Abstract
The function of cells in their native habitat often cannot be reliably predicted from genomic data or from physiology studies of isolates. Traditional experimental approaches to study the function of taxonomically and metabolically diverse microbiomes are limited by their destructive nature, low spatial resolution, or low throughput. Recently developed technologies can offer new insights into cellular function in natural and human-made systems and how microorganisms interact with and shape the environments that they inhabit. In this Review, we provide an overview of these next-generation physiology approaches and discuss how the non-destructive analysis of cellular phenotypes, in combination with the separation of the target cells for downstream analyses, provide powerful new, complementary ways to study microbiome function. We anticipate that the widespread application of next-generation physiology approaches will transform the field of microbial ecology and dramatically improve our understanding of how microorganisms function in their native environment.
ToC blurb
In this Review, Hatzenpichler et al. introduce next-generation physiology, which is a suite of new techniques that enable to investigate the phenotypes of individual cells in a non-destructive manner. Next-generation physiology complements genomics and culturing and provides new insights into microbiome function.
Introduction
Microorganisms dominate every ecosystem on our planet. They are the main drivers of global biogeochemical cycling, control the levels of many climate-active gases, and associate with virtually all multicellular lifeforms, including plants, animals, and humans. The microbiome [G] of each human is estimated to contain 1013-1015 microbial cells from 103-104 bacterial, archaeal and fungal species1 and recent predictions suggest a total number of more than 1030 microbial cells and 1031 viruses in the biosphere2. High-throughput sequencing technologies have revolutionized microbial community studies and led to a more complete view of the diversity of life on Earth3–5. However, in order to understand how microorganisms function and interact with their biotic and abiotic environment, experiments targeting the phenotype [G] of cells in their native habitat must complement cultivation-based and sequencing-based work. Physiology, the functioning of a cell at a given time and set of physiochemical conditions, is an emergent property that cannot be reliably predicted from genomic data or metabolic reconstructions alone. Rather, these approaches formulate valuable hypotheses that require experimental testing before definitive conclusions can be drawn about the physiology of a specific microorganism.
The realization that heterogeneity of gene expression and as a result changes in cellular phenotype are observed in synchronized, clonal cultures6,7 led microbiologists to study physiology at the level of the individual cell (Fig. 1). In natural systems, the need to work at such high resolution is more pronounced; most DNA-sequencing and bioinformatic methods cannot differentiate between strains of the same species, and microorganisms sometimes have dramatically different genotypes [G] 8 and in situ phenotypes9 despite indistinguishable or near-identical 16S ribosomal RNA (rRNA) gene sequences (the most commonly used taxonomic marker gene for bacteria and archaea). Although many powerful approaches exist to study microbial physiology, most of these techniques are only applicable to genetically amenable model archaea and bacteria that can be grown in pure culture. Commonly, these techniques depend on genetically encoded fluorescent reporters [G], the creation of deletion mutants to causally link genotype and phenotype, and/or the ability to reliably grow microorganisms under tightly controlled conditions, such as in bioreactors or microfluidic devices10–14. Outside of laboratory-based experiments, however, microorganisms live as members of spatially structured, taxonomically diverse, and metabolically interdependent communities, which are exposed to varying physicochemical conditions. These complexities are an important reason why most taxa have so far proven recalcitrant to cultivation15,16. Even if representatives of environmentally and medically relevant taxa can be isolated, it is sometimes unclear to what extent laboratory findings can inform us about the ecophysiology [G] of a microorganism and the way it functions in its native habitat.
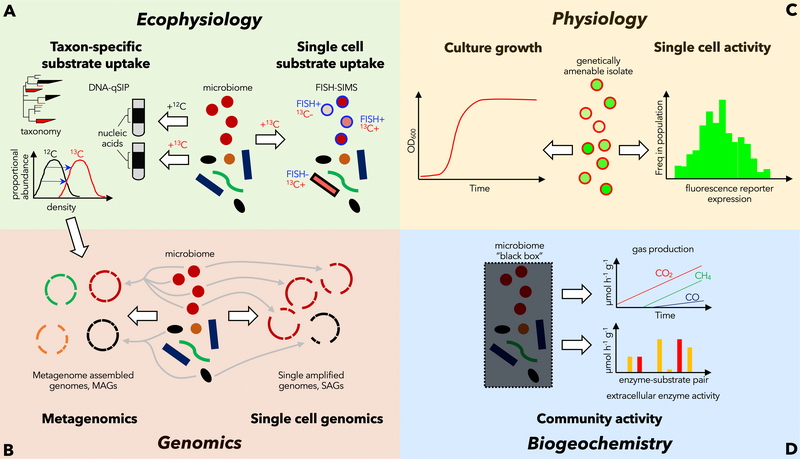
Figure 1. Examples of traditional approaches to study microbial physiology.
Most physiology-targeted techniques in the microbiome field depend on the availability of genetically amenable pure cultures, rely on destructive analyses that cannot directly link genotype with phenotype, or are unable to study functional activity at the level of single cells. (A) Stable isotope probing (SIP) can be coupled to secondary ion mass spectrometry (SIMS) and fluorescence in situ hybridization (FISH) to link cell function and identity.Isotopically heavy DNA can be separated from light DNA via buoyant density centrifugation. In quantitative stable isotope probing (qSIP), multiple density fractions are collected and analyzed by 16S rRNA gene sequencing or metagenomics. (B) The genetic makeup of entire microbial communities or individual cells can be studied by metagenomics or single cell genomics. Whereas single cell genomics typically captures only the most abundant members of a microbial community, metagenomics integrates the genomic information obtained from many individual cells into population genomes, that is, metagenome-assembled genomes (MAGs). (C) If genetically tractable microorganisms are available, they can be studied using reporter-gene constructs, which enables direct insights into variation of metabolic and anabolic activity between cells. (D) Many biogeochemical approaches treat microbiome samples as an undefined ‘black box’ but provide highly sensitive and precise measurements of overall community activity.
Ecophysiology experiments typically target phenotypes of populations or cells based on predictions of their metabolic potential from sequencing of enzyme marker genes, metagenomics [G], or single cell genomics [G]. All of these methods require the destruction of the original sample (through cell lysis), thereby preventing subsequent analyses. Metabolic predictions are tested using experimental approaches that also destroy cells. For example, microautoradiography (MAR) [G] and nano-scale secondary ion mass spectrometry (nanoSIMS) [G] are arguably the most successfully applied ecophysiology techniques capable of single cell resolution9,17–20 but are incompatible with downstream applications such as cultivation or genome sequencing. Quantitative stable isotope probing (qSIP) [G] 21 provides a complementary and more high-throughput approach to study microbial physiology and can provide a direct link between cell taxonomy and substrate uptake. Although qSIP has led to fascinating discoveries in microbial ecology and is particularly powerful when combined with meta-omics22–24, it cannot distinguish between individual cells. Similarly, many biogeochemistry-targeted approaches, such as extracellular enzyme assays, gas production measurements, or metabolome profiling are sensitive and easily replicable but currently cannot be applied at a scale relevant to microorganisms (μm to mm; with the notable exception of microsensors). Because these methods are either destructive, incompatible with correlative methods or have limited spatial resolution, one frequently has to determine the genotype of a cell first before subsequently characterizing the phenotype of a different cell.
In the past 15 years several new techniques have been developed in the fields of microbial ecology, chemical engineering, and analytical chemistry that radically break from the above approach. They enable studying the function of cells informing about, for example, their role in biogeochemical cycling, biotechnological potential, or medical relevance, irrespective of cell identity or genotype25,26. To distinguish these novel approaches from traditional methodologies, we introduce the term next-generation physiology [G]. Next-generation physiology approaches are independent of the need for prior knowledge about the genetic makeup of a microbial community and focus on cellular function. They do not require laboratory cultivation and are non-destructive, thus enabling microbiologists to bridge the gap between historically separated fields in microbiome research (Fig. 1). While cultivation, omics, and traditional physiology techniques are central components of microbiology research, next-generation physiology approaches provide a novel, complementary, and highly resolved view into the lives of microorganisms.
In this Review, we first discuss the general concept of next-generation physiology approaches before describing in detail the currently available techniques for studying cellular phenotypes without destruction of studied cells. We discuss how these approaches can be combined with cell sorting techniques and a suite of powerful downstream applications, including genetic characterization and cultivation-based experimentation.
Concept of next-generation physiology
We define a next-generation physiology approach as any combination of techniques that analyses the phenotype of an individual cell in a microbiome in a non-destructive way, which enables the physical separation of this cell based solely on its phenotype for subsequent, downstream applications. Ideally, these approaches can be applied in high-throughput (103–107 cells per hour).
Next-generation physiology approaches can be either label-free or label-dependent. Label-free approaches target native and inherent cellular properties and provide valuable information about the phenotype of a cell under non-invasive conditions. Label-based approaches introduce a chemical reporter into the cell that can provide a more comprehensive or complementary view of dynamic cellular processes. Before we discuss label-free and label-dependent approaches in detail, we provide an outline of the three steps of every next-generation physiology approach: (i) non-destructive phenotype observation, (ii) sorting of the observed cell based on its phenotype, and (iii) downstream analyses (Fig. 2).
Figure 2. Next-generation physiology workflow to study microorganisms.
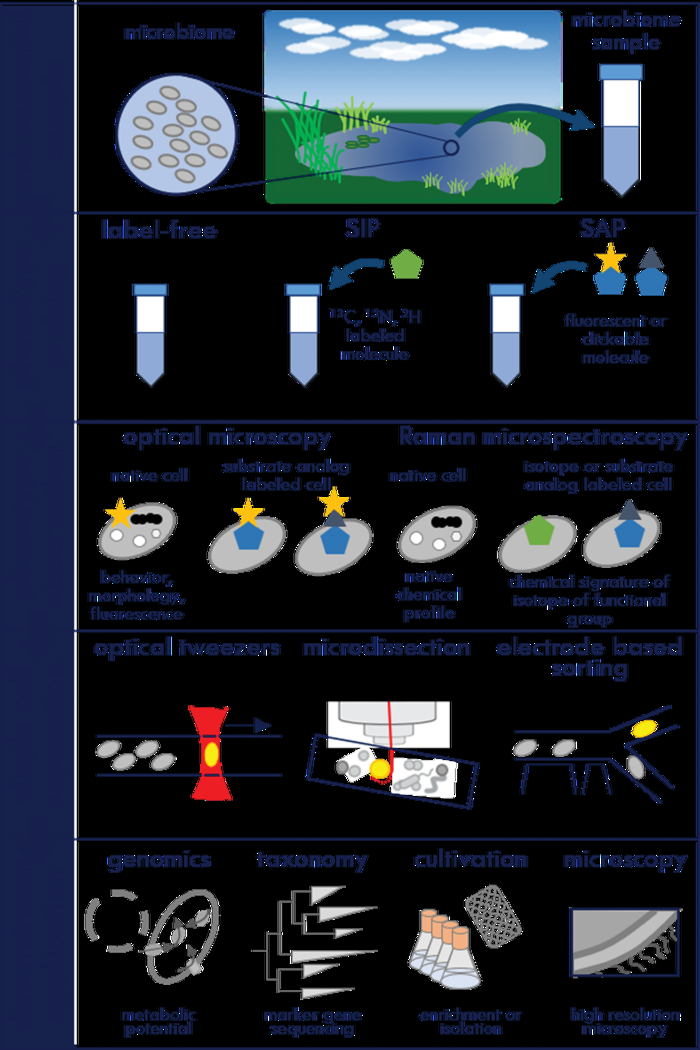
A microbiome sample is obtained using minimally invasive protocols and a phenotype of interest is detected using non-destructive methodology, for example via light or fluorescence microscopy or Raman microspectroscopy. Label-free approaches are directed at intrinsic properties of a cell, including chemotactic behavior, the expression of cofactors or pigments, or the presence of storage compounds. Label-based approaches introduce a chemical reporter into the cell that provides information about dynamic processes. Stable isotope probing (SIP) in combination with Raman microspectroscopy reveals substrate assimilation. Substrate analog probing (SAP) employs molecules that carry either a fluorescence tag or a side-group amenable to azide-alkyne click chemistry to obtain information on the overall biosynthetic activity or specific enzymatic function of the cell. After identifying a cell expressing the phenotype of interest that same cell is separated from the sample using, for example, optical tweezers, laser microdissection, or electrostatic deflection. The unaltered, sorted cell is then committed to downstream applications, which could include whole-genome sequencing, targeted cultivation, or complementary microscopic analyses. Different reporters used in next-generation physiology are described in table 1 and figure 3.
Non-destructive phenotype observation
The phenotype of an organism is defined by its observable characteristics in a given environment. Microscopy-based imaging is essential for studying the phenotype of individual microbial cells and is ideally coupled with molecular analysis to obtain taxonomic information. Microscopy uses transmitted light to visualize morphological features and optical properties or detects fluorescence characteristics upon excitation with light of specific wavelengths. Coupling microscopy with spectral analysis by Raman spectroscopy (Raman microspectroscopy) provides high-resolution (submicron spatial scale) spectral information. Raman spectroscopy measures the vibrational energy of molecular bonds after excitation with monochromatic light, which is informative of the molecular and, to some extent, isotopic composition of a cell (Box 1). The Raman spectrum of a cell typically consists of over 1,000 Raman bands (data points), each representing specific biochemical properties. Measurements are rapid (0.1–10 seconds per measurement) and can be non-destructive, thus enabling monitoring of living cells over time. There are reports of detrimental effects of laser irradiation on microorganisms and cell exposure to laser beams can have a range of outcomes from no observable effects to physical disintegration of the cell. However, negative effects are typically only observed after long-term exposure to intense laser light27.
Box 1. The chemistry underlying many nextgeneration physiology approaches.
Raman microspectroscopy
Raman spectroscopy is a classical technique in analytical chemistry that measures the vibrational energy of molecular bonds. In Raman microspectroscopy analyses, the molecules in a sample are excited with monochromatic light and inelastically scattered (re-emitted) photons are analyzed. Following excitation, most molecules return to their ground vibrational state and emit photons with the same wavelength as the incident light, an effect referred to as Rayleigh scattering (figure part a). In very rare cases (one in every 106-108 photons) the wavelength of a scattered photon is shifted compared to the incident light by either Stokes or anti-Stokes inelastic scattering. Stokes scattering, the more common form, occurs when an excited molecule returns to a state of elevated vibrational energy compared to the ground state, resulting in increased vibrational energy and emission of photons with lower energy. Alternatively, a molecule that is already in an excited state can be further excited and return to its vibrational ground state, emitting a photon with higher energy compared to the incoming light (anti-Stokes scattering). The detection of these scattered photons can be used to study the chemical composition of a sample.
Spontaneous Raman scattering, the most commonly used Raman microspectroscopy method, is limited by inherently low signal intensities. Low signal intensity can be problematic when analyzing cells with high levels of autofluorescence. Several techniques are available for signal enhancement and faster acquisition times, including surface- or tip-enhanced Raman spectroscopy (SERS or TERS), stimulated Raman scattering (SRS), coherent anti-Stokes Raman spectroscopy (CARS), and resonance Raman spectroscopy67,163–168. Although these advanced Raman microspectroscopy techniques have sporadically been applied to microbial isolates161,164,165,169–171 and hold great promise for microbial ecophysiology, they are currently absent from the microbiome literature. The acquisition of a Raman spectrum is relatively fast and easy, although Raman spectra can be very complex and their interpretation requires robust data analysis and reliable reference databases.
Azide-alkyne click chemistry
Click chemistry refers to any reaction that creates heteroatom links and that is modular and easy to perform, features fast kinetics, high chemo- and stereo-selectivity, as well as very high yields172. Although many reaction types fulfill these criteria81,83,173, the widely used azide-alkyne [3+2] cycloaddition reaction yielding a triazole conjugate has become the gold standard and is often simply referred to as the ‘click reaction’. Two types of labeling reactions yield triazole conjugates through azide-alkyne click chemistry (figure part b): a Cu(I)-catalyzed version that ligates an azide with a terminal alkyne, and a metal-free, strain-promoted reaction that links a highly reactive (strained) cyclooctyne-containing molecule (for example, dibenzocyclooctyne) with a reporter azide81,83,174,175.
In Cu(I)-catalyzed click reactions, chelating ligands for copper (such as tris[(1-hydroxypropyl-1H-1,2,3-triazol-4-yl)methyl]amine, THPTA) improve reaction kinetics and protect the cell from oxidative damage, whereas addition of the reductant sodium ascorbate maintains copper in the catalytically active Cu(I) state. To avoid protein crosslinking by byproducts of ascorbate oxidation, aminoguanidine is added to the reaction mix. Fluorescent dyes containing Cu-chelating picolyl motifs raise the effective concentration of Cu(I) at the reaction site176, which permits the use of lower metal concentrations and thus lowers the risk of copper cytotoxicity for downstream analyses that require viable cells, such as cultivation attempts.
Exploiting the reactivity of cyclooctyne-containing molecules with azides provides a metal-free alternative to Cu(I)-catalyzed click reactions. However, strain-promoted click chemistry can be accompanied by nonspecific reactions with free thiols (for example, the thiol group of reduced cysteine). Hence, free thiols must be blocked prior to the click reaction to avoid nonspecific labeling, which is typically achieved by incubation with a haloacetamide (for example, 2-chloroacetamide).
Azide-alkyne click chemistry reactions to fluorescently label cells are simple to perform because they involve cheap reagents (totaling ~$500 for the clickable substrate analog and dye as well as all necessary reagents 118) and a small number of working steps. Labeling and washing protocols are well established and can be completed in one (copper-catalyzed click) to three hours (strain-promoted click)118. Both types of click reactions are solvent- and pH-independent and are not affected by the presence of complex organic or inorganic matrices (for example, the extracellular polymeric substance of a biofilm, sediment particles or minerals), ensuring a low level of background noise when applied to microbiome samples. New generations of clickable fluorophores, including picolyl dyes176 and fluorogenic ‘turn-on’ azide probes177, which only become fluorescent upon reaction with an alkyne, are particularly well suited for complex sample types. The low molecular weight of all reagents (<1 kDa) makes it possible to click-stain cells without the ethanol-dehydration or permeabilization steps (such as treatment with lysozyme or proteinase K) required for successful fluorescence in situ hybridization. Click chemistry-mediated fluorescence-staining can be achieved on formaldehyde-fixed42,46,112–115,118, ethanol-fixed46, or intact, not chemically fixed42,46,112 cells.
The Raman spectrum of a cell is a unique fingerprint of its chemical composition and contains information on its taxonomic identity and physiological state28–30. Label-based phenotype studies use introduced reporters (that is, stable isotopes, functional groups, or fluorophores) to detect unique chemical signatures or fluorescence properties using Raman microspectroscopy or fluorescence microscopy, respectively.
Cell sorting
Cells can be separated from complex samples based on morphological, optical, fluorescence, or Raman spectral properties. Optical microscopy and cell separation via optical tweezers or laser microdissection are manual and often tedious processes with limited throughput (10–100 cells per hour). By contrast, fluorescence activated cell sorting (FACS) automates separation and can sort 103-104 cells per second by combining fluorescence detection of individual cells with flow cytometry or microfluidics-based separation. Furthermore, cells with unique chemical signatures in their Raman spectrum (for example, compound-specific bands or peak shifts due to isotope incorporation; Fig. 2) can be separated by Raman-activated cell sorting (RACS) [G]. RACS techniques (reviewed in 31) combine single cell Raman spectral acquisition with cell separation via optical tweezers32, microfluidic sorting33–36 or cell ejection36–39. Although a recently developed automated RACS platform that combined optical tweezers, microfluidics, and Raman spectral acquisition provided improved sorting efficiency (200–500 cells per hour33), Raman signal acquisition times of 0.1–10 seconds per spectrum currently limit the throughput of RACS compared to FACS. Future modifications of Raman microspectroscopy signal enhancement (Box 1) could theoretically achieve spectral acquisition rates over 100 times faster than classical Raman microspectroscopy.
A potential bias associated with all cell sorting is that the initial separation of cells from the sample matrix depends on the specific sample and can lead to preferential cell recovery. Proper cell extraction particularly important for samples with high structural complexity or high numbers of particle-attached or otherwise immobilized cells. To achieve maximal cellular yields at minimal risk of preferential recovery, cell extraction protocols typically require optimisation for each sample type and thorough testing by comparing the in situ community composition to the extracted cell fraction25,40,41. Although no single protocol works for all sample types, a combination of washes with mild detergents, sonication, and density gradient centrifugation with or without filtration has been reported to yield the best results for complex samples, including sediments and soils41–46. Finding the appropriate cell extraction protocol often is the most time-consuming step in any next-generation physiology workflow.
Downstream analyses
After separation and sorting of individual cells with a desired phenotype they can be used for subsequent investigation. The main applications in microbiome research identify taxa through rRNA-targeted fluorescence in situ hybridization (FISH) [G], taxonomic marker gene sequencing, genotype characterization through single cell or metagenome sequencing, or further phenotypic characterization with different microscopy techniques (for example, electron microscopy or atomic force microscopy47). Because chemical fixation can dramatically decrease DNA quality (for example, formaldehyde cross-links proteins and DNA), intact cells [G] (cells that have not been chemically fixed) are desired for DNA-targeted downstream applications40,48. Genome amplification from ultra-low biomass samples, including single cells, is commonly achieved by multiple displacement amplification (MDA). MDA can lead to uneven genome coverage, genome rearrangements including chimera formation, or erroneous nucleotide incorporation. Most of these biases, however, can be overcome through long mate-pair libraries, high sequence coverage, and post-sequencing normalization40,41,48–50.
Alternatively, intact, sorted cells can be used as inoculum for cultivation, which enables in-depth culture-dependent physiology, biochemistry, and systems biology studies51,52. These downstream investigations complement initial phenotype characterization and lead to a more comprehensive understanding of the ecophysiology of a microorganism. To the best of our knowledge, high-throughput axenic cultures of cells separated from a sample based on their phenotype has not been achieved yet. However, a study demonstrated that cells separated from lake sediment by FACS based on their activity response to methane addition, could be regrown in enrichment media52.
Label-free approaches
Non-invasive optical microscopy and Raman microspectroscopy observe the behavior and native chemical composition of individual cells. This is mostly informative of the presence of transient traits, but in the case of time-resolved analyses of living cells, it also provides insights into dynamic cellular processes. Phenotypic observations by optical microscopy include the formation of spores, storage compounds, cellular segmentation, the behavioral responses of cells to external stimuli (for example, aero-, chemo-, magneto, or photo-taxis), or the occurrence of intrinsic autofluorescence from cofactors, pigments, or vitamins. Similarly, compounds with known Raman bands can be identified in the Raman profile of a cell based on database comparisons. Cells with specific characteristics can be separated based on their optical properties53 (for example, cell volume or refractory index) or their chemical composition, such as the presence of auto-fluorescent compounds54 or compound-specific Raman bands31. For example, RACS of a functional guild was elegantly demonstrated in a recent study38, which separated uncultured bacteria from the Red Sea based on distinctive Raman bands of their carotenoid pigments. RACS-separated cells were further characterized by single cell genomics, revealing novel insights into carotenoid biosynthesis and previously unknown phototrophs38. Table 1 provides an extensive list of reporters available for label-free imaging and sorting of individual microbial cells.
Table 1.
Next-generation physiology approaches to study microorganisms.
| Reporter | Phenotype of single cells characterized by light or fluorescence microscopy | Phenotype of single cells characterized by Raman microspectroscopy | Phenotype-based cell separation and downstream application |
|---|---|---|---|
| Label-free approaches | |||
| Behavioral reaction to external stimuli | Aerotaxis, chemotaxis, magnetotaxis, or phototaxis | N.A. | Magnetic enrichment and single cell genomics of magnetotactic bacteria151 |
| Cofactors | Cofactor F420 in methanogenic pure and enrichment cultures54 | Cofactor F420 in an ammonia-oxidizing archaeon152 | FACS based on cofactor F420 autofluorescence and sequencing of marker gene of methanogens54 |
| Pigments | Chlorophyll | Carotenoid-containing bacteria38 | RACS and single-cell genomics of carotenoid-containing bacteria38 |
| Spores | Endospores detection by differential interference contrast microscopy | Bacillus cereus spores153 | * |
| Extracellular polymeric substance | Stains for extracellular DNA, proteins, or polysaccharides145,146 | Proteins and polysaccharides in biofilm matrix59,60 | * |
| Carbon storage | N.A. | Glycogen, poly-hydroxyalkanoate, and poly-hydroxybutanoate in wastewater sludge bacteria154,155 | * |
| Cytochromes | N.A. | Cytochrome C in nitrifiers156, anammox bacteria156 and Beggiatoa spp.157; cytochrome redox potential158 | automated RACS of cytochrome C rich cells from a marine enrichment culture33 |
| Magnetosomes | N.A. | Magnetotactic bacteria containing magnetite and greigite159 | * |
| Phosphate storage | N.A. | Orthophosphate and poly-phosphate in cultured159 and environmental154,155,159 bacteria | * |
| Sulfur inclusions | N.A. | Polysulfides in sulfur-oxidizing Beggiatoa spp.157; cyclo-octasulfur in pure cultures159 and bacterial symbionts of flatworms160 | * |
| Stable isotope probing (with substrate or heavy water) | |||
| 2H | N.A. | Naphthalene and glucose degradation by Pseudomonas sp. and Escherichia coli 57 | * |
| 13C | N.A. | Naphthalene-degraders in groundwater59,60; phenylalanine uptake by extracellular Chlamydiae62; marine autotrophs37; degraders of cyanobacterial necromass65 | Raman-activated cell ejection and single cell genomics of marine autotrophs37 |
| 15N | N.A. | 15N2 fixers in soil63; ammonia, nitrite and N2 assimilation in freshwater bacteria161 | * |
| 2H2O | N.A. | Mucin degraders in mouse gut microbiome33; cellulose degraders129, detection of antibiotic-resistant bacteria in freshwater36; degraders of organic matter in groundwater162 | Manual sorting using optical tweezers, followed by 16S rRNA gene sequencing55; automated sorting using optical tweezers on a microfluidic platform, followed by metagenomics33 |
| H218O | N.A. | * | * |
| Substrate analog probing | |||
| Fluorescent analogs | Uptake of glucose in rumen76; xylan and lamarin uptake by bacterioplankton77; fluorescent amino acids75 | * | FACS and 16S rRNA gene sequencing and single cell genomics of cells taking up fluorescent glucose76 or polysaccharides77, respectively |
| Non-canonical substrates | Clickable nucleosides130,131,138, L-amino acids 46,112–115,119, D-amino acids80, sugars117,137, fatty acids132,133 | Alkyne containing amino acids, nucleosides, sugars, and fatty acids visualized by SERS67,68,88 | FACS followed by 16S rRNA gene sequencing42,46 |
| Activity- and affinity-based protein profiling | Ammonia monooxygenases, antibiotic-reactive proteins, ATP-ases, ATP-binding proteins, cellulases, cytochromes, fatty acid synthases, glycoside hydrolases, lipases, redox-reactive proteins, vitamin transporters139,141,142 | * | FACS separation and 16S rRNA gene sequencing of β-glucuronidase active cells144 |
application feasible but not yet demonstrated; anammox, anaerobic ammonium oxidation; FACS, fluorescence-activated cell sorting; N.A., not applicable; RACS, Raman-activated cell sorting; rRNA, ribosomal RNA; SERS, surface-enhanced Raman scattering.
As these label-free approaches to phenotypic characterization detect inherent cellular properties, they have limited application in studying metabolically active [G] cells, which requires the incorporation of chemical reporters to be tracked on a single cell level.
Isotope probing
Isotope probing approaches involve the incubation of a microbial sample with an isotopically labeled reactant (substrate or water) and track its incorporation into cellular components, identifying anabolically active [G] microorganisms. Incubation with an isotopically labeled substrate (for example, 13C-glucose or 15NH4+) enables substrate incorporation into biomass and tracking of the flow of intermediates within a system. Alternatively, incubation with heavy water (2H2O or H218O) provides a labeling strategy in which all anabolically active cells are detected independently of assimilatory capacities36,55–57.
Single cell resolved isotope probing, such as MAR or nanoSIMS combined with FISH, has seen wide application in microbial ecophysiology studies9,17–20. However, MAR and nanoSIMS destroy cells and thus preclude subsequent downstream analysis. Raman microspectroscopy is a non-destructive analysis strategy. Isotopically labeled cells are identified by characteristic peak shifts in their Raman spectrum due to the replacement of a light isotope by a heavy isotope, which changes the vibrational energy of a molecule through the increased molecular mass introduced by the heavy isotope (for example, shift of the C-H peak from 2,935 cm−1 to 2,178 cm−1 due to 2H incorporation58; Fig. 3). The intensity of this shift towards smaller wavenumbers correlates with the amount of assimilated heavy isotope55,59. Detection limits of isotope uptake depend on the specific capabilities of the Raman microspectroscopy system but typically are ~10% 13C, ~10% 15N, and ~0.2% 2H replacement of cellular carbon, nitrogen, and hydrogen, respectively55,58,59.
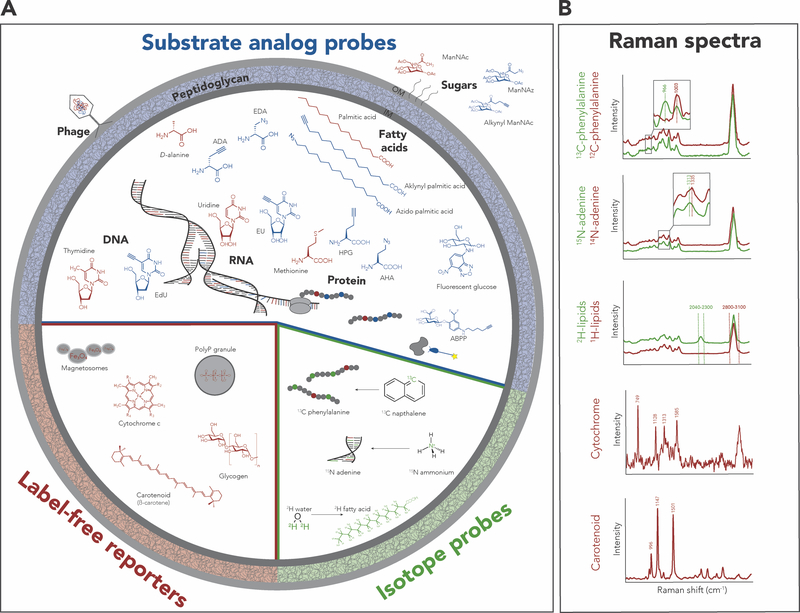
Figure 3. Reporters and their associated Raman spectral fingerprints in microbial next-generation physiology.
(A) Label-free reporters (red) are produced by the cell and do not require addition by the researcher. Substrate analog probes (blue) are traceable compounds that are amenable to biorthogonal labelling and that are incorporated into biomass by the cell of interest after addition to the microbiome sample. Finally, isotope probes (green) can be added to a sample to trace the uptake and incorporation of isotopically labeled compounds. Many substrate analog probes contain azide or terminal alkyne groups, which can be conjugated to a fluorescent dye for detection by click chemistry. (B) Incorporation of stable isotopes into biomass leads to spectral shifts towards lower wavenumbers in the spectrum of labeled cell compared to unlabeled cells and some of these shifts are pronounced enough to be detectable by Raman microspectroscopy58. The figure shows examples for the most commonly used indicator peak shifts used to trace isotope incorporation into single cells, including from top down, the symmetric ring breathing effect by phenylalanine (13C), C–H stretching of adenine (15N), and C–H stretching of lipids and proteins (2H). Other reporters, including alkyne-labeled substrate analogs and some label-free compounds, have distinct Raman spectral fingerprints that also can be used for phenotype detection38,68,88. Table 1 includes detailed information on the application of all depicted reporters and references to the primary literature.
Substrate stable isotope probing
Substrates labeled with heavy isotopes can be used to identify all members of a microbial community that can specifically assimilate the substrate. In addition, isotopically labeled metabolic intermediates (degradation products of the initially added substrate) can reveal cross-feeding within a community and metabolic interactions between cells. However, some isotope-containing compounds, in particular those of high structural or compositional complexity (such as cellulose, lignin, or mucin) often are prohibitively expensive or commercially unavailable. In addition, substrate SIP requires amendment of a microbial sample with an isotopically labeled compound, which could alter natural substrate concentrations and change the composition of the incubated communities [.
Detection of 13C and 15N labeled cells has been achieved by Raman microspectroscopy in multi-species communities and has been successfully combined with FISH59,60, genome sequencing37,60 or cultivation60. An initial study detected labeled cells after incubation with 13C-glucose based on peak shifts in the Raman spectrum due to incorporation of the heavy isotope61. Subsequently, substrate SIP-Raman microspectroscopy was used to investigate the niche differentiation of naphthalene degraders in ground water communities59,60, uptake of phenylalanine in an amoeba-Chlamydiae symbiont system62, and assimilation of different carbon and nitrogen sources in environmental communities60,63–66. Since the first successful separation of individual isotope labeled cells from a cell mixture into sterile capillaries using optical tweezers32, different types of RACS have been used to sort 13C or 15N labeled cells from complex samples37,39, demonstrating the importance of this approach to ecophysiology research (Tab. 1).
Using stimulated Raman scattering (SRS) microscopy, 2H-containing nucleosides, amino acids, and fatty acids can be used to visualize DNA-, RNA-, protein-, and membrane-synthesizing cells67,68; however, this application has, to the best of our knowledge, not yet been demonstrated for microbial samples.
Heavy water SIP
Isotope labeling approaches with heavy water (2H2O or H218O) aim to identify all members of a community that are anabolically active36,55–57, rather than those members involved in specific transformations. Heavy water SIP enables testing cellular activity under either close to in situ conditions or a specific physicochemical condition or substrate amendment. Heavy water has gained increasing interest in environmental microbiology as it generally can be used without prior knowledge of the growth substrate(s) of a microorganism, it does not interfere with the natural substrate pool, and it is inexpensive. SIP with heavy water commonly requires an experimental setup in which a portion of regular water (ideally 30–50%55) is replaced with heavy water to achieve high enough labeling percentages for reliable detection, a feat that can be challenging to achieve in certain sample types (for example, soil and aqueous samples). In addition, the effect of heavy water on the growth rates of physiologically diverse and taxonomically distinct cells has not been evaluated and the molecular underpinnings of this effect are not yet understood, demanding further investigation55,69,70.
For Raman microspectroscopy analyses, labeling of cells with 2H2O is superior to H218O labeling because H from water readily exchanges with the NADPH pool of the cells, the main source of H for lipid synthesis71–73. The introduction of 2H-C bonds is easily detectable in the Raman spectrum of a cell by a characteristic peak shift of the abundant C-H peak into the silent region [G] of the cellular chemical profile (Fig. 3). 2H2O SIP-Raman microspectroscopy has been combined with FISH to detect targeted taxa and with RACS to select functionally active cells for 16S rRNA gene or whole-genome sequencing33,36,55 (Tab. 1).
Substrate analog probing
An alternative approach to SIP is to incubate a sample with a synthetic compound that is a structural and/or functional analog of a naturally occurring molecule. Such experiments are either directed towards anabolic processes, such as non-canonical substrate labeling, or towards metabolic reactions catalyzed by specific enzymes or enzyme families, such as fluorescent substrate analog labeling or activity- and affinity-based protein profiling. To contrast these approaches from SIP, we here introduce the term substrate analog probing (SAP). An important advantage of SAP compared to most SIP and some label-free approaches is that SAP uses infrastructure that is readily available to most laboratories, that is, standard fluorescence microscopes and FACS instruments. Therefore, many SAP approaches, in particular those that use azide-alkyne click chemistry [G], are comparatively easy to set up.
Fluorescent SAP
Fluorophore-tagged derivatives of natural compounds can be used to track the uptake of molecules on a cellular level. This provides a powerful approach for determining specific substrate uptake capabilities of individual microorganisms in multi-species communities. Examples of fluorescent SAP include the use of fluorescent cobalamin analogs to demonstrate the uptake of this vitamin into bacteria, worms, and plants74, or the use of fluorescently labeled D-amino acids to visualize regions of active peptidoglycan-synthesis in cell walls of different bacterial pure cultures75. Furthermore, the combination of fluorescent substrate analog probing with FACS and subsequent marker gene and whole genome sequencing enabled the identification of diverse but low abundance degraders of glucose in the rumen76,of xylan and laminarian in bacterioplankton77, and cellulose degraders in a geothermal spring78.
Fluorescent SAP specifically detects cells that take up the fluorescent substrate under the assumption that there is no transfer of the fluorescent group to other metabolites. The broader implementation of fluorescent SAP is limited by the development of fluorescent labeling techniques that target different molecule classes. Furthermore, the addition of a fluorescent tag directly to the substrate might interfere with enzyme-substrate binding and recognition. Newer, click chemistry-based approaches, such as non-canonical substrate labeling and activity- and affinity-based protein profiling, overcome these problems by making the detection of these molecules (for example, by dye staining) independent of the labeling chemistry by using substrate analogs. Examples of this are the use of clickable [G] vitamin B1279 or D-amino acids80 rather than fluorescently labeled vitamins or D-amino acids.
Non-canonical SAP
Non-canonical molecules are synthetic structural analogs of biological molecules that are incorporated into biomass due to enzyme promiscuity. Many non-canonical molecules contain a reporter group that can be specifically traced within the complex environment of the cell through a bioorthogonal reaction [G]. These reactions are chemical transformations that do not interact with functional groups present in naturally occurring molecules, have no or only minimal byproducts, and do not interfere with cellular processes81–83. Azides and terminal alkynes are particularly attractive reporter groups because they rarely occur in biology, are biocompatible, and can be fluorescently detected by azide-alkyne click chemistry conjugation reactions (Box 1). To our knowledge, only one natural azide-containing molecule (a secondary metabolite produced by a dinoflagellate) has been identified84. Terminal alkynes, as functional groups of amino acids and fatty acids, are more common but still restricted to only a few lineages85–87. An alternative to detecting azides or alkynes through a bioorthogonal fluorescence labeling reaction is to use SRS to trace them inside the cell68,88.
Biorthogonal labeling approaches are well established in the study of bacterial89–93 and eukaryotic94–97 model organisms. In multi-species systems, however, they have mainly been used to study de novo protein synthesis. Indeed, proteins are the most promising target for in situ studies because they constitute the largest proportion of cellular dry weight (50–65%)98–101. This results in a higher sensitivity for proteins than other molecules, as the cellular dry weights of DNA (1–3%), RNA (10–20%), and lipids (10–25%) are much lower (Fig. 3).
Bioorthogonal non-canonical amino acid tagging
Labeling of newly translated proteins with synthetic amino acids can be accomplished through bioorthogonal non-canonical amino acid tagging (BONCAT)102–104. BONCAT achieves the co-translational labeling of proteins by exploiting the substrate promiscuity of amino-acyl tRNA synthetases, which are enzymes responsible for catalyzing the esterification of amino acids with their cognate tRNAs. Only two clickable amino acids, L-azidohomoalanine (AHA) and L-homopropargylglycine (HPG), which both replace L-methionine (Met) during translation (Fig. 3), can be incorporated without genetic modification104–106. Since its inception103, BONCAT has been used to study protein synthesis in a range of microbial pathogens89,107–111 and was recently applied in several complex samples, including marine and freshwater sediments46,112, surface113,114 and deep115 seawater, soil42, and an oral biofilm112. In these studies, BONCAT was applicable to cultured and uncultured members of at least 20 archaeal and bacterial phyla42,46,112–118 as well as bacteriophages119 and eukaryotic viruses119,120. Because of their structural similarity to Met and their low activation rate by methionyl-tRNA synthetase104, HPG and AHA have only small effects on rates of protein synthesis and degradation in E. coli121 and mammalian cells95,103,121, as well as on protein tertiary structure122. BONCAT correlates well with other independent proxies of growth, such as the incorporation of 15NH3 into single cells visualized by nanoSIMS112, 35S-Met uptake as measured by MAR113, or incorporation of 3H-leucine into bulk biomass measured by scintillation counting115. In a study on deep-sea methane seeps, no measurable effect on either microbial community composition or rates of sulfide production and methane oxidation was observed when sediment samples were incubated with HPG46. When AHA or HPG are used at levels that resemble the intracellular concentration of Met (~100 μM)123 or over more than two generations, growth rates of some bacterial cultures are negatively affected112. Therefore, low concentrations of AHA or HPG (nM to μM range) and no-addition (blank) controls are required to compare and minimize effects on growth rates as well as unwanted reactions with naturally occurring azides or terminal alkynes. Incubation times should also be optimized (ideally to less than one to a few cell generations46,114,118) to avoid excessive substitution of Met, which could lead to nonfunctional proteins. It is still unknown how non-canonical amino acids enter the cell and interact with the translational machinery, which currently limits the ability to directly quantify, on a single cell level, newly made proteins in complex communities (Box 2). It is also unknown whether AHA or HPG are misrecognized for Met by enzymes other than Met-tRNA-synthase; if so, the azide and alkyne functional groups could be transferred to other molecules.
Box 2. Limitations of single cell BONCAT studies.
Several unknowns currently limit our ability to absolutely quantify protein synthesis rates in individual cells, which challenge the use of bioorthogonal non-canonical amino acid tagging (BONCAT) in quantifying activity rates of single cells (figure part a). (i) The routes by which non-canonical amino acids enter a cell are unknown, and the roles of facilitated diffusion and/or transporters could differ between species. (ii) In addition, although the catalytic efficiency of methionyl-tRNA synthetase of Escherichia coli for L-homopropargylglycine (HPG) and L-azidohomoalanine (AHA) is known (1:500 for Met:HPG and 1:390 for Met:AHA104), the extent of this substrate promiscuity might differ between organisms. Varying promiscuity would lead to differences in the substitution rate of Met in new proteins and ultimately labeling intensity. Furthermore, (iii) variations in the Met content of proteins and (iv) the rate at which proteins are expressed might compound interpretations. Heterogeneity in gene expression rates is observed even in clonal cultures and is likely amplified in multi-species samples6,7,178. Lastly, (vi) variability in click staining efficiency as a result of differences in the rate of dye entry into the cell could also lead to differences in cell labeling intensity. Similar limitations probably exist for other non-canonical substrate analog probing approaches capable of labeling DNA, lipids, or peptidoglycan but are currently untested.
Analysis of genomes deposited in NCBI RefSeq reveals a range in the use of Met in proteins (figure part b; Narchaea=1,561,087 proteins; Nbacteria=14,597,681 proteins). On average, predicted bacterial and archaeal proteins have a Met content of 2.49% and 2.19%, respectively. >99.9% of these proteins contain Met, suggesting that virtually all proteins are in principle amenable to labeling by AHA or HPG. However, possible modifications to the start Met (for example, N-formyl-Met, which uses a separate tRNA) could render some proteins unamenable to replacement by AHA and HPG, which depends on the promiscuity of methionyl-tRNA synthetase. If the starting amino acid is ignored, 5.70% and 10.88% of predicted bacterial and archaeal proteins do not contain Met (figure part c). For these calculations, only one genome from each species was analyzed and only complete genomes were considered for bacteria. Average values for archaea and bacteria are shown in each plot. The number of archaeal and bacterial bins for drawing plots were 410 and 270 in part b and 550 and 350 in part c, respectively.
Intact or chemically fixed cells identified by BONCAT can be stained with clickable fluorophores (Box 1) that serve as reporter groups in fluorescence microscopy studies. When coupled to rRNA-targeted FISH or catalyzed reporter deposition FISH (BONCAT-FISH112, BONCAT-CARD-FISH46), active cells can be identified, thus revealing taxonomy-function relationships and co-localization patterns of taxonomically identified active cells46,112,114,115. BONCAT-FISH has been used to visualize the cell organization of protein-synthesizing methane-oxidizing archaeal-bacterial consortia in deep-sea sediments46. In the same study, BONCAT was, for the first time, combined with FACS of both ethanol-fixed and intact (chemically unaltered) cells (BONCAT-FACS) for subsequent whole genome amplification and gene sequencing. Recently, the same approach was used to study the active cell fraction in soil42, an ecosystem that is notoriously difficult to investigate due to its structural complexity and high microbial diversity. The study revealed that a large fraction (20–70%) of soil-extractable cells was translationally active and that a high diversity of bacterial taxa was labeled with BONCAT42. This result was in stark contrast to previous, more labor-intensive studies, such as DNA-SIP124 or labeling with the thymidine surrogate 5-bromo-2’-deoxyuridine (BrdU)125, which suggested that up to 95% of cells in soil are inactive at a given time. Recent studies that employed qSIP-methodology are consistent with findings by BONCAT-FACS126. This discrepancy can be explained by the inherent biases associated with BrdU-labeling125,127,128.
The ability to combine bioorthogonal labeling incubations with other compounds enables designing experiments to screen for physicochemical factors (such as temperature, pH, or O2 levels in the headspace) or growth substrates that drive cellular, population, or community activity46,112. BONCAT is particularly useful for studying non-assimilatory pathways or if isotope labeled substrates are not available. Accordingly, BONCAT-FISH and BONCAT-FACS combined with marker gene or whole-genome sequencing can be used to monitor microbial community dynamics or identify specific taxa with changing activity after substrate changes46,112. This approach is conceptually similar to tracking the growth response of cells to substrate addition in the presence of heavy water and separating 2H2O-labeled cells by RACS33,36,55,129. Neither BONCAT nor 2H2O-Raman microspectroscopy can disentangle whether cell labeling is due to direct substrate uptake or metabolic cross-feeding, but measuring multiple samples over the course of an incubation may help reconstructing metabolic interactions and population dynamics within communities.
Targeting non-proteinaceous cell components and viruses
BONCAT is arguably the most sensitive non-canonical substrate labeling approach due to the large contribution of proteins to cellular biomass; however, many other biomolecules can be targeted, including nucleic acids, lipids, and polysaccharides (Fig. 3). The introduction of (deoxy)ribonucleoside surrogates amenable to click chemistry, for example, provides a straight-forward approach for detecting cells that synthesize RNA and DNA. A recent proof-of-concept study demonstrated the applicability of the alkyne-carrying thymidine surrogate 5-ethynyl-2’-deoxyuridine (EdU) to studying DNA synthesis in individual marine microorganisms by azide-dye staining130. Click chemistry-based detection of EdU can be performed in an hour and yields cell labeling rates comparable to the more biased and experimentally more complex BrdU-labeling approach125,127,128. By contrast, the alkyne-carrying uridine analog 5-ethynyl-uridine (EU) is incorporated into RNA due the promiscuity of RNA polymerase131 but has not yet been used on complex samples.
Other bioorthogonal labeling approaches use azide- or alkyne-modified fatty acids132,133, D-amino acids 80,134,135, or sugars117,136,137 to label the lipid membrane, peptidoglycan layer, or cell surface polysaccharides, respectively (Fig. 3; Tab. 1). Because pathways for lipid and cell wall biosynthesis, the use of peptidoglycan, and cell wall modifications differ widely across the tree of life, these approaches lack the general applicability of protein labeling via BONCAT. Although some of these substrate analogs have been used in studies targeting specific microorganisms, they have yet to be tested on taxonomically and physiologically diverse pure cultures and their effect on cellular activity remains unclear. Thus, researchers interested in applying these activity proxies in their research should proceed carefully before applying them to diverse samples.
Recent successful application of bioorthogonal labeling to cultured virus-host models of pathogenic120 and environmental relevance are also very promising119,138. In 2012, a study demonstrated that EdU-modified T4 phages can infect E. coli and that T4-containing cells stained with a clickable dye can be separated by FACS from an artificial waste water community138. Furthermore, BONCAT was recently used to quantify in situ marine viral production rates by fluorescence staining119. These pioneering studies demonstrated that non-canonical SAP approaches have strong potential to increase our understanding of the turnover rates of viruses in single cells as well as microbial communities, the viral impacts on elemental cycling through the release of nutrients from lysed cells, and might help to identify new virus-host relationships119,138.
Activity- and affinity-based protein profiling
A complementary set of SAP techniques targets catabolic rather than anabolic functions of the cell. Activity-based protein profiling (ABPP) is arguably the most broadly applicable catabolism-targeted approach that identifies active enzymes. Most importantly, in contrast to all other methods discussed in this Review, ABPP enables researchers to reveal the function of open reading frames in microbial genomes lacking functional prediction. ABPP achieves this objective with catalytic mechanism-based, electrophilic reactive groups (‘warheads’) that covalently label the active site of specific enzymes or enzyme classes139–142 (Fig. 3). The bound enzyme is later detected by a functionalizable reporter attached to the warhead by a spacer group. Although other reporter groups are available143, terminal azides and alkynes are the most commonly used and adaptable reporter tags owing to their biocompatibility and small molecular size, which guarantees minimal interference with substrate binding and reactivity and improves cell permeability.
Affinity-based substrate analogs are similar to their counterparts used in ABPP but rather than relying on enzymatic activity, the analogous substrates interact with proteins based on structural mimicry of the substrate rather than by bond creation with the active site of the enzyme. Thus, affinity-based protein profiling cannot resolve catalytically active from inactive enzymes. To achieve irreversible covalent linking of the affinity-based substrate analog to the enzyme, photoactivatable groups can be used141,142.
Activity- and affinity-based protein profiling are well-established approaches for identifying new enzymes in cultured microorganisms but, to our knowledge, have only once been applied to complex microbiomes144. Their potential importance for single cell ecophysiology studies, however, cannot be overstated. In an approach called ABP-FACS, a recent study used activity-based probes (ABP) to fluorescently detect, separate by FACS, and taxonomically identify β-glucuronidase active members of the mouse gut microbiome144. They also demonstrated that treating mice with vancomycin drastically affects glucuronidase activity and leads to strong shifts in the taxonomic composition of glucuronidase-active cells separated by ABP-FACS.
The limitation of activity- and affinity-based protein profiling lies in the challenge to design a substrate analog that reacts and binds to only one particular enzyme or enzyme class; however, substrate analogs are already available for a wide variety of enzyme classes139,141,142 (Tab. 1). In the future, microbiologists will need to more frequently and effectively collaborate with analytical chemists, chemical engineers, protein biochemists, and synthetic organic chemists to identify the most promising targets for functional studies and develop specific reporters for probing the activities of specific enzymes as well as intact cells.
Although other fluorescence-based tracers of enzyme activity, cell integrity, or cell structure are in use, most of them suffer from limitations that currently restrict their widespread application in microbial ecology. Many stains used for staining extracellular matrixes or cell internal structures are class-specific (for example, DNA, polysaccharides, or protein) but not compound-specific, and their specificity has not been validated using independent methods145,146. Furthermore, most commercially available stains of metabolic activity have been shown to be inapplicable to complex samples for a variety of reasons (Box 3).
Box 3. Alternative cell-staining approaches.
‘Vitality’ and ‘viability’ dyes
Advertised as ‘vitality’ and ‘viability’ stains (table), commercially available redox stains and mixes of membrane-permeable and impermeable dyes have lately seen use in microbiome studies to identify supposedly ‘living’ or ‘active’ cells. However, all these stains have some limitations that restrict their use in many complex samples resulting in rough estimates of vital or viable microorganisms at best179,180. Nevertheless, these stains can be useful in mixed-species samples, but only after extensive testing, including with pure cultures relevant to the specific study system. RedoxSensor™ Green has been successfully applied in combination with substrate stimulation and fluorescence-activated cell sorting (FACS) to investigate metabolically active methane oxidizers in Lake Washington52,181. Although such targeted applications are possible, researchers should apply caution when using these dyes.
Genome-inferred antibody engineering
An exciting new approach at the interface of phenotypic and taxonomy-based cell separation is ‘reverse genomics’51. In this workflow, antibodies are raised against proteins predicted to be located in the outer membrane or cell wall and FACS is used to sort fluorescent antibody-stained cells from a sample for subsequent single cell cultivation. The power of this approach was elegantly demonstrated by a study that used it to culture individual cells of ‘Saccharibacteria’ (formerly known as TM7) and the candidate phyla ‘Absconditabacteria’ (SR1) from human saliva51. Genome-inferred antibody engineering depends on the availability of genomes from cells of interest and cannot differentiate between metabolically active and inactive cells. However, if suitable cell surface antigens can be identified and specific antibodies targeting them can be developed, reverse genomics could be a promising tool to bring new microorganisms into culture.
| Type of stain | Working principle | Method-specific limitations | Dye-specific limitations | General limitations of all ‘viability’ and ‘vitality’ dyes |
|---|---|---|---|---|
| Redox stains (for example, 5-cyano-2,3-ditolyltetrazolium chloride (CTC) or RedoxSensor™ Green (RSG)) | Redox dyes that depend on activity of electron transport chain | Not useful for tracking activity of microorganisms that lack an electron transport chain (for example, strict fermenters) | CTC suppresses cellular activity182,183; counts of CTC positive cells were 2–100 times lower than microautoradiography counts184–186 | Practically unsuitable for structurally complex sample types (such as sediments, soils, or biofilms) because cell extraction reduces cell activity; general applicability to physiologically and taxonomically diverse communities is unknown; dyes are typically tested only on a small subset of clinically relevant, easy to culture, heterotrophic bacteria adapted to high nutrient conditions; rarely compared to independent measures of activity or cell growth other than the formation of colony-forming units |
| Live-dead stains (for example, LIVE/DEAD™ BacLight™, SYTOX Red Dead, FUN®−1, ReadyProbes) | Mixture of a cell-permeable (for example, SYTO TM9) and membrane-impermeable DNA-stain (for example, propidium iodide) | Not useful or yield inaccurate results for cells with hard to permeate cell walls or membranes (for example, spores; Gram positive versus Gram negative bacteria)179,187,188 | Background fluorescence, bleaching, fluorescence resonance energy transfer between dyes, double staining, and decrease in vitality during staining179,187,189 |
Outlook
The non-destructive nature of next-generation physiology approaches enables crucial downstream analyses of individual cells that express a phenotype of interest. These unique, phenotype-targeted approaches complement more established methodologies including cultivation, enzyme characterization, and meta-omics.. Once appropriate instrumentation becomes more widely available and experimental protocols more broadly adapted by the research community, the concepts we have described will enable highly parallelized characterization of microbiome function. For example, we expect that BONCAT-FACS and 2H2O-RACS will soon be widely applied to study the activity response of microbial communities to substrate addition or environmental changes, thus allowing physiological characterization of uncultured microbes at a hitherto unprecedented speed33,42,46,55. These and other single cell targeted approaches will be aided by the anticipated expansion of droplet microfluidics to employ culture-independent assays. Most currently available microfluidics approaches still depend on the ability to grow microorganisms on-chip, exploit genetically encoded fluorescence reporters, or are targeted at the genotype rather than the phenotype10,13,147–150.
To reach these goals, microbiologists are encouraged to work hand-in-hand with researchers outside the microbiome sciences, including analytical chemists, synthetic organic chemists, and biological and chemical engineers. Tremendous opportunities exist for non-microbiologists who are willing to go outside their comfort zone and break into the realm of living systems. Examples for their potential impact on microbiome sciences include the synthesis of new probes to interrogate cellular and enzyme function under non-invasive conditions, the adaptation of lab-on-the-chip designs to characterize uncultured microbial cells extracted from complex samples, or the development of new high-speed phenotype-based cell-sorting devices. Whereas fluorescence microscopes and FACS instruments are already widely available to most microbiome researchers, university core facilities are now beginning to incorporate advanced microscopy techniques (such as Raman microspectroscopy and cryo-electron tomography), microfluidics, and nanofabrication equipment.
We predict that once broadly applied, next-generation physiology approaches will greatly help with the transition of microbiome research from correlative studies to a causal understanding of microbial activity and function.
Acknowledgements
We are grateful to Anthony Kohtz for generating the Raman data depicted in figure 3. We thank Robin Gerlach and Heidi Smith (Montana State University), James Hemp (University of Utah), members of the Hatzenpichler lab – Anthony Kohtz, Mackenzie Lynes, and Nicholas Reichart – as well as the three reviewers for critical comments that improved the manuscript. Next-generation-physiology research in the Hatzenpichler lab is supported through grants by the Gordon and Betty Moore Foundation (GBMF5999), the National Science Foundation (MCB award 1817428 and RII Track-2 FEC award 1736255), as well as an Early Career Fellowship by the National Aeronautics and Space Administration to R. H. (80NSSC19K0449). Montana State University’s Confocal Raman microscope was acquired with support by the National Science Foundation (DBI- 1726561) and the M. J. Murdock Charitable Trust (SR-2017331).
Glossary
- Microbiome
Synonymous with microbial community; all microscopic organisms, including archaea, bacteria, unicellular eukaryotes, and their viruses, within a sample
- Phenotype
An observable characteristic of an organism that is manifested on molecular, cellular, or population level. A phenotype of a cell varies over time and with changing physicochemical conditions.
- Genotypes
A genotype is the set of genes or the entire genome of an organism
- Reporters
Molecules or chemical motifs that can be specifically traced within the cell; ideally, the reporter group is entirely absent from the target cell under natural conditions
- Ecophysiology
The functioning of a cell in its native habitat under a given set of conditions, including interactions with other cells and the abiotic environment
- Metagenomics
Random shotgun sequencing of DNA from a sample containing more than one genotype
- Single cell genomics
An individual cell is separated from a microbiome and its genome is amplified and sequenced
- Microautoradiography (MAR)
A method that detects uptake of radioactively labeled substrates into cells through formation of silver grains after exposure to a photographic emulsion. MAR is limited in its wide-spread application because of its dependency on isotopes with a suitable half-life, its low throughput, and its destructive nature.
- Nano-scale secondary ion mass spectroscopy (nanoSIMS)
A technique that expels secondary ions from a sample surface through a focused ion beam in high vacuum, extracts them by an electric field, and analyzes them by time of flight mass spectrometry. nanoSIMS provides unrivaled sensitivity and spatial resolution but has very low throughput and destroys the sample
- Quantitative SIP (qSIP)
A technique that separates isotopically heavy biomolecules (for example, 13C-containing DNA) from unlabeled molecules by buoyant density centrifugation. By collecting multiple density fractions and determining their taxonomic and genetic makeup, taxon-specific isotope enrichments can be calculated.
- Next-generation physiology
Any approach enabling to study the physiology of an individual cell in a microbiome in a non-destructive way, thus enabling physical separation of this cell based on its phenotype for further downstream applications
- Raman activated cell sorting (RACS)
A set of techniques that combines Raman spectral acquisition with single cell separation
- Fluorescence in situ hybridization (FISH)
A technique that uses single-stranded DNA probes and fluorescence microscopy to visualize cells based on their taxonomic identity (rRNA FISH) or gene expression (mRNA FISH)
- Intact cells
cells that have not been exposed to a chemical fixative (such as formaldehyde or ethanol) that might interfere with downstream analyses (such as cultivation or DNA-sequencing)
- Metabolically active
A cell carrying out specific metabolic function (such as redox activity or activity of a specific enzyme); this term is agnostic about whether this activity leads to the build-up of new biomass (that is, anabolic activity)
- Anabolically active
Performing de novo synthesis of specific macromolecules (e.g. DNA, RNA, proteins, and lipids)
- Silent region
Area in the Raman spectrum of a cell that is free of background interference from cellular vibrations (~1,800–2,700 cm−1)
- Click chemistry
Summary term for a range of reactions with a high thermodynamic driving force and extremely high yields and reaction efficiencies. The term is often used synonymously for azide-alkyne cycloaddition reactions, which are the most commonly used type of click chemistry reactions in biology
- Clickable
Used here to characterize a molecule carrying a functional group that is amenable to azide-alkyne click chemistry
- Bioorthogonal reaction
A reaction that does not interfere with biological processes; can be used to label a cell or molecule with a reporter
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Microbiology thanks Wei Huang, Aaron Wright and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gilbert JA et al. Current understanding of the human microbiome. Nat Med 24, 392–400, doi: 10.1038/nm.4517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flemming HC & Wuertz S Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol 17, 247–260, doi: 10.1038/s41579-019-0158-9 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Castelle CJ & Banfield JF Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell 172, 1181–1197, doi: 10.1016/j.cell.2018.02.016 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Karst SM et al. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol 36, 190–195, doi: 10.1038/nbt.4045 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hug LA et al. A new view of the tree of life. Nat Microbiol 1, 16048, doi: 10.1038/nmicrobiol.2016.48 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ackermann M A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13, 497–508, doi: 10.1038/nrmicro3491 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Lidstrom ME & Konopka MC The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol 6, 705–712, doi: 10.1038/nchembio.436 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Ansorge R et al. Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nat Microbiol, doi: 10.1038/s41564-019-0572-9 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Gruber-Dorninger C et al. Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9, 643–655, doi: 10.1038/ismej.2014.156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasdekis AE & Stephanopoulos G Review of methods to probe single cell metabolism and bioenergetics. Metab Eng 27, 115–135, doi: 10.1016/j.ymben.2014.09.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenthal K, Oehling V, Dusny C & Schmid A Beyond the bulk: disclosing the life of single microbial cells. FEMS Microbiol Rev 41, 751–780, doi: 10.1093/femsre/fux044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nai C & Meyer V From Axenic to Mixed Cultures: Technological Advances Accelerating a Paradigm Shift in Microbiology. Trends Microbiol 26, 538–554, doi: 10.1016/j.tim.2017.11.004 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Oomen PE, Aref MA, Kaya I, Phan NTN & Ewing AG Chemical Analysis of Single Cells. Anal Chem 91, 588–621, doi: 10.1021/acs.analchem.8b04732 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Taheri-Araghi S, Brown SD, Sauls JT, McIntosh DB & Jun S Single-Cell Physiology. Annu Rev Biophys 44, 123–142, doi: 10.1146/annurev-biophys-060414-034236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd KG, Steen AD, Ladau J, Yin J & Crosby L Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 3, doi: 10.1128/mSystems.00055-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen AD et al. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J, doi: 10.1038/s41396-019-0484-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen JL, Christensen D, Kloppenborg M & Nielsen PH Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ Microbiol 5, 202–211 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lee N et al. Combination of fluorescent in situ hybridization and microautoradiography-a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol 65, 1289–1297 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlynn SE, Chadwick GL, Kempes CP & Orphan VJ Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Kopf SH et al. Heavy water and 15N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ Microbiol 17, 2542–2556, doi: 10.1111/1462-2920.12752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hungate BA et al. Quantitative microbial ecology through stable isotope probing. Appl Environ Microbiol 81, 7570–7581, doi: 10.1128/AEM.02280-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziels RM, Sousa DZ, Stensel HD & Beck DAC DNA-SIP based genome-centric metagenomics identifies key long-chain fatty acid-degrading populations in anaerobic digesters with different feeding frequencies. ISME J 12, 112–123, doi: 10.1038/ismej.2017.143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eyice O et al. SIP metagenomics identifies uncultivated Methylophilaceae as dimethylsulphide degrading bacteria in soil and lake sediment. ISME J 9, 2336–2348, doi: 10.1038/ismej.2015.37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortunato CS & Huber JA Coupled RNA-SIP and metatranscriptomics of active chemolithoautotrophic communities at a deep-sea hydrothermal vent. ISME J 10, 1925–1938, doi: 10.1038/ismej.2015.258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doud DFR & Woyke T Novel approaches in function-driven single-cell genomics. FEMS Microbiol Rev 41, 538–548, doi: 10.1093/femsre/fux009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer E, Wagner M & Woyke T Capturing the genetic makeup of the active microbiome in situ. The ISME Journal 11, 1949–1963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan X et al. Effect of Laser Irradiation on Cell Function and Its Implications in Raman Spectroscopy. Appl Environ Microbiol 84, doi: 10.1128/AEM.02508-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Wang X, Ma B & Xu J Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol Adv 37, 107388, doi: 10.1016/j.biotechadv.2019.04.010 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Harrison JP & Berry D Vibrational Spectroscopy for Imaging Single Microbial Cells in Complex Biological Samples. Front Microbiol 8, 675, doi: 10.3389/fmicb.2017.00675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorenz B, Wichmann C, Stockel S, Rosch P & Popp J Cultivation-Free Raman Spectroscopic Investigations of Bacteria. Trends Microbiol 25, 413–424, doi: 10.1016/j.tim.2017.01.002 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q et al. Towards high-throughput microfluidic Raman-activated cell sorting. Analyst 140, 6163–6174, doi: 10.1039/c5an01074h (2015). [DOI] [PubMed] [Google Scholar]
- 32.Huang WE, Ward AD & Whiteley AS Raman tweezers sorting of single microbial cells. Environ Microbiol Rep 1, 44–49, doi: 10.1111/j.1758-2229.2008.00002.x (2009). [DOI] [PubMed] [Google Scholar]
- 33.Lee KS et al. An automated Raman-based platform for the sorting of live cells by functional properties. Nat Microbiol 4, 1035–1048, doi: 10.1038/s41564-019-0394-9 (2019).This study describes the development and application of the first microfluidic platform for automated Raman-activated sorting of isotope-labelled microorganisms. SIP-RACS and metagenomics are used to characterize mucin-degrading bacteria from a mouse colon.
- 34.McIlvenna D et al. Continuous cell sorting in a flow based on single cell resonance Raman spectra. Lab on a chip 16, 1420–1429, doi: 10.1039/c6lc00251j (2016). [DOI] [PubMed] [Google Scholar]
- 35.Wang X et al. Raman-Activated Droplet Sorting (RADS) for Label-Free High-Throughput Screening of Microalgal Single-Cells. Anal Chem 89, 12569–12577, doi: 10.1021/acs.analchem.7b03884 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Song Y et al. Raman-Deuterium Isotope Probing for in-situ identification of antimicrobial resistant bacteria in Thames River. Sci Rep 7, 16648, doi: 10.1038/s41598-017-16898-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing X et al. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean. Environ Microbiol 20, 2241–2255, doi: 10.1111/1462-2920.14268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb Biotechnol, doi: 10.1111/1751-7915.12420 (2016).This study uses label-free Raman-activated cell sorting and single cell genomics to characterize yet uncultured carotenoid-containing microorganisms.
- 39.Wang Y et al. Raman activated cell ejection for isolation of single cells. Anal Chem 85, 10697–10701, doi: 10.1021/ac403107p (2013). [DOI] [PubMed] [Google Scholar]
- 40.Rinke C et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 9, 1038–1048, doi: 10.1038/nprot.2014.067 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Rinke C et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437, doi: 10.1038/nature12352 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Couradeau E et al. Probing the active fraction of soil microbiomes using BONCAT-FACS. Nat Commun 10, 2770, doi: 10.1038/s41467-019-10542-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morono Y, Terada T, Kallmeyer J & Inagaki F An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environ Microbiol 15, 2841–2849, doi: 10.1111/1462-2920.12153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eichorst SA et al. Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol Ecol 91, doi: 10.1093/femsec/fiv106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lunau M, Lemke A, Walther K, Martens-Habbena W & Simon M An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ Microbiol 7, 961–968 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Hatzenpichler R et al. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci U S A 113, E4069–E4078 (2016).This study uses BONCAT-FISH and BONCAT-FACS in combination with 16S rRNA gene sequencing to characterize translationally active methane-oxidizing microbial consortia in deep sea sediments.
- 47.Hao L et al. Novel prosthecate bacteria from the candidate phylum Acetothermia. ISME J 12, 2225–2237, doi: 10.1038/s41396-018-0187-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clingenpeel S, Schwientek P, Hugenholtz P & Woyke T Effects of sample treatments on genome recovery via single-cell genomics. ISME J 8, 2546–2549, doi: 10.1038/ismej.2014.92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clingenpeel S, Clum A, Schwientek P, Rinke C & Woyke T Reconstructing each cell’s genome within complex microbial communities-dream or reality? Front Microbiol 5, 771, doi: 10.3389/fmicb.2014.00771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowers RM et al. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16, 856, doi: 10.1186/s12864-015-2063-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cross KL et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37, 1314–1321, doi: 10.1038/s41587-019-0260-6 (2019).This study employs genome-informed antibody engineering to sort individual TM7 and SR1 cells from oral microbiome samples and regrows them in cultivation media.
- 52.Kalyuzhnaya MG, Lidstrom ME & Chistoserdova L Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J 2, 696–706, doi: 10.1038/ismej.2008.32 (2008).This study demonstrates that, if carefully applied, redox sensing dyes can be used to sort metabolically active methylotrophic bacteria via FACS and bring sorted cells into enrichment culture.
- 53.Koch C, Gunther S, Desta AF, Hubschmann T & Muller S Cytometric fingerprinting for analyzing microbial intracommunity structure variation and identifying subcommunity function. Nat Protoc 8, 190–202, doi: 10.1038/nprot.2012.149 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Lambrecht J et al. Flow cytometric quantification, sorting and sequencing of methanogenic archaea based on F420 autofluorescence. Microb Cell Fact 16, 180, doi: 10.1186/s12934-017-0793-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berry D et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A 112, E194–203, doi: 10.1073/pnas.1420406112 (2015).This study for the first time combines heavy water labeling, Raman-activated cell sorting and 16S rRNA gene sequencing and uses this workflow to identify glucosamine- and mucin-degrading bacteria from mouse cecal samples.
- 56.Kopf SH et al. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113, E110–116, doi: 10.1073/pnas.1512057112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J et al. Raman Deuterium Isotope Probing Reveals Microbial Metabolism at the Single-Cell Level. Anal Chem 89, 13305–13312, doi: 10.1021/acs.analchem.7b03461 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Huang WE, Cui L & Wagner M Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr Opin Biotechnol 41, 34–42, doi: 10.1016/j.copbio.2016.04.018 (2016).This review provides an excellent overview of the principle and potential applications of single cell targeted SIP-Raman studies.
- 59.Huang WE et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol 9, 1878–1889 (2007).This study combines Raman microspectroscopy and FISH for the first time and uses it to identify 13C-naphtalene-degraders in groundwater and quantifies isotope incoportation into individual cells.
- 60.Huang WE et al. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell raman-fluorescence in situ hybridization. Appl Environ Microbiol 75, 234–241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang WE, Griffiths RI, Thompson IP, Bailey MJ & Whiteley AS Raman microscopic analysis of single microbial cells. Anal Chem 76, 4452–4458 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Haider S et al. Raman microspectroscopy reveals long-term extracellular activity of Chlamydiae. Mol Microbiol 77, 687–700, doi: 10.1111/j.1365-2958.2010.07241.x (2010). [DOI] [PubMed] [Google Scholar]
- 63.Angel R et al. Application of stable-isotope labelling techniques for the detection of active diazotrophs. Environ Microbiol 20, 44–61, doi: 10.1111/1462-2920.13954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M et al. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J 6, 875–885, doi: 10.1038/ismej.2011.150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller AL et al. Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ Microbiol 20, 2927–2940, doi: 10.1111/1462-2920.14297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor GT et al. Single-Cell Growth Rates in Photoautotrophic Populations Measured by Stable Isotope Probing and Resonance Raman Microspectrometry. Front Microbiol 8, 1449, doi: 10.3389/fmicb.2017.01449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu F, Shi L & Min W Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nat Methods 16, 830–842, doi: 10.1038/s41592-019-0538-0 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Wei L et al. Live-Cell Bioorthogonal Chemical Imaging: Stimulated Raman Scattering Microscopy of Vibrational Probes. Acc Chem Res 49, 1494–1502, doi: 10.1021/acs.accounts.6b00210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crespi HL, Conard SM, Uphaus RA & Katz JJ Cultivation of microorganisms in heavy water. Ann N Y Acad Sci 84, 648–666, doi: 10.1111/j.1749-6632.1960.tb39098.x (1960). [DOI] [PubMed] [Google Scholar]
- 70.Kselikova V, Vitova M & Bisova K Deuterium and its impact on living organisms. Folia Microbiol (Praha) 64, 673–681, doi: 10.1007/s12223-019-00740-0 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Gillespie AL & Sessions AL Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc Natl Acad Sci U S A 106, 12580–12586, doi: 10.1073/pnas.0903030106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valentine DL, Sessions AL, Tyler SC & Chidthaisong A Hydrogen isotope fractionation during H2/CO2 acetogenesis: Hydrogen utilization efficiency and the origin of lipid-bound hydrogen. Geobiology 2, 179–188 (2004). [Google Scholar]
- 73.Sessions AL, Jahnke LL, Schimmelmann A & Hayes JM Hydrogen isotope fractionation in lipids of the methane-oxidizing bacterium Methylococcus capsulatus. Geochimica Et Cosmochimica Acta 66, 3955–3969 (2002). [Google Scholar]
- 74.Lawrence AD et al. Construction of Fluorescent Analogs to Follow the Uptake and Distribution of Cobalamin (Vitamin B12) in Bacteria, Worms, and Plants. Cell Chem Biol 25, 941–951 e946, doi: 10.1016/j.chembiol.2018.04.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuru E et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 51, 12519–12523, doi: 10.1002/anie.201206749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tao J et al. Use of a Fluorescent Analog of Glucose (2-NBDG) To Identify Uncultured Rumen Bacteria That Take Up Glucose. Appl Environ Microbiol 85, doi: 10.1128/AEM.03018-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Garcia M et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 7, e35314, doi: 10.1371/journal.pone.0035314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doud DFR et al. Function-driven single-cell genomics uncovers cellulose-degrading bacteria from the rare biosphere. ISME J, doi: 10.1038/s41396-019-0557-y (2019).This study uses fluorescent substrate analog probing, FACS of active cells, and mini-metagenomics to identify thermophilic cellulose degraders.
- 79.Rosnow JJ et al. A Cobalamin Activity-Based Probe Enables Microbial Cell Growth and Finds New Cobalamin-Protein Interactions across Domains. Appl Environ Microbiol 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liechti GW et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510, doi: 10.1038/nature12892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Best MD Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 48, 6571–6584, doi: 10.1021/bi9007726 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Devaraj NK The Future of Bioorthogonal Chemistry. ACS Cent Sci 4, 952–959, doi: 10.1021/acscentsci.8b00251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sletten EM & Bertozzi CR Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl 48, 6974–6998, doi: 10.1002/anie.200900942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Griffin RJ The medicinal chemistry of the azido group. Prog Med Chem 31, 121–232 (1994). [DOI] [PubMed] [Google Scholar]
- 85.Marchand JA et al. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–424, doi: 10.1038/s41586-019-1020-y (2019). [DOI] [PubMed] [Google Scholar]
- 86.Zhu X, Liu J & Zhang W De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol 11, 115–120, doi: 10.1038/nchembio.1718 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Zhu X & Zhang W Terminal Alkyne Biosynthesis in Marine Microbes. Methods Enzymol 604, 89–112, doi: 10.1016/bs.mie.2018.01.040 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Wei L et al. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat Methods 11, 410–412, doi: 10.1038/nmeth.2878 (2014).This study combines non-canonical substrate analog probing and Raman microspectroscopy to visualize alkyne-containing nucleoside, amino acid, and fatty acid analogs in a variety of eukaryotic cells.
- 89.Sinai L, Rosenberg A, Smith Y, Segev E & Ben-Yehuda S The molecular timeline of a reviving bacterial spore. Mol Cell 57, 695–707, doi: 10.1016/j.molcel.2014.12.019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shieh P, Siegrist MS, Cullen AJ & Bertozzi CR Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci U S A 111, 5456–5461, doi: 10.1073/pnas.1322727111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bagert JD et al. Time-resolved proteomic analysis of quorum sensing in Vibrio harveyi. Chem Sci 7, 1797–1806, doi: 10.1039/C5SC03340C (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Babin BM et al. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113, E597–605, doi: 10.1073/pnas.1514412113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mahdavi A et al. Engineered Aminoacyl-tRNA Synthetase for Cell-Selective Analysis of Mammalian Protein Synthesis. J Am Chem Soc 138, 4278–4281, doi: 10.1021/jacs.5b08980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Glenn WS et al. BONCAT enables time-resolved analysis of protein synthesis in native plant tissue. Plant Physiol 173, 1543–1553, doi: 10.1104/pp.16.01762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Calve S, Witten AJ, Ocken AR & Kinzer-Ursem TL Incorporation of non-canonical amino acids into the developing murine proteome. Sci Rep 6, 32377, doi: 10.1038/srep32377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuet KP et al. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc Natl Acad Sci U S A 112, 2705–2710, doi: 10.1073/pnas.1421567112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dieterich DC et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13, 897–905, doi: 10.1038/nn.2580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taymaz-Nikerel H, Borujeni AE, Verheijen PJ, Heijnen JJ & van Gulik WM Genome-derived minimal metabolic models for Escherichia coli MG1655 with estimated in vivo respiratory ATP stoichiometry. Biotechnol Bioeng 107, 369–381, doi: 10.1002/bit.22802 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Beck AE, Hunt KA & Carlson RP Measuring Cellular Biomass Composition for Computational Biology Applications. processes 6, 1–27 (2018). [Google Scholar]
- 100.Zavrel T et al. Quantitative insights into the cyanobacterial cell economy. Elife 8, doi: 10.7554/eLife.42508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beck AE, Hunt KA & Carlson RP Measuring Cellular Biomass Composition for Computational Biology Applications. Processes 6, doi: 10.3390/pr6050038 (2018). [DOI] [Google Scholar]
- 102.Beatty KE et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl 45, 7364–7367, doi: 10.1002/anie.200602114 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Dieterich DC, Link AJ, Graumann J, Tirrell DA & Schuman EM Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A 103, 9482–9487, doi: 10.1073/pnas.0601637103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kiick KL, Saxon E, Tirrell DA & Bertozzi CR Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A 99, 19–24, doi: 10.1073/pnas.012583299 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lang K & Chin JW Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev 114, 4764–4806, doi: 10.1021/cr400355w (2014). [DOI] [PubMed] [Google Scholar]
- 106.Ngo JT & Tirrell DA Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc Chem Res 44, 677–685, doi: 10.1021/ar200144y (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chakrabarti S, Liehl P, Buchon N & Lemaitre B Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12, 60–70, doi: 10.1016/j.chom.2012.06.001 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Sherratt AR et al. Rapid Screening and Identification of Living Pathogenic Organisms via Optimized Bioorthogonal Non-canonical Amino Acid Tagging. Cell Chem Biol 24, 1048–1055 e1043, doi: 10.1016/j.chembiol.2017.06.016 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Mahdavi A et al. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc Natl Acad Sci U S A 111, 433–438, doi: 10.1073/pnas.1301740111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ouellette SP, Dorsey FC, Moshiach S, Cleveland JL & Carabeo RA Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS One 6, e16783, doi: 10.1371/journal.pone.0016783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siegrist MS et al. (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 8, 500–505, doi: 10.1021/cb3004995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hatzenpichler R et al. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol 16, 2568–2590, doi: 10.1111/1462-2920.12436 (2014).This study for the first time uses bioorthogonal labeling and click chemistry on complex, multi-species samples and demonstrates that BONCAT-FISH can be used to link the identify and in situ function of uncultured microorganisms.
- 113.Samo TJ, Smriga S, Malfatti F, Sherwood BP & Azam F Broad distribution and high proportion of protein synthesis active marine bacteria revealed by click chemistry at the single cell level. Front Mar Sci 1, doi:doi: 10.3389/fmars.2014.00048 (2014). [DOI] [Google Scholar]
- 114.Leizeaga A, Estrany M, Forn I & Sebastian M Using Click-Chemistry for Visualizing in Situ Changes of Translational Activity in Planktonic Marine Bacteria. Front Microbiol 8, 2360, doi: 10.3389/fmicb.2017.02360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sebastian M et al. High Growth Potential of Long-Term Starved Deep Ocean Opportunistic Heterotrophic Bacteria. Front Microbiol 10, 760, doi: 10.3389/fmicb.2019.00760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kjeldsen KU et al. On the evolution and physiology of cable bacteria. Proc Natl Acad Sci U S A 116, 19116–19125, doi: 10.1073/pnas.1903514116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Geva-Zatorsky N et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med 21, 1091–1100, doi: 10.1038/nm.3929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hatzenpichler R & Orphan VJ in Hydrocarbon and Lipid Microbiology Protocols Vol. Vol. 7: Single-cell and single-molecule methods (ed McGenity TJ) 145–157 (Springer, 2015). [Google Scholar]
- 119.Pasulka AL et al. Interrogating marine virus-host interactions and elemental transfer with BONCAT and nanoSIMS-based methods. Environ Microbiol 20, 671–692, doi: 10.1111/1462-2920.13996 (2018).This study applies BONCAT to microscopically quantify viral production rates in model systems and seawater and nanoSIMS to quantify carbon and nitrogen transfer rates between viruses and their microbial hosts.
- 120.Muller TG, Sakin V & Muller B A Spotlight on Viruses-Application of Click Chemistry to Visualize Virus-Cell Interactions. Molecules 24, 1–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bagert JD et al. Quantitative, Time-Resolved Proteomic Analysis by Combining Bioorthogonal Noncanonical Amino Acid Tagging and Pulsed Stable Isotope Labeling by Amino Acids in Cell Culture. Mol Cell Proteomics 13, 1352–1358, doi: 10.1074/mcp.M113.031914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lehner F et al. The Impact of Azidohomoalanine Incorporation on Protein Structure and Ligand Binding. Chembiochem 18, 2340–2350, doi: 10.1002/cbic.201700437 (2017). [DOI] [PubMed] [Google Scholar]
- 123.Bennett BD et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5, 593–599, doi: 10.1038/nchembio.186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Neufeld JD et al. DNA stable-isotope probing. Nat Protoc 2, 860–866 (2007). [DOI] [PubMed] [Google Scholar]
- 125.Urbach E, Vergin KL & Giovannoni SJ Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl Environ Microbiol 65, 1207–1213 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Papp K et al. Quantitative stable isotope probing with H2(18)O reveals that most bacterial taxa in soil synthesize new ribosomal RNA. ISME J 12, 3043–3045, doi: 10.1038/s41396-018-0233-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pernthaler A, Pernthaler J, Schattenhofer M & Amann R Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl Environ Microbiol 68, 5728–5736, doi: 10.1128/aem.68.11.5728-5736.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hamasaki K Comparison of Bromodeoxyuridine Immunoassay with Tritiated Thymidine Radioassay for Measuring Bacterial Productivity in Oceanic Waters. Journal of Oceanography 62, 793–799 (2006). [Google Scholar]
- 129.Olaniyi OO, Yang K, Zhu YG & Cui L Heavy water-labeled Raman spectroscopy reveals carboxymethylcellulose-degrading bacteria and degradation activity at the single-cell level. Appl Microbiol Biotechnol 103, 1455–1464, doi: 10.1007/s00253-018-9459-6 (2019). [DOI] [PubMed] [Google Scholar]
- 130.Smriga S, Samo TJ, Malfatti F, Villareal J & Azam F Individual cell DNA synthesis within natural marine bacterial assemblages as detected by ‘click’ chemistry. Aquat Microb Ecol 72, 269–280 (2014). [Google Scholar]
- 131.Jao CY & Salic A Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105, 15779–15784, doi: 10.1073/pnas.0808480105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kho Y et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A 101, 12479–12484, doi: 10.1073/pnas.0403413101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Neef AB & Schultz C Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed Engl 48, 1498–1500, doi: 10.1002/anie.200805507 (2009). [DOI] [PubMed] [Google Scholar]
- 134.Garcia-Heredia A et al. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7, doi: 10.7554/eLife.37243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bublitz DC et al. Peptidoglycan Production by an Insect-Bacterial Mosaic. Cell 179, 703–712 e707, doi: 10.1016/j.cell.2019.08.054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saxon E et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J Am Chem Soc 124, 14893–14902 (2002). [DOI] [PubMed] [Google Scholar]
- 137.Siegrist MS, Swarts BM, Fox DM, Lim SA & Bertozzi CR Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev 39, 184–202, doi: 10.1093/femsre/fuu012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ohno S et al. A method for evaluating the host range of bacteriophages using phages fluorescently labeled with 5-ethynyl-2’-deoxyuridine (EdU). Appl Microbiol Biotechnol 95, 777–788, doi: 10.1007/s00253-012-4174-1 (2012). [DOI] [PubMed] [Google Scholar]
- 139.Liu Y et al. Advancing understanding of microbial bioenergy conversion processes by activity-based protein profiling. Biotechnol Biofuels 8, 156, doi: 10.1186/s13068-015-0343-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jehmlich N, Vogt C, Lunsmann V, Richnow HH & von Bergen M Protein-SIP in environmental studies. Curr Opin Biotechnol 41, 26–33, doi: 10.1016/j.copbio.2016.04.010 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Sadler NC & Wright AT Activity-based protein profiling of microbes. Curr Opin Chem Biol 24, 139–144, doi: 10.1016/j.cbpa.2014.10.022 (2015). [DOI] [PubMed] [Google Scholar]
- 142.Whidbey C & Wright AT Activity-Based Protein Profiling-Enabling Multimodal Functional Studies of Microbial Communities. Curr Top Microbiol Immunol 420, 1–21, doi: 10.1007/82_2018_128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Willems LI, Overkleeft HS & van Kasteren SI Current developments in activity-based protein profiling. Bioconjugate chemistry 25, 1181–1191, doi: 10.1021/bc500208y (2014). [DOI] [PubMed] [Google Scholar]
- 144.Whidbey C et al. A Probe-Enabled Approach for the Selective Isolation and Characterization of Functionally Active Subpopulations in the Gut Microbiome. J Am Chem Soc 141, 42–47, doi: 10.1021/jacs.8b09668 (2019).This study demonstrates the high potential of activity-based protein profiling for single cell physiology studies. Activity-based protein profiling is combined with fluorescence-actviated cell sorting and 16S rRNA gene sequencing to identify β-glucuronidase active members of the mouse gut microbiome.
- 145.Flemming HC & Wingender J The biofilm matrix. Nat Rev Microbiol 8, 623–633, doi: 10.1038/nrmicro2415 (2010). [DOI] [PubMed] [Google Scholar]
- 146.Schlafer S & Meyer RL Confocal microscopy imaging of the biofilm matrix. J Microbiol Methods 138, 50–59, doi: 10.1016/j.mimet.2016.03.002 (2017). [DOI] [PubMed] [Google Scholar]
- 147.Kehe J et al. Massively parallel screening of synthetic microbial communities. Proc Natl Acad Sci U S A 116, 12804–12809, doi: 10.1073/pnas.1900102116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lan F, Demaree B, Ahmed N & Abate AR Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol 35, 640–646, doi: 10.1038/nbt.3880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Spencer SJ et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J 10, 427–436, doi: 10.1038/ismej.2015.124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Terekhov SS et al. Ultrahigh-throughput functional profiling of microbiota communities. Proc Natl Acad Sci U S A 115, 9551–9556, doi: 10.1073/pnas.1811250115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kolinko S et al. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ Microbiol 14, 1709–1721, doi: 10.1111/j.1462-2920.2011.02609.x (2012). [DOI] [PubMed] [Google Scholar]
- 152.Spang A et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14, 3122–3145, doi: 10.1111/j.1462-2920.2012.02893.x (2012). [DOI] [PubMed] [Google Scholar]
- 153.Chan JW et al. Reagentless identification of single bacterial spores in aqueous solution by confocal laser tweezers Raman spectroscopy. Anal Chem 76, 599–603, doi: 10.1021/ac0350155 (2004). [DOI] [PubMed] [Google Scholar]
- 154.Fernando EY et al. Resolving the individual contribution of key microbial populations to enhanced biological phosphorus removal with Raman-FISH. ISME J 13, 1933–1946, doi: 10.1038/s41396-019-0399-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Majed N, Chernenko T, Diem M & Gu AZ Identification of functionally relevant populations in enhanced biological phosphorus removal processes based on intracellular polymers profiles and insights into the metabolic diversity and heterogeneity. Environ Sci Technol 46, 5010–5017, doi: 10.1021/es300044h (2012). [DOI] [PubMed] [Google Scholar]
- 156.Pätzold R et al. In situ mapping of nitrifiers and anammox bacteria in microbial aggregates by means of confocal resonance Raman microscopy. Journal of Microbiological Methods 72, 241–248 (2008). [DOI] [PubMed] [Google Scholar]
- 157.Berg JS, Schwedt A, Kreutzmann A-C, Kuypers MMM & Milucka J Polysulfides as Intermediates in the Oxidation of Sulfide to Sulfate by Beggiatoa spp. Appl Environ Microbiol 80, 629–636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bjerg JT et al. Long-distance electron transport in individual, living cable bacteria. Proc Natl Acad Sci U S A 115, 5786–5791, doi: 10.1073/pnas.1800367115 (2018).This study uses Raman microspectroscopy to visualize gradients in cytochrome redox states along living cable bacteria.
- 159.Eder SH, Gigler AM, Hanzlik M & Winklhofer M Sub-micrometer-scale mapping of magnetite crystals and sulfur globules in magnetotactic bacteria using confocal Raman micro-spectrometry. PLoS One 9, e107356, doi: 10.1371/journal.pone.0107356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gruber-Vodicka HR et al. Paracatenula, an ancient symbiosis between thiotrophic Alphaproteobacteria and catenulid flatworms. Proc Natl Acad Sci U S A 108, 12078–12083, doi: 10.1073/pnas.1105347108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cui L, Yang K, Zhou G, Huang WE & Zhu YG Surface-Enhanced Raman Spectroscopy Combined with Stable Isotope Probing to Monitor Nitrogen Assimilation at Both Bulk and Single-Cell Level. Anal Chem 89, 5793–5800, doi: 10.1021/acs.analchem.6b04913 (2017). [DOI] [PubMed] [Google Scholar]
- 162.Taubert M et al. Tracking active groundwater microbes with D2O labelling to understand their ecosystem function. Environ Microbiol 20, 369–384, doi: 10.1111/1462-2920.14010 (2018). [DOI] [PubMed] [Google Scholar]
- 163.Ando J, Palonpon AF, Sodeoka M & Fujita K High-speed Raman imaging of cellular processes. Current Opinion in Chemical Biology 33, 16–24, doi: 10.1016/j.cbpa.2016.04.005 (2016). [DOI] [PubMed] [Google Scholar]
- 164.Chisanga M, Muhamadali H, Ellis DI & Goodacre R Surface-Enhanced Raman Scattering (SERS) in Microbiology: Illumination and Enhancement of the Microbial World. Appl Spectrosc 72, 987–1000, doi: 10.1177/0003702818764672 (2018). [DOI] [PubMed] [Google Scholar]
- 165.Ivleva NP, Kubryk P & Niessner R Raman microspectroscopy, surface-enhanced Raman scattering microspectroscopy, and stable-isotope Raman microspectroscopy for biofilm characterization. Anal Bioanal Chem 409, 4353–4375, doi: 10.1007/s00216-017-0303-0 (2017).This review discusses the potential of advanced Raman microspectroscopy application, with a focus on the characterization of the extracellular polymeric substances and cells in microbial biofilms.
- 166.Cicerone M Molecular imaging with CARS micro-spectroscopy. Curr Opin Chem Biol 33, 179–185, doi: 10.1016/j.cbpa.2016.05.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Camp CH & Cicerone MT Chemically sensitive bioimaging with coherent Raman scattering. Nat Photonics 9, 295–305, doi: 10.1038/Nphoton.2015.60 (2015). [DOI] [Google Scholar]
- 168.Opilik L, Schmid T & Zenobi R Modern Raman imaging: vibrational spectroscopy on the micrometer and nanometer scales. Annu Rev Anal Chem (Palo Alto Calif) 6, 379–398, doi: 10.1146/annurev-anchem-062012-092646 (2013). [DOI] [PubMed] [Google Scholar]
- 169.Ivleva NP, Wagner M, Horn H, Niessner R & Haisch C Raman microscopy and surface-enhanced Raman scattering (SERS) for in situ analysis of biofilms. J Biophotonics 3, 548–556, doi: 10.1002/jbio.201000025 (2010). [DOI] [PubMed] [Google Scholar]
- 170.Kubryk P et al. Exploring the Potential of Stable Isotope (Resonance) Raman Microspectroscopy and Surface-Enhanced Raman Scattering for the Analysis of Microorganisms at Single Cell Level. Anal Chem 87, 6622–6630, doi: 10.1021/acs.analchem.5b00673 (2015). [DOI] [PubMed] [Google Scholar]
- 171.Weiss R et al. Surface-enhanced Raman spectroscopy of microorganisms: limitations and applicability on the single-cell level. Analyst 144, 943–953, doi: 10.1039/c8an02177e (2019). [DOI] [PubMed] [Google Scholar]
- 172.Kolb HC, Finn MG & Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl 40, 2004–2021 (2001). [DOI] [PubMed] [Google Scholar]
- 173.Moses JE & Moorhouse AD The growing applications of click chemistry. Chem Soc Rev 36, 1249–1262, doi: 10.1039/b613014n (2007). [DOI] [PubMed] [Google Scholar]
- 174.Codelli JA, Baskin JM, Agard NJ & Bertozzi CR Second-generation difluorinated cyclooctynes for copper-free click chemistry. J Am Chem Soc 130, 11486–11493, doi: 10.1021/ja803086r (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agard NJ, Prescher JA & Bertozzi CR A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc 126, 15046–15047, doi: 10.1021/ja044996f (2004). [DOI] [PubMed] [Google Scholar]
- 176.Uttamapinant C et al. Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew Chem Int Ed Engl 51, 5852–5856, doi: 10.1002/anie.201108181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shieh P et al. CalFluors: A Universal Motif for Fluorogenic Azide Probes across the Visible Spectrum. J Am Chem Soc 137, 7145–7151, doi: 10.1021/jacs.5b02383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zimmermann M et al. Phenotypic heterogeneity in metabolic traits among single cells of a rare bacterial species in its natural environment quantified with a combination of flow cell sorting and NanoSIMS. Front Microbiol 6, 243, doi: 10.3389/fmicb.2015.00243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Netuschil L, Auschill TM, Sculean A & Arweiler NB Confusion over live/dead stainings for the detection of vital microorganisms in oral biofilms - which stain is suitable? BMC Oral Health 14, 1–12 (2014).This review provides an in-depth discussion of the short-comings of viability and vitality stains.
- 180.Emerson JB et al. Schrodinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86, doi: 10.1186/s40168-017-0285-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Konopka MC et al. Respiration response imaging for real-time detection of microbial function at the single-cell level. Appl Environ Microbiol 77, 67–72, doi: 10.1128/AEM.01166-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ullrich S, Karrasch B, Hoppe H-G, Jeskulke K & Mehrens M Toxic Effects on Bacterial Metabolism of the Redox Dye 5-Cyano-2,3-Ditolyl Tetrazolium Chloride. Appl Environ Microbiol 62, 4587–4593 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hatzinger PB, Palmer P, Smith RL, Penarrieta CT & Yoshinari T Applicability of tetrazolium salts for the measurement of respiratory activity and viability of groundwater bacteria. J Microbiol Methods 52, 47–58 (2003). [DOI] [PubMed] [Google Scholar]
- 184.Karner M & Fuhrman JA Determination of Active Marine Bacterioplankton: a Comparison of Universal 16S rRNA Probes, Autoradiography, and Nucleoid Staining. Appl Environ Microbiol 63, 1208–1213 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Servais P, Agogue H, Courties C, Joux F & Lebaron P Are the actively respiring cells (CTC+) those responsible for bacterial production in aquatic environments? FEMS Microbiol Ecol 35, 171–179, doi: 10.1111/j.1574-6941.2001.tb00801.x (2001). [DOI] [PubMed] [Google Scholar]
- 186.Nielsen JL, Aquino de Muro M & Nielsen PH Evaluation of the redox dye 5-cyano-2,3-tolyl-tetrazolium chloride for activity studies by simultaneous use of microautoradiography and fluorescence in situ hybridization. Appl Environ Microbiol 69, 641–643, doi: 10.1128/aem.69.1.641-643.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Stiefel P, Schmidt-Emrich S, Maniura-Weber K & Ren Q Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiology 15, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Berney M, Hammes F, Bosshard F, Weilenmann HU & Egli T Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl Environ Microbiol 73, 3283–3290, doi: 10.1128/AEM.02750-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nocker A, Cheung CY & Camper AK Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67, 310–320, doi: 10.1016/j.mimet.2006.04.015 (2006). [DOI] [PubMed] [Google Scholar]