Abstract
Rapidly developing technologies have recently fueled an exciting era of discovery in the field of chromosome structure and nuclear organization. In addition to chromosome conformation capture (3C) methods, new alternative techniques have emerged to study genome architecture and biological processes in the nucleus, often in single or living cells. This sets an unprecedented stage for exploring the mechanisms that link chromosome structure and biological function. Here we review popular as well as emerging approaches to study chromosome organization, focusing on the contribution of complementary methodologies to our understanding of structures revealed by 3C methods and their biological implications, and discuss the next technical and conceptual frontiers.
Introduction
The three-dimensional (3D) structure of mammalian genomes has emerged as an important player in fundamental processes occurring in the cell nucleus. Transcription, replication, DNA damage and repair have all been shown to be intimately connected to the way chromosomes are folded in the 3D space. Current knowledge on 3D chromosome structure derives from two complementary classes of techniques. The first (and historically earlier) is microscopy, which revealed many principles of nuclear organization including the existence of sub-nuclear organelles such as the nucleolus (Pederson 2011), nuclear speckles (Spector and Lamond 2011) and polycomb bodies (Pirrotta and Li 2012). Fluorescence RNA and DNA in situ hybridization (FISH) have also revealed that chromosomes occupy distinct chromosome territories (Bolzer et al. 2005; Cremer and Cremer 2001; Stack et al. 1977), and that nuclear positioning can correlate with gene expression levels (Finlan et al. 2008). Live-cell fluorescence microscopy has given insights into the dynamic properties of chromosome organization (Chubb et al., 2002; Heun et al., 2001; Janicki et al., 2004; Marshall et al., 1997; Masui et al., 2011; Robinett et al., 1996), and has begun to reveal how this organization relates to transcription (Alexander et al. 2019; Chen et al. 2013; Germier et al. 2017; Gu et al. 2018). The key advantage of fluorescence microscopy is the direct visualization of the position and arrangement of chromosomes in the nucleus. However, although recent developments have considerably shifted the boundaries of possibilities (see Section II below), it has been traditionally limited in throughput as well as genomic and spatial resolution. Electron microscopy has further expanded our understanding of the fine-scale structure of the chromatin fiber (Belmont 2014; Ou et al. 2017), but despite its exquisite spatial resolution it remains incompatible with sequence determination. These limitations have been circumvented by a second complementary class of methods, which we refer to collectively here as chromosome conformation capture (3C) techniques.
In 3C methods, digestion and subsequent re-ligation of crosslinked chromatin in cell nuclei allows the detection of spatial proximity between DNA sequences (Figure 1A). Method variants such as 4C, 5C, and Hi-C (described in several excellent reviews such as (Denker and de Laat 2016; de Wit and de Laat 2012)) are based on high-throughput sequencing and have revolutionized the field of nuclear structure by providing high-resolution, genome-wide measurements of physical proximity events within and across chromosomes, which are generally interpreted in terms of chromosomal ‘contacts’ or ‘interactions’ (see Section I below). 3C experiments have revealed that each chromosome is folded into complex structural patterns emerging at different scales. Active and inactive genomic sequences tend to mutually exclusively associate into A and B compartments (Figure 1B, left) (Lieberman-Aiden et al. 2009) which appear to be formed by attractive interactions of unclear origin, but possibly mediated by mechanisms involving phase separation of chromatin-associated proteins (Wang et al. 2019b; Strom et al. 2017; Sanulli et al. 2015; Strom et al. 2017; Falk et al. 2019), (Figure 1C). At shorter scales (<1 megabase), chromosomes fold into topologically associating domains (TADs) (Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012) (Figure 1B, middle). Although the exact definition of TADs is ambiguous due to the high complexity of sub-megabase interaction patterns and the multiple mechanisms they arise from (see below), they can be operatively defined as the domains whose boundaries are most conserved during cell differentiation (Zhan et al. 2017). The complex folding patterns at the scale of TADs are in part mediated by compartmental interactions between active and inactive genes, as well as polycomb-mediated interactions (Bonev et al. 2017; Rowley et al. 2017; Schoenfelder et al. 2015). However the predominant structural features at the TAD level are point-like focal interactions that connect sequences bound by the DNA-binding factor CTCF (de Wit et al. 2015; Guo et al. 2015; Rao et al. 2014; Vietri Rudan et al. 2015) (Figure 1B, right). CTCF-bound sites also occasionally interact across entire domains forming ‘stripe’-like structures (Vian et al. 2018). Formation of interactions associated with CTCF requires the cohesin complex (Hadjur et al. 2009; Rao et al. 2017; Wutz et al. 2017), which has been proposed to extrude DNA loops until it is arrested by CTCF bound to DNA in a certain orientation or other barrier proteins (Fudenberg et al. 2016; Nichols and Corces 2015; Sanborn et al. 2015) (Figure 1C). Although this might require interactions with RNA (Saldaña-Meyer et al. 2019; Hansen et al. 2019), the mechanisms by which CTCF interact with cohesin remain largely unclear. Together with constraints provided by the nuclear lamina and sub-nuclear compartments such as speckles and nucleoli, interactions mediated by CTCF-cohesin, compartmental interactions, and polycomb-coated sequences appear to shape the complex folding of mammalian chromosomes.
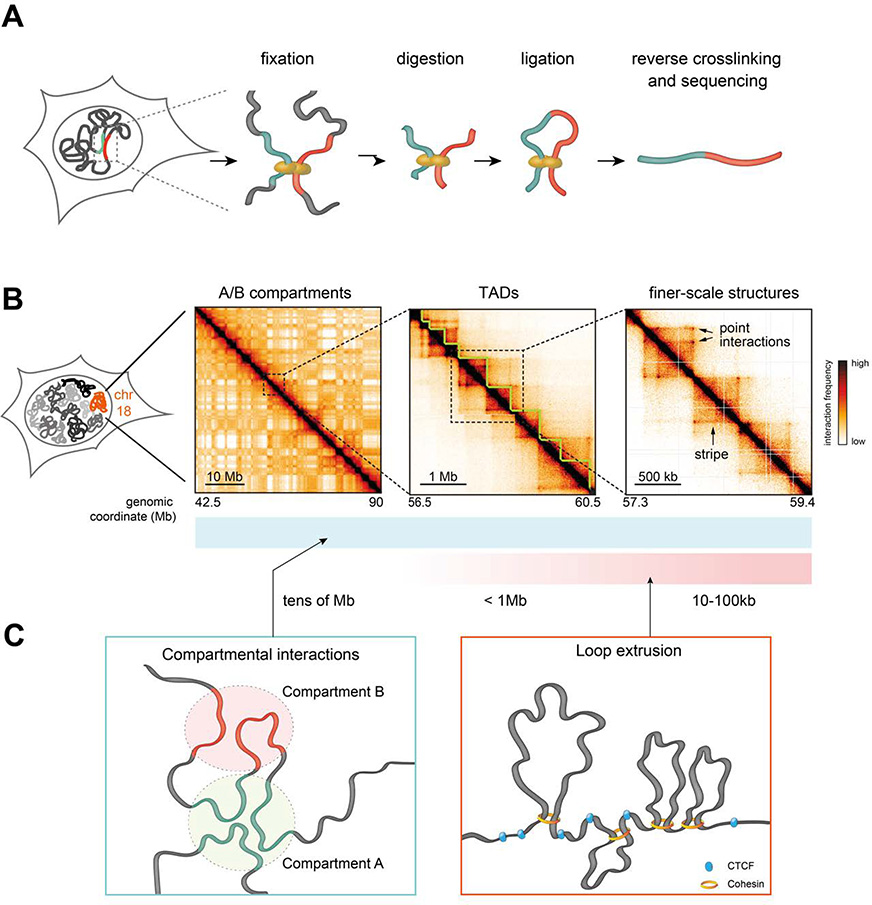
Figure 1.
A. Scheme of the core steps in chromosome conformation capture (3C) methods. Chromatin is crosslinked in cell nuclei and digested with a restriction enzyme (or endonuclease in the case of Micro-C), followed by ligation and decrosslinking. This results in the formation of hybrid DNA molecules that can be identified by high-throughput sequencing.
B. Hi-C contact maps illustrating that mammalian chromosomes are folded into checkerboard-like A/B compartments (left panel), TADs (middle panel), and shorter-scale structures. Sub-TAD structures include CTCF-related point interactions and stripes, as well as other interactions e.g. compartmental and polycomb-associated interactions (not shown in this example). Hi-C data were obtained mouse embryonic stem cells, from Ref. (Redolfi et al. 2019). Colormap saturation and scaling were modified across the three examples to emphasize structural features.
C. Hierarchies observed in chromosome folding are mainly driven by compartmental interactions involving attractions between active and inactive chromatin regions, manifesting at all genomic length scales; and CTCF/cohesin-mediated interactions likely originating from the loop extrusion activity of cohesin that is arrested by CTCF bound to DNA in defined orientations.
These discoveries have sparked enormous interest in the field due to experimental evidence suggesting that chromosome structure might play an important role in scaffolding the physical contacts between regulatory sequences, which are thought to help instruct gene expression during development and homeostasis (Galupa and Heard 2018; Spitz 2016) and to participate in other nuclear processes such as DNA damage repair (McCord and Balajee 2018; Fabre and Zimmer 2018) and replication (Marchal et al. 2019). This view is however challenged by some contradicting evidence suggesting that in some cases at least gene expression might not depend on chromosome structure (Williamson et al. 2019; Benabdallah et al. 2019; Ghavi-Helm et al. 2019) and despite our increasing ability to characterize chromosome structure, many fundamental questions concerning the causal links between chromosome interactions and fundamental nuclear processes remain completely open. Do chromosomal contacts have a direct causal impact on transcription? How dynamic are chromosomal interactions? How are CTCF/cohesin loops created? Are cooperative interactions between multiple regulatory sequences functionally relevant? Are CTCF loops and TAD boundaries causally linked to the accumulation of DNA damage, and how? Does the 3D structure of chromosomes play a causal role in DNA replication timing? How do properties of genome folding relate to the structural properties and shape of the nucleus? Fundamental questions also remain open on whether structure/function relationships are shared across evolution. Gene expression in yeast and Drosophila seems to be poorly responsive to massive rearrangements of chromosome structure (Shao et al. 2019; Luo et al. 2018; Ghavi-Helm et al. 2019). These results notably question the role of chromosome organization in Drosophila, where TAD-like domains do not result from CTCF looping but rather might arise as a consequence of compartmental interactions (Rowley et al. 2017).
Recent considerable technological advances give unprecedented opportunities to explore the mechanistic connections between chromosome structure and nuclear biology. Coupling existing methods (notably 3C) with newly developed techniques will expand the current paradigm, allow better measurement of dynamic molecular processes, and address the key open questions. In this review, we discuss 3C methods and the findings they enabled in the light of recent results obtained with new and rapidly developing orthogonal techniques. We focus on the strengths, limitations, and possible synergy of these methods and what they can reveal about the physical details and biological implications of chromosome structure.
Section I: Inferring 3D structure from chromosome contacts: considerations, limitations, and extensions made possible by orthogonal approaches
What do 3C methods actually measure?
A widespread interpretation of 3C data (and notably Hi-C) is that the number of ligation products detected is directly proportional to actual contact probabilities between genomic sequences. This has important implications for how data are interpreted (Fudenberg and Mirny 2012) and normalized (Imakaev et al. 2012). But what does ‘contact’ mean? 3C methods rely on formaldehyde crosslinking and ligation, two molecular processes that have been criticized as potential sources of bias (Belmont 2014; Williamson et al. 2014; Gavrilov et al. 2015, 2013). Crosslinking might capture contacts that are not mediated by direct molecular interactions of the chromatin fiber but rather by indirect crosslinking events through intervening nuclear proteins, and even organelles. Ligation might favor the formation of nonspecific molecular hybrids between crosslinked and partially solubilized chromatin. Orthogonal measurements of chromosomal interactions that are not based on crosslinking and ligation are thus important for providing context and comparison to 3C methods.
DNA FISH experiments use crosslinking but not ligation, and have largely confirmed that 3C-based data at various genomic scales are correlated with colocalization frequencies of FISH probes and/or inversely proportional to their spatial distances (Nora et al. 2012; Hakim et al. 2011; Giorgetti et al. 2014; Finn et al. 2019; Wang et al. 2016; Bintu et al. 2018; Crane et al. 2015; Gizzi et al. 2019; Giorgetti et al. 2016) (Figure 2A). Significant discrepancies have been however occasionally reported (Williamson et al. 2014), and even in concordant studies the correlation between FISH and 3C is high but not perfect, with a fraction of the variance in either technique that cannot be explained by the other (Finn et al. 2019; Wang et al. 2016) (Figure 2A). FISH and 3C methods are indeed affected by different technical and detection biases (Giorgetti and Heard 2016). Of note, signals in 3C methods arise from a subpopulation of cells where two loci occur in physical proximity, which can be compared with the fraction of cells where the two loci are closer than an arbitrary threshold distance in DNA FISH. This generally inversely correlates with the mean spatial separation between probes (Figure 2B), but deviations from this behavior are possible (e.g. in presence of bimodal distance distributions characterized by more than one peak, or in the presence of non-trivial dynamic processes such as loop extrusion (Fudenberg and Imakaev 2017)). 3C signals should thus better correlate with FISH when they are compared with the fraction of cells where probes are close to each other, rather than with their mean spatial separation (as discussed in detail in Refs. (Fudenberg and Imakaev 2017; Giorgetti and Heard 2016; Dekker 2016)). In addition, in DNA FISH physical proximity is defined using arbitrary distance or overlap thresholds that do not necessarily represent actual crosslinking radii in 3C (which incidentally might vary along the genome e.g. depending on the identity and concentration of protein complexes bound to DNA). Reassuringly, irrespective of these biases contact maps inferred from super-resolution DNA FISH are in excellent agreement with Hi-C data when spatial thresholds of 120–150 nm are imposed on distance distributions (Wang et al. 2016; Gizzi et al. 2019; Mateo et al. 2019; Bintu et al. 2018). Thus, DNA FISH and Hi-C concordantly interpret spatial proximity between two genomic sequences as their localization within a radius of a few hundred nanometers. It is interesting to notice that this distance range is still arguably one order of magnitude larger than distances required for protein complexes bound to DNA to be in reciprocal molecular contact. However the rapid pace of technical development in both classes of methods promises to soon give insight into these shorter-range interactions (Hsieh et al. 2019; Krietenstein et al. 2019; Mateo et al. 2019).
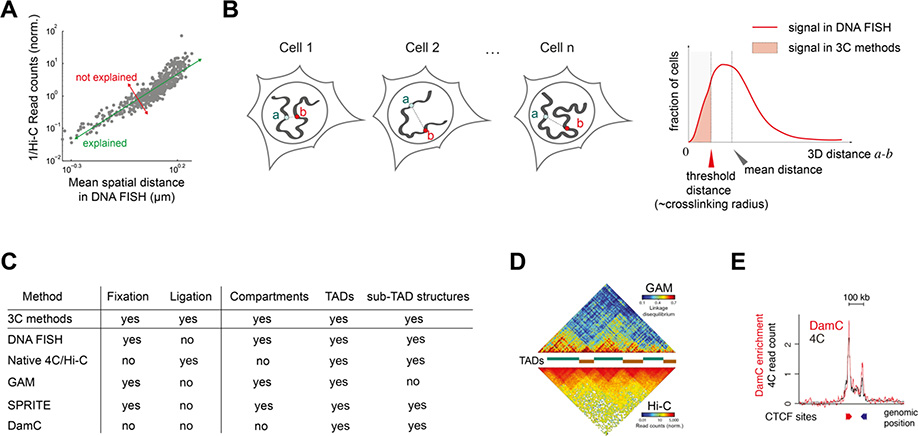
Figure 2.
A. 3C-based counts (Hi-C in this case) and spatial distances between loci measured in DNA FISH are generally well correlated, with a fraction of the variability that cannot be explained in terms of each other technique. Adapted from Ref. (Wang et al. 2016).
B. DNA FISH measures 3D distances between genomic loci (a and b here) and their distribution across the cell population. Signals in 3C methods arise from the fraction of cells where a and b can be crosslinked, which is usually (but not always) correlated with their average distance.
C. A summary of recently developed methods that are orthogonal to 3C and FISH and the structures they detected.
D. Ligation-free Genome Architecture Mapping (GAM) detects TADs and compartments in agreement with Hi-C. Adapted from Ref. (Beagrie et al. 2017)
E. Crosslinking and ligation-free DamC detects TAD boundaries and loops between convergent CTCF sites, in agreement with 4C-seq. Adapted from Ref. (Redolfi et al. 2019).
Several methods orthogonal to both 3C and FISH have recently provided evidence that 3C methods faithfully detect chromosomal contacts occurring within short spatial ranges, and confirmed the existence of compartments, TADs and sub-TAD structures (Figure 2C). ‘Native’ 4C and Hi-C performed in the absence of crosslinking were able to detect CTCF-mediated interactions as well as TAD boundaries, suggesting that these patterns do not arise as a mere consequence of an artifactual crosslinking of certain interactions (Brant et al. 2016). Two ligation-free techniques have further shown that interactions detected in 3C originate from short-range proximity (≲200 nm) between chromosomal sequences. In genome architecture mapping (GAM) (Beagrie et al. 2017) TADs, compartments and long-range inter-TAD interactions were recovered by measuring physical proximity (~200 nm) through ultrathin nuclear cryosectioning followed by high-throughput sequencing of DNA extracted from nuclear slices (Figure 2D). In split-pool recognition of interactions by tag extension (SPRITE) (Quinodoz et al. 2018), split-pool barcoding of DNA molecules within the same crosslinking complex discriminates different classes of interactions. Short-range SPRITE interactions quantitatively correlate with Hi-C data, contrary to long-range indirect crosslinking events that may possibly be mediated by subnuclear compartments. Finally, a modified version of DNA adenine methyltransferase identification (DamID) named DamC has emerged as a simultaneously crosslinking- and ligation-free assay which detects chromosomal interactions in living cells using DNA adenine methylation (Redolfi et al. 2019). Interactions detected by DamC show excellent agreement with 4C and Hi-C data (Figure 2E). In DamC DNA is methylated only by direct proximity with Dam molecules recruited to genomic viewpoints. Quantitative similarity with 3C-based measurements thus implies that a substantial fraction of chromosomal interactions measured by 3C methods involve nanometer-scale distances between genomic sequences.
Results from these orthogonal approaches collectively argue that 3C-based methods do not significantly distort the detection of chromosomal interactions. They also show that 3C data (and Hi-C in particular) are directly proportional to the fraction of cells in the population where a certain contact occurs at the moment of crosslinking. Far from being a nuance, this concept underlies all physical models of chromosome folding, including the highly influential loop extrusion model (see Section II below), in which simulated contact frequencies are compared with those inferred from Hi-C experiments.
One remaining challenge is that 3C-based data can only be converted to relative contact probabilities, given the absence of internal normalization criteria. This information is however required to address fundamental questions such as how mild differences in relative contact frequencies across TAD boundaries (~2-fold) manage to functionally insulate neighboring regulatory sequences. Lack of internal normalization also limits the ability to definitively compare contact frequencies at given loci between conditions and cell types. Precise conversion of experimental data into mechanistic models will require developing methods that are able to count 3D genomic interactions using quantitative molecular readouts.
How does contact frequency relate to spatial distance?
Contact frequencies alone cannot fully specify the physical shape and arrangement of the chromosome within the nucleus. This is a key challenge for comparing 3C-based data with microscopic observations (where physical distances, but not contacts are directly observed) and for modeling approaches that use Hi-C data to deduce 3D models of a chromosome or the whole nucleus. Complementary approaches are needed to measure physical distances between genomic sequences and their proximity to nuclear landmarks such as the nuclear envelope and nucleoli. DamID can identify regions of DNA that often contact the nuclear lamina (Lamin Associated Domains; LADs) (Peric-Hupkes et al. 2010) but again measures contact frequency instead of quantitating distance. One recent advance in this direction is TSA-seq (Tyramide signal amplification followed by high throughput sequencing) (Chen et al. 2018b), where antibody-coupled enzymes are targeted to nuclear features of interest (e.g. nuclear speckles) then generate diffusing biotinylated molecules that label nearby DNA in a distance-dependent manner. This approach enables the quantitative measurement of distances between genomic DNA and subnuclear compartments.
Remarkable improvements in microscopy techniques are also enabling better distance measurements within nuclei. Traditionally limited to small numbers of chromosomal loci and low genomic resolution, DNA FISH has been recently revamped by super-resolution chromosome tracing approaches employing highly multiplexed FISH probes, allowing distance measurements between thousands of chromosome loci in single cells at unprecedented scale. Concordant with earlier lower-throughput measurements (Nora et al. 2012; Giorgetti et al. 2014), these methods have shown that single TADs spatially extend over a few hundred nanometers in Drosophila and in mammals (Bintu et al. 2018; Gizzi et al. 2019; Boettiger et al. 2016), with active domains being larger/more decompacted than inactive domains. Strikingly, TADs belonging to A and B compartments spatially segregate, significantly overcoming their linear genomic arrangement (Wang et al. 2016). DNA FISH measures absolute spatial distances within the nucleus and reveals sub-population conformations that cannot be deconvolved out of population-averaged Hi-C data. In addition, super-resolution imaging can help clarify ambiguities in what Hi-C contact frequencies mean in terms of physical distance. For example, when a region is classified by Hi-C as weakly associated with the A compartment, it is equally possible that this region fluctuates between strong associations with A and B in different cells or that it consistently sits at an intermediate distance between the strongly separated A and B-type regions of the chromosome. Chromosome tracing clarifies that these “weak compartment” regions are in fact positioned in between the stronger A and B regions in most cells, rather than alternating between extremes (Wang et al. 2016).
Which interactions are simultaneous or cooperative?
Biologically relevant chromosome interactions may occur not only between pairs of loci, but also within clusters of cooperative contacts. Standard 3C methods are however unable to reveal whether multiple regions are interacting simultaneously or mutually exclusively. For example, when Hi-C reports that interactions occur pairwise between regions a-b, b-c and a-c, it is not clear whether a and b interact cooperatively with c or if a and b alternately contact c in different cells (Figure 3A). Distinguishing between simultaneous and alternate contacts is however necessary to answer a variety of biological questions. Is gene expression governed by the cooperative activity of several enhancers or by the mutually exclusive use of different enhancers in different sub-populations? Do nested interactions between several CTCF-bound regions represent an average of many alternate loops or cooperative interactions bringing together multiple loops? Measuring simultaneous contacts is also necessary to investigate whether groups of active genes cluster together in transcription factories (Sutherland and Bickmore 2009), to observe clustering of repressed regions potentially mediated by polycomb or HP1 (Schoenfelder et al. 2015; Strom et al. 2017), and to clarify the role of phase separation in the collective spatial partitioning of chromosome regions.
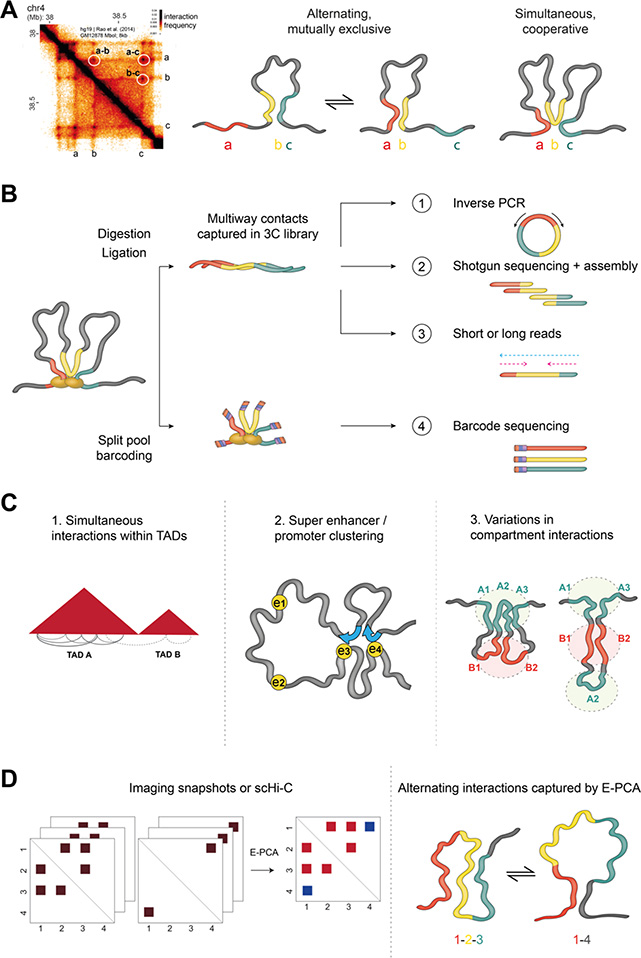
Figure 3.
A. Hi-C contacts between pairs of genomic loci (e.g. a, b and c here) can either correspond to mutually exclusive or to simultaneous (and possibly cooperative) interactions in single cells.
B. Simultaneous multi-way interactions can be identified using modified 3C methods that implement alternative strategies to sequence concatamers that exist in all 3C libraries, such as inverse PCR in MC-4C (scheme 1), shotgun sequencing and assembly in C-walks (2) or short-read sequencing as in Tri-C and similar methods (3). Multi-way contacts are also retrieved in SPRITE (4) using split-pool barcoding followed by sequencing of barcodes to identify genomic regions that were captured in the same cluster.
C. Multi-way chromosome conformation capture methods, as well as SPRITE and GAM, have shown overall that simultaneous contacts occur more frequently within TADs than across TAD boundaries (left), simultaneous interactions can occur between promoters and clusters of super-enhancers (middle), and that different subsets of A and B compartments cluster together in different subsets of cells (right).
D. E-PCA uses sets of contact matrices and reports patterns of correlated or anticorrelated contacts. Left: simplistic example where two possible contact patterns are found across single cells. Middle: The E-PCA result reports that the red set of interactions occur together and in opposition to the blue interactions, as in the schematic on Right.
3C libraries can contain long DNA concatemers in which several chromatin regions that were in close proximity in individual nuclei are ligated together. Modified versions of 3C methods have been developed to detect these simultaneous interactions using either shotgun sequencing (Olivares-Chauvet et al. 2016), short-read (Oudelaar et al. 2018; Ay et al. 2015; Jiang et al. 2016; Zheng et al. 2019) or ultra-long read sequencing (Allahyar et al. 2018) (Figure 3B). These methods have revealed common cooperative interactions between multiple loci, but also multi-contact configurations occurring in a small subset of cells and would be missed in population-averaged pairwise contact maps. Detecting simultaneous contacts in cells lacking the cohesin-unloading factor WAPL also revealed that CTCF-loop anchors collide with each other if cohesin is not efficiently unloaded from DNA (Allahyar et al. 2018). Overall, multi-way contacts are more likely to occur between regions within TADs (Figure 3C, left), and it is very rare to find cooperative interactions between multiple loci on different chromosomes, making it unlikely that such cooperative interchromosomal contacts are essential for key biological processes in most cases (Olivares-Chauvet et al. 2016).
While they have given important insights, proximity ligation-based methods are inherently limited in their ability to capture simultaneous contacts. One restriction fragment can only ligate directly to two other sequences in a single cell, regardless of how many other genomic loci were nearby. As such, while Hi-C maps from single cells (discussed in more detail in the next section) inherently represent sets of interactions that occur simultaneously (Nagano et al. 2013; Ramani et al. 2017; Stevens et al. 2017; Tan et al. 2018), any single map cannot sample all the interactions occurring in that cell due to substantial technical subsampling in current versions of single-cell Hi-C protocols. Computational approaches may help detect likely simultaneous or exclusive interactions from large sets of such data. In the protein structure field, the correlated forming and breaking of contacts has been analyzed by principal components analysis (PCA) across a large set of contact matrices from dynamic protein structure snapshots (Doshi et al. 2016) [Johnson J. Comput. Chem 2018]. A similar technique (termed E-PCA) has recently been applied to sets of chromosome contact maps from single cell imaging or Hi-C across different cell types (Lindsay et al. 2018) (Figure 3D). ). This approach has revealed certain compartment associations that tend to form and break together across single cells in a population and has identified the sets of compartment interactions that best distinguish blood cell type chromosome folding patterns. Future applications of this approach across single cell Hi-C contact maps may allow the inference of cooperative or mutually exclusive sets of contacts, even when these contacts cannot all be captured simultaneously in the same cell.
The limitations of 3C-based methods to capture multi-contact interactions have also driven the development of complementary approaches. The SPRITE approach described earlier (Quinodoz et al. 2018) does not require ligation and thus can identify multiple loci that simultaneously interact in a single cluster. SPRITE data suggest that certain long-distance interactions within the A compartment may tend to occur together in the same cell while other A compartment regions contact one another alternately, resulting in average high contact frequencies between all A compartment regions in a Hi-C map (Figure 3C, right). The GAM approach (Beagrie et al. 2017) also captures complex multi-way contacts and has identified numerous 3-way interactions between TADs containing highly transcribed regions or TADs containing super-enhancers (Figure 3C, middle). Super-resolution microscopic chromosome tracing has also revealed locations of cooperative, multi-way interactions that occur simultaneously in single cells (Bintu et al. 2018). Finally, single-cell applications of DamID have revealed extensive evidence for cooperative coordinated contacts between the nuclear lamina and chromosomes in individual cells (Kind et al. 2015). It should be noted that none of these approaches can definitively determine whether simultaneous interactions are essential for regulatory communication between loci. As described further below, perturbations of specific combinations of interactions will be needed to determine their function and how they are established.
Section II: Capturing dynamics and heterogeneity in chromosome conformation
Chromosome structure in single cells
With the exception of single-cell Hi-C and some multi-contact approaches, 3C data generally represent snapshots of chromosome conformations at a given time point, averaged over an entire cell population. But how do they relate to the actual conformations of the chromatin fiber in single cells, and how do they evolve in time? Well before the advent of super-resolution chromosome tracing, DNA FISH and live-cell imaging experiments in yeast, Drosophila and mammalian cells revealed that chromosome conformation is highly variable across cell populations or tissues (Jhunjhunwala et al. 2008; Amano et al. 2009; Shopland et al. 2006; Marshall et al. 1997; Heun et al. 2001; Berger et al. 2008). With the development of single-cell Hi-C, it has become clear that single pairwise contacts occur as stochastic events in single cells (Nagano et al. 2013; Ramani et al. 2017; Stevens et al. 2017; Flyamer et al. 2017; Tan et al. 2018). Interaction motifs observed in population Hi-C maps such as TADs and compartments only become evident when averaging over many single cells, indicating that they reflect preferential interactions in a highly diverse ensemble of structures (Figure 4A). The genomic resolution of single-cell Hi-C is currently limited, preventing short-range contacts such as single enhancer/promoter interactions (typically below 200kb within single TADs) to be quantitatively assessed. Polymer models and super-resolution DNA FISH have however shown that single conformations of the chromatin fiber inside single TADs are also highly variable (Nora et al. 2012; Giorgetti et al. 2014; Bintu et al. 2018; Gizzi et al. 2019; Mateo et al. 2019; Boettiger et al. 2016) (Figure 4A). Only a subset of interactions actually occur in a single cell, implying that enhancer-promoter interactions and CTCF loops are stochastic events (Giorgetti et al. 2014). TAD boundaries themselves emerge from the superposition of conformations where stochastic contacts within TADs are favored compared to contacts across boundaries (Giorgetti et al. 2014; Bintu et al. 2018). In the absence of cohesin, the disappearance of TAD boundaries from population Hi-C matrix is most likely a result of loss of preferential contacts within TADs and increased randomness in interactions (Bintu et al. 2018). This is consistent with the idea that contacts within TADs in individual cells represent the captured position of loop extrusion complexes, and thus reflect not static loops but a captured moment of a dynamic process (Hansen et al. 2018). Super-resolution DNA FISH experiments have additionally revealed that compartments emerge from the superposition of highly stochastic but mutually exclusive interactions between A- and B-type chromatin regions (Wang et al. 2016). Single-cell DamID experiments also showed high cell-to-cell variability in associations of LADs to the nuclear periphery (Kind et al. 2015). Experimental evidence thus agrees on the existence of extensive conformational variability at all genomic length scales, from sub-TAD structures (Nora et al. 2012; Giorgetti et al. 2014; Mateo et al. 2019; Boettiger et al. 2016) to TAD-scale interactions (Finn et al. 2019; Gizzi et al. 2019; Cattoni et al. 2017) all the way up to A/B compartments (Wang et al. 2016; Kind et al. 2015) and even multi-Mb regions on the very special case of inactive X chromosomes (Giorgetti et al. 2016). Pervasive cell-to-cell structural variability might have important implications for transcriptional regulation, since stochastic interactions between regulatory sequences are likely to result in the stochastic transfer of regulatory information. Methods that will help define the functional role of structure variation are discussed further in Section III.
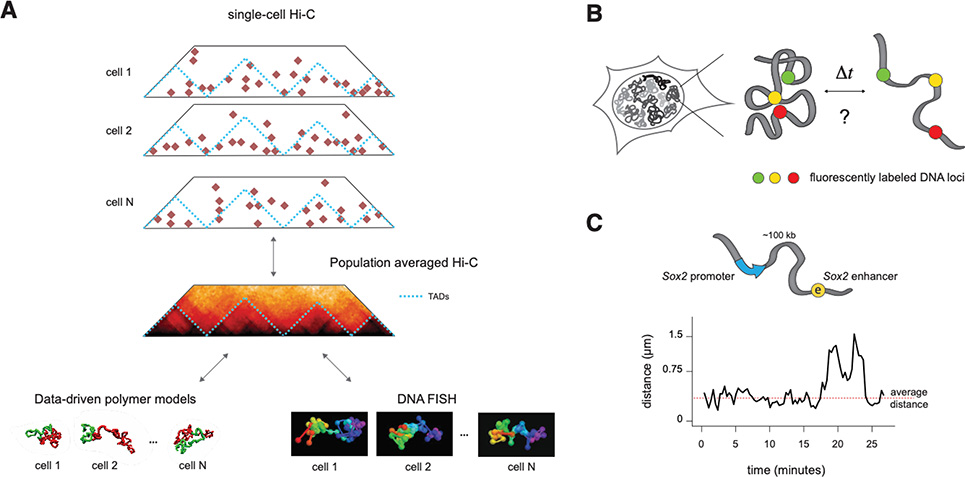
Figure 4.
A. Single-cell Hi-C (top), polymer simulations (bottom left) and super-resolution DNA FISH (bottom right) show that population-averaged signals in 3C methods arise from highly variable structures that occur simultaneously in single cells.
B. To measure the temporal dynamics of chromosome structures, experiments are needed where the spatial positions of two or more chromosomal locations in cis can be measured in time, in living cells.
C. Simultaneous live-cell imaging of genomic locations flanking the Sox2 promoter and its (super-) enhancer in mESC shows that their distances fluctuate around average values over an experimental timescale of two hours, with sporadic stochastic larger changes. Adapted from. (Alexander et al. 2019).
Chromosome structure in time
Very little is known about the timescales over which cell-to-cell structural differences are established and how they might relate to transcription and other nuclear processes. In one extreme hypothetical scenario, a conformational state is inherited at the exit from mitosis, maintained throughout the cell cycle, and transmitted with some degree of shuffling to daughter cells. In this scenario, regulatory interactions are stable during the cell cycle, with a subset of contacts maintained for several hours and others never taking place in this timespan. In an opposite extreme scenario, every cell would experience a large number of different conformations within a single cell cycle. In this case, exchange of regulatory information would occur as a highly stochastic and dynamic event, with contacts assembling and disassembling on a timescale of seconds or minutes. Considering that the time interval separating consecutive encounters between two loci on a chromosome is expected to rapidly increase as a function of their genomic distance (Amitai et al. 2010; Zhang and Dudko 2016), both scenarios could actually take place. Contacts between regulatory sequences separated by small genomic distances could assemble and disassemble rapidly, whereas the spatial distance between sequences located hundreds of kilobases away might change on slower time scales (Gibcus and Dekker 2013).
Several approaches have enabled imaging of chromosome dynamics at the scale of the whole nucleus, without detecting specific sequences. Labeling histone proteins with a photoactivatable fluorophore has revealed that the positions of chromosome territories are stable throughout interphase (Strickfaden et al. 2010), but that large-scale rearrangements happen after each mitosis. A method to visualize the dynamics of lamina-associated domains (LADs) (Kind et al. 2013), where a fluorescent tracer of N6-methylated adenines marks regions that have been associated with the laminB1 fused with Dam, came to similar conclusions. Shorter-scale intra-chromosomal dynamics have been probed using Dense Flow Reconstruction and Correlation (Shaban et al. 2018), which revealed localized micron-sized domains of correlated chromosome dynamics within transcriptionally active nuclei.
Measuring temporal fluctuations in chromosome structure in a sequence-specific manner requires to simultaneously visualize two or more genomic regions in living cells using combinations of orthogonal fluorescent proteins (Figure 4B). Classically, researchers have visualized single chromosomal loci via homologous recombination-mediated insertion of arrays of ectopic transcription factor binding sites such as Lac or Tet operators, which can be localized as diffraction-limited spots upon binding of the corresponding repressors fused to fluorescent proteins (Marshall et al. 1997; Heun et al. 2001; Chuang et al. 2006; Masui et al. 2011; Chubb et al. 2002; Kumaran and Spector 2008). More recently, visualization of single loci has also been achieved by recruiting multiple copies of catalytically inactive Cas9 (dCas9) fused to GFP to repetitive (Ma et al. 2018; Stanyte et al. 2018) or non-repetitive (Chen et al. 2013; Gu et al. 2018) DNA. The bacterial ParB/parS system, in which the ParB protein multimerizes upon binding to a single parS sequence on DNA, has also been successfully used to image single chromosomal locations (Germier et al. 2017; Saad et al. 2014). These strategies however present conceptual and technical hurdles which have limited their application to simultaneous multi-color imaging: Repetitive operator arrays are prone to recombining and generating DNA damage (Dubarry et al. 2011). Insertion of two or more orthogonal arrays in cis requires sequential targeting and cumbersome screening for genomically stable subclones. Multi-color CRISPR approaches require simultaneous expression of multiple gRNAs arrays and multiple orthogonal catalytically inactive Cas proteins. Additionally, imaging cells in physiological conditions over timescales ranging from seconds to hours demands microscopes that are able to deliver low photon numbers (to minimize photobleaching and phototoxicity), while recovering high-quality signals from dim sub-diffraction light sources.
Two recent studies have overcome these limitations and measured the time evolution of two genomic loci in cis in mouse cells using orthogonal operator arrays (Alexander et al. 2019; Khanna et al. 2019). In mouse embryonic stem cells, locations flanking the Sox2 gene and its super-enhancer, which are located ~100 kb apart, were found to have different average positions in each individual cell (Alexander et al. 2019). While their instantaneous positions fluctuated significantly around these averages over time, the average positions remained stable across an experimental time window of twenty-five minutes, with sporadic drastic changes (Figure 4C). The situation is similar in B-cell progenitors where the spatial positions of the VH and DHJH regions at the immunoglobulin locus (separated by approx. 1 Mb) were found to fluctuate around stable average values on a 10-minute timescale, which can change over longer times in a subset of cells (Khanna et al. 2019). Whether and how this behavior is linked to cell cycle progression and/or potential loop extrusion by cohesin is not clear. Based on experimental estimations (Cattoglio et al. 2019; Holzmann et al. 2019) numerical simulations (Fudenberg et al. 2016) and recent in vitro evidence (Golfier et al. 2019; Davidson et al. 2019), cohesin could extrude DNA loops at rates as high as ~100 kb/min. The loop extrusion process should hence affect the dynamics of loci separated by hundreds of kb on timescales of minutes. Shorter-timescale (<1 minute) fluctuations around stable positions may instead reflect extrusion-independent polymer behavior. In any case, these results overall suggest that enhancer-promoter interactions might occur as unstable and transient stochastic events several times in a cell cycle.
Another study monitored the dynamics of an ectopic even-skipped (eve) promoter and the endogenous eve locus located 100 kb apart in living Drosophila embryos (Chen et al. 2018a). Looping between the two loci was enforced through an ectopic insulator known to be able to pair with its endogenous copy at the eve locus. Interestingly, physical interactions between the two loci lasted for several minutes, in stark contrast with experiments in mouse cells (Alexander et al. 2019), and were further stabilized by transcription at the reporter gene and the looped state.
Advances in genomic engineering and imaging are opening exciting possibilities to extend and generalize these observations to other systems and conditions. Besides CRISPR-based knock-in strategies, transposon-based and recombinase-mediated genomic engineering allow efficient insertion of operator arrays (Alexander et al. 2019; Redolfi et al. 2019). Synthetic arrays with non-repetitive linkers (Lau et al. 2003) minimize risks of recombination, whereas new or modified repressor/operator systems (Alexander et al. 2019; Khanna et al. 2019) extend possibilities while minimizing side effects due to DNA damage (Dubarry et al. 2011). An alternative dCas9 targeting strategy where ribonuclear particles of dCas9 and fluorescently labeled sgRNAs are delivered to cells offers improved signal-to-noise ratio and easiness of use (Wang et al. 2019a). New imaging technologies such as highly inclined and laminated optical sheet (HILO) (Tokunaga et al. 2008) or lattice light-sheet microscopy (Chen et al. 2014) enable high-resolution, low-phototoxicity multicolor measurements of living samples. Coupled with advances in super-resolution live-cell microscopy and labeling strategies (Zheng and Lavis 2017), these tools will allow addressing fundamental questions such as if and how CTCF loops, loop extrusion and topological constraints modulate chromosome contact dynamics; and how structural dynamics relates to the dynamic exchange of regulatory information and ultimately impacts transcription.
Modeling approaches translate 3C information into a context of distance, cooperativity, heterogeneity, and dynamics
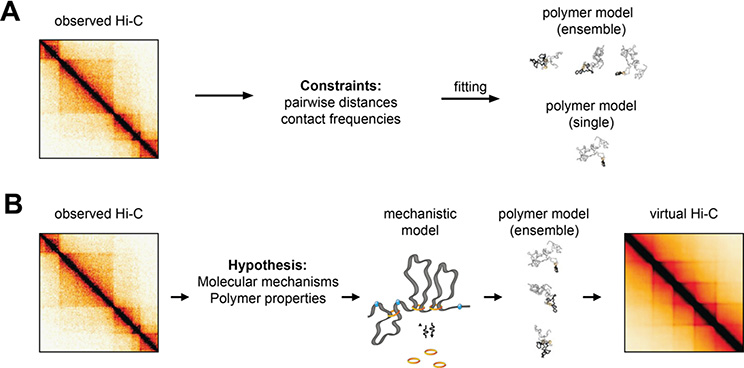
Computational modeling of 3C information and chromosome structure provides an important avenue to address many of the challenges in chromosome structure data interpretation presented thus far: translating contacts into distances, inferring simultaneous contacts, and representing heterogeneity in structures between cells and across time. There are numerous reasons that physical models of chromatin have proven to be powerful tools in studying spatial genome organization. First, since both microscopy and 3C-based methods do not yet allow a direct high-resolution large-scale view of chromatin structure, physical models are necessary to infer the underlying structure. Second, models provide a quantitative framework to integrate information from different types of experimental systems. Third, modelling approaches often allow us to directly test hypotheses regarding underlying molecular mechanisms, and to generate predictions that can be tested experimentally.
Since 3C-based methods measure interaction frequencies rather than produce actual structures, the most obvious goal of physical chromatin models is to discover the underlying structures (Figure 5A). The simplest approach is to seek a single structure (Duan et al. 2010; Lesne et al. 2014; Varoquaux et al. 2014; Baù et al. 2011). This process makes two critical assumptions: that one can convert interaction frequencies to spatial distances, and that a single structure is sufficient to represent the entire distribution of structures measured by bulk genomic methods. While the first assumption may be reasonable (as discussed previously), the second assumption is problematic, since genome structure is highly variable.
Figure 5.
A. Data-driven polymer models use parameter fitting to determine consensus 3D structures that satisfy contact or distance constrains inferred from Hi-C maps. They can result in either single consensus structures, or more realistic structural ensembles reflecting to some extent the cell-to-cell variability observed experimentally. Adapted from (Fudenberg et al. 2017).
B. Mechanistic polymer models implement specific hypotheses concerning the mechanisms that drive 3D folding, and result in ensembles of structures that can be used to predict the outcome of virtual Hi-C experiments.
Due to the limitations of using a single genome structure, modelling approaches have been extended to identify a set of structures whose average matches measured population-average interaction frequencies (Serra et al. 2017; Kalhor et al. 2012; Tjong et al. 2016; Paulsen et al. 2017). While this is closer to biological reality, a new challenge is introduced. Since these methods try to reconstruct a distribution by considering only its average, the problem is highly under-constrained, as many different sets of structure distributions may be consistent with measured population-averaged interaction frequencies. Some approaches constrain the set of solutions using energy landscapes and molecular dynamics simulations (Di Pierro et al. 2016) while others use Bayesian inference, shortest path calculations, or other assumptions to calculate an optimal ensemble (Lesne et al. 2014). Some approaches add constraints from microscopy data, either constraining conformations by visually measured landmarks or by detailed chromosome tracing results (Stevens et al. 2017; Nir et al. 2018). In addition to validation by in silico simulations, it is critical that these methods yield testable hypotheses that can be validated otherwise. For example, microscopy can be used to test the distribution of distances of specific pairs of loci across the population, and these can be compared to distribution of distances produced by models (Baù et al. 2011; Giorgetti et al. 2014). Despite anecdotal validations, it remains unclear whether this type of approach provides accurate descriptions of genome structure and quantitative predictions. While single-cell Hi-C experiments bypass the limitation of population averaging, their low yield per cell seriously limits the resolution of models based on single cell Hi-C maps.
An alternative class of modelling approaches start from simple physical or molecular mechanisms and attempt to produce structures consistent with 3C-based interaction data (Figure 5B). While these will generally reproduce the data less accurately, they have the benefit of directly testing hypotheses regarding molecular mechanisms and the dynamic processes underlying genome structure. Several models based on biophysical principles have been proposed to explain features of Hi-C data: interaction frequency is inversely proportional to genomic distance in human cells, consistent with a fractal globule model (Lieberman-Aiden et al. 2009; Grosberg et al. 1988); volume-exclusion and chromosome tethering have been used to model S. cerevisiae Hi-C (Wong et al. 2012; Tjong et al. 2012); supercoiling has been used to model TADs (Benedetti et al. 2014); supercoiled plectonemes have been used to model Caulobacter Hi-C (Le et al. 2013); block copolymers have been used to model TADs (Jost et al. 2014); binders and switchers have been used to model TADs and compartments (Barbieri et al. 2012). One modelling achievement has been the loop extrusion model which provides a parsimonious mechanism that explains both mitotic chromosome structure and TAD formation (Fudenberg et al. 2016; Goloborodko et al. 2016; Naumova et al. 2013). Notably, the model produced testable hypotheses, some of which were verified in genetic perturbations such as CTCF, NIPBL and WAPL deletions (Haarhuis et al. 2017; Nora et al. 2017; Schwarzer et al. 2017; Rao et al. 2017), as well as microscopy evidence demonstrating loop extrusion by yeast condensin (Ganji et al. 2018). Still, some details of the model remain unresolved such as extrusion speed, dependence on ATP, uni- vs. bi-directionality of extrusion and the molecular mechanism underlying insulation. An outstanding challenge will be consolidating mechanisms underpinning interaction patterns at different scales (Nuebler et al. 2018), e.g. how loop extrusion and phase separation could work together to produce loop, TAD, and compartment structures. Additionally, new methods for systematically integrating 3C data with microscopy data will be required (Abbas et al. 2019). Finally, in addition to highlighting the features which models explain well, it will be equally useful to study features of the data that are left unexplained or are poorly reproduced by models.
Section III: Connecting Chromosome Structure to Function
Which interactions are functionally relevant, and how can they be identified?
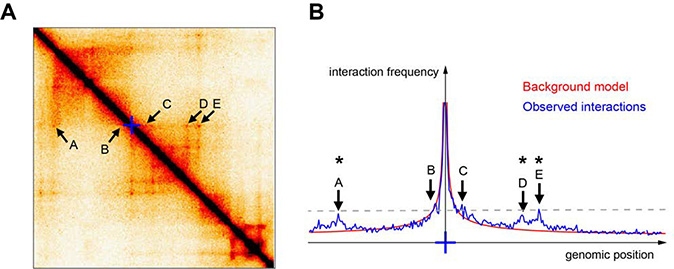
Precise measurements of interactions between genomic loci, even if translated into sophisticated polymer models or confirmed with high-resolution imaging, cannot report which interactions are linked with (or give rise to) functional events. Contact frequencies between two or more genomic loci presumably inform about whether those loci are likely to communicate functionally: a tendency in the field has thus been to develop computational methods to identify ‘specific’, ‘significant’ or ‘looping’ interactions within Hi-C or 4C data. Although the nomenclature varies slightly between studies, these are usually defined as pairs of sequences whose interaction frequencies stand out from the interaction background generated by random ‘collisions’ across the chromatin polymer (Figure 6A). Many competing methods have been proposed to detect these interactions using a variety of statistical approaches. These approaches can vary widely in their results, from identifying enriched point interactions (Rao et al. 2014; Geeven et al.) to finding stripes of enrichment (Schmitt et al. 2016) to detecting entire TADs (Dixon et al. 2012) or hierarchies of interaction domains (Zhan et al. 2017; Norton et al. 2018). Further, the apparent properties of these interactions in a contact map can vary somewhat depending on the details of the experimental approach. For example, 3C techniques that shorten the length of DNA molecules before ligation, such as micro-C, detect arrays of specific interaction dots where other Hi-C datasets find more homogeneous lines (Hsieh et al. 2019; Krietenstein et al. 2019).
Figure 6.
A. Example of a 1Mb-by-1Mb region in a Hi-C matrix illustrating various potentially ‘significant’ (A-E). Blue cross: viewpoint used for panel B.
B. Blue line: Relative contact frequencies measured from the blue cross viewpoint are shown as a virtual 4C plot derived from the Hi-C matrix shown in A. Red line: background model derived from the same map by genome-wide averaging Hi-C counts from pairs of loci separated by the same genomic distances. Algorithms that determine ‘significant’ or ‘specific’ interactions would typically identify interactions with A, D and E (indicated with an asterisk) because they stand over background, but not B and C. However, B and C interact as frequently with the viewpoint as A, D and E, as indicated by the dashed line.
Though attempts have been made (Forcato et al. 2017), determining the best loop- or domain-calling algorithm (or the best loop- or domain-resolving experimental method) is impossible without a clear reference for the biological implications of different classes of ‘significant’ interactions. Above-background interactions are certainly significant with respect to a null model where only random collisions occur (Figure 6B), and very often correlate with CTCF loops, interactions between active genes, or interactions between genes and their potential regulatory sequences (Bonev et al. 2017; Hsieh et al. 2019; Krietenstein et al. 2019; Andrey et al. 2016). However, a sizeable fraction of regulatory sequences do not show particularly enriched interactions with surrounding partners. Does this necessarily imply that these interactions are non-functional? Enrichment levels observed at ‘specific’ interactions are at most 2–3 fold higher than the random collision background, and only 0.5-fold higher on average. Do such small differences in interaction probabilities matter for transcriptional regulation or other biological processes? Conversely, is it correct to discount as non-functional the high-frequency interactions at short genomic distances that are not detected as significant, but occur as background polymer collisions (Figure 6B)?
Similar considerations might apply to contacts between genomic sequences located far in cis (≫1 Mb on the same chromosome) or in trans (i.e. on different chromosomes). Due to the power-law decay of contact probabilities in cis, and the near-random arrangement of chromosome territories in trans, these interactions are constitutively much rarer than those between sequences located nearby in cis (< 1 Mb), and as such are underrepresented in 3C ligation libraries. Due to their rarity, far-cis and trans interactions are preferentially detected using target-enrichment approaches such as 4C (van de Werken et al. 2012), ChIA-PET (Fullwood et al. 2009) or capture 3C/HiC (Schoenfelder and Fraser 2019; Hughes et al. 2014; and other methods reviewed in Denker and de Laat 2016). Specific interactions between loci separated by multiple Mb or on different chromosomes have been identified between polycomb-bound locations (Schoenfelder et al. 2015; Bonev et al. 2017), active genes and regulatory sequences (Javierre et al. 2016; Fullwood et al. 2009; Joshi et al. 2015; Bonev et al. 2017; Noordermeer et al. 2011) or DNA double-strand breaks (Aymard et al. 2017). Notably, a recent study has shown the emergence of long range cis and trans interactions in olfactory neurons, directly mediating their function by selection of individual olfactory receptors (Monahan et al. 2019). Despite being detected at high levels of statistical significance compared to background collisions or generic trans interactions, and despite notable exceptions (Maass et al. 2018), in most cases far-cis or trans interactions are very rare (in the order of a fraction of % of cells based on DNA FISH and using distance thresholds of few hundred nm, see e.g. (Schoenfelder et al. 2015; Noordermeer et al. 2011)).This raises the question as to whether single interactions, or rather (potentially cooperative) synergies between different combinations of them in different cells might be functionally relevant.
Addressing these questions requires developing experimental techniques that go beyond the identification of significantly enriched contacts, and rather use molecular readouts to detect ‘productive’ functional interactions. Global perturbations such as depletion of CTCF and cohesin have suggested that overall, the ‘significant’ structures associated to CTCF/cohesin are important for long range gene regulation of at least a subset of genes (Rao et al. 2017; Schwarzer et al. 2017; Nora et al. 2017). However, when so many interactions are affected at once, it is still difficult to pinpoint local causality and importance of particular interactions. Targeted genomic rearrangements affecting TAD boundaries or CTCF-CTCF interactions lead to rewiring of chromosomal contacts, and have been shown to result in gene misregulation or decreased developmental robustness (Despang et al. 2019; Symmons et al. 2016), but occasionally no effect at all on gene expression, at least in the studied context and conditions (Williamson et al. 2019; Ghavi-Helm et al. 2019). New techniques such as the CLOuD9 system have also been recently developed to ectopically introduce new CTCF-independent interactions and monitor their consequences (Morgan et al. 2017). Such induced loops have helped dissect the complex influence of chromatin looping on transcription at specific loci. Supporting the idea that chromosome interactions can regulate gene expression, interactions forced to occur between enhancers and gene promoters were found to increase gene transcription, but only in cell types normally permissive to activation of the studied gene (Morgan et al. 2017. Further development allowing light-activatable loop induction (Kim et al. 2019) has shown modest effects on gene expression. Other approaches such as CRISPR-GO have also enabled to investigate whether the inducible repositioning of chromosome loci to a variety of nuclear compartments alters gene expression (Wang et al. 2018). (Wang et al. 2018). This technique has shown, for example, that artificially localizing genomic regions to the periphery or Cajal bodies represses gene transcription. A key caveat to such approaches, however, is that artificially stabilized loops may not mimic the dynamics of endogenous contacts measured by 3C-based approaches. Additionally, such approaches assume that the stable, highly enriched point interaction is the key functional unit, rather than addressing whether less isolated, more stochastic, but high frequency interactions (such as those typically dismissed and normalized away as characteristic of random polymer collisions) could also have biological implications.
Developing molecular assays that will detect productive functional interactions will also require defining the full spectrum of molecular events and chromatin-based processes that can be affected by chromosome interactions. Transcription in interphase nuclei is an obvious process to focus on, but other biological events such as DNA translocations, DNA repair and replication, and the physical properties of the nucleus must also be considered as candidate processes and properties that can be directly affected by chromosome structure. Approaches to consider these connections between structure and function and recent results in these investigations are described in the following sections.
Chromosome structure and transcription: what causes what and how?
Although perplexing reports have sporadically challenged this view in mammals (Williamson et al. 2019; Benabdallah et al. 2019) and Drosophila (Ghavi-Helm et al. 2019), modulations in chromosome structure are thought to alter transcriptional activity and/or robustness in gene expression by altering interaction frequencies between regulatory regions: Enhancer activity (defined in terms of chromatin accessibility and/or enrichment in active histone modifications) generally correlates with increased spatial proximity to their target genes (Carter et al. 2002; Tolhuis et al. 2002); TADs overlap with genomic regions of concordant tissue-specific expression of randomly integrated transgenes in mice (Symmons et al. 2014); Deletion, inversion or duplication of TAD boundaries, and single CTCF sites to a lesser extent, can result in ectopic enhancer-promoter contacts that can cause oncogene activation (Hnisz et al. 2016), aberrant gene expression leading to malformation syndromes (Lupiáñez et al. 2015; Franke et al. 2016) or loss of developmental robustness (Despang et al. 2019); Distal quantitative trait loci also tend to occur at regulatory locations that interact frequently in space (Grubert et al. 2015; Delaneau et al. 2019).
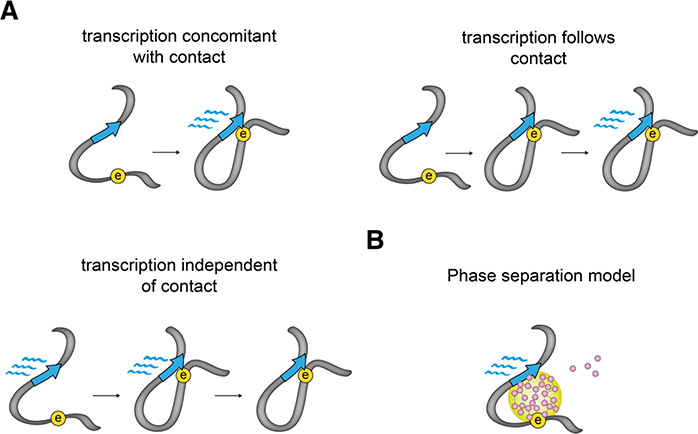
There is nonetheless a profound lack of understanding of the mechanistic principles that could link chromosome interactions to transcriptional regulation. Accumulating evidence suggests that enhancers can modulate promoter bursting frequency (Senecal et al. 2014; Bartman et al. 2016; Fukaya et al. 2016; Larsson et al. 2019; Rodriguez et al. 2019) or burst duration (Diez-Roux et al. 2011). It is however unknown if this is related to physical encounters, in the absence of information on whether transcription occurs concomitant with, subsequent to, or independent of physical contacts (Figure 7A). Live-cell imaging studies that aim at correlating enhancer-promoter spatial positions with RNA production levels in single cells (Bertrand et al. 1998; Chao et al. 2008) have just begun to shed some light on these questions. In Drosophila, ectopically enforced physical contacts between the endogenous eve enhancers and a minimal reporter gene causally lead to transcription of the reporter (Chen et al. 2018a), which in turn stabilizes the looped state. In mouse embryonic stem cells however, the physical proximity of the Sox2 gene locus with a region flanking its super-enhancer does not appear to temporally correlate with transcription levels (Alexander et al. 2019). Interestingly, insulator-induced loops in Drosophila are rare and stable, which might facilitate the detection of correlations between physical proximity and transcription. By contrast interactions at the Sox2 locus are frequent and transient. It will thus be interesting to test if engineered stable interactions in mammals would show more correlation with transcription states in single cells. As a note of caution however, it should be noted that to avoid interfering with transcriptional regulation, bacterial operator arrays in (Alexander et al. 2019) were inserted few kb outside the Sox2 and super-enhancer regions proper. Their spatial positions might thus not precisely reproduce the positions of the regulatory sequences, considering that the chromatin fiber might bend considerably in the 2–10 kb range (Redolfi et al. 2019; Mateo et al. 2019) and that the array marking the Sox2 gene is separated by a CTCF-bound region which might function as an insulator. Similarly, in (Chen et al. 2018a), MS2- and PP7-labelled RNA signals are used as proxies for the spatial positions of the endogenous and reporter genes, introducing some uncertainty on the latter.
Figure 7.
A. Transcription of RNA could either occur together with, or follow, or be completely unrelated to enhancer-promoter interactions. This could possibly depend on the specific enhancer-promoter pair, or the stability of their interactions.
B. Sub-micrometer sized phase-separated compartments could alternatively lead to functional communication and RNA production without the need of actual physical interactions between an enhancer and its target promoter.
An alternative and influential view of how enhancers might transfer regulatory information to promoters is that communication might happen without the strict requirement of a short-range (nanometer-range) physical contact. In this model regulatory sequences, even if not in direct contact, would be able to nucleate the formation of dynamic phase-separated transcription factor condensates spanning several hundred nanometers (Figure 7B). This is proposed to happen through non-specific interactions mediated by low-complexity regions present on many transcription factors. Nuclear phase-separated condensates have been recently observed (Boija et al. 2018; Cho et al. 2018; Sabari et al. 2018) and are proposed to mediate transcriptional activation by locally increasing the concentration of transcription factors in the immediate spatial neighborhood of promoters. This provocative concept, which presents analogies to previously proposed ‘transcription factory’ model (Sutherland and Bickmore 2009), might also explain why transcriptional activities at the Sox2 locus are uncorrelated to enhancer distance. Other modes of communication, including ‘tracking’ mechanisms where regulatory information is transmitted by 1D diffusion of transcription factors along the chromatin fiber, have also been proposed (Vernimmen and Bickmore 2015) and might be notably relevant for promoter-proximal enhancers.
To clarify the rules that govern regulatory communication mediated by 3D chromosome structure, an ideal experiment would measure promoter transcriptional output accurately and correlate it with its 3D distance to enhancers separated by diverse genomic distances and across topological boundaries. However, this remains a daunting task given the number of genomic manipulations required and the regulatory and structural complexity of transcribed genomic regions in eukaryotic genomes. Experiments in E.coli, where genomic engineering is not a major rate-limiting step, have revealed that transcription driven by enhancer-like bacterial elements is directly proportional to their contact probabilities with a promoter, and decreases like a power law with increasing genomic distances (Hao et al. 2019). Engineered loops induced through LacI tetramerization are able to insulate or stimulate transcription depending on their relative position with respect to the ‘enhancer’-promoter pair. It will be exciting to see similar experiments performed in higher eukaryotes.
Finally, interaction frequencies between enhancers and promoters might not be the main determinant of a promoter’s transcriptional level or robustness, or at least not at all genomic locations. Other factors have a major impact on gene activity, notably a gene’s nuclear position and proximity to repressive compartments such as the nuclear lamina (Akhtar et al. 2013), the sequence composition of its proximal and distal regulatory sequences, as well as the identity and concentrations of transcription factors that bind them (Spitz and Furlong 2012; Haberle et al. 2019). Advances in genome engineering, inducible protein degradation systems, imaging and sequencing-based approaches now make it possible to design experiments that disentangle the various dependencies of transcription on sequence composition, nuclear localization and chromosome structure. Further developments of recent methods allowing the simultaneous detection of chromatin states or nuclear positioning and transcription in single cells (Rooijers et al. 2019; Cao et al. 2018) might soon enable the measurement of chromosomal contacts together with transcription factor binding or transcription in the same cell. Together with single-molecule studies in living cells, these methods will increase our understanding of how molecular events occurring over a disparate range of timescales, from transcription factor binding to loop extrusion dynamics, are integrated to reach defined transcriptional outputs.
Chromosome structure and other nuclear processes: DNA damage/repair, replication timing, and nuclear mechanics
While the impact of genome folding on transcription often dominates the conversation, chromosome architecture may also play important roles in other nuclear processes, notably DNA damage, repair, and translocation formation; replication timing; and the physical properties of the nucleus. 3D genome folding influences DNA damage and repair in numerous ways. Results from FISH, translocation sequencing and Hi-C have shown correlations between translocation frequency and the pre-existing 3D proximity between chromosome regions on average in the cell population (Parada et al. 2004; Roix et al. 2003; Zhang et al. 2012). Image tracking of fluorescently labelled DNA breaks in live cells has confirmed in individual cells that DNA double strand break (DSB) pairing most often occurs between breaks that were initially proximal, but has also revealed that large movements of DSBs sometimes occur (Roukos et al. 2013). Such variations in DNA repair progression at different sites are also influenced by chromosome structure: DSBs that occur in different nuclear locations and in heterochromatin vs. euchromatin exhibit different degrees of motion and choose different repair pathways (Lemaître and Soutoglou 2014). Spatial clustering of DSBs is revealed by monitoring genome folding with Capture-C after breaks are induced in euchromatin (Aymard et al. 2017), but inducing and monitoring specific breaks in heterochromatin has thus far been technically challenging.
DNA repair and chromosome folding pathways strikingly share important protein factors. CTCF and cohesin are emerging as key players in genome architecture, but have long been recognized as responding to DNA damage (Birkenbihl and Subramani 1992). Cohesin and CTCF are both recruited to DSBs downstream of the DNA damage response protein ATM (Luo et al. 2008) (Lang et al. 2017). With new experimental approaches, more connections between these genome architecture factors and DNA damage are emerging. Precise mapping of DNA DSBs with END-Seq revealed that CTCF TAD boundary sites are vulnerable to breakage due to topological strain (Canela et al. 2017, 2019; Gothe et al. 2019). Increasingly powerful superresolution microscopy has further shown that the domains of ɣH2AX DNA damage marks are flanked by CTCF sites and are the same size as TADs (Natale et al. 2017). The process of DNA repair can also have major consequences for chromosome structure. If the repair of DNA damage disrupts a TAD boundary, enhancers and promoters that are normally segregated may become juxtaposed, causing misregulation of oncogenes (Hnisz et al. 2016). It is possible that the links between DNA repair factors and chromosome architecture proteins promote the integrity of 3D genome structure during DNA damage repair. Indeed, as measured by Hi-C across a cell population, there is increased insulation between TADs associated with DNA damage repair (Sanders et al. 2019). This might serve to prevent mis-repair across TAD boundaries.
Further work will be needed to explore potential connections between loop extrusion, TADs, and DNA damage repair in the future. Population measurements cannot monitor genome contacts and DNA damage at the same loci in the same cell, but monitoring individual DSBs often requires artificial sequences to be introduced into the genome. Linked single cell profiling by several different genomic techniques would allow a better assessment of the connections between DNA DSBs and genome structure changes. Thus far, single cell multi-omics have measured ATAC-Seq and RNA-Seq in the same cells (Rooijers et al. 2019; Cao et al. 2018), and this concept might be extended to include techniques like END-Seq in the future.
Replication timing (RT) strongly correlates with A/B compartment identity, with early replicating regions in the A compartment and late replicating regions in the B compartment. Correspondingly, boundaries of RT domains correlate with TAD boundaries, and RT switches between cell types involve changes of whole TADs (Pope et al. 2014). In differentiated cells, these RT domains are more strongly correlated with A/B compartment structure than individual histone modifications (Dixon et al. 2012; Ryba et al. 2010). As such, RT and chromosome contact measurements are two orthogonal approaches that both reveal biological significance of the same type of domain structure. Replication timing profiles across patients and controls have identified local RT switches that may reveal biomarkers of disease. In this sense, the strong correlation between replication timing and chromosome structure allows RT profiling to be an efficient, high resolution approach to read out potential differences in local chromosome state and structure (Rivera-Mulia et al. 2017).
Future work will be needed to clarify what the correlation between RT and spatial compartments tells us about the mechanisms and implications of chromosome structure formation. Recent work reveals that neither RT domains nor A/B compartments are disrupted by deletion of their local boundary regions, but both RT and compartments are disrupted when certain sites (termed Early replicating control elements; ERCEs) inside domains are deleted (Sima et al. 2019). These regions appear to act on both RT and compartmentalization by affecting the active histone marks and gene expression of the broader region. In contrast, at very early time points of cell fate determination and differentiation, RT changes occur that do not coincide with A/B compartment changes (Dileep et al. 2019). More work is needed to identify how chromatin marks and other factors can sometimes specify RT change without spatial contact changes, when most often these two are closely related.
In addition to carrying, copying, and expressing genetic information, chromosomes are large physical objects that influence the mechanical properties of the cell (Bustin and Misteli 2016). Cellular manipulation and imaging approaches are revealing ways in which chromosome structure may impact the biophysical function of cells. Stretching individual nuclei has shown that chromatin state influences nuclear elasticity: increasing heterochromatin stiffens nuclei, while increasing euchromatin makes nuclei more elastic (Banigan et al. 2017). Conversely, physical forces can affect genome structure: nuclei aspirated through narrow micropipettes experience stretching of chromatin domains that is sometimes irreversible (Irianto et al. 2017), and external forces on cells can deform chromatin and directly affect gene expression (Tajik et al. 2016). Relatedly, changes to B compartment interactions have been observed after neutrophil nuclei squeeze through tight spaces in confined migration (Jacobson et al. 2018).
These observations suggest that certain chromosome structures could play a larger role in maintaining nuclear mechanical integrity than in specific gene regulation, or that there may be coupling between gene regulatory and physical roles. Additional work and new approaches are needed to bridge the gap between the sequence-specific contact information of chromosome conformation capture techniques and measurements of single cell mechanical properties and nucleus deformations. Approaches are emerging to mechanically manipulate large numbers of cells or nuclei simultaneously, making it possible to link these perturbations with bulk genomic measurements (Earle et al. 2019). But more work will be needed to address the heterogeneity of initial chromosome structures in individual cells and how these influence the effect of mechanical perturbation on the genome.
Conclusions and Outlook
Debate around chromosome architecture has sensibly shifted from how to obtain increasingly resolved and realistic measurement of 3D structures, to how to understand the forces that create them and how to determine their functionality in the broader context of nuclear processes. Given the explosion of studies that employ 3C methods and Hi-C in particular, it will be important to keep in mind which aspects these different methods can capture. As discussed in this review, some caution is required because population-averaged 3C-based measurements cannot represent all the structural heterogeneity in space and time: signals in 3C methods cannot fully represent 3D distances, and vice versa. In addition pairwise contacts measured by 3C do not necessarily capture simultaneous interactions; and importantly, interactions identified with high levels of confidence over background polymer behavior do not necessarily correlate with biological function. Despite these technical and conceptual limitations, however, recently developed orthogonal techniques have shown that population-averaged 3C methods capture realistic aspects of chromosome folding. Notably, the fact that 3C data can be interpreted quantitatively in terms of contact probabilities paves the way for a truly mechanistic understanding of chromosome organization and the forces that drive it. Key steps ahead in this direction are likely to emerge from approaches that combine increasingly refined and quantitative 3C methods with targeted perturbation approaches and mechanistic physical models.
As the search for the links between chromosome structure and biological function ramps up, it is interesting to note that the most statistically striking features that can be observed in 3C data may not always be the most biologically relevant. Transcriptional changes, for example, may be induced by brief contacts rather than only stable interactions, making correlations with structural changes hard to detect, or through more distant interactions mediated by microphase-separated condensates rather than locus-specific colocalization events. In the same way that systems biologists have moved from simple deterministic signaling pathways to emergent properties of complex regulatory networks, future work in defining the impact of chromosome contacts on function will likely involve models integrating complex subtle and dynamic changes into overall changes in phenotype. Increasingly sophisticated live-cell microscopy techniques will certainly play a major role in this direction. But discovery might also be enabled by emerging technologies such as cryo-electron tomography, which allows to visualize macromolecular complexes in situ, inside cells (Bäuerlein et al. 2017). We look forward to seeing future work addressing clear associations between chromosome structures and biological outputs, as well as the molecular mechanisms by which chromatin properties such as proximity, stiffness, compaction, affect biological processes inside and perhaps outside the nucleus.
Acknowledgements
We would like to apologize to all colleagues whose work could not be cited because of space constraints. We thank members of the Giorgetti lab and McCord lab for useful discussions and feedback on the text, Tongye Shen, Rafael Galupa and Tim Pollex for critically reading the manuscript, and Véronique Juvin for help with figures. We are grateful to the two anonymous reviewers for sharing insightful comments and helping improve the manuscript. Research in the Giorgetti lab is supported by the Novartis Research Foundation and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation (grant agreement no. 759366, ‘BioMeTre’). Research in the McCord lab is supported by the NIH NIGMS grant R35GM133557. Research in the Kaplan Lab is supported by the Taub Family Foundation, the Azrieli Faculty Fellows program and Israel Science Foundation Personal Research Grant 1479/18.
REFERENCES
- Abbas A, He X, Niu J, Zhou B, Zhu G, Ma T, Song J, Gao J, Zhang MQ, Zeng J. 2019. Integrating Hi-C and FISH data for modeling of the 3D organization of chromosomes. Nat Commun 10: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B. 2013. Chromatin Position Effects Assayed by Thousands of Reporters Integrated in Parallel. Cell 154: 914–927. [DOI] [PubMed] [Google Scholar]
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. https://elifesciences.org/articles/41769 (Accessed September 10, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen MJAM, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, et al. 2018. Enhancer hubs and loop collisions identified from single-allele topologies. Nature Genetics 50: 1151. [DOI] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal Dynamics at the Shh Locus: Limb Bud-Specific Differential Regulation of Competence and Active Transcription. Developmental Cell 16: 47–57. [DOI] [PubMed] [Google Scholar]
- Amitai A, Kantor Y, Kardar M. 2010. First-passage distributions in a collective model of anomalous diffusion with tunable exponent. Phys Rev E 81: 011107. [DOI] [PubMed] [Google Scholar]
- Andrey G, Schöpflin R, Jerković I, Heinrich V, Ibrahim DM, Paliou C, Hochradel M, Timmermann B, Haas S, Vingron M, et al. 2016. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. http://genome.cshlp.org/content/early/2017/01/20/gr.213066.116 (Accessed July 27, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F, Vu TH, Zeitz MJ, Varoquaux N, Carette JE, Vert J-P, Hoffman AR, Noble WS. 2015. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics 16: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, et al. 2017. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nature Structural & Molecular Biology 24: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan EJ, Stephens AD, Marko JF. 2017. Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Biophysical Journal 113: 1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Chotalia M, Fraser J, Lavitas L-M, Dostie J, Pombo A, Nicodemi M. 2012. Complexity of chromatin folding is captured by the strings and binders switch model. PNAS 109: 16173–16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CC-S, Raj A, Blobel GA. 2016. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Molecular Cell 62: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baù D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. 2011. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäuerlein FJB, Saha I, Mishra A, Kalemanov M, Martínez-Sánchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, et al. 2017. In Situ Architecture and Cellular Interactions of PolyQ Inclusions. Cell 171: 179–187.e10. [DOI] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DCA, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas L-M, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS. 2014. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Current Opinion in Cell Biology 26: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. 2019. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Molecular Cell 0 https://www.cell.com/molecular-cell/abstract/S1097-2765(19)30593-3 (Accessed September 5, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Dorier J, Burnier Y, Stasiak A. 2014. Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucl Acids Res 42: 2848–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, Olivo-Marin J-C, Gadal O, Zimmer C. 2008. High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods 5: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA Particles in Living Yeast. Molecular Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su J-H, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. 1992. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res 20: 6605–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu C, Zhuang X. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 0 https://www.cell.com/cell/abstract/S0092-8674(18)31398-9 (Accessed November 16, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. 2005. Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLOS Biology 3: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cohen NM, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. 2017. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171: 557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant L, Georgomanolis T, Nikolic M, Brackley CA, Kolovos P, van Ijcken W, Grosveld FG, Marenduzzo D, Papantonis A. 2016. Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Molecular Systems Biology 12: 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Misteli T. 2016. Nongenetic functions of the genome. Science 352: aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Huang SN, Wutz G, Tang W, Zagnoli-Vieira G, Callen E, Wong N, Day A, Peters J-M, et al. 2019. Topoisomerase II-Induced Chromosome Breakage and Translocation Is Determined by Chromosome Architecture and Transcriptional Activity. Molecular Cell 75: 252–266.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon K-R, Pekowska A, Zhang H, Rao SSP, et al. 2017. Genome Organization Drives Chromosome Fragility. Cell 170: 507–521.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline-Figueroa JL, Packer JS, Christiansen L, et al. 2018. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361: 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai Y, Fraser P. 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626. [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Pustova I, Walther N, Ho JJ, Hantsche-Grininger M, Inouye CJ, Hossain MJ, Dailey GM, Ellenberg J, Darzacq X, et al. 2019. Determining cellular CTCF and cohesin abundances to constrain 3D genome models eds. Sherratt DJ, Struhl K, and Sherratt DJ. eLife 8: e40164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoni DI, Gizzi AMC, Georgieva M, Stefano M, Valeri A, Chamousset D, Houbron C, Déjardin S, Fiche J-B, González I, et al. 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nature Communications 8: 1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Almo SC, Singer RH. 2008. Structural basis for the coevolution of a viral RNA–protein complex. Nat Struct Mol Biol 15: 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z, et al. 2014. Lattice Light Sheet Microscopy: Imaging Molecules to Embryos at High Spatiotemporal Resolution. Science 346: 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. 2018a. Dynamic interplay between enhancer–promoter topology and gene activity. Nature Genetics 50: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018b. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol jcb.201807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science eaar 4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-Range Directional Movement of an Interphase Chromosome Site. Current Biology 16: 825–831. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. 2002. Chromatin Motion Is Constrained by Association with Nuclear Compartments in Human Cells. Current Biology 12: 439–445. [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2: 292–301. [DOI] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters J-M. 2019. DNA loop extrusion by human cohesin. Science. https://science.sciencemag.org/content/early/2019/11/20/science.aaz3418 (Accessed November 22, 2019). [DOI] [PubMed] [Google Scholar]
- Dekker J 2016. Mapping the 3D genome: Aiming for consilience. Nature Reviews Molecular Cell Biology 17: 741–742. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zazhytska M, Borel C, Giannuzzi G, Rey G, Howald C, Kumar S, Ongen H, Popadin K, Marbach D, et al. 2019. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science 364: eaat8266. [DOI] [PubMed] [Google Scholar]
- Denker A, de Laat W. 2016. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev 30: 1357–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan W-L, Timmermann B, Wittler L, Vingron M, et al. 2019. Functional dissection of the Sox9 – Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nature Genetics 1. [DOI] [PubMed] [Google Scholar]
- de de Wit E, de Laat W. 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev 26: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. 2015. CTCF Binding Polarity Determines Chromatin Looping. Molecular Cell 60: 676–684. [DOI] [PubMed] [Google Scholar]
- Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. 2016. Transferable model for chromosome architecture. Proc Natl Acad Sci USA 113: 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. 2011. A High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo. PLoS Biol 9: e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Wilson KA, Marchal C, Lyu X, Zhao PA, Li B, Poulet A, Bartlett DA, Rivera-Mulia JC, Qin ZS, et al. 2019. Rapid Irreversible Transcriptional Reprogramming in Human Stem Cells Accompanied by Discordance between Replication Timing and Chromatin Compartment. Stem Cell Reports 13: 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi U, Holliday MJ, Eisenmesser EZ, Hamelberg D. 2016. Dynamical network of residue–residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. PNAS 113: 4735–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry M, Loïodice I, Chen CL, Thermes C, Taddei A. 2011. Tight protein–DNA interactions favor gene silencing. Genes Dev 25: 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, Iruvanti S, Moore SA, Bonne G, Wallrath LL, Lammerding J. 2019. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. bioRxiv 364778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Zimmer C. 2018. From dynamic chromatin architecture to DNA damage repair and back. Nucleus 9: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. 2008. Recruitment to the Nuclear Periphery Can Alter Expression of Genes in Human Cells. PLOS Genetics 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton A-L, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 176: 1502–1515.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcato M, Nicoletti C, Pal K, Livi CM, Ferrari F, Bicciato S. 2017. Comparison of computational methods for Hi-C data analysis. Nat Methods 14: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan W-L, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Abdennur N, Imakaev M, Goloborodko A, Mirny LA. 2017. Emerging Evidence of Chromosome Folding by Loop Extrusion. Cold Spring Harb Symp Quant Biol 82: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M. 2017. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nature Methods 14: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports 15: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Mirny LA. 2012. Higher-order chromatin structure: bridging physics and biology. Current Opinion in Genetics & Development 22: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. 2016. Enhancer Control of Transcriptional Bursting. Cell 166: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2018. Topologically Associating Domains in Chromosome Architecture and Gene Regulatory Landscapes during Development, Disease, and Evolution. Cold Spring Harb Symp Quant Biol 035030. [DOI] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. 2018. Real-time imaging of DNA loop extrusion by condensin. Science eaar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov A, Razin SV, Cavalli G. 2015. In vivo formaldehyde cross-linking: it is time for black box analysis. Briefings in Functional Genomics 14: 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov AA, Gushchanskaya ES, Strelkova O, Zhironkina O, Kireev II, Iarovaia OV, Razin SV. 2013. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res 41: 3563–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeven G, Teunissen H, de Laat W, de Wit E. peakC: a flexible, non-parametric peak calling package for 4C and Capture-C data. Nucleic Acids Res. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gky443/5003460 (Accessed August 13, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germier T, Kocanova S, Walther N, Bancaud A, Shaban HA, Sellou H, Politi AZ, Ellenberg J, Gallardo F, Bystricky K. 2017. Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophysical Journal 113: 1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 51: 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The Hierarchy of the 3D Genome. Molecular Cell 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. 2014. Predictive Polymer Modeling Reveals Coupled Fluctuations in Chromosome Conformation and Transcription. Cell 157: 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Heard E. 2016. Closing the loop: 3C versus DNA FISH. Genome Biology 17: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi AMC, Cattoni DI, Fiche J-B, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G, et al. 2019. Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Molecular Cell 74: 212–222.e5. [DOI] [PubMed] [Google Scholar]
- Golfier S, Quail T, Kimura H, Brugués J. 2019. Cohesin and condensin extrude loops in a cell-cycle dependent manner. bioRxiv 821306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. 2016. Compaction and segregation of sister chromatids via active loop extrusion ed. A.M. Van Oijen. eLife 5: e14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothe HJ, Bouwman BAM, Gusmao EG, Piccinno R, Petrosino G, Sayols S, Drechsel O, Minneker V, Josipovic N, Mizi A, et al. 2019. Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Molecular Cell 75: 267–283.e12. [DOI] [PubMed] [Google Scholar]
- Grosberg AY, Nechaev SK, Shakhnovich EI. 1988. The role of topological constraints in the kinetics of collapse of macromolecules. J Phys France 49: 2095–2100. [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. 2015. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell 162: 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. 2017. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169: 693–707.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Arnold CD, Pagani M, Rath M, Schernhuber K, Stark A. 2019. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung M-H, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. 2011. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res 21: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Cattoglio C, Darzacq X, Tjian R. 2018. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 9: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hsieh T-HS, Cattoglio C, Pustova I, Saldaña-Meyer R, Reinberg D, Darzacq X, Tjian R. 2019. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Molecular Cell. http://www.sciencedirect.com/science/article/pii/S1097276519305945 (Accessed October 16, 2019). [DOI] [PMC free article] [PubMed]
- Hao N, Shearwin KE, Dodd IB. 2019. Positive and Negative Control of Enhancer-Promoter Interactions by Other DNA Loops Generates Specificity and Tunability. Cell Reports 26: 2419–2433.e3. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. 2001. Chromosome Dynamics in the Yeast Interphase Nucleus. Science 294: 2181–2186. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Politi AZ, Nagasaka K, Hantsche-Grininger M, Walther N, Koch B, Fuchs J, Dürnberger G, Tang W, Ladurner R, et al. 2019. Absolute quantification of cohesin, CTCF and their regulators in human cells ed. D.J. Sherratt. eLife 8: e46269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T-HS, Slobodyanyuk E, Hansen AS, Cattoglio C, Rando OJ, Tjian R, Darzacq X. 2019. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. bioRxiv 638775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, Gobbi MD, Taylor S, Gibbons R, Higgs DR. 2014. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature Genetics 46: 205–212. [DOI] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Meth 9: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Greenberg RA, Discher DE. 2017. As a Nucleus Enters a Small Pore, Chromatin Stretches and Maintains Integrity, Even with DNA Breaks. Biophysical Journal 112: 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson EC, Perry JK, Long DS, Olins AL, Olins DE, Wright BE, Vickers MH, O’Sullivan JM. 2018. Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biology 16: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, et al. 2016. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167: 1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJM, Grosveld FG, Knoch TA, Murre C. 2008. The 3D Structure of the Immunoglobulin Heavy-Chain Locus: Implications for Long-Range Genomic Interactions. Cell 133: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Raviram R, Snetkova V, Rocha PP, Proudhon C, Badri S, Bonneau R, Skok JA, Kluger Y. 2016. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Res 44: 8714–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi O, Wang S-Y, Kuznetsova T, Atlasi Y, Peng T, Fabre PJ, Habibi E, Shaik J, Saeed S, Handoko L, et al. 2015. Dynamic Reorganization of Extremely Long-Range Promoter-Promoter Interactions between Two States of Pluripotency. Cell Stem Cell 17: 748–757. [DOI] [PubMed] [Google Scholar]
- Jost D, Carrivain P, Cavalli G, Vaillant C. 2014. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucl Acids Res 42: 9553–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotech 30: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Zhang Y, Lucas JS, Dudko OK, Murre C. 2019. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nature Communications 10: 2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Rege M, Valeri J, Dunagin MC, Metzger A, Titus KR, Gilgenast TG, Gong W, Beagan JA, Raj A, et al. 2019. LADL: light-activated dynamic looping for endogenous gene expression control. Nat Methods 16: 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide Maps of Nuclear Lamina Interactions in Single Human Cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh T-HS, Parsi KM, Yang L, Maehr R, Mirny LA, et al. 2019. Ultrastructural details of mammalian chromosome architecture. bioRxiv 639922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. The Journal of Cell Biology 180: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Li X, Zheng W, Li Z, Lu D, Chen G, Gong D, Yang L, Fu J, Shi P, et al. 2017. CTCF prevents genomic instability by promoting homologous recombination-directed DNA double-strand break repair. PNAS 114: 10912–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, Segerstolpe Å, Rivera CM, Ren B, Sandberg R. 2019. Genomic encoding of transcriptional burst kinetics. Nature 565: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Søballe B, Økstad O-A, Barre F-X, Sherratt DJ. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Molecular Microbiology 49: 731–743. [DOI] [PubMed] [Google Scholar]
- Le TBK, Imakaev MV, Mirny LA, Laub MT. 2013. High-Resolution Mapping of the Spatial Organization of a Bacterial Chromosome. Science 342: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Soutoglou E. 2014. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair 19: 163–168. [DOI] [PubMed] [Google Scholar]
- Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J. 2014. 3D genome reconstruction from chromosomal contacts. Nat Meth 11: 1141–1143. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay RJ, Pham B, Shen T, McCord RP. 2018. Characterizing the 3D structure and dynamics of chromosomes and proteins in a common contact matrix framework. Nucleic Acids Res 46: 8143–8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Li Y, Mu J-J, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. 2008. Regulation of Intra-S Phase Checkpoint by Ionizing Radiation (IR)-dependent and IR-independent Phosphorylation of SMC3. J Biol Chem 283: 19176–19183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sun X, Cormack BP, Boeke JD. 2018. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 560: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Tu L-C, Naseri A, Chung Y-C, Grunwald D, Zhang S, Pederson T. 2018. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging. Nat Methods 15: 928–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Weiner CL, Rinn JL. 2018. Inter-chromosomal Contact Properties in Live-Cell Imaging and in Hi-C. Molecular Cell 69: 1039–1045.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Sima J, Gilbert DM. 2019. Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol. http://europepmc.org/abstract/med/31477886 (Accessed October 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Current Biology 7: 930–939. [DOI] [PubMed] [Google Scholar]
- Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupé P, Barillot E, Belmont AS, Heard E. 2011. Live-Cell Chromosome Dynamics and Outcome of X Chromosome Pairing Events during ES Cell Differentiation. Cell 145: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RP, Balajee A. 2018. 3D Genome Organization Influences the Chromosome Translocation Pattern. Adv Exp Med Biol 1044: 113–133. [DOI] [PubMed] [Google Scholar]
- Monahan K, Horta A, Lomvardas S. 2019. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu J-F, et al. 2017. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale F, Rapp A, Yu W, Maiser A, Harz H, Scholl A, Grulich S, Anton T, Hörl D, Chen W, et al. 2017. Identification of the elementary structural units of the DNA damage response. Nat Commun 8: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the Mitotic Chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MH, Corces VG. 2015. A CTCF Code for 3D Genome Architecture. Cell 162: 703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir G, Farabella I, Estrada CP, Ebeling CG, Beliveau BJ, Sasaki HM, Lee SD, Nguyen SC, McCole RB, Chattoraj S, et al. 2018. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLOS Genetics 14: e1007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. 2011. Variegated gene expression caused by cell-specific long-range DNA interactions. Nature Cell Biology 13: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169: 930–944.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton HK, Emerson DJ, Huang H, Kim J, Titus KR, Gu S, Bassett DS, Phillips-Cremins JE. 2018. Detecting hierarchical genome folding with network modularity. Nature Methods 15: 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. PNAS 115: E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Chauvet P, Mukamel Z, Lifshitz A, Schwartzman O, Elkayam NO, Lubling Y, Deikus G, Sebra RP, Tanay A. 2016. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540: 296–300. [DOI] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. 2017. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar AM, Davies JOJ, Hanssen LLP, Telenius JM, Schwessinger R, Liu Y, Brown JM, Downes DJ, Chiariello AM, Bianco S, et al. 2018. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat Genet 50: 1744–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. 2004. Tissue-specific spatial organization of genomes. Genome Biol 5: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J, Sekelja M, Oldenburg AR, Barateau A, Briand N, Delbarre E, Shah A, Sørensen AL, Vigouroux C, Buendia B, et al. 2017. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biology 18: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 2011. The Nucleolus. Cold Spring Harb Perspect Biol 3: a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Molecular Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Li H-B. 2012. A view of nuclear Polycomb bodies. Current Opinion in Genetics & Development 22: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174: 744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J. 2017. Massively multiplex single-cell Hi-C. Nat Meth 14: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin Loss Eliminates All Loop Domains. Cell 171: 305–320.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redolfi J, Zhan Y, Valdes-Quezada C, Kryzhanovska M, Guerreiro I, Iesmantavicius V, Pollex T, Grand RS, Mulugeta E, Kind J, et al. 2019. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nat Struct Mol Biol 26: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Desprat R, Trevilla-Garcia C, Cornacchia D, Schwerer H, Sasaki T, Sima J, Fells T, Studer L, Lemaitre J-M, et al. 2017. DNA replication timing alterations identify common markers between distinct progeroid diseases. PNAS 114: E10972–E10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Ren G, Day CR, Zhao K, Chow CC, Larson DR. 2019. Intrinsic Dynamics of a Human Gene Reveal the Basis of Expression Heterogeneity. Cell 176: 213–226.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291. [DOI] [PubMed] [Google Scholar]
- Rooijers K, Markodimitraki CM, Rang FJ, de Vries SS, Chialastri A, de Luca KL, Mooijman D, Dey SS, Kind J. 2019. Simultaneous quantification of protein–DNA contacts and transcriptomes in single cells. Nat Biotechnol 37: 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. 2013. Spatial Dynamics of Chromosome Translocations in Living Cells. Science 341: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Molecular Cell 67: 837–852.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad H, Gallardo F, Dalvai M, Tanguy-le-Gac N, Lane D, Bystricky K. 2014. DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells. PLOS Genet 10: e1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, et al. 2019. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Molecular Cell. http://www.sciencedirect.com/science/article/pii/S1097276519306549 (Accessed October 16, 2019). [DOI] [PMC free article] [PubMed]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. PNAS 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JT, Freeman TF, Xu Y, Golloshi R, Stallard MA, Martin RS, Balajee AS, McCord RP. 2019. Radiation-Induced DNA Damage and Repair Effects on 3D Genome Organization. bioRxiv 740704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST, Offer J, et al. 2015. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Molecular Cell 57: 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. 2016. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Reports 17: 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Fraser P. 2019. Long-range enhancer–promoter contacts in gene expression control. Nat Rev Genet 20: 437–455. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre B-M, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. 2015. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering C H, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecal A, Munsky B, Proux F, Ly N, Braye FE, Zimmer C, Mueller F, Darzacq X. 2014. Transcription Factors Modulate c-Fos Transcriptional Bursts. Cell Reports 8: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra F, Baù D, Goodstadt M, Castillo D, Filion GJ, Marti-Renom MA. 2017. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLOS Computational Biology 13: e1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Shaban HA, Barth R, Bystricky K. 2018. Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res 46: e77–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Lu N, Cai C, Zhou F, Wang S, Zhao Z, Zhao G, Zhou J-Q, Xue X, Qin Z. 2019. A single circular chromosome yeast. Cell Res 29: 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, von Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, et al. 2019. Identifying cis Elements for Spatiotemporal Control of Mammalian DNA Replication. Cell 176: 816–830.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear Speckles. Cold Spring Harb Perspect Biol 3: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F 2016. Gene regulation at a distance: From remote enhancers to 3D regulatory ensembles. Seminars in Cell & Developmental Biology 57: 57–67. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Stack SM, Brown DB, Dewey WC. 1977. Visualization of interphase chromosomes. Journal of Cell Science 26: 281–299. [DOI] [PubMed] [Google Scholar]
- Stanyte R, Nuebler J, Blaukopf C, Hoefler R, Stocsits R, Peters J-M, Gerlich DW. 2018. Dynamics of sister chromatid resolution during cell cycle progression. J Cell Biol 217: 1985–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 2010. 4D Chromatin dynamics in cycling cells. Nucleus 1: 284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. 2009. Transcription factories: gene expression in unions? Nat Rev Genet 10: 457–466. [DOI] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, Spitz F. 2016. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Developmental Cell 39: 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. 2014. Functional and topological characteristics of mammalian regulatory domains. Genome Res 24: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nature Materials 15: 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Xing D, Chang C-H, Li H, Xie XS. 2018. Three-dimensional genome structures of single diploid human cells. Science 361: 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Gong K, Chen L, Alber F. 2012. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res 22: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, Zhou Y, Li H, Zhou XJ, Gros MAL, et al. 2016. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. PNAS 113: E1663–E1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature Methods 5: 159–161. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. 2002. Looping and Interaction between Hypersensitive Sites in the Active β-globin Locus. Molecular Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- van de Werken HJG, Landan G, Holwerda SJB, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Öz Y, Bouwman BAM, et al. 2012. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Meth 9: 969–972. [DOI] [PubMed] [Google Scholar]
- Varoquaux N, Ay F, Noble WS, Vert J-P. 2014. A statistical approach for inferring the 3D structure of the genome. Bioinformatics 30: i26–i33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, Bickmore WA. 2015. The Hierarchy of Transcriptional Activation: From Enhancer to Promoter. Trends in Genetics 31: 696–708. [DOI] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon K-R, Jung S, Baranello L, Huang S-C, Khattabi LE, Dose M, Pruett N, et al. 2018. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 173: 1165–1178.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. 2015. Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. Cell Reports 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nakamura M, Abbott TR, Zhao D, Luo K, Yu C, Nguyen CM, Lo A, Daley TP, Russa ML, et al. 2019a. CRISPR-mediated live imaging of genome editing and transcription. Science 365: 1301–1305. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu X, Nguyen CM, Liu Y, Gao Y, Lin X, Daley T, Kipniss NH, La Russa M, Qi LS. 2018. CRISPR-Mediated Programmable 3D Genome Positioning and Nuclear Organization. Cell 175: 1405–1417.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, et al. 2019b. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Molecular Cell. http://www.sciencedirect.com/science/article/pii/S1097276519306586 (Accessed November 14, 2019). [DOI] [PubMed]
- Wang S, Su J-H, Beliveau BJ, Bintu B, Moffitt JR, Wu C, Zhuang X. 2016. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. 2014. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev 28: 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Kane L, Devenney PS, Flyamer IM, Anderson E, Kilanowski F, Hill RE, Bickmore WA, Lettice LA. 2019. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146: dev179523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Marie-Nelly H, Herbert S, Carrivain P, Blanc H, Koszul R, Fabre E, Zimmer C. 2012. A Predictive Computational Model of the Dynamic 3D Interphase Yeast Nucleus. Current Biology 22: 1881–1890. [DOI] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. The EMBO Journal 36: 3573–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Mariani L, Barozzi I, Schulz EG, Blüthgen N, Stadler M, Tiana G, Giorgetti L. 2017. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res 27: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dudko OK. 2016. First-Passage Processes in the Genome. Annual Review of Biophysics 45: 117–134. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho Y-J, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial Organization of the Mouse Genome and Its Role in Recurrent Chromosomal Translocations. Cell 148: 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Tian SZ, Capurso D, Kim M, Maurya R, Lee B, Piecuch E, Gong L, Zhu JJ, Li Z, et al. 2019. Multiplex chromatin interactions with single-molecule precision. Nature 566: 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Lavis LD. 2017. Development of photostable fluorophores for molecular imaging. Current Opinion in Chemical Biology 39: 32–38. [DOI] [PubMed] [Google Scholar]