Abstract
Background
Although analysis of cardiac magnetic resonance (CMR) images provides accurate and reproducible measurements of left ventricular (LV) volumes, these measurements are usually not performed throughout the cardiac cycle because of lack of tools that would allow such analysis within a reasonable timeframe. A fully-automated machine-learning (ML) algorithm was recently developed to automatically generate LV volume-time curves. Our aim was to validate ejection and filling parameters calculated from these curves using conventional analysis as a reference.
Methods
We studied 21 patients undergoing clinical CMR examinations. LV volume-time curves were obtained using the ML-based algorithm (Neosoft), and independently using slice-by-slice, frame-by-frame manual tracing of the endocardial boundaries. Ejection and filling parameters derived from these curves were compared between the two techniques. For each parameter, Bland-Altman bias and limits of agreement (LOA) were expressed in percent of the mean measured value.
Results
Time-volume curves were generated using the automated ML analysis within 2.5 ± 0.5 min, considerably faster than the manual analysis (43 ± 14 min per patient, including ~10 slices with 25–32 frames per slice). Time-volume curves were similar between the two techniques in magnitude and shape. Size and function parameters extracted from these curves showed no significant inter-technique differences, reflected by high correlations, small biases (< 10%) and mostly reasonably narrow LOA.
Conclusion
ML software for dynamic LV volume measurement allows fast and accurate, fully automated analysis of ejection and filling parameters, compared to manual tracing based analysis. The ability to quickly evaluate time-volume curves is important for a more comprehensive evaluation of the patient’s cardiac function.
Keywords: Left ventricle, Time-volume curves, Artificial intelligence
1. Introduction
To date, most cardiac parameters derived from cardiovascular magnetic resonance (CMR) images are obtained using manual measurements, including chamber volumes that rely on tracing of endocardial boundaries. Automated identification of cardiac chambers followed by accurate measurements without time-consuming and experience-dependent user input would be a major development in clinical cardiac imaging. Over the past decades, several techniques geared toward automation of CMR analysis have been reported with mixed results [1–4]. With the recent surge of artificial intelligence approaches, such as machine learning (ML) [5–10], a novel ML-based CMR technique was developed to automatically detect left ventricular (LV) boundaries and thus allow fully automated measurements of LV volume not only at end-systole and end-diastole, but for every frame throughout the cardiac cycle.
This approach allows, in addition to automated measurement of commonly used indices of LV function, such as ejection fraction (EF), quantification of dynamic volume changes that can be used to obtained potentially clinically valuable information regarding pathophysiology of disease, especially as it relates to changes in diastolic function and remodeling [11–16]. Despite the growing understanding of such dynamic indices, their use has been mostly limited to research studies, because their derivation required dedicated tools for off-line analysis that was time-consuming to a degree that hindered its use in clinical routine. The new ML approaches offer an opportunity for a more comprehensive evaluation of LV function, based on several parameters of ejection/filling, such as ejection and filling rates or percent ejection or filling at certain phases of the cardiac cycle, that are currently not used clinically. This study was designed to test this new automated algorithm by comparing LV time-volume curves and parameters of ventricular size and function derived from them to those obtained using conventional CMR volumetric analysis based on manual tracing of the LV endocardial boundaries.
2. Methods
2.1. Population
We prospectively studied 21 consecutive patients (age 53 ± 18, 13 females, BSA 1.9 ± 0.2 m2) undergoing clinical CMR examinations for various indications. Patients with pacemaker or defibrillator leads, significant arrhythmias or incomplete datasets were excluded. This study complied with ethical standards, was approved by the Institutional Review Board and each patient signed an informed consent.
2.2. Study design
LV time-volume curves were obtained from CMR cine images using a ML-based algorithm, and independently using frame-by-frame hand tracing of short-axis stacks in the same datasets. Analysis was performed by an experienced reader with level III CMR training, who was blinded to all prior measurements. Time curves obtained using the automated ML-based analysis and the manual methodology were automatically analyzed to derive LV volumes and ejection/filling parameters.
2.3. Image acquisition
CMR imaging was performed using a 1.5-T system (Achieva, Philips Healthcare) with a 5-channel cardiac coil. Steady-state free precession dynamic gradient-echo sequence with retrospective ECG gating and parallel imaging sensitivity encoding during ~5 second breath-holds (TR 2.9 ms, TE 1.5 ms, flip-angle 60°, and temporal resolution ~3040 ms) was used to obtain cine loops of 6-mm thick short-axis slices with 2-mm gaps and 2.0 × 2.0-mm in-plane spatial resolution from above the mitral valve to below the LV apex. The resultant frame rate varied between 25 and 32 frames per cardiac cycle.
2.4. ML image analysis
Automated analysis was performed using the SuiteHEART software (Neosoft, Pewaukee, WI), which uses a ML-based approach to measure LV volumes frame-by-frame throughout the cardiac cycle. The initial segmentation process identifies anatomical features, including LV cavity, and creates contours, which are fit into these anatomical structures. This ML algorithm utilizes training information from contours validated by experts in a large number of images [17]. With this training information, the ML algorithm effectively learns where an expert user would position boundaries between structures in actual clinical images, resulting in robust contour identification even on images of suboptimal quality, where less sophisticated approaches are likely to produce unsatisfactory results [17]. Using this approach, LV time-volume curves were derived by applying the method of disks to each consecutive frame of the cardiac cycle. No manual editing of endocardial boundaries was performed in this study, although the software allows such editing if deemed necessary.
2.5. Manual image analysis
The same CMR images were analyzed conventionally using QMass software (Medis, Leiden, Netherlands). Analysis included tracing of the circumference of the LV cavity that appeared surrounded by myocardial tissue from LV base to the apical tip of the LV cavity. Long-axis views were used to confirm the basal and apical slices. Manual boundary tracing was performed frame-by-frame throughout the cardiac cycle, while including the papillary muscles and trabeculae in the LV cavity. LV time-volume curve were generated by calculating LV volume for each frame, using the method of disks. With the average of 10 slices with 25–30 frames per slice, the total number of manual tracings was 250–300 per patient.
2.6. Analysis of time-volume curves
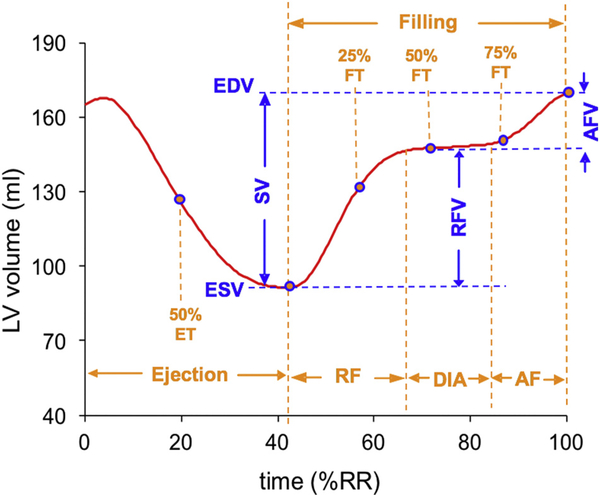
LV time-volume curves obtained using the two techniques (ML and manual tracing) were analyzed using Microsoft Excel worksheet that was designed to calculate several LV volume and timing parameters. The various phases of the cardiac cycle were detected using peaks and troughs of the volume curve and its time-derivative, which first underwent temporal smoothing using weighted averaging in a moving 3-point window. The resultant LV ejection and filling metrics included: end-diastolic and end-systolic volumes (EDV, ESV), ejection fraction (EF), volume at 50% ejection time (ET), volumes at 25, 50 and 75% filling time (FT), volume at diastasis, rapid and atrial filling volumes (RFV and AFV) (Fig. 1).
Fig. 1.
Analysis of left ventricular volume-time curves, resulting in dynamic ejection and filling parameters: end-diastolic and end-systolic volumes (EDV, ESV) and stroke volume (SV), volume at 50% ejection time (ET), volumes at 25, 50 and 75% filling time (FT), volume at diastasis (DIA), rapid filling volume (RFV) and atrial filling volume (AFV).
2.7. Statistical analysis
For each calculated parameter, comparisons between automated and manual techniques included paired two-tailed student’s t-tests, linear regression with Pearson correlation coefficients and Bland-Altman analyses to assess the bias and limits of agreement (LOA, namely 2SD around the mean). For each parameter, bias and LOA were expressed in percent of the mean measured value, in order to put the magnitude of the differences in perspective of what is measured. Values of p < 0.05 by t-tests were considered significant.
3. Results
The ML algorithm was successfully applied to all datasets, and the time needed to generate time-volume curves was significantly shorter (2.5 ± 0.5 min per patient), compared to manual frame-by-frame tracing of every slice from base to apex (43 ± 14 min, p < 0.0001).
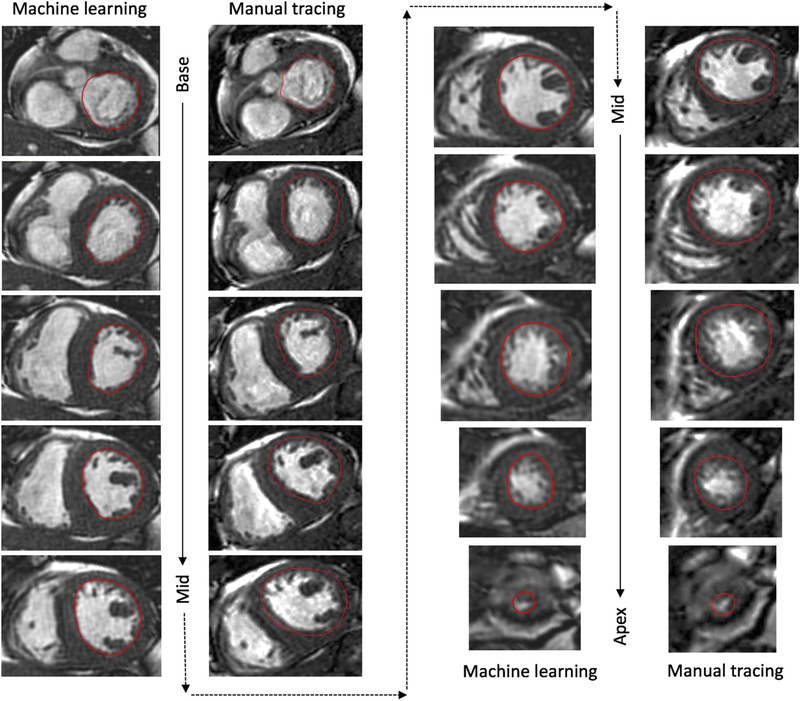
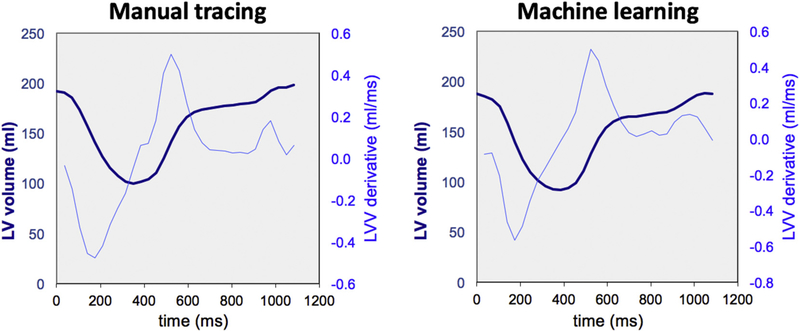
Fig. 2 shows an example of short-axis images of the left ventricle from base to apex with endocardial boundaries automatically detected by the ML algorithm side-by-side with those manually traced for comparison. Fig. 3 shows an example of LV time-volume curve and its time-derivative, obtained using the automated ML-based technique and manual tracing, where notably, the curves are very similar in their shape and magnitude. LV time-volume curves clearly differentiated systolic contraction from the biphasic filling with the rapid active LV relaxation separated by diastasis from the slower passive LV filling driven by atrial contraction. Importantly, the time-derivative curve clearly depicted peaks and troughs that were easy to detect despite the signal noise. This included a first peak corresponding to the maximum rate of systolic contraction, followed by zero crossing, indicating the end of systole, and then followed by two troughs: the first reflecting peak rate of rapid filling during active ventricular relaxation and the second corresponding to peak passive filling driven by atrial contraction.
Fig. 2.
Example of end-systolic short-axis images from left-ventricular base to apex with endocardial boundaries traced manually (left) and detected automatically by the machine learning algorithm (right).
Fig. 3.
Example of left ventricular volume-time curves (dark-blue, thick lines) and their time-derivatives (light-blue, thin lines), obtained in one study subject using the two analysis techniques: manual tracing (left), and machine learning (right). See text for details. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
These features of the curves allowed the determination of the cardiac cycle phases and derivation of the above dynamic indices of LV ejection and filling. Table 1 shows the summary of LV ejection and filling parameters measured by the automated ML algorithm and by manual tracing. None of the measured parameters showed significant inter-technique differences. The Table also shows the results of the detailed analysis of inter-technique agreement for the LV parameters, including correlation coefficients and Bland-Altman biases and LOA for all measured parameters. Generally, ML-derived indices were in good agreement with the manual reference technique, as evidenced by high correlations and small biases (≤10% of the measured values for all indices, and ≤ 5% for 14/16 of them). Of note, however, the LOA were rather wide for some of the parameters relying on the time derivatives, e.g. rapid and atrial filling volumes and volume fractions, indicating considerable inter-technique differences in individual subjects in these particular parameters.
Table 1.
Left ventricular ejection and filling parameters derived from volume-time curves obtained using the automated machine learning (ML) algorithm, and by using conventional CMR analysis in 21 study subjects (volumes are expressed in units of ml; fractions are in percent), as well as inter-technique agreement assessed by linear regression and Bland-Altman analysis (bias and limits of agreement (LOA) are expressed in percent of the corresponding measured value).
| Manual | ML | r | Bias ± LOA (%) | ||
|---|---|---|---|---|---|
| Ejection indices | End-systolic volume | 76 ± 27 | 77 ± 30 | 0.93 | 2 ± 29 |
| Stroke volume | 100 ± 32 | 93 ± 29 | 0.85 | −7 ± 35 | |
| Ejection fraction | 57 ± 9 | 55 ± 8 | 0.70 | −4 ± 25 | |
| Volume (50% ejection time) | 118 ± 39 | 118 ± 43 | 0.96 | 0 ± 20 | |
| Filling indices | End-diastolic volume | 176 ± 52 | 170 ± 53 | 0.97 | −3 ± 14 |
| Filling duration (%RR) | 63 ± 6 | 62 ± 7 | 0.64 | −1 ± 19 | |
| Volume (25% filling time) | 102 ± 41 | 98 ± 41 | 0.95 | −5 ± 26 | |
| Volume (50% filling time) | 137 ± 51 | 132 ± 41 | 0.99 | −4 ± 12 | |
| Volume (75% filling time) | 151 ± 50 | 145 ± 49 | 0.98 | −4 ± 15 | |
| Volume at diastasis | 144 ± 52 | 140 ± 51 | 0.98 | −3 ± 14 | |
| Rapid filling volume | 68 ± 34 | 63 ± 27 | 0.88 | −9 ± 51 | |
| Rapid filling fraction | 0.67 ± 0.17 | 0.66 ± 0.15 | 0.88 | −1 ± 24 | |
| Atrial filling volume | 31 ± 16 | 30 ± 15 | 0.86 | −3 ± 53 | |
| Atrial filling fraction | 0.33 ± 0.17 | 0.34 ± 0.15 | 0.88 | 1 ± 48 |
4. Discussion
The emergence ML techniques in cardiac imaging will undoubtedly have major impact on how we diagnose and manage patients with cardiovascular disease. Specifically, in CMR imaging, ML-based algorithms promise to provide automated chamber measurements that are accurate, improve efficiency, and are likely to yield clinically relevant information that has the potential to aid with diagnosis and evaluate prognosis in individual patients. In this study, an automated ML algorithm was used to demonstrate that: 1) ML can measure LV volume throughout the cardiac cycle, lending itself to automated analysis of potentially clinically meaningful ejection/filling parameters, and 2) these measurements are accurate, when compared to conventional CMR analysis, but require considerably less time. These findings indicate that incorporation of ML into daily practice of CMR analysis of LV size and function in the near future may be a realistic expectation.
The concept of computer aided diagnosis is not new within the realm of cardiac imaging in the context of nuclear stress testing, where automated quantification of myocardial perfusion is routinely used to facilitate the interpretation of images. Recent advances in machine learning, such as deep learning and advanced convolutional neural network architectures have revolutionized the performance of tasks such as image classification and segmentation. These developments have enabled computer programs to learn complex relationships or patterns from empirical data and generate accurate decisions [18]. Unlike chest radiography or even computed tomography, which usually require interpretation of static images, CMR images are dynamic. As a consequence, the incorporation of artificial intelligence and ML into CMR has been lagging behind. Nevertheless, the recent decade has witnessed significant efforts geared toward the realization of the potential of ML algorithms for analysis of CMR images. However, previous applications of ML were limited in their ability to accurately assess ventricular volume throughout the cardiac cycle.
Recent technological developments in artificial intelligence, such as ML and deep learning techniques, have facilitated the development of a new, fully-automated algorithm for dynamic quantification of LV size and function, which until recently might have seemed unrealistic. Our current study focused on a new ML algorithm that yields LV volume curves and allows analysis of potentially useful indices of LV ejection/ filling parameters that cannot be derived from static volume measurements at select phases of the cardiac cycle. This is similar to the recently developed ML-based dynamic LV analysis from 3D echocardiographic images [19], except the latter requires manual editing of endocardial boundaries, which was not part of the current CMR study. The additional parameters of LV ejection/filling may become part of future more comprehensive analysis of LV function, which may be particularly useful in the evaluation of patients with diastolic dysfunction. Our study was designed to test the accuracy of this methodology, by comparing the automatically derived indices to the reference derived from curves obtained using conventional manual analysis. This study showed that the automatically derived dynamic LV ejection/filling indices agreed well with the above reference. It is true that the agreement between the ML-derived parameters and the manual reference values was not perfect, as reflected by non-zero biases. However, the biases were ≤ 5% of the measured value for the majority of parameters, which is similar to or even below intrinsic inter-measurement variability and likely not clinically significant. The relatively wide limits of agreement indicate that differences can be large at times in individual patients, mostly as a result of differences in boundary detection in the basal slices, where the differentiation between ventricular and atrial cavities may be challenging. These differences can probably be minimized by manual corrections of the automatically determined boundaries, which the software provides an opportunity for. Such corrections were not part of our study, which was designed to test the feasibility and accuracy of the fully automated approach.
Not surprisingly, the ML technique was considerably faster than the conventional analysis, due to its automated nature. In fact, it is conceivable that this algorithm could provide accurate measurements when used in the background, before a cardiologist even starts reviewing the patient’s images.
4.1. Future directions
CMR is a multi-parametric technique capable of providing a large variety of information beyond cine imaging. In addition to more completely assessing dynamic changes in chamber size over the cardiac cycle, it is conceivable that in the future ML may also be useful to identify perfusion defects on stress perfusion images or to detect the presence of myocardial damage on late gadolinium enhancement images [10,20,21]. Such applications of ML could help harmonize the variation in image interpretation that currently exists.
4.2. Limitations
Our sample size was small, which raises the question whether insufficient statistical power was the reason for our measurements not being significantly different between techniques. However, our detailed analysis of inter-technique agreement, which included linear regression and Bland-Altman analyses, showed good correlation and only small biases for most parameters, even if limits of agreement were rather wide for some of the indices, suggesting that in some patients the agreement was not perfect. Further studies are needed to further validate this new methodology. Specifically, this is important for patients with more complex cardiac anatomy.
Additionally, ejection and filling indices were calculated in this study using custom Excel worksheet from time-volume curves and their time-derivative curves, which had varying degrees of signal noise. Because small changes in volumes are amplified in the derivative curves, their random nature might have affected the calculated parameters. Additional algorithmic refinements are needed to improve the confidence in the accuracy of the derived functional parameters.
5. Conclusions
In this study, we demonstrated that a new ML-based approach to analysis of CMR images allows fully automated, dynamic measurement of LV volume throughout the cardiac cycle, lending itself to accurate derivation of potentially useful ejection and filling indices, which characterize LV function more comprehensively than the traditional, isolated end-systolic and end-diastolic volumes and ejection fraction. Importantly, the automated nature of this analysis offers a pathway to solving the workflow limitations and may eventually result in integration of the additional parameters into clinical practice. One may anticipate that following further validation of such techniques, physicians will be starting their evaluation of clinical CMR examinations with an array of automatically measured functional parameters, that they will see with the images as part of their diagnostic interpretation.
Acknowledgments
Declaration of competing interest
ARP has received research support from Philips Healthcare and Neosoft. Akhil Narang was funded by a T32 Cardiovascular Sciences Training Grant (5T32HL7381) from the National Institutes of Health (USA).
Abbreviations
- AFV
atrial filling volume
- CMR
cardiovascular magnetic resonance
- EF
ejection fraction
- EDV, ESV
end-diastolic and end-systolic volumes
- ET
ejection time
- FT
filling time
- LV
left ventricular
- ML
machine learning
- RFV
rapid filling volume
References
- [1].Afshin M, Ben Ayed I, Islam A, Goela A, Peters TM, Li S. Global assessment of cardiac function using image statistics in MRI. Med Image Comput Comput Assist Interv 2012;15:535–43. [DOI] [PubMed] [Google Scholar]
- [2].Tan LK, Liew YM, Lim E, McLaughlin RA. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Med Image Anal 2017;39:78–86. [DOI] [PubMed] [Google Scholar]
- [3].Oksuz I, Mukhopadhyay A, Dharmakumar R, Tsaftaris SA. Unsupervised myocardial segmentation for cardiac BOLD. IEEE Trans Med Imaging 2017;36:2228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mantilla J, Paredes J, Bellanger JJ, Donal E, Leclercq C, Medina R, et al. Classification of LV wall motion in cardiac MRI using kernel Dictionary Learning with a parametric approach. Conf Proc IEEE Eng Med Biol Soc 2015;2015:7292–5. [DOI] [PubMed] [Google Scholar]
- [5].Emad O, Yassine IA, Fahmy AS. Automatic localization of the left ventricle in cardiac MRI images using deep learning. Conf Proc IEEE Eng Med Biol Soc 2015;2015:683–6. [DOI] [PubMed] [Google Scholar]
- [6].Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformablemodel approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016;30:108–19. [DOI] [PubMed] [Google Scholar]
- [7].Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal 2017;35:159–71. [DOI] [PubMed] [Google Scholar]
- [8].Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 2018;20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Samad MD, Wehner GJ, Arbabshirani MR, Jing L, Powell AJ, Geva T, et al. Predicting deterioration of ventricular function in patients with repaired tetralogy of Fallot using machine learning. Eur Heart J Cardiovasc Imaging 2018;19:730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Larroza A, Lopez-Lereu MP, Monmeneu JV, Gavara J, Chorro FJ, Bodi V, et al. Texture analysis of cardiac cine magnetic resonance imaging to detect nonviable segments in patients with chronic myocardial infarction. Med Phys 2018;45:1471–80. [DOI] [PubMed] [Google Scholar]
- [11].Poutanen T, Ikonen A, Jokinen E, Vainio P, Tikanoja T. Transthoracic three-dimensional echocardiography is as good as magnetic resonance imaging in measuring dynamic changes in left ventricular volume during the heart cycle in children. Eur J Echocardiogr 2001;2:31–9. [DOI] [PubMed] [Google Scholar]
- [12].Zeidan Z, Erbel R, Barkhausen J, Hunold P, Bartel T, Buck T. Analysis of global systolic and diastolic left ventricular performance using volume-time curves by real-time three-dimensional echocardiography. J Am Soc Echocardiogr 2003;16:29–37. [DOI] [PubMed] [Google Scholar]
- [13].Tashiro H, Aoki T, Sadamatsu K, Ooe K, Yamawaki T, Sagara S. Evaluation of the left ventricular diastolic function using three-dimensional echocardiography. Echocardiography 2008;25:968–73. [DOI] [PubMed] [Google Scholar]
- [14].Kort S, Mamidipally S, Madahar P, Dave S, Brown DL. Real time three-dimensional stress echocardiography: a new approach for assessing diastolic function. Echocardiography 2011;28:676–83. [DOI] [PubMed] [Google Scholar]
- [15].Yodwut C, Lang RM, Weinert L, Ahmad H, Mor-Avi V. Three-dimensional echocardiographic quantitative evaluation of left ventricular diastolic function using analysis of chamber volume and myocardial deformation. Int J Cardiovasc Imaging 2013;29:285–93. [DOI] [PubMed] [Google Scholar]
- [16].Nakanishi K, Fukuda S, Watanabe H, Seo Y, Mahara K, Hyodo E, et al. The utility of fully automated real-time three-dimensional echocardiography in the evaluation of left ventricular diastolic function. J Cardiol 2015;66:50–6. [DOI] [PubMed] [Google Scholar]
- [17].Backhaus SJ, Staab W, Steinmetz M, Ritter CO, Lotz J, Hasenfuss G, et al. Fully automated quantification of biventricular volumes and function in cardiovascular magnetic resonance: applicability to clinical routine settings. J Cardiovasc Magn Reson 2019;21:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16:933–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Narang A, Mor-Avi V, Prado A, Volpato V, Prater D, Tamborini G, et al. Machine learning based automated dynamic quantification of left heart chamber volumes. Eur Heart J Cardiovasc Imaging 2018;0:1–9. 10.1093/ehjci/jey137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Larroza A, Materka A, Lopez-Lereu MP, Monmeneu JV, Bodi V, Moratal D. Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur J Radiol 2017;92:78–83. [DOI] [PubMed] [Google Scholar]
- [21].Baessler B, Mannil M, Maintz D, Alkadhi H, Manka R. Texture analysis and machine learning of non-contrast T1-weighted MR images in patients with hypertrophic cardiomyopathy-preliminary results. Eur J Radiol 2018;102:61–7. [DOI] [PubMed] [Google Scholar]