Abstract
The deregulation of exosomal microRNAs (miRNAs) plays an important role in the progression of hepatocarcinogenesis. In this study, we highlight exosomes as mediators involved in modulating miRNA profiles in liver cancer cells after induction of the epithelial-mesenchymal transition (EMT) and metastasis. Initially, we induced EMT in a hepatocellular carcinoma cell (HCC) line (Hep3B) by stimulation with transforming growth factor-β (TGF-β) and confirmed by western blot detection of EMT markers such as vimentin and E-cadherin. Exosomes were then isolated from the cells and identified by nanoparticle tracking analysis (NTA). The isolated exosomal particles from unstimulated Hep3B cells (Hep3B exo) or TGF-β-stimulated EMT Hep3B cells (EMT-Hep3B exo) contained higher levels of exosome marker proteins, CD63 and TSG101. After incubation with EMT-Hep3B exo, Hep3B cell proliferation increased. EMT-Hep3B exo promoted the migration and invasion of Hep3B and 7721 cells. High-throughput sequencing of miRNAs and mRNA within the exosomes showed 119 upregulated and 186 downregulated miRNAs and 156 upregulated and 166 downregulated mRNA sequences in the EMT-Hep3B exo compared with the control Hep3B exo. The most differentially expressed miRNAs and target mRNA sequences were validated by RT-qPCR. Based on the known miRNA targets for specific mRNA sequences, we hypothesized that GADD45A was regulated by miR-374a-5p. Inhibition of miR-374a-5p in Hep3B cells resulted in exosomes that inhibited the proliferation, migration, and invasion of HCC cells. These results enhance our understanding of metastatic progression of liver cancer and provide a foundation for the future development of potential biomarkers for diagnosis and prognosis of hepatic cancer.
Keywords: Exosome, microRNA, epithelial-mesenchymal transition, liver cancer, metastasis, TGF-β
Introduction
Hepatocellular carcinoma (HCC) remains the sixth most common lethal malignancy and is considered to be the third highest cause of mortality in cancer-related deaths worldwide. Due to its poor prognosis and a high risk of recurrence with metastatic complications, it is mandatory to identify effective, noninvasive, and specific biomarkers that could provide early predictive potential in diagnosis of HCC [1]. Metastasis of liver cancers including HCC is a multistep process including an epithelial-mesenchymal transition (EMT), which is characterized by the loss of epithelial-like characteristics and the gain of mesenchymal-like attributes including tumor cell migration and invasion. The establishment of the mesenchymal state and metastasis is facilitated by intercellular crosstalk, which can occur in many different ways between neighboring and distant tumor cells as well as immune and stromal cells in the tumor microenvironment [2].
Extracellular vesicles called exosomes carry a concentrated group of functional molecules, provide protection to the transported molecules, and serve as intercellular communicators that can regulate many cellular processes while promoting angiogenesis, invasion, and proliferation in recipient cells to support tumorigenesis. Most prokaryotic and eukaryotic cells release exosomes, and this quality is even retained in cancers such as HCC [3,4], colorectal [5] lung, breast, ovarian cancers, glioblastoma (GBM), and melanoma [6]. Exosomes are nanosized microvesicles (30-100 nm in diameter) produced by many cell types, including T cells, B cells, dendritic cells, epithelial cells, and tumor cells [7-9]. Emerging studies have shown that tumor cells can secrete exosomes into the extracellular space, which then migrate to a distant place to transfer their functional effectors (RNAs, microRNAs (miRNAs), and proteins) to recipient cells and exert pleiotropic roles in regulating tumor progression, metastasis, and immune dysregulation [10]. Exosomes provide a novel way to transfer effector messages between cells including RNA, proteins, miRNAs, long noncoding RNAs (lncRNAs), or DNA fragments [11,12]. A number of studies have demonstrated that abundant cell-free miRNAs from exosomes can induce miRNA-mediated repression of target genes and function effectively in various pathological processes as either oncogenes or tumor suppressors in recipient normal or tumor cells [13,14].
MiRNAs, a family of non-coding RNAs of 20-25 nt, play pivotal roles during this remodeling process by regulating target genes at the post-transcriptional level [15]. Recent studies have shown that exosomal miRNAs have greater stability than free miRNAs, and the dysregulated expression of miRNAs in exosomes can accelerate HCC progression by promoting cell proliferation and metastasis via alteration of a network of genes [1,16]. As post-transcriptional regulators, miRNAs negatively regulate gene expression by binding directly to the 3’-untranslated regions (UTRs) of target mRNAs, thereby inducing mRNA degradation or repression of protein translation [10].
Previously validated exosomal-mediated pathogenic mechanisms include activation of target cells through the transfer of ligands promoting oncogenic activity and transfer of receptors resulting in oncogenic activity via activation of growth factor signaling pathways in recipient cells. In addition, a novel mechanism of endocytosis of exosomes delivering miRNAs, which can functionally repress target genes in recipient cells, has been demonstrated [17]. Tumor-derived exosomes usually circulate systemically to interact with surrounding stromal tissue and promote tumor progression by activating proliferative and angiogenic pathways [18-21]. Because exosomes carry their parental cell-type specific proteins and other regulatory molecules including miRNAs, they are explored as prognostic indicators of several types of malignancies and serve as biomarkers in assessing tumor types and stages [22-24]. A growing number of miRNAs have been implicated in the regulation of EMT-related pathways in cancer [25-28]. Although exosomes have been studied for many years, biological roles of exosomal miRNAs in mediating liver cancers are just beginning to be understood.
In this study, we have performed high-throughput sequencing of miRNAs in exosomes derived from hepatic cancer cells after stimulating EMT by transforming growth factor-β (TGF-β). The miRNAs specifically upregulated in sensitized cells were validated and subjected to gene target prediction, gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, which showed possible functions associated with these signature miRNAs. In addition, we studied the functional roles of the screened miRNAs in hepatic cancer cells. The results of this study may help in developing potential biomarkers for diagnosis and prognosis of hepatic cancer progression.
Materials and methods
Cell culture and treatment with TGF-β
Human hepatocellular carcinoma cells (Hep3B) and SMMC-7721 cells (7721) were obtained from Sun Yat-Sen University Laboratory Animal Center (Guangdong, China). Hep3B cells were grown in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS, SBI System Biosciences, Palo Alto, CA, USA), 100 units/mL of penicillin, and 100 μg/mL of streptomycin in a 37°C incubator with a humidified atmosphere containing 5% CO2.
EMT differentiation in Hep3B cells was performed by treatment with 10 ng/mL TGF-β1 for three days. Stimulated cells and 300 mL of cell supernatant were collected to extract exosomes.
Exosome isolation and identification
Exosomes were isolated by differential centrifugation of conditioned media collected from Hep3B cells with or without TGF-β. Cells were grown in medium containing 10% exosome-depleted FBS. After three days’ incubation, the conditioned medium was initially cleared of cellular debris, and the dead cells were removed with two sequential centrifugation steps at 2500×g for 10 min at 4°C. The supernatants were then ultracentrifuged at 100,000×g for 70 min at 4°C. The pellets were washed with phosphate-buffered saline (PBS), and the ultracentrifugation protocol was repeated. The final exosome pellets were re-suspended in PBS and quantified by nanoparticle tracking system (LM10-HS instrument, NanoSight, Amesbury, UK).
Characterization of exosome and western blotting analysis
Total protein was extracted from cells and exosomes using an extraction buffer with a protease inhibitor cocktail (Thermo Scientific), and equal amounts of protein (50 μg) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad). PVDF membranes were blocked in Tris-buffered saline containing 5% albumin bovine V and incubated with specific antibodies to exosomal markers including CD63 (Abcam, ab134045, 1:3000), and tumor susceptibility gene 101 (TSG101; Abcam, ab125011, 1:3000). Cells were detected by using anti-E-Cadherin (Cell Signaling Technologies, 14472, 1:2000), anti-vimentin (Santa Cruz Biotechnology, 6260, 1:1000), growth arrest and DNA damage 45-alpha (GADD45A) (abcam, ab180768, 1:500) and GAPDH (Life Technologies, A-1310016, 1:4000) as an internal control. After several washes, membranes were incubated in blocking buffer with polymerization horse radish peroxidase (HRP)-conjugated anti-rabbit IgG (Boster, Wuhan, China) coupled to horseradish peroxidase for 2 h at room temperature. The complexes were detected by ECLplus (Amersham Biosciences/GE Healthcare, Velizy, France).
Immunofluorescence staining
Pretreated sterile slides were laid into the wells of a 12-well plate, then 5×104 cells were plated in each well and incubated for 12 h at 37°C in 5% CO2. Exosomes were labeled with carboxy fluorescein succinimidyl ester (CFSE) (System Biosciences, USA) according to the manufacturer’s instructions and then pre-incubated with the cells. Nuclei were stained with DAPI, and the cells were photographed under a fluorescence microscope (Nikon, Tokyo, Japan) at 200× magnification.
MTS assay
Hepatic cancer cells at a density of 1×106/mL were seeded into 96-well plates (100 μL/well), treated with exosomes, and examined after 24, 48, and 72 h. The MTS/PMS mixture (Promega, USA) was then added to the cells and incubated for 3 h. The optical density of the cells was measured at 490 nm.
Migration and invasion assays
For the migration assay, Hep3B and 7721 cells were plated into 24-well Boyden chambers with an 8 µm pore polycarbonate membrane. For the invasion assay, Hep3B and 7721 cells were plated in chambers pre-coated with Matrigel. In both assays, the cells were plated in medium with exosomes in the lower chamber. After 48 h, the cells that did not migrate or invade through the pores were removed. The membrane inserts were fixed and stained, and then counted under a microscope.
Small RNA and RNA library construction and high-throughput sequencing
Purified exosomes from Hep3B cells with or without TGF-β stimulation separately underwent extraction of total RNA, including the small RNA fraction, with Trizol (Invitrogen, Carlsbad, CA, USA). The quality and quantity of the isolated RNA was determined by the ND100 Nanodrop (Thermo Fisher), while RNA integrity was evaluated based on the RIN value using the Agilent 2200 TapeStation (Agilent Technologies, CA, USA). Two small RNA libraries were constructed and sequenced with Illumina TruSeq deep sequencing technology. RNAs were ligated with 3’ RNA adapter, followed by a 5’ adapter ligation. Subsequently, the adapter-ligated RNAs were subjected to RT-PCR and amplified from single-stranded cDNA templates to double-stranded cDNA. PCR products were size-selected by PAGE according to instructions of TruSeq® Small RNA Sample Prep Kit (Illumina, USA). The purified library products were evaluated using the Agilent 2200 TapeStation and diluted to 10 pM for cluster generation in situ on the HiSeq2500 single-end flow cell, followed by sequencing on the HiSeq 2500 platform. Image files generated by the sequencer were processed to produce digital quality data (raw FASTQ files).
Bioinformatic analyses
The resulting raw data was filtered to generate clean reads (18-30 nt), and then annotated by aligning to miRBase 21 (http://www.mirbase.org), Rfam11.0 (http://www.rfam.janelia.org), UCSC (http://www.gtrnadb.ucsc.edu), and pirnabank (http//pirnabank.ibab.ac.in/). The un-mapped RNA reads were used for prediction of novel miRNAs with Mireap software. Expression profiles of known miRNAs and mRNA sequences were identified and compared. Significant miRNA changes were selected based on the following criteria: (i) statistical significance of miRNA expression changes were identified using a P-value threshold of 0.01 and (ii) fold change expression with a minimum twofold difference in either direction was required. The expression of miRNA was normalized as the number of reads per million clean tags.
Target genes prediction of differentially expressed microRNAs
Four software tools (TargetScan, miRanda, CLIP, and miRDB) were employed to predict target genes for selected miRNAs. Only targets that were found by at least three of the four tools were identified to be the target genes of miRNAs.
Gene ontology and KEGG pathway analysis
Putative genes were subjected to GO enrichment and KEGG pathway analysis with DAVID 6.7 software (http://david.abcc.ncifcrf.gov/home.jsp). Fisher’s exact test and χ2 test were used to select the significant GO categories and signaling pathways. The threshold of significance was defined by the P-value, with P < 0.1 regarded as significant for GO and KEGG analyses, respectively.
Reverse transcriptase quantitative PCR (RT-qPCR) analysis
RNA was isolated using TRizol reagent (MRC, TR118-500) according to standard instructions. Total extracted RNA was used to synthesize cDNA by reverse transcription reaction using M-MLV Reverse Transcriptase (Promega, M1705). Target gene and GAPDH mRNA levels were detected using ChamQ SYBR RT-qPCR Master Mix, and miRNA levels were detected using TaqMan® miRNA assays (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions with a mature miRNA-specific forward primer and the universal reverse primer. The mean expression level of human endogenous control (U6) was used as an internal control for all miRNA experiments for comparative analysis. The relative mRNA and miRNA expression levels were then calculated by the comparative threshold cycle method (2-ΔΔCt). The primers used in this study are listed in Table 1.
Table 1.
Sequences of primers used for quantitative RT-PCR method
| Gene ID | Primer | Product Length |
|---|---|---|
| GAPDH | Forward primer | GAGTCAACGGATTTGGTCGT |
| Reverse primer | GACAAGCTTCCCGTTCTCAG | |
| GADD45A | Forward primer | GAAGGATCCTGCCTTAAGTCAAC |
| Reverse primer | CCTCTTTCCATCTGCAAAGTCATC | |
| HMOX1 | Forward primer | CTGGAGGAGGAGATTGAGCG |
| Reverse primer | TGGCACTGGCAATGTTGG | |
| E2F2 | Forward primer | TCCCAATCCCCTCCAGATC |
| Reverse primer | CAAGTTGTGCGATGCCTGC | |
| ARID3B | Forward primer | AGAGGACGGAGGTTTGGAAGA |
| Reverse primer | GCTGGAAGTAGATTGGACATGGG | |
| TUBB3 | Forward primer | GCAAGGTGCGTGAGGAGTAT |
| Reverse primer | GTCTGACACCTTGGGTGAGG | |
| U6 | Forward primer | CTCGCTTCGGCAGCACA |
| Reverse primer | AACGCTTCACGAATTTGCGT | |
| hsa-miR-125b-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAA |
| Forward primer | GCTCCCTGAGACCCTAACT | |
| hsa-miR-374a-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACTTA |
| Forward primer | GGCGGTTATAATACAACCTGAT | |
| hsa-miR-24-3p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGTTC |
| Forward primer | ATGGCTCAGTTCAGCAGG | |
| hsa-miR-200b-3p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATCA |
| Forward primer | GGCTAATACTGCCTGGTAATG | |
| hsa-miR-21-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA |
| Forward primer | GCCGTAGCTTATCAGACTGA | |
| Universe-R | GTGCAGGGTCCGAGGT |
Statistical analysis
The data with mean and standard deviation (SD) as presented here were prepared using GraphPad Prism software 5. Statistical significance between two groups was tested using unpaired Student’s t-test. P-values < 0.05 were considered statistically significant.
Results
TGF-β treatment of Hep3B cells induces EMT
Hep3B cells were treated with 10 ng/mL TGF-β1 as previously reported [29] for six to eight days for EMT differentiation. Figure S1A shows that the cells appeared square shaped before treatment and progressively changed to a distinct fusiform shape. We confirmed EMT differentiation after TGF-β1 stimulation by detecting the typical expression pattern of EMT markers such as vimentin and E-cadherin. During the EMT process, there was diminished expression of E-cadherin and increased expression of vimentin (Figure S1B). These results confirmed that Hep3B cells were induced to EMT differentiation by TGF-β1.
Isolation and identification of exosomes
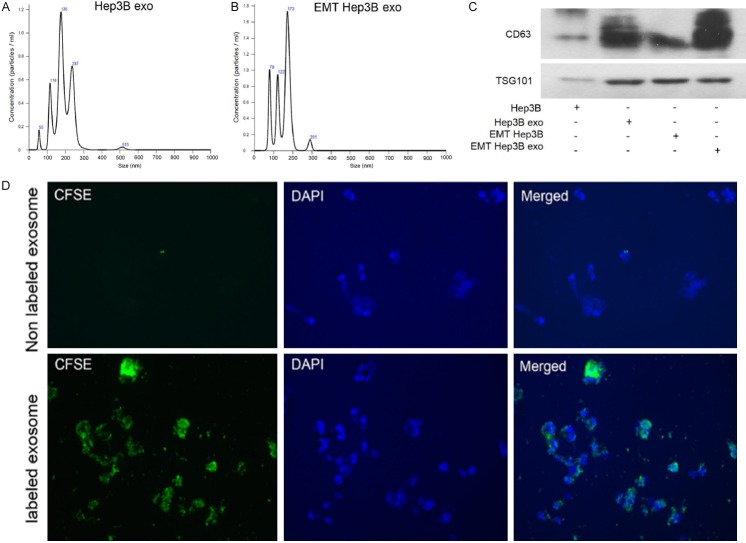
Exosomes were isolated from the Hep3B cells by sequential centrifugation, and their size distribution and relative concentration were determined by nanoparticle tracking analysis (NTA) (Figure 1A and 1B). The expression of the exosome markers CD63 and tumor susceptibility gene 101 (TSG101) were significantly enriched in Hep3B cell exosomes (Hep3B exo), with the highest expression found in the TGF-β1-stimulated Hep3B cell exosomes (EMT Hep3B exo) (Figure 1C). These results confirmed that the isolated exosomal particles from Hep3B cells induced for EMT by TGF-β expressed higher levels of exosome-associated marker proteins.
Figure 1.
Exosomes secreted by Hep3B and TGF-β1-induced Hep3B were taken up by Hep3B cells. A and B. Purified particles were analyzed by nanoparticle tracking analysis (NTA). Sizes of the particles were between 50 and 200 nm. C. Expression of exosome markers (CD63 and TSG101) was detected by western blot. The protein expression of exosome markers was upregulated in Hep3B-exo and EMT-Hep3B exo compared with their cells. D. CFSE-labeled Hep3B-exosomes showed green fluorescence in the cytoplasm of Hep3B cells. EMT Hep3B exo: exosomes secreted from Hep3B induced by TGF-β1.
To confirm whether cells could take up exosomes derived from Hep3B cells, exosomes were labeled with CFSE, pre-incubated with the cells, and analyzed under the fluorescence microscope. Exosomes accumulated in the labeled cells compared with the unlabeled control cells after observing under confocal microscope (Figure 1D).
EMT Hep3B cell-derived exosomes promote proliferation and metastasis of the hepatoma cells
To investigate the biological effects of EMT Hep3B exosomes on mediating cell proliferation and metastasis of hepatic liver cancer, we selected SMMC-7721 cells along with Hep3B cells and treated each separately with Hep3B exo or EMT-Hep3B exo and analyzed cell proliferation changes. Whereas EMT-Hep3B exo did not significantly affect the proliferation of 7721 cells (Figure 2A), EMT-Hep3B exo statistically significantly increased the rate of proliferation of Hep3B cells (Figure 2B). In addition, transwell migration and Matrigel invasion assays showed that EMT-Hep3B exo promoted migration and invasion of SMMC-7721 cells (Figure 2C) and Hep3B cells (Figure 2D) compared with control cells.
Figure 2.
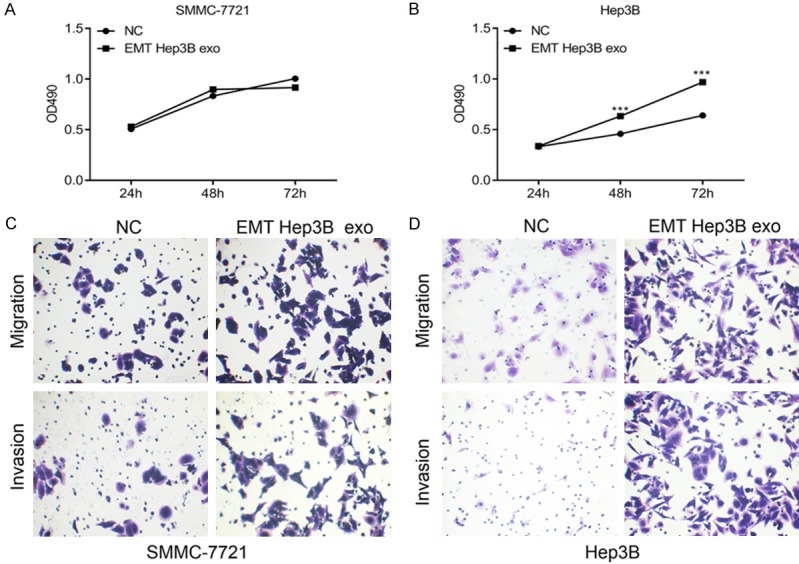
EMT Hep3B exo promoted proliferation, migration, and invasion of hepatic cancer cells. A and B. MTS assay was performed to detect 7721 and Hep3B cell viability. C and D. Cell migration and invasion were determined using Transwell chambers. ***P < 0.001.
High-throughput sequencing to detect differentially expressed miRNAs
To investigate the difference in miRNA expression profiles between Hep3B exo and EMT-Hep3B exo, we conducted small RNA (< 40 nt) sequencing. Following low-intensity filtering, hierarchical clustering was conducted to display the miRNA expression patterns (Figure 3A). After applying fold-change filtering and Student’s t-tests to identify differentially expressed miRNAs, we identified 119 significantly upregulated miRNAs and 186 downregulated miRNAs in the EMT-Hep3B exo. The miRNAs in exosomes released by these cells were ranked based on the clean reads data, and the top 20 prevalent miRNAs in each group were compared (Table 2).
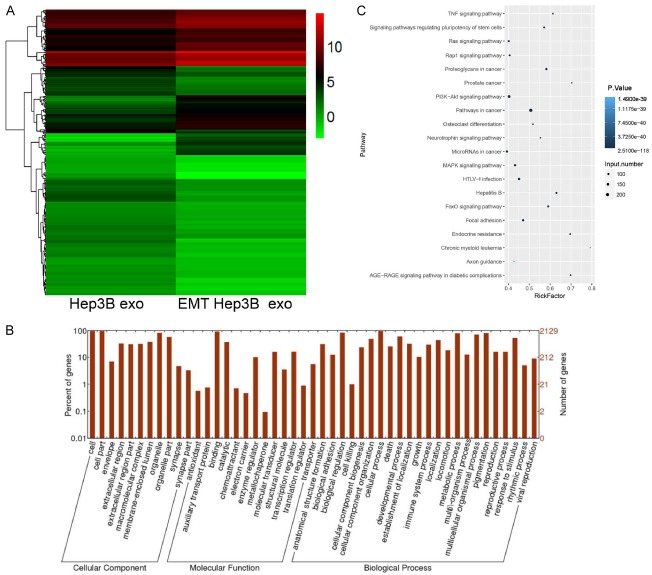
Figure 3.
Differentially expressed miRNAs were detected in EMT Hep3B exo and Hep3B exo. A. Cluster graph of differentially expressed miRNAs in EMT Hep3B exo and Hep3B exo. B. GO analysis of the differentially expressed miRNAs. C. KEGG pathway analysis of the differentially expressed miRNAs.
Table 2.
The top10 differentially expressed miRNAs in highest and lowest expression level in the EMT-Hep3B exo
| geneID | Hep3BNC-Expression | Hep3BTGF-Expression | Hep3BNC-RPKM | Hep3BTGF-RPKM | log2 Ratio (Hep3BTGF/Hep3BNC) | Up-Down-Regulation (Hep3BTGF/Hep3BNC) | P-value | FDR |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-656-3p | 2 | 122 | 0.180710427 | 13.38935792 | 6.211263217 | Up | 9.50E-17 | 1.41E-15 |
| hsa-miR-495-3p | 29 | 1355 | 2.62030119 | 148.709672 | 5.82662202 | Up | 5.30E-167 | 4.29E-165 |
| hsa-miR-758-3p | 3 | 128 | 0.27106564 | 14.04785093 | 5.695563379 | Up | 1.63E-16 | 2.36E-15 |
| hsa-miR-655-3p | 5 | 155 | 0.451776067 | 17.01106949 | 5.23472219 | Up | 2.11E-18 | 3.31E-17 |
| hsa-miR-543 | 7 | 200 | 0.632486494 | 21.94976708 | 5.117027147 | Up | 6.58E-23 | 1.19E-21 |
| hsa-miR-889-3p | 8 | 224 | 0.722841708 | 24.58373913 | 5.087880801 | Up | 2.27E-25 | 4.38E-24 |
| hsa-miR-410-3p | 4 | 111 | 0.361420854 | 12.18212073 | 5.074941746 | Up | 4.35E-13 | 5.37E-12 |
| hsa-miR-654-3p | 21 | 561 | 1.897459482 | 61.56909666 | 5.020065417 | Up | 8.98E-61 | 3.52E-59 |
| hsa-miR-376a-3p | 3 | 80 | 0.27106564 | 8.779906832 | 5.017491473 | Up | 1.32E-09 | 1.27E-08 |
| hsa-miR-409-3p | 28 | 566 | 2.529945977 | 62.11784084 | 4.6178292 | Up | 1.80E-55 | 6.64E-54 |
| hsa-miR-154-5p | 4 | 79 | 0.361420854 | 8.670157997 | 4.584306627 | Up | 1.19E-08 | 1.05E-07 |
| hsa-miR-3135b | 460 | 17 | 41.56339819 | 1.865730202 | -4.47750133 | Down | 4.57E-212 | 4.22E-210 |
| hsa-miR-383-3p | 26 | 1 | 2.34923555 | 0.109748835 | -4.419913839 | Down | 3.55E-13 | 4.40E-12 |
| hsa-miR-7156-3p | 531 | 27 | 47.97861834 | 2.963218556 | -4.017154669 | Down | 1.42E-235 | 1.47E-233 |
| hsa-miR-562 | 19 | 1 | 1.716749056 | 0.109748835 | -3.967401634 | Down | 1.17E-09 | 1.13E-08 |
| hsa-miR-4709-5p | 66 | 4 | 5.963444088 | 0.438995342 | -3.76386824 | Down | 6.51E-30 | 1.43E-28 |
| hsa-miR-4300 | 16 | 1 | 1.445683415 | 0.109748835 | -3.719474121 | Down | 3.67E-08 | 2.97E-07 |
| hsa-miR-548a-3p | 47 | 3 | 4.246695032 | 0.329246506 | -3.689100472 | Down | 1.54E-21 | 2.68E-20 |
| hsa-miR-1184 | 561 | 37 | 50.68927475 | 4.06070691 | -3.641877716 | Down | 1.78E-239 | 1.92E-237 |
| hsa-miR-4764-5p | 14 | 1 | 1.264972988 | 0.109748835 | -3.526829043 | Down | 3.60E-07 | 2.62E-06 |
| hsa-miR-5699-3p | 14 | 1 | 1.264972988 | 0.109748835 | -3.526829043 | Down | 3.60E-07 | 2.62E-06 |
In order to understand the function of the identified differentially expressed miRNAs, GO and KEGG analyses were performed. GO analysis showed the differentially expressed miRNAs were enriched in various biological process and molecular functions. Besides this, cellular component analysis showed that miRNAs were highly enriched in cells, cell parts, organelles, extracellular region, macromolecular complex, and membrane-enclosed lumen, as shown in Figure 3B. KEGG pathway analysis showed that differentially expressed miRNAs were mainly involved in Ras, Rap1, PI3K-Akt, and MAPK signaling, as well as other signaling pathways involved in cancer (Figure 3C).
Analysis of mRNA expression differences between Hep3B exo and EMT-Hep3B exo
A total of 322 mRNA sequences were differentially expressed, of which 156 were upregulated and 166 were downregulated in EMT-Hep3B exo. GO enrichment analysis of mRNA genes corresponding to the differentially expressed miRNAs showed the same enrichment pattern in percentage of gene expression corresponding to the GO enrichment of differentially expressed miRNAs (Figure 4A). KEGG pathway analysis revealed that metabolic pathways and pathways in cancer were the two highest expressed pathways (Figure 4B).
Figure 4.
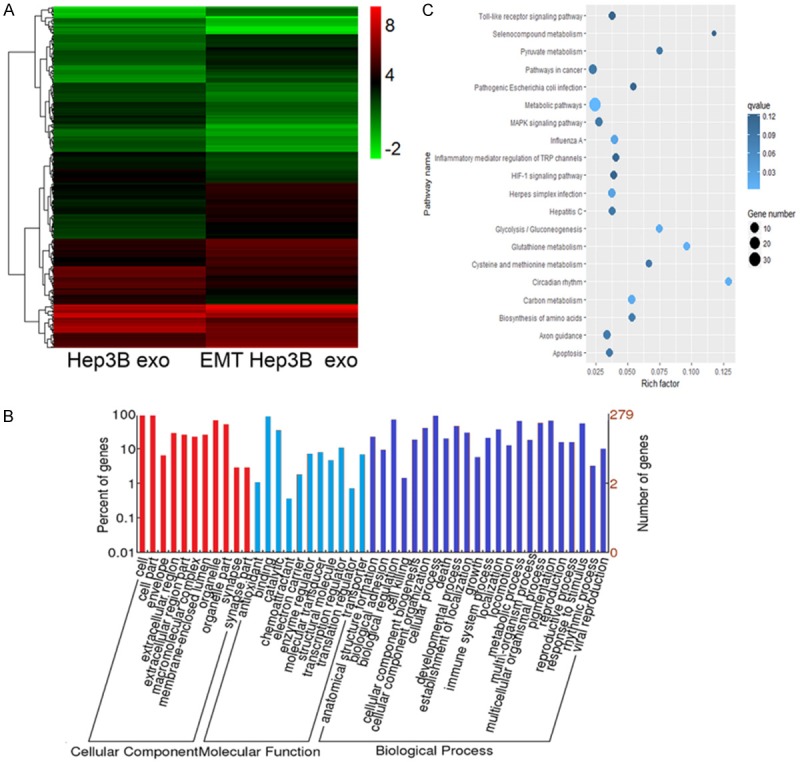
Differentially expressed mRNAs were detected in EMT Hep3B exo and Hep3B exo. A. Cluster graph of differentially expressed mRNAs in EMT Hep3B exo and Hep3B exo. B. GO analysis of the differentially expressed mRNAs. C. KEGG pathway analysis of the differentially expressed mRNAs.
Exosomal miRNA target genes mediate EMT in hepatic cancer
We selected EMT-related miRNAs from the sequencing results and verified their expression in exosomes and hepatic cells by RT-qPCR. These results showed that five miRNAs (hsa-miR-125b-5p, hsa-miR-374a-5p, hsa-miR-24-3p, hsa-miR-200b-3p, and hsa-miR-21-5p) were statistically upregulated in the EMT-Hep3B exo compared with the Hep3B exo, which was consistent with our sequencing results (Figure 5). Meanwhile, we found that hsa-miR-374a-5p, hsa-miR-200b-3p, and hsa-miR-21-5p were increased in the Hep3B cells after treatment with EMT-Hep3B exo compared with Hep3B exo.
Figure 5.
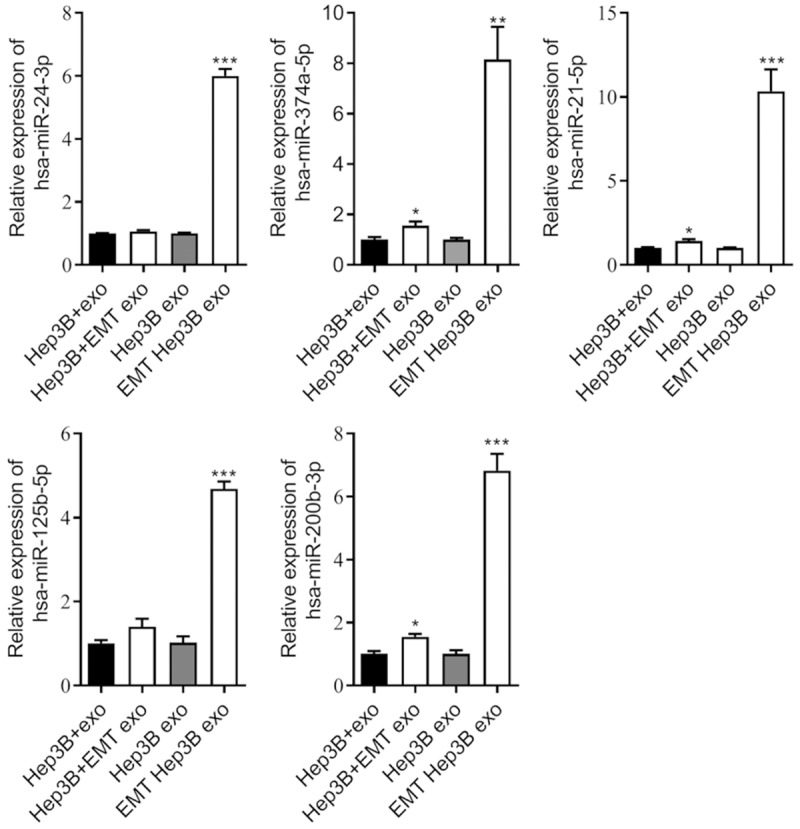
RT-qPCR validated differentially expressed miRNAs in EMT Hep3B exo and Hep3B exo. miR-125b-5p, hsa-miR-374a-5p, hsa-miR-24-3p, hsa-miR-200b-3p, and hsa-miR-21-5p were detected in the Hep3B exo, EMT-Hep3B exo, Hep3B cells, and Hep3B cells after treatment with EMT-Hep3B exo. *P < 0.05; **P < 0.01; ***P < 0.001.
Furthermore, several cancer-related signaling pathways were enriched, and multiple target genes of differentially expressed miRNAs were functionally regulated. Expression of the target genes of miRNAs hsa-miR-374a-5p, hsa-miR-200b-3p, and hsa-miR-21-5p were detected by RT-qPCR. Expression of target mRNAs including ARID3B, GADD45A, and HMOX1 was decreased, and the expression of two other target mRNAs, E2F2 and TUBB3, was increased in the Hep3B cells with EMT-Hep3B exo (Figure 6). Based on the relationship between miRNA targeting and regulated mRNA, we selected hsa-miR-374a-5p and GADD45A for further experiments.
Figure 6.
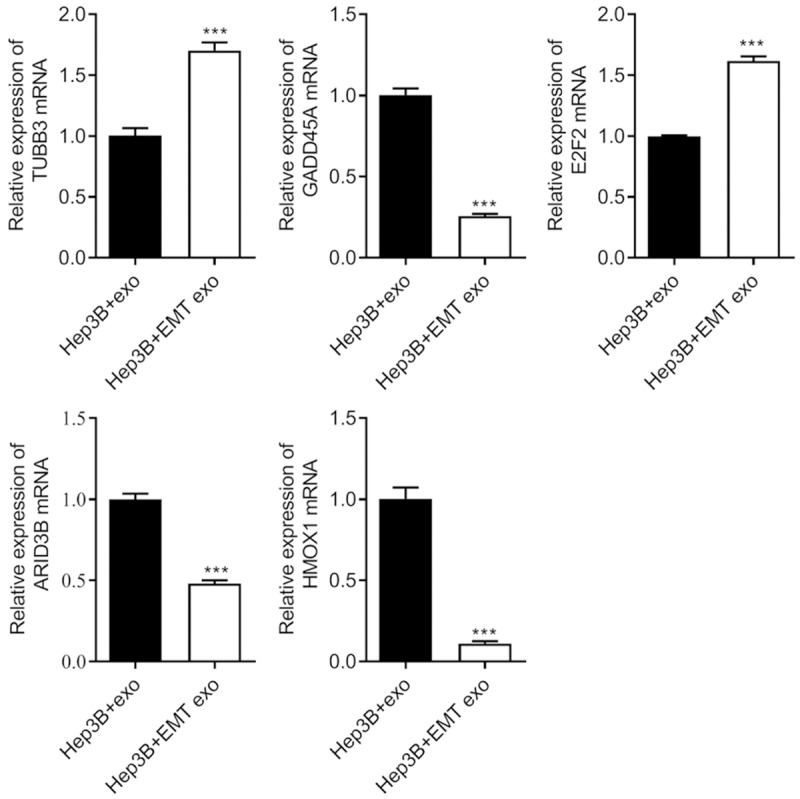
Regulation of expression of mRNA target genes by exosomal miRNA mediated EMT in hepatic cells. ARID3B, GADD45A, and HMOX1 were decreased, while the expression of E2F2 and TUBB3 were significantly increased in Hep3B cells after treatment with EMT-Hep3B exo. ***P < 0.001.
To clarify whether miRNAs contained within exosomes properly function, we interfered with the expression of miR-374a-5p in Hep3B cells with TGF-β-induced EMT and then collected exosomes, designated EMT Hep3B exo (In). Compared with the EMT-Hep3B exo group without interference (EMT Hep3B exo), EMT Hep3B exo (In) inhibited the expression of miR-374a-5p in cells (Figure 7A) and upregulated the expression of GADD45A mRNA and protein (Figure 7A and 7B). MTS results indicated that EMT Hep3B exo (In) significantly inhibited cell proliferation (Figure 7C). Figure 7D shows that EMT Hep3B exo (In) decreased migration and invasion of Hep3B cells compared with the EMT Hep3B exo group. These results suggested that interference of miR-374a-5p within Hep3B cell-derived exosomes might inhibit hepatocellular metastasis by regulating the expression of GADD45A.
Figure 7.
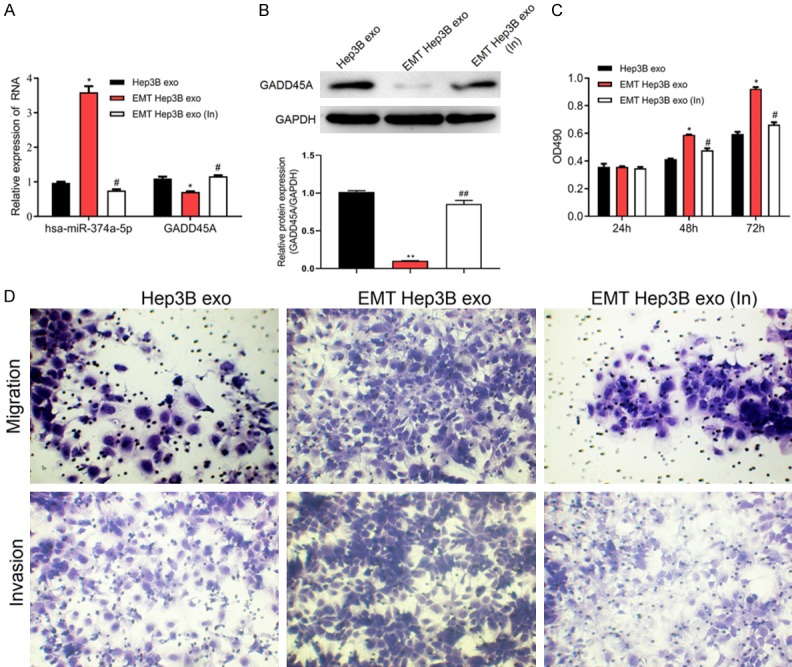
Interference of miR-374a-5p within Hep3B cell-derived exosomes inhibits proliferation and migration of Hep3B cells. A. The expression of miR-374a-5p and GADD45A were detected by RT-qPCR in Hep3B cell exo, EMT Hep3B exo, and EMT Hep3B exo (In). B. The protein expression of GADD45A was measured by western blot. C. MTS assay was performed to detect Hep3B cell proliferation. D. Hep3B cell migration was determined using Transwell analysis. *P < 0.05 vs. Hep3B exo; #P < 0.05 vs. EMT Hep3B exo. EMT Hep3B exo (In), Interference with miR-374a-5p in Hep3B cell-derived exosomes.
Discussion
Metastasis in hepatocellular carcinoma consists of multiple processes, including EMT, which is characterized by the loss of epithelial-like characteristics and the gain of mesenchymal-like attributes including cell migration and invasion. Exosomes carrying diverse groups of functional molecules mediate an intercellular crosstalk between tumor cells and neighboring cells and play a critical role in establishing a mesenchymal state and metastasis. A number of studies have demonstrated that abundant cell-free miRNAs from exosomes can induce miRNA-mediated repression of target genes and function effectively in various pathological processes as either oncogenes or tumor suppressors in recipient normal or tumor cells [13,14]. Moreover, substantial numbers of miRNAs have been implicated in the regulation of EMT-related pathways in cancer [25-28]. Recent studies have shown that the dysregulated expression of miRNAs in exosomes can accelerate HCC progression, including cell proliferation and metastasis, via alteration of a network of genes [16].
A previous study revealed that exosomal miRNAs are important molecules with significant functions in HCC and can reflect abnormal regulation in an intra-hepatic manner [16]. However, few studies have sought to characterize the role of HCC-derived exosomes in inducing EMT in tumor progression. In this study, we highlighted exosomes as mediators in modulating miRNA profiles in the HCC cell line Hep3B. Initially, we induced EMT in Hep3B cells by stimulating the cells with a multifunctional cytokine, TGF-β. We demonstrated that EMT-Hep3B exo promoted hepatic cancer cell proliferation, migration, and invasion. Our results are consistent with a previous study [30], which showed that incubation of SMMC-7721 cells and Hep3B cells with their self-derived exosomes caused a notable increase in cell growth, migration, and invasion. These findings indicate that HCC-cell-derived exosomes can promote the biological behavior of the parental HCC cells.
A large array of HCC-related genes is regulated by miRNAs, changes in the endogenous expression of these miRNAs may be a crucial mechanism in hepatocarcinogenesis [31-33]. Many studies have showed convincing results to support this hypothesis. miR-105 has been reported to function as a potential tumor suppressor and inhibit cell proliferation by regulating the PI3K/AKT signaling pathway in human HCC [31]. A set of serum miRNA panels including has-miR-206, 141-3p, 433-3p, 1228-5p, 199a-5p, 122-5p, 192-5p, and 26a-5p has shown considerable clinical value in HCC diagnosis [32]. miR-101 was implicated as a potential prognostic marker and therapeutic target for HBV-related HCC [33]. These findings are possible clues to highlight the potential role of miRNAs in the pathogenesis of hepatic cancers, and they also elucidate the prospects of developing novel therapeutic targets for HCC. However, the existing knowledge of miRNA interventions to achieve this goal is limited due to a lack of expertise and knowledge in the accurate delivery of miRNAs to hepatic cancer cells. Through this study, we have successfully unraveled the process of exosome-mediated delivery of miRNAs that could promote liver cancer EMT and metastasis in hepatic cancer cell lines. Moreover, we have performed high-throughput sequencing and detected differentially expressed miRNAs and their target genes; the expression of hsa-miR-125b-5p, hsa-miR-374a-5p, hsa-miR-24-3p, hsa-miR-200b-3p, and hsa-miR-21-5p were significantly higher in EMT-Hep3B exo than in Hep3B exo.
Recently, circulating miRNAs have been reported as promising biomarkers for various pathologic conditions including cancer, and certain miRNAs such as miR-125-5p were upregulated in CHB, HBV-positive cirrhosis, and HBV-positive HCC when compared with the control group [34]. Moreover, assessing tissue miR-125b-5p expression and intrahepatic metastasis was useful for stratifying patients at risk of early HCC recurrence after curative resection [35]. MiR-24-3p was previously reported to be significantly upregulated in HCC tumor tissues compared with non-tumor tissues and was shown to play an important role in the initiation and progression of HCC by targeting metallothionein [36]. A recent study reported that propofol inhibited cell proliferation, migration, and invasion while promoting cell apoptosis of HepG2 and SMMC-7721 cells through inhibiting the Wnt/β-catenin and PI3K/AKT pathways via downregulation of miR-374a [37]. Another study reported that overexpression of miR-374a activated the EGFR and AKT/ERK signaling pathways by regulation of MIG-6, and suggest that miR-374a could promote cell viability by targeting MIG-6 and activating the EGFR and AKT/ERK signaling pathways [38]. EV-associated hsa-miR-21-5p and hsa-miR-144-3p are markedly elevated in the serum of patients with HCC, and the ratio of miR-144-3p/miR-21-5p is correlated with the development of HCC [39]. The expression pattern of miRNAs in all these studies shows similar results with our findings, and the exact role of functional studies in development of hepatic cancer can be further explored. Moreover, our study suggests that interference with miR-374a-5p in Hep3B cell-derived exosomes inhibits proliferation and migration of HCC cells.
Our analysis of target genes of miRNAs showed that the expression of three target mRNAs, including ARID3B, GADD45A, and HMOX1 were decreased, and the expression of two target mRNAs, E2F2 and TUBB3, increased. GADD45A, the target gene of miR-374a-5p, is a nuclear protein involved in maintenance of genomic stability, DNA repair, and suppression of cell growth through interaction with nuclear elements, including cyclin-dependent kinase inhibitor 1A (CDKN1A) and PCNA. In a study conducted to assess GADD45A expression in 28 cases of HCC and matched cirrhosis tissues, GADD45A mRNA was downregulated in 20 of 26 HCCs with respect to matched cirrhosis [40]. Moreover, it was also reported that regulation of the expression of GADD45A could influence the phosphorylation level of Akt and further regulate the expression of Bcl-2 and Bax proteins involved in the mitochondrial apoptosis pathways [41]. Hence, we selected GADD45A for further experiments based on the targeted effects of selected miRNAs.
Conclusion
The potential role of these differentially expressed exosomal microRNAs in the pathogenesis of HCC is worth further study. Identification of signature miRNAs and further analyses on the multiple regulatory functions of their target genes could offer novel alternative biomarkers for diagnosis of liver cancer, and this could provide a promising prognostic and therapeutic strategy for HCC treatment.
Acknowledgements
Authors would like to thank the National Natural Science Foundation of China (81172193) for financial support.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Li S, Yao J, Xie M, Liu Y, Zheng M. Exosomal miRNAs in hepatocellular carcinoma development and clinical responses. J Hematol Oncol. 2018;11:54. doi: 10.1186/s13045-018-0579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vella LJ. The emerging role of exosomes in epithelial-mesenchymal-transition in cancer. Front Oncol. 2014;4:361. doi: 10.3389/fonc.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, Xi J, Yan L, Gu J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang JY, Zhang K, Chen DQ, Chen J, Feng B, Song H, Chen Y, Zhu Z, Lu L, De W, Wang R, Chen LB. MicroRNA-451: epithelial-mesenchymal transition inhibitor and prognostic biomarker of hepatocelluar carcinoma. Oncotarget. 2015;6:18613–18630. doi: 10.18632/oncotarget.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, Xue Y, Xu T, Zhu HJ, Simpson RJ. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 6.Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28–41. doi: 10.1016/j.ajpath.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 8.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 9.Wahlgren J, De L Karlson T, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, Li Y, Liu Y. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16:132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosaka N. Decoding the secret of cancer by means of extracellular vesicles. J Clin Med. 2016;5 doi: 10.3390/jcm5020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Valencia CA, Dong B, Chen M, Guan PJ, Pan L. Transfer of microRNAs by extracellular membrane microvesicles: a nascent crosstalk model in tumor pathogenesis, especially tumor cell-microenvironment interactions. J Hematol Oncol. 2015;8:14. doi: 10.1186/s13045-015-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Liu XS, Chopp M, Zhang RL, Zhang ZG. MicroRNAs in cerebral ischemia-induced neurogenesis. J Neuropathol Exp Neurol. 2013;72:718–722. doi: 10.1097/NEN.0b013e31829e4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Z, Wu JH, Wu JY, Ji AL, Qiang GH, Jiang Y, Jiang CP, Ding YT. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, Milosevic J, Tkacheva OA, Divito SJ, Jordan R, Lyons-Weiler J, Watkins SC, Morelli AE. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris DA, Patel SH, Gucek M, Hendrix A, Westbroek W, Taraska JW. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS One. 2015;10:e0117495. doi: 10.1371/journal.pone.0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gernapudi R, Yao Y, Zhang Y, Wolfson B, Roy S, Duru N, Eades G, Yang P, Zhou Q. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res Treat. 2015;150:685–695. doi: 10.1007/s10549-015-3326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 21.Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, Jiang YH, Yang XH, Liu YP. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell PJ, Welton J, Staffurth J, Court J, Mason MD, Tabi Z, Clayton A. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toth B, Nieuwland R, Liebhardt S, Ditsch N, Steinig K, Stieber P, Rank A, Gohring P, Thaler CJ, Friese K, Bauerfeind I. Circulating microparticles in breast cancer patients: a comparative analysis with established biomarkers. Anticancer Res. 2008;28:1107–1112. [PubMed] [Google Scholar]
- 25.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653–662. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E, Anderson KC, Scadden DT, Ghobrial IM. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. 2013;123:1542–1555. doi: 10.1172/JCI66517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, Rogatko A, Posadas EM, Haga CL, Chung LW. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 2015;34:2690–2699. doi: 10.1038/onc.2014.212. [DOI] [PubMed] [Google Scholar]
- 29.Miyazono K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:314–323. doi: 10.2183/pjab.85.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, Han QF, Liu PQ, Zhou G, Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, Zhou Q, Ji T, Huang S, Zhang J, Jia H. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35:2748–2755. doi: 10.1093/carcin/bgu208. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, Zou L, Li Z, Zhao J, Lin N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–4370. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 34.Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, Sezgin O, Altintas E, Tiftik EN. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep. 2014;41:4513–4519. doi: 10.1007/s11033-014-3322-3. [DOI] [PubMed] [Google Scholar]
- 35.Shimagaki T, Yoshizumi T, Harimoto N, Yoshio S, Naito Y, Yamamoto Y, Ochiya T, Yoshida Y, Kanto T, Maehara Y. MicroRNA-125b expression and intrahepatic metastasis are predictors for early recurrence after hepatocellular carcinoma resection. Hepatol Res. 2018;48:313–321. doi: 10.1111/hepr.12990. [DOI] [PubMed] [Google Scholar]
- 36.Dong X, Ding W, Ye J, Yan D, Xue F, Xu L, Yin J, Guo W. MiR-24-3p enhances cell growth in hepatocellular carcinoma by targeting metallothionein 1M. Cell Biochem Funct. 2016;34:491–496. doi: 10.1002/cbf.3213. [DOI] [PubMed] [Google Scholar]
- 37.Liu SQ, Zhang JL, Li ZW, Hu ZH, Liu Z, Li Y. Propofol inhibits proliferation, migration, invasion and promotes apoptosis through down-regulating mir-374a in hepatocarcinoma cell lines. Cell Physiol Biochem. 2018;49:2099–2110. doi: 10.1159/000493814. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Chen H, Wang H, Dong Y, Yin M, Zhang L, Wei J. MicroRNA-374a promotes hepatocellular carcinoma cell proliferation by targeting mitogen-inducible gene 6 (MIG-6) Oncol Res. 2018;26:557–563. doi: 10.3727/096504017X15000784459799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pu C, Huang H, Wang Z, Zou W, Lv Y, Zhou Z, Zhang Q, Qiao L, Wu F, Shao S. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol. 2018;9:930. doi: 10.3389/fphys.2018.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gramantieri L, Chieco P, Giovannini C, Lacchini M, Trere D, Grazi GL, Venturi A, Bolondi L. GADD45-alpha expression in cirrhosis and hepatocellular carcinoma: relationship with DNA repair and proliferation. Hum Pathol. 2005;36:1154–1162. doi: 10.1016/j.humpath.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Zou F, Zhong J, Yue L, Wang F, Wei H, Yang G, Jin T, Dong X, Li J, Xiu P. Secretory clusterin mediates oxaliplatin resistance via the Gadd45a/PI3K/Akt signaling pathway in hepatocellular carcinoma. J Cancer. 2018;9:1403–1413. doi: 10.7150/jca.23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.