Abstract
Chlorogenic acids (CGAs) and luteolin are active compounds in Lonicera japonica, a plant of high medicinal value in traditional Chinese medicine. This study provides a comprehensive overview of gene families involved in chlorogenic acid and luteolin biosynthesis in L. japonica, as well as its substitutes Lonicera hypoglauca and Lonicera macranthoides. The gene sequence feature and gene expression patterns in various tissues and buds of the species were characterized. Bioinformatics analysis revealed that 14 chlorogenic acid and luteolin biosynthesis-related genes were identified from the L. japonica transcriptome assembly. Phylogenetic analyses suggested that the function of individual gene could be differentiation and induce active compound diversity. Their orthologous genes were also recognized in L. hypoglauca and L. macranthoides genomic datasets, except for LHCHS1 and LMC4H2. The expression patterns of these genes are different in the tissues of L. japonica, L. hypoglauca and L. macranthoides. Results also showed that CGAs were controlled in the first step of biosynthesis, whereas both steps controlled luteolin in the bud of L. japonica. The expression of LJFNS2 exhibited positive correlation with luteolin levels in L. japonica. This study provides significant information for understanding the functional diversity of gene families involved in chlorogenic acid and the luteolin biosynthesis, active compound diversity of L. japonica and its substitutes, and the different usages of the three species.
Abbreviations: RACE, raid amplification of cDNA ends; cDNA, complementary DNA; DNA, deoxyribonucleic acids; Sp, Selaginella pulvinta; TPS, trehalose-6-phosphate synthase; TPS, trehalose-6-phosphate synthase gene; TPP, trehalose-6-phosphate phosphatase; RNA, ribonucleic acids; DP, degenerate primer; ORF, open reading frame; PCR, polymerase chain reaction
Keywords: Honeysuckle, Othologous genes, Gene expression, Chlorogenic acid, Luteolin
Highlights
► Twelve genes in Lonicera japonica were reported for the first time. ► All genes in L. hypoglauca and L. macranthoides were reported for the first time. ► The first step of biosynthesis controls the chlorogenic acid in the bud. ► Two steps control the luteolin content in the bud. ► A different medical usage of species was according to gene duplication.
1. Introduction
Phenylpropanoids (PDCs), as ubiquitous plant secondary metabolites, control flower color, pollination, and stress response (Ververidis et al., 2007). As constituents of plant-rich diets and an assortment of herbal medicinal agents, phenylpropanoids exhibit measurable cancer chemopreventive, antimitotic, estrogenic, antimalarial, antioxidant, and anti-asthmatic activities (Ferrer et al., 2008). In the last two decades, the molecular biochemistry and genomics of phenylpropanoid biosynthesis have attracted widespread interest (Fraser and Chapple, 2011). Due to the important role of PDCs in plant development and the potential value of metabolic engineering of PDC biosynthesis pathways, identification and characterization of genes encoding the enzymes involved in PDC biosynthesis have been carried out in various plant species, such as Arabidopsis (Fraser and Chapple, 2011) and tomato (Adato et al., 2009).
Lonicera japonica Thunb (LJ) and its substitutes are anti-inflammatory herbs that have been used in traditional Chinese medicine for thousands of years (Shang et al., 2011). This important Chinese medicine is used to prevent and treat severe acute respiratory syndromes, H1N1 influenza, and hand-foot-and-mouth disease. L. japonica extracts also possess other biological and pharmaceutical properties, including anti-bacterial, anti-inflammatory, anti-viral, liver protectant, anti-angiogenic, and antinociceptive activities (Ku et al., 2009). L. japonica substitutes that have been recorded as Chinese material medicine in China pharmacopoeia include L. hypoglauca Miquel (LH), L. macranthoides Hand.-Mazz (LM), L. confusa, and L. fulvotomentosa.
Genes for phenylpropanoid synthesis pathway are members of diverse gene families. Their isolation and characterization can provide important clues about enzyme evolution in plant secondary metabolism (Fraser and Chapple, 2011). For L. japonica and its substitutes, the gene families involved in phenylpropanoid biosynthesis exhibit expansion and diversification in different species, suggesting diversification of chemical compounds.
Chlorogenic acid is a major family of soluble plant phenolics (Clifford, 1999) commonly found in Asteraceae, Solanaceae, and Rubiaceae (Molgaard and Ravn, 1988). L. japonica and its substitutes belong to the Caprifoliaceae family, which exhibits accumulation of CGAs in their flowers, stems, and leaves (Chen et al., 2010, Zhang et al., 2009). Luteolin is a flavonoid commonly found in fruits and vegetables. This compound has anti-inflammatory, antioxidant, and anti-cancer properties (Lim et al., 2012). Both CGAs and luteolin-7-O-glucoside (L7G) are standard compounds used to evaluate the chemical quality of L. japonica. However, only chlorogenic acid is used to evaluate the chemical quality of L. hypoglauca, L. macranthoides, L. confusa, and L. fulvotomentosa (Chinese pharmacopoeia, 2010). The content of chlorogenic acid in buds of L. japonica is higher than in its leaves (Zhao et al., 2010), whereas in L. hypoglauca, buds have lower CGAs than leaves (Deng, 2007).
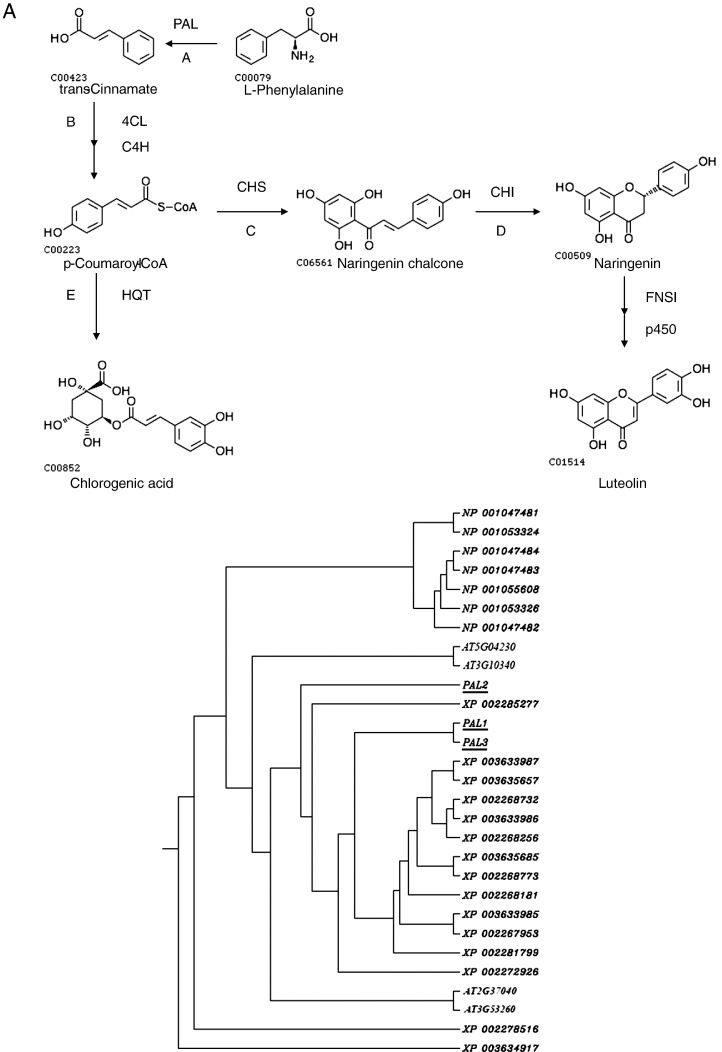
Generally, the biosynthesis of CGAs and luteolin is controlled by the phenylpropanoid pathway and can be divided into two stages (Fig. 1 ). The first stage leads to the synthesis of the universal precursor, p-Coumaroyl-CoA, under the catalysis of phenylalanine amononiar-lyase (PAL), 4-coumarate CoA ligase (4CL), and cinnamate 4-hydroxylase (C4H). In the second stage, luteolin is synthesized under the catalysis of chalcone synthase (CHS), chalcone isomerase (CHI), flavone synthase (FNS), and a P450 modifying enzyme. CGA is synthesized under the catalysis of hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase (HQT). Bai et al. (2010) isolated a LJHQT gene and showed that the tissue distribution of HQT is correlated with the pattern of CGA abundance. Yuan et al. (2012) completed L. japonica transcriptome and obtained genes in the biosynthetic pathway of CGA and luteolin.
Fig. 1.
Biosynthesis of chlorogenic acid and lutelin. PAL, phenylalanine amononiar-lyase; 4CL, 4-coumarate CoA ligase; C4H,cinnamate 4-hydroxylase; CHS, chalcone synthase, CHI, chalcone isomerase FNS, flavone synthase; HQT, hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase. A, Phylogenetic relationship of plant PALs. The rooted Neighbor–Joining tree was constructed using the ClustalW program. PALs included 4 Arabidopsis thaliana, 7 rice, 15 wine grape and 3 Lonicera japonica PALs. B, Phylogenetic relationship of plant 4CLs. The rooted neighbor-joining tree was constructed using the ClustalW program. 4CLs included 4 Arabidopsis thaliana, 10 rice, 2 wine grape and 2 Lonicera japonica 4CLs. C, Phylogenetic relationship of plant CHSs. The rooted neighbor-joining tree was constructed using the ClustalW program. CHSs included 1 Arabidopsis thaliana, 12 rice, 5 wine grape and 2 Lonicera japonica CHSs. D, Phylogenetic relationship of plant CHIs. The rooted neighbor-joining tree was constructed using the ClustalW program. CHIs included 5 Arabidopsis thaliana, 7 rice, 4 wine grape and 2 Lonicera japonica CHIs. E, Phylogenetic relationship of plant HQTs. The rooted neighbor-joining tree was constructed using the ClustalW program. HQTs included 1 Arabidopsis thaliana, 6 rice, 2 wine grape and 2 Lonicera japonica HQTs. The genes in this study were indicated by underline.
Recently, the genome sequencing program of L. japonica substitutes including L. hypoglauca and L. macranthoides, has been initiated. A working draft of the genome has been obtained (Yuan et al., unpublished data). The current assembly consists of 1,142,296 and 1,537,755 contigs representing ~ 85% of the protein-coding genes in the entire L. hypoglauca and L. macranthoides genome, respectively. The assembly allows a genome-wide identification and characterization of genes involved in chlorogenic acid and luteolin biosynthesis.
To understand the biosynthesis of chlorogenic acid and luteolin, we characterized the genes involved in the synthesis pathway. This study shows that almost all enzymes involved in the phenylpropanoid pathway of L. japonica and its substitutes are encoded by multigene families. The results in this study will be helpful for the modification of the accumulation of active components by biotechnological means in the future.
2. Materials and methods
2.1. Plant materials
Fresh buds and leaves of L. japonica, L. hypoglauca, and L. macranthoides were collected from Guangxi province, China in May 2011. During this month, buds were formed, and in pharmacological perspective, active components were rapidly accumulating. Six plants were sampled for each species. Samples were authenticated by Prof. Liying Yu (Guangxi Medical Plant Garden, China).
2.2. Sequence retrieval and gene prediction
We applied RNA-seq to L. japonica and generated over 100 million reads using Illumina's GA platform (Yuan et al., 2012). The current assembly of L. japonica transcriptome was investigated for homologs of chlorogenic acid and luteolin biosynthesis-related proteins from various plant species using BLASTx algorithm (Altschul et al., 1997). An e-value cut-off of 10− 5 was applied to the homolog recognition. L. hypoglauca and L. macranthoides genome sequences (Yuan et al., unpublished data) were searched for orthologs from L. japonica genes. An all-against-all sequence comparison was performed using BLAST (cutoff < 10− 20) to identify orthologs. They were determined from the best reciprocal hits (> 80% alignment length) (Tatusov et al., 1997). All retrieved sequences from the Genscan web server (http://genes.mit.edu/GENSCAN.html) were used for gene prediction (Burge and Karlin, 1998). The predicted gene models were further examined and corrected manually by comparison with related genes identified from other plant species.
2.3. Sequence feature analysis
Theoretical isoelectric point and molecular weight were predicted using Compute pI/MW tool on the ExPASy server (Bjellqvist et al., 1994) (http://web.expasy.org/compute_pi/). The localizations of deduced proteins were predicted on the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2007), iPSORT, and PSORT programs (ipsort.hgc.jp/). Conserved domains were searched against the Pfam protein family database using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) (Quevillon et al., 2005). The conserved amino acids were analyzed by protein alignment using tools such as ClustalW, and were checked manually (Thompson et al., 1994).
2.4. Phylogenetic analysis
Phylogenetic analyses of the alignments were performed using ClustalW (Thompson et al., 1994) and MEGA 5.0 (Tamura et al., 2011) for neighbor-joining (NJ) analysis. The reliability of these tree topologies was evaluated using bootstrap support with 1000 replicates (Kumar et al., 2008).
2.5. RNA extraction
Total RNA was extracted from plant tissues using Trizol Reagent (Invitrogen, USA) and was pre-treated with RNase-Free DNase (Promega, USA) to eliminate genomic DNA contamination. RNA integrity was analyzed on 1% agarose gel. RNA quantity was determined using NanoDrop 2000C Spectrophotometer (Thermo Scientific, USA).
2.6. Quantitative real-time reverse transcription-PCR
Total RNA was reverse-transcribed by Reverse Transcriptase MMLV (Takara, China). The polymerase chain reactions (PCRs) were performed according to the instructions in SYBR premix Ex Taq kit (TaKaRa, China) and were carried out in triplicate using 7500 real-time PCR system (ABI, USA). Gene-specific primers were designed using primer designing tools, particularly Primer3 (http://frodo.wi.mit.edu/primer3/) (Rozen and Skaletsky, 2000). The primer sequences found in orthologs of L. japonica, L. hypoglauca, and L. macranthoides are listed in Table 2 . The lengths of amplicons range between 100 bp and 250 bp. LJ18S was chosen as an endogenous control in studying gene expressions in buds and leaves of L. japonica, L. hypoglauca, and L. macranthoides. Standard deviations were calculated from the three PCR replicates. The specificity of amplification was assessed by melting curve analysis, and the relative abundance of genes was determined using the comparative Ct method as suggested by the 7500 Software v2.0.1 (ABI, USA).
Table 2.
Primer sequences for real-time RT-PCR.
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| PAL1 | GACTTGAGGCATTTGGAGGA | AGAACCTTGAGGGGTGGAGT |
| PAL2 | TGCCGAAAGAAGTCGAAAGT | TGTCAAATACTCGCCCTTCA |
| PAL3 | AAATGCGTCGATCTTCCAGA | AACCGGATTCTCACTCTCCA |
| 4CL1 | GCCAAACTTGGACAGGGTTA | ATAACGGCGTTTCTCACCAC |
| 4CL2 | CATTCCCCTCAACTCTCCAA | GAGGAGATAAGGGCGGTTTC |
| CHS1 | TTCGTGGTTAAGGACCATTTG | TGAAAGAAAAATAAACCCAAGACC |
| CHS2 | ATCCGGACAAGTCTGTCGAG | GCACGTCCTTCAACAAATGA |
| CHI1 | TCCCTGCAACTTCTTCCACT | TCCCACCCAAGTACCACTTC |
| CHI2 | AACGGGGCAACAATACTCAG | ATGAGCCAAGAGGTGATTGG |
| C4H1 | CGGTTCCATTCCCCTAAAAT | ATTTATAGAGGCGGGGGTTG |
| C4H2 | CTTTTCCCATCTGACCAACC | CTTTTCCCATCTGACCAACC |
| HQT1 | TGAGATCCTAGCTGCCCACT | TGGCTGTGAACACCACATTT |
| HQT2 | CAATCAAGTCCCAAGGCTGT | GGCAGCTAGGACCTCGTATG |
| FNS1 | AGGCTAGTGAGGGGGTGAAC | CACCTCACGTACCAATGTCCT |
| FNS2 | AGGGAAGCACAGGCTAGTCA | GGTGTTAGCCGAGATGTGGT |
| 18S | TGGTGGTCGTAACATTCC | TCCTTGTAAGTTTCTTTTCCT |
2.7. Enzyme activity assay
The activity of PAL was analyzed according to Brueske's method (Brueske, 1980). Approximately 500 mg of homogenized plant material in 5 mL 25 mM borate-HCl buffer (cold) with pH 8.8 and 5 mM mercaptoethanol (0.4 mL/L) was prepared. The homogenate was subjected to centrifugation at 12,000 g for 20 min. Then, the supernatant was used as an enzyme source. Reaction rate was expressed as micromole trans-cinnamic acid formed per mg protein per min.
For C4H and 4CL, 500 mg of plant material was extracted and mixed with 200 mM Tris–HCl buffer (pH 7.5) containing 25% (v/v) glycerol and 0.1 M DTT. The extracts were then homogenized and centrifuged at 10,000 g for 20 min at 4 °C. The supernatants were used for the enzyme assays. C4H activity was assayed using the method described by Cao et al. (2010) with slight modifications. The extract (0.2 mL) was added to 2-mercaptoethanol, 2 mM trans-cinnamic acid, and 0.5 mM NADPH, which was subsequently incubated for 1 h at 37 °C. The reaction was stopped with 6 M HCl. The absorbance values were measured at 290 nm. One unit of C4H activity was defined as the amount of enzyme causing an increase in A290 of 0.001 unit h− 1.
The method of Voo et al. (1995), which entails monitoring the increase in A 333 using p-coumarate, was used to assay 4CL activity. The 1 mL reaction mixture contained 50 μL crude enzyme, 0.2 mM p-coumarate, 0.8 mM ATP, 7.5 mM MgCl2, and 38 M CoA in 100 mM Tris–HCl buffer (pH 7.5). One unit of 4CL was defined as the amount of enzyme that causes a decrease in A333 of 0.01 units min− 1.
Protein concentration in the extracts was determined using Lowry method (Lowry et al., 1951).
2.8. Chemical analysis
As with our previous report, dried buds or leaves were separately comminuted with a miller. Each solid sample (40 mesh, 0.50 g) was accurately weighed and extracted with 50 mL of 70% aqueous ethanol by ultrasonication for 30 min. The extract was cooled to 25 °C, diluted to 50 mL with 70% aqueous ethanol, and filtered with 0.45 μm Millipore filter membrane. Afterward, 10 μL of the filtrate was injected into the HPLC system for analysis. The HPLC system was an Agilent 1200LC series (Agilent Technologies, Palo Alto CA, USA) composed of an online vacuum degasser (G1379B), a Bin pump SL (G1312B), an auto-sampler (GB67C), a thermostatic column compartment (G1316B), and a diode-array detector (DAD) (G1315C) coupled with an analytical workstation. The column configuration was an Agilent TC-C18 reserved-phase column (5 μm, 250 mm × 4.6 mm). The sample injection volume was 10 μL. The detection wavelength was set at 242 nm for the analysis with a flow rate of 1.0 mL min− 1 and 25 °C column temperature. The mobile phase was composed of deionized water–formic acid (A; 99:1, v/v) and methanol (B).
3. Results
3.1. Identification of 14 L. japonica chlorogenic acid and luteolin biosynthesis-related genes
Using systematic computational approach, 14 chlorogenic acid and luteolin biosynthesis-related genes were identified from the current L. japonica transcriptome assembly, of which 12 are novel (Table 1). These genes are members of six gene families which encode all the enzymes involved in the biosynthesis of chlorogenic acid and luteolin (Fig. 1). The deduced proteins demonstrated different lengths, isoelectric points (pI), molecular weights, subcellular localizations, and signal peptides. They also contained conserved domains and motifs (Table 1). The sequences of newly found genes generated from this study were submitted to the GenBank database and the accession numbers are JX068601–JX068613.
Table 1.
Biosynthesis genes of Chlorogenic acid and Luteoloside in Lonicera Japonica.
| Name | GenBank no. | Length (aa) |
Molecular weight(KD) | Isoelectric point | Subcellular localization | Signal peptide | Domains (# of domains) | Reference |
|---|---|---|---|---|---|---|---|---|
| PAL1 | JX068601 | 714 | 77.4 | 6.11 | Cytosol | – | Phenylalanine/histidine ammonia-lyase (IPR001106) | This study |
| PAL2 | JX068602 | 707 | 77.2 | 6.26 | Cytosol | – | This study | |
| PAL3 | JX068603 | 701 | 76.3 | 5.90 | Cytosol | – | This study | |
| 4CL1 | JX068604 | 538 | 58.7 | 5.73 | Cytosol | – | AMP-dependent synthetase/ligase (IPR000873) |
This study |
| 4CL2 | JX068605 | 540 | 58.7 | 8.49 | Cytosol | – | This study | |
| CHS1 | JX068606 | 351 | 38.2 | 6.10 | Cytosol | – | Chalcone/stilbene synthase, N-terminal (IPR001099) | This study |
| CHS2 | JX068607 | 393 | 42.9 | 6.34 | Cytosol | – | This study | |
| CHI1 | JX068608 | 208 | 23.2 | 4.76 | Cytosol | – | Chalcone isomerase, subgroup (IPR003466) | This study |
| CHI2 | JX068609 | 240 | 26.1 | 5.44 | Chloroplast | – | This study | |
| C4H1 | JX068610 | 505 | 58.1 | 9.09 | Cytosol | Yes | Cytochrome P450 (IPR001128) | This study |
| C4H2 | JX068611 | 535 | 61.1 | 8.73 | Cytosol | Yes | This study | |
| HQT1 | ACZ52698 | Peng et al., 2010 | ||||||
| HQT2 | JF261014 | |||||||
| FNS1 | JX068612 | Partial cDNA | This study | |||||
| FNS2 | JX068613 | Partial cDNA | This study | |||||
3.2. Characterization of full-length cDNA from L. japonica new genes
Eleven full-length cDNAs were obtained from L. japonica chlorogenic acid and luteolin biosynthesis-related genes. PAL is the first enzyme in flavonoid biosynthesis and is responsible for the conversion of l-phenylalanine to trans-cinnamic acid. As in many plants, the active PAL isoforms of Arabidopsis are encoded by a gene family designated as PAL1–PAL4 (Raes et al., 2003). In this paper, three new cytosolic PAL genes were uncovered in L. japonica. LJPAl1, LJPAL2, and LJPAL3 show 71.46% to 74.64% and 82.74% to 86.73% identity at the nucleotide and amino acid levels, respectively. All three genes could encode proteins with domains and motifs conserved among previously known PALs in the Interpro database. They include the consensus phenylalanine/histidine ammonia-lyase domain (IPR001106), suggesting that LJPALs have the same type of biochemical activity (Table 1).
In addition to PAL, cinnamate 4-hydroxylase (C4H) and 4-coumarate: coenzyme A ligase (4CL) are important enzymes in allocating significant amounts of carbon from phenylalanine into the biosynthesis of several important secondary metabolites. Two C4H (LJC4H1 and LJC4H2) and two 4CL (LJ4CL1 and LJ4CL2) exist in L. japonica. C4H is a P450-dependent monooxygenase cytochrome that catalyzes the conversion of cinnamate into 4-coumarate (also known as p-coumarate; Fig. 1) (Lu et al., 2006). LJC4H1 and LJC4H2 showed 53.95% and 60.40% identity at the nucleotide and amino acid levels. They included the consensus Cytochrome P450 domain (IPR001128), suggesting that LJC4Hs have the same type of biochemical activity. The importance of 4CL genes and the proteins they encode is especially evident when one considers the role of the activated thioester p-coumaroyl CoA in plant secondary metabolism. In higher plants, p-coumaroyl CoA represents the starting point for the biosynthesis of not only phenylpropanoid compounds, but a wide variety of other secondary metabolites as well (Vogt, 2010). LJ4CL1 and LJ4CL2 show 18.92% identity at the nucleotide level and 30.30% at the amino acid level. They include the consensus AMP-dependent synthetase/ligase (IPR000873), suggesting that LJ4CLs have the same type of biochemical activity.
3.3. Phylogenetic analysis of chlorogenic acid and luteolin biosynthesis-related genes
Phylogenetic analysis shows that these proteins are highly similar to those involved in the biosynthesis of flavonoids in Arabidopsis thaliana and wine grape (Figs. 1A–E). Gene duplication and divergence contribute to genetic novelty and adaptation (Hittinger and Carroll, 2007). As shown in Fig. 1A, LJPAL1 and LJPAL3 have similar functions compared with LJPAL2. Two LJ4CLs, two LJCHSs, and two LJCHIs may have different functions (Figs. 1B, C, D), whereas two LJHQTs may have similar functions (Fig. 1E).
3.4. Orthologous gene in L. hypoglauca and L. macranthoides
CHS1 and C4H2 were not detected in the genome of L. macranthoides and L. hypoglauca, respectively. Sequences of other genes were submitted to GenBank (JX068614–JX068636) and first reported in this paper.
3.5. Content of chlorogenic acid and luteoloside
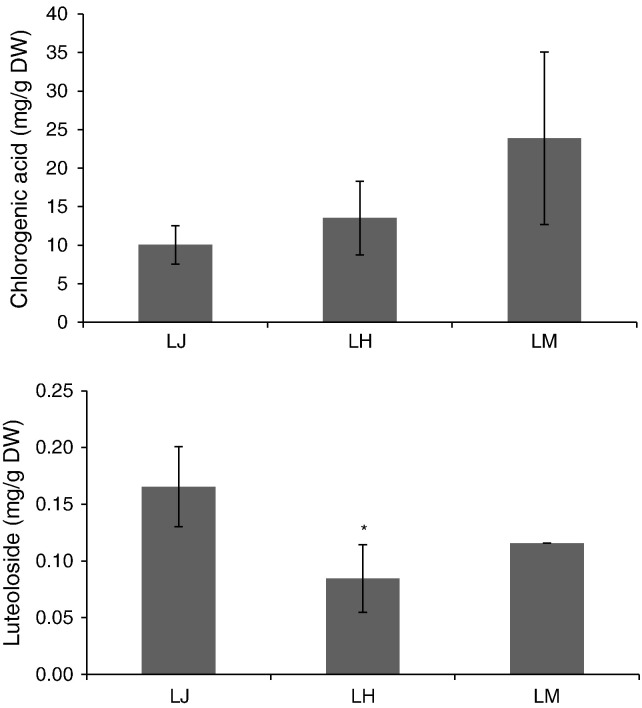
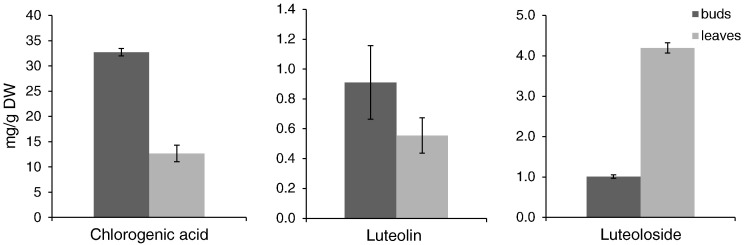
Chlorogenic acid and L7G are active compounds of L. japonica and its substitutes. The results of HPLC showed that chlorogenic acid content has no significant difference in the buds of the three species and the L7G content is lesser in buds of L. hypoglauca compared with those in L. japonica (Fig. 2 ). In L. japonica, chlorogenic acid in buds is higher than that in leaves; L7G in buds is lower than that in leaves, whereas luteolin content does not show significant difference (Fig. 3 ).
Fig. 2.
Content of chlorogenic acid and luteoloside in bud of Lonicera japonica Thunb (LJ), L. hypoglauca Miquel (LH), and L. macranthoides Hand.-Mazz (LM). The star means the significant difference at P < 0.05 level.
Fig. 3.
Content of chlorogenic acid, luteolin and luteoloside in bud and leaves of Lonicera japonica.
3.6. PAL gene family
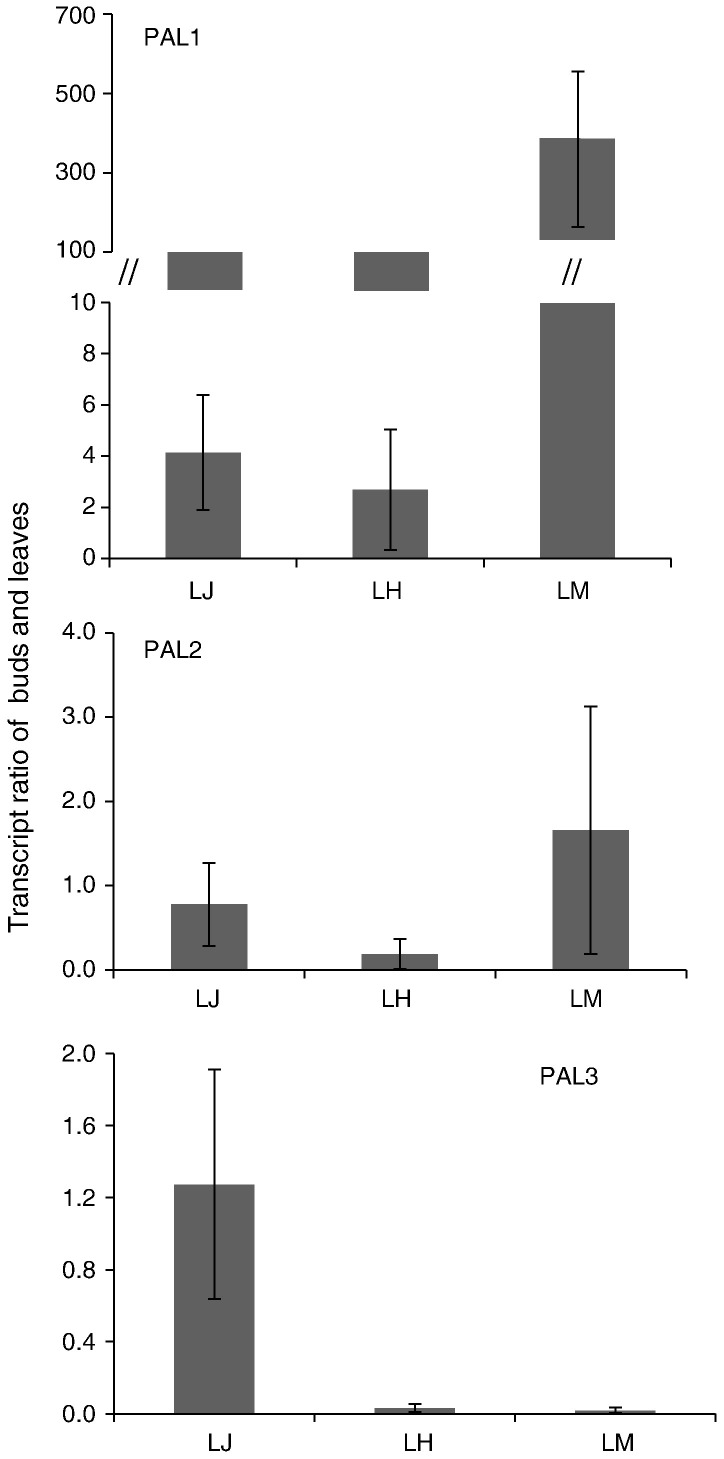
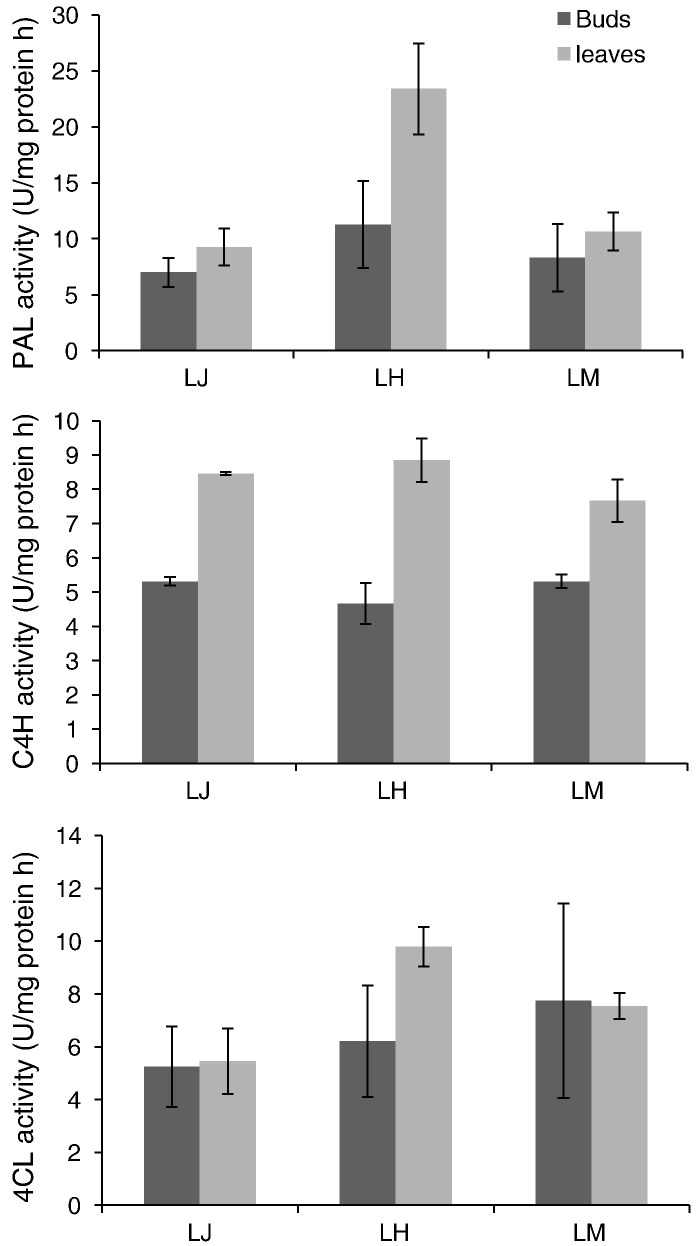
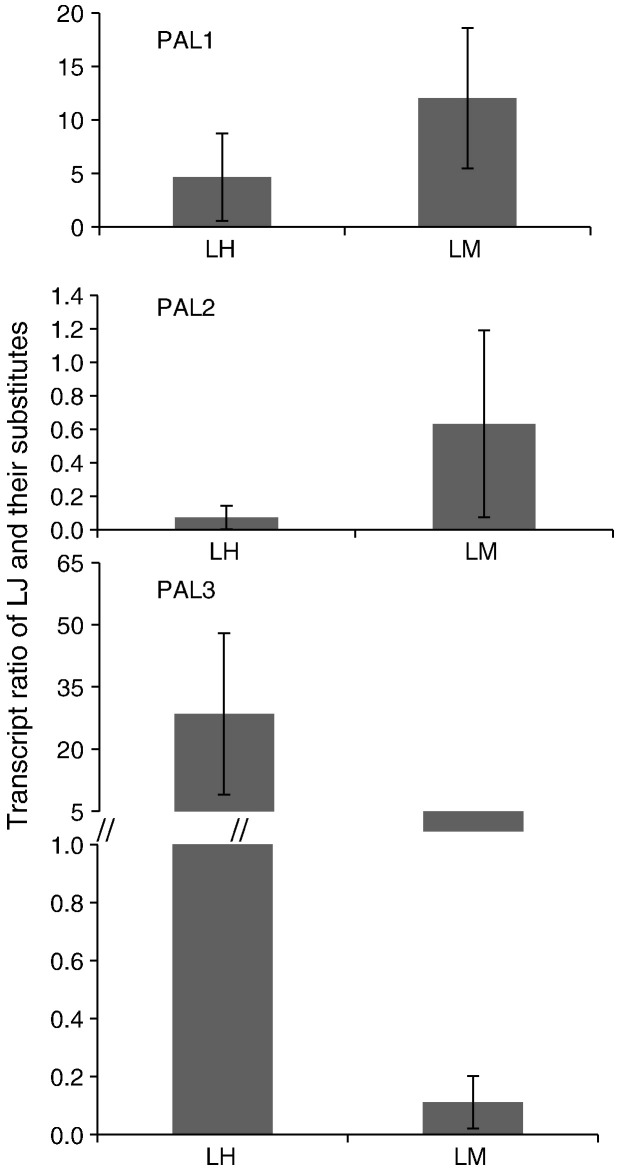
The transcript levels of three individual LJPAL and total enzyme activity were investigated in various tissues of L. japonica, L. hypoglauca, and L. macranthoides to ascertain the function of LJPALs on the accumulation of chlorogenic acid and luteolin. The investigation included the buds and leaves used in traditional Chinese medicine as recorded in Chinese pharmacopoeia. Gene expression analysis revealed that the transcripts of LJPAL1 and LMPAL1 are more abundant in buds than in leaves, whereas the transcripts of LHPAL2 and PAL3 (LHPAL3 and LMPAL3) are less abundant in buds than in leaves (Fig. 4 ). The total activity of PAL enzyme in buds is lower than that in leaves of L. hypoglauca and has no significant difference between buds and leaves of L. japonica and L. macranthoides (Fig. 5 ). The occurrence of chlorogenic acid in buds of L. hypoglauca is lower than that in leaves (Deng, 2007), suggesting that PALs could be important enzymes in the regulation of CGAs and luteolin content in L. hypoglauca. Although the transcripts of PAL orthologous in buds of various species differ (Fig. 6 ), their activities are not significantly different (Fig. 5).
Fig. 4.
The transcript ratio of PAL gene families in buds and leaves of Lonicera japonica Thunb (LJ), L. hypoglauca Miquel (LH), and L. macranthoides Hand.-Mazz (LM).
Fig. 5.
Enzymatic activity of PAL, C4H and 4CL in buds and leaves of Lonicera japonica Thunb (LJ), L. hypoglauca Miquel (LH), and L. macranthoides Hand.-Mazz (LM).
Fig. 6.
Fold changes of PAL gene families in buds of Lonicera japonica Thunb (LJ), L. hypoglauca Miquel (LH), and L. macranthoides Hand.-Mazz (LM). The expression level of PAL genes in of Lonicera japonica Thunb was arbitrarily set to 1.
3.7. C4H and 4CL gene family
The metabolic flux for the operation of the flavonoid pathway is maintained through the activities of C4H and 4CL (Kashmir et al., 2009), so chlorogenic acid and luteolin biosynthesis in L. japonica and its substitutes is critically dependent on the products of these enzymes. The expression of C4H and 4CL was also examined in the various tissues of the species used. Gene expression analysis revealed that the transcripts of LJC4H2 and LJ4CL2 were more abundant in buds than in leaves. Only LJC4H1 was less abundant in buds (Supplement Fig. 1). Orthologous genes in different species are function-divergent, whereas tissue-specific expression is related within species. In L. hypoglauca, only LH4CL2 is more abundant in buds than in leaves. In other species, particularly L. macranthoides, the transcripts of LMC4H2 are less abundant in buds than in leaves, whereas LM4CL1 and LM4CL2 are more abundant in buds. The activity of C4H in buds of the three species is lower than that in leaves, whereas 4CL does not show significant difference between buds and leaves (Fig. 5). Although transcripts of C4H and 4CL orthologous in buds of various species diverged (Supplement Fig. 2), their activities are not significantly different (Fig. 5).
3.8. CHS, CHI, and FNS gene family
Gene expression analysis revealed that the transcripts of LJCHS1, LJCHI2, LJFNS1, and LJFNS2 are more abundant in buds than in leaves. The difference in transcript level of CHS, CHI, and FNS orthologous in various species was analyzed. In L. hypoglauca, the transcripts of LHCHS1 are less abundant in buds than in leaves. In L. macranthoides, the transcripts of LHCHS2, LHCHI1, LHCHI2, LHFNS1, and LHFNS2 are more abundant in buds than in leaves (Supplement Fig. 3).
To understand the roles of individual CHS, CHI, and FNS members, comparison was made in terms of the expression of CHS, CHI, and FNS orthologous in buds of various species. Gene expression analysis revealed that the transcripts of LJFNS2 are higher than their orthologous genes in L. hypoglauca, and the transcript level of LJCHS2, LJCHI1, LJCHI2, LJFNS1, and LJFNS2 is lower than their orthologous genes in L. macranthoides (Supplement Fig. 4).
3.9. HQT gene family
Hydroxycinnamoyl-CoA quinate: hydroxycinnamoyl transferase (HQT) is an enzyme in chlorogenic acid biosynthesis that exhibits catalysis activity in the esterification of quinic acid with caffeoyl-CoA to generate CGA. Peng et al. (2010) reported that the tissue distribution of LJHQT1 is in accordance with the pattern of CGA content, based on the RT-PCR results they obtained. The transcripts of two HQTs in buds of L. japonica and L. hypoglauca have no significant difference in the leaves. The tissue distribution of LJHQTs is different in L. macranthoides, where both LMHQT1 and LMHQT2 in buds are higher than in leaves (Supplement Fig. 5). Meanwhile, the expression of HQT orthologous in buds of various species is not significantly different (Supplement Fig. 6).
4. Discussion
4.1. Organ preferential expression of gene families
For thousands of years, buds and leaves of L. japonica and its substitutes have been used as traditional Chinese medicine. The buds are used as Jin-Yin-Hua or Shan-Yin-Hua medicine, whereas the stems and leaves are used as Ren-Dong-Teng medicine. Previous studies have indicated that the active compound content differs among the plant parts (Deng, 2007; Zhao et al., 2010). This study confirmed that the chlorogenic acid and luteolin contents are significantly different between the buds and the leaves of L. japonica. Organ preferential expression of PAL, C4H, 4CL, CHS and CHI (Li et al., 2008, Rani et al., 2012, Roslan et al., 2012) is exhibited in the leaves, stem, and roots. PAL1, PAL2, and PAL4 are related to tissue-specific lignin synthesis in the leaves of A. thaliana (Olsen et al., 2008). Few papers have reported on the gene expression patterns in flowers. In this study, genes expressed in buds and leaves of L. japonica were analyzed. However, in the first step of biosynthesis, only the transcripted levels of LJPAL1, LJC4H2, and LJ4CL2 were higher in the buds than that in the leaves. The total activity of C4Hs in the buds was lower than that in the leaves, and PAL and 4CL did not change significantly. In the last step, LJHQTs did not show significant organ preferential expression, suggesting that LJPAL1, LJC4H2, and LJ4CL2 are among the critical genes that regulate CGA content within the organs. The activities of these individual enzymes in L. japonica need to be further analyzed. LJCHS1, LJCHI2, LJFNS1, and LJFNS2 in the last step exhibited bud preferential expression, suggesting that LJPAL1, LJC4H2, LJ4CL2, LJCHS1, LJCHI2, LJFNS1, and LJFNS2 are among the critical genes regulating the luteolin content of the organs. Therefore, only the first step of biosynthesis controls the CGAs, whereas the two steps control the luteolin content in the bud of L. japonica.
In the other two medical species, organ preferential expression of the gene families exhibited slight diversity. The two steps of biosynthesis could regulate the CGAs and luteolin in the bud of L. macranthoides. The critical genes include LMPAL1, LM4CL1, and LM4CL2 in the first step and LMHQT1, LMHQT2, LMCHS2, LMCHI1, LMCHI2, LMFNS1, and LMFNS2 in the last step. In L. hypoglauca, LHPAL2, LHPAL2, and LHCHS1 showed leaf preferential expression. The higher activity of LHPAL found in the leaves could be due to the presence of more active compounds in the leaves (Deng et al., 2007).
4.2. Species preferential expression of gene families
Although the three species are recorded in China pharmacopoeia, their active compounds and medicinal usages are diverse (Chu et al., 2011). The expression of orthologs genes differs in every species and could possibly induce diversity of active compound content. Previous reports have shown that PAL (Kovacik et al., 2012) and CHS (Pierantoni et al., 2010) have species or cultivar preferential expression and activity. In this paper, the transcripted level of orthologous genes related to CGAs and luteolin biosynthesis in the buds of the three medical species was analyzed. Compared with L. japonica, PAL1, 4CL1, and 4CL2 orthologs in the first step and CHS2, CHI1, CHI2, FNS1, and FNS2 orthologs in the last step showed preferential expression in the bud of L. macranthoides. For L. macranthoides, PAL3 and C4H2 orthologs exhibited preferential expression in the bud of L. japonica, whereas in L. hypoglauca, PAL2, 4CL2, and FNS2 orthologs showed preferential expression in the buds of L. japonica. However, the activity of PAL, C4H, and 4CL revealed no significant change in the buds of the three medical species (Fig. 5), similar to the CGA content (Fig. 2). The content of luteoloside in the bud of L. hypoglauca was lower than that in L. japonica (Fig. 2), suggesting that FNS2 orthologs could control luteoloside content in the buds of L. japonica and L. hypoglauca.
4.3. Gene and functional diversity of chlorogenic acid and luteolin biosyntheses
How gene duplication and divergence contribute to functional diversity has been of intense interest, but experimental evidence is limited. Based on the phylogenetic analysis, PAL, 4CL, CHS, and CHI paralogs were used to predict functional diversity. HQT paralogs could be the same function. According to the results of gene expression analysis, only HQT orthologs and paralogs have a similarly expressed pattern in different tissues among different species. This finding suggests that the complexity of orthologs and paralogs in the first step of CGA biosynthesis and the two steps in luteolin biosynthesis could induce functional diversity and chemical compound difference.
5. Conclusion
Fourteen genes in six gene families of CGAs, as well as luteolin biosynthesis in three medicinal plant species, were studied. Twelve genes in L. japonica and all the genes in L. hypoglauca and L. macranthoides were reported for the first time in this work. Results revealed that in biosynthesis, only the first step controls the CGAs, whereas both steps control luteolin in the bud of L. japonica. The expression of LJFNS2 exhibits positive correlation with luteolin levels in L. japonica. In addition, the higher activity of LHPAL may be related to the higher content of active compounds in the leaves of L. hypoglauca. The different usages of the three species of L. japonica and their substitutes were comprehensively studied according to expression diversity of orthologs and paralogs in metabolic pathways.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. (81001605). We thank Prof. Jun Yu for giving a revise to this paper.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gene.2012.09.051.
Appendix A. Supplementary data
Supplementary materials.
References
- Adato A. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 2009;5:e1000777. doi: 10.1371/journal.pgen.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G.B., Peng X.X., Li W.D., Wang W.Q. Cloning and characterization of a cDNA coding a hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase involved in chlorogenic acid biosynthesis in Lonicera japonica. Planta Med. 2010;76:1921–1926. doi: 10.1055/s-0030-1250020. [DOI] [PubMed] [Google Scholar]
- Bjellqvist B., Basse B., Olsen E., Celis J.E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- Brueske C.H. Phenylalanine ammonia lyase activity in tomatoroots infected and resistant to the root-knot nematode, Meloidogyne incognita. Physiol. Plant Pathol. 1980;16:409–414. [Google Scholar]
- Burge C.B., Karlin S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Cao S., Hu Z., Zheng Y., Lu B. Effect of BTH on anthocyanin content and activities of related enzymes in strawberry after harvest. J. Agric. Food Chem. 2010;58:5801–5805. doi: 10.1021/jf100742v. [DOI] [PubMed] [Google Scholar]
- Chen Q.Z., Lin R.C., Wang G.L., Li F.M. Studies on chemical constituents of the extract of Lonicera japonica. Zhong Yao Cai. 2010;33:920–922. [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission . 2010 ed. China Medical Science Press; Beijing: 2010. The Pharmacopoeia of the People’s Republic of China; pp. 205–206. [Google Scholar]
- Chu C., Liu H.J., Qi L.W., Liu E.H., Li P. Combination of normal light and fluorescence microscopy for authentication of five Lonicera species flower buds. Microsc. Res. Tech. 2011;74:133–141. doi: 10.1002/jemt.20882. [DOI] [PubMed] [Google Scholar]
- Clifford M.N. Chlorogenic acids and other cinnamates-nature, occurrence and dietary burden. J. Sci. Food Agric. 1999;79:362–372. [Google Scholar]
- Deng L.J., Zhao X.Z., Huang S.M., Huang S.J., Ou S.S., Wu Q.H. Chlorogenic acid content determination in branches, leaves and flowers of Lonricera japonica from Xincheng County. J. Guangxi Agric. 2007;22:34–36. [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Ferrer J.L., Austin M.B., Stewart C., Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M., Chapple C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book. 2011;9:e0152. doi: 10.1199/tab.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger C.T., Carroll S.B. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Kashmir S., Sanjay K., Arti R., Ashu G., Paramvir S. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct. Integr. Genomics. 2009;9(1):125–134. doi: 10.1007/s10142-008-0092-9. [DOI] [PubMed] [Google Scholar]
- Kovacik J., Klejdus B., Repčáková K. Phenolic metabolites in carnivorous plants: inter-specific comparison and physiological studies. Plant Physiol. Biochem. 2012;52:21–27. doi: 10.1016/j.plaphy.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Ku S. Effect of Lonicerae Flos extracts on reflux esophagitis with antioxidant activity. World J. Gastroenterol. 2009;15:4799–4805. doi: 10.3748/wjg.15.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Dudley J., Nei M., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ogita S., Keya C.A., Ashihara H. Expression of caffeine biosynthesis genes in tea (Camellia sinensis) Z. Naturforsch. C. 2008;63:267–270. doi: 10.1515/znc-2008-3-417. [DOI] [PubMed] [Google Scholar]
- Lim D., Cho H., Kim J., Nho C., Lee K., Park J. Luteolin decreases IGF-II production and downregulates insulin-like growth factor-I receptor signaling in HT-29 human colon cancer cells. BMC Gastroenterol. 2012;12:9. doi: 10.1186/1471-230X-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.R., Farr A.L. Protein measurement with the Folin-phenol reagent. J. Biol. Chem. 1951;193:265–269. [PubMed] [Google Scholar]
- Lu S., Zhou Y., Li L., Chiang V. Distinct roles of cinnamate 4-hydroxylase genes in populus. Plant Cell Physiol. 2006;47:905–914. doi: 10.1093/pcp/pcj063. [DOI] [PubMed] [Google Scholar]
- Molgaard P., Ravn H. Evolutionary aspects of caffeoyl ester distribution in dicotyledons. Phytochemistry. 1988;27:2411–2421. [Google Scholar]
- Olsen K.M., Lea U.S., Slimestad R., Verheul M., Lillo C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008;165:1491–1499. doi: 10.1016/j.jplph.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Peng X., Li W., Wang W., Bai G. Cloning and characterization of a cDNA coding a hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase involved in chlorogenic acid biosynthesis in Lonicera japonica. Planta Med. 2010;76:1921–1926. doi: 10.1055/s-0030-1250020. [DOI] [PubMed] [Google Scholar]
- Pierantoni L., Dondini L., De Franceschi P., Musacchi S., Winkel B.S., Sansavini S. Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear. Pyrus communis. Plant Physiol. Biochem. 2010;48:1020–1026. doi: 10.1016/j.plaphy.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Quevillon E. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani A., Singh K., Ahuja P.S., Kumar S. Molecular regulation of catechins biosynthesis in tea [Camellia sinensis (L.) O. Kuntze] Gene. 2012;495:205–210. doi: 10.1016/j.gene.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Roslan N.D. Flavonoid biosynthesis genes putatively identified in the aromatic plant polygonum minus via expressed sequences tag (EST) analysis. Int. J. Mol. Sci. 2012;13:2692–2706. doi: 10.3390/ijms13032692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Shang X., Pan H., Li M., Miao X., Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2011;138:1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ververidis F., Trantas E., Douglas C., Vollmer G., Kretzschmar G., Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol. J. 2007;2(10):1214–1234. doi: 10.1002/biot.200700084. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Voo K.S., Whetten R.W., O'Malley D.M., Sederoff R.R. 4-Coumarate: coenzyme A ligase from loblolly pine xylem (isolation, characterization, and complementary DNA cloning) Plant Physiol. 1995;108:85–97. doi: 10.1104/pp.108.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica Thunb. BMC Genomics. 2012;13:195. doi: 10.1186/1471-2164-13-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Chemical constituents from stems of Lonicera japonica. Zhongguo Zhong Yao Za Zhi. 2009;34:3051–3053. [PubMed] [Google Scholar]
- Zhao X., Ge Y., Wang D., Li S., Liu J., Wang X. Determination of extracted Chlorogenic acid and Luteolin-7-O-glucoside in Flos lonicerae by HPLC-DAD. Chin. J. Anal. Lab. 2010;29s:77–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.