Abstract
Immunotherapies can mediate regression of human tumors with high mutation rates, but responses are rarely observed in patients with common epithelial cancers. This raises the question of whether patients with these common cancers harbor T lymphocytes that recognize mutant proteins expressed by autologous tumors which may represent ideal targets for immunotherapy. Using high throughput immunologic screening of mutant gene products identified via whole exome sequencing, we identified neoantigen reactive tumor infiltrating lymphocytes (TIL) from 62 of 75 (83%) patients with common gastrointestinal cancers. In total, 124 neoantigen reactive TIL populations were identified, and all but one of the neoantigenic determinants were unique. The results of in vitro T cell recognition assays demonstrated that 1.6% of the gene products encoded by somatic non-synonymous mutations were immunogenic. These findings demonstrate that the majority of common epithelial cancers elicit immune recognition and open possibilities for cell based immunotherapies for patients bearing these cancers.
Introduction
Evidence from murine tumor models suggests that tumor rejection can be mediated by T lymphocytes reactive with antigenic determinants derived from tumor-associated mutations, also referred to as neoantigens (1,2). Subsequent studies in humans demonstrated that neoantigens represented the predominant targets of tumor reactive T cells in patients with melanoma (3). Support for the importance of neoantigen reactivity in immunotherapy responses in patients with cancer was provided by recent studies linking responses to therapy using antibodies directed against the inhibitory molecules CTLA-4 and PD-1 with mutation burden in patients with non-small-cell lung cancer (4) and with the total number of mutations, as well as the number of predicted neoepitopes identified using major histocompatibility complex (MHC) binding prediction algorithms in patients with melanoma (5,6). Clinical responses to checkpoint blockade have also been observed in over 50% of patients bearing mismatch repair (MMR) deficient tumors, primarily those of the gastrointestinal tract, that possessed high numbers of somatic mutations (4,7,8), whereas responses have rarely been seen in patients bearing MMR proficient GI tumors containing low numbers of mutations. Clinical responses in the latter study were also associated with peripheral expansion of T cells recognizing predicted HLA class I neoepitopes in three patients following checkpoint blockade (5,6). However, in studies where clinical responses were associated with the numbers of predicted neoepitopes, the ability of patient T cells to recognize these neoepitopes was either not evaluated or was only evaluated for a limited number of candidates (7,9), making it difficult to determine how many of the predicted targets were naturally processed and presented. Further evidence of immunologic reactivity of both CD8+ and CD4+ T lymphocytes against proteins encoded by somatic mutations comes from studies of individual patients with metastatic melanoma, cholangiocarcinoma (bile duct tumors), cervical cancer, and breast cancer in which the neoantigens associated with objective clinical responses following the adoptive transfer of autologous TIL were identified (10-16).
Here we analyzed the landscape of neoantigens arising from somatic mutations in 75 patients with MMR proficient gastrointestinal (GI) cancers.
Results
Identification of mutations in tumors
Using whole exome sequencing of tumor and normal tissue, we identified mutations present in tumors from 75 patients with GI cancers (originating in the rectum, colon, bile duct, pancreas, stomach, and esophagus), all of which were microsatellite stable and did not appear to be mis-match repair deficient (Supplementary Table 1). The number of mutations detected ranged from 22 to 928 with a median of 114. (Table 1 and Supplementary Tables 2,3). The number and range of somatic mutations were similar to those reported in a recent study of mutation frequencies in metastatic cancer (17). Approximately 94% of the mutations represented single nucleotide variants (SNVs) (Supplementary Table 3).
Table 1:
Neoantigens recognized by T lymphocytes from 75 patients with gastroinstinal cancers
| Patient ID* | Tumor type | Number of variant transcripts screened /total variant transcripts |
CD8 reactivities | CD4 reactivities |
|---|---|---|---|---|
| 3942 (32) | Colorectal | 131/150 | NUP98(A359D), KARS(D328H) | GPD2(E426K) |
| 3971 (32) | Colorectal | 51/116 | CASP8(F126V) | - |
| 3995 (32) | Colorectal | 57/67 | TUBGCP2(P293L), RNF213(N1702S), KRAS(G12D) | - |
| 4007 (32) | Colorectal | 124/142 | SKIV2L(R653H), H3F3B(A48T) | - |
| 4032 (32) | Colorectal | 92/112 | API5(R189Q), RNF10(E572K), PHLPP1(G566E) | - |
| 4060 | Colorectal | 56/86 | - | SMC1A(R192W) |
| 4071 | Colorectal | 53/188 | QSOX2(R524W) | - |
| 4072 | Colorectal | 45/93 | - | VPS51(Y348C), NEDD9(A728T) |
| 4081 | Colorectal | 93/110 | ALDOC(R69H) | - |
| 4090 | Colorectal | 80/84 | RPS15(R43Q) | MRPL39(L189V) |
| 4095 (20) | Colorectal | 55/118 | KRAS(G12D) | - |
| 4108 | Colorectal | 141/438 | - | SH3BP2(S130I) |
| 4115 | Colorectal | 111/162 | - | PTBP1(N112K), YWHAG(R86W) |
| 4141 | Colorectal | 111/134 | - | MPI(L408R) |
| 4151 | Colorectal | 73/88 | - | - |
| 4160 | Colorectal | 139/148 | APMAP(K195T), RNF43(P160L) | - |
| 4166 | Colorectal | 134/152 | NPLOC4(I473V) | - |
| 4171 | Colorectal | 114/133 | SIN3A(N520I) | PRMT3(I380S) |
| 4182 | Colorectal | 81/85 | - | ETV6(V265M) |
| 4196 | Colorectal | 164/223 | TP53(R175H) | GTF2E1(S334F) |
| 4207 | Colorectal | 29/42 | - | - |
| 4211 | Colorectal | 89/94 | - | RAB1A(I72S) |
| 4213 | Colorectal | 174/245 | DDX1(S362F), SMAD5(P268delinsPKH) | - |
| 4214 | Colorectal | 129/159 | - | NCK1(K24R), MCU(D123Y) |
| 4217 | Colorectal | 159/265 | MUC4(R4435S) | UEVLD(F191V), RAD51B(L321R), MAP3K2(S153F) |
| 4223 | Colorectal | 145/204 | - | - |
| 4232 | Colorectal | 33/58 | - | - |
| 4235 | Colorectal | 103/123 | GALK(A249V) | - |
| 4236 | Colorectal | 139/158 | - | ADAR(I723T) |
| 4238 | Colorectal | 93/102 | NCKAP1(D438Y), CYFIP1(I121V) | CSNK1E(R340W) |
| 4239 | Colorectal | 93/99 | - | - |
| 4241 | Colorectal | 76/80 | CCAR2(R417W), MED14(Q226R) | CCAR2(R417W), MED14(Q226R), ATM(D899H) |
| 4245 | Colorectal | 189/256 | NBEAL2(R371C) | FAM129B(R493W) |
| 4246 | Colorectal | 117/123 | ARMC9(L146F), MYO5B(K1410Q) | - |
| 4252 | Colorectal | 235/467 | ENDOG(V121L), NUP85(G349D), SLC2A1(I272V) | EXPH5(P1490T), SMU1(L346V) |
| 4255 | Colorectal | 86/89 | - | DDX21(R634H) |
| 4257 | Colorectal | 154/166 | - | PPL(T922I), NAV2(R496Q) |
| 4259 | Colorectal | 143/154 | TP53(Y220C) | TP53(Y220C) |
| 4262 | Colorectal | 212/218 | NAMPT(I178T) | GNG5(A21T) |
| 4263 | Colorectal | 120/130 | - | - |
| 4266 | Colorectal | 129/140 | TP53(R248W), ECI2(N352I) | NFASC(H1031Y), FAM208B(D677E) |
| 4268 | Colorectal | 85/91 | RNF149(D372H), USP37(D100Y) | KRAS(G12R) |
| 4271 | Colorectal | 117/119 | CPSF6(G178E), WDFY1(E44K), DHTKD1(V643I) | CHD2(K1351R), USP47(F1156L) |
| 4273 | Colorectal | 117/124 | - | TP53(R248W) |
| 4274 | Colorectal | 96/101 | - | TARS(R689C), ICAM2(E159K), BOD1(Q58K) |
| 4275 | Colorectal | 81/88 | GPATCH8(R954H) | WLS(R445G) |
| 4278 | Colorectal | 137/150 | COPS2(P42L,P308L) | - |
| 4283 | Colorectal | 154/161 | DNMT3A(P282S) | MUC6(V307M) |
| 4284 | Colorectal | 203/219 | CYP2E1(L101F) | RPL8(R241H), DDX47(E175K) |
| 4285 | Colorectal | 208/354 | - | TP53(R175H), ATP6VOB(Y88F) |
| 4289 | Colorectal | 86/105 | - | RAD21(D162V) |
| 3737 (32) | Bile duct | 22/29 | - | ERBB2IP(E805G) |
| 3812 (32) | Bile duct | 44/54 | - | - |
| 3978 (32) | Bile duct | 32/67 | - | ITGB4(S1002I) |
| 4077 | Bile duct | 34/41 | HIST1H2BE(E72V) | - |
| 4107 | Bile duct | 18/22 | - | - |
| 4110 | Bile duct | 121/208 | TES(K62N) | HYDIN(G4555V), ACLY(R794W) |
| 4112 | Bile duct | 205/931 | NBAS(C144S) | EIF4A3(D169Y) |
| 4200 | Bile duct | 36/45 | - | HIPK1(N66T) |
| 4203 | Bile duct | 54/74 | - | TUBGCP6(R611Q), ECE2(A40T) |
| 4220 | Bile duct | 94/208 | - | - |
| 4230 | Bile duct | 61/67 | - | - |
| 4272 | Bile duct | 145/155 | - | RSU1(P150T) |
| 4069 (32) | Pancreatic | 13/34 | ZFYVE27(R6H) | - |
| 4114 | Pancreatic | 23/30 | - | TP53(C135R), NTN1(596_597del) |
| 4145 | Pancreatic | 57/64 | - | IFI16(A577T) |
| 4231 | Pancreatic | 41/41 | - | - |
| 4247 | Pancreatic | 55/61 | - | - |
| 4265 | Pancreatic | 97/99 | PRPF6(S261T) | ATP11B(C705R) |
| 4270 | Pancreatic | 66/78 | MCAT(Q121K) | KRAS(G12R) |
| 4078 | Stomach | 84/107 | - | - |
| 4242 | Stomach | 151/177 | CDC42BPA(D428Y) | GOLGB1(E255G) |
| 4251 | Stomach | 143/161 | HNRNPU(F580I), RNF213(P4766H) | FMOD(S332N), TRAFD1(R11L) |
| 3948 (32) | Esophageal | 78/92 | - | PLEC(E1289K), XPO7(P274S), AKAP2(Q329K) |
| 4264 | Esophageal | 147/160 | CCNYL1(H84Y) | - |
Identification of neoantigen reactive TIL
Using high throughput immunologic testing, we evaluated whether autologous lymphocytes from these 75 patients could recognize neoantigens encoded by the identified mutations as previously described (18)(Table 1 and Supplementary Table 2). For several of the tumors, particularly those with mutational burdens of 200 or above, screening was limited using filters that accounted for expression of the variant transcript, the presence or absence of the variant in RNA-seq, and the presence or absence of the variant in multiple tumor samples (Supplementary Table 2). Algorithms for predicting MHC binding were not used for filtering variants, but we have included patient HLA typing data for reference (Supplementary Table 4). The overall frequency of screened variants was 73%. Here we defined “neoantigens” as proteins or 25 amino acid peptides that contained mutant amino acids that were recognized by T lymphocytes when loaded onto autologous antigen presenting cells. We distinguished this term from “neoantigenic determinants” which we refer to as shorter peptides containing mutant amino acids bound to specific HLA molecules that are recognized by T cells. We have mostly avoided the use of the term “neoepitopes” which has previously been used to describe peptides predicted to bind to particular MHC molecules but not necessarily determined to be immunogenic in vitro.
We generated multiple TIL cultures from different parts of the same metastatic tumor specimens on which whole exome sequencing was performed as previously described (19). To determine if any of the TIL cultures specifically recognized any of the mutant gene products present in the autologous tumor(s), we screened each variant for recognition of autologous dendritic cells (DCs) transfected with tandem minigenes (TMGs) encoding all the mutations or pulsed with synthetic peptide pools (PPs) containing the mutations as previously described (14,20). A schematic diagram outlining our overall screening approach has been previously published (18). TIL cultures that were positive in the preliminary screening assays were subsequently co-cultured with autologous DCs pulsed with individual peptides derived from the TMGs or PPs to identify the specific neoantigens being recognized. Finally, recognition of mutant peptides was compared to their wild type counterparts to determine if the recognition was truly neoantigen specific.
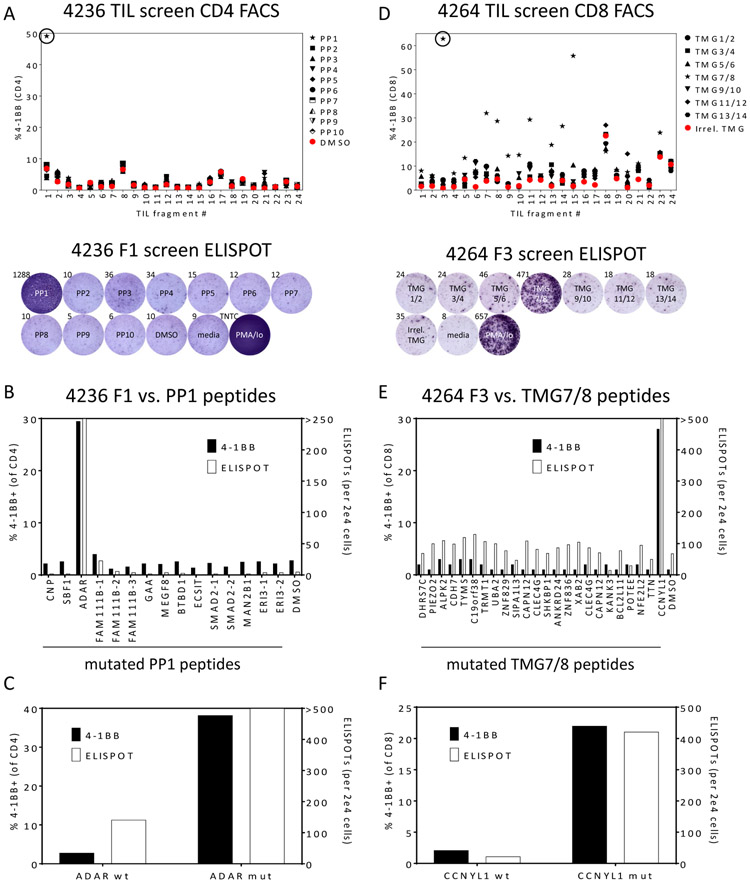
As one example of the identification of neoantigen reactive CD4+ T cells in a patient with colorectal cancer (patient 4236), we originally screened 24 TIL fragment cultures derived from five metastatic lung lesions for recognition of 10 PPs which comprised 138 variants unique to that patient. Reactive fragments were identified by interferon γ (IFNγ) secretion in an enzyme-linked immunospot (ELISPOT) assay and up-regulation of the T cell activation marker 4-1BB using flow cytometry. Only one of the 24 TIL cultures (fragment 1, F1) demonstrated reactivity against one PP (PP1) in both assays (Fig. 1A). Flow cytometric analysis of 4-1BB upregulation revealed this reactivity was mediated by CD4+ T cells (Fig. 1A). To identify the specific neoantigen being recognized, F1 was co-cultured with autologous DCs pulsed with each of the 15 individual peptides in PP1 (Fig. 1B). IFNγ secretion and expression of 4-1BB on CD4+ T cells indicated that F1 contained T cells which recognized mutant ADAR, a double-stranded RNA-specific adenosine deaminase. Of potential note is that loss of function of ADAR1 in tumor cells has previously been demonstrated to make them more susceptible to immunotherapy (21). To determine if this reactivity was neoantigen-specific, we evaluated recognition of the wild-type ADAR peptide in comparison to the mutant peptide (Fig. 1C), and both IFNγ ELISPOTs and 4-1BB on CD4+ T cells were significantly higher after co-culture with the mutant peptide.
Figure 1: Identification of neoantigen reactive CD4+ T cells from a patient with colon cancer (4236) and CD8+ T cells from a patient with esophageal cancer (4264).
(A) 24 TIL subcultures from patient 4236 were co-cultured overnight with autologous DCs pulsed with DMSO or the indicated peptide pool (PP) containing 15 or 16 peptides of 25 amino acids in length containing mutations identified by whole exome sequencing. TIL were also cultured without DCs (media) as negative controls or with PMA/Ionomycin (PMA/Io) as positive controls. T cell responses were measured by flow cytometric analysis of 4-1BB on CD4+ T cells (upper panel) and IFNγ ELISPOT (lower panel – one example showing the reactivity of the circled fragment in the upper panel).
(B) TIL fragment culture F1 from patient 4236 was co-cultured overnight with autologous DCs pulsed with individual peptides from PP1, and T cell responses were measured by flow cytometric analysis of 4-1BB on CD4+ T cells (left axis) and IFNγ ELISPOT (right axis). By both criteria, F1 contained CD4+ T cells that recognized mutant ADAR.
(C) 4236 TIL were co-cultured with autologous DCs pulsed with the indicated wild type and mutant peptides, and T cell responses were measured as in (B).
(D) 24 TIL subcultures from patient 4264 were co-cultured overnight with autologous DCs transfected with an irrelevant TMG or the indicated TMG encoding mutations identified by whole exome sequencing. TIL were also cultured without DCs (media) as negative controls or with PMA/Ionomycin (PMA/Io) as positive controls. T cell responses were measured by flow cytometric analysis of 4-1BB on CD8+ T cells (upper panel) and IFNγ ELISPOT (lower panel – one example showing the reactivity of the circled fragment in the upper panel).
(E) TIL fragment culture F3 from patient 4264 was co-cultured overnight with autologous DCs pulsed with individual 25 amino acid peptides derived from TMGs 7 and 8, and T cell responses were measured by flow cytometric analysis of 4-1BB on CD8+ T cells (left axis) and IFNγ ELISPOT (right axis). By both criteria, F3 contained CD8+ T cells that recognized mutant CCNYL1.
(F) 4264 TIL were co-cultured with autologous DCs pulsed with the indicated wild type and mutant peptides, and T cell responses were measured as in (E).
As one example of the identification of neoantigen reactive CD8+ T cells in a patient with esophageal cancer (patient 4264), we originally screened 24 TIL fragment cultures derived from three metastatic lung lesions for recognition of 14 TMGs which comprised 144 variants unique to that patient. In the preliminary screening assay, RNAs encoding two TMGs were pooled and electroporated into autologous DCs. Eleven of the 24 TIL cultures (F3, F5, F7, F8, F9, F10, F11, F13, F14, F15, and F23) demonstrated reactivity against one TMG pool (TMG7/8), and flow cytometric analysis of 4-1BB upregulation revealed this reactivity was mediated by CD8+ T cells (Fig. 1D). Seven of these TIL cultures (F3, F5, F7, F8, F9, F14, and F15) also recognized TMG7/8 as evaluated by IFNγ ELISPOT, and one example is presented in Fig. 1D (lower panel). To identify the specific neoantigen being recognized, F3 was co-cultured with autologous DCs pulsed with each of the 24 individual peptides encoded by TMG7/8 (Fig. 1E). IFNγ secretion and expression of 4-1BB on CD8+ T cells indicated that F3 contained T cells which specifically recognized mutant CCNYL1 (Cyclin Y Like 1) compared to the wild-type peptide (Fig. 1F). Thus, our screening approach enabled the identification of both CD4+ and CD8+ neoantigen reactive T cells.
We used this same approach to determine if neoantigen reactive T cells could be identified from the additional 73 patients (Tables 1 and 2, Supplementary Fig. 1A, and Supplementary Table 2), and in total, we identified neoantigen reactive T cells in 62 of the 75 (83%) patients (Table 2 and Supplementary Fig. 1B). Nearly all of the neoantigen-reactive T cells demonstrated minimal or no reactivity to the corresponding wild type peptides, but in cases where substantial recognition of the wild type peptide was observed, equivalent responses were observed against at least a 10-fold lower concentration of the mutant than the wild type peptide (22).
Table 2:
Summary of neoantigen reactivies identified in 75 patients with gastroinstinal cancers
| Cancer | Number of patients screened |
Number (%) of patients with neoantigen reactive TIL |
Median number of non- synonymous mutations |
Total number of variant transcripts screened / total number of variant transcripts (%) |
Total number of immunogenic neoantigens (% of variant transcripts screened) |
Number (%) CD8 reactivities |
Number (%) CD4 reactivities |
Number (%) of patients with TIL recognizing the following # of neoantigens: |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | >3 | ||||||||
| Colorectal | 51 | 45 (88%) | 126 | 5833/7710(76%) | 94 (1.6%)* | 47 (50%) | 47 (50%) | 6 (12%) | 17 (33%) | 16 (31%)* | 7 (14%) | 5 (10%)* |
| Bile duct | 12 | 8 (67%) | 59 | 866/1901(46%) | 12 (1.4%) | 3 (25%) | 9 (75%) | 4 (33%) | 5 (42%) | 2 (17%) | 1 (8%) | 0 (0%) |
| Pancreatic | 7 | 5 (71%) | 56 | 352/407(86%) | 8 (2.3%) | 3 (38%) | 5 (63%) | 2 (29%) | 2 (29%) | 3 (43%) | 0 (0%) | 0 (0%) |
| Stomach | 3 | 2 (67%) | 148 | 378/445(85%) | 6 (1.6%) | 3 (50%) | 3 (50%) | 1 (33%) | 0 (0%) | 1 (33%) | 0 (0%) | 1 (33%) |
| Esophageal | 2 | 2 (100%) | 119 | 225/252(89%) | 4 (1.8%) | 1 (25%) | 3 (75%) | 0 (0%) | 1 (50%) | 0 (0%) | 1 (50%) | 0 (0%) |
| Totals or Medians (GI) | 75 | 62 (83%) | 114 | 7654/10715(71%) | 124 (1.6%)* | 57 (46%) | 67 (54%) | 13 (17%) | 24 (32%) | 23 (31%) | 9 (12%) | 6 (8%) |
One patient (4259) had CD8+ and CD4+ T cells that recognized mutated TP53 epitopes that were counted as 2 separate antigens.
One patient (4241) had CD8+ and CD4+ T cells that recognized mutated CCAR2 and MED14 epitopes, both of which were counted as 2 separate antigens.
The median frequency of positive neoantigen reactive T cell cultures was 12.5%, and for 23 of the tumor samples where more than 20 cultures were available, only a single well was positive for the identified neoantigen (Supplementary Table 2). Mutational clustering analysis (described in the Supplemental Materials and Methods) indicated that 108 of the 121 (89%) somatic mutations encoding tumor neoantigens were clonal. Multiple factors that include variations between the infiltration and expansion of T cells within different tumor regions and stochastic expansion of T cells in vitro, rather than tumor heterogeneity, is likely to be responsible for these findings.
In these, we identified 124 T cell populations reactive with gene products encoded by nonsynonymous somatic mutations, representing 1.6% of the screened variants. All of the neoantigen-reactive TIL recognized products encoded by SNVs, with the exception of two TIL that recognized mutant proteins derived from non-frameshift indels, which represented 1.4% of the mutations found in these cancers. Frameshift mutations encoded 4.4% of the screened mutant transcripts but did not encode any of the identified neoantigens. Given the finding that 1.6% of SNVs encoded a neoantigen target, two or more frameshifted transcripts would also have been expected to encode a target neoantigen. Nonsense-mediated decay, which leads to a rapid decay of most frame-shifted products, may have influenced this observation, and only 135 frame-shifted transcripts were seen in RNA-seq libraries. The relatively small number of expressed frame-shifted products make it difficult to draw any conclusion other than the fact that these products do not appear to be more likely to encode neoantigen than SNVs.
TIL cultures from 53% of patients contained T lymphocytes that recognized more than one mutation (Table 2 and Supplementary Fig. 1C). For therapeutic application, it may be beneficial to target multiple mutations to overcome inter- and intra-tumoral heterogeneity (23,24).
The number of neoantigens identified for each patient was only weakly correlated with the number of variants screened (r2=0.15, Supplementary Fig. 1D). However, antigen expression, as inferred from RNA-seq data, was highly correlated with neoantigen recognition (Supplementary Table 5). RNA-seq data was available for 109 neoantigens identified in GI tumors, and 106 (97%) of those were encoded by transcripts in the top two expression quartiles.
Determination of whether neoantigen recognition was mediated by CD8+ or CD4+ T cells
The determination of whether a neoantigen was recognized by CD8+ or CD4+ T cells was usually clear based on the upregulation of 4–1BB in one cell type compared to the other in initial screening assays. For example, only CD4+ T cells in the F1 culture from patient 4236 expressed 4-1BB after stimulation with PP1 (Fig. 1A). Similarly, only CD8+ T cells from the positive fragment cultures from patient 4264 expressed 4-1BB after stimulation with TMG7/8 (Fig. 1A). However, in some cases, 4-1BB was expressed by both CD8+ and CD4+ T cells after stimulation. For example, 4-1BB upregulation was observed on both CD8+ and CD4+ T cells after stimulation of F5 from patient 4274 with PP7 (Supplementary Fig. 2A). In subsequent assays, we determined T cells in this fragment culture recognized mutant TARS (Threonyl-TRNA Synthetase) (Supplementary Fig. 2B). To determine whether this recognition was mediated by CD8+ T cells, CD4+ T cells, or both, we separated CD4+ T cells by positive selection using anti-CD4 coated magnetic beads and evaluated peptide recognition by both CD4+ and CD4- cell fractions (Supplementary Fig. 2C). In this assay, it was clear that only CD4+ T cells specifically recognized the mutant TARS peptide in comparison to its wild type counterpart.
In 2 patients, 4241 and 4259, we identified neoantigens that were recognized by both CD8+ and CD4+ T cells. For patient 4241, both purified CD4+ and CD8+ T cell populations from fragment cultures upregulated 4-1BB after stimulation with mutant CCAR2 and MED14 peptides (data not shown). For patient 4259, by pulsing peptides onto COS7 cells expressing different class I and class II HLA molecules, we identified 2 different restriction elements, HLA-A*02:01 and HLA-DRA1*01:01/DRB1*04:01:01, that were capable of presenting TP53 Y220C (25).
Overall, 46% of the identified neoantigens were recognized by CD8+ T cells and 54% were recognized by CD4+ T cells (Table 2). This may be a reflection the phenotype of the TIL fragment cultures which contained a median of 31.7% CD8+ T cells and 57.8 CD4+ T cells (Supplementary Fig. 3). The neoantigens recognized by CD8+ T cells were then analyzed using the NetMHCpan4.0 MHC class I prediction algorithm to identify candidate minimal epitopes and determine their predicted binding strength. The results indicated that 43 of the 56 class I neoepitopes ranked among the top 0.5% of predicted binders to an autologous MHC restriction element, whereas for eight of the neoantigens there were no predicted binders with a rank below 2 (Supplementary Table 6).
IFNγ ELISPOT vs. 4-1BB upregulation assays
The interpretation of preliminary screening assays was not always straightforward. In general, results from IFNγ ELISPOT and 4–1BB upregulation screening assays correlated well with each other as presented in Fig. 1. However, in some cases these results were discordant. It has previously been described that some T lymphocytes upregulate 4-1BB after stimulation and exhibit the ability to lyse target cells but are not capable of secreting IFNγ (26). Here we sometimes observed the opposite trend of potent IFNγ ELISPOT results with little or no apparent upregulation of 4-1BB on CD3+ T cells. In some cases, this appeared to be due to downregulation or complete loss of expression of CD3 by a few populations of activated T cells. For example, a TIL fragment F11 from patient 4257 was originally identified as recognizing mutant NAV2 in preliminary screening assays by IFNγ ELISPOT, but no expression of 4-1BB was observed on CD3+ CD4+ T cells (Supplementary Fig. 4A). Upon further investigation, by reanalyzing the FACS data, gating first on CD4+ cells and then subsequently evaluating 4-1BB expression on CD3+ and CD3- cells, we discovered that ~44% of CD4+ T cells completely lost surface expression of CD3 after stimulation with the mutant peptide, and ~86% of the CD3- cells expressed 4-1BB (Supplementary Fig. 4B).
Activation induced cell death (AICD) also caused discordant IFNγ ELISPOT and 4-1BB assay results, most notably for some CD4+ T cells. If the stimulus was too potent, as was often the case for CD4+ T cells stimulated with high concentrations of peptides, many of the reactive cells died during the overnight coculture. This resulted in an underestimate of the percentage of reactive cells by 4-1BB expression (Supplementary Fig. 5A-C).
In general, we attempted to use a thorough screening approach by evaluating recognition by both cytokine secretion and upregulation of activation markers to prevent missing reactivities that might only be present at low frequencies.
Screening of TMGs vs. PPs
We screened 62 of the 75 patients for neoantigen reactive TIL using both TMGs and PPs. The others were only screened using one of these two approaches due to lack of reagents during the preliminary screening. From these patients we identified 55 CD4 neoantigens and 43 CD8 neoantigens that were included in both TMGs and PPs in the preliminary screening assays (Supplementary Table 2). 26 (47%) of the CD4 neoantigens and 28 (65%) of the CD8 neoantigens were recognized in both TMG and PP formats. The majority of the remaining CD4 neoantigens were only detected using PPs, whereas the majority of the remaining CD8 neoantigens were only detected using TMGs in the preliminary screening assays (Supplementary Fig. 6). In subsequent assays, individual peptides were pulsed onto autologous DCs at ~10-20-fold higher concentrations than in the original screening assays, and all neoantigens could be detected, including the CD8 neoantigens that were only detected using TMGs in the preliminary screening assays. These observations may be accounted for by differences between class I and class II MHC antigen processing pathways. Class I MHC restricted epitopes are generally derived from intracellular proteins. Cross-presentation of class I MHC restricted epitopes from exogenously loaded 25-mer peptides is inefficient which may explain why higher peptide concentrations were necessary to visualize recognition by some CD8+ TIL. Class II MHC restricted epitopes are generally derived from exogenous proteins. Class II processing and presentation of epitopes derived from intracellular proteins is inefficient which may explain why some CD4 reactivities were not apparent using TMG RNAs electroporated into the cytoplasm of DCs (27).
Loss of neoantigen reactive T cells with prolonged in vitro culture
For 40 of the 75 patients, we selected fragments containing neoantigen reactive T cells, expanded them approximately 1000-fold in vitro with OKT3 and IL-2, and combined them. We analyzed the final expanded product for 26 of these patients by evaluating IFNγ ELISPOT and 4-1BB upregulation after coculture with mutant and wild type peptide pulsed autologous DCs. In 11 of these samples, we could not detect the presence of at least one neoantigen reactive T cell population in the expanded product (Supplementary Table 7). However, we could not accurately quantify the degree of loss since the expanded product and original fragments were not tested in the same assays. In addition, we were not able to compare actual percentages of reactive TCRs in the original fragments to those in the expanded products by TCR Vβ deep sequencing since we used all the cells from the original fragments in the expansion. Since nearly all the expanded products were made by combining fragments, some losses could have been accounted for by dilution. However, in some cases, we noted that the composition of the individual TIL fragment cultures changed dramatically over time during the in vitro culture period and that the neoantigen reactive T cells were sometimes lost or were present at very low frequencies in the expanded cultures (Supplementary Fig. 7A and B).
Reactivity of T cells transduced with neoantigen reactive T cell receptors
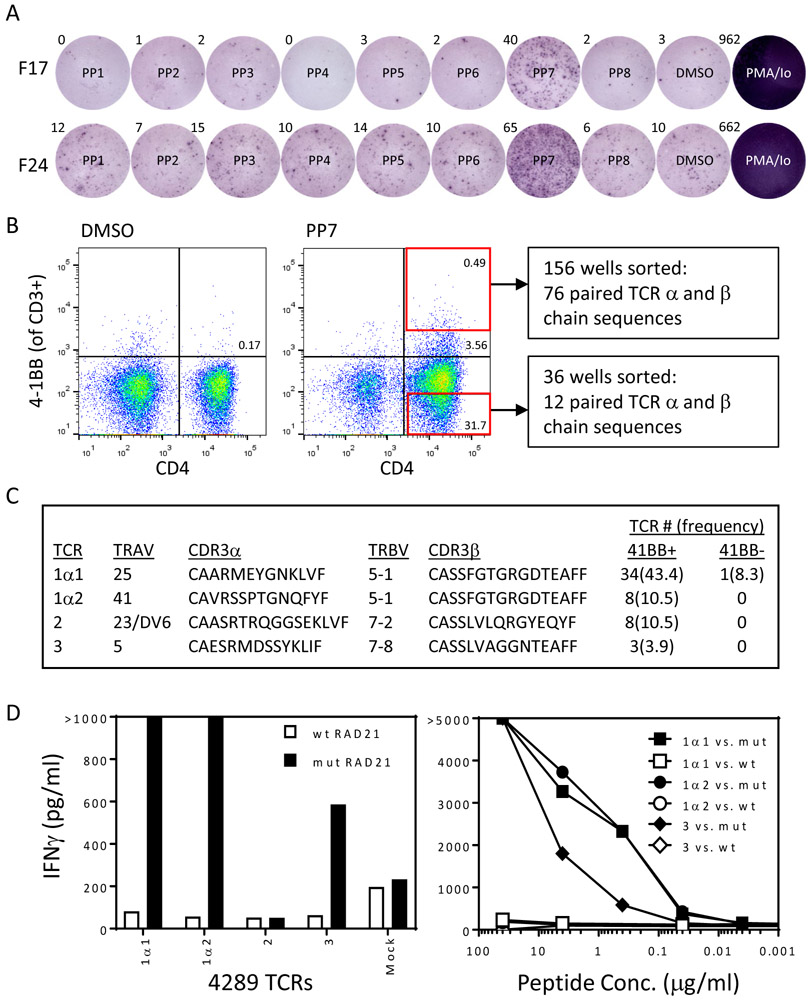
The finding that TIL from 83% of patients with metastatic common GI cancers contain T cells that recognize unique neoantigens expressed by the autologous cancer opens new possibilities for cell-based immunotherapies for patients with these cancers. However, as described above, we often encountered difficulties in consistently expanding enriched populations of these rare T cells. This is likely due to the lower proliferative potential of repeatedly stimulated neoantigen reactive T cells in the tumor microenvironment compared to the less differentiated naïve or central memory T cells in the original TIL cultures. To further validate the specific reactivities of the neoantigen reactive TIL, we developed strategies for rapidly identifying and testing the specificity of neoantigen reactive TCRs isolated from TIL (28). As one example, in patient 4289, we identified 3 TIL fragment cultures that weakly recognized PP7 in our original screening assay (F17, F18, and F24). ELISPOT data is presented in Fig. 2A for F17 and F24. Reactivity in F18 was only apparent by 4-1BB upregulation on CD4+ cells (data not shown). We combined these 3 fragments, stimulated them with autologous DCs pulsed with PP7, and sorted 156 individual cells expressing high levels of 4-1BB on CD4+ T cells (highest 0.49%) (Fig. 2B). We also sorted 36 individual cells that did not express 4-1BB as negative controls. We performed single-cell RT-PCR (29) to identify TCR α and β chains present in each of the wells, identified the 4 most frequent paired TCR α and β chain combinations (Fig. 2C), and constructed retroviral vectors encoding them as previously described (22). We transduced autologous T cells with these TCRs and measured IFNγ secretion by ELISA in response to all the mutant and wild type 25 amino acid peptides from PP7. Three of the 4 TCRs specifically mediated recognition of mutant, but not wild-type RAD21, a gene believed to be involved in the repair of DNA double-strand breaks (Fig. 2D). Interestingly, the β chains of the 2 TCRs with the highest avidities had identical amino acid sequences (Fig. 2C), but 3 silent base pair changes were present within non-germline-encoded CDR3 residues (Supplementary Fig. 8A-C). This may represent a case of convergent recombination, or it could be a set of naïve T cells that proliferated in the thymus after TCRβ, but before TCRα rearrangement. It is unclear if this is a “public” TCR β chain (30), but it has not been published in VDJdb, a curated database of TCR β chain sequences with known antigen specificities (https://vdjdb.cdr3.net/). Of potential interest is that a similar CDR3 is present in the VDJdb database, CASSFGGAGDTEAFF, which is reactive with p24 of HIV-1, although it is associated with a different TCRAV. Also, one of the identified TCR α chain CDR3 regions (CAARMEYGNKLVF) was previously reported in a study aimed at defining T cell states associated with response to checkpoint immunotherapy in melanoma (31), although it was associated with a different TCRBV.
Figure 2: Identification of neoantigen reactive TCRs from a patient with colon cancer (4289).
(A) 24 TIL subcultures from patient 4289 were co-cultured overnight with autologous DCs pulsed with DMSO or the indicated peptide pool (PP) containing 12 or 13 peptides of 25 amino acids in length containing mutations identified by whole exome sequencing. T cell responses were measured by flow cytometric analysis of 4-1BB on CD4+ T cells and IFNγ ELISPOT. ELISPOT results for 2 positive fragment cultures (F17 and F24) are shown.
(B) T cells from F17, F24, and F18 (which recognized PP7 by flow cytometric analysis but not ELISPOT) were combined and co-cultured with autologous DCs pulsed with PP7. After gating on CD3+ cells, 156 individual CD4+ T cells that expressed high levels of 4-1BB, as indicated by the red box in the upper right hand panel of the FACS plot, were sorted into 96 well plate wells, and single cell RT-PCR was performed on the contents of each well to amplify TCR α and β chains. In addition, as negative controls, 36 individual CD4+ T cells that did not express 4-1BB, as indicated by the red box in the lower right hand panel of the FACS plot, were sorted into 96 well plate wells, and single cell RT-PCR was performed on the contents of each well to amplify TCR α and β chains.
(C) Abbreviated sequences of the 5 most frequent TCRs identified by analysis of single-cell RT-PCR from CD4+ 4-1BB+ sorted cells in comparison to the frequencies of those TCRs in the CD4+ 4-1BB- population.
(D) Autologous open-repertoire PBL were genetically modified via retroviral transduction to express the TCRs described in (C) and were then co-cultured overnight with DCs pulsed with all the wild type and mutant peptides from PP7. T cell responses were measured by IFNγ ELISA, and recognition of wild type and mutant RAD21 by T cells expressing each of the 5 TCRs is shown (left panel). Recognition of titrated amounts of wild type and mutant RAD21 25mers by T cells expressing the 3 reactive TCRs was evaluated by IFNγ ELISA (right panel).
Isolation of neoantigen reactive TCRs was occasionally complicated by downregulation or complete loss of expression of CD3 by some populations of activated T cells. For example, as previously noted, ~44% of CD4+ T cells from patient 4257 TIL completely lost expression of surface CD3 after stimulation with the mutant NAV2 peptide, but ~86% of the CD3- cells expressed 4-1BB (Supplementary Fig. 4A). To identify which of these populations contained neoantigen reactive TCRs, we set up a second coculture and first gated on CD4+ cells. Subsequently we FACS sorted 48 individual cells with each of the following phenotypes for TCR analyses: CD3+ 4-1BB+, CD3-4-1BB+, and CD3-4-1BB- cells. We constructed retroviruses encoding the most frequently paired TCR α and β chain combinations from each of these populations. We transduced allogeneic PBL with these TCRs and measured IFNγ secretion by ELISA in response to mutant and wild type NAV2 25 mer peptides. Only the TCRs derived from the CD3- populations (both 4-1BB+ and 4-1BB-) mediated neoantigen recognition (Supplementary Fig. 9A-C).
Using this technique, we identified neoantigen reactive TCRs from 32 of the 75 patients with GI cancers targeting a total of 57 neoantigens. In all cases, the TCR transduced cells exhibited specific recognition of neoantigens in comparison to their wild type counterparts (examples in Fig. 2D and Supplementary Fig. 9).
Discussion
Immunotherapies can mediate regression of human tumors with high mutation rates, but responses are rarely observed in patients with common epithelial cancers. The lack of susceptibility of these cancers to immunotherapies such as IL-2, checkpoint modulators, and adoptive cell transfer (ACT), has raised questions concerning whether patients with these common cancers harbor T lymphocytes that can recognize neoantigens encoded by mutations found in autologous tumors. We previously reported the identification of neoantigen reactive T cells from 9 of 10 patients with GI cancers (32). Here we extended those findings to a cohort of 75 patients with a variety of gastrointestinal cancer diagnoses. We identified neoantigen reactive T lymphocytes from 83% of patients.
One advantage to the screening approach described here is that it does not rely on the use of MHC binding algorithms for the prediction of potential neoepitopes. Instead, by electroporating DCs with TMG RNAs encoding the mutations or pulsing DCs with long peptides containing the mutant amino acid(s), we allowed all potential neoantigenic determinants to be presented in the context of all of a patient’s class I and class II HLA molecules. In most cases, we did not specify the minimal neoantigenic determinant. Nonetheless, it appeared as though all were unique to each patient with one exception: CD8+ T cells from patients 3995 and 4095 recognized KRAS(G12D) in the context of HLA-C*08:02 as previously reported (20,32). CD4+ T cells from patients 4268 and 4270 recognized KRAS (G12R) albeit with different class II MHC restriction elements. T cells from six patients (4114, 4196, 4259, 4266, 4273, and 4285) each recognized unique neoantigenic determinants in TP53. T cells that recognized the R175H mutant gene product were identified from two patients (4196 and 4285), but recognition was mediated by CD8+ T cells in patient 4196 and CD4+ T cells in patient 4285. Similarly, T cells from two other patients recognized TP53 (R248W) (4266 and 4273), but recognition was mediated by CD8+ T cells in patient 4266 and CD4+ T cells in patient 4273. Thus, from the 124 neoantigen reactive T cell populations identified in the 75 patients we screened, 99% of the neoantigenic determinants appeared to be unique and not shared between any two patients.
Twenty-five (20%) of the neoantigen reactive T cell populations recognized mutant proteins encoded by genes listed in the Cancer Gene Census (CGC), a compilation of genes that are implicated to play a role in cancer when mutated (Supplementary Table 8). The functional significance of the majority of mutations in CGS gene products described in this report has not been experimentally verified, as the majority of these have not previously been identified in other tumors; however, 11 (9%) of the neoantigen reactive T cell populations recognized mutant proteins encoded by frequent mutations in TP53 and KRAS, while 99 (80%) of the neoantigen reactive T cell populations recognized mutant proteins resulting from somatic mutations in genes not listed in the CGC. These findings indicate that genes in the CGC were more likely to be recognized by T cells than non-CGC genes (p<0.001, Fisher’s exact test, Supplementary Table 9). The association between expression and T cell recognition described above is likely to play a significant role in these findings, and driver genes such as TP53 and KRAS are more likely to be expressed than passenger genes. Some of the additional mutations in CGC genes targeted by T cells that were not previously identified may nevertheless promote tumorigenesis which, given the association between expression and T cell recognition, could also provide a potential explanation for this association. Interestingly, 1.6% of the somatic cancer mutations were immunogenic (Table 2). However, this may be an underestimate because the T cells used for screening were obtained from individual tumor fragments that may not have represented the entire T cell pool from the patient.
All of the 75 patients described here were prospectively screened for the presence of neoantigen reactive TIL. All of these patients had previously received at least one systemic therapy (Supplementary Table 1) and had progressive disease at the time of tumor resection. Of the 62 patients in whom we found neoantigen reactive TIL, 40 were treated with autologous cell products selected on the basis of neoantigen reactivity. For these patients, individual TIL fragment cultures that contained neoantigen reactive T cells were selected, expanded approximately 1000-fold in vitro, and combined prior to ACT. In addition, 5 of these patients were treated with bulk, unselected TIL. 17 patients had a positive screen but were not treated: 10 had rapidly progressive disease, and the other 7 had very low frequencies of neoantigen reactive TIL as estimated on the basis of 4-1BB or OX40 upregulation after stimulation so we opted not to treat them. There were five partial responses by standard RECIST criteria.
We were not able to compare the compositions of expanded cell products to those of primary fragment cultures by TCR Vβ deep sequencing due to lack of starting material. However, we noted that the composition of individual TIL fragment cultures can change dramatically over time during the in vitro culture period and that the neoantigen reactive T cells can sometimes be lost or be present at very low frequencies in expanded cultures (Supplementary Table 7 and Supplementary Fig. 7). Therefore, to better evaluate the clinical effectiveness of targeting neoantigens, we are now developing techniques to enrich neoantigen reactive T cells by identifying neoantigen reactive TCRs and retrovirally introducing them into autologous lymphocytes with high transduction efficiencies for ACT. We are also developing methods to sort and selectively expand neoantigen reactive T cells based on upregulation of activation markers following coculture with neoantigen loaded autologous DCs.
The primary goal of the work presented here was to determine if patients with common epithelial cancers harbor T lymphocytes that can specifically recognize proteins encoded by somatic mutations uniquely expressed by autologous tumor cells. By evaluating TIL fragment cultures from 75 consecutive patients, we identified neoantigen reactive T cells in 83% of patients with common gastrointestinal cancers, and 99% of the neoantigenic determinants appeared to be unique to the autologous patient. Although we observed a few objective clinical responses after treating patients with autologous TIL, we can not draw any conclusions about the clinical efficacy of targeting neoantigens until we conduct clinical trials in which patients are treated with highly enriched populations of such T cells or with T cells genetically modified to express high levels and percentages of neoantigen reactive TCRs. Nonetheless, our observations demonstrate that most epithelial cancers, generally considered to be non-immunogenic, do elicit in vivo immune reactions and provide a rationale for developing new personalized cell-based immunotherapeutic treatments for patients with common epithelial cancers by targeting the unique tumor associated mutations expressed by those cancers.
Materials and Methods
Study Design
The primary objective of the study described here was to determine if tumor infiltrating lymphocytes (TIL) from patients with metastatic GI cancers can recognize neoantigens encoded by mutations expressed by autologous tumor cells. We performed prospective analyses of 75 patients with metastatic GI cancers (originating in the colon, rectum, bile duct, pancreas, stomach, and esophagus). Samples were derived from patients enrolled on a clinical protocol (NCT01174121) approved by the institutional-review board (IRB) of the National Cancer Institute (NCI). We obtained informed written consent from all patients, and all studies were conducted in accordance with The Declaration of Helsinki, The Belmont Report, and the U.S. Common Rule.
Whole Exome Sequencing, variant calling, andRNA-seq
Whole exome sequencing (WES) was performed by Personal Genome Diagnostics, the Broad Institute, and the Surgery Branch, NCI on tumor tissue and normal peripheral blood cells as previously described (33). The data was processed, and variants were called as described in the Supplementary Materials and Methods section. BLAT analysis of peptide Nmers against the ucsc database resulted in the identification of 27 transcripts, representing less than 0.3% of the total variant calls, that showed identity with germline segments that were potentially mis-mapped using the DNA alignment pipeline (Supplementary Table 10).
Identification of neoantigens recognized by TIL
TIL were generated from patients by dissecting resected lesions into small fragments and allowing the infiltrating T lymphocytes to migrate out of the fragments and expand individually in vitro in the presence of IL-2 as previously described (32) and as further detailed in the Supplementary Materials and Methods section. All resections and peripheral lymphocyte collections were performed at least four weeks after the last dose of cytotoxic chemotherapy or radiation to minimize the potential impact of those treatments on lymphocytes. To determine if any of the TIL cultures specifically recognized any of the potential neoantigens encoded by mutations present in the autologous tumor(s), we evaluated recognition of autologous dendritic cells (DCs) transfected with tandem minigenes (TMGs) encoding all the mutations or pulsed with synthetic peptide pools containing mutant peptides. TMGs and peptide pools were constructed as described in the Supplementary Materials and Methods section. Briefly, each TMG comprised a string of minigenes, each of which was a genetic construct encoding an identified mutation flanked on each side by the 12 wild-type amino acids from the parent protein. In the case of frameshift mutations, the cDNA was translated until the next stop codon. RNAs encoding these TMGs were synthesized by in vitro transcription. Alternatively, peptides of 25 amino acids in length were synthesized containing the mutated amino acid flanked on each side by the 12 wild-type amino acids and were mixed equally to form peptide pools (PPs) containing between 8 and 24 individual peptides. For frameshift mutations, peptides of 25 amino acids in length were synthesized, overlapping by 15 amino acids to cover the translated sequence until the next stop codon. Autologous DCs were electroporated with individual TMG RNAs and were pulsed with individual PPs, allowing for processing and presentation of all potential neoantigenic determinants by any of the patient’s class I or class II MHC molecules. Each TIL fragment population that had been expanded in vitro for 2-4 weeks with IL-2 was then co-cultured with RNA-electroporated and/or peptide-pulsed autologous DCs and evaluated for recognition via cytokine release and expression of activation markers as described in the Supplementary Materials and Methods section.
Identification and evaluation of neoantigen reactive T cell receptors
From some neoantigen reactive TIL cultures we attempted to isolate and characterize the TCRs that mediated recognition as described in the Supplementary Materials and Methods section. Briefly, individual T cells that upregulated an activation marker upon co-culture with mutant TMGs or peptides were sorted into by FACS, and TCRs were sequenced. Alternatively, in cases where there was a dominant reactivity that correlated with a dominant TCR-Vβ clonotype, the dominant TCR-Vβ expressing T cells were FACS purified, and TCRs were sequenced. DNAs encoding the TCR α and β chains separated by a cleavable picornavirus ribosomal skip element were constructed and cloned into retroviral vectors. Retroviral particles were then used to transduce autologous or allogeneic PBL, and recognition of relevant target cells was evaluated via cytokine release and expression of activation markers as described in the Supplementary Materials and Methods section.
Supplementary Material
Statement of Significance.
Tumor infiltrating lymphocytes cultured from 62 of 75 (83%) patients with gastrointestinal cancers recognized neoantigens encoded by 1.6% of somatic mutations expressed by autologous tumor cells, and 99% of the neoantigenic determinants appeared to be unique and not shared between patients.
Acknowledgements
We would like to acknowledge the generous support of the Center for Clinical Research, National Cancer Institute, and the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov), which was used to analyze the WES and RNA-seq data.
Footnotes
Conflict of interest disclosure statement
The authors have no conflicts of interest to disclose.
References
- 1.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity 1995;2(1):45–59. [DOI] [PubMed] [Google Scholar]
- 2.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482(7385):400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A 2005;102(44):16013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018. May 14;33(5):843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351(6280):1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017;551(7681):517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357(6349):409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359(6377):801–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevanovic S, Pasetto A, Helman SR, Gartner JJ, Prickett TD, Howie B, et al. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 2017;356(6334):200–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol 2013;190(12):6034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 2013;19(6):747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 2014;20(13):3401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344(6184):641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prickett TD, Crystal JS, Cohen CJ, Pasetto A, Parkhurst MR, Gartner JJ, et al. Durable Complete Response from Metastatic Melanoma after Transfer of Autologous T Cells Recognizing 10 Mutated Tumor Antigens. Cancer immunology research 2016;4(8):669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 2018;24(6):724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, et al. Integrative clinical genomics of metastatic cancer. Nature 2017;548(7667):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran E, Robbins PF, Rosenberg SA. ‘Final common pathway’ of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol 2017;18(3):255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother 2003;26(4):332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med 2016;375(23):2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019;565(7737):43-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, et al. Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression. Clin Cancer Res 2017;23(10):2491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27(1):15–26. [DOI] [PubMed] [Google Scholar]
- 25.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 2019;129(3):1109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turcotte S, Gros A, Tran E, Lee CC, Wunderlich JR, Robbins PF, et al. Tumor-reactive CD8+ T cells in metastatic gastrointestinal cancer refractory to chemotherapy. Clin Cancer Res 2014;20(2):331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, Noe F, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Frontiers in immunology 2017;8:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR, et al. High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol 2003;171(6):3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasetto A, Gros A, Robbins PF, Deniger DC, Prickett TD, Matus-Nicodemos R, et al. Tumor- and Neoantigen-Reactive T-cell Receptors Can Be Identified Based on Their Frequency in Fresh Tumor. Cancer immunology research 2016;4(9):734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol 2008;8(3):231–8. [DOI] [PubMed] [Google Scholar]
- 31.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018;175(4):998–1013 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350(6266):1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin J, Sabatino M, Somerville R, Wilson JR, Dudley ME, Stroncek DF, et al. Simplified method of the growth of human tumor infiltrating lymphocytes in gas-permeable flasks to numbers needed for patient treatment. J Immunother;35(3):283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.