Abstract
Lung, breast, colorectal, and prostate cancers are the most incident worldwide. Optimal population-based cancer screening methods remain an unmet need, since cancer detection at early stages increases the prospects of successful and curative treatment, leading to a lower incidence of recurrences. Moreover, the current parameters for cancer patients’ stratification have been associated with divergent outcomes. Therefore, new biomarkers that could aid in cancer detection and prognosis, preferably detected by minimally invasive methods are of major importance. Aberrant DNA methylation is an early event in cancer development and may be detected in circulating cell-free DNA (ccfDNA), constituting a valuable cancer biomarker. Furthermore, DNA methylation is a stable alteration that can be easily and rapidly quantified by methylation-specific PCR methods. Thus, the main goal of this review is to provide an overview of the most important studies that report methylation biomarkers for the detection and prognosis of the four major cancers after a critical analysis of the available literature. DNA methylation-based biomarkers show promise for cancer detection and management, with some studies describing a “PanCancer” detection approach for the simultaneous detection of several cancer types. Nonetheless, DNA methylation biomarkers still lack large-scale validation, precluding implementation in clinical practice.
Keywords: lung cancer, breast cancer, colorectal cancer, prostate cancer, DNA methylation, biomarker, detection, prognosis, liquid biopsy, cell-free DNA
1. Introduction
During the last decades, efforts have scaled up worldwide to develop more effective biomarkers as an approach to reduce cancer mortality [1]. A cancer biomarker can be defined as an objectively measurable biomolecule, such as a protein, metabolite, RNA, DNA, or an epigenetic alteration, found in body fluids or tissues, that indicates the presence of cancer or provides information on cancer’s expected future behavior [2,3].
Tissue biopsy sampling has been the gold-standard approach for patients’ diagnosis and prognostication. However, several drawbacks have been pointed out over the years to this approach [4,5]. Firstly, and mainly, tissue samples might not fully represent tumor heterogeneity, constituting a limitation for accurate outcome prediction and treatment efficacy [5,6]. Moreover, early-stage tumor, residual disease, and early recurrence detection might be difficult, since tissue biopsy sampling requires a highly invasive intervention with risk of complications, and, depending on the tumor anatomical location, may be extremely difficult to obtain [4,7]. Therefore, a minimally invasive method that allows for cancer detection at an early stage and patients’ follow-up is essential. Recently, liquid biopsies obtained from easily assessable body fluids, including blood, urine or sputum, have surfaced as a viable alternative to overcome these challenges [8].
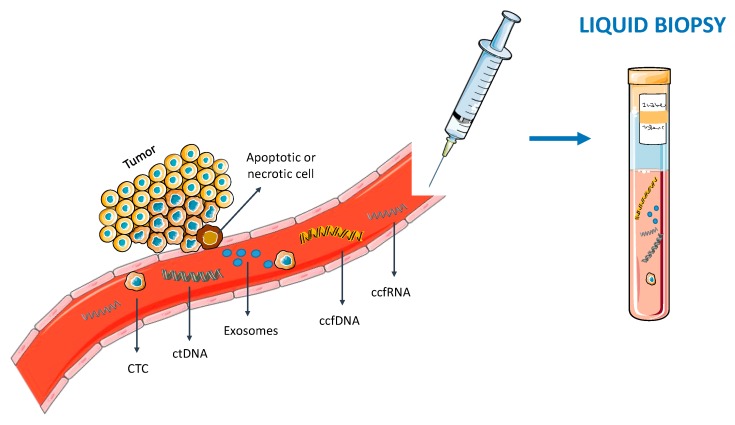
Liquid biopsies, mainly based on circulating cell-free DNA (ccfDNA), circulating tumor cells (CTCs), circulating cell-free RNA (ccfRNA) and exosomes [8] are a fast, reliable, cost-effective and minimally invasive approach [9] (Figure 1). Hence, owing to these features, they may allow for a real time monitoring of the cancer evolution, while better representing the heterogeneous genetic profile of all tumor sub clones [8,10]. Furthermore, it has been shown that these molecules contain alterations present in the tumor itself, including mutations [11] and epigenetic alterations [12]. Indeed, due to its early onset, cancer specificity, biological stability, and accessibility in bodily fluids, aberrant DNA methylation has called attention as epigenetic-based biomarker, making it an attractive target to be studied in liquid biopsies [3].
Figure 1.
Blood-based liquid biopsy. Circulating tumor cells (CTC), circulating cell-free DNA (ccfDNA) [including circulating tumor DNA (ctDNA)], circulating cell-free RNA (ccfRNA) and exosomes are released from tumor cells to the bloodstream. Hence, blood can be collected and analyzed in the context of a liquid biopsy.
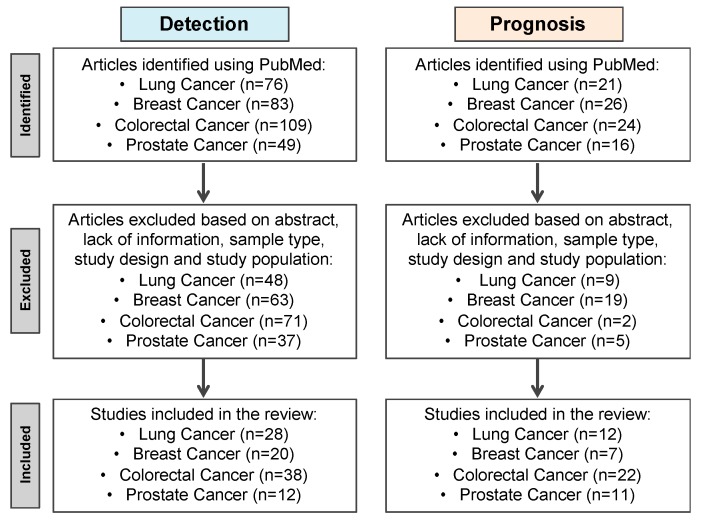
Lung (LC), breast (BrC), colorectal (CRC) and prostate (PCa) cancers are the most incident and among the deadliest worldwide, despite the efforts for early cancer detection and the emergence of new therapies [13]. Given the shortcomings of current screening methods and prognostic biomarkers, the development and implementation of effective biomarkers for cancer detection at curative, early stages, and for better patients’ stratification, is crucial. Thus, for the purposes of this review, a detailed and extensive literature review was conducted aimed to explore the state of the art of ccfDNA methylation blood-based biomarkers for cancer screening, diagnosis, prognosis, prediction, and monitoring of the four most incident cancers worldwide. On a Pubmed database search, the key words “Lung Cancer/Breast Cancer/Colorectal Cancer/Prostate Cancer”, “DNA methylation”, “Diagnosis/Detection/Prognosis” and “Serum/Plasma” were used (Figure 2).
Figure 2.
Flow diagram of Pubmed available studies’ selection procedure using the key words “Lung Cancer/Breast Cancer/Colorectal Cancer/Lung Cancer”, “DNA methylation”, “Diagnosis/Detection/Prognosis” and “Serum/Plasma”.
2. Circulating Cell-Free DNA Liquid Biopsies
CcfDNA was firstly described in 1948 when extracellular nucleic acids were found in human blood from healthy individuals by Mandel and Métais [14]. Later on, it was found that, in cancer patients, circulating tumor DNA (ctDNA) fragments between 150 and 1000 base pairs could also be detected due to their release into the bloodstream, either by cell death (apoptosis or necrosis) or active secretion by the release of extracellular vesicles, such as exosomes [10,15]. Depending on several factors including tumor burden, metastatic sites or cellular turnover, ctDNA might account for 0.01% to 90% of the total ccfDNA in the blood of cancer patients [15]. Owing to the fact that ctDNA might represent tumor-specific genetic and epigenetic alterations of all tumor’s sub clones present, ccfDNA is an ideal candidate for blood-based liquid biopsies by offering the possibility to test for the presence of cancer and the discrimination of lethally aggressive cancer [16].
3. DNA Methylation
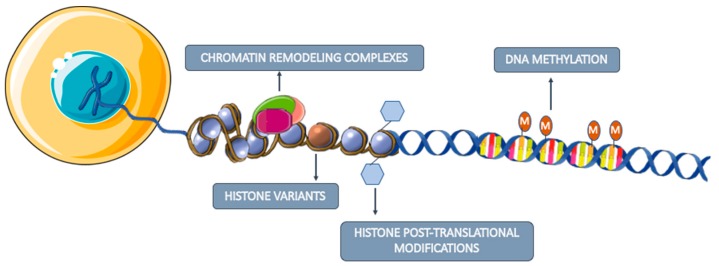
Although the study of tumor mutations was the focus of biomarker research for a long time, their wide diversity has been a challenge for the development of effective diagnostic biomarkers since very large proportions of the genome would need to be examined in order to provide adequate sensitivity [17]. Contrarily, epigenetic alterations seem to be more stable and homogenous in cancer, representing a good alternative for biomarker development [18]. The main studied epigenetic mechanisms are DNA methylation, histone post-translational modifications, histone variants and chromatin remodeling complexes (Figure 3) [19]. Although these mechanisms are crucial for normal cell development and regulation of specific gene expression patterns, epigenetic dysregulation often leads to inappropriate activation or inhibition of several signaling pathways, which may trigger the development of several pathologies, including cancer [20].
Figure 3.
Major studied epigenetic mechanisms involved in gene expression regulation. DNA methylation consists in the addition of a methyl group to a cytosine present in a cytosine-phosphate-guanine (CpG). Histone post-translational modifications refer to the addition of biochemical modifications on histone tails, such as methylation, acetylation, phosphorylation, ubiquitylation and SUMOylation that regulate gene expression. Histone variants differ a few amino acids from canonical histones and regulate chromatin remodeling and histone post-translational modifications. Chromatin remodeling complexes regulate the nucleosome structure by removing, relocate and shifting histones.
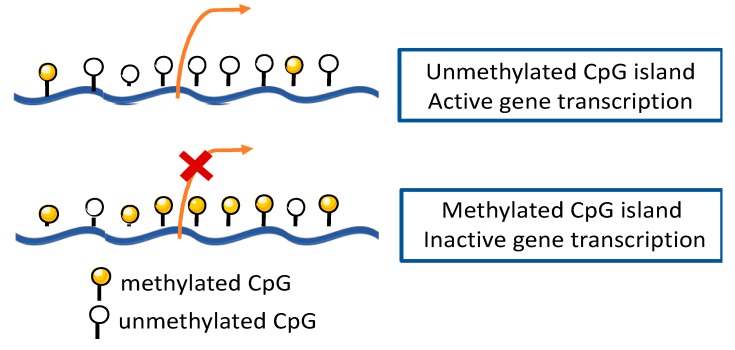
DNA methylation, the most widely studied epigenetic modification in humans, was also the first to be identified in cancer [20,21]. This epigenetic mechanism consists of covalent addition of a methyl group, donated by S-adenosylmethionine (SAM), to the 5-position carbon of a cytosine ring to form 5-methylcytosine (5mC) [22,23]. This modification is catalyzed by DNA methyltransferase enzymes (DNMTs), namely, DNMT3a and DNMT3b that catalyze de novo DNA methylation during embryonic development, establishing tissue-specific DNA methylation, and DNMT1 that is often associated with maintenance of methylation patterns during replication [23]. Typically, this process occurs on cytosine residues present at CpG dinucleotides commonly found in large clusters named CpG islands, which are predominantly located at the 5′ end of genes, occupying approximately 60% of human gene promoter regions [20,23,24]. Although gene promoter hypermethylation is associated with transcription repression of the nearby gene (Figure 4), depending on DNA methylation genomic location, it can display different functions [22,25]. Epigenetic gene silencing by DNA promoter methylation may occur either directly, by blocking transcription factors to prevent binding to target sites in or near the promoter, or indirectly, through binding of methyl-CpG-binding proteins (MBP), which can recruit other enzymes like DNMTs and histone deacetylases (HDAC), leading to chromatin conformation changes that further repress gene transcription [20,22].
Figure 4.
DNA methylation within a gene promoter region. Unmethylated CpG islands enable gene transcription. When CpG island is methylated, gene transcription is repressed.
As aforementioned, DNA methylation is crucial for multiple cellular processes, thus it is understandable that its deregulation has been linked to cancer. Indeed, normal and cancer cells display different methylomes. Usually, a global hypomethylation pattern, which contributes to genomic instability and activation of silenced oncogenes, is observed in cancer [23,26]. Alongside, tumor suppressor genes (TGS) frequently undergo inactivation due to focal promoter hypermethylation [23,26]. Currently, the latter process is considered a major contributor to neoplastic transformation [27].
Interestingly, aberrant DNA methylation is thought to occur at very early stages of cancer development and specific genes seem to be methylated at different tumor stages [23]. Moreover, since these alterations can be assessed in several body fluid samples [23], it is widely accepted that DNA methylation-based liquid biopsies are a promising approach, not only for premalignant/early cancer detection, but also for prognostic assessment. Furthermore, since some genes seem to acquire tissue-specific DNA methylation, it may also be possible to discriminate between different cancer types in the context of metastatic tumors [23] or in liquid biopsies.
4. Cell-Free DNA Methylation-Based Biomarkers
4.1. Lung Cancer
4.1.1. Screening and Diagnosis
Despite advances in new treatment options over the years, the high mortality rate observed in LC patients is mainly related to the fact that more than 75% of LC patients are diagnosed with advanced stage disease [28]. Hence, effective screening options aiming to shift LC diagnosis from advanced to curative early stages are crucial to change the fate of this disease [29]. Currently, low-dose computed tomography (LD-CT) is considered the best LC screening method available [30]. Nonetheless, despite The National Lung Screening Trial has shown a 20% decrease in LC-related mortality rate among high-risk smokers with LD-CT screening comparing to chest X-ray [corroborated by the largest European trial (Dutch-Belgian Lung Cancer Screening Trial)] from the 24% positive test results in this trial, 96.4% were deemed as false-positive [31,32]. Hence, due to the risks related to LD-CT screening, namely, overdiagnosis, radiation exposure, and false positive results leading to unnecessary anxiety and costs [29], the development of specific and accurate screening tools is urgently needed to improve LC survival.
In 2002, Usadel et al. and Bearzatto et al. reported, for the first time, APCme and p16INK4ame, respectively, in ccfDNA as putative minimally-invasive biomarkers for LC detection [33,34]. Thenceforth, numerous other methylated gene promoters detected in ccfDNA have been proposed for LC detection either individually or in panel (Table 1). RASSF1Ame and p16INK4ame represent the two most frequently reported genes in blood-based liquid biopsies displaying 22–66% sensitivity and 57–100% specificity for LC detection, individually [34,35,36].
Table 1.
CcfDNA-based methylation biomarkers for lung cancer (LC) detection.
| Lung Cancer | ||||||
|---|---|---|---|---|---|---|
| Genes | Number of Cases/Controls | Sensitivity (%) | Specificity (%) | Sources | Methods | References |
| APCme | 89 LC/50 AC | 47 | 100 | Serum/Plasma | qMSP | [33] |
| p16INK4ame | 35 NSCLC/15 AC | 34 | 100 | Plasma | F-MSP | [34] |
| MGMTme/p16INK4ame/RASSF1Ame/DAPKme/RARβme | 91 LC/109 BPD | 50 | 85 | Serum | MSP | [47] |
| p16INK4ame/CDH13me | 61 NSCLC/15 BPD | 39 | 100 | Serum | MSP | [48] |
| RASSF1Ame | 80 LC/50 AC a | 34 | 100 | Serum | MSP | [49] |
|
CDH13me/p16INK4ame/FHITme/
RARβme /RASSF1Ame/ZMYND10me |
63 NSCLC/36 BPD | 73 | 83 | Plasma | Two-step MSP | [50] |
| KLK10me | 78 NSCLC/50 AC a | 38 | 96 | Plasma | MSP | [51] |
| SFRP1me | 78 NSCLC/50 AC a | 28 | 96 | Plasma | MSP | [52] |
| DLEC1me | 78 NSCLC/50 AC a | 36 | 96 | Plasma | MSP | [53] |
| Kif1ame/DCCme/RARβ2me/NISCHme | 70 LC/80 BPD | 73 | 71 | Plasma | qMSP | [54] |
|
APCme/RASSF1Ame/CDH13me/
KLK10me/DLEC1me |
110 NSCLC b/50 AC a | 84 | 74 | Plasma | MSP | [55] |
| APCme/CDH1me/MGMTme/DCCme RASSF1Ame/AIM1me | 76 LC/30 AC | 84 | 57 | Serum | qMSP | [56] |
| SHOX2me | 188 LC/155 AC a,c | 60 | 90 | Plasma | qMSP | [37] |
| TMEFF2me | 316 NSCLC/50 AC | 9 | 100 | Serum | Two-step MSP | [57] |
| RARβ2me | 60 NSCLC/32 AC | 72 | 62 | Plasma | qMSP | [35] |
| RASSF1Ame | 66 | 57 | ||||
| SEPT9me | 70 LC/100 AC | 44 | 92 | Plasma | qMSP | [43] |
| p14ARFme | 107 NSCLC/20 BPD | 25 | 95 | Plasma | Two-step MSP | [58] |
| DCLK1me | 65 LC/95 AC | 49 | 92 | Plasma | qMSP | [42] |
| SOX17me | 48 Operable NSCLC/49 AC | 56 | 98 | Plasma | qMSP | [59] |
| 74 Advanced NSCLC/49 AC | 36 | |||||
| SHOX2me | 38 LC/31 BPD | 81 | 79 | Plasma | qMSP | [38] |
| SHOX2me/PTGER4me | 50 LC/122 AC a | 67 90 |
90 73 |
Plasma | Multiplex qMSP | [39] |
| CDO1me/TAC1me/SOX17me | 150 NSCLC b/60 AC | 93 | 62 | Plasma | qMSP | [60] |
|
MARCH11me/HOXA9me/CDO1me/
UNCXme/PTGDRme/AJAP1me |
43 LUAD d/42 AC | 72 | 71 | Plasma | qMSP | [44] |
| 40 LUSC d/42 AC | 60 | |||||
| NID2me | 46 NSCLC/30 BPD | 46 | 80 | Plasma | qMSP | [61] |
| APCme | 73 LC e/103 AC e | 36 | 94 | Plasma | Multiplex qMSP | [36] |
| FOXA1me | 72 | 74 | ||||
| RARβ2me | 25 | 95 | ||||
| RASSF1Ame | 22 | 98 | ||||
| SOX17me | 38 | 95 | ||||
|
CDH13me/WT1me/CDKN2Ame/HOXA9me/
PITX2me/CALCAme/RASSF1Ame/DLEC1me |
39 LC/11 BPD | 72 | 91 | Plasma | qMSP | [62] |
| APCme/RASSF1Ame | 129 LC/28 BDP | 38 | 93 | Plasma | qMSP | [45] |
| FOXA1me/RARβ2me/RASSF1Ame/SOX17me | 102 LC f/136 AC f | 66 | 70 | Plasma | qMSP | [46] |
a Included Benign pulmonary diseases; b Only stage I/II; c Included other cancer types; d Only included stage I; e Only included females; f Only included males; Abbreviations: AC—Asymptomatic Controls; BPD—Benign Pulmonary Diseases; F-MSP—Fluorescent methylation-specific PCR; LC—Lung Cancer; LUAD—Lung Adenocarcinoma; LUSC—Lung Squamous Cell Carcinoma; MSP—Methylation-specific PCR; NSCLC—Non-Small Cell Lung Cancer; qMSP—Quantitative methylation-specific PCR.
After the commercialization in Europe of a test based on SHOX2me assessment in bronchial aspirates, the methylation of this gene was also evaluated as an LC detection biomarker in plasma. Indeed, plasma SHOX2me discriminated LC from control samples with 60% sensitivity and 90% specificity, although higher sensitivity was found in stages II (72%), III (55%) and IV (83%) compared with stage I (27%) patients [37]. In line with these results, another study reported that SHOX2me discriminated LC in subjects undergoing bronchoscopy with 81% sensitivity and 79% specificity [38]. Later on, Weiss et al. reported that SHOX2me and PTGER4me panel distinguished LC patients with 67% sensitivity for a fixed specificity of 90%, and with 73% specificity for a fixed sensitivity of 90% [39]. Remarkably, in the end of 2017, the “Epi proLung®” assay, developed by Epigenomics AG, based on these two genes received the Conformité Européenne (CE) mark for In Vitro Diagnostic (IVD) test. According to their validation study comprising 360 patients from the US and Europe, of which 152 were diagnosed with LC, depending on the Epi proLung® test score threshold chosen, 85% sensitivity was achieved for 50% specificity, whereas sensitivity decreased to 59% if 95% specificity was considered [40,41].
Although the majority of these studies have focused on detection of non-small cell lung carcinoma (NSCLC) (Table 1) (the most diagnosed LC subtype), different detection frequencies of genes’ methylation have been reported among the different subtypes. Indeed, SHOX2me detected with higher sensitivity small cell lung cancer (SCLC) (80%) and lung squamous cell carcinoma (LUSC) (63%) than lung adenocarcinoma (LUAD) (39%) [37]. Similarly, DCLK1me was more frequent in SCLC than NSCLC [42], and our research team recently reported higher APCme and RARβ2me levels in SCLC compared to LUAD, in females [36]. Conversely, SEPT9me was more frequent in NSCLC (53%) than in SCLC (26%) [43]. A serum-based gene panel (MARCH11me, HOXA9me, CDO1me, UNCXme, PTGDRme and AJAP1me) detected stage I LUAD and LUSC with 71% specificity and 72% and 60% sensitivity, respectively [44]. Interestingly, HOXA9me, and RASSF1Ame were able to discriminate SCLC from NSCLC with 64% and 52% sensitivity and 84% and 96% specificity respectively [45], whereas HOXD3me and RASSF1Ame panel reached 75% sensitivity and 88% specificity in male samples [46].
4.1.2. Prognosis, Prediction, and Monitoring
Besides LC histological subtype, TNM prognostic stage groups remain the most important prognostic feature to predict recurrence and survival, followed by tumor histological grade, gender and age [63,64]. Recently, molecular subtypes have also emerged to provide more personalized genetic information, which may improve prognostic estimates and therapy response prediction [63].
Thus far, only a few small-scaled studies reported the prognostic, predictive and monitoring potential of ccfDNA methylation for LC, most of them performed in patients with advanced disease. DCLK1me and SOX17me levels were associated with reduced overall survival (OS) in advanced LC and NSCLC, respectively [42,59]. Likewise, higher SHP1P2me levels in advanced NSCLC associated with reduced progression-free survival (PFS) and OS [65], whereas BRMS1me associated with both reduced disease-free survival (DFS) and OS in operable NSCLC, and reduced PFS and OS in advanced NSCLC [66].
Interestingly, after neoadjuvant chemotherapy and surgery with intraoperative radiation therapy, NSCLC patients showed decreased RASSF1Ame and RARβ2me levels, similar to levels in healthy subjects. Moreover, methylation levels increase in at least one of these genes, up to the levels detected before treatment, was observed in all five patients, which disclosed evidence of disease progression [35]. Increased APCme and/or RASSF1Ame levels within 24h after cisplatin-based chemotherapy also associated with increased OS [67]. Detectable circulating levels of APCme and RASSF1Ame panel at diagnosis was also found to be an independent predictor of increased disease-specific mortality in LC patients, displaying a 3.9-fold risk of dying from LC comparing to those lacking methylation [46].
Remarkably, advanced LC patients that clinically responded to chemo/radiotherapy demonstrated a decrease in SHOX2me plasma levels, observed at 7–10 days after therapy initiation [68]. Additionally, higher SHOX2me levels, both before and 7–10 days after therapy beginning were indicative of shorter OS [68]. Similar results were obtained after two cycles of chemotherapy or TKI-based targeted therapy, being SHOX2me levels before therapy again predictive of OS [69]. Contrarily, 14-3-3σme levels in stage IV NSCLC patients before treatment with cisplatin-gemcitabine associated with longer survival [70]. Stage IV NSCLC patients with unmethylated CHFR depicted longer OS when treated with EGFR-TKI compared to those treated with chemotherapy, as second-line therapy [71].
4.2. Breast Cancer
4.2.1. Screening and Diagnosis
BrC is estimated to be the most incident and deadly cancer in females, worldwide [13]. BrC incidence increased after the implementation of mammography-based screening [72]. Screening mammography endures a sensitivity around 82% and 91% specificity [73]; however, sensitivity decreases with high breast density [74]. Furthermore, although BrC screening features several benefits, it presents some important disadvantages, including overdiagnosis and false-positive results. Indeed, about 11% of cases detected in a population invited to screening would probably not be clinically relevant in the woman’s lifetime, still are treated [75]. Additionally, false-positive results lead to extra imaging exams and eventually biopsy procedures, which can cause discomfort and anxiety to the subjects [75,76]. The diagnosis of BrC comprises clinical examination and biopsy procedures either by image-guided core needle biopsy or fine-needle aspiration, to confirm the diagnosis by histopathological analysis [72]. Since these are invasive procedures, advances in BrC detection are needed, specifically in pre-screening methods, which might select patients to invasive/costly screening tests, avoiding overdiagnosis and unnecessary exams.
Several methylated genes have been proposed as tumor biomarkers for BrC detection [77,78,79,80]. Nevertheless, the sensitivity for cancer detection of one methylated gene in ccfDNA is limited, hence several studies attempted to assemble gene methylation panels to increase the test sensitivity [77,78,79,80]. The currently reported ccfDNA methylation biomarkers for BrC are displayed on Table 2. One of the first studies reported 62% sensitivity and 87% specificity for BrC detection in plasma samples by assessing APCme, GSTP1me, RARβ2me and RASSF1Ame [77]. Nevertheless, a panel including DAPK1me and RASSF1Ame showed the highest sensitivity, i.e., 96% for BrC detection using methylation-specific PCR (MSP) [81]. Other studies attempted to assemble panels for BrC detection using quantitative methylation-specific PCR (qMSP), a quantitative method, achieving sensitivities above 80% [78,82]. Furthermore, several panels reached 100% specificity for BrC detection [83,84,85]. Altogether, APCme, RARβ2me and RASSF1Ame are the most reported genes for BrC detection [77,83,86].
Table 2.
CcfDNA-based methylation biomarkers for breast cancer (BrC) detection.
| Breast Cancer | ||||||
|---|---|---|---|---|---|---|
| Genes | Number of Cases/Controls | Sensitivity (%) | Specificity (%) | Sources | Methods | References |
| APCme/DAPkinaseme/RASSF1Ame | 34 BrC/20 AC + 8 Benign | 94 | 100 | Serum | MSP | [83] |
| ATMme/RASSF1Ame | 50 BrC/14 AC | 36 | 100 | Plasma | qMSP | [84] |
| RARβ2me/RASSF1Ame | 20 BrC/10 AC | 95 | 100 | Plasma a | MSP | [85] |
| APCme/GSPT1me/RARβ2me/RASSF1Ame | 47 BrC/38 AC | 62 | 87 | Plasma | qMSP | [77] |
| 14-3-3-σme/ESR1meb | 106 BrC/74 AC | 81 | 55 | Serum | qMSP | [80] |
| APCme/ESR1me/RASSF1Ame | 79 BrC/19 AC | 53 | 84 | Serum | qMSP | [86] |
| RASSF1Ame | 61 BrC/29 AC | 18 | 100 | Plasma | MSP | [88] |
| DAPK1me/RASSF1Ame | 26 BrC/16 AC 26 BrC/12 Benign |
96 | 92 | Serum | MSP | [81] |
| 57 | ||||||
| APCme/BIN1me/BRCA1me/CST6me/GSTP1me/p16 (CDKN2A)me/p21 (CDKN1A)me/TIMP3me | 36 BrC/30 AC | 91.7 | - | Plasma | Mass Spectrometry | [89] |
| RARβme/RASSF1Ame | 119 BrC/125 AC | 94.1 | 88.8 | Serum | Two-step qMSP | [78] |
| GSTP1me/RARβ2me/RASSF1Ame | 101 BrC c/87 AC | 22 | 93 | Serum | One-step MSP | [90] |
| SOX17me | 114 BrC/49 AC | 38 | 98 | Plasma | qMSP | [91] |
| ITIH5me/DKK3me/RASSF1Ame | 138 BrC/135 AC | 67 | 69 | Serum | qMSP | [79] |
| 138 BrC/39 Benign | 82 | |||||
|
APCme
RARβ2me |
121 BrC/66 AC + 79 Benign | 93.4 | 95.4 | Serum | MSP | [92] |
| 95.6 | 92.4 | |||||
| SFNme/p16me/hMLH1me/HOXD13me/PCDHGB7me/RASSF1Ame | 125 BrC/104 Benign | 82.4 | 78.1 | Serum | qMSP | [87] |
| 125 BrC/104 AC | 79.6 | 72.4 | ||||
| CDH1me/RASSF1Ame | 50 BrC/25 AC | 76 | 90 | Serum | MSP | [93] |
| NBPF1me | 52 BrC c/30 AC | 67.1 d | 59.1 d | Plasma | MSP | [94] |
| APCme | 108 BrC/103 AC | 32.4 | 94.2 | Plasma | Multiplex qMSP | [36] |
| FOXA1me | 38.9 | 79.6 | ||||
| RASSF1Ame | 19.4 | 100 | ||||
| SCGB3A1me | 21.3 | 92.2 | ||||
| APCme/FOXA1me/RASSF1Ame | 44 BrC/39 AC | 81.8 | 76.9 | Plasma | Multiplex qMSP | [82] |
| PER1me/NKX2-6me/SPAG6me | 111 BrC/14 Benign | 58 | 79 | Plasma | Pyrosequencing | [95] |
a Total circulating DNA, including cell-bound circulating DNA; b Considering ER-αme or 14-3-3-σme; c Stages I-III; d For one gene site. Abbreviations: AC—Asymptomatic Controls; BrC—Breast cancer; MSP—Methylation-specific PCR; qMSP—Quantitative methylation-specific PCR.
Interestingly, Shan et al. reported a six-gene panel that detects BrC with a tumor size ≤ 1cm with higher sensitivity (85.82%) than mammography (81.82%) [87]. Even though numerous studies report the feasibility of DNA methylation to detect BrC, its validation and transfer to the clinical setting is still overdue. Nonetheless, DNA methylation shows promise to complement BrC standard screening and diagnosis methods.
4.2.2. Prognosis, Prediction, and Monitoring
In addition to staging, several biomarkers are used to predict BrC patients’ response to therapy and prognosis, including histological grade—a high histological grade is associated with poorly differentiated tumors and worse prognosis [96]. Furthermore, 75% of BrC cases are estrogen receptor (ER)-positive, being less aggressive, and associated with a better prognosis [97]. Accordingly, progesterone receptor (PR)-positive tumors are frequently ER-positive. On the contrary, ER-negative and PR-negative tumors are frequently grade 3, often recur, and do not respond to hormonotherapy [96]. Erb-B2 receptor tyrosine kinase 2 (ERBB2) [or human epidermal growth factor receptor 2 (HER2)] is a growth factor overexpressed in 15% of all BrC cases [97]. Hence, therapies targeting ERBB2 such as trastuzumab are currently available for BrC patients, improving patient outcome [97]. Moreover, gene expression profiles have emerged to assist in adjuvant treatment decision [64] Oncotype DX®, MammaPrint® and PAM50 (Prosigna) are examples of gene expression profiles that allow for BrC classification and prognostic stratification [64]. Nonetheless, their usefulness in clinical practice is limited and therefore their implementation remains restricted [98]. Thus, new and reliable biomarkers to aid in defining a BrC patient prognosis is of major importance.
Chimonidou et al. analyzed CST6me in ccfDNA from plasma samples and, during follow-up time, CST6 was methylated in 13 of the 25 BrC patients that relapsed and in 3 of the 9 patients which died, however not reaching statistical significance [99]. Furthermore, RASSF1Ame and PITX2me were found to be independent biomarkers of poor OS and RASSF1Ame together with lymph node and ER status indicated poor distant DFS in BrC patients’ ccfDNA [100]. Recently, a study analyzing methylation patterns of several genes in ccfDNA showed that BrC patients with positive SOX17me and WNT5Ame displayed shorter OS, whereas KLK10me associated with higher number of relapses and shorter disease-free interval [101]. Apart from being a detection biomarker, SOX17me associated with lymph node metastasis, poor DFS and OS in plasma samples from BrC’s patients [102]. Visvanathan et al. described a cumulative methylation index (CMI) comprising the methylation of 6 genes (AKR1B1me, HOXB4me, RASGRF2me, RASSF1me, HIT1H3Cme, and TM6SF1me) that associates with PFS and OS, i.e., PFS and OS were significantly shorter in metastatic BrC patients with high CMI [103]. Additionally, 14-3-3-σme methylation was significantly associated with the response to metastatic BrC treatment [104]. APCme and RASSF1Ame were also associated with poor outcome, with a relative risk of death of 5.7 [105].
4.3. Colorectal Cancer
4.3.1. Screening and Diagnosis
Due to its slow progression time and the opportunity to easily remove precancerous and early stage cancerous lesions, if caught on time, CRC entail minimal risk to the patient with about 90% long-term survival [106,107]. Therefore, CRC screening programs are of great interest. Currently, analysis of trace blood in stool by fecal occult blood test (FOBT)/fecal immunochemical test (FIT), and internal imaging of the colon by colonoscopy are the main screening options available for CRC early detection [106,108]. Additionally, biopsy samples during colonoscopy are mandatory for histological diagnosis [109]. Nevertheless, fecal screening tests have limited sensitivity to detect precancerous lesions, whereas colonoscopy is very precise and can be used to remove the lesion during the examination, although it is a costly and invasive procedure with low patient compliance [110]. Hence, despite being recognized that screening reduces CRC incidence and mortality, the availability and compliance to the current screening tests remain suboptimal [111].
The increasing knowledge of the influence of epigenetic alterations in malignant transformation in the gut gave rise to an opportunity for development of sensitive and specific minimally invasive epigenetic-based biomarkers for CRC. Hence, plentiful studies have investigated the detection value of these biomarkers (Table 3). SEPT9me is the mostly reported methylated gene in blood from CRC patients. Remarkably, this marker was the first blood-based IVD assay for detection of occult cancer based on an epigenetic alteration, approved by the US Food and Drug Administration (FDA) in 2016, under the designation “Epi ProColon® 2.0” (Epigenomics AG) [40]. Additionally, this CE-IVD marked test is also commercially available in Europe and China [112]. A meta-analysis published in 2017 reported that SEPT9me sensitivity for CRC detection varies between 73–78% depending on the algorithm used to consider a positive result, while specificity varies between 84–96% [113]. Nevertheless, when the biomarker performance of this gene’s methylation was assessed in a multicenter screening setting (PRESEPT clinical trial) with asymptomatic individuals older than 50 years old, the results from 53 CRC cases and 1457 subjects without CRC yielded 48% sensitivity and 92% specificity [114].
Table 3.
CcfDNA-based methylation biomarkers for colorectal cancer (CRC) detection.
| Colorectal Cancer | ||||||
|---|---|---|---|---|---|---|
| Genes | Number of Cases/Controls | Sensitivity (%) | Specificity (%) | Sources | Methods | References |
| p16INK4ame | 52 CRC/44 AC a | 27 | 100 | Serum | MSP | [122] |
| APCme/hMLH1me/HLTFme | 49 CRC/41 AC | 57 | 90 | Serum | qMSP | [123] |
| ALX4me | 30 CRC/30 AC | 83 | 70 | Serum | qMSP | [124] |
| HPP1me | 38 CRC/20 AC | 49 | 100 | Serum | qMSP | [125] |
| HLTFme | 67 | |||||
| hMLH1me | 47 | |||||
| SEPT9me | 133 CRC/179 AC | 69 | 86 | Plasma | qMSP | [126] |
| TMEFF2me | 65 | 69 | ||||
| NGFRme | 51 | 84 | ||||
| RASSF1Ame | 45 CRC/30 AC | 29 | 100 | Serum | MSP | [127] |
| VIMme | 81 CRC/110 AC | 59 | 93 | Plasma | Methyl BEAMing | [128] |
|
APCme/MGMTme/RASSF2Ame/
WIF1me |
243 CRC b/276 AC | 87 | 92 | Plasma | MSP | [118] |
| 64 Adenoma/276 AC | 75 | |||||
| ALX4me/SEPT9me/TMEFF2me | 182 CRC/170 AC | 81 | 90 | Plasma | Multiplex qMSP | [129] |
| NEUROG1me | 97 CRC b/45 AC | 61 | 91 | Serum | qMSP | [130] |
| TFPI2me | 215 CRC/20 AC | 18 | 100 | Serum | qMSP | [131] |
| DLC1me | 85 CRC/45 AC | 42 | 91 | Serum | MSP | [132] |
|
CYCD2me/HIC1me/PAX5me/
RASSF1Ame/RB1me/SRBCme |
30 CRC b/30 AC | 84 | 68 | Plasma | Microarray | [133] |
| HIC1me/MDG1me/RASSF1Ame | 30 Adenoma/30 AC | 55 | 65 | |||
| SMAD4me | 60 CRC/100 AC a | 52 | 64 | Plasma | MSP-SSCP | [134] |
| FHITme | 50 | 84 | ||||
| DAPK1me | 50 | 74 | ||||
| APCme | 57 | 86 | ||||
| CDH1me | 60 | 84 | ||||
| SDC2me | 131 CRC/125 AC | 87 | 95 | Serum | qMSP | [135] |
| TAC1me/SEPT9me | 26 CRC c/26 AC | 73 | 92 | Serum | qMSP | [136] |
| NPYme/PENKme/WIF1me | 32 CRC/161 AC | 87 59 |
80 95 |
Serum | Multiplex qMSP | [137] |
| CAHMme | 73 CRC/74 AC | 55 | 93 | Plasma | qMSP | [138] |
| 73 Adenoma/74 AC | 4 | |||||
| PPP1R3Cme/EFHD1me | 120 CRC/96 AC | 53 (2 genes) | 96 (2 genes) | Plasma | MSP | [139] |
| 90 (at least 1 gene) | 64 (at least 1 gene) | |||||
| SYNE1me/FOXE1me | 66 CRC/140 AC | 58 | 91 | Plasma | Multiplex qMSP | [140] |
| GATA5me/SFRP2me | 57 CRC/47 AC | 43 | 91 | Plasma | MSP | [141] |
| 30 Adenoma/47 AC | 27 | |||||
| BCAT1me/IKZF1me | 74 CRC/144 AC | 77 | 92 | Plasma | qMSP | [142] |
| BCAT1me/IKZF1me | 129 CRC/450 AC | 66 | 95 | Plasma | qMSP | [120] |
| 338 Advanced Adenoma/450 AC | 6 | |||||
| 346 Non-Advanced Adenoma/450 AC | 7 | |||||
| BCAT1me/IKZF1me | 66 CRC/1315 AC a | 62 | 92 | Plasma | qMSP | [121] |
| 170 Advanced Adenoma | 9 | --- | ||||
| 278 Non-Advanced Adenoma | 9 | --- | ||||
| ALX4me | 25 CRC/25 AC | 68 | 88 | Serum | MSP | [143] |
| FGF5me | 20 CRC/40 AC | 85 | 82 | Plasma | qMSP | [144] |
| GRASPme | 44 CRC/44 AC | 55 | 93 | Plasma | qMSP | [145] |
| IRF4me | 22 CRC/24 AC | 59 | 96 | |||
| PDX1me | 20 CRC/20 AC | 45 | 70 | |||
| SDC2me | 44 CRC/44 AC | 59 | 84 | |||
| SEPT9me | 44 CRC/44 AC | 59 | 95 | |||
| SOX21me | 20 CRC/20 AC | 85 | 50 | |||
| SPG20me | 37 CRC/37 AC | 81 | 97 | |||
| SEPT9me | Meta-analysis | 78 (1/3) | 84 (1/3) | Plasma/ Serum |
--- | [113] |
| 73 (2/3) | 96 (2/3) | |||||
| ALX4me/BMP3me/NPTX2me/RARβme/SDC2me/SEPT9me/VIMme | 193 CRC/102 AC | 91 | 73 | Plasma | Two-step qMSP | [146] |
|
SFRP1me/SFRP2me/SDC2me/
PRIMA1me |
47 CRC/37 AC | 92 | 97 | Plasma | qMSP | [119] |
| 37 Adenoma/37 AC | 89 | 87 | ||||
| BMP3me | 45 CRC/50 AC | 40 | 94 | Plasma | BS-HRM | [147] |
| TWIST1me | 18 CRC/25 AC | 44 | 92 | Serum | Multiplex ddMSP | [117] |
| 70 Advanced Adenoma/25 AC | 30 | |||||
| 25 Non-Advanced Adenoma/25 AC | 36 | |||||
| SEPT9me | 98 CRC/253 AC | 61 | 98 | Plasma | qMSP | [117] |
| 101 Adenoma/253 AC | 8 | |||||
| APCme | 72 CRC d/103 AC d | 21 | 94 | Plasma | Multiplex qMSP | [36] |
| FOXA1me | 50 | 88 | ||||
| RARβ2me | 17 | 95 | ||||
| RASSF1Ame | 14 | 99 | ||||
| SCGB3A1me | 26 | 90 | ||||
| SEPT9me | 11 | 100 | ||||
| SOX17me | 24 | 90 | ||||
| SFRP2me | 62 CRC/55 AC | 69 | 87 | Serum | qMSP | [148] |
| SEPT9 me/SDC2me | 117 CRC/166 AC | 88.9 | 92.9 | Plasma | qMSP | [149] |
| C9orf50me/KCNQ5me/CLIP4me | 143 CRC/91 AC | 91 | 99 | Plasma | ddMSP | [150] |
| SEPT9me/SOX17me | 100 CRC e/136 AC e | 12 | 100 | Plasma | Multiplex qMSP | [46] |
a Included patients with adenomatous polyps; b Only included stages I/II; c Only included stage I; d Only included females; e Only included males; Abbreviations: AC – Asymptomatic Controls; Adenoma – Adenomatous polyps; BS-HRM – Bisulfite specific high-resolution melting analysis; CRC – Colorectal Cancer; ddMSP – Digital droplet methylation-specific PCR; MSP – Methylation-specific PCR; MSP-SSCP – Methylation-Specific PCR – single strand conformation polymorphism; qMSP – Quantitative methylation-specific PCR.
Given the importance of detecting pre-malignant conditions, several studies have not only studied the CRC detection performance, but also the ability to detect adenomas. Disappointingly, SEPT9me performance to detect advanced adenomas ranged between 8–31% [115,116,117], being reported to be 11% in the previously mentioned screening setting study [114]. Thus, the usefulness of this gene for population-based screening is questionable.
As expected, gene panels have improved the performance to detect both adenomas and CRC. Remarkably, APCme, MGMTme, RASSF2Ame and WIF1me panel discriminated adenomas and early stage CRC with 75% and 87% sensitivity, respectively, and 92% specificity [118], whereas SFRP1me, SFRP2me, SDC2me, and PRIMA1me panel detected adenomas with 89% sensitivity and 87% specificity, and CRC with 92% sensitivity and 97% specificity [119]. Nevertheless, the performance of these panels in a large screening setting remains to be elucidated. Interestingly, BCAT1me and IKZF1me panel performance has been evaluated in large multicenter studies, displaying 62–66% sensitivity and 92–95% specificity for CRC detection, although with a limited 6–9% sensitivity for adenoma detection [120,121].
4.3.2. Prognosis, Prediction, and Monitoring
TMN prognostic stage groups, in addition to cancer location, patient’s age and comorbidities are the basis for CRC prognostication and patient management [64,109]. Regarding ccfDNA methylation-based biomarkers, higher RUNX3me levels associated with lymphatic invasion, advanced pathological stage and tumor recurrence [151]. TFPI2me and SFRP2me levels associated with poorly differentiated carcinoma, deep invasion, lymph node and distant metastasis [131,152], the former also associating with tumor size [131], and the latter with shorter OS [152]. SSTme was independently associated with higher recurrence risk and shorter DSS in patients who underwent curative surgical resection [153].
A small-scale study suggested that p16INK4ame could reflect the recurrence status during follow-up after surgery [154]. Interestingly, in a prospective cohort study including 150 stage I-III CRC patients from whom serum samples were obtained 1 week before, as well as 6 months and 1 year after surgery, high levels of TAC1me after 6 months and SEPT9me after 1 year were independent predictors of tumor recurrence and shorter DSS [155]. Additionally, the increment of TAC1me and SEPT9me levels independently predicted disease recurrence, whereas NELL1me at both 6 months and 1 year associated with disease-specific survival (DSS) [155]. In the same line, SEPT9me was suggested as follow-up marker for recurrence and metastasis detection, also associating with tumor size, histological grade and histological type [117]. GATA5me, SFRP2me, ITGA4me, SHOX2me and SEPT9me levels were also associated with tumors’ histological grade, TNM stage and lymph node metastasis [141,156], whereas GATA5me also associated with large tumor size [141]. Furthermore, APCme, SEPT9me, SHOX2me and SOX17me levels were also shown to be significantly higher in female patients with metastatic disease [36], whereas higher RARB2me, SEPT9me and SOX17me levels were disclosed in metastatic CRC male patients [46].
Interestingly, various studies have demonstrated the prognostic value of HLTFme and HPP1me levels in CRC patients. Indeed, serum HLTFme, HPP1me and hMLH1me were significantly correlated with tumor size, and the two former genes (HLTFme and HPP1me) further associated with metastatic disease and tumor stage, as well as with worst outcome [125]. Later, Philipp et al., in a study involving 311 serum samples of CRC patients, reported that HLTFme and HPP1me associated with tumor size, stage, grade and metastatic disease, and HPP1me also associated with nodal status. Moreover, in stage IV patients, high levels of both these genes associated with reduced OS [157]. In patients curatively resected for CRC, pre-therapeutic serum HLTFme levels were associated with increased relative risk of disease recurrence [158]. In a clinical trial including 467 metastatic CRC patients treated with a combination therapy containing fluoropyrimidine, oxaliplatin and bevacizumab, patients with detectable plasmatic HPP1me before the start of treatment showed a significantly poorer OS [159]. Moreover, patients in which it reduced to undetectable levels 2–3 weeks after treatment, showed a better OS compared to patients that maintained detectable plasma HPP1me levels [159], suggesting the usefulness of HPP1me as a prognostic and early response biomarker. Recently, Barault et al. also suggested that plasmatic methylation changes of EYA4me, GRIA4me, ITGA4me, MAP3K14-AS1me and MSCme panel over time correlate with tumor response in metastatic CRC patients treated with chemo- or targeted therapy [160]. Interestingly, using the same panel, Amatu et al. reported that circulating methylated DNA normalized to the total amount of circulating DNA dynamics can reflect clinical response to treatment with regorafenib [161].
Since 2017, COLVERA™, a Laboratory Developed Test (LDT), based on a two-gene panel (IKZF1me and BCAT1me) is commercialized in the USA, for post-surgery residual disease detection and CRC patient surveillance [162]. The detection of these two genes in blood associated with stage [120] and showed a rapid reversion after surgical resection (35 of 47 positive patients at diagnosis, became negative after surgery) [163]. Moreover, in patients undergoing surveillance after primary CRC treatment, this panel was positive in 68% plasma samples of the 28 patients with clinically detectable recurrent CRC, whereas CEA was positive in only 32%, although specificity was similar in both tests (87% and 94%, respectively) [164]. Hence, this panel doubled the sensitivity of the current gold-standard marker for CRC monitoring. Recently, positivity of this panel after surgery also independently associated with increased risk of recurrence [165].
4.4. Prostate Cancer
4.4.1. Screening and Diagnosis
Curable PCa is mostly asymptomatic, and, thus, patients with clinically detected disease are mostly at advanced stage, resulting in worse outcome and limited treatment options [166]. Digital rectal examination (DRE), in combination with serum prostate-specific antigen (PSA) quantification, remains the gold standard PCa screening tools [167], however both methods present drawbacks. A DRE positive result is dependent on clinicians’ expertise and the majority of cancers detected by this method are at advanced stage [168], besides compliance is rather low. The widespread adoption of PSA screening since the late 1980s has facilitated the shift to detection of PCa at early stages [168]. Nonetheless, despite being highly sensitive, since benign prostatic hyperplasia (BPH) and other benign conditions also cause PSA elevation, the lack of cancer-specificity of this approach entails a high false-positive rate and overdiagnosis of non-life threatening PCa [167,169]. Indeed, only less than one third of the patients undergoing transrectal ultrasound-guided (TRUS) biopsy (standard diagnostic approach) due to elevated PSA levels or abnormal DRE are diagnosed with cancer [170]. In parallel, a negative result does not completely rule out the existence of cancer, leading to a large number of unnecessary invasive tissue biopsies that might be repeated due to the uncertainty of diagnosis if elevated PSA levels persist [170]. Thus, the introduction of more specific alternatives is urgently sought.
Besides blood-based liquid biopsies, aberrant DNA methylation in urological cancers can also be detected in urine, which is a non-invasive, easily accessible source of exfoliated cells and ccfDNA from diverse sites of the urinary system [171]. Hence, several studies have been carried out using this source [171]. Nonetheless, in PCa, higher sensitivities are achieved after manipulation of the prostate, either by prostate massage or DRE, increasing the invasiveness of this procedure [171]. Therefore, blood-based liquid biopsies might represent the better minimally invasive procedure for PCa detection. A summary of the currently reported ccfDNA methylation PCa detection biomarkers is depicted in Table 4.
Table 4.
CcfDNA-based methylation biomarkers for prostate cancer (PCa) detection.
| Prostate Cancer | ||||||
|---|---|---|---|---|---|---|
| Genes | Number of Cases/Controls | Sensitivity (%) | Specificity (%) | Sources | Methods | References |
| GSTP1me/PTGS2me/RPRMme/TIG1me | 168 PCa/42 BPH | 47 | 93 | Serum | qMSP | [172] |
| MDR1me | 192 PCa/35 AC a | 32 | 100 | Serum | qMSP | [181] |
| GSTP1me/RASSF1Ame/RARβ2me | 83 PCa/40 AC | 29 | 100 | Serum | MSP | [173] |
| GSTP1me | 80 PCa/51 AC a | 26 | 80 | Plasma | qMSP | [175] |
| RASSF2Ame | 28 | |||||
| HIST1H4Kme | 17 | |||||
| TFAP2Eme | 12 | |||||
| GSTP1me | Meta-analysis | 40 | 90 | Plasma/Serum | Non-qMSP | [174] |
| 36 | 96 | qMSP | ||||
| RARβ2m | 91 PCa/94 BPH | 93 | 89 | Serum | qMSP | [182] |
| GSTP1me | 31 PCa/44 BPH | 93 | 89 | Plasma | MSP | [176] |
| CDH13me | 98 PCa/47 AC b | 45 | 100 | Serum | MSP | [183] |
| GADD45ame | 34 PCa/48 BPH | 38 | 98 | Serum | Pyrosequencing | [180] |
| MCAMme/ERαme/ERβme | 84 PCa/30 AC | 75 | 70 | Serum | qMSP | [177] |
| CCDC181me/ST6GALNAC3me/HAPLN3me | 27 PCa/10 BPH | 67 | 100 | Serum | ddMSP | [178] |
| ZNF660me | 22 | |||||
| FOXA1me/RARβ2m/RASSF1Ame/GSTP1me | 121 PCa/136 AC | 72 | 72 | Plasma | Multiplex qMSP | [46] |
a Biopsy negative; b Included BPH; Abbreviations: AC—Asymptomatic Control; BPH—Benign Prostatic Hyperplasia; ddMSP—Digital droplet methylation-specific PCR; MSP—Methylation-specific PCR; PCa—Prostate Cancer; qMSP—Quantitative methylation-specific PCR.
GSTP1me is the most frequently described epigenetic alteration in ccfDNA of PCa patients due to its remarkably high specificity for PCa [172,173,174,175,176]. Indeed, a meta-analysis evaluating GSTP1me PCa detection performance in plasma/serum reported a pooled specificity of 90% (non-qMSP) and 96% (qMSP-based detection), although with a modest sensitivity of 40% (non-qMSP) and 36% (qMSP-based detection) [174].
Since epigenetic alterations are usually multiple and not necessarily overlapped, multigene panels are pivotal to increase the modest sensitivities of individual genes. Indeed, Ellinger et al. reported that using a gene panel comprising GSTP1me, PTGS2me, RPRMme and TIG1me, PCa diagnostic coverage increased from 42% (GSPT1me alone) to 47% (panel), maintaining 93% specificity [172]. Furthermore, Sunami et al. reported that GSTP1me, RASSF1Ame and RARβ2me were hypermethylated in 13%, 24% and 12% of serum samples from PCa patients, respectively, whereas the three gene panel increased the detection rate to 29%, with 100% specificity [173]. Remarkably, the addition of FOXA1me to the previous panel increased sensitivity to 72% although at the expense of lower specificity (72%) [46]. More recently, other panels without comprising GSTP1me have also been tested. Indeed, MCAMme, ERαme and ERβme panel disclosed 75% sensitivity and 70% specificity for early PCa detection [177]. Likewise, ZNF660me, CCDC181me, ST6GALNAC3me and HAPLN3me in serum displayed 22%, 26%, 31% and 44% sensitivity, respectively, and 100% specificity for PCa. Remarkably, the best gene panel (ST6GALNAC3me, CCDC181me and HAPLN3me) increased sensitivity to 67%, keeping 100% specificity [178].
Interestingly, given the modest sensitivities obtained with ccfDNA methylation even with gene panels, studies have also attempted to understand whether ccfDNA might have a better performance by complementing it with serum PSA levels. Indeed, in a Mexican cohort with biopsy-confirmed PCa, a panel comprising GSTP1me and RASSF1Ame allowed for cancer detection with 73% positive predictive value (PPV) and 59.6% negative predictive value (NPV), increasing to 81% and 66%, respectively, when serum PSA was also considered [179]. Likewise, serum GADD45ame increased sensitivity from 38% to 94% when PSA and free circulating DNA levels were also considered, although specificity decreased from 98% to 88% [180].
4.4.2. Prognosis, Predicition, and Monitoring
PCa is a very heterogeneous disease, ranging from small, low grade, clinically indolent tumors to large, lethally aggressive ones [184]. Thus, the main goal after establishing the presence of cancer is to evaluate its extension and aggressiveness through grading and staging, to assess prognosis and plan the treatment strategy. Currently, PCa prognostic stage groups are based on nomograms combining TNM classification, preoperative serum PSA levels, and histological International Society of Urological Pathology (ISUP) Grade Group [185,186], which is based on the evaluation of the two most common differentiation patterns in a tumor [187]. Furthermore, it is estimated that 30-50% patients treated with curative intent may show rising serum PSA levels (biochemical recurrence) within 10 years after treatment, with clinical disease progression occurring in up to 40% of these [188,189]. Thus, considering these uncertainties, there is an urgent need for development and implementation of more accurate PCa biomarkers to assist clinicians and patients in decision-making.
Besides the study of its detection value, the potential of methylation-based biomarkers in ccfDNA to predict disease progression and therapy response have also been tackled. GSTP1me [181,190], MDR1me, EDNRBme and RARβ2me [181] were reported to be more frequent in castration-resistant prostate cancer (CRPC) patients than in early-stage PCa patients. GSTP1me levels also associated with Gleason score and presence of metastasis [190] as well as with reduced DSS along with APCme [191] in CRPC patients. Furthermore, GSTP1me and RASSF2Ame were more frequently detected in men with non-organ confined compared to organ-confined disease and both associated with increased Gleason score [175]. Interestingly, preoperative serum GSTP1me was also reported as an independent predictor of biochemical recurrence following radical prostatectomy [192]. Sunami et al. reported that GSTP1me, RASSF1Ame and RARβ2me associated with Gleason score and serum PSA levels, whereas GSTP1me and RARβ2me also associated with advanced stages of disease [173]. Moreover, individually, serum PCDH17me, PCDC10me and PCDH8me were also associated with advanced clinical stage, higher preoperative serum PSA, as well as lymph node metastasis and shorter biochemical recurrence-free survival [193,194,195]. Besides being also associated with Gleason score, advanced tumor stage and high PSA, CDH13me was further associated with shorter OS [183].
Remarkably, a recent phase III multicenter trial enrolling 600 CRPC patients showed that detectable serum GSTP1me levels prior and after two cycles of chemotherapy were both independently associated with decreased OS [196]. In the same study, undetectable serum GSTP1me after two cycles of docetaxel further associated with longer time to PSA progression [196].
5. Cell-Free DNA Methylation as a Candidate “PanCancer” Screening Biomarker
The screening of these four cancer types faces different challenges. On the one hand, LC and CRC demand an early cancer detection that is still not fully accomplished with the current screening tools available. On the other hand, BrC and PCa diagnosis should not only be focused on early disease detection, but also be restrictive to clinically significant disease detection. Hence, new screening biomarkers have been exhaustively searched (Table 1, Table 2, Table 3 and Table 4). One may wonder if a minimally invasive “PanCancer” detection approach based on liquid biopsies with good sensitivity and specificity for simultaneous cancer detection would increase screening effectiveness. In fact, a study performed in the context of CRC screening revealed that only 37% of the 172 subjects were compliant with screening colonoscopy [197]. Interestingly, 97% of the subjects who refused this modality accepted a non-invasive alternative, of which 83% selected SEPT9me blood-test and only 15% the stool test, being the primary reason for this choice the convenience of the procedure [197]. Regardless of being small scaled, this study clearly demonstrates that screening compliance can dramatically increase if a convenient minimally invasive option, such as a liquid biopsy, is offered. Nevertheless, the establishment of a “PanCancer” panel for a simultaneous detection of several cancer types is a challenge given their heterogeneity.
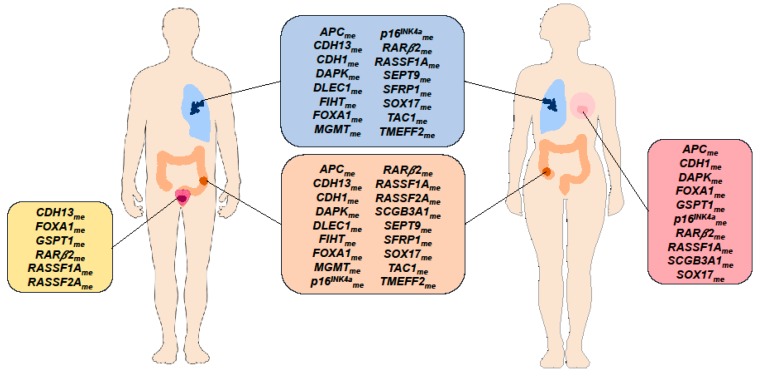
Regarding ccfDNA methylation-based biomarkers, although only RARβ2me, and RASSF1Ame levels were reported in all four cancer types (LC, BrC, CRC, and PCa), several other genes have already been reported in at least two of the four different cancer types (Figure 5).
Figure 5.
Circulating cell-free DNA methylation-based biomarkers described in the literature for cancer detection common to at least two cancer types [Breast Cancer (pink box), Lung Cancer (blue box), Prostate Cancer (yellow box), Colorectal Cancer (orange box)].
Interestingly, a study published in 2007 that included 70 serum samples from metastatic BrC, NSCLC, gastric, pancreatic, colorectal and hepatocellular carcinoma and 10 healthy serum controls demonstrated that a gene panel hypermethylation (RUNX3me, p16me, RASSF1Ame and CDH1me) detected cancer samples with 89% sensitivity and 100% specificity, using MSP [198], suggesting the putative value of using a single panel to detect several malignancies.
Remarkably, genome-wide DNA methylation studies have also been performed aiming to detect the presence of cancer and underlying cancer type [199,200,201]. Indeed, Li et al. and Kang et al. developed “CancerDetector” and “CancerLocator” that may detect cancer using probabilistic approaches based on ccfDNA methylation sequencing [200,201]. Moreover, Moss et al. using Illumina methylation arrays demonstrated that plasma methylation patterns can be used to identify cell type-specific ccfDNA in healthy and pathological conditions, including different types of cancer [202].
Interestingly, in 2018, the Laboratory for Advanced Medicine launched the LTD IvyGene® Test in the USA for detection of BrC, CRC, liver cancer and LC [203]. This test utilizes a multi-target approach derived from sequencing methods to detect ccfDNA methylation profile in 40mL of whole blood samples [203,204]. Notwithstanding, according to their website, the test detected cancer with 84% sensitivity and 90% specificity, it was only validated in 197 samples obtained from subjects with either no history of cancer or diagnosis of one of the four cancers [203]. Thus, the performance evaluation of IvyGene® Test in a large screening setting is still warranted.
Although epigenome-wide approaches allow for simultaneous screen of several hundreds of genes and might be advantageous to boost the discovery of new differentially methylated DNA regions, they are not still widely available in clinical laboratories and require high-level bioinformatics expertise, fast data processing and large data storage capabilities [205,206]. Additionally, depending on the number of samples and genes to be analyzed, targeted approaches, such as qMSP or droplet digital MSP (ddMSP) might be more cost-effective [207]. Indeed, two recent studies aimed to develop a “PanCancer” panel to detect the most incident malignancies, evaluated methylation levels of several genes in plasma samples using multiplex qMSP. Remarkably, APCme, FOXA1me and RASSF1Ame panel was able to detect BrC, CRC and LC in female patients with 72% sensitivity and 74% specificity [36], whereas FOXA1me and RARβ2me panel detected LC and PCa in males with 64% sensitivity and 70% sensitivity [46]. Although both studies included a “CancerType” panel to indicate the most likely cancer topography, reaching specificities above 80%, sensitivity to this end was rather limited [36,46].
Despite the significant advancements, standardization of technical procedures, such as amount of blood collected, type of samples to be used (plasma vs. serum), and DNA extraction and methylation analysis are still necessary to allow reproducibility between different studies and boost the translation of these markers into clinical practice. Nevertheless, although validation in large independent prospective cohorts by clinical trials in asymptomatic individuals to access the real clinical value of a screening method to detect several cancer types is still required, the data suggests that the hypothetical use of a “PanCancer” panel based on ccfDNA methylation is amenable to increase patient compliance to screening programs and decrease patient morbidity and mortality as well as healthcare systems costs.
6. Conclusions
In conclusion, these data indicate that LC, BrC, CRC, and PCa detection and patients’ stratification according to prognosis can be achieved by analyzing gene methylation levels in ccfDNA extracted from plasma or serum, constituting a minimally invasive approach. Methylation levels’ analysis in ccfDNA may not only complement current screening methods, but also aid in stratifying cancer patients according to recurrence risk and response to therapy. Nevertheless, these findings still lack validation in larger multicenter studies to enable their implementation in clinical practice.
Author Contributions
V.C. and S.P.N. reviewed the literature and drafted the manuscript. R.H. and C.J. supervised and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Research Center-Portuguese Oncology Institute of Porto (grant PI 74-CI-IPOP-19-2015). S.P.N. was supported by a fellowship from Liga Portuguesa Contra o Cancro/Pfizer and V.C. was supported by a fellowship from Liga Portuguesa Contra o Cancro/Fundação PT.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy M.J. Tumor markers in clinical practice: A review focusing on common solid cancers. Med. Princ. Pr. 2013;22:4–11. doi: 10.1159/000338393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa-Pinheiro P., Montezuma D., Henrique R., Jeronimo C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics. 2015;7:1003–1015. doi: 10.2217/epi.15.56. [DOI] [PubMed] [Google Scholar]
- 4.Marrugo-Ramirez J., Mir M., Samitier J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018;19:2877. doi: 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng F., Su L., Qian C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832–48841. doi: 10.18632/oncotarget.9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constancio V., Barros-Silva D., Jeronimo C., Henrique R. Known epigenetic biomarkers for prostate cancer detection and management: Exploring the potential of blood-based liquid biopsies. Expert Rev. Mol. Diagn. 2019 doi: 10.1080/14737159.2019.1604224. [DOI] [PubMed] [Google Scholar]
- 7.Poulet G., Massias J., Taly V. Liquid Biopsy: General Concepts. Acta. Cytol. 2019 doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 8.Han X., Wang J., Sun Y. Circulating Tumor DNA as Biomarkers for Cancer Detection. Genom. Proteom. Bioinf. 2017;15:59–72. doi: 10.1016/j.gpb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann M.H.D., Bender S., Krahn T., Schlange T. ctDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput Struct Biotechnol. J. 2018;16:190–195. doi: 10.1016/j.csbj.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Meo A., Bartlett J., Cheng Y., Pasic M.D., Yousef G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie K., Jia Y., Zhang X. Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol. 2015;36:7–19. doi: 10.1007/s13277-014-2758-3. [DOI] [PubMed] [Google Scholar]
- 12.Chan K.C., Jiang P., Chan C.W., Sun K., Wong J., Hui E.P., Chan S.L., Chan W.C., Hui D.S., Ng S.S., et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA. 2013;110:18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 14.Mandel P., Metais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc. Biol. Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 15.Elazezy M., Joosse S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol. J. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 17.Berdasco M., Esteller M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 18.Warton K., Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front. Mol. Biosci. 2015;2:13. doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeronimo C., Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer Lett. 2014;342:264–274. doi: 10.1016/j.canlet.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 20.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 22.Jeronimo C., Bastian P.J., Bjartell A., Carbone G.M., Catto J.W., Clark S.J., Henrique R., Nelson W.G., Shariat S.F. Epigenetics in prostate cancer: Biologic and clinical relevance. Eur. Urol. 2011;60:753–766. doi: 10.1016/j.eururo.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 23.Kulis M., Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg A.D., Allis C.D., Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Sweet T.J., Ting A.H. WOMEN IN CANCER THEMATIC REVIEW: Diverse functions of DNA methylation: Implications for prostate cancer and beyond. Endocr Relat. Cancer. 2016;23:T169–T178. doi: 10.1530/ERC-16-0306. [DOI] [PubMed] [Google Scholar]
- 26.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol. 2016:8. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zane L., Sharma V., Misteli T. Common features of chromatin in aging and cancer: Cause or coincidence? Trends Cell Biol. 2014;24:686–694. doi: 10.1016/j.tcb.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prosch H., Schaefer-Prokop C. Screening for lung cancer. Curr. Opin. Oncol. 2014;26:131–137. doi: 10.1097/CCO.0000000000000055. [DOI] [PubMed] [Google Scholar]
- 29.Wu G.X., Raz D.J. Lung Cancer Screening. Cancer Treat. Res. 2016;170:1–23. doi: 10.1007/978-3-319-40389-2_1. [DOI] [PubMed] [Google Scholar]
- 30.Oudkerk M., Devaraj A., Vliegenthart R., Henzler T., Prosch H., Heussel C.P., Bastarrika G., Sverzellati N., Mascalchi M., Delorme S., et al. European position statement on lung cancer screening. Lancet Oncol. 2017;18:e754–e766. doi: 10.1016/S1470-2045(17)30861-6. [DOI] [PubMed] [Google Scholar]
- 31.National Lung Screening Trial Research T., Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osarogiagbon R.U., Veronesi G., Fang W., Ekman S., Suda K., Aerts J.G., Donington J. Early-Stage NSCLC: Advances in Thoracic Oncology 2018. J. Thorac. Oncol. 2019;14:968–978. doi: 10.1016/j.jtho.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usadel H., Brabender J., Danenberg K.D., Jeronimo C., Harden S., Engles J., Danenberg P.V., Yang S., Sidransky D. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–375. [PubMed] [Google Scholar]
- 34.Bearzatto A., Conte D., Frattini M., Zaffaroni N., Andriani F., Balestra D., Tavecchio L., Daidone M.G., Sozzi G. p16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin. Cancer Res. 2002;8:3782–3787. [PubMed] [Google Scholar]
- 35.Ponomaryova A.A., Rykova E.Y., Cherdyntseva N.V., Skvortsova T.E., Dobrodeev A.Y., Zav’yalov A.A., Bryzgalov L.O., Tuzikov S.A., Vlassov V.V., Laktionov P.P. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81:397–403. doi: 10.1016/j.lungcan.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Nunes S.P., Moreira-Barbosa C., Salta S., Palma de Sousa S., Pousa I., Oliveira J., Soares M., Rego L., Dias T., Rodrigues J., et al. Cell-Free DNA Methylation of Selected Genes Allows for Early Detection of the Major Cancers in Women. Cancers. 2018;10:357. doi: 10.3390/cancers10100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kneip C., Schmidt B., Seegebarth A., Weickmann S., Fleischhacker M., Liebenberg V., Field J.K., Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J. Thorac. Oncol. 2011;6:1632–1638. doi: 10.1097/JTO.0b013e318220ef9a. [DOI] [PubMed] [Google Scholar]
- 38.Konecny M., Markus J., Waczulikova I., Dolesova L., Kozlova R., Repiska V., Novosadova H., Majer I. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma. 2016;63:246–253. doi: 10.4149/210_150419N208. [DOI] [PubMed] [Google Scholar]
- 39.Weiss G., Schlegel A., Kottwitz D., Konig T., Tetzner R. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 2017;12:77–84. doi: 10.1016/j.jtho.2016.08.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beltran-Garcia J., Osca-Verdegal R., Mena-Molla S., Garcia-Gimenez J.L. Epigenetic IVD Tests for Personalized Precision Medicine in Cancer. Front. Genet. 2019;10:621. doi: 10.3389/fgene.2019.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.AG E. Epi proLung®–Liquid Biopsy Test for Lung Cancer Detection. [(accessed on 10 July 2019)]; Available online: https://www.epigenomics.com/products/epi-prolung/
- 42.Powrozek T., Krawczyk P., Nicos M., Kuznar-Kaminska B., Batura-Gabryel H., Milanowski J. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin. Trans. Oncol. 2016;18:398–404. doi: 10.1007/s12094-015-1382-z. [DOI] [PubMed] [Google Scholar]
- 43.Powrozek T., Krawczyk P., Kucharczyk T., Milanowski J. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: Preliminary report. Med. Oncol. 2014;31:917. doi: 10.1007/s12032-014-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ooki A., Maleki Z., Tsay J.J., Goparaju C., Brait M., Turaga N., Nam H.S., Rom W.N., Pass H.I., Sidransky D., et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non-Small Cell Lung Cancer and Serum DNA. Clin. Cancer Res. 2017;23:7141–7152. doi: 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 45.Nunes S.P., Diniz F., Moreira-Barbosa C., Constâncio V., Silva A.V., Oliveira J., Soares M., Paulino S., Cunha A.L., Rodrigues J., et al. Subtyping Lung Cancer Using DNA Methylation in Liquid Biopsies. J. Clin. Med. 2019;8:1500. doi: 10.3390/jcm8091500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Constâncio V., Nunes S.P., Moreira-Barbosa C., Freitas R., Oliveira J., Pousa I., Oliveira J., Soares M., Dias C.G., Dias T., et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin. Epigenetics. 2019;11:175. doi: 10.1186/s13148-019-0779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara K., Fujimoto N., Tabata M., Nishii K., Matsuo K., Hotta K., Kozuki T., Aoe M., Kiura K., Ueoka H., et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin. Cancer Res. 2005;11:1219–1225. [PubMed] [Google Scholar]
- 48.Ulivi P., Zoli W., Calistri D., Fabbri F., Tesei A., Rosetti M., Mengozzi M., Amadori D. p16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J. Cell Physiol. 2006;206:611–615. doi: 10.1002/jcp.20503. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Yu Z., Wang T., Zhang J., Hong L., Chen L. Identification of epigenetic aberrant promoter methylation of RASSF1A in serum DNA and its clinicopathological significance in lung cancer. Lung Cancer. 2007;56:289–294. doi: 10.1016/j.lungcan.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Hsu H.S., Chen T.P., Hung C.H., Wen C.K., Lin R.K., Lee H.C., Wang Y.C. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110:2019–2026. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Song H., Miao Y., Wang R., Chen L. Frequent transcriptional inactivation of Kallikrein 10 gene by CpG island hypermethylation in non-small cell lung cancer. Cancer Sci. 2010;101:934–940. doi: 10.1111/j.1349-7006.2009.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y.W., Miao Y.F., Yi J., Geng J., Wang R., Chen L.B. Transcriptional inactivation of secreted frizzled-related protein 1 by promoter hypermethylation as a potential biomarker for non-small cell lung cancer. Neoplasma. 2010;57:228–233. doi: 10.4149/neo_2010_03_228. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Miao Y., Yi J., Wang R., Chen L. Frequent epigenetic inactivation of deleted in lung and esophageal cancer 1 gene by promoter methylation in non-small-cell lung cancer. Clin. Lung Cancer. 2010;11:264–270. doi: 10.3816/CLC.2010.n.034. [DOI] [PubMed] [Google Scholar]
- 54.Ostrow K.L., Hoque M.O., Loyo M., Brait M., Greenberg A., Siegfried J.M., Grandis J.R., Gaither Davis A., Bigbee W.L., Rom W., et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin. Cancer Res. 2010;16:3463–3472. doi: 10.1158/1078-0432.CCR-09-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Wang R., Song H., Huang G., Yi J., Zheng Y., Wang J., Chen L. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–28. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Begum S., Brait M., Dasgupta S., Ostrow K.L., Zahurak M., Carvalho A.L., Califano J.A., Goodman S.N., Westra W.H., Hoque M.O., et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin. Cancer Res. 2011;17:4494–4503. doi: 10.1158/1078-0432.CCR-10-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S.M., Park J.Y., Kim D.S. Methylation of TMEFF2 gene in tissue and serum DNA from patients with non-small cell lung cancer. Mol. Cells. 2012;34:171–176. doi: 10.1007/s10059-012-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L., Shen Y., Wang M., Tang D., Luo Y., Jiao W., Wang Z., Yang R., Tian K. Identification of the methylation of p14ARF promoter as a novel non-invasive biomarker for early detection of lung cancer. Clin. Transl. Oncol. 2014;16:581–589. doi: 10.1007/s12094-013-1122-1. [DOI] [PubMed] [Google Scholar]
- 59.Balgkouranidou I., Chimonidou M., Milaki G., Tsaroucha E., Kakolyris S., Georgoulias V., Lianidou E. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin. Chem Lab. Med. 2016;54:1385–1393. doi: 10.1515/cclm-2015-0776. [DOI] [PubMed] [Google Scholar]
- 60.Hulbert A., Jusue-Torres I., Stark A., Chen C., Rodgers K., Lee B., Griffin C., Yang A., Huang P., Wrangle J., et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin. Cancer Res. 2017;23:1998–2005. doi: 10.1158/1078-0432.CCR-16-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng X., Xie X., Zheng B., Peng C., Zhou H., Qin J. The more potential performance of nidogen 2 methylation by tissue or plasma DNA over brichoalveolar lavage DNA in diagnosis of nonsmall cell lung cancer. J. Cancer Res. 2018;14:S341–S346. doi: 10.4103/0973-1482.235352. [DOI] [PubMed] [Google Scholar]
- 62.Yang Z., Qi W., Sun L., Zhou H., Zhou B., Hu Y. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv. Clin. Exp. Med. 2019;28:355–360. doi: 10.17219/acem/84935. [DOI] [PubMed] [Google Scholar]
- 63.Woodard G.A., Jones K.D., Jablons D.M. Lung Cancer Staging and Prognosis. Cancer Treat. Res. 2016;170:47–75. doi: 10.1007/978-3-319-40389-2_3. [DOI] [PubMed] [Google Scholar]
- 64.Amin M.B., Edge S., Greene F., Byrd D.R., Brookland R.K., Washington M.K., Gershenwald J.E., Compton C.C., Hess K.R., Sullivan D.C., et al. AJCC Cancer Staging Manual. 8th ed. Springer International Publishing; Chicago, IL, USA: American Joint Commission on Cancer; Chicago, IL, USA: 2017. pp. 276–726. [Google Scholar]
- 65.Vinayanuwattikun C., Sriuranpong V., Tanasanvimon S., Chantranuwat P., Mutirangura A. Epithelial-specific methylation marker: A potential plasma biomarker in advanced non-small cell lung cancer. J. Thorac. Oncol. 2011;6:1818–1825. doi: 10.1097/JTO.0b013e318226b46f. [DOI] [PubMed] [Google Scholar]
- 66.Balgkouranidou I., Chimonidou M., Milaki G., Tsarouxa E.G., Kakolyris S., Welch D.R., Georgoulias V., Lianidou E.S. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br. J. Cancer. 2014;110:2054–2062. doi: 10.1038/bjc.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H., Zhang B., Chen D., Xia W., Zhang J., Wang F., Xu J., Zhang Y., Zhang M., Zhang L., et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin. Epigenetics. 2015;7:119. doi: 10.1186/s13148-015-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt B., Beyer J., Dietrich D., Bork I., Liebenberg V., Fleischhacker M. Quantification of cell-free mSHOX2 Plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS ONE. 2015;10:e0118195. doi: 10.1371/journal.pone.0118195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng X., Liu X., Xu L., Li Y., Wang H., Song L., Xiao W. The mSHOX2 is capable of assessing the therapeutic effect and predicting the prognosis of stage IV lung cancer. J. Thorac. Dis. 2019;11:2458–2469. doi: 10.21037/jtd.2019.05.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramirez J.L., Rosell R., Taron M., Sanchez-Ronco M., Alberola V., de Las Penas R., Sanchez J.M., Moran T., Camps C., Massuti B., et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J. Clin. Oncol. 2005;23:9105–9112. doi: 10.1200/JCO.2005.02.2905. [DOI] [PubMed] [Google Scholar]
- 71.Salazar F., Molina M.A., Sanchez-Ronco M., Moran T., Ramirez J.L., Sanchez J.M., Stahel R., Garrido P., Cobo M., Isla D., et al. First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer. 2011;72:84–91. doi: 10.1016/j.lungcan.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 72.Senkus E., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rutgers E., Zackrisson S., Cardoso F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26 doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 73.Kemp Jacobsen K., O’Meara E.S., Key D., Buist D.S.M., Kerlikowske K., Vejborg I., Sprague B.L., Lynge E., von Euler-Chelpin M. Comparing sensitivity and specificity of screening mammography in the United States and Denmark. Int. J. Cancer. 2015;137:2198–2207. doi: 10.1002/ijc.29593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holland R., Mravunac M., Hendriks J.H., Bekker B.V. So-called interval cancers of the breast. Pathologic and radiologic analysis of sixty-four cases. Cancer. 1982;49:2527–2533. doi: 10.1002/1097-0142(19820615)49:12<2527::AID-CNCR2820491220>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 75.The benefits and harms of breast cancer screening: An independent review. Lancet. 2012;380:1778–1786. doi: 10.1016/S0140-6736(12)61611-0. [DOI] [PubMed] [Google Scholar]
- 76.Shapiro S., Strax P., Venet L. Periodic breast cancer screening in reducing mortality from breast cancer. Jama. 1971;215:1777–1785. doi: 10.1001/jama.1971.03180240027005. [DOI] [PubMed] [Google Scholar]
- 77.Hoque M.O., Feng Q., Toure P., Dem A., Critchlow C.W., Hawes S.E., Wood T., Jeronimo C., Rosenbaum E., Stern J., et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J. Clin. Oncol. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 78.Kim J.H., Shin M.H., Kweon S.S., Park M.H., Yoon J.H., Lee J.S., Choi C., Fackler M.J., Sukumar S. Evaluation of promoter hypermethylation detection in serum as a diagnostic tool for breast carcinoma in Korean women. Gynecol. Oncol. 2010;118:176–181. doi: 10.1016/j.ygyno.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 79.Kloten V., Becker B., Winner K., Schrauder M.G., Fasching P.A., Anzeneder T., Veeck J., Hartmann A., Knüchel R., Dahl E. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15:R4. doi: 10.1186/bcr3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martinez-Galan J., Torres B., Del Moral R., Munoz-Gamez J.A., Martin-Oliva D., Villalobos M., Nunez M.I., Luna Jde D., Oliver F.J., Ruiz de Almodovar J.M. Quantitative detection of methylated ESR1 and 14-3-3-sigma gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol. 2008;7:958–965. doi: 10.4161/cbt.7.6.5966. [DOI] [PubMed] [Google Scholar]
- 81.Ahmed I.A., Pusch C.M., Hamed T., Rashad H., Idris A., El-Fadle A.A., Blin N. Epigenetic alterations by methylation of RASSF1A and DAPK1 promoter sequences in mammary carcinoma detected in extracellular tumor DNA. Cancer Genet. Cytogenet. 2010;199:96–100. doi: 10.1016/j.cancergencyto.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Salta S., Nunes P., Fontes-Sousa M., Lopes P., Freitas M., Caldas M., Antunes L., Castro F., Antunes P., Palma de Sousa S., et al. A DNA Methylation-Based Test for Breast Cancer Detection in Circulating Cell-Free DNA. J. Clin. Med. 2018;7:420. doi: 10.3390/jcm7110420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dulaimi E., Hillinck J., Ibanez de Caceres I., Al-Saleem T., Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin. Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 84.Papadopoulou E., Davilas E., Sotiriou V., Georgakopoulos E., Georgakopoulou S., Koliopanos A., Aggelakis F., Dardoufas K., Agnanti N.J., Karydas I., et al. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Ann. N. Y. Acad. Sci. 2006;1075:235–243. doi: 10.1196/annals.1368.032. [DOI] [PubMed] [Google Scholar]
- 85.Skvortsova T.E., Rykova E.Y., Tamkovich S.N., Bryzgunova O.E., Starikov A.V., Kuznetsova N.P., Vlassov V.V., Laktionov P.P. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br. J. Cancer. 2006;94:1492–1495. doi: 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van der Auwera I., Elst H.J., Van Laere S.J., Maes H., Huget P., van Dam P., Van Marck E.A., Vermeulen P.B., Dirix L.Y. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br. J. Cancer. 2009;100:1277–1286. doi: 10.1038/sj.bjc.6605013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shan M., Yin H., Li J., Li X., Wang D., Su Y., Niu M., Zhong Z., Wang J., Zhang X., et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7:18485–18494. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yazici H., Terry M.B., Cho Y.H., Senie R.T., Liao Y., Andrulis I., Santella R.M. Aberrant methylation of RASSF1A in plasma DNA before breast cancer diagnosis in the Breast Cancer Family Registry. Cancer Epidemiol. Biomark. Prev. 2009;18:2723–2725. doi: 10.1158/1055-9965.EPI-08-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Radpour R., Barekati Z., Kohler C., Lv Q., Bürki N., Diesch C., Bitzer J., Zheng H., Schmid S., Zhong X.Y. Hypermethylation of Tumor Suppressor Genes Involved in Critical Regulatory Pathways for Developing a Blood-Based Test in Breast Cancer. PLoS ONE. 2011;6:e16080. doi: 10.1371/journal.pone.0016080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamamoto N., Nakayama T., Kajita M., Miyake T., Iwamoto T., Kim S.J., Sakai A., Ishihara H., Tamaki Y., Noguchi S. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARbeta2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res. Treat. 2012;132:165–173. doi: 10.1007/s10549-011-1575-2. [DOI] [PubMed] [Google Scholar]
- 91.Chimonidou M., Strati A., Malamos N., Georgoulias V., Lianidou E.S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin. Chem. 2013;59:270–279. doi: 10.1373/clinchem.2012.191551. [DOI] [PubMed] [Google Scholar]
- 92.Swellam M., Abdelmaksoud M.D., Sayed Mahmoud M., Ramadan A., Abdel-Moneem W., Hefny M.M. Aberrant methylation of APC and RARbeta2 genes in breast cancer patients. Iubmb Life. 2015;67:61–68. doi: 10.1002/iub.1346. [DOI] [PubMed] [Google Scholar]
- 93.Nandi K., Yadav P., Mir R., Khurana N., Agarwal P., Saxena A. The Clinical Significance of Rassf1a and Cdh1 Hypermethylation in Breast Cancer Patients. Int. J. Sci. Res. 2008 doi: 10.36106/ijsr. [DOI] [Google Scholar]
- 94.Li D., Li P., Wu J., Yi J., Dou Y., Guo X., Yin Y., Wang D., Ma C., Qiu L. Methylation of NBPF1 as a novel marker for the detection of plasma cell-free DNA of breast cancer patients. Clin. Chim. Acta. 2018;484:81–86. doi: 10.1016/j.cca.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 95.Mijnes J., Tiedemann J., Eschenbruch J., Gasthaus J., Bringezu S., Bauerschlag D., Maass N., Arnold N., Weimer J., Anzeneder T., et al. SNiPER: A novel hypermethylation biomarker panel for liquid biopsy based early breast cancer detection. Oncotarget. 2019;10:6494–6508. doi: 10.18632/oncotarget.27303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.WHO . Classification of Tumours of the Breast. 4th ed. International Agency for Research on Cancer; Lyon, France: 2012. [Google Scholar]
- 97.Dai X., Xiang L., Li T., Bai Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer. 2016;7:1281–1294. doi: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Verma A., Kaur J., Mehta K. Molecular oncology update: Breast cancer gene expression profiling. Asian J. Oncol. 2015;1:65. doi: 10.4103/2454-6798.173282. [DOI] [Google Scholar]
- 99.Chimonidou M., Tzitzira A., Strati A., Sotiropoulou G., Sfikas C., Malamos N., Georgoulias V., Lianidou E. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clin. Biochem. 2013;46:235–240. doi: 10.1016/j.clinbiochem.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 100.Gobel G., Auer D., Gaugg I., Schneitter A., Lesche R., Muller-Holzner E., Marth C., Daxenbichler G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res. Treat. 2011;130:109–117. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 101.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., Karamitrousis E., Nena E., Tsamardinos I., Kolios G., Lianidou E., et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38:3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 102.Fu D., Ren C., Tan H., Wei J., Zhu Y., He C., Shao W., Zhang J. Sox17 promoter methylation in plasma DNA is associated with poor survival and can be used as a prognostic factor in breast cancer. Medicine. 2015;94:e637. doi: 10.1097/MD.0000000000000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Visvanathan K., Fackler M.S., Zhang Z., Lopez-Bujanda Z.A., Jeter S.C., Sokoll L.J., Garrett-Mayer E., Cope L.M., Umbricht C.B., Euhus D.M., et al. Monitoring of Serum DNA Methylation as an Early Independent Marker of Response and Survival in Metastatic Breast Cancer: TBCRC 005 Prospective Biomarker Study. J. Clin. Oncol. 2017;35:751–758. doi: 10.1200/JCO.2015.66.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zurita M., Lara P.C., del Moral R., Torres B., Linares-Fernandez J.L., Arrabal S.R., Martinez-Galan J., Oliver F.J., Ruiz de Almodovar J.M. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer. 2010;10:217. doi: 10.1186/1471-2407-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muller H.M., Widschwendter A., Fiegl H., Ivarsson L., Goebel G., Perkmann E., Marth C., Widschwendter M. DNA methylation in serum of breast cancer patients: An independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 106.Rawson J.B., Bapat B. Epigenetic biomarkers in colorectal cancer diagnostics. Expert Rev. Mol. Diagn. 2012;12:499–509. doi: 10.1586/erm.12.39. [DOI] [PubMed] [Google Scholar]
- 107.Payne S.R. From discovery to the clinic: The novel DNA methylation biomarker (m)SEPT9 for the detection of colorectal cancer in blood. Epigenomics. 2010;2:575–585. doi: 10.2217/epi.10.35. [DOI] [PubMed] [Google Scholar]
- 108.Issa I.A., Noureddine M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017;23:5086–5096. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 110.Simon K. Colorectal cancer development and advances in screening. Clin. Interv. Aging. 2016;11:967–976. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Molnar B., Toth K., Bartak B.K., Tulassay Z. Plasma methylated septin 9: A colorectal cancer screening marker. Expert Rev. Mol. Diagn. 2015;15:171–184. doi: 10.1586/14737159.2015.975212. [DOI] [PubMed] [Google Scholar]
- 112.Lamb Y.N., Dhillon S. Epi proColon((R)) 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol. Diagn. 2017;21:225–232. doi: 10.1007/s40291-017-0259-y. [DOI] [PubMed] [Google Scholar]
- 113.Song L., Jia J., Peng X., Xiao W., Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci. Rep. 2017;7:3032. doi: 10.1038/s41598-017-03321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Church T.R., Wandell M., Lofton-Day C., Mongin S.J., Burger M., Payne S.R., Castanos-Velez E., Blumenstein B.A., Rosch T., Osborn N., et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ahlquist D.A., Taylor W.R., Mahoney D.W., Zou H., Domanico M., Thibodeau S.N., Boardman L.A., Berger B.M., Lidgard G.P. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin. Gastroenterol. Hepatol. 2012;10:272–277.e271. doi: 10.1016/j.cgh.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Toth K., Wasserkort R., Sipos F., Kalmar A., Wichmann B., Leiszter K., Valcz G., Juhasz M., Miheller P., Patai A.V., et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS ONE. 2014;9:e115415. doi: 10.1371/journal.pone.0115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suehiro Y., Hashimoto S., Higaki S., Fujii I., Suzuki C., Hoshida T., Matsumoto T., Yamaoka Y., Takami T., Sakaida I., et al. Blood free-circulating DNA testing by highly sensitive methylation assay to diagnose colorectal neoplasias. Oncotarget. 2018;9:16974–16987. doi: 10.18632/oncotarget.24768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee B.B., Lee E.J., Jung E.H., Chun H.K., Chang D.K., Song S.Y., Park J., Kim D.H. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin. Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 119.Bartak B.K., Kalmar A., Peterfia B., Patai A.V., Galamb O., Valcz G., Spisak S., Wichmann B., Nagy Z.B., Toth K., et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics. 2017;12:751–763. doi: 10.1080/15592294.2017.1356957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pedersen S.K., Symonds E.L., Baker R.T., Murray D.H., McEvoy A., Van Doorn S.C., Mundt M.W., Cole S.R., Gopalsamy G., Mangira D., et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. doi: 10.1186/s12885-015-1674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Symonds E.L., Pedersen S.K., Baker R.T., Murray D.H., Gaur S., Cole S.R., Gopalsamy G., Mangira D., LaPointe L.C., Young G.P. A Blood Test for Methylated BCAT1 and IKZF1 vs. a Fecal Immunochemical Test for Detection of Colorectal Neoplasia. Clin. Transl. Gastroenterol. 2016;7:e137. doi: 10.1038/ctg.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zou H.Z., Yu B.M., Wang Z.W., Sun J.Y., Cang H., Gao F., Li D.H., Zhao R., Feng G.G., Yi J. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin. Cancer Res. 2002;8:188–191. [PubMed] [Google Scholar]
- 123.Leung W.K., To K.F., Man E.P., Chan M.W., Bai A.H., Hui A.J., Chan F.K., Sung J.J. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am. J. Gastroenterol. 2005;100:2274–2279. doi: 10.1111/j.1572-0241.2005.50412.x. [DOI] [PubMed] [Google Scholar]
- 124.Ebert M.P., Model F., Mooney S., Hale K., Lograsso J., Tonnes-Priddy L., Hoffmann J., Csepregi A., Rocken C., Molnar B., et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131:1418–1430. doi: 10.1053/j.gastro.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 125.Wallner M., Herbst A., Behrens A., Crispin A., Stieber P., Goke B., Lamerz R., Kolligs F.T. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin. Cancer Res. 2006;12:7347–7352. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 126.Lofton-Day C., Model F., Devos T., Tetzner R., Distler J., Schuster M., Song X., Lesche R., Liebenberg V., Ebert M., et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y.C., Yu Z.H., Liu C., Xu L.Z., Yu W., Lu J., Zhu R.M., Li G.L., Xia X.Y., Wei X.W., et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J. Gastroenterol. 2008;14:3074–3080. doi: 10.3748/wjg.14.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li M., Chen W.D., Papadopoulos N., Goodman S.N., Bjerregaard N.C., Laurberg S., Levin B., Juhl H., Arber N., Moinova H., et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He Q., Chen H.Y., Bai E.Q., Luo Y.X., Fu R.J., He Y.S., Jiang J., Wang H.Q. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet. Cytogenet. 2010;202:1–10. doi: 10.1016/j.cancergencyto.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 130.Herbst A., Rahmig K., Stieber P., Philipp A., Jung A., Ofner A., Crispin A., Neumann J., Lamerz R., Kolligs F.T. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am. J. Gastroenterol. 2011;106:1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 131.Hibi K., Goto T., Shirahata A., Saito M., Kigawa G., Nemoto H., Sanada Y. Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett. 2011;311:96–100. doi: 10.1016/j.canlet.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 132.Wu P.P., Zou J.H., Tang R.N., Yao Y., You C.Z. Detection and Clinical Significance of DLC1 Gene Methylation in Serum DNA from Colorectal Cancer Patients. Chin. J. Cancer Res. 2011;23:283–287. doi: 10.1007/s11670-011-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cassinotti E., Melson J., Liggett T., Melnikov A., Yi Q., Replogle C., Mobarhan S., Boni L., Segato S., Levenson V. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int. J. Cancer. 2012;131:1153–1157. doi: 10.1002/ijc.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pack S.C., Kim H.R., Lim S.W., Kim H.Y., Ko J.Y., Lee K.S., Hwang D., Park S.I., Kang H., Park S.W., et al. Usefulness of plasma epigenetic changes of five major genes involved in the pathogenesis of colorectal cancer. Int. J. Colorectal. Dis. 2013;28:139–147. doi: 10.1007/s00384-012-1566-8. [DOI] [PubMed] [Google Scholar]
- 135.Oh T., Kim N., Moon Y., Kim M.S., Hoehn B.D., Park C.H., Kim T.S., Kim N.K., Chung H.C., An S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J. Mol. Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 136.Liu Y., Tham C.K., Ong S.Y., Ho K.S., Lim J.F., Chew M.H., Lim C.K., Zhao Y., Tang C.L., Eu K.W. Serum methylation levels of TAC1. SEPT9 and EYA4 as diagnostic markers for early colorectal cancers: A pilot study. Biomarkers. 2013;18:399–405. doi: 10.3109/1354750X.2013.798745. [DOI] [PubMed] [Google Scholar]
- 137.Roperch J.P., Incitti R., Forbin S., Bard F., Mansour H., Mesli F., Baumgaertner I., Brunetti F., Sobhani I. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pedersen S.K., Mitchell S.M., Graham L.D., McEvoy A., Thomas M.L., Baker R.T., Ross J.P., Xu Z.Z., Ho T., LaPointe L.C., et al. CAHM, a long non-coding RNA gene hypermethylated in colorectal neoplasia. Epigenetics. 2014;9:1071–1082. doi: 10.4161/epi.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takane K., Midorikawa Y., Yagi K., Sakai A., Aburatani H., Takayama T., Kaneda A. Aberrant promoter methylation of PPP1R3C and EFHD1 in plasma of colorectal cancer patients. Cancer Med. 2014;3:1235–1245. doi: 10.1002/cam4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Melotte V., Yi J.M., Lentjes M.H., Smits K.M., Van Neste L., Niessen H.E., Wouters K.A., Louwagie J., Schuebel K.E., Herman J.G., et al. Spectrin repeat containing nuclear envelope 1 and forkhead box protein E1 are promising markers for the detection of colorectal cancer in blood. Cancer Prev Res. (Phila) 2015;8:157–164. doi: 10.1158/1940-6207.CAPR-14-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang X., Song Y.F., Lu H.N., Wang D.P., Zhang X.S., Huang S.L., Sun B.L., Huang Z.G. Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World J. Gastroenterol. 2015;21:2629–2637. doi: 10.3748/wjg.v21.i9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pedersen S.K., Baker R.T., McEvoy A., Murray D.H., Thomas M., Molloy P.L., Mitchell S., Lockett T., Young G.P., LaPointe L.C. A two-gene blood test for methylated DNA sensitive for colorectal cancer. PLoS ONE. 2015;10:e0125041. doi: 10.1371/journal.pone.0125041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Salehi R., Atapour N., Vatandoust N., Farahani N., Ahangari F., Salehi A.R. Methylation pattern of ALX4 gene promoter as a potential biomarker for blood-based early detection of colorectal cancer. Adv. Biomed. Res. 2015;4:252. doi: 10.4103/2277-9175.170677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitchell S.M., Ho T., Brown G.S., Baker R.T., Thomas M.L., McEvoy A., Xu Z.Z., Ross J.P., Lockett T.J., Young G.P., et al. Evaluation of Methylation Biomarkers for Detection of Circulating Tumor DNA and Application to Colorectal Cancer. Genes. 2016;7:125. doi: 10.3390/genes7120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rezvani N., Alibakhshi R., Vaisi-Raygani A., Bashiri H., Saidijam M. Detection of SPG20 gene promoter-methylated DNA, as a novel epigenetic biomarker, in plasma for colorectal cancer diagnosis using the MethyLight method. Oncol. Lett. 2017;13:3277–3284. doi: 10.3892/ol.2017.5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rasmussen S.L., Krarup H.B., Sunesen K.G., Johansen M.B., Stender M.T., Pedersen I.S., Madsen P.H., Thorlacius-Ussing O. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS ONE. 2017;12:e0180809. doi: 10.1371/journal.pone.0180809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rokni P., Shariatpanahi A.M., Sakhinia E., Kerachian M.A. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes Genom. 2018;40:423–428. doi: 10.1007/s13258-017-0644-2. [DOI] [PubMed] [Google Scholar]
- 148.Li H., Wang Z., Zhao G., Ma Y., Chen Y., Xue Q., Zheng M., Fei S. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int. J. Biol. Markers. 2019;34:54–59. doi: 10.1177/1724600818820536. [DOI] [PubMed] [Google Scholar]
- 149.Zhao G., Li H., Yang Z., Wang Z., Xu M., Xiong S., Li S., Wu X., Liu X., Wang Z., et al. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 2019;8:5619–5628. doi: 10.1002/cam4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jensen S.Ø., Øgaard N., Ørntoft M.-B.W., Rasmussen M.H., Bramsen J.B., Kristensen H., Mouritzen P., Madsen M.R., Madsen A.H., Sunesen K.G., et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin. Epigenetics. 2019;11:158. doi: 10.1186/s13148-019-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nishio M., Sakakura C., Nagata T., Komiyama S., Miyashita A., Hamada T., Kuryu Y., Ikoma H., Kubota T., Kimura A., et al. RUNX3 promoter methylation in colorectal cancer: Its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res. 2010;30:2673–2682. [PubMed] [Google Scholar]
- 152.Tang D., Liu J., Wang D.R., Yu H.F., Li Y.K., Zhang J.Q. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin. Investig. Med. 2011;34:E88–E95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 153.Liu Y., Chew M.H., Tham C.K., Tang C.L., Ong S.Y., Zhao Y. Methylation of serum SST gene is an independent prognostic marker in colorectal cancer. Am. J. Cancer Res. 2016;6:2098–2108. [PMC free article] [PubMed] [Google Scholar]
- 154.Nakayama G., Kodera Y., Ohashi N., Koike M., Fujiwara M., Nakao A. p16INK4a methylation in serum as a follow-up marker for recurrence of colorectal cancer. Anticancer Res. 2011;31:1643–1646. [PubMed] [Google Scholar]
- 155.Tham C., Chew M., Soong R., Lim J., Ang M., Tang C., Zhao Y., Ong S.Y., Liu Y. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer. 2014;120:3131–3141. doi: 10.1002/cncr.28802. [DOI] [PubMed] [Google Scholar]
- 156.Bergheim J., Semaan A., Gevensleben H., Groening S., Knoblich A., Dietrich J., Weber J., Kalff J.C., Bootz F., Kristiansen G., et al. Potential of quantitative SEPT9 and SHOX2 methylation in plasmatic circulating cell-free DNA as auxiliary staging parameter in colorectal cancer: A prospective observational cohort study. Br. J. Cancer. 2018;118:1217–1228. doi: 10.1038/s41416-018-0035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Philipp A.B., Stieber P., Nagel D., Neumann J., Spelsberg F., Jung A., Lamerz R., Herbst A., Kolligs F.T. Prognostic role of methylated free circulating DNA in colorectal cancer. Int. J. Cancer. 2012;131:2308–2319. doi: 10.1002/ijc.27505. [DOI] [PubMed] [Google Scholar]
- 158.Herbst A., Wallner M., Rahmig K., Stieber P., Crispin A., Lamerz R., Kolligs F.T. Methylation of helicase-like transcription factor in serum of patients with colorectal cancer is an independent predictor of disease recurrence. Eur. J. Gastroenterol. Hepatol. 2009;21:565–569. doi: 10.1097/MEG.0b013e328318ecf2. [DOI] [PubMed] [Google Scholar]
- 159.Herbst A., Vdovin N., Gacesa S., Ofner A., Philipp A., Nagel D., Holdt L.M., Op den Winkel M., Heinemann V., Stieber P., et al. Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer. Int. J. Cancer. 2017;140:2134–2144. doi: 10.1002/ijc.30625. [DOI] [PubMed] [Google Scholar]
- 160.Barault L., Amatu A., Siravegna G., Ponzetti A., Moran S., Cassingena A., Mussolin B., Falcomata C., Binder A.M., Cristiano C., et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Amatu A., Schirripa M., Tosi F., Lonardi S., Bencardino K., Bonazzina E., Palmeri L., Patanè D.A., Pizzutilo E.G., Mussolin B., et al. High Circulating Methylated DNA Is a Negative Predictive and Prognostic Marker in Metastatic Colorectal Cancer Patients Treated With Regorafenib. Front. Oncol. 2019;9:622. doi: 10.3389/fonc.2019.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.COLVERA: Because the Truth Matters. [(accessed on 20 July 2019)]; Available online: https://www.clinicalgenomics.com/colvera.html.
- 163.Symonds E.L., Pedersen S.K., Murray D.H., Jedi M., Byrne S.E., Rabbitt P., Baker R.T., Bastin D., Young G.P. Circulating tumour DNA for monitoring colorectal cancer-a prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin. Epigenetics. 2018;10:63. doi: 10.1186/s13148-018-0500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Young G.P., Pedersen S.K., Mansfield S., Murray D.H., Baker R.T., Rabbitt P., Byrne S., Bambacas L., Hollington P., Symonds E.L. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016;5:2763–2772. doi: 10.1002/cam4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Murray D.H., Symonds E.L., Young G.P., Byrne S., Rabbitt P., Roy A., Cornthwaite K., Karapetis C.S., Pedersen S.K. Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival. J. Cancer Res. Clin. Oncol. 2018;144:1741–1750. doi: 10.1007/s00432-018-2701-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hammerich K.H., Ayala G.E., Wheeler T.M. Anatomy of the prostate gland and surgical pathology of prostate cancer. Camb. Univ. Camb. 2009:1–10. [Google Scholar]
- 167.Roobol M.J., Carlsson S.V. Risk stratification in prostate cancer screening. Nat. Rev. Urol. 2013;10:38–48. doi: 10.1038/nrurol.2012.225. [DOI] [PubMed] [Google Scholar]
- 168.Hoffman R.M. Clinical practice. Screening for prostate cancer. N. Engl. J. Med. 2011;365:2013–2019. doi: 10.1056/NEJMcp1103642. [DOI] [PubMed] [Google Scholar]
- 169.Etzioni R., Penson D.F., Legler J.M., di Tommaso D., Boer R., Gann P.H., Feuer E.J. Overdiagnosis due to prostate-specific antigen screening: Lessons from U.S. prostate cancer incidence trends. J. Natl. Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 170.Rice M.A., Stoyanova T. Prostatectomy. IntechOpen; London, UK: 2018. Biomarkers for Diagnosis and Prognosis of Prostate Cancer. [DOI] [Google Scholar]
- 171.Larsen L.K., Lind G.E., Guldberg P., Dahl C. DNA-Methylation-Based Detection of Urological Cancer in Urine: Overview of Biomarkers and Considerations on Biomarker Design, Source of DNA, and Detection Technologies. Int. J. Mol. Sci. 2019;20:2657. doi: 10.3390/ijms20112657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ellinger J., Haan K., Heukamp L.C., Kahl P., Buttner R., Muller S.C., von Ruecker A., Bastian P.J. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate. 2008;68:42–49. doi: 10.1002/pros.20651. [DOI] [PubMed] [Google Scholar]
- 173.Sunami E., Shinozaki M., Higano C.S., Wollman R., Dorff T.B., Tucker S.J., Martinez S.R., Mizuno R., Singer F.R., Hoon D.S. Multimarker circulating DNA assay for assessing blood of prostate cancer patients. Clin. Chem. 2009;55:559–567. doi: 10.1373/clinchem.2008.108498. [DOI] [PubMed] [Google Scholar]
- 174.Wu T., Giovannucci E., Welge J., Mallick P., Tang W.Y., Ho S.M. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: A meta-analysis. Br. J. Cancer. 2011;105:65–73. doi: 10.1038/bjc.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Payne S.R., Serth J., Schostak M., Kamradt J., Strauss A., Thelen P., Model F., Day J.K., Liebenberg V., Morotti A., et al. DNA methylation biomarkers of prostate cancer: Confirmation of candidates and evidence urine is the most sensitive body fluid for non-invasive detection. Prostate. 2009;69:1257–1269. doi: 10.1002/pros.20967. [DOI] [PubMed] [Google Scholar]
- 176.Dumache R., Puiu M., Motoc M., Vernic C., Dumitrascu V. Prostate cancer molecular detection in plasma samples by glutathione S-transferase P1 (GSTP1) methylation analysis. Clin. Lab. 2014;60:847–852. doi: 10.7754/Clin.Lab.2013.130701. [DOI] [PubMed] [Google Scholar]
- 177.Brait M., Banerjee M., Maldonado L., Ooki A., Loyo M., Guida E., Izumchenko E., Mangold L., Humphreys E., Rosenbaum E., et al. Promoter methylation of MCAM, ERalpha and ERbeta in serum of early stage prostate cancer patients. Oncotarget. 2017;8:15431–15440. doi: 10.18632/oncotarget.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Haldrup C., Pedersen A.L., Ogaard N., Strand S.H., Hoyer S., Borre M., Orntoft T.F., Sorensen K.D. Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol. Oncol. 2018;12:545–560. doi: 10.1002/1878-0261.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sanchez B.E., Aguayo A., Martinez B., Rodriguez F., Marmolejo M., Svyryd Y., Luna L., Munoz L.A., Jimenez M.A., Sotomayor M., et al. Using Genetic and Epigenetic Markers to Improve Differential Diagnosis of Prostate Cancer and Benign Prostatic Hyperplasia by Noninvasive Methods in Mexican Patients. Clin. Genitourin Cancer. 2018;16:e867–e877. doi: 10.1016/j.clgc.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 180.Reis I.M., Ramachandran K., Speer C., Gordian E., Singal R. Serum GADD45a methylation is a useful biomarker to distinguish benign vs malignant prostate disease. Br. J. Cancer. 2015;113:460–468. doi: 10.1038/bjc.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bastian P.J., Palapattu G.S., Yegnasubramanian S., Rogers C.G., Lin X., Mangold L.A., Trock B., Eisenberger M.A., Partin A.W., Nelson W.G. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J. Urol. 2008;179:529–534; discussion 534–535. doi: 10.1016/j.juro.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dumache R., Puiu M., Minciu R., Bardan R., David D., Tudor A., Bumbacila B. Retinoic acid receptor beta2 (RARbeta2): Nonivasive biomarker for distinguishing malignant versus benign prostate lesions from bodily fluids. Chirurgia (Bucur) 2012;107:780–784. [PubMed] [Google Scholar]
- 183.Wang L., Lin Y.L., Li B., Wang Y.Z., Li W.P., Ma J.G. Aberrant promoter methylation of the cadherin 13 gene in serum and its relationship with clinicopathological features of prostate cancer. J. Int. Med. Res. 2014;42:1085–1092. doi: 10.1177/0300060514540631. [DOI] [PubMed] [Google Scholar]
- 184.Garzotto M. Prostate Cancer. Cambridge University Press; Cambridge, UK: 2008. The natural and treated history of prostate cancer. [Google Scholar]
- 185.Fine S.W. Evolution in Prostate Cancer Staging: Pathology Updates From AJCC 8th Edition and Opportunities That Remain. Adv. Anat. Pathol. 2018;25:327–332. doi: 10.1097/PAP.0000000000000200. [DOI] [PubMed] [Google Scholar]
- 186.Mottet N., Bellmunt J., Bolla M., Briers E., Cumberbatch M.G., De Santis M., Fossati N., Gross T., Henry A.M., Joniau S., et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 187.Gordetsky J., Epstein J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016;11:25. doi: 10.1186/s13000-016-0478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van den Bergh R.C., van Casteren N.J., van den Broeck T., Fordyce E.R., Gietzmann W.K., Stewart F., MacLennan S., Dabestani S., Bellmunt J., Bolla M., et al. Role of Hormonal Treatment in Prostate Cancer Patients with Nonmetastatic Disease Recurrence After Local Curative Treatment: A Systematic Review. Eur. Urol. 2016;69:802–820. doi: 10.1016/j.eururo.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 189.Shapiro D., Tareen B. Current and emerging treatments in the management of castration-resistant prostate cancer. Expert Rev. Anticancer. 2012;12:951–964. doi: 10.1586/era.12.59. [DOI] [PubMed] [Google Scholar]
- 190.Reibenwein J., Pils D., Horak P., Tomicek B., Goldner G., Worel N., Elandt K., Krainer M. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67:427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 191.Hendriks R.J., Dijkstra S., Smit F.P., Vandersmissen J., Van de Voorde H., Mulders P.F.A., van Oort I.M., Van Criekinge W., Schalken J.A. Epigenetic markers in circulating cell-free DNA as prognostic markers for survival of castration-resistant prostate cancer patients. Prostate. 2018;78:336–342. doi: 10.1002/pros.23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bastian P.J., Palapattu G.S., Lin X., Yegnasubramanian S., Mangold L.A., Trock B., Eisenberger M.A., Partin A.W., Nelson W.G. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin. Cancer Res. 2005;11:4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- 193.Lin Y.L., Deng Q.K., Wang Y.H., Fu X.L., Ma J.G., Li W.P. Aberrant Protocadherin17 (PCDH17) Methylation in Serum is a Potential Predictor for Recurrence of Early-Stage Prostate Cancer Patients After Radical Prostatectomy. Med. Sci. Monit. 2015;21:3955–3960. doi: 10.12659/MSM.896763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Deng Q.K., Lei Y.G., Lin Y.L., Ma J.G., Li W.P. Prognostic Value of Protocadherin10 (PCDH10) Methylation in Serum of Prostate Cancer Patients. Med. Sci. Monit. 2016;22:516–521. doi: 10.12659/MSM.897179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lin Y.L., Li Y.L., Ma J.G. Aberrant Promoter Methylation of Protocadherin8 (PCDH8) in Serum is a Potential Prognostic Marker for Low Gleason Score Prostate Cancer. Med. Sci. Monit. 2017;23:4895–4900. doi: 10.12659/MSM.904366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mahon K.L., Qu W., Lin H.M., Spielman C., Cain D., Jacobs C., Stockler M.R., Higano C.S., de Bono J.S., Chi K.N., et al. Serum Free Methylated Glutathione S-transferase 1 DNA Levels, Survival, and Response to Docetaxel in Metastatic, Castration-resistant Prostate Cancer: Post Hoc Analyses of Data from a Phase 3 Trial. Eur. Urol. 2018 doi: 10.1016/j.eururo.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 197.Adler A., Geiger S., Keil A., Bias H., Schatz P., deVos T., Dhein J., Zimmermann M., Tauber R., Wiedenmann B. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14:183. doi: 10.1186/1471-230X-14-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tan S.H., Ida H., Lau Q.C., Goh B.C., Chieng W.S., Loh M., Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol. Rep. 2007;18:1225–1230. doi: 10.3892/or.18.5.1225. [DOI] [PubMed] [Google Scholar]
- 199.Liu L., Toung J.M., Jassowicz A.F., Vijayaraghavan R., Kang H., Zhang R., Kruglyak K.M., Huang H.J., Hinoue T., Shen H., et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018;29:1445–1453. doi: 10.1093/annonc/mdy119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li W., Li Q., Kang S., Same M., Zhou Y., Sun C., Liu C.C., Matsuoka L., Sher L., Wong W.H., et al. CancerDetector: Ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 2018;46:e89. doi: 10.1093/nar/gky423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kang S., Li Q., Chen Q., Zhou Y., Park S., Lee G., Grimes B., Krysan K., Yu M., Wang W., et al. CancerLocator: Non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 2017;18:53. doi: 10.1186/s13059-017-1191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.IvyGene Diagnostics: Early Cancer Confirmation Test. [(accessed on 21 July 2019)]; Available online: https://www.ivygenelabs.com/ivygene-technology/core-test/
- 204.Hao X., Luo H., Krawczyk M., Wei W., Wang W., Wang J., Flagg K., Hou J., Zhang H., Yi S., et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl. Acad. Sci. USA. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Daber R., Sukhadia S., Morrissette J.J. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet. 2013;206:441–448. doi: 10.1016/j.cancergen.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 206.Soto J., Rodriguez-Antolin C., Vallespin E., de Castro Carpeno J., Ibanez de Caceres I. The impact of next-generation sequencing on the DNA methylation-based translational cancer research. Transl. Res. 2016;169:1–18.e11. doi: 10.1016/j.trsl.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 207.Sigalotti L., Covre A., Colizzi F., Fratta E. Quantitative Methylation-Specific PCR: A Simple Method for Studying Epigenetic Modifications of Cell-Free DNA. Methods Mol. Biol. 2019;1909:137–162. doi: 10.1007/978-1-4939-8973-7_11. [DOI] [PubMed] [Google Scholar]