PCS1 is a moonlighting protein that functions in abiotic stress response and is indispensable for proper extracellular immune responses and pathogen-triggered glucosinolate metabolism.
Abstract
Phytochelatin synthase (PCS) is a key component of heavy metal detoxification in plants. PCS catalyzes both the synthesis of the peptide phytochelatin from glutathione and the degradation of glutathione conjugates via peptidase activity. Here, we describe a role for PCS in disease resistance against plant pathogenic fungi. The pen4 mutant, which is allelic to cadmium insensitive1 (cad1/pcs1) mutants, was recovered from a screen for Arabidopsis mutants with reduced resistance to the nonadapted barley fungal pathogen Blumeria graminis f. sp. hordei. PCS1, which is found in the cytoplasm of cells of healthy plants, translocates upon pathogen attack and colocalizes with the PEN2 myrosinase on the surface of immobilized mitochondria. pcs1 and pen2 mutant plants exhibit similar metabolic defects in the accumulation of pathogen-inducible indole glucosinolate-derived compounds, suggesting that PEN2 and PCS1 act in the same metabolic pathway. The function of PCS1 in this pathway is independent of phytochelatin synthesis and deglycination of glutathione conjugates, as catalytic-site mutants of PCS1 are still functional in indole glucosinolate metabolism. In uncovering a peptidase-independent function for PCS1, we reveal this enzyme to be a moonlighting protein important for plant responses to both biotic and abiotic stresses.
As sessile organisms, plants have developed sophisticated mechanisms that allow them to adapt to and survive the biotic and abiotic stresses that they encounter in their environments. Different stress signaling pathways are connected to maximize plant fitness (Berens et al., 2019). For instance, one evolutionary conserved mechanism involves phytohormone signaling mediated by abscisic acid, which promotes abiotic stress tolerance and suppresses signaling of the biotic stress-related phytohormone salicylic acid (Berens et al., 2017). On the other hand, an overlapping enzymatic machinery is engaged to cope with xenobiotics and pathogen stress. The metabolism of xenobiotics, including plant metal tolerance, as well as the biosynthesis of sulfur-containing immunomodulatory and antimicrobial secondary metabolites such as glucosinolates and indole-type phytoalexins involves in Brassicaceae species the formation and processing of glutathione conjugates (Grill et al., 1989; Cobbett and Goldsbrough, 2002; Bednarek et al., 2009; Pastorczyk and Bednarek, 2016; Czerniawski and Bednarek, 2018).
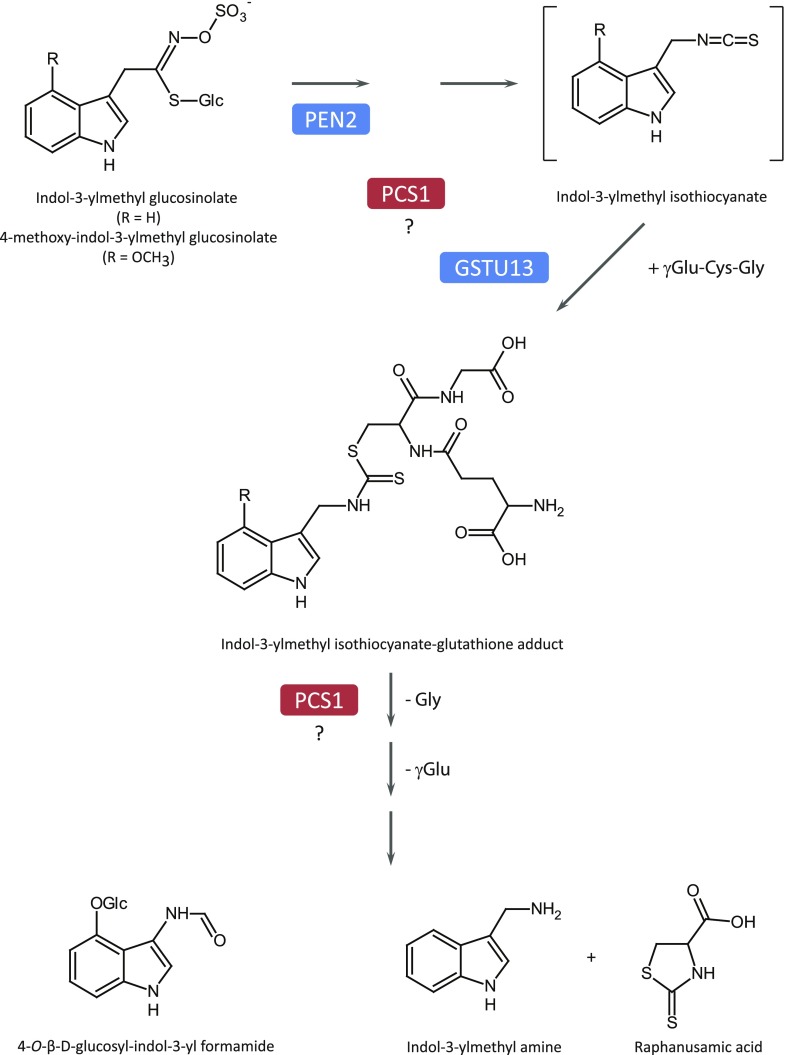
Invasive growth is an essential part of the pathogenesis of many fungal and oomycete plant pathogens, and extracellular defense mechanisms play a critical role in blocking their entry into plant cells (Underwood and Somerville, 2008; Hématy et al., 2009). For example, Arabidopsis (Arabidopsis thaliana) is an inappropriate or nonhost plant for the nonadapted barley powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh; Lipka et al., 2008). The ability of Arabidopsis cells to resist Bgh entry is partly dependent on the PENETRATION (PEN) proteins. For instance, mutants of the syntaxin PEN1 (SYP121; Collins et al., 2003), the myrosinase PEN2 (Lipka et al., 2005), and the ABC transporter PEN3 (ABCG36/PDR8; Stein et al., 2006) are more susceptible to Bgh entry and exhibit increased formation of the fungal feeding structure, the haustoria, in leaf epidermal cells. It has been shown that PEN2 hydrolyzes indole glucosinolates (IGs), a class of sulfur-containing secondary metabolites, to form products that could act as broad-spectrum toxins and confer antifungal defense in the extracellular space at pathogen contact sites (Bednarek et al., 2009). It has also been suggested that IG-derived compounds are signaling molecules for callose deposition following treatment with the microbe-associated molecular pattern flagellin (flg22), a peptide epitope derived from the bacterial motor protein (Clay et al., 2009). Recently, Arabidopsis glutathione-S-transferase class-τ member 13 (GSTU13) was identified as an indispensable component of the PEN2 immune pathway for IG metabolism (Piślewska-Bednarek et al., 2018), suggesting that this pathway involves conjugation of the tripeptide glutathione (ECG) with unstable isothiocyanates (ITCs) that are products of IG metabolism and further processing of the resulting adducts to biologically active molecules. Arabidopsis PAD2/CAD2, which encodes γ-glutamyl-Cys synthetase and catalyzes the first committed step of glutathione biosynthesis, is essential for biotic stress-induced accumulation of IGs and the PEN2 immune pathway (Bednarek et al., 2009). The same enzyme is also needed for heavy metal ion tolerance, as glutathione is the precursor of heavy metal-chelating polypeptides, called phytochelatins (PCs), which in Arabidopsis are predominantly produced by PC synthase1 (PCS1; Grill et al., 1985, 1989; Howden et al., 1995; Rea, 2006). Notably, pcs1 mutants display elevated cell death responses in leaves upon inoculation with the oomycete pathogen Phytophthora infestans, enhanced susceptibility to the bacterial phytopathogen Pseudomonas syringae DC3000, and reduced callose deposition upon treatment with the peptide-epitope flg22 derived from the motor protein of P. syringae, indicating that apart from a function in tolerance toward heavy metal ions, PCS1 has also a role in Arabidopsis immunity (Clay et al., 2009; Kühnlenz et al., 2015; De Benedictis et al., 2018). Moreover, given that a PCS from Caenorhabditis elegans was able to restore PC production, but not to revert the cell death phenotype in the pcs1 mutant background, it was suggested that PCS1 function in plant immunity is not dependent on PC accumulation (Kühnlenz et al., 2015). Finally, pen2 and pcs1 mutants were shown to share an aberrant pathogen-triggered accumulation and secretion of IGs and their metabolism products. This includes a hyperaccumulation of 4-methoxyindol-3-ylmethyl glucosinolate (4MI3G), reduced levels of indol-3-ylmethyl amine and raphanusamic acid (RA), as well as reduced secretion of 4-methoxyindol-3-yl methanol and S-(4-methoxyindol-3-ylmethyl) Cys (Matern et al., 2019).
We present here the isolation and characterization of Arabidopsis mutants with defects in a PENETRATION resistance gene (PEN) designated PEN4. These mutants are allelic with pcs1 and are sensitive to invasive powdery mildew pathogens. We show that PCS1 is translocated underneath pathogen contact sites, where it colocalizes with PEN2, which is known to accumulate on the surface of immobilized mitochondria (Fuchs et al., 2016). Mutagenesis of PCS1 residues abolishing PC synthesis affects tolerance to heavy metals but does not alter PCS1 function in IG metabolism or extracellular defense. Together our results show that PCS1 acts in the PEN2 pathway and is a multifunctional protein with two independent activities. In addition to its long-known function in the synthesis of the peptide PC for heavy metal tolerance, PCS1 also has an independent role in IG metabolism and immune responses.
RESULTS
Mutations in PCS1 Result in Enhanced Invasive Growth of Several Fungal Pathogens
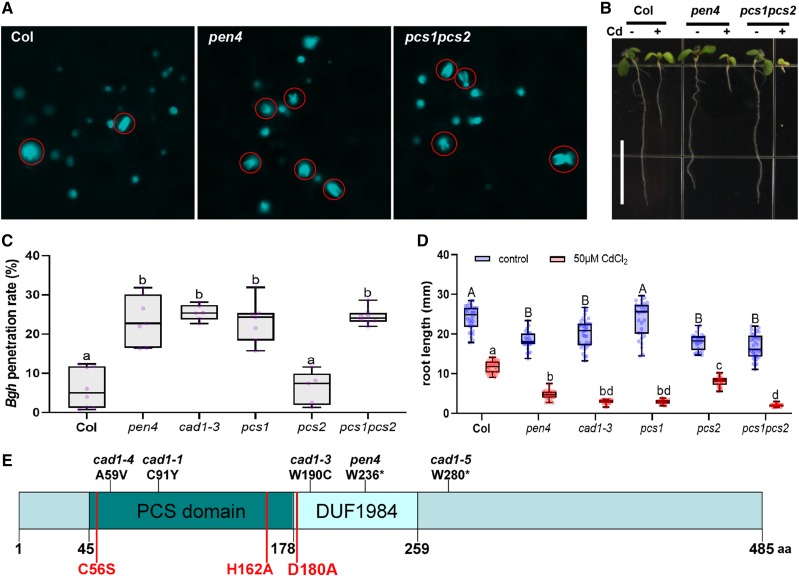
The pen4 mutant was isolated in a screen for Arabidopsis lines that allowed increased penetration of the nonadapted barley powdery mildew Bgh into leaf cells (Stein et al., 2006). Typically, 5% to 10% of germinated Bgh conidiospores successfully breach the plant cell wall and differentiate into fungal haustoria for nutrient uptake in Arabidopsis ecotype Columbia (Col-0; wild-type) leaf epidermal cells (Fig. 1A), while the remaining attempts at fungal entry culminate in de novo synthesized callose-rich deposits called papillae (Fig. 1A). The frequency of haustorium formation by Bgh increases to 20% to 25% on the leaves of pen4 plants (Fig. 1C).
Figure 1.
PEN4/PCS1 is important for both nonhost penetration resistance and heavy metal tolerance. A, Aniline blue-stained leaves of 3-week-old Arabidopsis plants 24 h after inoculation with Bgh. Aniline blue stains callose papillae symptomatic of failed penetration attempts and callose-containing encasements of haustorial necks formed after a successful penetration by the fungal appressorium (red circles). B, Seven-day-old Arabidopsis seedlings of the indicated genotype grown in the presence or absence of Cd (50 μm CdCl2; between 25 and 30 individual seedlings for each genotype/treatment grown on the same plate were used for the measurement). Scale bar = 1 cm. C, Box plots showing the penetration rate of Bgh spores on leaves of 3-week-old Arabidopsis plants. Measurements were done 24 h postinfection on the first true leaves of five to six individual plants (600–800 total penetration attempts counted per genotype). Whiskers represent minimum and maximum values. Lowercase letters show statistical differences according to Tukey’s honestly significant difference (HSD) mean-separation test following one-way ANOVA There was a significant genotype effect (P < 0.0001). D, Root length of 7-d-old seedlings grown in the presence or absence of Cd (50 μm CdCl2; between 25 and 30 individual seedlings for each genotype/treatment grown on the same plate were used for the measurement). Upper- and lowercase letters show statistical differences according to Tukey’s HSD mean-separation test following two-way ANOVA. There were significant main effects for Cd treatment and genotype (P < 0.001) and a significant interaction effect (P < 0.001). E, Domain organization of PCS1, showing the position of several mutant alleles (above) and residues important for PC synthesis (below in red).
Map-based cloning of PEN4 revealed that it encodes PCS1. PCS proteins are known for their glutathione γ-glutamylcysteinyltransferase activity (EC 2.3.2.15). The mutation in pen4 results in a premature stop codon that truncates the protein at amino acid residue 236 (Fig. 1E), in the middle of a DUF1984 domain (PF09328), which is always associated with the PC domain (PF05023; Rea, 2006). To confirm that the mutation in PCS1 is responsible for enhanced Bgh entry into pen4 leaves, we tested another pcs1 allele previously isolated in screens for plants with reduced tolerance to heavy metal cadmium (Cd) ions (cad1-3; Howden et al., 1995) and a null line carrying a transfer DNA (T-DNA) insertion in the second exon of PCS1 (Blum et al., 2007). All pcs1 lines were more susceptible than the wild-type Col-0 to Bgh haustorium formation (Fig. 1C). Like the parental lines, the F1 progeny of a cross between the cad1-3 and pen4 plants led to an increase of Bgh entry rates up to 20%, indicating that pen4 is allelic to cad1-3 (Supplemental Fig. S1).
Consistent with an impaired function of PCS1, the pen4 mutant also showed decreased tolerance to Cd (Fig. 1, B and D). PCS2, the second isoform of the PCS in Arabidopsis, shares 83% amino acid identity with PCS1. Although PCS2 can synthesize PCs in response to heavy metals, the gene appears to be dispensable for heavy metal tolerance (Fig. 1D; Blum et al., 2007). We asked whether PCS2, like PCS1, has a role in penetration resistance to fungal pathogens. A mutant line harboring a T-DNA insertion that abolishes PCS2 expression (pcs2; Blum et al., 2007) showed a frequency of cell entry by Bgh that was comparable to that in wild-type Col-0 plants (Fig. 1C). Furthermore, a line harboring T-DNA insertions in both PCS1 and PCS2 (pcs1 pcs2; Blum et al., 2007) did not consistently exhibit a higher incidence of cell entry by Bgh than the pcs1 single mutant (Fig. 1C). As with heavy metal tolerance, PCS2 has no detectable role in extracellular defense to the nonadapted pathogen. Furthermore, impaired disease resistance of the pen4 or pcs1 pcs2 double mutant line could be complemented by transforming these mutants with PCS1 expressed under the control of its native promoter or a 35S promoter and tagged with GFP at its N terminus (used later in this study). Together, these data indicate that PCS1 plays a role in limiting plant cell entry by Bgh.
To determine whether PCS1 is important for resistance responses to other fungal pathogens, we inoculated pen4 plants with the necrotrophic fungi Botrytis cinerea (Supplemental Fig. S2A) and Plectosphaerella cucumerina (Supplemental Fig. S2B; Sánchez-Rodríguez et al., 2009). Infection with both fungi was more severe in these mutant plants. Furthermore, pen4 mutants, like pen2 mutants, were hypersusceptible to Golovinomyces cichoracearum, a powdery mildew adapted for growth on Arabidopsis (Supplemental Fig. S2C). Together, these results show that PCS1 is important both in preventing the entry of nonadapted pathogens and in basal defense responses against multiple fungal pathogens.
Translocation of Cytosolic PCS1 to Immobile Mitochondria underneath Pathogen Contact Sites
Powdery mildew attack induces dramatic changes in cellular organization at the site of attempted fungal penetration (Underwood and Somerville, 2008). Several proteins that function in preventing host cell entry by Bgh show focal accumulation beneath fungal appressoria within epidermal cells at the site of attack, suggesting that these proteins act locally in extracellular defense responses (Koh et al., 2005; Underwood and Somerville, 2008). For instance, entry attempts by the fungus trigger local arrest of a subpopulation of mitochondria underneath Bgh appressoria, which is accompanied by transient aggregate formation of tail-anchored PEN2-GFP on the surface of these organelles (Fuchs et al., 2016). The plasma membrane-resident PEN3-GFP focally accumulates as disks, sometimes with bullseye-like “rings” around these contact sites (Koh et al., 2005; Stein et al., 2006).
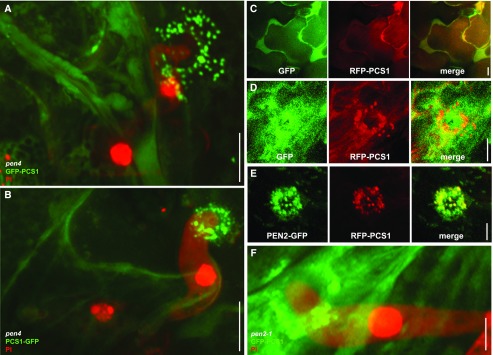
Given PCS1’s involvement in limiting host cell entry, we tested whether PCS1 localized to the site of fungal attack. To this end, we generated GFP fusions at either the amino- or carboxy terminus of PCS1 under control of the 35S promoter in the pen4 background (Fig. 2, A and B). In addition, we generated a transgenic pcs1 pcs2 double mutant line expressing red fluorescent protein (RFP)-PCS1 driven by the 35S promoter. In noninoculated leaf epidermal cells, the RFP-PCS1 fluorescence signal suggested a cytosolic localization similar to that of free GFP expressed under the control of the 35S promoter (EGAD line; Fig. 2C; Cutler et al., 2000). This is consistent with the absence of canonical organelle targeting sequences or predicted transmembrane domains in PCS1. In pathogen-challenged leaf epidermal cells, in addition to their cytosolic localization, GFP-PCS1 and PCS1-GFP localized to bright round aggregates underneath Bgh contact sites (Fig. 2, A and B); GFP alone did not exhibit this localization (Fig. 2D), excluding the possibility that the fluorescent tag is itself driving PCS1 aggregation underneath pathogen contact sites. Similarly, RFP-PCS1 also accumulated focally at incipient Bgh entry sites (Fig. 2, D and E). During the early stages of fungal entry, PCS1 tightly localized just beneath the tip region of the appressorium (Fig. 2, B and E), while at later stages the aggregates appeared more dispersed and excluded from the center of the penetration site (Fig. 2, A, D, and F). Furthermore, the signal recruiting PCS1 to attempted entry sites was able to cross cell frontiers (Fig. 2F, GFP-PCS1 from two adjacent cells organized as a ring around the fungal penetration site).
Figure 2.
Pathogen attack triggers translocation of cytoplasmic PCS1 and mitochondrial colocalization with PEN2 at fungal penetration sites. Three-week-old Arabidopsis seedlings were inoculated with the nonadapted powdery mildew Bgh. The epidermis of the first true leaves was visualized by confocal laser scanning microscopy 12–16 h after Bgh inoculation. The fungal structures are stained with propidium iodide except in GFP/RFP localization experiments. All pictures represent the average Z-projection of stacks of 30–50 sections, except in the case of C, where a single median section is shown. A and B, Localization at the attempted penetration site of GFP-PCS1 (A) or PCS1-GFP (B) in a pen4 mutant leaf epidermal cell. C Cytosolic localization of soluble GFP with RFP-PCS1 in noninfected epidermal cells. D, Translocation of RFP-PCS1, but not soluble GFP, into aggregates at the penetration site. E, Colocalization of RFP-PCS1 with mitochondria-associated PEN2-GFP at the penetration site. F, Localization of GFP-PCS1 in a pen2 mutant background after Bgh infection. Scale bars = 10 μm (A–C) or 5 μm (D–F).
As the PCS1 aggregation pattern at pathogen contact sites was reminiscent of PEN2 localization upon Bgh inoculation, we examined the localization of RFP-PCS1 in PEN2-GFP-expressing transgenic plants (Lipka et al., 2005). In pathogen-free plants, RFP-PCS1 showed a strong cytoplasmic fluorescence signal, similar to the GFP-tagged version described above, whereas PEN2-GFP was detectable in the cytoplasm and in the periphery of mobile membrane compartments (Supplemental Fig. S3, C–E) recently shown to represent peroxisomes and mitochondria (Fuchs et al., 2016). Pathogen challenge induced colocalization of RFP-PCS1 and PEN2-GFP in aggregate structures with enhanced fluorescence intensity (Fig. 2E). RFP-PCS1 did not translocate to all PEN2-GFP-tagged organelles surrounding attempted fungal entry sites, but was confined to a subset with increased PEN2-GFP fluorescence and aggregation (Supplemental Fig. S3E). These organelles were recently identified as a subpopulation of epidermal mitochondria, which become immobile at plant-pathogen contact sites (Fuchs et al., 2016).
To investigate a possible docking role of PEN2 for PCS1, we examined PCS1 relocalization after inoculation in a pen2-1 null mutant background (Lipka et al., 2005). Both GFP-PCS1 and RFP-PCS1 are able to aggregate underneath Bgh contact sites even in the absence of PEN2 (Fig. 2F; Supplemental Fig. S3F), suggesting that mitochondria-associated PEN2 is not necessary for the recruitment of PCS1 to these organelles.
PCS1 Catalytic Residues Are Not Required for Extracellular Defense
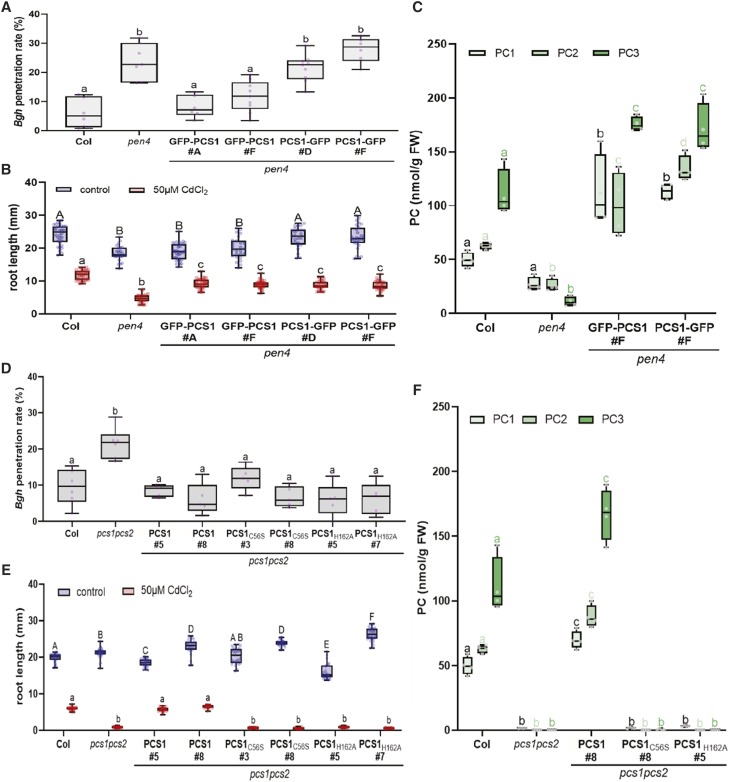
When preparing our fusion constructs to investigate the subcellular localization of PCS1 (GFP-PCS1 and PCS1-GFP), we found a striking difference that suggested the two functions of PCS1 in heavy metal tolerance and disease resistance could be uncoupled. Consistent with a previous study (Blum et al., 2010), both GFP fusions were soluble and were found to localize to the cytosol. To verify the functionality of the recombinant proteins, we tested their ability to complement the characteristic penetration resistance and Cd tolerance defects of the pen4 mutants. The Cd hypersensitivity of pen4 could be complemented by either GFP-PCS1 or PCS1-GFP fusions (Fig. 3B), which resulted, in both cases, in the accumulation of high levels of PC upon Cd treatment (Fig. 3C), suggesting that both fusions are functional. However, the penetration resistance defect of pen4 could only be complemented by the GFP-PCS1 fusion (Fig. 3A). Similar to GFP-PCS1, RFP-PCS1 was fully functional in both penetration resistance and heavy metal tolerance (Supplemental Fig. S3, A and B). Both GFP/RFP-PCS1 and PCS1-GFP relocalize as aggregates at focal sites of attempted fungal penetration (Fig. 2, A and B), indicating that the lack of complementation of entry resistance in pen4 by PCS1-GFP is not due to mislocalization of the PCS1-GFP fusion protein. The difference in complementation of the pen4 disease resistance phenotype by PCS1-GFP versus GFP-PCS1 is in accordance with an earlier study of Kühnlenz et al. (2015), who showed that the two functions of PCS1 in heavy metal tolerance and immunity can be uncoupled. This suggests that disease resistance conferred by PCS1 is independent of the ability of the enzyme to synthesize PC.
Figure 3.
Phytochelatin synthesis is required for heavy metal tolerance but not for nonhost resistance. A and D, Box plots showing the penetration rate of Bgh spores on leaves of 3-week-old Arabidopsis plants. Measurements were done 24 h postinfection on the first true leaves of five to nine individual plants (600–800 total penetration attempts counted per genotype). Different letters show statistical differences according to Tukey’s honestly significant difference (HSD) mean-separation test following one-way ANOVA. Lowercase letters indicate a significant genotype effect (P < 0.0001). B and E, Root length of 7-d-old seedlings grown in the presence or absence of Cd (50 µm CdCl2; between 25 and 30 individual seedlings for each genotype/treatment grown on the same plate were used for the measurement). Upper- and lowercase letters show statistical differences within treatment groups according to Tukey’s HSD mean-separation test following two-way ANOVA. There were significant main effects for Cd treatment and genotype (P < 0.001), and a significant interaction effect (P < 0.001). C and F, PC accumulation in 4-d-old seedlings transferred for 2 d onto 50 μm CdCl2. Lowercase letters show statistical differences within PC length groups according to Tukey’s HSD mean-separation test following two-way ANOVA. There were significant main effects for the genotype and the PC length factor (P < 0.001), and a significant interaction effect (P < 0.001).
To test this hypothesis, we generated catalytically inactive PCS1 mutants to investigate the importance of PC production for fungal resistance. The PCS activity of PCS1 is dependent on a highly conserved catalytic triad located in the N-terminal half of the protein, consisting of Cys-56, His-162 and Asp-180 (Romanyuk et al., 2006). Notably, the Cys at position 56 has been shown to be involved in the formation of an acyl-enzyme intermediate (Vatamaniuk et al., 2004). Thus, PCS1 proteins carrying C56A, C56S, or H162A substitutions lack PC synthesis activity in vitro (Vatamaniuk et al., 2004; Romanyuk et al., 2006). To test directly whether the function of PCS1 in fungal resistance is dependent on PC biosynthesis, we generated transgenic pcs1pcs2 Arabidopsis plants expressing, under its native promoter, the genomic coding sequence of wild-type PCS1 or constructs carrying either a C56S (PCS1C56S) or H162A (PCS1H162A) amino acid substitution in PCS1. As predicted by the inability of the mutated proteins to synthesize PC in vitro (Vatamaniuk et al., 2004; Romanyuk et al., 2006), transgenic lines expressing PCS1C56S or PCS1H162A were deficient in PC in vivo (Fig. 3F) and showed the same degree of root growth inhibition as pcs1pcs2 when grown on 50 μm Cd chloride (Fig. 3E). In contrast, in PCS1C56S or PCS1H162A lines, Bgh entry frequencies were similar to those seen in wild-type Col-0 plants or pcs1pcs2 complemented by PCS1 (Fig. 3D). These results confirm that the function of PCS1 in fungal resistance depends on a PCS1 activity or function that is distinct from PC synthesis.
PCS1, But Not PCS Activity, Is Involved in IG Metabolism to Restrict Powdery Mildew Entry in Arabidopsis Cells
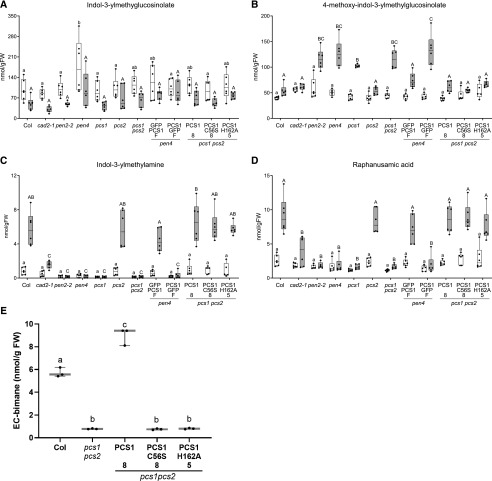
As indicated by the aberrant accumulation of 4MI3G and IG-metabolism products in pcs1 plants upon flg22 or Cd2+ treatment, or inoculation with P. infestans (Clay et al., 2009; De Benedictis et al., 2018; Matern et al., 2019), PCS1, like PEN2, is required for the pathogen-inducible metabolism of IGs. Similarly, upon inoculation with Bgh, pen2 and pen4 hyperaccumulate 4MI3G (Fig. 4B) and are not able to produce indol-3-ylmethylamine (I3A; Fig. 4C) and RA (Fig. 4D). Metabolism of IGs and their metabolic products is restored to wild-type levels when the pen4 or pcs1pcs2 mutants are complemented by either GFP-PCS1 or the mutated PCS1C56S or PCS1H162A, consistent with their wild-type-like disease resistance phenotype (Fig. 4, B–D). The ABC transporter PEN3/PDR8/ABCG36, although not deficient in I3A or RA (Bednarek et al., 2009), is thought to be involved in this IG metabolic pathway as an exporter of protoxins or toxic end products (Stein et al., 2006). This putative role would explain the proper accumulation of I3A/RA and hyperaccumulation of 4-O-β-d-glucosyl-indol-3-yl formamide (4OGlcI3F), a product presumably formed upon PEN2-mediated hydrolysis of 4MI3G, in pen3 leaves inoculated with Bgh (Lu et al., 2015). Our analysis of pcs1 mutants revealed that in addition to I3A and RA, PCS1 is also indispensable for 4OGlcI3F accumulation during the Arabidopsis response to pathogen inoculation (Supplemental Fig. S4A).
Figure 4.
PCS1 is involved in IG metabolism. A to D, Box plots showing accumulation of selected secondary metabolites, indicated as nmol/g of fresh tissue weight (FW), in Arabidopsis genotypes I3G (A), 4MI3G (B), I3A (C), and RA (D) 16 h after inoculation with Bgh conidiospores (gray bars; n = 6). Different letters show statistical differences according to Tukey’s honestly significant difference (HSD) mean-separation test following two-way ANOVA. We observed significant main effects for Bgh inoculation and genotype (P < 0.0001), and a significant interaction effect (P < 0.0001) for 4MI3G, I3A, and RA. In the case of I3G, significant main effects were observed for Bgh inoculation and genotype (P < 0.0001), but there was no apparent interaction effect. E, EC-bimane content in seedlings infiltrated with ECG-bimane measured by HPLC (n = 3). Lowercase letters show statistical differences according to Tukey’s HSD mean-separation test following one-way ANOVA. There was a significant genotype effect (P < 0.0001).
To directly test whether PEN2 and PEN4 operate in the same pathway, the two mutants were crossed. The pen2 pen4 double mutant supported a similar increase in entry frequency by Bgh as did the pen2 or pen4 single mutants (Supplemental Fig. S4B). Together with the colocalization with PEN2, these results suggest that PEN2 and PEN4 act together in one pathway that confers extracellular resistance to fungal attack. The correlation between the lack of pathogen-triggered accumulation of I3A and RA and defective entry resistance in pcs1 lines emphasizes the importance of PCS1 in IG metabolism.
Upon pathogen challenge, the pen4 mutant hyperaccumulates the physiological substrate of PEN2 myrosinase, 4MI3G, but not its unsubstituted precursor I3G (Fig. 4, A and B). In contrast, the pathogen-induced accumulation of both IG- and 4MI3G-derived hydrolysis products is essentially abolished (Fig. 4, C and D; Supplemental Fig. S4A). This yields a phenocopy of the metabolite profile seen in pen2 plants (Bednarek et al., 2009; Lu et al., 2015), suggesting that like PEN2, PCS1 may act downstream of 4MI3G biosynthesis to produce I3A, RA, and 4OGlcI3F, but upstream of the ABC transporter PEN3/PDR8.
Is PCS1 Peptidase Activity Required for IG Metabolism?
PCSs are known for their γ-glutamyl-Cys dipeptidyl transpeptidase activity (EC 2.3.2.15), which catalyzes the transfer of the dipeptide glutamyl-Cys (EC) from ECG onto another EC unit to generate the polypeptide PCs (EC)n-G. In addition, PCS has also been shown to be involved in the turnover of glutathione conjugates of xenobiotics by cleaving the Gly residue of their ECG moiety (Beck et al., 2003; Grzam et al., 2006; Blum et al., 2007).
It has been proposed that the IG-derived ITCs could be glutathionylated by GSTU13 and further processed by peptidases including PCS1 (Piślewska-Bednarek et al., 2018). To investigate whether the PC-deficient (pen4)-complementing mutants PCS1C56S and PCS1H162A still possessed peptidase activity, we assayed their deglycination activity on glutathionylated bimane (ECG-bimane; Blum et al., 2007). None of the PC-deficient lines could produce EC-bimane from ECG-bimane (Fig. 4E). This suggests that, unlike PCS1 metabolism of IG-derived compounds, both PC synthesis and the deglycination of glutathione conjugates depends on the residues C56 and H162 of the catalytic triad.
DISCUSSION
PCS is primarily known for its role in tolerance of heavy metals, notably Cd (Grill et al., 1989; Clemens et al., 1999). Here, we identified PEN4/AtPCS1 as a player in preinvasive immune responses against fungal pathogens. The second PCS paralogue, the weakly expressed protein AtPCS2, appears to be dispensable for both heavy metal tolerance and disease resistance. pcs1 mutants are hypersusceptible to adapted and nonadapted fungi, unveiling the importance of PCS1 for defense against fungal invasion. This impaired preinvasive immunity might also explain the previously reported strong cell death phenotype in Arabidopsis pcs1 plants in response to inoculation with the nonadapted oomycete pathogen P. infestans, because invasive growth of this oomycete in pen2 plants is also linked to a localized plant cell death response (Lipka et al., 2005; Kühnlenz et al., 2015). Like the other PEN proteins, PCS1 concentrates underneath the penetration site of fungal appressoria. Interestingly, in uninfected cells, PCS1 is cytosolic, and in response to infection it relocalizes to an endomembrane compartment(s) at incipient entry sites of attack. We did not observe this aggregation phenomenon after Cd treatment, further underlining the dual functions of PCS1. Colocalization of RFP-PCS1 and PEN2-GFP in plant cells under attack suggests that these compartments are mitochondria, as previously reported (Fuchs et al., 2016). Recent proteome studies localized PCS1 to the cytosol (Ito et al., 2011) and to the Arabidopsis mitochondrial complexome (Senkler et al., 2017), supporting its dual localization capacity.
Unlike the pen1 pen2 double mutant (Lipka et al., 2005), the susceptibility of the pen2 pen4 mutant to fungal infection is not more pronounced than that of either of the single mutants (Supplemental Fig. S4). This suggests that PEN2 and PEN4/PCS1 act in the same pathway. PEN2 has been shown to be an essential component of an IG metabolism pathway involved in broad-spectrum resistance to filamentous eukaryotic pathogens (Bednarek et al., 2009; Pastorczyk and Bednarek, 2016). PCS1 appears to be equally involved in this pathway, as the pcs1 mutants exhibit exactly the same I3A, RA, and 4OGlcI3F deficiencies as pen2 plants (Fig. 4, A and B; Supplemental Fig. S4A; Matern et al., 2019). The colocalization of both enzymes in response to pathogen attack could facilitate stimulus-induced metabolite channeling of these metabolites, which in turn could account for the lack of detection of the intermediates between I3G and I3A/RA or 4MI3G and 4OGlcI3F. However, this does not allow us to unambiguously determine whether PEN4 acts upstream of, downstream of, or in parallel with, PEN2.
The shared phenotype of the pen2 and pen4 mutants and the colocalization of PEN2 and PEN4 in the same compartment at the site of fungal ingress suggested a possible protein-protein interaction. However, we did not obtain any positive interactions between PEN2 and PEN4 in split-ubiquitin yeast two-hybrid experiments (Supplemental Fig. S5A), nor could we coimmunoprecipitate these two proteins from infected leaves (Supplemental Fig. S5B). The lack of physical interaction is confirmed by the fact that PCS1 is still able to relocalize in a null pen2 mutant background (Fig. 2F; Supplemental Fig. S3F). Focal accumulation of the PEN3 ABC transporter as disks in the plasma membrane and colocalization of PEN2 and PEN4 aggregates on mitochondria underneath pathogen contact sites rather points to pathogen-inducible macromolecular crowding as a potential alternative mechanism. Such an arrangement of PEN2 and PEN4 would facilitate metabolite channeling for targeted release of antimicrobials (Ellis, 2001; Stein et al., 2006; Fuchs et al., 2016; Guigas and Weiss, 2016). However, it remains to be tested whether enforced PEN4 mislocalization impairs its activity in extracellular defense.
Using mutated versions of PCS1, we showed that the function of PCS1 in IG metabolism does not require PCS activity sensu stricto, as we could generate PC-deficient mutants functional for IG metabolism. Indeed, the ability to metabolize IGs is retained in mutant versions of PCS1 (Fig. 4, A–D) that can neither produce any PCs (Fig. 3F) nor cleave the ECG bimane conjugate (Fig. 4E). Our findings confirm and extend a previous study in which heterologous expression of C. elegans PCS in the Arabidopsis pcs1 background complemented Cd hypersensitivity, but not a leaf cell death phenotype in response to P. infestans inoculation (Kühnlenz et al., 2015). Residue Cys-56 of the catalytic triad (Cys-56, His-162, and Asp-180) has been implicated in the formation of an acyl-enzyme intermediate with γ-EC. There appears to be an as yet unidentified second acylation site in the C-terminal part of PCS1, which shows a low level of sequence conservation across plant species (Vatamaniuk et al., 2004; Rea, 2012). This second site, which is absent in prokaryotic PCS, could potentially participate in IG metabolism. This hypothesis is supported by the fact that the PCS from C. elegans that was not able to complement the cell death phenotype in pcs1 plants has a significantly shorter C-terminal part compared to AtPCS1 (Rea et al., 2004; Kühnlenz et al., 2015). This assumed significance of the PCS1 C terminus could also explain why GFP-PCS1, but not the PCS1-GFP fusion, is nonfunctional in IG metabolism and extracellular defense despite its proper subcellular localization. Alternatively, rather than acting catalytically, PCS1 may instead stabilize a chemically labile intermediate in the IG metabolism pathway or a protein complex required for IG catabolism.
The chemical structures of RA and I3A, combined with the dependence of their formation on ECG supply via γ-glutamyl-Cys synthetase, suggested indol-3-ylmethyl-ITC-ECG conjugates as intermediates in pathogen-triggered IG metabolism in Brassicaceae (Bednarek et al., 2009). The existence of such conjugates in planta is supported by the identification of indolyl-dithiocarbamate glucoside during engineering of brassinin (an IG-derived phytoalexin from Brassica spp.) biosynthesis in Nicotiana benthamiana (Klein and Sattely, 2017). GSTU13 was recently found to be essential for pathogen-inducible formation of RA, I3A, and 4OGlcI3F, suggesting that it catalyzes the conjugation of unstable glucosinolate hydrolysis products, the indol-3-ylmethyl-ITCs, with ECG to form a dithiocarbamate adduct in planta (Piślewska-Bednarek et al., 2018). Although the exact mechanism of PCS1 in this pathway remains to be defined, our analysis combined with earlier work (Matern et al., 2019) suggests that PCS1 is a plausible candidate for further processing of this adduct to I3A and RA and the corresponding 4-methoxy adduct to 4OGlcI3F. However, as mentioned above, we cannot exclude the possibility that PCS1 acts upstream of GSTU13, or even in parallel to PEN2, or that it controls expression of the corresponding genes (Fig. 5). In conclusion, AtPCS1 is a multifunctional protein: it detoxifies xenobiotics via chelation of heavy metals by PC or via degradation of glutathione conjugates, and it participates in the production of antimicrobials formed from IGs in the PEN2 pathway.
Figure 5.
Function of PCS1 in pathogen-triggered IG metabolism. As indicated by a deficiency in IG hydrolysis products and hyperaccumulation of 4MI3G in pcs1 mutant plants, PCS1 acts downstream of intact IGs. PCS1 might contribute to the processing of the ITC-glutathione adduct, modulate the activity of other enzymes of the pathway, or control expression of the corresponding genes.
MATERIALS AND METHODS
Plant and Fungal Lines and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown for 15–20 d in growth chambers at 22°C with a 12-h photoperiod. All Arabidopsis mutants and transgenic lines were in the Col-0 background (Supplemental Table S2). Host powdery mildew (Golovinomyces cichoracearum UCSC1) was cultured on squash for 10–12 d and then applied to Arabidopsis using settling towers. Nonhost barley powdery mildew (Blumeria graminis f. sp. hordei CR3) was grown on the barley (Hordeum vulgare) line Algerian-S (CI-16138) and inoculated onto Arabidopsis using the methods described by Zimmerli et al. (2004).
Transgenic Arabidopsis lines were generated according to the floral dip method (Clough and Bent, 1998). T1 lines carrying single T-DNA insertion events were identified by screening their progeny for segregation of antibiotic resistance in the appropriate ratio. Resistant T2 seedlings were transferred to soil and used in pathogen assays, in which several representative independent lines were selected. T2 individuals homozygous for T-DNA insertions (based on 100% antibiotic resistance or 100% GFP fluorescence among their T3 progeny) were further used for pathogen resistance or heavy metal tolerance assays and metabolic analysis.
For assaying tolerance to Cd, Arabidopsis seeds were surface sterilized and sown onto agar plates containing one-half strength Murashige and Skoog medium and 50 μm Cd chloride (CdCl2). Plates were grown under continuous light at 22°C. After 7 d, plates were scanned and root lengths measured from scanned images using the software ImageJ. All experiments were repeated at least three times with similar results.
Map-Based Cloning of PEN4
The pen4 mutation was mapped to the bottom arm of chromosome 5 using an F2 mapping population generated from a cross between pen4 and Landsberg-erecta (Ler), according to the methods of Lukowitz et al. (2000). We narrowed down the region containing PEN4 to a genomic interval of ∼20 kb at the end of bacterial artificial chromosome (BAC) clone MRH10. Analysis of lists of genes that coexpress with either PEN1, PEN2, or PEN3 across publicly available microarray datasets (Obayashi et al., 2009) identified a gene highly correlated in expression with all three PEN genes (Supplemental Fig. S2) and that also resided in the 20 kb mapping interval. Sequencing of this gene, At5g44070, from the pen4 mutant showed that it harbored a G-to-A transition at nucleotide position 1,713 from the A of the translational start site of the genomic sequence. This mutation results in a predicted truncation of the AtPCS1 protein at residue 236.
Plasmids and DNA Constructs
PCS1 fusions to GFP or RFP were constructed using Gateway technology (Invitrogen). The Arabidopsis PCS1 (AtPCS1) genomic coding sequence was amplified by PCR using BAC clone MRH10 as template DNA and recombined into donor vector pDONR/Zeo (Invitrogen; Supplemental Table S1). The resulting entry vector was then recombined by a gateway LR reaction with the destination vectors pMDC43 and pMDC83 (Curtis and Grossniklaus, 2003) to produce expression clones GFP-PCS1 and PCS1-GFP, respectively. pDONR/Zeo AtPCS1 was recombined with a modified pEG106 (Gutierrez et al., 2009) containing a hemagglutinin-tagged mCherry to generate RFP-PCS1. GFP fusions were transformed into a pen4 mutant background (and selected on hygromycine 50 mg/L) and RFP-PCS1 into a pcs1pcs2 double mutant (selected on soil after Basta spray [BASF]; Blum et al., 2007). For complementation analyses, the entire PCS1 genomic coding sequence, in addition to 3 kb of promoter sequence upstream of the translational start site and 1 kb downstream of the translational stop codon, was amplified by PCR using BAC clone MRH10 as template DNA and recombined into donor vector pDONR/Zeo. The QuikChange mutagenesis kit (Stratagene) was used to introduce mutations in pDONR/Zeo PCS1, resulting in a change of Cys 56 to Ser (PCS1C56S) or of His 162 to Ala (PCS1H162A). The resulting entry vectors were then recombined with destination vector pGWB1 (conferring resistance to hygromycin; Nakagawa et al., 2007) to generate expression clone PCS1-wild type. These mutated PCS1 genomic clones were further recombined in the binary vector pGWB1. All expression clones were introduced into Agrobacterium tumefaciens (strain GV3101), and the resulting strains were used for stable transformation of Arabidopsis according to the floral dip method.
Generation of Double Mutants
Double mutants were generated by crossing the pen4 mutant to pen2-1, pen2-3, or pen3-1 mutants (Stein et al., 2006). F2 individuals homozygous at the mutated loci were identified using the cleaved amplified polymorphic sequence markers described. For the marker used to identify the pen4 mutation, the pen4 sequence was PCR-amplified with forward primer 5′taagacaaaacttgtggattgg3′ and reverse primer 5′cacccatctgatgaattgg3′, and the resulting DNA product was cleaved using the MboI restriction enzyme.
Microscopic Observations
Bgh penetration assays were carried out as described by Zimmerli et al. (2004), and stained with aniline blue according to Vogel and Somerville (2000). Successful penetrations (resulting in haustoria with extensive callose encasement) and failed penetrations (resulting in callosic papillae) were counted to determine the penetration rate (successful penetration/number of attempts). For imaging of fluorescent proteins in Arabidopsis epidermal cells, leaves from 2- to 3-week-old plants were mounted in a 0.01 mg/mL propidium iodide solution in water (to stain fungal structures). Leaves were examined between 12 and 18 h following fungal inoculation on a spinning-disc confocal microscope consisting of a Leica DMI 6000 B inverted microscope (Leica Microsystems) fitted with a Yokogawa CSU-10 spinning-disc confocal attachment (Yokogawa Electric) and a Photometrics QuantEM 512SC EM-CCD camera (Photometrics). Samples were mounted in water and observed with a 63× water immersion objective. Enhanced GFP was excited at 488 nm, and fluorescence was collected through a 525/50 nm band-pass filter and fungal structures stained with propidium iodide using a 620/60 band-pass filter (Chroma Technologies). RFP (mCherry) was excited at 561 nm, and fluorescence was collected through a 620/60 nm band-pass filter (Chroma Technologies). Microscope control and acquisition of images and z-series were accomplished using Metamorph software (Molecular Devices), and image processing was performed using ImageJ (W. Rasband, National Institutes of Health) software.
Metabolic Profiling
Analysis of PC biosynthesis and the bimane-conjugate hydrolysis assay were performed according to Blum et al. (2007). Extraction and analysis of IGs and their derivatives were performed according to Bednarek et al. (2011) and Lu et al. (2015).
Statistical Analysis
Statistical analysis was conducted with Graphpad Prism 8. Normality of samples was tested using the D’Agostino and Pearson test (P > 0.05). Normality was assumed for small size samples (n < 6). ANOVA was performed coupled to a Tukey’s honestly significant difference (HSD) mean-separation test in order to proceed to pairwise comparison between samples (confidence index, 95%). Statistical analyses between two samples were performed using Student’s t test (P-value = 0.05). In the box plots, the boxes indicate the first and third quartiles, the lines represent the median values, the whiskers indicate minimal and maximal values (excluding any outliers), and the points represent individual measurements.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. pen4 is allelic to cad1.
Supplemental Figure S2. pen4 is more susceptible to various fungal pathogens.
Supplemental Figure S3. Upon pathogen attack, the functional fusion protein RFP-PCS1 translocates from the cytosol to the surface of immobilized mitochondria tagged with PEN2-GFP.
Supplemental Figure S4. PEN2 and PCS1 act in the same biochemical and immune pathways.
Supplemental Figure S5. PCS1-PEN2 physical interaction assays.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. Mutant alleles used in this study.
Acknowledgments
We acknowledge the work of the late Ralph Blum (Technische Universtät München) for metabolite analysis and data processing shown in Figures 3, C and F, and 4E. We are grateful to Chris Cobbett (University of Melbourne) for providing the cad1 mutants and Phil Rea (University of Pennsylvania) for AtPCS1 complementary DNA; Ryan Guttierez (Carnegie Institution of Science) for the pEG106 mCherry vector; Chris Cobbett, Phil Rea, and Stephan Clemens (Universität Bayreuth) for helpful discussions; Bill Underwood (Energy Biosciences Institute) for advice and help in microscopy; Barbara Kracher (Max Planck Institute for Plant Breeding Research) for help with statistical analysis; and the Somerville lab for stimulating discussion.
Footnotes
This work was supported by the National Science Foundation and the Carnegie Institution for Science (S.S.), Stanford University (Graduate Fellowship to M.S.), the Max Planck Society (P.S.L.), the Deutsche Forschungsgemeinschaft (DFG; grants SPP1212 to P.S.L., GR 938 to E.G., and LI 1317/2-1 to V.L.), the Spanish Ministry of Economy and Competitiveness (MINECO; grants BIO2015-64077-R and BIO2012-32910 to A.M.), and the Polish National Science Centre (grant 2012/07/E/NZ2/04098 to P.B.).
Articles can be viewed without a subscription.
References
- Beck A, Lendzian K, Oven M, Christmann A, Grill E (2003) Phytochelatin synthase catalyzes key step in turnover of glutathione conjugates. Phytochemistry 62: 423–431 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Ver Loren van Themaat E, Maddula RK, Svatoš A, Schulze-Lefert P (2011) Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol 192: 713–726 [DOI] [PubMed] [Google Scholar]
- Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K (2017) Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol 55: 401–425 [DOI] [PubMed] [Google Scholar]
- Berens ML, Wolinska KW, Spaepen S, Ziegler J, Nobori T, Nair A, Krüler V, Winkelmüller TM, Wang Y, Mine A, et al. (2019) Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc Natl Acad Sci USA 116: 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Beck A, Korte A, Stengel A, Letzel T, Lendzian K, Grill E (2007) Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J 49: 740–749 [DOI] [PubMed] [Google Scholar]
- Blum R, Meyer KC, Wünschmann J, Lendzian KJ, Grill E (2010) Cytosolic action of phytochelatin synthase. Plant Physiol 153: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J 18: 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniawski P, Bednarek P (2018) Glutathione S-Transferases in the biosynthesis of sulfur-containing secondary metabolites in Brassicaceae plants. Front Plant Sci 9: 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis M, Brunetti C, Brauer EK, Andreucci A, Popescu SC, Commisso M, Guzzo F, Sofo A, Ruffini Castiglione M, Vatamaniuk OK, et al. (2018) The Arabidopsis thaliana knockout mutant for Phytochelatin synthase1 (cad1-3) is defective in callose deposition, bacterial pathogen defense and auxin content, but shows an increased stem lignification. Front Plant Sci 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. (2001) Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci 26: 597–604 [DOI] [PubMed] [Google Scholar]
- Fuchs R, Kopischke M, Klapprodt C, Hause G, Meyer AJ, Schwarzländer M, Fricker MD, Lipka V (2016) Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28: 130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Löffler S, Winnacker EL, Zenk MH (1989) Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific γ-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc Natl Acad Sci USA 86: 6838–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH (1985) Phytochelatins: The principal heavy-metal complexing peptides of higher plants. Science 230: 674–676 [DOI] [PubMed] [Google Scholar]
- Grzam A, Tennstedt P, Clemens S, Hell R, Meyer AJ (2006) Vacuolar sequestration of glutathione S-conjugates outcompetes a possible degradation of the glutathione moiety by phytochelatin synthase. FEBS Lett 580: 6384–6390 [DOI] [PubMed] [Google Scholar]
- Guigas G, Weiss M (2016) Effects of protein crowding on membrane systems. Biochim Biophys Acta 1858: 2441–2450 [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Howden R, Goldsbrough PB, Andersen CR, Cobbett CS (1995) Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol 107: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Batth TS, Petzold CJ, Redding-Johanson AM, Mukhopadhyay A, Verboom R, Meyer EH, Millar AH, Heazlewood JL (2011) Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J Proteome Res 10: 1571–1582 [DOI] [PubMed] [Google Scholar]
- Klein AP, Sattely ES (2017) Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proc Natl Acad Sci USA 114: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44: 516–529 [DOI] [PubMed] [Google Scholar]
- Kühnlenz T, Westphal L, Schmidt H, Scheel D, Clemens S (2015) Expression of Caenorhabditis elegans PCS in the AtPCS1-deficient Arabidopsis thaliana cad1-3 mutant separates the metal tolerance and non-host resistance functions of phytochelatin synthases. Plant Cell Environ 38: 2239–2247 [DOI] [PubMed] [Google Scholar]
- Lipka U, Fuchs R, Lipka V (2008) Arabidopsis non-host resistance to powdery mildews. Curr Opin Plant Biol 11: 404–411 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Lu X, Dittgen J, Piślewska-Bednarek M, Molina A, Schneider B, Svatoš A, Doubský J, Schneeberger K, Weigel D, Bednarek P, et al. (2015) Mutant allele-specific uncoupling of PENETRATION3 functions reveals engagement of the ATP-binding cassette transporter in distinct tryptophan metabolic pathways. Plant Physiol 168: 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible W-R (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern A, Böttcher C, Eschen-Lippold L, Westermann B, Smolka U, Döll S, Trempel F, Aryal B, Scheel D, Geisler M, et al. (2019) A substrate of the ABC transporter PEN3 stimulates bacterial flagellin (flg22)-induced callose deposition in Arabidopsis thaliana. J Biol Chem 294: 6857–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorczyk M, Bednarek P (2016) The function of glucosinolates and related metabolites in plant innate immunity In Kopriva S, ed, Glucosinolates. Academic Press Ltd-Elsevier Science Ltd, London, pp 171–198 [Google Scholar]
- Piślewska-Bednarek M, Nakano RT, Hiruma K, Pastorczyk M, Sanchez-Vallet A, Singkaravanit-Ogawa S, Ciesiołka D, Takano Y, Molina A, Schulze-Lefert P, et al. (2018) Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol 176: 538–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. (2006) Phytochelatin synthase, papain’s cousin, in stereo. Proc Natl Acad Sci USA 103: 507–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. (2012) Phytochelatin synthase: Of a protease a peptide polymerase made. Physiol Plant 145: 154–164 [DOI] [PubMed] [Google Scholar]
- Rea PA, Vatamaniuk OK, Rigden DJ (2004) Weeds, worms, and more. Papain’s long-lost cousin, phytochelatin synthase. Plant Physiol 136: 2463–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanyuk ND, Rigden DJ, Vatamaniuk OK, Lang A, Cahoon RE, Jez JM, Rea PA (2006) Mutagenic definition of a papain-like catalytic triad, sufficiency of the N-terminal domain for single-site core catalytic enzyme acylation, and C-terminal domain for augmentative metal activation of a eukaryotic phytochelatin synthase. Plant Physiol 141: 858–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I, Berrocal M, Marco Y, Somerville S, Molina A (2009) The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant Microbe Interact 22: 953–963 [DOI] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun H-P (2017) The mitochondrial complexome of Arabidopsis thaliana. Plant J 89: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W, Somerville SC (2008) Focal accumulation of defences at sites of fungal pathogen attack. J Exp Bot 59: 3501–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA (2004) Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with gamma-glutamylcysteine during catalysis: Stoichiometric and site-directed mutagenic analysis of Arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. J Biol Chem 279: 22449–22460 [DOI] [PubMed] [Google Scholar]
- Vogel J, Somerville S (2000) Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA 97: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40: 633–646 [DOI] [PubMed] [Google Scholar]