Abstract
Receptor-like protein kinase complexes regulate plant growth and development.
Cell-to-cell and cell-to-environment communications are essential for plant growth, development, and adaptation to various environmental conditions. Receptor-like protein kinases (RLKs) are critical plasma membrane-localized proteins transducing extracellular signals into the cell to coordinate cellular activities during these processes. ZmPK1, the first plant RLK isolated via screening a maize (Zea mays) root cDNA library, contains a transmembrane domain linking a cytoplasmic Ser/Thr protein kinase domain and an extracellular domain that is similar to the S-locus glycoproteins of Brassica (Walker and Zhang, 1990). Subsequently, a growing number of plant RLK genes were identified, such as S-LOCUS RECEPTOR KINASE from Brassica oleracea (Stein et al., 1991) as well as ARABIDOPSIS RECEPTOR KINASE1 (Tobias et al., 1992) and TRANSMEMBRANE KINASE1 from Arabidopsis (Arabidopsis thaliana; Chang et al., 1992). Since this class of plant protein kinases show structural similarity to the mammalian growth factor receptor protein kinases, but no corresponding ligands of these putative receptors were identified at the time, they were designated as RLKs (Walker, 1993, 1994). Xa21, an RLK conferring rice (Oryza sativa) pathogen recognition and response, was the first plant RLK with defined biological function (Song et al., 1995). Thereafter, bioinformatics analyses identified an RLK superfamily that contains more than 610 and 1,131 members in the genomes of Arabidopsis and rice, respectively (Shiu and Bleecker, 2001; Shiu et al., 2004). Among these, there are around 420 typical RLKs in Arabidopsis, and the rest were named receptor-like cytoplasmic kinase (RLCK) due to absence of the extracellular domain (Shiu and Bleecker, 2001). Henceforth, a broad avenue was opened for investigating the molecular mechanisms underlying cell signaling in response to various extracellular signals during plant growth and development.
In the past two decades, the biological functions of a number of RLKs have been revealed. Many of them mediate signals of canonical phytohormones or peptide hormones to control plant development. For example, BRASSINOSTEROID INSENSITIVE1 (BRI1) perceives brassinosteroids to regulate cell elongation and plant growth (Li and Chory, 1997; Clouse and Sasse, 1998; Wang et al., 2001). Sulfated peptide phytosulfokine (PSK) is perceived by PSK RECEPTOR1 (PSKR1) to regulate cell division and expansion (Matsubayashi and Sakagami, 1996; Matsubayashi et al., 2002, 2006). Some RLKs, including the Catharanthus roseus RLK1-LIKE (CrRLK1L) subfamily members FERONIA (FER), ANXUR1/2 (ANX1/2), and BUDDHA’S PAPER SEAL1/2 (BUPS1/2), regulate pollen tube-female gametophyte interactions during fertilization (Huck et al., 2003; Escobar-Restrepo et al., 2007; Boisson-Dernier et al., 2009; Miyazaki et al., 2009; Ge et al., 2017). In addition, RLKs are also involved in the recognition of molecular pattern elicitors to regulate plant immune responses. For instance, FLAGELLIN-SENSING2 (FLS2) is required for the perception of bacterial flagellin (Gómez-Gómez and Boller, 2000). ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR) is required for the EF-Tu-triggered response (Zipfel et al., 2006). PEP1 RECEPTOR1 (PEPR1) and PEPR2 sense endogenous danger signal Pep peptides (Yamaguchi et al., 2006, 2010; Krol et al., 2010). In addition, CHITIN ELICITOR RECEPTOR KINASE1 specifically interacts with chitin oligomers (Miya et al., 2007; Wan et al., 2008). To date, only a small portion of RLKs have been functionally characterized.
Most functionally defined RLK receptors require a different RLK as a coreceptor to recognize diverse extracellular signals. BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) was the first identified RLK coreceptor essential for brassinosteroid signaling (Li et al., 2002; Nam and Li, 2002). BAK1, also designated as SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 (SERK3), belongs to a small SERK subfamily that comprises five members in Arabidopsis (Hecht et al., 2001). Daucus carota SERK (DcSERK) was originally identified in carrot suspension cell cultures as a marker for competent and embryogenic cells during somatic embryogenesis (Schmidt et al., 1997). Arabidopsis SERKs were recently demonstrated to control stem cell division patterns during zygotic embryogenesis (Li et al., 2019). The extracellular domains of all five Arabidopsis SERKs contain five leucine-rich repeats (LRRs), which are relatively short compared with most known ligand-binding RLK receptors, such as BRI1 with 25 LRRs in its extracellular domain (Chinchilla et al., 2009). BAK1 and SERKs function as coreceptors that can directly interact with diverse ligand-binding RLK receptors to regulate a variety of biological processes (Li, 2010; Ma et al., 2016). For example, BAK1 is required by BRI1 to mediate brassinosteroid signaling (Li et al., 2002; Nam and Li, 2002). SERK1 and SERK2 are required for EXCESS MICROSPOROCYTES1 (EMS1)/EXTRA SPOROGENOUS CELLS (EXS)-regulated tapetum specification (Li et al., 2017). The pattern recognition receptors FLS2, EFR, and PEPR1/2 also recruit BAK1/SERKs as coreceptors to regulate plant immune responses (Chinchilla et al., 2007; Heese et al., 2007; Schulze et al., 2010; Roux et al., 2011; Antolín-Llovera et al., 2012; Böhm et al., 2014). More recently, CLV3 INSENSITIVE RECEPTOR KINASES (CIKs) were uncovered as another group of coreceptors that control anther cell specification and the homeostasis of the shoot apical meristem (SAM; Cui et al., 2018; Hu et al., 2018). Both SERK and CIK subfamilies belong to group II LRR-RLKs. Several recent studies revealed that a group of glycosylphosphatidylinositol (GPI)-anchored proteins, including LORELEI and its homologs LORELEI-like-GPI-anchored protein1 (LLG1), LLG2, and LLG3, are also required as coreceptors of CrRLK1L receptors FER, ANX1/2, and BUPS1/2 to control root growth, pollen cell wall integrity, and pollen tube reception (Li et al., 2015; Liu et al., 2016; Feng et al., 2019; Ge et al., 2019; Xiao et al., 2019). In this review, we focus on current advances in RLK-SERK and RLK-CIK complexes during plant growth and development.
THE BRI1-SERK COMPLEX REGULATES BRASSINOSTEROID SIGNALING
Brassinosteroids are a group of phytohormones essential for cell elongation and plant growth (Clouse and Sasse, 1998). BRI1, a typical RLK containing an LRR-type extracellular domain, was first identified as a putative brassinosteroid receptor (Li and Chory, 1997). A number of later analyses demonstrated that BRI1 is indeed the receptor of brassinosteroids. For example, photoaffinity cross-linking analyses indicated that LRR22 and the 70-amino acid island domain between LRR21 and LRR22 in the extracellular domain of BRI1 are required for brassinosteroid perception (Kinoshita et al., 2005). Structural analysis further demonstrated that the island domain and LRR21 to LRR25 form a surface pocket in the interior of the superhelical BRI1 extracellular domain for brassinosteroid binding (Hothorn et al., 2011; She et al., 2011).
Although the physical interaction of brassinolide, the most active form of brassinosteroids, with BRI1 does not require the coreceptor BAK1 (Kinoshita et al., 2005), genetic results strongly demonstrated that BAK1 and SERKs are absolutely required for brassinosteroid signaling (Li et al., 2002; Gou et al., 2012). BRI1 and BAK1 interact and transphosphorylate each other in a brassinosteroid-dependent manner in planta (Li et al., 2002; Wang et al., 2005, 2008). The dark-grown serk1 bak1 bkk1 triple mutant exhibits a typical deetiolation phenotype with opened cotyledons and deetiolated hypocotyls, which is similar to null bri1 mutants. The phosphorylation levels of BRI1 in serk1 bak1 bkk1 triple mutants cannot be induced upon exogenous application of brassinolide, indicating that phosphorylation and activation of BRI1 depend on functional BAK1 and SERKs. Consistent with the disruption of the brassinosteroid signaling, the downstream unphosphorylated active bri1-EMS-SUPPRESSOR1 (BES1) cannot be detected in the triple serk mutants with or without the treatment of exogenous brassinolide (Gou et al., 2012). Further structural analyses revealed that brassinosteroids act as molecular glue to mediate the physical interaction between the extracellular domains of BRI1 and BAK1. BAK1 is recruited to the interaction platform after the perception of brassinosteroids by BRI1 to stabilize the interaction and activate the downstream signaling pathway (Santiago et al., 2013; Sun et al., 2013). These results demonstrated that BAK1 and other SERK members function as essential coreceptors in BRI1-mediated brassinosteroid signaling (Fig. 1A). It was proposed that the interaction between BRI1 and BAK1 may stabilize the αC helix, which may be necessary for the activation of these two RLKs upon brassinosteroid binding (Moffett et al., 2017). Recent results demonstrated that the interaction between BRI1 and BAK1 is tightly regulated to appropriately transduce the brassinosteroid signal. For instance, BAK1-INTERACTING RECEPTOR-LIKE KINASE3 (BIR3), a member of LRR-RLK subgroup X, can directly interact with BRI1 and BAK1 in the absence of brassinosteroid to prevent the formation of the BRI1-BAK1 receptor complex, thereby negatively regulating brassinosteroid signaling (Imkampe et al., 2017; Großeholz et al., 2019). Consistent with this, structural analyses revealed that the elongated mutant of BAK1 contains a single amino acid substitution in the core of the SERK-BIR complex interface, which disrupts the interaction of BAK1 to BIRs, resulting in enhanced brassinosteroid signaling (Whippo and Hangarter, 2005; Hohmann et al., 2018a). By contrast, the immunophilin-like FK506-BINDING PROTEIN42/TWISTED DWARF1 promotes the interaction and phosphorylation of BRI1 and BAK1 in response to brassinosteroids (Chaiwanon et al., 2016; Zhao et al., 2016). The regulation of interaction and phosphorylation of the receptor and coreceptor provides an effective way to fine-tune early brassinosteroid signaling as well as integrate other external signals, such as phytohormones and pathogen stresses. Consistent with the vital and conserved functions of brassinosteroid signaling, homologous BRI1 and BAK1/SERKs have also been identified in crop plants, such as rice (Nakamura et al., 2006; Li et al., 2009; Park et al., 2011; Zhang et al., 2016a), maize (Kir et al., 2015), and tomato (Solanum lycopersicum; Bajwa et al., 2013; Peng and Kaloshian, 2014).
Figure 1.
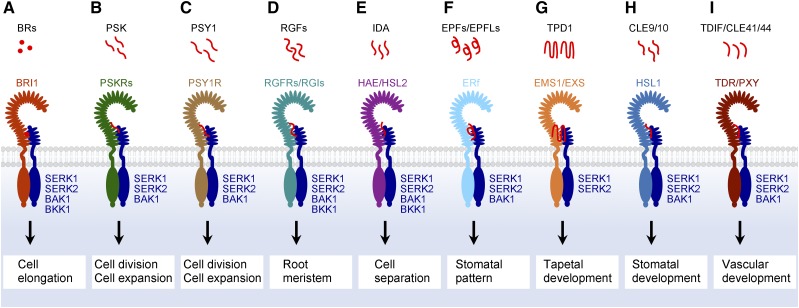
SERKs mediate a variety of signals to regulate plant growth and development. Extracellular signals, phytohormones or peptides, are perceived by RLK receptors with a large extracellular LRR domain. SERKs with a small extracellular LRR domain are recruited to form complete ligand-receptor-coreceptor complexes that transduce extracellular signals into intracellular responses for diverse biological functions. A, Phytohormone brassinosteroids (BRs) are perceived by the BRI1-SERK complex to regulate cell elongation. B and C, PSKR-SERK (B) and PSY1R-SERK (C) complexes perceive PSK and PSY1 peptides to control cell division and cell expansion, respectively. D, RGFs are perceived by the RGFR/RGI-SERK complex to regulate the root meristem. E, The HAE/HSL2-SERK complex senses IDA peptide to regulate cell separation processes. F, EPF/EPFL family peptides are perceived by the ERf-SERK complex to control the stomatal pattern. G, TPD1 peptide is perceived by the EMS1/EXS-SERK complex to specify tapetal cells. H, CLE9/10 is perceived by the HSL1-SERK complex to regulate stomatal development. I, The TDR/PXY-SERK complex perceives the TDIF/CLE41/44 peptide to regulate vascular development.
PSKR/PSY1R/RGI-SERK PAIRS PERCEIVE SULFATED PEPTIDES TO REGULATE CELL PROLIFERATION AND ROOT MERISTEM MAINTENANCE
Besides their roles in response to the canonical phytohormones, brassinosteroids, RLKs also mediate peptide hormones to regulate various processes in plant growth and development (Murphy et al., 2012; Hirakawa and Sawa, 2019). PSK is a posttranslationally sulfated pentapeptide that was identified from suspension cultures of Asparagus officinalis and rice and that can effectively promote cell proliferation at nanomolar concentrations (Matsubayashi and Sakagami, 1996; Matsubayashi et al., 1997). Preprophytosulfokine genes were identified in Arabidopsis and crop plants, such as rice, maize, and cotton (Gossypium hirsutum; Yang et al., 1999; Lorbiecke et al., 2005; Han et al., 2014; Sauter, 2015), suggesting that they possess conserved roles in regulating plant development. PSK acts as a non‐cell-autonomous signal to promote growth, which is dependent on brassinosteroid biosynthesis and signaling (Hartmann et al., 2013). PSK signaling is also required for pollen tube growth and funicular pollen tube guidance in Arabidopsis and fiber elongation in cotton (Han et al., 2014; Stührwohldt et al., 2015). PSK is perceived at the plasma membrane by PSKR1, an LRR-RLK containing 21 LRRs and a 36-amino acid island between the 17th and 18th LRRs (Matsubayashi et al., 2002, 2006). When PSKR1 and its closest paralog PSKR2 were both knocked out, the double mutant exhibited significantly shortened roots that were insensitive to PSK, indicating that the Arabidopsis genome possesses two PSK receptors to redundantly control cell proliferation (Amano et al., 2007). PSKR1 possesses kinase, guanylate cyclase, and Ca2+/calmodulin-binding activities that are required for PSK signaling (Kwezi et al., 2011; Hartmann et al., 2014; Kaufmann et al., 2017). Biochemical analyses indicated that PSKR1 interacts with BAK1. PSK-induced root growth and protoplast expansion are extremely impaired in both bak1-3 and bak1-4 (Ladwig et al., 2015). In addition, both the pskr1-3 pskr2-1 and serk1-3 serk3-1 serk4-1 mutants produced increased numbers of metaxylem cell compared with the wild type, indicating that PSKR1/2 and BAK1/SERKs suppress xylem identity. RLP44, a receptor-like protein (RLP), functions as a scaffold to promote the interaction between PSKR1 and BAK1 (Holzwart et al., 2018). These results suggested that BAK1 may function as a coreceptor to transduce the PSK signal similar to that in BRI1-mediated brassinosteroid signaling. PSK can effectively induce PSKR1 to form a heterodimer with BAK1 and other SERKs. However, structural results showed that PSK is not directly involved in the PSKR-SERK interface, which is different from the interaction of BRI1 and BAK1. PSK binding allosterically induces PSKR-SERK interaction, in which the completely disordered island domain of free PSKR is well stabilized for the subsequent recruitment of a SERK as the coreceptor to form a stable PSKR-SERK complex (Fig. 1B; Wang et al., 2015 ).
PLANT PEPTIDE CONTAINING SULFATED TYROSINE1 (PSY1) is another 18-amino acid Tyr-sulfated glycopeptide identified from Arabidopsis suspension cell culture, which can significantly promote cellular proliferation and expansion. PSY1R, an LRR-RLK paralogous to PSKR1, is the receptor perceiving PSY1 (Amano et al., 2007). Since both PSK and PSY1 promote cell proliferation and the pskr1 pskr2 psy1r triple knockout mutant showed significantly decreased cell size and cell number, it was proposed that the paralogous receptors PSKR1/2 and PSY1R integrate two distinct growth-promoting peptide signals to redundantly control cellular proliferation and plant growth (Amano et al., 2007). A recent study revealed that PSY1R and BAK1/SERKs can interact with and mutually transphosphorylate each other, suggesting that BAK1 and SERKs may also function as coreceptors of PSY1R to transduce the PSY1 signal and promote plant growth (Oehlenschlæger et al., 2017). Genetic and structural results are still required to test whether BAK1 and SERKs are bona fide coreceptors of PSY1R (Fig. 1C).
A tyrosylprotein sulfotransferase (TPST) was identified in Arabidopsis that can catalyze Tyr sulfation of both PSY1 and PSK (Komori et al., 2009). tpst mutants displayed similar but more severe phenotypes compared with the pskr1 pskr2 psy1r triple mutant (Amano et al., 2007; Komori et al., 2009), leading to the identification of another group of Tyr-sulfated peptides named ROOT MERISTEM GROWTH FACTORS (RGFs) that can rescue the extreme short-root phenotype of tpst when PSK and PSY1 are both supplied (Matsuzaki et al., 2010). RGFs are expressed in the quiescent center and columella stem cells. RGFs diffuse with a gradient into the root meristem to define the expression levels of PLETHORA (PLT) transcription factors and thereby to maintain the root stem cell niche. Loss of function of RGFs results in a short-root phenotype due to impaired root meristem (Aida et al., 2004; Matsuzaki et al., 2010). RGF peptides were also named CLE-LIKE (CLEL)/GOLVEN (GLV) because they contain a conserved CLEL/GLV domain responsible for root length, root growth direction, and lateral root initiation (Meng et al., 2012; Whitford et al., 2012).
Matsubayashi’s group expressed all 95 RLKs that could serve as ligand receptors with a large extracellular domain in tobacco (Nicotiana tabacum) BY-2 cells. Three LRR-RLKs named RGF RECEPTORs (RGFRs) were identified by exhaustive binding assay using photoaffinity-labeled RGF1 against membrane fractions of the transgenic lines (Shinohara et al., 2016). A total of five RGFRs, also designated as RGF1 INSENSITIVE (RGI) proteins, were independently identified by two other groups based on yeast two-hybrid screening and ligand recognition motif analysis, respectively (Ou et al., 2016; Song et al., 2016). The stability of these RGFR/RGI receptors are finely tuned by the ubiquitin-specific proteases UBP12 and UBP13 (An et al., 2018). The gradients of PLT1/2 in the root meristem were drastically impaired in the rgfr/rgi mutants that showed extremely shortened roots and were insensitive to exogenously applied RGF peptides, consistent with their role as the receptors of RGFs (Ou et al., 2016; Shinohara et al., 2016).
Two conserved motifs (RxR and RxGG) in the extracellular domains of RGFRs/RGIs were revealed to be crucial for the recognition of RGF1 by its receptors. Biochemical assays showed that RGF1 can induce the interaction between the extracellular LRR domains of RGFRs/RGIs and BAK1/SERKs, suggesting that SERK family members function as coreceptors of RGFRs/RGIs. This notion was further supported by genetic analyses with serk mutants that displayed short roots with smaller meristems (Song et al., 2016; Fig. 1D). In line with this, RGFRs/RGIs were also identified by a yeast two-hybrid screening using BAK1 as a bait (Ou et al., 2016). It was proposed that the new surface formed by the interaction of RGFRs/RGIs and RGFs could be used as a platform to recruit a SERK member to mediate RGF signaling (Song et al., 2016). In addition, the wavy growth of wild-type roots upon RGF1 treatment disappeared in the rgi1/2/3/4 quadruple mutants (Ou et al., 2016), indicating that the gravitropic response caused by RGF treatment may also be mediated by the RGFR/RGI receptors.
THE HAE-SERK COMPLEX SENSES THE HYDROXYLATED PEPTIDE IDA TO REGULATE CELL SEPARATION PROCESSES
INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and eight IDA-LIKE (IDL) homologs in Arabidopsis are a different type of peptide (Butenko et al., 2003). The mature active IDA is generated from the C terminus of its preproprotein and posttranslationally hydroxylated on its Pro residues (Vie et al., 2015). IDA is specifically expressed in floral abscission zones. ida mutants show altered abscission characteristics, and their senesced floral organs remain attached to the plant (Butenko et al., 2003). Consistent with this phenotype, constitutive and ectopic expression of IDA resulted in earlier abscission of floral organs and ectopic abscission of other organs, such as branches and cauline leaves (Stenvik et al., 2006). IDA encodes a secreted precursor protein containing a conserved motif named extended PIP (EPIP) that is sufficient to rescue the defective abscission phenotype seen in the ida mutants, and synthetic EPIP peptide induces abscission in the wild type and ida mutants, suggesting that proteolytic processing is required to produce a mature and bioactive form of IDA (Stenvik et al., 2008). Three subtilisin-like proteinases involved in processing the IDA precursor into the mature and bioactive 14-amino acid peptide were identified by biochemical approaches (Schardon et al., 2016). The central Pro residue in the minimal 12-amino acid IDA peptide must be hydroxylated for its activity (Butenko et al., 2014; Santiago et al., 2016).
HAESA (HAE) and its paralog HAESA-LIKE2 (HSL2), two LRR-RLKs, are also strongly expressed in the floral organ abscission zones (Jinn et al., 2000; Cho et al., 2008). Similar to the ida mutants, the hae hsl2 double mutants exhibit strong abscission defects and the floral organs fail to abscise (Cho et al., 2008; Stenvik et al., 2008). Overexpression of IDA cannot restore the abscission defects of the hae hsl2 double mutants, demonstrating that the function of IDA relies on HAE/HSL2, and IDA and HAE/HSL2 act in the same signaling pathway (Cho et al., 2008; Stenvik et al., 2008). The repressed expression of HAE by AGAMOUS-LIKE15 (AGL15) can be released through the abscission process when AGL15 is phosphorylated by a MKK4/5-MAPK3/6 cascade, which thus regulates the IDA-mediated floral organ abscission in a positive feedback manner (Cho et al., 2008; Patharkar and Walker, 2015). Higher-order serk mutants display defective floral organ abscission similar to that of the hae hsl2 and ida mutants, indicating that SERKs redundantly regulate floral organ shedding. IDA can induce heterodimerization via the extracellular domains of HAE/HSL2 and SERKs, and the IDA signal can be transduced by transphosphorylations between HAE/HSL2 and SERKs (Meng et al., 2016). Structural results demonstrated that SERKs function as coreceptors of HAE/HSL2. In the presence of SERK1, HAE perceives hydroxylated IDA with a much higher binding affinity (Santiago et al., 2016; Fig. 1E). Similar IDA/IDL1-HAE/HSL2 ligand-receptor pairs are also required to control other cell separation processes, including lateral root emergence and root cap sloughing (Aalen et al., 2013; Kumpf et al., 2013; Shi et al., 2018; Zhu et al., 2019). However, no coreceptors of HAE/HSL2 have been reported yet in regulating lateral root emergence and root cap sloughing. The IDA/IDL peptide signaling is evolutionarily conserved, and some key components of this pathway have been identified in all orders of flowering plants, including some important crops (Shi et al., 2019). Genetic breeding targeting these genes may generate crop varieties to decrease severe yield and quality losses caused by untimely organ abscission.
ERF/EMS1 (EXS)-SERK PAIRS MEDIATE CYS-RICH PEPTIDES TO REGULATE STOMATA PATTERN AND TAPETUM SPECIFICATION
The EPIDERMAL PATTERNING FACTOR (EPF) and EPF-LIKE (EPFL) peptide family in Arabidopsis comprises 11 small secreted Cys-rich peptides, including EPF1, EPF2, and nine EPFLs. Mature EPF peptides contain six or eight conserved Cys residues that form intramolecular disulfide bonds important for their functions (Silverstein et al., 2007; Hara et al., 2009; Ohki et al., 2011; Torii, 2012). EPF1 confers the one-cell-spacing stomatal pattern (stomata are not adjacent to each other). Overexpression of EPF1 resulted in decreased stomatal density while the epf1 mutant exhibited clustered stomata and moderately increased stomatal density (Hara et al., 2007). Different from EPF1, EPF2 inhibits protoderm cells from entering into the stomatal lineage, and excessive numbers of stomata and numerous small epidermal cells are produced in the epf2 mutant while most stomata still obey the one-cell-spacing rule (Hara et al., 2009; Hunt and Gray, 2009). By contrast, STOMAGEN, also named EPFL9, a mesophyll-derived inner-tissue EPF/EPFL family peptide, positively regulates stomatal development in the epidermis. Overexpression of STOMAGEN leads to numerous clustered stomata, whereas loss of function of STOMAGEN results in reduced stomatal density (Hunt et al., 2010; Kondo et al., 2010; Sugano et al., 2010). Three other EPFL peptides, EPFL6/CHALLAH, EPFL5/CHALLAH-LIKE1 (CLL1), and EPFL4/CLL2, are involved in region-specific regulation of stomatal production (Abrash and Bergmann, 2010; Abrash et al., 2011).
The ERECTA subfamily (ERf) LRR-RLKs, including ERECTA (ER), ERECTA-LIKE1 (ERL1), and ERL2, play overlapping but diverse roles as negative regulators to promote pavement cell differentiation and inhibit stomatal lineage formation. The triple er mutants generate high-density stomatal clusters (Shpak et al., 2005; Shpak, 2013). Overexpression of EPF1/2 and STOMAGEN in the er triple mutants cannot decrease stomatal density (Hara et al., 2007, 2009; Lee et al., 2015). On the other hand, the epf2 er double mutant exhibited stomatal density similar to the er single mutant (Hunt and Gray, 2009). The er erl1 erl2 epf1 epf2 and stomagen er erl1 erl2 mutants exhibit stomatal density similar to the er triple mutants (Hara et al., 2009; Lee et al., 2015). These results demonstrated that ERf members are necessary for EPF/EPFL peptides to regulate stomatal development.
Further protein interaction analyses indicated that ERf RLKs are the primary receptors of EPF1/2 and STOMAGEN (Lee et al., 2012, 2015). EPF2-ER and EPF1-ERL1 pairs initiate asymmetric entry divisions for stomatal lineage fate and enforce spacing divisions, respectively (Lee et al., 2012). EPF2 activates ER-mediated signaling to inhibit stomatal initiation, whereas STOMAGEN competes with EPF2 to bind to ER for blocking the signaling pathway, leading to optimal stomatal patterning (Lee et al., 2015). TOO MANY MOUTHS (TMM), an LRR-RLP, is also required to form a receptor complex with ERf for EPF binding in controlling stomatal development with distinct functions in different organs (Nadeau and Sack, 2002; Shpak et al., 2005; Hara et al., 2007, 2009; Hunt and Gray, 2009; Kondo et al., 2010; Sugano et al., 2010; Lee et al., 2012, 2015). However, structural results suggested that a coreceptor is still necessary for ERfs to transduce EPF signals, since EPFs cannot induce ERfs to dimerize no matter whether TMM is present or not (Lin et al., 2017).
The serk1 serk2 bak1 knockout mutants produce clustered stomata, similar to the er triple mutants. SERKs play unequal redundant roles in regulating stomatal patterning, as revealed by analyzing a series of higher-order serk mutants in the bak1-5 background, a semidominant allele of BAK1 without defects in brassinosteroid signaling and cell-death control. Application of EPF1/2 can enhance the interactions between ERf and SERKs, and in vitro kinase assays indicated that ER and BAK1 can transphosphorylate each other. In addition, the serk1 serk2 bak1 mutants were insensitive to exogenous treatment or induced expression of bioactive EPF1/2 peptides regarding the clustered stomata. Taken together, these results demonstrated that SERKs function as coreceptors for ERf-mediated EPF signaling in regulating stomatal patterning through transphosphorylation between SERKs and ERf RLKs (Fig. 1F; Meng et al., 2015 ). However, no structural data are currently available to further reveal the molecular mechanisms by which SERKs function together with TMM and ERf in regulating stomatal patterning.
TAPETUM DETERMINANT1 (TPD1) is another type of secreted Cys-rich peptide with four Cys residues after posttranslational processing that plays a crucial role in specifying the tapetal cells during anther development (Yang et al., 2003; Huang et al., 2016). TPD1 is expressed early in the archesporial cells and the descendant cells, and later predominantly in the sporogenous cells and microsporocytes. The tpd1 anthers produce excess microsporocytes and lack the tapetal cell layer, whereas overexpression of TPD1 delays the degeneration of the tapetal cells, indicating that TPD1 plays an important role in communicating between the tapetum and microsporocytes (Yang et al., 2003, 2005). Blocking the TPD1-mediated communication between the tapetum and microsporocytes by directing TPD1 into vacuoles resulted in defective anther development, and no pollen grains can be produced (Huang et al., 2016).
TPD1 is synthesized in the sporogenous cells/microsporocytes and secreted to the primary tapetum/tapetal cells, where it is perceived by EMS1/EXS, an LRR-RLK, to coordinate tapetal cell specification (Canales et al., 2002; Zhao et al., 2002). EMS1/EXS is predominantly expressed in the tapetal cells, exhibiting a partially complementary expression pattern compared with that of TPD1 (Yang et al., 2003). Similar to the tpd1 mutant, the ems1/exs mutant anthers cannot properly specify the tapetum (Canales et al., 2002; Zhao et al., 2002). Genetic analysis indicated that the tpd1, ems1/exs, and tpd1 ems1/exs mutants all show the same tapetal defects and that functional EMS1/EXS is required for TPD1 signaling, supporting that TPD1 and EMS1/EXS function in the same genetic pathway (Yang et al., 2003; Jia et al., 2008). Protein interaction assays demonstrated that TPD1 interacts with the extracellular LRR domain of EMS1/EXS, and TPD1 can induce the phosphorylation of EMS1/EXS in planta (Jia et al., 2008). Moreover, EMS1/EXS is required for proper TPD1 localization to the plasma membrane.
It was proposed that perception of the reproductive cell-secreted TPD1 by its somatic cell-localized receptor EMS1/EXS activates a signaling pathway that promotes the division of the secondary parietal cells and then specifies the fate of the tapetal cells that consequently suppress microsporocyte proliferation (Jia et al., 2008; Huang et al., 2016). MULTIPLE SPOROCYTE1, TPD1-LIKE1A/MICROSPORELESS2, and MULTIPLE ARCHESPORIAL CELLS1, the homologs of EMS1/EXS or TPD1 in rice and maize, function in a similar manner to regulate early anther cell specification (Zhao et al., 2008; Hong et al., 2012; Wang et al., 2012; Yang et al., 2016). Two SERKs, SERK1 and SERK2, function redundantly as the coreceptor of EMS1/EXS to control anther cell specification in Arabidopsis. Similar to the tpd1 and ems1/exs mutants, the serk1 serk2 mutant anthers cannot specify the tapetum (Albrecht et al., 2005; Colcombet et al., 2005). The ems1 serk1 serk2 mutant anthers exhibited tapetal and microsporocyte defects similar to tpd1, ems1/exs, and serk1 serk2. Ectopic expression of TPD1 in serk1 serk2 anther epidermis still exhibited similar defects to the serk1 serk2 mutant, whereas six somatic cell layers could be formed when TPD1 was ectopically expressed in wild-type anthers. Moreover, SERK1 physically interacts with and transphosphorylates EMS1/EXS to enhance EMS1/EXS kinase activity that is critical for EMS1/EXS to regulate anther cell specification. These results demonstrated that SERK1/2 function in the same genetic pathway as TPD1-EMS1/EXS and that the functions of SERK1/2 are required for TPD1-EMS1/EXS signaling (Fig. 1G; Li et al., 2017).
A conserved role of GhSERK1 in regulating anther development was also revealed in cotton (Shi et al., 2014). Very recently, it was revealed that BES1 and its homologs, a group of transcription factors, are employed as a common downstream signaling component by the BR-BRI1-SERK and TPD1-EMS1/EXS-SERK signaling pathways to regulate cell elongation and anther cell differentiation, respectively (Chen et al., 2019; Zheng et al., 2019).
HSL1/TDR (PXY)-SERK PAIRS PERCEIVE CLE PEPTIDES TO REGULATE STOMATAL DEVELOPMENT AND CAMBIUM CELL PROLIFERATION
CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION-related (CLE) family precursors contain a conserved CLE domain near their C termini that are proteolytically processed to produce mature CLE peptides with 12 to 13 amino acid residues. CLE peptides regulate a variety of biological processes in plant development and responses to stresses (Yamaguchi et al., 2016; Goad et al., 2017). For instance, upon dehydration stress, root-derived CLE25 moves to the leaves to modulate stomatal closure, enhancing drought resistance (Takahashi et al., 2018). As valves in the epidermis, stomata control gas exchange and water loss between plants and the ambient atmosphere. As aforementioned, the EPF-ERf peptide-receptor pairs strictly regulate critical steps of stomatal development, including spacing and density.
Recently, it was revealed that the CLE9 and CLE10 peptides, identical mature peptides produced by CLE9 and CLE10, also regulate stomatal development (Qian et al., 2018). CLE9/10 genes are expressed in stomatal lineage cells, and exogenous application of synthetic CLE9/10 peptide decreased the number of stomatal lineage cells. The cle9 single mutants showed increased number and density of stomatal lineage cells, similar to a cle9 cle10 double mutant, indicating that CLE9 plays a major role in repressing the meristemoid mother cell identity during stomatal development (Qian et al., 2018). Application of CLE9/10 peptide can still decrease the number of stomatal lineage cells in the er triple mutant, whereas EPF2 depends on the function of ERf (Hara et al., 2009), indicating that CLE9/10 functions independently of the EPF-ERf signaling pathway and that its corresponding receptor is required to control stomatal development (Qian et al., 2018).
By testing the sensitivity of mutants from group XI LRR-RLKs to CLE9/10 treatment in stomatal development, HSL1 was identified as the receptor of CLE9/10. Compared with wild-type plants, the hsl1 mutants produced more stomatal lineage cells, which were not decreased upon CLE9/10 treatment. Biochemical assays indicated that the extracellular LRR domain of HSL1 can directly and specifically bind to CLE9/10. However, HSL1 homodimerization cannot be induced upon CLE9/10 binding, suggesting that a coreceptor is needed for HSL1-mediated signaling during stomatal development. In fact, interactions between SERKs and HSL1 can be specifically induced by CLE9/10. On the other hand, SERK1 can significantly increase the binding affinity between HSL1 and CLE9/10 (Qian et al., 2018). These results indicated that SERKs are recruited as coreceptors of HSL1 for sensing CLE9/10 to regulate stomatal development. Future structural analysis will facilitate our understanding of this important biological process (Fig. 1H).
The TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF) dodecapeptide, a homolog of the Arabidopsis CLE41/44 peptide, which specifically suppresses the tracheary element differentiation, was originally identified in a Zinnia elegans xylogenic culture. When TDIF was added, the transition of procambial cells to tracheary elements was suppressed (Ito et al., 2006). Overexpression of CLE44 or exogenous application of TDIF resulted in discontinuous xylem vessels and stele enlargement caused by increased procambial cells that were suppressed to differentiate into xylem vessel cells in Arabidopsis (Hirakawa et al., 2008). Consistent with this, loss of function of CLE41/44 results in decreased numbers of procambial cells and some xylem vessel cells adjacent to the phloem cells (Hirakawa et al., 2010; Yamaguchi et al., 2017). Both CLE41 and CLE44 are expressed in the phloem and the pericycle. However, the CLE41/44 peptide was specifically detected in the apoplast surrounding the phloem precursors in root tips and in the procambial cells of hypocotyls, indicating that the CLE41/44 peptide is produced by the phloem cells and function in a non-cell-autonomous manner in the procambium.
CLE41/44 is perceived by TDIF RECEPTOR (TDR), an LRR-RLK also named PHLOEM INTERCALATED WITH XYLEM (PXY), to promote the maintenance of vascular stem cells and suppress their differentiation into xylem cells (Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010; Zhang et al., 2016c). TDR/PXY expression is restricted in procambial cells. TDR/PXY mutations result in more flattened vascular bundles with phloem adjacent to or interspersed with the xylem because of defective orientation of cell division during vascular development. Procambial cell proliferation in tdr hypocotyls cannot be enhanced by TDIF treatment (Fisher and Turner, 2007; Hirakawa et al., 2008, 2010; Etchells and Turner, 2010).
It was reported that SERK1 is expressed in vascular tissues and that TDIF induces the interaction between the extracellular LRR domains of TDR/PXY and SERKs (Kwaaitaal and de Vries, 2007; Zhang et al., 2016b, 2016c). The serk1 serk2 bak1 triple mutant cannot maintain the procambial cells, and the xylem is adjacent to the phloem. Moreover, the serk1 serk2 bak1 triple mutant showed impaired responses to TDIF treatment, similar to that in pxy (Fisher and Turner, 2007; Zhang et al., 2016c). Crystal structure results further supported that SERKs function as the coreceptors of TDR/PXY to regulate cell-to-cell communication during vascular stem cell maintenance and xylem differentiation (Fig. 1I; Zhang et al., 2016b).
CLV-CIK PAIRS MEDIATE CLV3 SIGNAL TO REGULATE THE SAM
Plants generate aerial organs, such as stems, branches, leaves, and flowers, during postembryonic development to establish appropriate architectures for survival in their life spans. The whole process is dependent on the SAM, which contains pluripotent stem cells that keep generating daughter cells to renew themselves and to replenish cells for new tissues or organs (Vernoux and Benfey, 2005; Williams and Fletcher, 2005; Aichinger et al., 2012; Wu et al., 2018; Kitagawa and Jackson, 2019). CLV3, the founding member of the CLE peptide family, is a secreted arabinosylated glycopeptide containing 13-amino acid residues and plays a crucial role in maintaining SAM homeostasis (Ohyama et al., 2009). Mutations of CLV3 resulted in an enlarged SAM and a fasciated stem (Fletcher et al., 1999; Rodriguez-Leal et al., 2019). A feedback regulation mediated by CLV3 and WUSCHEL (WUS), a critical transcription factor, delicately balances self-renewal and differentiation of the stem cells in the SAM (Brand et al., 2000; Schoof et al., 2000; Rojo et al., 2002; Lenhard and Laux, 2003; Yadav et al., 2011; Perales et al., 2016). Recent studies indicated that HAIRY MERISTEM (HAM) transcription regulators are required interacting cofactors of WUS, and the HAM concentration gradient spatially specifies the CLV3 expression domain (Zhou et al., 2015, 2018).
Loss of function of CLV1, a typical LRR-RLK, leads to an enlarged SAM and extra floral organs, similar to clv3 (Clark et al., 1993, 1997). CLV3 can directly bind to the extracellular LRR domain of CLV1 to repress the expression of WUS in the organizing center (Fletcher et al., 1999; Schoof et al., 2000; Ogawa et al., 2008). On the other hand, CLV3 promotes the membrane-localized CLV1 to move into lytic vacuoles for degradation, which is a possible mechanism to buffer CLV3 signaling in the maintenance of stem cells (Nimchuk et al., 2011). RECEPTOR-LIKE PROTEIN KINASE2 (RPK2), another typical LRR-RLK, and CLV2/CORYNE (CRN), a complex of an LRR-RLP and an RLCK without kinase activity, are also involved in sensing CLV3 (Kayes and Clark, 1998; Jeong et al., 1999; Müller et al., 2008; Kinoshita et al., 2010). Different from CLV1, however, CLV3 cannot directly bind to the LRR domain of RPK2 or CLV2 (Shinohara and Matsubayashi, 2015). Genetic evidence suggested that the CLV3 signal is possibly transduced by three parallel pathways mediated by CLV1, CLV2/CRN, and RPK2, respectively (Müller et al., 2008; Kinoshita et al., 2010).
When CLV1 function is compromised, BARELY ANY MERISTEMS (BAMs), the close homologs of CLV1, are ectopically expressed in the rib meristem, which partially compensates for the loss of CLV1 (Deyoung and Clark, 2008; Nimchuk et al., 2015). Consistent with this, photoaffinity labeling showed that CLV3 can directly bind to BAM1 and BAM2 (Shinohara and Matsubayashi, 2015). The CLV-WUS negative signaling pathway is conserved in higher plants and has also been extensively studied in crops, such as tomato, rice, and maize. The components in this pathway are potential targets for genetic manipulation to improve yield traits (Somssich et al., 2016; Fletcher, 2018; Kitagawa and Jackson, 2019).
Although SERKs function as critical coreceptors in various signaling pathways, no evidence supports that they are involved in the CLV-WUS signaling pathway to control stem cell homeostasis in the SAM. Recently, another group of coreceptors named CIKs were identified to function together with CLV1, CLV2/CRN, and RPK2 in mediating the CLV3 signal to maintain the SAM (Hu et al., 2018). The cik quadruple mutants exhibited severe SAM defects similar to clv3 and clv1 clv2 rpk2 triple mutants. Genetic analyses demonstrated that cik mutants were epistatic to the receptor mutants. Moreover, CIKs can interact with and be phosphorylated by CLV1 and RPK2. The cik quadruple mutant was insensitive to exogenous application of CLV3, and the phosphorylation level of CIKs cannot be elevated by CLV3 treatment in clv1 bam1 bam2. Taken together, these results suggest that CIKs function as shared coreceptors to integrate three parallel CLV signaling pathways mediated by CLV1, CLV2/CRN, and RPK2 (Fig. 2; Hu et al., 2018).
Figure 2.
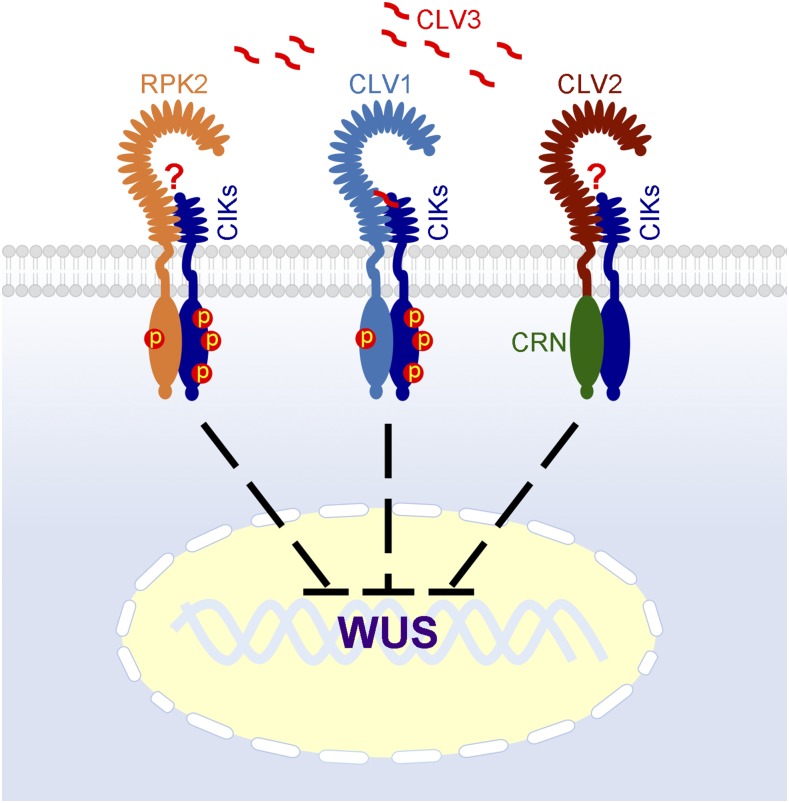
CIKs integrate RLK-mediated pathways to maintain stem cell homeostasis in the SAM. CIKs redundantly interact with CLV1, CLV2/CRN, and RPK2 to transduce the CLV3 signal into the nucleus while suppressing the expression of WUS. CIKs can at least be phosphorylated by CLV1 and RPK2 for signaling, according to the results from phosphorylation assays (Cui et al., 2018; Hu et al., 2018). Question marks indicate that no physical interaction between CLV3 and RPK2 or CLV2 has been confirmed.
BAM/RPK2-CIK PAIRS REGULATE ANTHER CELL SPECIFICATION
The Arabidopsis anther wall of each locule at stage 5 consists of four somatic cell layers, the epidermis, endothecium, middle layer, and tapetum from outside to inside, which surround the microspore mother cells (Sanders et al., 1999). Besides TPD1-EMS1/EXS (Jia et al., 2008; Huang et al., 2016; Li et al., 2017), no other peptide-RLK pairs have been characterized as specifying anther wall cells in Arabidopsis. Nevertheless, several other RLKs with long extracellular LRR domains have been identified as regulating anther cell specification. For instance, ERf members play important roles in promoting anther lobe formation and anther cell differentiation. The er triple mutants produced either antherless stamens or anthers with only defective abaxial lobes with disorganized somatic cell layers, especially abnormal tapetum and middle layer cells (Shpak et al., 2004; Hord et al., 2008). BAM1 and BAM2 play important roles in early anther development by specifying the identity of L2-derived somatic cells. The archesporial cells in the bam1 bam2 anthers exhibited defective asymmetric cell division and produced extra microspore mother cells but lacked the endothecium, middle layer, and tapetum (Hord et al., 2006). RPK2 promotes tapetal cell fate. The inner secondary parietal cells in rpk2 mutant anthers failed to differentiate into the middle layer cells and finally formed abnormally hypertrophic tapetal cells (Mizuno et al., 2007). It was recently reported that RPK2 also regulates the periclinal division of the archesporial cells and the primary parietal cells: disruption of RPK2 resulted in defective anthers lacking either two or three parietal cell layers (Cui et al., 2018). However, the ligands perceived by these RLKs during anther development have yet to be identified.
CIK RLKs play vital roles in redundantly regulating not only meristem homeostasis but also anther cell specification. The most severe defective anthers of cik mutants produced extra microspore mother cells surrounded only by the epidermis and lacked all other parietal cell layers because of abnormal anticlinal division of the archesporial cells, which is similar to that of bam1 bam2 and rpk2 mutants. The other defective cik anthers lacked either one or two parietal cell layers like rpk2 because of the failure of division of the inner secondary parietal cells or the primary parietal cells. CIKs can physically interact with and be phosphorylated by BAM1/2 and RPK2 and can function as coreceptors of BAM1/2 and RPK2 to regulate parietal cell differentiation during early anther development. The RPK2-CIK and EMS1-SERK complexes may coordinate to regulate cell division of the inner secondary parietal cells and specification of the middle layer and the tapetum (Cui et al., 2018).
CONCLUSION AND PERSPECTIVE
Significant progress has been made in revealing various biological functions of RLKs and the underlying mechanisms in past years, leading to numerous fundamental breakthroughs in plant biology. This knowledge has dramatically promoted our understanding of this key type of plasma membrane-localized signaling molecule. It has been widely accepted that ligand-binding RLKs require coreceptors to form receptor complexes for transducing external signals during plant growth and development. Numerous lines of genetic and biochemical evidence demonstrated that SERK receptor kinases function as pivotal coreceptors in regulating many developmental processes and responses to biotic and abiotic stresses, although structural results are still accumulating. Recent results showed that CIK receptor kinases function as coreceptors of CLV1, BAM1/2, and RPK2 to control SAM stem cell homeostasis and anther wall cell specification (Cui et al., 2018; Hu et al., 2018). CIK members, CIK1/NSP-INTERACTING KINASE3, CIK2/CLE-RESISTANT RECEPTOR KINASE, and CIK3/SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE, were reported to regulate antivirus response, root protophloem differentiation, and leaf senescence, respectively, although the cognate receptors have not yet been identified (Fontes et al., 2004; Xu et al., 2011; Anne et al., 2018). These results suggest that CIKs function as another group of RLK coreceptors in regulating plant growth, development, and responses to stresses, although more of the pairing receptors of CIKs need to be identified and further studied. See Table 1 for an overview of mentioned ligands, receptors, and coreceptors.
Table 1. Receptors, coreceptors, and peptides involved in plant development mentioned in this review.
| Ligand Name | Ligand Type | Receptor | Subfamily | Coreceptor | Function |
|---|---|---|---|---|---|
| Brassinosteroids | Phytohormone | BRI1 | LRR X | SERK1, SERK2, BAK1, BKK1 | Cell elongation and plant growth |
| PSK | Sulfated peptide | PSKR1, PSKR2 | LRR X | SERK1, SERK2, BAK1 | Cell division and cell expansion |
| PSY1 | Sulfated peptide | PSY1R | LRR X | SERK1, SERK2, BAK1 | Cell division and cell expansion |
| RGFs/CLELs/GLVs | Sulfated peptide | RGFR/RGI1 to RGI5 | LRR XI | SERK1, SERK2, BAK1, BKK1 | Root meristem size |
| IDA | Hydroxylated peptide | HAE, HSL2 | LRR XI | SERK1, SERK2, BAK1, BKK1 | Cell separation |
| EPF1, EPF2, EPFLs | Cys-rich peptide | ER, ERL1, ERL2 | LRR XIII | SERK1, SERK2, BAK1 | Stomatal development |
| TPD1 | Cys-rich peptide | EMS1/EXS | LRR X | SERK1, SERK2 | Tapetum specification |
| CLE9/10 | CLE peptide | HSL1 | LRR XI | SERK1, SERK2, BAK1 | Stomatal development |
| TDIF/CLE41/44 | CLE peptide | TDR/PXY | LRR XI | SERK1, SERK2, BAK1 | Cambium cell proliferation |
| CLV3 | CLE peptide | CLV1, BAM1, BAM2 | LRR XI | CIK1, CIK2, CIK3, CIK4 | SAM homeostasis |
| RPK2 | LRR X | ||||
| Unknown | Unknown | BAM1, BAM2 | LRR XI | CIK1, CIK2, CIK3, CIK4 | Anther development |
| Unknown | Unknown | RPK2 | LRR X | CIK1, CIK2, CIK3, CIK4 | Tapetum specification |
Efforts have been made to elucidate the signaling components after the perception of ligands by the receptor-coreceptor complexes. The BRI1-SERK complex-mediated brassinosteroid signaling recruits RLCKs, protein phosphatases, and GSK3/Shaggy-like kinases to activate the BES1 family transcription factors that directly regulate the expression of brassinosteroid-responsive genes and consequently modulate many aspects of plant growth and development (Planas-Riverola et al., 2019; Ackerman-Lavert and Savaldi-Goldstein, 2020). Recent reports indicated that both the BRI1-SERK and EMS1-SERK signaling pathways ultimately recruit the same BES1 family transcription factors to control cell elongation and anther development (Chen et al., 2019; Zheng et al., 2019). It is unknown, however, whether the EMS1-SERK pathway utilizes downstream signaling components similar to the BRI1-SERK pathway. Accumulating evidence indicates that RLCKs play essential roles in RLK-mediated signaling pathways, especially in plant immune responses (Liang and Zhou, 2018). Although RLCKs have been identified in the BRI1-SERK and HAE/HSL2-SERK pathways (Tang et al., 2008; Burr et al., 2011; Sreeramulu et al., 2013; Zhang et al., 2016a), it is still challenging to discover those possible RLCKs in other RLK-SERK pathways. The MAPK cascade is another signaling module found in many RLK-mediated signaling pathways, such as ERf-SERK-regulated stomatal patterning and HAE/HSL2-SERK-regulated cell separation processes (Wang et al., 2007; Cho et al., 2008; Aalen et al., 2013; Kumpf et al., 2013; Meng et al., 2015; Patharkar and Walker, 2015; Zhu et al., 2019). Whether other RLK-SERK pairs also employ a MAPK cascade is still elusive. As recently identified, however, the CIK-regulated signaling pathways are poorly understood with regard to their downstream signaling components.
In spite of the achievements that have been made in the RLK field, many interesting questions remain to be answered (see Outstanding Questions). SERKs and CIKs all belong to the LRR II-RLK family and have very similar structures (Cui et al., 2018). The current results support that the kinase domains of ligand-binding receptors determine their signaling specificity (Hohmann et al., 2018b). It is therefore fascinating to elucidate how a specific ligand-binding receptor selects a SERK or a CIK as its coreceptor. The specific phosphocode of SERKs or CIKs responding to diverse signals may be one of the solutions to determine the specificity (Perraki et al., 2018). Whether SERKs and CIKs employ similar mechanisms to transduce different signals is another open question. It is already known that the interaction between CIKs and CLV1 or RPK2 cannot be enhanced upon the application of CLV3, whereas the interaction between BAK1 and BRI1 can be dramatically elevated upon brassinosteroid application (Wang et al., 2005; Hu et al., 2018), suggesting that CIKs and SERKs may transduce signals differently, at least in some signaling pathways. Furthermore, the corresponding ligands and/or receptors of some SERK- or CIK-regulated biological processes have not been uncovered, such as the ligands involved in BAM-CIK and RPK2-CIK complex-controlled anther cell specification and the ligand and receptor in SERK-controlled stem cell division during embryogenesis. In addition, further studies are needed to uncover other unknown signaling pathways regulated by SERKs and CIKs and the unknown downstream components in SERK- and CIK-regulated signaling pathways.
Footnotes
This work was supported by National Natural Science Foundation of China (grant nos. 31970339, 31770312, 31470380, 31530005, and 31720103902), the 111 Project (grant no. B16022), and the Fundamental Research Funds for the Central Universities (grant nos. lzujbky-2019-kb05 and lzujbky-2020-kb05).
Articles can be viewed without a subscription.
References
- Aalen RB, Wildhagen M, Stø IM, Butenko MA(2013) IDA: A peptide ligand regulating cell separation processes in Arabidopsis. J Exp Bot 64: 5253–5261 [DOI] [PubMed] [Google Scholar]
- Abrash EB, Bergmann DC(2010) Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Abrash EB, Davies KA, Bergmann DC(2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman-Lavert M, Savaldi-Goldstein S(2020) Growth models from a brassinosteroid perspective. Curr Opin Plant Biol 53: 90–97 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Kornet N, Friedrich T, Laux T(2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B(2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S(2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y(2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Z, Liu Y, Ou Y, Li J, Zhang B, Sun D, Sun Y, Tang W(2018) Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci USA 115: 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne P, Amiguet-Vercher A, Brandt B, Kalmbach L, Geldner N, Hothorn M, Hardtke CS(2018) CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145: dev162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Binder A, Parniske M(2012) Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol 50: 451–473 [DOI] [PubMed] [Google Scholar]
- Bajwa VS, Wang X, Blackburn RK, Goshe MB, Mitra SK, Williams EL, Bishop GJ, Krasnyanski S, Allen G, Huber SC, et al. (2013) Identification and functional analysis of tomato BRI1 and BAK1 receptor kinase phosphorylation sites. Plant Physiol 163: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T(2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20: 47–54 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U(2009) Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R(2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ(2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB(2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G(2014) Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H(2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Garcia VJ, Cartwright H, Sun Y, Wang ZY(2016) Immunophilin-like FKBP42/TWISTED DWARF1 interacts with the receptor kinase BRI1 to regulate brassinosteroid signaling in Arabidopsis. Mol Plant 9: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Schaller GE, Patterson SE, Kwok SF, Meyerowitz EM, Bleecker AB(1992) The TMK1 gene from Arabidopsis codes for a protein with structural and biochemical characteristics of a receptor protein kinase. Plant Cell 4: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lv M, Wang Y, Wang PA, Cui Y, Li M, Wang R, Gou X, Li J(2019) BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat Commun 10: 4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B(2009) One for all: The receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T(2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC(2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM(1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM(1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM(1998) Brassinosteroids: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI(2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hu C, Zhu Y, Cheng K, Li X, Wei Z, Xue L, Lin F, Shi H, Yi J, et al. (2018) CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 30: 2383–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung BJ, Clark SE(2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U(2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Etchells JP, Turner SR(2010) The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Feng H, Liu C, Fu R, Zhang M, Li H, Shen L, Wei Q, Sun X, Xu L, Ni B, et al. (2019) LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 regulate pollen tube growth as chaperones and coreceptors for ANXUR/BUPS receptor kinases in Arabidopsis. Mol Plant 12: 1612–1623 [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S(2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fletcher JC.(2018) The CLV-WUS stem cell signaling pathway: A roadmap to crop yield optimization. Plants (Basel) 7: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM(1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J(2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Zhao Y, Liu MC, Zhou LZ, Wang L, Zhong S, Hou S, Jiang J, Liu T, Huang Q, et al. (2019) LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr Biol 29: 3256–3265.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad DM, Zhu C, Kellogg EA(2017) Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol 216: 605–616 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J(2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großeholz R, Feldman-Salit A, Wanke F, Schulze S, Glöckner N, Kemmerling B, Harter K, Kummer U(2019) Specifying the role of BAK1-interacting receptor-like kinase 3 in brassinosteroid signaling. J Integr Plant Biol doi:10.1111/jipb.12803. [DOI] [PubMed] [Google Scholar]
- Han J, Tan J, Tu L, Zhang X(2014) A peptide hormone gene, GhPSK promotes fibre elongation and contributes to longer and finer cotton fibre. Plant Biotechnol J 12: 861–871 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T(2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T(2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hartmann J, Fischer C, Dietrich P, Sauter M(2014) Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1. Plant J 78: 192–202 [DOI] [PubMed] [Google Scholar]
- Hartmann J, Stührwohldt N, Dahlke RI, Sauter M(2013) Phytosulfokine control of growth occurs in the epidermis, is likely to be non-cell autonomous and is dependent on brassinosteroids. Plant J 73: 579–590 [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC(2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP(2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H(2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Sawa S(2019) Diverse function of plant peptide hormones in local signaling and development. Curr Opin Plant Biol 51: 81–87 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H(2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Nicolet J, Moretti A, Hothorn LA, Hothorn M(2018a) The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat Plants 4: 345–351 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Santiago J, Nicolet J, Olsson V, Spiga FM, Hothorn LA, Butenko MA, Hothorn M(2018b) Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc Natl Acad Sci USA 115: 3488–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzwart E, Huerta AI, Glöckner N, Garnelo Gómez B, Wanke F, Augustin S, Askani JC, Schürholz AK, Harter K, Wolf S(2018) BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc Natl Acad Sci USA 115: 11838–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Tang D, Shen Y, Hu Q, Wang K, Li M, Lu T, Cheng Z(2012) MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol 196: 402–413 [DOI] [PubMed] [Google Scholar]
- Hord CLH, Chen C, Deyoung BJ, Clark SE, Ma H(2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CLH, Sun YJ, Pillitteri LJ, Torii KU, Wang H, Zhang S, Ma H(2008) Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J(2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zhu Y, Cui Y, Cheng K, Liang W, Wei Z, Zhu M, Yin H, Zeng L, Xiao Y, et al. (2018) A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat Plants 4: 205–211 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang T, Linstroth L, Tillman Z, Otegui MS, Owen HA, Zhao D(2016) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U(2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Hunt L, Bailey KJ, Gray JE(2010) The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186: 609–614 [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE(2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H(2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE(1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Liu X, Owen HA, Zhao D(2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acad Sci USA 105: 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC(2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Motzkus M, Sauter M(2017) Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J Exp Bot 68: 1411–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes JM, Clark SE(1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S(2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J(2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW(2015) RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol 169: 826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Jackson D(2019) Control of meristem size. Annu Rev Plant Biol 70: 269–291 [DOI] [PubMed] [Google Scholar]
- Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y(2009) Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA 106: 15067–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. (2010) Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol 51: 1–8 [DOI] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB(2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal MACJ, de Vries SC(2007) The SERK1 gene is expressed in procambium and immature vascular cells. J Exp Bot 58: 2887–2896 [DOI] [PubMed] [Google Scholar]
- Kwezi L, Ruzvidzo O, Wheeler JI, Govender K, Iacuone S, Thompson PE, Gehring C, Irving HR(2011) The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J Biol Chem 286: 22580–22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig F, Dahlke RI, Stührwohldt N, Hartmann J, Harter K, Sauter M(2015) Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27: 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Putarjunan A, Han SK, Avila J, Torii KU(2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU(2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Laux T(2003) Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130: 3163–3173 [DOI] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K(2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li H, Cai Z, Wang X, Li M, Cui Y, Cui N, Yang F, Zhu M, Zhao J, Du W, et al. (2019) SERK receptor-like kinases control division patterns of vascular precursors and ground tissue stem cells during embryo development in Arabidopsis. Mol Plant 12: 984–1002 [DOI] [PubMed] [Google Scholar]
- Li J.(2010) Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol 13: 509–514 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J(1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC(2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang Y, Huang J, Ahsan N, Biener G, Paprocki J, Thelen JJ, Raicu V, Zhao D(2017) Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol 173: 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou JM(2018) Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Lin G, Zhang L, Han Z, Yang X, Liu W, Li E, Chang J, Qi Y, Shpak ED, Chai J(2017) A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev 31: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Castro C, Wang Y, Noble J, Ponvert N, Bundy M, Hoel C, Shpak E, Palanivelu R(2016) The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28: 1035–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorbiecke R, Steffens M, Tomm JM, Scholten S, von Wiegen P, Kranz E, Wienand U, Sauter M(2005) Phytosulphokine gene regulation during maize (Zea mays L.) reproduction. J Exp Bot 56: 1805–1819 [DOI] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L(2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y(2006) Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol 142: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y(2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y(1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Takagi L, Sakagami Y(1997) Phytosulfokine-α, a sulfated pentapeptide, stimulates the proliferation of rice cells by means of specific high- and low-affinity binding sites. Proc Natl Acad Sci USA 94: 13357–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y(2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S(2012) CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA 109: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L(2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou J, Tang J, Li B, de Oliveira MVV, Chai J, He P, Shan L(2016) Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep 14: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N(2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M(2009) ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol 19: 1327–1331 [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K(2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50: 751–766 [DOI] [PubMed] [Google Scholar]
- Moffett AS, Bender KW, Huber SC, Shukla D(2017) Molecular dynamics simulations reveal the conformational dynamics of Arabidopsis thaliana BRI1 and BAK1 receptor-like kinases. J Biol Chem 292: 12643–12652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R(2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I(2012) Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 24: 3198–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD(2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J(2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM(2011) Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM(2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlenschlæger CB, Gersby LBA, Ahsan N, Pedersen JT, Kristensen A, Solakova TV, Thelen JJ, Fuglsang AT(2017) Activation of the LRR receptor-like kinase PSY1R requires transphosphorylation of residues in the activation loop. Front Plant Sci 8: 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y(2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohki S, Takeuchi M, Mori M(2011) The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat Commun 2: 512. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y(2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X, et al. (2016) RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res 26: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Ryu HY, Kim BH, Kim SY, Yoon IS, Nam KH(2011) A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Mol Cells 32: 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Walker JC(2015) Floral organ abscission is regulated by a positive feedback loop. Proc Natl Acad Sci USA 112: 2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HC, Kaloshian I(2014) The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS ONE 9: e93302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV(2016) Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc Natl Acad Sci USA 113: E6298–E6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, DeFalco TA, Derbyshire P, Avila J, Séré D, Sklenar J, Qi X, Stransfeld L, Schwessinger B, Kadota Y, et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI(2019) Brassinosteroid signaling in plant development and adaptation to stress. Development 146: dev151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Song W, Yokoo T, Minobe A, Wang G, Ishida T, Sawa S, Chai J, Kakimoto T(2018) The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat Plants 4: 1071–1081 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Xu C, Kwon CT, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, Van Eck J, et al. (2019) Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat Genet 51: 786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC(2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C(2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB(1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M(2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M(2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- Sauter M.(2015) Phytosulfokine peptide signalling. J Exp Bot 66: 5161–5169 [DOI] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A(2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Schmidt ED, Guzzo F, Toonen MA, de Vries SC(1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T(2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D(2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, Alling RM, Hammerstad M, Aalen RB(2019) Control of organ abscission and other cell separation processes by evolutionary conserved peptide signaling. Plants (Basel) 8: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, von Wangenheim D, Herrmann U, Wildhagen M, Kulik I, Kopf A, Ishida T, Olsson V, Anker MK, Albert M, et al. (2018) The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nat Plants 4: 596–604 [DOI] [PubMed] [Google Scholar]
- Shi YL, Guo SD, Zhang R, Meng ZG, Ren MZ(2014) The role of Somatic embryogenesis receptor-like kinase 1 in controlling pollen production of the Gossypium anther. Mol Biol Rep 41: 411–422 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y(2015) Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: Dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J 82: 328–336 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y(2016) Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci USA 113: 3897–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB(2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH(2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED.(2013) Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol 55: 1238–1250 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU(2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU(2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Silverstein KAT, Moskal WA Jr., Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA(2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D(2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143: 3238–3248 [DOI] [PubMed] [Google Scholar]
- Song W, Liu L, Wang J, Wu Z, Zhang H, Tang J, Lin G, Wang Y, Wen X, Li W, et al. (2016) Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res 26: 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, et al. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74: 905–919 [DOI] [PubMed] [Google Scholar]
- Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB(1991) Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA 88: 8816–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB(2006) Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA(2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Dahlke RI, Kutschmar A, Peng X, Sun MX, Sauter M(2015) Phytosulfokine peptide signaling controls pollen tube growth and funicular pollen tube guidance in Arabidopsis thaliana. Physiol Plant 153: 643–653 [DOI] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I(2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J(2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 23: 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, Shinozaki K(2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556: 235–238 [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY(2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CM, Howlett B, Nasrallah JB(1992) An Arabidopsis thaliana gene with sequence similarity to the S-locus receptor kinase of Brassica oleracea: Sequence and expression. Plant Physiol 99: 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU.(2012) Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci 17: 711–719 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Benfey PN(2005) Signals that regulate stem cell activity during plant development. Curr Opin Genet Dev 15: 388–394 [DOI] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Liu B, Winge P, Butenko MA, Hornslien KS, Kumpf R, Aalen RB, Bones AM, Brembu T(2015) The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: Phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J Exp Bot 66: 5351–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC.(1993) Receptor-like protein kinase genes of Arabidopsis thaliana. Plant J 3: 451–456 [DOI] [PubMed] [Google Scholar]
- Walker JC.(1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26: 1599–1609 [DOI] [PubMed] [Google Scholar]
- Walker JC, Zhang R(1990) Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345: 743–746 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G(2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ(2012) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S(2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Han Z, Zhang H, Wang T, Lin G, Chang J, Yang W, Chai J(2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD(2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD(2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J(2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP(2005) A brassinosteroid-hypersensitive mutant of BAK1 indicates that a convergence of photomorphogenic and hormonal signaling modulates phototropism. Plant Physiol 139: 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Pérez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, et al. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Williams L, Fletcher JC(2005) Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 8: 582–586 [DOI] [PubMed] [Google Scholar]
- Wu Q, Xu F, Jackson D(2018) All together now, a magical mystery tour of the maize shoot meristem. Curr Opin Plant Biol 45: 26–35 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J(2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Xu F, Meng T, Li P, Yu Y, Cui Y, Wang Y, Gong Q, Wang NN(2011) A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol 157: 2131–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV(2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA(2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA(2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S(2016) CLE peptides and their signaling pathways in plant development. J Exp Bot 67: 4813–4826 [DOI] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Yoshimura M, Imamura Y, Shimaoka C, Sawa S(2017) A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9-mediated gene targeting. Plant Cell Physiol 58: 1848–1856 [DOI] [PubMed] [Google Scholar]
- Yang H, Matsubayashi Y, Nakamura K, Sakagami Y(1999) Oryza sativa PSK gene encodes a precursor of phytosulfokine-α, a sulfated peptide growth factor found in plants. Proc Natl Acad Sci USA 96: 13560–13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Qian X, Chen M, Fei Q, Meyers BC, Liang W, Zhang D(2016) Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol 171: 2085–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Jiang L, Puah CS, Xie LF, Zhang XQ, Chen LQ, Yang WC, Ye D(2005) Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with excess microsporocytes1/extra sporogenous cells. Plant Physiol 139: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D(2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang X, Zhao Z, Wang R, Huang X, Zhu Y, Yuan L, Wang Y, Xu X, Burlingame AL, et al. (2016a) OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol 170: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin X, Han Z, Qu LJ, Chai J(2016b) Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res 26: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin X, Han Z, Wang J, Qu LJ, Chai J(2016c) SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol Plant 9: 1406–1414 [DOI] [PubMed] [Google Scholar]
- Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J(2016) TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol Plant 9: 582–592 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H(2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J(2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Bai Q, Wu L, Liu H, Liu Y, Xu W, Li G, Ren H, She X, Wu G(2019) EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat Commun 10: 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM(2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yan A, Han H, Li T, Geng Y, Liu X, Meyerowitz EM(2018) HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 361: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Shao Y, Ge S, Zhang M, Zhang T, Hu X, Liu Y, Walker J, Zhang S, Xu J(2019) A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat Plants 5: 414–423 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G(2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]