Abstract
Recent progresses made in structural analysis of plant PRRs and NLRs show the advancements in cryo-EM structural biology.
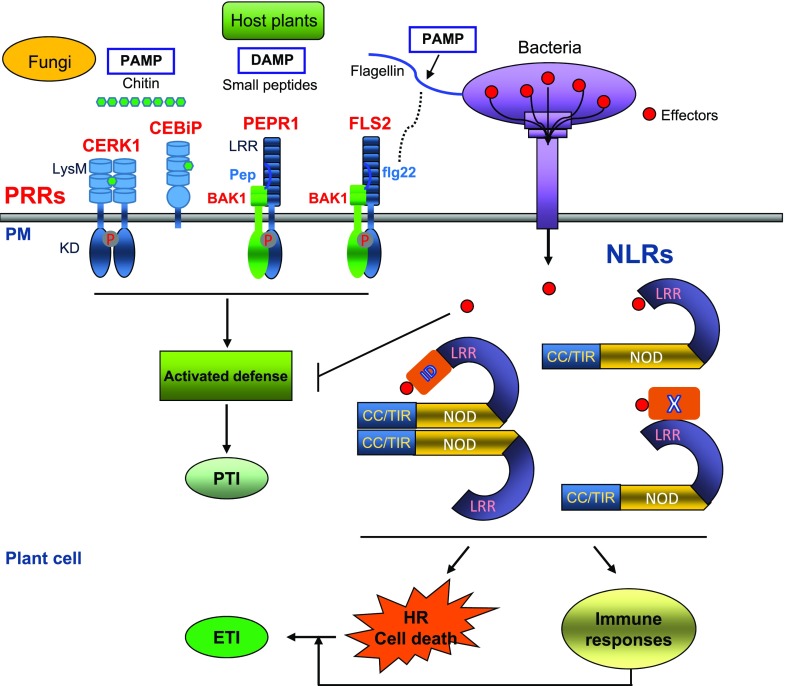
The plant immune system is composed of two classes of receptors—the membrane-anchored pattern recognition receptors (PRRs) and the intracellular nucleotide-binding and leucine-rich repeat receptors (NLRs; Jones and Dangl, 2006; Dangl et al., 2013). The PRR family consists of receptor kinases (RKs) and receptor-like proteins (RLPs) acting in the first tier of the plant immune system. PRRs contain a variable ectodomain that usually function to recognize either conserved microbial signatures known as pathogen-associated molecular patterns (PAMPs) or damage indicators known as danger-associated molecular patterns (DAMPs). PRR activation induces immune responses known as PAMP-triggered immunity (PTI) including expression of immune-related genes (Fig. 1) to ward off microbes (Macho and Zipfel, 2014). Many pathogenic microbes, however, can successfully deliver effector proteins into the plant cell to dampen PTI signaling by manipulating host targets. Plants hence have evolved NLRs as intracellular immune receptors to mediate the second level of surveillance (Fig. 1) through specific recognition of pathogen effectors (Chisholm et al., 2006). Upon perception of effectors, NLRs coordinate a rapid and robust immune signaling response termed effector-triggered immunity, which often leads to hypersensitive response (HR, local cell death at the infection site) and limitation of pathogenic microbes (Fig. 1; Jones et al., 2016).
Figure 1.
A schematic view of the two-tiered plant immune system. Recognition of PAMPs like bacterial flagellin and fungal chitin or DAMPs like secreted small peptides by PRRs induces intracellular signaling, leading to PTI. Successful pathogens deliver effector proteins (red dots) into the plant cell to dampen PTI. In some host plants, effector proteins are specifically recognized by the intracellular NLR immune receptors via different strategies, inducing effector-triggered immunity (ETI) that includes expression of immune-related genes and localized cell death referred to as HR. X, a host molecule guarded by NLRs; ID, integrated domain.
In the last two decades, tremendous advances have been made in functional and mechanistic dissection of plant PRRs and NLRs. There are many excellent reviews on these exciting achievements, mainly from the point view of genetics and physiology (Cui et al., 2015; Boutrot and Zipfel, 2017; Tang et al., 2017; Kourelis and van der Hoorn, 2018; Wan et al., 2019b; Zhang et al., 2017b). In this review, we highlight some of recent structural studies of PRRs and NLRs and discuss how they provided insights into their acting mechanisms.
STRUCTURAL MECHANISMS OF RECOGNITION, ACTIVATION, AND REGULATION OF PLANT PRRS
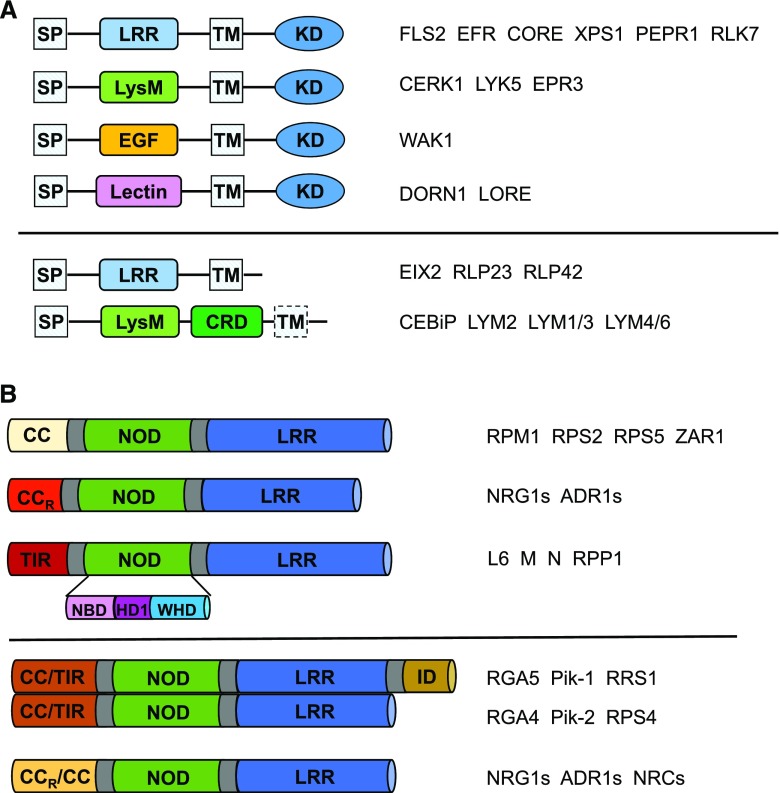
RK-PRRs contain a variable N-terminal extracellular domain (ECD), a transmembrane segment (TM), and a conserved cytoplasmic kinase domain (KD); whereas RLP-PRRs lack an obvious intracellular domain that is typically short (∼24 amino acids). Based on their ECDs, RK-PRRs can be categorized into several groups (Fig. 2A; Böhm et al., 2014; Macho and Zipfel, 2014; Zipfel, 2014). The largest one is leucine-rich repeat (LRR)-RKs and the well-known examples are FLAGELLIN-SENSITIVE2 (FLS2; Gómez-Gómez and Boller, 2000) and EF-TU RECEPTOR (Zipfel et al., 2006), sensing the PAMPs of peptide epitopes of flagellin and elongation factor, respectively. Other examples from this group include PEP RECEPTORs (PEPRs) and RLK7, which perceive the DAMPs of PLANT ELICITOR PEPTIDEs (Yamaguchi et al., 2006, 2010) and PAMP-INDUCED SECRETED PEPTIDEs (Hou et al., 2014), respectively. The Lys-motif (LysM) RK-PRRs such as CHITIN ELICITOR RECEPTOR KINASE1 (CERK1; Miya et al., 2007; Wan et al., 2008) and LYSIN MOTIF RECEPTOR KINASE5 (Cao et al., 2014) are the receptors of the polysaccharide PAMPs like chitin. WALL-ASSOCIATED KINASE1 perceiving the oligogalacturonide DAMP (Brutus et al., 2010) belongs to the epidermal growth factor-like RK-PRR. Another group of RK-PRRs contains extracellular lectin domains. Two newly identified members of this group are DOES NOT RESPOND TO NUCLEOTIDES1 (Choi et al., 2014) and LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (Kutschera et al., 2019), which recognize the extracellular ATP DAMP and the bacterial medium-chain 3-hydroxy fatty acids PAMP, respectively. Compared to RK-PRRs, fewer subgroups of RLP-PRRs have been characterized. One is the LRR-RLPs that usually sense PAMPs (Fig. 2A, lower). Another one is the LysM-RLPs, including the receptors of chitin, CHITIN OLIGOSACCHARIDE ELICITOR BINDING PROTEIN (CEBiP; Kaku et al., 2006) and LYSIN MOTIF DOMAIN-CONTAINING GPI-ANCHORED PROTEIN2 (LYM2; Faulkner et al., 2013). Other members from this group are LYM1 and LYM3, and LYSIN MOTIF–CONTAINING PROTEIN4 and LYSIN MOTIF–CONTAINING PROTEIN6. The former two are receptors of peptidoglycan (PGN; Willmann et al., 2011), while the latter two function to sense both PGN and chitin (Liu et al., 2012a).
Figure 2.
Plant PRRs and NLRs. A, Schematic diagrams depicting domain structures of different classes of plant PRRs. Upper, The RK-PRRs. Lower, The RLP-PRRs. The representatives in each class are shown on the right. SP, signal peptide; EGF, epidermal growth factor. B, Schematic diagrams of domain structures of different NLRs. The representatives in each class are shown on the right. Upper, Domain structures of plant NLRs based on their variable N-terminal domains. Lower, Domain structures of plant-paired NLRs and helper NLRs.
Ligand sensing by ECDs activates KDs for immune signaling. Because of the lack of KDs, RLP-PRRs generally function together with RKs. PAMP perception by single RK-PRRs such as Arabidopsis (Arabidopsis thaliana) AtCERK1 has been reported (Miya et al., 2007; Wan et al., 2008), but many RK-PRRs require a coreceptor for signaling. Several recent studies (Jaillais et al., 2011; Smakowska-Luzan et al., 2018; Xi et al., 2019) suggested that the size of ECD is crucial for ligand perception by LRR-RKs and could be used to predict whether they function as receptors or coreceptors. For instance, when the LRR-RKs containing large size of ECDs act as the ligand-binding receptors, another small LRR-RK is preferred for coreceptor, but not itself. The LRR-RK SOMATIC EMBRYOGENESIS RECEPTOR KINASE3 (SERK3, also called BRI1-ASSOCIATED KINASE1 [BAK1]) and its orthologs are commonly shared coreceptors by LRR-RKs involved in diverse signaling pathways including immunity (Ma et al., 2016). Additionally, BAK1/SERKs together with the LRR-RK SUPPRESSOR OF BIR-1 (SOBIR1) also function as coreceptors of multiple LRR-RLPs (Liebrand et al., 2014). Given their critical role in plant immunity, BAK1/SERKs are subjected to negative regulation by pathogen effectors (Ma et al., 2016) and by host components like the LRR-RK BAK1-INTERACTING RECEPTOR KINASEs (BIRs; Gao et al., 2009; Halter et al., 2014) in Arabidopsis. More recently, the Arabidopsis malectin-like RK FERONIA (FER) was shown to act as a scaffold for regulation of assembly of PRR-containing complexes (Shen et al., 2017; Stegmann et al., 2017).
PAMP-induced Homodimerization of PRRs for Activation
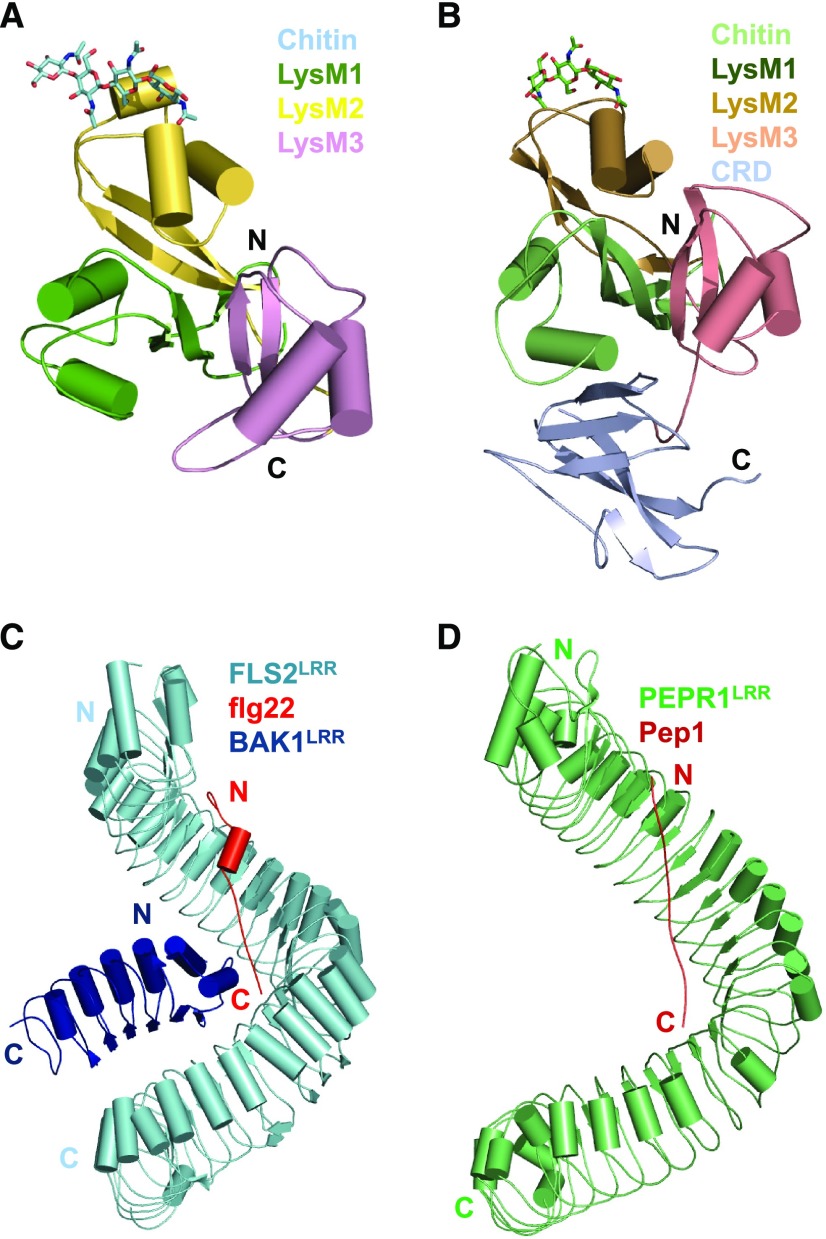
Chitin, a polymeric n-acetyl-glucosamine (NAG), is a well-characterized fungal PAMP. Different mechanisms are employed by Arabidopsis and rice (Oryza sativa) for perception of the PAMP. The LysM-RKs AtCERK1 (Miya et al., 2007; Wan et al., 2008; Liu et al., 2012b) and LYSIN MOTIF RECEPTOR KINASE5 (Cao et al., 2014) are direct receptors of chitin in Arabidopsis. In contrast, direct chitin recognition in rice is through the LysM-RLP OsCEBiP to activate OsCERK1 (Kaku et al., 2006; Shimizu et al., 2010; Liu et al., 2016a). The crystal structures of AtCERK1ECD and OsCEBiPECD in complex with a chito-pentamer and a chito-tetramer, respectively, revealed a conserved chitin recognition mechanism (Fig. 3, A and B; Liu et al., 2016a, 2012b). The structures of AtCERK1ECD and OsCEBiPECD are conserved in their LysMs, but only LysM2 was found to bind chitin. In the structures, chito-oligomers anchor to a shallow groove created by two loops of LysM2. Recognition of the chitin oligomers by AtCERK1 and OsCEBiP is mainly through a conserved set of residues of LysM2 that interacts with three NAG units.
Figure 3.
PAMP/DAMP recognition and activation of PRRs. A, Recognition of chitin by AtCERK1. Shown in the figure is the crystal structure of AtCERK1ECD in complex with a chito-pentamer (PDB: 4EBZ). AtCERK1ECD is shown in cartoon representation. Four chitin residues well defined in the structure are shown in stick representation. Color codes for three LysM domains are indicated. N, N terminus; C, C terminus. B, Recognition of chitin by OsCEBiPECD. Shown in the figure is the crystal structure of OsCEBiPECD in complex with a chito-tetramer (PDB: 5JCE). C, Flg22 acts as molecular glue to induce heterodimerization of FLS2 with its coreceptor BAK1. Shown is the overall structure of the FLS2LRR–flg22–BAK1LRR complex in cartoon representation (PDB: 4MN8). The color codes are indicated. D, Recognition of Pep1 by its receptor PERP1. The crystal structure of PERP1LRR in complex with AtPep1 is shown in cartoon representation (PDB: 5GR8).
Interaction of the N-acetyl groups of chitin with AtCERK1 and OsCEBiP can allow the two proteins to distinguish the PAMP from β‐1,3‐glucan (Glc; Liu et al., 2016a, 2012b). However, in addition to chitin, PGN (Willmann et al., 2011) and nonbranched Glc (Melida et al., 2018) also trigger AtCERK1-mediated immune responses in Arabidopsis. PGN perception by AtCERK1 is through the LysM-RLPs LYM1 and LYM3, although how AtCERK1 is activated remains elusive. Modeling studies suggested that β‐1,3‐Glc6 also binds to LysM2 of AtCERK1 but with a different orientation from the chitopentamer or the chitotetramer (Melida et al., 2018). AtCERK1 was suggested to recognize chitosan, a partially deacetylated chitin, to elicit immune responses in Arabidopsis (Cabrera et al., 2010; Petutschnig et al., 2010), which is further confirmed by a more recent study (Gubaeva et al., 2018).
Biochemical and functional data support chitin-induced AtCERK1 homodimerization for activation (Liu et al., 2012b). A longer chitin chain was proposed to act as a cross linker along which two AtCERK1 molecules bind for dimerization, known as the cross-linking model. A later modeling study of OsCEBiP dimerization suggested that hexachitin mediates homodimerization of OsCEBiP, with each LysM2 binding three NAGs in a “sliding mode” (Liu et al., 2016a), further supporting the cross-linking model. Two alternative models were recently suggested on chitin-induced OsCEBiP/AtCERK1 dimerization. One is called the sandwich-like model, where two CEBiP molecules simultaneously bind to one chitin chain from opposite sides (Hayafune et al., 2014). Analyses of chitin/chitosan oligosaccharides with varying degrees of polymerization or acetylation led to the slipped sandwich model (Gubaeva et al., 2018), in which two AtCERK1 molecules form an off-set chitin-binding groove for chitin or chitosan binding. This new model is a combination of the above models and provides an explanation for inhibition of chito-octamer-induced immunity by a chitosan octamer consisting of alternating GlcN and GlcNAc (Hayafune et al., 2014). Regardless of the mechanisms involved, chitin-induced homodimerization is required for AtCERK1 and OsCEBiP activation.
PAMPs and DAMPs Act as Molecular Glue To Induce Heterodimerization of PRRs with Their Co-receptors
The LRR-RK PRR FLS2 perceives flagellin by recognizing its highly conserved N-terminal epitope, a 22-residue peptide called flg22. BAK1 as a coreceptor is required for FLS2-mediated immune signaling (Chinchilla et al., 2007; Sun et al., 2013). The crystal structure of the ecto-LRR domain of FLS2 (FLS2LRR) in complex with flg22 and BAK1LRR revealed that flg22 adopts an elongated conformation interacting with the inner surface of FLS2LRR (Sun et al., 2013; Fig. 3C). Flg22 binding creates a novel surface on FLS2LRR for interaction with BAK1LRR. The C-terminal side of flg22 is sandwiched between FLS2LRR and BAK1LRR, indicating that flg22 acts as molecular glue to connect FLS2 with BAK1. In addition to the flg22-mediated interaction, BAK1LRR also anchors to the C-terminal portion of FLS2LRR. Both flg22-mediated and direct FLS2LRR–BAK1LRR contacts are important to form the FLS2–flg22–BAK2 complex.
The Arabidopsis PLANT ELICITOR PEPTIDES are classic DAMPs recognized by the LRR-RKs PEPR1 and PEPR2 (Yamaguchi et al., 2006, 2010). The crystal structure of PEPR1LRR bound by AtPep1 showed that their recognition mechanism is remarkably conserved with that for FLS2LRR recognition of flg22 (Tang et al., 2015; Fig. 3, C and D), although flg22 and AtPep1 are sequence-unrelated. Similarly, AtPep1 induced a heterodimeric PEPR1LRR–BAK1LRR complex. Modeling and binding studies indicated that the C-terminal side of AtPep1 is required for PEPR1LRR interaction with BAK1LRR, supporting AtPep1 as molecular glue to induce PEPR1LRR–BAK1LRR heterodimerization. Later biochemical and structural studies showed that many plant growth-promoting peptides such as CLAVATA3/ENDOSPERM SURROUNDING REGION-RELATED41 (Zhang et al., 2016), INFLORESCENCE DEFICIENT IN ABSCISSION (Santiago et al., 2016), and ROOT MERISTEM GROWTH FACTOR1 (Song et al., 2016) employ a similar mechanism to induce heterodimerization of their respective receptors with the BAK1/SERKs coreceptors.
Sequestering of BAK/SERKs by BIRs Negatively Regulates Plant Immunity
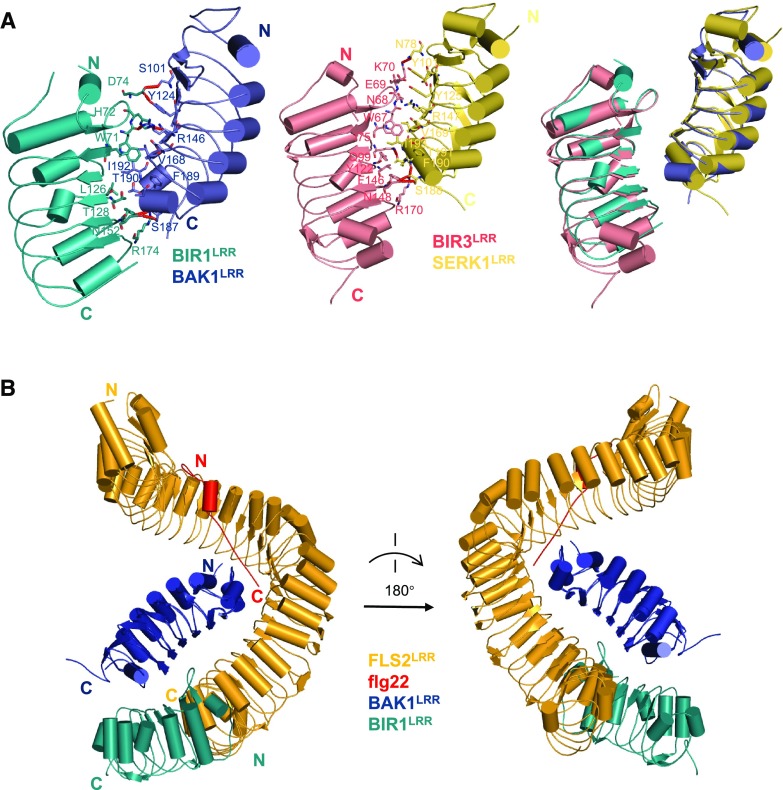
BIR1 was initially identified as a BAK1-interacting protein (Gao et al., 2009). Loss of BIR1 led to SOBIR1-dependent autoimmunity and cell death. There are four BIR members (BIR1, BIR2, BIR3, and BIR4) in Arabidopsis, and all of them interacted with BAK1 when expressed in Nicotiana benthamiana (Halter et al., 2014). Recent structural and biochemical studies (Ma et al., 2017; Hohmann et al., 2018) underlined that the ecto-domains of BAK1 and BIR1-4 are sufficient for their interaction. BIR1LRR–BAK1LRR interaction is mediated by packing of one lateral side of BIR1LRR against the C-terminal inner surface and the C-terminal capping domain of BAK1LRR (Fig. 4A). The BAK1-interacting residues are highly conserved among BIR1–BIR4, suggesting a conserved mechanism of BIR–BAK1 interaction as further confirmed by the structure of BIR3LRR–SERK1LRR (Fig. 4A; Hohmann et al., 2018). Importantly, structural comparison showed that the BIR1-contacting surface of BAK1LRR or the BIR3-contacting surface of SERK1LRR is also involved in interaction with other LRR-RKs such as FLS2 (Fig. 4B), suggesting that a BIR and these LRR-RKs may compete for interaction with BAK1/SERKs. Indeed, the FLS2LRR–flg22 complex efficiently outcompeted BIR1LRR for BAK1LRR binding (Ma et al., 2017). A similar observation was made for BRI1LRR–brassinolide (BRASSINOSTEROID INSENSITIVE1 [BRI1]) with BIR2LRR and BAK1LRR (Hohmann et al., 2018). These data support the idea that a BIR can negatively regulate BAK1/SERK signaling by sequestering them from their paired RKs, as suggested by Halter et al. (2014). A similar mechanism is applied to negative regulation of BR (brassinosteroid) signaling by BIR3 (Imkampe et al., 2017).
Figure 4.
Sequestering of the coreceptor of PRRs by BIRs. A, Crystal structures of the BIR1LRR–BAK1LRR complex (left) and the BIR3LRR–SERK1LRR complex (middle, PDB: 6FG8), and the structural alignment of these two complexes (right). Residues mediating detail interactions between BIR1LRR–BAK1LRR and BIR3LRR–SERK1LRR are shown in stick. B, Structural alignment between FLS2LRR–flg22–BAK1LRR complex and BIR1LRR–BAK1LRR. BAK1LRR was used as the template for the alignment.
Loss of BIR1 promotes BAK1-SOBIR1 interaction (Liu et al., 2016c), suggesting that BIR1 and SOBIR1 may interact with BAK1 in a competitive manner. This mechanism is consistent with the observation that overexpression of full-length or the ECD-TM of BAK1 in plants generated SOBIR1-dependent autoimmunity (Domínguez-Ferreras et al., 2015). On the other hand, overexpression of ECD-TM of BAK1 can interfere with immune signaling mediated by BAK1 and SOBIR1. Consistently, plants overexpressing the ECD-TM of BAK1 developed better than those overexpressing full-length BAK1 (Domínguez-Ferreras et al., 2015). Although both BIR1 and SOBIR1 interact with BAK1, the BIR1-binding region of BAK1 is less likely to completely overlap with the SOBIR1-interacting domain of BAK1, as transgenic plants expressing a BAK1 mutant protein with compromised binding to BIR1 were constitutively active in inducing immune responses (Ma et al., 2017). It currently remains unknown what signals relieve BIR1-inhibited SOBIR1 signaling when needed. Given the fact that cell death in bir-1 occurs even under sterile conditions (Gao et al., 2009), such signals, if present, appear to be endogenous.
Despite their conserved biochemical activities, BIR1–BIR4 have diversified functions. BIR1 is important to inhibit immunity mediated by BAK1 and SOBIR1 (Gao et al., 2009), whereas BIR2 and BIR3 have critical roles in negative regulation of PTI (Halter et al., 2014) and negative regulation of BR signaling (Imkampe et al., 2017; Hohmann et al., 2018), respectively. One possibility to reconcile the conserved biochemical activities of BIRs with their signaling specificity may be that they exist in distinct pools that are accessible to different RK-signaling complexes. It is of interest to note that a recent study using live-cell imaging showed that FLS2 and BRI1 localize to distinct plasma membrane (PM) nanodomains (Bücherl et al., 2017). But whether this is the case with BIRs remains undetermined.
GPI-Anchored Proteins as Co-receptors of RKs to Regulate Plant Immunity
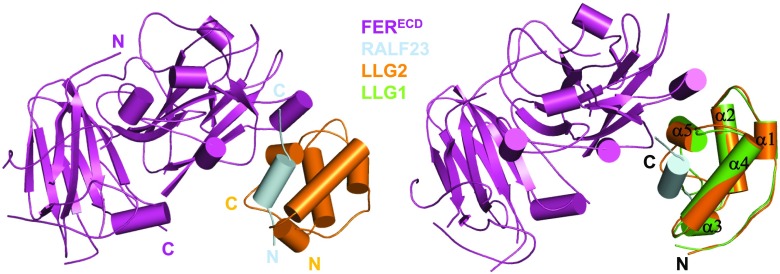
FER belongs to the Catharanthus roseus RLK1-like subfamily with 17 members in Arabidopsis (Franck et al., 2018) and plays pleiotropic roles in plant growth, development, and immunity. The endogenous Cys-rich peptides RAPID ALKALINIZATION FACTOR (RALFs; Pearce et al., 2001; Escobar-Restrepo et al., 2007; Haruta et al., 2014; Ge et al., 2017; Stegmann et al., 2017) and the glycosyl-phosphatidyl-inositol (GPI)-anchored proteins (GAPs) LORELEI and its homologs LLG1, LLG2, and LLG3 (Capron et al., 2008; Li et al., 2015; Liu et al., 2016b; Shen et al., 2017) are essential for FER-mediated signaling. FER negatively regulates PTI via recognition of RALF23 (Stegmann et al., 2017). The recently solved crystal structure of the RALF23–LLG2-FERECD complex revealed that RALF23 directly binds to LLG2 (Xiao et al., 2019; Fig. 5). A highly conserved N-terminal region is sufficient for RALF23 recognition by LLG1 and LLG2. Consistently, RALFs containing this region interact with LLG1, LLG2, and LLG3 and induce binding of the three LLG proteins to FERECD in vitro. Recognition of diverse RALFs via LLGs is consistent with the multitasking FER. Biochemical and functional data showed that recognition of RALF23 by LLG1 results in recruitment of FER through formation of a composite LLG1–RALF23 interface. Structural comparison between apo-LLG1 and the RALF23–LLG2–FERECD complex suggests that RALF23 binding induces no conformational change in LLG2 (Fig. 5). These and functional data established LLG1 as a coreceptor of FER to modulate plant immunity. Two more recent studies showed that LLG2 and LLG3 also function as coreceptors of the FER orthologs ANXUR/BUPS to regulate pollen tube growth and development (Feng et al., 2019; Ge et al., 2019) in response to RALF4 and RALF9. The emerging data suggest that LLGs function as coreceptors of different members of the CrRLK1-like subfamily for regulation of diverse signaling pathways.
Figure 5.
Structure of the RLF23–LLG2–FERECD complex. Left: Overall structure of the RLF23–LLG2–FERECD complex (PDB: 6A5E) shown in cartoon. The color codes are indicated. N, N terminus; C, C terminus. Right: Structural superposition of the RLF23–LLG2–FERECD complex and apo-LLG1 (PDB: 6A5D). The five α-helixes of LLG1 and LLG2 are indicated.
Around 250 GAPs are encoded in the genome of Arabidopsis (Zhou, 2019). The data discussed above suggest that other GAPs might also function as coreceptors to indirectly transmit signals from the PM by working in concert with RKs. OsCEBiP was initially thought to be an RLP, but was recently determined to be a GAP (Gong et al., 2017). Like RALF23 with LLG1 and FER, chitin binding induces OsCEBiP interaction with the RK OsCERK1 for defense signaling. The LysM-containing proteins LYM1 and LYM2 from Medicago truncatula were also shown to be GAPs (Fliegmann et al., 2011). Unlike OsCEBiP and BAK1/SERKs that act as coreceptors through homotypic interactions with other RKs, however, LLGs form ligand-induced complexes with the phylogenetically unrelated FER family members. Thus, the RALF-induced LLG-FER/ANXUR/BUPS complexes represent a novel type of ones for perception of plant peptides. The GAP GFRα in animals recognizes the glial-cell-line–derived neurotrophic factors and is consequently recruited to the receptor Tyr kinase RET (Paratcha and Ledda, 2008), forming complexes similar to those induced by RALFs.
In addition to LLGs, LRR-extensin (LRX) proteins also recognize RALF peptides. RALF4, RALF9–LRX1, and LRX2–LRX5 interaction was initially shown to be important for pollen tube growth (Mecchia et al., 2017). A recent structural study revealed the interaction mechanism between RALF and LRX (Moussu et al., 2019). Additionally, LRX3–LRX5 were also shown to associate with RALF22 and RALF23, and FER, to modulate plant salt tolerance in Arabidopsis (Zhao et al., 2018). More recently, Herger et al. (2019) indicated that FER, RALF1, and LRX1–LRX5 function together to coordinate plant growth. In future it will be worth testing whether the LRX proteins act as coreceptors of FER and its orthologs.
PLANT NLRS: INNATE IMMUNE RECEPTORS WITH HIGH SPECIFICITY IN PATHOGEN RECOGNITION
Conserved in animals and plants, NLRs have a modular domain architecture comprising a variable N-terminal domain, a conserved central nucleotide binding and oligomerization domain (NOD), and a C-terminal LRR domain (Maekawa et al., 2011b; Duxbury et al., 2016). A similar domain structure is present in the apoptotic protein APOPTOTIC PEPTIDASE ACTIVATING FACTOR1 (Apaf-1). The NOD module can be further divided into NB domain (NBD), helical domain1 (HD1), and winged helical domain (WHD; Fig. 2B). Both NLRs and Apaf-1 belong to the signal transduction ATPase with a numerous domain family (Lukasik and Takken, 2009). Depending on their N-terminal domains, plant NLRs can be broadly classified into coiled-coil (CC) and Toll/IL1 receptor/resistance proteins (TIR) NLRs (Fig. 2B). Among the CC-NLRs, one basal clade is distinguished by having CC domains resembling the resistance to powdery mildew8 protein, referred to as CCR-NLR (Collier et al., 2011). Despite their conserved domain structure, plant NLRs display highly diverse modes for perception of pathogen effectors. The most straightforward way for NLRs to detect pathogen effectors is through direct association. However, many NLRs indirectly recognize effectors by sensing effector-modified host components (guard model; Khan et al., 2016). Effector recognition in some cases requires two genetically linked NLRs (called paired NLRs) with one functioning as the sensor and the other as the executor (Fig. 2B; Césari et al., 2014a). A sensor NLR often contains an integrated domain responsible for effector binding as supported by structural and biochemical studies (Maqbool et al., 2015; Ortiz et al., 2017). Well-characterized paired NLRs include the CC-NLR pairs R-GENE ANALOG5 (RGA5)/RGA4 (Césari et al., 2013, 2014b) and Pik locus1/Pik locus2 (Zhai et al., 2011; Kanzaki et al., 2012) from rice and the Arabidopsis TIR-NLR pair RESISTANCE TO RALSTONIA SOLANACEARUM1 (RRS1)/RESISTANCE TO PSEUDOMONAS SYRINGAE4 (RPS4; Le Roux et al., 2015; Sarris et al., 2015). Recent studies showed that immune responses mediated by some NLRs require helper NLRs including N-REQUIRED GENE 1.1 (NRG1.1), NRG1.2, ACTIVATED DISEASE RESISTANCE GENE 1 (ADR1), ADR1-L1, and ADR1-L2 (Peart et al., 2005; Bonardi et al., 2011; Collier et al., 2011; Qi et al., 2018; Castel et al., 2019; Lapin et al., 2019; Wu et al., 2019) in Arabidopsis and tobacco (Nicotiana benthamiana) and NB-LRR PROTEIN REQUIRED FOR HR-ASSOCIATED CELL DEATH 2 (NRC2), NRC3, and NCR4 (Gabriëls et al., 2007; Wu et al., 2016, 2017) in Solanaceae. All helper NLRs belong to CC-NLRs (Fig. 2B). NRGs and/or ADR1/ADR-Ls are downstream components of TIR-NLR-mediated signaling but are differentially required for CC-NLRs, whereas NRCs are required for a subset of CC-NLRs from many families of angiosperms (Wu et al., 2017).
Cryo-Electron Microscopy Structures of ZAR1
The CC-NLR HOPZ-ACTIVATED RESISTANCE1 (ZAR1; Lewis et al., 2010) in Arabidopsis forms a constitutive complex with the RESISTANCE-RELATED KINASE1 (RKS1) pseudokinase to mediate immunity induced by the Xanthomonas campestris pathovar campestris effector AvrAC (Wang et al., 2015). AvrAC uridylylates the receptor-like cytoplasmic kinase PBS1-LIKE PROTEIN2 (PBL2), allowing the modified PBL2 (PBL2UMP) to be recognized by RKS1 in the preformed ZAR1–RKS1 complex. Recent cryo-electron microscopy (cryo-EM) structures of ZAR1 in resting, primed, and activated states revealed the mechanisms of autoinhibition, effector recognition, nucleotide exchange, and activation of the NLR (Wang et al., 2019a, 2019b).
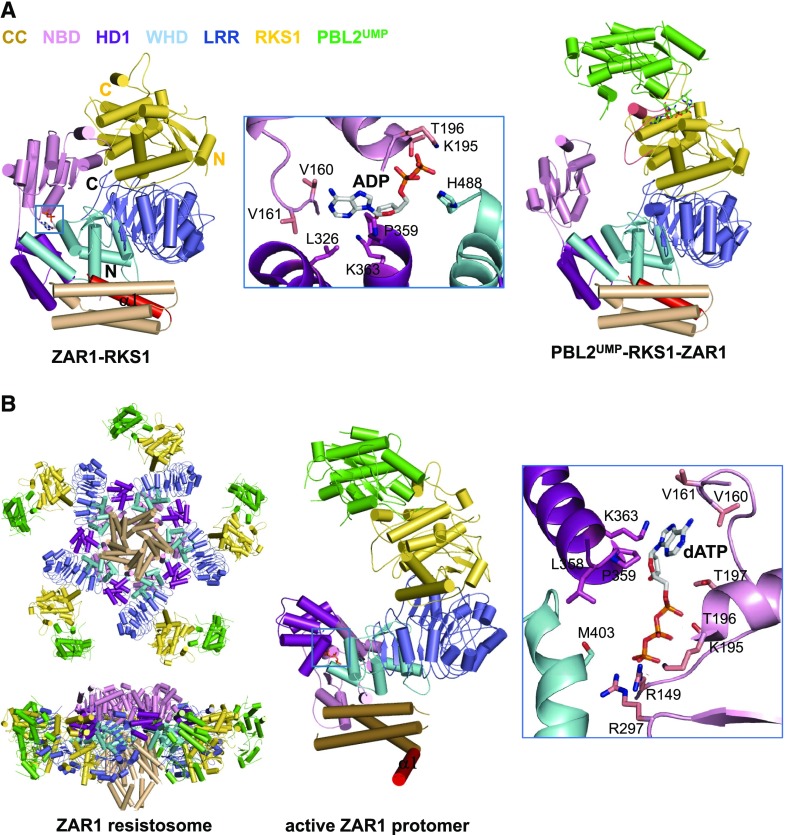
As found in the inactive NLRs NRC1 (Steele et al., 2019), NLR FAMILY CARD DOMAIN CONTAINING4 (NLRC4; Hu et al., 2013) and the NLR-like Apaf-1 (Riedl et al., 2005; Reubold et al., 2011), an ADP molecule, binds to a conserved pocket of inactive ZAR1 (Figs. 6A and 7). Multiple interdomain interactions within ZAR1 further stabilize the autoinhibited conformation (Wang et al., 2019b). One lateral surface of ZAR1LRR mediates specific ZAR1 interaction with RKS1, whereas PBL2UMP contacts exclusively RKS1 largely via the uridylated moieties of PBL2UMP. PBL2UMP binding stabilizes the activation segment of RKS1 that is unstructured in the inactive ZAR1–RKS1 complex, and induces ZAR1NBD rotation ∼60° outwards (Fig. 6A). Structural comparison further showed the PBL2UMP-stabilized activation region of RKS1 clashes with the inactive ZAR1NBD. These structural observations indicate that PBL2UMP binding allosterically induces conformational changes in ZAR1NBD to release ADP from the NLR (Fig. 6A).
Figure 6.
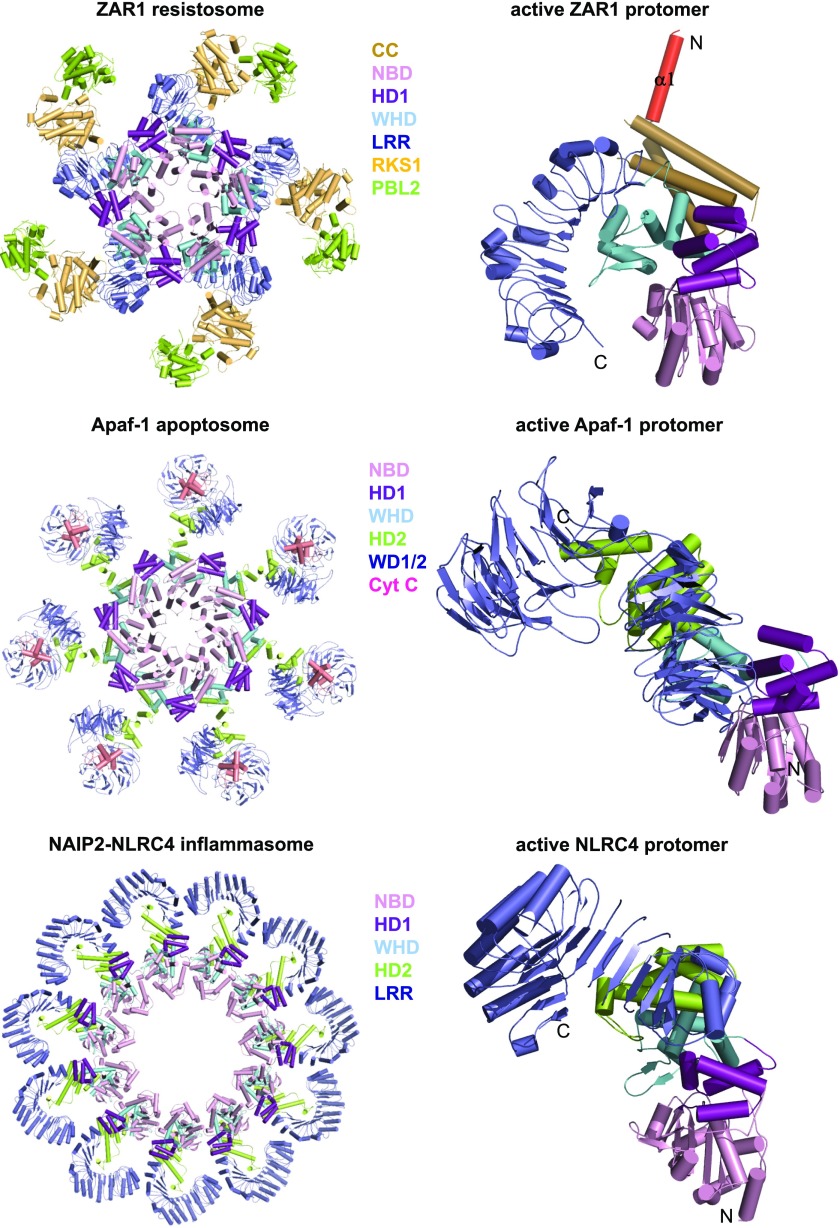
Auto-inhibition, priming, and activation of ZAR1. A, Overall structure of the ZAR1–RKS1 complex (PDB: 6J5W) shown in cartoon (left). The α1 is labeled in red. The bound ADP molecule is shown in stick and its detailed interactions with ZAR1 are shown in the middle representation.Overall structure of the PBL2UMP–RKS1–ZAR1 complex (PDB: 6J5V) shown in the same orientation as that in the left (right). The loop region that is disordered in the ZAR1–RKS1 complex is shown in salmon. B, Structure of the ZAR1 resistosome (PDB: 6J5T; left). Structure of an active ZAR1 protomer (middle). The α1 is labeled in red. The detailed interactions between dATP and ZAR1NOD (right)
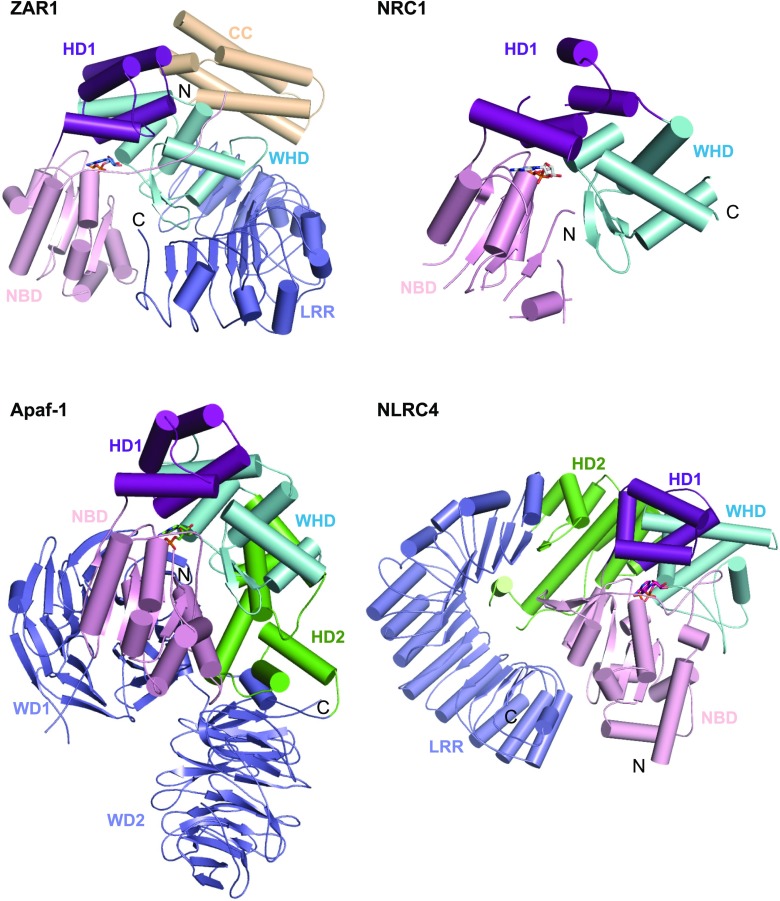
Figure 7.
Structural comparison of inactive ZAR1, NRC1, Apaf-1, and NLRC4. Structures indicated are shown with their NBD–HD1–WHD segments aligned. ZAR1 (PDB: 6J5W), NRC1 (PDB: 6S2P), Apaf-1 (PDB: 3SFZ), and NLRC4 (PDB: 4KXF).
The monomeric ZAR1–RKS1–PBL2UMP in the absence of (d)ATP is reminiscent of the monomeric Apaf-1–cytochrome c complex (Zhou et al., 2015). Like Apaf-1 assembly into the apoptosome (Zhou et al., 2015), (d)ATP induces formation of an oligomeric ZAR1–RKS1–PBL2UMP complex termed ZAR1 resistosome (Wang et al., 2019a). Cryo-EM analysis revealed a wheel-like pentamer of the ZAR1 resistosome, comparable to the structures of the Apaf-1 apoptosome (Zhou et al., 2015) and the NLRC4 inflammasome (Hu et al., 2015; Zhang et al., 2015). Formation of the ZAR1 resistosome is mediated by ZAR1 but not by RKS1 and PBL2UMP. dATP binding induces structural reorganization between ZAR1HD1 and ZAR1WHD (Fig. 6B), as demonstrated in Apaf-1 (Zhou et al., 2015) and NLRC4 (Hu et al., 2015; Zhang et al., 2015). Structural alignment revealed fold switching of ZAR1CC after activation. Interestingly, the very N-terminal α-helix (α1) largely buried in the inactive ZAR1 becomes completely exposed after ZAR1 activation, forming a funnel-shaped structure in the ZAR1 resistosome (Fig. 6B). These results support stepwise activation of the ZAR1 resistosome, first primed by AvrAC and then fully activated by (d)ATP (Wang et al., 2019a).
ZAR1CC is sufficient to induce HR cell death when expressed in N. benthamiana (Baudin et al., 2017). The oligomerized ZAR1CCs, however, are deeply buried in the ZAR1 resistosome except the funnel-shaped structure, suggesting that α1 is important for ZAR1 function. Indeed, N-terminal deletion mutants of ZAR1 lost AvrAC-induced HR cell death in protoplasts and resistance to X. campestris (Wang et al., 2019a). Remarkably, simultaneous mutation of Glu-11 and Glu-18 from the inner surface of the funnel-shaped structure substantially compromised the AvrAC-induced activities of ZAR1. Fractionation and mutagenesis assays showed that ZAR1 became PM-associated upon activation. These results suggest that the ZAR1 resistosome may directly function as a channel or a pore to mediate HR cell death and immune responses. Alternatively, it is also possible that recruitment to the membrane could bring ZAR1 into proximity with other yet-unidentified signaling proteins for further induction of cell death and resistance.
Autoinhibition and Ligand Sensing of NLRs
Although animal and plant NLRs are believed to have evolved independently (Jones et al., 2016), arrangement of NBD, HD1, and WHD is highly conserved in the inactive ZAR1, NRC1, NLRC4, and Apaf-1 (Fig. 7). Similar domain positioning is also found in the prototype NLR PH0952 from the hyperthermophilic euryarchaeota Pyrococcus horikoshii (Lisa et al., 2019). The C-terminal domains of ZAR1, NLRC4, Apaf-1, and PH0952 function to sequester these NLRs in a monomeric state, although they are differently positioned in the structures (Reubold et al., 2011; Hu et al., 2013; Lisa et al., 2019; Wang et al., 2019b). These structural observations suggest a conserved autoinhibition mechanism of NLRs.
The C-terminal LRR domain is widely hypothesized to act as the ligand sensor of an NLR. Indeed, some plant NLRs including RECOGNITION OF PERONOSPORA PARASITICA1 (Krasileva et al., 2010) from Arabidopsis and MILDEW-A (MLA; Lu et al., 2016) from barley (Hordeum vulgare) have been mapped to recognize their ligands through the variable C-terminal LRR region. While ZAR1LRR does not directly contact PBL2UMP, recognition of the AvrAC-modified PBL2 is through the LRR-bound RKS1. Thus, ZAR1LRR is the structural determinant for specific recognition of AvrAC. The integrated domains from several sensor NLRs are responsible for effector binding (Le Roux et al., 2015; Maqbool et al., 2015; Sarris et al., 2015; Ortiz et al., 2017). Additionally, the CC and TIR domains can also act as a sensor of effectors. For example, the TIR-only protein RESPONSE TO THE BACTERIAL TYPE III EFFECTOR PROTEIN HOPBA1 may act as a receptor of the effector protein HopB1 (Nishimura et al., 2017). Regardless of the recognition mechanisms, effector binding would function to trigger conformational changes in an NLR, promoting exchange of ADP with ATP/dATP to induce structural remodeling for full activation.
Activation and Oligomerization of NLRs
The conserved positioning of NBDs, HD1s, and WHDs in the inactive (Fig. 7) and active states of ZAR1, NLRC4, and Apaf-1 (Fig. 8) further solidifies the notion that structural remodeling generally accompanies NLR activation. The underlying mechanisms, however, can vary among different types of plant NLRs. Singleton NLRs could follow the mechanism demonstrated in ZAR1 (Wang et al., 2019a) and Apaf-1 (Zhou et al., 2015) for structural reorganization and activation. The NAIP–NLRC4 inflammasomes’ NEURONAL APOPTOSIS INHIBITOR PROTEIN (NAIP; Hu et al., 2015; Zhang et al., 2015) appears to be an attractive model for activation of paired NLRs. This model, however, needs formation of a substoichiometric complex between the sensor and the executor, which share a common promoter in an NLR pair. Furthermore, unlike the ligand-induced NAIP–NLRC4 complexes, constitutive heteromeric complexes have been shown for several paired NLRs (Césari et al., 2014b; Le Roux et al., 2015; Sarris et al., 2015). A more recent study showed that knockout of sensor NLRs from several NLR pairs in rice produced HR-like phenotypes (Wang et al., 2019c), supporting an inhibitory role of the sensors in activating the paired NLRs and agreeing with the model on activation of the paired NLRs RRS1/RPS4 (Le Roux et al., 2015; Sarris et al., 2015) and RGA5/RGA4 (Césari et al., 2014b).
Figure 8.
Structural comparison of active ZAR1, Apaf-1, and NLRC4. Shown in the left column are the structures of the ZAR1 resistosome (PDB: 6J5T), the Apaf-1 apoptosome (PDB: 3JBT), and the NLRC4 inflammasome (PDB: 3JBL). Protomers of these three complexes are shown in the right column with the NBD1–HD1–WHD segments aligned.
Less is known about how helper NLRs are activated. Signaling mediated by TIR-NLRs requires ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1)/PHYTOALEXIN DEFICIENT4 or EDS1/SENESCENCE-ASSOCIATED GENE101 (Wiermer et al., 2005) and the helper NLRs NRG1s and/or ADR1/ADR1-Ls (Peart et al., 2005; Roberts et al., 2013; Qi et al., 2018; Castel et al., 2019; Wu et al., 2019). Two recent studies (Horsefield et al., 2019; Wan et al., 2019a) showed that TIR-NLRs possess NADase activity. Thus, one plausible model on activation of helper NLRs might be that they sense a signaling molecule(s) generated by TIR-NLRs probably through EDS1. Identification of the putative signaling molecule(s) would be a key to understanding how NRG1s and/or ADR1/ADR1-Ls are activated. However, ADR1/ADR1-Ls are also required for some CC-NLRs (Bonardi et al., 2011; Roberts et al., 2013), raising the question of whether these helper NLRs can sense different signals. In contrast with paired NLRs, NRCs and NRC-dependent NLRs were proposed to act through positive regulation (Wu et al., 2017). In line with this model, overexpression of RxNBD of the NRC-dependent RESISTANCE TO POTATO VIRUS X in N. benthamiana induces HR-like cell death (Rairdan et al., 2008). This appears to suggest that activation of NRCs is comparable to that of NLRC4 in the NAIP–NLRC4 inflammasomes. However, whether the observed phenotype is dependent on an NRC was not tested. Furthermore, there has been no evidence showing effector-induced interaction between a sensor NLR and an NRC.
The active ZAR1, NLRC4, and Apaf-1 form wheel-like structures despite their different oligomerization status (Fig. 8; Hu et al., 2015; Zhang et al., 2015; Zhou et al., 2015; Wang et al., 2019a). Oligomerization functions to bring their N-terminal signaling domains into proximity, forming a funnel-like structure in the ZAR1 resistosome and caspase-recruiting platforms in the NLRC4 inflammasome and the Apaf-1 apoptosome. Oligomerization can be generally important for plant NLR function. Self-association of TIR domains required for effector-triggered immune response (Bernoux et al., 2011; Williams et al., 2014; Zhang et al., 2017a) supports such a model. However, it remains unknown whether TIR-NLRs form a structure similar to that of the ZAR1 resistosome. Notably, unclosed structures have been observed for some NLRs. For example, deletion of the N-terminal CARD domain of NLRC4 resulted in formation of helical NAIP5–NLRC4 structures (Diebolder et al., 2015; Matyszewski et al., 2018). Another example is the NLR-like protein MalT (the central activator of the mal genes) from Escherichia coli, which forms maltotriose-induced curved oligomers (Larquet et al., 2004).
ATP/dATP-Binding and Hydrolysis of NLRs
Structural studies support the notion that the ADP- and (d)ATP-bound forms of an NLR correspond to off- and on-states, respectively. As in the Apaf-1 apoptosome (Zhou et al., 2015), the γ-phosphate group of dATP functions to stabilize the active conformation of ZAR1 for oligomerization (Wang et al., 2019a). Supporting the important role of the γ-phosphate group, other triphosphate nucleosides such as GTP or the nonhydrolyzable AMP-PNP also support assembly of the Apaf-1 apoptosome (Reubold et al., 2009). However, activation of an NLR may not necessarily require (d)ATP binding. For example, flagellin binding alone stabilizes the active conformation of NAIP5 (Tenthorey et al., 2017; Yang et al., 2018), explaining the dispensability of the ATP-binding of this NLR (Halff et al., 2012). Similarly, the P-loop of NRG1.1, NRG1.2, and ADR1-L2 from Arabidopsis is dispensable for their helper function (Bonardi et al., 2011; Wu et al., 2019) downstream of the effector-activated sensor NLRs. Interestingly, however, the auto-activity of overexpressed N. benthamiana NRG1 (Peart et al., 2005) and the autoactive mutant ADR1-L2 (D484V) is P-loop–dependent (Roberts et al., 2013). The reason for this remains unclear, but it appears that different NLRs employ distinct mechanisms to stabilize their active conformation for oligomerization, although ATP/dATP binding is likely the most commonly used one.
Many NLRs are predicted to have the catalytic elements of an ATPase. Indeed, NLR proteins including M and L6 (Williams et al., 2011; Bernoux et al., 2016) from plants, and NLRC4 (Hu et al., 2013) from animals, exhibit ATP-hydrolyzing activity. ATP hydrolysis may function to switch the (d)ATP-bound active state back to the ADP-bound inactive state. However, whether the proteins tested for ATP hydrolysis were in active states was not reported. Notably, the catalytic pocket of NLRs is formed by an individual monomer and is not, as in the case for the canonical ATPASES ASSOCIATED WITH DIVERSE CELLULAR ACTIVITIES, a composite pocket formed by two neighboring monomers in the oligomer (Erzberger and Berger, 2006). One study appeared to argue against the model above by showing that only inactive Apaf-1 displayed low ATPase activity but not Apaf-1 from the apoptosome (Reubold et al., 2009). This agrees with the idea that activation of Apaf-1 apoptosome represents the point-of-no-return of programmed cell death pathways (Riedl and Salvesen, 2007).
Altered Subcellular Localization of ZAR1 upon Activation
In parallel to MIXED LINEAGE KINASE DOMAIN-LIKE PROTEIN (MLKL) oligomerization and translocation to the PM after activation (Cai et al., 2014; Chen et al., 2014; Wang et al., 2014), ZAR1 activation induced by AvrAC results in relocalization of the NLR from the cytosol to the PM to mediate cell death. Strong evidence for the altered localization of ZAR1 comes from the E11A/E18A mutation, which did not affect assembly of the ZAR1 resistosome but nearly abolished AvrAC-induced cell death in protoplasts. Because of the loss of cell death activity, the PM-association of the ZAR1 mutant was easily detected (Wang et al., 2019a). Identification of similar mutations in other CC-NLRs is possible, because a more recent study (Adachi et al., 2019) showed that the very N-terminal fragments of many singleton and helper CC-NLRs are also functionally important when tested in tobacco. Such mutations would be valuable in investigating cellular localization of NLRs, particularly because NLRs have been shown to function in different compartments including nucleus, endoplasmic reticulum, and Golgi apparatus (Cui et al., 2015). Additionally, because these mutations can arrest an activated form of NLRs, they might also be used to identify components regulating NLR complexes. Mutations of the conserved catalytically and functionally important glutamic residue in TIR-NLRs can serve similar purposes.
Pore-Forming Activity of ZAR1CC
Structural and biochemical data suggest that the funnel-shaped structure in the ZAR1 resistosome may function as a channel or pore in the PM. As noted in Burdett et al. (2019), the funnel-shaped structure bears striking similarity to the pore-forming protein MITOCHONDRIAL CALCIUM UNIPORTER from Caenorhabditis elegans (Oxenoid et al., 2016) and the calcium channel Orai from fruit fly (Drosophila melanogaster; Hou et al., 2012). Although many more investigations are needed to test this model, the pore-forming activity of a CC domain was demonstrated in other proteins. For example, the HeLo domain of fungal Het-S (a prion protein encoded by het-s locus of the nine het-loci), which is a four-helix bundle like a canonical CC domain, forms pores in the PM after activation to mediate cell death (Seuring et al., 2012). Induced pore formation by the N-terminal CC domain of MLKL in animals was also demonstrated (Huang et al., 2017). The very N-terminal α1 helix forming the funnel-shaped structure in the ZAR1 resistosome is conserved in many distantly related CC-NLRs (Adachi et al., 2019). Assays performed in N. benthamiana showed that the N-terminal fragments are functionally exchangeable among several CC-NLRs. Notably, when fused with Yellow Fluorescent Protein at the C terminus, the N-terminal 29 amino acids of NRC4 were sufficient to induce cell death. But whether the N-terminal fragment of NRC4 associates with PM and the NLR forms a ZAR1 resistosome-like structure remains unknown.
Formation of the funnel-like structure is remarkably similar to that of the hemolytic actinoporin fragaceatoxin (FraC; Tanaka et al., 2015), although ZAR1CC and FraC share little structural similarity. Interestingly, fold switching occurs to both ZAR1CC and FraC during assembly of the funnel-like structures. This is also true with the pore-forming protein Het-S (Daskalov et al., 2015). Fold plasticity of the CC domain appears to also exist in other CC-NLRs. The CC domains of the barley NLR Sr33 and wheat NLR MLA10 display different fold topologies when their structures were determined by nuclear magnetic resonance (Casey et al., 2016) and crystallography (Maekawa et al., 2011a; Casey et al., 2016), despite their highly conserved sequences.
FUTURE PERSPECTIVES
Despite the progress in structural studies of PRRs and NLRs, many open questions remain concerning these two families of proteins (see Outstanding Questions). Obtaining structures of full-length signaling-competent PRR complexes is one challenge for full understanding of how PAMPs/DAMPs activate them. Clustering of receptor Tyr kinases is important for their activation in animals (Kotani et al., 2008) and is now beginning to be appreciated as an important facet of RK activation in plants (Somssich et al., 2015; Bücherl et al., 2017). Structural and biochemical investigations will allow us to understand how this mechanism operates in PRR activation. Although several structures of LRR- and LysM-type PRRs have been solved, structural mechanisms of ligand recognition and activation of several types of PRRs remain to be elucidated. Similarly, how ligand recognition by LRR-RLPs, including those as resistance proteins such as the Cladosporium fulvum proteins (Postma et al., 2016; Wan et al., 2019b) activate their coreceptors BAK1/SERKs and/or SOBIR1, is still poorly understood. The fact that BAK1/SERKs function as coreceptors of many LRR-RKs, including PRRs, raises the question of how the loose specificity is achieved. It should be noted that the coreceptor RKs typically have diverse functions and can mediate different signaling than the ligand-binding ones. Assignment of the nonligand binding functions of these RKs would be a direction in the future studies.
Currently it remains unknown whether the ZAR1 resistosome functions as an executor or a trigger of immune responses. Many investigations will be required to test the model on the ZAR1 resistosome as a channel or a pore. Although oligomerization can be ingrained into the model of NLR activation, direct evidence for this from TIR-NLRs is still lacking. Whether other plant NLRs can form resistosome-like structures is another open question. Structural information of an active TIR-NLR is of particular interest, because it will not just help address this question but also may explain whether and why oligomerization is required for its potential NADase activity. Reconstitution of active complexes containing the helper NLRs NRG1s and ADR1/ADR1-Ls may critically depend on the molecule(s) produced by TIR-NLR as NADases or probably even other enzymes. Thus, identification of such a molecule(s) represents one major challenge to dissect the activation mechanisms of helper NLRs. In addition to effector sensing and negative regulation of immune response, RRS1 also contributes to RPS4-mediated signaling (Narusaka et al., 2009; Ma et al., 2018). How effector binding relieves the negative regulation by RRS1, and how RRS1 contributes to the activation of RPS4, remains elusive. Addressing these questions would provide a model on how other paired NLRs are activated. Emerging evidence suggested that NRCs may follow a similar mechanism to ZAR1 for signaling (Adachi et al., 2019). But how NRCs are activated remains enigmatic. The ZAR1 resistosome is just the tip of the NLR iceberg. With the evergrowing advance in cryo-EM, structural biology will reveal many more exciting mechanisms of NLR action.
Acknowledgments
We apologize to researchers whose relevant studies were not cited in this review due to page limitations.
Footnotes
The work was supported by the Alexander von Humboldt Foundation (Humboldt Professorship to J.C.).
Articles can be viewed without a subscription.
References
- Adachi H, Contreras MP, Harant A, Wu CH, Derevnina L, Sakai T, Duggan C, Moratto E, Bozkurt TO, Maqbool A, et al. (2019) An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 8: e49956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M, Hassan JA, Schreiber KJ, Lewis JD(2017) Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol 174: 2038–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Burdett H, Williams SJ, Zhang X, Chen C, Newell K, Lawrence GJ, Kobe B, Ellis JG, Anderson PA, et al. (2016) Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28: 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN(2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T(2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20: 47–54 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL(2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C(2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55: 257–286 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G(2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C(2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett H, Bentham AR, Williams SJ, Dodds PN, Anderson PA, Banfield MJ, Kobe B(2019) The plant “Resistosome”: Structural insights into immune signaling. Cell Host Microbe 26: 193–201 [DOI] [PubMed] [Google Scholar]
- Cabrera JC, Boland A, Cambier P, Frettinger P, Van Cutsem P(2010) Chitosan oligosaccharides modulate the supramolecular conformation and the biological activity of oligogalacturonides in Arabidopsis. Glycobiology 20: 775–786 [DOI] [PubMed] [Google Scholar]
- Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, Ward Y, Wu LG, Liu ZG(2014) Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 16: 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G(2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A, Gourgues M, Neiva LS, Faure JE, Berger F, Pagnussat G, Krishnan A, Alvarez-Mejia C, Vielle-Calzada JP, Lee YR, et al. (2008) Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey LW, Lavrencic P, Bentham AR, Césari S, Ericsson DJ, Croll T, Turk D, Anderson PA, Mark AE, Dodds PN, et al. (2016) The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci USA 113: 12856–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG(2019) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222: 966–980 [DOI] [PubMed] [Google Scholar]
- Césari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN(2014a) A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front Plant Sci 5: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T(2014b) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33: 1941–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li W, Ren J, Huang D, He WT, Song Y, Yang C, Li W, Zheng X, Chen P, et al. (2014) Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res 24: 105–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T(2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ(2006) Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G(2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P(2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE(2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ(2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Martinez D, Debets AJ, Sabaté R, Loquet A, Saupe SJ(2015) Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol 13: e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI(2015) Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure 23: 2349–2357 [DOI] [PubMed] [Google Scholar]
- Domínguez-Ferreras A, Kiss-Papp M, Jehle AK, Felix G, Chinchilla D(2015) An overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1-1–dependent manner. Plant Physiol 168: 1106–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z, Ma Y, Furzer OJ, Huh SU, Cevik V, Jones JD, Sarris PF(2016) Pathogen perception by NLRs in plants and animals: Parallel worlds. BioEssays 38: 769–781 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM(2006) Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 35: 93–114 [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U(2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ(2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Liu C, Fu R, Zhang M, Li H, Shen L, Wei Q, Sun X, Xu L, Ni B, et al. (2019) LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 regulate pollen tube growth as chaperones and coreceptors for ANXUR/BUPS receptor kinases in Arabidopsis. Mol Plant 12: 1612–1623 [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Uhlenbroich S, Shinya T, Martinez Y, Lefebvre B, Shibuya N, Bono JJ(2011) Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol Biochem 49: 709–720 [DOI] [PubMed] [Google Scholar]
- Franck CM, Westermann J, Boisson-Dernier A(2018) Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69: 301–328 [DOI] [PubMed] [Google Scholar]
- Gabriëls SH, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GC, Rainey DY, Martin GB, Takken FL, de Wit PJ, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Zhao Y, Liu MC, Zhou LZ, Wang L, Zhong S, Hou S, Jiang J, Liu T, Huang Q, et al. (2019) LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr Biol 29: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gong BQ, Xue J, Zhang N, Xu L, Yao X, Yang QJ, Yu Y, Wang HB, Zhang D, Li JF(2017) Rice chitin receptor OsCEBiP is not a transmembrane protein but targets the plasma membrane via a GPI anchor. Mol Plant 10: 767–770 [DOI] [PubMed] [Google Scholar]
- Gubaeva E, Gubaev A, Melcher RLJ, Cord-Landwehr S, Singh R, El Gueddari NE, Moerschbacher BM(2018) ‘Slipped sandwich’ model for chitin and chitosan perception in Arabidopsis. Mol Plant Microbe Interact 31: 1145–1153 [DOI] [PubMed] [Google Scholar]
- Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG(2012) Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J Biol Chem 287: 38460–38472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24: 134–143 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR(2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, et al. (2014) Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci USA 111: E404–E413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herger A, Gupta S, Kadler G, Franck CM, Boisson-Dernier A, Ringli C (2019) LRR-extensins of vegetative tissues are a functionally conserved family of RALF1 receptors interacting with the receptor kinase FERONIA. bioRxiv 783266, doi:10.1101/783266 [Google Scholar]
- Hohmann U, Nicolet J, Moretti A, Hothorn LA, Hothorn M(2018) The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat Plants 4: 345–351 [DOI] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX, et al. (2019) NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799 [DOI] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W(2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Pedi L, Diver MM, Long SB(2012) Crystal structure of the calcium release-activated calcium channel Orai. Science 338: 1308–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. (2013) Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341: 172–175 [DOI] [PubMed] [Google Scholar]
- Hu Z, Zhou Q, Zhang C, Fan S, Cheng W, Zhao Y, Shao F, Wang HW, Sui SF, Chai J(2015) Structural and biochemical basis for induced self-propagation of NLRC4. Science 350: 399–404 [DOI] [PubMed] [Google Scholar]
- Huang D, Zheng X, Wang ZA, Chen X, He WT, Zhang Y, Xu JG, Zhao H, Shi W, Wang X, et al. (2017) The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol Cell Biol 37: e00497-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J(2011) Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL(2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL(2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N(2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R(2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72: 894–907 [DOI] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D(2016) Of guards, decoys, baits and traps: Pathogen perception in plants by type III effector sensors. Curr Opin Microbiol 29: 49–55 [DOI] [PubMed] [Google Scholar]
- Kotani N, Gu J, Isaji T, Udaka K, Taniguchi N, Honke K(2008) Biochemical visualization of cell surface molecular clustering in living cells. Proc Natl Acad Sci USA 105: 7405–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL(2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ(2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schäffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364: 178–181 [DOI] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019) A coevolved EDS1–SAG101–NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31: 2430–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larquet E, Schreiber V, Boisset N, Richet E(2004) Oligomeric assemblies of the Escherichia coli MalT transcriptional activator revealed by cryo-electron microscopy and image processing. J Mol Biol 343: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D(2010) Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet 6: e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TW, van den Burg HA, Joosten MH(2014) Two for all: Receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci 19: 123–132 [DOI] [PubMed] [Google Scholar]
- Lisa MN, Cvirkaite-Krupovic V, Richet E, André-Leroux G, Alzari PM, Haouz A, Danot O(2019) Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor. Nucleic Acids Res 47: 3795–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. (2012a) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24: 3406–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang J, Han Z, Gong X, Zhang H, Chai J(2016a) Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 24: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. (2012b) Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164 [DOI] [PubMed] [Google Scholar]
- Liu X, Castro C, Wang Y, Noble J, Ponvert N, Bundy M, Hoel C, Shpak E, Palanivelu R(2016b) The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28: 1035–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang X, Li M, He P, Zhang Y(2016c) Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol 212: 637–645 [DOI] [PubMed] [Google Scholar]
- Lu X, Kracher B, Saur IM, Bauer S, Ellwood SR, Wise R, Yaeno T, Maekawa T, Schulze-Lefert P(2016) Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc Natl Acad Sci USA 113: E6486–E6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FL(2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Ma C, Liu Y, Bai B, Han Z, Tang J, Zhang H, Yaghmaiean H, Zhang Y, Chai J(2017) Structural basis for BIR1-mediated negative regulation of plant immunity. Cell Res 27: 1521–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L(2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Ma Y, Guo H, Hu L, Martinez PP, Moschou PN, Cevik V, Ding P, Duxbury Z, Sarris PF, Jones JDG(2018) Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc Natl Acad Sci USA 115: 10218–10227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C(2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011a) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P(2011b) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CE, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ(2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyszewski M, Zheng W, Lueck J, Antiochos B, Egelman EH, Sohn J(2018) Cryo-EM structure of the NLRC4CARD filament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly. J Biol Chem 293: 20240–20248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603 [DOI] [PubMed] [Google Scholar]
- Melida H, Sopena-Torres S, Bacete L, Garrido-Arandia M, Jorda L, Lopez G, Munoz-Barrios A, Pacios LF, Molina A(2018) Non-branched beta-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J 93: 34–49 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N(2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussu S, Broyart C, Santos-Fernandez G, Augustin S, Wehrle S, Grossniklaus U, Santiago J (2019) Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. bioRxiv 695874, doi:10.1101/695874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y(2009) A dual resistance gene system prevents infection by three distinct pathogens. Plant Signal Behav 4: 954–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung EH, El Kasmi F, et al. (2017) TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci USA 114: E2053–E2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, de Guillen K, Césari S, Chalvon V, Gracy J, Padilla A, Kroj T(2017) Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenoid K, Dong Y, Cao C, Cui T, Sancak Y, Markhard AL, Grabarek Z, Kong L, Liu Z, Ouyang B, et al. (2016) Architecture of the mitochondrial calcium uniporter. Nature 533: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratcha G, Ledda F(2008) GDNF and GFRα: A versatile molecular complex for developing neurons. Trends Neurosci 31: 384–391 [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr.(2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC(2005) NRG1, a CC-NB-LRR protein, together with N, a TIR–NB–LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15: 968–973 [DOI] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V(2010) The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Liebrand TW, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MH, Robatzek S(2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 210: 627–642 [DOI] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ(2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR–mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115: E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P(2008) The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S(2009) A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J Biol Chem 284: 32717–32724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S(2011) Crystal structure of full-length Apaf-1: How the death signal is relayed in the mitochondrial pathway of apoptosis. Structure 19: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y(2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434: 926–933 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS(2007) The apoptosome: Signalling platform of cell death. Nat Rev Mol Cell Biol 8: 405–413 [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V(2013) Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet 9: e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M(2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R(2012) The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10: e1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Bourdais G, Pan H, Robatzek S, Tang D(2017) Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc Natl Acad Sci USA 114: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grünwald K, et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Ma Q, Weidtkamp-Peters S, Stahl Y, Felekyan S, Bleckmann A, Seidel CA, Simon R(2015) Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci Signal 8: ra76. [DOI] [PubMed] [Google Scholar]
- Song W, Liu L, Wang J, Wu Z, Zhang H, Tang J, Lin G, Wang Y, Wen X, Li W, et al. (2016) Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res 26: 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele JFC, Hughes RK, Banfield MJ(2019) Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PLoS One 14: e0221226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C(2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J(2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Caaveiro JM, Morante K, González-Mañas JM, Tsumoto K(2015) Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat Commun 6: 6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM(2017) Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Han Z, Sun Y, Zhang H, Gong X, Chai J(2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res 25: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenthorey JL, Haloupek N, López-Blanco JR, Grob P, Adamson E, Hartenian E, Lind NA, Bourgeois NM, Chacón P, Nogales E, et al. (2017) The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358: 888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G(2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Osborne Nishimura E, DiAntonio A, Milbrandt J, Dangl JL, et al. (2019a) TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan WL, Fröhlich K, Pruitt RN, Nürnberger T, Zhang L(2019b) Plant cell surface immune receptor complex signaling. Curr Opin Plant Biol 50: 18–28 [DOI] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chabannes M, et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295 [DOI] [PubMed] [Google Scholar]
- Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, Wang X(2014) Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 54: 133–146 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J(2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: eaav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, et al. (2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364: aav5868. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao L, Zhang X, Zhang Q, Jia Y, Wang G, Li S, Tian D, Li WH, Yang S(2019c) Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc Natl Acad Sci USA 116: 18479–18487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE(2005) Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sornaraj P, deCourcy-Ireland E, Menz RI, Kobe B, Ellis JG, Dodds PN, Anderson PA(2011) An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact 24: 897–906 [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV, et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S(2017) NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci USA 114: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Belhaj K, Bozkurt TO, Birk MS, Kamoun S(2016) Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol 209: 1344–1352 [DOI] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X(2019) Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222: 938–953 [DOI] [PubMed] [Google Scholar]
- Xi L, Wu XN, Gilbert M, Schulze WX(2019) Classification and interactions of LRR receptors and co-receptors within the Arabidopsis plasma membrane: An overview. Front Plant Sci 10: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J(2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA(2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA(2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang F, Wang W, Lin G, Hu Z, Han Z, Qi Y, Zhang L, Wang J, Sui SF, et al. (2018) Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res 28: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q(2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189: 321–334 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin X, Han Z, Wang J, Qu LJ, Chai J(2016) SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol Plant 9: 1406–1414 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, et al. (2015) Cryo-EM structure of the activated NAIP2–NLRC4 inflammasome reveals nucleated polymerization. Science 350: 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernoux M, Bentham AR, Newman TE, Ve T, Casey LW, Raaymakers TM, Hu J, Croll TI, Schreiber KJ, et al. (2017a) Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci USA 114: E2046–E2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dodds PN, Bernoux M(2017b) What do we know about NOD-like receptors in plant immunity? Annu Rev Phytopathol 55: 205–229 [DOI] [PubMed] [Google Scholar]
- Zhao C, Zayed O, Yu Z, Jiang W, Zhu P, Hsu CC, Zhang L, Tao WA, Lozano-Durán R, Zhu JK(2018) Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci USA 115: 13123–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K.(2019) Glycosylphosphatidylinositol-anchored proteins in Arabidopsis and one of their common roles in signaling transduction. Front Plant Sci 10: 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Li Y, Hu Q, Bai XC, Huang W, Yan C, Scheres SH, Shi Y(2015) Atomic structure of the apoptosome: Mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev 29: 2349–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C.(2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G(2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]