Virtually all proteins at the plasma membrane are segregated in nanodomains, which are critical for signaling.
Abstract
Plasma membranes provide a highly selective environment for a large number of transmembrane and membrane-associated proteins. Whereas lateral movement of proteins in this lipid bilayer is possible, it is rather limited in turgid and cell wall-shielded plant cells. However, membrane-resident signaling processes occur on subsecond scales that cannot be explained by simple diffusion models. Accordingly, several receptors and other membrane-associated proteins are organized and functional in membrane nanodomains. Although the general presence of membrane nanodomains has become widely accepted as fact, fundamental functional aspects, the roles of individual lipid species and their interplay with proteins, and aspects of nanodomain maintenance and persistence remain poorly understood. Here, we review the current knowledge of nanodomain organization and function, with a particular focus on signaling processes involving proteins, lipids, and their interactions. Furthermore, we propose new and hypothetical aspects of plant membrane biology that we consider important for future research.
Together with the cell wall, the plasma membrane forms the frontier of the cell. As such, it acts as a physical barrier and allows the generation and maintenance of chemical gradients between the outside and inside of the cell. At the same time, the plasma membrane is a critical checkpoint for the perception and integration of extracellular signals prior to signal transduction in the cytoplasm. The fluid mosaic model initially predicted that biological membranes are fluids, with the underlying assumption that their protein and lipid constituents can laterally diffuse in the plane of the membrane without major restrictions (Singer and Nicolson, 1972). According to this view, membrane-embedded receptors would distribute uniformly throughout the cell surface and irrespective of the plasma-membrane proteome. However, the very opposite seems to be the case. Unequivocal evidence shows that the plasma membrane itself is highly compartmentalized into subdomains and that lateral segregation of proteins and lipids is a critical facet of cell surface signaling, modulating signal perception, specificity, and integration.
This view of a compartmentalized plasma membrane first arose from biochemical fractionation, which could separate biological membranes in a binary manner between so-called detergent-resistant (also referred to as detergent-insoluble) and detergent-sensitive membranes (Brown and Rose, 1992; Mongrand et al., 2004; Borner et al., 2005; Morel et al., 2006; Laloi et al., 2007; Lefebvre et al., 2007). However, fluorescent microscopy techniques with ever increasing resolution power have largely replaced biochemical fractionation, as it rapidly became clear that plasma membrane subdomains are not binary but rather a part of a large patchwork of many subdomains that coexist on various spatial and temporal scales. Such a view is supported by a wealth of data from colocalization analyses using confocal, total internal reflection fluorescence, and superresolution microscopy (Kleine-Vehn et al., 2011; Demir et al., 2013; Jarsch et al., 2014; Hosy et al., 2015; Bücherl et al., 2017; Martinière et al., 2019; Platre et al., 2019). Furthermore, the fact that different membrane constituents display varying diffusion patterns within the plane of the plasma membrane is also sustained by studies on the dynamics of protein/lipid lateral diffusion using fluorescence recovery after photobleaching (FRAP), single-molecule imaging (e.g. single particle tracking photoactivated localization microscopy), and fluctuation correlation spectroscopy (Li et al., 2011, 2016b, Martinière et al., 2012, 2019, Wang et al., 2013, 2015; Jarsch et al., 2014; Hosy et al., 2015; Gronnier et al., 2017; Cui et al., 2018; McKenna et al., 2019; Platre et al., 2019). Technically, receptor/scaffold complexes have also often been studied using distance-based imaging techniques such as Förster resonance energy transfer-fluorescence lifetime imaging (FRET-FLIM). However, a note of caution has recently been put forward concerning the use of bimolecular-fluorescence complementation, which can artificially stabilize membrane proteins in membrane contact sites of the endoplasmic reticulum and the plasma membrane (Tao et al., 2019).
In this update, we review the current evidence for the coexistence of a patchwork of membrane nanodomains in the plant plasma membrane and their functional importance and assess the roles of lipids, the cell wall, and the cytoskeleton in shaping this diverse plasma membrane landscape. Finally, we discuss plausible scenarios for the functional importance of protein nanoclustering in signal transduction.
THE PLASMA MEMBRANE AS A PATCHWORK OF COEXISTING FUNCTIONAL MEMBRANE NANODOMAINS
Receptor Scaffolding at the Nanoscale
Unequivocal evidence demonstrates that a significant number of membrane-resident proteins cluster in higher-order structures that have been termed “membrane nanodomains” or “membrane microdomains,” for which a nomenclature has been suggested recently (Ott, 2017). Briefly, nanodomains are submicron protein and/or lipid assemblies (∼20–300 nm and <1 μm), whereas microdomains are significantly larger assemblies (>1 μm, e.g. perimicrobial membranes, the Casparian strip domain, polar domains, plasmodesmata [PD], or membrane contact sites; Ott, 2017). Herein, we will specifically focus on plasma membrane nanodomains, which have often been termed “lipid rafts.”
Whereas the lipid raft model was mainly based on biochemical evidence, recent cell biological approaches revealed that a number of proteins distribute heterogeneously on plant cell membranes mostly labeling puncta-like structures (Kleine-Vehn et al., 2011; Demir et al., 2013; Jarsch et al., 2014; Bücherl et al., 2017; Martinière et al., 2019; Platre et al., 2019). Over time, members of the flotillin (FLOT) and plant-specific remorin protein families have been described as marker proteins for these membrane nanodomains (Fig. 1; Raffaele et al., 2009; Jarsch and Ott, 2011; Li et al., 2012; Marín et al., 2012; Perraki et al., 2012; Hao et al., 2014; Jarsch et al., 2014; Wang et al., 2015; Bücherl et al., 2017; Gronnier et al., 2017; Liang et al., 2018). This allowed comparative studies that revealed the coexistence of multiple nanodomains within the same cell (Jarsch et al., 2014). FLOTs comprise a small protein family with only two members in Arabidopsis (Arabidopsis thaliana; Daněk et al., 2016), whereas at least 16 remorins have been identified with an additional two legume-specific members (group 2; Fig. 1; Raffaele et al., 2007). Whereas the precise molecular function of most of these proteins remains unknown, most group 1 remorins were repeatedly shown to regulate viral spreading in leaves, possibly by modulating PD conductance (Raffaele et al., 2009; Perraki et al., 2012, 2018; Ishikawa et al., 2017). A recent preprint report described reduced numbers of PDs in a rem1.2 rem1.3 double knockout mutant, indicating a role of remorins in PD biogenesis (Wei et al., 2019).
Figure 1.
Remorin proteins and their targeting to plasma membrane nanodomains. Remorins have an N-terminal intrinsically disordered region, a central coiled-coil domain, and a REM-CA. The intrinsically disordered region is regulated by posttranslational modification, such as phosphorylation, and possibly modulates interaction with many different proteins. The coiled-coil domain is an oligomerization domain and mainly contributes to remorin trimer formation and interactions with other proteins. REM-CA is required for both plasma membrane and nanodomain targeting via interaction with inner leaflet lipids such as sterols and PI4P.
Additionally, remorins form higher-order oligomers leading to filamentous structures in vitro (Bariola et al., 2004; Marín et al., 2012; Martinez et al., 2019) and in vivo (Wei et al., 2019) and this may drive association of remorins with the plasma membrane (Legrand et al., 2019). Remorin oligomer formation itself is mainly mediated by the conserved C-terminal coiled-coil region, a hallmark feature of these proteins (Raffaele et al., 2009; Marín et al., 2012; Martinez et al., 2019), and further supported by the intrinsically disordered N-terminal region that harbors the majority of phosphorylation sites (Marín and Ott, 2012; Marín et al., 2012).
The effect of remorin phosphorylation on their association patterns with membrane nanodomains remains elusive. However, functionality of remorins in plasmodesmata is hampered in phospho-mutants (Perraki et al., 2018). Whereas remorin phosphorylation was mostly studied in vitro, several remorins are able to associate, or at least colocalize, with a number of soluble or membrane-associated kinases such as CALCIUM-DEPENDENT PROTEIN KINASE 3 (CPK3; Perraki et al., 2018), CPK21 (Demir et al., 2013), AVRPPHB SUSCEPTIBLE1 (PBS1; Albers et al., 2019), SNF1 RELATED KINASE (SnRK1; Son et al., 2014), and receptor-like kinases (RLKs) such as the Arabidopsis brassinosteroid receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) and innate immune receptor FLAGELLIN SENSING2 (FLS2; Bücherl et al., 2017), the rice (Oryza sativa) SOMATIC EMBRYOGENEIS RECEPTOR KINASE1 (SERK1) and OsBRI1 (Gui et al., 2016), and the Medicago truncatula RLKs NOD FACTOR PERCEPTION (NFP), LYSIN MOTIF KINASE3 (LYK3), and DOES NOT MAKE INFECTIONS2 (DMI2), as well as their corresponding homologs in Lotus japonicus (Lefebvre et al., 2010; Tóth et al., 2012; Liang et al., 2018). Increasing evidence suggests that remorins and FLOTs play versatile roles in nanodomain stabilization (Huang et al., 2019a) and receptor recruitment into these structures (Haney et al., 2011; Bücherl et al., 2017; Liang et al., 2018).
In rice, significant progress was made with respect to remorin functionality and the functional relevance of receptor recruitment into nanodomains, where ligand-induced phosphorylation of the remorin OsREM4.1 results in its dissociation from OsSERK1, which consequently allows the assembly of the signaling-competent OsSERK1/OsBRI1 receptor complex (Gui et al., 2016). A different mode of maintaining an active receptor complex has been proposed in the legume M. truncatula, where receptor-mediated ligand perception results in transcriptional activation of the group 2 remorin SYMREM1, which in turn physically associates with the entry receptor LYK3. This interaction results in stabilization and physical recruitment of LYK3 in a laterally stable and FLOT4-positive nanodomain. Genetic evidence further suggests that this process is required for receptor stabilization at the plasma membrane (Haney et al., 2011; Liang et al., 2018).
Nanodomain Targeting
Even though the evidence is still slightly scattered, a number of studies suggest that receptor nanoclusters colocalize with one or the other scaffold belonging to the FLOT and/or REM protein families. This implies that these proteins may act as organizing centers, as recently suggested for remorins (Gui et al., 2016; Liang et al., 2018; Huang et al., 2019a) and at least for the M. truncatula FLOT2/4 (Haney and Long, 2010; Haney et al., 2011; Liang et al., 2018).
Recruitment of remorins themselves to the plasma membrane and, in some cases, also to nanodomains is mediated by a C-terminal hydrophobic stretch called the remorin C-terminal anchor (REM-CA; Fig. 1). The REM-CA peptide specifically binds in a pH-dependent manner to sterols and phosphoinositides and requires the simultaneous presence of both β-sitosterol and phosphoinositides in the same nanodomain (Legrand et al., 2019). This demonstrates the tight link between specific lipid species and nanodomain recruitment of proteins.
In addition, remorin association with membranes is further supported by S-acylation of Cys residues, as well as protein-protein interactions (Raffaele et al., 2009; Perraki et al., 2012; Konrad et al., 2014; Gronnier et al., 2017; Legrand et al., 2019). It remains, however, still an open point of discussion whether S-acylation represents a major and general posttranslational modification supporting nanodomain targeting of remorins and other proteins. Among the >600 acylated proteins identified in Arabidopsis is also the immune receptor FLS2 (Hemsley et al., 2013). Whereas S-acylation on two Ser residues (S830 and S831) within the juxtamembrane domain of FLS2 have been mapped, these residues are not required for plasma membrane localization of the receptor per se (Hurst et al., 2019). However, a recent preprint suggests that they may contribute to the localization of FLS2 in nanodomains (Chen et al., 2019). This study also claims that S-acylation might be a hallmark of receptor targeting into nanodomains, as a posttranslational modification-dependent recruitment to FLOT1-labeled nanodomains was also observed for the receptors CERK1 and P2K1 (DORN1), which mediate perception of chitin and extracellular ATP, respectively (Chen et al., 2019). However, these results were obtained using transient expression in protoplasts, in which the formation of nanodomains is heavily impacted due to the absence of a cell wall (see below). Furthermore, nanodomains were visualized using deconvolution as an imaging postprocessing method, rather than microscopy techniques that can actually resolve diffraction-limited structures. Therefore, these results should be taken with caution until they have been verified in situ. In addition, Hurst and coworkers found that FLS2 mutants, in which C830 and C831 were substituted by serines, were fully functional, arguing that S-acylation at these sites is dispensable for FLS2 signaling function. Receptor tagging and expression levels are critical for FLS2 function and therefore need to be considered in functional studies (Hurst et al., 2018). Similar concerns have been raised for the FLS2 coreceptor BAK1 (Ntoukakis et al., 2011).
IMPORTANCE OF LIPIDS IN PLASMA MEMBRANE LATERAL SEGREGATION
Asymmetric Localization of Lipids in the Two Leaflets of the Plasma Membrane
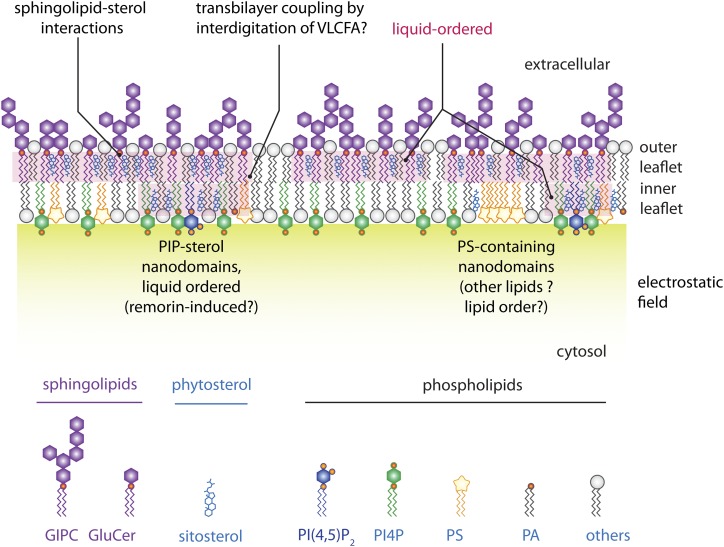
Like proteins, lipids are not uniformly localized in the plasma membrane. In animal cells, they display a strong asymmetry between the outer and inner membrane leaflets, which face the cell wall and the cytosol, respectively (Fig. 2). The outer membrane leaflet is rich in glycosphingolipids and sterols, as well as the phospholipid phosphatidylcholine. In plants, lipid asymmetry between the two membrane leaflets has not been extensively addressed experimentally, but was proposed to be largely similar to animal cells (Gronnier et al., 2018), with the main plant sphingolipids, glycosylinositol phosphorylceramides (GIPCs; Cacas et al., 2016), being enriched in the outer membrane leaflet (Fig. 2). By contrast, the inner leaflet is enriched in phospholipids, notably phosphatidylethanolamine and the minor anionic phospholipids, which include phosphatidic acid, phosphatidyl-Ser (PS), phosphatidylinositol (PI) and its phosphorylated derivatives phosphoinositides (Colin and Jaillais, 2019). These anionic lipids confer a strong electronegative property to the inner surface of the plasma membrane, which drives the identity of the plasma membrane and is crucial for the recruitment of many soluble or lipid-anchored proteins to this compartment (Fig. 2; Simon et al., 2016; Noack and Jaillais, 2017; Platre et al., 2018).
Figure 2.
Schematic representation of the lipid distribution within a plant plasma membrane. Note the asymmetric repartition of lipids across the bilayer as well as lateral segregation of lipids in both inner and outer membrane leaflets (Gronnier et al., 2018). PIP, phosphoinositide; GluCer, glucosylceramide; VLCFA, very long chain fatty acid; PA, phosphatidic acid.
Lipid Order in Plasma Membrane Compartmentalization
In addition to lipid segregation between the outer and inner plasma membrane leaflets, there is also lipid segregation laterally within each leaflet. The lipid raft hypothesis postulates that lipids in biological membranes may be in a liquid-disordered or liquid-ordered phase (Kusumi et al., 2012). The liquid-ordered phase is enriched in sterols and glycosphingolipids, which largely corresponds to the lipid composition of the outer leaflets. Sphingolipids, such as GIPC, interact with phytosterols to increase lipid order (Fig. 2; Cacas et al., 2016).
Evidence for different ordered phases at the plant plasma membrane recently emerged when using di-4-ANEPPDHQ (ANEPP), a probe sensitive to the lipid order (Roche et al., 2008; Frescatada-Rosa et al., 2014; Zhao et al., 2015a, 2015b; Gerbeau-Pissot et al., 2016; Gronnier et al., 2017; Grosjean et al., 2018; Huang et al., 2019a; Laurent et al., 2019; Pan et al., 2019). Its fluorescence emission spectrum is blue-shifted in liquid-ordered compared with liquid-disordered phases (Jin et al., 2005, 2006). Initially validated in vitro in model membranes, this dye is also amenable to live imaging, as it is easy to apply and can detect both the liquid-ordered and liquid-disordered lipid phases in living cells (Gerbeau-Pissot et al., 2016). ANEPP staining in vivo shows heterogeneous labeling of the plasma membrane (Gronnier et al., 2017; Pan et al., 2019). It is sensitive to methyl-β-cyclodextrin (mßcd), a sterol-depleting agent, which decreases the ANEPP-stained liquid-ordered phase at the expense of the liquid-disordered phase (Huang et al., 2019a). Similarly, a recent preprint suggested that the amount of staining of the liquid-ordered phases is reduced in the sterol biosynthesis mutant fackel-J79 (fk-J79; Pan et al., 2019).
Together, these data support the notion that sterols are required for the formation of liquid-ordered lipid phases in planta. However, these approaches also have limitations, and conclusions obtained with either ANEPP or mßcd should be taken with caution. Indeed, ANEPP is a membrane-intercalating dye and may therefore impact membrane properties. In addition, mßcd has a massive impact on membrane structure and integrity, and protein-lipid/lipid-lipid interactions.
Roles of Lipids in Protein Targeting in Nanodomains
Recent data imply that liquid-ordered phases are relevant for protein localization, since the ANEPP-stained liquid-ordered phases colocalize with the Solanum tuberosum group 1 remorin StREM1.3 (Gronnier et al., 2017). Interestingly, the localization of StREM1.3 in nanodomains is sensitive to fenpropimorph, an inhibitor that affects the plasma membrane sterol composition but not the total amount of sterols (Gronnier et al., 2017). Below, we will use StREM1.3 as an example of the role of protein/lipid interactions in nanodomain localization, as this is to date the best studied case in plants (Fig. 1). Although indirect, these data are consistent with the notion that sterols participate in the formation of liquid-ordered membrane domains, which themselves are required for localization of proteins, including remorins (Gronnier et al., 2017; Legrand et al., 2019). However, as mentioned above, sterols and sphingolipids mainly accumulate in the outer membrane leaflet, whereas remorins are inserted into the cytosolic leaflet (Cacas et al., 2016; Gronnier et al., 2017). This raises two questions: (1) Are there any lipids directly involved in remorin nanodomain localization in the cytosolic leaflet? And (2) if yes, how do the liquid-ordered domains formed in the outer leaflet influence the nanoscale organization of the plasma membrane at the inner leaflet?
The first question is partially understood, at least for StREM1.3. Indeed, biophysical and modeling approaches showed that StREM1.3 is recruited to plasma membrane nanodomains via its C-terminal anchor and that it directly binds to phosphoinositides (Fig. 1; Gronnier et al., 2017). Accordingly, immuno-electron microscopy on purified membranes showed that the phosphoinositide phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2) localizes to nanodomains ∼40 nm in size (Furt et al., 2010). Furthermore, inducible genetic perturbation indicates that phosphatidylinositol-4-phosphate (PI4P) is required for StREM1.3 plasma membrane localization (Gronnier et al., 2017). Whether the liquid-ordered phase induced by sterols and GIPCs at the outer leaflet modulates the formation of nanodomains inside the cell remains unknown. However, studies in animal cells suggest transbilayer coupling between outer- and inner-leaflet lipids via very long-chain fatty acids (Raghupathy et al., 2015). Indeed, GIPCs have very long-chain fatty acids, which would perfectly fit such a role in transbilayer coupling (Fig. 2).
From the cytosolic side, the most prominent phospholipid for transbilayer coupling is PS, which contains very long-chain fatty acids of 20–24 carbons in plants (Mamode Cassim et al., 2019). Single-molecule superresolution imaging of a PS biosensor suggests that, indeed, PS accumulates in nanodomains ∼50–70 nm in size in the inner plasma membrane leaflet (Fig. 2; Platre et al., 2019). However, there are still many unanswered questions concerning these PS-containing domains, as it is unclear whether (1) they also contain phosphoinositides or sterols, (2) their formation depends on sterols and/or GIPCs, (3) there is a coupling between liquid-ordered membrane domains in the outer leaflet and PS-containing domains in the inner leaflet, and (4) remorins themselves depend on PS for localization in nanodomains. In vitro analyses detected no interaction between PS and StREM1.3 (Legrand et al., 2019), but this does not exclude that PS could be indirectly involved in StREM1.3 localization in vitro and in vivo. Indeed, PS is required to stabilize RHO-OF-PLANTS6 (ROP6) in nanodomains in root epidermis (Platre et al., 2019). A recent preprint proposes that ROP6 nanoclustering in the leaf epidermis also depends on sterols (Pan et al., 2019). This suggests a combined action of PS and sterols for Rho GTPase clustering, although to be fully validated, it needs to be investigated further in the same developmental context.
Protein Feedback on the Lateral Segregation of Lipids
Lipid nano-patterning at the plasma membrane appears critical for the localization of proteins. However, the experimental evidence for the presence of lipids in different nanodomains is comparatively scarce and the full extent of lipid segregation in nanodomains is not fully appreciated. Although lipids alone may cluster into distinct domains in vitro depending on the composition, it is also important to note that proteins highly influence such lateral sorting of lipids. For example, remorins themselves affect the local membrane order (Huang et al., 2019a; Legrand et al., 2019).
The underlying molecular basis is not fully understood, but it is likely that remorin oligomerization is important to remodel membrane properties and perhaps also induce local phosphoinositide clustering (Gronnier et al., 2018; Legrand et al., 2019; Martinez et al., 2019). Indeed, if each C-terminal anchor of StREM1.3 binds PI4P, and considering that StREM1.3 is present as a trimer, this could induce local clustering of PI4P (Legrand et al., 2019). Similar mechanisms were proposed for the Sec14-Nodulin protein AtSfh1, which is critical for the focal accumulation of PI(4,5)P2 at the tip of growing root hairs (Ghosh et al., 2015). Local PI(4,5)P2 accumulation requires both PI(4,5)P2 binding by a polycationic peptide at the C terminus of AtSfh1 and the homo-oligomerization activities of its nodulin domain. In the case of AtSfh1, this could act as part of a self-organizing system, since AtSfh1 stimulates PI(4,5)P2 synthesis, either directly or by inducing PI4P production (Ghosh et al., 2015; Kf de Campos and Schaaf, 2017).
PLASMA MEMBRANE COMPARTMENTALIZATION: REVISITING THE ANCHORED PICKET FENCE CONCEPT
One of the most unifying models to explain the regulation of plasma membrane organization is the anchored picket fence model (Kusumi et al., 2012). In this model, the cortical cytoskeleton, made of actin microfilaments, defines membrane domains ∼40–300 nm in size by acting as a fence that restricts lateral diffusion of proteins and lipids within these domains (Fig. 3). As such, the cortical actin network is seen as a “membrane skeleton” that is key for plasma membrane organization (Fig. 3). Because lipids in both inner and outer leaflets are fenced by this membrane skeleton, the model additionally postulates the existence of transmembrane proteins that act as pickets. These pickets are then anchored either by the cytoskeleton in the cytosol or the extracellular matrix (Fig. 3; Kusumi et al., 2012). The diffusion of lipids, even on the outer leaflet, is thereby physically hindered by these anchored pickets and lipids may also transiently interact with them. Although the picket fence model is useful when thinking about the organization of the plant plasma membrane, it fails to include some plant-specific features that may have a profound impact on the organization of the cell surface.
Figure 3.
The picket fence model in animals and possible revision of the model in plants. A, Picket fence model for plasma membrane compartmentalization in animal cells, with a prominent influence of cortical actin as a membrane skeleton (Kusumi et al., 2012). B, The picket fence model in plants is likely to involve both intra- (actin and microtubules) and extracellular (cell wall) skeletons (i.e. fences), which may explain the limited diffusion of plasma membrane proteins. Lines with arrowheads represent examples of diffusion trajectories of the respective plasma membrane proteins.
The Role of the Cytoskeleton in Nanodomain Organization
In addition to cortical actin, plants also have cortical microtubules, and it is possible that microtubules could additionally act as a membrane skeleton (Fig. 3). Indeed, a number of membrane nanodomain proteins align with or directly bind to intact actin or microtubule filaments (Homann et al., 2007; Jarsch et al., 2014; Gui et al., 2015; Szymanski et al., 2015; Liang et al., 2018). In addition, a recent preprint suggested that stimulus-dependent microtubule fragmentation by a member of the M. truncatula DEVELOPMENTALLY REGULATED PLASMA MEMBRANE POLYPEPTIDE (DREPP) family occurs within SYMREM1/FLOT4-containing membrane nanodomains (Su et al., 2019).
In several cases, chemical disruption of these filaments was shown to cause a reduction or loss of nanodomain localization even though the used marker proteins themselves mostly resided at the plasma membrane (e.g. Raffaele et al., 2008; Jarsch et al., 2014; Konrad et al., 2014; Szymanski et al., 2015; Bücherl et al., 2017; Lv et al., 2017). This indicates an organizational role of the cytoskeleton during nanodomain and/or protein complex assembly, which is supported by studies of Arabidopsis HYPERSENSITIVE INDUCED REACTION 1 (HIR1; Lv et al., 2017). Nonetheless, it was recently shown that HIR1 as well as FLOT2-containing nanodomains, remain intact upon chemical disruption of the cytoskeleton (Daněk et al., 2020). In addition, FRAP and single-particle analyses showed a minor effect of the cytoskeleton on lateral diffusion of transmembrane proteins such as the FLS2 receptor (McKenna et al., 2019). Given the size of these protein assemblies in the nanometer range, the usual width of the cytoskeleton network, and the restricted data availability, a general mechanism of how the cytoskeleton supports or even mediates nanodomain assembly cannot be proposed to date. Overall, there is strong evidence that cortical cytoskeleton components participate in the dynamics and spatial repartitioning of plasma membrane nanodomains, but they may play a less prominent role in plasma membrane compartmentalization compared to animal cells.
The Cell Wall-Plasma Membrane Continuum in Shaping the Plasma Membrane Landscape
In addition to the cytoskeleton in the cytosol, a key component in understanding the nanoscale organization of the plasma membrane landscape in plants is the extracellular cell wall. Indeed, plant cells have a high turgor pressure that physically presses the plasma membrane to the rigid, yet porous, cell wall. This has a strong impact on the dynamic behavior of lipids and proteins at the plasma membrane-cell wall contacts and sets apart the plasma membrane from any other membranes of the cell. Most importantly, the cell wall is a barrier to the free diffusion of plasma membrane molecules that are sticking out into the wall (Fig. 3; Martinière et al., 2012). Using FRAP and particle tracking, it was demonstrated that synthetic reporters with an extracellular domain exhibit limited lateral diffusion at least on the minute timescale (Martinière et al., 2012; McKenna et al., 2019). This is in great contrast to soluble cytosolic synthetic reporters that interact with the inner leaflet of the plasma membrane and diffuse significantly faster (Martinière et al., 2012). Disruption of the plasma membrane/cell wall continuum, by digestion of the cell wall, induction of plasmolysis, or chemical perturbations of cell wall synthesis, have a profound impact on the diffusion of cell surface proteins and the formation of nanodomains (Martinière et al., 2012; McKenna et al., 2019; Daněk et al., 2020).
Whereas the full extent of the importance of the plasma membrane/cell wall continuum for organization of the cell surface is not understood and has barely been challenged experimentally, its existence points toward several predictions and hypotheses, as well as differences from the canonical view of membrane organization derived from animal systems. Animal transmembrane proteins that stably and directly interact with extracellular matrix components are excellent candidates as anchored pickets (Freeman et al., 2016, 2018), whereas proteins without interaction with the extracellular matrix are expected to diffuse within the boundary imposed by the membrane skeleton. However, in plants, the anchored picket situation seems to be pushed to the extreme, since virtually every protein with an extracellular domain could be seen as an anchored picket. Indeed, even the diffusion of proteins with few extracellular residues appears to be impacted by the presence of the cell wall (Martinière et al., 2012). Pushing this reasoning further, GIPCs, which have a very large head group sticking out of the outer leaflet of the plasma membrane, should also encounter limited diffusion because of the cell wall (Fig. 2; Cacas et al., 2016; Gronnier et al., 2018). Given their high abundance, both membrane proteins with an extracellular domain and GIPCs may have a very strong impact on the diffusion of plasma membrane constituents.
It is even possible that components of the outer membrane leaflets are barely diffusing. This could explain the observation in plants that membrane proteins are basically “fixed” (i.e. nondiffusing), a feature that is rarely seen in animal systems (Martinière et al., 2012). However, if receptors are fixed and unable to diffuse, one may wonder how protein complexes are formed upon ligand binding. It is possible that receptor complexes are preformed but inhibited, and that ligand binding triggers their activation rather than oligomerization. Such a view is supported by in vivo interaction data using FRET-FLIM between BRI1 and BAK1 receptors (Bücherl et al., 2013). Indeed, such analyses suggest that BRI1/BAK1 heterodimers are present even in the absence of brassinosteroid and that the proportion of dimers is not induced by exogenous treatment with this ligand (Bücherl et al., 2013). The results of these experiments have recently been confirmed using selective surface observation FILM, where the confocal spot is placed perpendicular to the surface of the observed cells to reduce signal detection from structures below the plasma membrane (Hutten et al., 2017). However, they contradict results obtained by coimmunoprecipitation, and also models based on crystallographic data, which indicate that brassinosteroids act as a molecular glue to bridge BRI1 and BAK1 extracellular domains (Belkhadir and Jaillais, 2015; Hohmann et al., 2017).
The idea that membrane proteins with an extracellular domain do not diffuse in plants because of the presence of the cell wall is perhaps too strong. Indeed, it is possible that we perceive an apparent lack of diffusion due to the resolution limits of the techniques used, notably FRAP and total internal reflection fluorescence microscopy. The plant cell wall is to some extent porous, so it is possible that proteins are able to diffuse within the boundaries of this porous material. In such a view, the cell wall could constitute a membrane skeleton, but it would act as an exoskeleton rather than a cytoplasmic cortical actin skeleton (Fig. 3).
From a regulatory point of view, this implies that modifications of the cell wall, rather than (or in parallel with) the cytoskeleton could impact protein diffusion. In addition, the cell wall may form membrane domains that could be much smaller than the 300 nm delimited by the actin membrane skeleton. If so, this could perhaps reconcile data obtained from FRET-FLIM and coimmunoprecipitation of receptor kinases (Wang et al., 2005, 2008; Bücherl et al., 2013; Hutten et al., 2017). Indeed, FRET is a proximity technique. Consequently, constitutive FRET can be observed even in the absence of physical interaction if receptors are enclosed in small membrane domains. If they exist, determining the size of such cell wall-delimited membrane domains and their composition would provide critical information for future studies.
According to the scenario highlighted above, receptor proximity could be achieved by preformation of nanodomains during the insertion of transmembrane proteins into the endoplasmic reticulum membrane and thus prior to their exocytosis. Here, but also subsequently, scaffolding could be executed by proteins like the peptide-binding receptor FERONIA, which facilitates complex formation of EFR and FLS2 with their coreceptor BAK1 (Stegmann et al., 2017). Interestingly, FER itself, or its protein partners, directly bind cell wall components (Li et al., 2016a; Feng et al., 2018; Dünser et al., 2019), which could be an additional way to integrate the cell wall information (i.e. composition, remodeling, peptide signaling, etc.) into nanodomain formation. However, whether FER also controls nanodomain patterning must be further tested. Interestingly, soluble scaffold proteins localizing to the inner membrane leaflet only access the receptor complex upon exocytotic vesicle fusion with the plasma membrane. This spatiotemporal association seems highly specific for scaffolds like remorins, as the great majority of studies on remorins did not observe any labeling of intracellular membrane, even though these proteins were often strongly overexpressed. In fact, remorins do not use the secretory pathway but are rather directly synthesized as soluble proteins in the cytosol and subsequently inserted in the inner plasma membrane leaflet via their REM-CA anchor (Fig. 1; Gronnier et al., 2017).
In addition, some proteins or lipids may act as real anchored pickets through direct interaction with wall components (Voxeur and Fry, 2014; Herger et al., 2019; Vaahtera et al., 2019; Rui and Dinneny, 2020). This is exemplified by extensin-like formins that bind the cell wall via a long extracellular domain or a comparably short Pro- and Ser-rich amino acid stretch in their N terminus, which resembles an extensin-like motif (Martinière et al., 2011; Herger et al., 2019). These domains are connected via a single-span transmembrane helix with an actin- or microtubule-binding formin homology domain. As such, members of the formin family were shown to act as actin nucleation facilitators in tip growing systems such as root hairs (Yi et al., 2005). Therefore, it is impossible to fully uncouple the cell wall from the cytoskeleton. Furthermore, there is a continuum between cortical microtubules inside the cell, cellulose synthases in the plasma membrane, and cellulose microfibrils in the cell wall. This three-way connection may impact membrane partitioning, but it has so far remained largely unexplored.
To conclude, the relative impact of the cytoskeleton and the cell wall on plasma membrane organization is poorly understood, but it is likely that they are different from animal systems.
THE PLASMA MEMBRANE AS A DIGITAL SCREEN FOR SIGNAL INTEGRATION: A TWO-DIMENSIONAL PERSPECTIVE
What is the function of plasma membrane compartmentalization in signal perception and integration? There is neither a single response to this question nor satisfying exhaustive answers. During signaling, extracellular information is transmitted through the plasma membrane inside the cell. The signals perceived by single activated receptors have to be amplified. It becomes increasingly evident that such amplification steps often happen directly at the plasma membrane. Indeed, whereas the initial view of signaling “cascades” would place receptors at the plasma membrane and their downstream components in the cytosol, this is often not the case. For example, in the brassinosteroid pathway, nearly all downstream components of BRI1 signaling, including the transcription factors BES1/BZR1, have recently been shown to localize to the plasma membrane at some point during signal transduction (Amorim-Silva et al., 2019; Ren et al., 2019). This is achieved through the action of scaffolds that transiently recruit kinases/phosphatases and other signaling molecules (Amorim-Silva et al., 2019). It is likely that the mechanisms of lateral membrane organization discussed above contribute to the formation and scaffolding of active receptor complexes.
In addition to signal amplification, clustering may also increase sensitivity. For example, clustered receptors are more likely to transduce information carried by ligands with weak binding properties. Furthermore, the localization of receptors in laterally segregated nanodomains may also increase signaling specificity by unmixing downstream signaling components. This is particularly relevant when different receptors share the same amplifying downstream components. For example, spatial separation of BRI1 and FLS2 in distinct nanoclusters may ensure robust signaling from these receptors, even though they share several kinases, such as BAK1 or BSKs (Bücherl et al., 2017). Another possible role of receptor clustering is their connection with intracellular trafficking, notably endocytosis, which is often associated with signal downregulation.
Modeling suggests that the plasma membrane may also act as a digital two-dimensional screen, allowing the integration of analog information and its conversion into digital pixels before this information is relayed with high fidelity inside the cell (Tian et al., 2007, 2010). This has been mainly studied at experimental and theoretical levels for Ras signaling in animal cells (Harding and Hancock, 2008). Ras is a small GTPase from the Rho/Ras superfamily that is involved in canonical mitogen-activated protein kinase (MAPK) signaling downstream of growth factor receptors (i.e. receptor Tyr kinase; Harding and Hancock, 2008). The idea behind digital signaling is that it relies on bistable switches that can only be in two states, namely off and on (Fig. 4). This is the opposite of analog circuits, which transmit continuous information (input) into a proportional continuous output. Most information in biology is of analog nature, including the varying amounts of growth factors experienced by the cell (Fig. 4). The challenge is to transmit such analog signal with high fidelity across the plasma membrane into a proportional output signal. Thus, Ras signaling works as an analog-digital-analog converter, which is one way to convey the signal across the membrane with high fidelity (Fig. 4; Tian et al., 2007). Indeed, membrane-localized Ras may be in two states, diffusing or in nanoclusters, which correspond to its inactive state (off) and active state (on), respectively (Prior et al., 2001; Tian et al., 2007). As such, each Ras nanocluster can be seen as a digital pixel at the membrane, which relays the information to downstream MAPK. Ultimately, this digital information is processed in an analog (i.e. continuous) output signal, which is the amount of MAPK phosphorylation (Fig. 4; Tian et al., 2007).
Figure 4.
The plasma membrane may act as an analog-digital-analog converter for high-fidelity signal transduction. A, During signaling both the input (i.e. ligand) and output (i.e. cellular response such as phosphorylation) signals are analog by nature. However, small GTPases, such as ROPs, act in a binary fashion akin to a digital signal by being OFF when they are freely diffusing, and being ON when they are immobilized in nanodomains (Harding and Hancock, 2008). B, As such, molecules (such as PS in roots; Platre et al., 2019) that favor or dampen the OFF (diffusing) or ON (nanodomains) state of the GTPase may act as a digital gain, which may modify the magnitude of the output signal even though the input signal is constant (Zhou and Hancock, 2015).
Whereas it has been validated mathematically for Ras signaling, it is likely that such analog-digital-analog relays are widespread in signal transduction processes across the plasma membrane. One of the prerequisites for such a system is that it indeed behaves as a binary system with nonclustered molecules that are fully unable to transduce the signal. This is the case for the plant Rho GTPase ROP6, which belongs to the same GTPase superfamily as Ras and, like Ras, is not able to signal when it is not stably localized in nanoclusters, even when it is in a constitutive active GTP-loaded conformation (Platre et al., 2019).
One of the interesting features of analog-digital-analog converters is that it is possible to adjust their “digital gain” (Harding and Hancock, 2008). In other words, it is possible to tune the sensitivity of the system by boosting or restricting the formation of the nanoclusters (Fig. 4). This is well exemplified for ROP6, whose nanoclustering is regulated by PS (Platre et al., 2019). In the absence of PS, ROP6 is not stabilized in nanoclusters and does not signal, whereas in the presence of low amounts of PS, it can be stabilized in nanoclusters but not without an increased input signal (i.e. auxin; Platre et al., 2019). By contrast, in plants with elevated PS levels, ROP6 nanoclustering is boosted and requires less input signal. Therefore, variations in PS level act as a molecular rheostat that adjusts the digital gain of the system and as such may modulate the strength of the output signal while keeping the input signal constant (Fig. 4; Platre et al., 2019). Given that PS levels at the plasma membrane vary during root epidermis differentiation, such variations are one way to modify cellular output according to the developmental context of the cell (Colin and Jaillais, 2019; Platre et al., 2019).
The comparison between the plasma membrane and a two-dimensional digital screen is useful, as it allows the conceptualization of how different membrane parameters may participate together in signaling. However, it should also be emphasized that this is a simplification, as the plasma membrane should not be seen as isolated from its environment, but rather as part of a three-dimensional continuum that includes the extracellular cell wall and the cytoplasm.
LIQUID-LIQUID PHASE SEPARATION IN PLASMA MEMBRANE NANO-ORGANIZATION: HOW MUCH THREE-DIMENSIONAL IS IN A NANODOMAIN?
At present, we define nanodomains purely as membrane compartments, which only extend into the cytosol by proteins interacting with the inner membrane leaflet or proteins within the complex. However, given the time scales of reactions, the lack of free diffusion in the cytosol due to tight packing, the high net charges of lipids and proteins, and the necessity to compartmentalize cellular processes in order to avoid unwanted cross talk between pathways, a concept that spatially extends nanodomains significantly into the cytosol is favored.
Recently, liquid-liquid phase separations (LLPSs) have been proposed as a novel organization concept (Hyman et al., 2014; Banani et al., 2017; McSwiggen et al., 2019). These LLPSs are dynamic cellular compartments that lack a membrane envelope and are also referred to as “membraneless organelles.” Similar to reactions inside the nucleolus, proteins and nucleic acids can be sequestered into LLPSs that have so far mostly been found in the nucleus or the cytosol and termed e.g. “stress granules” or “liquid droplets.” In contrast to lipid droplets, LLPSs rely on a reversible and protein-induced liquid unmixing providing almost viscoelastic properties to these compartments (Hyman et al., 2014). As such, four major characteristics of LLPSs have been proposed: (1) they are spheres; (2) they have a fluid content; (3) they fuse upon contact; and (4) they can be actively reshaped by shear flow (Cuevas-Velazquez and Dinneny, 2018).
They are most often organized by highly oligomeric proteins with long stretches of intrinsic disorder (ID). LLPSs only occur under specific conditions, and dissociation of proteins maintaining this phase separation results in total disintegration of the structure. Whereas theoretically most ID proteins could contribute to LLPS formation, some proteins have a higher tendency to self-coalesce at critical pH or salt concentrations or to bind nucleic acids or other proteins (Bergeron-Sandoval et al., 2016). In contrast to protein aggregates, LLPS-associated proteins can maintain native folding and activity upon liquid mixing and dissociation. These features are well exemplified by the nuclear Arabidopsis RNA-binding protein FCA that interacts with RNA processing components (Fang et al., 2019).
To date, there is no convincing experimental evidence demonstrating the existence and functional importance of LLPSs at or near the plant plasma membrane. However, the plasma membrane, as any membrane surface, restricts the molecular diffusion to a two-dimensional plane, which reduces the concentration threshold required for phase separation (Fig. 5; Case et al., 2019a; Snead and Gladfelter, 2019). In addition, recent advances in mammalian systems suggest that LLPSs may play a crucial role in cell surface signaling (Case et al., 2019a, 2019b; Martin and Mittag, 2019). Indeed, several examples were found in which multivalent interactions between transmembrane receptors and their downstream partners trigger phase transitions (Fig. 5; Case et al., 2019b; Huang et al., 2019b). In this scenario, the clustered receptor-effector complexes separate from the rest of the cytosol in a membraneless liquid compartment, which significantly enhances the residence time of the adaptors at the plasma membrane (Fig. 5). The induction of the enzymatic activity of effector molecules is slow, and their recruitment to unclustered receptors is too transient to allow signal transduction (Huang et al., 2019b).
Figure 5.
Liquid-liquid phase separation: concepts and roles in signaling at the plasma membrane/cytosol interface. A, Schematic representation of ideal liquids and solids. In liquids (left), molecules diffuse to distances greater than their size. In solids (right), molecules are confined by their neighbors. Furthermore, for crystalline solids, positional order exists over long distances and it is possible to draw straight lines along which particles are equally spaced (dashed lines; Hyman et al., 2014). In LLPS, two liquids are unmixed, similar to a water/oil solution. This limits the diffusion of molecules in and out of each liquid phase and allows specific reactions to occur in each of these “membraneless” compartments. B, LLPS in the cytosol may be coupled with lipid phase separation in membranes (Snead and Gladfelter, 2019). C, Phase separation may also occur upon clustering of receptors and adaptor molecules at the plasma membrane. The resulting biomolecular condensate will locally limit the diffusion of downstream signaling components, increasing their dwell time at the plasma membrane and thereby triggering signaling. This process was coined “kinetic proofreading” because it allows the filtering out of noise (i.e. uncontrolled recruitment of effectors at the plasma membrane) from real activation (i.e. induction of LLPS upon receptor nanoclustering; Huang et al., 2019b).
By contrast, prolonged residence time in the biomolecular condensate at the receptor nanocluster phase favors signal activation (Fig. 5). This model was coined “kinetic proofreading,” as it directly relates signaling activity to the dwell time of membrane association of downstream receptor components (Fig. 5). This dwell time is regulated by LLPS, which can be rapidly modulated by posttranslational modification such as Tyr phosphorylation (Huang et al., 2019b). Given that (1) the N-terminal regions of remorin proteins are intrinsically disordered (Marín et al., 2012; Marín and Ott, 2014), (2) their C-terminal regions are able to form higher-order oligomers (Bariola et al., 2004; Marín et al., 2012; Legrand et al., 2019; Martinez et al., 2019), (3) they actively bind polyanions, including anionic lipids (Gronnier et al., 2017; Legrand et al., 2019), and (4) they are phosphorylated (Perraki et al., 2018), remorin proteins are promising candidates to induce LLPSs at the plasma membrane interface and would support and extend a recently proposed model for liquid-like receptor clustering compartments (Cuevas-Velazquez and Dinneny, 2018).
Furthermore, multivalent association with the extracellular matrix in animal cells also potentiates LLPSs at the extracellular surface of animal cells (Case et al., 2019a). Accordingly, the extent of phase separations within the plant cell wall could also profoundly impact signaling, and reciprocally, plasma membrane organization could impact phase separation within the plant cell wall. For example, cellulose may be present in either of two states: amorphous or crystalline. In addition, cellulose microfibrils are embedded in a matrix composed of water, polysaccharides, proteins, and ions. This matrix is a biphasic mixture between a porous solid phase and a liquid phase (Ali and Traas, 2016). One may envision that a change in the equilibrium between these different states may impact lateral diffusion of plasma membrane proteins/lipids. Conversely, the presence of anchored pickets could locally impact the equilibrium between the cell wall porous solid phase and the liquid phase.
CONCLUDING REMARKS AND PERSPECTIVES
Altogether, accumulating evidence over the last years clearly demonstrates that the plant plasma membrane, like the plasma membrane in other eukaryotic cells, is highly compartmentalized. However, most of our knowledge originates either from biochemical fractionation of detergent-resistant membranes or from diffraction-limited microscopy techniques. It is increasingly clear that these techniques lack the resolution to properly address the challenges underlying the study of plasma membrane compartmentalization, as the size of nanodomains are typically below the resolution limit of light microscopy. Nonetheless, in the past few years, superresolution microscopy was used in several studies to address this problem in planta, and we expect that it will be used more frequently in future studies. So far, such techniques have mainly been used to localize one protein at a time, and an important future development of superresolution microscopy in plants will be to set up pipelines for colocalization analysis.
As described above, the cell wall plays a crucial role in plasma membrane organization and the diffusion of cell surface proteins and lipids. This highlights the importance of studying the cell in its native state inside its organism rather than in isolated cellular systems such as protoplasts. It also exemplifies the differences between plants and animals with respect to plasma membrane compartmentalization. A clear challenge for the coming years will be to better understand the potential coupling between the plasma membrane and cell wall organization (see Outstanding Questions). This will notably require experimental approaches to characterize their respective physical states and to monitor the evolution of these states in both space and time. In that respect, new developments in the field of atomic force microscopy, Raman microscopy, Brillouin microscopy, and mass spectrometry imaging may open new opportunities to challenge our current understanding of the cell wall/plasma membrane continuum. Since both systems are highly complex, we envision that such studies will likely require mathematical and/or physical modeling together with reconstitution experiments using minimal membrane/cell wall components.
Alongside extracellular matrices, the differences between plants and animals also include differences in their use of cytoskeleton components and the presence of different lipid species, notably sphingolipids, phytosterols, and very long-chain phosphatidyl-Sers. Therefore, although models based on animal or yeast studies are useful, they should not be taken at face value when thinking about the plant plasma membrane. It is likely that the plasma membrane greatly contributes to the phenotypic plasticity of plants. We therefore need to understand it on a functional level and address local and temporal specifications of the bilayer in response to the ever-changing environment (see Outstanding Questions). In comparison to many other model organisms, the ability to combine genetic, biochemical, and cell biological approaches at a tissue, organ, and even organismic level provides outstanding potential to mechanistically understand signal transduction across multiple scales.
Acknowledgments
We thank Julien Gronnier for his membrane lipid template and Alexandre Martinière and Matthieu Platre for critical comments on the manuscript.
Footnotes
This work was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft) under Germany’s Excellence Strategy (Centre for Integrative Biological Signalling Studies; EXC-2189 – Project ID 39093984 to T.O.) and by the French National Research Agency (Agence Nationale de la Recherche) caLIPSO (ANR-18-CE13-0025-02 to Y.J.) and STAYING-TIGHT (ANR-18-CE13-0016-02 to Y.J.) on lipids and ERA-NET Coordinating Action in Plant Sciences SICOPID on receptor kinase signaling (ANR-17-CAPS-0003-01 to Y.J.).
Articles can be viewed without a subscription.
References
- Albers P, Üstün S, Witzel K, Kraner M, Börnke F (2019) A remorin from Nicotiana benthamiana interacts with the Pseudomonas Type-III effector protein HopZ1a and is phosphorylated by the immune-related kinase PBS1. Mol Plant Microbe Interact 32: 1229–1242 [DOI] [PubMed] [Google Scholar]
- Ali O, Traas J (2016) Force-driven polymerization and turgor-induced wall expansion. Trends Plant Sci 21: 398–409 [DOI] [PubMed] [Google Scholar]
- Amorim-Silva V, García-Moreno Á, Castillo AG, Lakhssassi N, Esteban Del Valle A, Pérez-Sancho J, Li Y, Posé D, Pérez-Rodriguez J, Lin J, et al. (2019) TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in Arabidopsis. Plant Cell 31: 1807–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Retelska D, Stasiak A, Kammerer RA, Fleming A, Hijri M, Frank S, Farmer EE (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol Biol 55: 579–594 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 [DOI] [PubMed] [Google Scholar]
- Bergeron-Sandoval LP, Safaee N, Michnick SW (2016) Mechanisms and consequences of macromolecular phase separation. Cell 165: 1067–1079 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Rose JK (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68: 533–544 [DOI] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, van Esse GW, Kruis A, Luchtenberg J, Westphal AH, Aker J, van Hoek A, Albrecht C, Borst JW, de Vries SC (2013) Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol 162: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas JL, Buré C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F, Fouillen L, Germain V, et al. (2016) Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol 170: 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Ditlev JA, Rosen MK (2019a) Regulation of Transmembrane Signaling by Phase Separation. Annu Rev Biophys 48: 465–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK (2019b) Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ahsan N, Thelen JJ, Stacey G (2019) S-Acylation of plant immune receptors mediates immune signaling in plasma membrane nanodomains. bioRxiv 720482 doi:10.1101/720482 [Google Scholar]
- Colin LA, Jaillais Y (2019) Phospholipids across scales: Lipid patterns and plant development. Curr Opin Plant Biol 53: 1–9 [DOI] [PubMed] [Google Scholar]
- Cuevas-Velazquez CL, Dinneny JR (2018) Organization out of disorder: Liquid-liquid phase separation in plants. Curr Opin Plant Biol 45(Pt A): 68–74 [DOI] [PubMed] [Google Scholar]
- Cui Y, Yu M, Yao X, Xing J, Lin J, Li X (2018) Single-particle tracking for the quantification of membrane protein dynamics in living plant cells. Mol Plant 11: 1315–1327 [DOI] [PubMed] [Google Scholar]
- Daněk M, Angelini J, Malínská K, Andrejch J, Amlerová Z, Kocourková D, Brouzdová J, Valentová O, Martinec J, Petrášek J (2020) Cell wall contributes to the stability of plasma membrane nanodomain organization of Arabidopsis thaliana FLOTILLIN2 and HYPERSENSITIVE INDUCED REACTION1 proteins. Plant J 101: 619–636 [DOI] [PubMed] [Google Scholar]
- Daněk M, Valentová O, Martinec J (2016) Flotillins, erlins, and HIRs: From animal base camp to plant new horizons. Crit Rev Plant Sci 35: 191–214 [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D, et al. (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110: 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, Kleine-Vehn J (2019) Extracellular matrix sensing by FERONIA and leucine-rich repeat extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J 38: e100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Wang L, Ishikawa R, Li Y, Fiedler M, Liu F, Calder G, Rowan B, Weigel D, Li P, et al. (2019) Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569: 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SA, Goyette J, Furuya W, Woods EC, Bertozzi CR, Bergmeier W, Hinz B, van der Merwe PA, Das R, Grinstein S (2016) Integrins form an expanding diffusional barrier that coordinates phagocytosis. Cell 164: 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SA, Vega A, Riedl M, Collins RF, Ostrowski PP, Woods EC, Bertozzi CR, Tammi MI, Lidke DS, Johnson P, et al. (2018) Transmembrane pickets connect cyto- and pericellular skeletons forming barriers to receptor engagement. Cell 172: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescatada-Rosa M, Stanislas T, Backues SK, Reichardt I, Men S, Boutté Y, Jürgens G, Moritz T, Bednarek SY, Grebe M (2014) High lipid order of Arabidopsis cell-plate membranes mediated by sterol and DYNAMIN-RELATED PROTEIN1A function. Plant J 80: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F, König S, Bessoule JJ, Sargueil F, Zallot R, Stanislas T, Noirot E, Lherminier J, Simon-Plas F, Heilmann I, et al. (2010) Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol 152: 2173–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau-Pissot P, Der C, Grebe M, Stanislas T (2016) Ratiometric fluorescence live imaging analysis of membrane lipid order in Arabidopsis mitotic cells using a lipid order-sensitive probe. Methods Mol Biol 1370: 227–239 [DOI] [PubMed] [Google Scholar]
- Ghosh R, de Campos MK, Huang J, Huh SK, Orlowski A, Yang Y, Tripathi A, Nile A, Lee HC, Dynowski M, et al. (2015) Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol Biol Cell 26: 1764–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Crowet JM, Habenstein B, Nasir MN, Bayle V, Hosy E, Platre MP, Gouguet P, Raffaele S, Martinez D, et al. (2017) Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife 6: e26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Gerbeau-Pissot P, Germain V, Mongrand S, Simon-Plas F (2018) Divide and rule: Plant plasma membrane organization. Trends Plant Sci 23: 899–917 [DOI] [PubMed] [Google Scholar]
- Grosjean K, Der C, Robert F, Thomas D, Mongrand S, Simon-Plas F, Gerbeau-Pissot P (2018) Interactions between lipids and proteins are critical for organization of plasma membrane-ordered domains in tobacco BY-2 cells. J Exp Bot 69: 3545–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Zheng S, Liu C, Shen J, Li J, Li L (2016) OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev Cell 38: 201–213 [DOI] [PubMed] [Google Scholar]
- Gui J, Zheng S, Shen J, Li L (2015) Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front Plant Sci 6: 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CH, Long SR (2010) Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA 107: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CH, Riely BK, Tricoli DM, Cook DR, Ehrhardt DW, Long SR (2011) Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23: 2774–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Fan L, Chen T, Li R, Li X, He Q, Botella MA, Lin J (2014) Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26: 1729–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding A, Hancock JF (2008) Ras nanoclusters: Combining digital and analog signaling. Cell Cycle 7: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 197: 805–814 [DOI] [PubMed] [Google Scholar]
- Herger A, Dünser K, Kleine-Vehn J, Ringli C (2019) Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr Biol 29: R851–R858 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol 68: 109–137 [DOI] [PubMed] [Google Scholar]
- Homann U, Meckel T, Hewing J, Hütt MT, Hurst AC (2007) Distinct fluorescent pattern of KAT1:GFP in the plasma membrane of Vicia faba guard cells. Eur J Cell Biol 86: 489–500 [DOI] [PubMed] [Google Scholar]
- Hosy E, Martinière A, Choquet D, Maurel C, Luu DT (2015) Super-resolved and dynamic imaging of membrane proteins in plant cells reveal contrasting kinetic profiles and multiple confinement mechanisms. Mol Plant 8: 339–342 [DOI] [PubMed] [Google Scholar]
- Huang D, Sun Y, Ma Z, Ke M, Cui Y, Chen Z, Chen C, Ji C, Tran TM, Yang L, et al. (2019a) Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc Natl Acad Sci USA 116: 21274–21284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM, Biswas KH, Kuriyan J, Groves JT (2019b) A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363: 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CH, Turnbull D, Myles SM, Leslie K, Keinath NF, Hemsley PA (2018) Variable effects of C-terminal fusions on FLS2 function: Not all epitope tags are created equal. Plant Physiol 177: 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CH, Wright KM, Turnbull D, Leslie K, Jones S, Hemsley PA (2019) Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci Rep 9: 12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten SJ, Hamers DS, Aan den Toorn M, van Esse W, Nolles A, Bücherl CA, de Vries SC, Hohlbein J, et al. (2017) Visualization of BRI1 and SERK3/BAK1 nanoclusters in Arabidopsis roots. PLoS One 12: e0169905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F (2014) Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Hashimoto M, Yusa A, Koinuma H, Kitazawa Y, Netsu O, Yamaji Y, Namba S (2017) Dual targeting of a virus movement protein to ER and plasma membrane subdomains is essential for plasmodesmata localization. PLoS Pathog 13: e1006463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch IK, Konrad SS, Stratil TF, Urbanus SL, Szymanski W, Braun P, Braun KH, Ott T (2014) Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26: 1698–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch IK, Ott T (2011) Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol Plant Microbe Interact 24: 7–12 [DOI] [PubMed] [Google Scholar]
- Jin L, Millard AC, Wuskell JP, Clark HA, Loew LM (2005) Cholesterol-enriched lipid domains can be visualized by di-4-ANEPPDHQ with linear and nonlinear optics. Biophys J 89: L04–L06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Millard AC, Wuskell JP, Dong X, Wu D, Clark HA, Loew LM (2006) Characterization and application of a new optical probe for membrane lipid domains. Biophys J 90: 2563–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kf de Campos M, Schaaf G (2017) The regulation of cell polarity by lipid transfer proteins of the SEC14 family. Curr Opin Plant Biol 40: 158–168 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. (2011) Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad SS, Popp C, Stratil TF, Jarsch IK, Thallmair V, Folgmann J, Marín M, Ott T (2014) S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol 203: 758–769 [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG (2012) Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol 28: 215–250 [DOI] [PubMed] [Google Scholar]
- Laloi M, Perret AM, Chatre L, Melser S, Cantrel C, Vaultier MN, Zachowski A, Bathany K, Schmitter JM, Vallet M, et al. (2007) Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol 143: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent N, Der C, Simon-Plas F, Gerbeau-Pissot P (2019) Cell stage appears critical for control of plasma membrane order in plant cells. Plant Signal Behav 14: 1620058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, Rossignol M, Napier JA, Cullimore J, Bessoule JJ, et al. (2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144: 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand A, Martinez D, Grélard A, Berbon M, Morvan E, Tawani A, Loquet A, Mongrand S, Habenstein B (2019) Nanodomain clustering of the plant protein remorin by solidsState NMR. Front Mol Biosci 6: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wu HM, Cheung AY (2016a) FERONIA and her pals: Functions and mechanisms. Plant Physiol 171: 2379–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liu P, Wan Y, Chen T, Wang Q, Mettbach U, Baluska F, Samaj J, Fang X, Lucas WJ, et al. (2012) A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24: 2105–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J (2011) Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xing J, Qiu Z, He Q, Lin J (2016b) Quantification of membrane protein dynamics and interactions in plant cells by fluorescence correlation spectroscopy. Mol Plant 9: 1229–1239 [DOI] [PubMed] [Google Scholar]
- Liang P, Stratil TF, Popp C, Marín M, Folgmann J, Mysore KS, Wen J, Ott T (2018) Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc Natl Acad Sci USA 115: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Jing Y, Xiao J, Zhang Y, Zhu Y, Julian R, Lin J (2017) Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J 90: 3–16 [DOI] [PubMed] [Google Scholar]
- Mamode Cassim A, Gouguet P, Gronnier J, Laurent N, Germain V, Grison M, Boutté Y, Gerbeau-Pissot P, Simon-Plas F, Mongrand S (2019) Plant lipids: Key players of plasma membrane organization and function. Prog Lipid Res 73: 1–27 [DOI] [PubMed] [Google Scholar]
- Marín M, Ott T (2012) Phosphorylation of intrinsically disordered regions in remorin proteins. Front Plant Sci 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Ott T (2014) Intrinsic disorder in plant proteins and phytopathogenic bacterial effectors. Chem Rev 114: 6912–6932 [DOI] [PubMed] [Google Scholar]
- Marín M, Thallmair V, Ott T (2012) The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein-protein interactions. J Biol Chem 287: 39982–39991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Mittag T (2019) Dwelling at membranes promotes decisive signaling. Science 363: 1036–1037 [DOI] [PubMed] [Google Scholar]
- Martinez D, Legrand A, Gronnier J, Decossas M, Gouguet P, Lambert O, Berbon M, Verron L, Grélard A, Germain V, et al. (2019) Coiled-coil oligomerization controls localization of the plasma membrane REMORINs. J Struct Biol 206: 12–19 [DOI] [PubMed] [Google Scholar]
- Martinière A, Fiche JB, Smokvarska M, Mari S, Alcon C, Dumont X, Hematy K, Jaillais Y, Nollmann M, Maurel C (2019) Osmotic stress activates two ROS pathways with distinct impacts on protein nanodomains and diffusion. Plant Physiol 179: 1581–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Gayral P, Hawes C, Runions J (2011) Building bridges: formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J 66: 354–365 [DOI] [PubMed] [Google Scholar]
- Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu DT, Botchway SW, Webb SE, Mongrand S, Maurel C, et al. (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109: 12805–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna JF, Rolfe DJ, Webb SED, Tolmie AF, Botchway SW, Martin-Fernandez ML, Hawes C, Runions J (2019) The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc Natl Acad Sci USA 116: 12857–12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R (2019) Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Morel J, Claverol S, Mongrand S, Furt F, Fromentin J, Bessoule JJ, Blein JP, Simon-Plas F (2006) Proteomics of plant detergent-resistant membranes. Mol Cell Proteomics 5: 1396–1411 [DOI] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2017) Precision targeting by phosphoinositides: How PIs direct endomembrane trafficking in plants. Curr Opin Plant Biol 40: 22–33 [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Schwessinger B, Segonzac C, Zipfel C (2011) Cautionary notes on the use of C-terminal BAK1 fusion proteins for functional studies. Plant Cell 23: 3871–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T. (2017) Membrane nanodomains and microdomains in plant-microbe interactions. Curr Opin Plant Biol 40: 82–88 [DOI] [PubMed] [Google Scholar]
- Pan X, Fang L, Liu J, Senay-Aras B, Lin W, Zheng S, Zhang T, Manor U, Chen W, Yang Z (2019) Auxin-induced nanoclustering of membrane signaling complexes underlies cell polarity establishment in Arabidopsis. bioRxiv 734665 doi:10.1101/734665 [Google Scholar]
- Perraki A, Cacas JL, Crowet JM, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol 160: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Gronnier J, Gouguet P, Boudsocq M, Deroubaix AF, Simon V, German-Retana S, Legrand A, Habenstein B, Zipfel C, et al. (2018) REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14: e1007378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, Marquès-Bueno MDM, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62 [DOI] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018) A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell 45: 465–480 [DOI] [PubMed] [Google Scholar]
- Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF (2001) GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol 3: 368–375 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T (2007) Genome-wide annotation of remorins, a plant-specific protein family: Evolutionary and functional perspectives. Plant Physiol 145: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al. (2015) Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 161: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Willige BC, Jaillais Y, Geng S, Park MY, Gray WM, Chory J (2019) BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet 15: e1007904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Y, Gerbeau-Pissot P, Buhot B, Thomas D, Bonneau L, Gresti J, Mongrand S, Perrier-Cornet JM, Simon-Plas F (2008) Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J 22: 3980–3991 [DOI] [PubMed] [Google Scholar]
- Rui Y, Dinneny JR (2020) A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol 225: 1428–1439 [DOI] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud MC, Jaillais Y (2016) A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175: 720–731 [DOI] [PubMed] [Google Scholar]
- Snead WT, Gladfelter AS (2019) The control centers of biomolecular phase separation: How membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell 76: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Oh CJ, An CS (2014) Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to Beet Curly Top Virus and Beet Severe Curly Top Virus. Plant Pathol J 30: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Su C, Klein M-L, Hernández-Reyes C, Batzenschlager M, Ditengou FA, Keller J, Delaux P-M, Ott T (2019) The Medicago truncatula DREPP protein triggers microtubule fragmentation in membrane nanodomains during symbiotic infections. bioRxiv 794008 doi:10.1101/794008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski WG, Zauber H, Erban A, Gorka M, Wu XN, Schulze WX (2015) Cytoskeletal components define protein location to membrane microdomains. Mol Cell Proteomics 14: 2493–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Waletich JR, Arredondo F, Tyler BM (2019) Manipulating endoplasmic reticulum-plasma membrane tethering in plants through fluorescent protein complementation. Front Plant Sci 10: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF (2007) Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol 9: 905–914 [DOI] [PubMed] [Google Scholar]
- Tian T, Plowman SJ, Parton RG, Kloog Y, Hancock JF (2010) Mathematical modeling of K-Ras nanocluster formation on the plasma membrane. Biophys J 99: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, Stratil TF, Madsen EB, Ye J, Popp C, Antolín-Llovera M, Grossmann C, Jensen ON, Schüssler A, Parniske M, et al. (2012) Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS One 7: e30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtera L, Schulz J, Hamann T (2019) Cell wall integrity maintenance during plant development and interaction with the environment. Nat Plants 5: 924–932 [DOI] [PubMed] [Google Scholar]
- Voxeur A, Fry SC (2014) Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J 79: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li H, Lv X, Chen T, Li R, Xue Y, Jiang J, Jin B, Baluška F, Šamaj J, et al. (2015) Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol Plant 8: 1334–1349 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhao Y, Luo W, Li R, He Q, Fang X, Michele RD, Ast C, von Wirén N, Lin J (2013) Single-particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and internalization. Proc Natl Acad Sci USA 110: 13204–13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wei Z, Tan S-T, Liu T, Wu Y, Lei J-G, Xue H-W, Liao K (2019) Expressing remorin in animal cells forms filamentous structure unraveling the mechanism of plasmodesmata biogenesis. bioRxiv 791137 doi:10.1101/791137 [Google Scholar]
- Yi K, Guo C, Chen D, Zhao B, Yang B, Ren H (2005) Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol 138: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li R, Lu C, Baluška F, Wan Y (2015a) Di-4-ANEPPDHQ, a fluorescent probe for the visualisation of membrane microdomains in living Arabidopsis thaliana cells. Plant Physiol Biochem 87: 53–60 [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhang X, Qu Y, Li R, Baluška F, Wan Y (2015b) Mapping of membrane lipid order in root apex zones of Arabidopsis thaliana. Front Plant Sci 6: 1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hancock JF (2015) Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta 1853: 841–849 [DOI] [PubMed] [Google Scholar]