Abstract
The Automatic Rhodopsin Modeling (ARM) protocol has recently been proposed as a tool for the fast and parallel generation of basic hybrid quantum mechanics/molecular mechanics (QM/MM) models of wild type and mutant rhodopsins. However, in its present version, input preparation requires a few hours long user’s manipulation of the template protein structure, which also impairs the reproducibility of the generated models. This limitation, which makes model building semiautomatic rather than fully automatic, comprises four tasks: definition of the retinal chromophore cavity, assignment of protonation states of the ionizable residues, neutralization of the protein with external counterions, and finally congruous generation of single or multiple mutations. In this work, we show that the automation of the original ARM protocol can be extended to a level suitable for performing the above tasks without user’s manipulation and with an input preparation time of minutes. The new protocol, called a-ARM, delivers fully reproducible (i.e., user independent) rhodopsin QM/MM models as well as an improved model quality. More specifically, we show that the trend in vertical excitation energies observed for a set of 25 wild type and 14 mutant rhodopsins is predicted by the new protocol better than when using the original. Such an agreement is reflected by an estimated (relative to the probed set) trend deviation of 0.7 ± 0.5 kcal mol–1 (0.03 ± 0.02 eV) and mean absolute error of 1.0 kcal mol–1 (0.04 eV).
Graphical Abstract
1. INTRODUCTION
Vertebrate, invertebrate, and microbial rhodopsins constitute an ecologically widespread class of membrane photoresponsive proteins driving fundamental biological functions such as vision, photoentrainment, chromatic adaptation, ion-gating, and ion-pumping.1–3 The recent discovery of a new family of light-sensing microbial rhodopsins4–7 indicates that we do not still fully comprehend the vast distribution and functional diversity of these systems, which are likely to exploit, globally, an amount of sun-light energy larger than that harnessed by photosynthetic systems.
In spite of their functional diversity, rhodopsins display a remarkably common protein architecture featuring seven α-helices forming a cavity hosting a retinal protonated Schiff base (rPSB) chromophore covalently bound to a lysine located in the middle of helix VII (helix G for microbial rhodopsins).2 Furthermore, the protein functions are invariably initiated by the photoisomerization of the chromophore triggered by the absorption of light of a specific wavelength.8–12 The molecularlevel understanding of how variations in the amino acid sequence can modify the functionality of the rhodopsin molecular architecture appears to be not only central to photobiology13–20 but of importance for the rational design of synthetic mimics21–23 and artificial molecular devices.24–26
In the past, the investigation of how rhodopsin sequence variations modify the residue–chromophore interaction, and in turn, the protein light-response has been limited to a relatively, small number of cases.27–32 For instance, the comprehension of how such variation determines a change in the wavelength of absorption maximum () in tens of rhodopsins or rhodopsin mutants, was studied as the first stage in the understanding of functional variation.20 However, it is apparent that a solid comprehension of how different functions emerged would require the comparative investigation of entire arrays of rhodopsins, thus actively searching for common molecularlevel (e.g., residue type, placement, and conformation) traits associated with an observed property.
There is another reason for moving from the investigation of few rhodopsins to the investigation of larger rhodopsin arrays.
Rhodopsins are of central importance in the field of optogenetics.3,11,33–37 In optogenetics, specific microbial rhodopsins and/or their mutants are expressed in neurons, with the aim of activating, inhibiting, or visualizing neuronal activity through their interaction with light of a specific wavelength. In this context, the search for novel or better optogenetics tools (e.g., rhodopsins with specific values) requires the construction and screening of several sets of mutants of one or more rhodopsins.3,11,37–40 Indeed, red-shifted mutants, which minimize light scattering and absorption by biological tissues, are presently a target of great importance.39,41–46 As discussed above, both the understanding of function variability and the search for mutants with desired properties call for a comparative investigation of large arrays (e.g., hundreds, if not thousands) of rhodopsins with different amino acid sequences. In principle, this type of investigation could be carried out experimentally via expression and purification of rhodopsins from many organisms or, in the case of mutant screening, using directed evolution methods based on random mutagenesis. However, this appears to be an expensive and unpractical research effort to be carried out systematically. As we will now discuss, these issues can, in principle, be pursued through computational means, provided that novel and specialized protocols become available.
Arguably, a computational protocol suitable for the investigation of large arrays of photoresponsive proteins must be based on the construction of hybrid quantum mechanical/molecular mechanical (QM/MM) models.47–51 In fact, QM/MM models decrease the computational cost by limiting the size of the protein moiety treated at the expensive QM level. For instance, in the rhodopsin models considered here, the rPSB chromophore is treated at the QM level using a multiconfigurational quantum chemical method, whereas the protein itself is treated at the inexpensive MM level using a suitable force field. However, even though the application of such technology had, and still has, an important impact for rhodopsin studies, conventional QM/MM models are, almost regularly, computationally complex models which are built manually and feature different QM methods and MM force fields when designed by different research groups. For this reason, they are often (i) time-consuming to construct, (ii) non-congruous (e.g., not comparable), and (iii) error prone. Features i–iii impair the production of such models for extended rhodopsin arrays.
The recently proposed Automatic Rhodopsin Modeling protocol (from now on called ARM)52 represents a first attempt toward the automated and fast generation of congruous QM/MM models of rhodopsins. As illustrated in Figure 1, ARM models are specialized QM/MM models and, in general, would not be applicable to other (e.g., cytoplasmic) photoresponsive proteins. ARM is not designed to produce the most accurate QM/MM models possible (see, for instance, the models of refs 50 and 53 targeting accurate spectroscopic studies), but basic, gas-phase, and computationally fast models aimed at the rationalization and prediction of trends between sequence variability and function. Therefore, ARM aims to satisfy the following desirable features suitable for the generation of arrays of models: automation, so as to reduce building errors and avoid biased QM/MM modeling; speed, so as to deal with large sets of rhodopsins and/or rhodopsin mutants; documented accuracy, so as to be able to translate results into an experimentally assessable hypothesis; transferability, so as to treat rhodopsins with large differences in sequence (i.e., organism belonging to different life domains and kingdoms).
Figure 1.
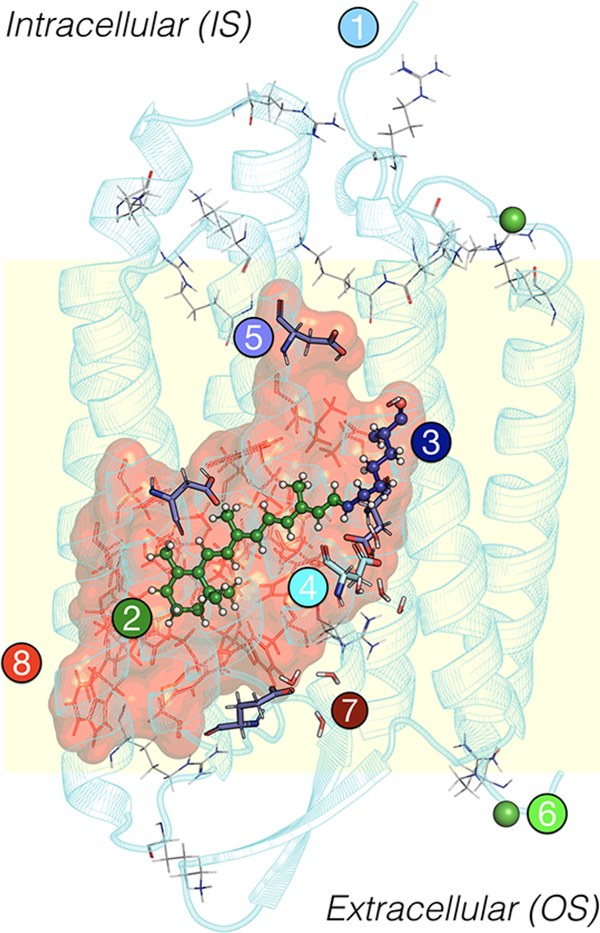
General scheme of a QM/MM ARM and a-ARM model, composed by (1) main chain (cyan cartoon), (2) chromophore rPSB (green ball-and-sticks), (3) Lys side chain covalently linked to the chromophore (blue ball-and-sticks), (4) main counterion MC (cyan tubes), (5) protonated residues GLH and ASH (violet tubes), (6) external Cl– (green balls) counterions, (7) water molecules (tubes), and the (8) residues of the chromophore cavity subsystem (red frames). Parts 1 and 6 form the environment subsystem. Parts 2 and 3 form the Lys-QM subsystem, which includes the H-link atom located along the only bond connecting blue and green atoms. Parts 4 and 8 form the cavity subsystem. Water molecules (Part 7) may be part of the environment or cavity subsystems. The external OS and IS charged residues are shown in frame representation. This figure, and all other protein structures presented in this work, were produced using PyMOL, version 1.2.59
The current version of ARM has been tested for the prediction of trends in of a limited set of wild type and mutant vertebrate, invertebrate, and microbial rhodopsins,10,20,52,54,55 showing good agreement with experimental data. The required input includes (A) an X-ray crystallographic structure or comparative model of the protein in PDB (Protein Data Bank) format,56,57 (B) a list of residues forming the chromophore cavity, (C) the protonation states of ionizable side chains, and (D) the position of extracellular (OS) and intracellular (IS) counterions. As we will detail below, the main drawback of ARM is that it is, substantially, only a semiautomatic (i.e., not fully automated) protocol, as the generation of its input is achieved through a manual manipulation of the template structure necessary to provide the information on points A–D.52 Furthermore, due to possible different user choices (e.g., during the placement of IS and OS counterions), the reproducibility of the results cannot be guaranteed. The latter is a worrisome aspect, since the produced ARM model and, consequently, the calculated properties may be user-dependent. Such limitations, added to the human error factor, represent a serious issue when the target is the generation of hundreds of rhodopsin models (see for instance ref 58 for an example where this would be the case).
In order to overcome the above-described limitations, here we report a novel version of ARM named a-ARM. We will show that, when adopting certain default choices/parameters, a-ARM is capable of performing automatically (i.e., avoiding user manipulation) the following four key steps: (A) definition of the chromophore cavity, (B) assignment of protonation states of ionizable residues side chain, (C) placement of OS and IS counterions, and (D) congruous generation of single or multiple point mutations, allowing in principle for a faster and parallel model building. Such an automated approach, called a-ARMdefault, adopts a set of default values for the choices determining how the QM/MM model is built. These are chain A, if different chains are present in the crystallographic data; chromophore cavity generation based on Voronoi tessellation and alpha spheres theory and including the lysine residue covalently linked to the rPSB, plus the main (MC) and secondary (SC) chromophore counterion residues; protonation states of the ionizable residues based on partial charges calculated at the crystallographic pH and using neutral His residues with the δ-nitrogen of the imidazole protonated (HID tautomer); OS and IS counterion (Na+/Cl–) positions optimized with respect to an electrostatic potential grid constructed around each charged OS and IS residue.
Based on a benchmark set of 25 wild type rhodopsins (including vertebrate, invertebrate, and microbial) and 14 mutants and providing 39 observed values, below we report that a-ARMdefault has a 32/39 success ratio in reproducing the observed trend. In the cases for which the fully automated protocol fails (i.e., produces ΔES1–S0 values far from the observed ones), we show that a semiautomatic approach called a-ARMcustomized can be employed, allowing for the construction of customized models, which display consistency with the observed trend.
Both a-ARMdefault and a-ARMcustomized not only have a high level of automation with respect to the original ARM, but also greatly reduced input preparation time, higher accuracy even when considering distant rhodopsins, and finally full reproducibility of the final results.
2. THEORY AND IMPLEMENTATION OF a-ARM
a-ARM is an improved version of the original ARM based on a Python subroutine, which allows for an automated production of QM/MM models of the type described in Figure 1. a-ARM is designed to generate the ARM input therefore avoiding, as much as possible, human manipulations. In a sense, a-ARM incorporates the original protocol but provides automatically (but also semiautomatically) all required input information. In order to facilitate the description of how a-ARM works, in section 2.1 we revise the main feature of the original input. The following sections 2.2 and 2.3 deal with the a-ARM structure and section 2.4 deals with a-ARM benchmarking.
2.1. ARM Input: Assets and Limitations
The original ARM is, substantially, a Bash shell script that links a series of publicly available computational packages, by automatically managing and passing information between them. The input (herein called ARM input) is constituted by two files containing the information described in points A–D of section 1. The PDBARM input file contains the protein structure in PDB format (from either crystallographic or comparative modeling data) with the assigned residue protonation states and positions of Na+/Cl– external counterions. Instead, the cavity input file contains a list of residues constituting the cavity where the chromophore resides.
In the workflow of the protocol shown in Figure S1 of the Supporting Information, the ARM input is treated sequentially to perform the following actions by a series of software packages: mutation and rotamer selection, using SCWRL4;60 addition of waters and hydrogens, employing DOWSER;61 MM energy minimization and simulated annealing (SA)/molecular dynamics (MD) relaxation, with GROMACS;62 geometry optimization and energy reevaluation at the CASSCF(12,12)/AMBER and CASPT2(12,12) levels, respectively,48 using a combination of the quantum chemical package MOLCAS63 and molecular mechanics package TINKER.64 The SA/MD procedure is performed starting with N = 10 different seeds that provide 10 independent sets of initial velocities for generating 10 independent QM/MM models. Therefore, the resulting output files include 10 replicas of the final equilibrated ARM model as well as the average vertical excitation energy, from now on called simply vertical excitation energy (ΔES1–S0), between ground state (S0) and the first singlet excited state (S1) computed at the CASPT2 level. From these 10 models, the output structure characterized by ΔES1–S0 values closest to the average (N = 10) is selected. As anticipated above, such models correspond to gasphase and globally uncharged models of a rhodopsin monomer, composed of three subsystems, i.e., environment, cavity, and Lys-QM (see Figure 1). The QM part of the Lys-QM subsystem is treated at the CASSCF level and corresponds to the rPSB chromophore, while the Lys part of the same subsystem as well as the environment and cavity subsystems correspond to the MM part of the model and are described at the AMBER level. The entire model construction and 3-root state-average CASPT2(12,12) vertical excitation energy calculation takes, after the input file preparation, ~36 h CPU time when running the 10 replicas in parallel on a modern workstation.
In spite of their elementary structure, ARM models have been shown to be able to reproduce trends in variation in a set of diverse rhodopsins.52 In addition, further studies have demonstrated that the same models are able to successfully simulate, thanks to CASSCF gradients, properties associated with rhodopsin fluorescence,20,55 and photoisomerization.65–67 However, as mentioned in section 1, a critical automation limit of ARM is related to the manual preparation of its input files. Such preparation takes time (see section S1 in the Supporting Information). In our experience, we found that a skilled user can complete the preparation of an ARM input for a new rhodopsin protein in not less than 3 h.
The first step in the manual preparation of an ARM input is the manipulation of the PDB file containing the original rhodopsin crystallographic structure (see point A of section 1), aimed at removing unnecessary information such as unwanted protein chains and subsequently adjusting atoms and residue numbering. This step will also deal with the possible presence of two alternate locations of selected side chains in the same PDB file, for which there is no established selection procedure. Related to this issue, also selection of the residue containing the retinal chromophore (i.e., the residue that will define the Lys-QM subsystem) has to be performed manually. The selected protein chain, side-chain rotamers, and chromophore residue are ultimately written in the PDBARM file.
ARM models are sensitive to the correct choice of protonation state of the protein ionizable residues52 (see point C of section 1). To perform such an assignment, one may use experimental data and/or execute the external program PROPKA68 (see also section 2.2.3) and analyze its output. In this way, residues with uncommon protonation states are identified and their threeletter code manually written in the PDBARM file.
The location of the residues belonging to the cavity surrounding the retinal chromophore (see point B of section 1) is also performed manually through an external Web-based tool (CASTp,69 see section 2.2.2). The user has then to manually prepare the cavity file containing the list of the selected cavity residues. Finally, the last step of the ARM input preparation is the neutralization of the protein environment, through the distribution of OS and IS counterions (see point D of section 1). This step is the most time-demanding and does not follow a well-defined procedure, since it requires the visual inspection of the protein structure and, therefore, has an impact on the reproducibility of the generated model. Again, the final type and coordinates of the selected counterions are added to the PDBARM file. For a more detailed description of the above steps see section S1 in the Supporting Information.
2.2. a-ARM
As already mentioned above, a-ARM has the ability to operate either as a fully automated tool or as an interactive system for the semiautomatic generation of the ARM input presented above. More specifically, in a-ARM the information required for generating complete PDBARM and cavity files may be provided via either default choices or by answering specific questions in the command line of terminal window. With such an input, the QM/MM model generated by the subsequent calculation is called a-ARM model.
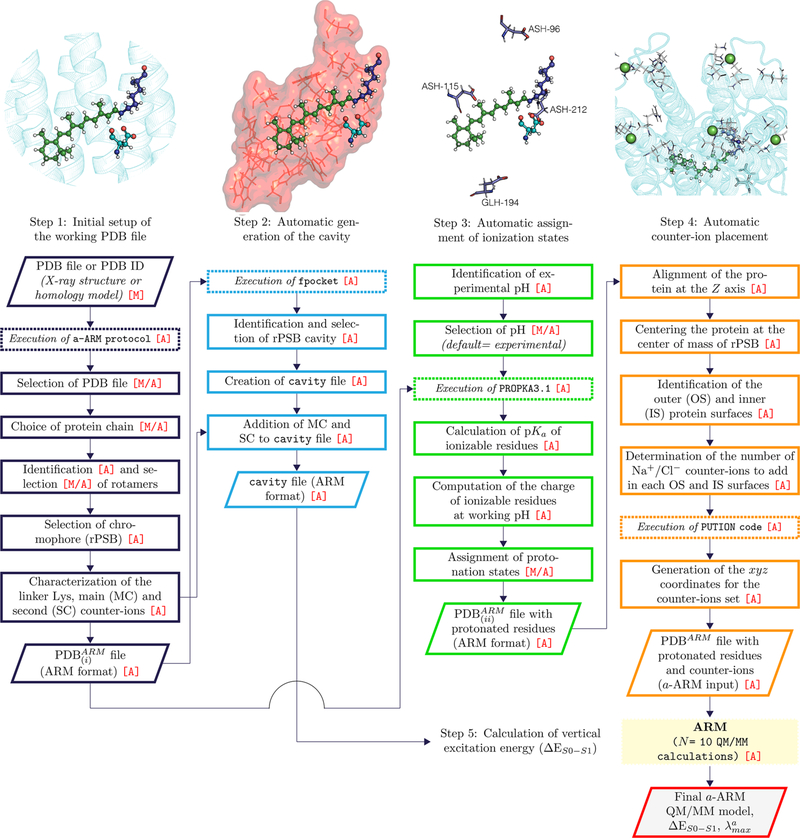
According to the general workflow of a-ARM (see Figure 2), the procedure starts with the selection of the rhodopsin structure of interest used to prepare the ARM input and ends with the generation of the QM/MM a-ARM model and the calculation of the ΔES1–S0 and corresponding (throughout this work, we assume that the vertical excitation energy provides a good approximation for the energy corresponding to the at the CASPT2 level of theory). The code behind the workflow reported in Figure 2 is driven by a modular, Python-based collection of routines and can be accessed upon request to the authors. In the following, we detail the four steps (see sections 2.2.1–2.2.4) of the a-ARM workflow. In section 2.2.5, we will instead report on an automatic mutant generation method also currently incorporated in a-ARM.
Figure 2.
a-ARM workflow. After the selection of the protein chain, a-ARM generates the ARM input files with complete information on the chromophore cavity, protonation states, and counterion placement (see Figure 1) corresponding to points B–D of section 1. The input is then used for the execution of the original ARM,52 obtaining as output 10 a-ARM models along with the calculated average vertical excitation energy (ΔES1–S0). The parallelograms represent input or output data, the continuous line squares refer to processes or actions, and the dashed lines mean software executions. The [A] mark symbolizes fully automation, whereas the [M] mark represent manual decision. Finally, the [M/A] mark indicates situation that may be either manual or automated (see text). Notice that the software execution labeled “QM/MM calculation” is the same as in the original ARM (see ref 52). In a-ARM the production of the PDBARM and cavity input files takes only a few minutes.
2.2.1. Step 1. Automatic Identification of Protein Chain, rPSB, Chromophore Bounded Lys, MC, and SC
In Step 1 of Figure 2 (see also Figure S2 in the Supporting Information) we display the workflow necessary to obtain the initial structure of the rhodopsin of interest. To begin with, the user has the option to provide a crystallographic structure or a comparative model in PDB format or type the PDB ID to download the file directly from the RCSB PDB.57 The program is then able to identify automatically the different protein chains, which may be present in the PDB file and select chain A by default (i.e., automatically or [A]) or, alternatively, let the user select the chain (i.e., manually [M]). Thus, the program generates a file , which contains information on the selected chain, residue conformations, chromophore, and water molecules.
Due to their local flexibility, certain residues may have two alternate side chains locations (i.e., conformations) in the protein crystallographic structure. The strategy adopted to assign a single rotamer, without the need to visualize the structure, is to analyze the atom occupancy number in the coordinate section of the file. This parameter, which takes values from 0 to 1, is used as a criteria to estimate the frequency of each conformation. Accordingly, a-ARM identifies the residues with atom occupancy different from 1.0, creates a list with residue name and sequential number, and the occupancy value of the alternate side chain locations and acts automatically [A] by selecting the rotamer with the largest occupancy or, alternatively, asks the user to select the wanted rotamer by typing the corresponding number [M].
The rPSB chromophore is automatically recognized and selected. For this purpose, the program identifies all residues which are not standard amino acids, waters, and membrane lipids and generates a list of possible chromophores. Once again, the chromophore can be selected automatically by default, which corresponds to the ordinary rPSB chromophore [A], or the user may select the correct option manually by typing the corresponding residue number [M]. Here, we should stress that, while this step is instrumental for a future generalization of a-ARM (e.g., for considering multiple chromophores), there is only a single rPSB chromophore in rhodopsins and therefore the user intervention is not needed. Moreover, although in the majority of rhodopsin coordinate files in the RCSB PDB,57 the retinal and the covalently linked lysine are two distinct residues (i.e., RET and LYS, respectively) in a minority of cases (e.g., 6EID70 and 6EIG70) retinal and lysine constitute a single residue (LYR). In principle, this LYR formatting is not compatible with ARM52 algorithms, which are designed to recognize the RET and linker LYS as distinct residues. To deal with that, a-ARM is now able to automatically recognize the LYR residue and subsequently split it into RET and LYS, respecting the standard format of residue and atom names (see section S4 in the Supporting Information).
Another important feature of the program is that, based on the geometrical parameters of the selected chromophore, the chromophore-linked Lys side chain and the potential MC and SC counterions are automatically identified. This is achieved by first locating the linked Lys as the residue geometrically closest to the chromophore, by computing the Euclidean distance between each atom in the chromophore and the coordinates of the nitrogen “NZ” of all the Lys residues present in the structure. Then, the MC and SC are identified as the two Asp and/or Glu and/or crystallographic Cl– residues geometrically closest to the chromophore-linked Lys side chain, by computing the distance between the coordinates of its nitrogen “NZ” and the coordinates of the oxygen “O” of each of the carboxylatebearing residues (or the chlorine atom). However, this selection is only preparatory to the ionization state assignment (see section 2.2.3) that determines if the SC and MC are indeed acting as negatively charged Schiff base counterions. The inclusion of the Cl– anions contained in the X-ray structure into the QM/MM model, even when not considered as MC or SC, is a new feature of a-ARM that allows a more realistic description of rhodopsin chloride pumps (i.e., 5B2N71 and 5G2872).
2.2.2. Step 2: Automatic Generation of the Chromophore Cavity
The identification and characterization of the chromophore cavity is a key step for the definition of congruous QM/MM models of rhodopsins (see Figure 2 and section 2.1). There are different algorithms for protein pocket detection.73 These are mainly available via Web server-based facilities, but a few are distributed as a code for local usage. The widely used Web servers include CASTp,69 employed in the original ARM protocol.52 Even though CASTp has proven to be effective, the fact that it is not available as a command line code makes it unsuitable for a full automation. Thus, we decided to use the Fpocket software,74 which can be integrated in a-ARM as illustrated in Figure 2 (see also Figure S3 in the Supporting Information). Fpocket detects the chromophore cavity based on Voronoi tessellation and alpha spheres built on top of the publicly available package Qhull.74 First, a-ARM receives as input the previously generated file and automatically executes the Fpocket software using the default options.74 As output, several protein pockets are obtained along with their scores. The selected chromophore cavity is the one that contains the Lys covalently linked to the rPSB and has the highest score. Finally, the previously identified MC and SC counterion residues are added to the cavity list (if not already present) and the updated list is written in the final cavity file.
2.2.3. Step 3: Automatic Assignment of Ionization States
Our procedure for the assignment of the protonation state of the ionizable residues at a given pH and in their specific protein environments is based on the assumption that such state is a function of the pKa value.75 Accordingly, each residue with a titratable group is associated with a model pKa value (),76 interpreted as the pKa displayed when the other protein side chains are in their neutral state. On the other hand, is affected by the interaction between the residue and its actual environment, causing a change from the model value to the real pKa value (see eq 1) called . The magnitude of this change, called shift value (ΔpKa), depends on the presence of hydrogen bonds, desolvation effects, and Coulomb interactions, all modulated through the degree to which the ionizable residue is “buried” within the protein.68,75
| (1) |
The adopted procedure is outlined in Step 3 of Figure 2 (see also Figure S4 in the Supporting Information), and it is initialized automatically after the detection of the and cavity files. In case that the crystallization conditions are available in the initial PDB structure file, the program identifies the experimental pH making the pH selection automatic [A]. Otherwise, the user is asked to insert the pH value and the pH selection is thus not automatic [M]. Once the working pH is assigned, the is obtained using the PROPKA software which also determines the burying percentage.68 A preliminary preparation of the file, consisting of completing the heavy missing atoms of chain residues (including hydrogen atoms), is needed to guarantee the correct operation of PROPKA.77 This requires using the PDB2PQR78,79 software, which operates under the following workflow: (i) check for missing heavy atoms, (ii) reconstruct heavy atoms, (iii) build and optimize hydrogens, and (iv) assign atomic parameters (for further details see ref 78). PDB2PQR is automatically launched using as input the file and as arguments the given pH and the AMBER force field. After that, PROPKA is launched and its output contains information on the calculated () values for each ionizable residue in the protein at the given pH.68 The subsequent assignment of the protonation states of the ionizable groups is carried out based on the above information.
According to a first approach (not reported in Figure 2) employed by Melaccio et al.52 for the construction of the original ARM models, the parameters used to identify the state of the ionizable residues are the burying percentage, which indicates how accessible the residue is from the surface (for further details see ref 68), and the ΔpKa shift calculated at pH 7.0 as shown in eq 1. In contrast, in a-ARM the parameter used to identify the state of the ionizable residues is the side-chain ionization equilibrium. Such equilibrium is estimated by inserting both the value and the established working pH in the Henderson–Hasselbalch equation,80 which describes the relationship between the pH and the pKa and the equilibrium concentrations of dissociated [A–] and non-dissociated acid [HA], respectively:80–82
| (2) |
The charges of the positive and negatively ionizable residues are then deduced from eq 2 using the following approximated rules:81
| (3) |
and
| (4) |
where ⌈Q+⌉ and ⌈Q−⌉ are integers obtained by rounding the decimals using the “round half to even” convention. Once ⌈Q+⌉ and ⌈Q−⌉ are obtained, the following criteria is used to assign the ionization (i.e., protonation) state:
| (5) |
The final result is added to the file to generate the file now also containing the ionization states.
There are two aspects that limit the confidence in the automation of the ionizable-state assignment described above. The first is that, due to the fact that the information provided by PROPKA68 is approximated, the computed value may, in certain cases, be not sufficiently realistic. The second aspect concerns the assignment of the correct tautomer of histidine. This amino acid has charge of +1 when both the δ-nitrogen and δ-nitrogen of the imidazole ring are protonated (HIP), while it is neutral when either the δ-nitrogen (HID) or the ϵ-nitrogen (HIE) are deprotonated. a-ARM uses as a default the HID tautomer for the automatic assignment [A] or allows the user to choose between the three tautomers for a non-automated selection [M]. Therefore, when possible, the user should collect the available experimental data and/or inspect the chemical environment of the ionizable residues including the histidines and propose the appropriate tautomer. Further details are given in section S8 in the Supporting Information.
2.2.4. Step 4: Automatic Counterion Placement
The procedure to select and place OS and IS counterions in the model represents a difficult automation problem (see section 2.1). Herein, we report a novel approach for automatically generating and placing such counterions and therefore avoiding user manipulation. The approach is documented in Step 4 of Figure 2 (see also Figure S5 of the Supporting Information). The initial task consists in determining the type (Cl– and/or Na+) and number of counterions needed to neutralize the protein environment. This calculation is carried out based on the actual charges of the OS and IS surfaces, which depend on the quantity of positively and negatively charged residues. Therefore, the output of Step 4 depends on the result of Step 3.
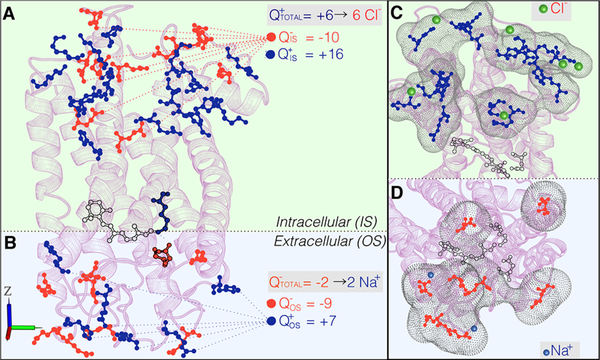
To define the OS and IS surfaces, the protein is oriented along the z axis, as illustrated in Figure 3. To this aim, the protein coordinates found in the file are first centered at the protein center of mass (xyzcm). The new set of coordinates are then rotated such as the main rotational axis is aligned with the z axis, using the Orient utility of the VMD83 software. Finally the coordinates are recentered at the center of mass of the chromophore. These coordinate transformations allow to define an imaginary plane orthogonal to the z-axis and containing the z coordinate of the NZ atom (zPSB) of the rPSB moiety. Such a plane divides the protein in two halves and establishes the OS and IS surfaces in terms of the z value: the ionized residues with a z value larger than zrPSB belong to the OS surface, whereas those residues with z lower or equal to zrPSB belong to the IS surface. The charge of each surface (QOS, QIS) is calculated as the difference between the number of positively charged (Arg, Lys, and His) and negatively charged (Asp, Glu, and crystallographic Cl– anions) residues. Once the surface charge is known, the protocol provides the type and number of counterions required to neutralize the net charge of each surface independently and, in turn, of the full protein. This procedure is illustrated in Figure 3A,B for the case of bovine rhodopsin (Rh).84 Accordingly, the net charge of the IS surface is QIS = +6, resulting from 16 positively charged and 10 negatively charged residues, whereas the net charge of the OS is QOS = –2, given by 7 positively charged and 9 negatively charged residues. As a consequence, 6 Cl– and 2 Na+ must be added to compensate the positive and negative charge of the IS and OS, respectively.
Figure 3.
External counterion placement. Schematic representation of the procedure for the definition of the number and type of external counterions needed to neutralize the IS (A) and OS (B) surfaces of bovine rhodopsin. We also illustrate the grid generated by the PUTION code to calculate the coordinates of the Cl– counterions in the IS (C) and the Na+ in the OS (D). The negatively and positively charged residues are illustrated as red and blue sticks, respectively, and the Na+ and Cl– counterions as blue and green spheres, respectively.
One main difference between the original ARM and the new a-ARM protocol is that, whereas the original version requires the visual inspection of the PDB file to manually identify the charged residues and calculate the number and identity of the counterions to be added on each surface, the new version performs these tasks automatically. The automatic location of ionized residues on OS and IS provides the basis to properly and automatically place the counterions.
As described for the original ARM,52 the user-defined OS and IS surfaces are neutralized using a set of counterions placed, semiautomatically, in the regions where the field generated by the charge of the ionized residues is stronger. In fact, while ARM employs a program called PUTION (described by Melaccio et al. as the ION Module52) that uses an energy minimization procedure to place the counterions, the user has to manually specify the target residues on the IS and OS surfaces, including number of residues, residue number identification, and the number and type of counterions to be added. With the aim of removing the above automation limits, a-ARM adopts a different strategy to assign the target residues and execute PUTION automatically. More specifically, PUTION optimizes the counterion positions on the basis of the Coulomb’s law,85 by computing an electrostatic potential grid constructed around all charged residues and excluding points whose distance is larger than 8.0 Å from the center of charge of a ionized residue and shorter than 2.0 Å from any residue atom.
As reported in Step 4 of Figure 2, the PUTION code is automatically launched right after the determination of the partial charges of each residue (see previous section). The program starts by placing a counterion on the surface with the highest net charge. The placement process is then alternated between the OS and IS surfaces, until both are neutralized. The energy of the Nth counterion is computed from the electrostatic interaction with the protein and the N – 1 preceding counterions. As an output, the geometry of all external counterions is generated as illustrated in Figure 3C,D and added to to generate the final PDBARM file, which is ready to be used as an input for the QM/MM model building.
2.2.5. Automatic Generation of Mutants: Redefinition of Cavity, Ionization States, and Counterion Placement
By exploiting the backbone-dependent rotamer library implemented in the software SCWRL4,60 the original ARM has the ability to perform amino acid substitutions on rhodopsin structures and generate QM/MM models of mutants.52 However, such calculation has serious limitations, since the generated mutants tend to preserve the chromophore environment (i.e., chromophore cavity, protonation states, and external counterions) of the wild type form (unless this information is manually modified). Therefore, although the method has been shown to be effective when a wild type residue is substituted with a residue with the same charge,52 it is unsuitable for replacements altering the residue charge or polarity, thus possibly affecting the protonation state of nearby residues and, in general, the distribution of OS and IS counterions. An additional problem with the original ARM is that when a mutated residue does not belong to the chromophore cavity, this is not relaxed but kept frozen.
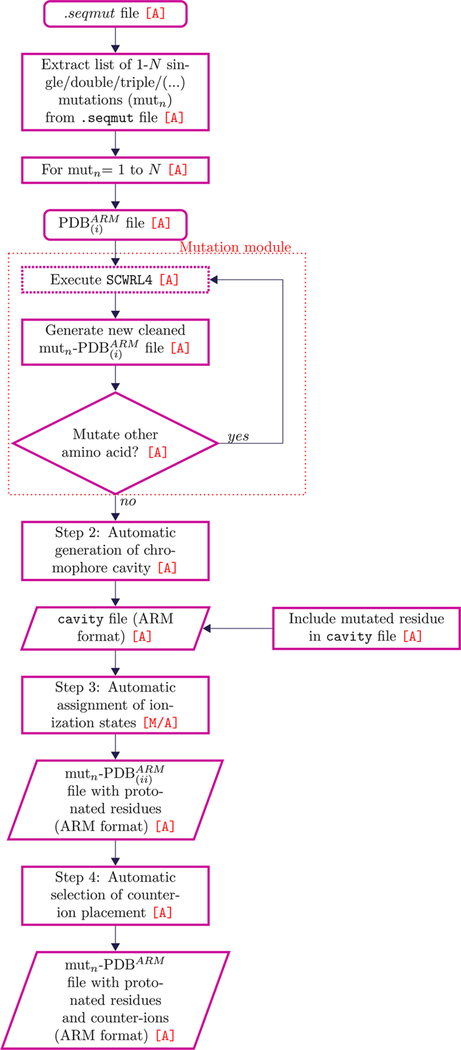
Given the importance of developing a suitable tool for the construction of congrous QM/MM models of mutants, we implemented in a-ARM a new mutation method that takes into account the effects of amino acid substitution on the protein environment (see the workflow in Figure 4). The method requires an additional input file with a .seqmut extension that contains the information on the type (single, double, triple, etc.) and number (N in the flowchart of Figure 4) of required mutations. After detecting the .seqmut file, a-ARM generates N lists with information on each mutation (mutn in the flowchart of Figure 4). Each list, along with the generated for the wild type structure in Step 1, provides the input for the automatic execution of the SCWRL4 software. In the case of multiple mutations, the SCWRL4 software is re-executed. When the mutation process is concluded, the mutant QM/MM models are built through generation of the cavity, assignment of protonation states, and selection of counterion placement carried out by following Steps 2–4, as described above for the wild type structure. Notice that in Step 2 the mutated residues are always included in the cavity subsystem (MM part) and, consequently, they are relaxed during the SA/MD procedure and subsequent QM/MM level geometry optimization.
Figure 4.
Automatic generation of mutants by using the SCWRL460 software. The code does not require any interaction with the user during execution.
2.3. a-ARMdefault and a-ARMcustomized Approaches
In Figure 2, we marked as automatic [A] or manual [M] the choices in Steps 1–4 described in sections 2.2.1–2.2.4. The [A] or [M] choices define two different approaches for the generation of a-ARM models. The first, named a-ARMdefault, is a fully automated approach that delivers maximum input preparation speed (see section 3.1) for the systematic building of a-ARM models and, therefore, useful for the generation of large arrays of wild type rhodopsins and of their mutants (as described in section 2.2.5). This is achieved by employing the default choices described in sections 2.2.1–2.2.4. Accordingly, these models are built on the basis of chain A and the side-chain rotamer with the highest occupancy, a chromophore cavity generated by Fpocket and including the Lys covalently linked to the rPSB and MC and SC residues, ionization states predicted at the crystallographic pH (or at physiological pH 7.4 in the case of no experimental information available) with a neutral HID tautomer of histidine and automatic counterion placement decided by the PUTION code.52 In addition to these choices, a default choice has to be taken for the rhodopsins displaying, for certain residues, alternate side chain locations with exactly the same top occupancy. As we will see below, this is found in two crystallographic structure of our benchmark set (see section 2.4) where two rotamers display a 50% probability (occupancy number 0.5) to contribute to the observed structure. In this situation, the default action of the automated a-ARMdefault approach is to generate one a-ARM model for each rotamer.
The second approach, named a-ARMcustomized, is semiautomatic and slower than a-ARMdefault but of increased accuracy (see section 3.2). In fact it allows, for instance, the construction of “customized” a-ARM models useful when the default choices give a poor result in terms of reproducing the experimental ΔES0–S1 trends (e.g., differences with experimental data larger than 3–4 kcal mol–1; 0.1–0.2 eV). a-ARMcustomized requires user manipulation during Steps 1 and 3, which consists of selecting the protein chain (in case of multi-chain rhodopsins), typing the number identifier of ionizable residues with neutral charge (based on chemical criteria or experimental data), and selecting the tautomer of the histidine. Instead, Steps 2 and 4 are performed as in the a-ARMdefault approach. Notice that even though the semiautomatic procedure requires user manipulation, the resulting models are always replicated even when different users select the options.
2.4. Benchmark Set for a-ARM
In sections 2.2 and 2.3, we have mainly dealt with the automation, speed, and reproducibility of a-ARM. However, no information is provided on the protocol accuracy in predicting property trends and, at the same time, on the transferability of the a-ARM model between rhodopsins with diverse (i.e., non-homologous) sequences. Information on both accuracy and transferability requires a benchmark study that, here, we limit to the calculation of ΔES1–S0. In order to compare this computed quantity with the experimental data, we assume that the observed ΔES1–S0 values can be derived from the observed via the equation ΔES1–S0 = hc/. As mentioned above, the calculated values are obtained via single-point 3-roots state-average CASPT2(12,12)//CASSCF(12,12)/AMBER calculations yielding the potential energy of the S0, S1, and S2 states. The fact that ΔES1–S0 corresponds to an allowed electronic transition is supported via oscillator strength (f Osc) calculations.
A benchmark data set comprising a pool of observed (expressed in terms of ΔES0–S1) values for 25 wild type and 14 mutant rhodopsins was employed for testing a-ARM. From these mutant rhodopsins, only 2 have an available X-ray structure (i.e., ASRAT-D217E and ChR2-C128T), while the other 12 were generated by the procedure described in section 2.2.5. The data set incorporates the set employed by Melaccio et al.52 for testing the original ARM (m-set), an additional set of rhodopsins (a-set), and a set of Rh mutants (Rh mutants). The full set, which includes vertebrate, invertebrate, and microbial rhodopsins is presented in Table 1 and features values ranging from 430 to 575 nm. The number of observed values will provide information on the method accuracy while the diversity (e.g., microbial vs vertebrate) of rhodopsins will provide information on the transferability of the generated models.
Table 1.
Benchmark Set of Wild Type and Mutant Rhodopsinsa
| proteinb | code | PDB ID | RET-Cc | chain(s) | ΔES0–S1 | |
|---|---|---|---|---|---|---|
| m-Set | ||||||
| X-ray Crystallographic Structures | ||||||
| Anabaena sensory rhodopsin (M) | ASRAT | 1XIO86 | all-trans | A | 550 | 52.0 (2.25)86 |
| ASR13C | 1XIO86 | 13-cis | A | 537 | 53.2 (2.31)86 | |
| Bacteriorhodopsin (M) | bRAT | 6G7H87 | all-trans | A | 568 | 50.3 (2.18)2 |
| bR13C | 1X0S88 | 13-cis | A | 548 | 52.2 (2.26)2 | |
| Bathorhodopsin (V) | bathoRh | 2G8789 | all-trans | A, B | 529 | 54.0 (2.34)90,91 |
| Blue proteorhodopsin (M) | BPR | 4JQ692 | all-trans | A, B, C | 490 | 58.3 (2.52)93 |
| Bovine rhodopsin (V) | Rh | 1U1984 | 11-cis | A, B | 498 | 57.4 (2.49)32,91 |
| Chimaera channelrhodopsin (M) | ChRC1C2 | 3UG994 | all-trans | A | 458 | 62.4 (2.71)40 |
| Squid rhodopsin (I) | SqRh | 2Z7395 | 11-cis | A, B | 489 | 58.5 (2.54)96 |
| Comparative Models | ||||||
| Human melanopsin (V) | hMeOp | 2Z73e | 11-cis | A | 473 | 60.4 (2.62)d |
| a-Set | ||||||
| X-ray Crystallographic Structures | ||||||
| Anabaena sensory rhodopsin D217E (M) | ASRAT-D217E | 4TL397 | all-trans | A, B | 552 | 51.8 (2.25)98 |
| Archaerhodopsin-1 (M) | Arch1 | 1UAZ99 | all-trans | A, B | 568 | 50.3 (2.18)100 |
| Archaerhodopsin-2 (M) | Arch2 | 3WQJ101 | all-trans | A | 550 | 52.0 (2.25)101 |
| Channelrhodopsin-2 (M) | ChR2 | 6EID70 | all-trans | A, B | 470 | 60.8 (2.64)70 |
| Channelrhodopsin-2 N24Q/C128T (M) | ChR2-C128T | 6EIG70 | all-trans | A, B | 485 | 59.0 (2.56)70 |
| Krokinobacter eikastus rhodopsin 2 (M) | KR2 | 3X3C102 | all-trans | A | 525 | 54.5 (2.36)102 |
| Nonlabens marinus rhodopsin-3 (M) | NM-R3 | 5B2N71 | all-trans | A | 517 | 55.3 (2.40)71 |
| C1R | 5G2872 | all-trans | A | 517 | 55.3 (2.40)72 | |
| Sensory rhodopsin II (M) | SRII | 1JGJ103 | all-trans | A | 497 | 57.5 (2.49)103 |
| Squid bathorhodopsin (I) | SqbathoRh | 3AYM104 | all-trans | A, B | 530 | 53.9 (2.34)104 |
| Comparative Models | ||||||
| Ancestral archosaur rhodopsin (V) | AARh | 1U19e | 11-cis | A | 508 | 56.3 (2.44)19 |
| Human blue cone (V) | BCone | 1U19e | 11-cis | A | 430 | 66.5 (2.88)105 |
| Human green cone (V) | GCone | 1U19e | 11-cis | A | 535 | 53.4 (2.32)105 |
| Human red cone (V) | RCone | 1U19e | 11-cis | A | 575 | 49.7 (2.15)105 |
| Mouse melanopsin (V) | mMeOp | 2Z73e | 11-cis | A | 467 | 61.2 (2.65)106 |
| Parvularcula oceani Xenorhodopsin (M) | PoXeRAT | 4TL397,e | all-trans | B | 568 | 50.3 (2.18)107 |
| PoXeR13C | 4TL3e | 13-cis | B | 549 | 52.1 (2.26)107 | |
| Rh Mutants Set | ||||||
| Bovine rhodopsin mutation (V) | A292S | 1U19e | 11-cis | A | 491 | 58.2 (2.52)32 |
| A269T | 1U19e | 11-cis | A | 514 | 55.6 (2.41)43 | |
| E113D | 1U19e | 11-cis | A | 510 | 56.1 (2.43)108 | |
| E122Q | 1U19e | 11-cis | A | 480 | 59.6 (2.58)109 | |
| F261Y | 1U19e | 11-cis | A | 510 | 56.0 (2.43)43 | |
| G90S | 1U19e | 11-cis | A | 489 | 58.5 (2.54)110 | |
| T94S | 1U19e | 11-cis | A | 494 | 57.9 (2.51)111 | |
| T118A | 1U19e | 11-cis | A | 484 | 59.1 (2.56)112 | |
| W265F | 1U19e | 11-cis | A | 480 | 59.6 (2.58)113 | |
| W265Y | 1U19e | 11-cis | A | 485 | 59.0 (2.56)110 | |
| D83N-E122Q | 1U19e | 11-cis | A | 475 | 60.2 (2.61)109 | |
| A292S-A295S-A299C | 1U19e | 11-cis | A | 484 | 59.1 (2.56)110 |
Experimental maximum absorption wavelength, , in nm, and first vertical excitation energy, ΔES0–S1, in kcal mol−1. Values of ΔES0–S1 in eV are also provided in parentheses.
Vertebrate (V), invertebrate (I), and microbial (M).
Retinal conformation.
X-ray structure template model.
See the details of the comparative model construction in section S6 of the Supporting Information.
In our benchmark study, we initially use the a-ARMdefault approach to obtain, in a fully automated fashion, the ΔES0–S1 trend. However, as reported in the previous section, default choices do not always generate a single a-ARM model. As we will document in section 3.2, this happens for the ASRAT, ASR13C, and KR2 rhodopsins. In these cases, the selection of a single representative rotamer is only possible when the corresponding observed ΔES0–S1 value is available (as for our benchmarks). The selected a-ARM model will be the one yielding the computed ΔES0–S1 value closest to the observed one.
3. RESULTS AND DISCUSSION
We are interested in answering the question of whether the a-ARM models generated using the input files PDBARM, cavity, and seqmut are suitable for predicting trends of ΔES1–S0 of wild type and mutant rhodopsins. For this purpose, we first compute the trend generated using the fully automated a-ARMdefault approach. Then, we describe some specific models that do not produce values consistent with the observed trend (i.e., with deviations larger than ~3–4 kcal mol–1), for which the use of a-ARMcustomized is needed. We recall that, in all cases, the computed ΔES1–S0 values are averages over 10 replicas of the final equilibrated a-ARM model (see section 2.1). The S0 and S1 energies, for each of the 10 replicas, are reported in Table S3 in the Supporting Information.
3.1. a-ARMdefault
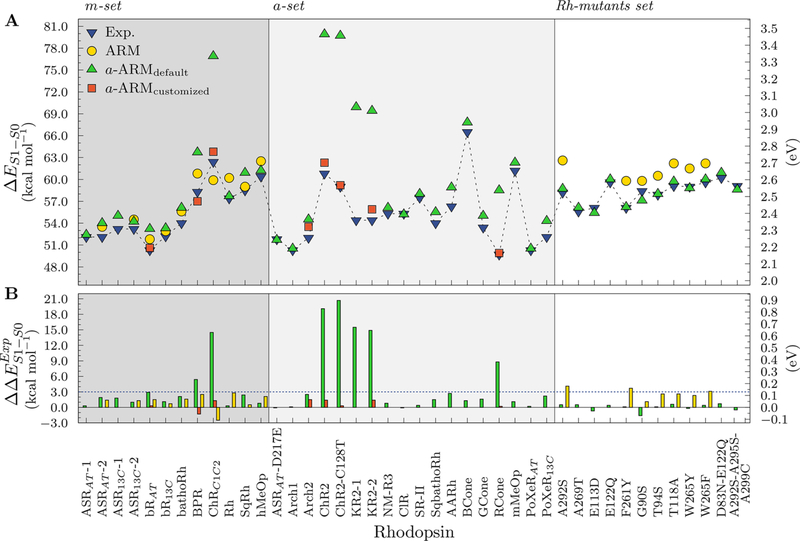
Figure 5A displays the ΔES1–S0 values for the 25 wild type and 14 mutant rhodopsins of the benchmark set (see Table 1), using the a-ARMdefault approach described in section 2.3 (green up triangles), whereas Figure 5B displays their differences calculated with respect to experimental data . The numerical values together with the corresponding and transition oscillator strength (f Osc) values are given in Table 2 and demonstrate that the S1 state is indeed a strongly absorbing state.
Figure 5.
(A) Vertical excitation energies (ΔES1–S0) computed with a-ARMdefault (up triangles) and a-ARMcustomized (squares), along with reported ARM52 (circles) and experimental data (down triangles). S0 and S1 energy calculations were performed at the CASPT2(12,12)//CASSCF(12,12)/AMBER level of theory using the 6–31G(d) basis set. The calculated ΔES1–S0 values are the average of 10 replicas (see Table S3 in the Supporting Information). (B) Differences between calculated and experimental ΔES1–S0 (). Values presented in kcal mol–1 (left vertical axis) and eV (right vertical axis).
Table 2.
Vertical Excitation Energies (ΔES1–S0, kcal mol−1 and eV in Italic and Parentheses), Maximum Absorption Wavelengths (, nm), and Oscillator Strength (fOsc)a
| experimental |
calculatedb |
error |
|||||
|---|---|---|---|---|---|---|---|
| model | ΔES1–S0 | fOsc | |||||
| a-ARMdefault | |||||||
| m-Set | |||||||
| ASRAT-1 | 52.0 (2.25) | 550 | 52.31.4 (2.27) | 547 | 1.29 | 0.3 (0.01) | −3 |
| ASRAT-2d | 52.0 (2.25) | 550 | 54.02.3 (2.34) | 529 | 1.25 | 2.0 (0.09) | −21 |
| ASR13C-1d | 53.2 (2.31) | 537 | 55.00.5 (2.38) | 520 | 1.04 | 1.8 (0.08) | −17 |
| ASR13C-2 | 53.2 (2.31) | 537 | 54.20.4 (2.35) | 528 | 1.08 | 1.0 (0.04) | −9 |
| bRAT | 50.3 (2.18) | 568 | 53.20.1 (2.30) | 537 | 1.25 | 2.9 (0.12) | −31 |
| bR13C | 52.2 (2.26) | 548 | 53.30.1 (2.31) | 536 | 0.94 | 1.1 (0.05) | −12 |
| bathoRh | 54.0 (2.34) | 529 | 56.20.3 (2.44) | 509 | 0.96 | 2.2 (0.10) | −20 |
| BPR | 58.3 (2.53) | 490 | 63.70.2 (2.76) | 449 | 0.57 | 5.4 (0.24) | −41 |
| ChRC1C2 | 62.4 (2.71) | 458 | 76.92.4 (3.33) | 372 | 0.88 | 14.5 (0.63) | −86 |
| Rh | 57.4 (2.49) | 498 | 57.70.7 (2.50) | 496 | 0.87 | 0.3 (0.01) | −2 |
| SqRh | 58.5 (2.54) | 489 | 60.90.2 (2.64) | 469 | 0.80 | 2.4 (0.10) | −20 |
| hMeOp | 60.4 (2.62) | 473 | 61.21.7 (2.65) | 467 | 0.72 | 0.8 (0.04) | −6 |
| a-Set | |||||||
| ASRAT-D217E | 51.8 (2.25) | 552 | 52.01.1 (2.25) | 550 | 1.29 | 0.2 (0.01) | −2 |
| Arch1 | 50.3 (2.18) | 568 | 50.51.2 (2.19) | 567 | 1.23 | 0.1 (0.01) | −1 |
| Arch2 | 52.0 (2.25) | 550 | 54.50.6 (2.36) | 525 | 1.23 | 2.5 (0.11) | −25 |
| ChR2 | 60.8 (2.64) | 470 | 79.92.3 (3.46) | 358 | 0.70 | 19.1 (0.83) | −112 |
| ChR2-C128T | 59.0 (2.56) | 485 | 79.71.1 (3.45) | 360 | 0.65 | 20.7 (0.90) | −126 |
| KR2-1 | 54.5 (2.36) | 525 | 69.90.9 (3.03) | 409 | 1.46 | 15.5 (0.67) | −116 |
| KR2-2d | 54.5 (2.36) | 525 | 69.60.7 (3.02) | 411 | 1.41 | 14.9 (0.66) | −114 |
| NM-R3 | 55.3 (2.40) | 517 | 56.10.1 (2.44) | 509 | 1.01 | 0.8 (0.04) | −8 |
| C1R | 55.3 (2.40) | 517 | 55.20.2 (2.39) | 518 | 1.05 | −0.2 (−0.01) | +1 |
| SRII | 57.5 (2.49) | 497 | 58.00.9 (2.51) | 493 | 1.11 | 0.5 (0.02) | −4 |
| SqbathoRh | 53.9 (2.34) | 530 | 55.50.2 (2.41) | 515 | 1.08 | 1.6 (0.07) | −15 |
| AARh | 56.3 (2.44) | 508 | 59.00.8 (2.56) | 485 | 0.79 | 2.7 (0.12) | −23 |
| BCone | 66.5 (2.88) | 430 | 67.80.1 (2.94) | 430 | 0.63 | 1.3 (0.06) | −8 |
| GCone | 53.4 (2.32) | 535 | 55.01.3 (2.39) | 519 | 0.90 | 1.6 (0.07) | −16 |
| RCone | 49.7 (2.16) | 575 | 58.61.6 (2.54) | 486 | 0.78 | 8.8 (0.38) | −87 |
| mMeOp | 61.2 (2.65) | 467 | 62.50.2 (2.71) | 457 | 0.76 | 1.3 (0.06) | −10 |
| PoXeRAT | 50.3 (2.18) | 568 | 50.50.5 (2.19) | 566 | 1.48 | 0.2 (0.01) | −2 |
| PoXeR13C | 52.1 (2.26) | 549 | 54.40.2 (2.36) | 525 | 1.07 | 2.2 (0.10) | −24 |
| Rh Mutants Set | |||||||
| Rh-A292S | 58.2 (2.54) | 491 | 58.70.3 (2.54) | 487 | 0.86 | 0.5 (0.01) | −1 |
| Rh-A269T | 55.6 (2.41) | 514 | 56.10.6 (2.44) | 510 | 0.91 | 0.5 (0.02) | −4 |
| Rh-E113D | 56.1 (2.43) | 510 | 55.40.4 (2.40) | 516 | 0.91 | −0.7 (−0.03) | +6 |
| Rh-E122Q | 59.6 (2.58) | 480 | 60.00.5 (2.60) | 477 | 0.81 | 0.4 (0.02) | −3 |
| Rh-F261Y | 56.1 (2.43) | 510 | 56.20.8 (2.44) | 509 | 0.86 | 0.1 (0.01) | −1 |
| Rh-G90S | 58.4 (2.53) | 489 | 56.80.7 (2.46) | 503 | 0.90 | −1.6 (−0.07) | +14 |
| Rh-T94S | 57.9 (2.51) | 494 | 58.00.7 (2.51) | 493 | 0.86 | 0.1 (0.00) | −1 |
| Rh-T118A | 59.1 (2.56) | 484 | 59.70.4 (2.59) | 479 | 0.86 | 0.6 (0.03) | −5 |
| Rh-W265Y | 59.0 (2.57) | 485 | 58.80.6 (2.55) | 486 | 0.88 | −0.2 (−0.02) | +1 |
| Rh-W265F | 59.6 (2.58) | 480 | 60.00.4 (2.60) | 476 | 0.83 | 0.5 (0.02) | −4 |
| Rh-D83N-E122Q | 60.2 (2.61) | 475 | 61.10.6 (2.65) | 468 | 0.76 | 0.9 (0.04) | −7 |
| Rh-A292S-A295S-A299C | 59.1 (2.56) | 484 | 58.60.7 (2.54) | 488 | 0.86 | −0.5 (−0.02) | +4 |
| ADmax | 20.7 (0.89) | ||||||
| MAE ± MAD of | 3.0 ± 3.4 (0.13 ± 0.15) | ||||||
| MAE ± MAD of ||Trend Dev.|| | 2.5 ± 1.2 (0.11 ± 0.05) | ||||||
| a-ARMcustomized | |||||||
| KR2-2(c) | 54.5 (2.36) | 525 | 55.90.4 (2.43) | 511 | 0.91 | 1.5 (0.06) | −14 |
| BPR(c) | 58.3 (2.53) | 490 | 57.20.3 (2.48) | 500 | 0.75 | −1.1 (−0.05) | 10 |
| RCone(c) | 49.7 (2.16) | 575 | 49.91.3 (2.16) | 572 | 1.12 | 0.2 (0.01) | −3 |
| 50.3 (2.18) | 568 | 50.70.3 (2.20) | 564 | 1.43 | 0.3 (0.02) | −4 | |
| 50.3 (2.18) | 568 | 50.40.5 (2.18) | 567 | 1.37 | 0.1 (0.00) | −1 | |
| 62.4 (2.71) | 458 | 63.80.8 (2.76) | 449 | 0.88 | 1.3 (0.06) | −9 | |
| ChR2(c) | 60.8 (2.64) | 470 | 63.31.2 (2.70) | 459 | 0.77 | 1.4 (0.06) | −11 |
| ChR2-C128T(c) | 59.0 (2.56) | 485 | 59.20.3 (2.57) | 483 | 0.95 | 0.3 (0.01) | −2 |
| ADmaxc | 2.7 (0.12) | ||||||
| MAE ± MAD of c | 0.9 ± 0.7 (0.04 ± 0.03) | ||||||
| MAE ± MAD of ||Trend Dev.||c | 0.7 ± 0.5 (0.03 ± 0.02) | ||||||
Calculated using the a-ARMdefault and the a-ARMcustomized approaches. Differences between calculated and experimental data (, ) are also presented.
Average value of 10 replicas, along with the corresponding standard deviation given as subindex.
For BPR, bRAT, ChRC1C2, ChR2, ChR2-C128T, RCone, and KR2-2 a-ARMcustomized are considered.
ASRAT-2, ASR13C-1, KR2-1, and are excluded from the statistical analysis.
Before discussing the performance of the fully automated approach, it is necessary to explain why Figure 5 shows, for certain rhodopsins, results from more than one model.
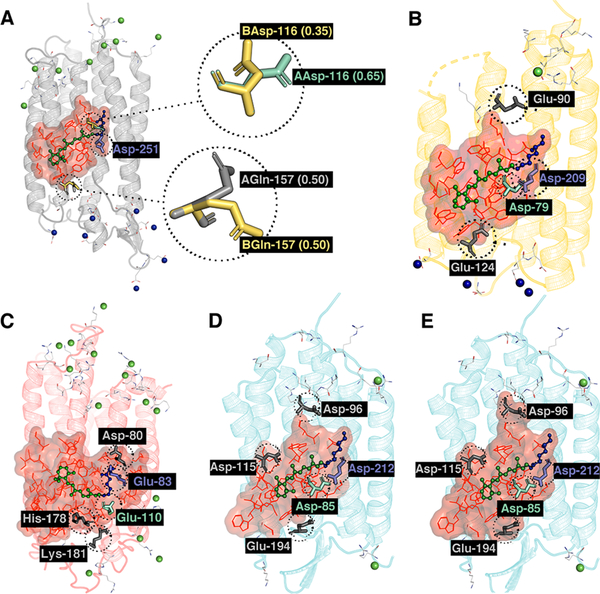
According to the a-ARMdefault approach (see section 2.3), this occurs for rhodopsins whose crystallographic data contain two alternate locations of some side chains. Multiple rotamers are found in the 1XIO,86 3X3C,102 6G7H,87 and 6EID70 crystallographic structures. In the 1XIO structure, corresponding to Anabaena sensory rhodopsin (ASR), two possible conformations were identified for both residues Lys-310 (ALys and BLys) and RET-301 (all-trans and 13-cis rPSB) that form the Lys-QM subsystem. Each rotamer in each pair exhibits 50% probability (occupancy number 0.5) to contribute to the observed structure.86,11 Therefore, the favored rotamer cannot be selected based on their occupancy, and thus, a-ARMdefault generates four models: the all-trans (ASRAT) models using ALys (ASRAT-1) and BLys (ASRAT-2) and the 13-cis (ASR13C) models with, again, ALys (ASR13C-1) and BLys (ASR13C-2), as also done manually in previous studies.52,54,55,86,115 The final models are then assigned to be those yielding a ΔES0–S1 value closest to the ones observed experimentally. More specifically, for ASRAT, we have selected model ASRAT-1 since (i) both the error and the standard deviation are lower than that observed for the second model (ASRAT-2), while (ii) the oscillator strengths are practically the same (see Table 2). A similar argument applies to the case of ASR13C where, however, the selected model is ASR13C-2.
In the case of the 3X3C structure, corresponding to Krokinobacter eikastus rhodopsin 2 (KR2), two alternate conformations (AAsp and BAsp) are present for the MC residue Asp-116 with occupancy numbers 0.65 and 0.35, respectively, and two alternate conformations (AGln and BGln) for Gln-157, both with occupancy number 0.5 (see Figure 6A). Given their occupancy numbers, a-ARMdefault uses AAsp-116 and generates two models relative to Gln-157: KR2–1, which includes AAsp-116 and AGln-157, and KR2–2, which includes AAsp-116 and BGln-157. KR2–2 is the chosen model, after comparing the computed and observed ΔES0–S1 values.
Figure 6.
a-ARM models. Conformational (the occupancy factor of the rotamers Asp-116 and Gln-157 are presented in parentheses) and ionization state variability for KR2 [PDB ID 3X3C] (A), BPR [PDB ID 4JQ6] (B), RCone (C) [PDB ID template 1U19], bRAT [PDB ID 6G7H] with standard (D) and modified cavity (E). MC and SC are presented as cyan and violet tubes, respectively.
The 6G7H structure, corresponding to Bacteriorhodopsin (bR), contains alternate locations for Asp-104, Leu-109, and Leu-15. However, the default choice leads to the generation of a single model with the rotamers AAsp-104, ALeu-109, and Aleu-15, since the occupancy numbers of these specific rotamers are 0.80, 0.54, and 0.57, respectively.
Furthermore, in the case of 6EID structure, corresponding to Channelrhodopsin-2 (ChR2), two alternate locations exist for the rPSB LYR: ALYR and BLYR, with occupancy numbers of 0.70 and 0.30, respectively. Therefore, the default model was generated using the conformation ALYR. This choice is consistent with the all-trans configuration of the rPSB presented in the resting conformation of ChR2.70
In the following sections, when discussing the results of ASRAT, ASR13C, and KR2, we will solely consider the models ASRAT-1, ASR13C-2, and KR2–2, respectively.
We now discuss the performance of the fully automated approach in predicting experimental , expressed in terms of ΔES1–S0 trends. As observed in Figure 5A, the general trend for wild type and Rh mutants models is qualitatively reproduced, mostly displaying blue-shifted absorption similar to the results of the original ARM.20,52,54,55 Actually, as can be seen in Figure 5B, 30 out of the 39 studied rhodopsins (77%) exhibit blue-shifted errors lower than 3 kcal mol–1, 6 (15%) higher than 5 kcal mol–1, and only 3 (8%) present red-shifted values of just few (0.5–1.6) kcal mol–1. More specifically, among the m-set, BPR and ChRC1C2 shows deviation of 5.4 kcal mol–1 and 14.5 kcal mol–1, respectively, which are larger than the more acceptable 3–4 kcal mol–1 difference. Among the a-set, ChR2, ChR2-C128T, KR2, and RCone are off the observed value, with deviations around 9 and 21 kcal mol–1.
The ability of a-ARMdefault models to predict rhodopsin functions can be estimated by using the data in Table 2. The analysis of these data reveals a mean absolute error (MAE) of 3.0 kcal mol–1, a mean absolute deviation (MAD) of 3.4 kcal mol–1, and a maximum absolute deviation (ADmax) of 20.7 kcal mol–1. Clearly, these large statistical values are due to the fact that models created for BPR, ChR2, ChR2-C128T, KR2, and RCone with default parameters are insufficient to provide an acceptable description. For such cases, we employ the a-ARMcustomized approach, as detailed in the next section.
3.2. a-ARMcustomized
We now employ the a-ARMcustomized approach to generate improved models for the KR2, BPR, ChRC1C2, ChR2, ChR2-C128T, and RCone outliers identified in the previous section. Indeed, we show that it is possible to construct a-ARM models (sections 3.2.1–3.2.5) yielding ΔES1–S0 values in good agreement with the observed quantities in all cases (see the orange squares and bars in Figure 5A,B, respectively). Moreover, in section 3.2.6 we deepen the study of bRAT, given its intrinsic importance and the debate surrounding the protonation state of Asp-85 and Asp-212, linked to which of the two residues constitutes the actual MC.87,116,117
3.2.1. KR2
Since the KR2 models generated using a-ARMdefault (KR2–1 and KR2–2) are unable to reproduce the experimental ΔES1–S0, we explored other possible protonation states, although without changing the other default choices (e.g., the default rotamer choices AAsp-116 and BGln-157), as shown in Figure 6A. The hypothesis we followed is that, in certain cases, a-ARMdefault does not correctly assign the residue charge (i.e., through Steps 3 and 4). According to the default model, the charge of the rPSB is stabilized by a counterion complex comprising two aspartic acid residues, Asp-116 and Asp-251. Based on distance analysis (see section 2.2.1) and the experimental evidence,102,118 Asp-116 and Asp-251 are identified as the MC and SC residues, respectively. The a-ARMdefault approach suggests that, at the crystallographic pH 8.0, both residues are deprotonated and therefore negatively charged. However, this seems questionable as two negative charges would outbalance the rPSB chromophore single positive charge (see Figure 6A). We propose that Asp-251 could be, instead, protonated (i.e., neutral). Accordingly, we generated a new model (KR2–2(c)) with the same features of KR2–2 (i.e., the default selected rotamers) but with a protonated Asp-251 residue. As observed in Figure 5 (orange square) and Table 2, this model successfully reproduces the observed data. Thus, KR2 indicates a possible limit of the default protocol for the assignment of protonation states residue and shows how the a-ARMcustomized approach may be used to explore different choices based on chemical reasoning and/or experimental evidence, so as to achieve a model with better agreement with experimental data.
We used KR2 also for testing the performance of the rotamer default assignment. As described in section 2.2.1, the assigned rotamer is the one with the highest occupancy number. To test this choice, we generated the models for all possible rotamers (see Figure 6A and Table 3) reported in the crystallographic data (keeping the ASH-251 customized choice). As reported in Table 3, we found that both models generated using the rotamer BAsp-116 with occupancy number 0.35 (KR2–3(c) for AGln-157 and KR2–4(c) for BGln-157) produce a ΔΔES1–S0 of ~15 kcal mol–1, whereas those with AAsp-116 with occupancy factor of 0.65 (KR2–1(c) for AGln-157 and KR2–2(c) for BGln-157) produce ΔΔES1–S0 of ~2 kcal mol–1. As discussed above, the choice of the Gln-157 rotamer, being relatively far from the Schiff base, does not have a significant effect on ΔES1–S0, but the conformer BGln (corresponding to the KR2–2(c) model discussed above) has a slightly smaller value and may be selected as the KR2 representative rotamer.
Table 3.
a-ARMcustomized Models for KR2 [PDB ID 3X3C] Testing All the Possible Combinations of Rotamers for Both Residues Asp-116 and Gln-157a
| rotamers |
||||||
|---|---|---|---|---|---|---|
| model | Asp-116 | Gln-157 | ΔES1–S0 | () | () | |
| KR2-1(c) | A (0.65) | A (0.50) | 56.9 | (503) | 2.4 | (−22) |
| KR2-2(c)b | A (0.65) | B (0.50) | 55.9 | (511) | 1.5 | (−14) |
| KR2-3(c) | B (0.35) | A (0.50) | 70.0 | (408) | 15.6 | (−117) |
| KR2-4(c) | B (0.35) | B (0.50) | 69.2 | (413) | 14.8 | (−112) |
Occupancy factor in parentheses. ΔES1–S0 in kcal mol−1 and in nm.
Best model, presented in Figure 5 as orange square.
3.2.2. BPR
Blue Proteorhodopsin has a structure (and a function) close to that of bR.119 Whereas the a-ARMdefault approach suggests to protonate both residues Glu-90 and Glu-124, within the a-ARMcustomized approach (see Figure 6B), we propose to keep the residue Glu-124 deprotonated and to protonate only the residue Glu-90. This choice was based on the protonation states found when imposing a pH of 7.4, as later shown in section 3.4. As observed in Figure 5 (orange square) and Table 2, such a choice has a favorable effect reducing the ΔΔES1–S0 from 5.4 kcal mol–1 to –1.1 kcal mol–1.
3.2.3. ChRC1C2
Similar to the case of KR2 explained above, the a-ARMdefault model for the Chimaera channelrhodopsin ChRC1C2 suggests that at the crystallographic pH 6.0, both MC and SC residues (Asp-292 and Glu-162) are deprotonated and therefore negatively charged. At a first glance, this seems to be the cause of its largely blue-shifted (14.5 kcal mol–1) computed ΔES1–S0 value. This hypothesis is supported by the fact that the default models for other microbial rhodopsins in the benchmarking set provide accurate results when one of the counterions is protonated (see Table S2 in the Supporting Information). Since the protonation states in a-ARM are defined by the pH choice, we compared the crystallographic pH values for KR2 and ChRC1C2 (8.0 and 6.0, respectively) with those corresponding to the other microbial rhodopsins in the benchmarking set (i.e., ASR, bR, Arch1, SR-II). Remarkably, the range of crystallographic pH of such rhodopsins is 5.2–5.6, suggesting that one should calculate the charges for microbial rhodopsins using a low pH. To test this hypothesis, we generated an a-ARMcustomized model for ChRC1C2 at pH 5.2. As a result, besides the protonated residues predicted in the default model (see Table S2 in the Supporting Information), the SC Glu-162 as well as Glu-140 are protonated. This customized model provides a decrease in the ΔΔES1–S0 from 14.5 kcal mol–1 to 1.3 kcal mol–1, highlighting the importance of ensuring a proper balance to the rPSB chromophore single positive charge.
3.2.4. ChR2 and ChR2-C128T
In the X-ray structures for Channelrhodopsin-2 and its C128T mutant, there is no available information on their experimental crystallographic pH, and therefore, the default model reverts to use the physiological pH value of 7.4. In such default models, both MC and SC (Glu-123 and Asp-253) are deprotonated and therefore negatively charged. As observed in Table 2 and Figure 5, these default models present large deviations of 19.1 and 20.7 kcal mol–1, respectively, with respect to experimental data. However, these rhodopsins represent a good case for testing the above presented hypothesis concerning the generation of customized models at low pH. To this aim, we generated a-ARMcustomized models at pH 5.2 for ChR2 (6EID) and ChR2-C128T (6EIG), with a protonated SC Asp-253, obtaining ΔΔES1–S0s of 1.4 and 0.3 kcal mol–1, respectively, which are in good agreement with experimental values (see Figure 5).
3.2.5. RCone
Starting from the comparative model of the human red cone generated using as a template the crystallographic structure of Rh (PDB ID 1U19),84 we generated a default model (RCone) that displays a large deviation from the experimental data, as opposed to the related green and blue cone models. For this reason, we also built a customized model with protonation states that better reproduce the observed ΔES1–S0 values. Specifically, considering that the pair Glu-83 and Glu-110 are the two negatively charged residues closest to the rPSB chromophore single positive charge and may play a role in its stabilization (see Figure 6C), we switched their protonation states, which in the default model are predicted to be protonated and unprotonated, respectively. As documented in Figure 5 (orange square) and Table 2, the resulting customized model (RCone(c)) produces a ΔES1–S0 value in good agreement with the experimental data, decreasing the ΔΔES1–S0 from 8.8 to 0.2 kcal mol–1.
3.2.6. bRAT
The structure corresponding to bRAT, the all-trans conformation of bacteriorhodopsin, has been recently structurally elucidated at a resolution high enough to detect hydrogen atoms (PDB ID 6G7H87). We used such a structure, after removing all hydrogen atoms for consistency with the building process, for generating the a-ARM model (see Figure 6D) at pH 5.6, as listed in Table 4. As observed in Figure 5 and Table 2, the ΔΔES1–S0Exp produced by the default model (bRAT) is smaller than 3.0 kcal mol–1, which is within the expected error range. However, since the experimental evidence does not establish the role of Asp-85 and Asp-212 as MC or SC,87,116,117 we propose a customized model in which Asp-85 is assumed to be the MC residue and it is therefore deprotonated, whereas Asp-212 is protonated. Using this model (bRAT (c)) we obtained results in even better agreement with experimental data, showing a of 0.3 kcal mol–1. Furthermore, considering the compelling importance of having a high quality model for bR, we found that the default cavity does not include the Asp-96, Asp-115, and Glu-194 residues, which are crucial for the proton pump function,87 and may therefore sensibly interact with the surrounding and even the rPSB chromophore. When we include these residues in a customized cavity (see Figure 6E), we get ΔES1–S0 values in consistent agreement with experimental data, showing a ΔES1–S0 of 50.4 kcal mol–1 and a of 0.1 kcal mol−1 ( in Table 2). These results show that not only the state of the ionizable residues (possibly the most relevant) but also the definition of the chromophore cavity may affect the quality of a-ARM models.
Table 4.
Effect of the pH on the State of Ionizable Residues for the Rhodopsins of the m-Seta
| modelb | chain | pH | neutral residuesd | ΔES1–S0e | e,f |
|---|---|---|---|---|---|
| m-Set | |||||
| ASRAT-1 | A | 5.6c | D(198,217), E(36), H(8,69) | 52.3 (2.27) | 0.3 (0.01) |
| ASRAT-1(c-pH) | A | 7.4 | E(36), H(8,69) | 58.9 (2.46) | 6.9 (0.29) |
| ASR13C-2 | A | 5.6c | D(198,217), E(36), H(8,69) | 54.2 (2.26) | 1.0 (0.04) |
| ASR13C-2(c-pH) | A | 7.4 | E(36), H(8–69) | 59.2 (2.47) | 6.0 (0.25) |
| bR13C | A | 5.2c | D(85,96,115), E(194) | 53.3 (2.22) | 1.1 (0.05) |
| A | 7.4 | D(96), E(194) | 63.2 (2.64) | 11.0 (0.48) | |
| bRAT | A | 5.6c | D(85,96,115), E(194) | 53.2 (2.30) | 2.9 (0.13) |
| A | 7.4 | D(96), E(194) | 64.2 (2.78) | 13.9 (0.60) | |
| bathoRh | A | 6.0c | D(83), E(122,181,249), H(211) | 56.2 (2.44) | 2.2 (0.09) |
| bathoRh(c-pH) | A | 7.4 | D(83), E(122,181), H(211) | 57.1 (2.48) | 3.1 (0.13) |
| bathoRhc | B | 6.0c | D(83), E(122,181), H(211) | 54.0 (2.34) | 0.0 (0.00) |
| bathoRh(c-pH-2) | B | 7.4 | D(83), E(122,181), H(211) | 54.0 (2.34) | 0.0 (0.00) |
| BPR | A | 4.5c | E(90,124) | 63.7 (2.76) | 5.4 (0.23) |
| BPR(c) | A | 7.4 | E(90) | 57.2 (2.48) | −1.1 (−0.05) |
| BPR(c-2) | B | 4.5c | E(90,124) | 63.7 (2.76) | 5.4 (0.23) |
| BPR(c-pH-2) | B | 7.4 | E(90) | 57.0 (2.47) | −1.3 (0.06) |
| Rh | A | 6.0c | D(83), E(122,181), H(211) | 57.7 (2.50) | 0.3 (0.01) |
| Rh(c-pH) | A | 7.4 | D(83), E(122,181), H(211) | 57.7 (2.50) | 0.3 (0.01) |
| Rhc | B | 6.0c | D(83), E(122,181), H(211) | 55.8 (2.42) | −1.6 (−0.07) |
| Rh(c-pH-2) | B | 7.4 | D(83), E(122), H(211) | 65.0 (2.82) | 7.6 (0.33) |
| SqRh | A | 6.4c | D(80), H(319) | 60.9 (2.64) | 2.4 (0.10) |
| SqRh(c-pH) | A | 7.4 | D(80), H(319) | 60.9 (2.64) | 2.4 (0.10) |
| SqRh(c) | B | 6.4c | D(80), H(319) | 59.4 (2.57) | 0.9 (0.04) |
| SqRh(c-pH-2) | B | 7.4 | D(80), H(319) | 59.4 (2.57) | 0.9 (0.04) |
| hMeOp | A | 6.4c | D(50), H(288,279) | 61.2 (2.65) | 0.8 (0.03) |
| hMeOp(c-pH) | A | 7.4 | D(50), H(288,279) | 61.2 (2.65) | 0.8 (0.03) |
Residues with neutral charge at physiological (7.4) and experimental crystallographic pH.
Customized models at physiological pH (c-pH).
Experimental crystallographic pH.
One letter code: Aspartic acid (D), Glutamic acid (E) and Histidine (H).
Values in kcal mol−1 and in eV in parentheses.
Experimental values are reported in Table 2.
The results presented in sections 3.2.1, 3.2.3, and 3.2.4 provide a first clue to deal with the rational design of customized models for microbial rhodopsins. In summary, for the models with high crystallographic pH (≥6.0), in which bothMCand SC are deprotonated, one can try producing new customized models at lower pH (5.2). In case the SC is still deprotonated, the next step sOkhould be to protonate it, to balance the charges around the rPSB. Finally, considering that not always the MC is the one closest to the rPSB as suggested by a-ARM, one can attempt to identify the role of the MC and SC by switching the pair predicted by a-ARM, as shown for the case of bR and RCone in the benchmarking set.
3.3. Models Comparison
When we consider for KR2–2, ChRC1C2, ChR2, ChR2-C128T, BPR, RCone, and bRAT rhodopsins, the a-ARMcustomized values rather than the corresponding a-ARMdefault values, our benchmark result analysis yields a calculated MAE of 0.9 kcal mol–1, a MAD of 0.7 kcal mol–1, and an ADmax of 2.7 kcal mol–1 (see Table 2) and thus show a substantial agreement with the experimental data.
Comparing the results for a subset constituted by the m-set and Rh mutants (excluding E122Q, A269T, E113D, D83NE122Q, and A292S-A295S-A299C) with the corresponding values reported by Melaccio et al.52 using the original ARM protocol (gold circles in Figure 5), one sees an improvement in the accuracy of the predicted trend (see Figure 5). In fact, the agreement between computed and observed quantities for such a subset is improved not only in terms of trend but also in terms of individual errors. For instance, the MAE ± MAD for this subset is reduced from 2.1 ± 0.8 kcal mol–1 (see values in Tables 1 and 2 of ref 52) to 0.9 ± 0.6 kcal mol–1 (see values in Table 2) when using a-ARM with respect to using the original ARM. Notice also that the X-ray structure-based and comparative model-based a-ARM models show a similar quality.
With the aim of quantifying the parallelism between the computed and experimental trends inΔES1–S0 and thus compare the performance of the a-ARMdefault and a-ARMcustomized approaches, we defined the trend deviation factor (‖Trend Dev.‖). This ‖Trend Dev.‖ describes the ability of the a-ARM models to predict the changes in ΔES1–S0 observed experimentally from one rhodopsin to another, with respect to a selected reference rhodopsin. For our benchmark set, we selected Rh as the reference. To compute ‖Trend Dev.‖, we first calculated the change in experimental ΔES1–S0 produced for each of the x = 37 rhodopsins with respect to Rh, as the absolute difference . Then, we performed a similar procedure but this time considering the calculated ΔES1–S0 of Rh as reference to be compared with the calculated value of the other x = 37 rhodopsins . Once obtained and for each rhodopsin, we computed the difference between these two quantities and, finally, the ‖Trend Dev.‖ value as the corresponding MAE and MAD. Further information on the complete data for the calculation is provided in Table S4 in the Supporting Information.
The results of ‖Trend Dev.‖ for the 37 rhodopsins in the benchmark set, expressed as MAE±MAD, are reported in Table 2. As observed, there is a significant improvement when we consider the a-ARMcustomized values for KR2–2, ChRC1C2, ChR2, ChR2-C128T, BPR, RCone, and bRAT instead of the a-ARMdefault values. More specifically, ‖Trend Dev.‖ changed from 1.3 ± 1.2 kcal mol–1 for the a-ARMdefault to 0.7 ± 0.5 kcal mol–1 for the a-ARMcustomized approach. The latest data validates the excellent agreement between our calculated and the available experimental values.
3.4. Effect of the Chain and pH on ΔES1–S0
As previously discussed by Melaccio et al.,52 and discussed above, when a different ionization state is assigned to a chromophore cavity residue, significant variations in the predictedΔES1–S0 have to be expected. The KR2, ChRC1C2, ChR2, ChR2-C128T, BPR, bR, and RCone cases indicate that the method used in a-ARMdefault for predicting the state of the ionizable residues of rhodopsins should be mainly used as a guideline. In fact, the change in protonation state of specific residues also have a direct effect on its global charge and, consequently, on the number of counterions needed to neutralize its OS and IS surfaces which, in turn, also affects the ΔES1–S0.
Another way of changing the ionization states of certain residues in an a-ARM model is through a pH change. In this last section, we document the effect of specific pH changes, namely, from crystallographic to physiologic pH, which shows that, in certain cases, the default choice of using the crystallization pH may not lead to a satisfactory result. In fact, such change may determine the change in the residues charge, as seen in eqs 3 and 4. To explore this potential issue, we look at the a-ARM model change in protonation state induced by a pH variation for the rhodopsins of the m-set. In particular, we selected two pH values: physiological (7.4) and experimental (imposed during crystallization) pH and compute the corresponding charges. Concurrently, we show that the charge variation can also be a function of the selected protein chain when the crystallographic data includes more than one chain.
Table 4 reports the list of ionizable residues which are calculated to be neutral for the m-set. Therefore, with the aim of evaluating the effect of the pH on the predicted ΔES1–S0, we generated a a-ARM model for each pH value, as specified in the last column of Table 4 and detailed in Table S2 of the Supporting Information. The table shows that the crystallization pH of animal rhodopsins fall in the 6.0–6.4 range, whereas for microbial rhodopsins fall in the 4.5–6.0 range. It can be seen that in most of the table entries the pH change has an influence on the calculated charges and therefore on the ionization state and, ultimately, ΔES1–S0.
Inside the explored pH range, SqRh residues do not change their protonation state, irrespective of the employed chain. The difference in computed ΔES1–S0 between SqRh(A) and SqRh(B) is, evidently, due to other factors. hMeOp is also insensitive to the change in pH, while bathoRh and Rh have different behaviors depending on the employed chain: bathoRh-(B) and Rh(A) residues do not change protonation state when varying pH, as opposed to bathoRh(A) and Rh(B). Conversely, for BPR there is no significant variation on ΔES1–S0 when chains A and B are considered, and the same residues protonation state change is found due to the pH.
Finally, it should be noted that for both bathoRh and SqRh we found a better agreement of ΔES1–S0 with respect to experimental data when chain B is considered. These results are consistent with previous studies in which some authors recommend the use of chain B in bathoRh120,121 and SqRh,122 because this is more compact than chain A and the retinal included in chain B takes a closer form to the 11-cis-conformation than the retinal included in chain A.
4. CONCLUSIONS
The possibility of automatically building QM/MM models of rhodopsins rather than via user manipulation opens up new perspectives in diverse fields, including the engineering of light-responsive proteins. In fact, automation is an unavoidable prerequisite for the production of sizable arrays (from hundreds to thousands) of rhodopsin models and, therefore, for the design of novel optogenetic tools through the in silico screening of mutant rhodopsins or for following evolutionary steps along the branches of a phylogenetic tree. However, to be useful, automation has to be accompanied by other properties such as speed in preparing the model building input and reproducibility of the final model when different users operate. Furthermore, the resulting models has to show a suitable accuracy in reproducing property trends as well as transferability to rhodopsins of very different sequence. In fact, one of the most appealing features of a-ARM is that the generation of the input for the QM/MM construction and ΔES0–S1 calculation is reduced from ~3 h to less than 5 min with respect to the original ARM protocol. This time reduction is a consequence of the automation of points A–D (see section 2.2), for which the user does not need to directly manipulate text files or visualize chemical structures anymore.
Above we have introduced and benchmarked a-ARM, a protocol designed to automatically build QM/MM models using a multiconfigurational QM level suitable for electronically excited state computations, including spectroscopy and photochemical reactivity. With respect to the previous semiautomatic version, a-ARM features an automated assignment of the residues defining the chromophore cavity, including the chromophore linker and counterions, of the state of ionizable residues and, finally, the unambiguous placement of cytoplasmic and extracellular counterions. These steps ensure automation, speed in input preparation for the QM/MMmodel building, and reproducibility.
While, presently, the benchmarking of a-ARM has been limited to a relatively small set of rhodopsins and to a single property (i.e., ), our study has revealed that (1) when used in a fully automated mode (a-ARMdefault) the protocol has a relatively high rate of success in predicting/simulating the trend in vertical excitation energies obtained from the corresponding values, (2) the automatically constructed models which do not follow the trend can be analyzed and improved using a semiautomatic version of the protocol (a-ARMdefault) to modify parameters such as the ionization states of specific residues, and (3) the trend seems to hold not only for homologous proteins (like mutants) but also for distant rhodopsins displaying severely different sequences and even chromophore isomers. These results indicate useful levels of accuracy and transferability.
In spite of the encouraging outcome of our studies, additional work has to be done for moving to a systematic applications of a-ARM to the production of sizable rhodopsin arrays. More specifically, since rhodopsin structural data are rarely available, it would be important to investigate the possibility of building, automatically, the corresponding comparative models. With such an additional tool one could achieve a protocol capable of producing QM/MM models starting directly from the constantly growing repositories of rhodopsin amino acid sequences. This target is currently pursued in our lab.
Finally, we have to stress that the structure of the a-ARM tool discussed in this manuscript could, in principle, be replicated for other biologically or technologically important photoresponsive proteins (e.g., the natural photoactive yellow proteins or the synthetic rhodopsin mimics). Therefore, our research effort can also be considered a first step toward a more general photobiological tool applicable outside the rhodopsin area.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Dr. Alessio Valentini and Dr. Federico Melaccio for fruitful discussions during the writing of the code. The code of a-ARM behind the workflow reported in Figure 2 is available upon request to the authors.
Funding
The authors acknowledge a MIUR (Ministero dell’Istruzione, dell’ Universitàe della Ricerca) grant “Dipartimento di Eccellenza 2018–2022”. M.O. is grateful for Grants NSF CHECLP-1710191 and NIH GM126627 01. M.O. is also grateful for a USIAS 2015 grant.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jctc.9b00061.
Details on the automation limits of ARM in terms of the ARM input preparation (section S1); general workflow of the a-ARM protocol, including the procedure for the QM/MM calculations (Figure S1); detailed workflow of each of the steps for the automatic input generation in a-ARM (Figures S2–S5); automatic format conversion from LYR to RET plus LYS residues (section S4); detailed information about the main features of the a-ARMdefault and a-ARMcustomized QM/MM models presented in Table 2 (Table S2); summary of the 10 a-ARM QM/MM calculations performed for each of the models presented in Table 2; computed total ground state (S0) and first excitation (S1) energies, transition oscillator strength (f Osc), first vertical excitation energy (ΔES1–S0), and maximum absorption wavelength ; statistical parameters such as average , difference between calculated and experimental data , and standard deviation (Table S3); trend deviation factor (‖Trend Dev.‖) for the a-ARMdefault and a-ARMcustomized approaches, expressed as mean absolute error (MAE) and mean absolute deviation (MAD) of the 38 rhodopsins of the benchmark set (Table S4); details of the employed comparative modeling protocol for AARh, PoXeR, hMeOp, mMeOp, BCone, GCone, and RCone (section S7); and further details on the assignment of ionizable residues protonation state (section S8) (PDF)
REFERENCES
- (1).Nilsson D-E Photoreceptor Evolution: Ancient Siblings Serve Different Tasks. Curr. Biol 2005, 15, R94–R96. [DOI] [PubMed] [Google Scholar]
- (2).Ernst OP; Lodowski DT; Elstner M; Hegemann P; Brown LS; Kandori H Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev 2014, 114, 126–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gushchin I; Shevchenko V; Polovinkin V; Borshchevskiy V; Buslaev P; Bamberg E; Gordeliy V Structure of the Light-Driven Sodium Pump KR2 and its Implications for Optogenetics. FEBS J. 2016, 283, 1232–1238. [DOI] [PubMed] [Google Scholar]
- (4).Pushkarev A; Inoue K; Larom S; Flores-Uribe J; Singh M; Konno M; Tomida S; Ito S; Nakamura R; Tsunoda SP; Philosof A; Sharon I; Yutin N; Koonin EV; Kandori H; Beéjaà OA Distinct Abundant Group of Microbial Rhodopsins Discovered using Functional Metagenomics. Nature 2018, 558, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Otomo A; Mizuno M; Singh M; Shihoya W; Inoue K; Nureki O; Beéjaà O; Kandori H; Mizutani Y Resonance Raman Investigation of the Chromophore Structure of Heliorhodopsins. J. Phys. Chem. Lett 2018, 9, 6431–6436. [DOI] [PubMed] [Google Scholar]
- (6).Singh M; Inoue K; Pushkarev A; Beéjaà O; Kandori H Mutation Study of Heliorhodopsin 48C12. Biochemistry 2018, 57, 5041–5049. [DOI] [PubMed] [Google Scholar]
- (7).Flores-Uribe J; Hevroni G; Ghai R; Pushkarev A; Inoue K; Kandori H; Beéjaà O Heliorhodopsins are Absent in Diderm (Gramnegative) Bacteria: Some Thoughts and Possible Implications for Activity. Environ. Microbiol. Rep 2019, DOI: 10.1111/1758-2229.12730. [DOI] [PubMed] [Google Scholar]
- (8).Kandori H; Shichida Y; Yoshizawa T Photoisomerization in Rhodopsin. Biochemistry (Moscow) 2001, 66, 1197–1209. [DOI] [PubMed] [Google Scholar]
- (9).Sinicropi A; Andruniow T; De Vico L; Ferré N; Olivucci M Toward a Computational Photobiology. Pure Appl. Chem 2005, 77, 977–993. [Google Scholar]
- (10).Gozem S; Luk HL; Schapiro I; Olivucci M Theory and Simulation of the Ultrafast Double-Bond Isomerization of Biological Chromophores. Chem. Rev 2017, 117, 13502–13565. [DOI] [PubMed] [Google Scholar]
- (11).Govorunova EG; Sineshchekov OA; Li H; Spudich JL Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem 2017, 86, 845–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Luk HL; Melaccio F; Rinaldi S; Gozem S; Olivucci M Molecular Bases for the selection of the Chromophore of Animal Rhodopsins. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 15297–15302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Herwig L; Rice AJ; Bedbrook CN; Zhang RK; Lignell A; Cahn JKB; Renata H; Dodani SC; Cho I; Cai L; Gradinaru V; Arnold FH Directed Evolution of a Bright Near-Infrared Fluorescent Rhodopsin Using a Synthetic Chromophore. Cell Chem. Biol 2017, 24, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bedbrook CN; Rice AJ; Yang KK; Ding X; Chen S; LeProust EM; Gradinaru V; Arnold FH Structure-Guided SCHEMA Recombination Generates Diverse Chimeric Channelrhodopsins. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E2624–E2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).McIsaac RS; Engqvist MKM; Wannier T; Rosenthal AZ; Herwig L; Flytzanis NC; Imasheva ES; Lanyi JK; Balashov SP; Gradinaru V; Arnold FH Directed Evolution of a Far-Red Fluorescent Rhodopsin. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bickelmann C; Morrow JM; Du J; Schott RK; van Hazel I; Lim S; Müller J; Chang BS The Molecular Origin and Evolution of Dim-Light Vision in Mammals. Evolution 2015, 69, 2995–3003. [DOI] [PubMed] [Google Scholar]
- (17).Morrow JM; Castiglione GM; Dungan SZ; Tang PL; Bhattacharyya N; Hauser FE; Chang BS An Experimental Comparison of Human and Bovine Rhodopsin Provides Insight into the Molecular Basis of Retinal Disease. FEBS Lett. 2017, 591, 1720–1731. [DOI] [PubMed] [Google Scholar]
- (18).Castiglione GM; Chang BS Functional Trade-Offs and Environmental Variation Shaped Ancient Trajectories in the Evolution of Dim-Light Vision. eLife 2018, 7, e35957–e35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chang BS; Jönsson K; Kazmi MA; Donoghue MJ; Sakmar TP Recreating a Functional Ancestral Archosaur Visual Pigment. Mol. Biol. Evol 2002, 19, 1483–1489. [DOI] [PubMed] [Google Scholar]
- (20).Marín M. d. C.; Agathangelou D; Orozco-Gonzalez Y; Valentini A; Kato Y; Abe-Yoshizumi R; Kandori H; Choi A; Jung K-H; Haacke S; Olivucci M Fluorescence Enhancement of a Microbial Rhodopsin Via Electronic Reprogramming. J. Am. Chem. Soc 2019, 141, 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Vasileiou C; Vaezeslami S; Crist RM; Rabago-Smith M; Geiger JH; Borhan B Protein Design: Reengineering Cellular Retinoic Acid Binding Protein II into a Rhodopsin Protein Mimic. J. Am. Chem. Soc 2007, 129, 6140–6148. [DOI] [PubMed] [Google Scholar]
- (22).Huntress MM; Gozem S; Malley KR; Jailaubekov AE; Vasileiou C; Vengris M; Geiger JH; Borhan B; Schapiro I; Larsen DS; Olivucci M Toward an Understanding of the Retinal Chromophore in Rhodopsin Mimics. J. Phys. Chem. B 2013, 117, 10053–10070. [DOI] [PubMed] [Google Scholar]
- (23).Ghanbarpour A; Nairat M; Nosrati M; Santos EM; Vasileiou C; Dantus M; Borhan B; Geiger JH Mimicking Microbial Rhodopsin Isomerization in a Single Crystal. J. Am. Chem. Soc 2019, 141, 1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lumento F; Zanirato V; Fusi S; Busi E; Latterini L; Elisei F; Sinicropi A; Andruniów T; Ferré N; Basosi R; Olivucci M Quantum Chemical Modeling and Preparation of a Biomimetic Photochemical Switch. Angew. Chem., Int. Ed 2007, 46, 414–420. [DOI] [PubMed] [Google Scholar]
- (25).Gueye M; Manathunga M; Agathangelou D; Orozco Y; Paolino M; Fusi S; Haacke S; Olivucci M; Léonard J Engineering the Vibrational Coherence of Vision into a Synthetic Molecular Device. Nat. Commun 2018, 9, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sinicropi A; Martin E; Ryazantsev M; Helbing J; Briand J; Sharma D; Léonard J; Haacke S; Cannizzo A; Chergui M; Zanirato V; Fusi S; Santoro F; Basosi R; Ferré N; Olivucci M An Artificial Molecular Switch that Mimics the Visual Pigment and Completes its Photocycle in Picoseconds. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 17642–17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Choi AR; Kim SY; Yoon SR; Bae K; Jung KH Substitution of Pro206 and Ser86 Residues in the Retinal Binding Pocket of Anabaena Sensory Rhodopsin is not Sufficient for Proton Pumping Function. J. Microbiol. Biotechnol 2007, 17, 138–145. [PubMed] [Google Scholar]
- (28).Kim SY; Waschuk SA; Brown LS; Jung KH Screening and Characterization of Proteorhodopsin Color-Tuning Mutations in Escherichia coli with Endogenous Retinal Synthesis. Biochim. Biophys. Acta, Bioenerg 2008, 1777, 504–513. [DOI] [PubMed] [Google Scholar]
- (29).Lee S; Geiller T; Jung A; Nakajima R; Song Y-K; Baker BJ Improving a Genetically Encoded Voltage Indicator by Modifying the Cytoplasmic Charge Composition. Sci. Rep 2017, 7, 8286–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Melaccio F; Ferré N; Olivucci M Quantum Chemical Modeling of Rhodopsin Mutants Displaying Switchable Colors. Phys. Chem. Chem. Phys 2012, 14, 12485–12495. [DOI] [PubMed] [Google Scholar]
- (31).Hunt DM; Dulai KS; Partridge JC; Cottrill P; Bowmaker JK The Molecular Basis for Spectral Tuning of Rod Visual Pigments in Deep-Sea Fish. J. Exp. Biol 2001, 204, 3333–3344. [DOI] [PubMed] [Google Scholar]
- (32).Altun A; Yokoyama S; Morokuma K Spectral Tuning in Visual Pigments: An ONIOM(QM:MM) Study on Bovine Rhodopsin and its Mutants. J. Phys. Chem. B 2008, 112, 6814–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang F; Vierock J; Yizhar O; Fenno LE; Tsunoda S; Kianianmomeni A; Prigge M; Berndt A; Cushman J; Polle J; Magnuson J; Hegemann P; Deisseroth K The Microbial Opsin Family of Optogenetic Tools. Cell 2011, 147, 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Deisseroth K Optogenetics. Nat. Methods 2011, 8, 26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Rein ML; Deussing JM The Optogenetic (R)evolution. Mol. Genet. Genomics 2012, 287, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Berndt A; Lee SY; Ramakrishnan C; Deisseroth K Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel. Science 2014, 344, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kandori H; Inoue K; Tsunoda SP Light-Driven Sodium-Pumping Rhodopsin: A New Concept of Active Transport. Chem. Rev 2018, 118, 10646–10658. [DOI] [PubMed] [Google Scholar]
- (38).Zhang F; Prigge M; Beyrière F; Tsunoda SP; Mattis J; Yizhar O; Hegemann P; Deisseroth K Red-Shifted Optogenetic Excitation: A Tool for Fast Neural control Derived from Volvox carteri. Nat. Neurosci. 2008, 11, 631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mattis J; Tye KM; Ferenczi EA; Ramakrishnan C; O’Shea DJ; Prakash R; Gunaydin LA; Hyun M; Fenno LE; Gradinaru V; Yizhar O; Deisseroth K Principles for Applying Optogenetic Tools Derived from Direct Comparative Analysis of Microbial Opsins. Nat. Methods 2012, 9, 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schneider F; Grimm C; Hegemann P Biophysics of Channelrhodopsin. Annu. Rev. Biophys 2015, 44, 167–186. [DOI] [PubMed] [Google Scholar]
- (41).Govorunova EG; Spudich EN; Lane CE; Sineshchekov OA; Spudich JL New Channelrhodopsin with a Red-Shifted Spectrum and Rapid Kinetics from Mesostigma viride. mBio 2011, 2, e00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Lin JY; Knutsen PM; Muller A; Kleinfeld D; Tsien RY ReaChR: A Red-Shifted Variant of Channelrhodopsin Enables Deep Transcranial Optogenetic Excitation. Nat. Neurosci 2013, 16, 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Chan T; Lee M; Sakmar T Introduction of Hydroxyl-Bearing Amino Acids Causes Bathochromic Spectral Shifts in Rhodopsin. Amino Acid Substitutions Responsible for Red-Green Color Pigment Spectral Tuning. J. Biol. Chem 1992, 267, 9478–9480. [PubMed] [Google Scholar]
- (44).Inagaki HK; Jung Y; Hoopfer ED; Wong AM; Mishra N; Lin JY; Tsien RY; Anderson DJ Optogenetic Control of Drosophila using a red-Shifted channelrhodopsin Reveals Experience-Dependent Influences on Courtship. Nat. Methods 2014, 11, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Klapoetke NC; Murata Y; Kim SS; Pulver SR; Birdsey-Benson A; Cho YK; Morimoto TK; Chuong AS; Carpenter EJ; Tian Z Independent Optical Excitation of Distinct Neural Populations. Nat. Methods 2014, 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).McIsaac RS; Bedbrook CN; Arnold FH Recent Advances in Engineering Microbial Rhodopsins for Optogenetics. Curr. Opin. Struct. Biol 2015, 33, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Altun A; Yokoyama S; Morokuma K Quantum Mechanical Molecular Mechanical Studies on Spectral Tuning Mechanisms of Visual Pigments and other Photoactive Proteins. Photochem. Photobiol 2008, 84, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Andruniów T; Ferré N; Olivucci M Structure, Initial Excited-State Relaxation, and Energy Storage of Rhodopsin Resolved at the Multi-Configurational Perturbation Theory Level. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 17908–17913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Demoulin B; Altavilla SF; Rivalta I; Garavelli M Fine Tuning of Retinal Photoinduced Decay in Solution. J. Phys. Chem. Lett 2017, 8, 4407–4412. [DOI] [PubMed] [Google Scholar]
- (50).Valsson O; Campomanes P; Tavernelli I; Rothlisberger U; Filippi C Rhodopsin Absorption from First Principles: Bypassing Common Pitfalls. J. Chem. Theory Comput 2013, 9, 2441–2454. [DOI] [PubMed] [Google Scholar]
- (51).Watanabe HC; Welke K; Schneider F; Tsunoda S; Zhang F; Deisseroth K; Hegemann P; Elstner M Structural Model of Channelrhodopsin. J. Biol. Chem 2012, 287, 7456–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Melaccio F; Marín M. d. C.; Valentini A; Montisci F; Rinaldi S; Cherubini M; Yang X; Kato Y; Stenrup M; Orozco-González Y; Ferre N; Luk HL; Kandori H; Olivucci M Toward Automatic Rhodopsin Modeling as a Tool for High-Throughput Computational Photobiology. J. Chem. Theory Comput 2016, 12, 6020–6034. [DOI] [PubMed] [Google Scholar]
- (53).Campomanes P; Neri M; Horta BAC; Rohrig UF; Vanni S; Tavernelli I; Rothlisberger U Origin of the Spectral Shifts among the Early Intermediates of the Rhodopsin Photocycle. J. Am. Chem. Soc 2014, 136, 3842–3851. [DOI] [PubMed] [Google Scholar]
- (54).Orozco-Gonzalez Y; Manathunga M; Marín M. d. C.; Agathangelou D; Jung K-H; Melaccio F; Ferré N; Haacke S; Coutinho K; Canuto S; Olivucci M An Average Solvent Electrostatic Configuration Protocol for QM/MM Free Energy Optimization: Implementation and Application to Rhodopsin Systems. J. Chem. Theory Comput 2017, 13, 6391–6404. [DOI] [PubMed] [Google Scholar]
- (55).Agathangelou D; Orozco-Gonzalez Y;Marín M. d. C.; Roy PP; Brazard J; Kandori H; Jung K-H; Léonard J; Buckup T; Ferré N; Olivucci M; Haacke S Effect of Point Mutations on the Ultrafast Photo-Isomerization of Anabaena Sensory Rhodopsin. Faraday Discuss. 2018, 207, 55–75. [DOI] [PubMed] [Google Scholar]
- (56).Bernstein FC; Koetzle TF; Williams GJ; Meyer EF Jr; Brice MD; Rodgers JR; Kennard O; Shimanouchi T; Tasumi M The Protein Data Bank: A Computer-Based Archival File for Macromolecular Structures. Eur. J. Biochem 1977, 80, 319–324. [DOI] [PubMed] [Google Scholar]
- (57).Berman HM; Westbrook J; Feng Z; Gilliland G; Bhat TN; Weissig H; Shindyalov IN; Bourne PE The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Karasuyama M; Inoue K; Nakamura R; Kandori H; Takeuchi I Understanding Colour Tuning Rules and Predicting Absorption Wavelengths of Microbial Rhodopsins by Data-Driven Machine-Learning Approach. Sci. Rep 2018, 8, 15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Schrödinger, L. L. C. PyMOL Molecular Graphics System, Version 1.2r1; 2010.
- (60).Krivov GG; Shapovalov MV; Dunbrack RL Improved Prediction of Protein Side-Chain Conformations with SCWRL4. Proteins: Struct., Funct., Genet 2009, 77, 778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhang L; Hermans J Hydrophilicity of Cavities in Proteins. Proteins: Struct., Funct., Genet 1996, 24, 433–438. [DOI] [PubMed] [Google Scholar]
- (62).Pronk S; Páll S; Schulz R; Larsson P; Bjelkmar P; Apostolov R; Shirts MR; Smith JC; Kasson PM; Van Der Spoel D; Hess B; Lindahl E GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Aquilante F; Autschbach J; Carlson RK; Chibotaru LF; Delcey MG; De Vico L; Fdez Galván I; Ferré N; Frutos LM; Gagliardi L; Garavelli M; Giussani A; Hoyer CE; Li Manni G; Lischka H; Ma D; Malmqvist PÅ; Müller T; Nenov A; Olivucci M; Bondo Pedersen T; Peng D; Plasser F; Pritchard B; Reiher M; Rivalta I; Schapiro I; Segarra-Martí J; Stenrup M; Truhlar DG; Ungur L; Valentini A; Vancoillie S; Veryazov V; Vysotskiy VP; Weingart O; Zapata F; Lindh R Molcas8: New capabilities for multiconfigurational quantum chemical calculations across the periodic table. J. Comput. Chem 2016, 37, 506–541. [DOI] [PubMed] [Google Scholar]
- (64).Rackers JA; Wang Z; Lu C; Laury ML; Lagardére L; Schnieders MJ; Piquemal J-P; Ren P; Ponder JW Tinker 8: Software Tools for Molecular Design. J. Chem. Theory Comput 2018, 14, 5273–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Frutos LM; Andruniów T; Santoro F; Ferré N; Olivucci M Tracking the Excited-State Time Evolution of the Visual Pigment with Multiconfigurational Quantum Chemistry. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 7764–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).El-Tahawy M; T, M.; Nenov A; Weingart O; Olivucci M; Garavelli M Relationship between Excited State Lifetime and Isomerization Quantum Yield in Animal Rhodopsins: Beyond the One-Dimensional Landau-Zener Model. J. Phys. Chem. Lett 2018, 9, 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Schnedermann C; Yang X; Liebel M; Spillane KM; Lugtenburg J; Fernandez I; Valentini A; Schapiro I; Olivucci M; Kukura P; Mathies RA Evidence for a Vibrational Phase-Dependent Isotope Effect on the Photochemistry of Vision. Nat. Chem 2018, 10, 449–455. [DOI] [PubMed] [Google Scholar]
- (68).Olsson MH; Søndergaard CR; Rostkowski M; Jensen JH PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput 2011, 7, 525–537. [DOI] [PubMed] [Google Scholar]
- (69).Tian W; Chen C; Lei X; Zhao J; Liang J CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Volkov O; Kovalev K; Polovinkin V; Borshchevskiy V; Bamann C; Astashkin R; Marin E; Popov A; Balandin T; Willbold D; Büldt, G.; Bamberg, E.; Gordeliy, V. Structural insights into ion conduction by channelrhodopsin 2. Science 2017, 358, No. eaan8862. [DOI] [PubMed] [Google Scholar]
- (71).Hosaka T; Yoshizawa S; Nakajima Y; Ohsawa N; Hato M; DeLong EF; Kogure K; Yokoyama S; Kimura-Someya T; Iwasaki W; Shirouzu M Structural Mechanism for Light-Driven Transport by a New Type of Chloride Ion Pump, Nonlabens marinus Rhodopsin-3. J. Biol. Chem 2016, 291, 17488–17495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Kim K; Kwon S-K; Jun S-H; Cha JS; Kim H; Lee W; Kim JF; Cho H-S Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an NTQ Motif. Nat. Commun 2016, 7, 12677–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Verma R; Schwaneberg U; Roccatano D Computer-Aided Protein Directed Evolution: A Review of Web Servers, Databases and Other Computational Tools for Protein Engineering. Comput. Struct. Biotechnol. J 2012, 2, No. e201209008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Le Guilloux V; Schmidtke P; Tuffery P Fpocket: An Open Source Platform for Ligand Pocket Detection. BMC Bioinf. 2009, 10, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Davies MN; Toseland CP; Moss DS; Flower DR Benchmarking pKa Prediction. BMC Biochem 2006, 7, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Warshel A Calculations of Enzymatic Reactions: Calculations of pKa, Proton Transfer Reactions, and General Acid Catalysis Reactions in Enzymes. Biochemistry 1981, 20, 3167–3177. [DOI] [PubMed] [Google Scholar]
- (77).Hediger MR; Jensen JH; De Vico L BioFET-SIM Web Interface: Implementation and Two Applications. PLoS One 2012, 7, No. e45379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Dolinsky TJ; Nielsen JE; McCammon JA; Baker NA PDB2PQR: An Automated Pipeline for the Setup of Poisson–Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Dolinsky TJ; Czodrowski P; Li H; Nielsen JE; Jensen JH; Klebe G; Baker NA PDB2PQR: Expanding and Upgrading Automated Preparation of Biomolecular Structures for Molecular Simulations. Nucleic Acids Res. 2007, 35, W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Po HN; Senozan N The Henderson-Hasselbalch Equation: its History and Limitations. J. Chem. Educ 2001, 78, 1499–1503. [Google Scholar]
- (81).Moore DS Amino Acid and Peptide Net Charges: A Simple Calculational Procedure. Biochem. Educ 1985, 13, 10–11. [Google Scholar]
- (82).Reijenga J; Van Hoof A; Van Loon A; Teunissen B Development of Methods for the Determination of pKa Values. Anal. Chem. Insights 2013, 8, 53–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (84).Okada T; Sugihara M; Bondar AN; Elstner M; Entel P; Buss V The Retinal Conformation and its Environment in Rhodopsin in Light of a New 2.2 Å crystal structure. J. Mol. Biol 2004, 342, 571–583. [DOI] [PubMed] [Google Scholar]
- (85).McLaughlin S The Electrostatic Properties of Membranes. Annu. Rev. Biophys. Biophys. Chem 1989, 18, 113–136. [DOI] [PubMed] [Google Scholar]
- (86).Vogeley L; Sineshchekov OA; Trivedi VD; Sasaki J; Spudich JL; Luecke H Anabaena Sensory Rhodopsin: A Photochromic Color Sensor at 2.0 Å. Science 2004, 306, 1390–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Nogly P; Weinert T; James D; Carbajo S; Ozerov D; Furrer A; Gashi D; Borin V; Skopintsev P; Jaeger K; Nass K; Bath P; Bosman R; Koglin J; Seaberg M; Lane T; Kekilli D; Brunle S; Tanaka T; Wu W; Milne C; White T; Barty A; Weierstall U; Panneels V; Nango E; Iwata S; Hunter M; Schapiro I; Schertler G; Neutze R; Standfuss J Retinal Isomerization in Bacteriorhodopsin Captured by a Femtosecond X-Ray Laser. Science 2018, No. eaat0094. [DOI] [PubMed] [Google Scholar]
- (88).Nishikawa T; Murakami M; Kouyama T Crystal Structure of the 13-cis Isomer of Bacteriorhodopsin in the Dark-Adapted State. J. Mol. Biol 2005, 352, 319–328. [DOI] [PubMed] [Google Scholar]
- (89).Nakamichi H; Okada T Crystallographic Analysis of Primary Visual Photochemistry. Angew. Chem 2006, 118, 4376–4379. [DOI] [PubMed] [Google Scholar]
- (90).Stuart JA; Birge RR Biomembranes: A Multi-Vol. Treatise; Elsevier, 1996; Vol. 2; pp 33–139. [Google Scholar]
- (91).Nickle B; Robinson P The Opsins of the Vertebrate Retina: Insights from Structural, Biochemical, and Evolutionary Studies. Cell. Mol. Life Sci 2007, 64, 2917–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Ran T; Ozorowski G; Gao Y; Sineshchekov OA; Wang W; Spudich JL; Luecke H Cross-Protomer Interaction with the Photoactive Site in Oligomeric Proteorhodopsin Complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr 2013, 69, 1965–1980. [DOI] [PubMed] [Google Scholar]
- (93).Béja O; Spudich EN; Spudich JL; Leclerc M; DeLong EF Proteorhodopsin Phototrophy in the ocean. Nature 2001, 411, 786–789. [DOI] [PubMed] [Google Scholar]
- (94).Kato HE; Zhang F; Yizhar O; Ramakrishnan C; Nishizawa T; Hirata K; Ito J; Aita Y; Tsukazaki T; Hayashi S; Hegemann P; Maturana AD; Ishitani R; Deisseroth K; Nureki O Crystal structure of the channelrhodopsin light-gated cation channel. Nature 2012, 482, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Murakami M; Kouyama T Crystal Structure of Squid Rhodopsin. Nature 2008, 453, 363–367. [DOI] [PubMed] [Google Scholar]
- (96).Shichida Y; Tokunaga F; Yoshizawa T Squid Hypsorhodopsin. Photochem. Photobiol 1979, 29, 343–351. [Google Scholar]
- (97).Dong B; Sánchez-Magraner L; Luecke H Structure of an Inward Proton-Transporting Anabaena Sensory Rhodopsin Mutant: Mechanistic Insights. Biophys. J 2016, 111, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Kawanabe A; Furutani Y; Jung KH; Kandori H Engineering an Inward Proton Transport from a Bacterial Sensor Rhodopsin. J. Am.Chem. Soc 2009, 131, 16439–16444. [DOI] [PubMed] [Google Scholar]
- (99).Enami N; Yoshimura K; Murakami M; Okumura H; Ihara K; Kouyama T Crystal Structures of Archaerhodopsin-1 and –2: Common Structural Motif in Archaeal Light-driven Proton Pumps. J. Mol. Biol 2006, 358, 675–685. [DOI] [PubMed] [Google Scholar]
- (100).Ming M; Lu M; Balashov SP; Ebrey TG; Li Q; Ding J pH Dependence of Light-Driven Proton Pumping by an Archaerhodopsin from Tibet: Comparison with Bacteriorhodopsin. Biophys. J 2006, 90, 3322–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Kouyama T; Fujii R; Kanada S; Nakanishi T; Chan SK; Murakami M Structure of Archaerhodopsin-2 at 1.8 Å Resolution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2014, 70, 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Kato HE; Inoue K; Abe-Yoshizumi R; Kato Y; Ono H; Konno M; Hososhima S; Ishizuka T; Hoque MR; Kunitomo H; Ito J; Yoshizawa S; Yamashita K; Takemoto M; Nishizawa T; Taniguchi R; Kogure K; Maturana AD; Iino Y; Yawo H; Ishitani R; Kandori H; Nureki O Structural Basis for Na+ Transport Mechanism by a Light-Driven Na+ Pump. Nature 2015, 521, 48–53. [DOI] [PubMed] [Google Scholar]
- (103).Luecke H; Schobert B; Lanyi JK; Spudich EN; Spudich JL Crystal Structure of Sensory Rhodopsin II at 2.4 Angstroms: Insights Into Color Tuning and Transducer Interaction. Science 2001, 293, 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Murakami M; Kouyama T Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin. J. Mol. Biol 2011, 413, 615–627. [DOI] [PubMed] [Google Scholar]
- (105).Merbs SL; Nathans J Absorption Spectra of Human Cone Pigments. Nature 1992, 356, 433–435. [DOI] [PubMed] [Google Scholar]
- (106).Matsuyama T; Yamashita T; Imamoto Y; Shichida Y Photochemical Properties of Mammalian Melanopsin. Biochemistry 2012, 51, 5454–5462. [DOI] [PubMed] [Google Scholar]
- (107).Inoue K; Ito S; Kato Y; Nomura Y; Shibata M; Uchihashi T; Tsunoda SP; Kandori H A Natural Light-Driven Inward Proton Pump. Nat. Commun 2016, 7, 13415–13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Sakmar TP; Franke RR; Khorana HG The Role of the Retinylidene Schiff Base Counterion in Rhodopsin in Determining Wavelength Absorbance and Schiff Base pKa. Proc. Natl. Acad. Sci. U. S. A 1991, 88, 3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Fahmy K; Jäger F; Beck M; Zvyaga TA; Sakmar TP; Siebert F Protonation States of Membrane-Embedded Carboxylic Acid Groups in Rhodopsin and Metarhodopsin II: A Fourier-Transform Infrared Spectroscopy Study of Site-Directed Mutants. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 10206–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Lin SW; Kochendoerfer GG; Carroll KS; Wang D; Mathies RA; Sakmar TP Mechanisms of Spectral Tuning in Blue Cone Visual Pigments Visible and Raman Spectroscopy of Blue-Shifted Rhodopsin Mutants. J. Biol. Chem 1998, 273, 24583–24591. [DOI] [PubMed] [Google Scholar]
- (111).Ramon E; del Valle LJ; Garriga P Unusual Thermal and Conformational Properties of the Rhodopsin Congenital Night Blindness Mutant Thr-94 →Ile. J. Biol. Chem 2003, 278, 6427–6432. [DOI] [PubMed] [Google Scholar]
- (112).Janz JM; Farrens DL Engineering a Functional Blue-Wavelength-Shifted Rhodopsin Mutant. Biochemistry 2001, 40, 7219–7227. [DOI] [PubMed] [Google Scholar]
- (113).Nakayama T; Khorana HG Mapping of the Amino Acids in Membrane-Embedded Helices that Interact with the Retinal Chromophore in Bovine Rhodopsin. J. Biol. Chem 1991, 266, 4269–4275. [PubMed] [Google Scholar]
- (114).Walker MT; Brown RL; Cronin TW; Robinson PR Photochemistry of Retinal Chromophore in Mouse Melanopsin. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 8861–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Strambi A; Durbeej B; Ferré N; Olivucci M Anabaena sensory Rhodopsin is a Light-Driven Unidirectional Rotor. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 21322–21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Nogly P; Panneels V; Nelson G; Gati C; Kimura T; Milne C; Milathianaki D; Kubo M; Wu W; Conrad C; Coe J; Bean R; Zhao Y; Bath P; Dods R; Harimoorthy R; Beyerlein KR; Rheinberger J; James D; DePonte D; Li C; Sala L; Williams GJ; Hunter MS; Koglin JE; Berntsen P; Nango E; Iwata S; Chapman HN; Fromme P; Frank M; Abela R; Boutet S; Barty A; White TA; Weierstall U; Spence J; Neutze R; Schertler G; Standfuss J Lipidic Cubic Phase Injector is a Viable Crystal Delivery System for Time-Resolved Serial Crystallography. Nat. Commun 2016, 7, 12314–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Nango E; Royant A; Kubo M; Nakane T; Wickstrand C; Kimura T; Tanaka T; Tono K; Song C; Tanaka R; Arima T; Yamashita A; Kobayashi J; Hosaka T; Mizohata E; Nogly P; Sugahara M; Nam D; Nomura T; Shimamura T; Im D; Fujiwara T; Yamanaka Y; Jeon B; Nishizawa T; Oda K; Fukuda M; Andersson R; BÅth P; Dods R; Davidsson J; Matsuoka S; Kawatake S; Murata M; Nureki O; Owada S; Kameshima T; Hatsui T; Joti Y; Schertler G; Yabashi M; Bondar AN; Standfuss J; Neutze R; Iwata S A Three-Dimensional Movie of Structural Changes in Bacteriorhodopsin. Science 2016, 354, 1552–1557. [DOI] [PubMed] [Google Scholar]
- (118).Tomida S; Ito S; Inoue K; Kandori H Hydrogen-bonding network at the cytoplasmic region of a light-driven sodium pump rhodopsin KR2. Biochim. Biophys. Acta, Bioenerg 2018, 1859, 684. [DOI] [PubMed] [Google Scholar]
- (119).Bamann C; Bamberg E; Wachtveitl J; Glaubitz C Proteorhodopsin. Biochim. Biophys. Acta, Bioenerg 2014, 1837, 614–625. [DOI] [PubMed] [Google Scholar]
- (120).Schreiber M; Sugihara M; Okada T; Buss V Quantum Mechanical Studies on the Crystallographic Model of Bathorhodopsin. Angew. Chem., Int. Ed 2006, 45, 4274–4277. [DOI] [PubMed] [Google Scholar]
- (121).Sandberg MN; Amora TL; Ramos LS; Chen M-H; Knox BE; Birge RR Glutamic Acid 181 is Negatively Charged in the Bathorhodopsin Photointermediate of Visual Rhodopsin. J. Am. Chem. Soc 2011, 133, 2808–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Nagai Y; Kito K; Maddess T Spacing Between Retinal and Amino Acid Residues in Squid Rhodopsin. Mem. Kokushikan U Cent. Inf. Sci 2012, 33, 1–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.