Abstract
Antibiotic resistance is on the rise, leading to an increase in morbidity and mortality due to infectious diseases. Klebsiella pneumoniae is a Gram-negative bacterium that causes bronchopneumonia, abscesses, urinary tract infection, osteomyelitis, and a wide variety of infections. The ubiquity of this microorganism confounds with the great increase in antibiotic resistance and have bred great concern worldwide. K. pneumoniae sequence type (ST) 307 is a widespread emerging clone associated with hospital-acquired infections, although sporadic community infections have also been reported. The aim of our study is to describe the first case of Klebsiella pneumoniae (ST) 307 harboring the blaOXA-48-like gene in Ecuador. We characterized a new plasmid that carry OXA-48 and could be the source of future outbreaks. The strain was recovered from a patient with cancer previously admitted in a Ukrainian hospital, suggesting that this mechanism of resistance could be imported. These findings highlight the importance of programs based on active molecular surveillance for the intercontinental spread of multidrug-resistant microorganisms with emergent carbapenemases.
Keywords: Klebsiella pneumoniae, OXA-48 carbapenemase, carbapenemase-producing Enterobacterales
1. Introduction
Antibiotic resistance is considered a XXI century pandemic [1], and several federal and governmental organisms, such as the World Health Organization (WHO), are calling for an urgent action to halt the spread the multidrug resistance microorganisms [2]. Carbapenem-resistant Enterobacterales are included in the list of urgent threats provided by the CDC (https://www.cdc.gov/drugresistance/biggest-threats.html [3]), and its dissemination throughout the world is a major public health worry. Carbapenemases are enzymes capable of hydrolyzing almost all β-lactam antibiotics and they are commonly located in plasmids, allowing to be spread throughout a great diversity of Gram-negative bacteria [4,5]. Serine carbapenemases (KPC) and metallo-β-lactamases (NDM, IMP, VIM) are the most prevalent carbapenemases reported all around the globe [6], KPC being the first to be described in 2001 in an isolate of Klebsiella pneumoniae in USA [7]. Carbapenem-resistant isolates within K. pneumoniae have been spread globally [8,9,10,11], including clone 307, which has significantly increased in the mid-1990s and is associated with several resistance genes, such as OXA-48 [12,13].
OXA-48-like carbapenemase was first described in Turkey in 2004 in a K. pneumoniae strain [8], and since then it has been reported in Europe, North Africa, and the USA [14,15,16,17]. Nowadays, several outbreaks have been registered in many South America countries, including Colombia [18], Argentina [19], and Brazil [20]. There are 11 allelic variants of OXA-48-like that have been described up to now [21]. The gene blaOXA-48 is located in 60- to 70-kb conjugative plasmids, generally associated with the transposon Tn1999 and its variants Tn1999.2 and Tn1999.3, and in the majority of the published data it has been associated with a nearby copy of IS1R [18,22,23]. Interestingly, OXA-48-like β-lactamases hydrolyzes penicillins and carbapenems but spares extended-spectrum cephalosporins. Here we report the first case of Klebsiella pneumoniae ST 307 carrying the blaOXA-48-like carbapenemase gene in Ecuador, and we use whole-genome sequencing (WGS) to characterize the carrying plasmid.
2. Materials and Methods
2.1. Patient History
In August 2016, a woman with colorectal cancer status—post two operations—in Ukraine was admitted to the Eugenio Espejo Hospital in Quito, Ecuador, for palliative treatment. Twenty days later, an ascitic fluid sample was processed at the microbiology laboratory of the hospital. An isolate of Klebsiella pneumoniae was identified using the VITEK®2 GN ID card (bioMerieux Inc., France).
2.2. Antimicrobial Susceptibility
Drug susceptibility was tested using the Vitek-2 compact system (bioMerieux Inc., France) utilizing the AST N272 card. Furthermore, the isolated sensititre (Trek Diagnostic Systems, UK) GNX2F microdilution broth plates and Modified Carbapenem Inactivation Methods (mCIM) and EDTA-modified carbapenem inactivation method (eCIM) were performed to differentiate metallo-β-lactamases from serine carbapenemases [24]. The results were interpreted according to the Clinical and Laboratory Standards Institute [24].
The isolate was sent to the National Reference Laboratory of Antimicrobial Resistance (NRLAR) “Dr. Leopoldo Izquieta Pérez”, Quito, for further investigation. At the NRLAR, we utilized molecular approaches to determine the mechanism of resistance.
2.3. DNA Extraction and Polymerase Chain Reaction
DNA was extracted following the recommendations of the manufacturer DNA Wizard (Promega). Polymerase chain reaction (PCR) was used to identify carbapenem resistance genes (blaKPC, blaIMP, blaNDM, blaVIM, and blaOXA-48-like) [25]. Multilocus sequence typing (MLST) analysis was performed using seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) [26], as recommended by the Pasteur Institute scheme (http://www.pasteur.fr).
2.4. Conjugation Assays
To demonstrate the horizontal gene transference, conjugation assays were performed using the broth-mating technique with an azide-resistant Escherichia coli J53 as a recipient [27,28,29]. The plasmid incompatibility group was determined using a PCR-based replicon typing (PBRT) kit (Diatheva, Fano, Italy) [30]. The IS1999 element was amplified and linked with the blaOXA-48-like gene using the primers reported previously [31].
2.5. Whole Genome Sequencing and Bioinformatic Analysis
Whole Genome Sequencing (WGS) was executed at the Institute Malbran using Illumina Miseq (https://www.illumina.com) for paired-end reads; the quality of reads was evaluated using a FastQC version [32], and adapters were eliminated with PEAT [33]. The reads below a quality 25 Phred score were eliminated with Trim Galore [34]. The assembly was performed with Spades [35], and the quality of the assembly was evaluated with Quast [36]; our statistics are based on contigs of size ≥ 500 bp. The identity of the strain was verified using the standard 16s ribosomal, groEL, rpoB, and recA genes in the Genome Peek online tool [37]. The functional annotation was made with Prokka [38] and the NCBI Prokaryotic Genomes Automatic Annotation Pipeline [39] (http://www.ncbi.nlm.nih.gov/genome/annotation_prok) and eggnog mapper [40].
From the assembly, we search for the presence of plasmids using Plasmidfinder [41]. To identify the OXA-48-like plasmid sequence, we mapped our sequence against the reference sequence CP034283.1, both sequenced with PacBio RSII and Illumina NextSeq. Coverage was evaluated using BBmap [42], and the consensus sequence was extracted using UGENE [29]. The plasmid map was obtained with BLAST Genome Atlas [43].
3. Results and Discussion
3.1. OXA-48 was Identified a K. pneumoniae MLST ST307 Isolate
The Ecuadorian surveillance network aims to provide solid data regarding epidemiological observation of antibacterial resistance. A Klebsiella pneumoniae strain with a high-resistance profile to imipenem, meropenem (intermediate), ertapenem (resistance), and piperacillin/tazobactam (intermediate), and susceptible to the third generation of cephalosporins (Table 1) was sent to the reference laboratory in order to determine the molecular mechanism of resistance associated with this phenotype. When studying inhibition to determine possible mechanisms of resistance, the mCIM resulted positive and eCIM was negative [24], suggesting that a carbapenemase Class D might be responsible for the identified phenotype. To further explore this hypothesis, Polymerase Chain Reaction (PCR) of several carbapenemases encoding genes revealed the presence of the blaOXA-48-like gene (Genebank accession number: KY609322.1).
Table 1.
Minimal concentration inhibitory of the Klebsiella pneumoniae OXA-48 producer and Escherichia coli J53 transconjugant.
| MIC (μg/mL) * | K. pneumoniae OXA-48-like | E. coli J 53 Transconjugant |
|---|---|---|
| IMI | 2 (Intermediate) | 2 (Intermediate) |
| MER | 2 (Intermediate) | <1 (Susceptible) |
| ERT | 4 (Resistant) | 4 (Resistant) |
| CAZ | <1 * (Susceptible) | <1 (Susceptible) |
| CTX | <1 * (Susceptible) | <1 (Susceptible) |
| FEP | <2 * (Susceptible) | <2 (Susceptible) |
| ATM | <2 (Susceptible) | <2 (Susceptible) |
| PIP/TAZ | >64/4 (Intermediate) | >64/4 (Intermediate) |
| CIP | >2 (Resistant) | <0.25 (Susceptible) |
| COL | 0.5 (Susceptible) | 0.25 (Susceptible) |
| AK | <4 (Susceptible) | <4 (Susceptible) |
| GEN | 8 (Intermediate) | 8 (Intermediate) |
| TGC | <0.25 (Susceptible) | <0.25 (Susceptible) |
| mCIM | POS | POS |
* Susceptible to third and four generation of cephalosporins. IMI: imipenem; MER: meropenem; ERT: ertapenem; CAZ: Ceftazidime; CTX: Cefotaxime; FEP: cefepime; ATM: aztreonam; PIP/TAZ: piperacillin/tazobactam; CIP: ciprofloxacin; COL: colistin; AK: amikacin; GEN: gentamicin; TGC: Tigecyclin; mCIM: modified carbapenem inactivation method.
Next, we wanted to investigate if this strain belongs to a previously described epidemic clone. Multilocus sequence typing (MLST) analysis using the Pasteur Institute scheme (http://www.pasteur.fr) as reference, revealed that the isolate belongs to the Sequence Type (ST) 307 (gapA: 4; infB: 1; mdh: 2; pgi:52; phoE:1; tomb: 7; rpoB:1). Interestingly, this clone was not previously reported in Ecuador, suggesting that this could be the first of many isolates. Importantly, one of the main characteristics of OXA-48 is its mobility, and this particular clone has been correlated with the presence of mobile elements.
3.2. pOXA-48 is Found on the HEEJev_01 Plasmid
To investigate if the mechanism of resistance was located in the plasmid or in the chromosome, conjugation assays were performed [44], revealing that the blaOXA-48-like gene was in the transconjugant strains suggesting that a plasmid was carrying the blaOXA-48-like resistance gene. The results were further corroborated using phenotypic (antimicrobial susceptibility) and molecular (PCR) tests (Table 1).
To determine if the plasmid also contained mobile elements that would allow to more strikingly disseminate, PCR assays were performed. PCR-based replicon typing (PBRT) from the recipient E. coli J53 (12) showed the presence of IncF, IncH, and IncL plasmid groups.
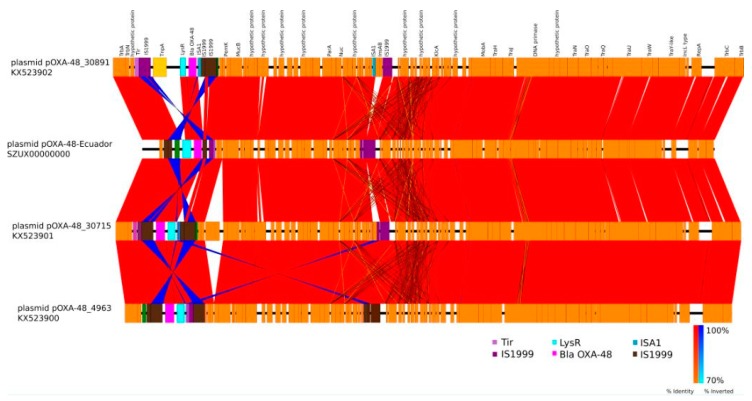
Although sequencing results were somehow limited, we were able to identify the presence of three independent plasmids, namely IncFIB (Mar), IncHI1B, and IncL/M(pOXA-48), further verifying the results obtained with PCR-based replicon typing (Table 1). Since it is the first isolate of Klebsiella pneumoniae ST 307 containing the pOXA-48 plasmid isolated in our country (Genebank accession number: SZUX00000000), we aimed to reconstruct the pOXA-48 plasmid using fragment recruitment, and the reference sequence CP034283.1 (Figure 1) plasmid that was obtained completely by sequencing in PacBio. Subsequently, our plasmid was compared with the reference plasmids KX523902, KX523901, and KX523900. We chose to compare it with these plasmids because according to a phylogenetic analysis (data not shown), the OXA-Ecuador plasmid was grouped together with these sequences. Thus, we were able to identify the sequence of the blaOXA-48-like gene and the IS transposons. Interestingly, the IS transposons were inverted when comparing the strains (Figure 2), suggesting that this plasmid is still evolving, and that the genes are rearranging to maybe increase its transmission and fitness.
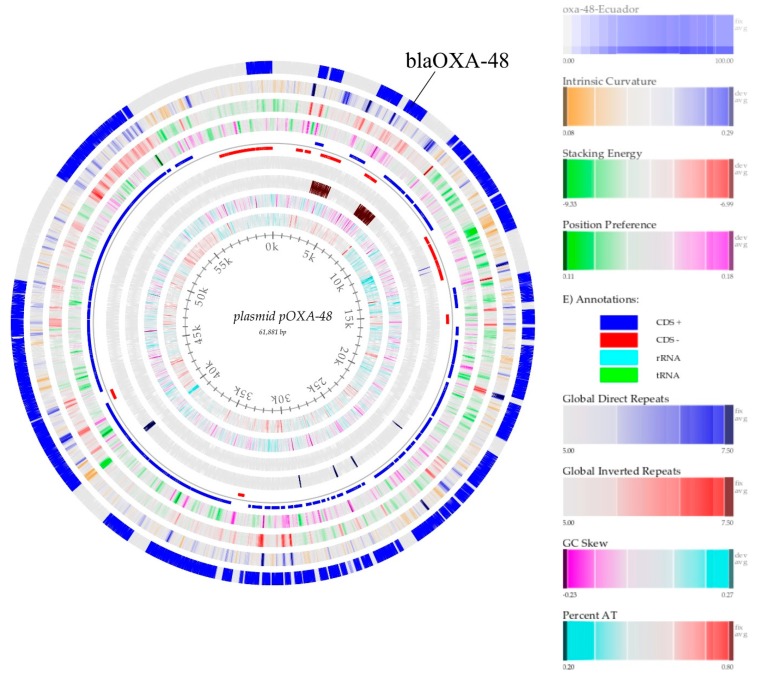
Figure 1.
From center to outside, the circular map of plasmid pOXA-48 reference CP034283.1 outside in blue plasmid, pOXA-48 from Ecuador, intrinsic curvature, stacking energy, position preference, CDS+, CDS-, global inverted repeats, GC content, GC skew, and percent AT.
Figure 2.
Plasmid OXA-48 of K. pneumoniae of Ecuador. Whole sequence shows a high degree of identity with plasmids from Czech Republic.
4. Discussion
With the rapid increase of sophisticated mechanisms of resistance, the need for developing new methods to allow their identification is imperative [45,46]. Although Enterobacterales are sensitive to third and fourth generation cephalosporins and with intermediate sensitivity to imipenem/meropenem and resistance to ertapenem are uncommon and difficult to identify [47], the mCIM and eCIM appear to be suitable methods for carbapenemase screening in healthcare centers where molecular techniques are lacking [48,49]. There is evidence that K. pneumoniae ST307 has been described as a high-risk clone [50,51,52].
The ST307 genome contains some virulence genes, suggesting that this clone might harbor the necessary tools to confer advantages in the hospital environment [50]. For example, glycogen synthesis can provide long-term survival and growth in environments outside the host as it increases biofilm formation, which has been correlated with nosocomial pathogens [13,53,54,55]. Moreover, the presence of the IncF plasmid, which is spread among the Enterobacterales, was identified, suggesting that this may be involved in the diffusion of resistance determinants and, with a combination of certain high-risk clones, contribute to the dissemination of AMR genes [5,56], which has been previously reported for K. pneumoniae [57,58]. From a search of the Institute Pasteur database (https://bigsdb.pasteur.fr/klebsiella/; 3987 STs last updated 10 April 2019), 35 human and 1 environmental isolate around the world were ST307 and 12 isolates from Europe and 1 from Asia were found in human samples with carbapenemase blaOXA-48-like [50]. We recently reported a strain of Raoutella ornithinolytica that harbored the blaOXA-48-like gene with no plasmid characterization that was recovered after this strain [59], and to the best of our knowledge, this is the first description of carrying OXA-48-like carbapenemase in an IncL plasmid isolated in Ecuador. Interestingly, our patient came from Croatia were this mechanism of resistance was reported previously [60,61,62], indicating that contagion with this strain most likely happened in Europe. The implications of globalization and free travel are now being revealed with the fast spread of the SARS-CoV-19, but in reality, this is nothing new and it has long been the case with mechanisms of resistance. Overall, these results highlight the implications of globalization, supporting previously published data from here and other countries [59,63,64,65,66]. Altogether this suggests that global guidelines for antibiotic stewardship might need to be implemented in order to better control the dissemination of the current increasing antibiotic resistance. This type of emergent resistance mechanism has an important risk as it can be easily spread between different environments, especially hospital settings. Further research will be performed in order to monitor this strain and others that can carry this and other mechanisms of resistance.
5. Conclusions
Early detection and establishment of immediate control measures are key to control dissemination. From a public health perspective, the existence of an active national surveillance system for multidrug-resistant organisms would allow for early warnings and a subsequent quick ascertaining of the magnitude of the problem at the national level. Programs based on molecular surveillance, infection control, and antibiotic stewardship are required in hospitals of this country and all other countries, to detect emerging resistance genes as well as high-risk clones, and halt them.
Acknowledgments
We thank Carolina Satán and Rafael Tamayo for their excellent technical assistance in performing replicon typing and antimicrobial testing. We thank Denise Galvan Rosales for her comments and revision of English.
Author Contributions
J.E.V., J.A.R., M.A.L., L.A.B. and A.W. designed the study; H.G.C.-S., S.D.-R. and L.M. defined the methodology; Y.T., C.N. and J.C. performed the experiments; J.E.V., J.A.R., M.G.G., H.G.C.-S. and M.C.G. wrote the first draft of the manuscript and developed the soty; J.E.V., H.G.C.-S. and M.C.G. worked and edited the revised version; all the authors reviewed and edited the manuscript and approved the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Instituto Nacional de Investigación en Salud Pública “Dr. Leopoldo Izquieta Pérez”, Quito, Ecuador. WGS were supported by Instituto Carlos G. Malbrán Buenos Aires, Argentina.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance 2014. The Organization; Geneva, Switzerland: 2014. [(accessed on 5 April 2019)]. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en. [Google Scholar]
- 2.World Health Organization . Global Action Plan on Antimicrobial Resistance 2015. The Organization; Geneva, Switzerland: 2014. [(accessed on 5 April 2019)]. Available online: https://www.who.int/ [Google Scholar]
- 3.CDC . Antibiotic Resistance Threats in the United States, 2019. U.S. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2019. [Google Scholar]
- 4.Bonomo R.A. β-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2016;7:a025239. doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathers A.J., Peirano G., Pitout J. The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tangden T., Giske C.G. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: Clinical perspectives on detection, treatment and infection control. J. Intern. Med. 2015;277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 7.Yigit H., Queenan A.M., Anderson G.J., Doménech-Sánchez A., Biddle J.W., Steward C.D., Alberti S., Bush K., Tenover F.C. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008;52:809. doi: 10.1128/AAC.01445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers J.R., Kitchel B., Driebe E.M., MacCannell D.R., Roe C., Lemmer D., De Man T., Rasheed J.K., Engelthaler D.M., Keim P., et al. Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic. PLoS ONE. 2015;10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escandón-Vargas K., Reyes S., Gutiérrez S., Villegas M.V. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infective Ther. 2016;15:277–297. doi: 10.1080/14787210.2017.1268918. [DOI] [PubMed] [Google Scholar]
- 10.Nordmann P., Naas T., Poirel L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canton R., Akova M., Carmeli Y., Giske C., Glupczynski Y., Gniadkowski M., Livermore D., Miriagou V., Naas T., Rossolini G.M., et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012;18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowe M., Kock M.M., Coetzee J., Hoosien E., Peirano G., Strydom K.-A., Ehlers M.M., Mbelle N.M., Shashkina E., Haslam D.B., et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg. Infect. Dis. 2019;25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyres K.L., Hawkey J., Hetland M.A.K., Fostervold A., Wick R.R., Kupfer S.S., Hamidian M., Howden B.P., Löhr I.H., Holt K.E. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J. Antimicrob. Chemother. 2019;74:577–581. doi: 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djahmi N., Dunyach-Rémy C., Pantel A., Dekhil M., Sotto A., Lavigne J.-P. Epidemiology of Carbapenemase-Producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean Countries. BioMed Res. Int. 2014;2014:305784. doi: 10.1155/2014/305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L., Potron A., Nordmann P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 16.Skálová A., Chudějová K., Rotová V., Medvecky M., Študentová V., Chudáčková E., Lavicka P., Bergerova T., Jakubu V., Zemlickova H., et al. Molecular Characterization of OXA-48-Like-Producing Enterobacteriaceae in the Czech Republic and Evidence for Horizontal Transfer of pOXA-48-Like Plasmids. Antimicrob. Agents Chemother. 2017;61:e01889-16. doi: 10.1128/AAC.01889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mairi A., Pantel A., Sotto A., Lavigne J.-P., Touati A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 2017;37:587–604. doi: 10.1007/s10096-017-3112-7. [DOI] [PubMed] [Google Scholar]
- 18.Vanegas J.M., Ospina W.P., Higuita-Gutiérrez L.F., Jiménez-Quiceno J.N., Higuita L.F. First reported case of an OXA-48-producing isolate from a Colombian patient. J. Glob. Antimicrob. Resist. 2016;6:67–68. doi: 10.1016/j.jgar.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Lascols C., Peirano G., Hackel M., Laupland K.B., Pitout J. Surveillance and Molecular Epidemiology of Klebsiella pneumoniae Isolates That Produce Carbapenemases: First Report of OXA-48-Like Enzymes in North America. Antimicrob. Agents Chemother. 2012;57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampaio J.L.M., Ribeiro V.B., Campos J.C., Rozales F.P., Magagnin C.M., Falci D.R., Da Silva R.C.F., Dalarosa M.G., Luz D.I., Vieira F.J., et al. Detection of OXA-370, an OXA-48-Related Class D β-Lactamase, in Enterobacter hormaechei from Brazil. Antimicrob. Agents Chemother. 2014;58:3566–3567. doi: 10.1128/AAC.02510-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitout J., Peirano G., Kock M.M., Strydom K.-A., Matsumura Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019;33:e00102-19. doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giani T., Conte V., Di Pilato V., Aschbacher R., Weber C., Larcher C., Rossolini G.M. Escherichia coli from Italy Producing OXA-48 Carbapenemase Encoded by a Novel Tn1999 Transposon Derivative. Antimicrob. Agents Chemother. 2012;56:2211–2213. doi: 10.1128/AAC.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beyrouthy R., Robin F., Delmas J., Gibold L., Dalmasso G., Dabboussi F., Hamzé M., Bonnet R. IS1R-Mediated Plasticity of IncL/M Plasmids Leads to the Insertion of blaOXA-48 into the Escherichia coli Chromosome. Antimicrob. Agents Chemother. 2014;58:3785–3790. doi: 10.1128/AAC.02669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. CLSI Supplement M100. [Google Scholar]
- 25.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Diancourt L., Passet V., Verhoef J., Grimont P.A.D., Brisse S. Multilocus Sequence Typing of Klebsiella pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber T.M., Göttig S., Mark L., Christ S., Kempf V.A.J., Wichelhaus T.A., Hamprecht A. Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J. Antimicrob. Chemother. 2014;70:1026–1030. doi: 10.1093/jac/dku482. [DOI] [PubMed] [Google Scholar]
- 28.Bou G., Cartelle M., Tomas M., Canle D., Molina F., Moure R., Eiros J.M., Guerrero A. Identification and Broad Dissemination of the CTX-M-14 β-Lactamase in Different Escherichia coli Strains in the Northwest Area of Spain. J. Clin. Microbiol. 2002;40:4030–4036. doi: 10.1128/JCM.40.11.4030-4036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández A., Gil E., Cartelle M., Pérez A., Beceiro A., Mallo S., Tomás M.M., Pérez-Llarena F.J., Villanueva R., Bou G. Interspecies spread of CTX-M-32 extended-spectrum beta-lactamase and the role of the insertion sequence IS1 in down-regulating bla CTX-M gene expression. J. Antimicrob. Chemother. 2007;59:841–847. doi: 10.1093/jac/dkm030. [DOI] [PubMed] [Google Scholar]
- 30.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Poirel L., Héritier C., Tolün V., Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 24 October 2019)];2011 Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 33.Li Y.-L., Weng J.-C., Hsiao C.-C., Chou M.-T., Tseng C.-W., Hung J.-H. PEAT: An intelligent and efficient paired-end sequencing adapter trimming algorithm. BMC Bioinform. 2015;16(Suppl. 1):S2. doi: 10.1186/1471-2105-16-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger F. Trim Galore. A Wrapper Tool around Cutadapt and fastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. [(accessed on 24 October 2019)]; Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 35.Bankevich A., Sergey N., Dmitry A., Alexey G., Mikhail D., Alexander K., Lesin V.M., Nikolenko S., Pham S., Prjibelski A.D., et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNair K., Edwards R.A. GenomePeek—An online tool for prokaryotic genome and metagenome analysis. PeerJ. 2015;3:e1025. doi: 10.7717/peerj.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., Von Mering C., Bork P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Boil. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carattoli A., Zankari E., García-Fernández A., Larsen M.V., Lund O., Villa L., Aarestrup F.M., Hasman H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushnell B. BBMap Short Read Aligner. University of California; Berkeley, CA, USA: 2016. [(accessed on 24 October 2019)]. Available online: http://sourceforgenet/projects/bbmap. [Google Scholar]
- 43.Golosova O., Henderson R., Vaskin Y., Gabrielian A., Grekhov G., Nagarajan V., Oler A.J., Quinones M., Hurt D., Fursov M., et al. Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ. 2014;2:e644. doi: 10.7717/peerj.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero-Alvarez D., Reyes J., Quezada V., Satán C., Cevallos N., Barrera S., Trueba G., Escobar L.E., Villacís J.E. First case of New Delhi metallo-β-lactamase in Klebsiella pneumoniae from Ecuador: An update for South America. Int. J. Infect. Dis. 2017;65:119–121. doi: 10.1016/j.ijid.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Boutal H., Vogel A., Bernabeu S., Devilliers K., Creton E., Cotellon G., Plaisance M., Oueslati S., Dortet L., Jousset A., et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:909–915. doi: 10.1093/jac/dkx521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jing X., Min X., Zhang X., Gong L., Wu T., Sun R., Chen L., Liu R., Zeng J. The Rapid Carbapenemase Detection Method (rCDM) for Rapid and Accurate Detection of Carbapenemase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Front. Microbiol. 2019;9:371. doi: 10.3389/fcimb.2019.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans B., Amyes S.G.B. OXA -Lactamases. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce V.M., Simner P.J., Lonsway D.R., Roe-Carpenter D.E., Johnson J.K., Brasso W.B., Bobenchik A.M., Lockett Z.C., Charnot-Katsikas A., Ferraro M.J., et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017;55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tijet N., Patel S.N., Melano R. Detection of carbapenemase activity in Enterobacteriaceae: Comparison of the carbapenem inactivation method versus the Carba NP test: Table 1. J. Antimicrob. Chemother. 2015;71:274–276. doi: 10.1093/jac/dkv283. [DOI] [PubMed] [Google Scholar]
- 50.Villa L., Feudi C., Fortini D., Brisse S., Passet V., Bonura C., Endimiani A., Mammina C., Ocampo A.M., Jimenez J.N., et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 2017;3:e000110. doi: 10.1099/mgen.0.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domokos J., Damjanova I., Kristof K., Ligeti B., Kocsis B., Szabo R. Multiple Benefits of Plasmid-Mediated Quinolone Resistance Determinants in Klebsiella pneumoniae ST11 High-Risk Clone and Recently Emerging ST307 Clone. Front. Microbiol. 2019;10:157. doi: 10.3389/fmicb.2019.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long S.W., Olsen R.J., Eagar T.N., Beres S.B., Zhao P., Davis J.J., Brettin T., Xia F., Musser J.M. Population Genomic Analysis of 1,777 Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Isolates, Houston, Texas: Unexpected Abundance of Clonal Group 307. mBio. 2017;8:e00489-17. doi: 10.1128/mBio.00489-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salipante S.J., Sengupta D.J., Cummings L.A., Land T.A., Hoogestraat D.R., Cookson B.T. Application of Whole-Genome Sequencing for Bacterial Strain Typing in Molecular Epidemiology. J. Clin. Microbiol. 2015;53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bocanegra-Ibarias P., Garza-González E., Padilla-Orozco M., Mendoza-Olazarán S., Perez-Alba E., Flores-Treviño S., Garza-Ramos U., Silva-Sanchez J., Camacho-Ortiz A. The successful containment of a hospital outbreak caused by NDM-1-producing Klebsiella pneumoniae ST307 using active surveillance. PLoS ONE. 2019;14:e0209609. doi: 10.1371/journal.pone.0209609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haller S., Kramer R., Becker K., Bohnert J.A., Eckmanns T., Hans J.B., Hecht J., Heidecke C.-D., Hübner N.-O., Kramer A., et al. Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, north-eastern Germany, June to October 2019. Euro Surveill. 2019;24:1900734. doi: 10.2807/1560-7917.ES.2019.24.50.1900734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahérault A.-C., Kemble H., Magnan M., Gachet B., Roche D., Le Nagard H., Tenaillon O., Denamur E., Branger C., Landraud L. Advantage of the F2:A1:B- IncF Pandemic Plasmid over IncC Plasmids in In Vitro Acquisition and Evolution of blaCTX-M Gene-Bearing Plasmids in Escherichia coli. Antimicrob. Agents Chemother. 2019;63:e01130-19. doi: 10.1128/AAC.01130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peirano G., Bradford P.A., Kazmierczak K.M., Chen L., Kreiswirth B.N., Pitout J. Importance of Clonal Complex 258 and IncFK2-like Plasmids among a Global Collection of Klebsiella pneumoniae with blaKPC. Antimicrob. Agents Chemother. 2017;61:e02610-16. doi: 10.1128/AAC.02610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villa L., García-Fernández A., Fortini D., Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 59.Reyes J., Villavicencio F., Villacís J.E., Pavón E., Campoverde N., Espinel M., Núñez B., Trueba G. First report of a clinical isolate of blaOXA-48- carbapenemase producing Raoultella ornithinolytica in South America. Revista Argentina de Microbiología. 2020;52:82–83. doi: 10.1016/j.ram.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Bedenić B., Slade M., Starcevic L.Z., Sardelic S., Vranić-Ladavac M., Benčić A., Atalić V.Z., Bogdan M., Bubonja-Sonje M., Tomić-Paradzik M., et al. Epidemic spread of OXA-48 beta-lactamase in Croatia. J. Med Microbiol. 2018;67:1031–1041. doi: 10.1099/jmm.0.000777. [DOI] [PubMed] [Google Scholar]
- 61.Khajuria A., Praharaj A.K., Kumar M., Grover N. Emergence of Escherichia coli, Co-Producing NDM-1 and OXA-48 Carbapenemases, in Urinary Isolates, at a Tertiary Care Centre at Central India. J. Clin. Diagn. Res. 2014;8:DC01–DC04. doi: 10.7860/JCDR/2014/7952.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jelić M., Škrlin J., Bejuk D., Koscak I., Butic I., Guzvinec M., Andrasević A.T. Characterization of Isolates Associated with Emergence of OXA-48-Producing Klebsiella pneumoniae in Croatia. Microb. Drug Resist. 2018;24:973–979. doi: 10.1089/mdr.2017.0168. [DOI] [PubMed] [Google Scholar]
- 63.Salloum T., Arabaghian H., Alousi S., Abboud E., Tokajian S. Genome sequencing and comparative analysis of an NDM-1-producing Klebsiella pneumoniae ST15 isolated from a refugee patient. Pathog. Glob. Heal. 2017;111:166–175. doi: 10.1080/20477724.2017.1314069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munoz-Price L.S., Poirel L., Bonomo R.A., Schwaber M.J., Daikos G.L., Cormican M., Cornaglia G., Garau J., Gniadkowski M., Hayden M.K., et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee C.-R., Lee J.H., Park K.S., Kim Y.B., Jeong B.C., Lee S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Machuca J., López-Cerero L., Fernández-Cuenca F., Mora-Navas L., Mediavilla-Gradolph C., López-Rodríguez I., Pascual Á. OXA-48-Like-Producing Klebsiella pneumoniae in Southern Spain in 2014–2015. Antimicrob. Agents Chemother. 2018;63:e01396-18. doi: 10.1128/AAC.01396-18. [DOI] [PMC free article] [PubMed] [Google Scholar]