Abstract
Hfq regulates bacterial gene expression post-transcriptionally by binding small RNAs and their target mRNAs, facilitating sRNA-mRNA annealing, typically resulting in translation inhibition and RNA turnover. Hfq is also found in the nucleoid and binds double-stranded (ds) DNA with a slight preference for A-tracts. Here, we present the crystal structure of the Escherichia coli Hfq Core bound to a 30 bp DNA, containing three 6 bp A-tracts. Although previously postulated to bind to the ‘distal’ face, three statistically disordered double stranded DNA molecules bind across the proximal face of the Hfq hexamer as parallel, straight rods with B-DNA like conformational properties. One DNA duplex spans the diameter of the hexamer and passes over the uridine-binding proximal-face pore, whereas the remaining DNA duplexes interact with the rims and serve as bridges between adjacent hexamers. Binding is sequence-independent with residues N13, R16, R17 and Q41 interacting exclusively with the DNA backbone. Atomic force microscopy data support the sequence-independent nature of the Hfq-DNA interaction and a role for Hfq in DNA compaction and nucleoid architecture. Our structure and nucleic acid-binding studies also provide insight into the mechanism of sequence-independent binding of Hfq to dsRNA stems, a function that is critical for proper riboregulation.
INTRODUCTION
Hfq is a pleiotropic post-transcriptional regulator found in approximately 50% of all sequenced bacteria (1–3). Hfq acts as an RNA chaperone and regulates gene expression by binding U-tracts and A-rich regions of small noncoding RNAs (sRNAs) and target mRNAs, respectively, to effect their annealing (4–9). The resulting sRNA–mRNA complex typically inhibits translation of the target mRNA and ultimately leads to its degradation as well as that of the sRNA, although some mRNAs, including rpoS, which encodes the stress response sigma factor σS, require Hfq for efficient translation (5,10–11). Studies on Δhfq mutant strains reveal Hfq has a dramatic effect on the virulence and robustness/fitness of bacterial pathogens: Δhfq mutants in certain Gram-negative pathogens such as Vibrio cholerae and Pseudomonas aeruginosa are highly attenuated in mouse model infections (12,13). Additionally, Hfq is a virulence factor in a number of pathogens including uropathogenic Escherichia coli (Ec), Neisseria meningitidis and Salmonella typhimurium (14–16). Ec Hfq has also been implicated in antibiotic resistance via its post-transcriptional regulation of the multidrug efflux transporter AcrB (17).
Hfq is a member of the Sm/LSm superfamily of RNA-binding proteins (6,8,18–19). These proteins have a common structural core comprised of an N-terminal α-helix followed by a twisted five-stranded β sheet (20) (Supplementary Figure S1). The core contains two Sm motifs, Sm1, encompassing the first three β strands and Sm2, which encompasses the last two strands (21–23). The Sm1 motif is highly conserved among all family members, whilst the sequence of Sm2 motifs differs between bacterial, archaeal and eukaryotic proteins (20,24–26). The Sm proteins assemble into a variety of quaternary structures ranging from pentamers to octamers, although those of eubacteria are only hexamers (19,24–27).
The first structures of an Hfq protein, those of the ligand-free Staphylococcus aureus (Sa) Hfq and Sa Hfq in complex with AU5G RNA, revealed a toroidal hexameric protein with RNA bound in a circular fashion encircling the pore of the ‘proximal’ face of the protein (28) (Supplementary Table S1). By contrast, a structure of Ec Hfq bound to an A-rich (A15) RNA sequence revealed this oligoribonucleotide bound in a circular conformation on the opposite face, the so-named ‘distal’ face of the hexamer (29). These structures confirmed the presence of two non-overlapping RNA binding sites on the Hfq protein and revealed the roles of key residues in single-stranded RNA recognition. Indeed, a subsequent Ec Hfq structure showed A7 and AU6A bound simultaneously on the distal and proximal faces, respectively (30). Additional studies have identified a third lateral surface binding site, located on the rim of the proximal face and consisting of three highly conserved arginine residues (31,32). Individual studies on both Ec Hfq and the Gram-positive Listeria monocytogenes (Lm) Hfq demonstrated that the affinity of U16 RNA for Hfq is greater than that of U6 RNA, as the longer RNA is able to simultaneously bind in the proximal pore and to the lateral rim of Hfq (33,34). Mutation of these arginine residues to alanine weakens but does not abolish binding to sRNA. However, these mutants do not exhibit the chaperone activity of wild-type Hfq, suggesting they are critical for sRNA-mRNA annealing (32).
In addition to its ability to bind RNA, Hfq is also known to interact with DNA (35–40), with the lateral rim-binding site and the apparently unstructured C-terminal region (CTR) of Ec Hfq (residues 64–102) implicated in this interaction (38–40). Although most Hfq is found either in the cytoplasm and associated with ribosomes, up to 20% of Hfq is contained within the bacterial nucleoid and is the third most prevalent protein in nucleoid fractions of exponentially growing cells (36,41–43). This observation led to the discovery that Hfq can bind with physiologically relevant affinity to double-stranded (ds) DNA (35–36,38). Additional studies revealed that Hfq has a slight preference for DNA containing 6 base pair (bp) A-tract repeats, which are found at regular intervals throughout the bacterial genome (36,38). These regions have been hypothesized to be important to DNA packaging in the nucleoid (44). The available data indicate that this DNA binding activity is likely affecting either transcriptional or translational control over target genes, but may also involve RNA interactions (45).
Multiple questions remain concerning the nature of Hfq–nucleic acid interactions. For example, our understanding of how Hfq interacts with structured nucleic acids, in particular the hairpin regions of RNA that are widespread in both sRNA and mRNA, is limited. There are no crystal structures of an Hfq–mRNA complex, and the first crystal structure of Hfq bound to an sRNA (RydC from Salmonella sp.) was only recently determined (46). RydC, an sRNA responsible for the regulation of a cyclopropane fatty acid synthase, has a characteristic pseudo-knot structure and is observed to bind simultaneously in the proximal-face pore and to residues R16 and R17 of the lateral rim. The uridine-tract binding observed in the pore is consistent with that seen in the Sa Hfq–AU5G (28), Salmonella typhimurium (St) Hfq–U6 (47), Ec Hfq–U6 (48) and Lm Hfq–U6 (34) complexes. The RNA also makes crystal contacts with other Hfq hexamers, which the authors suggest represent the full range of interactions the RNA is capable of making with a single hexamer. Given the variance in structures of RNA and the possibility of their structural rearrangement upon binding to Hfq, additional studies with other RNA would help strengthen the current proposed model of the sRNA–mRNA–Hfq ternary complex. Whilst significant attention has been given to Hfq–RNA interactions, much less is known about the ability of Hfq to interact with dsDNA. An early study using analytical ultracentrifugation (AUC), electrophoretic mobility shifts, and site-directed mutagenesis studies indicated that dsDNA was interacting only with the distal side of the Hfq hexamer (38). A more recent series of investigation utilizing nanofluidics, atomic force microscopy (AFM), electrophoretic mobility shift assays, and isothermal titration calorimetry has revealed that the Hfq–CTR not only interacts with duplex DNA, but can self-assemble into amyloid-like structures leading to compaction and condensation of the DNA (37,39–40).
In order to elucidate more fully the mechanism of dsDNA binding by Hfq, we carried out the structure determination of the Ec Hfq Core (residues 2–69) bound to a dsDNA ligand containing three 6-bp A-tracts separated by CGGC tracts, which had been demonstrated previously to bind this riboregulator with high affinity (36). The structure reveals an unanticipated binding mode and surface whereby the dsDNA lays laterally across the proximal face of the protein including one duplex that occludes the RNA-binding proximal pore. Interactions between the protein and DNA are made entirely with the phosphate backbone, via hydrogen bonds and electrostatic interactions. Thus, Hfq appears to bind dsDNA in a sequence-independent manner much like some other nucleoid-associated proteins (NAP) including the ubiquitous NAPs HU and H-NS (36,49–50). Additionally, we show that Sa Hfq, which contains neither the DNA-binding residues nor the elongated CTR as found in Ec Hfq, binds dsDNA 1000-fold less tightly. We also tested the ability of this A-tract dsDNA to interact with Hfq that has a polyA RNA pre-bound to its distal face and discovered the affinity of this interaction is unaltered by the presence of this RNA, supporting the validity of the observed dsDNA, proximal-face binding mode. Structure-guided fluorescence polarization and tryptophan fluorescence quenching studies examined the ability of structured RNAs, such as the stems of hairpin RNAs, to interact with the Ec Hfq Core at this newly observed dsDNA-binding site. In sum, our structural and biochemical studies reveal not only the dsDNA binding mode of the Ec Hfq Core but also provide a plausible binding mechanism of the ds stems of the rho-dependent terminators found in at the 3′-UTR of most sRNAs. Our results also support a role for Hfq in nucleoid DNA compaction or packaging.
MATERIALS AND METHODS
Protein expression, purification and site-directed mutagenesis
Escherichia coliHfq was purified using the IMPACT-CN system as described previously (8). Briefly, E. coli Hfq (residues 2–69) was overexpressed in E. coli strain ER2566 Δhfq using the pTYB11 vector. The cells were grown in Luria Broth (LB) containing 50 μg/ml ampicillin at 37°C to an OD600 between 0.4 and 0.6. Expression was induced with 0.5 mM IPTG for 20 h at 15°C. Cells were harvested at 4°C and stored at −80°C or lysed immediately using a microfluidizer. To remove contaminating RNA and DNA, 10 μg/ml DNAse and 10 μg/ml RNAse were added to crude lysate and stirred at 4°C for 2 h before clarification by centrifugation at 17 500 rpm for 30 min at 4°C. Mutations in the WT Hfq 2–69 plasmid were generated using standard protocols. WT Hfq and mutants were buffer exchanged into 20 mM HEPES pH 8.0, 150 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA) and concentrated to ∼230 μM hexamer.
Crystallization, data collection, structure determination and refinement
Nucleic acids were purchased from Oligos etc., Inc. and Integrated DNA Technologies. Crystals of Hfq Core (residues 2–69)-DNA crystals were grown by the hanging drop vapor diffusion method from equal volumes of 230 μM Hfq and 280 μM DNA (sequence; d(CGGCA6)3:d(T6GCCG)3) in 10 mM sodium cacodylate pH 6.5 (0.5 μl each) and an equal volume (1 μl) of crystallization buffer (28–38% MPD and 0.1 M Tris pH 7.5–7.9). Data were collected at 100K at the Advanced Photon Source (Argonne National Laboratory) using the SER-CAT beamline (22-ID). The complex crystallized in the P1 space group with cell constants of a = 65.75 Å, b = 65.80 Å, c = 82.00 Å, α = 105.9°, β = 92.4°, γ = 119.9° and diffracted nominally to 2.5 Å resolution. The structure was determined to 3.0 Å resolution by molecular replacement with Phaser using E. coli Hfq from the Hfq–A15 RNA complex (PDB ID: 3GIB) as a search model (51). The Hfq–DNA complex was built iteratively in Coot and refined with Phenix and CNS, to Rwork and Rfree values of 21.0 and 26.0%, respectively (52–54). Selected data collection and refinement statistics are listed in Table 1.
Table 1.
Selected crystallographic data and refinement statistics
| Unit Cell [abc, Å; αβγ, °] | a = 65.7, b = 65.8, c = 82.0 |
| α = 105.9, β= 92.3, γ = 119.9 | |
| Space Group | P1 |
| Wavelength [Å] | 1.000 |
| X-ray Source | APS SER-CAT 22-BM |
| Resolution [Å] (Highest shell) | 50.0–3.00 (3.05–3.00) |
| No. of Reflections [unique] | 18 997 |
| Completeness [%] | 83.8 (53.1) |
| Rsyma [%] | 10.6 (35.0) |
| I/(σI) | 10.3 (2.4) |
| Redundancy | 1.8 (1.5) |
| Refinement Summary | |
| Resolution [Å] (Highest shell) | 25.69–3.00 (3.05–3.00) |
| Hfq protomers/DNA base pairs/Unit Cell | 12/40 |
| No. of non-solvent atoms/Unit Cell | 8055 |
| No. of waters/Unit Cell | 59 |
| Rworkb [%] | 21.1 (23.4) |
| Rfreec [%] | 26.0 (27.9) |
| Average B factor [Å2] | |
| Protein | 48.6 |
| DNA | 81.6 |
| RMS deviations | |
| Bonds [Å] | 0.006 |
| Angles [o] | 1.13 |
| Ramachandran analysis [%] | |
| Favored, allowed, disallowed | 94.9, 5.1, 0.0 |
| PDB ID: code | 5UK7 |
a R sym = Σ|I- 〈I〉|/ Σ|I|, where I is the observed intensity and is the average intensity of several symmetry-related observations.
b R work = Σ||Fo|-|Fc||/ Σ|Fo, where Fo and Fc are the observed and calculated structure factors, respectively.
c R free = Σ||Fo|-|Fc||/ Σ|Fo for 5% of the data not used at any stage of the structural refinement.
Fluorescence polarization
Fluorescence polarization-based Hfq–nucleic acid binding measurements were performed with a PanVera Beacon 2000 instrument (Invitrogen, Madison, WI, USA) at 295 K. Hfq was serially diluted into 100 μl of binding buffer containing 20 mM HEPES pH 8.0, 0.5 mM EDTA, 75 mM NaCl and 1 nM 5′-fluorescein-labeled RNA or DNA (IDT DNA) or 20 mM HEPES pH 8.0, 0.5 mM EDTA, 150 mM NaCl and 1 nM 5′-fluorescein-labeled RNA or DNA (IDT DNA). Samples were excited at 490 nm and emission was detected at 530 nm. Data were analyzed assuming a 1:1 binding stoichiometry between one Hfq hexamer and one molecule of RNA or DNA. The data were plotted using KaleidaGraph (Synergy Software) and the generated curves were fit using non-linear least squared analysis, assuming a bimolecular model such that the Kd values represent the protein concentration at half maximal nucleic acid binding. The binding isotherms were fit to the equation, P = {(Pbound – Pfree)[protein]/(Kd +[protein])}, with Pbound being the maximum polarization at saturation, P is the polarization at a given protein concentration, Pfree is the polarization of free fluorescein-labeled nucleic acid and Kd is the equilibrium dissociation constant (28,29). All values were independently determined in triplicate.
Tryptophan fluorescence quenching (TFQ)
Tryptophan fluorescence quenching (TFQ) measurements were performed using an RF-5301PC spectrofluorophotometer (Shimadzu, Nakagyo-ku, Kyoto, Japan) at 298 K as described previously (33). Briefly, quenching was accomplished by exciting the single Hfq Trp residue at 298 nm and scanning the emission fluorescence from 320 – 400 nm. A 1 ml sample containing 1 μM of each Hfq protein in binding buffer (20 mM HEPES pH 8.0, 200 mM NaCl, 0.5 mM EDTA) was scanned and followed by a titration with 1 or 4 μM RNA. Each titration was done at least three times. Data were analyzed using Microsoft Excel. Quenching was determined using the experimentally observed fluorescence maximal height for each Trp mutant. Quenching percentage was calculated using the following equation: (1 – ((FR-FB) ÷ (F0-FB))) x 100, where FR is the fluorescence value after addition of RNA to the Hfq solution, F0 is the initial fluorescence value of the Hfq solution without RNA and FB is the fluorescence of buffer without RNA or Hfq (33).
Atomic force microscopy (AFM)
Hfq was added to a 500 bp DNA solution of the hipBA operon resulting in two final concentrations of 158 and 19.8 μM, respectively. For sample deposition, specially modified mica surfaces (APS mica) were used. The APS mica was obtained by incubation of freshly cleaved mica in 167 nM 1-(3-aminopropyl)silatrane as previously described (55). Data collection was carried out using a sample (5–10 μl), which was deposited on the APS mica immediately after dilution 30 times with 1 × binding buffer (100 mM NaCl, 20 mM Tris pH 7.5). After a 2-min incubation on the surface, excess sample was washed off with deionized water and the resultant sample dried with argon gas. AFM images in air were acquired using a MultiMode AFM NanoScope IV system operating in tapping mode. Regular tapping mode silicon probes with a spring constant of ∼42 N/m and a resonant frequency between 300 and 320 kHz were used.
RESULTS AND DISCUSSION
Ec Hfq Core–DNA complex structure reveals a novel nucleic acid binding mode
The Ec Hfq Core (residues 2–69) was crystallized in the presence of the 30-bp DNA, d(CGGCA6)3/d(T6GCCG)3. Although it has been reported that this truncated Hfq, which is missing residues 70–102, disrupts rpoS binding (56), this form of the protein is still capable of binding DNA with high affinity (Table 2). The DNA sequence used for crystallization was chosen on the basis of previous work that suggested Ec Hfq preferentially bound DNA with interspersed 6-bp A-tracts (36). The Hfq Core–dsDNA complex crystallized in the space group P1 and diffracted anisotropically to 2.5 Å resolution with data scaled and refined to 3.0 Å resolution. The structure was determined by molecular replacement using Hfq from the Ec Hfq–A15 RNA complex (PDB ID: 3GIB) as a search model, and refined to Rwork and Rfree values of 21.1 and 26.0%, respectively (Table 1). The asymmetric unit contains two hexamers and two statistically disordered dsDNA segments of 20 bp each (Supplementary Figure S2). Additional molecules in the unit cell generate the 30-bp DNA deoxyoligonucleotide that was used in crystallization, as the DNA forms a pseudo-continuous double helix throughout the crystal lattice (Supplementary Figure S3). The protein is similar to previously determined Hfq structures whereby each subunit contains an N-terminal α-helix and a twisted 5-stranded β-sheet and together form a toroidal hexamer. The protein superimposes well on the Ec Hfq apo structure (PDB ID: 3QHS) with a root mean square deviation (RMSD) of 1.1 Å over all atoms.
Table 2.
Ec Hfq binding to select DNA sequences
| Hfq | Kd (nM) |
|---|---|
| 150 mM NaCl binding buffer | |
| Full length, WT Hfq-xtal* | 175.0 ± 4.5 |
| Full length, dA16 | >3000 |
| Hfq(2-69)-xtal | 94.0 ± 8.0 |
| Hfq(2-69)-dA16 | >3000 |
| 75 mM NaCl binding buffer | |
| Hfq(2-69)-xtal | 4.9 ± 2.4 |
| Hfq(2-69)-3base A-tract DNA | 19.4 ± 6.0 |
| Hfq(2-69)-RacA DNA | 16.2 ± 3.7 |
| Hfq(2-69:N13A)-xtal | 6.0 ± 2.3 |
| Hfq(2-69:R16A)-xtal | 34.1 ± 7.0 |
| Hfq(2-69:R16A/R17A)-xtal | 66.8 ± 10.6 |
| Hfq(2-69:Q41E)-xtal | 47.1 ± 4.7 |
| Hfq(2-69:Q41A)-xtal | 299.6 ± 33.6 |
*xtal indicates the DNA sequence used for crystallization.
Hfq Core–dsDNA binding is sequence independent
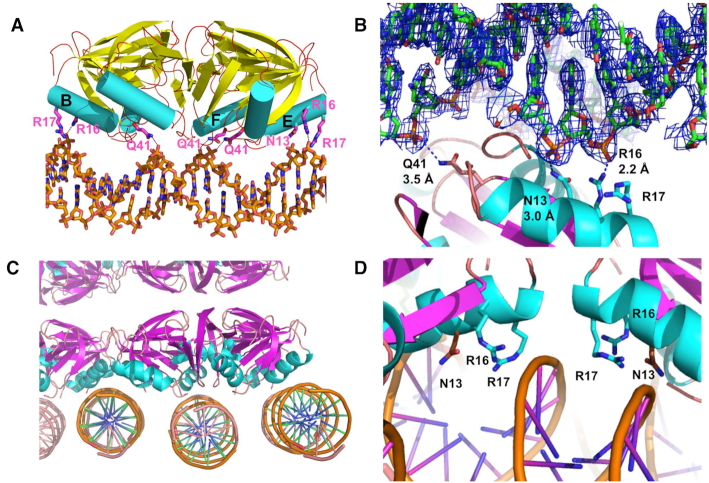
The structure reveals three dsDNA fragments bound to the proximal side or face of the Hfq hexameric ring, in sharp contrast to the previous hypothesis that suggested DNA binding takes place only on the distal face of the protein (Figure 1A and C; Supplementary Figure S3). Interestingly, the distal face of one Hfq hexamer stacks on top of the distal face of a vertically adjacent Hfq hexamer, precluding the possibility of that face binding DNA (Figure 1C and Supplementary Figure S3). Within the unit cell ∼20 bp span the width of the hexamer across the central pore whilst the other two dsDNA fragments bind to the proximal-face edges of two adjacent hexamers (Figure 1C and D). Interactions between the protein and the DNA are sequence-independent and occur only with the phosphate backbone of the DNA. Previous cloning and sequencing of 41 DNA sequences that co-purified with Hfq identified 24 different core sequences, reinforcing the sequence-independent nature of the interaction (38). Residues from multiple protomers are observed to make contacts with this DNA with residues R16 and R17 from 2-fold related subunits forming an electrostatic clamp at their respective lateral surfaces (Figure 1C and D). Interactions are also made with residues N13 and Q41, which form hydrogen bonds with the phosphate backbone (Figure 1A and B). These four residues are the only protein–DNA contacts in the unit cell, including those, which interact with the parallel-running DNA fragments that are offset from the centrally located DNA. These offset strands run along the interface between adjacent hexamers, making contact with residues N13, R16 and R17, stabilizing the lateral spread of Hfq hexamers (Figure 1C and D; Supplementary Figure S3). Intriguingly, each of these residues are also observed to be important for sRNA binding to Hfq in the structure of the St Hfq–RydC complex (Figure 2D) (46). Further, previous mutation of R16 was shown via gel shift to disrupt full length Hfq binding to DNA (38).
Figure 1.
The structure of Hfq bound to double stranded DNA. (A) The DNA that lies laterally occluding the proximal pore, shown from the ‘edge’ view of the hexamer. Hfq subunits contact the DNA exclusively through the phosphate backbone. Residues R16 and R17 make electrostatic interactions whilst Q41 and N13 form hydrogen bonds. The lone alpha helices of each subunits are shown as cylinders and colored cyan and the beta strands of all subunits are shown as arrows and colored magenta. Three alpha helices from the B, E and F subunits are labeled with residues from B and E contributing the electrostatic clamp. (B) Close up of one Hfq protomer interacting with DNA stretched across the pore. The DNA is shown within the 2FO-FC electron density map (1.0 σ). Interacting protein residues are labeled with distances measured between the nitrogen atom of each side chain and the most proximal oxygen atom of the DNA phosphate backbone. The lone alpha helices of each subunits are shown as ribbons and colored cyan and the beta strands of all subunits are shown as arrows and colored magenta. (C) Crystal packing reveals additional DNA strands bridge adjacent Hfq hexamers. (D) Hfq contacts the bridging DNA molecules utilizing the same residues that interact with the DNA that is bound across the pore. Again, the Hfq subunits contact the DNA exclusively through the phosphate backbone. Residues R16 and R17 make electrostatic interactions whilst N13 and Q41 form hydrogen bonds.
Figure 2.
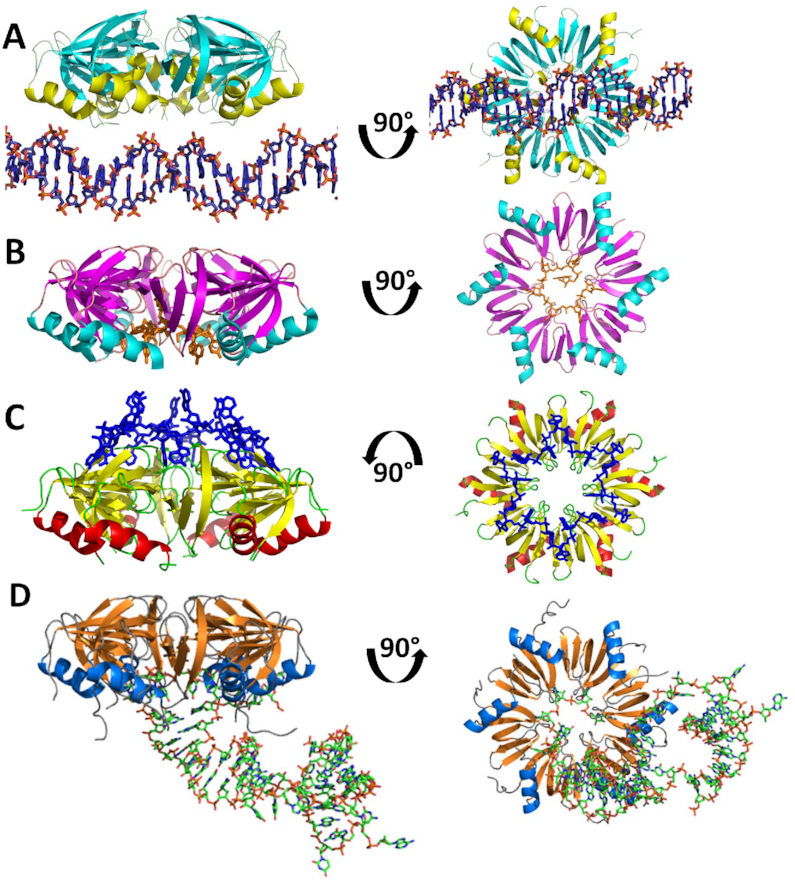
Hfq has three distinct nucleic acid binding sites. (A) Side view of the Hfq–DNA complex. Ninety-degree rotation shows view looking into the proximal face. (B) Side view of AU5G RNA (orange) shown bound in the proximal pore of Staphylococcus aureus Hfq. Ninety-degree rotation shows view into the proximal face. (C) Side view of A15 RNA (blue) shown bound to the distal face of Escherichia coli Hfq. Ninety-degree rotation shows view into the distal face. (D) Side view of RydC RNA (green) bound to the proximal face of E. coli Hfq. Ninety-degree rotation shows view into the proximal face.
The Hfq–DNA interaction observed in this crystal structure visualizes a novel double stranded nucleic acid binding site on Hfq (Figure 2A). Other structures of Hfq bound to RNA reveal two distinctly different binding surfaces: U-rich RNAs bind circularly in the pore of the proximal side of the hexamer (Figure 2B), whereas A-rich sequences bind circularly on the distal side but do not interact with the distal pore (Figure 2C) (28,29). Solution studies also indicate the importance of a lateral surface binding site for ds regions of sRNA on the proximal side of the protein (31). sRNA RybB binding to St Hfq is disrupted using lateral surface point mutants on the proximal side of the protein; two of these mutants are R16S and R17A, residues which are homologous to the E. coli R16 and R17 residues. Moreover, the recent RydC-Hfq structure, confirmed the lateral rim binding site interacts with sRNA (46) (Figure 2D) and its importance in nucleic acid binding is further underscored by this Hfq–DNA structure, in which many of the same residues that interact with RydC constitute the DNA binding site of the Hfq–DNA complex.
Compared to other NAP–DNA complexes, the Hfq–DNA interaction is unique. For example, integration host factor (IHF) interacts with DNA predominantly through the phosphate backbone; however binding to the DNA is sequence-specific, with recognition occurring indirectly through a narrow minor groove that is characteristic of A-tract DNA (57). IHF also plays an architectural role by introducing a sharp bend of greater than 160° in its nucleic acid sites, enabling higher order protein complexes to assemble on the DNA. This type of recognition mechanism is similar to the recently described DNA binding protein, BldC, a monomeric member of the MerR superfamily from the Gram-positive bacteria Streptomyces that binds hundreds of promoter regions (58). Specifically, BldC recognizes and coats a number of pseudo-degenerate sequences of key promoters in a head-to-tail manner to regulate global gene expression through DNA conformational distortion, thereby controlling the entry of Streptomyces into development. By contrast, the histone-like protein HU, which is 40% identical to IHF, shows very little sequence specificity but has a strong preference for certain DNA structures, such as supercoiled DNA, and nicks and junctions found in recombination intermediates (59). Contacts between HU and DNA consist mainly of charged interactions between basic residues and the phosphate backbone of the substrate. HU also induces a sharp bend in the DNA substrate (105°–140°). IHF, BldC and HU play key roles in either replication initiation or transcriptional regulation. We observe by sharp contrast, that Hfq binds a moderately compacted B-form DNA, with an average helical twist of 37.0° and average helical rise of 3.25 Å, resulting in 9.73 bp per turn and displaying essentially no helical curvature. When the curvature of the aforementioned 24 core sequences that co-purified with Hfq was assessed by two separate models, five sequences scored below the cutoff for curved DNA in both models whilst an additional seven scored below the cutoff in one or the other of the test models (38). This suggest that whilst there appears to be a small preference for Hfq binding to intrinsically curved DNA, the protein is certainly capable of binding straight DNA sequences in vivo as well as in vitro.
Hfq binds the ds DNA used in our crystallization studies tightly (Table 2). Indeed, fluorescence polarization-based binding experiments revealed full length Ec Hfq (residues 2–102) and the Ec Hfq Core construct (residues 2–69) used in crystallization bind in buffer containing 150 mM NaCl with a Kd of 175.0 ± 4.5 and 94.0 ± 8.0 nM, respectively (Supplementary Figure S4AB). When the experimental NaCl concentration was reduced to 75 mM, Hfq (2–69) bound the same DNA with a Kd of 4.9 ± 2.4 nM supporting the idea that ionic interactions are important, but certainly not solely responsible, for Hfq Core-DNA binding (Table 2 and Supplementary Figure S4C). A previous study had suggested that DNA binding by the Hfq–CTR binding is also dominated by electrostatic interactions again pointing toward a sequence-independent binding mechanism by Hfq (39).
In order to assess the contributions of each DNA-binding residue to the affinity of this complex, a series of site-directed mutants were tested using a fluorescence polarization-based DNA binding assay. The lower salt concentration was used in these binding experiments to allow for better comparison of the contribution of binding by individual protein residues. Interestingly, the Hfq mutant N13A has little effect on affinity, maintaining near wild type affinity (Table 2). The single R16A mutant and a double arginine mutant, R16A/R17A, however, lead to 7- and 13-fold decreases in affinity, respectively. Loss of the phosphate–backbone interaction provided by residue Q41 has the greatest single impact on binding affinity whereby a Q41E mutation reduces affinity nearly 10-fold. Surprisingly, the Q41A mutant bound DNA with a Kd of 299.6 nM, a >60-fold decrease in affinity (Table 2). Although this later result is somewhat surprising, perhaps the loss of the long side chain of glutamine might result in a local conformational change in the loop on which residue 41 resides, accounting for the larger loss of affinity. Strikingly, neither the DNA sequence nor the structure of the DNA appears to have a large effect on wild-type Hfq Core binding. The crystallization DNA contains 30 bp, with three 6-bp A-tracts spaced throughout the double helix. The junctions between A-tract DNA and other sequences can induce curvature to the helical axis; this bend has been estimated to be 10°–30° for 6 bp A-tracts (60–62). Previous studies suggest Hfq exhibits a very modest preference for A-tract-induced curved DNA, consistent with other nucleoid-associated proteins (Supplementary Figure S5A) (36). However, modification of the 6 bp A-tracts to 3 bp A-tracts in the context of a 30 bp oligodeoxynucleotide has only a small, four-fold effect on DNA binding affinity (Table 2 and Supplementary Figure S5B). Additionally, Hfq Core seems to exhibit little preference for particular dsDNA sequences or length. A GC-rich, 14 bp DNA site (RacA) with no A-tracts bound to Hfq with a Kd of 16.2 nM (Table 2 and Supplementary Figure S5C).
Interestingly, A-tracts of at least 4 bp are more prevalent in the E. coli genome than random probability would dictate. Further, they are clustered with the curvature of the A-tract DNA reducing the energetic cost of DNA looping (44). We posit that even a slight preference for these sequences should increase the local concentration of Hfq in these locations emphasizing a potential role in DNA compaction and nucleoid architecture. On the other hand, the single-stranded deoxyA-tract, (dA16), which would be predicted to bind to the distal face of Hfq, shows weak binding (Kd > 3 μM) under our assay conditions to both the full-length Hfq and the Hfq Core (Table 2). Hence, we did not test binding of dA16 to the Q41 mutant. Although our mutational analyses and binding data support proximal-face dsDNA binding, we cannot rule out alternative binding modes for A-tract containing sequences under different environmental conditions, including binding to the distal face, as has been proposed recently by Geiguenaud et al. (39,40).
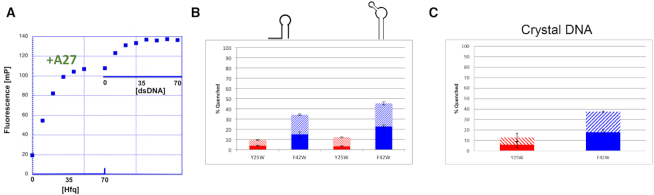
We also employed fluorescence polarization to show that dsDNA can bind Hfq after pre-binding a polyA RNA to the protein. The structure of A15 RNA in complex with E. coli Hfq shows the polyA ligand bound in a circular conformation on the distal face of the hexamer (29). We hypothesized that this binding should have little effect on protein interaction with the 30-mer dsDNA, which is bound on the opposite face of the protein. After saturating the distal binding site of Hfq with a fluoresceinated A27 RNA, addition of dsDNA produced another distinct binding isotherm that fit with a similar Kd to that of dsDNA and Hfq Core in the absence of A27 (Figure 3A). These solution studies are completely consistent with the binding modes observed in the individual crystal structures.
Figure 3.
Hfq uses the same interface to bind dsDNA and dsRNA but a different interface to bind distal-face binding polyA. (A) Fluorescence polarization demonstrates that A27 RNA and dsDNA can simultaneously bind to Hfq on distal and proximal faces, respectively. Hfq was first titrated into fluorescent A27 RNA until saturation was reached. DNA was then titrated into the same mixture and a second binding event was observed. (B) Tryptophan Fluorescence quenching reveals that hairpin RNAs (Left) site A, sequence AUUUUUUCGAAUCGAAAGGUUCA, (Right) Hairpin 2, sequence (CAUGAUUCUUAUACGUACGACGGAAGAUGAGAAUUAUGGU) preferentially bind to and quench the proximal face (F42W) over the distal face (Y25W). The solid portion of each column is quenching by the addition of 1 μM protein and the hatched portion of each column is quenching by the addition of 4 μM protein. (C) Tryptophan fluorescence quenching reveals that the dsDNA oligodeoxynucleotide, which was used in crystallization, preferentially binds to and quenches proximal face residue F42W and not distal face residue Y25W.
Hfq also binds dsRNA in a sequence-independent manner
The non-specific nature of the Hfq Core-DNA interaction led us to postulate that the DNA interaction surface should also act as the site for dsRNA binding. Previous studies have shown that Hfq can interact with a piece of hairpin RNA (site A) located in the 5′ UTR of hfq mRNA (Supplementary Figure S5D) (63). We also observe tight binding of Hfq to other RNA hairpins (Table 3 and Supplementary Figure S5E-F). To assess contributions of DNA-binding residues to hairpin RNA binding, we used the Q41E mutant; we observe that whilst binding affinity to A15 RNA (distal side) is unaffected, binding to the Hfq mRNA hairpin displays reduced affinity and is non-saturable. At last, tryptophan fluorescence quenching (33) reveals that multiple hairpin RNA substrates (site A and Hairpin 2, Supplementary Figure S5D-F) bind to the proximal face of the Hfq hexamer. The processed data for Site A were reported previously, in part, in Robinson et al. (33), but are included here to demonstrate the same proximal-face quenching phenomenon of yet a second, novel dsRNA hairpair (Hairpin 2). In both cases we observe a significant increase in fluorescence quenching of a tryptophan mutation on the proximal side of the protein (F42W) upon titration with our dsRNA hairpins (Figure 3B and Supplementary Figure S6A-B). This contrasts sharply with the tryptophan quenching results, i.e. little to no quenching, that are seen when identical titrations are carried out using the distal face-located Hfq–mutant Y25W (Figure 3B and Supplementary Figure S6C-D). Using this approach, we also observed robust tryptophan quenching of the F42W Hfq mutant upon titration with the dsDNA, which was used in our crystallization experiments, but no significant quenching of the Y25W Hfq mutant (Figure 3C and Supplementary Figure S6E-F). These results provide strong evidence of hairpin RNA binding and the binding of our dsDNA on the proximal face of Ec Hfq Core.
Table 3.
Ec Hfq binding to select RNA
| Hfq | K d (nM) |
|---|---|
| WT+ site A | 22.7 ± 1.5 |
| WT+ Hairpin 1 | 4.2 ± 1.7 |
| WT+ Hairpin 2 | 61.5 ± 18.3 |
| WT+ A15 | 3.4 ± 1.1 |
| Hfq(2-69:Q41E) + A15 | 1.3 ± 1.1 |
| Hfq(2-69:Q41E) + site A | Non-specific binding |
A model for Hfq–sRNA–mRNA interaction
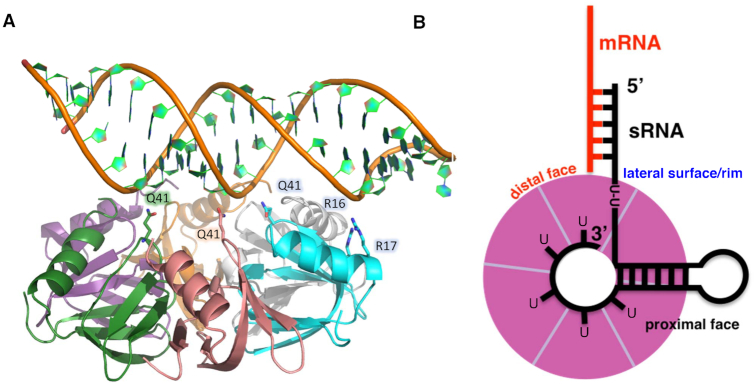
Our studies indicate the DNA-binding surface that we observe in our crystal structure is a ds nucleic acid binding site, which includes dsRNA regions such as those found in myriad sRNAs (64). Indeed, an A-form RNA hairpin can be readily docked on this surface of Hfq such that most of the residues that contact B-form DNA in our crystal structure can make similar interactions with the A-form hairpin phosphate backbone (Figure 4A). We performed this dsRNA modeling using an RNA hairpin derived from the pre-mRNA of the glutamate receptor GluR-2 (65). Its structure was determined as part of a complex with the adenosine deaminase ADAR2 (PDB ID: 2L2K) via NMR. This ds nucleic acid binding site hypothesis is corroborated by studies that reveal residue R16 contributes to the binding of both dsDNA (38) as well as the 38 nt domain II of the sRNA DsrA, which contains a 12 nt single stranded region followed by a stem loop (38,66).
Figure 4.
Hfq is uses the same residues to bind either dsDNA or dsRNA. (A) Residues that bind to a B-form like DNA can be rotated to interact with the dsRNA stem of an RNA hairpin. (B) A model of Hfq–sRNA–mRNA binding and annealing. The U-rich 3′ end of the sRNA wraps around the inside of the proximal pore whilst the dsRNA stem of the hairpin lies across the proximal face of the protein interacting with residues revealed by the Hfq–dsDNA structure. Single stranded regions of the RNA containing a U-U dinucleotide sequence may also bind to the lateral rim, whilst the remainder of the sRNA is free to anneal to mRNA bound on the distal face (colored red), forming an mRNA–sRNA duplex.
Additional studies have shown that the conserved arginines. R16 and R17, are integral to nucleic acid binding. Mutation of four St Hfq residues, including two that correspond to R16 and R17 of E. coli Hfq, abrogate RybB sRNA binding (31). This study further demonstrated that altering the structure of the hairpin that resides adjacent to the 3′ poly-U tail of RybB can negatively impact the binding of this sRNA to Hfq, suggesting the importance of this double stranded region in binding. In addition, an R16A mutation in E. coli Hfq was shown to eliminate rapid binding and release of short RNAs from the hexamer, and this residue was shown to contribute to binding of native sRNAs (32). In that study, the authors suggest that R16 is specifically interacting with the ds region of sRNAs, and that the conserved arginines may be playing a role in mRNA-sRNA annealing and hence translational regulation.
Notably, recent studies on Sa Hfq suggest that sRNA-mRNA interactions in this organism do not require Hfq as a chaperone; additional studies suggest that this Hfq does not bind to sRNA at all (67,68). These results are particularly interesting in light of our observation that Sa Hfq binds dsDNA extremely poorly (Supplementary Figure S4D). Sa Hfq lacks three of the four conserved DNA binding residues observed in our crystal structure as well as an extended CTR, and binds the 30-bp DNA ligand with ∼1000-fold lower affinity than E. coli Hfq (Kd = 6.4 μM) (Supplementary Figure S4D). It is tempting to speculate that the lack of conserved DNA binding residues is also responsible for deficiency or absence of binding to sRNA ligands.
The importance of these lateral rim residues in binding sRNA was confirmed by the crystal structure of Ec Hfq and RydC sRNA in which two separate U-U di-nucleotide stretches were observed to bind to Hfq lateral-rim residues N13, R16, R17 and F39. These U-U stretches, however, reside in single stranded regions of the RydC pseuodknot, rather than a double stranded portion (46). Given the variable structures of sRNA and the wide variety of RNA that require Hfq as a chaperone, it is likely that both double stranded and single stranded segments of a given sRNA can bind simultaneously to different lateral binding sites within a single hexamer. Indeed, studies of the sRNA SgrS led Ishikawa et al. to propose that the ‘functional Hfq binding module of bacterial sRNAs consists of either a double or single hairpin preceded by a U-rich sequence and followed by a 3′-poly(U) tail’ (64).
Taken together with recent solution studies, our structural characterization of the duplex nucleic acid binding site enables us to construct a model for Hfq–sRNA interaction, similar to the one proposed by Sauer et al. (31) and Dimastrogiovanni et al. (46) (Figure 4B). Our results confirm the presence of three independent nucleic acid binding sites, which are all likely to involve interaction with different regions of sRNA and mRNA substrates. In our model, U-rich sequences at the 3′ untranslated region of sRNA bind in the proximal pore, as observed in the Sa Hfq AU5G and St Hfq–U6 structures, whereas a hairpin region in the sRNA interacts with the conserved DNA-binding residues and lays flat against the proximal surface of the hexamer. This arrangement stabilizes the sRNA and places no conformational restriction on bases upstream of the hairpin, enabling single stranded U-U sequences to position the seed region proximal to the rim, thereby facilitating interaction with distal-face bound cognate mRNA substrates. Further, the model is structurally consistent with all known Hfq functions in translational regulation and mRNA degradation.
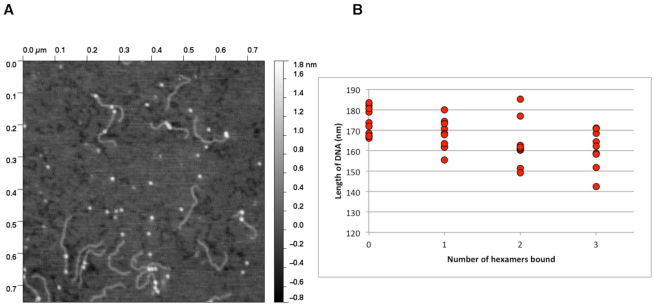
Potential DNA wrapping highlights role for Hfq in DNA compaction
AFM has been used to identify DNA wrapping around proteins (69–72). In addition to the proximal-face DNA binding observed in our crystal structure, AFM shows that longer pieces of DNA may possibly wrap around Hfq and interact simultaneously with multiple binding sites beyond the proximal face. We observe, when combined in an 8:1 ratio (Hfq:DNA) with 500 bp DNA fragments containing the E. coli hipBA promoter, up to four Hfq hexamers bound per DNA strand (Figure 5A), further indicating the sequence-nonspecific nature of the Hfq–DNA interaction. Using Gwyddion (73) and ImageJ (74) we measured the DNA length when 0, 1, 2 or 3 Hfq molecules are bound and observed shortening of the DNA as the number of Hfq hexamers increased (Figure 5B). This is consistent with our Hfq–dsDNA crystal structure complex whereby the average number of base pairs per turn is 9.7. Recently, a separate study using nanofluidics and AFM also observed compaction of DNA with increasing concentrations of full-length Ec Hfq (37). Additionally, SANS experiments suggest Hfq-mediated bridging of two DNA molecules via the Hfq–CTR (37). The same group later observed, again using nanofluidics, that increasing concentrations of Hfq (residues 1–72) resulted in shortening of the DNA, consistent with our AFM data, but could not induce condensation. However, they found that the Hfq–CTR alone (residues 64–102), whilst displaying lower binding affinity for DNA than Hfq-Core, could shorten the DNA ultimately resulting in an abrupt condensation of the DNA (39). This condensation was later attributed to the ability of Hfq to self-assemble on DNA, generating amyloid-like fibers consisting of the Hfq–CTR and DNA (40). These studies, along with our results, strongly suggest the DNA–Hfq interaction plays a role in genomic architecture and potentially in transcriptional regulation. However, our crystal structure and subsequent biochemical and biophysical studies suggest that the proximal face, not the distal face, is the initial high-affinity DNA binding site. Indeed, recent ChIP-seq data from the Dove laboratory have shown that Pseudomonas aeruginosa Hfq associated with 656 different regions of the chromosome suggesting a nascent mRNA–Hfq–DNA complex plays a role in transcription with the proximal face of Hfq binding DNA whilst the distal face binds the elongating mRNA (75). Intriguingly, to the best of our knowledge, it would appear that the archael Sm proteins are unlikely to bind dsDNA using the mechanism described herein, as these proteins appear to lack some key residues, e.g. the RRER motif, that are found in the Hfq proteins of most Gram-negative bacteria. Regardless, we expect the discovery of this novel binding site and binding mode taken in conjunction with the evidence of DNA condensation will allow the identification of additional conserved residues involved in anchoring of both sRNA and DNA, as well as enable further biological studies on the potential role of Hfq in DNA compaction, nucleoid architecture and transcription regulation.
Figure 5.
The binding of multiple Hfq(2-69) hexamers compacts dsDNA. (A) A representative AFM topograph demonstrating a 500 bp DNA (the Escherichia coli hipBA promoter region) bound by one or more Hfq hexamers (bright spots). (B) Representative scatter plot of DNA measurements made in the presence and absence of Hfq from a series of AFM topographs. Each point is the average of three measurements of the length of DNA molecules where the number of Hfq hexamers bound was obvious, i.e. Hfq was not bound at the ends or multiple hexamers were clustered together preventing delineation of the number bound to the DNA.
DATA AVAILABILITY
The coordinates and structure factors for the Ec Hfq–dsDNA complex structure have been deposited in the RSBC Protein Data Bank with accession code 5UK7 are already available to the public. Plasmids and strains used in this work are available from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beamlines and their support staff at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy Office of Science and Basic Energy Sciences. We also acknowledge Dr Luda Shlyakhtenko at the University of Nebraska Nanoimaging Core Facility for help with the AFM data collection.
Notes
Present address: Jillian Orans, United Therapeutics Corporation, Durham, NC 27709, USA.
Present address: Kirsten E. Hoff, Fujifilm Diosynth Biotechnologies, U.S.A., Durham, NC 27709, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R21AI115438 to R.G.B.]; U.S. Department of Energy Office of Science and Basic Energy Sciences [W-31-109-Eng-38]. Funding for open access charge: Duke University School of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sun X., Zhulin I., Wartell R.M.. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002; 30:3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valentin-Hansen P., Eriksen M., Udesen C.. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004; 51:1525–1533. [DOI] [PubMed] [Google Scholar]
- 3. Tsui H.C., Leung H.C., Winkler M.E.. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994; 13:35–49. [DOI] [PubMed] [Google Scholar]
- 4. Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G.. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998; 17:6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Storz G., Opdyke J.A., Zhang A.. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004; 7:140–144. [DOI] [PubMed] [Google Scholar]
- 6. Zhang A., Wassarman K.M., Ortega J., Steven A.C., Storz G.. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002; 9:11–22. [DOI] [PubMed] [Google Scholar]
- 7. Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S.. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001; 15:1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P.. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002; 9:23–30. [DOI] [PubMed] [Google Scholar]
- 9. Gorski S.A., Vogel J., Doudna J.A.. RNA-based recognition and targeting: sowing the seeds of specificity. Nat. Rev. Mol. Cell Biol. 2017; 18:215–228. [DOI] [PubMed] [Google Scholar]
- 10. Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004; 58:303–328. [DOI] [PubMed] [Google Scholar]
- 11. Muffler A., Traulsen D.D., Fischer D., Lange R., Hengge-Aronis R.. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1997; 179:297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding Y., Davis B.M., Waldor M.K.. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004; 53:345–354. [DOI] [PubMed] [Google Scholar]
- 13. Sonnleitner E., Hagens S., Rosenau F., Wilhelm S., Habel A., Jager K.E., Blasi U.. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003; 35:217–228. [DOI] [PubMed] [Google Scholar]
- 14. Kulesus R.R., Diaz-Perez K., Slechta E.S., Eto D.S., Mulvey M.A.. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 2008; 76:3019–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fantappie L., Metruccio M.M., Seib K.L., Oriente F., Cartocci E., Ferlicca F., Giuliani M.M., Scarlato V., Delany I.. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun. 2009; 77:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sittka A., Pfeiffer V., Tedin K., Vogel J.. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007; 63:193–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamada J., Yamasaki S., Hirakawa H., Hayashi-Nishino M., Yamaguchi A., Nishino K.. Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli. J. Antimicrob. Chemother. 2010; 65:853–858. [DOI] [PubMed] [Google Scholar]
- 18. Brennan R.G., Link T.M.. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007; 10:125–133. [DOI] [PubMed] [Google Scholar]
- 19. Mura C., Randolph P.S., Patterson J., Cozen A.E.. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013; 10:636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kambach C., Walke S., Young R., Avis J.M., de la Fortelle E., Raker V.A., Luhrmann R., Li J., Nagai K.. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999; 96:375–387. [DOI] [PubMed] [Google Scholar]
- 21. Cooper M., Johnston L.H., Beggs J.D.. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995; 14:2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hermann H., Fabrizio P., Raker V.A., Foulaki K., Hornig H., Brahms H., Luhrmann R.. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995; 14:2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995; 14:2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mura C., Cascio D., Sawaya M.R., Eisenberg D.S.. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toro I., Thore S., Mayer C., Basquin J., Seraphin B., Suck D.. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001; 20:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collins B.M., Harrop S.J., Kornfeld G.D., Dawes I.W., Curmi P.M., Mabbutt B.C.. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001; 309:915–923. [DOI] [PubMed] [Google Scholar]
- 27. Walke S., Bragado-Nilsson E., Seraphin B., Nagai K.. Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol. 2001; 308:49–58. [DOI] [PubMed] [Google Scholar]
- 28. Schumacher M.A., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G.. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002; 21:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Link T.M., Valentin-Hansen P., Brennan R.G.. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19292–19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W., Wang L., Wu J., Gong Q., Shi Y.. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013; 41:5938–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sauer E., Schmidt S., Weichenrieder O.. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9396–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Panja S., Schu D.J., Woodson S.A.. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013; 41:7536–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson K.E., Orans J., Kovach A.R., Link T.M., Brennan R.G.. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014; 42:2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kovach A.R., Hoff K.E., Canty J.T., Orans J., Brennan R.G.. Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA. 2014; 20:1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takada A., Wachi M., Kaidow A., Takamura M., Nagai K.. DNA binding properties of the hfq gene product of Escherichia coli. Biochem. Biophys. Res. Commun. 1997; 236:576–579. [DOI] [PubMed] [Google Scholar]
- 36. Azam T.A., Ishihama A.. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999; 274:33105–33113. [DOI] [PubMed] [Google Scholar]
- 37. Jiang K., Zhang C., Guttula D., Liu F., van Kan J.A., Lavelle C., Kubiak K., Malabirade A., Lapp A., Arluison V. et al.. Effects of Hfq on the conformation and compaction of DNA. Nucleic Acids Res. 2015; 43:4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Updegrove T.B., Correia J.J., Galletto R., Bujalowski W., Wartell R.M.. E. coli DNA associated with isolated Hfq interacts with Hfq's distal surface and C-terminal domain. Biochim. Biophys. Acta. 2010; 1799:588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malabirade A., Jiang K., Kubiak K., Diaz-Mendoza A., Liu F., van Kan J.A., Berret J.F., Arluison V., van der Maarel J.R.C.. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017; 45:7299–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malabirade A., Partouche D., El Hamoui O., Turbant F., Geinguenaud F., Recouvreux P., Bizien T., Busi F., Wien F., Arluison V.. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018; 8:16792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A.. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999; 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kajitani M., Kato A., Wada A., Inokuchi Y., Ishihama A.. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J. Bacteriol. 1994; 176:531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Azam T.A., Hiraga S., Ishihama A.. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000; 5:613–626. [DOI] [PubMed] [Google Scholar]
- 44. Tolstorukov M.Y., Virnik K.M., Adhya S., Zhurkin V.B.. A-tract clusters may facilitate DNA packaging in bacterial nucleoid. Nucleic Acids Res. 2005; 33:3907–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Le Derout J., Boni I.V., Regnier P., Hajnsdorf E.. Hfq affects mRNA levels independently of degradation. BMC Mol. Biol. 2010; 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dimastrogiovanni D., Frohlich K.S., Bandyra K.J., Bruce H.A., Hohensee S., Vogel J., Luisi B.F.. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife. 2014; 3:e05375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauer E., Weichenrieder O.. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13065–13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang W., Wang L., Zou Y., Zhang J., Gong Q., Wu J., Shi Y.. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011; 25:2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johansson J., Eriksson S., Sonden B., Wai S.N., Uhlin B.E.. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 2001; 183:2343–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 51. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 53. Afonine P.V., Grosse-Kunstleve R.W., Adams P.D.. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D. Biol. Crystallogr. 2005; 61:850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al.. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 55. Shlyakhtenko L.S., Gall A.A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y.L.. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003; 97:279–287. [DOI] [PubMed] [Google Scholar]
- 56. Vecerek B., Rajkowitsch L., Sonnleitner E., Schroeder R., Blasi U.. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008; 36:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rice P.A., Yang S., Mizuuchi K., Nash H.A.. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996; 87:1295–1306. [DOI] [PubMed] [Google Scholar]
- 58. Schumacher M.A., den Hengst C.D., Bush M.J., Le T.B.K., Tran N.T., Chandra G., Zeng W., Travis B., Brennan R.G., Buttner M.J.. The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development. Nat. Commun. 2018; 9:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A.. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003; 22:3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Calladine C.R., Drew H.R., McCall M.J.. The intrinsic curvature of DNA in solution. J. Mol. Biol. 1988; 201:127–137. [DOI] [PubMed] [Google Scholar]
- 61. Ulanovsky L., Bodner M., Trifonov E.N., Choder M.. Curved DNA: design, synthesis, and circularization. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:862–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haran T.E., Mohanty U.. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009; 42:41–81. [DOI] [PubMed] [Google Scholar]
- 63. Vecerek B., Moll I., Blasi U.. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005; 11:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ishikawa H., Otaka H., Maki K., Morita T., Aiba H.. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012; 18:1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B. et al.. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010; 143:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee Y., Chin R.K., Christiansen P., Sun Y., Tumanov A.V., Wang J., Chervonsky A.V., Fu Y.X.. Recruitment and activation of naive T cells in the islets by lymphotoxin beta receptor-dependent tertiary lymphoid structure. Immunity. 2006; 25:499–509. [DOI] [PubMed] [Google Scholar]
- 67. Rochat T., Bouloc P., Yang Q., Bossi L., Figueroa-Bossi N.. Lack of interchangeability of Hfq-like proteins. Biochimie. 2012; 94:1554–1559. [DOI] [PubMed] [Google Scholar]
- 68. Bohn C., Rigoulay C., Bouloc P.. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gaczynska M., Osmulski P.A., Jiang Y., Lee J.K., Bermudez V., Hurwitz J.. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17952–17957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shin M., Song M., Rhee J.H., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E.. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005; 19:2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rivetti C., Guthold M., Bustamante C.. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 1999; 18:4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clarey M.G., Botchan M., Nogales E.. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J. Struct. Biol. 2008; 164:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Necas D., Klapetek P.. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012; 10:181–188. [Google Scholar]
- 74. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kambara T.K., Ramsey K.M., Dove S.L.. Pervasive Targeting of Nascent Transcripts by Hfq. Cell Rep. 2018; 23:1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for the Ec Hfq–dsDNA complex structure have been deposited in the RSBC Protein Data Bank with accession code 5UK7 are already available to the public. Plasmids and strains used in this work are available from the corresponding author.