Abstract
U3 snoRNA and the associated Rrp9/U3-55K protein are essential for 18S rRNA production by the SSU-processome complex. U3 and Rrp9 are required for early pre-rRNA cleavages at sites A0, A1 and A2, but the mechanism remains unclear. Substitution of Arg 289 in Rrp9 to Ala (R289A) specifically reduced cleavage at sites A1 and A2. Surprisingly, R289 is located on the surface of the Rrp9 β-propeller structure opposite to U3 snoRNA. To understand this, we first characterized the protein-protein interaction network of Rrp9 within the SSU-processome. This identified a direct interaction between the Rrp9 β-propeller domain and Rrp36, the strength of which was reduced by the R289A substitution, implicating this interaction in the observed processing phenotype. The Rrp9 R289A mutation also showed strong synergistic negative interactions with mutations in U3 that destabilize the U3/pre-rRNA base-pair interactions or reduce the length of their linking segments. We propose that the Rrp9 β-propeller and U3/pre-rRNA binding cooperate in the structure or stability of the SSU-processome. Additionally, our analysis of U3 variants gave insights into the function of individual segments of the 5′-terminal 72-nt sequence of U3. We interpret these data in the light of recently reported SSU-processome structures.
INTRODUCTION
Eukaryotic ribosome biogenesis is a highly complex process initiated in the nucleolus within a large macromolecular complex, the SSU-processome or 90S pre-ribosomal particle (1). Production of the 40S and 60S subunits follows two independent pathways. It begins with the transcription by RNA polymerase I of a pre-ribosomal RNA (pre-rRNA) containing the 18S, 5.8S and 25/28S ribosomal RNA (rRNA) sequences (35S pre-rRNA in Saccharomyces cerevisiae). An ordered series of endo- and exo-ribonucleolytic cleavages then generates the mature rRNAs. Here, we focus on the model organism S. cerevisiae. However, as pre-rRNA processing is highly conserved from yeast to human, insights gained in this study are of general interest.
In yeast, the 35S pre-rRNA is initially cleaved at sites A0, A1 and A2. This generates the 20S pre-rRNA, a precursor to the 18S rRNA, and the 27SA2 pre-rRNA intermediate, a precursor to the 5.8S and 25S rRNAs (2,3). The released 20S pre-rRNA is assembled into pre-40S particles, while pre-60S particles assemble on the 27S pre-rRNA. Early during this maturation process, the pre-rRNA is 2′-O methylated and pseudouridylated at many positions by small nucleolar ribonucleoprotein particles (snoRNPs). Box C/D snoRNPs generally catalyze ribose 2′-O-methylation and contain a small nucleolar RNA (snoRNA) together with four core proteins Snu13, Nop56, Nop58 and Nop1 (4–8). In contrast, the U3 box C/D snoRNP lacks a known role in RNA methylation but is essential for pre-rRNA cleavage at sites A0, A1 and A2 (9,10). Compared to other box C/D snoRNPs, the U3 snoRNP also contains an additional protein, designated Rrp9 in yeast and U3-55K in humans (11–13), which is required for A0, A1 and A2 cleavages (12–15).
U3 is highly conserved and strongly expressed throughout eukaryotes, including human cells (16,17). Similar to other box C/D snoRNAs, U3 contains two ‘C/D like’ motifs, named the C’/D and B/C motifs, each forming a K-turn structure recognized by Snu13 (18,19). While the C’/D motif binds the canonical set of box C/D proteins (15,18), the B/C motif additionally recruits the specific Rrp9 (yeast) or U3-55K (human) protein (11–14,20,21). Rrp9 and U3-55K each contain a WD40 domain (11), comprised of seven WD repeats folded into a β-propeller structure (22–24). WD40 domains generally participate in protein–protein interactions and the SSU-processome contains several proteins of this family (25–27). Compared to other C/D snoRNAs, U3 has a long 5′-terminal region (72 nts in yeast). This protrudes from the C’/D box core region (10) and has been the subject of a large number of functional analyses (9,10,17,28–33). The U3 5′-terminal sequence was proposed to form five evolutionarily conserved base-paired interactions with the pre-rRNA, designated as helices I, II, III, V and VI (Figure 1 top) (9,10,28,30–32,34–36).
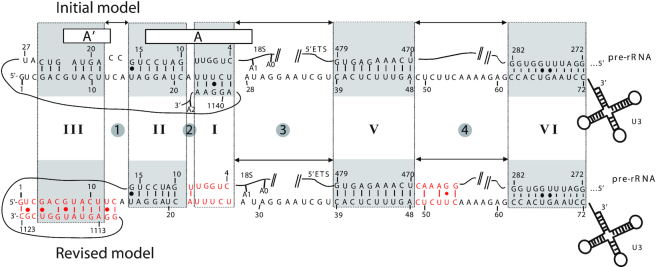
Figure 1.
Previous and new models for the yeast U3 snoRNA/pre-ribosomal RNA interaction. (Top) The 5′-terminal sequence of U3 snoRNA was previously proposed to form five base-pair interactions with the pre-rRNA. They were respectively designated as helices I, II, III, V and VI in yeast (9,10,28,30–32,34–36). (Bottom) Based on cryo-EM studies, an alternative model including three of the previously proposed interactions (helices II, V with an extension represented in red and VI) and an alternative helix III (represented in red) was established (40,44).
The U3 snoRNP is needed for assembly of the SSU-processome (1), which contains more than 70 proteins and coordinates pre-rRNA processing, RNA modification and pre-ribosome assembly (1,7,9,28,29,34,37–39). Published SSU-processome structures include the U3 snoRNP with Rrp9 (24,40–45). The two specific features of the U3 snoRNPs, namely, the specific Rrp9 protein and the extended U3 snoRNA 5′-terminal sequence are expected to contribute to the specific activity of the U3 snoRNP within the processome. However, while the U3 5′ region is located in the vicinity of the pre-rRNA cleavage sites (Figure 1), this is not the case for Rrp9. Moreover, some SSU-processome proteins remain unmapped, including Rrp36 and Sgd1 for which no 3D structures are established (46). In consequence, the protein densities visualized around Rrp9 have not yet been identified (46–49), indicating the need for further analyses. In addition to snoRNPs, various protein sub-complexes participate to the SSU-processome (including UTP-A/t-UTPs, UTP-B, UTP-C and the Mpp10 complex) (41,45,50–55). The U3 snoRNP is incorporated together with the UTP-B complex, after UTP-A complex formation onto the pre-rRNA. The UTP-C complex and protein Rrp5 are associated in a parallel pathway (52,53,55).
Based on phylogenetic analysis, yeast genetics experiments, in vivo chemical probing in yeast and Xenopus oocyte microinjections, a structure including five base-paired interactions formed between the 5′ region of U3 and pre-rRNA sequences in the 5′-ETS and 18S segments has been proposed. Ordered from the 5′ end of U3, these are designated III to I, V and VI, and are separated by spacer regions designated 1–4 (Figure 1, top). In this model, helices V and VI are formed with the 5′-ETS region of the pre-rRNA and these were shown to be essential for cleavages at sites A0–A2 by compensatory mutation assays (9,10,30). Helices I, II and III were proposed to base-pair with 18S rRNA segments implicated in formation of the central pseudoknot, a long-range interaction essential for 40S subunit function (56,57). Helix VI binds the trimeric Mpp10–Imp3–Imp4 complex, which is also needed for cleavages at sites A0, A1 and A2 (10,30,58–61). Imp3 functions to open internal structures in U3 and the pre-rRNA to facilitate intermolecular helix II formation (62,63). The functional importance of helix II could be demonstrated by compensatory mutations (29,34), but this could not be done for helices I and III. However, U3 mutations expected to block formation of helices II as well as helix III prevent cleavage at sites A1 and A2 but not A0, leading to accumulation of the aberrant 22S RNA cleaved at sites A0 and A3 (28,29,34). The U3 segments forming heterologous helices are separated by linker segments. Previous analyses in yeast and Xenopus laevis oocytes highlighted possible roles of these segments in pre-rRNA processing (28,31,32), and we therefore also performed functional analyses on these regions.
Cryo-EM structures from S. cerevisiae and Chaetomium confirmed the occurrence of helices V, VI and II in the SSU-processome. However, helix V appeared to be more extended than anticipated and an alternative form of helix III was proposed, while helix I was not detected (24,44) (Figure 1, bottom).
Here, we determined the functional interactions between Rrp9, other SSU-processome components and the 5′-terminal region of U3 in yeast pre-rRNA processing. The results identified a crucial amino acid at the surface of the Rrp9 β-propeller, a protein–protein interaction network and roles for key segments of the U3 5′-terminal region. Based on these results and re-analysis of cryo-EM structural data, we propose a new model for U3 binding to the yeast pre-rRNA.
MATERIALS AND METHODS
Plasmids
The pACT2, pGBKT7 and pAS2 plasmids (Clontech) were used for the two-hybrid assays. Plasmid pG1::protA (Addgene) was used to clone Rrp9 mutants with a N-ter Protein A (ProtA) tag. As previously described, the U3 snoRNA variants produced by site-directed mutagenesis were cloned into the pASZ11 plasmid to produce pASZ11::snoRNA U3A variant plasmids (10,28). The pDONR™ 207, pDONR™ 221, pDEST™ 15 (GST-tag) and pDEST™ 17 (6-His-tag) plasmids were used to clone SSU-processome proteins or proteins sub-domains using the GATEWAY technology (Invitrogen). Plasmids pnEA-3CH (6-His-tag) and pnCS (64) were modified to become compatible with the GATEWAY cloning technology (Invitrogen). Detailed plasmids information is available upon request.
Strains used and growth conditions
The S. cerevisiae W303 strain (Mata trp1-Δ63; his3-Δ200; ura3–52; lys2–801; ade2– 101; leu2-Δ1) was used to generate the S. cerevisiae W303-pGal-Rrp9 strain needed to test the functionality of mutated forms of Rrp9. In this strain, the S. cerevisiae galactose promoter was inserted upstream of the endogenous Rrp9 ORF, so that endogenous Rrp9 is only expressed if Galactose is present in the medium.
In the S. cerevisiae JH84 strain (Mata snr17a.Gald:URA3 snr17b::LEU2 his3 ade2 can1) (7,28), the endogenous U3A snoRNA is only expressed in the presence of Galactose, while the U3B snoRNA is not expressed at all. This strain was used to generate the S. cerevisiae JH84-pGal-Rrp9, using the same approach as for the W303-pGal-Rrp9 strain. This new strain can be used to test the functionality of mutated yeast U3 snoRNAs in the presence of the Rrp9 R289 mutant, as both endogenous Rrp9 and U3 snoRNA are only expressed in the presence of Galactose, and repressed in the presence of glucose.
One colony of transformed yeast cells was pre-cultured in 10 ml of YPG medium (10 g/l yeast extract, 20 g/l bactopeptone, 2% galactose) for 48 h at 30°C under shaking. From the saturating pre-culture, a culture was launched by inoculating 50 ml of YPD medium (10 g/l yeast extract, 20 g/l bactopeptone, 2% glucose) at 0.1 A600nm/ml, and incubation at 20, 30 or 37°C until the culture reaches an A600nm/ml comprised between 0.5 and 0.8 (for 8 h on average at 30 and 37°C, and 24 h at 20°C). Cells were then diluted at 0.1 A600 nm/ml in 100 ml (30 and 37°C cultures) or 200 ml (20°C cultures) of YPD medium and grown over night at the respective temperature. After 24 h, depending on the strain considered, endogenous U3 and/or endogenous Rrp9 were not detectable anymore.
Test of U3 snoRNA variants functionality
The S. cerevisiae strain JH84 was transformed with plasmid pASZ11 (65) or its derivatives using the Li-acetate method (66). Selection of transformants was made on minimum medium plates. Recombinant JH84 cells grown in liquid medium, as described above, were spread on YPD plates. Growth was tested at three temperatures (20, 30 and 37°C). Sizes of colonies were examined after 48 h of incubation at 30 and 37°C, or after 72 h of incubation at 20°C.
PCR mutagenenesis of Rrp9
A 50 μl incubation mixture made up in Pfu DNA polymerase buffer (ThermoFisher Scientific), containing 200 ng of plasmid to be modified, 2.5 U of Pfu DNA polymerase (ThermoFisher Scientific), 0.2 mM of dNTPS and 40 nM of each of the two overlapping complementary mutagenesis primers was used for PCR amplification using the following incubation conditions: 96°C for 10 min; 30 cycles : 96°C for 30 s, 55°C for 30 s, 72°C for 30 s; final incubation at 72°C for 10 min. Incubation time and temperature were adapted to the plasmid and primers used. Next, 10 μl of mixture were digested with 1 μl of DpnI restriction enzyme (ThermoFisher Scientific) for 10 min at 37°C in a final volume of 30 μl and Escherichia coli DH5α competent cells were transformed with 3 μl of the mixture.
Production of pASZ11 derivatives with U3 gene variants
Phage M13 mp9::T7-snR17A (67,68) containing the coding sequence of U3 snoRNA under the control of T7 promoter was used as the matrix for site-directed mutagenesis of the U3 gene (64), using the oligonucleotides listed in Supplementary Table S1. Plasmids pASZ11::U3 with the mutated forms of U3 were obtained by substitution of the SalI-EcoRI fragment of plasmid pASZ11::U3 encoding the WT U3 snoRNA by their mutated SalI-EcoRI counterparts.
Test of the U3 allele expression and stability
Total RNA was extracted from recombinant JH84 cells grown for 24 h in YPD liquid medium, using the procedure of (7). For each assays, 20 μg of total RNA were fractionated on a denaturing 6% polyacrylamide gel. Northern-blot analyses were performed as previously described (19), using a 5′-end labeled oligonucleotide (Supplementary Table S1), complementary to the 3′-end of the yeast U3 snoRNA as the probe, and the 5′-end labeled U6 oligonucleotide (Supplementary Table S1), complementary to U6 snRNA, for standardization of the data. The radioactivity of the hybridization bands were quantified with a phosphorimager (GE Healthcare) using the Image Quant software. The ratio between the radioactivity measured for the U3 snoRNA hybridization signals and the total radioactivity measured in U3 snoRNA and U6 snRNA hybridization signals was estimated for each recombinant strain.
Analysis of pre-rRNA processing
Ten micrograms of yeast total RNA were dissolved in 20 μl of MOPS buffer (20 mM MOPS, 5 mM NaAc, 1 mM EDTA) and were fractionated on a 1.5% agarose gel run in MOPS buffer for 24 h at 2 V/cm. The gel was washed for 10 min in water and then twice in 75 mM NaOH for 20 min. RNAs were transferred onto a Nylon membrane (Amersham Hybond™-N+, GE Healthcare) using a vacuum system for 30 min in transfer buffer 1 (0.5 M Tris–HCl, 1.5 M NaCl) and next for 3 h in transfer buffer 2 (10× SSC buffer, 1.5 M NaCl, 150 mM citrate sodium pH 7). After transfer, the membrane was UV-cross-linked for 5 min and northern-blotting was performed as previously described (69), using U3 and U6 specific probes (Supplementary Table S1).
Primer extension analysis
Two μg of total RNA were incubated for 2 min at 96°C with 200 000 cpm of labeled RT-primer in a final volume of 2.5 μl of hybridization buffer (50 mM Tris–HCl pH 8.3, 40 mM KCl). The hybridization material was immediately cooled on ice for 5 min. Next, 100 μM dNTPs were added together with 0.5 U of AMV-Reverse Transcriptase (MP Biomedicals) in a final volume of 5 μl of RT buffer (50 mM Tris–HCl pH 8.3, 50 mM KCl, 10 mM MgCl2, 0.5 mM spermidine, 10 mM DTT), and the mixture was incubated at 42°C for 30 min. After addition of 4 μl of formamide loading dye (10 mM EDTA, pH 8, 0.05% bromophenol blue, 0.05% xylene cyanol in deionized formamide), samples were incubated for 2 min at 96°C and loaded on a 7% acrylamide/bisacrylamide (38:2) denaturing gel (45 mM Tris–HCl pH 8, 45 mM H3BO3, 0.5× TBE, 1 mM EDTA, pH 8, 8 M urea). Migration was performed for 4 h at 2 V/cm in 0.5× TBE buffer, before phosphorimager analysis.
In vivo analysis of mutated U3 snoRNA variant structures
The procedure used was previously described by Méreau et al. (28). Recombinant JH84 cells were grown at 30°C in 20 ml of YPD medium until the A600 reached a value comprised between 0.5 and 1.2. Then, they were gently rocked at room temperature for 2 min in the presence of DMS at a final concentration of 30 or 60 mM. The reaction was stopped by addition of 0.7 M ice-cold 2β-mercaptoethanol and 5 ml of cold water-saturated isoamyl alcohol, followed by shaking and centrifugation. The cell pellet was first washed with 10 mM MgCl2, 3 mM CaCl2, 10 mM Tris–HCl pH 7.5 containing 250 mM sucrose and 0.7 M 2β-mercaptoethanol, then washed with 5 ml of sterile water, and ground with acid-washed beads in the presence of 200 μl extraction buffer (100 mM NaCl, 10 mM EDTA, 50 mM Tris, pH 7.5). The mixture was successively shaken for 1 min, put on ice for 2 min, and shaken again for 1 min. Then, 200 μl of extraction buffer and 50 μl SDS 10% were added, and total RNA was phenol extracted and ethanol precipitated. The RNA pellet was washed with ethanol (70%), dried and dissolved in milliQ water to a final concentration of 5 μg/μl, as estimated by UV absorption at 260 nm. Sites of DMS modifications were mapped by reverse transcriptase analysis with the 5′-end labeled 112 primer (Supplementary Table S1) complementary to U3 snoRNA nucleotides from position 92–106. The primer was annealed to 5 μg of total RNA. cDNA synthesis was made in the presence of four deoxynucleotide triphosphates and the AMV reverse transcriptase, as previously described by Mougin et al. (70). For comparison, total RNA from untreated cells was prepared in the same way than DMS treated cells. Positions of chemical modifications were then identified by reference to a sequencing ladder made with unmodified RNA and the same 5′-end labeled primer.
Yeast two hybrid assays
Yeast two-hybrid assays were performed at 30°C as previously described, using increasing 3-amino-1,2,4-Triazole (3-AT) concentrations from 10 to 50 mM to test for the stability of the detected interactions (71).
Immunoprecipitation
Cell extracts were incubated with Rabbit IgG-Agarose beads (SIGMA) in IPP-150 buffer (10 mM Tris–HCl, pH 8, 150 mM NaCl, 0.1% NP-40) for 3 h at 4°C. Beads were washed three times for 5 min in IPP-150 buffer. For analysis of immunoprecipitated RNAs, the pelleted agarose beads were suspended in 300 μl of IPP-150 buffer containing 0.5% SDS and 150 μg of proteinase K (Roche) and incubated for 30 min at 37°C and next for 10 min at 65°C. RNAs were phenol extracted twice and ethanol precipitated (72) before northern-blotting.
GST pull-down
GST-tagged proteins were expressed in E. coli BL21(DE3) pRARE2 cells (Novagen) and purified by affinity chromatography on glutathione-Sepharose beads (GE healthcare). [35S]-methionine-labeled proteins were prepared using the E. coli T7 S30 Extract System for Circular DNA (Promega). One microgram of GST or GST-tagged proteins bound to glutathione-sepharose beads was incubated with 5 μl of [35S]-methionine-labeled proteins for 2 h at 20°C in 400 μl of RSB 200 buffer (10 mM Tris–HCl, pH 7.4, 2.5 mM MgCl2, 200 mM NaCl, 0.01% NP40). Beads were washed three times in RSB 200 buffer and proteins were eluted in loading buffer (45 mM Tris–HCl pH 6.8; 8% SDS; 0.02% bromophenol blue; glycerol, 20% 2-mercapto-ethanol) and analyzed by SDS-PAGE followed by autoradiography.
RESULTS
The β-propeller surface residue R289 of Rrp9 is important for cell growth
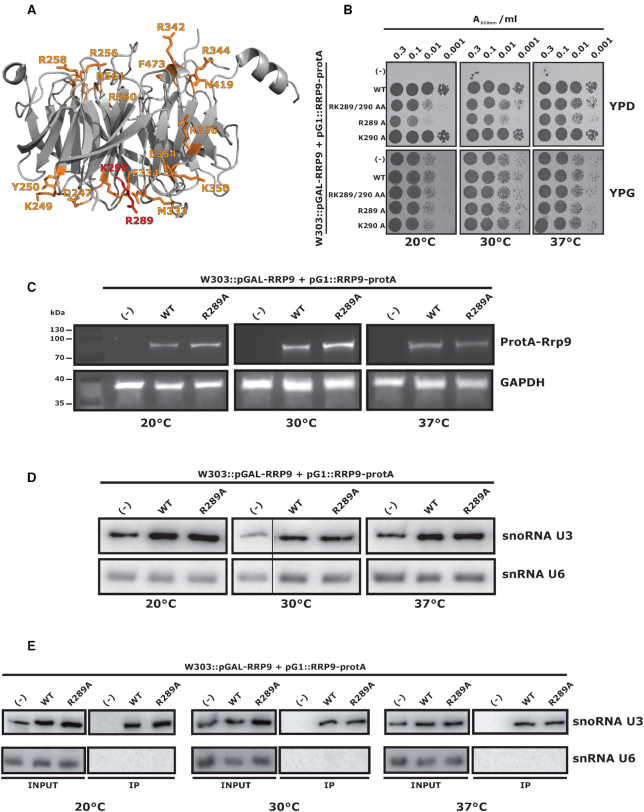
The β-propeller domain of Rrp9 is formed by seven WD subdomains and was predicted to mediate protein contacts within the SSU-processome. To test this hypothesis, we mutated β-propeller surface residues (Figure 2A) (26). We preferentially mutated amino-acids present both in Rrp9 and human U3-55K but omitted residues that were likely to play structural roles. We substituted nineteen residues with alanine, either individually or in combination, generating nine Rrp9 mutants (Figure 2A, B; Supplementary Figure S1). Since Rrp9 is essential, we inserted a regulated PGAL10 promoter upstream of the endogenous RRP9 open-reading frame in yeast strain W303. In the resulting strain (W303::pGAL-RRP9), Rrp9 is produced in galactose medium (YPG), but repressed in glucose (YPD). Cells were transformed either with an empty pG1::protA plasmid as control, or with a pG1::Rrp9-protA plasmid derivative expressing the WT or a mutant Rrp9 protein carrying a N-terminal protein A tag. Serial dilutions of transformed cell cultures were plated on both YPG and YPD solid media at three different temperatures (20, 30 and 37°C) (Figure 2B, Supplementary Figure S1). As expected, the empty plasmid did not restore cell growth on YPD medium, while expression of WT Rrp9-protA fully restored cell growth at all temperatures. In this initial screen, a clear growth defect was observed for cells expressing the R289A/K290A double mutant (Figure 2B, Supplementary Figure S1), which was exacerbated at 20°C. Deconvoluting the mutated residues of this double mutant showed that R289 was important for normal growth (Figure 2B). Indeed, the growth defect in cells expressing Rrp9–R289A was more severe than upon expression of the double mutant Rrp9-R289A/K290A. This observation was unexpected, as both consecutive residues are polar, but could reflect some compensatory effect of the K290A mutation.
Figure 2.
The R289A substitution in the Rrp9 β-propeller domain is deleterious for cell growth but alters neither Rrp9 / U3 stabilities, nor U3 snoRNP assembly. (A) Tridimensional representation of the Rrp9 protein structure (44). For simplicity, only the β-propeller domain of Rrp9 is shown. Surface residues that were mutated in this study are depicted in orange. Residues R289 and K290 are highlighted in red (44). (B) Effects of Rrp9 mutations on growth of yeast W303::pGAL-RRP9 cells. Growth of cells expressing mutant or wild-type Rrp9 proteins was monitored under permissive galactose (YPG) or repressive glucose medium conditions (YPD) at 3 different temperatures (20°C, 30°C and 37°C), using 4 increasing dilutions (indicated as A600 values: from 0.3 to 0.001). Identities of the amino-acid substitutions are indicated on the left of the panel. (–) Cells transformed with an empty plasmid. Growth defect is reinforced at low temperature. (C) Comparison by Western-blotting of the stabilities of Rrp9-Protein A and R289A Rrp9-Protein A fusion proteins expressed from plasmid pG1. Cells were grown on repressive YPD medium at 20°C, 30°C and 37°C. Antibodies were directed against the Protein A tag. GAPDH was used as a control. (–) corresponds to the negative control experiment performed with cells transformed with an empty pG1 plasmid. (D) Comparison of U3 snoRNA stability upon expression of wild-type Rrp9 (WT) or mutant Rrp9 (R289A) from plasmid pG1, and in the absence (–) of Rrp9 expression from plasmid pG1. Northern-blots on total RNA of cells grown on YPD medium at 20, 30 and 37°C were performed with the U3 specific labeled probe 41. U6 snRNA was used as a loading control (see Supplementary Table S1 for the probes). (E) RNA pull-down assays in the view to compare association of U3 snoRNA with the WT and the mutated Rrp9 (R289A) proteins. Pull-downs were performed with an antibody directed against the Protein A tag on extracts of cells transformed with pG1 plasmids expressing the wild-type (WT), the R289A Rrp9 protein or no Rrp9 protein (–). Immunoselection of U3 and U6 was monitored by northern-blotting using the same specific oligonucleotide probes as in D.
Western-blot analysis showed that Rrp9–R289A–protA was expressed at the same levels as WT Rrp9–protA at all temperatures (Figure 2C). To test effects of the R289A mutation on U3 association and stability, we performed northern-blot analyses using a U3 specific probe and affinity-purification with the protA-tag (Figure 2D, E). U3 levels were the same for cells expressing Rrp9–R289A or WT Rrp9 (Figure 2D). Recovery of co-purified U3 was also similar for Rrp9–R289A–protA and WT Rrp9–protA (Figure 2E).
We conclude that the R289A substitution is detrimental to Rrp9 function, particularly at low temperature, possibly reflecting altered protein interactions.
An aberrant 22S RNA species accumulates in cells expressing Rrp9-R289A
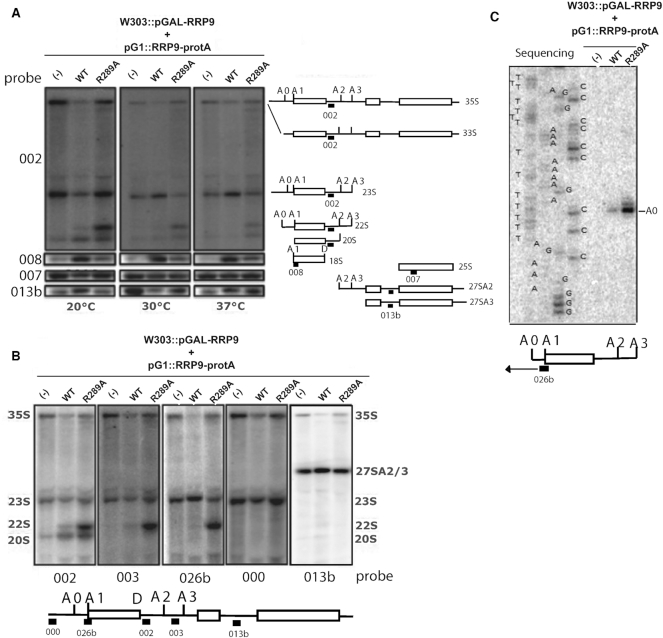
Rrp9 is required for pre-rRNA cleavage (12) and we therefore tested whether 35S pre-rRNA processing is defective in the Rrp9–R289A mutant (Figure 3). As the efficiency of pre-rRNA cleavage cannot be directly assessed from the steady levels of intermediates, but must be determined from the accumulation of precursors, we evaluated the levels of these intermediates by northern-blot assays and by reverse transcriptase analysis. Following Rrp9 depletion in the empty vector control (pG1), the 35S pre-rRNA was accumulated together with the 23S RNA. This aberrant intermediate results from cleavage at site A3 located in internal transcribed spacer 1 (ITS1), in the absence of prior cleavage at sites A0-A2 (Figure 3A, lanes (–)). Site A3 is cleaved by RNase MRP and does not require the U3 snoRNP (73). In addition, the 20S pre-rRNA and 18S rRNA were depleted consistent with the loss of cleavage at sites A0, A1 and A2 (12) (compare lanes (–) to WT, Figure 3A). Rrp9 depleted cells expressing Rrp9–R289A–protA showed an accumulation of 35S pre-rRNA and 23S RNA. This was more marked following growth at 20°C, correlating with the observed growth defects (lane R289A, Figure 3A). However, accumulation of 18S rRNA was markedly decreased at all tested temperatures. This was explained by a marked increase in abundance of the aberrant 22S RNA, predicted to result from cleavage at sites A0 and A3 in the absence of processing at sites A1 and A2. This was especially marked at 20°C, the temperature corresponding to the strongest growth defect. At this temperature 22S RNA is only detected in trace amounts when WT Rrp9 is expressed. This identification was verified by northern-blot assays, using probes targeting pre-rRNA sequences located between sites A0 and A1 (026b), A1 and A2 (002), A2 and A3 (003), as well as upstream of site A0 (000) and downstream of A3 (013b) (Figure 3B, Supplementary Table S1). As a further confirmation, primer extension with reverse transcriptase (RT) was performed with a primer (026b) hybridizing 36 nucleotides downstream from site A0 (the 5′-end of 22S). This revealed a marked increase of the RT signal at site A0 when Rrp9–R289A was expressed (Figure 3C). Since all pre-rRNA is considered to be cleaved at site A0 in a wild-type strain, this demonstrated the accumulation of RNA intermediates cleaved at site A0 in cells expressing the mutated Rrp9 protein. Comparison of different growth temperatures (Figure 3A) showed stronger accumulation of 35S pre-RNA and the 23S and 22S intermediates at 20°C, with a concomitantly greater decrease in 18S rRNA production.
Figure 3.
The Rrp9-R289A mutation affects pre-rRNA early cleavages at sites A1 and A2 leading to strong accumulation of the aberrant 22S RNA at 20°C. (A) The different RNA species produced from the 35S pre-rRNA at 20, 30 and 37°C were detected by northern-blotting using specific probes, indicated on the left side of panel A (Supplementary Table S1) (002 downstream of the 18S sequence, 008 close to the 5′ extremity of the 18S, 007 in the 5′-terminal part of the 25S, and 013b between the 5.8S and 25S sequence, in the ITS2 segment, as drawn right of panel A). Cells were transformed with an empty pG1 plasmid or pG1 plasmids expressing WT- or R289A–Rrp9. The cleavage steps generating the 18S rRNA from the 35S pre-rRNA are schematized on the right side of the panel. Positions of the probes are indicated on the schematic representations of the various intermediates. Upon expression of Rrp9 R289A, an accumulation of 22S RNA is detected, especially at 20°C and 18S production is decreased at all the tested temperatures. (B) The identities of cleavage sites occurring at 20°C in the pre-rRNA were determined by northern-blotting using five discriminating probes shown under the panel (Supplementary Table S1). Briefly, the binding site of probe 002 was located between sites A2 and D enabling detection of the 35S pre-rRNA and the 23S, 22S and 20S RNA intermediates. The probe 003 binding sequence was located between sites A2 and A3. This probe was dedicated to the detection of intermediates that were not cleaved at site A2, namely, the 35S, 23S and 22S RNAs, but not the 20S intermediate. Probe 026b hybridized between sites A0 and A1 and revealed the intermediates that were not cleaved at site A1, thus detecting the 35S, 23S and 22S pre-rRNAs. Probe 000 was binding upstream of site A0, revealing only the 35S pre-rRNA and the 23S intermediate that is not cleaved at site A0. Finally, probe 013b was binding downstream from site A3, and was only detecting the 35S pre-rRNA and intermediates involved in the 5.8S and 25S maturation. The absence of binding of probes 000 and 013b and the binding of probes 002, 003 and 026b to the abnormally accumulated intermediate species confirmed that it was the 22S intermediate cleaved at sites A0 and A3 without cleavage at sites A1 and A2. (C) To define the 5′ extremity of the 22S RNA intermediate, a primer extension experiment was performed with oligonucleotide 026b binding upstream of site A1 as depicted in the cartoon at the bottom of the panel. Products of the primer extension were fractionated in parallel with the products of sequencing reactions generated with the same primer (026b). Site +1, the 5′ end of 35S and 23S RNAs, is too far from the primer to be detected.
Our results show that in cells expressing Rrp9–R289A, cleavage efficiency at sites A1 and A2 is strongly reduced, while cleavages at sites A0 and A3 are less affected. The reduced accumulation of 18S likely explains the observed growth phenotype.
Rrp9 belongs to a large protein–protein interaction network within the SSU-processome
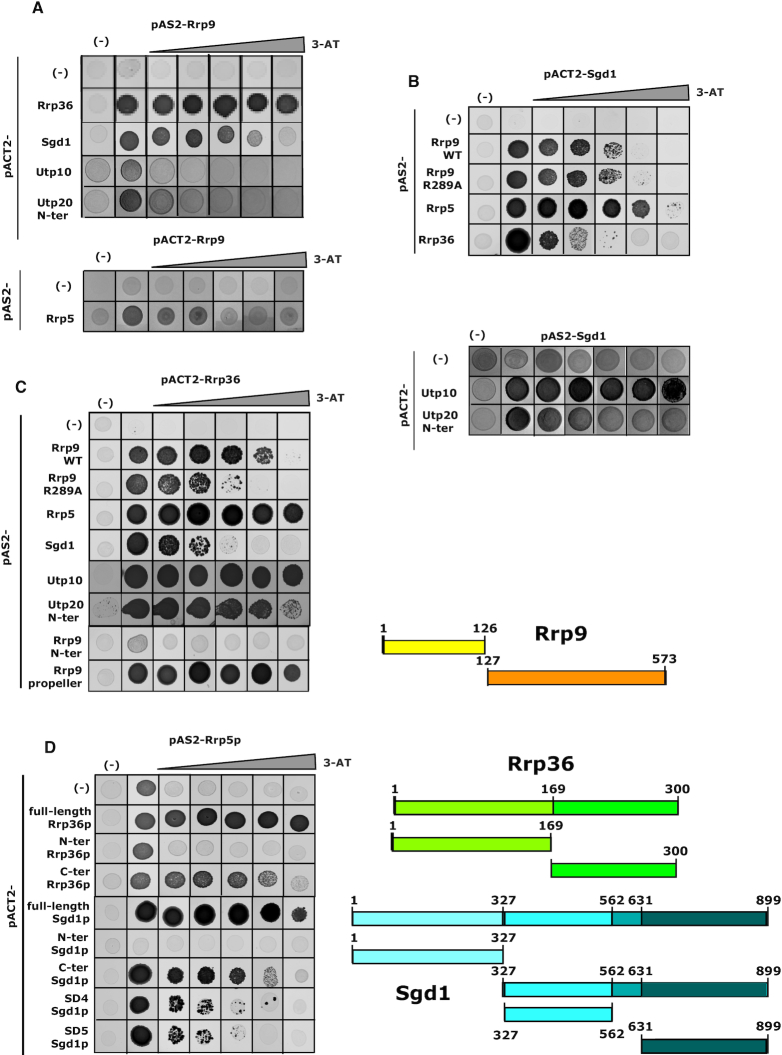
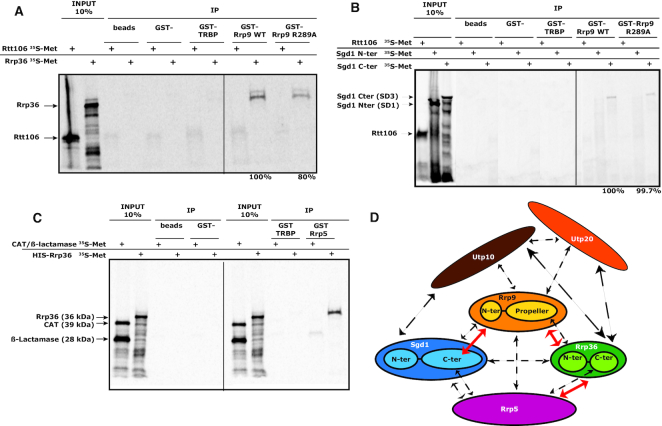
β-propeller domains are involved in many protein–protein interactions (22,23), suggesting that the R289A substitution might disturb interactions between Rrp9 and other components of the SSU-processome. To identify such interactions, we performed a candidate-based yeast two-hybrid (Y2H) screen focused on Rrp9. The proteins included in this screen were selected on the basis of previous SSU-processome interactome studies and structural analyses (24,40–45,74,75). A positive Y2H interaction between the two test proteins drives expression of the HIS3 gene and confers the ability to grow on medium lacking histidine (plates in Figure 4 and in Supplementary Figure S3). The strength of the interaction can be assessed by competition with increasing concentrations of the HIS3 inhibitor 3-amino-1,2,4-triazole (3-AT). Consistent with previous reports (74–76), among the numerous combinations tested, strong positive interactions were observed for Rrp9 with Rrp36 and Sgd1 (Figure 4A-C and Supplementary Table S2). In addition to Rrp36 and Sgd1, our candidate-based Y2H screen only identified positive interactions at lower intensities with Rrp5, Utp10 and the N-terminal domain of Utp20 (Figure 4A and Supplementary Table S2). Rrp36 is required for the early cleavages at sites A0, A1 and A2 (46), while Rrp5 is involved in both SSU and LSU maturation (77). Utp10 and Utp20 are two large HEAT-repeat-containing proteins forming arch-like structures on the surface of the SSU-processome, thus bridging distant regions of the SSU-particle (24,40,42–44).
Figure 4.
Y2H assays reveal that Rrp9 is involved in a protein interaction network including Rrp36, Sgd1, Rrp5, Utp10 and Utp20. Y2H assays were performed as described in Materials and Methods and increasing 3-AT concentrations were used to test the stabilities of the detected interactions (0, 2, 5, 10, 20 and 40 mM 3-AT). Here, we illustrate the strongest and more reproducible interactions that we detected (results of the interaction tested in the opposite orientation are given in Supplementary Table S2). (A) A strong positive interaction between Rrp9 and both Rrp36 and Sgd1 (up to 40 mM), an interaction of medium stability (up to 4 mM 3-AT) between Rrp9 and Rrp5 and weak interactions of Rrp9 with Utp10 and the N-ter part of Utp20 (up to 2 mM 3-AT) are detected. (B) Sgd1 interacts with Rrp9, Rrp36, Sgd1, Utp10 and the N-ter domain of Utp20. Stronger interactions are detected between Sgd1 and both Rrp5 and Utp10. The R289A mutation in Rrp9 does not alter the Rrp9-Sgd1 Y2H interaction. (C) Rrp36 shows strong Y2H interactions with Rrp9, Rrp5, Utp10 and the N-ter part of Utp20 and a medium interaction with Sgd1. The Rrp9 R289A mutation destabilizes the Rrp36-Rrp9 Y2H interaction. The Rrp9-Rrp36 interaction takes place through the Rrp9 C-ter domain. (D) Interactions of Rrp5 were tested in Y2H assays with Rrp36 and Sgd1 domains defined on the basis of protein 2D structure predictions with the IUPred (79) and the PSIPRED V3.3 (78) web servers as illustrated in Supplementary Figure S2. Domains are schematically represented on the right side of the panel. The C-terminal domains of both Rrp36 and Sgd1, but not their N-terminal domains, interact with Rrp5. Two subfragments of the Sgd1 C-terminal domain (SD4 and SD5) are also able to interact with Rrp5. However, the stability of the interaction is reduced according to 3-AT resistance.
To assess the functional importance of the R289 residue of Rrp9, Y2H assays were performed by co-expression of Rrp9-R289A with Sgd1 (Figure 4B), Rrp36 (Figure 4C), Utp10 and the N-terminal part of Utp20, which are the only proteins with which we observed a significant interaction with WT Rrp9 (Supplementary Table S2). The Y2H interaction between Rrp9 and Rrp36 was significantly destabilized by the R289A mutation, as indicated by a marked reduction in resistance to 3-AT (Figure 4C), while that with Sgd1 was unaffected (Figure 4B). The weak interactions between WT Rrp9 and Utp10 or the N-terminal part of Utp20 were completely abolished for Rrp9–R289A (Supplementary Table S2C). Therefore, the R289A substitution markedly decreases the strength of the Rrp9-Rrp36 interaction, likely contributing to the observed pre-rRNA processing defects.
One reasonable hypothesis is that the Rrp9–Rrp36 interaction constitutes a link within a broader protein-protein interaction network. Therefore, we also tested whether the Rrp9 interaction partners interact with each other. In agreement with previous data, a positive Y2H assay was observed for the Rrp36–Sgd1 pair (Figure 4B, C), and both Rrp36 and Sgd1 gave positive Y2H results with Utp10. For the N-terminal part of Utp20, a strong positive signal was only observed for Rrp36 (Figure 4C, Supplementary Table S2C) (76). In addition, interestingly, Y2H assays also indicated the association of Rrp5 with both Sgd1 and Rrp36 (Figure 4B, C). The interaction of Rrp36 and Sgd1 with Rrp5 appeared stronger than their binding to Rrp9, as judged by 3-AT resistance.
We therefore propose that Rrp9 functions in the context of a protein interaction network, including Sgd1, Rrp36, Rrp5, Utp10 and Utp20, within the SSU-processome.
The Rrp9 β-propeller domain alone interacts with Rrp36 while intact Rrp9 is required for Sgd1 interaction
To further characterize this protein network, we used Y2H assays to map the protein domains involved (Figure 4C, D, Supplementary Figures S2 and S3). Based on its structure, Rrp9 was subdivided into its N-terminal and β-propeller domains. Sgd1 and Rrp36 lack reported 3D structures, so subdomains to be tested were delineated using protein-structure prediction programs, as well as predictions of structure flexibility (Supplementary Figure S2) (78,79). Two and four subdomains were defined for Rrp36 and Sgd1, respectively (Figure 4C, D, Supplementary Figures S2 and S3). Significantly, the C-terminal β-propeller domain of Rrp9, but not the N-terminal region, interacted strongly with Rrp36 (Figure 4C). However, the intact Rrp9 protein was required for Y2H interaction with Sgd1 (Supplementary Figure S3A, Figure 4B). The C-terminal region of Rrp36 was sufficient for Y2H interaction with Rrp5 (Figure 4D), while the full-length protein was necessary for Y2H interaction with Rrp9, as well as with Sgd1 (Supplementary Figure S3B, C). The C-terminal part of Sgd1 was sufficient for Y2H interaction with Rrp5 (Figure 4D). No clear Sgd1 interacting sub-domain could be identified for the Sgd1–Rrp9 and Sgd1–Rrp36 interactions (Supplementary Figure S3D, E). We failed determining specific regions of the very large, multi-domain Rrp5 protein that are responsible for the Rrp5–Rrp36 and Rrp5–Sgd1 interactions (data not shown).
Demonstration of the direct interaction of Rrp9 with Rrp36 and Sgd1, and of Rrp36 with Rrp5
To test whether the Y2H assays identified direct or indirect interactions, we performed GST-pull down assays, using purified GST-tagged proteins and in vitro translated proteins labeled with [35S]-methionine (Figure 5A–C).
Figure 5.
Some of the detected Y2H interactions correspond to direct interactions. (A–C) GST-tagged wild-type Rrp9, GST-tagged R289A mutant Rrp9, a GST-tagged non-relevant protein (GST-TRBP) or GST alone were bound to glutathione sepharose beads and incubated in the presence of Rrp36, Sgd1 domains or control proteins (Rtt106, CAT and β-Lactamase) labeled with [35S]-methionine. Protein–protein interactions were revealed by autoradiography. As shown by GST pull-down assays, interactions of Rrp36 (A) and the C-terminal domain of Sgd1 (B) with Rrp9 are direct interactions. Rrp36 also interacts directly with Rrp5 (C). The strength of the Rrp9-Rrp36 interaction is decreased by the Rrp9 R289A mutation by 20% (A), which is confirmative of Y2H results. (D) Schematic representation of the interaction network characterized for proteins Rrp9 and Sgd1, Rrp36, Rrp5, Utp10 and Utp20. Dashed arrows represent the Y2H interactions detected between proteins and/or protein sub-fragments (Figure 4, Supplementary Table S2). Plain red arrows represent direct interactions. Due to solubility problems, only three direct interactions were tested. Therefore, direct interactions of Rrp9, Rrp36, Rrp5 and Sgd1 with Utp10 and Utp20 were not tested.
GST-Rrp9 co-precipitated labeled Rrp36 but not the Rtt106 control protein. In the same conditions, no recovery was seen for GST alone or the control GST-tagged protein TRBP (Figure 5A). Treatment with RNase demonstrated that the Rrp9–Rrp36 interaction does not depend on the presence of RNA (data not shown). Full-length Sgd1 was poorly expressed in vitro, so we generated separate N-terminal and C-terminal fragments (Figure 5B). The C-terminal Sgd1 domain was recovered with GST-Rrp9, whereas binding was not seen for GST or GST-TRBP controls. The N-terminal domain of Sgd1 and the control Rtt106 protein were not recovered. Purified GST-Rrp9-R289A interacted with both Rrp36 and the Sgd1 C-terminal fragment (Figure 5A, B). However, recovery of Rrp36 with GST–Rrp9–R289A was reduced by 20% relative to WT Rrp9 protein (Figure 5A), consistent with the negative effect of the Rrp9 R289A mutation on the strength of the Rrp9–Rrp36 Y2H interaction (Figure 4C). A direct interaction was also observed between GST-Rrp5 and Rrp36, which was well recovered relative to CAT and β-lactamase proteins (Figure 5C). Due to solubility problems, we could not test whether the interactions detected for Utp10 and the Utp20 N-ter domain are direct and whether the Utp10 and Utp20 proteins interact through domains showing some structural homologies.
We conclude that Rrp9 interacts directly with both the C-terminal region of Sgd1 and with Rrp36 (the interaction destabilized by the Rrp9 R289A mutation). In addition, Rrp36 directly interacts with Rrp5, pointing to potential roles for these direct interactions in the efficiency of cleavage at sites A1 and A2 (Figure 5D).
Testing for synergistic negative interactions between Rrp9–R289A and mutations in U3
Association of the U3 snoRNP with the SSU-processome involves base-pairing between multiple sequences in the 5′ region of U3 and the pre-rRNA (Figure 1, Supplementary Table S3). These interactions are designated helices I–III, V and VI. We predicted that the Rrp9 protein network may reinforce the stability of these RNA–RNA interactions. To test this, we looked for synthetic lethality or synergistic negative interactions in strains expressing both Rrp9–R289A and U3 carrying mutations that alter binding to the pre-rRNA (Figures 6 and 8, panel A, Supplementary Table S3). We also investigated the effects of mutations in the U3 segments linking intermolecular helices, as these regions could be directly involved in RNA-protein interaction and/or SSU-processome architecture.
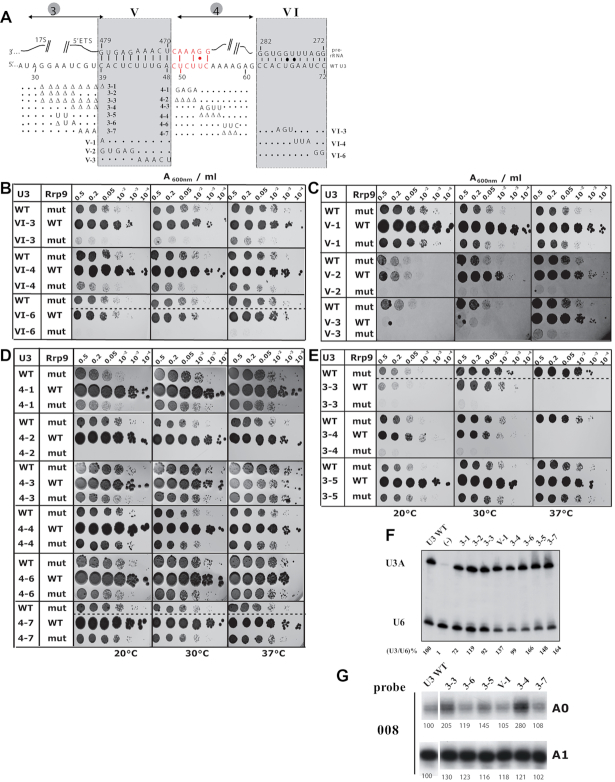
Figure 6.
The Rrp9–R289A mutation is synthetic lethal with several mutations destabilizing the heterologous interactions between U3 and the 5′-ETS sequence or reducing the size of the linker sequences (10,17,28,30). (A) Representation of the U3 snoRNA/pre-rRNA base-pair interaction. Helix V and VI and segments 3 and 4 are represented (10,44). The recently proposed extension of helix V (40,44) is shown in red. The mutations tested in the synthetic lethality screen are indicated. For the complete set of studied mutations, see Supplementary Table S3. (B–E) Yeast cells were transformed with plasmid expressing wild type Rrp9 (WT) or the R289A Rrp9 mutant (mut) and U3 snoRNA (Wild-type WT or mutants 3-3 to 3–5, 4-1 to 4–7, V-1 to V-3 and VI-3, VI-4 and VI-6) and grown on YPD medium after serial dilutions (2, 5, 20, 102, 103 or 104 X) at 20, 30 and 37°C (left, middle and right panels, respectively; for details see methods section). As a control, a comparison of cell growth on YPD and on YPG is shown for WT and all mutant cells in Supplementary Figure S4. (F) Northern-blot analysis of the in vivo stability of the different U3 mutants, expressed in JH84 yeast cells. Identities of the variant U3 RNAs are indicated on top of the autoradiogram. About 20 μg of total RNA extracted from the transformed JH84 cells grown at 30°C in YPD medium were fractionated by electrophoresis on a 6% denaturing polyacrylamide gel. After electrotransfer on a nylon membrane, the U3 snoRNA variants were detected by hybridization of the membrane with the 5′-end labeled U3 probe and the U6 oligonucleotide, complementary to U6 snRNA, was used as a control (Supplementary Table S1). The ratios between the U3 and U6 signals for each U3 mutant were calculated and expressed as a percentage of the ratio obtained for WT U3, arbitrary taken as 100%. (G) Primer extension analysis of total RNAs extracted from JH84 cells expressing various U3 mutants. Cells were grown on YPD medium and primer extensions were performed as described in Materials and Methods using equal amounts of total RNA (2 μg) and the labeled 008 probe complementary to 18S rRNA (Figure 3A). The autoradiogram signals corresponding to the A0- and A1-ending products are shown for cells expressing WT U3 or variant U3 as indicated on top of the autoradiogram. Intensities of the A0 and A1 bands were quantified and results are expressed as percentages of the intensities obtained for WT U3 arbitrary taken as 100%.
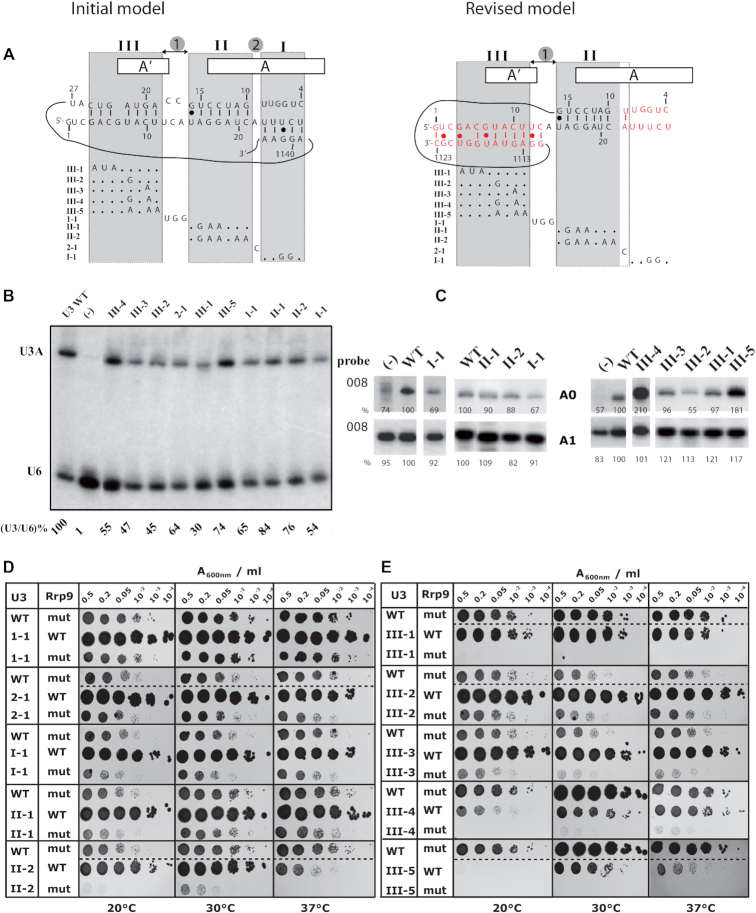
Figure 8.
The Rrp9-R289A mutation is synthetic lethal with some of the U3 snoRNA mutations destabilizing the formation of the helices formed between U3 and the 18S pre-rRNA (10,17,28,30). (A) Representation of the initial and new models for U3 snoRNA/pre-rRNA base-pair interactions. In the initial model, the three helices (I, II, III) and segments (1,2) are represented. In the new model, helix II which is unchanged and the new version of helix III (in red) are shown, as well as the opened helix I (in red) (40,44). The mutations tested in the synthetic lethality screen are indicated below each model. For the complete set of mutations, see Supplementary Table S3. (B) Northern-blot analysis of the in vivo stability of the different U3 mutants indicated on top of the autoradiogram. They were expressed in JH84 yeast cells grown at 30°C in YPD medium. About 20 μg of total RNA were fractionated by electrophoresis on a 6% denaturing polyacrylamide gel. After electrotransfer on a nylon membrane, the U3 snoRNA variants were detected by hybridization of the membrane with the 5′-end labeled U3 probe and for internal control, with the U6 oligonucleotide, complementary to U6 snRNA (Supplementary Table S1). For each U3 variant, the ratio between the U3 and U6 signals of the autoradiogram were calculated and expressed as a percentage of the value obtained for WT U3 arbitrary taken as 100%. (C) Primer extension analysis of total RNA extracted from JH84 cells expressing WT or variant U3 snoRNA, grown on YPD medium, The labeled 008 primer, complementary to 18S rRNA (Figure 3A) was used as the probe. The signals corresponding to the A0- and A1-ending products are shown and their relative amounts were expressed as percentages of the amount obtained for WT U3 arbitrary taken as 100%. (D, E) Yeast cells were transformed with plasmids expressing wild-type Rrp9 (WT) or R289A Rrp9 mutant (mut) and U3 snoRNA (Wild-type WT or mutants 1-1, 2-1, I-1, II-1, II-2, III-1 to III-5) and grown on YPD medium after serial dilutions (2, 5, 20, 102, 103 or 104 X) at 20, 30 and 37°C (left, middle and right panels, respectively; for details see methods section). A comparative analysis of cell growth on YPD and YPG media is shown in the Supplementary Figure S5.
Each of the selected U3 snoRNA variants was tested for effects on cell growth in the PGAL-Rrp9 strain following transfer to glucose medium (YPD) at 20, 30 and 37°C in the presence of WT Rrp9 (WT rows in Figures 6 and 8, Supplementary Figures S4 and S5, Supplementary Table S3). To test for synergistic effects, the U3 mutations were combined with Rrp9-R289A under the same conditions (mut rows in Figures 6 and 8, Supplementary Figures S4 and S5, Supplementary Table S3).
Mutations in the 5′-ETS interaction region of U3
The U3 region from position +30 to 72 forms helices V and VI, with the 5′-ETS region of the pre-rRNA. Both interactions were experimentally validated and are required for the early pre-rRNA cleavages at sites A0-A2 (10,30,40,44).
Deletion of helix VI is lethal, so we used previously described U3 mutations (U3 VI-3, U3 VI-4 and U3 VI-6) with more limited effects on growth (Figure 6A, B, Supplementary Figure S4A, B, Supplementary Table S3) (10). U3 VI-6 variant had the strongest growth phenotype alone, very similar to that found upon expression of the Rrp9–R289A protein alone, and combination of these two mutations led to synthetic lethality. Variant VI-3 alone had a weaker negative effect on cell growth, but markedly increased the negative effect of the Rrp9–R289A mutation. Finally, while the U3 VI-4 mutation had very limited effect on cell growth, it also reinforced the negative effect of the Rrp9 mutation. Therefore, the three mutations tested in segment VI either had a complete synthetic lethal effect or markedly increased the sickness of mutants carrying the Rrp9–R289A mutation (Figure 6B, Supplementary Figure S4B), suggesting some functional relationships between helix VI and Rrp9 during processome assembly or activity.
Substitution of a large region of helix V blocks pre-rRNA processing and growth (9,17), so we generated new mutations (U3 V-1, U3 V-2 and U3 V-3). The single nucleotide mutation in U3 V-1 had no effect on cell growth alone and did not significantly increase the negative effect of Rrp9–R289A (Figure 6C, Supplementary Figure S4C, Supplementary Table S3). In contrast, 5 nt substitutions of the 5′- or 3′-halves of segment V (U3 V-2 and U3 V-3, respectively) had marked effects on cell growth. U3 V-3 was almost inviable at 20 or 30°C, but supported robust growth at 37°C (Figure 6C, Supplementary Figure S4C, Supplementary Table S3), revealing a strong cryo-sensitive phenotype. Both U3 V-2 and U3 V-3 showed synergistic negative growth defects with Rrp9-R289A at all tested temperatures.
Segment 4 is located between helices V and VI and was proposed to play an important role in SSU-processome assembly (45). We therefore tested three substitutions (U3 mutants 4-1, U3 4-3, U3 4–6) as well as three short deletions that we previously described (U3 4-2, U3 4-4, U3 4-7) (Figure 6A and D) (10). All mutations had very limited effects on cell growth in the presence of the WT or mutated Rrp9, except for the U3 4-2 deletion that was fully synthetic lethal with Rrp9–R289A (Figure 6D and Supplementary Figure S4D). No synergistic negative effects were detected for the other mutants. We conclude that, in combination with Rrp9-R289A, the presence of the four 5′-terminal nucleotides of segment 4 proximal to intermolecular helix V is crucial, but this is independent of the nucleotide sequence.
Segment 3 lies 5′ to helix V and large deletions between +28 and +38 (renamed here as U3 3-1 and U3 3-2 for homogeneity) abolish growth (Supplementary Table S3) (28). We therefore generated five new mutations with short deletions (U3 3-3 and U3 3-4), tracts of base substitutions (U3 3–5 and U3 3-7) or both deletion and substitutions (U3 3–6) (Figure 6A). The 2 nt substitution in U3 3–5, showed no clear effects in the presence of Rrp9 or Rrp9–R289A (Figure 6E, Supplementary Figure S4E). In contrast, the 4 nt deletion in U3 3-4 was thermosensitive lethal at 37°C (Supplementary Table S3) and synergistically negative with Rrp9-R289A at other temperatures. An overlapping 8 nt deletion U3 3-3 was both cold-sensitive and thermosensitive and synergistically negative with Rrp9-R289A at 30°C (Figure 6E, Supplementary Figure S4E, Supplementary Table S3). All of the mutant U3 constructs were stably expressed (Figure 6F, Supplementary Table S3). Primer extension analyses were performed with primer 008 on 2 μg total RNA extracted from cells grown at 30°C. This primer allows detection of pre-rRNA intermediates cleaved at site A0 (22S and A0-A2 intermediates), as well as intermediates cleaved at site A1 (20S and mature 18S rRNA) (Figure 6G and 7B). Interestingly, U3 truncations in segment 3 (variants 3-1, 3-2, 3-3 and 3-4) markedly affected 18S processing with unusual amounts of processing intermediates cleaved at site A0 (Figures 6G and 7B). In contrast, the segment 3 substitutions U3 3-5, U3 3-6 and U3 3-7 did not induce clear defects, suggesting that the length of segment 3 may be more important for correct processing than its sequence.
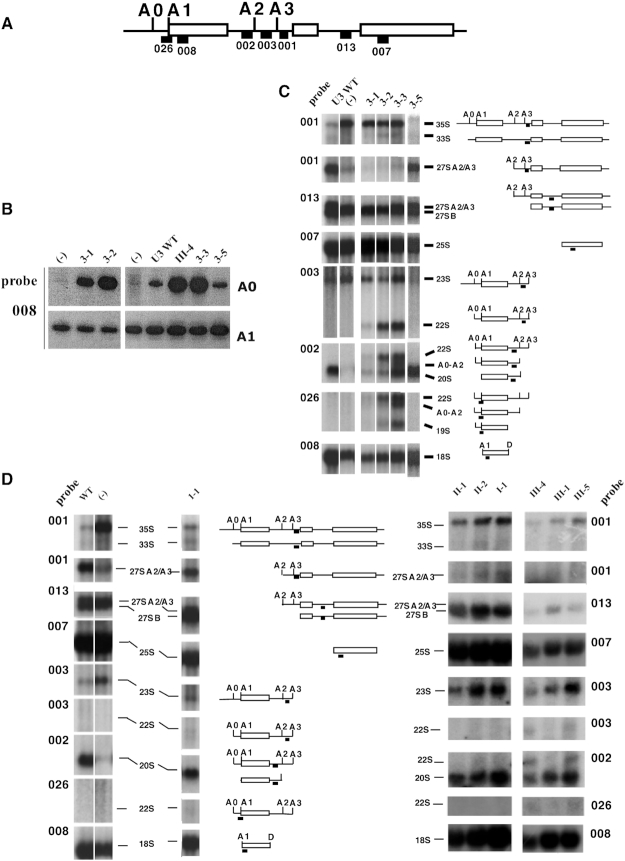
Figure 7.
Primer extension analysis and Northern-blot probing of pre-rRNA maturation intermediates for U3 variants mutated in helices I, II, III and their linking segments. (A) Target sites on the pre-rRNA of oligonucleotides used for northern-blotting. The positions of the probe target sequences are indicated with black squares. Probe 001 was used to detect the entire 35S pre-rRNA, the 33S RNA only cleaved at site A0 and the 27S A2 and 27S A3 intermediates containing the pre-rRNA sequences upstream of sites A2 or A3. Probe 013 was used for RNAs 27A2/A3 and 27B which 3′ extremities corresponds to sites A2, A3 and B, respectively. Probe 007 allowed evaluation of the amount of mature 25S rRNA. Probe 003 was used to detect 23S RNA cleaved at site A3 without cleavage at A0 and the aberrant accumulation of 22S RNA cleaved at site A0 and A3 without cleavage at sites A1 and A2. Probe 002 also detected the accumulation of 22S RNA, the aberrant apparition of an A0-A2 intermediates, and the 20S RNA, the normal last intermediate before 18S production. Probe 026 detected the aberrant 22S and A0-A2 intermediates as well as an aberrant 19S RNA in extracts of cells expressing the variant 3-2 and 3-3 U3 RNAs. Finally, probe 008 was used to detect mature 18S rRNA in Northern-blot experiments and to detect 5′-termini A0 and A1 in primer extension analyses, as described in Figure 6G. (B) Primer extension analyses with primer 008 performed on 2 μg of total RNA extracted from JH84 cells expressing WT or mutant U3 snoRNAs: (–) control assays with an empty plasmid; for other lanes, the identity of the U3 snoRNA expressed in the cells is indicated above the autoradiograms. Longer exposure time was used for detection of pre-rRNA intermediates cleaved at site A0 (22S and A0–A2 intermediates) than for detection of products cleaved at A1 (20S intermediates and mature 18S rRNA). (C, D) Autoradiograms of Northern-blot analyses performed on total RNA extracted from JH84 cells expressing WT or mutant U3 snoRNAs after growth on YPD medium. RNA were fractionated by electrophoresis in 1.2% agarose-6% formaldehyde gels. The parts of interest in the autoradiogram obtained for each probes are shown as distinct sub-panels, identified by the number of the probe used, indicated on the side (see Supplementary Table S1 for probe sequences). The identities of the U3 snoRNAs variants used in the experiments are given on top of the series of autoradiograms. Panel C corresponds to U3 mutated in segment 3 (variants 3-1, 3-2, 3-3 and 3–5). The first two lanes correspond to WT U3 and no U3 expression (–), respectively. Panel D corresponds to U3 mutated in linking segment 1 (1-1) and mutants of segments III, II and I, the latter previously proposed to form helix I. The pre-rRNA maturation intermediates identified by Northern-blot and their schematic representation are indicated on the right (C) or between (D) the panels.
The effects of the newly generated mutations in segment 3 were further analyzed for U3 3-1, U3 3-2, U3 3-3 and U3 3–5 (Figure 7). Northern analyses of the pre-rRNA maturation intermediates (Figure 7C) showed that transfer of the JH84-PGAL-U3 strain to glucose medium without complementing U3 expression had the expected phenotype, with loss of the products of the normally closely coupled cleavages at sites A0, A1 and A2: the 27SA2 and 20S pre-rRNAs. Both low levels of these pre-rRNAs intermediates, as well as mature 18S rRNA, were also observed in cells complemented with the U3 mutant U3 3-1, explaining its severe slow growth. The U3 3-2 and U3 3-3 strains showed significant accumulation of the 22S species. This observation is in agreement with data in Figures 6G and 7B showing an increased level of products cleaved at site A0. We also detected a 19S species that is apparently cleaved at sites A1 and A3 without A2 cleavage. This was unexpected, as the cleavages at sites A1 and A2 are generally tightly coupled (by an unknown mechanism) such that A1 is always cleaved prior to A2. These results suggest that U3, and specifically segment 3, plays a role in coupling cleavage at sites A1 at the 5′ end of 18S and A2 within ITS1, which are far distant in the primary sequence. In contrast, in agreement with data in Figure 6G, the pattern of maturation intermediates for the short U3-3-5 substitution closely resembled WT U3, supporting the conclusion that the length of segment 3 is more important than its actual sequence.
Mutations in the 18S rRNA-interaction region of U3
To assess the interactions between Rrp9 and U3-18S base-pairing, we generated a large collection of U3 mutations over the 5′ 27 nts of U3, including the segments previously proposed to form helices I, II and III (Figure 1 top, Figure 8A left), as well as a newly proposed version of helix III (Figure 1, bottom, Figure 8A, right) (40,44). All of the mutant U3 constructs were stably expressed, although some were less abundant than wild-type U3 (Figure 8B, Supplementary Table S3).
In helix I, we mutated the two central residues (U3 I-1; U25 and C26) proposed to base pair with the 18S rRNA on the 3′ side of the central pseudoknot (Figure 8A). This mutation would be expected to abolish helix I formation, but had no clear effect on cell growth (Figure 8D, Supplementary Table S3) or pre-rRNA processing (Figures 7D and 8C). In combination with Rrp9-R289A, U3 I-1 conferred a slight negative effect on growth (Figure 8D, Supplementary Figure S5B), possibly related to reduced U3 abundance (Figure 8B). The data demonstrate that helix I is not important for ribosome synthesis and are consistent with cryo-EM data in which it was not detected. This region of U3 is conserved in evolution, but appears to be more important for the U3 stability than pre-rRNA processing.
U3 residue A22 forms the short segment 2 in the original model (Figure 8A left), located between helices I and II, but was visualized base-paired with the 18S U9 residue by cryo-EM (Figure 8A right) (40,44). However, its substitution by a C residue (variant U3 2-1) had no effect on cell growth (Figure 8D, Supplementary Table S3) and did not increase the Rrp9–R289A growth defect (Figure 8D, Supplementary Figure S5B).
The importance of helix II is supported by genetic and structural analyses. However, a 3nt substitution (U3 II-1) had no detectable effect on cell growth (Figure 8D, Supplementary Table S3, Supplementary Figure S5B) and did not reinforce the Rrp9–R289A phenotype. A 5 nt substitution (U3 II-2) conferred a thermosensitive phenotype alone and was synthetic lethal with Rrp9 R289A at 20°C or 37°C and synergistically impaired at 30°C (Figure 8D, Supplementary Table S3, Supplementary Figure S5B). Primer extension revealed no accumulation of products cleaved at A0 in U3 II-1 or U3 II-2 (Figure 8C). However, northern analysis showed accumulation of 23S RNA, especially for U3 II-2 variant reflecting delayed cleavage at sites A0-A2 (Figure 7D).
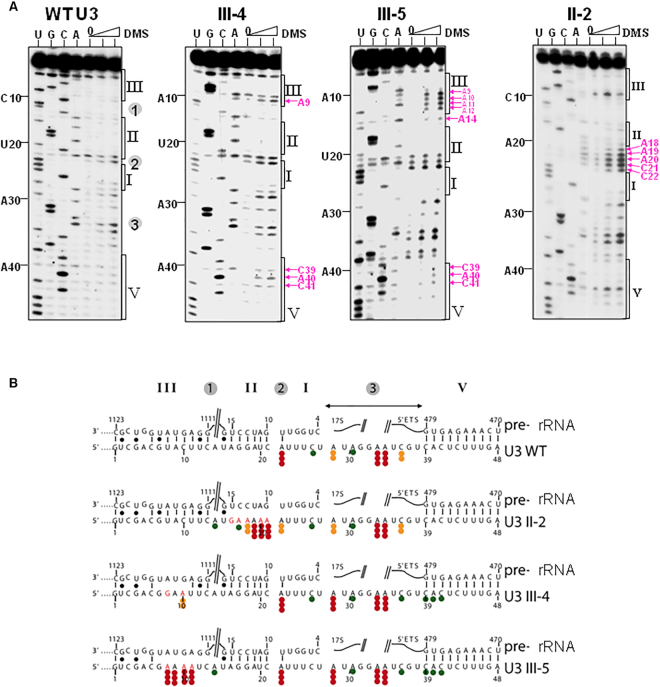
The structure of helix II was assessed in vivo by modification with DMS (Figure 9), which introduces methyl groups that act as blocks to primer extension in single stranded regions. Comparison of U3 with U3 II-2 confirmed the opening of the helix II structure in this variant with the appearance of numerous DMS modifications indicating non-base paired nucleotides. These analyses show that disruption of helix II is synthetic lethal with the Rrp9–R289A mutation, revealing the interdependence of helix II formation and protein Rrp9 integrity in SSU-processome activity.
Figure 9.
In vivo structural analysis with DMS of U3 snoRNA in cells expressing the U3 WT or the II-2, III-4 and III-5 variants supports the new model of U3/pre-rRNA interaction. (A) Transformed JH84 cells grown in YPD medium were modified in vivo with two DMS concentrations (30 and 60 mM) (28). The increase of DMS concentration (30–60 mM of DMS) is indicated by a triangle above the lanes. A control experiment was made in the absence of DMS (lane 0 in each panel). Methylated adenosines and cytidines in the extracted RNAs were identified by extension of primer 112 (Supplementary Table S1) with reverse transcriptase. Lanes U, G, C, A correspond to sequencing ladders obtained with the same primer. Position of nucleotides within U3 snoRNA or the pre-rRNA are indicated as well as the positions of segments involved in intermolecular helices I, II, III and V. Pink arrows indicate nucleotides, which are specifically methylated in the mutated U3 snoRNAs. (B) Positions of modified residues are shown on the new model of U3/pre-rRNA interaction. The intensities of DMS modifications are indicated by dots: three red dots, two orange dots, and one green dot represent strong, medium and low level of modification, respectively.
Segment 1 of U3 comprises the U12C13A14 trinucleotide joining helix II to helix III. Its substitution by a UGG sequence (U3 1-1) had no effect on cell growth (Figure 8D, Supplementary Table S3) and did not increase the phenotype of Rrp9 R289A (Figure 8D, Supplementary Figure S5B). This U3 1-1 mutation did neither clearly affect U3 levels (Figure 8B, Supplementary Table S3), nor accumulation of A0 cleaved pre-rRNAs or 18S rRNA (Figure 8C), nor other steps in pre-rRNA processing (Figure 7D). A base pair could be formed between U3 C13 and 18S G1111 in the revised version of helix III, but this seems to be dispensable for growth and pre-rRNA processing.
Five substitution mutations were generated in the helix III region of U3. Little effect on growth was seen for substitution of the 3 residues at the 5′-extremity (U3 III-1) or single nucleotide mutations in the 3′ region (U3 III-2 and U3 III-3) (Figure 8E, Supplementary Table S3, Supplementary Figure S5C). In contrast, double and triple base substitutions in the 3′ part of segment III (U3 III-4 and U3 III-5, respectively) generated clear growth defects, which were exacerbated at 20°C for both mutations and at 37°C for U3 III-5 (Figure 8E, Supplementary Table S3, Supplementary Figure S5C). Co-expression of Rrp9 R289A was synthetic lethal with U3 III-1, U3 III-4 or U3 III-5, but not with the single substitutions U3 III-2 and U3 III-3 (Figure 8E, Supplementary Figure S5C).
The mutants U3 III-1, U3 III-2, U3 III-3 and U3 III-4 all had a low in vivo stability compared to WT RNA (comprised between 55% for variant III-4 and 30% for variant III-1) (Figure 8B, Supplementary Table S3). In addition, primer extension analysis of RNAs extracted from cells expressing U3 III-4 or U3 III-5 (Figure 8C), showed accumulation of pre-rRNA cleaved at site A0, although the 18S 5′ end at A1 was not clearly altered (Figure 8C). Northern-blot analysis detected increased levels of 22S RNA, cleaved at sites A0 and A3 (Figure 7D). Altogether, this reflects a processing defect for variants U3 III-4 and U3 III-5 carrying base substitutions between positions 7 and 11, which explains the growth phenotypes (Figure 8E, Supplementary Figure S5C).
In recent cryo-EM structures, the 5′-terminal region of helix III is poorly base-paired (Figure 8A, right). In this structure, the 3 nt substitutions in U3 III-1 are not expected to dramatically change the stability of helix III. In contrast, these substitutions would be expected to dramatically reduce the stability of the previous version of helix III, with the disappearance of three Watson-Crick base-pairs (Figure 8A, left). In contrast, mutations U3 III-4 and U3 III-5 are predicted to destabilize helix III for both the initial and the revised versions of helix III (Figure 8A). In vivo structure probing of helix III formation with DMS showed that U3 III-5 mutation completely destabilized helix III, with the appearance of several DMS modification sites (Figure 9). More limited destabilization was observed for U3 III-4, consistent with its milder growth phenotype.
DISCUSSION
R289 in the Rrp9 β-propeller is required for efficient cleavages at sites A1 and A2
The SSU-processome UTP-A and UTP-B complexes contain a remarkable set of 19 proteins with β-propeller structures which functions in SSU-processome assembly (41,42,45,53). However, Rrp9 is the only U3 snoRNP protein with a β-propeller domain (26). Here, we report that the R289A substitution in this domain specifically impairs cleavage at two of the early pre-rRNA cleavage sites A1 and A2, while residue K290A does not seem to be important. The compensatory effect observed for the double mutant RK289/290AA might be explained by a redistribution of charges, which has often been observed in similar situations. Interestingly, this defect is more marked at lower temperature (20°C) and, consistently, growth of the strain is cold-sensitive. Interestingly, no clear defect was seen for cleavage at site A0, which is abolished by depletion of Rrp9 (12–14). Additionally, the R289A mutation interferes with the interaction between Rrp9 and Rrp36, and is synthetic lethal with mutations in the U3 snoRNA 5′-terminal region, which by themselves are not highly critical for growth. Mutation of other tested amino acids on the surface of the Rrp9 β-propeller domain generated no marked growth phenotype, neither individually nor in combination. This points to the high importance of the evolutionarily conserved R289 residue of Rrp9. Interestingly, the phenotype found for this Rrp9 R289A mutation is similar to those found for a dominant negative mutation of the Rrp3 helicase and the deletion of the Dhr1 helicase gene, which are both involved in pre-rRNA remodeling prior to early pre-rRNA cleavages (80–82). This suggests an involvement of Rrp9 R289A in the SSU-processome structural determinants required for efficient cleavages at sites A1 and A2.
R289 in Rrp9 is important to establish a protein-protein network within the SSU-processome
The Rrp9 R289 residue is located far from the A1 and A2 cleavage sites and the Utp24 RNase responsible for their cleavages, opening the question for a possible long distance effect through protein-protein interaction (Supplementary Figure S6). Accordingly, our Y2H assays and GST-pull down assays revealed an intricate protein network, involving the processome proteins Rrp9, Rrp36, Sgd1, Utp10, Utp20 as well as Rrp5, one of the few proteins involved in both 40S and 60S subunit production (74–77). We also demonstrated the direct interaction of Rrp9 with Rrp36 and with Sgd1. While the entire Rrp9 protein is required for direct interaction with the Sgd1 C-terminal domain, the β-propeller domain of Rrp9 was sufficient for direct interaction with the Rrp36 N-terminal domain. Importantly, the Rrp9 R289A mutation decreases the stability of the Rrp9-Rrp36 interaction. Little is known on Rrp36 except that it is required for pre-rRNA cleavage at sites A0–A2 (46). Of note, while in the Y2H map previously proposed by the Baserga team, Rrp9 gave positive signals with a limited number of processome proteins, Rrp36 gave a positive signal with a larger number of proteins, such as Krr1 required to maintain a critical SSU-processome conformation, or the DEAD-box helicase Dbp4 (76). In addition, we show here the direct interaction of the Rrp36 C-terminal domain with Rrp5. Therefore, the decreased Rrp9-Rrp36 interaction may play a crucial role in the Rrp9 R289A phenotype.
Mutations at Rrp9 R289 and in U3 segments base-paired with the pre-rRNA are synergistically negative
Formation of both helices V and VI between U3 and the 5′-ETS region of the pre-rRNA is essential at site A0, A1 and A2, likely reflecting key roles in processome assembly (9,10,30,40–42,44,45). Base substitutions in the 5′ or the 3′ half of the U3 stem V (U3 V-2 and U3 V-3, respectively, Supplementary Table S3) do not impair growth, whereas combination of these mutations with Rrp9 R289A mutation completely abolishes growth at any of the tested temperatures (Figure 6C, Supplementary Table S3). Similarly, partial substitutions in U3 stem VI (U3 VI-3 and U3 VI-6, Figure 6B, Supplementary Table S3) do not drastically affect cell growth, while their combination with the Rrp9 R289A mutation is lethal (Figure 6B). These data support a role for the protein interaction network including Rrp9 in formation and/or stability of the intermolecular helices V and VI.
In previous models (28,33,34), helix I was proposed to interact with the 3′ side of the 18S rRNA central pseudoknot. However, in contrast to helices II and III, helix I was neither observed in recent SSU-processome cryo-EM structures (40,44), nor supported by complementary mutations experiments (29). Consistent with these findings, U3 I-1 abolishes the possibility of forming helix I but did not result in any clear growth or pre-rRNA processing defects (Figure 8D, Supplementary Table S3) and its combination with the Rrp9 R289A mutation had almost no effect.
Helix II, formed between U3 (nts 10–16) and a 5′ region of 18S rRNA (nts 14–21), was detected in the cryo-EM structures (40,44) and supported by compensatory mutations (29). Substantial base substitutions in the U3 II-1 that were expected to lead to the formation of an alternative helix II and in the U3 II-2 constructs that led to a complete opening of helix II (variant U3 II-2) as demonstrated by DMS modification in vivo, did not have a drastic effect on cell growth at 30°C. Nevertheless, the marked thermosensitive phenotype observed for variant U3 II-2 reveals a functional importance of helix II at high growth temperature (Figure 8D, Supplementary Table S3). In addition, combination of the Rrp9 R289A mutation with U3 II-2 was synthetic lethal at 20 or 37°C and conferred very slow growth at 30°C, consistent with a much stricter requirement for the helix II sequence in the presence of weaker protein interactions.
In the revised interaction model (Figure 1 bottom), helix III is formed between the 5′ end of U3 and a region located downstream of the 18S rRNA (nts 1111–1123) (40,44). Notably, this 18S region is brought into proximity with the 5′ end of 18S in the mature 40S subunit, due to formation of the long-range, central pseudoknot. Substitutions in helix III had dramatic effects on cell growth (Figure 8E, Supplementary Table S3), which correlated with very high phylogenetic conservation of this region (box A’). Cells expressing the U3 III-4 and U3 III-5 variants accumulated the aberrant 22S maturation intermediate cleaved at sites A0 and A3. This indicates a greater inhibition of A1 and A2 cleavage by the Utp24 activity, relative to the (unidentified) endonuclease responsible for A0 cleavage. Notably, Rrp9 R289A which also reduces A1 and A2 cleavages relative to A0 was synthetic lethal with these U3 mutations. Rrp9 R289A was also synthetic lethal with the mutation in the more poorly base-paired, 5′ region of helix III (U3 III-1, Figure 8E). This may be explained by the observed lower in cellulo levels of U3 variants mutated in the helix III region, presumably linked to some defect in the U3 maturation or stability (Figure 8B, Supplementary Table S3) (70,83,84).
The spacing of U3 intermolecular helices is functionally important and their reduction is synthetic lethal with R289A in Rrp9
In vivo analysis of U3 snoRNA structure using DMS indicated that disruption of helix II (U3 II-2) or of helix III (U3 III-5) had limited effects on the flanking helices, showing that they can be formed independently (Figure 9). We only observed a very slight increase of accessibility of residues 39, 40 and 41 in helix V upon drastic mutation in helix III. However, the spacing between helices emerged as an important feature.
Segment 3 links the U3 regions that base-pair with the 5′-ETS and 18S regions, respectively. Truncation mutations U3 3-1 to U3 3–4 that shortened this distance reduced cell growth and impaired cleavage at sites A1 and A2 (Figure 6). Moreover, U3 3-3 and U3 3-4 were synthetic lethal in the presence of the Rrp9 R289A (Figure 6E). The importance of the length of U3 spacer sequences was also reported in Xenopus laevis (31,32,36,85). In cryo-EM structures, the SSU-processome 3D structure appears to be highly constraint by some rigid protein-protein interactions, as well as by RNA-protein interactions and the U3/pre-rRNA interaction (40,42,44). The truncations in segment 3 probably modify the availability of the U3 sequence for base-pair interaction with the pre-rRNA and for interaction with proteins. A highly surprising and interesting observation was made for U3 3-3. A large number of different mutations (>50) have been reported to create defects in pre-40S maturation. In all previous cases, loss of cleavage at site A1 was accompanied by loss of A2 cleavage. It was therefore quite unexpected that aberrant intermediates cleaved at site A0 and A2 were detected in U3 3-3 strains (Figure 7). The mechanism of coupling between A1 and A2 remains unclear, but is clearly influenced by the structure of the U3 snoRNP.
U3 segment 4 links helices V and VI, and deletion of the 5′ section of the spacer in variant U3 4-2, which alters the possibility to extend helix V according to EM data (40,44) was synthetic lethal with Rrp9 R289A (Figure 6D). Interestingly, none of the substitution or deletion mutants in segment 4 had an effect on cell growth in the presence of WT Rrp9 (Figure 6D). However, U3 variant 4-2, with a 4 nt deletion at the 5′ terminus of segment 4, was synthetic lethal with the Rrp9 R289A substitution. This indicates that the presence, but not the identity, of the 4 residues proximal to helix V is important for U3 function in pre-rRNA maturation.
Integrating the new data into the cryo-EM processome structure
The large heat domain proteins Utp10 and Utp20 very likely join the 90S complex before the A0 cleavage (86). Based on the SSU-processome structure (44,87), the U3 snoRNP is located inside a horseshoe formed by the extended Utp10 protein (Figure 10). Utp20 interacts with Utp10 and forms another smaller horseshoe on the opposite side (Figure 10). The β-propeller domain of Rrp9 faces the Utp10-Utp20 connection, with residue R289 pointing toward this junction (Figure 10). Neither Rrp36 nor Sgd1 were localized in the current 3D structures of processome complexes (24,40,42–44,87). Based on our interaction data, we predict the formation of an Rrp9–Rrp36–Sgd1 complex. Inspecting the published structures, there is sufficient space to accommodate Sgd1 and Rrp36 directly bound to Rrp9, between Rrp9 and the Utp10-Utp20 junction (Figure 10, Supplementary Figure S6). The Rrp5 protein chain is only partially identified in the established structures (40,44,87) (Figure 10). Its length and the fragments already localized, would be consistent with interactions between Rrp5 and Rrp36 associated with Rrp9.
Figure 10.
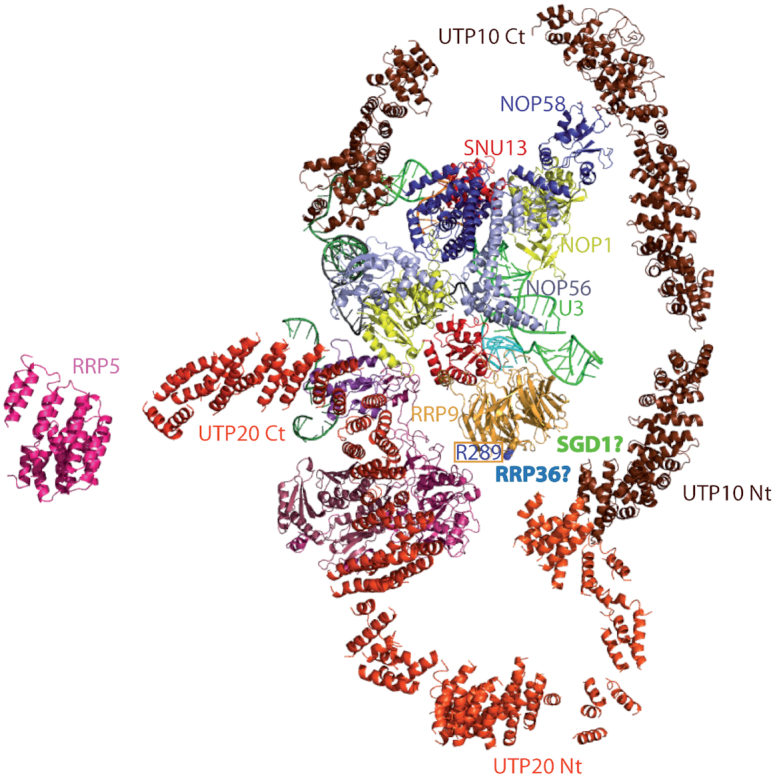
Putative localization of the Rrp36 and Sgd1 proteins, directly interacting with Rrp9 within the SSU-processome 3D structure (44). In addition to the U3 snoRNP (Snu13, Nop1, Nop56, Nop58), only the proteins Rrp9, Utp10, Utp20 are represented, together with a small part of Rrp5. Complete representation of all protein structures would otherwise mask the putative localization for the Rrp36 and Sgd1 proteins that we propose (44). The U3 snoRNA is colored in green, the pre-rRNA in gray.
U3 helix V is in close vicinity to proteins Utp7, Utp10, one of the Rrp9 partners, and protein Imp3 of the Mpp10-Imp3-Imp4 complex, stabilized by Utp3, which gave a positive Y2H signal with both Rrp36 and Sgd1 (10,30,63,76) (Figure 10, Supplementary Figures S6 and S7). Therefore, intricate connections between both RNA-protein interactions and long-distance protein-protein interactions involving Rrp9 might collaborate in formation and/or stabilization of the base-pair interaction of U3 with the 5′-ETS.
In conclusion, this study highlights the interplay between the protein-protein and RNA-RNA interaction networks required during formation of the SSU-processome and its function in pre-rRNA processing.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Yves Henry, Natacha Dreumont and members of the IMoPA team 1 for helpful discussions.
Notes
Present address: Kufel Joanna, Institute of Genetics and Biotechnology, Faculty of Biology, University of Warsaw, Warsaw, Poland.
Present address: Méreau Agnès, Université de Rennes, CNRS, Institut de Génétique et Développement de Rennes (IGDR), UMR6290, Rennes, France.
Present address: Senty-Ségault Véronique, Centre des Sciences du Goût et de l'Alimentation (CSGA), Université de Bourgogne Franche-Comté, UMR1324, INRAE, CNRS, Dijon, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Université de Lorraine; Bioengineering project of the Pôle de Recherche Scientifique et Technologique (PRST) Ingénierie Moléculaire et Thérapeutique-Santé (IMTS) of the Plan Etat Région Lorraine; N.M.G., N.R. and G.C. were fellows from the French Ministry of Education and Research; Research of M.R. was supported by the Université de Lorraine and the CNRS (‘Chaire d’Excellence’ program); Research in the Lab of D.L.J.L. is supported by the Belgian Fonds de la Recherche Scientifique (F.R.S./FNRS), the Université Libre de Bruxelles (ULB), the Région Wallonne, the Fonds Jean Brachet, the Internationale Brachet Stiftung, and the Epitran COST action (CA16120); D.T. is supported by a Wellcome Fellowship [077248] and work in the Wellcome Centre for Cell Biology is supported by a Centre Core grant [203149]. Funding for open access charge: Université de Lorraine, CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dragon F., Gallagher J.E.G., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al.. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brand R.C., Planta R.J.. The molecular weights of yeast ribosomal precursor RNAs. Mol. Biol. Rep. 1975; 2:321–325. [DOI] [PubMed] [Google Scholar]
- 3. Granneman S., Baserga S.J.. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004; 296:43–50. [DOI] [PubMed] [Google Scholar]
- 4. Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001; 20:3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachellerie J.P., Cavaillé J.. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997; 22:257–261. [DOI] [PubMed] [Google Scholar]
- 6. Ganot P., Bortolin M.L., Kiss T.. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997; 89:799–809. [DOI] [PubMed] [Google Scholar]
- 7. Hughes J.M., Ares M.. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991; 10:4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerbi S.A. Small nucleolar RNA. Biochem. Cell Biol. Biochim. Biol. Cell. 1995; 73:845–858. [DOI] [PubMed] [Google Scholar]
- 9. Beltrame M., Tollervey D.. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995; 14:4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marmier-Gourrier N., Cléry A., Schlotter F., Senty-Ségault V., Branlant C.. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011; 39:9731–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pluk H., Soffner J., Lührmann R., van Venrooij W.J.. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol. Cell Biol. 1998; 18:488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Venema J., Vos H.R., Faber A.W., van Venrooij W.J., Raué H.A.. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA. 2000; 6:1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cléry A., Senty-Ségault V., Leclerc F., Raué H.A., Branlant C.. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell Biol. 2007; 27:1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lukowiak A.A., Granneman S., Mattox S.A., Speckmann W.A., Jones K., Pluk H., van Venrooij W.J., Terns R.M., Terns M.P.. Interaction of the U3-55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55k. Nucleic Acids Res. 2000; 28:3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Lührmann R.. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004; 16:789–798. [DOI] [PubMed] [Google Scholar]
- 16. Weinberg R.A., Penman S.. Small molecular weight monodisperse nuclear RNA. J. Mol. Biol. 1968; 38:289–304. [DOI] [PubMed] [Google Scholar]
- 17. Beltrame M., Henry Y., Tollervey D.. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994; 22:4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watkins N.J., Ségault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Lührmann R.. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000; 103:457–466. [DOI] [PubMed] [Google Scholar]
- 19. Marmier-Gourrier N., Cléry A., Senty-Ségault V., Charpentier B., Schlotter F., Leclerc F., Fournier R., Branlant C.. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003; 9:821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Granneman S., Pruijn G.J.M., Horstman W., van Venrooij W.J., Luhrmann R., Watkins N.J.. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J. Biol. Chem. 2002; 277:48490–48500. [DOI] [PubMed] [Google Scholar]
- 21. Knox A.A., McKeegan K.S., Debieux C.M., Traynor A., Richardson H., Watkins N.J.. A weak C’ box renders U3 snoRNA levels dependent on hU3-55K binding. Mol. Cell. Biol. 2011; 31:2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wall M.A., Coleman D.E., Lee E., Iñiguez-Lluhi J.A., Posner B.A., Gilman A.G., Sprang S.R.. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995; 83:1047–1058. [DOI] [PubMed] [Google Scholar]
- 23. Gardner K.H., Anderson S.F., Coleman J.E.. Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain. Nat. Struct. Biol. 1995; 2:898–905. [DOI] [PubMed] [Google Scholar]
- 24. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 25. Smith T.F., Gaitatzes C., Saxena K., Neer E.J.. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999; 24:181–185. [DOI] [PubMed] [Google Scholar]
- 26. Zhang L., Lin J., Ye K.. Structural and functional analysis of the U3 snoRNA binding protein Rrp9. RNA. 2013; 19:701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang C., Lin J., Liu W., Chen X., Chen R., Ye K.. Structure of Utp21 tandem WD domain provides insight into the organization of the UTPB complex involved in ribosome synthesis. PLoS One. 2014; 9:e86540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Méreau A., Fournier R., Grégoire A., Mougin A., Fabrizio P., Lührmann R., Branlant C.. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997; 273:552–571. [DOI] [PubMed] [Google Scholar]
- 29. Sharma K., Tollervey D.. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell Biol. 1999; 19:6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dutca L.M., Gallagher J.E.G., Baserga S.J.. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011; 39:5164–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borovjagin A.V., Gerbi S.A.. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA N Y N. 2004; 10:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borovjagin A.V., Gerbi S.A.. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005; 33:4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beltrame M., Tollervey D.. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992; 11:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hughes J.M. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996; 259:645–654. [DOI] [PubMed] [Google Scholar]
- 35. Gerbi S.A., Borovjagin A.V., Ezrokhi M., Lange T.S.. Ribosome biogenesis: role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harb. Symp. Quant. Biol. 2001; 66:575–590. [DOI] [PubMed] [Google Scholar]
- 36. Borovjagin A.V., Gerbi S.A.. Xenopus U3 snoRNA GAC-Box A’ and Box A sequences play distinct functional roles in rRNA processing. Mol. Cell Biol. 2001; 21:6210–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phipps K.R., Charette J.M., Baserga S.J.. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA. 2011; 2:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D. et al.. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002; 10:105–115. [DOI] [PubMed] [Google Scholar]
- 39. Bernstein K.A., Gallagher J.E.G., Mitchell B.M., Granneman S., Baserga S.J.. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell. 2004; 3:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S.. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017; 24:944–953. [DOI] [PubMed] [Google Scholar]
- 41. Chaker-Margot M., Hunziker M., Barandun J., Dill B.D., Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015; 22:920–923. [DOI] [PubMed] [Google Scholar]
- 42. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R. et al.. Architecture of the 90S Pre-ribosome: A structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 43. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 44. Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., Chen R., Wang B., Zheng S., Zhang C. et al.. Molecular architecture of the 90S small subunit pre-ribosome. eLife. 2017; 6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang L., Wu C., Cai G., Chen S., Ye K.. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016; 30:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gérus M., Bonnart C., Caizergues-Ferrer M., Henry Y., Henras A.K.. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol. Cell Biol. 2010; 30:1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bleichert F., Granneman S., Osheim Y.N., Beyer A.L., Baserga S.J.. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9464–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tomecki R., Labno A., Drazkowska K., Cysewski D., Dziembowski A.. hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0. RNA Biol. 2015; 12:1010–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C.. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016; 44:5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krogan N.J., Peng W.-.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P. et al.. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004; 13:225–239. [DOI] [PubMed] [Google Scholar]
- 51. Dosil M., Bustelo X.R.. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004; 279:37385–37397. [DOI] [PubMed] [Google Scholar]
- 52. Pérez-Fernández J., Román A., De Las Rivas J., Bustelo X.R., Dosil M.. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007; 27:5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pérez-Fernández J., Martín-Marcos P., Dosil M.. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011; 39:8105–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wery M., Ruidant S., Schillewaert S., Leporé N., Lafontaine D.L.J.. The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009; 15:406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hunziker M., Barandun J., Buzovetsky O., Steckler C., Molina H., Klinge S.. Conformational switches control early maturation of the eukaryotic small ribosomal subunit. eLife. 2019; 8:e45185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gutell R.R., Larsen N., Woese C.R.. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994; 58:10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Neefs J.M., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R.. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993; 21:3025–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dunbar D.A., Wormsley S., Agentis T.M., Baserga S.J.. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell Biol. 1997; 17:5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gallagher J.E.G., Baserga S.J.. Two-hybrid Mpp10p interaction-defective Imp4 proteins are not interaction defective in vivo but do confer specific pre-rRNA processing defects in Saccharomyces cerevisiae. Nucleic Acids Res. 2004; 32:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wormsley S., Samarsky D.A., Fournier M.J., Baserga S.J.. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001; 7:904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee S.J., Baserga S.J.. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell Biol. 1999; 19:5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shah B.N., Liu X., Correll C.C.. Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing. RNA. 2013; 19:1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gérczei T., Correll C.C.. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15301–15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Diebold M.-.L., Fribourg S., Koch M., Metzger T., Romier C.. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011; 175:178–188. [DOI] [PubMed] [Google Scholar]
- 65. Stotz A., Linder P.. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990; 95:91–98. [DOI] [PubMed] [Google Scholar]
- 66. Gietz D., St Jean A., Woods R.A., Schiestl R.H.. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992; 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ségault V., Mougin A., Grégoire A., Banroques J., Branlant C.. An experimental study of Saccharomyces cerevisiae U3 snRNA conformation in solution. Nucleic Acids Res. 1992; 20:3443–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kramer W., Drutsa V., Jansen H.W., Kramer B., Pflugfelder M., Fritz H.J.. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984; 12:9441–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rederstorff M., Bernhart S.H., Tanzer A., Zywicki M., Perfler K., Lukasser M., Hofacker I.L., Hüttenhofer A.. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010; 38:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mougin A., Grégoire A., Banroques J., Ségault V., Fournier R., Brulé F., Chevrier-Miller M., Branlant C.. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996; 2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 71. Rothé B., Back R., Quinternet M., Bizarro J., Robert M.-.C., Blaud M., Romier C., Manival X., Charpentier B., Bertrand E. et al.. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res. 2014; 42:2015–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clerget G., Bourguignon-Igel V., Rederstorff M.. Alcoholic precipitation of small non-coding RNAs. Methods Mol. Biol. 2015; 1296:11–16. [DOI] [PubMed] [Google Scholar]
- 73. Schmitt M.E., Clayton D.A.. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1993; 13:7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lim Y.H., Charette J.M., Baserga S.J.. Assembling a protein-protein interaction map of the SSU processome from existing datasets. PLoS One. 2011; 6:e17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tarassov K., Messier V., Landry C.R., Radinovic S., Serna Molina M.M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S.W.. An in vivo map of the yeast protein interactome. Science. 2008; 320:1465–1470. [DOI] [PubMed] [Google Scholar]
- 76. Vincent N.G., Charette J.M., Baserga S.J.. The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA. 2018; 24:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khoshnevis S., Askenasy I., Johnson M.C., Dattolo M.D., Young-Erdos C.L., Stroupe M.E., Karbstein K.. The DEAD-box protein Rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from Pre-40S ribosomes to allow for 60S maturation. PLoS Biol. 2016; 14:e1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T.. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013; 41:W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dosztányi Z., Csizmok V., Tompa P., Simon I.. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinforma Oxf. Engl. 2005; 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- 80. Roychowdhury A., Joret C., Bourgeois G., Heurgué-Hamard V., Lafontaine D.L.J., Graille M.. The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis. Nucleic Acids Res. 2019; 47:7548–7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhu J., Liu X., Anjos M., Correll C.C., Johnson A.W.. Utp14 recruits and activates the RNA helicase Dhr1 To undock U3 snoRNA from the Preribosome. Mol. Cell Biol. 2016; 36:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. O’Day C.L., Chavanikamannil F., Abelson J.. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996; 24:3201–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Brulé F., Grégoire A., Ségault V., Mougin A., Branlant C.. Secondary structure conservation of the U3 small nucleolar RNA introns in Saccharomyces. C. R. Acad. Sci. III. 1995; 318:1197–1206. [PubMed] [Google Scholar]
- 84. Rothé B., Manival X., Rolland N., Charron C., Senty-Ségault V., Branlant C., Charpentier B.. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 2017; 45:7455–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Borovjagin A.V., Gerbi S.A.. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000; 300:57–74. [DOI] [PubMed] [Google Scholar]
- 86. Dez C., Dlakić M., Tollervey D.. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007; 13:1516–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Heuer A., Thomson E., Schmidt C., Berninghausen O., Becker T., Hurt E., Beckmann R.. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. eLife. 2017; 6:e30189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.