Abstract
Some transcription factors that specifically bind double-stranded DNA appear to also function as RNA-binding proteins. Here, we demonstrate that the transcription factor Sox2 is able to directly bind RNA in vitro as well as in mouse and human cells. Sox2 targets RNA via a 60-amino-acid RNA binding motif (RBM) positioned C-terminally of the DNA binding high mobility group (HMG) box. Sox2 can associate with RNA and DNA simultaneously to form ternary RNA/Sox2/DNA complexes. Deletion of the RBM does not affect selection of target genes but mitigates binding to pluripotency related transcripts, switches exon usage and impairs the reprogramming of somatic cells to a pluripotent state. Our findings designate Sox2 as a multi-functional factor that associates with RNA whilst binding to cognate DNA sequences, suggesting that it may co-transcriptionally regulate RNA metabolism during somatic cell reprogramming.
INTRODUCTION
Multi-functionality and cell context specific activities are features of many nucleic acid-binding proteins. Global nucleic acid interaction assays revealed that ∼8.1% of human nucleic acid-binding proteins are dual DNA- and RNA- binding proteins (DRBPs) (1). These DRBPs are widespread, and are implicated in biological processes such as mRNA processing, transcriptional regulation, DNA replication, DNA repair, stress response and apoptosis (1). DRBPs primarily act by initiating mRNA synthesis and by regulating alternative splicing (AS). Several transcriptional and chromatin regulators have been reported to bind RNA with diverse consequences on gene regulation. RNAs transcribed from cis-regulatory elements contribute to enhanced YY1 occupancy at these sites and thus consolidate gene expression programs (2). PARP1 associates with nucleosomes harbouring exonic H3K4me3 marks, binds nascent RNA and co-transcriptionally regulates splicing by recruiting splicing factors (3,4). The growth arrest-specific 5 (Gas5) noncoding RNA competitively binds to the DNA-binding domain of the glucocorticoid receptor and modulates its transcriptional activity (5). However, there are few concrete examples as to the mechanism how dual RNA/DNA binders affect RNA metabolism and chromatin regulation, and the impact of DRBPs on gene regulation in a biologically relevant context is still largely unknown.
Sox transcription factors (TFs) are well-established regulators of cell fate determination during embryonic development and cellular reprogramming (6,7). They are characterized by a highly conserved 79-amino-acid-long high-mobility group (HMG) box domain that binds short and degenerate DNA sequences 5′-(A/T)(A/T)CAA(A/T)G-3′ through the minor groove of the DNA (8). Mammalian species contain 20 Sox TFs that are classified into nine groups (A, B1, B2, C, D, E, F, G, H) based on their amino acid sequences. The SoxB1 group includes Sox1, Sox2 and Sox3. SoxB1 group members are composed of short N-terminal regions, HMG boxes, and long C-terminal sequences including a conserved ‘group B homology’ comprised mainly of basic amino acids immediately adjacent to the HMG box (9). Sox2 is the best studied SoxB1 protein for its prominent roles in a large array of cell and tissue types including pluripotent and neural stem cells (10). The HMG box not only mediates DNA binding but also doubles as a protein-protein interaction interface for different cofactors including the Pit-Oct-Unc (POU) family factor Oct4 (11,12). The formation of Oct4/Sox2 complexes on the enhancers of pluripotency gene loci is essential for the induction and maintenance of pluripotent stem cells (PSC) (13–15). The staggeringly diverse and often seemingly opposing regulatory roles of Sox2, i.e. in the maintenance of pluripotency as well as in the regulation of neural differentiation, are believed to rely on dynamically changing and cell-context dependent molecular interactions.
Several Sry-related box (Sox) proteins have been reported to interface with RNA. In oocytes of Xenopus tropicalis, Sox9 interacts with the ribonucleoprotein matrix of the lampbrush chromosomes lateral loops in an RNA-dependent manner and regulates post-transcriptional processes (16). Sry and Sox6 colocalize with splicing factors in the nucleus and block pre-mRNA splicing in HeLa cells (17). Recent work suggested that Sox2 is also a node of the RNA network. Ng et al. reported that the lncRNA_ES1 and lncRNA_ES2 play a role in the maintenance of pluripotency in human embryonic stem cells (hESCs) in a Sox2-dependent manner (18). The lncRNA RMST was found to be necessary for Sox2 to bind to a subset of promoter regions of neurogenesis-relevant TFs, leading to the regulation of genes critical for neural stem cell development (19). Bioinformatic analyses predicted that Sox2 interacts with the 5′-end of lncRNA_ES1 (18,20). RNA immunoprecipitation with hESC and neural stem cell lysate revealed Sox2 in the interactome with several RNAs (18,19).
Given the implications that Sox2 links regulatory networks involving DNAs and RNAs, we sought to elucidate whether Sox2 is a DRBP and how its DNA- and RNA-binding activities coordinate somatic cell reprogramming to pluripotency. We set out to scrutinise the proposed RNA-binding function of Sox2 to tackle several unresolved questions. Does Sox2 bind RNA directly? Which domains of Sox2 mediate the interaction? How DNA and RNA binding functions can be reconciled in the process of cellular reprogramming? To address these, we used chemilumilescent photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP), systematic evolution of ligands by exponential enrichment (SELEX), electrophoretic mobility shift assays (EMSA) and RNA immunoprecipitation assays (RIP) to show that Sox2 is an RNA-binding protein with a preference for G/C-rich sequences. In the C-terminal region of Sox2, we identify a 60-residue-long RNA binding motif (RBM) encompassing the group B homology domain that directs the preference for RNA sequences. We show that Sox2 employs the RBM and HMG domain to associate with RNA and DNA simultaneously. Deletion of the RBM significantly reduces the ability of Sox2 to induce pluripotency. Co-immunoprecipitation assays show that Sox2 interacts with Zcchc8, Skiv2l2, SFPQ and hnRNP K, which are involved in RNA processing/splicing, and deletion of the RBM does not perturb this association. Nevertheless, deletion of the RBM causes a series of pluripotency-related AS changes. This study highlights a hitherto unknown interplay of distinct Sox2 domains with RNA during somatic cell reprogramming.
MATERIALS AND METHODS
Preparation of wild-type and truncated Sox2 proteins
Mouse Sox2 (Gene ID: 20674), Sox2-ΔHMG, Sox2(1–180), Sox2(1–120), Sox2(120–319), Sox2(180–319), Sox2-HMG, Sox2-ΔRBM, Sox11 (Gene ID: 20666) and Sox11N2C cDNAs were amplified with Phusion® High-Fidelity DNA Polymerase (NEB, Cat#M0530S) and cloned into the pET28a (Novagen, Cat# 69864-3) vector with NdeI and XhoI restriction sites. Primers used are shown in Supplementary Table S4. pETG20a-Sox1 (Gene ID: 20664) was kindly provided by Dr Calista Keow Leng Ng. All recombinant proteins were expressed in Escherichia coli Rosetta (DE3) cells (Tiangen, Cat#CB108-02). The transformed cells were cultivated at 37°C until reaching OD600 = 0.4–0.6 and protein expression was induced with 0.2 mM IPTG for Sox2 variants and Sox11 or 0.5 mM IPTG for Sox1 at 30°C for 4 h. Cells were harvested by centrifugation, resuspended in a buffer containing 8 M urea, 50 mM Tris–HCl pH 7.4, 150 mM NaCl and 1 mM PMSF and disrupted by ultrasonification (Xinzhi JY92 Ultrasonic homogenizer; 400 W, 20 min for 30 ml suspension). The proteins were captured with Ni-NTA resin (ThermoFisher Scientific, Cat#88222) under denaturing condition. Purified proteins were subsequently refolded by gradually dialyzing to 6, 4 and 2 M urea followed by buffer exchange with PD-10 desalting columns (GE Healthcare, Cat#17-0851-01) and refolding buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl (Sox2 variants and Sox11) or 500 mM NaCl (Sox1), 1 mM PMSF). The His6-TrxA was removed from Sox1 by digestion with TEV (tobacco etch virus) protease (w/w = 1:5) at 4°C for 16 h and purification with a RESOURCE™ S column (GE Healthcare, Cat#17-1180-01) followed by desalting with PD-10 columns (GE Healthcare, Cat#17-0851-01). Protein concentrations were determined by measuring the UV absorbance at 280 nm or by measuring the intensity of bands after SDS-PAGE.
Systematic evolution of ligands by exponential enrichment (SELEX)
Oligonucleotides containing a random 25 bp sequence flanked by two primer binding sites [5′-TGGGCACTATTTATATCAAC(N)25AATGTCGTTGGTGGCCC-3′] were procured from BGI and amplified by nine cycles of PCR as described (21,22) with primers listed in Supplementary Table S5 followed by gel extraction (GeneJET gel extraction kit, ThermoFisher Scientific, Cat#K0691). The RNA library was transcribed using the T7 RNA polymerase (ThermoFisher Scientific, Cat#EP0111) and purified from a 10% denaturing polyacrylamide gel after DNase treatment. Binding reactions were performed in RNA binding buffer (10 mM Tris–HCl (pH 7.4), 1 mM MgCl2, 10 mM ZnCl2, 1 mM DTT, 10 mM KCl, 5% glycerol, 1 unit/μl RNase inhibitor). Before binding, the purified RNA library was first incubated with anti-His conjugated Dynabeads® Protein G (ThermoFisher Scientific, Cat#10004D) in RNA binding buffer at 4°C for 1 h to eliminate RNA un-specifically binding to the beads. Around 1.4 nmol unbound RNA was collected and gently mixed with 41.6–55.6 pmol recombinant His6-Sox2 immobilized to anti-His antibody conjugated Dynabeads® Protein G at 4°C for 2 h in a 200 μl reaction volume. After six times of washing with RNA binding buffer, the Sox2 bound RNA was eluted with TRIzol reagent (ThermoFisher Scientific, Cat#15596026), precipitated with 2.5 volumes of ice-cold 100% ethanol and resuspended into RNase- and DNase-free water (Tiangen, Cat# RT121-01). The RNA was then reverse-transcribed with M-MLV Reverse Transcriptase (ThermoFisher Scientific, Cat#28025013) and amplified by nine cycles of PCR using primers encoding a T7 promoter (underlined) in the forward primer (5′-CGCGGATCCTAATACGACTCACTATAGGGGCCACCAACGACATT-3′) and a reverse primer (5′-CCCGACACCCGCGG ATCCATGGGCACTATTTATATCAAC-3′). The DNA was subjected to another round of SELEX. Selection stringency was increased in the last six rounds by increasing the amount of RNA from 1.4 to 7.7 nmol. Counterselections were done in Round 5 and Round 8. After the 12th round of SELEX, the PCR products were subcloned into vector pTZ57R/T and transformed into chemically competent Top10 E. coli cells (Tiangen, Cat#CB104-02). Seventy-two colonies were picked for plasmid DNA extraction and Sanger sequencing.
Mouse induced pluripotent stem cell (iPSC) generation
OG2 mouse embryonic fibroblasts (OG2 MEFs) were obtained from E13.5 mouse embryos carrying the Oct4-GFP transgene (JAX ID #004654) following standard procedures (23). MEFs were maintained in DMEM/high glucose (Hyclone, Cat#SH30022-2B) containing 10% fetal bovine serum (FBS) (Natocor, Cat#SFBE), 1× GlutaMax (Gibco, Cat#35050079), 1× MEM nonessential amino acids (Corning, Cat#25-025-CI), and 0.5× penicillin/streptomycin (Hyclone, Cat#SV30010). Virus producing Plat-E cells were maintained in DMEM/high glucose containing 10% FBS. Around 15 000 OG2 MEFs in a well of 12-well plates were transduced twice with 0.5 ml of each viral supernatants in the presence of polybrene. Each reprogramming condition was assessed with at least three biological repeats for each individual experiment. After the second transduction, the medium was changed to mouse embryonic stem cells (mESCs) medium (DMEM/high glucose (Hyclone, Cat#SH30022-2B) containing 15% FBS (Natocor, Cat#SFBE), 1× GlutaMax (Gibco, Cat#35050079), 1× MEM nonessential amino acids (Corning, Cat#25-025-CI), 1× sodium pyruvate (Corning, Cat#25-000-CI), 0.055 mM 1× 2-Mercaptoethanol (Gibco, Cat#21985023), 0.5× penicillin/streptomycin (Hyclone, Cat#SV30010) with the presence of 1000 U/ml leukemia inhibitory factor (LIF, produced at the GIBH core facility)) at day 1 post-infection and renewed daily. At day 12 post-infection, GFP+ colonies were counted.
Mouse embryonic stem cell (mESC) culture
The OG2 mESCs were plated at feeder-free and gelatin-coated 6-cm plates, and were grown in standard mouse ESC medium containing DMEM (Hyclone, Cat#SH30022-2B), 15% FBS (Natocor, Cat#SFBE), 1× recombinant leukemia inhibitory factor (LIF, produced at the GIBH core facility), 0.055 mM β-mercaptoethanol (Gibco, Cat#21985023), 0.5× penicillin/streptomycin (Hyclone, Cat#SV30010), 1× GlutaMax (Gibco, Cat#35050079), 1× pyruvate sodium (Corning, Cat#25-000-CI) and 1× MEM non-essential amino acids (Corning, Cat#25-025-CI). To avoid differentiation, we also added two inhibitors: 3 μM ChIR099021 (Selleck; #S2924-25mg) and 1 μM PD0325901 (Selleck; #S1036-25mg). Cells used for different analyses were harvested in exponential growth phase (∼70% confluent).
PAR-CLIP-biotin chemiluminescent nucleic acid detection and PAR-CLIP sequencing
The experiment was performed as described (24). In brief, HEK293T cells were transfected with pMX-FLAG-Sox2 plasmid with 1 μg/μl polyethylenimine (Polyscience, Cat#23966). Twenty-four hours after transfection, the cells were treated with 0.2 mM 4-thiouridine (4-SU) (Sigma-Aldrich, Cat#T4509) for 16 h. OG2 mESCs and reprogramming cells were treated with 0.2 mM 4-SU for 16 h when cells grew to 50% confluence or on day 5 after transfected with pMX-FLAG3-Sox2, pMX-Oct4, pMX-c-Myc and pMX-Klf4 plasmids. Cells were crosslinked with 0.4 J/cm2 of 365 nm UV light and harvested by centrifugation at 500 g for 5 min at 4°C. Lysis was performed in 300 μl (for each plate) NP40 lysis buffer (50 mM HEPES pH 7.5, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% NP40, 0.5 mM DTT, RNase inhibitor (ThermoFisher Scientific, Cat#EO0384) and protease inhibitor cocktail (Roche, Cat#11873580001)) by 10 min incubation on ice and followed homogenization with a 0.4 mm syringe needle. The cleared lysates were digested with RNase T1 (Sigma-Aldrich, Cat#R1003) at a final concentration of 1 U/ml at 22°C for 15 min and then were incubated with Anti-FLAG® M2 Magnetic Beads (Sigma, Cat#F3165) (10 μl for each plate) for 2 h at 4°C. The beads were washed with washing buffer (50 mM HEPES–KOH pH 7.5, 300 mM KCl, 0.05% NP40, 0.5 mM DTT and protease inhibitor cocktail) at 4°C and incubated in washing buffer with RNase T1 at a final concentration of 10 U/ml for 15 min at 22°C. After washing with high-salt washing buffer (50 mM HEPES–KOH pH 7.5, 500 mM KCl, 0.05% NP40, 0.5 mM DTT and protease inhibitor cocktail) at 4°C, beads were dephosphorylated in dephosphorylation buffer (50 mM Tris–HCl, pH 7.9, 100 mM NaCl, 10 mM MgCl2 and 1 mM DTT supplemented with calf intestinal alkaline phosphatase at a final concentration of 0.5 U/ml) at 37°C for 10 min. The beads were then washed with phosphatase washing buffer (50 mM Tris–HCl, pH 7.5, 20 mM EGTA and 0.5% NP40) and with T4 polynucleotide kinase buffer without DTT (50 mM Tris–HCl pH 7.5, 50 mM NaCl and 10 mM MgCl2). For PAR-CLIP-biotin chemiluminescent nucleic acid detection, RNAs co-immunoprecipitated with the proteins were labelled with biotin according to the instructions of the Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFisher Scientific, Cat#89880). After labelling, beads were washed with washing buffer and incubated with SDS-PAGE loading buffer. The beads were heated at 95°C for 5 min and the released RNA–protein complexes were separated in NuPAGE® Novex 4–12% Bis–Tris Gel in Novex™ NuPAGE™ MOPS SDS Running Buffer followed by western blotting or chemiluminescence nucleic acid detection. For PAR-CLIP sequencing, RNAs co-immunoprecipitated with proteins were phosphorylated with 1 U/μl T4 polynucleotide kinase (NEB, Cat#M0201L) for 1 h at 37°C. Samples were washed, eluted and separated in NuPAGE® Novex 4–12% Bis–Tris Gel in Novex™ NuPAGE™ MOPS SDS Running Buffer. RNA–protein complexes were then transferred onto nitrocellulose membranes. One membrane was applied for PAR-CLIP-biotin chemiluminescent nucleic acid detection to identify the position of the captured RNAs. The other membrane was cut into small slices and processed with 4 μg/μl of proteinase K (ThermoFisher Scientific, Cat#25530049) at 37°C for 20 min. Thereafter, equal volume of urea buffer (100 mM Tris–HCl pH 7.4, 50 mM NaCl, 10 mM EDTA and 7 M urea) was added and incubated for 20 min at 37°C. RNAs were extracted with acid phenol/CHCl3 (pH 4.3–4.7) and was precipitated in 75% ethanol supplemented with glycogen and NaAcO3 (pH 5.5) at −20°C overnight. The extracted RNAs were first ligated with 3′ RNA adapter and subsequent 5′ RNA adapter. cDNAs were synthesized with SuperScript™ III Reverse Transcriptase (ThermoFisher Scientific, Cat#18080093) and amplified by 18 cycles of PCR. PCR products were screened based on size using from PAGE gels according to instructions of TruSeq® Small RNA Sample Prep Kit (Illumina, Cat#15019892). The purified library products were evaluated using the Agilent 2200 TapeStation and diluted to 10 pM for cluster generation and high-throughput sequencing (50 bp single end) on HiSeq 2500 (Illumina).
Electrophoretic mobility shift assay (EMSA)
RNA binding reactions were carried out at 4°C for 2 h in a 20 μl system containing 10 mM Tris–HCl pH 7.4, 1 mM MgCl2, 10 mM ZnCl2, 1 mM DTT, 10 mM KCl, 5% glycerol, 1 U/μl RNase inhibitor, 50 nM Cy3- or Cy5-labelled RNA and indicated amounts of proteins with or without oligonucleotide competitors. DNA binding reactions were performed as previously described (25,26). Reaction samples were then resolved on a 8% native polyacrylamide gel in dark in 0.5× TAE at 4°C, 200 V. Signals were visualized with a FLA-7000 image reader (FUJIFILM/GE Healthcare) and quantified using the ImageQuant TL software (GE Healthcare).
RNA immunoprecipitation assay (RIP)
In vitro RIP was performed with purified His6-Sox2. After washing with buffer 1 (10 mM Tris–HCl pH 7.4, 1 mM MgCl2, 10 mM ZnCl2, 1 mM DTT, 10 mM KCl, 5% glycerol, 1 unit/μl RNase inhibitor) for three times, 250 μl Dynabeads® Protein G (Lifetech, Cat#10004D) was incubated with 50 μl of anti-His antibodies (CW0083, 1 mg/ml) with gentle rotating at room temperature (RT) for 1 h. Wash with 1 ml buffer 1 for three times and then incubate with Sox2 with gentle rotating at 4°C for 2 h. Repeat three times of washing with buffer 1. Combined fluorescently-labelled RNAs with the beads and incubated with gentle rotating at 4°C for 2 h. After six times of washing with buffer 1, the bound RNAs were eluted with 10 mM EDTA pH 8.2 and 95% formamide at 90°C for 10 min. The elution fractions were then resolved on a 8% native polyacrylamide gel in dark in 0.5× TAE at 4°C, 200 V. Signals were visualized with a FLA-7000 image reader (FUJIFILM/GE Healthcare) and quantified using the ImageQuant TL software (GE Healthcare).
RIP using cell lysate was performed as previously described (27). Briefly, cells were detached with 0.25% Trypsin (Gibco, Cat#25200114). The cell pellet was resuspended in equal volume of lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), 0.5% NP40, 1 mM DTT, 100 U/ml RNase Out, 400 μM VRC, Protease inhibitor cocktail), kept on ice for 5 min and then immediately used for immunoprecipitation or transfer to liquid nitrogen for storage. The lysate was thawed on ice and cell debris were removed by centrifugation at 4°C, precleared with Dynabeads® Protein G (Lifetech, Cat#10004D) before adding anti-Sox2 antibodies (Abcam, Cat #ab97959) pre-bound to Dynabeads® Protein G for 4 h at 4°C. In all, 5 μg of antibodies was used for each RNA-IP. Beads were then washed five times in ice-cold NT2 buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP40). RNAs were released from ribonucleoprotein complexes with Proteinase K (ThermoFisher Scientific, Cat#25530049) at 55°C for 30 min. RNA was isolated with Trizol reagent (ThermoFisher Scientific, Cat#15596026) and precipitated in 75% ethanol, resuspended into RNase- and DNase-free water (Tiangen, Cat#RT121-01) for further analysis.
Co-immunoprecipitation (co-IP)
Around 107 cells expressing FLAG3-Sox2 or FLAG3-Sox2-ΔRBM at day 6 of reprogramming were lysed in TNE lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% NP40, 0.1% SDS and 1 mM EDTA) containing protease inhibitor cocktail on ice for 15 min. Lysates were homogenized with a 0.4 mm needle and centrifuged at 13 000 g for 15 min at 4°C. Supernatants were incubated with 20 μl of Anti-FLAG® M2 Magnetic Beads (Sigma, Cat#F3165) or Dynabeads® Protein G (Lifetech, Cat#10004D) covered with anti-IgG antibody (Abcam, Cat#ab124055) and rotated overnight at 4°C. The next day, beads were washed five times with wash buffer (20 mM Tris–HCl, pH 7.4, 137 mM NaCl, 0.05% NP40 and protease inhibitor cocktail) and eluted by boiling in 50 μl SDS loading buffer and followed by for western blotting with antibodies (anti-Sox2, Abcam, Cat#ab97959; anti-hnRNP K, Abcam, Cat#ab52600; anti-Zcchc8, Proteintech, Cat#23374-1-AP; anti-FLAG, Sigma, Cat#F3165; anti-Skiv2l2, Abcam, Cat#ab70551; anti-SFPQ, Abcam, Cat# ab11825).
Chromatin immunoprecipitation assay (ChIP)
The experiment was performed as described (28). Briefly, ∼107 reprogramming cells at day 4 were crosslinked with 1% formaldehyde and then were disrupted on ice successively with Lysis Buffer 1 (50 mM HEPES–KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100, 1× protease inhibitors), Lysis Buffer 2 (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× protease inhibitors) and Lysis Buffer 3 (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1× protease inhibitors). The cell suspension was sonicated at high amplitude for 35 × 30 s cycles using a Bioruptor sonicator to yield DNA fragments ∼200–300 bp. Cell debris were pelleted in 1% Triton X-100 by spinning at 20 000 g for 10 min at 4°C. The supernatant was then incubated with 2–5 μg of anti-FLAG (CST, Cat#14793S) antibodies overnight at 4°C with gently rotating. The next day, 20– 50 μl Dynabeads® Protein G (Lifetech, Cat#10004D) was added to the mixture and incubated for 4–5 h. The beads were washed with Low Salt Buffer (0.1% SDS, 1% Triton-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0 and 150 mM NaCl), High Salt Buffer (0.1% SDS, 1% Triton-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0 and 500 mM NaCl), LiCl Buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA and 10 mM Tris–HCl pH 8.0) and TE Buffer (10 mM Tris–HCl and 1 mM EDTA pH 8.0). Protein–DNA complexes were eluted with TE buffer at 65°C for 30 min. Coeluted RNA was cleaned with Rnase A (ThermoFisher Scientific, Cat#EN0531) at 37°C for 1 h and bound proteins were eliminated with Proteinase K (ThermoFisher Scientific, Cat#25530049) at 55°C for 2 h. DNA fragments were extracted and precipitated in isopropanol for further analyses. qPCR was performed using iTaq™ Universal SYBR® Green Supermix (BIO-RAD, Cat#172-5124) following the manufacturer's instructions. Primer sequences are listed in Supplementary Table S6.
mRNA-sequencing (mRNA-seq)
RNA-seq samples were isolated using the TRIzol™ Reagent (ThermoFisher Scientific, Cat#15596026) from reprogramming OG2 MEFs at day 12 post-infection. A total amount of 1 μg RNA per sample was used as input material for the library preparation. The sequencing libraries were generated using the VAHTS mRNA-seq v2 Library Prep Kit for Illumina® (Vazyme, Cat#NR601) following the manufacturer's recommendations and sequenced on an Illumina Hiseq X Ten platform with 6G base 150 bp paired end reads (Vazyme Biotech Co., Ltd).
Bioinformatics analysis
For PAR-CLIP sequencing analysis, adapters and reads shorter than 15 nucleotides were removed. PCR duplicates were ignored using the FASTQ/A Collapser. Data were then aligned to hg19 human genome assembly using Bowtie (v1.1.2) (29) with the settings ‘-v 2 -m 1 -best -strata’. Aligned reads were used for peak calling with PARalyzer (v1.5) (30), requiring a minimum cluster (equal as ‘peak’) size of 10 nucleotides, a minimum T>C conversion count of 2 and a minimum of 5 reads for cluster inclusion (BANDWIDTH=3, CONVERSION=T>C, MINIMUM_READ_ COUNT_PER_GROUP=10, MINIMUM_READ_COUNT_PER_CLUSTER=5, MINIMUM_READ_COUNT_FOR_KDE=5, MINIMUM_CLUSTER_SIZE=15, MINIMUM_CONVERSION_LOCATIONS_FOR_CLUSTER=2, MINIMUM_ CONVERSION_COUNT_FOR_CLUSTER=1, MINIMUM_READ_COUNT_FOR _CLUSTER_INCLUSION=5, MINIMUM_READ_LENGTH=15, MAXIMUM_ NUMBER_OF_NON_CONVERSION_MISMATCHES=3). Peaks were used for de novo motif finding using DREME (MEME suite) (31) and annotated according to GENCODE v19. For splicing analysis, all the samples were first aligned to mm10 using Tophat (v2.1.0) (32) with default settings. Differentially spliced genes were detected using rMATS (v4.0.1) (33) according to GENCODE vM15 transcript models with false discovery rate (FDR) lower than 0.05. The G/C content of the 5′ splice site was analysis for 400 nt windows centered at the 5′ splice sites of the 1102 AS-affected exons. The control set contains corresponding regions for 22 752 exons. GO analysis was performed using metascape (34). Based on the alternative splicing information produced from rMATS, all AS events in the same cluster were pooled and the AS events with FDR <0.05 were included for downstream analysis. Duplicated events were identified manually according to the flanking sequences and the exon sequences. The number of AS events or genes in different AS types was recorded and plotted with the Venn function in gplotss. To perform the correlation analysis between Sox2-ΔRBM and Sox2-ΔHMG, gene ID, gene name, FDR, IncLevel difference information of AS events were collected from rMATS analysis results. Correlation analysis between Sox2-ΔRBM and Sox2-ΔHMG was performed by gene symbol with the pearson method (cor R package). For ChIP-seq anlaysis, Sox2 ChIP-seq were taken from Gene Expression Omnibus database (GSE90895) (35). Data were first aligned to the mm10 mouse genome assembly using Bowtie2 with the settings ‘-very-sensitive’. Low quality reads were removed using Samtools with the settings ‘-q 30’. Binding peaks were called using MACS2 (36) with default settings and further annotated by ChIPSeeker (for promoter, –3 kb to 3 kb to TSS; for intergenic binding events, peaks are assigned to nearest gene) (37). DESeq2 (38) with log2(fold change) > 2 and adjusted P-value < 0.05 criteria was used for differential gene expression analyses.
RNA isolation, RT-PCR and qRT-PCR
Total RNA was extracted from cells using TRIzol™ Reagent (ThermoFisher Scientific, Cat#15596026) according to the manufacturer's instructions. cDNA was synthesized by ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Cat#FSQ-301) from 2 μg of total RNA. Low-cycle PCR was then performed with Cy5-labelled forward primers and unlabelled reverse primers with DreamTaq PCR Master Mix (ThermoFisher Scientific, Cat#K1071) (39). Primers sequences are listed in Supplementary Table S7. Gapdh and Tada2a were used as controls. PCR products were analyzed on 8% PAGE gel and visualized using with a FLA-7000 image reader (FUJIFILM/GE Healthcare) and quantified using the ImageQuant TL software (GE healthcare). For qPCR, iTaq™ Universal SYBR® Green Supermix (BIO-RAD, Cat#172-5124) was used following the manufacturer's instructions.
RESULTS
Sox2 directly binds to RNA
Previous studies showed that Sox2 functionally synergizes with lncRNAs to control pluripotency and neuronal differentiation, unveiling potential roles of Sox2/RNA complexes in cell fate transitions and development (18,19). However, it remained unclear whether the interaction of Sox2 with RNAs was direct or indirect. To investigate this, we first performed a chemilumilescent photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) assay in HEK293T cells using overexpressed FLAG3-Sox2 (Figure 1A) (40). Sox2 showed robust binding to RNA, similar to a FLAG3-hnRNP K positive control whilst a FLAG3-GFP negative control does not bind RNA. We verified the Sox2-RNA interaction with retrovirally expressed FLAG3-Sox2 during the reprogramming of mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells (iPSCs) as well as with endogenous Sox2 in mouse embryonic stem cell (mESCs) (Figure 1B). To probe the RNA binding activity of purified Sox2 in vitro, we performed systematic evolution of ligands by exponential enrichment (SELEX) with an RNA library transcribed from DNAs containing 25 random nucleotides and bacterially-expressed full-length Sox2 with N-terminal hexahistidine tag (His6-Sox2) (Supplementary Figure S1). To verify the complexity of the library, we sequenced 17 randomly selected elements and observed non-redundant sequences with similar overall base composition (A = 20.47%, C = 26.12%, T = 28.94%, G = 24.47%; Supplementary Table S1). After 12 rounds of selection, 51 unique sequences were identified from 72 sequenced clones (Supplementary Table S2). Motif discovery using MEME (41) identified two G/C-rich consensus motifs in all 51 selected RNAs: a core motif defined as ‘GCCCY (Y: A or U) and a side motif with consensus ‘CGCG’ sequences as Sox2-binding motifs (Figure 1C and Supplementary Table S2). Consistent with this, FLAG3-Sox2 binding sites obtained from PAR-CLIP sequencing in HEK293T cells are also enriched for a similar ‘CCCY’ motif, which is often found in intronic regions (Figure 1D and Supplementary Figure S2).
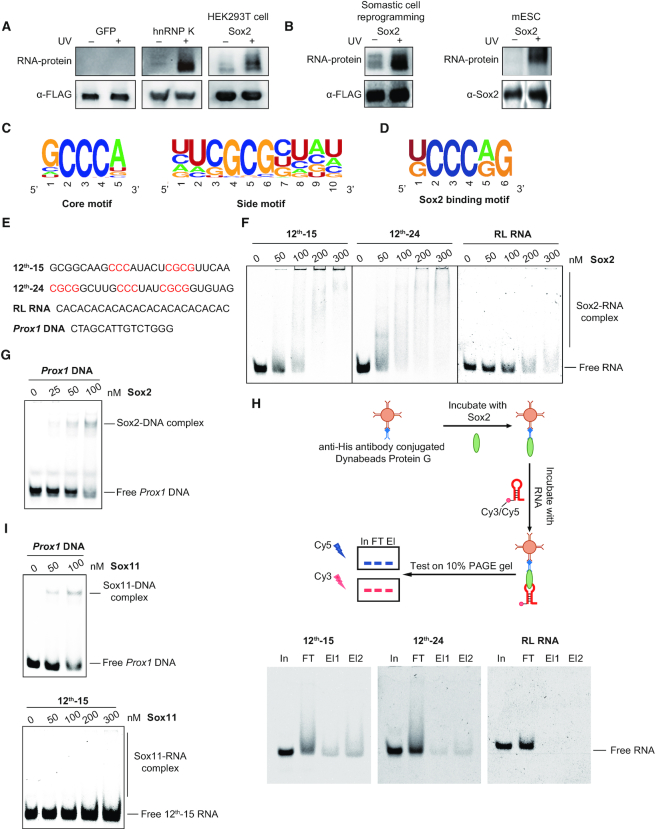
Figure 1.
Sox2 preferentially binds G/C-rich RNA sequences. (A) RNA binding activity of Sox2 was detected by PAR-CLIP. The upper panel shows the protein-bound RNA detected by biotin chemiluminescence. Antibody against FLAG tag was used to confirm the protein size and as loading control. FLAG-tagged Sox2, GFP (negative control) and hnRNP K (positive control) were expressed in HEK293T cells. (B) Same as in (A), FLAG-tagged Sox2 was expressed in MEFs at day 6 of pluripotency reprogramming, and endogenous Sox2 antibody was used in mESCs. Antibody against Sox2 or FLAG tag was used to confirm the protein size and as loading control. (C) Consensus motifs discovered by MEME in sequences enriched after 12 rounds of RNA SELEX with full length His6-Sox2 as bait (Supplementary Table S2). (D) Consensus motifs found by MEME in PAR-CLIP sequencing data for Sox2 expressed in HEK293T cells. (E) Nucleic acid ligands used for the EMSAs in (F–I). Bases highlighted in red are the SELEX consensus. 12th-15 RNA and 12th-24 RNA are Cy3- or Cy5-labelled RNA selected by SELEX after 12-round enrichment; RL RNA is a randomly chosen CA-rich RNA from the original RNA library; Prox1 DNA is a Cy5-labelled DNA with Sox2-binding motif. (F) EMSAs for the binding of 0–300 nM Sox2 to 50 nM fluorescently-labelled RNAs. The concentrations of Sox2 are as indicated above each panel. Shift patterns of free RNA and RNA bound by Sox2 (Sox2-RNA complex) are labelled to the right. (G) EMSAs of Cy5-labelled Prox1 DNA (50 nM) with increasing concentrations (0–100 nM) of Sox2. (H) In vitro RIP with 100 nM Sox2 and 100 nM fluorescently-labelled RNAs. In: input; FT: flow-through; El: elution. (I) EMSAs of Cy5-labelled Prox1 DNA (50 nM) (left panel) or Cy3-labelled 12th-15 RNA (50 nM) (right panel) with increasing concentrations of Sox11.
We next tested the RNA-binding activity of Sox2 using electrophoretic mobility shift assays (EMSA) with two enriched sequences (12th-15 RNA, 12th-24 RNA), one control sequence (RL RNA) from the initial RNA library and a cognate double-stranded DNA (dsDNA) (Figure 1E-G and Supplementary Table S3). The 12th-15 RNA contains one side motif and one core motif, and 12th-24 RNA consists of two side motifs flanking one core motif. RL RNA is a CA-rich RNA harbouring neither of the selected motifs. Strong binding was observed between Sox2 and both enriched RNAs as well as the cognate dsDNA control (left panel and middle panel in Figure 1F and G). In contrast, Sox2 only poorly binds to RL RNA (right panel in Figure 1F). This difference was then verified using an in vitro RIP assays in which RNA–protein complexes were immunoprecipitated with antibody specific to Sox2 (Figure 1H). The RIP assay includes a wash with binding buffer that removes low-affinity RNAs from immobilized Sox2 and is therefore more stringent than EMSAs. We observed 12th-15 RNA and 12th-24 RNA in the elution fraction but the RL RNA was not detectable. By contrast, the homologous Sox11 protein solely bound dsDNA but does not exhibit detectable affinity for RNA (Figure 1I). Collectively, these data establish Sox2 as a bona fide RNA binding protein with a preference for G/C-rich sequences.
Sox2 possesses a novel RNA binding module
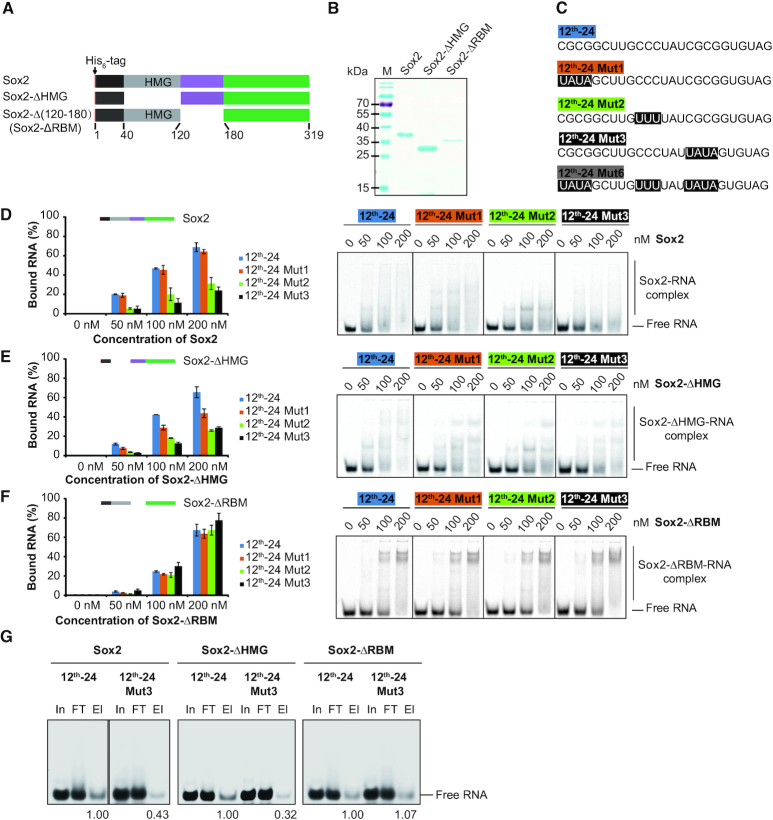
We next sought to identify the domains responsible for the RNA binding by Sox2. To this end, we constructed Sox2 proteins with internal domain deletions and purified them to homogeneity (Figure 2A and B) (42,43). We selected the 12th-24 RNA for these assays and designed three mutants where each of the three motifs was mutated (Figure 2C and Supplementary Table S3). EMSAs showed that binding of full-length Sox2 to 12th-24 Mut2, 12th-24 Mut3 and 12th-24 Mut6 was reduced (Figure 2D and Supplementary Figure S3A). Recent studies showed that the HMG box proteins, such as Hmgb1 and Hmgb2, are enriched in the proteins of the HeLa and mESC mRNA interactome, and the HMG box could act as an RNA-binding domain (44,45). However, surprisingly, deletion of the HMG box of Sox2 (Sox2-ΔHMG) did not disrupt RNA-binding, indicating that regions outside the HMG box contribute to RNA binding (Figure 2E). We hypothesized that a 60 amino acids motif C-terminal of the HMG box might be an RNA-binding motif (RBM) because this region is rich of amino acids highly favoured in RNA–protein interfaces (46). Removal of the 60-amino-acid motif (Sox2-ΔRBM) next to the HMG box retained RNA-binding activity but could not discriminate between original and mutated RNA elements (Figure 2F and Supplementary Figure S3A). To confirm the function of the RBM, we performed in vitro RIP assays with 12th-24 RNA and 12th-24 Mut3 RNA. Sox2 and Sox2-ΔHMG showed preferential binding to 12th-24 RNA, whilst Sox2-ΔRBM showed similar binding to 12th-24 RNA and mutants thereof (Figure 2G).
Figure 2.
Identification of a novel RNA binding motif of Sox2. (A) Schematic representation of the His6-Sox2 constructs used for binding assays. (B) Coomassie stained 12% SDS-PAGE with purified His6-Sox2 constructs. M: protein marker and sizes are given in kDa. (C) RNA ligands used for the EMSAs. Bases shaded in black denote sequences that are altered between the SELEX consensus (12th-24) and mutant versions (12th-24 Mut1, 12th-24 Mut2, 12th-24 Mut3 and 12th-24 Mut6). (D–F) Comparison of the binding activities of Sox2 constructs to the 12th-24 RNA and mutants thereof. Fractions of bound RNA (left panels) were determined by EMSAs (right panels) using densitometric analysis using the ImageQuantTL software. Barplots represent the mean ± SD (n = 3). (G) In vitro RIP with 100 nM Sox2 construct and 100 nM fluorescently-labelled RNAs. The names of Sox2 constructs and RNA substrates are as indicated above each panel. In: input; FT: flow-through; El: elution. Relative levels of eluted 12th-24 and 12th-24 Mut3 are noted.
To further clarify whether the RBM is sufficient for RNA binding, we next generated Sox2 truncations containing the short N-terminal sequence followed by the HMG domain with (Sox2(1–180)) or without (Sox2(1–120)) the RBM (Supplementary Figure S3B and C). Sox2(1–120) bound the SELEX consensus and mutants thereof with indistinguishable affinity, while addition of the RBM (Sox2(1–180)) restored the sequence selectivity (Supplementary Figure S3D and E). To exclude that the extended C-terminus also contributes to RNA binding, we generated N-terminal deletions in the absence of presence of the RBM (Sox2(120–319) and Sox2(180–319); Supplementary Figure S3B and C). The construct without RBM abolished the RNA-binding capacity of Sox2(180–319) in contrast to Sox2(120–319) (Supplementary Figure S3F). Collectively, these analyses identified a 60-amino-acid RNA binding motif (RBM) C-terminally of the HMG box that endows Sox2 with a preference for GC-rich RNA sequences.
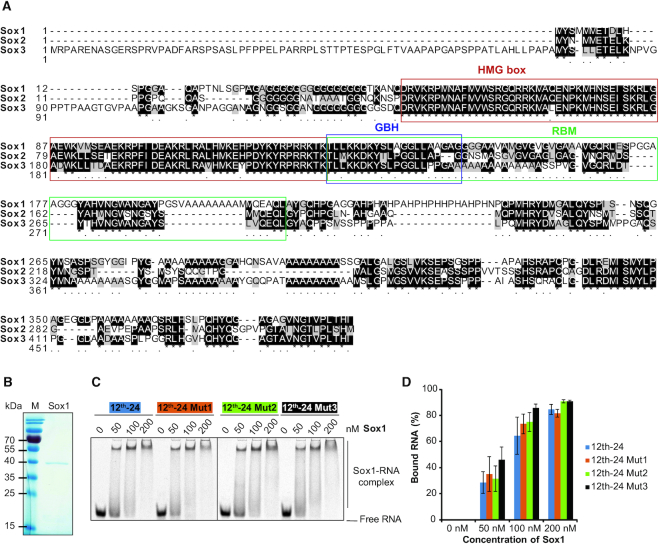
The SoxB1 subfamily members- Sox1, Sox2 and Sox3 share a highly conserved HMG box immediately followed by a ‘group B homology (GBH)’ domain comprised largely of basic amino acids (Figure 3A) (47). To test whether the RNA binding of Sox2 is directed by the GBH and shared by other SoxB1 factors, we performed binding assays with recombinant Sox1 and the 12th-24 RNAs. Sox1 is able to bind RNA with high affinity but the binding is not weakened upon mutations of consensus elements (Figure 3B–D). These results suggest that the G/C directed RNA binding is not shared within the SoxB1 subfamily and likely driven by features encoded outside the GBH.
Figure 3.
The group B homology domain does not confer sequence selective RNA binding. (A) Multiple sequence alignment of mouse SoxB1 group done with ClustalX 2.0.9 (http://www.clustal.org/download/2.0.9/). The DNA-binding HMG domains are framed in red. The RNA-binding motif (RBM) of Sox2 and its N-terminal part corresponding to the group B homology domain are boxed in green or blue, respectively. (B) Recombinant Sox1 resolved by SDS-PAGE and visualized by Coomassie staining. M: protein size markers in kDa. (C, D) Comparison of the binding of Sox1 to 12th-24 RNA and its mutants. Representative EMSAs are shown in (C) and the fractional RNA binding is plotted in (D). The concentrations of Sox1 are as indicated and complexes are labelled to the right. Data are represented as mean ± SD (n = 2).
Next, we asked whether the Sox2-RNA interaction is driven by dedicated structural units. To this end, we predicted the RNA structures using Mfold (48), and generated mutants predicted to either retain or disrupt the stem-loop architecture of the 12th-24 RNA (Supplementary Figure S4A–G). The ‘CCC’ core motif is predicted to map to a loop region and its conversion into ‘UUU’, ‘GGG’ or ‘AAA’ does not change the secondary structure (Supplementary Figure S4A, C, E and F). All three mutations reduced the binding to Sox2 (Supplementary Figure S4H). Mutations predicted to perturb the stem-loop architecture have varying consequences, suggesting that structural changes pleiotropically influence binding modalities (12th-24 Mut1, Mut3, Mut6; Supplementary Figures S4B, D, G, S3A and Figure 2D). The Sox2-ΔRBM construct indistinguishably bound to the 12th-24 RNA and both classes of mutants (Figure 2F and Supplementary Figure S4I). We conclude that the RBM mediated binding of Sox2 to RNA can occur in the context of alternatively structured RNA scaffolds with a contextual preference for the ‘CCC’ element.
Sox2 is capable of binding DNA and RNA simultaneously via distinct domains
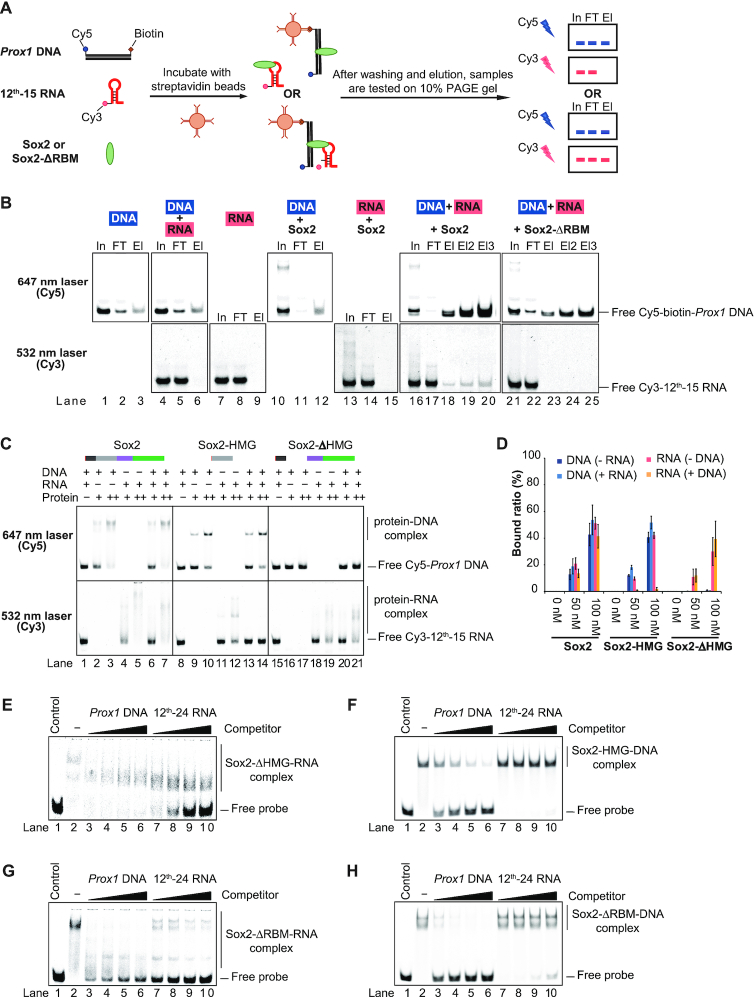
Sox2 is constantly exposed to both DNA and RNA in a cellular or physiological environment. To mimic the simultaneous exposure of Sox2 to both types of nucleic acids, we decided to dissect the binding of Sox2 with DNA and RNA in single tube reactions. We established RNA fishing assays with a Sox consensus dsDNA element carrying both 5′-Cy5 and 3′-biotin labels and 5′-Cy3-labelled 12th-15 RNA (Figure 4A and Supplementary Table S3). The protein/RNA/DNA complexes were assembled, captured with streptavidin beads, washed, and then electrophoresed on a 10% PAGE gel and scanned twice with different lasers (532 nm laser line for Cy3; 647 nm laser line for Cy5) to successively detect DNA and RNA in input, flow through and elution fractions. We verified that the streptavidin beads capture biotin- and Cy5-labelled DNA efficiently (Figure 4B, upper panel). Neither the biotin-labelled DNA nor Sox2 alone can capture the 5′-Cy3-labelled 12th-15 RNA (lanes 4–9 and 13–15 in Figure 4B, bottom panel). Yet, if the DNA and Sox2 were present simultaneously, the co-elution of RNA was observed (lanes 16–20 in Figure 4B), demonstrating that Sox2 can bind DNA and RNA simultaneously. However, when the RBM was deleted the RNA did no longer co-elute with the Sox2/DNA complex, showing that the formation of ternary RNA/Sox2/DNA complexes critically relies on the RBM (lanes 21–25 in Figure 4B).
Figure 4.
Sox2 forms a ternary complex with DNA and RNA. (A) Schematic representation of the RNA fishing experiment to test for the simultaneous binding of RNA and DNA to Sox2. (B) After immobilization of biotinylated DNA on magnetic streptavidin beads and washing, the eluates were analysed by native 10% PAGE gels. Components of the reaction mixtures are labelled on the top of each gel. The lasers used to sequentially detect Cy5-labelled DNA and Cy3-labelled RNA are indicated to the left. Results shown in the same column in upper or lower panels were from identical gels but scanned with different lasers. In: input; FT: flow-through; El: elution. (C) EMSAs of Sox2 constructs with Cy5-DNA and Cy3-RNA. Concentrations of DNA, RNA and protein are labelled with ‘–’, ‘+’ and ‘++’ indicating 0 nM, 50 nM and 100 nM, respectively. In each column, the upper panel and bottom panel are results from identical gels but scanned with the Cy5 (top) or the Cy3 excitation wavelength (bottom). (D) Quantification of the results shown in (C). The barplot shows the mean ± SD (n = 3). (E–H) Competition EMSAs using Sox2-ΔHMG-Cy5-RNA (E), Sox2-HMG-Cy5-DNA (F), Sox2-ΔRBM-RNA (G) and Sox2-ΔRBM-DNA (H) complexes with increasing concentrations of unlabelled Prox1 DNA or unlabelled 12th-24 RNA (from 500 to 4000 nM). Control: free Cy5-Prox1 DNA or Cy5-12th-24 RNA only; ‘-’, Cy5-Prox1 DNA or Cy5-12th-24 RNA with Sox2 constructs but without competitor.
We also performed EMSAs that excluded the biotin capture step using 50 nM of Cy5-DNA/Cy3-RNA and 50 nM or 100 nM of full-length Sox2, Sox2-HMG and Sox2-ΔHMG constructs followed by sequential detection of the DNA and RNA in the same gel (Figure 4C and D). Sox2 bound to RNA and DNA with similar affinities (lanes 2–5 in Figure 4C). Moreover, we observed no reduction of the binding when Sox2 was incubated with an RNA/DNA mixture as compared to incubations with DNA or RNA alone (lanes 6–7 in Figure 4C), confirming the simultaneous binding of DNA and RNA. The HMG domain of Sox2 (Sox2-HMG) was able to interact with both DNA and RNA in separate binding reactions (lanes 9–12 in Figure 4C). However, when the Sox2-HMG was incubated with the DNA/RNA mixture it did not bind to RNA at all but was exclusively directed to its dsDNA consensus element (lanes 13–14 in Figure 4C). Conversely, a Sox2 construct lacking the HMG box completely lost DNA binding activity but RNA binding remained unaffected (lanes 16–21 in Figure 4C).
We next verified the binding preference of the RBM and the HMG box using competition assays. Without competitor, 50 nM of 5′-Cy5-labelled 12th-24 RNA was retarded by the Sox2-ΔHMG (lane 2 in Figure 4E). Competition with 80 × molar excess of unlabelled Prox1 DNA (lanes 3–6 in Figure 4E) failed to displace Sox2-ΔHMG from RNA, whereas 80 × excess of unlabelled 12th-24 RNA displaced the Sox2-ΔHMG from the labelled RNA (lanes 7–10 in Figure 4E). Conversely, a pre-bound Sox2-HMG/DNA complex can be effectively disrupted with DNA competitors but not with RNA competitors (Figure 4F). Thus, different binding preferences of non-HMG domain and HMG domain contribute to the simultaneous interaction of Sox2 with RNA and DNA. The RNA binding of Sox2-ΔRBM was strongly diminished by excess amount of both DNA and RNA (Figure 4G). Yet, similarly as the Sox2-HMG, Sox2-ΔRBM/DNA complexes can be disrupted by DNA but not by RNA competitors (Figure 4H). Collectively, in the presence of DNA, Sox2 employs its HMG box exclusively to bind cognate DNA sequences and concurrently uses a novel RBM to mediate the formation of ternary RNA/Sox2/DNA complexes in vitro.
Deletion of the Sox2 RBM impairs iPSC generation
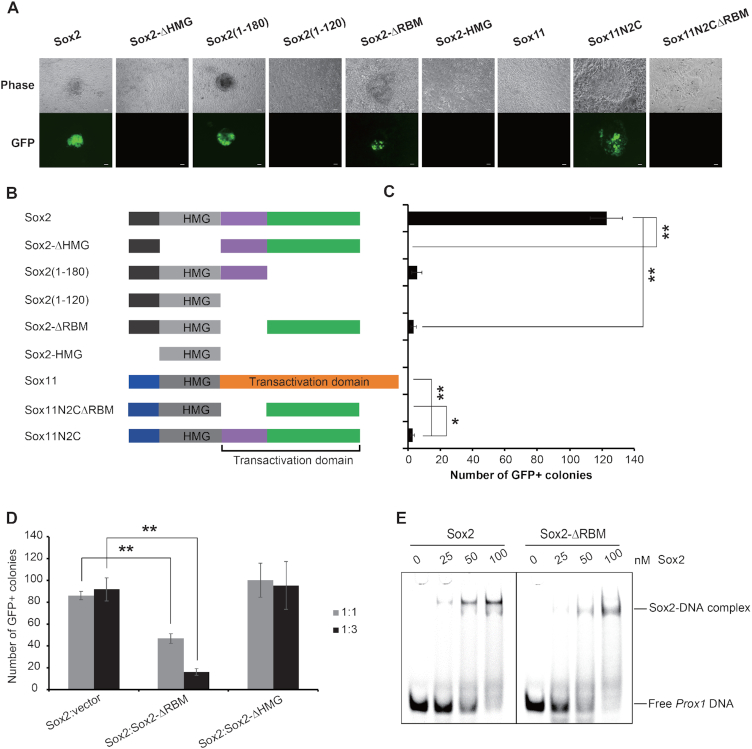
RNA-binding proteins are dynamically regulated at the initial phase of iPSC generation, implicating important roles of co-transcriptional or post-transcriptional processes during pluripotency reprogramming (44). Given that Sox2 is a core component of pluripotency reprogramming cocktails, we used iPSC generation as a functional read-out to assess the biological importance of the RBM domain. We aimed to compare the efficiency of iPSC generation with 4-factor cocktails including Oct4, Klf4 and c-Myc (OKM) as well as different Sox2 variants (Figure 5A, B and Supplementary Figure S5A). We introduced retroviruses into MEFs carrying a GFP transgene under the control of an Oct4 promoter (OG2 MEFs) cultured in Serum/LIF conditions (23). Efficiency was scored 12 days after the transduction by counting GFP positive colonies. Neither the Sox2-HMG box alone nor a Sox2 construct lacking the HMG box (Sox2-ΔHMG) was able to induce pluripotency alongside OKM (Figure 5A–C and Supplementary Figure S5B). The Sox2-ΔRBM produced 36 times less GFP-positive colonies than wild-type Sox2, indicating a relevance of the RBM for efficient iPSC generation (Figure 5A–C and Supplementary Figure S5B). In addition, deletion of the C-terminal 139 residues succeeding the RBM (Sox2(1–180)) also led to notable reduction of the reprogramming efficiency (Figure 5A–C and Supplementary Figure S5B). Reprogramming activity is completely lost when the RBM and the C-terminal 139 residues are both deleted (Sox2(1–120), Figure 5A–C), suggesting functional synergies between both domains. Sox11 shares over 70% sequence similarity with Sox2 in the HMG domain, while their C-terminal sequences are devoid of homologous sequences. Sox11 cannot convert somatic cells into iPSCs (49). When the C-termini of Sox2 and Sox11 are interchanged, the chimeric Sox11N2C protein could produce GFP-positive colonies (Figure 5A–C). However, deletion of the RBM led to a reprogramming incapacitated construct (Sox11N2CΔRBM). To assess the relative significance of RBM compared to the HMG box, we next performed reprogramming experiments using mixtures of intact Sox2 and the deletion mutants Sox2-ΔRBM or Sox2-ΔHMG at a 1:1 or 1:3 ratio of viral supernatants. Interestingly, we observed that the presence of Sox2-ΔRBM mitigates the ability of Sox2 to reprogram cells to pluripotency whilst Sox2-ΔHMG has no effect (Figure 5D and Supplementary Figure S6). The deletion of the RBM does not affect DNA binding in vitro (Figure 5E), or the recognition of cognate chromatin binding sites in reprogramming MEFs whilst the deletion of the HMG abrogates chromatin association (Supplementary Figure S7A). Co-immunoprecipitation showed that the lack of the RBM had no effect on the interaction of Sox2 with Oct4 (Supplementary Figure S7B). This suggests that deletion of the HMG prevent the association of Sox2 with its target genes. By contrast, Sox2-ΔRBM remains able to bind cognate chromatin targets but fails to execute subsequent regulatory processes. Therefore, Sox2-ΔRBM acts as a dominant negative and interferes with normal Sox2 function during the reprogramming. Altogether, these findings suggest that the RBM of Sox2 imparts activities relevant for somatic cell reprogramming in the context of chromatin bound Sox2 and imply a role of ternary Sox2/RNA/DNA complexes in the context of cell fate changes.
Figure 5.
Deletion of the Sox2 RBM impairs iPSC generation. (A) Morphology of mouse iPSCs generated by Sox2 or Sox11 constructs in combination with Oct4, c-Myc and Klf4 under LIF/Serum conditions generated from OG2 MEFs expressing a transgenic Oct4-GFP reporter. Colonies were photographed using a fluorescence microscope 12 days after the viral infection. The scale bar indicates 100 μm. (B) A schematic representation of Sox2 and Sox11 constructs used for iPSCs generation. (C) The iPSC generation efficiency of these constructs is presented by the absolute number of GFP-positive colonies in a well of a 12-well cell culture. Barplot shows the mean ± SD of three biological replicates and differences between selected samples were compared using ANOVA (* and ** indicate P-values of < 0.05 and < 0.01, respectively). (D) The numbers of iPSC colonies generated by Sox2 plus empty vector, Sox2-ΔRBM or Sox2-ΔHMG at 1:1 or 1:3 ratio of viral supernatants from three independent experiments are shown. Data are represented as mean ± SD, and ANOVA was used to assess significance (** indicates P-value of < 0.01). (E) Comparison of the binding activities of Sox2 and Sox2-ΔRBM to Prox1 DNA. The reactions were analyzed by EMSAs in 10% native PAGE gels. Concentrations of Sox2 constructs are labelled above each lane. Free DNA and protein-DNA complex are marked.
The Sox2-RBM regulates splicing during reprogramming
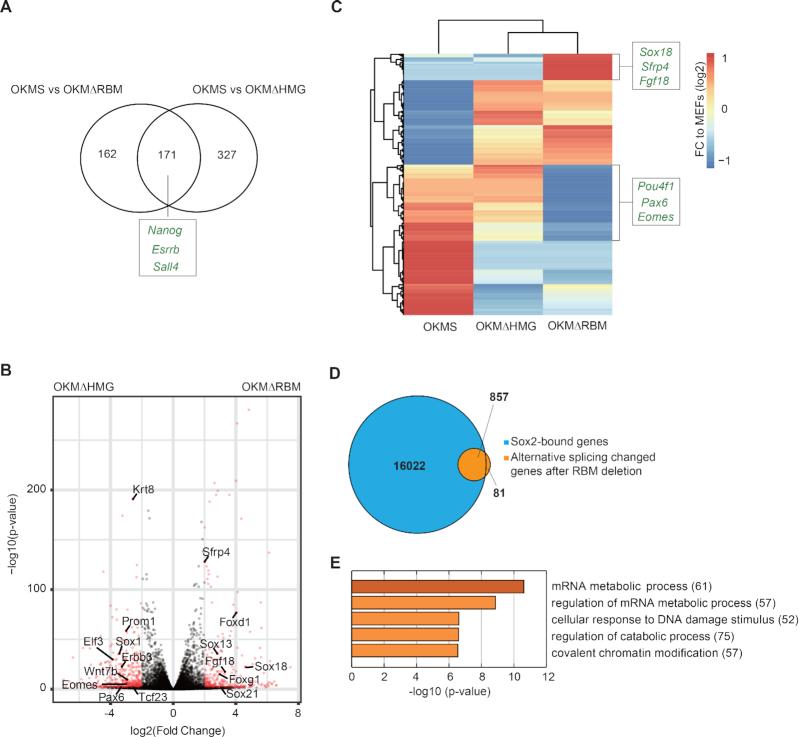
We next sought to explore the mechanism by which the RBM contributes to reprogramming. To this end, we first collected RNA-sequencing (RNA-seq) data at day 12 of reprogramming from cells transduced with Oct4-Klf4-c-Myc-Sox2 (OKMS), Oct4-Klf4-c-Myc-Sox2-ΔRBM (OKMΔRBM) and Oct4-Klf4-c-Myc-Sox2-ΔHMG (OKMΔHMG). qRT-PCR showed that all transduced factors were expressed at similar levels (Supplementary Figure S5A). Pluripotency genes (e.g. Nanog, Esrrb and Sall4) were downregulated in OKMΔRBM as well as the OKMΔHMG control compared to OKMS (Figure 6A and Supplementary Figure S8A). This is consistent with the observation that deletion of HMG or RBM reduces reprogramming efficiency (Figure 5B, C). We also found that the set of differentially regulated genes differs in Sox2-ΔRBM versus Sox2-ΔHMG expressing cells (e.g. Sfrp4, Fgf18, Sox18, Pax6 and Eomes) indicating that both deletions have non-redundant functional consequences (Figure 6B and C). This suggests that the HMG and the RBM impart different functional modalities which in the case of the RBM is not tied to target gene selection and transactivation.
Figure 6.
Deletion of the Sox2-RBM and Sox2-HMG have different effects on gene expression and splicing. (A) The Venn diagram compares gene sets differentially upregulated (determined with DESeq2 (38)) in OKMS versus OKMΔRBM with OKMS and OKMΔHMG. Examples of genes related to pluripotency that are depleted in both Sox2 deletion mutants compared to the OSKM control are marked. (B) Volcano plot highlighting differentially expressed genes determined with DESeq2 from OKMΔRBM and OKMΔHMG RNA-seq data. Genes at –log10(P-value) > 2 and absolute log2(fold change) value >0.5 are in red. Representative genes related to differentiation in OKMΔRBM are labelled on the right. (C) Differentially expressed genes upon OKSM, OKMΔHMG and OKMΔRBM are shown. The names of representative genes related to differentiation are depicted. (D) 938 transcripts were detected to undergo alternative splicing under Sox2-ΔRBM compared to Sox2 control conditions at day 12 of iPSC generation using rMATS (33). The Venn diagram shows the overlap genes bound by Sox2 (blue) at early reprogramming stages (35) and genes affected by alternative splicing after RBM deletion (orange). (E) The top five GO terms provided by metascape (http://www.metascape.org) of 857 genes shown in (D). The number of genes in each GO term is labelled in brackets.
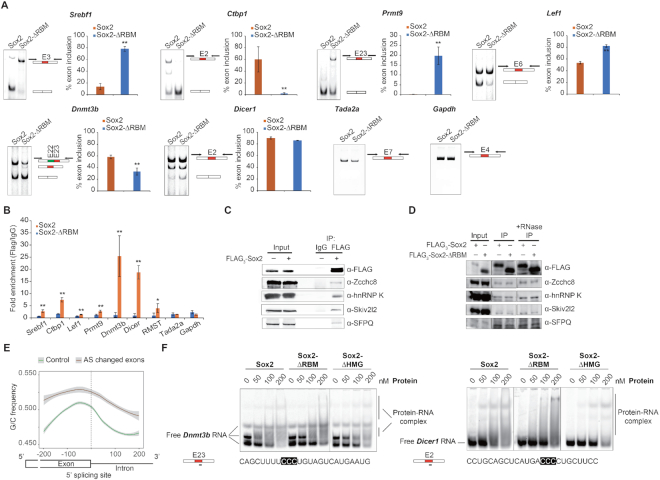
RNA-binding proteins are involved in all steps of RNA metabolism, including transcription, splicing, RNA modification and turnover (50). Regulation of alternative splicing (AS) has recently emerged as a critical event in controlling stem cell pluripotency, differentiation and somatic cell reprogramming (51,52). As splicing can occur co-transcriptionally (53) and a growing number of TFs has been reported to concurrently regulate transcription and splicing (54), we surmised that Sox2 may also regulate splicing. To test this hypothesis, we first analysed the differential splicing events using rMATS (33) for OKMΔRBM compared to OKMS. This led to an identification of 1176 alternative splicing events of 938 genes. Interestingly, we also identified 1219 alternative splicing events in 975 genes for OKMΔHMG compared to OKMS. However, the majority of the genes alternatively spliced in OKMΔRBM and OKMΔHMG are unique for the respective condition (Supplementary Figure S8B and C). This indicates that perturbations to different regulatory pathways derail the Sox2-dependent reprogramming activity after excision of the RBM or the HMG box, respectively. Since the RBM does not affect the association of Sox2 with chromatin or its partner factor Oct4 (Supplementary Figure S7), we next filtered for transcripts that could be concurrently regulated on the transcriptional and post-transcriptional level. To this end, we intersected the list of alternatively spliced genes with genes having a proximal Sox2 binding at early stages of reprogramming cells (35) and found evidence for direct binding for 91% (857) of the AS genes (Figure 6D). Gene ontology (GO) analysis for these 857 genes revealed an enrichment of terms associated with RNA metabolism and chromatin modification (Figure 6E). The AS genes include known regulators of cellular reprogramming such as Lef1 which blocks the reprogramming at early stage (55), Srebp1 which facilitates reprogramming through interacting with c-Myc (56) and the histone methyltransferases Ehmt2 which modulates the efficiency of reprogramming (57,58). We validated a series of AS exons for a panel of pluripotency-related genes using RT-PCR including Srebf1, Lef1, Dnmt3b, Ctbp1, Dicer1 and Prmt9 as well as for two unaffected genes (Tada2a, Gapdh) (Figure 7A) (55–63). Altogether, these results indicate that the RBM of Sox2 might regulate pluripotency reprogramming through modifying splicing.
Figure 7.
Deletion of the Sox2-RBM affects exon selection and RNA binding. (A) RT-PCR assays examining mRNA splicing levels of a group of pluripotency-related genes (Srebf1, Ctbp1, Lef1, Prmt9, Dnmt3b and Dicer1) and control genes (Tada2a, Gapdh) at day 12 of iPSC generation under Sox2-ΔRBM or Sox2 control conditions. Histograms show quantifications of each RT-PCR measurements. Error bars represent the mean ± SD from two biological replicates. Differences were compared using ANOVA (** indicates P-value of < 0.01). (B) RIP of FLAG3-Sox2 and FLAG3-Sox2-ΔRBM using anti-FLAG. RIP enrichment was measured by qRT–PCR, and values were normalized to background immunoprecipitation measured by isotype IgG. ANOVA was used to assess statistical significance (* and ** indicate P-values of <0.05 and <0.01, respectively). (C) FLAG3-Sox2 was transfected into MEF cells along with Oct4, Klf4 and c-Myc. Co-immunoprecipitations were performed with IgG or anti-FLAG antibody and immunoblots with anti-FLAG, anti-Zcchc8, anti-hnRNP K, anti-Skiv2l2 and anti-SFPQ. (D) FLAG3-Sox2 or FLAG3-Sox2-ΔRBM was transfected into MEF cells along with Oct4, Klf4 and c-Myc. Co-immunoprecipitations were performed with anti-FLAG antibody and immunoblots with anti-FLAG, anti-Zcchc8, anti-hnRNP K, anti-Skiv2l2 and anti-SFPQ. (E) G/C content around the 5′ splice sites of exons affected by AS (red, n = 749) and exons not affected by AS (control, green, n = 14 342). The dotted black line highlights the 5′ splicing sites. (F) Comparison of the binding activities of Sox2 constructs to Dnmt3b and Dicer1 RNA. The locations and sequences of RNAs are marked under each panel. Fractions of bound RNA were determined by EMSAs on 10% native gels.
To test whether the effect of Sox2 on splicing is direct or indirect, we performed RIP using reprogramming MEFs at day 12 and found that many of the alternatively spliced transcripts co-immunoprecipitated with intact Sox2 but the deletion of the RBM mitigates the association (Figure 7B). As an additional control we performed RIP for the lncRNA RMST that was reported to physically interact with SOX2 in human neural and embryonic stem cells (19). Our RIP data in reprogramming MEFs verified the physical association of Sox2 and RMST in mouse cells during reprogramming and shows that the deletion of the RBM alleviates this interaction (Figure 7B). A subset of the AS transcripts is also bound by Sox2 in mESCs (Supplementary Figure S9). Because splicing is extensively regulated by a plethora of proteins, we next used co-immunoprecipitation to test whether Sox2 interacts with splicing factors. We validated the interaction of Sox2 with several splicing factors including hnRNP K, Skiv2l2, SFPQ and the spliceosome associated protein Zcchc8 (64–66) at day 6 of pluripotency reprogramming (Figure 7C and D). These protein-protein interactions of Sox2 were not affected by the deletion of RBM or treatment with RNase A.
As the Sox2-RBM prefers G/C-rich RNA sequences and the 5′ splicing site is essential for spliceosome assembly, we compared the G/C contents around the 5′ splicing site for exons affected under Sox2-ΔRBM with unaffected exons. Intriguingly, we found an enrichment of G/C rich sequences around the 5′ splice site for exons subject to AS but not for controls (Figure 7E and Supplementary Figure S10). The G/C content in exons and introns was previously reported to contribute to exon selection by the splicing machinery (67). To test whether Sox2 can directly bind splice sites of AS transcripts, we carried out EMSAs with two 25 nucleotide RNA fragments which flank the GC-rich 5′ splicing site of AS exons in Dnmbt3b and Dicer1 mRNA. Removal of the RBM (Sox2-ΔRBM and Sox2(180–319)) impairs the Sox2–RNA interaction (Figure 7F and Supplementary Figure S11A), while removal of the HMG (Sox2-ΔHMG and Sox2(120–319)) largely maintains association with RNA (Figure 7F and Supplementary Figure S11B). Removal of both RBM and HMG completely abolished RAN binding (Supplementary Figure S11A). The homologous Sox11 protein does not exhibit detectable affinity for Dicer1 and Dnmt3b RNA (Supplementary Figure S11C). These findings suggest that the Sox2-RBM may contribute to exon selection via interacting with the 5′ splice site during reprogramming.
DISCUSSION
Multi-functionality and cell context specific activities are features of many nucleic acid-binding proteins. A list of transcription factors, such as TFIIIA and p53, have been defined as dual DNA and RNA binding proteins, participating in multiple, sometimes unrelated biological pathways (68–72). Here, we provide direct evidence that for the interaction between Sox2 and RNA. We first identified this interaction in a cellular context using PAR-CLIP. Next, we used purified Sox2 to verify this interaction in vitro by RNA SELEX. Both assays indicated that Sox2 preferentially targets a ‘CCCY’ core motif and a secondary motif with consensus ‘CGCG’. EMSA (Figure 2D and Supplementary Figure S4) emphasized the importance of the ‘CCCY’ motif in Sox2-RNA interaction whilst the role of the side motif is less certain. Splicing regulatory elements (SRE) are short cis motifs that generally associate with splicing factors that regulate the spliceosome assembly on an adjacent potential splice site (73). Some SREs are contained in introns. RNA binding protein Nova-1 regulates AS by binding to intronic SREs, such as YCAY, and enhancing the downstream splicing (74,75). Since the ‘CCCY’ core motif of Sox2 is enriched in intron regions (Supplementary Figure S2), we propose that it constitutes a SRE targeted by Sox2 to regulate AS. Interestingly, the Sox2 interaction partner PARP1 also binds G/C-rich RNA sequences and plays a role in the co-transcriptional AS regulation (3,41), suggesting that Sox2 might cooperate with PARP1 to regulate AS co-transcriptionally during reprogramming.
The HMG domain is primarily deemed as a sequence-specific DNA-binding domain. Recent studies revealed that it could also interact with RNA. In mESCs, Hmgb1 and Hmgb2, which belong to the class of non-sequence specific HMG box proteins, were identified as RNA-binding proteins (44). In HeLa cells, a series of HMG box proteins were also detected in mRNA interactomes (45). Consistently, our results show that the Sox2-HMG domain is able to bind to RNA but without sequence specificity (Supplementary Figure S3E). Moreover, in the presence of cognate DNA elements the HMG box is readily removed from RNA to bind DNA (Figure 4C and D). The region outside HMG box of Sox2 exhibits RNA-, rather than DNA-, binding activity (Figure 4C and D). We found that a group B homology-containing motif (RBM) mediates binding to GC-rich RNA elements (Figures 2 and 3A). This RBM shares no similarity with known RNA-binding domains (76). We hypothesized that the G/C-rich sequence-biased RNA binding is a common activity of the SoxB1 group. However, Sox1 does not discriminate between mutants and the SELEX enriched 12th-24 RNA variant, suggesting that this activity is unique for Sox2 (Figure 3). Indeed, although there is some degree of functional redundancy, individual SoxB1 members still have their own unique roles. For example, Sox1 is required during the differentiation and migration of ventricular zone neurons (77). Sox2 and Sox3 sequential act during neurogenesis from ESC to neural progenitor cells (78). Furthermore, an RNA-binding domain succeeding the DNA-binding domain seems a common modular organization for dual activity DNA- and RNA-binding proteins. In PARP1, the DNA-binding domain is composed of the first two Zinc fingers, and the following third Zinc finger is responsible for RNA binding (3,79). ADAR family members pose one to three copies of RNA-binding domains after the N-terminal DNA-binding domain (80). p53 has a centrally-located DNA-binding domain and a C-terminal RNA-binding domain (81). These could be important for orchestrating functions associated with DNA as well as RNA binding.
Co-operativity of Sox2 with Oct4 is critical to convert somatic cells into iPSCs and to maintain pluripotency and mediated by the HMG box of Sox2 and the POU domain of Oct4 (15,82,83). Yet, the C-terminus of Sox2 confers the potency of reprogramming. Chimeric Sox2 with the C-terminal region of Sox17 enhances its potency in iPSC generation, while chimeric Sox17 with the C-terminus of Sox2 is not able to activate the pluripotency network reminiscent to wild-type Sox17 (49). In agreement with this, the removal of the C-terminus of Sox2 impairs reprogramming (Figure 5). Our observation that the replacement of the TAD of Sox11 with the intact C-terminal region of Sox2 inclusive of the RBM converts Sox11 into an iPSC inducer prompted us to further study how the RBM of Sox2 regulates the pluripotency network. The efficiency of iPSC generation sharply decreased in the absence of the RBM (Figure 5A–C). Yet, deletion of the RBM does not influence DNA and chromatin association. This is consistent with a recent report showing that the interaction of Sox2 with the non-coding RNA 7SK does not significantly affect binding to regulatory chromatin regions (84). Consistently, Sox2-ΔRBM acts as dominant negative and compromises the ability of Sox2 to reprogram whilst the expression of the Sox2-ΔHMG does not interfere with the activity of Sox2 as its unable to bind its target genes (Figure 5D and Supplementary Figure S6B). Apparently, Sox2-ΔRBM occupies the same genomic locations as Sox2 but fails to fulfill functions subsequent to target gene selection that require an intact RBM. A proteomic study in mESCs revealed that 9% of Sox2-interacting proteins take part in RNA processing and two of them (Zcchc8, Skiv2l2) are prominent RNA splicing-associated factors (64). Spliceosome components, such as SFPQ, hnRNP K, were also detected in Sox2 interactome (65,66). Indeed, we found that Sox2 interacts with splicing factors (hnRNP K, SPFQ, Skiv2l2 and Zcchc8) independently of its RBM, suggesting a role in splicing (Figure 7C and D). In addition, Zcchc8 is also a component of the nuclear exosome complex, implicating that Sox2 may be a regulator of 3′ processing after splicing (85). Comparison of the RNA-seq data from OKSM and OKMΔRBM samples uncovered AS in 857 Sox2-bound genes, many of which have roles in RNA metabolism and chromatin modification (Figure 6D and E). Amongst the proteins linked to RNA metabolism, some play important roles in cell fate determination such as transcription factors (Srebf1, Lef1), m6A binding proteins (YTHDF1, YTHDF2, YTHDF3) and methyltransferases (Ehmt2). Chromatin modifications, mainly DNA methylation and histone modification, are pivotal in regulating higher-ordered chromatin structure during early embryonic development, and are reported as major barriers to iPSC generation (86,87). We validated AS for the chromatin modifiers (Ctbp1, Dnmt3b, Dicer1) and RNA metabolism participators by RT-PCR (Figure 7A). During the late stage of iPSC induction, the expression of Dnmt3b is upregulated to ensure full pluripotency maturation and the full-length isoform predominates (88). A Lef1 variant lacking exon 6 (Lef1Δ6) is the predominant isoform in PSC whilst the isoform including the exon is present in MEF cells (89). Therefore, full-length Dnmt3b and Lef1Δ6 can be treated as hallmarks of pluripotency. At day 12 of reprogramming, dominant amount of full-length Dnmt3b and Lef1Δ6 were detected in cells transduced with OKMS but not in OKMΔRBM-induced cells, suggesting that OKMS-induced cells are in the maturation phase, whereas the OKMΔRBM-induced cells are still in the transition of MEF cells to iPSCs. Yet, further studies are needed to clarify how Sox2 and splicing factors synergistically regulate alternative splicing during somatic cell reprogramming. Since AS is often regulated by the selection of 5′ splicing site, we studied the sequence composition of the 857 genes exhibiting AS in Sox2-ΔRBM versus Sox2 wild-type conditions. The AS-affected exons possess higher G/C content in contrast to the unaffected ones (Figure 7E and Supplementary Figure S10). As the Sox2-RBM targets G/C-rich sequences and deletion of this region attenuates binding of Sox2 to transcripts subject to alternative splicing during pluripotency reprogramming (Figure 7A, B, F and Supplementary Figure S11), we surmised that Sox2 directly regulates their exon selection. A connection between G/C content and splicing site selection has previously been posited for other splicing modifiers (67,90). Taken together, our data suggest that the RBM of Sox2 might influence reprogramming via splice site selection. Moreover, Sox2 associates with RNA and DNA simultaneously (Figure 4). This observation implies that Sox2 links RNA and chromatin. In human cells, pre-mRNA splicing is initiated during transcription (53), and factors regulating chromatin and transcription affect the splicing process (91). Hirsch et al. found that Gcn5 associates with Myc to induce AS in the early stages of somatic cell reprogramming (92). Therefore, we hypothesize that Sox2 regulates splicing co-transcriptionally employing its RBM whilst bound to cognate enhancers through its HMG box. Further work should explore the crosstalk between Sox2-regulated transcription and splicing at different stages of reprogramming.
DATA AVAILABILITY
The RNA-seq and PAR-CLIP sequencing data reported in this paper have been deposited with the NCBI GEO under accession number GSE115452.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Rory Johnson (Universität Bern) for advice and comments on the manuscript. We thank Dr Calista Keow Leng Ng (Institute of Medical Biology, Singapore) for the kind gift of the pETG20a-Sox1 plasmid.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Young Scientists Fund of the National Natural Science Foundation of China [31501058 to L.H.]; National Natural Science Foundation of China [31771454, 31471238, 31611130038 to R.J.]; National Key Research and Development Program of China Stem Cell and Translational Research [2017YFA0105103, 2017YFA0105101 to R.J.]; Ministry of Science and Technology of China [2013DFE33080, 2016YFA0100701 to R.J.]; 100 talent award of the Chinese Academy of Sciences [to R.J.]; Science and Technology Planning Projects of Guangdong Province, China [2014B030301058, 2016A050503038 to R.J.]; Youth Innovation Promotion Association of the Chinese Academy of Sciences [2015294 to X.B.]; Natural Science Foundation of Guangdong Province [2018B030306042 to X.B.]. Funding for open access charge: National Science Foundation for Young Scientists of China [31501058]; Ministry of Science and Technology of China [2016YFA0100701].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hudson W.H., Ortlund E.A.. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014; 15:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A.. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015; 350:978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Melikishvili M., Chariker J.H., Rouchka E.C., Fondufe-Mittendorf Y.N.. Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov. 2017; 3:17043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matveeva E., Maiorano J., Zhang Q., Eteleeb A.M., Convertini P.. Involvement of PARP1 in the regulation of alternative splicing. Cell Discov. 2016; 2:15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P.. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010; 3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamachi Y., Kondoh H.. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013; 140:4129–4144. [DOI] [PubMed] [Google Scholar]
- 7. Sarkar A., Hochedlinger K.. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013; 12:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hou L., Srivastava Y., Jauch R.. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017; 63:2–12. [DOI] [PubMed] [Google Scholar]
- 9. Kamachi Y., Uchikawa M., Kondoh H.. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000; 16:182–187. [DOI] [PubMed] [Google Scholar]
- 10. Reiprich S., Wegner M.. Sox2: a multitasking networker. Neurogenesis (Austin). 2014; 1:e962391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Remenyi A., Lins K., Nissen L.J., Reinbold R., Scholer H.R., Wilmanns M.. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003; 17:2048–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ng C.K., Li N.X., Chee S., Prabhakar S., Kolatkar P.R., Jauch R.. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012; 40:4933–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dailey L., Basilico C.. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell Physiol. 2001; 186:315–328. [DOI] [PubMed] [Google Scholar]
- 14. Lodato M.A., Ng C.W., Wamstad J.A., Cheng A.W., Thai K.K., Fraenkel E., Jaenisch R., Boyer L.A.. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLos Genet. 2013; 9:e1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jerabek S., Ng C.K., Wu G., Arauzo-Bravo M.J., Kim K.P., Esch D., Malik V., Chen Y., Velychko S., MacCarthy C.M. et al.. Changing POU dimerization preferences converts Oct6 into a pluripotency inducer. EMBO Rep. 2017; 18:319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Penrad-Mobayed M., Perrin C., L’Hote D., Contremoulins V., Lepesant J.A., Boizet-Bonhoure B., Poulat F., Baudin X., Veitia R.A.. A role for SOX9 in post-transcriptional processes: insights from the amphibian oocyte. Sci. Rep. 2018; 8:7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohe K., Lalli E., Sassone-Corsi P.. A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ng S.Y., Johnson R., Stanton L.W.. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012; 31:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ng S.Y., Bogu G.K., Soh B.S., Stanton L.W.. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013; 51:349–359. [DOI] [PubMed] [Google Scholar]
- 20. Bellucci M., Agostini F., Masin M., Tartaglia G.G.. Predicting protein associations with long noncoding RNAs. Nat. Methods. 2011; 8:444–445. [DOI] [PubMed] [Google Scholar]
- 21. Sakashita E., Sakamoto H.. Characterization of RNA binding specificity of the Drosophila sex-lethal protein by in vitro ligand selection. Nucleic Acids Res. 1994; 22:4082–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Loughlin F.E., Mansfield R.E., Vaz P.M., McGrath A.P., Setiyaputra S., Gamsjaeger R., Chen E.S., Morris B.J., Guss J.M., Mackay J.P.. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S. et al.. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010; 6:71–79. [DOI] [PubMed] [Google Scholar]
- 24. Li A., Chen Y.S., Ping X.L., Yang X., Xiao W., Yang Y., Sun H.Y., Zhu Q., Baidya P., Wang X. et al.. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017; 27:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klaus M., Prokoph N., Girbig M., Wang X., Huang Y.H., Srivastava Y., Hou L., Narasimhan K., Kolatkar P.R., Francois M. et al.. Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction. Nucleic Acids Res. 2016; 44:3922–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y.H., Jankowski A., Cheah K.S., Prabhakar S., Jauch R.. SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci. Rep. 2015; 5:10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keene J.D., Komisarow J.M., Friedersdorf M.B.. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006; 1:302–307. [DOI] [PubMed] [Google Scholar]
- 28. Lee T.I., Johnstone S.E., Young R.A.. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006; 1:729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011; 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen S., Park J.W., Lu Z.X.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C. et al.. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015; 18:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K.. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017; 168:442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 38. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heyd F., Lynch K.W.. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol. Cell. 2010; 40:126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.C., Munschauer M. et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Z., Kraus W.L.. Catalytic-Independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell. 2017; 65:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cox J.L., Mallanna S.K., Luo X., Rizzino A.. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLoS One. 2010; 5:e15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gagliardi A., Mullin N.P., Ying Tan Z., Colby D., Kousa A.I., Halbritter F., Weiss J.T., Felker A., Bezstarosti K., Favaro R. et al.. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J. 2013; 32:2231–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwon S.C., Yi H., Eichelbaum K., Fohr S., Fischer B., You K.T., Castello A., Krijgsveld J., Hentze M.W., Kim V.N.. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1122–1130. [DOI] [PubMed] [Google Scholar]
- 45. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 46. Terribilini M., Lee J.H., Yan C., Jernigan R.L., Honavar V., Dobbs D.. Prediction of RNA binding sites in proteins from amino acid sequence. RNA. 2006; 12:1450–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pevny L.H., Lovell-Badge R.. Sox genes find their feet. Curr. Opin. Genet. Dev. 1997; 7:338–344. [DOI] [PubMed] [Google Scholar]
- 48. Galarneau A., Richard S.. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005; 12:691–698. [DOI] [PubMed] [Google Scholar]
- 49. Aksoy I., Jauch R., Eras V., Chng W.B., Chen J., Divakar U., Ng C.K., Kolatkar P.R., Stanton L.W.. Sox transcription factors require selective interactions with Oct4 and specific transactivation functions to mediate reprogramming. Stem Cells. 2013; 31:2632–2646. [DOI] [PubMed] [Google Scholar]
- 50. Guallar D., Wang J.. RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol. (Beijing). 2014; 9:389–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O’Hanlon D., Sung H.K., Alvarez M., Talukder S., Pan Q., Mazzoni E.O. et al.. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011; 147:132–146. [DOI] [PubMed] [Google Scholar]
- 52. Lu Y., Loh Y.H., Li H., Cesana M., Ficarro S.B., Parikh J.R., Salomonis N., Toh C.X., Andreadis S.T., Luckey C.J. et al.. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014; 15:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brody Y., Shav-Tal Y.. Transcription and splicing: when the twain meet. Transcription. 2011; 2:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G.. Multiple links between transcription and splicing. RNA. 2004; 10:1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye S., Zhang T., Tong C., Zhou X., He K., Ban Q., Liu D.. Depletion of Tcf3 and Lef1 maintains mouse embryonic stem cell self-renewal. Biol. Open. 2017; 6:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu Y., Chen K., Liu X., Huang L., Zhao D., Li L., Gao M., Pei D., Wang C., Liu X.. Srebp-1 interacts with c-Myc to enhance somatic cell reprogramming. Stem Cells. 2016; 34:83–92. [DOI] [PubMed] [Google Scholar]
- 57. Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. et al.. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012; 483:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M. et al.. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat. Cell Biol. 2013; 15:872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pawlak M., Jaenisch R.. De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev. 2011; 25:1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu J.Q., Habegger L., Noisa P., Szekely A., Qiu C., Hutchison S., Raha D., Egholm M., Lin H., Weissman S. et al.. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bodak M., Cirera-Salinas D., Yu J., Ngondo R.P., Ciaudo C.. Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Biol. 2017; 7:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu G., Zheng H., Ai W.. C-terminal binding proteins (CtBPs) attenuate KLF4-mediated transcriptional activation. FEBS Lett. 2009; 583:3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jain K., Warmack R.A., Debler E.W.. Protein arginine methyltransferase product specificity is mediated by distinct active-site architectures. J. Biol. Chem. 2016; 291:18299–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mallanna S.K., Ormsbee B.D., Iacovino M., Gilmore J.M., Cox J.L., Kyba M., Washburn M.P., Rizzino A.. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010; 28:1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fang X., Yoon J.G., Li L., Tsai Y.S., Zheng S., Hood L., Goodlett D.R., Foltz G., Lin B.. Landscape of the SOX2 protein-protein interactome. Proteomics. 2011; 11:921–934. [DOI] [PubMed] [Google Scholar]
- 66. Saud K., Canovas J., Lopez C.I., Berndt F.A., Lopez E., Maass J.C., Barriga A., Kukuljan M.. SFPQ associates to LSD1 and regulates the migration of newborn pyramidal neurons in the developing cerebral cortex. Int. J. Dev. Neurosci. 2017; 57:1–11. [DOI] [PubMed] [Google Scholar]
- 67. Amit M., Donyo M., Hollander D., Goren A., Kim E., Gelfman S., Lev-Maor G., Burstein D., Schwartz S., Postolsky B. et al.. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012; 1:543–556. [DOI] [PubMed] [Google Scholar]
- 68. Rawlings S.L., Matt G.D., Huber P.W.. Analysis of the binding of Xenopus transcription factor IIIA to oocyte 5 S rRNA and to the 5 S rRNA gene. J. Biol. Chem. 1996; 271:868–877. [DOI] [PubMed] [Google Scholar]
- 69. Nolte R.T., Conlin R.M., Harrison S.C., Brown R.S.. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mosner J., Mummenbrauer T., Bauer C., Sczakiel G., Grosse F., Deppert W.. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995; 14:4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ewen M.E., Oliver C.J., Sluss H.K., Miller S.J., Peeper D.S.. p53-dependent repression of CDK4 translation in TGF-beta-induced G1 cell-cycle arrest. Genes Dev. 1995; 9:204–217. [DOI] [PubMed] [Google Scholar]
- 72. Miller S.J., Suthiphongchai T., Zambetti G.P., Ewen M.E.. p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol. Cell Biol. 2000; 20:8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Carmel L., Chorev M.. The function of introns. Front. Genet. 2012; 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M. et al.. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005; 37:844–852. [DOI] [PubMed] [Google Scholar]
- 75. Dredge B.K., Darnell R.B.. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell Biol. 2003; 23:4687–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ekonomou A., Kazanis I., Malas S., Wood H., Alifragis P., Denaxa M., Karagogeos D., Constanti A., Lovell-Badge R., Episkopou V.. Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 2005; 3:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bergsland M., Ramskold D., Zaouter C., Klum S., Sandberg R., Muhr J.. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011; 25:2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huambachano O., Herrera F., Rancourt A., Satoh M.S.. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 2011; 286:7149–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Barraud P., Allain F.H.. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 2012; 353:35–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yoshida Y., Izumi H., Torigoe T., Ishiguchi H., Yoshida T., Itoh H., Kohno K.. Binding of RNA to p53 regulates its oligomerization and DNA-binding activity. Oncogene. 2004; 23:4371–4379. [DOI] [PubMed] [Google Scholar]
- 82. Tapia N., Reinhardt P., Duemmler A., Wu G., Arauzo-Bravo M.J., Esch D., Greber B., Cojocaru V., Rascon C.A., Tazaki A. et al.. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012; 3:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Malik V., Glaser L.V., Zimmer D., Velychko S., Weng M., Holzner M., Arend M., Chen Y., Srivastava Y., Veerapandian V. et al.. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat. Commun. 2019; 10:3477–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Samudyata, Amaral P.P., Engstrom P.G., Robson S.C., Nielsen M.L., Kouzarides T., Castelo-Branco G.. Interaction of Sox2 with RNA binding proteins in mouse embryonic stem cells. Exp. Cell Res. 2019; 381:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Falk S., Finogenova K., Melko M., Benda C., Lykke-Andersen S., Jensen T.H., Conti E.. Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors. Nat. Commun. 2016; 7:13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002; 3:662–673. [DOI] [PubMed] [Google Scholar]
- 87. Wang G., Weng R., Lan Y., Guo X., Liu Q., Liu X., Lu C., Kang J.. Synergetic effects of DNA methylation and histone modification during mouse induced pluripotent stem cell generation. Sci. Rep. 2017; 7:39527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gopalakrishna-Pillai S., Iverson L.E.. A DNMT3B alternatively spliced exon and encoded peptide are novel biomarkers of human pluripotent stem cells. PLoS One. 2011; 6:e20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. He S., Pant D., Schiffmacher A., Meece A., Keefer C.L.. Lymphoid enhancer factor 1-mediated Wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells. 2008; 26:842–849. [DOI] [PubMed] [Google Scholar]
- 90. Roca X., Krainer A.R., Eperon I.C.. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013; 27:129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Braunschweig U., Gueroussov S., Plocik A.M., Graveley B.R., Blencowe B.J.. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013; 152:1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hirsch C.L., Coban Akdemir Z., Wang L., Jayakumaran G., Trcka D., Weiss A., Hernandez J.J., Pan Q., Han H., Xu X. et al.. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 2015; 29:803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and PAR-CLIP sequencing data reported in this paper have been deposited with the NCBI GEO under accession number GSE115452.