Abstract
Rediscovered as a potential eukaryotic epigenetic mark, DNA N6-adenine methylation (6mA) varies across species in abundance and its relationships with transcription. Here we characterize AMT1—representing a distinct MT-A70 family methyltransferase—in the ciliate Tetrahymena thermophila. AMT1 loss-of-function leads to severe defects in growth and development. Single Molecule, Real-Time (SMRT) sequencing reveals that AMT1 is required for the bulk of 6mA and all symmetric methylation at the ApT dinucleotides. The detection of hemi-methylated ApT sites suggests a semi-conservative mechanism for maintaining symmetric methylation. AMT1 affects expression of many genes; in particular, RAB46, encoding a Rab family GTPase involved in contractile vacuole function, is likely a direct target. The distribution of 6mA resembles H3K4 methylation and H2A.Z, two conserved epigenetic marks associated with RNA polymerase II transcription. Furthermore, strong 6mA and nucleosome positioning in wild-type cells is attenuated in ΔAMT1 cells. Our results support that AMT1-catalyzed 6mA is an integral part of the transcription-associated epigenetic landscape. AMT1 homologues are generally found in protists and basal fungi featuring ApT hyper-methylation associated with transcription, which are missing in animals, plants, and true fungi. This dichotomy of 6mA functions and the underlying molecular mechanisms may have implications in eukaryotic diversification.
INTRODUCTION
First identified and characterized as a DNA modification in bacteria, N6-adenine methylation (6mA) was for a long time only known in a few eukaryotes, including the ciliate Tetrahymena thermophila (1–5). More recently, 6mA has been identified in the genomic DNA of a wide range of eukaryotic organisms, from protists and basal fungi (5–7) to animals and plants (8–17). Nevertheless, DNA 6mA is far from universal in eukaryotes. Furthermore, in animals such as worms, flies, mice, and humans (11,12,14,15,18), 6mA levels are orders of magnitude lower than those in unicellular eukaryotes such as Tetrahymena (1,19) and Chlamydomonas (6). Even more complicated are the functions of 6mA. It is implicated as an epigenetic mark for either transcription activation or repression, depending on the organism under study (6,10,13,14,17). It is also reported to regulate development in the fly (16), carry heritable epigenetic information in the worm (8,20), respond dynamically to stress in the mouse brain (21), and participate in carcinogenesis of human glioblastoma (15,18). The patchy distribution, varying abundance, and divergent functions of DNA 6mA in eukaryotes suggest a complex evolutionary history.
Comprehensive phylogenomic analyses guide the search for DNA 6mA methyltransferases (MTases) in eukaryotes. Prominent among potential candidates are members of the MT-A70 family, which evolved from the bacterial M.MunI-like DNA 6mA MTase (22). Eukaryotic MT-A70 family members METTL3 and METTL14 form a heterodimer and deposit m6A in mRNA—facilitated by the single-stranded RNA binding CCCH domain of METTL3 (23). It is extrapolated that the MT-A70 MTase domain, tethered to structural domains with an alternative substrate preference, may catalyze DNA 6mA. Indeed, recent studies support that METTL4 orthologues, representing a separate subclade of the eukaryotic MT-A70 family widespread in animals and plants, are likely DNA 6mA MTases (8). Still, other subclades of the eukaryotic MT-A70 family—each with their own distinct domain architectures—are poorly studied. Their functional analyses promise to reveal the molecular mechanisms underpinning divergent 6mA behaviors and substantiate their independent origins.
DNA 6mA in Tetrahymena, discovered >40 years ago (1), is highly abundant (∼1% of adenine). It is enriched in linker DNA of stereotypical nucleosome arrays downstream of transcription start sites, as an integral part of the epigenetic landscape (1,19,24,25). These findings strongly suggest that DNA 6mA in Tetrahymena is deposited by specific MTases rather than by random uncatalyzed reactions. As putative DNA 6mA MTases, there are several MT-A70 family members in the Tetrahymena genome (Figure 1). Here, we identify AMT1 (adenine methyltransferase 1), belonging to a distinct and previously uncharacterized eukaryotic MT-A70 subclade, as the one required for the bulk DNA 6mA in general, and for symmetric ApT methylation in particular. We also provide detailed functional analysis of AMT1-dependent 6mA, supporting its role for regulating cell growth and development, as an active epigenetic mark associated with RNA polymerase II (Pol II) transcription. Despite their absence in animals, plants and true fungi, AMT1 orthologues are present in all the eukaryotic super-groups. Their phylogenetic distribution coincides with abundant 6mA in genomic DNA, especially symmetric ApT methylation, supporting AMT1 homologues as prototypical DNA 6mA MTases in eukaryotes.
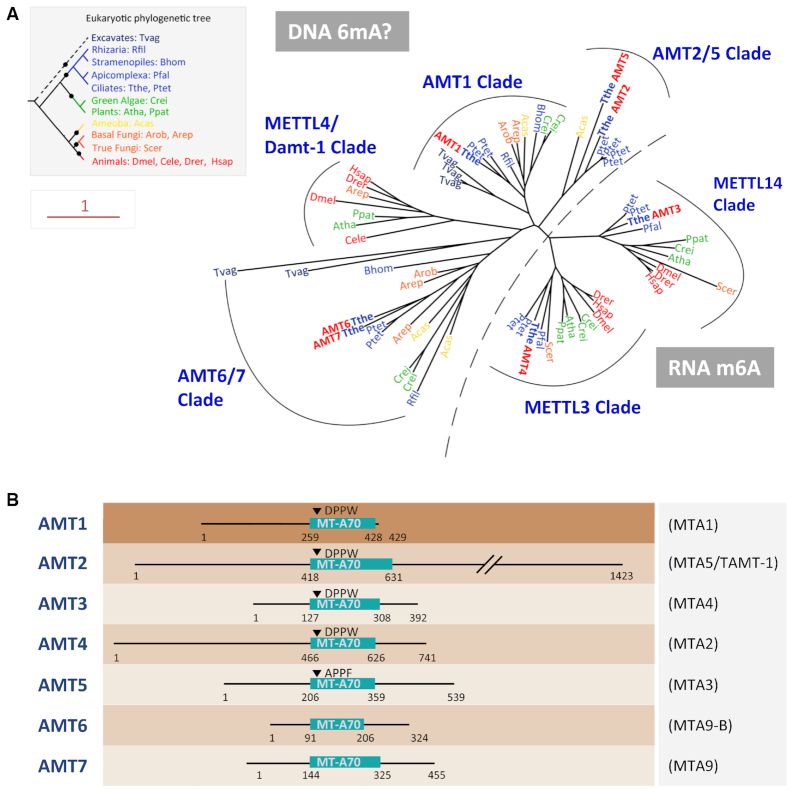
Figure 1.
Phylogenetic analysis and domain structure comparison of AMTs 1–7. (A) Phylogenetic analysis of MT-A70 proteins. DNA 6mA (subclades AMT2/5, AMT1, METTL4/DAMT1 and AMT6/7) and RNA m6A (subclades METTL3 and METTL14) methyltransferase candidates are separated by a dotted line. Species are marked by different colors based on their phylogenetic position in the eukaryotic tree (inset). AMTs 1–7 of Tetrahymena are in bold plus red. The scale bar corresponds to 1 expected amino acid substitution per site. See Supplementary Table S9 for details (species full name and NCBI GI number). (B) Conserved domains and motifs in AMTs 1–7. Gene names used in Luo et al. (70) and Beh et al. (75) are shown in parentheses. MT-A70 domains of AMTs 1–5 were predicted by CD-Search (69), while domain structures of AMT6 and AMT7 were inferred from sequence alignment with AMTs 1–5.
MATERIALS AND METHODS
Cell culture
Tetrahymena thermophila wild-type strains (SB210 and CU428) were obtained from the Tetrahymena Stock Center (http://tetrahymena.vet.cornell.edu). ΔAMT1 was a homozygous homokaryon strain generated in this study. Cells were grown in SPP medium at 30°C (26,27).
Generation of Tetrahymena strains
To generate the ΔAMT1 construct, the neo4 cassette (28,90) was flanked with the 5′ and 3′ flanking regions of AMT1 (TTHERM_00704040) (Supplementary Figure S1A, B and Table S8). Starved WT cells of two different mating types (SB210 and CU428) were mated and transformed with the ΔAMT1 construct at 3 h post-mixing. Homozygous heterokaryon strains that were AMT1 knockout in the MIC, while retaining the intact AMT1 gene in the MAC, were generated by germline transformation and standard genetic manipulations (29,30). By crossing two homozygous heterokaryon strains, homozygous homokaryon strains (ΔAMT1 cells) were generated that are AMT1 knockout in both the MIC and MAC. This was confirmed by the SMRT sequencing and by the amplification of the AMT1 transcript using RT-PCR (Supplementary Figure S1C, S8A–C). ΔAMT2-5 and point mutation (DPPW to APPA, AMT1-APPA) cells (Supplementary Figure S9A–C) were generated using the same strategy.
We initially introduced the hemagglutinin (HA) tag to the N-terminus of the endogenous AMT1 gene, but failed to detect any immunofluorescence signal, probably due to the low expression level (31). Instead, we generated the AMT1 overexpression (OE) construct (AMT1-NHA-OE), by placing AMT1 ORF, with an N-terminal HA-tag under the cadmium-inducible MTT1 (Supplementary Figure S5A, B). The AMT1-NHA-OE construct was transformed into SB210 cells and selected in increasing concentration of Paromomycin to generate a somatic AMT1-overexpression strain or was transformed into mating pairs of SB210 and CU428 to generate a germline AMT1-overexpression strain.
To generate the AMT1 rescue (AMT1-RS) construct, AMT1 ORF and its flanking regions were cloned and the bsr2 cassette was inserted into the 5′ UTR (32) (Supplementary Figure S7A). The AMT1-RS construct was transformed into ΔAMT1 cells and selected by increasing the concentration of blasticidin S (Supplementary Figure S7B).
To generate the RAB46 knockdown (RAB46-KD) construct (33,34), a fragment containing 203bp RAB46 CDS was inserted in opposite orientations downstream of the cadmium-inducible MTT1 promoter, to produce hairpin RNA for RNAi (Supplementary Figure S16A). The RAB46-KD construct was transformed into WT cells and selected in increasing concentration of paromomycin (Supplementary Figure S16B). Cells were treated with 4 μg/ml CdCl2 for 24 h to induce the large contractile vacuole phenotype.
Phylogenetic analysis
The AMTs 1–7 amino acid sequences (gi numbers are listed in Supplementary Table S9) were queried against the NCBI nr database using PSI-BLAST (35–37) (maximum E-value = 1e–4). Retrieved hits were collapsed using CD-HIT (38) (-c 0.97) to remove redundant sequences. Only sequences corresponding to the taxa in Figure 1A were retained. Sequences were aligned using the MUSCLE program (39,40). A Phylogenetic tree was obtained using the approximate maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (41). All protein sequences used for phylogenetic analysis are listed in Supplementary Table S9.
Immunofluorescence staining
Immunofluorescence (IF) staining experiments followed previously described procedures (19,42–44). For the single antibody staining, the primary antibodies were α -6mA (Synaptic Systems, 202003, 1:2000) (19) and α -HA (Cell Signaling, #3724, 1:200) and the secondary antibody was Goat anti-Rabbit IgG (H+L) (Invitrogen, A-21428, 1:4000). For the co-staining, cells were first incubated with α -HA (Covance, MMS-101P, 1:500, mouse) and its secondary antibody (goat anti-mouse IgG (H+L), Alexa Fluor 555, Invitrogen, A-21127, 1:4000). After crosslink by 3% PFA, α -6mA (Synaptic Systems, 202003, 1:2000) and its secondary antibody (goat anti-rabbit IgG (H+L), Alexa Fluor 488, Invitrogen, A-11008, 1:4000) were used. Digital images were collected using an Olympus BX43 microscope with an Olympus DP72 camera.
Preparation of Tetrahymena DNA and RNA samples
Genomic DNA was collected from vegetative Tetrahymena log-phase cells (∼2 × 105 cells/ml) using Wizard® Genomic DNA Purification Kit (Promega, A1120). RNA samples were extracted from vegetative log-phase cells (∼2 × 105 cells/ml) by the RNeasy Plus Mini kit (Qiagen, 74134). The quality and concentration of DNA and RNA samples were analyzed by agarose gel electrophoresis and Qubit®3.0 Fluorometer (Thermo Fisher Scientific).
UHPLC–QQQ–MS/MS Analysis
500 ng genomic DNA of Tetrahymena thermophila was denatured in 20 μl of ddH2O at 95°C for 5 min, chilled on ice rapidly for 5 min. After cooling, genomic DNA was digested into mononucleosides using a mixture of enzymes, including DNase I (1 U, NEB, M0303L), calf intestinal phosphatase (1 U, NEB, M0290L), and snake venom phosphodiesterase I (0.005 U, Sigma, P4506), at 37°C for 12 h. Digested DNA was diluted and then purified by 0.22 μm filter (Millipore, SLGVR04NL). Samples were analyzed by ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC–QQQ–MS/MS) on an Acquity BEH C18 column (75 mm × 2.1 mm, 1.7 μm, Waters, MA, USA), using a Xevo TQ-S triple quadrupole mass spectrometer (Waters, Milford, MA, USA) (16,45). The mass spectrometer was set to multiple reaction monitoring (MRM) in positive electrospray mode. The selective MRM transitions were detected under m/z 266/150 for 6mA, m/z */* for 1 mA and m/z 252/136 for dA. The ratio of 6mA/A was quantified by the calibration curves of nucleoside standards running at the same time.
Phenotypic analysis of vegetative ΔAMT1 cells
To monitor the growth rate, ΔAMT1 progeny and WT (SB210) cells were cultured in SPP medium at 30°C by starting at the concentration of 0.6–0.8 × 105 cells/ml. Beckman Coulter Z2 Particle Counter was used to count the cell density at indicated time points for 32 h. Statistical analysis was performed by GraphPad Prism5 (46).
Movies of swimming cells were captured in rates of 500 frames per second (fps) using an Olympus BX53F microscope with an Olympus DP74 camera. Image processing steps were performed with ImageJ (National Institutes of Health, Bethesda, MD, USA).
For the osmotic pressure test, ΔAMT1 and WT (SB210) cells were cultured in SPP medium (-glucose) with different concentrations of sucrose (5%, 4%, 3%, 2%, 1%, 0%) until reaching the log-phase (2–3 × 105 cells/ml). The numbers of cells with or without large contractile vacuoles were then calculated.
Conjugation progress analysis and viability test
Starved ΔAMT1-1-10 and ΔAMT1-1-11 cells (2 × 105 cells/ml) were mixed to induce conjugation. Conjugating WT cells (SB210 × CU428) were used as control setups. To monitor the progress of conjugation, cells were collected at different stages of conjugation (3, 4, 6, 10 h after mixing) and fixed with 1% PFA and 100 μg/ml DAPI.
For the viability test, individual mating pairs were isolated and placed into drops with SPP medium at 10 h post-mixing. After 48 h incubation in 96-well plates at 30°C, the number of wells with viable cells was counted. To distinguish true progeny (exconjugants) from aborted pairs (non-conjugants), viable cells were starved to induce conjugation with WT cells (SB210 or CU428 respectively). Cells that were unable to mate with two WT strains of different mating types were characterized as true conjugation progeny.
Reverse transcription polymerase chain reaction (RT-PCR) and quantitative reverse transcription PCR (qRT-PCR)
Total RNA after DNase treatment (Invitrogen, AM1907) was reverse-transcribed using an oligo-dT primer and M-MLV Reverse Transcriptase (Invitrogen, 28025013). RT-PCR was performed by using Premix Taq (TaKaRa, RR901A). The housekeeping gene JMJ1 (TTHERM_00185640) was used for loading control and normalization. For qRT-PCR analysis of AMT1 expression levels in growing, starved, and conjugating cells, the housekeeping gene RPL5 (TTHERM_00736480) was used for loading control and normalization. All PCR primers are listed in Supplementary Table S8.
DpnI/DpnII digestion and quantitative PCR (qPCR) analysis
DpnI/DpnII digestion experiment followed previously described procedures (19). DpnI/DpnII digested and non-digested DNA (4 ng) were loaded into the qPCR analysis using EvaGreen Express 2 × qPCR MasterMix-Low ROX (Abm, MasterMix-LR). Primers flanking selected GATC sites are listed in Supplementary Table S6. Primers in the CDS of JMJ1 (TTHERM_00185640) were used as internal controls. The methylation level (6mA/A) is reflected by fold difference between DpnI- and DpnII-digested samples (ΔΔCt = ΔCtDpnI – ΔCtDpnII). ΔCtDpnI and ΔCtDpnII were normalized respectively between digested and undigested samples (ΔCtDpnI = CtDpnI – Ctundigested and ΔCtDpnII = CtDpnII – Ctundigested). As DpnI and DpnII cut methylated and unmethylated GATC sequences respectively, the following results are expected. For highly methylated sites, the methylation level (6mA/A) should be larger than zero in WT and AMT1-RS cells, but below zero in ΔAMT1 and AMT1-APPA cells. For unmethylated sites, the methylation level (6mA/A) should be substantially below zero in all four strains.
Nuclei purification and MNase sequencing analysis
Nuclei purification was carried out following established protocols (47). Approximately 5 × 107 purified MACs from WT and ΔAMT1 cells were digested by Micrococcal Nuclease (400 U/ml MNase, NEB, M0247S) at 25°C for 15 min and mono-nucleosome-sized DNA was collected by phenol–chloroform extraction for sequencing.
Sequencing reads were mapped to the latest MAC genome assembly in the Tetrahymena genome database (TGD) (http://ciliate.org) (48,49). The mapping results were visualized using GBrowse 2.0 (50). Only mono-nucleosome-sized fragments (120–260 bp) were analyzed. To calculate nucleosome distribution around TSS and the distance between 6mA sites and nucleosomes dyads, a dyad was defined as the midpoint of a fragment. Nucleosome frequency was counted by dyads accumulated in every base from 1000-nt upstream to 2000-nt downstream TSS. Nucleosomes were called using the NucPosSimulator (51) which identifies non-overlapping nucleosomes using fragment centers. The degree of translational positioning for each called nucleosome was described previously, defined as the number of fragment centers within ±20 bp of the called nucleosome dyad, relative to the number of all fragment centers within the 147 bp called nucleosome footprint (52). +1 to +5 nucleosomes with positioning degree ≥0.7 were selected according to their distance to TSS. 6mA between dyads was defined as all 6mA sites between dyads of nucleosome N and nucleosome (N + 1).
SMRT sequencing and data analysis
Genomic DNA prepared for SMRT sequencing libraries was extracted from WT (SB210) and ΔAMT1 cells using Wizard® Genomic DNA Purification Kit (Promega, A1120), and sequencing was carried out by Novogene Co. Ltd (Beijing, China). Even though SMRT sequencing does not discriminate 6mA versus 1mA, we could call 6mA with confidence from SMRT sequencing results as 1mA was not detected in the Tetrahymena genome by mass spectrometry (Supplementary Figure S3A–C).
The latest SB210 MAC genome downloaded from TGD (http://ciliate.org) was used as a reference for reads mapping. 6mA was identified using the Base Modification and Motif Analysis protocol with default parameters in the SMRT link v5.10 (Pacific Biosciences). Considering the different sequencing depths between SB210 and ΔAMT1, all data were normalized to 100× while using strict cut-off (Qv >30 and coverage >25×) to filter out unauthentic modifications.
The two longest chromosomes (scf_8254667 and scf_8254697) were used for SMRT Circular Consensus Sequences (CCS) analysis. The single molecules were extracted from the raw sequencing data to calculate the IPD ratio of every base in both strands by customized Perl scripts.
To calculate 6mA density across scaffolds, the MAC scaffolds were mapped onto MIC chromosomes using NUCmer (53). Note that five scaffolds (scf_8255058, scf_8255661, scf_8255483, scf_8255586, scf_8255776) were unmapped in MIC, scaffolds without 6mA and scf_8254915 with extremely high 6mA density were also removed from the analysis. Smooth curves were plotted by ggplot2 in R (54).
For composite analysis and motif identification, 6mA was divided into groups based on their methylation level (low 10–20%, intermediate 20–80%, high 80–100%) or motifs (symmetric/asymmetric/non-AT). The number and percentage of sites of different 6mA groups were calculated by customized Perl scripts and plotted using GraphPad Prism 6 (46).
The genome-wide distribution of 6mA groups on scaffolds was counted by customized Perl scripts and the 6mA density was calculated as the number of methylated adenine sites divided by the total number of adenine sites (6mA/A) in each bin (bin size = 1 kb). The circle diagrams were generated using circlize 0.4.4 (55). Among these, the longest scaffold (scf_8254803) was shown individually.
For analysis of 6mA, H2A.Z and H3K4me3 distribution among genes, 15 841 well-modeled genes (13 854 of which are longer than 1 kb) that are strongly supported by deep RNA-seq sequencing results were selected (52,56,57). The gene body length was scaled to 1 unit and one-unit length was extended to each side. Customized Perl scripts were used for locus statistic (bin size = 0.05). To calculate distribution of these marks around the transcription start sites (TSS), the numbers of 6mA sites and midpoints of ChIP-Seq fragments were accumulated in every base from 1000-nt upstream to 2000-nt downstream TSS. The 6mA amount was defined as the number of methylated adenine sites combined with their methylation level.
The correlation matrix of H2A.Z, H3K4me3 and 6mA frequency was accomplished by corrplot (58), of which H2A.Z (19) and H3K4me3 were defined as midpoints of ChIP fragments accumulated in the 1 kb downstream of TSS.
To identify conserved motifs around the methylated adenines, sequences between 20-nt upstream and 20-nt downstream of 6mA sites were extracted. Local motifs nearby 6mA were illustrated by WebLogo3 (59) and GraphPad Prism 6.
To determine the correlation between changes in the number and methylation level of 6mA sites and gene expression, one-factor analysis of variance (one-way ANOVA), one sample t-test and independent sample t-test were all carried out by SPSS v. 22.0 (60).
To validate the dramatic change in knockout cells, we generated another ΔAMT1 strain and subjected it to SMRT sequencing. The two strains shared 69 154 methylated adenine sites, corresponding to ∼50% overlap (Supplementary Figure S17A). More strikingly, a similar pattern of change (preferential loss of symmetric, highly methylated 6mA in the genic region) was observed in this independently generated strain (Supplementary Figure S17B, C), indicating that AMT1 indeed modulates genome-wide 6mA distribution.
RNA sequencing and data analysis
A total of twelve RNA samples of T. thermophila were sequenced, three replicates each for WT (SB210), AMT1-RS (AMT1-RS-14 and AMT1-RS-17) and ΔAMT1 respectively. After trimming sequencing adapters and filtering low quality reads according to Trimmomatic (61) (TruSeq3-PE.fa: 2:30:10, leading: 3, trailing: 3, sliding window: 4:15, minlen: 80), the numbers of reads mapped to the genome were determined using the HTSeq2 software (62). Stringtie 1.3.4 (63) was used for assembling potential transcripts as the reference, and featureCounts (64) was implemented for counting reads to genomic features with the assembled transcripts as reference. Effective expression levels (FPKM > 1) were calculated based on RNA-Seq coverage of these strains for counting the Pearson's correlation coefficients in gene expression with DEsq2 (65). Differentially expressed genes (DEGs) were also identified by DEsq2 (log2FoldChange > 1 or < -1, P < 0.05). The selected genes were annotated using Interproscan 5.29 (66) and Gene Ontology (GO) analysis was performed by WEGO (67). Pathway analysis of DEGs was carried out on the KEEG web server (https://www.genome.jp/kegg/pathway.html). RNA-Seq data for starved cells were acquired from Feng et al. (68).
RESULTS
DNA 6mA in Tetrahymena mainly depends on AMT1
We identified in the genome of Tetrahymena thermophila seven genes with strong homology to the MT-A70 domain of methyltransferases (MTases), and named them adenine MTase 1–7 (AMTs 1–7) (Figure 1A, B; Supplementary Table S1). AMT1 belongs to a distinct eukaryotic subclade, with orthologues widely distributed in protists and basal fungi, but missing in animals, plants, and true fungi. AMT2 and AMT5 are grouped into another subclade with more limited distribution in protists. AMT3 is the METTL14 orthologue, while AMT4, with the conserved N-terminal CCCH domain, is the Tetrahymena orthologue of METTL3. AMT6 and AMT7 form a divergent subclade; the phylogenetic distribution of their orthologues mirrors that of AMT1. Tetrahymena AMTs 1–4, but not 5–7, have the 6mA MTase signature motif ([DNSH]PP[YFW]) required for substrate recognition and catalytic activity (Supplementary Table S1) (22,69). None of them are METTL4 homologues, which are represented by the putative DNA MTase DAMT-1 in Caenorhabditis elegans (8), but are not found in any protists.
To determine whether any AMTs are responsible for DNA 6mA in Tetrahymena, we generated knockout strains deleting each of AMTs 1–5, mainly focusing on genes with potential catalytic activity. Immunofluorescence staining showed that compared with wild-type (WT) cells, DNA 6mA levels in the macronucleus (MAC) of growing cells were greatly reduced in ΔAMT1 cells, but not in AMTs 2–5 knockout cells (Figures 2A, B, S2A–C). This was corroborated by mass spectrometry analysis, showing that global DNA 6mA levels in ΔAMT1 cells were reduced to about 30%–40% of WT levels (Figures 2B, C, S2D, S3A–C). No significant change in RNA m6A levels was observed in ΔAMT1 cells (Supplementary Figure S3D), indicating that DNA is the specific substrate for AMT1, consistent with a recently published finding that AMT1 is selective for DNA over RNA in vitro (75). We therefore focused our analysis on AMT1, the main DNA 6mA MTase in Tetrahymena.
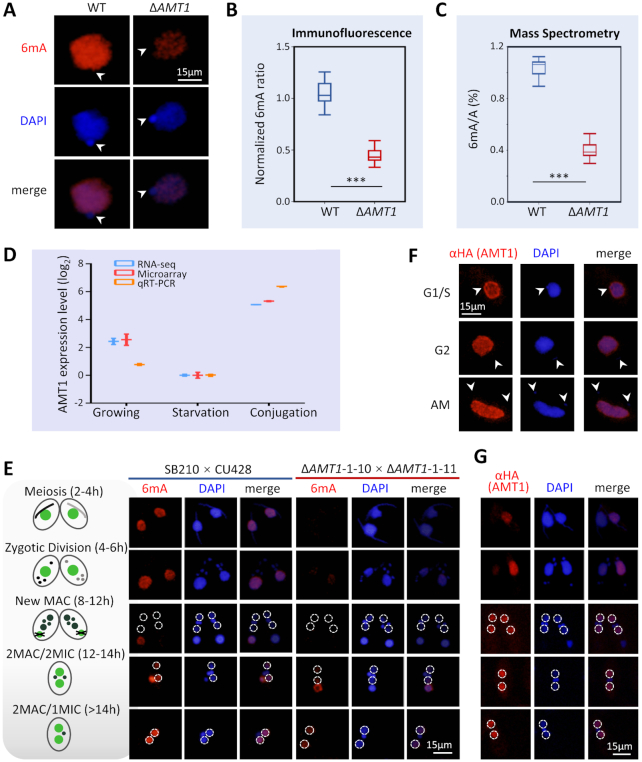
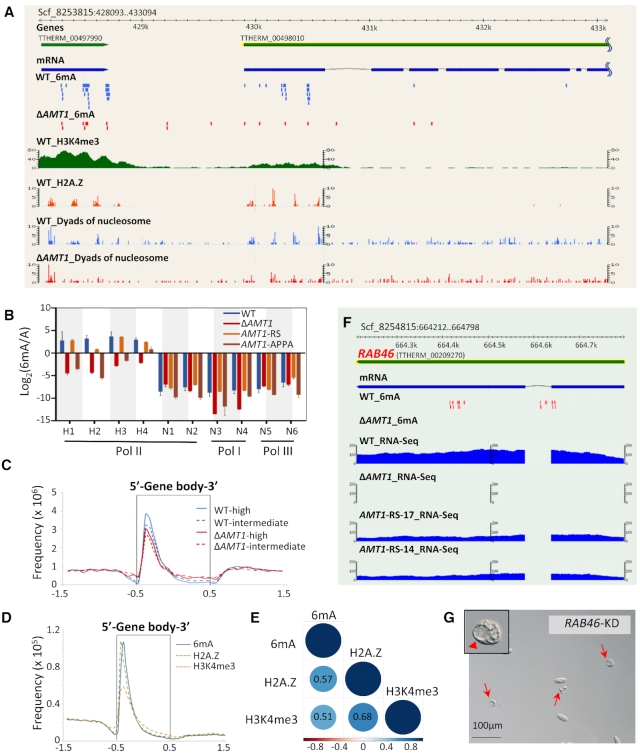
Figure 2.
Germline knockout of AMT1 (ΔAMT1) dramatically reduced the DNA 6mA level. (A) Immunofluorescence (IF) staining of DNA 6mA in logarithmically growing WT and ΔAMT1 cells. Note the absence of 6mA signals in the MIC (arrowheads). (B) Statistical analysis of 6mA IF signal intensity in A. Cell images were randomly selected (WT: n = 263, ΔAMT1: n = 450) and processed by ImageJ. Data are presented as box plots (from top to bottom: max, first quartile, median, third quartile, and min). Student's t-test was performed. ***P < 0.001. (C) Mass spectrometry analysis of 6mA, performed on five biological replicates for WT and 39 for ΔAMT1. Data are presented as box plots. Student's t-test was performed. ***P < 0.001. (D) Expression profile of AMT1 during growth (mid-log phase, ∼3.5 × 105 cells/ml), starvation (15 h after starvation) and conjugation (4 h post-mixing). Expression levels are represented by normalized RNA-seq reads numbers (http://tfgd.ihb.ac.cn/) (88), microarray signals (31) and quantitative RT-PCR data. All expression values (in logarithm scale) were compared with values during starvation (set at 0). (E) IF staining of DNA 6mA in conjugating WT and ΔAMT1 cells. Nuclear events are used to ascertain conjugation stages (see schematics on the left). The new MAC is outlined (dotted circles). Note that de novo occurrence of 6mA in the new MAC was dramatically reduced in ΔAMT1 cells. (F) IF staining of AMT1 in growing cells. AMT1 was HA-tagged at the N-terminus (HA-AMT1) and over-expressed under the control of the MTT1 promoter. After induction by cadmium chloride (1.5μg/ml, 30min), HA-AMT1 was detected by an α-HA antibody. Note the absence of AMT1 signals in the MIC (arrowheads). (G) IF staining of AMT1 in conjugating cells. Nuclear events are used to ascertain conjugation stages (see schematics on the left). Note that AMT1 signals appeared in the new MAC (dotted circles), before buildup of 6mA signals.
AMT1 was expressed at high levels during growth and at low levels during starvation (Figure 2D). AMT1 expression levels increased dramatically during conjugation, the sexual reproduction stage of the Tetrahymena life cycle (Figure 2D). Consistent with this expression profile, we detected abundant 6mA in WT cells during conjugation (Figure 2E) as well as during growth (Figure 2A). 6mA was exclusively detected in the parental MAC during early conjugation, while it was established de novo in the new MAC at late conjugation (Figure 2E). In comparison, 6mA signals were markedly lower in the parental MAC during early conjugation and in the new MAC during late conjugation in ΔAMT1 cells (Figure 2E, S4). Taken together, these results strongly argue that DNA 6mA levels in Tetrahymena are mainly dependent on AMT1 throughout its life cycle. Our conclusion is consistent with Beh et al. identifying AMT1 as a major source of the DNA MTase activity in Tetrahymena (referred to as MTA1 therein) (75), but directly contradicts another publication by Luo et al. claiming that DNA 6mA levels in Tetrahymena are mainly dependent on AMT2 (referred to as TAMT1 therein) (70).
We next analyzed the cellular localization of AMT1 by tracking an ectopically expressed version (AMT1-NHA-OE). In growing cells, AMT1 was detected only in the MAC but not in the micronucleus (MIC) (Figure 2F, S5A–B), which is consistent with the exclusive presence of 6mA in the MAC (Figure 2A). During conjugation, AMT1 was detected in the parental MAC at early time points and in the new MAC at late time points (Figure 2G). This pattern is again consistent with the distribution of 6mA at different developmental stages (Figure 2E).
Defects in growth and development of ΔAMT1 cells
Deleting AMT1 severely impaired cell growth, with the doubling time increased to ∼22.8 h (versus ∼5.1 h in WT cells) (Figure 3A). ΔAMT1 cells swam very slowly: instead of the rapid spiraling movement typical of WT cells, the knockout cells showed dramatically reduced rotational and translational speeds (Figure 3B and C). We also noticed that many knockout cells contained an abnormally large contractile vacuole (Figure 3D), which took much longer to complete a systole/diastole cycle for water expulsion (Supplemental video). As expected, far fewer cells with an abnormally large contractile vacuole were found when cultured in hypertonic media, which eliminates the need for water expulsion (Supplementary Figure S6).
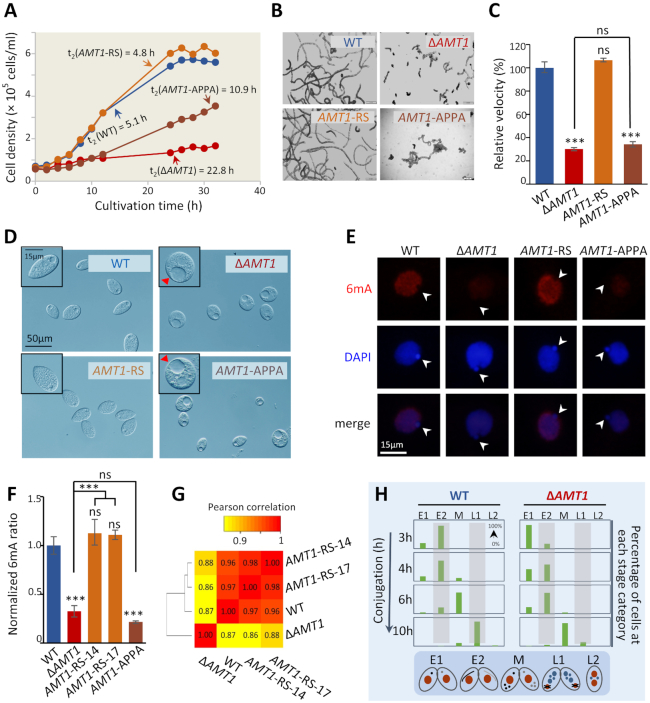
Figure 3.
Growth and development were severely impaired in AMT1 loss-of-function mutants. (A) Growth rates of WT (SB210), ΔAMT1, AMT1-APPA and AMT1-rescued cells (AMT1-RS). Cells were enumerated using a Coulter counter at the indicated time points. Doubling time (h) were calculated based on the log-phase data. (B) Trajectory analysis of the swimming ability. Swimming paths of indicated cells in half a second were recorded and converted to illustrations by Image J. ΔAMT1 and AMT1-APPA cells swam much slower than WT cells. The swimming ability was restored in AMT1-RS cells. (C) Quantification of the translational swimming velocity. All values are normalized with that of WT cells. Data are presented as mean ± standard deviations. Student's t-test was performed. ***P < 0.001; ns: not significant (P > 0.05); compared with WT, unless indicated otherwise. (D) Contractile vacuoles (CVs) observed by phase-contrast microscopy. Abnormal CVs (red arrowheads) were observed in a large portion of ΔAMT1 and AMT1-APPA cells, but not in WT and AMT1-RS cells. (E) IF staining of 6mA in WT, ΔAMT1, AMT1-APPA and AMT1-RS cells. Note the absence of 6mA signal in the MIC (white arrowheads). (F) Mass spectrometry analysis of 6mA in WT, ΔAMT1, AMT1-APPA and AMT1-RS cells (two independent strains). Data are presented as mean ± standard deviations. Student's t-test was performed. ***P < 0.001; ns: not significant (P > 0.05); compared with WT, unless indicated otherwise. (G) Correlations in gene expression profiles. Pearson's correlation coefficients (PCC) were calculated for pair-wise comparison and represented by the color scale. Cluster analysis, based on PCC, is shown on the left. Note the similarities between the gene expression profiles of WT and AMT1-rescued cells (AMT1-RS-14 and AMT1-RS-17) and their distinction from that of ΔAMT1 cells. (H) Conjugation progress (h) of WT (SB210 × CU428) and ΔAMT1 (1–10 × 1–11) cells. Cells at each time point (n > 200) were classified into different developmental stages by their nuclear morphology: pre-meiosis (E1), meiosis (E2), mitosis (M), new MAC development (L1) and pair separation (L2) (89).
To test whether the aforementioned phenotypes were solely attributable to the lack of AMT1, we transformed ΔAMT1 cells with a DNA fragment containing the WT AMT1 gene (Supplementary Figure S7A, B). RT-PCR and qRT-PCR confirmed that AMT1 mRNA expression was restored to WT levels in rescued (AMT1-RS) cells (Supplementary Figure S8A, B). Immunofluorescence staining showed that 6mA levels in rescued cells were comparable to WT levels (Figures 3E, S8C); this was further confirmed by mass spectrometry analysis (Figure 3F). Remarkably, restoring AMT1 alone was sufficient to rescue the aforementioned defects of ΔAMT1 cells: both slow growth (Figure 3A) and poor swimming (Figure 3B, C) were rectified, abnormally large contractile vacuoles eliminated, and the normal systole/diastole cycle restored (Supplemental video). RNA-seq analysis further confirmed that the transcription profile in rescued cells had been largely restored to that of WT cells (Figure 3G).
To determine whether the MTase activity of AMT1 underpins its functions, we mutated the conserved DPPW motif to APPA (Supplementary Figure S9A–C). The AMT1 APPA mutant phenocopied ΔAMT1 cells: it had reduced 6mA level, slow growth, poor swimming, and large contractile vacuoles (Figure 3A–F), demonstrating that AMT1 catalyzes DNA 6mA, which in turn is required for normal cell growth and development. Intriguingly, the AMT1 mRNA expression level in AMT1-APPA cells was lower than that in WT cells (Supplementary Figure S8B). A likely scenario is that AMT1 expression is promoted by DNA 6mA levels, and is thus dependent on the MTase activity of AMT1. This scenario is consistent with high levels of 6mA sites on the gene body of AMT1 (14.52, compared with 9.90 for genes on average).
Functions of AMT1 in conjugation were investigated by crossing two germline knockout cells. Conjugation progress was poorly synchronized and suffered major delays at key stages, including cell pairing, meiotic MIC elongation, and new MAC formation (Figure 3H). Many cells aborted conjugation, likely due to defective meiosis and gametogenesis, with only a small percentage reaching the final developmental stage and giving rise to viable conjugation progeny (ΔAMT1 6.3% versus WT 92.4%). These strong conjugation phenotypes in ΔAMT1 cells are consistent with high expression levels of AMT1 during conjugation (Figure 2G), underscoring the functional significance of AMT1.
Collectively, our results directly link AMT1 loss-of-function to severe defects with high fitness cost, which is potentially mediated by the transcriptional effect of AMT1-catalyzed 6mA.
AMT1 promotes 6mA with high methylation levels
We next performed Single Molecule, Real-Time (SMRT) sequencing of genomic DNA, generating 1 684 630 and 1 851 283 reads for WT and ΔAMT1 cells, corresponding to 137× and 160× average coverage of the Tetrahymena MAC genome (Supplementary Figure S10A, Table S2). We focused our analysis on 431 346 and 123 935 6mA sites called with high confidence (normalized coverage > 25×, Qv > 30), representing 0.54% and 0.16% of the total adenines in WT and ΔAMT1 cells (Figure 4A; Supplementary Table S3). We calculated 6mA density across the genome, which was dramatically and evenly reduced in ΔAMT1 relative to WT cells (Figures 4A, S10B). Consistent with the immunofluorescence staining and mass spectrometry results (Figure 2B, C), the SMRT sequencing result corroborates our conclusion of global 6mA reduction upon AMT1 loss.
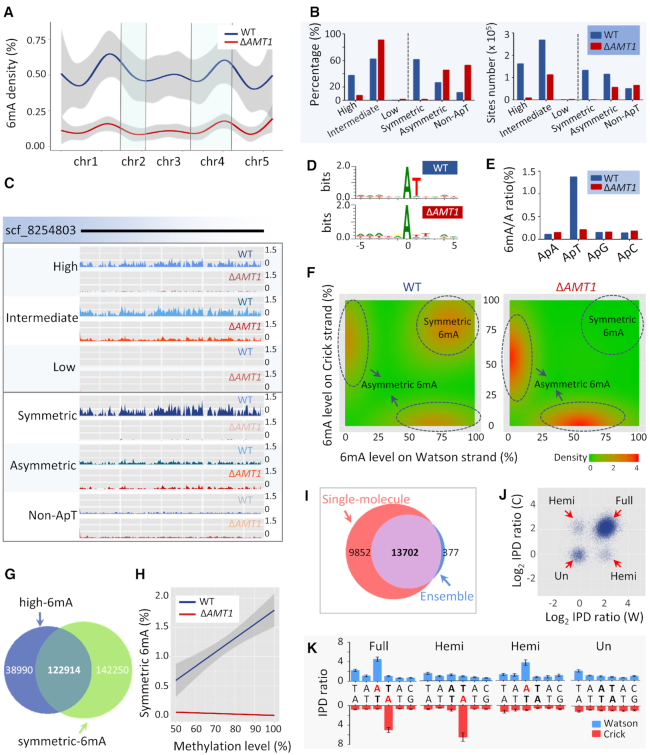
Figure 4.
Deletion of AMT1 affected the genome-wide distribution of 6mA. (A) 6mA density (6mA/A) in WT (blue) and ΔAMT1 cells (red), across the assembled Tetrahymena MAC genome, which are concatenated according to their positions in the MIC chromosomes. (B) Classification of 6mA sites according to their methylation levels (left half) and symmetry (right half), in WT (blue) and ΔAMT1 cells (red). The left and right panels represent percentage (a particular class of 6mA/all 6mA) and the site numbers for the classes, respectively. See Supplementary Table S3 for details. (C) Normalized 6mA distribution on scaffold_8254803 (the longest in the Tetrahymena MAC genome assembly) in WT and ΔAMT1 cells. Classification of 6mA sites are the same as in (B). (D) Sequence logos for 6mA sites (at position 0) in WT and ΔAMT1 cells. (E) Relative distribution of 6mA in ApN (ApA/ApT/ApG/ApC) dinucleotides in WT (blue) and ΔAMT1 cells (red). Note the even distribution of 6mA on all four ApN dinucleotides in ΔAMT1 cells. (F) Density plot of 6mA distribution, according to methylation levels on Watson (x-axes) or Crick strands (y-axes) in WT (left panel) and ΔAMT1 cells (right panel). (G) Area-proportional Venn diagram representing highly methylated 6mA and symmetrically methylated 6mA in WT cells. (H) Relationship between methylation levels (x-axis) and proportion of symmetric 6mA sites (y-axis: symmetric 6mA/total 6mA) in WT (blue) and ΔAMT1 cells (red). The linear regression trendlines and 95% confidence intervals (very narrow for ΔAMT1 cells) are shown. (I) Area-proportional Venn diagram representing symmetrically methylated AT sites identified in the ensemble and 6mA-containing AT sites in single molecules. The analysis was limited to the two longest chromosomes in Tetrahymena (scf_8254667 and scf_8254697). (J) Different methylation states revealed by SMRT CCS. IPD ratios were calculated for A’s in symmetrically methylated AT sites (the intersection in (I)) of every single molecule. In the scatterplot, the strand-specific IPD ratios (log2 transformed) is explicitly denoted in the x- (Watson strand) and y-axes (Crick strand). Note the four distinct clusters in the plot representing different methylation states: The cluster near the origin represents the unmethylated state (IPD ratios≈1); the cluster in the upper-right corner represents the full methylation state (IPD ratios≈4 for both Watson and Crick strands); two other clusters represent the hemi-methylation state (IPD ratios ≈ 4 for either Watson or Crick strand). (K) Four single molecules representing different methylation states for the same AT site (scf_8254697: 1 830 057–1 830 062). For each DNA molecule, the mean IPD ratios and their dispersions for Watson and Crick strands are plotted for the 6-bp region containing the AT site.
In the polyploid MAC, methylation levels varied almost continuously from saturation to depletion at different 6mA sites. We therefore distinguished 6mA sites with high (80–100%), intermediate (20–80%), or low methylation levels (10–20%). In WT cells, we found 37.6%, 62.3% and 0.2% of 6mA sites with high, intermediate, and low methylation levels, respectively (Figure 4B, left panel; Supplementary Table S3). We observed a downward shift of methylation levels in ΔAMT1 cells, with 7.4%, 91.0% and 1.6% of 6mA at corresponding levels (Figure 4B, left panel; Supplementary Table S3). When the overall reduction of 6mA sites in ΔAMT1 cells was taken into account, the reduction of highly methylated 6mA was even more conspicuous (Figure 4B, right panel). As illustrated in a representative genomic region, highly methylated 6mA was greatly reduced in ΔAMT1 relative to WT cells, while 6mA with intermediate and low levels were only moderately affected (Figure 4C, top panel). We conclude that even though AMT1 is not the only MTase for 6mA, it strongly promotes high level methylation.
AMT1 is required for symmetric 6mA at ApT dinucleotides
In WT cells, 6mA was highly enriched at the sequence 5′-ApT-3′ (ApT: 88.2%; non-ApT: 11.8%); this effect was diminished in ΔAMT1 cells (ApT: 47.2%, non-ApT: 52.8%) (Figure 4B, left panel; Supplementary Table S3). Indeed, the AT motif was obvious in the 6mA consensus sequence of WT cells, but was obscured in ΔAMT1 cells (Figure 4D). In contrast to the strong preference for the ApT dinucleotides in WT cells, 6mA was distributed with similar probabilities in all four ApN dinucleotides in ΔAMT1 cells (Figure 4E; Supplementary Table S4). These results strongly suggest that AMT1 preferentially targets the AT motif.
The palindromic AT sequence can carry 6mA on the Watson strand and/or the Crick strand. In an ensemble of DNA molecules, if 6mA is found on both the Watson and the Crick strands of a particular ApT site, this site is defined as symmetrically methylated; if 6mA is only found on either the Watson strand or the Crick strand, it is asymmetrically methylated. In WT cells, 53.5% of 6mA-containing ApT sequences were symmetrically methylated (61.5% of total 6mA), while 46.5% were asymmetrically methylated (26.7% of total 6mA) (Figure 4B, left panel; Supplementary Table S3). In ΔAMT1 cells, symmetrically methylated ApT sequences were minimal (1.5% of total 6mA), relative to 6mA at asymmetric ApT and non-ApT sites (45.6% and 52.8% of total 6mA) (Figure 4B, left panel; Supplementary Table S3). When the overall reduction of 6mA sites in the mutant was taken into account, symmetric 6mA was all but eliminated (Figure 4B, right panel; Supplementary Table S3). As shown in a representative genomic region, there was almost no symmetric 6mA in ΔAMT1 cells relative to WT cells, along with moderately reduced levels of asymmetric 6mA and slightly increased levels of non-ApT 6mA (Figure 4C, bottom panel).
The distribution of methylated ApT sites was plotted against methylation levels on the Watson strand and the Crick Strand. In WT cells, symmetrically and asymmetrically methylated 6mA were distributed in isolated peaks, with no obvious transition between them (Figure 4F). Symmetrically methylated ApT sites distributed along the diagonal line, indicating that they generally have similar methylation levels between the Watson and Crick strands (Figure 4F). In ΔAMT1 cells, the peak corresponding to symmetric 6mA distribution disappeared, while the asymmetric 6mA peaks shifted towards the origin point—reflecting reduced methylation levels (Figure 4F). Collectively, these results indicate that AMT1 is specifically required for symmetric 6mA.
Symmetric 6mA was linked to 6mA with high methylation levels. In WT cells, there was significant overlap between the two sets of 6mA (28.5% of total 6mA; representation factor: 1.2) (Figure 4G). We also found a strong positive correlation between methylation levels and symmetric 6mA ratios (Figure 4H). In ΔAMT1 cells, highly methylated 6mA was greatly reduced, accompanied by virtual elimination of symmetric 6mA (Figure 4B). A parsimonious interpretation of these results is that AMT1 drives symmetric 6mA to high methylation levels.
While symmetric and asymmetric methylation are defined at the ensemble level, full methylation and hemi-methylation are defined at the single molecule level. In an individual DNA molecule, if both adenines in the palindromic ApT site are methylated, this site is in the full methylation state; if only one of the adenines is methylated, this site is the hemi-methylation state. In WT Tetrahymena cells, the high levels of symmetric 6mA make it likely that at least some — if not most — of them are in the full methylation state. Nonetheless, SMRT sequencing in continuous long read (CLR) mode, used to generate the aforementioned results, is most informative about DNA 6mA at the ensemble level, but not at the single molecule level. To address this issue, we performed SMRT sequencing of WT cells in circular consensus sequencing (CCS) mode, in which both strands of a DNA single molecule are read multiple times, generating strand-specific and highly accurate calling of 6mA.
Analysis of the SMRT CCS data showed that almost all symmetrically methylated AT sites called at the ensemble level were also called as containing 6mA at the single molecule level (Figure 4I). Indeed, all four possible methylation states were revealed at the single molecule level (Figure 4J, K): The vast majority (77.3%) of symmetric methylated AT sites were found in the full methylation state, with 6mA on both strands. 10.5% were in one of the two hemi-methylation states, representing the transitory state of symmetrically methylated AT, after DNA replication splitting parental strands in the full methylation state. Another minor group (12.2%) was in the unmethylated state, revealing the presence of the alternative epigenetic state in these sites. These results strongly support the involvement of AMT1 in maintaining symmetric 6mA as an epigenetic mark, by effective conversion of the hemi-methylation state to the full methylation state, as proposed in our previous publication (19). Our hypothesis is further supported by a recent study reporting that the hemi-methylated AT, rather than the unmethylated site, is the preferred substrate for AMT1 (75).
AMT1 affects Pol II-transcribed genes
SMRT sequencing showed that 6mA, particularly symmetric methylation at ApT, was prominently associated with RNA polymerase II (Pol II)-transcribed genes in WT cells (Figure 5A), consistent with our previous work (19). By contrast, a dramatic reduction was observed in ΔAMT1 cells (Figure 5A). With increased sequencing coverage (Supplementary Table S2), we identified more putative 6mA sites on Pol I- and Pol III-transcribed genes (Supplementary Table S5). However, most are likely false positives as they differed significantly from 6mA associated with Pol II-transcribed genes in their kinetic signature, methylation levels, and sequence characteristics (Supplementary Figure S11A–D). Even if real, these 6mA sites still only occurred at near background levels.
Figure 5.
AMT1 affects Pol II-transcribed genes. (A) Distributions of 6mA, H3K4me3, H2A.Z, and nucleosomes. In this GBrowse snapshot of a representative genomic region, tracks from top to bottom are: gene models, mRNA transcripts, 6mA (WT in blue, ΔAMT1 in red), H3K4me3 (X-ChIP coverage), dyads of nucleosomes containing H2A.Z, and dyads of nucleosomes (WT in blue, ΔAMT1 in red). Note the biased distribution of 6mA toward the 5′ end of a long gene (TTHERM_00498010). (B) Validating the methylation states of 10 GATC sites by DpnI/DpnII digestion. The sites were selected for their location on genes transcribed by different RNA polymerases (Pol I, II and III), and their methylation levels calculated from SMRT sequencing data (H1-H4: high methylation; N1-N6: no methylation; all in WT cells). Genomic DNA was digested with DpnI or DpnII; qPCR was performed with primers flanking the GATC sites to quantify undigested DNA (Supplementary Table S6). Y-axis represents the ratios between the methylated and unmethylated state (Log2 transformed), deduced from differential digestion by DpnI and DpnII. See Materials and Methods for details. (C) Composite analysis of 6mA distribution on the gene body of WT (blue) and ΔAMT1 cells (red). Genes are scaled to unit length and is extended to each side by unit length. Distribution frequency was calculated as ‘6mA amount at a certain position/total 6mA amount’. Solid lines: high methylation levels (80–100%); dashed lines: intermediate methylation levels (20–80%). Note that the remaining 6mA in ΔAMT1 cells was similarly accumulated downstream of transcription start sites (TSS), towards the 5′ end of the gene body. (D) Distribution profiles of 6mA, H2A.Z, H3K4me3 on the gene body of WT cells. Genes are scaled to unit length and is extended to each side by unit length. Note that all of them were accumulated downstream of TSS, towards the 5′ end of the gene body. (E) Correlation matrix of H2A.Z, H3K4me3, and 6mA frequency in 1 kb of the gene body downstream of TSS. Correlation coefficients and correlation color dots are shown. (F) GBrowse snapshot of the RAB46 locus (TTHERM_00209270). Tracks from top to bottom are: gene model, mRNA transcripts, 6mA (WT and ΔAMT1 cells), and RNA-seq coverage in WT, ΔAMT1 and AMT1-RS cells (two replicates, AMT1-RS-17 and AMT1-RS-14). Note that both 6mA and RAB46 expression were eliminated in ΔAMT1 cells. (G) Extraordinarily large contractile vacuoles (CV) were observed upon RAB46 knockdown (KD). Red arrowheads: large CV; red arrows: cells with large CV.
Restriction enzyme digestion was employed to validate the SMRT sequencing results. We compared DpnI-digested (recognizing GATC with 6mA), DpnII-digested (recognizing unmethylated GATC), and undigested genomic DNA, and determined 6mA/A ratios of several GATC sites by qPCR with flanking primers (Figure 5B). Four symmetrically and highly methylated sites and two unmethylated sites, determined by SMRT sequencing, were selected for Pol II-transcribed genes (Supplementary Table S6). Random sites were also selected for Pol I- and Pol III-transcribed genes (two each) (Supplementary Table S6). The four methylated sites all had high 6mA/A ratios in WT and AMT1-rescued cells, but much reduced 6mA/A ratios in ΔAMT1 cells and the AMT1-APPA mutant. The six unmethylated sites all had very low 6mA/A ratios in all four strains. This provides an independent verification of the SMRT sequencing results and the specified 6mA distribution pattern.
6mA was mapped to about 88.33% (20 973 genes, 381 027 sites) and 76.29% (18 895 genes, 94 547 sites) of Pol II-transcribed genes in WT and ΔAMT1 cells, respectively. As reported previously (19), 6mA was preferentially localized at the gene body in WT cells (Figure 5C). Despite a global reduction of 6mA levels in ΔAMT1 cells, the distribution preference was not significantly changed (P > 0.05) (Supplementary Figure S12A; Table S7). Composite analysis of 6mA distribution in 13 854 well-annotated long genes (>1 kb) revealed moderate reductions in 6mA accumulation immediately downstream of transcription start sites (TSS) in ΔAMT1, relative to WT cells (Figures 5C, S12B and C). Enrichment at the 5′ end of the gene body was observed for 6mA of high and intermediate methylation levels, but not low methylation levels (Figures 5C, S12B). These results are likely underpinned by recruitment of AMT1 as well as other Tetrahymena 6mA MTases to promoters of Pol II-transcribed genes.
The biased distribution of 6mA toward the 5′ end of the gene body is reminiscent of distribution patterns of the histone modification H3K4 methylation and the histone variant H2A.Z (Figure 5D), two well-established epigenetic marks associated with Pol II transcription (71–73). Indeed, we found strong correlations between distributions of 6mA, H3K4 methylation, and H2A.Z (Figure 5E). RNA-seq analysis of WT and ΔAMT1 growing cells revealed a large number of genes differentially expressed upon AMT1 deletion (6,047 out of 26,996 well-annotated genes), including 3,205 upregulated and 2,842 downregulated genes (Padj < 0.05; at least 2-fold increase or decrease) (Supplementary Figure S13A, B). Among thousands of genes differentially expressed in WT and ΔAMT1 cells, many were associated with high levels of 6mA in WT cells, and with substantially reduced 6mA levels in ΔAMT1 cells (Supplementary Figure S13C); these genes are potentially under direct regulation of AMT1-deposited 6mA. However, many genes with divergent 6mA levels were not differentially expressed in WT and ΔAMT1 cells, suggesting that they may be mainly controlled by mechanisms other than 6mA, particularly H3K4 methylation and H2A.Z, which overlap with 6mA in their distributions and potentially also in their functions (19).
Gene ontology analysis of differentially expressed genes revealed many conserved pathways affected in ΔAMT1 cells, with potential connections to its phenotypes (Supplementary Figure S13D). Here we focused on one gene, RAB46, encoding a Rab family GTPase involved in membrane trafficking (74). In WT cells, RAB46 was associated with high levels of 6mA, particularly symmetric ApT methylation, which was eliminated in ΔAMT1 cells. RAB46 was robustly expressed in WT cells, but not expressed in ΔAMT1 cells (Figure 5F). Importantly, RNAi knockdown of RAB46 led to abnormal contractile vacuoles, phenocopying ΔAMT1 cells (Figure 5G). These results link AMT1-catalyzed 6mA with RAB46 transcription, which in turn accounts for the most dramatic phenotype of ΔAMT1 cells.
Attenuation of strong 6mA and nucleosome positioning relative to each other in ΔAMT1 cells
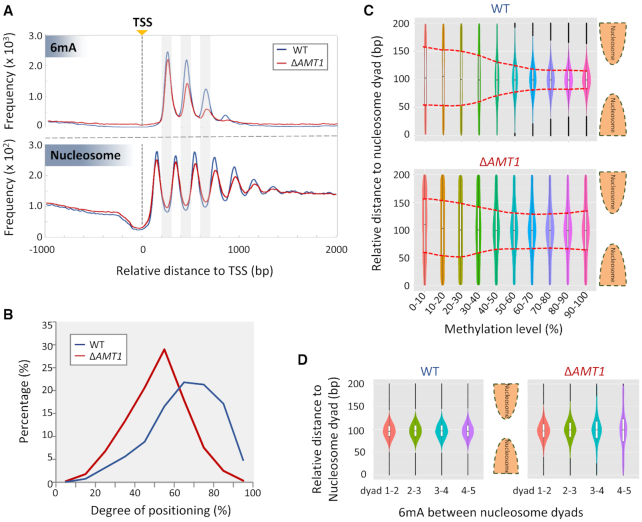
In WT cells, 6mA was present mostly in linker DNA (∼50 bp) between adjacent nucleosomes, mapped by micrococcal nuclease digestion (19) (Figure 6A). Despite dramatic reduction, 6mA was still preferentially found in linker DNA in ΔAMT1 cells (Figure 6A). This is best illustrated by composite analysis of 6mA and nucleosome distributions downstream of TSS of Pol II-transcribed genes, revealing two damped oscillations with the same periodicity of ∼200 bp, but in opposite phases (Figure 6A). The strong anti-correlation was obvious in both WT and ΔAMT1 cells (Figure 6A). Nonetheless, the amplitude (peak-to-trough distance) for both 6mA and nucleosome distributions was reduced in ΔAMT1 relative to WT cells (Figure 6A). We also calculated the degree of nucleosome positioning (Figure 6B), measuring the dispersion of nucleosome distribution (52). This revealed a global reduction in the degree of nucleosome positioning in ΔAMT1 relative to WT cells (Supplementary Figure S14A–C). This trend was even more accentuated for the +1, +2 and +3 nucleosomes (as part of stereotypical nucleosome arrays downstream of TSS), which, in general, were strongly positioned in WT cells, but were weakly positioned in ΔAMT1 cells (Figures 6B, S14D). These results support that the chromatin landscape of the gene body—especially the strongly positioned stereotypical nucleosome arrays—is shaped by AMT1-deposited 6mA.
Figure 6.
Attenuation of strong 6mA and nucleosome positioning relative to each other in ΔAMT1 cells. (A) Distribution profiles of 6mA (top panel) and nucleosome (bottom panel) around TSS in WT (blue) and ΔAMT1 cells (red). (B) Nucleosome positioning in WT (blue) and ΔAMT1 (red) cells. Degrees of positioning were calculated for +1, +2 and +3 nucleosomes in the gene body. See Materials and Methods for details. (C) 6mA distribution relative to the nucleosome dyad in WT (top panel) and ΔAMT1 cells (bottom panel). The violin plots show the density of 6mA between neighboring nucleosome dyads, grouped by methylation levels. The box plots within represent the median and the interquartile range of each group. Red dotted lines mark the trend for 6mA with high methylation levels to be enriched in linker DNA in WT (top panel) but not ΔAMT1 cells (bottom panel). (D) Dispersions of 6mA increase further downstream of TSS in ΔAMT1 cells (right panel) but not WT cells (left panel). The violin/box plots show 6mA distribution between neighboring nucleosomes, grouped by their positions in the gene body (+1/+2, +2/+3, +3/+4, +4/+5).
The remaining 6mA sites in ΔAMT1 cells, the vast majority of which were not symmetric methylated, were more dispersed relative to adjacent nucleosomes (Figure 6C, D). In WT cells, 6mA sites were progressively enriched in linker DNA as their methylation levels increased, and highly enriched therein upon methylation saturation (Figure 6C, top panel). In ΔAMT1 cells, many more 6mA sites were found outside linker DNA, and the dispersion did not decrease monotonically with increasing methylation levels (Figure 6C, bottom panel). Dispersion of 6mA sites was also prominently observed in the gene body, as the oscillation amplitudes of 6mA distribution downstream of TSS were damped much faster in ΔAMT1 than in WT cells (Figure 6A, top panel). Focusing on 6mA sites in between stereotypical nucleosome arrays, we found that while they were highly enriched in linker DNA in WT cells (Figure 6D, left panel), they were more dispersed into the nucleosomal DNA in ΔAMT1 cells, and increasingly so further downstream of TSS (Figure 6D, right panel). We argue that 6mA and nucleosome distributions reinforce each other to establish the epigenetic landscape for proper expression of Pol II-transcribed genes.
DISCUSSION
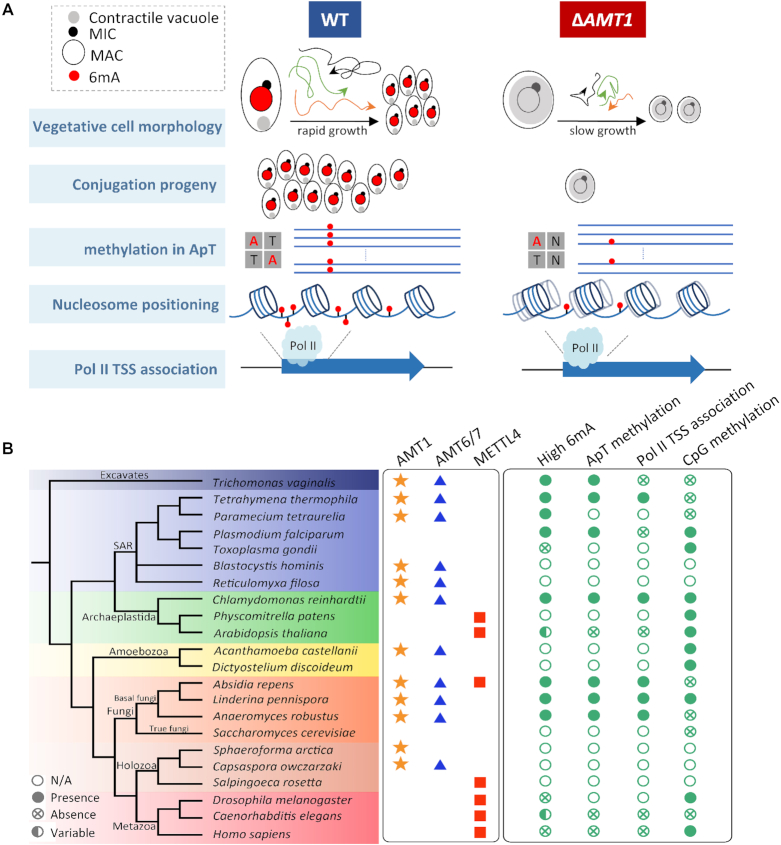
Here we delineate the roles of the MT-A70 homologue AMT1 as an epigenetic regulator (Figure 7A): (i) AMT1 is the DNA 6mA methyltransferase (MTase) required for symmetric methylation at the AT motif, which constitutes the majority of 6mA in Tetrahymena; (ii) 6mA, especially AMT1-dependent 6mA, accumulates toward the 5′ end of Pol II-transcribed genes, orders stereotypical nucleosome arrays therein and, together with H3K4 methylation and H2A.Z, shapes the epigenetic landscape for Pol II transcription; (iii) AMT1 is required for normal growth and development while affecting expression of thousands of genes, in particular RAB46, which is linked to the abnormal contractile vacuole phenotype in ΔAMT1 cells.
Figure 7.
AMT1 represents a new class of DNA 6mA methyltransferases and epigenetic regulators in eukaryotes. (A) A diagram summarizing AMT1′s roles as an epigenetic regulator. (B) Phyletic distribution of MT-A70 family genes (AMT1, AMT6/7, and METTL4 clades) and features of DNA methylation (high 6mA, ApT methylation, Pol II TSS association, and CpG methylation). High 6mA is defined as 6mA/A ratio higher than 0.1%. Star, triangle and square denote presence of AMT1, AMT6/7, and METTTL4 homologues in corresponding species. Solid circle, cross in circle, hollow circle, and hemi-solid circle denote presence, absence, no available data (N/A), variability/uncertainty in corresponding features of DNA methylation. See Supplementary Table S10 for details.
This study is a logical extension of our recently published work (19), describing the first genome-wide 6mA distribution map of Tetrahymena and its coordination with other epigenetic players. While our manuscript was in preparation, a related study was published (75). Told from different perspectives (the present study focusing on genetics and in vivo cellular functions, while Beh et al. (75) focused on enzymology), these two studies are broadly consistent in areas where they overlap, and are complementary in many aspects in identifying AMT1, referred to by Beh et al. as MTA1 (75), as the major 6mA MTase in ciliates (Supplementary Table S1) (Figure 1B).
AMT1 represents a new family of DNA 6mA methyltransferases in eukaryotes
AMT1 represents a divergent subclade of eukaryotic MT-A70 MTases, which are distributed in all the super-groups of eukaryotic classification, despite evident lacunae in many branches (Figure 7B). AMT1 orthologues are found in all three major clades—Stramenopiles, Alveolata (containing ciliates), and Rhizaria—of the SAR super-group, though missing in Apicomplexa, a sub-group of Alveolata represented by parasitic protists such as Plasmodium and Toxoplasma. For the neighboring Archaeplastida super-group, AMT1 orthologues are widespread in green algae, but absent in plants. They are also present in some basal eukaryotes in the Excavates super-group. On the other main branches of eukaryotic divergence, AMT1 orthologues are distributed in Amoebozoa, basal (early-diverging) fungi, and some Holozoa (unicellular relatives of animals), but are conspicuously absent in true fungi (represented by budding and fission yeasts) and animals. The broad but patchy distribution of AMT1 orthologues suggests that they arose early in eukaryotic evolution, but experienced lineage-specific gene loss.
AMT6 and AMT7 share remarkably similar expression profiles with AMT1 in Tetrahymena, with two peaks at early and late conjugation respectively (Supplementary Figure S15). More strikingly, AMT6 and AMT7 mirror the distribution of AMT1 in early-branching eukaryotes (Figure 7B). Only AMT1, but not AMT6 and AMT7, contains the catalytic motif (DPPW). A likely scenario is that AMT6 and AMT7 are the heterodimeric partner(s) for AMT1, analogous to the heterodimeric RNA m6A MTase subunits METTL3 (containing the catalytic site) and METTL14 (no enzymatic activity) (76). Indeed, Beh et al. identified AMT7 (referred to therein as MTA9) as an AMT1-associated protein required for its MTase activity (75). The in vivo functions of AMT6 and AMT7 await further investigation.
Intriguingly, the MT-A70 MTases of the METTL4 subclade, also present in a wide range of eukaryotes, show a contrasting phylogenetic distribution pattern (Figure 7B): they are widespread in animals and plants, they overlap with AMT1 homologues in some basal fungi and their presence in protists is obscure. The largely mutually exclusive distributions of AMT1 and METTL4 homologues coincide with the two alternative modes of DNA 6mA in eukaryotes: the former is associated with abundant 6mA in genomic DNA, symmetric methylation at ApT, and Pol II transcription; the latter corresponds to low 6mA abundance, shows no preference for the AT motif, and is generally targeted to non-genic regions. The phylogenetic distribution of METTL4 homologues bears a resemblance to that of DNA methylation at 5-methylcytosine (5mC) and its cognate DNMT MTases, which are involved in transcriptional gene silencing and heterochromatin formation (77). This suggests that METTL4-dependent 6mA may have co-evolved with DNMT-dependent 5mC, especially in animals and plants, whose genomes are generally more transcriptionally repressive and heterochromatic than those of unicellular eukaryotes (78). We argue that AMT1 homologues and METTL4 homologues represent two deep branches of eukaryotic DNA 6mA MTases, which are functionally antagonistic to each other. Their lineage-specific loss, often in a complementary pattern, may be a driver for divergent evolution of eukaryotes.
AMT1-dependent symmetric 6mA is an epigenetic mark for Pol II-transcribed genes
A strong role for AMT1-dependent 6mA in transcription regulation emerges from our results. As symmetrically and highly methylated 6mA is broadly distributed in the Tetrahymena genome and associated with a majority of Pol II-transcribed genes, its effects are not limited to specific pathways, as reported for animals (15,18). Instead, 6mA in unicellular eukaryotes like Tetrahymena and Chlamydomonas is more likely to be integrated into transcription in general. Here we posit that symmetrically and highly methylated 6mA shapes the epigenetic landscape of transcription, analogous to, and perhaps partially redundant with, other transcription-associated epigenetic markers such as histone H3K4 methylation and histone H2A.Z variant (80,81). Indeed, 6mA, H3K4 methylation and H2A.Z are strongly correlated with one another in Tetrahymena, with the same biased distribution towards the 5′ end of the gene body (19,52). This bias further suggests that AMT1 is recruited to the promoter, similar to the H3K4 methylation-depositing MLL/SET1 complex and H2A.Z-depositing SWR1 complex (82,83), supporting functional interplays in transcription regulation among one another (19,52).
Distributions of 6mA and the nucleosome are mutually dependent on each other. On the one hand, AMT1 preferentially deposits 6mA in linker DNA, likely precluded by steric hindrance of the nucleosome. A similar observation is made in Oxytricha (another ciliate)(84), in which 6mA is located in linker DNA downstream of TSS (75). On the other hand, AMT1-deposited 6mA also affects the chromatin landscape of transcription by promoting nucleosome positioning, especially for the +1, +2, and +3 nucleosomes, which are flanked by 6mA. Consistent with this, Beh et al. reports that the nucleosome distribution becomes fuzzier when the flanking 6mA is depleted in Oxytricha (75). Furthermore, in vitro nucleosome assembly—using the Tetrahymena genomic DNA with 6mA (85)—can recapitulate the in vivo nucleosome array downstream of TSS, more so than using unmethylated DNA (70). Similarly, by comparing in vitro nucleosome occupancy on identical DNA sequences, with or without 6mA, Beh et al. finds that 6mA disfavors nucleosome occupancy (75). This may be attributed to intrinsic structural rigidity of DNA containing 6mA, which hinders its wrapping around the histone octamer (70). Alternatively, symmetric 6mA in linker DNA may increase the affinity for ATP-dependent chromatin remodeling complexes, or promote their activities otherwise, which are essential for ordering nucleosomes in the gene body. We envision a positive feedback loop between 6mA and nucleosome positioning for maintaining the epigenetic landscape of transcription.
Molecular mechanism for epigenetic inheritance of symmetric 6mA at ApT dinucleotides
A striking parallel can be drawn between AMT1-deposited symmetric 6mA at the AT motif in Tetrahymena and DNMT1-deposited symmetric 5mC at the CG motif in mammals (86), suggesting that symmetric 6mA is also maintained by a semi-conservative mechanism, on top of the specific recruitment of AMT1 to the promoter. Indeed, in asynchronously growing Tetrahymena cells, most symmetric 6mA sites have methylation levels near saturation; most AT sites are in the full methylation state, while only a minority is in the hemi-methylation state. These observations indicate that after replication splitting full into hemi-methylation, the latter is rapidly and efficiently converted to the former in an AMT1-dependent manner. This is corroborated by the recent finding that AMT1 preferentially methylates hemi-methylated sites (75). The two AMT1-associated proteins identified by Beh et al., featuring Myb-like domains in their N-terminal regions, are potentially involved in DNA binding (75). In support, Myb binding occurs at the major groove (87) and 6mA also occurs at the major groove. Structural studies are needed to elucidate the underlying molecular mechanism for specific recognition of hemi-methylated AT and propagation of symmetric 6mA as an epigenetic mark.
Other potential regulators of 6mA in Tetrahymena
6mA levels are reduced, but not abolished, in ΔAMT1 cells. The complete removal of AMT1 provides tantalizing glimpses into features of other potential 6mA regulators. By carefully checking the kinetic signatures, we are confident that the remaining 6mA in ΔAMT1 cells are not false positives and are enzymatically catalyzed. These other DNA N6-adenine MTase activities are distinguished from that of AMT1 in several aspects: (i) random targeting of the ApN dinucleotides, instead of specific targeting of ApT; (ii) no symmetric methylation on the AT motif and (iii) diminished enrichment in linker DNA. On the other hand, they still preferentially deposit 6mA at the 5′ end of Pol II-transcribed genes, suggesting that the corresponding MTases, similar to AMT1, are recruited to the promoter.
AMT2 and AMT5 are distinguished by several ZZ-type zinc fingers at the C-terminus (69), which might be involved in interacting with AMT1 or binding the target DNA molecule (79). Of note, a recent study reported that somatic knockout of AMT2 (referred to therein as TAMT1) suppresses 6mA levels in Tetrahymena (70). However, this is directly contradicted by our finding of no significant 6mA reductions in either somatic or germline knockout strains of AMT2. The grouping of AMT2 and AMT5 in the same subclade raised the possibility that they are functionally redundant, hence the lack of obvious phenotypes in the single knockout strain (70). AMT2 and AMT5 are likely involved in asymmetric AT methylation and non-AT methylation that are AMT1-independent. The mechanism to maintain epigenetic information carried by these 6mA sites is apparently distinct from the semi-conservative mechanism for symmetric AT methylation, and provides an intriguing direction for future studies.
DATA AVAILABILITY
The latest SB210 MAC genome can be found at the Tetrahymena genome database (TGD) (http://ciliate.org). Accession number for publicly available Tetrahymena datasets: BioProject PRJNA510457 (NCBI).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the following people for assistance with this study: Dr. Lifang Feng (Zhejiang Gongshang University, China) for sharing protocols of somatic rescue and hypertonic media culture; Borong Lu (Ocean University of China, China) for taking ‘contractile vacuole’ pictures and videos; Haibo Xie (OUC, China) for helping with the swimming velocity experiment and Dr. Alan Warren (Natural History Museum, UK) for English editing. Our special thanks are given to Prof. Weibo Song (OUC) for his helpful suggestions during drafting the manuscript.
Author contributions: Y.W. and S.G. conceived the study; Y.W., Y.L., T.C. and L.D. performed the experiments; Y.S. performed the bioinformatics analysis; B.P. performed the phylogenetic analysis; W.Z. performed the mass spectrometry analysis; Y.Q. performed phenotypic analysis for RAB46-KD cells; Y.W., Y.L. and S.G. wrote the paper. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) [2018SDKJ0406-2]; Natural Science Foundation of Shandong Province [JQ201706]; Fundamental Research Funds for the Central Universities [201841005]; Blue Life Breakthrough Program of LMBB of Pilot National Laboratory for Marine Science and Technology (Qingdao) [MS2018NO04]. Funding for open access charge: Natural Science Foundation of Shandong Province [JQ201706].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gorovsky M.A., Hattman S., Pleger G.L.. [6N] Methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol. 1973; 56:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ammermann D., Steinbrück G., Baur R., Wohlert H.. Methylated bases in the DNA of the ciliate Stylonychia mytilus. Eur. J. Cell Biol. 1981; 24:154–156. [PubMed] [Google Scholar]
- 3. Cummings D.J., Tait A., Goddard J.M.. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1974; 374:1–11. [DOI] [PubMed] [Google Scholar]
- 4. Rae P.M., Steele R.E.. Modified bases in the DNAs of unicellular eukaryotes: an examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978; 10:37–53. [DOI] [PubMed] [Google Scholar]
- 5. Salvini M., Barone E., Ronca S., Nobili R.. DNA methylation in vegetative and conjugating cells of a protozoan ciliate: Blepharisma japonicum. Dev. Genet. 1986; 7:149–158. [Google Scholar]
- 6. Fu Y., Luo G.-Z., Chen K., Deng X., Yu M., Han D., Hao Z., Liu J., Lu X., Doré L.C.. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015; 161:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mondo S.J., Dannebaum R.O., Kuo R.C., Louie K.B., Bewick A.J., Labutti K., Haridas S., Kuo A., Salamov A., Ahrendt S.R.. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017; 49:964–968. [DOI] [PubMed] [Google Scholar]
- 8. Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizábal-Corrales D., Hsu C.-H., Aravind L., He C., Shi Y.. DNA methylation on N6-adenine in C. elegans. Cell. 2015; 161:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koziol M.J., Bradshaw C.R., Allen G.E., Costa A.S., Frezza C., Gurdon J.B.. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2016; 23:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang Z., Shen L., Cui X., Bao S., Geng Y., Yu G., Liang F., Xie S., Lu T., Gu X.. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell. 2018; 45:406–416. [DOI] [PubMed] [Google Scholar]
- 11. Liu J., Zhu Y., Luo G.-Z., Wang X., Yue Y., Wang X., Zong X., Chen K., Yin H., Fu Y.. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016; 7:13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schiffers S., Ebert C., Rahimoff R., Kosmatchev O., Steinbacher J., Bohne A.V., Spada F., Michalakis S., Nickelsen J., Müller M.. Quantitative LC–MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. Int. Ed. 2017; 56:11268–11271. [DOI] [PubMed] [Google Scholar]
- 13. Wang X., Li Z., Zhang Q., Li B., Lu C., Li W., Cheng T., Xia Q., Zhao P.. DNA methylation on N6-adenine in lepidopteran Bombyx mori. Biochim. Biophys. Acta, Gene Regul. Mech. 2018; 1861:815–825. [DOI] [PubMed] [Google Scholar]
- 14. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M.. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao C.-L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.-Q., Luo F.. N6-Methyladenine DNA modification in the human genome. Mol. Cell. 2018; 71:306–318. [DOI] [PubMed] [Google Scholar]
- 16. Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J.. N6-methyladenine DNA modification in Drosophila. Cell. 2015; 161:893–906. [DOI] [PubMed] [Google Scholar]
- 17. Zhou C., Wang C., Liu H., Zhou Q., Liu Q., Guo Y., Peng T., Song J., Zhang J., Chen L.. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants. 2018; 4:554–563. [DOI] [PubMed] [Google Scholar]
- 18. Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U.. N6-methyladenine DNA modification in glioblastoma. Cell. 2018; 175:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y.Y., Chen X., Sheng Y.L., Liu Y.F., Gao S.. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 2017; 45:11594–11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma C., Niu R., Huang T., Shao L.-W., Peng Y., Ding W., Wang Y., Jia G., He C., Li C.-Y.. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019; 21:319–327. [DOI] [PubMed] [Google Scholar]
- 21. Yao B., Cheng Y., Wang Z., Li Y., Chen L., Huang L., Zhang W., Chen D., Wu H., Tang B.. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017; 8:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iyer L.M., Abhiman S., Aravind L.. Progress in Molecular Biology and Translational Science. 2011; 101:Elsevier; 25–104. [DOI] [PubMed] [Google Scholar]
- 23. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y.Y., Sheng Y.L., Liu Y.Q., Pan B., Huang J.B., Warren A., Gao S.. N6-methyladenine DNA modification in the unicellular eukaryotic organism Tetrahymena thermophila. Eur. J. Protistol. 2017; 58:94–102. [DOI] [PubMed] [Google Scholar]
- 25. Karrer K.M., VanNuland T.A.. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 2002; 30:1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorovsky M.A., Yao M.C., Keevert J.B., Pleger G.L.. Isolation of micro-and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975; 9:311–327. [DOI] [PubMed] [Google Scholar]
- 27. Sweet M., Allis C.. Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual. 1998; 1:NY: Cold Spring Harbor Laboratory press. [Google Scholar]
- 28. Mochizuki K., Novatchkova M., Loidl J.. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J. Cell Sci. 2008; 121:2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cassidy-Hanley D., Bowen J., Lee J.H., Cole E., VerPlank L.A., Gaertig J., Gorovsky M.A., Bruns P.J.. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997; 146:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hai B., Gorovsky M.A.. Germ-line knockout heterokaryons of an essential α-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Nat. Acad. Sci. U.S.A. 1997; 94:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miao W., Xiong J., Bowen J., Wang W., Liu Y., Braguinets O., Grigull J., Pearlman R.E., Orias E., Gorovsky M.A.. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009; 4:e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Talsky K.B., Collins K.. Strand-asymmetric endogenous Tetrahymena small RNA production requires a previously uncharacterized uridylyltransferase protein partner. RNA. 2012; 18:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Howard-Till R.A., Yao M.-C.. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol. Cell Biol. 2006; 26:8731–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao X.L., Xiong J., Mao F.B., Sheng Y.L., Chen X., Feng L.F., Dui W., Yang W.T., Kapusta A., Feschotte C.. RNAi-dependent Polycomb repression controls transposable elements in Tetrahymena. Genes Dev. 2019; 33:348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schäffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F.. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001; 29:2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen X., Wang Y., Sheng Y., Warren A., Gao S.. GPS it: an automated method for evolutionary analysis of nonculturable ciliated microeukaryotes. Mol. Ecol. Resour. 2018; 18:700–713. [DOI] [PubMed] [Google Scholar]
- 38. Huang Y., Niu B., Gao Y., Fu L., Li W.. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010; 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Wang C., Jiang Y., Katz L.A., Gao F., Yan Y.. Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci. China Life Sci. 2019; 62:203–214. [DOI] [PubMed] [Google Scholar]
- 41. Price M.N., Dehal P.S., Arkin A.P.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao S., Xiong J., Zhang C.C., Berquist B.R., Yang R.D., Zhao M., Molascon A.J., Kwiatkowski S.Y., Yuan D.X., Qin Z.H. et al.. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev. 2013; 27:1662–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu J., Li X., Song W., Wang W., Gao S.. Cyclin Cyc2p is required for micronuclear bouquet formation in Tetrahymena thermophila. Sci. China Life Sci. 2019; 62:668–680. [DOI] [PubMed] [Google Scholar]
- 44. Zhao X.L., Wang Y.Y., Wang Y.R., Liu Y.F., Gao S.. Histone methyltransferase TXR1 is required for both H3 and H3.3 lysine27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci. China Life Sci. 2017; 60:264–270. [DOI] [PubMed] [Google Scholar]
- 45. Yin R., Mao S.-Q., Zhao B., Chong Z., Yang Y., Zhao C., Zhang D., Huang H., Gao J., Li Z.. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013; 135:10396–10403. [DOI] [PubMed] [Google Scholar]
- 46. Swift M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comp. Sci. 1997; 37:411–412. [Google Scholar]
- 47. Chen X., Gao S., Liu Y.F., Wang Y.Y., Wang Y.R., Song W.B.. Enzymatic and chemical mapping of nucleosome distribution in purified micro-and macronuclei of the ciliated model organism, Tetrahymena thermophila. Sci. China Life Sci. 2016; 59:909–919. [DOI] [PubMed] [Google Scholar]
- 48. Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M. et al.. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006; 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stover N.A., Punia R.S., Bowen M.S., Dolins S.B., Clark T.G.. Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database. 2012; 2012:bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stein L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinf. 2013; 14:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schöpflin R., Teif V.B., Müller O., Weinberg C., Rippe K., Wedemann G.. Modeling nucleosome position distributions from experimental nucleosome positioning maps. Bioinformatics. 2013; 29:2380–2386. [DOI] [PubMed] [Google Scholar]
- 52. Xiong J., Gao S., Dui W., Yang W., Chen X., Taverna S.D., Pearlman R.E., Ashlock W., Miao W., Liu Y.. Dissecting relative contributions of cis-and trans-determinants to nucleosome distribution by comparing Tetrahymena macronuclear and micronuclear chromatin. Nucleic Acids Res. 2016; 44:10091–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delcher A.L., Adam P., Jane C., Salzberg S.L.. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002; 30:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wickham H., Grolemund G.. R for Data Science: Import, Tidy, Transform, Visualize Andmodel Data. 2016; USA: O’Reilly Press. [Google Scholar]
- 55. Gu Z., Gu L., Eils R., Schlesner M., Brors B.. circlize implements and enhances circular visualization in R. Bioinformatics. 2014; 30:2811–2812. [DOI] [PubMed] [Google Scholar]
- 56. Xiong J., Lu X., Zhou Z., Chang Y., Yuan D., Tian M., Zhou Z., Wang L., Fu C., Orias E.. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS One. 2012; 7:e30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen X., Jiang Y., Gao F., Zheng W., Krock T.J., Stover N.A., Lu C., Katz L.A., Song W.. Genome analyses of the new model protist Euplotes vannus focusing on genome rearrangement and resistance to environmental stressors. Mol. Ecol. Resour. 2019; 19:1292–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wei T., Simko V., Levy M., Xie Y., Jin Y., Zemla J.. Package ‘corrplot’. Statistician. 2017; 56:316–324. [Google Scholar]
- 59. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Norusis M. SPSS 16.0 Statistical Procedures Companion. 2008; Prentice Hall Press. [Google Scholar]
- 61. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 65. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L.. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006; 34:W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Feng L., Wang G., Hamilton E.P., Xiong J., Yan G., Chen K., Chen X., Dui W., Plemens A., Khadr L.. A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res. 2017; 45:9481–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Iyer L.M., Zhang D., Aravind L.. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays. 2016; 38:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Luo G.-Z., Hao Z., Luo L., Shen M., Sparvoli D., Zheng Y., Zhang Z., Weng X., Chen K., Cui Q.. N6-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 2018; 19:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stargell L.A., Bowen J., Dadd C.A., Dedon P.C., Davis M., Cook R.G., Allis C.D., Gorovsky M.A.. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993; 7:2641–2651. [DOI] [PubMed] [Google Scholar]
- 72. Guillemette B., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L.. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. Plos Biol. 2005; 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Strahl B.D., Ohba R., Cook R.G., Allis C.D.. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Park H. Structural basis of membrane trafficking by Rab family small G protein. Int. J. Mol. Sci. 2013; 14:8912–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Beh L.Y., Debelouchina G.T., Clay D.M., Thompson R.E., Lindblad K.A., Hutton E.R., Bracht J.R., Sebra R.P., Muir T.W., Landweber L.F.. Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell. 2019; 177:1781–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Richards E.J., Elgin S.C.R.. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002; 108:489–500. [DOI] [PubMed] [Google Scholar]
- 78. Driel R.V., Fransz P.. Nuclear architecture and genome functioning in plants and animals: what can we learn from both. Exp. Cell Res. 2004; 296:86–90. [DOI] [PubMed] [Google Scholar]
- 79. Ponting C.P., Blake D.J., Davies K.E., Kendrick-Jones J., Winder S.J.. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 1996; 21:11–13. [PubMed] [Google Scholar]
- 80. Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T.. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004; 6:73–77. [DOI] [PubMed] [Google Scholar]
- 81. Schübeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J.. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004; 18:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mizuguchi G., Shen X., Landry J., Wu W.-H., Sen S., Wu C.. ATP-driven exchange of histone H2A.Z variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004; 303:343–348. [DOI] [PubMed] [Google Scholar]
- 83. Van Nuland R., Smits A.H., Pallaki P., Jansen P.W., Vermeulen M., Timmers H.M.. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell Biol. 2013; 33:2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Swart E.C., Bracht J.R., Magrini V., Minx P., Chen X., Zhou Y., Khurana J.S., Goldman A.D., Nowacki M., Schotanus K.. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PloS Biol. 2013; 11:e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M.. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PloS Biol. 2006; 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bashtrykov P., Ragozin S., Jeltsch A.. Mechanistic details of the DNA recognition by the Dnmt1 DNA methyltransferase. Febs. Lett. 2012; 586:1821–1823. [DOI] [PubMed] [Google Scholar]
- 87. Jaiswal R., Choudhury M., Zaman S., Singh S., Santosh V., Bastia D., Escalante C.R.. Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe. Proc. Nat. Acad. Sci. U.S.A. 2016; 113:E2267–E2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xiong J., Yuan D., Fillingham J.S., Garg J., Lu X., Chang Y., Liu Y., Fu C., Pearlman R.E., Miao W.. Gene network landscape of the ciliate Tetrahymena thermophila. PLoS One. 2011; 6:e20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aronica L., Bednenko J., Noto T., DeSouza L.V., Siu K.M., Loidl J., Pearlman R.E., Gorovsky M.A., Mochizuki K.. Study of an RNA helicase implicates small RNA–noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008; 22:2228–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008; 425:79–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The latest SB210 MAC genome can be found at the Tetrahymena genome database (TGD) (http://ciliate.org). Accession number for publicly available Tetrahymena datasets: BioProject PRJNA510457 (NCBI).