Abstract
Liquid–liquid phase separation plays an important role in a variety of cellular processes, including the formation of membrane-less organelles, the cytoskeleton, signalling complexes, and many other biological supramolecular assemblies. Studies on the molecular basis of phase separation in cells have focused on protein-driven phase separation. In contrast, there is limited understanding on how RNA specifically contributes to phase separation. Here, we described a phase-separation-like phenomenon that SHORT ROOT (SHR) RNA undergoes in cells. We found that an RNA G-quadruplex (GQ) forms in SHR mRNA and is capable of triggering RNA phase separation under physiological conditions, suggesting that GQs might be responsible for the formation of the SHR phase-separation-like phenomenon in vivo. We also found the extent of GQ-triggered-phase-separation increases on exposure to conditions which promote GQ. Furthermore, GQs with more G-quartets and longer loops are more likely to form phase separation. Our studies provide the first evidence that RNA can adopt structural motifs to trigger and/or maintain the specificity of RNA-driven phase separation.
INTRODUCTION
Liquid–liquid phase separation (LLPS) phenomena, with two liquid phases coexisting in one system, have been recently observed in the cells and shown to be responsible for organization of many complex biochemical reactions temporally and spatially (1). Thus, LLPS is of great importance for many biological processes, including the formation of membrane-less organelles (2,3), the cytoskeleton (4,5), signalling complexes (6,7), and many other biological supramolecular assemblies (8,9).
Previous studies mainly focused on protein-driven phase separation through multivalent interactions. This multivalence can be attributed to at least one of three different specific interactions: (a) proteins with well-defined domains interact with stereospecific interaction surfaces to form oligomers (10); (b) proteins with linker motifs string together to generate linear multivalent interactions (11); (c) proteins featured with intrinsically disordered regions (IDRs) can serve as scaffolds to act with distinctive short linear motifs to form multiple interactions (12–14). Apart from these protein–protein interactions, the specificity for the protein-driven phase separation could also be achieved by the properties of their interacting RNAs including their amount (15), sequence (16) and secondary structure (16,17). For example, the secondary structures of both CLN3 and BIN2 RNAs determine the identity of droplets formed by RNA binding protein, whi3, by uncovering or masking its interaction regions (16). Furthermore, ALS/FTD-associated C9ORF72 RNA containing G4C2 repeat sequences is also involved in the phase separation of G-quadruplex-binding proteins (17).
RNA has generally been considered as a regulatory component in protein-driven phase separation (16). However, there is evidence that RNA itself is also capable of forming phase separation, potentially through multivalent RNA–RNA interactions (18). Examples of these include RNA tandem repeat sequences (19) and RNA homopolymers (20). However, these do not provide any specificity in the formation and/or maintenance of RNA-driven phase separation. Hence, it is still unclear which properties of RNA assure the specificity of RNA-driven phase separation.
In our study, we investigated the properties of SHR mRNA, an RNA important in plant development, that was observed to form an RNA phase-separation-like phenomenon in vivo. We found that SHR contains a G-quadruplex (GQ), a specific RNA tertiary structure motif. G-quadruplexes are composed of G-tetrads formed by Hoogsteen hydrogen bonding and stabilized by π-π stacking interactions and the coordination of cations, especially K+ (21). The SHR-GQ structure is capable of triggering RNA-driven phase separation. Furthermore, the stability of this GQ, when modified by different conditions, correlated with the extent of phase separation formation. Our study suggests that RNA may adopt specific RNA structure motifs such as G-quadruplexes, to trigger and/or maintain RNA-driven phase separation.
MATERIALS AND METHODS
Single molecule FISH (smFISH)
Probes for SCARECROW (At3g54220) and SHORT ROOT (At4g37650) were designed using the online program Stellaris Probe Designer version 2.0 from LGC Biosearch Technologies (see Supplementary Table S3). smFISH experiments were carried out for Arabidopsis roots as previously described (22). Briefly, seedlings were removed from the media and the root tips were cut and fixed in 4% paraformaldehyde for 30 min. The roots were washed twice with nuclease free 1 × PBS (Thermo Fisher Scientific) and then placed onto a poly-l-lysine slide (Thermo Fisher Scientific) and covered by a glass coverslip (R&L Slaughter, Upminster, UK). The meristems were then squashed under the coverslip, before being submerged in liquid nitrogen until frozen. The coverslips were removed using a razor blade and the roots were left on the slide to dry at room temperature for 30 min. Tissue permeabilization was then carried out by immersing the samples in 70% ethanol for a minimum of one hour. Probe hybridization following removal from ethanol, slides were left at room temperature for 5 min before two washes were carried out with wash buffer (10% formamide and 2× saline-sodium citrate buffer; SSC). 100 ml of hybridization solution (10% dextran sulphate, 2× SSC and 10% formamide) containing SHR/SCR probes (at a final concentration of 250 nM) was added to each slide. Coverslips were placed over the samples to prevent evaporation and the probes were left to hybridize at 37°C overnight in the dark. Excess hybridization solution (containing unbound probes) was pipetted off the following morning. Each sample was washed twice with wash buffer, with the second wash left to incubate for 30 min at 37°C in the dark. After wash buffer removal, 100 ml of the nuclear stain DAPI (4′,6-diamidino-2-phenylindole, 100 ng/ml) was added to each slide and left to incubate at 37°C for 30 min. Following DAPI removal, a 100 ml 2× SSC wash was carried out before 100 ml GLOX buffer (0.4% glucose in 10 mM Tris, 2× SSC) was added to each slide and left to equilibrate at room temperature for 2 min. This was pipetted off and replaced with an anti-fade solution containing 100 ml of GLOX buffer, 1 ml glucose oxidase (Sigma) and 1 ml catalase (Sigma). The samples were then covered by 22 mm × 22 mm No. 1 coverslips (R&L Slaughter, Upminster, UK), sealed with CoverGrip sealant (Biotium, UK) and imaged using a Delta Vision Elite Deconvolution microscope with a ×100 oil immersed objective (NA 1.46).
Preparation of in vitro transcribed RNAs
DNA Templates (see Supplementary Table S3) were amplified and purified, RNAs were in vitro transcribed using Hiscribe T7 High Yield RNA Synthesis Kit (New England Biolabs, NEB). Each RNA was purified using pre-casted 6% denaturing polyacrylamide gel (Thermo Fisher Scientific) and the gel band with desired RNA product was cut and soaked in 1× TEL800 buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 800 mM LiCl) overnight, RNA was then purified with ZYMO RNA purification kit (ZYMO S1016).
Reverse transcription stalling assay
Reverse transcription stalling assay was performed as previously described (23). 500 ng (about 0.3 pmol) of SHR-GQ or SHR-GQm RNA was mixed with folding buffer (10 mM Tris–Cl, pH 7.5, 150 mM Li+ or K+) and denatured at 95°C for 1.5 min. For the reaction with PDS, PDS was added to a final concentration of 5 μM. For reverse transcription, 5× RT buffer (200 mM Tris–HCl, pH 8.3, 750 mM KCl/LiCl, 2.5 mM dNTPs, 12.5 mM MgCl2 and 5 mM DTT), 1 μl of 5 μM Cy5-labelled primer and 0.5 μl SuperScript III reverse transcriptase (200 U/μl) (Thermo Fisher Scientific) were added. After reverse transcription, 0.5μl NaOH (1M) was added to the reaction and incubate at 95°C to eliminate RNA template. Finally, 10 μl of 2 × stopping dye solution containing 20 mM Tris–HCl, pH 7.5, 20 mM EDTA, 94% deionized formamide and Orange G was added to the reaction mixture (Orange G dye as tracker). The cDNAs were size fractionated by 8 M urea 8% denaturing polyacrylamide gel. The gel was scanned with typhoon phosphorimager and analyzed by ImageQuant 5.2.
Circular dichroism (CD)
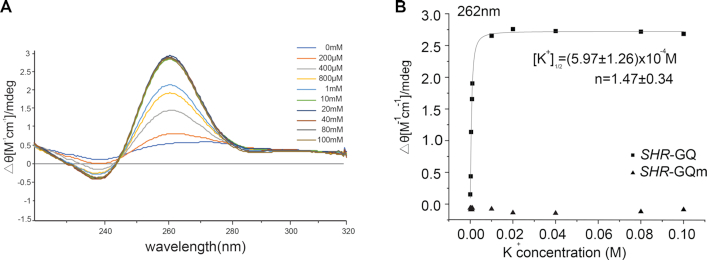
RNA oligos SHR-GQ, SHR-GQm, SHR-GQsc (see Supplementary Table S3) were purchased from Gentech Inc. 10 μM RNA in 10 mM Li cacodylate, pH 7.0, was denatured at 95°C for 3 min, cooled to room temperature at the rate of 1°C/min and then incubated at room temperature for 15 min for equilibration. The CD measurements were performed using Applied Photophysics Chirascan-plus with the pathlength of 1 mm, spectra were acquired at each wavelength from 220 to 320 nm at 20°C. Spectrum is an average of four scans with a response time 0.5 s/nm. For data fitting, the K+ titration was performed to determine the amount of K+ needed for GQ formation. To determine the [K+]1/2, the CD values at 262 nm as a function of potassium concentration were fit with Origin 8.1 according to the Hill equation.
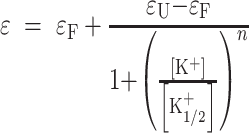 |
where ϵF is the normalized ellipticity of fully folded CD signal, and ϵu is the normalized ellipticity of unfolded CD signal, [K+]1/2 is the concentration of potassium ion needed for 50% of the RNA to fold into GQ, and n is the Hill coefficient.
RNA structure prediction
To predict the RNA secondary/tertiary structure, SHR sequence was subjected to ViennaRNA web server (24), with adjustment of temperature to 22°C to match the growth temperature of Arabidopsis. For the GQ prediction, GQRS mapper (25) prediction was used with the default settings.
Phase separation assay
RNAs and K+ was mixed with concentration shown on the phase diagram (Figure 6). Before the experiment, RNA, K+ and 10% PEG8000 were mixed thoroughly, the mixture was then heated to 95°C for 3 min and cooled down at 0.5°C/min to room temperature and imaged immediately. RNA was stained by nucleic acid dye SYTO 17 (Sigma, S7579). Samples were visualized by a confocal microscope (ZEISS LSM 880) with ×40 oil immersion objective.
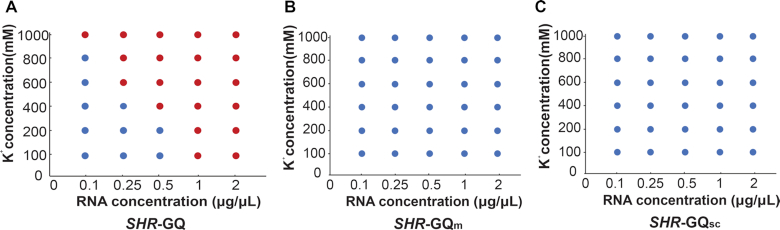
Figure 6.
Phase diagrams of SHR-GQ (A), SHR-GQm (B) and SHR-GQsc (C) under different RNA and K+ concentrations. Red dots indicate where liquid–liquid-phase-separation occurs; blue dots indicate a lack of liquid–liquid-phase-separation.
Quantification of droplets in ROI (region of interest)
The extent of phase separation was quantified by the index of dispersion (σ2/μ) of fluorescence intensity per pixel (pixel size 43 nm × 43 nm). In brief, for each ROI, the variance of fluorescence intensity was determined, and normalized by the mean RNA fluorescence intensity in the solution phase. 10 independent imaging areas (∼2500 μm2 each) were analyzed for each condition.
RESULTS AND DISCUSSION
SHR mRNA displays a phase separation like phenomenon in cells
Initially our work focused on visualizing the mRNA distribution of SHORT ROOT (SHR) and SCARECROW (SCR), two key genes for Arabidopsis root development (26,27), using single molecule fluorescence in situ hybridization (smFISH). This is a technique which utilizes fluorescence-labelled antisense-probes to visualize individual RNA molecules in vivo (28). We used DAPI (4′,6-diamidino-2-phenylindole) to stain the nuclei (Figure 1A, Supplementary Movie S1). We found that the signals for SCR RNA were evenly distributed in cells, with each cytosolic green spot of a uniform signal intensity representing a single SCR RNA molecule (Figure 1B, Supplementary Movie S1). This even distribution of RNAs is typical for mRNA localization (29,30). Surprisingly, in the same cells, we observed aggregated, phase separation-like signals for SHR RNAs (Figure 1C, Supplementary Movie S1). After merging the nuclei and RNA images, we determined that SHR signals were mainly localized to the cytoplasm (Figure 1D, Supplementary Movie S1).
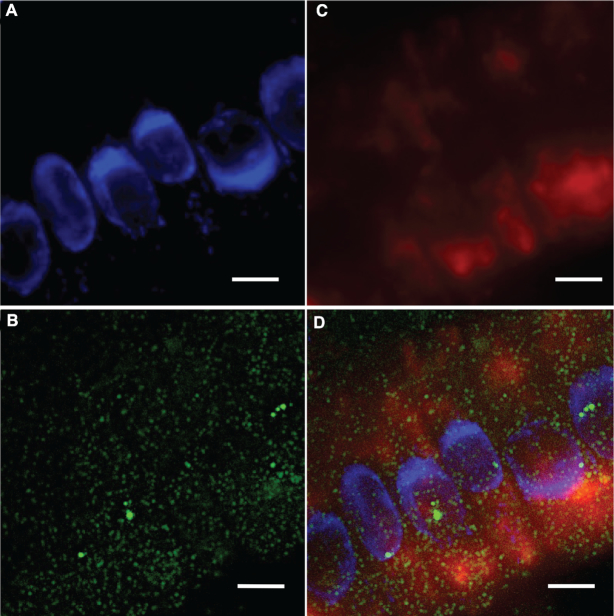
Figure 1.
Phase-separation-like SHR and punctate SCR RNA labelling observed in plant root cells, images generated by smFISH. (A) Nuclei stained with DAPI (blue), SCR RNA (green) is shown in (B) and SHR RNA (red) in (C). Merged image in (D). Scale bar, 10 μm.
To further investigate this phase-separation-like phenomena in cells, we performed another smFISH experiment on a transgenic line with inducible SHR (shr-2 pSHR::SHR:GR), where the expression of SHR was induced by dexamethasone(2). Following the dexamethasone induction, we observed that the big globular-shape signals of SHR RNA appeared in cells after 2 h (Supplementary Figure S1, Supplementary Movie S2). These big globular-shaped signals were very different from the dot-shaped signals (such as SCR), indicating a phase separation-like phenomenon and this phase separation-like phenomenon is specifically associated with SHR expression.
Our smFISH results indicate that SHR mRNA displays a phase-separation-like phenomenon in plant cells. As a critical gene in ground tissue patterning and identity maintenance, SHR plays important roles in root development (26,31,32). During root pattern formation, SHR proteins are translated in the stele and move from the stele into the endodermis where they interact with SCR. This interaction leads to restriction of SHR movement to a single cell layer of endodermis (26,31,32). While the focus to date has been on SHR protein movement and activity, the cellular concentration and localization of SHR mRNA may also affect SHR function. The phase-separation-like phenomenon of SHR we observed might result in the sequestering of SHR mRNA enhancing or suppressing its translational efficacy. Thus, our observations lead to a new hypothesis that plant root cells might organize the cellular SHR mRNA levels in space and time to assure the accurate regulation of root development.
A G-quadruplex forms in SHR mRNA
To identify the molecular properties of SHR RNAs capable of producing this phase-separation-like phenomenon, we investigated both SHR mRNA sequence and secondary/tertiary structures. Previous studies showed that multivalent RNA–RNA interactions through RNA tandem repeat sequences are able to form phase separation (19). Therefore, we searched the SHR sequence using tandem repeat finder (33) but found no tandem repeats, suggesting RNA phase separation was caused by an alternative trigger. Single stranded regions of RNA have also been implicated as the basis for multivalent interactions (16). Additionally, stable secondary structures might inhibit the phase separation by masking the flanking single-strandedness, which could contribute to multivalent interactions (16). Using ViennaRNA (24) to predict secondary/tertiary structures formed in SHR RNA, we found a quite stable RNA structure without any long single-stranded loop regions or predicted pseudoknot structures (Figure 2, Supplementary Table S1).
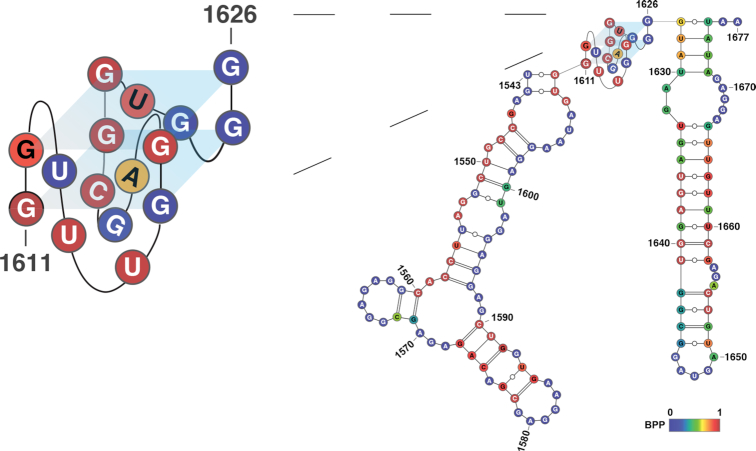
Figure 2.
Formation of GQ in SHR mRNA. RNA structure model of SHR, showing the fragment with GQ structure, predicted by ViennaRNA (24). A parallel topology is shown (supported by circular dichroism studies), nucleotides are color-coded according to base-pairing probability (BPP), where 1 represent 100% double-stranded, 0 represent 100% single-stranded. GQ structure is enlarged for better visualization.
Interestingly, we found a putative G-quadruplex (GQ) motif, was predicted at position 1611–1626 (Figure 2). An RNA G-quadruplex is one specific tertiary structure motif that could form in a special sequence context of GxLaGxLbGxLcGx (G stands for guanine; L stands for loop), where x ≥2, and a, b, c are ≥1 nt and ≤7 (34). This putative GQ motif was also predicted using QGRS mapper, a program designed to predict GQ forming sequences (25). We found that the GQ motif predicted by ViennaRNA had the highest confidence score among all the putative GQs found by QGRS mapper in the sequence (Supplementary Table S2). This putative GQ motif predicted in SHR is a two-guanine-quartet (G2) GQ, with the sequence motif as G2L3G2L3G2L2G2 (Figure 2).
To probe the presence of this GQ in SHR mRNA, we performed a reverse transcription (RT) stalling assay (35). The GQ formation is typically stabilized by potassium (K+) but not lithium (Li+), and GQs may be preferentially stabilized by using both potassium and the G-quadruplex stabilizing ligand pyridostatin (K++PDS) (36). If GQ is stably formed, the reverse transcriptase will stall during reverse transcription at positions where there is a folded RNA G-quadruplex (23). We observed stronger RT stalling signals in both K+ and K++PDS as compared to Li+ conditions (lanes 1–3, Figure 3), confirming the formation of this GQ in SHR mRNA under the K+ and K+ + PDS conditions.
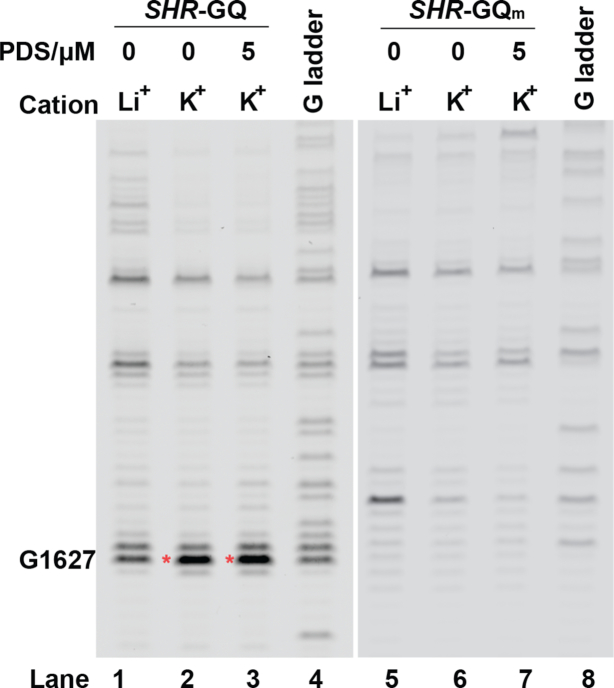
Figure 3.
Reverse transcription stalling assays of SHR-GQ RNA shows K+ dependent stalling (red asterisk) at one nucleotide after the folded GQ (lane 2 and lane 3), SHR-GQm do not have stalling at the corresponding position (lane 6 and lane 7). G-ladders (lane 4 and lane 8) show the sequencing lanes of G for SHR-GQ (lane 4) and SHR-GQm (lane 8), respectively.
To further confirm that the stalling was caused by GQ folding, we used a mutated SHR-GQm RNA, in which the Gs that contribute to G-quartets were mutated to As (Supplementary Table S3). No RT stalling was observed for the SHR-GQm RNA (lanes 5–7, Figure 3), providing further evidence that RT stalling is due to GQ formation in SHR.
SHR G-quadruplex forms under physiological conditions
To determine the type of GQ formation in SHR we used circular dichroism (CD) (37). A titration with K+ was performed and we observed a spectrum with a positive signal at 262 nm and a negative signal at 242 nm, indicative of a parallel-type GQ structure (Figure 4A) (38,39). The K+ titration CD profile fitted well to a two-state model with a [K+]1/2 value of (5.96 ± 1.26) × 10−4 M (Figure 4B).
Figure 4.
SHR-GQ forms a parallel GQ structure. (A) Circular Dichroism profile of SHR-GQ RNA with potassium ion titration. (B) Circular Dichroism signal (ellipticity monitored at 262 nm) of SHR-GQ (squares) and SHR-GQm (triangles) are shown as a function of K+ concentration. Data were fitted with Hill equation. The [K+]1/2 and Hill coefficients (n) are provided in the plot.
The formation of GQ reached saturation at ∼10 mM, indicating strong GQ formation (Figure 4B). In contrast, the mutated SHR-GQm RNA did not show any GQ spectrum signature at any concentration of K+, only giving a spectrum consistent with a random coil, suggesting a complete abolition of GQ formation (Figure 4B, Supplementary Figure S2A). These CD results combined with the RT stalling experiments confirmed the formation of a parallel-type GQ structure in SHR mRNA. Notably, this GQ could be triggered and stabilized at very low potassium level ([K+]1/2 is ∼0.6 mM) (Figure 4B). Since the physiological K+ concentration is ∼100 mM (40), it is most likely that this GQ forms and is stable in plant cells.
SHR G-quadruplex can trigger phase separation
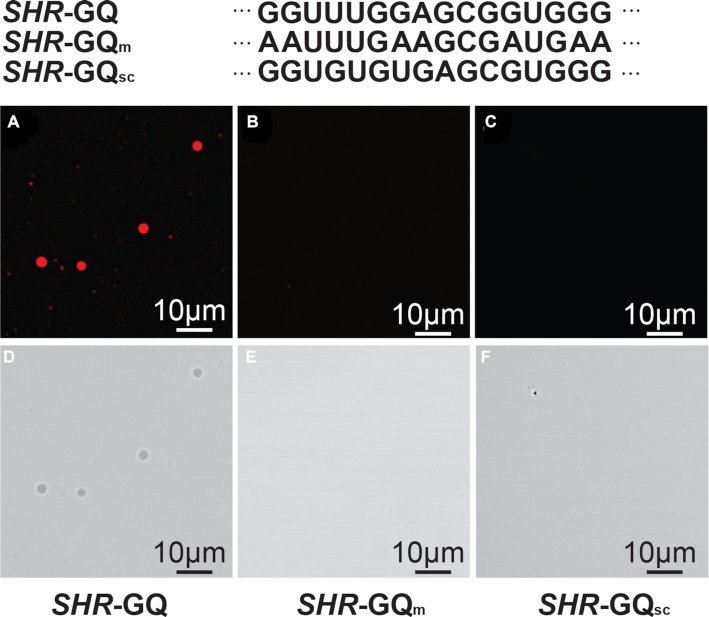
We next asked if this GQ motif is able to trigger phase separation of SHR mRNA. We performed a phase separation assay on SHR-GQ and SHR-GQm RNAs at the physiological K+ concentration (100 mM K+) (40) and found that SHR-GQ formed clear droplets (Figure 5A, D) whereas the mutant (SHR-GQm) was not able to trigger phase separation (Figure 5B, E).
Figure 5.
GQ is able to trigger the liquid–liquid phase separation. Fluorescence micrographs of SHR-GQ (A, D), GQ-mutation (SHR-GQm) (B, E) and GQ-scramble (SHR-GQsc) (C, F) RNAs at the physiological K+ concentration. Scale bars = 10 μm
This result provided evidence that this SHR-GQ is capable of triggering the formation of phase separation (Figure 5A, D). We further generated another mutated GQ (SHR-GQsc), in which the GQ sequence was scrambled to avoid intramolecular GQ formation (Supplementary Table S3). As expected, SHR-GQsc did not show the same responsiveness to K+ (Supplementary Figure S2B), producing only very weak GQ signals at high K+ concentration. This suggests that SHR-GQsc is not able to form intramolecular GQ but might be able to form weak GQ through intermolecular interactions (Supplementary Figure S2B). The phase separation assay for SHR-GQsc showed SHR-GQsc that was unable to trigger phase separation (Figure 5C, F). In the following systematic assessment of the capability of SHR-GQ to form phase separation under different K+ and RNA concentrations, we found that the extent of phase separation triggered by SHR-GQ increased with the rise of RNA concentration as well as with K+ concentration (Figure 6A, Supplementary Figure S3).
In contrast, neither the SHR-GQm nor SHR-GQsc mutants were able to trigger phase separation, even at high K+ concentration (1000 mM) and RNA concentration (2 μg/μl) (Figure 6B, C). These results indicate that the specificity of RNA-driven phase separation is provided by the SHR intramolecular GQ structure. We next investigated the physical properties of the SHR-GQ triggered droplets, these droplets were found to undergo rapid rearrangement and fusion events (Figure 7, Supplementary Movie S3), indicating that the droplets were in a relaxed, liquid-like state.
Figure 7.
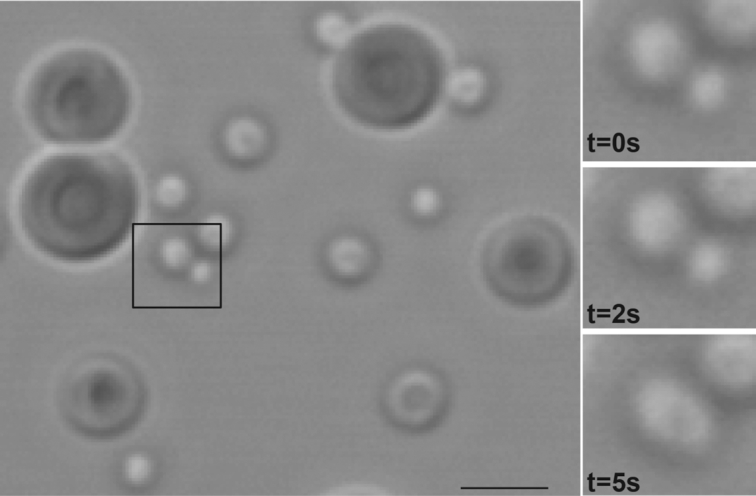
The quick rearrangement of GQ-triggered droplets. Left, droplets formed by SHR-GQ. Right, droplets formed by SHR-GQ undergo fusion over time. Scale bar = 5 μm.
To further determine whether GQ structure act as a general trigger of phase separation when embedded within any RNA sequence, we generated another two mutated sequences (SHRscramble1-GQ and SHRscramble2-GQ), in which the flanking sequences of GQ were scrambled (Supplementary Table S3). We found that both RNAs were able to trigger RNA phase separation (Supplementary Figures S4 and S5), suggesting that GQ serves as a general trigger of RNA phase separation. To further confirm the full-length SHR is capable of triggering RNA phase separation at the physiological K+ concentration, we performed the in vitro phase separation assay on the RNAs with full-length SHR and full-length SHR GQ mutants (Supplementary Table S3). We found that the full-length SHR, similar to the short-length SHR, was able to trigger RNA phase separation at physiological K+ concentration (Supplementary Figure S6), while the full-length SHR GQ mutant was not capable of triggering RNA phase separation (Supplementary Figure S6). By increasing the potassium concentration up to 400 mM, we observed RNA phase separation of the full-length SHR GQ mutant (Supplementary Figure S7), which might be due to non-specific helical stacking (18). Overall, the full-length SHR is capable of triggering RNA phase separation at physiological K+ concentration, further confirming that SHR is likely to trigger RNA phase separation in cells.
Taken together, our results indicate that SHR-GQ is capable of triggering liquid–liquid phase separation through intramolecular GQ structure formation. Notably, this RNA G4 located within SHR coding region. It is possible that SHR-GQ driven RNA phase separation might act as a regulator/factor for SHR translation.
Properties of GQ affect the extent of GQ-triggered phase separation
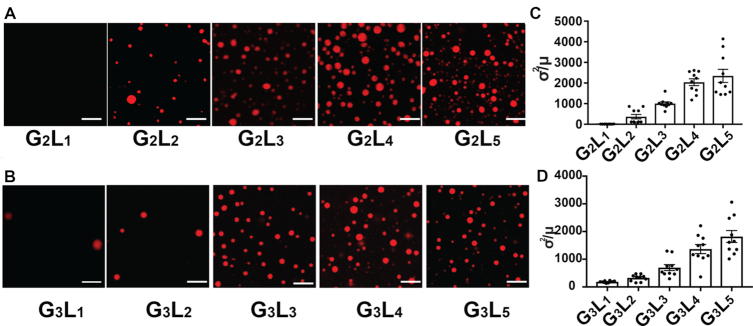
Previous studies indicate that the stability of GQ forming sequences is generally determined by the number of G-quartets and the length of the loops (39,41). To probe the relationship between phase separation and stability of the GQ forming sequence, we determined the capacity for phase separation of sequences with different numbers of quartets (G = 2 or 3) and different loop lengths (L = 1 to 5) (see Supplementary Table S3 for full list). We found that the G3 GQs tend to trigger phase separation under lower K+ concentrations and lower RNA concentrations as compared to G2 GQs (Figure 8A, B, Supplementary Figures S8 and S9).
Figure 8.
The capability for GQ-triggered-phase-separation is affected by the number of G-quartets and loop lengths. (A) Fluorescence micrographs of phase separation of G2 GQ RNAs with different loop lengths (1 μg/μl RNA under 800mM K+). (B) Fluorescence micrographs of phase separation of G3 GQ RNAs with different loop lengths (1 μg/μl RNA under 400 mM K+). (C) Quantitation of inhomogeneity as normalized variance (σ2/μ) of phase separation for G2 GQ RNAs with different loop lengths. (D) Quantitation of inhomogeneity as normalized variance (σ2/μ) of phase separation for G3 GQ RNAs with different loop lengths. Data are shown as mean ± SE.
Our results indicate that the G3 GQs provide a better trigger for phase separation as compared to G2 GQs and provides further evidence that the GQ structure is responsible for the phase separation formation. Interestingly, we also found that the GQs with longer loop lengths tend to form more droplets as compared to those with shorter loop lengths (Figure 8A, B, Supplementary Figures S8 and S9). The index of dispersal (σ2/μ) in Region Of Interest (ROI) was used to quantify the extent of phase separation (19). Consistently, longer loop lengths resulted in an increased extent of droplets formation (Figure 8C, D). The longer loops might allow more multivalent interactions among GQs. Overall, our results indicate that GQs with more G-quartets and longer loops have a greater extent of triggering phase separation. From a previous prediction of GQs across Arabidopsis genome, G2s with longer loops (G2L3–4) are shown to be more dominant among all the GQ types (42). Given that in our experiments G2 with longer loops (L = 3 to 5) triggered phase separation more easily (Supplementary Figure S8), this suggested that these G2 longer-loop GQs in Arabidopsis are likely to form similar RNA-driven phase separation as SHR-GQ under physiological conditions.
RNA GQs have been suggested to influence translation in the plant development (34,43). In plants, the cellular levels of K+ can vary between different developmental stages and stress conditions (44,45). For instance, under drought conditions, plants increase cytosolic K+ concentrations from 100 mM under normal cellular conditions up to 700 mM in order to avoid cellular dehydration (46). This dramatic increase of K+ concentration in the cell is likely to promote more GQ to form strongly, subsequently inducing more GQ-triggered RNA-driven phase separation. This GQ-triggered RNA-driven phase separation might be a regulatory mechanism for the cells to protect and/or store RNAs under the stress conditions such as drought.
In conclusion, our results provide the first demonstration of RNA adopting a specific RNA structure, e.g. GQ, to allow the formation of RNA-driven phase separation. This RNA-structure-triggered phase separation might be responsible for our observation of the phase-separation-like phenomenon of SHR in vivo, providing the possibility of a new regulatory role in plant root development. It also reveals a new functional role for GQ structures, serving as a trigger for RNA-driven phase separation.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council [BB/L025000/1 and BB/N022572/1 to Y.Z., M.Y., Z.W., Y.D.]; European Commission Horizon 2020 European Research Council (ERC) Starting Grant [680324 to Y.Z., M.Y., Y.D.]; Company of Biologists Travelling Fellowship (to S.D.); Howard Hughes Medical Institute (to P.N.B.). Funding for open access charge: Biotechnology and Biological Sciences Research Council and European Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hyman A.A., Weber C.A., Jülicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 2. Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A.. Germline p granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009; 324:1729. [DOI] [PubMed] [Google Scholar]
- 3. Brangwynne C.P., Mitchison T.J., Hyman A.A.. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A.. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017; 169:1066–1077. [DOI] [PubMed] [Google Scholar]
- 5. Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., Alberti S., Diez S., Hyman A.A.. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017; 20:2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D.. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016; 352:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chong P.A., Forman-Kay J.D.. Liquid–liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 2016; 41:180–186. [DOI] [PubMed] [Google Scholar]
- 8. Burke Kathleen A., Janke Abigail M., Rhine Christy L., Fawzi Nicolas L.. Residue-by-residue view of in vitro fus granules that bind the c-terminal domain of RNA polymerase II. Mol. Cell. 2015; 60:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conicella Alexander E., Zerze Gül H., Mittal J., Fawzi Nicolas L.. ALS mutations disrupt phase separation mediated by α-helical structure in the tdp-43 low-complexity c-terminal domain. Structure. 2016; 24:1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J.. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003; 160:823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Kato M., Han Tina W., Xie S., Shi K., Du X., Wu Leeju C., Mirzaei H., Goldsmith Elizabeth J., Longgood J., Pei J. et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee C., Occhipinti P., Gladfelter A.S.. PolyQ-dependent RNA–protein assemblies control symmetry breaking. J. Cell Biol. 2015; 208:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y., Protter David S.W., Rosen Michael K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F. et al.. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012; 483:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M. et al.. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018; 360:918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M. et al.. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018; 360:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fay M.M., Anderson P.J., Ivanov P.. ALS/FTD-Associated C9ORF72 repeat rna promotes phase transitions in vitro and in cells. Cell Rep. 2017; 21:3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Treeck B., Parker R.. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell. 2018; 174:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jain A., Vale R.D.. RNA phase transitions in repeat expansion disorders. Nature. 2017; 546:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D., Parker R.. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hardin C.C., Watson T., Corregan M., Bailey C.. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry. 1992; 31:833–841. [DOI] [PubMed] [Google Scholar]
- 22. Duncan S., Olsson T.S.G., Hartley M., Dean C., Rosa S.. Single molecule RNA FISH in Arabidopsis root cells. Bio-protocol. 2017; 7:e2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwok C.K., Balasubramanian S.. Targeted detection of G-quadruplexes in cellular RNAs. Angew. Chem. Int. Ed. 2015; 54:6751–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms for Molecular Biology. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kikin O., D’Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakajima K., Sena G., Nawy T., Benfey P.N.. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001; 413:307–311. [DOI] [PubMed] [Google Scholar]
- 27. Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.-T., Benfey P.N.. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000; 101:555–567. [DOI] [PubMed] [Google Scholar]
- 28. Duncan S., Olsson T.S.G., Hartley M., Dean C., Rosa S.. A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods. 2016; 12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zenklusen D., Larson D.R., Singer R.H.. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008; 15:1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zenklusen D., Singer R.H.. Methods in Enzymology. 2010; 470:Academic Press; 641–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N.. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007; 316:421. [DOI] [PubMed] [Google Scholar]
- 32. Petricka J.J., Winter C.M., Benfey P.N.. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012; 63:563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwok C.K., Ding Y., Shahid S., Assmann S.M., Bevilacqua P.C.. A stable RNA G-quadruplex within the 5′-UTR of Arabidopsis thaliana ATR mRNA inhibits translation. Biochem. J. 2015; 467:91. [DOI] [PubMed] [Google Scholar]
- 35. Kwok C.K., Sahakyan A.B., Balasubramanian S.. Structural analysis using shalipe to reveal RNA g-quadruplex formation in human precursor microRNA. Angew. Chemie Int. Ed. 2016; 55:8958–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez R., Müller S., Yeoman J.A., Trentesaux C., Riou J.-F., Balasubramanian S.. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008; 130:15758–15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vorlíčková M., Kejnovská I., Sagi J., Renčiuk D., Bednářová K., Motlová J., Kypr J.. Circular dichroism and guanine quadruplexes. Methods. 2012; 57:64–75. [DOI] [PubMed] [Google Scholar]
- 38. Zhang A.Y.Q., Bugaut A., Balasubramanian S.. A sequence-independent analysis of the loop length dependence of intramolecular RNA g-quadruplex stability and topology. Biochemistry. 2011; 50:7251–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pandey S., Agarwala P., Maiti S.. Effect of loops and G-quartets on the stability of RNA G-quadruplexes. J. Phys. Chem. B. 2013; 117:6896–6905. [DOI] [PubMed] [Google Scholar]
- 40. Wang Y.i, Wu W.-H.. Plant sensing and signaling in response to K+-deficiency. Mol. Plant. 2010; 3:280–287. [DOI] [PubMed] [Google Scholar]
- 41. Palumbo S.L., Ebbinghaus S.W., Hurley L.H.. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009; 131:10878–10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mullen M.A., Olson K.J., Dallaire P., Major F., Assmann S.M., Bevilacqua P.C.. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: prevalence and possible functional roles. Nucleic Acids Res. 2010; 38:8149–8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho H., Cho H.S., Nam H., Jo H., Yoon J., Park C., Dang T.V.T., Kim E., Jeong J., Park S. et al.. Translational control of phloem development by RNA G-quadruplex–JULGI determines plant sink strength. Nature Plants. 2018; 4:376–390. [DOI] [PubMed] [Google Scholar]
- 44. Greenway H., Munns R.. Mechanisms of Salt Tolerance in Nonhalophytes. Annual Review of Plant Physiology. 1980; 31:149–190. [Google Scholar]
- 45. Zhang H.-X., Blumwald E.. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nature Biotechnology. 2001; 19:765–768. [DOI] [PubMed] [Google Scholar]
- 46. Levi A., Paterson A.H., Cakmak I., Saranga Y.. Metabolite and mineral analyses of cotton near-isogenic lines introgressed with QTLs for productivity and drought-related traits. Physiologia Plantarum. 2011; 141:265–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.