Abstract
During meiosis, telomere attachment to the inner nuclear envelope is required for proper pairing of homologous chromosomes and recombination. Here, we identified F-box protein 47 (FBXO47) as a regulator of the telomeric shelterin complex that is specifically expressed during meiotic prophase I. Knockout of Fbxo47 in mice leads to infertility in males. We found that the Fbxo47 deficient spermatocytes are unable to form a complete synaptonemal complex. FBXO47 interacts with TRF1/2, and the disruption of Fbxo47 destabilizes TRF2, leading to unstable telomere attachment and slow traversing through the bouquet stage. Our findings uncover a novel mechanism of FBXO47 in telomeric shelterin subunit stabilization during meiosis.
INTRODUCTION
Disorders of meiosis often result in dire consequences, such as infertility (1–7). During meiotic prophase I, associations between homologous chromosomes (homologs) are established to ensure the exchange of genetic information and the correct division of homologs in the subsequent cell division steps. To this end, chromosomes are tethered to the inner nuclear membrane (INM) through telomeres and move along the INM throughout meiotic prophase I (8,9). In mammals, meiotic telomeres connect to the cytoskeleton through the transmembrane linker of the nucleoskeleton and cytoskeleton (LINC) complex, which is composed of SUN-KASH domain proteins. The SUN domain protein SUN1 interacts with telomeres at the INM, whereas the KASH domain proteins interact with cytoplasmic motors at the outer nuclear membrane (ONM) (10–12). During meiotic prophase I, telomere attachment to the nuclear membrane is achieved through the formation of a chimeric complex of TERB1/2-MAJIN and telomere shelterin. By releasing shelterin, the chimeric complex matures into DNA-bound TERB1/2-MAJIN, forming a direct link between telomeric DNA and the INM (13,14).
These transmembrane linkages conduct cytoskeletal forces to telomeres, and this process drives chromosome movement (15,16). The telomeres attach to the INM during the late-preleptotene stage, followed by moving and transiently clustering adjacent to the centrosome, forming a structure termed ‘bouquet’ (17). The telomere bouquet is thought to directly facilitate homologous chromosome pairing, synapsis and homologous recombination by bringing the ends of chromosomes into close proximity and coalignment, and an aberrant bouquet is always related to the failure of meiosis (18–26). Mammalian telomeres are composed of repetitive TTAGGG DNA sequences and are bound by a six-protein shelterin complex consisting of TRF1, TRF2, RAP1, TIN2, TPP1 and POT1 (27). While shelterin components, such as TRF1, are reportedly degraded by ubiquitin-dependent proteolysis (28), the molecular mechanism underlying the dynamic changes in the telomere-bound shelterin complex during meiotic prophase I remains largely elusive.
Ubiquitination by the ubiquitin proteasome system (UPS) is a post-translational modification that governs diverse cellular processes, such as cell proliferation, cell cycle progression, transcription and apoptosis. The UPS exerts its biological functions through a cascade of enzymatic reactions, which are catalyzed by the ubiquitin-activating E1 enzyme, the ubiquitin-conjugating E2 enzyme and the ubiquitin–protein E3 ligase. Crucially, the ubiquitin–protein E3 ligase determines the specific substrate targeted for ubiquitination and subsequent degradation (29,30). We identified a meiosis-specific member of the F-box protein family (31), FBXO47 (F-box only protein 47). F-box proteins consist of at least two major functional domains: an F-box motif and a carboxy-terminal domain. First identified in F-box only 1 (FBXO1) (32), the F-box motif is a protein-protein interaction domain that recruits F-box proteins to the SKP1-cullin1-F-box protein (SCF) E3 ligase complex via direct binding to the adaptor protein SKP1 (33). The carboxy-terminal domain binds to specific substrates.
While mutation of a limited homolog of FBXO47 in Caenorhabditis elegans, prom-1, has been shown to impair homologous synapsis during meiotic prophase I (34), the role of FBXO47 in mammalian meiosis remains to be explored. In this study, we found that FBXO47 interacts with TRF1/2 and prevents ubiquitin-dependent proteolysis of TRF2. Loss of FBXO47 caused the failure of chromosomal synapsis near telomeres, aberrations in telomere-protecting proteins and arrest of telomere movement during the ‘bouquet stage’, highlighting the importance of FBXO47 in the protection of telomere ends.
MATERIALS AND METHODS
Animals
Mice were maintained in a controlled environment at 20–22°C with a 12 h light/dark cycle, 50–70% humidity, and food and water ad libitum. Fbxo47 knockout mice were originally transferred from the Knockout Mouse Project (KOMP) consortium and were bred at the animal center of the Animal Core Facility of Nanjing Medical University. To evaluate the reproductive performance of different males, the mice were individually housed with different females for 9 days, and the males were then paired with different females for an additional 9 days. Females with the presence of copulation plugs were observed for pregnancy and litter size.
Generation of Fbxo47-flag mice by using CRISPR/Cas9
Cas9 mRNA was produced and purified as described previously (35). In brief, the Cas9 plasmid (Addgene No. 44758) was linearized with AgeI, and the digestion product was purified using the MinElute PCR Purification Kit (QIAGEN, 28004). Cas9 mRNA was produced by transcription in vitro using the mMESSAGEmMACHINE® T7 Ultra Kit (Ambion, AM1345) and purified using the RNeasy Mini Kit (QIAGEN, 74104) according to the manufacturer's instructions. The sgRNA was designed in proximity to the Fbxo47 gene stop codon. The target sequence of sgRNA was 5′-ACGCTATCTCTTCCTAAGTCAGG-3′. The two complementary DNA oligos were annealed and ligated to the BsaI-digested pUC57-T7-sgRNA vector. The sgRNA plasmid was linearized with DraI and purified using the MinElute PCR Purification Kit (QIAGEN, 28004). sgRNA was produced using the MEGAshortscript™ Kit (Ambion, AM1354) and purified using the MEGAclear Kit (Ambion, AM1908) according to the manufacturer's instructions. The donor ssDNA sequence is shown in Supplementary Table S1. The donor ssDNA sequence was 5′-CGAACTTCCATAAGGAGGTGCTGTATCTGACCATGAACGCTATCTCTTCCGGATCCGATTACAAGGATGACGACGATAAGTAAGTCAGGAAGCTTGTGTCCCTCTGGACTGGCATTCAGGGGAGTGATGC-3′, included a 56 bp left arm and a 58 bp right arm (including a stop codon) at the position of the stop codon and 1 *flag sequence with 6 linked GGATCC nuclease sites. Mouse zygotes were co-injected with Cas9 mRNA (25 ng/μl), sgRNA (15 ng/μl) and donor ssDNA (1 μM). The injected zygotes were transferred into pseudo-pregnant recipients. The newborn mice (7 days old) were tagged by a toe cut, and DNA was extracted using the Mouse Direct PCR Kit (Biotool, B40013). PCR amplification was carried out with primers (Forward: ATATGTTCCTGAGCCGCATGTTAA, Reverse: ACCAAGACCTGAAGAGCCAGAA) using PrimeSTAR HS DNA Polymerase (Takara, DR010A) under the following conditions: 95°C for 5 min; 10 cycles of 95°C for 30 s, 68°C (–1°C/cycle) for 30 s, and 72°C for 30 s; 25 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s; and a final step of 72°C for 5 min. The PCR products were subjected to Sanger sequencing.
Antibodies
Rabbit antibodies specific for SYCP3 (ab15093), RPA (ab87272), SUN1 (ab103021), H2A (ab18255), H2B (ab1790) and LAMIN B1 (ab16048) and mouse antibodies specific for SYCP3 (ab97672), γH2AX (ab26350) and TRF1 (ab10579) were purchased from Abcam. A rabbit antibody specific for DMC1 (sc-22768) was purchased from Santa Cruz Biotechnology. Rabbit antibodies specific for TRF2 (NB110-57130) and SYCP1 (NB300-229) were purchased from Novus Biologicals (Lihhleton, CO, USA). Mouse antibodies specific for α-tubulin (T9026) and FLAG (F1804) were purchased from Sigma-Aldrich. The rabbit antibody specific for the DDDDK-tag (PM020) that was used for co-immunoprecipitation was purchased from Medical & Biological Laboratories (Nagoya, JP).
The generation of the antibody specific for SHCBP1L has been previously described (36). The anti-FBXO47 antibody recognizes the peptide corresponding to the amino acid sequence DLDLPGTKEETALLE-C of FBXO47.
Real-time quantitative PCR (qPCR) analyses
Total RNA was extracted from mouse tissues using the RNeasy Plus Micro Kit with on-column DNase digestion (Qiagen Ltd., 74034). Samples were homogenized with a heat-sterilized Teflon micropestle in 350 μl of RLT buffer and 4 μl of β-mercaptoethanol. RNA was extracted following the manufacturer's protocol with on-column DNase I treatment. To avoid RNA degradation, all centrifugation steps were carried out at 4°C, and the samples were kept on ice during the entire procedure. The RNA was eluted in 14 μl of RNase-free water. cDNA synthesis was carried out on 1 μg of total RNA, using the PrimeScript RT reagent Kit (TaKaRa Bio Inc., RR037A). The tube was incubated for 15 min at 37°C, followed by heat-inactivation of the reverse transcriptase for 5 s at 85°C. SYBR green-based qPCR was performed using a qPCR machine (StepOnePlus, Applied Biosciences). Each cDNA amplification was performed in triplicate. The results were analyzed using StepOnePlus Real-Time PCR Systems. The primers used for qPCR analyses are listed in Supplementary Table S1, and 18S rRNA was used as an internal control.
Histological analysis
For H&E staining, testes were fixed in modified Davidson's Fluid (30% of a 37–40% stock solution of formaldehyde, 15% ethanol, 5% glacial acetic acid and 50% distilled H2O) and embedded in paraffin. Sections were cut at a thickness of 5 μm. The sections were then deparaffinized, rehydrated, stained with H&E, dehydrated, and mounted.
Ultrastructural examination has been described previously (37). Briefly, 4% (vol/vol) glutaraldehyde-fixed tissues were postfixed with 2% (w/v) OsO4 and embedded in araldite. Ultrathin sections (80 nm) were stained with uranyl acetate and lead citrate and analyzed by EM (JEM. 1010, JEOL).
For focused ion beam scanning electron microscopy (FIB-SEM; Carl Zeiss AG), the tissues embedded in araldite were coated with a thin layer of gold using a Cressington 108Auto system. FIB-SEM was used to collect 3D data. The focused ion beam milled the cross-section, and the scanning electron beam imaged the newly milled cross-section. The XY pixel size was 10 nm, and the Z interval was 20 nm.
Western blot analysis
Western blotting was performed as described previously with a modification (38). Briefly, protein extracts were prepared using lysis buffer (7 M urea, 2 M thiourea, 2% (w/v) DTT) in the presence of 1% (v/w) protease inhibitor mixture (Pierce Biotechnology). The proteins were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk in a TBST solution for 2 h at room temperature and incubated overnight at 4°C with primary antibodies. The membranes were washed with TBST buffer three times and incubated at room temperature for 2 h with secondary antibodies. The signals of the detected proteins were visualized with the SuperSignal West Femto Chemiluminescent Substrate Western Blotting detection system (Thermo Scientific).
Immunofluorescence staining of paraffin-embedded sections, spermatocytes and nuclear spread
For paraffin-embedded sections, the samples were deparaffinized and rehydrated. After three 10 min washes with PBS, heat-induced antigen retrieval was carried out by boiling the slides in 10 mM citrate buffer (pH 6.0) with a microwave oven for 10 min. After three 10 min washes with PBST (0.1% Tween-20 in PBS), the slides were incubated with 5% BSA diluted in PBST for 1 h and then with the indicated antibodies overnight at 4°C.
Immunostaining of spermatocytes was performed based on a previous study with modifications (39). Briefly, testes were digested with collagenase IV at 37°C for 10 min and a trypsin-EDTA solution at 37°C for 15 min and then washed briefly in PBS. After washed several times with PBS, The cell pellets were resuspended in hypotonic buffer (30 mM Tris, pH 7.5, 17 mM Tris sodium citrate, 5 mM EDTA and 50 mM sucrose) for 5 min at room temperature and then fixed by adding the same volume of fixation buffer with detergent (1% PFA, with 0.1% Triton X-100). The fixed cell suspensions were placed on slides and air-dried.
For nuclear spread analysis, testicular tubules were extracted in a hypotonic treatment buffer (30 mM Tris–Cl pH 8.5, 50 mM sucrose, 17 mM sodium citrate, 5 mM EDTA, 50 mM DTT, 10 mM PMSF) for 30 min at roomtemperature. Cells were resuspended in 0.1 M sucrose and were then spread on a thin layer of paraformaldehyde solution containing Triton X-100 for 3 h. After fixation, the slides were air-dried at room temperature.
For immunofluorescence, the samples were washed three times with PBST (0.1% Tween-20 in PBS) for 10 min each. The slides were incubated with 1% BSA diluted in PBST for 1 h and then with the indicated antibodies overnight at 4°C. After incubation with the secondary antibody at room temperature for 2 h, the slides were incubated with Hoechst 33342 for 5 min. Finally, the slides were washed in PBS and then mounted with glycerol. The slides were viewed with an LSM700 confocal microscope (Carl Zeiss AG) driven by ZEN2013 software (Carl Zeiss AG).
TUNEL assays
TUNEL assays were performed with the TUNEL BrightGreen Apoptosis Detection Kit (Vazyme, A112-03) and the In Situ Cell Death Detection Kit (Roche Diagnostics, 11684795910), as described previously (40).
Germ cell purification
Germ cells purification was performed following a published protocol (41) with modifications. Briefly, testes were collected from male mice at postnatal day 10 and 14. Testes tissue was digested with 1 mg/ml collagenase type IV (Invitrogen, 17104-019) for 15 min, followed by 0.25% trypsin/1 mM EDTA digestion (Gibco, 25200-072) for 10 min. Dispersed cells were cultured in cell culture dishes for 24 h. During this time, Sertoli cells attach to the bottom of the dish (42), such that cells in suspension cells were enriched for germ cells (41). Cells in suspension were collected, and washed by DPBS (Gibco, 14190-144) for the follow-up tests.
Transmission electron microscopy
The adult mouse testes were dissected and fixed with 2.5% (v/v) glutaraldehyde in 0.2 M cacodylate buffer (50 mM cacodylate, 50 mM KCl, and 2.5 mM MgCl2, pH 7.2) overnight. After washing in cacodylate buffer, the tissues were cut into small pieces of approximately 1 mm3 and immersed in 1% OsO4 in 0.2 M cacodylate buffer for 2 h at 4°C. Next, the samples were washed and submerged in 0.5% uranyl acetate overnight, dehydrated through a graded ethanol series, and embedded in resin (Low Viscosity Embedding Media Spurr's Kit, EMS, 14300). Ultrathin sections were cut on an ultramicrotome and mounted on copper grids. The sections then were stained with uranyl acetate and lead citrate for 10 min and observed using a JEM-1400 transmission electron microscope (JEOL).
Yeast two-hybrid assays
Yeast two-hybrid assays were performed as described previously (43). Mouse Fbxo47 and Sun1 C (residues 458–913) were subcloned into the pGBT9 plasmid as the bait. Mouse Skp1 and Kash LR (residues 627–648) were subcloned into the pGAD424 plasmid as the prey. The bait and prey plasmids were co-transformed into PJ69-4a and selected on an SD-Leu-Trp-His plate.
Co-immunoprecipitation
Co-immunoprecipitation was performed on Fbxo47-flag mouse testes with anti-FLAG antibodies. Briefly, testes (200 mg) were homogenized in 2 ml of lysis buffer (25 mM Tris, pH 7.4, 500 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol) with 1× protease inhibitor cocktail (Selleckchem, B14002). The lysate was incubated on ice for 40 min, followed by centrifugation at 12 000 g for 20 min at 4°C. The supernatant was transferred to a new tube and used for immunoprecipitation followed by western blotting.
pCS2-Myc-Fbxo47 and pRK-Flag-Trf1 or pRK-Flag-Trf2 were co-transfected into HEK293T cells. Transfected cells were lysed in TAP lysis buffer (50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% NP-40 10 mM NaF, 0.25 mM Na3VO4 and 50 mM β-glycerolphosphate) plus protease inhibitors (Roche, 04693132001) for 30 min on ice and centrifuged at 13 000 g for 15 min. For immunoprecipitation, cell lysates were incubated with anti-FLAG or anti-MYC antibodies overnight at 4°C and then incubated with protein A-Sepharose (GE, 17-1279-03) for 2 h at 4°C. Thereafter, the precipitants were washed two times with IP buffer (20 mM Tris, pH 7.4, 2 mM EGTA and 1% NP-40), and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 95°C and analyzed by immunoblotting.
Ubiquitination assay
pCS2-Myc-Fbxo47, pRK-Flag-Trf1 or pRK-Flag-Trf2 and pCMV-HA-Ubiquitin were co-transfected into HEK293T cells. The transfected cells were lysed in TAP lysis buffer plus protease inhibitors for 30 min on ice and centrifuged at 13 000 g for 15 min. The cell lysates were incubated with an anti-FLAG antibody overnight at 4°C and then incubated with protein A-Sepharose (GE, 17-1279-03) for 2 h at 4°C. Then, the precipitants were washed 4 times with TAP lysis buffer, and the immune complexes were analyzed by immunoblotting. For in vitro ubiquitination assays, pCS2-Myc-Fbxo47 was transfected into HEK293T cells and immunoprecipitated using an anti-MYC antibody. E1, UbcH5c, ad GST-TRF1 or FLAG-TRF2, and FLAG-Ub (10 nM) were incubated with immune complexes of FBXO47 at 37°C in buffer containing 25 mM Tris–HCl (pH 7.4), 2 mM magnesium/ATP, and 0.1 mM DTT for 1 h. Ubiquitination of TRF1 or TRF2 was detected by immunoblotting.
Cycloheximide chase assay
Cycloheximide experiments were performed as described below. HEK293T cells transfected with pCS2-Myc-Fbxo47, pRK-Flag-Trf1 or pRK-Flag-Trf2 were treated with CHX (10 mg/ml) for 1 to 12 h to inhibit de novo protein synthesis; subsequently, the degradation of TRF1 or TRF2 was examined using immunoblotting.
Quantitative co-localization analysis
The quantitative co-localization analysis was performed by means of the ImageJ software, in particular by using the Just Another Co-localization Plugin (JACoP) plugin (44).
Statistical analysis
Numerical data are presented as mean ± SEM. The statistical significance of the difference between the mean values for the different genotypes was examined using Student's t-test with a paired two-tailed distribution. The data were considered significant when P < 0.05 (*), 0.01(**), 0.001(***) or 0.0001(****).
RESULTS
FBXO47 is predominantly expressed in male germlines with specific localization in the spermatocytes
FBXO47 belongs to the F-box family of proteins, of which most members are components of Skp1-Cul1-F-box protein-type (SCF-type) ubiquitin ligases (45,46). Fbxo47 is an evolutionarily conserved gene (Supplementary Figure S1), and the conservation of this F-box motif among species suggests it might have a conserved function.
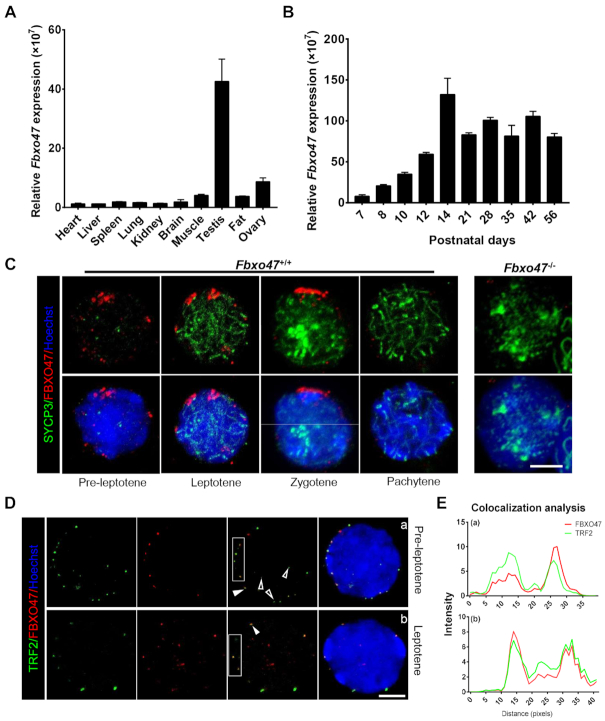
The expression pattern of Fbxo47 transcripts in mouse tissues was analyzed by real-time q-PCR. Fbxo47 transcripts were found to be abundantly expressed in testis (Figure 1A). In the mouse testis, the first round of spermatogenesis is fairly synchronous. To determine the temporal expression of Fbxo47, we performed q-PCR from testis tissues collected at different postnatal days (P) to capture the leading edge of the first wave of spermatogenesis. The analysis showed that transcripts of Fbxo47 were first detected at approximately P8, with a continual increase occurring from P10 to P14; which is the developmental stage corresponding to the emergence of meiotic spermatocytes in mouse testes (Figure 1B) (17,47–50).
Figure 1.
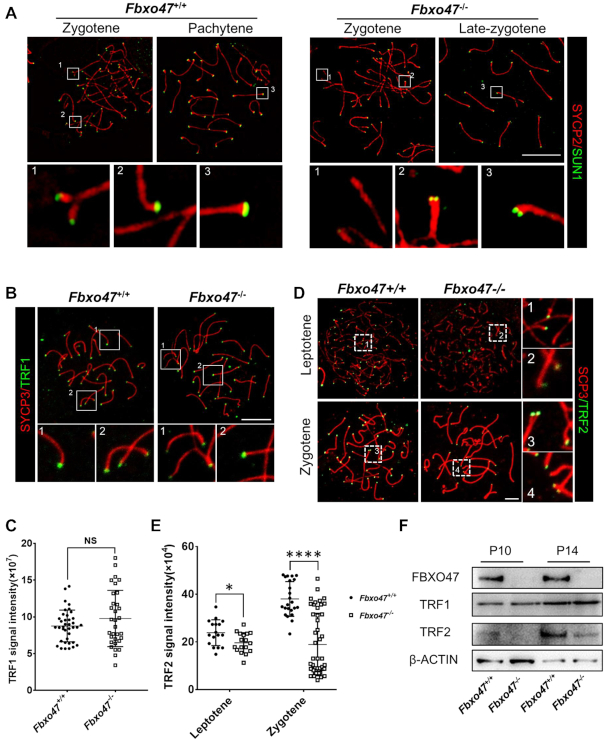
Distribution pattern of FBXO47 in the testis. (A) Real-time q-PCR for Fbxo47 in various mouse tissues with 18S rRNA as a control. Error bars, SEM (n = 3). (B) Real-time q-PCR of testis samples at various postnatal days with 18S rRNA as a control. Error bars, SEM (n = 3). (C) Orthogonal z-stack projections of spermatocytes stained with the indicated antibodies. Testis cells in suspension were prepared with a mild hypotonic treatment and then fixed in Triton X-100 (39). Bars, 5 μm. (D) Nuclear colocalization of TRF2 and FBXO47 (nuclear equator). TRF2 co-localizes with FBXO47 at the INM. Arrowheads mark TRF2 located at the nuclear periphery. Hollow arrowheads mark TRF2 at the nuclear internal domain. Asterisks indicate FBXO47 at the nuclear internal domain. Bars, 5 μm. (E) Quantitative co-localization analysis of TRF2 and FBXO47 of the cell shown in panel (D).
We generated an antibody specific for mouse FBXO47. The expression of FBXO47 in mouse spermatocytes was verified by immunofluorescence (IF) with hypotonic treatment and Triton X-100 permeabilization (39). FBXO47 foci were distributed on the nuclear surface during the early stage of meiotic prophase I (Figure 1C, D and Supplementary Figure S2). Using the expression pattern of SYCP3 to define meiotic stages, we found FBXO47 is expressed at the onset of meiosis (Figure 1C). During this stage, meiotic telomeres have been attached to the INM and the chromosome ‘bouquet’ formed. We next evaluated the potential interaction of FBXO47 with other meiosis-related proteins and found that FBXO47 co-localized with TRF2 (Figure 1D and E). More importantly, FBXO47 only accumulated at TRF2 protein when telomeres relocate to the nuclear periphery, but not at TRF2 located in the internal domain of the nucleus at preleptotene (Figure 1D and E). FBXO47 co-localized with of TRF2 at the nuclear periphery at the preleptotene and leptotene (Figure 1D). During these stages, only 25% FBXO47 foci were localized in the internal domain of nucleus and not co-localized with TRF2 (Figure 1D). The function of FBXO47 in non-telomeric regions remains unclear, but we speculate that the internal localization may reflect stages of FBXO47 transport prior to binding to TRF2. In summary, we observe FBXO47 is present in male germ cells at the early stage of meiotic prophase I and co-localizes with TRF2 at the nuclear periphery (Figure 1D). We therefore conclude that FBXO47 is a telomere-associating protein and hypothesize that FBXO47 may be involved in telomere attachment and bouquet formation.
Fbxo47 is indispensable for spermatogenesis in mice
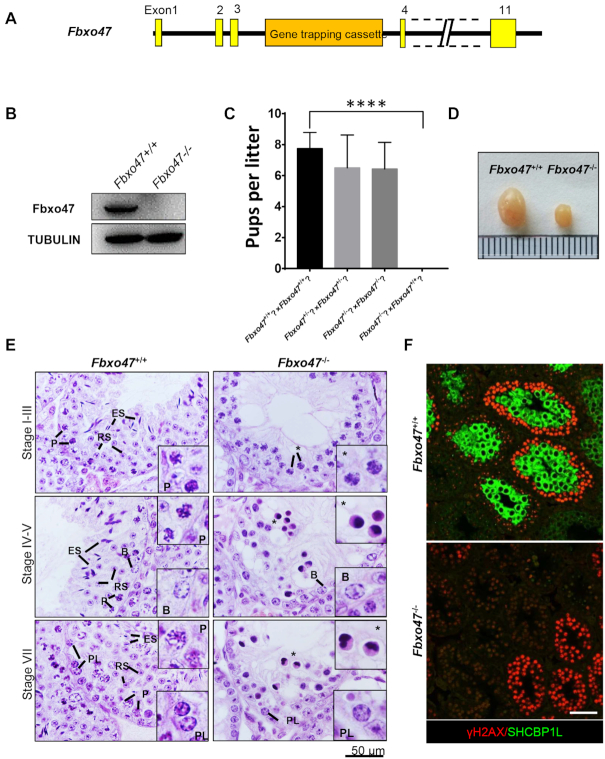
To determine the function of Fbxo47, we obtained mice that were heterozygous for a knockout mutation of the Fbxo47 allele (Fbxo47tm1a(EUCOMM)Wtsi; herein referred to as Fbxo47−/−) from the Wellcome Trust Sanger Institute (Figure 2A). The Fbxo47−/− mice had been generated as part of the Knockout Mouse Project (KOMP) (51). We intercrossed Fbxo47+/− males with Fbxo47+/− females to obtain homozygous males (Fbxo47−/−) and their control littermates (Fbxo47+/+). Western blot analysis confirmed the absence of the FBXO47 protein in Fbxo47−/− testes (Figure 2B). Homozygous Fbxo47−/− males showed no overt abnormality in development or behavior. To test their fertility, Fbxo47−/− and Fbxo47+/+ males were mated to wild-type females for 2 months. The number of offspring per litter was recorded. Eight Fbxo47+/+ males had 8 ± 1.03 pups per litter, whereas Fbxo47−/− males failed to sire any offspring despite copulation plugs being observed. However, homozygous females (Fbxo47−/−) were fertile (Figure 2C). These results demonstrated that Fbxo47 is required for male fertility in mice.
Figure 2.
Fbxo47 null males display azoospermia and arrest of meiosis. (A) Schematic of the Fbxo47tm1a(KOMP)Wtsi (Fbxo47−) allele from the Wellcome Trust Sanger Institute. (B) Western blot analysis of Fbxo47+/+ and Fbxo47−/− testes with a mouse polyclonal antibody raised against FBXO47. (C) Breeding results from Fbxo47 homozygous (Fbxo47−/−) and heterozygous mice (Fbxo47+/−). Error bars, SEM (n = 8); ****P ≤ 0.0001. (D) Testes from 8-week-old mice with the indicated genotypes. (E) Hematoxylin and eosin-stained histological cross-sections of testes from adults. B, type B spermatogonia; PL, preleptotene; P, pachytene; RS, round spermatids; ES, elongated spermatids; Asterisks, loss of germ cells. Bar, 50 μm. (F) Immunofluorescence staining of γH2AX and SHCBP1L in Fbxo47+/+ and Fbxo47−/− histological cross-sections. Scale bar, 50 μm.
Causes of male infertility range from defects in spermatogenesis to impairments in fertilization once the spermatozoa reach the egg (52). To investigate the functional role of FBXO47 in spermatogenesis, we carried out histological analyses on testes from Fbxo47−/− mice. The testes of adult Fbxo47−/− mice were much smaller in size and weight than those of their Fbxo47+/+ littermates (Figure 2D). Only abnormal spermatocyte-like cells were observed in some seminiferous tubules, and round spermatids and spermatozoa were completely absent in Fbxo47−/− testes, suggesting that Fbxo47 is essential for meiotic progression (Figure 2E) (31). TUNEL assays revealed that extensive apoptotic events in the Fbxo47−/− seminiferous tubules may account for the elimination of round spermatids and spermatozoa in the Fbxo47−/− testis (Supplementary Figure S3). The expression of a mid-pachytene marker, SHCBP1L (36), in Fbxo47+/+ and Fbxo47−/- testes was then evaluated by immunofluorescence. Although the seminiferous tubules in Fbxo47−/− testes in stage IV contained dying spermatocytes (Figure 2E), SHCBP1L could not be detected in the remaining germ cells of Fbxo47−/− testes (Figure 2F), confirming that the Fbxo47−/− spermatocytes died before the mid-pachytene stage. To investigate the starting point for apoptosis in Fbxo47−/− spermatocytes, we performed TUNEL assays on testes from Fbxo47+/+ and Fbxo47−/− mice at P14. Yet no differences in apoptosis were observed at P14 (Supplementary Figure S4). We therefore conclude that FBXO47 plays a crucial role in meiosis and that deletion of this protein causes spermatocyte apoptosis before the mid-pachytene stage.
Impaired DNA double-strand break repair in Fbxo47 knockout mice
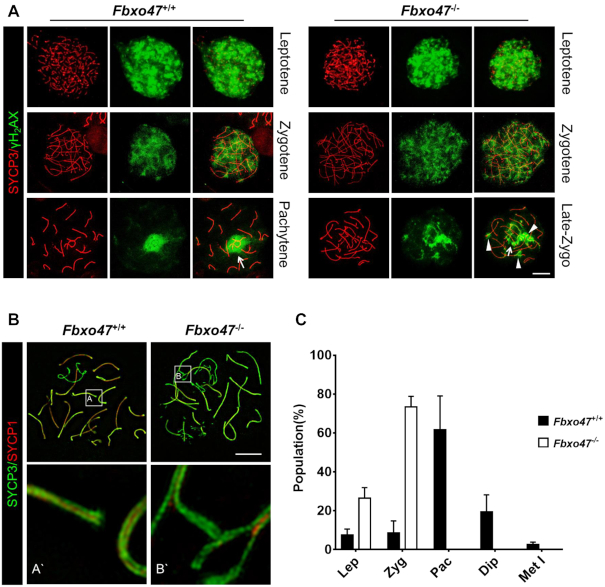
To monitor meiotic recombination in Fbxo47−/− spermatocytes, we evaluated the formation of DNA double-strand breaks (DSBs). During meiosis, following chromatin changes at recombination hotspots mediated by PRDM9, the SPO11 protein catalyzes the formation of DSBs, eliciting the DNA damage response (53–57) and leading to activation of the ATM kinase and subsequent phosphorylation of H2AX (termed γH2AX). During the leptotene and zygotene stages, γH2AX is present on autosomal chromatin and distributes widely throughout the nucleus (Figure 3A). The presence of strong γH2AX signals suggested that DSBs were formed in Fbxo47−/− leptotene and zygotene spermatocytes (Figure 3A). In normal spermatocytes, γH2AX disappears from the autosomes after meiotic DSB repair and becomes restricted to the XY chromatin during the pachytene and diplotene stages, concomitant with meiotic sex chromatin inactivation (Figure 3A). However, late-zygotene arrest Fbxo47−/− spermatocytes remained γH2AX-positive in some sections of the autosomes, especially the terminus (Figure 3A), suggesting that DNA double-strand repair was compromised in Fbxo47−/− spermatocytes.
Figure 3.
Uncompleted double-strand repair and synapsis. (A) Spread spermatocytes from Fbxo47+/+ and Fbxo47−/− males were immunostained with anti-SYCP3 and anti-γ-H2AX antibodies. Arrows indicate the sex body; Arrowheads indicate impaired double-strand repair. Scale bars, 10 μm. (B) Spread spermatocytes from Fbxo47+/+ and Fbxo47−/− males were immunostained for synaptonemal complex proteins (SYCP3 and SYCP1). Scale bars, 10 μm. (C) Frequencies of meiotic stages in testes from 8-week-old mice with the indicated genotypes. SYCP3-positive spermatocytes (889 cells were counted) are classified into the following substages: Lep, leptotene (no SYCP1); Zyg, zygotene (partially assembled SYCP1); Pac, pachytene (fully assembled SYCP1); Dip, diplotene (disassembled SYCP1); Met I, metaphase I (no SYCP1 and SYCP3 accumulation at centromeres). Mean values of three independent experiments from three different mice are shown. Error bars, SEM (n = 3).
Meiotic recombination is executed by the coordinated actions of a large number of DNA repair proteins (58,59). We examined the single-stranded DNA-binding proteins RPA1 and DMC1 (Supplementary Figure S5). These DNA repair proteins form distinct foci on meiotic chromosomes. The RPA heterotrimer consists of RPA1, RPA2 and RPA3, which bind to the ssDNA ends of the meiotic DSBs (60). DMC1 is a recombinase that recruits filaments on RPA-coated ssDNA and directs strand invasion into homologous chromosomes, which is required for crossover formation, homologue pairing, and chromosomal synapsis (61). The number of RPA and DMC1 foci was similar between Fbxo47+/+ and Fbxo47−/− at the zygotene stage (Supplementary Figure S5B, D). These data suggested that the initialization of meiotic recombination was not influenced by FBXO47 depletion.
Disruption of FBXO47 causes spermatogenesis arrest at the late-zygotene stage
We assessed the impact of FBXO47 disruption on chromosomal synapsis by immunofluorescence of spermatocyte spreads using antibodies derived against SYCP1, a component of transverse elements in the synaptonemal complex (SC) (62,63). SC axial elements are formed during the leptotene stage, initiate synapsis through physical juxtaposition during the zygotene stage, achieve full synapsis on autosomes during the pachytene stage, and are subsequently separated during the diplotene stage (58,64). Fbxo47+/+ pachytene spermatocytes contained fully synapsed autosomes, whereas the most advanced spermatocytes from Fbxo47−/− males were at a late-zygotene stage, characterized by apparent chromosome pairing and formation of SC axial elements (SYCP3) without complete chromosomal synapsis (Figure 3B and 3C). We observed the synapsis complex of the Fbxo47−/− late-zygotene arrest cells by super-resolution microscopy (SIM). The unaccomplished synapsis was always found near the telomeres (Figure 3B), which is a mark of late-zygotene arrest.
Loss of FBXO47 leads to arrest of chromatin remodelling in spermatocytes
Telomeres are central players in meiotic prophase I, and although the underlying mechanisms remain poorly understood, the initial steps of meiotic prophase I are strongly linked to telomeres (9,65). At the onset of meiosis, telomeres are presumed to be lengthened in two ways, alternative lengthening of telomeres by telomerase (ALT) (66–68) and mechanical fortification by the addition of cohesins (69). Successful lengthening prepares the telomeres for anchoring to the nuclear envelope (NE), and anchored telomeres are then pulled along the NE to form ‘bouquet’ clusters.
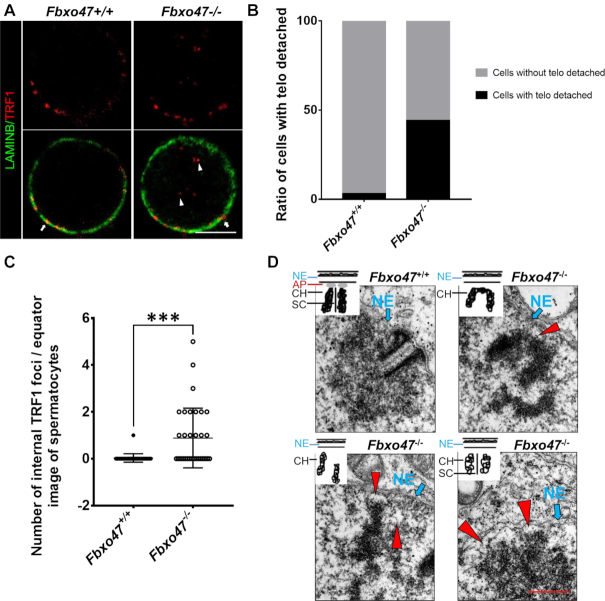
To investigate the function of telomeres, we performed TRF1/LAMIN B staining on the testis suspension. TRFI foci were observed at the edge of the nucleus in both Fbxo47+/+ and Fbxo47−/- spermatocytes. Although the density of TRF1 immunofluorescence was not decreased in Fbxo47−/− spermatocytes compared with controls, an increased number of spermatocytes with clustered telomeres indicated a possible temporal extension of the ‘bouquet stage’ in Fbxo47−/− cells (Supplementary Figure S6A and B). Interestingly, a subset of TRF1 foci remained in the nuclear internal domain at the bouquet stage, although attachment to the nuclear membrane should have been completed at the preleptotene stage (Supplementary Figure S6A). We speculate that the telomere attachment plate may be defective in Fbxo47−/− spermatocytes. To quantify the localization of telomeres to the nuclear periphery, we counted the number of TRF1 foci located in the nuclear internal domain of spermatocytes (Figure 4C). TRF1 foci were largely absent from the internal compartment of the nuclei of Fbxo47+/+ spermatocytes, indicating that most telomeres were anchored at the INM. In contrast, Fbxo47−/− contained a significantly larger number of TRF1 foci in the internal nuclear compartment, indicating that the telomeres of Fbxo47−/− spermatocytes had not attached to the INM (Figure 4A−C). To explore the telomere attachment status, we examined telomere attachment by investigating the ultrastructures of meiotic telomeres using electron microscopy (EM). While conical electron-dense thickenings of synapsed telomeres and capping structures attached to the INM were observed in Fbxo47+/+ spermatocytes, an increased number of telomeres were detached in Fbxo47−/− spermatocytes (Figure 4D). Our results suggest that FBXO47 may regulate telomere attachment.
Figure 4.
FBXO47 is indispensable for telomere attachment. (A) Equator images of spermatocytes at zygotene, stained with the indicated antibodies. Arrowheads indicate internal telomeres and arrows indicate telomeres at the nuclear periphery. Scale bars, 10 μm. (B) Graphs showing the ratio of spermatocytes with telomeres detached from the INM. (C) Graphs showing the number of internal TRF1 foci per image (n > 30 cells for each genotype). Error bars, SEM, ***P < 0.001. (D) Electron microscopic images of telomeres (arrowheads) and INMs (arrows) in spermatocytes of the indicated genotypes, with schematics representing the structures. NE, nuclear envelope; AP, attachment plate; CH, chromatin; SC, synaptonemal complex. Scale bars, 200 nm.
Thus, we proposed that there must be developmental defects in bouquet stage spermatocytes due to FBXO47 depletion. We then investigated the 3D structure of chromatin in ‘bouquet stage’ spermatocytes with a Carl Zeiss CrossBeam 550 scanning electron microscope (SEM) equipped with a focused ion beam (FIB). During the bouquet stage, a dense structure was formed along the INM with a jagged interspace in Fbxo47+/+ spermatocytes (Supplementary Figure S7). Additionally, meiotic telomeres attached to the INM through the dense structure (Supplementary Figure S8). In contrast, in Fbxo47−/− spermatocytes, the dense structures were incomplete and remained close to the NE without an obvious interspace. The function of this dense structure remains to be studied. Our data suggest that deficiency of FBXO47 impairs the structure of the bouquet.
Disruption of FBXO47 causes aberrations in telomere-protecting proteins
In recent years, mounting evidence from diverse mammalian species has indicated that the progression of meiotic prophase I depends on the telomere-led rapid movement of chromosomes along the NE (20,70,71). A prerequisite for this rapid chromosome movement is the attachment of telomeres to the NE, where the transmembrane linker of the nucleoskeleton and cytoskeleton (LINC) complex, SUN1-KASH5 (72), serves as a structural bridge to connect telomeres to the cytoskeleton and conducts forces generated in the cytoplasm to the ends of the chromosomes (71,73). The SUN1 was detected by immunofluorescence. Although SUN1 accumulation at telomeres was observed in Fbxo47−/− spermatocytes, the intensity of signal was much weaker in Fbxo47−/− cells. Magnified views of telomeres revealed that in Fbxo47+/+ spermatocytes, SUN1 foci on homologous chromosomes seemed to merge with each other after telomere synapsis. However, in Fbxo47−/− late-zygotene cells, the SUN1 foci remained separated (Figure 5A).
Figure 5.
Telomere-binding protein aberrations in FBXO47-deficient spermatocytes. (A) Spread spermatocytes from Fbxo47+/+ and Fbxo47−/− males were stained with anti-SYCP3 and anti-SUN1 antibodies. Scale bars, 10 μm. (B) Spread spermatocytes from Fbxo47+/+ and Fbxo47−/− males were stained with anti-SYCP3 and anti-TRF1 antibodies. Scale bars, 10 μm. (C) Graphs showing the TRF1 intensities per cells (n > 30 cells for each genotype). (D) Spread spermatocytes from Fbxo47+/+ and Fbxo47−/− males were stained with anti-SYCP3 and anti-TRF2 antibodies. Scale bars, 10 μm. (E) Graphs showing the TRF2 intensities per cells (n > 30 cells for each genotype). TRF1 signal intensity: Fbxo47+/+, 8.769 ± 0.351; Fbxo47−/−, 9.786 ± 0.6768; TRF2 signal intensity, leptotene: Fbxo47+/+, 23.873 ± 5.414; Fbxo47−/−, 19.660 ± 4.207; Zygotene: Fbxo47+/+, 38.046 ± 7.281; Fbxo47−/−, 18.953 ± 13.110. (F) FBXO47 is expressed in murine germ cells at P10 and P14. TRF2 is destabilized in Fbxo47-deficient germ cells. β-ACTIN served as a loading control.
In addition to SUN1, several telomere-protecting proteins also play important roles in mediating telomere movement, such as the shelterin complex (74–81). We examined TRF1 and TRF2, which are core subunits of the shelterin complex involved in anchoring telomeres to the NE (69). Through the immunofluorescence, we found TRF2 protein expression was remarkably reduced in Fbxo47−/- spermatocytes, while TRF1 protein expression was not notably influenced (Figure 5B–E). We next determined the levels of TRF1 and TRF2 in Fbxo47-deficient germ cells. As the onset of spermatogenesis is fairly synchronous in the mice testis, we selected testes from mice at P10 and P14, during which time the most advanced germ cells have passed preleptotene and zygotene respectively (17,47–50). We performed Western blot analysis of cell extracts from purified germ cells. The expression level of the TRF1 protein was not dramatically affected. However, TRF2 was significantly down regulated in Fbxo47 deficient germ cells, which might be related to the loss of FBXO47 (Figure 5F). Our results suggest that FBXO47 may regulate telomere attachment through SUN1 and TRF2.
FBXO47 interacts with TRF1 and TRF2
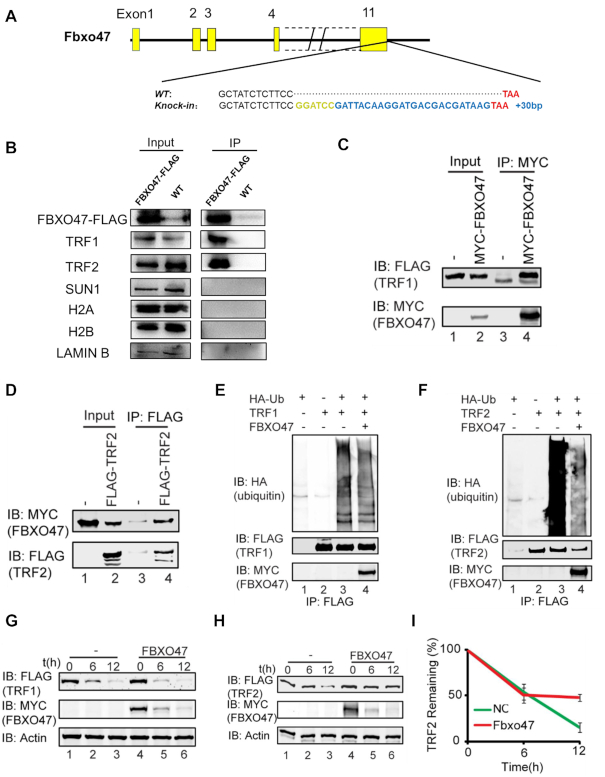
To further investigate the molecular function of FBXO47, we employed CRISPR/Cas9 to create a FBXO47 C-terminal FLAG-tagged knock-in mouse line (Figure 6A). To confirm successful expression of the FBXO47-FLAG fusion protein in vivo, we performed immunoprecipitation (IP) with a commercial anti-FLAG antibody. To avoid non-specific antibody binding, we used a FLAG antibody produced by a different company for Western blot analysis. A band of ∼55 kDa, which was consistent with the theoretical molecular weight of the FBXO47-FLAG fusion protein, was detected (Figure 6B). To determine whether FBXO47 regulates the expression of TRF1/TRF2 and SUN1 directly, we performed co-immunoprecipitation (co-IP) with testis lysates from these Fbxo47-flag knock-in mice using an anti-FLAG antibody. Results of co-IP analysis indicated that FBXO47 bound TRF1 and TRF2 directly but did not bind SUN1 (Figure 6B).
Figure 6.
Disruption of Fbxo47 destabilizes TRF2. (A) Schematic of FLAG-tagged alleles of endogenous Fbxo47 generated using CRISPR/Cas9. A 2-aa (Gly-Ser) spacer was added. (B) Co-IP analysis of FBXO47-FLAG and indicated proteins from Fbxo47-flag testicular protein extracts. (C and D) FBXO47 interacts with TRF1 and TRF2. pCS2-Myc-Fbxo47 and pRK-Flag-Trf1 or pRK-Flag-Trf2 were co-transfected into HEK293T cells. After 24 h, the cells were collected for immunoprecipitation (IP) with an anti-MYC (C) or anti-FLAG (D) antibody and analyzed with an anti-MYC and anti-FLAG antibodies. (E) The over-expression of FBXO47 has no influence on TRF1 ubiquitination. pCS2-Myc-Fbxo47, pRK-Flag-Trf1 and pCMV-HA-Ubiquitin were co-transfected into HEK293T cells. After 24 h, the cells were collected for immunoprecipitation (IP) with an anti-FLAG antibody and analyzed with anti-HA, anti-FLAG and anti-MYC antibodies. (F) FBXO47 impairs the ubiquitination of TRF2. pCS2-Myc-Fbxo47, pRK-Flag-Trf2 and pCMV-HA-Ubiquitin were co-transfected into HEK293T cells. After 24 h, cells were collected for immunoprecipitation (IP) with an anti-FLAG antibody and analyzed with anti-HA, anti-FLAG and anti-MYC antibodies. (G and H) FBXO47 stabilizes TRF2, rather than TRF1. Cycloheximide chase (CHX) assays of TRF1 (G) and TRF2 (H) were performed in FBXO47-over-expressing or empty vector-transfected samples. (I) Quantification of the relative TRF2 levels in (I).
Disruption of Fbxo47 destabilizes TRF2
As FBXO47 belongs to the F-box protein family, whose members are key recognition subunits of Skp1-Cullin1-F-Box protein (SCF) E3 ubiquitin ligase complexes (82), we speculated that FBXO47 might regulate telomere-IMN integration during meiosis by recognizing and promoting specific substrates for proteasome degradation. To test this hypothesis, we first evaluated the interaction between FBXO47 and SKP1, which links F-box proteins and other subunits of SCF complexes. As shown in Supplementary Figure S9A–C, FBXO47 indeed interacted with SKP1, indicating that FBXO47 likely cooperates with other components to form SCF complexes. Next, we examined the E3 ubiquitin ligase activity of FBXO47 immunoprecipitants and found that FBXO47 functioned as a component of SCF complexes to catalyze the formation of ubiquitin chains (Supplementary Figure S9D). Therefore, FBXO47 should be a subunit of the SCF complex, which contains an E3 ubiquitin ligase.
Since FBXO47 can directly bind TRF1 and TRF2, these two proteins might be substrates of SCFFBXO47. We carried out co-IP on HEK293T cells with simultaneous over-expression of pCS2-Myc-Fbxo47 and pRK-Flag-Trf1 or pRK-Flag-Trf2, and we found FBXO47 indeed interacted with TRF1 and TRF2 (Figure 6C and D). Further investigation showed that SCFFBXO47 could not add ubiquitin chains to TRF1 either in vitro or in vivo (Figure 6E and Supplementary Figure S9E), and the over-expression of FBXO47 had no influence on the degradation of the TRF1 protein (Figure 6G), indicating that TRF1 might not be a substrate of SCFFBXO47 during meiosis. However, over-expression of FBXO47 dramatically impaired the ubiquitination of TRF2 (Figure 6F), and SCFFBXO47 could not assemble ubiquitin chains on TRF2 in vitro (Supplementary Figure S9F). Furthermore, CHX assays of TRF2 showed that the over-expression of FBXO47 could stabilize TRF2 (Figure 6H and I). This result was consistent to the deficiency of TRF2 in the Fbxo47−/− germ cells (Figure 5D, E and F). Thus, the disruption of Fbxo47 destabilizes TRF2 during meiosis. Although FBXO47 may not function as a recognition subunit of the SCF complex, it may function as a ubiquitination inhibitor by binding to TRF2 to prevent its degradation (Figure 7).
Figure 7.
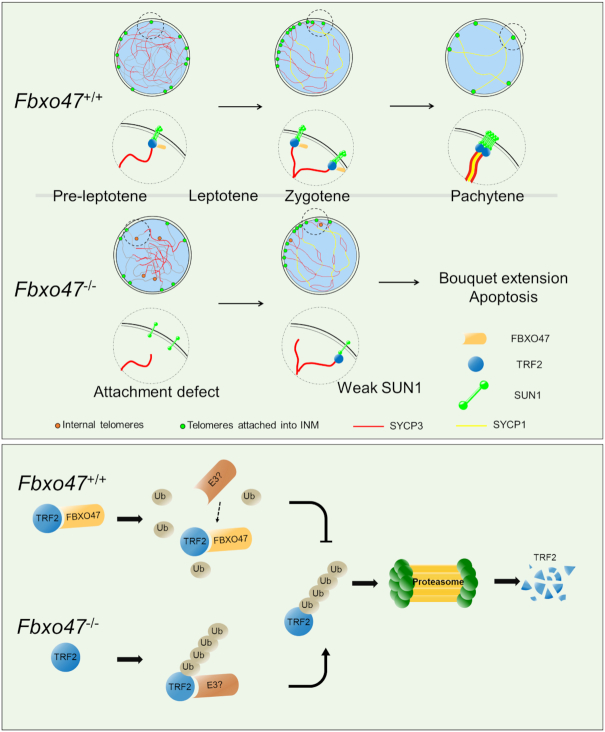
Schematic of meiosis-specific telomere shelterin protection in mammals. FBXO47 prevents TRF2 degradation as a ubiquitination inhibitor during meiosis and maintains telomere shelterin integrity for initial telomere attachment. The absence of FBXO47 destabilizes TRF2 during meiosis and disrupts telomere shelterin protein localization and attachment to the inner nuclear membrane.
DISCUSSION
Meiosis is an evolutionarily conserved process that is crucial to gamete formation for zoogamy. During meiosis, rapid chromosome movement along the nuclear envelope and clustering of chromosome ends during the bouquet stage are essential for the pairing and recombination of homologous chromosomes. Over the past decade, a number of telomere-associated proteins have been reported (13,72,81,83,84). However, the molecular architectures that drive dynamic changes in meiotic telomeres have not been elucidated. In this study, we identified a novel testis-enriched protein named FBXO47. FBXO47 is localized at the telomeres attached to the INM and clustered on the spermatocyte nuclear envelope during the bouquet stage, and male Fbxo47 knockout mice showed an arrest of meiosis before the early pachytene stage with failure of double-strand repair and synapsis complex formation (Figure 2 and 3). Fbox47−/− testes contain abundant bouquet stage spermatocytes and a subset of the telomeres are detached from the NE (Figure 4). Taken together, our results indicate that FBXO47 is a specific regulator that is essential for telomere function during the bouquet stage.
Telomere attachment is regulated by an extensive protein network. Sun1−/− mice were the first genetic model to provide functional insights into telomere-NE attachment during meiosis in mammals. Deficiency of SUN1 caused a failure of telomere attachment, recombination and synapsis (72). TRF1 and TRF2 are core components of the shelterin complex. TRF1/TRF2 were found to be essential for telomeres attachment during meiosis (8,85,86). Here, we demonstrate that FBXO47 is required for the telomere localization of SUN1 and TRF2 (Figure 5), which may be the cause of the defect of telomere attachment (Figure 4).
FBXO47 belongs to the F-box protein family, whose members are the key recognition subunits of Skp1-Cullin1-F-Box protein (SCF) E3 ubiquitin ligase complexes (82). Our data show that FBXO47 could work as a component of SCF to catalyze ubiquitin chain formation (Supplementary Figure S9). However, FBXO47 directly interacts with TRF2 to protect it from ubiquitination, thus stabilizing the shelterin complex (Figure 6). TRF1 connects to TRF2 through TIN2 (78,87), and depletion of TRF1 affects the telomeric association of TRF2 (88). In our study, FBXO47 interacted directly with TRF1 but did not affect the ubiquitination of TRF1. Furthermore, the immunoblotting results showed that the level of TRF1 was not dramatically affected in Fbxo47-deficient testis. In addition, TRF2 is usually recognized as a telomere-protecting protein; however, the destabilization of TRF2 leads to spermatocyte arrest without telomere fusion. Therefore, TRF2 might have a specific function during meiosis.
Our identification and characterization of TRF2 stability highlights the elaborate regulatory processes that occur during telomere attachment. In past decades, multiple proteins and complexes have been identified. The SUN1–KASH complex proteins were the earliest proteins identified as connecting structures between telomeres and the NE (72). Dernburg et al. discovered that cytoskeletal forces coordinate meiotic chromosome pairing and synapsis (16). Watanabe and co-workers identified the multi-subunit TERB1/2-MAJIN complex as a driving force of telomere cap exchange during meiotic progression (13). In addition, a drastic reformation of INM proteins takes place specifically during meiosis, such as the expression of the meiosis-specific short isoform of the lamina protein lamin C2 (89–91) and the reorganization of the nuclear pore complex (92). Furthermore, chromosome movement during meiosis is regulated by multiple protein modifications, especially meiosis-specific phospho-modifications by CDK2 in mammals (65,84,93) and by CHK2 and PLK2 in nematodes (94–96). F-box proteins are involved in meiotic cell cycle by governing the ubiquitination and subsequent degradation of proteins, for example, ZYGO1 functioned as an E3 ligase during the ‘bouquet’ stage in rice (97). To our knowledge, FBXO47 is the first protein that functions as TRF2 protector during meiosis. And we expect to identify which E3 ligase has a function on TRF2 ubiquitination in the near future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jiahao Sha for participating in the experimental design. Zibin Wang and Chaohao Wu for sample preparation and FIB-SEM analysis; Shuqin Zhao for zygote injuctions and transplantation and Sigrid Eckardt for assistance with manuscript preparation.
Author contributions: R.H., H.W. and C.L. performed most of the experiments; Y.Z., S.L., Y.C., Y.G., X.G. and M.L. performed some of the experiments; R.H., H.W., W.L. and M.L. analyzed the data; M.L., and W.L. initiated the project and designed the experiments; R.H., H.W., Y.Z., C.L. and M.L. wrote the paper. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2017YFA0103803]; National Natural Science Foundation of China [31890784]; National Key Research and Development Program of China [2016YFA0500902 to W.L. and M.L.); Natural Science Foundation of China [31571536, 31771654 and 31530047 to M.L.]; Natural science fund for colleges and universities in Jiangsu Province [16KJA310003 to M.L.]; The Natural Science Foundation of Jiangsu Province [BK20190081 to M.L.]; Qing Lan Project (to M.L.). Funding for open access charge: Qing Lan Project (to M.L.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Page S.L., Hawley R.S.. Chromosome choreography: the meiotic ballet. Science. 2003; 301:785–789. [DOI] [PubMed] [Google Scholar]
- 2. Nagaoka S.I., Hassold T.J., Hunt P.A.. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012; 13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adelfalk C., Janschek J., Revenkova E., Blei C., Liebe B., Gob E., Alsheimer M., Benavente R., de Boer E., Novak I. et al.. Cohesin SMC1beta protects telomeres in meiocytes. J. Cell Biol. 2009; 187:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Novak I., Wang H., Revenkova E., Jessberger R., Scherthan H., Hoog C.. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J. Cell Biol. 2008; 180:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., Jessberger R.. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 2004; 6:555–562. [DOI] [PubMed] [Google Scholar]
- 6. Handel M.A., Schimenti J.C.. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010; 11:124–136. [DOI] [PubMed] [Google Scholar]
- 7. Bolcun-Filas E., Handel M.A.. Meiosis: the chromosomal foundation of reproduction. Biol. Reprod. 2018; 99:112–126. [DOI] [PubMed] [Google Scholar]
- 8. Alsheimer M. The dance floor of meiosis, evolutionary conservation of nuclear envelope attachment and dynamics of meiotic telomeres. Genome Dyn. 2009; 5:81–93. [DOI] [PubMed] [Google Scholar]
- 9. Scherthan H. Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci.: CMLS. 2007; 64:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Link J., Jahn D., Alsheimer M.. Structural and functional adaptations of the mammalian nuclear envelope to meet the meiotic requirements. Nucleus. 2015; 6:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Starr D.A., Fridolfsson H.N.. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010; 26:421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M.. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007; 12:863–872. [DOI] [PubMed] [Google Scholar]
- 13. Shibuya H., Hernandez-Hernandez A., Morimoto A., Negishi L., Hoog C., Watanabe Y.. MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell. 2015; 163:1252–1266. [DOI] [PubMed] [Google Scholar]
- 14. Hu C., Rai R., Huang C., Broton C., Long J., Xu Y., Xue J., Lei M., Chang S., Chen Y.. Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex. Cell Res. 2017; 27:1485–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scherthan H., Wang H., Adelfalk C., White E.J., Cowan C., Cande W.Z., Kaback D.B.. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. PNAS. 2007; 104:16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F.. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009; 139:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scherthan H., Weich S., Schwegler H., Heyting C., Härle M., Cremer T.. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 1996; 134:1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomita K., Cooper J.P.. The telomere bouquet controls the meiotic spindle. Cell. 2007; 130:113–126. [DOI] [PubMed] [Google Scholar]
- 19. Scherthan H., Sfeir A., de Lange T.. Rap1-independent telomere attachment and bouquet formation in mammalian meiosis. Chromosoma. 2011; 120:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koszul R., Kleckner N.. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections. Trends Cell Biol. 2009; 19:716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Storlazzi A., Gargano S., Ruprich-Robert G., Falque M., David M., Kleckner N., Zickler D.. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010; 141:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pandita T.K., Westphal C.H., Anger M., Sawant S.G., Geard C.R., Pandita R.K., Scherthan H.. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol. Cell Biol. 1999; 19:5096–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fernandez-Capetillo O., Liebe B., Scherthan H., Nussenzweig A.. H2AX regulates meiotic telomere clustering. J. Cell Biol. 2003; 163:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liebe B., Petukhova G., Barchi M., Bellani M., Braselmann H., Nakano T., Pandita T.K., Jasin M., Fornace A., Meistrich M.L. et al.. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp. Cell Res. 2006; 312:3768–3781. [DOI] [PubMed] [Google Scholar]
- 25. Roig I., Robles P., Garcia R., Martinez-Flores I., Cabero L., Egozcue J., Liebe B., Scherthan H., Garcia M.. Chromosome 18 pairing behavior in human trisomic oocytes. Presence of an extra chromosome extends bouquet stage. Reproduction. 2005; 129:565–575. [DOI] [PubMed] [Google Scholar]
- 26. Trelles-Sticken E., Dresser M.E., Scherthan H.. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 2000; 151:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 28. Zeng Z., Wang W., Yang Y., Chen Y., Yang X., Diehl J.A., Liu X., Lei M.. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev. Cell. 2010; 18:214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richburg J.H., Myers J.L., Bratton S.B.. The role of E3 ligases in the ubiquitin-dependent regulation of spermatogenesis. Semin. Cell Dev. Biol. 2014; 30:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Z., Liu P., Inuzuka H., Wei W.. Roles of F-box proteins in cancer. Nat. Rev. Cancer. 2014; 14:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., Wang Q., Xie N., Hua R., Liu M. et al.. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018; 28:879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J.W., Elledge S.J.. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996; 86:263–274. [DOI] [PubMed] [Google Scholar]
- 33. Nakayama K.I., Nakayama K.. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006; 6:369–381. [DOI] [PubMed] [Google Scholar]
- 34. Jantsch V., Tang L., Pasierbek P., Penkner A., Nayak S., Baudrimont A., Schedl T., Gartner A., Loidl J.. Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol. Biol. Cell. 2007; 18:4911–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen B., Zhang J., Wu H., Wang J., Ma K., Li Z., Zhang X., Zhang P., Huang X.. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013; 23:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu M., Shi X., Bi Y., Qi L., Guo X., Wang L., Zhou Z., Sha J.. SHCBP1L, a conserved protein in mammals, is predominantly expressed in male. Mol. Hum. Reprod. 2014; 20:463–475. [DOI] [PubMed] [Google Scholar]
- 37. Jiang M., Gao M., Wu C., He H., Guo X., Zhou Z., Yang H., Xiao X., Liu G., Sha J.. Lack of testicular seipin causes teratozoospermia syndrome in men. PNAS. 2014; 111:7054–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng B., Zhao D., Zhang P., Shen C., Guo Y., Zhou T., Guo X., Zhou Z., Sha J.. Quantitative proteomics reveals the essential roles of stromal interaction molecule 1 (STIM1) in the testicular cord formation in mouse testis. Mol. Cell Proteomics. 2015; 10:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shibuya H., Morimoto A., Watanabe Y.. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014; 10:e1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y.J., Liu C., Chang Z., Wadas B., Brower C.S., Song Z.H., Xu Z.L., Shang Y.L., Liu W.X., Wang L.N. et al.. Degradation of the separase-cleaved Rec8, a meiotic cohesin subunit, by the N-end rule pathway. J. Biol. Chem. 2016; 291:7426–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T.. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 2003; 69:612–616. [DOI] [PubMed] [Google Scholar]
- 42. Bernardino R.L., Alves M.G., Oliveira P.F.. Evaluation of the purity of sertoli cell primary cultures. Methods Mol. Biol. 2018; 1748:9–15. [DOI] [PubMed] [Google Scholar]
- 43. Guo H., Wen R., Wang Q., Datla R., Xiao W.. Three Brachypodium distachyon Uev1s promote Ubc13-mediated Lys63-linked polyubiquitination and confer different functions. Front. Plant Sci. 2016; 7:1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mandolesi G., Vanni V., Cesa R., Grasselli G., Puglisi F., Cesare P., Strata P.. Distribution of the SNAP25 and SNAP23 synaptosomal-associated protein isoforms in rat cerebellar cortex. Neuroscience. 2009; 164:1084–1096. [DOI] [PubMed] [Google Scholar]
- 45. Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J.W., Elledge S.J.. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-Box. Cell. 1996; 86:263–274. [DOI] [PubMed] [Google Scholar]
- 46. Bai C., Richman R., Elledge S.J.. Human cyclin F. EMBO J. 1994; 13:6087–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goetz P., Chandley A.C., Speed R.M.. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J. Cell Sci. 1984; 65:249–263. [DOI] [PubMed] [Google Scholar]
- 48. Bellvé A.R., Cavicchia J.C., Millette C.F., O’Brien D.A., Bhatnagar Y.M., Dym M.. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1997; 74:68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kluin P.M., de Rooij D.G.. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int. J. Androl. 1981; 4:475–493. [DOI] [PubMed] [Google Scholar]
- 50. Vergouwen R.P., Jacobs S.G., Huiskamp R., Davids J.A., de Rooij D.G.. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J. Reprod. Fertil. 1991; 93:233–243. [DOI] [PubMed] [Google Scholar]
- 51. Bradley A., Anastassiadis K., Ayadi A., Battey J.F., Bell C., Birling M.C., Bottomley J., Brown S.D., Burger A., Bult C.J. et al.. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012; 23:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matzuk M.M., Lamb D.J.. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008; 14:1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baudat F., Manova K., Yuen J.P., Jasin M., Keeney S.. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000; 6:989–998. [DOI] [PubMed] [Google Scholar]
- 54. Romanienko P.J., Camerini-Otero R.D.. The mouse Spo11 gene is required for meiotic chromosome symapsis. Mol. Cell. 2000; 6:975–987. [DOI] [PubMed] [Google Scholar]
- 55. Bellani M.A., Romanienko P.J., Cairatti D.A., Camerini-Otero R.D.. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J. Cell Sci. 2005; 118:3233–3245. [DOI] [PubMed] [Google Scholar]
- 56. Parvanov E.D., Petkov P.M., Paigen K.. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010; 327:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., Przeworski M., Coop G., de Massy B.. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010; 327:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Handel M.A., Schimenti J.C.. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010; 11:124–136. [DOI] [PubMed] [Google Scholar]
- 59. Rockmill B., Engebrecht J.A., Scherthan H., Loidl J., Roeder G.S.. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995; 141:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ribeiro J., Abby E., Livera G., Martini E.. RPA homologs and ssDNA processing during meiotic recombination. Chromosoma. 2015; 125:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng K., Xiol J., Reuter M., Eckardt S., Leu N.A., McLaughlin K.J., Stark A., Sachidanandam R., Pillai R.S., Wang P.J.. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. PNAS. 2010; 107:11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de Vries F.A., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A.. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005; 19:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yuan L., Liu J.G., Hoja M.R., Wilbertz J., Nordqvist K., Höög C.. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science. 2002; 296:1115–1118. [DOI] [PubMed] [Google Scholar]
- 64. Bisig C.G., Guiraldelli M.F., Kouznetsova A., Scherthan H., Hoog C., Dawson D.S., Pezza R.J.. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLos Genet. 2012; 8:e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mikolcevic P., Isoda M., Shibuya H., del Barco Barrantes I., Igea A., Suja J.A., Shackleton S., Watanabe Y., Nebreda A.R.. Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat. Commun. 2016; 7:11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de Lange T., Shiue L., Myers R.M., Cox D.R., Naylor S.L., Killery A.M., Varmus H.E.. Structure and variability of human chromosome ends. Mol. Cell Biol. 1990; 10:518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu L., Franco S., Spyropoulos B., Moens P.B., Blasco M.A., Keefe D.L.. Irregular telomeres impair meiotic synapsis and recombination in mice. PNAS. 2004; 101:6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Siderakis M., Tarsounas M.. Telomere regulation and function during meiosis. Chromosome Res. 2007; 15:667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shibuya H., Ishiguro K., Watanabe Y.. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 2014; 16:145–156. [DOI] [PubMed] [Google Scholar]
- 70. Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S.. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008; 133:1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee C.Y., Horn H.F., Stewart C.L., Burke B., Bolcun-Filas E., Schimenti J.C., Dresser M.E., Pezza R.J.. Mechanism and regulation of rapid telomere prophase movements in mouse meiotic chromosomes. Cell Rep. 2015; 11:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M.. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007; 12:863–872. [DOI] [PubMed] [Google Scholar]
- 73. Hiraoka Y., Dernburg A.F.. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 2009; 17:598–605. [DOI] [PubMed] [Google Scholar]
- 74. Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M.. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007; 445:506–510. [DOI] [PubMed] [Google Scholar]
- 75. O’Connor M.S., Safari A., Xin H., Liu D., Songyang Z.. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11874–11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li B., de Lange T.. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell. 2003; 14:5060–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E.. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997; 17:236–239. [DOI] [PubMed] [Google Scholar]
- 78. O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 80. Bianchi A., Smith S., Chong L., Elias P., deLange T.. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997; 16:1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang L., Tu Z., Liu C., Liu H., Kaldis P., Chen Z., Li W.. Dual roles of TRF1 in tethering telomeres to the nuclear envelope and protecting them from fusion during meiosis. Cell Death Differ. 2018; 25:1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Skaar J.R., Pagan J.K., Pagano M.. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013; 14:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tu Z., Bayazit M.B., Liu H., Zhang J., Busayavalasa K., Risal S., Shao J., Satyanarayana A., Coppola V., Tessarollo L., Singh M., Zheng C., Han C., Chen Z., Kaldis P., Gustafsson J.Å., Liu K.. Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. PNAS. 2017; 114:592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cohen P.E., Martinerie L., Manterola M., Chung S.S.W., Panigrahi S.K., Weisbach M., Vasileva A., Geng Y., Sicinski P., Wolgemuth D.J.. Mammalian E-type cyclins control chromosome pairing, telomere stability and CDK2 localization in male meiosis. PLos Genet. 2014; 10:e1004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cooper J.P., Watanabe Y., Nurse P.. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998; 392:828–831. [DOI] [PubMed] [Google Scholar]
- 86. Nimmo E.R., Pidoux A.L., Perry P.E., Allshire R.C.. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998; 392:825–828. [DOI] [PubMed] [Google Scholar]
- 87. Maciejowski J., de Lange T.. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017; 18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Iwano T., Tachibana M., Reth M., Shinkai Y.. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004; 279:1442–1448. [DOI] [PubMed] [Google Scholar]
- 89. Alsheimer M., von Glasenapp E., Hock R., Benavente R.. Architecture of the nuclear periphery of rat pachytene spermatocytes distribution of nuclear envelope proteins in relation to synaptonemal complex attachment sites. Mol. Biol. Cell. 1999; 10:1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jahn D., Schramm S., Benavente R., Alsheimer M.. Dynamic properties of meiosis-specific lamin C2 and its impact on nuclear envelope integrity. Nucleus. 2010; 1:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Link J., Jahn D., Schmitt J., Gob E., Baar J., Ortega S., Benavente R., Alsheimer M.. The meiotic nuclear lamina regulates chromosome dynamics and promotes efficient homologous recombination in the mouse. PLoS Genet. 2013; 9:e1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Scherthan H., Jerratsch M., Li B., Smith S., Hultén M., Lock T., de Lange T.. Mammalian meiotic telomeres protein composition and redistribution in relation to nuclear pores. Mol. Biol. Cell. 2000; 11:4189–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Viera A., Alsheimer M., Gomez R., Berenguer I., Ortega S., Symonds C.E., Santamaria D., Benavente R., Suja J.A.. CDK2 regulates nuclear envelope protein dynamics and telomere attachment in mouse meiotic prophase. J. Cell Sci. 2015; 128:88–99. [DOI] [PubMed] [Google Scholar]
- 94. Harper N.C., Rillo R., Jover-Gil S., Assaf Z.J., Bhalla N., Dernburg A.F.. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell. 2011; 21:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Labella S., Woglar A., Jantsch V., Zetka M.. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell. 2011; 21:948–958. [DOI] [PubMed] [Google Scholar]
- 96. Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y. et al.. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009; 139:920–933. [DOI] [PubMed] [Google Scholar]
- 97. Zhang F., Tang D., Shen Y., Xue Z., Shi W., Ren L., Du G., Li Y., Cheng Z.. The F-Box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell. 2017; 29:2597–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.