Abstract
The inflammatory cytokine tumor necrosis factor alpha (TNFα) is considered to play a key role in the pathogenesis of intervertebral disc disease. To evaluate the importance of this cytokine we examined the inflammatory environment and spinal phenotype of 9-month-old hTNFα overexpressing (hTNFα-TG) mice. The mice evidenced increased circulating levels of IL-1β, IL-2, KC/GRO, and MCP-1 along with thinning of the cortical and trabecular vertebral bone. Surprisingly, while the nucleus pulposus (NP) of these mice was intact and healthy, the caudal annulus fibrosus (AF) evidenced robust cell death and immune cell-infiltration. Despite these differences, there were no obvious alterations in the collagen or aggrecan content in the NP and AF. However, there was a reduction in cartilage oligomeric matrix protein (COMP) suggesting destabilization of the AF matrix. Microarray analysis of the NP from hTNFα-TG mice cells revealed minimal changes in global gene expression. These findings lend support to the notion that NP tissue is isolated from systemic inflammation. In contrast, the severe AF phenotype suggests that systemic inflammation interferes with AF health, predisposing discs to herniation as opposed to directly causing NP degeneration.
Keywords: Cytokines, genetic animal models, collagen
INTRODUCTION
Low back pain (LBP) and associated intervertebral disc degeneration is a widespread, costly, and complex medical condition affecting a huge proportion of the population (1-4). As the disc degenerates with age or disease, the nucleus pulposus (NP) becomes more fibrotic, and cells transition from a vacuolated notochordal phenotype to one that resembles hypertrophic chondrocytes (5-7). In concert with these cellular and extracellular matrix changes, the dramatic increase in interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNFα) expression is correlated with severity of disc degeneration (8). These cytokines are thought to further promote disc degeneration by activating matrix metalloproteinases (MMPs) (9-12).
Despite the association between cytokine expression and disc degeneration, our recently published work shows that global hTNFα overexpression in a murine model (Tg197), results in healthier and more cellular discs than the wild type (WT) controls (13,14). This observation was very surprising considering both the large body of literature linking cytokine expression to disc degeneration and the finding that the synovial joints of Tg197 animals are severely arthritic (13,15-17). In contrast to the healthier NP compartments of these animals, global hTNFα overexpression predisposed these Tg197 mice to caudal disc herniation (14). These findings raised two interdependent questions: first, would the NP compartment remain healthy after long-term TNFα challenge; second, would disc herniation of the TNFα transgenic animals would become more prevalent over a longer time period. To address these questions, we explored the effect of global hTNFα overexpression on disc health over an extended period of time.
To address these questions, we studied the intervertebral disc health of a longer living hTNFα transgenic mouse model with a less severe arthritic phenotype (18). We measured cytokine concentrations in the blood and performed comprehensive histological and microCT (μCT) analyses to characterize effects of the hTNFα driven systemic inflammation. We confirmed that systemic inflammation leads to dramatic vertebral bone erosion and found that TNFα overexpression lead to AF cell death, immune cell infiltration, and AF matrix erosion. Despite these osseous and AF changes, the NP remains healthy. These findings support the view that the NP is a closed immune-privileged compartment isolated from systemic inflammation and immune cell infiltration.
MATERIALS AND METHODS
Mice and organ culture
All animal care procedures, housing, breeding, and the collection of animal tissues were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Thomas Jefferson University. hTNFα transgenic (hTNFα-TG) mice and age- and sex- matched C57BL6/N wild type (WT) mice were purchased from Taconic Farms (Hudson, NY). These mice contain a 2.8 kb fragment of the human TNFα gene comprising the promoter and coding region fuse d to the human β-globin 3’ untranslated region (UTR) replacing the endogenous TNFα 3’UTR (13). This alteration results is deregulated overexpression of hTNFα (19). Organ culture was performed using previously described methods (20). Briefly, dissected vertebra-disc-vertebra motion segments from WT (C57BL6) mice were cultured with or without hTNFα (10 ng/mL) in DMEM (1g/L glucose) supplemented with 10% FBS for 24 hours (n=3 mice/group, 6 lumbar and 9 caudal discs/animal). Discs were dissected under stereo microscope (Zeiss, Stemi 503) and RNA extraction as described below.
Blood collection and analysis
Blood from 9-month-old mice (n=5) was collected immediately post-mortem by intra-cardiac puncture using heparinized needles. Cells were separated from the plasma using centrifugation. Cytokine concentrations were assayed using the V-PLEX Mouse Cytokine 19-Plex Kit (Meso Scale Diagnostics) according to manufacturer’s specifications.
Micro-computed tomography analysis
Micro-computed tomography scans (MicroCT40, SCANCO Medical, Switzerland) were performed on the lumbar and caudal hTNFα-TG and WT spines fixed with 4% PFA. Five mice per genotype were used (2–3 spinal levels/mouse); graphs show all measured levels. Caudal and lumbar segments were scanned with an energy of 70 kVp, a current of 114 mA, and a 200-ms integration time resulting in 16 μm3 voxel resolution. The region of interest for trabecular bone analysis was identified using hand contouring and included the entire vertebral trabecular bone excluding cortical bone and the growth plate. Three-dimensional reconstructions of these trabecular bone scans were compiled using a Gaussian filter (σ = 1.0, support = 1) and converted to binary images with a fixed grey-scale threshold of 200. The data sets were then assessed using software supplied by the system manufacturer. To measure cortical thickness, spines were first aligned in the x-y plane using bony landmarks. Mean cortical thickness at the midpoint of each vertebra was manually quantified by averaging measurements at four points separated by 90 degrees (21). Disc height index (DHI) was calculated by dividing average disc height by the height of adjacent vertebral bodies (22).
Histological analysis
Processing of histological samples began with 48 hours of 4% paraformaldehyde fixation before decalcification with 20% ethylenediaminetetracetic acid (EDTA) for 15 days at 4°C. After decalcification, spinal motion segments were dissected and embedded in paraffin. Mid-coronal 7 μm sections were used for staining. Xylene deparaffinization followed by graded ethanol rehydration preceded all protocols. Safranin O/Fast Green/Hematoxylin stained slides were imaged using an Axio Imager 2 microscope, 5x/0.15 N-Achroplan or 10x/0,3 EC Plan-Neofluar objectives, Axiocam 105 color camera,and Zen2TM software (Carl Zeiss). Five blinded observers performed the scoring using a Modified Thompson grading scale (23,24). Five mice per genotype with 4 discs per mouse for both caudal and lumbar levels were analyzed. AF width was quantified from Safranin O/Fast Green/Hematoxylin stained slides using ImageJ length measurement tool, each plotted value is the average of the midpoint length of both sides of the AF normalized to WT Safranin O/Fast Green/Hematoxylin stained slides (n=5, and 4 levels per animal).
Picrosirius Red™ Analysis
Picrosirius Red™ staining visualized localization and quality of the collagen fibrils (25,26). Stained sections were imaged on a polarizing microscope (Eclipse LV100 POL, Nikon). High magnification AF images were used for the analysis of the area occupied by green, yellow, or red pixels. Threshold levels for the colors remained constant.
Cell number quantification
DAPI (Thermo Fisher Scientific, P36934) stained mid-coronal 7μm sections were used to quantify cell number in the NP and AF. Three sections per animal (n=5) were used, and the NP area was used for analysis. Using ImageJ software (NIH), images were converted to 32-bit, then the background was subtracted using rolling=50. Next the images were auto-thresholded, made binary, and then cell number was calculated using the analyze particles function (27). Cell band percent area was calculated by hand contouring the cell band and NP compartment on Safranin O/Fast Green/Hematoxylin stained slides using ImageJ.
TUNEL assay
TUNEL assay was performed on disc tissue sections using an “In situ cell death detection” Kit (Roche Diagnostic) 7. Sections were permeabilized with Proteinase K (20 μg/mL) for 15 min at room temperature before the TUNEL assay and imaged as described above.
Immunofluorescence microscopy
Mid-coronal 7 μm sections were used for all immunofluorescence studies. Antigen retrieval was accomplished in an antibody-specific manner including: 20 minutes in heated citrate buffer, 10 min incubation with proteinase K, 30 min in Chondroitinase ABC at 37 °C, or TRIS/EDTA. Sections were blocked in 5% normal serum (Thermo Fisher Scientific, 10000C) in PBS-T (0.4% Triton X-100 in PBS), and incubated with primary antibody. Mouse on Mouse Kit (Vector laboratories, BMK-2202) was used for blocking and primary antibody incubation. The primary antibodies used were: Aggrecan (1:50, Millipore, AB1031), Collagen I detecting COL1A1 (1:100, Abcam, ab34710), Collagen II detecting COL2A1 (1:400, Fitzgerald, 70R-CR008), COMP (1:200, Abcam, ab231977), CA3 (1:150, Santa Cruz, sc-50715), in blocking buffer at 4 °C overnight. For GLUT-1 (1:200, Abcam, ab40084), ARGxx (1:200, Abcam, ab3773), and CS (1:300, Abcam, ab11570). Sections were incubated for 1h at room temperature with the appropriate Alexa Fluor®−594 conjugated secondary antibody (1:700;Jackson ImmunoResearch) before washing and mounting with ProLong® Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, P36934). All mounted slides were allowed to set before visualization with Axio Imager 2 using 5x/0.15 N-Achroplan or 10x/0,3 EC Plan-Neofluar objectives, AxioCam MRm camera, and Zen2TM software (Carl Zeiss). Exposure settings remained constant (28). Staining percent area quantification of three levels from five animals of each genotype was performed using ImageJ software (NIH); thresholds remained constant.
RNA isolation and microarray analysis
NP tissue was manually micro-dissected under a stereo microscope (Zeiss, Stemi 503) and immediately placed in RNAlater® Reagent (Invitrogen) as previously described (29). Seven mice per genotype were sacrificed for RNA isolation and NP tissue pooled from single animal served as an individual sample. NP tissue was collected in RNAlater® Reagent and homogenized with a Pellet Pestle Motor (Sigma Aldrich, Z359971). RNA was extracted from the lysates using RNeasy® Mini kit (Qiagen). Half of the DNA-free RNA converted to cDNA using EcoDry™ Premix (Clontech) and the other half was used in the Clariome™ S Assay, mouse (ThermoFisher). RNA was quantified on a Nanodrop ND-100 spectrophotometer, followed by RNA quality assessment analysis on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). For subsequent rtPCR, template cDNA and gene-specific primers were added to Power SYBR Green master mix (Applied Biosystems) and mRNA expression was quantified and normalized to GAPDH using the Step One Plus Real-time PCR System (Applied Biosystems). Melting curves were analyzed to verify the specificity of the RT-PCR and the absence of primer dimer formation. Thermal cycle was programmed for 20 s at 95 °C as initial denaturation, followed by 40 cycles of 30 s at 95 °C, and 30 s at 60 °C, with final melt curve and extension for 15 s at 95 °C, 1 min. at 60 °C, and 15 s at 95°C. Custom PCR primers specific to murine genes are described in Supplementary Figure 1.
Microarray data analysis
Fragmented biotin labeled cDNA was synthesized according to ABI using the GeneChip WT Plus kit (Thermo Fisher Scientific). Mouse Clariom S gene chips were hybridized with fragmented and biotin-labeled cDNA in 100 μl of hybridization cocktail. Arrays were washed and stained with GeneChip hybridization wash & stain kit using Gene chip Fluidic Station 450. Chips were scanned on an Affymetrix Gene Chip Scanner 3000 7G, using Command Console Software. Quality Control of the experiment was performed by Expression Console Software v 1.4.1. Chp files were generated by sst-rma normalization from Affymetrix cel file using Expression Console Software. A heat map was generated using MeV 4_8. The normalized values were Log2 transformed and mean centered and a t-test (p < 0.001) was used to isolate significant genes for further analysis. Significant genes and samples were then both clustered using Pearson Correlation. SAM analysis was performed in MeV with delta value of 0.74 for a FDR of 5%. Volcano plot was generated in R studio.
Antigen Presentation
Primary rat NP cells were isolated as previously described (30). For particle phagocytosis image, cells were incubated for 24 hours with Fluoro-Max Dyed Green Aqueous Florescent Particles (ThermoFisher) before trypsinization and live cell flow cytometer sorting (BD FACSCelesta). Florescent positive cells were plated on p-lysine treated coverslips, fixed with methanol, and imaged as described above. For particle internalization quantification, NP cells were initially plated on p-lysine treated coverslips before incubation with particles for specified period. At least 5 fields of view were counted per sample. Additionally, cells treated with DQ™ Ovalbumin (Thermo Fisher) and quantified using a microplate reader (Tecan Infinite M100).
Statistics
Five animals per genotype were used for analysis (n=5), and data are presented as mean ± SD. Differences between genotypes were analyzed using the Student’s t test when only two groups were presented on graph, or one-way ANOVA with a Sidak’s multiple comparison test between groups when more than two groups were presented. Three lumbar or tail levels per mouse were combined and averaged for both μCT and histological analysis. At least five independent blinded individuals performed histological grading. Significance between collagen fiber distributions was determined using a χ2 test. All statistical analyses were performed using Prism7 (GraphPad Software). P ≤ 0.05 was considered statistically significant.
RESULTS
hTNFα transgenic mice show elevated systemic inflammatory cytokine levels.
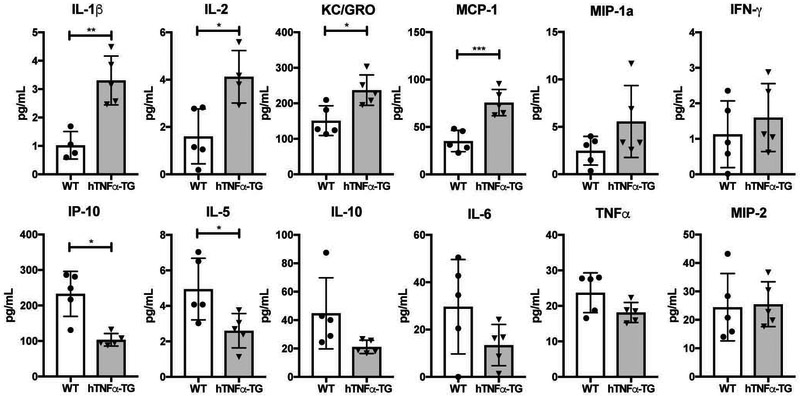
To investigate the alterations in circulating inflammatory mediators resulting from global hTNFα overexpression, we used the Mesoscale Discovery multiplex ELISA platform to measure the concentrations of cytokines in the blood. The transgenic TNFα overexpressing (hTNFα-TG) mice evidenced significant increases of IL-1β (p = 0.0022), IL-2 (p = 0.0131), KC/GRO (p = 0.0126), and monocyte chemoattractant protein-1 (MCP-1) (p = 0.0009). Interestingly, there was a significant decrease in both IL-5 (p = 0.0298) and interferon gamma-induced protein 10 (IP-10) (p = 0.023) compared to WT controls. There were no significant differences between the genotypes in the circulating levels of macrophage inflammatory protein (MIP)-1a, IFN-γ, IL-10, IL-6, TNFα and MIP-2 (Fig. 1). Additionally, there was no change in TNFα concentration between hTNFα-TG and WT mice. This is expected, as the ELISA specifically measures murine TNFα not the human protein. These results show that hTNFα causes alterations in circulating inflammatory factors.
Figure 1: Circulating cytokine levels in hTNFα-TG mice showed an overall increase of inflammatory markers compared to WT controls.
IL-1β, IL-2, KC/GRO, and MCP-1 show a significant increase in concentration in the hTNFα-TG mice. IP-10 and IL-5 both show a significant decrease in the hTNFα-TG mice. Scatter plots show all data points plotted as mean ± SD. t-test was used to determine significance between groups. (n = 5) * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
TNFα transgenic vertebrae show cortical and trabecular thinning.
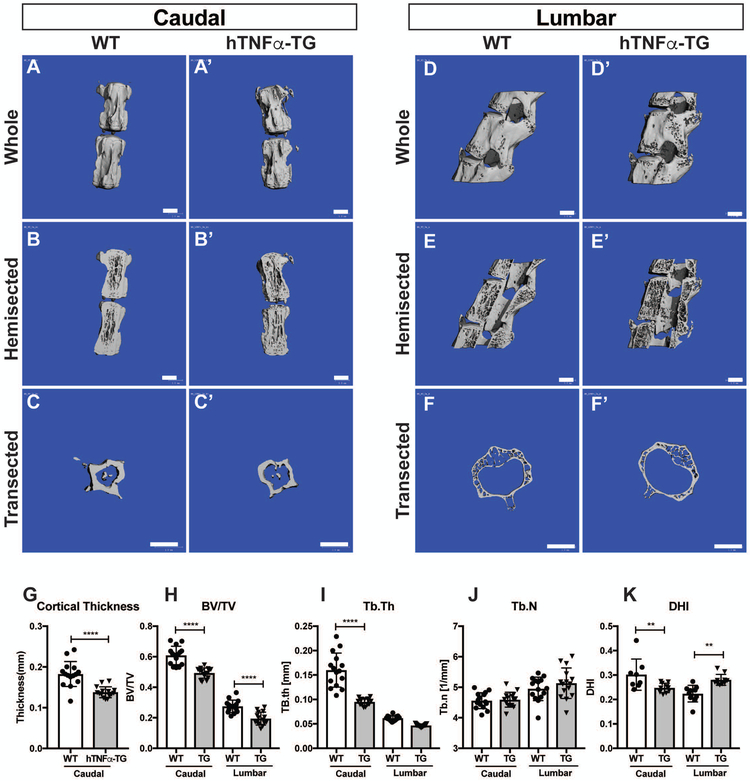
Micro-computed tomography (μCT) analysis showed that the morphology of the caudal cortical bone of the WT and hTNFα-TG mice were similar (Fig. 2A, A’). However, sagittal optical hemisectioning reveals marked thinning of the caudal cortical and trabecular vertebral bone in hTNFα-TG mice (Fig. 2B, B’). Analysis of the midpoint transection of the caudal vertebrae provided further details of the cortical thinning of the hTNFα-TG caudal levels (Fig. 2C, C’). While the cortical shell is much thinner in the lumbar spine than caudal spine in both genotypes, the hTNFα-TG lumbar vertebrae showed similar bone changes as the caudal, with robust trabecular thinning in the hTNFα-TG vertebrae (Fig. 2 D-F’).
Figure 2: hTNFα-TG vertebrae showed cortical and trabecular thinning.
(A-B’) Representative μCT scans of caudal motion segments of 9-month-old spines showing cortical thinning in hTNFα-TG vertebrae. (C-C’) Cross-section of a representative caudal vertebrae showing a robust cortical thinning of hTNFα-TG bone. (D-F’) Representative μCT scans lumbar vertebrae (D-D’) whole, (E-E’) optical hemi-section, and (F-F’) cross section. (G) Quantification of cortical thickness in caudal vertebrae. (H-K) Bone volume/trabecular volume (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and disc height index (DHI) of WT and hTNFα-TG mice (mean ± SD) (n = 5 mice per genotype with 3 consecutive vertebrae/animal). Scatter plots show all data points and plotted as mean ± SD. Significance was determined using ANOVA and Sidak’s multiple comparison test. Scale bar = 1mm ** p ≤ 0.01, **** p ≤ 0.0001.
Quantitative analysis of the μCT studies showed that there was a robust decrease in cortical thickness (p < 0.0001) (Fig. 2 G). Thinning of vertebral bone was not limited to the cortical shell as the hTNFα-TG mice had significantly less trabecular bone in both the caudal (p < 0.0001) and lumbar (p < 0.0001) regions of the spine (bone volume/total volume; BV/TV) (Fig. 2 H). Together with the reduction in BV/TV there was a reduction in trabecular thickness (Tb.Th) in the hTNFα-TG caudal levels (p < 0.0001); however, while the lumbar Tb.Th was reduced the difference was not significant (Fig 2 I). While there was no change in either the caudal or lumbar trabecular number (Fig 2 J), there is a significant decrease in DHI of the caudal levels (p = 0.0081), but a significant increase (p = 0.0034) in the lumbar levels (Fig. 2 K). Together, our results clearly show that the hTNFα-TG vertebral bone showed marked cortical and trabecular thinning.
TNFα transgenic lumbar discs are healthier than wild type controls, but the caudal annulus fibrosus shows cell infiltration of the outer lamellae.
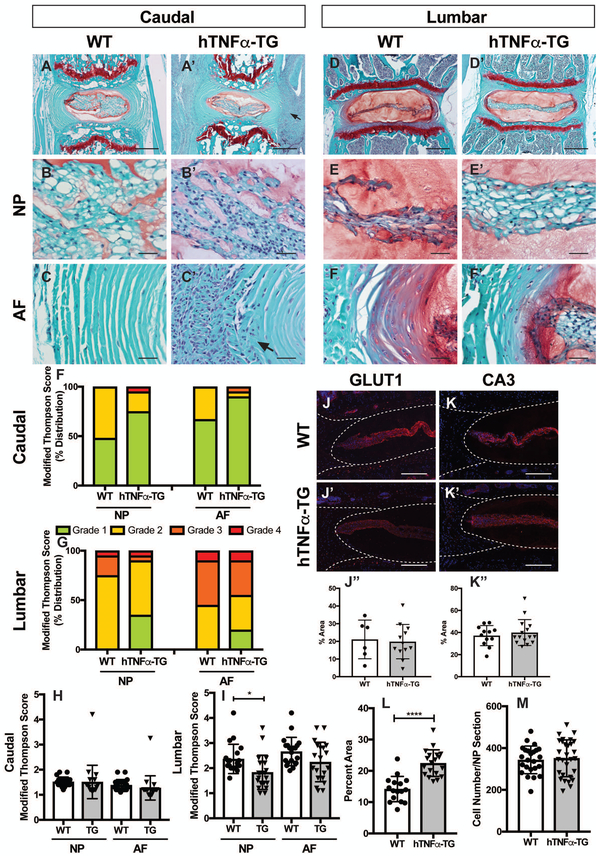
First, we confirmed using RT-PCR analysis that hTNFα was expressed by the NP tissue of hTNFα-TG mice and that the murine NP cells responded to hTNFα stimulation (S Fig. 2A and B). There was significantly higher expression of hTNFα in NP tissue of hTNFα-TG mice, with little to no signal detected in WT controls (S Fig. 2A). Importantly, when treated with hTNFα for 24 hours, organ cultured murine discs responded by increasing expression of mmp3 and ccl2 mRNA with a concomitant decrease in Col2a1 levels (S Fig. 2B). These results showed and validated that murine discs in hTNFα-TG mice not only produce hTNFα but similar to other skeletal tissues are responsive to hTNFα. To investigate whether the systemic and bony inflammatory changes translated to alterations in disc health, we performed an in-depth histological analysis of 9-month-old hTNFα-TG mice and age matched wild type controls. Safranin O/fast green and hematoxylin staining of caudal discs showed comparable microanatomy between the hTNFα-TG mice and their WT controls (Fig. 3 A, A’). Despite the overall similarity, there was an accumulation of sequestered cells outside the AF of the hTNFα-TG caudal discs (Fig. 3 A’, arrow). NP cells of the caudal discs were scattered throughout the tissue, and the hTNFα-TG NP cells appeared slightly smaller than their WT controls (Fig. 3 B, B’). A high magnification image of the caudal AF revealed that the cell agglomeration noted above infiltrated the outer lamella of the hTNFα-TG AF causing fraying of the organized collagen matrix (Fig. 3 C’, arrow). The NP compartment in lumbar discs of both genotypes had a vacuolated cell band within, and surrounded by, an abundant proteoglycan-rich matrix (Fig. 3 D, D’). The cell band of the hTNFα-TG discs was wider than the WT controls, and the NP cells appeared larger and more vacuolated (Fig. 3 E, E’). In both genotypes, the junction between NP and AF compartments was well demarcated, and the AF exhibited a well-organized collagenous lamellae interspersed with fibroblastic cells (Fig. 3 F, F’).
Figure 3: TNFα transgenic lumbar discs are healthier than wild type controls, but the caudal annulus fibrosus shows cell infiltration of the outer lamellae.
(A-F’) Safranin O/Fast Green/Hematoxylin staining of coronal sections of WT and hTNFα-TG mouse intervertebral discs (A, A’, D, D’; scale bars = 200μm; B-C’ and E-F’, scale bars = 20μm). (F, G) Distribution of histological grades of (F) caudal and (G) lumbar discs using the modified Thomson scale. (H, I) Average modified Thompson scores for (K) caudal and (L) lumbar intervertebral discs of 9-month-old WT and hTNFα-TG mice. (J-K”) Representative images and quantification showed similar expression of NP cell markers glucose transporter 1 (GLUT1) (n=4; 1–2 levels per animal for WT, n = 5; 3 levels per animal for hTNFα-TG)(J-J”) and carbonic anhydrase 3 (CA3) (n=5; 3 levels per animal) (K-K”) in WT and hTNFα-TG lumbar discs (J-K’; scale bar = 200 μm). (L) The area occupied by the NP cell band in lumbar discs is significantly larger in the hTNFα-TG animals. However, the NP compartment showed comparable cell number between WT and hTNFα-TG mice (M). Histological grading data was collected from 4 caudal and 4 lumbar discs per mouse (n=5 mice/genotype). Scatter plots show all data points and plotted as mean ± SD. Significance was determined using ANOVA and Sidak’s multiple comparison test. * p ≤ 0.05, **** p ≤ 0.0001.
The health of the discs in both the caudal (Fig. 3 H), and lumbar (Fig. 3 I) regions of the hTNFα-TG and WT animals were systematically graded and the average Modified Thompson Score determined. In both regions of the spine, the hTNFα-TG mice showed a healthier distribution of scores in both the NP and AF compartment (Fig. 3 F, G). The average scores plotted for each disc are shown for caudal (Fig. 3 H) and lumbar (Fig. 3 I) levels. Although hTNFα-TG mice showed a healthier distribution of scores in both the NP and AF compartments of the caudal spine, this did not translate to a statistically significant reduction in average Modified Thompson Score (Fig. 3 H). However, comparison of the average lumbar grade scores indicated a significant (p = 0.0249) reduction in degenerative changes (Fig. 3 I).
To further characterize the effects of hTNFα on cell phenotype, immunofluorescent staining of NP cell markers was performed. There were no discernable no differences in the expression of glucose transporter 1 (GLUT1) (Fig. 3 J-J”) or carbonic anhydrase III (CA3)(Fig. 3 K-K”). Further, there was no difference in NP cell number between the genotypes (Fig. 3 M). However, careful examination of the histology of the NP indicated that compared to WT discs, NP cells of the hTNFα-TG lumbar region occupied a larger proportion of the NP compartment than those of the WT levels (p < 0.0001) (Fig. 3 L). Additionally, we found no difference in the distribution of aggrecan or chondroitin sulfate between genotypes (S Fig. 3 A-B”). In summary, the NP and AF of the hTNFα-TG mice are comparable to, or healthier than, the WT controls, and while the NP cells of both genotypes are phenotypically similar, subtle cellular differences are noted.
hTNFα-TG mice evidence a vigorous inflammatory response adjacent to the caudal AF accompanied by AF cell loss and reduced AF width.
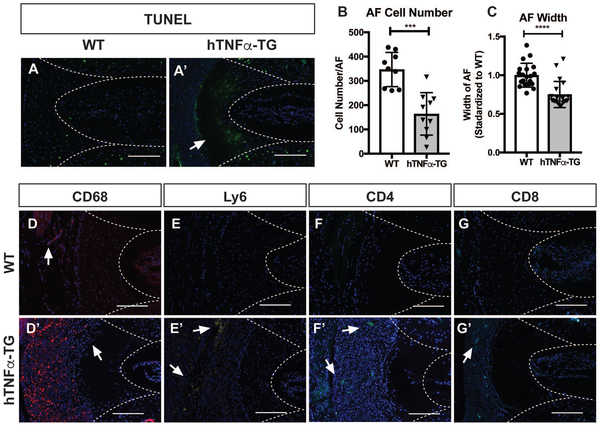
Safranin O/fast green and hematoxylin staining of hTNFα-TG caudal discs revealed a dense cellular response directly outside and infiltrating the outer lamella of the AF that was not present in the WT controls. To characterize the changes in AF, sections were TUNEL stained and DAPI positive nuclei of AF cells counted. There was a small amount of cell death identified by TUNEL positivity in the WT levels scattered throughout the endplate (EP), AF, and NP (Fig. 4 A). In the hTNFα-TG levels, there was robust cell death in the AF forming a border between what was now an acellular outer AF lacking DAPI stained nuclei and the inner AF, which included a population of DAPI and TUNEL positive nuclei (Fig 4 A’). There was no difference in NP or EP TUNEL staining between the WT and hTNFα-TG mice. Since TUNEL staining only captures actively dying cells, we chose to quantify the number of nuclei in the AF to provide a more complete picture of AF cellularity. There was a significant reduction in cell number in the hTNFα-TG AF compared to the WT controls (p = 0.0001) (Fig. 4 B). Additionally, we measured the width of the AF compartment. Compared with WT controls, the AF width of the hTNFα-TG animals was significantly reduced (p < 0.0001) (Fig. 4 C).
Figure 4: hTNFα-TG mice evidence a vigorous inflammatory response adjacent to the caudal AF accompanied by AF cell loss and reduced AF width.
TUNEL staining of (A) WT and (A’) hTNFα-TG caudal levels. (B) Quantification of AF cellularity by counting the number of nuclei on DAPI stained sections. (C) Quantification of AF width measured from Safranin O/Fast Green/Hematoxylin stained images. (D-G’) Representative immunofluorescence staining of caudal discs from WT and hTNFα-TG mice showing elevated immune cell staining of (D,D’) CD68 a phagocytic macrophage marker, (E,E’) Ly6 a neutrophil marker, and (F-G’) both CD4 and CD8 T-cell markers. Quantitative data was plotted as mean ± SD and differences between groups were analyzed using t-test. *** p ≤ 0.001, **** p ≤ 0.0001 Staining was performed on 5 animals/genotype and representative images are shown. Scale bar = 200μm.
We next interrogated the types of cells that were present outside of the caudal AF; cells were stained for immune cell markers: CD68 a phagocytic macrophage marker, lymphocyte antigen-6 (Ly6) a neutrophil marker, and both T-cell markers CD4 and CD8. In comparison with the AF of WT discs, which display one or two CD68 positive cells (Fig. 4 D, arrow), the hTNFα-TG discs exhibit robust CD68 positive cell staining adjacent to the AF (Fig. 4 D’). Widespread AF cell loss was particularly apparent on this image as was the loss of DAPI positive cells in the AF, and the nuclei adjacent to the AF were more dispersed and organized in a linear pattern giving the impression that they were immune cells infiltrating the organized AF collagen matrix (Fig. 4 D’, arrow).
As for the other cell markers, while there were no Ly6 immunopositive cells in the WT discs, there were pockets of Ly6 positive cells adjacent to the hTNFα-TG AF (Fig. 4 E, E’ arrows). Likewise, the WT AF sections were negative for both CD4 and CD8 positive cells, but some CD4 or CD8 positive cells were present in the hTNFα-TG AF cell infiltrate (Fig. 4 F-G’, arrows). The vigorous inflammatory response adjacent to the caudal AF of hTNFα-TG mice is primarily composed of phagocytic macrophages. While we did not establish a causation between immune activation and AF changes in this model, the immune response described herein occurred concurrently with AF cell loss and reduced AF width.
hTNFα-TG mice show no difference in AF collagens but have a reduction in cartilage oligomeric matrix protein
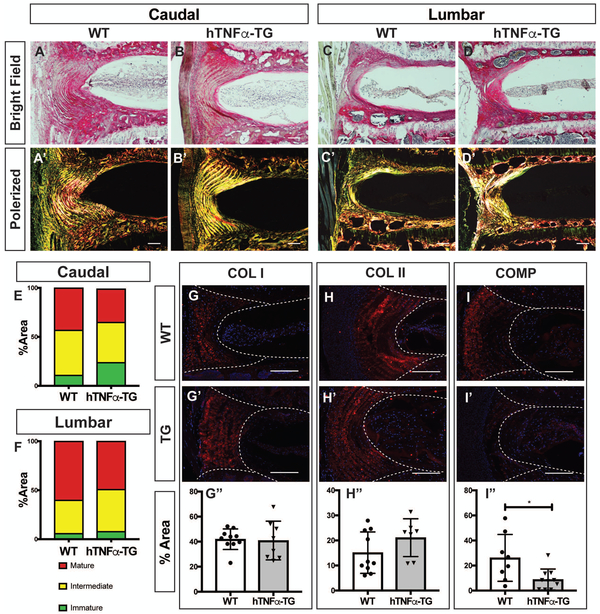
To determine the collagen content and fiber diameter of the fibrillar collagens, sections were stained with Picrosirius red and visualized under both bright field and polarized microscopy. Both caudal and lumbar discs showed strong collagen localization in the AF with a very weak NP pericellular staining (Fig. 5 A-D). Polarized light images of the AF exhibited strong red, yellow and green birefringence, an indicator of collagen fiber maturity, with no variation between genotypes (Fig. 5 A’-D’). To quantify the fiber maturity, the percent area occupied by green, yellow, or red fibers was evaluated to confirm that there was no difference in caudal or lumbar discs between the genotypes (Fig. 5E, F). We observed that both collagen I and collagen II were primarily localized in the AF in a fibril-morphology in the caudal discs of both WT and hTNFα-TG mice (Fig. 5 G-H’). There were no differences in the levels of collagen I or II (Fig. 5 G” and H”). Additionally, we measured the expression of cartilage oligomeric matrix protein (COMP), which helps maintain the structural integrity of the collagen fibrils. There was a significant decrease in COMP content in the AF of the hTNFα-TG discs (p = 0.0185) (Fig. 5 I-I”). There was also a significant decrease in aggrecan cleavage product ARGxx (SFig. 3 C-C”). While there are no changes in the collagen I or II content or maturity in the hTNFα-TG mouse AF compartment, the reduction in COMP suggests a potential reduction in matrix stability.
Figure 5: hTNFα-TG mice show no difference in AF collagens but have reduction in cartilage oligomeric matrix protein.
(A-D’) Picrosirius red staining of (A-B’) caudal and (C-D’) lumbar discs showing collagen organization in the annulus fibrosus. Collagen fibers visualized under polarized light (A’, B’, C’, and D’) show organized lamellae (scale bar 50 μm). (E, F) Quantification of the fiber content distribution in (E) caudal and (F) lumbar levels showing no significant difference in fiber maturity distribution (n = 5 animals/genotype). Representative images of immunofluorescence staining of disc and quantification showed comparable expression of (G) caudal collagen I (COL I), (H) caudal collagen II (COL II), and a reduction in (I) cartilage oligomeric matrix protein (COMP) levels of hTNFα-TG mice. Staining was performed on 5 animals/genotype and 1-2 levels per animal; A-D’ scale bar = 100μm; G-I’ scale bar = 200μm. Significance between fiber distribution was determined using χ2 test and differences between immunofluorescence staining data plotted as mean ± SD was analyzed using t-test. * p ≤ 0.05
Microarray analysis reveals minimal gene expression changes in the NP
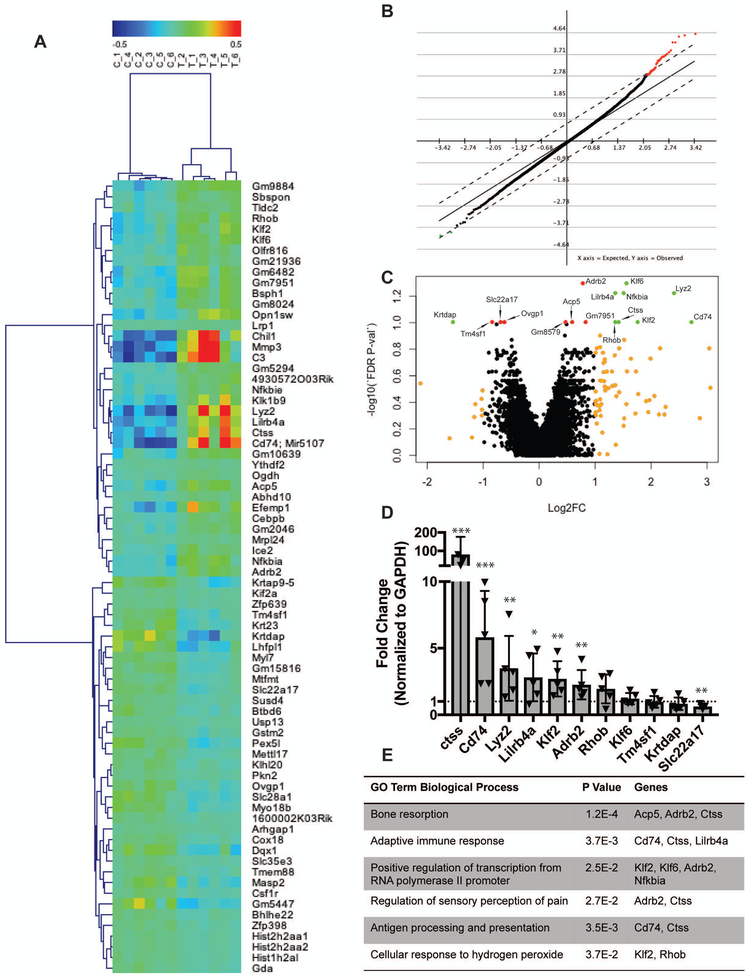
Microarray analysis of hTNFα-TG and WT NP showed only subtle changes in NP cell gene expression. Even with 6 animals/genotype analyzed, no genes were significantly different between the groups using a false discovery rate adjusted p-value <0.05. Taking this into consideration, we used multiple data analysis methods to get a sense of the subtle gene expression changes that did not rise to the level of statistical significance when factoring in adjustments due to false discovery rate. A heat map of the 74 genes with an unadjusted p-value <0.001 showed the clustering of the WT samples and hTNFα-TG samples into two distinct groups with more sample variation in the hTNFα-TG mice apparent in the dendrogram at the top of the heat map (Fig. 6 A). Both significance analysis of microarrays (SAM) with a delta value set to 0.74 for a FDR of 5% and using a FDR adjusted p-value <0.1, the volcano plot identified a similar small family of genes (Fig. 6 B and C). Along the phenotypes, PCR analysis confirmed that the following genes are significantly different: kruppel like factor 2 (klf2), cathepsin S (ctss), adrenoceptor beta 2 (adrb2), leukocyte immunoglobulin-like receptor, subfamily B (Lilrb4a), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha (Nfkbia), lysozyme C-2 precursor (Lyz2), cluster of differentiation 74 (Cd74), and solute carrier family 22 member 17 (Slc22a17) (Fig. 6 D). DAVID analysis of the gene list determined from FDR p-value <0.1, revealed a number of associated GO terms including bone resorption, adaptive immune response, positive regulation of transcription, regulation of sensory perception of pain, antigen processing and presentation, and cellular response to hydrogen peroxide (Fig. 6 E). While terms like bone resorption and sensory perception of pain were expected functions of TNFα, NP cells expressing genes for type II antigen processing and presentation was a unique finding. We confirmed the finding that NP cells are highly phagocytic and can internalize and cleave DQ Ovalbumin, a necessary step for type II antigen presentation (S Fig. 4). Additionally, vacuole was the top GO CC direct term, which echoes the vacuolated NP cells of the hTNFα-TG animals. Together microarray analysis confirmed that TNFα overexpression has little effect on the NP compartment, and the minimal effect is consistent with the changes shown in this model.
Figure 6: Microarray analyses reveals minimal gene expression changes in the NP of hTNFα-TG mice.
(A) A heat map showing gene and sample clustering of the genotypes in to two distinct groups with more distance between the hTNFα-TG samples than the WT controls. (B) Significance analysis of microarrays (SAM) with a delta value of 0.74 for an FDR of 5%. (C) Volcano plot showing genes with Log2 (fold change) greater than 1 or less than −1 in orange, genes with Log2 (fold change) greater than 1 or less than −1 and adjusted p value < 0.1 in green, and genes with adjusted p value <0.1 in red. (D) PCR verification of genes identified by microarray analysis with significance determine by t-test. (E) A table showing the DAVID identified significant GO biological terms associated with the gene expression changes identified by the microarray. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
DISCUSSION
There is a vast body of literature linking inflammatory cytokines with disc disease (12,15,31,32). However, our recent studies of a hTNFα overexpressing mouse model revealed no signs of disc degeneration after 16 weeks; in fact, the NP of these hTNFα overexpressing mice appeared healthier and more cellular than WT controls (14). However, it is unclear whether the nutrient limited NP compartment could support this increased cellularity and consequent metabolic demands over a longer time course (33). To ascertain whether a long-term TNFα challenge would sustain both a healthy NP and AF compartments we evaluated structural changes in discs of TNFα transgenic animals with less severe arthritic disease burden and a relatively longer lifespan than the Tg197 strain (34). This study showed that longer-term exposure to hTNFα caused changes in the AF characterized by cell death, altered collagen fibril structure, immune cell infiltration and macrophage-mediated loss of outer lamellae. Importantly, even with extensive AF damage, the NP maintained its cellularity, tissue morphology and matrix composition.
Systemic overexpression of hTNFα resulted in an increase in numerous blood cytokine concentrations: IL-1β, IL-2, KC/GRO, and MCP-1. Notably absent from this list was TNFα itself. The multiplex platform used to measure blood cytokine concentrations is mouse-specific and consequently does not reflect hTNFα levels in these mice, which should be around 2.5 pg/mL (34). Suppression of endogenous TNFα in the context of TNFα overexpression is expected based on previously published data examining the TNFα concentration of these mice in response to lipopolysaccharide (LPS) stimulation (34). Increased systemic cytokine concentrations, in particular TNFα, characterize conditions like obesity and smoking that are implicated in increasing the risk of disc disease associated LBP (35-38). Establishing a severe model of systemic inflammation thus allowed the evaluation of its effect on disc health.
In addition to the altered systemic cytokine profile, the overexpression of hTNFα resulted in alterations in vertebral bone structure including cortical and trabecular thinning. There is a well-established link between systemic inflammatory conditions and reduced bone mass for both smoking and diabetes (39,40). The state of the neighboring vertebral bone inflammation is pertinent to the discussion of degenerative disc disease: bone marrow inflammation with Modic changes, is highly correlated with painful disc degeneration (41,42). However, despite vertebral bone inflammation there were no corresponding changes in disc health. This finding reinforces the view that the disc is largely isolated from the neighboring bony inflammation (14).
Despite the robust and significant changes in both systemic cytokine profile and vertebral bone phenotype, the intervertebral discs in the hTNFα-TG mice remained largely unaffected, with comparable or healthier Modified Thompson scores in both regions of the spine. The NP cells in the lumbar hTNFα-TG levels occupied a larger proportion of the NP compartment than WT controls, however this trend did not appear to hold true in the caudal levels. Caudal NP cells of hTNFα-TG mice did not retain the vacuolated phenotype seen in the lumbar region. The change in apparent cell size mirrors the small changes of DHI described by μCT; there is a small decrease in DHI and apparent cell size in the caudal spine, and a small increase in DH and cell size in the lumbar spine. This result suggests that NP cell vacuoles may contribute to the DHI measurement. Cell size is particularly relevant to degenerative disc disease, because smaller chondrocyte-like cells are associated with aging and degeneration (6,7). The increase in cell band size in the lumbar region is consistent with a recent study involving overexpression of TNFα. Our previous studies showed that the cell band area was elevated together with increased size and vacuolar morphology of NP cells (14). The lack of vacuolar change in the caudal NP cells is likely secondary to the robust changes in the neighboring AF. While the basal TNFα overexpression does not affect NP cell health, the presence of this immune response could result in a more inflammatory environment inducing this phenotypic change.
Despite the lack of change in AF grade, it is important to note that the Modified Thompson Grading system only considers NP cellularity and neglects AF cellularity, focusing instead on AF clefts, buckling, and the mucinous infiltration characteristic of the loss of demarcation between the AF and the NP (43). Consequently, this grading system misses the most striking disc-related phenotypic effect of systemic hTNFα overexpression, AF cell death and thinning. AF cell death progression appeared to proceed from the outside to the inner layers of the tissue. Hence, there were few DAPI stained nuclei at the periphery of the AF; within the AF, a zone of TUNEL positive cells was evident, and deep to this zone, the cells are TUNEL negative. This out-to-in progression of cell death suggests that the AF cell death is in response to the invading immune cells and not nutrient deprivation.
The lack of this immune reaction in the lumbar discs is puzzling. Most likely, this reflects the difference in mechanical stress experienced by the caudal and lumbar component of the axial skeleton (44). Increased displacement in the caudal spine translates to elevated stress on the caudal discs leading to micro-fractures and an immune response in the caudal levels of the TNFα transgenic mice. The observation that COMP expression is low in the hTNFα-TG animals is of particular interest since this molecule has been shown to be protective against collagen-induced arthritis (45). Thus, decreased COMP would be expected to increase the susceptibility of the collagen-rich matrix to immune mediated degradation. Although there was clear loss of cellularity and loss of AF width, the fibrillar collagen content in the remaining AF was structurally similar to WT AF collagen. This similarity suggests that the immune response is not due to an intrinsic issue with the collagen itself, but it may be due to proteins organizing the AF collagen matrix.
The limited changes in hTNFα-TG NP gene expression identified by microarray analysis further reinforces the view that cytokine overexpression has a minimal effect on the health of the NP compartment. While increased cytokine expression within the NP compartment is typically considered pathological, it is important to acknowledge that TNFα is expressed in the juvenile human NP indicating that this cytokine may play a physiological role in disc homeostasis (46). TNFα may be involved in maintaining cell viability and matrix homeostasis in the physiologically challenging disc niche that is hypoxic, hyperosmolar, and under mechanical stress (47). Earlier work from our group found that TNFα expression is maintained in the post-natal NP cells by tonicity-responsive binding-protein (TonEBP); this transcription factor controls cell adaptive responses in the hyperosmolar disc environment likely in part by regulating the expression of pro-survival and anabolic matrix genes (20,48,49). Additionally, complete removal of IL-1β, another cytokine closely related to disc degeneration, renders mice more susceptible to age-related disc degeneration, further reinforcing the importance of the physiological role that certain cytokines play in the preservation of disc health (50). Of the genes that did change in the hTNFα-TG animals, two were related to antigen presentation. This is a particularly intriguing finding considering that MHC type II antigen presentation is typically restricted to a small population of immune cells. However, NP cells possess the ability of internalize and cleave external material as reported before and by our present study. This process could represent a novel way for NP cells to communicate with each other or keep the disc space free of cellular debris; however, much more work is necessary to adequately explore this idea.
In summary, findings from this investigation confirm that AF health and integrity is pivotal to disc health. Clinical research supports the idea; a longitudinal study of adults looking at the association between AF tears and degenerative disc disease found that annular tears likely occur early in the course of disc disease and speed NP degeneration (51). A second study of a pediatric population found that only 30 percent of discs with radial annular tears had intact discs while only 3 percent of degenerate discs had an intact AF (52). While systemic cytokine expression did not directly induce NP degeneration, the loss of vertebral bone mass and thinning of the AF suggest that systemic inflammation may weaken the integrity of the AF-EP junction predisposing these discs to herniation (53). The findings presented herein further support the hypothesis that the NP compartment is isolated from systemic inflammation and the immune system; and importantly, any procedure that compromises AF integrity should be avoided.
Supplementary Material
Supplementary Figure 1: Details of the PCR primers used in this study.
Supplementary Figure 2: RT-PCR results confirming the presence of and responsiveness to hTNFα in murine NP cells. (A) RT-PCR analysis of mTNFα and hTNFα in NP mRNA isolated from WT control and hTNFα-TG mice. Presence of hTNFα mRNA was seen only in the hTNFα-TG mice. TNFα expression was measured from 3 mice/genotype. (B) RT-PCR analysis of NP tissue isolated from organ cultured disc motion segments from WT mice treated with hTNFα for 24h. hTNFα treatment resulted in upregulation of MMP3 and CCL2 mRNA along with marked downregulation of COL2A1 confirming responsiveness of mouse disc tissue to hTNFα. Organ culture experiment was repeated 3 independent times, n =3 mice/group, 6 lumbar and 9 caudal discs/mouse. The data are plotted as mean ± SD and analyzed using t-test, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
Supplementary Figure 3: hTNFα-TG and WT mice showed comparable proteoglycan content, but a reduction in ARGxx. (A-C”) Representative immunofluorescence staining images of discs from WT and hTNFα-TG mice and their quantification showing comparable expression levels of (A-A”) aggrecan (ACAN) and (B-B”) Chondortin sulfate (CS), but a hTNFα-TG mice show a reduction (p = 0.0424) in aggrecan cleavage product (C-C”) ARGxx. (n = 5 animals/genotype and at least 2 levels per animal; scale bar = 200 μm). Differences between immunofluorescence staining data plotted as mean ± SD and analyzed using t-test. * p ≤ 0.05
Supplementary Figure 4: Preliminary data suggesting NP cells phagocytose and process protein antigens. (A) Images showing DAPI stained rat NP cells with internalized florescent beads after 24h of incubation. (B) Quantification of the number of beads-per-cell. (C) Emission intensity of cleaved DQ Ovalbumin internalized and cleaved by NP cells in vitro. Data plotted as mean ± SD. Differences between experimental groups were analyzed by one-way ANOVA* p ≤ 0.05, ** p ≤ 0.01, **** p ≤ 0.0001.
ACKNOWLEDGMENTS
This study was funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health under Award Numbers AR055655, AR064733, AR074813, T32 AR052273, and F30AR071256. We thank Sidney Kimmel Cancer Center Cancer Genomics Facility of Thomas Jefferson University, Philadelphia for help with generating microarray data.
Footnotes
Conflicts of Interest: None for all authors.
REFERENCES
- 1.Hoy D, Bain C, Williams G, March L, Brooks P, Blyth F, et al. A systematic review of the global prevalence of low back pain. Arthritis Rheum. 2012;64(6):2028–37. [DOI] [PubMed] [Google Scholar]
- 2.Freburger JK, Holmes GM, Agans RP, Jackman AM, Darter JD, Wallace AS, et al. The rising prevalence of chronic low back pain. Arch Intern Med. 2009;169(3):251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deyo RA, Mirza SK, Martin BI. Back Pain Prevalence and Visit Rates. Spine. 2006;31(23):2724–7. [DOI] [PubMed] [Google Scholar]
- 4.Arnbak B, Jensen TS, Egund N, Zejden A, Hørslev-Petersen K, Manniche C, et al. Prevalence of degenerative and spondyloarthritis-related magnetic resonance imaging findings in the spine and sacroiliac joints in patients with persistent low back pain. Eur Radiol. 2015;26(4):1191–203. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Xiong C, Kudelko M, Li Y, Wang C, Wong YL, Tam V, Rai MF, Cheverud J, Lawson HA, Sandell L, Chan WCW, Cheash KSE, Sham PC, Chan D. Early onset of disc degeneration in SM/J mice is associated with changes in ion transport systems and fibrotic events. Matrix Biol. 2018; 70:123–139. [DOI] [PubMed] [Google Scholar]
- 6.Choi H, Tessier S, Silagi ES, Kyada R, Yousefi F, Pleshko N, et al. A novel mouse model of intervertebral disc degeneration shows altered cell fate and matrix homeostasis. Matrix Biol. 2018;70:102–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohanty S, Pinelli R, Pricop P, Albert TJ, Dahia CL. Chondrocyte-like nested cells in the aged intervertebral disc are late-stage nucleus pulposus cells. Aging Cell 2019; 18(5):e13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther. 2007;9(4):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson ZI, Doolittle AC, Snuggs JW, Shapiro IM, Le Maitre CL, Risbud M V. TNF-α promotes nuclear enrichment of the transcription factor TonEBP/NFAT5 to selectively control inflammatory but not osmoregulatory responses in nucleus pulposus cells. J Biol Chem. 2017;292(42):17561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wang H, Yang H, Li J, Cai Q, Shapiro IM, et al. Tumor Necrosis Factor-α- and Interleukin-1β-Dependent Matrix Metalloproteinase-3 Expression in Nucleus Pulposus Cells Requires Cooperative Signaling via Syndecan 4 and Mitogen-Activated Protein Kinase-Nuclear Factor κB Axis: Implications in Inflammatory D. Am J Pathol. 2014;184(9):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Tian Y, Phillips KLE, Chiverton N, Haddock G, Bunning RA, et al. Tumor necrosis factor α- and interleukin-1β-dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis Rheum. 2013;65(3):832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Risbud M V, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keffer J, Probert L, Cazlaris H, Georgopoulos S, Kaslaris E, Kioussis D, et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 1991;10(13):4025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorth DJ, Shapiro IM, Risbud M V. Transgenic mice overexpressing human TNF-α experience early onset spontaneous intervertebral disc herniation in the absence of overt degeneration. Cell Death Dis. 2019;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips KLELE, Cullen K, Chiverton N, Michael ALRLR, Cole AAA, Breakwell LMM, et al. Potential roles of cytokines and chemokines in human intervertebral disc degeneration: interleukin-1 is a master regulator of catabolic processes. Osteoarthr Cartil. 2015;23(7):1165–77. [DOI] [PubMed] [Google Scholar]
- 16.Markova DZ, Kepler CK, Addya S, Murray HB, Vaccaro AR, Shapiro IM, et al. An organ culture system to model early degenerative changes of the intervertebral disc II: profiling global gene expression changes. Arthritis Res Ther. 2013;15(5):R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purmessur D, Walter BA, Roughley PJ, Laudier DM, Hecht AC, Iatridis J. A role for TNFα in intervertebral disc degeneration: a non-recoverable catabolic shift. Biochem Biophys Res Commun. 2013;433(1):151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward MD, Jones BK, Saparov A, Hain HS, Trillat A-C, Bunzel MM, et al. An extensive phenotypic characterization of the hTNFalpha transgenic mice. BMC Physiol. 2007;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob CO, Lee SK, Strassmann G. Mutational analysis of TNF-alpha gene reveals a regulatory role for the 3’-untranslated region in the genetic predisposition to lupus-like autoimmune disease. J Immunol. 1996;156(8):3043–50. [PubMed] [Google Scholar]
- 20.Johnson ZI, Shapiro IM, Risbud MV. RNA Sequencing Reveals a Role of TonEBP Transcription Factor in Regulation of Pro-inflammatory Genes in Response to Hyperosmolarity in Healthy Nucleus Pulposus Cells. J Biol Chem. 2016;291(52):26686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox KM, Kimura S, Powell-Threets K, Plato CC. Radial and ulnar cortical thickness of the second metacarpal. J Bone Miner Res. 2009;10(12):1930–4. [DOI] [PubMed] [Google Scholar]
- 22.Tajerian M, Alvarado S, Millecamps M, Dashwood T, Anderson KM, Haglund L, et al. DNA methylation of SPARC and chronic low back pain. Mol Pain. 2011;7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IK, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine. 1990;15(5):411–5. [DOI] [PubMed] [Google Scholar]
- 24.McCann MR, Patel P, Pest MA, Ratneswaran A, Lalli G, Beaucage KL, et al. Repeated exposure to high-frequency low-amplitude vibration induces degeneration of murine intervertebral discs and knee joints. Arthritis Rheumatol. 2015;67(8):2164–75. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker P, Kloner RA, Boughner DR, Pickering JG. Quantitative assessment of myocardial collagen with picrosirius red staining and circularly polarized light. Basic Res Cardiol. 1994;89(5):397–410. [DOI] [PubMed] [Google Scholar]
- 26.Steplewski A, Fertala J, Beredjiklian PK, Abboud JA, Wang MLY, Namdari S, et al. Blocking collagen fibril formation in injured knees reduces flexion contracture in a rabbit model. J Orthop Res. 2017;35(5):1038–46. [DOI] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novais EJ, Diekman BO, Shapiro IM, Risbud M V. p16Ink4a deletion in cells of the intervertebral disc affects their matrix homeostasis and senescence associated secretory phenotype without altering onset of senescence. Matrix Biol. 2019;82:54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessier S, Tran VA, Ottone OK, Novais EJ, Doolittle A, DiMuzio MJ, Shapiro, IM, Risbud MV. TonEBP-deficciency accelerates intervertebral disc degeneration underscored by matrix remodeling, cytoskeletal rearrangements, and changes in proinflammatory gene expression. Matrix Biol. 2019. (Accepted). S0945–053X(19)30387–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, et al. Nucleus pulposus cells express HIF-1α under normoxic culture conditions: A metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98(1):152–9. [DOI] [PubMed] [Google Scholar]
- 31.Phillips KLE, Jordan-Mahy N, Nicklin MJH, Le Maitre CL. Interleukin-1 receptor antagonist deficient mice provide insights into pathogenesis of human intervertebral disc degeneration. Ann Rheum Dis. 2013;72(11):1860–7. [DOI] [PubMed] [Google Scholar]
- 32.Johnson ZI, Schoepflin ZR, Choi H, Shapiro IM, Risbud M V. Disc in flames: Roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104–16; discussion 116–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urban JPG, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5(3):120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayward MD, Jones BK, Saparov A, Hain HS, Trillat A-C, Bunzel MM, et al. An extensive phenotypic characterization of the hTNFalpha transgenic mice. BMC Physiol. 2007;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dario A, Ferreira M, Refshauge K, Harmer A, Sánchez-Romera J, Pérez-Riquelme F, et al. Mapping the association between back pain and type 2 diabetes: A cross-sectional and longitudinal study of adult Spanish twins. PLoS One. 2017;12(4):e0174757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased Adipose Tissue Expression of Tumor Necrosis Factor-α in Human Obesity and Insulin Resistance. J Clin Invest. 1995;95:2409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battié MC, Videman T, Kaprio J, Gibbons LE, Gill K, Manninen H, et al. The Twin Spine Study: Contributions to a changing view of disc degeneration. Spine J. 2009;9(1):47–59. [DOI] [PubMed] [Google Scholar]
- 38.Petrescu F, Voican SC, Silosi I. Tumor necrosis factor-alpha serum levels in healthy smokers and nonsmokers. Int J Chron Obstruct Pulmon Dis. 2010;5:217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghadimi R, Hosseini S, Asefi S, Bijani A, Heidari B, Babaei M. Influence of smoking on bone mineral density in elderly men. Int J Prev Med. 2018;9(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundararaghavan V, Mazur MM, Evans B, Liu J, Ebraheim NA. Diabetes and bone health: latest evidence and clinical implications. Ther Adv Musculoskelet Dis. 2017;9(3):67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modic MT, Steinberg PM, Ross JS, Masaryk TJ, Carter JR. Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology. 1988;166(1 Pt 1):193–9. [DOI] [PubMed] [Google Scholar]
- 42.Jensen TS, Karppinen J, Sorensen JS, Niinimäki J, Leboeuf-Yde C. Vertebral endplate signal changes (Modic change): A systematic literature review of prevalence and association with non-specific low back pain. Eur Spine J. 2008;17(11):1407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutges JPHJ, Duit RA, Kummer JA, Bekkers JEJ, Oner FC, Castelein RM, et al. A validated new histological classification for intervertebral disc degeneration. Osteoarthr Cartil. 2013;21(12):2039–47. [DOI] [PubMed] [Google Scholar]
- 44.Sarver JJ, Elliott DM. Mechanical differences between lumbar and tail discs in the mouse. J Orthop Res. 2005;23(1):150–5. [DOI] [PubMed] [Google Scholar]
- 45.Geng H, Carlsen S, Nandakumar K, Holmdahl R, Aspberg A, Oldberg Å, et al. Cartilage oligomeric matrix protein deficiency promotes early onset and the chronic development of collagen-induced arthritis. Arthritis Res Ther. 2008;10(6):R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiler C, Nerlich AG, Bachmeier BE, Boos N. Expression and distribution of tumor necrosis factor alpha in human lumbar intervertebral discs: a study in surgical specimen and autopsy controls. Spine. 2005;30(1):44–53. [DOI] [PubMed] [Google Scholar]
- 47.Risbud MV, Schipani E, Shapiro IM. Hypoxic Regulation of Nucleus Pulposus Cell Survival. Am J Pathol. 2010;176(4):1577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai T-T, Danielson KG, Guttapalli A, Oguz E, Albert TJ, Shapiro IM, et al. TonEBP/OREBP Is a Regulator of Nucleus Pulposus Cell Function and Survival in the Intervertebral Disc. J Biol Chem. 2006;281(35):25416–24. [DOI] [PubMed] [Google Scholar]
- 49.Choi H, Chaiyamongkol W, Doolittle AC, Johnson ZI, Gogate SS, Schoepflin ZR, et al. COX-2 expression mediated by calcium-TonEBP signaling axis under hyperosmotic conditions serves osmoprotective function in nucleus pulposus cells. J Biol Chem. 2018;293(23):8969–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorth DJ, Shapiro IM, Risbud M V. A new understanding of the role of IL‐1 in age‐related intervertebral disc degeneration in a murine model. J Bone Miner Res. 2019;34(8):1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma A, Pilgram T, Wippold FJ, Wippold Ii FJ. Association between Annular Tears and Disk Degeneration: A Longitudinal Study. Am J Neuroradiol. 2009;30(3):500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma A, Parsons MS, Pilgram TK. Temporal association of annular tears and nuclear degeneration: lessons from the pediatric population. AJNR Am J Neuroradiol. 2009;30(8):1541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajasekaran S, Bajaj N, Tubaki V, Kanna RM, Shetty AP. ISSLS Prize Winner. Spine. 2013;38(17):1491–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Details of the PCR primers used in this study.
Supplementary Figure 2: RT-PCR results confirming the presence of and responsiveness to hTNFα in murine NP cells. (A) RT-PCR analysis of mTNFα and hTNFα in NP mRNA isolated from WT control and hTNFα-TG mice. Presence of hTNFα mRNA was seen only in the hTNFα-TG mice. TNFα expression was measured from 3 mice/genotype. (B) RT-PCR analysis of NP tissue isolated from organ cultured disc motion segments from WT mice treated with hTNFα for 24h. hTNFα treatment resulted in upregulation of MMP3 and CCL2 mRNA along with marked downregulation of COL2A1 confirming responsiveness of mouse disc tissue to hTNFα. Organ culture experiment was repeated 3 independent times, n =3 mice/group, 6 lumbar and 9 caudal discs/mouse. The data are plotted as mean ± SD and analyzed using t-test, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
Supplementary Figure 3: hTNFα-TG and WT mice showed comparable proteoglycan content, but a reduction in ARGxx. (A-C”) Representative immunofluorescence staining images of discs from WT and hTNFα-TG mice and their quantification showing comparable expression levels of (A-A”) aggrecan (ACAN) and (B-B”) Chondortin sulfate (CS), but a hTNFα-TG mice show a reduction (p = 0.0424) in aggrecan cleavage product (C-C”) ARGxx. (n = 5 animals/genotype and at least 2 levels per animal; scale bar = 200 μm). Differences between immunofluorescence staining data plotted as mean ± SD and analyzed using t-test. * p ≤ 0.05
Supplementary Figure 4: Preliminary data suggesting NP cells phagocytose and process protein antigens. (A) Images showing DAPI stained rat NP cells with internalized florescent beads after 24h of incubation. (B) Quantification of the number of beads-per-cell. (C) Emission intensity of cleaved DQ Ovalbumin internalized and cleaved by NP cells in vitro. Data plotted as mean ± SD. Differences between experimental groups were analyzed by one-way ANOVA* p ≤ 0.05, ** p ≤ 0.01, **** p ≤ 0.0001.