Abstract
Radiomics has potential for reflecting the differences in glioma perfusion heterogeneity between arterial spin labeling (ASL) and dynamic susceptibility contrast (DSC) imaging. The aim of this study was to compare radiomic features of ASL and DSC imaging-derived parameters (cerebral blood flow, CBF) and assess radiomics-based classification models for low-grade gliomas (LGGs) and high-grade gliomas (HGGs) using their parameters. The ASL-CBF and DSC-relative CBF of 46 glioma patients were normalized (ASL-nCBF and DSC-nrCBF) for data analysis. For each map, 91 radiomic features were extracted from the tumor volume. Seventy-five radiomic features were significantly different (P < 0.00055) between ASL-nCBF and DSC-nrCBF. Positive correlations were observed in 75 radiomic features between ASL-nCBF and DSC-nrCBF. Even though ASL imaging underestimated CBF compared with DSC imaging, there were significant correlations (P < 0.00055) in the first-order-based mean, median, 90th percentile, and maximum. Texture analysis showed that ASL-nCBF and DSC-nrCBF characterized similar perfusion patterns, while ASL-nCBF could evaluate perfusion heterogeneity better. The areas under the curve of the ASL-nCBF and DSC-nrCBF radiomics-based classification models for gliomas were 0.888 and 0.962, respectively. Radiomics in ASL and DSC imaging is useful for characterizing glioma perfusion patterns quantitatively and for classifying LGGs and HGGs.
Subject terms: CNS cancer, Tumour heterogeneity, Magnetic resonance imaging
Introduction
Angiogenesis, the formation of new blood vessels for tumor growth, plays a key role in gliomas1. Gliomas rely on angiogenesis to maintain an adequate blood supply for the delivery of nutrients and oxygen, thereby consisting of a complex and heterogeneous vasculature1,2. Therefore, high-grade gliomas (HGGs) are characterized by heterogeneous and relatively higher perfusion compared with low-grade gliomas (LGGs)3. These characteristics are critical elements in the determination of preoperative grade, treatment strategy, and prognosis of gliomas.
Magnetic resonance (MR) perfusion-weighted imaging can provide information about tumor vascularity. Cerebral blood flow (CBF) and cerebral blood volume (CBV) are representative perfusion parameters that can be measured by MR perfusion-weighted imaging. Dynamic susceptibility contrast (DSC) imaging is an established MR perfusion-weighted imaging4. DSC imaging requires a bolus injection of a gadolinium-based (exogenous) contrast agent. A deconvolution method is used for the quantification analysis of relative CBF (rCBF) and mean transit time (MTT)4. Relative CBV (rCBV) is related to rCBF and MTT via the equation MTT = rCBV/rCBF, and can be estimated from the area of the time–concentration curve in tissue4. DSC imaging-derived parameters (rCBF and rCBV) are known to correlate with histological findings of glioma angiogenesis5,6 and glioma grade7–9.
On the other hand, arterial spin labeling (ASL) imaging, which is another MR perfusion-weighted imaging, does not require an exogenous contrast agent10. ASL imaging allows absolute quantification of CBF by using magnetically labeled arterial blood water as an endogenous tracer10. Some studies have compared ASL with DSC imaging in brain tumors11–14. These studies concluded that ASL imaging can be an alternative method to DSC imaging for evaluating brain tumors. The usefulness of ASL imaging for differentiation between LGGs and HGGs has also been reported15–18. Thus, ASL imaging is establishing itself as a non-invasive perfusion measurement without a contrast agent and X-ray exposure, making it valuable for renal failure19 and pediatric patients20.
Radiomics can describe tumor phenotypic characteristics using various quantitative features based on histogram and texture in medical images21,22. Recently, several studies have reported that radiomic features in MR imaging reflect tumor heterogeneity and have potential for predicting glioma grading23,24. We assumed that radiomics has the potential for quantitative assessment of glioma perfusion heterogeneity in ASL and DSC imaging. However, because ASL and DSC imaging are based on different techniques, it is unclear whether they exhibit similar perfusion patterns (e.g., heterogeneity) or how their perfusion patterns differ. In order to use ASL imaging as an alternative to DSC imaging, it is necessary to investigate this issue. Therefore, the purpose of this study was to compare radiomic features of ASL and DSC imaging in gliomas. Additionally, we constructed radiomics-based classification models for glioma grading using ASL and DSC imaging and assessed their diagnostic performance.
Results
Glioma patients
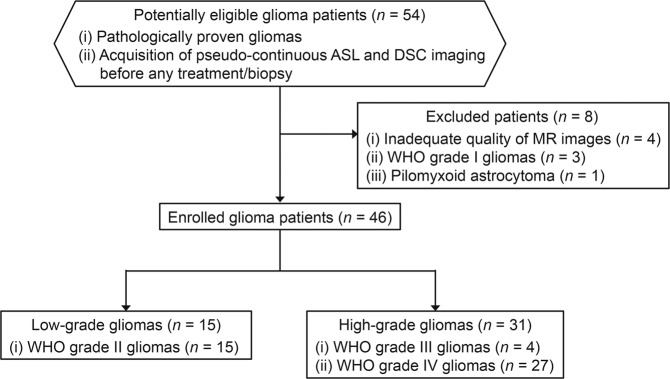
Figure 1 shows the flow diagram for glioma patient selection in this study. Among the 46 gliomas enrolled, 15 gliomas were LGGs (World Health Organization (WHO) grade II; 11 males and 4 females; mean age, 41.5 ± 12.6 years), and 31 gliomas were HGGs (WHO grade III/IV; 25 males and 6 females; mean age, 57.7 ± 16.7 years). All LGGs were WHO grade II (diffuse astrocytoma = 11, oligodendroglioma = 4). The HGGs included were WHO grade III (anaplastic astrocytoma = 3, anaplastic oligodendroglioma = 1) and IV (glioblastoma = 26, diffuse midline glioma = 1).
Figure 1.
Flow diagram for glioma patient selection. ASL, arterial spin labeling; DSC, dynamic susceptibility contrast; MR, magnetic resonance; WHO, World Health Organization.
Comparisons of radiomic features between ASL and DSC imaging
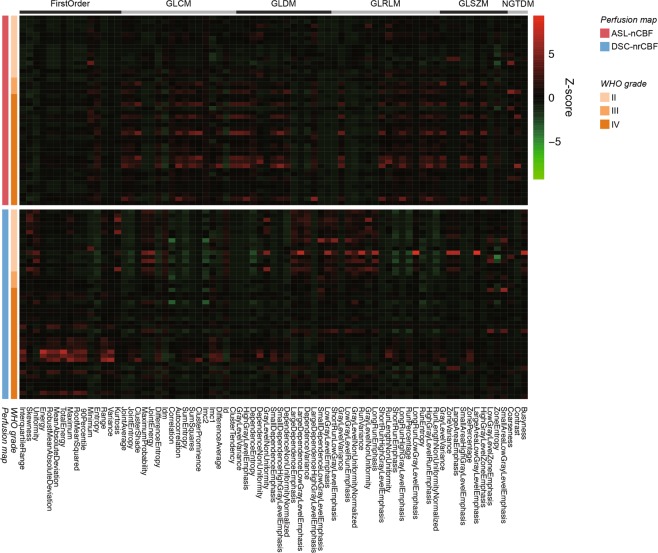
All 91 radiomic features (Supplementary Table S1) in all gliomas were compared between ASL-normalized CBF (ASL-nCBF) and DSC-normalized rCBF (DSC-nrCBF) using the paired t-test or Wilcoxon signed-rank test as appropriate. Figure 2 shows the heatmap of the 75 radiomic features that were significantly different (P < 0.00055). As shown in Fig. 2 and Supplementary Table S2, 15 out of the 18 first-order-based features (83.3%), including the minimum (ASL-nCBF, 0.49 ± 0.31; DSC-nrCBF, 0.13 ± 0.26), 90th percentile (ASL-nCBF, 3.15 ± 1.51; DSC-nrCBF, 5.64 ± 4.75), and maximum (ASL-nCBF, 5.30 ± 2.83; DSC-nrCBF, 24.41 ± 21.77), had significant differences (P < 0.00055) between ASL-nCBF and DSC-nrCBF. Sixty out of the 73 texture features (82.2%), including features from all 5 texture classes, had significant differences (P < 0.00055) between ASL-nCBF and DSC-nrCBF. The mean and standard deviation of all radiomic features and statistical results are shown in Supplementary Table S2.
Figure 2.
Heatmap of radiomic features for comparing between ASL-nCBF and DSC-nrCBF. All the 75 radiomic features shown in the heatmap were found to be significantly different (P < 0.00055) between ASL-nCBF and DSC-nrCBF. Rows correspond to patients, and columns correspond to Z-score normalized radiomic features. The heatmap is grouped by perfusion map, glioma grade, and radiomic feature class. ASL-nCBF, arterial spin labeling normalized cerebral blood flow; DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow; WHO, World Health Organization; GLCM, gray-level co-occurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size-zone matrix; NGTDM, neighboring gray-tone difference matrix.
Correlations of radiomic features between ASL and DSC imaging
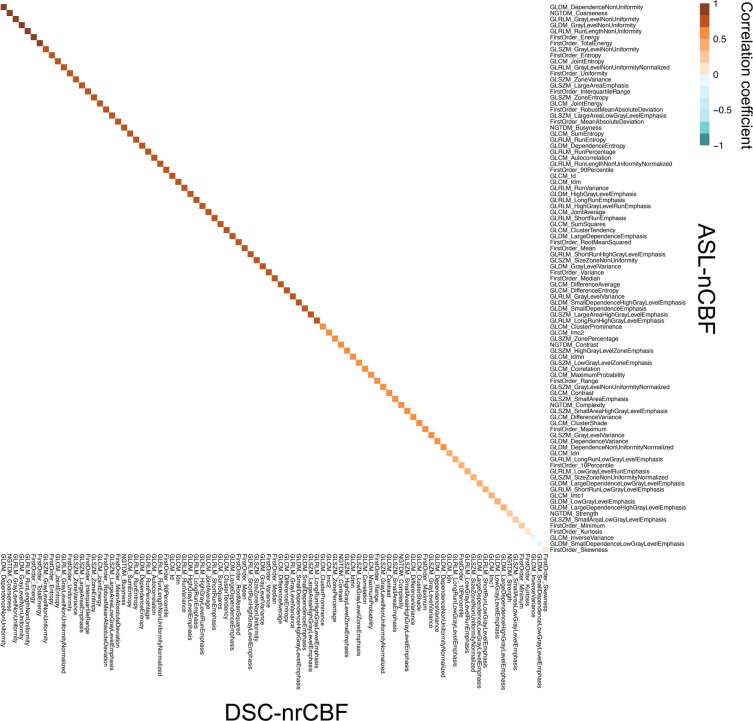
Figure 3 shows the heatmap of Pearson’s product-moment correlation coefficients (r) or Spearman’s rank-order correlation coefficients (ρ) of all radiomic features between ASL-nCBF and DSC-nrCBF in all gliomas. Significant correlations (P < 0.00055) were observed in 14 first-order-based features (14/18, 77.8%) and 61 texture features (61/73, 83.6%). The detailed results of the correlation analysis can be found in Supplementary Table S3.
Figure 3.
Heatmap of correlation coefficients between ASL-nCBF and DSC-nrCBF radiomic features. Brown and blue colors represent positive and negative correlations, respectively. The correlation coefficients are arranged in order from the highest significant correlation in the upper left corner, and the corresponding features are shown in the x- and y-axes. Significant correlations (P < 0.00055) were observed in the top 75 radiomic features. ASL-nCBF, arterial spin labeling normalized cerebral blood flow; DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow; GLCM, gray-level co-occurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size-zone matrix; NGTDM, neighboring gray-tone difference matrix.
Radiomics-based classification models for LGGs and HGGs
Logistic regression models were constructed using the radiomic features selected by least absolute shrinkage and selection operator (LASSO) and Eq. (3) in the Materials and methods section, as follows:
| 1 |
| 2 |
where GLCM, GLDM, GLRLM, and GLSZM are the gray-level co-occurrence matrix, gray-level dependence matrix, gray-level run-length matrix, and gray-level size-zone matrix, respectively.
According to the receiver operating characteristic (ROC) analysis (Fig. 4 and Table 1), the DSC-nrCBF model (AUC, 0.962; sensitivity, 89.3%; specificity, 92.9%) showed better diagnostic performance for differentiating HGGs from LGGs than the ASL-nCBF model (AUC, 0.888; sensitivity, 85.7%; specificity, 85.7%). However, no significant differences (P = 0.133) were found between the areas under the curve (AUCs) of the models.
Figure 4.
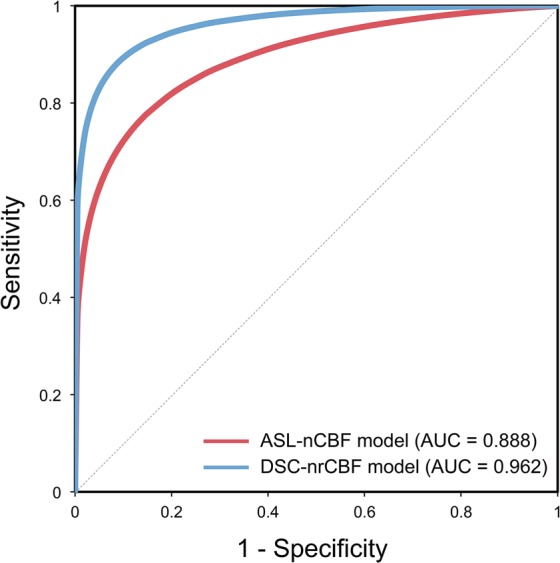
Receiver operating characteristic curves for the radiomics-based classification models for low- and high-grade gliomas. The red and blue solid lines represent the curves of the ASL-nCBF and DSC-nrCBF models, respectively. ASL-nCBF, arterial spin labeling normalized cerebral blood flow; DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow; AUC, area under the curve.
Table 1.
Receiver operating characteristic analysis results of radiomics-based classification models for low- and high-grade gliomas.
| AUC (95% confidence interval) | Significance (P-value) | Cut-off | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|
| ASL-nCBF model | 0.888 (0.783–0.993) | <0.0001 | 0.517 | 85.7 | 85.7 |
| DSC-nrCBF model | 0.962 (0.913–1.000) | <0.0001 | 0.556 | 89.3 | 92.9 |
AUC, area under the curve; ASL-nCBF, arterial spin labeling normalized cerebral blood flow; DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow.
The performance of the radiomics-based classification models was further evaluated using the test set (n = 4). Both the ASL-nCBF and DSC-nrCBF models correctly predicted the class (LGG or HGG) of all patients included in the test set (Tables 2 and 3). Furthermore, the cut-off values determined from the ROC analysis could also correctly differentiate the LGG and HGG patients.
Table 2.
Prediction results of the ASL-nCBF classification model for low- and high-grade gliomas.
| Test data # (n = 4) | #1 (Diffuse astrocytoma, grade II) | #2 (Anaplastic oligodendroglioma, grade III) | #3 (Glioblastoma, grade IV) | #4 (Glioblastoma, grade IV) |
|---|---|---|---|---|
| logit(p)ASL-nCBFa | −0.411 | 0.673 | 1.897 | 1.371 |
| Probability (p)b | 0.399 | 0.662 | 0.870 | 0.797 |
| Predicted class | LGG | HGG | HGG | HGG |
aCut-off value: 0.517; bp: probability of a patient with high-grade glioma (0 < p < 1); ASL-nCBF, arterial spin labeling normalized cerebral blood flow; LGG, low-grade glioma; HGG, high-grade glioma.
Table 3.
Prediction results of the DSC-nrCBF classification model for low- and high-grade gliomas.
| Test data # (n = 4) | #1 (Diffuse astrocytoma, grade II) | #2 (Anaplastic oligodendroglioma, grade III) | #3 (Glioblastoma, grade IV) | #4 (Glioblastoma, grade IV) |
|---|---|---|---|---|
| logit(p)DSC-nrCBFa | −2.379 | 0.829 | 5.154 | 1.106 |
| Probability (p)b | 0.085 | 0.696 | 0.994 | 0.751 |
| Predicted class | LGG | HGG | HGG | HGG |
aCut-off value: 0.556; bp: probability of a patient with high-grade glioma (0 < p < 1); DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow; LGG, low-grade glioma; HGG, high-grade glioma.
Discussion
This study compared 91 radiomic features in gliomas between ASL-nCBF and DSC-nrCBF. To our knowledge, this is the first study focusing on the comparison between ASL- and DSC-derived perfusion parameters in gliomas using radiomics. Compared with DSC imaging, ASL imaging underestimates CBF and better characterizes perfusion heterogeneity. However, there are a great number of radiomic features with positive correlations between ASL-nCBF and DSC-nrCBF. Moreover, the radiomics-based classification models showed high diagnostic performance for differentiating between LGGs and HGGs.
The main first-order statistics, including the mean, median, 90th percentile, and maximum values of ASL-nCBF tended to be lower than those of DSC-nrCBF. This could be explained from the fact that ASL tends to underestimate CBF due to increased MTT (e.g., complex and heterogeneous vasculature in tumors or low blood flow in white matter regions)11,13. Our ASL-CBF and DSC-rCBF maps were normalized to normal-appearing white matter (NAWM). Previous studies also reported that ASL-CBF tends to be lower than DSC-rCBF when normalized to NAWM11,12. Additionally, DSC imaging tends to overestimate rCBF because of its high sensitivity to large vessels25. In this study, large vessels were excluded from the tumor volume by using a semi-automated segmentation method. However, intratumoral microvessels might not have been removed completely. This could explain the hyperperfusion observed in DSC imaging, particularly in the maximum values.
The second- or higher-order statistics has been used for the assessment of texture pattern in brain tumors23,24,26. The texture analysis in our study included the GLCM-, GLDM-, GLRLM-, GLSZM-, and neighboring gray-tone difference matrix (NGTDM)-based features. The 82.2% texture features that were significantly different (P < 0.00055) between ASL-nCBF and DSC-nrCBF mainly point that ASL-nCBF is more heterogeneous (GLCM-based Idm, GLCM-based Id, GLDM-based gray-level variance, GLDM-based large dependence emphasis, and GLRLM-based run entropy) with greater variations in neighboring voxel values (GLCM-based joint entropy and GLCM-based sum entropy) than DSC-nrCBF. Furthermore, the GLDM-based high gray-level emphasis and GLRLM-based high gray-level run emphasis suggest that ASL-nCBF consists of a greater proportion of high voxel values in the segmented tumor region, while the GLDM-based low gray-level emphasis, GLRLM-based low gray-level run emphasis, and GLSZM-based low gray-level zone emphasis suggest that ASL-nCBF consists of a smaller proportion of low voxel values. These findings are consistent with the trend observed in our first-order-based features, such as entropy, uniformity, and skewness.
Intratumoral heterogeneity is an important predictor of tumor prognosis27. Preclinical studies in ASL imaging have demonstrated that CBF exhibits intratumoral heterogeneity in rat and mouse glioma models28,29. The present study revealed that ASL-nCBF is more heterogeneous than DSC-nrCBF. The discrepancy in perfusion patterns between ASL and DSC imaging may be attributed to their principles and methodology. ASL measures CBF by using labeled arterial water as a freely diffusible tracer, which distributes across capillary membranes throughout the brain tissue11 and is not affected by the destruction of the blood–brain barrier (BBB)10. On the contrary, DSC imaging utilizes an exogenous contrast agent as a non-diffusible tracer and is sensitive to the BBB destruction30. ASL imaging more accurately reflects the physiological perfusion of tissues and can objectively evaluate the degree of angiogenesis and malignancy in brain tumors due to the different diffusivity of the tracers14,31. These suggest that ASL-nCBF reflects glioma perfusion heterogeneity better than DSC-nrCBF. Therefore, ASL imaging has potential for characterizing perfusion heterogeneity in gliomas.
MR perfusion-weighted imaging studies have reported that ASL-CBF is positively correlated with DSC-rCBF and DSC-rCBV in brain tumors11–13. For the first-order-based mean in this study, there was a strong positive correlation between ASL-nCBF and DSC-nrCBF (ρ = 0.73, P < 0.00055) in gliomas. This finding agrees with those reported previously11–13. Besides the mean, strong and moderate positive correlations were observed in the first-order based median (ρ = 0.70, P < 0.00055), 90th percentile (ρ = 0.75, P < 0.00055), and maximum (ρ = 0.57, P < 0.00055). This suggests that even though ASL imaging underestimates CBF compared with DSC imaging, ASL imaging could be an alternative method to DSC imaging for evaluating glioma perfusion. On the other hand, very strong positive correlations (r ≥ 0.8 or ρ ≥ 0.8) between ASL-nCBF and DSC-nrCBF were found in texture features. Some features with significant correlations were, for example, the GLCM-based joint entropy and GLCM-based sum entropy, which are associated with randomness of voxel values, and the GLDM-, GLRLM-, GLSZM-based gray-level non-uniformity, and NGTDM-based coarseness, which are associated with heterogeneous texture. These relationships between ASL and DSC imaging imply that both techniques may evaluate similar perfusion patterns (e.g., randomness and heterogeneity) in gliomas.
The ROC analysis showed that the DSC-nrCBF model has better diagnostic performance for differentiating HGGs from LGGs. Several studies have applied radiomics to MR imaging and reported diagnostic performance for distinguishing between LGGs and HGGs23,24,26. A study showed that a logistic regression model using radiomic features from ASL-CBF had an AUC, sensitivity, and specificity of 0.750, 71.4%, and 63.9%, respectively23. Combining radiomic features from various MR imaging techniques improved the model performance with an AUC, sensitivity, and specificity of 0.911, 85.2%, and 85%, respectively23. Another study reported that the AUC, sensitivity, and specificity of a support vector machine model using 30 texture features from multi-parametric MR imaging, including ASL imaging, were 0.987, 96.4%, and 97.3%, respectively24. Zacharaki et al. proposed a support vector machine model, including DSC-rCBV, with an AUC, sensitivity, and specificity of 0.896, 84.6%, and 95.5%, respectively26. Our models have a diagnostic performance comparable to that of these studies for distinguishing between LGGs and HGGs. Furthermore, our ASL-nCBF and DSC-nrCBF models have a relatively high performance, with their AUCs being not significantly different (P ≥ 0.05). These imply that preoperative glioma grading using radiomics may be feasible with only a single MR perfusion-weighted imaging-derived parameter, such as CBF. Additionally, most selected features for model construction were related to texture patterns. This suggests that texture analysis in MR perfusion-weighted imaging is useful for distinguishing HGGs from LGGs. The ASL-nCBF model is interchangeable to the DSC-nrCBF model for glioma grading and should be preferred, especially for patients who cannot undergo DSC imaging.
This study has several limitations. First, the sample size was small. To avoid any bias in patient selection, the glioma patients were enrolled consecutively. Even though there was this limitation, it was possible to identify differences and correlations between radiomic features of ASL and DSC imaging-derived parameters, which was the main purpose of our study, and to reach a conclusion about the perfusion patterns of ASL and DSC imaging. A future study with a larger sample size may yield improved ASL and DSC models for glioma grading. Second, post-labeling delay (PLD), one of the scan parameters of ASL imaging, was fixed at 1525 ms for all cases. If the PLD is shorter/longer than the arrival time of labeled arterial blood, CBF may not be assessed correctly. Ideally, PLD is set by considering the patient background (e.g., age and medical history). However, it is difficult to set a PLD for each individual patient with brain tumor. Third, the ASL and DSC imaging data were acquired at different resolutions (voxel spacing; ASL: 1.9 mm × 1.9 mm × 4.0 mm, DSC: 1.7 mm × 1.7 mm × 6.0 mm). This may have contributed to the increased glioma perfusion heterogeneity in ASL imaging and differences in the texture features between ASL and DSC imaging. Despite these limitations, our findings demonstrated that radiomics is useful for characterizing glioma perfusion in ASL and DSC imaging and for classifying LGGs and HGGs.
In conclusion, radiomics in ASL and DSC imaging showed that ASL imaging has lower CBF and reflects glioma perfusion heterogeneity better than DSC imaging, whereas both techniques characterize similar perfusion patterns. For classifying LGGs and HGGs, the DSC-nrCBF model showed higher diagnostic performance and was comparable to the ASL-nCBF model. Radiomics can provide glioma perfusion patterns quantitatively, which can be used to differentiate gliomas, and ASL imaging can be a non-invasive alternative to DSC imaging for evaluating glioma perfusion.
Materials and methods
Patient population
This retrospective observational study was approved by our institutional review board (Osaka University Ethics Committee, approval number 17225-2), and passive informed consent was obtained in the form of opt-out on the institutional website from all individual patients. All procedures performed were in accordance with the ethical standards of the institutional review board and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Fifty-four patients with pathologically proven gliomas who underwent pseudo-continuous ASL and DSC perfusion-weighted imaging during the same session in our institution prior to any treatment and/or biopsy between January 2014 and December 2019 were eligible for this study. All patients’ histopathological diagnoses fulfilled the 2007 or 2016 WHO classification criteria of the central nervous system tumors32,33. The flow diagram for patient selection is shown in Fig. 1. Eight patients were excluded due to the following reasons: (1) inadequate quality of MR images (e.g., motion and susceptibility artifacts) for tumor segmentation (n = 4), (2) WHO grade I gliomas (n = 3), and (3) pilomyxoid astrocytoma (n = 1). WHO grade I gliomas, such as pilocytic astrocytoma, are benign and are considered a separate entity34. A definite grading for pilomyxoid astrocytoma is not currently recommended according to the 2016 WHO classification criteria33. Finally, 46 glioma patients, including 15 LGGs and 31 HGGs, were enrolled in this study.
MR imaging
All patients underwent multi-parametric MR imaging using a 3-T scanner (Discovery MR750 3.0 T, GE Healthcare, Milwaukee, WI, USA) with a 32-channel head coil. The following 7 MR imaging sequences were acquired: (1) T1 fluid-attenuated inversion recovery (FLAIR) image, (2) T2 FLAIR image, (3) T2-weighted image (T2WI), (4) T2*-weighted image (T2*WI), (5) pseudo-continuous ASL imaging, (6) DSC imaging, and (7) contrast-enhanced T1-weighted image (CE-T1WI). The main parameters of ASL and DSC imaging were set as follows. Axial ASL imaging, acquired using a three-dimensional pseudo-continuous ASL with spiral fast spin-echo sequence: PLD = 1525 ms, repetition time (TR) = 4642 ms, echo time (TE) = 10.5 ms, flip angle (FA) = 111°, 512 sampling points on 8 spiral arms, in-plane matrix = 128 × 128, field of view (FOV) = 240 mm, slice thickness = 4 mm, band width (BW) = ±62.5 kHz, number of slices = 37, scan time = 2 min 1 s. Oblique-axial DSC imaging, acquired using a gradient-echo echo-planar imaging sequence after a bolus injection of 0.1 ml/kg of gadoterate meglumine (MAGNESCOPE, Guerbet, Tokyo, Japan) at a rate of 3 ml/s, followed by a 30 ml bolus of saline flush at the same rate: TR = 2000 ms, TE = 13.3 ms, FA = 60°, matrix size = 96 × 128, FOV = 220 mm, slice thickness = 5 mm, slice spacing = 1 mm, BW = ±250 kHz, number of slices = 22, scan time = 1 min 20 s. Before DSC imaging, a bolus injection of 0.1 ml/kg of gadoterate meglumine was used for dynamic contrast-enhanced imaging (preload leakage correction to minimize T1 effects for DSC imaging). The scan parameters of the anatomical MR imaging, T1 FLAIR, T2 FLAIR, T2WI, T2*WI, and CE-T1WI, are described in Supplementary Text S1.
Post-processing
The post-processing of the ASL and DSC imaging data was performed with 3DASL and BrainStatAIF applications implemented with FuncTool (version 14.3.01, GE Healthcare, Milwaukee, WI, USA), respectively. The ASL-CBF and DSC-rCBF maps for each patient were generated from ASL imaging and DSC imaging, respectively. Motion correction and arterial input function (AIF) deconvolution with singular value decomposition were applied to the DSC-derived map. The AIF locations were detected automatically.
All MR images of each patient were registered to the patient’s own oblique-axial CE-T1WI, which was set to align along the anterior commissure–posterior commissure line, using Functional MR Imaging of the Brain Software Library (version 5.0.9, FMRIB Analysis Group, University of Oxford, Oxford, UK). Then, these MR images were resliced to match the oblique-axial CE-T1WI.
Additionally, a 6.8-mm diameter spherical volume-of-interest (VOI) (162.8 mm3) was placed within NAWM, avoiding any abnormalities, in CE-T1WIs and T2 FLAIR images. The VOIs were transferred to all perfusion maps in order to normalize ASL-CBF and DSC-rCBF in each patient. The VOI measurements were performed using ITK-SNAP (version 3.6.0, Penn Image Computing and Science Laboratory, University of Pennsylvania, Philadelphia, PA, USA). All voxels in the ASL-CBF and DSC-rCBF maps were divided by the mean value of the NAWM VOI in the respective map, whereby the ASL-nCBF and DSC-nrCBF maps were generated.
Tumor segmentation
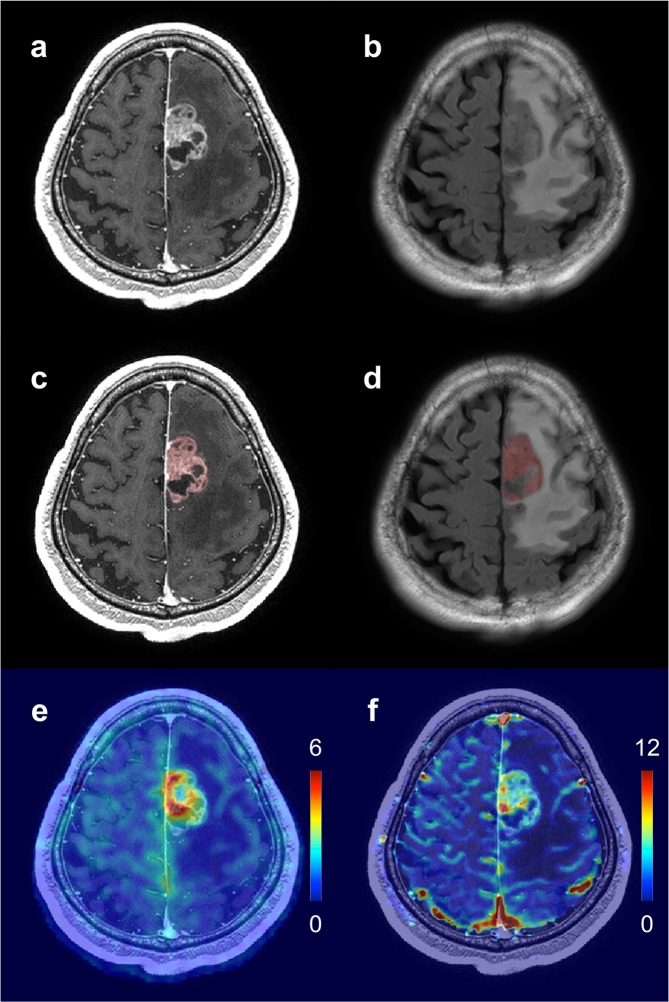
A supervised random forests machine-learning algorithm implemented with ITK-SNAP was used for semi-automatic tumor segmentation. CE-T1WIs and T2 FLAIR images were used to cross-reference the solid portion of the tumor in all patients, which was defined as the gadolinium-enhanced region on the CE-T1WIs and/or the signal abnormality region on the T2 FLAIR images. The machine-learning algorithm took into account the intensities of both MR images during segmentation35. T1 FLAIR images, T2WIs, and T2*WIs were used to exclude any cysts, calcifications, edema, hemorrhage, necrosis, and large vessels in the tumor volume. Figure 5 shows an example of tumor segmentation.
Figure 5.
Example of tumor segmentation for a patient with WHO grade IV glioblastoma (59-year-old male). (a) CE-T1WI. (b) T2 FLAIR image. (c) The segmented tumor volume (red color) overlaid on the CE-T1WI. (d) The segmented tumor volume (red color) overlaid on the T2 FLAIR image. (e) The ASL-nCBF map overlaid on the CE-T1WI. (f) The DSC-nrCBF map overlaid on the CE-T1WI. WHO, World Health Organization; CE-T1WI, contrast-enhanced T1-weighted image; FLAIR, fluid-attenuated inversion recovery; ASL-nCBF, arterial spin labeling normalized cerebral blood flow; DSC-nrCBF, dynamic susceptibility contrast normalized relative cerebral blood flow.
Radiomic feature extraction
The segmented tumor volume was used as a mask image for radiomic feature extraction. A total of 91 radiomic features (Supplementary Table 1) were extracted from each of the ASL-nCBF and DSC-nrCBF maps by using PyRadiomics (version 1.3.0, Computational Imaging and Bioinformatics Lab, Harvard Medical School, Boston, MA, USA)22. The extracted radiomic features can be divided into the following 6 classes: (1) first-order statistics (18 features), (2) GLCM (22 features), (3) GLDM (14 features), (4) GLRLM (16 features), (5) GLSZM (16 features), and (6) NGTDM (5 features). All radiomic features were extracted using the default parameters of PyRadiomics. The feature descriptions and mathematical definitions can be found elsewhere (http://www.radiomics.io/pyradiomics.html)22.
Statistical analysis
All values are expressed as the mean ± standard deviation. All statistical analyses were conducted using R software (version 3.5.0, R Foundation for Statistical Computing, Vienna, Austria). A P-value of < 0.05 was considered statistically significant.
The paired t-test or Wilcoxon signed-rank test was used to compare the radiomic features between ASL-nCBF and DSC-nrCBF. Pearson’s product-moment correlation coefficients (r) or Spearman’s rank-order correlation coefficients (ρ) were used to examine the correlations of the radiomic features between ASL-nCBF and DSC-nrCBF. The Shapiro–Wilk test was used for testing the data normality. The Bonferroni correction method for multiple comparisons was applied to adjust the significance level (α), resulting in P < 0.00055 (α = 0.05/91) to be considered statistically significant.
Before the construction of the radiomics-based classification models, the sample was randomly divided into a training set (n = 42) and a test set (n = 4). The test set included 1 patient with WHO grade II, 1 patient with WHO grade III, and 2 patients with WHO grade IV gliomas. The LASSO regression was used to select the radiomic features that best classify LGGs and HGGs in the ASL-nCBF and DSC-nrCBF maps (training set, n = 42). Leave-one-out cross validation was used for the tuning parameter (λ) selection. The radiomic features with non-zero coefficients were selected. The selected features were used to construct a logistic regression model for each perfusion map, as follows:
| 3 |
where p indicates the probability of a patient with HGG (0 < p < 1), and βi and xi indicate regression coefficients and explanatory variables, respectively. In this study, the xi variables corresponded to the selected radiomic features.
ROC analysis was performed to assess the performance of each model for differentiating between LGGs and HGGs. The empirical method by DeLong et al.36 was used to compare the AUCs of the ASL-nCBF and DSC-nrCBF models. Additionally, the test set was used to evaluate whether the models can correctly predict the class (LGG or HGG) of new patients.
Supplementary information
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (KAKENHI, No. JP19K08172) from the Japan Society for the Promotion of Science (JSPS).
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and data analysis were performed by T.H., S.S., and T.I. The first draft of the manuscript was written by T.H., and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Takashi Hashido and Shigeyoshi Saito.
Supplementary information
is available for this paper at 10.1038/s41598-020-62658-9.
References
- 1.Würdinger T, Tannous BA. Glioma angiogenesis: Towards novel RNA therapeutics. Cell Adh. Migr. 2009;3:230–235. doi: 10.4161/cam.3.2.7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain R, et al. Glioma Angiogenesis and Perfusion Imaging: Understanding the Relationship between Tumor Blood Volume and Leakiness with Increasing Glioma Grade. AJNR Am. J. Neuroradiol. 2015;36:2030–2035. doi: 10.3174/ajnr.A4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young R, Babb J, Law M, Pollack E, Johnson G. Comparison of region-of-interest analysis with three different histogram analysis methods in the determination of perfusion metrics in patients with brain gliomas. J. Magn. Reson. Imaging. 2007;26:1053–1063. doi: 10.1002/jmri.21064. [DOI] [PubMed] [Google Scholar]
- 4.Covarrubias DJ, Rosen BR, Lev MH. Dynamic magnetic resonance perfusion imaging of brain tumors. Oncologist. 2004;9:528–537. doi: 10.1634/theoncologist.9-5-528. [DOI] [PubMed] [Google Scholar]
- 5.Aronen HJ, et al. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology. 1994;191:41–51. doi: 10.1148/radiology.191.1.8134596. [DOI] [PubMed] [Google Scholar]
- 6.Sugahara T, et al. Correlation of MR imaging-determined cerebral blood volume maps with histologic and angiographic determination of vascularity of gliomas. AJR Am. J. Roentgenol. 1998;171:1479–1486. doi: 10.2214/ajr.171.6.9843274. [DOI] [PubMed] [Google Scholar]
- 7.Shin JH, et al. Using relative cerebral blood flow and volume to evaluate the histopathologic grade of cerebral gliomas: preliminary results. AJR Am. J. Roentgenol. 2002;179:783–789. doi: 10.2214/ajr.179.3.1790783. [DOI] [PubMed] [Google Scholar]
- 8.Law M, et al. Zagzag D. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am. J. Neuroradiol. 2003;24:1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 9.Hakyemez B, et al. High-grade and low-grade gliomas: differentiation by using perfusion MR imaging. Clin. Radiol. 2005;60:493–502. doi: 10.1016/j.crad.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Grade M, et al. A neuroradiologist’s guide to arterial spin labeling MRI in clinical practice. Neuroradiology. 2015;57:1181–1202. doi: 10.1007/s00234-015-1571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White CM, et al. Regional and voxel-wise comparisons of blood flow measurements between dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) and arterial spin labeling (ASL) in brain tumors. J. Neuroimaging. 2014;24:23–30. doi: 10.1111/j.1552-6569.2012.00703.x. [DOI] [PubMed] [Google Scholar]
- 12.Jiang J, et al. Comparative analysis of arterial spin labeling and dynamic susceptibility contrast perfusion imaging for quantitative perfusion measurements of brain tumors. Int. J. Clin. Exp. Pathol. 2014;7:2790–2799. [PMC free article] [PubMed] [Google Scholar]
- 13.Khashbat D, et al. Correlation of 3D Arterial Spin Labeling and Multi-Parametric Dynamic Susceptibility Contrast Perfusion MRI in Brain Tumors. J. Med. Invest. 2016;63:175–181. doi: 10.2152/jmi.63.175. [DOI] [PubMed] [Google Scholar]
- 14.Ma H, et al. Three-dimensional arterial spin labeling imaging and dynamic susceptibility contrast perfusion-weighted imaging value in diagnosing glioma grade prior to surgery. Exp. Ther. Med. 2017;13:2691–2698. doi: 10.3892/etm.2017.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warmuth C, Gunther M, Zimmer C. Quantification of blood flow in brain tumors: comparison of arterial spin labeling and dynamic susceptibility-weighted contrast-enhanced MR imaging. Radiology. 2003;228:523–532. doi: 10.1148/radiol.2282020409. [DOI] [PubMed] [Google Scholar]
- 16.Wolf RL, et al. Grading of CNS neoplasms using continuous arterial spin labeled perfusion MR imaging at 3 Tesla. J. Magn. Reson. Imaging. 2005;22:475–482. doi: 10.1002/jmri.20415. [DOI] [PubMed] [Google Scholar]
- 17.Chawla S, et al. Arterial spin-labeling and MR spectroscopy in the differentiation of gliomas. AJNR Am. J. Neuroradiol. 2007;28:1683–1689. doi: 10.3174/ajnr.A0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HS, Kim SY. A prospective study on the added value of pulsed arterial spin-labeling and apparent diffusion coefficients in the grading of gliomas. AJNR Am. J. Neuroradiol. 2007;28:1693–1699. doi: 10.3174/ajnr.A0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadowski EA, et al. Nephrogenic systemic fibrosis: risk factors and incidence estimation. Radiology. 2007;243:148–157. doi: 10.1148/radiol.2431062144. [DOI] [PubMed] [Google Scholar]
- 20.Dangouloff-Ros V, et al. Arterial Spin Labeling to Predict Brain Tumor Grading in Children: Correlations between Histopathologic Vascular Density and Perfusion MR Imaging. Radiology. 2016;281:553–566. doi: 10.1148/radiol.2016152228. [DOI] [PubMed] [Google Scholar]
- 21.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Griethuysen JJM, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su C, et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur. Radiol. 2019;29:1986–1996. doi: 10.1007/s00330-018-5704-8. [DOI] [PubMed] [Google Scholar]
- 24.Tian Q, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging. 2018;48:1518–1528. doi: 10.1002/jmri.26010. [DOI] [PubMed] [Google Scholar]
- 25.Reishofer G, et al. Automated macrovessel artifact correction in dynamic susceptibility contrast magnetic resonance imaging using independent component analysis. Magn. Reson. Med. 2011;65:848–857. doi: 10.1002/mrm.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zacharaki EI, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Silva, A. C., Kim, S. G. & Garwood, M. Imaging blood flow in brain tumors using arterial spin labeling. Magn. Reson. Med. 44, 169–173. 10.1002/1522-2594(200008)44:2<169::aid-mrm1>3.0.co;2-u (2000). [DOI] [PubMed]
- 29.Sun Y, et al. Perfusion MRI of U87 brain tumors in a mouse model. Magn. Reson. Med. 2004;51:893–899. doi: 10.1002/mrm.20029. [DOI] [PubMed] [Google Scholar]
- 30.Ferré JC, Shiroishi MS, Law M. Advanced techniques using contrast media in neuroimaging. Magn. Reson. Imaging Clin. N. Am. 2012;20:699–713. doi: 10.1016/j.mric.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YF, et al. Diagnostic significance of arterial spin labeling in the assessment of tumor grade in brain. J. Cancer Res. Ther. 2016;12:259–266. doi: 10.4103/0973-1482.155978. [DOI] [PubMed] [Google Scholar]
- 32.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 34.Larsen J, et al. ‘Low grade glioma’: an update for radiologists. Br. J. Radiol. 2017;90:20160600. doi: 10.1259/bjr.20160600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yushkevich PA, Gao Y, Gerig G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:3342–3345. doi: 10.1109/EMBC.2016.7591443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.