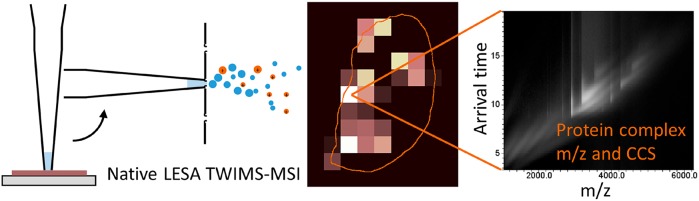
Abstract
We have previously demonstrated native liquid extraction surface analysis (LESA) mass spectrometry imaging of small intact proteins in thin tissue sections. We also showed calculation of collision cross sections for specific proteins extracted from discrete locations in tissue by LESA traveling wave ion mobility spectrometry (TWIMS). Here, we demonstrate an integrated native LESA TWIMS mass spectrometry imaging (MSI) workflow, in which ion mobility separation is central to the imaging experiment and which provides spatial, conformational, and mass information on endogenous proteins in a single experiment. The approach was applied to MSI of a thin tissue section of mouse kidney. The results show that the benefits of integration of TWIMS include improved specificity of the ion images and the capacity to calculate collision cross sections for any protein or protein complex detected in any pixel (without a priori knowledge of the presence of the protein).
Introduction
Native mass spectrometry enables the analysis of tertiary and quaternary protein structure in the gas phase.1 Noncovalent interactions such as hydrogen bonds and salt bridges that were present in solution are preserved in the gas phase through the use of mild electrospray ionization conditions including native-like solvents. Typically, native mass spectrometry results in lower charge states and a narrower charge state distribution than observed under denaturing conditions, due to more folded protein conformations with limited available protonation sites and solvent-accessible surface areas.2−4 Native mass spectrometry is often integrated with ion mobility spectrometry. In particular, traveling wave ion mobility spectrometry (TWIMS) has found widespread use for over a decade since the introduction of commercially available TWIMS-enabled mass spectrometers.5−9 Structural information may be inferred from collision cross section (CCS) values derived from drift tube ion mobility spectrometry (DTIMS) or calibrated TWIMS measurements.10 The CCS is a measure of the rotationally averaged 3D shape of a gas-phase ion and has been used as further evidence for the retention of solution phase structures in the gas phase.11,12 The CCS may be evaluated against theoretical CCS values predicted from X-ray crystallography structures, which can provide insight into the protein conformation, e.g., folded or unfolded.6,13,14 TWIMS may also serve as a filter, for example, enabling the separation of isobaric signals with differing size-to-charge ratios. TWIMS separation of singly charged species from multiply charged peptide and protein signals was recently demonstrated for desorption electrospray ionization.15
Liquid extraction surface analysis (LESA) MS is an ambient surface sampling technique that is particularly suited to the direct analysis of intact proteins from biological substrates.16,17 LESA mass spectrometry imaging (MSI) allows the spatial distribution of analytes, including proteins, to be mapped.18 As LESA MS makes use of electrospray ionization, the technique is suitable for native mass spectrometry through use of native-like solvents. We have previously demonstrated native LESA MS of folded proteins and protein complexes from dried blood spots and thin tissue sections.19−21 More recently, we demonstrated native LESA MSI of thin tissue sections of mouse brain and liver.21 In that work, discrete locations on adjacent tissue sections were sampled by native LESA coupled with TWIMS-MS enabling calculation of CCS of small proteins. Here, we have developed a LESA TWIMS-MSI workflow, in which ion mobility separation is integral to the imaging experiment and which provides spatial, conformational, and mass information for proteinaceous constituents of tissues within a single experiment. The advantages of TWIMS are, first, the filtering of interfering signals associated with direct sampling of tissue and consequent improved specificity of the ion images and improvement in the signal-to-noise ratio and, second, the capacity for calculation of the CCS for any protein detected in any pixel. We have applied native LESA TWIMS-MSI to a thin tissue section of mouse kidney. We demonstrate the measurement of the CCS values of proteins, including the 64 kDa tetrameric Hb complex, directly from tissue during MSI, and improved ion images, especially for proteins with low intensity signals that are obscured by isobaric species.
Methods
Materials
Kidney from wild-type mice (extraneous tissue from culled animals) was the gift of Dr. Caroline Chadwick (University of Birmingham). Tissue was frozen in liquid nitrogen and then stored at −80 °C until sectioned. Tissue was sectioned at −22 °C at a thickness of 10 μm with a CM1810 Cryostat (Leica Microsystems, Wetzlar, Germany) and thaw mounted to glass microscope slides. Sections were stored at −80 °C until use. The tissue was not subjected to washing prior to analysis.
Sodium iodide (2 μg/μL in 50% isopropanol) for TOF calibration was obtained from Waters Corporation (Manchester, UK). Ammonium acetate and the protein standards ubiquitin (bovine, U6253), cytochrome C (equine, C2506), and myoglobin (equine, M0630) were obtained from Sigma-Aldrich (Gillingham, UK). Argon (purity >99.998%), nitrogen (>99.995%), and helium (>99.996%) gases were obtained from BOC (Guildford, UK). MS grade water and methanol were obtained from Fisher Scientific (Loughborough, UK).
LESA TWIMS-MSI
“Contact” LESA was performed using a Triversa NanoMate (Advion Biosciences, Ithaca, NY). The diameter of the sampled area was approximately 600 μm (external diameter of pipet tip), with sampling locations separated by 1 mm, the smallest spacing available in the Triversa NanoMate control software ChipSoft (version 8.3.3) (see Figure 1). A 5 μL aliquot of extraction solvent (200 mM ammonium acetate + 5% methanol) was aspirated into the conductive pipet tip and moved to the location above the tissue. The tip was pressed into the tissue surface, and 2.1 μL of solvent was dispensed. After 1 min, a 2.2 μL volume was aspirated, and the tip was moved to the nanoESI chip. ESI was initiated with a potential of 1.8 kV and back pressure of 0.15 PSI.
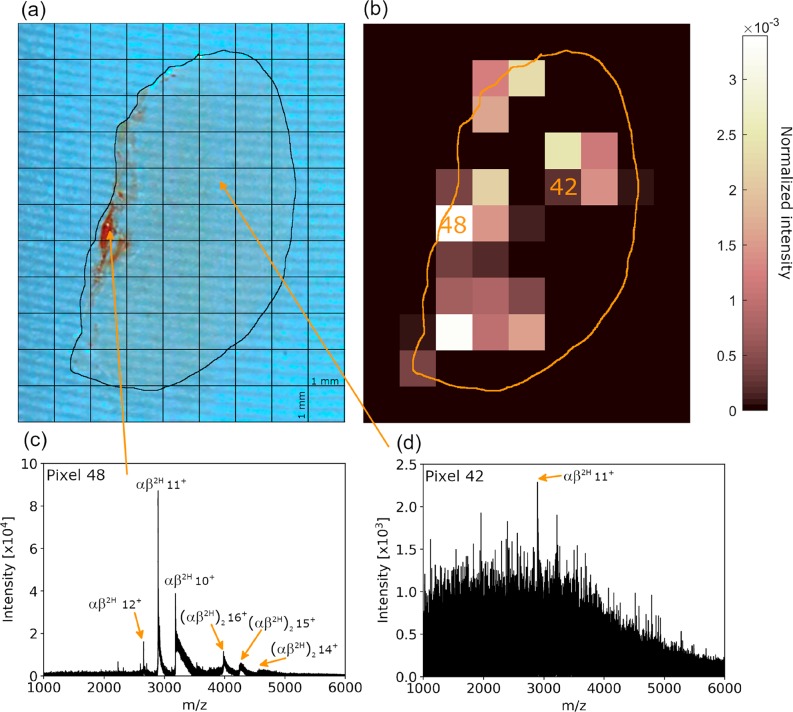
Figure 1.
(a) Kidney section with pixel grid overlaid. (b) Ion image for m/z 2894.7 (11+), corresponding to hemoglobin heterodimer. (c) The raw mass spectrum (i.e., without arrival time filtering) obtained from pixel 48. Peaks corresponding to a hemoglobin heterotetramer and heterodimer are observed. (d) The raw mass spectrum obtained from pixel 42 reveals low ion abundance.
IM-MS data were recorded with a Synapt G2-S HDMS (Waters Corporation). The instrument was operated in “mobility-TOF mode” and “sensitivity mode”. The backing pressure was raised to approximately 6.35 mBar using a Speedivalve on the roughing pump. The trap and transfer cells were provided with argon (5 mL/min) to give a pressure of approximately 3 × 10–2 mBar. Helium was introduced to the helium cell at 180 mL/min. Nitrogen was provided to the TWIMS cell at 90 mL/min to give a pressure of ∼3 mBar. The laboratory temperature was approximately 20 °C.
The source conditions were as follows: source temperature, 100 °C; sampling cone, 30–80 V; source offset, 30 V; cone gas flow, 10 L/h. The StepWave ion guide settings were left at default values. The trap DC entrance and DC bias were set to 3 and 42 V, respectively. The TWIMS device was operated with a fixed wave velocity of 500 m/s and a wave height of 25 V. Additionally, pixels 15, 43, 48, and 49 were sampled and analyzed a second and third time with wave heights of 24 and 26 V to assess CCS measurement errors. Transfer collision energy was set between 10 and 15 V to improve mass accuracy. A manual quadrupole profile was set to assist higher m/z transmission (see Table S1, Supporting Information). Waters .raw files were acquired with the discrete “Drift Time function” option checked in the MS Tune acquisition window. Data were acquired for 5 min per pixel over the m/z range 1000–8000 (pusher frequency, 138 μs; enhanced duty cycle coefficient, 1.57 ms).
Collision Cross Sections
IM-MS data were collected for ubiquitin, cytochrome C, and apo-myoglobin by direct infusion nanoESI under denaturing conditions (10 μM in water/MeOH/acetic acid 49:49:2 v:v:v). The cone voltage and source offset voltages were both set to 150 V to maximize unfolding of the calibrant ions. The m/z range was 700–8000 (pusher frequency; 138 μs, enhanced duty cycle coefficient; 1.57 ms). Each standard was analyzed at three TWIMS traveling wave heights (24, 25, 26 V). The arrival times for ions of each charge state were determined from the apex of the most intense arrival time peak. Mean smoothing (±1 scan, 1 smooth) of the arrival time distribution (ATD) plot was performed using MassLynx 4.1. Previously published drift tube ion mobility CCS values obtained in nitrogen (DTCCSN2) were used for generating a calibration curve for calculation of TWCCSN2→N2.10TWCCSN2→N2 was calculated by a previously published method adapted for N2 calibration values.21 For proteins detected in pixels analyzed at the three wave heights (pixels 15, 43, 48, and 49), the standard deviation of TWCCSN2→N2 was calculated. CCSN2 calculated from crystal structures were obtained using the trajectory model (TM) in IMoS v1.09 and .pdb files as inputs.22
Image Processing
Data files in the Waters .raw format were converted to mzML using MSConvert (Version 3.0, ProteoWizard Software Foundation).23 mzML files were imported into MATLAB (version R2019a, MathWorks, Natick, USA.) with the imzML converter using code adapted from Spectral Analysis software (downloaded from https://github.com/AlanRace/SpectralAnalysis on 21/03/2017).2425 In-house code from previous work was adapted to enable the extraction of mass spectra from specific regions of interest from the arrival time versus m/z plot (see Supplemental File S1).21 The software is freely available from http://www.biosciences-labs.bham.ac.uk/cooper/software.php.
The mass spectra from multiple scans were summed at each location to form a single spectrum (retaining arrival time information) per position. Broad filtering: selection rules (arrival time (tA), m/z) were defined in DriftScope (v2.9, Waters) and then exported as a text file and imported into the in-house software. Specific filtering: the arrival time window at approximately full width half-maximum for the peak (m/z) of interest was selected in Driftscope and exported as a text file and imported into the in-house software. After filtering (either broad or specific), baseline subtraction (max baseline corrected at 25% base peak ion intensity, with a median filter setting between 2000 and 8000) and normalization to total ion count were performed to account for variability in absolute signal intensity between pixels.
Results and Discussion
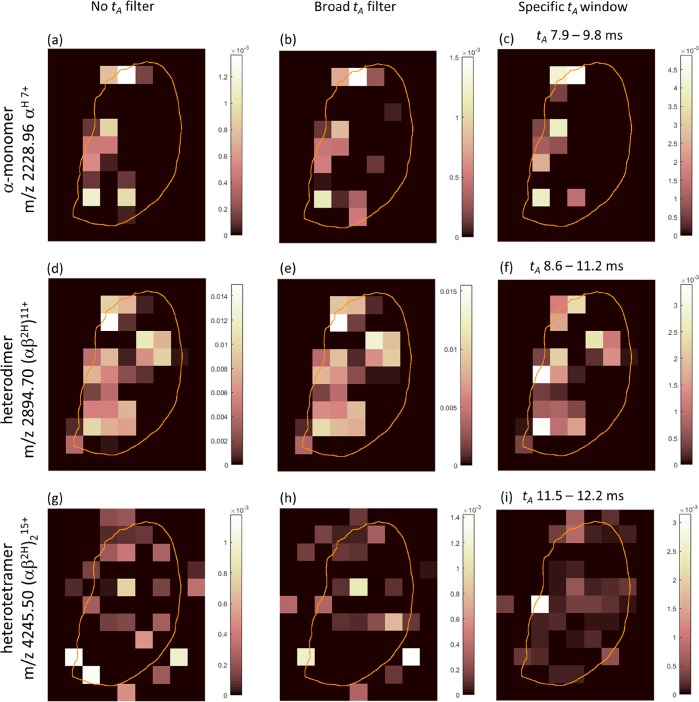
A thin tissue section of mouse kidney was subjected to LESA TWIMS-MSI. Figure 1a shows the kidney section with the 1 × 1 mm pixel grid overlaid. The major blood vessels are located toward the left side of the section at the renal pelvis. The renal pelvis is the entry and exit point for the major renal artery and vein, respectively. Figure 1b shows an arrival time (tA) filtered ion image for m/z 2894.7, the 11+ charge state of the hemoglobin heterodimer (αβ2H). The unfiltered mass spectrum obtained for pixel 48 reveals strong (αβ2H)11+ signals (Figure 1c), whereas the unfiltered mass spectrum of pixel 42 (Figure 1d) exhibits weak signals. Figure 2 shows ion images generated for the heme-bound hemoglobin (Hb) tetramer (αβ2H)2 (15+, MW 63.7 kDa), heterodimer αβ2H (11+, MW 31.8 kDa), and alpha-subunit monomer Hb (αH) (7+, MW 15.6 kDa). The images reveal that these hemoglobin-related ions were most abundant near the large vascular structures at the renal pelvis. The distributions of the monomer and dimer correlate reasonably well, with signals detected across the tissue, whereas the tetramer is confined to the renal pelvis vasculature. In part, this observation can be explained by the fact that tetrameric Hb is known to dissociate to dimers following hemolysis, i.e., outside of red blood cells, with subsequent clearance to the kidney.26 Another factor is the lower signal intensity of the tetramers. Three sets of ion images are shown: produced using no ion mobility filtering, with a broad filtering rule, and with filtering specific to the ion of interest. Broad filtering (see Figure S1, Supporting Information) involved selection of a protein-containing region of interest from the 2D heat-map plot of arrival time versus m/z. This selection rule was applied to all pixels within the data set, and ion images were generated from the resulting mass spectra. Specific filtering involved selection of an arrival time (tA) window set approximately at the full width half-maximum of the arrival time peak of ions of interest, followed by image generation for those ions. Additional ion images for small proteins are shown in Figure S2, Supporting Information. The images are generally comparable in appearance for all ion mobility filtering options; however, there are clear differences between the images for m/z 4245 (Figure 2g–i). The specific tA filtering results in an image with the most intense signal where the blood vessel is seen on the photograph of the kidney section, whereas this is not the case for the unfiltered images. This observation suggests that a lack of arrival time specificity is detrimental to the accuracy of the image. Arrival time filtered images for three charge states of each of the Hb-related ions are shown in Figure S3, Supporting Information. In each case, the middle charge state represents the most intense signal. Dimer ions exhibit comparable images for charge states 12+–10+. Images for the heme-bound α-monomer and tetramer ions show consistency between the 7+, 6+ and 16+, 15+ charge states respectively, while the 8+ and 14+ charge state images suffer from low signal intensity and are visually less consistent.
Figure 2.
Ion images for Hb αH 7+ monomer (a–c), αβ2H 11+ dimer (d–f), and (αβ2H)215+ tetramer (g–i) produced without arrival time (tA) filtering (a,d,g), with a broad selection rule that predominantly removed singly charged signals (b,e,h), and with a specific tA selected for each ion of interest (c,f,i). Color bars indicate normalized signal intensity after baseline subtraction.
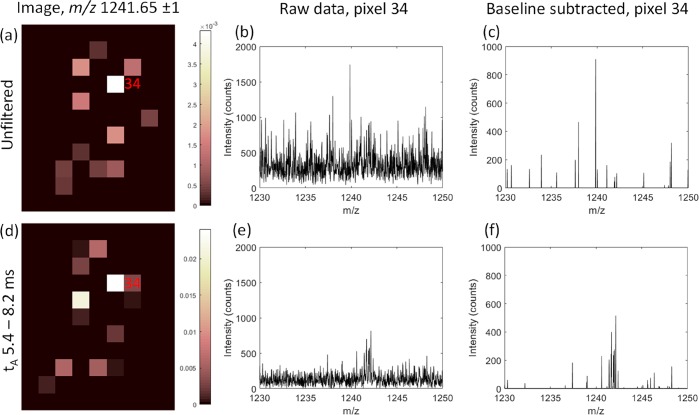
The use of specific arrival time (tA) filtering is especially useful where protein ion signals are of low intensity or otherwise obscured by isobaric signals. As an example, Figure 3 shows how, with specific tA filtering, peaks for β-thymosin 4 [M + 4H]4+ in pixel 34 are revealed from baseline signals and are thus incorporated into the ion image once further processed. Without filtering, real ion signals were subtracted as if they were part of the baseline, whereas tA filtering improved the S/N prior to baseline subtraction. Figure S4, Supporting Information, shows an alternative situation, where ubiquitin [M + 5H]5+ signal is apparently detected in pixel 32 for the unfiltered image (Figure S4a), but the signal results from the noise. tA filtering reduces the noise but does not reveal any peaks attributable to ubiquitin ions, and as such, the reduced pixel intensity is reported in the ion image (Figure S4d). It is important to note that some baseline signals may be retained, i.e., tA filtering results in improved rather than complete specificity.
Figure 3.
Comparison of ion images for m/z 1241.65 ± 1 where (a) includes signals at all arrival times (tA), whereas (d) is restricted to signals with tA between 5.4 and 8.2 ms. The raw spectrum for pixel 34 (b) shows low intensity peaks for β-thymosin 4 are lost within the baseline signals, but tA filtering increases the S/N ratio (e). With baseline subtraction during image processing, real ion signals within the baseline may be lost entirely (c). The increased S/N provided by specific tA filtering in (e) improved detection of β-thymosin signals in pixel 34 (f). Color bars in (a) and (d) indicate normalized intensity after baseline subtraction.
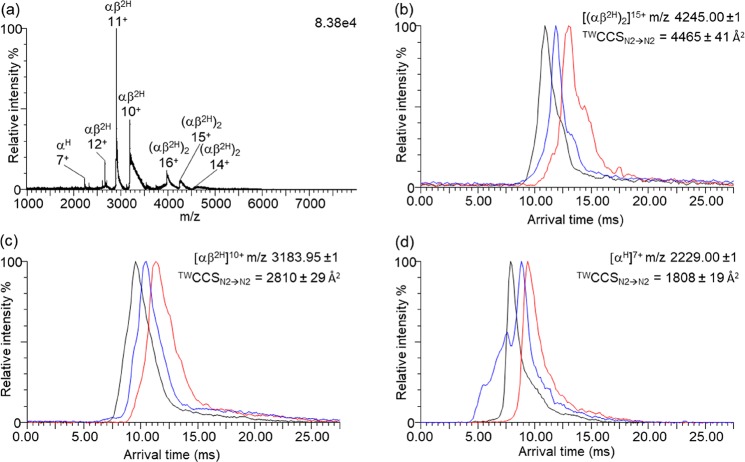
Figure 4a shows the mass spectrum obtained from pixel 48. Peaks corresponding to heme-coordinated hemoglobin ion signals, including the tetramer Hb (αβ2H)2, heterodimer Hb (αβ2H), and alpha-subunit monomer Hb (αH), are detected. The narrow charge state envelopes and presence of protein complex ions is characteristic of native analysis conditions. Arrival time distribution (ATD) plots obtained at three wave heights are shown for [(αβ2H)2]15+, (αβ2H)10+, and (αH)7+ (see Figure 4b–d, respectively). CCS values may be obtained at a single traveling wave height, but information from three different wave heights enables estimation of the measurement error. For each wave height, the same tissue location was resampled (see Methods). The TWCCSN2→N2 for the Hb tetramer was calculated for the 14+ (approximate m/z 4550, 4378 ± 89 Å2), 15+ (approximate m/z 4245, 4465 ± 41 Å2), and 16+ (approximate m/z 3980, 4503 ± 33 Å2) charge states. The CCSN2 of the Hb (αβ2H)2 crystal structure (3HRW) calculated using the TM was 4699 Å2, up to 7% larger than the experimentally determined TWCCSN2→N2 values. This discrepancy indicates slightly compacted ions, explained by a degree of gas-phase compaction of the tetramer as a consequence of the traveling wave height in the ion mobility device, and has been noted previously.9,27 A similar observation was made for human hemoglobin ions by Scarff et al. with the suggestion that polar side chains collapse in the gas phase.8,28 The TWCCSN2→N2 for Hb (αβ2H)2 ions is larger for higher charge states, as is often observed, due to Coulombic repulsion in the gas phase.29 The 16+ charge state also features a second arrival time peak (see Figure S5, TWCCSN2→N2 = 4714 ± 44 Å2), suggesting that there was also a more unfolded conformation present. Hemoglobin dimers, αβ2H, also exist naturally under physiological conditions and have been observed in previous native MS studies of hemoglobin.26,30 The presence of abundant dimer and monomer peaks may also be the result of natural dissociation due to low Hb concentration in the extraction solution, occurring in the interval between extraction and ionization.31 Peaks corresponding to αβ2H in the 10+ (TWCCSN2→N2 = 2810 ± 29 Å2), 11+ (TWCCSN2→N2 = 3030 ± 17 Å2), and 12+ (TWCCSN2→N2 = 3221 ± 40 Å2) charge states were detected. The 7+ charge state of αH had a TWCCSN2→N2 of 1808 ± 19 Å2. An unidentified protein (∼14 561 Da) was detected at m/z 2428 (6+, TWCCSN2→N2 = 1637 ± 4 Å2), m/z 2081 (7+, TWCCSN2→N2 = 1705 ± 10 Å2), and m/z 1821 (8+, TWCCSN2→N2 = 1809 ± 10 Å2). TWCCSN2→N2 for a further unidentified protein (∼14 866 Da) was similarly determined (7+, TWCCSN2→N2 = 1715 ± 10 Å2; 8+, 1791 ± 18 Å2). Ubiquitin (8560 Da (5+), TWCCSN2→N2 = 1089 ± 17 Å2) was found to have a comparable TWCCSN2→N2 to those reported in the literature.32,33 β-thymosin 4 (4960 Da (4+), TWCCSN2→N2 = 796 ± 9 Å2) was also detected—this is the first instance of its TWCCSN2→N2 being reported, although the TWCCSN2→He has been reported.21 It is important to note that as ion mobility measurements were performed in tandem with mass analysis, no additional experiments were necessary for determination of TWCCSN2→N2. As such, future native LESA MSI investigations could study protein–protein and protein–ligand complexes with location specificity on a single tissue section but without additional sample preparation or instrument time required.
Figure 4.
(a) Mass spectrum for pixel 48 after background subtraction (MassLynx function, polynomial order 15, 10% below curve) with labels indicating the peaks for heme-coordinated ions; hemoglobin tetramer (αβ2H)2, heterodimer (αβ2H), and monomer (αH). Arrival time distributions are shown for (b) [(αβ2H)2]15+, (c) [(αβ2H)]10+, and (d) [αH]7+. Errors indicate one standard deviation above and below the mean value of three measurements at TW heights of 24 (red), 25 (blue), and 26 V (black).
Conclusions
Mouse tissue was analyzed by contact LESA TWIMS-MSI under native conditions. The ability to collect spatial, conformational, and mass information for intact proteins in a single experiment allows protein structure to be associated with tissue features, e.g., the vascular regions here. IM-MS measurements revealed that Hb was retained as a tetrameric complex, among the other protein signals. The complex’s TWCCSN2→N2 was found to be similar to that predicted from the crystal structure. This finding demonstrates that room temperature native LESA sampling retains delicate noncovalent interactions. Ion images showed Hb-related ions distributed toward the renal pelvis, where the largest blood vessels are located. The ion mobility data was also used to filter ion signals in the m/z dimension by arrival time. Broad tA selection rules or specific tA selection may be used to further improve specificity of ion images, for example, by increasing signal-to-noise ratios for low intensity signals otherwise lost in a noisy baseline.
Acknowledgments
H.J.C. is an EPSRC Established Career Fellow. O.J.H. and H.J.C. are funded by EPSRC (EP/S002979/1). H.J.C. and R.L.G. were funded by EPSRC (EP/L023490/1). E.K.S. received funding from the EPSRC via the Centre for Doctoral Training in Physical Sciences for Health (Sci-Phy-4-Health) (EP/L016346/1). E.K.S.’ studentship is in collaboration with UCB Pharma. The Advion Triversa Nanomate and the Waters Synapt G2S mass spectrometer was funded by EPSRC (EP/K039245/1). Supplementary data supporting this research is openly available from https://doi.org/10.25500/edata.bham.00000448.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.9b00122.
Table S1: Manual quadrupole profile. Figure S1: Broad DriftScope selection rule. Figure S2: Ion images for small proteins detected by LESA TWIMS-MSI. Figure S3: Ion images for three charge states of hemoglobin-related ions. Figure S4: Unfiltered and tA filtered ion images for ubiquitin [M + 5H]5+. Figure S5: ATD for Hb tetramer 16+ (PDF)
File S1: Details of ion image generation and associated .m files (ZIP)
Author Present Address
# School of Pharmacy, University of Nottingham, Nottingham NG7 2RD, U.K. (R.L.G.)
The authors declare no competing financial interest.
Supplementary Material
References
- Leney A. C.; Heck A. J. Native Mass Spectrometry: What is in the Name?. J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. 10.1007/s13361-016-1545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de la Mora J. Electrospray ionization of large multiply charged species proceeds via Dole’s charged residue mechanism. Anal. Chim. Acta 2000, 406, 93–104. 10.1016/S0003-2670(99)00601-7. [DOI] [Google Scholar]
- Kaltashov I. A.; Mohimen A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 2005, 77, 5370–5379. 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijnenberg A.; Butterer A.; Sobott F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta, Proteins Proteomics 2013, 1834, 1239–1256. 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Giles K.; Pringle S. D.; Worthington K. R.; Little D.; Wildgoose J. L.; Bateman R. H. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004, 18, 2401–2414. 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- Ruotolo B. T.; Giles K.; Campuzano I.; Sandercock A. M.; Bateman R. H.; Robinson C. V. Evidence for macromolecular protein rings in the absence of bulk water. Science 2005, 310, 1658–1661. 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- Ruotolo B. T.; Benesch J. L.; Sandercock A. M.; Hyung S. J.; Robinson C. V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3, 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Scarff C. A.; Patel V. J.; Thalassinos K.; Scrivens J. H. Probing hemoglobin structure by means of traveling-wave ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2009, 20, 625–631. 10.1016/j.jasms.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Michaelevski I.; Kirshenbaum N.; Sharon M. T-wave ion mobility-mass spectrometry: basic experimental procedures for protein complex analysis. J. Visualized Exp. 2010, e1985. 10.3791/1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M. F.; Hall Z.; Giles K.; Hoyes J.; Robinson C. V.; Ruotolo B. T. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 2010, 82, 9557–9565. 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- Jurneczko E.; Barran P. E. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst 2011, 136, 20–28. 10.1039/C0AN00373E. [DOI] [PubMed] [Google Scholar]
- Lanucara F.; Holman S. W.; Gray C. J.; Eyers C. E. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014, 6, 281–294. 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- Scarff C. A.; Thalassinos K.; Hilton G. R.; Scrivens J. H. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun. Mass Spectrom. 2008, 22, 3297–3304. 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- van Duijn E.; Barendregt A.; Synowsky S.; Versluis C.; Heck A. J. Chaperonin complexes monitored by ion mobility mass spectrometry. J. Am. Chem. Soc. 2009, 131, 1452–1459. 10.1021/ja8055134. [DOI] [PubMed] [Google Scholar]
- Towers M. W.; Karancsi T.; Jones E. A.; Pringle S. D.; Claude E. Optimised Desorption Electrospray Ionisation Mass Spectrometry Imaging (DESI-MSI) for the Analysis of Proteins/Peptides Directly from Tissue Sections on a Travelling Wave Ion Mobility Q-ToF. J. Am. Soc. Mass Spectrom. 2018, 29, 2456–2466. 10.1007/s13361-018-2049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz V.; Ford M. J.; Van Berkel G. J. Automation of a surface sampling probe/electrospray mass spectrometry system. Anal. Chem. 2005, 77, 7183–7189. 10.1021/ac0510742. [DOI] [PubMed] [Google Scholar]
- Kertesz V.; Van Berkel G. J. Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform. J. Mass Spectrom. 2010, 45, 252–260. 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L.; Creese A. J.; Race A. M.; Bunch J.; Cooper H. J. LESA FAIMS Mass Spectrometry for the Spatial Profiling of Proteins from Tissue. Anal. Chem. 2016, 88, 6758–6766. 10.1021/acs.analchem.6b01060. [DOI] [PubMed] [Google Scholar]
- Martin N. J.; Griffiths R. L.; Edwards R. L.; Cooper H. J. Native Liquid Extraction Surface Analysis Mass Spectrometry: Analysis of Noncovalent Protein Complexes Directly from Dried Substrates. J. Am. Soc. Mass Spectrom. 2015, 26, 1320–1327. 10.1007/s13361-015-1152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. L.; Konijnenberg A.; Viner R.; Cooper H. J. Direct Mass Spectrometry Analysis of Protein Complexes and Intact Proteins up to > 70 kDa from Tissue. Anal. Chem. 2019, 91, 6962–6966. 10.1021/acs.analchem.9b00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. L.; Sisley E. K.; Lopez-Clavijo A. F.; Simmonds A. L.; Styles I. B.; Cooper H. J. Native mass spectrometry imaging of intact proteins and protein complexes in thin tissue sections. Int. J. Mass Spectrom. 2019, 437, 23–29. 10.1016/j.ijms.2017.10.009. [DOI] [Google Scholar]
- Larriba-Andaluz C.; Hogan C. J. Jr Collision cross section calculations for polyatomic ions considering rotating diatomic/linear gas molecules. J. Chem. Phys. 2014, 141, 194107. 10.1063/1.4901890. [DOI] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M. Y.; Paulse C.; Creasy D.; Flashner L.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race A. M.; Styles I. B.; Bunch J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 2012, 75, 5111–5112. 10.1016/j.jprot.2012.05.035. [DOI] [PubMed] [Google Scholar]
- Race A. M.; Palmer A. D.; Dexter A.; Steven R. T.; Styles I. B.; Bunch J. SpectralAnalysis: Software for the Masses. Anal. Chem. 2016, 88, 9451–9458. 10.1021/acs.analchem.6b01643. [DOI] [PubMed] [Google Scholar]
- Schaer D. J.; Buehler P. W.; Alayash A. I.; Belcher J. D.; Vercellotti G. M. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2013, 121, 1276–1284. 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine P. W. A.; Fisher H. C.; Calabrese A. N.; Whelan F.; Higazi D. R.; Potts J. R.; Lowe D. C.; Radford S. E.; Ashcroft A. E. Investigating the Structural Compaction of Biomolecules Upon Transition to the Gas-Phase Using ESI-TWIMS-MS. J. Am. Soc. Mass Spectrom. 2017, 28, 1855–1862. 10.1007/s13361-017-1689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelimov K. B.; Clemmer D. E.; Hudgins R. R.; Jarrold M. F. Protein Structurein Vacuo: Gas-Phase Conformations of BPTI and Cytochromec. J. Am. Chem. Soc. 1997, 119, 2240–2248. 10.1021/ja9619059. [DOI] [Google Scholar]
- Lermyte F.; Konijnenberg A.; Williams J. P.; Brown J. M.; Valkenborg D.; Sobott F. ETD allows for native surface mapping of a 150 kDa noncovalent complex on a commercial Q-TWIMS-TOF instrument. J. Am. Soc. Mass Spectrom. 2014, 25, 343–350. 10.1007/s13361-013-0798-3. [DOI] [PubMed] [Google Scholar]
- Griffith W. P.; Kaltashov I. A. Highly Asymmetric Interactions between Globin Chains during Hemoglobin Assembly Revealed by Electrospray Ionization Mass Spectrometry. Biochemistry 2003, 42, 10024–10033. 10.1021/bi034035y. [DOI] [PubMed] [Google Scholar]
- Manning L. R.; Jenkins W. T.; Hess J. R.; Vandegriff K.; Winslow R. M.; Manning J. M. Subunit dissociations in natural and recombinant hemoglobins. Protein Sci. 1996, 5, 775–781. 10.1002/pro.5560050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. P.; Knapman T. W.; Campuzano I.; Malham R. W.; Berryman J. T.; Radford S. E.; Ashcroft A. E. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur. J. Mass Spectrom. 2009, 15, 113–130. 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- May J. C.; Jurneczko E.; Stow S. M.; Kratochvil I.; Kalkhof S.; McLean J. A. Conformational Landscapes of Ubiquitin, Cytochrome c, and Myoglobin: Uniform Field Ion Mobility Measurements in Helium and Nitrogen Drift Gas. Int. J. Mass Spectrom. 2018, 427, 79–90. 10.1016/j.ijms.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.