Abstract
Objective
Previous work has revealed a genetic association between Takayasu arteritis and a non-coding genetic variant in an enhancer region within IL6 (rs2069837 A/G). The risk allele in this variant (allele A) has a protective effect against chronic viral infection and cancer. The goal of this study was to characterise the functional consequences of this disease-associated risk locus.
Methods
A combination of experimental and bioinformatics tools were used to mechanistically understand the effects of the disease-associated genetic locus in IL6. These included electrophoretic mobility shift assay, DNA affinity precipitation assays followed by mass spectrometry and western blotting, luciferase reporter assays and chromosome conformation capture (3C) to identify chromatin looping in the IL6 locus. Both cell lines and peripheral blood primary monocyte-derived macrophages were used.
Results
We identified the monocyte/macrophage anti-inflammatory gene GPNMB,~520 kb from IL6, as a target gene regulated by rs2069837. We revealed preferential recruitment of myocyte enhancer factor 2–histone deacetylase (MEF2–HDAC) repressive complex to the Takayasu arteritis risk allele. Further, we demonstrated suppression of GPNMB expression in monocyte-derived macrophages from healthy individuals with AA compared with AG genotype, which was reversed by histone deacetylase inhibition. Our data show that the risk allele in rs2069837 represses the expression of GPNMB by recruiting MEF2–HDAC complex, enabled through a long-range intrachromatin looping. Suppression of this anti-inflammatory gene might mediate increased susceptibility in Takayasu arteritis and enhance protective immune responses in chronic infection and cancer.
Conclusions
Takayasu arteritis risk locus in IL6 might increase disease susceptibility by suppression of the anti-inflammatory gene GPNMB through chromatin looping and recruitment of MEF2–HDAC epigenetic repressive complex. Our data highlight long-range chromatin interactions in functional genomic and epigenomic studies in autoimmunity.
INTRODUCTION
Takayasu arteritis is a granulomatous large vessel vasculitis, mainly affecting women of childbearing age.1 The disease primarily involves the aorta and its major branches, leading to thickening, stenosis and occlusion of involved vessels. Takayasu arteritis is relatively more prevalent in Asia and North Africa compared with Europe and North America.2
Although Takayasu arteritis occurs worldwide, the aetiology of this disease remains elusive, in part due to its rarity and indolent course. It is suggested that both genetic and environmental factors contribute to the development of Takayasu arteritis.2 Several genetic susceptibility loci have been identified and confirmed in Takayasu arteritis; however, there is no convincing evidence regarding the specific environmental factors that could be involved. The genetic association between Takayasu arteritis and HLA-B*52 has been confirmed in multiple cohorts and ethnicities.3 In addition, non-HLA susceptibility loci including FCGR2A/FCGR3A, IL12B, IL6, RPS9/LILRB3, PTK2B, LILRA3/LILRB2, DUSP22 and KLHL33 have been reported with a genome-wide level of significance.4–7
Similar to other autoimmune diseases, the majority of genetic susceptibility loci reported in Takayasu arteritis in genome-wide association studies are in non-coding regions. In addition, causal genetic variants in these loci and the target genes affected are largely unknown. Given the complex and three-dimensional nature of the human genome, genetic susceptibility loci do not necessarily affect genes within which they are located or to which they are closest but instead can potentially affect other target genes at a distance. Therefore, identifying causal variants and uncovering the potential regulatory effects within these genetic susceptibility loci and affected cell types are critical to revealing target genes and elucidating the mechanisms of disease susceptibility.
We have previously identified a genetic association between IL6 and Takayasu arteritis.4 Specifically, we reported a genetic association between a locus tagged by rs2069837(A/G) located within the second intron of the IL6 gene, with allele A being the disease risk allele in Takayasu arteritis. This same genetic variant has been reported to be associated with multiple other diseases and conditions. The Takayasu arteritis-associated risk allele in this variant is associated with longevity, spastic tetraplegia, cerebral palsy and antipsychotic-induced weight gain.8–11 In contrast, the Takayasu arteritis risk allele in rs2069837 was shown to be protective against late-onset alzheimer’s disease, cervical cancer, chronic hepatitis B virus infection, colorectal cancer and hepatocellular carcinoma.12–16
In the present study, we characterise the regulatory function of rs2069837 and reveal a distant target gene affected by this genetic polymorphism. The findings of this study mechanistically elucidate the genetic effect of this variant relevant to multiple disorders.
MATERIALS AND METHODS
Cell culture
HEK293 cell line (American Type Culture Collection (ATCC)) was cultured using Dulbecco’s Modified Eagle’s Mediumdium (DMEM, high glucose 4.5 g/L, Hyclone) supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin (100 U/mL). Cells were passaged at 80%–90% confluence. THP-1 cell line (ATCC) was cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 2 mM L-glutamine, 4.5 g/L glucose, 10% FBS (not heat inactivated) at a density between 0.5×106 and 1.0×106 cells/mL. For peripheral blood mononuclear cells or monocyte-derived macrophages, RPMI-1640 supplemented with 10% FBS was used.
Extraction and concentration detection of nuclear proteins
NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) were applied to extract nuclear protein according to the manufacturer’s manual. Next, Pierce BCA protein assay kit (Thermo Fisher Scientific) was used to measure the concentration of nuclear protein.
Electrophoretic mobility shift assay
Nuclear protein was extracted from HEK293 and THP-1 cell lines in different experiments. Thirty mer oligonucleotides flanking rs2069837 with A/G allele (5′-TGCCAGGCACTTTAA/GATAAATATTGTGTCT-3′) and their complementary oligonucleotides (5′-AGACACAATATTTATT/CTAAAGTGCCTGGCA-3′) were synthesised with or without 5′ end biotin label (Integrated DNA Technologies). The complementary oligomers were annealed into corresponding double-stranded DNA on Bio-Rad T100 Thermal Cycler (95°C for 5 min; step cooling (95°C (−1°C/cycle), 70 cycles; holding at 4°C). The biotin-labelled probes with A or G were incubated with nuclear extract in a binding buffer (50 ng/μl Poly (dI•dC), 2.5% glycerol, 0.05% nonidet P-40, 5 mM MgCl2, 10 mM EDTA, 20 μL system) using LightShift Chemiluminescent electrophoretic mobility shift assay (EMSA) Kit (Thermo Fisher Scientific) for 20 min at room temperature before separating on a 6% retardation gel (Thermo Fisher Scientific). For competitive EMSA, different fold excess (1–400×) of unlabeled DNA were added. After electrophoresis, the protein and DNA complexes were transferred to biodyne B nylon membrane (Thermo Fisher Scientific) and observed by chemiluminescent detection methods postultraviolet cross-linking.
Prediction of transcription factor binding
Different platforms were applied to analyse potential binding proteins to the IL6 locus, including HaploReg and Catalog of Inferred Sequence Binding Preferences (CIS-BP) database.17,18 HaploReg reports transcription factor binding experiments data from Encyclopedia of DNA Elements.17 CIS-BP database collects data from >25 sources, including other databases such as TRANSFAC, JASPAR, Homo sapiens Comprehensive Model Collection (HOCOMOCO) and FactorBook, and predicts binding proteins based on previous experiments or motif similarity across species. When 30 mer oligonucleotides used in EMSA were analysed, several proteins were predicted to bind to the flanking regions and were not related to the single-nucleotide polymorphism (SNP) of interest. Therefore, we narrowed down the input sequence to 16 oligonucleotides centering on rs2069837 with A or G (CACTTTAA/GATAAATAT) to focus on the proteins most likely affected by this SNP.
DNA affinity precipitation assay
Transcription factor purification was done using μMACS FactorFinder kit (Miltenyi Biotech, Germany). Protein–DNA binding reaction system was based on EMSA conditions and scaled up to purify enough proteins for further analysis. Specifically, 2–2.5 mg nuclear protein was incubated with 1 μg labelled probe with either A, G, mutated probe (5′- TGCCAGGCACTTT GTGC AAATATTGT GTC T-3′) or non-biotinylated oligos with A allele for 20 min at room temperature. Next, 100 μL μMACS streptavidin microbeads were added to the binding reactions and incubated for 15 min at room temperature. Protein–DNA–μMACS beads complexes were then loaded onto a column in a strong magnet separator. Washing and elution processes were done following the manufacturer’s instructions. The purified proteins were identified by mass spectrometry or validated by Western blotting. In addition, about 1.5% of the input nuclear extract in this experiment was stored and later used as loading control in Western blotting.
Identification of differential transcription factor binding by mass spectrometry
For the purified protein complex, complete reduction, alkylation and digestion were performed by dithiothreitol (10 mM, 30 min at room temperature), iodoacetamide (10 mM, dark for 30 min at 37°C) and trypsin (MS Grade, Promega, 12.5 ng/μl, 37°C overnight) sequentially before analysing by Thermo Scientific Orbitrap Fusion Tribrid Mass Spectrometer. The proteomics data were analysed by Proteome Discoverer software (Thermo Scientific). The number of peptide spectrum matches was used to compare the protein amount preliminarily. Proteins that were predicted to bind to the motif around rs2069837 and had twofold differences between A and G alleles were further validated by Western blotting.
Western blotting
Proteins purified from DNA affinity precipitation assay and the corresponding 1.5% input protein (loading control) as mentioned above were added to 4X Laemmli sample buffer and denatured by boiling for 10 min at 100°C on a heat block. Next, protein samples were resolved on a 4%–20% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose membranes. Various selected proteins were detected using mouse anti-GATA1, GATA2, myocyte enhancer factor 2A (MEF2A), MEF2C, CTCF, AT-rich interaction domain 3A (ARID3A), histone deacetylase (HDAC) 2, HDAC4, HDAC5, HDAC7 and TAFII (all purchased from Santa Cruz Biotechnology) and visualised using Super Signal West Dura substrate (Thermo Fisher Scientific) on the Omega Lum C imaging system (Gel Company).
Luciferase reporter assays
Luciferase reporter vectors were constructed using a promoterless pRMT-Luc vector (PR100001, Origene). A 315-nucleotide fragment of the human IL6 promoter (nucleotides –303 to +12, Ensembl ENSG00000136244)19 was inserted upstream of the luciferase gene with the restriction enzyme SpeI and MluI to construct an IL6 promoter-driven pRMT-Luc vector. In addition, a 251-nucleotide fragment flanking rs2069837 with A or G allele (hg19 chr7:22767927–22768177) was then inserted upstream of the IL6 promoter sequence using XbaI and EcoRI restriction sites.
For transfection, 3×104 HEK293 cells/well were seeded in a 96-well plate. Sixteen to twenty hours later, the cells were transfected with different luciferase vectors (100 ng) along with 0.5 ng renilla vector (pGL4.74, (hRluc/TK), Promega) using Lipofectamine 3000 reagent (Invitrogen). When THP-1 cells were transfected, SG Cell Line 4D- Nucleofector X Kit and Lonza Amaxa Nucleofection system were used (Program FF100) according to the manufacturer’s protocols. After 48 hours, the luminescence of firefly luciferase and renilla luciferase were detected by Synergy H1 multimode microplate reader (BioTek) using Dual-Glo luciferase reporter assay reagents (Promega). The activity of firefly luciferase was normalised by renilla luciferase. Each condition had six replicated wells, and the experiment was repeated three times. In the experiment using HDAC pan-inhibitor, Trichostatin A (TSA, 400 nM) or dimethyl sulfoxide (DMSO) was added 2 hours prior to transfection and throughout the cell culture until luminescence was assessed.
Real time PCR-based SNP genotyping
TaqMan SNP Genotyping Assay for rs2069837 and TaqMan genotyping master mix (Thermo Fisher Scientific) were used to detect rs2069837 genotype of 48 healthy subjects from whom peripheral blood mononuclear cells (PBMC) were collected and stored. Genomic DNA of 10 ng was used for each sample, and same volume of DNase-free water was used as negative control.
Peripheral blood monocytes isolation and differentiation into macrophages
Seven pairs of PBMC samples from healthy subjects with AA or AG genotypes at rs2069837 were used. The age, sex and ethnicities were matched between these two groups of subjects. Frozen PBMC were thawed quickly in a 37°C water bath and suspended in prewarmed RPMI-1640 medium supplemented with 10% FBS and 25 U/mL benzonase. After washing twice, PBMC were counted and transferred to 12-well plates in 3×106 cells/mL with RPMI-1640 supplemented with 10% FBS and 20 ng/mL macrophage colony-stimulating factor (M-CSF). Two days later, the floating cells were removed, while adherent cells were washed twice and incubated with fresh medium supplemented with M-CSF for another 2 days. On the fourth day, fibroblast-like macrophages were obtained and media were changed again. RNA was extracted after another 24 hours. In the experiment with HDAC inhibition, the media were supplemented with DMSO or HDAC pan-inhibitor TSA (100 nM) for the final 24 hours.
Extraction of messenger RNA and quantitative reverse transcription PCR
RNA was prepared using Direct-zol RNA MiniPrep kit (Zymo research) and converted to complementary DNA (cDNA) using Verso cDNA Synthesis kit (Thermo Fisher Scientific). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) on a ViiA 7 real-time PCR system. The primers used in the present study are listed in online supplementary table 1. Gene expression data were normalised to beta-actin.
Chromosome conformation capture (3C)
THP-1-or primary monocytes-derived macrophages were used in this experiment. 1×107 THP-1 cells were seeded in a T75 flask with RPMI-1640 medium supplemented with 10% FBS and treated with 120 ng/mL phorbol 12-myristate-13-acetate (PMA) for 24 hours. Next, RPMI-1640 without FBS and PMA was used to rest cells for 24 hours. For primary monocytes, 1×108 PBMC from healthy donors were cultured for 4 days with M-CSF (20 ng/mL) as mentioned above to obtain macrophages. For 3C experiments, we followed the protocol published by Hagège et alwith some modifications.20 Briefly, per 1×107 cells, 10 mL 2% formaldehyde was used to fix the cells, and glycine (final concentration 0.125M) was applied to quench the formaldehyde. Then the cells were lysed to obtain nuclei, which were digested using 100U BsrDI (NEB) at 65°C overnight. Ligase (NEB) of 2000U T4 was applied to ligate the chromatin fragments (4 hours at 16°C, followed with 0.5 hour at room temperature) after restriction enzyme inactivation (20 min at 80°C). Finally, 300 μg proteinase K was used to decross-link the chromatin (65°C, overnight), and the ligated fragments were purified by phenol-chloroform extraction. A control template from four bacterial artificial chromosome (BAC) clones (CH17–8M1, CH17–33O24, CH17–211I22, CH17–41A22, BACPAC Resources Center) which together cover 520 kb sequence spanning from IL6 to GPNMB with minimally overlapping sequences were used. After culturing the clones overnight in 400 mL LB Broth (Lennox, Sigma), DNA from each clone was extracted using nucleoBond Xtra BAC kit (Macherey Nagel). Then equimolar amounts of DNA from those clones (total 10 μg) were digested using BsrDI and ligated by the T4 ligase following the conditions as mentioned above. During this process, digestion efficiency was detected by quantitaive PCR (qPCR) using chromatin before and after digestion with pairs of primers targeting each restrictions site, and only samples that had over 65% digestion efficiency were used. In 3C-qPCR, a constant primer was designed upstream of a BsrDI site (871 bp downstream of rs2069837). Paired primers for 3C detection were designed. The loading control was evaluated with serial dilutions of genomic DNA from THP-1 cells or primary monocytes-derived macrophages using primers targeting a DNA sequence devoid of BsrDI restriction sites in GAPDH. The sequence of all primers used in this experiment are listed in online supplementary table 2. Relative cross-linking frequencies were normalised to both GAPDH loading control and BAC clones.
Statistical analysis
Results were presented as the mean±SD. The differences of luciferase signals between constructs with A and G alleles, and gene expression in macrophages between samples with AA and AG genotypes were compared by Student’s t-test. Paired t-test was used to compare AA or AG samples with or without TSA treatment. All the analyses were performed in GraphPad Prism V.7.03. P values less than 0.05 were considered significant.
RESULTS
We have previously revealed a genetic association between Takayasu arteritis and rs2069837 (OR=2.07, p=6.70×10−9), which is located in the second intron of IL6 (hg19, chr7: 22768026). No genetic variants in linkage disequilibrium with rs2069837 (r2≥0.6) were identified, suggesting that rs2069837 is the likely causal variant in this locus.4 DNase hypersensitivity and acetylation of histone H3 on lysine 27 (H3K27ac) data in various immune cells suggest that this genetic variant is near an enhancer region (figure 1A). To identify regulatory proteins that bind rs2069837, we first performed an EMSA using DNA oligos with the Takayasu arteritis risk allele (allele A) and the protective allele (allele G) in rs2069837. Using nuclear proteins extracted from HEK293 or THP-1 cells, we show that more DNA protein complexes were formed in the presence of allele A than allele G (figure 1B, online supplementary figure 1A). Both complexes can be competed out by their respective unlabeled probes. In addition, unlabeled probe with A allele was able to compete for binding and eliminate the shifted band formed in the presence of probe with G allele in both cell lines (figure 1B, online supplementary figure 1A). When we used different folds of unlabeled allele A probe to compete with labelled allele A probe, a 10-fold concentration (1.2 pmol) of unlabeled probe was sufficient to almost completely compete out the shifted band, suggesting that the binding affinity between the DNA sequence and nuclear proteins was high (figure 1C).
Figure 1.
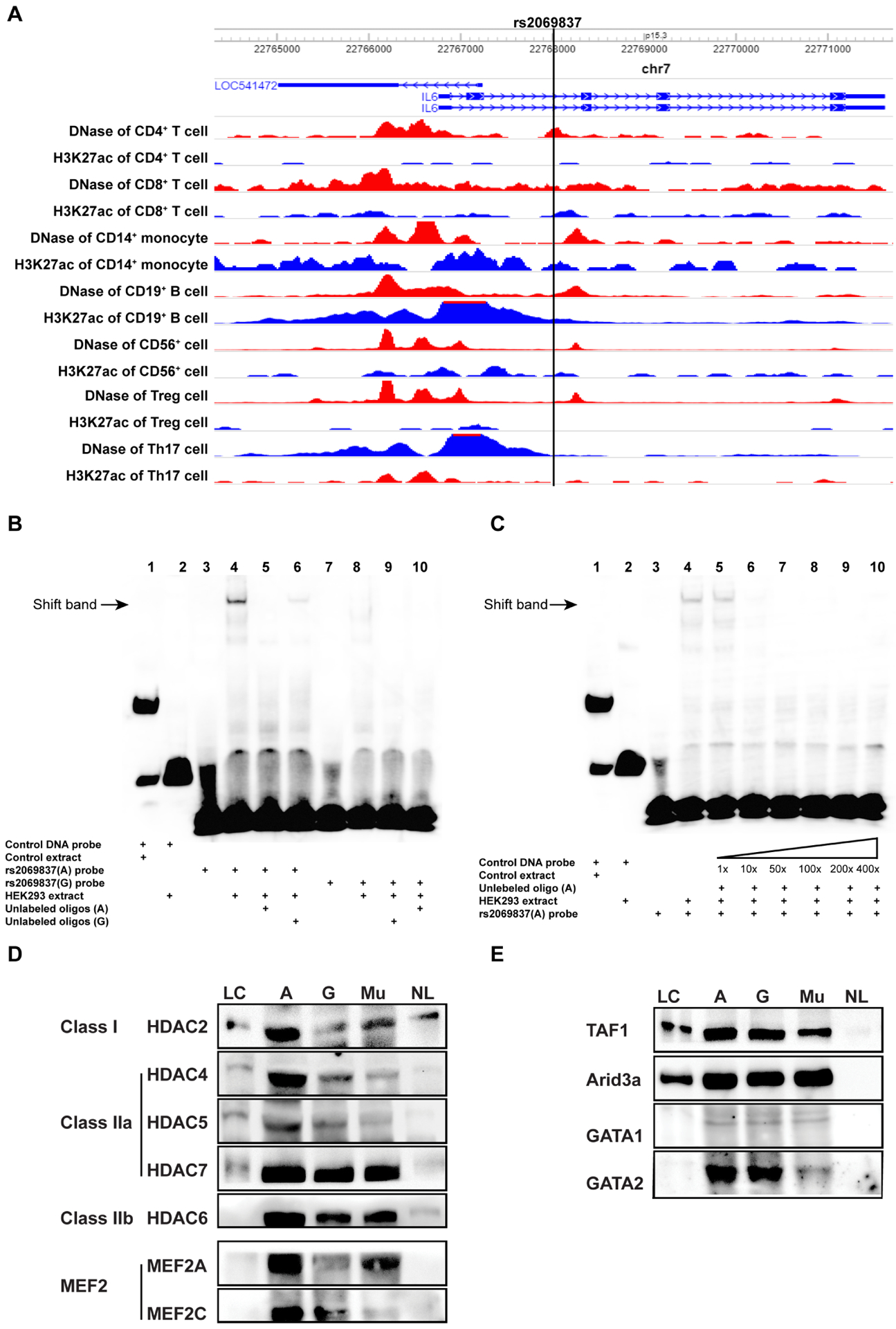
Characterisation of rs2069837 and detection of binding proteins. (A) rs2069837 (hg19, chr7: 22768026) is located in the second intron of IL6. Epigenetic markers (DNase and H3K27ac) in multiple immune cells indicate that this locus is close to a functional region. (B) EMSA assay using nuclear protein from HEK293 cells demonstrating that more DNA–protein complexes were formed with the probe with allele A (lane 4) than allele G (lane 8); both unlabeled probes with A allele or G allele (24 pmol, 200×) can totally compete out the corresponding shifted band separately (lane 5 and lane 9); unlabeled G allele probe cannot completely compete out the shifted band formed with the A allele probe (lane 6); unlabeled A allele probe was able to compete out the shifted band formed in the presence of the G allele probe (lane 10). (C) Different concentrations of the unlabeled A allele probe were applied in the competitive EMSA assay (1×, 10×, 50×, 100×, 200×, 400×). At 10× concentration, the band can be competed out (lane 6). (D, E) Validation using Western blotting. More HDAC and MFE2 proteins bound to the probe with A than with G allele, while no differences were detected in TAF1, ARID3A, GATA1 and GATA2 between probes with A and G alleles. When the four nucleotides around rs2069837 were mutated, the binding of those proteins decreased except for ARID3A. The protein binding in the unlabeled group was barely observed. A, probe with A allele; EMSA, electrophoretic mobility shift assay; G, probe with G allele; Mu, probe with mutated sequence; NL, non-biotinylated oligos with A allele; LC, loading control.
To investigate possible regulatory proteins that bind rs2069837, we first examined publicly available data using HaploReg and CIS-BP databases.17,18 Publicly available experimental data indicate that rs2069837 can alter motifs of several important transcription regulators (see online supplementary table 3). The changes of log-odds scores between reference allele A and alternative allele G indicated alterations of binding affinities for corresponding transcription regulators. These scores were higher for almost all the binding motifs in the presence of allele A compared with allele G, except TATA_known3, indicating that the risk allele A potentially binds more proteins, consistent with our EMSA results. The binding affinities of MEF2 and FOXJ1 increased the most in the presence of allele A, followed by ARID3A, FOXA, FOXD3, FOXF1 and FOXI1. When we used CIS-BP database to predict potential binding proteins to the rs2069837 locus, we identified ARID3 proteins, Forkhead family proteins, MEF2 and GATA proteins. Among these, ARID3 and MEF2 proteins were shown only in the sequence with A allele, while GATA proteins were predicted to bind only the sequence with G allele (see online supplementary table 4).
Next, we used DNA affinity precipitation followed by mass spectrometry to confirm proteins that differentially bind to the motif around rs2069837 in the presence of A and G alleles (see online supplementary table 5). MEF2 proteins were only detected in proteins bound to DNA with A allele, while more GATA proteins appeared in complexes with G allele, which is consistent with bioinformatics prediction and mining publicly available data. However, no remarkable differences were observed in the binding of ARID family, forkhead family and TATA binding proteins associated factors between A and G alleles. In addition, HDACs were also detected in these protein complexes, among which HDAC5 was detected in complex with allele A. These results were further validated by Western Blotting which showed increased MEF2 and HDAC binding in the presence of the A allele compared with the G allele, consistent with mass spectrometry results (figure 1D,E). It has been known that HDAC class IIa family such as HDAC4, HDAC5 and HDAC7 have a unique regulatory domain which can mediate an interaction with MEF2, causing the repression of MEF2-target genes. Taken together, our data indicate that the Takayasu arteritis risk allele (allele A) in rs2069837 induces transcriptional repression due to preferential recruitment of the MEF2–HDAC complex, thereby weakening the enhancer function in this locus.
To confirm that rs2069837 is in a regulatory region and that allele A is associated with transcriptional repression, we used a luciferase reporter assay in HEK293 cells (figure 2A). After normalising to renilla luciferase activity, both plasmids inserted with the intronic sequence containing A or G showed significantly higher luciferase activity compared with the basic vector or promoter-only vector (p<0.05, figure 2B), indicating that rs2069837 is within a regulatory region. When comparing luciferase activity between plasmids with A and G alleles, the plasmid with G allele demonstrated stronger luciferase activity (p<0.05, figure 2B). This was also confirmed in THP-1 cells (see online supplementary figure 1B). These data suggest that rs2069837 is in a regulatory region, which is significantly more repressive in the presence of the Takayasu arteritis risk allele (allele A), consistent with our findings above. To test the hypothesis that allele A induces transcriptional repression by preferentially recruiting MEF2–HDAC complex, we assessed the effect of HDAC inhibitor TSA on luciferase activity in the presence of the A and G alleles. With TSA treatment, the luciferase activity was increased in both constructs with A and G alleles, confirming recruitment of HDAC to rs2069837. Importantly, luciferase activity was significantly more increased with TSA treatment in the presence of the A allele, confirming a functional consequence of the preferential recruitment of MEF2–HDAC complex to allele A in rs2069837 (figure 2C).
Figure 2.
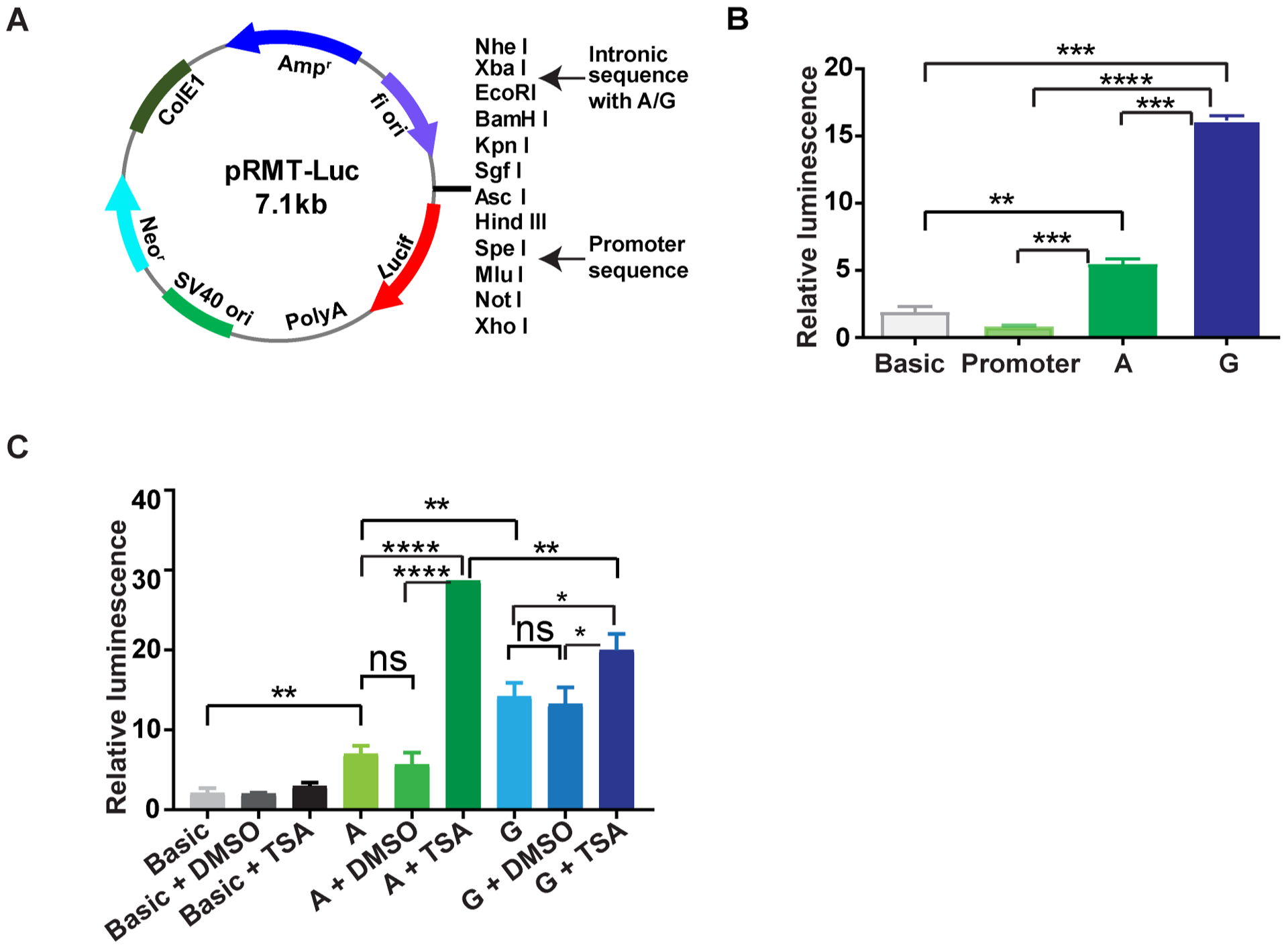
Luciferase reporter assay to assess enhancer function in rs2069837 and the effect of HDAC inhibition. (A) The intronic sequence with A or G (hg19 chr7:22767927–22768177) was inserted between XbaI and EcoRI, while the promoter sequence (nucleotides −303 to +12, Ensembl ENSG00000136244) was inserted between SPEI and MluI. (B) No increase in luciferase expression was observed in promoter-only vector. Significantly increased luciferase expression was detected in both vectors with intronic sequence (A/G), with significantly higher expression in the construct with G allele than that with A allele (p<0.05). (C) TSA treatment increased luciferase expression in both constructs with intronic sequence (A/G) compared with DMSO-treated samples. In the presence of TSA, the luciferase expression was significantly higher in the construct with A allele than that with G allele (p<0.05). DMSO, dimethyl sulfoxide; HDAC, histone deacetylase; TSA, Trichostatin A.
To identify target genes regulated by the rs2069837 locus, we examined chromatin interactions involving rs2069837 using DNAase hypersensitivity site (DHS) linkage patterns. In this analysis, distal DHSs (relative to transcription start sites of genes) were correlated using Pearson correlation coefficient with DHSs in gene promoters across multiple cell types.21,22 When a distal DHS highly correlates with another DHS in a gene promoter region, it is reasonable to assume a regulatory effect from the distal DHS on that gene. Multiple DHS linkages were observed between rs2069837 and other DHS loci (hg19, chr7: 22528026, chr7:22568026, chr7:22688026, chr7:22968026, chr7:23128026, chr7:23228026, chr7:23248–026, chr7:23288026). These potentially interacting regions covered several genes, including STEAP1B, LOC100506178, LOC541472, IL6, TOMM7, SNORD93, FAM126A, KLHL7, NUPL2 and GPNMB (figure 3A). Since CTCF-mediated chromatin looping is the most common mechanism for long-distance chromatin interaction,23 we also checked CTCF enrichment at this region using ChIP-seq data in immune cells through WashU EpiGenome browser. ChIP-seq data for CTCF were only available in monocytes and B cells, and the rs2069837 locus was enriched for CTCF binding in both cell types (figure 3A). CTCF binding site in multiple cell types in aggregate is shown in figure 3B. Binding of CTCF to this locus was further validated by Western blotting and was not different between alleles A and G in rs2069837, and the mutated sequence, indicating that the binding of CTCF occurs in this locus but is not affected by rs2069837 (figure 3C). Moreover, CTCF and units of cohesin complex, which is composed of multiple proteins that play important role in CTCF-mediated looping, were also detected in our mass spectrometry results (see online supplementary table 6).
Figure 3.
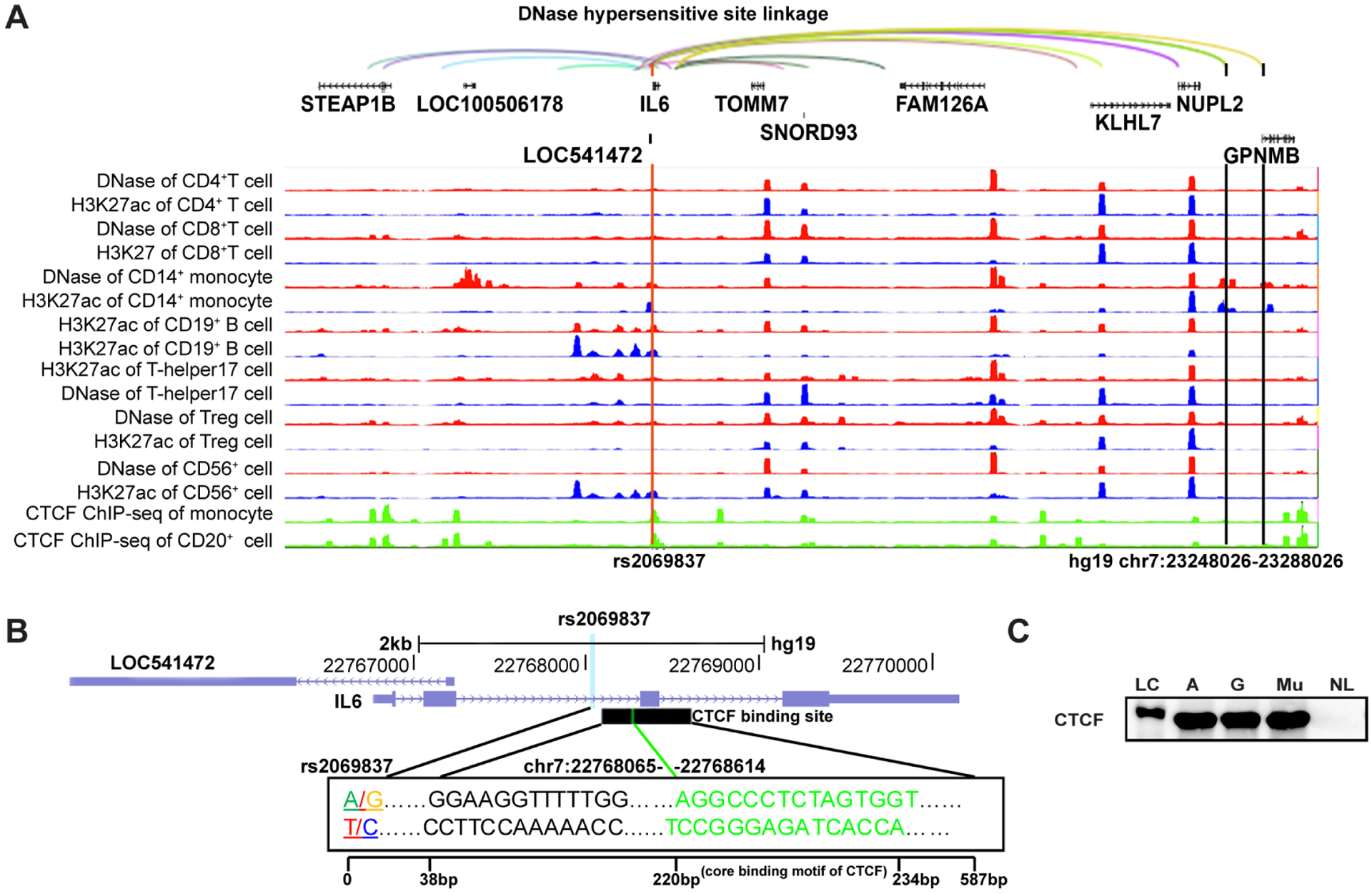
Potential interactions between rs2069837 and distant loci. (A) DNase hypersensitivity linkage (40 kb resolution) showed correlation of DNase hypersensitivities between rs2069837 and several distant loci. The interaction region (890 kb) included several genes such as STEAP1B, LOC100506178, LOC541472, IL6, TOMM7, SNORD93, FAM126A, KLHL7, NUPL2 and GPNMB from 5′ to 3′. The interaction sites from upstream to downstream were chr7: 22528026, chr7:22568026, chr7:22688026, chr7:22968026, chr7:23128026, chr7:23228026, chr7:23248–026, chr7:23288026 (hg19). Interaction sites chr7: 23248026 and chr7:23288026 overlapped with DNase hypersensitivity and H3K27ac peaks in monocyte, where GPNMB is the closest gene. In addition, CTCF binding is enriched at rs2069837 in monocytes and B cells. (B) CTCF binding site downstream of rs2069837 (position: chr7:22768065–22768614, 38–587 bp downstream of rs2069837; core binding motif: chr7:22768247–22768261, 220–234 bp downstream of rs2069837). The black bar/track shows regions of transcription factor binding derived from a large collection of ChIP-Seq experiments across multiple cell types, presented using genome browser. (C) Western blot analysis demonstrating CTCF binding to the probes with A or G alleles. No difference between A, G or mutated sequence was detected. A, probe with A allele; G, probe with G allele; LC, loading control; Mu, probe with mutated sequence; NL, non-biotinylated oligos with A allele.
Next, we explored whether the rs2069837 locus regulates IL6 or other genes indicated by chromatin interaction loops using DHS data (STEAP1B, LOC100506178, LOC541472, TOMM7, SNORD93, FAM126A, KLHL7, NUPL2, GPNMB). First, we performed functional annotation analysis of these genes using the Database for Annotation, Visualization and Integrated Discovery (DAVID). The results demonstrated that GPNMB has a close relationship with IL-6 and participates in multiple biological processes such as angiogenesis (see online supplementary table 7). No relevant significant finding was found for the other genes. By further literature review, we found that GPNMB is expressed by multiple cell types including monocytes, and it is increasingly expressed when monocytes differentiate into macrophages. Because monocytes/macrophages play an important role in the pathogenesis of Takayasu arteritis,24–27 and epigenetic patterns (DNase hypersensitivity and H3K27ac; figure 3A) also indicated that the rs2069837-interacting site in the GPNMB locus is a potential regulatory region in monocytes rather than other immune cells, we used primary monocyte-derived macrophages for our subsequent functional studies.
Forty-eight normal healthy individuals were genotyped for rs2069837 (see online supplementary figure 1C). Among them, seven were AG and the remaining subjects were AA genotypes. Thus, we selected seven pairs of age, sex and ethnicity-matched individuals with AG and AA genotypes for further studies (see online supplementary table 8). In monocyte-derived macrophages, we observed no differences in IL-6 mRNA expression, while GPNMB expression was significantly lower in macrophages derived from subjects with AA compared with AG genotype (figure 4A). No differential expression was observed in all other interacting genes between samples with AA and AG genotypes (see online supplementary figure 1D). Importantly, TSA treatment significantly enhanced GPNMB expression in individuals with AA genotype but not AG genotype (figure 4B,C). This is consistent with our data derived from the luciferase assays and confirms a role for preferential recruitment of MEF2–HDAC complex in regulating GPNMB expression in the presence of the A allele in rs2069837.
Figure 4.
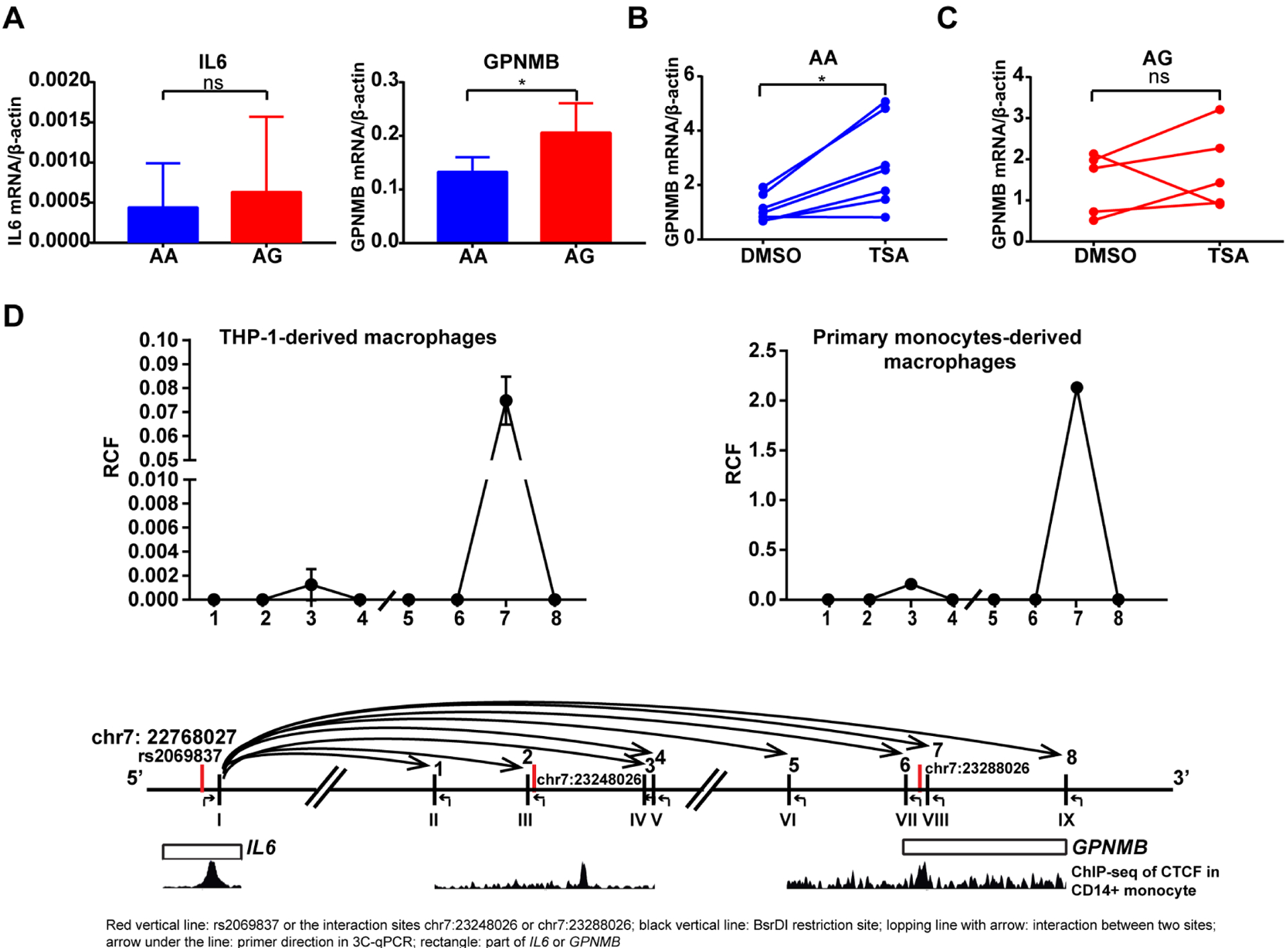
GPNMB is a target gene repressed by the Takayasu arteritis risk allele in rs2069837 through long-distance chromatin looping. (A) The mRNA expression level of GPNMB was significantly lower in monocyte-derived macrophages with AA compared with AG genotypes (p<0.05, n=7 in each group). No difference was observed in IL-6 expression. (B) TSA treatment (100 nM) significantly increased GPNMB expression in monocyte-derived macrophages with AA genotype (p<0.05, n=7). (C) TSA treatment (100 nM) did not affect GPNMB expression in monocyte-derived macrophages with AG genotypes (p>0.05, n=5). (D) 3C data revealed the interaction between rs2069837 and chr7:23288026 (GPNMB) in THP-1 cells (representative of two replicates) and primary monocytes-derived macrophages (representative of two independent samples). The interactions examined relative to chromosomal positions are shown in the bottom panel. Restriction sites are depicted using Latin numbers, and interaction loops tested are depicted in Arabic numbers. ChIP-Seq data of CTCF in CD14+ monocyte demonstrated that CTFC was strongly enriched at rs2069837 and chr7:23288026 (hg19) but weakly enriched at chr7:23248 026 (hg19). The relative positions of restriction sites are as follows: I: +871 bp of rs2069837 and +284 bp of CTCF core binding motif, II: −7797 bp of chr7:23248026, III: −29 bp of chr7:23248026, IV: +6906 bp of chr7:23248026, V: +7179 bp of chr7:23248026, VI: −9293 bp of chr7: 23288026, VII: −1933 bp of chr7:23288026; VIII:+578 bp of chr7:23288026; IX:+9224 bp of chr7:23288026. mRNA, messenger RNA; TSA, Trichostatin A.
Further examination of the interaction between rs2069837 and GPNMB based on DHS linkage indicates two interaction sites, one 38.29 kb upstream of GPNMB (hg19, chr7:23248026), while the other (hg19, chr7:23288026) is located in the first intron of GPNMB. Both loci are within DNase and H3K27ac peaks in monocytes, and CTCF ChIP-seq data also indicated peaks at these loci (figure 3A and 4D). Other positive epigenetic marks (H3K4me1 and H3K4me3) in monocytes further demonstrate that these two rs2069837-interacting loci, especially chr7:23288026, are within important regulatory regions in GPNMB (see online supplementary figure 1E). In addition, ChIP-seq data in several cell lines showed GATA and MEF2 enrichment at these loci which were also demonstrated by analysing the sequence motifs (see online supplementary figure 1E).
To confirm the interaction between rs2069837 and GPNMB, a 3C experiment was performed which demonstrated an interaction between the rs2069837 locus and a locus close to chr7:23288026 (figure 4D). Since the BsrDI restriction sites close to rs2069837 and chr7: 23288026 were also close to the regions enriched with CTCF, CTCF was assumed to mediate this interaction.
Taken together, our data indicate that the Takayasu arteritis risk allele in rs2069837 represses an enhancer function in this locus by recruiting MEF2–HDAC complex and inhibits GPNMB expression via long-distance intrachromatin looping likely mediated by CTCF (figure 5).
Figure 5.
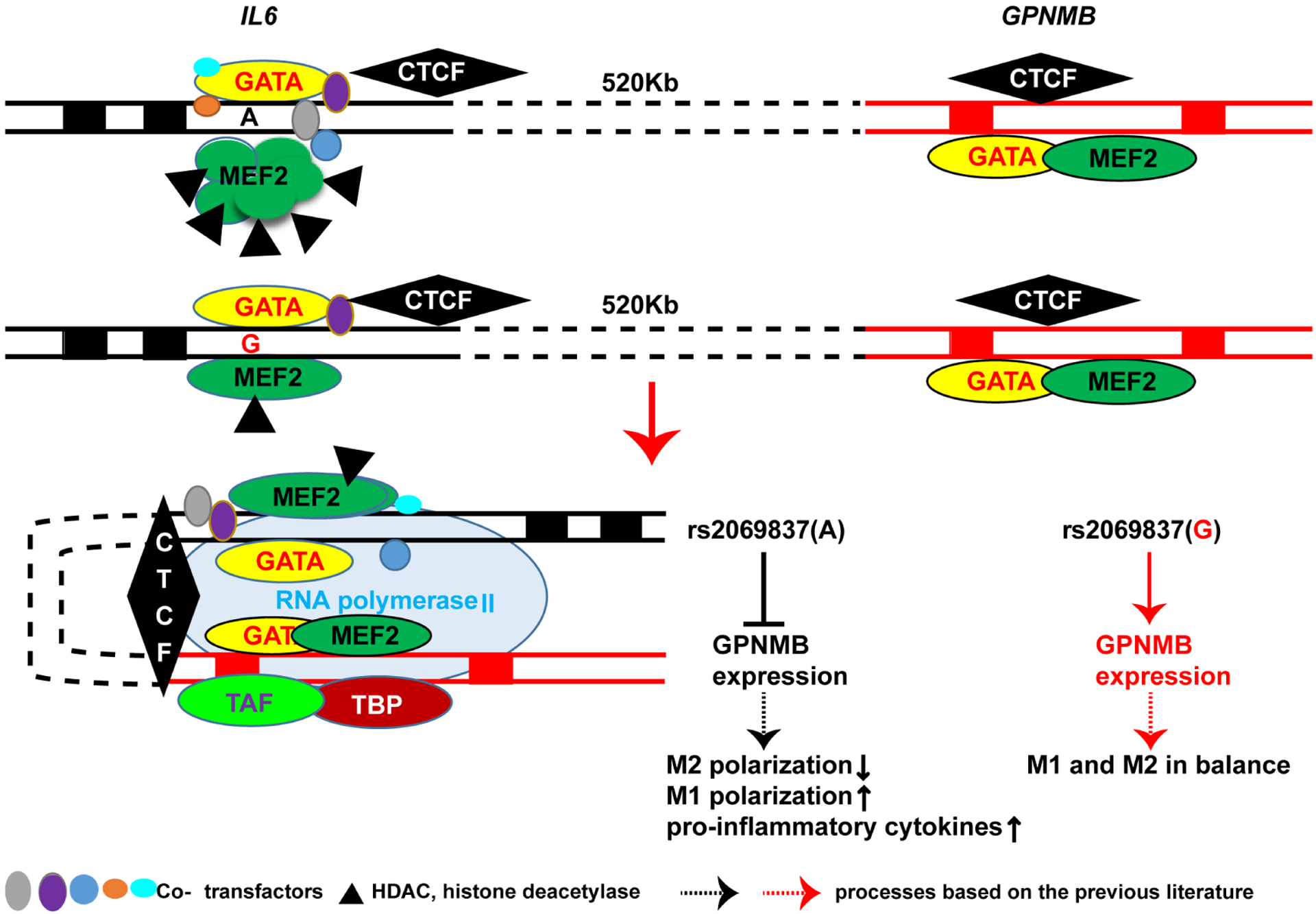
A proposed model for the regulatory mechanism of rs2069837 on the expression of GPNMB. The Takayasu arteritis risk allele A at rs2069837 preferentially recruits MEF2 and thereby HDAC proteins compared with the G allele resulting in a repressive effect by weakening the enhancer function at this locus. CTCF binding downstream of this SNP mediates the interaction between this locus and a regulatory locus in GPNMB, where a CTCF binding site also exists. The differential binding of MEF2–HDAC complex between A and G results in differential expression of GPNMB, with inhibited expression in the presence of risk allele A. Inhibiting GPNMB suppresses M2 macrophage polarisation and enhances M1 polarisation and overexpression of proinflammatory cytokines. HDAC, histone deacetylase; MEF2, myocyte enhancer factor 2; SNP, single-nucleotide polymorphism.
DISCUSSION
In this study, we demonstrate that rs2069837 is in an enhancer region that is preferentially suppressed in the presence of the Takayasu arteritis risk allele, and although located within IL6, it affects the expression of a distant anti-inflammatory gene in human monocyte-derived macrophages. We show that the risk allele A recruits a repressive MEF2–HDAC protein complex, weakening the enhancer function at this locus. We confirmed CTCF binding to this locus, and an interaction between rs2069837 and a regulatory region in GPNMB located ~520 kb downstream of this SNP, suggesting CTCF-mediated long-distance chromatin looping. Indeed, we demonstrated reduced expression of GPNMB in monocyte-derived macrophages with AA genotype, which can be restored by inhibiting HDAC. A reduction in GPNMB expression in the presence of the Takayasu arteritis risk allele in rs2069837 may enhance inflammatory responses, suggesting a mechanism for a pathogenic role of rs2069837 in Takayasu arteritis, while at the same time explaining a protective effect of this same allele in chronic viral infection and malignancy. Previously, studies have demonstrated an anti-inflammatory role for GPNMB by enhancing M2 and suppressing M1 macrophage differentiation.28 The importance of proinflammatory macrophages in Takayasu arteritis has been well established.
Using a combination of bioinformatics and experimental approaches, we revealed that MEF2 and HDAC proteins differentially bind to rs2069837 (more in A allele), which was highly likely to contribute to the differential regulatory function between A and G alleles. MEF2 is a transcription factor that regulates several cellular processes including differentiation, proliferation and apoptosis.29 The N-terminus of MEF2 contains a highly conserved MADS-box and an adjacent motif termed as the MEF2 domain, which together mediate dimerisation, DNA binding and cofactor interactions.30 The cofactors binding at MEF2 N-termini determine MEF2 functions. Binding of GATA proteins activates the expression of MEF2-target genes, while binding of HDAC IIa proteins represses the expression of MEF2-target genes.30 In the present study, the same amounts of GATA proteins were observed to bind at probes with A and G alleles, while multiple HDAC proteins were detected at higher levels in the presence of A allele, including HDAC IIa (HDAC4, 5 and 7). Thus, the enhancer function was weakened in this locus with A allele, as we also showed using a luciferase reporter assay with and without an HDAC inhibitor. It was reported that the deacetylase activity of HDAC IIa family is not strong, but that they can recruit other robust repressors such as HDAC I proteins.31 Indeed, we showed significantly enriched recruitment of HDAC2 (an HDAC I family member) in the presence of the A allele in rs2069837, probably resulting from the presence of HDAC IIa proteins which are recruited by MEF2.
The MEF2–HDAC axis plays a role in several differentiation pathways and numerous adaptive responses, including skeletal muscle differentiation, heart development and vascular integrity.32 In most conditions, due to the suppression mediated by HDAC, MEF2-target genes cannot be expressed, leading to pathophysiological alterations, as implicated in pulmonary arterial hypertension and oestrogen receptor positive breast tumours.33,34 Therefore, strategies to release MEF2 from HDAC proteins have been attempted. Recent findings demonstrated that inhibition of HDAC IIa or blocking the interactions of HDAC IIa and MEF2 were two potential therapeutic strategies.35,36
Existence of CTCF binding sites close to rs2069837 and CTCF enrichment at this region indicated that CTCF might mediate interactions between rs2069837 and other loci via chromatic looping. Our mass spectrometry data also showed CTCF and cohesin complex binding to this locus, further suggesting formation of chromatin loops involving this locus. Furthermore, no difference in CTCF binding between A allele and G allele was observed, implying that CTCF-mediated looping was not altered by rs2069837. Using DNase hypersensitivity maps followed by expression analysis in monocyte-derived macrophages, we identified GPNMB as target gene regulated by rs2069837. The 3C data further confirmed the interaction between the rs2069837 locus and a regulatory region in GPNMB. Of interest, we did not detect a direct effect from rs2069837 on IL-6 expression, despite that this polymorphism is located within IL6. These data suggest that in monocytes/macrophages, it does not appear that this genetic variant is directly relevant to the transcriptional regulation of IL6. It should be noted, however, that this does not rule out a possible direct transcriptional effect of rs2069837 on IL6 expression in other cell types. Further studies to examine the direct effect of rs2069837 alleles in homogeneous genetic backgrounds using CRISPR/Cas9 genetic editing are warranted.
GPNMB is a negative regulator of inflammatory responses in macrophages. In dextran sulfate sodium-induced colitis model, the deficiency of GPNMB resulted in more severe colitis characterised by higher levels of proinflammatory cytokines including IL-6.37 GPNMB is also involved in M2 polarisation of macrophages. In GPNMB-knockdown mice, M2 polarisation from bone marrow-derived macrophages was inhibited, while M1 polarisation and the secretion of IL-1β and TNF-α were enhanced.38 Therefore, it is possible that lower levels of GPNMB in the presence of allele A can indirectly contribute to the expression of multiple inflammatory cytokines including IL-6, which is important in the pathogenesis of Takayasu arteritis.
Previous genetic association studies have reported a protective effect of the A allele in rs2069837 in late-onset alzheimer’s disease, cervical cancer, chronic hepatitis B virus infection, colorectal cancer and hepatocellular carcinoma.12–16 Based on our data, the A allele in rs2069837 is associated with suppression of GPNMB expression, which is consistent with the findings that GPNMB expression is increased in Alzheimer’s disease, cancer and infections.39–41 GPNMB weakens the immune response against cancer cells and infections, contributing to cancer development or chronic infections.39–41 Since GPNMB has been implicated in multiple tumours, our data discovering genetic regulation of GPNMB by rs2069837 via MEF2–HDAC complex also shed light on mechanisms that can be potentially targeted in malignant disorders.
In conclusion, our study elucidated a pathogenic mechanism underlying the genetic association between rs2069837 and Takayasu arteritis. In addition, our findings are relevant to other disorders, including chronic infection and malignancy. Targeting GPNMB and the MEF2–HDAC complex might provide novel therapeutic strategies in Takayasu arteritis. Our work highlights long-range chromatin interactions involving regulatory variants in elucidating functional consequences of disease-associated genetic variants to pave the way for using genetic data in identifying novel therapeutic approaches.
Supplementary Material
Key messages.
What is already known about this subject?
Our previous work revealed a genetic association between Takayasu arteritis and a non-coding genetic variant in an enhancer region within IL6 (rs2069837 A/G).
The Takayasu arteritis-associated allele in this variant has a protective effect against chronic viral infection and cancer.
What does this study add?
This study characterised the functional consequences of this disease-associated risk locus.
The Takayasu arteritis-associated variant in this locus represses the anti-inflammatory gene GPNMB, which is located about 520 kb away, through chromatin looping and recruitment of myocyte enhancer factor 2–histone deacetylase repressive complex.
Our data highlight long-range chromatin interactions in functional genomic and epigenomic studies in autoimmunity.
How might this impact on clinical practice or future developments?
GPNMB is a novel target molecule that might play a role in the pathogenesis of Takayasu arteritis and can be considered for therapeutic intervention.
Funding
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health grant number R01AR070148.
Footnotes
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement All data relevant to the study are included in the article or uploaded as supplementary information.
REFERENCES
- 1.Seyahi E Takayasu arteritis: an update. Curr Opin Rheumatol 2017;29:51–6. [DOI] [PubMed] [Google Scholar]
- 2.Koster MJ, Matteson EL, Warrington KJ. Recent advances in the clinical management of giant cell arteritis and Takayasu arteritis. Curr Opin Rheumatol 2016;28:211–7. [DOI] [PubMed] [Google Scholar]
- 3.Renauer P, Sawalha AH. The genetics of Takayasu arteritis. La Presse Médicale 2017;46:e179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renauer PA, Saruhan-Direskeneli G, Coit P, et al. Identification of susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in Takayasu arteritis in a genome-wide association study. Arthritis Rheumatol 2015;67:1361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saruhan-Direskeneli G, Hughes T, Aksu K, et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am J Hum Genet 2013;93:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terao C, Yoshifuji H, Kimura A, et al. Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the Il12b and HLA-B regions in a Japanese population. Am J Hum Genet 2013;93:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terao C, Yoshifuji H, Matsumura T, et al. Genetic determinants and an epistasis of LILRA3 and HLA-B*52 in Takayasu arteritis. Proc Natl Acad Sci U S A 2018;115:13045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Y, Nie C, Min J, et al. Novel loci and pathways significantly associated with longevity. Sci Rep 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi D, Chen M, Zhang X, et al. The association between sex-related interleukin-6 gene polymorphisms and the risk for cerebral palsy. J Neuroinflammation 2014;11:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, Li T, Lin S, et al. Association of interleukin 6 gene polymorphisms with genetic susceptibilities to spastic tetraplegia in males: a case-control study. Cytokine 2013;61:826–30. [DOI] [PubMed] [Google Scholar]
- 11.Fonseka TM, Tiwari AK, Gonçalves VF, et al. The role of genetic variation across IL-1β, IL-2, IL-6, and BDNF in antipsychotic-induced weight gain. World J Biol Psychiatry 2015;16:45–56. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X-M, Gao Y-F, Zhou Q, et al. Relationship between interleukin-6 polymorphism and susceptibility to chronic hepatitis B virus infection. World J Gastroenterol 2013;19:6888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng X, Han C, Shan R, et al. Association of interleukin-6 polymorphisms with susceptibility to hepatocellular carcinoma. Int J Clin Exp Med 2015;8:6252–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S-Y, Chen T-F, Lai L-C, et al. Sequence variants of interleukin 6 (IL-6) are significantly associated with a decreased risk of late-onset alzheimer’s disease. J Neuroinflammation 2012;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsusaka S, Hanna DL, Cao S, et al. Prognostic impact of IL6 genetic variants in patients with metastatic colorectal cancer treated with bevacizumab-based chemotherapy. Clin Cancer Res 2016;22:3218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi T-Y, Zhu M-L, He J, et al. Polymorphisms of the interleukin 6 gene contribute to cervical cancer susceptibility in eastern Chinese women. Hum Genet 2013;132:301–12. [DOI] [PubMed] [Google Scholar]
- 17.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in encode TF binding experiments. Nucleic Acids Res 2014;42:2976–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weirauch MT, Yang A, Albu M, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014;158:1431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann E, Ashouri J, Wolter S, et al. Transcriptional regulation of Egr-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J Biol Chem 2008;283:12120–8. [DOI] [PubMed] [Google Scholar]
- 20.Hagège H, Klous P, Braem C, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2007;2:1722–33. [DOI] [PubMed] [Google Scholar]
- 21.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oti M, Falck J, Huynen MA, et al. Ctcf-Mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics 2016;17:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzi R, Bruno S, Stellacci C, et al. Takayasu’s arteritis: a cell-mediated large-vessel vasculitis. Int J Clin Lab Res 1999;29:8–13. [DOI] [PubMed] [Google Scholar]
- 25.Noris M Pathogenesis of Takayasu’s arteritis. J Nephrol 2001;14:506–13. [PubMed] [Google Scholar]
- 26.Noris M, Daina E, Gamba S, et al. Interleukin-6 and RANTES in Takayasu arteritis: a guide for therapeutic decisions? Circulation 1999;100:55–60. [DOI] [PubMed] [Google Scholar]
- 27.Tripathy NK, Sinha N, Nityanand S. Antimonocyte antibodies in Takayasu’s arteritis: prevalence of and relation to disease activity. J Rheumatol 2003;30:2023–6. [PubMed] [Google Scholar]
- 28.Yu B, Sondag GR, Malcuit C, et al. Macrophage-Associated Osteoactivin/GPNMB mediates mesenchymal stem cell survival, proliferation, and migration via a CD44-dependent mechanism. J Cell Biochem 2016;117:1511–21. [DOI] [PubMed] [Google Scholar]
- 29.McKinsey TA, Zhang CL, Olson EN. Mef2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci 2002;27:40–7. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Gao B, Ponnusamy M, et al. Mef2 signaling and human diseases. Oncotarget 2017;8:112152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischle W, Dequiedt F, Hendzel MJ, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 2002;9:45–57. [DOI] [PubMed] [Google Scholar]
- 32.Potthoff MJ, Olson EN. Mef2: a central regulator of diverse developmental programs. Development 2007;134:4131–40. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Hwangbo C, Hu X, et al. Restoration of impaired endothelial myocyte enhancer factor 2 function rescues pulmonary arterial hypertension. Circulation 2015;131:190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clocchiatti A, Di Giorgio E, Ingrao S, et al. Class IIa HDACs repressive activities on MEF2-depedent transcription are associated with poor prognosis of ER⁺ breast tumors. Faseb J 2013;27:942–54. [DOI] [PubMed] [Google Scholar]
- 35.Sofer A, Lee S, Papangeli I, et al. Therapeutic engagement of the histone deacetylase IIA-Myocyte enhancer factor 2 axis improves experimental pulmonary hypertension. Am J Respir Crit Care Med 2018;198:1345–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clocchiatti A, Di Giorgio E, Viviani G, et al. The MEF2-HDAC axis controls proliferation of mammary epithelial cells and acini formation in vitro. J Cell Sci 2015;128:3961–76. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki F, Kumagai K, Uto H, et al. Expression of glycoprotein nonmetastatic melanoma protein B in macrophages infiltrating injured mucosa is associated with the severity of experimental colitis in mice. Mol Med Rep 2015;12:7503–11. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Zhuo H, Ouyang H, et al. Glycoprotein non-metastatic melanoma protein B (Gpnmb) is highly expressed in macrophages of acute injured kidney and promotes M2 macrophages polarization. Cell Immunol 2017;316:53–60. [DOI] [PubMed] [Google Scholar]
- 39.Hüttenrauch M, Ogorek I, Klafki H, et al. Glycoprotein NMB: a novel Alzheimer’s disease associated marker expressed in a subset of activated microglia. Acta Neuropathol Commun 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Smith AJ, Schacker TW, et al. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol 2009;183:1975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onaga M, Ido A, Hasuike S, et al. Osteoactivin expressed during cirrhosis development in rats fed a choline-deficient, L-amino acid-defined diet, accelerates motility of hepatoma cells. J Hepatol 2003;39:779–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


