SUMMARY
Glioblastoma (GBM) is characterized by aberrant vascularization and a complex tumor microenvironment. The failure of anti-angiogenic therapies suggests pathways of GBM neovascularization, possibly attributable to glioblastoma stem cells (GSCs) and their interplay with the tumor microenvironment. It has been established that GSC-derived extracellular vesicles (GSC-EVs) and their cargoes are proangiogenic in vitro. To further elucidate EV-mediated mechanisms of neovascularization in vitro, we perform RNA-seq and DNA methylation profiling of human brain endothelial cells exposed to GSC-EVs. To correlate these results to tumors in vivo, we perform histoepigenetic analysis of GBM molecular profiles in the TCGA collection. Remarkably, GSC-EVs and normal vascular growth factors stimulate highly distinct gene regulatory responses that converge on angigenesis. The response to GSC-EVs shows a footprint of post-transcriptional gene silencing by EV-derived miRNAs. Our results provide insights into targetable angiogenesis pathways in GBM and miRNA candidates for liquid biopsy biomarkers.
In Brief
Extensive intercellular interactions occur within the notoriously heterogeneous tumor microenvironment of GBM. Lucero et al. identify distinct angiogenic gene regulatory responses of brain endothelial cells to growth factors and to extracellular vesicles (EVs) secreted by GBM stem-like cells. The response to EVs shows a footprint of EV-derived miRNAs.
Graphical Abstract
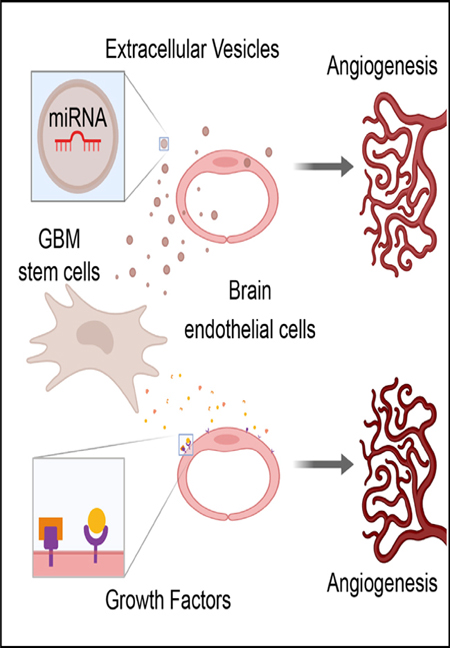
INTRODUCTION
Glioblastoma (GBM), the most common primary brain cancer in adults, is incurable, with 2- and 5-year survival rates of 16% and 5%, respectively (Ostrom et al., 2015). Aggressive diffuse growth, high tumor heterogeneity, vascular abnormalities, and a population of GBM stem-like cells (GSCs) are major factors that complicate treatment (Ramirez et al., 2013; Harder et al., 2018, Rooj et al., 2017; Garnier et al., 2019). Diffuse infiltrative growth of GBM, which arises in large part due to the perivascular migration of GSCs, precludes complete resection, sparing the GSCs that resist chemotherapy and radiation and revive the tumor (Hanif et al., 2017; Ramirez et al., 2013). The development of effective targeted therapies is hindered by the heterogeneity and plasticity of GBM cells, which provide the tumor with multiple paths of resistance, while GBM vasculature provides various obstacles to drug delivery (Ramirez et al., 2013; Zanders et al., 2019; Perrin et al., 2019; Kane, 2019).
Extensive molecular profiling of GBM tissues (Freije et al.,2004; Phillips et al., 2006; Verhaak et al., 2010), single cells (Patel et al., 2014; Rooj et al., 2017; Neftel et al., 2019; Ricklefs et al., 2016; Wang et al., 2019b), and secreted nanoparticles (Ricklefs et al., 2016; Wei et al., 2017; Spinelli et al., 2018; Zhang et al., 2019b) has driven recent developments in the identification of GBM tumor subtypes that support a coherent model of GBM heterogeneity. Gene expression-based tumor subtypes have been resolved to sample-specific mixtures of up to 4 dominant single-cell GBM expression signatures with unique underlying functional cell states that are governed by genetic and microenvironmental cues, but appear to be both plastic and commutable, which is consistent with other similarities to neural precursor cells (Neftel et al., 2019). Studies focused on GSCs identified two distinct functional states that match GBM molecular subtypes (Rooj et al., 2017; Wang et al., 2019b), as well as a microRNA (miRNA)-driven, possibly extracellular vesicle (EV)-mediated, bidirectional transition between distinct GSC sub-9+within the tumor may depend on the molecular subtype and functional state of the donor cells (Spinelli et al., 2018; Wei et al., 2017).
Heterotypic interactions of GSCs with immune, endothelial, and other cells within the tumor microenvironment (TME) are implicated in generating histological heterogeneity of GBM tumors, recurrent treatment failures, and high lethality (Perrin et al., 2019; Schiffer et al., 2018). Molecular profiles capturing the effects of GBM on these non-malignant cells are emerging (Sankowski et al., 2019; Venteicher et al., 2017), but the predominant intercellular agents are not known. GSCs communicate with the TME through several modes, including exchange of soluble molecules and EVs, as well as by cell-to-cell contacts (Broekman et al., 2018; Spinelli et al., 2019). The exchange of molecules and genetic information via EVs plays a critical role in GBM progression and tumor angiogenesis, which may involve reprogramming the epigenome and transcriptome of endothelial cells (ECs) (Aslan et al., 2019; Broekman et al., 2018; Godlewski et al., 2017; Nakano et al., 2015; Quezada et al., 2018; Rooj et al., 2016; Spinelli et al., 2018, 2019; Todorova et al., 2017; Treps et al., 2017). An increasing body of evidence suggests a major role for GSCs in non-conventional angiogenesis (Das and Marsden, 2013; Hardee and Zagzag, 2012; Kane, 2019), consistent with the failure of anti-angiogenic drugs that typically target the classic vascular endothelial growth factor (VEGF)-mediated angiogenic pathways (Geraldo et al., 2019; Ameratunga et al., 2018; Kane, 2019).
To further elucidate non-conventional angiogenic pathways in GBM, we here examine the GSC-EV-mediated transfer of extracellular RNAs (exRNAs) from human GSCs to human brain microvascular ECs (HBMVECs) in vitro by molecular profiling and to ECs in vivo via histoepigenetic analysis by computational deconvolution. EV-derived miRNAs are known to convey growth-promoting and angiogenic signaling in GBM (Beyer et al., 2017; Chen et al., 2019; Todorova et al., 2017; Wong et al., 2015); however, the molecular events controlling this process in HBMVECs are largely unknown. We hypothesized that GSC-derived ex-RNAs, along with more conventional vascular GFs, jointly modulate the gene-expression landscape of ECs to promote angiogenesis. To this end, we compared the effects of GFs and GSC-EVs on angiogenic pathways elicited in cultured HBMVECs, by associating changes in DNA methylome and total RNA profiles in ECs with microRNA (miRNA) content of GSC-EVs. The expression profiles obtained from ECs by histoepigenetic analysis of GBM molecular profiles in the The Cancer Genome Atlas (TCGA) collection (Cancer Genome Atlas Research Network, 2008) revealed a concordance of effects in vitro and in vivo. Finally, we identified candidate proangiogenic miRNAs that are transferred via GSC-EVs into HBMVECs.
RESULTS
GFs and GSC-Derived EVs Induce Visually Similar Vascularization Patterns but Divergent Transcriptional and Epigenomic Changes in HBMVECs
To investigate the potential of GSC-EVs to elicit an angiogenic response from brain microvasculature, we isolated EVs from the conditioned media of GBM8 human primary GBM stem-like cells (Wakimoto et al., 2012) and added them to the basal medium of HBMVECs cultured on a Matrigel substrate (Figure 1A, top panel). A standardized cocktail of angiogenic GFs added to HBMVECs in identical conditions served as a positive control. Cells cultured identically but without added stimulus served as the baseline for comparison. Vascularization metrics were quantified 16 h after application of the stimuli.
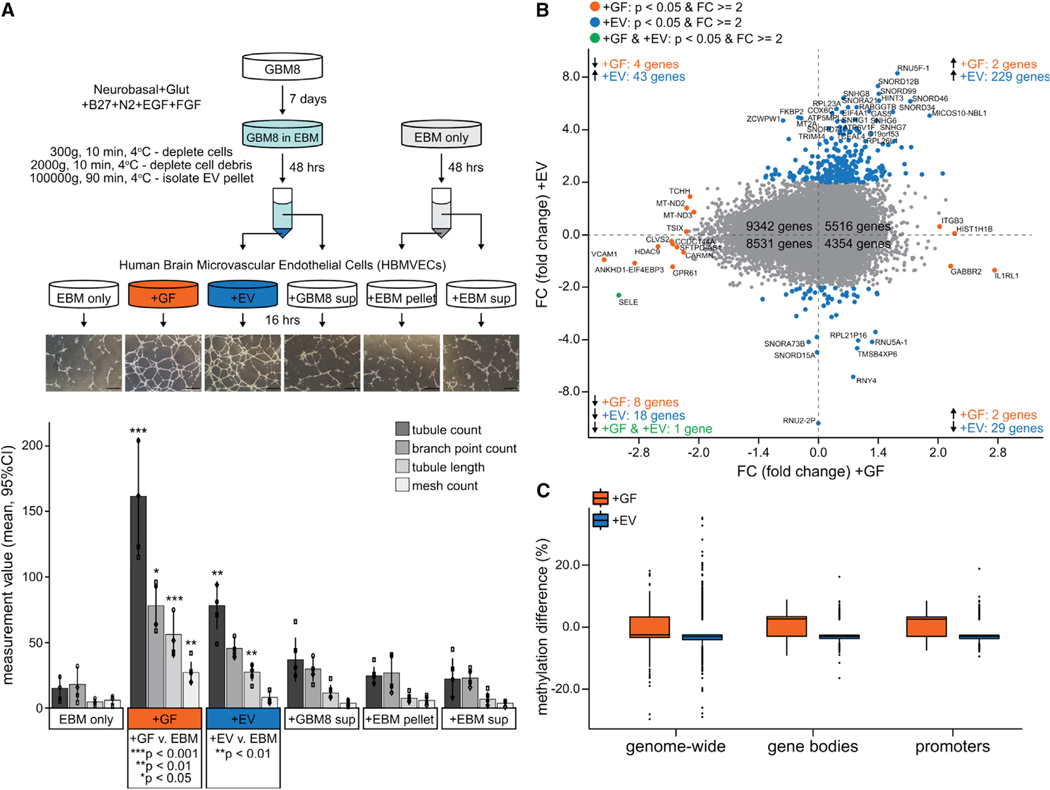
Figure 1. GFs and GSC-Derived EVs Induce Similar Vascularization Patterns but Divergent Transcriptional and Epigenomic Responses in HBMVECs.
(A) Schematic and results of in vitro tube-formation assay. (i) Pellet and supernatant fractions were isolated from media conditioned by GBM8 neurospheres (EV, GBM8 sup) or unconditioned media (EBM pellet, EBM sup). (ii) HBMVECs were cultured on Matrigel for 16 h under EBM containing angiogenic GFs or 1 of the 4 media fractions, then (iii) plates were photographed and harvested for molecular profiling. (iv) Bar plot shows tube-formation assay (n = 4) metrics (mean ± 95% confidence interval [CI]).
(B) Comparative transcript-level changes for +GF versus +EV (log2 fold change versus “EBM only”; n = 2) (quadrant I is top right and that quadrant numbering is counterclockwise).
(C) Comparative DNA methylation changes (log2 fold change versus EBM only; n = 3).
GSC-EV treatment (+EV) stimulated vascularization similar to that of the GF treatment (+GF), as indicated by increases in total tubule length and total counts of tubules, branch points, and meshes (Figure 1A, bar plot). No meaningful vascularization was observed when HBMVECs were treated with supernatant from the EV isolation procedure (+GBM sup), nor with the pellet or supernatant from a mock isolation of EVs from unconditioned endothelial basal medium (+EBM pellet, +EBM sup) (Figure 1A, bar plot).
The responses obtained from GBM8-conditioned media fractions (+EV, +GBM sup) and GFs could not be compared quantitatively because the concentrations in the conditioned media are not normalized to one another nor are they calibrated to physiologically relevant concentrations. These in vitro experiments were designed to detect broad qualitative differences in the EC response to EV and GF stimuli obtained according to well-established (+EV; Zaborowski et al. 2015) or standardized (+GF; tube-formation assay) protocols. Specifically, we asked whether the similar vascularization phenotypes of +EV and +GF were associated with similar or divergent transcriptional and epigenomic changes in HBMVECs. Over the set of synergic transcriptional changes (>2-fold), we detected, for +EV and +GF, respectively, the upregulation of 229 and 2 genes (Figure 1B, quadrant I, top right) and the downregulation of 18 and 8 genes (Figure 1B, quadrant III, bottom left). Only 1 gene (SELE: E-Selectin) showed a large concordant change (>2-fold decrease) in both treatments, indicative of divergent transcriptional responses. We therefore focused on the 78 genes showing opposite changes in transcript levels, 72 of which showed larger perturbations in +EV as compared to +GF (Figure 1B, quadrants II—top left and IV—bottom right). Specifically, +EV decreased the abundance of 29 genes, with just 4 genes reduced by +GF. DNA methylation over gene bodies and promoters also diverged (Figure 1C), with +GF increasing and +EV decreasing on average, which is consistent with the upregulation of more genes in +EV versus +GF. Methylation over 100-kb tiles taken genome-wide was less divergent, with demethylation dominating for both treatments, although the methylation gain was more pronounced with +GF, in accordance with the signal from promoters and gene bodies. The highly divergent transcriptomic and epigenomic responses to +GF and +EV belie the similar vascularization phenotypes in vitro and hint at different primary pathways of action.
Transcriptional and Epigenomic Perturbations Induced by GFs and EVs In Vitro in HBMVECs Largely Resemble Those within Human GBM Tumor ECs
To examine the relevance of our cell line experiments for tumor biology in vivo, we compared the transcriptomic and epigenomic signatures observed in vitro to those observed in ECs of human GBM tumors in vivo. Specifically, we exploited the transcriptomic divergence to determine whether changes in ECs in vivo correlated primarily with the in vitro responses of HBMVECs to +GF or +EV. GBM-associated changes in ECs in vivo were identified by the histoepigenetic analysis of glioma tumors from the TCGA collection (Brennan et al., 2013) using the Epigenomic Deconvolution (EDec) method (Onuchic et al., 2016). The TCGA GBM collection generally lacks molecular profiling data for matched normal non-cancerous samples, so we included lower-grade glioma (LGG) samples as a control group, given that microvascular structures of GBM and LGG are characteristically disparate (Guarnaccia et al., 2018; Louis et al., 2007; Bergers and Benjamin, 2003).
EDec estimated 5 cancer-cell epigenome profiles, all of which correspond to previously defined LGG and GBM molecular subtypes. In GBM tumors, 3 of the cancer-cell profiles (GBM 1, 2, and 3) were found in appropriately high proportions with in tumors of the Proneural+G-CIMP (glioma-CpG island methylator phenotype), classical, and proneural subtypes (Figure 2A). The remaining profiles (LGG1 and LGG2) were enriched within LGG tumors (Figure 2A). EDec also estimated proportions of 4 non-cancer cell types: neuronal, glial, immune, and endothelial. Normal adjacent tissue samples collected by TCGA were highly enriched for non-cancer profiles, although some cancer profiles could be detected in certain samples, consistent with the diffuse growth of gliomas (Figure 2A). The GBM8 epigenome revealed the greatest similarity to that of the estimated Proneural cancer epigenome (Figure 2B, GBM.3), consistent with the previous characterization of the GBM8 cell line as a Proneural-like, stem-like cell type with wild-type IDH1 (Teng et al., 2017). The results indicate successful deconvolution, warrant confidence in the inferred gene expression profiles, and validate the GBM8 cell line as an in vitro model for GBM in the TCGA collection. The differences in deconvoluted EC gene expression and methylation profiles between GBM and LGG should therefore reflect GBM-associated differences of the microvasculature in vivo. Differential expression analysis of GBM versus LGG ECs revealed GBM-associated perturbations (>2-fold change, false discovery rate [FDR] < 0.05) of 1,632 genes.
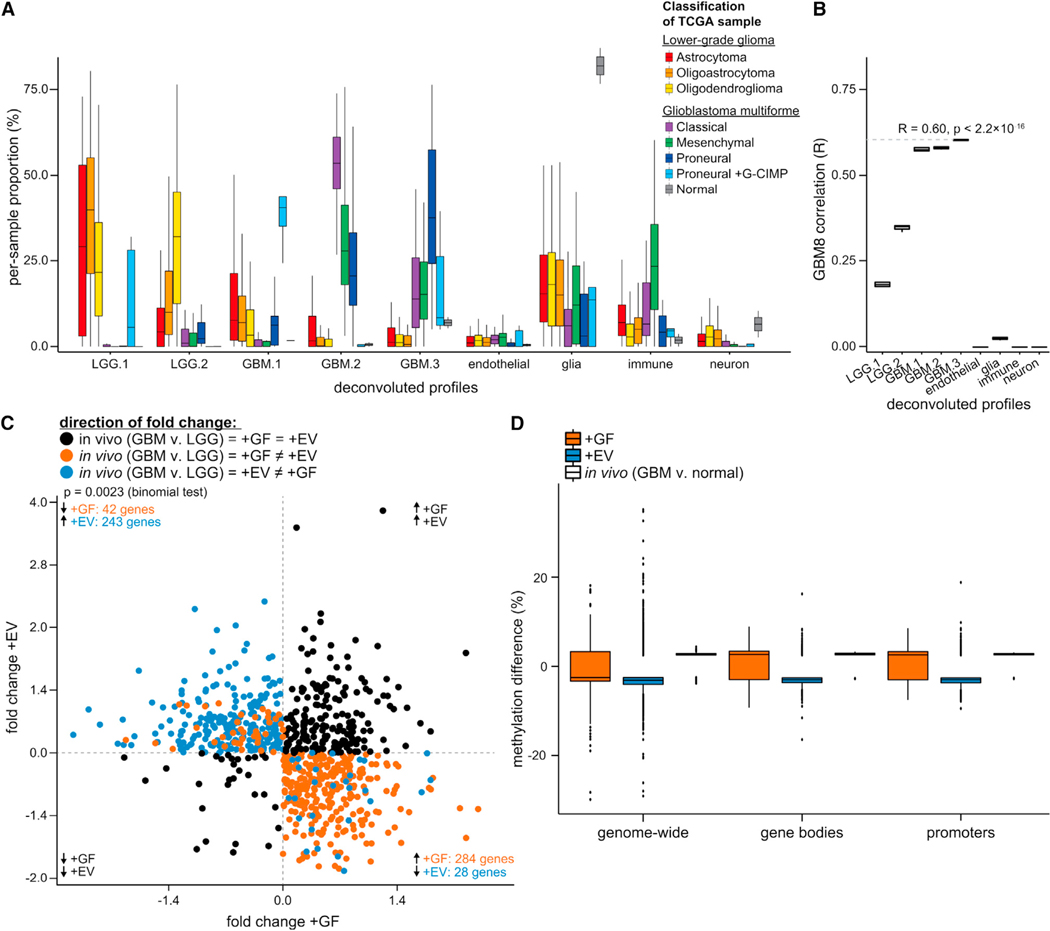
Figure 2. Transcriptional and Epigenomic Perturbations Induced In Vitro by GFs or GSC-Derived EVs in ECs Largely Resemble Those within Human GBM Tumors.
Histoepigenetic analysis of GBM and LGG tumors in the TCGA collection identified constituent cell types of in vivo GBM and LGG tumors (cancer, endothelial, immune, glial, and neuronal).
(A) Inferred cellular composition of GBM tumors (classical, mesenchymal, and proneural ± G-CIMP) and LGG tumors (astrocytoma, oligoastrocytoma, oligo-dendroglioma). The inferred cancer epigenomic profiles (LGG1 and −2 and GBM1, −2, and −3) are enriched in specific tumor subtypes. Non-cancer epigenomic profiles are named according to the highest correlation with normal reference profiles and expression of select marker genes.
(B) Correlation of deconvoluted profiles with GBM8 GSCs is consistent with the proneural origin of GBM8 (see A, GBM.3 profile).
(C) Intersection of expression changes of the EC fraction in vivo (GBM versus LGG) and in vitro (+GF or +EV versus EBM). Quadrants II and IV show genes with opposite changes in HBMVECs upon +GF and +EV treatments (color denotes treatment-specific concordance with the expression change in vivo; this panel shows a subset of genes from Figure 1B) (quadrant I is top right and that quadrant numbering is counterclockwise).
(D) Side-by-side view of in vivo and in vitro DNA methylation changes (this panel includes the data from Figure 1C, augmented by in vivo changes).
To determine whether GFs or EVs play a dominant role in vivo, we asked whether the GBM-associated transcriptomic perturbations in vivo mostly reflected the transcriptomic response of HBMVECs to +GF or +EV in vitro (Figure 2C). The treatments elicited divergently trending changes from 597 (42+243)+(284+28) of the 1,632 genes (Figure 2C, quadrants II—top left and IV—bottom right), and the directions of +GF-induced expression changes were concordant with 54.6% [(42 + 284)/597] (p = 0.0023, binomial test) of the GBM-associated perturbations (Figure 2C, orange dots). This suggests a somewhat larger transcriptional influence of GF compared to GSC-EV stimulation in vivo. Moreover, DNA methylation differences of GBM ECs in vivo were overwhelmingly more concordant with those observed in HBMVECs upon +GF treatment, especially over gene body and promoter regions (Figure 2D). These observations are consistent with a highly dominant effect of GFs on transcriptional regulation, leaving open the possibility of post-transcriptional influence of GSC-EVs, perhaps mediated by miRNAs.
Transcriptional Perturbations of Angiogenic Pathways in ECs Are Consistent with Post-transcriptional Silencing by GBM EV miRNAs Transferred into ECs
To explore the possibility that miRNAs delivered to ECs by GSC-EVs contribute to post-transcriptional downregulation, we focused on the 28 genes downregulated in vivo and in vitro by +EV only (Figure 2C, quadrant IV—bottom right blue dots). Asking whether the magnitude of changes in vitro correlate with those observed in vivo, we observed significant correlation (R = 0.57), supporting a gene-silencing role for GSC-EVs in vivo (Figure 3A). To explore this more broadly, we exploited the fact that transcriptionally modulated attenuation should leave “epigenomic footprints” at gene-regulatory regions, whereas most gene attenuation by miRNAs would be post-transcriptional and leave no such signature. Joint examination of transcriptomic and DNA methylation changes at genes with detectably altered promoter DNA methylation revealed that diminished expression was accompanied by increased promoter methylation with +GF (p = 1.77 × 10−5, chi-square test), as expected for transcriptionally mediated gene silencing (Figure 3B), whereas decreased expression did not accompany promoter methylation gain with +EV (p = 0.26, chi-square test) (Figure 3C), which is consistent with post-transcriptional regulation by miRNAs.
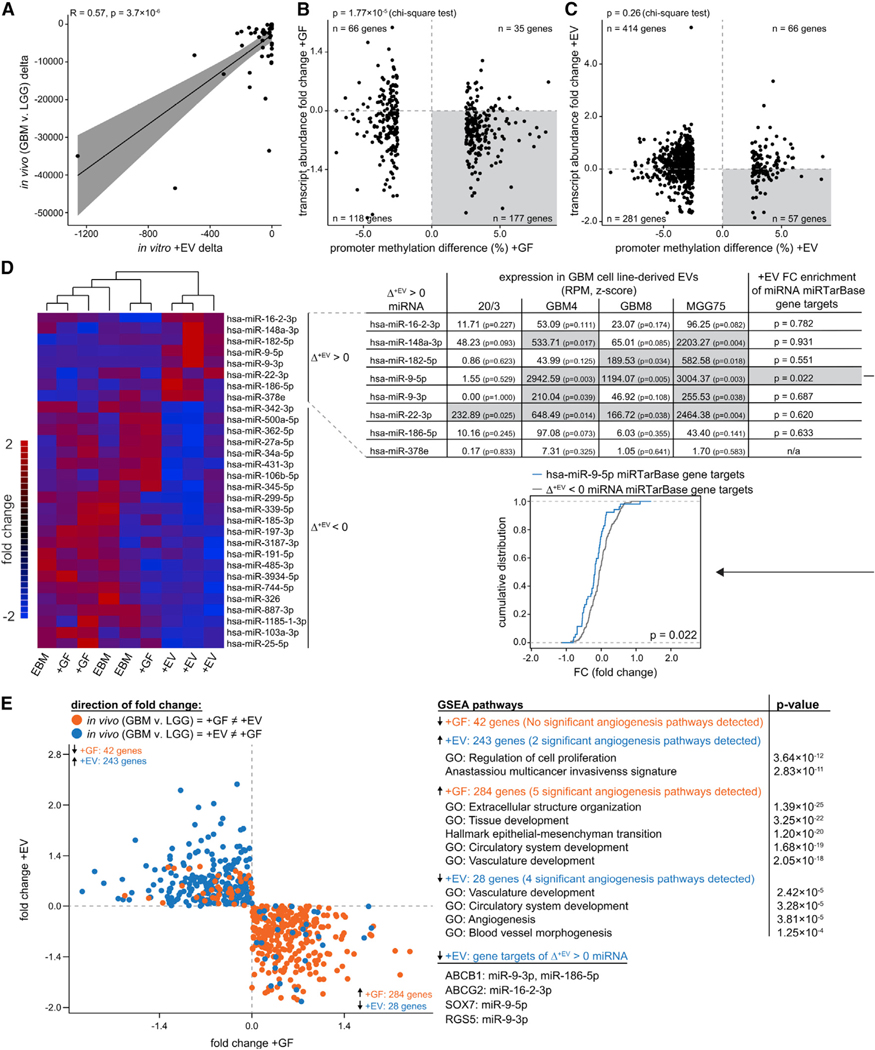
Figure 3. Identification of Candidate miRNAs that May Mediate EV-Induced Vascularization.
(A) The 28 genes downregulated in vitro upon +EV treatment (indicated by blue dots in E, quadrant IV) show concordant downregulation in vivo with correlated magnitudes (R = 0.57).
(B and C) Transcript depletion upon +GF treatment associates with DNA methylation gain over promoters (B), whereas transcript depletion upon +EV treatment does not associate with promoter methylation (C).
(D) Of the miRNAs that showed a notable increase in abundance in HBMVECs following +EV treatment (top 8 rows of heatmap, first column of table), 5 showed significantly high abundance within GSC-EVs (center columns of table, highlighted in gray). Downstream targets of miR-9 were significantly downregulated (last column of table).
(E) GSEA implicates 5 angiogenic pathways enriched for the 284 genes upregulated in vivo and upon +GF treatment (quadrant IV, orange dots) and 4 angiogenic pathways enriched for the 28 genes downregulated in vivo and upon EV treatment (quadrant IV, blue dots) (quadrant I is top right and that quadrant numbering is counterclockwise).
This prompted us to ask which miRNAs may be delivered by GSC-EVs to effect gene attenuation in HBMVECs. Small RNA sequencing (RNA-seq) of HBMVECs before and 16 h after +EV treatment revealed an increased abundance of 8 miRNAs (Figure 3D, upper right), consistent with the uptake of GSC-EV miRNAs. To determine the potential for EV-mediated delivery of these 8 miRNAs to HBMVECs, we analyzed small RNA-seq data from GSC-EVs of 4 GBM cell lines (20/3, GBM8, GBM4, and MGG75; Wei et al., 2017). The ubiquitous presence of the 8 miRNAs in GSC-EVs (Figure 3D, right panel) substantiates the inference that the miRNAs were not simply transcribed in HBMVECs as an effect of +EV treatment. Relative to all of the other miRNAs identified within GBM8 EVs, 3 of the 8 miRNAs— hsa-miR-9–5p, hsa-miR-22–3p, and hsa-miR-182–5p—were significantly enriched (Figure 3D, right panel). Similarly, GBM4 and MGG75 EVs were enriched in 4 and 5, respectively, of the candidate miRNAs, with both lines also showing strong enrichment for miR-9–5p.
We then asked whether some of the 8 miRNAs were associated with the downregulation of validated miRTarbase (Chou et al., 2018) target mRNAs in vitro (HBMVEC, +EV) or in vivo (GBM ECs). The strongest and the only statistically significant signal was obtained for hsa-miR-9–5p, with the following 3 target genes downregulated both in vitro and in vivo: RGS5, ABCB1, and SOX7 (Figure 3E, right panel, bottom).
We next tested the 28 genes (downregulated in vivo and in vitro by +EV only) for association with angiogenesis pathways (Figure 3E, right panel). Gene set enrichment analysis (GSEA) identified significant enrichment in 4 angiogenesis-related pathways. A comparable 5 angiogenesis pathways were returned when we queried the 284 genes upregulated in vivo and specifically in response to +GF treatment. In contrast, no angiogenesis pathways were enriched in the 42 genes uniquely downregulated in response to +GF treatment. These results suggest that the downregulation of angiostatic genes, which could be mediated by miRNAs, may be a unique feature of GSC-EV communication.
miR-9–5p in GBM Stem Cells Supports Metabolic Activity and May Influence Resistance to Therapeutic Intervention on LGGs
The signature associated with miR-9–5p is consistent with proangiogenic influence; therefore, we aimed to assess the degree to which it contributes to the proangiogenic effect of GSC-EVs. Therefore, we aimed to treat HBMVECs with EVs derived from GBM8 cells depleted of miR-9–5p. However, knockdown of miR-9–5p severely reduced the metabolic activity of the GBM8 neurosphere cultures as measured by the WST assay (Figures 4A and 4B), ending this line of investigation. This result suggested the possibility that the effects of the export of miR-9–5p from cancer cells may play a distinct and complementary role to its angiogenic effects in ECs.
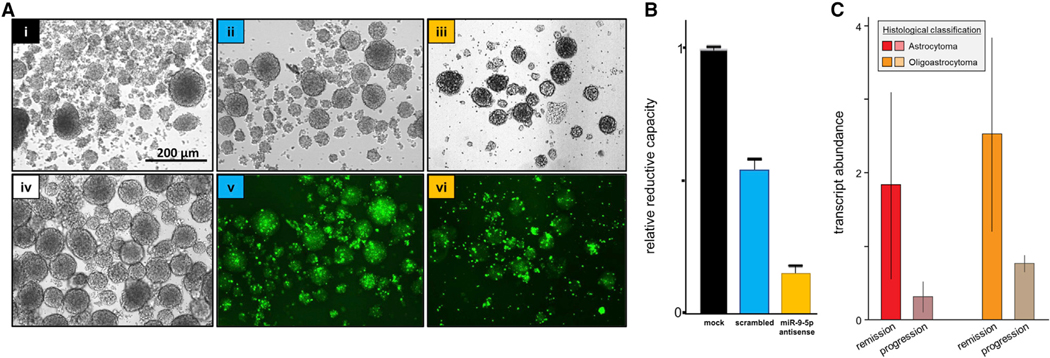
Figure 4. miR-9–5p Supports the Metabolic Activity of GBM Stem Cells and May Influence Resistance to Therapeutic Intervention on Gliomas.
(A) GBM8 neurosphere cultures: transfected with 50 nM lipofectamine plus no oligonucleotide (i, mock); FAM-labeled scrambled oligonucleotide (ii, v, scrambled); or FAM-labeled antagomir (iii, vi, miR-9–5p antisense); non-transfected GBM8 (iv). Micrographs taken 12 days after transfection (seeding at1 × 104 cells/ well). Scale bar, 200 μm.
(B) Metabolic activity (mean ± SD) of transfected cells measured by WST-1reduction assay.
(C) miR-9–5p levels (mean ± SD) in the cancer cell fraction of LGG tumors (astrocytomas or oligoastrocytomas), stratified by response to therapeutic intervention.
To identify other GSC-EV miRNAs that may drive vascular proliferation, we aimed to contrast miRNA compositions of LGG-derived EVs (LGG-EVs) and GSC-EVs hypothesizing that LGG-EVs would be less angiogenic and that comparison of miRNA compositions would highlight additional pro-angiogenic GSC-EV miRNAs. Due to constraints associated with the culture of LGG cell lines, we could not isolate enough LGG EVs for analysis. We therefore sought to use histoepigenetic analysis to facilitate comparison of the miRNA composition of ECs and cancer cells in LGG versus GBM as a proxy for direct investigation of miRNA levels in the respective isolated EVs. Unavailability of small RNA-seq expression data for GBM in the TCGA collection precluded this analysis. Small RNA-seq data are, however, available for the LGG samples. Thus, we performed additional histoepigenetic analysis to compare the in vivo expression patterns of miRNAs in the cancer and EC fractions of LGG tumors under the premise that therapy-resistant LGG may contain a significant undetected proportion of GSCs cells that survive treatment. We focused on LGG samples with a component of astrocytic neoplasia and grouped them by response to therapeutic intervention (progression or remission) and tumor molecular subtype (astrocytoma, oligoastrocytoma) for the analysis. We did not detect meaningful differences in the ECs or GSC-like cells based on this stratification, but we did observe that treatment-resistant LGG tumors tended to have lower miR-9–5p levels in the GSC-like fraction (Figure 4C), consistent with cancer resistance induced by the export of miR-9–5p from cancer cells.
DISCUSSION
We provide multiple lines of evidence from both in vitro experiments on cell lines and computational deconvolution of tumors in vivo that EV-mediated transfer of RNAs from GBM cells to brain ECs induces angiogenesis. Similar morphologic patterns of vascularization induced by in vitro treatment of HBMVECs with either GFs or GSC-EV stand in sharp contrast to highly divergent transcriptional and epigenomic changes observed in HBMVECs upon these 2 treatments. While the angiogenic pathway response to GFs is characherized by gene upregulation, downregulation dominates the angiogenic pathway response to GSC-EVs. Moreover, 28 of 29 genes downregulated by GSC-EVs in vitro were also downregulated in vivo without observable gains in promoter methylation, which is consistent with post-transcriptional downregulation by miRNAs delivered from GSCs to ECs.
Following up on the above results, we identified 8 candidate miRNAs that may mediate the EV-associated angiogenesis. The set includes miR-148a and miR-9–5p, both previously associated with glioma angiogenesis (Kim et al., 2014; Zhang et al., 2019a; Madelaine et al., 2017; Yi and Gao, 2019) and poor survival. Our results are concordant with previous studies (Chen et al., 2019) reporting that when miR-9 is delivered to human umbilical ECs (HUVEC) via GBM-derived EVs, its expression levels in HUVECs directly correlated with the resulting tubule formation count and length. Our results are also concordant with previous studies (Wong et al., 2015) reporting that the silencing of miR-148a normalizes the aberrant tumor vasculature in mouse models of GBM.
In search of downstream mediators of angiogenesis, we also examined transcript-level changes of validated mRNA targets of miR-9 (Chen et al., 2019). Three miR-9 targets (RGS5, SOX7, and ABCB1) were downregulated both in vivo in GBM ECs and in HBMVECs by GBM8-EV treatment in vitro. RGS5 plays a central role in vascular growth (Svensson et al., 2015); it has been shown to reduce endothelial growth (Wang et al., 2019a); and small interfering RNA (siRNA)-mediated knockdown of RGS5 stimulates endothelial growth (Jin et al., 2009). In a wide variety of cell types, both tumor and normal, SOX7 inhibits cell proliferation by antagonizing the Wnt/β-catenin signaling pathway, whereas miRNA-mediated knockdown of SOX7 restores proliferation (Zheng et al., 2016). SOX7 expression is critical for physiological angiogenesis, and its loss leads to weakly formed microvasculature (Kim et al. 2016).
Our results raise the possibility that miRNA export may also have important consequences for cancer cells. Specifically, xenobiotic efflux pumps such as ABCB1 appear to play roles in cancer therapy resistance. Speculatively, the depletion of miR-9–5p in glioblastoma cells may weaken the efficacy of chemotherapeutics such as temozolomide by de-repression of ABCB1, a miR-9 target, and consequent improvement of drug efflux. In that regard, we note that the overexpression of ABCG2, another xenobiotic efflux pump and target of miR-16–2-3p (found in GSC-EVs) can result in temozolomide resistance and poor clinical outcomes (Emery et al., 2017; de Gooijer et al., 2018). Moreover, reduction of the metabolic activity of GBM8 GSCs induced by the knockdown of miR-9–5p is consistent with previous reports of decreased proliferation in other GBM cell lines (Chen et al. 2019), suggesting that removal of miR-9–5p may also confer chemotherapy resistance by reducing proliferation.
The diversity of secreted nanoparticles that carry exRNA and the heterogeneity of many nanoparticle isolates are increasingly recognized sources of confounding in exRNA studies and thus require careful consideration. Recent reports highlight distinct nucleic acid compositions of vesicular and non-vesicular carriers of exRNA (Wei et al., 2017; Jeppesen et al., 2019), while others demonstrate that common EV isolation protocols often co-isolate non-vesicular carriers (Jeppesen et al., 2019; Murillo et al., 2019). Thus, clear evidence obtained using improved isolation methods is required before attributing RNA transfer to a specific carrier class. In that regard, the stringent studies by Wei et al. (2017) and Jeppesen et al. (2019) identified miR-9–5p as enriched in the vesicular fractions of 4 distinct GBM cell lines (GBM8, GBM4, MGG75, and Gli36), suggesting that miR-9–5p is a bona fide EV-derived miRNA in these systems. Jeppesen et al. (2019) reported enrichment of miR-22 miR-148a-3p, and miR-182–5p in the non-vesicular fraction of Gli36 (an established glioma cell line), although they were identified as enriched in EVs by Wei et al., 2017 in primary GBM GSC cultures. Because our EV isolation method was less stringent, we cannot completely rule out the effects of co-isolated non-vesicular miRNA on angiogenesis in our system.
While there are manifold mechanisms by which the transcriptome can be modified, the present analysis accounts only for variations in the transcript levels of miRNAs and messenger RNA (mRNA) resolved at the level of genes. Our study does not address subtler transcriptomic changes, including alternative transcript initiation, alternative splicing, or post-transcriptional nucleobase modifications, nor does it consider non-coding RNAs other than miRNAs. It will be of interest to determine in future studies whether the EV-mediated changes involve a greater degree of post-transcriptional modifications and whether exRNAs other than miRNAs may be involved as mediators.
The implied role of EV-miRNAs in GBM vascularization revealed in this study has both therapeutic and diagnostic implications. EV and GF signaling likely exhibits spatial and temporal heterogeneity within a tumor, and their relative contributions could account for substantive and clinically important differences in the types of vessels formed—for example, with respect to permeability and morphology. Because of the poor accessibility of brain tumors, liquid biopsy biomarkers that may distinguish highly malignant GBM from LGG have potentially high clinical utility. In that regard, previous studies detected GBM-associated EVs in patient plasma (Jones et al., 2019), elevated levels of miR-9 in GBM tissues (Wu et al., 2013) and in the serum-derived EVs (Ji et al. 2016) and cerebrospinal fluid (CSF) (Sørensen et al. 2017) of acute ischemic stroke patients, suggesting miR-9 as a potential liquid biopsy marker for GBM progression and brain damage. The prognostic value of miR-9 levels has already been endorsed with its inclusion in multi-marker prognostic panels for GBM (Yuan et al., 2017). Our study supports these results by providing a mechanistic role of miR-9 in GBM progression. By elucidating pathways of GBM vascularization that are distinct from the well-known GF pathway, our results open the way toward new types of anti-angiogenic therapies of these lethal tumors.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for other resources and reagents should be directed to the Lead Contact, Aleksandar Milosavljevic (amilosav@bcm.edu). This study did not generate new or unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
Established primary human glioblastoma cells (GBM8 (also called MGG8), kindly provided by H. Wakimoto through A. Krichevsky, Harvard Medical School, Boston, MA) were cultured as spheroids in 0.22um-filtered Neurobasal® medium (GIBCO Invitrogen Corporation, San Diego, CA) supplemented with Glutamax (GIBCO Invitrogen Corporation, San Diego, CA) (3mM), N2 (GIBCO Invitrogen Corporation, San Diego, CA) (0.5mL/100mL), B27 (GIBCO Invitrogen Corporation, San Diego, CA) (2mL/100mL), EGF (R&D system) (20ng/mL), FGF (PEPROTECH) (20ng/mL), and penicillin-streptomycin (10 IU/mL and 10 μg/mL, respectively, Sigma-Aldrich, St Louis, MO, USA). GBM8 were used at passages 23 to 25. Primary Human Brain Microvascular Endothelial Cells (HBMVEC) from Cell Systems (Catalogue #ACBRI-376, Kirkland, WA, USA) were cultured in EGM-2 MV (Lonza, Basel, Switzerland) supplemented with 5% EV-depleted FBS. FBS was depleted from EVs by 16 h of ultracentrifugation at 160,000 × g. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. HBMVECs were used at passages 5–8. Growth medium (EBM-2) from plates where no cells were seeded (unconditioned medium – UCM) was included in the experiments and used as a negative control. Cells were regularly checked for mycoplasma contamination using Mycoplasma PCR detection kit (ABM, Richmond, BC) and only negative cells were used for the experiments. In brief, conditioned media (cultured for at least 24 hours) was collected, centrifuged at 2,000 × g to get rid of cell debris and subjected for PCR. HBMVECs and GBM8 cells were confirmed to be female (by analysis of sex chromosome DNA methylation patterns).
Extracellular Vesicle Purification
GBM8 single cells were seeded at 10,000 cells/cm2 in four 150mm dishes and cultured as neurospheres for 8 days, adding fresh medium every 3 days. GBM8 neurospheres were then transferred on basal medium (EBM-2 Basal Medium, Lonza, Basel, Switzerland) for 48 hours before extracellular vesicle (EV) isolation.
The conditioned basal medium (100mL) from approximately 20 million GBM8 cells was harvested after 48 hr. The EVs were isolated by differential centrifugation. Briefly, conditioned and unconditioned media were centrifuged at 300 3 g for 10 min at 4°C to remove any cells/cell debris. The supernatant was transferred to a clean 50 mL tube and further centrifuged at 2,000 3 g for 10 minutes at 4°C to remove additional cell debris. The supernatant was then transferred to a clean ultracentrifuge tube (Beckman Coulter, Brea, CA, USA) and ultracentrifuged at 100,000 3 g for 90 min at 4°C (70.Ti Beckman rotor) to obtain EV-enriched pellet. The ultracentrifuged conditioned and unconditioned supernatants were removed and preserved at 4°C and the EV pellets and the UCM pellets were resuspended in 200uL of sterile double-filtered (df) (0.22um) PBS. EV pellets were measured for their nanoparticles content using Nanosight instrument technology (NanoSight NTA 2.2 software) (3×60sec videos/sample, detection threshold: 6). All of the EV pellet isolated from the three independent experiments were used to treat the HBMVEC cells. In parallel, we collected EV pellets from approximately 20 million GBM8 cells, and the purified EVs were characterized using NTA.
HBMVEC in vitro Angiogenesis Assay
HBMVECs (500,000/well) were cultured on Matrigel-coated (BD Matrigel 10mg/mL, BD Biosciences, Franklin Lakes, NJ, USA) wells in a 6-well plate in i) endothelial basal medium (EBM-2); ii) EBM-2 supplemented with a cocktail of angiogenic factors (EGM-2 SingleQuot Kit Suppl. & Growth Factors, Lonza); iii) EBM-2 with 200 ul GBM8-derived EV pellet (10×104EVs/cell); iv) EBM-2 with 200 ul GBM8-derived supernatant; v) EBM-2 with 200 ul UCM-derived pellet; and vi) EBM-2 with 200 ul UCM-derived supernatant. After 16 hours of exposure, 3 random pictures at 4X and 25 random pictures at 10X per well were taken. Angiogenesis was analyzed with the ImageJ software (NIH). Specifically, the tubules length (10X), the number of tubules (10X), the number of branching points (10X), and the mesh size (4X) were assessed. Experiments were conducted in triplicates.
Cell culture and transfection for RNA inhibition
Confluent GBM8 cells were dissociated into single cell suspension using the Neurocult Stem Cells chemical dissociation kit (Stem Cell Technologies). The cells were transfected via magnetofection with magnetic nanobeads (Neuromag, Oz Biosciences) either with fluorescently-labeled miRCURY LNA™ hsa-miR-9–5p inhibitor or scramble control at a final concentration of 50 nM. The mixture of transfection reagent and miRNA inhibitor was added directly to cells and seeded into 6-well plate at density of 1×104 cells/well. Growth of GBM8 neurospheres was monitored up to 12 days in vitro.
WST-1 Assay for Cell Proliferation and Viability
Cell viability was assessed by WST reduction assay (Cell counting kit, Dojindo), which detects dehydrogenase activity of viable cells. The cells were incubated with 10% WST reagent for 2 hours at 37°C. The absorbance of the culture medium was measured with a microplate reader at a test and reference wavelengths of 450 nm and 630 nm respectively. The cell viability was calculated as a comparison to the control/mock group (only transfection agent, no miRNA inhibitor). Data are represented as mean + SEM of 3 independent experiments. Statistical significance was evaluated using one-way ANOVA to compare among groups, with Tukey’s test for multiple comparisons. A p value of < 0.05 was considered to be statistically significant.
METHOD DETAILS
Total genomic DNA and RNA extraction
After 16 hours of exposure, the medium was gently removed, centrifuged at 2,000 × g for 15 minutes at 4°C to collect floating cells, and dry pellets preserved at 4°C. Equilibrated (37°C) Dispase (BD Biosciences, Franklin Lakes, NJ, USA) was added (0.2 mL per cm2) to Matrigel and incubated at 37°C for 1.5 h. After pipetting to carefully resuspend the cells, 3.6 mL EDTA (5mM sterile, pH = 8) was used to stop Dispase activity and cells were pelleted twice by centrifugation (2,000 × g for 15 minutes at 4°C). Cell pellets from medium and Matrigel were collected for each sample and washed in 600 ul sterile dfPBS (1X PBS, filtered through 0.22 um filter twice) at 2,000 × g for 15 minutes at 4°C and finally resuspended in 100 ul sterile dfPBS for either DNA or RNA extraction.
Selection of RNA isolation methods can greatly impact exosomal RNA yield and size distribution, and ultimately data interpretation (Eldh et al., 2012; Srinivasan et al., 2019). Total RNA for this study was isolated from samples using the QIAGEN (Exiqon) miRCURY RNA isolation kit. The miRCURY kit was chosen since it was one of the best performing kits in terms of cellular and exosomal RNA yield and size distribution (Eldh et al., 2012; Srinivasan et al., 2019).
Genomic DNAs for methylation analysis were purified (QIAGEN QIAamp® DNA Micro Kit, QIAGEN,Stanford, CA, USA) from HBMVECs cultures as above, and also from GBM8 cells cultured (in triplicate) for 48h both in supplemented Neurobasal® medium (GIBCO Invitrogen Corporation, San Diego, CA) and in basal medium (EBM-2 Basal Medium, Lonza Biologics Inc., Portsmouth, NH, USA).
RNA isolation and Sequencing
Long RNA sequencing
RNA-sequencing was performed using the Clontech/Takara SMARTer Stranded Total RNA-seq PICO v2 kit (Clontech/Takara 634414) for long RNA expression profiling. All libraries then had 75 bp paired end sequencing on the NextSeq500 using a 150 cycle high output kit. Raw Illumina reads were quality filtered as follows. First, ends of the reads were trimmed to remove N’s and bases with quality less than 20. After that the quality scores of the remaining bases were sorted and the quality at the 20th percentile was computed. If the quality at the 20th percentile was less than 15, the whole read was discarded. Also, reads shorter than 40 bases after trimming were discarded. If at least one of the reads in the pair failed the quality check and had to be discarded, we discarded the mate as well.
Transcript Abundance and Differential Expression Estimates for long RNA
Prior to mapping for transcript-level quantification, to assess sample integrity all fastq files from long-RNA sequencing runs were uploaded to the Genboree workbench and mapped to hg19 and all exogenous genomes using the exceRpt [v4.6.3] small RNA-seq pipeline (Kaczor-Urbanowicz et al., 2018; Rozowsky et al., 2019). Data quality assessment and read clipping was performed using TrimGalore [v0.4.1] with CutAdapt [v1.15] and FastQC[v0.11.6] (Martin, 2011). Paired-reads were mapped simultaneously to the GRCh37.83 cDNA and ncRNA transcriptomes (Kinsella et al., 2011) using Kallisto [v0.44.0] (Bray et al., 2016). Kallistos were imported to R with tximport. All reads mapping to ncRNA were excluded from the analysis, then transcript abundances were aggregated by Ensembl Gene ID. Genes with less than 10 reads across all samples were excluded from further analysis. Differential transcript abundance was characterized by DeSeq2 with EBM as the baseline for all treatments, and samples treated as paired within replicates. Threshold values for differential expression were set at 2 and 0.05, respectively, for fold change and adjusted p value.
Small RNaseq Library Preparation and Sequencing
Small RNaseq libraries were generated using the NEB Next SmallRNA library preparation kit according to the manufacturer’s protocol, with some minor modifications. The reaction volumes were reduced to 1/5thof the recommended volume. Due to the low amounts of RNA input, the 3° SR adaptor, SR RT primer and the 5° SR adaptors were diluted 1:6 to avoid excessive amounts of free adaptors and the formation of adaptor dimers. The final libraries were purified using the Zymo DNA clean and concentrator-5 kit and subjected to size selection on the Pippin Prep with a cut off between 117 and 135 bp to deplete adaptor dimers and most of larger RNA species such as tRNA fragments. The size selected libraries were then sequenced on a HiSeq 4000 sequencing system.
Small RNaseq Data Analysis
Condition2 Replicate1 was removed from analysis due to a markedly lower total miRNA count (273,468) compared to the other 8 samples (nearly 5x fewer counts than the sample with the next fewest counts). In the remaining samples, the samples with the fewest total miRNA counts (which was Condition3 Replicate2) had a total miRNA count of 1,338,649. Therefore, the scaled data for the remaining samples were filtered to remove miRNAs with fewer than 7.5 scaled counts (corresponding to a cutoff of 10 raw counts/ 1,338,649 total miRNA counts) in at least 2 samples; after this filtering, 430 miRNAs remained. The data were further filtered using the Qlucore data analysis and visualization software package (https://www.qlucore.com) by selecting for miRNAs that displayed a variance across the dataset of 0.01, leaving 29 miRNAs. Differential expression analysis was then performed among the three conditions, and using a p value < 0.03/q-value < 0.75, obtained 15 miRNAs.
QUANTIFICATION AND STATISTICAL ANALYSIS
Methylation analysis
HBMVECs and GBM8 genomic DNA 3-replicates for each sample were submitted for genomic methylation analysis to the Translational Genomics Core (65 Landsdowne Street, Cambridge, MA 02139). Methylation analysis on genomic DNAs were performed using the Illumina Human 450K Infinium Methylation BeadChip.
Differential Methylation Analysis
Illumina Human 450K Infinium Methylation BeadChip beta values were input to RnBeads [version 2.0.1] with annotations from RnBeads.hg19 [version 1.14.0] (Assenov et al., 2014) in R [version 3.5.1]. Filtering removed probes annotated as non-CpG, cross-reactive, snp-containing, or on sex chromosomes. Probes with variance of less than 0.05 across all samples were excluded. All comparisons were performed with EBM as the baseline. Differential methylation was determined over annotated gene body and promoter regions using limma.
Statistics
The statistical analyses for angiogenesis assay were performed using GraphPad software 6.0 (GraphPad software Inc., La Jolla, CA 92037 USA). Shapiro’s test was performed to check for normality. Either t test or Mann-Whitney were applied to analyze the in vivo angiogenesis assay. P threshold of significance was fixed as p < 0.05.
GSEA
Pathways analysis was performed using Gene Set Enrichment Analysis. The pathways were collated by running the candidate genes using the “Compute Overlap” feature. All presented pathways have FDR < 0.05.
Epigenomic Deconvolution
EDec R script (Onuchic et al., 2016) was utilized to deconvolute the GBM, LGG, and normal samples collected by TCGA. Following the EDec instructions, Stage 0 informative probes were collected by running a t test across the 450K array beta values of cell type references collected from GEO database. This included GBM8 cell lines, glia, neurons, immune, and endothelial cell profiles. For Stage 1, k = 9 was selected as the most stable model using the estimate stability function which uses 80% of the data over n = 3 iterations. Stage 1 results in the deconvoluted methylation profiles and per-sample proportions (Figure 2A). Each profile was correlated to the GEO references across the informative probes to determine its identity by selecting the highest correlation. Stage 2 was performed on GBM and LGG sample separately to determine the endothelial cell type specific gene-expression. Differential expression was calculated using the mean, standard errors and degrees of freedom calculated by EDec Stage 2. All reported changes are > 2 fold-change and FDR < 0.05.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical Commercial Assays | ||
| QIAGEN miRCURY RNA isolation kit | QIAGEN | 76743 |
| QIAGEN QIAamp DNA Micro Kit | QIAGEN | 56304 |
| Human 450K Infinium Methylation BeadChip | Illumina | WG-314–1003 |
| SMARTer Stranded Total RNA-seq PICO v2 kit | Clontech/Takara | 634414 |
| Deposited Data | ||
| reference cell molecular profiles | GEO | see GSE138115 for sample Identifiers |
| Glioma tumor molecular profiles | TCGA | see GSE138115 for sample Identifiers |
| DNA methylation profiles of HBMVECs | This Paper | see GSE138115 |
| RNA-seq profiles of HBMVECs | This Paper | see GSE138115 |
| Experimental Models: Cell Lines | ||
| GBM8 primary GBM cells | Dr. Hiroaki Wakimoto | NA |
| Primary Human Brain Microvascular Endothelial Cells | Cell Systems | ACBRI-376 |
| Oligonucleotides | ||
| Anti-miR-9: Cy5-TCATACAGCTAGATAACCAAAG | This Paper | NA |
| Software and Algorithms | ||
| exceRpt [v4.6.3] small RNA-seq pipeline | Rob Kitchen | https://github.com/rkitchen/exceRptgenboree.org |
| EDec | Onuchic et al., 2016 | https://github.com/BRL-BCM/EDec |
| RnBeads | Assenov et al., 2014 | rnbeads.org |
| GSEA | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Other | ||
| EGM-2 MV Microvascular Endothelial SingleQuots Kit | Lonza | CC-4147 |
| EBM-2 Endothelial Cell Growth Basal Medium-2 | Lonza | CC-3156 |
Highlights.
GBM EVs and growth factors promote angiogenesis by distinct pathways
EV-exposed endothelial cells show a footprint of gene regulation by EV miRNAs
The EV miRNA pathway explains failure of anti-angiogenic therapy for GBM
The results suggest an angiogenic pathway and liquid biopsy biomarkers for GBM
ACKNOWLEDGMENTS
This publication is part of the NIH Extracellular RNA Communication Consortium paper package and was supported by the NIH Common Fund exRNA Communication Program. This work was supported by a grant from the Common Fund of the National Institutes of Health (NIH) 5U54 DA036134 (to A.M.) and U19 CA179563 (to X.O.B. and A.M.K.). This work was also in part supported by the NIH/National Cancer Institute (NCI) grant 5RO1 CA163849 (to A.M.), NIH/NCI R01 CA215072 (to A.M.K.), and NIH/NCI P01 CA069246 and R35 CA232103 (to X.O.B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
The RNA-seq and DNA-methylation data produced in the course of this study are accessible via GEO archives at the NCBI accession GEO: GSE138115.
REFERENCES
- Ameratunga M, Pavlakis N, Wheeler H, Grant R, Simes J, and Khasraw M. (2018). Anti-angiogenic therapy for high-grade glioma. Cochrane Database Syst. Rev 11, CD008218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T, and Kharaziha P. (2019). Tumor-derived exosomes: implication in angiogenesis and antiangiogenesis cancer therapy. J. Cell. Physiol 234, 16885–16903. [DOI] [PubMed] [Google Scholar]
- Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, and Bock C. (2014). Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 11, 1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, and Benjamin LE (2003). Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 3, 401–410. [DOI] [PubMed] [Google Scholar]
- Beyer S, Fleming J, Meng W, Singh R, Haque SJ, and Chakravarti A. (2017). The Role of miRNAs in angiogenesis, invasion and metabolism and their therapeutic implications in gliomas. Cancers (Basel) 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. ; TCGA Research Network (2013). The somatic genomic landscape of glioblastoma. Cell 155, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, and Breakefield XO (2018). Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol 14, 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y, Zhang D, Huo Y, Zhang J, Yang A, and Wang T. (2019). MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J. Exp. Clin. Cancer Res 38, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, Huang W-C, Sun T-H, Tu S-J, Lee W-H, et al. (2018). miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 46 (D1), D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, and Marsden PA (2013). Angiogenesis in glioblastoma. N. Engl. J. Med 369, 1561–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gooijer MC, de Vries NA, Buckle T, Buil LCM, Beijnen JH, Boogerd W, and van Tellingen O. (2018). Improved Brain Penetration and Anti-tumor Efficacy of Temozolomide by Inhibition of ABCB1 and ABCG2. Neoplasia 20, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldh M, Lötvall J, Malmhäll C, and Ekström K. (2012). Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol 50, 278–286. [DOI] [PubMed] [Google Scholar]
- Emery IF, Gopalan A, Wood S, Chow KH, Battelli C, George J, Blas-zyk H, Florman J, and Yun K. (2017). Expression and function of ABCG2 and XIAP in glioblastomas. J. Neurooncol 133, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, and Nelson SF (2004). Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64, 6503–6510. [DOI] [PubMed] [Google Scholar]
- Garnier D, Renoult O, Alves-Guerra MC, Paris F, and Pecqueur C. (2019). Glioblastoma stem-like cells, metabolic strategy to kill a challenging target. Front. Oncol 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo LHM, Garcia C, da Fonseca ACC, Dubois LGF, de Sampaio E Spohr TCL, Matias D, de Camargo Magalhaães ES, do Amaral RF, da Rosa BG, Grimaldi I, et al. (2019). Glioblastoma Therapy in the age of molecular medicine. Trends Cancer 5, 46–65. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Ferrer-Luna R, Rooj AK, Mineo M, Ricklefs F, Takeda YS, Nowicki MO, Salińska E, Nakano I, Lee H, et al. (2017). MicroRNA signatures and molecular subtypes of glioblastoma: the role of extracellular transfer. Stem Cell Reports 8, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia L, Navone SE, Trombetta E, Cordiglieri C, Cherubini A, Crisa FM, Rampini P, Miozzo M, Fontana L, Caroli M, et al. (2018). Angiogenesis in human brain tumors: screening of drug response through a patient-specific cell platform for personalized therapy. Sci. Rep 8, 8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanif F, Muzaffar K, Perveen K, Malhi SM, and Simjee ShU (2017). Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev 18, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardee ME, and Zagzag D. (2012). Mechanisms of glioma-associated neovascularization. Am. J. Pathol 181, 1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder BG, Blomquist MR, Wang J, Kim AJ, Woodworth GF, Winkles JA, Loftus JC, and Tran NL (2018). Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma. Front. Oncol 8, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, et al. (2019). Reassessment of Exosome Composition. Cell 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Ji Y, Peng J, Zhou X, Chen X, Zhao H, Xu T, Chen L, and Xu Y. (2016). Increased Brain-Specific MiR-9 and MiR-124 in the Serum Exosomes of Acute Ischemic Stroke Patients. PLoS One 11, e0163645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, An X, Ye Z, Cully B, Wu J, and Li J. (2009). RGS5, a hypoxia-inducible apoptotic stimulator in endothelial cells. J. Biol. Chem 284, 23436–23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PS, Yekula A, Lansbury E, Small JL, Ayinon C, Mordecai S, Hochberg FH, Tigges J, Delcuze B, Charest A, et al. (2019). Characterization of plasma-derived protoporphyrin-IX-positive extracellular vesicles following 5-ALA use in patients with malignant glioma. EBioMedicine 48, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczor-Urbanowicz KE, Kim Y, Li F, Galeev T, Kitchen RR, Gerstein M, Koyano K, Jeong S-H, Wang X, Elashoff D, et al. (2018). Novel approaches for bioinformatic analysis of salivary RNA sequencing data for development. Bioinformatics 34, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JR (2019). The Role of Brain Vasculature in Glioblastoma. Mol. Neurobiol 56, 6645–6653. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, Schiff D, Purow B, Parsons S, Lawler S, and Abounader R. (2014). microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 74, 1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kim IK, Yang JM, Lee E, Koh BI, Song S, Park J, Lee S, Choi C, Kim JW, et al. (2016). SoxF transcription factors are positive feedback regulators of VEGF signaling. Circ. Res 119, 839–852. [DOI] [PubMed] [Google Scholar]
- Kinsella M, Harismendy O, Nakano M, Frazer KA, and Bafna V. (2011). Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics 27, 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, and Kleihues P. (2007). The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madelaine R, Sloan SA, Huber N, Notwell JH, Leung LC, Skariah G, Halluin C, Pasxca SP, Bejerano G, Krasnow MA, et al. (2017). MicroRNA-9 couples brain neurogenesis and angiogenesis. Cell Rep. 20, 1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, Jackson AR, Srinivasan S, Chung A, Laurent CD, et al. (2019). exRNA Atlas Analysis Reveals Distinct Extracellular RNA Cargo Types and Their Carriers Present across Human Biofluids. Cell 177, 463–477.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano I, Garnier D, Minata M, and Rak J. (2015). Extracellular vesicles in the biology of brain tumour stem cells–implications for inter-cellular communication, therapy and biomarker development. Semin. Cell Dev. Biol 40, 17–26. [DOI] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic V, Hartmaier RJ, Boone DN, Samuels ML, Patel RY, White WM, Garovic VD, Oesterreich S, Roth ME, Lee AV, and Milosavljevic A. (2016). Epigenomic deconvolution of breast tumors reveals metabolic coupling between constituent cell types. Cell Rep. 17, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, and Barnholtz-Sloan JS (2015). CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neurooncol. 17 (Suppl 4), iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin SL, Samuel MS, Koszyca B, Brown MP, Ebert LM, Oksdath M, and Gomez GA (2019). Glioblastoma heterogeneity and the tumour microenvironment: implications for preclinical research and development of new treatments. Biochem. Soc. Trans 47, 625–638. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. (2006). Molecular sub-classes of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173. [DOI] [PubMed] [Google Scholar]
- Quezada C, Torres A, Niechi I, Uribe D, Contreras-Duarte S, Toledo F, San Mart´ın R, Gutié rrez J, and Sobrevia L. (2018). Role of extracellular vesicles in glioma progression. Mol. Aspects Med 60, 38–51. [DOI] [PubMed] [Google Scholar]
- Ramirez YP, Weatherbee JL, Wheelhouse RT, and Ross AH (2013). Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals (Basel) 6, 1475–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs F, Mineo M, Rooj AK, Nakano I, Charest A, Weissleder R, Breakefield XO, Chiocca EA, Godlewski J, and Bronisz A. (2016). Extracellular Vesicles from High-Grade Glioma Exchange Diverse Pro-oncogenic Signals That Maintain Intratumoral Heterogeneity. Cancer Res. 76, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooj AK, Mineo M, and Godlewski J. (2016). MicroRNA and extracellular vesicles in glioblastoma: small but powerful. Brain Tumor Pathol. 33, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooj AK, Ricklefs F, Mineo M, Nakano I, Chiocca EA, Bronisz A, and Godlewski J. (2017). MicroRNA-Mediated Dynamic Bidirectional Shift between the Subclasses of Glioblastoma Stem-like Cells. Cell Rep. 19, 2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, Thistlethwaite W, Subramanian SL, Milosavljevic A, and Gerstein M. (2019). exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Syst. 8, 352–357.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, Seredenina T, Muhs A, Scheiwe C, Shah MJ, et al. (2019). Mapping mi-croglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci 22, 2098–2110. [DOI] [PubMed] [Google Scholar]
- Schiffer D, Annovazzi L, Casalone C, Corona C, and Mellai M. (2018). Glioblastoma: microenvironment and niche concept. Cancers (Basel) 11, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen SS, Nygaard A-BB, Carlsen AL, Heegaard NHH, Bak M, and Christensen T. (2017). Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomark. Res 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli C, Montermini L, Meehan B, Brisson AR, Tan S, Choi D, Nakano I, and Rak J. (2018). Molecular subtypes and differentiation programmes of glioma stem cells as determinants of extracellular vesicle profiles and endothelial cell-stimulating activities. J. Extracell. Vesicles 7, 1490144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli C, Adnani L, Choi D, and Rak J. (2019). Extracellular Vesicles as Conduits of Non-Coding RNA Emission and Intercellular Transfer in Brain Tumors. Noncoding RNA 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, Filant J, Laurent CD, Laurent LD, Magee R, et al. (2019). Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 177, 446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson A, Özen I, Genove G, Paul G, and Bengzon J. (2015). Endogenous brain pericytes are widely activated and contribute to mouse glioma microvasculature. PLoS One 10, e0123553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J, da Hora CC, Kantar RS, Nakano I, Wakimoto H, Batchelor TT, Chiocca EA, Badr CE, and Tannous BA (2017). Dissecting inherent intratumor heterogeneity in patient-derived glioblastoma culture models. Neuro-oncol. 19, 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorova D, Simoncini S, Lacroix R, Sabatier F, and Dignat-George F. (2017). Extracellular vesicles in angiogenesis. Circ. Res 120, 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treps L, Perret R, Edmond S, Ricard D, and Gavard J. (2017). Glioblastoma stem-like cells secrete the pro-angiogenic VEGF-A factor in extracellular vesicles. J. Extracell. Vesicles 6, 1359479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin M, Hovestadt V, Escalante L, Shaw M, Rodman C, et al. (2017). Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. ; Cancer Genome Atlas Research Network (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr., Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, et al. (2012). Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-oncol. 14, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Xu Y, Feng L, Yin P, Song SS, Wu F, Yan P, and Liang Z. (2019a). RGS5 decreases the proliferation of human ovarian carcinoma-derived primary endothelial cells through the MAPK/ERK signaling pathway in hypoxia. Oncol. Rep 41, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Babikir H, Müller S, Yagnik G, Shamardani K, Catalan F, Kohanbash G, Alvarado B, Di Lullo E, Kriegstein A, et al. (2019b). The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 9, 1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabi-novsky R, Balaj L, Chen CC, Hochberg F, et al. (2017). Coding and non-coding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HA, Fatimy RE, Onodera C, Wei Z, Yi M, Mohan A, Gowrisankaran S, Karmali P, Marcusson E, Wakimoto H, et al. (2015). The Cancer Genome Atlas analysis predicts microRNA for targeting cancer growth and vascularization in glioblastoma. Mol. Ther 23, 1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Wang L, Li G, Liu H, Fan F, Li Z, Li Y, and Gao G. (2013). Increased expression of microRNA-9 predicts an unfavorable prognosis in human glioma. Mol. Cell. Biochem 384, 263–268. [DOI] [PubMed] [Google Scholar]
- Yi J, and Gao ZF (2019). MicroRNA-9–5p promotes angiogenesis but in-hibits apoptosis and inflammation of high glucose-induced injury in human umbilical vascular endothelial cells by targeting CXCR4. Int. J. Biol. Macromol 130, 1–9. [DOI] [PubMed] [Google Scholar]
- Yuan GQ, Wei NL, Mu LY, Wang XQ, Zhang YN, Zhou WN, and Pan YW (2017). A 4-miRNAs signature predicts survival in glioblastoma multiforme patients. Cancer Biomark. 20, 443–452. [DOI] [PubMed] [Google Scholar]
- Zaborowski MP, Balaj L, Breakefield XO, and Lai CP (2015). Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience 65, 783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders ED, Svensson F, and Bailey DS (2019). Therapy for glioblastoma: is it working? Drug Discov. Today 24, 1193–1201. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li Y, Tan Y, Liu Q, Jiang S, and in … -D. L (2019a). MiR-9–5p Inhibits Glioblastoma Cells Proliferation Through Directly Targeting FOXP2 (fork-head box P2). Front. Oncol 9, 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang Y-P, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA, et al. (2019. b). Transfer of Functional Cargo in Exomeres. Cell Rep. 27, 940–954.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, Lin B, Chen T, Xing C, Liu Z, et al. (2016). MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/β-catenin signaling pathway. Oncotarget 7, 28000–28012. [DOI] [PMC free article] [PubMed] [Google Scholar]