Significance
Bacteria constantly transition between conditions of feast and famine. Colonization of the human intestine by Vibrio cholerae is associated with a surge in host-derived fatty acids, demanding rapid regulation of fatty acid metabolism. Here, we provide evidence for an RNA-based mechanism controlling the expression of central fatty acid metabolism genes in response to changing external fatty acid concentrations. We identified a small regulatory RNA, FarS, which is processed from the 3′UTR of the fabB fatty acid biosynthesis gene and inhibits the production of proteins required for fatty acid degradation. Tight control of fatty acid biosynthesis and degradation is vital for all bacteria, and, in V. cholerae, FarS plays an important role in balancing these processes.
Keywords: small RNA, feed-forward loop, fatty acid metabolism, RNase E, Vibrio cholerae
Abstract
Hfq (host factor for phage Q beta) is key for posttranscriptional gene regulation in many bacteria. Hfq’s function is to stabilize sRNAs and to facilitate base-pairing with trans-encoded target mRNAs. Loss of Hfq typically results in pleiotropic phenotypes, and, in the major human pathogen Vibrio cholerae, Hfq inactivation has been linked to reduced virulence, failure to produce biofilms, and impaired intercellular communication. However, the RNA ligands of Hfq in V. cholerae are currently unknown. Here, we used RIP-seq (RNA immunoprecipitation followed by high-throughput sequencing) analysis to identify Hfq-bound RNAs in V. cholerae. Our work revealed 603 coding and 85 noncoding transcripts associated with Hfq, including 44 sRNAs originating from the 3′ end of mRNAs. Detailed investigation of one of these latter transcripts, named FarS (fatty acid regulated sRNA), showed that this sRNA is produced by RNase E-mediated maturation of the fabB 3′UTR, and, together with Hfq, inhibits the expression of two paralogous fadE mRNAs. The fabB and fadE genes are antagonistically regulated by the major fatty acid transcription factor, FadR, and we show that, together, FadR, FarS, and FadE constitute a mixed feed-forward loop regulating the transition between fatty acid biosynthesis and degradation in V. cholerae. Our results provide the molecular basis for studies on Hfq in V. cholerae and highlight the importance of a previously unrecognized sRNA for fatty acid metabolism in this major human pathogen.
Many if not all microorganisms use posttranscriptional control mechanisms to regulate gene expression. Small regulatory RNAs (sRNAs) are frequently involved in these processes, and an overwhelming majority of sRNAs seem to function by base-pairing with either cis- or trans-encoded target transcripts. However, these sRNAs typically do not act in isolation but rather require the aid of RNA-binding proteins (1). One prime example of this type of proteins is the Hfq RNA chaperone. Hfq belongs to the family of Sm/Lsm proteins characterized by a multimeric, ring-like structure, which promotes the binding of nucleic acid molecules (2). Mechanistically, Hfq functions as a “molecular matchmaker” by facilitating the interaction of sRNAs with cognate target mRNAs. The protein also protects sRNAs from ribonucleolytic decay (3, 4). Hfq can make contact with RNA at four different sites—rim, distal face, proximal face, and C terminus—though not all Hfq homologs carry the C-terminal extension (5, 6).
Studies from bacterial model organisms such as Escherichia coli and Salmonella enterica showed that Hfq binds hundreds of mRNAs and several dozen sRNAs in vivo (7–10). Accordingly, deletions of hfq give rise to drastic phenotypic changes ranging from impaired stress responses to failure to engage collective cell functions, such as biofilm formation (11, 12). Significantly reduced infectivity is also observed for hfq mutants of pathogenic microorganisms (13), including the major human pathogen Vibrio cholerae (14). Here, activation of virulence gene expression relies on a complex pathway integrating signals from V. cholerae itself, other microorganisms, and the host (15, 16). Indeed, recent work on V. cholerae’s cholera toxin (CTX) has revealed that host-derived heme and fatty acids are central factors for efficient colonization of the intestine (17).
Fatty acids also modulate the activity of the major virulence transcription factor ToxT (18), which, among many other genes, controls the expression of the TarB sRNA (19). TarB is a posttranscriptional inhibitor of the secreted colonization factor TcpF (19), as well as the pathogenicity island-encoded transcription factor VspR (20). In addition, V. cholerae sRNAs controlling cell–cell communication, e.g. Qrr1-4 (21) and VqmR (22–24), as well as sRNAs responding to cell-envelope damage (25, 26), contribute to virulence gene expression.
Numerous other sRNAs exist in V. cholerae. In fact, transcriptomic approaches have reported hundreds of uncharacterized sRNAs, including a large group of sRNAs originating from the 3′ end of mRNAs (23, 27). Similar observations have been made for other Gram-negative bacteria (28); however, it is often unclear if and how these sRNAs participate in gene regulation. Knowledge about the interaction of an sRNA with an RNA-binding protein can provide strong hypotheses regarding their regulatory functions. For example, Hfq- and ProQ-dependent sRNAs are likely to engage base-pairing with other transcripts, whereas CsrA-dependent sRNAs typically act by protein sequestration (1). For V. cholerae, we currently lack this information.
In this work, we have performed RIP-seq (RNA immunoprecipitation followed by high-throughput sequencing) analysis of Hfq in V. cholerae. We discovered 603 mRNAs and 82 sRNAs interacting with Hfq. A total of 25 of these sRNAs were previously unknown, and 44 sRNAs mapped to the 3′ end of a coding sequence. One highly abundant and 3′-encoded sRNA was FarS (for fatty acid regulated sRNA; as detailed later), which we studied in more detail. We show that FarS is expressed from the 3′UTR of the fabB gene producing β-ketoacyl-ACP synthase, a key enzyme for initiating fatty acid biosynthesis. As the farS gene does not have its own promoter, expression depends on the FadR transcription factor (activating fabB), as well as RNase E-mediated processing of the fabB mRNA. Mature FarS base-pairs with and inhibits the expression of two paralogous fadE mRNAs encoding acyl-CoA dehydrogenase, which is the rate-limiting enzyme in fatty acid β-oxidation (29). Transcription of fadE is repressed by FadR (30), and, together, FadR, fabB-FarS, and fadE constitute a previously unknown type 3 coherent feed-forward loop (FFL) regulating the transition between fatty acid biosynthesis and degradation in V. cholerae.
Results
RIP-Seq Analysis of Hfq in V. Cholerae.
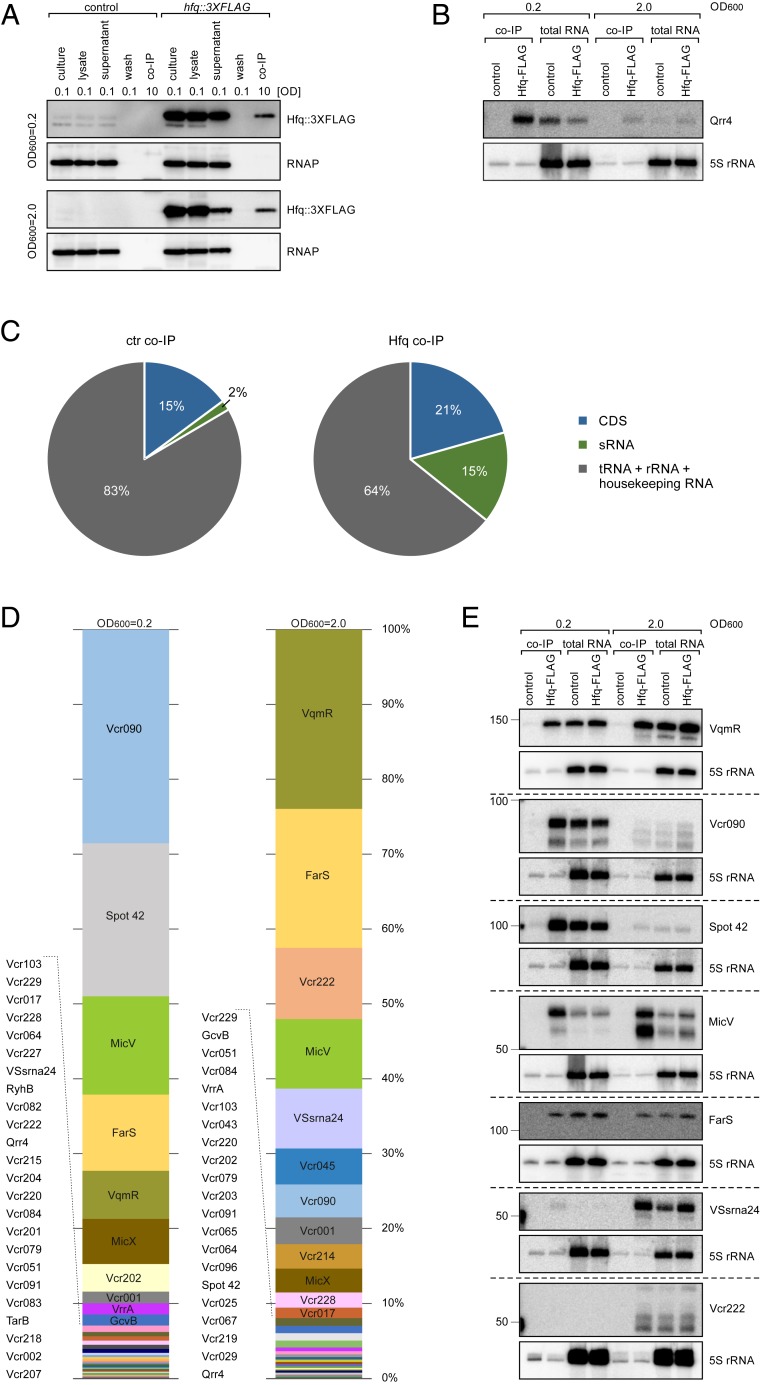
To identify the RNA ligands of Hfq in V. cholerae, we added the 3XFLAG epitope to the C terminus of the chromosomal hfq locus (vc0347) and tested protein production at various stages of growth. Hfq was produced under all tested conditions (SI Appendix, Fig. S1A), allowing us to employ a RIP-seq approach (9) to determine the set of Hfq-bound transcripts of V. cholerae cells cultivated to low (OD600 of 0.2) and high cell densities (OD600 of 2.0). Western blot analysis of the coimmunoprecipitated samples revealed specific enrichment of the Hfq::3XFLAG protein when compared to the negative control lacking the FLAG epitope (Fig. 1A). Likewise, the Hfq-dependent sRNA Qrr4 (21) was strongly enriched in the Hfq::3XFLAG samples, verifying our approach (Fig. 1B; note that Qrr4 is most strongly expressed at low cell densities). To obtain the full set of Hfq binding partners from both cell densities, we next converted the copurified RNAs (using the Hfq::3XFLAG strain as well as the untagged controls) into cDNA, followed by deep sequencing (31). We obtained 7.9 to 39.1 million reads for the individual libraries, of which 96.7 to 98.0% mapped to the V. cholerae N16961 genome (32) (SI Appendix, Table S1). As expected, the majority of reads (∼83%) obtained from the control libraries mapped to rRNAs, tRNAs, and housekeeping RNAs (tmRNA, 6S RNA, 4.5S RNA), whereas only 64% of reads mapped to this category in the Hfq::3XFLAG libraries (Fig. 1C). Instead, the fractions of bound mRNAs and sRNAs increased from 15 to 21% and from 2 to 15%, respectively. We detected a total of 82 sRNAs, 3 annotated riboswitches, and 603 mRNAs interacting with Hfq (>twofold enrichment over the untagged control sample; SI Appendix, Table S2). A total of 25 of these sRNAs (SI Appendix, Table S3) were discovered by our approach. As proof of concept, we confirmed that all previously reported Hfq-dependent sRNAs in V. cholerae, i.e., VqmR, Qrr1-4, MicV, VrrA, MicX, RyhB, TfoR, and TarA (23, 25, 33–37), were included in our dataset (SI Appendix, Table S2).
Fig. 1.
RIP-seq analysis of Hfq-binding sRNAs. (A) V. cholerae wild-type cells (control) and cells carrying a 3XFLAG epitope at the C-terminal end of the chromosomal hfq gene were cultivated in LB medium to low (OD600 of 0.2) and high cell densities (OD600 of 2.0) and subjected to coimmunoprecipitation. Protein samples were collected at different steps of the IP procedure and analyzed by Western blots. Culture refers to total protein before treatment, lysate refers to total protein after cell lysis, supernatant refers to remaining protein after incubation with anti-FLAG antibody and protein G Sepharose, wash refers to remaining protein in the lysis buffer after five washing steps, and co-IP indicates coimmunoprecipitated protein sample. The relative amount of cells loaded (OD600 units) is indicated. RNAP served as loading control. (B) RNA samples of co-IP and total RNA (lysate) fractions were loaded on a Northern blot and analyzed for Qrr4 levels. 5S rRNA served as loading control. (C) Pie charts of control and Hfq co-IP samples showing the relative fractions of the different RNA classes. The relative amount of total cDNA reads from each class in the control and Hfq co-IP libraries are shown. (D) Distribution of reads of significantly enriched sRNAs (fold enrichment > 2, P value ≤ 0.05) in Hfq co-IP libraries obtained from low (OD600 of 0.2) and high cell densities (OD600 of 2.0). Reads matching to a given sRNA were compared to all enriched sRNAs in the cDNA libraries. Shown are all sRNAs corresponding to at least 0.1% of the mapped reads. The relative amount of reads and enrichment factors for each sRNA are listed in SI Appendix, Table S2. (E) Co-IP and total RNA (lysate) fractions were obtained from V. cholerae wild-type and hfq::3XFLAG-tagged strains cultivated in LB medium to low (OD600 of 0.2) and high cell densities (OD600 of 2.0). The RNA was loaded on Northern blots and probed for the indicated sRNAs. 5S rRNA served as a loading control.
Patterns of Hfq-Binding sRNAs at Low and High Cell Densities.
Next, we sorted the Hfq-binding sRNAs by abundance, i.e., the relative number of reads obtained from the Hfq::3XFLAG samples (Fig. 1D). At low cell densities, the top five most abundant sRNAs were the yet-uncharacterized Vcr090 sRNA (23), the highly conserved Spot 42 (38), MicV (26), FarS [previously identified as Vcr076 (23)], and VqmR (23). At high cell densities, the relative levels of Vcr090, MicV, and Spot 42 decreased, while VqmR became the most abundant sRNA, followed by FarS. The top five sRNAs now also included the carbon controlled VSsrna24 (39) and the newly discovered Vcr222 (Fig. 1D and SI Appendix, Table S3). We verified direct Hfq binding of these and 13 additional sRNAs using coimmunoprecipitation followed by Northern blot analyses (Fig. 1E and SI Appendix, Fig. S1B). In addition, we performed electrophoretic mobility shift assays using purified Hfq and synthetic Vcr090, Spot 42, FarS, VqmR, MicV, and Vcr222 transcripts, which confirmed Hfq binding of these sRNAs in vitro (SI Appendix, Fig. S2 A–F). VqmR and Vcr090 displayed the highest affinity for Hfq in these assays (Kd of ∼5 to 10 nM), while MicV showed the weakest binding (Kd of ∼60 nM). These values are similar to Hfq-binding affinities of previously reported sRNAs, such as RybB and RydC from S. enterica (40, 41).
An Abundant Class of 3′UTR-Derived Hfq-Binding sRNAs.
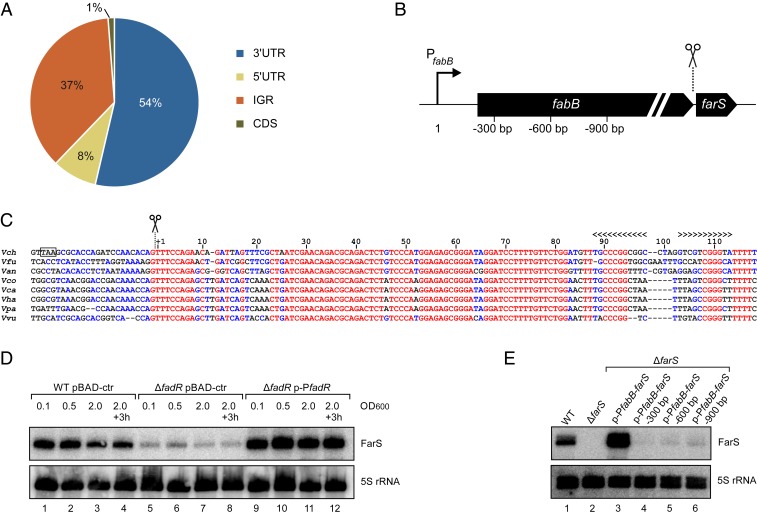
Our previous transcriptome analysis of V. cholerae cultivated under conditions of low and high cell densities indicated 44 possible 3′UTR-derived sRNAs (23); however, it remained unclear if these sRNAs were involved in posttranscriptional gene control and if Hfq would be required in this process. To address this question, we categorized the list of Hfq-binding sRNAs by their genomic location, i.e., intergenic, 5′UTR, CDS, and 3′UTR (Fig. 2A). In line with our previous hypothesis, we discovered that a large fraction of Hfq-binding sRNAs are expressed from the 3′UTR of mRNAs (54%), followed by intergenic sRNAs (37%) and sRNAs located in 5′UTRs (8%). Only one sRNA originated from an annotated coding sequence.
Fig. 2.
Identification and expression of the FarS sRNA. (A) Classification of Hfq-binding sRNAs according to their genomic location. The pie chart shows the relative fractions of Hfq-binding sRNAs (fold enrichment > 2, P value ≤ 0.05) originating from 3′UTRs, intergenic regions (IGRs), 5′UTRs, and coding sequences (CDSs). (B) Schematic representation of the fabB-farS genomic organization. Scissors indicate the processing site. Numbers correspond to the fabB promoter truncations tested in E. (C) Alignment of farS sequences in different Vibrio species. The sequences were aligned using the Multalign tool (76). The start of the sRNA and the Rho-independent terminator are indicated. The stop codon of fabB in V. cholerae is marked with a black box. Vch, Vibrio cholerae; Vfu, Vibrio furnissii; Van, Vibrio anguillarum; Vco, Vibrio coralliilyticus; Vca, Vibrio campbellii; Vha, Vibrio harveyi; Vpa, Vibrio parahaemolyticus; Vvu, Vibrio vulnificus. (D) V. cholerae wild-type and ΔfadR cells harboring either a control plasmid (pBAD-ctr) or a plasmid containing the fadR gene and its native promotor (p-PfadR) were cultivated in LB medium. Total RNA samples were collected at different stages of growth, and expression of FarS was analyzed on Northern blot. 5S rRNA was used as loading control. (E) V. cholerae wild-type and ΔfarS strains harboring different plasmids containing fabB-farS gene fragments (as indicated in B) were grown to stationary phase (OD600 of 2.0) in LB medium. Northern blot analysis was performed to determine FarS levels. Probing for 5S rRNA served as a loading control.
These results suggested that 3′UTR-derived sRNAs could have important regulatory roles in V. cholerae. To test this prediction, we focused on FarS, which was the most abundant 3′UTR-derived sRNA in our Hfq coimmunoprecipitation experiments (Fig. 1D). The farS gene is located in the 3′UTR of fabB (encoding β-ketoacyl-ACP synthase; Fig. 2B) and highly conserved among the Vibrio spp. (Fig. 2C). Northern analysis of V. cholerae cultivated in rich medium indicated that FarS is detectable at all stages of growth (Fig. 2D, lanes 1 to 4), and similar results were obtained for growth in minimal medium (SI Appendix, Fig. S3A, lanes 1 to 4). Expression of fabB has previously been reported to rely on the dual transcriptional regulator FadR (42), and we were able to confirm this result (SI Appendix, Fig. S3B). We therefore speculated that FadR might also affect the expression of FarS. Indeed, mutation of fadR resulted in approximately eightfold reduced farS levels in rich and minimal medium (Fig. 2D and SI Appendix, Fig. S3A, lanes 5 to 8), and expression was fully complemented by introduction of an FadR-producing plasmid (Fig. 2D and SI Appendix, Fig. S3A, lanes 9 to 12). These results are in agreement with a previous study suggesting that farS does not have its own promoter (23) and indicated that FarS expression strictly relies on transcriptional input signals integrated at the fabB promoter. To test this hypothesis, we first constructed a farS mutant strain by removing base pairs 1 to 85 of the farS sequence from the V. cholerae genome while keeping the Rho-independent terminator intact (Fig. 2C). Importantly, this mutation did not affect fabB mRNA stability (SI Appendix, Fig. S3C), avoiding possible secondary effects resulting from this mutation. We next introduced a plasmid containing the fabB-farS gene locus, as well as the fabB promoter, and monitored FarS production by Northern analysis. As expected, the fabB-farS plasmid fully restored FarS expression in the ΔfarS mutant (Fig. 2E, lanes 1 to 3). In contrast, deletion of the fabB promoter sequence in the fabB-farS plasmid strongly reduced FarS levels (>100-fold; Fig. 2E, lane 4), and expression remained low when we eliminated additional segments of the fabB coding sequence in the fabB-farS plasmid (Fig. 2E, lanes 5 and 6). Together, these results show that FarS is produced from the 3′UTR of fabB and that expression of the sRNA depends on the fabB promoter.
RNase E Is Required for FarS Production.
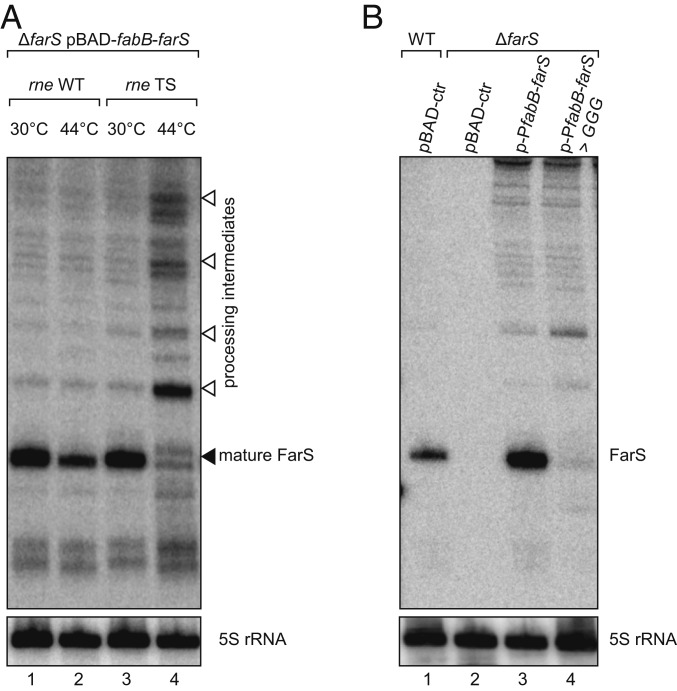
The class of 3′UTR-derived sRNAs has been divided into two groups: (i) sRNAs carrying their promotors [e.g., DapZ (9) and MicL (43)] and (ii) sRNAs requiring ribonuclease-dependent cleavage for full maturation [e.g., SdhX (44, 45) and CpxQ (46, 47)]. Our previous results indicated that FarS belongs to the second class (Fig. 2 D and E); however, the respective ribonuclease required for FarS maturation remained unclear. Inspection of the farS gene revealed a conserved sequence stretch located at the very 5′ end of the sRNA (Fig. 2C), matching the recently determined recognition motif for RNase E-mediated cleavage (48). To test a possible involvement of RNase E in FarS maturation, we transferred the farS mutation into a V. cholerae strain, producing a temperature-sensitive RNase E variant [rne encodes RNase E and is an essential gene in V. cholerae (49)], and transformed this strain with a plasmid allowing pBAD-inducible expression of the fabB-farS gene locus. We cultivated this strain under permissive (30 °C) and nonpermissive temperature (44 °C) and induced the pBAD promoter by addition of L-arabinose (0.2% final concentration). Total RNA samples were collected, and FarS expression was tested by Northern analysis. Mature FarS expression was readily detected at 30 °C but strongly reduced at 44 °C in the temperature-sensitive RNase E mutant (Fig. 3A, lanes 3 and 4). In addition, nonpermissive temperatures also resulted in the accumulation of various processing intermediates, suggesting inadequate degradation (maturation) of the fabB mRNA. This effect was specific to RNase E, as the relevant control strain (carrying the native rne gene) displayed accurate FarS maturation at 30 °C and 44 °C (Fig. 3A, lanes 1 and 2).
Fig. 3.
RNase E is required for FarS production. (A) V. cholerae ΔfarS carrying either a wild-type (rne WT) or temperature-sensitive RNase E (rne TS) allele and the pBAD-fabB-farS plasmid were cultivated at 30 °C in LB medium. When cells reached an OD600 of 1.0, cultures were split and kept at permissive temperature (30 °C) or shifted to nonpermissive temperature (44 °C) and incubated for 30 min. Next, expression of fabB-farS was induced using L-arabinose (0.2% final concentration, 30 min), and FarS levels were monitored by Northern blot. Probing for 5S rRNA served as loading control. (B) V. cholerae wild-type or ΔfarS cells harboring either a control plasmid (pBAD-ctr), a plasmid containing the fabB-farS gene locus and the fabB promotor, or a version of the plasmid where the first three base pairs of farS were mutated (TTT to GGG) were cultivated in LB medium. Total RNA samples were collected when cells reached an OD600 of 1.0. The Northern blot was probed for FarS, and 5S rRNA was used as a loading control.
To corroborate a direct role of RNase E in FarS production, we exchanged the first three base pairs of farS (TTT to GGG; mutating the predicted RNase E recognition motif) and tested FarS production. In line with our prediction, exchange of these critical residues almost completely abolished FarS production, while cleavage events located further upstream in the transcript remained functional (Fig. 3B). In summary, these data strongly suggest that FarS is produced by RNase E-mediated processing of the fabB mRNA.
FarS inhibits the Expression of Two Paralogous fadE mRNAs.
A hallmark of Hfq-dependent sRNAs is their ability to base-pair with trans-encoded target mRNAs, affecting transcript stability and translation initiation (3). This feature has been demonstrated for conventional sRNAs encoded by free-standing genes, as well as 3′UTR-derived sRNAs requiring ribonuclease-assisted maturation (28). To investigate if FarS functions as a trans-acting regulator in V. cholerae, we cloned the farS gene onto a plasmid downstream of the pBAD promoter (initiating transcription at the RNase E cleavage site; see Fig. 2C). Next, we cultivated V. cholerae wild-type cells carrying either pBAD-farS or a control plasmid to exponential phase (OD600 of 0.5) and induced pBAD expression for 15 min. Comparison of pBAD-controlled and endogenous FarS expression revealed that both transcripts migrated at the same size and that pBAD-driven FarS levels were ∼10-fold increased when compared to the control (SI Appendix, Fig. S4A). Next, we converted the total RNA obtained from these samples into cDNA and determined global transcriptome changes using deep sequencing (31). We identified only two differentially expressed transcripts upon FarS induction (both repressed, vc1740 and vc2231; Table 1), and both encoded homologs of the same enzyme, FadE (acyl-CoA dehydrogenase).
Table 1.
Genes differentially expressed in response to FarS pulse expression
| Gene | Description | Fold change |
| vc1740 | acyl-CoA dehydrogenase | −2.02 |
| vc2231 | acyl-CoA dehydrogenase | −2.54 |
Description is based on the annotation at KEGG (https://www.genome.jp/kegg/). Fold changes were obtained by transcriptomic analysis of pBAD-driven FarS expression using RNA-seq. Genes regulated >twofold with an FDR-adjusted P value ≤ 0.05 are listed.
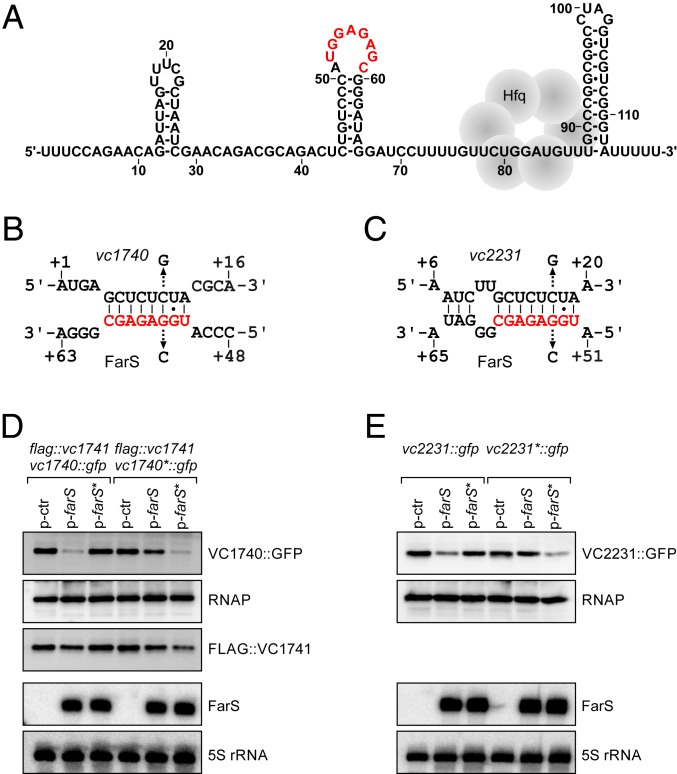
To explore the mechanistic details of FarS-mediated fadE repression, we first determined the secondary structure of FarS using chemical and enzymatic probing (SI Appendix, Fig. S4B). FarS contains three hairpin elements, the most distal of which most likely serves as a Rho-independent terminator (Fig. 4A). In addition, we identified a potential Hfq-binding site located between the second and third stem-loops of FarS, indicating that base-pairing will occur further upstream in the sRNA. Using the RNAhybrid algorithm (50), we were able to predict potential base-pairing sites of FarS with vc1740 and vc2231 involving an exposed loop in the second hairpin element of FarS (Fig. 4A) and the sequence encoding the N terminus of both FadE proteins (Fig. 4 B and C). To test these predictions, we used E. coli as a heterologous host (lacking the farS gene) and a reporter system with constitutive expression of the sRNA and a translational fusion of the target mRNA to gfp on two individual plasmids (51). Given that vc1740 constitutes the second gene in a di-cistronic operon with vc1741 (23) (encoding a transcriptional regulator), we employed a variant of this system in which the proximal gene (i.e., vc1741) carries an N-terminal FLAG epitope to monitor the effect of the sRNA on both genes. Using this setup, we discovered that FarS specifically repressed the distal part of this operon (producing FadE::GFP), whereas FLAG::VC1741 levels remained unchanged (Fig. 4D). We also found that FarS inhibits VC2231::GFP expression at the posttranscriptional level (Fig. 4E) and that a single point mutation in FarS (G54 to C) was sufficient to block regulation of both fadE targets (Fig. 4 D and E). Vice versa, mutation of vc1740 and vc2231 at the indicated positions (C10 to G and C17 to G, respectively; Fig. 4 B and C) inhibited regulation by FarS, while combination of the mutated FarS and fadE variants restored GFP repression (Fig. 4 D and E). FarS stability was reduced in hfq-deficient V. cholerae cells (SI Appendix, Fig. S5A), and, despite FarS accumulating to similar levels when expressed from a multicopy plasmid in wild-type and Δhfq cells, regulation of both targets was strictly dependent on Hfq (SI Appendix, Fig. S5 B and C). Our data suggest that FarS uses an exposed loop element to base-pair with the two paralogous fadE mRNAs and that this process requires Hfq. We note that, although the FadE protein sequence is highly divergent at the N terminus, the FarS base-pairing site is highly conserved at the DNA level (SI Appendix, Fig. S5 D and E). These results might suggest that, at the phylogenetic level, FarS-mediated repression of the fadE mRNA was established before the gene was duplicated.
Fig. 4.
Structure of FarS and base-pairing to fadE target mRNAs. (A) Secondary structure of FarS. The secondary structure was derived from chemical and enzymatic structure probing experiments (SI Appendix, Fig. S4B). The base-pairing site is marked in red, and the Hfq binding site is indicated. (B) Predicted base-pairing of FarS with vc1740. Arrows indicate the single nucleotide mutations tested in D. (C) Predicted base-pairing of FarS with vc2231. Arrows indicate the single nucleotide mutations tested in E. (D) Discoordinate regulation of FLAG::VC1741 and VC1740::GFP. E. coli cells carrying a reporter plasmid for FLAG::VC1741 and VC1740::GFP or VC1740*::GFP (C10 to G) were cotransformed with a control plasmid (p-ctr), p-farS, or p-farS* (G54 to C). Cells were grown in LB medium to stationary phase (OD600 of 2.0). GFP and FLAG levels were measured by Western blot, and FarS levels were determined by Northern blot. RNAP and 5S rRNA served as loading controls for the Western and Northern blots, respectively. (E) E. coli harboring a reporter plasmid for VC2231::GFP or VC2231*::GFP (C17 to G) and either a control plasmid (p-ctr), p-farS, or p-farS* (G54 to C) were grown in LB medium to OD600 of 2.0. GFP and FarS levels were monitored as in D.
FarS Restricts FadE Protein Production.
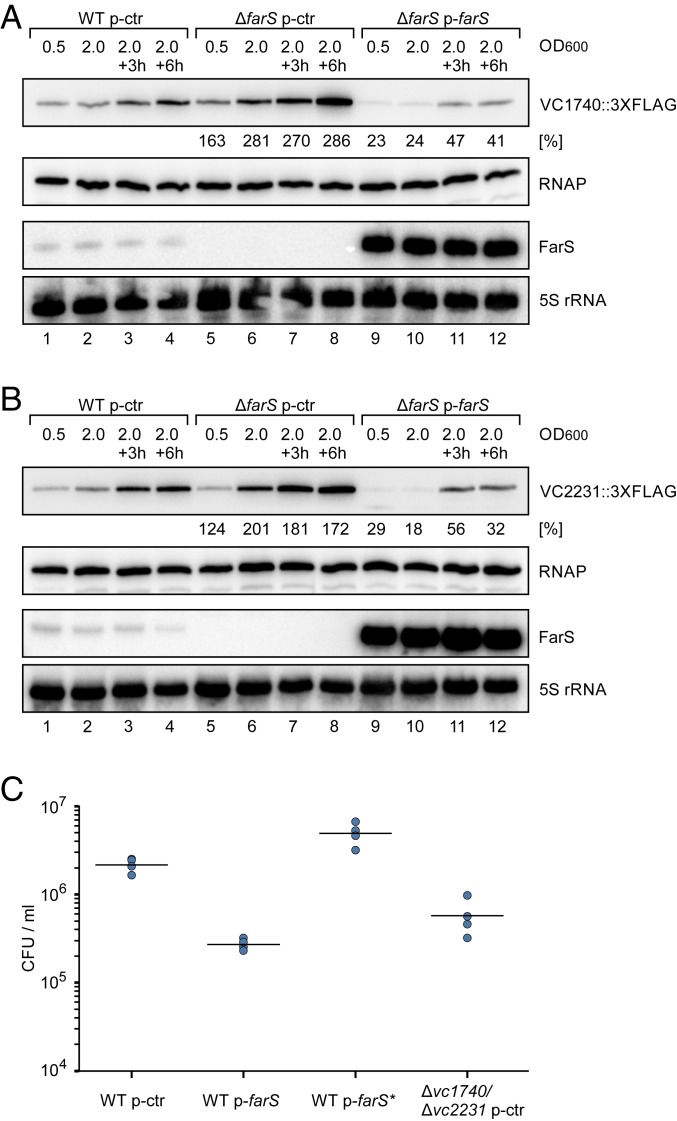
Repression of the two fadE::gfp fusions suggested that FarS also inhibits the synthesis of both FadE paralogs in V. cholerae. To test this hypothesis, we added a 3XFLAG epitope to the C termini of the chromosomal vc1740 and vc2231 genes and monitored protein production in wild-type and ΔfarS cells (both harboring a control plasmid). In agreement with our prediction, the production of both proteins was elevated in farS-deficient cells. For VC1740::3XFLAG, increased protein abundance was detected at all stages of growth accumulating to ∼2.5-fold higher levels in late stationary phase (6 h after cells reached an OD600 of 2.0; Fig. 5A, lanes 1 to 8, and SI Appendix, Fig. S6A). Similarly, VC2231::3XFLAG levels were elevated in ΔfarS, with the most pronounced differences in protein production (∼twofold) when cells reached an OD600 of 2.0 (Fig. 5B, lanes 1 to 8, and SI Appendix, Fig. S6B). In both cases (VC1740::3XFLAG and VC2231::3XFLAG), introduction of a FarS overexpression plasmid into ΔfarS cells strongly reduced FadE levels at all stages of growth (Fig. 5 A and B, lanes 9 to 12, and SI Appendix, Fig. S6 A and B).
Fig. 5.
FarS inhibits FadE protein production. (A and B) V. cholerae wild-type and ΔfarS strains carrying a chromosomal 3XFLAG epitope either at the vc1740 (A) or at the vc2231 (B) gene and harboring the indicated plasmids were cultivated in M9 minimal medium. Protein and total RNA samples were collected at the indicated OD600 readings. FadE::3XFLAG protein production (A, VC1740::3XFLAG; B, VC2231::3XFLAG) was analyzed on Western blots, and expression of FarS was monitored on Northern blots. RNAP and 5S rRNA served as loading controls for the Western and Northern blots, respectively. Percentages indicate the amount of protein relative to the wild-type level at the corresponding growth phase. A quantification of data obtained from three independent biological replicates is shown in SI Appendix, Fig. S6 A and B. (C) V. cholerae wild-type and Δvc1740/Δvc2231 strains carrying the indicated plasmids were cultivated for 10 h in M9 minimal medium containing fatty acid (sodium oleate) as sole carbon source. Serial dilutions were prepared and recovered on agar plates, and colony-forming units (CFU) per milliliter were determined. Dots represent individual replicates (n = 4), and lines indicate the mean CFU.
To test the effect of FarS-mediated FadE repression on fatty acid metabolism of V. cholerae, we cultivated wild-type V. cholerae carrying either a control or the FarS overexpression plasmid in minimal medium containing sodium oleate as sole carbon source. We discovered that, after 10 h of incubation under these conditions, V. cholerae cells expressing FarS from a plasmid displayed ∼10-fold decreased survival when counted on agar plates (Fig. 5C). This effect was specific to the repression of vc1740 and vc2231 by FarS, since plasmid-borne expression of mutated FarS (FarS*, e.g., see Fig. 4B) did not inhibit growth under these conditions, whereas V. cholerae cells deleted for vc1740 and vc2231 showed survival rates similar to the FarS overexpression strain. Together, we conclude that FarS down-regulates the synthesis of both FadE paralogs and thereby affects the fatty acid metabolism in V. cholerae.
FarS Is the Central Regulator of a Mixed Feed-Forward Loop.
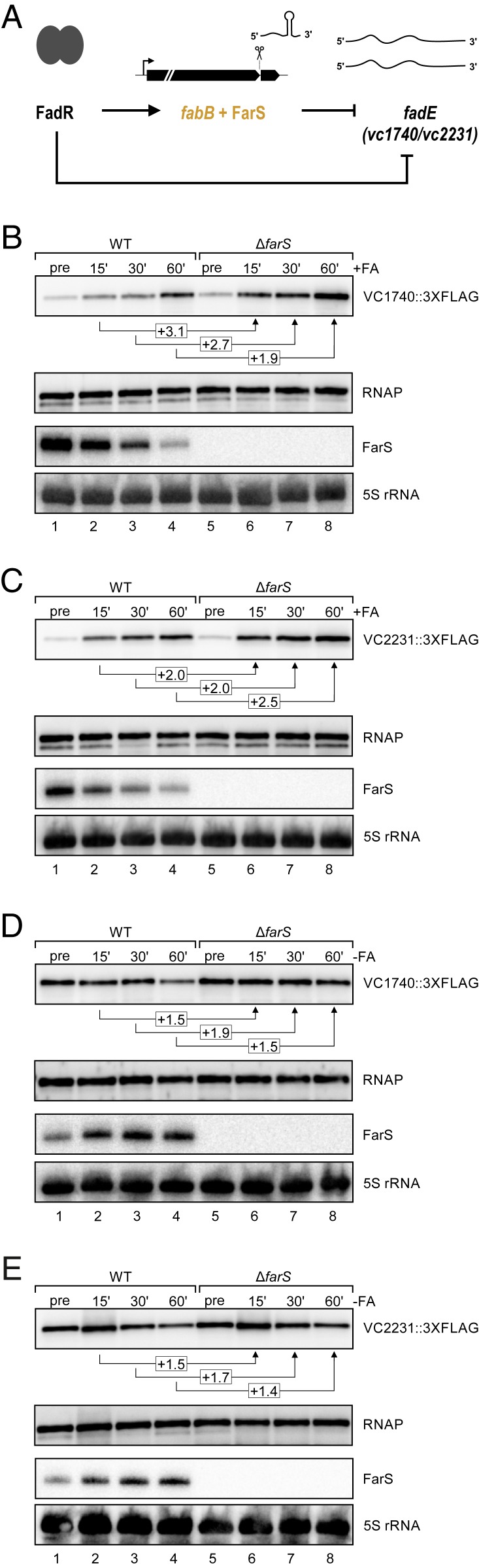
Previous reports have shown that FadR of V. cholerae functions as a dual transcriptional regulator inhibiting fadE and activating fabB (52). Our data now show that FarS is coexpressed from the fabB promoter (Fig. 2E) and represses the production of the FadE paralogs (Fig. 5 A and B and SI Appendix, Fig. S6 A and B). Thus, FadR and FarS both repress synthesis of the FadE proteins, establishing a mixed type 3 coherent FFL in which one regulator, FarS, is clipped off the 3′UTR of a functionally related mRNA product (Fig. 6A). The logic implied in this regulatory setup predicts two possible functions for FarS: (i) FarS acts as a delay element limiting FadE production when V. cholerae transitions from low to high external fatty acid concentrations and, (ii) in the reverse scenario (transition from high to low fatty acid concentrations), FarS accelerates the repression of FadE. To test this prediction, we examined the effects of adding or removing fatty acids. We first determined the effects on the sRNA levels by cultivating V. cholerae cells to early stationary phase (OD600 of 1.0) and then monitoring the expression of FarS in response to the addition or removal of fatty acids. Indeed, addition of sodium oleate (0.005% final concentration) efficiently repressed FarS production in V. cholerae (∼fivefold; SI Appendix, Fig. S6C). Removal of external fatty acids from the medium, on the contrary (by washing and reinoculation of V. cholerae cells into fatty-acid-free minimal medium), resulted in increased FarS expression (∼10-fold 60 min after reinoculation; SI Appendix, Fig. S6D). These results are in line with the expected patterns of FabB and FarS expression under conditions of low and high fatty acids.
Fig. 6.
FarS is part of a mixed feed-forward loop. (A) Schematic display of a mixed type 3 coherent feed-forward loop involving the transcription factor FadR, the fabB mRNA, FarS, and the two fadE mRNAs. (B and C) V. cholerae wild-type and ΔfarS strains carrying a chromosomal 3XFLAG epitope either at the vc1740 (B) or at the vc2231 (C) gene were cultivated in M9 minimal medium to stationary phase (OD600 of 2.0). Total protein and RNA samples were collected before and after addition of fatty acids (+FA; sodium oleate, 0.005% final concentration) at the indicated time points. Expression patterns of the VC1740 (B) and VC2231 (C) proteins were analyzed on Western blots, and expression of FarS was determined using Northern blot analysis. RNAP and 5S rRNA served as loading controls for the Western and Northern blots, respectively. (D and E) V. cholerae wild-type and ΔfarS strains carrying a chromosomal 3XFLAG epitope either at the vc1740 (D) or at the vc2231 (E) gene were cultivated in M9 minimal medium containing sodium oleate (0.005% final concentration) to an OD600 of 2.0. Cells were washed with PBS and resuspended in M9 minimal medium lacking fatty acids (-FA). Total protein and RNA samples were collected before and after removal of fatty acids at the indicated time points. Western and Northern blots show VC1740 (D) and VC2231 (E) protein and FarS levels, respectively. RNAP was used as loading control for Western blots; 5S rRNA for Northern blots.
We next tested the effect of fatty acid addition and removal on the expression of the two FadE paralogs in wild-type and ΔfarS V. cholerae using the experimental conditions established earlier. We discovered that addition of sodium oleate led to the increased production of the VC1740 and VC2231 proteins, albeit with slightly different kinetics. Accumulation of VC2231 was more rapid when compared to VC1740, and VC2231 showed a larger dynamic range (∼12-fold vs. ∼8-fold increased protein production comparing the preinduction and the 60-min time points; Fig. 6 B and C, lanes 1 to 4). V. cholerae cells lacking farS also showed elevated VC1740 and VC2231 levels upon fatty acid supplementation; however, the dynamics of the response were accelerated and resulted in ∼2 to 2.5-fold higher protein levels at the final time point of the experiment (Fig. 6 B and C, lanes 5 to 8, and SI Appendix, Fig. S6 E and F). Thus, we conclude that, as hypothesized earlier, FarS slows down FadE protein production when V. cholerae is exposed to sudden surges in fatty acid concentration.
FarS also inhibited FadE production when fatty acids were removed from the environment. Here, we observed that VC1740 and VC2231 levels decreased upon reinoculation of V. cholerae into fresh medium lacking fatty acids and that ΔfarS cells displayed ∼1.5 to 2-fold higher protein levels during the course of the experiment (Fig. 6 D and E and SI Appendix, Fig. S6 G and H). Of note, FarS only acts to down-regulate the existing vc1740 and vc2231 mRNAs, while transcription of these genes is simultaneously repressed by FadR (52). Together, both factors (transcriptional and posttranscriptional regulation) allow FadE repression when fatty acids become scarce.
Discussion
Bacterial sRNAs constitute a heterogeneous group of regulators that are produced from almost all segments of the genome. Traditionally, sRNAs from intergenic regions have been the focus of attention, which may well be explained by the design of early biocomputational screens scoring for conserved sequences associated with potential promoters and Rho-independent terminators that should be transcribed independent of both adjacent genes (53). Similarly, microarray-based approaches discovered a wealth of sRNAs in model organisms such as E. coli (8); however, due to the lacking resolution in microarray technologies, these analyses also favored the discovery of sRNAs from intergenic regions. The perception that sRNAs strictly originate from intergenic sequences was first challenged by shotgun cloning approaches (54) and further revised using deep-sequencing analyses (9, 48, 55).
In the well-studied Salmonella and E. coli models, 3′UTR-derived sRNAs constitute ∼20 to 30% of the Hfq-binding sRNAs, and, in this study, we reveal even higher numbers for V. cholerae (54%; Fig. 2A). The molecular determinants for this strong preference for 3′UTRs might well be explained by Hfq’s affinity toward Rho-independent terminators (56) and its relatively weak sequence specificity (57). However, the TransTerm algorithm (58) predicts a total of ∼760 high-confidence Rho-independent terminators for V. cholerae, suggesting that additional factors are required to guide Hfq to these 3′UTRs. It is interesting to note that initial FarS biogenesis seems to be independent of Hfq, as mature FarS is readily detectable in Δhfq cells (SI Appendix, Fig. S5A). This finding could also indicate an order of events for the synthesis of 3′UTR-derived sRNAs in which transcription is followed by RNase E-mediated mRNA decay, which is followed by binding of Hfq to the final degradation product. This process differs from the reported maturation of the ArcZ sRNA. Here, Hfq binding to the sRNA’s 3′ end is required to guide RNase E to the correct cleavage position (48). For 3′UTR-derived sRNAs such as MicL, CpxQ, and FarS, this function might be compensated by the presence of stem-loop elements inhibiting continued RNase E-mediated transcript decay (Fig. 4A, SI Appendix, Fig. S4B, and refs. 46 and 59).
Understanding the biogenesis and functions of 3′UTR-derived sRNAs could also make an interesting case for the study of sRNA evolution in bacteria (60). This is particularly evident for sRNAs that are produced by ribonucleolytic cleavage from mRNAs, as transcriptional control is already established by the promoter elements located upstream of coding sequence(s). Given that RNase E-mediated cleavage is pervasive in the enterobacteria (48, 61), one may speculate that de novo sRNA generation from 3′UTRs is driven by the affinity of Hfq for Rho-independent terminator elements (56) followed by potential base pair mutations that allow for the interaction with selected trans-encoded mRNAs. Other global RNA chaperones such as ProQ (55, 62, 63) could take Hfq’s position in this scenario as well and mediate target mRNA interactions. For example, RaiZ of S. enterica, still the single thoroughly characterized ProQ-dependent sRNA, is produced by cleavage of the raiA mRNA and base-pairs with the hupA mRNA to repress translation initiation (64). Interestingly, while FarS produced from the fabB 3′UTR of V. cholerae binds Hfq (Figs. 1 and 2), the fabB 3′UTR is a strong binding partner of ProQ in E. coli and Salmonella (65), indicating two possible analogous pathways (using Hfq or ProQ) to evolve functional sRNAs from the 3′ end of mRNAs. Finally, in contrast to their upstream coding sequences, the 3′ ends of mRNAs typically do not show conservation at the sequence level unless these are required to base-pair with mRNAs (Fig. 2C and refs. 9, 34, 43–45, and 48). This might be an exploitable feature for future bioinformatic searches aiming at 3′UTR-derived sRNAs in other microbes that have not yet been investigated for their Hfq–RNA interactions.
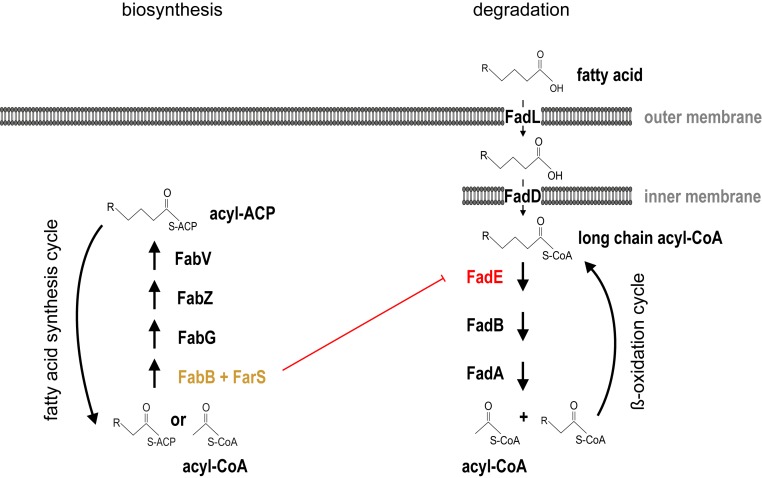
The expression of a regulatory RNA from the 3′ end of an mRNA using ribonucleolytic cleavage also adds an intriguing feature to the operon concept (54). Operons typically constitute a set of coding genes that are cotranscribed and together build a biologically relevant unit or pathway. This concept has now been extended to noncoding regulators, as these can provide a regulatory function to mRNAs, which would typically only produce an enzyme or a structural protein. How the regulatory role of these 3′ end-encoded sRNAs relates to the function of their upstream coding sequences has now been established in several cases. First, CpxQ is part of the CpxAR stress response system in Salmonella and produced from the 3′UTR of cpxP to reduce the translation of inner membrane proteins that trigger the same pathway (46). Second, SdhX, which is cotranscribed with the ∼10-kb-long sdhCDAB-sucABCD operon of the TCA cycle, down-regulates the synthesis of AckA (acetate kinase) and thereby adjusts TCA flux and acetate metabolism (44, 45). Third, s-SodF sRNA is expressed from the 3′UTR of the sodF mRNA (encoding a Fe-containing superoxide dismutase) under nickel starvation and limits the synthesis of the nickel-containing SodN superoxide dismutase (66). We showed here that FarS (produced from the 3′ end of the fabB fatty acid biosynthesis gene) inhibits the expression of two paralogous FadE proteins, which are involved in fatty acid degradation (Fig. 7). Thus, in the studied examples, the 3′ end-derived sRNAs provide a strong functional link between their origin of expression (i.e., their upstream mRNAs) and their targets. Importantly, s-SodF is produced in Gram-positive Streptomyces coelicolor, and regulation of sodN does not require Hfq (66), suggesting that this type of gene control is relevant beyond the enterobacterial clade. Stable 3′UTR RNA tails have now also been documented in mammalian cells (67), proposing an even broader regulatory concept.
Fig. 7.
Regulatory model for FarS-mediated fatty acid metabolism in V. cholerae. The fabB gene is part of the fatty acid biosynthesis regulon. It encodes β-ketoacyl-ACP synthase catalyzing the rate-limiting step in the synthesis of unsaturated fatty acid. The downstream reactions are catalyzed by FabG (3‐ketoacyl‐ACP reductase), FabZ (3‐hydroxyacyl‐ACP dehydratase), and FabV (enoyl‐ACP reductase). FarS is produced from the 3′UTR of fabB and posttranscriptionally inhibits the expression of two paralogous fadE mRNAs. The fadE genes encode acyl-CoA dehydrogenase catalyzing the initial step in fatty acid β-oxidation. Here, long‐chain fatty acids are transported across the outer and inner membranes by FadL and FadD, respectively. Following FadE activity, the remaining steps in fatty acid degradation are performed by a complex consisting of FadB and FadA.
One exciting future question related to the biological roles of 3′UTR-derived sRNAs is how they modulate the dynamics of their associated regulatory systems or pathways. We could show here that FarS is part of a type 3 coherent FFL that modifies the expression of two fadE genes (i.e., vc1740 and vc2231) in response to the availability of external fatty acids. Other sRNAs have recently been identified as part of so-called mixed regulatory circuits involving transcription factors and regulatory RNAs (68); however, none involved a 3′UTR-derived sRNA. Mixed FFLs come in two different designs with the sRNA working either as the top or middle regulator. However, only few mixed circuits have been analyzed for their regulatory dynamics. Two prime examples for sRNAs acting at the top and middle of the circuitry are RprA and Spot 42, respectively. RprA activates the production of rpoS and ricI at the posttranscriptional level, and RpoS is required for transcriptional activation of ricI. RicI inhibits plasmid conjugation in Salmonella, and, together, this system serves as a safety device to limit plasmid transfer under membrane-damaging conditions (69). The Spot 42 sRNA is repressed by the Crp transcriptional regulator and inhibits the expression of genes involved in the uptake and utilization of secondary carbon sources (constituting a type 4 coherent FFL). Spot 42 here modulates the dynamics of carbon utilization gene expression and reduces the overall leakiness of the system (70). Similarly, we discovered that FarS accelerates FadE repression when fatty acids are limited and serves as delay element when V. cholerae is transferred to high concentrations of fatty acids (Fig. 6 A–C). In addition, FarS inhibits FadE expression under regular growth conditions (Fig. 5 A and B), indicating a regulatory role when fatty acid concentrations are constant.
How this regulatory setup affects V. cholerae’s physiology is currently not fully understood, and, given various molecular mechanisms employed by Hfq-binding sRNAs (4), it is well possible that FarS regulates additional genes besides vc1740 and vc2231. Nevertheless, it is interesting to note that fabB (which also produces FarS) constitutes the first gene in fatty acid biosynthesis, while the FarS target genes, the two fadE paralogs, are the first genes required for fatty acid degradation (Fig. 7). FabB carries out the rate-limiting step for the biosynthesis of unsaturated fatty acids (71) and has recently been employed to artificially control membrane viscosity in E. coli (72). Conversely, FadE, i.e., acyl-CoA dehydrogenase, is the rate-limiting enzyme for one cycle of oxidation of acyl-CoA (73). It is well conceivable that V. cholerae limits the production of fatty acid degradation genes when fatty acid biogenesis is activated, as high levels of FadE could result in a futile cycle in which newly synthesized fatty acids are degraded by the cellular machinery. At the transcriptional level, switching between fatty acid biosynthesis and degradation is controlled by FadR (29), and our data suggest that FarS improves the robustness of this system through a posttranscriptional control mechanism (Figs. 5 and 6). Such tight regulation of fatty metabolism might be particularly relevant for V. cholerae’s lifestyle. Transcriptomic analysis of V. cholerae infecting infant rabbits revealed a strong activation of fatty acid degradation genes (including fadE), which can be explained by the influx of long-chain fatty acids in the cecal fluid of infected animals (74). Indeed, recent work focusing on the role of CTX during the infection process showed that the acquisition of host-derived long-chain fatty acids is necessary for V. cholerae’s survival and replication in the host (17). Further, fatty acids also directly modulate the activity of the major virulence transcription factor ToxT, which is required for CTX production (18). We therefore conclude that fatty acid metabolism is a key feature of V. cholerae’s pathogenic lifestyle and possibly requires dynamic regulatory mechanisms, including the mixed feed-forward loop identified here, to balance fatty acid biosynthesis and degradation.
Methods
Bacterial Strains and Growth Conditions.
All strains used in this study are listed in SI Appendix, Table S4. Details on strain construction are provided in SI Appendix, Materials and Methods. V. cholerae and E. coli cells were grown under aerobic conditions in LB (Lennox broth) or M9 minimal medium (0.4% glucose, 0.4% casamino acids) at 37 °C unless stated otherwise. Where appropriate, antibiotics were used at the following concentrations: 100 µg/mL ampicillin, 20 µg/mL chloramphenicol, 50 µg/mL kanamycin, 50 U/mL polymyxin B, and 5,000 µg/mL streptomycin. When cultivated in minimal medium with fatty acids (sodium oleate; 0.005% [wt/vol] final concentration; Sigma; O3880) as sole carbon source, V. cholerae cells were inoculated ∼1:1,000 from overnight cultures (M9 minimal medium with 0.4% glucose and 0.4% casamino acids) to the same starting OD600 and grown for 10 h at 37 °C (200 rpm shaking conditions). Serial dilutions were prepared and spotted on agar plates, and colony-forming units per milliliter were determined.
Plasmids and DNA Oligonucleotides.
Plasmids and DNA oligonucleotides are listed in SI Appendix, Tables S5 and S6, respectively. Details on plasmid construction are provided in SI Appendix, Materials and Methods.
Hfq Coimmunoprecipitation and cDNA Library Preparation.
V. cholerae wild-type (KPS-0014) and hfq::3XFLAG-tagged strains (KPS-0995) were cultivated in LB medium to low (OD600 of 0.2) and high cell densities (OD600 of 2.0). Cells equivalent to 50 OD600 units were collected and subjected to coimmunoprecipitation as described previously (9), with slight modifications. Briefly, cells were resuspended in lysis buffer (20 mM Tris⋅HCl [pH 8], 150 mM KCl, 1 mM MgCl2, 1 mM DTT) and disrupted with 0.3-mL glass beads (Roth; 0.1 mm diameter) using a Bead Ruptor 4 (Omni). Cleared lysates were incubated with monoclonal anti-FLAG antibody (Sigma; F1804) and protein G Sepharose (Sigma; P3296). After stringent washing with lysis buffer, RNA and protein fractions were isolated by phenol-chloroform-isopropanol extraction. The RNA was subjected to DNase I (Thermo Fisher Scientific) digestion, and RNA integrity was confirmed using a Bioanalyzer (Agilent). cDNA libraries were prepared using the NEBNext Small RNA Library Prep Set for Illumina (NEB; E7300S) according to the manufacturer’s instructions.
RIP-Seq Analysis.
cDNA libraries were sequenced on a HiSeq 1500 system in single-read mode with 100-nt read length. Demultiplexed raw reads were imported into CLC Genomics Workbench (Qiagen) and subjected to quality control and adaptor trimming. The trimmed reads were mapped to the V. cholerae reference genome [National Center for Biotechnology Information (NCBI) accession numbers NC_002505.1 and NC_002506.1, https://www.ncbi.nlm.nih.gov/assembly/GCF_000006745.1] with standard parameter settings. sRNA annotations were added manually based on previously identified sRNA candidates (23). Fold enrichment in the hfq::3XFLAG-tagged samples over the untagged control samples was calculated using the CLC “Differential Expression for RNA-Seq” tool.
Western Blot Analysis.
Western blot analysis of FLAG and GFP fusion proteins followed previously published protocols (24). Briefly, proteins were separated using SDS-PAGE and transferred to PVDF membranes. FLAG-tagged fusions were detected using anti-FLAG antibody (Sigma; F1804) and GFP-tagged fusions using anti-GFP antibody (Roche; no. 11814460001). RNAP served as loading control and was detected using anti-RNAP antibody (BioLegend; WP003). Signals were visualized on a Fusion FX imager (Vilber), and band intensities were quantified using the BIO-1D software (Vilber).
RNA Isolation and Northern Blot Analysis.
Total RNA was prepared and transferred as described previously (75). Membranes were hybridized in Roti-Hybri-Quick buffer (Roth) at 42 °C with [32P] end-labeled DNA oligonucleotides or at 63 °C for riboprobes. Riboprobes were prepared using the MAXIscript T7 Transcription Kit (Thermo Fisher Scientific; AM1312). Signals were visualized on a Typhoon PhosphorImager (Amersham), and band intensities were quantified using the GelQuant software (BioChemLabSolutions). Oligonucleotides for Northern blot analyses are listed in SI Appendix, Table S6.
Transcriptome Analysis.
V. cholerae wild-type cells harboring either pBAD-ctr or pBAD-farS were cultivated in triplicates to exponential phase (OD600 of 0.5). Expression of FarS was induced by adding L-arabinose (0.2% final concentration). After 15 min of arabinose treatment, transcription was stopped by adding 0.2 volumes of stop mix (95% ethanol, 5% [vol/vol] phenol) and cells were harvested. Total RNA was prepared and subjected to Turbo DNase (Thermo Fisher Scientific) digestion. After confirming RNA integrity using a Bioanalyzer (Agilent), ribosomal RNA was depleted using the Ribo-Zero rRNA Removal Kit (Epicentre) for Gram-negative bacteria. cDNA libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB; E7760) according to the manufacturer’s instructions. High-throughput sequencing was performed on a HiSeq 1500 system in single-read mode with 50-nt read length. Demultiplexed raw reads were trimmed for quality and adaptors and mapped to the V. cholerae reference genome (NCBI accession numbers NC_002505.1 and NC_002506.1) using CLC Genomics Workbench (Qiagen) with standard parameter settings. Reads mapped to annotated coding sequences were counted, and differential expression was calculated.
Fatty Acid Transition Assays.
V. cholerae cells were grown to the desired cell densities in M9 minimal medium. To study the effect of addition of fatty acids, sodium oleate (0.005% [wt/vol] final concentration; Sigma; O3880) was added to the cultures, and RNA and protein samples were collected at different time points (as indicated in figure legends). To analyze the reverse scenario, when fatty acids are removed, cultures were first cultivated in M9 minimal medium containing sodium oleate (0.005% [wt/vol] final concentration), washed at room temperature in 1× PBS, and resuspended for further growth in fresh M9 minimal medium lacking fatty acids. Again, RNA and protein samples were collected at the indicated time points. Expression of FarS was analyzed by Northern blots, and VC1740::3XFLAG and VC2231::3XFLAG protein levels were determined by Western blots.
Data Availability.
The sequencing data of the RIP-seq experiment and the transcriptome analysis are available at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database under the accession number GSE140516.
Supplementary Material
Acknowledgments
We thank Helmut Blum, Stefan Krebs, Nikolai Peschek, and Andreas Brachmann for help with the RNA sequencing experiments and Andreas Starick for excellent technical support. We thank Jörg Vogel, Gisela Storz, and Mona Hoyos for comments on the manuscript and all members of the Papenfort lab for insightful discussions and suggestions. This work was supported by the GIF (Grant G-2411-416.13/2016), the Human Frontier Science Program (Grant CDA00024/2016-C), Deutsche Forschungsgemeinschaft (EXC 2051, Project 390713860), and the European Research Council (StG-758212).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE140516).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920753117/-/DCSupplemental.
References
- 1.Holmqvist E., Vogel J., RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Santiago-Frangos A., Woodson S. A., Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9, e1475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Updegrove T. B., Zhang A., Storz G., Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kavita K., de Mets F., Gottesman S., New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schu D. J., Zhang A., Gottesman S., Storz G., Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34, 2557–2573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santiago-Frangos A., et al. , Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc. Natl. Acad. Sci. U.S.A. 116, 10978–10987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilusic I., Popitsch N., Rescheneder P., Schroeder R., Lybecker M., Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol. 11, 641–654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang A., et al. , Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50, 1111–1124 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Chao Y., Papenfort K., Reinhardt R., Sharma C. M., Vogel J., An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sittka A., et al. , Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4, e1000163 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobrero P., Valverde C., The bacterial protein Hfq: Much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 38, 276–299 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Vogel J., Luisi B. F., Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao Y., Vogel J., The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Ding Y., Davis B. M., Waldor M. K., Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53, 345–354 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Silva A. J., Benitez J. A., Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10, e0004330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler S. M., Camilli A., Going against the grain: Chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3, 611–620 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Chávez F., Mekalanos J. J., Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572, 244–248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowden M. J., et al. , Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. U.S.A. 107, 2860–2865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley E. S., Bodi K., Ismail A. M., Camilli A., A genome-wide approach to discovery of small RNAs involved in regulation of virulence in Vibrio cholerae. PLoS Pathog. 7, e1002126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies B. W., Bogard R. W., Young T. S., Mekalanos J. J., Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y., Feng L., Rutherford S. T., Papenfort K., Bassler B. L., Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 32, 2158–2171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzog R., Peschek N., Fröhlich K. S., Schumacher K., Papenfort K., Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae. Nucleic Acids Res. 47, 3171–3183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papenfort K., Förstner K. U., Cong J. P., Sharma C. M., Bassler B. L., Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 112, E766–E775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papenfort K., et al. , A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 13, 551–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song T., et al. , A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 70, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peschek N., Hoyos M., Herzog R., Förstner K. U., Papenfort K., A conserved RNA seed-pairing domain directs small RNA-mediated stress resistance in enterobacteria. EMBO J. 38, e101650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J. M., et al. , Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 37, e46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyakoshi M., Chao Y., Vogel J., Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 24, 132–139 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Fujita Y., Matsuoka H., Hirooka K., Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66, 829–839 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Campbell J. W., Cronan J. E. Jr, The enigmatic Escherichia coli fadE gene is yafH. J. Bacteriol. 184, 3759–3764 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber M., Papenfort K., Switching fatty acid metabolism by an RNA-controlled feed forward loop. National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140516. Deposited 17 November 2019.
- 32.Heidelberg J. F., et al. , DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenz D. H., et al. , The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Davis B. M., Waldor M. K., RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 65, 373–385 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis B. M., Quinones M., Pratt J., Ding Y., Waldor M. K., Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187, 4005–4014 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto S., et al. , Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J. Bacteriol. 193, 1953–1965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard A. L., Withey J. H., Beyhan S., Yildiz F., DiRita V. J., The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol. Microbiol. 78, 1171–1181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahagan B. G., Dahlberg J. E., A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. I. Nucleotide sequence analysis. J. Mol. Biol. 131, 573–592 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Hansen G. A., et al. , Expression profiling reveals Spot 42 small RNA as a key regulator in the central metabolism of Aliivibrio salmonicida. BMC Genomics 13, 37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papenfort K., et al. , SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimastrogiovanni D., et al. , Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife 3, e05375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi W., et al. , The 40-residue insertion in Vibrio cholerae FadR facilitates binding of an additional fatty acyl-CoA ligand. Nat. Commun. 6, 6032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo M. S., et al. , MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Mets F., Van Melderen L., Gottesman S., Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 1043–1052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyakoshi M., Matera G., Maki K., Sone Y., Vogel J., Functional expansion of a TCA cycle operon mRNA by a 3′ end-derived small RNA. Nucleic Acids Res. 47, 2075–2088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao Y., Vogel J., A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell 61, 352–363 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Grabowicz M., Koren D., Silhavy T. J., The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. MBio 7, e00312–e00316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao Y., et al. , In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65, 39–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cameron D. E., Urbach J. M., Mekalanos J. J., A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 105, 8736–8741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R., Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corcoran C. P., et al. , Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84, 428–445 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Kovacikova G., Lin W., Taylor R. K., Skorupski K., The fatty acid regulator FadR influences the expression of the virulence cascade in the El Tor biotype of Vibrio cholerae by modulating the levels of ToxT via two different mechanisms. J. Bacteriol. 199, e00762-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Argaman L., et al. , Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11, 941–950 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Vogel J., et al. , RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31, 6435–6443 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smirnov A., et al. , Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 113, 11591–11596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishikawa H., Otaka H., Maki K., Morita T., Aiba H., The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA 18, 1062–1074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmqvist E., et al. , Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kingsford C. L., Ayanbule K., Salzberg S. L., Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8, R22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Updegrove T. B., Kouse A. B., Bandyra K. J., Storz G., Stem-loops direct precise processing of 3′ UTR-derived small RNA MicL. Nucleic Acids Res. 47, 1482–1492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Updegrove T. B., Shabalina S. A., Storz G., How do base-pairing small RNAs evolve? FEMS Microbiol. Rev. 39, 379–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dar D., Sorek R., Extensive reshaping of bacterial operons by programmed mRNA decay. PLoS Genet. 14, e1007354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melamed S., Adams P. P., Zhang A., Zhang H., Storz G., RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 77, 411–425.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westermann A. J., et al. , The major RNA-binding protein ProQ impacts virulence gene expression in salmonella enterica serovar typhimurium. MBio 10, e02504-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smirnov A., Wang C., Drewry L. L., Vogel J., Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 36, 1029–1045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holmqvist E., Li L., Bischler T., Barquist L., Vogel J., Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell 70, 971–982.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Kim H. M., Shin J. H., Cho Y. B., Roe J. H., Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3'UTR of the sodF mRNA. Nucleic Acids Res. 42, 2003–2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malka Y., et al. , Post-transcriptional 3´-UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat. Commun. 8, 2029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nitzan M., Rehani R., Margalit H., Integration of bacterial small RNAs in regulatory networks. Annu. Rev. Biophys. 46, 131–148 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Papenfort K., Espinosa E., Casadesús J., Vogel J., Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 112, E4772–E4781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beisel C. L., Storz G., The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell 41, 286–297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng Y., Cronan J. E., Escherichia coli unsaturated fatty acid synthesis: Complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284, 29526–29535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Budin I., et al. , Viscous control of cellular respiration by membrane lipid composition. Science 362, 1186–1189 (2018). [DOI] [PubMed] [Google Scholar]
- 73.O’Brien W. J., Frerman F. E., Evidence for a complex of three beta-oxidation enzymes in Escherichia coli: Induction and localization. J. Bacteriol. 132, 532–540 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandlik A., et al. , RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fröhlich K. S., Haneke K., Papenfort K., Vogel J., The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res. 44, 10406–10422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corpet F., Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of the RIP-seq experiment and the transcriptome analysis are available at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database under the accession number GSE140516.