Significance
Clostridioides difficile is the most common cause of hospital-acquired diarrhea. It is responsible for more than half of all hospital enteric infections and >90% of mortalities resulting from these infections. Gut damage sustained during C. difficile infection is devastating, inducing epithelial damage deep into the gut tissue, to regions where stem cells responsible for tissue repair reside. Our work has shown that toxin-mediated damage during severe murine C. difficile infection alters the colonic epithelial protective barrier, which promotes colonic stem cell dysfunction, exacerbating disease and perturbing the repair capacity of the host. Furthermore, we have shown that this damage induces a chronically dysfunctional stem cell state, which prevents homeostatic cellular repair, possibly increasing susceptibility to subsequent infections or disease recurrence.
Keywords: Clostridium difficile, toxins, stem cell damage, infection, gut infection
Abstract
Gastrointestinal infections often induce epithelial damage that must be repaired for optimal gut function. While intestinal stem cells are critical for this regeneration process [R. C. van der Wath, B. S. Gardiner, A. W. Burgess, D. W. Smith, PLoS One 8, e73204 (2013); S. Kozar et al., Cell Stem Cell 13, 626–633 (2013)], how they are impacted by enteric infections remains poorly defined. Here, we investigate infection-mediated damage to the colonic stem cell compartment and how this affects epithelial repair and recovery from infection. Using the pathogen Clostridioides difficile, we show that infection disrupts murine intestinal cellular organization and integrity deep into the epithelium, to expose the otherwise protected stem cell compartment, in a TcdB-mediated process. Exposure and susceptibility of colonic stem cells to intoxication compromises their function during infection, which diminishes their ability to repair the injured epithelium, shown by altered stem cell signaling and a reduction in the growth of colonic organoids from stem cells isolated from infected mice. We also show, using both mouse and human colonic organoids, that TcdB from epidemic ribotype 027 strains does not require Frizzled 1/2/7 binding to elicit this dysfunctional stem cell state. This stem cell dysfunction induces a significant delay in recovery and repair of the intestinal epithelium of up to 2 wk post the infection peak. Our results uncover a mechanism by which an enteric pathogen subverts repair processes by targeting stem cells during infection and preventing epithelial regeneration, which prolongs epithelial barrier impairment and creates an environment in which disease recurrence is likely.
Colonic homeostasis, mediated by a functional stem cell compartment and with a normal turnover rate of 3 to 5 d (1, 2), is partly modulated by the 1014 bacteria in the colon (3). These microbes aid in the development and function of innate intestinal immunity and provide colonization resistance to opportunistic infections (4). Antibiotic treatment can alter this host–microbial balance, permitting colonization by pathogenic bacteria. In a recent survey of gastrointestinal illnesses in the United States, Clostridioides difficile was found to be responsible for more than half of all gastrointestinal infections in hospitals and was the causative agent in >90% of mortalities resulting from these infections (5). C. difficile infection (CDI) occurs in a polymicrobial environment and affects multiple cell types, inducing a spectrum of diarrheal diseases mediated by two exotoxins, TcdA and TcdB, which share sequence and structural homology (6, 7) but may contribute to disease severity unequally (8–11). Some clinical strains also produce binary toxin or C. difficile transferase (CDT), which alone does not induce severe disease in a hamster model of CDI (12) but seemingly aids in colonization (13) and partially enhances virulence (14). The prevalence of CDI and high disease relapse rate of ∼20 to 30% render C. difficile a clinically relevant and ideal candidate to study how infection and toxin-mediated damage may alter epithelial integrity and the colonic stem cell population.
Disease Outcome during CDI Is Dictated by Toxin Titer and Depth of Colonic Epithelium Damage
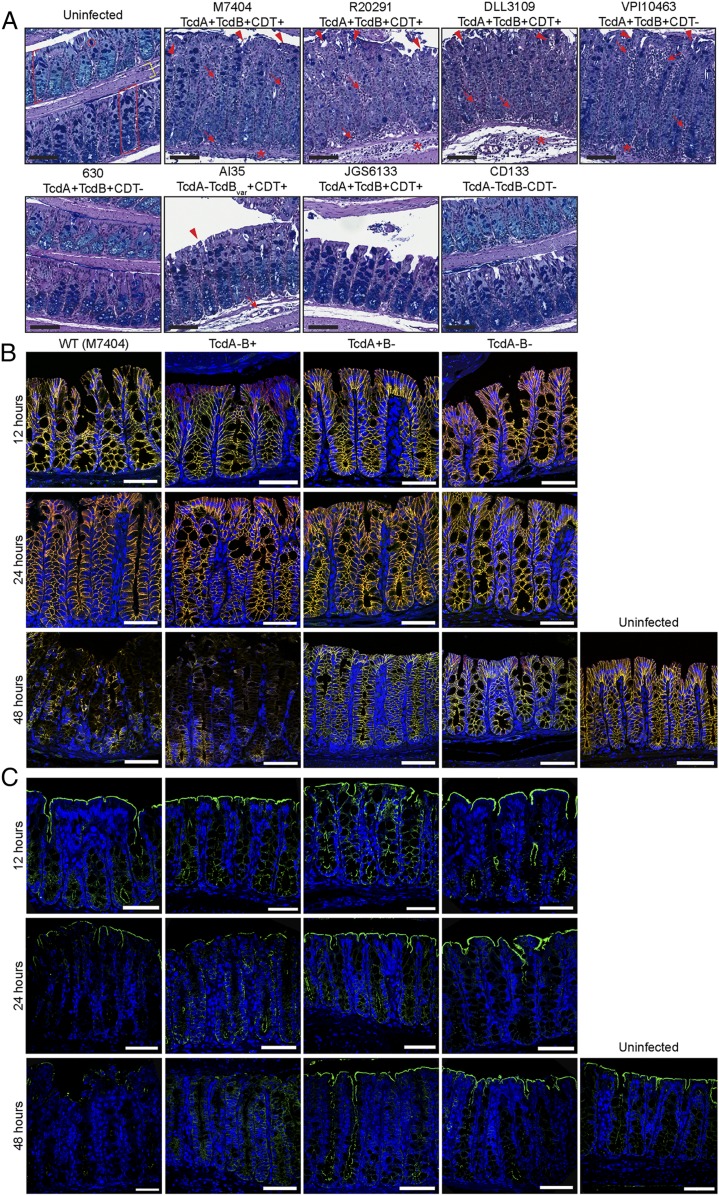
The changing epidemiology of CDI and diversity of strains, coupled with the heightened disease severity associated with ribotype (RT) 027, 017, 126, and 244 C. difficile strains, among others, has resulted in more CDI cases involving life-threatening complications and prolonged disease (15–20). Differences in disease severity are seemingly associated with C. difficile strains from different clades (21, 22); however, how genetically diverse C. difficile strains affect the host, and particularly the stem cell compartment, during infection is poorly characterized. Here, using a mouse model of CDI, we show that three genetically and geographically distinct RT027 strains (M7404, R20291, and DLL3109) and the RT003 strain VPI10463 are capable of inducing severe and devastating colonic damage that penetrates deep into the epithelium, characterized by damage to the base of the colonic crypts, severe inflammation, and edema (Fig. 1A). This deep colonic damage is associated with poor disease outcomes in these mice, limiting survival to 48 h of infection (SI Appendix, Fig. S1A). Interestingly, infection with the prototypical C. difficile strain 630 (23, 24), the pathogenicity locus (PaLoc) of which has 99% sequence identity to VPI10463 PaLoc (25–27), and strain AI35, a naturally occurring TcdA−TcdB+CDT+ strain which encodes a variant TcdB (28), as well as strain JGS6133, a RT078 animal isolate (28), was unable to induce damage beyond the surface of the colonic epithelium (Fig. 1A) and correlated with 100% survival and recovery from infection similar to CD133, a nontoxigenic clinical isolate (SI Appendix, Fig. S1 A and B). Indeed, the level of damage associated with each strain correlated with toxin production during infection (SI Appendix, Fig. S1 A, iii and iv). Strains capable of producing increased amounts of toxin during infection were able to induce damage deep into the epithelium. We suggest that higher toxin production during infection damages the epithelium at a rate higher than normal cellular turnover or repair, leading to progressive damage through the mucosa, which can then reach the crypt base. Thus, it appears that the capacity for a strain to induce a collapse in intestinal integrity and expose cells deep within the colonic epithelium is critical to poor disease outcome in CDI.
Fig. 1.
C. difficile induces severe and deep epithelial damage through TcdB alterations in adherens-junction formation and cellular polarity. (A) Mice were infected with a panel of genetically distinct C. difficile isolates and monitored for disease severity and colonic damage through periodic acid–Schiff (PAS)/Alcian blue staining. Representative images of swiss-rolled colonic tissue are shown. The colonic mucosa (red bracket) and, more specifically, the crypts of Lieberkühn (red box), comprised of colonic epithelial cells and goblet cells (red circle), sit above the submucosa and muscle layers (yellow bracket) of the colon. Arrow, inflammation; arrowhead, crypt damage/goblet cell loss; asterisk, edema. n ≥ 5. (Black scale bars: 100 µm.) (B and C) Colonic tissues were collected at 12, 24, and 48 h postinfection with M7404 (WT), and the isogenic toxin mutants of this strain (TcdA−B+, TcdA+B−, and TcdA−B− C. difficile), or from uninfected mice (48 h). A representative image of tissues stained for (B) β-catenin (green) and E-cadherin (red) (merge, yellow) or (C) ezrin (green) is shown, with nuclei stained with DAPI (blue). n ≥ 5. (White scale bars: 50 µm.) See also SI Appendix, Fig. S1.
TcdB Mediates Severe Intestinal Damage, Disrupting Intestinal Integrity and Exposing Stem Cells Deep within the Colonic Crypt to Intoxication
To gain a better understanding of the deep epithelial damage seen during CDI, we next focused on examining the disruption to tissue integrity over time during CDI. Junctional complexes, including tight junctions, adherens junctions, desmosomes, and gap junctions (29), play an important role in maintaining epithelial integrity and control the movement of molecules or microorganisms through the epithelial layer (30). Several pathogens, including Clostridium perfringens and Helicobacter pylori, target intercellular junctions to penetrate deeper into the epithelium and exacerbate disease (30). Previous in vitro work has shown that TcdA and TcdB alter tight-junction permeability (31, 32); however, toxin-mediated effects on β-catenin/E-cadherin adherens junctions (33) have not been reported. Cellular polarity is also essential for intestinal integrity. Ezrin, an ERM protein, assists in linking transmembrane proteins with the actin cytoskeleton and coordinating apical/basolateral receptor localization, among other functions in cytoskeletal stabilization and regulation (34). Ezrin also interacts with Rho-GTPases, which aid in actin cytoskeleton maintenance (35) and, importantly, are the primary targets of TcdA/TcdB (6, 35). Thus, to analyze changes in intestinal integrity during CDI in more detail, mice were infected with either M7404 (wild type [WT]; RT027), which produces TcdA, TcdB, and CDT, or an isogenic mutant strain lacking either TcdA (DLL3045, hereafter TcdA−B+), TcdB (DLL3101, hereafter TcdA+B−), or TcdA and TcdB (DLL3121, hereafter TcdA−B−) (8) and euthanized at either 12, 24, or 48 h postinfection, to track the progression of intestinal integrity collapse. Consistent with previous results (8), we showed that CDT alone (TcdA−B− infection) did not play a role in the induction of colonic pathology in this model and that infection with TcdB-producing strains (WT or TcdA−B+), but not TcdA+B−, induced rapid weight loss, severe diarrhea, and extensive colonic epithelial damage that progressively exposes deeper regions of the colonic epithelium until the peak of infection at 48 h (SI Appendix, Fig. S1 B–D). At 12 h postinfection, β-catenin/E-cadherin interactions and ezrin localization appeared intact and similar to those in uninfected tissues, regardless of the infecting strain (Fig. 1 B and C). At 24 h, β-catenin/E-cadherin immunostaining remained unchanged (Fig. 1B); however, the apical distribution of ezrin in the colon was altered in mice infected with TcdB-producing strains (Fig. 1C). This loss of cellular polarity staining seemingly initiates the collapse in intestinal integrity that begins to expose the deeper regions of the colonic epithelium since, by the 48-h time point, mice infected with WT or TcdA−B+ strains displayed dramatic TcdB-mediated changes to both the adherens junctions and cellular polarity (Fig. 1 B and C). Specifically, β-catenin/E-cadherin staining was severely disrupted, and ezrin was undetectable in tissues from mice infected with TcdB-producing strains, suggesting a collapse in adherens junctions and cellular polarity of differentiated cells within the colonic epithelium, that would otherwise function to protect the stem cell compartment at the base of the crypt from intoxication (Fig. 1 B and C). By contrast, β-catenin/E-cadherin and ezrin localization was indistinguishable between uninfected mice and those infected with TcdA+B− or TcdA−B− strains (Fig. 1 B and C), suggesting that TcdA and CDT do not play a role in altering the integrity of the intestinal barrier during CDI.
TcdB-Mediated Damage Deep into the Colonic Crypts Induces Stem Cell Injury
Under normal conditions, the crypts of Lieberkühn form the functional units of the colonic epithelium and are generated by stem cells at the crypt base that are maintained in an undifferentiated state through localized WNT signaling (36–38). Few studies have examined the effect of infection on intestinal stem cells, with some studies implicating effects on WNT signaling in the small intestine following infection (39, 40) and a study demonstrating that inflammation in the stomach could expand the stem cell pool (41). Others have used mature intestine-like organoids derived from human hair follicles (42) and mature colonic organoids to assess C. difficile toxin function (using purified toxins) and tissue damage (43). However, there are no published studies investigating the direct effect of infection on colonic stem cells and their function. WNT signaling is required for maintenance of intestinal stem cells where WNT ligands secreted from cells in the surrounding microenvironment interact with Frizzled (FZD) receptors on stem cells (44). A key marker of intestinal stem cells in both the small intestine and colon is LGR5 (36, 37), a seven-transmembrane receptor that binds R-spondin ligands (45–47). This complex intensifies WNT signaling in stem cells, stabilizing FZD/WNT receptors at the cell surface, which maintains stem cell function (45–47). Thus, damage to the stem cell population during intestinal infection is likely to have downstream consequences on disease severity or recovery. Given the level and depth of TcdB-mediated tissue damage associated with WT or TcdA−B+ infection, and that we detected significant increases in apoptosis at the crypt base, where stem cells reside, during infection with TcdB-producing strains (SI Appendix, Fig. S2), we investigated the effect of CDI on colonic stem cells and WNT-signaling activity.
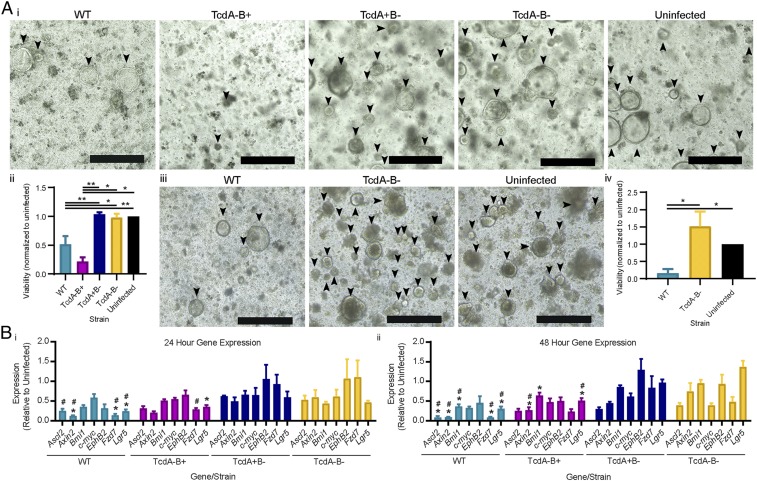
To characterize the effect of CDI on stem cell function and regenerative potential, crypts were isolated from mouse colonic tissues 24 and 48 h postinfection and assessed for their capacity to form viable colonic organoids (three-dimensional, multicellular stem cell derived structures) (48). Given the nature of this culture system, only functional colonic stem cells will develop into colonic organoids, providing a readout of stem cell function and regenerative capacity (49). At 24 h postinfection, fewer mature colonic organoids formed from mice infected with WT or TcdA−B+ strains, compared to uninfected mice or mice infected with TcdA+B− or TcdA−B− strains (Fig. 2 A, i). This was confirmed by quantitative assessment of viable cells in organoids from WT- or TcdA−B+-infected mice when compared to uninfected tissues (both P = 0.0079), TcdA−B− infection (P = 0.0317 and P = 0.0079, respectively), and TcdA+B− infection (P = 0.0268 and P = 0.0159, respectively) (Fig. 2 A, ii). These experiments provide direct evidence that toxin-mediated damage to the colon during infection alters the functional capacity of colonic stem cells, significantly reducing the establishment of organoids, and that this effect is mediated by TcdB and not TcdA. TcdA does not appear to alter stem cell function as similar numbers of organoids were formed from crypts isolated from TcdA+B−-infected mice or from uninfected mice, or those infected with the TcdA−B− strain (Fig. 2A). By 48 h postinfection, extensive damage to the colon and a decreased survival rate of mice infected with TcdB-producing strains, particularly the TcdA−B+ derivative, limited the numbers of mice from which viable crypts could be isolated for organoid culture. A comparison could therefore only be performed between WT-infected mice and uninfected or TcdA−B−-infected mice at the 48-h time point. Crypts isolated from WT-infected mice had a significantly decreased regenerative capacity, with very few mature colonic organoids forming compared to the TcdA−B−-infected and uninfected mice (P = 0.0286) (Fig. 2 A, iii and iv). This result again highlights that CDI induces significant cellular damage deep into the mucosa, which is severe enough to compromise the stem cell compartment and to alter the ability of the intestinal epithelium to be regenerated.
Fig. 2.
C. difficile TcdB alters murine colonic stem cell function and gene expression. Equal numbers of colonic crypts isolated from tissues collected postinfection with WT, TcdA−B+, TcdA+B−, and TcdA−B− C. difficile, or uninfected mice were used to derive murine colonic organoids, at 24 or 48 h postinfection. (A, i) Representative images of organoids 24 h postinfection are shown, (ii) with cell viability assessed via a PrestoBlue assay. (iii) Representative images of mature organoids after 48 h of infection are shown, (iv) with cell viability assessed via a PrestoBlue assay. (B) Quantitative ddPCR analysis of colonic tissue (i) 24 h and (ii) 48 h postinfection. Gene expression, as fold change relative to uninfected mice, was plotted (*, significant difference compared to TcdA−B−; #, significant difference compared to TcdA+B−). Data are represented as mean + SEM, n > 4. *P ≤ 0.05, **P ≤ 0.01. (Scale bars: 400 µm.) See also SI Appendix, Fig. S2.
Further investigation of this infection-mediated damage of the stem cell population via quantitative digital droplet-PCR (ddPCR) for seven genes that are representative markers of stem cells (Bmi1, Ascl2, Ephb2, and Lgr5), WNT-signaling (Axin2 and Fzd7), and cellular proliferation (c-myc) revealed that CDI, primarily through TcdB, perturbs the WNT-signaling pathway and alters the stem cell compartment, likely a consequence of fewer viable stem cells being present in the epithelium following TcdB-mediated intoxication. Alteration of the stem cell compartment was confirmed by the significant reduction in gene expression of several key stem cell genes, including Ascl2, Axin2, Bmi1, Fzd7, and Lgr5 (SI Appendix and Fig. 2B), when compared to TcdA+B−- and TcdA−B−-infected mice. TcdB-mediated intoxication, damage, or functional impairment of the LGR5+ cell population is likely to change the ability of the stem cells to replenish the colonic epithelium following damage, which may be compounded by a loss of viable stem cells within the colonic epithelium.
RT027 TcdB Induces Stem Cell Damage in a Frizzled Independent Manner
Since a number of TcdB receptors have previously been identified, we next examined their possible contributions to the disease phenotypes observed within the context of the infected gut. Recent work using purified TcdB, together with cell lines and mice, has identified FZD1/2/7 as potential TcdB receptors (50); however, the contribution of these interactions to stem cell damage during CDI has not been reported. Under normal conditions, expression of FZD1/2 in the colon is very low or undetectable (51), with increased expression seen in colonic cancers (51, 52). Importantly, as FZD7 is known to function as a WNT receptor in small-intestinal stem cells (44, 53), it may function as a TcdB receptor in the colon. As such, we wanted to confirm that FZD7 was expressed within the stem cell compartment of the colon. Using qPCR, we confirmed the expression of Fzd7 in colonic stem cells and immediate daughter cells from LGR5-GFP mice, where stem and progenitor cells can be isolated by levels of GFP expression (36), and showed that Fzd7 is highly expressed in LGR5+ stem cells and decreased in expression in progeny cells, at a similar level to Lgr5 (SI Appendix, Fig. S3). We also analyzed Lrp1 and Pvrl3 (NECTIN-3 hereafter) expression in colonic stem cells and daughter cells from LGR5-GFP mice (54, 55), both of which are putative TcdB receptors, with expression levels similar to Lgr5 seen for each receptor, across all cell types (SI Appendix, Fig. S3). Several attempts were also made to detect Cspg4 expression in both mouse colonic and small intestinal tissues; however, all were unsuccessful, suggesting that CSPG4 (another reported TcdB receptor) (56) is unlikely to act as a TcdB receptor on mouse intestinal epithelial and stem cells since it is not expressed in these tissues (SI Appendix, Fig. S3).
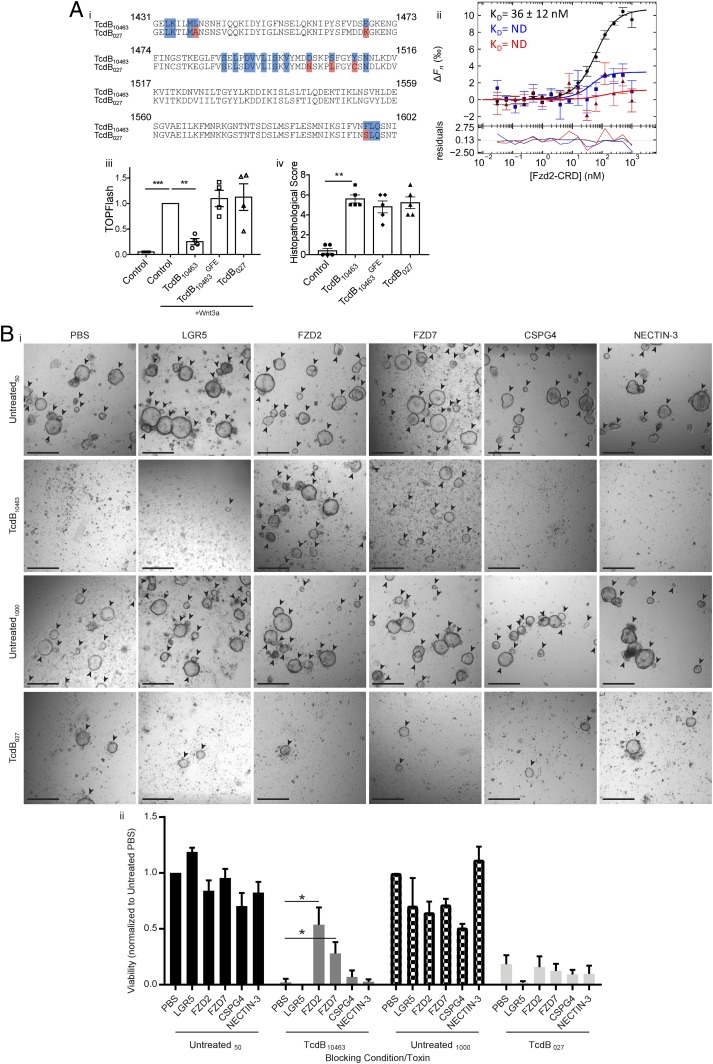
Given the loss of stem cell function and reduced capacity to regenerate the colonic epithelium following RT027 TcdB-mediated intestinal assault, we further characterized the underlying mechanism of stem cell damage during CDI and compared this to VPI10463 TcdB, which was also capable of inducing severe damage deep into the colonic mucosa in our in vivo modeling. As FZD7 is a well-characterized WNT receptor (44), we expected that RT027 TcdB binding to FZD proteins on colonic stem cells would be responsible for the observed stem cell dysfunction in vivo. Strikingly, examination of the key residues for TcdB–FZD2 interaction identified in a crystal structure of the VPI10463 TcdB delivery domain bound to the FZD2 cysteine-rich domain (CRD) (57) revealed that many of the TcdB contact residues are not conserved in the RT027 TcdB sequence from C. difficile M7404 (Fig. 3 A, i), confirming recent observations regarding TcdB sequence variations in binding regions (26). A phylogenic analysis of TcdB from annotated RT027 strains deposited onto the National Center for Biotechnology Information (NCBI) database revealed a complete clonal conservation of TcdB within RT027 strains, suggesting that these differences exist for all RT027 strains (SI Appendix, Fig. S4A).
Fig. 3.
RT027 C. difficile TcdB does not interact with FZD receptors but still induces stem cell death and dysfunction. (A, i) An alignment of the FZD binding domain of TcdB10463 and TcdB027 with TcdB10463 contact residues (blue) and nonconserved TcdB027 residues (red). (ii) MST binding curves of TcdB10463 (black), TcdB027 (blue), and TcdB10463GFE (red) to FZD2-CRD. The residuals with regard to the fit of the binding curve are depicted at the bottom (n = 3). ΔFn (‰) is the change in normalized fluorescence (parts per thousand). ND, not determined. (iii) TOPFlash assay performed in HEK293 STF cells incubated with 1:5 molar ratio of WNT3a and TcdB (n = 4). (iv) Mice were given 50 µg of TcdB intrarectally and were killed after 4 h postintoxication. The descending colon from each mouse was collected and stained with hematoxylin/eosin (H&E) (n = 5). (B) Equal numbers of colonic crypts were isolated from uninfected mice and then exposed to toxin, with or without recombinant receptor prior to organoid seeding. (i) Representative images of organoids (arrowheads) cultured from crypts incubated for 4 h with 5 nM TcdB10463 or 100 nM TcdB027. Blocking was conducted with 50 nM or 1,000 nM, respectively, of either recombinant LGR5, FZD2, FZD7, CSPG4, or NECTIN-3 (ii) with cell viability assessed via a PrestoBlue assay. n ≥ 3. Data are represented as mean + SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (Scale bars: 500 µm.) See also SI Appendix, Figs. S4 and S5.
As these sequence differences may affect the binding affinity of TcdB for FZD, or any of the other reported receptors, we purified TcdB toxins from VPI10463 (TcdB10463) (RT003) (previously used to identify and characterize the CSPG4, FZD1/2/7, and NECTIN-3 receptors) (50, 54–56, 58) and M7404 (TcdB027) and used microscale thermophoresis (MST) to determine the binding affinity of each toxin to the purified ectodomains of CSPG4, FZD2-CRD, and NECTIN-3. While the binding between TcdB10463 and FZD2-CRD could be reproducibly measured with a dissociation constant (KD) of 36 nM, the interactions of FZD2-CRD with TcdB027 or TcdB10463GFE (a TcdB10463 mutant lacking residues required for FZD2 binding) (57) were weak and did not permit the calculation of KD. The affinities of TcdB027 for CSPG4 and NECTIN-3 were similar to those with TcdB10463, suggesting that the amino acid differences at the FZD binding site do not prevent the interaction of TcdB027 with other reported receptors (Fig. 3 A, i and SI Appendix, Fig. S4B). As FZD1/2/7 are identical in the residues that contact TcdB10463 (57), these data suggest that TcdB027 does not interact with FZD1/2/7 but retains the capacity to interact with CSGP4 and NECTIN-3 (SI Appendix, Fig. S4B).
Although TcdB10463 was able to inhibit WNT signaling in a TOPFlash assay and TcdB027 and the FZD-binding mutant, TcdB10463GFE, could not (Fig. 3 A, iii), our in vivo data indicate that TcdB027 is still capable of altering WNT signaling in vivo, likely as a consequence of inducing cell death at the crypt base, where the stem cell compartment resides. To further test this hypothesis, we used a rectal instillation mouse toxicity model and found that there were no significant differences in histological scoring between toxins that could or could not interact with FZD, as measured by injury, edema, and inflammation (Fig. 3 A, iv and SI Appendix, Fig. S4C). These data indicate that TcdB can damage the colonic epithelium via receptors other than FZD and that there are likely multiple receptors that allow for TcdB entry into cells on the colonic epithelium and, more specifically, colonic stem cells, altering their regenerative capacity.
To examine this differential receptor binding in the context of our in vivo infection data, colonic crypts from uninfected mice were isolated and intoxicated with purified TcdB10463 and TcdB027 toxins (at 5 nM and 100 nM, respectively) (SI Appendix, Fig. S5A), to induce crypt death. These crypts were also pretreated with purified recombinant soluble CSPG4, FZD2, FZD7, NECTIN-3, and LGR5 (at 50 nM and 1,000 nM for each respective toxin), for 30 min, and used in organoid seeding to assess if stem cell intoxication could be prevented by blocking toxin binding. Only unintoxicated intestinal stem cells that survive will generate mature organoids. Thus, these assays serve as a functional readout of the potential for TcdB to kill stem cells and the ability of each purified receptor to prevent this. Treatment with each receptor alone did not significantly alter organoid seeding, with similar numbers of mature organoids in each control condition (Fig. 3B). TcdB10463 and TcdB027 treatment alone induced crypt death, with a lack of viable organoids following intoxication, and little to no cell viability detected (Fig. 3B). Pretreatment with FZD2 and FZD7 blocked TcdB10463-mediated stem cell intoxication, indicated by increased mature organoid formation and cell viability at a significantly higher level than phosphate-buffered saline (PBS)-treated TcdB10463-exposed crypts (P = 0.0286) (Fig. 3B). However, soluble CSPG4, NECTIN-3, and LGR5 had no effect in modifying TcdB10463 toxicity and may reflect low cell surface expression levels of these alternate receptors (Fig. 3B). As predicted, pretreatment with FZD2 and FZD7 did not block TcdB027-mediated stem cell intoxication, indicated by the similar levels of mature colonic organoids and cell viability between the FZD2/7-treated and PBS-treated TcdB027-exposed groups (Fig. 3B). Furthermore, soluble CSPG4, NECTIN-3, and LGR5 also had no effect in modifying TcdB027 toxicity, and, as such, TcdB027 may bind to a previously unknown receptor on colonic stem cells. Similar blocking experiments were performed on human colonic organoids, indicating that FZD7 can prevent TcdB10463 but not TcdB027 intoxication of colonic stem cells, evidenced by a significant increase in organoid viability and organoid numbers in FZD7-treated TcdB10463-intoxicated stem cells (P = 0.0021) (SI Appendix, Fig. S5B). However, similar assays performed on Vero cells indicated that, despite being protective in organoid culture, FZD7 could not prevent TcdB10463-mediated Vero cell rounding, with levels of cell rounding consistent across all test conditions (SI Appendix, Fig. S5C). We also measured the toxicity of TcdB10463, TcdB10463GFE, and TcdB027-in HeLa and Caco-2 cells, two cell lines that differ in their receptor profiles. HeLa cells are known to express high levels of CSPG4, but low levels of FZD1/2/7, and can differ in whether or not NECTIN-3 is present (58). Caco-2 cells express NECTIN-3 and low levels of FZD1/2/7 but have little to no CSPG4 (58). Cells were treated with 0.1, 1, 10, and 100 nM of each toxin, assessing viability after 2.5 h (HeLa) or 24 h (Caco-2). In HeLa cells, all of the toxins are equally active in the presence of the shared HeLa-dominant receptor CSPG4, implying that interaction with FZD1/2/7 is not required for this toxicity (SI Appendix, Fig. S5 D, i). In contrast, TcdB10463 had significantly higher cytotoxicity than TcdB10463GFE and TcdB027 in Caco-2 cells that correlated with the capacity to bind FZD1/2/7, evidenced by cells treated with TcdB10463GFE and TcdB027 exhibiting an increased viability over the concentration range of 0.1 nM to 10 nM TcdB (P < 0.05) (SI Appendix, Fig. S5 D, ii). Taken together, our data indicate TcdB027 is able to induce stem cell death during infection, but this does not result from binding to FZD. Additionally, these observations appear to be relevant in both human and mouse colonic stem cells.
Stem Cell Damage during CDI Results in a Delay in Intestinal Repair
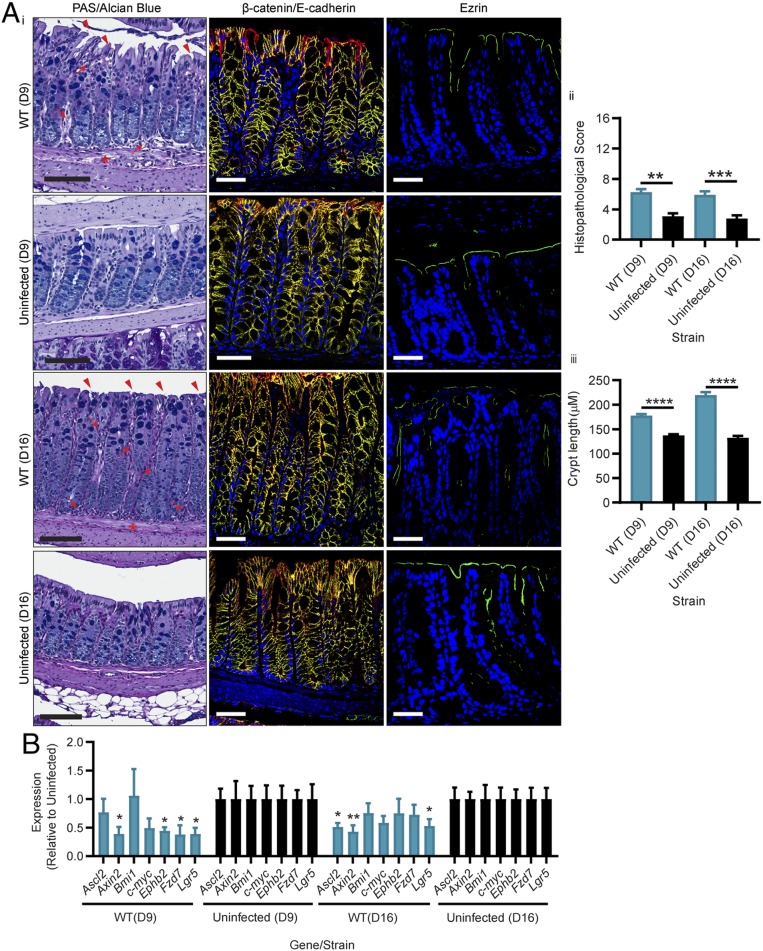
CDI-mediated damage and loss of viable stem cells is likely to have downstream effects for recovery; however, this aspect of the disease cycle has not been explored. To gain this understanding, we examined mice that had reached the peak of WT infection (losing between 10% and 15% of their body weight within 48 h) (SI Appendix) and which had then been allowed to recover for up to 2 wk. Strikingly, even after this long recovery period, colonic tissues from these mice did not resemble uninfected tissues, with regions of crypt erosion and hyperplasia, inflammation, and edema seen at both days 9 and 16 postinfection (P = 0.0012 and 0.0022, respectively) (Fig. 4 A, i and ii), as well as significant differences in crypt length and overall tissue pathology detected when compared to tissues from uninfected mice at both time points (SI Appendix and Fig. 4 A, i and iii; P < 0.001). This abnormal colonic pathology in the WT-infected recovery mice was coupled with disrupted ezrin staining, when compared to uninfected tissues at both days 9 and 16 postinfection, indicating that, although the mice had begun to clear the CDI and regenerate cell-to-cell contacts via β-catenin/E-cadherin, the capacity for the stem cells to repair the damage mediated by TcdB was significantly altered. This hypothesis was supported by ddPCR on the panel of stem cell genes used previously, which identified that several stem cell genes were expressed at significantly lower levels in the WT-infected recovery mice, when compared to uninfected tissues (Fig. 4B). Specifically, Axin2, Ephb2, Fzd7, and Lgr5 expression was significantly lower than uninfected tissues at day 9 (Fig. 4B) (P < 0.02), and Ascl2, Axin2, and Lgr5 expression was significantly lower than uninfected tissues at day 16, at levels less than half that detected for uninfected mice (Fig. 4B) (P < 0.01). Thus, our data show that CDI is capable of inducing significant stem cell damage, which consequently prolongs recovery and repair of the intestinal epithelium after infection.
Fig. 4.
C. difficile TcdB-mediated stem cell death and dysfunction hinders host repair for up to 2 wk. Mice were infected with M7404 (WT) C. difficile or left uninfected and allowed to recover after reaching a peak of infection (between 10% and 15% weight loss). Two weeks post the peak of infection, (A) colonic tissues were collected and assessed with (i) PAS/Alcian blue for overall pathology, β-catenin (green), and E-cadherin (red) (merge, yellow) for adherens junctions, or ezrin (green) for cellular polarity. (ii) Overall histopathology was scored and plotted, and (iii) crypt length was measured for 30 crypts per mouse across the entire length of the colon. Arrows, inflammation; arrowheads, crypt damage/goblet cell loss; asterisk, edema. (Scale bars: black, 100 µm; white, 50 µm.) (B) Quantitative ddPCR analysis of colonic tissue 9 d (D9) and 16 d (D16) postinfection (7 and 14 d post peak of infection, respectively). Gene expression, as fold change relative to uninfected mice, was plotted. n ≥ 5. Data are represented as mean + SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Discussion
Important clinical implications result from the capacity of infections to alter epithelial homeostasis, stem cell function, and regenerative capacity. Damage to the epithelium during infection requires repair to ensure that intestinal integrity is maintained and to prevent severe disease outcomes, including disease recurrence. A collapse in intestinal integrity may disrupt the repair process and may expose deeper tissues to damage, which is of particular importance for the stem cell population which resides deep in the base of the crypts within the intestinal epithelium, to protect it from damage. Enterococcus faecalis, enterotoxigenic Bacteroides fragilis, and H. pylori, for example, are able to disrupt homeostasis by cleaving E-cadherin, thereby promoting cellular adherence and deeper tissue penetration that induces inflammation and severe pathologies during infection (29). Here, we have shown that TcdB-mediated disruption of adherens junctions during CDI is linked with severe pathology, exposure of the protected stem cell compartment, and a poor disease outcome, likely through actin cytoskeleton collapse of the intoxicated cell, which is independent of TcdA activity. This toxin-mediated effect may also disrupt ezrin localization to the apical cell surface, which occurs prior to adherens-junction collapse, leading to a progressive exposure of deeper regions of the colonic epithelium as infection advances. One consequence of this cellular disorganization may be an alteration to the distribution of cell receptors, which is particularly intriguing in the context of previous studies in which TcdA and TcdB were administered to fetal-like human small-intestinal organoids, which showed that TcdB had higher cytotoxic capacity when applied to cells via the basolateral surface (59). It is possible that the TcdB-mediated alteration of cell polarity during early infection may redistribute basolateral receptors to the apical surface, thereby changing intestinal functional capacity and possibly amplifying toxin uptake and disease severity.
The results of the infection study presented here highlight the important role that host factors play in the outcomes that occur during CDI. Previous studies using purified TcdA and cultured cells showed that TcdA attenuates WNT signaling (60); however, a similar effect was not seen in mouse colonic tissue after infection with a TcdA+B− strain. Rather, the work presented here suggests that, during infection, TcdB induces significant colonic damage, which attenuates WNT signaling and the expression of key stem cell markers, and damages the stem cell population (50). Importantly, CDI diminishes the LGR5+ stem cell compartment, which drives epithelial regeneration (61), and this dysregulation of key signaling pathways and acute disruption of stem cell function affect the renewal and repair capacity of the colonic epithelium. In turn, this damage delays intestinal repair for up to 2 wk, which is consistent with the severe disease outcomes seen here with TcdB-producing strains.
The continual emergence of TcdA−B+ C. difficile variants (15, 16, 62, 63) provides an important clinical context for the work described here. Infection with the TcdA−B+ strain was more detrimental to stem cell functionality than TcdA+B− and TcdA−B− strains, demonstrated by the reduced capacity of tissue isolated from TcdA−B+-infected mice to form colonic organoids. There is accumulating evidence that naturally occurring clinical TcdA−B+ strains cause more severe disease, which highlights the compelling need to better understand toxin-mediated effects on the host (15, 16). TcdB is clearly capable of causing severe and potentially irreversible damage to the intestinal epithelium which is compounded by the ability of the toxin to target colonic stem cells and disrupt regeneration.
The work presented here, showing a reduced regenerative capacity of the gastrointestinal tract following infection and a decreased ability of the host to recover appropriately, brings a broader understanding to gastrointestinal infections and their effects on the host. Damage to intestinal stem cells resulting from infection may increase host susceptibility to reinfection and disease relapse. This outcome is particularly relevant for C. difficile, which thrives in a disrupted colonic niche. Relapse rates for CDI range from 18 to 32% for patients treated for primary CDI with vancomycin and metronidazole (64). Risk for subsequent recurrences following an initial relapse increases to between ∼45% and 65%, which is partly linked to continued disruption of the microbiota (64); however, the risk factors that contribute to infection relapse are likely to be complex. If the capacity of the colonic epithelium to repair and form a functional barrier is impaired, relapse risk may be heightened. For C. difficile, the outcome of TcdB-mediated disruption of adherens junctions, cellular polarity, and stem cell function is a reduction in epithelial integrity and capacity to repair damage, which will exacerbate epithelial impairment and intestinal inflammation. This work therefore expands our understanding of C. difficile pathogenesis and provides valuable insights into gastrointestinal infection and repair with respect to the stem cell compartment, which is a previously unrecognized target of infectious damage.
Materials and Methods
Bacterial Strains, Growth Conditions, Cell Lines, and Reagents.
Detailed descriptions of the bacterial culture, cloning, protein purification, and cell culture methods are included in SI Appendix.
Microscale Thermophoresis.
Microscale thermophoresis (MST) experiments were performed on a NanoTemper Monolith NT.115 (NanoTemper Technologies GmbH, Munich, Germany). VPI TcdB10463, TcdB027, VPI TcdB10463GFE, FZD2-CRD, CSPG4, and NECTIN-3 were equilibrated prior to labeling with size exclusion chromatography using either a Superdex 200 10/300 or Superdex 75 10/300 column (GE) with 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 8.0, 100 mM NaCl. Toxins, CSPG4, and NECTIN-3 were labeled with the Monolith NT His-tag Labeling Kit RED-Tris-NTA, following the manufacturer’s instructions. TcdB or receptor concentration was held constant at 50 nM. Serial dilutions of FZD2-CRD or TcdB were prepared using a 1:1 dilution from 3 to 10 µM to 90 to 300 pM in 20 mM Hepes, pH 8.0, 100 mM NaCl, 0.05% Tween 20, and protease inhibitor mixture for His-tagged proteins (Sigma). Samples were loaded into Monolith NT.115 capillaries (NanoTemper), and measurements were carried out at 21 °C with 40% MST power and 60% excitation power. MO.Control v1.6 was used for data collection, and MO.Affinity Analysis v2.3 and PALMIST were used for data analysis (65). The KD constants were calculated in PALMIST utilizing the saturation binding curve at equilibrium.
Animal Model of C. difficile Infection and Intoxication.
Detailed descriptions of the C. difficile infection and intoxication models are included in SI Appendix.
Immunohistochemistry and Immunofluorescence Analysis.
Paraffin-embedded colonic tissues were processed using standard procedures. Slides were dewaxed, and antigen retrieval was performed using 10 mM citrate (Sigma-Aldrich) buffer, with 0.05% Tween 20 (Amresco), pH 6.0. Slides were blocked for 60 min with CAS-Block (Thermo-Fisher Scientific) at room temperature. For immunofluorescence staining, slides were incubated with either mouse anti-ezrin (1:200 dilution in 1% bovine serum albumin [BSA] [Sigma Aldrich] in PBS; Thermo-Fisher Scientific) or mouse anti-β-catenin (1:200 dilution in 1% BSA; BD) and rabbit anti–E-cadherin (1:200 dilution in 1% BSA; Cell Signaling) overnight at 4 °C. Slides were rinsed three times in PBS before incubation with secondary antibodies for 60 min at room temperature. For ezrin and β-catenin, goat anti-mouse IgG, Alexa Fluor 488 (1:1,000 dilution in 1% BSA; Thermo-Fisher Scientific) was used; for E-cadherin, goat anti-rabbit IgG, Alexa Fluor 568 (1:1,000 dilution in 1% BSA; Thermo-Fisher Scientific) was used. Slides were washed three times in PBS before staining the nuclei with DAPI (300 nM; Thermo Fisher). Slides were mounted with ProLong Gold (Thermo Fisher), sealed, and imaged using a Leica SP8 Confocal Invert microscope on a 20×/1.0 oil objective with LasX Software (Leica).
For immunohistochemical staining, see SI Appendix.
Colonic Stem Cell Sorting and Quantitative PCR for Stem Cell and TcdB Receptors.
Detailed descriptions of the stem cell isolation, sorting, RNA extraction, and quantitative PCR for known TcdB receptors are included in SI Appendix.
RNA Extraction, Complementary DNA Preparation, and Digital Droplet PCR Analysis.
Colonic tissues collected from infected mice were placed in RNAlater (Ambion) prior to RNA extraction. The tissues were then homogenized and total RNA was extracted using the RNeasy mini kit (Qiagen). One microgram of RNA was used for complementary DNA (cDNA) synthesis using a QuantiTect Reverse Transcription kit (Qiagen). The cDNA was quantified using the QIAExpert prior to dilution for use in digital droplet PCR (ddPCR). The ddPCR was conducted as previously described (66) and is described in detail in SI Appendix.
Growth of Murine Colonic Organoids from C. difficile-Infected Mice.
Mouse colonic crypt isolation and organoid establishment were based on protocols previously described (67, 68). See SI Appendix for a detailed description.
TOPFlash Assay.
Five hundred thousand human embryonic kidney 293 (HEK293) Super TOPFlash (STF) cells were seeded in a 12-well dish for 18 h. The media was then replaced with 1 mL of prewarmed media with or without combinations of 1:5 molar ratio of human WNT3a (100 ng/mL, 2.67 nM; StemRD) to toxin (13.35 nM) for 20 h. Following the incubation, the media was removed, and cells were lysed in 110 µL of Passive Lysis Buffer (Promega) for 15 min while shaking. The solubilized supernatant was collected and immediately used for the determination of the TOPFlash luciferase activities with the Steady-Glo Luciferase assay (Promega) and CellTiter Glo (Promega).
Treatment of Colonic Crypts, with Purified Toxins and Receptor-Blocking Proteins, and Assessment of Organoid Formation.
Colonic crypts were isolated from C57BL/6J mouse tissues as described previously. The crypt pellets were resuspended in PBS containing 1% fetal bovine serum (FBS) and supplemented with 5 nM purified VPI10463TcdB (Abcam) or 100 nM purified RT-027 TcdB. Blocking of toxin variants was conducted with, respectively, 50 nM or 1,000 nM of either recombinant human FZD2 (see above), recombinant human FZD7 Fc chimera (R&D Systems), recombinant human CSPG4 (see above), recombinant human NECTIN-3 (see above), or recombinant human LGR5 Fc chimera (R&D Systems). After 4 h at 4 °C, the crypt pellets were washed twice with PBS and resuspended in Matrigel (BD), and 12 µL were seeded into each well of a 48-well plate (Nunc) and incubated for 10 min at 37 °C until solidified. The crypt culture media described above was added to each well. After 3 d, the medium was replaced with fresh culture medium without Y-27632 Rock inhibitor. After 4 d in culture, cell viability was measured using the PrestoBlue reagent (Life Technologies) and imaged using an EVOS FL Auto Cell Imaging System (Invitrogen). Viability was plotted using GraphPad Prism 7, with statistical significance assessed using a Mann–Whitney U test.
For human organoid establishment and intoxication, see SI Appendix for a detailed description.
Sequence Analysis and Phylogeny.
Fully annotated genomes of C. difficile strains deposited on the National Center for Biotechnology Information (NCBI) database were used as the source of sequences in this study. Detailed description of sequence analysis and phylogeny can be found in SI Appendix.
Data Availability Statement.
All data discussed in the paper will be made available to readers, through contact with the corresponding authors.
Supplementary Material
Acknowledgments
We thank the Monash Histology Platform, Department of Anatomy and Developmental Biology (Monash University, Clayton, Australia), Monash Biomedicine Discovery Institute Organoid Program, and Monash Micro Imaging (Monash University, Clayton, Australia) for their technical assistance with pathology and confocal imaging. We thank Jamie Rossjohn and Ben Kile for manuscript discussions, Kay Washington for pathology scoring, and Bliss Cunningham for assistance in animal handling. This work was supported by National Health and Medical Research Council Grant GNT1107812 (to D.L. and H.E.A.). Work in the D.B.L. laboratory was supported by NIH National Institute of Allergy and Infectious Diseases Grant AI95755, US Department of Veterans Affairs Grant BX002943, Molecular Biophysics Training Grant T32GM008320 (to K.O.C.), and Gastroenterology Training Grant T32DK007673 (to J.L.J. and M.J.S.). Collection of human tissue was supported by Monash Strategic Grant SGS15-0156 (to H.E.A., D.L., and P.J.M.) and, in part, by “Let’s Beat Bowel Cancer” (https://www.letsbeatbowelcancer.com.au/), a benevolent fundraising and public awareness foundation. This funder had no part in the design, conduct, outcomes, decision to publish, or the drafting of this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915255117/-/DCSupplemental.
References
- 1.van der Wath R. C., Gardiner B. S., Burgess A. W., Smith D. W., Cell organisation in the colonic crypt: A theoretical comparison of the pedigree and niche concepts. PLoS One 8, e73204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozar S., et al. , Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell 13, 626–633 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Sender R., Fuchs S., Milo R., Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkaid Y., Naik S., Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 14, 646–653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peery A. F., et al. , Burden of gastrointestinal, liver, and pancreatic diseases in the united states. Gastroenterol 149, 1731–1741.e3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jank T., Giesemann T., Aktories K., Rho-glucosylating Clostridium difficile toxins A and B: New insights into structure and function. Glycobiology 17, 15R–22R (2007). [DOI] [PubMed] [Google Scholar]
- 7.Chumbler N. M., et al. , Crystal structure of Clostridium difficile toxin A. Nat. Microbiol. 1, 15002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter G. P., et al. , Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. MBio 6, e00551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyras D., et al. , Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehne S. A., et al. , Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J. Infect. Dis. 209, 83–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuehne S. A., et al. , The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Geric B., et al. , Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 193, 1143–1150 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Schwan C., et al. , Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 5, e1000626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowardin C. A., et al. , The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat. Microbiol. 1, 16108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., Kim Y., Pai H., Clinical characteristics and treatment outcomes of Clostridium difficile infections by PCR ribotype 017 and 018 strains. PLoS One 11, e0168849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin B. M., Kuak E. Y., Yoo S. J., Shin W. C., Yoo H. M., Emerging toxin A-B+ variant strain of Clostridium difficile responsible for pseudomembranous colitis at a tertiary care hospital in Korea. Diagn. Microbiol. Infect. Dis. 60, 333–337 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Autenrieth D. M., Baumgart D. C., Toxic megacolon. Inflamm. Bowel Dis. 18, 584–591 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Lim S. K., et al. , Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin. Infect. Dis. 58, 1723–1730 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Stabler R. A., et al. , Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One 7, e31559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung Y. P., et al. , Clostridium difficile ribotype 126 in southern Taiwan: A cluster of three symptomatic cases. Anaerobe 30, 188–192 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Walker A. S. et al.; Infections in Oxfordshire Research Database , Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin. Infect. Dis. 56, 1589–1600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis B. B., et al. , Pathogenicity locus, core genome, and accessory gene contributions to Clostridium difficile virulence. MBio 8, e00885-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wüst J., Sullivan N. M., Hardegger U., Wilkins T. D., Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol. 16, 1096–1101 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wüst J., Hardegger U., Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother. 23, 784–786 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrigan M., et al. , Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192, 4904–4911 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Ureña D., et al. , Toxin B variants from Clostridium difficile strains VPI 10463 and NAP1/027 share similar substrate profile and cellular intoxication kinetics but use different host cell entry factors. Toxins (Basel) 11, E348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanis J. M., Barua S., Ballard J. D., Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 6, e1001061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Squire M. M., et al. , Novel molecular type of Clostridium difficile in neonatal pigs, Western Australia. Emerg. Infect. Dis. 19, 790–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa A. M., Leite M., Seruca R., Figueiredo C., Adherens junctions as targets of microorganisms: A focus on Helicobacter pylori. FEBS Lett. 587, 259–265 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Guttman J. A., Finlay B. B., Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 1788, 832–841 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Hecht G., Pothoulakis C., LaMont J. T., Madara J. L., Clostridium difficile toxin A perturbs cytoskeletal structure and tight junction permeability of cultured human intestinal epithelial monolayers. J. Clin. Invest. 82, 1516–1524 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nusrat A., et al. , Clostridium difficile toxins disrupt epithelial barrier function by altering membrane microdomain localization of tight junction proteins. Infect. Immun. 69, 1329–1336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartsock A., Nelson W. J., Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta 1778, 660–669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fehon R. G., McClatchey A. I., Bretscher A., Organizing the cell cortex: The role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276–287 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aspenström P., Fransson A., Saras J., Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 377, 327–337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Barker N., van de Wetering M., Clevers H., The intestinal stem cell. Genes Dev. 22, 1856–1864 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degirmenci B., Valenta T., Dimitrieva S., Hausmann G., Basler K., GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Lu R., Wu S., Sun J., Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett. 584, 911–916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed I., et al. , Critical roles of Notch and Wnt/β-catenin pathways in the regulation of hyperplasia and/or colitis in response to bacterial infection. Infect. Immun. 80, 3107–3121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata W., et al. , Helicobacter-induced gastric inflammation alters the properties of gastric tissue stem/progenitor cells. BMC Gastroenterol. 17, 145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z., et al. , The antibiotic bacitracin protects human intestinal epithelial cells and stem cell-derived intestinal organoids from Clostridium difficile toxin TcdB. Stem Cells Int. 2019, 4149762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., et al. , Cloning and variation of ground state intestinal stem cells. Nature 522, 173–178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flanagan D. J., et al. , Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5(+) stem cells. Stem Cell Reports 4, 759–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmon K. S., Gong X., Lin Q., Thomas A., Liu Q., R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Lau W., et al. , Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Peng W. C., et al. , Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 3, 1885–1892 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Sato T., et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Sato T., et al. , Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Tao L., et al. , Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holcombe R. F., et al. , Expression of Wnt ligands and Frizzled receptors in colonic mucosa and in colon carcinoma. Mol. Pathol. 55, 220–226 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gujral T. S., et al. , A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 159, 844–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gregorieff A., et al. , Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638 (2005). [DOI] [PubMed] [Google Scholar]
- 54.LaFrance M. E., et al. , Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 112, 7073–7078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schöttelndreier D., et al. , Expression and (Lacking) internalization of the cell surface receptors of Clostridioides difficile toxin B. Front. Microbiol. 9, 1483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan P., et al. , Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res. 25, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen P., et al. , Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360, 664–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta P., et al. , Functional defects in Clostridium difficile TcdB toxin uptake identify CSPG4 receptor-binding determinants. J. Biol. Chem. 292, 17290–17301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leslie J. L., et al. , Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bezerra Lima B., et al. , Clostridium difficile toxin A attenuates Wnt/β-catenin signaling in intestinal epithelial cells. Infect. Immun. 82, 2680–2687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barker N., Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Cairns M. D., et al. , Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London, England. J. Clin. Microbiol. 53, 3141–3147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hung Y. P., et al. , Predominance of Clostridium difficile ribotypes 017 and 078 among toxigenic clinical isolates in southern Taiwan. PLoS One 11, e0166159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelly C. P., Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin. Microbiol. Infect. 18 (suppl. 6), 21–27 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Scheuermann T. H., Padrick S. B., Gardner K. H., Brautigam C. A., On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 496, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvay K., et al. , Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelium. EMBO J. 34, 1319–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jardé T., et al. , In vivo and in vitro models for the therapeutic targeting of Wnt signaling using a Tet-OΔN89β-catenin system. Oncogene 32, 883–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nefzger C. M., et al. , A versatile strategy for isolating a highly enriched population of intestinal stem cells. Stem Cell Reports 6, 321–329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper will be made available to readers, through contact with the corresponding authors.