Significance
Persistent aquatic environmental reservoirs for Vibrio cholerae O1 are present in Asia; however, their existence in other parts of the world remains controversial. The single-source introduction of toxigenic V. cholerae O1 in Haiti provides a unique opportunity to assess the potential role played by aquatic reservoirs in subsequent seasonal outbreaks. Whole-genome sequence Bayesian phylogeography showed robust evidence of V. cholerae O1 evolution in riverine sites, through the establishment of reservoirs, during lull periods of the Haitian epidemic. Novel lineages emerged in the environment from sequential population bottlenecks, characterized by mutations in genes potentially involved in adaptive response. The data highlight the need for long-term investments in cholera control at both human and environmental levels.
Keywords: cholera, reservoir, evolution, phylodynamics
Abstract
The spread of cholera in the midst of an epidemic is largely driven by direct transmission from person to person, although it is well-recognized that Vibrio cholerae is also capable of growth and long-term survival in aquatic ecosystems. While prior studies have shown that aquatic reservoirs are important in the persistence of the disease on the Indian subcontinent, an epidemiological view postulating that locally evolving environmental V. cholerae contributes to outbreaks outside Asia remains debated. The single-source introduction of toxigenic V. cholerae O1 in Haiti, one of the largest outbreaks occurring this century, with 812,586 suspected cases and 9,606 deaths reported through July 2018, provided a unique opportunity to evaluate the role of aquatic reservoirs and assess bacterial transmission dynamics across environmental boundaries. To this end, we investigated the phylogeography of both clinical and aquatic toxigenic V. cholerae O1 isolates and show robust evidence of the establishment of aquatic reservoirs as well as ongoing evolution of V. cholerae isolates from aquatic sites. Novel environmental lineages emerged from sequential population bottlenecks, carrying mutations potentially involved in adaptation to the aquatic ecosystem. Based on such empirical data, we developed a mixed-transmission dynamic model of V. cholerae, where aquatic reservoirs actively contribute to genetic diversification and epidemic emergence, which underscores the complexity of transmission pathways in epidemics and endemic settings and the need for long-term investments in cholera control at both human and environmental levels.
Cholera is a severe, acute dehydrating diarrheal disease caused by toxigenic Vibrio cholerae that harbors the CTX prophage promoting cholera toxin (CT) production. The disease has repeatedly manifested in global pandemics emanating from Asia, with seven pandemics formally designated since 1817 (1). It has demonstrated a remarkable ability to persist and spread in the modern world. The current seventh pandemic is associated with the V. cholerae El Tor biotype (1). El Tor strains are thought to have an increased ability to persist in aquatic reservoirs over the classical biotype (1). V. cholerae can survive in aquatic reservoirs in a variety of forms, and can also live in association with zooplankton, copepods, or other natural aquatic hosts. Toxigenic V. cholerae in epidemic serogroups such as O1 and O139 can also survive in these environmental reservoirs (2) and represent a source for recurrent annual epidemics on the Indian subcontinent (3, 4), with environmental triggers resulting in seasonal blooms of the microorganism followed by “spillover” into human populations and subsequent epidemic spread. On the other hand, the existence and potential role played by aquatic reservoirs of toxigenic V. cholerae and their existence outside of Asia remain controversial (5), though such information has profound repercussions for the development of effective public health methods for global cholera prevention, control, and elimination. If toxigenic V. cholerae O1 populations are fueling seasonal cholera outbreaks in areas with endemic cholera outside of the Indian subcontinent, eradication may be difficult, if not impossible.
Unlike the other American (6) and African (5) epidemics caused by multiple introductions of toxigenic V. cholerae O1 El Tor strains from Asia, the Haitian epidemic was the result of a single introduction of toxigenic V. cholerae O1 into Haiti’s Artibonite River (7, 8), with initial transmission of the infection to communes found along its lower course, followed by spread throughout the country (8). Moreover, due to its island location, the Haitian epidemic is at reduced risk of recurrent outside introductions. The cholera epidemic in Haiti provides a unique opportunity to assess the role of aquatic reservoirs in the evolution and persistence of cholera outside Asia. As V. cholerae is capable of growth and long-term survival in aquatic ecosystems (2), elucidating the role of aquatic environmental reservoirs has become critical to the development of elimination strategies.
Results
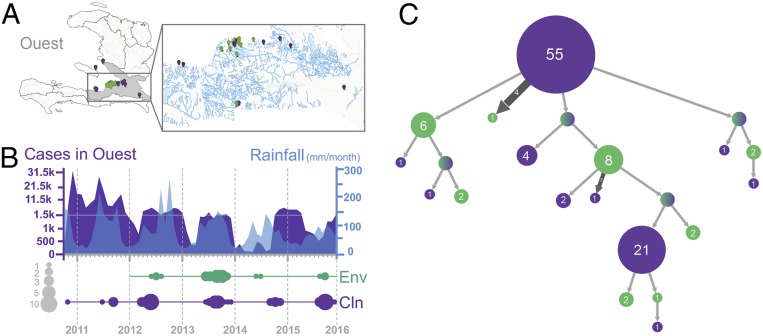
Four departments—Artibonite, Centre, Nord-Ouest, and Ouest—account for 90% of reported cholera cases in Haiti (9). We focused our efforts on the Ouest Department (Fig. 1A), a coastal region rich in aquatic ecosystems where we have isolated toxigenic V. cholerae O1 from fixed riverine and estuarine sites (Fig. 1A). The prevalence of toxigenic aquatic V. cholerae O1 correlated with increased temperature and rainfall but not with fecal coliform counts, suggesting that the presence was not due to fecal contamination alone (10). Monthly case counts in Ouest, oscillating yearly but decreasing during the dry season and increasing during the rainy season (April to October), correlated with increased temperature and rainfall (Fig. 1B). This is in agreement with patterns of endemic V. cholerae O1 in Bangladesh and Peru, where seasonal cholera outbreaks correlate with increased V. cholerae O1 in the environment and increases in water temperature or rainfall (11, 12).
Fig. 1.
Cholera epidemic in the Ouest Department, Haiti, 2010 to 2015. (A) Geographical distribution of environmental (n = 27) and clinical (n = 89) isolates sampled from the Haitian toxigenic V. cholerae O1 lineage in the Ouest Department between 2010 and 2015. (B, Upper) Monthly case counts of cholera infections in the Ouest Department and mean monthly precipitation (mm/mo) between 2010 and 2015. (B, Lower) Temporal distribution of V. cholerae genomes sampled in Haiti and of the number of cases reported monthly to the World Health Organization from October 2010, the beginning of the epidemic, until December 2015 (ticks correspond to the month of January for each year indicated on the x axis). The size of the circle is proportional to the number of environmental (green) and clinical (violet) genomes sampled in our study for each year. (C) Ancestral-state reconstruction using maximum likelihood inferred with PastML from a rooted ML phylogenetic tree inferred with IQ-TREE (SI Appendix, Fig. S1E). ML tree clades are merged vertically (clade circles contain the number of tips of the initial tree contained in them) and horizontally (branch size corresponds to the number of times its subtree is found in the initial tree) to cluster independent events of the same kind.
To test the hypothesis that V. cholerae established aquatic reservoirs in the Haitian river system, contributing to recurrent seasonal epidemics, we carried out whole-genome sequencing of 27 aquatic environmental toxigenic V. cholerae O1 strains, isolated in Gressier between 2012 and 2015, and 89 clinical strains collected in Ouest and Artibonite between 2010 and 2015 (Fig. 1B and Datasets S1 and S2). We investigated their phylogenetic relationship by maximum-likelihood (ML) (SI Appendix, Fig. S1C) and ML ancestral-state reconstruction (Fig. 1C) based on high-quality single-nucleotide polymorphisms (hqSNPs) (Dataset S3), after confirming the presence of a robust phylogenetic signal (SI Appendix, Fig. S1 A and B). Environmental strains are intermixed with clinical strains in each major phylogenetic clade, including isolates from seasonal epidemic waves. Connection of epidemic waves through environmental lineages suggests that such lineages played a substantial role in promoting the epidemic, rather than simply providing an occasional contribution.
Sufficient temporal signal was present (SI Appendix, Fig. S1 D and E) to calibrate a reliable molecular clock (SI Appendix, Table S1) and infer time-scaled phylogenies. We reconstructed the spatiotemporal spread of the epidemic and quantified the potential contribution of environmental V. cholerae to this spread using Bayesian phylogeography (13). In agreement with previous findings (14), the maximum clade credibility (MCC) time-scaled tree obtained by discrete trait analysis (DTA) (Fig. 2A) and by Bayesian structured coalescent approximation (BASTA) (SI Appendix, Fig. S2A) showed a staircase phylogeny typical of evolution driven by repeated selective sweeps through sequential population bottlenecks with a major surviving lineage leading, over time, to each subsequent wave (15). The surviving lineage is represented by 4 preenvironmental contribution unambiguous and unique SNPs (uuSNPs): 98 C→T (gene Vch1786_II0360), 32 G→A (gene Vch1786_I1120), 51 T→G (gene exeA), and 53 T→C (upstream of gene rseA); there are 10 uuSNPs linked to environmental contribution: 1 C→T (gene Vch1786_I0012), 3 C→T (gene Vch1786_I0051), 30 G→A (gene Vch1786_I0998), 44 A→G (gene Vch1786_I1539), 45 T→G (gene Vch1786_I1601), 52 G→A (gene rseA), 61 A→C (gene epsG), 69 C→A (gene Vch1786_I2482), 105 G→A (upstream of gene Vch1786_II0538), and 107 C→A (gene Vch1786_II0794); and there is 1 postenvironmental contribution uuSNP: 110 C→T (gene Vch1786_II0977) (SI Appendix, Fig. S3). Eleven waning lineages defined by uuSNPs, shared in both inferences, are summarized in SI Appendix, Table S2 (SI Appendix, Fig. S3).
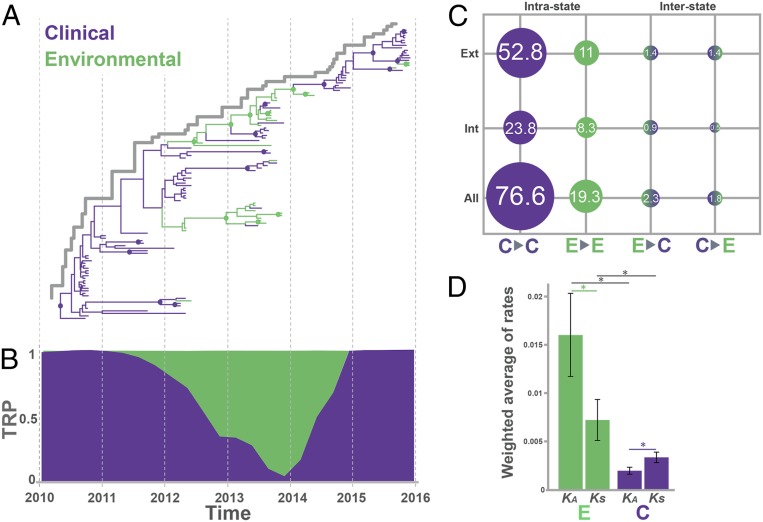
Fig. 2.
Contribution of toxigenic V. cholerae O1 environmental isolates to the evolution of the cholera epidemic in Haiti between 2012 and 2014. (A) MCC phylogeny for 116 environmental and clinical toxigenic V. cholerae O1 isolates collected between October 2010 and December 2015 inferred from genome-wide hqSNP data using the Bayesian phylogeography framework implemented in BEAST package version 1.8.4. Branch lengths are scaled in time by enforcing a relaxed molecular clock. Environmental and clinical states are indicated in green and violet, respectively. Circles at internal nodes indicate high posterior probability (PP) support (PP > 0.9). (B) Trunk reward proportion (TRP) at each ancestral location state estimated over time inferred using the continuous-time Markov chain model. Green- and purple-shaded areas represent the trunk proportions over time for environmental and clinical transitions, respectively. (C) Number of jumps (%) of all possible intrastate (clinical-to-clinical, environmental-to-environmental) and interstate transitions (clinical-to-environmental, environmental-to-clinical) normalized by total numbers of transitions obtained from the MCC phylogeny. Internal refers to all internal branches including the backbone path, and external to terminal branches of the tree. (D) Weighted averages of synonymous substitution rate estimates for environmental toxigenic V. cholerae O1 isolates were based on 200 randomly sampled trees from the posterior distribution of molecular clock-calibrated Bayesian phylogenies. Internal refers to estimates based on all internal branches of the tree, while external refers to estimates based on terminal branches. An asterisk indicates a significant (P < 0.001) difference between rate estimates. Error bars indicate standard deviation.
The classic source–sink cholera population dynamic, where each epidemic wave is essentially propagated by ongoing transmissions within the human population (source) followed by spillover into the environment (sink), predicts that the backbone path (trunk) of the tree, that is, the surviving lineate successfully propagated through time (Fig. 2A), would be occupied only by lineages of clinical origin. Therefore, we used DTA and BASTA to assess the origin (clinical or environmental) of the internal branches (ancestral cholera lineages) in the tree through ancestral-state reconstruction. Both methods consistently inferred aquatic environmental V. cholerae lineages along the trunk of the tree giving rise, in turn, to clinical isolates circulating during different epidemic waves (Fig. 2A and SI Appendix, Fig. S2A). The presence of environmental ancestors directly connected along the trunk of the tree was highly supported (posterior probability > 0.9) between May 2012 (95% highest posterior density [HPD] November 2011 to August 2012) and January 2014 (95% HPD November 2013 to April 2014) (Fig. 2A and SI Appendix, Fig. S2 A and B). This finding provides robust evidence of sustained replication and evolution of V. cholerae in the aquatic environment followed by reintroduction into the human population, that is, environmental isolates playing an active role in epidemic spread rather than contributing solely to dead-end transmission chains. Moreover, we observed the presence of a monophyletic clade in the phylogeny, which arose in 2012, dominated by environmental isolates with few intermixed clinical ones (Fig. 2A). Although we cannot exclude that the clade is an artifact of sampling bias, its presence in the tree suggests a concurrent scenario where independent evolution of V. cholerae in aquatic reservoirs can occasionally spill over to humans without necessarily contributing to an epidemic wave. Previous genetic studies suggest that the first cholera outbreak in the Port-au-Prince area, after the lull period in 2014, was propagated by autochthonous local transmission in the south rather than introduction from the north (16). Such observations, together with our findings, reinforce a scenario of undetected environmental reservoirs serving as a source for a later wave of clinical cases.
To investigate further, we quantified the contribution of V. cholerae environmental ancestors to the backbone path of the tree by calculating the proportion of time spent by isolates in environmental (river/estuarine) or clinical (human host) periods (Fig. 2B). The estimates support the concept that aquatic reservoirs were a main source of V. cholerae from June 2012 to May 2014 (60 to 80% of the trunk, P < 0.0001) (Fig. 2B and SI Appendix, Fig. S2C). Since Bayesian phylogeography reconstruction can be sensitive to unequal sampling size, all analyses were repeated on 10 additional datasets, each one including all 27 aquatic isolates as well as a random subsample of 27 clinical isolates. Results were in agreement with the findings for the full dataset (SI Appendix, Fig. S4). The majority of the transitions (connections between two neighboring nodes) in the MCC tree (76.6%) occurred between isolates of clinical origin, representing a large number of human-to-human transmission events throughout the epidemic (Fig. 2C). Yet, a proportion of all transitions (19.3%) occurred between aquatic isolates, with about half of these (8.3%) located in internal branches (i.e., connections between ancestral neighboring nodes). These data support, again, a scenario of independent replication/evolution in the aquatic environment. Clinical-to-environmental and environmental-to-clinical transitions were also present at a low proportion in the tree, indicating spillover between the environment and humans (Fig. 2C). Spillover between host and environment, as well as host colonization (17) or host-to-host transmission cycles (14), can cause bottlenecks that dramatically reduce the bacterial (population genetic measure) effective population size (Ne). Indeed, V. cholerae demographic history, inferred from the Bayesian time-scaled phylogeny (18), showed two population bottlenecks in mid-2012 and 2014 (SI Appendix, Fig. S2D). The demographic history also showed that, despite the low prevalence of clinical cases observed in 2014, bacterial diversification continued in the aquatic environment, as indicated by the Ne increase in 2012 to 2013 after the first bottleneck.
V. cholerae is capable of surviving in aquatic environments due to its varied adaptive responses to stressors, including nutrient deprivation (19), changes in salinity, and temperature and predation by bacteriophages and protists (20). Persistence in the environment has been linked to a variety of phenotypes and mechanisms (21, 22), including vps-dependent or vps-independent biofilm formation (23). To explore the rate at which toxigenic V. cholerae isolates replicated, we calculated the weighted average of synonymous substitution rates (KSs) as a proxy for bacterial replication rate. To control for the potential bias of transient polymorphisms, KS was separately estimated along internal and external branches, of either environmental or clinical isolates, of Bayesian phylogenies, after testing for phylodynamic quality and structure of the datasets and calibrating the best evolutionary model (SI Appendix, Fig. S5 and Tables S3 and S4). The environmental KS estimate for internal branches was about twice higher than the clinical (P < 0.001) (Fig. 2D and SI Appendix, Fig. S6 and Table S5), evidence of faster replication in isolates circulating in aquatic environmental sources. While not completely in keeping with prior findings that the human gut is the “optimal” environment for V. cholerae (24) replication, it is known that stress-induced mutations contribute to adaptive evolution (25) and bacteria increase their mutation rates in response to environmental stressors such as starvation (25). A faster replication of environmental isolates is expected to lead to accumulation of deleterious mutations in the population, with a large number of transient polymorphisms segregating on external branches of the phylogeny. Indeed, while KS in external branches of environmental phylogenies, that is, along lineages that have not propagated successfully, is significantly higher than in internal branches (Fig. 2D and SI Appendix, Table S5), the contrary is observed for clinical isolates (Fig. 2D and SI Appendix, Fig. S6 and Table S5).
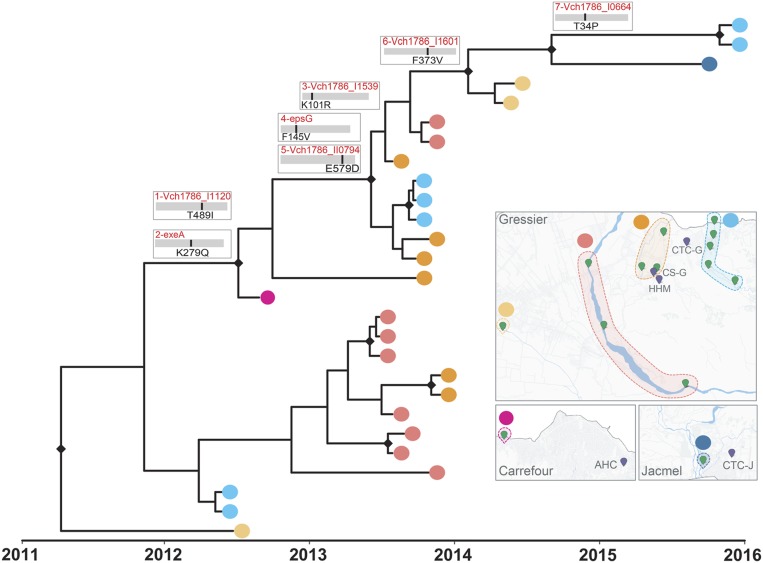
To distinguish between evolution under selective pressure (adaptation) or genetic drift, we calculated the nonsynonymous (dN) and synonymous substitution (dS) rate ratio (dN/dS) (SI Appendix, Fig. S6 and Table S5). The evolutionary dynamic of the clinical isolates was mainly driven by purifying selection (dN/dS = 0.6, P < 0.001), consistent with a negative Tajima’s D (−2.3, P < 0.005) and a population experiencing bottlenecks and/or selective sweeps. Negative selection is expected to drive the already-established epidemic in humans that has reached a peak in the adaptive landscape during an early epidemic stage driven by diversifying selection (14). In contrast, environmental isolates showed greater nonsynonymous rates (dN/dS = 2.1, P < 0.001), characteristic of a heterogeneous microbial population recently introduced to a new environment and undergoing fixation of new variant-driven diversifying selection. In fact, we found environment-specific molecular imprints in the genomes of V. cholerae that successfully replicated and persisted in the aquatic environment. A total of seven nonsynonymous mutations in coding regions, potentially granting a selective advantage to the environmental population of V. cholerae, emerged along the backbone of the environmental isolate phylogeny between 2012 and 2015 (Fig. 3 and SI Appendix, Table S7). Three environmental-specific mutations, two early in the backbone bifurcation (Fig. 3) and one in an internal branch (SI Appendix, Table S7), affected genes in the type II secretion system (TSSII): general secretion pathway proteins A and G, and K (SI Appendix, Table S7). The TSSII plays a pivotal role in the virulence and survival of V. cholerae in different niches, such aquatic reservoirs or human hosts (26). Other mutations found along internal branches affected genes related to environmental stress response, chemotaxis/motility of V. cholerae in response to fluctuating environmental cues, flagellum hook-length control, biofilm formation in the extraintestinal environment, and exponential growth in response to available nutrients (SI Appendix, Fig. S7 and Table S7). Environmental isolates that contributed successfully to the evolving environmental lineage were collected not only near hospitals and cholera treatment centers located in Gressier but also in the nearby Goave and Carrefour regions (Fig. 3).
Fig. 3.
Evolution and adaptation of V. cholerae in Haitian aquatic reservoirs. Phylogenetic relationship and geographical distribution of 27 environmental isolates collected between 2012 and 2015 in Haiti. Nonsynonymous mutations acquired during the evolution of the environmental population, reconstructed by Bayesian inference of ancestral states, are indicated along the backbone of the tree. The SNPs detected along the trunk (surviving lineage) of the tree were sequentially numbered from 1 to 7 to facilitate comparison with the additional information reported in SI Appendix, Table S7. Maps show the sampling sites, grouped by aquatic source, labeled with yellow, red, orange, and cyan dots in the Gressier region, purple for Carrefour, and blue in the Jacmel region.
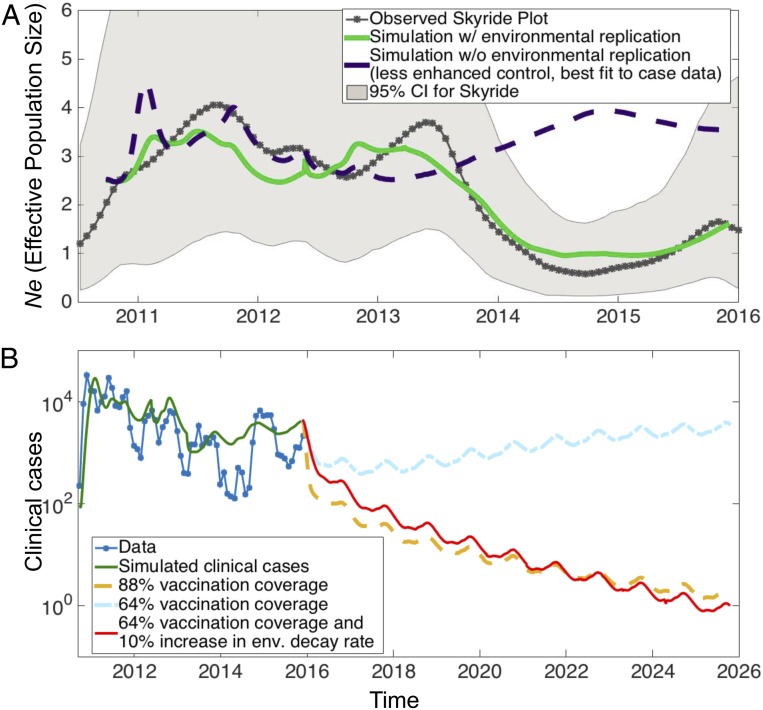
The evidence of epidemiological and environmental evolutionary forces driving the Haitian epidemic motivated us to consider a dynamic model of cholera tracking transmission and population genetics through aquatic environments and hosts. As in prior models of cholera from our group (27), our underlying epidemiological model is based on the susceptible–infected–recovered with reservoir (SIRW) framework, an extension of the classic susceptible–infected–recovered (SIR) framework with an added compartment for pathogen concentration in an aquatic reservoir (W) (Fig. 4 and SI Appendix, Figs. S8 and S9). In keeping with Kirpich et al. (27), we allowed for replication outside of the host, along with seasonality in environmental transmission, shedding, and decay, as suggested by the significant correlation in case counts with precipitation. In addition to this epidemiological formulation, we considered an evolutionary component in our model. Both neutral drift and selection pressures are accounted for in our multilocus framework, where we measure Ne by calculating genetic diversity and utilizing coalescent approaches (SI Appendix, Text). Consistent with other studies, we assumed a tradeoff between environmental survivability and fitness in host transmission for loci under selection. The results from model simulations recapitulate qualitative features of Ne and monthly case counts (Fig. 4A and SI Appendix, Fig. S8).
Fig. 4.
Mixed-transmission model of cholera and vaccination prediction. (A) Ne (effective population size) observed from data (gray) and in simulation with environmental replication (green) and Ne in simulation without environmental replication (violet). (B) Distinct scenarios of vaccination and control for the model simulation with environmental replication, in particular vaccination coverages of 64% (rate of 0.01 d−1), 88% (rate 0.04 d−1), and 64% with a 10% increase in environmental decay rate. Here we define vaccination coverage as the percent reduction in susceptible individuals (upon reaching an approximate steady state after 2 to 3 mo of vaccination). As opposed to Kirpich et al. (27) and simulations without environmental replication (SI Appendix, Fig. S10) in which 64% vaccination coverage eradicates the pathogen within a year, here either more vaccination (88% coverage) or increased environmental decay (10% increase) is needed to control cholera in the presence of environmental replication.
First, the model output and data both display a rise and drop in Ne initially reflecting rapid expansion followed by negative selection after the initial outbreak (Fig. 4A). Then came the observation of a second growth in Ne in 2013, larger in extent than would be expected by host transmission alone, produced by a preceding seasonal flare-up and extending into a period of reducing incidence. Our hypothesis of diversification within the aquatic reservoir offers an explanation for this relative upsurge in Ne, and is supported by reasonable model simulations where inclusion of environmental replication allows for this evolutionary exploration in contrast to cases without environmental replication (Fig. 4A and SI Appendix, Fig. S8). In particular, during the late 2012-to-mid 2013 peak, uuSNPs were detected in the following genes of the surviving environmental lineage: Vch1786_I0012 (1 C→T), Vch1786_I0051 gene (3 C→T), Vch1786_I0998 (30 G→A), Vch1786_I1539 (44 A→G), rseA (52 G→A), epsG (61 A→C), and Vch1786_II0794 (107 C→A) (SI Appendix, Fig. S3). Mutations in these genes may have contributed to increased adaptation of environmental strains in the aquatic ecosystem, although future in vitro studies will be needed to investigate their contribution to V. cholerae fitness. The peak in environmental replication is followed by a substantial decline in Ne beginning prior to the major lull period in 2014. Again, the case incidence does not fully explain the magnitude of this drop. While the population bottleneck induces a decrease in Ne, simulations suggest the environmental replication is necessary to accelerate fixation of genes adapted to persisting in the aquatic reservoir. At the end of 2014, environmental reservoirs spill over to hosts with a spike in clinical cases, possibly enhanced by a beneficial host adaptation and/or loss of immunity in the host population. Extensive simulations calibrated to the case data suggest that in order to additionally fit the observed Ne, environmental replication should be included in the model. A major ramification of our findings is that the replication and adaptation within the aquatic reservoir may make control strategies targeting host transmission more difficult. Indeed, projecting forward from 2015 in simulations, we observed that vaccination can readily clear the pathogen in the absence of environmental replication (SI Appendix, Fig. S10). However, for environmental adaptation and replication consistent with the observed Ne, vaccinating a much larger portion of the population or adding an intervention affecting the aquatic reservoir is necessary to eliminate cholera in our dynamic mixed-transmission model (Fig. 4B).
Discussion
Our findings have direct applicability to the development of control and elimination strategies for this ancient and devastating disease. Assumptions that transmission outside of the Indian subcontinent occurs only at a direct person-to-person level have resulted in a major focus on “quick response” teams that go to case households to provide disinfection combined with a local emphasis on water, sanitation, and hygiene. Using Haiti as an example, our data underscore a greater complexity of the transmission process, illustrating the key role that aquatic reservoirs play in evolution, dissemination, and maintenance of the disease in an area well-removed from its Asian homeland. This translates into a recognition that elimination of toxigenic V. cholerae may be difficult, if not impossible, so long as environmental reservoirs are present. This, in turn, emphasizes the need for careful and ongoing environmental studies in areas where cholera cases have occurred and the need for a long-term control strategy, including long-term investments in water, sanitation, and hygiene infrastructure, even if cholera cases are not being identified. It also plays into the question of vaccination. There has been a recent focus on local, “ring” vaccination for cholera to try to limit spread from identified human cases. Our data, in keeping with earlier models published by our group, suggest that population-based vaccination may be more effective, recognizing the potential for broad distribution of the disease through environmental sources.
There has been a well-recognized link between water and cholera epidemics (28). However, recent genetic studies based on clinical isolates from global sources hypothesized that environmental reservoirs, while important in the persistence of the disease on the Indian subcontinent, have been playing a minimal role in cholera outbreaks in Africa and the Americas (5, 6, 29). Such studies lacked an analysis of aquatic environmental toxigenic V. cholerae O1 isolates. Our findings show the critical role played by aquatic isolates in the transmission and evolution of cholera in Haiti. We propose a dynamic mixed-transmission model that assumes bacterial dissemination into an aquatic reservoir, evolution and adaptation to the new condition, and spillover to humans with subsequent transmission within human populations. Ultimate control of cholera is possible, but requires a recognition of the complexity of the transmission process and the need for long-term investments in cholera control at both the human and environmental levels.
Materials and Methods
Sample Collection.
Between 2010 and 2017, we have isolated and characterized ∼800 toxigenic V. cholerae O1 strains from cholera patients attended in diverse cholera treatment centers and clinics run by nongovernmental agencies and Haitian national clinics. For environmental monitoring of toxigenic V. cholerae O1 strains, we collected surface water samples monthly from 17 sentinel sites in the Gressier/Leogane regions in Haiti effective May 2012. Environmental survey resulted in the isolation of 27 toxigenic V. cholerae O1 strains between 2012 and 2015. Both clinical and environmental V. cholerae O1 isolates were confirmed by standard microbiological, biochemical, serological, and genetic analysis as described previously (10, 30). Of 800 clinical isolates, 205 strains (116 from the Ouest Department) as well as all 27 environmental O1 strains were analyzed in this study. Information about the samples is available in SI Appendix, Tables S1 and S2.
Monthly case counts in the Ouest Department were obtained from the Pan American Health Organization (https://new.paho.org/hq/images/Atlas_IHR/CholeraHispaniola/atlas.html) (SI Appendix, Table S2), and mean monthly precipitation and average water surface temperature at 10 m above displacement height in Haiti were obtained from the NASA Goddard Distributed Active Archive Center (https://disc.gsfc.nasa.gov/). We tested if the variables used in a model to predict the case precipitations and temperatures were relevant to the outcome (cases) using a stepwise selection using the Akaike information criterion. The F test was used to compare this model with the one with the intercept only (no variables).
Whole-Genome Mapping and HqSNP Calling.
Our dataset was composed of 116 toxigenic V. cholerae strains: 27 environmental isolates were obtained in the Ouest Department, 23 isolates were sequenced in this study, and 4 were previously sequenced by our group (14); and 89 clinical isolates, of which one is the reference strain EL-1786 for the Haitian epidemic (31): 37 isolates were sequenced in this study, 32 isolates were previously sequenced by our group and collected between 2010 and 2012 (14), and 20 isolates also added to ensure correct calibration of the root of the tree were collected in the Artibonite and Ouest departments at the beginning of the epidemic between 2010 and 2011 (32). The newly sequenced toxigenic V. cholerae strains were confirmed by serology and PCR (33). After subculture, genomic DNA extraction was performed using the Qiagen DNeasy Blood and Tissue Kit. Genomic DNA from all isolates was cultured and extracted from bacterial pellets. Sample library construction using the Nextera XT DNA Library Preparation Kit was performed (Illumina). Whole-genome sequencing on all isolates was executed on the Illumina MiSeq for 500 cycles (Illumina). Adapter and raw sequence reads were filtered by length and quality by using the program Trimmomatic (34). After quality filtering, Bowtie 2 (35) was used to map the sequence reads to the reference genome, V. cholerae O1 strain 2010EL-1786 (GenBank accession nos. NC_016445.1 and NC_016446.1) (31). This reference sequence was isolated in Haiti and is generally used for reference mapping of the samples that were collected in Haiti. After the reads were mapped to the reference genome, duplicate reads were marked and the reads were realigned using the program Picard (http://broadinstitute.github.io/picard/). The reference-based mapping alignment was then verified and fixed accordingly, if needed. FreeBayes (36) was used to create a custom genome-wide SNP calling database (dbSNP) from all isolates in the dataset to perform base quality score recalibration (BQSR), outlined in GATK’s best practice guidelines for germline variation (https://gatk.broadinstitute.org/hc/en-us). When the variant call format (VCF) file was obtained, hard filtering of called SNPs was performed and the subsequent VCF file was used as a dbSNP for BQSR. When the BQSR step was completed, the alignment files were recalibrated and FreeBayes (36) was then used again to call variants on the recalibrated alignment files. Afterward, the newly created VCF file was filtered only for SNPs. After filtering, the VCF file was normalized using BCFtools (http://www.htslib.org/doc/bcftools.html). Normalization simplifies the represented variants in the VCF file by showing as few bases as possible at a particular SNP site in the genome. SNPs were then filtered by depth of coverage, quality, and genotype likelihood, as described in Azarian et al. (14). A SNP alignment in FASTA format was extracted from the VCF file from a custom python script. The SNP alignment was then filtered by site, leaving only sites with greater than 75% SNPs at that particular site, making a high-quality SNP alignment (SI Appendix, Table S3). The hqSNP alignment was annotated using the program SnpEff (37). HqSNPs found in protein-coding regions of the V. cholerae O1 genome that were identified by annotation were used to produce a codon alignment that was extracted by in-home scripts (available upon request). All bioinformatic analysis was performed using this optimized pipeline implemented on the University of Florida’s HiPerGator cluster (https://www.rc.ufl.edu/services/hipergator/).
Phylogenetic Quality Assessments and Maximum-Likelihood Analysis.
All datasets used in this study passed phylogenetic quality checks as follows: in order to evaluate the presence of sufficient phylogenetic signal to resolve the phylogenetic relationship among V. cholerae O1 isolates, we performed likelihood mapping analysis with IQ-TREE (http://www.iqtree.org/) that calculates and maps the likelihood of all possible sequence quartets using the best-fitting nucleotide substitution model (38). Absence of substitution saturation was assessed by plotting pairwise transition/transversion vs. genetic distance with DAMBE6 (http://dambe.bio.uottawa.ca/DAMBE/) (39) (SI Appendix, Fig. S2).
For each dataset, the presence of temporal signal was assessed with TempEst version 1.5 (http://tree.bio.ed.ac.uk/software/tempest/) (40) on ML phylogenies inferred by IQ-TREE (41, 42) (http://iqtree.cibiv.univie.ac.at) using the best-fitting nucleotide substitution model according to the Bayesian information criterion (41, 42) and ultrafast bootstrap approximation (1,000 replicates) to assess robustness of the phylogeny internal branches (43) (SI Appendix, Fig. S2). In order to test whether or not the spread of sampling times was sufficient to allow the substitution rate to be estimated accurately, that is, the presence of temporal structure within each of the time-structured datasets, we also performed the date randomization test on all phylogeographic datasets using TipDatingBeast implemented in the R package (44) (SI Appendix, Fig. S2). Ancestral-state reconstruction using maximum likelihood inferred with PastML (45) from a rooted maximum likelihood phylogenetic tree inferred with IQ-TREE, and the tree was compressed vertically and horizontally: clades have been vertically (clade circles contain the number of tips of the initial tree contained in them) and horizontally (the branch size corresponds to the number of times its subtree is found in the initial tree) merged to cluster independent events of the same kind.
Bayesian Coalescent Inference: Discrete Phylogeographic Reconstruction, Markov Jumps, and Markov Rewards.
To test our hypothesis of long-term aquatic reservoirs of toxigenic V. cholerae O1 being established in Haiti, we used the Bayesian phylogeographic (46) coalescent-based method (15) implemented in the BEAST version 1.8.4 (47) software package. The reconstruction of V. cholerae spatiotemporal spread in different environments through Bayesian phylogeography requires calibration of a molecular clock. Evolutionary rates were estimated implementing the HKY nucleotide substitution model (48) with empirical base frequencies, gamma distribution of site-specific rate heterogeneity, and ascertainment bias correction (49, 50), testing a constant demographic prior against nonparametric demographic models, Gaussian Markov random-field Skyride (51), and Bayesian Skyline (52) to rule out spurious changes in effective population size inferred by a nonparametric model that would in turn impact the timing of divergence events (53). Additionally, for each demographic model, we compared strict and relaxed uncorrelated (lognormal distribution among branches) molecular clocks (18, 54). The best molecular clock and demographic model were chosen by estimating the marginal likelihood of each model, with path sampling and stepping-stone methods, followed by Bayes factor comparisons (47, 55) (SI Appendix, Tables S1, S3, and S4). Markov chain Monte Carlo samplers were run for a number of generations (50 to 200 million) sufficient to achieve proper mixing of the Markov chain, which was evaluated by calculating the effective sampling size (ESS) of each parameter estimate under a given model. ESS values >200 for all parameter estimates were considered as evidence of proper mixing. The origin of each isolate, environmental or clinical, was used as a trait for “location” in order to reconstruct the evolution of V. cholerae O1 and bacterial flow (migration) events between environmental and/or clinical sources. Transitions between discrete states (environmental and clinical) were estimated using the continuous-time Markov chain model using the asymmetric migration model with Bayesian stochastic search variable selection (46). In our reconstruction of ancestral states, we assume migration occurs at the tree nodes.
Bayesian phylogeographic reconstructions are sensitive to disproportional sample sizes across subpopulations. We tested for sampling bias using two approaches: 1) using the less sensitive to sampling biases Bayesian structured coalescent approximation (56) implemented in BEAST2 version 2.5.2 for the Ouest Department dataset (SI Appendix, Fig. S2); and 2) using 10 resampled datasets on clinical (187) and environmental strains (27) isolated in 2012 to 2015, since no environmental strains were collected in 2016 to 2017, and all clinical strains sampled during these 2 y were monophyletic (i.e., representing a new epidemic wave) not connected to any previously sampled environmental strain. Each dataset was generated by randomly subsampling clinical strains to match the same number of sequences available for environmental isolates collected between 2012 and 2015, and by adding the isolates collected at the beginning of the epidemic (2010) to allow correct rooting of the tree (31). The analyses described below were carried out in parallel on each one of the resampled datasets and are shown in SI Appendix, Fig. S4.
The maximum clade credibility trees were obtained from the posterior distribution of trees with TreeAnnotator version 1.8.4 after 10% burn-in. The MCC phylogeny was manipulated in R using the package GGTREE (57) for publishing purposes.
Within the same phylogeographic inference, we also calculated the number of transitions between environmental and clinical states (Markov jump) (58) and the time between state changes (Markov reward) (58, 59). Markov jumps (or transitions) were plotted as the total number of state counts for the migration in and out of each location (environment, clinical) among all internal and external branches. Markov rewards can be estimated along the phylogenetic tree trunk to calculate the length of time that an ancestral population occupies a particular location (environmental or clinical) (60–62). We defined the backbone path, or trunk of the tree, following Lemey et al. (63), as the internal subset of branches that connects the root node of a time-scaled phylogeny to the most recent common ancestor of the sequences sampled at the latest time point. In other words, the trunk represents the successful lineage that persists through time. Trunk proportions for time and ancestral locations were calculated after 10% burn-in, slicing time in 0.5-y sections. If the tree trunk is occupied by a single location, cholera would exhibit source–sink population dynamics (60), and if the trunk is occupied by multiple locations, cholera would emerge from a population migrating between states (61, 62). Markov rewards were compared against a null distribution of 10 replicates obtained by randomization of the tip states performing binomial tests (64) to assess whether the mean of the posterior probability at every time point differed. Markov jumps and rewards were plotted using the R package ggplot2 (65).
Calculation of the Weighted Average of Synonymous Substitution Rates and Selection Analysis.
In order to investigate replication rates among environmental versus clinical strains and to assess the selective pressure driving their evolution, we filtered the genome-wide hqSNP data to hqSNPs located in protein-coding regions. Using hqSNP in codon format, a subset of 200 Bayesian MCC genealogies was randomly obtained from the posterior distribution of trees for each subsampled data set and used for selection analysis. The weighted average of synonymous substitution rates (KSs) and nonsynonymous substitution rates (KAs) in the protein-coding regions of the V. cholerae O1 genome for all, internal and external, branches were obtained from a subset of 200 Bayesian MCC trees randomly obtained from the posterior distribution of trees, as described by Lemey et al. (63). The ratio of nonsynonymous and synonymous substitution rates (dN/dS)—providing information on whether evolution is occurring mainly through random genetic drift (dN/dS ∼ 1) or positive (dN/dS > 1) or negative (dN/dS < 1) selection—was obtained from a subset of 200 Bayesian MCC trees randomly obtained from the posterior distribution of trees, as described by Lemey et al. (63). For the calculation of Tajima’s D (66) and P values, we used the tajima.test function in the R package pegas.
Statistical Analyses for dN and dS Substitution Accumulation.
The null hypothesis of equality of two means H0:μX = μY, against the alternative H1:μX ≠ μY, was assessed through a Welch’s two-sample location test (67). It is an adaptation of the Student’s t test and is applicable when the two samples have unequal variances. The resulting P value is used as evidence against the hypothesis that the two population means are equal. Testing the null hypothesis that the ratio of the two means is equal to 1 can be stated as H0:φ = 1, where φ = μX∕μY against the alternative H1:φ ≠ 1, which means that φ = μx∕μy = 1 = μx∕μy = 1 μx∕μy*μY = 1*μY μY = μY, and the null hypothesis on the ratio can be restated in terms of equality in means, that is, H0:μX = μY, against H1:μX ≠ μY. CIs are computed using the bias-corrected and accelerated bootstrap method. It adjusts for both bias and skewness in the bootstrap distribution (68).
SIRW Mathematical Model.
See SI Appendix for a mathematical model for the ecoevolutionary dynamics of cholera.
Additional Data.
R scripts and xml files are available at the GitHub page: https://github.com/salemilab/environmental_cholera_haiti. The genomic sequences have been deposited with the National Center for Biotechnology Information (NCBI) in the Sequence Read Archive under BioProject ID code PRJNA510624. All other data are available in SI Appendix or upon request.
Supplementary Material
Acknowledgments
We thank Philippe Lemey for his help with Markov rewards analysis, and Nicola De Maio for his help with BASTA analysis. This work was entirely supported by NIH Grants R01 AI097405, R01 AI128750, and R01 AI123657. C.B. was supported by US NSF Grant DMS-1815095. E.J.N. was supported by NIH Director’s Early Independence Award DP5OD019893.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic sequences reported in this paper have been deposited with the National Center for Biotechnology Information (NCBI) in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject ID code PRJNA510624). R scripts and xml files are available on GitHub (https://github.com/salemilab/environmental_cholera_haiti).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918763117/-/DCSupplemental.
References
- 1.Cvjetanovic B., Barua D., The seventh pandemic of cholera. Nature 239, 137–138 (1972). [DOI] [PubMed] [Google Scholar]
- 2.Colwell R. R., Huq A., Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann. N. Y. Acad. Sci. 740, 44–54 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Stine O. C., et al. , Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerg. Infect. Dis. 14, 831–833 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam M., et al. , Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 72, 4096–4104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weill F. X., et al. , Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Domman D., et al. , Integrated view of Vibrio cholerae in the Americas. Science 358, 789–793 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Hendriksen R. S., et al. , Population genetics of Vibrio cholerae from Nepal in 2010: Evidence on the origin of the Haitian outbreak. MBio 2, e00157-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piarroux R., et al. , Understanding the cholera epidemic, Haiti. Emerg. Infect. Dis. 17, 1161–1168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UN Office for the Coordination of Humanitarian Affairs, Haiti: Cholera figures (2018). https://reliefweb.int/report/haiti/haiti-cholera-figures-31-january-2019. Accessed 6 June 2019.
- 10.Alam M. T., et al. , Increased isolation frequency of toxigenic Vibrio cholerae O1 from environmental monitoring sites in Haiti. PLoS One 10, e0124098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco A. A., et al. , Cholera in Lima, Peru, correlates with prior isolation of Vibrio cholerae from the environment. Am. J. Epidemiol. 146, 1067–1075 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Alam M., et al. , Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72, 2849–2855 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faria N. R., Suchard M. A., Rambaut A., Lemey P., Toward a quantitative understanding of viral phylogeography. Curr. Opin. Virol. 1, 423–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azarian T., et al. , Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from Haiti reveals diversification driven by positive selection. MBio 5, e01824-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grenfell B. T., et al. , Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303, 327–332 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Rebaudet S., et al. , Epidemiological and molecular forensics of cholera recurrence in Haiti. Sci. Rep. 9, 1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levade I., et al. , Vibrio cholerae genomic diversity within and between patients. Microb. Genom. 3, e000142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummond A. J., Rambaut A., Shapiro B., Pybus O. G., Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Nelson E. J., et al. , Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 4, e1000187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutz C., Erken M., Noorian P., Sun S., McDougald D., Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 4, 375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jubair M., Morris J. G. Jr, Ali A., Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS One 7, e45187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colwell R. R., Viable but nonculturable bacteria: A survival strategy. J. Infect. Chemother. 6, 121–125 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Sinha-Ray S., Ali A., Mutation in flrA and mshA genes of Vibrio cholerae inversely involved in vps-independent biofilm driving bacterium toward nutrients in lake water. Front. Microbiol. 8, 1770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson E. J., Harris J. B., Morris J. G. Jr, Calderwood S. B., Camilli A., Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7, 693–702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjedov I., et al. , Stress-induced mutagenesis in bacteria. Science 300, 1404–1409 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Sikora A. E., Proteins secreted via the type II secretion system: Smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog. 9, e1003126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirpich A., et al. , Cholera transmission in Ouest Department of Haiti: Dynamic modeling and the future of the epidemic. PLoS Negl. Trop. Dis. 9, e0004153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colwell R. R., Global climate and infectious disease: The cholera paradigm. Science 274, 2025–2031 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Weill F. X., et al. , Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature 565, 230–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam M. T., et al. , Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerg. Infect. Dis. 20, 356–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimer A. R., et al. ; V. cholerae Outbreak Genomics Task Force , Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg. Infect. Dis. 17, 2113–2121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz L. S., et al. , Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio 4, e00398-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali A., et al. , Recent clonal origin of cholera in Haiti. Emerg. Infect. Dis. 17, 699–701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabor M., Garrison E., Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (17 July 2012).
- 37.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt H. A., Strimmer K., Vingron M., von Haeseler A., TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Xia X., Xie Z., DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 92, 371–373 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trifinopoulos J., Nguyen L. T., von Haeseler A., Minh B. Q., W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minh B. Q., Nguyen M. A., von Haeseler A., Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duchêne S., Duchêne D., Holmes E. C., Ho S. Y., The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol. Biol. Evol. 32, 1895–1906 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa S. A., Zhukova A., Iwasaki W., Gascuel O., A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol. Biol. Evol. 36, 2069–2085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemey P., Rambaut A., Drummond A. J., Suchard M. A., Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond A. J., Rambaut A., BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasegawa M., Kishino H., Yano T., Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985). [DOI] [PubMed] [Google Scholar]
- 49.Leaché A. D., Banbury B. L., Felsenstein J., de Oca A. N., Stamatakis A., Short tree, long tree, right tree, wrong tree: New acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 64, 1032–1047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis P. O., A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Minin V. N., Bloomquist E. W., Suchard M. A., Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25, 1459–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strimmer K., Pybus O. G., Exploring the demographic history of DNA sequences using the generalized skyline plot. Mol. Biol. Evol. 18, 2298–2305 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Hall M. D., Woolhouse M. E., Rambaut A., The effects of sampling strategy on the quality of reconstruction of viral population dynamics using Bayesian skyline family coalescent methods: A simulation study. Virus Evol. 2, vew003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gill M. S., et al. , Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 30, 713–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baele G., et al. , Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29, 2157–2167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Maio N., Wu C. H., O’Reilly K. M., Wilson D., New routes to phylogeography: A Bayesian structured coalescent approximation. PLoS Genet. 11, e1005421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu G. C., Smith D. K., Zhu H. C., Guan Y., Lam T. T. Y., GGTREE: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 58.Minin V. N., Suchard M. A., Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 56, 391–412 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Lemey P., et al. , Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 10, e1003932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell C. A., et al. , The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Bedford T., Cobey S., Beerli P., Pascual M., Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog. 6, e1000918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bahl J., et al. , Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc. Natl. Acad. Sci. U.S.A. 108, 19359–19364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lemey P., et al. , Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 3, e29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clopper C. J., Pearson E. S., The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26, 404–413 (1934). [Google Scholar]
- 65.Ginestet C., ggplot2: Elegant graphics for data analysis. J. R Stat. Soc. Ser. A Stat. Soc. 174, 245–246 (2011). [Google Scholar]
- 66.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Welch B. L., The generalisation of Student’s problems when several different population variances are involved. Biometrika 34, 28–35 (1947). [DOI] [PubMed] [Google Scholar]
- 68.Efron B., Better bootstrap confidence-intervals. J. Am. Stat. Assoc. 82, 171–185 (1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.