Significance
While understanding metabolic change of tumor cells is crucial to improve cancer therapy, current 2D cell culture condition may not fully recapitulate in vivo metabolic environment of tumors. Our metabolomics analysis demonstrates that proline metabolism is critical for the KSHV-transformed cell growth in 3D culture, not in 2D culture. Specifically, the KSHV K1 oncoprotein activates PYCR proline biosynthesis enzyme, increasing intracellular proline concentration for tumor cell growth in in vitro 3D spheroid culture and in vivo tumorigenesis. This study describes an oncogenic strategy of KSHV to enhance proline synthesis for virus-induced transformation, adding the proline metabolic pathway as a potential target for KS treatment.
Keywords: cancer metabolism, proline metabolism, pyrroline-5-carboxylate reductase (PYCR), Kaposi's sarcoma-associated herpesvirus (KSHV), K1
Abstract
Three-dimensional (3D) cell culture is well documented to regain intrinsic metabolic properties and to better mimic the in vivo situation than two-dimensional (2D) cell culture. Particularly, proline metabolism is critical for tumorigenesis since pyrroline-5-carboxylate (P5C) reductase (PYCR/P5CR) is highly expressed in various tumors and its enzymatic activity is essential for in vitro 3D tumor cell growth and in vivo tumorigenesis. PYCR converts the P5C intermediate to proline as a biosynthesis pathway, whereas proline dehydrogenase (PRODH) breaks down proline to P5C as a degradation pathway. Intriguingly, expressions of proline biosynthesis PYCR gene and proline degradation PRODH gene are up-regulated directly by c-Myc oncoprotein and p53 tumor suppressor, respectively, suggesting that the proline-P5C metabolic axis is a key checkpoint for tumor cell growth. Here, we report a metabolic reprogramming of 3D tumor cell growth by oncogenic Kaposi’s sarcoma-associated herpesvirus (KSHV), an etiological agent of Kaposi’s sarcoma and primary effusion lymphoma. Metabolomic analyses revealed that KSHV infection increased nonessential amino acid metabolites, specifically proline, in 3D culture, not in 2D culture. Strikingly, the KSHV K1 oncoprotein interacted with and activated PYCR enzyme, increasing intracellular proline concentration. Consequently, the K1-PYCR interaction promoted tumor cell growth in 3D spheroid culture and tumorigenesis in nude mice. In contrast, depletion of PYCR expression markedly abrogated K1-induced tumor cell growth in 3D culture, not in 2D culture. This study demonstrates that an increase of proline biosynthesis induced by K1-PYCR interaction is critical for KSHV-mediated transformation in in vitro 3D culture condition and in vivo tumorigenesis.
Cells are often statically cultured as monolayers on flat surfaces, and these conditions do not faithfully reflect the biological situation in vivo given that proper tissue architecture and cell–cell contacts are often lost in such two-dimensional (2D) cell culture. Three-dimensional (3D) cell culture is well documented to regain intrinsic properties and to better mimic the in vivo physiologic environment than 2D cell culture (1, 2). Intriguingly, proline metabolism has been recently identified as one of the metabolic pathways that is regulated differently in 3D cell culture than 2D cell culture (3). Proline is one of the nonessential amino acids that shows emerging roles in stress response, cell survival, aging, and development in many different organisms (4–6). Proline protects cells against reactive oxygen species (ROS) like hydrogen peroxide, tert-butyl hydroperoxide, and carcinogenic oxidative stress inducer (7). Proline depletion reduces clonogenic potential of cancer cells with unresolved endoplasmic reticulum stress, therefore proving the importance of proline in cancer cell growth (8). Furthermore, proline and hydroxyproline are major components of collagen that constitute the scaffold of the tumor microenvironment. Indeed, global metabolic profiling demonstrates a crucial role of proline and hydroxyproline metabolism in hepatocellular carcinoma (HCC) (9). A recent study has also identified that collagen-derived proline is metabolized to fuel the tricarboxylic acid (TCA) cycle and contribute to cancer cell survival under restrictive nutrient conditions (10). This indicates that proline metabolism is critical for 3D tumor formation.
Pyrroline-5-carboxylate (P5C) is a central intermediate that is synthesized from glutamate by P5C synthase (P5CS) and then converted to proline by P5C reductase (PYCR/P5CR). The degradation step from proline to P5C is catalyzed by proline dehydrogenase (PRODH). Overexpression of PRODH in HEK293 cells resulted in a sixfold lower intracellular proline content and decreased cell survival relative to vector-alone transfected HEK293 cells (7). Indeed, the ProDH has been identified as a tumor suppressor p53-induced gene 6 (PIG6) (11). In contrast, overexpression of the proline biosynthetic enzymes P5CS and PYCR in HEK293 cells resulted in a twofold higher proline content, significantly lower ROS levels, and increased cell survival relative to vector-alone transfected HEK293 cells (7). Correspondingly, c-Myc oncogene markedly up-regulates PYCR expression, whereas it suppresses ProDH expression (12, 13). Thus, the proline metabolic axis can serve as a scaffold on which the oncogene and tumor suppressor are tightly integrated, suggesting that proline metabolism may be a good target for adjunctive cancer therapy.
Tumor cells alter cellular metabolism to achieve survival and growth advantages in an unfavorable environment (14). Metabolic reprogramming has been explored for its contribution to cancer cell growth in addition to the initial identification of the Warburg effect where cancer cells increase glycolysis and lactate fermentation. However, the impact of oncogenic viruses on metabolic phenotype is less well understood. Kaposi’s sarcoma-associated herpesvirus (KSHV) is an etiologic agent of Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL). As an oncogenic virus, KSHV induces angiogenesis, proliferation, and metabolic reprogramming. Previous studies showed that KSHV infection targets metabolic regulators such as HIF1α and AMPK in addition to affecting metabolic pathways like glycolysis and lipogenesis (15–19). However, cell types and culture conditions as well as testing model systems can drastically affect tumor virus-induced metabolic phenotypes (20).
Similar to other herpesvirus, KSHV undergoes latent or lytic phase upon infection, and both latent and lytic gene products contribute to viral transformation (21). One of the lytic oncoproteins is a type I transmembrane glycoprotein called K1, encoded by the first open reading frame (ORF) in the KSHV genome (22). K1 has an extracellular domain that demonstrates a regional homology with the Ig family and an immunoreceptor tyrosine-based activation motif (ITAM) in the cytoplasmic domain. The phosphorylated ITAM of K1 subsequently interacts with cellular Src homology 2 (SH2)-containing signaling proteins Lyn, Syk, p85, PLCγ2, RasGAP, Vav, SHIP 1/2, and Grab2 (23), which increases intracellular tyrosine phosphorylation, calcium mobilization, and transcription factor activity (24, 25). Consequently, K1 expression inhibits apoptosis, promotes cell survival, and produces inflammatory cytokines and proangiogenic factors (26, 27).
In this study, we performed the unbiased metabolomics analysis of KSHV-infected cells in 3D culture to better mimic the in vivo environment than standard 2D culture. Therefore, we identified that proline amino acid metabolism was one of the most significantly altered metabolic pathways of KSHV-infected cells in 3D culture. Moreover, we discovered that the KSHV K1 oncoprotein interacted with host PYCR and enhanced its enzyme activity via its carboxyl (C)-terminal ITAM-independent manner, increasing intracellular proline concentration, and promoting 3D cell growth as well as in vivo tumorigenesis. This study describes an oncogenic strategy of KSHV to enhance proline synthesis for virus-induced pathogenesis, adding the proline metabolic pathway as a potential target for KS treatment.
Results
KSHV Induces Nonessential Amino Acid Metabolism in 3D Cell Culture.
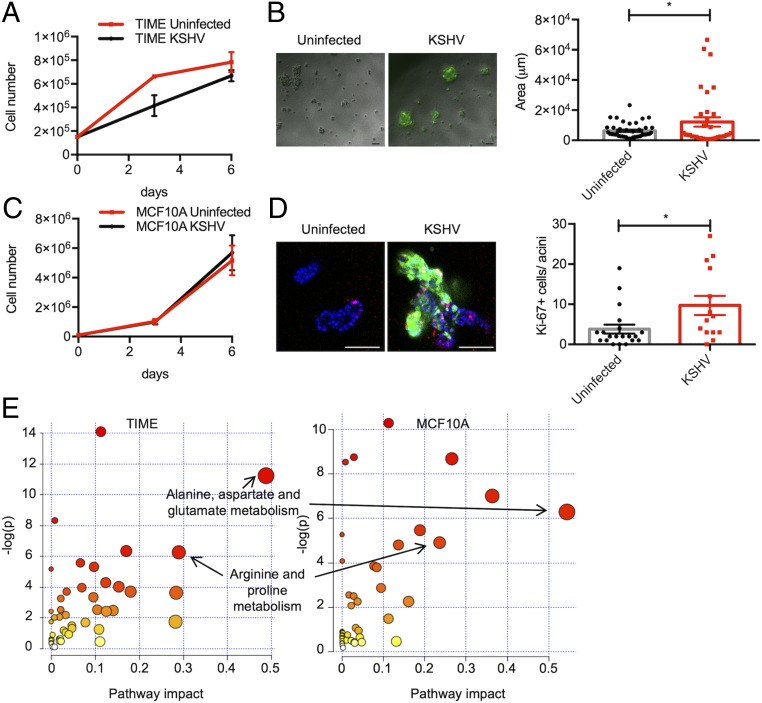
To study the KSHV-mediated metabolic reprogramming, we cultured telomerase-immortalized microvascular endothelial (TIME) cells and MCF10A nontumorigenic epithelial cells in 2D monolayer or in 3D matrigel culture. While KSHV-infected TIME or MCF10A cells proliferated similarly to the uninfected cells in 2D culture (Fig. 1 A and C), KSHV-infected TIME cells formed larger spheroids than uninfected TIME cells (Fig. 1B). It is well known that MCF10A cells in 3D culture are able to form growth-arrested and hollow lumen containing polarized acini and it is a good model to analyze the tumorigenesis and oncogenicity (28). For example, MCF10A cells expressing human papillomavirus E7 form larger acini and result in excessive proliferation within acini instead of growth arrest (29). KSHV-infected MCF10A cells in 3D culture also showed hyperproliferation and filling of the luminal space within acini compared to uninfected cells in 3D culture (Fig. 1D). To elucidate metabolic changes, we performed liquid chromatography-mass spectrometric analysis (LC-MS) and detected 176 metabolites in TIME cells, and 165 metabolites in MCF10A cells (SI Appendix, Fig. S1A). Principal component analysis (PCA) plots showed that there were more obvious distinctions between uninfected cells and KSHV-infected cells in 3D culture than in 2D culture, indicating intracellular metabolism of KSHV-infected cells is considerably altered in 3D culture compared to that in 2D culture (SI Appendix, Fig. S1B). The KSHV-induced metabolites were analyzed using Metaboanalyst 4.0 to identify metabolic pathways that were altered by the virus infection only in 3D culture. The metabolic pathways with impact >0.15 and P value <0.05 were considered to be significantly influenced pathways in TIME and MCF10A cells (SI Appendix, Tables S1 and S2). Among these pathways, nonessential amino acid metabolic pathways, including alanine, aspartate, and glutamate metabolisms and arginine and proline metabolisms, were commonly induced by KSHV infection of both TIME and MCF10A cells in 3D culture (Fig. 1E). Taken together, targeted metabolomics analysis shows that nonessential amino acid metabolism is significantly altered by KSHV in 3D culture.
Fig. 1.
KSHV induces nonessential amino acid metabolism in 3D culture conditions. (A) Proliferation rate of uninfected or KSHV-infected TIME cells in 2D monolayer. Proliferation was measured by counting the cell numbers. Data are presented as the mean ± SEM. (B) Representative pictures (Left) and size quantification (Right) of 3D spheroids of uninfected or KSHV-infected TIME cells cultured in ultralow attachment condition with 4% Matrigel for 8 d. (Scale bars: 100 μm.) Data are presented as the mean ± SEM *P < 0.05, by Student’s t test. (C) Proliferation rate of uninfected or KSHV-infected MCF10A cells in 2D monolayer. Proliferation was measured by counting the cell numbers. Data are presented as the mean ± SEM. (D) Representative images of uninfected or KSHV-infected MCF10A cells cultured on Matrigel for 14 d. Acini were stained with the nuclei counterstained with Hoechst 33342 (blue) and Ki67 antibody (red). GFP signal was from the BAC16 KSHV genome. (Scale bars: 100 μm, Left.) Quantitation of Ki67 positive cells in acini as box-and-whisker plot. The number of biological replicates for each experiment was n ≥ 3 (Right). *P < 0.05, by Student’s t test. (E) Scatterplots of KSHV-induced metabolic pathways in TIME cells and MCF10A cells. KSHV-induced metabolic pathways in TIME and MCF10A cell types are annotated in the graph. Node color is based on its P value, and the node size is based on pathway impact score.
KSHV K1 Binds to Proline Synthesis Enzyme PYCR via Its C-Terminal ITAM-Independent Manner.
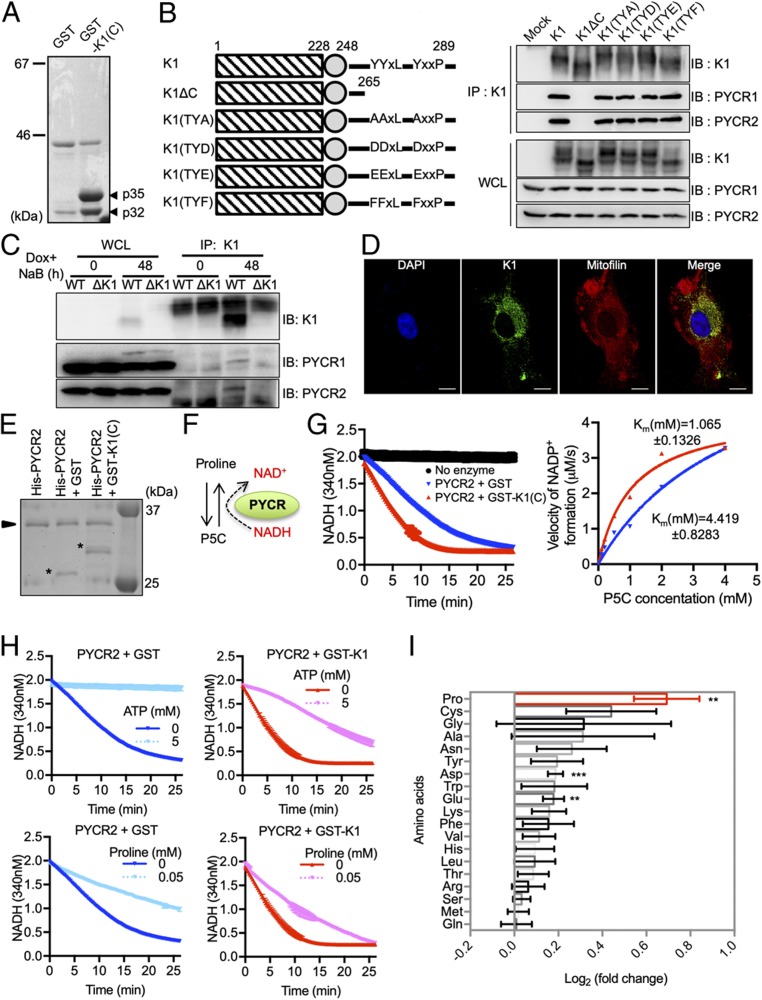
As cancer cells demand higher proline level compared to normal cells, metastatic cancer cells induce de novo proline synthesis (30). For instance, not only is PYCR transcriptionally up-regulated by c-MYC oncogene (12), but its gene copy is also amplified in many types of cancers, suggesting that the PYCR expression is an unfavorable prognostic marker of various cancers (SI Appendix, Fig. S2 A and B) (31). Furthermore, proline metabolism is differently regulated in 3D culture from 2D culture and critically contributes to tumor cell growth in 3D culture but not in 2D culture (3). During the study of which KSHV induced nonessential amino acid metabolisms in 3D culture, we coincidentally identified the robust interaction between KSHV K1 oncoprotein and host PYCR. Bacterially purified GST fusion protein containing the K1 cytoplasmic region [GST-K1(C)] was mixed with lysates of human Burkitt lymphoma BJAB cells, followed by GST pull-down (GST-PD) and mass spectrometry analysis. This showed that K1 strongly bound to endogenous PYCR1 and PYCR2 through its cytoplasmic domain (Fig. 2A and SI Appendix, Table S3). To further confirm the interaction, HEK293T cells were transfected with expression vectors of full-length K1 (K1), the cytoplasmic domain truncated mutant (ΔC) or the TYA, TYD, TYE, or TYF mutants carrying the replacements of three tyrosine residues with alanine, aspartate, glutamate, or phenylalanine, respectively. Cell lysates were used for immunoprecipitation (IP) with an anti-K1 antibody that reacted with its amino (N)-terminal extracellular domain. This showed that K1 wild type (WT) and all tyrosine residue mutants (TYA, TYD, TYE, or TYF), but not the cytoplasmic domain truncated K1 ΔC mutant, interacted with endogenous PYCR1 and PYCR2 (Fig. 2B). GST fusions containing a series of the truncated cytoplasmic domain revealed that the cytoplasmic 24 amino acids of K1 were sufficient to interact with endogenous PYCR1 and PYCR2 (SI Appendix, Fig. S3A). A series of GST-PYCR2 truncation mutants also showed that the NAD(P)H-binding and dimerization domains, but not the C-terminal short region, of PYCR2 were required for K1 binding (SI Appendix, Fig. S3B).
Fig. 2.
K1 interacts with and enhances PYCR enzyme activity. (A) Purification of K1(C) binding proteins. Arrowheads denote the bands that were excised and subjected to MS-based analysis for protein identification. (B) A series of K1 expression constructs (Left) were transfected into HEK293 cells and cell extracts were immunoprecipitated with an anti-K1 antibody, followed by immunoblotting with the indicated antibodies (Right). (C) iSLK-BAC16 KSHV WT and iSLK-BAC16 ΔK1 (ΔK1) cells were stimulated with doxycyline (Dox, 1 μg/mL) and sodium butyrate (NaB, 1 mM) to induce viral reactivation. Cell extracts were immunoprecipitated with an anti-K1 antibody, followed by immunoblotting with the indicated antibodies. (D) Representative confocal fluorescence images of K1. HeLa cells expressing K1 were stained with anti-K1 and anti-mitofilin antibodies. Merged images show K1 (green), mitofilin (red), and nucleus (blue). (Scale bars: 10 μm.) (E) Purification of recombinant PYCR2 complex from E. coli. The arrowhead indicates the His-PYCR2 and the asterisks indicate GST and GST-K1. As a control, the purified recombinant GST protein was added to His-PYCR2. (F) PYCR2 enzymatic reaction. (G) Enzyme activity plots of purified His-PYCR2 with GST or GST-K1. Right graph shows Michaelis–Menten model for His-PYCR2 enzyme activity with GST or GST-K1. Data are presented as the mean ± SD. (H) Enzyme activity plots of purified His-PYCR2 with GST or GST-K1 in presence of proline and ATP. Data are presented as the mean ± SD. (I) MS-based metabolomics analysis of intracellular amino acids. HEK293T cells were collected 48 h after transfection with mock or K1 expression vector, and the amounts of intracellular amino acid were determined by LC-MS (n = 5). Log2 fold change values (K1/mock) data are presented as the mean ± SEM. **P < 0.01 and ***P < 0.001, by Student’s t test.
GST-K1(C) and His-tagged PYCR2 (His-PYCR2) protein were bacterially purified for in vitro binding to demonstrate the direct interaction. Unphosphorylated GST-K1(C) was purified from Escherichia coli BL21 strain, and tyrosine-phosphorylated GST-K1(C)pY was purified from E. coli TKX1 strain carrying promiscuous protein tyrosine kinase. Immunoblotting analysis showed that over 90% of GST-K1(C)pY protein purified from TKX1 strain underwent tyrosine phosphorylation (SI Appendix, Fig. S3 C, Upper). Either GST-K1(C) or GST-K1(C)pY directly interacted with His-PYCR2 in vitro at an equivalent level (SI Appendix, Fig. S3 C, Lower). Upon viral lytic reactivation, K1 interaction with endogenous PYCR1 and PYCR2 was readily detected in iSLK-BAC16 KSHV WT cells, but not in iSLK-BAC16 KSHV ΔK1 (K1 deletion mutant virus) cells (Fig. 2C). All subtypes of K1, including A (North America), B (Africa), C (Europe and Asia), and D (Pacific island) KSHV subtypes, based on their geographic sequence variation, bound endogenous PYCR1 and PYCR2 at similar levels (SI Appendix, Fig. S3D) (32). In contrast, other ITAM-containing herpesviral oncoproteins, Epstein-Barr virus (EBV) latent membrane protein 2 (LMP2) and Rhesus monkey rhadinovirus (RRV) R1, showed no interaction with PYCR1/2 (SI Appendix, Fig. S3E).
As it has been reported that a majority of PYCR1/2 were detected in mitochondria, we analyzed the subcellular location of K1 and PYCR complex. A majority of K1 localizes at the plasma membrane, but it is also detected in the cytosol (33, 34). Indeed, the subcellular fractionations showed that while half of K1 was localized in the plasma membrane fractions, the other half of K1 was detected in the mitochondrial fractions where PYCR1 and PYCR2 were primarily localized (SI Appendix, Fig. S4A). Confocal analysis further showed four different subtypes of K1 also colocalizing with PYCR2 at the mitochondria (Fig. 2D and SI Appendix, Fig. S4B). Collectively, these results demonstrate the specific interaction of KSHV K1 and cellular PYCR1/2 at the mitochondria.
K1 Enhances PYCR Enzyme Activity and Proline Synthesis.
To examine the role of K1 in PYCR enzymatic activity, His-tagged PYCR2 was copurified with GST or GST-K1(C) in E. coli and subjected to in vitro enzyme assay with P5C substrate. PYCR enzymatic activity was monitored by measuring the reduction of NADH (Fig. 2 E and F). This showed that the presence with K1 WT considerably increased PYCR2 enzymatic activity—decreasing KM = 4.419 to KM = 1.065 (Fig. 2G). Furthermore, the proline-mediated feedback inhibition or ATP-mediated competitive inhibition of PYCR2 enzymatic activity was considerably lower in the presence of GST-K1 than in the presence of GST (Fig. 2H). These results suggest that K1 interaction not only enhances PYCR2 enzymatic activity but also renders PYCR2 less sensitive to proline- or ATP-mediated inhibition. Finally, LC-MS indeed showed that K1 expression significantly increased intracellular proline the most with marginally significant changes in other amino acid levels (Fig. 2I).
Given that KSHV infection induced the proline metabolism pathway in 3D cell culture, we investigated the possibility that K1-PYCR interaction regulates proline metabolism in KSHV-infected cells. Although K1 is significantly expressed in lytic phase, a low level of K1 mRNA has been detected in latently infected cells and Kaposi’s sarcoma (35, 36). We wanted to determine the expression of K1 in KSHV-infected cells in 2D and 3D culture. Interestingly, we found higher expression of K1 transcript in TIME and MCF10A 3D culture than 2D culture (SI Appendix, Fig. S5 A and B). Next, to evaluate the effect of K1 on proline metabolism in KSHV-infected cells, we utilized TIME cells infected by KSHV mutant lacking cytoplasmic domain of K1 (KSHV K1 ΔC). Metabolomics analysis revealed that most metabolites in the proline metabolism pathway that were induced by KSHV WT were significantly decreased in KSHV K1 ΔC-infected cells in 3D culture (SI Appendix, Fig. S6). These data indicate that K1 interacts with and activates PYCR activity to increase intracellular proline synthesis.
K1 Enhances Cell Growth in 3D Spheroid Culture in a PYCR Binding-Dependent Manner.
Previous studies have shown that PYCR-deficient cells proliferate normally in 2D monolayer culture in vitro, but they show impaired growth in 3D spheroid culture and fail to develop tumors in vivo (3). This suggests that 3D spheroid cultures are more physiologically relevant than 2D monolayer cultures, as they more closely represent the metabolic changes, microenvironments, cell-to-cell interactions, and biological processes that occur in vivo. Given that KSHV specifically alters proline metabolism in 3D cell culture and K1 enhances proline synthesis by interacting with PYCR, we tested whether K1 expression affected the growth of red fluorescence protein (RFP)-positive TIME (R-TIME) cells in 3D spheroidal culture conditions. R-TIME cells expressing empty vector (mock), K1 WT, K1 ΔC, or K1 TYF were cultured in 2D monolayer or cocultured with human foreskin fibroblasts (HFF) in 3D spheroid conditions (SI Appendix, Fig. S7A). While none of the R-TIME cells showed growth difference in 2D monolayer conditions (SI Appendix, Fig. S7B), R-TIME cells expressing K1 WT or K1 TYF displayed considerably enhanced growth in 3D spheroid conditions compared to R-TIME cells expressing vector or K1 ΔC (Fig. 3A). Besides 3D spheroid culture, we used bioprinted 3D hydrogel scaffolds that were fabricated as organized hollow vessel-like structures where endothelial cells were guided to grow into 3D vascular network (SI Appendix, Fig. S8A). Overtime, the endothelial cells extended lumen-like structure lining within the hollow microchannels of the scaffold and continuously grew along the hollow microchannels of scaffold, recapitulating in vivo endothelial structures (SI Appendix, Fig. S8B). When R-TIME cells were seeded into 3D-printed hydrogel scaffolds, all R-TIME cells grew into 3D vascular lumen-like structures. However, R-TIME-K1 or R-TIME-K1 TYF cells grew much faster and more extensively along the hollow scaffold and denser into the hollows of scaffold than R-TIME vector or R-TIME K1 ΔC cells. The average volumes of R-TIME K1 or R-TIME K1 TYF cells inside the hollow microchannels were 1.5-fold higher than those of R-TIME vector or R-TIME K1 ΔC cells (Fig. 3B).
Fig. 3.
K1 promotes cell growth in 3D spheroid culture system. (A) Representative pictures and size quantification of 3D spheroids of mock-, K1 WT-, or mutant-expressing TIME and HFF cells cocultured in ultralow attachment condition. (Scale bars: 200 μm.) *P < 0.05, by one-way ANOVA. (B) A 3D confocal microscopy image of R-TIME-cultured hydrogel scaffold. Bottom shows the average of total fluorescence intensity at day 9 normalized to day 2 (seeding amount). *P < 0.05 and **P < 0.01, by one-way ANOVA. The 3D reconstruction and analysis of fluorescence intensity from multiple z-series images were performed using ImageJ. (C) Representative pictures and size quantification of 3D spheroids of mock-, K1 WT-, or mutant-expressing MCF10A cells. (D) MDA-MB-231 cells cultured in ultralow attachment condition. (Scale bars: 200 μm.) *P < 0.05; **P < 0.01; ****P = 0.0001, by one-way ANOVA. (E) ATP production of MCF10A cells expressing mock, K1 WT, or mutant. *P < 0.05, by one-way ANOVA. (F) Representative pictures (Upper) and size quantification (Lower) of 3D spheroids of uninfected, KSHV WT-infected, or KSHV K1 ΔC-infected or KSHV K1 TYF-infected TIME cells cultured in Matrigel for 12 d. (Scale bars: 100 μm.) The number of biological replicates for each experiment was n ≥ 3. **P = 0.0001, by one-way ANOVA. (G) Representative images of uninfected, KSHV WT-infected, or KSHV K1 ΔC-infected or KSHV K1 TYF-infected MCF10A cells cultured on Matrigel for 21 d. (Upper) Acini were stained with the nuclei counterstained with Hoechst 33342 (blue) and Ki67 antibody (red). GFP is from the BAC16 KSHV genome. (Scale bars: 100 μm.) (Lower) Quantification of Ki67 positive cells in acini. The number of biological replicates for each experiment was n ≥ 3. **P < 0.01 and ****P = 0.0001; ns, not significant, by one-way ANOVA.
As PYCR has been extensively characterized in epithelial cells (37) and epithelial cells are the primary cell type for KSHV infection (38), we included nontumorigenic MCF10A and tumorigenic MDA-MB-231 epithelial cells expressing vector, K1 WT, K1 ΔC, or K1 TYF for growth transformation in 3D culture. As seen with R-TIME cells, MCF10A or MDA-MB-231 cells expressing vector, K1 WT, or its mutant showed no growth difference in 2D monolayer cultures (SI Appendix, Fig. S7 C–F). In contrast, K1 WT- or K1 TYF-expressing MCF10A or MDA-MB-231 cells displayed considerably larger spheroids in 3D cells cultures than vector- or K1 ΔC-expressing MCF10A or MDA-MB-231 cells (Fig. 3 C and D). The enhanced spheroid growth was also evidenced by higher ATP production in MCF10A cells expressing K1 or K1 TYF than in MCF10A cells expressing vector or K1 ΔC (Fig. 3E). To further investigate K1 activity in the context of viral genome, we examined the 3D spheroid and acini formation of KSHV-infected TIME and MCF10A cells. KSHV WT- or KSHV K1 TYF-infected TIME cells showed lager spheroids formation in 3D matrigel culture than uninfected or KSHV K1 ΔC-infected TIME cells (Fig. 3F). Similarly, KSHV WT- or KSHV K1 TYF-infected MCF10A cells showed enhanced proliferation in 3D matrigel culture than uninfected or KSHV K1 ΔC-infected MCF10A cells (Fig. 3G). KSHV K1 ΔC-infected MCF10A acini are also larger in size compared to uninfected acini, but retain a low proliferation rate. It suggests other oncogenic factors than K1 might contribute to elicit distinct morphological phenotypes in KSHV-infected MCF10A acini. These results collectively demonstrate that expression of K1 WT or K1 TYF mutant individually or in the context of viral genome leads to enhanced growth in 3D culture conditions.
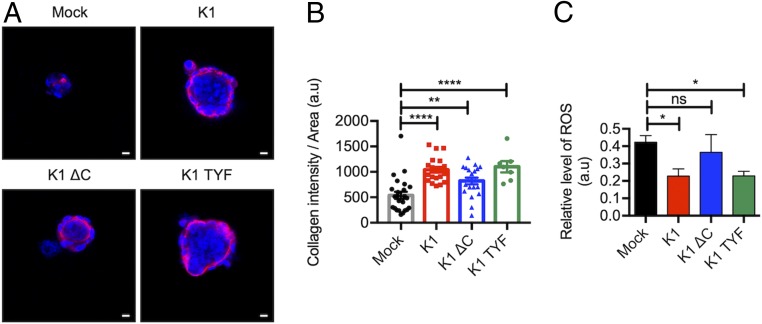
It has been well studied that proline is stored as collagen in extracellular matrix (39). To investigate the role of K1-induced proline production in collagen synthesis, the collagen accumulation was examined in 3D spheroids. We found that K1- and K1 TYF-expressing spheroids induced higher accumulation of collagen type I compared to vector or K1 ΔC-expressing spheroids (Fig. 4 A and B). Proline has also been shown to modulate the intracellular redox environment and protect mammalian cells by reducing ROS. When intracellular ROS level was measured by CellROX staining, K1- and K1 TYF-expressing spheroids showed detectably reduced ROS levels compared to vector or K1 ΔC-expressing spheroids (Fig. 4C). These results suggest a potential role of K1-induced proline production in the regulation of collagen synthesis and ROS homeostasis in 3D culture conditions.
Fig. 4.
K1 induces collagen synthesis and reduces ROS in 3D spheroids. (A) Representative confocal image of collagen I (red) and K1 (green) immunofluorescence on mock-, K1 WT-, or mutant-expressing TIME and HFF cell cocultured spheroids in ultralow attachment condition. Nuclei were stained with Hoechst 33342 (blue). (Scale bars: 10 μm.) (B) Quantification of collagen I signal intensity in 3D spheroids. Collagen intensity of spheroids was divided by areas of spheroids. Data are mean ± SEM. **P < 0.01 and ****P = 0.0001, by one-way ANOVA. (C) Intracellular ROS level in mock-, K1 WT-, or mutant-expressing TREX 293T cell spheroids. a.u., arbitrary units. Data are mean ± SEM. *P < 0.05; ns, not significant, by one-way ANOVA.
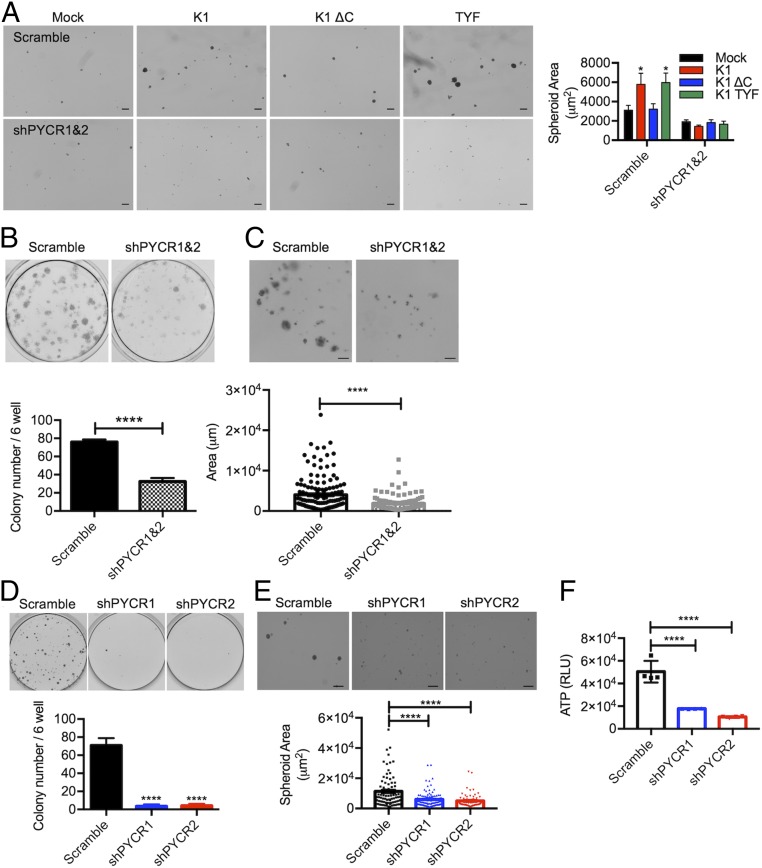
To test a direct role of PYCR in K1- or K1 TYF-induced growth transformation of R-TIME cells in 3D spheroid cultures, we depleted expression of PYCR1 and PYCR2 by their specific shRNAs (SI Appendix, Fig. S7G). This showed that depletion of the PYCR1/2 expression abrogated 3D spheroid growth of R-TIME K1 or R-TIME K1 TYF cells, whereas R-TIME vector or R-TIME K1 ΔC cells showed only slightly reduced growth upon depletion of the PYCR1/2 expression (Fig. 5A). On the other hand, depletion of the PYCR1/2 expression led to no detectable effect on the growth of R-TIME cells in 2D culture conditions (SI Appendix, Fig. S7H). Next, we aimed to identify the functions of PYCR in the context of KSHV infection; thus, we depleted expression of PYCR1 and PYCR2 in KSHV-infected TIME cells (SI Appendix, Fig. S7I). Decreasing PYCR1 and PYCR2 expression significantly reduced the number of foci and the size of spheroids in low-density culture and 3D culture of KSHV-infected TIME cells (Fig. 5 B and C). Finally, as KSHV efficiently transformed primary rat embryonic metanephric mesenchymal precursor (MM) cells (40), KSHV-transformed MM (KMM) cells efficiently formed a number of foci and spheroids in low-density culture and 3D culture, respectively. However, the shRNA-mediated depletion of PYCR1 or PYCR2 expression (SI Appendix, Fig. S7J) drastically depleted the foci and spheroid-forming activity of KMM cells in low-density culture and 3D culture (Fig. 5 D–F). These results indicate that PYCR1 and PYCR2 play a critical role in K1-induced growth transformation in 3D culture.
Fig. 5.
PCYR expression and interaction are essential for K1-mediated transformation. (A) Representative pictures of mock-, K1 WT-, or mutant-expressing TIME cells treated with scramble shRNA (scramble) or PYCR-specific shRNA (shPYCR1/2) in ultralow attachment condition. Right graph shows size quantification of the spheroids. (Scale bars: 200 μm.) *P < 0.05, by two-way ANOVA. (B) Scramble- or shPYCR-treated KSHV-infected TIME cells were subjected to low-density culture (1,500 cells per six-well plates) for clonogenic assay. Representative pictures for colony growth are shown (Upper). Quantification of the number of colonies is shown (Lower). Data are presented as the mean ± SD. ****P < 0.0001, by Student’s t test. (C) Representative pictures and size quantification of 3D spheroids of scramble- or shPYCR-treated KSHV-infected TIME cells cultured in ultralow attachment condition with 4% Matrigel. (Scale bars: 100 μm.) Data are presented as the mean ± SEM. ****P < 0.0001, by Student’s t test. (D) Scramble- or shPYCR-treated KMM cells were subjected to low-density culture (1,000 cells per six-well plates) for clonogenic assay. Representative pictures for colony growth are shown (Upper). Quantification of the number of colonies is shown (Lower). Data are presented as the mean ± SD. ****P = 0.0001, by one-way ANOVA. (E) Representative pictures and size quantification of 3D spheroids of scramble control- or shPYCR-treated KMM cells cultured in ultralow attachment condition. (Scale bars: 400 μm.) Data are presented as the mean ± SEM. ****P = 0.0001, by one-way ANOVA. (F) Intracellular ATP levels of scramble control or shPYCR KMM cells are seeded in ultralow attachment condition. Intracellular ATP levels were measured as an indicator of cell viability using CellTiter-Glo reagents after 4 d of shRNA treatment. Data are presented as the mean ± SD. The number of biological replicates for each experiment was n ≥ 3. ****P = 0.0001, by one-way ANOVA.
K1-Induced Proline Metabolism within In Vivo Tumors.
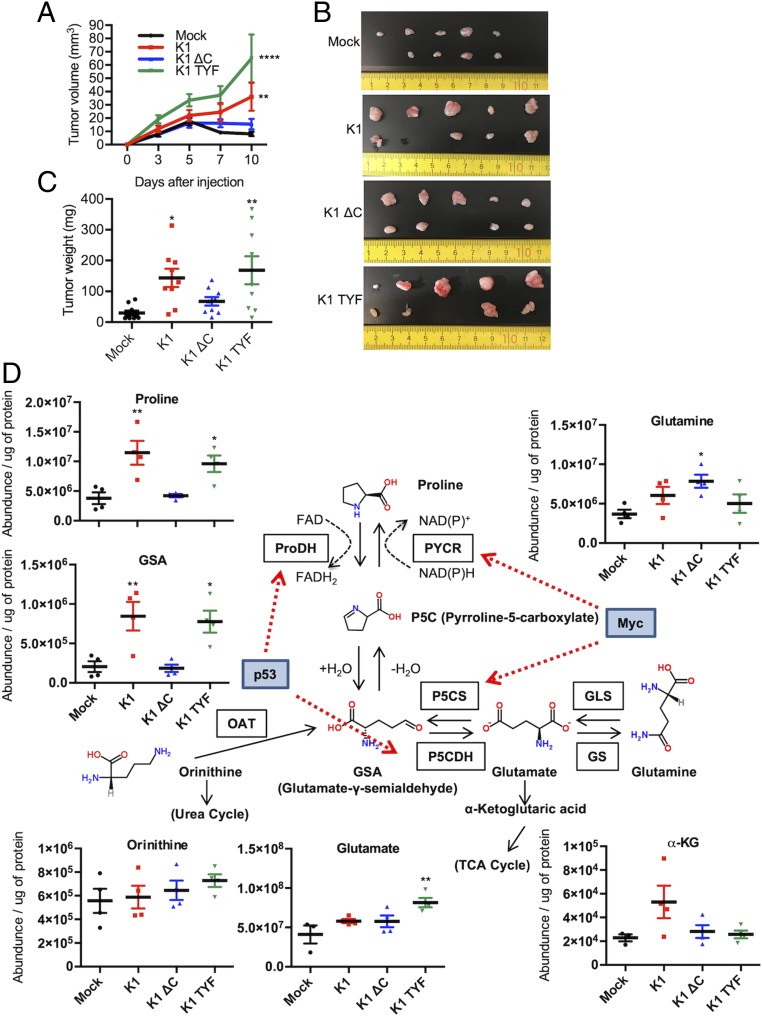
As MDA-MB-231 cells are tumorigenic in nude mice, MDA-MB-231 cells expressing vector, K1 WT, K1 ΔC, or K1 TYF were s.c. injected into nude mice, followed by measuring tumor development, volume, weight, and metabolites. The results showed that K1 WT- or K1 TYF-expressing MDA-MB-231 cells formed significantly larger tumors in volume and weight compared to vector- or K1 ΔC-expressing MDA-MB-231 cells in nude mice (Fig. 6 A–C). The metabolomics profile of each tumor revealed that the levels of nonessential amino acids, including proline, glycine, and arginine, were significantly elevated in K1 WT- or K1 TYF-induced tumors as compared to those in vector- or K1 ΔC-induced tumors, indicating the contribution of K1-mediated proline biosynthesis activity to the tumorigenesis in vivo (SI Appendix, Fig. S9). As aggressive tumors rely on glycine and arginine for their growth (41, 42), K1 WT and K1 TYF tumors may also increase glycine and arginine pools along with proline for their growth. Additionally, glycine can be derived from increased collagen deposit due to enhanced proline metabolism, since glycine is one of the components of collagen along with proline and hydroxyl proline. Metabolite analysis of the proline synthesis pathway also revealed that intratumor proline and glutamate-γ-semialdehyde (GSA) levels were markedly higher in K1- or K1 TYF-induced tumors than in vector- or K1 ΔC-induced tumors (Fig. 6D). As GSA is a nonenzymatically interconvertible form of P5C, K1-induced proline synthesis and subsequent proline catabolism appeared to contribute to the increase of in vivo tumor formation. Thus, these results demonstrate that K1-induced growth transformation is evident only in 3D spheroid culture and in vivo tumor formation, but not in 2D monolayer culture.
Fig. 6.
Proline metabolic differences between in vivo tumors. (A) MDA-MB-231 cells (1 × 106 cells) expressing mock, K1 WT, or mutants were injected s.c. into nude mice (n = 9 to 10). Tumor volume was plotted as indicated. Data are presented as the mean ± SEM. **P < 0.01 and ****P = 0.0001, by two-way ANOVA. At 11 d after injection, xenograft tumors were (B) harvested and (C) weighed. Data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01, by one-way ANOVA. (D) Metabolite abundance of proline metabolism pathway in mock-, K1 WT-, or mutant-expressing MDA-MB-231 tumors measured by LC-MS. Levels of metabolites are related to proline synthesis. Data are presented as mean ± SEM. *P < 0.05 and **P < 0.01, by one-way ANOVA.
Discussion
Metabolic reprogramming plays a major role in cancer cell growth, maintenance, and metastasis. Therefore, a better understanding of metabolic changes in cancer cells may contribute to the therapy targeting tumor metabolism. However, in vitro cancer metabolism is not only readily affected by cell types and culture conditions, but also fundamentally different from in vivo cancer metabolism (43). While 2D monolayer cells are exposed to abundant and even amounts of nutrients, in vivo tumors expose gradients in nutrient, oxygen, and pressure, which eventually leads to differences in cell proliferation rate within tumors (44). Besides the biological gradient, 2D monolayer cells interact with both plastic surface of culture dish and nearby equivalent cells, but the microenviroment of in vivo tumors consists of extracellular matrix and heterogeneous stromal cells that release a number of signals, nutrients, and metabolic wastes (45). These characteristics ultimately generate different metabolic phenotypes and drug responses (46). In fact, culturing KSHV-infected cells in 3D conditions has been shown to carry different gene expression profiles compared to those in 2D conditions (47). To minimize the difference between in vitro and in vivo tumor metabolic phenotype, we conducted untargeted metabolomics of KSHV-infected cells in a 3D culture model that better represents the physical microenviroment of tumors than 2D monolayer culture. KSHV latent infection has been previously shown to be associated with the increases of glucose and glutamine uptake, lactic acid production, and fatty acid synthesis in 2D culture (18, 48, 49). However, metabolic reprogramming of KSHV-infected cells has not been studied in 3D culture. We found that alanine, aspartate, and glutamate metabolism, and arginine and proline metabolism, were highly up-regulated in both KSHV-infected TIME and MCF10A cells compared to uninfected TIME and MCF10A cells when they were grown in 3D culture. This suggests that those nonessential amino acids play specific roles in KSHV-infected cell growth in 3D culture conditions but not in 2D culture conditions.
The proline biosynthesis pathway includes P5C as an intermediate synthesized from glutamine or ornithine. P5C is converted to proline by PYCR while producing NAD(P)+. The proline catabolism pathway is initiated by PRODH with formation FADH2 that serves as a substrate of the electron transport chain. Overexpression of PRODH generates ROS and induces apoptosis (50), while knockdown of PYCR decreases cancer cell growth and proliferation (51). There are three isoforms of PYCR in human: PYCR1 and PYCR2 are very similar (84% similarity) and localized in mitochondria, whereas PYCRL has a 45% similarity with the other two forms and cytosol localization. Among isoforms of PYCR, PYCR1 has been studied as the most overexpressed metabolic enzyme in cancer (52), but expression level of both PYCR1 and PYCR2 are high in various types of cancers according to the Human Protein Atlas database (https://www.proteinatlas.org). Indeed, recent studies have shown that PYCR1 and PCYR2 both protect cells from oxidative stress and support tumorigenesis (53, 54). Moreover, cells that lack PYCR proliferate normally in 2D monolayer culture, while showing impaired growth in 3D spheroid culture and failing to develop tumors in vivo (3, 51), indicating that PYCR-mediated proline metabolism is critical for tumorigenesis. Indeed, expressions of the proline biosynthesis PYCR gene and proline degradation ProDH gene are up-regulated directly by c-Myc oncoprotein and p53 tumor suppressor (11, 13), respectively. This collectively demonstrates that the proline-P5C metabolic axis is a key checkpoint for tumor cell growth and a potential target for adjunctive cancer therapy (55).
We identified the proline synthesis enzyme PYCR1/2 as the K1-binding protein in an ITAM-independent manner. Consequently, K1 interaction not only activated PYCR activity to increase intracellular proline synthesis, but also renders PYCR less sensitive to proline- or ATP-mediated inhibition. The proline metabolism pathway is involved in supporting ATP production, protein, and nucleotide synthesis, anaplerosis, and redox homeostasis in cancer cells (3, 12, 56). Especially, proline functions as a stress scavenger under conditions of metabolic, oxidative, and nutrient stresses that are similar to intrinsic properties of the tumor environment (57). Indeed, the disruption of the proline synthesis pathway in cancer cells prevents 3D spheroid formation and tumorigenesis in nude mice, while PYCR-deficient cells proliferate normally in 2D monolayer culture (3, 51). We also observed that K1-PYCR interaction enhanced in vitro 3D spheroidal growth and in vivo tumorigenesis, while it had no effect on cell growth in in vitro 2D culture. Thus, our study further emphasizes the difference of tumor metabolisms between 3D conditions and 2D conditions, and the importance of proline metabolism in 3D conditions that reflects actual metabolic challenges of KSHV-induced tumor cells.
Proline metabolism is critically important for tumor metastasis since PYCR is highly overexpressed in various metastasized tumors. There are several host proteins, DJ-1, ORAOV1, and Kindlin-2 that carry oncogenic characters that have also been shown to cooperate with PYCR to promote cancer cell survival (53, 58, 59). This study shows that KSHV K1 is a viral oncoprotein that interacts with and activates PYCR enzyme activity, which ultimately enhances cell growth in 3D spheroid culture as well as tumorigenesis in nude mice. This suggests that K1 and PYCR interaction may be a potential therapeutic target against KSHV-derived tumorigenesis.
Materials and Methods
Cell Lines.
Cells were maintained at 37 °C in a humidified incubator with 5% CO2. HEK293T, iSLK, HeLa, TIME, HFF, MCF10A, and MDA-MB-231 cell lines were purchased from ATCC. MM and KMM cells were kindly provided by Shou-Jiang Gao, University of Pittsburgh, Pittsburgh, PA. HEK293T, iSLK, HeLa, HFF, MM, KMM, and MDA-MB-231 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Seradigm), and 1% penicillin/streptomycin (Gibco). For KMM cells, 250 μg/mL of hygromycin was used to maintain the KSHV genome. iSLK cells were cultured in the presence of 1 μg/mL of puromycin and 250 μg/mL of G418. BAC16, BAC16 ΔK1, BAC16 K1ΔC, and BAC16 K1 TYF were introduced into iSLK cells via transfection with Fugene HD reagent (Roche) and then selected with 200 μg/mL of hygromycin (Invitrogen). TIME cells were cultured in VascuLife vascular endothelial growth factor endothelial medium (LIFELINE). TIME cells infected with KSHV were selected and maintained with 10 μg/mL of hygromycin. MCF10A cells were cultured in DMEM-F12 supplemented with 5% horse serum (Thermo Fisher), EGF 20 ng/mL (Peprotech), insulin 10 μg/mL (Sigma), hydrocortisone 0.5 mg/mL (Sigma), cholera toxin 100 ng/mL (Sigma), and 1% penicillin/streptomycin (Gibco). MCF10A cells infected with KSHV were selected and maintained with 100 μg/mL of hygromycin.
Mice.
All experiments were approved and done according to the guidelines of the Institutional Animal Care and Use Committee at the University of Southern California. NCr nude mice were purchased from Taconic. Tumors were initiated by s.c. injection of K1 and K1 mutants expressing MDA-MB-231 cells (1 × 106 cells) expressing K1 or K1 mutants into the flank of NCr nude mice. Mice were killed after 11 d after the injection of tumor cells.
Plasmids.
The constructs encoding GST-K1(C) and a series of GST-PYCR truncates for GST pull-down were generated in pEBG vector to fuse GST to the N-terminal of the K1 cytoplasmic domain and PYCR truncates. For in vitro PYCR enzyme assay, full-length PYCR2 was introduced to the MCS1 site of pRSFDuet-1 vector (Novagen) and GST or GST-K1(C) was introduced to the MCS2 site after removing S-tag from the MCS2 site. The full-length of K1, K1 ΔC, and K1 TYF constructs were made in pCDH-hygro vector. K1 TYF mutant was obtained by site-directed mutagenesis with traditional PCR. For knockdown of PYCR1 and PYCR2, shRNAs that target human PYCR1 (5′-TGTCCTTGAGCTGGCCTGG-3), human PYCR2 (5′-CCATGCCAGCTTAAGGACAAT-3), rat PYCR1 (5′-CATTGAGGACAGGCACATTGT-3), rat PYCR2 (5′-CTGTCGGCTCACAAGATAATA-3), or scrambled sequence were introduced into pLKO vectors.
Protein Expression and Purification.
GST fusion proteins were purified from either E. coli strain BL21 or TKX1 transformed with pGEX-K1(C) plasmid. Protein expression was induced by adding 0.25 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 37 °C and cells were further incubated for 4 h. Cell pellets were resuspended in binding buffer (20 mM Hepes [pH 7.4], 100 mM NaCl, 1% Nonidet P-40, protease inhibitors), lysed by adding lysozyme, followed by sonication. Lysate was cleared by centrifugation and then applied to GST4B beads (GE Healthcare) at 4 °C for 2 h. GST4B beads were washed four times with binding buffer, and the proteins associated with the beads were eluted by 10 mM glutathione. Recombinant His-PYCR2 protein was purified from E. coli strain BL21 transformed with pRSFDuet-PYCR2 plasmid. The protein expression was induced by the addition of 0.25 mM IPTG at 37 °C and cells were further incubated for 4 h. Cell pellets were resuspended in binding buffer (50 mM NaH2PO4 [pH 7.4], 0.5 M NaCl, 10 mM imidazole), lysed by adding lysozyme, followed by sonication. Lysates were cleared by centrifugation. The cleared lysate was applied to Ni-NTA agarose beads (Life Technologies) for 4 °C for 4 h. Ni-NTA agarose beads were washed four times with wash buffer (50 mM NaH2PO4 [pH 7.4], 0.5 M NaCl, 20 mM imidazole), and the proteins associated with the beads were eluted by elution buffer (50 mM NaH2PO4 [pH 7.4], 0.5 M NaCl, 250 mM imidazole).
GST Pull-Down Assay.
GST fusion proteins purified from either E. coli strain BL21 or mammalian cell (BJAB, HEK293T) lysates were incubated with GST4B beads (GE Healthcare) in binding buffer (20 mM Hepes [pH 7.4], 100 mM NaCl, 1% Nonidet P-40, protease inhibitors) at 4 °C for 2 h. GST4B beads were then washed four times with binding buffer, and the proteins associated with the beads were analyzed by SDS/PAGE and subjected to Coomassie blue staining, Western blot assay and peptide sequencing was performed at the Harvard University mass spectrometry facility.
Immunoprecipitation and Western Blotting.
For immunoprecipitation, cells were harvested 48 h after polyethylenimine (PEI) transfection and lysed in RIPA buffer (500 mM Tris⋅HCl, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 1 mM EDTA, protease inhibitor). Cell extracts were precleared with protein A/G agarose for 1 h at 4 °C and subsequently incubated with anti-K1 (3H4) (24) for 4 h at 4 °C, followed by incubation with protein A/G agarose (Thermo Fisher) for 1 h at 4 °C. Immunoprecipitates were washed with RIPA lysis buffer and resuspended in SDS sample buffer, boiled at 95 °C, resolved on SDS/PAGE gels, and transferred onto polyvinylidene difluoride (PVDF) membranes. Antibody concentrations were as follows: anti-K1(3H4), 1:1,000; anti-PYCR1 (Abcam), 1:1,000; anti-phosphotyrosine antibody (Millipore), 1:1,000; anti-PYCR2 (LSBio), 1:2,000; anti-GST (Santa Cruz), 1:1,000; anti-mitofilin (Proteintech), 1:1,000; anti-β-actin (Santa Cruz), 1:1,000; anti-histone H3 (Santa Cruz), 1:1,000; and secondary antibodies, affinity-purified with horseradish peroxidase conjugate, 1:5,000. Images were developed with ECL reagent (Thermo Scientific) and imaged on a Bio-Rad ChemiDoc-Touch.
Subcellular Membrane Fractionation.
HEK293T cell lines were grown in 5 × 15 cm tissue culture plates and transfected with empty vector (mock) or full-length K1 (K1) using PEI. At 48 h after transfection, the medium was aspirated, and cells were washed twice with phosphate-buffered saline (PBS). Cells were scraped in cold fractionation buffer (0.25 M sucrose, 1 mM EDTA, 1 mM EGTA, 10 mM KCl, 1.5 mM MgCl2, 20 mM Hepes [pH 7.4], protease inhibitor, 1 mM dithiothreitol [DTT]). Cells were then homogenized by passing through a 25-gauge syringe 10 times. After incubation on ice for 20 min, the homogenate was spun at 3,000 × g for 10 min at 4 °C, and the supernatant were transferred into a fresh tube. The nuclear pellet was washed with fractionation buffer again. The pellet was dispersed with a pipette and passed through a 25-gauge syringe 10 more times, and the nuclear pellet was resuspended in nuclear buffer (standard lysis buffer with 10% glycerol and 0.1% SDS added). Supernatant was centrifuged at 8,000 rpm (10,000 × g) and the supernatant was used as cytosolic and membrane fraction. For mitochondrial fraction, the saved pellet was resuspended in the nuclear buffer as above. For membrane fraction, the supernatant was centrifuged in an ultracentrifuge at 40,000 rpm (100,000 × g) for 1 h. A total of 400 μL of the fractionation buffer was added to this pellet, resuspended by passing through a 25-gauge syringe as above, and then recentrifuged for 45 min. The resulting membrane pellet was resuspended in the same buffer as the nuclei and mitochondria.
Cell Culture in 3D Spheroid.
To generate 3D spheroid, MCF10A or MDA-MB-231 or TIME cells with 4% Matrigel (Corning) or TIME and HFF cell coculture (1:1 ratio) or TREX 293T cells were seeded at 3 × 104 cells per six-well ultralow attachment plates (Corning). Spheroid areas were measured by Fiji software. Cell viability was measured using CellTiter-Glo reagents (Promega). To detect intracellular ROS, spheroids were incubated with CellROX Green Reagent (Life Technologies) for 30 min and fluorescence was measured using a plate reader (Molecular Device). The ROS amount was normalized as to number of viable cells (luminescent value from CellTiter-Glo). For 3D spheroid culture of MCF10A cell on Matrigel, 5,000 MCF10A cells were resuspended in assay medium (DMEM-F12 supplemented with 2% horse serum, 10 μg/mL insulin, 100 ng/mL cholera toxin, 0.5 μg/mL hydrocortisone, 50 U/mL penicillin, 50 μg/mL streptomycin, 5 ng/mL EGF, and 2% Matrigel) and seeded on an eight-chamber slide coated with Matrigel. Cultures were fed every 3 to 4 d with assay medium.
Immunofluorescence.
HeLa cells were plated on coverslips overnight and transfected with full-length K1 by using FugeneHD. At 48 h after transfection, cells were stained with MitoTracker Deep Red FM (100 nM) for 45 min. The 3D spheroid of MCF10A cells on Matrigel were cultured in eight-well chamber slides for immunofluoresence. TREx293 cell spheroids obtained by culturing in ultralow attachment plates (Corning) were collected and transferred to 1.5-mL tubes for further staining procedures. After washing with PBS, cells were fixed using 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. After blocking with 10% goat serum in PBS, cells were stained with anti-K1(2H5) (24), anti-Flag, anti-PYCR2, anti-Ki67 (Thermo Fisher), anti-collagen I (Genetex), and anti-mitofilin diluted in 1% BSA-PBS for 2 h. Appropriate fluorescence-conjugated secondary antibodies from goat were incubated for 1 h. DNA was stained with 1 μg/mL Hoechst 33342 and coverslips were mounted with Fluoromount-G (Thermo Scientific) mounting media. Stained spheroids were transferred to µ-Dish 35 mm (ibidi) for imaging. Cells and spheroids were examined under a Nikon Eclipse Ti confocal microscope.
PYCR In Vitro Enzyme Assay.
Recombinant purified His-PYCR and GST/GST-K1(C) were dialyzed in enzyme reaction buffer (300 mM Tris, pH 8.0 containing 0.01% Brij 35) and used for in vitro enzyme assay. Enzymatic assays were measured by continuous monitoring of NADH consumption (Sigma) at 340 nm in the conversion reaction of Δ1-Pyrroline-5-carboxylic acid (P5C, Angene International Limited) to proline by a FiterMax F5 plate reader (Molecular Devices).
Metabolite Extraction and LC-MS Analysis.
Transfected HEK293T cells, TIME, MCF10 2D cultured cells, or collected 3D spheroid cells were washed twice with cold PBS and fixed in 80% methanol (Fisher Chemical, LC-MS grade) precooled in dry ice. Cells were detached by scraping and then transferred to a 2-mL screw cap tube. For tumors, 1 mL of 80% methanol per 100 mg of tumor was added right after isolation, then homogenized with TissueLyser II (QIAGEN). Lysate of cells or tumors was quantified by bicinchoninic acid (BCA) assay and pelleted by centrifugation (12,000 rpm for 10 min at 4 °C). The supernatant was transferred into a new 2-mL screw cap tube and dried using a vacuum concentrator. Dried metabolites were resuspended in acetonitrile/methanol/water (40:40:20). Before applying LC-MS, metabolites were mixed with acetonitrile containing 0.2% folic acid solution and spun down at 12,000 rpm for 10 min at 4 °C. The supernatant was injected into Agilent Accurate Mass 6230 time-of-flight (TOF) coupled with Agilent 1290 LC system. Detected ions were deemed metabolites on the basis of unique accurate mass-retention time identifiers for masses exhibiting the expected distribution of accompanying isotopologues. The abundance of metabolites was extracted using Agilent Qualitative Analysis B.07.00 and Profinder B.08.00 software (Agilent Technologies) with a mass tolerance of <0.005 Da. Metabolism pathway analyses were carried out using Metaboanalyst 4.0 (https://www.metaboanalyst.ca/).
Lentivirus Induction and Making Stable Cell Lines.
Lentiviruses were produced in HEK293T cells using the pCDH-hygro system or pLKO shRNA system with PEI transfection. MCF10A, TIME, MM, and KMM cells were infected with lentiviruses containing 8 µg/mL polybrene. The inoculum was exchanged for fresh media after a 4-h incubation. To make stable cell lines, MCF10A cells were selected using 100 µg/mL of hygromycin, and TIME cells were selected using 20 µg/mL of hygromycin. We generated MCF10A and TIME cells expressing K1, K1 ΔC, K1 TYF, as well as control cells expressing empty pCDH-hygro vector (System Biosciences) by lentivirus transduction.
Quantitative RT-qPCR.
RNA was isolated using TRIzol reagent (Invitrogen), according to manufacturer’s instructions. Total RNA used for cDNA synthesis was obtained using iScript (Bio-Rad), according to manufacturer’s instructions. RT-qPCR using SyberGreen (Bio-Rad) was performed on CFX96 Real Time PCR (Bio-Rad), according to manufacturer’s instructions. Samples were run in triplicate and normalized to the housekeeping gene, beta-actin. Relative expression was calculated using the ΔΔCT method. Primer sequences are found in SI Appendix, Table S3.
Three-Dimensional Printing of the Hydrogel Scaffolds.
The 3D-printed hydrogel scaffolds were designed to feature the shape of a hexagonal prism (diagonal length: 3 mm; height: 3 mm) consisting of parallel microchannels (diameter: 150 µm) to study the 3D growth and vascular formation of seeded endothelial cells. The prepolymer solution for 3D printing was prepared by dissolving and mixing 10% poly(ethylene glycol) diacrylate (vol/vol), 7.5% gelatin methacrylate (wt/vol), and 0.25% lithium phenyl-2,4,6-trimethylbenzoylphosphinate (wt/vol) in sterile PBS. The hydrogel scaffolds were 3D printed using a digital light processing (DLP)-based rapid bioprinter described in previously published work (60). Briefly, the 3D bioprinter consists of the following major components: 1) a UV LED light source (365 nm) for photopolymerization; 2) a digital micromirror array device (DMD) for modulating the optical patterns projected to the fabrication plane for selective photopolymerization; 3) projection optics for projecting the optical patterns from the DMD chip to the fabrication plane; 4) a motorized stage; and 5) a computer controlling the light source, the DMD chip, and the motorized stage. The DMD chip is composed of ∼2 million micromirrors (1,920 × 1,080) which can be controlled by user-defined computer-aided design (CAD) models to produce 3D scaffolds with customized geometries and dimensions. DLP-based bioprinters print by projecting an entire plane of the optical pattern to the prepolymer without scanning line-by-line or drop-by-drop like in conventional nozzle-based bioprinters, which significantly reduces the printing time. Thus, a total time of 0.6 s was needed to print a single hydrogel scaffold used in this study. In addition, by focusing the UV light with the proper projection optics, microscale printing resolution can be achieved. To print the hydrogel scaffolds, the prepolymer solution was loaded into the fabrication stage and exposed to the UV optical pattern derived from the 3D CAD design. The 3D-printed hydrogel scaffolds were transferred to a 24-well plate and rinsed with PBS. The 1.5 × 105 R-TIME cells were loaded to the hydrogel scaffold and the vascular formation was tracked by RFP.
Quantification and Statistical Analysis.
The statistical tests were calculated using GraphPad Prism 6. Details of the specific statistical analysis are indicated in the figure legends.
Data and Software Availability.
PYCR1 and PYCR2 gene expression data in various type cancers were obtained from The Cancer Genome Atlas databank, through the cBioportal web-based utility (cbioportal.org/) (61, 62).
Supplementary Material
Acknowledgments
We thank Dr. Bok-Soo Lee for initiating the current work. We also thank Drs. Richard Longnecker and Blossom Damania for providing reagents. This work was partly supported by NIH grants (CA200422, AI073099, AI116585, AI129496, AI140718, AI140705, DE023926, DE027888, and DE028521); the Fletcher Jones Foundation grant (J.U.J.); NIH grants (DE027556 and CA221521) (P.F.); NIH grants (R01CA197153 and R01DE025465) (S.-J.G.); NIH grants (AR074763 and EB021857) (S.C.); the Wright Foundation award; and start-up funding from Department of Molecular Microbiology and Immunology, Keck School of Medicine, University of Southern California (H.E.).
Footnotes
Competing interest statement: J.U.J. is a scientific advisor of Vaccine Stabilization Institute, a California corporation.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918607117/-/DCSupplemental.
References
- 1.Langhans S. A., Three-Dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pampaloni F., Reynaud E. G., Stelzer E. H., The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Elia I., et al. , Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 8, 15267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantilla B. S., et al. , Proline metabolism is essential for trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog. 13, e1006158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyoshima K., et al. , Increased plasma proline concentrations are associated with sarcopenia in the elderly. PLoS One 12, e0185206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G., et al. , Proline metabolism in the conceptus: Implications for fetal growth and development. Amino Acids 35, 691–702 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Krishnan N., Dickman M. B., Becker D. F., Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 44, 671–681 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu N., et al. , Proline starvation induces unresolved ER stress and hinders mTORC1-dependent tumorigenesis. Cell Metab. 24, 753–761 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Tang L., et al. , Global metabolic profiling identifies a pivotal role of proline and hydroxyproline metabolism in supporting hypoxic response in hepatocellular carcinoma. Clin. Cancer Res. 24, 474–485 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Olivares O., et al. , Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat. Commun. 8, 16031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B., A model for p53-induced apoptosis. Nature 389, 300–305 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Liu W., Hancock C. N., Fischer J. W., Harman M., Phang J. M., Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci Rep 5, 17206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W., et al. , Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. U.S.A. 109, 8983–8988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L., et al. , cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 25, 429–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders P. M., Zhang Z., Bhende P. M., Giffin L., Damania B., The KSHV K1 protein modulates AMPK function to enhance cell survival. PLoS Pathog. 12, e1005985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Q., et al. , Kaposi’s sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1 alpha to upregulate RTA expression during hypoxia: Latency control under low oxygen conditions. J. Virol. 80, 7965–7975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado T., et al. , Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 10696–10701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado T., Sanchez E. L., Camarda R., Lagunoff M., Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 8, e1002866 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., et al. , An oncogenic virus promotes cell survival and cellular transformation by suppressing glycolysis. PLoS Pathog. 12, e1005648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson S. M., et al. , Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mesri E. A., Feitelson M. A., Munger K., Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 15, 266–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H., et al. , Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat. Med. 4, 435–440 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Lee B. S., et al. , Characterization of the Kaposi’s sarcoma-associated herpesvirus K1 signalosome. J. Virol. 79, 12173–12184 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee B. S., Connole M., Tang Z., Harris N. L., Jung J. U., Structural analysis of the Kaposi’s sarcoma-associated herpesvirus K1 protein. J. Virol. 77, 8072–8086 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sousa-Squiavinato A. C. M., Silvestre R. N., Elgui De Oliveira D., Biology and oncogenicity of the Kaposi sarcoma herpesvirus K1 protein. Rev. Med. Virol. 25, 273–285 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson C. C., Damania B., The K1 protein of Kaposi’s sarcoma-associated herpesvirus activates the Akt signaling pathway. J. Virol. 78, 1918–1927 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., et al. , The Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) K1 protein induces expression of angiogenic and invasion factors. Cancer Res. 64, 2774–2781 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Debnath J., Muthuswamy S. K., Brugge J. S., Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Debnath J., et al. , The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Richardson A. D., Yang C., Osterman A., Smith J. W., Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res. Treat. 110, 297–307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding J., et al. , Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 38, 519–531 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zong J. C., et al. , High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J. Virol. 73, 4156–4170 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B. S., Alvarez X., Ishido S., Lackner A. A., Jung J. U., Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi’s sarcoma-associated herpesvirus K1. J. Exp. Med. 192, 11–21 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlinson C. C., Damania B., Critical role for endocytosis in the regulation of signaling by the Kaposi’s sarcoma-associated herpesvirus K1 protein. J. Virol. 82, 6514–6523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samaniego F., Pati S., Karp J. E., Prakash O., Bose D., Human herpesvirus 8 K1-associated nuclear factor-kappa B-dependent promoter activity: Role in Kaposi’s sarcoma inflammation? J. Natl. Cancer Inst. Monogr., 15–23 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Wang L., Dittmer D. P., Tomlinson C. C., Fakhari F. D., Damania B., Immortalization of primary endothelial cells by the K1 protein of Kaposi’s sarcoma-associated herpesvirus. Cancer Res. 66, 3658–3666 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Debnath J., et al. , The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 (2002). Correction in: Cell 111, 757 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Webster-Cyriaque J., Duus K., Cooper C., Duncan M., Oral E. B. V., Oral EBV and KSHV infection in HIV. Adv. Dent. Res. 19, 91–95 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Phang J. M., Pandhare J., Liu Y., The metabolism of proline as microenvironmental stress substrate. J. Nutr. 138, 2008S–2015S (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones T., et al. , Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest. 122, 1076–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain M., et al. , Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patil M. D., Bhaumik J., Babykutty S., Banerjee U. C., Fukumura D., Arginine dependence of tumor cells: Targeting a chink in cancer’s armor. Oncogene 35, 4957–4972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir A., Danai L. V., Vander Heiden M. G., Microenvironmental regulation of cancer cell metabolism: Implications for experimental design and translational studies. Dis. Model. Mech. 11, dmm035758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin R. Z., Chang H. Y., Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 3, 1172–1184 (2008). Correction in: Biotechnol J.3, 1285 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Bissell M. J., Radisky D., Putting tumours in context. Nat. Rev. Cancer 1, 46–54 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riedl A., et al. , Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 130, 203–218 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Cheng F., et al. , KSHV-initiated notch activation leads to membrane-type-1 matrix metalloproteinase-dependent lymphatic endothelial-to-mesenchymal transition. Cell Host Microbe 10, 577–590 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Sanchez E. L., Carroll P. A., Thalhofer A. B., Lagunoff M., Latent KSHV infected endothelial cells are glutamine addicted and require glutaminolysis for survival. PLoS Pathog. 11, e1005052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tso F. Y., et al. , RNA-Seq of Kaposi’s sarcoma reveals alterations in glucose and lipid metabolism. PLoS Pathog. 14, e1006844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Borchert G. L., Surazynski A., Hu C. A., Phang J. M., Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: The role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene 25, 5640–5647 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Loayza-Puch F., et al. , Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530, 490–494 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Nilsson R., et al. , Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5, 3128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Togashi Y., et al. , Frequent amplification of ORAOV1 gene in esophageal squamous cell cancer promotes an aggressive phenotype via proline metabolism and ROS production. Oncotarget 5, 2962–2973 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo M. L., et al. , PYCR1 and PYCR2 interact and collaborate with RRM2B to protect cells from overt oxidative stress. Sci. Rep. 6, 18846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanner J. J., Fendt S. M., Becker D. F., The proline cycle as a potential cancer therapy target. Biochemistry 57, 3433–3444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandhare J., Donald S. P., Cooper S. K., Phang J. M., Regulation and function of proline oxidase under nutrient stress. J. Cell. Biochem. 107, 759–768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang X., Zhang L., Natarajan S. K., Becker D. F., Proline mechanisms of stress survival. Antioxid. Redox Signal. 19, 998–1011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda T., et al. , DJ-1 cooperates with PYCR1 in cell protection against oxidative stress. Biochem. Biophys. Res. Commun. 436, 289–294 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Guo L., et al. , Kindlin-2 links mechano-environment to proline synthesis and tumor growth. Nat. Commun. 10, 845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu W., et al. , Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture. Biomaterials 124, 106–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). Correction in: Cancer Discov. 2, 960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.