Abstract
Coronaviruses infect many species of animals including humans, causing acute and chronic diseases. This review focuses primarily on the pathogenesis of murine coronavirus mouse hepatitis virus (MHV) and severe acute respiratory coronavirus (SARS-CoV). MHV is a collection of strains, which provide models systems for the study of viral tropism and pathogenesis in several organs systems, including the central nervous system, the liver, and the lung, and has been cited as providing one of the few animal models for the study of chronic demyelinating diseases such as multiple sclerosis. SARS-CoV emerged in the human population in China in 2002, causing a worldwide epidemic with severe morbidity and high mortality rates, particularly in older individuals. We review the pathogenesis of both viruses and the several reverse genetics systems that made much of these studies possible. We also review the functions of coronavirus proteins, structural, enzymatic, and accessory, with an emphasis on roles in pathogenesis. Structural proteins in addition to their roles in virion structure and morphogenesis also contribute significantly to viral spread in vivo and in antagonizing host cell responses. Nonstructural proteins include the small accessory proteins that are not at all conserved between MHV and SARS-CoV and the 16 conserved proteins encoded in the replicase locus, many of which have enzymatic activities in RNA metabolism or protein processing in addition to functions in antagonizing host response.
Keywords: Nidovirus, Coronavirus, Replicase protein, Pathogenesis, Interferon
I. Introduction
Coronaviruses, a family of viruses within the Nidovirus superfamily, were divided into three groups (1, 2, 3), originally based on antigenic reactivity, later confirmed by genome sequencing. Recently, a new taxonomic nomenclature was adapted by the International Committee on Taxonomy of Viruses (2009) (http://talk.ictvonline.org/media/g/vertebrate-2008/default.aspx). As such, coronaviruses are divided into three genera (alpha, beta and gammacoronaviruses), corresponding to groups 1, 2, 3, within the subfamily coronavirinae, within the family of coronaviridae, and within the order or superfamily of nidovirales. Coronaviruses cause diseases in a variety of domestic and wild animals as well as in humans. Probably the most well-studied coronavirus is the betacoronavirus, murine coronavirus (MuCoV), mouse hepatitis virus (commonly referred to as MHV) that has long provided model systems for the study of central nervous system (CNS) diseases such as encephalitis and multiple sclerosis (MS) and acute hepatitis. While most coronavirus infections cause the common cold in humans, the emergence of the agent for severe acute respiratory syndrome (SARS), the SARS-associated coronavirus (SARS-CoV), also a betacoronavirus, demonstrated the potential for further significant human diseases to result from coronavirus infections. Indeed, shortly after the identification of the SARS-associated human coronavirus (HCoV), new coronavirus were identified in association with more severe infections in humans, NL63 an alphacoronavirus, believed to cause bronchiolitis in children, and HKU1, a betacoronavirus, associated with chronic respiratory disease in the elderly (Pyrc et al., 2007). This review will concentrate on the model MuCoV and the human SARS-CoV.
II. Genome and Virion
Coronaviruses are enveloped positive strand RNA viruses with the largest known RNA genomes, of 30–32 kb (Fig. 1 ). All coronavirus genomes are arranged similarly with the replicase locus encoded within the 5′ end and the structural proteins encoded in the 3′ third of the genome arranged in the order hemagglutinin esterase (HE), if present (HE is only present in some betacoronaviruses), spike (S), small membrane (E), membrane (M) and nucleocapsid (N) and internal (I) protein, encoded within the N gene (Fig. 1). The nucleocapsid protein complexes with the genome RNA to form a helical capsid structure found within the viral envelope. Trimers of the spike protein form the peplomers embedded in the envelope giving the virion its corona or crown-like morphology. In some coronavirus virions, the HE protein forms smaller spikes on the membrane. M and E are also transmembrane proteins involved in virus assembly (Fig. 2 ).
Figure 1.
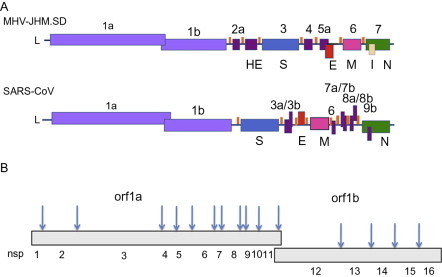
Genome organization and replicase encoded nonstructural proteins. (A) The genomes of MHV-JHM.SD and SARS-CoV are diagrammed. L, leader; ORF1a/1b, replicase; structural genes: HE, hemagglutinin-esterase; S, spike; E, small membrane envelope; M, membrane; N, nucleocapsid; I, internal. orfs encoding accessory genes are designated with numbers. (B). Arrows indicate cleavage sites for orf1a, orf1ab encoded polypeptides and numbers indicate individual nsp cleavage products.
Figure 2.
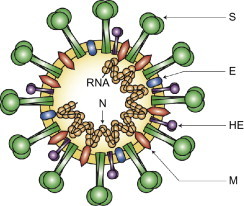
Coronavirus virion structure. The genome RNA is complexed with the N protein to form a helical cased within the viral membrane, HE, hemagglutinin-esterase; S, spike; E, small membrane envelope; M, membrane are all transmembrane proteins.
(Reproduced from Finlay and Hancock, 2004).
The 5′ end of the coronavirus genome encodes the replicase gene, containing two very large open reading frames (orfs), orf1a and orf1b, encompassing about 20 kb or two-thirds of the genome. The replicase is translated as two large polyproteins (pp) 1a and 1ab, with pp1ab expressed via a translational frame shift encoded near the end of orf1a. These replicase polyproteins are cotranslationally cleaved into 16 proteins, many of which have enzymatic activities, including two or three proteases, several RNA modification enzymes as well as a polymerase and helicase, as will be discussed below. Intermingled with the structural genes are a variable number of accessory nonstructural genes encoding usually small, accessory proteins not essential for replication in cell culture. These proteins differ in number, sequence, and function among coronavirus groups and between MHV and SARS-CoV. It has been widely speculated that these proteins mediate virus host interactions, and there are some new data suggesting important functions for some of these proteins, as will be discussed below.
III. Coronavirus-Induced Diseases
A. MHV pathogenesis
The MHV is a collection of strains with different organ tropisms. MHV strains may be divided into two major biotypes, based on general patterns of tropism. One group is enterotropic and includes MHV-D, -Y, -RI, -S/CDC, LIVIM, and DVIM; these viruses are the frequent cause of MHV outbreaks in housed rodent colonies (Homberger et al., 1998). The other biotype, the polytropic strains are those generally studied as models of human disease. Various strains from this group provide model systems for diseases of several organ systems. Neurotropic MHV strains induce acute encephalitis and chronic demyelinating diseases, serving as one of the few recognized mouse models for MS. Hepatotropic strains provide one of the few small animal models for viral hepatitis, and the pneumotropic MHV-1 strain induces severe pneumonitis and reproduces the pathology of SARS. Curiously, despite the very different organ tropisms, all MHV strains use the same cellular receptor, carcinoembryonic antigen molecule (CEACAM)-1, with no known requirements for coreceptors, suggesting that MHV tropism is in part determined by postviral entry events.
1. Central nervous system disease
The most frequently studied MHV strains are the neurotropic ones, primarily JHM and A59. The original JHM isolate, recovered from a paralyzed mouse, was highly neurovirulent, inducing encephalomyelitis with extensive demyelination (Bailey et al., 1949, Cheever et al., 1949). It was subsequently passaged multiple times through mouse brains (Lavi et al., 1984a, Weiner, 1973, Weiner et al., 1973). From this mouse brain-adapted stock, various clones with very different pathogenic phenotypes were isolated and used in many labs, all under the name JHM, causing confusion as to the actual phenotype of JHM. More recently, attempts have been made to differentiate among the JHM isolates, as described further below (Table I ). Among the various JHM isolates, some induce severe encephalitis and high mortality and others induce more mild acute disease followed by chronic demyelination; the origins and pathogenic phenotypes of the various strains has been reviewed recently (Bender and Weiss, 2010, Weiss and Leibowitz, 2008). The A59 strain is a relatively neuroattenuated, yet moderately hepatovirulent strain that was isolated in 1961 from a mouse with leukemia (Manaker et al., 1961).
Table I.
Neurotropic MHV strains
| MHV strain | Pathogenesis | Tropism | Spike/spread | References |
|---|---|---|---|---|
| JHM.SD (MHV-4) | Highly lethal; severe encephalitis | Neurons, glial cells | Gly310; Leu1114; CEACAM1-independent spread | Dalziel et al. (1986) |
| V5A13.1 (mAb escape mutant of JHM.SD) | Neuroattenuated; spreads more slowly in CNS | Neurons, glial cells | HVR deletion (142 aa) | Fazakerley et al. (1992) |
| OBLV60 (variant of JHM.SD isolated from persistently infected OBL21A cells) | Neuroattenuated | Olfactory bulb neurons | L1114R; CEACAM1-dependent spread | Gallagher et al., 1991, Pearce et al., 1994 |
| JHM-DL | Highly lethal | Neurons, glial cells | Leu1114 | Stohlman et al., 1982, Wang et al., 1992 |
| 2.2-V-1 (mAb escape mutant of JHM-DL) | Neuroattenuated; subacute demyelination | Glial cells, primarily oligodendrocytes | L1114F; CEACAM1-dependent spread | Fleming et al., 1986, Wang et al., 1992 |
| JHM cl-2 | Highly lethal | Neurons, glial cells | Gly310; Leu1114; CEACAM1-independent spread | Taguchi et al. (1985) |
| srr7 (soluble receptor-resistant mutant of JHM cl-2) | Neuroattenuated | Macrophages/microglia (in vitro) | L1114F; CEACAM1-dependent spread | Matsuyama et al., 2001, Nakagaki et al., 2005 |
| JHM.IA | Highly lethal, but less than JHM.SD | Neurons, glial cells | S310; Leu1114; CEACAM1-dependent spread | Ontiveros et al. (2003) |
| rJHM.IA.S310G (mutant of JHM.IA) | Highly lethal; more than JHM.IA | Neurons, glial cells | S310G; CEACAM1-independent spread | Ontiveros et al. (2003) |
| JHM. WU (MHV Wb3) | Highly neurovirulent, highly hepatotropic | Not determined | CEACAM1-dependent; HVR deletion (140 aa) | Schwarz et al., 1990, Zhao et al., 2011 |
| A59 | Neuroattenuated; mild encephalitis; subacute demyelination; hepatitis | Neurons, glial cells | HVR deletion (52 aa); CEACAM1-dependent spread | Lavi et al., 1984a, Lavi et al., 1984b, Phillips et al., 2002 |
The general paradigm for neurotropic MHV infection can be summarized as follows. Following intracranial or intranasal inoculation, neurotropic MHV infects all of the major CNS cell types including neurons, the most frequently infected cell type, and glial cells, astrocytes, oligodendrocytes, and microglia. Viral titers typically peak in the CNS at day 5 postinfection and then begin to decline (Leparc-Goffart et al., 1998), with infectious virus becoming undetectable by approximately 2 weeks postinfection (Matthews et al., 2002). Infected mice develop mild to severe encephalomyelitis, characterized by infiltration of a variety of inflammatory cells. Innate immune responses are detectable within the first few days postinfection, followed by the development of an adaptive immune response (Bergmann et al., 2006, Savarin and Bergmann, 2008). Virus is cleared primarily by CD8+ T-cells with help from CD4+ T-cells (Williamson et al., 1991). However, despite clearance of infectious virus, viral RNA, both genome and mRNA persist in the CNS and demyelination, largely immune-mediated, develops, peaking at approximately 1 month postinfection (Lavi et al., 1984a, Lavi et al., 1984b, Marten et al., 2001).
Among the highly neurovirulent isolates are JHM.SD (San Diego, formerly called MHV-4; Dalziel et al., 1986, Ontiveros et al., 2003), JHM.IA (Iowa), JHM.WU (Wurzburg, previously called Wb3; Schwarz et al., 1990), JHM-DL (Stohlman et al., 1982, Wang et al., 1992), and JHM-cl2 (Taguchi et al., 1995). These isolates kill weanling mice with a lethal dose (LD)50 of < 10 pfu following intracranial inoculation. There are subtle phenotypic differences among these isolates which map to the spike gene as well as to other viral genes, as discussed further below. The most neurovirulent strains (e.g., JHM.SD, JHM-cl2) are able to spread cell to cell in the absence of the only known MHV receptor, CEACAM1a (Gallagher and Buchmeier, 2001). JHM 2.2-V-1 (Fleming et al., 1986, Wang et al., 1992), an attenuated monoclonal antibody escape variant, is glialtropic and nonlethal in immunocompetent mice; however, JHM 2.2-V-1 infection along with A59 infection provides useful models to demyelination, in that mice do not die of acute encephalitis (Bergmann et al., 2001, Lavi et al., 1984a, Lavi et al., 1984b). JHM.IA infection of suckling mice, passively immunized, provides another model that has been used to study MHV-induced demyelination (Pewe et al., 1996).
2. Hepatitis
MHV-induced hepatitis has been studied using several strains, including highly hepatovirulent MHV-3 and MHV-2 and the more moderately hepatotropic A59. The MHV-3 strain, most commonly used to study the pathogenesis of MHV-induced hepatitis, was isolated from a VS weanling mouse that developed acute hepatitis after inoculation with serum from a patient with acute hepatitis (Dick et al., 1956). A liver homogenate from this initial isolate produced no clinical signs when inoculated into naive mice. However, as with the neurovirulence of JHM, following serial passage of MHV3 in suckling or weanling mice, a virus emerged that caused fulminant hepatitis that was lethal for weanling VS mice. This virus was primarily hepatotropic, producing massive hepatic necrosis and has been called MHV-3 (Dick et al., 1956).
The extent of liver pathology induced by MHV-3 is dependent on the age and the strain of the mouse (Le Prevost et al., 1975). Most strains, including DBA/2, BALB/c, and C57BL/6 are highly susceptible to lethal disease. However, A/J mice are highly resistant and C3H mice are semisusceptible (Le Prevost et al., 1975). Pathology, characterized by necrotic foci and inflammatory infiltrates of neutrophils and mononuclear cells (Dick et al., 1956), develops quickly after infection of susceptible mice and peaks at 3–4 days postinfection, coinciding with the peak of viral replication, with death occurring 4–7 days after infection. MHV-3 induced hepatitis is characterized by abnormalities in blood flow, including the development of micro thrombi in the liver sinusoids (Bloch et al., 1975, Levy et al., 1983). Levy et al. (1981) observed that MHV-3 infection of peripheral blood mononuclear cells (PBMCs) from susceptible mice induces a procoagulant activity (PCA) and that induction of PCA expression in monocytes in response to MHV-3 infection correlated with susceptibility to disease. Furthermore, the PCA activity is encoded by the fgl2 gene, which is induced at the transcriptional level during MHV infection, specifically by the MHV-3 nucleocapsid and not by nucleocapsids from nonhepatotropic strains (Ning et al., 2003, Ning et al., 1999). MHV-3 infection failed to induce the expression of PCA in macrophages from fgl2 null mice in vitro and in vivo, supporting an important role for fgl2 encoded PCA in the pathogenesis of MHV-3 induced hepatitis. This loss of PCA was reflected by an almost complete absence of fibrin deposition in the liver and hepatocellular necrosis at 3 days postinfection. Interestingly, it is not clear that induction of the fg2 gene is a common feature of hepatitis induced by the other MHV strains, such as A59 (data not shown).
The adaptive immune response to MHV-3 differs between susceptible and resistant mouse strains. In susceptible, but not resistant mouse strains, MHV-3 infection results in necrosis and destruction of splenic and lymphoid follicles (Hirano and Ruebner, 1965, Lamontagne et al., 1989, Virelizier et al., 1975, Yamada et al., 1979). T and B cells from susceptible, but not resistant, mice infected with MHV-3 in vitro were permissive to viral replication and underwent cell lysis (Lamontagne et al., 1989). In addition, antibody responses to MHV-3 were undetectable in BALB/c mice up to death at 5 days postinfection, in contrast to A/J mice that began to mount a robust antibody response by that time (Levy et al., 1984). Like JHM.SD infection of the CNS, high virulence is associated with an inability to induce a robust T-cell response.
Using well-characterized moderately hepatotropic A59, nonhepatotropic JHM, and severely hepatotropic MHV-2 strains, reverse genetics was used to map the viral genes that influence the induction of hepatitis, as will be discussed in detail below. These studies showed that spike protein is a major determinant of hepatovirulence but that one or more background genes in the 3′ end of the genome are also influential. Furthermore, the ns2 protein, an interferon antagonist encoded in the genomes of all known MHV strains, is necessary but in the case of JHM not sufficient for the induction of hepatitis (Zhao et al., 2011).
3. Pneumonitis
The MHV-1 strain is primarily pneumovirulent, different from the previously discussed strains. MHV-1-induced pneumonitis is highly mouse strain dependent; A/J mice, resistant to MHV-3 induced hepatitis, are the most susceptible. While Balb/c and C57Bl/6 mice are resistant to MHV-1-induced pulmonary disease, MHV-1 infection of A/J mice provides a mouse model for the pathogenesis of SARS-CoV in humans (De Albuquerque et al., 2006). Following intranasal infection of MHV-1, A/J mice develop consolidated pneumonitis characterized by hyaline membranes, fibrin deposition and lymphocytic and macrophage infiltration and die by 7 days postinfection. Virions are found mostly localized to pulmonary macrophages. C3H/HeJ mice exhibit an intermediate pattern of resistance/susceptibility, developing chronic pulmonary fibrosis and bronchial hyperplasia with 40% of the mice dying by day 28. MHV-1 replicated in all mouse strains, regardless of susceptibility to disease, suggesting that the development of pneumonitis was a result of the host immune responses. One of the striking differences between infection of susceptible A/J mice and resistant Balb/c and C57Bl/6 mice was the less robust type I interferon response in A/J mice. In contrast to the type I IFN response, A/J mice respond to infection with higher levels of cytokines including macrophage chemo-attractant protein 1(MCP-1/CCL2), IFN-γ, and TNF-α. In addition, in A/J the expression of fgl2 and fibrin deposition were markedly increased (De Albuquerque et al., 2006, Leibowitz et al., 2010). Thus mice susceptible to MHV-3 induced hepatitis bear some similarities in cytokine response to A/J mice infected with MHV-1.
B. SARS-CoV pathogenesis
SARS is a novel infectious disorder that was first diagnosed in China in November 2002 and subsequently spread worldwide (Booth et al., 2003, Dwosh et al., 2003, Holmes, 2003, Ksiazek et al., 2003, Lee et al., 2003, Peiris et al., 2003b, Poutanen et al., 2003, Tsang et al., 2003, Varia et al., 2003, WHO, 2003). SARS was documented in over 8000 persons with 778 deaths (WHO, 2003) before the outbreak was extinguished. In 2004, laboratory-associated cases in Singapore, Taiwan, and Beijing were reported, as were four nonlaboratory associated cases in Guandong Province, P.R.C. (WHO, 2004), underlining the possibility of reemergence of SARS. Spread of SARS was via airborne droplets and through fomites (Donnelly et al., 2003). Electron microscopy, virus isolation, cloning, and sequencing studies demonstrated that a novel coronavirus was the etiologic agent of SARS (Drosten et al., 2003, Ksiazek et al., 2003, Marra et al., 2003, Peiris et al., 2003b, Rota et al., 2003). Shortly thereafter, the coronavirus etiology of SARS was confirmed when Koch's postulates were fulfilled using cynomolgus macaques (Macaca fascicularis) (Fouchier et al., 2003). Although the SARS-CoV was initially thought to represent a novel coronavirus subgroup (Marra et al., 2003, Rota et al., 2003), subsequent more extensive phylogenetic analyses place it as an early branch of the betacoronaviruses, the genus that includes the MHV (Eickmann et al., 2003, Snijder et al., 2003, Zhu and Chen, 2004).
Clinically, patients with SARS had a triphasic pattern of disease (Peiris et al., 2003a). Patients most frequently initially presented with fever, a nonproductive cough, sore throat, and myalgia, with dyspnea often not becoming a prominent feature until days 7–14 of the illness. During the second phase of the illness, dyspnea and hypoxia, with continued fever and frequently accompanied by diarrhea, became more prominent. Some patient's respiratory status continued to deteriorate and they developed acute respiratory distress syndrome often requiring mechanical respiration by the third week. Deaths occurred as early as day 4 and as late as 108 days after onset. Virus shedding from the respiratory tract generally peaked around day 10 and subsequently declined. Virus excretion from the GI tract was frequently present. IgG antibodies were detected 10–15 days after onset and their development was associated with decreased virus load. The severity of the disease was correlated with increasing age, with mortality reaching 50% for patients over 60 (Booth et al., 2003, Chan et al., 2003, Donnelly et al., 2003, Lee et al., 2003, Peiris et al., 2003a, Peiris et al., 2003c, Tsui et al., 2003).
The primary pathology observed at autopsy of patients that succumbed to infection was diffuse alveolar damage (Ding et al., 2003, Franks et al., 2003, Hwang et al., 2005, Nicholls et al., 2003). The lungs of patients that died in the early phases of the disease contained hyaline membranes, edema, fibrin exudates, small vessel thrombi, loss and sloughing of pneumocytes, and a mixed cellular infiltrate of lymphocytes, macrophages, and polymorphonuclear leukocytes. Multinucleated giant cells that carried markers for macrophages and pneumocytes were frequently present. At later phases of the disease, a histologic picture of an organizing pneumonitis and consolidation, with type II pneumocyte hyperplasia, squamous metaplasia, and bronchiolitis obilterans, was found. The association of worsening clinical progression with declining virus loads and the onset of an immunological response, plus the presence of markedly elevated cytokines levels suggested that severe lung damage was largely immunopathological in nature (Beijing Group of National Research Project for SARS, 2003, Cameron et al., 2007, He et al., 2006, Nicholls et al., 2003, Peiris et al., 2003a, Wong et al., 2004a).
The zoonotic origin of the SARS outbreak has recently been reviewed (Graham and Baric, 2010, Yip et al., 2009). The earliest cases of SARS in Guandong, P.R.C., were disproportionally in workers at wild animal markets. Subsequent studies of wild caught animals in these markets detected evidence of SARS-CoV infection in Himalayan palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) suggesting these species as possible sources for human infections (Guan et al., 2003). Attention was focused on civets because of their longer time of virus excretion, and epidemiological studies, including finding additional cases of SARS among food handlers in restaurants that served civet meat (Wang et al., 2005, Xu et al., 2004). Sequence comparisons of SARS-CoVs isolated from civets and patients supported the transmission from civets to humans (Hu et al., 2003, Song et al., 2005, Yeh et al., 2004). Elegant work demonstrating that ACE2 is the SARS-CoV receptor, characterizing the receptor-binding domain (RBD) including a determination of its structure bound to ACE2, and characterization of the key residues involved in adaptation of the SARS-CoV RBD to the human ACE2 has illuminated the structural changes that evolved to enable efficient human infection (Li et al., 2005a, Li et al., 2005c, Li et al., 2003, Wong et al., 2004b) to be further discussed below. However, civets in the wild did not have evidence of current or past infection with SARS-CoV making them unlikely as the natural host for SARS-CoV (Kan et al., 2005). The discovery of SARS-like bat coronaviruses with approximately 90% sequence identity with SARS-CoV in Chinese horseshoe bats (Rhinolophus sinicus) suggests that this or a related species of bat is likely origin of SARS-CoV (Lau et al., 2005, Li et al., 2005b).
A number of animal models for SARS were developed during and after the SARS outbreak. Three excellent reviews of these models are available (Nagata et al., 2010, Roberts et al., 2007b, Subbarao and Roberts, 2006); thus these models will be only be briefly reviewed here. These include nonhuman primate models employing SARS-CoV isolates from later phases of the epidemic and cynomolgus macaques (M. fascicularis) (Fouchier et al., 2003, Kuiken et al., 2003, Rowe et al., 2004), rhesus macaques (Macaca mulatta) (McAuliffe et al., 2004, Qin et al., 2005, Rowe et al., 2004) African Green monkeys (Cercopithecus atheiops or Chlorocebus sabeus) (McAuliffe et al., 2004), and in marmosets (Callithrix jacchus)(Greenough et al., 2005). Although all of these animals support the replication of SARS-CoV in their respiratory tracts, most develop relatively mild disease. In addition, the degree of the severity of the disease and pathology observed in cynomolgus macaques by different workers was variable. SARS-CoV is able to infect cats (Felis domesticus) and ferrets (Mustela furo) (Martina et al., 2003) and although infection does not produce severe disease in either species, the ferret model has been utilized in protection studies and to study the host response to infection (Chu et al., 2008, Czub et al., 2005, Danesh et al., 2011, ter Meulen et al., 2004, Weingartl et al., 2004).
Multiple rodent models of SARS have been developed. Young BALB/c mice can be infected by SARS-CoV but develop minimal pathologic changes and no disease but virus does replicate for a short period of time in the respiratory tract, reaching substantial titers in the lung (Subbarao et al., 2004). Similar to the case with human SARS-CoV infections, aged mice develop more severe disease than young mice with greater viral replication in the lungs, evidence of clinical illness, and the histologic changes of interstitial pneumonitis with alveolar damage, similar to that observed in human SARS (Roberts et al., 2005a). In contrast to 4–6-week-old mice, BALB/c mice 12–14 months of age had elevated levels of IFN-α, IFN-γ, and TNF-α early in infection, suggesting that high levels of proinflammatory cytokines contribute to the more severe disease observed with increased age. Infection of Syrian Golden hamsters with SARS-CoV results in an acute interstitial pneumonitis and lymphocytic inflammatory lesions in the liver (not typically seen in human SARS) without clinical symptoms, and virus was completely cleared by day 14 (Roberts et al., 2005b). Several mouse-adapted strains of SARS-CoV have been developed (Day et al., 2009, Nagata et al., 2008, Roberts et al., 2007a) with the MA15 strain developed by the Baric lab being the most extensively studied. These viruses produce severe lethal disease resembling SARS in young (Day et al., 2009, Roberts et al., 2007a) or aged mice (Nagata et al., 2008, Roberts et al., 2007a). Two of these viruses have been sequenced and both carry identical Y436H mutations in the RBD of the S protein as well as other mutations in replicase (nsp3, nsp 5, nsp9, nsp13), structural (M and additional S mutations), and accessory (3b) proteins (Day et al., 2009, Roberts et al., 2007a). In addition, a rat-adapted strain of SARS-CoV has also been developed that replicates more efficiently in rats than the parental Frankfort1 strain from which it was derived and produces clinical disease and pneumonitis with diffuse alveolar damage in 6-month-old rats (Nagata et al., 2007). This virus contains a mutation in the RBD of the S protein, allowing it to bind more efficiently to rat ACE2, the SARS-CoV receptor (Nagata et al., 2007). Thus adaptation of the S protein to rodent ACE2 appears to be a significant element of the evolution to strains pathogenic for rodents. Transgenic mouse models in which human ACE2 was expressed in the respiratory tract as well as other tissues have been developed (McCray et al., 2007, Tseng et al., 2007). In both of these models, the transgenic mice rapidly succumb after intranasal inoculation of SARS-CoV due to infection of the CNS, limiting their usefulness in the pathogenesis of SARS (Netland et al., 2008, Tseng et al., 2007).
Genetic knockout mice have been extensively employed to identify important elements of host immunity that contribute to the pathogenesis of SARS. Infection of beige, CD1−/−, and RAG1−/− mice resulted in minimal pulmonary pathology, and virus grew to similar titers in the lungs and was cleared with similar kinetics to that observed in wild-type C57Bl/6 mice, demonstrating that NK cells and adaptive cellular immunity are not required for viral clearance (Glass et al., 2004). However, recent experiments using adoptive transfer have demonstrated that virus-specific T-cells derived from immunized mice ameliorate the development of disease and pulmonary pathology and decrease mortality in mice challenged with mouse-adapted MA15 (Zhao and Perlman, 2010) Infection of type I, type II, or type III interferon receptor knockout mice on a strain 129 background, with SARS-CoV (Urbani strain) or the mouse-adapted MA15 virus, resulted in clinical disease and pathologic changes identical to that observed in wild-type strain 129 mice (Frieman et al., 2010). This contrasted with the results obtained with a STATI knockout where genetic ablation of STAT1 increased the severity of disease with both the MA15 and the Urbani strains of SARS-CoV. This suggests that STAT1 may contribute to SARS-CoV pathogenesis by an interferon independent mechanism and it has been speculated that this is related to its role in regulating cell proliferation (Frieman et al., 2010). Microarray analysis of lungs harvested from IFNRA1−/− mice demonstrated strong expression of interferon-stimulated genes in spite of the lack of type I interferon receptors (Zornetzer et al., 2010). In contrast, STAT1−/− mice exhibited a defect in the expression of interferon-stimulated genes and were unable to clear the infection, resulting in a lethal outcome (Frieman et al., 2010). Microarray data suggested dysregulation of T-cell and macrophage differentiation, with a TH2-biased immune response and a profibrotic environment within the lung (Zornetzer et al., 2010). Infection of mice in which MyD88 was genetically ablated with MA15 resulted in increased mortality and pulmonary pathology with higher viral loads in lung, compared to MA15 infection of wild-type mice (Sheahan et al., 2008). In spite of the high viral loads the transcription of proinflammatory cytokine and chemokine genes in lung, and recruitment of macrophages to the lung were severely impaired. Mice in which the CCR1, CCR2, or CCR5 chemokine receptors had been genetically ablated also had more severe disease (Sheahan et al., 2008), suggesting a role for macrophage recruitment in controlling the disease.
Other host factors have also been implicated in the pathogenesis of SARS, primarily from work on murine models. Multiple SARS-CoV proteins have been reported to interact with components of the innate immune system to evade an antiviral interferon response, and these are discussed below with the individual proteins that have been implicated in this process. The expression of ACE2, the SARS-CoV receptor, on the surface of cells is downregulated after infection with SARS-CoV (Kuba et al., 2005). The mechanism of this downregulation appears to be due to internalization of ACE2 during SARS-CoV entry (Wang et al., 2008) and by induction of tumor necrosis factor alpha converting enzyme activity or Adams family metalloproteases which cleave the ACE2 extracellular domain from its transmembrane domain, resulting in shedding of this domain into the media (Haga et al., 2008). ACE2 has a pneumoprotective effect on acute lung injury induced by acid injury (Imai et al., 2005), and instillation of a recombinant fusion protein containing the SARS S protein RBD increased acute lung injury by acid (Kuba et al., 2005). These results have led to the hypothesis that the binding of SARS-CoV S protein is a virulence factor for SARS above and beyond its role in viral attachment and entry. Furthermore, in a mouse model, SARS-CoV replication in myocardium during pulmonary infection correlated with downregulation of ACE2 in the heart (Oudit et al., 2009). This data combined with the detection of inflammatory lesions and viral replication in myocardial tissue of patients that died of SARS suggests that downregulation of ACE2 and cardiac infection could contribute to SARS mortality (Oudit et al., 2009). As described in more detail in Section V.A.2 below, several different proteases, including cathepsin L (Simmons et al., 2005) and the serine protease TMPRSS2 (Matsuyama et al., 2010, Shulla et al., 2011) have been reported to affect SARS-CoV entry through cleavage of the spike protein and activation of its membrane fusion activity. A large number of noncoding RNAs have also been demonstrated to be differentially regulated during infection of mice with MA15 (Peng et al., 2010). About 40% of these noncoding RNAs are similarly regulated during in vitro infection of mouse embryonic fibroblasts with mouse-adapted influenza virus and by interferon treatment, suggesting that these noncoding RNAs may play a role in regulating the host response to virus infection, particularly the innate immune response.
IV. Coronavirus Reverse Genetics
The development of coronavirus reverse genetic systems has greatly enhanced our understanding of coronavirus replication and pathogenesis. This is particularly true in regard to illuminating the functions of viral proteins that interact with host proteins that are part of the host response to infection. We briefly review the various reverse genetic systems that are available for the different coronaviruses. For more detailed information about the various approaches that have been employed, the reader can consult several excellent reviews (Baric and Sims, 2005, Enjuanes et al., 2005, Masters, 2006, Masters and Rottier, 2005, Thiel and Siddell, 2005) and the primary literature on each reverse genetic system.
The large size of the coronavirus genome, 27–32 kb, presented serious obstacles to developing reverse genetic systems similar to those used for smaller positive sense RNA viruses, where a cDNA clone of the genome is transcribed in vitro and the RNA product is transfected into permissive cells to regenerate infectious virus. These obstacles were due to both the large size of cDNAs corresponding to complete coronavirus genomes, and to the instability of various portions of coronavirus replicase genes when cloned into conventional E. coli plasmid vectors (Almazan et al., 2000, Yount et al., 2000). This delayed the development of reverse genetic systems for these viruses for a number of years following the completion of the first coronavirus sequence (Boursnell et al., 1987) and resulted in the development of alternatives to more conventional plasmid-based approaches.
A. Targeted recombination
The first coronavirus reverse genetic system that was developed was targeted recombination for MHV, strain A59 (MHV-A59), and took advantage of the phenotype of a particular temperature-sensitive mutant virus, Alb4, that contained a small in-frame deletion of the N gene that rendered it both much more sensitive to thermal inactivation at 40 °C than wild-type MHV-A59, and conferred a temperature-sensitive phenotype in that it produced a lesser number of very small plaques at 39 °C that were easily distinguished from those formed by wild-type virus under identical conditions (Masters et al., 1994). In this system, a synthetic defective interfering (DI) RNA consisting of the first 467 nucleotides of the MHV genome fused to 48 nucleotides derived from the VSV N gene, fused in turn to the MHV N coding sequence followed by the MHV 3′UTR and a poly (A) tail functioned as an RNA replicon that could recombine with Alb 4, thus introducing mutations engineered into the N gene or 3′UTR of the resulting recombinant viruses. This system allowed the efficient recovery of viruses that contained mutations introduced into the N gene or the 3′ UTR, but was less efficient in introducing mutations into genes 5′ of the N gene.
Subsequently, the targeted recombination methodology was significantly advanced by taking advantage of the fact that the host range of coronaviruses is largely controlled at the entry step, thus providing a powerful means of selecting recombinant viruses (Kuo et al., 2000). In this system a cDNA clone representing a synthetic DI RNA has been enlarged to now contain all of the MHV sequences from codon 28 of the HE gene to the 3′UTR (pMH54, see Fig. 3 ). To create an appropriate acceptor virus, Kuo et al. (2000) created a donor DI RNA in which the sequences encoding the ectodomain of the S protein were replaced by the corresponding sequences of feline infectious peritonitis virus (FIPV). After transfection with this donor DI RNA into cells infected with MHV-A59, recombinant viruses (fMHV) in which the MHV S ectodomain coding sequences replaced by their FIPV counterparts were selected by their ability to grow in feline cells but not murine cells. FMHV can then be used as the acceptor virus in the reverse process, where feline cells are infected with fMHV and transfected with pMH54 or mutation-containing derivatives, and recombinants viruses containing the introduced mutations selected on murine cells. The powerful host range selection enables manipulation of the sequences extending from the S gene through the 3′UTR, a region that includes all of the essential MHV structural genes and is sufficiently efficient to allow the isolation of mutants with very crippled phenotypes (Kuo and Masters, 2003). Although the targeted recombination system was created for MHV-A59, it was extended to the JHM strain of MHV (MHV-JHM) (Ontiveros et al., 2001) and to MHV-1 (B.M. McGruder and J.L. Leibowitz, unpublished). In principle, a system similar to this can be created for any coronavirus that replicates in cultured cells.
Figure 3.
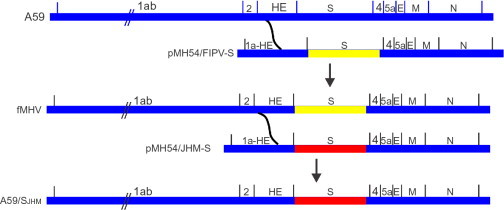
Targeted recombination. A schematic representation of targeted recombination. A59 sequences are shown in blue, FIPV S ectodomain sequences are shown in yellow, and JHM sequences are shown in red. Based on Kuo et al. (2000).
B. Reverse genetic systems that regenerate virus from cDNA
A significant limitation of the targeted recombination system is the inability to easily genetically manipulate genes upstream of the S gene, and for introducing mutations or extensive chimeric sequences downstream of the S gene the need to screen recombinant viruses by sequencing to avoid selecting viruses in which a double crossover has occurred to produce a virus that lacks the desired genotype. To overcome this limitation, reverse genetic systems that regenerate virus from cDNA copies of the genome have been developed. As alluded to above, the large size of the coronavirus genome and the instability of portions of the coronavirus replicase gene posed significant obstacles to the development of reverse genetic systems for coronaviruses (Almazan et al., 2000, Baric and Yount, 2000).
1. Transmissible gastroenteritis virus
The first full-length infectious cDNA clone of a coronavirus was created for transmissible gastroenteritis virus (TGEV) by Almazan et al. (2000). Initial attempts at assembling cDNAs representing the entire TGEV genome by stepwise ligation in a plasmid vector failed due to instability that could be remedied by omitting one 5.2 kbp fragment derived from the replicase. This difficulty was circumvented by transferring the cDNA lacking the 5.2 kbp replicase fragment to a bacterial artificial chromosome (BAC) and then ligating the missing replicase fragment into the BAC to reconstruct the complete TGEV genome. The cDNA was under the control of a cytomegalovirus (CMV) promoter, and the hepatitis delta virus ribozyme is down stream of a poly (A) tail to ensure that the transcript contains the correct 3′ end. Virus is regenerated by transfection into a TGEV-permissive cell, where after translocation of the BAC to the nucleus, transcription of the cDNA regenerates the TGEV genome, which is subsequently exported to the cytoplasm. Once the genome reaches the cytoplasm, viral replication resumes normally. Although there are several potential splice sites in the TGEV genome, splicing appeared to occur at only low levels, allowing efficient recovery of virus. Although the BAC TGEV clone was stable in E. coli, additional stabilization was subsequently obtained by inserting an intron into the regions of the ORF1 gene that are associated with cDNA instability in bacteria (Gonzalez et al., 2002).
Almost simultaneously with the development of the BAC-based reverse genetic system for TGEV, a second reverse genetic system for TGEV was created using an in vitro cDNA assembly approach (Yount et al., 2000). This approach molecularly cloned the TGEV genome as six cDNAs, which together spanned the entire TGEV genome. These cDNAs were created by RT-PCR such that the cDNA which represented the 5′ end of the genome was immediately downstream of a T7 promoter, and the cDNA ends contained restriction sites for a subset of restriction enzymes (such as BglI, recognition site GCCNNNN↓NGGC) that leave sticky ends that are arbitrary in sequence and which will occur very infrequently, making it possible to assemble these cDNAs by in vitro ligation. As was the case with the BAC-based system described above, plasmid instability made it necessary to divide one replicase cDNA fragment into two separate clones in order to stably maintain the cDNAs in E. coli. The separately cloned cDNAs were then excised from plasmids by restriction enzyme digestion and ligated together to assemble a cDNA that corresponds to the complete TGEV genome. The assembled TGEV cDNA was used as template for in vitro transcription to generate capped high molecular weight RNAs. To enhance the recovery of infectious virus, transcripts of the TGEV N gene were separately synthesized in vitro and mixed with the full-length cDNA transcripts, and the mixture electroporated into BHK cells. Although BHK cells do not express the porcine amino peptidase N receptor for TGEV and thus cannot support secondary rounds of infection, they are fully permissive for transfected RNA and were used because of their high electroporation efficiency. The electroporated cells were then seeded with the fully TGEV-permissive and infectable swine testicle (ST) cell line to recover infectious TGEV.
Each of these two approaches to generating a reverse genetic system has its advantages and disadvantages. The BAC approach using a CMV promoter is simpler in that the viral cDNA is propagated as a single clone in E. coli, and because regenerating virus relies on host cell transcription rather than in vitro transcription to regenerate virus genomes, it is considerably less expensive than other approaches utilizing in vitro transcription withT7 RNA polymerase. However, a disadvantage of the BAC system is that genetic manipulation of the cDNA clone in the BAC is not as facile compared to the in vitro cDNA assembly system where the separation of the cDNA into multiple smaller fragments facilitates the introduction of mutations.
2. Human coronavirus 229E
Thiel et al., 2001a, Thiel et al., 2001b used a vaccinia virus-based approach to develop a reverse genetic system for human coronavirus 229E (HCoV-229E). This was necessary due to the instability of a region within orf1a (approximately nts 5200–7000) when cloned into plasmid vectors in E. coli. A series of cloned cDNAs were ligated with an RT-PCR amplicon containing the unstable region to assemble a cDNA, which together represented the entire replicase region of HCoV-229E. To obviate the problem of plasmid instability, the assembled replicase cDNAs were ligated to NotI-cleaved vNotI/tk vaccinia virus DNA (Merchlinsky and Moss, 1992), and a recombinant vaccinia virus containing this HCoV-229E cDNA under the control of a T7 promoter was then recovered by transfecting the ligation products into fowlpoxvirus (FPV)-infected cells (Thiel et al., 2001b). DNA from the recovered recombinant vaccinia virus was subsequently extracted and ligated in vitro to a second cDNA clone representing the remaining 3′ portion of the genome. The resulting cDNA which corresponded to the complete HCoV-229E genome was then ligated to NotI-cleaved vNotI/tk vaccinia virus DNA (Merchlinsky and Moss, 1992), and a second recombinant vaccinia virus was then recovered by transfecting the ligation products into fowlpoxvirus-infected cells. The vaccinia virus DNA was then extracted and digested with ClaI (ClaI sites are absent in the HCoV-229E cDNA, but one is present just 3′ to the HCoV-229E cDNA insert), and capped RNA corresponding to the HCoV-229E genome was synthesized by in vitro transcription with T7 RNA polymerase in the presence of cap analog. Transfection with this RNA regenerated infectious HCoV-229E. Mutations can be introduced into the HCoV-229E cDNA by vaccine virus-mediated homologous recombination with transfected plasmid DNA carrying the desired mutation and the E. coli gpt gene by sequential selection for and against gpt-containing recombinant vaccinia viruses (Falkner and Moss, 1988, Thiel et al., 2001a) or by trans-dominant selection (Falkner and Moss, 1990).
3. Mouse hepatitis virus
Two different approaches have been employed to develop reverse genetic systems for MHV. Both systems allow relatively facile genetic manipulation of MHV-A59. The Baric lab utilized a cDNA fragment assembly approach similar to the one they utilized for TGEV to develop a plasmid-based system in which the MHV-A59 genome is cloned in seven fragments (Yount et al., 2002). One improvement made in this system was the incorporation of type IIS restriction enzyme recognition sites (such as Esp3I) at the ends of the amplified and cloned fragments. Type IIS restriction enzymes recognize a strand-specific sequence rather than a palindromic sequence and cleave double-stranded DNA such that the restriction enzyme recognition site is cleaved from the DNA, but leave an overhang that can be an arbitrary sequence (lower case sequences in Fig. 4 ), permitting the ligation of any two cDNAs in which compatible overhangs have been engineered. This allowed the joining of the seven MHV-A59 cDNAs at points of the investigators’ choosing rather than having to either rely on the existence of rare unique restriction sites or to have to engineer coding silent mutations to create unique sites. The assembled MHV cDNA was used as template for in vitro transcription to generate capped high molecular weight RNAs. As described above, the recovery of infectious virus was enhanced by adding in vitro synthesized transcripts of the MHV N gene to the full-length cDNA transcripts, and the mixture electroporated into BHK-R cells (Dveksler et al., 1991), a cell line that has been transformed with Ceacam1a, the receptor for MHV. The electroporated cells are then overlaid onto fully permissive DBT cells to increase the yield of virus. The initial recombinant virus generated by this approach, MHV-1000, did not produce disease in mice after intracranial inoculation (Sperry et al., 2005). This attenuated phenotype was mapped to two mutations, one in nsp14, and the second in the ns2 gene. Neither mutation was in the predicted active sites of the two proteins. Correction of these mutations allows regeneration of recombinant MHV-A59 that is as virulent as the nonrecombinant virus. The Weiss lab has created a similar reverse genetic system for the widely studied JHM.SD strain of MHV (T.J. Cowley et al., unpublished).
Figure 4.
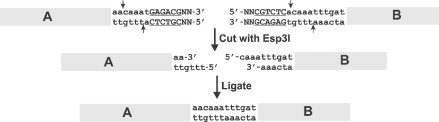
Assembly strategy of full-length coronavirus genomes. The use of the type II S restriction enzyme Esp3I to ligate two cDNAs of arbitrary sequence. The Esp3I recognition site is shown in underlined upper case text. The arbitrary sequence at which the two cDNAs are joined is shown in lower case text. Based on Yount et al. (2002).
The second reverse genetic system for MHV-A59 employed vaccinia virus as a eukaryotic cloning vector (Coley et al., 2005). Four cDNAs representing the entire MHV-A59 genome were assembled from smaller cDNA clones that were stable in E. coli and from RT-PCR products with the 5′ end of the genome immediately downstream of a T7 promoter. These four cDNAs were then ligated in vitro, and the resulting ligation product which contained NotI compatible ends and corresponded to full-length MHV-A59 cDNA was subsequently ligated to NotI-cleaved vNotI/tk vaccinia virus DNA (Merchlinsky and Moss, 1992). Recombinant vaccinia virus containing the MHV cDNA was recovered by transfecting the ligation products into fowlpoxvirus-infected cells, and the recombinant virus subsequently plaque cloned as described previously (Thiel et al., 2001a). Sequence errors inadvertently introduced into the MHV cDNA during the RT-PCR and cloning steps were corrected stepwise by four cycles of vaccinia virus-mediated homologous recombination with transfected plasmids carrying the corrected sequence and the E. coli gpt gene with selection for and against gpt as described previously (Falkner and Moss, 1988, Thiel et al., 2001a). Large amounts of DNA can then be prepared from purified recombinant vaccinia virus, and the purified DNA containing the MHV-A59 cDNA digested with EagI to provide a template for T7 transcription. The MHV cDNA was transcribed in vitro to generate capped high molecular weight RNAs. Recovery of infectious virus was enhanced by adding in vitro synthesized transcripts of the MHV N gene prior to electroporation into BHK-21 cells, which are subsequently incubated with the MHV permissive murine fibroblast cell line, 17Cl-1, to regenerate recombinant MHV-A59. This virus was virulent in mice. Mutant strains of recombinant MHV can be generated by employing vaccinia-mediated recombination as described above for correcting RT-PCR mutations.
4. Avian infectious bronchitis virus
Two reverse genetic systems have been developed for avian infectious bronchitis virus (IBV) (Casais et al., 2001, Youn et al., 2005). Casais et al. (2001) utilized a vaccinia-based approach similar to those described above to ligate three cloned cDNAs into a single large cDNA representing the entire IBV genome under the control of a T7 promoter. Recombinant vaccinia virus containing the IBV genome was recovered as described above for HCoV-229E. Rather than using in vitro transcription to regenerate recombinant IBV, permissive chicken kidney cells were infected with rFPV-T7 to provide cytoplasmic T7 RNA polymerase and the poxvirus guanylyltransferase to cap any T7 transcripts, and at 1 h p.i., the cells were transfected with SalI- or AscI-digested recombinant vaccinia virus DNA containing the IBV full-length cDNA to generate IBV genomes in vivo, which subsequently replicated to generate recombinant IBV. Youn et al. (2005) utilized the in vitro cDNA fragment assembly strategy followed by in vitro transcription to regenerate infectious recombinant IBV from seven discrete cloned cDNAs. Both reverse genetic systems have been used to study various facets of IBV pathogenesis and replication, including the development of avian vaccines.
5. SARS-coronavirus and related Bat-SARS-like coronavirus
Two reverse genetic systems have been developed to study SARS-CoV, one based on the in vitro cDNA assembly approach (Yount et al., 2003); in the second reverse genetic system, a cDNA corresponding to the complete SARS-CoV genome was cloned into a BAC under the control of a CMV promoter and followed by the hepatitis delta virus ribozyme to create a correct 3′ end during transcription from the transfected BAC (Almazan et al., 2006). These systems have been widely utilized to investigate the replication and pathogenesis of SARS. Using synthetic biology and the cDNA assembly approach, Becker et al. (2008) created a series of cloned cDNAs corresponding to the consensus sequence of several bat SARS-like CoVs (bat-SCoV) and subsequently attempted to recover this previously uncultivated virus. This effort failed although evidence for viral replication in the electroporated cells was detected by RT-PCR, most likely due to not having a fully permissive bat cell line containing the cognate receptor (likely bat ACE2) for this virus. When the putative bat-SCoV RBD of the S protein was replaced by the homologous SARS-CoV RBD, the authors were able to recover and characterize infectious virus. This work illustrates the power of the combination of synthetic biology and coronavirus reverse genetics to generate coronavirus species that are only known from sequence information, but have not been successfully grown in cell culture.
6. Human coronaviruses OC43 and NL63
A reverse genetic system for a mouse neurovirulent strain of the human coronavirus OC43 (HCoV-OC43) was created using a BAC system similar to that used for TGEV and SARS-CoV (St-Jean et al., 2006). Like the TGEV and SARS-CoV systems described above, the HCoV-OC43 reverse genetic system relies on transcription from a CMV promoter to transcribe HCoV-OC43 genome RNAs from the transfected BAC containing a cDNA clone of the complete HCoV-OC43 genome. The recovered virus was neurovirulent for mice after intracranial injection, as was the parental virus.
A reverse genetic system for HCoV-NL63 was created using a cDNA assembly approach in which five cDNAs representing the complete HCoV-NL63 genome were ligated in vitro and transcribed from a T7 promoter to regenerate viral genomes to recover recombinant virus by electroporation (Donaldson et al., 2008). This system was applied to demonstrate that HCoV-NL63 ORF3 was not essential for virus replication in cell culture and could be replaced by GFP to create a virus containing this marker.
7. Feline coronavirus
A reverse genetic system for type I feline coronavirus (FCoV) strain Black has been developed using a vaccinia-based approach (Tekes et al., 2008) similar to those described above for HCoV-229E, IBV, and MHV. This system was utilized to create two recombinant FCoVs in which the nonessential 3abc genes in the FCoV genome were replaced by GFP or Renilla luciferase genes to create recombinant viruses that are suitable for both in vivo and cell culture studies of FCoV.
V. Structural Proteins
We review the coronavirus structural proteins, which have important functions in pathogenesis as well as virion assembly and structure. These include the membrane-spanning proteins found in all coronavirus virions, spike, membrane, small membrane, and the HE, expressed by a subset of coronaviruses. We then discuss the nucleocapsid protein complexes with virion RNA to form a helical encased structure and the I protein of unknown function.
A. Spike protein (S)
1. MHV spike protein
The spike protein is a type I membrane protein that is inserted in the viral envelope to form the peplomers that both give the virions their characteristic crown-like morphology (Fig. 2) and interact with viral receptors to mediate viral entry as well as cell to cell spread, through their ability to induce membrane fusion. Spike is synthesized as an approximately 120 kDa precursor that is cotranslationally glycosylated to obtain its final 180 kDa molecular weight. The S proteins of most MHV strains (with the notable exception of MHV-2) are cleaved by a cellular furan-like protease into two noncovalently associated approximately 90 kDa subunits, the N-terminal S1 and C-terminal S2 (Frana et al., 1985, Sturman et al., 1985) (Fig. 5 ). Spike is assembled on the membrane as a trimer in which the S1 subunits form a globular head structure and the S2 subunits form a transmembrane stalk. During infection, S attaches to the MHV receptor CEACAM1a and mediates viral entry, usually directly at the plasma membrane (Gallagher et al., 1991, Qiu et al., 2006), but MHV may also employ an endosomal route of entry (Eifart et al., 2007) and this may be cell type dependent. The spike protein of MHV-2 like that of SARS-CoV (see below) is not cleaved during synthesis; MHV-2 entry occurs via an endosomal route and requires cleavage by cathepsin in a low pH environment (Qiu et al., 2006).
Figure 5.
Structures of the JHM.SD and SARS-CoV spike glycoproteins. RBD, receptor-binding domain; HVR, hypervariable region; HR, heptad repeat domain; TM, transmembrane domain. Arrow indicates cleavage site yielding S1 and S2 subunits in JHM.SD spike. Mutations/deletions found in other neurotropic MHV strains and SARS-CoV variants are indicated below structures and discussed in the text.
Recently, the crystal structure of the MHV spike N-terminal domain (NTD), the RBD, complexed with CEACAM1a has been solved (Peng et al., 2011). Interestingly, the core structure of the NTD contains the same β-sandwich fold as in human galectins, suggesting binding activity to carbohydrates, as well an additional structural motif that binds to the N-terminal Ig-like CEACAM1 domain. Interestingly, while the MHV spike does not bind to sugars and uses only the protein receptor CEACAM1a, other coronavirus spikes, for example, those of BoCV, HCoV-OC43, TGEV (Krempl et al., 2000), and IBV (Niesters et al., 1987), bind to cells through a lectin-like activity. Peng et al. (2011) speculated that coronavirus NTDs were derived originally from human galectins, which evolved over time to lose carbohydrate-binding ability and to gain the ability to interact with CEACAM1a, while other coronaviruses spikes remained dependent on carbohydrate binding for cell attachment.
The coronavirus spike has a major influence on viral tropism and pathogenic phenotype. In the case of MHV, selection by reverse genetic techniques and characterization of chimeric and mutant viruses has been extremely useful for the mapping of pathogenic properties. Exchange of the spike genes between the highly neurovirulent JHM.SD strain and the weakly neurovirulent, yet hepatotropic, A59 strain demonstrated that the spike is a major (but not sole) determinant of the high neurovirulence of JHM.SD (Navas and Weiss, 2003, Navas et al., 2001, Phillips et al., 2002). However, the role of spike in determining liver tropism is more complicated. When A59 background viruses differing only in spike were compared, the level of virulence was determined by the spike protein expressed (Navas et al., 2001). Thus, a chimeric virus expressing the spike of the highly hepatotropic MHV-2 strain from within the A59 background genes (rA59/SMHV-2) was significantly more hepatotropic than A59, while a chimeric virus expressing the spike from the nonhepatotropic JHM.SD within the A59 background (rA59/SJHM.SD) was less hepatotropic than A59. However, perhaps surprisingly, rA59/SJHM.SD was able to replicate to a significant extent in the liver, albeit only at high dose; this is in contrast to JHM.SD that replicates at or near the limit of detection even when inoculated at very high doses. A recombinant virus expressing the hepatotropic A59 spike within the JHM background (rJHM/SA59) failed to infect the liver even at high doses of virus, demonstrating that JHM genetic background eliminates A59 spike-determined hepatotropism. These data imply that liver tropism is at least in part determined by postentry events (Navas and Weiss, 2003).
Comparison of the spike proteins of the many JHM isolates has been informative in elucidating the viral determinants of high neurovirulence. As a general rule, the most neurovirulent JHM isolates (e.g., JHM.SD, JHM-cl2) are able to spread cell-to-cell in a CEACAM1a-independent manner, a process referred to as “receptor independent spread” or “RIS.” There are several functional domains in the MHV spike that have been demonstrated to effect the ability to carry out RIS and to contribute to the corresponding highly neuropathogenic phenotype. These are: (1) the N-terminal RBD, originally defined as the first 330 amino acid of spike (Kubo et al., 1994), which encompasses the NTD described above (Peng et al., 2011), (2) the hypervariable domain (HVR) within S1, and (3) the two the heptad repeat domains (HR1 and HR2) within S2 (Fig. 5). The RBD binds to the NTD of the MHV receptor, CEACAM1, a member of the IgG superfamily. Single amino acid substitutions within the RBD have major effects on neurovirulence and organ tropism. The enhanced neurovirulence of JHM.SD over JHM.IA was mapped to a S310G substitution within the JHM.SD spike protein, conferring the ability to carry out RIS (Ontiveros et al., 2003). Characterization of chimeric JHM.SD/A59 recombinant viruses with exchanges of the RBDs demonstrated that CEACAM1a-independent spread and the very high neurovirulence that accompanies RIS require both the RBD and the rest of the spike to be derived from JHM.SD (Tsai et al., 2003b). In addition to modulating neurovirulence, the RBD clearly also plays a role in hepatotropism as a single Q159L amino acid substitution eliminates the ability of A59 to infect the liver and induce hepatitis while having no effect on neurovirulence (Leparc-Goffart et al., 1997, 1998). Interestingly, the crystal structure of MHV spike NTD in complex with CEACAM1a would predict that while Q159 does not directly interact with receptor, it would influence the binding of R20 to CEACAM1a receptor (Peng et al., 2011). A recent study demonstrated that the MHV-1 spike gene expressed within the A59 genome conferred pneumovirulence; however, other genes both within the 3′ and 5′ portions of the genome were required for full pneumovirulence of MHV-1 (Leibowitz et al., 2010).
The highly neurovirulent JHM.SD (Dalziel et al., 1986, Ontiveros et al., 2003), JHM cl-2 (Taguchi et al., 1985), and JHM-DL (Wang et al., 1992), all capable of carrying out RIS, express spikes with relatively long HVRs. The neuroattenuated phenotypes of a group of monoclonal antibody escape variants of JHM.SD, for example, V5A13.1 (Fazakerley et al., 1992), are associated with single site mutations or deletions within the HVR (Dalziel et al., 1986, Gallagher and Buchmeier, 2001, Phillips and Weiss, 2001) and the spike of the neuroattenuated A59 strain contains a large deletion (52 amino acids) within the HVR. As with the RBD, the long HVR of JHM.SD is, however, not alone sufficient to confer high neurovirulence in that replacement of the HVR of A59 with that of JHM.SD did not confer a highly neurovirulent phenotype to the virus (Phillips and Weiss, 2001). In addition, a spike containing S1 of JHM.SD and S2 of A59 was unable to mediate RIS. These observations indicate that cooperation among several regions of spike, including RBD, the long HVR and S2, is likely required for the high neurovirulence conferred by the JHM.SD spike. Recent data suggest, however, that the long HVR is not required for high neurovirulence as JHM.WU has a large (approximately 400) nucleotide deletion in the HVR spike gene relative to that of JHM.SD (data not shown) and despite its inability to mediate RIS, is highly neurovirulent.
Finally, mutations within the HR domains, which undergo conformational changes during the process of membrane fusion, also effect virulence and the ability to perform RIS. Most notably, substitution of amino acid 1114, L1114R or L1114F has been associated with neuroattenuation in several mutants. The spike protein of the OBLV60 mutant of JHM.SD, which is restricted in replication to the olfactory bulbs, contains three amino acid substitutions within HR1 that have been associated with the requirement for low pH for induction of fusion. However, the L1114R, alone is sufficient to confer neuroattenuation and restriction of viral replication to the olfactory bulbs (Gallagher et al., 1991, Tsai et al., 2003a). An L1114F substitution has been identified both in the spike of the 2.2-V-1 glial-tropic variant of JHM-DL (Wang et al., 1992) and in the spike of a highly attenuated soluble receptor-resistant mutant srr7, derived from JHM-cl2 (Saeki et al., 1997, Saeki et al., 1998). This substitution is associated with an inability to induce RIS as well as with neuroattenuation and the restriction of infection to glial cells in the CNS (Matsuyama and Taguchi, 2002a, Matsuyama and Taguchi, 2002b, Taguchi and Matsuyama, 2002). It is curious that viruses expressing the JHM spike with a L1114F substitution have lost their tropism for neurons while the OBLV60 mutant, expressing a spike with the L1114R substitution, can readily infect neurons of the olfactory bulb in vivo. Thus, small changes within the HR domains, even different substitutions of the same residue, may result in alterations in spike/receptor interaction and subsequent virus entry and pathogenesis in vivo.
High neurovirulence and the associated ability to carry out RIS is associated with less stable association of S1 and S2 as compared with spike proteins that are CEACAM1a dependent in order to mediate fusion, such that the conformational changes that lead to fusion are more easily triggered, even in the absence of CEACAM1a (Gallagher and Buchmeier, 2001, Krueger et al., 2001). Characterization of chimeric A59/JHM.SD viruses in which the S1 and S2 subunits have been exchanged demonstrated that S1 of JHM.SD was not alone sufficient to confer high neurovirulence, underscoring the notion that the cooperation of many domains within spike are required for the full virulence. Further evidence for cooperation among spike domains, noncontiguous in the primary structure comes from the observation that an E1035D substitution within HR1 of S2 may overcome the Q159L substitution in the RBD, since a spike with both of these substitutions confers hepatotropism upon a recombinant A59 (Navas-Martin et al., 2005). Escape mutants selected by resistance to a monoclonal antibody mapping to the RBD had point mutations in the region of HR2 (Grosse and Siddell, 1994), providing further support for the interaction of these domains. Thus, the high neurovirulence conferred by the JHM.SD spike can be thought of as a perfect storm. Very small changes in the sequence can significantly reduce its virulence.
In an effort to understand whether the ability of MHV to perform RIS is truly independent of receptor or whether there is an alternative to CEACAM1a, particularly in the brain, a tissue very poor in expression of CEACAM1a, there have been attempts at identifying additional receptors. The most notable perhaps was the report that PSG16, a protein identified by expression from a cDNA isolated from a mouse brain library, when expressed in COS cells could mediate MHV entry (Chen et al., 1995). We recently confirmed that psg16 mRNA is indeed expressed in the brain, more highly in neurons as compared to glial cells (Bender et al., 2010). However, the PSG16 isoform expressed by Chen et al. (1995), as well as other known isoforms are N-terminally truncated relative to other PSG family proteins (Zebhauser et al., 2005) and thus lacked the sequences that interact with the MHV spike. We have recently cloned a novel full-length isoform of psg16 that is also expressed in the brain, placenta, and retina but, like the truncated form, lacks MHV receptor activity when expressed on the surface of 293T cells (Phillips et al., submitted), suggesting that PSG16 does not mediate CEACAM1a-independent spread of MHV.
2. The SARS-CoV spike
The interaction of SARS-CoV S protein with its cellular receptor, angiotensin-converting enzyme (ACE)-2 is the major determinant of SARS-CoV host range. In contrast to MHV, which infects only mice, and to a limited extent rats, SARS-CoV isolates can infect a variety of species of animals other than humans, including palm civets and raccoon dogs in nature and in the laboratory mice and ferrets as well as nonhuman primates. The RBD of the SARS-CoV spike is not at the amino terminus of spike as it is for MHV; rather, the SARS-CoV RBD is a 192-amino acid region spanning residues 319–510 (Fig. 5). While the core domain of SARS-CoV spike is homologous to a similar region in other betacoronavirus spikes, a loop from residues 424–494, distinct from betacoronaviruses, is the so-called receptor-binding motif (RBM) that contacts ACE-2 directly. It was speculated that this binding loop may have been acquired from a human alphacoronavirus such as NL-63 which also uses ACE-2 as its receptor (Li et al., 2006). Comparisons of the sequences of highly pathogenic human isolates from the 2002–2003 SARS epidemic (e.g., TOR-2 or Urbana), viruses isolated from humans with milder infections in 2003–2004, viruses isolated from civets and raccoon dogs early in the epidemic and more recently bat SARS-like coronaviruses demonstrated that one or two amino acid substitutions in spike can have large effects on the interaction of SARS-CoV spike with human ACE-2 receptor. Such changes were probably responsible for the adaptation of SARS-CoV into humans. The crystal structure of the SARS RBD with ACE-2 has been used to predict how spike variants interact with ACE-2. Two important residues, within the RBM of the spikes of SARS-CoV isolates from humans during the 2002–2003 epidemic, that make contact with the receptor, are N479 and T487 (Fig. 5). Most viruses isolated from palm civets encode K479, which is compatible with the palm civet ACE2; while the human ACE-2 prefers N479, the palm civet ACE-2 can equally accommodate K479 of the civet isolates or N479 of human isolates. Viruses isolated from more mild human cases of SARS in 2003–2004 encode S487 as do the palm civet isolates; these spikes bind less effectively to human ACE-2 than the T487 containing spike, associated with the more pathogenic human isolates. These types of data have lead to the belief that the civet was the intermediate species of transfer for the SARS-CoV from its animal reservoir into humans (Li et al., 2006). As discussed above, many SARS-CoV like viruses have been isolated from bats, leading to the belief that the reservoir for SARS-CoV is the bat (Lau et al., 2005, Li et al., 2005b).
Unlike the spikes of most betacoronaviruses, the spike of SARS-CoV is not cleaved into S1 and S2 subunits during synthesis. However, an endosomal low pH requiring cleavage by cathepsin L takes place during viral entry, similar to that of MHV-2. The exact sites of cleavage and even the number of cleavage events required for viral entry and/or cell-to-cell fusion events have been elusive. However, it was recently reported that cathepsin was required for fusion during viral entry and a second leupeptin-sensitive-like cleavage by a cellular protease was required for activation of cell to cell fusion (Simmons et al., 2011). Whittaker and coworkers proposed that there are two critical cleavage events, one at the S1/S1 boundary and the other within S2 at R797 which act in concert to mediate membrane fusion and virus infectivity (Belouzard et al., 2009, Belouzard et al., 2010). Two other labs reported that a transmembrane protease/serine subfamily member 2 (TMPRSS2) was shown to be colocalized with ACE2 on the cell surface and to enhance SARS-CoV entry (Matsuyama et al., 2010, Shulla et al., 2011). Since TMPRSS2 family proteases are found in the lung, these findings suggest that cleavage by this protease may be a determinant of viral tropism and pathogenesis during the initiation of SARS-CoV infection in vivo. Thus, the precise processing steps needed to activate the SARS-CoV are still not well understood.
In addition to mediating virus entry, the SARS-CoV spike also has effects on regulation of the rennin angiotensin system, which are mediated by the downregulation of ACE2 expression on the plasma membrane, resulting from SARS-CoV infection (Inoue et al., 2007) (Haga et al., 2008, Rockx et al., 2009, Wang et al., 2008). The rennin–angiotensin system regulates blood pressure and fluid balance; this system is widely studied in the kidney, while little is known about regulation in the lung. ACE2 has been shown to be pneumoprotective in multiple models of lung injury, likely through its effect on degrading angiotensin II, a proinflammatory mediator, synthesized by ACE-1 (Hamming et al., 2007, Imai et al., 2005, Kuba et al., 2005, Wosten-van Asperen et al., 2008, Zhang and Sun, 2005). There are many known inhibitors of ACE-1 and the angiotensin II receptor that may have potential to ameliorate the effects of SARS-CoV induced lung pathology, a strategy yet to be explored.
B. Small membrane (E) protein
Coronavirus E proteins are small, 76–109 amino acid, integral transmembrane proteins and are minor components of purified virus particles (Arbely et al., 2004, Corse and Machamer, 2000, Godet et al., 1992, Liu and Inglis, 1991, Raamsman et al., 2000, Yu et al., 1994). Rather than being expressed from a subgenomic mRNA solely dedicated to its expression as it is for the SARS-CoV, the E orf may be downstream of one (i.e., MHV; Leibowitz et al., 1988) or two (i.e., IBV; Liu et al., 1991) orfs encoding accessory genes that are expressed from the same mRNA as E. For IBV translation of the E protein, encoded in orf3c, downstream of the 3a and 3b orfs, from subgenomic mRNA 3 has been shown to be mediated by an IRES that facilitates its translation (Liu and Inglis, 1992). It is not known if other coronaviruses also use this strategy to translate E protein from downstream orfs. The E protein contains three domains, a short N-terminal domain, an unusually long transmembrane domain (see below for discussion of topology), and a hydrophilic C-terminal domain. The C-terminal domain of E protein is palmitoylated (Boscarino et al., 2008, Liao et al., 2006, Yu et al., 1994) and ubiquitinated (Alvarez et al., 2010), and palmitoylation is required for proper virus assembly.
The E protein plays an important role in assembly. Coexpression of E and M proteins is sufficient to direct the assembly of virus-like particles for most coronaviruses that have been examined (Baudoux et al., 1998, Bos et al., 1996, Corse and Machamer, 2000). Cross-linking experiments have further demonstrated an interaction between the E and M proteins (Corse and Machamer, 2003). This interaction appears to be largely mediated by their cytoplasmic tails, although there is also a role for the E protein alpha-helical transmembrane domain in proper assembly and release of virus (Ye and Hogue, 2007). Some, but not all, investigators have shown that for SARS-CoV, VLP assembly may require expression of N protein (Hsieh et al., 2005, Huang et al., 2004, Siu et al., 2008), but not E protein (Huang et al., 2004). Interestingly, although the E protein plays an important role in assembly of virus particles (Fischer et al., 1998), the E protein is not absolutely required for virion assembly for all coronaviruses. Kuo and Masters employed targeted recombination to isolate an MHV mutant that carried a deletion in the E gene (Kuo and Masters, 2003). This virus was viable, and although it produced tiny plaques and replicated to a much lower titer than wild-type virus, it was stable through several passages in cell culture. A similar result was obtained for SARS-CoV; a recombinant SARS-CoV lacking the E gene was viable, reaching titers only 1–2 logs lower than wild-type virus in cell culture, consistent with the observation that E may not be required for SARS-CoV VLP formation (DeDiego et al., 2007). This contrasts with results obtained for TGEV, where deletion of the E gene was lethal (Ortego et al., 2007).
Only a small fraction of the intracellular pool of E protein is assembled into virions. The data on intracellular localization of E protein and the topology of E protein in membranes are conflicting. Immunofluorescence studies in MHV (Raamsman et al., 2000, Yu et al., 1994), SARS-CoV (Liao et al., 2006, Nieto-Torres et al., 2011), IBV (Corse and Machamer, 2000), and TGEV (Godet et al., 1992) infected cells demonstrate that the majority of the E protein localizes to juxtanuclear membranes. The precise origin of these juxtanuclear membranes containing E appears to vary somewhat from virus to virus and from study to study, with E being reported to colocalize with Golgi markers (Corse and Machamer, 2000) and with ER markers for IBV (Lim and Liu, 2001); SARS-CoV E protein has been reported to colocalize with Golgi (Cohen et al., 2011, Liao et al., 2006), ER (Nal et al., 2005), or ERGIC markers (Nieto-Torres et al., 2011); MHV E protein colocalizes with ER and ERGIC markers (Raamsman et al., 2000); TGEV E protein colocalizes with the ERGIC markers (Ortego et al., 2007). Although most studies examining E protein localization did not report E as being present on plasma membranes but rather in an intracellular membranous compartment (see above), several studies reported that a small fraction of E protein could also be detected on plasma membranes (Godet et al., 1992, Pervushin et al., 2009, Yuan et al., 2006a). However, a recent careful study using four different methods failed to detect SARS-CoV E protein at the plasma membrane in infected cells (Nieto-Torres et al., 2011). Some of the differences in the results obtained in different studies on the same E protein may be attributable to the use of N- or C-terminal tags that have the potential for interfering with proper targeting of the E protein when overexpressed from plasmids. A Golgi-targeting sequence in the C-terminal cytoplasmic tail has also been identified for the SARS-E protein (Cohen et al., 2011) and a dilysine-like ER retention signal was identified in the C-terminal 6 amino acids of IBV E (Lim and Liu, 2001). The dilysine-like motif is not conserved in other coronaviruses, whereas the Golgi-targeting signal is conserved in beta and gammacoronaviruses. A chimeric protein containing the VSV transmembrane ectodomain and transmembrane domain fused to the E protein C-terminal domain was retained in the Golgi rather than transported to the plasma membrane. The targeting signal, two predicted beta-strands flanking a conserved proline residue, was identified by mutagenesis. This signal is conserved in the beta and gammacoronaviruses but not the alpha coronaviruses and is functional in both the IBV and MHV E proteins. The N-terminal half of the E protein appears to contain an additional Golgi-targeting signal (Cohen et al., 2011).
A variety of E protein topologies in the membrane have been described for different coronaviruses; these include topologies with a somewhat longer than usual single transmembrane segment and those that describe a hairpin transmembrane domain. TGEV E has been reported to have its C-terminus oriented toward the lumen of intracellular membranes with its N-terminus exposed to the cytoplasm (Godet et al., 1992). IBV E protein (Corse and Machamer, 2000) has been reported to take up the opposite orientation, with the C-terminus exposed to the cytoplasm and the N-terminus of the protein in the luminal position. The MHV E protein has been reported to have both the C-terminus (Maeda et al., 2001, Raamsman et al., 2000) and its N-terminus (Maeda et al., 2001) oriented toward the cytoplasm in a hairpin-like topology. Two different topologies have been reported for the SARS protein, one a hairpin topology with both the N- and C-termini oriented toward the cytoplasm (Arbely et al., 2004, Khattari et al., 2005, Yuan et al., 2006a), and a second topology with a single membrane-spanning domain with the C-terminus in the cytoplasm and the N-terminus oriented toward the lumen (Nieto-Torres et al., 2011, Yuan et al., 2006a).
In addition to its important role in virus assembly, the E protein has several additional effects on infected cells. Overexpression of MHV (An et al., 1999) and SARS-CoV (Yang et al., 2005) E proteins resulted in apoptosis. Overexpression of Bcl-2 (An et al., 1999) or Bcl-xL (Yang et al., 2005) inhibited MHV and SARS-CoV-induced apoptosis, respectively, and the SARS-CoV E protein was shown to bind Bcl-xL through a BH3-like region located in its C-terminal cytosolic domain. The precise mechanism by which E protein triggers apoptosis has not been determined. Caution should be observed in interpreting studies in which E protein is overexpressed since the level of E protein in infected cells is quite low relative to other coronavirus structural proteins; thus infected cells might not show all of the biologic effects observed in overexpression studies. Teoh et al. (2010) demonstrated that the SARS-E protein interacts with PALS1, a protein that is essential for the development and maintenance of epithelial tight junctions, through the C-terminal four amino acids which interact with the PALS1 PZD domain. This redistributes PALS1 to the Golgi and interferes with tight junction formation and thus may contribute to the acute alveolar damage that characterizes SARS-CoV infection of humans. Coronavirus E proteins also have a cation-selective ion channel activity (Liao et al., 2004, Parthasarathy et al., 2008, Pervushin et al., 2009, Wilson et al., 2006, Wilson et al., 2004). The E protein transmembrane domain forms an amphipathic alpha-helix which assembles into pentameric bundles in model lipid bilayers to form functional ion channels (Pervushin et al., 2009, Torres et al., 2006). Although most studies of the E protein ion channel activity have focused on the SARS-CoV E protein, the human coronavirus 229E (HCoV-229E), MHV, and IBV E proteins have also been shown to exhibit ion channel activity (Wilson et al., 2006). This ion channel activity is inhibited by hexamethylene amiloride at doses comparable to those which inhibit the replication of MHV and HCoV-229E (Wilson et al., 2006), suggesting that the ion channel activity plays an important, but as yet unknown, role in coronavirus replication. The E protein transmembrane domain also appears to alter the host secretory machinery to slow down transport of cargo proteins to the plasma membrane (Ruch and Machamer, 2011). It has not as yet been determined if this effect is mediated by the E protein ion channel activity.
A SARS-CoV mutant that carried a deletion of the E gene (SARS-CoVΔE) had an attenuated phenotype in several rodent models of SARS. (DeDiego et al., 2007, Dediego et al., 2008, Lamirande et al., 2008, Netland et al., 2010). After infection with SARS-CoVΔE, animals were protected from a subsequent challenge with wild-type SARS-CoV, making SARS-CoVΔE a potential vaccine candidate. The attenuated phenotype of SARS-CoVΔE raises the possibility that E has a specific but as yet unknown role in pathogenesis.
C. Membrane (M) protein
The coronavirus M protein (formerly called E1) is a multiple membrane-spanning protein containing 221–262 amino acids and it is the most abundant protein in the virus envelope (Cavanagh, 1983, Escors et al., 2001a, Godet et al., 1992). M protein consists of a short (∼ 25 amino acids for MHV) hydrophilic glycosylated N-terminal domain that is exposed on the external surface of the virion, followed by three transmembrane domains followed by a long C-terminal tail that is positioned in the interior of the virus (Armstrong et al., 1984, Rottier et al., 1984). The C-terminal tail contains two domains: an amphipathic domain adjacent to the third transmembrane domain followed by a short hydrophilic region. The N-terminal domain is O-glycosylated for the majority of the betacoronaviruses (Holmes et al., 1981, Lapps et al., 1987, Niemann and Klenk, 1981, Niemann et al., 1984), with the SARS-CoV (Nal et al., 2005, Oostra et al., 2006, Voss et al., 2006) and the MHV-2 strain of MHV (Yamada et al., 2000) being notable exceptions, having an N-glycosylated N-terminal domain. For alphacoronaviruses and gammacoronaviruses, the N-terminal domain is N-glycosylated (Cavanagh and Davis, 1988, Garwes et al., 1984, Jacobs et al., 1986, Stern and Sefton, 1982). The N-terminal domain is exposed on the virus surface and is protease sensitive; it is translocated to the lumen of the ER after in vitro translation of a cDNA encoding the M protein in the presence of microsomes (Cavanagh and Davis, 1988, Rottier et al., 1984, Rottier et al., 1986). The N-terminal ectodomain can be recognized by monoclonal antibodies which are able to neutralize viral infectivity in the presence of complement (Fleming et al., 1989). The majority of the amphipathic domain in the C-terminal tail is thought to be associated with the viral envelope or with the cytoplasmic face of the vesicular compartment where virus assembly and budding occurs, based upon its relative resistance to protease digestion after in vitro translation in the presence of microsomes of a cDNA encoding the M protein (Rottier et al., 1984).
When expressed alone, M is localized to the Golgi (Krijnse-Locker et al., 1994, Machamer and Rose, 1987, Machamer et al., 1990, Nal et al., 2005). The first transmembrane domain from IBV M protein appears to contain the signals that retain this protein in the cis-Golgi and is sufficient to retain otherwise plasma membrane-exposed proteins in the Golgi (Machamer et al., 1993, Swift and Machamer, 1991). Interestingly, for the MHV M protein, it appears that deletion of either of the first two transmembrane domains, or of the cytoplasmic tail, results in failure of the M protein to be retained in the Golgi (Locker et al., 1994). In the infected cell, the M protein is localized to intracellular membranes where virus budding takes place, the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) (Krijnse-Locker et al., 1994, Tooze and Tooze, 1985, Tooze et al., 1984). The localization to the ERGIC in IBV infected cells appears to be dependent upon coexpression of the E protein (Lim and Liu, 2001).
M protein plays a crucial role in the assembly and budding of virus particles (reviewed in de Haan and Rottier, 2005). M proteins interact with each other, and with the other virion proteins, N, E, S and in some betacoronaviruses, HE (de Haan et al., 1998, de Haan et al., 1999, de Haan et al., 2000, Escors et al., 2001b, Fang et al., 2005, Kuo and Masters, 2002, Nguyen and Hogue, 1997, Opstelten et al., 1995). Reverse genetic approaches (Arndt et al., 2010, de Haan et al., 1998, Hurst et al., 2005, Kuo and Masters, 2002, Verma et al., 2006, Verma et al., 2007), experiments utilizing virus-like particles (VLPs) (Corse and Machamer, 2003, de Haan et al., 1998, Huang et al., 2004, Nakauchi et al., 2008, Siu et al., 2008, Vennema et al., 1996), and biochemical and two-hybrid studies of protein–protein interactions (de Haan et al., 1999, He et al., 2004, Hsieh et al., 2008, McBride and Machamer, 2010, Narayanan et al., 2000, Nguyen and Hogue, 1997, Opstelten et al., 1995) have been used to study the role of M protein in the assembly of coronaviruses. M:M interactions are mediated by the transmembrane domains (de Haan et al., 2000, Hida et al., 2000) and by a highly conserved stretch of 12 amino acids, SWWSFNPETNNL, that immediately follow the third transmembrane domain (Arndt et al., 2010). M:S interactions are largely mediated by residues in the cytoplasmic tail although there is evidence that residues in the N-terminal half of the molecule contribute to this association as well (de Haan et al., 1999, McBride and Machamer, 2010, Voss et al., 2009). M:N interactions are also mediated by the M cytoplasmic domain (Escors et al., 2001b, Fang et al., 2005, Hirano and Ruebner, 1965, Hsieh et al., 2008, Hurst et al., 2005, Kuo and Masters, 2002, Verma et al., 2006, Verma et al., 2007). In addition to its interaction with the N protein, the MHV M protein has been demonstrated to interact directly with the MHV RNA packaging signal in an N protein-independent manner to direct packaging of genomes into virions (Narayanan and Makino, 2001, Narayanan et al., 2003). An interaction between the IBV M protein cytoplasmic domain and β-actin is essential for virus budding and assembly (Wang et al., 2009).
A role of M protein in the induction of an interferon response to coronavirus infection was first demonstrated for TGEV, where the induction of interferon-α in PBMCs by glutaraldehyde-fixed purified virus or virus-infected cells could be blocked by monoclonal antibodies that recognized the N-terminal exposed M protein ectodomain (Bernard and Hubert, 1988). Studies with VLPs containing M proteins from representatives of the alpha, beta, and gammacoronaviruses indicated that the interferogenic property of M protein was not confined to TGEV but extended to other coronaviruses as well (Baudoux et al., 1998). Analysis of the interferogenic activity of a panel of escape mutants generated with monoclonal antibodies directed against the N-terminal ectodomain of the M protein implicated N-linked glycans in this region to the induction of interferon by TGEV (Laude et al., 1992). Genetic studies with recombinant MHV-A59 mutants in which the ectodomain of the MHV M protein had been altered to either abolish glycosylation, or to replace the normal O-glycosylation sites with N-glycosylation sites also affected their interferogenic capacity in that viruses-containing N-glycosylated M protein were better inducers of interferon than those containing O-glycosylated M protein (de Haan et al., 2003). Mutants with unglycosylated M proteins were poor interferon inducers. In vivo challenge with these viruses demonstrated that their abilities to replicate in the liver, but not brain, correlated with their in vitro interferogenic capacity (de Haan et al., 2003). This correlation may be the result of binding of virus to lectins, such as the mannose receptor, which are abundantly, expressed in the liver but also play a role in the induction of interferon-α in dendritic cells. In contrast, overexpressed SARS M protein associated with RIG-I, TBK1, IKKε, and TRAF3, thus inhibiting activation of IRF3 and IRF7, leading to significant suppression of the induction of the interferon-β promoter by dsRNA (Siu et al., 2009). It is not clear if this difference in the effects of M protein on the induction (or inhibition) of an interferon response reflects real differences between the different coronaviruses used in these experiments or rather differences between virus infection and overexpression studies.
D. Hemagglutinin-esterase (HE)
HE forms a second, smaller spike on the envelope of some betacoronaviruses (Kienzle et al., 1990, Smits et al., 2005, Yokomori et al., 1991, Yokomori et al., 1989). HE is synthesized as a 42 kDa polypeptide, glycosylated to 65 kDa and disulfide linked to form a homodimer. The MHV HE was observed to have 30% sequence homology to the HA1 subunit of the hemagglutinin esterase fusion (HEF) protein of influenza C virus (Luytjes et al., 1988), leading to the speculation that HE was obtained via nonhomologous RNA recombination involving a betacoronavirus after the split off of SARS-CoV, which does not encode an HE protein (Snijder et al., 2003). HE protein has sialic acid binding and acetyl esterase (or receptor destroying) activities (Brian et al., 1995, Kienzle et al., 1990), which could potentially contribute to viral entry and/or release from the cell surface via interaction with sialic acid-containing moieties. HE is an essential protein for viruses within the betacoronavirus-1 species, including bovine coronavirus (BCoV) and HCoV-OC43 (Kienzle et al., 1990, Vlasak et al., 1988a, Vlasak et al., 1988b). In contrast, for MHV, HE is a nonessential protein expressed by some JHM isolates in addition to MHV-S and the enteropathogenic strains, including MHV-DVIM (Yokomori et al., 1991). It has been speculated that, for MHV, HE may function as an initial or additional binding molecule while spike mediates binding to a specific glycoprotein receptor on the cell surface, in addition to supplying receptor destroying activity to remove the virus when attached to nonsusceptible cells or aiding in virus release. More recently, it was demonstrated that for the betacoronavirus-1 species, it is the spike protein that binds to sialic acid residues which implies that the major function of HE, for these viruses, would be release from glycans via esterase activity (Wurzer et al., 2002). Thus, for these coronaviruses, it has been speculated that during evolution, the spike protein extended its receptor specificity or even shifted its specificity to glycan residues (Langereis et al., 2010). Consistent with this, a specific protein receptor for this species of coronaviruses has not been identified. An alternative evolutionary history in which the S protein of an ancestor of all of the coronavirus genera had a hemaggulinating (lectin) activity that was lost in the MuCoVs sometime after the acquisition of HE from influenza C is also possible (Fig. 6 ).
Figure 6.
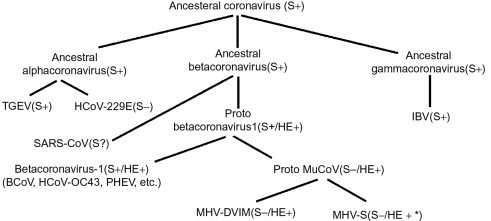
A speculative evolutionary history of the lectin activity of HE and S proteins. S proteins that maintain the ability to hemaggultinate and bind to glycans are denoted S+; those that have lost this ability as S−. For SARS-CoV, hemagglutinating activity has not been reported and to our knowledge has not been tested for; this is denoted as S? HE proteins with a requirement or a strong preference for 9-O-acetyl sialic acid as their ligand (and esterase substrate) are depicted as HE+. HE proteins from strains of MHV where the preferred ligand (substrate) of HE is 4-O-acetyl sialic acid are depicted as HE + *. This schema is supported by the presence of hemagglutinin (lectin-like) activity of TGEV (an alphacoronavirus) (Krempl et al., 2000) and IBV (a gammacoronavirus) (Niesters et al., 1987), and the presence of a galectin fold in the receptor-binding domain of MHV (Peng et al., 2011).
A role for HE in infection and/or pathogenesis has not yet been defined. While HE is clearly dispensable during replication in cell culture, it is conserved among enteropathogenic field strains, suggesting an important function in the wild. The genome of the tissue culture-adapted A59 strain has multiple mutations in the HE orf as well as in the transcriptional regulatory sequences (TRS), resulting in an inactive HE pseudogene and HE (of strain MHV-S) expression from a chimeric A59/MHV-S was lost during passage in vitro. It had long been speculated that HE may play a role in acute and/or chronic MHV disease, either as a determinant of organ and/or cellular tropism (Yokomori et al., 1995, Yokomori et al., 1992, Yokomori et al., 1991) or to aid in spread of the virus (Kienzle et al., 1990, Smits et al., 2005). More recently, a role for HE in spread of MHV in the CNS was demonstrated by comparison of isogenic recombinant viruses expressing a wild-type HE protein, an HE protein in which the esterase activity had been eliminated, and a virus expressing a truncated HE polypeptide (Kazi et al., 2005). The viruses that expressed full-length HE polypeptides (with or without a functional esterase activity) were more virulent when inoculated intracranially into mice and spread more extensively in the CNS compared to viruses expressing a truncated HE polypeptide. Thus, perhaps surprisingly, enhanced virulence does not require an intact esterase activity, suggesting that HE may instead enhance virus attachment and/or spread via binding to sialic acid-containing molecules. Since expression of the MHV receptor CEACAM1a is relatively low in the brain, we speculate that HE interaction with cell surface molecules may enhance attachment to one or more neural cell types. However, isogenic recombinant JHM strains that differ only in expression of HE showed no differences in neurovirulence, suggesting that in this context HE had no major effects on neuropathogenesis. A caveat to this result is that expression of HE was at a considerably lower level than that in the chimeric viruses expressing the HE protein of the MHV-S strain (T.J. Cowley and Weiss, data not shown). The MHV esterase activity may be more important in other organs such as the gastrointestinal tract where the virus may need to pass through mucous or have the ability to detach cells that may not be productively infected, both believed to be functions of neuraminidases. In the case of the influenza neuraminidase, the sialic acid specificity of the enzymatic activity was shown to determine the cell subtype infected within the respiratory track and hence the pathogenic outcome (Matrosovich et al., 2004).
E. Nucleocapsid protein (N) and Internal (I) proteins
The nucleocapsid protein (N), a basic RNA-binding protein (Armstrong et al., 1983) encoded in the most 3′ portion of the MHV genome, plays both structural and nonstructural roles in infection. N complexes with genome RNA to form the viral capsid (Sturman et al., 1980) and interacts with the viral membrane protein (M) during assembly (Hurst et al., 2005) as described above. In addition, N associates with genomic and subgenomic messenger RNA, binding specifically to the TRS (Baric et al., 1988, Grossoehme et al., 2009) and significantly enhances recovery of infectious virus from transfected genome length synthetic RNA (Yount et al., 2002). It was recently demonstrated that N protein is associated with replication-transcription complexes in infected cells and that recruitment of N to these complexes requires the C-terminal N2b domain, which interacts with other N proteins (Verheije et al., 2010). Interestingly, N is unique among MHV structural proteins in that it is partially localized to the nucleus of infected cells (Wurm et al., 2001).
There are data suggesting that N has several potential roles in pathogenesis of CNS disease as well as in MHV-induced hepatitis. Analysis of chimeric viruses in which the N genes of A59 and JHM.SD have been exchanged demonstrated that expression of the JHM N protein from within the A59 background confers increased neurovirulence characterized by increased spread of viral antigen in the brain compared to A59 (Cowley et al., 2010). In addition, N has been reported to associate with microtubules (Pasick et al., 1994), suggesting a possible role in trafficking and axonal transport in neurons; however, there are no further reports following up these intriguing data and in the context of JHM/A59 chimeras N of strain JHM.SD did not enhance spread in cultures of primary hippocampal neurons (Cowley et al., 2010).
N has been reported to antagonize type I interferon by blocking RNase L activity; this activity was demonstrated in 17Cl-1 cells, a cell type in which MHV does not induce type I IFN and may be due to the ability of N to bind RNA and thus sequester it from detection by pattern recognition receptors and the consequent induction of type I interferon (Ye et al., 2007). Furthermore, N proteins from hepatotropic MHV-3 and A59, but not JHM, were shown to be responsible for the induction of fibrinogen-like protein 2 (fgl2), a multifunctional protein that has both procoagulant and immunosuppressive activities and leads to enhanced liver damage during MHV infection (Ding et al., 1998, McGilvray et al., 1998, Ning et al., 1999). The N protein of SARS N was also reported to be an interferon antagonist. When overexpressed in 293T cells, N protein inhibited production of IFN by inhibiting activation of IRF-3 and also inhibited activation of expression from an NF-κB-responsive promoter (Kopecky-Bromberg et al., 2007). These experiments were carried out with overexpressed N, which may have acted through its RNA-binding properties.
The internal protein (I) is a 23 kDa hydrophobic viral membrane-associated structural protein of unknown function. The I gene is encoded within the + 1 reading frame of the N orf (Fischer et al., 1997). An I gene negative recombinant A59 virus displayed no major differences in replication in vitro or in vivo in the brain or liver compared to its isogenic control wild type. Interestingly, all MHV strains express an I gene with the notable exception of JHM (Fischer et al., 1997, Parker and Masters, 1990). Thus, the lack of expression of the I protein could possibly be responsible for the inability of JHM to induce hepatitis. However, this is unlikely as expression of the I gene from within the JHM genome was not sufficient to confer the ability to induce hepatitis (Cowley et al., 2010).
VI. Replicase Proteins
The coronavirus replicase locus is expressed via pp1a and pp1ab, which are processed into 16 nonstructural proteins, some of which provide essential functions such as the RNA-dependent RNA polymerases, the enzymes that modify the 5′ end of the genome with a methylated cap structure and two or three proteases that process precursor proteins. Other replicase proteins provide nonessential functions in virus–host interaction. We discuss these proteins below.
A. Nsp12 polymerase and Nsp8 primase
Nsp12 is encoded in orf1b, just downstream of and containing the orf1a/1b translational frame shift region. Nsp12 contains the RNA-dependent RNA polymerase (RdRp) core activity that is responsible for replication of the viral genome via a negative strand intermediate as well as carrying out transcription of the multiple subgenomic mRNAs containing 5′ termini derived from the 5′ end of the genome, also via negative strand intermediates. Based on structures that have previously characterized RdRps, Xu et al. (2003) built a three-dimensional model of the catalytic domain and located conserved motifs that are shared by all RdRps. There has been little characterization of this activity as a result of difficulties of expression of the proteins. However, recently nsp12 was expressed in E. coli with its natural N-terminus and was shown to have primer dependent activity in vitro on RNA substrates similar to enzymes from poliovirus and Hepatitis C (te Velthuis et al., 2010). Nsp8 has RdRp activity that prefers internal 5′-(G/U)CC-3′ sequences to initiate synthesis of oligonucelotides of less than six residues. In addition, the C-terminus of nsp8 has homology with the catalytic palm domain of RNA viral polymerases. These data lead to the suggestion that nsp8 may serve as a primase to synthesize primers for nsp12 dependent coronavirus RNA synthesis (Imbert et al., 2006). There are no data regarding how the polymerase mediates the joining of noncontiguous sequences in the genesis of negative stranded subgenomic RNAs that serve as templates for mRNAs or which, if any, replicase proteins participate in these processes.
B. Nsp13 helicase
Nsp13 is a 66 kDa protein containing an N-terminal zinc finger structure linked to a C-terminal superfamily 1 helicase domain. A histidine-tagged form of the alphacoronavirus 229E protein was expressed using baculovirus vectors in insect cells (Ivanov and Ziebuhr, 2004) and a SARS-CoV nsp13-maltose binding protein (MBP)-fusion was expressed in E. coli (Ivanov et al., 2004b). The purified recombinant proteins had both RNA and DNA duplex 5′-to-3′ unwinding activities. This is the opposite direction from the well-characterized flaviviral helicases and it has been speculated that this may reflect the synthesis of the multiple subgenomic mRNAs and their negative polarity templates. More recently, Sarafianos and colleagues (personal communication) expressed a glutathione S-transferase-tagged SARS-CoV nsp13 (GST-nsp13) via recombinant baculovirus in insect cells and demonstrated that it unwinds nucleic acids at rates comparable to other helicases, two to three orders of magnitude faster than His tagged or MBP-fusion nsp13 proteins. These data also demonstrated that nsp12 complexes with nsp13 and the complex unwinds nucleic acids twice as fast as nsp13 alone. Nsp13 also has NTPase, dNTPase and 5′ triphosphatase activities. The association of triphosphatase activity with the helicase has led to the suggestion that it may carry out the first step in the capping of genome and mRNAs.
C. Nsp1 protein
Coronavirus nsp1 protein is the N-terminal protein in the orf1a polyprotein (pp1a), and is cotranslationally cleaved from pp1a by a papain-like protease (PLP), also contained in orf1a (nsp3) (Baker et al., 1989, Bonilla et al., 1995, Denison and Perlman, 1987, Denison et al., 1992, Dong and Baker, 1994, Hughes et al., 1995, Perlman, 1986, Soe et al., 1987). In viruses with two PLP domains, PLP-1 carries out this cleavage; in viruses that have only a single PLP domain, that domain catalyzes the proteolytic cleavage of nsp1 from the pp1a precursor. The proteolytic cleavage of this protein from its precursor produces nsp1s of varying sizes depending upon the coronavirus genus. While nsp1s of most of the betacoronaviruses are about 245 amino acids long (also called p28), the SARS-CoV nsp1 is 179 amino acids long. Nsp1s from alphacoronaviruses are shorter, about 110 amino acids in length. The sequences of nsp1s from different coronaviruses are highly divergent with a very low level of sequence similarity when comparing members of different genera. Only 20% sequence similarity can be detected between the MHV and SARS-CoV nsp1s, and even within the less divergent alphacoronaviruses only 20–50% sequence identity is found in pairwise comparisons (Almeida et al., 2007). The avian gammacoronaviruses do not have an nsp1 homologue (Almeida et al., 2007). For the SARS-CoV nsp1, NMR studies of recombinant nsp1 has shown that residues 13–128 contain a novel alpha/beta fold formed by a six stranded beta barrel with an alpha-helix covering one end of the barrel and another helix alongside the barrel (Almeida et al., 2007). No biochemical activity could be ascribed to this protein from this structure. Molecular modeling of HCoV-229E and HCoV-NL63 nsp1 (two alphacoronaviruses) suggests that this beta barrel structure is also found in the alphacoronavirus nsp1 (Wang et al., 2010). These authors also found evidence for functional conservation of nsp1 functional activities among these three viruses (see below).
Immunofluorescence studies have shown that nsp1 colocalizes with proteins found in replication complexes during early times after infection, but at later times, the protein colocalizes with the M protein at the site of virus budding and assembly (Brockway et al., 2004). Biochemical fractionation studies of infected cells suggest that MHV p28 is also present in the soluble fraction of the cytosol at late times postinfection (Bi et al., 1998). Yeast two-hybrid and coimmunoprecipitation experiments demonstrated potential interactions with nsp7 (P10) and nsp10 (P15) (Brockway et al., 2004). Reverse genetic experiments to assess the function of nsp1 demonstrated that for MHV, deletion of the C-terminal half (residues 124–245) of nsp1 was tolerated and gave rise to viable virus, whereas deletion of the N-terminal half of the protein was lethal (Brockway and Denison, 2005). Two clustered changed-to-alanine mutants (R64A/E69A and R78A/D79A) were also lethal, although other changed-to-alanine mutations in the N-terminal half of the molecule were viable. The interpretation of these studies is complicated by the presence of cis-acting replication signals in the MHV RNA genome that extend from the 5′UTR into the first half of the nsp1 coding region (Brown et al., 2007, Kim et al., 1993), raising the possibility that the lethality observed was due to changes made to these cis-acting sequences. Brian and coworkers have demonstrated that purified recombinant BCoV nsp1 binds in vitro to several cis-acting stem-loop structures present in BoCV 5′ and 3′ UTRs (Gustin et al., 2009). Furthermore, expression of nsp1 from a DI RNA-encoded subgenomic mRNA resulted in a reduction in the replication of the DI RNA relative to a control DI construct, but there was only a slight transient reduction in helper virus RNA replication. These results suggest that nsp1 is an RNA-binding protein that may function to regulate viral genome translation or replication.
Infection with MHV leads to cell cycle arrest in the G0/G1 phase associated with a reduction in the amounts of G1 cyclin–Cdk complexes, active Cdk and insufficient phosphorylation of retinoblastoma protein (pRb) (Chen and Makino, 2004). The expression of the MHV nsp1 protein from plasmid vectors in uninfected cells resulted in a similar cell cycle arrest in G0/G1 and inhibition of cell proliferation, suggesting that this effect is due at least in part to nsp1 (Chen et al., 2004). Examination of cell cycle regulatory proteins demonstrated that p28 expression resulted in hypophosphorylation of pRb, increased levels of tumor suppressor p53 and the cyclin-dependent kinase (Cdk) inhibitor p21Cip1. This suggests that p28 expression stabilizes p53, either directly or indirectly, and that accumulated p53 causes upregulation of p21Cip1 with subsequent inhibition of cyclin E/Cdk2 activity, resulting in the inhibition of pRb phosphorylation and thus cell cycle arrest in G0/G1. It should be noted that another MHV protein, nsp15, also interacts with Rb and thus also influences the cell cycle (see nsp15 below). Exogenous expression of SARS-CoV nsp1 also results in decreased cell proliferation with an accumulation of cells in the G0/G1 phase of the cell cycle (Wathelet et al., 2007), suggesting that this effect on the cell cycle may be exerted by many coronavirus nsp1 proteins.
Work by the Makino and Baric labs suggests that nsp1 has a role in pathogenesis by blocking both the synthesis of type I interferons in SARS-CoV infected cells and the induction of interferon-dependent anti-viral proteins such as ISG15 and ISG56 (Kamitani et al., 2006, Narayanan et al., 2008a, Wathelet et al., 2007). There is conflicting data concerning the mechanism(s) by which SARS-CoV nsp1 inhibits the interferon response. Wathelet et al. reported that nsp1 expression inhibits the activation of IRF3, NF-KB, and c-Jun, three transcription factors that are required for activation of the interferon-β promoter. They also report that nsp1 expression blunts the phosphorylation of STAT1 in response to type 1 interferon. In contrast, Kamitani et al. failed to detect an effect of nsp1 expression on IRF3 activation using IRF3 dimerization as a readout for activation (Kamitani et al., 2006). Their data demonstrates that exogenously expressed SARS-CoV nsp1 increases the rate of mRNA degradation, thus inhibiting the accumulation of interferon-β mRNA (and other mRNAs as well) and protein that is normally observed after infection by Sendai virus. Further biochemical studies demonstrated that recombinant nsp1 inhibited in vitro translation reactions, bound to the 40S ribosomal subunit, inhibiting 80S ribosome formation but permitting the formation of a ternary 48S complex with mRNA (Kamitani et al., 2009). Primer extension analysis of the mRNA in the ternary complex indicated that nsp1 resulted in a modification of the 5′ region of the mRNA, rendering it translationally incompetent. Interestingly, Wathelet et al. found that expression of nsp1 had variable effects on protein synthesis depending upon the protein examined, although it should be noted that the overall protein content of cells transfected with nsp1 expressing plasmids was significantly lower than that in cells transfected with control plasmid suggesting that overall protein synthesis (or degradation) may be affected (Wathelet et al., 2007). The differences in the mechanisms by which nsp1 blocks the induction of interferon by virus infection reported by the two groups may be due to differences in the cell lines employed by the two groups, as well as differences in the methodologies they employed. It should be emphasized that both groups are in agreement that nsp1 antagonizes the induction of an interferon response.
Both groups have employed reverse genetic approaches to examine the role of nsp1 in the context of SARS-CoV infection. Mutagenesis experiments demonstrated that two basic residues (K164 and H165) were crucial for stimulating the degradation of mRNA and inhibition of host protein synthesis in cells transfected with nsp1 expressing plasmids (Narayanan et al., 2008a). A recombinant SARS-CoV encoding a mutant nsp1 (K164A/H165A) grew as well as the wild-type virus but was not as efficient as wild-type SARS-CoV in promoting host mRNA degradation and in inhibiting host cell protein synthesis. In contrast to the minimal amounts of type I interferon that were observed after infection with wild-type SARS-CoV, infection of cells with the SARS-CoV nsp1 mutant resulted in the induction of type I interferons at levels similar to those observed after infection with Sendai virus, a strong inducer of interferon. Wathelet et al. employed two different mutations (R124S/K125E and N128S/K129E) that rendered nsp1 much less effective than wild-type nsp1 in blocking the induction of the interferon-β promoter and of interferon responsive genes (Wathelet et al., 2007). A recombinant SARS-CoV encoding the nsp1 R124S/K125E mutation replicated as efficiently as wild-type virus in cells with a defective interferon response but its replication was significantly decreased relative to wild virus in cells with an intact interferon response. The results from both groups suggest that the introduction of mutations into nsp1 might be effective in attenuating the virulence of SARS-CoV by increasing its sensitivity to interferon.
Studies with nsp1s from bat coronaviruses belonging to the betacoronavirus genus showed that they were also able to inhibit the induction of interferon and interferon responsive genes (Tohya et al., 2009), suggesting that the activity of nsp1 in blocking the development of an interferon-mediated antiviral state in the infected cells is not unique to the SARS-CoV. The alphacoronavirus TGEV nsp1, like the SARS-CoV nsp1, also inhibits host protein synthesis. The mechanism by which TGEV nsp1 accomplishes this may differ between the two viruses, since TGEV nsp1 did not bind to 40S ribosomal subunits, nor did it promote host mRNA degradation (Huang et al., 2011). Thus it appears that nsp1 from diverse coronaviruses may have some common functions in spite of their sequence divergence.
D. Nsp3 protein
Nsp3 is a 180–200 kDa multifunctional protein, encoded within coronavirus orf1a, containing multiple functional domains, of which the two most characterized are a PLP domain, described above, and a macrodomain (ADP-ribose 1″-phosphatase or ADRP), also referred to as the “X” domain, both of which have been shown to be virulence factors for MHV and will be discussed. Interestingly. Neuman et al. (2008) using mass spectrometry and kinase-profiling techniques identified nsp3 as a virion component and based on both bioinformatics analyses and characterization of E. coli expressed proteins, identified two additional RNA-binding domains, a chaperone-like domain encoded immediately downstream of the PLP domain and a cysteine-coordinated metal ion-binding domain (Fig. 7 ). The PLP of SARS-CoV and the analogous PLP-2 of MHV have deubiquitinating activity in addition to protease activity, and it has been suggested that this activity could confer type I IFN antagonism (Barretto et al., 2005, Zheng et al., 2008). Indeed, several studies have demonstrated that PLP of SARS-CoV inhibits both the IRF3 and NF-κB pathways (Devaraj et al., 2007, Frieman et al., 2009). However, there are conflicting data regarding the role of MHV PLP-2 as a type I IFN antagonist (Frieman et al., 2009, Zheng et al., 2008), and it is possible that the MHV- and SARS-CoV-encoded proteases may differ in this activity.
Figure 7.

A schematic diagram of the domain structure of MHV nsp3. The domains depicted are: ubiquitin-related domains (UB1 and UB2), papain-like proteases (PLP-1 and PLP-2), ADP-ribose 1″-phosphatase (ADRP), metal binding domain (MBD), betacoronavirus-specific nucleic acid binding (NAB) and marker (G2M) domains, a putative metal-binding region containing a zinc finger (ZF), and three subdomains forming part of the Y region (Y1–Y3). Transmembrane domains are depicted by vertical bars. Based on Neuman et al., 2008.
Macrodomains are ubiquitous and highly conserved among many viral groups and throughout all eukaryotic organisms, bacteria, and archae. The best characterized macro domain is the histone-associated MacroH2A, which plays a role in cell type-specific regulation of transcription (Changolkar et al., 2008). The ADRPs of several coronaviruses (including SARS-CoV, HCoV-229E, and porcine TGEV) were demonstrated to have phosphatase activity, converting ADP-ribose 1″ phosphate, into ADP-ribose and inorganic phosphate (Putics et al., 2005, Putics et al., 2006). Mutation of a conserved residue within the MHV ADRP domain resulting in loss of enzymatic activity conferred loss of hepatitis and decreases in inflammatory cytokine induction (Eriksson et al., 2008). More recently, it was concluded that the ADRP domains of SARS-CoV and 229E conferred resistance to type I IFN treatment (Kuri et al., 2011). Because MHV, as well as some other group II coronaviruses, encode a putative cyclophosphodiesterase activity within the ns2 protein, encoded in ORF2a, just downstream of the replicase gene (Fig. 1), it was predicted that together, the CPD and ADRP activities could participate in a pathway of nucleotide processing (Snijder et al., 2003) in which the CPD would convert ADP-ribose-1″, 2″-cyclic phosphate into ADP-ribose 1″ phosphate and the ADRP would convert the product of the CPD, ADP-ribose 1″ phosphate, into ADP-ribose and inorganic phosphate (Putics et al., 2005, Putics et al., 2006). It seems unlikely now that this pathway is utilized during coronavirus infection in that most coronaviruses do not express ns2 and in addition there are data indicating that MHV ns2 acts as a virulence factor via 2′, 5′ phosphodiesterase activity by inhibition of RNase L pathway, not through a CPD activity (L. Zhao and S.R. Weiss, unpublished data described further below). In addition, the ADRP domains of SARS-CoV, hepatitis E virus and Sindbis virus were shown to have only weak enzymatic activity. However, these macrodomains also have binding activity to mono- and poly-ADP-ribose, implying that they may participate in ribosylation of host cell proteins, which may promote apoptosis or necrosis and interfere with numerous host pathways (Egloff et al., 2006). Finally, studies of the crystal structure of SARS-CoV and the gammacoronavirus IBV ADRP domains indicated that the ADRP of IBV fails to bind ADP-ribose implying that ADRP functions may differ among viruses (Piotrowski et al., 2009). Thus, the function of the ADRP during infection in vivo is still an open question.
E. Nsp14 protein
The nsp14 protein is encoded in the coronavirus 1b orf and is synthesized as pp1ab precursor from which the mature nsp14 protein of approximately 60 kDa is proteolytically released by the CoV 3CLpro protease (Hegyi and Ziebuhr, 2002, Prentice et al., 2004, Xu et al., 2001) [reviewed in (Ziebuhr, 2005)]. The N-terminal half of nsp14 contains a domain predicted by a comparative genomics approach to be a member of a 30-to-50 member exonuclease (ExoN) family belonging to the DEDD superfamily of proteins (Snijder et al., 2003). It was speculated that this putative enzymatic activity could be part of a nucleic acid (RNA) modification pathway also involving nsp15 (NendoU, a predicted endonuclease) and nsp16 (a predicted 2′-O-methyltransferase) (Snijder et al., 2003). A biochemical and genetic (complementation) analysis of a panel of MHV-A59 temperature-sensitive (ts) mutants identified two ts mutants in the nsp14 gene which were unable to carry out RNA synthesis at the nonpermissive temperature, implying an essential role for nsp14 in these processes (Sawicki et al., 2005). The predicted 3′ → 5′ exoribonuclease activity of nsp14 has been characterized biochemically, including identification of residues required for enzyme activity (Chen et al., 2007, Minskaia et al., 2006). The introduction of putative active site mutations into the ExoN coding sequence in the HCoV-229E genome by reverse genetics was lethal (Minskaia et al., 2006). These mutations had a major effect on viral RNA synthesis, greatly decreasing the amount of virus-specific RNA in cells transfected by mutant genomes relative to those observed with wild-type genomes and significantly altered the amounts and the electrophoretic mobility of the HCoV-229E subgenomic mRNA, indicating that the exonuclease activity of nsp14 has an important role in both transcription and RNA replication. The introduction of similar mutations into both the SARS-CoV and MHV genomes had a much less dramatic effect on replication, and viable viruses were recovered, although they replicated more poorly than the wild-type viruses (Eckerle et al., 2007, Eckerle et al., 2010).
The sequence relationships between the CoV ExoN domain and DNA polymerase-associated 3′ → 5′ exonuclease domains and its greater activity with dsRNA substrates than with single stranded RNAs, suggests a possible role of nsp14 in proofreading during CoV RNA synthesis (Minskaia et al., 2006, Snijder et al., 2003). Consistent with this hypothesis, for MHV and SARS-CoV, alanine replacement of conserved ExoN active-site residues yielded viable mutant viruses that accumulated 15–21 fold more mutations than wild-type virus during passage (Eckerle et al., 2010, Eckerle et al., 2007). The estimated mutation rate for these ExoN mutants was similar to that reported for other RNA viruses, approximately 1–3 × 10− 5 substitutions per nucleotide per replication cycle, whereas that of the wild-type viruses was about 10-fold less, suggesting that ExoN contributes to an unusually high fidelity of RNA synthesis for coronaviruses. Recently, a yeast genetic functional screen for the cap-forming enzymes encoded by SARS-CoV identified a second RNA modification activity in nsp14, a (guanine-N7)-methyltransferase (N7-MTase) (Chen et al., 2009). This N7-MTase activity mapped to the C-terminal half of nsp14. Functional studies of mutants introduced into nsp14 in a replicon system showed that the N7-MTase activity is important for SARS-CoV replication and transcription (Almazan et al., 2006, Chen et al., 2009). Interestingly, a mammalian two-hybrid screen for interactions among the SARS-CoV nsps indicated that nsp14 interacted with nsp10, a protein which also interacted with nsp16 (Bouvet et al., 2010, Pan et al., 2008). The interaction of nsp14 with nsp10 had little effect on nsp14 N7-MTase activity (Bouvet et al., 2010). An earlier yeast two-hybrid screen revealed additional interactions: nsp14 with nsp8 (a putative RdRp primase) and with the SARS-CoV 9a accessory protein (von Brunn et al., 2007). A screen directed toward identifying cellular interacting partners identified an interaction with a cellular protein, DDX1, an RNA helicase in the DExD/H helicase family (Xu et al., 2010). siRNA knockdown of DDXI modestly decreased viral replication suggesting that this interaction makes a contribution to efficient coronavirus replication.
Nsp14 also may have one or more pathogenesis-related activities. Sperry et al. identified a Y414H mutation within the MHV-A59 nsp14 N7-MTase domain that resulted in the complete attenuation of lethal disease after intracranial challenge with this virus while having no effect on replication in cell culture (Sperry et al., 2005). This mutation is located in a predicted β-strand that does not contain putative active-site residues for the N7-MTase activity (Minskaia et al., 2006). The mechanism for attenuation is unknown, nor is the effect, if any, of this mutation on the N7-MTase and ExoN activities of nsp14.
F. Nsp15 protein
The nsp15 protein is encoded in the coronavirus 1b orf and is synthesized as part of the pp1ab precursor polyprotein from which the mature nsp15 protein of approximately 38 kDa is proteolytically released by the 3CLpro protease (Hegyi and Ziebuhr, 2002, Prentice et al., 2004, Xu et al., 2001) (reviewed in Ziebuhr, 2005). Nsp15 contains a domain (NendoU) predicted to be related to a Xenopus laevis U specific endonuclease (XendoU) that functions in small nucleolar RNA processing (Snijder et al., 2003). Among RNA viruses, this domain is unique to nidoviruses and is present in members of the arterivirus family as well as in the coronavirus family. Purified recombinant nsp15 from four different coronaviruses representing the three coronavirus genera exhibited an endoribonuclease activity that required divalent cations with a strong preference for Mn++ and preferentially cleaved at uridine nucleotides in both single- and double-stranded RNA substrates leaving a 2′–3′ cyclic phosphate end (Bhardwaj et al., 2004, Ivanov et al., 2004a). The enzyme cleaves 3′ of the recognition uridylate residues and although it cleaves preferentially at U, it also has a much slower reactivity with C containing substrates (Bhardwaj et al., 2006). A 2′-O-methylated RNA substrate was reported to be resistant to cleavage by nsp15 (Ivanov et al., 2004a), suggesting a possible role for the 2′-OMT activity of nsp16 (see below) in regulating nsp15 activity. Purified recombinant nsp15 exists in equilibrium between monomeric and hexameric forms, but only the hexameric form is enzymatically active (Guarino et al., 2005) and binds RNA (Bhardwaj et al., 2006). Cryoelectron microscopy studies and subsequent crystallographic determination of the SARS-CoV (Bhardwaj et al., 2006, Bhardwaj et al., 2008, Ricagno et al., 2006) and MHV nsp15 structures confirmed the hexameric structure for nsp15 and demonstrated that the hexamer was a dimer of trimers (Xu et al., 2006). Mutagenesis studies based on the alignment of coronavirus nsp15s with the Xenopus XendoU sequence suggested that the catalytic center contained two histidines and a lysine that were completely conserved and were essential for enzyme activity (Guarino et al., 2005, Ivanov et al., 2004a). The determination of the nsp15 structure confirmed this assignment and further determined that these catalytic residues were arranged in space virtually identically to the catalytic residues of RNase A, suggesting a similar mechanism of action for that enzyme (Bhardwaj et al., 2008, Ricagno et al., 2006, Xu et al., 2006). Each monomer in the hexamer has a three-domain structure with a small N-terminal domain followed by two larger domains and contains an enzymatically active site. Determination of the structure of a monomeric form of nsp15 showed how oligomerization stabilized the formation of the active site and provided an explanation as to why the monomers are catalytically inactive (Joseph et al., 2007).
Functional studies on the role of nsp15 in the coronavirus life cycle were initially performed in the TGEV replicon (Almazan et al., 2006) and HCoV-229E (Ivanov et al., 2004a) reverse genetic systems. An alanine replacement mutation of a TGEV catalytically active histidine residue reduced RNA synthesis of the TGEV replicon N RNA to levels below 1% of those observed with the wild-type replicon. A mutation in a conserved aspartate that also abrogated NendoU activity of the HCoV-229E enzyme also prevented viral RNA accumulation when tested in the reverse genetic system, suggesting that nsp15 NendoU activity is required for viral replication. However, subsequent structural studies demonstrated that this particular mutation likely interfered with hexamer formation (Ricagno et al., 2006). A reverse genetic analysis of the related arterivirus nsp11 NendoU domain revealed a more complex pattern, with mutation of catalytic residues giving rise to viable viruses that were impaired for virus growth and directed decreased levels of viral RNA synthesis, particularly subgenomic RNA synthesis, whereas replacement of two conserved aspartate residues, one of which corresponded to the aspartate mutated in HCoV-229E (Ivanov et al., 2004a), rendered viral RNA synthesis and virus production undetectable (Posthuma et al., 2006). Generally, similar results were obtained with the MHV reverse genetic system (Kang et al., 2007). Taken together the data suggest that NendoU enzymatic activity plays a role in facilitating maximal viral RNA synthesis, but it is not essential for coronavirus replication. The fact that mutations in a conserved aspartate residue required for nsp15 hexamer formation are lethal suggests that nsp15 has an as yet undiscovered essential role in coronavirus replication that is likely dependent on the formation of nsp15 hexamers.
Examination of betacoronavirus nsp15 amino acids sequence revealed that they contained a retinoblastoma (pRb)-binding motif (LXCXE/D) located on the surface of the protein near the NendoU active site (K. Bhardwaj, C.C. Kao et al., unpublished results). The addition of pRb to recombinant nsp15 stimulated endonuclease activity in vitro and the two proteins coimmunoprecipitated from cellular extracts. Expression of nsp15 in cells shifted the cellular distribution of pRb toward the cytoplasm, increased ubiquitination of pRb, decreased pRb levels, increased the fraction of cells in the S phase of the cell cycle, and increased the number of foci of proliferating 3T3 cells in a transfection assay. Mutation of the LXCXE/D-motif in MHV resulted in a viable virus that exhibits a modestly reduced growth phenotype. Together these data suggest that nsp15 and pRb interact, and that this interaction alters the regulation of cellular proliferation and has subtle effects on coronavirus replication in culture.
G. Nsp16 protein
The nsp16 protein is encoded in the coronavirus 1b orf and is synthesized as pp1ab precursor from which the mature nsp16 protein of approximately 33 kDa is proteolytically released by the CoV 3CLpro protease (Hegyi and Ziebuhr, 2002, Prentice et al., 2004, Xu et al., 2001) [reviewed in (Ziebuhr, 2005)]. Using a comparative genomic approach, Snijder et al. identified a conserved S-adenosylmethionine-dependent ribose 2′-O-methyltransferase (2′-OMT) domain that contains the conserved K-D-K-E catalytic tetrad characteristic of the RrmJ family of methyltransferases (Decroly et al., 2008, Snijder et al., 2003). A biochemical analysis of recombinant FCoV nsp16 demonstrated that nsp16 did carry the predicted 2'-OMT activity (Decroly et al., 2008). Recombinant nsp16 selectively binds short-capped RNAs that have previously undergone N7-methylation of the guanosine cap, a reaction that is carried out by nsp14 (see Section VI.E). Nsp16 then catalyzes the transfer of a methyl group from S-adenosylmethionine to the 2′-hydroxyl group of the first transcribed nucleotide, thereby converting a cap-0 structure to a cap-1 structure (Decroly et al., 2008). Mutagenesis studies demonstrated that the presumptive catalytic amino acids were essential for significant enzymatic activity. Nsp16 has been shown to interact with nsp10, an RNA-binding protein (Pan et al., 2008). Surprisingly, purified SARS-CoV nsp16 had virtually no 2′-OMT activity in the absence of nsp10; however, binding of SARS-CoV nsp16 to nsp10 greatly enhanced 2′-OMT activity to levels higher than that observed with FCoV nsp16 (Bouvet et al., 2010, Decroly et al., 2011) and a mixture of nsp10, nsp14, and nsp16 proteins efficiently converted cap-0 containing RNAs to 2’-O-methylated cap-1 RNAs (Bouvet et al., 2010). Guided by the crystal structure of nsp10 (Joseph et al., 2006) Lugari et al. were able to perform a series of studies of mutant recombinant SARS-CoV nsp10 to define the binding surface that interacted with nsp16 (Lugari et al., 2010). Mutations that abolished the nsp10:nsp16 interaction also abrogated nsp16's 2′-OMT activity. Determination of the structure of the SARS-CoV nsp10:nsp16 heterodimer by X-ray crystallography enabled a series of biochemical experiments with nsp16 mutants that defined key nsp16 residues for binding nsp10 (Decroly et al., 2011). Studies with nsp16 mutants also identified key residues for the highly specific binding of N7-methylated capped RNAs (Decroly et al., 2011). Since nsp10 also interacts with nsp14 as well as nsp16 (Pan et al., 2008), and nsp10 is a non-specific RNA-binding protein (Joseph et al., 2006), it is likely that these three proteins act coordinately and we speculate that they may act as a multicomponent complex.
To better understand the biologic function(s) of nsp16, viruses-containing mutations in nsp16 have been generated by forward and reverse genetic methods. A biochemical and genetic (complementation) analysis of a panel of MHV-A59 ts mutants identified two viruses with nsp16 mutations, and these viruses exhibited defects in RNA synthesis under nonpermissive conditions, implicating an important role for nsp16 in viral RNA synthesis (Sawicki et al., 2005). Consistent with this result, a reverse genetic study employing a SARS-CoV replicon demonstrated reductions in RNA synthesis of 90% (Almazan et al., 2006). However, a subsequent reverse genetic study with active-site mutants of HCoV-229E and MHV-A59 demonstrated that viruses-containing mutations that completely abrogated 2′-OMT activity (D129A for HCoV-229E, D130A for MHV) were viable (Zust et al., 2011). Although the HCoV-229E D129A mutant had slower growth kinetics and reached a peak titer about 10-fold lower than wild-type virus in MRC9 fibroblasts, the MHV-A59 D130 mutant grew identically to wild type in 17Cl-1 cells, a transformed fibroblast cell line. Both the HCoV-299E D129A mutant and the MHV D130A mutant elicited consistently higher levels of interferon-β than wild-type virus when infecting primary macrophages and the replication of both mutants was dramatically more sensitive to interferon-α treatment than wild-type virus. The vigorous induction of interferon-β by the MHV D130 mutant was completely dependent on MDA-5, suggesting that 2′-O-methylation interferes with the sensing of coronavirus RNA by MDA-5, the major sensor of coronavirus infection and induction of an interferon response in macrophages (Roth-Cross et al., 2008). The 2′-OMT activity of nsp16 was also demonstrated to play a key role in avoiding the antiviral effect of IFIT-2 (ISG54) and IFIT-1 (ISG56), two members of the IFIT family of proteins that play an important role in the development of an antiviral state after interferon treatment (Daffis et al., 2010, Zust et al., 2011). Infection of C57Bl/6 mice by intraperitoneal injection showed that although wild-type MHV-A59 replicated to high titer in liver and spleen, the nsp16 D130A mutant failed to replicate and spread in wild-type mice (Zust et al., 2011). This phenotype was dependent upon an intact type I interferon response; IFNAR−/− mice failed to restrict replication of the mutant. These data suggest that a major function of the nsp16 2′-OMT activity is to allow coronaviruses to evade restriction of viral replication by interferon.
VII. MHV Accessory Proteins
The genomes of all coronaviruses have small accessory proteins, not essential for replication in cell culture. Such proteins, encoded in the genomes of other virus families, have been shown to have important functions in virus host interaction, many of which antagonize the host type I interferon response. Accessory proteins are distinct among the three coronavirus groups and are completely distinct between MHV and SARS-CoV, perhaps indicative of the early evolutionary split off of SARS-CoV from the other betacoronaviruses (Snijder et al., 2003), and another reason to place them in separate subgroups of betacoronaviruses. We will review the accessory proteins of MHV and SARS and their roles in antagonizing the host response.
MHV encodes three such accessory proteins, ns2 (orf2a), ns4 (orf4) [or ns4a,4b in some strains (orf4,b)] and ns5a (orf5a). Early on it was shown that naturally occurring viruses not expressing ns2 (Schwarz et al., 1990) or ns4, 5a (Yokomori and Lai, 1991) were replication competent in vitro, confirming that expression of these proteins was nonessential for replication in vitro in transformed cell lines (de Haan et al., 2002).
A. ns2 protein
The most well-studied MHV accessory protein, ns2 is a 30 kDa cytoplasmic protein, encoded in orf2a, just downstream of the replicase gene and expressed from a distinct mRNA from HE (encoded in orf2b) (Schwarz et al., 1990, Zoltick et al., 1990). ns2 contains a domain with high homology to a superfamily of 2H phosphoesterases and was predicted to have a 1″, 2″-cyclophosphodiesterase (CPD) activity. The structure of ns2 has been predicted based on the cellular phosphoesterase AKAP18, and includes two His-x-Thr/Ser motifs for ns2 (Roth-Cross et al., 2009). While expression of ns2 is nonessential for replication in tissue culture cell lines, it is necessary for A59-induced hepatitis in vivo. Introduction of amino acid substitutions into the predicted catalytic His residues (Roth-Cross et al., 2009) attenuates A59 replication in the liver, and reduces hepatitis to a minimal level, without affecting viral replication in the brain or encephalitis (Roth-Cross et al., 2009). Thus ns2 is a tissue-specific virulence factor and in addition has been shown to antagonize type I interferon signaling. Replication of ns2 mutant viruses, in which either of the predicted catalytic His residues has been replaced is attenuated in bone marrow-derived macrophages (BMM) from wild-type (wt) mice but not in BMM derived from type I interferon receptor knockout (IFNAR−/−) mice, and in addition, ns2 mutants are more sensitive than wt virus to pretreatment of BMM. Consistent with these in vitro data, ns2 mutants replicate to nearly the same titers as wt virus after depletion of macrophages in vivo (Zhao et al., 2011). Recently, we found that ns2 mutants replicate to wild-type titers in BMM isolated from RNase L deficient mice and we have obtained evidence, albeit indirect, that ns2, as predicted from sequence, has a phosphodiesterase activity (Mazumder et al., 2002) that cleaves 2-5A, the activator of RNase L and thus, most likely does not act as an IFN antagonist via a cyclophosphodiesterase activity. These data imply that the ability of MHV to replicate in macrophages is a prerequisite for replication in the liver and the induction of hepatitis, but not for CNS replication and disease, highlighting the importance of IFN signaling in macrophages in vivo for the protection of the host from hepatitis. We suggest the Kupffer cells, macrophages of the liver, serve as a gateway to the live parenchyma and restrict viruses through their robust IFN signaling. These ns2 studies point out the cell type and organ type specificity of the IFN response and the interaction of MHV with that response and furthermore, underscore the importance of studying virus–host interaction in primary cells rather than or in addition to transformed cell lines.
B. ns4 protein(s)
The ns4 gene of the JHM strain of MHV encodes a 14 kDa protein that has not yet been detected in infected cells. Comparison of a recombinant JHM.IA ablated for ns4 expression with its isogenic wt parent demonstrated that ns4 was not essential for high neurovirulence (Ontiveros et al., 2001). The ns4 orf of A59 genome has a premature termination codon, converting orf4 into two smaller orfs 4a and 4b; there are no data regarding expression of these proteins or a role in virulence for either one. Thus orf4 has been used a site for expression of foreign genes (Chua et al., 2004, MacNamara et al., 2005, Zhou and Perlman, 2006). It has, however, not been unambiguously shown that the gene products of orf4 do not have an as yet undetected role in virus host interaction, possibly in an organ specific way as has been observed for ns2.
C. ns5a protein
Orf5a encodes ns5a, a protein of approximately 13 kDa, encoded in an upstream orf on the same mRNA as the E protein (orf5b). In vitro translation studies suggested that ns5b is translated from an internal ribosome-binding site (Budzilowicz and Weiss, 1987, Skinner et al., 1985) and orf5a was less efficiently translated than orf5b in an in vitro system and has not yet been detected in infected cells. Orf5a has been reported to encode a type I IFN antagonist activity. A recombinant chimeric virus expressing the 3′ end of MHV-A59 and the replicase gene of MHV-S was strikingly more sensitive to IFN pretreatment of L2 cells than parental A59, similar to that of mildly virulent MHV-S (Koetzner et al., 2010). An A59 mutant abrogated for expression of orf5a displayed intermediate sensitivity to type I IFN pretreatment of L2 cells, supporting the finding that orf5a is an IFN antagonist and also implying that other genes encoded in more 5′ regions within the A59 genome contribute to IFN antagonist activity as well (Koetzner et al., 2010), consistent with the ns2 findings described above. Further unpublished studies supporting these data showed that the ns5a mutant was more sensitive than wt A59 in replication in BMM, but that replication is recovered in BMM derived from IFNAR−/− mice (data not shown). The virulence conferred by ns5 is through a different mechanism from that of ns2 in that the ns5a mutant was attenuated for replication in the CNS as well as in the liver (data not shown). Thus the mechanism by which ns5a confers IFN antagonism is not yet understood.
VIII. SARS Accessory Proteins
The SARS-CoV genome encodes a number of accessory proteins with no identified homologies to those of MHV or other known host cell proteins. SARS-CoV accessory proteins are encoded in orfs 3a, 3b; 7a, 7b; 8a, 8b; 9b. The orf3b,7b, 8b encoded proteins are translated via internal downstream initiation codons from the same mRNAs as 3a,7a,8a, respectively. Of these, proteins encoded in orfs3a, 6, 7a, and 7b have all been found in virus particles (Narayanan et al., 2008b). Systematic deletions individually and in combination of orfs 3a, 3b, 6, 7a, or 7 in recombinant viruses demonstrated that none of these orfs is essential for replication in cell culture, demonstrating that like ns4 and ns5a of MHV, these are nonessential proteins. However, there was some loss efficiency in replication particularly for a virus with the deletion of orf3a (Yount et al., 2005). Importantly, there are data, however, showing that the orf3b and 6 have type I interferon induction and signaling antagonizing activities while orfs3a and 7a have roles in interfering with signaling pathways including apoptosis, as described below. Functions for the proteins encoded in the other accessory orfs are as yet not known.
A. orf6
SARS-CoV orf6 encodes a 63-amino acid endoplasmic reticulum (ER)/Golgi membrane-associated protein, which is expressed in cell culture and in the lung and intestines of infected patients (Narayanan et al., 2008b). The orf6 encoded protein was shown to be a virulence factor; when expressed ectopically from within an MHV genome, orf6 protein conferred lethality upon a nonlethal JHM isolate (Tangudu et al., 2007). The orf6a protein was further demonstrated to function as a virulence factor within the SARS-CoV genome. The orf6 protein inhibits nuclear import and as such inhibits interferon signaling by preventing import of ISGF3 (STAT1/STAT2/IRF-9), the transcription factor that mediates expression of type I interferon-stimulated genes or STAT1/STAT2 complexes following IFN-χ treatment. The C-terminal tail of orf6 binds karyopherin alpha (KPNA)2, which recruits KPNB1 a component of the classical nuclear import complex, thus blocking proteins with classical import signals (Frieman et al., 2007, Hussain et al., 2008). Interestingly Ebola virus, another human pathogen from a different virus family encodes VP24 which inhibits host nuclear import, illustrating that this strategy is utilized by multiple viruses in modulating host responses to viral infection. When expressed in the absence of other viral proteins, orf6 induces the formation of membranous structures, similar to double membrane vesicles involved in virus replication and in addition partially colocalizes with nonstructural protein 3 (nsp3) (Zhou et al., 2010), leading to the suggestion that orf6 protein is also involved in virus replication.
B. orf3b
Several functions have been reported for orf3b encoded protein, which is expressed during infection of patients (Chan et al., 2005). Overexpressed orf3b protein was localized primarily to the nucleus in A549 cells in culture and was shown to inhibit both interferon induction and signaling (Kopecky-Bromberg et al., 2007). Other reports concluded that expressed orf3b also induced cell growth arrest (Yuan et al., 2005) or apoptosis and necrosis (Khan et al., 2006). Most of the data available regarding orf3b protein activities come from overexpression studies, and there is little information on the activities of orf3b during infection.
C. orf7a
The orf7a-encoded protein is a 122 amino acid type I transmembrane protein, localized to perinuclear regions in SARS-infected cells (Nelson et al., 2005) through interactions with M and E (Huang et al., 2006). The precise subcellular localization of the orf7a protein has been disputed (ER, ERGIC Fielding et al., 2006), trans Golgi (Nelson et al., 2005). Likewise, several biological activities have been reported for the orf7a encoded protein, including induction of apoptosis through a caspase-dependent pathway (Tan et al., 2004), inhibition of cellular protein synthesis, activation of p38 mitogen activated protein kinase (Kopecky-Bromberg et al., 2006) and cell cycle arrest at Go/G1 (Yuan et al., 2006b). As with the orf3a encoded protein, most of the data has been obtained from overexpression studies rather than from infected cells.
IX. Conclusions and Future Directions
The coronavirus field, viral replication, cell biology and pathogenesis has advanced quickly in part due to the availability of reverse genetics systems but also, in the past 7 or 8 years due to the increased interest in this class of viruses following the SARS epidemic and resources added to the study of these viruses. The impressive speed with which the SARS-associated virus was identified and the genome sequenced was made possible by the data accumulated previously on the other members of the coronavirus family, illustrating the value of basic science research. There are still some important and intriguing questions to be addressed about coronaviruses, a few of them being the following.
What determines the varied organ tropisms among MHV strains? One of the remaining puzzling aspects of MuCoV pathogenesis is how MHV strains have different organ tropisms despite the observations that they all use the same cellular receptor, CEACAM1a, and the lack of evidence for an alternative receptor. This is in part explained by the contributions of other virus genes and postviral entry events to the determination of tropism; however, the mechanisms underlying differential organ tropism are not at all understood.
How the very different cell type and organ type specific levels of CEACAM1a receptor expression influence tropism? While the liver expresses detectable levels of CEACAM1a protein, CEACAM protein is undetectable in the brain and the levels of mRNA are expressed in the brain are approximately 100-fold less than in the liver. Neurons are the most frequently infected cell type and express levels of CEACAM1a mRNA at or near the level of detection, yet despite this observation, the brain remains a major target of MHV infection. Thus there are still unanswered questions regarding the requirements for CEACAM1a expression and other potential receptors for efficient infection and spread of MHV in vivo.
What are all the replicase proteins/activities for? The coronavirus replicase coding region is longer (approximately 21 kb) than most RNA viruses. Granted that the Coronavirus discontinuous mode of mRNA synthesis is more complex than that of other RNA viruses, the closely related Arteriviruses are also members of the Nidovirus family and synthesize their mRNAs by the same general mechanism but do so using much less genetic information. This raises the question as to what functionalities this additional genetic potential encodes. As exemplified by nsp1 and nsp3, it is likely that at least some of this additional genetic potential is directed toward manipulation of the host environment to directly further virus replication or toward aiding immune evasion. A number of the nsps or domains within nsps (e.g., nsp3) have no biochemical activity associated with them, or if a biochemical activity has been demonstrated, an in vivo correlate of that activity has not yet been demonstrated. Unraveling these functions will continue to be fruitful area of coronavirus research.
Will SARS or another HCoV emerge from its reservoir? The data suggest that SARS adapted to humans by only a few mutations into the viral spike proteins. It seems like this could happen again given the identification of numerous bat SARS-like viruses and the finding of SARS-like virus in animal such as the civet.
Acknowledgments
This work was supported in part by NIH grants AI-060021, AI-080797 and NS-054695 (SRW) and AI-078148 and AI067416 (JLL).
References
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuniga S., Alonso S., Moreno J.L., Nogales A., Capiscol C. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wuthrich K. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Marcos-Villar L., Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Chen C.J., Yu X., Leibowitz J.L., Makino S. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 1999;73:7853–7859. doi: 10.1128/jvi.73.9.7853-7859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341:769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Smeekens S., Rottier P. Sequence of the nucleocapsid gene from murine coronavirus MHV-A59. Nucleic Acids Res. 1983;11:883–891. doi: 10.1093/nar/11.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature (London) 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey O.T., Pappenheimer A.M., Sargent F., Cheever M.D., Daniels J.B. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. II. Pathology. J. Exp. Med. 1949;90:195–212. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.C., Shieh C.K., Soe L.H., Chang M.F., Vannier D.M., Lai M.M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Sims A.C. Development of mouse hepatitis virus and SARS-CoV infectious cDNA constructs. In: Enjuanes L., editor. Coronavirus Replication and Reverse Genetics. Springer; New York: 2005. pp. 229–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N. Interactions between coronavirus nucleocapsid protein and viral RNAs: Implications for viral transcription. J. Virol. 1988;62:4280–4427. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. USA. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijing Group of National Research Project for SARS Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin. Med. J. (Engl) 2003;116:1283–1287. [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010;285:22758–22763. doi: 10.1074/jbc.M110.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S.J., Weiss S.R. Pathogenesis of murine coronavirus in the central nervous system. J. Neuroimmune Pharmacol. 2010;5:336–354. doi: 10.1007/s11481-010-9202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S.J., Phillips J.M., Scott E.P., Weiss S.R. Murine coronavirus receptors are differentially expressed in the central nervous system and play virus strain-dependent roles in neuronal spread. J. Virol. 2010;84:11030–11044. doi: 10.1128/JVI.02688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C.C., Marten N.W., Hinton D.R., Parra B., Stohlman S.A. CD8 T cell mediated immunity to neurotropic MHV infection. Adv. Exp. Med. Biol. 2001;494:299–308. doi: 10.1007/978-1-4615-1325-4_46. [DOI] [PubMed] [Google Scholar]
- Bergmann C.C., Lane T.E., Stohlman S.A. Coronavirus infection of the central nervous system: Host-virus stand-off. Nat. Rev. Microbiol. 2006;4:121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C., Hubert L. Induction of alpha interferon by transmissible gastroenteritis coronavirus: Role of transmembrane glycoprotein E1. J. Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J.M., Yi L.L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Pinon J.D., Hughes S., Bonilla P.J., Holmes K.V., Weiss S.R., Leibowitz J.L. Localization of mouse hepatitis virus open reading frame 1A derived proteins. J. Neurovirol. 1998;4:594–605. doi: 10.3109/13550289809114226. [DOI] [PubMed] [Google Scholar]
- Bloch E.H., Warren K.S., Rosenthal M.S. In vivo microscopic observations of the pathogenesis of acute mouse viral hepatitis. Br. J. Exp. Pathol. 1975;56:256–264. [PMC free article] [PubMed] [Google Scholar]
- Bonilla P.J., Pinon J.L., Hughes S., Weiss S.R. Characterization of the leader papain-like protease of MHV-A59. Adv. Exp. Med. Biol. 1995;380:423–430. doi: 10.1007/978-1-4615-1899-0_68. [DOI] [PubMed] [Google Scholar]
- Booth C.M., Matukas L.M., Tomlinson G.A., Rachlis A.R., Rose D.B., Dwosh H.A., Walmsley S.L., Mazzulli T., Avendano M., Derkach P., Ephtimios I.E., Kitai I. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Hogue B.G., Kienzle T.E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 165–179. [Google Scholar]
- Brockway S.M., Denison M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005;340:209–223. doi: 10.1016/j.virol.2005.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Lu X.T., Peters T.R., Dermody T.S., Denison M.R. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 2004;78:11551–11562. doi: 10.1128/JVI.78.21.11551-11562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.G., Nixon K.S., Senanayake S.D., Brian D.A. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 2007;81:7716–7724. doi: 10.1128/JVI.00549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzilowicz C.J., Weiss S.R. In vitro synthesis of two polypeptides from a nonstructural gene of coronavirus, mouse hepatitis virus strain A59. Virology. 1987;157:509–515. doi: 10.1016/0042-6822(87)90293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Willey B.M., DeVries M.E. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. coronavirus ibv: Further evidence that the surface projections are associated with two glycoproteins. J. Gen. Virol. 1983;64:1787–1791. doi: 10.1099/0022-1317-64-8-1787. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Evolution of avian coronavirus IBV: Sequence of the matrix glycoprotein gene and intergenic region of several serotypes. J. Gen. Virol. 1988;69:621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- Chan J.W., Ng C.K., Chan Y.H., Mok T.Y., Lee S., Chu S.Y., Law W.L., Lee M.P., Li P.C. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 2003;58:686–689. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar L.N., Singh G., Pehrson J.R. macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol. Cell. Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever F.S., Daniels J.B., Pappenheimer A.M., Baily O.T. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. I. Isolation and biological properties of the virus. J. Exp. Med. 1949;90:181–194. doi: 10.1084/jem.90.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 2004;78:5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S., Asanaka M., Yokomori K., Wang F., Hwang S.B., Li H.P., Lai M.M. A pregnancy-specific glycoprotein is expressed in the brain and serves as a receptor for mouse hepatitis virus. Proc. Natl. Acad. Sci. USA. 1995;92:12095–12099. doi: 10.1073/pnas.92.26.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Sugiyama K., Kubo H., Huang C., Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J. Virol. 2004;78:10410–10419. doi: 10.1128/JVI.78.19.10410-10419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Jiang M., Hu T., Liu Q., Chen X.S., Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 2007;40:649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.K., Ali G.D., Jia F., Li Q., Kelvin D., Couch R.C., Harrod K.S., Hutt J.A., Cameron C., Weiss S.R., Jonsson C.B. The SARS-CoV ferret model in an infection-challenge study. Virology. 2008;374:151–163. doi: 10.1016/j.virol.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua M.M., MacNamara K.C., San Mateo L., Shen H., Weiss S.R. Effects of an epitope-specific CD8+ T-cell response on murine coronavirus central nervous system disease: Protection from virus replication and antigen spread and selection of epitope escape mutants. J. Virol. 2004;78:1150–1159. doi: 10.1128/JVI.78.3.1150-1159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85:5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley S.E., Lavi E., Sawicki S.G., Fu L., Schelle B., Karl N., Siddell S.G., Thiel V. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 2005;79:3097–3106. doi: 10.1128/JVI.79.5.3097-3106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312:25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley T.J., Long S.Y., Weiss S.R. The murine coronavirus nucleocapsid gene is a determinant of virulence. J. Virol. 2010;84:1752–1763. doi: 10.1128/JVI.01758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel R.G., Lampert P.W., Talbot P.J., Buchmeier M.J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J. Virol. 1986;59:463–471. doi: 10.1128/jvi.59.2.463-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh A., Cameron C.M., Leon A.J., Ran L., Xu L., Fang Y., Kelvin A.A., Rowe T., Chen H., Guan Y., Jonsson C.B., Cameron M.J. Early gene expression events in ferrets in response to SARS coronavirus infection versus direct interferon-alpha2b stimulation. Virology. 2011;409:102–112. doi: 10.1016/j.virol.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C.W., Baric R., Cai S.X., Frieman M., Kumaki Y., Morrey J.D., Smee D.F., Barnard D.L. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Albuquerque N., Baig E., Ma X., Zhang J., He W., Rowe A., Habal M., Liu M., Shalev I., Downey G.P., Gorczynski R., Butany J. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J. Virol. 2006;80:10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: Primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Smeets M., Vernooij F., Vennema H., Rottier P.J. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: Homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., de Wit M., Kuo L., Montalto-Morrison C., Haagmans B.L., Weiss S.R., Masters P.S., Rottier P.J. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology. 2003;312:395–406. doi: 10.1016/S0042-6822(03)00235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2'O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2'-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Alvarez E., Almazan F., Rejas M.T., Lamirande E., Roberts A., Shieh W.J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. Epub 2006 Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Perlman S. Identification of a putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157:565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Zoltick P.W., Hughes S.A., Giangreco B., Olson A.L., Perlman S., Leibowitz J.L., Weiss S.R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.-T.K., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick G.A., Niven J.F., Gledhill A.N. A virus related to that causing hepatitis in mice. Br. J. Exp. Pathol. 1956;37:90–98. [PMC free article] [PubMed] [Google Scholar]
- Ding J.W., Ning Q., Liu M.F., Lai A., Peltekian K., Fung L., Holloway C., Yeger H., Phillips M.J., Levy G.A. Expression of the fgl2 and its protein product (prothrombinase) in tissues during murine hepatitis virus strain-3 (MHV-3) infection. Adv. Exp. Med. Biol. 1998;440:609–618. doi: 10.1007/978-1-4615-5331-1_79. [DOI] [PubMed] [Google Scholar]
- Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., Lu Y., Wu D. The clinical pathology of severe acute respiratory syndrome (SARS): A report from China. J. Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82:11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Baker S.C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C.A., Ghani A.C., Leung G.M., Hedley A.J., Fraser C., Riley S., Abu-Raddad L.J., Ho L.M., Thach T.Q., Chau P., Chan K.P., Lam T.H. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Dveksler G.S., Pensiero M.N., Cardellichio C.B., Williams R.K., Jiang G.S., Holmes K.V., Dieffenbach C.W. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwosh H.A., Hong H.H., Austgarden D., Herman S., Schabas R. Identification and containment of an outbreak of SARS in a community hospital. CMAJ. 2003;168:1415–1420. [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-Ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Eifart P., Ludwig K., Bottcher C., de Haan C.A., Rottier P.J., Korte T., Herrmann A. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 2007;81:10758–10768. doi: 10.1128/JVI.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Sola I., Alonso S., Escors D., Zuniga S. Coronavirus reverse genetics and development of vectors for gene expression. In: Enjuanes L., editor. Coronavirus Replication and Reverse Genetics. Springer; New York: 2005. pp. 161–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K.K., Cervantes-Barragan L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1''-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F.G., Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F.G., Moss B. Transient dominant selection of recombinant vaccina viruses. J. Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Ye L., Timani K.A., Li S., Zen Y., Zhao M., Zheng H., Wu Z. Peptide domain involved in the interaction between membrane protein and nucleocapsid protein of SARS-associated coronavirus. J. Biochem. Mol. Biol. 2005;38:381–385. doi: 10.5483/bmbrep.2005.38.4.381. [DOI] [PubMed] [Google Scholar]
- Fazakerley J.K., Parker S.E., Bloom F., Buchmeier M.J. The V5A13.1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology. 1992;187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Gunalan V., Tan T.H., Chou C.F., Shen S., Khan S., Lim S.G., Hong W., Tan Y.J. Severe acute respiratory syndrome coronavirus protein 7a interacts with hSGT. Biochem. Biophys. Res. Commun. 2006;343:1201–1208. doi: 10.1016/j.bbrc.2006.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.B., Hancock R.E. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2004;2:497–504. doi: 10.1038/nrmicro908. [DOI] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S.T., Weiss S.R., Masters P.S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.O., Trousdale M.D., El-Zaatari F.A., Stohlman S.A., Weiner L.P. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J. Virol. 1986;58:869–875. doi: 10.1128/jvi.58.3.869-875.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.O., Shubin R.A., Sussman M.A., Casteel N., Stohlman S.A. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology. 1989;168:162–167. doi: 10.1016/0042-6822(89)90415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., Van Amerongen G., Van Doornum G.J., Van Den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: Host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.J., Chong P.Y., Chui P., Galvin J.R., Lourens R.M., Reid A.H., Selbs E., McEvoy C.P., Hayden C.D., Fukuoka J., Taubenberger J.K., Travis W.D. Lung pathology of severe acute respiratory syndrome (SARS): A study of 8 autopsy cases from Singapore. Hum. Pathol. 2003;34:743–748. doi: 10.1016/S0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. SARS coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NFkB signaling. J. Virol. 2009;83(13):6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Escarmis C., Buchmeier M.J. Alteration of the pH dependence of coronavirus-induced cell fusion: Effect of mutations in the spike glycoprotein. J. Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwes D.J., Bountiff L., Millson G.C., Elleman C.J. Defective replication of porcine transmissible gastroenteritis virus in a continuous cell line. Adv. Exp. Med. Biol. 1984;173:79–93. doi: 10.1007/978-1-4615-9373-7_7. [DOI] [PubMed] [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- Godet M., L'Haridon R., Vautherot J.F., Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Penzes Z., Almazan F., Calvo E., Enjuanes L. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by insertion of an intron. J. Virol. 2002;76:4655–4661. doi: 10.1128/JVI.76.9.4655-4661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough T.C., Carville A., Coderre J., Somasundaran M., Sullivan J.L., Luzuriaga K., Mansfield K. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am. J. Pathol. 2005;167:455–463. doi: 10.1016/S0002-9440(10)62989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse B., Siddell S.G. Single amino acid changes in the S2 subunit of the MHV surface glycoprotein confer resistance to neutralization by S1 subunit-specific monoclonal antibody. Virology. 1994;202:814–824. doi: 10.1006/viro.1994.1403. [DOI] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd, Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L. Isolation and characterization of viruses related to the sars coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: Identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. Epub 2005 Oct 03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.M., Guan B.J., Dziduszko A., Brian D.A. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 2009;83:6087–6097. doi: 10.1128/JVI.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Cooper M.E., Haagmans B.L., Hooper N.M., Korstanje R., Osterhaus A.D., Timens W., Turner A.J., Navis G., van Goor H. The emerging role of ACE2 in physiology and disease. J. Pathol. 2007;212:1–11. doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J., Deng Y., Yang L. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Hida S., Ogasawara K., Sato K., Abe M., Takayanagi H., Yokochi T., Sato T., Hirose S., Shirai T., Taki S., Taniguchi T. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity. 2000;13:643–655. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- Hirano T., Ruebner B.H. The effect of murine hepatitis virus infection on lymphatic organs. Lab. Invest. 1965;14:488–500. [PubMed] [Google Scholar]
- Holmes K.V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Holmes K.V., Doller E.W., Sturman L.S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: Demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberger F.R., Zhang L., Barthold S.W. Prevalence of enterotropic and polytropic mouse hepatitis virus in enzootically infected mouse colonies. Lab. Anim. Sci. 1998;48:50–54. [PubMed] [Google Scholar]
- Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.C., Li H.C., Chen S.C., Lo S.Y. Interactions between M protein and other structural proteins of severe, acute respiratory syndrome-associated coronavirus. J. Biomed. Sci. 2008;15:707–717. doi: 10.1007/s11373-008-9278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.D., Zheng G.Y., Jiang H.S., Xia Y., Zhang Y., Kong X.Y. Mutation analysis of 20 SARS virus genome sequences: Evidence for negative selection in replicase ORF1b and spike gene. Acta Pharmacol. Sin. 2003;24:741–745. [PubMed] [Google Scholar]
- Huang Y., Yang Z.Y., Kong W.P., Nabel G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: Implications for assembly and vaccine production. J. Virol. 2004;78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.A., Bonilla P.J., Weiss S.R. Identification of the murine coronavirus p28 cleavage site. J. Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Perlman S., Gallagher T.M. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J. Virol. 2008;82:7212–7222. doi: 10.1128/JVI.02406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.M., Chamberlain D.W., Poutanen S.M., Low D.E., Asa S.L., Butany J. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod. Pathol. 2005;18:1–10. doi: 10.1038/modpathol.3800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. Epub 2006 Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: Characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5'-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs L., van der Zeijst B.A., Horzinek M.C. Characterization and translation of transmissible gastroenteritis virus mRNAs. J. Virol. 1986;57:1010–1015. doi: 10.1128/jvi.57.3.1010-1015.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Brooun A., Griffith M., Moy K., Yadav M.K., Velasquez J., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi L., Lissenberg A., Watson R., de Groot R.J., Weiss S.R. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 2005;79:15064–15073. doi: 10.1128/JVI.79.24.15064-15073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Fielding B.C., Tan T.H., Chou C.F., Shen S., Lim S.G., Hong W., Tan Y.J. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattari Z.Y., Brotons G., Akkawi M., Arbely E., Arkin I.T., Salditt T. SARS coronavirus E protein in phospholipid bilayers: A X-ray study. Biophys. J. 2005;90(6):2038–2050. doi: 10.1529/biophysj.105.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienzle T.E., Abraham S., Hogue B.G., Brian D.A. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J. Virol. 1990;64:1834–1838. doi: 10.1128/jvi.64.4.1834-1838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N., Jeong Y.S., Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner C.A., Kuo L., Goebel S.J., Dean A.B., Parker M.M., Masters P.S. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J. Virol. 2010;84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Ballesteros M.L., Zimmer G., Enjuanes L., Klenk H.D., Herrler G. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J. Gen. Virol. 2000;81:489–496. doi: 10.1099/0022-1317-81-2-489. [DOI] [PubMed] [Google Scholar]
- Krijnse-Locker J., Ericsson M., Rottier P.J.M., Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: Evidence that transport from the RER to the golgi complex requires only one vesicular transport step. J. Cell Biol. 1994;125:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger D.K., Kelly S.M., Lewicki D.N., Ruffolo R., Gallagher T.M. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 2001;75:2792–2802. doi: 10.1128/JVI.75.6.2792-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri T., Eriksson K.K., Putics A., Zust R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V., Ziebuhr J., Weber F. The ADP-ribose-1"-monophosphatase domains of SARS-coronavirus and Human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Lamirande E.W., DeDiego M.L., Roberts A., Jackson J.P., Alvarez E., Sheahan T., Shieh W.J., Zaki S.R., Baric R., Enjuanes L., Subbarao K. A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden Syrian hamsters. J. Virol. 2008;82:7721–7724. doi: 10.1128/JVI.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne L., Descoteaux J.P., Jolicoeur P. Mouse hepatitis virus 3 replication in T and B lymphocytes correlate with viral pathogenicity. J. Immunol. 1989;142:4458–4465. [PubMed] [Google Scholar]
- Langereis M.A., van Vliet A.L., Boot W., de Groot R.J. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J. Virol. 2010;84:8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Gelfi J., Lavenant L., Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi E., Gilden D.H., Highkin M.K., Weiss S.R. Persistence of mouse hepatitis virus A59 RNA in a slow virus demyelinating infection in mice as detected by in situ hybridization. J. Virol. 1984;51:563–566. doi: 10.1128/jvi.51.2.563-566.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi E., Gilden D.H., Wroblewska Z., Rorke L.B., Weiss S.R. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology. 1984;34:597–603. doi: 10.1212/wnl.34.5.597. [DOI] [PubMed] [Google Scholar]
- Le Prevost C., Levy-Leblond E., Virelizier J.L., Dupuy J.M. Immunopathology of mouse hepatitis virus type 3 infection. Role of humoral and cell-mediated immunity in resistance mechanisms. J. Immunol. 1975;114:221–225. [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Leibowitz J.L., Perlman S., Weinstock G., DeVries J.R., Budzilowicz C., Weissemann J.M., Weiss S.R. Detection of a murine coronavirus nonstructural protein encoded in a downstream open reading frame. Virology. 1988;164:156–164. doi: 10.1016/0042-6822(88)90631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J.L., Srinivasa R., Williamson S.T., Chua M.M., Liu M., Wu S., Kang H., Ma X.Z., Zhang J., Shalev I., Smith R., Phillips M.J. Genetic determinants of mouse hepatitis virus strain 1 pneumovirulence. J. Virol. 2010;84:9278–9291. doi: 10.1128/JVI.00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I., Hingley S.T., Chua M.M., Jiang X., Lavi E., Weiss S.R. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239:1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I., Hingley S.T., Chua M.M., Phillips J., Lavi E., Weiss S.R. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J. Virol. 1998;72:9628–9636. doi: 10.1128/jvi.72.12.9628-9636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy G.A., Leibowitz J.L., Edgington T.S. Induction of monocyte procoagulant activity by murine hepatitis virus type 3 parallels disease susceptibility in mice. J. Exp. Med. 1981;154:1150–1163. doi: 10.1084/jem.154.4.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy G.A., MacPhee P.J., Fung L.S., Fisher M.M., Rappaport A.M. The effect of mouse hepatitis virus infection on the microcirculation of the liver. Hepatology. 1983;3:964–973. doi: 10.1002/hep.1840030614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy G.A., Shaw R., Leibowitz J.L., Cole E. The immune response to mouse hepatitis virus: Genetic variation in antibody response and disease. Adv. Exp. Med. Biol. 1984;173:345–364. doi: 10.1007/978-1-4615-9373-7_35. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lescar J., Tam J.P., Liu D.X. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 2004;325:374–380. doi: 10.1016/j.bbrc.2004.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349:264–275. doi: 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Cavanagh D., Green P., Inglis S.C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker J.K., Klumperman J., Oorschot V., Horzinek M.C., Geuze H.J., Rottier P.J. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J. Biol. Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J.C., Debarnot C., Borg J.P., Bouvet M., Canard B., Morelli X., Lecine P. Molecular mapping of the RNA Cap 2'-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 2010;285:33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P.J., Noten A.F., Horzinek M.C., Spaan W.J. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C.E., Rose J.K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C.E., Mentone S.A., Rose J.K., Farquhar M.G. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. USA. 1990;87:6944–6948. doi: 10.1073/pnas.87.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C.E., Grim M.G., Esquela A., Chung S.W., Rolls M., Ryan K., Swift A.M. Retention of a cis Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol. Biol. Cell. 1993;4:695–704. doi: 10.1091/mbc.4.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNamara K.C., Chua M.M., Nelson P.T., Shen H., Weiss S.R. Increased epitope-specific CD8+ T cells prevent murine coronavirus spread to the spinal cord and subsequent demyelination. J. Virol. 2005;79:3370–3381. doi: 10.1128/JVI.79.6.3370-3381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda J., Repass J.F., Maeda A., Makino S. Membrane topology of coronavirus E protein. Virology. 2001;281:163–169. doi: 10.1006/viro.2001.0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaker R.A., Piczak C.V., Miller A.A., Stanton M.F. A hepatitis virus complicating studies with mouse leukemia. J. Natl. Cancer Inst. 1961;27:29–51. [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Marten N.W., Stohlman S.A., Bergmann C.C. Role of viral persistence in retaining CD8(+) T cells within the central nervous system. J. Virol. 2001;74:7903–7910. doi: 10.1128/jvi.74.17.7903-7910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Rottier P.J. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 2005;287:133–159. doi: 10.1007/3-540-26765-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Koetzner C.A., Kerr C.A., Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Communication between S1N330 and a region in S2 of murine coronavirus spike protein is important for virus entry into cells expressing CEACAM1b receptor. Virology. 2002;295:160–171. doi: 10.1006/viro.2002.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 2002;76:11819–11826. doi: 10.1128/JVI.76.23.11819-11826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Watanabe R., Taguchi F. Neurovirulence in mice of soluble receptor-resistant (srr)mutants of mouse hepatitis virus: intensive apoptosis caused by less virulent srr mutant. Arch. Virol. 2001;146(9):1643–1654. doi: 10.1007/s007050170053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.E., Weiss S.R., Paterson Y. Murine hepatitis virus—A model for virus-induced CNS demyelination. J. Neurovirol. 2002;8:76–85. doi: 10.1080/13550280290049534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J., Butler E., Zaki S., St Claire M., Murphy B., Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Machamer C.E. A single tyrosine in the severe acute respiratory syndrome coronavirus membrane protein cytoplasmic tail is important for efficient interaction with spike protein. J. Virol. 2010;84:1891–1901. doi: 10.1128/JVI.02458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., Meyerholz D.K., Kirby P. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGilvray I.D., Lu Z., Wei A.C., Dackiw A.P., Marshall J.C., Kapus A., Levy G., Rotstein O.D. Murine hepatitis virus strain 3 induces the macrophage prothrombinase fgl-2 through p38 mitogen-activated protein kinase activation. J. Biol. Chem. 1998;273:32222–32229. doi: 10.1074/jbc.273.48.32222. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: Recombination-independent selectable cloning vectors. Virology. 1992;190:522–526. doi: 10.1016/0042-6822(92)91246-q. [DOI] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwata N., Hasegawa H., Fukushi S., Yokoyama M., Harashima A., Sato Y., Saijo M., Morikawa S., Sata T. Participation of both host and virus factors in induction of severe acute respiratory syndrome (SARS) in F344 rats infected with SARS coronavirus. J. Virol. 2007;81:1848–1857. doi: 10.1128/JVI.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwata N., Hasegawa H., Fukushi S., Harashima A., Sato Y., Saijo M., Taguchi F., Morikawa S., Sata T. Mouse-passaged severe acute respiratory syndrome-associated coronavirus leads to lethal pulmonary edema and diffuse alveolar damage in adult but not young mice. Am. J. Pathol. 2008;172:1625–1637. doi: 10.2353/ajpath.2008.071060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwata-Yoshikawa N., Taguchi F. Studies of severe acute respiratory syndrome coronavirus pathology in human cases and animal models. Vet. Pathol. 2010;47:881–892. doi: 10.1177/0300985810378760. [DOI] [PubMed] [Google Scholar]
- Nakagaki K., Taguchi F. Receptor-independent spread of a highly neurotropic murine coronavirus JHMV strain from initially infected microglial cells in mixed neural cultures. J. Virol. 2005;79(10):6102–6110. doi: 10.1128/JVI.79.10.6102-6110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakauchi M., Kariwa H., Kon Y., Yoshii K., Maeda A., Takashima I. Analysis of severe acute respiratory syndrome coronavirus structural proteins in virus-like particle assembly. Microbiol. Immunol. 2008;52:625–630. doi: 10.1111/j.1348-0421.2008.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75:9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas S., Weiss S.R. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J. Virol. 2003;77:4972–4978. doi: 10.1128/JVI.77.8.4972-4978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas S., Seo S.H., Chua M.M., Das Sarma J., Lavi E., Hingley S.T., Weiss S.R. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 2001;75:2452–2457. doi: 10.1128/JVI.75.5.2452-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Martin S., Hingley S.T., Weiss S.R. Murine coronavirus evolution in vivo: Functional compensation of a detrimental amino acid substitution in the receptor binding domain of the spike glycoprotein. J. Virol. 2005;79:7629–7640. doi: 10.1128/JVI.79.12.7629-7640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure (Camb) 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., DeDiego M.L., Zhao J., Fett C., Alvarez E., Nieto-Torres J.L., Enjuanes L., Perlman S. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399:120–128. doi: 10.1016/j.virol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.P., Hogue B.G. Protein interactions during coronavirus assembly. J. Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H., Klenk H.D. Coronavirus glycoprotein E1, a new type of viral glycoprotein. J. Mol. Biol. 1981;153:993–1010. doi: 10.1016/0022-2836(81)90463-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H. The carbohydrates of mouse hepatitis virus(mhv) a59: Structures of the o-glycosidically linked oligosaccharides of glycoprotein E1. EMBO J. 1984;3:665–670. doi: 10.1002/j.1460-2075.1984.tb01864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H.G., Bleumink-Pluym N.M., Osterhaus A.D., Horzinek M.C., van der Zeijst B.A. Epitopes on the peplomer protein of infectious bronchitis virus strain M41 as defined by monoclonal antibodies. Virology. 1987;161:511–519. doi: 10.1016/0042-6822(87)90145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Dediego M.L., Alvarez E., Jimenez-Guardeno J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Q., Liu M., Kongkham P., Lai M.M., Marsden P.A., Tseng J., Pereira B., Belyavskyi M., Leibowitz J., Phillips M.J., Levy G. The nucleocapsid protein of murine hepatitis virus type 3 induces transcription of the novel fgl2 prothrombinase gene. J. Biol. Chem. 1999;274:9930–9936. doi: 10.1074/jbc.274.15.9930. [DOI] [PubMed] [Google Scholar]
- Ning Q., Lakatoo S., Liu M., Yang W., Wang Z., Phillips M.J., Levy G.A. Induction of prothrombinase fgl2 by the nucleocapsid protein of virulent mouse hepatitis virus is dependent on host hepatic nuclear factor-4 alpha. J. Biol. Chem. 2003;278:15541–15549. doi: 10.1074/jbc.M212806200. [DOI] [PubMed] [Google Scholar]
- Ontiveros E., Kuo L., Masters P.S., Perlman S. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology. 2001;289:230–238. doi: 10.1006/viro.2001.1167. [DOI] [PubMed] [Google Scholar]
- Ontiveros E., Kim T.S., Gallagher T.M., Perlman S. Enhanced virulence mediated by the murine coronavirus, mouse hepatitis virus strain JHM, is associated with a glycine at residue 310 of the spike glycoprotein. J. Virol. 2003;77:10260–10269. doi: 10.1128/JVI.77.19.10260-10269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., de Groot R.J., Rottier P.J. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80:2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M.J., Wolfs K., Horzinek M.C., Rottier P.J. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Ceriani J.E., Patino C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368:296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.M., Masters P.S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy K., Ng L., Lin X., Liu D., Pervushin K., Gong X., Torres J. Structural flexibility of the pentameric sars coronavirus envelope protein ion channel. Biophys. J. 2008;95:L39–L41. doi: 10.1529/biophysj.108.133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J.M., Kalicharran K., Dales S. Distribution and trafficking of JHM coronavirus structural proteins and virions in primary neurons and the OBL-21 neuronal cell line. J. Virol. 1994;68:2915–2928. doi: 10.1128/jvi.68.5.2915-2928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce B.D., Hobbs M.V., McGraw T.S., Buchmeier M.J. Cytokine induction during T-cell-mediated clearance of mouse hepatitis virus from neurons in vivo. J. Virol. 1994;68(9):5483–5495. doi: 10.1128/jvi.68.9.5483-5495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Yuen K.Y., Osterhaus A.D., Stohr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio. 2010;1:e00206–e00210. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman M.D.S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Wu G.F., Barnett E.M., Castro R.F., Perlman S. Cytotoxic T cell-resistant variants are selected in a virus-induced demyelinating disease. Immunity. 1996;5:253–262. doi: 10.1016/s1074-7613(00)80320-9. [DOI] [PubMed] [Google Scholar]
- Phillips J.J., Weiss S.R. MHV neuropathogenesis: The study of chimeric S genes and mutations in the hypervariable region. Adv. Exp. Med. Biol. 2001;494:115–119. doi: 10.1007/978-1-4615-1325-4_18. [DOI] [PubMed] [Google Scholar]
- Phillips J.J., Chua M.M., Rall G.F., Weiss S.R. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology. 2002;301:109–120. doi: 10.1006/viro.2002.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J. M., Kuo, I., Richardson, C., Weiss, S. R., submitted. A novel full-length isoform of murine pregnancy-specific glycoprotein 16 (PSG16) expressed in the brain does not mediate mouse hepatitis virus (MHV) entry. Manuscript submitted [DOI] [PMC free article] [PubMed]
- Piotrowski Y., Hansen G., Boomaars-van der Zanden A.L., Snijder E.J., Gorbalenya A.E., Hilgenfeld R. Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 2009;18:6–16. doi: 10.1002/pro.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma C.C., Nedialkova D.D., Zevenhoven-Dobbe J.C., Blokhuis J.H., Gorbalenya A.E., Snijder E.J. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1"-monophosphatase: A conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1''-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Pyrc K., Berkhout B., van der Hoek L. The novel human coronaviruses NL63 and HKU1. J. Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Wang J., Wei Q., She M., Marasco W.A., Jiang H., Tu X., Zhu H., Ren L., Gao H., Guo L., Huang L. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J. Pathol. 2005;206:251–259. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsman M.J., Locker J.K., de Hooge A., de Vries A.A., Griffiths G., Vennema H., Rottier P.J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Paddock C., Vogel L., Butler E., Zaki S., Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 2005;79:5833–5838. doi: 10.1128/JVI.79.9.5833-5838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Vogel L., Guarner J., Hayes N., Murphy B., Zaki S., Subbarao K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79:503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., Zaki S.R., Baric R. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Lamirande E.W., Vogel L., Jackson J.P., Paddock C.D., Guarner J., Zaki S.R., Sheahan T., Baric R., Subbarao K. Animal models and vaccines for SARS-CoV infection. Virus Res. 2007;133:20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., Dyer M.D., Teal T.H., Proll S., van den Brand J., Baric R., Katze M.G. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Stokes H., Chang G., Chua M.M., Thiel V., Weiss S.R., Gorbalenya A.E., Siddell S.G. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 2009;83:3743–3753. doi: 10.1128/JVI.02203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P., Brandenburg D., Armstrong J., van der Zeijst B., Warren G. Assembly in vitro of a spanning membrane protein of the endoplasmic reticulum: The E1 glycoprotein of coronavirus mouse hepatitis virus A59. Proc. Natl. Acad. Sci. USA. 1984;81:1421–1425. doi: 10.1073/pnas.81.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J., Welling G.W., Welling-Wester S., Niesters H.G., Lenstra J.A., Van der Zeijst B.A. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- Rowe T., Gao G., Hogan R.J., Crystal R.G., Voss T.G., Grant R.L., Bell P., Kobinger G.P., Wivel N.A., Wilson J.M. Macaque model for severe acute respiratory syndrome. J. Virol. 2004;78:11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki K., Ohtsuka N., Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 1997;71:9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki K., Ohtsuka N., Taguchi F. Isolation and characterization of murine coronavirus mutants resistant to neutralization by soluble receptors. Adv. Exp. Med. Biol. 1998;440:11–16. doi: 10.1007/978-1-4615-5331-1_2. [DOI] [PubMed] [Google Scholar]
- Savarin C., Bergmann C.C. Neuroimmunology of central nervous system viral infections: The cells, molecules and mechanisms involved. Curr. Opin. Pharmacol. 2008;8:472–479. doi: 10.1016/j.coph.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B., Routledge E., Siddell S.G. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4:e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Bertram S., Glowacka I., Steffen I., Chaipan C., Agudelo J., Lu K., Rennekamp A.J., Hofmann H., Bates P., Pohlmann S. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology. 2011;413:265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Kok K.-H., Ng M.-H.J., Poon V.K.M., Yuen K.-Y., Zheng B.-J., Jin D.-Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3·TANK·TBK1/IKKϵ complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M.A., Ebner D., Siddell S.G. Coronavirus MHV-JHM mRNA 5 has a sequence arrangement which potentially allows translation of a second, downstream open reading frame. J. Gen. Virol. 1985;66:581–592. doi: 10.1099/0022-1317-66-3-581. [DOI] [PubMed] [Google Scholar]
- Smits S.L., Gerwig G.J., van Vliet A.L., Lissenberg A., Briza P., Kamerling J.P., Vlasak R., de Groot R.J. Nidovirus sialate-O-acetylesterases: Evolution and substrate specificity of corona- and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe L.H., Shieh C.K., Baker S.C., Chang M.F., Lai M.M.C. Sequence and translation of the murine coronavirus 5'-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J. Virol. 1987;61:3968–3976. doi: 10.1128/jvi.61.12.3968-3976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry S.M., Kazi L., Graham R.L., Baric R.S., Weiss S.R., Denison M.R. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F., Sefton B.M. Coronavirus proteins: Structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J. Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Desforges M., Almazan F., Jacomy H., Enjuanes L., Talbot P.J. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 2006;80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Brayton P.R., Fleming J.O., Weiner L.P., Lai M.M. Murine coronaviruses: Isolation and characterization of two plaque morphology variants of the JHM neurotropic strain. J. Gen. Virol. 1982;63:265–275. doi: 10.1099/0022-1317-63-2-265. [DOI] [PubMed] [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Ricard C.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: Activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 1985;56:904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., Roberts A. Is there an ideal animal model for SARS? Trends Microbiol. 2006;14:299–303. doi: 10.1016/j.tim.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift A.M., Machamer C.E. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Matsuyama S. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J. Virol. 2002;76:950–958. doi: 10.1128/JVI.76.3.950-958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Siddell S.G., Wege H., ter Meulen V. Characterization of a variant virus selected in rat brains after infection by coronavirus mouse hepatitis virus JHM. J. Virol. 1985;54:429–435. doi: 10.1128/jvi.54.2.429-435.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Suzuki H., Takahashi H., Kubo H. Neurovirulence for rats of the JHMV variants escaped from neutralization with the S1-specific monoclonal antibodies. Adv. Exp. Med. Biol. 1995;380:185–187. doi: 10.1007/978-1-4615-1899-0_31. [DOI] [PubMed] [Google Scholar]
- Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.-J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2008;82:1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh K.-T., Siu Y.-L., Chan W.-L., Schluter M.A., Liu C.-J., Peiris J.S.M., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W., Gelderblom H.R., Goudsmit J. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Springer; New York: 2005. Reverse Genetics of Coronaviruses Using Vaccinia Virus Vectors, Coronavirus Genome Structure and Replication. pp. 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohya Y., Narayanan K., Kamitani W., Huang C., Lokugamage K., Makino S. Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 2009;83:5282–5288. doi: 10.1128/JVI.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S.A. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: Virus budding is restricted to the Golgi region. Eur. J. Cell Biol. 1985;37:203–212. [PubMed] [Google Scholar]
- Tooze J., Tooze S., Warren G. Replication of coronavirus MHV-A59 in sac- cells: Determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- Torres J., Parthasarathy K., Lin X., Saravanan R., Kukol A., Liu D.X. Model of a putative pore: The pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.C., de Groot L., Pinon J.D., Iacono K.T., Phillips J.J., Seo S.H., Lavi E., Weiss S.R. Amino acid substitutions within the heptad repeat domain 1 of murine coronavirus spike protein restrict viral antigen spread in the central nervous system. Virology. 2003;312:369–380. doi: 10.1016/S0042-6822(03)00248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.C., Zelus B.D., Holmes K.V., Weiss S.R. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 2003;77:841–850. doi: 10.1128/JVI.77.2.841-850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C., Lam B. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348 doi: 10.1056/NEJMoa030666. 1977–1985. [DOI] [PubMed] [Google Scholar]
- Tseng C.T., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui P.T., Kwok M.L., Yuen H., Lai S.T. Severe acute respiratory syndrome: Clinical outcome and prognostic correlates. Emerg. Infect. Dis. 2003;9:1064–1069. doi: 10.3201/eid0909.030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varia M., Wilson S., Sarwal S., McGeer A., Gournis E., Galanis E., Henry B. Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. CMAJ. 2003;169:285–292. [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S., de Haan C.A. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Bednar V., Blount A., Hogue B.G. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 2006;80:4344–4355. doi: 10.1128/JVI.80.9.4344-4355.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Lopez L.A., Bednar V., Hogue B.G. Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein. J. Virol. 2007;81:5339–5348. doi: 10.1128/JVI.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virelizier J.L., Dayan A.D., Allison A.C. Neuropathological effects of persistent infection of mice by mouse hepatitis virus. Infect. Immun. 1975;12:1127–1140. doi: 10.1128/iai.12.5.1127-1140.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Leider J., Spaan W., Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss D., Kern A., Traggiai E., Eickmann M., Stadler K., Lanzavecchia A., Becker S. Characterization of severe acute respiratory syndrome coronavirus membrane protein. FEBS Lett. 2006;580:968–973. doi: 10.1016/j.febslet.2006.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss D., Pfefferle S., Drosten C., Stevermann L., Traggiai E., Lanzavecchia A., Becker S. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol. J. 2009;6:79. doi: 10.1186/1743-422X-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.I., Fleming J.O., Lai M.M. Sequence analysis of the spike protein gene of murine coronavirus variants: Study of genetic sites affecting neuropathogenicity. Virology. 1992;186:742–749. doi: 10.1016/0042-6822(92)90041-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E., Wang H., Ye J. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Guo F., Liu K., Wang H., Rao S., Yang P., Jiang C. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Fang S., Xiao H., Chen B., Tam J.P., Liu D.X. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS One. 2009;4:e4908. doi: 10.1371/journal.pone.0004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shi H., Rigolet P., Wu N., Zhu L., Xi X.G., Vabret A., Wang X., Wang T. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect. Genet. Evol. 2010;10:919–924. doi: 10.1016/j.meegid.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: Role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner L.P. Pathogenesis of demyelination induced by a mouse hepatitis. Arch. Neurol. 1973;28:298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]
- Weiner L.P., Johnson R.T., Herndon R.M. Viral infections and demyelinating diseases. N. Engl. J. Med. 1973;288:1103–1110. doi: 10.1056/NEJM197305242882106. [DOI] [PubMed] [Google Scholar]
- Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Pathogenesis of murine coronavirus infection. In: Perlman S., Gallagher T.M., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2008. [Google Scholar]
- WHO . 2003. Severe Acute Respiratory Syndrome (SARS): Multi-country outbreak. [Google Scholar]
- WHO . 2004. WHO calls for urgent investigation into sources of infection in recent SARS cases in Guangdong Province, after a fourth case is announced. Press Release, January 31. [Google Scholar]
- Williamson J.S.P., Sykes K.C., Stohlman S.A. Characterization of brain-infiltrating monuclear cells during infection with mouse hepatitis virus strain JHM. J. Neuroimmunol. 1991;32:199–207. doi: 10.1016/0165-5728(91)90189-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten-van Asperen R.M., Lutter R., Haitsma J.J., Merkus M.P., van Woensel J.B., van der Loos C.M., Florquin S., Lachmann B., Bos A.P. ACE mediates ventilator-induced lung injury in rats via angiotensin II but not bradykinin. Eur. Respir. J. 2008;31:363–371. doi: 10.1183/09031936.00060207. [DOI] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzer W.J., Obojes K., Vlasak R. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: A proposal to reorganize group 2 Coronaviridae. J. Gen. Virol. 2002;83:395–402. doi: 10.1099/0022-1317-83-2-395. [DOI] [PubMed] [Google Scholar]
- Xu H.Y., Lim K.P., Shen S., Liu D.X. Further identification and characterization of novel intermediate and mature cleavage products released from the orf 1b region of the avian coronavirus infectious bronchitis virus 1a/1b polyprotein. Virology. 2001;288:212–222. doi: 10.1006/viro.2001.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: Implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31:7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.F., Wang M., Zhang Z.B., Zou X.Z., Gao Y., Liu X.N., Lu E.J., Pan B.Y., Wu S.J., Yu S.Y. An epidemiologic investigation on infection with severe acute respiratory syndrome coronavirus in wild animals traders in Guangzhou. Zhonghua Yu Fang Yi Xue Za Zhi. 2004;38:81–83. [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Khadijah S., Fang S., Wang L., Tay F.P., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A., Taguchi F., Fujiwara K. T lymphocyte-dependent difference in susceptibility between DDD and C3H mice to mouse hepatitis virus, MHV-3. Jpn. J. Exp. Med. 1979;49:413–421. [PubMed] [Google Scholar]
- Yamada Y.K., Yabe M., Ohtsuki T., Taguchi F. Unique N-linked glycosylation of murine coronavirus MHV-2 membrane protein at the conserved O-linked glycosylation site. Virus Res. 2000;66:149–154. doi: 10.1016/S0168-1702(99)00134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F. Bcl-xL inhibits T cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81:3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hauns K., Langland J.O., Jacobs B.L., Hogue B.G. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S.H., Wang H.Y., Tsai C.Y., Kao C.L., Yang J.Y., Liu H.W., Su I.J., Tsai S.F., Chen D.S., Chen P.J. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: Molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C.W., Hon C.C., Shi M., Lam T.T.-Y., Chow K.Y.-C., Zeng F., Leung F.C.-C. Phylogenetic perspectives on the epidemiology and origins of SARS and SARS-like coronaviruses. Infect. Genet. Evol. 2009;9:1185–1196. doi: 10.1016/j.meegid.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Lai M.M. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J. Virol. 1991;65:5605–5608. doi: 10.1128/jvi.65.10.5605-5608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., La Monica N., Makino S., Shieh C.K., Lai M.M. Biosynthesis, structure, and biological activities of envelope protein gp65 of murine coronavirus. Virology. 1989;173:683–691. doi: 10.1016/0042-6822(89)90581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Banner L.R., Lai M.M. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991;183:647–657. doi: 10.1016/0042-6822(91)90994-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Baker S.C., Stohlman S.A., Lai M.M. Hemagglutinin-esterase-specific monoclonal antibodies alter the neuropathogenicity of mouse hepatitis virus. J. Virol. 1992;66:2865–2874. doi: 10.1128/jvi.66.5.2865-2874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Asanaka M., Stohlman S.A., Makino S., Shubin R.A., Gilmore W., Weiner L.P., Wang F.I., Lai M.M. Neuropathogenicity of mouse hepatitis virus JHM isolates differing in hemagglutinin-esterase protein expression. J. Neurovirol. 1995;1:330–339. doi: 10.3109/13550289509111022. [DOI] [PubMed] [Google Scholar]
- Youn S., Leibowitz J.L., Collisson E.W. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology. 2005;332:206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: Transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Bi W., Weiss S.R., Leibowitz J.L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Liao Y., Torres J., Tam J.P., Liu D.X. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580:3192–3200. doi: 10.1016/j.febslet.2006.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B., Chen B., Zhao Z., Wang S., Chen J., Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebhauser R., Kammerer R., Eisenried A., McLellan A., Moore T., Zimmermann W. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang H., Sun G.Y. LPS induces permeability injury in lung microvascular endothelium via AT(1) receptor. Arch. Biochem. Biophys. 2005;441:75–83. doi: 10.1016/j.abb.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Rose K.M., Elliott R., Van Rooijen N., Weiss S.R. Cell type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J. Virol. 2011;85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Perlman S. Preferential infection of mature dendritic cells by mouse hepatitis virus strain JHM. J. Virol. 2006;80:2506–2514. doi: 10.1128/JVI.80.5.2506-2514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ferraro D., Zhao J., Hussain S., Shao J., Trujillo J., Netland J., Gallagher T., Perlman S. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J. Virol. 2010;84:3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Chen H.W. Monophyletic relationship between severe acute respiratory syndrome coronavirus and group 2 coronaviruses. J. Infect. Dis. 2004;189:1676–1678. doi: 10.1086/382892. Epub 2004 Apr 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltick P.W., Leibowitz J.L., Oleszak E.L., Weiss S.R. Mouse hepatitis virus ORF 2a is expressed in the cytosol of infected mouse fibroblasts. Virology. 1990;174:605–607. doi: 10.1016/0042-6822(90)90114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zornetzer G.A., Frieman M.B., Rosenzweig E., Korth M.J., Page C., Baric R.S., Katze M.G. Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection. J. Virol. 2010;84:11297–11309. doi: 10.1128/JVI.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


