INTRODUCTION
The advances discussed in this chapter should be relevant to a broad and heterogeneous readership because of their significance, novelty, or wide applicability of viral chip technology in genomic medicine. These advances are making an outstanding contribution to the development of an important field of research. The state-of-the-field, particularly as it has evolved over the last years, is surveyed. As the field is already too expansive for a single article, I apologize in advance for the many omissions, but hope that this chapter captures the excitement of recent achievements in viral chip technology for genomic medicine.
Viruses are a heterogeneous group of infectious agents composed mainly of nucleic acids, either DNA or RNA, packed tightly inside the protein coat. The outer shell mainly protects virus from physical, chemical, or enzymatic damage. Following entry into the cell by specific receptors (Table 48.1 ), the virus uses the host cellular regulatory system and the host cells synthetic machinery to produce enzymes required for its replication, and in many cases the viral genome encodes proteins, which act, for example, as RNA-polymerases, transcriptases that are needed in replication. These components are then assembled and released as viral particles. Viruses also use host cell ribosomes to translate viral proteins. Viruses can only replicate within cells. Thus, viruses are an exception to what is understood as the concept of “life.”
TABLE 48.1.
Representative viruses and viral binding receptors
| Virus | Cell surface receptora |
|---|---|
| Influenza | Glycophorin A |
| Rhinovirus | Adhesion molecule ICAM-1 |
| Reovirus | β-adrenergic hormone receptor |
| Rabies | Acetylcholine receptor |
| Vaccinia | Epidermal growth factor receptor |
| Epstein-Bar | B cell complement receptor |
| HIV | T cell, macrophage CD4 receptor |
All viruses need to bind to a specific cell surface molecule in order to enter into the cell. The binding can be blocked by antibody, but there are cases in which the antibody helps the virus to get in.
ROLE OF VIRUSES IN HUMAN INFECTIOUS DISEASE
Viruses cause diseases ranging from acute infections (e.g., poliomyelitis) to chronic infections that are relatively benign (e.g., herpes) to life-threatening chronic infections (e.g., AIDS). As shown in Table 48.2 and in the recommended general references, different viruses lead to markedly different diseases, reflecting the diverse processes by which they damage human tissues and cells. Some common viruses are listed with typical examples of diseases caused in humans. The variety of viruses has required potential hosts to develop two crucial features of adaptive immunity. First, the need to recognize a wide range of different DNA and RNA pathogens had driven the development of receptors on B and T cells of equal or greater diversity. Second, the distinct habitants and life cycles have to be countered by a range of different effector mechanisms. Characteristic features of each virus are its mode of transmission, its mechanism of replication, its pathogenesis or the means by which it causes disease, and the immune response it elicits.
TABLE 48.2.
Some common viruses that cause diseases in humansa
| Virus genome | Virus family | Virus Genus with type species | Human disease |
|---|---|---|---|
| DNA(ds) | Herpesvirida | Simplex virus with human herpesvirus 1 and 2 or herpes simplex type 1and 2 (HSV-1, HSV-2) | Cold sores, genital herpes, encephalitis |
| Varicellovirus with human herpesvirus 3 or varicella-zoster virus (VZV) | Chickenpox, shingles | ||
| Cytomegalovirus with human herpesvirus 5 (CMV) | Mononucleosis | ||
| Lymphocryptovirus with human herpesvirus 4 or Epstein-Bar virus (EBV) | Mononucleosis, Burkitt's lymphoma | ||
| Roseolovirus with human herpesvirus 6 (HHV-6) | Erythema subitum, roseola infantum | ||
| (ds) | Adenoviridae | Mastadenovirus with human adenoviruses types 3 (HAdV-A), 4 (HAdV-E), and 7 (HAdV-B) | Acute respiratory disease (ARD) |
| (ds) | Poxviridae | Orthopoxvirus with vaccinia virus | Cowpox, vaccinia, smallpox |
| (ss) | Parvoviridae | Erythrovirus with human parvovirus B-19 | Erythema infectiosium, seronegative arthritis, hydrops fetalis |
| (ds) | Papillomaviridae | Human papilloma virus with human papilloma virus types (strains) 16, 18, and 31 | Cervical cancer |
| (ds) | Hepadnaviridae | Orthohepadnavirus with human hepatitis B virus | Hepatitis B |
| RNA(ss) | Orthomyxoviridae | Influenza virus A with types H1N1, H2N2, H3N2, and H5N1 | Spanish flu, Asian flu, Hong Kong flu, or pandemic threat in 2006-7 flu season |
| (ss) | Paramyxoviridae | Morbillivirus with measles virus | Measles |
| Rubulavirus with Mumps virus | Mumps | ||
| (ss) | Coronaviridae | Coronavirus with SARS coronavirus | Severe acute respiratory syndrome (SARS) |
| (ss) | Picornaviridae | Hepatovirus with hepatitis A virus | Hepatitis A |
| Enterovirus with polioviruses and coxsackieviruses | Poliomyelitis and infectious myocarditis by coxsackievirus B | ||
| Rhinovirus with human rhinovirus A | Common cold (acute viral nasopharyngitis) | ||
| (ds) | Reoviridae | Rotavirus with rotavirus A, and B | Severe diarrhea among infants and children, or adult diarrhea by rotavirus B |
| Coltivius with Colorado tick fever virus | Colorado tick fever | ||
| (ss) | Togaviridae | Rubivirus with rubella virus | Rubella |
| (ss) | Flaviviridae | Flavivirus with yellow fever virus, or Dengue virus | Yellow fever, or Dengue hemorrhagic fever found in the tropics |
| Hepacivirus with hepatitis C virus | Hepatitis C | ||
| (ss) | Arenaviridae | Arenavirus with lymphocytic choriomenigitis virus | Lymphocytic choriominigitis |
| Arenavirus with Lassa virus | Lassa fever | ||
| (ss) | Rhabdoviridae | Lyssavirus with rabies virus | Rabies |
| (ss) | Retroviridae | Deltaretrovirus with human T-lymphotropic virus | T-cell leukaemia and T-cell lymphoma in adults |
| Lentivirus with human immunodeficiency virus 1 and 2 (HIV-1 and HIV-2) | Acquired immunodeficiency syndrome (AIDS) |
For general information, see: Rich, R.R., Fleisher, T.A., Shearer, W.T., Schroeder, H.W., Frew, A.J. and Weyand, C.M. (eds.) (2008). Clinical Immunology: Principles and Practice, 3rd edition. Mosby Ltd., New York.
Elimination of a viral infection generally requires the destruction of virally infected cells by osmotic lysis, which follows natural killer (NK) or cytotoxic T-cell (CTL) activation. Activation of these immune cells involves cytokines secreted by antigen presenting cells such as macrophages and dendritic cells, or Type I cytokine producing CD4+ T cells. NK and CTL cells differ in killing virus–infected cells mainly at the recognition site. NK and CTL cells active in these processes recognize unique surface features of virally infected cells, which are not present on non-infected cells. NK cells can use several different receptors that signal them to kill, including lectin-like receptors that recognize carbohydrate on self cells, killing inhibitory receptors (KIRs) that recognize “free” major histocompatibility complex (MHC) class I molecules encoded by genes of the human leukocyte antigen (HLA) C region and overrule the actions of the killing activation receptors (KARs). Once recognition has occurred, the cytotoxic mechanisms of the NK and CTL cells are similar.
During the initial phase of viral infection and when the viruses bud from the cell, the particles can be targeted by IgM and IgG antibodies, leading to opsonin-mediated phagocytosis and/or neutralization. Phagocytosis and neutralization by antibodies are components of the natural and adaptive immune system in response to viral infection when the virus is disseminated via lymph, blood, or interstitial fluid, rather sequestered within a cell. Immune response to viruses also varies according to the site where the virus penetrates the human organisms and whether the infection is primary or secondary.
General Problem of Needing to Identify the Relevant Virus
Because viruses are sequestered within host cells and replicate rapidly, viruses produce mutations rapidly. When these mutations occur at the antigenic epitope, previously active immune effector cells may no longer recognize the altered virus. Immunological resistance develops. Many disease states have been associated with immune dysfunction of varying degrees of severity and significance, for example HIV/AIDS, flu, or colds. Antigenic diversity is the result of extensive antigenic variation in the past.
Viruses interfere with the proper functioning of the cytokine network. For example, adenoviruses and rotaviruses inhibit interferon formation, hepatitis B virus does not induce interferon, and the EBV releases inhibitory cytokines suchas IL-10.
Not all virus infections cause disease. The objective of the virus is to survive and be transmitted. Sometimes this may cause pathology; for example, diarrhea helps the rotaviruses (see Table 48.2) to get into water and to infect another host. Some viruses are obliged to destroy cells in order to spread, but many viruses inhabit the human body without causing disease symptoms for a long time, and these could be regarded as the most successful viruses. EBV in the throat and HSV-1 in the ganglia are examples of this.
How Were Viruses Identified Prior to the Development of Chip Technology?
Diagnosis depends on the detection of antiviral antibody and/or viral antigen, not on immunological markers. The nature of investigations undertaken should be selected accordingly. For example, immune cell proliferation is often abnormal in viral infections but does not help in identification of the relevant virus. Accurate monitoring of viral load is now performed by quantitative real-time PCR.
In general, there are three main and well-established approaches to identify a virus in the human (for overview, see Striebel et al., 2003):
-
(i)
Isolation and cultivation techniques
These techniques have some disadvantages, in particular long cultivation times from 3 up to 30 days, lack of sensitivity, and the rather limited number of viruses that can be identified in comparison to the broad spectrum of possible viruses that cause infection and/or disease. Identification of isolates from cell cultures occurs by titration with different serum pools. This is additionally hampered by constant evolving of new viral subtypes. Alternatively, type specific detection may directly be performed using monoclonal antibodies, immunofluorescence, or enzyme immunoassay (EIA).
-
(ii)
Indirect virus detection by virus-specific antibodies
Indirect virus detection methods include serological determination of specific antibodies by immunofluorescence assay, enzyme immunoassay, or by titration. After infection, only a few viruses release antigens in amounts sufficient to be detectable in body fluids by antibody assays (ELISAs). The presence of antibodies indicates immunity of the human organism in most virus infections, whereas re-infections do not show any symptoms. Antibody generation is detectable shortly after disease breaks out (e.g., in patients with viral hepatitis), but not during the highly infectious incubation time. Diagnosis of infections by cytopathogenic viruses, such as influenza or respiratory infections, may be easily performed in late states of these diseases, but are hardly possible during early disease states. While serotyping of viruses is mostly done by conventional immunological methods, many clinical isolates remain unclassifiable due to the limited number of antibodies against virus surface proteins, for example, of enteroviruses. Array-based assays are able to detect several serotypes with high accuracy. Highly sensitive array-based assay may become a useful alternative in clinical diagnostics.
-
(iii)
Direct detection of viral antigen or genomic features
Nucleic acid detection methods in virology may be separated into two basic groups: in vitro nucleic acid-amplifying techniques such as PCR including reverse transcriptase (RT)-PCR and non-amplifying techniques. Analysis of virus specific nucleic acids without in vitro amplification may be performed best by hybridization of gene specific probes, followed by radioactive or immunological detection methods (i.e., ELISA, or chemiluminescence). Practical applications of these direct methods are rare, as sensitivities often are too low in comparison to quantitative real-time PCR or RT-PCR by LightCycler or TaqMan. However, PCR and RT-PCR bear increased risks of contamination by non-specific amplification products but the main limitation resides in the difficulty of designing compatible multiplex primer sets for large numbers of viruses to be screened in a single assay, being restricted to a limited number of viral families. By contrast, serial analysis of gene expression and differential display is a powerful high-throughput methodology (Conejero-Goldberg et al., 2005).
Chip Technology
Modern DNA microarray technology started in the mid-1990's when Pat Brown and his colleagues at Stanford University first reported a high-capacity system in which DNA sequences specific to individual transcripts were immobilized at defined locations in an array format on a solid surface (Brown et al., 1999; DeRisi and Iyer, 1999; DeRisi et al., 1996; Schena et al., 1995), either through mechanical spotting (Lipshutz et al., 1995) or some implementation of in situ synthesis (Fodor et al., 1991). With the original DNA chip technology, microarrays were prepared by high-speed robotic printing of complementary DNAs on glass. Because of the small format and high density of the arrays, hybridization volumes of 2 μL could be used that enabled detection of rare transcripts in probe mixtures derived from 2 μg of total cellular messenger RNA (Schena et al., 1995). Differential expression measurements were performed by means of simultaneous, two-color fluorescence hybridization in parallel. This microarray-based genome-wide expression analysis of 45 genes first provided an unbiased view of the transcriptional state of a cell population (from Arabidopsis).
DNA chips/microarrays are the biotechnology tool of our decade. Pathogen detection, resequencing, comparative genome analysis, and gene expression – they do it all. The amount of data they are generating is overwhelming. The expression levels of thousands of viral genes as well as many mutant variants are documented for numerous states of growth and disease. It is inevitable that DNA chips/microarrays will drive genomic medicine to new horizons.
In the early 1990's, we purified DNA, ligated it, transformed it, and the cloned gene had to be somewhere. We screened thousands of clones by hand with an oligonucleotide just to find one insert. Of course, positive selection vectors facilitated screening, but still we had to do the hard stuff by hand. Today's DNA chip/array technology reverses that early approach. Instead of screening an array of unknowns with a defined probe like a synthetic oligonucleotide, PCR product or cloned gene, now a defined probe occupies the chip and then the chip is probed with the unknown sample. Today, identification of disease-relevant genes or proteins often starts with transcription profiling experiments on DNA chips and/or proteomics approaches. The interpretation of the measured data is largely based upon the complete sequences of the human genome and the most important model organisms, respectively.
The overall objective of this chapter is twofold. First, the objective is to increase awareness in clinical medicine of the challenges and opportunities presented by DNA microarray technology and the emerging and rapidly changing field of genomic medicine. Second, the aim is to publicize to a broader research community additional challenges associated with the use of microarray technology in medically relevant genome research.
MICROFABRICATION
Probe selection and microarray/chip design are central to the reliability, sensitivity, specificity, and robustness of viral arrays. Because common viral microarray/chip manufacturing technology synthesizes probes with defined sequences, positions, and lengths, array performance can be optimized using data collected from multiple databases, bioinformatics tools, and computer models. Viral oligonucleotide sequences can be designed according to open reading frames sequences obtained from Entrez Genome and Nucleotide Databases (http://www.ncbi.nlm.nih.gov/). The probe selection should be based on the criteria for specificity, sensitivity, and uniformity (Hughes et al., 2001; Wright and Church, 2002). Arrays can be designed with Array Designer 2.02 software (http://www.premierbiosoft.com). Recently, a bioinformatic platform and database were developed with specific probes of all known viral genome sequences to facilitate the design of diagnostic chips (Lin et al., 2006: http://www.bioinfo.csie.ncu.edu.tw/).
Some of the key elements of viral chip microfabrication are shown in Figure 48.1 and are common to the production of all microarrays/chips. Probe concentration, probe DNA length, and printing buffer need to be optimized for high quality chip performance depending on the surface chemistry. Slide autofluorescence, spot morphology, reproducibility of arraying, binding efficiency and purity of probes are crucial for the accuracy and reliability of chip data analysis. The manufacturing process ends with a comprehensive series of quality control tests (Földes-Papp et al., 2004). Additionally, a sampling of arrays from every wafer is used to test the batch by running control hybridizations. A quantitative test of hybridization is performed using standardized control probes. For example, the results of 12,900 hybridization reactions on about 150 configured human herpes virus microarrays for the parallel detection of HSV-1 and HSV-2, VZV, EBV, CMV, and HHV-6 showed that the established microarray/chipbased typing procedure was reproducible, virus-specific and sufficiently sensitive with a lower limit of 100 viral copies per mL sample (Földes-Papp et al., 2004, 2005b). A core element of array design, the perfect match/mismatch probe strategy, is also universally applied to the production of viral chips. For each probe designed to be perfectly complementary to a target sequence, a partner probe is generated that is identical except for a single base mismatch in its center. These probe pairs, which are called the perfect match probe and the mismatch probe, allow the quantization and subtraction of signals caused by nonspecific cross-hybridization. The difference in hybridization signals between the partners, and their intensity ratios, serve as indicators of specific target abundance.
Figure 48.1.
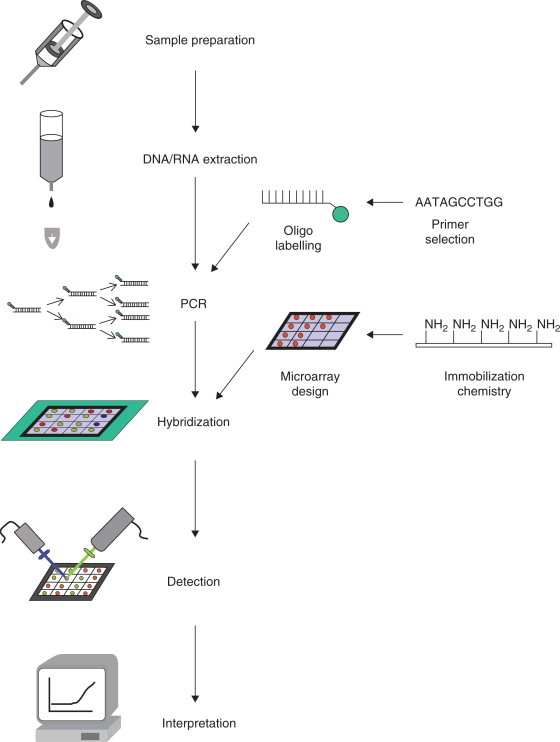
Some key elements of viral chip microfabrication (see Striebel et al., 2003).
The efficiency with which RNA target molecules are captured by, or hybridized to, surface-immobilized oligonucleotides depends upon secondary and tertiary structure of the RNA target strand. To overcome this limitation, RNA is often fragmented to reduce structural effects. The hybridization efficiency of the resulting fragments was determined as a function of fragment length and the amount of RNA captured was evaluated qualitatively by fluorescence intensity normalized to an internal standard (Mehlmann et al., 2005). Optimized conditions for influenza RNA were determined to include a fragmentation time of 20–30 m in at 75°C. These conditions resulted in a maximum concentration of fragments between 38 and 150 nucleotides in length and a maximum in the capture and label efficiency.
In the case of viral chips, for example, an online platform and database that provide users with specific probe sequences of all known viral genome sequences was established to facilitate the design of diagnostic chips (Lin et al., 2006). A user can select any number of different viruses and set the experimental conditions such as melting temperature and length of probe. The system then returns the optimal sequences from the database. The experimental design of a microarray determines the confidence that can be assigned to the data. Specificity, sensitivity, reproducibility, and robustness of the experimental data are essential to a successful use of a viral DNA chip (Földes-Papp et al., 2005b). The microarray-chip platform to be chosen has high value for “multiplex” array diagnostics, that is the parallel determination of several parameters from one sample. There are several platform categories available, such as amplified cDNAs, oligonucleotides chemically synthesized from known sequences, and one-color and two-color samples labeled and hybridized to a chip. The selected chip platform has an effect on the complexity and flexibility of the generated data. For example, the analysis of one-color chips is more straightforward than the analysis of two-color chips. Data formats like CEL, CHP, MAS 5, RMA GenePix, or ImaGene affect the signal intensity and quality of data for downstream analysis. In differential gene expression studies, thousands of genes are simultaneously interrogated, and this in turn determines the statistics to be applied (Draghici, 2002) and the use of real and simulated chip data for testing multiple hypotheses (Reiner et al., 2003). The application of z-score statistics to gene ontology terms is described in Doniger et al. (2003). Because microarray experiments produce tremendous amounts of data, data management by databases efficiently organizes and retrieves the raw data. Sharing the data both within the lab and with collaborators requires web-based system access.
Several aspects of probe selection and array design are dictated by an array s intended use. To select probes for viral microarrays/chips, sequence and annotation data from multiple databases are integrated. Custom arrays can be designed for subsets of known genes.
Structured Substrates
Traditional hybridization assays used nitrocellulose and nylon membranes, and measurements were carried out by autoradiography. Microarrays and other chip assays are based on solid supports, usually glass. Measurements are commonly performed by fluorescence labeling and detection. In contrast to membrane-based macroscopic arrays, microarray/chip supports (substrates) are non-porous, and therefore prevent absorption of reagents and samples on the substrate matrix. Non-porosity permits fast removal of organic and fluorescence substances during chip microfabrication and application. Microarray/chip supports allow highly parallel reactions, as well as significant increase in accuracy of measured data (Schena, 2001). These advantages maintain high sample concentrations and fast hybridization kinetics.
In general, glass substrates are characterized by high reproducibility of the fluorescence signal during hybridization of immobilized oligonucleotides with fluorescence-labeled samples due to low autofluorescence and thus high signal-to-noise ratio. They also show high reaction stability over extended time periods. Borofluorate glass substrates are particularly suitable because of their low alkali content and therefore low autofluorescence.
Spotters, Growers, and Grabbers
Spotting
Probe immobilization can be achieved using spotters for creating the arrays. The spatial resolution of the robotics determines the density of the array. Mechanical pins (contact printing) and ink-jetting usually gives spots of 100 p,m in diameter. The pin diameter and shape, solution viscosity, and substrate characteristics lead to some variation in spot size, shape, and concentration of solution transferred. A number of companies offer robotic systems for spotting arrays, but only commercially available DNA chips will pack as many spots as possible onto a chip. Modern inkjet-like printers reduce the spot volume into the picoliter range. Efficient covalent immobilization of biological molecules on solid supports is a crucial step during microarray production (Pirrung, 2002). Biomolecules need to be immobilized firmly enough to prevent their replacement during reaction and hybridization steps, but flexibly enough to allow conformational changes and binding of target molecules.
An advantage of spotting of prefabricated oligonucleotides onto solid supports (substrates) followed by chemical immobilization is the independence from the length of oligonucleotides to be immobilized. Oligonucleotides of 20–70 nucleotides in lengths are usually aminomodified and covalently bound onto the support. For glutaraldehyde immobilization, aminated slides and 5′-aminated oligonucleotides may be used (Pease et al., 1994). Glutaraldehyde-mediated coupling of biomolecules to solid supports is the classical chemical immobilization method. In order to covalently attach oligonucleotides to the chip, preactivated or surface-modified solid supports, homo- or hetero-bifunctional crosslinkers, and modified oligonucleotides are needed. Table 48.3 surveys common immobilization techniques (see also Wittmann 2005).
TABLE 48.3.
A selection of DNA immobilization techniques on microarrays/chips (see Striebel et al., 2003)
| Immobilization | Examples |
|---|---|
| Non-covalent complexation | Biotin-avidine/streptavidine |
| Self assembled monolayer | Alkanethioles, mercapto- or epoxysilane-compounds |
| Inclusion/crosslinking | NH2-dextrane matrix, aminoethylcellulose, polyacrylamide |
| Polymerization | Co-polymerization of pyrrole and oligo-nucleotides with pyrrole residue at 5′-ends, pyrimidine pyrimidine-dimers, vinylsubstituted nucleotides |
| Silanization by bifunctional reagents | Aminopropyltriethoxysilane (aminofunction with succinic anhydride convertible into carboxic acid function), 3-mercaptopropyl-triethoxysilane (disulfide bridge/thioether), glycidoxypropyltriethoxysilane, p-amino-phenyltrimethoxysilane (ATMS)/diazotation chemistry and unmodified oligonucleotides |
| Covalently by linkerphotoactivated crosslinking | 3′-Hydroxyfunction, 5′phosphate residue, bromcyan, cyanurchloride, carboxylfunction, aldehyde, primary amine, DNA coupling by thymidines, DNA coupling via alkylamines, photolinkage (i.e., psoralen, p-nitrophenyl-3-diazopyruvate) |
To immobilize modified DNA, glass slides with reactive aldehyde, amino, mercapto, or epoxy groups are commercially available. Another covalent and direct immobilization of DNA on glass with oxidic surface (Jung et al., 2001) requires positioning of DNA droplets on a heated surface with an efficiency of 150–300 fmol/mm2 during the coupling reaction. Lindroos et al. (2001) compared six different commercial slides with respect to fluorescence background, turnover efficiency and signal-to-noise ratio. They found that attachment chemistry affects genotyping accuracy when mini-sequencing is used for genotyping on micro-arrays. Genotyping results were best when mercaptosilane-coated slides were used to attach disulfide-modified oligonucleotides. Diazotized chip surfaces are described for immobilization of unmodified oligonucleotides (Dolan et al., 2001). Using p-aminophenyltrimethoxysilane (ATMS), diazotization chemistry was developed, and microarrays were fabricated and analyzed. The method produced uniform spots containing equivalent or even larger amounts of DNA than those obtained by commercially available immobilization techniques Oligonucleotides with hairpin stem-loop structure and multiple phosphorothioate moieties in the loop were used to anchor the oligonucleotide to glass slides that are pre-activated with bromoacetamidopropylsilane (Zhao et al., 2001). Consolandi et al. (2002) describe two robust procedures for oligonucleotide microarray preparation based on polymeric coating. Chemical procedures include a glass functionalization step with γ-aminopropyltriethoxysilane-APTES, or poly-L-lysine, or polyacrylic acid-polyacrylamide co-polymer, which is covalently bound to the modified glass. A surface activation step allows attachment of amino-modified oligonucleotides. Results show high loading capacity, good uniformity, ready availability of immobilized DNA to hybridization targets, and stability to thermal cycles.
The AR Chip Epoxy obtained from two competitors (3D-Link™ and Easy Spot) was compared with respect to slide autofluorescence, spacer length and signal-to-noise ratio (Preininger and Sauer, 2003). The two chip surfaces were assayed by hybridizations with the same targets, and under the same hybridization and scanning conditions. When polyamide (PAMAM) dendrimers containing 64 primary amino groups were used as linkers, signal intensities could be increased significantly compared with amino and epoxy silanized surfaces (Benters et al., 2002). It was shown that dendritic PAMAM linker systems reveal high immobilization efficiencies for amino-modified DNA-oligomers. This was used to assay the performance of dendrimer-based DNA microarrays for discrimination of SNPs.
Aminated glass slides are the most common functionalized supports available. The slides may be used for biomolecule immobilization by reaction of the amino groups with glutaraldehyde, which in turn is coupled to amino groups of target molecules in a sandwich-like manner. Resulting Schiff bases need to be reduced to secondary amines. This may be performed by sodium cyanoborohydride (Birch-Hirschfeld et al., 2002). The method has disadvantages such as complex storage conditions for chemically fully active glutaraldehyde and low biomolecule binding capacity. In addition, glutaraldehyde coatings often do not meet the criteria for low fluorescence background due to impurities and artifacts acquired during production.
Another immobilization chemistry is based on epoxycoated slides. For preparation, activated glass slides are coated with 3-glycido-oxypropyltrimethoxysilane (GOPS), dissolved in toluene. When low-fluorescence glass substrates are used, the resulting microarray supports show greatly reduced fluorescence background, combined with superior DNA immobilization properties (Földes-Papp et al., 2004; Striebel et al., 2003, 2004).
A hetero-bifunctional photoreactive cross-linking reagent, 4-nitrophenyl 3-diazopyruvate (DAPpNP), reacts with glass slides bearing amino groups (Földes-Papp et al., 2004; Striebel et al., 2003). After design and spotting of NH2-modified probe oligonucleotides, the DAPpNP-coated glass slides are irradiated at 360 nm. This UV-radiation leads to conversion of diazogroups into reactive ketene groups, which react with amino groups of the DNA (Goodfellow et al., 1989; Harrison et al., 1989; Kalachikov et al., 1992). In aqueous solutions, ketene groups formed by UV-irradiation of diazopyruvic acid are transformed into carboxyl groups, which then may be used to react with 5′-amino-modified 2′-deoxynucleotide oligomers in the presence of carbodiimide (Penchovsky et al., 2000).
A further light-driven oligonucleotide immobilization technique applies psoralen-mediated covalent coupling of complementary DNA strands (Kittler and Löber, 1995; Pieles and Englisch, 1989). In a first step, an oligonucleotide of 15 alternating adenine and thymidine nucleotides is synthesized directly on a glass support. The sequence carries an amino group at its 3′-end and a psoralen group at its 5′-end. Psoralen may be added using 2-[4′-(hydroxymethyl)-4,5′,8-trimethylpsoralen]-hexyl-1-O-[(2-cyanoethyl)-(N,N-diiso-propyl)]phosphoramidite. A strand, which is complementary to the synthesized support oligonucleotide carrying a stem of five thymidines plus the favored cDNA sequence at its 3′-end, is then hybridized to the support. In an irradiation step (2 m in at 280 nm), the psoralen function of the support oligonucleotide is bound covalently to the complementary sequence. Microarrays generated that way may be hybridized to single- or double-stranded, Cy3-labeled probe DNAs. Advantages of the psoralen-based technique are: oligonucleotides may be synthesized completely in the standard 3′ to 5′ direction; densities of immobilized oligonucleotides are optimally suited for subsequent hybridizations, and the light-driven coupling process may be performed in an aqueous environment without impairing bases or other parts of the DNA carrying the cDNA sequence (Földes-Papp et al., 2004).
Growing
In addition to spotting, oligonucleotide/DNA sequences can be grown directly on the microarray/chip. Photolithography opened the way for in situ fabrication of defined oligonucleotide arrays (Fodor et al., 1991). The oligonucleotide sequence grows on the surface of a glass wafer in a manner similar to conventional solid-phase oligonucleotide synthesis but modified to include a light-sensitive deprotection step. Masks are used to add photospecific bases to selected points on the chip to create a series of oligo-nucleotides with a variety of different sequences. The process, which comprises solid-phase synthesis, photolithography, and affinity labeling, allows the synthesis of up to 250,000 different oligonucleotides (or oligopeptides) per square centimeter on a glass slide by light-directed, spatially addressable chemical synthesis with spot sizes of about 20 μm in diameter. Optimal spacer lengths are crucial for hybridization behavior and fluorescence yields. Different spacer compositions to reduce steric interference of immobilized oligonucleotides with the support were studied in order to improve hybridization behavior (Shchepinov et al., 1997). Optimal spacer length was determined to be at least 40 atoms in length, yielding up to 150-fold increase in hybridization. These spacers are composed of a variety of monomeric units that are synthesized using phosphoramidite chemistry, by condensation onto an amino-functionalized polypropylene support (Matson et al., 1994). Steric hindrance during hybridization may also become problematic, if the immobilized oligonucleotides are too close to each other. Surface coverage was varied using a combination of cleavable and stable linkers. Results showed that highest hybridization yields were obtained from surfaces containing approximately 50% of the maximally possible oligonucleotide concentration.
High-density oligonucleotide arrays are one of these tools for large-scale hybridization. With their ability to produce global views of genome sequences and activity, they have emerged as a key analytical research tool. A vast amount of data must be collected and analyzed to understand biological functions. Affymetrix Inc. creates breakthrough tools for genomic applications. The unique combination of photolithography and combinatorial chemistry eliminates the need for individual laboratories to produce and test their own chips. Using technologies adapted from the semiconductor industry, GeneChip™ manufacturing begins with a 5-inch square quartz wafer. Initially the quartz is washed to ensure uniform hydroxylation across the surface. Because quartz glass is naturally hydroxylated, glass provides a very good substrate for the attachment of chemicals such as linker molecules that are later used to position the probes on the chips. The wafer is placed in a bath of silane that reacts with the hydroxyl groups of the quartz glass surface, and forms a matrix of covalently linked molecules. The distance between these silane molecules determines the probes' packing density, allowing arrays to hold over 500,000 probe locations, or features, within a mere 1.28 cm2. Linker molecules attached to the silane matrix provide a surface that may be spatially activated by UV light. Probe synthesis occurs in parallel and results in the addition of an A, C, T, or G nucleotide to multiple growing chains simultaneously. To define which oligonucleotide chain will receive a nucleotide in each coupling (growing, propagation step), photolithographic masks, which carry 18–20 square micron windows corresponding to the dimensions of individual features, are placed over the coated wafer. The windows are distributed over the mask based on the desired sequence of each probe. When UV light is shone over the mask in the initial step of oligonucleotide synthesis, the exposed linkers become deprotected and are then available for nucleotide coupling. Critical to this step is the precise alignment of the mask with the wafer. To ensure that this critical step is accurately completed, chrome marks on the wafer and on the mask are perfectly aligned. Once the desired features have been activated, a solution containing a single type of deoxynucleotide with a removable protection groups is flushed over the wafer) surface. The nucleotide attaches to the activated linkers, initiating the synthesis process. Although the synthesis process is highly efficient, some activated molecules failed to attach the new nucleotide. To prevent these “outliers” from becoming probes with missing nucleotides, a capping step truncates them in each round of oligonucleotide synthesis. In addition, the side chains of the nucleotides are protected to prevent the formation of branched oligonucleotides. In the next round of synthesis, another mask is placed over the wafer to allow the next cycle of deprotection, coupling and capping. The cyclic process is repeated until the probes reach their full length, usually 25 nucleotides. Although each position in the sequence of an oligonucleotide can be occupied by one of four nucleotides resulting in an apparent need for 25 × 4 = 100 different masks per wafer, situations are identified when the same mask can be used repeatedly. Once the synthesis is complete, the wafers are deprotected and diced. Disadvantages of this method are oligonucleotide lengths that are limited to approximately 30 nucleotides, as well as an unfavorable oligonucleotide orientation, where the 3′-end is bound to the solid support. This is important since hybridization efficiencies depend on orientation and accessibility of the immobilized probe (Southern et al., 1999). A further disadvantage of photolithographic array generation is the accumulation of truncated sequences that cannot be removed (fidelity of the synthesis process: Földes-Papp et al., 1994). For optimal performance, for example, of a photolithographically generated microarray, requirements are therefore (Földes-Papp et al., 1994): (i) Minimization of failure sequences during chemical synthesis of oligonucleotides. This may be achieved by keeping side reactions of chemical synthesis low. Side reactions comprise clustering, depurination in the detritylation reaction, substitution of nucleotides at nucleobases, or breaking of internucleotide bonds. (ii) Minimization of homologous sequences. Oligonucleotides become increasingly more similar in their physico-chemical properties with increasing chain lengths. Therefore, sequences of the type N-1, N-2, N-3, and so forth (N stands for the number of nucleotides in the projected probe oligonucleotide/DNA sequence) are to be minimized by optimization of the coupling and capping reactions. (iii) Suppression of incomplete reactions during chemical cycles. Incomplete reactions are due to masking of non-converted ends by degradation of protection groups. (iv) High reproducibility of chemical syntheses within single steps.
The chemical criteria “yield” (e.g., trityl yield) and “coupling and capping efficiency” used in practice do not meet the demands of the quantitative characterization of chemical oligonucleotide/DNA/RNA/peptide syntheses. Yields, coupling and capping efficiencies only relate to single-reaction steps, but do not consider the length N of projected probe sequences; therefore, these parameters do not say anything about the frequency distribution of failure (truncated, false, erroneous) sequences, and they do not express the dynamics with multi-cyclic syntheses. If syntheses of different nucleotide lengths are compared regarding the optimization of external and internal synthesis conditions, then a lower yield of achieved full length sequences does not mean poorer synthesis conditions. Synthesis conditions leading to more probe sequences of lengths N are better optimized if fewer false sequences are synthesized (Földes-Papp et al., 1995a, b, 1996, 1997a, b, 1998). From theoretical considerations, exact measures of cyclic multistep syntheses, such as chemical oligonucleotide/DNA/RNA/peptide syntheses, were derived. The studies showed that fractal dimensions are such measures for oligonucleotide/DNA/RNA/peptide syntheses. They are superior to commonly used parameters like trityl yield and coupling efficiencies, and they can be applied to the experimental separation of crude synthesis products by high-resolution ion-exchange HPLC, capillary electrophoresis, and gel electrophoresis. Based on experiments, the dynamics of the syntheses on solid support was modeled by stochastic processes with nucleotide chain length N, propagation/elongation (coupling, oxidation), and termination (capping) probabilities as the basic parameters. The following Eqn. 48.1 provides an exact measure for quantitative assessment and comparison of multi-cyclic syntheses of different target length, such as chemical oligonucleotide/DNA/RNA/peptide syntheses:
| (48.1) |
Here, N is the projected length of the probe sequence, and d0 the average (constant) coupling (propagation/elongation) probability that can be expressed, for example, as averaged trityl yield in the case of chemical oligonucleotide/DNA syntheses (Földes-Papp et al., 1995a, b). Oligonucleotide syntheses of different projected probe lengths are equally well optimized regarding their synthesis conditions, if the fractal dimensions D = D(N, d 0) are equal; syntheses showed less propagation and accumulation of erroneous sequences with respect to their projected target lengths N, which is formulated by Eqn. 48.1 and experimentally tested and validated (Földes-Papp et al., 1995a, b, 1996, 1997a, b, 1998).
Grabbing
A grabber is an electronic addressing technology. A chip is flooded with a solution of a probe, and a charge is introduced onto the surface by electrical activation of a row or spot on the chip. The oligonucleotide/DNA probe concentrates close to the charge and is then chemically bonded in place. The chip is washed and another probe solution is introduced. One mm2 of the microarray/chip contains 25 up to 400 wire-bonded electrodes, or even 10,000 sites.
Labeling Strategies and Fluidics Workstations
Once the DNA microarray/chip is fabricated, some form of chemistry has to occur between the sample and the array. The simplest approach is direct labeling of the viral target DNA with fluorophores. Common methods employ enzymatic incorporation of fluorescent nucleotides, or PCR amplification with fluorescent primer pairs. RNA probes are usually prepared from cloned DNA by incorporation of fluorescent nucleotides via RNA polymerase. Selection of fluorescence dyes for primer labeling depends on the filter system of the microarray reader and scanner, respectively. As fluorescence dyes are large hydrophobic molecules, care has to be taken to avoid intercalation of these dyes during PCR (Földes-Papp et al., 2001a, b). Suitable dyes that provide a good long-time stability, such as cyanide dyes Cy3 or Cy 5, are commercially available. Cyanide dyes show an intensive fluorescence and are all water-soluble. This property, combined with their good quantum yield, makes them superior to many other fluorescence dyes (Wessendorf and Brelje, 1992).
Usually, the dyes are attached to the respective DNA sequences in the form of their N-hydroxy-succinimide (NHS) esters. The attachment requires an amino group at the 3′ or 5′-ends of the DNA, which may be introduced via an amino link. Additionally, the method allows introduction of spacers of different lengths (Földes-Papp et al., 2001a, b). Optimal spacer lengths were obtained using C6-amino link reagents.
If solvents and pH values are appropriately adjusted (reactions of some fluorescence dyes require heterogeneous phases), covalent fluorescence dye coupling may be achieved within 1 h. Subsequently, an HPLC purification step is required.
An alternative to NHS ester coupling is coupling of some fluorescence dyes in the form of their phosphoramidite derivatives. Dye-phosphoramidites are easily attached covalently to DNA 5′-ends in computer-assisted automatic oligonucleotide synthesis without any difficulties. After cleavage from the support and chemical removal of protection groups, purification is carried out by preparative Rp18-HPLC.
Incorporation of fluorescent dyes, such as Cy3, Cy5, or fluorescein into nucleic acids, can also be performed with their appropriate 2′-desoxyuridine-5′-triphosphates. These commercially available compounds carry the respective dyes that are attached to spacers at nucleotide 5′-positions. With this internal incorporation of the dye-labeled nucleotides, false base pairing cannot be excluded during PCR reactions. In particular, this holds true for multiplex PCR reactions. Additionally, these dye-tagged nucleotides need to be added in excess, since they are less well inserted by DNA-polymerases than naturally occurring nucleotides (Földes-Papp et al., 2001a, b).
A serious problem of viral diagnostics is the variable, often small quantity of viral particles in patient samples. The amount may vary between one and several thousands of virus copies per milliliter. Signal intensities may be amplified by adding dendrimer building blocks at 5′-ends of PCR primers, allowing multiple coupling of fluorescence dyes per primer (Striebel et al., 2004). The review article of Caminade et al. (2006) focuses on the improvements that hyperbranched and perfectly defined dendrimers also called nanomolecules can provide to DNA microarrays. Two main uses of dendrimers for such purpose have been described up to now (Caminade et al., 2006). Either the dendrimer is used as linker between the solid surface and the probe oligonucleotide, or the dendrimer is used as a multilabeled entity linked to the target oligonucleotide. In the first case the dendrimer generally induces a higher loading of probes and an easier hybridization, due to moving away the solid phase. In the second case, the high number of localized labels (generally fluorescent) induces an increased sensitivity, allowing the detection of small quantities of biological entities.
Using dendrimer technology for multilabeling of viral DNA, Striebel et al. (2004) incorporated dendrimer building blocks at DNA ends during oligonucleotide synthesis, and then fluorescence dyes coupled to amino groups at dendrimers' branches via NHS ester chemistry. In this way, two, four, or eight fluorescence dye molecules may be attached covalently to a primer, using “doubler” dendrimer building blocks. By using “trebler” dendrimer building blocks, three or nine fluorescence dyes may be linked to a primer. In oligonucleotide chemistry, branched oligonucleotides are already being used for signal amplification in hybridization assays. Branched oligonucleotides allow detection of a concentration below 100 human CMV copies per milliliter sample (template) in the case of 5′-(Cy3)3-dendrimer-labeled CMV DNAs (Striebel et al., 2004). The fluorescence signal was enhanced via the dendrimers up to 30 times compared with the quenched single Cy3-labeled human HSV-1 DNA. The on-chip signal-amplifying effect depended upon the number of branches and the concentration of fluorophore-labeled pathogenic DNAs.
Treblers were superior to doublers, as trebler-labeled nucleic acids had fluorescence-signal-enhancing effects over a broad range of labeled DNA concentrations exemplified for the quenched single Cy3-labeled HSV-1 and non-quenched single Cy3-labeled CMV DNAs.
Fluorescence dye labeling invariably involves hybridization of the sample to the oligonucleotides/DNA present on the array. Because chip-bound DNA is often quite short and varied in sequence, a single stringent hybridization condition that is optimal for every spot on the chip is impossible. However, it is possible to find temperature and salt conditions that gives acceptably strong signals for the desired hybridization products and much weaker signals for mismatches. Unbound viral target DNA is then washed away, and the chip is ready to be scanned. Fluidics workstations are available to automate some or all of the wet working steps.
The traditional array detection strategy was autoradiography of radiolabeled samples, but other options are available including electronic signal transduction. With bioelectronic detection, electron transfer reactions from the DNA to the substrate allow fast and probe-cell specific detection systems. The ability of mass spectroscopy to identify and, increasingly, to precisely quantify genes and genome sequences (as well as proteins) from complex samples can be expected to impact broadly on biology and medicine. The systematic analysis of the much larger number of genes expressed in a cell, an explicit goal of functional genomics, is now also rapidly advancing, due mainly to the development of new experimental approaches. Using MALDI-TOF, DNA fragments are analyzed with the SpectroChip™ from Sequenom Inc.
A piezoelectric gene sensor microarray for HBV quantification in clinical samples was constructed using crystal units that oscillate independently (Chen et al., 2005a, b).
Imaging and Scanning, Data Processing
Scanning a fluorescence-labeled DNA array is conceptually quite simple. A light source excites the labeled samples and a detector system measures and records the emitted fluorescence. However, the instrumentation requirements differ according to the precise nature of the array. Most image capture instruments and microarray readers, respectively, use a scanning detector similar to line-scanning detector systems for DNA sequencing instruments. The number of discrete points that the detector can sample across the array and the row-to-row step interval determine the size of the features that the detector can image. It is important that the detector pixels are sufficiently small to gather data from enough significant pixels in each probe cell. Confocal laser-scanning systems for fluorescent microarray biochips should have a spatial resolution of a detector on the order of 1/8 to 1/10 of the diameter of the smallest element in the microarray. While the absolute number of DNA molecules that can be crammed into a small space will probably prevent the field from reaching the 0.2 μm features found in semiconductors, 5 μm probe cells appear attainable. This would put about four million features on a 1 cm2 chip.
Image analysis is performed by placing a grid over the array to allow integration of the signal from each probe cell. If the signals from the arrays are strong and fairly consistent, a threshold algorithm is usually adequate, but weak signals demand a computationally intensive approach. The background signal or baseline is the fluorescence from the support matrix. Raw fluorescence intensity data are background subtracted. Hybridization outcomes can be represented as log2 fluorescence intensity ratio of test versus reference samples. To virtually enrich viral specific concordant probes, data filtration can be further filtered by elimination of spots below signal intensity and a predefined absolute log2 ratio; this eliminates outlier data from further analysis due to labeling bias. The processing improves data correlation. Hierarchical clustering and visualization of data can be performed using Gene Cluster and Tree View software (Stanford University, USA). High-density DNA chips with thousands of probe cells need special managing software for microarray probe tracking and data analysis like Affymetrix's LIMS workstation, BioDiscovery's Clone Tracker software or GeneSpring, a software package offered by Silicon Genetics. Based on observed microarray hybridization patterns an algorithm called E-Predict is reported for microarray-based viral species identification. E-Predict compares observed hybridization patterns with theoretical energy profiles representing different viral species in a set of clinical samples but its relevance to other metagenomic applications is discussed (Urisman et al., 2005).
Viral Genotyping
Regardless of how the microarrays and chips, respectively, are made, viral chip technology is superior to conventional PCR methods for the fast, sensitive, specific, and parallelized diagnostics of heterogeneous populations of different viral species and strains (Li et al., 2001; Striebel et al., 2003). Viral genotyping is an approach made possible by the availability of viral gene and genome sequence databases and technical and conceptual advances in chip technology. Interest exists in providing high-throughput platforms and readout technologies that can selectively examine changes in the transcript levels of specific genes in response to multiple treatments. Although conventional microarrays (DeRisi et al., 1996; Schena et al., 1995; Wang et al., 2002) have, in principle, the necessary throughput, they were limited with respect to cost and dynamic range and therefore are not well suited for the task of producing genotyping data for multiple viruses from which accurate reliable information can be calculated. For routine clinical testing, it is not possible to test thousands of genes on a microarray. Therefore, some researchers have called for “less” genes to be put on the array, because most of the other data points are inconsequential (Wooster, 2000) or even detrimental to the usefulness of a set of classifier genes (Draghici et al., 2003). Can array data be used to identify a handful of critical genes that will lead to a more-detailed taxonomy and can this or similar array data be used to predict clinical outcome (Wooster, 2000)?
In genotyping applications, one looks for sequence variation that has previously been characterized. Oligonucleotides of known sequence variants of a viral gene or collections of viral genes are represented in the chip. Genotyping arrays that are designed to examine viral DNA sequences are listed in Table 48.4 . The viral chip has the enormous strength that only one chip is needed to analyze a mixture of different viral DNA sequences.
TABLE 48.4.
Representative examples of viral genotyping studies with microarrays/chips
In practice, there may be some pitfalls, as exemplified by long-term variability studies of the human herpes simplex virus type 1 (HSV-1) genome. The HSV-1 genome consists of iterative DNA sequences of variable amounts. The HSV-1 genome from a HSV-1 DNA pool has an isomolar genome organization with L and S components. Genome complexity is due to tetra-isomers derived from the rolling circle-like intracellular replication process. In the L-S region, transient variabilities are formed by insertions or deletions of the repetitive a-sequences. The genomic changes are more or less strain-specific. Outside the L-S junction the virus progeny under study showed no or negligible deviations from the parent strain. The viral DNA material of three HSV-1 strains F (Roizman), Mst (Ulm) and AK (Ulm) was sampled after intracellular self-assembled virus particles were formed (Földes-Papp et al., 1997a; Klauck et al., 1995). The DNA replication was carried out in Vero cell infections and the purified viral DNA was digested with BamHI restriction endonuclease (Földes-Papp et al., 1997a). The resulting BamHI-K2-fragments of HSV-1 were extracted from agarose gels by electroelution, ligated into plasmid pBR322 or pAT153 and cloned in E.coli K 12 C600 cells for hybridization in Southern blots. The restriction patterns obtained by endonucleases KpnI, and SalI were analyzed by hybridization with K2-fragments of HSV-1 (F) labeled by the random priming method with [32P]α-dCTP. Blots were autoradiographed and the bands quantified by scanning densiometry. We found that the nucleotide sequence pattern of the HSV-1 genome in the L-S region can be approximated by distribution profiles of digested components expressed in powers of nucleotide length N of the largest component in the region. The largest components of interest studied had nucleotide length N of 13,500 base pairs obtained with KpnI of HSV-1 strains F, Mst, AK and nucleotide length N of 9730 base pairs obtained with SalI of HSV-1 strains F, Mst, AK. We obtained the fractal dimensions D = 1.63 (strain F), D = 1.66 (strain Mst), and D = 1.66 (strain AK) with restriction endonuclease KpnI. With restriction endonuclease SalI, we found the fractal dimensions D = 1.36 (strain F), D = 1.36 (strain Mst), and D = 1.39 (strain AK). The fractal dimension D is here a measure for producing internal repeats in the L-S region of strains F, Mst, AK during replication at the viral genome level (Földes-Papp et al., 1997a). Here, a lower value of the fractal dimension D indicates a distribution of sequence variability with lower amounts of internal repeats. The fractal dimension D was obtained from the scaling to the largest nucleotide length of interest, N, and is interpreted as typical long-term DNA sequence variability.
Viral Sequence Determination
At its most complex, viral genotyping using a DNA chip involves complete determination of the nucleotide order via resequencing by hybridization. Viral resequencing discovers novel sequence variations (see Table 48.5 ). The longer the target DNA, the longer the oligonucleotides must be to eliminate ambiguities. For example, a viral chip was designed after dividing each viral genome into overlapping 70-mer oligonucleotides using sequencing analysis tools BLASTn (Wang et al., 2003; Wong et al., 2004). The 70-mers were ranked by the number of viral genomes to which significant homology were found by alignments. Virus resequencing of the genomes in infected humans was performed with the Affymetrix™ SARS resequencing GeneChips (Sulaiman et al., 2006, 2007). High-density resequencing GeneChips have potential biodefense applications and may be used as an alternate tool for rapid identification of smallpox virus in the future.
TABLE 48.5.
Representative examples of viral resequencing studies with microarrays/chips
| Viral species and strains | Examples |
|---|---|
| SARS coronavirus | Rota et al. (2003); Ksiazek et al. (2003); Wong et al. (2004); Sulaiman et al. (2006) |
| Smallpox viruses (variola virus) | Sulaiman et al. (2007) |
| Coccolithovirus | Allen et al. (2007) |
| Human influenza virus | Wang et al. (2006) |
For sequencing of any viral DNA/RNA, a gene of unknown sequence can be rapidly screened. In this approach, oligonucleotide probes represent all possible combinations of sequence in a given length. The number of probe “cells” can be calculated by four to the power of the oligonucleotide length, that is 65,536 8-mers, 262,144 9-mers, and so forth. The viral target sequence to be sequenced is broken into small pieces, fluorescently labeled, and hybridized with the immobilized oligonucleotides on the chip. However, the random sequencing procedure is weak for non-random sequences such as direct and inverted repeats.
Viral Gene Expression
There is a diverse range of experimental objectives and uses for viral gene expression microarray data, which makes the areas of experimental design and data analysis quite broad in scope (see Table 48.6 ). As such, there are many ways to design expression profiling experiments, as well as many ways to analyze and mine data functional genomics expression profiling experiments, including transcriptional analysis of normal biological processes of viral and host replication, discovery and validation of viral drug targets, and studies into the mechanism of action and toxicity of pharmaceutical compounds.
TABLE 48.6.
Representative examples of viral gene expression studies with microarrays/chips
| Viral species and strains | Examples |
|---|---|
| Pan-viral | Wang et al. (2002, 2003) |
| Human and mammalian retroviruses | Seifarth et al. (2003) |
| Choristoneura fumiferana nucleopolyhedrovirus | Yang et al. (2007) |
| Human immunodeficiency virus type 1, Human T cell leukemia virus types 1 and 2, Hepatitis C virus, Epstein-Barr virus, Human herpesvirus 6A and 6B, and Kaposi's sarcomaassociated herpesvirus | Ghedin et al. (2004) |
| HSV-1 and HSV-2 | Aguilar et al. (2006) |
| HPV18 (E2 mutants) | Thierry et al. (2004) |
Chambers et al. (1999) described the first viral DNA micro-array for the temporal profiling of viral gene expression human cytomegalovirus, CMV Basically, viral DNA microarrays/chips that apply a genomic approach from known open reading frames allow analysis of many virally encoded genes at the steady-state level of mRNA in the cell for a single (Kellam, 2001) or multiple pathogen detection (Wang et al., 2002).
Single Pathogen Detection
Viral RNA expression monitoring studied for a single pathogen identified patterns of immediate early, early, and late gene expression for some herpes viruses (Aguilar et al., 2004; Chambers et al., 1999; Stingley et al., 2000). Lytic expression profiles of viral herpes genes are established following reactivation from latency (Jenner et al., 2001; Leenman et al., 2004; Paulose-Murphy et al., 2001). The predicted open reading frames of the human CMV and an established protocol for simultaneously measuring the expression of all CMV genes have been studied by Yang et al. (2006); In order to study the global pattern of VZV gene transcription, VZV microarrays using 75-base oligomers to 71 VZV ORFs were designed and validated (Kennedy et al., 2005). A human herpes virus chip for six latent EBV genes (EBNA1, EBNA2, EBNA3A, EBNA3C, LMP1, LMP2), and four lytic EBV genes (BZLF1, BXLF2, BKRF2, BZLF2) allowed Bernasconi et al., 2006 to accurately measure EBV gene transcription changes triggered by treatment interventions. The EBV infection rate and the gene expression profile of EBV in tumor biopsies were determined using EBV genome chips (Li et al., 2006). By using Affymetrix™ human gene chip and NetAffx analysis through the Affymetrix™ website, the gene expression profiles of variant core proteins were implicated in HCV replication, pathogenesis, or oncogenesis in the Huh-7 cell line, which is useful for our understanding of HCV variant core protein biological function and its pathogenic mechanism (Dou et al., 2005).
Multiple Pathogen Detection
A viral detection DNA microarray composed of oligonucleotides corresponding to the most conserved sequences of all known viruses identified the presence of gammaretroviral sequences in cDNA samples that impair function of RNase L, particularly R462Q, from seven of 11 R462Q-homozygous cases, and in one of eight heterozygous and homozygous wild-type cases in human prostata tumors (Urisman et al., 2006). The novel virus, named XMRV, is closely related to xenotropic murine leukemia viruses (MuLVs), but its sequence is clearly distinct from all known members of this group. Attempts to identify differentially expressed transcripts in complex neuropsychiatric disorders, such as schizophrenia, have shown the necessity of screening a large sample set in order to rule out medication effects and normal individual variation when attempting to determine disease association (Conejero-Goldberg et al., 2005; Wang et al., 2002). The genome wide gene expression profile aids in the understanding of genes that may be regulated in a particular pathological condition-like cardiovascular disease and how it pertains to viral and parasitic infections of the heart (Mukherjee et al., 2006).
Viral Chips for Several Diagnostic Purposes
Recent successes with viral chips illustrate their role as an indispensable tool for molecular and cellular biology and for the emerging field of systems biology. Studies focusing on the analysis of viral populations from cells or tissues typically pose challenges owing to the high degree of complexity of a mixture of numerous viral species and the low abundance of many of the viruses, which necessitates highly parallelized analysis. The ability of viral chips to identify thousands of viruses from complex samples can be expected to impact broadly on biology and medicine (Wang et al., 2002). However, the use of viral chips for a diagnostic purpose is still limited because there is little agreement as to which portion of the viral genome is specific for diagnostics and allows establishment of intra- and inter-species relationships (Földes-Papp et al., 1997a; Striebel et al., 2003).
One application is the “forecast” of the efficacy of drugs for therapy (Chambers et al., 1999; Murphy, 2002). Identification of mutations responsible for resistances of bacterial and viral particles (i.e., mycobacteria, HIV), including the proof of special resistance genes, plays an important role (Gunthard et al., 1998; Hamels et al., 2001; Ramaswamy et al., 2003; Rogers and Barker, 2002; Schembri et al., 2002). Studying pathogen–host relations contributes to the understanding of disease pathogenesis and helps develop therapeutic strategies for new drugs (Cummings and Relman, 2000; Haselbeck et al., 2000; Kato-Maeda et al., 2001; Mahony, 2002; Peek et al., 2001). Since phenotype-based testing of resistance with virus isolation and growth on cell cultures is very time and labor consuming, genotype-based testing of resistance represents a meaningful alternative. In hepatitis C, duration and success of an interferon therapy mainly depends upon the HCV genotype (Zein, 2000), apart from other factors such as age, duration of infection and height of the HCV RNA titer before specific therapy starts.
So far, there have been very few applications of microarrays/chips for quantitative estimations of viral loads. Preliminary studies are reported for quantification of hepatitis B virus (HBV) DNA in serum (Kawaguchi et al., 2003). Because of the high clinical importance of virus quantification, in particular for estimation of virus replication and therapy monitoring, there is a great potential for chip application. For example, a quarter of the chronically infected people suffer from SNPs in the precore/basal core promoter region of the HBV genome, causing the loss of HBeAg expression. These HBeAg-negative patients often remain asymptomatic and viremic until the alanine aminotransferase level in serum is persistently elevated (Lai et al., 2003). It would therefore be important to develop methods to detect the precore/basal core promoter mutations of the HBV genome at an earlier stage of infection for large-scale clinical and diagnostic analysis. The conventional and common methods for detecting mutations such as single-strand conformation polymorphisms, denaturing gradient gel electrophoresis, direct sequencing and chemical cleavage are not practical for detecting multiple SNPs in high-throughput format. The arrayed primer extension method (APEX), which is based on a hybridization of short single-stranded DNA templates with an array of immobilized oligonucleotide probes on a glass surface followed by a polymerase-assisted incorporation of different fluorescently tagged dideoxynucleotides (Tonisson et al., 2000), enables large-scale SNP detection. All methods of mutant detection so far described can only identify mutants in the serum, and cannot determine the proportion of those mutants. Li et al. (2005) reported the development of a novel technique that can quantify the relative proportion of mutants in serum utilizing gene microarray technology.
Clinical applications of microarray technology predominantly refer to collections of virus variants and to differentiation of subtypes. A pan-virus DNA microarray (Virochip) was used to detect a human metapneumovirus (hMPV) strain associated with a critical respiratory tract infection in an elderly adult with chronic lymphocytic leukemia (Chiu et al., 2007). Using the Virochip, human parainfluenzavirus 4 (HPIV-4) infection was detected in an immunocompetent adult presenting with a life-threatening acute respiratory illness (Chiu et al., 2006). So far, viral chip technology has no practical applications in clinical diagnostics due to its high costs and insufficient standardization; it so cannot be compared to biological chip experiments. This technology thus only can be a useful supplement to other diagnostic methods (Wang et al., 2002).
The main focus of clinical diagnostics with viral DNA chips will be to find answers to a handful of questions about viruses safely, quickly and inexpensively. Clinical diagnostics approaches using viral DNA chip technology simply ask the question whether particular genes, which are indicative of particular viruses, are present in a sample or not. For example, the knowledge-based, low-density “focused” microarray is based on the molecular diagnostics of six human herpesvirus types: HSV-1, HSV-2, VZV, EBV, CMV, HHV-6 (Földes-Papp et al., 2004, 2005b; Striebel et al., 2004). These studies attempted to optimize parameters of chip design, surface chemistry, oligonucleotide probe spotting, sample labeling, and DNA hybridization. The well-designed and tailored chip platform developed utilizes low-fluorescence background coverslips, epoxy surface chemistry, standardized oligonucleotide probe spotting, PCR-labeling with Cy3 of isolated viral DNA, array hybridization, and detection of specific spot fluorescence by an automatic microarray reader. Korimbocus et al. (2005) reported on the simultaneous detection of major CNS pathogens HSV-1, HSV-2, CMV; all serotypes of human enteroviruses, and five flaviviruses (West Nile virus, dengue viruses, and Langat virus) using amplification by PCR and detection of amplified products by a DNA microarray. Consensus primers for the amplification of all members of each genus and sequences that are specific for the identification of each virus species were selected from the sequence alignments of each target gene. A multiplex PCR-based DNA array for EBV, CMV, and Kaposi's sarcoma-associated herpesvirus (KSHV) may serve as a rapid and reliable diagnostic tool for clinical applications (Fujimuro et al., 2006).
The macroarray test of Fitzgibbon and Sagripanti (2006) for capturing generic members of the orthopox- or alphavirus families and a collection of additional oligonucleotides to bind specifically nucleic acids from five individual alphaviruses, including Venezuelan equine encephalitis, or DNA from each of four orthopoxviruses, including variola virus (VAR) is easy to perform, inexpensive, relatively fast, uncomplicated to interpret, and its end point is read visually without the need of additional equipment. This nucleic acid hybridization assay onto nylon membranes in macroarray format can help in detecting or excluding the presence of threat viruses in environmental samples and appears promising for a variety of biodefense applications. A microarray-in-a-tube system, for example, in an eppendorf-tube format, was first suggested by Földes-Papp et al. (2005a, b). It is a solution to complicated handling and expensive microarry equipment. Recently, a microarray-in-a-tube system with a 5 × 5 oligonucleotide “microarray” was developed by Liu et al. (2007), for detecting four respiratory tract viruses (severe acute respiratory syndrome-associated coronavirus, influenza A virus, influenza B virus, and enterovirus) in a specially designed Eppendorf cap with a flat, optically transparent window.
NANOFABRICATION
Nanofabrication combines different metals, semiconductors, or carbon in spherical particles as quantum dots and in linear particles as nanowires, nanotubes, or nanorods. Nanofabrication reduces the amount of biological samples and reagents for the detection of viruses. A number of assays of metal nanoparticles for DNA detection and analysis and their basic principle, advantages, disadvantages and detection limits are very recently reviewed (Möller and Fritzsche, 2007). The detection schemes can aptly be divided into optical category such as quantum dots, stripped nanovirus and electrical/electrochemical category such as semiconducting nanowires and carbon nanotubes (Rosi and Mirkin, 2005). However, I distil the salient features of nanofabrication for viral detection.
Quantum Dots
Quantum dots are also known as nanocrystals or artificial atoms. They consist of clusters of a few hundred to a few thousand atoms. Their diameter is in the nanometer range. Quantum dots (QD) are made from metals such as gold, silver, cobalt, and semiconductor materials such as cadmium sulfide, cadmium selenide, or cadmium telluride (Gerion et al., 2003). QDs are characterized by broad absorption spectra from the ultraviolet to the far infrared, good photostability, and a narrow emission spectrum. These properties could improve the sensitivity of biological detection and imaging by 10- to 100-fold. Conjugated gold nanoparticles with nucleic acids were used as labels for DNA microarrays (Taton et al., 2000). With this method, target nucleic acids at concentrations of 500 femtomolar were detected (Bao et al., 2005; Liang et al., 2005; Park et al., 2002). QDs have also been applied to the detection of viral species (Perez et al., 2003).
To detect specific DNA sequences without first performing an amplification step, Wang and colleagues developed an ultrasensitive nanosensor that uses QDs linked to DNA probes (Zhang et al., 2005). The nanosensor can detect less than 50 copies of DNA and has much less background fluorescence than conventional fluorescence resonance energy transfer (FRET) systems. The nanosensor includes two oligonucleotide probes that bind to separate regions of an assayed strand of DNA (target DNA). The reporter probe is labeled with a Cy5 fluorophore, and the capture probe is labeled with biotin. The probes bind the target DNA, and a streptavidin-coated QD binds to the capture probe. This process brings the Cy5 molecule close to the QD for FRET to occur. Because QDs have broad absorption and narrow emission spectra, the QD-based nanosensor system can be finely tuned so that background fluorescence is negligible. QDs also concentrate the FRET signal by binding many target–probe complexes. To detect FRET signals, the researchers developed a novel confocal fluorescence microscopy platform. The QD–target–probe complexes were continuously flowed through a microcapillary past two detectors that were specific for signals from either FRET donors or acceptors. When the system was tested with a single-stranded target DNA, the fluorescence was observed with both detectors. However, when the target was absent or when a non-complementary DNA strand was used, only donor fluorescence was observed. Wang and colleagues also compared the performance of the QD nanosensor with that of a molecular beacon, which is typically used in FRET assays. The QD nanosensor was about 100 times more sensitive. Finally, the researchers combined the nanosensor with an oligonucleotide ligation assay. This method allowed the discrimination of point mutations in DNA samples from ovarian cancer patients.
Stripped Nanowires
These can be made of metals such as Au, Pt, Ni, Co, or Cu by chemical vapor deposition or more commonly by electrodeposition. The length of the stripped nanowires depends upon the electrochemical deposition time for each metal band (Nicewarner-Pena et al., 2001). Nanowires consisting of two metals and 13 strips are metallic barcodes in biological assays with up to 4160 distinguishable patterns. Stripped nanowires of three metals yield 8.0 × 105 different tags (Keating, 2003). On the surface of nanorods, the barcoded nanowire molecular beacon approach with a quenched fluorophore was used to detect DNA binding of human immunodeficiency virus (HIV), hepatitis B (HBV) and hepatitis C (HCV) virus sequences.
Semiconducting Nanowires
They are building blocks for nanoscale electronics. For example, they are set up for field-effect transistors for the real-time detection of DNA. The hybridized nucleic acid is used as substrate for the deposition of metal and construction of a nanowire. There are several nucleic acid metallization chemistries available. Hybridization between target DNA and the probe bound to the nanowire or a support followed by metallization of nucleic acid changes conductance. The hybridized and metallized nucleic acid is electrocatalytically detected by applying voltage to one of the two electrodes in each test structure and measuring the increased current of a redox indicator or the changes in conductivity or capacity (Jianrong et al., 2004). Semiconductor-based oligonucleotide microarrays were used for rapidly identifying influenza A virus hemagglutinin subtypes 1 through 15 and neuraminidase subtypes 1 through 9 (Lodes et al., 2006).
Silicon Nanowires
Silicon semiconducting nanowires transport electrons and holes (Cui and Lieber, 2001; Cui et al., 2001). They can be synthesized by chemical vapor deposition, electrochemical size reduction, and pulsed laser vaporation. Slicon nanowires with 20 nm gold clusters as catalysts, silane as reactant, and diboran as p-type dopant are label-free, electrochemical biosensors that were used for the detection of herpesvirus type 1 and 2 DNA (Patolsky et al., 2004). With an antibody functionalized nanowire, single influenza A viruses could also be detected.
Carbon Nanowire
The graphitic surface chemistry and electronic properties depend upon the tube diameter and chirality. Carbon nanowires can be metallic or semiconducting and yield semiconductor– semiconductor and semiconductor–metal junctions. They can be functionalized with nucleic acids, proteins and antibodies (Cai et al., 2003).
ARE THERE ADDITIONAL ALTERNATIVES TO DIAGNOSTIC MICROARRAYS?
For phenotype screening in functional genomics, live-cell chips are powerful new tools. For example, a cell-based microarray on primary and cancer cells based on the localized reverse infection by retroviruses (Carbone et al., 2007). Viral vectors are immobilized on a nanostructured titanium dioxide (ns-TiO2) film obtained by depositing a supersonic beam of titania clusters on a glass substrate. We validated the retroviral cell array by overexpression of Green Fluorescence Protein reporter genes in primary and cancer cells, and by RNA interference of p53 in primary cells by analyzing effects in cell growth.
The technical solutions for high mutation and recombination rate studies are power-low fluidic chips. The state-of-the-art in microfluidics is that different systems at different scales are available; almost any macrosystem can be micro. This novel direction in chemical/biochemical analytics offers the possibility of using nanofluidic functional elements (Mulvaney et al., 2007) and electrodynamic separation in nanochannels (Pal et al., 2005; Schwartz et al., 2004; Ye et al., 2005). However, technologies in the labs are not available in a cheap-and-easy form (Zaytseva et al., 2005).
A further trend is single molecule detection. It has the advantages of small volume platform, digital analysis, elimination of processing steps, approach for real-time measurements, and automation of sample preparation. This is definitely good work and is likely the trigger for further investigations on the behavior of single molecule. Current technologies can only measure biological mechanisms as an average of a population of molecules, as only their combined effect can be detected. The simplification ignores the fact that biological macromolecules oscillate between different activity and conformational states. Figure 48.2 summarizes a new physically grounded approach in single-molecule fluorescence fluctuation spectroscopy and imaging that is based upon the meaningful-time concept (Földes-Papp, 2006, 2007a, c). If we want to perform a single-molecule measurement in solution without immobilization or hydrodynamic flow, or within a live cell then we do not want to collect (integrate) data longer than we have to. The minimum time that we need for the measurement with one single molecule is:
| (48.2) |
Figure 48.2.
Synopsis of a new physically grounded technology of fluorescence fluctuation spectroscopy for observing single molecules at longer time scales than currently available. (See also Földes-Papp, 2001c, 2002, 2005, 2006, 2007a, b, c; Földes-Papp et al., 2001, 2005a) In many respects, such methods are an alternative to DNA chip technology (Földes-Papp et al., 2005a, b). They detect the molecular Brownian movement of fluorescent particles in a very tiny volume of the laser focus. Quantitative understanding of molecular interactions at the level of single molecules within single cells is the next step in basic and applied biomedical research for the analysis of the dynamics and localization of molecules in a variety of physiological and pathophysiological processes.
where Tm is the meaningful time that one can study the selfsame molecule, for example the viral nucleic acid, τdiff is the measurable diffusion time of the single fluorescent viral nucleic acid molecule at the measurable number of viral molecules N < 1 in the confocal probe volume ΔV of about 0.2 fl= 2 × 10−16 L and less, cm is the molar concentration of viral nucleic acid molecules of the same kind in the bulk solution (sample), and NA is Avogadro's number of [mol−1].
CONCLUSIONS
DNA microarrays/chips stand out for their simplicity, comprehensiveness, robustness, data consistency and high-throughput. They can be devided for form s sake into viral chips and host chips (Piersanti et al., 2004; Livingston et al., 2005), but the majority of applications can inherently be categorized as geno-typing, resequencing, or gene expression. Viral microarrays/chips provide a particularly powerful tool to reap these benefits. Their ability to assess the contribution of non-specific signals in a probe-specific manner allows the detection and quantitation of low abundance viral species and strains. In addition, analysis software can be used to adjust the balance between sensitivity and specificity to meet the particular requirements of an application. The microarray/chip experiment begins with well-defined goals, anticipated pitfalls, and minimized costs. It paves the way to the use of all kinds of molecular and clinical information to optimize diagnosis, treatment and health outcomes for individual patients. Today, we have entered into an era where biological and medical research can make inquiries about the entire genome of an organism. Investigators now have a unique opportunity to ask fundamental biological questions at an unexpected scale and depth. Viral microarrays/chips are one of the few technological tools available to take advantage of this explosion of genetic and sequence information. Each breakthrough that either allows a new type of measurement or improves the quality of data expands the range of potential applications of viral chips in genomic medicine.
ACKNOWLEDGEMENTS
Zeno Földes-Papp, who is principal investigator, is supported in part by the FWF Austrian Science Fund through his research project P20454-N13. Within this research project, Z.F.-P. has received visiting professorships at ISS Inc., Champaign, IL61822, USA, and at the Department of Molecular Biology and Immunology, Health Science Center, University of North Texas in Dallas-Fort Worth, U.S.A.
References
- Aguilar J.S., Ghazal P., Wagner E.K. Design of a herpes simplex virus type 2 long oligonucleotide-based microarray: Global analysis of HSV-2 transcript abundance during productive infection. Meth Mol Biol. 2004;292:423–448. doi: 10.1385/1-59259-848-x:423. [DOI] [PubMed] [Google Scholar]
- Aguilar J.S., Devi-Rao G.V., Rice M.K., Sunabe J., Ghazal P., Wagner E.K. Quantitative comparison of the HSV-1 and HSV-2 transcriptomes using DNA microarray analysis. Virology. 2006;348(1):233–241. doi: 10.1016/j.virol.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Albrecht V., Chevallier A., Magnone V., Barbry P., Vandenbos F., Bongain A., Lefebvre J.C., Giordanengo V. Easy and fast detection and genotyping of high-risk human papillomavirus by dedicated DNA microarrays. J Virol Meth. 2006;147(2):236–244. doi: 10.1016/j.jviromet.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Allen M.J., Martinez-Martinez J., Schroeder D.C., Somerfield P.J., Wilson W.H. Use of microarrays to assess viral diversity: From genotype to phenotype. Environ Microbiol. 2007;9(4):971–982. doi: 10.1111/j.1462-2920.2006.01219.x. [DOI] [PubMed] [Google Scholar]
- An H.J., Cho N.H., Lee S.Y., Kim I.H., Lee C., Kim S.J., Mun M.S., Kim S.H., Jeong J.K. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray method. Cancer. 2003;97(7):1672–1680. doi: 10.1002/cncr.11235. [DOI] [PubMed] [Google Scholar]
- Bao Y.P., Huber M., Wei T.F., Marla S.S., Storhoff J.J., Muller U.R. SNO identification in unamplified human genomic DNA with gold nanoparticle probes. Nucl Acids Res. 2005;33:e15. doi: 10.1093/nar/gni017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean P., Wilson J. HIV genotyping by chip technology. Am Clin Lab. 2000;19(4):16–17. [PubMed] [Google Scholar]
- Benters R., Niemeyer C.M., Drutschmann D., Blohm D., Wöhrle D. DNA microarrays with PAMAN dendritic linker systems. Nucl Acids Res. 2002;30(2):e10. doi: 10.1093/nar/30.2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi M., Berger C., Sigrist J.A., Bonanomi A., Sobek J., Niggli F.K., Nadal D. Quantitative profiling of housekeeping and Epstein-Barr virus gene transcription in Burkitt lymphoma cell lines using an oligonucleotide microarray. Virol J. 2006;3:43. doi: 10.1186/1743-422X-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Hirschfeld E., Egerer R., Striebel H.-M., Stelzner A. New Ways to immobilize Oligonucleotides on DNA-Chips. In: Chemistry of Nucleic Acid Components, XIIth Symposium, Czech Republic, September 3–8, 2. Collection Symposium Series. 2002;5:299–303. [Google Scholar]
- Boriskin Y.S., Rice P.S., Stabler R.A., Hinds J., Al Ghusein H., Vass K., Butcher P.D. DNA microarrays for virus detection in cases of central nervous system infection. J Clin Microbiol. 2004;42:5811–5818. doi: 10.1128/JCM.42.12.5811-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.O., Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21(Suppl. 1):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Cai H., Cao X., Jiang Y., He P., Fang Y. Carbon nanotube-enhanced electrochemical DNA biosensor for DNA hybridization detection. Anal Bioanl Chem. 2003;375:287–293. doi: 10.1007/s00216-002-1652-9. [DOI] [PubMed] [Google Scholar]
- Caminade A.-M., Padie C., Laurent R., Maravel A., Majoral J.-P. Uses of dendrimers for DNA microarrays. Sensors. 2006;6:801–914. [Google Scholar]
- Carbone R., Giorgetti L., Zanardi A., Marangi I., Chierici E., Bongiorno G., Fiorentini F., Faretta M., Piseri P., Pelicci P.G., et al. Retroviral microarray-based platform on nanostructured TiO2 for functional genomics and drug discovery. Biomaterials. 2007;28(13):2244–2253. doi: 10.1016/j.biomaterials.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Chambers J., Angulo A., Amaratunga D., Guo H., Jiang Y., Wan J.S., Bittner A., Frueh K., Jackson M.R., Peterson P.A., et al. DNA microarrays of the complex human cytomegalovirus genome: Profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73(7):5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., Huang J., Zhang X.P., Qiao P., Zhang W., Yang N.M., Liu H.J., Geng Y.Y., Qiu J.M., Wang S.Q. Clinical evaluation of oligonucleotide microarrays for the detection of HBV mutants associated with lamivudine resistance. Pharacogenomics. 2005;6(7):721–730. doi: 10.2217/14622416.6.7.721. [DOI] [PubMed] [Google Scholar]
- Chen M., Liu M., Yu L., Cai G., Chen Q., Wu R., Wang F., Zhang B., Jiang T., Fu W. Construction of a novel peptide nucleic acid piezoelectric gene sensor microarray detection system. J Nanosci Nanotechnol. 2005;5(8):1266–1272. doi: 10.1166/jnn.2005.223. [DOI] [PubMed] [Google Scholar]
- Cherkasova E., Laassari M., Chizhikov V., Korotkova E., Dragunsky E., Agol V.I., Chumakov K. Microarray analysis of evolution of RNA viruses: Evidence of circulation of virulent highly divergent vaccine-derived polioviruses. Proc Natl Acad Sci USA. 2003;100:9398–9403. doi: 10.1073/pnas.1633511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.Y., Rouskin S., Koshy A., Urisman A., Fischer K., Yagi S., Schnurr D., Eckburg P.B., Tompkins L.S., Blackburn B.G., et al. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43(8):e71–e76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.Y., Alizadeh A.A., Rouskin S., Merker J.D., Yeh E., Yagi S., Schnurr D., Patterson B.K., Ganem D., DeRisi J.L. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J Clin Microbiol. 2007;45(7):2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov V., Wagner M., Ivshina A., Hoshino Y., Kapikian A.Z., Chumakov K. Detection and genotyping of human group a rotaviruses by oligonucleotide microarray hybridization. J Clin Microbiol. 2002;40:2398–2407. doi: 10.1128/JCM.40.7.2398-2407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N.H., An H.J., Jeong J.K., Kang S., Kim J.W., Kim Y.T., Park T.K. Genotyping of 22 human papillomavirus types by DNA chip in Korean women: Comparison with cytologic diagnosis. Am J Obstet Gynecol. 2003;188(1):56–62. doi: 10.1067/mob.2003.120. [DOI] [PubMed] [Google Scholar]
- Conejero-Goldberg C., Wang E., Yir C., Goldberg T.E., Jones-Brando L., Marincola F.M., Webster M.J., Torrey E.F. Infectious pathogen detection arrays: Viral detection incell lines and postmortem brain tissue. BioTechniques. 2005;39(5):741–751. doi: 10.2144/000112016. [DOI] [PubMed] [Google Scholar]
- Consolandi C., Castiglioni B., Bordoni R., Busti E., Battaglia C., Bernar L.R., De Bellis G. Two efficient polymeric chemical platforms for oligonucleotide microarrays preparation. Nucleos Nucleot Nucleic Acids. 2002;21(8-9):561–580. doi: 10.1081/NCN-120015069. [DOI] [PubMed] [Google Scholar]
- Cui Y., Lieber C.M. Functional nanoscale electronic devices assembled using silicon nanowires building blocks. Science. 2001;291:851–853. doi: 10.1126/science.291.5505.851. [DOI] [PubMed] [Google Scholar]
- Cui Y., Wei Q., Park H., Lieber C.M. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- Cummings C.A., Relman D.A. Using DNA micoarrays to study host-microbe interactions. Emerg Infect Dis. 2000;6(5):513–525. doi: 10.3201/eid0605.000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiba A., Inaba N., Ando S., Kajiyama N., Yatsuhashi H., Terasaki H., Ito A., Ogasawara M., Abe A., Yoshioka J., et al. A low-density cDNA microarray with a unique reference RNA: Pattern recognition analysis for IFN efficacy prediction to HCV as a model. Buichem Biophys Res Commun. 2004;315(4):1088–1096. doi: 10.1016/j.bbrc.2004.01.160. [DOI] [PubMed] [Google Scholar]
- Dankbar D.M., Dawson E.D., Mehlmann M., Moore C.L., Smagala J.A., Shaw M.W., Cox N.J., Kuchta R.D., Rowlen K.L. Diagnostic microarray for influenza B viruses. Anal Chem. 2007;79(5):2084–2090. doi: 10.1021/ac061960s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson E.D., Moore C.L., Dankbar D.M., Mehlmann M., Townsend M.B., Smagala J.A., Smith C.B., Cox N.J., Kuchta R.D., Rowlen K.L. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal Chem. 2007;79(1):378–384. doi: 10.1021/ac061920o. [DOI] [PubMed] [Google Scholar]
- Delrio-Lafreniere S.A., Browning M.K., McGlennen R.C. Low-density addressable array for the detection and typing of the human papillomavirus. Diagn Microbiol Infect Dis. 2004;48(1):23–31. doi: 10.1016/j.diagmicrobio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer V.R. Genomics and array technology. Curr Opin Oncol. 1999;11:76–79. doi: 10.1097/00001622-199901000-00015. [DOI] [PubMed] [Google Scholar]
- DeRisi J., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Dolan P.L., Wu Y., Ista L.K., Metzenberg R.L., Nelson M.A., Lopez G.P. Robust and efficient synthetic method for forming DNA microarrays. Nucl Acids Res. 2001;29(21):e107. doi: 10.1093/nar/29.21.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger S.W., Salomonis N., Dahlquist K.D., Vranizan K., Lawlor S.C., Conklin B.R. MAPPFinder: Using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J., Liu P., Zhang X. Cellular response to gene expression profiles of different hepatitis C virus core proteins in the Huh-7 cell line with microarray analysis. J Nanosci Nanotechnol. 2005;5(88):1230–1235. doi: 10.1166/jnn.2005.209. [DOI] [PubMed] [Google Scholar]
- Draghici S. Statistical intelligence: Effective analysis of high-density microarray data. Drug Discov Today. 2002;7(11 Suppl):55–63. doi: 10.1016/s1359-6446(02)02292-4. [DOI] [PubMed] [Google Scholar]
- Draghici S., Khatri P., Bhavsar P., Shah A., Krawetz S.A., Tainsky M.A. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucl Acids Res. 2003;31(13):3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duburcq X., Olivier C., Malingue F., Desmet R., Bouzidi A., Zhou F., Auriault C., Gras-Masse H., Melnyk O. Peptide-protein microarrays for the simultaneous detection of pathogen infections. Bioconjug Chem. 2004;15:307–316. doi: 10.1021/bc034226d. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J.E., Sagripanti J.L. Simultaneous identification of orthopoxviruses and alphaviruses by oligonucleotide macroarray with special emphasis on detection of variola and Venezuelan equine encephalitis viruses. J Virol Meth. 2006;131(12):160–167. doi: 10.1016/j.jviromet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Fodor S.P., Read J.L., Pirrung M.C., Stryer L., Lu A.T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z. Ultrasensitive detection and identification of fluorescent molecules by FCS: Impact for immunobiology. Proc Natl Acad Sci U S A. 2001;98(20):11509–11515. doi: 10.1073/pnas.181337998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földes-Papp Z. Theory of measuring the selfsame single fluorescent molecule in solution suited for studying individual molecular interactions by SPSM-FCS. Pteridines. 2002;13:73–82. [Google Scholar]
- Földes-Papp Z. How the molecule number is correctly quantified in two-color fluorescence cross-correlation spectroscopy: Corrections for cross-talk and quenching in experiments. Curr Pharm Biotechnol. 2005;6(6):437–444. doi: 10.2174/138920105775159296. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z. What it means to measure a single molecule in solution by fluorescence fluctuation spectroscopy. Exp Mol Pathol. 2006;80(3):209–218. doi: 10.1016/j.yexmp.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z. “True” single-molecule molecule observations by fluorescence correlation spectroscopy and two-color fluorescence cross-correlation spectroscopy. Exp Mol Pathol. 2007;82(2):147–155. doi: 10.1016/j.yexmp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z. Exploring the biomedical applications of microscopy and spectroscopy. Exp Mol Pathol. 2007;82(2):103. [Google Scholar]
- Földes-Papp Z. Fluorescence fluctuation spectroscopic approaches to the study of a single molecule diffusing in solution and a live cell without systemic drift or convection: a theoretical study. Curr Pharm Biotechnol. 2007;8(5):267–273. doi: 10.2174/138920107782109930. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Herold A., Seliger H., Kleinschmidt A.K. In: Nonnenmacher T.F., Losa G.A., Weibel E.R., editors. Birkhauser; Boston: 1994. Error propagation theory of chemically solid phase synthesizedoligonucleotides and DNA sequences for biomedical application; pp. 165–173. (Fracals in Biology and Medicine Volume I). [Google Scholar]
- Földes-Papp Z., Birch-Hirschfeld E., Rösch R., Hartmann M., Kleinschmidt A.K., Seliger H. Efficiency of chemical oligonucleotide synthesis evaluated by ion-exchange high-performance liquid chromatography. J Chromatogr A. 1995;706:405–419. doi: 10.1016/0021-9673(96)00060-x. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Peng W.-G., Seliger H., Kleinschmidt A.K. Fractal dimension of error sequences dynamics in quantitative modeling of syntheses of short oligonucleotide and single stranded DNA sequences. J Theor Biol. 1995;174:391–408. doi: 10.1006/jtbi.1995.0107. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Birch-Hirschfeld E., Eickhoff H., Baumann G., Biber Th., Seydel R., Kleinschmidt A.K., Seliger H. Fractals for multicyclic synthesis conditions of biopolymers: Examples of oligonucleotide synthesis measured by high-performance capillary electrophoresis and ion-exchange high-per-formance liquid chromatography. J Chromatogr A. 1996;739:431–447. doi: 10.1016/0021-9673(96)00060-x. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Angerer B., Ankenbauer W., Baumann G., Birch-Hirschfeld E., Björling S., Conrad S., Hinz M., Rigler R., Seliger H., et al. In: Fractals in Biology and Medicine Volume II. Losa G.A., Merlini D., Nonnenmacher T.F., Weibel E.R., editors. Birkauser; Boston: 1997. Modeling the dynamics of nonenzymatic and enzymatic nucleotide processes by fractal dimension; pp. 238–254. [Google Scholar]
- Földes-Papp Z., Birch-Hirschfeld E., Seliger H., Kleinschmidt A.K. In: Spurenanalytische Bestimmung von Ionen: Ionenchromatographie und Kapillarelektrophorese. Kettrup A., Weiss J., Jensen D., editors. Munich, Ecomed. Publ; Landsberg: 1997. Modeling chromatographic and electrophoretic data in the separation of crude products of multicyclic oligonucleotide synthesis; pp. 158–172. [Google Scholar]
- Földes-Papp Z., Baumann G., Birch-Hirschfeld E., Eickhoff E., Greulich K.O., Kleinschmidt A.K., Seliger H. The analysis of oligonucleotide preparations by fractal measures. Biopolymers. 1998;45:361–379. [Google Scholar]
- Földes-Papp Z., Angerer B., Ankenbauer W., Rigler R. Fluorescent high-density labeling of DNA: Error-free substitution for a normal nucleotide. J Biotechnol. 2001;86:237–253. doi: 10.1016/s0168-1656(00)00416-8. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Angerer B., Thyberg P., Hinz M., Wennmalm S., Ankenbauer W., Seliger H., Holmgren A., Rigler R. Fluorescently labeled model DNA sequences for exonucleolytic sequencing. J Biotechnol. 2001;86:203–224. doi: 10.1016/s0168-1656(00)00414-4. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Egerer R., Birch-Hirschfeld E., Striebel H.-M., Wutzler P. Detection of multiple human herpes viruses by DNA microarray technology. Mol Diagn. 2004;8(1):1–9. doi: 10.1007/BF03260041. [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z., Baumann G., Kinjo M., Tamura M. In: Encyclopedia of Medical Genomics & Proteomics. Fuchs J., Podda M., editors. Marcel Dekker; New York: 2005. Single-phase single-molecule fluorescence correlation spectroscopy (SPSM-FCS). Distinguished article entry. [Google Scholar]
- Földes-Papp Z., Egerer R., Birch-Hirschfeld E., Striebel H.-M., Wutzler P. In: Encyclopedia of Medical Genomics & Proteomics. Fuchs J., Podda M., editors. Marcel Dekker; New York: 2005. Human herpes virus detection by DNA microarray technology. Distinguished article entry. [Google Scholar]
- Földes-Papp Z., Kinjo M., Tamura M., Birch-Hirschfeld E. An ultrasensitive way to circumvent PCR-based allele distinction: Direct probing of genomic DNA by solution-phase hybridization down to femtomolar allele concentrations and less using two-color fluorescence crosscorrelation spectroscopy. Exp Mol Pathol. 2005;78(3):177–189. doi: 10.1016/j.yexmp.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Nakaso K., Nakashima K., Sadanari H., Hisanori I., Teishikata Y., Hayward S.D., Yokosawa H. Multiplex PCR-based DNA array for simultaneous detection of three human herpesviruses, EVB, CMV and KSHV. Ecp Mol Pathol. 2006;80(2):124–131. doi: 10.1016/j.yexmp.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gerion D., Chen F., Kannan B., Fu A., Parak. W.J., Chen D.J., Majumdar A., Alivisatos A.P. Room-temperaturesingle-nucleotide polymorphism and multiallele DNA detection using fluorescent nanocrystals and microarrays. Anal Chem. 2003;75:4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Pumfery A., De La Fuente C., Yao K., Miller N., Lacoste V., Quackenbush J., Jacobson S., Kashanchi F. Use of a multi-virus array for the study of human viral and retroviral pathogens: Gene expression studies and ChIP-chip analysis. Retrovirology. 2004;1:10. doi: 10.1186/1742-4690-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheit T., Landi S., Gemignani F., Snijders P.J., Vaccarella S., Franceschi S., Canzian F., Tommasino M. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J Clin Microbiol. 2006;44(6):2025–2031. doi: 10.1128/JCM.02305-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Masquelier B., Fleury H., Lacroix B., Troesch A., Vernet G., Telles J.N. Detection of human immunodeficiency virus type 1 antiretroviral resistance mutations by high-density DNA probe arrays. J Clin Microbiol. 2004;42(87):2907–2912. doi: 10.1128/JCM.42.7.2907-2912.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow V.S., Settineri M., Lawton R.G. P-Nitrophenyldiazopyruvate and diazopyruvamides, a new family of photoactivatable cross-linking bioprobes. Biochemistry. 1989;28(15):6346–6360. doi: 10.1021/bi00441a030. [DOI] [PubMed] [Google Scholar]
- Günthard H.F., Wong J.K., Ignacio C.C., Havlir D.V., Richman D.D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res Human Retrovir. 1998;14:869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- Hamels S., Gala J.L., Dufour S., Vannuffel P., Zammatteo N., Remacle J. Consensus PCR and microarray for diagnosis of the genus Staphylococcus, species, and methicillin resistance. Biotechniques. 2001;31(6):1364–1366. doi: 10.2144/01316md04. [DOI] [PubMed] [Google Scholar]
- Harrison J.K., Lawton R.G., Gnegy M.E. Development of a novel photoreactive calmoduline derivate: Cross-linking of purified adenylate cyclase from bovine brain. Biochemistry. 1989;28(14):6023–6027. doi: 10.1021/bi00440a045. [DOI] [PubMed] [Google Scholar]
- Haselbeck R., Wall D., Jiang B., Ketela T., Zyskind J., Bussey H., Foulkes J.G., Roemer T. Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Curr Pharm Des. 2000;8(13):1155–1172. doi: 10.2174/1381612023394818. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Lohrey C., Hunziker A., Kahn T., Schwarz E. Human papillomavirus type 16 E6 and E7 genotypes in head-and-neck carcinomas. Oral Oncol. 2004;40(5):520–524. doi: 10.1016/j.oraloncology.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Ivshina A.V., Vodeiko G.M., Kuznetsov V.A., Volokhov D., Taffs R., Chizhikov V.I., Levandowski R.A., Chumakov K.M. Mapping of genomic segments of influenza B virus strains by an oligonucleotide microarray method. J Clin Microbiol. 2004;42(12):5793–5801. doi: 10.1128/JCM.42.12.5793-5801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen A.J., Maunula L. Applicability of microarray technique for the detection of noro- and astroviruses. J Virol Meth. 2006;136(1-2):210–216. doi: 10.1016/j.jviromet.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Alba M.M., Boshoff C., Kellam P. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jianrong M., Yuqing M., Nongyue H., Xiaohua W., Sijiao L. Nanotechnology and biosensors. Biotechnol Adv. 2004;22:505–518. doi: 10.1016/j.biotechadv.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Jung A., Stemmler I., Brecht A., Gauglitz G. Covalent strategy for immobilization of DNA-microspots suitable for microarrays with label-free and time-resolved optical detection of hybridization Fresenius. J Anal Chem. 2001;371:128–136. doi: 10.1007/s002160101001. [DOI] [PubMed] [Google Scholar]
- Kalachikov S.M., Adarichev V.A., Dymshits G.M. Immobilization of DNA on microporous membranes using UV-irradiation. Bioorganicheskaia Khimia. 1992;18(1):56–62. [PubMed] [Google Scholar]
- Kato-Maeda M., Gao Q., Small P.M. Microarray analysis of pathogens and their interactions with hosts. Cell Microbiol. 2001;3(11):713–719. doi: 10.1046/j.1462-5822.2001.00152.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K., Kaneko S., Honda M., Kawai H.F., Shirota Y., Kobayashi K. Detection of hepatitis B virus DNA in sera from patients with chronic hepatitis B virus infection by DNA microarray method. J Clin Microbiol. 2003;41(4):1701–1704. doi: 10.1128/JCM.41.4.1701-1704.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating C.D.N. Striped metal nanowires as building blocks and optical tags. Adv Mater. 2003;15:451–454. [Google Scholar]
- Kellam P. Post-genomic virology: The impact of bioinformatics, microarrays and proteomics on investigating host and pathogen interactions. Rev Med Virol. 2001;11:313–329. doi: 10.1002/rmv.328. [DOI] [PubMed] [Google Scholar]
- Kennedy P.G., Grinfeld E., Craigon M., Vierlinger K., Roy D., Forster T., Ghazal P. Transcriptomal analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J Gen Virol. 2005;86:2673–2684. doi: 10.1099/vir.0.80946-0. [DOI] [PubMed] [Google Scholar]
- Kessler N., Ferraris O., Palmer K., Marsh W., Steel A. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J Clin Microbiol. 2004;42(5):2173–2185. doi: 10.1128/JCM.42.5.2173-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Yoon M.S., Na Y.J., Park C.S., Oh M.R., Moon W.C. Development and evaluation of a highly sensitive human papillomavirus genotyping DNA chip. Gynecol Oncol. 2006;100(1):38–43. doi: 10.1016/j.ygyno.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Kittler K., Löber G. Sequence specificity of DNA-psoralen photoproduct formation in supercoiled plasmid DNA (puc 19) Photobiochem Photobiophys. 1995;27:161–166. doi: 10.1016/1011-1344(94)07071-u. [DOI] [PubMed] [Google Scholar]
- Klaassen C.H., Prinsen C.F., de Valk H.A., Horrevorts A.M., Jeunink M.A., Thunnissen F.B. DNA microarray format for detection and subtyping of human papillomavirus. J Clin Microbiol. 2004;42:2152–2160. doi: 10.1128/JCM.42.5.2152-2160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck S.M., Hampl W., Kleinschmidt A.K. HSV type 1 genome variants from persistently productive infections in RAJI and BJAB cell lines. Arch Virol. 1995;140:1195–1213. doi: 10.1007/BF01322746. [DOI] [PubMed] [Google Scholar]
- Korimbocus J., Scaramozzino N., Lacroix B., Crance J.M., Garin D., Vernet G. DNA probe array for the simultaneous identification of herpesviruses, enteroviruses, and flaviviruses. J Clin Microbiol. 2005;43(8):3779–3787. doi: 10.1128/JCM.43.8.3779-3787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. SARS Working Group. [DOI] [PubMed] [Google Scholar]
- Lai C.L., Ratziu V., Yuen M.F. Viral hepatitis B. Lancet. 2003;362:2089–2094. doi: 10.1016/S0140-6736(03)15108-2. [DOI] [PubMed] [Google Scholar]
- Leenman E.E., Panzer-Grumayer R.E., Fischer S., Leitch H.A., Horsman D.E., Lion T., Gadner H., Ambros P.F. Rapid determination of Epstein-Bar virus latent or lytic infection in single human cells using in situ hybridisation. Mod Pathol. 2004;17:1564–1572. doi: 10.1038/modpathol.3800228. [DOI] [PubMed] [Google Scholar]
- Li C., Chen R.S., Hung S.K., Lee Y.T., Yen C.Y., Lai Y.W., Teng R.H., Huang J.Y., Tang Y.C., Tung C.P., et al. Detection of Epstein-Barr virus infection and gene expression in human tumors by microarray analysis. J Virol Meth. 2006;133(2):158–166. doi: 10.1016/j.jviromet.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Li J., Chen S., Evans D.H. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J Clin Micorbiol. 2001;39:696–704. doi: 10.1128/JCM.39.2.696-704.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.G., Chen L.Y., Huang J., Qiao P., Qiu J.M., Wang S.Q. Quantification of the relative levels of wild-type and lamivudine-resistant mutant virus in serum of HBV-infected patients using microarray. J Viral Hepat. 2005;12(2):168–175. doi: 10.1111/j.1365-2893.2005.00562.x. [DOI] [PubMed] [Google Scholar]
- Liang R.Q., Li W., Li Y., Tan C.Y.Y., Li J.X., Jin Y.X., Ruan R.C. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Vora G.J., Thach D., Walter E., Metzgar D., Tibbetts C., Stenger D.A. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J Clin Microbiol. 2004;42:3232–3239. doi: 10.1128/JCM.42.7.3232-3239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Chen H.C., Lin R.W., You S.L., You C.M., Chuang L.C., Pan M.H., Lee M.H., Chou Y.C., Chen C.J. Quality assurance of genotyping array for detection and typing of human papillomavirus. J Virol Meth. 2007;140(1-2):1–9. doi: 10.1016/j.jviromet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Lin F.M., Huang H.D., Chang Y.C., Tsou A.P., Chan P.L., Wu L.C., Tsai M.F., Horng J.T. Database to dynamically aid probe design for virus identification. IEEE Trans Inf Technol Biomed. 2006;10(4):705–713. doi: 10.1109/titb.2006.874202. [DOI] [PubMed] [Google Scholar]
- Lindroos K., Liljedahl U., Raitio M., Syvänen A.Ch. Minisequencing on oligonucleotide microarrays: Comparison of immobilisation chemistries. Nucl Acids Res. 2001;29(13):e69. doi: 10.1093/nar/29.13.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz R.J., Morris D., Chee M., Hubbell E., Kozal M.J., Shah N., Shen N., Yang R., Fodor S.P. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- Liu Q., Bai Y., Ge Q., Zhou S., Wen T., Lu Z. Microarray-in-a-tube for detection of multiple viruses. Clin Chem. 2007;53(2):188–194. doi: 10.1373/clinchem.2006.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston A.D., Campbell C.J., Wagner E.K., Ghazal P. Biochip sensors for the rapid and sensitive detection of viral disease. Genome Biol. 2005;6(6):112. doi: 10.1186/gb-2005-6-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes M.J., Suciu D., Elliott M., Stover A.G., Ross M., Caraballo M., Dix K., Crye J., Webby R.J., Lyon W.J., et al. Use ofsemiconductor-based oligonucleotide microarrays for influenza a virus subtype identification and sequencing. J Clin Microbiol. 2006;44(4):1209–1218. doi: 10.1128/JCM.44.4.1209-1218.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmar L., Fock C., Espinoza F., Bucardo F., Syvanen A.C., Bondeson K. Microarrays for genotyping human group a rotavirus by multiplex capture and type-specific primer extension. J Clin Microbiol. 2003;41:5153–5158. doi: 10.1128/JCM.41.11.5153-5158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.W., Roan C.H., Liu C.J. Human papillomaviruses in oral squamous cell carcinoma and pre-cancerous lesions detected by PCR-based gene-chip array. Int. J Oral Maxillofac Surg. 2007;36(2):153–158. doi: 10.1016/j.ijom.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Mahony J.B. Chlamydiae host cell interactions revealed using DNA microarrays. Ann NY Acad Sci. 2002;975:192–201. doi: 10.1111/j.1749-6632.2002.tb05952.x. [DOI] [PubMed] [Google Scholar]
- Martell M., Briones C., de Vicente A., Piron M., Esteban J.I., Esteban R., Guardia J., Gómez J. Structural analysis of hepatitis C RNA genome using DNA microarrays. Nucl Acids Res. 2004;32(11):e90. doi: 10.1093/nar/gnh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson R.S., Rampal J.B., Coassin P.J. Biopolymer synthesis on polypropylene supports. I. Oligonucleotides. Anal Biochem. 1994;217:306–310. doi: 10.1006/abio.1994.1123. [DOI] [PubMed] [Google Scholar]
- Mehlmann M., Townsend M.B., Stears R.L., Kuchta R.D., Rowlen K.L. Optimization of fragmentation conditions for microarray analysis of viral RNA. Anal Biochem. 2005;347(2):316–323. doi: 10.1016/j.ab.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Mehlmann M., Dawson E.D., Townsend M.B., Smagala J.A., Moore C.L., Smith C.B., Cox N.J., Kuchta R.D., Rowlen K.L. Robust sequence selection method used to develop the FluChip diagnostic microarray for influenza virus. J Clin Micobiol. 2006;44(8):2857–2862. doi: 10.1128/JCM.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzasoma L., Bacarese-Hamilton T., Di Cristina M., Rossi R., Bistoni F., Crisanti A. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem. 2002;48:121–130. [PubMed] [Google Scholar]
- Min W., Wen-Li M., Bao Z., Ling L., Zhao-Hui S., Wen-Ling Z. Oligonucleotide microarray with RD-PCR labeling technique for detection and typing of human papillomavirus. Curr Microbiol. 2006;52(3):204–209. doi: 10.1007/s00284-005-0212-x. [DOI] [PubMed] [Google Scholar]
- Möller R., Fritzsche W. Metal nanoparticle-based detection for DNA analysis. Curr Pharm Biotechnol. 2007;8(5):274–285. doi: 10.2174/138920107782109967. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Belbin T.J., Spray D.C., Mukhopadhyay A., Nagajyothi F., Weiss L.M., Tanowitz H.B. Microarray technology in the investigation of diseases of myocardium with special reference to infection. Front Biosci. 2006;11:1802–1813. doi: 10.2741/1924. [DOI] [PubMed] [Google Scholar]
- Mulvaney S.P., Cole C.L., Kniller M.D., Malito M., Tamanaha C.R., Rife J.C., Stanton M.W., Whitman L.J. Bioelectron; Biosens: 2007. Rapid, femtomolar bioassays in complex matrices combining microfluidics and magnetoelectronics. in press, doi:10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- Murphy D. Gene expression studies using microarrays: principles, problems, and prospects. Advan Physiol Edu. 2002;26:256–270. doi: 10.1152/advan.00043.2002. [DOI] [PubMed] [Google Scholar]
- Neverov A.A., Riddell M.A., Moss W.J., Volokhov D.V., Rota P.A., Lowe L.E., Chibo D., Smit S.B., Griffin D.E., Chumakov K.M., et al. Genotyping of measles virus in clinical specimens on the basis of oligonucleotide microarray hybridization patterns. J Clin Microbiol. 2006;44(10):3752–3759. doi: 10.1128/JCM.00998-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicewarner-Pena S.R., Freeman R.G., Reiss B.D., He L., Pena D.J., Walton I.D., Cromer R., Keating C.D., Natan M.J. Submicrometer metallic barcodes. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- Nordstrom H., Johansson P., Li Q.G., Lundkvist A., Nilsson P., Elgh F. Microarray technology for identification and distinction of hantaviruses. J Med Virol. 2004;72:646–655. doi: 10.1002/jmv.20041. [DOI] [PubMed] [Google Scholar]
- Nordstrom H., Falk K.I., Lindegren G., Mouzavi-Jazi M., Waldén A., Elgh F., Nilsson P., Lundkvist A. DNA microarray technique for detection and identification of seven flaviviruses pathogenic for man. J Med Virol. 2005;77(4):528–540. doi: 10.1002/jmv.20489. [DOI] [PubMed] [Google Scholar]
- Oh T.J., Kim C.J., Woo S.K., Kim T.S., Jeong D.J., Kim M.S., Lee S., Cho H.S., et al. Development and clinical evaluation of a highly sensitive DNA microarray for detection and genotyping of human papillomaviruses. J Clin Microbiol. 2004;42:3272–3280. doi: 10.1128/JCM.42.7.3272-3280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R., Yang M., Lin R., Johnson B.N., Srivastava N., Razzacki S.Z., Chomistek K.J., Heldsinger D.C., Haque R.M., Ugaz V.M., et al. An integrated microfluidic device for influenza and other genetic analyses. Lab Chip. 2005;5(10):1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- Park T.C., Kim C.J., Koh Y.M., Lee K.H., Yoon J.H., Kim J.H., Namkoong S.E., Park J.S. Human papillomavirus genotyping by the DNA chip in the cervical neoplasia. DNA Cell Biol. 2004;23(2):119–125. doi: 10.1089/104454904322759939. [DOI] [PubMed] [Google Scholar]
- Park S.J., Taton T.A., Mirkin C.A. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- Patolsky F., Zheng G., Hayden O., Lakadamyali M., Zhuang X., Lieber C.M. Electrical detection of single viruses. Proc Natl Acad Sci U S A. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose-Murphy M., Ha N.K., Xiang C., Chen Y., Gillim L., Yarchoan R., Meltzer P., Bittner M. Transcription program of human herpesvirus 8 (kaposi's sarcoma-associated herpesvirus) J Virol. 2001;75:4843–4853. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease A.C., Solas D., Sullivan E.J., Cronin M.T., Holmes C.P., Fodor S.P.A. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994;91(11):5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek R.M., Moss S.F., Ando T., Camorlinga M., Wirth H.P., Moss S.F., Ando T., Arnold C.N., Falkow S., Tham K.T., et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 2001;107(5):611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penchovsky R., Birch-Hirschfeld E., McCaskill J.S. End-specific covalent photo-dependent immobilisation of synthetic DNA to paramagnetic beads. Nucl Acids Res. 2000;28:e98. doi: 10.1093/nar/28.22.e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J.M., Simeone F.J., Saeki Y., Josephson L., Weissleder R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J Am Chem Soc. 2003;125:10192–10193. doi: 10.1021/ja036409g. [DOI] [PubMed] [Google Scholar]
- Perrin A., Duracher D., Perret M., Cleuziat P., Mandrand B. A combined oligonucleotide and protein microarray for the codetection of nucleic acids and antibodies associated with human immunodeficiency virus, hepatitis B virus, and hepatitis C virus. Anal Biochem. 2003;322:148–155. doi: 10.1016/j.ab.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Pieles U., Englisch U. Psoralen covalently linked to oligodeoxyribonucleotides: synthesis, sequence specific recognition of DNA and photo-cross-linking to pyrimidine residues of DNA. Nucl Acids Res. 1989;17:285–299. doi: 10.1093/nar/17.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersanti S., Martina Y., Cherubini G., Avitabile D., Saggio I. Use of DNA microarrays to monitor host response to virus and virus-derived gene therapy vectors. Am J Pharmacogenomics. 2004;4(6):345–356. doi: 10.2165/00129785-200404060-00002. [DOI] [PubMed] [Google Scholar]
- Pirrung M.C. How to make a DNA Chip. Angew Chem. 2002;41:1276–1289. doi: 10.1002/1521-3773(20020415)41:8<1276::aid-anie1276>3.0.co;2-2. Int. Ed. [DOI] [PubMed] [Google Scholar]
- Preininger C., Sauer U. Quality control of chip manufacture and chip analysis using epoxy-chips as a model. Sens Actu B: Chem. 2003;90(1):98–103. [Google Scholar]
- Ramaswamy S.V., Reich R., Dou S.J., Jasperse L., Pan X., Wanger A., Quitugua T., Graviss E.A. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47(4):1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–373. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rogers P.D., Barker K.S. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob Agents Chemother. 2002;46(11):3412–3417. doi: 10.1128/AAC.46.11.3412-3417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N.L., Mirkin C.A. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1377–1378. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ryabinin V.A., Shundrin L.A., Kostina E.B., Laassri M., Chizhikov V., Shchelkunov S.N., Chumakov K., Sinyakov A.N. Microarray assay for detection and discrimination of Orthopox-virus species. J Med Viol. 2006;78(10):1325–1340. doi: 10.1002/jmv.20698. [DOI] [PubMed] [Google Scholar]
- Schembri M.A., Ussery D.W., Workman C., Hasman H., Klemm P. DNA microarray analysis of fim mutations in Escherichia coli. Mol Genet Genomics. 2002;267(6):721–729. doi: 10.1007/s00438-002-0705-2. [DOI] [PubMed] [Google Scholar]
- Schena M. Oxford University Press; 2001. DNA Microarrays, A Practical Approach. [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schwartz J.A., Vykoukal J.V., Gascoyne P.R. Droplet-based chemistry on a programmable micro-chip. Lab Chip. 2004;4(1):11–17. doi: 10.1039/b310285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifarth W., Spiess B., Zeilfelder U., Speth C., Hehlmann R., Leib-Mosch C. Assessment of retroviral activity using a universal retrovirus chip. J Virol Meth. 2003;112:79–91. doi: 10.1016/s0166-0934(03)00194-0. [DOI] [PubMed] [Google Scholar]
- Sengupta S., Onodera K., Lai A., Melcher U. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J Clin Microbiol. 2003;41:4542–4550. doi: 10.1128/JCM.41.10.4542-4550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.S., Song Y.S., Kim J.W., Park N.H., Kang S.B., Lee H.P. Good correlation of HPV DNA test between self-collected vaginal and clinician-collected cervical samples by the oligonucleotide microarray. Gynecol Oncol. 2006;102(1):67–73. doi: 10.1016/j.ygyno.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Sergeev N., Rubtcova E., Chizikov V., Schmid D.S., Loparev V.N. New mosaic subgenotype of varicella-zoster virus in the USA: VZV detection and genotyping by oligonucleotide-microarray. J Virol Meth. 2006;136(1-2):8–16. doi: 10.1016/j.jviromet.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Shchepinov M.S., Case-Green S.C., Southern E.M. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucl Acids Res. 1997;25(6):1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82(4):693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- Son S., Noh H.T., An S. Human papillomavirus status in cervical scrapes and biopsy specimens using the HPV genotyping DNA microarray Int. J Gynaecol Obstet. 2006;93(3):258–259. doi: 10.1016/j.ijgo.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Song Y., Dai E., Wang J., Liu H., Zhai J., Chen C., Du Z., Guo Z., Yang R. Genotyping of hepatitis B virus (HBV) by oligonucleotides microarray. Mol Cell Probes. 2006;20(2):121–127. doi: 10.1016/j.mcp.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Southern E., Mir K., Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21(1):5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- Stingley S.W., Ramirez J.J., Aguilar S.A., Simmen K., Sandri-Goldin R.M., Ghazal P., Wagner E.K. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J Virol. 2000;74:9916–9927. doi: 10.1128/jvi.74.21.9916-9927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel H.M., Birch-Hirschfeld E., Egerer R., Földes-Papp Z. Virus diagnostics on microarrays. Curr Pharm Biotechnol. 2003;4:401–415. doi: 10.2174/1389201033377274. [DOI] [PubMed] [Google Scholar]
- Striebel H.M., Birch-Hirschfeld E., Egerer R., Földes-Papp Z., Stelzer A. Enhancing sensitivity of human virus diagnosis with DNA microarrays using dendrimers. Exp Mol Pathol. 2004;77:89–97. doi: 10.1016/j.yexmp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Sulaiman I.M., Liu X., Frace M., Sulaiman N., Olsen-Rasmussen M., Neuhaus E., Rota P.A., Wohlhueter R.M. Evaluation of affymetrix severe acute respiratory syndrome resequencing GeneChips in characterization of the genomes of two strains of coronavirus infecting humans. Appl Environ Microbiol. 2006;72(1):207–211. doi: 10.1128/AEM.72.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Tang K., Osborne J., Sammons S., Wohlhueter R.M. GeneChip resequencing of the smallpox virus genome can identify novel strains: A biodefense application. J Clin Microbiol. 2007;45(2):358–363. doi: 10.1128/JCM.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton T.A., Mirkin C.A., Letsinger R.L. ScanometricDNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Thierry F., Benotmane M.A., Demeret C., Mori M., Teissier S., Desaintes C. A genomic approach reveals a novel mitotic pathway in papillomavirus carcinogenesis. Cancer Res. 2004;64(3):895–903. doi: 10.1158/0008-5472.can-03-2349. [DOI] [PubMed] [Google Scholar]
- Tönisson N., Kung A., Kaasik K., Löhmussaar E., Metspalu A. Unravelling genetic data by arrayed primer extension. Clin Chem Lab Med. 2000;38(2):165–170. doi: 10.1515/CCLM.2000.025. [DOI] [PubMed] [Google Scholar]
- Townsend M.B., Dawson E.D., Mehlmann M., Smagala J.A., Dankbar D.M., Moore C.L., Smith C.B., Cox N.J., Kuchta R.D., Rowlen K.L. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J Clin Microbiol. 2006;44(8):2863–2871. doi: 10.1128/JCM.00134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N., Berne R., Chann R., Gauthier M., Martin D., Armand M.A., Ollivet A., Teo C.G., Ijaz S., Flichman D., et al. European multicenter evaluation of high-density DNA probe arrays for detection of hepatitis B virus resistance mutations and identification of genotypes. J Clin Microbiol. 2006;44(8):2780–2792. doi: 10.1128/JCM.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A., Fischer K.F., Chiu C.Y., Kistler A.L., Beck S., Wang D., DeRisi J.L. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 2005;6(9):R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A., Molinaro R.J., Fischer N., Plummer S.J., Casey G., Klein E.A., Malathi K., Magi-Galluzzi C., Tubbs R.R., Ganem D., et al. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PloS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wallace J., Woda B.A., Pihan G. Facile, comprehensive, high-throughput genotyping of human genital papillomaviruses using spectrally addressable liquid bead microarrays. J Mol Diagn. 2005;7(1):71–80. doi: 10.1016/S1525-1578(10)60011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99(24):15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek Y.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1(2):E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. Identifying influenza viruses with resequencing microarrays. Emerg Infect Dis. 2006;12(4):638–646. doi: 10.3201/eid1204.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessendorf M.W., Brelje T.C. Which fluorophore is brightest? A comparison of the staining obtained using fluorescein, tetramethylrhodamine, lissamine rhodamine, Texas read and cyanine 3. Histochemistry. 1992;98(2):81–85. doi: 10.1007/BF00716998. [DOI] [PubMed] [Google Scholar]
- Wittmann, C. (ed.) (2005). Immobilization of DNA on chips I, II. Topics Curr Chem 260–261.
- Wong C.W., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond L.W., Stanton L.W., Liu E.T., Miller L.D. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R. Cancer classification with DNA microarrays is less more? Trend Genet. 2000;16(8):327–329. doi: 10.1016/s0168-9525(00)02064-3. [DOI] [PubMed] [Google Scholar]
- Wright M.A., Church G.M. An open-source oligomicroarray standard for human and mouse. Nature Biotechnol. 2002;20:1082–1083. doi: 10.1038/nbt1102-1082. [DOI] [PubMed] [Google Scholar]
- Yamagishi J., Isobe R., Takebuchi T., Bando H. DNA microarrays of baculovirus genomes: Differential expression of viral genes in two susceptible insect cell lines. Arch Virol. 2003;148:587–597. doi: 10.1007/s00705-002-0922-3. [DOI] [PubMed] [Google Scholar]
- Yang D.H., Barari M., Arif B.M., Krell P.J. Development of an oligonucleotide-based DNA microarray for transcriptional analysis of Choristoneura fumiferana nucleopolyhedrovirus (CfMNPV) genes. J Virol Meth. 2007;143m(2):175–185. doi: 10.1016/j.jviromet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Yang S., Ghanny S., Wang W., Galante A., Dunn W., Liu F., Soteropoulos P., Zhu H. Using DNA microarray to study human cytomegalovirus gene expression. J. Virol. Meth. 2006;131(2):202–208. doi: 10.1016/j.jviromet.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Ye M.Y., Yin X.F., Fang Z.L. DNA separation with low-viscosity sieving matrix on microfabricated polycarbonate microfluidic chips. Anal Bioanl Chem. 2005;381(4):820–827. doi: 10.1007/s00216-004-2988-0. [DOI] [PubMed] [Google Scholar]
- Zhang C.-Y., Yeh H.-C., Kuroki M.T., Wang T.-H. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4(11):826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- Zaytseva N.V., Montagna R.A., Baeumner A.J. Microfluidic biosensor for the serotype-specific detection of dengue virus RNA. Anal Chem. 2005;77(23):7520–7527. doi: 10.1021/ac0509206. [DOI] [PubMed] [Google Scholar]
- Zein N.N. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13(2):223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Nampalli S., Serino A.J. Kumar,S. Immobilization of oligonucleotides with multiple anchors to microchips. Nucl Acids Res. 2001;29(4):955–959. doi: 10.1093/nar/29.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Wan J.M., Liu W., Liu Q.J., Zhang L., Zhou Z.X., Liu X.J., Zhang H.R. Hepatitis gene chip in detecting HBV DNA, HCV RNA in serum and liver tissue samples of hepatitis patients. Hepatobiliary Pancreas Dis Int. 2003;2:234–241. [PubMed] [Google Scholar]
Recommended Resources
- Chiu C.Y., Alizadeh A.A., Rouskin S., Merker J.D., Yeh E., Yagi S., Schnurr D., Patterson B.K., Ganem D., DeRisi J.L. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J Clin Microbiol. 2007;45(7):2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földes-Papp Z. “True” single-molecule molecule observations by fluorescence correlation spectroscopy and two-color fluorescence cross-correlation spectroscopy. Exp Mol Pathol. 2007;82(2):147–155. doi: 10.1016/j.yexmp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Földes-Papp, Z. (ed.) (2007b). Half special issue exploring the biomedical applications of microscopy and spectroscopy. Exp Mol Pathol 82(2), 103–189.
- Földes-Papp Z., Enderlein J., Widengren J., Kinjo M. Special Edition. The way down from single genes and proteins to single molecules: Nucleic acid analyses in many-molecule systems (first part of the first two-part special issue) Curr Pharm Biotechnol. 2003;4(6):351–484. [Google Scholar]
- Földes-Papp Z., Enderlein J., Widengren J., Kinjo M. Special Edition. The way down from single genes and proteins to single molecules: Nucleic acid and protein analyses in many-molecule systems (second part of the first two-part special issue) Curr Pharm Biotechnol. 2004;5(1):1–126. [Google Scholar]
- Földes-Papp Z., Enderlein J., Widengren J., Kinjo M. Special Edition. The way down from single genes and proteins to single molecules: fluorescence correlation spectroscopy (auto- and two-color cross-correlation mode) in single-molecule systems (first part of the second two-part special issue) Curr Pharm Biotechnol. 2004;5(2):135–241. [Google Scholar]
- Földes-Papp Z., Enderlein J., Widengren J., Kinjo M. Special Edition. The way down from single genes and proteins to single molecules: Applying single-molecule analyses (second part of the second two-part special issue) Curr Pharm Biotechnol. 2004;5(3):243–319. [Google Scholar]
- Földes-Papp Z., Egerer R., Birch-Hirschfeld E., Striebel H.-M., Wutzler P. In: Encyclopedia of Medical Genomics & Proteomics. Fuchs J., Podda M., editors. Marcel Dekker; New York: 2005. Human herpes virus detection by DNA microarray technology. Distinguished article entry. [Google Scholar]
- Livingston A.D., Campbell C.J., Wagner E.K., Ghazal P. Biochip sensors for the rapid and sensitive detection of viral disease. Genome Biol. 2005;6(6):112. doi: 10.1186/gb-2005-6-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich R.R., Fleisher T.A., Shearer W.T., Schroeder H.W., Frew A.J., Weyand C.M., editors. Clinical Immunology: Principles and Practice. 3rd edition. Mosby Ltd; New York: 2008. [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Seliger H. In: Microarrays. Synthesis Methods Volume 1. 2nd edition. Rampal J.B., editor. Academic Press; 2007. Introduction: Array technology – an overview. [Google Scholar]
- Urisman A., Fischer K.F., Chiu C.Y., Kistler A.L., Beck S., Wang D., DeRisi J.L. E-Predict: A computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 2005;6(9):R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99(24):15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
National Center for Biotechnology Information and resource for molecular biology information. NCBI creates public databases, conducts research in computational biology, develops software tools for analyzing genome data, and disseminates biomedical information: http://www.ncbi.nlm.nih.gov/5
Software to accelerate research in molecular biology: http://www.premierbiosoft.com
DeRisi Lab: E-Predict Download. derisilab.ucsf.edu/epredict/
Stanford microarray database: genome-www5.stanford.edu/
ArrayExpress of EMBL-EBI: http://www.ebi.ac.uk/arrayexpress/
Microscale products and services for Biochemistry and Molecular Biology at the Open Directory Project: http://dmoz.org/dmoz.org/Science/Biology/Biochemistry_and_Molecular_Biology/Products_and_Services/Micro_Scale/


