Abstract
Shotgun lipidomics provides sensitive and fast lipid identification without the need for chromatographic separation. Challenges faced by shotgun analysis of glycerophospholipids (GPs) include the lack of signal uniformity across GP classes and the inability to determine the carbon-carbon double bond (C=C) location within the fatty acyl chains of an unsaturated species. Two distinct derivatization strategies were employed to both enhance the ionization of GPs, via trimethylation enhancement using 13C-diazomethane (13C-TrEnDi), as well as determine location of double bonds within fatty acyl chains, employing an in-solution photochemical reaction with acetone (via the Paternò–Büchi reaction). The modified GPs were then subjected to positive ion mode ionization via electrospray ionization, producing uniform ionization efficiencies for different classes of GP species. The GPs were charge inverted via gas-phase ion/ion reactions and sequentially fragmented using ion trap collision-induced dissociation (CID). The CID of the species led to fragmentation producing diagnostic ions indicative of C=C bond location. The approach enabled enhanced ionization and the identification of phosphatidylcholine and phosphatidylethanolamine species at the C=C level in a bovine lipid extract.
Keywords: Lipidomics, Mass Spectrometry, Charge Inversion, Paternò–Büchi Reaction, 13C-TrEnDi
Graphical Abstract
1. INTRODUCTION
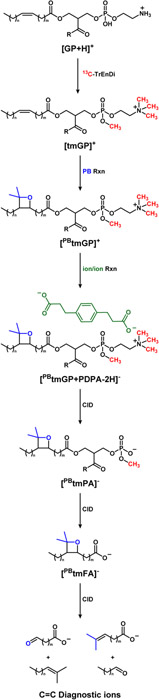
Mass spectrometry (MS) is a core tool in lipidomics research due to its high sensitivity and specificity. In favorable cases, lipid ions may be efficiently generated using electrospray ionization (ESI) and selectively fragmented via tandem MS (MS2) to obtain structural details and enable unambiguous identification. Shotgun lipidomics is an MS-based approach that involves the direct-infusion of complex lipid extracts without prior separation, offering both speed and efficient coverage for lipid analysis.1,2 Glycerophospholipids (GPs) are the most abundant lipid class by mass in eukaryotic cells.1 GP analysis benefits from different MS approaches that seek to improve the characterization of individual lipid species.3 Although relatively low in molecular weight when compared to other biologically relevant molecules, GPs are structurally diverse. For example, over 100 species can exist within a single sample, each potentially possessing unique biological activity.4 GPs, such as phosphatidylethanolamine (PE) and phosphatidylcholine (PC), are structurally characterized by a glycerol backbone with a polar head group attached at the sn3 position1,3 and acyl chains attached at the sn1 and/or sn2 positions. One of the challenges with the shotgun approach is that the ionization efficiency of GPs depends on the identity of the polar headgroup. For example, the headgroup of PE consists of a primary amine that is readily protonated but susceptible to matrix effects during the ESI process leading to variation in sensitivity between samples. Several approaches to address this issue have been made including the incorporation of a fixed positive charge using sulfonium ions5,6 or through the generation of a fixed quaternary ammonium and methylated phosphate using trimethylation enhancement using diazomethane (TrEnDi).7,8 Depending upon the headgroup identity, TrEnDi can improve ionization yields greater than 11-fold compared to unmodified lipids in the positive ion mode7 and is compatible with the use of isotopically labelled diazomethane to differentiate modified lipid classes and enable quantification strategies (13C-TrEnDi).8 However, TrEnDi modification generally inhibits ionization in the negative mode due to the complete methylation of functional groups with pKa values less than 11. For this reason, fatty acyl analysis of TrEnDi modified ions is severely compromised as traditional negative ion ESI-MS2 produces essentially no ions and positive ion tandem mass spectra are strongly dominated by head group product ions. Gas-phase charge inversion of the TrEnDi modified cations followed by MS2 has been shown to be an effective solution to this issue and enables full determination of fatty acyl chain composition.9-11 This gas-phase charge inversion strategy indicates the number of carbon-carbon double bonds (C=C) that are present in a given lipid but does not yield any information as to their location. Several methods currently exist to probe the C=C location within fatty acyl chains.12-22 However, most of them require instrument modifications or high energy collisions. Methods involving in-solution modification prior to ionization have been developed as means of obtaining C=C information without instrument modification23-25 including the Paternò–Büchi (PB) reaction.26-30 This UV-initiated reaction utilizes acetone as a reagent for a [2+2] cycloaddition to the unsaturation site(s) on a fatty acyl chain. Upon low-energy CID, signature fragments are formed that indicate C=C bond location. Charge inversion of PB reacted GPs has been applied for improved structural coverage of PEs and PCs allowing for C=C location to be determined.31 Herein, we demonstrate the utility of combining 13C-TrEnDi, PB chemistry and gas-phase charge inversion to extend PE and PC lipid structural characterization to also include the determination of C=C bond location.
2. EXPERIMENTAL
2.1. Materials.
All the GP samples were purchased as chloroform solutions from Avanti Polar Lipid, Inc. (Alabaster, AL). The analytes were 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and bovine liver polar lipid extract (BLE). 1,4-phenylenedipropionic acid (PDPA) was purchased from Sigma Aldrich (St. Louis, MO). All solvents were HPLC-grade. 13C-labeled N-methyl-N-nitroso-p-toluenesulfonamide and 13C-diazomethane were prepared according to procedures by Shields et al.32
2.2. The 13C-TrEnDi Modification of GPs.
The modification has previously been reported.8 In short, 1 μL of freshly prepared tetrafluoroboric acid diethyl ether complex in diethyl ether was added to a solution of lipid standard or BLE in ethanol. To the lipid solution approximately 1 mL of freshly distilled ethereal 13C-diazomethane was added, and allowed to react at room temperature for 5 min. Once the reaction was complete, the light-yellow solution was carefully dried down under a gentle stream of N2(g) and sealed under an atmosphere of N2(g) before shipping over dry ice.
2.3. The PB reactions in an offline flow microreactor.
The PB reaction was conducted using an offline flow microreactor as previously discussed.31 The flow cell of the device was a UV transparent fused silica capillary (fluoropolymer-coated, 100 μm i.d. 363 μm o.d.; Polymicro Technologies/Molex; Phoenix, AZ). In order to control the reaction and as a safety precaution, the exposure of the cell to UV emission was limited to a 4 cm portion within an aluminum-coated box. A syringe pump was used to facilitate the flow of solution through the microreactor. The optimal yield of the product was obtained by a 4-5 second reaction time. Thus, the flowrate was maintained between 4 to 6 μL/min. After UV exposure, about 10 μL of the solution was placed into a pulled nanoESI borosilicate capillary and used for mass spectrometric analysis.
2.4. Mass Spectrometry.
Standard solutions were analyzed on a TripleTOF 5600 triple quadrupole/time of flight instrument (Sciex, Concord, ON, Canada) and the polar lipid extract was analyzed on a QTRAP 4000 system (Sciex, Concord, ON, Canada) both modified to allow for mutual storage of cations and anions, as reported.33 Spray voltage was held at 1300 V for all experiments. Precursor ion scans (PIS) and neutral loss scans (NLS) (QTRAP) were obtained for the mass range 700 Da to 900 Da with a collision energy of 35 eV. PIS at m/z 184, 199, and 202 profiled unmodified PC, tmPC, and tmPE, respectively, while NLS of 141 Da profiled unmodified PE.
The PDPA anions ([PDPA − 2H]2−) were mass selected with Q1 and injected for 0.2 s into the q2 collision cell. The modified lipid cations ([PBtmGP]+) subsequently followed the same procedure joining the anions in q2 where both ion populations were mutually stored for 0.5 s. LMCO for q2 to Q3 transfer was set as m/z 600 or 700, depending on the mass of the species of interest, to preclude lower mass ions. A q-value of 0.2 was used to perform single frequency resonance excitation of all generations of ions produced throughout the experiments.
3. RESULTS AND DISCUSSION
3.1. Structural characterization of synthetic GPs.
Ionization enhancement of GPs is an attractive feature for lipidomics because it allows for the detection of less abundant species and improvement in identifying species that require protonation. The Smith group published methods of enhancing sensitivity of PE analysis via methylation using diazomethane and 13C-labeled diazomethane, TrEnDi and 13C-TrEnDi, respectively.7,8 In both methods, functional groups with pKa values 11 or less will be readily permethylated and, in the case of PE, the primary amine and phosphate functional groups become a quaternary ammonium and phosphoester, respectively. The resulting tetramethylated species contains a fixed charge and does not have signal splitting due to cationization competition between H+, Na+, and K+. These new features combine to give a sensitivity gain of about 11-fold compared to unmodified PE species. However, TrEnDi using unlabeled diazomethane on a complex lipid matrix will convert all PEs to PCs, which precludes the distinction of these lipid classes. 13C-TrEnDi modification of GPs (tmGPs) uses labeled diazomethane to differentiate modified PEs and from the corresponding PCs by a mass difference of 3 Da, due to a difference of three labeled methyl groups installed on PE, in contrast with the corresponding PC lipid (i.e. PC 36:2 vs PE 36:2).
Structural characterization beyond subclass is not readily achieved using 13C-TrEnDi modification alone. Positive ion mode fragmentation leading to the losses of neutral ketenes that are normally in low abundance for unmodified species are absent using 13C-TrEnDi. The phosphotriester formed during the modification prevents deprotonation and diminishes negative ion mode ionization using an anion adduct. Charge inversion of tmGP cations via gas phase ion/ion reactions has been described as a means of determining fatty acyl chain information in negative ion mode.11 The modified lipid is introduced to the mass spectrometer and mutually stored with a dianion, doubly-deprotonated 1,4-phenylenedipropionic acid (PDPA), which is also used in this study. The ion/ion reaction results in an electrostatically bound complex, with an overall negative charge. After charge inversion of the tmGP, sequential CID steps yield the product ions corresponding to the sum composition of the fatty acyl chains. The combined methods, TrEnDi and charge inversion, show increased PE coverage in a complex sample as well as fatty acid composition. However, location of alkenes in unsaturated GP fatty acyl chains is not forthcoming using the above approach. Structural characterization of lipid species is important for determining their biophysical properties,34 thus probing C=C location is of great interest. Recently, the PB reaction was used to localize the C=C within charge inverted unsaturated PEs and PCs.31 The PB reaction is a classic [2+2] photocycloaddition that forms a bond between an olefin and a carbonyl compound. This reaction is utilized for lipid structure characterization by selectively modifying an alkene with photoactivated acetone to give a four membered oxetane ring, which, upon CID, decomposes to dimethyl-alkene and aldehyde (Scheme 1). Both dimethyl-alkene and aldehyde product ions can be used as diagnostic ions for double bond localization based on a neutral loss when compared to the original m/z of [PBtmFA]−. Combining gas-phase charge inversion with the PB reaction allowed for specific structural elucidation of unsaturated GPs. The method is efficient at characterizing PC; however, PEs ionization in mixtures can be suppressed in the positive ion mode leading to many species being left unidentified. Herein, we report the combination of in-solution modifications followed by charge inversion for sensitive and specific characterization of PEs and PCs with a focus on 13C-TrEnDi modified PE species.
Scheme 1.
Sequence of events for lipid structure identification.
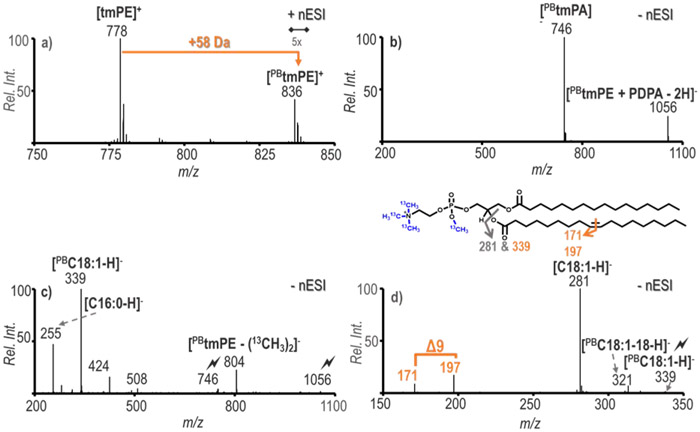
13C-TrEnDi modification was applied to PE 16:0/18:1(9Z) for proof of concept experiments. The PB reaction was facilitated by an offline flow microreactor, which had a product yield of approximately 15%.31 The reaction yield was calculated by normalizing the relative ion intensity of the PB product to that of the species before the photo-reaction. Figure 1a displays the PB reaction spectrum for the 13C-TrEnDi modified PE. The tmPE species appeared at m/z 778 and the PB reaction yielded product ions with a neutral gain of 58 Da (m/z 836). The PB reaction was also applied to tmPC (m/z 775), which yielded a product at m/z 833 (PBtmPC, Figure S1). The results provided in Figure S1 show that PC lipid species also benefit from this derivatization and charge inversion strategy. The [PBtmPE]+ ion was isolated and activated via ion trap CID and shows an exclusive product ion at m/z 202 [(13CH3)4C2H2NO4P]+, confirming the PE headgroup loss, (Figure S2). Product ions from fragmentation at the oxetane ring for positive ion mode C=C location were not present; therefore, all structural information was collected post charge inversion.
Figure 1.
Combination of PB reaction and charge inversion of tmPE 16:/18:1(9Z). a) The PB reaction spectrum of 5 μM tmPE 16:0/18:1(9Z) after 5 seconds of UV exposure, b) Gas-phase ion/ion reaction of [PBtmPE]+ and [PDPA − 2H ]2−, c) Ion trap CID of [PBtmPE + PDPA − 2H]− followed by ion trap CID of [PBtmPA]−, d) Ion trap CID of the PB product of C18:1(9Z) verifying the C=C position of PC 16:0/18:1(Δ9). CID of a target ion is depicted by a lightning bolt (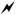 ).
).
The in-solution modified PE cation, [PBtmPE]+, was subjected to ionization via direct nESI and mutually stored in q2 with doubly deprotonated PDPA, [PDPA − 2H]2−. The mutual storage ion/ion reaction resulted in two major products, a complex formed between the two ions at m/z 1056 and an anionic 13C-methylated phosphatidic acid ([tmPA]−) at m/z 746, as shown in Figure 1b. The tmPA formation was due to the loss of (13CH3)3NCH2CH2 from the phosphate headgroup of the tmPE species leaving a 13C-methylated phosphatidic acid. These results are similar to what was observed for tmPE without PB reaction, further suggesting that the addition of the acetone does not affect the overall pathway for gas-phase charge inversion. The ion trap CID of the complex [PBtmPE + PDPA − 2H]− resulted in the increase of [tmPA]− and also the minor loss of two 13C-methyl groups ([PBtmPE - (13CH3)2]− (Figure S3). The sequential fragmentation of the precursor at m/z 746 generates the spectrum in Figure 1c. As detailed in Figure 1b, abundant product ions corresponding to unmodified C16:0 and PB reacted C18:1 at m/z 255 and 339, respectively, are obtained. Also present in lower abundance are product ions from the neutral ketene losses (NL ketene) of both acyl chains at m/z 424 and m/z 508. The relatively more abundant NL ketene loss of PBC18:1 at m/z 424 corresponds with the C18:1 being at the sn2 position relative to the C16:0 at the sn1 position. Ultimately, the series of experiments verified that the method can be used for determining the species as PE 16:0/18:1. To further probe the structure of the tmPE, fragmentation of the ion population at m/z 339 was carried out as shown in Figure 1d. Diagnostic ions at m/z 171 and m/z 197, arising from the respective NL of 168 Da and 142 Da from [PBtmPA]−, can be readily identified by the difference of 26 Da, which indicates the location of C=C to be Δ9 in the synthetic PE 16:0/18:1 standard.
Analysis of PC 16:0/18:1 was conducted in a similar fashion, yielding comparable results (Figure S4). The primary difference between PC and PE analysis is the 3 Da mass differences in the products due to the PC being 13C-TrEnDi modified only at the phosphate group whereas the PE is modified with four 13C-methyl groups (one on the phosphate and 3 on the primary amine). Based on these results, the method summarized in Scheme 1, which combines 13C-TrEnDi and PB modifications with ion/ion charge inversion, is demonstrated to be capable of simultaneously realizing the advantages of all three measures for the structural characterization of GP lipids.
3.2. Application of in-solution modification and gas-phase charge inversion.
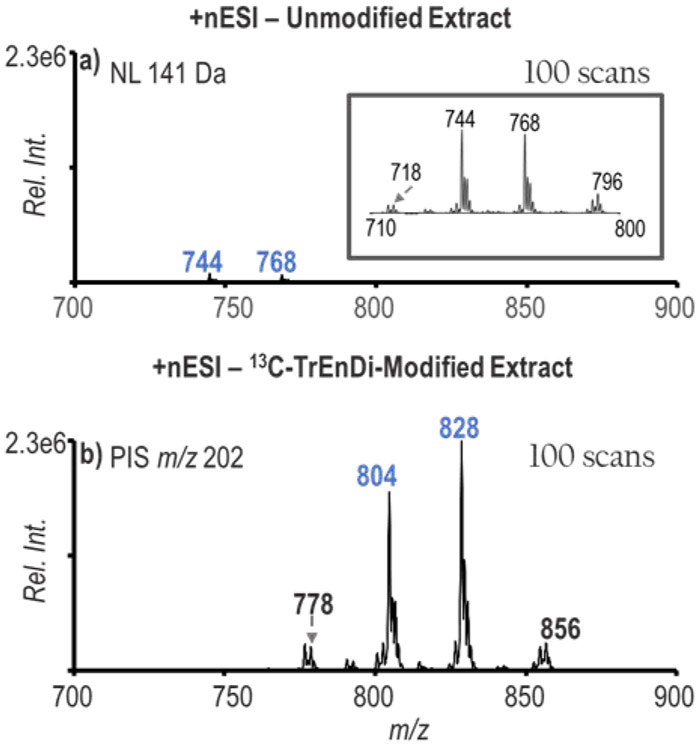
The PB reaction paired with charge inversion via ion/ion reactions is a method that provides sensitive and selective characterization, particularly for PCs due to their inherently high ionization yields in the positive mode. By improving ionization efficiency of PEs, the number of identified PE species in the sample can theoretically increase. A NLS for 141 Da for detection of PEs in unmodified bovine liver polar lipid extract is shown in Figure 2a with the most abundant ions in the spectrum labelled as m/z 744 (PE 36:2) and m/z 768 (PE 38:4). 13C-TrEnDi-modified bovine liver extract was also analyzed for PEs using a PIS of m/z 202 as shown in Figure 2b. The findings were consistent with 13C-TrEnDi studies in the past that show an increase in species identified with greater than 10-fold signal increase obtained with the modification. By comparing an equivalent number of scans, Figure 2 shows the potential for enhanced identification of PEs using a combination of derivatization and charge inversion.
Figure 2.
Bovine liver polar lipid extract (88 μg/mL) mass spectra for PE detection. a) Neutral loss scan for 141 of unmodified extract. Inset: Zoom-in of spectrum from m/z 710-800. b) Precursor ion scan for 202 of the modified extract.
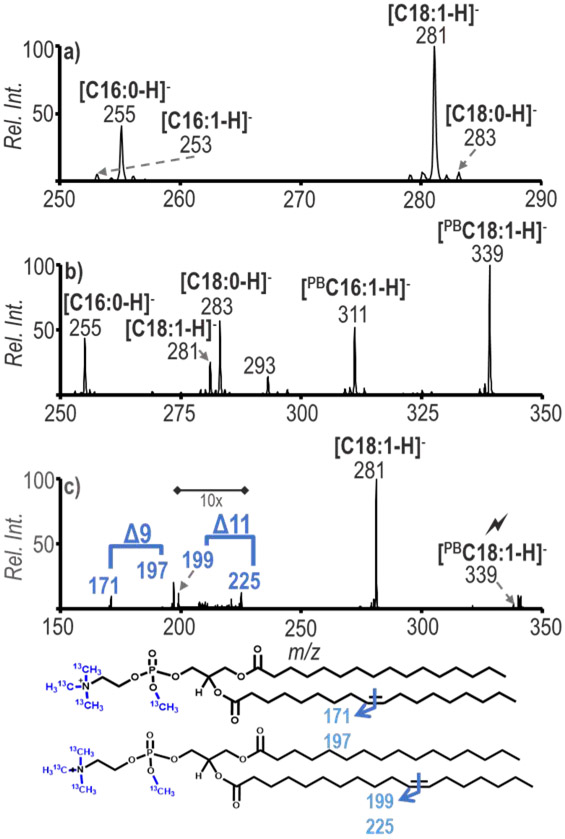
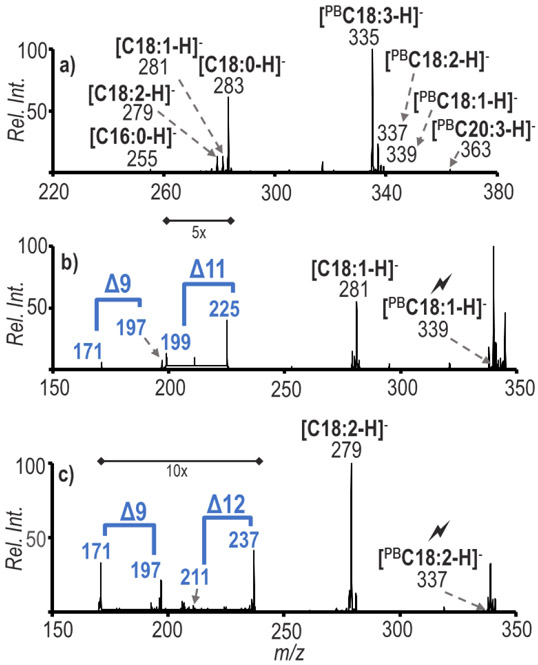
Bovine liver polar lipid extract was used to illustrate the utility of these combined methods for a complex mixture derived from a biological source. A primary goal for undertaking this work was to determine structural information for GPs present in biological extracts. Before the PB reaction was applied in the positive ion mode, the ion population at m/z 778 (tmPE 34:1) was isolated and subsequently transformed in the gas-phase via the ion/ion charge inversion chemistry to generate a [tmPE + PDPA − 2H]− complex. CID of the [tmPE +PDPA − 2H]− ion followed by CID of [tmPA]− generated anions representative of the fatty acyl chains present in the selected lipid, as demonstrated in Figure 3a. After the sequential steps of CID just described, fragmentation products were formed at m/z 253, 255, 281 and 283, which correspond to fatty acyl chains C16:1, C16:0, C18:1 and C18:0, respectively. The appearance of these peaks indicates the presence of the isomers PE 16:0_18:1 and PE 18:0_16:1 in the ion population at m/z 778. The PB reaction of the sample was conducted producing a post-reaction product for ions at m/z 778, 58 Da greater in mass at m/z 836, which, following the analogous set of experiments described above, resulted in the fatty acyl chain ions shown in Figure 3b. The fatty acyl chains present in the spectrum are consistent with those of Figure 3a, which also suggests the presence of PE 16:0_18:1 and PE 18:0_16:1 after the addition of a 58 Da acetone modification to the unsaturated fatty acyl species. The abundant peak at m/z 339 represents [PB18:1 - H]−. The [PB18:1 - H]− ion was subjected to ion trap CID to generate further structural information and the resulting spectrum is shown in Figure 3c. The major fragment ion at m/z 281 arises from the loss of acetone. The diagnostic ions at m/z 171/197 and 199/225 correspond to double bonds being present at the Δ9 and Δ11 positions of C18:1. The fragmentation of [PB16:1 - H]− ions produced diagnostic ions at 171/197 corresponding to the C=C position at the Δ9 position and identifies PE 18:0_16:1(Δ9) (Figure S5). The data show that the methods are capable of being combined for structural characterization down to the C=C location of lipids in a biological sample and allows for the distinction between C=C positional isomers.
Figure 3.
a) Charge inversion of positive ion mode m/z 778 followed by multiple steps of ion trap CID to obtain fatty acyl chain diagnostic ions, b) Charge inversion of positive ion mode m/z 836 (PB product of m/z 778) to obtain the PB reacted fatty acyl chain ions, c) Sequential CID spectrum of the ion population at m/z 339 from b) for C=C localization. ID: PE 16:0_18:1(Δ9) and PE 16:0_18:1 (Δ11)
3.3. Specific characterization of unsaturated fatty acyl chain isomers.
The structural complexity of GPs can include unsaturation on both of the fatty acyl chains at the sn1 and sn2 glycerol positions. Use of the PB reaction alone in the positive ion mode without recourse to the charge inversion MSn process can lead to ambiguities in interpretation regarding C=C bond location due to overlap in diagnostic ions from different fatty acyl chains.28 Charge inversion of the PB reacted species followed by generation and isolation of each fatty acyl chain allows for their independent characterization thereby avoiding this ambiguity.
An example of species present at the lower end of our dynamic range (which is 1.7) without 13C-TrEnDi modification was the subclass PE 36:3 (m/z 802 after 13C-TrEnDi modification). In previous work, PE 36:3 was not structurally characterized because of its low abundance.31 The tmPE 36:3 ions at m/z 802 were isolated, charge inverted and subjected to multiple steps of CID thereby generating the fatty acyl chains C16:0, C18:0, C18:1, C18:2, C18:3, and C20:3 (Figure S6). The PE 36:3 species were identified as PE 18:0_18:3, PE 18:1_18:2, and PE 16:0_20:3, exemplifying the structural diversity of the lipidome, particularly regarding the presence of GP species including two unsaturated fatty acyl chains. The sample underwent photochemical reaction and was subjected to ion/ion reaction followed by sequential CID steps resulting in Figure 4a showing that each unsaturated chain was modified. By fragmenting each individual chain separately, specific fatty acyl chain C=C localization can be achieved. As shown in Figure 4b, the least abundant chain [PBC18:1-H]− was probed and shows that there were C=C isomers present at Δ9 and Δ11 positions (i.e., isomers PE 18:1 (Δ9)_18:2 and PE 18:1 (Δ11)_18:2). Figure 4c shows results for the analogous experiments performed on [PBC18:2-H]− where the abundant fragment is the loss of the acetone (m/z 279) and there exist two sets of diagnostic ions that identify the species as C18:2(Δ9, Δ12). Identifying fatty acyl chains permits in-depth structural elucidation, as PE 36:3 can further be assigned as PE 18:1(Δ9)_18:2(Δ9, Δ12) and PE 18:1(Δ11)_18:2(Δ9, Δ12). The improved ionization yield resulting from 13C-TrEnDi and the structural information made accessible via PB modification with ion/ion charge inversion allowed for the characterization of these heretofore unidentified components in bovine liver polar lipid extract.
Figure 4.
a) Charge inversion of positive ion mode ions at m/z 860 (the PB product of 802 from Figure 2b) to obtain the PB reacted fatty acyl chains, b) Sequential CID spectrum of the ion population at m/z 339 from a) for C=C localization, c) Sequential CID spectrum of ions at m/z 337 from a) for C=C localization. ID: PE 18:1(Δ9)_18:2(Δ9, Δ12), PE 18:1(Δ11)_18:2(Δ9, Δ12)
3.4. Determining C=C location isomers for polyunsaturated fatty acyl chains.
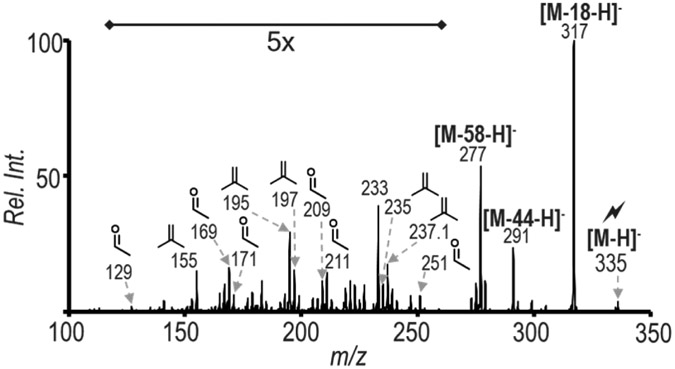
In 2017, Murphy and co-workers applied the PB reaction to polyunsaturated fatty acids and conducted subsequent MS/MS analysis in the negative ion mode.30 Novel diagnostic ions were found after fragmentation, permitting C=C location distinction for polyunsaturated fatty acids possessing 18 or 20 carbons and three to four degrees of unsaturation. We have used insights provided in that work for interpreting results from the workflow described herein for polyunsaturated species found in the complex lipid sample.
The fragment ion at m/z 335 in Figure 4a was subjected to ion trap CID to determine the ability to distinguish the location of unsaturation along C18:3. The resulting spectrum is provided in Figure 5 and shows results matching Murphy’s findings. The diagnostic ions at 129/155, 169/195, 209/235 indicate the presence of C18:3(Δ6, Δ9, Δ12), whereas the ions at m/z 171/197, 211/237, and 251/277 show the presence of C18:3(Δ9, Δ12, Δ15). The combination of TrEnDi/PB modification and charge inversion clearly shows that PE 36:3 bovine liver exists as a minimum of 4 species {PE 18:0_18:3(Δ6, Δ9, Δ12), PE 18:0_18:3(Δ9, Δ12, Δ15), PE 18:1(Δ9)_18:2(Δ9, Δ12), and PE 18:1(Δ11)_18:2(Δ9, Δ12)}. When the sample was analyzed directly after PB reaction without combining the other methods, 6 PE species could be characterized at the level of C=C bond position.31 The characterization of 18 PE species, listed in Table 1, was achieved herein, which shows a 3-fold increase in the number of species that could be characterized at the level of C=C bond location. When analyzing complex mixtures, typical MS/MS spectra used for characterizing the structures of GP anions may contain information for multiple isobaric species from different GP classes. Charge inversion of the modified species improved specificity because cations specific to GP class could be mass selected and probed for confident fatty acyl chain analysis. The development of data collection and analysis software would help move this method toward large-scale lipidomic analysis.
Figure 5.
Charge inversion of positive ion mode m/z 860 (the PB product of 802 from Figure 2b) followed by sequential CID spectrum of the ion population m/z 335 from Figure 4a in order to localize the C=C. ID: PE 18:0_18:3(Δ6, Δ9, Δ12), PE 18:0_18:3(Δ9, Δ12, Δ15).
Table 1:
Unsaturated GPs in bovine liver polar lipid extract characterized to C=C bond location level
| m/z | PB product |
PB Fatty Acyl Diagnostic Ions |
PB Reaction Diagnostics | Identified Species |
|---|---|---|---|---|
| 747 | 805 | 225; 339 | 171, 197 | PC 14:0_18:1(Δ9) |
| 747 | 805 | 225; 339 | 199, 225 | PC 14:0_18:1(Δ11) |
| 747 | 805 | 255; 311 | 171, 197 | PC 16:0_16:1(Δ9) |
| 773 | 831 | 255; 337 | 171, 197 ; 211, 237 | PC 16:0_18:2(Δ9,Δ12) |
| 775 | 833 | 255; 339 | 171, 197 | PC 16:0_18:1(Δ9) |
| 775 | 833 | 255; 339 | 199, 225 | PC 16:0_18:2(Δ11) |
| 775 | 833 | 283; 311 | 171, 197 | PC 16:1(Δ9)_18:0 |
| 801 | 859 | 283; 337 | 171, 197 ; 211, 237 | PC 18:0_18:2(Δ9, Δ12) |
| 801 | 859 | 339 | 171, 197 | PC 18:1(Δ9)_18:1(Δ9) |
| 801 | 859 | 339 | 199, 225 | PC 18:1(Δ11)_18:1(Δ11) |
| 801 | 859 | 255; 365 | 199, 225; 239, 265 | PC 16:0_20:2(Δ11, Δ14) |
| 803 | 861 | 283; 339 | 171, 197 | PC 18:0_18:1(Δ9) |
| 803 | 861 | 283; 339 | 199, 225 | PC 18:0_18:1(Δ11) |
| m/z | PB product |
PBFatty Acyl Diagnostic Ions |
PB Reaction Diagnostics | Identified Species |
| 764 | 822 | 339; 241 | 171, 197 | PE 18:1(Δ9)_15:0 |
| 774 | 832 | 255; 335 | 171, 197; 211, 237; 251, 277 | PE 16:0 18:3(Δ9, Δ12, Δ15) |
| 774 | 832 | 255; 335 | 129, 155; 169, 195; 209, 235 | PE 16:0_18:3(Δ6, Δ9, Δ12) |
| 776 | 834 | 255 ; 337 | 171, 197 ; 211, 237 | PE 16:0_18:2(Δ9, Δ12) |
| 778 | 836 | 255; 339 | 171, 197 | PE 16:0_18:1(Δ9) |
| 778 | 836 | 255; 339 | 199, 225 | PE 16:0_18:1(Δ11) |
| 778 | 836 | 253; 283 | 171, 197 | PE 16:1(Δ9)_18:0 |
| 790 | 848 | 337; 269 | 171, 197; 211, 237 | PE 17:0_18:2(Δ9, Δ12) |
| 792 | 850 | 269; 339 | 171, 197 | PE 17:0_18:1(Δ9) |
| 802 | 860 | 339; 337 | 171, 197; 171, 197; 211, 237 | PE 18:1(Δ9)_18:2(Δ9, Δ12) |
| 802 | 860 | 339; 337 | 199, 225; 171, 197; 211, 237 | PE 18:1(Δ11)_18:2(Δ9, Δ12) |
| 802 | 860 | 335; 283 | 171, 197; 211, 237; 251, 277 | PE 18:0_18:3(Δ9, Δ12, Δ15) |
| 802 | 860 | 335; 283 | 129, 155; 169, 195; 209, 235 | PE 18:0_18:3(Δ6, Δ9, Δ12) |
| 804 | 862 | 337; 283 | 171, 197 ; 211, 237 | PE 18:0_18:2(Δ9, Δ12) |
| 804 | 862 | 339; 281 | 171, 197 | PE 18:1(Δ9)_18:1(Δ9) |
| 804 | 862 | 339; 281 | 199, 225 | PE 18:1(Δ11)_18:1(Δ11) |
| 806 | 864 | 339; 283 | 171, 197 | PE 18:0_18:1(Δ9) |
| 806 | 864 | 339; 283 | 199, 225 | PE 18:0_18:1(Δ11) |
4. CONCLUSION
This work demonstrates the combination of three techniques to improve the sensitivity and specificity in characterizing the structures of PEs and PCs using a single ionization polarity and solvent composition. 13C-TrEnDi provides high and relatively uniform signal responses for multiple GP classes in the positive ion mode and allows for lipid class identification. Gas-phase charge inversion of TrEnDi modified lipid cations allows for the generation of structurally informative fatty acyl anions for subsequent tandem mass spectrometry. The PB reaction allows for the location of double bonds within the fatty acyl chain. This novel combination of the three techniques enables detailed structural characterization for lipid species that may not ionize efficiently in shotgun lipidomics (e.g., PEs) while retaining the ability to generating detailed fatty acid information (i.e., composition and double bond location). Consistent with previous TrEnDi studies, a roughly 10-fold enhancement in ionization yield for PE lipids was noted in this work. A three-fold increase in the number of PE lipids that could be characterized to the level of C=C bond location was noted relative to the use of PB derivatization and ionization in the negative mode for a polar lipid extract.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (GM R01-118484 and GM R37-45372), the Natural Sciences and Engineering Research Council of Canada, the Canadian Foundation for Innovation, the Ontario Research Fund and Sciex.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Additional information is provided on the positive ion mode analysis of [PBtmGPs]+, charge inversion [PBtmPE]+ with a sequential CID step, charge inversion [PBtmPC]+ with a sequential CID steps for C=C localization, MS/MS spectrum of m/z 836 (the PB product of m/z 778) for the localization of 16:1(Δ9), and fatty acyl chain determination of the ions at m/z 802.
REFERENCES
- (1).Han X; Gross RW Shotgun Lipidomics: Electrospray Ionization Mass Spectrometric Analysis and Quantitation of Cellular Lipidomes Directly from Crude Extracts of Biological Samples. Mass Spectrom. Rev 2005, 24 (3), 367–412. 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- (2).Han X; Gross RW Shotgun Lipidomics: Multidimensional MS Analysis of Cellular Lipidomes. Expert Rev. Proteomics 2005, 2 (2), 253–264. 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- (3).Han X Lipidomics: Comprehensive Mass Spectrometry of Lipids; Wiley Series on Mass Spectrometry; John Wiley & Sons, 2016. [Google Scholar]
- (4).Quehenberger O; Armando AM; Brown AH; Milne SB; Myers DS; Merrill AH; Bandyopadhyay S; Jones KN; Kelly S; Shaner RL; et al. Lipidomics Reveals a Remarkable Diversity of Lipids in Human Plasma,. J. Lipid Res 2010, 51 (11), 3299–3305. 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fhaner CJ; Liu S; Ji H; Simpson RJ; Reid GE Comprehensive Lipidome Profiling of Isogenic Primary and Metastatic Colon Adenocarcinoma Cell Lines. Anal. Chem 2012, 84 (21), 8917–8926. 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ryan E; Reid GE Chemical Derivatization and Ultrahigh Resolution and Accurate Mass Spectrometry Strategies for “Shotgun” Lipidome Analysis. Acc. Chem. Res 2016, 49 (9), 1596–1604. 10.1021/acs.accounts.6b00030. [DOI] [PubMed] [Google Scholar]
- (7).Wasslen KV; Canez CR; Lee H; Manthorpe JM; Smith JC Trimethylation Enhancement Using Diazomethane (TrEnDi) II: Rapid In-Solution Concomitant Quaternization of Glycerophospholipid Amino Groups and Methylation of Phosphate Groups via Reaction with Diazomethane Significantly Enhances Sensitivity in Mass Spectrometry Analyses via a Fixed, Permanent Positive Charge. Anal. Chem 2014, 86 (19), 9523–9532. 10.1021/ac501588y. [DOI] [PubMed] [Google Scholar]
- (8).Canez CR; Shields SWJ; Bugno M; Wasslen KV; Weinert HP; Willmore WG; Manthorpe JM; Smith JC Trimethylation Enhancement Using 13 C-Diazomethane ( 13 C-TrEnDi): Increased Sensitivity and Selectivity of Phosphatidylethanolamine, Phosphatidylcholine, and Phosphatidylserine Lipids Derived from Complex Biological Samples. Anal. Chem 2016, 88 (14), 6996–7004. 10.1021/acs.analchem.5b04524. [DOI] [PubMed] [Google Scholar]
- (9).Stutzman JR; Blanksby SJ; McLuckey SA Gas-Phase Transformation of Phosphatidylcholine Cations to Structurally Informative Anions via Ion/Ion Chemistry. Anal. Chem 2013, 85 (7), 3752–3757. 10.1021/ac400190k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rojas-Betancourt S; Stutzman JR; Londry FA; Blanksby SJ; McLuckey SA Gas-Phase Chemical Separation of Phosphatidylcholine and Phosphatidylethanolamine Cations via Charge Inversion Ion/Ion Chemistry. Anal. Chem 2015, 87 (22), 11255–11262. 10.1021/acs.analchem.5b02243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Betancourt SK; Canez CR; Shields SWJ; Manthorpe JM; Smith JC; McLuckey SA Trimethylation Enhancement Using 13 C-Diazomethane: Gas-Phase Charge Inversion of Modified Phospholipid Cations for Enhanced Structural Characterization. Anal. Chem 2017, 89 (17), 9452–9458. 10.1021/acs.analchem.7b02271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tomer KB; Crow FW; Gross ML Location of Double-Bond Position in Unsaturated Fatty Acids by Negative Ion MS/MS. J. Am. Chem. Soc 1983, 105 (16), 5487–5488. 10.1021/ja00354a055. [DOI] [Google Scholar]
- (13).Klein DR; Brodbelt JS Structural Characterization of Phosphatidylcholines Using 193 Nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2017, 89 (3), 1516–1522. 10.1021/acs.analchem.6b03353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pham HT; Maccarone AT; Thomas MC; Campbell JL; Mitchell TW; Blanksby SJ Structural Characterization of Glycerophospholipids by Combinations of Ozone- and Collision-Induced Dissociation Mass Spectrometry: The next Step towards “Top-down” Lipidomics. The Analyst 2014, 139 (1), 204–214. 10.1039/C3AN01712E. [DOI] [PubMed] [Google Scholar]
- (15).Brown SHJ; Mitchell TW; Blanksby SJ Analysis of Unsaturated Lipids by Ozone-Induced Dissociation. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids 2011, 1811 (11), 807–817. 10.1016/j.bbalip.2011.04.015. [DOI] [PubMed] [Google Scholar]
- (16).Thomas MC; Mitchell TW; Harman DG; Deeley JM; Nealon JR; Blanksby SJ Ozone-Induced Dissociation: Elucidation of Double Bond Position within Mass-Selected Lipid Ions. Anal. Chem 2008, 80 (1), 303–311. 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- (17).Pham HT; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid Differentiation of Isomeric Lipids by Photodissociation Mass Spectrometry of Fatty Acid Derivatives: Photodissociation of Derivatized Fatty Acids. Rapid Commun. Mass Spectrom 2013, 27 (7), 805–815. 10.1002/rcm.6503. [DOI] [PubMed] [Google Scholar]
- (18).Pham HT; Julian RR Radical Delivery and Fragmentation for Structural Analysis of Glycerophospholipids. Int. J. Mass Spectrom 2014, 370, 58–65. 10.1016/j.ijms.2014.06.022. [DOI] [Google Scholar]
- (19).Campbell JL; Baba T Near-Complete Structural Characterization of Phosphatidylcholines Using Electron Impact Excitation of Ions from Organics. Anal. Chem 2015, 87 (11), 5837–5845. 10.1021/acs.analchem.5b01460. [DOI] [PubMed] [Google Scholar]
- (20).Baba T; Campbell JL; Le Blanc JCY; Baker PRS Structural Identification of Triacylglycerol Isomers Using Electron Impact Excitation of Ions from Organics (EIEIO). J. Lipid Res 2016, 57 (11), 2015–2027. 10.1194/jlr.M070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Deimler RE; Sander M; Jackson GP Radical-Induced Fragmentation of Phospholipid Cations Using Metastable Atom-Activated Dissociation Mass Spectrometry (MAD-MS). Int. J. Mass Spectrom 2015, 390, 178–186. 10.1016/j.ijms.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li P; Hoffmann WD; Jackson GP Multistage Mass Spectrometry of Phospholipids Using Collision-Induced Dissociation (CID) and Metastable Atom-Activated Dissociation (MAD). Int. J. Mass Spectrom 2016, 403, 1–7. 10.1016/j.ijms.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Stinson CA; Zhang W; Xia Y UV Lamp as a Facile Ozone Source for Structural Analysis of Unsaturated Lipids Via Electrospray Ionization-Mass Spectrometry. J. Am. Soc. Mass Spectrom 2018, 29 (3), 481–489. 10.1007/s13361-017-1861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Harris RA; May JC; Stinson CA; Xia Y; McLean JA Determining Double Bond Position in Lipids Using Online Ozonolysis Coupled to Liquid Chromatography and Ion Mobility-Mass Spectrometry. Anal. Chem 2018, 90 (3), 1915–1924. 10.1021/acs.analchem.7b04007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cao W; Ma X; Li Z; Zhou X; Ouyang Z Locating Carbon–Carbon Double Bonds in Unsaturated Phospholipids by Epoxidation Reaction and Tandem Mass Spectrometry. Anal. Chem 2018, 90(17), 10286–10292. 10.1021/acs.analchem.8b02021. [DOI] [PubMed] [Google Scholar]
- (26).Ma X; Xia Y Pinpointing Double Bonds in Lipids by Paternò-Büchi Reactions and Mass Spectrometry. Angew. Chem. Int. Ed 2014, 53(10), 2592–2596. 10.1002/anie.201310699. [DOI] [PubMed] [Google Scholar]
- (27).Stinson CA; Xia Y A Method of Coupling the Paternò-Büchi Reaction with Direct Infusion ESI-MS/MS for Locating the C=C Bond in Glycerophospholipids. The Analyst 2016, 141 (12), 3696–3704. 10.1039/C6AN00015K. [DOI] [PubMed] [Google Scholar]
- (28).Ma X; Chong L; Tian R; Shi R; Hu TY; Ouyang Z; Xia Y Identification and Quantitation of Lipid C=C Location Isomers: A Shotgun Lipidomics Approach Enabled by Photochemical Reaction. Proc. Natl. Acad. Sci 2016, 113 (10), 2573–2578. 10.1073/pnas.1523356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ren J; Franklin ET; Xia Y Uncovering Structural Diversity of Unsaturated Fatty Acyls in Cholesteryl Esters via Photochemical Reaction and Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom 2017, 28(7), 1432–1441. 10.1007/s13361-017-1639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Murphy RC; Okuno T; Johnson CA; Barkley RM Determination of Double Bond Positions in Polyunsaturated Fatty Acids Using the Photochemical Paternò-Büchi Reaction with Acetone and Tandem Mass Spectrometry. Anal. Chem 2017, 89 (16), 8545–8553. 10.1021/acs.analchem.7b02375. [DOI] [PubMed] [Google Scholar]
- (31).Franklin ET; Betancourt SK; Randolph CE; McLuckey SA; Xia Y In-Depth Structural Characterization of Phospholipids by Pairing Solution Photochemical Reaction with Charge Inversion Ion/Ion Chemistry. Anal. Bioanal. Chem 2019, 411 (19), 4739–4749. 10.1007/s00216-018-1537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Shields SWJ; Manthorpe JM Efficient, Scalable and Economical Preparation of Tris(Deuterium)- and 13 C-Labelled N -Methyl- N -Nitroso- p -Toluenesulfonamide (Diazald®) and Their Conversion to Labelled Diazomethane: Efficient Preparation of Bis(Deuterium)- and 13 C-Labelled Diazomethane. J. Label. Compd. Radiopharm 2014, 57 (12), 674–679. 10.1002/jlcr.3231. [DOI] [PubMed] [Google Scholar]
- (33).Xia Y; Chrisman PA; Erickson DE; Liu J; Liang X; Londry FA; Yang MJ; McLuckey SA Implementation of Ion/Ion Reactions in a Quadrupole/Time-of-Flight Tandem Mass Spectrometer. Anal. Chem 2006, 78 (12), 4146–4154. 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dowhan W Molecular Genetic Approaches to Defining Lipid Function. J. Lipid Res 2009, 50 (Supplement), S305–S310. 10.1194/jlr.R800041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.