Abstract
Background
Heat waves can critically influence maize crop yields. Plant heat shock transcription factors (HSFs) play a key regulating role in the heat shock (HS) signal transduction pathway.
Method
In this study, a homologous cloning method was used to clone HSF gene ZmHsf01 (accession number: MK888854) from young maize leaves. The transcript levels of ZmHsf01 were detected using qRT-PCR in different tissues and treated by HS, abscisic acid (ABA), hydrogen peroxide (H2O2), respectively, and the functions of gene ZmHsf01 were studied in transgenic yeast and Arabidopsis.
Result
ZmHsf01 had a coding sequence (CDS) of 1176 bp and encoded a protein consisting of 391 amino acids. The homologous analysis results showed that ZmHsf01 and SbHsfA2d had the highest protein sequence identities. Subcellular localization experiments confirmed that ZmHsf01 was localized in the nucleus. ZmHsf01 was expressed in many maize tissues. It was up-regulated by HS, and up-regulated in roots and down-regulated in leaves under ABA and H2O2treatments. ZmHsf01-overexpressing yeast cells showed increased thermotolerance. In Arabidopsis seedlings, ZmHsf01 compensated for the thermotolerance defects of mutant athsfa2, and ZmHsf01-overexpressing lines showed enhanced basal and acquired thermotolerance. When compared to wild type (WT) seedlings, ZmHsf01-overexpressing lines showed higher chlorophyll content and survival rates after HS. Heat shock protein (HSP) gene expression levels were more up-regulated in ZmHsf01-overexpressing Arabidopsis seedlings than WT seedlings. These results suggest that ZmHsf01 plays a vital role in response to HS in plant.
Keywords: Thermotolerance, Heat shock transcription factors, Maize, ABA, Subcellular localization, H2O2, Adversity stresses, Signal transduction pathway, Functional analysis, ZmHsf01
Introduction
High temperature is an abiotic stress factor with a great impact on crop yields and quality. The North China Plain’s typical temperature and monsoonal climate means that agricultural production is often subjected to the frequency and intensity of heat waves (Li et al., 2018; Suchul & Eltahir, 2018). High temperature and drought stress often cause declines in the production and quality of maize, particularly during the early stages of the growth period (Cairns et al., 2012). Controllable and appropriate heat acclimation can induce related gene expression and the synthesis of heat shock proteins (HSPs) and protective enzymes (Nover et al., 1996), so as to help plants obtain thermotolerance and adjust to the even more extreme high temperatures. Studies have shown that heat shock transcription factors (HSFs) have a regulatory role in the thermotolerance-formation process. HSFs can bind to heat shock elements (HSEs) on the promoter regions of HSPs or other related genes and can activate downstream genes to generate a heat shock (HS) response (Nover et al., 2001). Therefore, HSFs have been proved to be important regulators in transcriptional activation, especially in the signal transduction pathway activated by heat and other stresses (Aranda et al., 1999).
Several HSFs have been cloned from different species since the yeast HSF was first isolated in the 1980s (Baniwal et al., 2004; Bharti et al., 2004; Czarnecka-Verner et al., 2004; Kotak et al., 2004; Zhu et al., 2006; Guo et al., 2008; Yang et al., 2016). The first plant HSF was cloned from tomato in 1990 (Yokotani et al., 2008). The HSF multi-gene family is divided into three classes (A, B, and C) based on gene structures, and each class contains different subclasses. The gene number of the HSF family also varies. There is only one HSF gene in the fruit fly and yeast, and vertebrates have at least four HSFs (Nakai, 2016). Plants possess more HSFs than other organism. To date, 21 HSFs in Arabidopsis and 24 HSFs in tomato have been identified (Scharf et al., 2012), and at least 56 HSFs in wheat have been predicted (Xue et al., 2014).
Few functional analyses were reported on plant HSFs, and previous studies were limited to analysis of the A1 and A2 subclasses of Arabidopsis and tomato. Previous studies showed that the tomato HsfA1a is constitutively expressed and that the deduced protein is localized in both nucleus and cytoplasm under normal growth conditions (Scharf et al., 1998). In tomato, HsfA2 and HsfB1 synthesis, which induces HSPs expression, is regulated by HsfA1a for HS resistance (Scharf et al., 1998; Liu et al., 2013). Strong and stable HsfA2 is up-regulated by heat induction, and a considerable HsfA2 accumulation appears during late and recovery periods after heat stress in tomato (Heerklotz, Döring & Bonzelius, 2001). Because of strong cytoplasmic localization signals, the nuclear localization of HsfA2 has to rely on the heterooligomer formed by the bond between HsfA2 and HsfA1 (Heerklotz, Döring & Bonzelius, 2001). In Arabidopsis, AtHsfA2 can be induced by HS. Basal and acquired thermotolerances, as well as the resistance to salt and the osmotic stresses can be enhanced in AtHsfA2-overexpressing Arabidopsis seedlings (Ogawa, Yamaguchi & Nishiuchi, 2007). Basal and acquired thermotolerances and antioxidant capacity are consistently reduced in mutant athsfa2 (Li et al., 2005).
As a key regulator in the response to environmental stresses, AtHsfA2 plays an important role in the regulating response to multiple abiotic stresses including heat (Busch, Wunderlich & Schöffl, 2005; Schramm et al., 2006; Nishizawa et al., 2006; Charng et al., 2007). It can directly bind to HSEs in the promoter region of target genes or interact with AtHsfA1s to form a heterogenic complex and regulate the expression of downstream genes, particularly HSPs (Liu & Charng, 2013; Nishizawa-Yokoi et al., 2011). AtHsfA1s function as key regulators in AtHsfA2 expression (Liu & Charng, 2013). AtHsfA1s and AtHsfA2 have distinct but overlapping functions in response to abiotic stresses (Liu & Charng, 2013; Nishizawa-Yokoi et al., 2011).
In maize, four HSF of A2 subclass, ZmHsf01, ZmHsf04, ZmHsf05, and ZmHsf17, have been identified (Lin et al., 2011). Ectopic overexpression of ZmHsf04 and ZmHsf05 improved the thermotolerance of transgenic Arabidopsis seedlings (Li et al., 2019; Jiang et al., 2018). In our study, we cloned and analyzed ZmHsf01, another member of A2 subclass, and observed the subcellular localization of ZmHsf01 protein, based on these, ZmHsf01 functions in response to HS were detected and discussed further.
Material and Methods
Plant materials and culture conditions
Maize (Zea mays L.) inbred line H21 was used in this study. Healthy seeds were surface-sterilized with 0.1% HgCl2 for 10 min and rinsed repeatedly with double distilled water. The seeds germinated in the dark and were planted in a controlled environment greenhouse at 28 °C with conditions of 16/8 h of day/night (100 µmol m−2 s−1) and 60% relative humidity (RH). Seedlings with two leaves were used for stress experiments and young roots, shoots, and leaves were sampled, respectively. Mature leaves, pollen, and ears were separated during the blooming period. Immature embryos were separated two weeks after pollination. All tissues and organs were frozen in liquid nitrogen for gene expression analysis.
Stress treatment
Uniform maize seedlings with two leaves were selected and subjected to treatment methods described by Li et al., with some modifications (Li et al., 2019). For HS treatment, the Hoagland nutrient solution was pre-heated at 42 °C before immersing into seedlings. Leaves and roots were sampled at treatment times of 0, 10, 20, 30, 40, 50, 60, and 120 min. For abscisic acid (ABA) treatment, the seedlings were treated with Hoagland nutrient solution with a concentration of 200 µM ABA. The treatment times were 0, 2, 4, 6, 12, 24, and 36 h. For H2O2 treatment, the seedlings were treated Hoagland nutrient solution with a final concentration of 10 mM H2O2. The treatment times were 0, 15, 30, 60, 90, 120, and 240 min. For qRT-PCR testing of HSPs, 5-day-old transgenic line and wild type (WT) seedlings were selected and the leaves both WT and overexpressing line were harvested at 2 h after HS treatment at 45 °C for 50 min for the basal thermotolerance and the the following treatment for the acquired thermotolerance: pre-treatment at 37 °C for 60 min, a recovery under the normal conditions for 2 days respectively, and then retreatment at 46 °C for 2 h. We carried out three biological experiments. The samples were collected and quickly frozen in liquid nitrogen.
Gene cloning and sequencing
A total RNA extraction from leaves was conducted using the RNArose Reagent Systems Kit (SBS, Beijing, China). The genomic DNA was digested using DNase I (TaKaRa, Dalian, China) for 30 min at 37 °C. 1 µg total RNA was used for the first standard synthesis of cDNA using a Reverse Transcription Kit (Invitrogen, USA). The quantity of RNA samples was checked using a NanoDrop 2000 (Thermo Scientific, USA).
The primers (forward primer 5′-CGTGGCGAGATGGACCTGATGC-3′ and reverse primer 5′-TTAACGCGATCATCTCTACTTC-3′) were designed to amplify the open reading frame of ZmHsf01. We submitted the full ZmHsf01 coding sequences (CDS) to GenBank under accession number MK888854. High-fidelity enzyme Pyrobest (TaKaRa, Dalian, China) was used for PCR amplification. The PCR reaction system consisted of 1 × Pyrobest buffer, 0.2 mM dNTP mixture, 200 ng first strand cDNA, 0.2 µM forward primer, 0.2 µM reverse primer, and 1.25 units Pyrobest DNA polymerase. The reaction procedure was 30 cycles of: 98 °C for 10 s, 55 °C for 15 s, and 72 °C for 2 min. The PCR products were ligated into T-Vector (TransGen Biotech, Beijing, China) and sequenced by the Shanghai Sangon Biotech Company.
Quantitative real time PCR (qRT-PCR) analyses
The PCR reaction mixtures contained 1 × SYBR Premix Ex TaqII (Takara, Dalian, China), 0.4 µM forward primer, 0.4 µM reverse primer, and 1 µg cDNA for a final volume of 20 µL. We used a 7500 Real-Time PCR system (Applied Biosystems, USA) in this experiment. The reaction procedure was as follows: pre-denaturation at 95 ° C for 10 min, 40 cycles of denaturation at 95 °C for 5 s, and annealing/extension at 60 °C for 1 min. After the reaction, we analyzed the data using the 2−ΔΔCt method. Three biological replicates were performed in each group of experiments. The data was analyzed with Microsoft Excel 2010. For the statistical analyses, each dataset was repeated at least three times. We set the expression level of young roots as 1 during the expression analysis of tissues and organs, and expression levels at 0 min were set as 1 for the expression analysis of different stress treatments. Maize gene β-Actin was used as an endogenous control, and AtActin8 (At1g49240) was used for Arabidopsis. The primers of ZmHsf01 and other relevant genes were listed in Table S1.
Subcellular localization of ZmHsf01 protein in tobacco epidermal cells
Using a ClonExpress II kit (Vazyme, Nanjing, China), we constructed a recombinant vector pCAMBIA1300-ZmHsf01-GFP driven by a CaMV 35S promoter containing the ZmHsf01 CDS amplified by gene-specific primers (forward primer: 5′-GAGAACACGGGGGACTCTAGAATGGACCTGATGCTG-3′; reverse primer: 5′-GCCCTTGCTCACCATGGATCCCTTCGCCGTGGTGTT-3′) and the GFP gene. The constructs were transformed into Agrobacterium tumefaciens EHA105 competent cells by the freeze-thaw method. ZmHsf01-GFP was expressed in the epidermal cells of tobacco leaf using the infiltration method described by Runions, Hawes & Kurup (2007) with EHA105 cells. We raised the tobacco seedlings in a glasshouse (12/12 h of day/night, 150 µmol m - 2 s - 1 50% RH, 19−23 °C temperature) for 72 h, and harvested the leaves and stained them with DAPI (a nuclei-special dye) (10 µg mL−1) for 5 min. After being rinsed three times with physiological saline, the tobacco epidermal cells were observed under a laser-scanning confocal LSM 710 microscope (Zeiss Microsystems, Germany).
Semi quantitative RT-PCR (semi qRT-PCR) assay
The transcription abundance of ZmHsf01 in transgenic Arabidopsis was tested using semi qRT-PCR method. RNA extraction and cDNA synthesis were carried out according to the protocols mentioned above. Based on the coding region sequence of ZmHsf01, we synthesized a pair of primers (forward primer, 5′-GTGACGGTAAAGGAGGAGTGGCCT-3′; reverse primer, 5′-GCCATAGGTGTTCAGCTGGCGGAC-3′). AtActin2 (At3g18780) (forward primers 5′-CAATCGTGTGTGACAATGG-3′ and reverse 5′-AACCCTCGTAGATTGGCA-3′) was used as a loading control.
Construction and transformation of yeast expression vectors and thermotolerance assays
The pYES2 vector (Invitrogen, USA) was used to detect the target protein expression in Saccharomyces cerevisiae. Using the ClonExpress II recombination system (Vazyme, Nanjing, China), we amplified the PCR products of ZmHsf01 CDS using a pair of specific primers (5′-GGGAATATTAAGCTTGGTACCATGGACCTGATGCTGCCG-3′and 5′- TGATGGATATCTGCAGAATTCCTACTTCGCCGTGGTGTT-3′) inserted into the pYES2 vector. The recombinants were transformed into INVSc1 competent yeast cells described by Gietz et al. (1992), and the cells were diluted and plated on a SC/Glu/Ura− agar screening plate at 30 °C. After 2 to 3 days, the positive clones were selected and verified by colony PCR.
For the thermotolerance assays, the positive clones were cultured using a liquid SC/Glu/Ura− medium in a shaking incubator (250 rpm min−1). When the OD600 of cells reached 0.6∼0.7, the cells were diluted to an OD600 of 0.2 with a SC/Glu/Ura− liquid medium, and were cultured by shaking for 2∼3 h. Cells were collected at an OD600 of 0.4∼0.8, eluted twice with sterile water, and serially diluted to OD600 levels of 0.1, 0.05, and 0.01. Separated the two samples into two groups: one group subjected to HS treatment in a 50 °C water bath for 15 min followed by culturing at 30 °C, and the control group with no intervention. 8 µL treated yeast cells was plated on SC/Gal/Ura− agar to be grown at 30 °C. Yeast colony formation was examined and photographed after 2∼3 days.
Plasmid construction and transformation in Arabidopsis
Special primers (forward: 5′- GAGAACACGGGGGACTCTAGAATGGACCTGATGCTG-3′; reverse: 5′- CGATCGGGGAAATTCGAGCTCCTACTTCGCCGTGGTGTT -3′) were designed to amplify the coding sequence of ZmHsf01 by RT-PCR. Using a ClonExpress II kit, we inserted the PCR products into the pCAMBIA1300 vector, digested in advance by Xba I and Sac I and driven by a CaMV 35S promoter. After the construct infected Agrobacterium GV3101, transformed them into the wild Arabidopsis and deletion mutants using the vacuum dipping method. The MS medium, containing 25 µg ml−1 hygromycin, was used to screen the progeny plants until the homozygous seeds were harvested. We used the transgenic T3 homozygous lines to identify thermotolerance.
Thermotolerance assays in Arabidopsis
The sterilized WT, athsfa2, and transgenic line seeds of Arabidopsis thaliana (Ecotype, Columbia) were placed in 1/2 MS solid plates containing 0.8% agar. The plates were then placed in an incubator at 22 °C (during the day) and 18 °C (at night) with conditions of 16/8 h of day/night (100 µmol m−2 s−1). We evaluated the basal thermotolerance using the following protocol: 5-day-old seedlings of WT and three overexpressed lines were subjected to 45 °C HS for 50 min, then recovered under normal growth conditions for 8 days. The acquired thermotolerance was evaluated through following treatment: 5-day-old seedlings of WT and three overexpressed lines were subjected to 37 °C temperatures for 60 min, recovered under normal conditions for 2 h (short-time recovery) and 2 days (long-time recovery), respectively, then re-heated at 46 °C for 60 min, and recovered under normal conditions for 8 days. The complementary thermotolerance was assayed after 5-day-old WT, athsfa2 mutant, and transgenic line seedlings were treated at 44 °C for 70 min and recovered under normal conditions for 8 days. We observed and photographed all phenotypes, measured the survival rates, and measured the chlorophyll content of the leaves (Li et al., 2015a). At least 30 seedlings for each line were analyzed, and all experiments were repeated for three times.
Yeast transcriptional activation analysis
Transcription activation activity assays of ZmHsf01 was performed according to the Y2H system instructions (TaKaRa, Dalian, China). Using the CloneExpress II cloning kit, we created the recombinants with the pGBKT7 vector and the full ZmHsf01 CDS amplified by specific primers (5′-ATGGCCATGGAGGCCGAATTCATGGACCTGATGCTG-3′ an 5′-CCGCTGCAGGTCGACGGATCCCTACTTCGCCGTGGT-3′). We used LiAc and PEG3350 in the yeast transformation assay. The pGBKT7, pGBKT7-53, and pGADT7-T vectors and the pGBKT7-ZmHsf01 fusion vectors were transformed into the yeast cell AH109. Transformed yeast cells of different concentrations were successively cultured in SD/Trp− and SD/Trp−/His−/Ade−/X- α-gal for 5 days.
Results
Cloning of ZmHsf01 and protein subcellular localization
Using the RT-PCR method, we cloned the ZmHsf01 CDS from H21 seedlings. Sequence analysis showed that the ZmHsf01 CDS were 1,176 bp in length and encoded a deduced protein with 391 amino acid residues. The CDS and amino acid sequence are shown in Fig. S1. The conserved DNA-binding domain (DBD), oligomerization A/B (HR-A/B), nuclear localization signal (NLS), nuclear export signal (NES), and C-terminal activator motif (AHA) domains are marked with red lines (Fig. 1). Amino acid sequence alignment results showed that ZmHsf01 shared 86%, 70%, 69%, 42%, and 43% identities with SbHsfA2d from sorghum bicolor (XP_002468465), DoHsfA2d from Dichanthelium oligosanthes (OEL38242), SiHsfA2d from Setaria italica (XP_004985605), AtHsfA2 from Arabidopsis thaliana (AEC07800), and AtHsfA6b from Arabidopsis thaliana (AEE76681), respectively (Fig. 1). These results verify that ZmHsf01 is a HSF gene of the A2 subclass.
Figure 1. Amino acid sequence alignment between ZmHsf01 and homologs from different species.
DBD, the conserved DNA binding domain; HR-A/HR-B, two hydrophobic heptad repeats; NLS, nuclear localization signal; NES, nuclear export signal; AHA, aromatic, large hydrophobic and acidic amino residues. The protein accession numbers in NCBI are as follows: SbHsfA2d from Sorghum bicolor, XP_002468465.1; DoHsfA2d from Dichanthelium oligosanthes, OEL38242.1; SiHsfA2d from Setaria italica, XP_004985605.1; AtHsfA2 from Arabidopsis thaliana, AEC07800; and AtHsfA6b from Arabidopsis thaliana, AEE76681.
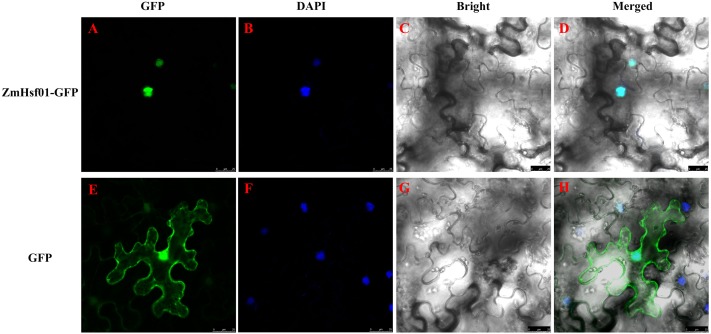
The ZmHsf01-GFP fusion protein were observed during the subcellular localization of ZmHsf01. The ZmHsf01 CDS were connected to the N-terminal of the green fluorescent protein (GFP) gene driven by a CaMV 35S promoter. The tobacco epidermal cells expressed the ZmHsf01-GFP fusion protein by an Agrobacterium-mediated transformation. After cultured for 3 days, stained the tobacco epidermal cells with DAPI. The laser confocal microscopy examination found individual GFP proteins throughout the control group’s entire cell, but ZmHsf01-GFP fusion protein was only detected in the nuclei, and was co-localized using DAPI florescence (Fig. 2). These results suggest that ZmHsf01 is localized in the nucleus.
Figure 2. Subcellular localization of ZmHsf01.
(A and E) Green fluorescence of GFP; (B and F) blue fluorescence of DAPI; (C and G) bright field; (D and H) merged images.
Expression analysis of ZmHsf01
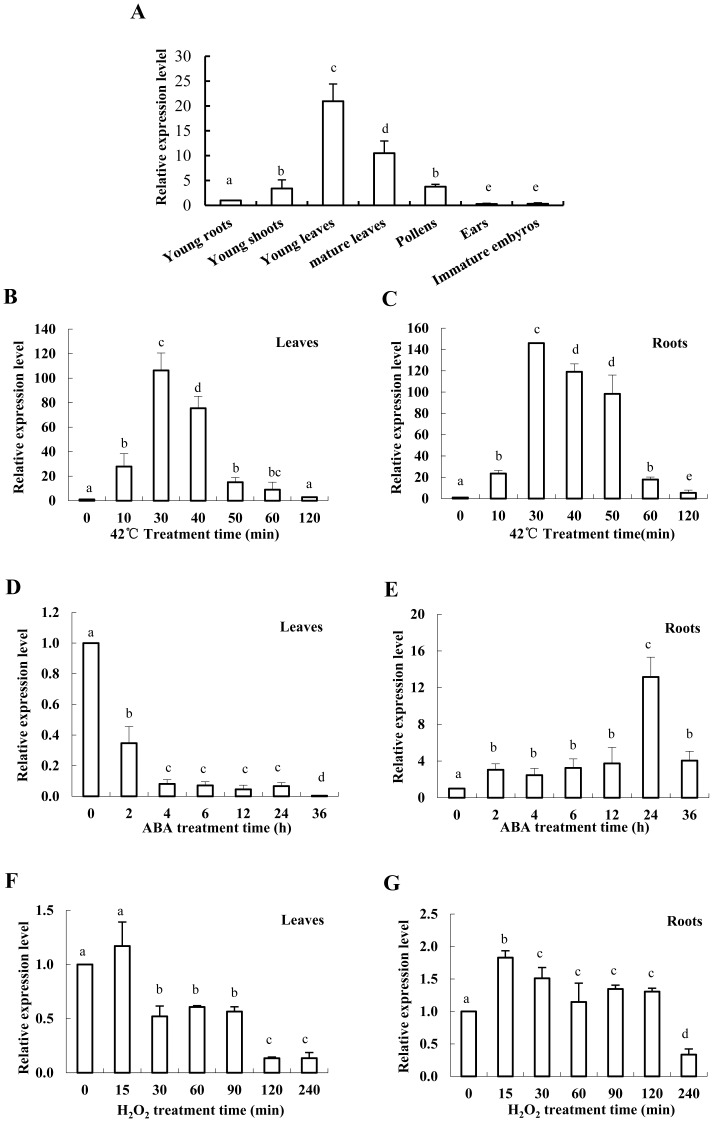
Under the normal growth conditions, ZmHsf01 was expressed in all detected maize organs: young roots, young shoots, young leaves, mature leaves, pollen, ears, and immature embryos (Fig. 3A). The expression level of ZmHsf01 was highest in young leaves and lowest in ears. ZmHsf01 was expressed 20 times more in young leaves than in roots.
Figure 3. Expression patterns of ZmHsf01 in different tissues and abiotic stresses include HS, ABA, and H2O 2.
(A) Expression levels of ZmHsf01 in different tissues and organs under normal growth conditions. (B and C) Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 42 °C HS. (D and E) Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 200 μ M ABA treatment. (F and G) Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 10 mM H2O 2 treatment. There were three biological repeats for each sample and the data mean ± standard error.
ZmHsf01 expression was significantly up-regulated in both roots and leaves after 42 °C HS. Expression levels reached their peak at 30 min after HS, and then gradually decreased (Figs. 3B, 3C). After ABA treatment, the expression of ZmHsf01 in roots was up-regulated and reached peak value at 24 h, but in leaves the ZmHsf01 expression showed a tendency to be down-regulated (Fig. 3E). The expression level of ZmHsf01 increased in roots and decreased in leaves under H2O2 treatment (Figs. 3F, 3G). These results suggest that ZmHsf01 can be up-regulated by HS, ABA, and H2O 2in roots(All the raw data of Fig. 3 were listed in Table S2).
Improving thermotolerance through ZmHsf01 expression in yeast cell
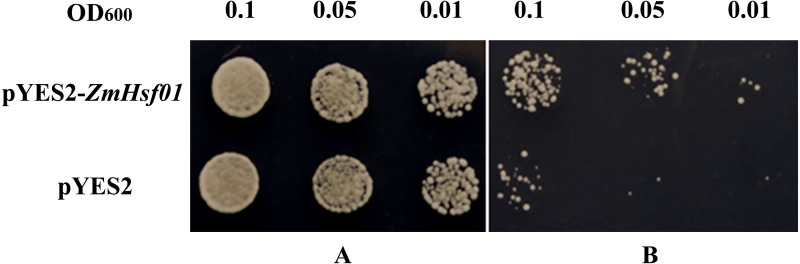
To further analyze the function of ZmHsf01, we used the pYES2-ZmHsf01 yeast expression vector to identify the genetic transformation and thermotolerance of yeast positive strains. Under the normal conditions, no significant phenotype difference was found between the two types of transgenic yeast cells (pYES2-ZmHsf01 and pYES2 control). The cellular growth of the two groups was inhibited after heat treatment at 50 °C for 15 min, but the growth potential of ZmHsf01-expressing cells was better than that of the control cells (Fig. 4). These results demonstrate that ZmHsf01 improves the thermotolerance of transgenic yeast cells.
Figure 4. Thermotolerance assays of yeast cell harboring pYES2 or pYES2-ZmHsf01 after 50 °C HS.
(A) Culture under normal conditions; (B) culture after HS at 50 °C for 15 min.
Yeast transcription activation activity of ZmHsf01
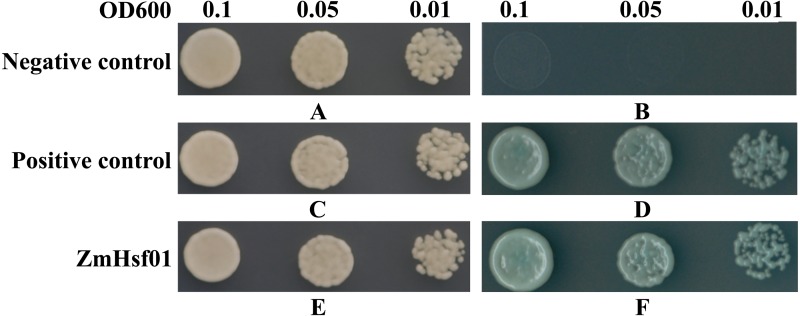
Based on our domain analysis, ZmHsf01 contains the AHA domain existing in class A members. We constructed ZmHsf01 into a pGBKT7 vector. The yeast strains transformed with fusion vector pGBKT7-ZmHsf01 grew well and turned blue similarly to the positive control groups in the SD/Trp-/His-/Ade-/X-α-gal culture medium (Fig. 5). Our results show that ZmHsf01 is active in yeast cell transcription activation.
Figure 5. Transcription activation analysis of ZmHsf01 in yeast.
Yeast cells transformed with pGBKT7 vector were set as the negative control, and yeast cells co-transformed with pGBKT7-53 and pGADT7-T vectors were set as the positive control. The ZmHsf01 group were the yeast cells transformed with the fusion vector pGBKT7-ZmHsf01. (A) The negative control in the SD/Trp- culture medium. (B) The negative control in the SD/Trp-/His-/Ade-/X-α-gal culture medium. (C) The positive control in the SD/Trp- culture medium. (D) The positive control in the SD/Trp-/His-/Ade-/X-α-gal culture medium. (E) The fusion vector pGBKT7-ZmHsf01 in the SD/Trp- culture medium. (F) The fusion vector pGBKT7-ZmHsf01 in the SD/Trp-/His-/Ade-/X-α-gal culture medium.
ZmHsf01 compensated for the thermotolerance defects of mutant Arabidopsisathsfa2
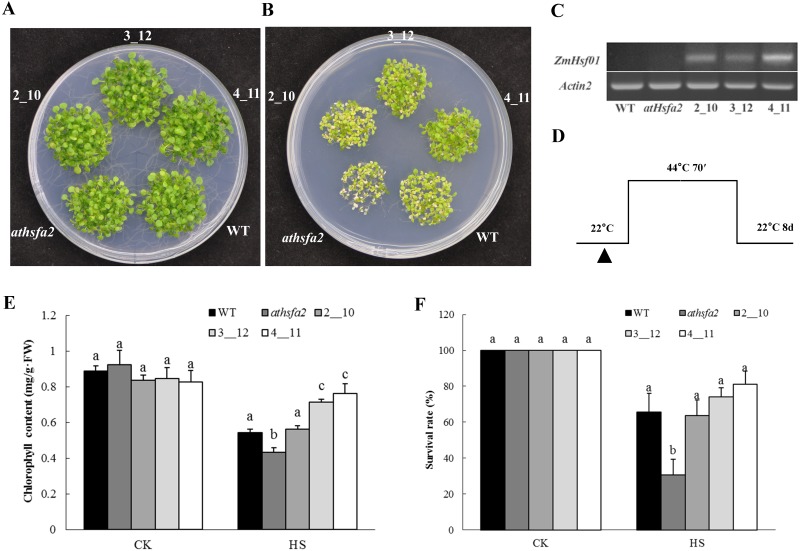
Charng et al. (2007) first reported the thermotolerance defects of Arabidopsis mutant athsfa2 (SALK_008978). We used the mutant athsfa2 to investigate the thermotolerance of ZmHsf01. Three ZmHsf01/athsfa2-complemented lines (2_10, 3_1, and 4_11) that expressed different levels of ZmHsf01 were selected using semi-quantitative RT-PCR (Fig. 6C). The seedlings of the three ZmHsf01/athsfa2-complemented lines, athsfa2 and WT were photographed, and the chlorophyll content were measured under normal conditions and HS (Fig. 6). The thermotolerances of ZmHsf01/athsfa2 were better than that both athsfa2 and WT (Fig. 6B). The chlorophyll contents of ZmHsf01/athsfa2 in lines 3_12 and 4_11 were higher than those of athsfa2 and WT (Fig. 6E). The survival rates of all complemented lines were higher than the mutant athsfa2, but not WT (Fig. 6F). The results showed that ZmHsf01 can partially or completely compensate for the thermotolerance defects of athsfa2.
Figure 6. The thermotolerant phenotypes of the deletion mutant and three restoration lines of Arabidopsis seedlings.
(A) Seedlings of WT, deletion mutant, and three restoration lines (2_10, 3_12, and 4_11) growing on the MS plate under normal conditions (control). (B) Seedlings of all genotypes under HS at 44 °C for 70 min and recovered under normal conditions for 8 days. (C) Chlorophyll content of seedlings under normal conditions and HS treatments. (D) Semi-qRT-PCR assay of the ZmHsf01 transcript. (E) Schematic representation of the HS regimes.
Functionally identifying ZmHsf01 thermotolerance in Arabidopsis
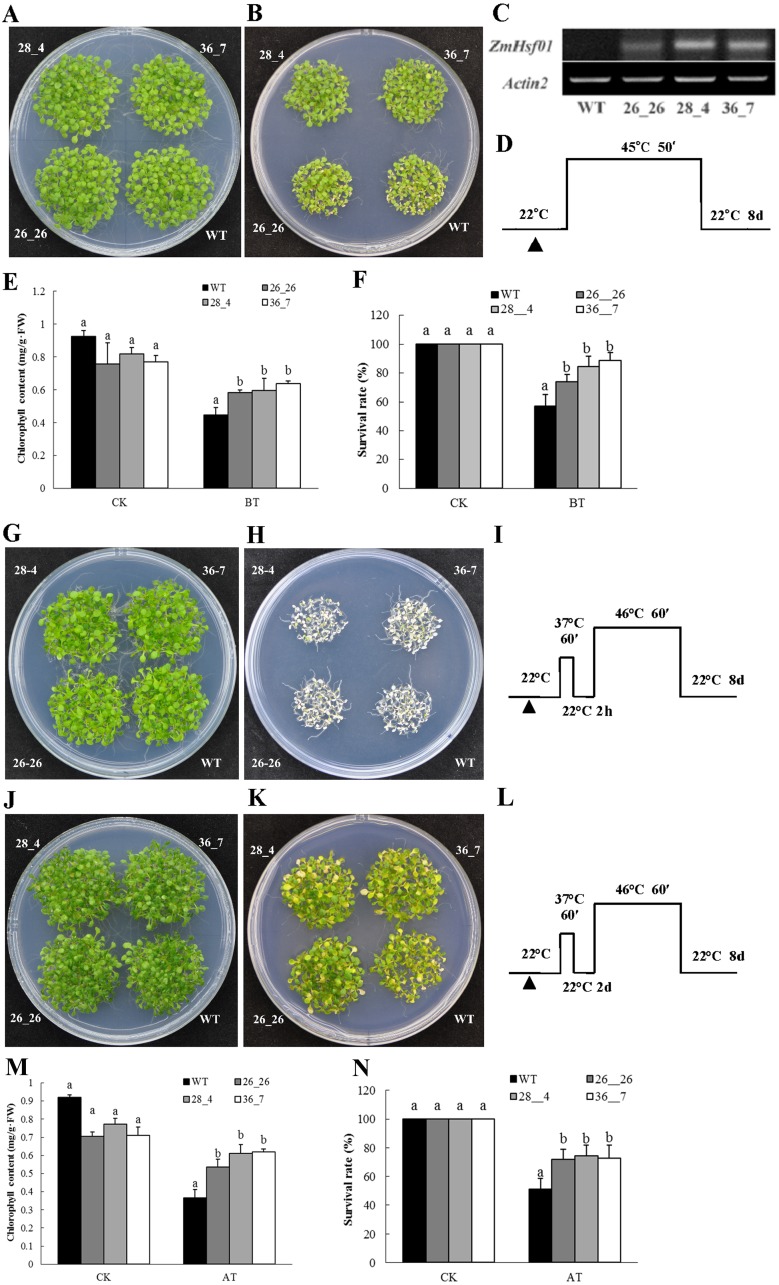
Three ZmHsf01-overexpressing lines (26_26, 28_4, and 36_7) were used to analyze the basal and acquired thermotolerances with different expression levels detected by semi-quantitative RT-PCR (Fig. 7C). Five-day-old seedlings of WT and three ZmHsf01-overexpressing lines growing on the same MS medium were exposed to special HS regimes (Fig. 7B), then recovered at 22 °C for 8 days (Fig. 7H). Under the normal conditions, no obvious phenotypic changes could be observed between the ZmHsf01-overexpressing lines and WT (Figs. 7A, 7G and 7J). After basal HS treatment and acquired HS treatment with a long-time recovery, the WT seedlings wilted, but the three ZmHsf01 over-expressing lines remained green (Figs. 7B and 7K). However, after acquired HS treatment with a short-time recovery, the seedlings of WT and three ZmHsf01 over-expressing lines all died (Fig. 7H). Further, we measured the chlorophyll contents and survival rates of different lines. Without HS treatment, no remarkable difference in chlorophyll content was found between the WT and three transgenic lines (Figs. 7E and 7M). After basal and acquired HS treatment, the chlorophyll contents of all genotypes decreased, but the chlorophyll contents of the three ZmHsf01-overexpressing lines were higher than that of WT (Figs. 7E and 7M), and the survival rates of the three over-expressing lines were higher than WT (Figs. 7F and 7N).
Figure 7. Overexpression of ZmHsf01 improved the basal and acquired thermotolerance of Arabidopsis seedlings.
(A) Seedlings of WT and three overexpressing lines (26_26, 28_4, and 36_7) growing on an MS plate under normal conditions used as the basal thermotolerance control. (B) Seedlings of all genotypes under basal HS at 45 °C for 50 min and recovered under normal conditions for 8 days. (C) Semi-qRT-PCR analysis of the ZmHsf01 transcript in different lines. The expression levels of AtActin2 were used as the control. (D) Schematic representation of the basal HS regimes. (E) Chlorophyll content of seedlings under normal conditions and basal HS treatments. (F) The survival rates of seedlings under normal conditions and basal HS treatments. (G) Seedlings of WT and three overexpressing lines (26_26, 28_4, and 36_7) growing on an MS plate under normal conditions used as the acquired thermotolerance control. (H) 5-day-old seedlings of all genotypes under acquired HS at 37 °C for 60 min, recovered under normal conditions for 2 h, treated at 46 °C for 60 min, and recovered under normal conditions for 8 days. (I) Schematic representation of the acquired HS regimes. (J) Seedlings of WT and three overexpressing lines (26_26, 28_4, and 36_7) growing on an MS plate under normal conditions used as the acquired thermotolerance control. (K) 5-day-old seedlings of all genotypes under acquired HS at 37 °C for 60 min, recovered under normal conditions for 2 days, treated at 46 °C C for 60 min, and recovered under normal conditions for 8 days. (L) Schematic representation of the acquired HS regimes. (M) Chlorophyll content of seedlings under normal conditions and acquired HS treatments. (N) The survival rates of seedlings under normal conditions and acquired HS treatments.
Affection of overexpression of ZmHsf01 in Arabidopsis on AtHsps expression
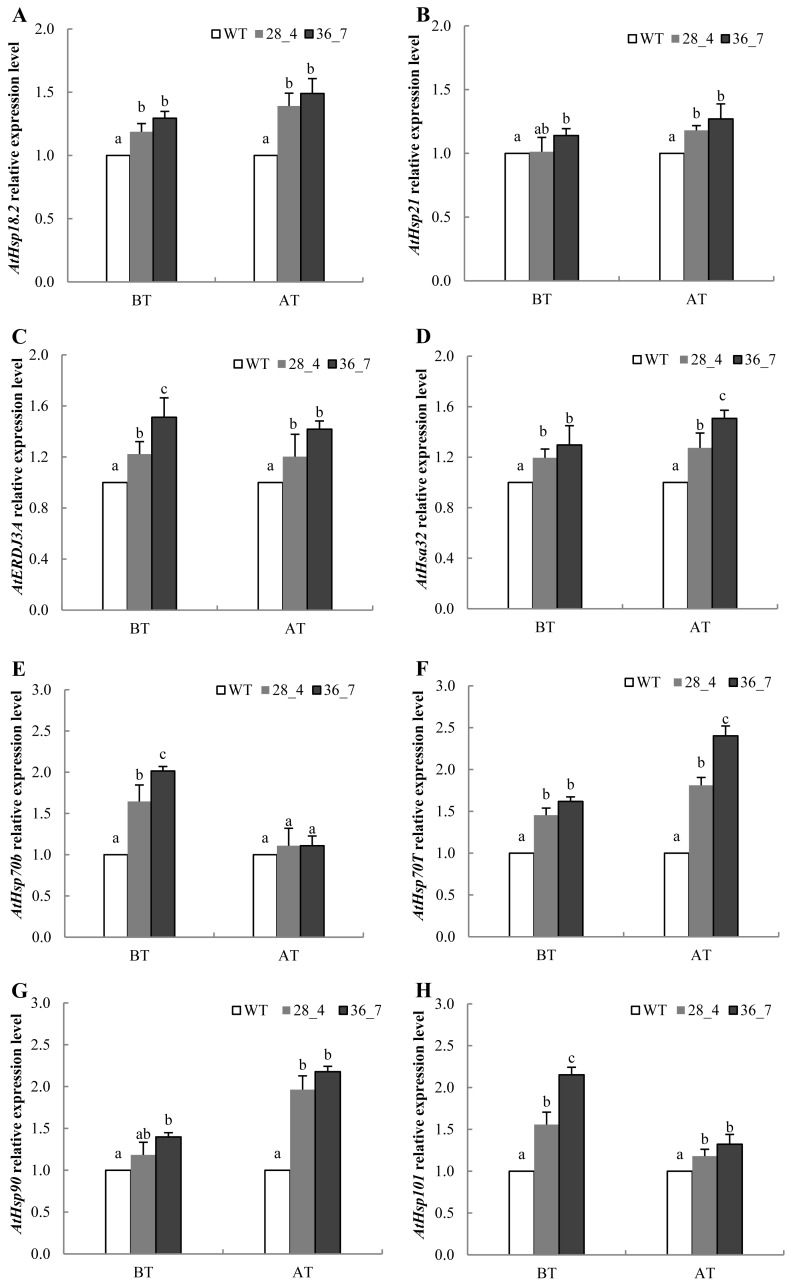
It has been shown that HSP genes can be induced and accumulate in cells so as to enhance the resistance of Arabidopsis plants under HS (Wan et al., 2016). To test the regulating role of ZmHsf01 in HSPs expression, we performed qRT-PCR using ZmHsf01 over-expressing lines 28_4 and 36_7. Five-day-old seedlings of WT and three ZmHsf01-overexpressing lines growing on the same MS medium were exposed to the HS treatment described in the Methods section. Such AtHsps, including AtHsp18.2, AtHsp21, AtHsa32, AtERDJ3A, AtHsp70b, AtHsp70T, AtHsp90.1, and AtHsp101, were detected after the basal and acquired heat treatments (Fig. 8). The results showed that the expression levels of AtHsps in ZmHsf01-overexpressing line 36_7 were 1.1 to 2.5 times higher than that in WT after HS treatment. The up-regulation of AtHsp70b and AtHsp101 only existed in BT, and the up-regulation of AtHsp70T and AtHsp90 was more obvious in AT. These results indicate that ZmHsf01 can increase the expression of AtHsps to enhance the thermotolerance of transgenic Arabidopsis (The raw data were listed in Table S3).
Figure 8. The expression levels of heat-related HSP genes in WT and ZmHsf01 over-expressing lines 28_4 and 36_7 after HS treatment.
qRT-PCR was performed on the Arabidopsis genes AtHsp18.2 (A), AtHsp21 (B), AtERDJ3A (C), AtHsfa32 (D), AtHsp70b (E), AtHsp70T (F), AtHsp90 (G), and AtHsp101 (H) under BT and AT HS. We set the expression level of WT samples as 1. The reference gene Atactin8 (At1g49240) was used as an internal control to normalize the loading of different samples. Data means ± SD from three biological experiments.
Discussion
In the HSF studies of Arabidopsis and tomato, the A1 and A2 subclasses were treated as the main research focus (Nakai, 2016; Scharf et al., 1998; Ogawa, Yamaguchi & Nishiuchi, 2007; Liu & Charng, 2013). With the recent developing in crop genome sequencing, HSF family members in more and more species have been speculated and genetically analyzed. The functional analysis of other subclass members have also been gradually developed (Ma et al., 2016; Hu et al., 2018). Research on HSFs in field crops started relatively late. Many transcription factor (TF) families, such as WRKY, MYB, DREB, and HSF, have enormous potential for improving the resistance of maize (Kimotho, Baillo & Zhang, 2019). In 2011, 25 HSFs were identified in maize, and total 30 ZmHsfs in 2016 (Lin et al., 2011; Guo et al., 2016). Xue et al. reported that at least 56 HSFs exist in wheat (Xue et al., 2014), but our analysis shows a more larger number family (Duan et al., 2019), suggesting the complexity and diversity of HSF family.
In Arabidopsis, A1 subclass HSFs are considered the master regulators in sensing stress and downstream gene activation (Liu et al., 2013). Unlike A1 subclass, the genes of A2 subclass are always induced after HS (Schramm et al., 2006; Mittal et al., 2009). ZmHsf01 expression after 30 min of HS increased and then decreased in leaves and roots. ZmHsf01 contains the typical domains of the HsfA class, such as DBD, HR-A/HR-B, NLS, and AHA motifs. ZmHsf01-GFP fusion protein is localized in the nuclei, which is consistent with the structure of the NLS motif within ZmHsf01 protein. This expression level trend in ZmHsf01 corresponds with previously reported ZmHsfA2s (Li et al., 2019; Jiang et al., 2018). The conserved domain among these class A members suggests similarities among gene functions. ABA, low temperatures, and NaCl treatments significantly induced the up-regulation of ZmHsf04 (Li et al., 2019). However, ZmHsf01 was only up-regulated after ABA or H2O2 treatment in the roots, and the ZmHsf01 expression levels were lower in leaves. It has been known that ABA can regulate tolerance to various abiotic stress, and exogenous ABA could induce the expression of HsfA2 and enhance the heat tolerance of Arabidopsis and tall fescue (Wang et al., 2017b). However, the expression levels of SbHsfA2 genes in sorghum leaves remained stable after 100 µM of ABA for 4 h (Nagaraju et al., 2015). H2O2 was also involved in the regulation of some stress responses. The ZmHsf06 expression could be up-regulated by 42 °C HS and H2O2, and this expression was H2O2-dependent (Li, Li & Guo, 2015b). In the process of acquired thermotolerance, the regulation of salicylic acid (SA) on the AtHsfA2 also depend on the existence of H2O2 signaling pathway(Gaffney et al., 1993; Snyman & Cronje, 2008). ABA up-regulated the OsHsfA2 and OsHsfA4a expression by inducing H2O2 in rice (Dang, Jiang & Lin, 2010), and the up-regulation of ABA on ZmHsf06 also partly depended on H2O2 (Li, Li & Guo, 2015b). The expression of OsHsfA2d increased 4∼6 folds under salt, PEG, and cold stress (Liu et al., 2010). The abiotic stress assays suggest that HsfA2s are induced by various environmental stresses, but showed different response pattern.
Previous studies on Arabidopsis have shown that AtHsfA2 can sustain the expression of HSP genes during the recovery stage and the acquired thermotolerance (Charng et al., 2007). There is only one HsfA2 in Arabidopsis, compared with WT, mutant athsfa2 is more sensitive to HS (Li et al., 2005; Charng et al., 2007). FaHsfA2c from tall fescue can improve the heat tolerance of mutant Arabidopsis athsfa2 (Wang et al., 2017a). ZmHsf05, another member of class A2, can also complement the lack of thermotolerance of mutant athsfa2 mutant (Li et al., 2019). In this study, ZmHsf01 rescued the thermotolerant phenotypes of the athsfa2 mutant, and ZmHsf01-overexpressing Arabidopsis seedlings improved the basal and acquired thermotolerance when compared with WT. These results demonstrate that ZmHsf01 can improve thermotolerance, playing a similar role to ZmHsf05. However, the other roles of ZmHsf01 such as involving in salinity and drought stresses have not been reported, it’s necessary to be studied further. Wheat TaHsfA2d has been proved to improve tolerance to high temperatures, salinity, and drought stresses in transgenic Arabidopsis plants (Chauhan et al., 2013).
Working as molecular chaperones, HSPs belong to multigene families and participate in various biological processes: protein folding, refolding, co-degradation of denatured proteins, and normal growth development (Kuang et al., 2017; Queitsch et al., 2000). The recognized “refolding machines”, Hsp70 and ERDJ3A, can refold denatured proteins under HS and alleviate stress damage (Ma et al., 2015). Both the regulation of HSFs on HSPs and the feedback inhibition of HSPs on HSFs play vital role in stress regulation (Frangkostefanakis et al., 2015). In both ZmHsf04 and Zmhsf05 over-expressing Arabidopsis, the expression levels of all detected AtHsps were higher than those in WT after HS (Li et al., 2019; Jiang et al., 2018). After heat treatment, the transcript levels of some HSPs, such as AtHsp70b, AtHsp70T, AtHsp90, and AtHsp101, in ZmHsf01 over-expressing lines were higher than those of WT. At the same time, the up-regulation of AtHsp70b and AtHsp101 only existed in BT, while the up-regulation of AtHsp70T and AtHsp90 was more obvious in AT. These results indicate that ZmHsf01 may improve the different thermotolerance of plants by regulating different HSPs expression.
Our analysis proved that heat treatment may help plants accumulate various HSPs to improve their thermotolerance. Different member of the HSF family plays different role in HS signal transduction and downstream gene expression. Further tests of the interactions of different HSFs and the gene regulatory mechanism in transgenic maize should be conducted.
Conclusion
We cloned ZmHsf01 from maize inbred line H21. ZmHsf01 was highly conserved compared to its homologs in other plants. ZmHsf01 compensated for the thermotolerant defects of athsfa2 mutant of Arabidopsis thaliana. The Arabidopsis seedlings of overexpressing ZmHsf01 had more stronger thermotolerance than WT seedlings.
Supplemental Information
The accession numbers of cDNA sequences of genes from Arabidopsis available at NCBI. The forward and reverse primers were designed with the software Primer Premier 5.
Abiotic stresses include heat stress, ABA, and H2O2. A, Expression levels of ZmHsf01 in different tissues and organs under normal growth conditions. B and C, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 42 °C heat shock. D and E, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 200μ M ABA treatment. F and G, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 10 mM H2O2 treatment. There were three biological repeats for each sample and the data mean ± standard error.
qRT-PCR was performed on the Arabidopsis AtHsp18.2, AtHsp21, AtERDJ3A, AtHsfa32, AtHsp70b, AtHsp70T, AtHsp90, and AtHsp101 under BT and AT HS. We set the expression level of WT samples as 1. The reference gene Atactin8 (At1g49240) was used as an internal control to normalize the loading of different samples. Data means ± SD from three biological experiments.
The β-actin blots of WT, athsfa2 and three over-expressing lines are tagged with white words.
The ZmHsf01 blots of WT, athsfa2 and three over-expressing lines are tagged with white words.
The β-actin blots of WT and three over-expressing lines are tagged with white words.
The ZmHsf01 blots of WT and three over-expressing lines are tagged with white words.
Acknowledgments
We thank Dr. Yeeyung Charng (Agricultural Biotechnology Research Center, Academia Sinica, Taipei, Taiwan) for providing athsfa2 mutant seeds, and Dr. Rongmin Chen (School of Medicine, Yale University, United States) for polishing the language.
Abbreviations
- HSFs
heat shock transcription factors
- HSE
heat shock element
- HS
heat shock
- HSPs
heat shock proteins
- RH
relative humidity
- CDS
coding sequence
- DBD
DNA-binding domain
- OD
oligomerization domain
- NLS
nuclear localization signal
- NES
nuclear export signal
- AHA motifs
C-terminal activator motifs
- ABA
abscisic acid
- SA
salicylic acid
- H2O2
hydrogen peroxide
- PEG
polyethylene glycol
- GFP
green fluorescence protein
- WT
wild type
- BT
basal thermotolerance
- AT
acquired thermotolerance
Funding Statement
This work was supported by the National Key Research and Development Program of China (2018YFD0300504), the Natural Science Foundation of Hebei Province (C2017301065), the Technological Innovation Basal Research Fund of Hebei Academy of Agriculture and Forestry Sciences (2018110101), the Technological Innovation Project of Modern Agriculture of Hebei Province (494-0402-JBN-C7GQ) and Excellent Scientists Plan of JAAS, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Hongbo Shao, Email: shaohongbochu@126.com.
Xiulin Guo, Email: myhf2002@163.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Huaning Zhang, Guoliang Li and Yuanyuan Zhang conceived and designed the experiments, performed the experiments, prepared figures and/or tables, and approved the final draft.
Dong Hu analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Yujie Zhang, Lina Zhao and Ruiping Yang performed the experiments, prepared figures and/or tables, and approved the final draft.
Hongbo Shao conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Xiulin Guo conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files and the ZmHsf01 CDS and amino acid sequences are available at GenBank: MK888854.
References
- Aranda et al. (1999).Aranda MA, Escaler M, Thomas CL, Maule AJ. A heat shock transcription factor in pea is differentially controlled by heat and virus replication. The Plant Journal. 1999;20(2):153–161. doi: 10.1046/j.1365-313x.1999.00586.x. [DOI] [PubMed] [Google Scholar]
- Baniwal et al. (2004).Baniwal SK, Bhaerti K, Chan KY, Fauth M, Ganguli A. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences. 2004;29(4):471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- Bharti et al. (2004).Bharti K, Von KoskullDöring P, Bharti S, Kumar P, Tintschlkörbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. The Plant Cell. 2004;16(6):1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, Wunderlich & Schöffl (2005).Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal. 2005;41(1):1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- Cairns et al. (2012).Cairns JE, Sonder K, Zaidi PH, Verhulst N, Mahuku G, Babu R, Nair1 SK, Das B, Govaerts B, Vinayan MT, Rashid Z, Noor JJ, Devi P, SanVicente F, Prasanna BM. Maize production in a changing climate: impacts, adaptation and mitigation strategies. Advances in Agronomy. 2012;114:1–58. doi: 10.1016/B978-0-12-394275-3.00006-7. [DOI] [Google Scholar]
- Charng et al. (2007).Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology. 2007;143(1):251–262. doi: 10.2307/40065230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan et al. (2013).Chauhan H, Khurana N, Agarwal P, Khurana JP, Khurna P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLOS ONE. 2013;8(11):e79577. doi: 10.1371/journal.pone.0079577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka-Verner et al. (2004).Czarnecka-Verner E, Pan S, Salem T, Gurley WB. Plant class B Hsfs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Molecular Biology. 2004;56(1):57–75. doi: 10.1007/s11103-004-2307-3. [DOI] [PubMed] [Google Scholar]
- Dang, Jiang & Lin (2010).Dang J, Jiang MY, Lin F. ABA up-regulate the expression of OsHSF gene in leaves of rice plants. Journal of Nanjing Agricultural University. 2010;33(1):11–15. doi: 10.7685/j.issn.1000-2030.2010.01.003. [DOI] [Google Scholar]
- Duan et al. (2019).Duan SN, Liu BH, Zhang YY, Li GL, Guo XL. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genomics. 2019;20(1):257. doi: 10.1186/s12864-019-5617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangkostefanakis et al. (2015).Frangkostefanakis S, Fragkostefanakis S, Röth S, Schleiff E, Scharf KD. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environment. 2015;38(9):1881–1895. doi: 10.1111/pce.12396. [DOI] [PubMed] [Google Scholar]
- Gaffney et al. (1993).Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261(5122):754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Gietz et al. (1992).Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for high transformation of intact yeast cells. Nucleic Acids Research. 1992;20(6):1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2008).Guo JK, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. Journal of Genetics and Genomics. 2008;35(2):105–118. doi: 10.1016/S1673-8527(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2016).Guo M, Liu HJ, Ma X, Luo DX, Gong ZH, Lu MH. The plant heat stress transcription factors (Hsfs): structure, regulation and function in response to abiotic stresses. Frontiers in Plant Science. 2016;7:114. doi: 10.3389/fpls.2016.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz, Döring & Bonzelius (2001).Heerklotz D, Döring P, Bonzelius F. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Molecular and Cellular Biology. 2001;21(5):1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2018).Hu XJ, Chen D, Mclntyre CL, Dreccer MF, Zhang ZB, Drenth J, Kalaipandian S, Chang H, Xue GP. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA- mediated regulatory pathway. Plant Cell Environment. 2018;41(1):79–98. doi: 10.1111/pce.12957. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2018).Jiang YL, Zheng QQ, Chen L, Liang YN, Wu JD. Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiologiae Plantarum. 2018;40(1):9–21. doi: 10.1007/s11738-017-2587-2. [DOI] [Google Scholar]
- Kimotho, Baillo & Zhang (2019).Kimotho RN, Baillo EH, Zhang Z. Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ. 2019;7:e7211. doi: 10.7287/peerj.preprints.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak et al. (2004).Kotak S, Port M, Ganguli A, Bicker F, Von Koskull-Doüring P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. The Plant Journal. 2004;39(1):98–112. doi: 10.1111/j.1365-313X.2004.02111.x. [DOI] [PubMed] [Google Scholar]
- Kuang et al. (2017).Kuang J, Liu JZ, Mei J, Wang CC, Hu HT, Zhang YJ, Sun MH, Ning X, Xiao LT, Yang L. A Class II small heat shock protein OsHsp180 plays positive roles in both biotic and abiotic defense responses in rice. Scientific Reports. 2017;7(1):11333. doi: 10.1038/s41598-017-11882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2005).Li CG, Chen QJ, Gao XQ, Qi BS, Chen NZ, Xu SM, Chen J, Wang XC. Heat shock transcription factor AtHsfA2 regulating genes expression related to stresses and increase endurance to heat and oxidation stress in Arabidopsis. Science in China Series C-Life Sciences. 2005;35(5):398–407. doi: 10.3969/j.issn.1674-7232.2005.05.003. [DOI] [Google Scholar]
- Li et al. (2019).Li GL, Zhang HN, Shao HB, Wang GY, Zhang YY, Zhang YJ, Zhao LN, Guo XL, Mohamed SS. ZmHsf05, a new heat shock transcription factor from Zea mays L improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Science. 2019;283:375–384. doi: 10.1016/j.plantsci.2019.03.002. [DOI] [PubMed] [Google Scholar]
- Li, Li & Guo (2015b).Li HC, Li GL, Guo XL. Cloning, expression characteristics and subcellular-location of heat shock transcription factor ZmHsf06 in Zea mays. Journal of Agricultural Biotechnology. 2015b;23(1):41–51. doi: 10.3969/j.issn.1674-7968.2015.01.005. [DOI] [Google Scholar]
- Li et al. (2015a).Li HC, Zhang HN, Li GL, Liu ZH, Zhang YM, Guo XL. Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Functional Plant Biology. 2015a;42(11):1080–1090. doi: 10.1071/FP15080. [DOI] [PubMed] [Google Scholar]
- Li et al. (2018).Li LY, Yang HM, Liu P, Ren WB, Wu XH, Huang F. Combined impact of heat stress and phosphate deficiency on growth and photochemical activity of sheepgrass (Leymus chinensis) Journal of Plant Physiology. 2018;231:271–276. doi: 10.1016/j.jplph.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Lin et al. (2011).Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics. 2011;12(1):76–89. doi: 10.1186/1471-2164-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2010).Liu AL, Zou J, Zhang XW, Zhou XY, Wang WF, Xiong XY, Chen LY, Chen XB. Expression profiles of class A rice heat shock transcription factor genes under abiotic stresses. Journal of Plant Biology. 2010;53(2):142–149. doi: 10.1007/s12374-010-9099-6. [DOI] [Google Scholar]
- Liu & Charng (2013).Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiology. 2013;163(1):276–290. doi: 10.1104/pp.113.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2013).Liu YF, Zhang CX, Chen J, Guo LH, Li XL, Li WP, Yu ZF, Deng JS, Zhang PY, Zhang KQ, Zhang LM. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiology Biochemistry. 2013;64(5):92–98. doi: 10.1016/j.plaphy.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2016).Ma H, Wang CT, Yang B, Cheng HY, Wang Z, Mijiti A, Ren C, Qu GH, Zhang H, Ma L. CarHSFB2, a Class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.) Plant Molecular Biology Reporter. 2016;34(1):1–14. doi: 10.1007/s11105-015-0892-8. [DOI] [Google Scholar]
- Ma et al. (2015).Ma ZX, Leng YJ, Chen GX, Zhou PM, Ye D, Chen LQ. The thermosensitive male sterile 1 interacts with the BiPs via DnaJ domain and stimulates their ATPase enzyme activities in Arabidopsis. PLOS ONE. 2015;10(7):e0132500. doi: 10.1371/journal.pone.0132500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal et al. (2009).Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiology and Biochemistry. 2009;47(9):785–795. doi: 10.1016/j.plaphy.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Nagaraju et al. (2015).Nagaraju M, Sudhakar Reddy P, Anil Kumar S, Srivastava RK, Kavi Kishor P, Rao DM. Genome-wide scanning and characterization of Sorghum bicolor L. heat shock transcription factors. Current Genomics. 2015;16:279–291. doi: 10.13140/RG.2.2.13051.72485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai (2016).Nakai A. Heat shock factor. 1st edition Springer; Japan: 2016. [Google Scholar]
- Nishizawa et al. (2006).Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. The Plant Journal. 2006;48(4):535–547. doi: 10.1111/j.1365-313x.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi et al. (2011).Nishizawa-Yokoi A, Nosaka R, Hayashi H, Tainaka H, Maruta T, Tamoi M, Ikeda M, Ohme-Takagi M, Yoshimura K, Yabuta Y, Shigeoka S. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiology. 2011;52(5):933–945. doi: 10.1093/pcp/pcr045. [DOI] [PubMed] [Google Scholar]
- Nover et al. (2001).Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need. Cell Stress Chaperones. 2001;6(3):177–189. doi: 10.2307/1601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover et al. (1996).Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The HSF world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1(4):215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, Yamaguchi & Nishiuchi (2007).Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. Journal of Experimental Botany. 2007;58(12):3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- Queitsch et al. (2000).Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. The Plant Cell. 2000;12(4):479–492. doi: 10.2307/3871063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions, Hawes & Kurup (2007).Runions J, Hawes C, Kurup S. Fluorescent protein fusions for protein localization in plants. Methods in Molecular Biology. 2007;390:239–256. doi: 10.1007/978-1-59745-466-7_16. [DOI] [PubMed] [Google Scholar]
- Scharf et al. (2012).Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica et Biophysica Acta. 2012;1819(2):104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Scharf et al. (1998).Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Molecular and Cellular Biology. 1998;18(4):2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm et al. (2006).Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, Von Koskull-Doring P. The heat stress transcription factor HSFA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Molecular Biology. 2006;60(5):759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- Snyman & Cronje (2008).Snyman M, Cronje MJ. Modulation of heat shock factors accompanies salicylic acid-mediated potentiation of Hsp70 in tomato seedlings. Journal of Experimental Botany. 2008;59(8):2125–2132. doi: 10.1093/jxb/ern075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchul & Eltahir (2018).Suchul K, Eltahir EAB. North China plain threatened by deadly heatwaves due to climate change and irrigation. Nature Communications. 2018;9(1):1–9. doi: 10.1038/s41467-018-05252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan et al. (2016).Wan XL, Yang J, Li XB, Zhou Q, Guo C, Bao MZ, Zhang JW. Over-expression of PmHSP179 in transgenic Arabidopsis thaliana confers thermotolerance. Plant Molecular Biology Reporter. 2016;34(5):899–908. doi: 10.1007/s11105-016-0974-2. [DOI] [Google Scholar]
- Wang et al. (2017a).Wang X, Huang W, Liu J, Yang Z, Huang B. Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnology Journal. 2017a;15(2):237–248. doi: 10.1111/pbi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2017b).Wang X, Zhuang L, Shi Y, Huang B. Up-Regulation of HSFA2c and HSPs by ABA Contributing to Improved Heat Tolerance in Tall Fescue and Arabidopsis. International Journal of Molecular Sciences, 2017. 2017b;18(9):1981. doi: 10.3390/ijms18091981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue et al. (2014).Xue GP, Sadat S, Drenth J, Mclntyre CL. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. Journal of Experimental Botany. 2014;65(2):539–557. doi: 10.1093/jxb/ert399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016).Yang X, Zhu W, Zhang H, Liu N, Tian S. Heat shock factors in tomatoes: genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. PeerJ. 2016;4:e1961. doi: 10.7717/peerj.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani et al. (2008).Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K. Expression of rice heat s transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta. 2008;227(5):957–967. doi: 10.2307/23389920. [DOI] [PubMed] [Google Scholar]
- Zhu et al. (2006).Zhu BG, Ye CJ, Lü HY, Chen XJ, Chai GH, Chen JN, Wang C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max) Journal of Plant Research. 2006;119(3):247–256. doi: 10.1007/s10265-006-0267-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The accession numbers of cDNA sequences of genes from Arabidopsis available at NCBI. The forward and reverse primers were designed with the software Primer Premier 5.
Abiotic stresses include heat stress, ABA, and H2O2. A, Expression levels of ZmHsf01 in different tissues and organs under normal growth conditions. B and C, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 42 °C heat shock. D and E, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 200μ M ABA treatment. F and G, Expression levels of ZmHsf01 in leaves and roots of maize seedlings after 10 mM H2O2 treatment. There were three biological repeats for each sample and the data mean ± standard error.
qRT-PCR was performed on the Arabidopsis AtHsp18.2, AtHsp21, AtERDJ3A, AtHsfa32, AtHsp70b, AtHsp70T, AtHsp90, and AtHsp101 under BT and AT HS. We set the expression level of WT samples as 1. The reference gene Atactin8 (At1g49240) was used as an internal control to normalize the loading of different samples. Data means ± SD from three biological experiments.
The β-actin blots of WT, athsfa2 and three over-expressing lines are tagged with white words.
The ZmHsf01 blots of WT, athsfa2 and three over-expressing lines are tagged with white words.
The β-actin blots of WT and three over-expressing lines are tagged with white words.
The ZmHsf01 blots of WT and three over-expressing lines are tagged with white words.
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files and the ZmHsf01 CDS and amino acid sequences are available at GenBank: MK888854.