Abstract
Psoriasis is characterized by hyperproliferation and defective differentiation of keratinocytes (KCs). Patients with psoriasis are at a high risk of developing diabetes and cardiovascular diseases. The debate on the genetic origin of psoriasis pathogenesis remains unresolved due to lack of suitable in vitro human models mimicking the disease phenotypes. In this study, we provide the first human induced pluripotent stem cell (iPSC) model for psoriasis carrying the genetic signature of the patients. iPSCs were generated from patients with psoriasis (PsO-iPSCs) and healthy donors (Ctr-iPSCs) and were efficiently differentiated into mature KCs. RNA sequencing of KCs derived from Ctr-iPSCs and PsO-iPSCs identified 361 commonly upregulated and 412 commonly downregulated genes. KCs derived from PsO-iPSCs showed dysregulated transcripts associated with psoriasis and KC differentiation, such as HLA-C, KLF4, chemokines, type I interferon-inducible genes, solute carrier family, IVL, DSG1, and HLA-DQA1, as well as transcripts associated with insulin resistance, such as IRS2, GDF15, GLUT10, and GLUT14. Our data suggest that the KC abnormalities are the main driver triggering psoriasis pathology and highlights the substantial contribution of genetic predisposition in the development of psoriasis and insulin resistance.
Keywords: induced pluripotent stem cells, genetic predisposition, keratinocytes, insulin resistance, skin disorder, transcriptome profiling
Introduction
Psoriasis is an immune-mediated chronic inflammatory skin disorder, characterized by hyperproliferation and defective differentiation of epidermal keratinocytes (KCs). Psoriasis affects multiple organs beyond the skin and has been associated with metabolic dysfunction (eg, insulin resistance) and cardiovascular diseases. It has been hypothesized that psoriasis arises due to environmental assaults on a background of genetic predisposition. Psoriasis can aggregate in families with estimated heritability of up to 80% [1,2]. Previous genome-wide association studies (GWAS) have identified more than 60 psoriasis susceptibility regions, including loci associated with regulation of T cell function, macrophage activation, nuclear factor-kB signaling, and interferon (IFN)-mediated antiviral responses [3–9]. Substantial work has been done to identify genes and pathways altered in psoriatic lesions compared with normal skin using different molecular and genomic approaches [4,9–17]. Genetic alterations in the signaling pathways that activate inflammatory immune responses in KCs can change skin homeostasis and induce psoriasis.
Although several hereditary and environmental factors are known to be involved in the development of psoriasis, the distinction between genetic and acquired factors implicated in the KC abnormalities during psoriasis development and its progression remain unknown. Mouse models have been used to understand psoriasis pathogenesis [18,19]; however, human psoriasis phenotypes are difficult to reproduce in animal models. Therefore, there is a need to establish a human model that can recapitulate the patient KC-specific genetic signature. Induced pluripotent stem cells (iPSCs) have allowed for the generation of in vitro models of human diseases. Patient-specific iPSCs can provide genetically relevant cells, recapitulating the disease phenotype in vitro to understand genetic alterations associated with psoriasis. To our knowledge, there are no reports studying KC abnormalities in psoriasis using human iPSC-based model. Therefore, in this study, we have established the first in vitro iPSC-based model to study genetic alterations in KCs derived from patient-specific iPSCs.
Materials and Methods
Patient samples
The study has been approved by the appropriate Institutional Research Ethics Committee and has been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Blood samples were obtained from two subjects with psoriasis and two healthy individuals from Hamad Medical Corporation (HMC) hospital with full informed consent. The protocol was approved by the Institutional Review Board (IRB) of HMC (no. 16260/16) and Qatar Biomedical Research Institute (QBRI) (no. 2016-003), Qatar. Both patients were insulin resistant and had a family history of psoriasis. Patient 1 (PsO1) was a 32 years old female; prediabetes (HBA1c of 6.4%) confirmed the presence of insulin resistance. Patient 2 (PsO2) was a 45 years old male; insulin resistance was confirmed based on a measured HOMA-IR of 2.55. Both patients with psoriasis do not smoke and both of them have family history of psoriasis. Both of them are on topical treatment for psoriasis. Both healthy controls had no family history of psoriasis or diabetes. Control 1 (Ctr1) was a 27 years old male and control 2 (Ctr2) was a 28 years old female.
Generation of human iPSCs
The peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples using Ficoll-Paque Premium according to the manufacturer's instructions (Sigma-Aldrich). The PBMCs were cultured in StemPro-34 SFM Complete Medium (Gibco) for at least 4 days before reprogramming. The cells were transduced with the CytoTune-iPS 2.0 Sendai reprogramming kit (Thermo Fisher Scientific). At day 3 after reprogramming, the cells were seeded on Matrigel-coated plates and cultured in StemPro medium without cytokines. At day 7, half of the medium was replaced with ReproTeSR medium (Stem Cell Technologies) and the medium was completely changed to ReproTeSR at day 8. The iPSCs colonies generated between days 15 and 30 were manually picked, expanded, and maintained in mTESR-1 medium (Stem Cell Technologies) (Fig. 1A). The established hiPSC lines were registered in (https://hpscreg.eu) website and were assigned unique stem cell line names as follows: QBRIi001-A (Ctr1-iPSCs-C1), QBRIi001-B (Ctr1-iPSCs-C2), QBRIi001-C (Ctr1-iPSCs-C3), QBRIi002-A (Ctr2-iPSCs-C1), QBRIi002-B (Ctr2-iPSCs-C2), QBRIi002-C (Ctr2-iPSCs-C3), QBRIi005-A (PsO1-iPSCs-C1), QBRIi005-B (PsO1-iPSCs-C2), QBRIi005-C (PsO1-iPSCs-C3), QBRIi006-A (PsO2-iPSCs-C1), QBRIi006-B (PsO2-iPSCs-C2), and QBRIi006-C (PsO2-iPSCs-C3).
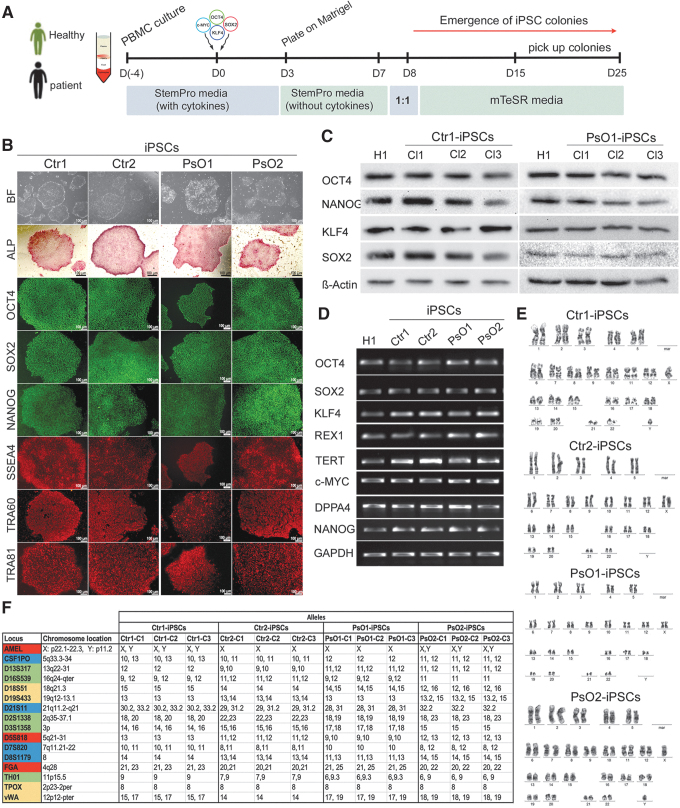
FIG. 1.
Generation and characterization of iPSC lines. (A) Schematic diagram of the protocol used for iPSCs generation. PBMCs were reprogrammed using cytotune 2.0 Sendai reprogramming kit. (B) Representative images showing the characterization of iPSCs generated from healthy controls (Ctr-iPSCs) and patients (PsO-iPSCs). The generated iPSCs showed hESC-like morphology, strong alkaline phosphatase activity (ALP), and expressed the pluripotency markers (OCT4, SOX2, NANOG, SSEA4, TRA-60, and TRA-81). (C) Western blotting showing the expression of pluripotency markers OCT4, NANOG, KLF4, and SOX2. (D) RT-PCR analysis of the generated iPSCs showing the expression of the pluripotency markers, OCT4, SOX2, NANOG, KLF4, REX1, TERT, DPPA4, and c-MYC. H1-hESCs were used as control. (E) Representative iPSC lines (Ctr1-iPSCs, Ctr2-iPSCs, PsO1-iPSCs, and PsO2-iPSCs) showed normal karyotype by G-banding analysis. (F) Genetic profiling of the generated iPSCs from healthy and psoriatic patients through the STR analysis showed that the iPSCs from each individual are typically identical. Scale bars = 100 μm. hESCs, human embryonic stem cells; iPSC, induced pluripotent stem cell; PBMCs, peripheral blood mononuclear cells; RT-PCR, reverse transcription polymerase chain reaction; STR, short tandem repeat.
Differentiation of iPSCs toward KCs
KC differentiation protocol was performed based on previously published protocols [20,21] with minor modifications. The commercially available H1-human embryonic stem cell (hESC) line was obtained from WiCell Research Institute (Madison, WI) and was used as a control. In brief, H1-hESCs, Ctr-iPSCs (Ctr1-iPSCs and Ctr2-iPSCs), and PsO-iPSCs (PsO1-iPSCs and PsO2-iPSCs) were seeded as small clumps on Geltrex (1:100; Thermo Fisher Scientific)-coated plates in mTeSR medium. Differentiation was started using unconditioned medium (UCM) composed of Knockout Dulbecco's modified Eagle's medium-F12 (DMEM/F12) (Thermo Fisher Scientific), 20% knockout serum replacement medium, 1 mM l-glutamine, 1% nonessential amino acids (NEAA), and 0.1 mM beta mercaptoethanol supplemented with 1 μM retinoic acid (RA) (Sigma-Aldrich) and 20 ng/mL bone morphogenic protein 4 (BMP4) (Thermo Fisher Scientific) for 1 week. At day 5 of differentiation, the cells were replated using ReLeSR (Stem Cell Technologies) and the medium was switched stepwise from UCM to N2 medium (1:1) containing knockout DMEM/F12, 1% N2-supplement (Thermo Fisher Scientific), 1% l-glutamine, and 1% NEAA supplemented with RA and BMP4. At day 7 of differentiation, the cultured medium was switched completely to N2 medium containing 10 ng/mL EGF and the cells were grown until day 14 of differentiation. At day 14 of differentiation, the cells were detached using Tryple E express (Thermo Fisher Scientific) and plated at high density in N2 medium containing 10 ng/mL EGF until day 30 of differentiation. The cells were treated for the last 8 days with high calcium (1.2 mM) to induce KC maturation (Fig. 3A).
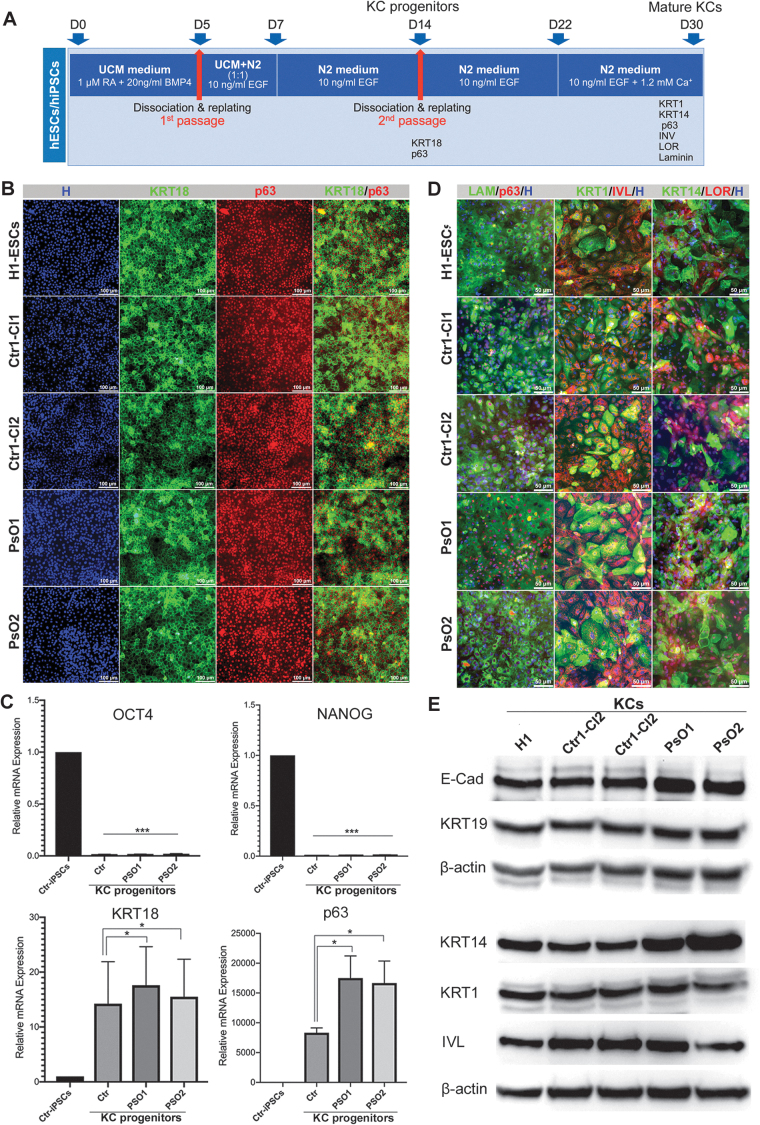
FIG. 3.
Differentiation of patient-specific iPSCs into KCs. (A) A schematic overview of iPSC differentiation into KCs. At day 14 of differentiation, the KC progenitor markers (KRT18 and p63) and pluripotency markers (OCT4 and NANOG) were examined using immunostaining (B) and qRT-PCR (C). (D) Immunostaining images showing the coexpression of mature KC markers, KRT14 and LOR, KRT1 and IVL, and p63 and LAM in KCs derived from H1-ESCs, Ctr-iPSCs and PsO-iPSCs at day 30 of differentiation. The nuclei were stained with Hoechst staining. (E) Western blotting showing the expression of mature KC markers, KRT19, KRT14, and IVL in mature KCs at day 30 of differentiation. *P < 0.05, ***P < 0.001. Scale bars = 100 μm. BMP4, bone morphogenetic protein 4; H, Hoechst; EGF, epidermal growth factor; IVL, involucrin; KCs, keratinocytes; LAM, laminin; LOR, loricrin; qRT-PCR, quantitative reverse transcription-PCR; RA, retinoic acid.
Immunostaining
Immunostaining was performed as previously reported [22,23]. The antibody details are described and listed in Supplementary Table S1.
Western blotting
Total protein was extracted from the undifferentiated iPSCs or differentiated KCs using RIPA (Thermo Fisher Scientific). The protein concentration was measured using Pierce™ BCA Protein Concentration Assay Kit (Thermo Fisher Scientific). The proteins were dissolved in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) buffer and transferred to polyvinylidene fluoride membranes. The membranes were incubated overnight at 4°C with primary antibodies and with secondary antibodies for 1 h (see Supplementary Table S1 for the antibody list). Membranes were developed using SuperSignal West Pico Chemiluminescent substrate (Pierce, Loughborough, UK) and visualized using iBright™ CL 1000 Imaging System (Invitrogen).
Alkaline phosphatase assay
For alkaline phosphatase assay, the iPSCs colonies were washed once with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde for 2 min. The colonies were stained using Alkaline Phosphatase Kit SCR004, Merck Millipore) according to the the manufacturer's instructions.
In vitro spontaneous differentiation
Spontaneous differentiation was induced by embryoid bodies (EBs) formation. Undifferentiated iPSCs of the generated clones were dissociated using TrypLE™ (12604013; Gibco) and cultured in low-attachment six-well plates in mTeSR media with 10 μM Rock inhibitor Y27632 (04001201; Stemgent) for the first 24 h followed by transition to EB differentiation medium (DMEM/F12, 20% KOSR, 1% pen–strep, 1 mM l-glutamine, 1% NEAA, and 0.1 mM β-mercaptoethanol). After 4 days in suspension culture, the formed EBs were transferred on Matrigel-coated plates in the same media with changing the media every day. After 10–15 days, the differentiated EBs were examined using immunostaining and reverse transcription polymerase chain reaction (RT-PCR) for the expression of the markers of the three germ layers (ectoderm, mesoderm, and endoderm).
Direct differentiation of the generated hiPSCs into the three germ layers
For ectodermal differentiation, the iPSCs were differentiated using EB formation method. In brief, the dissociated cells were cultured in suspension using the differentiation media consisting of DMEM/F12 with 20% KOSR, 1 mM l-glutamine, 1% NEAA, 1% pen–strep, 0.1 mM β-mercaptoethanol) supplemented with 1 μM of RA (R2626; Sigma-Aldrich) and 0.25 mM vitamin C (A4544; Sigma-Aldrich) for successive 2 to 3 days. For mesodermal differentiation, the cells were cultured for 2 days in MCDB media (Thermo Fisher Scientific) supplemented with 50 ng/mL activin A (338-AC; R&D Systems), 5 μM CHIR99021 (04-0004; Stemgent), and 0.25 mM vitamin C (A4544; Sigma-Aldrich). For endodermal differentiation, the cells were cultured for 1 day in MCDB media (Thermo Fisher Scientific) supplemented with 100 ng/mL activin A (338-AC; R&D Systems), 2 μM CHIR99021 (04-0004; Stemgent), and 0.25 mM vitamin C (A4544; Sigma-Aldrich) then followed by 2 days culture with 100 ng/mL activin A (338-AC; R&D Systems) and 5 ng/mL of bFGF (Stem Cell Technologies).
hPSC ScoreCard assay
EBs were formed as discussed previously, and 1 μg of RNA was used to prepare the template cDNA using the High Capacity cDNA Reverse Transcription kit (4374966; Applied Biosystems, CA). TaqMan® hPSC Scorecard™ Kit 96w fast assays (A15876; Life Technologies) was run on a QuantStudio7 Flex Real-Time PCR system (Applied Biosystems) using the “hpsc-ScoreCard-template-QuantStudio7-96-well” template according to manufacturer's instructions. Data analysis was performed through the cloud-based TaqMan hPSC Scorecard analysis software, which is available online at www.lifetechnologies.com/scorecarddata.
Karyotyping
The karyotyping status and chromosome spread in the generated iPSC clones was investigated using the Giemsa banding technique. In brief, the 60%–70% confluent cells were cultured for 2 h in KaryoMAX colcemid solution (Thermo Fisher Scientific) with a final concentration of 100 ng/mL to obtain a large number of metaphase spreads. Cells were dissociated with trypsin, followed by treatment with hypotonic solution; KaryoMAX 75 mM potassium chloride solution (Thermo Fisher Scientific) for 20 min at 37°C, then fixed with solution composed of methanol/glacial acetic acid (3:1). Ice-cold cell suspension was dropped onto cleaned slides and aged for 1 h at 90°C. Slides were stained with Giemsa after trypsin treatment. Twenty metaphases each were analyzed with a mean resolution of 200–300 bands per haploid chromosome set. The karyotype formula is given according to ISCN 2016 [24] and indicated as composite karyotype [cp20] reflecting the sum of 20 metaphases analyzed.
Short tandem repeat profiling
DNA were extracted from all the generated iPSC clones using DNA purification kit (NORGEN, BIOTEK). The genetic signature of the samples was detected using AmpFlSTR® Identifiler® Plus PCR Amplification Kit (Applied Biosynthesis Life Technologies) based on the multiplex assay of 15 tetranucleotide repeat loci and the Amelogenin gender-determining marker. PCR was amplified following the manufacture's instruction. The polymerase chain reaction (PCR) products were run in the 3,500/3,500 × L genetic analyzer (Applied Biosystems, Life Technologies) and analyzed using the GeneMapper® ID Software (Applied Biosystems, Life Technologies) following the manufacturer's recommendations.
RNA extraction, reverse transcription polymerase chain reaction, and real-time PCR
The total RNA was extracted from iPSCs and H1-hESCs using RNeasy Plus Mini Kit (QIAGEN) or Direct-zol RNA Extraction Kit (Zymo Research) following the manufacturer's instructions. cDNAs were synthesized from 1 μg of RNA using superscript IV, First-Strand Synthesis System (Thermo Fisher Scientific). PCR-Master mix (Thermo Fisher Scientific) used for amplification of the specified genes by conventional PCR. Real-time RT-PCR was performed using GoTaq qPCR Master Mix (Promega) and amplification was detected using Quant Studio 7 system (Applied Biosystems). The primer details were listed in Supplementary Table S2.
RNA sequencing and data analysis
RNA was purified from at least two biological replicates for each sample of mature KCs using Direct-zol RNA Extraction Kit (Zymo Research). One micrograms of total RNA was used to capture mRNA using NEBNext (Poly A) mRNA Magnetic Isolation Kit (NEB, E7490) according to manufacturer's instructions. RNA-sequencing (RNA-seq) libraries were generated using NEBNext Ultra Directional RNA Library Prep Kit (NEB, E7420L) and sequenced on an Illumina Hiseq 4000 system. Basic trimming and quality control were performed during the conversion of raw data to fastq using Illumina BCL2Fastq Conversion Software v2.20. The STAR aligner used in the following steps performs local as opposed to end-to-end alignment and thus automatically trims adaptor sequences. The FASTQ files of the RNA-seq reads were aligned to the UCSC hg38 reference genome using the STAR aligner with default parameters [25]. STAR produced uniquely mapped reads into a BAM file, which was then passed to Regtools to extract exon–exon junctions. The average length of mapped reads for the samples was between 289 and 292 nucleotides. The deep coverage of the sequencing experiment per sample are summarized in Supplementary Table S3.
Cuffdiff was then used with default settings for differential expression analysis of genes [26]. Cuffdiff allows for comparing samples with varying replicate numbers and compute significance of the observed change between samples, and outputs a P value for uncorrected test statistics and q value for false discovery rate (FDR)-adjusted P value using Benjamini-Hochberg correction for multiple testing. For normalization purposes, Cuffdiff computes fragment per kilobase per million mapped reads (FPKM), which takes into account local and global differences in the distribution of mapped read. For identifying differentially expressed genes (DEGs), we only considered genes with FPKM >0.5 and P value <0.05. For visualization purposes (Venn diagrams and Heatmaps), the replicates were combined.
Gene ontology analysis was performed using the Molecular Signature Data Base (MSigDB 6.2) software.
Glucose uptake assay
The differentiated KC progenitors at day 14 were dissociated and plated on 96-well plates at 1–2 × 104 cells/well and glucose uptake by these cells was monitored at day 30 using the fluorescent D-glucose analog (2-NBDG) according to the glucose uptake cell-based assay (Cat. no. 600470; Cayman, Ann Arbor, MI). The cells were glucose-starved with Krebs buffer for 4 h. Then one group of cells was treated with final concentration of 200 μg/mL of 2-NBDG and the other group was treated with 100 nM insulin along with 200 μg/mL of 2-NBDG for 3 h. After washing twice with the buffer, the retained fluorescence was measured with FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany) at excitation/emission wavelengths of 485 and 535 nm. Fold increase in glucose uptake in response to insulin was compared to its basal uptake in nontreated cells.
Proliferation assay
The proliferation assay was performed as previously reported [22]. In brief, the KC progneitors (at day 14) and the mature KCs (at day 30) of differentiation were treated with BrdU (1:100; Thermo Fisher Scientific) for 6 h and then were dissociated using TrypLE before fixation with 70% ethanol overnight. The fixed cells were denatured with 2 M HCl containing 0.5% Triton X-100 and neutralized by 0.1 M sodium borate for 10 min. Cells were incubated for 2 h at room temperature with anti-BrdU antibody, Alexa Fluor 488 (1:100, Cat. no. B35130; Thermo Fisher Scientific) in 2% bovine serum albumin in PBS. The results were acquired on BD Accuri™ C6 flow cytometer (BD Biosciences) and the data were analyzed using FlowJo software.
Statistical analysis
The results are expressed as mean ± standard deviation, as indicated in the figure legends. Statistical significance was examined by two-tailed Student's t-tests. Values of P < 0.05 were considered significant.
Data availability
The RNA-seq datasets generated and used in the present study are available on the Zenodo repository at https://doi.org/10.5281/zenodo.3484611.
Results
Generation and characterization of iPSCs from patients with psoriasis
iPSCs were generated from PBMCs isolated from two healthy donor controls (Ctr1-iPSCs and Ctr2-iPSCs) and two patients with psoriasis who also had insulin resistance and family history of psoriasis (PsO1-iPSCs and PsO2-iPSCs) (Fig. 1A). From each sample, several iPSC lines were generated, and only three fully reprogrammed iPSC lines were maintained from each sample. In the current study, we used six hiPSC lines from patients with psoriasis (PsO1 and PsO2) and six hiPSC lines from healthy controls (Ctr1 and Ctr2). Morphologically, the iPSC clones showed similar characteristics to hESCs (Fig. 1B). All iPSC lines were positive for alkaline phosphatase (ALP) activity and expressed pluripotency markers, including OCT4, SOX2, NANOG, SSEA4, TRA1-60, TRA1-81, KLF4, DPPA4, REX-1, and TERT, as examined using immunostaining, western blotting, and RT-PCR (Fig. 1B–D, Supplementary Figs. S1 and S2). The iPSC clones showed a normal karyotype using the G-banding technique (Fig. 1E) and lost the expression of transduced pluripotency markers and Sendai virus backbone at passage 10–15 (Supplementary Figs. S1 and S2). Short tandem repeat (STR) analysis showed that the genetic profiling of all the three hiPSC lines established from the same subject were identical (Fig. 1F).
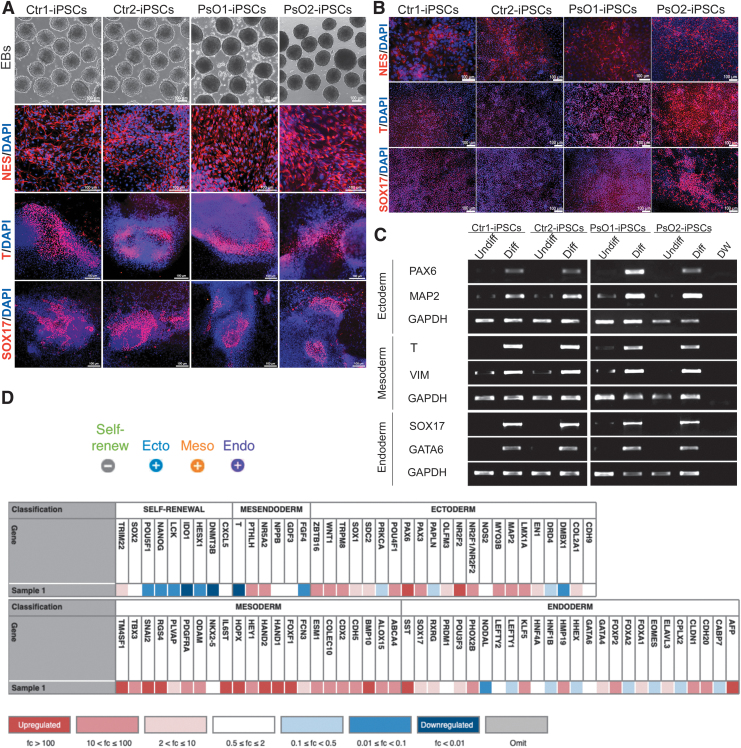
To further confirm the ability of the generated iPSCs to differentiate into the three germ layers, spontaneous and direct differentiation were performed, using the EB technique (Fig. 2A) and direct differentiation protocols (Fig. 2B), respectively. Immunostaining results showed the expression of ectodermal (NESTIN), mesodermal (BRACHYURY), and endodermal (SOX17) markers (Fig. 2A, B, Supplementary Figs. S1 and S2). RT-PCR analysis showed the expression of MAP2 and PAX6 (ectoderm), BRACHYURY and VIMENTIN (mesoderm), and SOX17 and GATA6 (endoderm) (Fig. 2C).
FIG. 2.
Characterization of the generated iPSC lines. Phase contrast of the spontaneously generated EBs from representative Ctr-iPSCs and PsO-iPSCs. Immunostaining showed the expression of the three germ layer markers, NESTIN, BRACHYURY (T), and SOX17 after spontaneous differentiation (A) and direct differentiaiton (B). (C) RT-PCR analysis of the three germ layer markers as indicated. The undifferentiated iPSCs of each sample used as a control. (D) Scorecard analysis of the EBs derived from iPSCs showing a represetnative heatmap of the indicated genes (self-renewal, ectodermal, mesodermal, and endodermal genes). Note that the expression of pluripotency markers were significantly downregulated and the trilineage markers were significantly upregulated in the representative images of Ctr1-iPSCs and PsO2-iPSCs. Scale bars = 100 μm. DW, distal water; EBs, embryoid bodies.
The pluripotent ability of the generated iPSCs was confirmed through a comprehensive real-time PCR TaqMan hPSC Scorecard Panel, consisting of 94 individual q-PCR assay including a combination of lineage-specific, self-renewal, control, and housekeeping genes. The scorecard results showed that the iPSCs were able to differentiate spontaneously into the three germ layers with loss of pluripotency marker expression (Fig. 2D).
Differentiation of patient-specific iPSCs into KCs
To identify the genetic defects in KCs associated with psoriasis, we sought to differentiate Ctr-iPSCs and PsO-iPSCs into KCs (Fig. 3A). H1-hESCs, Ctr-iPSCs, and PsO-iPSCs were differentiated into KC progenitors (Fig. 3B, C) and mature KCs (Fig. 3D, E). At day 14 of differentiation (KC progenitors), the cells were examined using immunostaining, real-time PCR (Fig. 3B, C), and RT-PCR (Supplementary Fig. S3). The results showed robust downregulation in the expression of the pluripotency markers in KC progenitors in comparison to the undifferentiated Ctr-iPSCs (Fig. 3C), confirming the loss of pluripotency. In contrast, all the differentiated cells showed significant increased expression of epithelial marker keratin 18 (KRT18) and KC marker p63 as confirmed by immunostaining and real-time PCR (Fig. 3B, C), indicating the generation of cell populations committed to ectodermal lineage resembling KC progenitors. Interestingly, we noticed a significant upregulation in the expression of p63 mRNA in KC progenitors derived from both PsO-iPSCs (KCs-PsO) in comparison to those derived from Ctr-iPSCs (KCs-Ctr) (Fig. 3C).
At day 30 of differentiation, the cells were examined for the expression of mature KC markers. The immunostaining results showed that the differentiated cells expressed markers of mature KCs, including keratin 14 (KRT14), loricrin (LOR), keratin 1 (KRT1), involucrin (IVL), laminin, and p63 (Fig. 3D, Supplementary Figs. S4–S6). Furthermore, western blot analysis showed the expression of E-Cadherin (E-Cad), KRT19, KRT14, and KRT1 (Fig. 3E). These results confirm the efficient differentiation of different iPSC lines into mature KCs. We noticed that E-Cad and KRT14 were upregulated in KCs-PsO1 and KCs-PsO2 in comparison to KCs-Ctr and KCs-H1 (Fig. 3E). Furthermore, the KCs successfully constructed three-dimensional (3D) skin equivalents and expressed markers of epidermal KCs (Supplementary Data and Supplementary Fig. S7). These results suggested that 3D skin equivalents can be generated from the KCs derived from iPSCs.
Transcriptomic analysis of iPSC-derived KCs
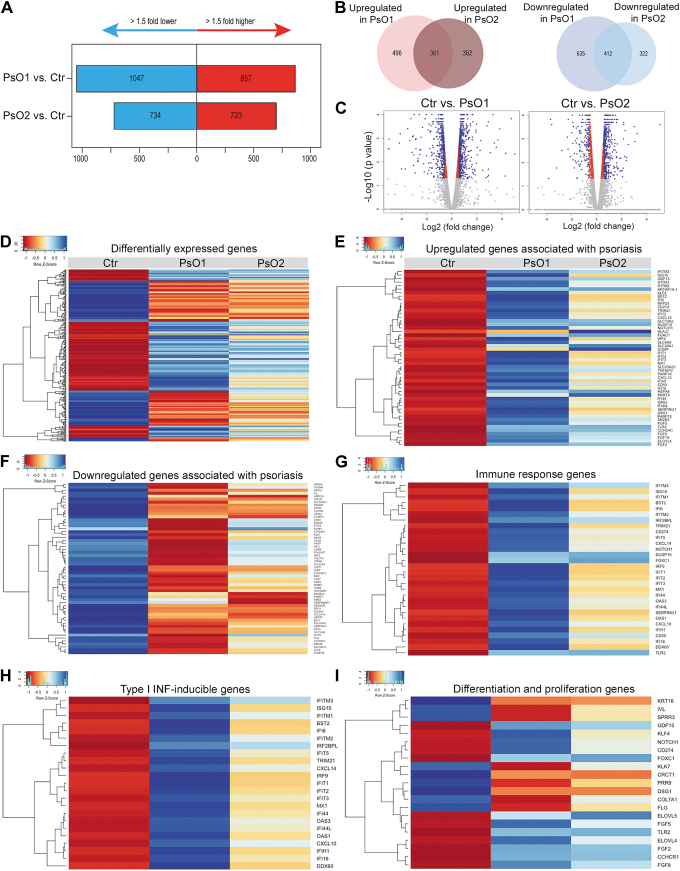
To investigate the differences in the gene expression profiles of KCs-PsO and KCs-Ctr, we performed transcriptomic analysis using RNA-seq. The RNA-seq analysis identified DEGs based on two comparisons (KCs-Ctrl vs. KCs-PsO1 and KCs-Ctrl vs. KCs-PsO2). Comparison of KCs-Ctrl with KCs-PsO1 identified 1,904 DEGs with P value <0.05, of which 857 gene were upregulated [fold change (FC) >1.5] and 1,047 genes were downregulated (FC <0.5) (Fig. 4A, B). In KCs-PsO2, 1,457 genes were differentially expressed with P value <0.05, of which 723 genes were upregulated (FC >1.5), while 734 genes were downregulated (FC <0.5) (Fig. 4A, B).
FIG. 4.
Transcriptomics alterations in KCs derived from PsO-iPSCs. Graphs (A) and Venn diagram (B) showing the number of significantly upregulated and downregulated genes in KCs-PsO1 and KCs-PsO2 in comparison to KCs-Ctr. (C) Volcano plot ploting the log2 fold change and the adjusted P value for all the detected transcripts. (D) A heatmap showing DEGs in KCs-PsO1 and KCs-PsO2 compared to KCs-Ctr. Heatmaps showing upregulated (E) and downregulated (F) genes assoiciated with psoriasis. Heatmaps showing important significantly upregulated genes related to immune response (G) and type I IFN-induced genes (H). (I) A heatmap of genes involved in the differentiation and proliferation of KCs. The relative value for each gene is depicted by color intensity, with blue indicating upregulated and red indicating downregulated genes. DEG, differentially expressed gene; IFN, interferon.
Next, we performed pathway analysis to identify biological functions/pathways enriched among DEGs that are common in KCs derived from both patients; KCs-PsO1 and KCs-PsO2. Among these, 361 genes were commonly upregulated (FC >1.5), while 412 genes were commonly downregulated in both KCs-PsO1 and KCs-PsO2 (FC <0.5) (Fig. 4B). The GO terms for these commonly upregulated genes showed their role in immune response, type I IFN signaling pathway, response to IFN-gamma, and cytokine-mediated signaling pathway, while the commonly downregulated genes were involved with GO terms epidermis development, response to external stimulus, KC differentiation, cell–cell adhesion, and glucose transport (Fig. 4D–I) (Table 1). Although we have identified large number of common DEGs in both PsO-iPSCs, the results showed that the phenotypes were more severe in KCs-PsO1 in comparision to KCs-PsO2 (Fig. 4D–I).
Table 1.
Top Upregulated and Downregulated Genes in KC-PsO1-iPSCs and KC-PsO2-iPSCs Compared with KCs Derived from Ctr-iPSCs (P < 0.05>
| Upregulated genes | |||||
|---|---|---|---|---|---|
| Gene symbol | Gene title | PsO1 vs. Ctr |
PsO2 vs. Ctr |
||
| Log2 FC | P | Log2 FC | P | ||
| IFITM1 | IFN-induced transmembrane protein 1 | 6.82 | 5.00E-05 | 3.95 | 5.00E-05 |
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | 6.73 | 5.00E-05 | 3.39 | 5.00E-05 |
| BST2 | Bone marrow stromal cell antigen 2 | 6.02 | 5.00E-05 | 2.02 | 5.00E-05 |
| IFIT2 | IFN-induced protein 44 | 5.92 | 5.00E-05 | 2.32 | 5.00E-05 |
| IFI44 | IFN-induced protein 44 like | 5.85 | 5.00E-05 | 2.67 | 5.00E-05 |
| IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 | 5.81 | 5.00E-05 | 2.03 | 5.00E-05 |
| IFI44L | IFN-induced protein 44 like | 5.64 | 5.00E-05 | 2.92 | 5.00E-05 |
| OAS3 | 2′-5′-oligoadenylate synthetase | 5.44 | 5.00E-05 | 3.01 | 5.00E-05 |
| MX1 | MX dynamin like GTPase 1 | 5.28 | 5.00E-05 | 2.11 | 5.00E-05 |
| OAS1 | 2′-5′-oligoadenylate synthetase 1 | 5.11 | 5.00E-05 | 2.17 | 5.00E-05 |
| IFI6 | IFN alpha inducible protein 6 | 4.72 | 5.00E-05 | 1.73 | 5.00E-05 |
| IFITM2 | IFN-induced transmembrane protein 2 | 4.45 | 5.00E-05 | 2.67 | 5.00E-05 |
| IFITM3 | IFN-induced transmembrane protein 3 | 4.42 | 5.00E-05 | 3.46 | 5.00E-05 |
| ISG15 | ISG15 ubiquitin-like modifier | 4.20 | 5.00E-05 | 1.62 | 5.00E-05 |
| IFI16 | IFN gamma inducible protein 16 | 2.19 | 5.00E-05 | 1.09 | 5.00E-05 |
| IFIH1 | IFN-induced with helicase C domain 1 | 1.91 | 5.00E-05 | 0.77 | 5.00E-05 |
| IRF9 | IFN regulatory factor 9 | 1.48 | 5.00E-05 | 0.55 | 0.0036 |
| IFIT5 | IFN-induced protein with tetratricopeptide repeats 5 | 2.69 | 5.00E-05 | 1.42 | 5.00E-05 |
| CXCL14 | C-X-C motif chemokine ligand 14 | 2.54 | 5.00E-05 | 1.44 | 5.00E-05 |
| CXCL10 | C-X-C motif chemokine ligand 10 | 2.32 | 0.00035 | 1.41 | 0.00345 |
| SERPING1 | Serpin family G member 1 | 4.37 | 5.00E-05 | 1.44 | 5.00E-05 |
| SH2D5 | SH2 domain containing 5 | 3.93 | 5.00E-05 | 2.77 | 5.00E-05 |
| PARP12 | Poly (ADP-ribose) polymerase family member 12 | 3.71 | 5.00E-05 | 1.99 | 5.00E-05 |
| PARP14 | Poly (ADP-ribose) polymerase family member 14 | 2.82 | 5.00E-05 | 1.35 | 5.00E-05 |
| KRTAP19–1 | Keratin-associated protein 19–1 | 3.27 | 5.00E-05 | 3.70 | 5.00E-05 |
| GDF15 | Growth differentiation factor 15 | 3.26 | 5.00E-05 | 2.80 | 5.00E-05 |
| HSPA6 | Heat shock protein family A (Hsp70) member 6 | 2.58 | 0.0001 | 4.37 | 5.00E-05 |
| RPP25 | Ribonuclease P and MRP subunit p25 | 2.32 | 5.00E-05 | 1.24 | 0.0003 |
| HLA-C | MHC, class I, C | 1.10 | 0.0082 | 4.55 | 5.00E-05 |
| CD274 | CD274 molecule | 2.13 | 5.00E-05 | 1.24 | 5.00E-05 |
| CD55 | CD55 molecule | 1.65 | 5.00E-05 | 1.21 | 5.00E-05 |
| TNFAIP2 | TNF-alpha-induced protein 2 | 2.04 | 5.00E-05 | 1.53 | 5.00E-05 |
| TRIM21 | Tripartite motif containing 21 | 2.02 | 5.00E-05 | 0.81 | 0.00015 |
| SLC6A8 | Solute carrier family 6 member 8 | 1.74 | 5.00E-05 | 1.05 | 5.00E-05 |
| SLC25A25 | Solute carrier family 25 member 25 | 1.71 | 5.00E-05 | 1.18 | 5.00E-05 |
| SLC20A1 | Solute carrier family 20 member 1 | 1.33 | 5.00E-05 | 1.89 | 5.00E-05 |
| SLC19A2 | Solute carrier family 19 member 2 | 1.28 | 5.00E-05 | 1.27 | 5.00E-05 |
| KLF4 | Kruppel like factor 4 | 1.66 | 5.00E-05 | 0.88 | 5.00E-05 |
| PRRT4 | Proline rich transmembrane protein 4 | 1.50 | 5.00E-05 | 2.78 | 5.00E-05 |
| FOXC1 | Forkhead box C1 | 1.30 | 5.00E-05 | 3.87 | 5.00E-05 |
| S100P | S100 calcium-binding protein P | 1.15 | 5.00E-05 | 2.88 | 5.00E-05 |
| S100A14 | S100 calcium-binding protein A14 | 0.73 | 0.00015 | 0.69 | 5.00E-05 |
| DUSP10 | Dual specificity phosphatase 10 | 1.07 | 5.00E-05 | 1.19 | 5.00E-05 |
| SOCS1 | Suppressor of Cytokine Signaling 1 | 1.01 | 0.00505 | 1.30 | 0.00005 |
| NOTCH1 | Notch receptor 1 | 0.99 | 5.00E-05 | 0.61 | 5.00E-05 |
| Downregulated genes | |||||
|---|---|---|---|---|---|
| |
|
PsO1 vs. Ctr |
PsO2 vs. Ctr |
||
| Gene symbol | Gene title | Log2 FC | P | Log2 FC | P |
|
SLC38A11 |
Solute carrier family 38 member 11 |
−6.15 |
5.00E-05 |
−2.93 |
5.00E-05 |
|
SLC22A3 |
Solute carrier family 22 member 3 |
−3.29 |
5.00E-05 |
−1.18 |
5.00E-05 |
|
SLC12A8 |
Solute carrier family 12 member 8 |
−3.16 |
5.00E-05 |
−2.45 |
5.00E-05 |
|
SLC2A14 |
Solute carrier family 2 member 14 |
−1.94 |
5.00E-05 |
−2.55 |
5.00E-05 |
|
SLC6A1 |
Solute carrier family 6 member 1 |
−2.06 |
5.00E-05 |
−2.12 |
5.00E-05 |
|
SLC9A3R1 |
SLC9A3 regulator 1 |
−1.10 |
5.00E-05 |
−0.64 |
5.00E-05 |
|
UCN2 |
Urocortin 2 |
−5.02 |
0.00465 |
−1.41 |
5.00E-05 |
|
ABCA4 |
ATP-binding cassette subfamily A member 4 |
−4.75 |
5.00E-05 |
−1.78 |
5.00E-05 |
|
PAEP |
Progestagen-associated endometrial protein |
−4.74 |
0.00015 |
−0.93 |
5.00E-05 |
|
GFI1 |
Growth factor-independent 1 transcriptional repressor |
−4.66 |
5.00E-05 |
−1.21 |
5.00E-05 |
|
KLK7 |
Kallikrein-related peptidase 7 |
−4.54 |
5.00E-05 |
−1.94 |
5.00E-05 |
|
CYP26A1 |
Cytochrome P450 family 26 subfamily A member 1 |
−4.29 |
0.0043 |
−2.33 |
5.00E-05 |
|
CFTR |
Cystic fibrosis transmembrane conductance regulator |
−7.95 |
5.00E-05 |
−0.60 |
0.0001 |
|
ICOS |
Inducible T cell costimulator |
−4.25 |
5.00E-05 |
−2.71 |
5.00E-05 |
|
PRR9 |
Proline rich 9 |
−3.67 |
0.0012 |
−2.28 |
5.00E-05 |
|
PITX2 |
Paired-like homeodomain 2 |
−3.56 |
5.00E-05 |
−0.91 |
5.00E-05 |
|
COL7A1 |
Collagen type VII alpha 1 chain |
−3.51 |
5.00E-05 |
−1.50 |
5.00E-05 |
|
IL2RB |
Interleukin 2 receptor subunit beta |
−3.46 |
5.00E-05 |
−0.91 |
5.00E-05 |
|
HLA-DQA1 |
MHC, class II, DQ alpha 1 |
−3.39 |
5.00E-05 |
−1.83 |
5.00E-05 |
|
KCNF1 |
Potassium voltage-gated channel modifier subfamily F member 1 |
−3.37 |
5.00E-05 |
−2.41 |
5.00E-05 |
|
CYP24A1 |
Cytochrome P450 family 24 subfamily A member 1 |
−3.28 |
5.00E-05 |
−2.26 |
5.00E-05 |
|
S100A9 |
S100 calcium-binding protein A9 |
−3.14 |
5.00E-05 |
−1.29 |
5.00E-05 |
|
CGB8 |
Chorionic gonadotropin subunit beta 8 |
−3.07 |
0.0041 |
−2.22 |
0.00045 |
|
ANXA8 |
Annexin A8 |
−3.04 |
5.00E-05 |
−0.73 |
5.00E-05 |
|
STRA6 |
Stimulated by retinoic acid 6 |
−3.03 |
5.00E-05 |
−1.24 |
5.00E-05 |
|
IVL |
Involucrin |
−3.01 |
5.00E-05 |
−1.54 |
5.00E-05 |
|
SERPINA1 |
Serpin family A member 1 |
−2.97 |
5.00E-05 |
−1.53 |
5.00E-05 |
|
NEFL |
Neurofilament ligh |
−2.91 |
0.0002 |
−0.68 |
0.0197 |
|
PGLYRP4 |
Peptidoglycan recognition protein 4 |
−2.85 |
5.00E-05 |
−1.99 |
5.00E-05 |
|
KRT3 |
Keratin 3 |
−2.80 |
0.0003 |
−2.05 |
5.00E-05 |
|
KRT16 |
Keratin 16 |
−1.69 |
5.00E-05 |
−1.56 |
5.00E-05 |
|
CD52 |
CD52 molecule |
−2.72 |
0.046 |
−1.59 |
0.00165 |
|
NR0B1 |
Nuclear receptor subfamily 0 group B member 1 |
−2.70 |
5.00E-05 |
−1.16 |
0.0002 |
|
EBI3 |
Epstein-Barr virus induced 3 |
−2.58 |
5.00E-05 |
−0.88 |
5.00E-05 |
|
PCDHB13 |
Protocadherin beta 13 |
−2.53 |
5.00E-05 |
−1.67 |
5.00E-05 |
|
PCDHGC5 |
Protocadherin gamma subfamily C, 5 |
−1.21437 |
0.03365 |
−1.0337 |
0.03975 |
|
EDN1 |
Endothelin 1 |
−2.51 |
5.00E-05 |
−0.70 |
0.00025 |
|
SPRR3 |
Small proline rich protein 3 |
−2.48 |
5.00E-05 |
−1.29 |
5.00E-05 |
|
PLA2G2F |
Phospholipase A2 group IIF |
−2.39 |
5.00E-05 |
−0.89 |
0.0002 |
|
GJB4 |
Gap junction protein beta 4 |
−2.35 |
5.00E-05 |
−2.08 |
5.00E-05 |
|
BST1 |
Bone marrow stromal cell antigen 1 |
−2.32 |
5.00E-05 |
−1.96 |
5.00E-05 |
|
C2orf54 |
Mab-21 like 4 |
−2.14 |
5.00E-05 |
−1.99 |
5.00E-05 |
|
CRCT1 |
Cysteine rich C-terminal 1 |
−2.13 |
5.00E-05 |
−2.19 |
5.00E-05 |
|
UCP2 |
Uncoupling protein 2 |
−1.81 |
5.00E-05 |
−1.54 |
5.00E-05 |
|
UCHL1 |
Ubiquitin C-terminal hydrolase L1 |
−1.75 |
5.00E-05 |
−1.90 |
5.00E-05 |
|
RWDD2B |
RWD domain containing 2B |
−1.72 |
5.00E-05 |
−1.95 |
5.00E-05 |
|
GPR78 |
G protein-coupled receptor 78 |
−1.67 |
0.0002 |
−1.96 |
5.00E-05 |
|
DSG1 |
Desmoglein 1 |
−1.63 |
5.00E-05 |
−1.56 |
5.00E-05 |
|
PROM2 |
Prominin 2 |
−1.57 |
5.00E-05 |
−1.50 |
5.00E-05 |
|
CYSRT1 |
Cysteine rich tail 1 |
−1.35 |
0.00105 |
−1.90 |
5.00E-05 |
| PAMR1 | Peptidase domain containing associated with muscle regeneration 1 | −1.29 | 5.00E-05 | −1.96 | 5.00E-05 |
FC, fold change; IFN, interferon; iPSC, induced pluripotent stem cell; KC, keratinocyte; MHC, major histocompatibility complex.
We observed significant upregulation in several antiviral (type I IFN-indcible) genes, including IFITM1, IFIT3, IFIT2, IFI44, IFI44L, IFIT1, IFI6, IFITM2, IFITM3, IFIT3, IFIT5, IFI16, IFIH1, IRF9, IRF2BPL, BST2, OAS3, OAS1, MX1, ISG15, IFIH1, DDX60, TRIM21, CXCL14, and CXCL10, in the differentiated KCs derived from both PsO-iPSCs (Fig. 4H) (Table 1). In addition to the antiviral genes, several immune response and chemokine genes were significantly upregulated, including CD55, CD274, SERPING1, TRIM21, IRF9, FOXC1, DUSP10, NOTCH1, IRF2BPL, CXCL10, and CXCL14 (Fig. 4G) (Table 1).
Among the signifcantly upregulated genes, there are genes reported by previous studies to be associated with psoriasis, such as immune response genes, HLA-C, KLF4, SERPING1, TNFAIP2, S100 calcium-binding proteins (S100P and S100A14) , CD274, NOTCH1, SOCS1, FOXC1, SLC20A1, SLC19A2, SLC25A25, SLC6A8, SLC20A1, and KRTAP19-1, while the downregulated genes, such as solute carrier genes (SLC38A11, SLC22A3, SLC12A8, SLC2A14, and SLC6A1), PRR9, and HLA-DQA1 (Fig. 4F) (Table 1). We also found several downregulated genes associated with KC differentiation, such as IVL, DSG1, KRT3, KRT16, KLK7, STRA6, PCDHB13, PCDHGC5, and PROM2 (Fig. 4I). Interestingly, some DEGs associated with insulin resistance and type 2 diabetes (T2D) were observed to be upregulated, such as IRS2 and GDF15, or downregulated, such as SLC2A14 and SLC2A10 (Table 1) (Supplementary Table S4).
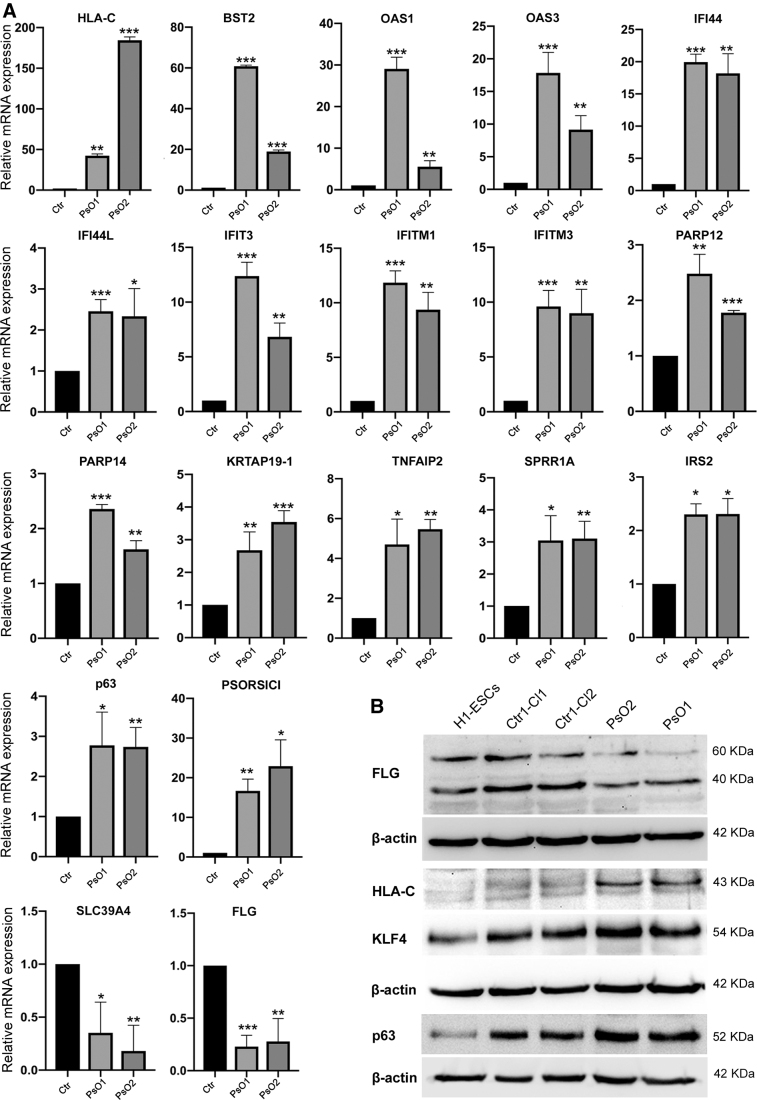
To confirm the RNA-seq data, we validated selected genes dysregulated in KCs-PsO of both subjects using quantitative reverse transcription-PCR (qRT-PCR) and western blotting (Fig. 5A, B), which were consistent with the RNA-seq data. Our qRT-PCR confirmed the significant upregulation of HLA-C, BST2, OAS1, OAS3, IFI44, IFI44L, IFIT3, IFITM1, IFITM3, PARP12, PARP14, KRTAP19-1, TNFAIP2, SRR1A, IRS2, p63, and PSORS1C1. In contrast, SLC39A4 and FLG were significantly downregulated (Fig. 5A)), as observed by RNA-seq. At the protein level, western blotting confirmed the upregulation of HLA-C, KLF4 and p63 and the downregulation of FLG (Fig. 5B).
FIG. 5.
Validation of RNA-seq data using real-time PCR (qRT-PCR) and western blot. (A) qRT-PCR for the main genes deregulated in the RNA-seq results. Graphs show mean with SEM of three independent replicates and data were statistically analyzed using unpaired t-test. (B) Validation of gene expression using western blotting. *P < 0.05, **P < 0.01, ***P < 0.001. RNA-seq, RNA-sequencing.
Alterations in the proliferation and glucose uptake of KCs derived from PsO-iPSCs
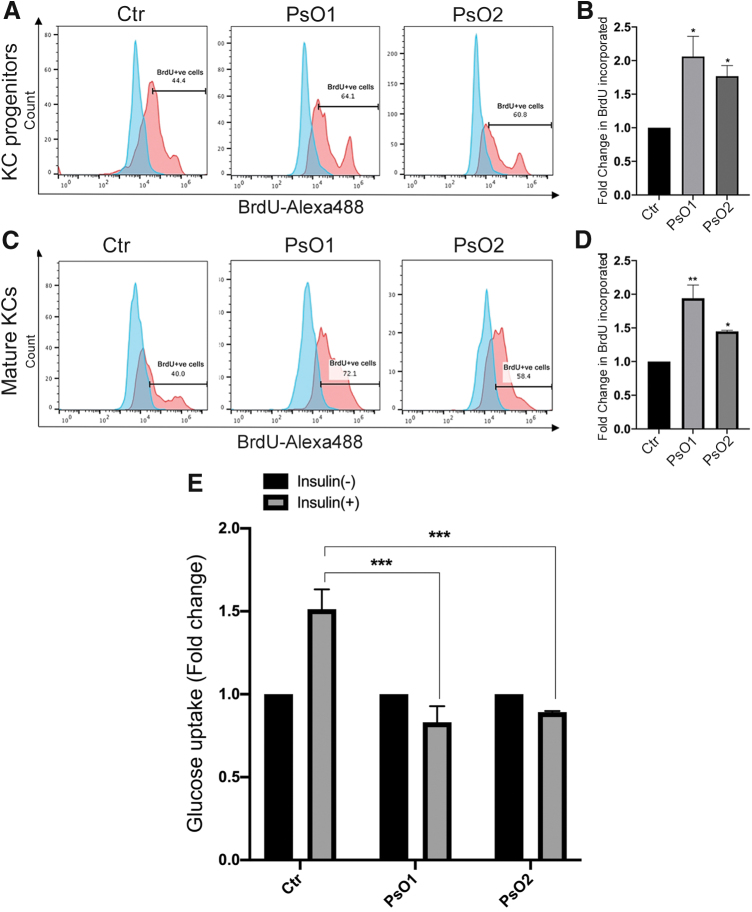
Psoriaisis is associated with hyperproliferation; therefore, we examined the proliferation capacity of the differentiated KCs derived from PsO-iPSCs in comparision to those of Ctr-iPSCs. As expected, the number of BrdU-positive cells increased significantly in KC progenitors (day 14) and mature KCs (day 30) derived from PsO-iPSCs compared to those derived from Ctr-iPSCs reflecting an increase in cell proliferation (Fig. 6A–D). These results indicate a genetic cause for the hyperproliferation of psoriatic KCs.
FIG. 6.
Alterations in the proliferation and glucose uptake of KCs derived from PsO-iPSCs. Flow cytometry analysis of BrdU incorporation showing increased cell proliferation (BrdU+cells) in KC progenitors (A, B) and mature KCs (C, D) derived from PsO-iPSCs in comparision to those derived from Ctr-iPSCs (n = 2). (E) Glucose uptake assay showing no change in glucose uptaken by the mature KCs-PsO in response to insulin treatment; however, KCs-Ctr showed a significant increase in insulin-induced glucose uptake at day 30 of differentiation (n = 2). Fluorescence units for glucose uptake for each sample induced with insulin was normalized to the basal. *P < 0.05, **P < 0.01, ***P < 0.001. Color images are available online.
To confirm our RNA-seq results showing defects in genes involved in insulin resistance, and to correlate the insulin-resistant phenotype observed in these psoriatic patients, we sought to test the insulin sensitivity in the generated KCs. A glucose uptake assay was performed at day 30 of differentiation. Our results showed no change in glucose uptaken by the mature KCs-PsO in response to insulin treatment; however, KCs-Ctr showed a significant increase in insulin-induced glucose uptake (Fig. 6E). These results indicate that psoriatic KCs possess genetic defects that confer insulin resistance.
Discussion
There has been an ongoing debate regarding the role of KCs in psoriasis being a secondary event, triggered by immune stimulation. Our iPSC model established here resolves this by illustrating that the hallmarks of psoriasis disease, hyperproliferation and abnormal differentiation of the KCs, and inflammatory response, are primarily triggered due to the genetic defects in the KCs. These findings suggest that psoriasis is not purely a T cell-dependent disorder, but genetic alterations of KCs are likely to play a major role in psoriasis pathogenesis. Furthermore, our results also uncover a genetic link between insulin resistance and psoriasis.
Genetic defects in the KCs could disrupt epidermal homeostasis in psoriasis. Our RNA-seq analysis revealed that genes involved in appropriate epidermis stratification and granular layer were dysregulated in the psoriatic KCs. Specifically, p63, a major player in differentiation of the ectodermal-specified cells to epidermal progenitors [27,28], was significantly upregulated in the KCs-PsO, which is in line with high expression of p63 in psoriatic skin as previously demonstrated [29,30]. In addition, we found that NOTCH1, which is downstream of p63 [29], was also significantly activated in KCs-PsO. Interestingly, Notch signaling is activated in the epidermis by cilia for balancing proliferation and differentiation of the epidermal cells [31], and has been previously shown to be upregulated in the psoriatic skin [32,33]. Another psoriasis susceptibility locus, KLF4 [34], which is required for differentiation and specification of the skin epithelium [35], was found to be increased in the KCs-PsO compared to KCs-Ctr, consistent with previous reports detailing its increased expression in psoriatic KCs [36,37]. In addition, the granualar layer was shown to be dramatically affected in the psoriatic skin [38]. Our results indicate that this defect originates due to decreased expression of FLG, IVL, and DSG1, responsible for granular layer differentiation, in KCs-PsO. Therefore, our results demonstrate that signaling pathways affected in the psoriatic skin according to previous reports are due to the genetic defects in KCs. Nonetheless, these genetic defects in KCs may be the cause of its hyperproliferation and inappropriate differentiation pathology in psoriasis. Nonetheless, these genetic defects in KCs may be the cause of its hyperproliferation, as indicated by our results showing an increased proliferation of KCs-PsO compared with KCs-Ctr and inappropriate differentiation pathology in psoriasis.
While psoriasis is a multifactorial disease, it is noteworthy that multiple transcriptome studies on psoriatic lesional and nonlesional skin biopsies have made it possible to comprehensively study the pathogenesis of psoriasis [39–41]. One of the limitations to prior transcriptome studies is that the lesions analysed contain multiple types of cells constituting and infiltrating the epidermis, such as fibroblasts and immune cells, which has hindered determination of genes exclusively associated with KC dysfunction [14,41–44]. Our study is uniquely different from these as KCs from psoriatic iPSCs, unadulterated by other cell types, were analysed to obtain DEGs. Indeed, analysis of pure population of sorted epidermal cells in lesional and nonlesional psoriasis has shed light on cell type-specific gene expression alterations [45]; however, it is still limited in concluding if these changes are primary or if they are cytokine-induced transcriptional responses. Our iPSC model resolves this debate as any DEGs identified are due to genetic inheritance and not a secondary transcriptional response to immune system activation and KC hyperplasia. Hence, our model distinguishes between genetic and acquired factors involved in psoriasis development.
The sequence of the pathogenic events triggering psoriasis development is not completely understood, however, it is thought to be initiated by defects in the immune response. It is debated that there occurs a defective T cell response stimulating proliferation that further activates normal KCs to hyperproliferate [46–49], and the resulting crosstalk between them sustains psoriasis progression [14,50–52]. It has been thought that in psoriatic patients, the inflammation and T cell activation occur before the hyperproliferation of the epidermis [49]. Our study showed that the class I major histocompatibility complex (MHC) allele HLA-C was significantly upregulated in the KCs derived from subjects with psoriasis. This finding is consistent with a previous study that reported the upregulation of HLA-C in the epidermis of patients with psoriasis [6,10,53]. HLA-C encodes a MHC class I receptor that participates in immune responses via antigens presentation to CD8+ T lymphocytes and has been previously reported as the strongest susceptibility factor for psoriasis [10,54]. Our results, therefore, suggest that genetically defective KCs in psoriasis might lead to T cell activation in the epidermis in contrast to the conventional hypothesis that genetic defects in the immune cells stimulate abnormal immune response.
Studies on profiling of psoriatic lesions have consistently highlighted the detrimental role of upregulated antiviral and chemokine genes in activating and attracting immune cells to the epidermis [45,55–62]. We show here that the origin of this inflammation in the psoriatic lesions is in the genetically defected KCs; as KCs generated from PsO-iPSCs showed significant upregulation of type I INF-inducible, immune response, and chemokine genes. These results agree with the accumulating evidences suggesting that KCs have a substantial influence on skin immunity, because they express multiple proinflammatory cytokines and chemokines indicating the existance of the immune system in the epidermal KCs [63,64]. In addition, it is likely that targeting of these genes could attenuate the progression of psoriasis as blocking of INF-alpha pathway in a xenograft model of psoriasis has suppressed the development of this disease [65]. Therefore, our KC-PsO-iPSCs carrying the genetic defects that cause the disease are an excellent model for studies on drug screening for psoriasis treatment.
Interestingly, several association and clinical studies have eluded to a potential link between altered gene transcription in the psoriatic skin and comorbid conditions, such as insulin resistance, T2D, and cardiovascular dieases [66]. In KCs, insulin-signaling pathway regulate proliferation and differentiation processes. KC differentiation is enhanced by insulin and is inhibited by IGF-1 [67]. The disruption in the balance between these two pathways may result in insulin resistance, leading to defects in the epidermal KCs. We found that IRS2 was significantly upregulated in KCs-PsO1 and KCs-PsO2 in comparision to KCs-Ctr. IRS2 deletion in vivo (IRS2-knockout mice model) and in vitro in KCs showed that the inhibition of this gene increases glucose transport in epidermal KCs [68], which is opposite to its function in other tissues [69,70]. Furthermore, IRS2 overexpression in KCs leads to a dramatic reduction in glucose uptake in KCs stimulated with insulin [68]. In contrast, it has been reported that inhibition of insulin receptor and IRS1 in the KCs leads to a reduction in the glucose transport rate [68,71]. We found that insulin was unable to stimulate uptake of glucose in mature KCs-PsO when compared to KCs-Ctrl, thereby validating the genetic origin of insulin resistance in the psoriatic KCs. Taken together, these results indicate a unique role of IRS2 in the KCs and suggest that the upregulation of IRS2 in the current study may be associated with the inhibition of the glucose transport.
Interestingly, the main glucose transporter in KCs, GLUT1 [72], was not differentially expressed in our study. However, other glucose transporters, including SLC2A10 (GLUT10) and SLC2A14 (GLUT14), were significantly downregulated. A previous study hypothesized that genetic variations in SLC2A14 may be associated with inflammatory disease [73]. Also, SLC2A10 was found to be associated with T2D [74,75]. Interestingly, we found that GDF15, known as a macrophage inhibiting cytokine, was significantly upregulated in KCs of both patients. Increased GDF15 level has been shown to be associated with insulin resistance, T2D, and cardiovascular diseases [76,77]. Taken together, these findings suggest that the insulin resistance in KCs associated with psoriasis may, in part, be due to genetic alterations in the expression of IRS2, GDF15, SLC2A10, and SLC2A14 in KCs of patients with psoriasis. Also, these data suggest that IRS-2 and GDF15 can be classified as candidate genes for psoriasis. Further investigations are needed to discover how these genetic variations causing psoriasis also induce insulin resistance and T2D.
Although we have identified large number of common DEGs in both PsO-iPSCs, we noticed more severe phenotypes in KCs derived from PsO1-iPSCs in comparision to those derived from PsO2-iPSCs. It could be argued that the severity of the genetic predisposition would have differences in the expression levels of certain genes. Further studies using the iPSC-based model established here are needed to gain deeper insight into signaling pathways and key genetic factors involved in the pathogenesis of psoriasis.
Although our results showed that the iPSCs can generate 3D skin equivalents, which expressed some key markers, the organotypic differentiation protocol needs further optimization to generate all layers of the stratified epithelium of the epidermis. Therefore, we are currently working on the generation of 3D epidermis equivalent and optimizing the condition to establish a reproducible protocol in our laboratory for the generation of epidermis equivalent. Also, in our future study, we will compare the 3D skin equivalents generated from PsO-iPSCs with those generated from Ctr-iPSCs and normal skin. Using 3D skin model derived from PsO-iPSCs would further help in understating the mechanism of psoriasis progression and testing drugs for psoriasis treatment.
In conclusion, we have established in this study the first human iPSC-based model to study genetic defects in KCs carrying the same genetic signatures of patients with psoriasis (Fig. 7). Our data showed that KCs derived from PsO-iPSCs harbor psoriasis-associated genetic defects, suggesting that the genetic alterations in KCs are the main drivers in the development of psoriasis. These findings are consistent with the concept that the KC abnormalities trigger psoriasis [78]. Our iPSC model established in this study can be used in future studies focusing on the understanding of the pathophysiology of psoriasis and associated comorbidities as well as developing novel therapeutics.
FIG. 7.
Schematic overview of establishing iPSCs carrying the genetic signature of patient with psoriasis. Patient iPSCs were differentiated into KCs and compared to those of healthy controls to identify psoriasis-associated genetic defects in KCs. Color images are available online.
Supplementary Material
Acknowledgments
We thank Dr. Manale Karam (QBRI/HBKU) for her critical reading of the manuscript. We thank the Genomic Core Facility of QBRI; Mrs. Khaoula Errafii, Ms. Hanan Abunada, and Dr. Richard Thompson for their assistance. We thank Ms. Samira Saleh, histocompatibility and immunogenetics laboratory, HMC.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was funded by grants from Qatar National Research Fund (QNRF) (grant no. NPRP9-283-3-056) and QBRI/HBKU (IGP ID 2014 009).
Supplementary Material
References
- 1. Bowcock AM and Cookson WO (2004). The genetics of psoriasis, psoriatic arthritis and atopic dermatitis. Hum Mol Genet 13 Spec No 1:R43–R55 [DOI] [PubMed] [Google Scholar]
- 2. Nestle FO, Kaplan DH and Barker J (2009). Psoriasis. N Engl J Med 361:496–509 [DOI] [PubMed] [Google Scholar]
- 3. Capon F. (2017). The genetic basis of psoriasis. Int J Mol Sci 18:2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellinghaus E, Ellinghaus D, Stuart PE, Nair RP, Debrus S, Raelson JV, Belouchi M, Fournier H, Reinhard C, et al. (2010). Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet 42:991–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huffmeier U, Uebe S, Ekici AB, Bowes J, Giardina E, Korendowych E, Juneblad K, Apel M, McManus R, et al. (2010). Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat Genet 42:996–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, et al. (2009). Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 41:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riveira-Munoz E, He SM, Escaramis G, Stuart PE, Huffmeier U, Lee C, Kirby B, Oka A, Giardina E, et al. (2011). Meta-analysis confirms the LCE3C_LCE3B deletion as a risk factor for psoriasis in several ethnic groups and finds interaction with HLA-Cw6. J Invest Dermatol 131:1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stuart PE, Nair RP, Ellinghaus E, Ding J, Tejasvi T, Gudjonsson JE, Li Y, Weidinger S, Eberlein B, et al. (2010). Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet 42:1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang XJ, Huang W, Yang S, Sun LD, Zhang FY, Zhu QX, Zhang FR, Zhang C, Du WH, et al. (2009). Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet 41:205–210 [DOI] [PubMed] [Google Scholar]
- 10. Elder JT, Bruce AT, Gudjonsson JE, Johnston A, Stuart PE, Tejasvi T, Voorhees JJ, Abecasis GR and Nair RP (2010). Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol 130:1213–1226 [DOI] [PubMed] [Google Scholar]
- 11. Sun LD, Cheng H, Wang ZX, Zhang AP, Wang PG, Xu JH, Zhu QX, Zhou HS, Ellinghaus E, et al. (2010). Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet 42:1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suarez-Farinas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C and Krueger JG (2012). Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol 132:2552–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gudjonsson JE, Ding J, Li X, Nair RP, Tejasvi T, Qin ZS, Ghosh D, Aphale A, Gumucio DL, et al. (2009). Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol 129:2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR and Elder JT (2010). Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol 130:1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swindell WR, Xing X, Stuart PE, Chen CS, Aphale A, Nair RP, Voorhees JJ, Elder JT, Johnston A and Gudjonsson JE (2012). Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One 7:e34594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ainali C, Valeyev N, Perera G, Williams A, Gudjonsson JE, Ouzounis CA, Nestle FO and Tsoka S (2012). Transcriptome classification reveals molecular subtypes in psoriasis. BMC Genomics 13:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reischl J, Schwenke S, Beekman JM, Mrowietz U, Sturzebecher S and Heubach JF (2007). Increased expression of Wnt5a in psoriatic plaques. J Invest Dermatol 127:163–169 [DOI] [PubMed] [Google Scholar]
- 18. Gudjonsson JE, Johnston A, Dyson M, Valdimarsson H and Elder JT (2007). Mouse models of psoriasis. J Invest Dermatol 127:1292–1308 [DOI] [PubMed] [Google Scholar]
- 19. Wagner EF, Schonthaler HB, Guinea-Viniegra J and Tschachler E (2010). Psoriasis: what we have learned from mouse models. Nat Rev Rheumatol 6:704–714 [DOI] [PubMed] [Google Scholar]
- 20. Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, Leung TL, Siprashvili Z, Tichy A, et al. (2014). Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med 6:264ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metallo CM, Azarin SM, Moses LE, Ji L, de Pablo JJ and Palecek SP (2010). Human embryonic stem cell-derived keratinocytes exhibit an epidermal transcription program and undergo epithelial morphogenesis in engineered tissue constructs. Tissue Eng Part A 16:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Memon B, Karam M, Al-Khawaga S and Abdelalim EM (2018). Enhanced differentiation of human pluripotent stem cells into pancreatic progenitors co-expressing PDX1 and NKX6.1. Stem Cell Res Ther 9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aigha II, Memon B, Elsayed AK and Abdelalim EM (2018). Differentiation of human pluripotent stem cells into two distinct NKX6.1 populations of pancreatic progenitors. Stem Cell Res Ther 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGowan-Jordan JSA and M. Schmid. 2016. An International System for Human Cytogenetic Nomenclature. Karger, Basel [Google Scholar]
- 25. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truong AB, Kretz M, Ridky TW, Kimmel R and Khavari PA (2006). p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20:3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Senoo M, Pinto F, Crum CP and McKeon F (2007). p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129:523–536 [DOI] [PubMed] [Google Scholar]
- 29. Okuyama R, Ogawa E, Nagoshi H, Yabuki M, Kurihara A, Terui T, Aiba S, Obinata M, Tagami H and Ikawa S (2007). p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene 26:4478–4488 [DOI] [PubMed] [Google Scholar]
- 30. Okuyama R, Tagami H and Aiba S (2008). Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J Dermatol Sci 49:187–194 [DOI] [PubMed] [Google Scholar]
- 31. Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE and Fuchs E (2011). A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 145:1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ota T, Takekoshi S, Takagi T, Kitatani K, Toriumi K, Kojima T, Kato M, Ikoma N, Mabuchi T and Ozawa A (2014). Notch signaling may be involved in the abnormal differentiation of epidermal keratinocytes in psoriasis. Acta Histochem Cytochem 47:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdou AG, Maraee AH, Sharaf A and Elnaidany NF (2012). Up-regulation of Notch-1 in psoriasis: an immunohistochemical study. Ann Diagn Pathol 16:177–184 [DOI] [PubMed] [Google Scholar]
- 34. Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, et al. (2012). Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44:1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Segre JA, Bauer C and Fuchs E (1999). Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22:356–360 [DOI] [PubMed] [Google Scholar]
- 36. Madonna S, Scarponi C, Sestito R, Pallotta S, Cavani A and Albanesi C (2010). The IFN-gamma-dependent suppressor of cytokine signaling 1 promoter activity is positively regulated by IFN regulatory factor-1 and Sp1 but repressed by growth factor independence-1b and Kruppel-like factor-4, and it is dysregulated in psoriatic keratinocytes. J Immunol 185:2467–2481 [DOI] [PubMed] [Google Scholar]
- 37. Kim KJ, Park S, Park YH, Ku SH, Cho EB, Park EJ and Kim KH (2014). The expression and role of kruppel-like factor 4 in psoriasis. Ann Dermatol 26:675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lowes MA, Bowcock AM and Krueger JG (2007). Pathogenesis and therapy of psoriasis. Nature 445:866–873 [DOI] [PubMed] [Google Scholar]
- 39. Jabbari A, Suarez-Farinas M, Dewell S and Krueger JG (2012). Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol 132:246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W and Bowcock AM (2011). Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 20:4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swindell WR, Johnston A, Voorhees JJ, Elder JT and Gudjonsson JE (2013). Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics 14:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suarez-Farinas M, Lowes MA, Zaba LC and Krueger JG (2010). Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA). PLoS One 5:e10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lovendorf MB, Mitsui H, Zibert JR, Ropke MA, Hafner M, Dyring-Andersen B, Bonefeld CM, Krueger JG and Skov L (2015). Laser capture microdissection followed by next-generation sequencing identifies disease-related microRNAs in psoriatic skin that reflect systemic microRNA changes in psoriasis. Exp Dermatol 24:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mitsui H, Suarez-Farinas M, Belkin DA, Levenkova N, Fuentes-Duculan J, Coats I, Fujita H and Krueger JG (2012). Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol 132:1615–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pasquali L, Srivastava A, Meisgen F, Das Mahapatra K, Xia P, Xu Landen N, Pivarcsi A and Sonkoly E (2019). The keratinocyte transcriptome in psoriasis: pathways related to immune responses, cell cycle and keratinization. Acta Derm Venereol 99:196–205 [DOI] [PubMed] [Google Scholar]
- 46. Kess D, Peters T, Zamek J, Wickenhauser C, Tawadros S, Loser K, Varga G, Grabbe S, Nischt R, et al. (2003). CD4+ T cell-associated pathophysiology critically depends on CD18 gene dose effects in a murine model of psoriasis. J Immunol 171:5697–5706 [DOI] [PubMed] [Google Scholar]
- 47. Gottlieb AB, Chamian F, Masud S, Cardinale I, Abello MV, Lowes MA, Chen F, Magliocco M and Krueger JG (2005). TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol 175:2721–2729 [DOI] [PubMed] [Google Scholar]
- 48. Sweeney CM, Tobin AM and Kirby B (2011). Innate immunity in the pathogenesis of psoriasis. Arch Dermatol Res 303:691–705 [DOI] [PubMed] [Google Scholar]
- 49. Baadsgaard O, Fisher G, Voorhees JJ and Cooper KD (1990). The role of the immune system in the pathogenesis of psoriasis. J Invest Dermatol 95:32S–34S [DOI] [PubMed] [Google Scholar]
- 50. Lowes MA, Suarez-Farinas M and Krueger JG (2014). Immunology of psoriasis. Annu Rev Immunol 32:227–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winge MC, Ohyama B, Dey CN, Boxer LM, Li W, Ehsani-Chimeh N, Truong AK, Wu D, Armstrong AW, et al. (2016). RAC1 activation drives pathologic interactions between the epidermis and immune cells. J Clin Invest 126:2661–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, Wong WH and Bowcock AM (2003). Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics 13:69–78 [DOI] [PubMed] [Google Scholar]
- 53. Carlen L, Sakuraba K, Stahle M and Sanchez F (2007). HLA-C expression pattern is spatially different between psoriasis and eczema skin lesions. J Invest Dermatol 127:342–348 [DOI] [PubMed] [Google Scholar]
- 54. Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NVC, Jenisch S, Weichenthal M, Abecasis GR, Lim HW, et al. (2006). Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet 78:827–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nedoszytko B, Sokolowska-Wojdylo M, Ruckemann-Dziurdzinska K, Roszkiewicz J and Nowicki RJ (2014). Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: atopic dermatitis, psoriasis and skin mastocytosis. Postepy Dermatol Alergol 31:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB and Leung DY (2003). Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol 112:1195–1202 [DOI] [PubMed] [Google Scholar]
- 57. van der Fits L, van der Wel LI, Laman JD, Prens EP and Verschuren MC (2003). Psoriatic lesional skin exhibits an aberrant expression pattern of interferon regulatory factor-2 (IRF-2). J Pathol 199:107–114 [DOI] [PubMed] [Google Scholar]
- 58. Raposo RA, Gupta R, Abdel-Mohsen M, Dimon M, Debbaneh M, Jiang W, York VA, Leadabrand KS, Brown G, et al. (2015). Antiviral gene expression in psoriasis. J Eur Acad Dermatol Venereol 29:1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolk K, Witte K, Witte E, Raftery M, Kokolakis G, Philipp S, Schonrich G, Warszawska K, Kirsch S, et al. (2013). IL-29 is produced by T(H)17 cells and mediates the cutaneous antiviral competence in psoriasis. Sci Transl Med 5:204ra129. [DOI] [PubMed] [Google Scholar]
- 60. Swindell WR, Xing X, Voorhees JJ, Elder JT, Johnston A and Gudjonsson JE (2014). Integrative RNA-seq and microarray data analysis reveals GC content and gene length biases in the psoriasis transcriptome. Physiol Genomics 46:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Keermann M, Koks S, Reimann E, Prans E, Abram K and Kingo K (2015). Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics 16:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, et al. (2014). Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 134:1828–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boyman O, Conrad C, Tonel G, Gilliet M and Nestle FO (2007). The pathogenic role of tissue-resident immune cells in psoriasis. Trends Immunol 28:51–57 [DOI] [PubMed] [Google Scholar]
- 64. Boyman O, Hefti HP, Conrad C, Nickoloff BJ, Suter M and Nestle FO (2004). Spontaneous development of psoriasis in a new animal model shows an essential role for resident T cells and tumor necrosis factor-alpha. J Exp Med 199:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ and Gilliet M (2005). Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med 202:135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davidovici BB, Sattar N, Prinz J, Puig L, Emery P, Barker JN, van de Kerkhof P, Stahle M, Nestle FO, Girolomoni G and Krueger JG (2010). Psoriasis and systemic inflammatory diseases: potential mechanistic links between skin disease and co-morbid conditions. J Invest Dermatol 130:1785–1796 [DOI] [PubMed] [Google Scholar]
- 67. Wertheimer E, Trebicz M, Eldar T, Gartsbein M, Nofeh-Moses S and Tennenbaum T (2000). Differential roles of insulin receptor and insulin-like growth factor-1 receptor in differentiation of murine skin keratinocytes. J Invest Dermatol 115:24–29 [DOI] [PubMed] [Google Scholar]
- 68. Sadagurski M, Weingarten G, Rhodes CJ, White MF and Wertheimer E (2005). Insulin receptor substrate 2 plays diverse cell-specific roles in the regulation of glucose transport. J Biol Chem 280:14536–14544 [DOI] [PubMed] [Google Scholar]
- 69. Fasshauer M, Klein J, Ueki K, Kriauciunas KM, Benito M, White MF and Kahn CR (2000). Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J Biol Chem 275:25494–25501 [DOI] [PubMed] [Google Scholar]
- 70. Kido Y, Burks DJ, Withers D, Bruning JC, Kahn CR, White MF and Accili D (2000). Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J Clin Invest 105:199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stachelscheid H, Ibrahim H, Koch L, Schmitz A, Tscharntke M, Wunderlich FT, Scott J, Michels C, Wickenhauser C, et al. (2008). Epidermal insulin/IGF-1 signalling control interfollicular morphogenesis and proliferative potential through Rac activation. EMBO J 27:2091–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gherzi R, Melioli G, de Luca M, D'Agostino A, Distefano G, Guastella M, D'Anna F, Franzi AT and Cancedda R (1992). “HepG2/erythroid/brain” type glucose transporter (GLUT1) is highly expressed in human epidermis: keratinocyte differentiation affects GLUT1 levels in reconstituted epidermis. J Cell Physiol 150:463–474 [DOI] [PubMed] [Google Scholar]
- 73. Amir Shaghaghi M, Zhouyao H, Tu H, El-Gabalawy H, Crow GH, Levine M, Bernstein CN and Eck P (2017). The SLC2A14 gene, encoding the novel glucose/dehydroascorbate transporter GLUT14, is associated with inflammatory bowel disease. Am J Clin Nutr 106:1508–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Andersen G, Rose CS, Hamid YH, Drivsholm T, Borch-Johnsen K, Hansen T and Pedersen O (2003). Genetic variation of the GLUT10 glucose transporter (SLC2A10) and relationships to type 2 diabetes and intermediary traits. Diabetes 52:2445–2448 [DOI] [PubMed] [Google Scholar]
- 75. Bento JL, Bowden DW, Mychaleckyj JC, Hirakawa S, Rich SS, Freedman BI and Segade F (2005). Genetic analysis of the GLUT10 glucose transporter (SLC2A10) polymorphisms in Caucasian American type 2 diabetes. BMC Med Genet 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kempf T, Guba-Quint A, Torgerson J, Magnone MC, Haefliger C, Bobadilla M and Wollert KC (2012). Growth differentiation factor 15 predicts future insulin resistance and impaired glucose control in obese nondiabetic individuals: results from the XENDOS trial. Eur J Endocrinol 167:671–678 [DOI] [PubMed] [Google Scholar]
- 77. Adela R and Banerjee SK (2015). GDF-15 as a target and biomarker for diabetes and cardiovascular diseases: a translational prospective. J Diabetes Res 2015:490842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ippagunta SK, Gangwar R, Finkelstein D, Vogel P, Pelletier S, Gingras S, Redecke V and Hacker H (2016). Keratinocytes contribute intrinsically to psoriasis upon loss of Tnip1 function. Proc Natl Acad Sci U S A 113:E6162–E6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets generated and used in the present study are available on the Zenodo repository at https://doi.org/10.5281/zenodo.3484611.