Introduction
The upper airways play a critical role in the respiratory system by conditioning and clearing contaminants from the inspired airstream before it accesses the lower respiratory system.1 Large particulate matter is removed from inhaled air in the anterior naris or nasal vestibule, a relatively dry environment lined by skin-like squamous epithelial cells and containing sebaceous glands and vibrissae. Smaller particulate matter including bacteria and hydrophilic aerosolized compounds are trapped in a flowing mucus blanket covering the sinonasal mucosa deeper in the nasal cavity and sinuses. Sinonasal mucociliary function is a key host defense mechanism that clears the inhaled particulate matter. Characterized by impaired mucociliary clearance (MCC), bacterial colonization may play some role in the initiation or sustenance of the inflammatory process in chronic rhinosinusitis (CRS).2
In recent years, growing understanding of the fundamental role of the microbiome in the initiation, adaptation, and function of the human immune system has revolutionized the field of mucosal immunology.3 While each inflammatory disease can be differentiated by exclusive genetic and biological mechanisms, many inflammatory diseases, including CRS, are associated with significant shifts in the resident microbiota from a ‘healthy’ to a ‘diseased’ state.3 The dysbiosis hypothesis--alteration of microbial composition associated with perturbation of the local ecological landscape--has been widely suggested as a mechanism involved in CRS pathogenesis. This hypothesis is supported by several studies identifying a healthy local environment with particular “keystone species,” or microbes that normally maintain a stable and interactive community.4-6 Yet, sinus microbiome studies are in their infancy; many findings have not been replicated due to small study cohorts and variable experimental methods. In addition, results across studies are difficult to interpret in aggregate, and observed associations do not establish causality between the presence of certain microbial communities in the airways and the development of CRS.7 Many of these difficulties are intrinsically related to the broad diagnostic parameters of CRS and lack of a universally appropriate animal model.
High-impact microbiome studies from other organ systems (e.g., gut) have been conceptually applied to the respiratory field.8 Although seemingly reasonable, nucleic-acid based surveys of airway microbial communities and their proposed role(s) in CRS remain to be addressed using appropriate model systems. Should an etiological of specific community structures hold true, opportunities will arise for novel therapeutic interventions with potential for personalized, microbiome-based treatment strategies. In this review we will discuss major concepts that highlight the complex role of the microbiota in sinus health and disease and explore future directions for study.
Considerations in Sinus Microbiome Investigation
Sampling locations vary between studies, reflecting subtly different microenvironments throughout the upper airway, and making cross-study meta-analyses a challenge.9 It is clear that the anterior nasal cavity microbiome is distinct from the middle meatus and sphenoethmoid recess in the healthy state,10 but the most representative single sampling site in the nasal cavity is often argued.4,11 How can a single site encompass the complexities of the many anatomic niches and account for differences in local immune and disease properties? The middle meatus is often used as a representative sampling site for the deeper sinuses, given:
its high agreement in culture comparison studies with the maxillary sinus,12
its location as a common drainage pathway of the three major (maxillary, anterior ethmoid, and frontal) sinuses, and
In the gastrointestinal (GI) microbiome field, stool is often studied as a convenient single sample that overrepresents the cecal contribution, acknowledging there are likely biogeographical differences between the upper GI tract, duodenum, jejunum, ileum, and large intestine. To address this concern in the context of CRS, we compared 12 sites from 8 subjects with CRS at the time of surgical intervention and found a fair concordance between the middle meatus and underlying sinuses. These data suggested that if one were interested in single site representation that the middle meatus would be a reasonable proxy for the entire upper airway.14
In terms of bacterial detection, it is clear that molecular methods are superior to traditional culture-based approaches in CRS, as identification of even the most fastidious of organisms can be achieved with DNA-based detection and classification by variable regions with the 16S rRNA gene.15-17 Hauser et al. demonstrated that bacterial detection using 16S rRNA gene sequencing allows for greater sensitivity and provides more information on bacterial diversity than standard clinical swab culture in CRS.16 Though clinical laboratory culture has been the gold standard for decades and offers useful information, these techniques are unique to institutional laboratories and may miss bacteria that are present in disease. However, the true clinical utility of culture-independent molecular techniques remains to be determined. The ability to more accurately detect bacteria that are present may allow for more effective treatment regimens and allow for an improved basis for clinical and laboratory research into CRS. Nevertheless, there are some shortcomings of culture-independent molecular techniques. 16S rRNA gene sequencing measures total or relative abundance of bacterial DNA and does not differentiate between actively growing, dormant, or dead biomass.18 As with all tests, it is important to be aware of such biases. To better understand in vivo bacterial activity, culture-independent approaches must be improved and new innovative techniques should continue to be integrated, for instance, by separating active cells from extracellular DNA and inactive microbial subpopulations.19,20
Currently, there are two main gene sequencing approaches used for studying microbial communities:
targeted sequencing of specific marker genes (i.e., 16S rRNA gene for bacteria and 18S rRNA or internal transcribed spacer (ITS) regions for fungi) and
shotgun sequencing of the metagenome.
16S rRNA gene sequencing is currently the most widely used approach for characterizing bacterial community membership and comparing phylogeny between samples. This method is based on the premise that nine hypervariable regions within the 16S rRNA gene harbor sufficient sequence diversity to differentiate bacterial taxa down to the genus or species level. Flanking these regions are highly conserved sequences across bacteria and archaea that facilitate the use of universal PCR primer sets.21,22 Though costlier, shotgun sequencing methods are useful for characterizing microbial communities more broadly, including viruses and fungi that have also been implicated in the development of upper airway disease. Shotgun metagenomics, the study of whole-community DNA extracted directly from samples, has increasingly been used in various settings, particularly as sequencing costs decrease and output increases.23,24 Furthermore, relative to targeted amplicon assays (e.g., 16S rRNA gene sequencing), shotgun metagenomics offers potential for both higher-resolution identification of organisms and the study of microbial communities without introduction of sequencing bias due to unequal amplification of the target gene.24,25 Moreover, shotgun approaches capture details of the microbial metagenome (i.e., antibiotic resistance, virulence factors) not provided using single marker gene studies.22,26 Sequencing technologies have undergone rapid advances during the past several years to attempt to resolve biases associated with current methods and to obtain a better balance between data yield, read length, and cost.22 These efforts have resulted in third generation sequencing technologies (e.g., Oxford Nanopore and PacBio platforms), which are single-molecule and real-time technologies that reduce amplification bias, as well as short-read length limitations.22,27,28 Reduction in cost and time presented by these sequencing methods are valuable assets, and certainly future incorporation of new technologies and bioinformatics is expected.
Based on the anatomic location and local disease environment, viruses and fungi have hypothesized interactions with the bacterial community, which have been borne out in prior study.29 The relative absence of fungal and viral study at the current time may be a simple lag behind the bacterial microbiome research explosion, as the early microbial detection techniques focused primarily on numerically dominant bacteria. Multiple studies have demonstrated the presence of viruses and fungi in CRS.30-35 Virus replication can result in epithelial damage and increase bacterial mucosal adhesion, whereas fungi may act synergistically with pathogenic bacteria to play a role in the pathogenesis of CRS.32,36 The precise roles of these organisms in the pathogenesis of CRS and etiological importance remains poorly understood.32
Dysbiosis of Sinus Microbiota in CRS
Analysis of the normal state of the microbiome in sinus cavities is crucial, as there is a clear role for commensals in pathogen exclusion and in the modulation of the healthy host-microbial immune response.37 The deeper nasal cavity and sinuses have unique local microenvironments (pO2, pH, etc.) and host immune properties.11,38,39 While Yan et al. recently examined deeper anatomical subsites in healthy human nasal cavities, and Ramakrishnan et al. compared upper airway subsites and sinuses in CRS, there has been no thorough comparison within normal sinus cavities to date, perhaps owing to the requirement of a more invasive approach.10,14
In the healthy state, commonly identified bacterial genera from the upper airways include Staphylococcus, Corynebacterium, Peptoniphilus and Propionibacterium.2,6,32,36,40,41 Interestingly, total bacterial load present in healthy and diseased sinuses appear to be surprisingly alike across adults. Further, high inter-individual microbiome variation is often observed in healthy controls and CRS patients.42,43 Many opportunistic pathogens are found at low abundance in healthy sinuses and, therefore, have the potential to create disease after an acute alteration in the stable baseline microbial community (i.e. dysbiosis).2,32
Disruption of stable microbiota may contribute to the exacerbation of chronic inflammatory disease in the absence of acute infection.11,44 Dysbiosis can lead to benign microbial communities becoming pro-inflammatory, invasive or allowing overgrowth of pathogens. There is also growing evidence that dysbiosis of the sinus microbiota is associated with CRS pathogenesis.45 Human studies have revealed that the CRS microbiome is characterized by loss of diversity compared to healthy controls,5,43,46 indicating the opportunity for prosperity of pathogens.47 Results from these and other sequence-based studies have transformed our understanding of the role of microbial community composition and dynamics in CRS pathogenesis.
Disruption of healthy commensal interactions with the local immune system appears to be a critical determinant of CRS progression. Linear discriminant analysis identified the genus Corynebacterium as a potential biomarker that was significantly increased in abundance in CRS patients, however this genus was also omnipresent in healthy subjects from other studies.4,11 Using a murine model challenged with C. tuberculostearicum after antibiotic-mediated microbial depletion, Abreu et al. demonstrated goblet cell hyperplasia and mucin hyper-secretion, two important histologic hallmarks of CRS.48 However, in this study there were only 7 samples from CRS patients, and another study subsequently reported opposing findings that CRS patients with enriched C. tuberculostearicum colonization at the time of endoscopic sinus surgery showed improved surgical outcomes.6,49 Regarding host interaction with local immune system, another group found that nasal lavage samples of microbiota collected from CRS patients stimulated the induction of proinflammatory cytokines such as IL-5 in peripheral leukocytes isolated from healthy controls.11,50 Together, these data suggest that the CRS state represents an altered ecological landscape interacting with an aberrant immune response. To this concept, a recent cross-sectional study of CRS and non-CRS patients who underwent endoscopic sinus surgery demonstrated a correlation between the loss of bacterial species richness and diversity and the severity of inflammation and tissue eosinophilia.51 Whether dysbiosis is causative or a result of the disrupted local immune system remains to be determined.
A preponderance of anaerobes has been consistently observed in studies of CRS, which may be explained by:
selective pressure of antimicrobial agents enabling anaerobic organisms to flourish,52 and
from the existence of conditions appropriate for anaerobic growth (i.e., sinus hypoxia).53
Anaerobic taxa such as Peptoniphilus, Anaerococcus, and Prevotella have been reported as abundant taxa in multiple CRS studies.4,42,54-56 Ambient conditions within the sinus cavities may not be hypoxic, especially after endoscopic sinus surgery has opened the cavities. However, expansion of anaerobes in CRS may be indicative of underlying tissue hypoxia, or may suggest that discrete micro-environments within mucus or bacterial biofilms in CRS can also be oxygen limited, allowing anaerobes to thrive .4,53,57 It is likely that, similar to mucus plugs in the lower airways of individuals with cystic fibrosis, oxygen levels within sinus mucus are dynamic and driven by both host and microbial processes.58 Whether anaerobic bacteria have an etiological role in CRS disease progression has been only marginally addressed and is an emerging area of research in chronic airway disease.
Microbial Interactions in CRS
Understanding the complexity and dynamics of interspecies and interkingdom relationships represents a major challenge in microbiome research, but has the potential to help clarify effects in several chronic respiratory diseases including CRS.8 Symbiosis in healthy microbial ecosystems allows for efficient nutrient utilization and results in decreased pathogen colonization.47 Most microorganisms face a constant battle for resources and there are diverse mechanisms by which bacterial species can coexist with, or dominate, other organisms competing for the same pool of resources.59 Understanding of microbial interactions will be crucial in establishing the function of microbial communities in CRS and implementing new therapeutic strategies.
Yan et al. studied the interaction between S. aureus and Corynebacterium in the healthy human nasal cavity and showed that Corynebacterium sp. are involved in both mutualistic and inhibitory interactions with S. aureus. C. accolens and S. aureus appear to be adapted to each other and mutually promote each other’s growth in vitro, whereas C. pseudodiphtheriticum may interfere with colonization of S. aureus and was observed to inhibit S. aureus growth in vitro10 Within the nasal cavity, these reciprocal interactions suggest the possibility for niche competition and possible protection against S. aureus nasal colonization.
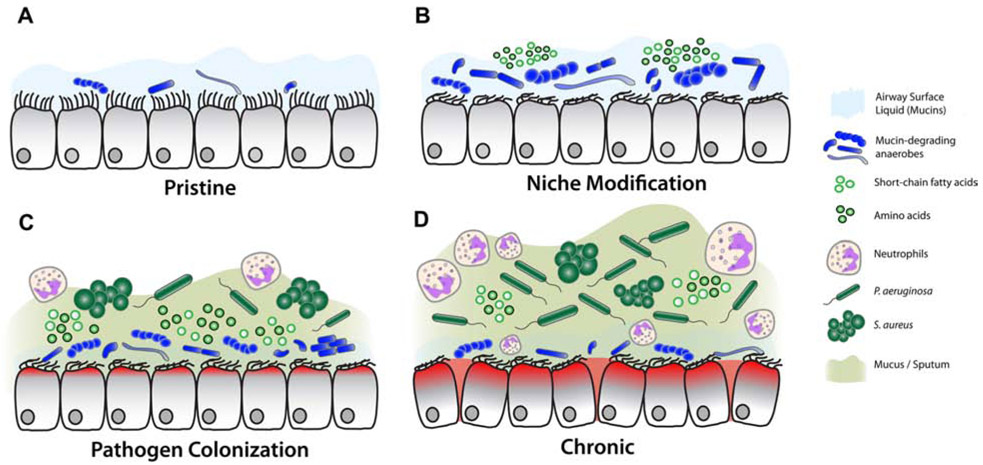
P. aeruginosa is also an important respiratory pathogen, and often carries intrinsic and/or acquired resistance to many classes of antibiotics. Its appearance and recalcitrance in a portion of CRS subjects is an ongoing clinical challenge. Flynn et al. investigated the role of airway mucins as the microbial carbon source in the cystic fibrosis (CF) airway and characterized their potential to stimulate the growth of Pseudomonas.60 Their group demonstrated that co-culture of P. aeruginosa with an anaerobic bacterial consortium facilitates robust growth of P. aeruginosa using mucins as a sole nutritional carbon source. These data support an ecological role for anaerobes in shaping the landscape of the human airway for progression of chronic disease (e.g., CRS), and proposed a model for the role of anaerobes in disease pathogenesis.60 In this model, potential pathogens that cannot degrade mucins (e.g. P. aeruginosa, S.aureus) do not establish an airway infection until mucin-fermenting bacteria (anaerobes) have colonized (Figure 1). Numerous 16S rRNA gene sequencing studies in CRS have demonstrated a previously unrecognized abundance of anaerobes in the disease state.6,48 Based on this hypothesis, chronic airway disease could develop through a defined series of dependent events:
Figure 1. Model for the role of mucin fermenting bacteria in the progression of CF lung disease, as applied to CRS.
(A) In early life, airway surface liquid harbors a low number of bacteria. Numerous factors allow for establishment of personal local microbiota. (B) Local insult resulting in impaired mucociliary clearance and defective immune responses results in hypoxic environment ideal for expansion of anaerobes. In turn, their ability to degrade and ferment respiratory mucins further modifies the airway environment for secondary colonizers. (C) The abundance of fermentation byproducts facilitates pathogen colonization, heightened inflammation, neutrophil recruitment and further hypoxia. (D) In late stages of disease, host inflammatory responses and epithelial damage increases the abundance of pathogens, while healthy commensals are eliminated by the host and via broad spectrum antibiotic therapies.
Data from Flynn JM, Niccum D, Dunitz JM, et al. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS Pathog 2016;12(8):e1005846.
impaired mucus clearance,
generation of anaerobic microenvironments,
dysbiosis with mucin-fermenting anaerobes,
mucin degradation to carbon source nutrients (e.g., short chain fatty acids (SCFAs)), and
proliferation of sinus pathogens.
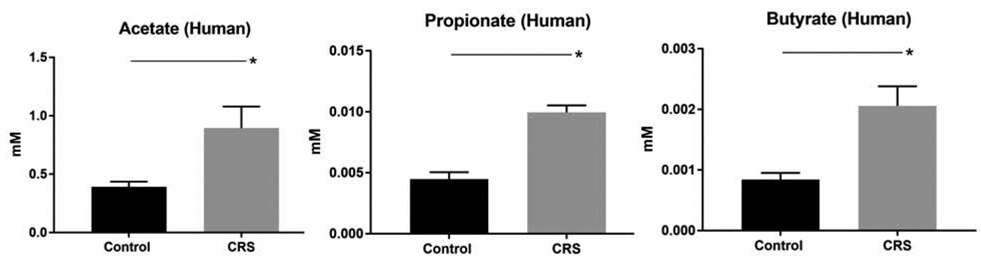
In this context, we preliminarily tested whether there is evidence of mucin fermentation in human CRS by analyzing the presence of SCFAs in the mucus of subjects during acute exacerbations. Using gas chromatography-mass spectrometry, three SCFAs (acetate, propionate, and butyrate) were quantified in human mucus samples collected from 6 controls and 9 CRS patients during acute exacerbation episodes. SCFAs were found at millimolar concentrations in all mucus samples, and at significantly higher concentrations in CRS compared to healthy subjects (Figure 2). Given that SCFAs are predominately derived from bacterial fermentation, this evidence suggests that mucin-fermenting bacteria are able to generate carbon-source nutrients for pathogenic bacteria in CRS, similar to proposed mechanisms of disease progression in the lower airways.60 Based on these data, it is intriguing to consider that the growth of canonical airway pathogens (e.g. S. aureus, P. aeruginosa) might be inhibited by targeting co-colonizing microbiota that potentiate their growth and virulence.
Figure 2. Concentrations of short chain fatty acids (SCFA) in human mucus samples from CRS with acute exacerbation vs healthy controls.
All 3 SCFAs were significantly higher in CRS (n = 9) compared to control (n = 6): 1) acetate = 0.89 +/− 0.19 versus 0.39 +/ 0.04 mM (p < 0.05); 2) propionate = 0.01 +/− 0.00 versus 0.0045 +/− 0.00 (p < 0.0001); 3) butyrate = 0.002 +/− 0.00 versus 0.0008 +/− 0.00 (p < 0.01).
Developing preclinical models
The study of dysbiosis in human CRS is especially challenging because medical therapies used in disease treatment are likely to affect resident bacterial communities.45,61,62 Observed alterations in CRS local microbiota in cross-sectional studies have been unable to account for the repeated and prolonged medical therapies that are common in study subjects. Unfortunately, small animals are not universally accepted in CRS as they do not develop upper airway phenotypes (e.g., CF murine models), possibly from absence of submucosal glands,63 and their small size precludes thorough examination of sinus pathology.64 Thus, there remains a need for a robust preclinical model of CRS for longitudinal sampling prior to and during disease initiation. Although there are some limitations when applying animal findings to human pathophysiology, preclinical models have played a significant role in the process of understanding CRS pathophysiology.65-67
Many different animals (e.g., murine, rabbit, sheep, pigs) have been used to establish acute and chronic sinus inflammation in prior studies. Small animal models used in CRS microbiome research include the murine model of sinusitis described earlier in investigation of C. tuberculostearicum as a potential pathogen on the sinus microbiota. By inoculating C. tuberculostearicum into the nasal cavity with and without preceding antibiotic treatment, this study showed the capability of C. tuberculostearicum to induce a CRS phenotype, particularly in conjunction with a depleted host commensal community. Co-inoculation of C. tuberculostearicum with Lactobacillus sakei, a putative probiotic, resulted in a reduced abundance of C. tuberculostearicum.48 In addition, mice have been used to understand the dynamics of sinonasal infection and the role of the mucosal microbiome in short- and long- term responses after topical inoculation of human pathogens (e.g. P. aeruginosa).68 Mice are easy to work with in the laboratory and carry many advantages of experimental application that have been extensively documented. However, murine CRS models are limited due to animal size, unclear similarity of commensal microbes to human counterparts, poorly defined ecological properties of stability and resilience, and that mice do not reproduce key aspects of human airway physiology. They do not have true sinuses, for instance, essential for the analysis of the pathophysiological mechanisms of CRS.69 Furthermore, immune responses in mice are notably different from those in humans.70 Compared to mice, rat models are much larger, which makes acquiring larger tissue specimens easier and ameliorates the technical limitations of smaller models.71 However, transgenic rat models useful for CRS are rare, and the cystic fibrosis transmembrane conductance regulator (CFTR) knockout (KO) rat model (Rattus norvegicus; SD-CFTRtm1sage) does not develop spontaneous sinusitis.64
As an alternative small animal model, the in vivo rabbit sinusitis model is established and may be well-suited for studies of therapeutic intervention. The rabbit sinusitis model:
can recapitulate histopathological features of sinusitis,
is of sufficient size to study spatial and temporal microbial changes, and
has been used to explore experimental ostial obstruction and/or microbial inoculation in the development of the disease.72
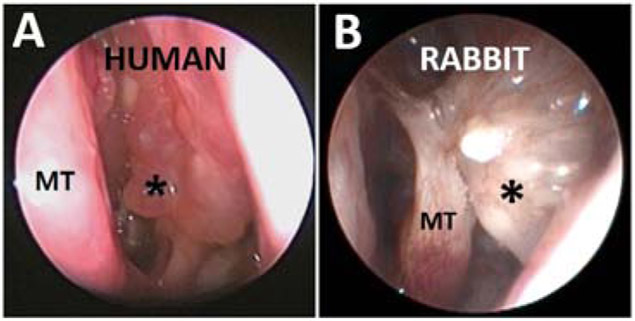
Cho et al. developed a rabbit model of sinusitis by blocking the maxillary sinus ostium for 2 weeks in the absence of infection to create an anaerobic environment with decreased MCC, resulting in the infiltration of sinus epithelium with acute inflammatory cells (neutrophils).73 When followed for another 12 weeks after removal of ostial obstruction, those rabbits exhibited a chronic inflammatory phenotype at week 14 (Figure 3). In this model, the mucin fermenting anaerobic phyla Firmicutes and Bacteroidetes dominated at week 2, but were followed by a significant microbial shift to pathogenic Proteobacteria (e.g. Burkholderiales and Pseudomonadales) during the development of chronic inflammation by week 14. Such a model provides the opportunity to study microbial host interactions with a level of experimental control that is not achievable in mouse or humans, and also permits multiple longitudinal samplings because the nasal cavity is accessible by nasal endoscopy.
Figure 3. Middle meatus (*) of Human (A) vs Rabbit (B) CRS.
Similar significant polypoid mucosal changes (asterisk). MT: Middle turbinate
Future Directions in CRS Microbiome Research
Standardization in sampling procedures
Many protocols have been utilized and advocated for different reasons. The “best” sampling protocol depends on the question being addressed. Mucus swab of the middle meatus or ethmoid cavity may be the simplest approach for longitudinal study of the sinus microbiome, considering that it can be obtained from a wide range of subjects and does not require invasive procedures. Whether microbes are sampled by swab, brush, or tissue biopsy, sequencing provides a general picture of the composition of the bacterial community, whereas as an accompanying clinically meaningful and functional physiologic approach is still required.
Healthy microbiome patterns
What is the healthy sinus microbiome consortium and what defines “normal”? How do healthy microbiota protect against potential pathogens, either passively through niche competition or actively through or metabolic processes or secretion of antimicrobial compound? Are these organisms susceptible to changes that occur in the sinus environment as a result of the CRS disease process, or iatrogenic manipulation? Normality patterns for viruses and fungi still need to be defined in the upper respiratory system.
Further characterization of non-cultivable and/or non-pathogenic bacteria
16S rRNA gene analyses have shown discrete patterns of non-cultivable microorganisms obtained from patients with CRS. However, conventional sequencing methods do not differentiate between actively growing, dormant, or dead biomass, nor do they capture in situ activity at the transcriptional and/or protein level. Detailed characterization of CRS-associated microbial communities therefore requires further innovative assessment. As an example, bioorthogonal non-canonical amino acid tagging (BONCAT) can be used to fluorescently label actively growing bacteria within samples prior to gene sequencing, and has the potential to enhance traditional sequencing methods by characterizing bacterial activity at the protein level.19 Other methods such as stable isotope probing or single cell transcriptional analyses coupled with in situ imaging also carry potential for generating unprecedented insights into the microbial basis of CRS disease progression.74,75
Local vs systemic microbial interactions
Future studies may need to address the contributions of both local and systemic microbial communities (i.e., local and GI occupants). New studies including bacteriophage, viral, and fungal contributions to functional host immune processes are eagerly anticipated.
Interventions
Bacterial supplementation and modulation of the microbiota through pre- or pro-biotics and equivalents are opportunities for thoughtful and ethical clinical research. Whether probiotics directly target inflammatory processes within the sino-nasal epithelium or aim to restore normal upper airway microbiota by mucus transfer, novel strategies to address pathogens in CRS are needed.
Key Points:
The dysbiosis hypothesis (alteration of microbial composition associated with perturbation of the local ecological landscape) has been widely implicated in CRS.
CRS might develop through a defined series of temporally dependent events: Impaired mucus clearance → anaerobic microenvironments → anaerobe proliferation → increased nutrient availability for sinus pathogens.
There remains a need for continued CRS research with longitudinal sampling prior to and during disease initiation, and application of robust preclinical models.
Synopsis.
Chronic rhinosinusitis (CRS) is defined as persistent inflammation and/or infection of the nasal cavity and paranasal sinuses. Recent advancements in culture-independent molecular techniques have enhanced our understanding of interactions between sinus microbiota and upper airway microenvironment. The dysbiosis hypothesis--alteration of microbiota associated with perturbation of the local ecological landscape--has been widely suggested as a mechanism involved in CRS pathogenesis. In this review, the authors discuss concepts that highlight the complex role of the microbiota in health and CRS and emphasize: 1) Considerations in sinus microbiome investigation; 2) dysbiosis of sinus microbiota in CRS; 3) microbial interactions in CRS; and 4) development of preclinical models. The authors conclude with future directions for CRS-associated microbiome research.
Acknowledgements
Research reported in this publication was supported by the National Institute on Deafness and Other Communication Disorders of the National Institutes of Health under award number K23DC014747 (VRR), National Institutes of Allergy and Infectious Disease K08AI146220 (DYC), Flight Attendants Medical Research Institute grant CIA130066 (DNF and VRR), American Rhinologic Society New Investigator Award (DYC), Cystic Fibrosis Foundation Research Development Pilot grant (ROWE15R0) to DYC, Cystic Fibrosis Foundation Postdoctoral Fellowship (FLYNN16F0) and National Center for Advancing Translational Sciences Grant (UL1TR000114) to RCH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shusterman D. The effects of air pollutants and irritants on the upper airway. Proc Am Thorac Soc. 2011;8(1):101–105. [DOI] [PubMed] [Google Scholar]
- 2.Ramakrishnan VR, Feazel LM, Gitomer SA, Ir D, Robertson CE, Frank DN. The microbiome of the middle meatus in healthy adults. PLoS One. 2013;8(12):e85507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copeland E, Leonard K, Carney R, et al. Chronic Rhinosinusitis: Potential Role of Microbial Dysbiosis and Recommendations for Sampling Sites. Front Cell Infect Microbiol. 2018;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner Mackenzie B, Waite DW, Hoggard M, Douglas RG, Taylor MW, Biswas K. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol. 2017;19(1):381–392. [DOI] [PubMed] [Google Scholar]
- 6.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol. 2015;136(2):334–342 e331. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan VR, Frank DN. Microbiome in patients with upper airway disease: Moving from taxonomic findings to mechanisms and causality. J Allergy Clin Immunol. 2018;142(1):73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faner R, Sibila O, Agusti A, et al. The microbiome in respiratory medicine: current challenges and future perspectives. Eur Respir J. 2017;49(4). [DOI] [PubMed] [Google Scholar]
- 9.Ramakrishnan VR, Hauser LJ, Frank DN. The sinonasal bacterial microbiome in health and disease. Curr Opin Otolaryngol Head Neck Surg. 2016;24(1):20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan M, Pamp SJ, Fukuyama J, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14(6):631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proctor DM, Relman DA. The Landscape Ecology and Microbiota of the Human Nose, Mouth, and Throat. Cell Host Microbe. 2017;21(4):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubin MG, Ebert CS, Coffey CS, Melroy CT, Sonnenburg RE, Senior BA. Concordance of middle meatal swab and maxillary sinus aspirate in acute and chronic sinusitis: a meta-analysis. Am J Rhinol. 2005;19(5):462–470. [PubMed] [Google Scholar]
- 13.Lund VJ, Stammberger H, Fokkens WJ, et al. European position paper on the anatomical terminology of the internal nose and paranasal sinuses. Rhinol Suppl. 2014;24:1–34. [PubMed] [Google Scholar]
- 14.Ramakrishnan VR, Gitomer S, Kofonow JM, Robertson CE, Frank DN. Investigation of sinonasal microbiome spatial organization in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7(1):16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feazel LM, Frank DN, Ramakrishnan VR. Update on bacterial detection methods in chronic rhinosinusitis: implications for clinicians and research scientists. Int Forum Allergy Rhinol. 2011;1(6):451–459. [DOI] [PubMed] [Google Scholar]
- 16.Hauser LJ, Feazel LM, Ir D, et al. Sinus culture poorly predicts resident microbiota. Int Forum Allergy Rhinol. 2015;5(1):3–9. [DOI] [PubMed] [Google Scholar]
- 17.Rhoads DD, Cox SB, Rees EJ, Sun Y, Wolcott RD. Clinical identification of bacteria in human chronic wound infections: culturing vs. 16S ribosomal DNA sequencing. BMC Infect Dis. 2012;12:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis AL, Calton JB, Carr TF, Chiu AG, Chang EH. Dead or alive: Deoxyribonuclease I sensitive bacteria and implications for the sinus microbiome. Am J Rhinol Allergy. 2016;30(2):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couradeau E, Sasse J, Goudeau D, et al. Probing the active fraction of soil microbiomes using BONCAT-FACS. Nat Commun. 2019;10(1):2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson MT, Pope CE, Marsh RL, et al. Human and Extracellular DNA Depletion for Metagenomic Analysis of Complex Clinical Infection Samples Yields Optimized Viable Microbiome Profiles. Cell Rep. 2019;26(8):2227–2240 e2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bent SJ, Pierson JD, Forney LJ, et al. Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl Environ Microbiol. 2007;73(7):2399–2401; author reply 2399-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malla MA, Dubey A, Kumar A, Yadav S, Hashem A, Abd Allah EF. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front Immunol. 2018;9:2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manichanh C, Chapple CE, Frangeul L, Gloux K, Guigo R, Dore J. A comparison of random sequence reads versus 16S rDNA sequences for estimating the biodiversity of a metagenomic library. Nucleic Acids Res. 2008;36(16):5180–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Noyes NR, Doster E, et al. Use of Metagenomic Shotgun Sequencing Technology To Detect Foodborne Pathogens within the Microbiome of the Beef Production Chain. Appl Environ Microbiol. 2016;82(8):2433–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah N, Tang H, Doak TG, Ye Y. Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics. Pac Symp Biocomput. 2011:165–176. [DOI] [PubMed] [Google Scholar]
- 26.Tessler M, Neumann JS, Afshinnekoo E, et al. Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Sci Rep. 2017;7(1):6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fichot EB, Norman RS. Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome. 2013;1(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93(24):13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson CM, Pfeiffer JK. Viruses and the Microbiota. Annu Rev Virol. 2014;1:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao B, Hu CY, Liu T, Liu Z. Respiratory viral infection in the chronic persistent phase of chronic rhinosinusitis. Laryngoscope. 2014;124(4):832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramadan HH, Farr RW, Wetmore SJ. Adenovirus and respiratory syncytial virus in chronic sinusitis using polymerase chain reaction. Laryngoscope. 1997;107(7):923–925. [DOI] [PubMed] [Google Scholar]
- 32.Sivasubramaniam R, Douglas R. The microbiome and chronic rhinosinusitis. World J Otorhinolaryngol Head Neck Surg. 2018;4(3):216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AJ, Antoszewska H, Fraser J, Douglas RG. Is chronic rhinosinusitis caused by persistent respiratory virus infection? Int Forum Allergy Rhinol. 2011;1(2):95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao YC, Bassiouni A, Tanjararak K, Vreugde S, Wormald PJ, Psaltis AJ. Role of fungi in chronic rhinosinusitis through ITS sequencing. Laryngoscope. 2018;128(1):16–22. [DOI] [PubMed] [Google Scholar]
- 35.Zhang I, Pletcher SD, Goldberg AN, Barker BM, Cope EK. Fungal Microbiota in Chronic Airway Inflammatory Disease and Emerging Relationships with the Host Immune Response. Front Microbiol. 2017;8:2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gevers D, Knight R, Petrosino JF, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JT, Frank DN, Ramakrishnan V. Microbiome of the paranasal sinuses: Update and literature review. Am J Rhinol Allergy. 2016;30(1):3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshadri S, Rosati M, Lin DC, et al. Regional differences in the expression of innate host defense molecules in sinonasal mucosa. J Allergy Clin Immunol. 2013;132(5): 1227–1230 e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White LC, Weinberger P, Coulson H, et al. Why sinonasal disease spares the inferior turbinate: An immunohistochemical analysis. Laryngoscope. 2016;126(5):E179–183. [DOI] [PubMed] [Google Scholar]
- 40.Kaspar U, Kriegeskorte A, Schubert T, et al. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 2016;18(7):2130–2142. [DOI] [PubMed] [Google Scholar]
- 41.Mahdavinia M, Engen PA, LoSavio PS, et al. The nasal microbiome in patients with chronic rhinosinusitis: Analyzing the effects of atopy and bacterial functional pathways in 111 patients. J Allergy Clin Immunol. 2018;142(1):287–290 e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas RG. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol. 2015;9:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoggard M, Biswas K, Zoing M, Wagner Mackenzie B, Taylor MW, Douglas RG. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7(3):230–239. [DOI] [PubMed] [Google Scholar]
- 44.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384(9944):691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlandi RR, Kingdom TT, Hwang PH, et al. International Consensus Statement on Allergy and Rhinology: Rhinosinusitis. Int Forum Allergy Rhinol. 2016;6 Suppl 1:S22–S209. [DOI] [PubMed] [Google Scholar]
- 46.Wilson MT, Hamilos DL. The nasal and sinus microbiome in health and disease. Curr Allergy Asthma Rep. 2014;14(12):485. [DOI] [PubMed] [Google Scholar]
- 47.Cardinale BJ, Duffy JE, Gonzalez A, et al. Biodiversity loss and its impact on humanity. Nature. 2012;486(7401):59–67. [DOI] [PubMed] [Google Scholar]
- 48.Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4(151):151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chalermwatanachai T, Vilchez-Vargas R, Holtappels G, et al. Chronic rhinosinusitis with nasal polyps is characterized by dysbacteriosis of the nasal microbiota. Sci Rep. 2018;8(1):7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, Sanford T. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2013;139(12):1328–1338. [DOI] [PubMed] [Google Scholar]
- 51.Rom D, Bassiouni A, Eykman E, et al. The Association Between Disease Severity and Microbiome in Chronic Rhinosinusitis. Laryngoscope. 2019;129(6):1265–1273. [DOI] [PubMed] [Google Scholar]
- 52.Barenfanger J, Drake CA, Lawhorn J, Kopec C, Killiam R. Outcomes of improved anaerobic techniques in clinical microbiology. Clin Infect Dis. 2002;35(Suppl 1):S78–83. [DOI] [PubMed] [Google Scholar]
- 53.Brook I. The role of anaerobic bacteria in sinusitis. Anaerobe. 2006;12(1):5–12. [DOI] [PubMed] [Google Scholar]
- 54.Cleland EJ, Bassiouni A, Vreugde S, Wormald PJ. The bacterial microbiome in chronic rhinosinusitis: Richness, diversity, postoperative changes, and patient outcomes. Am J Rhinol Allergy. 2016;30(1):37–43. [DOI] [PubMed] [Google Scholar]
- 55.Stephenson MF, Mfuna L, Dowd SE, et al. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J Otolaryngol Head Neck Surg. 2010;39(2):182–187. [PubMed] [Google Scholar]
- 56.Ivanchenko OA, Karpishchenko SA, Kozlov RS, et al. The microbiome of the maxillary sinus and middle nasal meatus in chronic rhinosinusitis. Rhinology. 2016;54(1):68–74. [DOI] [PubMed] [Google Scholar]
- 57.Kim YJ, Cho HJ, Shin WC, Song HA, Yoon JH, Kim CH. Hypoxia-mediated mechanism of MUC5AC production in human nasal epithelia and its implication in rhinosinusitis. PLoS One. 2014;9(5):e98136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. Pediatric Cystic Fibrosis Sputum Can Be Chemically Dynamic, Anoxic, and Extremely Reduced Due to Hydrogen Sulfide Formation. MBio. 2015;6(4):e00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flynn JM, Niccum D, Dunitz JM, Hunter RC. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS Pathog. 2016;12(8):e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu CM, Soldanova K, Nordstrom L, et al. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis. Int Forum Allergy Rhinol. 2013;3(10):775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wine JJ, Joo NS. Submucosal glands and airway defense. Proc Am Thorac Soc. 2004;1(1):47–53. [DOI] [PubMed] [Google Scholar]
- 64.Tipirneni KE, Cho DY, Skinner DF, et al. Characterization of primary rat nasal epithelial cultures in CFTR knockout rats as a model for CF sinus disease. Laryngoscope. 2017;127(11):E384–E391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia M, Chen Z, Du X, Guo Y, Sun T, Zhao X. A simple animal model of Staphylococcus aureus biofilm in sinusitis. Am J Rhinol Allergy. 2014;28(2):e115–119. [DOI] [PubMed] [Google Scholar]
- 66.London NR Jr., Lane AP. Innate immunity and chronic rhinosinusitis: What we have learned from animal models. Laryngoscope Investig Otolaryngol. 2016;1(3):49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin HW. Animal Models in CRS and Pathophysiologic Insights Gained: A Systematic Review. Laryngoscope Investig Otolaryngol. 2016;1(5):116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cope EK, Goldberg AN, Pletcher SD, Lynch SV. A chronic rhinosinusitis-derived isolate of Pseudomonas aeruginosa induces acute and pervasive effects on the murine upper airway microbiome and host immune response. Int Forum Allergy Rhinol. 2016;6(12):1229–1237. [DOI] [PubMed] [Google Scholar]
- 69.Lindsay R, Slaughter T, Britton-Webb J, et al. Development of a murine model of chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2006;134(5):724–730; discussion 731-722. [DOI] [PubMed] [Google Scholar]
- 70.Liang KL, Jiang RS, Wang J, et al. Developing a rabbit model of rhinogenic chronic rhinosinusitis. Laryngoscope. 2008;118(6):1076–1081. [DOI] [PubMed] [Google Scholar]
- 71.Mashimo T, Serikawa T. Rat resources in biomedical research. Curr Pharm Biotechnol. 2009;10(2):214–220. [DOI] [PubMed] [Google Scholar]
- 72.Al-Sayed AA, Agu RU, Massoud E. Models for the study of nasal and sinus physiology in health and disease: A review of the literature. Laryngoscope Investig Otolaryngol. 2017;2(6):398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho DY, Mackey C, Van Der Pol WJ, et al. Sinus Microanatomy and Microbiota in a Rabbit Model of Rhinosinusitis. Front Cell Infect Microbiol. 2017;7:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DePas WH, Starwalt-Lee R, Van Sambeek L, Ravindra Kumar S, Gradinaru V, Newman DK. Exposing the Three-Dimensional Biogeography and Metabolic States of Pathogens in Cystic Fibrosis Sputum via Hydrogel Embedding, Clearing, and rRNA Labeling. MBio. 2016;7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kopf SH, Sessions AL, Cowley ES, et al. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 2016;113(2):E110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]