Abstract
An in-depth annotation of the newly discovered coronavirus (2019-nCoV) genome has revealed differences between 2019-nCoV and severe acute respiratory syndrome (SARS) or SARS-like coronaviruses. A systematic comparison identified 380 amino acid substitutions between these coronaviruses, which may have caused functional and pathogenic divergence of 2019-nCoV.
An in-depth annotation of the newly discovered coronavirus (2019-nCoV) genome has revealed differences between 2019-nCoV and severe acute respiratory syndrome (SARS) or SARS-like coronaviruses. A systematic comparison identified 380 amino acid substitutions between these coronaviruses, which may have caused functional and pathogenic divergence of 2019-nCoV.
Main Text
A novel coronavirus (CoV) named “2019 novel coronavirus” or “2019-nCoV” by the World Health Organization (WHO) is responsible for the recent pneumonia outbreak that started in early December, 2019 in Wuhan City, Hubei Province, China (Huang et al., 2020, Zhou et al., 2020, Zhu et al., 2020). This outbreak is associated with a large seafood and animal market, and investigations are ongoing to determine the origins of the infection. To date, thousands of human infections have been confirmed in China along with many exported cases across the globe (China CDC, 2020).
Coronaviruses mainly cause respiratory and gastrointestinal tract infections and are genetically classified into four major genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Li, 2016). The former two genera primarily infect mammals, whereas the latter two predominantly infect birds (Tang et al., 2015). Six kinds of human CoVs have been previously identified. These include HCoV-NL63 and HCoV-229E, which belong to the Alphacoronavirus genus; and HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV), which belong to the Betacoronavirus genus (Tang et al., 2015). Coronaviruses did not attract worldwide attention until the 2003 SARS pandemic, followed by the 2012 MERS and, most recently, the 2019-nCoV outbreaks (China CDC, 2020, Song et al., 2019). SARS-CoV and MERS-CoV are considered highly pathogenic (Cui et al., 2019), and it is very likely that both SARS-CoV and MERS-CoV were transmitted from bats to palm civets (Guan et al., 2003) or dromedary camels (Drosten et al., 2014), and finally to humans (Cui et al., 2019).
The genome of coronaviruses, whose size ranges between approximately 26,000 and 32,000 bases, includes a variable number (from 6 to 11) of open reading frames (ORFs) (Song et al., 2019). The first ORF representing approximately 67% of the entire genome encodes 16 non-structural proteins (nsps), while the remaining ORFs encode accessory proteins and structural proteins (Cui et al., 2019). The four major structural proteins are the spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N). The spike surface glycoprotein plays an essential role in binding to receptors on the host cell and determines host tropism (Li, 2016, Zhu et al., 2018). The spike proteins of SARS-CoV and MERS-CoV bind to different host receptors via different receptor-binding domains (RBDs). SARS-CoV uses angiotensin-converting enzyme 2 (ACE2) as one of the main receptors (Ge et al., 2013) with CD209L as an alternative receptor (Jeffers et al., 2004), whereas MERS-CoV uses dipeptidyl peptidase 4 (DPP4, also known as CD26) as the primary receptor. Initial analysis suggested that 2019-nCoV has a close evolutionary association with the SARS-like bat coronaviruses (Zhou et al., 2020). Here, based on the first three determined genomes of the novel coronavirus (2019-nCoV), namely Wuhan/IVDC-HB-01/2019 (GISAID accession ID: EPI_ISL_402119) (HB01), Wuhan/IVDC-HB-04/2019 (EPI_ISL_402120) (HB04), and Wuhan/IVDC-HB-05/2019 (EPI_ISL_402121) (HB05), an in-depth genome annotation of this virus was performed with a comparison to related coronaviruses, including 1,008 human SARS-CoV, 338 bat SARS-like CoV, and 3,131 human MERS-CoV, whose genomes were published before January 12, 2020 (release date: September 12, 2019) from Virus Pathogen Database and Analysis Resource (ViPR) (http://www.viprbrc.org/) and NCBI.
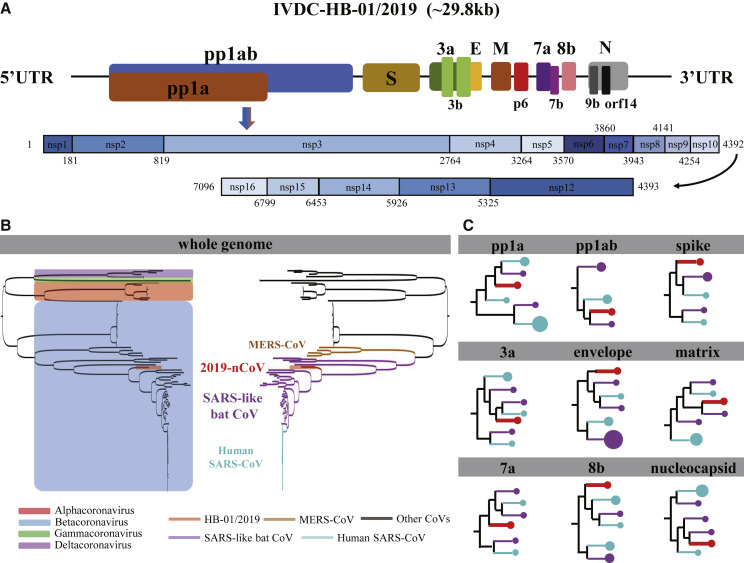
Comparison of genomes of these three strains showed that they are almost identical, with only five nucleotide differences in the genome of ~29.8 kb nucleotides (Figure S1). The 2019-nCoV genome was annotated to possess 14 ORFs encoding 27 proteins (Figure 1 A and Tables S1A and S1B). The orf1ab and orf1a genes located at the 5′-terminus of the genome respectively encode the pp1ab and pp1a proteins, respectively. They together comprise 15 nsps including nsp1 to nsp10 and nsp12 to nsp16 (Figure 1A and Table S1B). The 3′-terminus of the genome contains four structural proteins (S, E, M, and N) and eight accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14). At the amino acid level, the 2019-nCoV is quite similar to that of SARS-CoV, but there are some notable differences. For example, the 8a protein is present in SARS-CoV and absent in 2019-nCoV; the 8b protein is 84 amino acids in SARS-CoV, but longer in 2019-nCoV, with 121 amino acids; the 3b protein is 154 amino acids in SARS-CoV, but shorter in 2019-nCoV, with only 22 amino acids (Table S1A). Further studies are needed to characterize how these differences affect the functionality and pathogenesis of 2019-nCoV.
Figure 1.
Genome composition and phylogenetic tree for 2019-nCoV
(A) Schematic diagram of the genome organization and the encoded proteins of pp1ab and pp1a for the IVDC-HB-01/2019 (HB01) strain. The largest gene, namely the orf1ab, encodes the pp1ab protein that contains 15 nsps (nsp1-nsp10 and nsp12-nsp16). The pp1a protein encoded by the orf1a gene also contains 10 nsps (nsp1-nsp10). Structural proteins are encoded by the four structural genes, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes. The accessory genes are distributed among the structural genes. The protein-encoding genes of the genome of 2019-nCoV were predicted by the online servers of GeneMarkS (http://exon.gatech.edu/GeneMark/genemarks.cgi) and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) with manual check.
(B) Phylogenetic relationship based on the whole genome for the HB01 strain and other coronaviruses. All viral strains were classified by the genus and the type, which are presented on the left and right schematic phylogenetic trees, respectively. The four genera of the coronaviruses, including Alphacoronavirus (red), Betacoronavirus (blue), Gammacoronavirus (green), and Deltacoronavirus (violet) are blocked in the left phylogenetic tree. The MERS coronavirus (brown), the SARS-like bat coronavirus (violet), human SARS coronavirus (light blue), and the HB01 strain (red) are highlighted by lines of different colors in the right phylogenetic tree.
(C) Schematic phylogenetic trees of individual genes for the HB01 strain. The coronavirus species were colored in the same way as (B). The amount of the strains in the phylogenetic clade is denoted by the area of the circles.
As shown in a phylogenetic tree based on whole genomes (Figures 1B and S2) with the Molecular Evolutionary Genetics Analysis (MEGA) (version 7.0), the 2019-nCoV is in the same Betacoronavirus clade as MERS-CoV, SARS-like bat CoV, and SARS-CoV. The phylogenetic tree falls into two clades. The Betacoronavirus genus constitutes one clade, while the Alphacoronavirus, Gammacoronavirus, and Deltacoronavirus genera constitute the other clade. The 2019-nCoV is parallel to the SARS-like bat CoVs, while the SARS-CoVs are descended from the SARS-like bat CoVs, indicating that 2019-nCoV is closer to the SARS-like bat CoVs than the SARS-CoVs in terms of the whole genome sequence. Tables S1C and S1D also show that the genome of 2019-nCoV has the highest similarity with that of a SARS-like bat CoV (MG772933). In comparison, 2019-nCoV is distant from and less related to the MERS-CoVs. In terms of the encoded proteins of pp1ab, pp1a, envelope, matrix, accessory protein 7a, and nucleocapsid genes, phylogenetic analyses showed that the 2019-nCoV is closest to the SARS-like bat CoVs (Figure 1C and Table S1D). Regarding the spike gene, the 2019-nCoV is closest to the bat CoVs, while the 3a and 8b accessory genes are both closest to the SARS-CoVs. Although phylogenetic analyses for the whole genome and individual genes clearly show that the 2019-nCoV is most closely related to SARS-like bat viruses (Figures 1B and 1C), we did not find a single strain of a SARS-like bat virus that harbors all proteins with the most similarity to counterparts of the 2019-nCoV (Figures 1B and 1C).
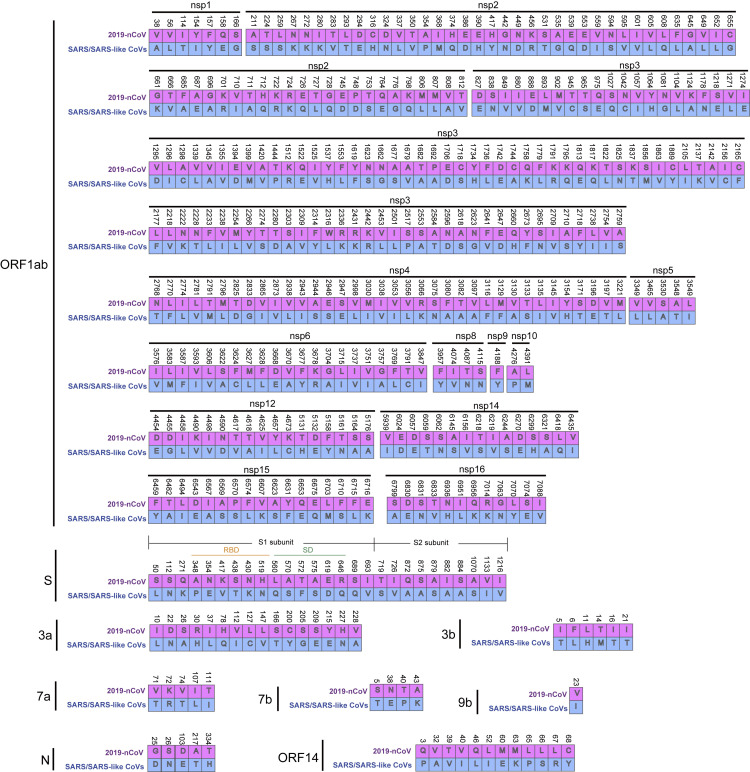
Given the close relationship between 2019-nCoV and SARS-CoVs or SARS-like bat CoVs (Figures 1B and 1C), an examination of the amino acid substitutions in different proteins could shed light into how 2019-nCoV differs structurally and functionally from SARS-CoVs. In total, there were 380 amino acid substitutions between the amino acid sequences of 2019-nCoV (HB01) and the corresponding consensus sequences of SARS and SARS-like viruses (Figure 2 and Tables S1E and S1F). No amino acid substitutions occurred in nonstructural protein 7 (nsp7), nsp13, envelope, matrix, or accessory proteins p6 and 8b (Table S1F). Respectively, 102 and 61 amino acid substitutions are located in nsp3 and nsp2. In addition, 27 amino acid substitutions were found in the spike protein with a length of 1,273 amino acids, including six substitutions in the RBD at amino acid region 357-528 and six substitutions in the underpinning subdomain (SD) at amino acid region 569-655. Moreover, four substitutions (Q560L, S570A, F572T, and S575A) in the C-terminal of the receptor-binding subunit S1 domain (Figure 2) are situated in two peptides previously reported to be antigens for SARS-CoV (Guo et al., 2004).
Figure 2.
Amino Acid Substitutions of 2019-nCoV against SARS and SARS-like Viruses
All 27 proteins encoded by 2019-nCoV have been aligned against SARS-CoVs and SARS-like bat CoVs using the FFT-NS-2 algorithm in MAFFT (version v7.407) (The number of aligned proteins were listed in Table S1E). An amino acid substitution was defined as an absolutely conserved site in the group of SARS and SARS-like CoVs but different from that of 2019-nCoV. In total, 380 amino acid substitutions have been identified between the amino acid sequences of 2019-nCoV (HB01) and the corresponding consensus sequences of SARS and SARS-like CoVs.
Due to very limited knowledge of this novel virus, we are unable to give reasonable explanations for the significant number of amino acid substitutions between the 2019-nCoV and SARS or SARS-like CoVs. For example, no amino acid substitutions were present in the receptor-binding motifs that directly interact with human receptor ACE2 protein in SARS-CoV (Ge et al., 2013), but six mutations occurred in the other region of the RBD. Whether these differences could affect the host tropism and transmission property of the 2019-nCoV compared to SARS-CoV is worthy of future investigation.
Acknowledgments
This work was supported by the National Key Plan for Scientific Research and Development of China (2016YFD0500301 and 2016YFC1200200), CAMS Initiative for Innovative Medicine (CAMS-I2M and 2016-I2M-1-005), the National Natural Science Foundation of China (U1603126), the Central Public-Interest Scientific Institution Basal Research Fund (2016ZX310195, 2017PT31026, and 2018PT31016), and NIH R01AI069120 (United States).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.02.001.
Supplemental Information
(A) The genome annotation of 2019-nCoV strain IVDC-HB-01/2019 (EPI_ISL_402119 in GISAID).
(B) The non-structural proteins included in orf1ab.
(C) Information on the most similar strains with the 2019-nCoV in terms of the whole genome and individual genes.
(D) The sequence similarity between the 2019 nCoV and the strains that had the blast hit and the closest human SARS CoVs in terms of individual genes.
(E) The number of examined protein sequences for Bat SARS-CoV, Human SARS-CoV, and Human MERS-CoV.
(F) The number of amino acid substitutions for 2019-nCoV against different reference sets of SARS and MERS strains.
References
- China CDC Tracking the Epidemic. 2020. http://weekly.chinacdc.cn/news/TrackingtheEpidemic.htm?from=timeline#Beijing%20Municipality%20Update
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Kellam P., Memish Z.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;371:1359–1360. doi: 10.1056/NEJMc1409847. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guo J.P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11:E59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Song Y., Shi M., Cheng Y., Zhang W., Xia X.Q. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci. Rep. 2015;5:17155. doi: 10.1038/srep17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 doi: 10.1101/2020.01.22.914952. [DOI] [Google Scholar]
- Zhu Z., Zhang Z., Chen W., Cai Z., Ge X., Zhu H., Jiang T., Tan W., Peng Y. Predicting the receptor-binding domain usage of the coronavirus based on kmer frequency on spike protein. Infect. Genet. evol. 2018;61:183–184. doi: 10.1016/j.meegid.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The genome annotation of 2019-nCoV strain IVDC-HB-01/2019 (EPI_ISL_402119 in GISAID).
(B) The non-structural proteins included in orf1ab.
(C) Information on the most similar strains with the 2019-nCoV in terms of the whole genome and individual genes.
(D) The sequence similarity between the 2019 nCoV and the strains that had the blast hit and the closest human SARS CoVs in terms of individual genes.
(E) The number of examined protein sequences for Bat SARS-CoV, Human SARS-CoV, and Human MERS-CoV.
(F) The number of amino acid substitutions for 2019-nCoV against different reference sets of SARS and MERS strains.