Summary
In Arabidopsis, the ASYMMETRIC LEAVES2 (AS2) protein plays a key role in the formation of flat symmetric leaves via direct repression of the abaxial gene ETT/ARF3. AS2 encodes a plant‐specific nuclear protein that contains the AS2/LOB domain, which includes a zinc‐finger (ZF) motif that is conserved in the AS2/LOB family. We have shown that AS2 binds to the coding DNA of ETT/ARF3, which requires the ZF motif. AS2 is co‐localized with AS1 in perinucleolar bodies (AS2 bodies). To identify the amino acid signals in AS2 required for formation of AS2 bodies and function(s) in leaf formation, we constructed recombinant DNAs that encoded mutant AS2 proteins fused to yellow fluorescent protein. We examined the subcellular localization of these proteins in cells of cotyledons and leaf primordia of transgenic plants and cultured cells. The amino acid signals essential for formation of AS2 bodies were located within and adjacent to the ZF motif. Mutant AS2 that failed to form AS2 bodies also failed to rescue the as2‐1 mutation. Our results suggest the importance of the formation of AS2 bodies and the nature of interactions of AS2 with its target DNA and nucleolar factors including NUCLEOLIN1. The partial overlap of AS2 bodies with perinucleolar chromocenters with condensed ribosomal RNA genes implies a correlation between AS2 bodies and the chromatin state. Patterns of AS2 bodies in cells during interphase and mitosis in leaf primordia were distinct from those in cultured cells, suggesting that the formation and distribution of AS2 bodies are developmentally modulated in plants.
Keywords: zinc‐finger motif, perinucleolar body, chromocenter, nucleolus, epigenetic factor AS2, 45S ribosomal RNA genes, ETTIN/AUXIN RESEPONSE FACTOR3
Significance Statement
Development of the abaxial and adaxial domains of leaves requires the cooperative actions of the ASYMMETRIC LEAVES2 (AS2) gene and genes for many nucleolus‐localized proteins. We report that formation of perinucleolar bodies (AS2 bodies) is important for normal leaf development and requires the zinc‐finger DNA‐binding motif in AS2, and propose that interactions among AS2, its target DNA, heterochromatin states of ribosomal RNA genes, and nucleolar proteins are essential for normal leaf development.
Introduction
In Arabidopsis thaliana, leaf development, with the establishment of adaxial–abaxial polarity, is regulated epigenetically by a perinucleolar repressor complex that consists of ASYMMETRIC LEAVES2 (AS2) and AS1 (the AS2–AS1 complex) (Guo et al., 2008; Yang et al., 2008; Iwasaki et al., 2013; Lodha et al., 2013) and this process provides a model for studies on the activation of a next genetic program, with repression of a previously functioning program (Machida et al., 2015).
Leaf primordia are formed initially as lateral organs, without adaxial–abaxial polarity, from the shoot apical meristem (a group of stem cells) that is maintained by some class 1 KNOTTED‐like HOMEOBOX (KNOX) genes (Long et al., 1996; Long and Barton, 1998; Barton, 2001). The leaf primordia become elongated and patterned along the proximal–distal axis in parallel with the repression of the KNOX genes via direct binding of AS2–AS1 to the promoter region of KNOX genes (Lin et al., 2003; Guo et al., 2008). During the growth of these primordia to form flat, symmetric leaves with adaxial and abaxial domains, expression of abaxial‐determining genes, such as ETTIN/AUXIN‐RESPONSE‐FACTOR3 (ETT/ARF3) and ARF4 (a functionally redundant version of ARF3), appears to precede the expression of adaxial‐determining genes (Eshed et al., 1999, 2001, 2004). The adaxial domain develops upon the direct repression of these abaxial genes via co‐ordinated binding of AS2–AS1 to the promoter region of ETT/ARF3 (Iwasaki et al., 2013), as well as upon the expression of adaxial‐determining genes in the presumptive adaxial domain (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). Levels of transcripts of ARF4 and ARF3 also decrease as a consequence of degradation of these transcripts via activation by AS2 of tasiR‐ARF‐mediated gene silencing (Iwasaki et al., 2013). Transcripts of the AS2 gene are specifically detected in the adaxial domains of primordia of cotyledons in embryos and leaves (Iwakawa et al., 2002, 2007), this is in agreement with the expression patterns of genes involving the adaxial–abaxial establishment of leaves as mentioned above. Then the proliferation of cells might be induced in the boundary region between the adaxial and abaxial domains, and such proliferation might result in balanced expansion of the leaf primordia along the medial–lateral axis to form flat symmetric leaves (Waites et al., 1998; Hudson, 2000; Semiarti et al., 2001; Tsukaya, 2006; Bowman and Floyd, 2008; Szakonyi et al., 2010; Nakata and Okada, 2013; Machida et al., 2015).
The AS2–AS1 complex also acts as an epigenetic regulator for leaf adaxial specification. This complex directly represses transcription of some genes in two families, as follows. Levels of transcripts of some genes in the class 1 KNOX family are downregulated via histone modification (Phelps‐Durr et al., 2005; Guo et al., 2008; Lodha et al., 2013). AS1 and AS2 are also involved in maintaining METHYLTRANSFERASE1 (MET1)‐mediated CpG methylation in exon 6 of the ETT/ARF3 locus (Iwasaki et al., 2013). In the shoot apices of met1 plants, levels of the ETT/ARF3 transcript are elevated. Therefore, the level of the ETT/ARF3 transcript is inversely correlated with the extent of methylation at CpG sites in this gene. In addition, AS2–AS1 and many other factors, including various nucleolar proteins that mediate biogenesis of ribosomal RNAs, act co‐operatively to repress levels of transcripts of target genes, suggesting critical roles for these nucleolar factors as modifiers of the actions of AS2 and AS1 (Ueno et al., 2007; Pinon et al., 2008; Yao et al., 2008; Szakonyi et al., 2010; Horiguchi et al., 2011; Szakonyi and Byrne, 2011; Machida et al., 2015; Matsumura et al., 2016). We recently reported that, in addition to AS2, genes for two nucleolar proteins, NUCLEOLIN1 (NUC1) and RNA HELICASE 10 (RH10), are also involved in repressing the transcription of ETT/ARF3, the target gene of AS2, and CpG methylation in exon 6 of ETT/ARF3 (Vial‐Pradel et al., 2018). Moreover, AS2 binds to the CpG repeat in exon 1 of the ETT/ARF3 gene. These findings lend further support to the hypothesis that the epigenetic repression of ETT/ARF3 involves CpG methylation mediated by AS2 and AS1.
AS1 encodes a myb‐domain protein (Byrne et al., 2000). AS2 encodes a nuclear protein of 199 amino acid residues and has a plant‐specific AS2/LOB (ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARY) domain of 100 amino acid residues (residues 10 to 109) near its N‐terminus, and this domain is conserved in all 42 known members of the family of AS2‐LIKE/LOB DOMAIN proteins (ASL/LBD proteins) within the AS2/LOB family (Iwakawa et al., 2002; Shuai et al., 2002; Matsumura et al., 2009). The AS2/LOB domain of AS2 contains a zinc‐finger‐like (ZF‐like) motif that includes four cysteine residues (formerly designated the C‐motif or C‐block; Iwakawa et al., 2002; Shuai et al., 2002) at its N‐terminus. This motif in a wheat member of the AS2/LOB family binds a zinc ion, which is required for binding to DNA (Chen et al., 2019), an observation that implies that members of this family can form zinc‐finger (ZF) structures. The ZF motif is essential for binding of AS2 to the CpG repeat in exon 1 of its target ETT/ARF3 gene (Vial‐Pradel et al., 2018). The AS2/LOB domain includes two additional conserved regions: an internal‐co‐nserved‐glycine (ICG) region and a leucine‐zipper‐like (LZL) region (Iwakawa et al., 2002; Matsumura et al., 2009). Both these regions are essential for the functions of AS2 in the normal development of leaf polarity (Semiarti et al., 2001; Iwakawa et al., 2002; Matsumura et al., 2009; Lee et al., 2013).
AS2 is localized as nuclear bodies, known as AS2 bodies, in regions adjacent to the nucleolus (Ueno et al., 2007). AS1 has also been found in AS2 bodies (Ueno et al., 2007). The AS2/LOB domain is sufficient for the formation of AS2 bodies (Luo et al., 2012a). Taken together with the co‐operation with nucleolus‐localized proteins in the repression of target gene expression, these observations suggest a potential role for AS2 in the repression of gene expression via nucleolus‐related events, such as the formation of AS2 bodies. The amino acid signals in the AS2/LOB domain that are required for its nuclear localization and the formation of AS2 bodies at the perinucleolar region remain, however, to be elucidated.
The results of the present study show that the ZF motif of AS2, which is required for binding to the coding DNA of ETT/ARF3 (Vial‐Pradel et al., 2018), is essential for the formation of AS2 bodies in adaxial epidermal cells but not for nuclear localization. Furthermore, the regions encompassing the ICG region and the LZL region are essential for the nuclear localization of AS2. Mutant AS2 proteins that were unable to form the AS2 bodies did not rescue the as2‐1 mutation, demonstrating a correlation between the formation of AS2 bodies and their functions in development. AS2 bodies partially overlapped with perinucleolar chromocenters and condensed rRNA genes. Therefore, there appears to be a correlation between AS2 bodies and heterochromatin states of rRNA genes in chromocenters. The distribution of AS2 bodies during the M phase of the cell cycle in intact plants was different from that of AS2 bodies in cultured cells, suggesting the developmental control of the formation of AS2 bodies and their distribution in plants.
Results
Detection of fluorescent signals due to AS2‐YFP in epidermal cells of developing cotyledons and leaf primordia
We examined the expression of an estradiol‐inducible recombinant AS2 gene in transgenic Arabidopsis plants. We fused the entire length of AS2 cDNA to the YFP sequence (which encodes yellow fluorescent protein (YFP)), linking the fusion construct to the estradiol‐inducible XVE promoter (XVE‐AS2‐YFP). Then we transformed wild‐type Arabidopsis (Col‐0) with the XVE‐AS2‐YFP construct. We determined the optimal conditions for induction of expression of each transgene by estradiol in the transgenic plants (Figure S1 and Appendix [Link], [Link], [Link]). When cotyledons of 7‐day‐old Arabidopsis seedlings, which had been transformed with an AS2‐YFP construct, were incubated with 0.05 µm 17β‐estradiol for 16 h, bands of protein with a molecular mass of c. 48 kDa (corresponding to the mass of AS2‐YFP) were visualized by western blotting with AS2‐specific antibodies (lanes 5–9 in Figure S1a). Endogenous AS2 protein in protein extracted from flower buds had a molecular mass of c. 25 kDa (lane 1 in Figure S1a). The level of AS2‐YFP induced by 0.05 µm 17β‐estradiol (lane 5) was higher than that of endogenous AS2 from flower buds. Under the same conditions, we observed signals due to YFP in many cells within the adaxial surface of cotyledons (see typical optical sections of the epidermis in Figures 1a, S2a and Appendix S1) and of leaf primordia (Figures 1b, S2b) around the shoot apex. These locations are consistent with observations of the expression of the endogenous AS2 gene reported by Iwakawa et al. (2002 and 2007). When we incubated plants with 0.005 µm 17β‐estradiol for 16 h or longer, we did not detect signals due to YFP in cells at these locations. We examined nuclei and chromosomes after staining with 4′6‐diamidino‐2‐phenylindole (DAPI). Analysis of signals due to DAPI revealed that almost all cells in cotyledons and leaf primordia had entered interphase or the G0 (differentiated) stage and only a few cells had undergone mitosis (examples are indicated by arrows in Figure 1(a,b), with magnified views in Figure 1(c,d), respectively). Although nuclear bodies were visible in cells in these sections, they were not clearly apparent in most cells at low magnification (Figure 1(a,b). Patterns of fluorescent signals from these cells are discussed in detail below. Levels of accumulated AS2‐YFP continued to rise during incubation for 48 h (Figure S1b). We chose to monitor cells in cotyledons and leaf primordia of transgenic seedlings that had been incubated in the presence of 0.05 µm 17β‐estradiol for 16 h in subsequent experiments. As confirmed below (see Figure 4), the AS2‐YFP construct was functional.
Figure 1.
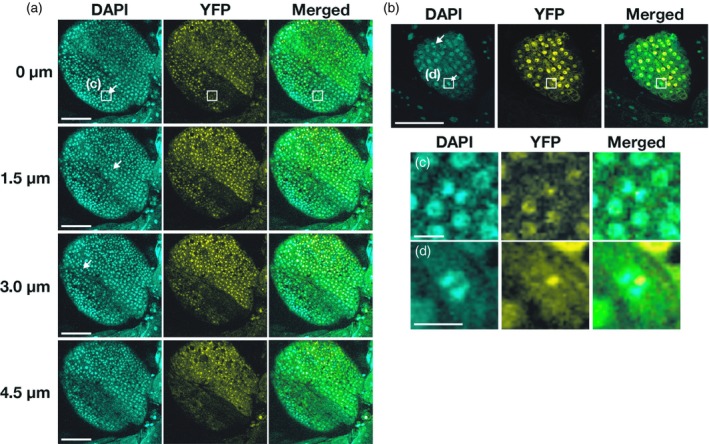
Observations of AS2‐YFP at the adaxial surfaces of cotyledons and leaf primordia. Expression of AS2‐YFP was induced by incubating 7‐day‐old transgenic Arabidopsis plants (7 days after sowing) with 0.05 µm 17β‐estradiol for 16 h. Aboveground parts of seedlings were fixed in 3.7% paraformaldehyde and chromosomes and nuclei were stained with DAPI. Cells at mitosis, with condensed chromosomes, are indicated by white arrows.
(a) Fluorescence due to DAPI (cyan) and YFP (yellow) in cells at the adaxial surface of a cotyledon was monitored at 0.5‐µm intervals by confocal fluorescence microscopy. Merged images are also shown. Optical sections of a given cotyledon at the indicated depths are shown as typical examples. Bars, 100 µm. (b) Signals due to DAPI (cyan) and YFP (yellow) in cells at the adaxial surface of a leaf primordium are shown in one typical optical section. Bar, 100 µm. Magnified views of boxed regions in panels (a) and (b) are shown in (c) and (d), respectively. Bars, 10 µm.
The ZF motif plays a role in the formation of AS2 bodies and the ICG and LZL regions are required for nuclear localization of AS2
As depicted schematically in Figure 2(a) (top left), we introduced deletions into four regions of the AS2 cDNA sequence, as follows: the N‐terminal region (residues 2–7; as2‐∆N2–7); the N‐terminal region plus the ZF motif (residues 2–24; as2‐ΔNΔZFL2–24); the ICG region (residues 25–80; as2‐ΔICG25–80), and the LZL region (residues 81–109; as2ΔLZL81–109). Although the N‐terminal region might not be expected to form a structurally distinct domain, the 18‐base‐pair sequence that corresponds to the amino acid sequence from the first to the sixth residue of AS2 (Table S1) is essential for the expression of AS2 (Iwakawa et al., 2007). Therefore, it seemed reasonable to examine the role of the N‐terminal region in the formation of AS2 bodies and the development of leaves. We fused each of the four deletion constructs to the YFP sequence and linked each of the four fusion constructs to the estradiol‐inducible XVE promoter. We transformed wild‐type Arabidopsis plants (Col‐0) with these constructs and established several independent transgenic lines of plants that expressed the respective constructs, as depicted in Figure 2(a).
Figure 2.
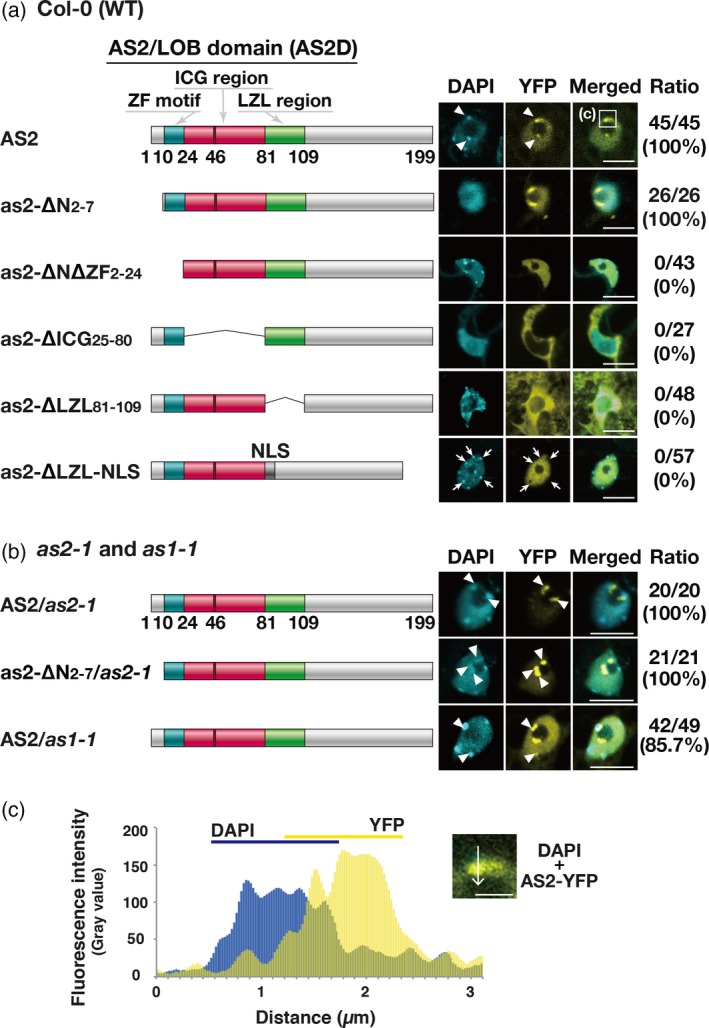
The zinc‐finger motif is required for localization of AS2 to AS2 bodies and the ICG and LZL regions are required for nuclear localization of AS2.
(a. left) Schematic representation of AS2 and variant proteins with deletions in the AS2/LOB domain. DNA constructs encoding mutant proteins were fused to the sequence encoding to the N‐terminus of YFP and these fused constructs were linked to the estrogen‐inducible promoter. See text for details. (a. right) Images showing the signals from YFP fusion proteins in Arabidopsis plants (Col‐0) transformed with these DNA constructs. Three to five independent transgenic lines were established for analysis of AS2‐YFP and each as2‐variant‐YFP: four lines for AS2‐YFP; five lines for as2‐∆N2‐7‐YFP; and three lines each for as2‐∆N∆ZF2–24‐YFP, as2‐∆ICG25–80‐YFP, as2‐∆LZL81–109‐YFP and as2‐∆LZL‐NLS‐YFP. Expression of these genes was induced as described in the legend to Figure 1. Four to 27 cells that were YFP‐positive were observed in the adaxial epidermis of cotyledons of each transgenic line, as described in the legend to Figure 1. Signals due to DAPI (cyan), those due to YFP (yellow) and merged images in representative cells are shown. Numbers on the right side of the images show the ratios of total numbers of AS2 body‐positive cells to total numbers of YFP‐positive cells, which were obtained by adding the numbers of cells from the analysis of each transgenic line. Arrows in the sixth row (as2‐∆LZL‐NLS) show DAPI‐positive granules overlapping YFP‐negative areas. Bars, 10 µm. (b, left) Schematic representation of AS2 and as2‐∆N2–7. (b, right) Images showing the signals from YFP fusion proteins in as2‐1 and as1‐1 plants. Three transgenic lines were selected for analysis of fluorescence, which was visualized as described above. Signals due to DAPI (cyan) and YFP (yellow) and merged images are shown. Numbers on the right show the ratios of total AS2 body‐positive cells to total YFP‐positive cells in each transgenic line, counted as described above. Chromocenters and AS2 bodies that partially overlapped one another are indicated by white arrowheads. In (a) and (b), bars, 10 µm. (c) Partial overlap of the DAPI signal from the chromocenter with the YFP signal from an AS2 body formed by AS2‐YFP. The inset is a magnified view of the boxed region in panel a (top right, labeled c). Bar, 3 µm. Intensities of fluorescence signals due to YFP and DAPI were measured along the white arrow with ImageJ software (https://imagej.nih.gov/ij/).
We examined whether the mutant proteins could support nuclear localization and the formation of AS2 bodies by observing signals due to YFP in cells at adaxial surfaces of cotyledons from transgenic seedlings. Nuclei were visualized by staining with DAPI. The nucleoplasm and the nucleolus were detected as DAPI‐positive and DAPI‐negative (dark) areas, respectively (Ueno et al., 2007; Matsumura et al., 2016). As shown in Figure 2(a) (top right), signals due to YFP in AS2‐YFP‐expressing cells were mainly detected in the nucleoplasm and nuclear bodies, as reported previously for AS2 bodies themselves (Ueno et al., 2007; Luo et al., 2012a) in all YFP‐positive cells (Figures 1, 2a and 3a). Nuclear bodies were detected adjacent to nucleoli and, sometimes, slightly inside nucleoli. As shown in Figure 2(b), AS2 bodies were also formed on as2‐1 and as1‐1 mutant backgrounds when AS2‐YFP was introduced into the respective mutants, suggesting that neither endogenous AS2 nor endogenous AS1 is required for the formation of AS2 bodies.
Figure 3.
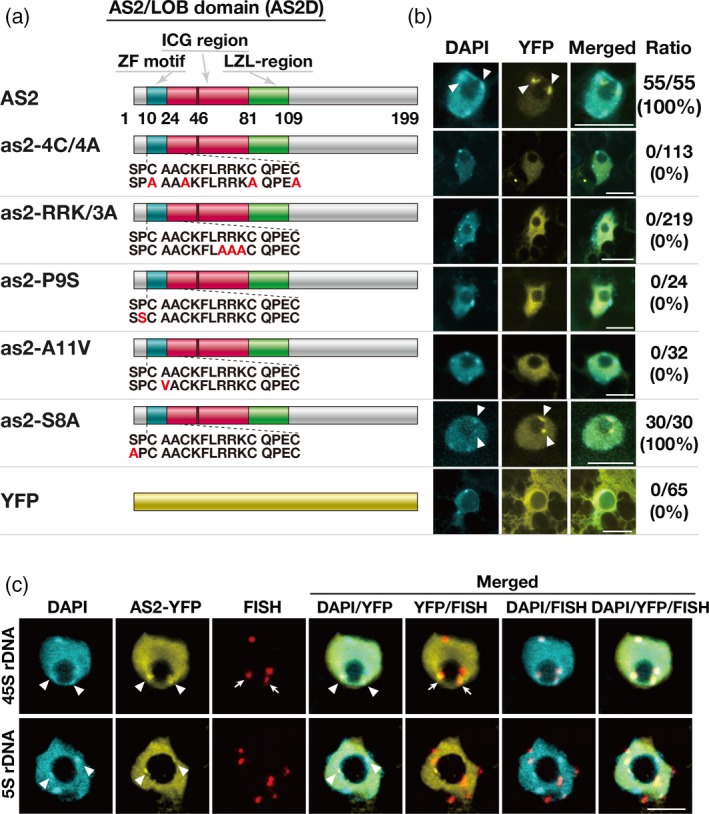
Amino acid residues in the zinc‐finger motif are critical for the formation of AS2 bodies.
(a) Schematic representation of AS2 and variant proteins with amino acid substitutions in the ZF motif. Numbers below the wild‐type representation indicate positions of amino acid residues in AS2. Amino acid sequences in and around the ZF motif of the wild‐type are shown and mutated residues are depicted in red. The DNA constructs were linked to an estrogen‐inducible promoter. Details of the DNA constructs that encoded AS2‐YFP and as2‐variant‐YFP proteins can be found in the text. YFP is shown by the yellow bar at the bottom. Arabidopsis plants (Col‐0) were transformed with indicated constructs. (b) Images showing the signals from YFP fusion proteins in Arabidopsis plants (Col‐0) that had been transformed with the respective DNA constructs. Three independent transgenic lines were established for analysis of AS2‐YFP and each as2‐variant‐YFP protein. Expression of these genes was induced as described in the legend to Figure 1. Eight to 75 cells that were YFP‐positive were observed in the adaxial epidermis of cotyledons of each transgenic line, as described in the legend to Figure 2. Signals due to DAPI (cyan), those due to YFP (yellow) and merged images are shown. Numbers on the right side of the fluorescence images show the ratios of total numbers of AS2 body‐positive cells to total numbers of YFP‐positive cells, obtained by adding numbers of cells from the analysis of each transgenic line. Chromocenters and AS2 bodies that partially overlapped one another are indicated by white arrowheads. Bars, 10 µm. (c) Observation of 45S rDNA and 5S rDNA loci in cells of the aerial part of the AS2‐YFP‐expressing transgenic plant, as described above. DNA‐FISH was performed using 45S (top row) and 5S rRNA gene‐specific probes (bottom row). Signals due to DAPI (cyan), anti‐GFP (AS2‐YFP; yellow), FISH (red), and the merged images are shown. Chromocenters and AS2 bodies that partially overlapped one another are indicated by white arrowheads. FISH signals against 45S rDNA loci that partially overlapped with AS2 bodies are indicated by white arrows. Bars, 10 μm.
Signals due to YFP in as2‐∆N2–7‐YFP cells were detected in AS2 bodies and the nucleoplasm in all the YFP‐positive cells in wild‐type (Figure 2a, second row) and as2‐1 mutant plants (Figure 2b, second row). These results suggested that the short N‐terminal region was not required for the formation of AS2 bodies. Fluorescence due to YFP in as2‐∆N∆ZF2–24‐YFP cells was detected as disperse signals in the nucleoplasm, but none was detected in AS2 bodies (Figure 2a, third row), suggesting that the ZF motif plays an essential role in the formation of AS2 bodies in Arabidopsis plants and that the region from the N‐terminus to the ZF motif is not involved in the nuclear localization of AS2, consistent with previous observations in the tobacco BY‐2 cultured cells (Luo et al., 2012a).
As shown in Figure 2(a) (fourth row), signals due to YFP in as2‐∆ICG25–80‐YFP cells were evident exclusively in the cytoplasm and there was no detectable fluorescence in the nuclei, implying that the ICG region is involved in the nuclear accumulation of the AS2 protein and that regions other than the ICG region play a role in retaining the AS2 protein in the cytoplasm. Fluorescent signals in as2‐∆LZL81–109‐YFP cells were detected both in the nucleus and in the cytoplasm (Figure 2a, fifth row), and this pattern of localization was similar to that of the localization of YFP alone (Figure 3, bottom row). Although some signals were detected in nuclei, no signals were associated with nuclear bodies. Therefore, the LZL region also appeared to be involved in the nuclear localization of AS2.
We examined the effects of the SV40 nuclear localization signal (NLS) on the subcellular localization of as2‐∆LZL‐YFP and the formation of AS2 bodies. We replaced the sequence that corresponded to the LZL region with the NLS sequence to generate as2‐∆LZL‐NLS‐YFP (Figure 2a, bottom row). Signals due to YFP in as2‐∆LZL‐NLS‐YFP cells were detected exclusively in the nucleoplasm but, notably, not as AS2 body‐like aggregates (Figure 2a, bottom). Therefore, loss of the LZL‐mediated ability to form AS2 bodies was not overcome by replacement of the LZL region by the SV40 NLS. The pattern of YFP signals was not homogeneous: many YFP‐negative (dark) areas were visible in the nucleoplasm, as indicated by arrows. Many chromocenter‐like granules were also visualized by DAPI staining in the nucleoplasm (DAPI in Figure 2a, bottom row). Some DAPI‐positive granules overlapped YFP‐negative areas, as also indicated by arrows.
We confirmed that the fusion proteins described above remained intact, for the most part, in transgenic plants (Figure S3 and Appendix [Link], [Link], [Link]). Taken together, our results suggest that the amino acid sequence encompassing the ICG and LZL regions is involved in the nuclear localization of AS2 and that the ZF motif is required for the formation of AS2 bodies.
AS2 bodies that were formed by wild‐type AS2‐YFP and as2‐∆N2–7‐YFP were in contact with the nucleolus and at least partially within the nucleolus. Moreover, they partially overlapped chromocenters that were densely stained with DAPI and localized to the periphery of nucleoli (arrowheads in Figure 2a, top row; Figure 2b, top, second and bottom rows; Figure 2c and Movie S1).
Amino acid residues conserved in and around the ZF motif are critical for the formation of AS2 bodies
The amino acid sequence in the ZF motif of AS2 (residues 10–24) is strongly conserved among at least 25 members of the class 1a subfamily of the AS2/LOB protein family (Table S1; AS2 and ASL/LBD proteins from ASL1 through ASL25), in which 73% of amino acid residues are identical, on average (Iwakawa et al., 2002; Table S1). We introduced various point mutations into the AS2 cDNA sequence to replace amino acid residues that were especially strongly conserved within this subfamily (Table S1 and Figure 3a). We replaced the four cysteine residues at positions 10, 13, 20 and 24 in the ZF motif, which are perfectly conserved in all members of the subfamily, with alanine residues to generate mutant as2‐4C/4A proteins. In all 25 members of the subfamily, the amino acid sequences on the N‐terminal side of the fourth cysteine residue consist of RRK or RRR sequences, which have been proposed to be part of nucleolar localization signals (NOLS; Song and Wu, 2005; Musinova et al., 2011; de Melo et al., 2013; Earley et al., 2015). The RRK sequence in AS2 was replaced with a cluster of three alanine residues to generate mutant as2‐RRK/3A. We also replaced the proline and alanine residues at positions 9 and 11 of AS2, which are adjacent to the first cysteine residue in the ZF motif, by serine (P9S) and valine (A11V) residues, respectively, because these proline and alanine residues are strongly conserved among AS2 and ASL/LBD proteins (Table S1 and Figure 3a). We also introduced a single mutation to replace the conserved serine residue at position 8 of AS2 with an alanine residue (S8A; Table S1 and Figure 3a), because SP and TP sequences are predicted sites of phosphorylation and are strongly conserved in the class 1a subfamily (77%; Table S1). We fused the respective mutated cDNAs to YFP cDNA and linked the fusion constructs to the inducible promoter. We transformed wild‐type Arabidopsis plants (Col‐0) with recombinant AS2 DNA and the various derivatives described above.
We selected three transgenic lines that expressed each construct and examined patterns of YFP signals in cells, as described in the previous section. As shown in Figure 3(b) (second row), YFP fluorescence in all cells that expressed as2‐4C/4A‐YFP was dispersed throughout the entire nucleoplasm and no nuclear bodies were observed, suggesting essential roles for the cysteine residues in the formation of AS2 bodies. In the cells that expressed as2‐RRK/3A‐YFP, the YFP signal was also dispersed in the nucleoplasm and none was seen in AS2 bodies (Figure 3b, third row), suggesting that the RRK sequence is essential for the formation of AS2 bodies but is not required for the nuclear localization of AS2.
Signals due to as2‐P9S‐YFP and as2‐A11V‐YFP were dispersed in the nucleoplasm and no signals were detected in AS2 bodies (Figure 3b, fourth and fifth rows). These results suggest an essential role for the proline and alanine residues adjacent to the cysteine residue in the formation of AS2 bodies. The replacement of the serine residue at position 8 by an alanine residue (as2‐S8A) did not affect the formation of AS2 bodies (Figure 3b, sixth row), suggesting that the serine residue is not critical for the formation of AS2 bodies. The partial overlap of AS2 bodies with perinucleolar chromocenters was observed in cells of transgenic Arabidopsis plants expressing the as2‐S8A‐YFP (Figure 3b, sixth row) as well as the wild‐type AS2‐YFP (Figure 3b, first row). We confirmed that the fusion proteins described above remained intact, for the most part, in the respective transgenic plants (Figure S3). Our observations indicate that the strongly conserved amino acid residues within the ZF motif of AS2 and the proline residue at position 9, adjacent to the ZF motif, are essential for the formation of AS2 bodies.
In leaf cells, inactive rDNA repeats are condensed into heterochromatin, are localized to the external region of the nucleolus, and have loci that are co‐incident with chromocenters (Pontes et al., 2003, 2007; Pontvianne et al., 2007). Using the DNA sequence coding for 45S pre‐ribosomal RNA (45S rDNA) of A. thaliana as a probe for fluorescence in situ hybridization (FISH), we examined the relationship between the 45S rDNA repeats and the AS2 bodies mentioned in earlier sections (Figure 3c, top row). In addition, we observed chromocenters by DAPI staining. As described in the Experimental Procedures section, the cells of the aerial part of a 6‐day‐old AS2‐YFP plant were stained with anti‐green fluorescent protein (GFP) antibodies, which also recognize YFP, to detect AS2‐YFP. Fluorescent signals from anti‐GFP antibody staining were detected in the nucleoplasm and nuclear bodies, as observed in the case of AS2 bodies that were detected as fluorescent signals due to AS2‐YFP. The fluorescent signals due to the AS2 bodies overlapped those observed due to the chromocenters (indicated by arrowheads in Figure 3c, top row). Using FISH staining for 45S rDNA repeats, four fluorescent signals (red) were observed, as expected, and at least two (indicated by arrows) of these four signals were found to partially overlap the fluorescent signals due to AS2‐YFP that were localized at the perinucleolar regions. We examined five cells to identify any possible patterns of the fluorescent signals and found these cells to exhibit patterns similar, in terms of the numbers of YFP and FISH signals, to those presented in Figure 3(c). When the DNA sequence for 5S ribosomal RNA was used as a FISH probe for a negative control, FISH signals at six chromosomal sites were detected, but no FISH signals were found to overlap with those due to AS2‐YFP (Figure 3c, bottom row). These results suggest an interaction of AS2‐YFP bodies with 45S rDNA repeats, which might correspond to that of chromosome 2 (inactive NOR2) or chromosome 4 (active NOR4) (Chandrasekhara et al., 2016).
A correlation between the ability of the AS2 protein to form AS2 bodies and the role of AS2 in leaf development
We examined the ability of the above‐described YFP‐fused AS2 derivatives to normalize the abnormal leaf phenotype of as2‐1. We used AS2‐YFP as a positive control to judge how efficiently these derivatives could rescue the as2‐1 mutation. In as2‐1 plants, the leaf margin is curls downward to a significant extent, as compared with that of wild‐type plants (Semiarti et al., 2001). When exogenous AS2 cDNA is expressed in as2‐1 plants, leaves have four distinct phenotypes: normal (Type I); partially upwardly curled (Type II); and upwardly curled phenotype (Type III; Iwakawa et al., 2007) in addition to the non‐rescued as2‐1 phenotype. Upward curling is typical, and the extent of curling corresponds to the level of expression of exogenous AS2 (Nakazawa et al., 2003; Iwakawa et al., 2007; Chen et al., 2013).
In the present study, we introduced DNA constructs that encoded YFP‐fused AS2 and derivatives of AS2 into as2‐1 mutant plants and, for each construct, we selected three to five independent transgenic lines in which fluorescence due to YFP was apparent. We used five independent transgenic lines that harbored AS2‐YFP, and we induced expression of the gene by germinating seeds and growing plants on Murashige and Skoog (MS) plates supplemented with 0.5 µm estradiol for 14 days. Figure 4(a) shows the typical phenotype of one representative transgenic line (#6) that harbored AS2‐YFP, and the leaf phenotypes are classified as Type I, Type II and Type III, as defined above. Two (1.7%) of 119 transgenic plants exhibited the wild‐type phenotype (Type I; Figure 4a,i), eight (6.7%) of 119 plants exhibited the Type II phenotype, having partially upwardly curled leaves (Figure 4i), and 56 (47.1%) plants exhibited the Type III phenotype, having clearly upwardly curled leaves (Figure 4i). The remaining 53 plants exhibited the as2‐1 phenotype. Therefore, the phenotypes of 55.5% of the as2‐1 transgenic plants might have been the result of complementation by the wild‐type AS2‐YFP construct under our experimental conditions (Figure 4i). The Type III phenotype might have been generated by overexpression of AS2‐YFP (Iwakawa et al., 2007).
Figure 4.
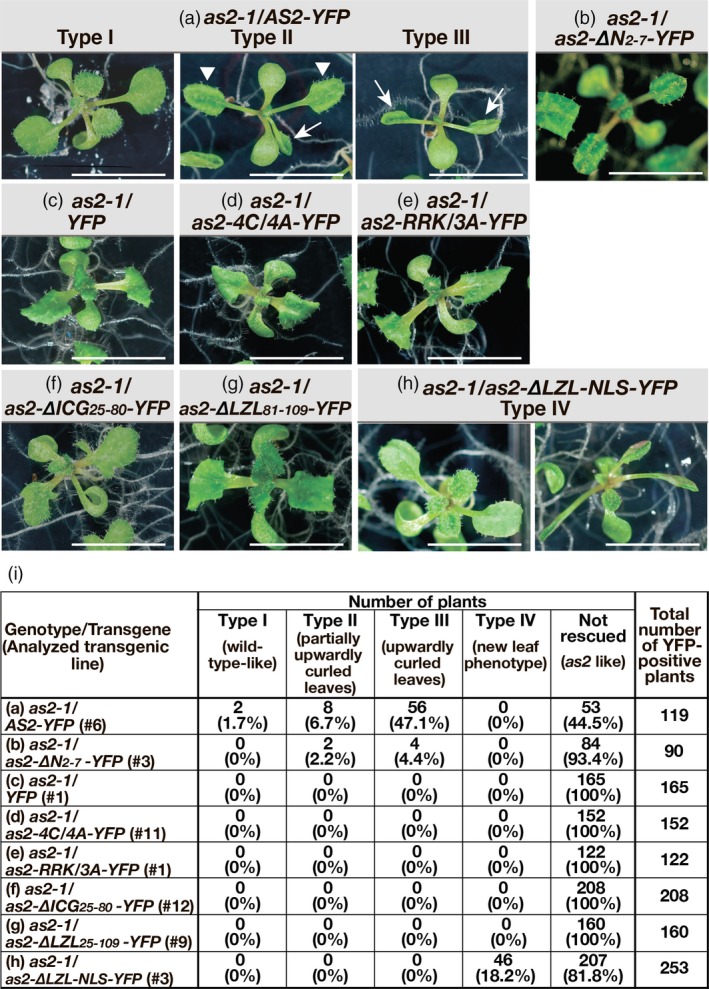
Variant as2 proteins that failed to form AS2 bodies did not rescue the as2‐1 mutation.
Transgenic as2‐1 seeds harboring AS2‐YFP, as2‐variant‐YFPs and YFP, as indicated above the images, were grown on MS plates supplemented with 0.5 µm 17β‐estradiol under white light for 16 h and in darkness for 8 h daily, at 22ºC, as described in Experimental Procedures. The gross morphology of 14‐day‐old transgenic plants was analyzed and representative morphology is shown for each transgenic line in panels (a–h). On the basis of phenotypes of transgenic as2‐1 plants that harbored AS2‐YFP plants were classified into four types (Types I, II, III and non‐rescued as2‐like plants). Panel (i) shows numbers of plants belonging to five types (Types I to IV and non‐rescued as2‐like plants) and corresponding percentages for YFP signal‐positive plants of individual transgenic lines, as denoted by a number with # in parenthesis in the left‐hand column. Arrowheads and arrows in panel (a) indicate partially upwardly curled leaves and upwardly curled leaves, respectively. Transgenic as2‐1 plants harboring as2‐∆LZL‐NLS‐YFP [panels (h)] exhibited an unexpected phenotype (Type IV) in addition to the non‐complemented as2‐like phenotype. Bars, 1 cm.
We selected five transgenic lines that harbored as2‐∆N2–7‐YFP. In one representative transgenic line (#3) harboring as2‐∆N2–7‐YFP, 6 of 90 plants (6.6%) exhibited Type II and Type III phenotypes (Figure 4b,i). We obtained similar results with three other lines. In one other transgenic line, no complemented plants were obtained. Therefore, as2‐∆N2–7‐YFP had very weak rescue ability. Clearly, the short deleted region must be required for full activity of AS2 in the development of wild‐type leaf morphology.
We selected three to five transgenic lines of the other variants. As shown in Figure 4(d–g), the as2‐4CA‐YFP, as2‐RRKA‐YFP, as2‐∆ICG25–80‐YFP, and as2‐∆LZL81–109‐YFP transgenes did not rescue the as2‐1 mutation: the transgenic plants all resembled YFP‐transformed as2‐1 plants (Figure 4c,i). Therefore, many amino acid residues in the ZF motif and the ICG and LZL regions, which are critical for the formation of AS2 bodies, are required for wild‐type leaf development.
When as2‐∆LZL‐NLS‐YFP was expressed in plants, 81.8% of transgenic plants exhibited phenotypes that were similar to that of as2‐1 plants (Figure 4h,i). However, 18.2% of transgenic plants (16 + 30 = 46 of 253 plants examined in groups represented by Figure 4h, Type IV) did have abnormal phenotypes that were somewhat different from the as2‐1 phenotypes. These results suggest that this variant might have a limited capacity to participate in leaf development even though it appeared unable to form AS2 bodies.
Western blotting analysis demonstrated that the chimeric proteins remained intact, for the most part, in the transgenic plants (Figure S3). We concluded that the mutant AS2 proteins that were unable to form AS2 bodies also failed to rescue the as2‐1 mutation. Our observations (Figures 2 and 4) suggest that the ability of AS2 to form AS2 bodies is a pre‐requisite for the normal development of leaves but is not, by itself, sufficient.
The behavior of AS2 bodies in cells of leaf primordia differs from that of AS2 bodies in cultured cells
During the present study, we noticed that the behavior of AS2 bodies in cells of intact Arabidopsis plants was very different from that of AS2 bodies in cells of the tobacco cultured cell line BY‐2 (Luo et al., 2012a). The number of AS2 body‐forming cells relative to the total number of YFP‐positive interphase BY‐2 cells was very low (4.7%; Luo et al., 2012a), but the proportion of AS2 body‐forming cells relative to all interphase (or G0 stage) cells in leaf primordia of Arabidopsis plants was very high (Figure 5). Moreover, the pattern of separation of AS2 bodies during the M phase of the cell cycle in BY‐2 cells (Luo et al., 2012a) was also different from that of separation of AS2 bodies during progression of the M phase in leaf primordial cells of Arabidopsis plants (Figure 5). Therefore, we examined whether a similar difference might be observed between Arabidopsis plants and the Arabidopsis cultured cell line MM2d.
Figure 5.
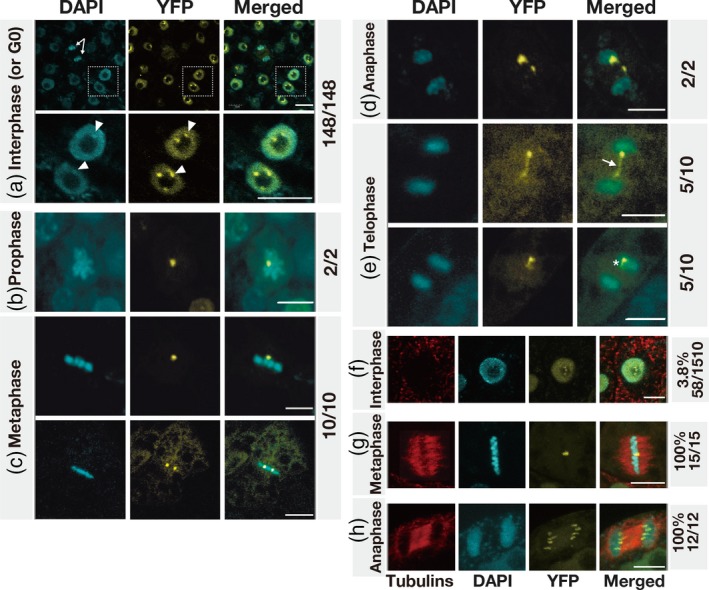
Subcellular localization of AS2‐YFP in interphase and M phase cells in leaf primordia and cultured cell line MM2d.
Expression of AS2‐YFP was induced by incubating 7‐day‐old seedlings of transgenic Arabidopsis plants with 0.05 µm 17β‐estradiol for 16 h (see Experimental Procedures). Seedlings were fixed in 3.7% paraformaldehyde, and chromosomes and nuclei were stained with DAPI. Fluorescence due to DAPI (cyan) and that due to YFP (yellow) was visualized by confocal fluorescence microscopy. Merged images of fluorescence due to DAPI and YFP are also shown. Stages of interphase (or G0, the differentiated phase) (a) and mitotic phases [panels (b–e)] were defined in terms of patterns of staining with DAPI. Panel (a) (top row) shows many interphase (or G0) cells and one anaphase cell (indicated by the white arrows) and magnified views of the boxed nuclei are shown in the lower row. Chromocenters and AS2 bodies that partially overlapped one another are indicated by white arrowheads. Images in (b–e) are seven‐image stacks from an individual sample or cell. The white arrow and the asterisk in (e) indicate a bridge‐like and a tail‐like structures, respectively (see text). The subcellular localization of AS2‐YFP in cells of the MM2d cultured cell line was analyzed similarly, as shown in (f–h). Cells were incubated for 16 h in the presence of 0.05 µm 17β‐estradiol and fixed as described above. Phases of the cell cycle were determined by fluorescence immunostaining of microtubules with antibodies raised in mouse against α‐tubulin from chicken and staining of chromosomal DNA with DAPI. Fluorescence from tubulins, DAPI and YFP was visualized by fluorescence microscopy as described in Experimental Procedures. Merged images (tubulin, red; DAPI, cyan; YFP, yellow) are shown on the right. Phragmoplast microtubules were observed between separating chromosomes (h). Numbers on the right represent ratios of AS2‐body‐containing cells to total YFP‐positive cells at each mitotic phase. Bars, 5 μm.
We monitored the subcellular localization of signals due to YFP in cells at interphase and/or the G0 (differentiated) phase in leaf primordia that harbored XVE‐AS2‐YFP (Figure 5a). Nuclei and mitotic chromosomes were visualized by staining with DAPI. As shown in Figure 5(a) (top), signals due to YFP were mainly detected in the nucleoplasm and in AS2 bodies in all YFP‐positive cells except one, as indicated by white arrows. Although AS2 bodies were not detected in the indicated cell in this optical section, two AS2 bodies were clearly visible in other optical sections of the same cell (Figure 5d). In other cells boxed (the upper panel in Figure 5a), two or three nuclear bodies and single chromocenters were detected adjacent to nucleoli (see also the magnified view at the lower panel of Figure 5a). In addition, at least one of these AS2 bodies was partially overlapped with the perinucleolar chromocenter also in these cells of leaf primordia. In all, we examined 148 YFP‐positive cells in leaf primordia and we detected AS2 bodies in all of the YFP‐positive cells at interphase (and/or the G0 stage) in the leaf primordia. The average number of AS2 bodies per YFP‐positive cell at interphase (and/or the G0 stage) was calculated as 1.9 (n = 148).
We examined 24 cells that were at the M phase in leaf primordia (Figure 5b–e). AS2 bodies were found in all the YFP‐positive cells at M phase, and condensed chromosomes and sets of separating chromosomes were also apparent. Most cells at stages from prophase to metaphase contained one or two AS2 bodies (1.5 on average; n = 12; Figure 5b,c). We observed 12 cells that were progressing from anaphase to telophase (Figure 5d, e): seven of the 12 cells contained two AS2 bodies, while the remaining five cells contained one AS2 body each (1.6 on average; n = 12; Figure 5e, bottom). Therefore, in a slight majority of cells (7 of 12) after metaphase in leaf primordia, there were two AS2 bodies, but the intensities of signals from the two AS2 bodies were different (Figure 5d). In addition, bridge‐like structures, which resembled ultra‐fine DNA bridges that are not detected by DAPI staining (Liu et al., 2014; Nielsen and Hickson, 2016), appeared to link the two AS2 bodies (indicated by the white arrow in Figure 5e, top). In the other five above‐mentioned cells, with only one AS2 body each, a faint signal due to YFP was observed as a tail‐like structure (indicated by the asterisk in Figure 5e, bottom). These observations are suggestive of the possible asymmetric separation or division of individual AS2 bodies during mitosis of primordial cells of leaves.
In BY‐2 cells, the proportion of AS2 body‐positive interphase cells is very low (3–5%), but all cells at M phase contain AS2 bodies and each AS2 body splits into two bodies after metaphase and the two bodies are symmetrically distributed to daughter cells (Luo et al., 2012a). To pursue this issue, we studied the subcellular localization of AS2‐YFP in the Arabidopsis cultured cell line MM2d (Figures 5f–h, S4 and Appendix S1). We found that the proportion of AS2 body‐containing interphase MM2d cells (3.8%) was similar to that of the corresponding BY‐2 cells and that the distribution of numbers of pairs of AS2 bodies was consistent during chromosome separation in anaphase–telophase cells (Figures 5h, S4 and Movie S2), suggesting the non‐random distribution of AS2 bodies, as observed in tobacco BY‐2 cells (Movies S3 and S4). The average number of AS2 bodies in MM2d cells from prophase to metaphase was 1.2. The average number of AS2 bodies in cells during the progression from anaphase to telophase was 2.3. These results suggest that the mechanisms involved in the formation and maintenance of AS2 bodies during leaf development might be different from those during progression of the cell cycle in cultured cells.
AS2 bodies are distinct from Cajal bodies
The shape of AS2 bodies around the nucleolus resembles that of the perinucleolar bodies known as Cajal bodies (Gall, 2000; Liu et al., 2009; Ohtani, 2018). To compare these two types of cell body, we generated transgenic Arabidopsis plants that expressed the AS2‐YFP gene and the chimeric gene U2B"‐GFP, in which the coding sequence for U2B", a marker protein for Cajal bodies, was fused to GFP DNA (Boudonck et al., 1999; Collier et al., 2006). The signals due to AS2‐YFP did not overlap those due to U2B‐GFP (Figure S5 and Appendix [Link], [Link], [Link]). Mitotic Cajal bodies disintegrate during early telophase; Cajal bodies are reformed in daughter cells (Carmo‐Fonseca et al., 1993; Ferreira et al., 1994; Dundr and Misteli, 2010). These observations indicate that AS2 bodies are distinct from Cajal bodies.
Discussion
The ZF motif is essential for the formation of AS2 bodies and subcellular localization of AS2 appears to be subject to multiple controls, including transport from the cytoplasm to the nucleolar periphery
The results of the present study are summarized in Figure 6. We found that the ZF motif is essential for the formation of AS2 bodies around the peripheral region of the nucleolus and, also, for the development of proper leaf morphology (Figures 2, 3, 4 and 6). The four conserved cysteine residues, the cluster of basic amino acid residues (RRK) in the ZF motif, and the proline residue immediately adjacent to the ZFL motif are required for the formation of AS2 bodies (Figures 3 and 6). The RRK sequence is found within proposed nucleolar localization signals (NOLSs; Song and Wu, 2005; Musinova et al., 2011; de Melo et al., 2013; Earley et al., 2015), and it is likely that this cluster participates in the perinucleolar localization of AS2. AS2 bodies also overlapped, to some extent, the chromocenters at the periphery of nucleoli (Figures 2, 3 and 5a). In leaf cells, inactive rDNA repeats are condensed as heterochromatin, are found outside the nucleolus, and are co‐incident with chromocenters (Pontes et al., 2003, 2007; Pontvianne et al., 2007). In addition, AS2 bodies were found to overlap with perinucleolar rDNA repeats (Figure 3c), suggesting the interaction of these bodies with the rDNA repeats.
Figure 6.
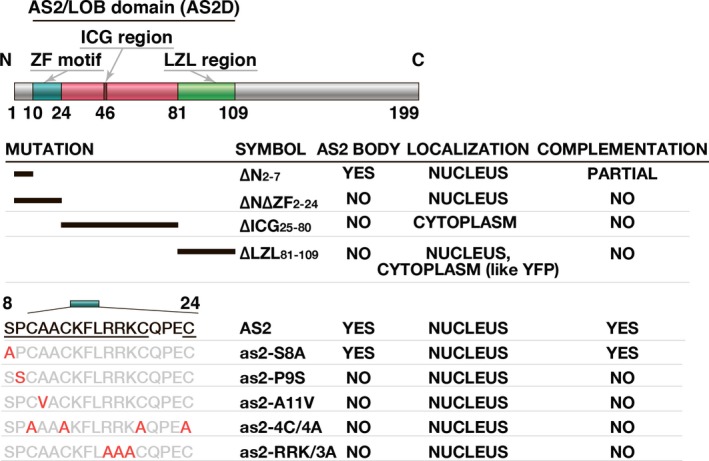
Summary of the association of mutations in the AS2/LOB domain with the ability to form AS2 bodies and the results of complementation tests with the AS2 variants used in the present study. Amino acid residues indicated in red were tested for their importance in the formation of AS2 bodies and development of normal leaf morphology, such as establishment of adaxial–abaxial polarity and development of proximal–distal polarity. Underlined residues are conserved in many ASL members of the AS2/LOB family (see Table S1) and were required for the formation of AS2 bodies and the functions of AS2 in leaf formation.
It was apparent that the ZF motif is not involved in the nuclear localization of AS2 since fluorescent signals due to YFP‐fused as2‐∆N∆ZF2‐24 and to fused derivatives with point mutations in this motif were localized in the nucleus (Figures 2 and 6; Luo et al., 2012b). Both the ICG and LZL regions, encompassing 85 amino acid residues, are responsible for the nuclear localization of AS2: the mutant derivative of AS2 lacking either the ICG or the LZL region failed to appear in the nucleus (Figures 2 and 6). The absence of an obvious NLS sequence in both of these regions suggests the involvement of some unidentified mechanism(s) in the nuclear localization of AS2. The mutant protein lacking the ICG region was found exclusively in the cytoplasm. Therefore, the LZL region and/or the ZF motif might encode a signal(s) for retention of AS2 in the cytoplasm and the ICG region might mask the capacity for the cytoplasmic retention of wild‐type AS2, allowing its nuclear localization. The nuclear transfer of AS2 protein as a consequence of protein modification, such as phosphorylation of the serine residue at position 8 (Table S1), is one conceivable mechanism. However, since the as2‐S8A variant retained the ability to form AS2 bodies, a role for phosphorylation of AS2 in its function remains to be demonstrated. The presence of a leucine‐rich sequence that is somewhat similar to a nuclear exporting signal (NES) (Iwakawa et al., 2002) might explain the export of nuclear AS2 protein to the cytoplasm. Ye et al. (2015) reported that AS2 was exported to the cytoplasm via the action of the geminivirus‐encoded nuclear shuttle protein. Therefore, the AS2 protein might be subject to multiple subcellular localization controls, depending on its interactions with other proteins. The pattern of formation of AS2 bodies in leaf primordia was distinct from that of the formation of AS2 bodies in cultured cells (Figure 5), an observation that suggests that the formation of AS2 bodies is developmentally modulated in the intact plant.
Importance of AS2 bodies in the normal development of leaves
Our observation that as2 derivatives that do not form AS2 bodies fail to rescue the as2‐1 phenotype (Figure 4i) suggests that the formation of AS2 bodies is a pre‐requisite for the normal development of leaves. The observation that as2‐∆N2‐7‐YFP formed AS2 bodies in all cells but had only limited ability with respect to rescue of the as2‐1 phenotype (Figure 4b,i) suggests, however, that the formation of AS2 bodies is, by itself, insufficient for the normal development of leaves.
AS2 bodies might be related to epigenetic repression of the target ETT/ARF3 gene and NUC1 might be involved in their formation
As discussed above, expression of the ETT/ARF3 gene is influenced by factors that are found in the nucleolus. For example, the AS2–AS1 complex binds directly to the upstream region of the leaf abaxial gene ETT/ARF3 to repress its transcription (Iwasaki et al., 2013; Takahashi et al., 2013). Such transcriptional repression is further reduced by mutations in various genes for nucleolus‐localized proteins, such as NUCLEOLIN1 (NUC1) and RNA‐HELICASE10 (RH10), which are involved in the biogenesis of ribosomal RNAs and the formation of nucleoli with normal morphology (Pontvianne et al., 2007; Matsumura et al., 2016). In eukaryotes, nucleolin is one of the major non‐ribosomal proteins in the nucleolus (Ginistry et al., 1999) and part of the small subunit processome (SSUP) complex that mediates ribosome biogenesis (Phipps et al., 2011; You et al., 2015). Similarly, in Arabidopsis, it is proposed that two orthologs of nucleolin, NUC1 and NUC2, are also abundant nucleolar proteins (Durut et al., 2014) and might act as components of the putative SSUP‐like complex (Matsumura et al., 2016). In this context, it is worth noting that NUC1, as well as the AS2–AS1 complex, is involved in maintaining methylated CpG dinucleotides in exon 6 in the target ETT/ARF3 gene (Vial‐Pradel et al., 2018). Such methylation depends on the activity of MET1, which is an Arabidopsis homolog of the vertebrate Dnmt1 methyltransferase that mediates maintenance of CpG methylation during DNA replication (Long et al., 2013; Iwasaki et al., 2013; Vial‐Pradel et al., 2018). Since the ZF motif, required for the formation of AS2 bodies in the peripheral region of the nucleolus, is essential for binding to a specific CpG cluster within exon 1 in ETT/ARF3 (Vial‐Pradel et al., 2018), binding of AS2 to DNA might be part of the process of formation of AS2 bodies. Therefore, it is plausible that AS2 bodies might play a role in epigenetic repression of the expression of the target gene ETT/ARF3, which is expressed and functions only at the early stages of leaf development. The binding of AS2 to exon 1 of the ETT/ARF3 gene and its relationship to maintenance of methylated CpG sequences in exon 6 remain to be investigated.
Perinucleolar regions might provide the molecular architecture, such as nucleolar‐associated domains (NADs; Padeken and Heun, 2014), that function in the regulation of gene transcription. In Arabidopsis, most 45S rDNA loci and transposable elements are condensed as heterochromatin at the periphery of the nucleolus in NADs (Neumüller et al., 2013; Pontvianne et al., 2016). MET1, NUC1, and HISTONE‐DEACETYLASE6 (HDA6) are localized at the periphery of the nucleolus, being responsible for repression of the expression of rDNA in the corresponding perinucleolar compartment (Pontvianne et al., 2007, 2010, 2013). AS1 and AS2 are also associated with HDA6 (Luo et al., 2012b), which interacts physically and genetically with MET1 (To et al., 2011; Liu et al., 2012). The above observations suggest that AS2–AS1 might interact with these putative epigenetic factors in peripheral regions of the nucleolus to control the expression of ETT/ARF3.
NUC1 is involved in the spatial organization of chromosomes, telomere nucleolar clustering (Pontvianne et al., 2016; Picart and Pontvianne, 2017), and maintenance of the repressive states of rRNA gene loci (Pontvianne et al., 2010). We suggest a role of NUC1 in the development of flat, symmetric leaves and the involvement of a co‐operative action between AS2–AS1 and NUC1 in the repression of the target gene ETT/ARF3, which triggers the development of the adaxial domain of leaves (Matsumura et al., 2016). The molecular mechanism behind the co‐operative action between these proteins leading to leaf development has yet to be demonstrated. The results of our analyses show that the number of AS2 bodies increased in nuc1‐1 mutants, and some AS2 bodies did not co‐localize with the perinucleolar chromocenters (Figure S6 and Appendix [Link], [Link], [Link]), indicating that NUC1 might be involved in the formation and/or maintenance of AS2 bodies. Pontvianne et al. (2007) showed that, in the nuc1‐1 mutant, chromocenters co‐localize with decondensed (compared with wild‐type) NORs. Taken together with this observation, our results imply that the decondensation of NORs might alter distribution patterns of AS2 bodies in nuc1‐1 mutants. Further molecular analyses are required to clearly elucidate the relationship between NUC1 and AS2 bodies.
Distribution of AS2 bodies to daughter cells during mitosis in leaf primordia is different from that of AS2 bodies during mitosis in cultured cell lines
AS2 bodies were distributed to daughter cells during progression of the M phase from anaphase to telophase both in cultured cell lines and in leaf primordia (Figure 5 and Movies [Link], [Link], [Link]; Luo et al., 2012a). However, the details of the distribution of AS2 bodies in leaf primordia differed from those of the distribution of AS2 bodies in cultured cells. In both Arabidopsis and tobacco cultured cells, individual AS2 bodies split into two bodies, which segregate into two daughter cells (Figure 5 and Movies [Link], [Link], [Link]; Luo et al., 2012a), suggesting the operation of a nonrandom mechanism for the separation of AS2 bodies in cultured cell lines. By contrast, such division of AS2 bodies was not clearly apparent in 40% of leaf primordial cells after metaphase and the AS2 bodies were distributed asymmetrically to daughter cells (Figure 5). Regardless of cell type, the separating AS2 bodies appeared to be physically associated, somehow, with the separating chromosomes, although no close co‐localization of signals due to YFP and of signals due to DAPI was observed (Figure 5). Signals derived from GFP‐fused RS2 (a maize homolog of AS1) strongly overlapped the separating chromosomes after metaphase in root epidermal cells of maize (Theodoris et al., 2003). The asymmetric mode of distribution of AS2 bodies in leaf primordial cells might reflect the distinct developmental states of two daughter chromosomes in dividing cells in leaf primordia. The role of the unequal distribution of AS2 bodies in leaf development merits further investigation.
Relationship between AS2 bodies and other nuclear bodies
We have shown that AS2 bodies are distinct from Cajal bodies (Figure S5), and finally, will compare with other known nuclear bodies.
Dicer‐like 1 (DCL1), which are involved in the biogenesis of miRNAs in Arabidopsis, form nuclear bodies that have been referred to as dicing bodies (D‐bodies; Fujioka et al., 2007; Fang and Spector, 2007). Some D‐bodies (c. 60%) are found close to the nucleolus, but they do not merge with it (Fang and Spector, 2007; Fujioka et al., 2007; Yu et at., 2017). All the AS2 bodies that we observed were, by contrast, localized adjacent to contact with and sometimes slightly inside the nucleolus (Figures 2, 3b and 5a and Movie S1). The number of D‐bodies is variable (zero to four) in cells of Arabidopsis (Fang and Spector, 2007; Song et al., 2007; Li et al., 2016; Yu et al., 2017). In the present study, our results showed that the number of AS2 bodies in cells of Arabidopsis was smaller and constant (one to two).
The AtMORC4 and AtMORC7 proteins of Arabidopsis are found as nuclear bodies adjacent to chromocenters in perinucleolar regions but they do not overlap the chromocenters (Harris et al., 2016). By contrast, at least one AS2 body partially overlapped the chromocenters in the cells that we examined, and sometimes some AS2 was detected inside the nucleolus. Moreover, global analyses of patterns of gene expression downstream of AS2 and AtMORC4/AtMORC7 showed that these two proteins regulate different sets of genes (Takahashi et al., 2013; Harris et al., 2016). For example, AS2 directly represses transcription of class 1 KNOX genes (Guo et al., 2008) and ETT/ARF3 (Iwasaki et al., 2013; see also Introduction). These genes are, however, not regulated downstream of AtMORC4/AtMORC7 (Harris et al., 2016). These data suggest that AS2 bodies are distinct from D‐bodies and AtMORC‐related bodies. Although GDS1 (growth, development and splicing 1), another ZF finger protein of Arabidopsis, also forms nuclear speckles (Kim et al., 2016), the pattern of these speckles is different from that of AS2 bodies.
Therefore, AS2 bodies are distinct from other nuclear bodies and the co‐ordinated roles of all these distinct entities require further detail analyses.
Experimental Procedures
Plant materials and growth conditions
Arabidopsis thaliana ecotype Col‐0 and the as2‐1 and as1‐1 mutants have been described previously (Semiarti et al., 2001). We performed three outcrosses of as2‐1 and as1‐1, separately, with Col‐0 and used the progeny for our experiments (Kojima et al., 2011). Seeds were sown on a 1‐cm‐thick layer of Golden peat (Sakata, Yokohama, Japan) spread over a mixture (1:1; v/v) of vermiculite and peat moss or on gellan gum‐solidified MS medium. After 2 days at 4°C in darkness, plants were transferred to a regimen of white light at 50 μmol m−2 sec−1 for 16 h and darkness for 8 h daily at 22°C, as described previously (Semiarti et al., 2001). Ages of plants are given in terms of numbers of days after sowing. Arabidopsis seeds containing U2B‐GFP were kindly provided by Peter J. Shaw of the John Innes Centre (Norwich, UK; Collier et al., 2006).
Chemicals
4′6‐Diamidino‐2‐phenylindole (DAPI) and 17β‐estradiol were purchased from Sigma (St. Louis, MO, USA). These chemicals were prepared as a 20 mm stock solution in dimethyl sulfoxide (DMSO) and a 20 mg/ml stock solution in distilled water, respectively. Solutions were stored at −20°C and 4°C, respectively, in darkness.
Construction of AS2‐fused YFP DNA and as2 variant DNAs
Full‐length AS2 cDNA and truncated fragments of this cDNA were amplified by PCR with appropriate pairs of primers (Table S2). Point mutations were introduced by site‐directed mutagenesis with the QuikChange Kit (Agilent Technologies, Santa Clara, CA, USA) with appropriate sets of primers for PCR (Table S2). Plasmids for expression of YFP‐fused AS2 and its variant proteins were made by cloning cDNA fragments into the YFP fusion vector pEYFP (Clontech, Mountain View, CA, USA). Accurate generation of all constructs was confirmed by sequencing. The resulting AS2‐EYFP (EYFP is referred to herein as YFP) and as2‐variant‐YFP cDNA fragments were subcloned into the binary vector pER8 (Zuo et al., 2000) and introduced into cells of Agrobacterium tumefaciens (GV3101).
Plant transformation and selection of transgenic plants
To introduce AS2‐YFP and as2‐variant‐YFPs into A. thaliana (Col‐0), as2‐1 and as1‐1 plants, we used the floral‐dip method for Agrobacterium‐mediated transformation (Clough and Bent, 1998). T1 seeds were grown on gellan gum‐solidified (0.2%) plates of MS medium supplemented with carbenicillin (300 µg/ml) and hygromycin (15 µg/ml). Hygromycin‐resistant T1 seedlings, 7 days after antibiotic selection, were picked out, transferred to MS plates supplemented with 17β‐estradiol (0.05 µm) and incubated for 16 h. Roots were non‐destructively monitored for YFP fluorescence under a stereofluorescence microscope. YFP fluorescence‐positive T1 plants were transplanted to soil and cultivated for production of T2 seeds. For complementation tests of the ability of wild‐type AS2‐YFP and as2‐variant‐YFPs to rescue the as2‐1 mutant phenotype, we established three to five transgenic plant lines that expressed AS2‐YFP and each as2‐variant‐YFP.
Observation of fluorescence
To examine the formation of AS2 bodies, we established three to five lines of transgenic Arabidopsis plants for analysis of the subcellular localization of AS2‐YFP and as2‐variant‐YFPs. Patterns of fluorescence due to YFP and 4,6‐diamidino‐2‐phenylindole (DAPI) were analyzed in 4 to 75 cells of each line of transgenic plants after induction of expression of transgenes. For DAPI staining, Arabidopsis cotyledons and shoot apices containing leaf primordia were separated, incubated in 3.7% paraformaldehyde for 30 min and stained with 0.2 µg/ml DAPI. For DAPI staining of cultured MM2d and BY‐2 cells, cells were fixed in 3.7% paraformaldehyde in phosphate‐buffered saline (PBS; pH 7.2) for 14 min and stained with 0.2 µg/ml DAPI. Images were recorded by confocal laser scanning fluorescence microscopy with a ×40 magnification 1.3 NA plan apochromat oil immersion objective (FV1000 confocal laser scanning fluorescence microscope; OLYMPUS, Tokyo, Japan).
Immunostaining and FISH analysis
Six‐day‐old Arabidopsis seedlings from AS2‐YFP plants were fixed for 15 min at 25°C in 4% formaldehyde in PBS after the induction of gene expression with 0.05 µm 17β‐estradiol for 16 h. After incubation for 10 min in the presence of 200 mm glycine and washing with PBS for 10 min, whole aerial parts of seedlings were minced in lysis solution (15 mm Tris–HCl pH 7.5, 2 mm EDTA, 0.5 mm spermine–4HCl, 80 mm KCl, 20 mm NaCl, and 0.1% Triton X‐100) using a razor blade. The homogenate was suspended in four volumes of nuclei suspension solution (100 mm Tris–HCl pH 7.5, 50 mm KCl, 2 mm MgCl2, 5% sucrose, and 0.05% Tween 20) and filtered through 30‐µm nylon mesh to isolate the nuclei. The nuclei suspension was placed on a glass microscope slide and air dried. Immunostaining with anti‐GFP (ab290; Abcam, Cambridge, UK), which also recognizes YFP, and Alexa‐Fluor‐488‐conjugated anti‐rabbit antibody (A21206; Thermo Fisher Scientific, Waltham, MA, USA) as a primary and secondary antibody, respectively, was used to detect AS2‐YFP signals. DNA‐FISH was performed as described previously (Kurita et al., 2019) following the detection of immunostaining signals. Probes recognizing 45S rDNA and 5S rDNA were synthesized by nick translation using DIG‐Nick Translation Mix (Roche, https://www.roche.com/). The primers for the probe synthesis are listed in Table S2. The DIG‐labeled probes were visualized using anti‐DIG‐rhodamine Fab fragments (Roche). The analysis of fluorescence signals was performed using a confocal fluorescence microscope (Olympus FV1200). Images were captured separately for each fluorochrome using appropriate excitation and emission filters and merged using imagej software. Details are described in Methods S1.
Complementation analysis
Transgenic as2‐1 plant lines harboring AS2‐YFP and as2‐variant‐YFPs, which had been established as described above, were grown on MS plates supplemented with 0.5 µm estradiol under white light for 16 h and in darkness for 8 h daily, at 22ºC. The gross morphology of 14‐day‐old transgenic plants was recorded.
Culture and transformation of Arabidopsis cultured cells
Arabidopsis cultured MM2d cells were maintained in suspension at 26ºC in darkness, with weekly subculturing, in modified Linsmaier and Skoog medium (LS medium; Banno et al., 1993). For transformation of MM2d cells, we used Agrobacterium EHA105 that harbored the above‐mentioned binary vector constructs that included AS2‐YFP or as2‐variant‐YFP cDNA. Details of transformation of MM2d cells are described in Methods S1.
Induction of expression of transgenes in cultured cell lines
For induction of gene expression in cultured MM2d and BY‐2 cells, independent transformants were separately subcultured in liquid LS medium that contained 0.05 µm 17β‐estradiol for 16 h at 26°C in darkness. Several lines expressing specific YFP fusion constructs were selected and analyzed.
Immunofluorescence analysis of MM2d cells
MM2d cells were fixed in 3.7% paraformaldehyde in PBS (pH 7.2) for 15 min. Cells were collected by gravity sedimentation. Fixed cells were washed four times in PBS that contained 0.02% Tween 20 (PBS‐T). Cell walls were digested in a solution of 0.5% cellulase Onozuka R‐10 (Yakult Pharmaceutical Industry, Tokyo, Japan), 0.05% pectolyase Y‐23 (Kyowa Chemical Products, Osaka, Japan), 0.25 m mannitol, and 1 mm phenylmethylsulfonyl fluoride (pH 5.5) at 30°C for 3 min. After two washes in PBS‐T, cells were permeabilized by a 15‐min incubation with PBS‐T that contained 0.5% Triton X‐100. After two washes in PBS‐T and one wash in PBS‐T that contained 1% BSA (PBS‐TB), cells were incubated with a 1:2000 dilution of mouse antibodies against chicken α‐tubulin (DM1A; Sigma) in PBS‐TB overnight at 4ºC. After two more washes with PBS‐T and one in PBS‐TB, cells were incubated with a 1:1000 dilution of tetramethylrhodamine isothiocyanate‐conjugated antibodies raised in goat against mouse IgG (Life Technologies, Eugene, OR, USA) at room temperature for 1 h. Cells were washed once in PBS‐T that contained 0.2 µg/ml DAPI and twice in PBS‐T. The suspension of cells was then mixed with an equal volume of mounting buffer (20 mm Na2HPO4, 150 mM NaCl, and 50% glycerol) and stored at 4ºC until used for observations.
Fluorescence images were captured with a fluorescence microscope (AxioImager2; Carl Zeiss, Oberkochen, Germany) equipped with a high‐end charge coupled device (CCD) camera system (Axiocam 503 mono; Carl Zeiss).
Conflict of Interest
The authors declare no conflict of financial interest.
Author Contributions
YM, MS, CM and SM designed research. LL, SA, YS, and DK performed research; YS, SM, NI, DK and KTY contributed new reagents/analytic tools, and MS, YM, CM, DK, TH, TS, SK and SM analyzed data. YM, MS, CM, YS, SM and HT wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Appendix S1. Legends to supporting figures and movies.
Figure S1. Induction of AS2‐YFP by 17β‐estradiol.
Figure S2. Views of the adaxial surfaces of a cotyledon and a leaf primordium.
Figure S3. Confirmation of the presence of full‐sized fusion proteins in transgenic as2‐1 plants.
Figure S4. (a) The subcellular localization of AS2‐YFP in cells of the MM2d cultured cell line. (b) Time‐lapse images of AS2 bodies from metaphase (0 min) to telophase (51 min) in an MM2d cell.
Figure S5. AS2‐YFP was not co‐localized with U2B‐GFP.
Figure S6. The number of AS2‐body signals increased in the nuc1‐1 mutant.
Methods S1. Supporting experimental procedures.
Movie S1a. Reconstruction of the 3D image of a nucleus in a cell that expressed AS2‐YFP in the adaxial epidermis of the cotyledon of the as2‐1 mutant.
Movie S2. Time‐lapse imaging of an AS2 body during mitosis, from metaphase to telophase, in a cell of the MM2d cultured cell line that harbored the AS2‐YFP construct.
Movie S3. Time‐lapse imaging of AS2 bodies during mitosis, from metaphase to anaphase, in a cell of the tobacco BY‐2 cultured cell line that harbored the AS2‐YFP construct.
Movie S4. Time‐lapse imaging of AS2 bodies during mitosis, from metaphase to telophase, in a cell of the tobacco BY‐2 cultured cell line that harbored the AS2‐YFP construct.
Table S1. Alignment of amino acid sequences in the N‐terminal regions, including the zinc‐finger motif, of proteins in Class Ia of the AS2/LOB family.
Table S2. Primers used for construction of AS2‐fused YFP DNA and AS2 variant DNAs and for FISH probes in this study.
Acknowledgements
The authors thank Peter J. Shaw at the John Innes Centre (Norwich, UK) for providing Arabidopsis seeds that harbored U2B‐GFP. We also thank Yoko Matsumura, Ayami Nakagawa and Takumi Ogawa for fruitful discussions on the role of AS2 bodies and experimental techniques. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant nos. JP26291056, JP16K14574, JP17K07432, JP18K06297, JP18H03330, JP18K14743 and JP19K06730); the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI (grant nos. JP19060008, JP15H01223, JP15H05962, JP16H01246 and JP17H05659). Microscopy for supporting Movie S4 was performed at the Institute of Transformative Bio‐Molecules at Nagoya University, which is supported by the Japan Advanced Plant Science Network.
Contributor Information
Sachihiro Matsunaga, Email: sachi@rs.tus.ac.jp.
Chiyoko Machida, Email: cmachida@isc.chubu.ac.jp.
Michiko Sasabe, Email: msasabe@hirosaki-u.ac.jp.
Yasunori Machida, Email: yas@bio.nagoya-u.ac.jp.
Data Availability Statement
Data supporting the findings of this work are provided in the main text and the supporting information files. All data and materials used in this study will be available from the corresponding authors.
References
- Banno, H. , Hirano, K. , Nakamura, T. , Irie, K. , Nomoto, S. , Matsumoto, K. and Machida, Y. (1993) NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11, and Byr2 protein kinases. Mol. Cell Biol. 13, 4745–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M.K. (2001) Leaving the meristem behind: regulation of KNOX genes. Genome Biol. 2, 1002.1–1002.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck, K. , Dolan, L. and Shaw, P.J. (1999) The movement of coiled Bodies visualized in living plant cells by the green fluorescent protein. Mol. Biol. Cell, 10, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L. and Floyd, S.K. (2008) Patterning and polarity in seed plant shoots. Annu. Rev. Plant Biol. 59, 67–88. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E. , Barley, R. , Curtis, M. , Arroyo, J.M. , Dunham, M. , Hudson, A. and Martienssen, R.A. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature, 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Carmo‐Fonseca, M. , Ferreira, J. and Lamond, A.I. (1993) Assembly of snRNP‐containing coiled bodies is regulated in interphase and mitosis – evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 120, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhara, C. , Mohannath, G. , Blevins, T. , Pontvianne, F. and Pikaard, C.S. (2016) Chromosome‐specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 30, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Wang, H. , Li, J. , Huang, H. and Xu, L. (2013) Quantitative control of ASYMMETRIC LEAVES2 expression is critical for leaf axial patterning in Arabidopsis . J. Exp. Bot. 64, 4895–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.F. , Wei, X.B. , Rety, S. , Huang, L.Y. , Liu, N.N. , Dou, S.X. and Xi, X.G. (2019) Structural analysis reveals a “molecular calipers” mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat. J. Biol. Chem. 294, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collier, S. , Pendle, A. , Boudonck, K. , van Rij, T. , Dolan, L. and Shaw, P. (2006) A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis . Mol. Biol. Cell, 17, 2942–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M. and Misteli, T. (2010) Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000711 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durut, N. , Mohamed, A.‐E. , Pontvianne, F. et al . (2014) A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis . Plant Cell, 26, 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, L.F. , Kawano, Y. , Adachi, K. , Sun, X.‐X. , Dai, M.‐S. and Nakai, H. (2015) Identification and characterization of nuclear and nucleolar localization signals in the Adeno‐associated virus serotype 2 assembly‐activating protein. J. Virol. 89, 3038–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F. , Floyd, S.K. , Alvarez, J. , Eshed, Y. , Hawker, N.P. , Izhaki, A. , Baum, S.F. and Bowman, J.L. (2003) Radial patterning of Arabidopsis shoots by Class III HD‐ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Eshed, Y. , Baum, S.F. and Bowman, J.L. (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis . Cell, 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y. , Baum, S.F. , Perea, J.V. and Bowman, J.L. (2001) Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y. , Izhaki, A. , Baum, S.F. , Floyd, S.K. and Bowman, J.L. (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development, 131, 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fang, Y. and Spector, D.L. (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 17, 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, J.A. , Carmo‐Fonseca, M. and Lamond, A.I. (1994) Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 126, 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, Y. , Utsumi, M. , Ohba, Y. and Watanabe, Y. (2007) Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis . Plant Cell Physiol. 48, 1243–1253. [DOI] [PubMed] [Google Scholar]
- Gall, J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Ginistry, H. , Sicard, H. , Roger, B. and Bouvet, P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Thomas, J. , Collins, G. and Timmermans, M.C.P. (2008) Direct repression of KNOX; loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis . Plant Cell, 20, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, C.J. , Husmann, D. , Liu, W. et al . (2016) Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein‐coding genes. PLoS Genet. 12, e1005998 10.1371/journal.pgen.1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi, G. , Mollá‐Morales, A. , Pérez‐Pérez, J.M. , Kojima, K. , Robles, P. , Ponce, M.R. , Micol, J.L. and Tsukaya, H. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736. [DOI] [PubMed] [Google Scholar]
- Hudson, A. (2000) Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 349–370. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H. , Ueno, Y. , Semiarti, E. et al . (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H. , Iwasaki, M. , Kojima, S. , Ueno, Y. , Soma, T. , Tanaka, H. , Semiarti, E. , Machida, Y. and Machida, C. (2007) Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51, 173–184. [DOI] [PubMed] [Google Scholar]
- Iwasaki, M. , Takahashi, H. , Iwakawa, H. et al (2013) Dual regulation of ETTIN (ARF3) gene expression by AS1‐AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial–abaxial partitioning in Arabidopsis . Development, 140, 1958–1969. [DOI] [PubMed] [Google Scholar]
- Kim, D.W. , Jeon, S.J. , Hwang, S.M. , Hong, J.C. and Bahk, J.D. (2016) The C3H‐type zinc finger protein GDS1/C3H42 is a nuclear‐speckle‐localized protein that is essential for normal growth and development in Arabidopsis . Plant Sci. 250, 141–153. [DOI] [PubMed] [Google Scholar]
- Kojima, S. , Iwasaki, M. , Takahashi, H. , Imai, T. , Matsumura, Y. , Fleury, D. , Van Lijsebettens, M. , Machida, Y. and Machida, C. (2011) Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana . Plant Cell Physiol. 52, 1259–1273. [DOI] [PubMed] [Google Scholar]
- Kurita, K. , Sakamoto, Y. , Naruse, S. et al . (2019) Intracellular localization of histone deacetylase HDA6 in plants. J. Plant Res. 132, 629–640. 10.1007/s10265-019-01124-8. [DOI] [PubMed] [Google Scholar]
- Lee, H.W. , Kim, M.‐J. , Kim, N.Y. , Lee, S.H. and Kim, J. (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis . Plant J. 73, 212–224. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Wang, S. , Cheng, J. et al . (2016) Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis . PLoS Genet. 12, e1006422 10.1371/journal.pgen.1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.‐C. , Shuai, B. and Springer, P.S. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES‐Domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. Plant Cell, 15, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.‐L. , Wu, Z.A. , Nizami, Z. , Deryusheva, S. , Rajendra, T.K. , Beumer, K.J. , Gao, H. , Matera, A.G. , Carroll, D. and Gall, J.G. (2009) Coilin is essential for Cajal body organization in Drosophila melanogaster . Mol. Biol. Cell, 20, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Yu, C.‐W. , Duan, J. , Luo, M. , Wang, K. , Tian, G. , Cui, Y. and Wu, K. (2012) HDA6 directly interacts with dna methyltransferase MET1 and maintains transposable element silencing in Arabidopsis . Plant Physiol. 158, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Nielsen, C.F. , Yao, Q. and Hickson, I.D. (2014) The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet Dev. 26, 1–5. [DOI] [PubMed] [Google Scholar]
- Lodha, M. , Marco, C.F. and Timmermans, M.C.P. (2013) The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb‐repressive complex2. Genes Dev. 27, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A. and Barton, M.K. (1998) The development of apical embryonic pattern in Arabidopsis . Development, 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A. , Moan, E.I. , Medford, J.I. and Barton, M.K. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis . Nature, 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Long, H.K. , Blackledge, N.P. and Klose, R.J. (2013) ZF‐CxxC domain‐containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 41, 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. , Ando, S. , Sasabe, M. , Machida, C. , Kurihara, D. , Higashiyama, T. and Machida, Y. (2012a) Arabidopsis ASYMMETRIC LEAVES2 protein required for leaf morphogenesis consistently forms speckles during mitosis of tobacco BY‐2 cells via signals in its specific sequence. J. Plant Res. 125, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Yu, C.W. , Chen, F.F. , Zhao, L.M. , Tian, G. , Liu, X.C. , Cui, Y.H. , Yang, J.Y. and Wu, K.Q. (2012b) Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8, e1003114 10.1371/journal.pgen.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, C. , Nakagawa, A. , Kojima, S. , Takahashi, H. and Machida, Y. (2015) The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis . Wiley Interdiscip. Rev. Dev. Biol. 4, 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, Y. , Iwakawa, H. , Machida, Y. and Machida, C. (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, Y. , Ohbayashi, I. , Takahashi, H. et al . (2016) A genetic link between epigenetic repressor AS1‐AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis . Biol. Open, 5, 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R. and Barton, M.K. (1998) Leaf polarity and meristem formation in Arabidopsis . Development, 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R. , Emery, J. , Eshed, Y. , Bao, N. , Bowman, J. and Barton, M.K. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature, 411, 709–713. [DOI] [PubMed] [Google Scholar]
- de Melo, I.S. , Jimenez‐Nuñez, M.D. , Iglesias, C. , Campos‐Caro, A. , Moreno‐Sanchez, D. , Ruiz, F.A. and Bolívar, J. (2013) NOA36 protein contains a highly conserved nucleolar localization signal capable of directing functional proteins to the nucleolus, in mammalian cells. PLoS One, 8, e59065 10.1371/journal.pone.0059065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musinova, Y.R. , Lisitsyna, O.M. , Golyshev, S.A. , Tuzhikov, A.I. , Polyakov, V.Y. and Sheval, E.V. (2011) Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim. Biophys. Acta. 1813, 27–38. [DOI] [PubMed] [Google Scholar]
- Nakata, M. and Okada, K. (2013) The leaf adaxial–abaxial boundary and lamina growth. Plants, 2, 174–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, M. , Ichikawa, T. , Ishikawa, A. , Kobayashi, H. , Tsuhara, Y. , Kawashima, M. , Suzuki, K. , Muto, S. and Matsui, M. (2003) Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34, 741–750. [DOI] [PubMed] [Google Scholar]
- Neumüller, R.A. , Gross, T. , Samsonova, A.A. et al . (2013) Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci. Signal. 6, ra70 10.1126/scisignal.2004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, C.F. and Hickson, I.D. (2016) PICH promotes mitotic chromosome segregation: identification of a novel role in rDNA disjunction. Cell Cycle, 15, 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani, M. (2018) Plant snRNP biogenesis: a perspective from the nucleolus and Cajal bodies. Front. Plant Sci. 8, 2184 10.3389/fpls.2017.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken, J. and Heun, P. (2014) Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 28, 54–60. [DOI] [PubMed] [Google Scholar]
- Phelps‐Durr, T.L. , Thomas, J. , Vahab, P. and Timmermans, M.C.P. (2005) Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell, 17, 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps, K.R. , Charette, J.M. and Baserga, S.J. (2011) The small subunit processome in ribosome biogenesis‐progress and prospects. Wiley Interdiscip. Rev. RNA, 2, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart, C. and Pontvianne, F. (2017) Plant nucleolar DNA: Green light shed on the role of Nucleolin in genome organization. Nucleus, 8, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon, V. , Etchells, J.P. , Rossignol, P. , Collier, S.A. , Arroyo, J.M. , Martienssen, R.A. and Byrne, M.E. (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development, 135, 1315–1324. [DOI] [PubMed] [Google Scholar]
- Pontes, O. , Lawrence, R.J. , Neves, N. , Silva, M. , Lee, J.H. , Chen, Z.J. , Viegas, W. and Pikaard, C.S. (2003) Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis . Proc. Natl Acad. Sci. USA, 100, 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes, O. , Lawrence, R.J. , Silva, M. , Preuss, S. , Costa‐Nunes, P. , Earley, K. , Neves, N. , Viegas, W. and Pikaard, C.S. (2007) Postembryonic establishment of megabase‐scale gene silencing in nucleolar dominance. PLoS One, 2, e1157 10.1371/journal.pone.0001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne, F. , Matía, I. , Douet, J. , Tourmente, S. , Medina, F.J. , Echeverria, M. and Sáez‐Vásquez, J. (2007) Characterization of AtNUC‐L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC‐L2 gene in Arabidopsis . Mol. Biol. Cell, 18, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne, F. , Abou‐Ellail, M. , Douet, J. et al . (2010) Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana . PLoS Genet. 6, e1001225 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne, F. , Blevins, T. , Chandrasekhara, C. et al (2013) Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 27, 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne, F. , Carpentier, M.‐C. , Durut, N. et al . (2016) Identification of nucleolus‐associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 16, 1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E. , Ueno, Y. , Tsukaya, H. , Iwakawa, H. , Machida, C. and Machida, Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem‐related homeobox genes in leaves. Development, 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai, B. , Reynaga‐Peña, C.G. and Springer, P.S. (2002) The Lateral Organ Boundaries gene defines a novel, plant‐specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z.Y. and Wu, M. (2005) Identification of a novel nucleolar localization signal and a degradation signal in Survivin‐deltaEx3: a potential link between nucleolus and protein degradation. Oncogene, 24, 2723–2734. [DOI] [PubMed] [Google Scholar]
- Song, L. , Han, M.H. , Lesicka, J. and Fedoroff, N. (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl Acad. Sci. USA, 104, 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi, D. and Byrne, M.E. (2011) Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana . Plant J. 65, 269–281. [DOI] [PubMed] [Google Scholar]
- Szakonyi, D. , Moschopoulos, A. and Byrne, M.E. (2010) Perspectives on leaf dorsoventral polarity. J. Plant Res. 123, 281–290. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Iwakawa, H. , Ishibashi, N. et al . (2013) Meta‐analyses of microarrays of Arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial–abaxial patterning and cell division during leaf development. Plant Cell Physiol. 54, 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris, G. , Inada, N. and Freeling, M. (2003) Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl Acad. Sci. USA, 100, 6837–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, T.K. , Kim, J.‐M. , Matsui, A. et al . (2011) Arabidopsis HDA6 regulates locus‐directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7, e1002055 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2006) Mechanism of leaf‐shape determination. Annu. Rev. Plant Biol. 57, 477–496. [DOI] [PubMed] [Google Scholar]
- Ueno, Y. , Ishikawa, T. , Watanabe, K. , Terakura, S. , Iwakawa, H. , Okada, K. , Machida, C. and Machida, Y. (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis . Plant Cell, 19, 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial‐Pradel, S. , Keta, S. , Nomoto, M. et al . (2018) Arabidopsis zinc‐finger‐like protein ASYMMETRIC LEAVES2 (AS2) and two nucleolar proteins maintain gene body DNA methylation in the leaf polarity gene ETTIN (ARF3). Plant Cell Physiol. 59, 1385–1397. 10.1093/pcp/pcy031. [DOI] [PubMed] [Google Scholar]
- Waites, R. , Selvadurai, H.R.N. , Oliver, I.R. and Hudson, A. (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell, 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Yang, J.‐Y. , Iwasaki, M. , Machida, C. , Machida, Y. , Zhou, X. and Chua, N.‐H. (2008) βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 22, 2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y. , Ling, Q. , Wang, H. and Huang, H. (2008) Ribosomal proteins promote leaf adaxial identity. Development, 135, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Ye, J. , Yang, J.Y. , Sun, Y.W. , Zhao, P.Z. , Gao, S.Q. , Jung, C. , Qu, J. , Fang, R.X. and Chua, N.H. (2015) Geminivirus Activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2‐mediated mRNA turnover and weakens RNA silencing in Arabidopsis . PLoS Pathog, 11, e1005196 10.1371/journal.ppat.1005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, K.T. , Park, J. and Kim, V.N. (2015) Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 29, 2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Jia, T. and Chen, X. (2017) The 'how' and 'where' of plant microRNAs. New Phytol. 216, 1002–1017. 10.1111/nph.14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J. , Niu, Q.W. and Chua, N.H. (2000) Technical advance: an estrogen receptor‐based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Legends to supporting figures and movies.
Figure S1. Induction of AS2‐YFP by 17β‐estradiol.
Figure S2. Views of the adaxial surfaces of a cotyledon and a leaf primordium.
Figure S3. Confirmation of the presence of full‐sized fusion proteins in transgenic as2‐1 plants.
Figure S4. (a) The subcellular localization of AS2‐YFP in cells of the MM2d cultured cell line. (b) Time‐lapse images of AS2 bodies from metaphase (0 min) to telophase (51 min) in an MM2d cell.
Figure S5. AS2‐YFP was not co‐localized with U2B‐GFP.
Figure S6. The number of AS2‐body signals increased in the nuc1‐1 mutant.
Methods S1. Supporting experimental procedures.
Movie S1a. Reconstruction of the 3D image of a nucleus in a cell that expressed AS2‐YFP in the adaxial epidermis of the cotyledon of the as2‐1 mutant.
Movie S2. Time‐lapse imaging of an AS2 body during mitosis, from metaphase to telophase, in a cell of the MM2d cultured cell line that harbored the AS2‐YFP construct.
Movie S3. Time‐lapse imaging of AS2 bodies during mitosis, from metaphase to anaphase, in a cell of the tobacco BY‐2 cultured cell line that harbored the AS2‐YFP construct.
Movie S4. Time‐lapse imaging of AS2 bodies during mitosis, from metaphase to telophase, in a cell of the tobacco BY‐2 cultured cell line that harbored the AS2‐YFP construct.
Table S1. Alignment of amino acid sequences in the N‐terminal regions, including the zinc‐finger motif, of proteins in Class Ia of the AS2/LOB family.
Table S2. Primers used for construction of AS2‐fused YFP DNA and AS2 variant DNAs and for FISH probes in this study.
Data Availability Statement
Data supporting the findings of this work are provided in the main text and the supporting information files. All data and materials used in this study will be available from the corresponding authors.


