Abstract
Odontogenic tumors show considerable morphologic heterogeneity and at times the diagnosis can be challenging. Ameloblastoma, the most common odontogenic tumor, can have morphological similarity to some salivary gland tumors and therefore we sought to identify biomarkers that might aid in the diagnosis by performing transcriptome wide gene expression profiling of 80 odontogenic and salivary gland neoplasms. These data identified the FOXP1+/SOX10− expression profile as characteristic of many odontogenic tumors including ameloblastoma but largely absent in salivary gland tumors. We then assessed 173 salivary gland tumors and 108 odontogenic tumors by immunohistochemistry for FOXP1 and SOX10 expression and found that 34/35 (97%) cases of ameloblastomas were diffusely positive for FOXP1 but completely negative for SOX10. None of the basaloid salivary neoplasms (basal cell adenoma, adenoid cystic carcinoma, polymorphous adenocarcinoma, and myoepitheloma) demonstrated FOXP1+/SOX10− expression pattern. Taken together, the results of this study suggest that the FOXP1+/SOX10− immunophenotype is common in odontogenic tumors including ameloblastoma and might be useful distinguishing these from similar appearing basaloid salivary gland tumors.
Introduction/background:
Odontogenic tumors arise from the tissues derived from the tooth forming apparatus and typically display a range of microscopic patterns and clinical behavior. The most common odontogenic tumor, ameloblastoma, is a locally infiltrative odontogenic tumor that on rare occasions may undergo malignant transformation. Surprisingly little is known about the molecular biology of most odontogenic tumors but recurrent genomic alterations have been identified in ameloblastomas,1,2 ghost cell odontogenic tumors,3-5 and odontogenic keratocysts.6-8
Accurate diagnosis of these tumors is important since different odontogenic tumors can require differing surgical management. Several odontogenic tumors can show morphological similarities to basaloid salivary gland neoplasms such that the distinction can be challenging in small biopsies and for tumors at unusual sites. For example, ameloblastomas arising in sinonasal mucosa or peripheral locations may closely resemble basaloid salivary gland neoplasms such as basal cell adenoma or adenoid cystic carcinoma. Conversely, intra-osseous salivary gland neoplasms may pose a diagnostic challenge with respect to odontogenic tumors. Malignant ameloblastoma that metastasizes to atypical locations (lung, liver, brain, etc) may cause significant diagnostic challenges and can lead to misdiagnosis and mismanagement. We hypothesize that by using a genomic approach new biomarkers for the diagnostic classification of odontogenic tumors can be generated.
Material and Methods:
Case selection:
Formalin fixed and paraffin embedded tissue blocks from 108 odontogenic tumors were retrieved from University California San Francisco Dermatopathology & Oral Pathology Service, Vancouver General Hospital, and Stanford University Hospital with IRB approval. These cases include 35 ameloblastomas, 5 ameloblastic fibromas (ABF), 2 ameloblastic carcinomas (ABC), 4 calcifying odontogenic cysts (COC), 3 adenomatoid odontogenic tumors (AOT), 2 glandular odontogenic cysts (GOC), 1 clear cell carcinoma (CCC), and 56 odontogenic keratocysts.
Formalin fixed and paraffin embedded tissue blocks from 173 salivary gland tumors were retrieved from the surgical pathology archives at Stanford University Hospital & Clinics with IRB approval. These cases include 9 basal cell adenomas, 63 adenoid cystic carcinomas, 7 polymorphous (low grade) adenocarcinomas, 9 myoepitheliomas, 43 pleomorphic adenomas, 7 acinic cell carcinomas, 3 salivary duct carcinomas, 9 oncocytomas, 22 mucoepidermoid carcinomas, and 1 low grade adenocarcinoma, NOS.
43 odontogenic tumors (28 ameloblastomas, 3 ameloblastic fibromas, 2 ameloblastic carcinomas, 4 calcifying odontogenic cysts, 3 adenomatoid odontogenic tumors, 2 glandular odontogenic cysts, 1 clear cell carcinoma) and 37 salivary gland tumors (6 basal cell adenomas, 4 polymorphous adenocarcinomas, 3 myoepitheliomas, 4 adenosquamous carcinomas, 20 adenoid cystic carcinomas) underwent laser capture microdissection. Smart-3SEQ9 protocol was performed to generate gene expression profiles.
Gene expression profiling
Slide preparation and Laser-Capture Microdissection
Consecutive sections of the FFPE block were cut at 7 μm thickness, mounted on glass slides with polyethylene naphthalate membranes (Thermo Fisher #LCM0522), dried for at least for 1 hour and then stored overnight in a nitrogen chamber. Slides were immersed for 20 seconds each in xylenes (3 times), 100% ethanol (3 times), 95% ethanol (2 times), 70% ethanol (2 times), water, hematoxylin (Dako #S3309), water, bluing reagent (Thermo Fisher #7301), water, 70% ethanol (2 times), 95% ethanol (2 times), 100% ethanol (3 times), and xylene (3 times) before performing laser-capture microdissection (LCM). Slides were dissected immediately after staining. The consecutive sections stained with hematoxylin and eosin as the reference. Epithelial component was dissected in most of the odontogenic tumors and salivary gland tumors. For the Ameloblastic fibroma, the stromal component and epithelial component were separately dissected. Around 500 Cells for each sample were captured on an Arcturus XT LCM System using both the ultraviolet (UV) laser to cut out each sample and the infrared laser to adhere it to a CapSure HS LCM Cap (Thermo Fisher #LCM0215). After LCM, the cap was sealed in an 0.5 mL tube (Thermo Fisher #N8010611) and stored at −80°C until library preparation.
Library preparation and sequencing
Sequencing libraries were prepared according to the Smart-3SEQ protocol (https://www.biorxiv.org/content/10.1101/207340v3; RNA extraction and preparation steps are built into the Smart-3SEQ protocol). Amplification of the cDNA was performed adding KAPA HiFi HotStart Ready mix (KAPA biosystems KK2602) and 2 μM primers with different barcodes in P7 primers. The number of PCR cycles was 22 cycles (15s at 98 °C, 30s at 60 °C, 10s at 72 °C). All of the libraries were purified by SPRI beads (Beckman Coulter #B23317) and then elute in the10ul 1xTE + 0.05% Tween20 buffer.
After bead purification, the concentration of the cDNA library was profiled for size distribution on an Agilent 2200 TapeStation system with the High Sensitivity D1000 reagent kits and quantified by a qPCR with a dual-labeled probe. The libraries were pooled together according to the qPCR measurements. Libraries were prepared according to the manufacturer’s instructions with a 1% spike-in of the PhiX control library (Illumina #FC-110-3002) and sequenced on an Illumina NextSeq 500 instrument with a High Output v2 reagent kit (Illumina #FC-404-2005), reading 76 nt for read 1 and 6 nt for the P7 index read.
Gene expression profile analysis
Raw sequencing data was converted to FASTQ using bcl2fastq. The raw reads were then aligned to the hg38 reference into sorted, indexed, duplicate-marked BAM files using a combination of STAR and samtools. We produced a read count matrix using the Subread R package and GENCODE release 25. Then unsupervised hierarchical clustering was performed using custom R scripts and differential gene expression analysis was performed with DESeq2.
Immunohistochemistry:
Immunohistochemical (IHC) staining was performed on paraffin-embedded tissue microarray (TMA) sections (4 um). Briefly, slides were deparaffinized and hydrated using a xylene-ethanol series. Antigen retrieval was carried out at 116 C for 3 minutes in a decloacking chamber in Antigen Unmasking Solution 10mM Citrate pH6 (Cat No. S236984-2 Agilent, Santa Clara, CA). Slides were incubated by hand with primary antibody FOXP1clone JC12 (Cat No.ab32010 Abcam, Cambridge, MA) at 1:1000 dilution in PBS. Vectastain Elite ABC kit (Mouse Cat No. PK-6102, Vector Laboratories, Burlingame, CA) was used as per the manufacture protocol. Antigen-antibody reactions were revealed with DAKO Liquid DAB+ Substrate Chromogen System (Cat No. K346811-2 Agilent, Santa Clara, CA). Slides were counter-stained with hematoxylin.
SOX-10, Clone BC34, mouse monoclonal from Biocare, Cat# ACI 3099C, was performed on the Leica Bond III using Leica’s Bond DAB Refine Kit, Cat# DS9800. HIER was performed as part of the protocol on the Leica Bond III instrument using Leica’s proprietary ER2 solution (pH9.0), Cat# AR9640. HIER is done for 20 min at 100°C. Oligo dT, from Ventana/Roche, Cat# 780-2221, ALU Positive Control Probe II, was performed on the Ventana Ultra using Ventana’s ISH iView Blue Detection Kit. Slides were scanned with Leica Microscope slide scanner with Ariol Software (Leica Biosystems, CA). Immunohistochemical stain for SOX10 was scored using similar criteria described by Hsieh, et al.10 Specifically, staining results were categorized as negative (< 10% of tumor cells with nuclear staining) or positive (≥ 10% of tumor cells with nuclear staining; interpretated by Y.C.K and R.W.).
Immunohistochemical stain for FOXP1 was scored as negative (<50% of tumor cells with nuclear staining) or positive (≥50% of tumor cells with nuclear staining; interpretated by Y.C.K and R.W.). Cores that had substandard staining with an OligodT in situ hybridization probe (Ventana) were flagged. One case of ameloblastoma with negative FOXP1 immunolabeling was tested for RNA in situ hybridization studies for kappa and lambda immunoglobulin light chains (Ventana automated staining platform).
Results:
Hierarchical clustering analysis of the odontogenic tumors showed grouping of ameloblastoma, ameloblastic carcinoma, and the epithelial component of the ameloblastic fibroma (Figure 1). Calcifying odontogenic cyst, which may mimic ameloblastoma on histology, appeared to have a distinct gene expression profile when compared to ameloblastoma. Hierarchical clustering analysis of both odontogenic tumors and salivary gland tumors showed differential expression profile in multiple genes (Figure 2).
Figure 1:

Hierarchical clustering of odontogenic tumors.
Figure 2:
Hierarchical clustering of odontogenic tumors and salivary gland tumors.
To identify useful biomarkers for ameloblastoma, differential gene expression analysis was performed with DESeq2 for ameloblastoma versus COC, GOC, ameloblastic fibroma (epithelial), ameloblastic fibroma (stromal), AOT, ameloblastic carcinoma, clear cell carcinoma, basal cell adenoma, polymorphous adenocarcinoma, adenoid cystic carcinoma, adenosquamous carcinoma, and myoepithelioma (see Supplemental data 1, http://links.lww.com/PAS/A900). Among significant genes, we identified 6 positively expressed gene candidates: PITX2 (p value adj. 9.57E-29), FLRT2 (p value adj. 1.04E-20), MSX2 (p value adj. 1.30E-09), PTHLH (p value adj. 1.16E-12), EPHA7 (p value adj. 1.60E-09), and FOXP1 (p value adj. 6.93E-08). Interestingly, SOX10 had a significant negative gene expression in ameloblastoma (p adj. 7.53E-24) while having neutral or positive expression in basal cell adenoma, polymorphous adenocarcinoma, adenoid cystic carcinoma, and myoepithelioma.
To validate these findings and determine their topographic distribution in tumors, FOXP1 and SOX10 immunohistochemical studies were performed on a second independent set of neoplasms comprising 173 salivary gland tumors and 108 odontogenic tumors. The results of FOXP1 and SOX10 protein expression in odontogenic tumors and salivary gland tumors are summarized (Table 1 and 2).
Table 1:
FOXP1/SOX10 immunophenotypes in 108 odontogenic tumors.
| FOXP1+/SOX10− | FOXP1+/SOX10+ | FOXP1−/SOX10− | FOXP1−/SOX10+ | |
|---|---|---|---|---|
| Ameloblastoma | 34/35 (97%) | 0/35 (0%) | 1/35 (3%) | 0/35 (0%) |
| Ameloblastic carcinoma | 2/2 (100%) | 0/2 (0%) | 0/2 (0%) | 0/2 (0%) |
| Ameloblastic fibroma | 5/5 (100%) | 0/5 (0%) | 0/5 (0%) | 0/5 (0%) |
| Calcifying odontogenic cyst | 4/4 (100%) | 0/4 (0%) | 0/4 (0%) | 0/4 (0%) |
| Adenomatoid odontogenic tumor | 1/3 (33%) | 0/3 (0/%) | 2/3 (67%) | 0/3 (0/%) |
| Glandular odontogenic cyst | 2/2 (100%) | 0/2 (0/%) | 0/2(0/%) | 0/2 (0/%) |
| Clear cell odontogenic carcinoma | 1/1 (100%) | 0/1 (0/%) | 0/1 (0/%) | 0/1 (0/%) |
| Odontogenic Keratocyst | 53/56 (95%) | 0/56 (0/%) | 3/56 (5/%) | 0/56 (0/%) |
Table 2:
FOXP1/SOX10 immunophenotypes in 173 salivary gland tumors. *This case was subsequently re-classified as ameloblastoma after consensus review.
| FOXP1+/SOX10− | FOXP1+/SOX10+ | FOXP1−/SOX10− | FOXP1−/SOX10+ | |
|---|---|---|---|---|
| Basal cell adenoma (9 cases) | 0/9 (0%) | 8/9 (89%) | 1/9 (11%) | 0 (0%) |
| Adenoid cystic carcinoma (63) | 0/63 (0%) | 47/63 (75%) | 2/63 (3%) | 14/63 (22%) |
| Polymorphus adenocarcinoma (7) | 0/7 (0%) | 3/7 (43%) | 1/7 (14%) | 3/7 (43%) |
| Myoepithelioma (9 cases) | 0/9 (0%) | 3/9 (33%) | 4/9 (44%) | 2/9 (22%) |
| Pleomorphic adenoma (43) | 2/43 (4%) | 14/43 (33%) | 8/43 (19%) | 19/43 (44%) |
| Acinic cell carcinoma (7) | 0/7 (0%) | 6/7 (86%) | 0/7 (0%) | 1/7 (14%) |
| Salivary duct carcinoma (3) | 0/3 (0%) | 0/3 (0%) | 3/3 (100%) | 0/3 (0%) |
| Oncocytoma (9 cases) | 0/9 (0%) | 0/9 (0%) | 9/9 (100%) | 0/9 (0%) |
| Mucoepidermoid carcinoma (22 cases) | 0/22 (0%) | 0/22 (0%) | 19/22 (86%) | 3/22 (14%) |
| Low grade adenocarcinoma, NOS* (1 case) | 1/1 (100%) | 0/1 (0%) | 0/1 (0%) | 0/1 (0%) |
FOXP1 was positive in 97% (34/35) ameloblastomas, 100% (2/2) ameloblastic carcinomas, 100% (5/5) ameloblastic fibromas, 100% (4/4) calcifying odontogenic cysts, 33% (1/3) adenomatoid odontogenic tumors, 100% (2/2) clear cell odontogenic cysts, 100% (1/1) clear cell odontogenic carcinomas, and 95% (53/56) odontogenic keratocysts. No correlation was identified between location (sinonasal, maxillary, mandibular, or peripheral) and histologic subtype of ameloblastoma and FOXP1 expression. Intense peripheral basal columnar cells staining was observed with variable protein expression in the central stellate reticulum. SOX10 was consistently negative in all of the odontogenic tumors tested.
SOX10 expressions in salivary gland tumors were consistent with previous reports.10-11 SOX10 was positive in 89% (8/9) basal cell adenomas, 97% (61/63) adenoid cystic carcinomas, 86% (6/7) polymorphous adenocarcinomas, 56% (5/9) myoepitheliomas, 77% (33/43) pleomorphic adenomas, 100% (7/7) acinic cell carcinomas, 0% (0/3) salivary duct carcinoma, 0% (0/9) oncocytoma, and 14% (3/22) mucoepidermoid carcinomas. Consistent with prior descriptions, expression of SOX10 was consistently seen in acinic cell carcinoma and strong abluminal component expression was observed in adenoid cystic carcinoma and basal cell carcinoma. All ameloblastomas are negative for SOX10 (0/35).
Interestingly, co-expression of FOXP1 and SOX10 are seen in basal cell adenoma (89%), adenoid cystic carcinoma (75%), polymorphous (low grade) adenocarcinoma (43%), myoepithelioma (33%), and acinic cell carcinoma (86%). No cases of FOXP1+/SOX10− immunophenotype were identified in basal cell adenoma, polymorphous (low grade) adenocarcinoma, adenoid cystic carcinoma, and myoepithelioma. One case of low grade adenocarcinoma, NOS demonstrated strong FOXP1 expression and complete absence of SOX10 expression. This case was subsequently re-classified as ameloblastoma after consensus review.
Discussion:
While many odontogenic tumors have a characteristic morphologic appearance, few sensitive and specific immunohistochemical markers have been described in the literature.12 Ameloblastoma, the most common odontogenic tumor, has been shown to harbor BRAF V600E mutation in 48-63% of mandibular cases.1-2 However, positive BRAFV600E expression by immunohistochemistry is only observed in a subset of ameloblastomas, thereby limiting its use as a diagnostic maker. A portion of calcifying odontogenic cysts and ghost cell odontogenic tumors have been shown to harbor CTNNB1 mutation with resultant nuclear localization of beta-catenin and the expression of LEF1 (Lymphoid Enhancer Binding Factor 1) by immunohistochemistry.4, 13 However, a recent comparative analysis of beta-catenin and LEF1 expression in odontogenic and salivary tumors showed low frequency of expression in odontogenic tumors, thereby argue against their utility as diagnostic markers.13
Distinction between ameloblastoma and basaloid salivary gland tumors can be particularly challenging, especially in intra-oral, sinonasal, and atypical locations. Ameloblastoma may be indistinguishable from basal cell adenoma, adenoid cystic carcinoma, or polymorphous (low grade) adenocarcinoma. In small incision biopsies, the specific features of these lesions may be absent or obscured. Misdiagnosis may lead to inappropriate treatment and management. For example, ameloblastoma is a locally aggressive tumor that may require wider resection than basal cell adenoma. Adenoid cystic carcinoma is known for its propensity for perineural invasion and may require advanced surgical planning and elective neck dissection. A sensitive and specific useful marker for odontogenic tumor may resolve this diagnostic dilemma.
FOXP1 (transcription factor forkhead box protein P1) belongs to subfamily P of the forkhead box (FOX) transcription factor family. Forkhead box transcription factors play important roles in the regulation of tissue- and cell type-specific gene transcription during both development and adulthood. Expression of FOXP1 has been shown to be associated with key regulatory development of the aboral and posterior regions of the jaw.14 SOX10 (Transcription factor SRY-related MMG-box10) is a sensitive and relatively specific marker for myoepithelial and basaloid salivary gland tumors.
We examine for the first time the expression of FOXP1 and SOX10 in odontogenic tumors and salivary gland tumors. Strong and diffuse nuclear FOXP1 expression is observed in 97% cases of ameloblastomas, including sinonasal ameloblastomas (Figure 3). FOXP1 was low in only 1 of 35 ameloblastomas (equivocal expression; <50% of tumor cells are staining). We postulate that this tumor may have lost some of its antigenicity due to decalcification. Indeed, this ameloblastoma showed a complete loss of expression on oligo DT probe ISH suggesting that the tissue preservation was poor. We observe that none of the tested odontogenic tumors express SOX10, suggesting that SOX10 could be a clinically useful negative marker for excluding salivary gland neoplasm in difficult and unusual locations (Figure 4). Two of the three adenomatoid odontogenic tumors have FOXP1−/SOX10− profile. We postulate that this unexpected finding could be due to limited tissue sampling (sampling bias) on the tissue microarray. The expression profile of FOXP1 and SOX10 in other tumors are currently unknown.
Figure 3:
Strong nuclear expressions for FOXP1 in maxillary ameloblastomas (A, B), mandibular ameloblastomas (C, D), and sinonasal ameloblastomas (E, F).
Figure 4:
Ameloblastoma (H&E, A) showed diffuse positivity for FOXP1 (B) but complete negativity for SOX10 (C). In contrast, most of the adenoid cystic carcinomas (H&E, D) and basal cell adenomas (H&E, G) demonstrated negative to variable patchy expression for FOXP1 (E, H) and strong expression for SOX10 (F, I) in the peripheral/myoepithelial cells.
Examining 173 salivary tumors, we saw similar results to previously reported SOX10 staining profiles for basal cell adenoma, adenoid cystic carcinoma, and polymorphous adenocarcinoma.10 For the first time, we also report the expression profile of FOXP1 in salivary gland tumors. Many of the salivary gland neoplasms have co-expression of FOXP1 and SOX10. More importantly, a FOXP1+/SOX10− immunoprofile is largely absent in basal cell adenoma (0/9), polymorphous adenocarcinoma (0/7), adenoid cystic carcinoma (0/63), and myoepithelioma (0/9). Two cases of FOXP1+/SOX10− pleomorphic adenomas demonstrate FOXP1staining only in the mesenchymal areas and absence of staining in the glandular areas. Negative expression of SOX10 in pleomorphic adenomas is unusual, however, SOX10 negative pleomorphic adenoma has been reported in the literature.15
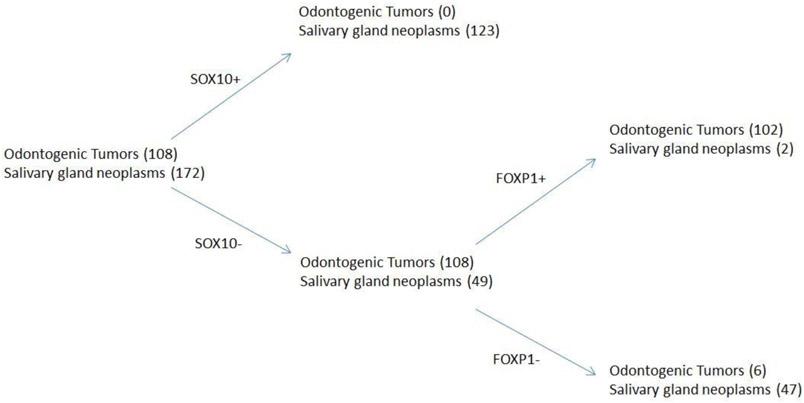
Given the strong negative predictive value of SOX10 expression in odontogenic tumors, an algorithmic based approach of SOX10 and FOXP1 immunohistochemical panel could be helpful in distinguishing odontogenic tumors from salivary gland neoplasms. Overall, a SOX10 negative and FOXP1 positive immunophenotype is 94% sensitive and 96% specific for odontogenic tumors (Figure 5). When restricting the algorithm to only ameloblastoma and basaloid salivary neoplasms such as basal cell adenoma, adenoid cystic carcinoma, polymorphous adenocarcinoma, and myoepithelioma, a FOXP1 positive and SOX10 negative immunophenotype is 97% sensitive and 100% specific for ameloblastoma.
Figure 5:
Using SOX10 and FOXP1 in separating odontogenic tumors and salivary gland neoplasms.
In summary, we established a unique FOXP1/SOX10 expression profile in ameloblastoma and salivary gland neoplasms. FOXP1+ and SOX10− immunophenotype can reliably distinguish ameloblastoma from basal cell adenoma, adenoid cystic carcinoma, polymorphous adenocarcinoma, and myoepithelioma. While unexpected FOXP1+/SOX10− pattern may occur in a small number of pleomorphic adenomas, especially in the mesenchymal chondromyxoid component, morphologic separation is possible by restricting analysis of the expression profile to the cellular ductal epithelium. A FOXP1/SOX10 immunohistochemical panel can provide a useful diagnostic adjunct in separating challenging odontogenic tumors basaloid salivary gland neoplasm in the head and neck region. Future studies to further characterize FOXP1 and SOX10 expressions in other basaloid neoplasms (i.e. basaloid variant of squamous cell cariconma, sinonasal undifferentiated carcinoma, etc) may allow clinically meaningful and diagnostically useful classification of tumors of the head and neck.
Supplementary Material
References:
- 1.Sweeney RT, McClary AC, Myers BR, et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet. 2014;46:722–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurppa KJ, Catón J, Morgan PR, et al. High frequency of BRAF V600E mutations in ameloblastoma. J Pathol. 2014;232:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekine S, Sato S, Takata T, et al. Beta-catenin mutations are frequent in calcifying odontogenic cysts, but rare in ameloblastomas. Am J Pathol. 200;163:1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yukimori A, Oikawa Y, Morita KI, et al. Genetic basis of calcifying cystic odontogenic tumors. PLoS One. 2017. June 28;12:e0180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Arruda JAA, Monteiro JLGC, Abreu LG, et al. Calcifying odontogenic cyst, dentinogenic ghost cell tumor, and ghost cell odontogenic carcinoma: A systematic review. J Oral Pathol Med. 2018;47:721–730. [DOI] [PubMed] [Google Scholar]
- 6.Li TJ. The odontogenic keratocyst: a cyst, or a cystic neoplasm? J Dent Res. 2011;90:133–142. [DOI] [PubMed] [Google Scholar]
- 7.Bresler SC, Padwa BL, Granter SR. Nevoid Basal Cell Carcinoma Syndrome (Gorlin Syndrome). Head Neck Pathol. 2016;10:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu J, Yu F, Hong Y, et al. Underestimated PTCH1 mutation rate in sporadic keratocystic odontogenic tumors. Oral Oncol. 2015;51:40–45. [DOI] [PubMed] [Google Scholar]
- 9.Foley JF, Zhu C, Jolivet P, et al. Gene-expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. BioRxiv. doi: 10.1101/207340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh MS, Lee YH, Chang YL. SOX10-positive salivary gland tumors: a growing list, including mammary analogue secretory carcinoma of the salivary gland, sialoblastoma, low-grade salivary duct carcinoma, basal cell adenoma/adenocarcinoma, and a subgroup of mucoepidermoid carcinoma. Hum Pathol. 2016;56:134–142. [DOI] [PubMed] [Google Scholar]
- 11.Ohtomo R, Mori T, Shibata S, et al. SOX10 is a novel marker of acinus and intercalated duct differentiation in salivary gland tumors: a clue to the histogenesis for tumor diagnosis. Mod Pathol. 2013;26:1041–1050. [DOI] [PubMed] [Google Scholar]
- 12.DeVilliers P, Liu H, Suggs C. Calretinin expression in the differential diagnosis of human ameloblastoma and keratocystic odontogenic tumor. Am J Surg Pathol. 2008;32:256–260. [DOI] [PubMed] [Google Scholar]
- 13.Bilodeau EA, Acquafondata M, Barnes EL, et al. A comparative analysis of LEF-1 in odontogenic and salivary tumors. Hum Pathol. 2015;46:255–259. [DOI] [PubMed] [Google Scholar]
- 14.Cesario JM, Almaidhan AA, Jeong J. Expression of forkhead box transcription factor genes Foxp1 and Foxp2 during jaw development. Gene Expr Patterns. 2016;20:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miettinen M, McCue PA, Sarlomo-Rikala M, et al. Sox10--a marker for not only schwannian and melanocytic neoplasms but also myoepithelial cell tumors of soft tissue: a systematic analysis of 5134 tumors. Am J Surg Pathol. 2015;39:826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.