Abstract
Head and neck cancer is the sixth most common malignancy around the world, and 90% of cases are squamous cell carcinomas. In this study, we performed a systematic investigation of the immunogenomic landscape to identify prognostic biomarkers for head and neck squamous cell carcinoma (HNSCC). We analyzed the expression profiles of immune‐related genes (IRGs) and clinical characteristics by interrogating RNA-seq data from 527 HNSCC patients in the cancer genome atlas (TCGA) dataset, including 41 HPV+ and 486 HPV− samples. We found that differentially expressed immune genes were closely associated with patient prognosis in HNSCC by comparing the differences in gene expression between cancer and normal samples and performing survival analysis. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to annotate the biological functions of the differentially expressed immunogenomic prognosis-related genes. Two additional cohorts from the Oncomine database were used for validation. 65, 56 differentially expressed IRGs was associated with clinical prognosis in total and HPV- samples, respectively. Furthermore, we extracted 10, 11 prognosis-related IRGs from 65, 56 differentially expressed IRGs, respectively. They were significantly correlated with clinical prognosis and used to construct the prognosis prediction models. The multivariable ROC curves (specifically, the AUC) were used to measure the accuracy of the prognostic models. These genes were mainly enriched in several gene ontology (GO) terms related to immunocyte migration and receptor and ligand activity. KEGG pathway analysis revealed enrichment of pathways related to cytokine−cytokine receptor interactions, which are primarily involved in biological processes. In addition, we identified 63 differentially expressed transcription factors (TFs) from 4784 differentially expressed genes, and 16 edges involving 18 nodes were formed in the regulatory network between differentially expressed TFs and the high-risk survival-associated IRGs. B cell and CD4 T cell infiltration levels were significantly negatively correlated with the expression of prognosis-related immune genes regardless of HPV status. In conclusion, this comprehensive analysis identified the prognostic IRGs as potential biomarkers, and the model generated in this study may enable an accurate prediction of survival.
Subject terms: Cancer, Computational biology and bioinformatics, Genetics, Immunology, Biomarkers, Oncology
Introduction
Head and neck cancer is a common malignancy accounting for 5–10% of cancers worldwide. Head and neck squamous cell carcinoma constitutes 90% of head and neck cancers, which arises from the pharynx, the oral cavity and lip, the ear, the larynx, the nasal cavity, the salivary glands and the paranasal sinuses1–3. The tobacco, alcohol use and human papillomavirus (HPV) infection are important causes of head and neck squamous cell carcinoma4. HNSCC represents a biologically complex disease process and a heterogeneous collection of tumors in which multiple pathways are altered, leading to the development of HNSCC, and the mechanisms leading to this disease are still not clearly understood5. The main treatments for HNSCC include surgery, radiotherapy, and chemotherapy, either alone or in combinations. Despite a multimodal approach, the majority of patients with locally advanced HNSCC develop recurrence or distant metastases, such that the 5-year overall survival does not usually exceed 60%6. The locoregionally advanced and distantly metastatic HNSCC cases that are unsuitable for surgery or radiotherapy are significantly associated with a poor prognosis, with an expected survival on the order of 6–10 months7.
Extracted from TCGA data, protein-coding genes that paint a molecular portrait of the disease can be helpful tools and can be tested as biomarkers. Many studies analyzing the cellular landscape indicated that several genes were differentially expressed between tumor and healthy tissues. However, the differential expression status of the IRGs in HNSCC has not been revealed by comprehensive analysis.
In recent years, it has been well established that the immune system plays a pivotal role in the control of tumor growth, and it has been suggested that potentially invading cancer cells are held in equilibrium via the immune system8. Cutting-edge immunotherapy treatments, which are beneficial to recurrent/metastatic (R/M) HNSCC patients, have recently revolutionized the treatment of multiple cancers9. Some clinical trials have demonstrated that immune checkpoint therapy is effective for R/M HNSCC and has less toxicity than other therapies10. Interestingly, one of the major advantages of immunotherapy over other forms of systemic cancer therapy is that responses can be quite durable—with clinical benefit sometimes measured in years. Long-term follow-up data for survival outcomes following immunotherapy for R/M HNSCC demonstrated that pembrolizumab exhibited durable antitumor activity and a high survival rate in advanced HNSCC patients. The overall response rate was 18%, while 85% of responses lasted 6 months or more, and 71% of responses lasted a year or more11. Many studies have emphasized that more immunological biomarkers need to be discovered to provide prognostic information and facilitate clinical decision-making. Therefore, identifying immune-related biomarkers to manipulate the immune response and achieve greater clinical benefit for R/M HNSCC patients is key.
In this study, we combined the expression profiles of immune-related genes (IRGs) with clinical information by comparing the differences in gene expression between normal and tumor tissues and performing Cox regression analysis. Furthermore, bioinformatics analysis was utilized to explore the intrinsic regulatory mechanisms of immune-related genes. Our findings reveal the potential clinical application of IRGs as biomarkers in prognosis prediction, and IRGs may represent therapeutic targets for HNSCC immunotherapy.
Results
Identification of differentially expressed IRGs
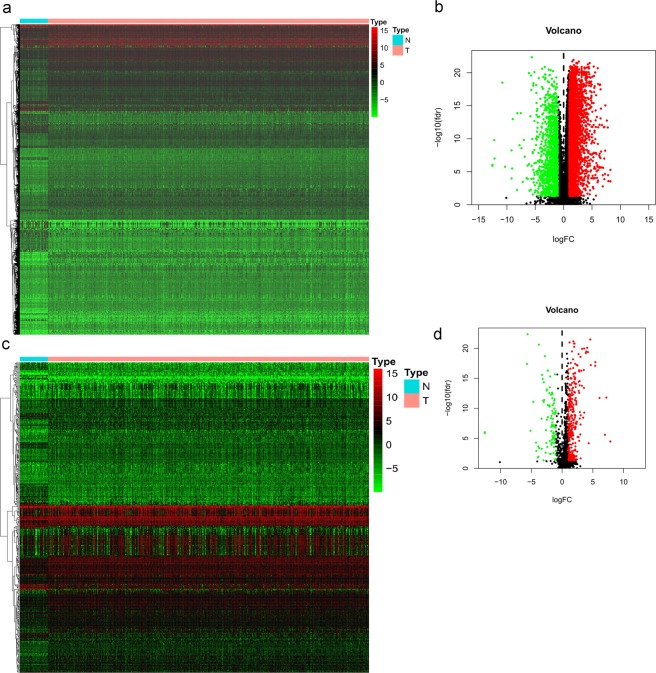
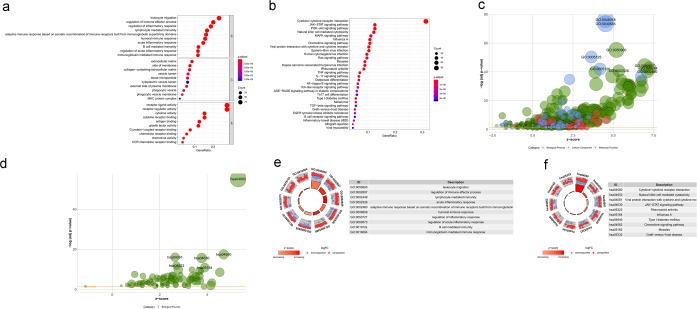
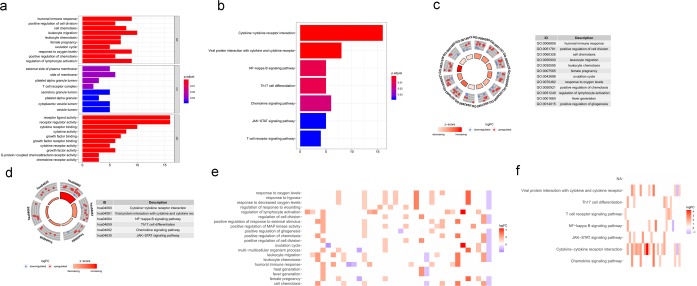
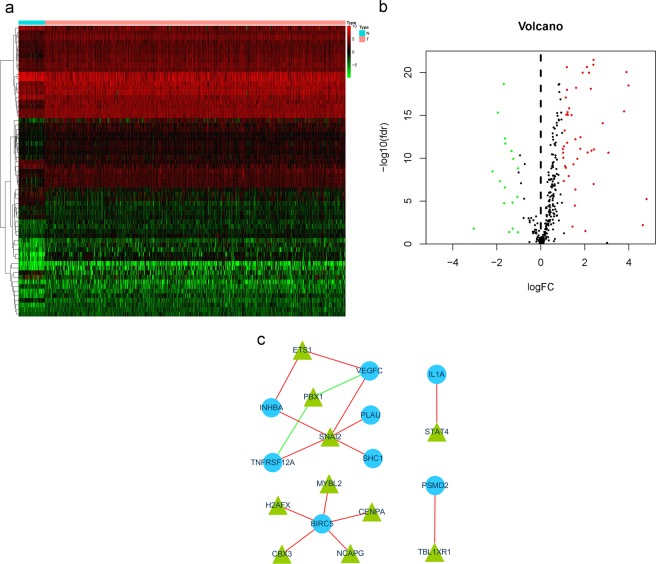
In the present study, we downloaded a total of 56753 genes information from TCGA database including tumor and normal samples. 4784 differentially expressed genes were identified, 3603 of which were upregulated and 1181 of which were downregulated in HNSCC patients (Fig. 1a,b). Furthermore, 399 differentially expressed IRGs were identified from a subset of the 4784 genes, 304 of which were upregulated and 95 of which were downregulated in HNSCC patients (Table 1; Fig. 1c,d). In addition, the functions of the intersecting genes (399 differentially expressed IRGs) were predicted. In the Gene Ontology (GO) and KEGG pathway analyses, 1550 terms and 94 pathways were identified. The top three GO terms were “leukocyte migration (GO:0050900)”, “regulation of immune effector process (GO:0002697)” and “regulation of inflammatory response (GO:0050727)” in terms of biological processes; “extracellular matrix (GO:0030198)”, “side of membrane (GO:0098552)” and “collagen-containing extracellular matrix (GO:0062023)” in terms of cellular components; and “receptor ligand activity (GO:0048018)”, “ receptor regulator activity (GO:0030545)” and “cytokine activity (GO:0005125)” in terms of molecular functions (Fig. 2a,c,e). In the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis, cytokine-cytokine receptor interaction (hsa04060) was the pathway most often enriched by the differentially expressed IRGs (Fig. 2b,d,f).
Figure 1.
Comparison of gene expression profiles between head and neck squamous cell carcinoma and nontumor tissues. (a) Heatmap and (b) volcano plot demonstrating differentially expressed genes. (c) Heatmap and (d) volcano plot demonstrating differentially expressed immune genes. Red represents upregulated differentially expressed genes, blue represents downregulated differentially expressed genes, and black dots (volcano) represent genes that were not differentially expressed.
Table 1.
Identification of the differentially expressed immune-related genes (The logFC means log2|fold change | , log F/C > 1 indicated that the expression of genes were up-regulated in HNSCC patients, while log F/C < 1 indicated that expression of genes were down-regulated in HNSCC patients).
| Gene | Normal mean | Tumor mean | LogFC | P value | FDR |
|---|---|---|---|---|---|
| AZGP1 | 140.761501 | 7.469646968 | −4.23607 | 2.14E-05 | 4.61E-05 |
| FCER1G | 11.2589868 | 26.17050568 | 1.216865 | 1.25E-09 | 5.33E-09 |
| HLA-A | 254.911533 | 613.2837215 | 1.266558 | 3.09E-13 | 2.44E-12 |
| HLA-B | 326.464724 | 877.4184076 | 1.426338 | 3.61E-13 | 2.81E-12 |
| HLA-C | 236.984649 | 555.0795393 | 1.227901 | 1.21E-12 | 8.58E-12 |
| HLA-DOB | 0.990856 | 2.369508193 | 1.25784 | 1.27E-08 | 4.54E-08 |
| HLA-F | 10.370698 | 30.66749407 | 1.564197 | 1.31E-12 | 9.22E-12 |
| HLA-G | 1.39598199 | 6.074506878 | 2.121487 | 3.54E-10 | 1.64E-09 |
| HLA-H | 22.4554075 | 49.17635987 | 1.130902 | 1.44E-09 | 6.03E-09 |
| HSPA2 | 9.76782464 | 19.61169521 | 1.005605 | 0.000272 | 0.000498 |
| ICAM1 | 11.2802693 | 25.07587424 | 1.152498 | 1.17E-08 | 4.23E-08 |
| IFNG | 0.1438456 | 0.654326879 | 2.18549 | 4.87E-08 | 1.58E-07 |
| KIR2DL4 | 0.1522916 | 0.529638836 | 1.798173 | 5.58E-06 | 1.32E-05 |
| KLRC1 | 0.11234459 | 0.279755016 | 1.316233 | 0.001152 | 0.001918 |
| LTA | 0.21982314 | 0.521766513 | 1.247061 | 1.24E-06 | 3.23E-06 |
| MICB | 0.89624206 | 3.786667263 | 2.078968 | 4.86E-17 | 9.00E-16 |
| PSMD2 | 39.4400045 | 91.67299729 | 1.216837 | 5.96E-25 | 9.64E-22 |
| RELB | 7.44585989 | 15.18863955 | 1.028482 | 2.08E-11 | 1.18E-10 |
| TAP1 | 24.7771851 | 74.2062648 | 1.582529 | 2.56E-14 | 2.51E-13 |
| TAP2 | 5.75960252 | 13.95711697 | 1.27696 | 8.25E-16 | 1.12E-14 |
| IFI30 | 0.18085808 | 0.521648619 | 1.52822 | 5.21E-17 | 9.54E-16 |
| PROCR | 6.49854461 | 24.59981521 | 1.920459 | 2.15E-16 | 3.40E-15 |
| ULBP3 | 1.29958122 | 2.880656553 | 1.148351 | 2.54E-08 | 8.61E-08 |
| ULBP2 | 3.1334658 | 12.91807779 | 2.04356 | 5.31E-18 | 1.31E-16 |
| ULBP1 | 0.13572911 | 1.001890626 | 2.883923 | 7.45E-11 | 3.83E-10 |
| RAET1E | 13.2144682 | 3.93391552 | −1.74808 | 3.46E-08 | 1.15E-07 |
| PDIA2 | 0.05969777 | 1.199043433 | 4.328063 | 2.80E-05 | 5.92E-05 |
| CXCL14 | 122.268502 | 372.1008438 | 1.605641 | 0.000748 | 0.00128 |
| SLPI | 2610.15495 | 830.9769458 | −1.65126 | 1.48E-08 | 5.24E-08 |
| CXCL10 | 15.196852 | 124.3922942 | 3.033053 | 2.83E-10 | 1.33E-09 |
| CXCL9 | 9.24006951 | 49.69692373 | 2.427181 | 4.01E-09 | 1.57E-08 |
| CXCL11 | 2.02329743 | 24.56421975 | 3.601778 | 9.19E-11 | 4.66E-10 |
| CXCL12 | 15.0119696 | 5.452086988 | −1.46123 | 4.96E-07 | 1.38E-06 |
| CXCL13 | 2.64456939 | 19.00938842 | 2.845607 | 1.00E-15 | 1.34E-14 |
| CXCL2 | 9.72385442 | 4.565688464 | −1.0907 | 0.017695 | 0.024321 |
| XCL1 | 0.28104112 | 1.248440254 | 2.151274 | 5.61E-13 | 4.22E-12 |
| DEFB1 | 176.677144 | 53.88491009 | −1.71316 | 3.51E-10 | 1.63E-09 |
| TMSB10 | 1092.64225 | 2821.300725 | 1.368539 | 6.92E-19 | 2.19E-17 |
| LCN2 | 447.352172 | 159.2711221 | −1.48993 | 2.39E-08 | 8.15E-08 |
| S100A9 | 15432.446 | 6667.03957 | −1.21085 | 3.22E-06 | 7.86E-06 |
| S100A8 | 6148.25986 | 2377.261991 | −1.37088 | 2.42E-06 | 6.04E-06 |
| HTN3 | 1113.90311 | 0.186136307 | −12.547 | 2.91E-07 | 8.39E-07 |
| MMP12 | 2.4760227 | 38.28779427 | 3.950788 | 2.99E-22 | 3.14E-20 |
| PTGDS | 20.3392441 | 7.284519543 | −1.48136 | 1.26E-09 | 5.36E-09 |
| TMSB15A | 0.31540397 | 2.161507024 | 2.776765 | 1.03E-05 | 2.34E-05 |
| DEFB126 | 0.05038056 | 0.226744114 | 2.170126 | 0.020469 | 0.027794 |
| S100A5 | 0.22413643 | 0.486194597 | 1.117157 | 0.008856 | 0.012838 |
| S100A1 | 28.6642829 | 2.833404063 | −3.33865 | 1.13E-09 | 4.85E-09 |
| HTN1 | 234.309738 | 0.038339574 | −12.5773 | 4.20E-07 | 1.18E-06 |
| S100A14 | 1032.71172 | 454.45533 | −1.18423 | 1.25E-06 | 3.25E-06 |
| TINAGL1 | 7.20176813 | 19.21614587 | 1.415896 | 4.63E-10 | 2.11E-09 |
| WFDC2 | 427.5378 | 29.38590271 | −3.86286 | 0.006801 | 0.010059 |
| TGFB1 | 12.6913796 | 45.44074526 | 1.840138 | 3.54E-25 | 6.30E-22 |
| MMP9 | 4.68250673 | 79.57169061 | 4.086902 | 3.60E-23 | 7.44E-21 |
| APOBEC3G | 1.42821266 | 3.614852766 | 1.339726 | 2.69E-07 | 7.79E-07 |
| FABP6 | 1.10315656 | 3.75998828 | 1.769091 | 2.90E-05 | 6.12E-05 |
| RBP1 | 7.59289009 | 37.30236814 | 2.296546 | 5.35E-16 | 7.61E-15 |
| PLAU | 17.1784923 | 110.1589636 | 2.680912 | 1.02E-21 | 8.42E-20 |
| IL1B | 3.71074115 | 13.10607739 | 1.820457 | 1.08E-06 | 2.85E-06 |
| PAEP | 0.03142237 | 1.32057542 | 5.393231 | 2.05E-19 | 7.58E-18 |
| MX1 | 11.8010874 | 33.38941718 | 1.500471 | 2.19E-08 | 7.53E-08 |
| DDX58 | 4.17161375 | 14.29679051 | 1.777014 | 5.30E-14 | 4.91E-13 |
| SFTPA2 | 0.76667193 | 0.336438314 | −1.18827 | 1.59E-10 | 7.76E-10 |
| LBP | 1.64869075 | 0.736595416 | −1.16238 | 0.024851 | 0.033252 |
| RBP4 | 2.27829793 | 0.912345169 | −1.3203 | 1.56E-08 | 5.49E-08 |
| NOX4 | 0.21387194 | 0.915830479 | 2.098333 | 7.43E-17 | 1.30E-15 |
| LTF | 917.674427 | 36.01701431 | −4.67123 | 1.10E-12 | 7.87E-12 |
| FABP7 | 5.56457715 | 0.157924359 | −5.13897 | 1.63E-07 | 4.88E-07 |
| FABP3 | 44.1770711 | 6.908420351 | −2.67687 | 0.00131 | 0.002165 |
| OASL | 2.62095302 | 17.0465449 | 2.701316 | 2.19E-16 | 3.45E-15 |
| DUOX1 | 28.8369572 | 14.15939772 | −1.02616 | 4.70E-08 | 1.53E-07 |
| FABP12 | 0.52989973 | 0.256180969 | −1.04856 | 0.000629 | 0.001092 |
| PI15 | 0.49099368 | 1.362655272 | 1.472644 | 3.64E-09 | 1.43E-08 |
| APOD | 46.1811346 | 8.244769222 | −2.48575 | 5.73E-15 | 6.43E-14 |
| ORM2 | 0.0788061 | 0.225477574 | 1.516605 | 4.84E-05 | 9.88E-05 |
| CTSG | 4.33606479 | 1.330628377 | −1.70428 | 2.12E-05 | 4.57E-05 |
| PML | 7.27390668 | 15.30806882 | 1.07349 | 5.06E-13 | 3.84E-12 |
| CYBB | 4.16431981 | 8.686060183 | 1.060621 | 0.000129 | 0.000248 |
| ISG20 | 2.49483593 | 5.24894123 | 1.073082 | 1.48E-06 | 3.83E-06 |
| TFRC | 11.9428901 | 37.92448056 | 1.666977 | 3.11E-13 | 2.46E-12 |
| IFIH1 | 5.25357518 | 15.36513689 | 1.548289 | 8.48E-12 | 5.16E-11 |
| IDO1 | 3.55528778 | 16.98751225 | 2.256436 | 5.29E-09 | 2.02E-08 |
| ADIPOQ | 4.43687949 | 0.090920784 | −5.60879 | 2.32E-27 | 4.12E-23 |
| STAT1 | 29.7280212 | 89.78982168 | 1.594728 | 6.47E-14 | 5.87E-13 |
| TNFSF10 | 37.0843358 | 85.95761174 | 1.212815 | 4.04E-08 | 1.32E-07 |
| CCL20 | 6.80143542 | 22.5198056 | 1.727283 | 1.20E-06 | 3.13E-06 |
| SOCS1 | 2.54808466 | 11.06232065 | 2.118169 | 8.88E-20 | 3.59E-18 |
| IL15 | 0.39916272 | 0.874389501 | 1.131299 | 5.20E-06 | 1.23E-05 |
| CHIT1 | 0.20032363 | 2.170568122 | 3.437668 | 7.40E-10 | 3.26E-09 |
| VEGFA | 4.16606964 | 10.34122599 | 1.311648 | 2.61E-12 | 1.75E-11 |
| ISG15 | 21.9969809 | 318.5958413 | 3.85635 | 6.27E-22 | 5.66E-20 |
| DHX58 | 2.5898532 | 5.375063939 | 1.053412 | 6.25E-11 | 3.25E-10 |
| TNFAIP3 | 10.9562653 | 24.03152342 | 1.133172 | 2.79E-09 | 1.12E-08 |
| TFR2 | 0.17390547 | 0.546720142 | 1.652499 | 4.38E-06 | 1.05E-05 |
| MUC4 | 10.5167619 | 3.130648606 | −1.74816 | 5.09E-05 | 0.000104 |
| F2R | 4.76270265 | 12.43949049 | 1.385075 | 4.74E-12 | 3.03E-11 |
| MAPT | 2.22286807 | 0.283387734 | −2.97157 | 2.79E-16 | 4.27E-15 |
| LYZ | 1480.74523 | 112.8877688 | −3.71336 | 0.00021 | 0.000391 |
| CCL5 | 11.7542346 | 49.97424577 | 2.088004 | 7.64E-10 | 3.36E-09 |
| ITGAV | 11.7548612 | 28.92172488 | 1.298896 | 5.51E-14 | 5.08E-13 |
| TLR8 | 0.30481515 | 0.7045093 | 1.208684 | 0.000726 | 0.001247 |
| GNLY | 1.87734623 | 7.933403211 | 2.079245 | 4.88E-13 | 3.71E-12 |
| EIF2AK2 | 3.79883745 | 10.20996947 | 1.426349 | 5.71E-20 | 2.46E-18 |
| BST2 | 37.5941515 | 254.1483336 | 2.757091 | 1.38E-18 | 4.07E-17 |
| PLA2G2A | 38.820626 | 7.929581253 | −2.29151 | 1.63E-18 | 4.67E-17 |
| ADAR | 23.6630778 | 47.86163445 | 1.016232 | 4.46E-18 | 1.14E-16 |
| MX2 | 1.7639436 | 5.455379419 | 1.628875 | 9.28E-10 | 4.03E-09 |
| MSR1 | 0.87519781 | 1.910406496 | 1.126199 | 5.59E-08 | 1.80E-07 |
| SLC11A1 | 0.34450751 | 1.21576439 | 1.819256 | 5.33E-19 | 1.75E-17 |
| DMBT1 | 65.6409166 | 3.617668986 | −4.18146 | 0.000676 | 0.001168 |
| DES | 728.159849 | 123.9189978 | −2.55486 | 2.51E-05 | 5.35E-05 |
| TNFRSF10B | 7.03680752 | 15.75748764 | 1.163045 | 4.48E-15 | 5.19E-14 |
| APOBEC3H | 0.24598789 | 0.706353938 | 1.521804 | 4.06E-07 | 1.14E-06 |
| SPINK5 | 354.298193 | 44.99588064 | −2.9771 | 1.07E-12 | 7.66E-12 |
| TNFSF11 | 0.10591856 | 0.530948712 | 2.325617 | 1.65E-12 | 1.14E-11 |
| CLDN4 | 114.276557 | 40.59751217 | −1.49307 | 1.23E-10 | 6.10E-10 |
| CCL28 | 24.4864213 | 1.584078293 | −3.95027 | 0.010034 | 0.014398 |
| IRF7 | 10.2207731 | 24.55095099 | 1.264275 | 6.17E-13 | 4.60E-12 |
| IL7R | 2.27764192 | 8.021914076 | 1.816406 | 1.44E-11 | 8.46E-11 |
| IL1A | 4.34170044 | 22.08057361 | 2.346446 | 9.64E-09 | 3.52E-08 |
| PTX3 | 7.05171955 | 1.821522249 | −1.95283 | 0.00205 | 0.003284 |
| PTGS2 | 2.97256793 | 10.74320113 | 1.853642 | 0.000581 | 0.001014 |
| PROC | 0.18503733 | 1.331676726 | 2.847356 | 1.62E-16 | 2.66E-15 |
| NDRG1 | 91.6022532 | 241.7426842 | 1.400017 | 3.15E-11 | 1.72E-10 |
| IRF9 | 0.48277151 | 1.48917545 | 1.625101 | 3.63E-16 | 5.40E-15 |
| ABCC4 | 1.19897173 | 2.407151667 | 1.005529 | 3.41E-10 | 1.58E-09 |
| PLSCR1 | 12.0066205 | 24.15015248 | 1.008202 | 1.79E-10 | 8.71E-10 |
| RSAD2 | 1.99533893 | 12.91988771 | 2.694888 | 1.03E-14 | 1.09E-13 |
| PDGFRB | 6.27611021 | 17.8452768 | 1.5076 | 6.19E-12 | 3.87E-11 |
| PDCD1 | 0.7684706 | 1.57420676 | 1.034563 | 0.021445 | 0.029014 |
| AQP9 | 0.59654601 | 1.638747522 | 1.457888 | 2.73E-11 | 1.51E-10 |
| FASLG | 0.32324901 | 0.724487013 | 1.164314 | 0.024377 | 0.03267 |
| BIRC5 | 5.05117624 | 20.54924905 | 2.024394 | 2.61E-24 | 1.53E-21 |
| OAS1 | 11.2092353 | 23.35089274 | 1.05879 | 7.05E-07 | 1.91E-06 |
| TNFSF4 | 0.17675193 | 1.549628874 | 3.132125 | 1.75E-18 | 4.99E-17 |
| NOS1 | 2.23283169 | 0.689385368 | −1.69549 | 2.59E-08 | 8.76E-08 |
| ACTA1 | 1981.05982 | 135.3248897 | −3.87177 | 0.000104 | 0.000202 |
| CCL26 | 1.01014639 | 4.561715584 | 2.175012 | 0.000101 | 0.000198 |
| CCR8 | 0.17349441 | 0.734053595 | 2.080996 | 2.32E-10 | 1.11E-09 |
| CCL2 | 34.9686388 | 11.61449106 | −1.59014 | 5.29E-09 | 2.02E-08 |
| CCL7 | 0.18927901 | 0.657953341 | 1.797471 | 1.96E-09 | 8.08E-09 |
| CCL3 | 2.14816002 | 4.902955744 | 1.19055 | 7.68E-08 | 2.42E-07 |
| CCL11 | 0.55157223 | 2.904377118 | 2.396607 | 1.38E-19 | 5.33E-18 |
| CCL23 | 1.12326388 | 0.273743401 | −2.0368 | 6.30E-12 | 3.93E-11 |
| CXCR4 | 9.22252573 | 19.16025406 | 1.054883 | 0.000386 | 0.000692 |
| LTBP1 | 6.62487458 | 25.99529833 | 1.972286 | 1.68E-14 | 1.71E-13 |
| PPARG | 2.88842354 | 0.756024662 | −1.93378 | 2.16E-17 | 4.49E-16 |
| MIF | 26.9699739 | 57.37594282 | 1.089092 | 1.10E-13 | 9.54E-13 |
| CD86 | 1.67553761 | 3.655170224 | 1.125315 | 6.99E-10 | 3.09E-09 |
| OLR1 | 0.72531218 | 3.224972232 | 2.152613 | 3.30E-10 | 1.54E-09 |
| RNASE2 | 0.34609099 | 0.725574867 | 1.067973 | 8.35E-07 | 2.24E-06 |
| CD79A | 3.16021095 | 10.3146119 | 1.706597 | 0.023568 | 0.031671 |
| BLNK | 10.4408627 | 4.286057645 | −1.28452 | 1.20E-10 | 5.99E-10 |
| VAV2 | 3.81897612 | 14.93115698 | 1.967068 | 2.07E-23 | 5.32E-21 |
| RAC2 | 8.0638533 | 28.71382317 | 1.832204 | 7.02E-18 | 1.67E-16 |
| RAC3 | 3.49005231 | 11.12945643 | 1.673063 | 2.03E-08 | 7.03E-08 |
| FOS | 338.296862 | 136.589998 | −1.30844 | 1.96E-12 | 1.34E-11 |
| CARD11 | 1.25857938 | 3.46194499 | 1.459787 | 2.19E-06 | 5.50E-06 |
| CD19 | 0.23053819 | 0.740556011 | 1.683603 | 0.001518 | 0.002482 |
| PIK3R1 | 8.78076848 | 4.252691387 | −1.04597 | 6.98E-07 | 1.89E-06 |
| PIK3CD | 2.3298192 | 6.209533165 | 1.414267 | 4.14E-17 | 7.84E-16 |
| AKT3 | 1.09039538 | 2.369801878 | 1.119915 | 6.20E-07 | 1.69E-06 |
| CD22 | 0.25129293 | 0.539412325 | 1.102018 | 0.012393 | 0.017507 |
| CD72 | 0.35093164 | 0.830922486 | 1.243524 | 3.41E-08 | 1.13E-07 |
| IFITM1 | 60.1765068 | 172.7061256 | 1.521047 | 2.55E-10 | 1.21E-09 |
| IGHE | 0.2414842 | 0.771029039 | 1.674856 | 0.001735 | 0.002811 |
| IGHG1 | 86.8715439 | 698.6877831 | 3.007692 | 1.24E-08 | 4.44E-08 |
| IGHG2 | 144.085491 | 642.3323018 | 2.156395 | 2.53E-05 | 5.39E-05 |
| IGHG3 | 30.1498131 | 193.1365392 | 2.6794 | 2.20E-06 | 5.52E-06 |
| IGHG4 | 71.4587322 | 410.6246363 | 2.522638 | 2.13E-06 | 5.38E-06 |
| IGHM | 25.724065 | 113.9007551 | 2.146587 | 0.000581 | 0.001014 |
| IGHV1-18 | 28.0447833 | 116.6576854 | 2.056477 | 0.006306 | 0.009362 |
| IGHV1-24 | 9.20112685 | 50.8718685 | 2.466986 | 0.000875 | 0.001484 |
| IGHV1-69 | 4.30104966 | 21.78746724 | 2.340738 | 0.005635 | 0.008425 |
| IGHV2-26 | 5.85552155 | 20.18384448 | 1.785331 | 0.03092 | 0.04069 |
| IGHV2-5 | 2.25448061 | 8.638621574 | 1.938006 | 0.010052 | 0.014422 |
| IGHV2-70 | 4.16961628 | 17.08320458 | 2.034592 | 0.007378 | 0.010847 |
| IGHV3-11 | 13.5472501 | 48.65714917 | 1.844652 | 0.012999 | 0.018306 |
| IGHV3-15 | 25.8863216 | 62.98550917 | 1.28283 | 0.033187 | 0.043404 |
| IGHV3-20 | 0.91442044 | 4.904326758 | 2.423126 | 0.020563 | 0.027907 |
| IGHV3-21 | 15.7078204 | 56.99584537 | 1.859374 | 0.003813 | 0.005856 |
| IGHV3-23 | 41.3186634 | 126.6624015 | 1.616123 | 0.007178 | 0.010576 |
| IGHV3-30 | 20.7333208 | 62.45990567 | 1.590979 | 0.031063 | 0.04086 |
| IGHV3-33 | 12.4224505 | 33.34431749 | 1.424491 | 0.021303 | 0.028845 |
| IGHV3-48 | 7.11143582 | 14.96467857 | 1.073349 | 0.009273 | 0.013399 |
| IGHV3-53 | 3.91411303 | 11.4157559 | 1.544269 | 0.024646 | 0.033 |
| IGHV3-64 | 0.8308217 | 6.716016837 | 3.014995 | 0.013989 | 0.019583 |
| IGHV3-7 | 1.23334926 | 2.669893683 | 1.114201 | 0.035765 | 0.046491 |
| IGHV4-34 | 10.5658183 | 42.39739749 | 2.004571 | 0.004522 | 0.00686 |
| IGHV4-39 | 30.0219714 | 104.3604529 | 1.797484 | 0.013882 | 0.019448 |
| IGHV4-59 | 15.9127994 | 60.28010631 | 1.921494 | 0.028581 | 0.037839 |
| IGHV5-51 | 42.2463818 | 136.4318714 | 1.691281 | 0.020611 | 0.027966 |
| IGHV6-1 | 1.13432516 | 2.954858673 | 1.381255 | 0.012314 | 0.017404 |
| IGKC | 310.677637 | 922.6267517 | 1.570329 | 0.00422 | 0.006436 |
| IGKJ5 | 1.28525265 | 4.612672177 | 1.843551 | 0.003279 | 0.005084 |
| IGKV1-12 | 0.99550699 | 2.573234524 | 1.37008 | 0.009952 | 0.014292 |
| IGKV1-16 | 11.9177292 | 41.34892335 | 1.79474 | 0.018027 | 0.024752 |
| IGKV1-5 | 61.956309 | 179.5210393 | 1.53483 | 0.017269 | 0.023778 |
| IGKV2D-29 | 3.9638841 | 13.4372983 | 1.761256 | 0.018362 | 0.025169 |
| IGKV3-11 | 49.2704227 | 151.7922943 | 1.623305 | 0.003912 | 0.005994 |
| IGKV3-15 | 25.5356016 | 77.71487129 | 1.605681 | 0.017013 | 0.023458 |
| IGKV3-20 | 85.2825162 | 278.3799343 | 1.706733 | 0.004424 | 0.006724 |
| IGKV4-1 | 68.2739321 | 202.0626519 | 1.565396 | 0.0065 | 0.009636 |
| IGLC2 | 151.274685 | 406.896779 | 1.427492 | 0.003367 | 0.005211 |
| IGLC3 | 104.742295 | 275.0896662 | 1.393058 | 0.005651 | 0.008449 |
| IGLJ2 | 0.30590016 | 0.916362063 | 1.582857 | 0.002942 | 0.004593 |
| IGLV1-40 | 47.7857769 | 132.2710552 | 1.468844 | 0.01631 | 0.022585 |
| IGLV1-44 | 26.0153565 | 81.27468738 | 1.643443 | 0.014452 | 0.020177 |
| IGLV1-47 | 25.4370506 | 66.27028303 | 1.381431 | 0.019104 | 0.026086 |
| IGLV1-50 | 0.44212857 | 1.185104284 | 1.422476 | 0.033796 | 0.044112 |
| IGLV1-51 | 36.3794548 | 117.9062168 | 1.696444 | 0.019806 | 0.026966 |
| IGLV2-23 | 34.5489759 | 97.452528 | 1.496057 | 0.028726 | 0.038017 |
| IGLV3-1 | 15.9420869 | 61.57060709 | 1.949401 | 0.007619 | 0.011173 |
| IGLV3-10 | 11.2153179 | 46.57915361 | 2.054214 | 0.029644 | 0.039118 |
| IGLV3-19 | 31.8797257 | 119.9531001 | 1.911759 | 0.009225 | 0.013333 |
| IGLV3-21 | 45.4931231 | 177.2342444 | 1.961937 | 0.005405 | 0.0081 |
| IGLV3-25 | 48.9199575 | 111.6293824 | 1.190222 | 0.022367 | 0.030162 |
| IGLV4-69 | 11.5982515 | 49.31871381 | 2.088228 | 0.031139 | 0.040953 |
| CMA1 | 1.98769994 | 0.403261295 | −2.30131 | 5.89E-10 | 2.64E-09 |
| CXCL17 | 117.062679 | 28.39879541 | −2.04338 | 4.71E-12 | 3.01E-11 |
| EDN3 | 6.75141587 | 0.130648725 | −5.69143 | 9.14E-20 | 3.66E-18 |
| SAA2 | 36.8129049 | 4.564767621 | −3.0116 | 0.031376 | 0.041211 |
| SEMA3C | 7.07077779 | 15.95431922 | 1.174006 | 8.65E-07 | 2.31E-06 |
| SEMA3G | 2.70000023 | 1.064567734 | −1.34269 | 3.80E-07 | 1.07E-06 |
| SEMA4F | 0.66486563 | 2.27662203 | 1.77576 | 1.15E-20 | 6.26E-19 |
| SEMA5B | 0.08279814 | 0.385262347 | 2.218171 | 2.10E-13 | 1.71E-12 |
| SEMA6D | 0.40114622 | 0.951156363 | 1.245554 | 0.009777 | 0.01406 |
| SEMA7A | 1.76833125 | 4.814156071 | 1.444894 | 6.52E-14 | 5.91E-13 |
| TNC | 23.5188264 | 115.8420979 | 2.300272 | 3.24E-12 | 2.13E-11 |
| TYMP | 45.4861884 | 141.5798149 | 1.638115 | 1.01E-14 | 1.06E-13 |
| CCRL2 | 0.34955428 | 0.735407853 | 1.073028 | 2.81E-08 | 9.44E-08 |
| CX3CR1 | 1.37980915 | 0.457133277 | −1.59378 | 4.56E-11 | 2.42E-10 |
| CXCR3 | 1.03249804 | 2.172631282 | 1.073304 | 0.018825 | 0.025735 |
| CYSLTR1 | 0.81979704 | 0.336677522 | −1.2839 | 1.74E-11 | 1.00E-10 |
| CYSLTR2 | 0.09923963 | 0.326614036 | 1.718598 | 6.95E-11 | 3.59E-10 |
| EDNRA | 2.36183216 | 5.059874066 | 1.099195 | 1.45E-06 | 3.74E-06 |
| EDNRB | 1.9785425 | 0.952446752 | −1.05473 | 0.003007 | 0.004688 |
| FPR2 | 0.1349368 | 0.443136924 | 1.715469 | 3.12E-07 | 8.93E-07 |
| PLAUR | 6.59774454 | 16.01458766 | 1.279342 | 2.78E-13 | 2.22E-12 |
| PLXNA1 | 6.91315204 | 21.17578982 | 1.615 | 2.84E-22 | 3.03E-20 |
| PLXNA3 | 1.55276495 | 3.273184314 | 1.075855 | 2.85E-12 | 1.89E-11 |
| PLXNB3 | 1.02087802 | 2.067987833 | 1.018417 | 1.02E-08 | 3.72E-08 |
| PLXND1 | 4.05003807 | 8.33955808 | 1.042035 | 9.74E-12 | 5.87E-11 |
| ROBO2 | 0.46743827 | 0.230123469 | −1.02237 | 4.91E-11 | 2.59E-10 |
| ADM | 23.1966792 | 46.9101623 | 1.015982 | 8.56E-07 | 2.29E-06 |
| AGT | 5.78163796 | 2.070608106 | −1.48142 | 3.33E-06 | 8.11E-06 |
| AMH | 0.07988846 | 0.62480892 | 2.967356 | 7.61E-07 | 2.05E-06 |
| ANGPTL7 | 1.68002292 | 0.201015181 | −3.0631 | 5.83E-13 | 4.37E-12 |
| APLN | 0.86235123 | 3.900377306 | 2.177266 | 4.54E-16 | 6.58E-15 |
| ARTN | 0.43390071 | 5.247002259 | 3.596057 | 5.63E-23 | 9.83E-21 |
| BMP1 | 3.33853928 | 12.80559466 | 1.939485 | 2.61E-24 | 1.53E-21 |
| BMP2 | 3.92212365 | 9.153133226 | 1.222631 | 1.08E-07 | 3.33E-07 |
| BMP3 | 1.72390668 | 0.339824676 | −2.34282 | 3.91E-12 | 2.54E-11 |
| BMP8A | 0.06636327 | 0.526073118 | 2.986807 | 3.79E-21 | 2.45E-19 |
| BMP8B | 0.5313324 | 1.349579758 | 1.344824 | 9.28E-12 | 5.61E-11 |
| BTC | 1.78168079 | 0.579803476 | −1.6196 | 2.21E-14 | 2.19E-13 |
| CD70 | 0.30839157 | 2.87821674 | 3.22234 | 1.26E-15 | 1.65E-14 |
| CGB5 | 0.00244445 | 0.360701481 | 7.205152 | 1.82E-13 | 1.50E-12 |
| CGB7 | 0.0621182 | 0.274841474 | 2.145512 | 4.00E-17 | 7.62E-16 |
| CGB8 | 0.00479987 | 0.327709573 | 6.093277 | 2.09E-13 | 1.70E-12 |
| CLEC11A | 2.71065169 | 9.312385266 | 1.780511 | 2.82E-15 | 3.43E-14 |
| CMTM1 | 0.19283448 | 0.524554219 | 1.443729 | 4.22E-12 | 2.72E-11 |
| CMTM3 | 4.81675736 | 10.42202404 | 1.113501 | 6.69E-12 | 4.15E-11 |
| CSF2 | 0.17931906 | 4.564203615 | 4.669762 | 1.47E-19 | 5.65E-18 |
| CSPG5 | 0.10920882 | 0.314350353 | 1.525284 | 0.000161 | 0.000305 |
| DKK1 | 2.00139197 | 8.598564529 | 2.103092 | 8.49E-08 | 2.65E-07 |
| EGF | 1.40614405 | 0.353739232 | −1.99099 | 1.08E-05 | 2.43E-05 |
| EPO | 0.11762463 | 0.588822801 | 2.323643 | 1.83E-12 | 1.25E-11 |
| ESM1 | 0.88518558 | 2.453007684 | 1.4705 | 5.86E-17 | 1.06E-15 |
| FAM3B | 16.2966982 | 3.222150502 | −2.33848 | 1.60E-18 | 4.62E-17 |
| FAM3D | 96.035144 | 6.988279824 | −3.78055 | 5.91E-24 | 2.29E-21 |
| FGF18 | 0.7907508 | 0.26729618 | −1.56478 | 1.54E-06 | 3.96E-06 |
| FGF19 | 0.00665932 | 1.529811499 | 7.843763 | 1.35E-05 | 3.00E-05 |
| FGF7 | 1.90308134 | 0.546490517 | −1.80007 | 9.11E-10 | 3.96E-09 |
| GAST | 0.12363756 | 5.276926614 | 5.415509 | 4.03E-20 | 1.82E-18 |
| GDF10 | 1.98146718 | 0.265373097 | −2.90048 | 4.92E-13 | 3.74E-12 |
| GDF6 | 0.03060174 | 0.223556987 | 2.868957 | 1.08E-09 | 4.63E-09 |
| GNRH1 | 0.18077715 | 0.466431407 | 1.367453 | 1.45E-07 | 4.38E-07 |
| GREM1 | 1.0455893 | 4.727065721 | 2.176629 | 5.57E-16 | 7.89E-15 |
| GREM2 | 1.58860774 | 0.179265326 | −3.14759 | 6.58E-17 | 1.17E-15 |
| GRP | 0.41557375 | 1.600923684 | 1.945728 | 1.75E-09 | 7.28E-09 |
| IFNE | 0.15189015 | 0.691375503 | 2.186441 | 2.92E-08 | 9.79E-08 |
| IFNK | 0.19121068 | 0.647341699 | 1.759364 | 1.60E-08 | 5.62E-08 |
| IL11 | 0.13642248 | 3.280347335 | 4.587695 | 1.24E-25 | 3.15E-22 |
| IL12A | 0.5559761 | 0.223027363 | −1.3178 | 0.000136 | 0.00026 |
| IL17C | 0.09787588 | 0.213580834 | 1.125757 | 0.016171 | 0.022405 |
| IL17D | 1.32095958 | 0.238188898 | −2.47141 | 1.67E-09 | 6.95E-09 |
| IL1F10 | 0.32539757 | 0.783656027 | 1.268017 | 0.002822 | 0.00442 |
| IL1RN | 308.866731 | 92.42516434 | −1.74063 | 2.00E-07 | 5.90E-07 |
| IL24 | 0.83979892 | 9.87783144 | 3.556079 | 2.98E-15 | 3.61E-14 |
| IL33 | 13.98023 | 4.709156551 | −1.56985 | 4.15E-14 | 3.92E-13 |
| IL34 | 6.63270625 | 2.842884765 | −1.22224 | 1.73E-11 | 9.96E-11 |
| INHA | 0.07616644 | 0.214585107 | 1.494323 | 1.13E-06 | 2.98E-06 |
| INHBA | 1.02075508 | 18.84559329 | 4.206519 | 4.04E-22 | 4.04E-20 |
| INHBE | 0.11804892 | 0.278753559 | 1.239605 | 4.59E-05 | 9.41E-05 |
| JAG1 | 18.6109461 | 44.3967847 | 1.254304 | 4.03E-14 | 3.81E-13 |
| JAG2 | 7.3803013 | 17.14258704 | 1.215833 | 7.38E-15 | 8.07E-14 |
| LHB | 0.08880278 | 0.6175139 | 2.797795 | 2.43E-18 | 6.56E-17 |
| LTBP2 | 8.59170092 | 17.58345766 | 1.033203 | 8.72E-08 | 2.72E-07 |
| MDK | 26.910812 | 60.19590711 | 1.161479 | 3.14E-06 | 7.68E-06 |
| MIA | 1.98022809 | 0.548150514 | −1.85302 | 0.002231 | 0.003555 |
| NGF | 0.89179839 | 2.087460938 | 1.22696 | 1.10E-05 | 2.48E-05 |
| NMB | 4.46159827 | 13.90247985 | 1.63971 | 1.82E-12 | 1.24E-11 |
| NRG1 | 0.9112157 | 4.468160248 | 2.293816 | 7.30E-13 | 5.39E-12 |
| NTF3 | 0.90704007 | 0.390411652 | −1.21617 | 3.42E-13 | 2.68E-12 |
| OGN | 3.59963628 | 0.867043828 | −2.05367 | 7.47E-12 | 4.60E-11 |
| OSM | 0.91005352 | 2.769073982 | 1.60538 | 8.63E-08 | 2.70E-07 |
| PDGFA | 2.97864939 | 7.432678127 | 1.319224 | 9.82E-14 | 8.64E-13 |
| PDGFB | 3.10490671 | 6.567244174 | 1.080738 | 1.28E-09 | 5.42E-09 |
| PDGFD | 2.00947478 | 0.801335171 | −1.32634 | 2.44E-05 | 5.22E-05 |
| PGF | 2.29505443 | 8.732569631 | 1.927878 | 2.04E-17 | 4.26E-16 |
| PTHLH | 5.48040688 | 63.6062284 | 3.536813 | 1.25E-18 | 3.74E-17 |
| PTN | 26.0423208 | 12.62390556 | −1.0447 | 1.06E-10 | 5.29E-10 |
| SCG2 | 0.56913811 | 1.344286679 | 1.23999 | 0.009749 | 0.014022 |
| SCGB3A1 | 269.255231 | 27.49958988 | −3.29149 | 0.000269 | 0.000494 |
| SLURP1 | 405.893276 | 66.07402152 | −2.61895 | 1.32E-07 | 4.00E-07 |
| SPP1 | 15.3697514 | 106.1032656 | 2.787303 | 2.42E-14 | 2.39E-13 |
| STC1 | 3.23967231 | 7.683465944 | 1.245909 | 6.80E-07 | 1.85E-06 |
| STC2 | 0.51361732 | 4.88917222 | 3.250824 | 7.15E-23 | 1.14E-20 |
| TGFB3 | 3.54746977 | 8.099078106 | 1.190967 | 9.70E-09 | 3.54E-08 |
| TNFSF13B | 1.24240408 | 2.870095712 | 1.207964 | 5.96E-06 | 1.40E-05 |
| TNFSF18 | 0.44962232 | 1.398115287 | 1.636698 | 0.013091 | 0.01842 |
| TNFSF9 | 2.76838158 | 5.596849403 | 1.015572 | 2.57E-07 | 7.46E-07 |
| UCN | 0.21565604 | 0.742440911 | 1.783544 | 2.41E-11 | 1.36E-10 |
| UCN2 | 0.72836626 | 4.120332153 | 2.500025 | 1.80E-19 | 6.73E-18 |
| VEGFC | 2.4144167 | 10.2701672 | 2.088713 | 3.42E-08 | 1.14E-07 |
| VGF | 0.14444269 | 0.441491762 | 1.611889 | 5.04E-08 | 1.63E-07 |
| ACVR1C | 0.18085289 | 0.522859525 | 1.531607 | 9.15E-13 | 6.62E-12 |
| ANGPTL1 | 4.0469674 | 0.423402202 | −3.25674 | 1.11E-17 | 2.51E-16 |
| AR | 0.84142851 | 0.279458225 | −1.59021 | 2.28E-13 | 1.84E-12 |
| BMPR1B | 0.42433224 | 0.97757387 | 1.204011 | 3.72E-07 | 1.05E-06 |
| CNTFR | 3.5781603 | 0.853604306 | −2.06758 | 2.78E-14 | 2.71E-13 |
| EGFR | 15.5484376 | 39.46628352 | 1.343851 | 5.59E-07 | 1.54E-06 |
| EPOR | 0.4418559 | 1.051799214 | 1.251211 | 9.80E-15 | 1.03E-13 |
| FGFR4 | 0.40046898 | 1.471692526 | 1.877714 | 3.35E-12 | 2.20E-11 |
| IGF1R | 4.97994875 | 9.993087189 | 1.0048 | 7.96E-12 | 4.88E-11 |
| IL12RB1 | 0.52176071 | 1.430100231 | 1.454656 | 1.35E-07 | 4.07E-07 |
| IL12RB2 | 0.55232543 | 2.540589041 | 2.201573 | 8.83E-09 | 3.25E-08 |
| IL15RA | 2.65784681 | 5.826970161 | 1.132488 | 1.44E-12 | 1.00E-11 |
| IL17RD | 1.96227708 | 0.814741728 | −1.26811 | 2.67E-05 | 5.67E-05 |
| IL21R | 0.32308714 | 1.142605044 | 1.822332 | 6.22E-09 | 2.35E-08 |
| IL22RA2 | 0.1338516 | 0.453174929 | 1.759434 | 6.36E-07 | 1.74E-06 |
| IL27RA | 3.5496236 | 9.703778415 | 1.450881 | 8.17E-09 | 3.03E-08 |
| IL2RA | 0.74667359 | 2.439247542 | 1.707887 | 7.69E-12 | 4.72E-11 |
| IL2RG | 5.99842991 | 13.36207169 | 1.155487 | 0.002085 | 0.003336 |
| IL31RA | 0.28666596 | 0.791027258 | 1.464357 | 4.68E-07 | 1.31E-06 |
| LEPR | 2.10085142 | 0.903359942 | −1.2176 | 6.96E-06 | 1.62E-05 |
| LGR5 | 0.56930049 | 1.348318763 | 1.243899 | 0.002398 | 0.0038 |
| MC1R | 0.22552167 | 0.629522882 | 1.480993 | 9.02E-16 | 1.22E-14 |
| MET | 6.4272196 | 16.07767268 | 1.322792 | 2.07E-13 | 1.69E-12 |
| NR2E1 | 0.06145965 | 0.268358343 | 2.126449 | 0.00037 | 0.000665 |
| NR3C2 | 2.06675998 | 0.367105007 | −2.49311 | 9.20E-15 | 9.80E-14 |
| NR4A1 | 23.7674383 | 7.62137528 | −1.64086 | 6.87E-06 | 1.60E-05 |
| NR4A3 | 4.13466038 | 1.581359678 | −1.3866 | 0.001584 | 0.002583 |
| NR5A1 | 0.01405336 | 1.762426165 | 6.970504 | 8.63E-07 | 2.31E-06 |
| PTGER3 | 1.01346032 | 0.389626298 | −1.37913 | 3.12E-07 | 8.95E-07 |
| PTGFR | 1.32754176 | 0.439742298 | −1.59403 | 2.32E-05 | 4.98E-05 |
| RORC | 4.24330743 | 0.607971833 | −2.80311 | 2.66E-21 | 1.90E-19 |
| RXRG | 0.48690854 | 0.202918239 | −1.26275 | 5.87E-06 | 1.38E-05 |
| SORT1 | 18.0520794 | 8.066695852 | −1.16212 | 8.58E-18 | 2.00E-16 |
| SSTR2 | 0.10550506 | 0.406339649 | 1.945374 | 4.01E-08 | 1.32E-07 |
| TACR1 | 0.68462766 | 0.172061105 | −1.9924 | 2.36E-15 | 2.91E-14 |
| TGFBR3 | 4.78706853 | 1.379474384 | −1.79502 | 5.43E-14 | 5.01E-13 |
| TNFRSF11A | 1.49016855 | 0.523554639 | −1.50906 | 4.12E-12 | 2.66E-11 |
| TNFRSF12A | 17.7723627 | 56.19964292 | 1.660925 | 4.14E-15 | 4.85E-14 |
| TNFRSF18 | 4.3003038 | 16.11444457 | 1.905844 | 5.18E-11 | 2.73E-10 |
| TNFRSF19 | 6.77463912 | 2.906295251 | −1.22096 | 3.60E-08 | 1.19E-07 |
| TNFRSF25 | 1.76759089 | 4.574161764 | 1.371723 | 4.06E-12 | 2.62E-11 |
| TNFRSF4 | 0.74157962 | 3.237799059 | 2.12634 | 4.82E-18 | 1.21E-16 |
| TNFRSF8 | 0.23116456 | 0.716018319 | 1.631076 | 3.15E-10 | 1.48E-09 |
| TNFRSF9 | 0.21987028 | 1.011939319 | 2.202398 | 2.09E-15 | 2.60E-14 |
| TUBB3 | 0.35949321 | 1.485323276 | 2.046741 | 4.84E-16 | 6.97E-15 |
| FCGR3A | 3.9329154 | 14.23552539 | 1.855825 | 6.68E-11 | 3.46E-10 |
| FCGR3B | 0.33728536 | 0.76681787 | 1.184914 | 0.000267 | 0.000491 |
| CD247 | 0.83658503 | 1.750058596 | 1.064819 | 0.000918 | 0.001553 |
| ZAP70 | 0.650674 | 1.400126297 | 1.10555 | 0.000721 | 0.001239 |
| SHC1 | 21.3436652 | 43.46714575 | 1.026117 | 5.53E-17 | 1.00E-15 |
| SH2D1B | 1.30987534 | 0.248778859 | −2.39649 | 0.000128 | 0.000245 |
| SH2D1A | 0.48679012 | 1.030303921 | 1.081698 | 0.01699 | 0.023428 |
| GZMB | 3.38306871 | 9.73932144 | 1.525489 | 1.80E-07 | 5.36E-07 |
| PRF1 | 2.17381804 | 6.330644293 | 1.542121 | 1.15E-06 | 3.03E-06 |
| BID | 3.94088989 | 9.343639602 | 1.245463 | 6.89E-16 | 9.59E-15 |
| TEC | 1.46982345 | 0.60201631 | −1.28777 | 6.31E-13 | 4.69E-12 |
| ICOS | 0.44116172 | 1.427023302 | 1.693629 | 6.12E-11 | 3.19E-10 |
| CTLA4 | 0.50396065 | 2.170800093 | 2.106844 | 2.15E-14 | 2.15E-13 |
| CBLB | 1.28623151 | 3.026043292 | 1.234282 | 8.12E-16 | 1.11E-14 |
| CDK4 | 14.1156616 | 28.71289684 | 1.024402 | 1.04E-18 | 3.14E-17 |
| PDK1 | 1.06635448 | 2.243918541 | 1.073333 | 1.46E-11 | 8.57E-11 |
| TRAV2 | 0.22091724 | 0.51174568 | 1.211921 | 0.005292 | 0.007947 |
| TRAV4 | 0.22383663 | 0.516858401 | 1.207323 | 0.003459 | 0.005347 |
| TRAV8-3 | 0.24255669 | 0.56073964 | 1.209009 | 0.003018 | 0.004704 |
| TRAV8-4 | 0.18845464 | 0.418056464 | 1.149481 | 0.008897 | 0.012887 |
| TRAV8-6 | 0.22023349 | 0.467775712 | 1.086783 | 0.004075 | 0.006228 |
| TRAV24 | 0.08147574 | 0.226358104 | 1.474164 | 0.02406 | 0.032283 |
| TRAV26-1 | 0.14875179 | 0.338231849 | 1.185105 | 0.000173 | 0.000326 |
| TRAV26-2 | 0.10735569 | 0.241319312 | 1.168545 | 0.008334 | 0.01214 |
| TRAV29DV5 | 0.14335924 | 0.3344734 | 1.222257 | 0.00784 | 0.011471 |
| TRBJ2-3 | 0.36593755 | 0.882313743 | 1.269694 | 0.005042 | 0.007595 |
| TRBV4-1 | 0.18012141 | 0.478476686 | 1.409479 | 0.020608 | 0.027965 |
The FDR means the false discovery rate and the FDR value is less than 0.05 as the filter criterion. Tumor mean represented the average gene expression in tumor samples and normal mean represented the average gene expression in normal samples.
Figure 2.
Gene functional enrichment of differentially expressed immune-related genes. Differentially expressed IRGs identified via Gene Ontology (GO) (a,c) and KEGG pathway (b,d) analyses. In the bubble plot (c), green, red and blue bars represent biological processes, cellular components and molecular functions, respectively. The top 10 most significant GO terms (e) and Kyoto Encyclopedia of Genes and Genomes pathways (f). Nodes in the concentric circle graph represent co-expressed genes clustered in specific biological process terms. The inner sectors with larger size and darker color represent more significant enrichment.
Identification of survival-associated IRGs
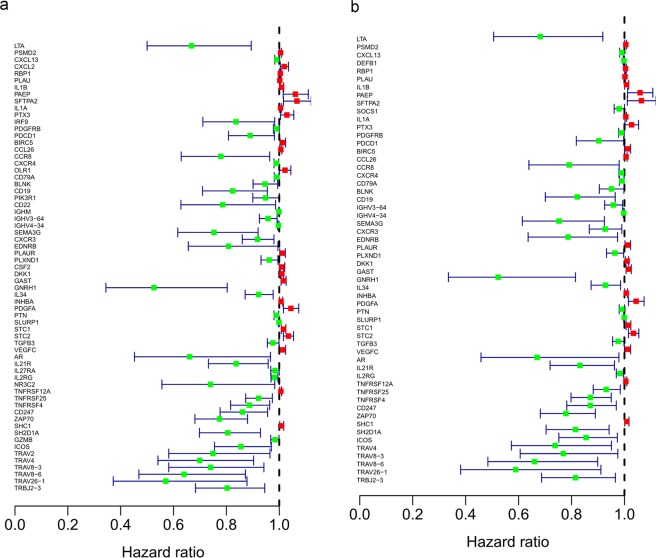
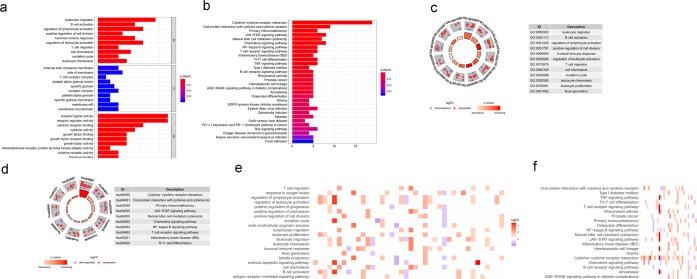
To explore the relationship between IRGs and prognosis, we identified 65, 56 IRGs that were significantly associated with overall survival (OS) from the total and HPV- HNSCC patients, respectively (P < 0.05; Table 2A,B; Fig. 3a,b). Moreover, we divided 65 IRGs into 23 high-risk (HR > 1) genes and 42 low-risk (HR < 1) genes (Fig. 3a). We also identified 20 high-risk (HR > 1) genes and 36 low-risk (HR < 1) genes from 56 IRGs (Fig. 3b). Similar to the results from the previous enrichment analysis of differentially expressed IRGs, 65 survival-associated IRGs were mainly involved in several Gene Ontology (GO) terms related to immunocyte migration and the activity of receptors and ligands (Fig. 4a,c,e). Additionally, KEGG pathway analysis indicated that survival-associated IRGs were enriched in “cytokine-cytokine receptor interaction (hsa04060)” (Fig. 4b,d,f). Interestingly, the results of 56 survival-associated IRGs enrichment analysis were almost consistent with our earlier research. GO terms immunocyte migration and the activity of receptors and ligands were mainly enriched (Fig. 5a,c,e). KEGG pathway “cytokine-cytokine receptor interaction (hsa04060)” was also enriched (Fig. 5b,d,f).
Table 2.
Relationships between the expression of immune-related genes and overall survival in head and neck squamous cell carcinoma.
| Gene | HR | HR95%CI | HR95%CI | P value |
|---|---|---|---|---|
| LTA | 0.668496 | 0.499696 | 0.894317 | 0.006684 |
| PSMD2 | 1.004766 | 1.001218 | 1.008327 | 0.008437 |
| CXCL13 | 0.990465 | 0.982905 | 0.998082 | 0.014244 |
| CXCL2 | 1.019352 | 1.003773 | 1.035172 | 0.014722 |
| RBP1 | 1.003664 | 1.000355 | 1.006985 | 0.029978 |
| PLAU | 1.0024 | 1.000922 | 1.00388 | 0.001448 |
| IL1B | 1.008894 | 1.001188 | 1.01666 | 0.023602 |
| PAEP | 1.061454 | 1.015316 | 1.10969 | 0.00853 |
| SFTPA2 | 1.067483 | 1.016221 | 1.12133 | 0.0093 |
| IL1A | 1.005049 | 1.002307 | 1.007798 | 0.000302 |
| PTX3 | 1.029444 | 1.004951 | 1.054535 | 0.018179 |
| IRF9 | 0.835539 | 0.710554 | 0.982507 | 0.029748 |
| PDGFRB | 0.9891 | 0.978489 | 0.999825 | 0.046403 |
| PDCD1 | 0.890434 | 0.807518 | 0.981864 | 0.019967 |
| BIRC5 | 1.013461 | 1.002572 | 1.024469 | 0.015264 |
| CCL26 | 1.005943 | 1.001596 | 1.010309 | 0.00733 |
| CCR8 | 0.778939 | 0.629352 | 0.96408 | 0.021665 |
| CXCR4 | 0.98885 | 0.980251 | 0.997524 | 0.011864 |
| OLR1 | 1.021934 | 1.000659 | 1.043662 | 0.043248 |
| CD79A | 0.990108 | 0.981287 | 0.999008 | 0.02945 |
| BLNK | 0.946274 | 0.90121 | 0.993591 | 0.026538 |
| CD19 | 0.823527 | 0.710098 | 0.955076 | 0.010232 |
| PIK3R1 | 0.948401 | 0.899575 | 0.999877 | 0.04947 |
| CD22 | 0.786499 | 0.627544 | 0.985716 | 0.037082 |
| IGHM | 0.998821 | 0.99779 | 0.999852 | 0.025075 |
| IGHV3-64 | 0.957713 | 0.924915 | 0.991675 | 0.015091 |
| IGHV4-34 | 0.99686 | 0.99438 | 0.999347 | 0.013377 |
| SEMA3G | 0.753086 | 0.616466 | 0.919983 | 0.005494 |
| CXCR3 | 0.918157 | 0.860136 | 0.980092 | 0.010355 |
| EDNRB | 0.807825 | 0.65587 | 0.994985 | 0.044723 |
| PLAUR | 1.012167 | 1.001904 | 1.022535 | 0.02003 |
| PLXND1 | 0.96239 | 0.93088 | 0.994967 | 0.024004 |
| CSF2 | 1.010432 | 1.000419 | 1.020545 | 0.041111 |
| DKK1 | 1.009998 | 1.003671 | 1.016365 | 0.001918 |
| GAST | 1.016163 | 1.006507 | 1.025912 | 0.000997 |
| GNRH1 | 0.525898 | 0.344358 | 0.803142 | 0.002933 |
| IL34 | 0.922265 | 0.870564 | 0.977036 | 0.005974 |
| INHBA | 1.007063 | 1.00146 | 1.012697 | 0.013411 |
| PDGFA | 1.044258 | 1.015614 | 1.073709 | 0.002275 |
| PTN | 0.988966 | 0.979748 | 0.998271 | 0.02022 |
| SLURP1 | 0.99868 | 0.997466 | 0.999896 | 0.033426 |
| STC1 | 1.016164 | 1.00751 | 1.024892 | 0.000238 |
| STC2 | 1.034862 | 1.015672 | 1.054415 | 0.000333 |
| TGFB3 | 0.976459 | 0.95481 | 0.9986 | 0.037301 |
| VEGFC | 1.012665 | 1.00125 | 1.024211 | 0.02956 |
| AR | 0.660928 | 0.451694 | 0.967084 | 0.032982 |
| IL21R | 0.83736 | 0.732213 | 0.957607 | 0.009523 |
| IL27RA | 0.983581 | 0.967608 | 0.999818 | 0.047502 |
| IL2RG | 0.981909 | 0.968983 | 0.995008 | 0.006932 |
| NR3C2 | 0.739584 | 0.55636 | 0.983148 | 0.037804 |
| TNFRSF12A | 1.005603 | 1.001962 | 1.009258 | 0.002538 |
| TNFRSF25 | 0.92232 | 0.873419 | 0.97396 | 0.003623 |
| TNFRSF4 | 0.887354 | 0.816476 | 0.964384 | 0.004896 |
| CD247 | 0.861279 | 0.775609 | 0.956411 | 0.005211 |
| ZAP70 | 0.773955 | 0.680878 | 0.879756 | 8.87E-05 |
| SHC1 | 1.009126 | 1.001624 | 1.016684 | 0.017021 |
| SH2D1A | 0.805145 | 0.697566 | 0.929315 | 0.003059 |
| GZMB | 0.983635 | 0.968192 | 0.999325 | 0.040998 |
| ICOS | 0.855448 | 0.754597 | 0.969777 | 0.014709 |
| TRAV2 | 0.749408 | 0.581806 | 0.965292 | 0.025518 |
| TRAV4 | 0.698698 | 0.540568 | 0.903086 | 0.00617 |
| TRAV8-3 | 0.739954 | 0.581527 | 0.941541 | 0.014286 |
| TRAV8-6 | 0.639837 | 0.46938 | 0.872197 | 0.004727 |
| TRAV26-1 | 0.571343 | 0.371677 | 0.87827 | 0.010721 |
| TRBJ2-3 | 0.803312 | 0.682749 | 0.945165 | 0.008298 |
| Gene | HR | HR95%CI | HR95%CI | P value |
| LTA | 0.681712 | 0.506441 | 0.917639 | 0.011511 |
| PSMD2 | 1.004734 | 1.001177 | 1.008303 | 0.009049 |
| CXCL13 | 0.989825 | 0.981869 | 0.997845 | 0.012991 |
| DEFB1 | 0.997616 | 0.995385 | 0.999852 | 0.036633 |
| RBP1 | 1.004001 | 1.000537 | 1.007476 | 0.023552 |
| PLAU | 1.002342 | 1.000849 | 1.003836 | 0.002091 |
| IL1B | 1.008056 | 1.000107 | 1.016068 | 0.046975 |
| PAEP | 1.058761 | 1.012306 | 1.107348 | 0.012622 |
| SFTPA2 | 1.063654 | 1.010685 | 1.119399 | 0.017896 |
| SOCS1 | 0.98023 | 0.961002 | 0.999843 | 0.048214 |
| IL1A | 1.004939 | 1.002162 | 1.007723 | 0.000483 |
| PTX3 | 1.027288 | 1.002224 | 1.05298 | 0.032662 |
| PDGFRB | 0.988423 | 0.977725 | 0.999239 | 0.035982 |
| PDCD1 | 0.903214 | 0.817886 | 0.997445 | 0.04438 |
| BIRC5 | 1.011754 | 1.000279 | 1.02336 | 0.044647 |
| CCL26 | 1.005815 | 1.001453 | 1.010196 | 0.008926 |
| CCR8 | 0.791404 | 0.639114 | 0.979982 | 0.031921 |
| CXCR4 | 0.988743 | 0.979416 | 0.998159 | 0.019232 |
| CD79A | 0.989859 | 0.980521 | 0.999287 | 0.035085 |
| BLNK | 0.950602 | 0.905354 | 0.998112 | 0.041761 |
| CD19 | 0.822477 | 0.701205 | 0.964723 | 0.01634 |
| IGHV3-64 | 0.958206 | 0.925079 | 0.992519 | 0.017394 |
| IGHV4-34 | 0.997012 | 0.994536 | 0.999494 | 0.018331 |
| SEMA3G | 0.753482 | 0.614441 | 0.923987 | 0.006537 |
| CXCR3 | 0.92655 | 0.867316 | 0.989829 | 0.023621 |
| EDNRB | 0.787011 | 0.635774 | 0.974225 | 0.027821 |
| PLAUR | 1.012231 | 1.002034 | 1.022531 | 0.018602 |
| PLXND1 | 0.963663 | 0.93165 | 0.996776 | 0.03177 |
| DKK1 | 1.0097 | 1.003305 | 1.016136 | 0.002901 |
| GAST | 1.015899 | 1.006204 | 1.025687 | 0.001263 |
| GNRH1 | 0.522748 | 0.335136 | 0.815388 | 0.00424 |
| IL34 | 0.927659 | 0.873885 | 0.984741 | 0.013715 |
| INHBA | 1.006638 | 1.000945 | 1.012363 | 0.022231 |
| PDGFA | 1.043827 | 1.014846 | 1.073636 | 0.002829 |
| PTN | 0.990012 | 0.980399 | 0.999719 | 0.043749 |
| SLURP1 | 0.998546 | 0.997284 | 0.999809 | 0.024046 |
| STC1 | 1.015356 | 1.006415 | 1.024377 | 0.000733 |
| STC2 | 1.034271 | 1.015038 | 1.053869 | 0.000434 |
| TGFB3 | 0.976175 | 0.954551 | 0.998288 | 0.034869 |
| VEGFC | 1.011847 | 1.00034 | 1.023486 | 0.043569 |
| AR | 0.669584 | 0.457741 | 0.979467 | 0.038746 |
| IL21R | 0.831783 | 0.719299 | 0.961857 | 0.012971 |
| IL2RG | 0.982533 | 0.968926 | 0.99633 | 0.013261 |
| TNFRSF12A | 1.005413 | 1.001712 | 1.009127 | 0.004113 |
| TNFRSF25 | 0.931332 | 0.881721 | 0.983734 | 0.01086 |
| TNFRSF4 | 0.870867 | 0.798383 | 0.949931 | 0.001818 |
| CD247 | 0.870532 | 0.78222 | 0.968814 | 0.01107 |
| ZAP70 | 0.779149 | 0.68226 | 0.889798 | 0.00023 |
| SHC1 | 1.009006 | 1.001361 | 1.016708 | 0.02086 |
| SH2D1A | 0.814511 | 0.704142 | 0.942179 | 0.005751 |
| ICOS | 0.855312 | 0.752063 | 0.972737 | 0.017261 |
| TRAV4 | 0.738136 | 0.573219 | 0.9505 | 0.018599 |
| TRAV8-3 | 0.76872 | 0.605943 | 0.975226 | 0.030265 |
| TRAV8-6 | 0.659702 | 0.483854 | 0.89946 | 0.008541 |
| TRAV26-1 | 0.589333 | 0.381387 | 0.910658 | 0.017244 |
| TRBJ2-3 | 0.81451 | 0.68679 | 0.965983 | 0.01839 |
(A) The prognosis-related genes in total HNSCC patients, (B) The prognosis-related genes in HPV- HNSCC patients. Immune-related genes were divided into high risk and low risk the prognosis-related genes via HR value (HR > 1 indicated high risk and HR < 1 indicated low risk).
Figure 3.
Forest plot of the hazard ratios showing the prognosis-related immune genes. (a) The prognosis-related genes in total HNSCC patients, (b) The prognosis-related genes in HPV- HNSCC patients. Red dots represent high-risk genes (HR > 1), and green dots represent low-risk genes (HR < 1).
Figure 4.
Functional enrichment of survival-associated IRGs. Survival-associated IRGs identified via Gene Ontology (GO) analysis (a) and KEGG pathway analysis (b). Circle graphs representing the most significant Gene Ontology terms (c) and Kyoto Encyclopedia of Genes and Genomes pathways (d). Nodes in the concentric circle graph represent co-expressed genes clustered in specific biological process terms. The inner sectors with larger size and darker color represent more significant enrichment. Heat maps representing the overall expression levels of 20 enriched Gene Ontology terms (e) and Kyoto Encyclopedia of Genes and Genomes pathways (f).
Figure 5.
Functional enrichment of HPV- HNSCC patients survival-associated IRGs. Survival-associated IRGs identified via Gene Ontology (GO) analysis (a) and KEGG pathway analysis (b). Circle graphs representing the most significant Gene Ontology terms (c) and Kyoto Encyclopedia of Genes and Genomes pathways (d). Nodes in the concentric circle graph represent co-expressed genes clustered in specific biological process terms. The inner sectors with larger size and darker color represent more significant enrichment. Heat maps representing the overall expression levels of 20 enriched Gene Ontology terms (e) and 7 enriched Kyoto Encyclopedia of Genes and Genomes pathways (f).
Transcription factor (TF) regulatory network
We found 63 differentially expressed transcription factors (TFs) within the genes that were differentially expressed between HNSCC patients and normal patients, 46 of which were upregulated and 17 of which were downregulated (Fig. 6a,b). To investigate the relationship between the differentially expressed TFs and 23 high-risk survival-associated IRGs, we constructed a regulatory network based on them. In this module (Fig. 6c), 16 edges involving 18 nodes were formed. SNA12 was remarkable for having the most connections with other high-risk genes, while BIRC5 was remarkable for having the most connections with other TFs.
Figure 6.
Differentially expressed transcription factors (TFs) in the differentially expressed genes between HNSCC and nontumor patients. (a) Heat map and (b) volcano plot demonstrating differentially expressed TFs. Red represents upregulated differentially expressed TFs, blue represents downregulated differentially expressed TFs, and black dots (volcano) represent TFs that were not differentially expressed. (c) Differentially expressed TFs and the high-risk survival-associated IRG interaction network. The red line represents positive regulation, and the green line represents negative regulation.
Immune gene-related prognosis model
To establish a prognosis prediction model based on survival-associated IRG expression, prognostic genes for HNSCC were identified by multivariable Cox regression analysis. Finally, to avoid false positive results, 10 genes were proven to be predictive of clinical outcomes via multivariable Cox regression analysis (P < 0.01) and constructed the optimal model according to Akaike information criterion. The model corresponding to minimum AIC value (AIC = 1,859.31) represented the target model. SEMA3G, GNRH1 and ZAP70 were positively correlated with OS. PLAU, SFTPA2, CCL26, DKK1, GAST, PDGFA and STC1 were negatively correlated with OS (Table 3A). Thus, the expression data of these prognostic genes and their coefficients were used to develop a gene-based prognosis prediction model, with a formula as follows: [expression level of PLAU * (0.0013)] + [expression level of SFTPA2 * (0.0590)] + [expression level of CCL26 * (0.0081)] + [expression level of SEMA3G * (−0.1523)] + [expression level of DKK1 * (0.0059)] + [expression level of GAST * (0.0173) + [expression
Table 3.
The coefficients and HR values of prognosis-associated IRGs included in the prognosis prediction model.
| Gene | coef | HR | HR95%CI | HR95%CI | P value |
|---|---|---|---|---|---|
| PLAU | 0.001272 | 1.001273 | 0.999585 | 1.002964 | 0.139493 |
| SFTPA2 | 0.059045 | 1.060823 | 1.008968 | 1.115344 | 0.020938 |
| CCL26 | 0.008102 | 1.008135 | 1.003507 | 1.012784 | 0.000558 |
| SEMA3G | −0.15228 | 0.858744 | 0.705155 | 1.045787 | 0.129856 |
| DKK1 | 0.005884 | 1.005902 | 0.998694 | 1.013162 | 0.108773 |
| GAST | 0.017251 | 1.0174 | 1.007052 | 1.027855 | 0.000942 |
| GNRH1 | −0.47171 | 0.623937 | 0.389605 | 0.99921 | 0.049617 |
| PDGFA | 0.026347 | 1.026697 | 0.994729 | 1.059693 | 0.102572 |
| STC1 | 0.01324 | 1.013328 | 1.003745 | 1.023003 | 0.006312 |
| ZAP70 | −0.12972 | 0.878341 | 0.758798 | 1.016718 | 0.082239 |
| Gene | coef | HR | HR95%CI | HR95%CI | P value |
| PLAU | 0.001459 | 1.00146 | 0.999768 | 1.003154 | 0.090764 |
| CCL26 | 0.008962 | 1.009002 | 1.004356 | 1.013669 | 0.000141 |
| SEMA3G | −0.15543 | 0.856043 | 0.694721 | 1.054827 | 0.144578 |
| DKK1 | 0.005887 | 1.005904 | 0.99853 | 1.013332 | 0.116845 |
| GAST | 0.017418 | 1.017571 | 1.007484 | 1.027759 | 0.000611 |
| GNRH1 | −0.53635 | 0.584881 | 0.359709 | 0.951008 | 0.03058 |
| PDGFA | 0.03171 | 1.032218 | 1.000327 | 1.065127 | 0.047663 |
| STC1 | 0.011712 | 1.011781 | 1.001787 | 1.021874 | 0.02075 |
| TNFRSF4 | −0.07703 | 0.925858 | 0.829668 | 1.0332 | 0.168698 |
| ZAP70 | −0.18953 | 0.827345 | 0.645459 | 1.060486 | 0.134569 |
| SH2D1A | 0.174511 | 1.190664 | 0.947321 | 1.496515 | 0.134644 |
(A) Total HNSCC patients, (B) HPV- HNSCC patients.
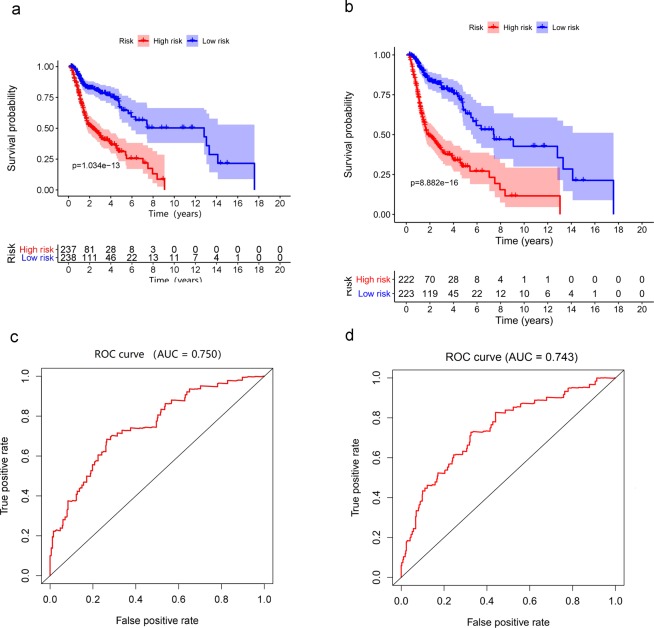
level of GNRH1 * (−0.4717)] + [expression level of PDGFA * (0.0263)] + [expression level of STC1 * (0.0132)] + [expression level of ZAP70 * (−0.1297)]. The 10-gene-based model was used to calculate a risk score for each sample as described above (Fig. 7a,c,e). All patients were divided into a high-risk group (n = 237) or a low-risk group (n = 238) according to the median risk score after removing some patients with missing clinical characteristics data. According to the Kaplan-Meier survival analysis, the overall survival time was significantly different between the high-risk and low-risk groups, and the five-year survival rates were 31.2% and 66.8%, respectively (Fig. 8a). The area under the curve (AUC) of the receiver operating characteristic (ROC) curve was 0.750, suggesting that the model could predict the survival outcomes of HNSCC patients (Fig. 8c).
Figure 7.
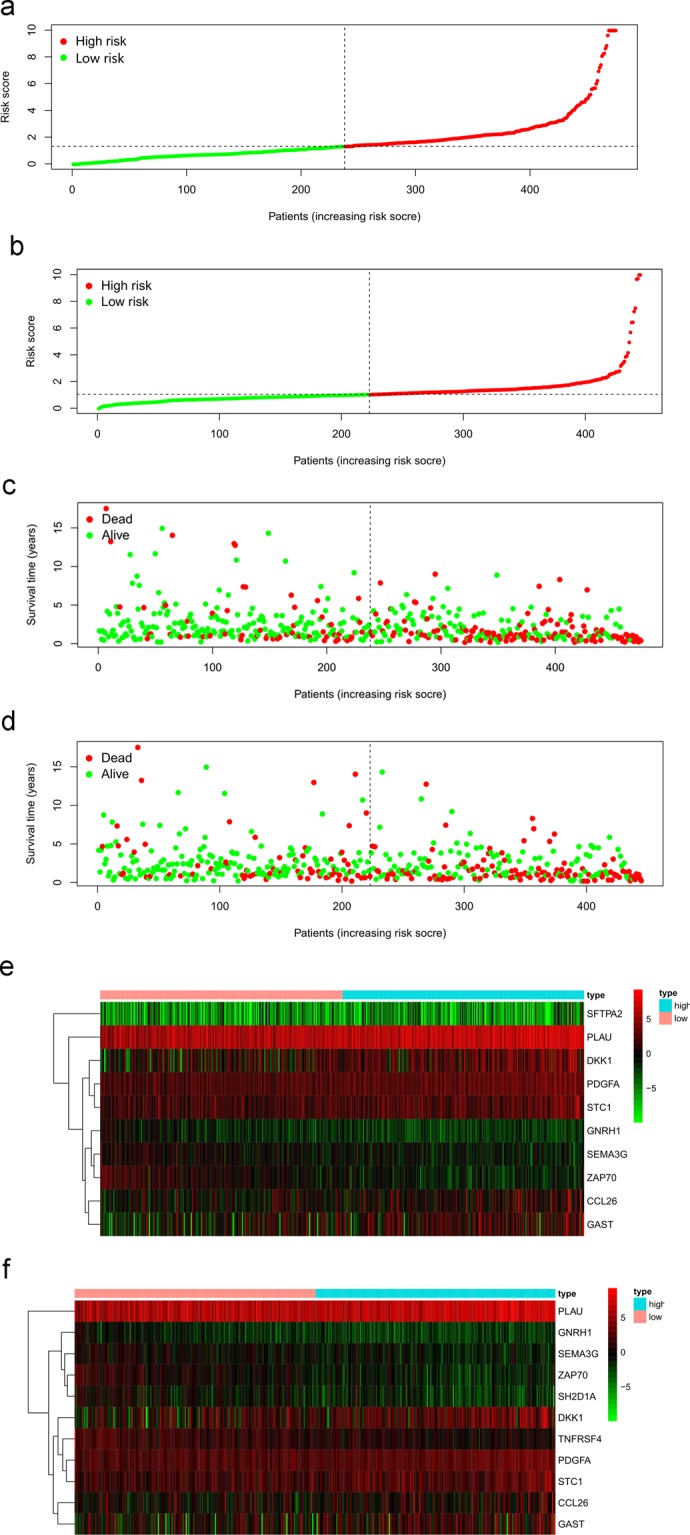
Development of a risk score based on survival-associated genes in total HNSCC patients (a,c,e) and HPV- HNSCC patients (b,d,f). (a,b) Association between risk score and the distribution of high (red) and low (green) risk groups. (c,d) Survival status of the HNSCC patients in different groups; red and green dots represent deceased and surviving patients, respectively. (e,f) Heat map representing the expression level of the prognostic genes based on the high-risk group and the low-risk group.
Figure 8.
Survival analysis in different groups of HNSCC patients. (a) The overall survival rates of different groups of HNSCC patients. (b) The overall survival rates of different groups of HPV-HNSCC patients. Verification of the accuracy of the prognostic model via analysis of the area under curve (AUC) of the receiver operating characteristic (ROC) curve (c). Total HNSCC patients, (d) HPV- HNSCC patients).
For HPV- HNSCC patients, we also identified 11 genes to predict clinical outcomes and establish the prognosis predict model via multivariable Cox regression analysis (P < 0.01). The same as the above methods and standards, the model corresponding to minimum AIC value (AIC = 1814.05) represented the optimal model. SEMA3G, GNRH1, TNFRSF4 and ZAP70 were positively correlated with OS. PLAU, SH2D1A, CCL26, DKK1, GAST, PDGFA and STC1 were negatively correlated with OS (Table 3B). Then, the 11-gene-based model was also used to calculate a risk score for each sample with an alike formula (Fig. 7b,d,f). Similarly, HPV- patients were divided into a high-risk group (n = 222) or a low-risk group (n = 223) and the five-year survival rates were 28.5% and 63.1%, respectively (Fig. 8b). The area under the curve (AUC) of the receiver operating characteristic (ROC) curve was 0.743, suggesting that the model could predict the survival outcomes of HPV- HNSCC patients (Fig. 8d).
The relationship between IRGs and clinical factors
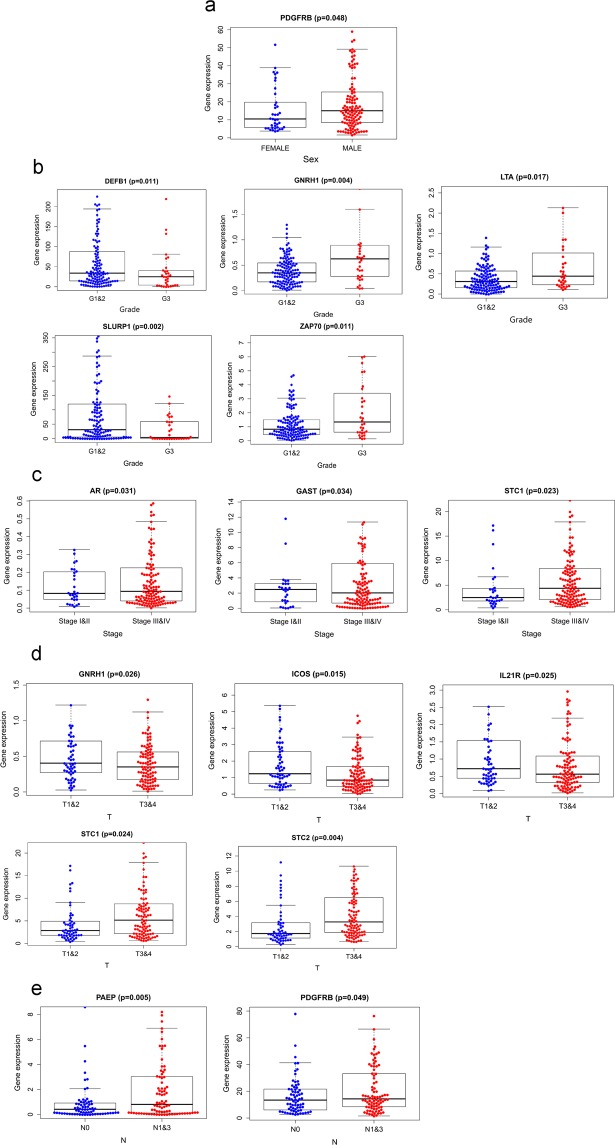
The relationships between the single genes and clinical factors were analyzed. The results indicated that the gene expression level of PDGFRB was significantly higher in males than in females whether total HNSCC patients (Fig. 9a) or HPV- HNSCC patients (Fig. 10a). AR was significantly higher in stage III and IV disease than in stage I and II disease, ICOS was significantly higher in T1–2 disease than in T3–4 disease whether total HNSCC patients (Fig. 9c,d) or HPV- HNSCC patients (Fig. 10c,d). Among the total HNSCC patients, NR3C2 was significantly higher in grade 1 and 2 disease than in grade 3 disease (Fig. 9b). Among the HPV- HNSCC patients, DEFB1 was significantly higher in grade 1 and 2 disease than in grade 3 disease, PAEP was significantly higher in N0 disease than in N1–3 disease (Fig. 10b,e).
Figure 9.
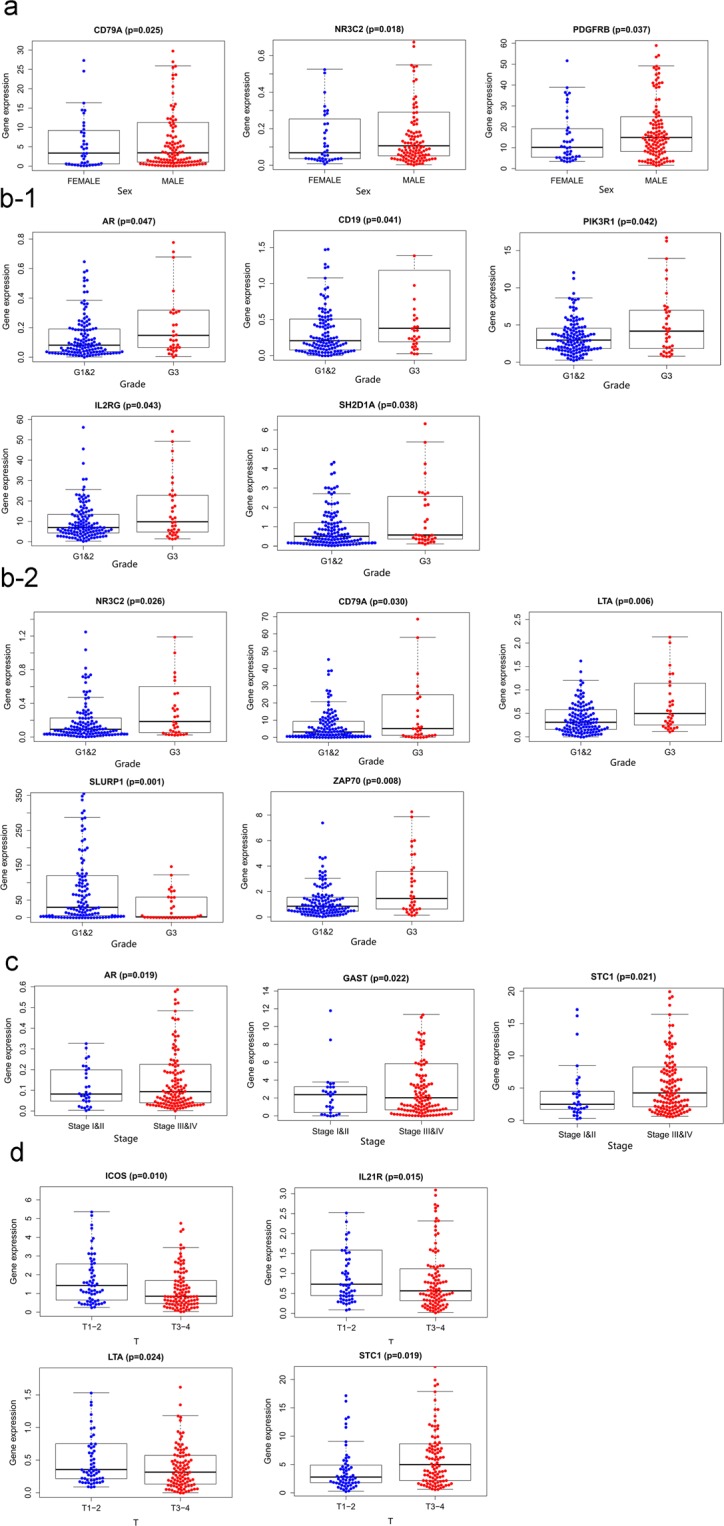
The relationships between prognosis-related IRG expression and sex (a), tumor grade (b), tumor stage (c), and T stage (d) in the high-risk (red) and low-risk (blue) groups of total HNSCC patients. The relationships between the expression of genes (SLURP1, ZAP70, CD79A, LTA, and NR3C2) (B2) and tumor grade were more significant than AR, CD19, IL2RG, PIK3R1 and SH2D1A (B1) according to p value.
Figure 10.
The relationships between prognosis-related IRG expression and sex (a), tumor grade (b), tumor stage (c), T stage (d) and N stage (e) in the high-risk (red) and low-risk (blue) groups of the HPV- HNSCC patients.
The relevance analysis of risk score and immune cell infiltration
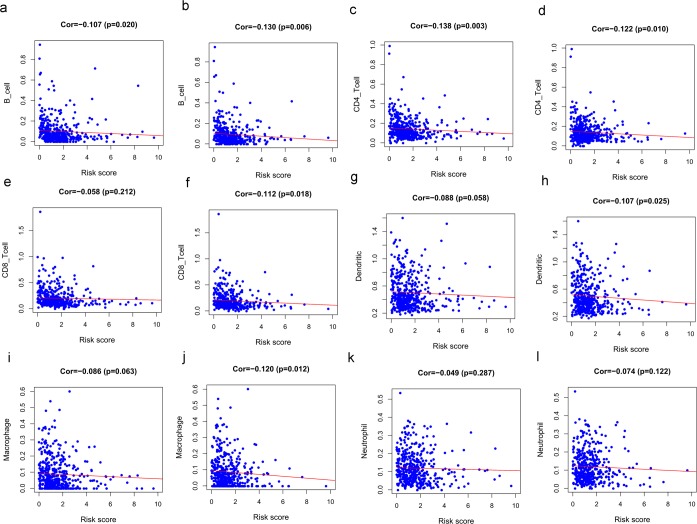
To reveal whether the immune-related genome can alter the tumor immune microenvironment, we analyzed the relationship between the risk score of IRGs and immune cell infiltration. B cell and CD4 T cell infiltration levels were significantly negatively correlated with the risk score in both total HNSCC patients (Fig. 11a,c) and HPV- patients (Fig. 11b,d) (P < 0.05). In total HNSCC patients, the infiltration of CD8 T cells, dendritic cells and macrophages were not statistically significant (P > 0.05) (Fig. 11e,g,i), while CD8 T cells, dendritic cells and macrophages infiltration levels were significantly negatively correlated with the risk score in the HPV- patients (P < 0.05) (Fig. 11f,h,j). The infiltration of neutrophils was not statistically significant in both total and HPV- HNSCC patients (P > 0.05) (Fig. 11k,l).
Figure 11.
The relationships between infiltration abundances of six types of immune cells and the risk score in different groups of HNSCC patients. B cell and CD4 T cell infiltration levels were significantly negatively correlated with the risk score in both total HNSCC patients (a,c) and HPV- patients (b,d). In total HNSCC patients, the infiltration of CD8 T cells (e), dendritic cells (g) and macrophages (i) were not statistically significant (P > 0.05) while CD8 T cells (f), dendritic cells (h) and macrophages (j) infiltration levels were significantly negatively correlated with the risk score of the HPV- patients. The infiltration of neutrophils was not statistically significant (P > 0.05) in both total (k) and HPV- (l) HNSCC patients.
Independent survival analysis
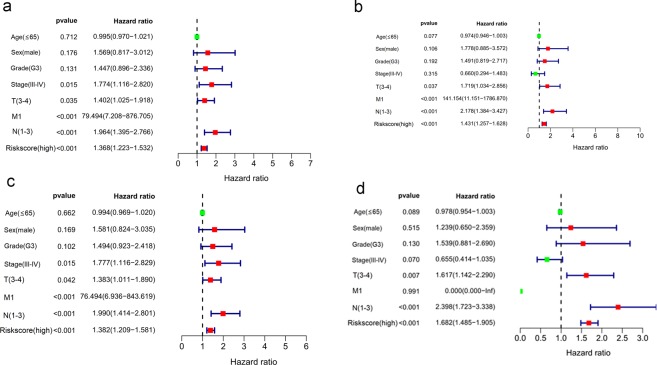
The univariable and multivariable Cox regression analyses suggested that the risk score could become an independent predictor for OS after adjusting for other parameters, including TN stage and risk score whether total HNSCC patients (Fig. 12a,b) or HPV- HNSCC patients (Fig. 12c,d) (P < 0.05).
Figure 12.
Univariable (a,c) and multivariable (b,d) independent subgroup analysis in terms of overall survival for the total HNSCC patients (a,b) and HPV- HNSCC patients (c,d).
Validation in the oncomine database
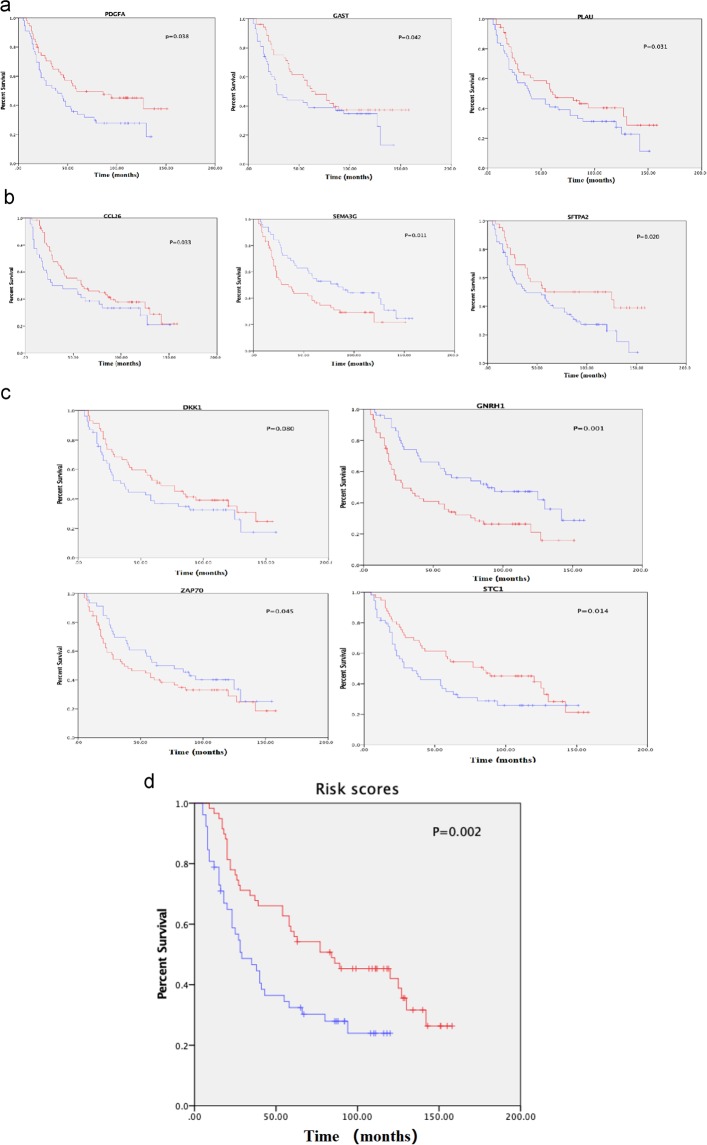
To verify the above results, we selected available cohorts of HNSCC samples from the Oncomine database, including Rickman’s cohort (81 samples) and Cormer’s cohort (31 samples). We extracted the expression levels of the survival-related IRGs, relevant clinical characteristics and follow-up times, and we performed survival analysis and created Kaplan-Meier survival curves (Fig. 13). As a result, 10 of the survival-related IRGs we identified were significantly associated with clinical prognosis according to the survival curves. SEMA3G, GNRH1 and ZAP70 were negatively correlated with OS, whereas PLAU, SFTPA2, CCL26, DKK1, GAST, PDGFA and STC1 were positively correlated with OS. Similarly, we also calculated the risk scores of 112 samples and created survival curves (Fig. 13). The results from this verification cohort were consistent with the results we obtained before, indicating their reliability and repeatability.
Figure 13.
Validation of TCGA results with Oncomine data. Kaplan-Meier survival curves were used to verify the relationship between genes (a). PDGFA, GAST and PLAU; (b) CCL26, SEMA3G and SFTPA2; (c) DKK1, GNRH1, ZAP70 and STC1) and risk scores (d) with overall survival. Red represents the high-risk group, and blue represents the low-risk group.
Discussion
The head and neck squamous cell carcinoma (HNSCC) is a common cancer which has attracted considerable and increasing attention12. While many patients with locally advanced HNSCC could be treated by surgery, radiation, chemotherapy and the combination application, those who develop recurrent/metastatic disease (R/M) has a median overall survival of less than a year13. Therefore, because second-line treatment options for advanced HNSCC are limited, immunotherapy has attracted increasing attention. However, depending on the different patterns of tumor-infiltrating immunocytes, individuals respond significantly differently after immunotherapy.
A number of studies have found that differential expression of IRGs affects tumor prognosis and response to immunotherapy, probably because these genes cause different levels of infiltration of various immune cell subtypes in tumors14–17. The interaction of the tumor with its microenvironment is crucial in the development and progression of the tumor. A comprehensive analysis of the expression of immune-related genes and the functional roles of different subsets of tumor-infiltrating cells in the tumor immune microenvironment could improve our knowledge of immunology and define subgroups of patients who are more likely to respond to immunotherapy. Therefore, in this study, we identified the survival-associated IRGs that were significantly related with the development and progression of HNSCC. In the functional enrichment analysis, one significant pathway identified was related to cytokine-cytokine receptor interactions in HNSCC, and the main enriched GO terms were immunocyte migration and activity of the receptor and ligand regardless of HPV status. The immune cell infiltration was mainly mediated by chemokine/receptor expression networks and cancer genetic alterations in tumor tissue via a systematic analysis of multiple cancers18. As expected, biological functions of the identified genes were significantly associated with inflammation progression. The results suggested that these differentially expressed genes were mostly enriched in inflammation-related terms and pathways.
Based on this result, we selected 10 genes (PLAU, SFTPA2, CCL26, SEMA3G, DKK1, GAST, GNRH1, PDGFA, ZAP70, and STC1) and 11 genes (SEMA3G, GNRH1, TNFRSF4, ZAP70, PLAU, SH2D1A, CCL26, DKK1, GAST, PDGFA and STC1) that were closely related to clinical prognosis to construct two prognostic prediction models to assess potential clinical outcomes for the total HNSCC patients and HPV- HNSCC patients. Notably, GNRH1 has been suggested as a marker of the metastatic spread of gynecological cancer19. PDGFA, a member of the platelet-derived growth factor (PDGF) family, may play a crucial role in the composition of the immune microenvironment20. PLAU, one of the major proteolytic enzymes involved in the degradation of extracellular matrix, has been demonstrated to play a critical role in tissue remodeling and migration in the developmental of cancer and in tumorigenesis21. Some studies indicated that PLAU was a marker to predict OS in HNSCC patients22. Stanniocalcin-1 (STC1) is a secreted glycoprotein implicated in several pathologies, including inflammation and cancer. Several studies have shown that STC1 is associated with cancer development23,24. It was verified that STC1 could accelerate tumor growth and reduce disease-free survival in mice. SEMA3G is a member of the class 3 semaphorin family originally characterized in axonal guidance25. Semaphorins have been shown to play multiple roles in normal and pathologic angiogenesis by acting on their receptors, plexins and neuropilins26. Methylation of the SFTPA2 promoter represents a potential biomarker for lung cancer diagnosis. The SFTPA2 DNA methylation profile was used as a potential tool to monitor disease progression and immunity27. The immune-related gene ZAP70 was associated with an increased risk of developing virally mediated head and neck squamous cell carcinoma. Mutations in many of these genes have previously been implicated in cancer risk, viral host-response, or epithelial immunity28. Gastrin is a growth factor of the gastrointestinal mucosa, and its role in gastrointestinal tumorigenesis is well studied. High levels of gastrin have been correlated with the poor prognosis of lung cancer patients. Gastrin-releasing peptide (GRP) signaling appears to mediate the autocrine growth of human squamous cell carcinoma of the head and neck29,30. Dickkopf-1 (DKK-1) is a secreted protein, and the expression and DKK-1 is different in various cancers. The methylation of DKK1 may be considered a prognostic marker in oral cancer31. DKK1 knockdown increased cellular migration and invasiveness in oral cancer cells32. High expression of DKK-1 was associated with poor prognosis, and this suggests that DKK-1 may be a useful molecular marker in breast cancer33. OChemokine ligand 26 (CCL26) levels were elevated in and positively correlated with stage III and IV colorectal cancer (CRC) tissues and were associated with a poor prognosis in CRC patients34. TNFRSF4 (also known as OX40 or CD134) is a member of the tumor necrosis factor receptor superfamily expressed on activated T cells35. TNFRSF4 has also been shown to promote T cell survival, proliferation, and memory, enhance cytokine secretion, thus further enhance antitumor immunity36. Therefore, the quality of T cells is as important as T cell number in response to HMSCC immunotherapy. SH2 domains are commonly found in adapter proteins that aid in the signal transduction of receptor tyrosine kinase pathways37. SH2D1A is also a SH2 domain-containing protein, which mutations caused the X-linked lymphoproliferative syndrome (XLP) and was associated with B-cell lymphomas38. These findings indicate the potential clinical application of IRGs as biomarkers in prognosis prediction and the promise of HNSCC immunotherapy. Our study will also provide a bioinformatics evidence for the prognosis prediction of HPV- HNSCC patients.
In univariable and multivariable Cox regression analyses, interestingly, we found that the expression of prognostic immune-related genes (CD79A, NR3C2, and PDGFRB) was significantly correlated with the sex of HNSCC patients. The results indicated that the immune microenvironment and the levels of infiltration by immune cells may be different in male and female HNSCC patients. Thus, male and female patients respond differently to immunotherapy. Recent findings reported that immune checkpoint inhibitors were twice as effective as standard cancer therapies in the treatment of men with advanced solid tumors compared to their female counterparts39,40. Immune checkpoint inhibitors can improve overall survival for patients of both sexes with some types of advanced cancers, but men have a greater treatment effect from these drugs versus control treatments than do women41. Sex differences in congenital and adaptive immune responses are known, and women generally have a stronger immune response than men. Therefore, different treatment strategies are adopted for patients of different sexes. The immune microenvironment should be improved in male cancer patients, while tumor antigenicity should be enhanced in female cancer patients42,43. However, Wallis et al. demonstrated no statistically significant association of patient sex with the efficacy of immunotherapy in the treatment of advanced cancers using overall survival as the outcome via meta-analysis44.
HNSCC represents a group of tumors occurring at various sites, including the oral mucosa and the palatine tonsils. Adding to this diversity is the recent observation that a proportion of these cancers, notably tonsillar carcinomas, are associated with human papillomavirus (HPV) infection41. HPV has been implicated in the etiology of a subset of HNSCCs, which often arise in younger patients without a history of alcohol or tobacco use42. Approximately 30% of HNSCC tumors are HPV+, and despite late-stage presentation, they often have a better prognosis and response to therapy45. A high level of CD8+ T-cell infiltration might be an important factor contributing to the improved survival of HPV+ HNSCC patients46. In current study, we also identified 13 differentially expressed IRGs that were significantly associated with overall survival (OS) of the HPV + HNSCC patients (Tables S2, 3; Fig. S1). However, the number of HPV + HNSCC samples are not enough for prognosis and immune cell infiltration analyses.
Tumor-infiltrating lymphocytes (TILs) are generally thought to represent a host immune response directed against antigens expressed on tumor cells, and a high density of TILs has been identified as a favorable marker in HNSCC47. Studies of HNSCC and other cancers have suggested a beneficial effect of B cells on outcome48–50. However, little is known about the role of TILs in the development of head and neck squamous cell carcinoma. No research has reported the relationship between the expression of IRGs and TIL status. Thus, knowledge of immune infiltration is important for an in-depth understanding of tumor and immune interactions. We investigated the relationship between prognosis-related immune gene expression and immune cell infiltration to determine the status of the HNSCC immune microenvironment and found that B cell and CD4 T cell infiltration levels were significantly negatively correlated with the expression of prognosis-related immune genes in total HNSCC patients. However, B cell, CD4 T cell, CD8 T cell, dendritic cell and macrophage infiltration levels were significantly negatively correlated with the risk score of the HPV- patients. IRGs were significantly associated with B cell infiltration in lung adenocarcinoma (LUAD), and these genes are involved in tumor immunity and may play an important role in the prognosis of patients51. Lin et al. demonstrated that an IRG-based risk score was significantly negatively correlated with the infiltration of CD4 + T cells, B cells and macrophages52. Similarly, our results confirmed a low level of infiltrating B cells and CD4 + T cells in high-risk patients who had a poor prognosis regardless of HPV status.
This study comprehensively analyzed IRGs and the TME based on data from TCGA. Differentially expressed IRGs were proposed to determine the abundance of infiltrating immune cells and assess potential clinical outcomes. We will aim to validate the molecular mechanism and further in-depth analysis the impact of different HPV status on the prognosis of HNSCC patients in future studies, particularly HPV + HNSCC patients.
Inevitably, there were several limitations in our study. The current results lack prognostic analysis of HPV + HNSCC patients due to small number of HPV + samples, and the lack of validation in a prospective clinical trial is also a limitation of the study. Moreover, the mechanism by which the expression of the prognosis-related IRGs affects the outcome of HNSCC patients remains unclear.
In conclusion, we arrived at a more comprehensive understanding of the TME and created a list of prognostic immune‐related genes with the potential to become prognostic biomarkers.
Materials and Methods
Clinical samples and immune gene data
All data in this study were obtained from a public database. The transcriptome profiling data of HNSCC samples were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/), which contained data from 501 primary HNSCC and 44 nontumor samples. The clinical information of 527 HNSCC patients was extracted and downloaded for further analysis. We also identified immune-related genes via the Immunology Database and Analysis Portal (ImmPort) (https://www.immport.org/) (Table S1). ImmPort is an archival repository and dissemination platform for clinical and molecular datasets. ImmPort is an important source of raw data and protocols from clinical trials, mechanistic studies, and novel methods for cellular and molecular measurements53. In addition, ImmPort is also a database that updates real-time data accurately and provides a list of IRGs that are actively involved in tumor immunological processes for cancer research.
Differential gene analysis
We performed differential gene analysis on all transcriptome profiling data using the R software limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html), with a false discovery rate (FDR) < 0.05 and log2 |fold change | > 1 as the cutoff values. Heatmaps were generated using the pheatmap package in R. Then, we extracted differentially expressed IRGs from the intersection of immune genes and all differentially expressed genes. To explore the potential molecular mechanisms of differentially expressed IRGs, functional enrichment analysis was performed via GO and KEGG pathway analyses54–56 using the R software clusterprofiler package57.
Survival-related IRGs and survival analysis
We used Perl software to analyze the clinical characteristics and follow-up data downloaded from TCGA and chose overall survival (OS) as the primary endpoint. Survival-related genes were selected by univariable COX regression analysis (FDR < 0.05). We made proportional hazards assumptions based on Schoenfeld residuals (phtest) for the COX regression model. The significance value for the overall test of proportional hazards is less than 0.05 (P < 0.05). Hazards ratio (HR) is the ratio of tumor samples and normal samples IRGs expression. We defined the high-risk IRGs (HR > 1) and low-risk IRGs (HR < 1) with HR = 1 as a cutoff. The Kaplan-Meier survival curve was drawn using the R software survival package according to a significance filter of P < 0.05. Functional enrichment analysis was also performed on survival-related IRGs that were significantly associated with OS.
Construction of the immune gene-related prognostic model
The expression data of these survival-related genes and their coefficients were used to develop a gene-based prognosis prediction model, and Akaike information criterion (AIC) was used to build the model in order to avoid the data of overfitting, with the minimum AIC value representing the target model58. The formula used is as follows: 59. Multivariable Cox regression analysis was used to illustrate the correlation of risk scores with patient overall survival (OS) and to identify potential prognostic genes. Patients were divided into high- and low-risk groups based on the median risk score value.
Correlation analysis of the clinical data
Relationships were analyzed between single genes and clinical factors via the R software beeswarm package. In addition, we also used clinical characteristics for univariable and multivariable Cox regression analyses with the R software survival package. The risk scores were divided into high- and low-risk groups based on median risk score, and ages were divided into ≤65 group and >65 group. The grades were divided into G1–2 and G3, stages were divided into I-II and III-IV groups, stage T were divided into T1–2 and T3–4 groups, stage N were divided into N0 and N1–3 groups and stage M were divided into M0 and M1 groups. We selected age (>65), sex (female), grade (G1–2), stage (I-II), T (1–2), M (M0) and N (N0) as the reference for each group. Risk score P-values of less than 0.001 were considered statistically significant.
Clinical relevance of tumor immune infiltration
We downloaded the immune infiltration levels of HNSCC patients using the Tumor IMmune Estimation Resource (TIMER) (http://cistrome.org/TIMER/), which is a web resource for the systematic evaluation of the clinical impact of different immune cells in diverse cancer types. We used this resource to investigate possible associations between the abundance of six subtypes of tumor-infiltrating immune cells (B cells, CD4 T cells, CD8 T cells, macrophages, neutrophils, and dendritic cells) and patient prognosis. Based on the immune gene-related prognostic model, we performed a correlation analysis combining the risk score and immune cell infiltration level of each sample via R software.
Transcription factor (TF) regulatory network
To explore the interactions between high-risk survival-associated IRGs and transcription factors (TFs), we obtained data on 318 TFs from the Cistrome online database (https://cistrome.org/). Cistrome is a comprehensive database for cancer transcription factor (TF) targets and provides regulatory links between TFs and transcriptomes. A protein–protein interaction (PPI) network was constructed based on data gleaned and displays many direct and indirect interactions with genes. The resulting PPI network was analyzed with Cytoscape software version 3.7.2. We extracted differential TFs to construct the regulatory network of the high-risk survival-associated IRGs (HR > 1) and potential TFs.
Statistical analysis
Differential analysis of immune genes, function enrichment analysis, COX regression analysis and survival analysis were performed using R software (version 3.6.1) and R packages. The Kaplan–Meier curve was created with the R survival and survminer packages. Based on the survival ROC R software package, the AUC of the survival ROC curve was calculated to verify the accuracy of the prognosis prediction model. Differences between clinical characteristics and prognosis-related IRGs were tested using an independent Student’s t test. P-values of less than 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (Grant No.81660448 and 81360401), the Special Health Technical Personnel Training program of Yunnan, China (Grant No. L-201612) and the Natural Science Foundation of Yunnan, China (Grant No. 2017FE468–006).
Author contributions
Conception and design: Yong-Wen He. Collection and assembly of data: Lei Li, Xiao-Li Wang. Data analysis and interpretation: Chuan-Zheng Sun, Yan Xi. Manuscript writing: Qian Lei, Ran Chen. Final approval of manuscript: All authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lei Li and Xiao-Li Wang.
Supplementary information
is available for this paper at 10.1038/s41598-020-63148-8.
References
- 1.Petr, S. & Vermorken, J. B. Treatment of Elderly Patients with Squamous Cell Carcinoma of the Head and Neck. Frontiers in Oncology610.3389/fonc.2016.00199 (2016). [DOI] [PMC free article] [PubMed]
- 2.Kish, J. A. et al. The effect of age on outcome in prospective, phase III NRG Oncology/RTOG trials of radiotherapy (XRT) +/− chemotherapy in locally advanced (LA) head and neck cancer (HNC). Journal of Clinical Oncology6003, 10.1200/jco.2015.33.15_suppl.6003 (2015).
- 3.Shield KD, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA: A Cancer Journal for Clinicians. 2017;67(1):51–64. doi: 10.3322/caac.21384. [DOI] [PubMed] [Google Scholar]
- 4.Chi MT, Tsao SW. The role of Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Virologica Sinica. 2015;30(2):107–121. doi: 10.1007/s12250-015-3592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Economopoulou, P. et al. Diagnostic Tumor Markers in Head and Neck Squamous Cell Carcinoma (HNSCC) in the Clinical Setting. Frontiers in Oncology10.3389/fonc.2019.00827 (2019). [DOI] [PMC free article] [PubMed]
- 6.Bledsoe TJ, Noble AR. Split-Course Accelerated Hypofractionated Radiotherapy (SCAHRT): A Safe and Effective Option for Head and Neck Cancer in the Elderly or Infirm. Anticancer Research. 2016;36(3):933. [PubMed] [Google Scholar]
- 7.Szturz, P. & Vermorken, J. B. Systemic Treatment of Recurrent/Metastatic Squamous Cell Carcinoma of the Head and Neck// Head and Neck Cancer. Springer New York, 10.1007/978-3-319-27601-4_42 (2016).
- 8.Economopoulou P, et al. The promise of immunotherapy in head and neck squamous cell carcinoma. Ann Oncol. 2016;27:1675–85. doi: 10.1136/esmoopen-2016-000122. [DOI] [PubMed] [Google Scholar]
- 9.Jazaeri AA. Immunotherapy in ovarian cancer: where are we now, and where are we going? Clinical Advances in Hematology & Oncology H & O. 2017;15(11):851. [PubMed] [Google Scholar]
- 10.Seiwert, T. Y. et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. The Lancet Oncology10.1016/S1470-2045(16)30066-3 (2016). [DOI] [PubMed]
- 11.Mehra, R. et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC): Pooled analyses after long-term follow-up in KEYNOTE-012. Journal of Clinical Oncology6012, 10.1200/JCO.2016.34.15_suppl.6012(2016).
- 12.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 13.Vermorken JB, et al. Platinum-Based Chemotherapy plus Cetuximab in Head and Neck Cancer. The New England Journal of Medicine. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 14.Kim JW, et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer. 2016;19(1):42–52. doi: 10.1007/s10120-014-0440-5. [DOI] [PubMed] [Google Scholar]
- 15.Vasaturo A, et al. Immune infiltrates impact on the prediction of prognosis and response to immunotherapy of melanoma patients. Journal of Translational Medicine. 2015;13:12–12. doi: 10.1186/1479-5876-13-S1-P12. [DOI] [Google Scholar]
- 16.Parikh F, et al. Chemoradiotherapy-induced upregulation of PD-1 antagonizes immunity to HPV-related oropharyngeal cancer. Cancer Research. 2014;74:7205–7216. doi: 10.1158/0008-5472.CAN-14-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bron, L. et al. Prognostic value of arginase-II expression and regulatory T‐cell infiltration in head and neck squamous cell carcinoma. International Journal of Cancer132, 10.1002/ijc.27728 (2013). [DOI] [PubMed]
- 18.Li, B. et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biology17, 10.1186/s13059-016-1028-7 (2016). [DOI] [PMC free article] [PubMed]
- 19.Andrusiewicz M, et al. CGB and GNRH1 expression analysis as a method of tumor cells metastatic spread detection in patients with gynecological malignances. Journal of Translational Medicine. 2011;9:130–130. doi: 10.1186/1479-5876-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartoschek M, Pietras K. PDGF family function and prognostic value in tumor biology. Biochemical and Biophysical Research Communications. 2018;503:984–990. doi: 10.1016/j.bbrc.2018.06.106. [DOI] [PubMed] [Google Scholar]
- 21.Yang K, et al. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. International Journal of Clinical Oncology. 2019;24:1030–1041. doi: 10.1007/s10147-019-01435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, et al. Expression profiles analysis identifies a novel three-mRNA signature to predict overall survival in oral squamous cell carcinoma. American Journal of Cancer Research. 2018;8:450–461. [PMC free article] [PubMed] [Google Scholar]
- 23.Chang AC, et al. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clinical & Experimental Metastasis. 2015;32:15–27. doi: 10.1007/s10585-014-9687-9. [DOI] [PubMed] [Google Scholar]
- 24.Chen FZF, et al. Role of stanniocalcin-1 in breast cancer (Review) Oncology Letters. 2019;18:3946–3953. doi: 10.3892/ol.2019.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature 2005. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai A, et al. Semaphorin signaling in angiogenesis, lymphangiogenesis and cancer. Cell Research. 2012;22:23–32. doi: 10.1038/cr.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grageda M, et al. DNA Methylation Profile and Expression of Surfactant Protein A2 gene in Lung Cancer. Experimental Lung Research. 2015;41:93–102. doi: 10.3109/01902148.2014.976298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levovitz C, et al. Abstract 2988: Exploring the association between immune-related genetic variations and virally mediated head and neck cancer. Cancer Research. 2012;72:2988–2988. doi: 10.1158/1538-7445.AM2012-2988. [DOI] [Google Scholar]
- 29.Lango MN, et al. Gastrin-releasing peptide receptor-mediated autocrine growth in squamous cell carcinoma of the head and neck. JNCI Journal of the National Cancer Institute. 2002;94:375–383. doi: 10.1093/jnci/94.5.375. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, et al. Antitumor mechanisms of combined gastrin-releasing peptide receptor and epidermal growth factor receptor targeting in head and neck cancer. Molecular Cancer Therapeutics. 2007;6:1414–1424. doi: 10.1158/1535-7163.MCT-06-0678. [DOI] [PubMed] [Google Scholar]
- 31.Paluszczak J, et al. The negative regulators of Wnt pathwayin-releasing peptide receptor and epidermal growth factoryngeal cancer and WIF1 methylation predicts shorter survival. Tumor Biology. 2015;36:2855–2861. doi: 10.1007/s13277-014-2913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogoshi K, et al. Dickkopf-1 in human oral cancer. Int J Oncol. 2011;39:329–36. doi: 10.3892/ijo.2011.1046. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, S. J. et al. Serum Dickkopf-1 expression level positively correlates with a poor prognosis in breast cancer. Diagnostic Pathology9, 10.1186/s13000-014-0161-4 (2014). [DOI] [PMC free article] [PubMed]
- 34.Lan Q, et al. CCL26 Participates in the PRL-3-Induced Promotion of Colorectal Cancer Invasion by Stimulating Tumor-Associated Macrophage Infiltration. Molecular Cancer Therapeutics. 2018;17:276. doi: 10.1158/1535-7163.MCT-17-0507. [DOI] [PubMed] [Google Scholar]
- 35.Popovic A, et al. Emerging strategies for combination checkpoint modulators in cancer immunotherapy. Journal of Clinical Investigation. 2018;128(8):3209–3218. doi: 10.1172/JCI120775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaer D, et al. Targeting tumor-necrosis factor receptor pathways for tumor immunotherapy. Journal for ImmunoTherapy of Cancer. 2014;2(1):7–7. doi: 10.1186/2051-1426-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endo TA, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387(6636):921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 38.Sandlund, J. T. et al. Frequent mutations in SH2D1A (XLP) in males presenting with high-grade mature B-cell neoplasms. Pediatric Blood & Cancer60(9) 10.1002/pbc.24525 (2013). [DOI] [PMC free article] [PubMed]
- 39.Conforti, F. et al. Cancer immunotherapy efficacy and patients’ sex: a systematic review and meta-analysis. Lancet Oncology19, 10.1016/S1470-2045(18)30261-4 (2018). [DOI] [PubMed]
- 40.Lin PY, et al. B7-H1-dependent sex-related differences in tumor immunity and immunotherapy responses. Journal of Immunology. 2010;185(5):2747–2753. doi: 10.4049/jimmunol.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, Sturgis EM. The role of human papillomavirus in squamous carcinoma of the head and neck. Curr Oncol Rep. 2006;8:130–139. doi: 10.1007/s11912-006-0048-y. [DOI] [PubMed] [Google Scholar]
- 42.Goon PK, et al. HPV & head and neck cancer: a descriptive update. Head Neck Oncol. 2009;1:36–43. doi: 10.1186/1758-3284-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Cowley LA, Liu XS. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy. Molecules. 2019;24:3214–3226. doi: 10.3390/molecules24183214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallis CJ, et al. Association of Patient Sex With Efficacy of Immune Checkpoint Inhibitors and Overall Survival in Advanced Cancers: A Systematic Review and Meta-analysis. JAMA. Oncology. 2019;5:529–536. doi: 10.1001/jamaoncol.2018.5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dayyani F, et al. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC) Head Neck oncology. 2010;2:15. doi: 10.1186/1758-3284-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell S, et al. Immune cell infiltration patterns and survival in head and neck squamous cell carcinoma. Head and Neck Oncology. 2013;5:24. [PMC free article] [PubMed] [Google Scholar]
- 47.Brandwein GM, et al. Oral squamous cell carcinoma: histologic risk assessment, but not margin status, is strongly predictive of local disease-free and overall survival. The American journal of surgery pathology. 2005;29:167–178. doi: 10.1097/01.pas.0000149687.90710.21. [DOI] [PubMed] [Google Scholar]
- 48.Nzula S, Going JJ, Stott DI. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer research. 2003;63:3275–3280. [PubMed] [Google Scholar]
- 49.Schmidt M, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer research. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 50.Herpen CM, et al. Intratumoral rhIL-12 administration in head and neck squamous cell carcinoma patients induces B cell activation. International journal of cancer. 2008;123:2354–2361. doi: 10.1002/ijc.23756. [DOI] [PubMed] [Google Scholar]
- 51.Wei BY, Kong W, Mou X, Wang S. Comprehensive analysis of tumor immune infiltration associated with endogenous competitive RNA networks in lung adenocarcinoma. Pathology, research and practice. 2019;215:159–170. doi: 10.1016/j.prp.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 52.Lin P, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging. 2019;11:480–500. doi: 10.18632/aging.101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharya S, et al. ImmPort: disseminating data to the public for the future of immunology. Immunologic Research. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. KEGG: Kyoto Encyclopaedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa MYM, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Research. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Science. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu G, et al. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omics A. Journal of Integrative Biology. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David P, Buckley TR. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches over Likelihood Ratio Tests. Systematic Biology. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 59.Guo Q, et al. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Scientific Reports. 2016;5:17683–17683. doi: 10.1038/srep17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.