Uncovering the mechanisms of resistance is a required step for countering the looming antibiotic resistance crisis. In this communication, we show how universally conserved histidine-triad hydrolases provide resistance to microcin C, a potent inhibitor of bacterial protein synthesis.
KEYWORDS: HinT, RiPPs, antibiotics, histidine-triad proteins, microcin C, peptide-nucleotides
ABSTRACT
The Escherichia coli microcin C (McC) and related compounds are potent Trojan horse peptide-nucleotide antibiotics. The peptide part facilitates transport into sensitive cells. Inside the cell, the peptide part is degraded by nonspecific peptidases releasing an aspartamide-adenylate containing a phosphoramide bond. This nonhydrolyzable compound inhibits aspartyl-tRNA synthetase. In addition to the efficient export of McC outside the producing cells, special mechanisms have evolved to avoid self-toxicity caused by the degradation of the peptide part inside the producers. Here, we report that histidine-triad (HIT) hydrolases encoded in biosynthetic clusters of some McC homologs or by standalone genes confer resistance to McC-like compounds by hydrolyzing the phosphoramide bond in toxic aspartamide-adenosine, rendering them inactive.
INTRODUCTION
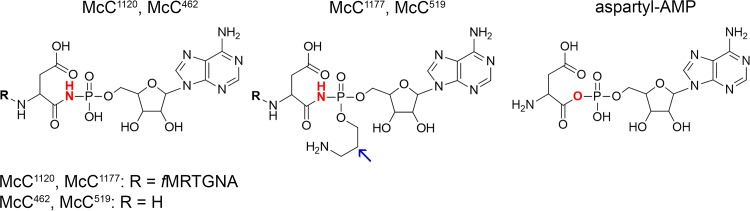
Microcin C (McC) is a ribosomally synthesized posttranslationally modified peptide (RiPP) antibiotic produced by some strains of Escherichia coli. Homologous compounds are encoded by gene clusters in numerous Gram-negative and Gram-positive bacteria (1). McC is produced by E. coli cells harboring a conjugative plasmid containing the mccABCDEF cluster (2). The mccA gene encodes a seven-amino-acid precursor peptide whose C-terminal residue is modified by the product of the mccB gene to yield a peptidyl-adenylate McC1120, in which the C-terminal aspartamide is linked to AMP through a nonhydrolyzable N-acyl phosphoramidate linkage (Fig. 1) (3). McC1120 is further modified by MccD and the N-terminal domain of MccE protein, whose joint action results in a fully matured microcin C, McC1177, harboring an aminopropyl decoration on the phosphate moiety (Fig. 1) (4). Both forms of McC are exported from the producing cell by a specialized transporter encoded by the mccC gene (5).
FIG 1.
Structures of nonaminopropylated (McC1120) and aminopropylated (McC1177) E. coli microcin C, processed forms (McC462 and McC519, respectively), and Asp-AMP, an intermediate of AspRS-catalyzed reaction. The aminopropyl group is indicated by an arrow.
McC acts through a Trojan horse mechanism. The peptide part facilitates uptake into the susceptible cell; once inside the cell, the peptide part is proteolytically degraded by aminopeptidases, releasing toxic “processed McC,” a nonhydrolyzable aspartamide-adenylate (Fig. 1), a structural mimic of intermediate of the reaction of aminoacylation of tRNAAsp catalyzed by aspartyl-tRNA synthetase (AspRS) (6, 7). Processed McC competitively inhibits AspRS, bringing protein biosynthesis to a halt (5).
Although most McC is efficiently exported outside the producing cell by the MccC pump, intracellular processing by aminopeptidases should inevitably lead to the accumulation of toxic nonhydrolyzable aspartamide-adenylate and self-intoxication of the producer, since MccC does not export processed McC. Many mcc-like clusters acquired additional genes whose products help avoid self-intoxication. In the case of E. coli, the C-terminal domain of MccE, a Gcn5-related N-acetyltransferase (GNAT)-type acetyltransferase, acetylates the α-amino group of processed McC, making it unable to bind to AspRS (8). In addition, MccF peptidase cleaves the carboxamide bond between the C-terminal aspartamide and AMP of both intact and processed McC (9).
In this work, we report a novel pathway of McC inactivation by histidine-triad (HIT) superfamily hydrolases encoded in some mcc-like biosynthetic clusters or by standalone genes located elsewhere in bacterial genomes. Proteins of the HIT superfamily form two separate functional groups: the first group includes nucleotide hydrolases, represented by HinT (10–13), Fhit (13), APTX (14), and Dcsp (15) enzymes, while the other group includes nucleotide transferases such as GalT (16). The most common members of the HIT superfamily, HinT proteins, were shown to possess phosphoramidase activity (17). We show that bacterial MccH, a product of a gene in an mcc-like cluster from Hyalangium minutum, as well as its homologs from Salmonella enterica, Nocardiopsis kunsanensis, and Pseudomonas fluorescens, are phosphoramidases that confer resistance to McC-like compounds by hydrolyzing the toxic aspartamide-adenylate that is produced after intracellular processing of peptidyl-nucleotides.
RESULTS
Bioinformatic prediction and experimental validation of an unusual mcc operon of Hyalangium minutum.
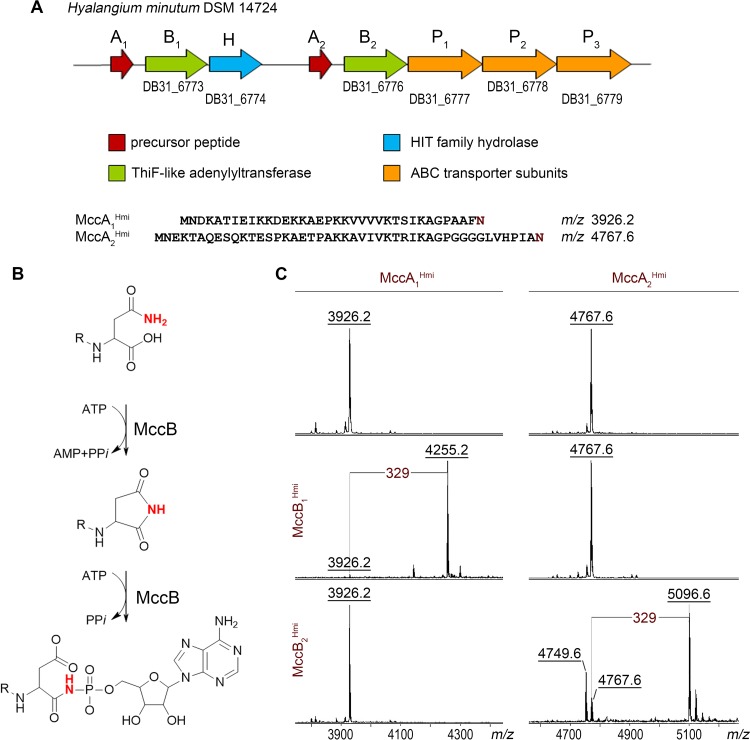
Bioinformatics analysis reveals a uniquely organized cluster in the genome of the Gram-negative bacterium Hyalangium minutum DSM 14724 that may determine the production of two putative McC-like compounds. The cluster contains two genes coding for putative precursor peptides, MccA1 and MccA2, two mccB genes, encoding THIF-like adenylyl transferases, and three genes whose products likely constitute a complex ABC-type transporter with integrated HlyD-like translocator and C39-like peptidase (18) (Fig. 2A). An additional gene, mccH, is located downstream of the mccB1 gene and encodes a protein belonging to a histidine-triad (HIT) superfamily (12).
FIG 2.
The mcc-like operon of H. minutum DSM 14724 and its products. (A) Organization of the mcc-like gene cluster from H. minutum DSM 14724. Genes are represented by colored arrows, with functional predictions corresponding to each color shown in the key beneath. (B) The mechanism of MccB-mediated adenylation of an MccA precursor peptide (3). (C) MALDI-TOF MS spectra of products of in vitro reactions between chemically synthesized H. minutum MccA1 and MccA2 precursor peptides and recombinant MccB1Hmi and MccB2Hmi enzymes in the presence of ATP. Spectra shown at the top are controls (no MccB enzyme added). The mass difference of 329 Da between MccA1 (m/z 3,926.2) and MccA2 (m/z 4,767.6) mass ions and the mass ions of the reaction products (m/z 4,255.2 and m/z 5,096.6, correspondingly) matches the modification with AMP; the MH+ ion at m/z 4,749.6 present in reaction mixtures containing MccA2Hmi and MccB2Hmi is a succinimide intermediate of the nucleotidyl transfer reaction.
To validate the predicted H. minutum mcc-like cluster, in vitro adenylation reactions of synthetic MccA1 and MccA2 peptides with recombinant MccB1 and MccB2 adenylyltransferases were performed and products analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). As can be seen from Fig. 2B and C, incubation of the 36-amino-acid-long MccA1 (MH+ at m/z 3,926.2) but not the 46-amino-acid-long MccA2 (MH+ at m/z 4,767.6) with MccB1 and an equimolar mixture of four nucleotide triphosphates led to the appearance of a mass ion at m/z 4,255.2. The 329-Da mass increase corresponds to the adenylated form of the peptide. Conversely, in the presence of MccB2, a prominent MH+ ion at m/z 5,096.6 was observed in reaction mixtures containing MccA2, corresponding to its adenylated form. An MH+ ion at m/z 4,749.6 with 18 mass units less than the original MccA2 ion was also detected. It corresponds to an MccB-catalyzed adenylation reaction intermediate, a succinimide derivative of the MccA2 peptide. MccA1 was not modified by MccB2. We conclude that the H. minutum mcc-like cluster directs the synthesis of two peptidyl-adenylates, each synthesized from separate precursors by dedicated MccB enzymes.
We next attempted to reconstruct the production of each of the H. minutum McC-like compounds in a heterologous E. coli host. Cognate mccA-mccB pairs were cloned on one expression plasmid, and the mccP1P2P3 genes encoding the putative transporter were cloned on a compatible plasmid. Under conditions of induction of plasmid-borne genes, E. coli cells or cellular extracts harboring both plasmid pairs did not inhibit the growth of an McC-sensitive E. coli tester strain, and no mass ions corresponding to H. minutum McC-like compounds were detected in cultured medium (see Fig. S1A in the supplemental material). To test if peptidyl-adenylates are synthesized but fail to export from the heterologous host, induced cells were subjected to MALDI-TOF MS. MH+ ions at m/z 4,255.2 and 5,096.6 corresponding to full-length adenylated MccA1 and MccA2, respectively, were identified in cells harboring plasmids producing MccA1-MccB1 and MccA2-MccB2 pairs. In addition, MH+ ions at m/z 4,124.2 and 4,965.6, corresponding to adenylated MccA1 and MccA2 peptides lacking the first methionine residue, were detected. Unmodified full-sized MccA1 and MccA2 polypeptides (MH+ ions at m/z 3,926.2 and 4,767.6, respectively) and their derivatives lacking methionine (MH+ ions at m/z 3,795.2 and 4,636.6) were also observed (Fig. S1B). Thus, two distinct products of the H. minutum mcc cluster are produced in the heterologous host but fail to be exported at a detectable level.
Production of McC1Hmi and McC2Hmi in a heterologous host. (A). E. coli cells harboring the plasmid-borne H. minutum mcc operon do not produce toxic compounds, as follows: mccA1B1, E. coli BL21(DE3) cells harboring pRSF_mccA1B1Hmi and pACYC_mccP1P2P3Hmi plasmids; mccA2B2, BL21(DE3) cells carrying pRSF_mccA2B2Hmi and pACYC_mccP1P2P3Hmi plasmids; and control, E. coli BL21(DE3) cells harboring empty pRSF and pACYC vectors. Cells were induced for 24 h at 30°С and then extracted as described in reference 37. Five microliters of 10× concentrated cell cultures (upper panel) or cellular extracts (lower panel) were deposited on the surface of McC-sensitive E. coli B cells lawn (upper panel). Two microliters of 0.5 μg/ml gentamicin solution was used as a control antibiotic. (B) MALDI-TOF MS analysis of E. coli BL21 cells harboring pRSF_mccA2B2Hmi and pACYC_mccP1P2P3Hmi (top) and pRSF_mccA1B1Hmi and pACYC_mccP1P2P3Hmi plasmids (bottom). At the top spectrum, MH+ at m/z 5,096.6 corresponding to adenylated MccA2Hmi, peptide-adenylate lacking N-terminal methionine (MH+ at m/z 4,965.6), full-length MccA2Hmi precursor peptide (MH+ at m/z 4,767.6), and MccA2Hmi lacking N-terminal methionine (MH+ at m/z 4,636.6) are labeled. MH+ ions at m/z 3,637.0 and 4,363.5 correspond to E. coli proteins. At the bottom spectrum, ions corresponding to adenylated MccA1Hmi (MH+ at m/z 4,255.2), peptide-adenylate lacking N-terminal methionine (MH+ at m/z 4,124.2), full-length MccA1Hmi precursor peptide (MH+ at m/z 3,926.2), and MccA1Hmi lacking N-terminal methionine (MH+ at m/z 3,795.2) are labeled. Download FIG S1, PDF file, 0.2 MB (200.4KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MccHHmi confers McC immunity when overproduced in E. coli.
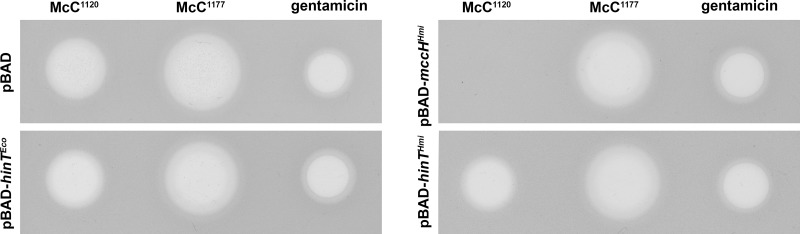
We hypothesized that MccHHmi is a HIT superfamily phosphoramidase that provides H. minutum with self-immunity through the inactivation of McC-like compounds. To check this conjecture, we cloned the mccHHmi gene in an arabinose-inducible E. coli expression vector. We also created plasmids overproducing HinTHmi, a product of a standalone H. minutum gene, and its E. coli homologue HinTEco. Since the final toxic aspartate-adenylate forms of McC produced by H. minutum and E. coli should be identical (Fig. 1, McC462), we tested the susceptibility of MccH- or HinT-expressing cells to E. coli McC1120. Additionally, we used the aminopropylated form of E. coli McC, McC1177. In the assay, the drops of solutions of two active forms of E. coli McC were deposited on lawns of E. coli B McC-sensitive cells producing HIT proteins or harboring control empty vector (Fig. 3). The results revealed that the size of the growth inhibition zones around drops of fully mature McC1177 solution on lawns of HIT protein-producing cells was the same as that on the control cell lawn. In contrast, E. coli cells overexpressing the mccHHmi gene were completely resistant to McC1120, an intermediate of the E. coli McC maturation process that does not contain the aminopropyl moiety. The expression of either hinTEco or hinTHmi had no effect on the size of growth inhibition zones produced by McC1120 (Fig. 3). All HIT proteins were produced in comparable amounts, as judged by SDS-PAGE (Fig. S2). We therefore conclude that the MccHHmi but not HinT proteins tested can provide resistance to externally added toxic peptidyl-adenylate.
FIG 3.
Overproduction of MccHHmi makes E. coli resistant to McC1120 but not to mature E. coli microcin C McC1177. Three microliters of 5 μM solutions of McC1177, McC1120, or 0.5 μg/ml gentamicin used as a control was deposited on lawns of E. coli cells harboring indicated plasmids. The results of overnight growth at 37°C under conditions of the induction of plasmid-borne genes are shown.
Coomassie-stained SDS polyacrylamide gel showing purified proteins used in the study. Lane L, PageRuler Plus prestained protein ladder; lane 1, MccHHmi; lane 2, MccHHmi H101N; lane 3, MccHHmi K103H; lane 4, MccHHmi F44H; lane 5, HinTEco; lane 6, HinTHmi. Download FIG S2, PDF file, 0.1 MB (122.8KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MccHHmi hydrolyzes the phosphoramide bond connecting the aminoacyl and nucleotide moieties of processed McC1120.
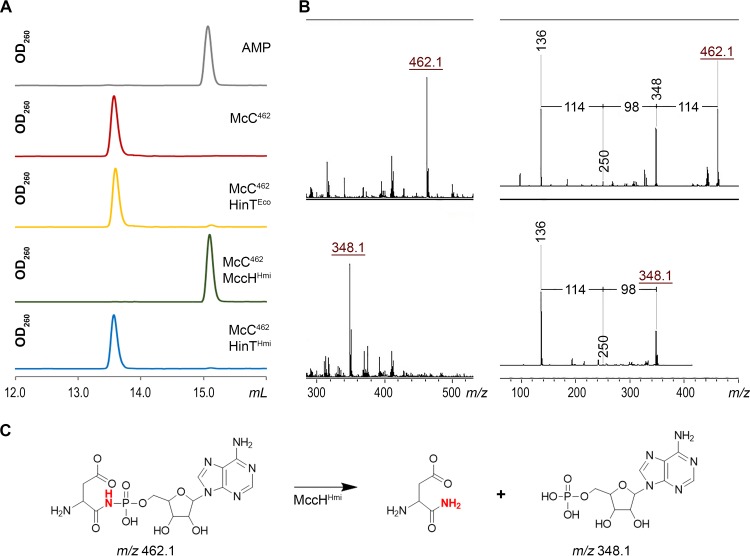
To determine the mechanism of MccHHmi-mediated resistance to toxic peptidyl-nucleotide, recombinant MccHHmi, HinTHmi, and HinTEco were purified and incubated with unprocessed McC1120 or McC1177, and the reaction products were analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) and MALDI-TOF MS. No changes were observed after a 1-h incubation (data not shown). Since no hydrolytic activity against either form of E. coli McC was detected, we considered whether the processed forms of McC1120 and McC1177 could be the substrates of MccHHmi. To this end, aspartamide-adenylates with (McC519) and without (McC462) the aminopropyl group were prepared by in vitro processing of McC1177 and McC1120 (see Materials and Methods). After incubation with MccHHmi, HinTHmi, or HinTEco, samples were analyzed by RP-HPLC and MALDI-TOF MS. Since the expected phosphoramidase activity of MccHHmi should result in the appearance of AMP or adenosine 5′-phosphoramidate, we used AMP as a marker (Fig. 4A). Upon a 1-h incubation with MccHHmi, McC462 was completely converted into a new compound with the same chromatographic mobility as AMP (Fig. 4A). The MH+ ion of this compound had m/z 348.1, matching that of AMP (Fig. 4B). The MS-MS fragmentation spectra confirmed this assignment (Fig. 4B). None of the enzymes was able to hydrolyze fully processed microcin with aminopropyl decoration (McC519) (Fig. S3).
FIG 4.
MccHHmi hydrolyzes aspartamide-adenylate McC462. (A) RP HPLC elution profiles of the products of incubation of aspartamide-adenylate McC462 with MccHHmi, HinTHmi, and HinTEco. (B) MALDI-TOF MS and MS-MS fragmentation analyses of McC462 and the product of its hydrolysis by MccHHmi. For a description of the mass ions, refer to the text. (C) Scheme of an aspartamide-adenylate hydrolysis reaction by MccHHmi.
Aminopropyl decoration of aspartamide-adenylate protects the compound from the phosphoramidase activity of MccHHmi, HinTEco, and HinTHmi. (A) MALDI-TOF MS spectra of McC519 incubated without the enzyme (top) and with HinTEco, MccHHmi, and HinTHmi (bottom). The MH+ ion at m/z 519.2 corresponds to aminopropylated aspartamide-adenylate. No MH+ ion at m/z 405.2 corresponding to hydrolyzed McC519 is observed. (B) RP-HPLC elution profile of products of incubation of McC519, processed aspartamide-adenylate with aminopropyl decoration, without the enzyme and with MccHHmi, HinTHmi, and HinTEco. Download FIG S3, PDF file, 0.1 MB (61.4KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
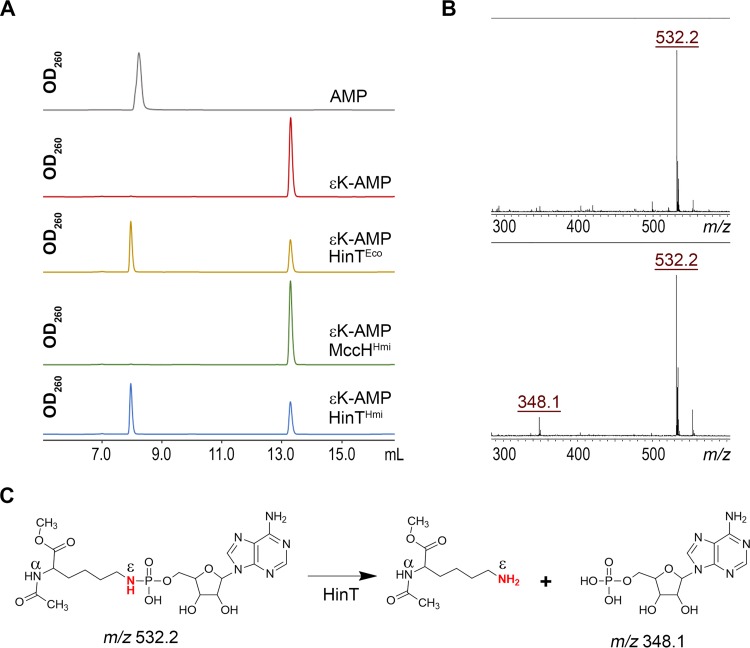
In the presence of HinTEco or HinTHmi, McC462 remained largely intact, with only trace amounts of AMP formed in the course of the reaction. We therefore decided to assess whether HinTHmi is an active phosphoramidase using AMP-N-ε-(N-α-acetyl-lysine methyl ester)-5ʹ-phosphoramidate (εK-AMP), a previously described HinT phosphoramidase model substrate (17). Incubation of εK-AMP with HinTHmi, HinTEco, or MccHHmi, followed by RP-HPLC and MALDI-TOF MS, revealed that both HinTHmi and HinTEco hydrolyzed it with the release of AMP, while MccHHmi did not (Fig. 5A to C). We therefore conclude that the MccHHmi hydrolase cleaves the P-N bond in aspartamide-adenylate but not in εK-AMP. HinTHmi and HinTEco have different specificities, where they hydrolyze εK-AMP well but are largely inactive toward processed McC1120 (Fig. 4A and 5A). These results explain why both HinT enzymes failed to provide resistance to McC in the antibiotic susceptibility test under our conditions in vivo (Fig. 3).
FIG 5.
Substrate specificity of MccHHmi, HinTHmi, and HinTEco phosphoramidases. (A) HPLC elution profiles of products of incubation of εK-AMP with HinTEco, HinTHmi, or MccHHmi. (B) MALDI-TOF MS analysis of the hydrolysis reaction of the εK-AMP by HinTHmi protein. (C) Scheme of a HinT-mediated hydrolysis reaction of εK-AMP (36).
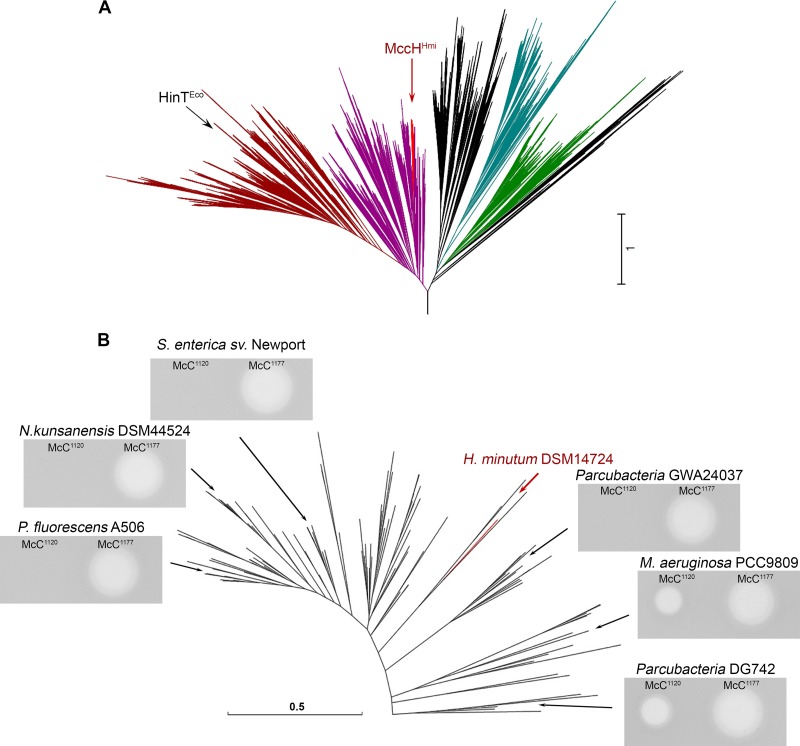
MccH homologs are present in diverse bacteria.
HIT domain-containing proteins are widespread among prokaryotes (see Materials and Methods for details on domain identification). A phylogenetic tree constructed using available HIT domain sequences revealed that MccHHmi belongs to a distinct clade, highlighted in red in Fig. 6A (see also Fig. S4). This clade also contains proteins from putative mcc-like clusters from Nocardiopsis, Pseudomonas, and Thermobifida spp., as well as multiple proteins encoded by standalone genes. Genes encoding MccH homologs from the mcc-like cluster from Nocardiopsis kunsanensis DSM 44524, as well as standalone genes from Pseudomonas fluorescens A506, Salmonella enterica serovar Newport, Microcystis aeruginosa PCC9809, and Parcubacteria sp. strains GWA24037 and DG742, were cloned into an E. coli expression vector, and the ability of the resulting plasmids to make E. coli cells resistant to the two forms of McC was tested. As can be seen from Fig. 6B, overexpression of most MccH-like genes led to resistance to McC1120 but not to McC1177. The apparently inactive MccH-like proteins from M. aeruginosa strain PCC9809 and Parcubacteria sp. strain DG742 are the earliest branching MccH homologs tested, and they may have evolved different substrate specificities.
FIG 6.
Diversity of McC-specific phosphoramidases. (A) Approximate maximum likelihood (ML) phylogenetic tree of the HIT domain proteins from completely sequenced genomes. Dark magenta and dark red indicate HinT-like proteins (dark red shows the protein kinase C interacting protein-related subgroup), bright red indicates the MccHHmi clade that is expanded in panel B, cyan indicates GalT, and green indicates FHIT. In black are clades without any clear profile signature. The arrows point to HinTEco (within the PKCI clade) and MccHHmi. (B) Approximate ML phylogenetic tree of proteins within the MccHHmi clade and growth inhibition zones formed by McC1120 and McC1177 on lawns of E. coli B cells transformed with plasmids expressing MccHHmi or homologs from the indicated positions on the tree. The MccHHmi branch is highlighted in red. sv., serovar.
Conservation of the amino acid sequence in the protein kinase C interacting protein-related clade of HIT proteins. Shown is a sequence alignment of the HinT clade of HIT proteins (HmiDSM14724a, NCBI RefSeq accession no. WP_044187632.1 of Hyalangium minutum DSM 14724; TteBAA798, GenBank accession no. ACZ41971.1, of Thermobaculum terrenum ATCC BAA-798; EcoNCTC9094, NCBI RefSeq accession no. WP_096759427.1 of Escherichia coli NCTC 9094; and SteATCC33386, GenBank accession no. ACZ09064.1 of Sebaldella termitidis ATCC 33386) and the MccH clade of HIT proteins (HmiDSM14724, NCBI RefSeq accession no. WP_044187428.1 of Hyalangium minutum DSM 14724; PflA506, GenBank accession no. AFJ55311.1 of Pseudomonas fluorescens A506; NkuDSM44524, NCBI RefSeq accession no. WP_017574753.1 of Nocardiopsis kunsanensis DSM 44524; SenNewport, ECU0367860.1 of Salmonella enterica subsp. enterica serovar Newport; ParGWA24037, GenBank accession no. KKR61370.1 of Parcubacteria sp. strain GW2011_GWA2_40_37; MaePCC9809, GenBank accession no. CCI22782.1 of Microcystis aeruginosa PCC 9809; and ParDG742, GenBank accession no. KPJ57467.1 of Parcubacteria sp. strain DG_74_2). Residues conserved in either of the two groups are shown in bold and underlined. The histidine-triad active-site region is indicated by a red-shaded box. Conserved and partially conserved hydrophobic and polar residues forming the nucleotide-binding pocket of HIT proteins are indicated by an asterisk (*). Substitutions of the active-site residues in MccH clade proteins are indicated by ‡. Red boxes mark residues of “inactive” MaePCC9809 and ParDG742 MccH-like proteins, which differ considerably from the MccH consensus. Download FIG S4, PDF file, 0.4 MB (414.3KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutational analysis of MccH active center.
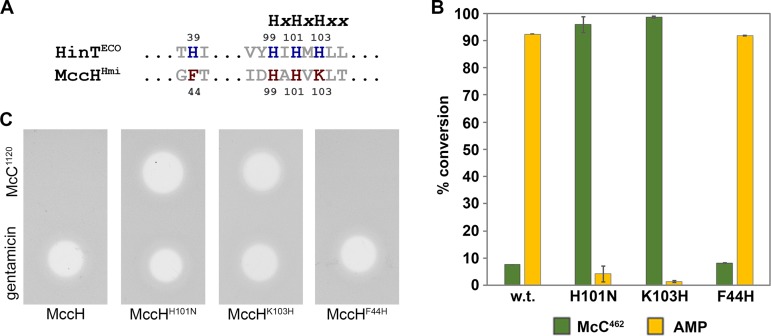
The mechanism of nucleotide phosphoramidate hydrolysis is best studied for the E. coli enzyme, HinTEco (19), and its human homologue, hHint1 (20, 21). The characteristic feature of the HIT superfamily proteins is a conserved histidine-triad motif, HxHxHxx, where H is a histidine, and x is a hydrophobic residue (12). The three essential catalytic histidines form a network of hydrogen bonds with the substrate that promotes proton transfer from the protonated C-terminal histidine of the triad (H103 in HinTEco or H102 in HinTHmi) to phosphoramidate unbridged oxygens and amide nitrogen and facilitate nucleophilic attack of the central histidine (HinTEco H101 or HinTHmi H100) on the phosphorus atom, resulting in P-N bond hydrolysis (19, 21, 22). Another conserved His residue, H39 in HinTEco (H38 in HinTHmi), located outside the triad motif closer to the N terminus of HIT enzymes, contributes to catalysis by stimulating the protonation of the third histidine of the triad by stabilizing its cationic state (21). Mutational analysis of human Hint1, a close homologue of HintEco, revealed that substitutions of conserved histidines equivalent to H39 and H103 in HinTEco substantially reduce the catalytic activity (22).
Interestingly, HIT proteins of the MccH clade contain a modified motif where the third histidine is substituted for lysine (K103 in MccHHmi). Together with this substitution, MccH-like proteins lost the N-terminal conserved histidine (H39 in HintEco), which is replaced by phenylalanine (F44 in MccHHmi) (Fig. 7A and S4). Structural modeling (see below) indicates that F44 makes hydrophobic contacts with the side chain of K103 in MccHHmi (Fig. 8A). In addition, all members of MccH clade acquired glutamate (E93 in MccHHmi) five residues away from the triad motif. In the structural model of MccHHmi, E93 makes a hydrogen bond (or a salt bridge) with K103, thus playing the same functional role as H39 in HinTEco (H38 in HinTHmi) (Fig. 8). Since this triple substitution should still allow phosphoramide bond hydrolysis, we speculate that the positively charged lysine occupying position of H103 in MccHHmi ultimately donates its stationary proton to the nitrogen of the phosphoramide bond (Fig. 8B). To test this conjecture and better understand the origin of MccH-like protein specificity, we prepared two MccHHmi mutants harboring the single substitutions K103H and F44H. An MccHHmi K103H-F44H double mutant was also engineered. As a key residue in the triad, H101 in MccHHmi is supposed to directly participate in catalysis. Therefore, a protein with substitution H101N was prepared and tested for phosphoramidase activity. As expected, the H101N mutant, which served as a control, was catalytically inactive, i.e., no hydrolysis of processed McC1120 was detected (Fig. 7B). The K103H substitution had also eliminated the hydrolytic activity of MccHHmi, confirming that a lysine characteristic of MccH-like proteins is essential for the catalytic function. The F44H mutant retained its activity, which is also an expected result. The K103H-F44H double mutant was insoluble, and thus, its enzymatic properties could not be assessed. To confirm the in vitro hydrolytic activity of the mutants, the E. coli cells harboring the corresponding plasmids were tested for their susceptibility to McC1120. As shown in Fig. 7C, the H101N and K103H substitutions completely abolished immunity to peptidyl-adenylates, while the phenotype of MccHHmi F44H-expressing cells was indistinguishable from that of the cells producing wild-type MccHHmi.
FIG 7.
Mutational analysis of active site residues of MccHHmi. (A) Sequence alignment of the conserved HIT motif of HinTEco and MccHHmi. (B) Phosphoramidase activity of MccHHmi active-site mutants. In vitro reaction mixtures containing aspartamide-adenylate McC462 were incubated with wild-type MccHHmi and the mutant proteins, containing the H101N or K103N substitution, and then analyzed by RP-HPLC. The conversion of McC462 was calculated as the percentage of McC462 and AMP absorption peak areas that remained after the reaction completion relative to the corresponding peak areas observed without the addition of enzymes. The bars represent the mean ± standard deviation (SD) conversion percentages calculated from three independent measurements. (C) Mutations in the active center of MccHHmi abolish immunity to McC1120. Growth inhibition of E. coli cells harboring pBAD plasmids encoding the indicated proteins. Solutions (5 μM) of McC1120 and 0.5 μg/ml gentamicin were deposited on freshly prepared lawns and allowed to grow overnight at 37°C under conditions of induction of plasmid-borne genes.
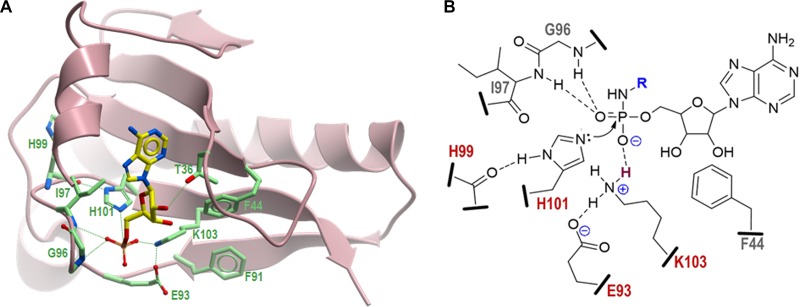
FIG 8.
Three-dimensional structural model of MccHHmi and proposed catalytic mechanism. (A) Model structure of MccHHmi generated by the SWISS-MODEL homology modeling server (23) using as the template the crystal structures of the HIT-like protein from Mycobacterium paratuberculosis (PDB 3P0T) (24) and human HINT1-AMP complex (PDB 3TW2) (28). Positions of catalytic residues in the MccHHmi active site in complex with AMP are shown as atom type-colored sticks: N, blue; O, red; P, orange. The C atoms in AMP are in yellow, and the C atoms in the side chains are in light green. (B) Schematic diagram representing the active site of MccHHmi with the bound substrate, aspartamide-adenylate (the aspartyl moiety is represented by R and shown in blue). Residues of the ExxxxxHxHxKxx motif conserved among all members of the MccH clade that are directly involved in the catalysis are shown in red. Potential hydrogen bonds between peptide side chains, backbone atoms, and phosphate oxygens are indicated by dashed lines. The proton to be transferred from the ε-amino group of K103 to the unbridged oxygen of the phosphoramide is depicted in dark red. Nucleophilic attack of the unprotonated nitrogen of H101 on electrophilic phosphorus atom is shown by a curved arrow.
Structural model of MccHHmi.
The loss of functional activity by MccHHmi mutant K103H suggested that this residue, which is specific to MccH clade proteins, is involved in substrate binding and/or catalysis. We also hypothesized that some other active-site residues might spatially constrain the catalytic pocket environment favoring the flexible aliphatic side chain of lysine over a more rigid imidazole ring of histidine. To explore the possible spatial organization of the active center of MccHHmi, we generated its three-dimensional (3D) model using the SWISS-MODEL homology modeling program (23). The resulting model of MccHHmi (Fig. 8A) is based on a top-ranked Mycobacterium paratuberculosis HIT-like protein structure (PDB 3P0T) (24) with a global model quality estimate (GMQE) quality score of 0.68, indicative of good reliability and accuracy. To reveal the potential interactions in the substrate-binding pocket, the model structure of MccHHmi was superimposed with the crystal structure of the human histidine-triad nucleotide-binding protein 1 (hHINT1) in complex with AMP (PDB 3TW2) (25).
As expected, the overall structure of MccHHmi and the spatial organization of its active site are very similar to those of other HinT and HIT-like proteins, with a notable exception of the C-terminal nonconserved 45 amino acids that model differently depending on the homology template used. In the model, MccHHmi forms a symmetric homodimer with each protein monomer capable of binding and hydrolyzing the substrate (Fig. S5). The nucleoside-binding pocket is formed mostly by conserved hydrophobic residues F11, F12, L15, F34, P37, V46, F38, and I97. The hydroxyl group of T36 makes a hydrogen bond with ribose 2′-OH in AMP, thus contributing to nucleotide recognition. The N atoms of the side chains of catalytic H101 and K103 are positioned (2.5 to 2.7 Å) to make strong hydrogen bonds with the P and unbridged O atoms of the phosphate moiety of AMP, respectively (Fig. 8A). The interatomic distances (2.9 to 3.6 Å) between the peptide backbone amide and carbonyl groups of residues G96, I97, and H99 and their interacting partners (unbridged oxygen and the protonated N atom of H101, respectively) are within a range that is optimal for hydrogen bonding and consistent with the proposed catalytic mechanism (Fig. 8B). Unexpectedly, the carbonyl group of E93, a conserved residue among members of the MccH clade, is in close proximity (2.3 Å) to the protonated N atom of K103, suggesting a strong hydrogen bond or salt bridge that would stabilize the charged state of K103 and facilitate the catalysis. Furthermore, consistent with the results of our mutagenesis experiments (Fig. 7), the bulky hydrophobic side chains of F44 and F91 replacing conserved H39 and I86 in HinTEco and HinTHmi, about the side chain of K103, stabilize its conformation and direct it toward the phosphate. Thus, the positioning of F44, F91, and E93 in the active center explains the observed preference for a catalytic lysine in MccH instead of histidine in the HinT clade.
Three-dimensional structural model of MccHHmi dimer in complex with AMP. The two monomers of MccH are depicted in light-green- and purple-colored ribbon diagrams. Residues of the active site that form the substrate-binding pocket are labeled and shown in a stick representation. Download FIG S5, PDF file, 0.1 MB (56.8KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this work, we uncover a novel mechanism of immunity to microcin C-like compounds by MccHHmi, a HIT-like phosphoramidase encoded in the mcc cluster of H. minutum. The cluster produces two separate peptide-adenylates that are analogous to McC1120, a toxic maturation intermediate of E. coli McC that lacks the aminopropyl decoration. As of today, the mcc operon from H. minutum is the only validated operon that produces two McC-like compounds with different peptide parts. Since peptide parts determine the specificity of antibacterial action by allowing selective import into sensitive cells (26), H. minutum DSM 14724 may target distinct, nonoverlapping sets of its competitors by the McC-like compounds it produces.
Like other McC-producing organisms, H. minutum should experience the buildup of toxic processed products inside the cell, which could lead to the cessation of protein biosynthesis. The MccHHmi enzyme, the product of the mcc operon, alleviates this problem by cleaving the bond between phosphorus and nitrogen in the toxic aspartamide-adenylate that is produced after proteolytic processing of either of the two McC-like compounds encoded by the operon, thus providing self-immunity to the producing cell. MccHHmi makes cells resistant to the maturation intermediate of E. coli McC that lacks the aminopropyl decoration but not to fully mature McC1177. The catalytic mechanism of phosphoramide hydrolysis requires a transient protonation of two unbridged oxygens (21, 22). The presence of an additional aminopropyl group on the phosphate in unprocessed McC1177 and processed McC519 precludes the proton transfer reaction and renders the phosphorus center inaccessible to nucleophilic attack by the catalytic histidine (HinTEco H101, HinTHmi H100, or MccHHmi H101). Thus, H. minutum and E. coli mcc operons use different strategies to overcome the self-intoxication of producers. The H. minutum mcc operon produces two peptidyl-adenylates without additional modifications, and the processing of both compounds leads to identical toxic aspartamide-adenylate. MccHHmi hydrolyzes the phosphoramide bond in aspartamide-adenylate with the formation of AMP and aspartamide. The absence of additional genes in the H. minutum mcc operon that may be involved in self-immunity suggests that MccHHmi is sufficient to counter the inhibitory effects caused by the buildup of the toxic product. In E. coli, the MccD/E enzyme complex installs the aminopropyl decoration at the phosphate of peptide-adenylate, which allows the potency of antibacterial action to be increased ∼10-fold by increasing the affinity of the processed compound to its target, AspRS (4). The presence of activity-enhancing decoration renders the MccHHmi enzyme inactive, necessitating another mechanism to overcome self-intoxication. MccE detoxifies both aminopropylated and nonaminopropylated aspartamide-adenylates by acetylating the amino group of the aspartate (8).
The structural model of MccHHmi built based on a crystal structure of homologous HIT-like protein from M. paratuberculosis (PDB 3P0T) (24) provides a plausible view on a spatial organization of the active center and offers clues to understanding the enzyme’s substrate specificity. Importantly, the model points to the functional role of the conserved hydrophobic (F44 and F91) and charged (E93) residues in the activation of catalytic K103 for the hydrolysis of aspartamide-adenylate that can be tested experimentally. It is also predicted that residues M95 and W115 of the C-terminal loop of one MccH monomer together with L110 from the adjacent C-terminal loop of the other MccH monomer form a tight aspartamide-binding site which would sterically occlude the binding of bulkier groups, such as the ε-lysine amide of εK-AMP. This view is consistent with the fact that HinTEco lacking the C-terminal extension present in MccHHmi was active toward εK-AMP but could not hydrolyze aspartamide-adenylate. It is also supported by previous observations that both deletion and swapping of the C-terminal loop between human HINT1 and HinTEco strongly affect both the catalytic activity and substrate specificity (19, 27, 28). The proposed model will be validated in our future genetic, biochemical, and structural studies of bacterial MccH and HinT proteins.
Previous studies have shown that bacterial and human HinT proteins exhibit a broad substrate specificity; they can accommodate both purine and pyrimidine nucleotides with various substitutions in the aminoacyl moiety, including d- and l-stereoisomers of tryptophan and sterically hindering N-ε-(N-α-acetyl-lysine methyl ester)-adenosine phosphoramidates (11, 27). Unlike HinT, the MccHHmi and its homologues from four diverse bacterial species characterized in this work apparently have evolved much more specialized enzymes that show a clear preference for aspartamide-adenylate (Fig. 4 and 5). Our results suggest that the fully processed McC is a bona fide substrate for MccH. We speculate that MccH-like proteins from M. aeruginosa PCC9809 and Parcubacteria sp. DG742, which are inactive toward E. coli McC, may recognize yet-unidentified species-specific McC-like compounds that carry different nucleoside or amino acid moieties.
MATERIALS AND METHODS
Molecular cloning.
E. coli DH5α was used for cloning. All primers were synthesized by Evrogen (Russia); their sequences are listed in Table S1. The H. minutum DSM 14724 or E. coli BW25113 genomic DNA was used as the template for PCR. Phusion DNA polymerase (Thermo Scientific) was used for PCR.
Primers used in the study. Download Tabl e S1, PDF file, 0.1 MB (109.9KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For MccB1Hmi and MccB2Hmi activity analysis, their coding sequences were amplified from the H. minutum DSM 14724 genomic DNA. The PCR products were digested with BamHI and SalI restriction endonucleases and inserted under the same sites into the pET22_MBP vector (26) to create an N-terminal fusion protein with a maltose-binding protein (MBP) tag.
For the heterologous mccHmi expression system, a DNA fragment spanning the mccP1Hmi, mccP2Hmi, and mccP3Hmi genes was amplified from genomic DNA, digested with EcoRI and KpnI, and inserted into pACYCDuet-1 vector (Novagen-Millipore, USA) linearized with the same restriction endonucleases. To construct MccA-MccB expression vectors, first, the phosphorylated self-complementary oligonucleotides containing sequences of the MccA1Hmi and MccA2Hmi open reading frames (ORFs) were inserted into the pRSFDuet-1 vector (Novagen-Millipore, USA) and digested with NcoI and HindIII, resulting in the pRSF-MccA1Hmi and pRSF-MccA2Hmi vectors, respectively. Then, mccB1Hmi and mccB2Hmi were PCR amplified, digested with NdeI and KpnI, and inserted into the pRSF_mccA1Hmi and pRSF_mccA2Hmi vectors linearized with NdeI and KpnI, resulting in pRSF_mccA1B1Hmi and pRSF_mccA2B2Hmi, respectively.
To obtain an arabinose-inducible vector for the heterologous expression of HIT proteins, the pBAD/His B vector (Invitrogen-Thermo Fisher, USA) was linearized by PCR with the appropriate primers that contained an introduced ribosomal binding site and SalI and HindIII restriction sites to generate pBAD30_SalRBS. Next, the mccHHmi, hinTHmi, and hinTEco genes were PCR amplified using genomic DNA as the template and the corresponding primers. Genes encoding the homologs of MccHHmi were purchased as synthetic DNA fragments from IDT, USA. All amplified PCR products and synthetic fragments were digested with SalI and HindIII and inserted between the same sites into the pBAD30 expression vector.
To create vectors for HIT protein fused with C-terminal His6 tags for protein purification, the mccHHmi, hinTHmi, and hinTEco genes were PCR amplified, digested with NdeI and XhoI, and inserted into the pET22(b) vector (Novagen-Millipore, USA). The site-directed mutagenesis of mccHHmi was carried out using overlap extension PCR (29), with appropriate primers.
Recombinant protein expression and purification.
Recombinant proteins were produced in E. coli BL21(DE3) transformed with the appropriate plasmid. The cells were grown in 500 ml of TB medium supplemented with ampicillin to an optical density at 600 nm (OD600) of ∼0.7 and induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After induction, the culture was transferred for overnight growth at 18°C and 180 rpm. The cells were harvested by centrifugation at 8,000 × g and 4°C for 20 min, resuspended in ice-cold resuspension buffer (20 mM Tris-HCl, 300 mM NaCl, 1 mM dithiothreitol [DTT] [pH 8.0]), supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), and disrupted by sonication. For the His-tagged proteins, imidazole was added up to 2 mM. The lysate was cleared by centrifugation at 30,000 × g and 4°C for 20 min. The cleared lysate was applied to a preequilibrated column with a tag-binding resin; depending on the tag present, either amylose resin (NEB) or Talon CellThru Co2+-chelating resin (TaKaRa-Clontech) was used. The resin was washed with 10 column volumes of resuspension buffer, followed by elution with 5 column volumes of the elution buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 10% glycerol) supplemented with either 10 mM maltose or 0.5 M imidazole.
Adenylation of MccA1Hmi and MccA2Hmi.
For validation of the MccA-MccB pairs of the H. minutum mcc-like gene cluster, in vitro modification of synthetic MccA1Hmi (MNDKATIEIKKDEKKAEPKKVVVVKTSIKAGPAAFN) and MccA2Hmi (MNEKTAQESQKTESPKAETPAKKAVIVKTRIKAGPGGGGLVHPIAN) (GenScript, USA) peptides using purified MccB1Hmi and MccB2Hmi reactions was performed. Reaction mixtures contained 50 μM synthetic peptides (either MccA1Hmi or MccA2Hmi), 5 μM recombinant MccB1Hmi or MccB2Hmi, and 2 mM each nucleoside triphosphate (NTP) in the reaction buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM MgCl2, 5 mM DTT). The reaction was carried out at 30°С for 16 h and then stopped by the addition of 0.1% trifluoroacetic acid (TFA) in water. The products of the reaction were analyzed by MALDI-TOF MS for the presence of adenylated MccA1Hmi and MccA2Hmi.
McC1Hmi and McC2Hmi production test.
E. coli BL21(DE3) cells harboring a combination of either pRSF_mccA1B1Hmi and pACYC_mccP1P2P3Hmi or pRSF_mccA2B2Hmi and pACYC_mccP1P2P3Hmi plasmids were grown in 25 ml of 2xYT medium (1.6% Tripton, 1% yeast extract, 0.5% NaCl) at 30°C with constant shaking at 180 rpm. Upon reaching an OD260 of ∼0.7, the cells were induced with 0.25 mM IPTG and grown for an additional 20 h at 30°C. After that, the cells were pelleted at 5,000 × g and resuspended in 250 μl of M9 medium. Aliquots of 15 μl were deposited on a freshly prepared M9 agar lawn of E. coli strain B harboring the pRSFduet-1, pETduet-1, and pACYCduet-1 vectors. Plates were incubated at 30°C overnight to allow formation of the lawn. The next day, the plates were inspected for the presence of growth inhibition zones. Additionally, the cells and the surrounding agar were analyzed for the presence of McC-like products using mass spectrometry.
In vivo immunity assay.
E. coli strain B was transformed with pBAD_SalRBS vectors containing the mccHHmi gene or its homologues. The overnight cultures of the E. coli cells with the appropriate vectors were diluted 1,000-fold in M9 agar medium supplemented with 1% glycerol, 0.02% yeast extract, 10 mM arabinose, and 100 μg/ml ampicillin. The sensitivity of cells carrying plasmid-borne genes encoding HIT-like proteins was measured by applying 5-μl drops of 5 μM McC1120 and 5 μM McC1177 on the surface of the plate and allowed to dry, and gentamicin (0.5 μg/ml) was used as a control growth inhibitor. Plates were incubated for 16 h at 30°C for a lawn to form. The next day, the growth inhibition zones were analyzed.
εK-AMP synthesis.
The synthesis of εK-AMP was performed as described elsewhere, with minor modifications (17). To synthesize εK-AMP, 0.12 g (6.25 mmol) of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide HCl (Sigma-Aldrich) was added to the flask containing 0.98 g (2.7 mmol) of AMP (Sigma-Aldrich), 0.1 g of (0.42 mmol) N-α-acetyl-l-lysine methyl ester HCl (Sigma-Aldrich), and 10 ml of ultrapure water, and the pH of the solution was adjusted with triethylamine to a value of 7.5. The mixture was incubated in the shaker at 65°C, with vigorous shaking at 250 rpm for 22 h. After allowing the reaction mix to cool down to room temperature, the mixture was lyophilized, redissolved in 0.1% TFA in water, and applied to a Luna 5-μm C18 100-Å, LC column (250 by 10 mm; Phenomenex). The purification of εK-AMP was performed in a linear gradient (5 to 25%) of acetonitrile. The fractions were analyzed for the presence of εK-AMP by MALDI-TOF mass spectrometry, and fractions containing εK-AMP were subjected to additional chromatographic purification on the same column in a linear gradient of acetonitrile (0 to 25%) in triethylammonium acetate (TEAA) buffer (pH 6.5).
In vitro phosphoramidase activity assay.
To test the phosphoramidase activity of MccHHmi, HinTHmi, HinTEco enzymes, and their respective variants, the processed forms of E. coli McC1177 and McC1120 (McC519 and McC462, respectively) and εK-AMP were used as the substrates. For production, purification, and processing of McC forms, refer to the study by Metlitskaya et al. (6). Fifty micromolar processed McC or εK-AMP was mixed with 5 μM the enzyme in the reaction buffer (20 mM HEPES [pH 7.2], 2.5 mM MgCl2, 2.5 mM MnCl2). The reaction mixture was incubated at 25°С for 30 min, terminated by the addition of 0.1% TFA, and analyzed for hydrolysis by HPLC and mass spectrometry.
Reverse-phase HPLC analysis of the products of the in vitro reactions.
All biochemical reactions were analyzed on 1220 Infinity II LC system (Agilent), and the peak separation occurred on a Zorbax Eclipse Plus C18 5-μm (4.6 by 250 mm) column (Agilent) in a 0.1 M TEAA buffer system (pH 6.0) in the varying linear gradient of acetonitrile.
The products of McC462 hydrolysis reactions were separated in a linear gradient of acetonitrile (0 to 20%) over a period of 15 min. After the incubation of McC519 with HIT enzymes, the reaction products were separated in the acetonitrile gradient (0 to 22%) for 15 min. After hydrolysis of εK-AMP by HIT enzymes, the reaction products were analyzed in the linear acetonitrile gradient (5 to 30%) lasting for 15 min. The chromatograms were processed with the use of the ChemStation software (Agilent), and elution profiles were exported in comma-separated values format.
Mass spectrometry analysis.
One to two microliters of the sample aliquots was mixed with 0.5 μl of matrix mix (Sigma-Aldrich) on a steel target. The mass spectra were recorded on an UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) equipped with a neodymium laser. The molecular MH+ ions were measured in reflector mode; the accuracy of the measured results was within 0.1 Da.
Sequence analysis.
Proteins containing the histidine catalytic triad were identified in 4,621 completely sequenced genomes available in 2016. Profiles belonging to the NCBI CDD (30) superfamily cl00228 were used as PSI-BLAST (31) queries to search the protein sequences encoded in this set. The resulting set of 10,580 proteins was clustered using UCLUST (32) and aligned using MUSCLE (33) (Fig. S6); alignments were iteratively compared to each other using HHSEARCH and aligned using the HHALIGN program (34). The approximate ML tree was reconstructed using the FastTree program (35) with a WAG evolutionary model and gamma-distributed site rates.
Alignment of cluster consensus sequences of the HIT domain proteins from completely sequenced genomes. The phylogenetic tree, constructed from the multiple alignment of 15,351 HIT sequences (Fig. 6A), was split into subtrees at the average depth of 1.5 from the tree tips, producing 292 clusters of 4+ sequences. Consensus sequences were derived for each cluster from the corresponding subsets of the alignment (positions less than 30% conserved denoted by “x”). Positions corresponding to active-site histidine residues are highlighted in yellow. Consensus sequences for the MccHHmi (CON.109), HinTEco (CON.19), and HinTHmi (CON.21) clades are shown in red font. Download FIG S6, PDF file, 0.3 MB (280.5KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by NIH RO1 grant AI117270 (to K.S. and Satish A. Nair) and Skoltech institutional funds to K.S., Russian Science Foundation grants RSF 16-14-10356 and RSF 19-14-00266 to S.D., and Rowan University departmental funds to S.B. Y.I.W. is supported through the intramural program of the U.S. National Institutes of Health. Bioinformatic analysis was partially supported by the Ministry of Science and Higher Education of the Russian Federation grant 075-15-2019-1661.
The MALDI-TOF MS facility was made available to us as part of the framework of the Moscow State University Development Program PNG 5.13.
Footnotes
This article is a direct contribution from Konstantin V. Severinov, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Michael Ibba, The Ohio State University; Sylvie Rebuffat, CNRS, Muséum National d'Histoire Naturelle; and Konstantinos Beis, Imperial College London.
Citation Yagmurov E, Tsibulskaya D, Livenskyi A, Serebryakova M, Wolf YI, Borukhov S, Severinov K, Dubiley S. 2020. Histidine-triad hydrolases provide resistance to peptide-nucleotide antibiotics. mBio 11:e00497-20. https://doi.org/10.1128/mBio.00497-20.
REFERENCES
- 1.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 2.Guijarro JI, González-Pastor JE, Baleux F, San Millán JL, Castilla MA, Rico M, Moreno F, Delepierre M. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem 270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 3.Roush RF, Nolan EM, Löhr F, Walsh CT. 2008. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc 130:3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 4.Kulikovsky A, Serebryakova M, Bantysh O, Metlitskaya A, Borukhov S, Severinov K, Dubiley S. 2014. The molecular mechanism of aminopropylation of peptide-nucleotide antibiotic microcin C. J Am Chem Soc 136:11168–11175. doi: 10.1021/ja505982c. [DOI] [PubMed] [Google Scholar]
- 5.Severinov K, Nair SK. 2012. Microcin C: biosynthesis and mechanisms of bacterial resistance. Future Microbiol 7:281–289. doi: 10.2217/fmb.11.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J Biol Chem 281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 7.Kazakov T, Vondenhoff GH, Datsenko KA, Novikova M, Metlitskaya A, Wanner BL, Severinov K. 2008. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J Bacteriol 190:2607–2610. doi: 10.1128/JB.01956-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novikova M, Kazakov T, Vondenhoff GH, Semenova E, Rozenski J, Metlytskaya A, Zukher I, Tikhonov A, Van Aerschot A, Severinov K. 2010. MccE provides resistance to protein synthesis inhibitor microcin C by acetylating the processed form of the antibiotic. J Biol Chem 285:12662–12669. doi: 10.1074/jbc.M109.080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tikhonov A, Kazakov T, Semenova E, Serebryakova M, Vondenhoff G, Van Aerschot A, Reader JS, Govorun VM, Severinov K. 2010. The mechanism of microcin C resistance provided by the MccF peptidase. J Biol Chem 285:37944–37952. doi: 10.1074/jbc.M110.179135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krakowiak A, Pace HC, Blackburn GM, Adams M, Mekhalfia A, Kaczmarek R, Baraniak J, Stec WJ, Brenner C. 2004. Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors. J Biol Chem 279:18711–18716. doi: 10.1074/jbc.M314271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Fang P, Schimmel P, Guo M. 2012. Side chain independent recognition of aminoacyl adenylates by the Hint1 transcription suppressor. J Phys Chem B 116:6798–6805. doi: 10.1021/jp212457w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner C. 2014. Histidine triad (HIT) superfamily. eLS John Wiley & Sons Ltd., Chichester, United Kingdom. doi: 10.1002/9780470015902.a0020545.pub2. [DOI] [Google Scholar]
- 13.Brenner C. 2002. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kijas AW, Harris JL, Harris JM, Lavin MF. 2006. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J Biol Chem 281:13939–13948. doi: 10.1074/jbc.M507946200. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Rodgers ND, Jiao X, Kiledjian M. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden HM, Rayment I, Thoden JB. 2003. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 17.Chou T-F, Bieganowski P, Shilinski K, Cheng JJ, Brenner C, Wagner CR. 2005. 31P NMR and genetic analysis establish hinT as the only Escherichia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J Biol Chem 280:15356–15361. doi: 10.1074/jbc.M500434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland IB, Peherstorfer S, Kanonenberg K, Lenders M, Reimann S, Schmitt L 13 December 2016, posting date. Type I protein secretion-deceptively simple yet with a wide range of mechanistic variability across the family. EcoSal Plus 2016 doi: 10.1128/ecosalplus.ESP-0019-2015. [DOI] [PubMed] [Google Scholar]
- 19.Bardaweel S, Pace J, Chou T-F, Cody V, Wagner CR. 2010. Probing the impact of the echinT C-terminal domain on structure and catalysis. J Mol Biol 404:627–638. doi: 10.1016/j.jmb.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 20.Shah R, Maize KM, Zhou X, Finzel BC, Wagner CR. 2017. Caught before released: structural mapping of the reaction trajectory for the sofosbuvir activating enzyme, human histidine triad nucleotide binding protein 1 (hHint1). Biochemistry 56:3559–3570. doi: 10.1021/acs.biochem.7b00148. [DOI] [PubMed] [Google Scholar]
- 21.Liang G, Webster CE. 2017. Phosphoramidate hydrolysis catalyzed by human histidine triad nucleotide binding protein 1 (hHint1): a cluster-model DFT computational study. Org Biomol Chem 15:8661–8668. doi: 10.1039/c7ob02098h. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Chou TF, Aubol BE, Park CJ, Wolfenden R, Adams J, Wagner CR. 2013. Kinetic mechanism of human histidine triad nucleotide binding protein 1. Biochemistry 52:3588–3600. doi: 10.1021/bi301616c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baugh L, Phan I, Begley DW, Clifton MC, Armour B, Dranow DM, Taylor BM, Muruthi MM, Abendroth J, Fairman JW, Fox D, Dieterich SH, Staker BL, Gardberg AS, Choi R, Hewitt SN, Napuli AJ, Myers J, Barrett LK, Zhang Y, Ferrell M, Mundt E, Thompkins K, Tran N, Lyons-Abbott S, Abramov A, Sekar A, Serbzhinskiy D, Lorimer D, Buchko GW, Stacy R, Stewart LJ, Edwards TE, Van Voorhis WC, Myler PJ. 2015. Increasing the structural coverage of tuberculosis drug targets. Tuberculosis (Edinb) 95:142–148. doi: 10.1016/j.tube.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolot R, Ozga M, Włodarczyk A, Krakowiak A, Nawrot B. 2012. A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5′-monophosphate at 1.38 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 68:883–888. doi: 10.1107/S1744309112029491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bantysh O, Serebryakova M, Zukher I, Kulikovsky A, Tsibulskaya D, Dubiley S, Severinov K. 2015. Enzymatic synthesis and functional characterization of bioactive microcin C-like compounds with altered peptide sequence and length. J Bacteriol 197:3133–3141. doi: 10.1128/JB.00271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T-F, Baraniak J, Kaczmarek R, Zhou X, Cheng J, Ghosh B, Wagner CR. 2007. Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and Escherichia coli histidine triad nucleotide binding proteins. Mol Pharm 4:208–217. doi: 10.1021/mp060070y. [DOI] [PubMed] [Google Scholar]
- 28.Chou T-F, Sham YY, Wagner CR. 2007. Impact of the C-terminal loop of histidine triad nucleotide binding protein1 (Hint1) on substrate specificity. Biochemistry 46:13074–13079. doi: 10.1021/bi701244h. [DOI] [PubMed] [Google Scholar]
- 29.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer AA. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113–119. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou T-F, Wagner CR. 2007. Lysyl-tRNA synthetase-generated lysyl-adenylate is a substrate for histidine triad nucleotide binding proteins. J Biol Chem 282:4719–4727. doi: 10.1074/jbc.M610530200. [DOI] [PubMed] [Google Scholar]
- 37.Zukher I, Pavlov M, Tsibulskaya D, Kulikovsky A, Zyubko T, Bikmetov D, Serebryakova M, Nair SK, Ehrenberg M, Dubiley S, Severinov K. 2019. Reiterative synthesis by the ribosome and recognition of the N-terminal formyl group by biosynthetic machinery contribute to evolutionary conservation of the length of antibiotic microcin C peptide precursor. mBio 10:e00768-19. doi: 10.1128/mBio.00768-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Production of McC1Hmi and McC2Hmi in a heterologous host. (A). E. coli cells harboring the plasmid-borne H. minutum mcc operon do not produce toxic compounds, as follows: mccA1B1, E. coli BL21(DE3) cells harboring pRSF_mccA1B1Hmi and pACYC_mccP1P2P3Hmi plasmids; mccA2B2, BL21(DE3) cells carrying pRSF_mccA2B2Hmi and pACYC_mccP1P2P3Hmi plasmids; and control, E. coli BL21(DE3) cells harboring empty pRSF and pACYC vectors. Cells were induced for 24 h at 30°С and then extracted as described in reference 37. Five microliters of 10× concentrated cell cultures (upper panel) or cellular extracts (lower panel) were deposited on the surface of McC-sensitive E. coli B cells lawn (upper panel). Two microliters of 0.5 μg/ml gentamicin solution was used as a control antibiotic. (B) MALDI-TOF MS analysis of E. coli BL21 cells harboring pRSF_mccA2B2Hmi and pACYC_mccP1P2P3Hmi (top) and pRSF_mccA1B1Hmi and pACYC_mccP1P2P3Hmi plasmids (bottom). At the top spectrum, MH+ at m/z 5,096.6 corresponding to adenylated MccA2Hmi, peptide-adenylate lacking N-terminal methionine (MH+ at m/z 4,965.6), full-length MccA2Hmi precursor peptide (MH+ at m/z 4,767.6), and MccA2Hmi lacking N-terminal methionine (MH+ at m/z 4,636.6) are labeled. MH+ ions at m/z 3,637.0 and 4,363.5 correspond to E. coli proteins. At the bottom spectrum, ions corresponding to adenylated MccA1Hmi (MH+ at m/z 4,255.2), peptide-adenylate lacking N-terminal methionine (MH+ at m/z 4,124.2), full-length MccA1Hmi precursor peptide (MH+ at m/z 3,926.2), and MccA1Hmi lacking N-terminal methionine (MH+ at m/z 3,795.2) are labeled. Download FIG S1, PDF file, 0.2 MB (200.4KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coomassie-stained SDS polyacrylamide gel showing purified proteins used in the study. Lane L, PageRuler Plus prestained protein ladder; lane 1, MccHHmi; lane 2, MccHHmi H101N; lane 3, MccHHmi K103H; lane 4, MccHHmi F44H; lane 5, HinTEco; lane 6, HinTHmi. Download FIG S2, PDF file, 0.1 MB (122.8KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Aminopropyl decoration of aspartamide-adenylate protects the compound from the phosphoramidase activity of MccHHmi, HinTEco, and HinTHmi. (A) MALDI-TOF MS spectra of McC519 incubated without the enzyme (top) and with HinTEco, MccHHmi, and HinTHmi (bottom). The MH+ ion at m/z 519.2 corresponds to aminopropylated aspartamide-adenylate. No MH+ ion at m/z 405.2 corresponding to hydrolyzed McC519 is observed. (B) RP-HPLC elution profile of products of incubation of McC519, processed aspartamide-adenylate with aminopropyl decoration, without the enzyme and with MccHHmi, HinTHmi, and HinTEco. Download FIG S3, PDF file, 0.1 MB (61.4KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conservation of the amino acid sequence in the protein kinase C interacting protein-related clade of HIT proteins. Shown is a sequence alignment of the HinT clade of HIT proteins (HmiDSM14724a, NCBI RefSeq accession no. WP_044187632.1 of Hyalangium minutum DSM 14724; TteBAA798, GenBank accession no. ACZ41971.1, of Thermobaculum terrenum ATCC BAA-798; EcoNCTC9094, NCBI RefSeq accession no. WP_096759427.1 of Escherichia coli NCTC 9094; and SteATCC33386, GenBank accession no. ACZ09064.1 of Sebaldella termitidis ATCC 33386) and the MccH clade of HIT proteins (HmiDSM14724, NCBI RefSeq accession no. WP_044187428.1 of Hyalangium minutum DSM 14724; PflA506, GenBank accession no. AFJ55311.1 of Pseudomonas fluorescens A506; NkuDSM44524, NCBI RefSeq accession no. WP_017574753.1 of Nocardiopsis kunsanensis DSM 44524; SenNewport, ECU0367860.1 of Salmonella enterica subsp. enterica serovar Newport; ParGWA24037, GenBank accession no. KKR61370.1 of Parcubacteria sp. strain GW2011_GWA2_40_37; MaePCC9809, GenBank accession no. CCI22782.1 of Microcystis aeruginosa PCC 9809; and ParDG742, GenBank accession no. KPJ57467.1 of Parcubacteria sp. strain DG_74_2). Residues conserved in either of the two groups are shown in bold and underlined. The histidine-triad active-site region is indicated by a red-shaded box. Conserved and partially conserved hydrophobic and polar residues forming the nucleotide-binding pocket of HIT proteins are indicated by an asterisk (*). Substitutions of the active-site residues in MccH clade proteins are indicated by ‡. Red boxes mark residues of “inactive” MaePCC9809 and ParDG742 MccH-like proteins, which differ considerably from the MccH consensus. Download FIG S4, PDF file, 0.4 MB (414.3KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Three-dimensional structural model of MccHHmi dimer in complex with AMP. The two monomers of MccH are depicted in light-green- and purple-colored ribbon diagrams. Residues of the active site that form the substrate-binding pocket are labeled and shown in a stick representation. Download FIG S5, PDF file, 0.1 MB (56.8KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in the study. Download Tabl e S1, PDF file, 0.1 MB (109.9KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of cluster consensus sequences of the HIT domain proteins from completely sequenced genomes. The phylogenetic tree, constructed from the multiple alignment of 15,351 HIT sequences (Fig. 6A), was split into subtrees at the average depth of 1.5 from the tree tips, producing 292 clusters of 4+ sequences. Consensus sequences were derived for each cluster from the corresponding subsets of the alignment (positions less than 30% conserved denoted by “x”). Positions corresponding to active-site histidine residues are highlighted in yellow. Consensus sequences for the MccHHmi (CON.109), HinTEco (CON.19), and HinTHmi (CON.21) clades are shown in red font. Download FIG S6, PDF file, 0.3 MB (280.5KB, pdf) .
Copyright © 2020 Yagmurov et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.