SUMMARY
Cellular identity is not driven by differences in genomic content but rather by epigenomic, transcriptomic and proteomic heterogeneity. Although regulation of the epigenome plays a key role in shaping stem cell hierarchies, differential expression of transcripts only partially explains protein abundance. The epitranscriptome, translational control and protein degradation have emerged as fundamental regulators of proteome complexity that regulate stem cell identity and function. Here, we discuss how post-transcriptional mechanisms enable stem cell homeostasis and responsiveness to developmental cues and environmental stressors by rapidly shaping the content of their proteome and how these processes are disrupted in pre-malignant and malignant states.
Introduction
Because of the expanded use of single cell nucleic acid sequencing technology, cell identity is being increasingly defined by transcriptional profiles. Transcriptional networks play a central role in governing stem cell function and fate. This is best exemplified by pluripotent stem cells, in which four transcription factors, MYC, OCT4, SOX2 and NANOG, are essential for driving the genetic programs that support pluripotency and self-renewal (Boyer et al., 2005) and are sufficient for reprogramming somatic cells into induced pluripotent stem cells (Park et al., 2008; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). A central question that consequently arose from these studies was whether cell-autonomous mechanisms shape cellular identity or vice versa. While initial work focused on epigenetic mechanisms, we now appreciate that transcriptional events do not entirely determine cellular identity. Recent studies have revealed that diverse post-transcriptional mechanisms influence the functional output of genetic programs (i.e. proteome content) required by stem cells (Nilsen and Graveley, 2010; van den Berg et al., 2017; Williamson et al., 2008).
In this Review, we will discuss how transcript sequence, stability and translational efficiency are regulated, at least in part, by a variety of biochemical modifications (defined in Table 1) to influence stem cell identity and function. We will then examine how protein synthesis and degradation influence proteome content and quality (Figure 1). In addition, we will discuss how defects in these post-transcriptional mechanisms deregulate tissue-specific stem cells and progenitors in human disease and stress conditions (Figure 2) and examine their potential as both diagnostic and therapeutic targets. Finally, we provide a summary of key publications investigating post-translational mechanisms and their effects on pluripotent, somatic, and malignant stem cells (Table S1).
Table 1.
Glossary of key terms
| Epitranscriptome | Collective term for biochemical changes that can modify RNA |
| Alternative splicing | Removal introns and joining of different exons to produce distinct mRNA isoforms |
| m6A | Methylation of adenosine at the nitrogen-6 position |
| m1A | Methylation of adenosine at the nitrogen-1 position |
| m5C | Methylation of adenosine at the carbon-5 position |
| Writer | Methyltransferase complex that deposits methyl groups onto RNA |
| Reader | Protein that recognize and binds methylated RNA |
| Eraser | Demethylase that removes methyl groups from RNA |
| Pseudouridylation | Conversion of uridine to pseudouridine (an isomer of uridine) on RNA |
| A-to-I editing | Deamination of adenosine on RNA into inosine |
| Messenger RNA (mRNA) | RNA molecule that specifies protein products |
| MicroRNA (miRNA) | Noncoding RNA involved in RNA silencing |
| Ribosomal RNA (rRNA) | Noncoding RNA that when processed is an essential component of the ribosome |
| tRNA-derived-stress-induced RNA (tiRNA) | tRNA “halves” produced by cleavage of tRNAs |
| Splicesomal small nuclear RNA (snRNA) | Non-coding RNA that participates in intron splicing |
| Small nucleolar RNA (snoRNA) | RNA that primarily modifies and processes rRNA |
| Protein homeostasis (proteostasis) | A network of physiological mechanisms and stress response pathways the maintain proteome content and quality |
| Eukaryotic initiation factors (eIF) | Proteins that are involved in the initiation of eukaryotic mRNA translation. E.g. eIF2A, eIF4E |
| eIF2α | Alpha subunit of translation initiation factor complex eIF2 that when phosphorylated typically suppresses global protein synthesis |
| Cap dependent translation | Translation of mRNA that requires binding of all canonical initiation factors to the 5’ mRNA cap structure |
| Cap independent translation | Translation of mRNA that does not require interaction of mRNA with initiation factors eIF4E and/or eIF4G but involves recruitment of the small ribosomal subunit to internal ribosome entry sites (IRES) |
| Poly(A)-binding protein (PABP) | RNA-binding protein that binds to the poly(A) tail of eukaryotic mRNA |
| eIF4 binding protein (4E-BP) | Binds and sequesters the cap-binding translation initiation factor eIF4E to suppress protein synthesis |
| Ribosomopathies | Diseases that are associated with mutations in ribosomal genes |
| ER unfolded protein response (UPRER) | Stress response pathway activated by the accumulation of unfolded/misfolded proteins in the endoplasmic reticulum |
| Mitochondrial unfolded protein response (UPRMT) | Stress response pathway activated by the accumulation of unfolded/misfolded proteins in the mitochondria |
| Heat shock response | Stress response pathway activated by the accumulation of unfolded/misfolded proteins in the cytoplasm |
| Ubiquitin-proteasome system | Cellular protein degradation system in which proteins destined for degradation are ubiquitinated and degraded by the proteasome |
| Cancer stem cells | A subset of cancer cells that can self-renew and differentiate into cells that comprise the tumor, thus contributing to tumorigenesis |
Figure 1. Post-transcriptional mechanisms influence proteome content.
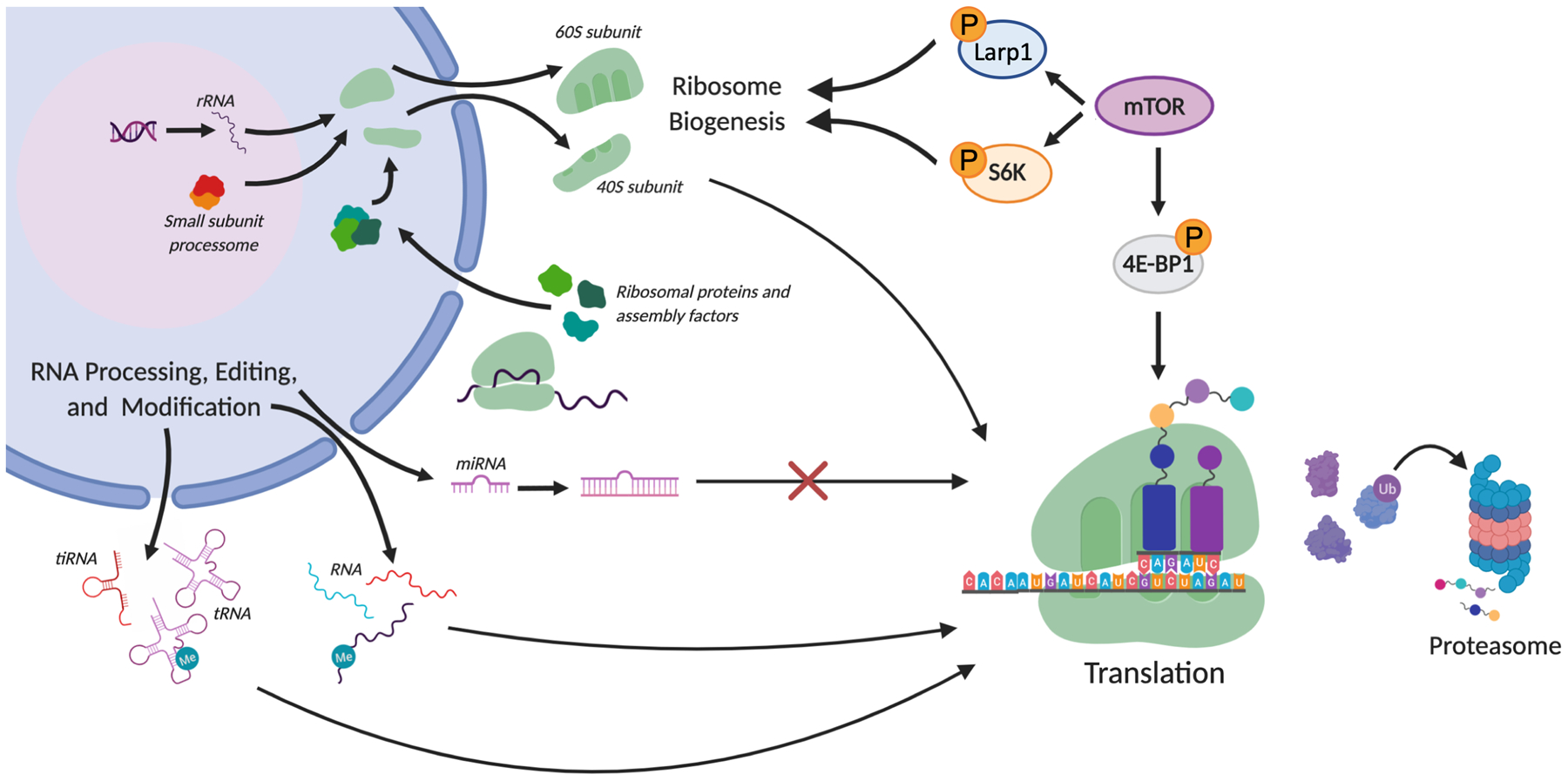
Proteome content can be regulated by RNA processing, ribosome biogenesis, signaling pathways and protein degradation. RNA splicing can produce mRNAs that code for distinct protein isoforms, introduce premature stop codons, or cause UTR variation that alters translational efficiency. mRNA methylation can alter transcript stability, localization and translational efficiency. Methylation and pseudouridylation of rRNA and tRNA can impact ribosome biogenesis, polysome assembly, translation fidelity and tRNA stability. RNA editing can alter miRNA biogenesis and alter mRNA coding or UTR sequence to alter translational efficiency. The mTOR signaling pathway can promote protein synthesis by enhancing the translation of ribosomal protein mRNAs via phosphorylation of Larp1, ribosome biogenesis via phosphorylation and activation of S6K, and translation initiation via phosphorylation and inhibition of 4E-BPs. The ubiquitin-proteasome system is a major cellular degradation system that contributes to maintaining proteostasis in stem cells by regulating both the content and quality of the proteome through the normal turnover of proteins and the degradation of misfolded proteins.
Figure 2. Post-transcriptional regulation of stem cell identity under conditions of stress.
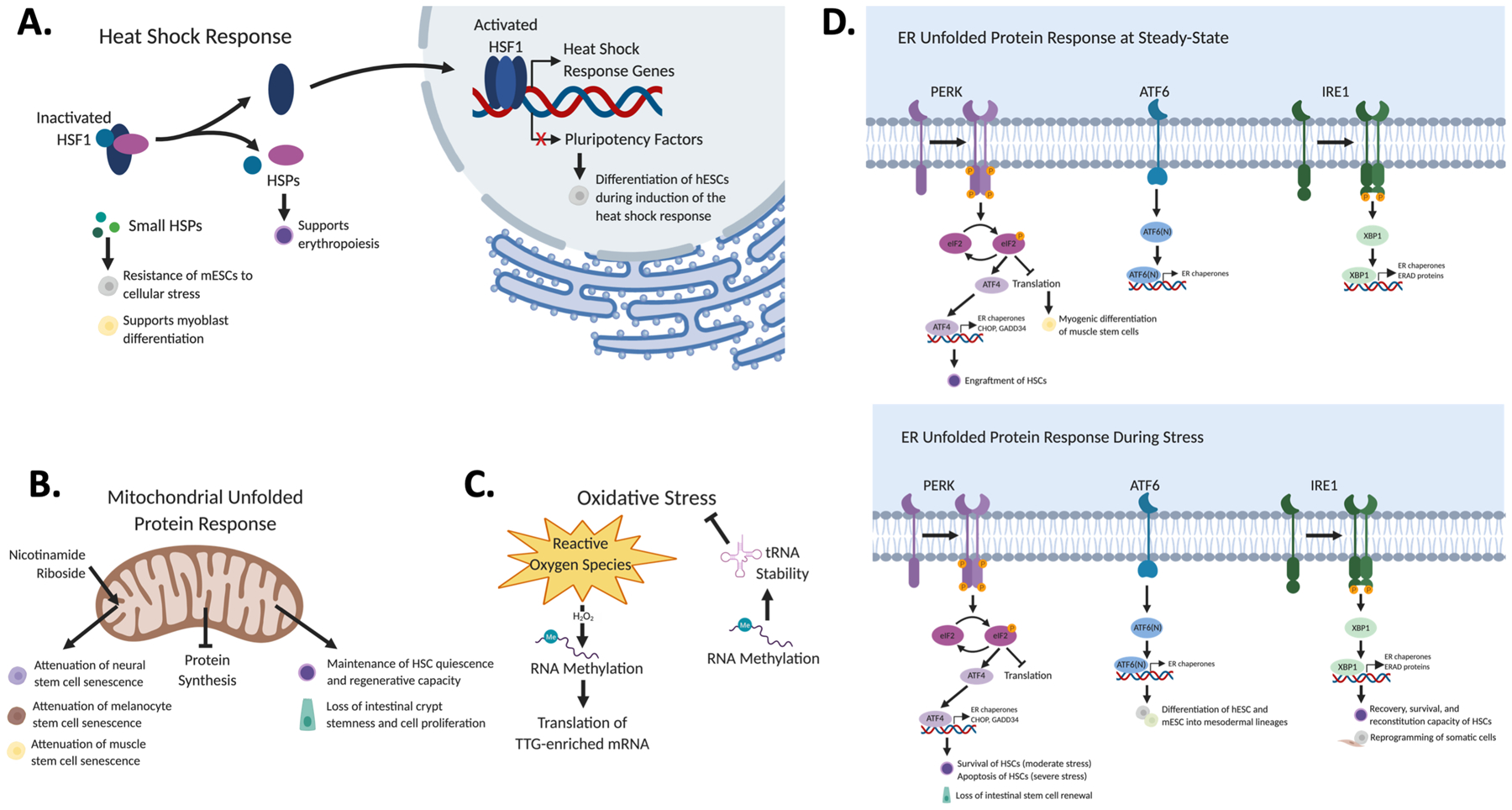
In response to proteotoxic insults, the cell mounts adaptive responses to maintain protein quality control. These stress response pathways also regulate stem cells. (A) The heat shock response induces ESC differentiation, protects ESCs from cellular stress, promotes myoblast differentiation, and supports erythropoiesis. (B) Activation of the UPRMT by nicotinamide riboside delays neural stem cell and melanocyte stem cell senescence, while dysregulation of the UPRMT also impairs hematopoietic and intestinal stem cell stemness and proliferation. (C) Post-transcriptional mechanisms of gene regulation including RNA methylation and protein synthesis can regulate the cellular response to oxidative stress. However, the precise nature of this relationship and its influence on stem cells is largely unknown. (D) The effects of UPRER activation on stem cells are tissue- and context-specific. Activation of the PERK branch regulates muscle satellite cell differentiation during homeostasis, but results in a loss of intestinal stem cell self-renewal during stress. Induction of ATF6 and IRE1 through pharmacological means supports mesodermal specification of ESCs and enhances the reprogramming efficiency of somatic stem cells. Activation of the PERK branch during homeostasis promotes the engraftment of HSCs, and during conditions of moderate stress promotes their survival. The IRE1 pathway provides a protective effect on HSCs experiencing stress. However, extreme stress induces HSC apoptosis.
RNA Processing
Post-transcriptional regulation begins with extensive processing and modification of RNA. Precursor messenger RNAs (pre-mRNAs) transcribed from coding genes may be capped, spliced, cleaved and polyadenylated to make them competent for translation into functional proteins. In addition to these processing events, coding and non-coding RNAs can be biochemically modified via methylation, pseudouridylation or editing. These epitranscriptomic modifications can alter RNA coding sequences, localization, stability and translational efficiency (Figure 1). Therefore, RNA processing and epitranscriptomic alterations play a key role in regulating proteome content and diversity (Kim et al., 2008; Kwon et al., 2013).
RNA Splicing
To date, alternative pre-mRNA splicing is the most extensively studied mRNA modification that influences protein composition (Chen and Manley, 2009). Splicing is the process of intron removal and exon joining that is necessary for converting intron containing pre-mRNAs into mRNAs that are competent for translation into functional proteins. In many cases, splicing takes place using alternative splice sites that can result in the production of distinct mRNAs that code for different protein isoforms that exhibit unique structural and functional properties. Many human splicing events are not conserved in mice thereby suggesting that splicing is essential for fine-tuning human gene regulation (Thanaraj et al., 2003; Yeo et al., 2005).
Splice isoform diversity is highest in human embryonic stem cells (hESCs) and decreases upon differentiation. This phenomenon, referred to as isoform specialization, is mediated in part by a number of splicing factors and other RNA binding proteins that are differentially expressed during development (Chen et al., 2015). Splice isoform expression patterns also distinguish human stem and progenitor cell fate, aging and malignant transforming potential (Cesana et al., 2018; Crews et al., 2016).
Alternative splicing can influence gene expression to either promote or impair stem cell function (Aaronson and Meshorer, 2013; Chen et al., 2015). For example, depletion of SRSF2, a member of the serine/arginine-rich pre-mRNA splicing factor family, was shown to decrease the expression of pluripotency factors OCT4 and NANOG, and disrupt self-renewal of hESCs (Lu et al., 2014). Interestingly, OCT4 itself can bind to the SRSF2 promoter, and depletion of OCT4 reduces SRSF2 expression, suggesting that there is reciprocal regulation of splicing and pluripotency factors (Lu et al., 2014). In contrast to SRSF2, muscleblind like splicing factors (MBNLs) negatively regulate stem cell self-renewal. MBNL proteins are more highly expressed by differentiated cells than hESCs and have been shown to repress stem cell specific splicing patterns (Han et al., 2015; Holm et al., 2015). One key splice variant whose production is repressed by MBNL proteins encodes a specific isoform of FOXP1, a transcription factor that is expressed by hESCs but absent during differentiation. This stem cell specific FOXP1 variant arises from inclusion of a stem cell specific exon that alters its DNA binding specificity to promote the expression of pluripotency factors, including OCT4, SOX2 and NANOG, and suppress the expression of differentiation factors (Gabut et al., 2011). Consistent with their role in suppressing pluripotency, knockdown of MBNL proteins enhances the reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Han et al., 2013). In addition, reprogramming has been associated with re-acquisition of a pluripotent alternative splicing profile (Ohta et al., 2013). Thus, alternative splicing has important roles in pluripotency as well as cellular reprogramming.
Alternative splicing can also directly affect the function of genes important for pluripotency, including OCT4 (Atlasi et al., 2008), TCF3 (Yamazaki et al., 2018), MBD2 (Lu et al., 2014), DNMT3B (Yeo et al., 2007) and SALL4 (Rao et al., 2010). The expression of OCT4A, a splice variant of OCT4, is specifically expressed in pluripotent stem cells and is necessary for their self-renewal (Atlasi et al., 2008). In contrast, OCT4B, another alternatively spliced isoform of OCT4, is expressed by both stem cells and somatic cells, but has no apparent role in regulating stem cell function (Atlasi et al., 2008). A third splice isoform, OCT4B1, is also specifically expressed in hESCs, and its expression is increased in response to various cellular stresses (Farashahi Yazd et al., 2011). Indeed, mRNA splicing efficiency is thought to be altered by stress, and may provide stem cells with a mechanism to rapidly alter the content of their proteome in response to environmental cues without requiring epigenetic and transcriptional changes (Kalsotra and Cooper, 2011). Overall, these studies indicate that alternative splicing is important for regulating pluripotency and cell fate specification.
In addition to generating functional variants, splicing can alter transcript stability and translational efficiency to influence stem cell development and function. Alternative splicing can introduce premature stop codons that can trigger nonsense mediated mRNA decay and can cause untranslated region (UTR) variation, which can affect translation efficiency, mRNA stability and subcellular localization (Kalsotra and Cooper, 2011; Licatalosi et al., 2012; Lou et al., 2014). One recent study identified temporal splicing changes in the 3’ UTR of HMGA2 during human hematopoietic stem cell (HSC) ontogeny (Cesana et al., 2018). Human fetal liver and cord blood HSCs were shown to express distinct isoforms of HMGA2. Fetal HSCs express a longer isoform of HMGA2 that contains a distinct terminal exon and a 3-fold longer 3’-UTR as compared to the short isoform which is highly expressed by neonatal HSCs. Although the function of both isoforms is similar, the shorter 3’UTR present in the short isoform of HMGA2 enables it to more effectively escape repression mediated by various microRNAs (miRNAs). This short isoform enables neonatal HSCs to sustain HMGA2 expression and self-renewal potential despite the increased presence of let-7 family miRNAs that can suppress HMGA2 (Cesana et al., 2018).
Given the importance of RNA splicing in regulating both transcriptome and proteome diversity in normal stem cells, recent attention has focused on cancer stem cells and their capacity to hijack splicing to support malignant growth. Cancer stem cells exhibit splicing patterns reminiscent of undifferentiated stem cells, and these splicing patterns are at least partly mediated by MBNL1 (Crews et al., 2016; Sebestyen et al., 2016). Furthermore, splicing factors, such as SRSF2, U2AF1 and SF3B1, have been found to be mutated or epigenetically modified in pre-leukemic and leukemic disorders (Ogawa, 2014; Yoshida and Ogawa, 2014). This dysregulation is coupled with observations that cancer stem cells express both stem cell regulatory and pro-survival splice variants of a number of genes. Chronic myeloid leukemia blast crisis stem cells have been shown to express high levels CD44v3, an isoform of CD44 that is typically expressed by hESCs (Holm et al., 2015). CD44 isoform switching also occurs in breast cancer stem cells (Zhang et al., 2019). Mis-splicing of GSK3β and concomitant activation of β-catenin was shown to be important for leukemia stem cell self-renewal, while pro-apoptotic splice variants of the BCL2 family promote leukemia stem cell survival (Abrahamsson et al., 2009; Goff et al., 2013). Overall, dozens of dysregulated spliceosome components, splice variants and splicing patterns have been identified in cancer stem cells, and much work remains to uncover the functional significance of these changes. However, these cancer stem cell specific splicing events are already revealing new opportunities to improve diagnostic and prognostic tools as well as to develop new targeted therapies (Crews et al., 2016). Thus, it is likely that we have only begun to uncover how cell-type and context specific differences in splicing enable stem cells to remodel their proteome for optimal function in response to developmental signals and environmental cues.
MicroRNA
miRNAs are small non-coding single stranded RNA molecules that repress gene expression through translational inhibition or by promoting degradation of mRNA. miRNA biogenesis is a stepwise process that starts with transcription of primary miRNAs (pri-miRNAs) in the nucleus. Pri-miRNAs are subsequently processed into stem-loop precursor miRNA (pre-miRNA) by a complex composed of DGCR8 and other factors (Gregory et al., 2004). Mature miRNAs are then cleaved by Dicer1 and incorporated into the RNA induced silencing complex (RISC) (Hammond, 2005). Base pairing between the RISC-bound miRNA and 3’ UTR of target mRNA triggers mRNA decay or translational repression (Gregory et al., 2005).
MicroRNA-mediated gene silencing is a mechanism that regulates stem cell pluripotency. Mouse ESCs deficient in Dgcr8 lose the ability to differentiate (Wang et al., 2007) and express the pluripotency genes Oct4, Sox2, and Nanog at high levels (Wang et al., 2017c). Additionally, Dicer1-null mouse ESCs have diminished expression of differentiation markers in vitro and in vivo (Kanellopoulou et al., 2005). Consistent with these observations, miR-134, miR-296, and miR-470 mediate mouse ESC differentiation by disrupting Oct4, Sox2, and Nanog expression (Tay et al., 2008). MicroRNAs similarly repress hESC pluripotency by targeting OCT4, SOX2, and KLF4 transcripts (Xu et al., 2009b). Studies on somatic cell reprogramming also point to the role of miRNA in regulating pluripotency. miRNAs can increase the reprogramming efficiency of mouse embryonic fibroblasts into iPSCs (Li et al., 2011) and human skin cancer cells to a pluripotent state (Lin et al., 2008).
miRNAs can also regulate the activation, proliferation, and differentiation of somatic stem cells. miR-128 and miR-181 maintain hematopoietic stem and progenitor cells by inhibiting their differentiation into mature hematopoietic lineages (Georgantas et al., 2007). In muscle stem cells Pax3, which controls stem cell activation, is subject to repression by miR-206 (de Morree et al., 2019). miRNAs can also promote myogenesis by targeting repressors of muscle-related transcription factors or can enhance myoblast proliferation by targeting transcripts essential for differentiation (Chen et al., 2006). Lastly, neuronal lineages and astrocytes differentially express miRNA species that influence lineage specification (Smirnova et al., 2005; Visvanathan et al., 2007).
Disruptions in the miRNA pathway have been implicated in several types of cancer. Germline and somatic mutations in DICER1 can predispose individuals to cancer (Foulkes et al., 2014) and impaired DICER1 function can promote colon cancer (Iliou et al., 2014) and endometrial cancer stemness (Wang et al., 2017b). Many cancer types show miRNA signatures characterized by a defect in miRNA biogenesis and global downregulation of miRNA production (Calin and Croce, 2006; Gaur et al., 2007; Lu et al., 2005). Downregulation of miR-34a has been observed in breast cancer, colon cancer, pancreatic cancer, neuroblastoma, hepatocellular carcinoma, and non-small-cell lung cancer (Asadzadeh et al., 2019) suggesting that it may function as a tumor suppressor. miR-34a also inhibits the proliferation of breast cancer stem cells (Ma et al., 2015) and prostate cancer stem cells by suppressing CD44 expression (Liu et al., 2011). Lastly, miRNAs have also been implicated in stem cell-related signal transduction pathways including Wnt, Notch, and Hedgehog (Asadzadeh et al., 2019). Given their role in cancer pathogenesis, miRNAs show potential as diagnostic and prognostic biomarkers and provide a new avenue for treating cancer.
RNA Methylation
RNA can undergo a variety of biochemical modifications that collectively are referred to as the epitranscriptome. The most prevalent mRNA modification is methylation of adenosine at the nitrogen-6 position (N6-methyl adenosine (m6A)) (Dominissini et al., 2013). m6A deposition is catalyzed by the m6A methyltransferase (“writer”) complex that consists of methyltransferase like protein 3 (METTL3) or METTL14 along with WTAP and VIRMA (KIAA1429) (Balacco and Soller, 2019; Bokar et al., 1997; Liu et al., 2014; Yue et al., 2018). Although RNA methylation has been known about for decades, only recently was it shown to be reversible through the discovery of m6A demethylases (“erasers”) such as FTO (Jia et al., 2011) and ALKBH5 (Zheng et al., 2013). The dynamic nature of m6A modifications has sparked tremendous interest in its biological function, which is mediated by RNA binding proteins that recognize and bind m6A modified RNA (“readers”).
m6A modifications can functionally alter mRNAs, pre-mRNAs, miRNAs and non-coding RNAs, such as rRNA and tRNA. The major effects of m6A on mRNA are mediated by the reader proteins YTHDF1 and YTHDF2. YTHDF1 can promote cap dependent translation of m6A modified mRNAs by enhancing interaction with translation initiation factors. In contrast, YTHDF2 typically promotes mRNA decay, thereby suppressing translation (Wang et al., 2014b; Wang et al., 2015b). m6A modification of pre-mRNAs can alter mRNA export or induce structural changes that promote interaction with different RNA binding proteins that in turn alter splicing or editing patterns (Dominissini et al., 2012; Geula et al., 2015; Liu et al., 2015; Peer et al., 2018; Xiao et al., 2016). m6A in pri-miRNAs promotes processing and miRNA biogenesis (Alarcon et al., 2015). Hence, m6A can influence the transcriptome and proteome through the regulation of diverse post-transcriptional mechanisms.
Patterns of m6A modifications vary dramatically in a temporal, tissue and cell type specific manner. Although these patterns are mediated in part by differential expression of writers, erasers and readers, how these context specific patterns of m6A are established remains largely unknown. In vivo, germline deletion of Mettl3 results in early (E5.5–7.5) embryonic lethality associated with impaired induction of cellular differentiation (Geula et al., 2015). In zebrafish embryos, morpholino-mediated knockdown of either mettl3 or wtap also cause widespread differentiation defects (Ping et al., 2014). Germline deletion of the erasers Fto or Alkbh5 in mice are not lethal, but the former causes severe growth defects and the latter impairs male fertility (Boissel et al., 2009; Zheng et al., 2013). Thus, m6A methylation is critical for normal development.
By regulating mRNA stability, m6A modifications have a striking impact on ESC self-renewal and differentiation. A wide range of transcripts, including the core pluripotency transcription factors SOX2 and NANOG, are marked by m6A. Genetic inactivation of Mettl3 in naïve mouse ESCs results in widespread loss of m6A modifications that enhance self-renewal and impair differentiation in vitro and in vivo (Batista et al., 2014). Since m6A modifications can promote mRNA degradation, the loss of m6A in ESCs stabilizes pluripotency-promoting transcripts such as NANOG (Geula et al., 2015). Conversely, overexpression of METTL3 enhances reprograming efficiency of human fibroblasts into iPSCs by stabilizing pluripotency factors (Chen et al., 2015). Interestingly, Mettl3 or Mettl14 knockdown within primed mouse ESCs also reduces m6A modifications, but has dichotomous effects on self-renewal and differentiation as compared to naïve ESCs. Knockdown of Mettl3 or Mettl14 impairs self-renewal and promotes differentiation of primed mouse ESCs (Wang et al., 2014c). This difference can be partially explained by the rebalancing of self-renewal and differentiation transcripts that occurs in primed but not naïve ESCs. In naïve ESCs, the loss of m6A enhances the stability of highly expressed self-renewal genes, while in primed ESCs the loss of m6A enhances the stability of highly expressed differentiation genes. This example demonstrates the importance and precision with which post-transcriptional mechanisms of gene regulation influence stem cell identity.
m6A also regulates the emergence, self-renewal and differentiation of somatic stem cells. Mettl3-deficient zebrafish embryos do not undergo the endothelial to hematopoietic transition and fail to produce early hematopoietic stem and progenitor cells (Zhang et al., 2017). This occurs in part because in the absence of m6A, YTHDF2, a reader protein that promotes mRNA decay, is delayed in binding to the arterial endothelial mRNAs for notch1a and rhoca. This results in sustained Notch signaling in endothelial cells, which suppresses hematopoietic specification (Zhang et al., 2017). Knockdown of either METTL3 or METTL14 in human cord blood derived hematopoietic stem and progenitor cells modestly impairs stem cell proliferation and promotes myeloid differentiation (Vu et al., 2017; Weng et al., 2018). However, conditional deletion of Mettl3 from adult mouse HSCs leads to HSC accumulation, reduced reconstituting activity and impaired differentiation due in part to a loss of m6A-mediated translation of c-Myc (Lee et al., 2019). Conditional deletion of Mettl14 from adult mouse HSCs also reduces long-term multilineage reconstituting activity in transplantation assays (Weng et al., 2018). In addition, conditional deletion of Mettl3 from mouse skeletal stem cells impairs osteogenic differentiation and bone development by regulating the translational efficiency of parathyroid hormone receptor 1 (Pthr1) (Wu et al., 2018). Thus, m6A exhibits exquisite context dependent regulation of gene expression that can contribute to a divergent transcriptome and proteome. Overall, our understanding of how m6A influences proteome content and cellular function is still in its infancy, but it clearly plays a key role in stem cell regulation and cell fate determination.
m6A methylation also influences cancer stem cells. Breast cancer stem cells exhibit reduced m6A methylation of NANOG and KLF4, which contributes to elevated expression of both pluripotency factors (Zhang et al., 2016a). In both cervical cancer and acute myeloid leukemia, high FTO expression has been reported to be important for cell survival (Li et al., 2017b; Wang et al., 2017a). High METTL3 expression has also been reported in acute myeloid leukemia, with subsequent methylation of MYC, MYB, PTEN, and BCL2, which could support leukemia stem cell survival (Barbieri et al., 2017; Vu et al., 2017; Weng et al., 2018). The finding that both m6A writers and erasers are highly expressed in cancer highlights the importance of determining the role of m6A methylation and subsequent binding of reader proteins on mRNA stability and translation efficiency in a temporal, tissue and cell type specific manner.
In addition to m6A, adenosines can be methylated at the nitrogen-1 position (m1A) (Dominissini et al., 2016; Li et al., 2017a; Safra et al., 2017; Zhang and Jia, 2018). m1A modifications were traditionally thought to regulate the stability of tRNAs and rRNAs. However, recent advances in sequencing technology have revealed tissue specific methylation of mRNAs, in the 5’ UTRs within the mRNA cap that enhances translational efficiency (Dominissini et al., 2016; Li et al., 2017a). To date, m1A modifications have been associated with both increased and suppressed protein synthesis (Dominissini et al., 2016; Li et al., 2017a; Safra et al., 2017). Moreover, m1A deposition occurs in a tissue and cell type specific manner and can be dynamically regulated in response to environmental stress. Because m1A methylation was shown to be highly conserved in mice, it is likely to be essential for gene regulation. However, the functional importance of m1A, particularly in stem cells, remains largely unknown.
RNA can also be methylated at the carbon-5 position of cytosine (m5C) (Schaefer et al., 2009). The m5C modification most commonly occurs on tRNAs and rRNAs (Agris, 2008; Schaefer et al., 2009). However, NSUN2, one of seven known cytosine-5 methylases, was recently shown to deposit m5C on some mRNAs as well (Khoddami and Cairns, 2013; Squires et al., 2012). There are at least six other enzymes capable of methylating cytosine 5, including NSUN1, NSUN3, NSUN4, NSUN5, NSUN6 and DNMT2 (Yang et al., 2017).
Dynamic changes in m5C deposition in rRNAs and tRNAs can impact ribosome biogenesis, polysome assembly, translation fidelity and tRNA stability (Blanco et al., 2016; Gigova et al., 2014; Schosserer et al., 2015; Sharma et al., 2013; Tuorto et al., 2015). This widespread influence on the translational apparatus enables m5C levels to modulate global protein synthesis and regulate specific translational programs (Roundtree et al., 2017). Loss of m5C is associated with suppression of global protein synthesis. Deletion or loss of function of NSUN2 leads to widespread loss of m5C in most tRNAs, leading to cleavage and the accumulation of tRNA-derived small non-coding RNAs, which can impair translation elongation and reduce protein synthesis (Blanco et al., 2016). In addition to dampening global protein synthesis, loss of m5C also increases translation of stress response genes (Chan et al., 2010), as well as genes regulating cell motility (Flores et al., 2017), morphogenesis and apoptosis (Blanco et al., 2014; Hussain et al., 2013).
Loss of m5C in tRNAs associated with Nsun2 and/or Dnmt2 deficiency impairs differentiation in multiple murine tissues, including the brain, blood, skin, testis, liver and fat (Blanco et al., 2011; Flores et al., 2017; Hussain et al., 2013; Rai et al., 2007; Tuorto et al., 2015). The specialized translational program associated with m5C loss is sufficient to maintain epidermal stem cells in their undifferentiated state, but does not enable normal differentiation. Increased m5C is required for epidermal stem cells to increase protein synthesis in response to cytotoxic stress (Blanco et al., 2016). These studies suggest that dynamic control of the epitranscriptome is required for stem cells to appropriately survive and promote regeneration in response to stress.
Pseudouridylation
Pseudouridine (Ψ, 5-ribosyluracil) is the most widespread RNA modification (Charette and Gray, 2000; Guzzi et al., 2018). Pseudouridine is present within mRNAs and non-coding RNAs such as rRNAs, tRNAs, splicesomal small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and telomerase RNA. Because pseudouridine contains an extra hydrogen bond donor, it promotes base stacking interactions that typically make RNA backbones more rigid (Charette and Gray, 2000). This modification thus primarily influences RNA structure, which can in turn influence interactions with other biomolecules. Pseudouridylation plays an important role in regulating protein synthesis by enhancing tRNA stability, influencing base pairing within the ribosome decoding center and altering translation termination (De Zoysa and Yu, 2017). While pseudouridylation is thought to promote translation (Roundtree et al., 2017), its effects are diverse thereby making it a complex modification to understand.
There are at least 13 pseudouridine synthases (PUSs). These PUSs can catalyze pseudouridylation in a guide RNA independent manner (Hamma and Ferre-D’Amare, 2006). Pseudouridylation can also be catalyzed in a guide RNA dependent manner by Dyskerin (DKC1), in a process that depends upon target sequence complementarity to a box H/ACA snoRNA in complex with several other proteins (Hamma and Ferre-D’Amare, 2006). Currently, pseudouridylation is thought to be irreversible as no readers or erasers have yet been identified.
The effects of pseudouridylation on stem cells were initially observed in the context of DKC1 mutations. DKC1 is mutated in the X-linked form of the human disease dyskeratosis congenita, which is a disorder characterized by short telomeres (Batista et al., 2011). TERC contains a highly conserved pseudouridylation site in a key region required for TERT binding, raising the possibility that defects in pseudouridylation impair telomerase activity. DKC1 promotes telomere elongation in iPSCs (Batista et al., 2011), and also regulates the expression of OCT4 and SOX2 (Fong et al., 2014). In agreement with a potential role in promoting pluripotency, DKC1 mutant fibroblasts exhibit impaired iPSC reprogramming (Agarwal et al., 2010). DKC1 mutations are also associated with widespread loss of rRNA modifications, and the catalytic activity of Dkc1 is required for normal HSC differentiation (Bellodi et al., 2013).
Guide independent pseudouridylation has been shown to be important for stem cells as well. PUS7 deficient hESCs exhibit impaired activation of tRNA derived small fragments that are required for translational control (Guzzi et al., 2018). PUS7 deficiency is associated with increased protein synthesis and impaired germ layer specification (Guzzi et al., 2018). Dysregulation of the Pus7 mediated translational program is also required for HSC commitment (Guzzi et al., 2018). Pseudouridylation is thus required for translational control and normal stem cell function.
RNA Editing
Another mechanism of RNA sequence modification that contributes to transcriptomic diversity is RNA editing. RNA editing is the most common post-transcriptional modification detected by whole transcriptome RNA sequencing in human cells (Peng et al., 2012). The most frequent type of RNA editing event in mammals involves deamination of adenosine into inosine (A-to-I) (Hartner et al., 2009; Tan et al., 2017; Zipeto et al., 2016). When A-to-I editing occurs within protein coding exons, inosine bases are read as guanosines by the translational apparatus (Tan et al., 2017; Zipeto et al., 2016; Zipeto et al., 2015). Although editing events can occur within coding regions, most RNA editing sites are located within non-coding regions, such as introns and UTRs (Peng et al., 2012). In humans, approximately 90% of these editing sites are located within primate-specific Alu sequences, which are transposable elements that represent approximately 11% of the human genome (Batzer and Deininger, 2002). Editing of Alu sequences within non-coding regions can cause the introduction of new splice sites (splicing machinery also recognizes inosines as guanosines) (Hsiao et al., 2018; Rueter et al., 1999; Solomon et al., 2013), promote RNA degradation, and induce sequestration of RNAs in discrete nuclear compartments (Mellis et al., 2017). Also, RNA editing can modulate gene expression by impairing miRNA biogenesis (Jiang et al., 2019; Zipeto et al., 2016).
Editing of RNA is catalyzed by members of the adenosine deaminase associated with RNA (ADAR) family. To date, three members of the ADAR family have been identified in vertebrate animals: ADAR (ADAR1), ADARB1 (ADAR2) and ADARB2 (ADAR3) (Tan et al., 2017; Zipeto et al., 2015). ADAR1 is ubiquitously expressed and is essential for embryonic development (Hartner et al., 2009). Adar1 deficiency causes embryonic lethality in mice as a consequence of defective erythropoiesis and is associated with hyperactive interferon signaling and widespread apoptosis (Hartner et al., 2009). Germline ADAR1 mutations in humans are associated with Aicardi–Goutières syndrome and dyschromatosis symmetrica hereditaria (Rice et al., 2012; Suzuki et al., 2005). ADAR2 is also widely expressed, but is not required for embryonic development (Jacobs et al., 2009). However, Adar2 deficiency is associated with neuronal death and seizures that cause postnatal lethality in the first few weeks of life (Higuchi et al., 2000; Yamashita and Kwak, 2019). Aberrant RNA editing profiles are accordingly associated with a number of human neurological and psychiatric disorders (Slotkin and Nishikura, 2013). ADAR3 expression is largely restricted to the brain and has not yet been shown to exhibit RNA editing activity (Whitney et al., 2008). Rather, ADAR3 has been shown to inhibit RNA editing (Oakes et al., 2017).
hESCs exhibit high levels of RNA editing (Osenberg et al., 2009). RNA editing in hESCs is enriched within non-coding regions of double stranded RNA marked by inverted Alu repeats (Osenberg et al., 2009). The global abundance of transcript editing is reduced during differentiation, particularly in the neural lineage (Osenberg et al., 2009; Tan et al., 2017). Knockdown of ADAR1 is associated with increased expression of genes associated with differentiation and developmental processes (Osenberg et al., 2009) and may be dispensable for hESCs (Chung et al., 2018). Human fibroblasts reprogrammed into iPSCs exhibit RNA editing profiles that more closely resemble hESCs than mature fibroblasts (Shtrichman et al., 2013), suggesting that there is reprogramming of the RNA editome. Furthermore, modulating ADAR1 expression influences the efficiency of reprogramming (Germanguz et al., 2014). Overall, these studies suggest that ADAR1-mediated RNA editing contributes to the establishment of pluripotency and cell fate determination.
RNA editing also regulates somatic stem and progenitor cell populations. Conditional deletion of Adar1 impairs the multi-lineage reconstituting activity of mouse HSCs (Orkin and Zon, 2008; XuFeng et al., 2009). Adar1 deficiency also increases hematopoietic progenitor cell apoptosis. This phenotype depends upon the RNA editing domain of Adar1 and is associated with upregulation of interferon signaling (XuFeng et al., 2009). Based on these phenotypes, it remains unclear whether Adar1 deficiency directly impairs HSC function or whether reconstitution is impaired because of defects in progenitor cells. In addition to regulating cell death, ADAR1 can also regulate quiescence and cell cycle entry of human hematopoietic stem and progenitor cells. Lentiviral overexpression of ADAR1 within human cord blood derived hematopoietic stem and progenitor cells increased expression of specific cell cycle and self-renewal regulatory transcripts that enhance the expansion of these cells in vitro (Jiang et al., 2019). At least some of the effects of ADAR1 on stem and progenitor cell expansion occur through an RNA editing dependent mechanism. ADAR1 mediated RNA editing of pri-miR-26a at the Drosha cleavage site impaired maturation of miR-26a. Subsequent reduction of miR-26a led to a cascade of gene expression changes that enhanced cell cycle transit. Expression of EZH2, a direct target of miR-26a, was increased in hematopoietic stem and progenitor cells following ADAR1 overexpression. Moreover, EZH2 subsequently repressed the expression of CDKN1A, a negative regulator of cell cycle entry that can promote HSC quiescence (Jiang et al., 2019).
ADAR1 also promotes intestinal homeostasis and stem cell maintenance. Adar1 is highly expressed by Lgr5+ cells in the intestine of adult mice. Conditional deletion of Adar1 results in rapid apoptosis of Lgr5+ stem cells in both the small and large intestine. In contrast to the Lgr5+ cells, Adar1 deficiency caused expansion of intestinal progenitors and Paneth cells, although enterocytes, goblet cells and enteroendocrine cells were all depleted (Qiu et al., 2013). Similar to the hematopoietic system, Adar1 deficiency in the intestine was associated with increased interferon signaling, but was also marked by endoplasmic reticulum (ER) stress and activation of the unfolded protein response (UPR) that at least partially contributed to crypt apoptosis (Qiu et al., 2013). Together, these studies support an essential role of ADAR1 and RNA editing in both tissue homeostasis and stem cell maintenance.
Though only a handful of editing sites have been identified in normal cells, editing events in cancer have been characterized in more detail. Editing of mRNAs encoding GLI1, GSK3β, AZIN1 and APOBEC3D have been identified and were found to be required for survival of leukemia stem and progenitor cells (Crews et al., 2015). ADAR1-mediated editing of the MDM2 3’UTR has also been shown to reduce the binding of mir-155 as well as other negative regulatory miRNAs (Jiang et al., 2019). As a consequence of hyper-editing, MDM2 mRNA is stabilized within leukemia stem cells, resulting in increased MDM2 protein and enhanced p53 degradation. In acute myeloid leukemia, editing of PTPN6 was found to abrogate splicing and is thought to be important for leukemogenesis (Beghini et al., 2000).
While only a small number of editing sites have been characterized, bioinformatics analyses have predicted A-to-I changes to be far more abundant. To date, A-to-I editing profiles of more than 6000 patient samples of 17 cancer types revealed a surprising increase in RNA editing events in tumor tissue relative to normal tissue (Han et al., 2015). Furthermore, it has been suggested that RNA editing could also affect the therapeutic response to immunotherapy. Loss of function of ADAR1 was found to improve response to PD-1 checkpoint blockade (Ishizuka et al., 2019), and PD-L1 expression is under significant translational control (Xu et al., 2019). Thus, posttranscriptional regulation appears to play a major role in resistance to immunotherapy. Overall, targets of RNA editing vary dramatically across species, tissues and cell types (Tan et al., 2017). Recent parallel analysis of RNA secondary structure sequencing (PARS-seq) reveals that ADAR1 regulates RNA topology and ribosomal occupancy resulting in cell type and context specific changes in protein turnover rates (Solomon et al., 2017). Consistent with these context dependent effects on gene expression, RNA editing exhibits distinct functional effects on stem and progenitor cells. Future studies must focus on identifying edited transcripts that further explain the emerging role of RNA editing in normal and malignant stem cell biology.
PROTEOSTASIS
The synthesis and degradation of proteins, while long appreciated to be fundamental processes, have been historically viewed as simple housekeeping functions performed similarly by most cells. Emerging research has challenged this notion and it is becoming increasingly clear that the unique proteomes that specify diverse cell types are assembled and regulated using various strategies to maximize tissue function. Transcriptional regulation of mRNA content plays a key role in defining the cellular proteome. However, numerous studies in eukaryotic systems have documented broad differences between mRNA and protein abundance for individual genes, suggesting that mRNA abundance itself cannot be used to predict proteome content (Jovanovic et al., 2015; Liu et al., 2019; Schwanhausser et al., 2011; Vogel and Marcotte, 2012). Rather, the combined actions of the transcriptional, translational and degradation machineries are required to shape cellular function through the integrated regulation of proteome complexity and protein homeostasis (proteostasis) (Figure 1) (Harper and Bennett, 2016).
Protein Synthesis
Gene expression models have long assumed that the protein synthesis machinery merely acts as a passive conduit connecting the dynamic transcriptome to the proteome. Research over the last decade has revealed these models to be overly simplistic, as the translation machinery and the nature of the mRNA sequences themselves can impart biologically meaningful control over protein production (Hershey et al., 2018; Hinnebusch, 2017; Jackson et al., 2010; Schuller and Green, 2018). Advances in systems biology, nucleic acid sequencing, mathematical modeling and single cell technologies have revealed important cell type specific differences in protein synthesis that influence proteome complexity. Cell-type specific differences in protein synthesis have emerged as a particularly important mechanism for regulating stem cells and tissue regeneration (Buszczak et al., 2014).
Several reports have demonstrated that translation increases during ESC differentiation. ESCs differentiated into embryoid bodies (EBs) by LIF withdrawal display increased Golgi apparatus and ER contents as well as a greater cytoplasm to nucleus ratio, presumably to accommodate for a rise in translational output. Consistent with this, mRNA abundance and steady-state levels of proteins are elevated, and the rate of protein synthesis was shown to be enhanced during differentiation (Sampath et al., 2008). Increased translational output is at least partly due to greater translation efficiency during differentiation. EBs display a greater abundance of polysomes than ESCs, indicating greater efficiency of ribosome loading onto mRNAs (Ingolia et al., 2011; Sampath et al., 2008). ESCs also exhibit an abundance of ribosome pausing, translation of unannotated products and translation initiation from upstream open reading frames and non-conventional translation start sites (Ingolia et al., 2011; Sampath et al., 2008). Precise control of the translational apparatus thus plays a complex role in regulating proteome content within ESCs.
In addition to global changes in translation during ESC differentiation, there are also changes in the translation of specific transcripts that may have important functional consequences. The transcriptional and translational efficiency of B-Myb, which promotes cell-cycle activation (Sampath et al., 2008), was shown to be elevated during differentiation. In contrast, Wnt1, which activates β-catenin and supports pluripotency and development, was shown to exhibit a decline in translational efficiency during differentiation (Sampath et al., 2008). Together, these studies suggest that global and specialized translational programs can influence ESC self-renewal and differentiation.
Precise control of protein synthesis is also essential in ESCs to maintain an open chromatin landscape and support hyper-transcription. Some regulators of euchromatin are relatively unstable proteins that are rapidly turned over within ESCs. Inhibition of protein synthesis mediated by mTOR inhibition, can lead to a loss of these euchromatin factors and a concomitant reduction in chromatin accessibility and transcription of key developmental genes (Bulut-Karslioglu et al., 2018). It has been postulated that this translational program protects ESCs from spurious differentiation in response to acute changes in nutrient availability.
In addition to ESCs, low protein synthesis is a broadly conserved feature of somatic stem cells present within adult tissues in vivo. This was first demonstrated in murine HSCs, which exhibit unusually low rates of protein synthesis as compared to restricted progenitors and differentiated cells (Signer et al., 2014). Subsequent studies have revealed that low protein synthesis is observed within mouse hair follicle stem cells (Blanco et al., 2016), quiescent neural stem cells (Llorens-Bobadilla et al., 2015), muscle satellite cells (Zismanov et al., 2016) and Drosophila germline stem cells (Sanchez et al., 2016).
Modest increases or decreases in protein synthesis impair somatic stem cell function (Cai et al., 2015; Goncalves et al., 2016; Signer et al., 2014; Signer et al., 2016). HSCs with a mutation in the ribosomal gene Rpl24 (Rpl24Bst/+) (Oliver et al., 2004) were shown to exhibit a ~30% reduction in protein synthesis and reduced regenerative activity (Signer et al., 2014). Deletion of Pten, a negative regulator of mTOR signaling (which promotes translation) (Laplante and Sabatini, 2012; Ma and Blenis, 2009), increases protein synthesis by ~30% in HSCs (Signer et al., 2014) and causes HSC depletion (Yilmaz et al., 2006; Zhang et al., 2006). In Pten−/−;Rpl24Bst/+ compound mutant mice, protein synthesis is restored to normal, and Pten−/− and Rpl24Bst/+ HSC function is largely rescued (Signer et al., 2014). This demonstrates that protein synthesis is tightly regulated in HSCs and that HSCs require low rates of protein synthesis.
Surprisingly, the low rate of protein synthesis within somatic stem cells is not simply a consequence of increased quiescence. Although activated HSCs or HSCs driven into cell cycle following treatment with cyclophosphamide and GCSF exhibit modestly increased protein synthesis as compared to their quiescent counterparts (Cabezas-Wallscheid et al., 2017; Signer et al., 2014), cycling and dividing HSCs still exhibit significantly lower protein synthesis than cycling and dividing progenitors in vivo (Signer et al., 2014). Furthermore, these differences in protein synthesis cannot be fully explained by differences in doubling time between stem and progenitor cells (Signer et al., 2016). Similar to HSCs, hair follicle stem cells consistently display lower levels of protein synthesis than more differentiated cells regardless of whether they are in the anagen, catagen, or telogen stage of the hair growth cycle (Blanco et al., 2016). Although the low rate of protein synthesis in stem cells is not attributed to quiescence, it may be influenced by additional secretory functions employed by stem cells. Stem and progenitor cells secrete cytokines and growth factors to modulate their environment and induce responses from nearby cells (Baraniak and McDevitt, 2010). It is possible that differences in secretory output contribute to differences in protein synthesis and its subsequent connection to cell growth and proliferation.
Initial observations on the lack of correlation between protein synthesis and proliferation in stem cells contrasts recent landmark studies utilizing bacterial systems that have successfully established quantitative and predictive models that describe how proteome flux governs cell proliferation (Basan et al., 2015; Dai et al., 2016; Hui et al., 2015; Klumpp and Hwa, 2014; Scott et al., 2010; Scott et al., 2014). These studies determined that because most ribosomes are engaged in translation, and that translational capacity is nearly saturated in exponentially growing bacterial cells, there is an obligatory constraint between protein synthesis and the rate of cell proliferation (Klumpp and Hwa, 2014; Mori et al., 2016). Despite protein synthesis being so highly conserved, these differences between prokaryotic and mammalian systems highlight the pressing need to identify how proteome flux and content influence mammalian cell proliferation, and to establish growth laws within complex tissues in vivo. Indeed, an intriguing possibility is that there are cell-type specific growth laws that govern mammalian cell proliferation, and that stem cells may have a unique set of constraints that limit their proliferation, self-renewal and differentiation as compared to restricted progenitors.
Similar to normal stem cells, low protein synthesis has also been reported as a feature of cancer stem cells. Tumor initiating cells present in squamous tumors that develop in K5-SOS mice, which have constitutive RAS activation in basal epidermal cells, exhibit low rates of protein synthesis as compared to more committed progenitors. Reducing the global rate of protein synthesis via deletion of Nsun2 increases tumor mass and number, and shortens the lifespan of K5-SOS mice (Blanco et al., 2016). Another study demonstrated that squamous cell carcinoma initiating cells that amplify and express Sox2 also exhibit low protein synthesis associated with a redistribution of translating ribosomes to upstream open reading frames. This shifting translational landscape, which at least partly depends upon the translation initiation factor eIF2A, enables a subset of cancer-related transcripts to be translated more efficiently in order to drive tumor progression (Sendoel et al., 2017). In the hematopoietic system, Runx1 mutations, which are frequently seen in patients with acute myeloid leukemia and myelodysplastic syndrome (Cai et al., 2011; Growney et al., 2005), reduce ribosome abundance and protein synthesis in hematopoietic stem and progenitor cells. In this context, reduced protein synthesis enables these pre-leukemic stem cells to better survive genotoxic stress, which provides a selective advantage that enables their persistence and expansion in the bone marrow (Cai et al., 2015).
The recent finding that low protein synthesis promotes cancer development and progression was rather surprising, since cancer had long been thought to depend upon elevated protein synthesis to sustain malignant growth. Indeed, several studies have demonstrated that reducing protein synthesis can impair cancer growth. Reducing protein synthesis with a mutation in the ribosomal gene Rpl24 has been shown to slow the development and progression of Myc driven B cell malignancies (Barna et al., 2008) as well as T cell neoplasms that arise from Pten deletion (Signer et al., 2014). Suppression of mTOR signaling can slow prostate cancer progression by inhibiting the translation of pro-invasion genes (Hsieh et al., 2012). Deletion of one allele of Eif4e reduces tumor burden in a mouse model of Kras driven lung cancer by reducing translation of proteins that reduce reactive oxygen species (Truitt et al., 2015).
It is not yet clear why reducing protein synthesis can have dichotomous effects on different cancers. One possibility is that it is dependent on the genetic landscape of the tumor. Another possibility is that the alterations on global protein synthesis are less therapeutically relevant than specific translational changes. Yet another intriguing possibility is that the therapeutic response is based on whether the tumor contains cancer stem cells; tumors driven by cancer stem cells may depend upon low protein synthesis while cancers that are not driven by cancer stem cells may depend more on higher protein synthesis. Indeed, much remains to be learned about how cell-type specific differences in protein synthesis contribute to cancer growth and progression. Such studies should enable improved context-dependent targeting of translational machinery to have significant therapeutic efficacy in a broad spectrum of cancers.
Mechanisms of Translational Control
One mechanism of translational control important for stem cells is mediated by cytoplasmic poly(A)-binding proteins (PABPs). PABPs drive translational activation by binding to poly(A) tails on mRNA and interacting with translation initiation factors. PABP1 can also mediate miRNA silencing by associating with the miRNA-induced silencing complex (Fabian et al., 2009). PABPs are essential for fertility and early development. Disruption of PABP-encoding genes or depletion of direct PABP binding partners can lead male to sterility in Drosophila (Blagden et al., 2009) and embryonic defects or lethality in various model organisms (Blagden et al., 2009; Gorgoni et al., 2011; Maciejowski et al., 2005). In Pabp1-depleted Xenopus, defects in embryonic structures have been attributed to a global reduction in protein synthesis, which is consistent with the role of PABPs in stimulating translation. DAZL, a protein that can enhance translation through interaction with PABPs, exhibits increased translational efficiency during ESC differentiation, revealing a possible mechanism through which global protein synthesis is elevated (Sampath et al., 2008). In addition, pab-1 mutations in C. elegans can reduce the proliferation of germline stem cells during the larval stages (Ko et al., 2010).
Another mechanism of translational control that may be particularly important for stem cells involves Eif4e binding proteins (4E-BPs), which can suppress cap-dependent translation. 4E-BPs support pluripotency by suppressing the translation of select transcripts such as Yy2, a negative regulator of mouse ESC self-renewal. Mouse ESCs depleted of 4E-BP1 and 4E-BP2 have increased abundance of Yy2, reduced expression of Nanog, c-Myc, and Oct4, and morphologically resemble differentiated cells. Thus, by suppressing the translation of Yy2, 4E-BPs can promote mouse ESC pluripotency (Tahmasebi et al., 2016). 4E-BPs also influence the efficiency of iPSC reprogramming. During reprogramming of mouse embryonic fibroblasts (MEFs) into iPSCs, 4E-BPs repress translation of p21, a known inhibitor of somatic cell reprogramming (Tahmasebi et al., 2014). Consistent with this, 4E-BP-deficient MEFs exhibit impaired iPSC generation in response to reprogramming factors. However, deletion of 4E-BPs also increases the translation of the reprogramming factors Sox2 and Myc. As a consequence, compound deletion of 4E-BPs and p53 enhances iPSC generation and enables reprogramming with ectopic expression of Oct4 alone (Tahmasebi et al., 2014).
4E-BPs also regulate somatic stem cells. Deletion of 4E-BP1 and 4E-BP2 modestly increases protein synthesis within HSCs and impairs their serial reconstituting activity (Signer et al., 2016). Similarly, 4E-BPs support neural stem cell self-renewal. Increasing mTORC1 activity in neural stem cells induces their differentiation into intermediate progenitors at the expense of self-renewal, an effect that is driven by inhibition of 4E-BP2 and subsequent activation of cap-dependent translation (Hartman et al., 2013).
Repression of protein synthesis within somatic stem cells can also be mediated through non-cell-autonomous mechanisms. Angiogenin (Ang), a secreted factor in the bone marrow microenvironment, non-cell autonomously restricts protein synthesis within HSCs by stimulating the biogenesis of tRNA-derived stress-induced small RNAs (tiRNAs) (Goncalves et al., 2016). tiRNAs can suppress protein synthesis through multiple mechanisms, including RNA interference and displacement of eIF4G from mRNA on ribosomes (Ivanov et al., 2011). Deletion of Ang increases protein synthesis within HSCs and is associated with increased cell cycle entry and diminished reconstituting activity (Goncalves et al., 2016).
Although the vast majority of cellular mRNA is translated in a cap-dependent manner, recent studies have also shown the importance of cap-independent translation in regulating stem cell differentiation. DAP5 (also known as NAT1) is a translation initiation factor that mediates IRES-dependent translation. DAP5-depleted human ESCs exhibit impaired differentiation due to a reduction in the translation efficiency of Hmgn3 and transcripts involved in mitochondrial and oxidative respiration pathways. Cap-independent translation in hESCs may therefore promote differentiation by supporting epigenetic changes mediated by the nucleosome binder HMGN3 and by maintaining oxidative respiration pathways needed during the initial stages of differentiation (Yoffe et al., 2016). Dap5 is also essential for mouse ESC differentiation (Yamanaka et al., 2000). Suppression of Dap5 reduces the levels of Map3k3 and Sos1, consequently repressing the Erk and Akt signaling pathways (Sugiyama et al., 2017). Forced expression of Map3k3 and Sos1 factors induces mouse ESC differentiation. Sugiyama et al. proposed that Dap5 mediates translation independent of eIF4E, which suggests that Dap5 may regulate protein synthesis required for mouse ESC differentiation in a cap-independent manner. However, further studies are needed to confirm cap-independent translation of Dap5 target mRNAs.
Ribosome Biogenesis
Ribosomes serve as the site for protein synthesis. The eukaryotic ribosome consists of the small 40S subunit and large 60S subunit. The 40S subunit is comprised of 18S rRNA along with 33 ribosomal proteins. The 60S subunit is composed of 25S, 5.8S, and 5S rRNA in addition to 46 additional proteins. Ribosome assembly requires the cooperation of assembly factors and various classes of enzymes to process pre-rRNA into mature rRNA and to export the assembled ribosomes from the nucleus into the cytoplasm (Pena et al., 2017).
Despite the ubiquitous requirement for ribosomes to produce proteins, mutations in ribosomal genes are associated with a number of human diseases, collectively referred to as ribosomopathies, which present with tissue and cell-type specific phenotypes. One of the most well-studied ribosomopathies is Diamond-Blackfan anemia (DBA), an inherited condition characterized primarily by a deficiency in erythrocytes. The underlying cause of DBA is often ribosomal haploinsufficiency due to a loss in one of several ribosomal protein genes (Narla and Ebert, 2010). Initial efforts in developing animal models for DBA focused on RPS19, a gene encoding a ribosomal protein that is mutated in approximately 25% of DBA patients (Draptchinskaia et al., 1999). However, deletion of both Rps19 alleles in mice resulted in embryonic lethality and deletion of a single allele produced no phenotype due to compensation by the wild-type allele (Matsson et al., 2004; Matsson et al., 2006). As an alternative approach, rps19-deficient zebrafish were generated using morpholino oligonucleotides. The morphant zebrafish recapitulated defects in ribosome biogenesis and hematopoiesis observed in DBA patients, such as reduced circulating red blood cells, impaired erythroid maturation, and decreased levels of hemoglobin (Danilova et al., 2008; Uechi et al., 2008). Rps19 mouse models have since been generated, including one expressing a dominant negative point mutation and another that silences Rps19 through an in vivo knockdown approach (Devlin et al., 2010; Jaako et al., 2011). Identification of pathogenic mutations in other ribosomal proteins have spurred the creation of additional animal models for DBA. These include Rps6, Rps19, Rps20, and Rps29 transgenic mouse and zebrafish lines, which also phenocopy some hematopoietic defects found in DBA (Keel et al., 2012; McGowan et al., 2008; Taylor et al., 2012).
It has been proposed that ribosomal mutations disrupt the stoichiometry required for ribosome biogenesis, and some free ribosomal proteins, such as Rpl5 and Rpl11 subsequently bind and inhibit Mdm2, a negative regulator of p53 (Lohrum et al., 2003; Marechal et al., 1994; Zhang et al., 2003). The pathogenesis of DBA has thus long been thought to be dependent on the accumulation of p53. However, recent studies have challenged this paradigm. Decreased translation of GATA1, a transcription factor essential for erythropoiesis, has been shown to contribute to DBA pathogenesis (Khajuria et al., 2018; Sankaran et al., 2012). Ribosomal haploinsufficiency preferentially reduces the translation of GATA1 (Ludwig et al., 2014).This has been shown to occur because the 5’ UTR of GATA1 is shorter and has a less complex secondary structure than the typical transcript, allowing GATA1 to be translated at high efficiency, and the most highly translated transcripts are inherently most sensitive to changes in ribosome abundance. Furthermore, when compared with other hematopoietic regulators, these properties of the 5’ UTR are quite specific to GATA1 (Khajuria et al., 2018). Thus, ribosome biogenesis influences lineage commitment by affecting the translation of select transcripts.
Stem cells are highly sensitive to changes in ribosome biogenesis factors. The small subunit processome (SSUP), which is responsible for processing pre-18S rRNA, is enriched in mouse ESCs as compared to EBs and is essential for maintaining pluripotency. Knockdown of SSUP components reduces small ribosomal subunit content and attenuates translation (You et al., 2015). Reducing protein synthesis has been proposed to impair ESCs by preventing the biogenesis of short-lived pluripotency factors that are rapidly lost by normal turnover.
One recent study has revealed a mechanism whereby a ribosome biogenesis factor, Htatsf1, is dynamically regulated to control protein synthesis and ESC fate determination. Htatsf1 is a RNA binding protein that promotes ribosome biogenesis in two ways; as a member of the U2 snRNP complex that mediates intron removal in a subset of ribosomal proteins, and by interacting with rRNA transcription and processing proteins to promote rRNA maturation (Corsini et al., 2018). Htatsf1 is highly expressed by mouse ESCs. Deletion of Htatsf1 reduces Nanog, Oct4 and Sox2 expression, and impairs ESC colony-forming potential. At the onset of differentiation, Htatsf1 expression transiently decreases before it increases again as a requirement for neuroectoderm specification. Consistent with its role in ribosome biogenesis, Htatsf1 expression during differentiation is mirrored by changes in the global rate of protein synthesis (Corsini et al., 2018). This study suggests that a transient reduction in ribosome production and protein synthesis is required to deplete pluripotency factors in ESCs to facilitate differentiation, before ultimately rising to promote neuroectoderm specification.
Defects in ribosome biogenesis also impair somatic stem cell function. Mice deficient in Notchless (Nle) suffer from hematopoietic defects including bone marrow cytopenia, a loss of HSC quiescence, and impaired HSC reconstitution capacity. The effects of Nle appear to be restricted to HSCs and immature progenitors, as Nle is dispensable for myeloid progenitor proliferation and differentiation, as well as B cell development. Nle shares homology with yeast ribosome assembly protein 4 (RSA4), which has been implicated in large ribosome subunit biogenesis. Mice deficient in Nle accumulate 60S ribosome subunit pre-rRNA species specifically in HSCs and multipotent progenitors, suggesting that defective ribosome maturation in those cell types is the underlying cause of the observed hematopoietic dysfunction (Le Bouteiller et al., 2013).
The observation that stem cells are sensitive to changes in ribosome biogenesis is difficult to reconcile with reports that protein synthesis does not appear to be constrained by ribosome abundance. ESCs appear to contain an abundance of free ribosomes, suggesting that translational efficiency is not limited by ribosome availability (Ingolia et al., 2011; Sampath et al., 2008). Adult HSCs do not synthesize or contain unusually low amounts of rRNA or ribosomal proteins (Jarzebowski et al., 2018; Signer et al., 2014) suggesting that a paucity of ribosomes is unlikely to underlie low protein synthesis within HSCs. One possible explanation for this apparent contradiction comes from studies in Drosophila germline stem cells. A complex consisting of Udd, TAF1B, and TAF1C-like factor promotes transcription of rRNA by RNA polymerase I. In Drosophila, it has been shown that germline stem cells exhibit high levels of rRNA transcription as compared to their differentiating daughters and are correspondingly enriched in Udd, a component of an RNA polymerase I regulatory complex (Sanchez et al., 2016; Zhang et al., 2014). Despite high levels of rRNA transcription, Drosophila germline stem cells, similar to mammalian stem cells, exhibit lower rates of protein synthesis than their differentiated progeny, and require increased ribosome biogenesis and translation for differentiation (Sanchez et al., 2016). Thus, it is possible that stem cells may synthesize ribosomal components to adequately prepare their differentiated progeny for their increased protein synthesis requirements, but that the stem cells actually contain a low abundance of translationally competent ribosomes. An alternative possibility is that stem and progenitor cells contain distinct types of specialized ribosomes (Xue and Barna, 2015) that facilitate cell type specific translation, and perturbations in ribosome biogenesis may preferentially affect a subset of ribosomes required by stem cells. Although there are not yet data to support this possibility, it has not yet been thoroughly examined. Overall, much remains to be determined about how the translational machinery regulates stem cell fate and function.
Protein Quality Control
In addition to regulating proteome content, translation can also regulate proteostasis by regulating proteome quality. Translation is the most error prone step in gene expression and approximately 18% of proteins include at least one missense amino acid substitution (Drummond and Wilke, 2009). Translational errors can lead to protein misfolding and the formation of potentially toxic aggregates. High rates of protein synthesis can increase amino acid misincorporation (Drummond and Wilke, 2009). Reducing translation decreases synthesis of defective translational products and promotes the clearance of misfolded proteins (Sherman and Qian, 2013). Interestingly, genetic or environmental interventions that reduce protein synthesis can extend organismal lifespan, at least in part by enhancing proteome quality (Taylor and Dillin, 2011). This raises the possibility that low protein synthesis could enhance proteome quality within stem cells, and could promote stem cell maintenance and longevity.
A recent study has indeed demonstrated a direct connection between protein synthesis and proteome quality in stem cells. Modest increases in protein synthesis lead to an accumulation of misfolded and unfolded protein within HSCs. The accumulation of defective translational products can overwhelm the capacity of the proteasome within HSCs, and can lead to changes in the HSC proteome, including accumulation of c-Myc protein, which drives increased proliferation and impairs HSC self-renewal (Hidalgo San Jose et al., 2020).
Proteotoxic stress can induce activation of stress response pathways that can exhibit important roles in stem cell regulation (Figure 2). There are three compartment specific proteotoxic stress response pathways; the ER unfolded protein response (UPRER), the mitochondrial unfolded protein response (UPRMT) and the heat shock response.
The ER is the major site of protein folding for secreted proteins and is thus a cellular compartment that is highly susceptible to the accumulation of unfolded proteins. An accumulation of unfolded proteins in the ER can trigger activation of the UPRER, which can attenuate protein synthesis, increase expression of folding chaperones, and in the case of severe or sustained stress, induce apoptosis (Walter and Ron, 2011). The UPRER is divided into three branches that work in parallel but are operated by distinct families of ER stress transducers: ATF6, IRE1 and PERK (Figure 2). These proteins are located on the ER membrane and initiate the UPRER by sensing protein-folding conditions. Activation of the ATF6 branch results in the transport of ATF6 into the Golgi apparatus where it is cleaved by S1P and S2P proteases. The resulting fragment, ATF6(N), translocates to the nucleus where it activates a transcriptional program that enhances the expression of chaperones such as BiP, PDI and GRP94 (HSP90B1). IRE1 promotes the splicing and translation of XBP1, a transcription factor that stimulates the expression of ER chaperones and ER-associated degradation proteins. PERK is a kinase that, upon activation, phosphorylates eIF2α. Phosphorylated eIF2α inactivates eIF2 to reduce overall protein synthesis, but also induces a specialized translational program that promotes the synthesis of downstream effector proteins including ATF4. ATF4 drives the expression of CHOP and GADD34, the former of which can promote apoptosis. Notably, eIF2α can also be phosphorylated by GCN2, PKR and HRI, which are kinases that are activated by the absence of essential amino acids, the presence of double stranded RNA, and a lack of heme, respectively. Thus, phosphorylation of eIF2α and suppression of protein synthesis is a common response to a variety of cellular stressors. Depending on the magnitude, duration and cellular context, UPRER activation can restore proteostasis, promote cell survival, and induce apoptosis (Walter and Ron, 2011).
The generation and differentiation of pluripotent stem cells requires significant proteome remodeling and can thus be influenced by the UPRER. Activation of the UPRER is predictive for successful somatic cell reprogramming, and genetic or pharmacologic interventions that transiently activate the IRE1 branch of the UPRER enhance reprogramming efficiency (Simic et al., 2019). Pharmacologic activation of the UPRER in ESCs activates Smad2 and β-catenin signaling and promotes endodermal differentiation (Xu et al., 2014). Activation of ATF6 also promotes differentiation of ESCs into mesoderm lineages (Kroeger et al., 2018; Wang et al., 2015a). Consistent with this, inhibition of ATF6 activation maintains ESCs in a pluripotent state and impedes mesodermal specification (Kroeger et al., 2018).
Activation of the UPRER also influences somatic stem cell fate in a tissue specific manner. Quiescent muscle satellite cells depend upon activation of the PERK branch of the UPRER to accumulate phosphorylated eIF2α, Atf4, and Chop. It has been shown that phosphorylated eIF2α enables satellite cells to sustain low levels of protein synthesis and suppress the translation of myogenesis-related transcripts. Failure to adequately phosphorylate eIF2α and suppress protein synthesis leads to the translational activation of a genetic program that drives satellite cells into cycle and promotes myogenic differentiation (Zismanov et al., 2016). Additionally, accumulation of Chop in satellite cells restricts myogenic differentiation by repressing transcription of MyoD (Alter and Bengal, 2011). In contrast, intestinal stem cells exhibit less UPRER activation than transit-amplifying progenitors and differentiated cells. UPRER signaling can have dichotomous effects on intestinal stem cells. Activation of PERK signaling is associated with a loss of self-renewal potential and is necessary for intestinal stem cell differentiation (Heijmans et al., 2013). In contrast, activation of Xbp1 reduces growth of Drosophila intestinal stem cells in vitro (Niederreiter et al., 2013; Wang et al., 2014a).
Similar to intestinal stem cells, distinct branches of the UPRER differentially regulate HSCs. Because of a low abundance of eIF2, human HSCs express high levels of ATF4 as compared to restricted progenitors, and increased translation of ATF4 enhances HSC engraftment in transplantation assays (van Galen et al., 2018). Moderate stress, such as nutrient depletion, enhances ATF4 activation to promote survival in primitive cord blood stem and progenitor cells (van Galen et al., 2018). Similarly, higher levels of Ire1 activation promote adult mouse HSC survival, reconstitution capacity (Liu et al., 2019) and recovery after irradiation (Chapple et al., 2018). Under more severe stress conditions, such as pharmacologic activation of the UPRER, human HSCs preferentially activate the PERK and ATF6 pathways, while restricted progenitors preferentially activate the IRE1 pathway (van Galen et al., 2014; van Galen et al., 2018). Increasing PERK activation as well as low Ire1 activity have been reported to preferentially induce apoptosis in HSCs, but promote the survival of restricted progenitors (Liu et al., 2019; van Galen et al., 2014). The induction of apoptosis in HSCs can eliminate damaged stem cells and preserve the overall integrity of the HSC pool.
Transformed cells are exposed to a variety of stressors that activate the UPRER, including hypoxia, nutrient shortage, oxidative stress, and genome instability (Vandewynckel et al., 2013). While activation of the UPRER sustains cell survival by reducing the burden of unfolded/misfolded proteins in the ER, chronic activation can also result in apoptosis via the IRE1-mediated JNK pathway (Tam et al., 2014). Cancer cells in which the UPRER has been activated must therefore balance the demands for cell survival with the risk of cell death. Preleukemic stem cells utilize the UPRER to enhance their survival and persistence. HSCs bearing the G12D mutation in Nras are highly resistant to ER stress, as they exhibit less apoptosis and higher reconstitution capacity following pharmacological induction of ER stress as compared to wild-type HSCs. NrasG12D hyperactivates IRE1 in pre-leukemic stem cells, resulting in the expression of DNAJB8, a target of XBP1. DNAJB8 serves as a molecular chaperone that increases protein folding capacity and ultimately enhances HSC reconstitution (Liu et al., 2019; van Galen et al., 2014). Although the UPRER can be protective for pre-malignant stem cells, enhanced activation of the UPRER has also been demonstrated to sensitize cancer stem cells to therapy. Induction of the UPRER by subtilase cytotoxin AB enhanced differentiation of colon cancer stem cells and made them more sensitive to chemotherapy (Wielenga et al., 2015). Due to the paradoxical effect of the UPRER on cancer cells, either inhibition or activation of the UPRER may have therapeutic potential in targeting cancer stem cells, but additional studies are needed to better understand the role of the pathway in a cell- and tissue-specific manner.
Translational stress in the mitochondria can also induce an unfolded protein response, and the UPRMT influences stem cell fate. HSCs exiting quiescence exhibit UPRMT activation, and mitochondrial integrity is monitored at the restriction point before HSCs proliferate (Mohrin et al., 2018). Dysregulation of the UPRMT impairs both HSCs (Mohrin et al., 2015) and intestinal stem cells (Berger et al., 2016). Treatment with nicotinamide riboside promotes UPRMT activation and can rejuvenate or delay senescence in muscle satellite cells, neural stem cells and melanocyte stem cells (Zhang et al., 2016b). These findings are consistent with studies demonstrating a dependence on mitochondrial health for optimal stem cell maintenance and function (Khacho et al., 2016; Vannini et al., 2019; Zhang et al., 2013).
Elevated temperatures, oxidative stress and toxic substances can cause the accumulation of misfolded proteins that disrupt proteostasis. To counteract this, the cell initiates the heat shock response. The heat shock response in eukaryotic cells is driven by heat shock factor 1 (HSF1), a highly-conserved, master transcription factor that induces the expression of heat shock proteins – molecular chaperones that coordinate protein folding, trafficking and degradation that enhance proteostasis buffering capacity and promote cell survival (Richter et al., 2010). Activation of the heat shock response in hESCs represses OCT4 expression and promotes differentiation (Byun et al., 2013). Mouse ESCs downregulate the expression of members of the Hsp70 family as well as small heat shock proteins during differentiation into EBs and neural precursor cells (Battersby et al., 2007; Saretzki et al., 2004). Similarly, in muscle, induction of myoblast differentiation is associated with elevated expression of several small heat shock proteins (Sugiyama et al., 2000). In the hematopoietic system, mutations in Hspa9b, a member of the Hsp70 family, reduce the number of hematopoietic progenitors in zebrafish (Craven et al., 2005) and depletion of HSP70 induces apoptosis in differentiating human erythroid precursors (Ribeil et al., 2007). However, Hsf1 appears dispensable for steady state young adult HSC function (Kourtis et al., 2018). Overall, little is yet known about how the heat shock response regulates stem cells either at steady state or when proteostasis is challenged.
Although each of these pathways is generally considered to be a stress responder, mounting evidence indicates that they also have important physiological function. A more thorough understanding of how these pathways influence stem cell behavior will require distinguishing their impact on the proteome and stem cell function at steady state and in response to developmental cues, regenerative demands and distinct stressors.
Protein Degradation
The ubiquitin-proteasome system is a major cellular protein degradation system. The covalent attachment of ubiquitin to proteins can mark them for recognition and destruction by the proteasome. The process of ubiquitination is orchestrated by a cascade of ubiquitin ligases and provides specificity to the degradation process. First, the ubiquitin activating enzyme (E1) activates ubiquitin before transferring it to the ubiquitin conjugating enzyme (E2). E2 then interacts with a ubiquitin ligase (E3) which recognizes and ubiquitinates specific substrates targeted to the proteasome (Zheng and Shabek, 2017). Although there are distinct assemblies of the proteasome, the 26S proteasome degrades damaged proteins. The 26S proteasome consists of the 20S core and 19S regulatory particle, and contains caspase-, trypsin-, and chymotrypsin-like activity that degrades substrates into 2–24 amino acid peptides (Bedford et al., 2010; Dikic, 2017). The proteasome contributes to maintaining proteostasis in stem cells by regulating both the content and quality of the proteome through the normal turnover of proteins and the degradation of misfolded proteins.
ESCs depend upon high levels of proteasome assembly and activity. NRF2, PSMD11, and PSMD14 have been identified as regulators of proteasome activity and assembly in ESCs (Buckley et al., 2012; Jang et al., 2014; Vilchez et al., 2012). All three proteins have elevated expression in pluripotent stem cells as compared to more differentiated cells. NRF2 governs proteasome activity partly through POMP, a chaperone that mediates 20S subunit assembly (Jang et al., 2014). Similarly, PSMD11 stabilizes the interaction between the 20S and 19S proteasome subunits. PSMD14 is a component of the 19S subunit, thereby facilitating 26S proteasome assembly (Buckley et al., 2012; Vilchez et al., 2012). Disrupted expression or activity of these factors each impairs normal ESC function. High levels of proteasome activity promote ESC function by regulating the abundance of pluripotency and differentiation factors (Buckley et al., 2012; Jang et al., 2014; Vilchez et al., 2012). The pluripotency factors Oct4 and Nanog are much more frequently ubiquitinated and degraded in ESCs as compared to differentiated cells, suggesting that their abundance is specifically fine-tuned in ESCs via the proteasome (Buckley et al., 2012).
In addition to having overall elevated levels of proteasome activity, proteasomal regulation of ESCs is supported through differential expression of E3 ubiquitin ligases. Wwp2, the E3 ubiquitin ligase that targets Oct4 for ubiquitination and degradation, is more highly expressed by ESCs than differentiated progeny enabling more flexible post-translational control of Oct4 abundance (Xu et al., 2009a; Xu et al., 2004). In contrast, the E3 ubiquitin ligase Fbxw7 is necessarily upregulated during ESC differentiation in order to ubiquitinate c-Myc and target it for degradation. The proteasome may also suppress the transcription of lineage specific genes by degrading components of the transcriptional pre-initiation complex at intergenic regions (Szutorisz et al., 2006). Inhibition of the proteasome within ESCs results in broad proteomic changes, marked by decreased abundance of proteins involved in ribosome biogenesis, nuclear transport of mRNA, carbohydrate metabolism, and telomere maintenance, which are all processes that influence ESC identity (Saez et al., 2018).
The proteasome also influences proteome content in adult stem cells to regulate self-renewal, differentiation and aging. In contrast to ESCs, Fbxw7 is required by HSCs to promote self-renewal and suppress differentiation by targeting c-Myc for degradation (Reavie et al., 2010; Thompson et al., 2008; Wilson et al., 2004). Similarly, the E3 ubiquitin ligase Huwe1 is required by HSCs to reduce the abundance of N-Myc via proteasome mediated degradation to prevent spurious HSC proliferation and exhaustion (King et al., 2016). In addition to Myc family members, the proteasome regulates protein abundance of other key regulators of HSC self-renewal, including p53, Hif1α, Notch and Stat5 (Moran-Crusio et al., 2012). In muscle, deletion of Rpt3 (Psmc4), a subunit of the 26S proteasome, results in muscle atrophy due to proteasome insufficiency (Kitajima et al., 2018). Rpt3-deficient muscle satellite cells exhibit a loss of quiescence, increased apoptosis, impaired differentiation and overall impaired regenerative activity in response to injury (Kitajima et al., 2018). Neural stem and progenitor cells exhibit an age-related decline in PSMB5, a catalytic subunit of the 20S proteasome, which results in a concomitant decline in proteasome activity. Treatment of neural stem and progenitor cells with a proteasome inhibitor or following knockdown of Psmb5 phenocopied some of the effects of aging, including diminished proliferation and impaired differentiation (Zhao et al., 2016). Overall, these studies indicate that proteasome mediated protein degradation is essential for dynamically regulating stem cell quiescence, activation and regeneration.
Several studies have demonstrated that low proteasome activity is a characteristic of cancer stem cells. Glioma and breast cancer cell lines that exhibit low levels of 26S proteasome activity exhibit higher tumorigenicity in vitro and in vivo as compared with cell lines with high proteasome activity (Vlashi et al., 2009). In lung carcinoma and colorectal cancer cell lines, low proteasome activity is also associated with a more stem cell like phenotype and high tumorigenic potential (Munakata et al., 2016; Pan et al., 2010). These studies raise the possibility that low proteasome activity may be a useful biomarker in identifying cancer stem cells.
Conclusion
Cumulative advances in whole genome, whole transcriptome and single cell RNA sequencing, in addition to murine and humanized model systems, have helped to shape our understanding of stem cell hierarchies in homeostatic, stressed and diseased states particularly in pre-malignancy and during malignant transformation. More recently, post-transcriptional (epitranscriptomic) RNA processing alterations, such as RNA editing and methylation, have been shown to alter transcript splicing, stability, and microRNA targeting that can cause recoding of transcripts or alter translational efficiency. The observation that many of these epitranscriptomic mechanisms are deregulated in cancer underscores the importance of determining their role in regulating proteome content in a cell type and tissue specific manner. Overall, transcriptomic and epitranscriptomic events that dictate alterations in translation can now be interrogated to predict stem cell function in both benign and malignant settings and can be developed as predictive biomarkers of stem cell fitness and may ultimately add new layers in the advancement and complexity of precision medicine.
As the most error-prone step in gene expression, translation not only regulates proteome content but also influences proteome quality. Highly regulated protein biogenesis and turnover has thus emerged as a primary regulator of stem cell identity, homeostasis and regenerative activity. In addition to regulating stem cell fitness, translational control also contributes to cancer progression. Disruptions in proteostasis have been linked to therapy resistant cancer stem cell generation in leukemia and are likely to play a role in invasion and metastasis of other malignancies. Altered proteasome activity may also fuel cancer stemness, and provide a therapeutic target for cancer stem cell eradication.
Ultimately, RNA processing, the epitranscriptome, translational control and protein degradation are key regulators of proteome complexity and play pivotal roles in regulating stem cell identity and function and when dysregulated contribute to cancer stem cell propagation (Table S1). The intersection of these nascent fields provides a fulcrum for developing clinically tractable methods to track stem cell fitness and cancer stem cell propagation for the benefit of patients with stem cell driven degenerative diseases and cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank E. Bennett for advice and discussions regarding the manuscript. Figures were created with BioRender.com. This research was supported by the Sanford Stem Cell Clinical Center, NIH/NIDDK (R01DK116951; RS), Scholar awards from the V Foundation for Cancer Research (RS) and the American Society of Hematology (RS) and NIH/NCI R01CA205944; NIH/NIDDK R01DK114468-01; NIH NCI 2P30CA023100-28; The Koman Family Foundation; the Strauss Family Foundation; and the Moores Family Foundation (CJ). We apologize to investigators whose studies we omitted from our discussion and references due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aaronson Y, and Meshorer E (2013). Stem cells: Regulation by alternative splicing. Nature 498, 176–177. [DOI] [PubMed] [Google Scholar]
- Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, et al. (2009). Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A 106, 3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. (2010). Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 464, 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF (2008). Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, and Tavazoie SF (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, and Bengal E (2011). Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS One 6, e29498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadzadeh Z, Mansoori B, Mohammadi A, Aghajani M, Haji-Asgarzadeh K, Safarzadeh E, Mokhtarzadeh A, Duijf PHG, and Baradaran B (2019). microRNAs in cancer stem cells: Biology, pathways, and therapeutic opportunities. J Cell Physiol 234, 10002–10017. [DOI] [PubMed] [Google Scholar]
- Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, and Andrews PW (2008). OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 26, 3068–3074. [DOI] [PubMed] [Google Scholar]
- Balacco DL, and Soller M (2019). The m(6)A Writer: Rise of a Machine for Growing Tasks. Biochemistry 58, 363–378. [DOI] [PubMed] [Google Scholar]
- Baraniak PR, and McDevitt TC (2010). Stem cell paracrine actions and tissue regeneration. Regen Med 5, 121–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, and Ruggero D (2008). Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, and Hwa T (2015). Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al. (2011). Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 474, 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby A, Jones RD, Lilley KS, McFarlane RJ, Braig HR, Allen ND, and Wakeman JA (2007). Comparative proteomic analysis reveals differential expression of Hsp25 following the directed differentiation of mouse embryonic stem cells. Biochim Biophys Acta 1773, 147–156. [DOI] [PubMed] [Google Scholar]
- Batzer MA, and Deininger PL (2002). Alu repeats and human genomic diversity. Nat Rev Genet 3, 370–379. [DOI] [PubMed] [Google Scholar]
- Bedford L, Paine S, Sheppard PW, Mayer RJ, and Roelofs J (2010). Assembly, structure, and function of the 26S proteasome. Trends Cell Biol 20, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, and Larizza L (2000). RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 9, 2297–2304. [DOI] [PubMed] [Google Scholar]
- Bellodi C, McMahon M, Contreras A, Juliano D, Kopmar N, Nakamura T, Maltby D, Burlingame A, Savage SA, Shimamura A, et al. (2013). H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep 3, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E, Rath E, Yuan D, Waldschmitt N, Khaloian S, Allgauer M, Staszewski O, Lobner EM, Schottl T, Giesbertz P, et al. (2016). Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat Commun 7, 13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, and Glover DM (2009). Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol 334, 186–197. [DOI] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. (2014). Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo j 33, 2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, and Frye M (2011). The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7, e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, et al. (2009). Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet 85, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, and Rottman FM (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. (2012). Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, Covarrubias S, Percharde M, Ku G, Diaz A, McManus MT, Burlingame AL, and Ramalho-Santos M (2018). The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell 22, 369–383.e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Signer RA, and Morrison SJ (2014). Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159, 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun K, Kim TK, Oh J, Bayarsaikhan E, Kim D, Lee MY, Pack CG, Hwang D, and Lee B (2013). Heat shock instructs hESCs to exit from the self-renewal program through negative regulation of OCT4 by SAPK/JNK and HSF1 pathway. Stem Cell Res 11, 1323–1334. [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, Pastor-Flores D, Roma LP, Renders S, Zeisberger P, et al. (2017). Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169, 807–823.e819. [DOI] [PubMed] [Google Scholar]
- Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, and Speck NA (2015). Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 17, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Gaudet JJ, Mangan JK, Chen MJ, De Obaldia ME, Oo Z, Ernst P, and Speck NA (2011). Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS One 6, e28430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, and Croce CM (2006). MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866. [DOI] [PubMed] [Google Scholar]
- Cesana M, Guo MH, Cacchiarelli D, Wahlster L, Barragan J, Doulatov S, Vo LT, Salvatori B, Trapnell C, Clement K, et al. (2018). A CLK3-HMGA2 Alternative Splicing Axis Impacts Human Hematopoietic Stem Cell Molecular Identity throughout Development. Cell Stem Cell 22, 575–588.e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, and Begley TJ (2010). A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6, e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple RH, Hu T, Tseng YJ, Liu L, Kitano A, Luu V, Hoegenauer KA, Iwawaki T, Li Q, and Nakada D (2018). ERalpha promotes murine hematopoietic regeneration through the Ire1alpha-mediated unfolded protein response. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, and Gray MW (2000). Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, and Wang DZ (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Dai X, and Wu J (2015). Alternative splicing: An important mechanism in stem cell biology. World J Stem Cells 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, and Manley JL (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, et al. (2018). Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 172, 811–824.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini NS, Peer AM, Moeseneder P, Roiuk M, Burkard TR, Theussl HC, Moll I, and Knoblich JA (2018). Coordinated Control of mRNA and rRNA Processing Controls Embryonic Stem Cell Pluripotency and Differentiation. Cell Stem Cell 22, 543–558.e512. [DOI] [PubMed] [Google Scholar]
- Craven SE, French D, Ye W, de Sauvage F, and Rosenthal A (2005). Loss of Hspa9b in zebrafish recapitulates the ineffective hematopoiesis of the myelodysplastic syndrome. Blood 105, 3528–3534. [DOI] [PubMed] [Google Scholar]
- Crews LA, Balaian L, Delos Santos NP, Leu HS, Court AC, Lazzari E, Sadarangani A, Zipeto MA, La Clair JJ, Villa R, et al. (2016). RNA Splicing Modulation Selectively Impairs Leukemia Stem Cell Maintenance in Secondary Human AML. Cell Stem Cell 19, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews LA, Jiang Q, Zipeto MA, Lazzari E, Court AC, Ali S, Barrett CL, Frazer KA, and Jamieson CH (2015). An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med 13, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, Williamson JR, Fredrick K, Wang YP, and Hwa T (2016). Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol 2, 16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, and Lin S (2008). Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112, 5228–5237. [DOI] [PubMed] [Google Scholar]
- de Morree A, Klein JDD, Gan Q, Farup J, Urtasun A, Kanugovi A, Bilen B, van Velthoven CTJ, Quarta M, and Rando TA (2019). Alternative polyadenylation of Pax3 controls muscle stem cell fate and muscle function. Science 366, 734–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zoysa MD, and Yu YT (2017). Posttranscriptional RNA Pseudouridylation. Enzymes 41, 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin EE, Dacosta L, Mohandas N, Elliott G, and Bodine DM (2010). A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood 116, 2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I (2017). Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem 86, 193–224. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, and Rechavi G (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8, 176–189. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. (1999). The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21, 169–175. [DOI] [PubMed] [Google Scholar]
- Drummond DA, and Wilke CO (2009). The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. (2009). Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35, 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farashahi Yazd E, Rafiee MR, Soleimani M, Tavallaei M, Salmani MK, and Mowla SJ (2011). OCT4B1, a novel spliced variant of OCT4, generates a stable truncated protein with a potential role in stress response. Cancer Lett 309, 170–175. [DOI] [PubMed] [Google Scholar]
- Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, Tailor J, Dietmann S, and Frye M (2017). Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Reports 8, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Ho JJ, Inouye C, and Tjian R (2014). The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Priest JR, and Duchaine TF (2014). DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14, 662–672. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. (2011). An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, and Israel MA (2007). Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67, 2456–2468. [DOI] [PubMed] [Google Scholar]
- Georgantas RW 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, and Civin CI (2007). CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A 104, 2750–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germanguz I, Shtrichman R, Osenberg S, Ziskind A, Novak A, Domev H, Laevsky I, Jacob-Hirsch J, Feiler Y, Rechavi G, et al. (2014). ADAR1 is involved in the regulation of reprogramming human fibroblasts to induced pluripotent stem cells. Stem Cells Dev 23, 443–456. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. [DOI] [PubMed] [Google Scholar]
- Gigova A, Duggimpudi S, Pollex T, Schaefer M, and Kos M (2014). A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. Rna 20, 1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff DJ, Court Recart A, Sadarangani A, Chun HJ, Barrett CL, Krajewska M, Leu H, Low-Marchelli J, Ma W, Shih AY, et al. (2013). A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell 12, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, Scadden DT, and Hu GF (2016). Angiogenin Promotes Hematopoietic Regeneration by Dichotomously Regulating Quiescence of Stem and Progenitor Cells. Cell 166, 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, Martins JP, Brook M, Sheets MD, and Gray NK (2011). Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A 108, 7844–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, and Shiekhattar R (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, and Shiekhattar R (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. [DOI] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, et al. (2005). Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M, et al. (2018). Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 173, 1204–1216.e1226. [DOI] [PubMed] [Google Scholar]
- Hamma T, and Ferre-D’Amare AR (2006). Pseudouridine synthases. Chem Biol 13, 1125–1135. [DOI] [PubMed] [Google Scholar]
- Hammond SM (2005). Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579, 5822–5829. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. (2013). MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HMJ, Eterovic AK, Yuan Y, et al. (2015). The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 28, 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, and Bennett EJ (2016). Proteome complexity and the forces that drive proteome imbalance. Nature 537, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, and Bordey A (2013). mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep 5, 433–444. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, and Orkin SH (2009). ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M, Ferrante M, Lee AS, Onderwater JJ, Paton JC, et al. (2013). ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 3, 1128–1139. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Sonenberg N, and Mathews MB (2018). Principles of Translational Control. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo San Jose L, Sunshine MJ, Dillingham CH, Chua BA, Kruta M, Hong Y, Hatters DM, and Signer RAJ (2020). Modest Declines in Proteome Quality Impair Hematopoietic Stem Cell Self-Renewal. Cell Rep 30, 69–80.e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, and Seeburg PH (2000). Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2017). Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem Sci 42, 589–611. [DOI] [PubMed] [Google Scholar]
- Holm F, Hellqvist E, Mason CN, Ali SA, Delos-Santos N, Barrett CL, Chun HJ, Minden MD, Moore RA, Marra MA, et al. (2015). Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc Natl Acad Sci U S A 112, 15444–15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YE, Bahn JH, Yang Y, Lin X, Tran S, Yang EW, Quinones-Valdez G, and Xiao X (2018). RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res 28, 812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Silverman JM, Chen SS, Erickson DW, Basan M, Wang J, Hwa T, and Williamson JR (2015). Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol Syst Biol 11, 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, and Frye M (2013). The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliou MS, da Silva-Diz V, Carmona FJ, Ramalho-Carvalho J, Heyn H, Villanueva A, Munoz P, and Esteller M (2014). Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene 33, 4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, Miller BC, Du PP, Yates KB, Dubrot J, et al. (2019). Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, and Anderson P (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, Henson A, Ellis S, Schambach A, Baum C, Richter J, et al. (2011). Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood 118, 6087–6096. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, and Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MM, Fogg RL, Emeson RB, and Stanwood GD (2009). ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev Neurosci 31, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J, Wang Y, Kim HS, Lalli MA, and Kosik KS (2014). Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells 32, 2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzebowski L, Le Bouteiller M, Coqueran S, Raveux A, Vandormael-Pournin S, David A, Cumano A, and Cohen-Tannoudji M (2018). Mouse adult hematopoietic stem cells actively synthesize ribosomal RNA. Rna 24, 1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Isquith J, Zipeto MA, Diep RH, Pham J, Delos Santos N, Reynoso E, Chau J, Leu H, Lazzari E, et al. (2019). Hyper-Editing of Cell-Cycle Regulatory and Tumor Suppressor RNA Promotes Malignant Progenitor Propagation. Cancer Cell 35, 81–94.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Rooney MS, Mertins P, Przybylski D, Chevrier N, Satija R, Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, et al. (2015). Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 347, 1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, and Cooper TA (2011). Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, and Rajewsky K (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19, 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel SB, Phelps S, Sabo KM, O’Leary MN, Kirn-Safran CB, and Abkowitz JL (2012). Establishing Rps6 hemizygous mice as a model for studying how ribosomal protein haploinsufficiency impairs erythropoiesis. Exp Hematol 40, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M, Clark A, Svoboda DS, Azzi J, MacLaurin JG, Meghaizel C, Sesaki H, Lagace DC, Germain M, Harper ME, et al. (2016). Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell 19, 232–247. [DOI] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. (2018). Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, and Cairns BR (2013). Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goren A, and Ast G (2008). Alternative splicing: current perspectives. Bioessays 30, 38–47. [DOI] [PubMed] [Google Scholar]
- King B, Boccalatte F, Moran-Crusio K, Wolf E, Wang J, Kayembe C, Lazaris C, Yu X, Aranda-Orgilles B, Lasorella A, et al. (2016). The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat Immunol 17, 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima Y, Suzuki N, Nunomiya A, Osana S, Yoshioka K, Tashiro Y, Takahashi R, Ono Y, Aoki M, and Nagatomi R (2018). The Ubiquitin-Proteasome System Is Indispensable for the Maintenance of Muscle Stem Cells. Stem Cell Reports 11, 1523–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, and Hwa T (2014). Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Curr Opin Biotechnol 28, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Park JH, Lee AR, Kim E, Jiyoung K, Kawasaki I, and Shim YH (2010). Two mutations in pab-1 encoding poly(A)-binding protein show similar defects in germline stem cell proliferation but different longevity in C. elegans. Mol Cells 30, 167–172. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Lazaris C, Hockemeyer K, Balandran JC, Jimenez AR, Mullenders J, Gong Y, Trimarchi T, Bhatt K, Hu H, et al. (2018). Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat Med 24, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger H, Grimsey N, Paxman R, Chiang WC, Plate L, Jones Y, Shaw PX, Trejo J, Tsang SH, Powers E, et al. (2018). The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, and Kim VN (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20, 1122–1130. [DOI] [PubMed] [Google Scholar]
- Laplante M, and Sabatini DM (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouteiller M, Souilhol C, Beck-Cormier S, Stedman A, Burlen-Defranoux O, Vandormael-Pournin S, Bernex F, Cumano A, and Cohen-Tannoudji M (2013). Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med 210, 2351–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, Hanna JH, and Ding L (2019). Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol 21, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al. (2017a). Base-Resolution Mapping Reveals Distinct m(1)A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol Cell 68, 993–1005.e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. (2017b). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, and Rana TM (2011). Small RNA-mediated regulation of iPS cell generation. Embo j 30, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, and Darnell RB (2012). Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev 26, 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, and Ying SY (2008). Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. Rna 14, 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhao M, Jin X, Ney G, Yang KB, Peng F, Cao J, Iwawaki T, Del Valle J, Chen X, et al. (2019). Adaptive endoplasmic reticulum stress signalling via IRE1alpha-XBP1 preserves self-renewal of haematopoietic and pre-leukaemic stem cells. Nat Cell Biol 21, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, and Pan T (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, and Martin-Villalba A (2015). Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329–340. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, and Vousden KH (2003). Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587. [DOI] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, and Wilkinson MF (2014). Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 6, 748–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834–838. [DOI] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. (2014). Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell 15, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, et al. (2014). Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med 20, 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang L, Fan S, Fan P, Hou Z, Li J, et al. (2015). Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget 6, 10432–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, and Blenis J (2009). Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10, 307–318. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Ahn JH, Cipriani PG, Killian DJ, Chaudhary AL, Lee JI, Voutev R, Johnsen RC, Baillie DL, Gunsalus KC, et al. (2005). Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169, 1997–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas JC, and Levine AJ (1994). The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol 14, 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, Forsberg E, Karlsson S, and Dahl N (2004). Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol 24, 4032–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson H, Davey EJ, Frojmark AS, Miyake K, Utsugisawa T, Flygare J, Zahou E, Byman I, Landin B, Ronquist G, et al. (2006). Erythropoiesis in the Rps19 disrupted mouse: Analysis of erythropoietin response and biochemical markers for Diamond-Blackfan anemia. Blood Cells Mol Dis 36, 259–264. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. (2008). Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet 40, 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellis IA, Gupte R, Raj A, and Rouhanifard SH (2017). Visualizing adenosine-to-inosine RNA editing in single mammalian cells. Nat Methods 14, 801–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, and Chen D (2015). Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Widjaja A, Liu Y, Luo H, and Chen D (2018). The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell 17, e12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie LB, and Aifantis I (2012). Regulation of hematopoietic stem cell fate by the ubiquitin proteasome system. Trends Immunol 33, 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Hwa T, Martin OC, De Martino A, and Marinari E (2016). Constrained Allocation Flux Balance Analysis. PLoS Comput Biol 12, e1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata K, Uemura M, Tanaka S, Kawai K, Kitahara T, Miyo M, Kano Y, Nishikawa S, Fukusumi T, Takahashi Y, et al. (2016). Cancer Stem-like Properties in Colorectal Cancer Cells with Low Proteasome Activity. Clin Cancer Res 22, 5277–5286. [DOI] [PubMed] [Google Scholar]
- Narla A, and Ebert BL (2010). Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreiter L, Fritz TM, Adolph TE, Krismer AM, Offner FA, Tschurtschenthaler M, Flak MB, Hosomi S, Tomczak MF, Kaneider NC, et al. (2013). ER stress transcription factor Xbp1 suppresses intestinal tumorigenesis and directs intestinal stem cells. J Exp Med 210, 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, and Graveley BR (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes E, Anderson A, Cohen-Gadol A, and Hundley HA (2017). Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma. J Biol Chem 292, 4326–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S (2014). Splicing factor mutations in AML. Blood 123, 3216–3217. [DOI] [PubMed] [Google Scholar]
- Ohta S, Nishida E, Yamanaka S, and Yamamoto T (2013). Global splicing pattern reversion during somatic cell reprogramming. Cell Rep 5, 357–366. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, and Glaser T (2004). Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, and Zon LI (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Dominissini D, Rechavi G, and Eisenberg E (2009). Widespread cleavage of A-to-I hyperediting substrates. Rna 15, 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Zhang Q, Wang Y, and You M (2010). 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One 5, e13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, and Daley GQ (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146. [DOI] [PubMed] [Google Scholar]
- Peer E, Moshitch-Moshkovitz S, Rechavi G, and Dominissini D (2018). The Epitranscriptome in Translation Regulation. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena C, Hurt E, and Panse VG (2017). Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 24, 689–699. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. (2012). Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30, 253–260. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Wang X, Buchanan M, He K, Sharma R, Zhang L, Wang Q, and Yu J (2013). ADAR1 is essential for intestinal homeostasis and stem cell maintenance. Cell Death Dis 4, e599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, and Cairns BR (2007). Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 21, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, and Orkin SH (2010). Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol 30, 5364–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavie L, Della Gatta G, Crusio K, Aranda-Orgilles B, Buckley SM, Thompson B, Lee E, Gao J, Bredemeyer AL, Helmink BA, et al. (2010). Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol 11, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeil JA, Zermati Y, Vandekerckhove J, Cathelin S, Kersual J, Dussiot M, Coulon S, Moura IC, Zeuner A, Kirkegaard-Sorensen T, et al. (2007). Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature 445, 102–105. [DOI] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. (2012). Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet 44, 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, and Buchner J (2010). The heat shock response: life on the verge of death. Mol Cell 40, 253–266. [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, and Emeson RB (1999). Regulation of alternative splicing by RNA editing. Nature 399, 75–80. [DOI] [PubMed] [Google Scholar]
- Saez I, Koyuncu S, Gutierrez-Garcia R, Dieterich C, and Vilchez D (2018). Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci Rep 8, 4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, and Schwartz S (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255. [DOI] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, and Murry CE (2008). A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, and Lehmann R (2016). Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 18, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, et al. (2012). Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest 122, 2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G, Armstrong L, Leake A, Lako M, and von Zglinicki T (2004). Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells 22, 962–971. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, and Lyko F (2009). RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. (2015). Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6, 6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, and Green R (2018). Roadblocks and resolutions in eukaryotic translation. Nat Rev Mol Cell Biol 19, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, and Hwa T (2010). Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Scott M, Klumpp S, Mateescu EM, and Hwa T (2014). Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol 10, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyen E, Singh B, Minana B, Pages A, Mateo F, Pujana MA, Valcarcel J, and Eyras E (2016). Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res 26, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. (2017). Translation from unconventional 5’ start sites drives tumour initiation. Nature 541, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kotter P, and Entian KD (2013). Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 41, 9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MY, and Qian SB (2013). Less is more: improving proteostasis by translation slow down. Trends Biochem Sci 38, 585–591. [DOI] [PubMed] [Google Scholar]
- Shtrichman R, Germanguz I, and Itskovitz-Eldor J (2013). Induced pluripotent stem cells (iPSCs) derived from different cell sources and their potential for regenerative and personalized medicine. Curr Mol Med 13, 792–805. [DOI] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Qi L, Zhao Z, Thompson D, Sigova AA, Fan ZP, DeMartino GN, Young RA, Sonenberg N, and Morrison SJ (2016). The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev 30, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic MS, Moehle EA, Schinzel RT, Lorbeer FK, Halloran JJ, Heydari K, Sanchez M, Jullie D, Hockemeyer D, and Dillin A (2019). Transient activation of the UPR(ER) is an essential step in the acquisition of pluripotency during reprogramming. Sci Adv 5, eaaw0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin W, and Nishikura K (2013). Adenosine-to-inosine RNA editing and human disease. Genome Med 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, and Wulczyn FG (2005). Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21, 1469–1477. [DOI] [PubMed] [Google Scholar]
- Solomon O, Di Segni A, Cesarkas K, Porath HT, Marcu-Malina V, Mizrahi O, Stern-Ginossar N, Kol N, Farage-Barhom S, Glick-Saar E, et al. (2017). RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat Commun 8, 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon O, Oren S, Safran M, Deshet-Unger N, Akiva P, Jacob-Hirsch J, Cesarkas K, Kabesa R, Amariglio N, Unger R, et al. (2013). Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). Rna 19, 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, and Preiss T (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H, Takahashi K, Yamamoto T, Iwasaki M, Narita M, Nakamura M, Rand TA, Nakagawa M, Watanabe A, and Yamanaka S (2017). Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc Natl Acad Sci U S A 114, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Suzuki A, Kishikawa M, Akutsu R, Hirose T, Waye MM, Tsui SK, Yoshida S, and Ohno S (2000). Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. J Biol Chem 275, 1095–1104. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Suzuki T, Inagaki K, Ito S, Kono M, Fukai K, Takama H, Sato K, Ishikawa O, Abe M, et al. (2005). Mutation analysis of the ADAR1 gene in dyschromatosis symmetrica hereditaria and genetic differentiation from both dyschromatosis universalis hereditaria and acropigmentatio reticularis. J Invest Dermatol 124, 1186–1192. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, and Dillon N (2006). The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell 127, 1375–1388. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, et al. (2014). Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14, 606–616. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos-Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li W, Atlasi Y, Caron M, et al. (2016). Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Natl Acad Sci U S A 113, 12360–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tam AB, Koong AC, and Niwa M (2014). Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep 9, 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. (2017). Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, and Rigoutsos I (2008). MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, and Zon LI (2012). Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol 40, 228–237.e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, and Dillin A (2011). Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Clark F, and Muilu J (2003). Conservation of human alternative splice events in mouse. Nucleic Acids Res 31, 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, and Aifantis I (2008). Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med 205, 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, and Ruggero D (2015). Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 162, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. Embo j 34, 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, and Kenmochi N (2008). Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet 17, 3204–3211. [DOI] [PubMed] [Google Scholar]
- van den Berg PR, Budnik B, Slavov N, and Semrau S (2017). Dynamic post-transcriptional regulation during embryonic stem cell differentiation. bioRxiv, 123497. [Google Scholar]
- van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, Xie S, Laurenti E, Hermans K, Eppert K, et al. (2014). The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272. [DOI] [PubMed] [Google Scholar]
- van Galen P, Mbong N, Kreso A, Schoof EM, Wagenblast E, Ng SWK, Krivdova G, Jin L, Nakauchi H, and Dick JE (2018). Integrated Stress Response Activity Marks Stem Cells in Normal Hematopoiesis and Leukemia. Cell Rep 25, 1109–1117.e1105. [DOI] [PubMed] [Google Scholar]
- Vandewynckel YP, Laukens D, Geerts A, Bogaerts E, Paridaens A, Verhelst X, Janssens S, Heindryckx F, and Van Vlierberghe H (2013). The paradox of the unfolded protein response in cancer. Anticancer Res 33, 4683–4694. [PubMed] [Google Scholar]
- Vannini N, Campos V, Girotra M, Trachsel V, Rojas-Sutterlin S, Tratwal J, Ragusa S, Stefanidis E, Ryu D, Rainer PY, et al. (2019). The NAD-Booster Nicotinamide Riboside Potently Stimulates Hematopoiesis through Increased Mitochondrial Clearance. Cell Stem Cell 24, 405–418.e407. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. (2012). Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, and Lee SK (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21, 744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M, Sayre JW, Stefani E, McBride W, and Pajonk F (2009). In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst 101, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, and Marcotte EM (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23, 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, and Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang L, Zeng X, Ryoo HD, and Jasper H (2014a). Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet 10, e1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Karamariti E, Simpson R, Wang W, and Xu Q (2015a). Dickkopf Homolog 3 Induces Stem Cell Differentiation into Smooth Muscle Lineage via ATF6 Signalling. J Biol Chem 290, 19844–19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Z, Kong B, Song C, Cong J, Hou J, and Wang S (2017a). Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget 8, 98918–98930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014b). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015b). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Jiang FZ, Tong H, Ke JQ, Li YR, Zhang HL, Yan XF, Wang FY, and Wan XP (2017b). Dicer1 dysfunction promotes stemness and aggression in endometrial carcinoma. Tumour Biol 39, 1010428317695967. [DOI] [PubMed] [Google Scholar]
- Wang XW, Hao J, Guo WT, Liao LQ, Huang SY, Guo X, Bao X, Esteban MA, and Wang Y (2017c). A DGCR8-Independent Stable MicroRNA Expression Strategy Reveals Important Functions of miR-290 and miR-183–182 Families in Mouse Embryonic Stem Cells. Stem Cell Reports 9, 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, and Zhao JC (2014c). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, and Blelloch R (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39, 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 22, 191–205.e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney NP, Peng H, Erdmann NB, Tian C, Monaghan DT, and Zheng JC (2008). Calcium-permeable AMPA receptors containing Q/R-unedited GluR2 direct human neural progenitor cell differentiation to neurons. Faseb j 22, 2888–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielenga MCB, Colak S, Heijmans J, van Lidth de Jeude JF, Rodermond HM, Paton JC, Paton AW, Vermeulen L, Medema JP, and van den Brink GR (2015). ER-Stress-Induced Differentiation Sensitizes Colon Cancer Stem Cells to Chemotherapy. Cell Rep 13, 489–494. [DOI] [PubMed] [Google Scholar]
- Williamson AJ, Smith DL, Blinco D, Unwin RD, Pearson S, Wilson C, Miller C, Lancashire L, Lacaud G, Kouskoff V, et al. (2008). Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol Cell Proteomics 7, 459–472. [DOI] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, and Trumpp A (2004). c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18, 2747–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, Li J, Sheng R, Deng P, Wang Y, et al. (2018). Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun 9, 4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- Xu H, Tsang KS, Wang Y, Chan JC, Xu G, and Gao WQ (2014). Unfolded protein response is required for the definitive endodermal specification of mouse embryonic stem cells via Smad2 and beta-catenin signaling. J Biol Chem 289, 26290–26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, and Jin Y (2009a). WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res 19, 561–573. [DOI] [PubMed] [Google Scholar]
- Xu HM, Liao B, Zhang QJ, Wang BB, Li H, Zhong XM, Sheng HZ, Zhao YX, Zhao YM, and Jin Y (2004). Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem 279, 23495–23503. [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, and Kosik KS (2009b). MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658. [DOI] [PubMed] [Google Scholar]
- Xu Y, Poggio M, Jin HY, Shi Z, Forester CM, Wang Y, Stumpf CR, Xue L, Devericks E, So L, et al. (2019). Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med 25, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, and Barna M (2015). Cis-regulatory RNA elements that regulate specialized ribosome activity. RNA Biol 12, 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, Yang Q, Wang Q, and Cheng T (2009). ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A 106, 17763–17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV Jr., Iwao H, and Innerarity TL (2000). Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. Embo j 19, 5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, and Kwak S (2019). Cell death cascade and molecular therapy in ADAR2-deficient motor neurons of ALS. Neurosci Res 144, 4–13. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Liu L, Lazarev D, Al-Zain A, Fomin V, Yeung PL, Chambers SM, Lu CW, Studer L, and Manley JL (2018). TCF3 alternative splicing controlled by hnRNP H/F regulates E-cadherin expression and hESC pluripotency. Genes Dev 32, 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, and Burge CB (2005). Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A 102, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, and Gage FH (2007). Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol 3, 1951–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, and Morrison SJ (2006). Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441, 475–482. [DOI] [PubMed] [Google Scholar]
- Yoffe Y, David M, Kalaora R, Povodovski L, Friedlander G, Feldmesser E, Ainbinder E, Saada A, Bialik S, and Kimchi A (2016). Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev 30, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, and Ogawa S (2014). Splicing factor mutations and cancer. Wiley Interdiscip Rev RNA 5, 445–459. [DOI] [PubMed] [Google Scholar]
- You KT, Park J, and Kim VN (2015). Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29, 2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. (2017). m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276. [DOI] [PubMed] [Google Scholar]
- Zhang C, and Jia G (2018). Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinformatics 16, 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, and Semenza GL (2016a). Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, et al. (2019). CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev 33, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. (2016b). NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. (2006). PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441, 518–522. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, and Buszczak M (2014). Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Marsboom G, Toth PT, and Rehman J (2013). Mitochondrial respiration regulates adipogenic differentiation of human mesenchymal stem cells. PLoS One 8, e77077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, and Xiong Y (2003). Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23, 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu X, He Z, Niu X, Shi W, Ding JM, Zhang L, Yuan T, Li A, Yang W, et al. (2016). Essential role of proteasomes in maintaining self-renewal in neural progenitor cells. Sci Rep 6, 19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, and Shabek N (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 86, 129–157. [DOI] [PubMed] [Google Scholar]
- Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, Pineda G, Morris SR, Mason CN, Geron I, et al. (2016). ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell 19, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipeto MA, Jiang Q, Melese E, and Jamieson CH (2015). RNA rewriting, recoding, and rewiring in human disease. Trends Mol Med 21, 549–559. [DOI] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, and Crist C (2016). Phosphorylation of eIF2alpha Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell 18, 79–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


