Abstract
LgDel mice, which model the heterozygous deletion of genes at human chromosome 22q11.2 associated with DiGeorge/22q11.2 deletion syndrome (22q11DS), have cranial nerve and craniofacial dysfunction as well as disrupted suckling, feeding and swallowing, similar to key 22q11DS phenotypes. Divergent trigeminal nerve (CN V) differentiation and altered trigeminal ganglion (CNgV) cellular composition prefigure these disruptions in LgDel embryos. We therefore asked whether a distinct transcriptional state in a specific population of early differentiating LgDel cranial sensory neurons, those in CNgV, a major source of innervation for appropriate oropharyngeal function, underlies this departure from typical development. LgDel versus wild-type (WT) CNgV transcriptomes differ significantly at E10.5 just after the ganglion has coalesced. Some changes parallel altered proportions of cranial placode versus cranial neural crest-derived CNgV cells. Others are consistent with a shift in anterior–posterior patterning associated with divergent LgDel cranial nerve differentiation. The most robust quantitative distinction, however, is statistically verifiable increased variability of expression levels for most of the over 17 000 genes expressed in common in LgDel versus WT CNgV. Thus, quantitative expression changes of functionally relevant genes and increased stochastic variation across the entire CNgV transcriptome at the onset of CN V differentiation prefigure subsequent disruption of cranial nerve differentiation and oropharyngeal function in LgDel mice.
Introduction
The potential relationship between transcriptional differences, anomalous neural circuit development and behavioral deficits has been explored for a number of neurodevelopmental and psychiatric disorders in older children and adults (1). Less attention, however, has been given to transcriptional distinctions that arise early in embryogenesis, perhaps modifying circuits for behaviors that must be in place by birth to ensure survival, growth and health. These behaviors include suckling, feeding and swallowing (S/F/S), whose disruption—perinatal dysphagia—results in substantial health challenges from birth onward, especially in infants and children with a broad range of developmental syndromes (2). We found that S/F/S is compromised in the LgDel mouse model of DiGeorge/22q11.2 deletion syndrome (22q11DS), a common genetic disorder (1/3000 live births) (3–5) with an enhanced incidence of perinatal dysphagia (5–7). We now ask whether transcriptional divergence during initial differentiation of key neurons and precursors that facilitate optimal S/F/S—those in the trigeminal ganglion (CNgV) (8) prefigures altered cranial nerve development and function in LgDel mice.
CNgV provides sensory innervation to the face, lips, oral cavity and anterior tongue critical for initiation of S/F/S and thus must develop appropriately prior to birth (5). CNgV, like most cranial ganglia, consists of mechanosensory neurons derived from neurogenic cranial placodes and nociceptive neurons derived from hindbrain neural crest (9). Our previous observations (6) show that patterning of the hindbrain neural crest, which gives rise to CNgV nociceptive neurons, is disrupted by the presumed 50% decrease in 22q11 gene dosage by E10.5. This disruption, as well as expression of several 22q11 orthologues in CNgV, and potential for altered interactions between distinct cell classes within and surrounding the ganglion (6,10), all suggest that the LgDel CNgV transcriptional state may differ from wild-type (WT). Such early differences in LgDel, and potentially in 22q11DS, may ultimately compromise sensory neuron identity and/or disrupt sensory innervation and function, resulting in sub-optimal CNgV-related oropharyngeal behaviors including dysphagia. Accordingly, we used RNA-Seq to define transcriptomes of microdissected LgDel and WT CNgV at embryonic day (E) 10.5, just after the ganglion coalesces (6,11), sensory neurogenesis accelerates and axons begin to grow. We analyzed multiple, pooled biological replicates from each genotype (12) to minimize contributions of individual variability, eliminate statistical ‘noise’ and securely identify expression differences due to 22q11 gene deletion with appropriate statistical power in a critical population of cranial sensory neurons.
We found that LgDel and WT E10.5 CNgV transcriptomes differ in parallel with differences in CNgV cellular composition in the two genotypes. These differences are distinct from those in transcriptomes of E10.5 LgDel or WT whole embryos, emphasizing the value of microdissection to accurately define transcriptional landscapes of developmentally critical cell populations. Differential expression levels of genes that may regulate neuronal differentiation suggest potential mechanisms that can lead to disrupted cranial nerve differentiation and function in LgDel mouse pups. The most striking quantitative distinction, however, is statistically verifiable increased expression variability across the entire LgDel CNgV transcriptome. Apparently, a stochastically variable transcriptome is a hallmark of LgDel cranial sensory neurons at the embryonic stage when cranial neural circuit development essential for optimal S/F/S diverges from the WT differentiation program.
Results
Developmental phenotypes suggest divergent transcriptomes for WT versus LgDel CNgV
We showed previously that CN V is dysmorphic in LgDel E10.5 embryos compared to WT (6). To visualize CNgV itself, we immunostained whole E10.5 embryos for βIII-tubulin, which detects early differentiating neurons as well as axons (Fig. 1A and B) (13). CNgV is dysmorphic in LgDel E10.5 embryos; its size and shape are altered, and the extension of axon fascicles from all three divisions (ophthalmic: op; maxillary: mx; mandibular: md; Fig. 1A and B, and inset) is aberrant. To determine if CNgV cellular composition changes similarly at E10.5, we quantified presumed placode derived (Six1 immunolabeled) and neural crest (Wnt1:Cre recombined, eGFP reporter-labeled) associated populations (Fig. 1C and D) (11). There is a decreased proportion of Wnt1:Cre/eGFP labeled cells in LgDel CNgV (29% Wnt1:Cre/eGFP in LgDel; 35% Wnt1:Cre/eGFP in WT; P < 0.02), as well as a corresponding increase in the proportion of Six1 labeled cells (34% Six1 in LgDel; 26% Six1 in WT; P < 0.02; Fig. 1E). Moreover, a non-Wnt1:Cre recombined, non-Six1 expressing, DAPI-labeled population, associated with Foxd3-expressing neural crest (11), also declines, similar to the Wnt1:Cre/eGFP population (21% in LgDel; 29% in WT; P < 0.04). The developmental origin of these changes in CNgV may reflect interactions with surrounding neural crest-derived mesenchyme (14,15), which in LgDel embryos may also be distinct due to its origin from ‘posteriorized’ hindbrain rhombomeres (see below) or hindbrain target cells as axons extend centrally. Nevertheless, CNgV differentiation diverges in LgDel versus WT by the time the ganglion has fully coalesced at E10.5.
Figure 1.
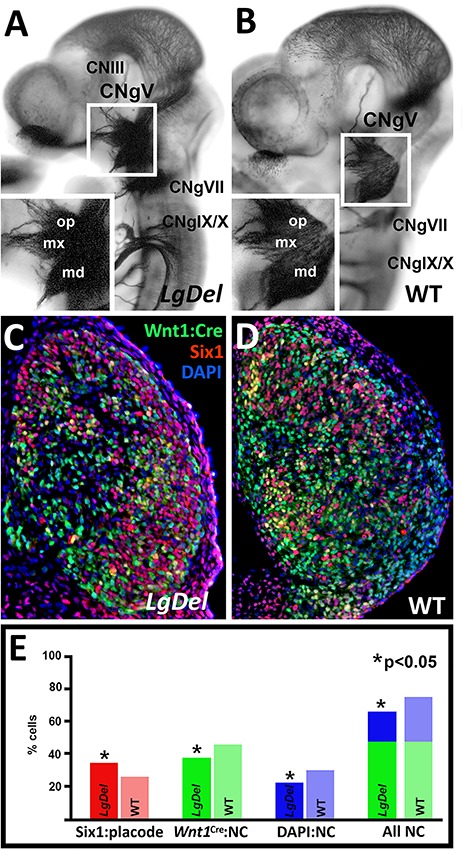
Divergent trigeminal nerve (CN V) differentiation and trigeminal ganglion (CNgV) cellular composition in the LgDel mouse model of 22q11.2 deletion syndrome (22q11DS). (A) A E10.5 LgDel embryo labeled for the early neuronal and axon marker βIII-tubulin, in which CN V axon fascicles extending peripherally from CNgV are sparse and dysmorphic. Inset: The differentiation of all three subdivisions of CNgV—op, mx and md—is compromised in concert with altered CN V axon growth. (B) A WT E10.5 embryo, the same gestational age as the LgDel in (A), in which there is robust, directed, fasciculated growth of CN V axons from a well-differentiated CNgV. Inset: The three subdivisions of CNgV and their associated axons are well defined, compared to the LgDel counterpart (A). (C) A transverse section through the LgDel E10.5 CNgV labeled immunohistochemically using Six1 (red), which distinguishes cranial placode-associated CNgV cells, Wnt1:Cre recombination lineage tracing (leading to constitutive eGFP reporter expression) for a subpopulation of CNgV neural crest cells (green); DAPI, which we have shown previously (11), identifies a second sub-population of primarily Foxd3-expressing neural crest cells (blue). There is a small population of cells identified by both Wnt1:Cre recombination and Six1 immunolabeling. Those cells have been excluded from the quantitative analysis summarized in the histogram in (D). (D) A section through the E10.5 WT CNgV, with cell classes labeled as described for (C). (E) Quantitative assessment of frequency of Six1 in LgDel (darker red) and WT (lighter red), Wnt1:Cre associated neural crest (LgDel: darker green, WT: lighter green), and DAPI presumed Foxd3 neural crest cells (LgDel: darker blue; WT: lighter blue); asterisks indicate significant differences, P < 0.05; (n = 11 WT, 9 LgDel ganglia from 8 WT, 5 LgDel embryos).
LgDel and WT CNgV transcriptome differences
Distinctions between LgDel and WT CNgV cellular composition suggest that altered 22q11.2 gene dosage due to heterozygous deletion may result in divergent transcriptional states at the earliest stages of gangliogenesis, leading to anomalous CNgV cell identity and differentiation. CNgV begins to form via local delamination and translocation of cranial ectodermal placode cells as well as arrival and accumulation of a small number of neural crest cells around E9.5 in the mouse; however, it is not fully defined cytologically until E10.5 (11). Thus, we microdissected and pooled CNgV from LgDel and WT E10.5 embryos and then sequenced and analyzed transcriptomes (n = 5 CNgV pools/genotype, 6 ganglia/pool from at least 3 different embryos/3 different litters; 1 pool = 1 biological replicate), the earliest stage when the ganglion can be confidently identified and isolated accurately from the whole embryo (Fig. 2A and B).
Figure 2.
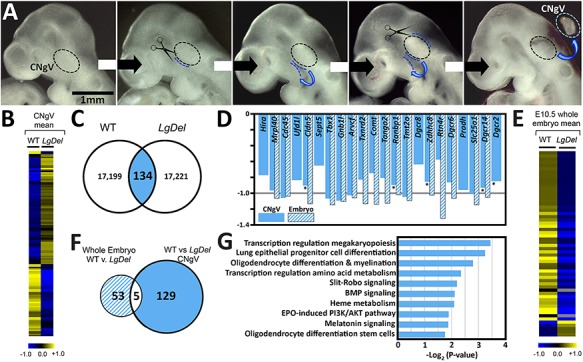
Quantitative characteristics of transcriptome divergence in LgDel and WT CNgV. (A) Images of E10.5 mouse embryos showing the step-wise microdissection approach used to harvest WT and LgDel CNgV. The dotted oval indicates the location of CNgV, the dotted blue lines indicate the incisions made by fine op dissecting scissors to isolate the ganglion. An isolated ganglion is shown at far right (dotted oval). (B) A heat map showing mean expression differences between WT and LgDel CNgV based on RNA-Seq transcriptome analysis of five biological replicates. The scale bar below the heat map (−1.0 to +1.0) is given on a log2 scale. (C) Venn diagram showing the distribution of differentially expressed genes in the WT versus LgDel CNgV. There are 134 genes with greater expression in either LgDel versus WT or WT versus LgDel. In addition, there are 9 transcripts in WT and 10 in LgDel that are apparently uniquely expressed. Each of these unique transcripts, in either genotype, is expressed at very low frequency and encodes either non-coding RNAs or other non-transcribed mRNAs. (D) Histogram showing detection (non-zero FPKM) and approximately 50% decreased expression levels (2-fold change) of 21 murine orthologues of human chromosome 22q11.2 genes deleted heterozygously in the LgDel CNgV (solid bars) and 15 of those orthologues detected non-zero values in the whole E10.5 embryo at a similar level of decreased expression (hatched bars). Of these genes, only five reach our criteria for expression and FDR levels in CNgV (asterisks). (E) A heat map showing average expression differences between whole E10.5 WT and LgDel whole embryos. The scale bar below the heat map (−1.0 to +1.0) is given on a log2 scale. (F) Venn diagram showing the minimal overlap of differentially expressed genes from the 134 identified in the comparison of WT versus LgDel CNgV and the 58 identified in the comparison of WT versus LgDel whole E10.5 embryos. There are only five of these genes, and four of them are 22q11 gene orthologues deleted heterozygously in the LgDel (see C). (G) A listing of the major GO terms associated with the RNA-Seq datasets for WT and LgDel CNgV.
This analysis of early developing WT versus LgDel CNgV sensory neurons and precursors identified 17 190 unique genes with non-zero FPKM values shared by both genotypes (Supplemental Table 2). There are 17 199 non-zero reads in WT and 17 221 non-zero reads in LgDel. Equally robust detection of similar transcripts in both genotypes indicates that E10.5 LgDel CNgV cells are transcriptionally active at levels approximating WT. Analysis of LgDel CNgV versus WT showed similar number of up- and downregulated genes (8677 versus 8553; Fig. 2B). From the total set of expressed CNgV genes, we found 134 genes differentially expressed at FDR q < 0.1 in LgDel versus WT CNgV (Fig. 2C and Supplemental Table 2). Of these 134 genes, 77 are upregulated and 57 downregulated in LgDel versus WT CNgV. The murine orthologues of the 22q11-deleted genes should be among the 134 differentially expressed genes, detected at an approximately 2-fold (i.e. 50%) decreased level (16) in LgDel CNgV. In CNgV, we detected 22 of the 28 murine 22q11 orthologues (17) with FPKMs greater than 1 for all but 1, Rtn4, a transmembrane protein involved in neurite outgrowth (18). Expression of these 22q11 genes in LgDel CNgV decreases by approximately 2-fold as expected (Fig. 2C, solid bars); however, the FDR of only five: Cldn5, Dgcr2, Dgcr14, Ranbp1 and Zdhhc8 met the q < 0.1 criterion of significance (asterisks, Fig. 2D).
To evaluate whether our microdissected CNgV samples enhance identification of transcriptome differences, we compared our CNgV results with RNA-Seq-measured transcriptomes of whole E10.5 WT and LgDel embryos (Fig. 2E). This comparison evaluates selectivity of the microdissection technique for CNgV enriched genes, and potential specificity of the CNgV dataset for detecting transcriptome changes associated with CNgV sensory progenitor, neuronal and glial differentiation rather than those broadly associated with 22q11 deletion at this stage of development. In transcriptomes from two replicates of four whole E10.5 embryos of each genotype, we found a total of 58 differentially expressed genes using the same significance threshold (FDR q < 0.1; Fig. 2F and Supplemental Table 3). Within these 58 differentially expressed genes, 17 are orthologues of the 22q11-deleted genes, and all decline in LgDel by approximately 2-fold (Fig. 2D, hatched bars). Thus, both datasets—E10.5 CNgV and E10.5 whole embryo—accurately represent the fundamental genotypic distinction: 50% (2-fold) decreased expression of murine 22q11 gene orthologues in LgDel. Of the 58 differentially expressed genes in the whole embryo transcriptome comparison, only 5 coincide with those identified as differentially expressed in CNgV transcriptomes (Fig. 2F). Four are 22q11 genes: Cldn5, Dgcr14, Ranbp1 and Zdhhc8. The fifth is Hsd3b6, which is expressed at relatively low levels (mean FPKM, WT: 2.1; LgDel:6.3), and apparently upregulated in LgDel versus WT CNgV and downregulated in LgDel versus WT E10.5 whole embryos. Hsd3b6 encodes hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 6 known to be localized to adrenal cortical cells (19). The minimal overlap of differentially expressed transcripts in CNgV with those in whole embryos confirms that RNA-Seq data obtained from the micro-dissected CNgV samples reflect its constituent cell classes and thus describe transcriptional states of cranial sensory neurons in the two genotypes.
Finally, we assessed the functional identities of LgDel versus WT CNgV differentially expressed genes. Half of the differentially expressed CNgV genes (84 of 134) could be assigned a known molecular and/or biological function based on Gene Ontology (GO, http://www.geneontology.org; Fig. 2G). The differentially expressed genes of known function were significantly over-represented (P < 0.05) in a number of broad cell-signaling and metabolic pathways, including several that might contribute to sensory neuron differentiation. These include Slit/Robo signaling (20–22), BMP signaling (23,24), oligodendrocyte myelination (25) and stem cell differentiation (Fig. 2G). Thus, a large but fairly well-defined set of differentially expressed genes, many potentially belonging to functional pathways associated with developmentally regulated signaling and neural differentiation processes, distinguishes the transcriptomes of LgDel versus WT CNgV.
Reliable detection of CNgV-associated gene expression
To assess the precision and reliability of our CNgV RNA-Seq data, we confirmed expression of several genes based on limited or enhanced CNgV localization of their encoded protein and then queried the RNA-Seq dataset for mRNA detection of the same genes. Hox1b protein is barely detected in CNgV but robustly and selectively localized in the adjacent hindbrain (Fig. 3A, left). There was no obvious difference in pattern or level of Hox1b protein detection in WT versus LgDel CNgV or hindbrain. The RNA-Seq data paralleled protein expression: Hox1b is detected at very low levels in CNgV from both genotypes (FPKM mean 0.37, WT; 0.55 LgDel, q = 0.995; Fig. 3A, right). We then evaluated localization of proteins that are selectively expressed at higher, but varying levels in CNgV at E10.5. Pax3, a transcription factor associated with a subset of CNgV neural progenitors, has no discernible difference in protein expression pattern or level in WT and LgDel CNgV. Similarly, Pax3 is detected at equivalent but relatively low levels in WT and LgDel CNgV in our RNA-Seq data (WT 22.1; LgDel 17.0, q = 0.99; Fig. 3B). Three additional neural progenitor-associated proteins—nestin, brain lipid-binding protein (blpb)/fatty acid-binding protein 7 (fabp7) and vimentin—are selectively expressed at low, intermediate and high levels, respectively, in WT and LgDel CNgV, based on apparent frequency of labeled cells (Fig. 3C–E, compare right-hand panels in each column). Their mRNAs are detected at similar relative levels with no significant differences between genotypes (Nes-94, WT; 74, LgDel, q = 0.99; Blpb/Fabp7–218.6, WT; 156.2, LgDel, q = 0.69; Vim-748.4, WT; 914.7, LgDel, q = 0.99; Fig. 3C–E, far right). Thus, based on comparison with an independent detection method for genes differentially expressed in CNgV, our RNA-Seq datasets appear robust and reliable.
Figure 3.
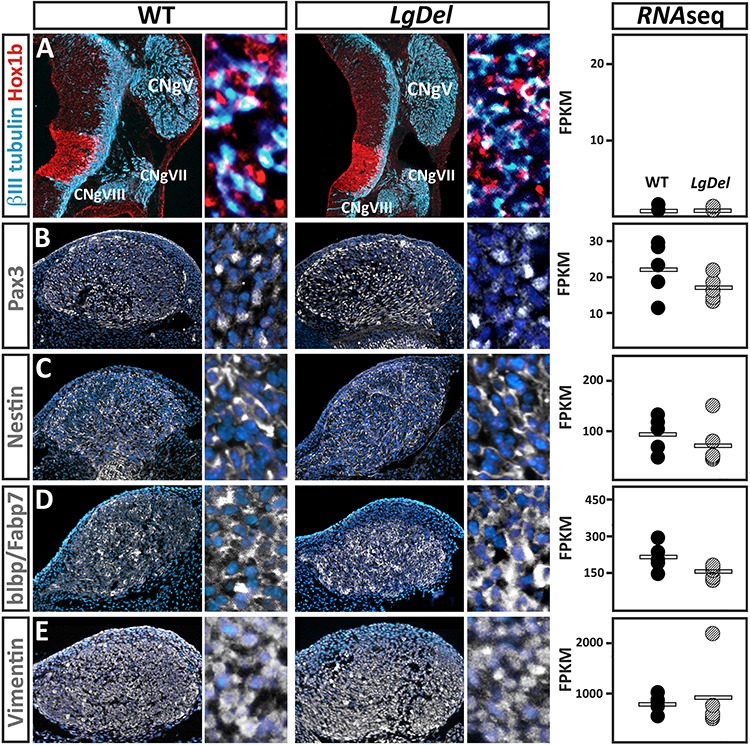
Registration of cellular CNgV protein expression localization at E10.5 and RNA-Seq mRNA detection from micro-dissected CNgV. For the images demonstrating protein expression, the first panel shows the expression pattern of each protein recognized immunocytochemically in CNgV in each genotype, and the adjacent second panel shows the distribution of individual labeled cells for the same protein. (A) Hox1b protein is expressed in a very limited population of CNgV cells but highly expressed in rhombomere 4 (r4) in the hindbrain immediately adjacent to CNgV in both WT and LgDel. In parallel, Hox1b mRNA is expressed at very low (FPKM < 0.1) but equivalent levels in the WT and LgDel CNgV RNA-Seq datasets. (B) Pax3 protein is detected in a subset of CNgV cells in both WT and LgDel. Right: Pax3 mRNA is detected at relatively low but statistically indistinguishable levels (FPKM < 5.0) in the WT and LgDel CNgV RNA-Seq datasets. (C) Left: Blpb/Fabp1 protein is localized in a somewhat broader subset of CNgV cells in both WT and LgDel. Right: Blpb/Fabp1 mRNA is detected at intermediate but statistically indistinguishable levels (FPKM < 100) in the WT and LgDel CNgV RNA-Seq datasets. (D) Left: nestin protein is more broadly localized in CNgV cells in both WT and LgDel. Right: Nes mRNA is detected at moderate (FPKM < 300) but statistically indistinguishable levels in the WT and LgDel CNgV RNA-Seq datasets. (E) Left: vimentin protein is localized to nearly all CNgV cells in both WT and LgDel. Right: Vim mRNA is detected at high (FPKM < 1000) but statistically indistinguishable levels in the WT and LgDel CNgV RNA-Seq datasets.
Validation of established, cell-class-selective CNgV transcriptional regulators
Four transcription factors are established markers for placode-derived/mechanosensory versus neural crest-derived/nociceptive neurons that constitute the two functionally distinct cell lineages in cranial sensory ganglia including CNgV: Six1, Brn3A, Sox10 and Foxd3 (11,26–28). Accordingly, each of these transcripts should be detected at substantial levels in both WT and LgDel CNgV samples, in register with cell-class-selective protein expression. In addition, these genes may be expressed at quantitatively distinct levels, in register with shifted proportions of placode- versus neural crest-derived cells in LgDel CNgV (see Fig. 1). We therefore compared the RNA-Seq values of these four genes with expression values measured by quantitative PCR (qPCR) in a parallel, independent set of 5 E10.5 microdissected, pooled CNgV samples.
We found robust expression levels in the RNA-Seq dataset for the placode-associated gene Six1, as well as Brn3a, a marker for CNgV neuronal differentiation (29), whose protein expression coincides with that of Six1 in the E10.5 CNgV (Fig. 4A and B, top) (11). We also found substantial expression levels in the RNA-Seq data of Sox10, whose protein expression nearly completely coincides with Wnt1:Cre-eGFP labeled neural crest-derived CNgV cells, and Foxd3, an established neural crest marker whose expression identifies Wnt1:Cre-negative, Six1 negative presumed CNgV neural crest cells (Fig. 4A, bottom) (11). Six1 and Brn3a expression levels appear to increase in the RNA-Seq and qPCR data for LgDel versus WT CNgV. Increased expression of both genes accords with the increased proportion of Six1-labeled cells in LgDel CNgV at E10.5 (see Fig. 1) that also express Brn3A. Statistical significance, however, was only reached for Six1 in the RNA-Seq data (P = 0.046) and for Brn3a in the qPCR data (P = 0.03; Fig. 4B and C, top). In contrast, even though we detected modest differences for the neural crest-associated transcription factors, only that for Sox10, measured by qPCR, reaches statistical significance (P = 0.037). The direction of change of transcript level, however, an increase, does not accord with the direction of change in neural crest cell frequency, a decrease.
Figure 4.
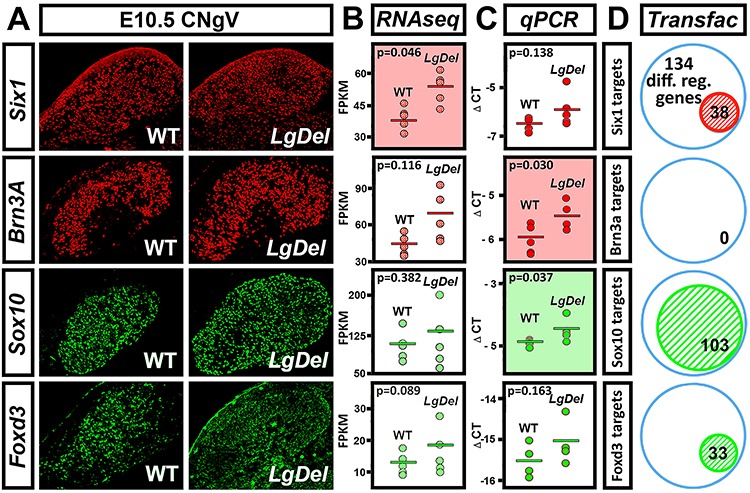
Validation of LgDel versus WT CNgV transcriptome comparison based on expression of transcription factors associated with placodal or neural crest-associated CNgV neurons or precursors. (A) Protein expression of Six1, Brn3a, Sox10 and Six1 in the E10.5 CNgV of WT and LgDel embryos. (B) RNA-Seq determined expression levels of diagnostic transcription factors associated with E10.5 cranial placode-derived CNgV cells—Six1 and Brn3a—and neural crest-derived CNgV cells—Sox10 and Foxd3—in LgDel and WT CNgV. Expression levels determined by RNA-Seq are presented for each of the five biological replicates of WT and LgDel CNgV as FPKM values. (C) Expression levels determined by qPCR in a parallel set of five biological replicates of WT and LgDel CNgV pooled samples are presented as delta CT (ΔCT) values. (D) TRANSFAC computational analysis identifies subsets of the 134 genes differentially expressed in WT versus LgDel CNgV (see Fig. 2) as potential transcriptional targets for the diagnostic transcription factors associated with either placode-derived or neural crest-derived CNgV cells.
Based on detection of these CNgV lineage/cell-class-selective markers in our RNA-Seq, qPCR and protein localization data, we asked whether the 134 differentially expressed genes detected by RNA-Seq might be potential regulatory targets of these four lineage-associated CNgV transcription factors. We conducted a gene promoter analysis to search for cis-acting binding sites of each transcription factor using the TRANSFAC (30) algorithm with a strict search criterion: 5% maximal matrix dissimilarity within 200 nucleotides upstream sequence from the start of transcription, to identify genes with cognate-binding elements for Six1, Brn3a, Sox10 or FoxD3. This analysis identified 38 of the 134 (28%) differentially regulated genes as potential Six1 targets, 0 of the 134 (0%) as potential Brn3a targets, 103 of the 134 (77%) as potential Sox10 targets and 33 of the 134 (25%) as potential FoxD3 targets (Fig. 4D; Supplemental Table 4). The lack of Brn3a targets is perhaps less surprising since it is expressed at the transition from neurogenesis to post-mitotic neuroblast to influence subsequent CNgV neuronal differentiation (31), which has not begun in earnest at E10.5. Together, these data indicate that our RNA-Seq analysis reliably detects established regulators of CNgV identity in both WT and LgDel. Moreover, expression changes of some of these genes parallel altered proportions of placode-associated CNgV cells—particularly for placode-associated genes. The relationship of these data to decreased neural crest cell frequency is less clear. Some expression level differences corresponding to quantitative cellular changes may be below the level of reliable detection of these methods. Alternately, the increased Sox10 expression in LgDel detected by qPCR but not RNA-Seq may reflect a real—yet quantitatively discordant—change in CNgV neural crest transcriptional state due to altered specification, migration, proliferation and cell–cell interactions associated with altered proportions of these cells.
Differentially expressed CNgV genes outside of the 22q11.2 deleted region
Our assessment of the 134 up- or downregulated genes identified several individual candidates that based on presumed functional significance for 22q11 deletion-associated phenotypes, cranial sensory neuron development or known roles in branchial arch and neural crest differentiation were chosen for further validation. We chose genes expressed at fairly high abundance based on FPKM values (>5) in our RNA-Seq dataset, and with substantial magnitude of expression changes (≥2-fold decrease or increase). Based on these criteria, we selected seven additional genes for qPCR analysis in a parallel set of five CNgV pooled samples from each genotype (Fig. 5). Four of these genes appear by RNA-Seq to decrease in expression (Fig. 5A–D): Josd2, a Josephin-domain containing presumed deubiquitinating enzyme (32); Lfng, a Notch signaling intermediate (33); Atoh1, a bHLH transcription factor known to regulate sensory and cerebellar neuronal development (34) and Epha7, an Ephrin receptor tyrosine kinase with putative functions in neuronal migration, axon growth and guidance (35). However, these four genes all appear to increase (although not statistically significantly) when assayed by qPCR. Two candidates increase significantly (Fig. 5E and F): Cited4, a transcriptional co-activator implicated in cardiac myocyte differentiation (36), and Icam4, an immunoglobulin/cell adhesion molecule implicated in cytokine signaling and immune cell adhesion (37). Expression levels of these genes also increase in the qPCR dataset, but only Cited4 reaches statistical significance. Thus, there is both divergence and convergence between expression levels measured in RNA-Seq and qPCR datasets generated from E10.5 microdissected WT and LgDel CNgV.
Figure 5.
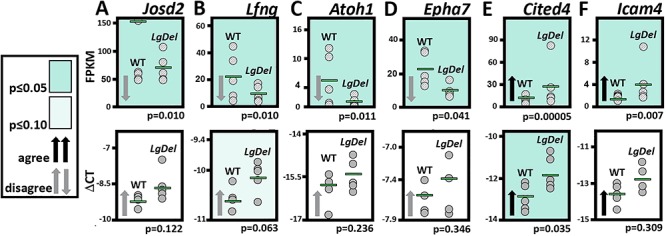
Comparison of expression differences for candidate genes identified by RNA-Seq using qPCR. (A–F) Differential expression validation for six candidate genes, chosen based on potential contributions to CNgV neuronal differentiation from among the full set of differentially expressed genes identified in the RNA-Seq dataset. Individual values (FPKM, top or ΔCT, bottom) are plotted for each gene. Green horizontal bars indicate mean expression values for each gene. Arrows indicate direction of expression change (up- or downregulated), and black versus gray shading indicates whether the qPCR and RNA-Seq mean expression differences are in agree (black) or do not agree (gray). The darker green shading indicates genes for which either the qPCR or RNA-Seq dataset identifies a statistically significant expression difference (P or FDR < 0.05). The lighter green shading indicates instances, where an apparent expression difference was detected and had a P value less than 0.1 in the qPCR validation.
CNgV transcriptome changes reflect altered hindbrain neural crest patterning
Our previous work suggests that patterning of anterior hindbrain rhombomeres (r1/r2), from which most CNgV neural crest progenitors originate, is altered in the LgDel embryo. This change, toward a more posterior identity, disrupts several aspects of CN V differentiation (6). To assess whether divergent transcriptional signatures in LgDel versus WT CNgV reflect this anterior to posterior (A/P) identity shift (Fig. 6A), we compared our transcriptome results to an existing RNA-Seq dataset (38) that catalogues transcripts associated with cranial neural crest of distinct A/P rhombomeric origins. To maximize compatibility of our data with that from the A/P neural crest transcriptomes, we used EdgeR (39) in addition to CuffDiff analysis to identify differentially expressed genes shared by the neural crest and CNgV datasets. We found 956 differentially expressed transcripts in LgDel versus WT CNgV using EdgeR and CuffDiff and confirmed 131 transcripts differentially expressed in the r1/r2 (CNgV-associated) versus r4 (posterior) neural crest.
Figure 6.
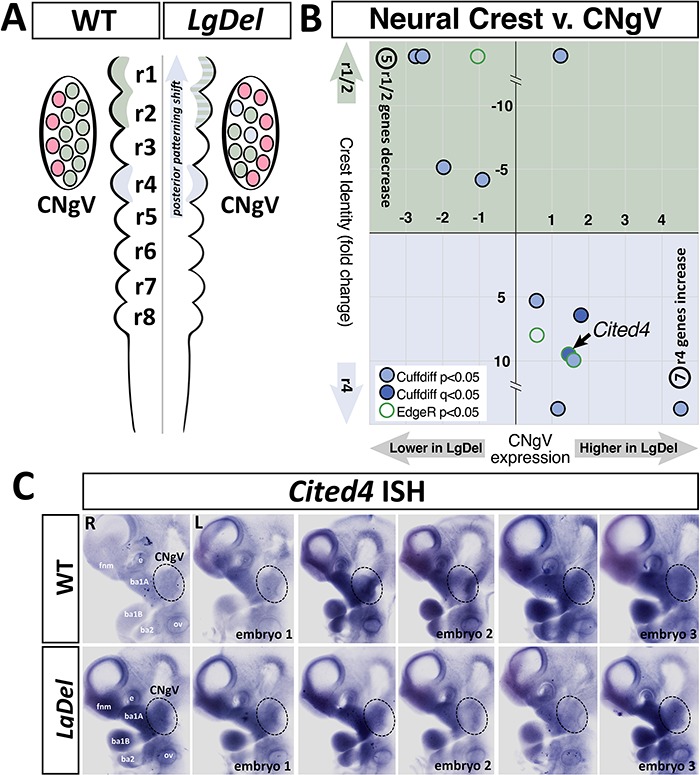
A posterior shift of neural crest-associated genes is detected in the LgDel CNgV transcriptome. (A) Schematic of the WT (left) and LgDel (right) hindbrain indicating the posterior shift of gene expression in rhombomeres (r)2 and r3 in the LgDel as well as the potential for this altered gene expression to be transferred to CNgV due to migration of neural crest progenitors from r2 and r3 into the coalescing CNgV (blue cell icons, far right). (B) A quadrant plot of gene identity (A/P) defined by an independent RNA-Seq analysis (38) and CNgV expression levels detected in our RNA-Seq analysis. Note that Cited4, whose expression is apparently increased in LgDel CNgV, by both RNA-Seq and qPCR (see Fig. 5), is one of the genes whose A/P expression is shifted. (C) ISH of Cited4 mRNA in E10.5 WT and LgDel embryos. The dotted oval indicates CNgV. These embryos were hybridized together, genotypes distinguished by tail clip.
Thirteen transcripts with significant expression differences, detected by CuffDiff (P < 0.05) and/or EdgeR analysis (P < 0.05), were common to both datasets. Those from the neural crest dataset whose expression levels are greater in r1/2 than r4 include Nkx2.9, Icam4, Ferd3l, Trh, Shh and Sox21; and those with greater expression levels in r4 versus r1/2 include Des, Acp5, Hox1b, Cited4, Krt14, Slc22a4 and Oc90. Of these, the expression levels of 12 LgDel versus WT CNgV transcripts had a shift consistent with altered A/P identity: five WT r1/r2-enhanced (anterior) transcripts had lower expression in LgDel CNgV: Nkx2.9, Ferd3l, Trh, Shh and Sox21 (Fig. 6B, upper left quadrant), whereas all seven WT r4-enhanced transcripts (posterior) had higher expression (Fig. 6B, lower right quadrant). These changes were statistically significant (P = 0.018; chi-square). Apparently, there is a shift in A/P gene expression consistent with a shift of A/P identity for the neural crest cells that contribute to CNgV.
Presumably, this distinction is established in hindbrain rhombomeres from which the neural crest directed to CNgV originates and is preserved in neural crest-derived progenitors that constitute a major component of the E10.5 CNgV. Consistent with this interpretation, the A/P shifted gene set includes Cited4, the one candidate gene from our RNA-Seq differential expression analysis that was validated as significantly differentially expressed by qPCR analysis (see Fig. 5E). The expression pattern of Cited4 in the developing head was not defined in the existing literature. Accordingly, we performed in situ hybridization (ISH) for Cited4 on WT and LgDel embryos as a further validation. WT (n = 4) and LgDel (n = 6) embryos were incubated in the same vial throughout hybridization, labeling and clearing, and imaged at standard illumination and exposure settings. We found that Cited4 is selectively expressed in CNgV in both WT and LgDel embryos (Fig. 6C). In these embryos, it was difficult to identify consistent expression differences between the two genotypes. Nevertheless, the selective expression of Cited4 in CNgV, as well as its association with the shift in A/P rhombomere identities confirms the sensitivity of our RNA-Seq—Cited4 has not been previously localized to CNgV—and detecting expression changes that accord with developmental disruption in LgDel versus WT embryos.
Non-parametric analysis of LgDel and WT transcriptome differences
Our analysis of mean expression level differences in E10.5 WT and LgDel CNgV replicates indicates changes in several genes that may influence divergent CN V differentiation in the LgDel. Nevertheless, validation of many of the differentially expressed genes in parallel samples of CNgV did not consistently confirm predictions based on mean expression comparisons from the RNA-Seq data. Indeed, we noted that a significant number of outliers in sample-by-sample expression levels of many transcripts, especially in the LgDel samples, seemed to drive mean expression differences, perhaps erroneously. Statistical assessment of mean expression differences, based on CuffDiff analysis, is known to be impacted by outliers, particularly when they deviate by an order of magnitude or more, as is often the case for LgDel values. This raised the question of whether the mean analysis had appropriate sensitivity to identify all significant expression level distinctions between WT and LgDel CNgV.
Accordingly, we evaluated our CNgV RNA-Seq dataset using a non-parametric method in which WT and LgDel expression values as a group were assigned ranks (1 through 10) based on FPKM detection levels (Fig. 7). Potential median differences between individual expression levels in the two genotypes identified by this analysis were compared using the Mann–Whitney U test (a rank-order test analogous to a two-tailed Student’s t test), which compares group differences based on median rather than average values. This approach, which minimizes the statistical effects of outliers, identified approximately eight times the number of significantly changed transcripts (1149) than by the CuffDiff analysis (134; Fig. 7A and Supplemental Table 5), and twice the number of transcripts even when no FDR correction is used (668 transcripts at CuffDiff P < 0.05). Nevertheless, there is relatively little overlap between the sets of transcripts identified by the two methods—only 178 of the 668 (26.6%) uncorrected transcripts (P < 0.05) and 27 of the 134 (20.1%) FDR corrected transcripts (FDR < 0.10) are also significant by the Mann–Whitney test (P < 0.05).
Figure 7.
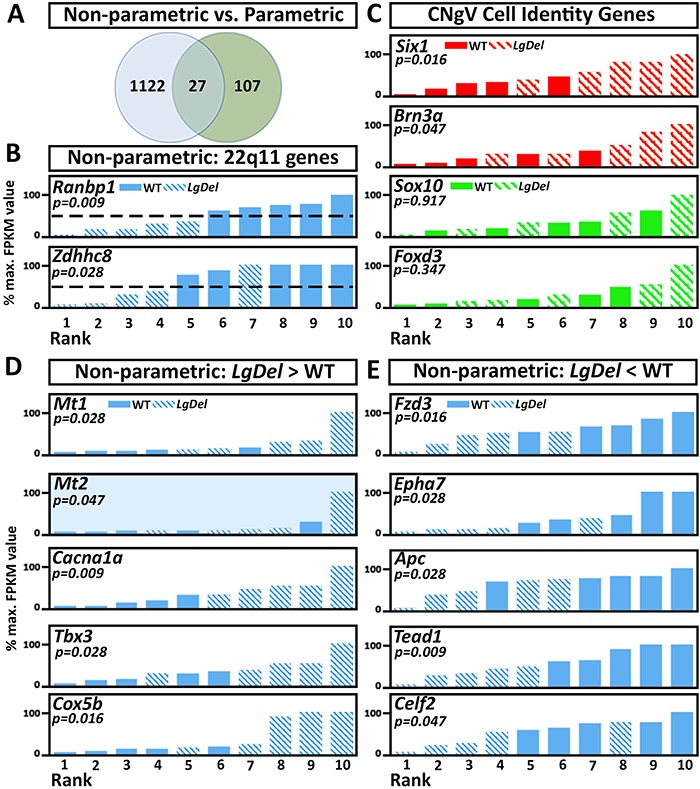
Non-parametric analysis of WT versus LgDel CNgV transcriptomes detects a greater number of significantly expression level differences in the two genotypes. (A) A summary of the number of significantly differentially expressed genes detected using non-parametric, rank-order based statistics and the Mann–Whitney U test of significance in the WT versus LgDel CNgV transcriptome. Nearly 10 times as many genes—1149—are detected at significance levels of P < 0.05 or less as compared with 134 detected using the previously described CuffDiff analysis, but only 27 transcripts are identified by both methods. (B) Non-parametric rank-order analysis detects significant 50% decrements in 22q11 gene expression with greater sensitivity. The most (Ranbp1) and least (Zdhhc8) abundant significantly different 22q11 transcripts are shown here. These values and all others in this figure are plotted in rank order, 1 through 10, left to right, and the expression values across both genotypes are displayed as percentage of the maximum expression value (100%). The dotted line indicates the 50% expression level. Mann–Whitney significance (P value) is given in italics. (C) Non-parametric analysis detects changes in expression levels of four diagnostic placode versus neural crest associated genes with accuracy similar to the parametric analysis. (D) Novel genes whose expression is detected as significantly increased based on non-parametric analysis in our RNA-Seq dataset. Mann–Whitney P value is given in italics. The shading (Mt2 non-parametric analysis histogram) indicates that qPCR assessment validated the RNA-Seq detected expression difference (P = 0.047; n = 5 pooled CNgV replicates/genotype). (E) Novel genes whose expression is detected as significantly decreased based on non-parametric analysis in our RNA-Seq dataset, presented as above. qPCR validation of transcripts shown in (D) and (E) is shown in Supplementary Table 5.
To validate whether this non-parametric approach identifies significantly changed transcripts with at least the same sensitivity as the parametric method, we first assessed the murine orthologues of 22q11 genes that are heterozygously deleted in the LgDel mouse. Of the 22 transcripts encompassed by the LgDel deletion that are robustly identified in the RNA-Seq dataset (see Fig. 2C), 5 have detectable reduced expression by CuffDiff parametric assessment; however, 20 are identified as significant by the non-parametric analysis (Fig. 7B and Supplemental Table 5). In addition, we assessed expression differences that reach significance or accord with CNgV cellular changes detected by the parametric approach: Six1 and Brn3a. The non-parametric analysis detected differential expression of the placode-associated Six1 and Brn3a and confirmed that the neural crest-associated genes Sox10 and Foxd3 are differentially expressed (Fig. 7C). Next, we asked whether a subset of transcripts identified by the non-parametric approach might be robustly replicated by qPCR from additional pooled samples. We assessed 10 differentially expressed genes with varying levels of significance based on our non-parametric analysis: five upregulated (Fig. 7D) and five downregulated (Fig. 7E); only 4/10 of these additional transcripts were significant by both parametric and non-parametric analyses of the RNA-Seq dataset. Our parallel qPCR analysis of these transcripts showed that the non-parametric approach is not more reliable; only one of the transcripts (Mt2; Fig. 7, shaded histogram) was significantly different (LgDel > WT; P = 0.047) in the independent qPCR analysis and the non-parametric analysis (Supplemental Table 5). Apparently, non-parametric statistics may be a useful adjunct for identifying differentially expressed genes, based on robust identification of LgDel deleted transcripts. Nevertheless, it appears that a high level of variability is a key feature of the LgDel transcriptome, and not merely measurement ‘noise’ that can be filtered out.
Significantly increased transcriptome variability in LgDel CNgV
The consensus of multiple clinical studies of 22q11DS patients (40–43) as well as analyses in mouse models (17,44), including our previous assessment of CN V anomalies that prefigure disrupted S/F/S in LgDel mouse pups (6), is that phenotypic variability is a hallmark of 22q11 deletion. Thus, it seemed possible that difficulties in confirming some—but not all—of apparent expression differences from the RNA-Seq dataset in independent samples with alternative methods might reflect a fundamental distinction between the transcriptomes of E10.5 CNgV cells from WT versus LgDel: there may be biologically established statistically verifiable greater variation in expression levels throughout the entire set of expressed genes in LgDel due to the apparent 50% change in 22q11 gene dosage. If this is the case, quantitative evidence of this variability should remain despite microdissection intended to limit cell classes being profiled and pooling intended to diminish or eliminate non-biological variability in expression levels.
We therefore compared variability of expression levels on a gene-by-gene basis in the five WT CNgV biological replicates with that in the five LgDel CNgV replicates (Fig. 8A). Inspection of the heat maps of each of the individual WT versus LgDel samples suggests that there may be greater variation of expression for each gene across the LgDel CNgV transcriptome. There seems to be far more frequent differences in magnitude of expression differences, for both over-expressed (Fig. 8B, top) and under-expressed genes (Fig. 8B, bottom), as well as those that are expressed at equivalent values. To verify this impression quantitatively across the entire 17 000-plus gene set, we did a 2 × 2 chi-square analysis for CNgV genes whose mean expression differed significantly in WT versus LgDel, as well as for genes whose mean expression did not differ between the two genotypes. We found that there is far greater variability in expression levels across all CNgV expressed genes in LgDel. Of the 17 190 expressed genes common to both genotypes, 14 996 have significantly greater variable expression levels in LgDel CNgV versus 2187 in WT with only 7 genes varying equally (Fig. 8C and D), based on the coefficient of variation calculated across the 5 biological replicates for each genotype. This difference in the number and level of variable expression between LgDel and WT CNgV is significant (P < 0.00001; chi-square).
Figure 8.
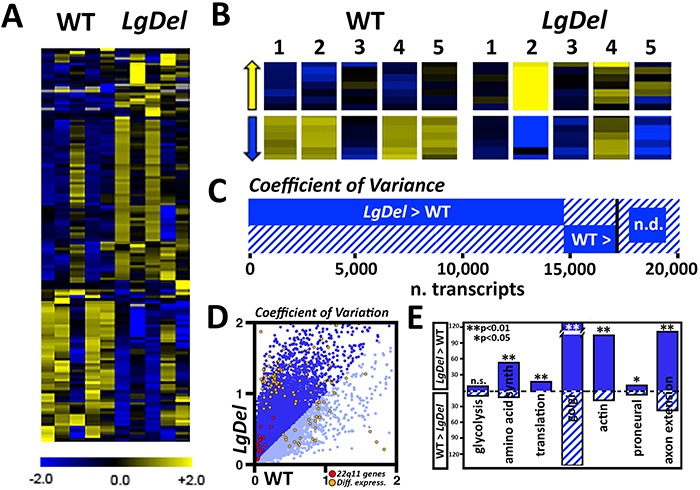
Transcriptome variation as a quantitative phenotype distinguishes LgDel and WT CNgV. (A) Heat maps for each of the five biological replicates of WT and LgDel CNgV assessed by RNA-Seq. Although there is some variation in the 5 WT replicates (e.g. compare replicate 3 with other WT samples), the variation in the 5 LgDel samples appears greater. The scale bar below the heat map is given on a log2 scale. (B) Higher resolution view of blocks from the heat map for both up- and downregulated genes confirms the impression of greater variability in the LgDel samples. (C) Summary plot of the proportion of 17 128 total genes with non-zero reads with a higher coefficient of variance (CV) in LgDel (14 996) than WT (2187), as detailed in Supplemental Table 2. (D) Scatterplot comparing CV in WT (x-axis) versus CV in LgDel (y-axis). Darker blue indicates higher CV in LgDel versus lighter blue for transcripts with higher CV in WT. 22q11 orthologues all have higher CV in LgDel and are shown in red; the subset of 72/134 significantly differentially expressed genes with higher CV in LgDel is shown in green. (E) Graphic representation of increased stochastic variation in LgDel versus WT (asterisks indicating significant differences, chi-square) among most, but not all of a subset of GO categories. These categories include gene sets associated with fundamental cellular (e.g. glycolysis, mitochondria) and neuronal differentiation mechanisms (e.g. proneural bHLH genes).
Greater variability extends across most classes of genes, including 22q11 orthologues: 20 of these genes had a higher coefficient of variation in LgDel CNgV (Fig. 8D, red dots), and this difference between LgDel and WT was also significant (P < 0.000007, chi-square). Similarly, the 134 genes found to be differentially expressed in LgDel versus WT CNgV (Fig. 8D, orange dots; P < 0.00009). In addition, we queried several additional GO categories and found that with the exception of genes associated with glycolysis, stochastic variation was greater in LgDel CNgV for several fundamental cellular pathways: amino acid synthesis, protein translation, Golgi apparatus structure and function, actin dynamics, as well as two additional neural development mechanisms: bHLH proneural genes expressed during neural differentiation and axon extension genes (Fig. 8E). Apparently, heterozygous deletion of mouse 22q11 gene orthologues results in a broad, significant increase in statistically verifiable transcriptome-wide transcriptional variability, extending to many—but not all—functional gene subsets expressed in CNgV. This suggests that an essential dimension of the distinctions between WT and LgDel CNgV transcriptomes, in addition to expression differences of particular genes, is increased stochastic variation of overall transcriptional state. Our RNA pooling strategy, which is thought to reduce biologic variability and increase detection power for differentially expressed genes (12,45), likely underestimates stochastic variation observed in LgDel. CNgV transcriptional instability, perhaps due to a cell-by-cell basis, may contribute to phenotypic change and may also indicate a potential general mechanism for aspects of phenotypic variability seen in 22q11DS mouse models (16) as well as individuals with 22q11DS (41).
Discussion
The transcriptomes of differentiating CNgV progenitors and sensory neurons are quantitatively distinct in E10.5 LgDel versus WT embryos, shortly after the ganglion has coalesced as a distinct structure, in parallel with altered CN V differentiation and CNgV cellular composition. Our transcriptome comparison provides enhanced tissue specificity and statistical resolution of changes that ultimately must reflect the 50% diminished expression level of 22q11 genes in CNgV and related structures that contribute cells to the ganglion: the cranial placodes and hindbrain. Additional quantitative analysis of candidate genes whose expression differs in LgDel versus WT CNgV validates some but not all of the RNA-Seq comparisons. Among the validated genes, changes in Six1 and Cited4 expression levels accord with cell biological changes in LgDel CNgV or in the hindbrain from where CNgV neural crest progenitors originate. In addition, we identified statistically robust increased variability in expression levels of the majority of the over 17 000 genes expressed in common in LgDel and WT CNgV as an essential divergent feature of the LgDel versus WT CNgV transcriptomes. We suggest that 22q11 deletion-related CN V differentiation phenotypes in LgDel originate in part from this increased stochastic variation across much of the LgDel CNgV transcriptome. Such divergence may de-stabilize programs for CNgV progenitor division or sensory neuron differentiation and disrupt initial oropharyngeal sensory-motor circuit development, including that essential for S/F/S, in 22q11.2DS.
Comparing embryonic CNgV transcriptomes
Our micro-dissection approach for isolating embryonic CNgV mRNA is novel. Several observations reinforce the robustness of the data generated using this approach. First, the number of expressed genes detected consistently in five biological replicates from each genotype is substantial and approximately equivalent: 17 221 in LgDel and 17 199 in WT. Apparently, CNgV cells in both genotypes are viable and actively transcribing a large number of genes. Second, we confirmed our previous validation of microdissected LgDel and WT CNgV (6) based on parallel detection of selective protein expression in E10.5 CNgV sections and mRNA in our RNA-Seq data. Third, the approximate 50% expression decrement of deleted 22q11 genes in LgDel CNgV versus the same genes in WT adds confidence. Fourth, quantitative cell biological changes in proportions of placode- versus neural crest-derived CNgV cells in the two genotypes are paralleled by expression level changes of relevant cell-class associated genes. Finally, an independent RNA-Seq dataset of a related subpopulation of cranial neural crest cells—derived from r1/2, which generates most of the neural crest component of CNgV—detects expression changes of several genes in our CNgV RNA-Seq dataset. Thus, our approach has precision, specificity and reliability to generate robust data to test specific hypotheses of transcriptional distinctions underlying early divergent CNgV differentiation in LgDel versus WT embryos.
Transcriptome divergence, candidate genes and independent validation
Our RNA-Seq analysis distinguishes the transcriptomes of early CNgV sensory neurons or their precursors in LgDel versus embryos at a critical stage of early CN V differentiation. The number of non-zero reads shared by these samples, 17 190, suggests that 68% of the approximately 25 059 mouse genes with protein sequence data (46) are expressed at some level in CNgV by E10.5. Our microdissection-based approach enriches for genes that are selectively expressed in CNgV, and indeed, we identify several significantly enriched pathways associated with trigeminal sensory neuron differentiation. Slit/Robo (20–22) and BMP signaling (23,24) are both established regulators of neural crest progenitors as well as trigeminal axon growth and branching. In addition, the enrichment of pathways associated with oligodendrocyte progenitor proliferation and differentiation may reflect commonalities of cranial neural crest cells with Schwann cell precursors, which are also of the neural crest lineage and generate peripheral glial cells among their many progeny (24). Thus, the informatics comparison of LgDel and WT CNgV transcriptomes yields an informative framework for assessing molecular foundations of trigeminal sensory neuron development and its divergence in LgDel embryos.
We tested several hypotheses generated by our biological observations of LgDel CN V developmental divergence. First, we found that differentially expressed genes include transcriptional regulators of CNgV cranial placode—Six1 and Brn3a—as well as neural crest—Sox10 and Foxd3—derived populations. These differences accord with a developmental/cellular distinction between WT and LgDel CNgV: a proportionate increase in Six1 mRNA and Six1-expressing placode-associated cells in LgDel. Moreover, analysis of regulatory sequences of the differentially expressed genes shows that significant subsets of these 134 genes are potential targets of three of the four transcriptional regulators that are ‘cardinal’ markers for placode and neural crest components of CNgV: Six1, Sox10 and Foxd3 (11,26–28).
Our evaluation of additional hypotheses yielded mixed results. We assessed RNA-Seq-identified candidate genes by qPCR, based on potential roles in progenitor proliferation, neuronal differentiation, axon growth and guidance. For each gene we analyzed, qPCR analysis detected expression at roughly comparable levels in parallel pooled samples collected and processed identically to those used to generate the RNA-Seq datasets. Thus, we found no false positive signals in our RNA-Seq data. Nevertheless, increased expression of only one these genes, Cited4, was confirmed at P < 0.05 by both RNA-Seq and qPCR. In addition, Cited4 was further validated as a novel CNgV-expressed gene by ISH in both WT and LgDel embryos. Thus, although our RNA-Seq comparison, with multiple biological replicates, provides a high resolution ‘screen’ for total transcriptome differences that underlie LgDel versus WT CNgV developmental divergence, it does not do so with absolute resolution on a gene-by-gene basis. Nevertheless, as demonstrated by our data on Cited4, RNA-Seq comparisons can be used to identify novel CNgV-expressed genes and generate testable hypotheses of altered expression of relevant candidates in LgDel versus WT. Such transcriptome-based hypotheses can be tested further by rigorous complimentary experiments.
Despite some limitations, our RNA-Seq data confirmed differential regulation of two genes that identify CNgV just after the ganglion has coalesced from placode and neural crest-derived cell populations. Six1 and Cited4 may contribute to distinctions in LgDel versus WT CNgV lineage and subsequent sensory neuron differentiation. Six1 is a key transcriptional regulator of cranial placode specification and differentiation of placode-derived cranial mechanosensory neurons. Our RNA-Seq analysis detected a significant increase in Six1 mRNA, parallel to a significant proportionate increase in Six1-expressing LgDel CNgV cells. This agreement between transcriptome and cell biological change is further reinforced by the concentration of Six1 transcriptional targets among the 134 genes that are significantly differentially expressed in LgDel versus WT CNgV. This apparently enhanced regulation may reflect the increased number of placode-derived presumed CNgV cells, or altered differentiation due to increased Six1 expression in LgDel. Less is known about primary developmental functions of Cited4, a CBP/P300 interacting transcriptional cofactor expressed in multiple embryonic and adult tissues including heart and mammary glands (47). Complete loss of Cited4 function is embryonic lethal; nevertheless, there is evidence of disrupted early skin differentiation (48). In addition, Cited4 inactivation is associated with oligodendroglial tumorigenesis (49). Thus, increased Cited4 levels as well as anomalous A-P expression in LgDel (6) might modify proliferation, neurogenesis or gliogenesis in LgDel CNgV neural crest-derived progenitors, especially since our GO analysis identifies several pathways related to gliogenesis as highly enriched in the E10.5 WT and LgDel CNgV transcriptomes. Thus, the two validated expression changes in our RNA-Seq analysis suggest potential downstream consequences for the differentiation of two key classes of CNgV cranial sensory neurons: placode-derived mechanosensors and neural crest-derived nociceptors.
Stochastic transcriptional variation is a key 22q11 deletion phenotype
The most robust distinction between the LgDel and WT CNgV transcriptomes is greater variation of LgDel gene expression levels. These results parallel one other transcriptome analysis that compared 22q11DS model mice (Df(16)A+/−: a deletion that parallels LgDel) with WT mice (50). In this microarray analysis, only one candidate with a modest transcriptional change was identified and validated in the developing cerebral cortex in the context of substantial additional variability. Similarly, an analysis of 22q11-deleted humans carrying either the 1.5 MB minimal critical or typical 3 MB deletion identified variability as an essential transcriptome characteristic in 22q11 deleted individuals (51). This common feature is unlikely to reflect ‘noise’ in the datasets. Instead, we suggest that transcriptome-wide stochastic expression variability due to 22q11 deletion results in cell-by-cell and embryo-by-embryo differences in development and function. In embryos, these differences amplified by local distinctions in cell signaling states and divergent gestational experience (maternal diet, stress, environmental exposures) destabilize optimal developmental programs for LgDel CNgV cell fates.
The quantitative expression variability we detect in CNgV may be similar to ‘fate drift’ described recently based on stochastic expression level variation identified by single-cell transcriptome analysis for pancreatic cell classes as a function of aging (52). In this instance, an ‘environmental’ factor—aging—apparently introduces instability in regulation of expression levels of multiple genes for which there are presumably two genomic copies. Copy number variation may increase stochastic variability. The kinetics of expression initiation and termination, which are thought to be buffered by normal copy numbers, may enhance expression level variation when copy number is greater or less than 2, potentially on a cell-by-cell basis (53). In the context of hChr.21 duplication, combined effects of allelic selection and variable transcription kinetics lead to cell-by-cell differences in expression levels of duplicated Chr.21 genes as well as diploid loci (54). The variation we report here may reflect similar mosaic expression regulation of heterozygously deleted 22q11 genes as well as other genes—individual cells may be effectively null for some loci, haploinsufficient or diploid for others. Such mosaicism could lead to variable, divergent differentiation across the entire population of early differentiating CNgV cells and provide an initial pathogenic departure for anomalous circuit development underlying perinatal dysphagia and other oropharyngeal dysfunction in 22q11DS.
The increased frequency of stochastic transcription variation we identify in LgDel CNgV is substantial; however, differences in expression levels of individual genes, though statistically significant, are far more modest and in some cases difficult to validate using additional methods. Quantitative measures of expression in tissue samples, including those we identify in carefully microdissected CNgV, may be complicated by such instability of individual cell-by-cell expression levels. Thus, there may be even greater distinctions in transcriptional state from cell to cell in LgDel CNgV. Nevertheless, across the entire relatively small population of CNgV cells at E10.5, there are quantitatively verifiable differences in gene expression as well as statistically significantly increased overall stochastic variability in LgDel, presumably reflecting the consequences of 50% diminished expression of heterozygously deleted 22q11 gene orthologues. Little is known of the deleterious—or perhaps beneficial—effects of locally variable gene expression in individual cells for any cell class in the developing or mature nervous system. Our findings highlight the need for further assessment of this quantitative feature of transcriptional states in genetically typical as well as mutant individuals during development and in the adult. Future analysis of single-cell sequencing data combined with highly sensitive, quantifiable expression localization in cells within their native tissues may further delineate the nature, mechanisms and potential developmental or functional significance of normal stochastic gene expression as well as that due to 22q11 deletion as well as additional single gene or gene dosage disorders.
Materials and Methods
Embryo microdissection, sample collection and RNA extraction
C57Bl/6N WT dams, obtained directly from the animal vendor (Charles River), carried timed pregnancies sired by LgDel males, also C57Bl/6N. The LgDel males from our colony descend from a lineage of over 20 generations. Each generation of LgDel studs inherits the deletion paternally, via matings with C57Bl/6N WT dams from the vendor to avoid inbreeding. For this study, pregnant dams were sacrificed by rapid cervical dislocation on gestational (Embryonic) day E10.5 (vaginal plug = E0.5) and embryos quickly collected in RNase-free HEPES buffer. In C57Bl/6 mice, the average gestational time in days is 19.26 ± 0.04 days (55). Thus, the stage at which we collect embryos can be considered late mid-gestation if one defines the mid-point gestational day as E9.6, and the mid-gestation period as ±1.0 days around this mid-point. CNgV was harvested by microdissection from these E10.5 WT and LgDel embryos. The two ganglia from each embryo were placed into RNAlater (Ambion) and stored separately until genotyping from additional tissue from that embryo was complete.
Once genotypes were established, pooled CNgV samples were generated to yield five biological replicates for each genotype (WT, LgDel). One biological replicate consisted of a pool of at least six ganglia from at least three embryos of the same genotype over several litters. The numbers of cells in mouse CNgV at E10.5–E11.0, the earliest age when such estimates have been reported in the literature, is between 5000 and 6000 (56,57), a number confirmed by our own quantitative estimates. We see no size difference or obvious cell loss in LgDel CNgV at E10.5 (see Fig. 1). Thus, 30 000–36 000 cells, at most, comprise each pooled sample for each genotype. Accordingly, we used all of the RNAs isolated from each pool for subsequent transcriptome sequencing. Total RNA from each pool was extracted using the single-step acid guanidinium thiocyanate-phenol-chloroform method (10) and subjected to RNA quality control (QC) with RIN scores ≥8 (Agilent 2100 Bioanalyzer, Agilent Tech., Santa Clara, CA, USA) using standard protocols (6). Similar methods were used for whole embryo samples (two pools of WT and LgDel, consisting of four embryos each).
Embryo immunocytochemical analysis
A separate set of E10.5 WT and LgDel embryos was collected to assess cranial ganglion and nerve developmental phenotypes and cell-class-selective protein expression within CNgV. One set of embryos was genotyped and then labeled whole for the early neuronal marker βIII-tubulin (mouse anti-βIII tubulin, Covance), which identifies both peripheral sensory ganglion neurons and their growing axons. A second set of LgDel and WT embryos also carried Wnt1:Cre recombinase and eGFP nuclear reporter alleles heterozygously (10). These embryos were processed for cryomicrotomy and immunolabeled for Six1, a marker of placode-derived cranial sensory neurons (rabbit anti-Six1, Proteintech) (11) and an antibody against eGFP (chicken anti-GFP, Abcam) to amplify the reporter signal established by Wnt1-mediated recombination in CNgV cells presumed to originate from the neural crest. Sections were imaged with a Leica tiling video microscope. Six1 (placode-derived) and Wnt1:Cre (neural crest derived) labeled cells were counted blind to genotype on digitized micrographic images to estimate relative frequencies (11). Primary antibodies directed against Brn3a (mouse, Millipore), Sox10 (goat, Santa Cruz) and Foxd3 (mouse, Thermo Scientific) as well as Hox1b (rabbit, Covance), Pax3, blpb/fabp7 (both mouse monoclonal antibodies, Developmental Studies Hybridoma Bank), nestin (mouse, BD Sciences) and vimentin (rabbit, Enzo Life Sciences) were used to label E10.5 sections of CNgV using the same methods. Primary antibody labeling was visualized using Alexa Fluor 488-, 546- and 647-conjugated secondary antibodies (Molecular Probes).
Library construction and sequencing
Paired-end libraries were constructed according to the Illumina protocol for the HiSeq2000 platform. Each pooled CNgV RNA sample, defined as a single biological replicate, was fragmented prior to cDNA conversion to ensure transcript coverage. WT (n = 5 replicates) and LgDel (n = 5 replicates) were subjected to sequencing (~100 million paired-end reads/replicate, 100 bp read length) on the Illumina HiSeq2000 platform. For WT RNA-Seq, 111–123 million paired-end reads were generated after QC filtering, and 97.3–97.9% of the reads aligned to the mouse genome reference sequence. For LgDel RNA-Seq, 77–137 million paired-end reads were generated after QC filtering, and 94.3–97.8% of the reads aligned to the mouse genome reference sequence.
Data processing
Raw sequence data were processed using Illumina’s RTA and CASAVA pipeline software, which include image analysis, base calling, sequence quality scoring and QC.
Sequencing datasets were processed through an in-house transcriptome analysis pipeline, employing HISAT/Cufflinks and SAMtools packages, including recent sub-developments (58–62), as described previously (63,64). The quality-filtered sequencing reads are aligned against the latest version of the mouse reference genome (GRCm38/mm10) using HISAT2 (v.2.1.0). These alignments were indexed using SAMtools (v.1.4) and aligned reads assembled into transcripts using Cufflinks (v.2.2.1) (58). The minimum possible spliced transcripts that explain all aligned read pairs were assembled; abundance was calculated as fragments per kilobase of exon per million fragments mapped (FPKM). Differential expression analysis was performed using two strategies in parallel: the CuffDiff utility of Cufflinks (58) and the outlier-resistant EdgeR-robust package (39,65). The expression profiles and the final transcript predictions of interest were examined individually using Integrated Genome Viewer (v.2.5) (66).
Functional and enrichment analyses
GO categories, pathway enrichment and network analysis were assessed using MetaCore (Clarivate Analytics), Ingenuity Pathway Analysis (Qiagen Bioinformatics) and the AmiGO 2 platform (http://amigo.geneontology.org/amigo).
Statistics
Quantitative estimation of the expression levels of the different isoforms between and within the genotype groups was performed using the Cufflinks package, CuffDiff, which contains normalization modules supporting across-sample comparisons. In addition, ANOVA was applied to define significant differentially regulated transcripts, and P values were corrected for multiple testing using False Discovery Rate (FDR) (67). Unless otherwise noted, FDR of 10% (q < 0.1) was considered significant for all analyses.
qPCR and ISH validation
We validated the quantitative expression changes identified by RNA-Seq using qPCR and ISH for mRNAs significantly up- or downregulated in LgDel versus WT CNgV. qPCR primers were designed for all of the genes (Supplemental Table 1). Parallel sets of 5 WT and 5 LgDel microdissected, pooled CNgV samples were prepared for qPCR analysis. For ISH, digoxigenin-labeled probes were synthesized using plasmids containing the full-length Cited4 open-reading frame sequences (OriGene). Whole-mount ISH was performed using E10.5 WT and LgDel embryos fixed in 4% paraformaldehyde overnight and stored in 100% methanol. Embryos were re-hydrated into PBS + Tween 20 buffer before probe hybridization overnight at 65°C. After several washes, embryos were incubated in 10% sheep serum in TBS + Tween 20 (TBS-T) buffer followed by overnight incubation at 4°C with anti-digoxigenin-AP antibody (Roche) in blocking solution to detect bound probe. Embryos were washed several times with TBS-T buffer and developed in BM-Purple (Roche). Embryos were photographed using a Leica M420 stereoscope fitted with a Leica DFC480 camera. ISH experiments were performed on a minimum of five control and five mutant embryos.
Supplementary Material
Acknowledgements
We gratefully acknowledge Dr Lisa Sadzewicz and Luke Tallon of the Institute for Genome Sciences for their assistance in coordinating the RNA-Seq study.
Funding
National Institute of Child Health and Human Development (P01 HD083157).
Conflict of Interest statement. None declared.
References
- 1. Sullivan P.F. and Geschwind D.H. (2019) Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell, 177, 162–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kohl J., Autry A.E. and Dulac C. (2017) The neurobiology of parenting: a neural circuit perspective. Bioessays, 39, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooper-Brown L., Copeland S., Dailey S., Downey D., Petersen M.C., Stimson C. and Van Dyke D.C. (2008) Feeding and swallowing dysfunction in genetic syndromes. Dev. Disabil. Res. Rev., 14, 147–157. [DOI] [PubMed] [Google Scholar]
- 4. Kakodkar K. and Schroeder J.W. Jr. (2013) Pediatric dysphagia. Pediatr. Clin. North Am., 60, 969–977. [DOI] [PubMed] [Google Scholar]
- 5. LaMantia A.S., Moody S.A., Maynard T.M., Karpinski B.A., Zohn I.E., Mendelowitz D., Lee N.H. and Popratiloff A. (2016) Hard to swallow: developmental biological insights into pediatric dysphagia. Dev. Biol., 409, 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karpinski B.A., Maynard T.M., Fralish M.S., Nuwayhid S., Zohn I.E., Moody S.A. and LaMantia A.S. (2014) Dysphagia and disrupted cranial nerve development in a mouse model of Digeorge (22q11) deletion syndrome. Dis. Model. Mech., 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Bryan C., LaMantia A.S. and Mendelowitz D. (2017) Altered neurobiological function of brainstem hypoglossal neurons in Digeorge/22q11.2 deletion syndrome. Neuroscience, 359, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maynard T.M., Zohn I.E., Moody S.A. and LaMantia A.S. (2020) Suckling, feeding, and swallowing: behaviors, circuits, and targets for neurodevelopmental pathology. Annu. Rev. Neurosci., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamburger V. (1961) Experimental analysis of the dual origin of the trigeminal ganglion in the chick embryo. J. Exp. Zool., 148, 91–123. [DOI] [PubMed] [Google Scholar]
- 10. Steventon B., Mayor R. and Streit A. (2014) Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol., 389, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karpinski B.A., Bryan C.A., Paronett E.M., Baker J.L., Fernandez A., Horvath A., Maynard T.M., Moody S.A. and LaMantia A.S. (2016) A cellular and molecular mosaic establishes growth and differentiation states for cranial sensory neurons. Dev. Biol., 415, 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kendziorski C., Irizarry R.A., Chen K.S., Haag J.D. and Gould M.N. (2005) On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. U. S. A., 102, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moody S.A., Quigg M.S. and Frankfurter A. (1989) Development of the peripheral trigeminal system in the chick revealed by an isotype-specific anti-beta-tubulin monoclonal antibody. J. Comp. Neurol., 279, 567–580. [DOI] [PubMed] [Google Scholar]
- 14. LaMantia A.S., Bhasin N., Rhodes K. and Heemskerk J. (2000) Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron, 28, 411–425. [DOI] [PubMed] [Google Scholar]
- 15. Bhasin N., Maynard T.M., Gallagher P.A. and LaMantia A.S. (2003) Mesenchymal/epithelial regulation of retinoic acid signaling in the olfactory placode. Dev. Biol., 261, 82–98. [DOI] [PubMed] [Google Scholar]
- 16. Meechan D.W., Tucker E.S., Maynard T.M. and LaMantia A.S. (2009) Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/Digeorge syndrome. Proc. Natl. Acad. Sci. U. S. A., 106, 16434–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Motahari Z., Moody S.A., Maynard T.M. and LaMantia A.S. (2019) In the line-up: deleted genes associated with Digeorge/22q11.2 deletion syndrome: are they all suspects? J. Neurodev. Disord., 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Josephson A., Trifunovski A., Widmer H.R., Widenfalk J., Olson L. and Spenger C. (2002) Nogo-receptor gene activity: cellular localization and developmental regulation of mRNA in mice and humans. J. Comp. Neurol., 453, 292–304. [DOI] [PubMed] [Google Scholar]
- 19. Yamamura K., Doi M., Hayashi H., Ota T., Murai I., Hotta Y., Komatsu R. and Okamura H. (2014) Immunolocalization of murine type vi 3beta-hydroxysteroid dehydrogenase in the adrenal gland, testis, skin, and placenta. Mol. Cell. Endocrinol., 382, 131–138. [DOI] [PubMed] [Google Scholar]
- 20. Ma L. and Tessier-Lavigne M. (2007) Dual branch-promoting and branch-repelling actions of Slit/Robo signaling on peripheral and central branches of developing sensory axons. J. Neurosci., 27, 6843–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozdinler P.H. and Erzurumlu R.S. (2002) Slit2, a branching-arborization factor for sensory axons in the mammalian CNS. J. Neurosci., 22, 4540–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiau C.E., Lwigale P.Y., Das R.M., Wilson S.A. and Bronner-Fraser M. (2008) Robo2-Slit1 dependent cell-cell interactions mediate assembly of the trigeminal ganglion. Nat. Neurosci., 11, 269–276. [DOI] [PubMed] [Google Scholar]
- 23. Hodge L.K., Klassen M.P., Han B.X., Yiu G., Hurrell J., Howell A., Rousseau G., Lemaigre F., Tessier-Lavigne M. and Wang F. (2007) Retrograde bmp signaling regulates trigeminal sensory neuron identities and the formation of precise face maps. Neuron, 55, 572–586. [DOI] [PubMed] [Google Scholar]
- 24. Ji S.J. and Jaffrey S.R. (2012) Intra-axonal translation of Smad1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron, 74, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simons M. and Nave K.A. (2015) Oligodendrocytes: myelination and axonal support. Cold Spring Harb. Perspect. Biol., 8, a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barembaum M. and Bronner-Fraser M. (2005) Early steps in neural crest specification. Semin. Cell Dev. Biol., 16, 642–646. [DOI] [PubMed] [Google Scholar]
- 27. Pandur P.D. and Moody S.A. (2000) Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech. Dev., 96, 253–257. [DOI] [PubMed] [Google Scholar]
- 28. Schlosser G., Awtry T., Brugmann S.A., Jensen E.D., Neilson K., Ruan G., Stammler A., Voelker D., Yan B., Zhang C. et al. (2008) Eya1 and Six1 promote neurogenesis in the cranial placodes in a Soxb1-dependent fashion. Dev. Biol., 320, 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Artinger K.B., Fedtsova N., Rhee J.M., Bronner-Fraser M. and Turner E. (1998) Placodal origin of Brn-3-expressing cranial sensory neurons. J. Neurobiol., 36, 572–585. [DOI] [PubMed] [Google Scholar]
- 30. Knuppel R., Dietze P., Lehnberg W., Frech K. and Wingender E. (1994) TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol., 1, 191–198. [DOI] [PubMed] [Google Scholar]
- 31. Lanier J., Dykes I.M., Nissen S., Eng S.R. and Turner E.E. (2009) Brn3a regulates the transition from neurogenesis to terminal differentiation and represses non-neural gene expression in the trigeminal ganglion. Dev. Dyn., 238, 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Todi S.V. and Paulson H.L. (2011) Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci., 34, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Semerci F., Choi W.T., Bajic A., Thakkar A., Encinas J.M., Depreux F., Segil N., Groves A.K. and Maletic-Savatic M. (2017) Lunatic fringe-mediated notch signaling regulates adult hippocampal neural stem cell maintenance. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamada M., Seto Y., Taya S., Owa T., Inoue Y.U., Inoue T., Kawaguchi Y., Nabeshima Y. and Hoshino M. (2014) Specification of spatial identities of cerebellar neuron progenitors by Ptf1a and Atoh1 for proper production of GABAergic and glutamatergic neurons. J. Neurosci., 34, 4786–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clifford M.A., Athar W., Leonard C.E., Russo A., Sampognaro P.J., Van der Goes M.S., Burton D.A., Zhao X., Lalchandani R.R., Sahin M. et al. (2014) Epha7 signaling guides cortical dendritic development and spine maturation. Proc. Natl. Acad. Sci. U. S. A., 111, 4994–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miake J., Notsu T., Higaki K., Hidaka K., Morisaki T., Yamamoto K. and Hisatome I. (2017) Cited4 is related to cardiogenic induction and maintenance of proliferation capacity of embryonic stem cell-derived cardiomyocytes during in vitro cardiogenesis. PLoS One, 12, e0183225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee G., Spring F.A., Parsons S.F., Mankelow T.J., Peters L.L., Koury M.J., Mohandas N., Anstee D.J. and Chasis J.A. (2003) Novel secreted isoform of adhesion molecule Icam-4: potential regulator of membrane-associated Icam-4 interactions. Blood, 101, 1790–1797. [DOI] [PubMed] [Google Scholar]
- 38. Lumb R., Buckberry S., Secker G., Lawrence D. and Schwarz Q. (2017) Transcriptome profiling reveals expression signatures of cranial neural crest cells arising from different axial levels. BMC Dev. Biol., 17, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson M.D., McCarthy D.J. and Smyth G.K. (2010) Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swillen A. and McDonald-McGinn D. (2015) Developmental trajectories in 22q11.2 deletion. Am. J. Med. Genet. C Semin. Med. Genet., 169, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonald-McGinn D.M., Sullivan K.E., Marino B., Philip N., Swillen A., Vorstman J.A., Zackai E.H., Emanuel B.S., Vermeesch J.R., Morrow B.E. et al. (2015) 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers., 1, 15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schneider M., Debbane M., Bassett A.S., Chow E.W., Fung W.L., Bree M., Owen M., Murphy K.C., Niarchou M., Kates W.R. et al. (2014) Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. Am. J. Psychiatry, 171, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y., Diacou A., Johnston H.R., Musfee F.I., McDonald-McGinn D.M., McGinn D., Crowley T.B., Repetto G.M., Swillen A., Breckpot J. et al. (2020) Complete sequence of the 22q11.2 allele in 1,053 subjects with 22q11.2 deletion syndrome reveals modifiers of conotruncal heart defects. Am. J. Hum. Genet., 106, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meechan D.W., Maynard T.M., Tucker E.S., Fernandez A., Karpinski B.A., Rothblat L.A. and LaMantia A.S. (2015) Modeling a model: mouse genetics, 22q11.2 deletion syndrome, and disorders of cortical circuit development. Prog. Neurobiol., 130, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajkumar A.P., Qvist P., Lazarus R., Lescai F., Ju J., Nyegaard M., Mors O., Borglum A.D., Li Q. and Christensen J.H. (2015) Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-Seq. BMC Genomics, 16, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bult C.J., Eppig J.T., Blake J.A., Kadin J.A., Richardson J.E. and Mouse Genome Database Group (2016) Mouse genome database 2016. Nucleic Acids Res., 44, D840–D847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yahata T., Takedatsu H., Dunwoodie S.L., Braganca J., Swingler T., Withington S.L., Hur J., Coser K.R., Isselbacher K.J., Bhattacharya S. et al. (2002) Cloning of mouse Cited4, a member of the cited family P300/CBP-binding transcriptional coactivators: induced expression in mammary epithelial cells. Genomics, 80, 601–613. [DOI] [PubMed] [Google Scholar]
- 48. Sundberg J.P., Dadras S.S., Silva K.A., Kennedy V.E., Garland G., Murray S.A., Sundberg B.A., Schofield P.N. and Pratt C.H. (2017) Systematic screening for skin, hair, and nail abnormalities in a large-scale knockout mouse program. PLoS One, 12, e0180682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tews B., Roerig P., Hartmann C., Hahn M., Felsberg J., Blaschke B., Sabel M., Kunitz A., Toedt G., Neben K. et al. (2007) Hypermethylation and transcriptional downregulation of the Cited4 gene at 1p34.2 in oligodendroglial tumours with allelic losses on 1p and 19q. Oncogene, 26, 5010–5016. [DOI] [PubMed] [Google Scholar]
- 50. Xu B., Hsu P.K., Stark K.L., Karayiorgou M. and Gogos J.A. (2013) Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell, 152, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jalbrzikowski M., Lazaro M.T., Gao F., Huang A., Chow C., Geschwind D.H., Coppola G. and Bearden C.E. (2015) Transcriptome profiling of peripheral blood in 22q11.2 deletion syndrome reveals functional pathways related to psychosis and autism spectrum disorder. PLoS One, 10, e0132542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Enge M., Arda H.E., Mignardi M., Beausang J., Bottino R., Kim S.K. and Quake S.R. (2017) Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell, 171, 321–330.e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cook D.L., Gerber A.N. and Tapscott S.J. (1998) Modeling stochastic gene expression: implications for haploinsufficiency. Proc. Natl. Acad. Sci. U. S. A., 95, 15641–15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stamoulis G., Garieri M., Makrythanasis P., Letourneau A., Guipponi M., Panousis N., Sloan-Bena F., Falconnet E., Ribaux P., Borel C. et al. (2019) Single cell transcriptome in aneuploidies reveals mechanisms of gene dosage imbalance. Nat. Commun., 10, 4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murray S.A., Morgan J.L., Kane C., Sharma Y., Heffner C.S., Lake J. and Donahue L.R. (2010) Mouse gestation length is genetically determined. PLoS One, 5, e12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elshamy W.M. and Ernfors P. (1996) Requirement of neurotrophin-3 for the survival of proliferating trigeminal ganglion progenitor cells. Development, 122, 2405–2414. [DOI] [PubMed] [Google Scholar]
- 57. Wilkinson G.A., Farinas I., Backus C., Yoshida C.K. and Reichardt L.F. (1996) Neurotrophin-3 is a survival factor in vivo for early mouse trigeminal neurons. J. Neurosci., 16, 7661–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim D., Langmead B. and Salzberg S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langmead B., Trapnell C., Pop M. and Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pertea M., Kim D., Pertea G.M., Leek J.T. and Salzberg S.L. (2016) Transcript-level expression analysis of RNA-Seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc., 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L. and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat. Protoc., 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Movassagh M., Alomran N., Mudvari P., Dede M., Dede C., Kowsari K., Restrepo P., Cauley E., Bahl S., Li M. et al. (2016) RNA2DNAlign: nucleotide resolution allele asymmetries through quantitative assessment of RNA and DNA paired sequencing data. Nucleic Acids Res., 44, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mudvari P., Movassagh M., Kowsari K., Seyfi A., Kokkinaki M., Edwards N.J., Golestaneh N. and Horvath A. (2015) SNPlice: variants that modulate intron retention from RNA-sequencing data. Bioinformatics, 31, 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McCarthy D.J., Chen Y. and Smyth G.K. (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res., 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thorvaldsdottir H., Robinson J.T. and Mesirov J.P. (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Benjamini Y. and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc., 57, 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


