Flaviviruses include many important human pathogens. The production of subgenomic flaviviral RNAs (sfRNAs) is important for viral pathogenicity as a common feature of flaviviruses. sfRNAs are formed through the incomplete degradation of viral genomic RNA by the cytoplasmic 5ʹ–3ʹ exoribonuclease Xrn1 halted at the Xrn1-resistant RNA (xrRNA) structures within the 3ʹ-UTR. The 3ʹ-UTRs of the flavivirus genome also contain distinct short direct repeats (DRs), such as RCS3, CS3, RCS2, and CS2. However, the biological functions of these ancient primary DR sequences remain largely unknown. Here, we found that DR sequences are involved in sfRNA formation and viral virulence and provide novel targets for the rational design of live attenuated flavivirus vaccine.
KEYWORDS: sfRNA, direct repeats, West Nile virus, Flavivirus
ABSTRACT
Mosquito-borne flaviviruses consist of a positive-sense genome RNA flanked by the untranslated regions (UTRs). There is a panel of highly complex RNA structures in the UTRs with critical functions. For instance, Xrn1-resistant RNAs (xrRNAs) halt Xrn1 digestion, leading to the production of subgenomic flaviviral RNA (sfRNA). Conserved short direct repeats (DRs), also known as conserved sequences (CS) and repeated conserved sequences (RCS), have been identified as being among the RNA elements locating downstream of xrRNAs, but their biological function remains unknown. In this study, we revealed that the specific DRs are involved in the production of specific sfRNAs in both mammalian and mosquito cells. Biochemical assays and structural remodeling demonstrate that the base pairings in the stem of these DRs control sfRNA formation by maintaining the binding affinity of the corresponding xrRNAs to Xrn1. On the basis of these findings, we propose that DRs functions like a bracket holding the Xrn1-xrRNA complex for sfRNA formation.
IMPORTANCE Flaviviruses include many important human pathogens. The production of subgenomic flaviviral RNAs (sfRNAs) is important for viral pathogenicity as a common feature of flaviviruses. sfRNAs are formed through the incomplete degradation of viral genomic RNA by the cytoplasmic 5ʹ–3ʹ exoribonuclease Xrn1 halted at the Xrn1-resistant RNA (xrRNA) structures within the 3ʹ-UTR. The 3ʹ-UTRs of the flavivirus genome also contain distinct short direct repeats (DRs), such as RCS3, CS3, RCS2, and CS2. However, the biological functions of these ancient primary DR sequences remain largely unknown. Here, we found that DR sequences are involved in sfRNA formation and viral virulence and provide novel targets for the rational design of live attenuated flavivirus vaccine.
INTRODUCTION
The Flavivirus genus includes many important human pathogens, such as West Nile virus (WNV), dengue virus (DENV), Zika virus (ZIKV), Japanese encephalitis virus (JEV), Murray Valley encephalitis virus (MVEV), and yellow fever virus (YFV), which have placed a large burden on public health worldwide. The flaviviruses are small, enveloped viruses with a single-stranded positive-sense RNA genome of around 11 kb that is divided into a 5ʹ untranslated region (5ʹ-UTR), an open reading frame (ORF), and a 3ʹ-UTR. The ORF encodes a polyprotein that can be processed into three structural and seven nonstructural proteins for viral particle formation and replication, respectively. The UTRs comprise highly complex structures involving well-conserved RNA sequences, which play multiple roles in different aspects of the viral life cycle (1–4), such as viral translation, replication, transmission, and pathogenesis. The 3ʹ-UTR of flaviviruses can be sequentially divided into three domains, namely, the stem-loop (SL) domain, the dumbbell (DB) domain, and the 3ʹ-stem-loop (sHP-3ʹ-SL) domain (5, 6). Besides these secondary and tertiary structures, flavivirus genomes also contain a few short direct repeats (DRs) in the 3ʹ-UTR (7). These DRs were originally identified as conserved sequence 3 (CS3) and CS2 and repeated conserved sequence 3 (RCS3) and RCS2 in the mosquito-borne flaviviruses (MBFV) (8). It has been proposed that DRs represent the evolutionary remnants of long repeated sequences from slowly evolving tick-borne flaviviruses (9) and may function during viral replication and transmission (10, 11). However, the biological functions of these ancient primary DR sequences in the flavivirus genome remain largely unknown (12), except for a report from Kieft’s group showing that RCS3 may be involved in subgenomic flaviviral RNA 1 (sfRNA1) production (13).
The accumulation of sfRNA has been well documented both in vitro and in vivo (4). Although the exact lengths and patterns of sfRNAs differ, all flaviviruses have the capacity to produce sfRNAs. sfRNAs function in viral pathogenicity (4), immune escape (3, 14, 15), and host adaptation (1, 16), and their production is attributed to incomplete degradation of viral genomic RNA by the cytoplasmic 5ʹ–3ʹ exoribonuclease Xrn1 (4, 6, 17, 18), which is halted at the Xrn1-resistant RNA (xrRNA) structures within the 3ʹ-UTR (1, 17). The xrRNA1 structures of MVEV and ZIKV have been characterized previously (13, 19), and it has been shown that RNA secondary structures combined with pseudoknot (PK) interactions within the RNA structure of xrRNAs are required for sfRNA formation (4, 17, 19). According to nomenclature of xrRNA1 used by Kieft’s laboratory, the DR sequence of RCS3 was recognized as the P4/L4 stem-loop region of xrRNA1. The docking of the crystal structure of ZIKV xrRNA1 with Xrn1 indicated the putative interaction between the stem (P4) of RCS3 and Xrn1 (13), which may serve to stabilize the xrRNA’s pseudoknot interaction and thus enhance resistance to Xrn1 cleavage. However, the biological function of RCS3 for sfRNA1 formation still needs to be explored.
In this study, we discovered that the specific DR elements (RCS3/CS3, RCS2/CS2) within the 3ʹ-UTR are involved in production of different sfRNA species through mutagenesis analysis in the context of the infectious clones of WNV, DENV2, and ZIKV. These DRs were found to form stem-loop structures, and the base pairings in the stem of each DR element are important for sfRNA production within virus-infected cells. Although an in vitro cleavage assay showed that RCS3 is dispensable for the cleavage of xrRNA1, an isothermal titration calorimetry (ITC) assay demonstrated that RCS3 is important for the binding of the exoribonuclease Xrn1 to xrRNA1.
RESULTS
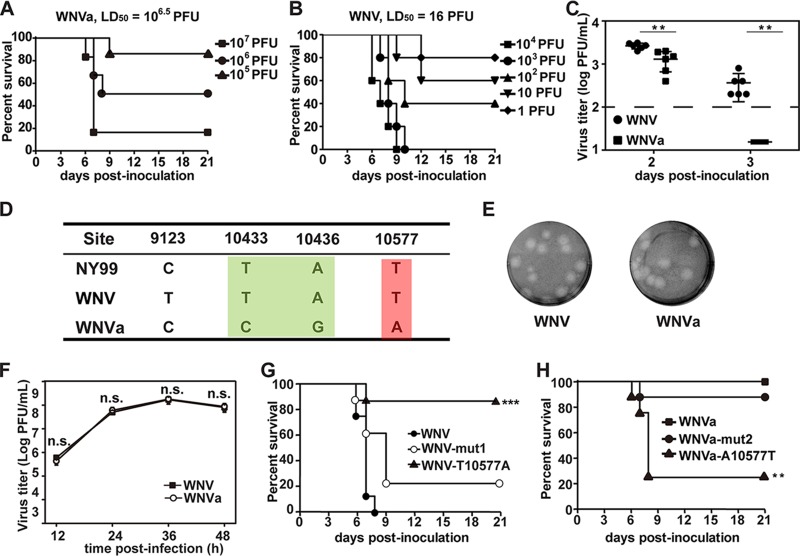
The viral pathogenicity of different WNV strains in mice correlates with a single nucleotide change (T10577A) within the 3ʹ-UTR.
During our initial plaque purification of the WNV infectious clone of strain 3356 from New York City (20), we accidently obtained a WNV variant (WNVa) with an excellent attenuation phenotype in mouse virulence (Fig. 1A). The 50% lethal dose (LD50) in adult BALB/c mice was found to have decreased by over 100,000-fold compared with that of the wild-type (WT) WNV strain (Fig. 1B). WNV produced significantly higher levels of viremia than WNVa on day 2 and day 3 (Fig. 1C). The viremia of WNVa dropped to almost undetectable levels on days 3 after infection. Whole-genome sequencing revealed only four nucleotide differences between these two viruses (Fig. 1D): one substitution at position 9123 within the NS5 coding region and three substitutions within the 3ʹ-UTR at positions 10433, 10436, and 10577. To confirm which nucleotide change resulted in the attenuation phenotype, T10577A or T10433C plus A10436G substitutions were first introduced into the infectious clone of WNV (Fig. 1G). The rescued virus harboring the T10433C plus A10436G double mutation (WNV-mut1) was similar to the WT virus with regard to viral growth properties as well as virulence in mice. Interestingly, the single T10577A substitution significantly attenuated the virulence of WNV (Fig. 1G). Meanwhile, complementary A10577T in the infectious clone of WNVa restored the virulence phenotype of WNVa, and C10433T plus G10436A substitutions had no effect on the viral virulence of WNVa (Fig. 1H). These results demonstrate that position 10577 in the 3ʹ-UTR is critical for WNV virulence.
FIG 1.
A single nucleotide change (T10577A) within RCS3 of the 3ʹ-UTR leads to attenuation of WNV virulence in mice. (A and B) Survival analysis of mice infected with WNV. Four-week-old BALB/c mice (n = 5 to 9) were i.p. inoculated with the indicated dose of (A) WNVa or (B) WNV and then monitored for clinical symptoms and mortality over 21 days. LD50, 50% lethal dose. (C) Viremia differences between WNV and WNVa. Four-week-old of BALB/c mice (n = 6) were intraperitoneally (i.p.) inoculated with 105 PFU indicated viruses. Blood was taken at the second and third days after infection, and virus titer was determined by plaque assay. The asterisks denote the statistical significance of differences between results from the indicated groups. **, P < 0.01. (D) Four nucleotide differences between WNV and WNVa were identified at nucleotide positions 9123, 10433, 10436, and 10577 through complete-genome sequence alignment. Corresponding nucleotides from the isolated virulent NY99 strain are also listed. (E) Plaque morphologies of WNV and WNVa in BHK-21 cells. (F) Growth kinetics of WNV and WNVa in BHK-21 cells. WNV and WNVa showed no significant differences in viral titers at each time point. n.s., no significant statistical differences. (G) Survival analysis of mice infected with WNV, WNV-mut1, and WNV-T10577A. Four-week-old of BALB/c mice (n = 8) were intraperitoneally (i.p.) inoculated with 105 PFU of the indicated viruses and monitored for clinical symptoms and mortality over 21 days. ***, P < 0.001. (H) Survival analysis of mice infected with WNVa, WNVa-mut2, and WNVa-A10577T. Four-week-old BALB/c mice (n = 8) were i.p. inoculated with 105 PFU of the indicated viruses and monitored for clinical symptoms and mortality over 21 days. **, P < 0.01.
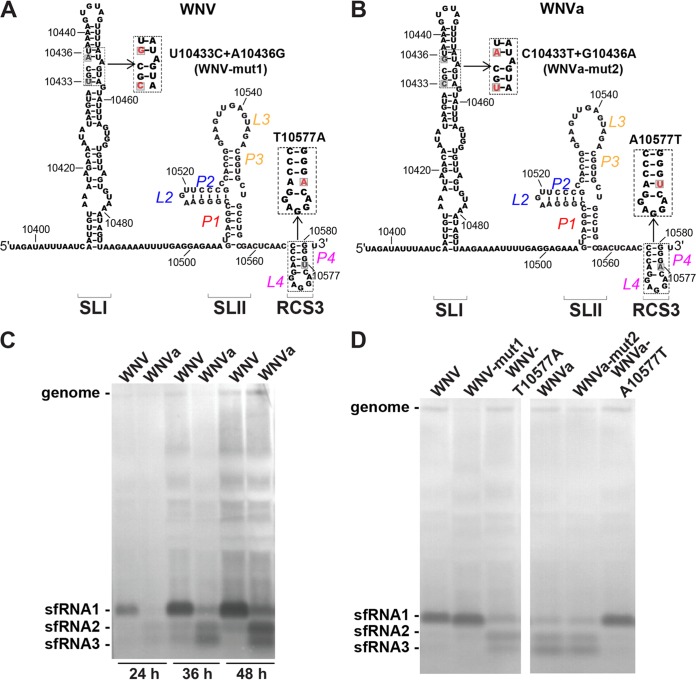
A single nucleotide change (T10577A) within RCS3 of the 3ʹ-UTR alters sfRNA production patterns.
On the basis of the known RNA secondary and tertiary structure model (Fig. 2A), T10577 resides in the P4 stem of RCS3 within the xrRNA1 RNA structure (13, 19). The putative interaction between P4 of RCS3 and Xrn1 (13) indicated a possible function of RCS3 for sfRNA production as mentioned above. We then first examined sfRNA production of WNV and WNVa. As shown in Fig. 2C, the patterns of sfRNA production for WNV and WNVa were distinct although the two viruses demonstrated similar growth curves and plaque morphologies (Fig. 1E and F) in BHK-21 cells. Specifically, abundant sfRNA1 was observed in WNV-infected BHK-21 cells, which agrees with previous studies (4), whereas lower levels of sfRNA2 and sfRNA3 were found to have accumulated in WNVa-infected BHK-21 cells (Fig. 2C). In the background of WNV, T10577A altered sfRNA1 production and T10433C plus A10436G substitutions (WNV-mut1) had no effect on sfRNA1 formation (Fig. 2A and D). At the same time, A10577T restored WNVa sfRNA1 production and C10433T plus G10436A substitutions had no effect on sfRNA production patterns of WNVa (Fig. 2B and D). These results demonstrate that position 10577 in the 3ʹ-UTR is critical for WNV sfRNA1 production and indicate that RCS3 may be involved in sfRNA1 production.
FIG 2.
A single nucleotide change (T10577A) within RCS3 of the 3ʹ-UTR alters sfRNA production patterns. (A) Diagram of putative secondary structure of WNV xrRNA1. RNA sequences represent nucleotide positions 10396 to 10581, and the location of the 10577 site within RCS3 is labeled. The inset shows the mutation used to test the importance of identified nucleotides in viral virulence in the context of the WNV infectious clone. (B) Putative secondary structure diagram and RNA sequence of nucleotide positions 10396 to 10581. The inset shows mutations in the context of the WNVa infectious clone. (C) Northern blot analysis of total RNA extracted from BHK-21 cells infected with WNV and WNVa at 24, 36, and 48 h postinfection. (D) Northern blot of total RNA extracted from BHK-21 cells infected with WNV or WNVa or the indicated mutant at 36 h postinfection. The sfRNA signals are indicated as sfRNA1, sfRNA2, and sfRNA3 in descending size order.
RCS3 determines sfRNA1 production of WNV.
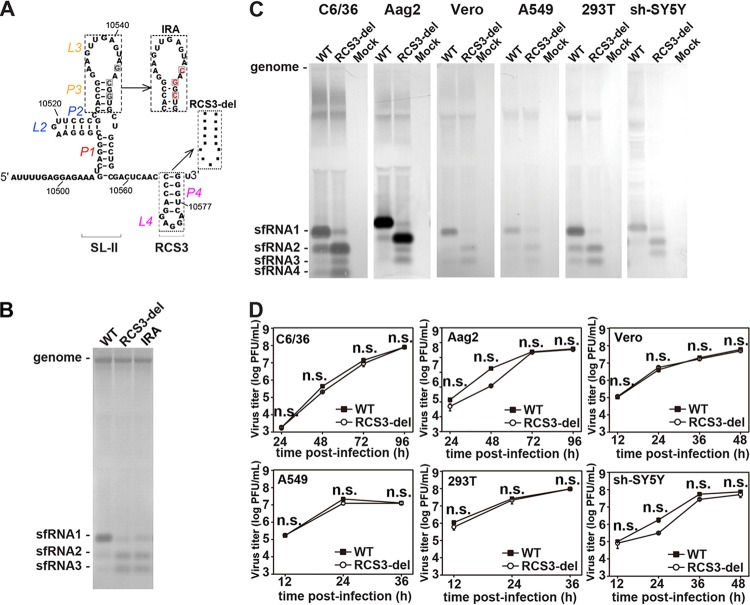
We next sought to further confirm the role of RCS3 in sfRNA1 formation. Initially, a recombinant WNV with a deletion of full-length RCS3 (RCS3-del) was constructed (Fig. 3A). In parallel, an inverted repeat (IRA) mutation (Fig. 3A) within the SL of SL-II containing a 3-nucleotide (nt) reversed complementary substitution (4) was prepared as a control for the attenuation of sfRNA1 production. Notably, the deletion of RCS3 dramatically impaired sfRNA1 production and led to increased sfRNA2 and sfRNA3 production in a manner similar to that seen with the IRA mutation (Fig. 3B). Further validation in C6/36, Aag2, Vero, A549, 293T, and sh-SY5Y cells showed sfRNA production patterns similar to those observed in BHK-21 cells (Fig. 3C), indicating that RCS3 determines sfRNA formation in a cell type-independent manner. Moreover, similar viral growth curves in these cells (Fig. 3D) were observed for the WT and RCS3-del, further indicating that the different sfRNA production profiles were not due to viral replication efficiency. Overall, our results demonstrate that RCS3, an RNA element downstream of xrRNA1, determines the production of upstream sfRNA1.
FIG 3.
RCS3 determines sfRNA1 production of WNV. (A) Secondary structure of WNV SLII and RCS3. The various mutations within SLII and RCS3 are indicated in the inset. (B) sfRNA detection through Northern blotting of total RNA extracted from BHK-21 cells infected with recombinant WNV containing mutations of IRA or RCS3 deletion. (C) sfRNA production profiles of WT WNV and RCS3-del in different cell lines. Data represent results of Northern blot analysis of total RNA purified from WNV-infected mosquito C6/36 and Aag2, Vero, human A549 and 293T, and human neuroblastoma sh-SY5Y cell lines. Total viral RNAs were collected at peak titers in the infected cells. (D) Growth kinetics of WT WNV and mutant WNV with the RCS3-del mutation in different cell lines. The RCS3-del and WT viruses showed no significant differences in viral titers at any time point in all the tested cell lines. n.s., no statistically significant differences.
The stem-loop structure of RCS3 is essential for sfRNA1 production.
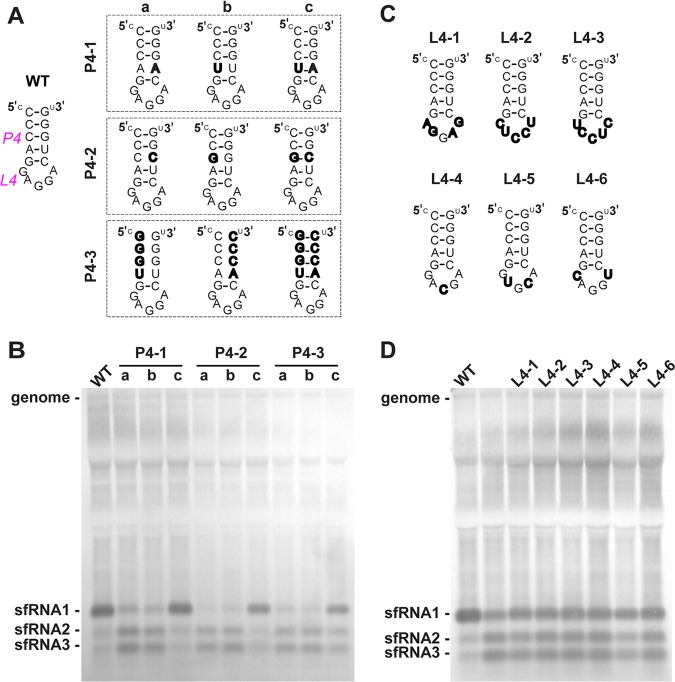
RCS3 contains a stem-loop structure that has been confirmed using RNase digestion in WNV (6). In accordance with the nomenclature of the secondary structure of xrRNA1 from Kieft’s laboratory (13, 17, 19), the stem and loop structures of RCS3 are identified as P4 and L4 (Fig. 4A), respectively. To determine which RNA sequences and/or structures of RCS3 are important for WNV sfRNA1 production, we designed several panels of mutants targeting the stem (P4) and loop (L4) of RCS3 (Fig. 4A) to test their functional importance in sfRNA1 production. Disruption of the base parings of the P4 stem (P4-1a/1b, P4-2a/2b, and P4-3a/3b) dramatically impaired sfRNA1 production and increased sfRNA2 and sfRNA3 levels (Fig. 4B). Reconstitution of P4 complementarity (P4-1c, P4-2c, and P4-3c) restored sfRNA1 production. In contrast, all six loop mutations favored sfRNA1 production (Fig. 4C and D), albeit with increased levels of sfRNA2 and sfRNA3 production. Thus, the base pairing of the P4 stem, rather than the primary sequence of RCS3, is critical for sfRNA1 production.
FIG 4.
The stem-loop structure of RCS3 is essential for sfRNA1 production. (A) Secondary structure of WNV RCS3. The various mutations within P4 are indicated in the inset. (B) sfRNA detection through Northern blotting of total RNA extracted from BHK-21 cells infected with recombinant WNV containing various mutations within stem of RCS3, recognized as P4. (C) Different loop mutations within the RCS3 loop, recognized as L4. To minimize structural changes, most mutations maintained the same number of nucleotides within the loop. L4-1 contains the complete reversed sequences of the loop. L4-2 and L4-3 have the complete complementary and reversed complementary loop sequences, respectively. L4-4, L4-5, and L4-6 have one or two complementary nucleotides altered at the corresponding position of the loop sequence. (D) Northern blot analysis of RNA purified from BHK-21 cells infected with recombinant WNVs containing the different L4 mutations at 36 h postinfection. Lane 2 represents an unrelated mutant.
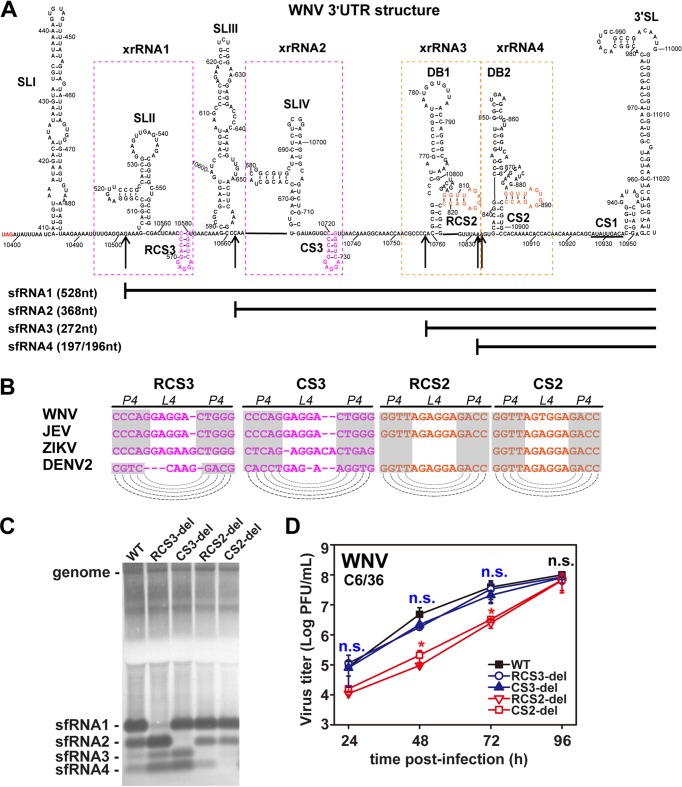
Conserved DRs of WNV are involved in the production of various sfRNA species.
In addition to RCS3, there are several other DRs within the 3ʹ-UTR of WNV (CS3, RCS2, and CS2) that are located downstream of their corresponding xrRNAs (Fig. 5A and B). Therefore, we further explored whether these DRs following the known xrRNAs also contribute to upstream sfRNA production in the manner in which RCS3 does. Typically, four species of sfRNAs (sfRNA1, sfRNA2, sfRNA3, and sfRNA4) are produced during WNV and DENV2 infection (5, 16) at the corresponding xrRNA structures (xrRNA1, xrRNA2, xrRNA3, and xrRNA4) (17) within the 3ʹ-UTRs (Fig. 5A). According to the classification of xrRNAs, xrRNA1 and xrRNA2 are SL-type structures whereas xrRNA3 and xrRNA4 are DB-type structures (2). WNV mutants with CS3, RCS2, or CS2 deletions were constructed and recovered. Remarkably, Northern blot analysis demonstrated that CS3, RCS2, and CS2 deletions impaired the production of sfRNA2, sfRNA3, and sfRNA4, respectively, in C6/36 cells (Fig. 5C and D) without altering sfRNA1 production. Further mutagenesis indicated that the SL structures of CS3 and RCS2 are also important for the production of their corresponding sfRNAs (sfRNA2 and sfRNA3). Together, these results demonstrate that the DR of each xrRNA is involved in the production of its upstream sfRNA.
FIG 5.
Conserved DRs of WNV control production of various sfRNA species. (A) Secondary structure of WNV 3ʹ-UTR. The various secondary structures formed within the 3ʹ-UTR are indicated. The putative SLs of RCS3/CS3 and RCS2/CS2 are marked in pink and orange, respectively. The 5′ ends of sfRNA1, sfRNA2, sfRNA3, and sfRNA4 are indicated by arrows. The sizes of different sfRNA species are also indicated. xrRNA1, xrRNA2, xrRNA3, and xrRNA4 are depicted by dotted boxes. (B) Conserved repeated sequences from various flaviviruses. Dashed lines indicate putative base pairing with the SL structures of conserved repeated sequences as indicated in the diagram. (C) Northern blot of total RNA extracted from C6/36 cells infected with WT or mutant WNVs with RCS3, CS3, RCS2, or CS2 deletions. sfRNA signals are marked as sfRNA1, sfRNA2, sfRNA3, and sfRNA4 in descending size order. (D) Growth kinetics of recombinant WNVs with individual RCS3, CS3, RCS2, and CS2 deletions in C6/36 cells. RCS3-del, CS3-del, and WT virus titers showed no significant differences at different time points. n.s., no statistically significant differences. RCS2-del and CS2-del virus titers showed statistically significant differences from the WT virus titers at 24, 48, and 72 h postinfection (hpi) (*, P < 0.05), and there were no statistically significant differences at 96 hpi.
DR sequences also control production of sfRNAs of DENV2 and ZIKV.
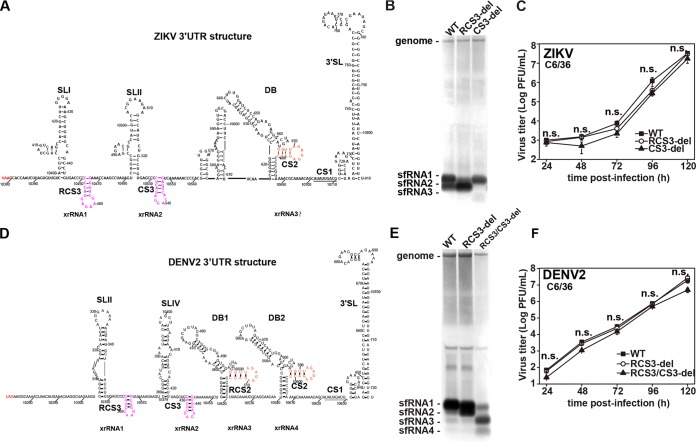
Since DRs are present in all flaviviruses (12), we further expanded our examination to ZIKV and DENV. Similarly, recombinant ZIKV with RCS3 and CS3 deletions were constructed and recovered on the basis of the infectious clone of ZIKV strain SZ-WIV01 (21) (Fig. 6A). Consistent with the WNV results, RCS3 and CS3 deletions also blocked formation of sfRNA1 (Fig. 6B, lane 2) and sfRNA2 (Fig. 6B, lane 3) of ZIKV, respectively, without dramatically impairing viral replication (Fig. 6C). Since the 3ʹ-UTR of ZIKV does not contain RCS2 sequence, we believed that the weak sfRNA3 band shown in Fig. 6B was derived from DB containing CS2 sequence. Similarly, we also demonstrated that RCS3 deletion in the context of DENV (22) impaired the pattern of sfRNA production with increasing amounts of sfRNA2 and that double deletions of RCS3 plus CS3 blocked the production of sfRNA1 and sfRNA2 (Fig. 6D to F). Overall, our results suggest that these DRs may play similar roles in the production of sfRNAs of flaviviruses.
FIG 6.
DR sequences also control production of DENV2 and ZIKV sfRNAs. (A) Secondary structure of ZIKV 3′-UTR. The different RNA secondary structures and CS sequences are indicated. (B) Northern blot of total RNA extracted from C6/36 cells infected with recombinant ZIKV with RCS3 or CS3 deletions. (C) Growth kinetics of recombinant ZIKVs with individual RCS3 and CS3 deletions in C6/36 cells. RCS3-del, CS3-del, and WT virus titers showed no significant differences at different time points. n.s., no statistically significant differences. (D) Secondary structure of DENV2 3′-UTR. The different RNA secondary structures and CS sequences are indicated. (E) Northern blot analysis of total RNA extracted from C6/36 cells infected with recombinant DENV2 containing RCS3 and RCS3/CS3 deletions. (F) Growth curves of recombinant DENV2 viruses in C6/36 cells. RCS3-del, RCS3/CS3-del, and WT virus titers showed no significant differences at different time points. n.s., no statistically significant differences.
RCS3 controls the binding affinity of xrRNA1 to Xrn1.
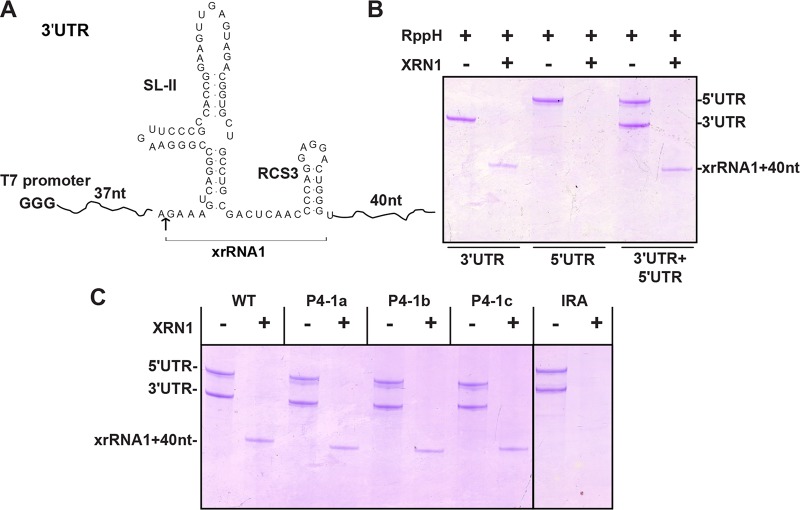
We then performed Xrn1 resistance cleavage assay of xrRNA1 to decipher the underlying mechanisms accounting for how these DR elements control sfRNA production. Surprisingly, xrRNA1 with P4-1a and P4-1b mutations within RCS3, which dramatically impaired sfRNA1 production in the cell culture (Fig. 4B), still resisted Xrn1 cleavage similarly to the WT (Fig. 7C). The discrepancies between the in vitro and in vivo assay results are discussed below (see Discussion).
FIG 7.
Xrn1 resistance assay performed in vitro for analysis of sfRNA RNA. (A) Briefly, a 158-nt uncapped WNV 3′-UTR RNA corresponding to nt 10464 to 10621 containing SLII plus RCS3 (nt 10502 to 10580; resistance to XRN1 cleavage) was prepared for Xrn1 resistance assay. (B) The production of shorter xrRNA1 of the input 3′-UTR was used to demonstrate Xrn1 cleavage resistance of 3′-UTR (lanes 1 and 2). Additionally, WNV 5′-UTR RNA corresponding to nt 1 to 190 was prepared as a control for complete cleavage by Xrn1 (lanes 3 and 4). (C) Xrn1 resistance assay of various RNAs with P4 mutations or IRA mutation.
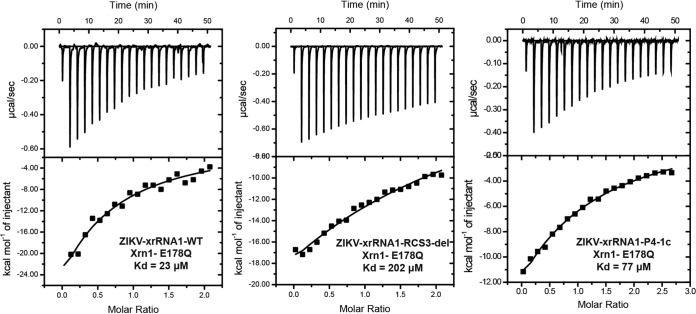
To generate sfRNA, host Xrn1 is first loaded onto the 5′ end of viral genomic RNA, which then begins to degrade the viral genome, halting at the xrRNAs within the 3′-UTR (17). Structural remodeling of Xrn1 and ZIKV xrRNA1 (13) revealed that the fully folded ZIKV xrRNA1 closely matches the contours of an electropositive patch of Xrn1, with P4 contacting a conserved charged region on the winged helix domain of Xrn1 (13). Thus, the binding affinity between Xrn1 and various xrRNAs may be of critical importance. We performed isothermal titration calorimetry (ITC) to assay the binding affinity of Xrn1 with xrRNA1 containing different RCS3 mutations (Fig. 8). To focus on the binding affinity, the Xrn1-E781Q mutant, which aborts the cleavage activity of Xrn1 (23), was used. As expected, WT ZIKV xrRNA1 bound to the Xrn1-E178Q protein with strong affinity (dissociation constant [Kd] = 23 μM). In contrast, the binding affinity for the ZIKV RCS3-del mutant was approximately 10 times lower than that for WT xrRNA1. Interchanging the bilateral sequence of the P4 helix (P4-1c), which maintains the base pairing of the stem, restored the binding affinity greatly (at three times higher than that seen with RCS3-del) (Fig. 8).
FIG 8.
RCS3 controls Xrn1 binding affinity to xrRNA. Data represent results of analyses of binding affinity of Xrn1 to the xrRNA1 of ZIKV measured by ITC assay. The dissociation constant (Kd) values were calculated to be 23 μM, 202 μM, and 77 μM for ZIKV-xrRNA1-WT, ZIKV-xrRNA1-RCS3-del, and ZIKV-xrRNA1-P4-1c, respectively.
The stem-loop structure of RCS3 stabilizes the three-dimensional structure of xrRNA.
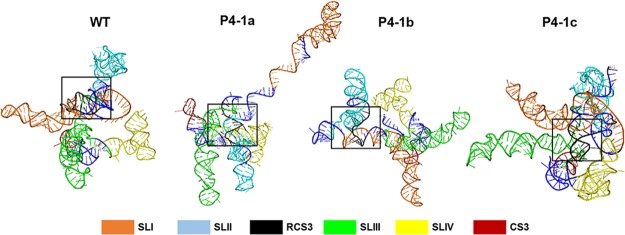
To clarify the mechanistic explanation of why the DR mutations destabilize interactions between xrRNAs and Xrn1, we performed computational modeling of xrRNAs with and without RCS3 mutations using RNAComposer (Fig. 9). P4-1a and P4-1b mutations resulted in relatively loose tertiary folding of xrRNA1, preventing the interaction between P4 and Xrn1 that is crucial for resistance to Xrn1. In contrast, P4-1c was able to greatly restore the stem-loop structure that displays a tight tertiary folding. Similarly to WT RCS3, it enabled P4 to bind to Xrn1 to stabilize xrRNA’s pseudoknot interaction for Xrn1 resistance (13).
FIG 9.
Three-dimensional RNA structure model of sfRNA. The first 350 nt of 3′-UTR from SLI to CS3 was used for three-dimensional (3D) RNA structure model analysis performed with the RNAComposer Web server with standard settings. The color of each structure matches the color of the labeled box. RCS3 is depicted as black in a square box. For WT and P4-1c, a complete pairing of RCS3 stem resulted in very tight tertiary folding, whereas for P4-1a or P4-1b mutant RNA, impaired base pairing of RCS3 stem resulted in loose tertiary folding.
DISCUSSION
Production of sfRNAs represents a tactic used by flaviviruses for productive infection (24). sfRNAs are the products of incomplete degradation of the viral genome by cellular Xrn1 (25). The xrRNA structures within the 3ʹ-UTR are the RNA elements required for stalling Xrn1 and are therefore crucial for sfRNA formation (4, 6, 17, 18). Two types of xrRNA structures (the SL type and the DB type) and the PK interactions stabilizing these structures have been demonstrated to play a significant role in the production of sfRNAs of different lengths (2, 6). In this study, we demonstrated that conserved repeat sequences of DRs (RCS3, CS3, RCS2, and CS2), which are associated with all of the known duplicated SL-based and DB-based stalling structures, are involved in the production of corresponding sfRNA species within infected cells and thus represent novel determinants for sfRNA formation. Our results confirmed the previously published hypothesis that RCS3 may play an important role in sfRNA1 formation (13). Previous analyses of the crystal structure of ZIKV xrRNA1 docking with Xrn1 showed that there are extensive contacts between the enzyme and xrRNA1 and that P4 of RCS3 contacts a conserved charged region on the winged helix domain of Xrn1 (13). It was proposed that this putative P4-Xrn1 interaction could serve to stabilize xrRNA’s pseudoknot interaction and thus enhance resistance to Xrn1 (13). The interaction between P4 of RCS3 and Xrn1 may also prevent conformational changes in the enzyme that are important for processive Xrn1 function (13). In the present study, we found that RCS3 did enhance xrRNA1 resistance to Xrn1 by interacting with Xrn1 through its stem (Fig. 8), contributing to sfRNA1 formation (Fig. 3; see also Fig. 4).
No role of DRs (RCS3/CS3, RCS2/CS2) in production of sfRNAs was observed in previous studies. One major reason is that RCS3 mutations exhibited no effect on xrRNA1-resistant cleavage of Xrn1 in biochemical assays (17, 19), which is consistent with our in vitro cleavage assay of Xrn1 (Fig. 7C). It is possible that the discrepancies between the in vitro and in vivo assay results can be accounted for by the mechanism involved in the binding affinity between Xrn1 and xrRNA1 mentioned above. Alternatively, it is possible that the discrepancies resulted from the choice of the source organism of Xrn1. The Xrn1 exonucleases used in our study were from Kluyveromyces lactis (23, 26) and may not completely mimic the properties of Xrn1 for sfRNA biogenesis in the case of viral infection within mammalian and mosquito cells.
DRs of 3′-UTRs are conserved among flaviviruses (8). Various numbers of DR elements have been found within the 3′-UTR of flaviviruses. Japanese encephalitis virus (JE) subgroups contain the greatest numbers of DRs, which are RCS3, CS3, RCS2, CS2, and CS1 in the 5′ to 3′ direction. With the exception of CS1, CS and RCS are not essential for virus replicative capacity or viability (11, 27, 28). The evolutionary analysis indicates that DRs may function in the transmission of flaviviruses (10, 11). For DENV2, it was demonstrated previously that duplicated RNA structures of SLI/SLII or DB1/DB2 are relevant to viral host adaptation (2, 25, 29), and a host-specific pattern of sfRNA generation was observed previously in DENV2-infected vertebrate and invertebrate cells (1). As the current study showed that DRs are required for production of sfRNAs, we speculated that conserved DRs may be involved in flavivirus transmission among different hosts through modulating the formation of sfRNAs. These hypotheses require further investigation in future study.
Many viral factors are involved in flavivirus virulence through different mechanisms. There is a growing body of evidence showing that sfRNA formation is related to viral pathogenesis through modulating the innate immunity responses (3, 4, 14). Here, we report that DRs are responsible for sfRNA production, which also explains the attenuation mechanism of live attenuated DENV and ZIKV vaccines containing the CS2 deletions (30, 31) and indicates that DRs could be considered an attractive target for development of live attenuated vaccines.
MATERIALS AND METHODS
Cell culture.
BHK-21, Vero, A549, 293T, and sh-SY5Y cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (PS) and incubated in 5% CO2 at 37°C. C6/36 cells were propagated in RPMI 1640 medium supplemented with 10% FBS and PS at 28°C. Aag2 cells were grown in Schneider’s Drosophila medium supplemented with 5% FBS and PS at 5% CO2 and 28°C. Infection of different cell lines was performed in the corresponding medium with 2% FBS and PS as indicated.
Plasmid construction.
We initially used the infectious clone of WNV—containing the T7 promoter (WNV and WNVa)—as the backbone (20) for introducing 3ʹ-UTR mutations. Briefly, SpeI and AflII restriction sites and MluI and XbaI restriction sites were used to engineer WNV and WNVa mutations by fusion PCR, respectively. To facilitate recombinant virus production, the cytomegalovirus (CMV) promoter was introduced to replace the T7 promoter of the infectious clone of WNVa-A10577T at BamHI and EcoNI sites via overlapping PCR, and the resulting cDNA clone was designated pCMV-WNV-WT. RCS3, CS3, RCS2, and CS2 deletions and multiple-location mutations were introduced into full-length pCMV-WNV-WT via the use of MluI and XbaI restriction sites and fusion PCR. ZIKV 3ʹ-UTR mutations were introduced into the F4 subclone through overlapping PCR, and then four ZIKV fragments were digested at the BglI site and ligated in vitro to construct full-length ZIKV cDNA as described before (21). DENV2 3ʹ-UTR mutations were constructed in the full-length pACYC-DENV2-NGC infectious clone (32) via the use of MluI and XbaI restriction sites and fusion PCR. All constructs were verified by DNA sequencing.
Virus production.
Recombinant wild-type (WT) and mutant WNV, ZIKV, and DENV2 with the T7 promoter were generated by electroporation of BHK-21 cells with in vitro-transcribed viral genome RNAs from linearized infectious cDNA clones (20, 21). A T7 mMESSAGE mMACHINE kit (Ambion, Waltham, MA) was used for in vitro transcription. WT and mutant WNV with the CMV promoter were obtained by transfection of the cDNA clone. BHK-21 cells were transfected with 1 μg DNA using 3 μl FuGENE HD transfection reagent (Promega, Madison, WI) in 35-mm-diameter dishes. The supernatants of transfected cells were harvested 72 h posttransfection.
Plaque assay and viral growth kinetics.
Virus titer and morphology were determined by the monolayer plaque assay. Briefly, a series of 1:10 dilutions were prepared by diluting 15 μl virus stock with 135 μl DMEM containing 2% FBS, and 100 μl of each dilution was seeded onto 24-well plates containing confluent BHK-21 cells (1 × 105 cells/well; plated 1 day in advance). Infected cells were incubated at 37°C and 5% CO2 for 1 h before the medium containing 2% methylcellulose was overlaid. After 3 or 4 days of incubation at 37°C and 5% CO2, the cells were fixed in 3.7% formaldehyde and then stained with 1% crystal violet. The viral titer was calculated as the number of PFU per milliliter. For determination of viral growth curves, BHK-21, Vero, A549, 293T, sh-SY5Y, mosquito C6/36, and Aag2 cells were infected at a multiplicity of infection (MOI) of 0.1 and the cell cultures were harvested at the indicated time points postinfection. Viral titers were then determined by the plaque assay. One-way analysis of variance was used with multiple comparisons to compare virus titers from all time points.
Northern blot analysis.
For WNV, ZIKV, and DENV2 genome and sfRNA detection, total RNAs from infected cells were obtained using TRIzol reagent (Invitrogen, Carlsbad, CA) at the indicated time points postinfection. Approximately 20 μg RNA was separated on a 1.5% agarose with 2% formaldehyde gel, followed by transfer onto Hybond-N+ membranes (GE Healthcare, Chicago, IL) using capillary migration. Next, the blots were UV cross-linked and hybridized with digoxigenin (DIG)-ddUTP-labeled 3ʹ-UTR DNA probes. Membrane blocking and visualization were performed using DIG Northern starter kit I (Roche, Basel, Switzerland). The probe sequence for WNV detection consisted of nt 10894 to 11029, the probe sequence for detection of labeled ZIKV was nt 10699 to 10808, and the labeled probe sequence for DENV2 RNA detection was nt 10622 to 10723 of the genome. Each assay was performed in triplicate, and similar results were obtained.
Sequencing of sfRNAs.
To identify the exact 5′-end sequence of different sfRNAs, TRIzol-extracted RNAs from infected C6/36 cells were used. RNAs from WT WNV, RCS3-del, RCS3/CS3-del, and RCS3/CS3/RCS2-del were used for sfRNA1, sfRNA2, sfRNA3, and sfRNA4 sequencing, respectively. The 5′ phosphorylated RNAs were subjected to 5′–3′ end ligation by the use of T4 RNA ligase (Ambion, Waltham, MA) at 4°C overnight and used as the templates for reverse transcription performed using SuperScript III reverse polymerase (Invitrogen) with primer 5′-TCCCAGGTGTCAATATGCTG-3ʹ. The cDNA was then used as the template for PCR with different sets of internal primers. Products were gel purified and sequenced, and then the 5′–3′-end junction was identified.
Expression and purification of recombinant proteins.
A plasmid encoding 6×His-tagged Xrn1 from Kluyveromyces lactis (residues 1 to 1245) was kindly provided by Liang Tong of Columbia University. Mutant Xrn1-E178Q was constructed using a fast mutagenesis system kit (Transgen Biotech, Beijing, China) and confirmed by sequencing. A plasmid encoding RppH from Bdellovibrio bacteriovorus was subjected to total gene synthesis by Wuxi Qinglan Biotechnology Inc. (Wuxi, China) and further subcloned into the pET28a expression vector. The expression and purification procedures were similar to those previously described (23, 26). Purified Xrn1-E178Q in buffer containing 20 mM Tris (pH 7.5), 150 mM KCl, 4 mM MgCl2, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was flash frozen with liquid nitrogen and stored at −80°C. Purified BdRppH was stored in buffer containing 20 mM Tris (pH 7.5), 200 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 50% (vol/vol) glycerol at −80°C.
In vitro transcription and purification of RNA.
Plasmids encoding an upstream T7 promoter and WNV 3′UTR corresponding to nt 10464 to 10621 and WNV 5′UTR corresponding to nt 1 to 190, as an internal control, were subjected to Xrn1 resistance assay. Plasmids encoding the T7 promoter and WNV xrRNA1 (nt 10501 to 10580) or ZIKV xrRNA1 (nt 10392 to 10466) were subjected to ITC assay. RNA was transcribed in vitro using T7 RNA polymerase and then purified by passage through a size exclusion chromatography Superdex 75 preparation-grade (pg) column (GE Healthcare, Chicago, IL) with running buffer (20 mM Tris [pH 7.5], 100 mM KCl, and 4 mM MgCl2).
In vitro Xrn1 resistance assay.
The purified 2 μg 3′-UTR and 2 μg 5′-UTR RNAs were first refolded (90°C, 2 min; 20°C, 5 min; cooling at 4°C) and then treated with RppH for 30 min at 37°C to remove 5′-triphosphate. 5′-Monophosphate RNA was split between two tubes. One tube was added into purified Xrn1, while the other served as a negative (-) Xrn1 control in 1× EC3 buffer (100 mM NaCl, 10 mM MgCl2, 50 mM Tris [pH 7.9], 1 mM DTT). RNA species were resolved by 10% polyacrylamide–7 M urea gel electrophoresis and visualized by staining with Stains-All (Sigma-Aldrich).
Isothermal titration calorimetry (ITC).
For ITC assay, purified RNAs were directly digested with RppH. After digestion, the RppH protein was removed by phenol-chloroform extraction and the RNA was further purified by size exclusion chromatography in a running buffer containing 20 mM Tris (pH 7.5), 150 mM KCl, 4 mM MgCl2, and 1 mM TCEP. Purified Xrn1-E178Q and RNA samples were extensively exchanged into a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM KCl, 1 mM TCEP, and 4 mM MgCl2 by the use of a Superdex 75 10/300 GL column (GE Healthcare, Chicago, IL). ITC measurements were performed with a MicroCaliTC 200 Calorimeter (GE Healthcare, Chicago, IL) at 25°C. Background data obtained from the buffer sample were subtracted before data analysis, and then the data were fitted using the Origin7 software package (MicroCal). Measurements were repeated twice, and similar results were obtained.
Mouse studies.
All experiments involving animals were approved by and carried out in strict accordance with the guidelines of the Institutional Experimental Animal Welfare and Ethics Committee of Beijing Institute of Microbiology and Epidemiology, Beijing, China (13-2016-001). BALB/c mice used in this study were purchased from the Laboratory Animal Center (AMMS; Beijing, China). For the virulence test, groups of 4-week-old female BALB/c mice (n = 6 to 9) were inoculated intraperitoneally (i.p.) with the corresponding WT and mutant viruses. Animals were monitored daily for 21 days after inoculation. Any mice found in a moribund condition were euthanatized and scored as dead. Viremia in infected mice was quantified by plaque assay as mentioned above.
RNA structure modeling.
Secondary structures were folded using the Mfold Web server with standard settings and the flat exterior loop type. Tertiary-structure folding of WNV 3′-UTR (1 to 350 nt; SLI to CS3) was performed using the RNAComposer Web server with standard settings (33).
Statistical analysis.
Student's t test was used to determine significant differences (P < 0.05) between all tested viruses. Statistical analyses were performed using IBM SPSS Statistics v18 (IBM, Armonk, NY). Kaplan-Meier survival curves were analyzed by the log rank test. All data were analyzed using GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA).
ACKNOWLEDGMENTS
We thank Liang Tong of Columbia University for providing the key reagents, Andrea V. Gamarnik for the protocol used for 5’ end determination of sfRNA, Wu-Xiang Guan for the protocol used for Northern blot analysis, Core Facility and Technical Support staff members (Pei Zhang, An-na Du, and Juan Min), and staff members of the Center for Animal Experiment (Xue-fang An, Fan Zhang, He Zhao and Li Li) and the biosafety level 3 (BSL-3) laboratory (Hao Tang) at Wuhan Institute of Virology and Wuhan Key Laboratory of Special Pathogens and Biosafety for their helpful support during the course of the work.
This work was supported by the National Key Research and Development Program of China (2016YFD0500400, 2016YFD0500304, and 2016YFC1200400), the National Natural Science Foundation of China (81572003, 31770190, 81621005, and 81522025) and the National Science and Technology Major Project of China (2018ZX09711003, 2018ZX10101004, and 2017ZX10304402).
We have no competing interests to declare.
REFERENCES
- 1.Filomatori CV, Carballeda JM, Villordo SM, Aguirre S, Pallares HM, Maestre AM, Sanchez-Vargas I, Blair CD, Fabri C, Morales MA, Fernandez-Sesma A, Gamarnik AV. 2017. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog 13:e1006265. doi: 10.1371/journal.ppat.1006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. 2016. RNA structure duplications and flavivirus host adaptation. Trends Microbiol 24:270–283. doi: 10.1016/j.tim.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. 2014. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gritsun TS, Gould EA. 2006. The 3’ untranslated regions of Kamiti River virus and cell fusing agent virus originated by self-duplication. J Gen Virol 87:2615–2619. doi: 10.1099/vir.0.81950-0. [DOI] [PubMed] [Google Scholar]
- 8.Gritsun TS, Gould EA. 2006. The 3’ untranslated region of tick-borne flaviviruses originated by the duplication of long repeat sequences within the open reading frame. Virology 350:269–275. doi: 10.1016/j.virol.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Gritsun TS, Gould EA. 2007. Direct repeats in the flavivirus 3’ untranslated region; a strategy for survival in the environment? Virology 358:258–265. doi: 10.1016/j.virol.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Gritsun TS, Gould EA. 2007. Origin and evolution of 3’UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv Virus Res 69:203–248. [DOI] [PubMed] [Google Scholar]
- 11.Lo MK, Tilgner M, Bernard KA, Shi PY. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3’ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol 77:10004–10014. doi: 10.1128/jvi.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinton MA, Basu M. 2015. Functions of the 3’ and 5’ genome RNA regions of members of the genus Flavivirus. Virus Res 206:108–119. doi: 10.1016/j.virusres.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi PY, Nix JC, Beckham JD, Kieft JS. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354:1148–1152. doi: 10.1126/science.aah3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. 2012. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol 86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang RY, Hsu TW, Chen YL, Liu SF, Tsai YJ, Lin YT, Chen YS, Fan YH. 2013. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol 166:11–21. doi: 10.1016/j.vetmic.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 16.Kieft JS, Rabe JL, Chapman EG. 2015. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: conservation, folding, and host adaptation. RNA Biol 12:1169–1177. doi: 10.1080/15476286.2015.1094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife 3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. 2010. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol 84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. 2014. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol 76:5847–5856. doi: 10.1128/jvi.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng CL, Zhang QY, Chen DD, Liu SQ, Qin CF, Zhang B, Ye HQ. 2017. Recovery of the Zika virus through an in vitro ligation approach. J Gen Virol 98:1739–1743. doi: 10.1099/jgv.0.000862. [DOI] [PubMed] [Google Scholar]
- 22.Zou G, Xu HY, Qing M, Wang QY, Shi PY. 2011. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res 91:11–19. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. 2011. Structural and biochemical studies of the 5’–>3’ exoribonuclease Xrn1. Nat Struct Mol Biol 18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gokhale NS, Horner SM. 2017. Knotty Zika virus blocks exonuclease to produce subgenomic flaviviral RNAs. Cell Host Microbe 21:1–2. doi: 10.1016/j.chom.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. 2015. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog 11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messing SA, Gabelli SB, Liu Q, Celesnik H, Belasco JG, Pineiro SA, Amzel LM. 2009. Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus. Structure 17:472–481. doi: 10.1016/j.str.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. 2005. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Wei Y, Qin C, Jiang T, Li X, Zhao H, Liu Z, Deng Y, Liu R, Chen S, Yu M, Qin E. 2009. Translational regulation by the 3’ untranslated region of the dengue type 2 virus genome. Am J Trop Med Hyg 81:817–824. doi: 10.4269/ajtmh.2009.08-0595. [DOI] [PubMed] [Google Scholar]
- 29.de Borba L, Villordo SM, Marsico FL, Carballeda JM, Filomatori CV, Gebhard LG, Pallares HM, Lequime S, Lambrechts L, Sanchez Vargas I, Blair CD, Gamarnik AV. 2019. RNA structure duplication in the dengue virus 3’ UTR: redundancy or host specificity? mBio 10:e02506-18. doi: 10.1128/mBio.02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehead SS. 2016. Development of TV003/TV005, a single dose, highly immunogenic live attenuated dengue vaccine; what makes this vaccine different from the Sanofi-Pasteur CYD vaccine? Expert Rev Vaccines 15:509–517. doi: 10.1586/14760584.2016.1115727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan C, Muruato AE, Nunes BT, Luo H, Xie X, Medeiros DB, Wakamiya M, Tesh RB, Barrett AD, Wang T, Weaver SC, Vasconcelos PF, Rossi SL, Shi PY. 2017. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med 23:763–767. doi: 10.1038/nm.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goertz GP, Fros JJ, Miesen P, Vogels CBF, van der Bent ML, Geertsema C, Koenraadt CJM, van Rij RP, van Oers MM, Pijlman GP. 2016. Noncoding subgenomic flavivirus RNA is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by Culex pipiens mosquitoes. J Virol 90:10145–10159. doi: 10.1128/JVI.00930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]