Abstract
We report herein a novel pipet-based “ELISA in a tip” as a new versatile diagnostic tool featuring better sensitivity, shorter incubation time, accessibility, and low sample and reagent volumes compared to traditional ELISA. Capture and analysis of data by a cell phone facilitates electronic delivery of results to health care providers. Pipette tips were designed and 3D printed as adapters to fit most commercial 50–200 μL pipettes. Capture antibodies (Ab1) are immobilized on the inner walls of the pipet tip, which serves as the assay compartment where samples and reagents are moved in and out by pipetting. Signals are generated using colorimetric or chemiluminescent (CL) reagents and can be quantified using a cell phone, CCD camera, or plate reader. We utilized pipet-tip ELISA to detect four cancer biomarker proteins with detection limits similar to or lower than microplate ELISAs at 25% assay cost and time. Recoveries of these proteins from spiked human serum were 85–115% or better, depending slightly on detection mode. Using CCD camera quantification of CL with femto-luminol reagent gave limits of detection (LOD) as low as 0.5 pg/mL. Patient samples (13) were assayed for 3 biomarker proteins with results well correlated to conventional ELISA and an established microfluidic electrochemical immunoassay.
Graphical Abstract
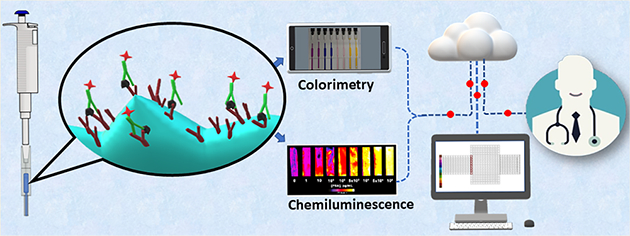
Enzyme linked immunosorbent assay (ELISA) has long been the gold standard for measuring proteins, pathogens, antibodies, and other biomolecules in complex matrices.1–3 Recent research has focused on developing innovative and accessible systems in order to decrease cost and assay time and increase sensitivity. Many ELISA systems use microplates with different volumes and sizes as assay compartments to immobilize antibodies that specifically capture target molecules followed by labor intensive, time-consuming incubation and washing steps. Recent approaches have included design of new labels to replace enzymes,4,5 utilizing magnetic beads6,7 and signal enhancement by nanomaterials8 as well as developing more accessible plate readers.9,10 3D printing has been utilized to increase microwell surface area, improving the performance of microplate ELISA.11 Remaining challenges that hinder the use of ELISA for low cost diagnostics include limited surface area for antibody immobilization, fluid handling, multiplexing, and sample size.12
Commercial alternatives for biomolecule detection include multiplexed microbead immunoassay technologies with optical or electrochemiluminescent (ECL) readout with LODs for proteins similar to standard ELISA at 1–10 pg mL−1.13–15 The Simoa HD-116 single protein counting device detects proteins with much better LODs (e.g., 4–70 fg mL−1).17 However, instrument and assay costs for these methods are high and prohibitive for many applications.
Incorporation of microfluidics into molecular diagnostic devices facilitates fluid control, decreases sample and reagent volumes, decreases assay time and cost, and can enable remote sensing.18 Microfluidics has improved immunoassay performance by enhancing reaction kinetics, providing multiplexing, and decreasing assay time and cost.19,20 ELISA has been integrated into PDMS chips,21,22 centrifugal assay platforms,23,24 disposable patterned discs,25,26 and microfluidic chips with an optical sensor27 to offer fast, cost-effective alternatives to microplate ELISA. However, there are as yet few off-the-shelf systems available, so for widespread lab use, these approaches will require in-house fabrication that will not be familiar or desirable to many users.28,29
3D printing provides fast, low cost, readily accessible fabrication of microfluidic devices.30–32 Desktop 3D printing allows laboratories around the world to reproduce bioanalytical diagnostic microfluidic devices at low cost.33 3D printed microfluidic electrochemical immunoarrays have been developed for determining proteins,34–38 nucleic acids,39–41 and small molecules.42,43 Other detection modes are also possible, and we have reported 3D printed microfluidic immunoarrays to detect up to eight proteins utilizing electrochemiluminescence (ECL) or chemiluminescence (CL).44–46
In the present paper, we report the first example of ELISA in 3D printed pipet tips. Our goal was to develop a simple, easy-to-use, low cost methodology for sandwich ELISA assays for virtually any protein. The tips were designed to fit to most commercially available 50–200 μL single and multichannel pipettes. The inner fluidic chamber of the tip serves as a solid support to strongly adsorb chitosan hydrogel, allowing covalent immobilization of capture antibodies (Ab1). The microfluidic nature of the assay chamber decreases assay time because of larger volume/surface area ratios than microwells. Sample and reagents are drawn into the tip and incubated there, and measurement of resulting absorbance or CL is done with a CCD camera, cell phone, or plate reader. Four protein biomarkers, prostate specific antigen (PSA), vascular endothelial growth factor (VEGF), insulin-like growth factor-1 (IGF-1), and cluster of differentiation-14 (CD-14), were detected simultaneously using tetramethylbenzidine (TMB) reagent and an eight-channel pipet-tip adaptor. Limits of detection (LOD) were 5 pg/mL for PSA, 25 pg/mL for VEGF, 2.5 pg/mL for IGF-1, and 0.5 pg/mL for CD-14, which are lower than or similar to microplate ELISA in <25% of the assay time. Cell phone image capture and analysis was demonstrated for VEGF with an LOD of 50 pg/mL and for IGF-1 with an LOD of 20 pg/mL. Integration with chemiluminescence (CL) detection was shown by measuring PSA using West Femto luminol with an LOD of 1 pg/mL. Recoveries from spiked human serum samples were between 85–115%. In addition, proteins PSA, CD-14, and IGF-1 were quantified in 13 patient samples and data correlated well with both conventional ELISA and an established electrochemical immunoassay. Cell-phone image capture was also used to quantify IGF-1 levels in 13 patient samples with good correlation to conventional ELISA.
RESULTS AND DISCUSSION
Design of ELISA Tip Adapter.
We used two different designs for the pipet tips. The first was a single channel tip and the second was an eight-channel tip that fits eight-channel pipettes (Figure 1A). Tips were designed in 123D design software (Autodesk) and transferred to a Form2 stereolithographic 3D printer where they were printed using Formlabs clear resin. The pipet tip (SI, Figure S1) has three main compartments, the pipet housing, immunoassay chamber, and inlet cylinder. The inlet cylinder has an outer diameter of 3 mm and 15 mm length, suitable for drawing samples and reagents into the immunoassay chamber from ordinary microcentrifuge tubes. Total fill volume of the immunoassay chamber and inlet cylinder is 50 ± 3 μL (45 μL immunoassay chamber and 5 μL inlet cylinder). The pipet housing compartment was designed as a conical cylinder to facilitate fitting onto different commercially available pipettes and was equipped with an overflow feature near the immunoassay chamber to prevent possible contamination of the pipet with reagents or samples (SI, Figure S2). Four different commercially available pipettes, Finnpipette II, Eppendorf, V-Pette, and Acumax, were tested. Designed tips snuggly fitted into these pipettes by applying less than 3 kg of force with pipetted volume variation <±1%.
Figure 1.
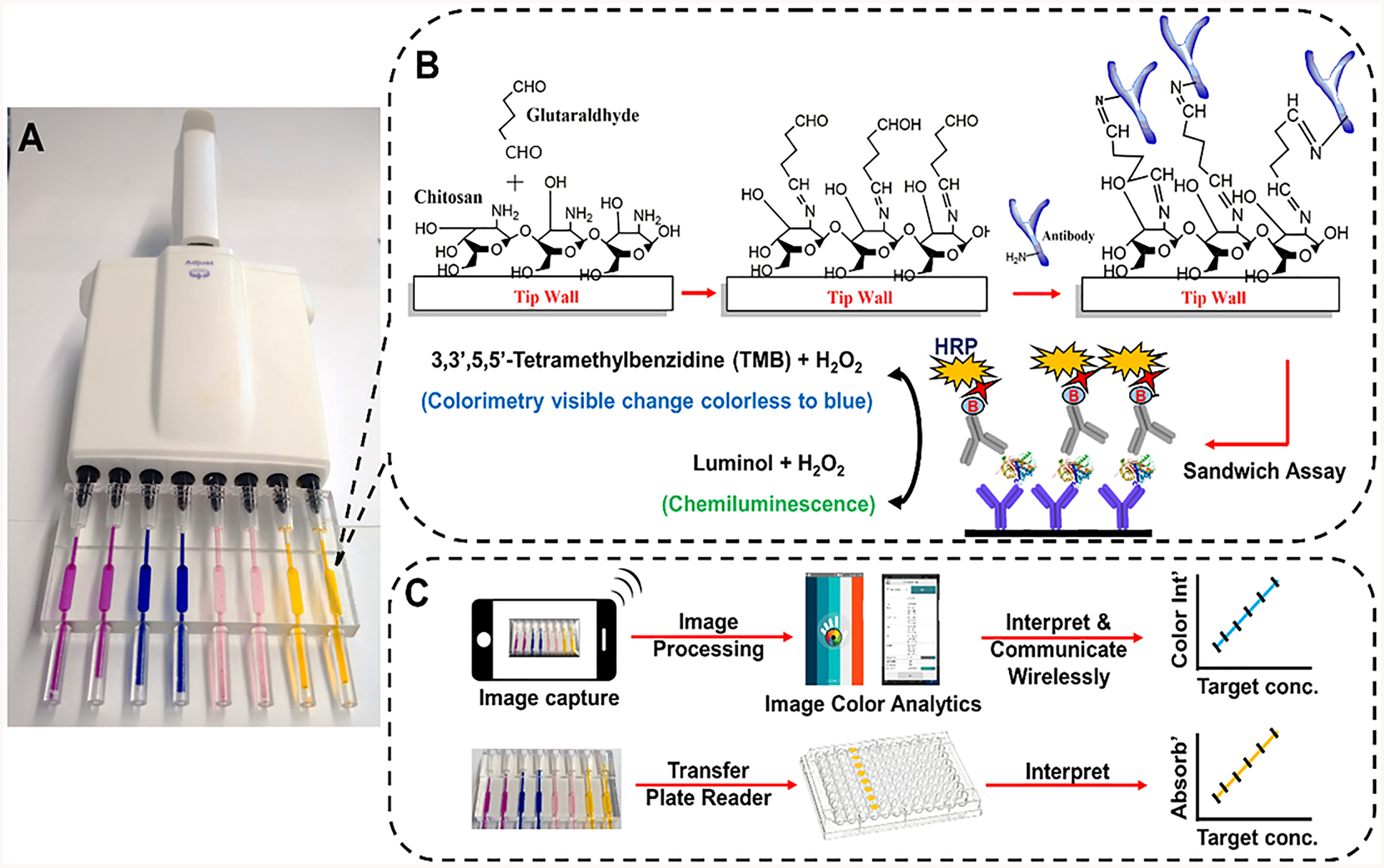
Representation of sandwich immunoassays in ELISA tips with protocols: (A) fully transparent eight-pipet 3D printed array loaded with food dyes; (B) precoating steps showing immobilization of capture antibodies on immunoarray tip wall coated with chitosan followed by sandwich immunoassay and signal measurement for colorimetry and CL. (C) Signal capture and processing flow using smartphone or microplate reader.
Hydrogel Film Characterization.
Chitosan is a naturally occurring polysaccharide that we found to adsorb strongly and irreversibly to the 3D-printed polyacrylate. A flat 1 × 1 cm 3D printed chip was coated with chitosan and used to estimate dry hydrogel film mass/cm2, swelling ratio %, and antibody coverage/cm2 in the ELISA tips. White light interferometry analysis of 3D printed surface coated with chitosan on showed large peaks of varying heights (5–33 μm) intercalated with small valleys that occupied ~10% of the chip area (Figure 2).
Figure 2.
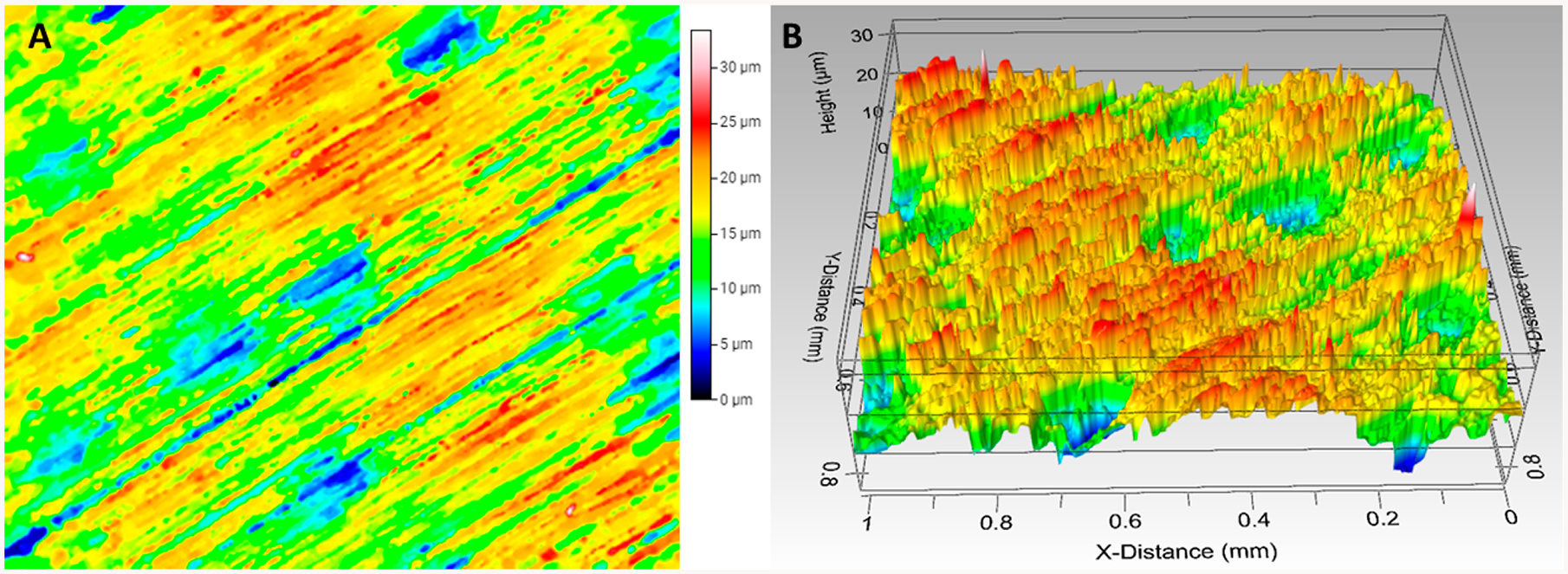
Surface profile of 3D printed chip. (A) Top view and (B) 3D view showing surface roughness with peaks of 5–33 μm intercalated by valleys that occupy ~10% of the 3D printed surface
It is likely that this roughness and chitosan swelling contributes to performance of the tip ELISA, as it increases surface area for chitosan–Ab1 immobilization. Dry film loading capacity of the 3D printed chip was 0.10 ± 0.01 mg chitosan/cm2. Film mass after hydration with water in a humidity chamber was 1.1 ± 0.05 mg, which indicated a swelling ratio % of 1100% from eq 1.47
| (1) |
where Ws is weight of the swollen film, and Wd is weight of dry chitosan film. Ab1 was anchored onto the chitosan film using glutaraldehyde as a cross-linker for covalent immobilization (Figure 1B). Ab1 coverage calculated from the BCA total protein assay was 10.0 ± 0.5 μg/cm2, which is approximately 1.0 × 1013 antibodies/cm2 (SI, Figure S4). We also estimated swelling of the chitosan film after immobilizing Ab1. Dried chitosan films with Ab1 had a mass of 0.9 ± 0.03 mg, and the mass of the chitosan–Ab1 film after hydration increased to 5.5 ± 0.04 mg for a swelling ratio of 600%.
Single Protein Assays.
Assay precision was evaluated at several analyte concentrations (n ≥ 3) in calf serum (Figure 3), which is an effective human serum surrogate for immune assays.48 Figure 3A shows the reproducibility of colorimetry from four different tips at 1.3 and 12 ng/mL VEGF in calf serum, Figure 3D shows images of colorimetric assays from three different tips at 1.3 and 5.5 ng/mL IGF-1 in calf serum, and standard deviations for each concentration were reproducible within ±5%. Day-to-day standard deviations were less than ±7%. The assay was also done in cell-phone-assisted mode. We utilized a simple photograph and instant signal integration with a commercial color analytics app, Color Grab (SI, Figure S3). Figure 3B,E shows colorimetric signals imaged after 10 min of color reaction with increasing concentrations of VEGF and IGF-1. Images were captured and analyzed with the Color Grab mobile application. VEGF had a detection limit of 50 pg/mL and a dynamic range of 50 pg/mL to 10 ng/mL. Figure 3C shows calibration curves obtained by plotting %K values as a measure of color intensity vs VEGF concentration. IGF-1 had an LOD of 20 pg/mL and a dynamic range of 20 pg/mL to 5.5 ng/mL. Figure 3F shows calibration curves obtained by plotting %K values measuring color intensity vs IGF-1 concentration. Percent recoveries of VEGF from spiked human serum samples were found to validate accuracy of the cell-phone-based assay and were 90–110% (Table 1). Stability of antibodies immobilized in the tip were estimated by measuring the same concentration of IGF-1 (625 pg/mL) daily for tips stored at 4° C over 7 days. Antibodies lost 5% activity after 3 days and maintained 80% of initial activity after 7 days of storage (SI, Figure S5).
Figure 3.
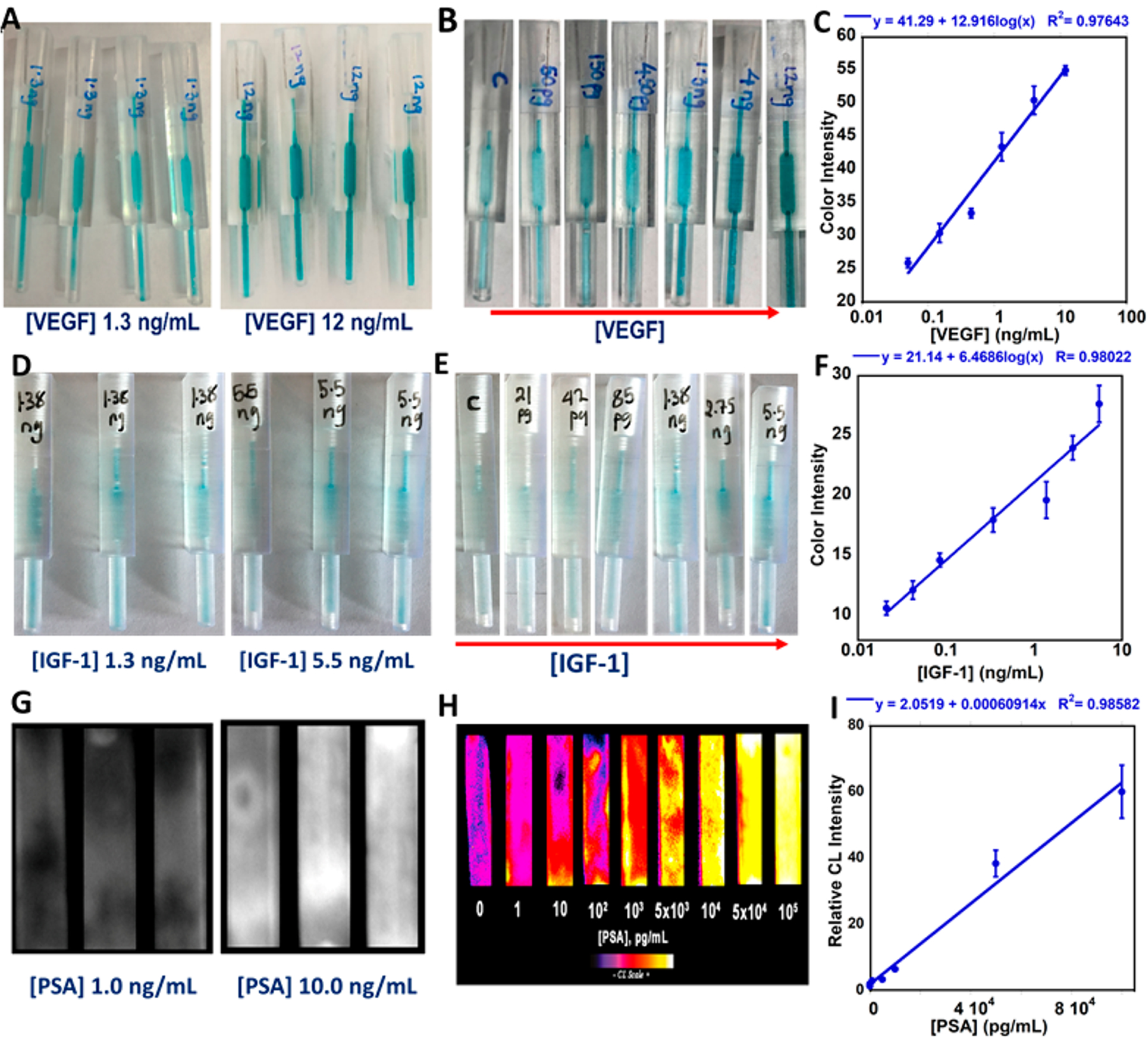
Single tip assays in calf serum using iPhone and CCD camera imaging: (A) reproducibility of colorimetric detection (n = 4) for VEGF; (B) change in colorimetric signal with increasing concentration of VEGF; (C) calibration curve obtained via iPhone imaging color intensity (K%) vs [VEGF]; (D) reproducibility of colorimetric detection (n = 3) for IGF-1; (E) change in colorimetric signal with increasing concentration of IGF-1; (F) calibration curve obtained via iPhone K% vs [IGF-1]; (G) raw CL images captured by CCD camera for PSA; (H) recolorized images showing change in CL intensity vs [PSA]; (I) calibration curve of relative CL vs [PSA].
Table 1.
Spike-Recovery Results from Single and Multiplex Immunoassays in Human Serum
| method | biomarker protein | spiked concentrations (pg/mL) | found concentrationsa (pg/mL) | (%) recovery (±SD) |
|---|---|---|---|---|
| cell-phone-assisted single tip assay | VEGF | 100 | 107 (±6) | 107 (±7) |
| 1000 | 950 (±84) | 95 (±9) | ||
| 4000 | 4133 (±454) | 103(±7) | ||
| colorimetric eight-chamber tip assay | PSA | 20 | 21 (±1.5) | 105 (±7) |
| 300 | 296 (±38) | 98 (±13) | ||
| 2000 | 2397 (±144) | 119 (±6) | ||
| VEGF | 125 | 122 (±15) | 98 (±12) | |
| 500 | 436 (±42) | 87 (±9) | ||
| 2000 | 2051 (±287) | 102 (±14) | ||
| IGF-1 | 125.0 | 121 (±16) | 100 (±13) | |
| 800 | 925 (±138) | 98.5 (±15) | ||
| 2000 | 2134 (±320) | 93 (±15) | ||
| CD-14 | 10 | 11 (±0.89) | 113 (±7) | |
| 500 | 450 (±43) | 90 (±9) | ||
| 5000 | 5402 (±486) | 108 (±9) |
After subtraction of the control human serum concentration.
CL capture from tips by CCD camera was demonstrated using PSA as the test protein, Here, HRP oxidized femto-luminol to an excited state that decays and emits light that was captured and integrated over 120 s using a CCD camera in a dark box. Figure 3H shows recolorized CL images that showing increased CL intensity with increasing PSA concentration. Figure 3I is a calibration curve obtained by plotting CL intensities against PSA concentration. The CL assay for PSA on calf serum had an LOD of 1 pg/mL and a dynamic range from 1 pg/mL to 100 ng/mL.
Multiplexed ELISA in a Tip.
The colorimetric assay was tested to simultaneously quantify four proteins in undiluted calf serum. A multichannel pipet-tip adapter (Figure 1) was used with two channels for each protein. Here we used a plate reader for absorbance detection to illustrate versatility of the approach. Figure 4 shows calibration curves for the proteins, where standard deviation for each concentration was less than 15%. The LODs were 5 pg/mL for PSA, 25 pg/mL for VEGF, 2.5 pg/mL for IGF-1, and 0.5 pg/mL for CD-14. Dynamic ranges were 5 to 10 000 pg/mL for PSA, 50 to 10 000 pg/mL for VEGF, 4 to 8000 pg/mL for IGF-1, and 1 to 10 000 pg/mL for CD-14.
Figure 4.
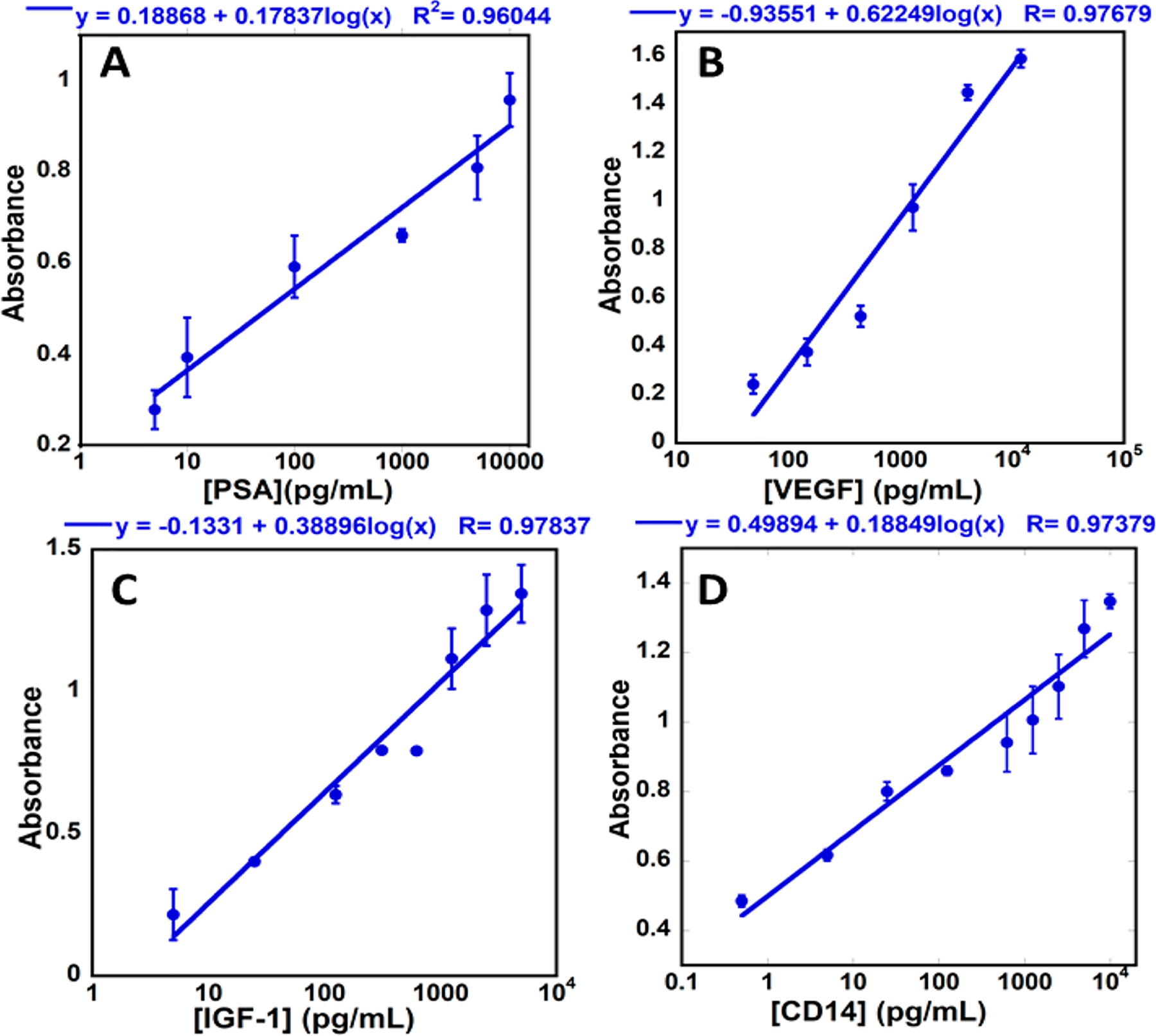
Calibration curves for ELISA tip colorimetric multiplexed detection of (A) PSA, (B) VEGF, (C) IGF-1, and ((D) CD-14. Absorbance was measured at 450 nm in a plate reader (n = 4).
Recoveries from spiked, diluted human serum were measured to validate accuracy of multiplexed ELISA in a tip. Human serum was diluted 100-fold before spiking to bring native protein levels below or equal to the protein LODs. Calibration curves in calf serum were used to estimate recoveries from spiked human serum. Recoveries were estimated after subtracting the signals from control serum and were in an analytically acceptable range49 of 100 ± 20% (Table 1).
Patient Sample Analyses.
PSA, IGF-1, and CD-14 represent promising biomarkers for prostate cancer diagnostics50–52 and were measured in 13 patient serum samples by multiplexed analysis. The sample cohort consisted of three normal cancer-free individuals, five samples collected from patients with indolent prostate cancer (Gleason score < 7), and five samples from patients with aggressive prostate cancer (Gleason score > 7). Samples were analyzed by colorimetric tip ELISA, and results were compared to both microwell plate ELISA (SI, Figure S7 and Table S2–S4) and an established validated microfluidic electrochemical assay (SI, p. S11, Table S6). Tip ELISA gave excellent correlation with microwell ELISA and the electrochemical assay as demonstrated by correlation coeffcients and slopes near 1.0 and intercepts near zero. Correlation coeffcients with the electrochemical assay were 0.999 for PSA, 0.991 for IGF-1, and 0.992 for CD-14, and slopes were 1.17 for PSA, 0.87 for IGF-1, and 1.23 for CD-14 (SI, Figure S8). Correlation coeffcients with microwell plate ELISA were 0.999 for PSA, 0.986 for IGF-1, and 0.981 for CD-14, and slopes were 0.775 for PSA, 0.949 for IGF-1, and 0.917 for CD-14 (Figure 5). IGF-1 concentration in 10× diluted serum samples was also estimated using iPhone imaging colorimetric assay and had a correlation coecient of 0.973 vs conventional ELISA with a slope of 0.978 (SI, Figure S9 and Table S5). Linear correlations of tip ELISA with the referee methods gave correlation coeffcients for the three proteins >0.90, slopes close to unity, and intercepts close to zero. Insets show correlations in the lower concentration range.
Figure 5.
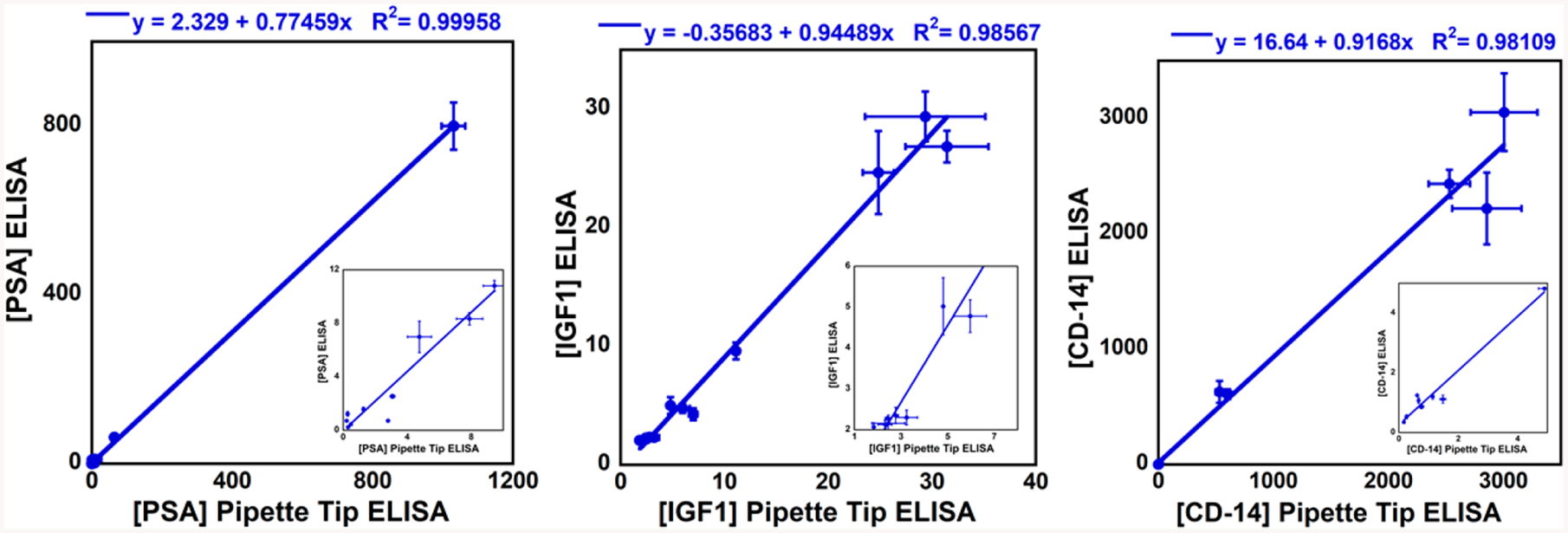
Linear correlations for pipet-tip ELISA vs microwell plate ELISA for (A) PSA, (B) IGF1, and (C) CD14, quantified in ng/mL (n = 3) in patient samples. Insets show low concentration ranges.
Although the number of samples is too small for definitive conclusions, receiver operator characteristics (ROC) were analyzed for preliminary diagnostic predictions (SI, Figure S6). PSA shows moderate increases for cancer and aggressive cancer samples, whereas CD-14 shows a very large increase for aggressive prostate cancer. In addition, clustered multiple variables box plots53 show the found expressions of the three biomarkers in the human serum samples (SI, Figure S10)
The results above demonstrate a novel ELISA in a tip strategy utilizing 25% less reagent and sample volumes and a 25% shorter assay time than traditional ELISA (Table 2). ELISA in a tip offers opportunities that can allow decreases in instrumentation cost and size (e.g., by using cell-phone data capture and analysis). The latter approach also facilitates electronic reporting to relevant health care providers and the patients themselves. We demonstrated sensitive detection of four cancer biomarkers proteins simultaneously with ELISA in a tip (Figures 3 and 4). Users with pipet use knowledge or training should be able to run the assays effortlessly, facilitating use in areas with limited resources. Pipetting speed and accuracy may slightly affect the performance of the assay, and we did not notice significant variation between users. We believe developing future automated systems engineered for the pipet-tip assay may eliminate user-to-user variation.
Table 2.
Comparison between ELISA in a Tip vs. Microplate ELISA
| comparison type | tip ELISA (colorimetry) | tip ELISA (chemiluminescence) | tip ELISA (cell phone) | traditional ELISA |
|---|---|---|---|---|
| assay time | ||||
| sample incubation | 30 min | 30 min | 30 min | 150 min |
| detection antibody | 20 min | 20 min | 20 min | 60 min |
| streptavidin–HRP | 15 min | 15 min | 15 min | 45 min |
| development and quench | 12 min | 12 min | 12 min | 32 min |
| detection | 2 min | 2 min | 3–5 s | 2 min |
| approx. total wash time | 6 min | 6 min | 6 min | 9 min |
| total assay time | 85 min | 85 min | 83 min | 298 min |
| reagent/sample volume | 50 μL for sample and wash | 50 μL for sample and wash | 50 μL for sample and wash | 150 μL for sample and 300 μL for wash |
| LOD (pg/mL) and dynamic range (pg/mL) | PSA - 5 and 5 – 10 000 VEGF - 25 and 50 – 10 000 |
PSA 1 and 1 – 100 000 | IGF-1 20 and 20 – 5500 | PSA - 900 and 900 – 60 000 VEGF - 25 and 35 – 2000 |
| IGF-1 4 and 4 – 8000 CD-14 - 0.5 and 1 – 10 000 |
VEGF 50 and 50 – 10 000 | IGF-1 - 100 and 100 – 8000 CD-14 - 62 and 62 – 4000 |
||
| assay cost per microwell | $1.20/single tip | $1.20/single tip | $1.20/single tip | $4.14/microwell (Human PSA ELISA Kit – $398) |
| required instrument | plate reader | CCD camera | smart phone | no |
| operator | professional | simple training | simple training | professional |
| multiplexity | yes | yes | yes | no |
Antigen–antibody reactions at a solid–liquid interface as in microplate ELISA are limited by diffusion and antigens, available to interact with immobilized antibodies, are depleted with time.54,55 Reagent volume/surface area ratios are 131 μL/cm2 for microwells and 31 μL/cm2 for the the pipet-tip chamber. This 4-time lower volume/surface area ratio improves Ag–Ab interaction kinetics, reflected in better sensitivities and shorter assay times.
The roughness of the 3D printed surface (Figure 2) of the tip and the use of a highly swelled hydrogel also contribute to effective surface area, which increases the antibody surface loading capacity.56 Antibody loading in 3D printed tips was 10.0 ± 0.5 μg/cm2, which is 15–50 times higher than reported capacities for commercial microplates (200–650 ng/cm2).57 This high surface loading helped reduce assay time while maintaining similar sensitivity to conventional ELISA. In addition, covalent immobilization of antibodies onto chitosan hydrogel maintains high activity compared to a documented loss of activity for passively adsorbed antibodies on polymer surfaces that induce antibody denaturation.58–60
The transparent tips 3D printed with a low-cost stereolithographic printer were shown to be applicable to colorimetric (Figure 3) and CL detection. CL assays detected PSA from complex serum matrix with detection limits of 1 pg/mL (Figure 3). No additional steps or modifications are required for CL detection compared to colorimetric assays. CL arrays can be easily integrated with a cell phone, making them available for resource-limited settings.61,62 The “ELISA in a tip” platform was also customized to multiplex multiple analytes and multisample assays simultaneously (Figure 2A). CAD files for ELISA tip printing can be found at https://rusling.research.uconn.edu/research/3d-printing-designs/. The simple design facilitates extension for multichannel pipettes with 12, 16, 48, and 64 channels.
Cell-phone interfacing can enable 3D printed tip arrays for resource-limited settings. Using free, commercially available, software “Color Grab”, we demonstrated sensitive detection of colorimetric signals right directly from the tip array and validated accuracy of detection with spiked human serum (Table 1) and by analyzing real serum samples (SI, Table S5). Interfacing cell phones could also help in telemedicine to establish networks among assay technician, centralized laboratories, and physicians. At rural sites in developing countries where availability of resources for traditional ELISA may be limited, cell-phone analysis of tip ELISA can be used at a fraction of the cost and time of standard ELISA kits.
In summary, we describe and demonstrate above an “ELISA in a tip” immunoassay tool designed by 3D printing immunoassay tips for conventional pipettes. “ELISA in a tip” can provide better LODs and dynamic range than microwell ELISA kits, at lower cost, sample volume, and assay time. Multiple signal detection strategies and multiplexed detection are accommodated. Cell-phone integration enables the prospect of low-cost detection diagnostics and telemedical communication in normal and low resource environments. This new approach offers a versatile, sensitive, multiplexing immunoassay with tips that any lab with a stereolithographic 3D printer can access, with capability that is faster and cheaper than conventional ELISA and may be particularly useful in resource-limited venues.
MATERIALS AND METHODS
Materials.
All reagents and chemicals were of analytical grade. Chitosan (low molecular weight) and glutaraldehyde were from Sigma-Aldrich. Blocker casein in PBS buffer was from Thermo Fisher. ELISA kits for prostate specific antigen (PSA) (DY1344), vascular endothelial growth factor (VEGF) (DY293B), insulin-like growth factor-1 (IGF-1) (DY291), and cluster of differentiation-14 (CD-14) (DY383) were from R&D Systems. Colorimetric signals were generated using R&D Systems’ substrate reagent pack with tetramethylbenzidine (TMB) color reagent and peroxide mixed immediately before use. Chemiluminescence (CL) was generated using Thermo Fisher Supersignal West Femto chemiluminescent substrate, with luminol and peroxide mixed right immediately before use. All colorimetric measurements were performed after transferring the solutions after color was developed to a microplate, and absorbance was measured using a Synergy HTX plate reader. CL was measured in pipet tips using a Syngene dark box with a CCD camera. Images were processed using GeneSnap software. Mobile phone image processing was done using an android/iPhone compatible application Loomatix ColorGrab. Human prostate cancer patient serum samples and controls were from George Washington University (GWU) Hospital under IRB ethical approval.
Antibody Immobilization.
Inner walls of tips were coated with a polymeric hydrogel layer of chitosan by dispensing 50 μL of 0.25% chitosan in 0.05 M HCl (pH.4.0) solution into the pipet tip and incubating for 3 h. Solution was then removed, and pipet tips were allowed to dry under vacuum at room temperature for 3 h, forming a thin, stable film of chitosan onto the inner walls. Amine groups of the hydrogel layer were then activated by filling the tip with 50 μL of 3% glutaraldehyde in phosphate buffer (PBS, pH.7.4) and were then incubated for another 3 h. Glutaraldehyde solution was then removed, and tips were washed with PBS. Aliquots of antibody solution (50 μL) at desired concentrations were pipetted into the tips, which were then incubated overnight (Figure 1B). To compare to microwell plate ELISA, all antibodies were used at concentrations recommended by ELISA kit suppliers (SI, Table S1). After incubation, antibody solutions were removed, and tips were washed with phosphate buffer–0.05% Tween 20 solution (PBS–T20, pH.7.4) to remove unbound antibodies and inhibit nonspecific binding (NSB).
Chitosan layers were characterized by coating a 1 × 1 cm 3D printed chip with chitosan using white light interferometry and a Filmetrics Profilm3D profilometer equipped with a 20× Mirau objective and Profilm software. Antibody (Ab1) coverage on the flat 3D printed chip was estimated using the bicinchoninic acid (BCA) total protein assay.63 Chitosan chips were incubated with 100 μL of 3% glutaraldehyde, washed, and incubated with 100 μL of 10 μg/mL anti-PSA antibodies overnight. Chips were then washed with PBS–T20 and incubated with 100 μL of BCA reagent (Thermo Fisher) at 37° C for 2 h. Reagent was then transferred to a microplate where absorbance was measured at 562 nm. The absorbance was then used to estimate total amount of antibodies on the chip from a calibration curve for standard anti-PSA (SI, Figure S4).
Colorimetric Assay.
Tips decorated with Ab1 were filled with 50 μL of standard protein or sample solution and incubated for 30 min. Tips were then washed 3× with PBS–T20, and 50 μL of biotinylated detection antibody (Ab2) solution in 1% casein in PBS solution was pipetted in and incubated for 20 min. Unbound Ab2 was removed by washing 3× with PBS–T20, tips were filled with 50 μL of streptavidin horseradish peroxidase (ST-HRP) diluted according to vendor specifications in 1% BSA in PBS, and incubated 15 min. Tips were washed 3× with PBS–T20 and filled with 50 μL of TMB reagent prepared by mixing equal volumes of TMB substrate and peroxide solution according to vendor specifications and incubated for 12 min. Finally, solutions with developed color were transferred from tips into a 96 well microplate and filled with 50 μL of 2N sulfuric acid stop solution, and absorbance at 450 nm was measured using a microplate reader (Figure 1C).
Chemiluminescence (CL) Assay.
Procedures were the same as for colorimetry except for the CL development and detection. Supersignal West Femto reagent was used to develop CL that was recorded with a CCD camera in a dark box. Tips with sandwich immunocomplex decorated with HRP were filled with luminol reagent prepared by mixing equal volumes of luminol/enhancer substrate and hydrogen peroxide solution according to vendor specifications. Tips with CL reagent were then immediately placed into the dark box, and images were captured over 120 s (Figure 1C). Images were analyzed using GeneTools software (Syngene) to convert to relative CL intensity.
Cell-Phone-Assisted Image Capture.
Using the colorimetric assay procedure, images of the developed blue color after incubation of TMB in each tip were captured using an Android cell-phone camera with 12 megapixel resolution. Images of each tip were analyzed individually using free mobile app Color Grab from Loomatix available for both Android cell phones and iPhones. Color Grab uses image digitizer software to convert color intensity into a set of numbers based on the color composition and intensity of the specific image (Figure 1C). K% values from analysis of each captured image were used to describe the intensity of the blue color developed, because it showed the best correlation with color development (SI, Figure S3).
Assay Validation and Patient Samples Analysis.
Assays were validated by measuring recoveries of biomarker proteins in spiked human serum samples. Filtered human serum, 100× diluted sterile, from human male AB plasma (Sigma) was spiked with different concentrations of protein biomarkers. Colorimetric, CL, or cell-phone-assisted quantification of spiked concentrations were done in triplicate, and concentrations were calculated after subtracting the signal from diluted human serum control. In addition, 13 human samples (3 controls, 5 indolent prostate cancer patients, and 5 aggressive prostate cancer patients) were analyzed for 3 prostate cancer biomarker proteins, PSA, IGF-1, and CD-14, using the colorimetric assay. Patient samples were diluted 10× in PBS buffer for analysis of PSA and IGF-1 and 100× for analysis of CD-14 in order to bring biomarkers into the dynamic range of the assays. Diluted samples were incubated in pipet tips for 30 min, washed 3× with PBS–T20, followed by incubation with 50 μL of optimized Ab2 solution. The same procedure described for colorimetric assays was followed in patient sample analysis using the same R&D ELISA kits for tip assay as well as electrochemical and microwell plate ELISA. The same patient sample dilutions were used for traditional microwell plate ELISA (SI, S9). Assay results were well correlated with those obtained from microwell ELISA and a well-established electrochemical immunoassay developed in house and similar to a published procedure. (SI, Figure S8).35,36
Supplementary Material
ACKNOWLEDGMENTS
This work was supported financially by an Academic Plan Grant from The University of Connecticut and in part by Grant no. EB016707 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH. The authors thank James F. Elman (Filmetrics Application Lab, Rochester, NY) for 3D printed surface profiling.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b01284.
Pipette tip 3D design and dimensions; photographs of the single and multichannel printed tips; optimized capture and detection antibody concentrations; Color Grab on Android smartphone image analysis; bicinchoninic acid assay calibration for PSA antibodies; stability study of immobilized antibodies; receiver operating characteristics (ROC) curve analysis; patient sample analysis/microwell ELISA; electrochemical assay procedure; IGF-1 smart phone imaging colorimetric tip– ELISA assay correlation to conventional ELISA; box-and-whisker plots (PDF)
The authors declare no competing financial interest.
CAD files for printing tips at https://rusling.research.uconn.edu/research/3d-printing-designs/.
REFERENCES
- (1).Sato S; Murakami A; Kuwajima A; Takehara K; Mimori T; Kawakami A; Mishima M; Suda T; Seishima M; Fujimoto M; et al. PLoS One 2016, 11, e0154285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gan SD; Patel KR J. Invest. Dermatol 2013, 133, 1–3. [DOI] [PubMed] [Google Scholar]
- (3).Lequin RM Clin. Chem 2005, 51, 2415–2418. [DOI] [PubMed] [Google Scholar]
- (4).Miao L; Zhu C; Jiao L; Li H; Du D; Lin Y; Wei Q Anal. Chem 2018, 90, 1976–1982. [DOI] [PubMed] [Google Scholar]
- (5).Chen X; Liang Y; Zhang W; Leng Y; Xiong Y Talanta 2018, 186, 29–35. [DOI] [PubMed] [Google Scholar]
- (6).Rissin DM; Kan CW; Campbell TG; Howes SC; Fournier DR; Song L; Piech T; Patel PP; Chang L; Rivnak AJ; et al. Nat. Biotechnol 2010, 28, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vidal JC; Bertolín JR; Ezquerra A; Hernández S; Castillo JR Anal. Methods 2017, 9, 3602–3611. [Google Scholar]
- (8).Liu D; Li X; Zhou J; Liu S; Tian T; Song Y; Zhu Z; Zhou L; Ji T; Yang C Biosens. Bioelectron 2017, 96, 332–338. [DOI] [PubMed] [Google Scholar]
- (9).Berg B; Cortazar B; Tseng D; Ozkan H; Feng S; Wei Q; Chan RY-L; Burbano J; Farooqui Q; Lewinski M; et al. ACS Nano 2015, 9, 7857–7866. [DOI] [PubMed] [Google Scholar]
- (10).Wang L-J; Chang Y-C; Sun R; Li L Biosens. Bioelectron 2017, 87, 686–692. [DOI] [PubMed] [Google Scholar]
- (11).Singh H; Shimojima M; Shiratori T; Van An L; Sugamata M; Yang M Sensors 2015, 15, 16503–16515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tan X; Khaing Oo MK; Gong Y; Li Y; Zhu H; Fan X Analyst 2017, 142, 2378–2385. [DOI] [PubMed] [Google Scholar]
- (13).Rusling J; Kumar C; Gutkind J; Patel V Analyst 2010, 135, 2496–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Srinivas P; Kramer B; Srivastava S Lancet Oncol. 2001, 2, 698–704. [DOI] [PubMed] [Google Scholar]
- (15).Quansys Biosciences. Multiplex Assays http://www.quansysbio.com/multiplex/multiplex-assays (Accessed January 18, 2019).
- (16).Rissin DM; Kan CW; Campbell TG; Howes SC; Fournier DR; Song L; Piech T; Patel PP; Chang L; Rivnak AJ; et al. Nat. Biotechnol 2010, 28, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rissin DM; Kan CW; Song L; Rivnak AJ; Fishburn MW; Shao Q; Piech T; Ferrell EP; Meyer RE; Campbell TG; et al. Lab Chip 2013, 13, 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jung W; Han J; Choi J-W; Ahn CH Microelectron. Eng 2015, 132, 46–57. [Google Scholar]
- (19).Han KN; Li CA; Seong GH Annu. Rev. Anal. Chem 2013, 6, 119–141. [DOI] [PubMed] [Google Scholar]
- (20).Das T; Chakraborty S Biomicrofluidics 2013, 7, 011811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Costantini F; Sberna C; Petrucci G; Manetti C; de Cesare G; Nascetti A; Caputo D Sens. Bio-Sens. Res 2015, 6, 51–58. [Google Scholar]
- (22).Yu L; Ming Li C; Liu Y; Gao J; Wang W; Gan Y Lab Chip 2009, 9, 1243–1247. [DOI] [PubMed] [Google Scholar]
- (23).Aeinehvand MM; Ibrahim F; Harun SW; Djordjevic I; Hosseini S; Rothan HA; Yusof R; Madou MJ Biosens. Bioelectron 2015, 67, 424–430. [DOI] [PubMed] [Google Scholar]
- (24).Burger R; Amato L; Boisen A Biosens. Bioelectron 2016, 76, 54–67. [DOI] [PubMed] [Google Scholar]
- (25).Hoegger D; Morier P; Vollet C; Heini D; Reymond F; Rossier JS Anal. Bioanal. Chem 2006, 387, 267–275. [DOI] [PubMed] [Google Scholar]
- (26).Wang G; Das C; Ledden B; Sun Q; Nguyen C; Kumar S Biomicrofluidics 2017, 11 (1), 014115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Weng X; Gaur G; Neethirajan S Biosensors 2016, 6 (2), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ho CMB; Ng SH; Li KHH; Yoon Y-J Lab Chip 2015, 15, 3627–3637. [DOI] [PubMed] [Google Scholar]
- (29).He Y; Wu Y; Fu J; Gao Q; Qiu J Electroanalysis 2016, 28, 1658–1678. [Google Scholar]
- (30).Waheed S; Cabot JM; Macdonald NP; Lewis T; Guijt RM; Paull B; Breadmore MC Lab Chip 2016, 16, 1993–2013. [DOI] [PubMed] [Google Scholar]
- (31).Yazdi AA; Popma A; Wong W; Nguyen T; Pan Y; Xu J Microfluid. Nanofluid 2016, 20, 50. [Google Scholar]
- (32).Sharafeldin M; Jones A; Rusling JF Micromachines 2018, 9, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rackus DG; Shamsi MH; Wheeler AR Chem. Soc. Rev 2015, 44, 5320–5340. [DOI] [PubMed] [Google Scholar]
- (34).Sharafeldin M; Bishop GW; Bhakta S; El-Sawy A; Suib SL; Rusling JF Biosens. Bioelectron 2017, 91, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Krause CE; Otieno BA; Bishop GW; Phadke G; Choquette L; Lalla RV; Peterson DE; Rusling JF Anal. Bioanal. Chem 2015, 407, 7239–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Otieno BA; Krause CE; Latus A; Chikkaveeraiah BV; Faria RC; Rusling JF Biosens. Bioelectron 2014, 53, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tang CK; Vaze A; Shen M; Rusling JF ACS Sens 2016, 1, 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sánchez-Tirado E; Salvo C; González-Corteás A; Yáñez-Sedeño P; Langa F; Pingarrón JM Anal. Chim. Acta 2017, 959, 66–73. [DOI] [PubMed] [Google Scholar]
- (39).Das J; Ivanov I; Montermini L; Rak J; Sargent EH; Kelley SO Nat. Chem 2015, 7, 569–575. [DOI] [PubMed] [Google Scholar]
- (40).Malecka K; Stachyra A; Góra-Sochacka A; Sirko A; Zagórski-Ostoja W; Radecka H; Radecki J Sens. Actuators, B 2016, 224, 290–297. [Google Scholar]
- (41).Conde J; Edelman ER; Artzi N Adv. Drug Delivery Rev 2015, 81, 169–183. [DOI] [PubMed] [Google Scholar]
- (42).Schoukroun-Barnes LR; Wagan S; White RJ Anal. Chem 2014, 86, 1131–1137. [DOI] [PubMed] [Google Scholar]
- (43).Somerson J; Plaxco KW Molecules 2018, 23, 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kadimisetty K; Malla S; Bhalerao KS; Mosa IM; Bhakta S; Lee NH; Rusling JF Anal. Chem 2018, 90, 7569–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kadimisetty K; Spak AP; Bhalerao KS; Sharafeldin M; Mosa IM; Lee NH; Rusling JF Anal. Methods 2018, 10, 4000–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tang C; Vaze A; Rusling J Lab Chip 2017, 17, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Park H; Guo X; Temenoff JS; Tabata Y; Caplan AI; Kasper FK; Mikos AG Biomacromolecules 2009, 10, 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yu X; Munge B; Patel V; Jensen G; Bhirde A; Gong JD; Kim S-N; Gillespie J; Gutkind JS; Papadimitrakopoulos F; Rusling JF J. Am. Chem. Soc 2006, 128, 11199–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Shah VP; Midha KK; Findlay JW; Hill HM; Hulse JD; McGilveray IJ; McKay G; Miller KJ; Patnaik RN; Powell ML; et al. Pharm. Res 2000, 17, 1551–1557. [DOI] [PubMed] [Google Scholar]
- (50).Prensner JR; Rubin MA; Wei JT; Chinnaiyan AM Sci. Transl. Med 2012, 4, 127rv3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Butkus de Aguiar B; Girardi I; D’Avila Paskulin D; de Franca E; Dornelles C; Suparregui Dias F; Bonorino C; Sampaio Alho C Immunol. Invest 2008, 37, 752–769. [DOI] [PubMed] [Google Scholar]
- (52).Svensson J; Carlzon D; Petzold M; Karlsson MK; Ljunggren Ö; Tivesten A; Mellström D; Ohlsson CJ Clin. Endocrinol. Metab 2012, 97, 4623–4630. [DOI] [PubMed] [Google Scholar]
- (53).Mcgill R; Tukey J; Larsen W Am. Stat 1978, 32, 12–16. [Google Scholar]
- (54).Stenberg M; Nygren HJ Immunol. Methods 1988, 113, 3–15. [DOI] [PubMed] [Google Scholar]
- (55).Nygren H; Werthen M; Stenberg M J. Immunol. Methods 1987, 101, 63–71. [DOI] [PubMed] [Google Scholar]
- (56).Malhotra R; Papadimitrakopoulos F; Rusling JF Langmuir 2010, 26, 15050–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Esser P Activity of Adsorbed Antibodies; 11b; Application note; Thermo Scientific, 2014. [Google Scholar]
- (58).Butler JE; Ni L; Brown WR; Joshi KS; Chang J; Rosenberg B; Voss EW Mol. Immunol 1993, 30, 1165–1175. [DOI] [PubMed] [Google Scholar]
- (59).Butler JE; Ni L; Nessler R; Joshi KS; Suter M; Rosenberg B; Chang J; Brown WR; Cantarero LA J. Immunol. Methods 1992, 150, 77–90. [DOI] [PubMed] [Google Scholar]
- (60).Isobe N; Lee D-S; Kwon Y-J; Kimura S; Kuga S; Wada M; Kim U-J Cellulose 2011, 18, 1251. [Google Scholar]
- (61).Zangheri M; Cevenini L; Anfossi L; Baggiani C; Simoni P; Di Nardo F; Roda A Biosens. Bioelectron 2015, 64, 63–68. [DOI] [PubMed] [Google Scholar]
- (62).Roda A; Michelini E; Cevenini L; Calabria D; Calabretta MM; Simoni P Anal. Chem 2014, 86, 7299–7304. [DOI] [PubMed] [Google Scholar]
- (63).Smith PK; Krohn RI; Hermanson GT; Mallia AK; Gartner FH; Provenzano MD; Fujimoto EK; Goeke NM; Olson BJ; Klenk DC Anal. Biochem 1985, 150, 76–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


