Abstract
The situation of infectious diseases and biothreats all over the world remains serious. The effective identification of such diseases plays a very important role. In recent years, gold nanoparticles have been widely used in biosensor design to improve the performance for the detection of infectious diseases and biothreats. Here, recent advances of gold‐nanoparticle‐based biosensors in this field are summarized.
Keywords: biosensors, biodetection, infectious diseases, gold nanoparticles
Gold‐nanoparticle‐based biosensors play an important role in the identification of infectious diseases and biothreats. Gold nanoparticles can be used as signal producers and signal amplifiers to improve the performance of biosensors. Based on this approach, colorimetric, electrochemical, and fluorescent biosensors with high sensitivity and specificity are designed to fight infectious diseases and biothreats.
1. Introduction
It's a small world: Following the emergence of Severe Acute Respiratory Syndrome (SARS) in south China in November, 2002, the SARS virus rapidly spread to over 30 countries within months1 (Figure 1 ). Besides SARS, there are many kinds of infectious diseases, such as tuberculosis, cholera, and influenza, that challenge our vulnerable world frequently. According to data from World Health Organization (WHO), about 1 billion people worldwide are affected by one or more infectious diseases. Each year, almost 9 million people are killed, many of whom are children under five; furthermore, some infectious diseases cause life‐long disability.2 The situation is serious for developed and developing countries alike. In addition to naturally occurring diseases, biothreats and biochemical warfare agents also pose a threat,3 as exemplified by the anthrax spores attacks through the US mail system in 2001. Unfortunately, our knowledge of infectious diseases is still inadequate in terms of fully predicting and controlling epidemic events. One of the important factors hampering the control of infectious disease is a lack of effective diagnosis; in order to stop the infectious diseases and biothreats before they spread, it is of critical important to develop diagnostic tests that are fast, cheap, miniaturized, accurate, and suitable for the direct use in developing countries, remote areas, and for global surveillance.4 In the past decades, much progress has been made to address this problem; most serious epidemics have been contained through global alerts and cooperation.
Figure 1.
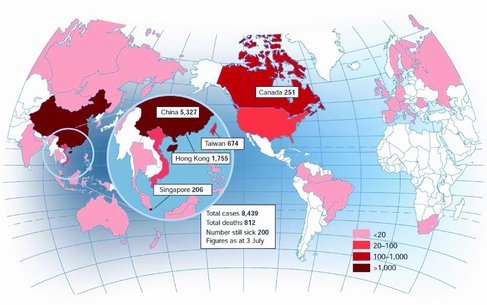
The global epidemic of SARS and the distribution of SARS cases worldwide in July, 2002. Reproduced with permission.1 Copyright 2003, Nature Publishing Group.
It's a “nano world”: When we look into infectious diseases, it is not difficult to find that their origins are related to mutated genes, abnormal proteins, and virulence factors from viruses or bacteria of nanometer scale or confined in nanometer‐ sized compartments. Coincidentally, nanotechnology provides an excellent platform in the nanometer range (1–100 nm) to improve the detection and identification of infectious diseases and biothreats. In recent years, various nanomaterials with unique properties have been synthesized and applied in many research areas, such as biosensors, bio‐imaging, nanomedicine, drug delivery, and nanofabrication.5 Among these nanomaterials, gold nanoparticles have attracted intense interests with their unique optical and electrical properties, high stability, and good biocompatibility. Based on these properties, gold nanoparticles have been used as biomolecular carriers, signal producers, and signal amplifiers in biosensor design.
Based on the characteristics of infectious diseases, such as rapid epidemic spread, high mutation rate, and frequent outbreaks in remote areas, only those detection strategies that are fast, cheap, stable, and miniaturized are suitable to fight these diseases effectively. In this article, we aim to summarize the applications of gold nanoparticles in such strategies. There are two detection modes in this research area, one of which is the detection of relevant molecular biomarkers (including DNA/RNA, proteins, and small molecules) of infectious diseases, while the other is the direct identification of intact pathogens.
2. The Applications of Gold Nanoparticles in Molecular Assays
2.1. Gold Nanoparticles as Signal Producers
2.1.1. Color Changes of Gold Nanoparticles
The color changes of gold nanoparticles tuned by interparticle plasmon coupling have been widely used in the biosensor field. The color of gold nanoparticle solution is red when the nanoparticles are very well dispersed and it turns blue (or purple) when the nanoparticles aggregate. With this intrinsic property of gold naanoparticles, a serials of biosensors based on predictable color changes have been designed to detect DNA (or RNA), proteins, and metal ions.6
Usually, two patterns of aggregation have been used in biosensor design. One pattern is based on the self‐assembly of recognition biomolecules (DNA, antibodies, and aptamers) on gold nanoparticles. After introducing the target into the system, the gold nanoparticles are brought together because of the formation of target biomolecule complexes (sometimes, this aggregation process is reversible). As a result, a color change from red to purple (or blue) occurs which can be easily observed by the naked eye; thus, complicated instrumentation is not required in this detection mode.
Mirkin's group were first to invent a method for the selective detection of DNA based on the distance‐dependent color changes of gold nanoparticles.7 This creative method is quite generalizable to any other DNA or RNA sequence. Then, Storhoff's group expanded this method to the detection of unamplified mecA in methicillin‐resistant Staphylococcus aureus (MRSA; the first outbreaks of infection occurred in Europe in early 1960s, and it has since spread worldwide)8 genomic DNA samples.9 Just like Mirkin's colorimetric assay, two types of DNA probes were immobilized on gold nanoparticles and used to hybridize to adjacent regions on the mecA sequence. In the presence of mecA target, a red‐to‐purple color change was observed because of the target‐induced aggregation of gold nanoparticles. This result confirmed that this method is selective enough to identify the specific target in the presence of total genomic DNA isolated from MRSA. While, as a negative control, the total genomic DNA from methicillin‐sensitive S. aureus (MSSA) strains did not bring any color change. An impressive detection limit was obtained and as few as 20 000 targets could be detected specifically in total bacterial genomic DNA without any signal amplification. Lai's group developed a similar method to identify Mycobacterium tuberculosis (MTB) and MTB complex (MTBC). Impressively, they collected 600 clinic sputum specimens and detected the relative DNA targets. Through comparing the results to the traditional culture and biomedical methods, their assay showed a 96.6% sensitivity and 98.9% specificity towards detection of MTBC, and a 94.7% sensitivity and 99.6% specificity for detection of MTB.10 After developing the gold nanoparticle‐based assay for tuberculosis, Baptista et al. estimated the overall cost was less than $0.35 per sample, which is much cheaper than for other molecular methods.11
Importantly, this strategy can be further developed and integrated with some other emerging technologies. As a successful integration, Chan et al. combined DNAzyme amplification with colorimetric coupling of surface plasmons of gold nanoparticles for point‐of‐care detection of infectious diseases.12 This method is based on a catalytic DNAzyme which is much cheaper and more stable than its protein counterparts. Furthermore, the simple colorimetric readout ensures this method is instrument‐free. As shown in Figure 2 , without the genetic target, the DNAzyme is disassembled into two parts (A and B) and has no cleavage activity on its substrate DNA strands (linker DNA); then, the gold nanoparticles conjugated with complementary strands to linker DNA can be bridged together and form aggregates, so the color turns blue. If the target is present, the components of DNAzyme assemble together and catalyze the cleavage of multiple linker DNAs; then, the gold nanoparticles disperse well because of the absence of the linker DNAs, so the color remains red. By rational design of the recognition arms and linker DNA, this method can be easily generalized to other DNA targets. They detected multiple DNA targets for infectious diseases (gonorrhea, syphilis bacteria, malaria parasite, and hepatitis B virus) in a multiplexed manner. This method inherently requires no complex equipment, expensive reagents, or complicated operations, making it suitable for point‐of‐care diagnosis.
Figure 2.
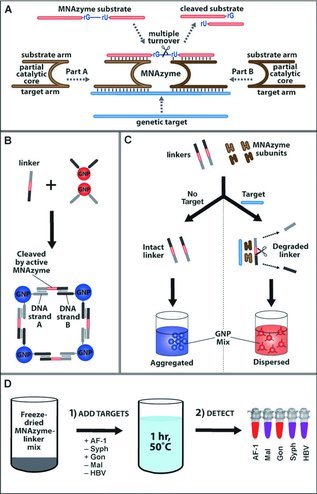
The integration of gold nanoparticles and DNAzyme technologies for multiplexed detection. A) The DNAzyme assembles together and catalyzes the cleavge of subatrate DNA. Reproduced with permission, Copyright 2009, American Chemical Society. B) Functionalized gold nanoparticles form aggregates with a color change in the prescence of linker DNA. C) With genetic target DNA, the DNAzyme is actived and catalytically digest the linker DNA; then, the functionalized gold nanoparticles remain red. Without target DNA, the linker DNA remains intact and brings aggregation of gold nanoparticles. D) This detection strategy can be generalized to many other targets for multiplexed detection. B–D) Reproduced with permission.12 Copyright 2013, Wiley‐VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
Interestingly, this detection mode is not limited to the detection of nucleic acids. Based on similar mechanisms, some biosensors for non‐nucleic acids targets such as proteins have been rationally designed.13 Russell's group developed a cholera toxin assay by using gold nanoparticles conjugated with specifically synthesized lactose derivative.13c They found that a lactose derivative with a thiolated anchor can form a stabilizing self‐assembled monolayer on the surface of gold nanoparticles. In an initial state, the stabilized gold nanoparticles dispersed well in solution and had red color. In the present of cholera toxin, the binding of cholera toxin and lactose induced the aggregation of gold nanoparticles, which broadened the surface plasmon adsorption band; as a result, the color changed to deep purple. The whole process was completed in 10 min and the detection limit was as low as 54 nm. A series of potential interferent ions and proteins had no effect on this assay.
The other pattern is based on the irreversible aggregation of gold nanoparticles without the self‐assembly of recognition biomolecules. After introducing the target, changes in the electrostatic, steric, or electrosteric properties of gold nanoparticles drive the aggregation and thus the color changes from red to purple or blue. Because the relatively complicated self‐assembly of biomolecules is not required, this pattern is simple in operation and does not need any specially trained personnel.
Ray's group has developed a sensitive and selective detection of hepatitis C virus (HCV), which is an enveloped RNA virus with a diameter of about 50 nm. They pointed out the limitations of traditional enzyme‐linked immunosorbent assays (ELISA) for HCV detection, which include false‐negative results in early stage and for immunosuppressed patients. First, single‐stranded RNA probes with flexible backbones were incubated with gold nanoparticles and adsorbed on the surface of gold nanoparticles, which can protect gold nanoparticles from aggregation. Then, HCV RNA targets were added into the solution. The hybridized double helices of target RNA and probes are relatively rigid and expose their negatively charged phosphate backbone to the gold nanoparticles. Thus, the double helices were separated from the gold nanoparticles due to electrostatic repulsion. As a result, the aggregation of gold nanoparticles and a color change were observed.14 A similar mechanism was later used by the same group to detect human immunodeficiency virus (HIV); however this time they used gold nanorods instead and achieved a detection limit of 100 pm.15
Most recently, a direct and rapid detection of MTB was developed using unmodified gold nanoparticles to detect unamplified genomic DNA extracted from MTB clinical samples.16 This method can eliminate the PCR amplification steps and reduce the turnaround time effectively. It was found that genomic DNA is difficult to denature because of its high G–C rate. Some physical methods, such as vortexing with glass beads and passing it through a fine syringe needle, did not work; an effective method was to use an enzyme (Bam HI) to digest the genomic DNA, which finally allowed detect of amounts of genomic DNA as low as 40 ng without any amplification.
When using this detection mode directly in complex matrices such as serum, we should note that there may be some difficulties. For example, many non‐specific proteins in serum may adsorb on the surface of gold nanoparticles that can stabilize the gold nanoparticles. This problem can be addressed by using filtered or diluted serum.
2.1.2. Gold Ions Dissolved from Gold Nanoparticles
Other than the color change, gold nanoparticles are an assembly of thousands to millions of gold atoms which can produce a much greater signal than a single atom. For example, a 20 nm spherical gold nanoparticle contains, in theory, 2.3 × 105 gold atoms.17 These gold atoms can be dissolved and transferred into Au3+ which can be easily detected by electrochemical methods. Based on this mechanism, electrochemical biosensors for infectious diseases have been developed in recent years.18
A 406‐base‐pair DNA sequence of human cytomegalovirus (HCMV, which is an important pathogen in transplant recipients and immunosuppressed individuals) was detected by Brossier's group. They extracted HCMV DNA from cells and amplified the DNA by PCR. Then they immobilized the amplified DNA on polystyrene microwell and allowed the hybridization of signaling probe‐gold nanoparticle conjugates. At last, the gold nanopartiles were dissolved by acidic bromine–bromide solution and quantified through anodic stripping voltammetry (ASV).17 A detection limit of 5 pm was achieved, which is equal to 10 amol of 406‐base pair DNA sequence per microwell. A further amplification could be realized by an enlargement process of gold nanoparticles using gold‐ion solution and formaldehyde reductant.19 After this, much more Au3+ could be generated for the electrochemical detection.
2.2. Gold Nanoparticles as Signal Amplifiers
Just like other nanomaterials, the high surface‐area‐to‐volume ratio is another property of gold nanoparticles that makes them excellent signal amplifiers for thousands of recognition biomolecules and signaling molecules. Recently, the self‐assembly of DNA (or RNA), aptamers, antibodies, and enzymes on the surface of gold nanoparticles has been extensively investigated.20
2.2.1. Antibody–Gold Nanoparticle Conjugates
The double‐antibody sandwich assay has become a mainstay of clinic diagnostics; however, a limitation of the traditional assay is that one target molecule binds only a single capture antibody and signaling probe, which limits the final signal gain and sensitivity. The combination of double‐antibody sandwich assay and gold nanoparticles provides a versatile platform for sensitive detection of antigens. Usually the antibodies are physically adsorbed onto the surface of gold nanoparticles without losing their binding activity. Sia and co‐workers demonstrated a useful and reliable diagnostic device based on antibody–gold nanoparticle conjugates21 (Figure 3 ). First, they immobilized the capture antibodies on a biochip. Then, HIV antigen, the antibody–gold nanoparticle conjugates, silver reagents, and washing buffer were delivered serially through a microfluidic device. The silver solution was used as signal amplification (silver reduction produces amplified signal on gold nanoparticles). Thus, an assay was completed within 20 min. Impressively, they pointed out that many technologies work well in laboratory but not in the field. To further prove the success of their detection, they tested their device in Rwanda (the HIV prevalence is about 3% nationally and even higher – 8% – in some areas such as Kigali). A small volume of unprocessed whole blood (less than 1 μL) from patient was used and 70 specimens were collected. They obtained sensitivity of 100% and specificity of 96%. Other than this, they tried to test the HIV in 101 archived specimens, many of which were also infected by hepatitis B, hepatitis C, or both. They obtained a high sensitivity of 100% and specificity of 94%. These results showed the excellent accuracy of their detection in complicated samples with different infection statuses. The antibody–gold nanoparticle conjugates are widely used for signal amplification in some other infectious factors and diseases,22 such as HIV type 1 in plasma,23 avian influenza virus,24 and respiratory infection.25
Figure 3.
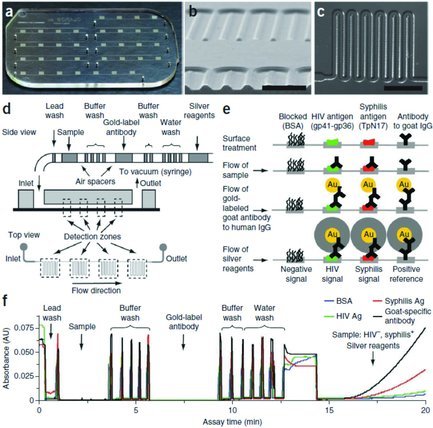
a) Microfluidic chip for multiplexed detection. b) Cross‐sectional image of microchannels through scanning electron microscope. c) The design of a meandering microchannel. d) Various reagents required are seperated by air spacers in a tube and then automatically and serially deliverd to the microchips. e) The detection steps and the signal amplification by the gold nanoparticles and silver stain. f) Various reagents passed through the detection zone; the signals are monitored in real time. a–f) Reproduced with permission.21 Copyright 2011, Nature Publishing Group.
2.2.2. DNA–Gold Nanoparticle Conjugates
DNA is recognized as an excellent component of biosensor design.20e, 26 With the development of DNA synthesis and modification, DNA can be synthesized at large scale and modified with many chemical groups (such as fluorophores and electrochemical moieties). The self‐assembly of DNA on gold nanoparticles was developed very well by Mirkin's group, who also invented an ultra‐sensitive protein detection called the nanoparticle‐based bio‐barcode.27 In this detection platform, they co‐immobilized DNA and signal‐antibody on the surface of gold nanoparticles to amplify the signal. Then, after the double‐antibody sandwich complex formed, a single target protein binding can bring a gold nanoparticle conjugated with antibodies and DNAs. Then, by quantifying the large amount of DNA released from the gold nanoparticle, highly sensitive protein detection was achieved with detection limit of 30 am concentration. Thus, a protein binding event can be transferred to the detection of a large number of DNA sequences. Based on the original mode, they further applied it to the sensitive detection of HIV‐1 p24 Gag protein.28 They found that commonly used methods, such as ELISA, are specific but not sensitive enough for the earliest stage of HIV infection: of 112 HIV‐positive plasma specimens, only 23 specimens were identified by ELISA, while using the nanoparticle based bio‐barcode method, 111 specimens were detected as HIV positive. A total of 34 uninfected specimens were not identified by both approaches. As a result, the specificity of the nanoparticle based bio‐barcode was 100% and the sensitivity was 99%.
DNA–gold nanoparticle conjugates are widely used in DNA detection, amplifying the signal and overcoming the limitation of the traditional DNA‐sandwich assay (capture DNA probe–DNA target–signaling DNA probe architecture). In the traditional sandwich assay, one DNA target can only hybridize one signaling probe, which brings a relatively weak signal from one signaling molecule (fluorophore or other signaling group). On the other hand, using DNA–gold nanoparticle conjugates, one hybridization event can bring an amplified signal from thousands of DNA sequences conjugated on the gold nanoparticles. Mirkin's group obtained a detection sensitivity of 500 zm using this method.20a, 29 A similar method was used to selectively detect mecA and tuf genes in the purified genomic DNA of MRSA; a success rate of 100% was obtained, comparing results with those from bacterial culturing.20d
2.2.3. Artificial Ligand–Gold‐Nanoparticle Conjugates30
Synthetic carbohydrate and protein interaction is widely used in biorecognition, but the binding affinity is relatively weak (k a is 103 –104 m −1).30a To improve this, Lin and co‐workers assembled globotriose (pk) on the surface of gold nanoparticles through Au–S bonding to interrogate the multivalent interaction of pk‐gold nanoparticle conjugates and B5 subunit of Shiga‐like toxin (B‐Slt). By doing so, the binding affinity was improved greatly and presented a ca. 108‐fold increase compared to the monovalent pk ligand. Basd on this, they developed a robust sandwich assay for the B‐Slt and obtained a detection limit of 1 μg mL−1.
2.2.4. Enzyme–Gold‐Nanoparticle Conjugates
The ease of assembly of biomolecules on gold nanoparticles has attracted intensive interest in biosensor design. Besides the ligands (antibodies, DNAs, and synthetic ligands) mentioned above, some enzymes (such as horse radish peroxidase (HRP) and alkaline phosphatase (ALP)) commonly used in ELISA can also be easily attached to gold nanoparticles. Comparing to traditional ELISA, which brings a signal from one enzyme for one binding event, an amplified signal from a large number of enzymes conjugated on gold nanoparticles is presented. We have demonstrated sensitive DNA detection through this mechanism.31 We co‐immobilized DNA signal probe (for the recognition of target DNA), HRP (for signal producing), and bovine serum albumin (BSA) (for the blocking of non‐specific adsorption). By using enzyme‐based multi‐component nanoprobes, a 100 pM target can easily be detected by the naked eye through the color generated from the enzyme catalysis reaction. A dual functional gold nanoparticle conjugated with antibodies and ALPs was used to detect Cryptosporidium parvum.32 By combining with an immunodot blot readout, a visually detection limit of 10 oocysts mL−1 was obtained, which represents a 500‐fold improvement over the conventional method.
However, a problem of concern is the activity and stability of the enzymes on gold nanoparticles, which may be affected by steric effects, structural changes, and crowding density. Instead of physical adsorption of enzymes on gold nanoparticles, Brust's group developed a covalent conjugation of enzymes and gold nanoparticles using “click” chemistry. This method may provide an efficient way to maintain the activity of the enzymes.33
3. The Application of Gold Nanoparticles in Identification and Inhibition of Pathogens
In molecular assays, the application of gold nanoparticles has led to many sensitive, selective, and specific biodetections for infectious diseases that are useful not only in routine laboratory tests but also in the field. In these molecular assays, the targets usually are biomarkers in pathogens; thus, to obtain these biomarkers, multiple extraction and separation steps are required that slow down the speed of detection and limit widespread use in remote settings. To overcome this, a new trend to directly identify the intact pathogens is emerging and recently has been attracting increasing interest. With a simplified process, the direct identification of intact pathogens is rapid and quite straightforward.
The perfect biocompatibility of gold nanoparticles and the well‐established surface modification of gold nanoparticles (the modification with antibodies, aptamers, and other ligands) means that they can be easily employed to the identification of pathogens. By using the excellent light‐scattering of gold nanoparticles, Li's group developed a simple and rapid counting method based on dark‐field imaging of E. coli. The whole detection process could be finished within 15–30 min which is impressive, and the detection limit was 2 × 104 colony forming units (CFU) per mL.34 Specifically, they used anti‐DH5α strain polyclonal antibody modified gold nanoparticles as probes (the antibody and the gold nanoparticle were covalently bridged through 11‐mercaptoundecanoic acid.). These probes directly bound to the target E. coli DH5α with satisfactory selectivity (Figure 4 ). As a control, another strain of E. coli BL21 and Rosetta were used. Through manual counting, they found that 77% of E. coli DH5α was targeted by the gold nanoparticles probes, while less than 15% was targeted for the other two strains. The possible reason for the non‐specific binding to other strains may be the relatively poor specificity of the polycolonal antibody. If some monoclonal antibodies or aptamers could be used, this method may be improved in specificity.
Figure 4.
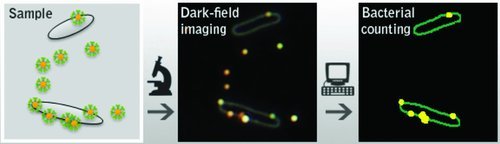
The antibody functionalized gold nanoparticles are used to image the E. coli directly based on the light‐scattering of gold nanoparticles. Through this simple method, the E. coli can be easily counted. Reproduced with permission.34 Copyright 2012, American Chemical Society.
After the specific identification of the pathogens, as a further step the pathogens could be destroyed by using the photothermal property of gold nanoparticles. Sabo‐attwood et al. covalently modified gold nanorods with primary antibodies specific to Pseudomonas aeruginosa obtained from the upper respiratory tract of sinusitis patients.35 The gold nanorod–antibody conjugates specifically bind to the target Pseudomonas aeruginosa. In this process, the nanorods that covalently conjugated with antibodies have higher binding efficiency than those modified with physically adsorbed antibodies. Near‐infrared radiation (NIR) was used to produce localized heating that can cause cellular damage. A 75% decrease in cell viability was observed. A similar method was used to identify the Gram‐positive bacterium Staphylococcus aureus with subsequent laser radiation.36 We should mention that aptamers, as promising recognition elements, could be used in these kinds of application to further improve selectivity and stability.
In the process of direct identification of pathogens using gold nanoparticles, the phenomenon of color change of gold nanoparticles is obvious and can be used as a direct readout. Because of the relatively large size of pathogen, a large number of functionalized gold nanoparticles can be adsorbed on the surface of pathogen and aggregate together, which causes obvious color change.37
Other than pathogen identification through the specific binding of antibody–surface antigen, electrostatic interactions can be used to detect whole pathogens. Rotello et al. used quaternary ammonium ligands to functionalize gold nanoparticles. Then, the functionalized gold nanoparticles were adsorbed on β‐galactosidase (β‐Gal) and thus the enzyme was inhibited. In the presence of target pathogen E. coli, competitive binding of the anionic surface of E. coli to the cationic gold nanoparticles occurred. Then the β‐Gal was free to catalyze its substrate to initialize a enzymatic reaction. A detection limit of 100 cells mL−1 was achieved and the detection could be complete in minutes. Furthermore, this method can be translated into a test strip with a visual sensitivity of 104 cells mL−1.38
Other than the direct identification of pathogens, there is an indirect way to inhibit the pathogens through gold nanoparticle‐mediated oligonucleotide delivery. As we mentioned above, the immobilization of oligonucleotides on the surface of gold nanoparticles is very well developed, allowing control of density and direction. We created a polyvalent immunostimulatory nanoagents by conjugating CpG oligonucleotides to gold nanoparticles, which can deliver the CpG oligonucleotides into cells and bring immunological effects (unmethylated CpG motifs are widely present in the genomic DNA of invading bacteria and viruses).39 It was noted that drug applications of oligonucleotides are largely limited by delivery approaches. Naked oligonucleotides cannot penetrate through the cell membrane and are prone to being cleared by nucleases in serum or cytoplasm. By using gold nanoparticles as a vehicle, the successful intracellular delivery of CpG was realized.
In order to inhibit pandemic H1N1 influenza viral replication, Prasad et al. used gold nanorods to enhance the delivery of a ssDNA immune activator, through which the expression of INF‐β and other INF‐stimulated genes was up‐regulated and thus viral replication was reduced.5c In their method, they modified gold nanorode with ssDNA through electrostatic binding; therefore the density, uniformity, and direction of ssDNA on the surface of gold nanorods needed to be improved. A possible way to enhance the efficiency further is covalent immobilization of ssDNA on gold nanorods with controlled density and direction.
4. Conclusions
The effective control of infectious diseases all over the world remains a challenge. For some infectious diseases, the lack of fast, cheap, sensitive, and specific biosensors is still a major problem. With the development of nanotechnology, we are happy to see recent advancements in the detection of infectious diseases. The application of color changes of gold nanoparticles provides a quite straightforward way to identify antigens and nucleic acids just using the naked eyes. This is potentially important for the screening of infectious diseases all over the world. In some resource‐limited countries and remote areas, this is equally important because it is an instrument‐free method and a power source is not required. On the other hand, the signal amplification through gold nanoparticles improves the sensitivity of biosensors which can be used effectively in the earliest stage of the infection. The direct idendification of pathogens provides a quite straightforward way to detect infectious diseases, which is very important for the control of diseases before they spread. Some concerns need to be raised, include the practical applications of biosensors in complicated matrices, the stability of biomolecule–gold nanoparticle conjugates, and reduction of the number of operation steps involved in detection.
Acknowledgements
The authors are grateful for financial support from the 100 Talent Project of the Chinese Academy of Sciences, National Basic Research Program of China (973 program, 2012CB932600, and 2013CB933800).
Contributor Information
Chunhai Fan, Email: fchh@sinap.ac.cn.
Xiaolei Zuo, Email: zuoxiaolei@sinap.ac.cn.
References
- 1. Pearson H., Clarke T., Abbott A., Knight J., Cyranoski D., Nature 2003, 424, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Global Report for Research on Infectious Diseases of Poverty, http://www.who.int/tdr/stewardship/global_report, accessed: April 2013.
- 3. a) Petrenko V. A., Sorokulova I. B., J. Microbiol. Methods 2004, 58, 147–168; [DOI] [PubMed] [Google Scholar]; b) Upadhyayula V. K. K., Anal. Chim. Acta 2012, 715, 1–18. [DOI] [PubMed] [Google Scholar]
- 4. a) Pang T., Peeling R. W., Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 856–857; [DOI] [PubMed] [Google Scholar]; b) Mabey D., Peeling R. W., Ustianowski A., Perkins M. D., Nat. Rev. Microbiol. 2004, 2, 231–240; [DOI] [PubMed] [Google Scholar]; c) Banoo S., Bell D., Bossuyt P., Herring A., Mabey D., Poole F., Smith P. G., Sriram N., Wongsrichanalai C., Linke R., O'Brien R., Perkins M., Cunningham J., Matsoso P., Nathanson C. M., Olliaro P., Peeling R. W., Ramsay A., Nat. Rev. Microbiol. 2010, S17–S27; [PubMed] [Google Scholar]; d) Forum Global Health Diagnostics, Nature 2006, 444, 681–681.17151639 [Google Scholar]
- 5. a) Kaittanis C., Santra S., Perez J. M., Adv. Drug Delivery Rev. 2010, 62, 408–423; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Arvizo R., Bhattacharya R., Mukherjee P., Expert Opin. Drug Delivery 2010, 7, 753–763; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chakravarthy K. V., Bonoiu A. C., Davis W. G., Ranjan P., Ding H., Hu R., Bowzard J. B., Bergey E. J., Katz J. M., Knight P. R., Sambhara S., Prasad P. N., Proc. Natl. Acad. Sci. USA 2010, 107, 10172–10177; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Li H., Huang J., Lv J., An H., Zhang X., Zhang Z., Fan C., Hu J., Angew. Chem. Int. Ed. 2005, 44, 5100–5103; [DOI] [PubMed] [Google Scholar]; e) Rosi N. L., Giljohann D. A., Thaxton C. S., Lytton‐Jean A. K. R., Han M. S., Mirkin C. A., Science 2006, 312, 1027–1030; [DOI] [PubMed] [Google Scholar]; f) Dubertret B., Calame M., Libchaber A. J., Nat. Biotechnol. 2001, 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 6. a) Li H. X., Rothberg L., Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li H. X., Rothberg L. J., Biophys. J. 2004, 86, 597a–597a; [Google Scholar]; c) Xia F., Zuo X. L., Yang R. Q., Xiao Y., Kang D., Vallee‐Belisle A., Gong X., Yuen J. D., Hsu B. B. Y., Heeger A. J., Plaxco K. W., Proc. Natl. Acad. Sci. USA 2010, 107, 10837–10841; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Pei H., Li F., Wan Y., Wei M., Liu H. J., Su Y., Chen N., Huang Q., Fan C. H., J. Am. Chem. Soc. 2012, 134, 11876–11879; [DOI] [PubMed] [Google Scholar]; e) Wang L. H., Liu X. F., Hu X. F., Song S. P., Fan C. H., Chem. Commun. 2006, 3780–3782; [DOI] [PubMed] [Google Scholar]; f) Zhang J., Wang L. H., Pan D., Song S. P., Boey F. Y. C., Zhang H., Fan C. H., Small 2008, 4, 1196–1200. [DOI] [PubMed] [Google Scholar]
- 7. a) Mirkin C. A., Letsinger R. L., Mucic R. C., Storhoff J. J., Nature 1996, 382, 607–609; [DOI] [PubMed] [Google Scholar]; b) Giljohann D. A., Seferos D. S., Daniel W. L., Massich M. D., Patel P. C., Mirkin C. A., Angew. Chem. Int. Ed. 2010, 49, 3280–3294; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rosi N. L., Mirkin C. A., Chem. Rev. 2005, 105, 1547–1562; [DOI] [PubMed] [Google Scholar]; d) Elghanian R., Science 1997, 277, 1078–1081. [DOI] [PubMed] [Google Scholar]
- 8. Chambers H. F., Clin. Microbiol. Rev. 1997, 10, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Storhoff J. J., Lucas A. D., Garimella V., Bao Y. P., Muller U. R., Nat. Biotechnol. 2004, 22, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soo P.‐C., Horng Y.‐T., Chang K.‐C., Wang J.‐Y., Hsueh P.‐R., Chuang C.‐Y., Lu C.‐C., Lai H.‐C., Mol. Cell. Probes 2009, 23, 240–246. [DOI] [PubMed] [Google Scholar]
- 11. Baptista P. V., Koziol‐Montewka M., Paluch‐Oles J., Doria G., Franco R., Clin. Chem. 2006, 52, 1433–1434. [DOI] [PubMed] [Google Scholar]
- 12. Zagorovsky K., Chan W. C. W., Angew. Chem. Int. Ed. 2013, 52, 3168–3171. [DOI] [PubMed] [Google Scholar]
- 13. a) Huang C. C., Huang Y. F., Cao Z., Tan W., Chang H. T., Anal. Chem. 2005, 77, 5735–5741; [DOI] [PubMed] [Google Scholar]; b) Chen S. J., Huang Y. F., Huang C. C., Lee K. H., Lin Z. H., Chang H. T., Biosens. Bioelectron. 2008, 23, 1749–1753; [DOI] [PubMed] [Google Scholar]; c) Schofield C. L., Field R. A., Russell D. A., Anal. Chem. 2007, 79, 1356–1361. [DOI] [PubMed] [Google Scholar]
- 14. Griffin J., Singh A. K., Senapati D., Lee E., Gaylor K., Jones‐Boone J., Ray P. C., Small 2009, 5, 839–845. [DOI] [PubMed] [Google Scholar]
- 15. Darbha G. K., Rai U. S., Singh A. K., Ray P. C., Chemistry 2008, 14, 3896–3903. [DOI] [PubMed] [Google Scholar]
- 16. Hussain M. M., Samir T. M., Azzazy H. M. E., Clin. Biochem. 2013, 46, 633–637. [DOI] [PubMed] [Google Scholar]
- 17. Authier L., Grossiord C., Brossier P., Anal Chem 2001, 73, 4450–4456. [DOI] [PubMed] [Google Scholar]
- 18. Saha K., Agasti S. S., Kim C., Li X., Rotello V. M., Chem. Rev. 2012, 112, 2739–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. a) Wang J., Xu D., Kawde A. N., Polsky R., Anal. Chem. 2001, 73, 5576–5581; [DOI] [PubMed] [Google Scholar]; b) Liao K. T., Cheng J. T., Li C. L., Liu R. T., Huang H. J., Biosens. Bioelectron. 2009, 24, 1899–1904. [DOI] [PubMed] [Google Scholar]
- 20. a) Nam J.‐M., Stoeva S. I., Mirkin C. A., J. Am. Chem. Soc. 2004, 126, 5932–5933; [DOI] [PubMed] [Google Scholar]; b) Boisselier E., Astruc D., Chem. Soc. Rev. 2009, 38, 1759–1782; [DOI] [PubMed] [Google Scholar]; c) Daniel M. C., Astruc D., Chem. Rev. 2004, 104, 293–346; [DOI] [PubMed] [Google Scholar]; d) Ramakrishnan R., Buckingham W., Domanus M., Gieser L., Klein K., Kunkel G., Prokhorova A., Riccelli P. V., Clin. Chem. 2004, 50, 1949–1952; [DOI] [PubMed] [Google Scholar]; e) Zhang J., Song S., Zhang L., Wang L., Wu H., Pan D., Fan C., J. Am. Chem. Soc. 2006, 128, 8575–8580. [DOI] [PubMed] [Google Scholar]
- 21. Chin C. D., Laksanasopin T., Cheung Y. K., Steinmiller D., Linder V., Parsa H., Wang J., Moore H., Rouse R., Umviligihozo G., Karita E., Mwambarangwe L., Braunstein S. L., van de Wijgert J., Sahabo R., Justman J. E., El‐Sadr W., Sia S. K., Nat. Med. 2011, 17, 1015–U1138. [DOI] [PubMed] [Google Scholar]
- 22. Bailes J., Mayoss S., Teale P., Soloviev M., in Nanoparticles in Biology and Medicine, Methods in Molecular Biology, Vol. 906, Humana Press, New York: 2012, pp. 45–55. [DOI] [PubMed] [Google Scholar]
- 23. Lee K.‐B., Kim E.‐Y., Mirkin C. A., Wolinsky S. M., Nano Lett. 2004, 4, 1869–1872. [Google Scholar]
- 24. Mu B., Huang X., Bu P., Zhuang J., Cheng Z., Feng J., Yang D., Dong C., Zhang J., Yan X., J. Virological Methods 2010, 169, 282–289. [DOI] [PubMed] [Google Scholar]
- 25. Halfpenny K. C., Wright D. W., Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. a) Fan C. H., Plaxco K. W., Heeger A. J., Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu G., Wan Y., Gau V., Zhang J., Wang L. H., Song S. P., Fan C. H., J. Am. Chem. Soc. 2008, 130, 6820–6825; [DOI] [PubMed] [Google Scholar]; c) Liu G., Sun C. F., Li D., Song S. P., Mao B. W., Fan C. H., Tian Z. Q., Adv. Mater. 2010, 22, 2148–2150. [DOI] [PubMed] [Google Scholar]
- 27. Nam J. M., Science 2003, 301, 1884–1886. [DOI] [PubMed] [Google Scholar]
- 28. Kim E.‐Y., Stanton J., Korber B. T. M., Krebs K., Bogdan D., Kunstman K., Wu S., Phair J. P., Mirkin C. A., Wolinsky S. M., Nanomedicine 2008, 3, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey R. C., Nam J.‐M., Mirkin C. A., Hupp J. T., J. Am. Chem. Soc. 2003, 125, 13541–13547. [DOI] [PubMed] [Google Scholar]
- 30. a) Chien Y. Y., Jan M. D., Adak A. K., Tzeng H. C., Lin Y. P., Chen Y. J., Wang K. T., Chen C. T., Chen C. C., Lin C. C., ChemBioChem 2008, 9, 1100–1109; [DOI] [PubMed] [Google Scholar]; b) Phillips R. L., Miranda O. R., You C. C., Rotello V. M., Bunz U. H., Angew. Chem. Int. Ed. Engl. 2008, 47, 2590–2594. [DOI] [PubMed] [Google Scholar]
- 31. a) Li J., Song S. P., Liu X. F., Wang L. H., Pan D., Huang Q., Zhao Y., Fan C. H., Adv. Mater. 2008, 20, 497–500; [Google Scholar]; b) Li J., Song S. P., Li D., Su Y., Huang Q., Zhao Y., Fan C. H., Biosens. Bioelectron. 2009, 24, 3311–3315. [DOI] [PubMed] [Google Scholar]
- 32. Thiruppathiraja C., Kamatchiammal S., Adaikkappan P., Alagar M., Biosens. Bioelectron. 2011, 26, 4624–4627. [DOI] [PubMed] [Google Scholar]
- 33. Brennan J. L., Hatzakis N. S., Tshikhudo T. R., Dirvianskyte N., Razumas V., Patkar S., Vind J., Svendsen A., Nolte R. J., Rowan A. E., Brust M., Bioconjugate Chem. 2006, 17, 1373–1375. [DOI] [PubMed] [Google Scholar]
- 34. Xu X., Chen Y., Wei H. J., Xia B., Liu F., Li N., Anal. Chem. 2012, 84, 9721–9728. [DOI] [PubMed] [Google Scholar]
- 35. Norman R. S., Stone J. W., Gole A., Murphy C. J., Sabo‐Attwood T. L., Nano Lett. 2008, 8, 302–306. [DOI] [PubMed] [Google Scholar]
- 36. Zharov V. P., Mercer K. E., Galitovskaya E. N., Smeltzer M. S., Biophys. J. 2006, 90, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. a) Singh A. K., Senapati D., Wang S. G., Griffin J., Neely A., Candice P., Naylor K. M., Varisli B., Kalluri J. R., Ray P. C., ACS Nano 2009, 3, 1906–1912; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang S. G., Singh A. K., Senapati D., Neely A., Yu H. T., Ray P. C., Chem. Eur. J. 2010, 16, 5600–5606; [DOI] [PubMed] [Google Scholar]; c) Khan S. A., Singh A. K., Senapati D., Fan Z., Ray P. C., Chem. Commun. 2011, 47, 9444–9446. [DOI] [PubMed] [Google Scholar]
- 38. Miranda O. R., Li X. N., Garcia‐Gonzalez L., Zhu Z. J., Yan B., Bunz U. H. F., Rotello V. M., J. Am. Chem. Soc. 2011, 133, 9650–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei M., Chen N., Li J., Yin M., Liang L., He Y., Song H. Y., Fan C. H., Huang Q., Angew. Chem. Int. Ed. 2012, 51, 1202–1206. [DOI] [PubMed] [Google Scholar]


