Abstract
In Belize, shallow populations (10 and 16 m) of the coral species Montastraea cavernosa from the back reef and reef crest are genetically differentiated from deeper populations on the fore reef and reef wall (25 and 35 m). Like many species of scleractinian corals, M. cavernosa has an obligate symbiosis with dinoflagellate microalgae from the family Symbiodiniaceae. Here, we describe the Symbiodiniaceae taxa found within previously sampled and genotyped M. cavernosa populations along a depth gradient on the Belize Barrier Reef by implementing high-throughput sequencing of the ITS2 region of Symbiodiniaceae ribosomal DNA and the SymPortal analysis framework. While Symbiodiniaceae ITS2 type profiles across all sampling depths were almost entirely (99.99%) from the genus Cladocopium (formerly Symbiodinium Clade C), shallow (10 and 16 m) populations had a greater diversity of ITS2 type profiles in comparison to deeper (25 and 35 m) populations. Permutational multivariate analysis of variance (PERMANOVA) confirmed significant differences in ITS2 type profiles between shallow and deep sample populations. Overall Symbiodiniaceae communities changed significantly with depth, following patterns similar to the coral host’s population genetic structure. Though physiological differences among species in the cosmopolitan genus Cladocopium are not well-described, our results suggest that although some members of Cladocopium are depth-generalists, shallow M. cavernosa populations in Belize may harbor shallow-specialized Symbiodiniaceae not found in deeper populations.
Keywords: mesophotic coral ecosystems, dinoflagellate, amplicon sequencing, ITS2, symbiosis, Cladocopium
Introduction
The association between scleractinian corals and their endosymbiotic dinoflagellate microalgae (family Symbiodiniaceae) supports the growth and persistence of important coral reef habitats worldwide (Muscatine and Cernichiari, 1969; Hatcher, 1988). Algal symbionts are sheltered and provided inorganic nitrogen, phosphorus, and carbon; consequently, as much as 95% of their photosynthetically produced saccharides are translocated to their coral host (Muscatine and Cernichiari, 1969; Muscatine et al., 1984; Rahav et al., 1989; Venn et al., 2008; Weis, 2008). This normally mutualistic symbiosis is also susceptible to breakdown when corals are exposed to external stressors, especially thermal anomalies (Goreau, 1964; Jokiel and Coles, 1977; Glynn and D’Croz, 1990; Brown et al., 1995; Glynn, 1996). Although corals are capable of heterotrophy, they are particularly reliant upon the photosynthate produced by algal symbionts to fully meet their energetic requirements. For most coral species in close association with Symbiodiniaceae, the coral host will ultimately perish if symbiosis is not re-established within a few weeks to months after bleaching (Glynn and D’Croz, 1990; Douglas, 2003).
The high diversity of algal symbiont communities within and among coral host species contributes to the intricacies of the coral-algal symbiosis. Morphometric differences observed among symbiotic dinoflagellates provided evidence of multiple genera and species within the initially described genus Symbiodinium (Blank and Trench, 1985). Recent work led to the revision of the former clades within the genus Symbiodinium sensu lato into distinct genera within the newly re-described family Symbiodiniaceae (LaJeunesse et al., 2018). These genera consist of numerous algal symbiont “types” or “strains” which align with species-level differences (LaJeunesse et al., 2012, 2018). Presently, there is still much focus on describing these species and evaluating their diverse genetic, physiological, and ecological characteristics.
Symbiodiniaceae species may exhibit varying tolerances to environmental conditions and stressors (Baker et al., 2004; LaJeunesse et al., 2010; Wham et al., 2017). For example, many species within the genus Durusdinium (formerly Clade D) exhibit higher thermal tolerance than most species in other genera, making them less likely to be expelled from their coral host in times of thermal anomaly (Stat and Gates, 2011; Cunning et al., 2015a; Silverstein et al., 2015). Corals may harbor different species of Symbiodiniaceae depending upon host species, geographic location, solar irradiance levels, or water depth (van Oppen et al., 2011; Bongaerts et al., 2015b; Cunning et al., 2015b, 2017). Corals often form symbioses with one Symbiodiniaceae species, yet some corals may harbor multiple Symbiodiniaceae taxa simultaneously (Thornhill et al., 2009; Silverstein et al., 2012; Baums et al., 2014; Serrano et al., 2014; Cunning et al., 2015a). These associations may also be ephemeral, with the numerically dominant Symbiodiniaceae taxa switching following environmental stress and disturbances, particularly after thermally induced coral bleaching events (Silverstein et al., 2012, 2015).
High-throughput sequencing of the internal transcribed spacer 2 (ITS2) region of the ribosomal DNA operon is one of the most useful molecular methods for describing Symbiodiniaceae communities within corals (LaJeunesse, 2001; Baker, 2003; Correa and Baker, 2009; Arif et al., 2014; LaJeunesse et al., 2018). ITS2 sequencing has been implemented to characterize Symbiodiniaceae community structure within and among coral colonies and to identify community profile shifts across environmental gradients, habitats, and temporal scales (Stat et al., 2009; Quigley et al., 2014; Klepac et al., 2015; Cunning et al., 2017; Polinski and Voss, 2018). While the ITS2 marker is widely used in studies characterizing Symbiodiniaceae communities and provides comparisons among studies, ITS2 is known to be multicopy and it is unclear how copy number varies among Symbiodiniaceae species (Thornhill et al., 2007). This can impact interpretations of inter- and intragenomic variation in bioinformatic analyses (Thornhill et al., 2007; Sampayo et al., 2009). Many studies collapse sequences into operational taxonomic units at 97% similarity threshold, as is common with many prokaryotic 16S amplicon analyses (Klepac et al., 2015; Cunning et al., 2017; Kenkel and Bay, 2018). This approach can be confounded by the intragenomic variation of Symbiodiniaceae ITS2 leading to an inability to resolve biologically relevant taxa (Smith et al., 2017; Hume et al., 2019). To overcome these hurdles, other studies have used additional markers (e.g., psbAncr) in conjunction with ITS2, allowing more robust analysis and interpretation of in hospite Symbiodiniaceae (LaJeunesse and Thornhill, 2011). Hume et al. (2019) recently developed and validated the SymPortal analysis framework to deal with issues of resolving Symbiodiniaceae taxa based only on ITS2 sequences. SymPortal identifies defining intragenomic variants (DIVs) within samples of ITS2 sequencing data. Combinations of DIVs are then used to determine ITS2 type profiles which are representative of putative Symbiodiniaceae taxa. This approach achieves finer resolution of inter- and intragenomic variation of Symbiodiniaceae ITS2 without the use of additional markers (Hume et al., 2019).
Coral reefs globally are imperiled by a number of anthropogenic influences, most notably climate change (Hughes, 1994; Glynn, 1996; Gardner et al., 2003; Mumby et al., 2006; De’ath et al., 2012). Sea surface temperature models and future emission scenarios project that the majority of the world’s coral reefs will experience harmfully frequent thermal stress events in the coming decades (Hoegh-Guldberg et al., 2008; Donner, 2009; Eakin et al., 2010; van Hooidonk et al., 2016; Hughes et al., 2017; Skirving et al., 2019), which may have devastating consequences for the delicate mutualism between corals and Symbiodiniaceae and the ecosystems it supports. With the present deterioration of coral reefs and the continued threat of decline, there has been an increased focus on mesophotic coral ecosystems (MCEs; Lesser et al., 2009, 2018; Bongaerts et al., 2010). Located at 30–150 m depths, MCEs experience different thermal regimes, light spectra, and irradiance as compared to shallow coral ecosystems (Lesser et al., 2000, 2009; Leichter et al., 2006; Kahng et al., 2010; Smith et al., 2016). Despite these differences, many scleractinian species in the Tropical Western Atlantic (TWA) co-occur on both shallow reefs and MCEs (as much as 25–40%; Bongaerts et al., 2010, 2017). Due in part to the species overlap with shallow reefs, MCEs are hypothesized to be potential refuges for shallow reefs (i.e., the Deep Reef Refugia Hypothesis; Glynn, 1996; Bongaerts et al., 2010; Lesser et al., 2018; Bongaerts and Smith, 2019). Multiple studies have examined the potential for “reseeding” of shallow reefs with larvae from MCE coral counterparts using molecular methods to quantify levels of genetic connectivity between these habitats (Brazeau et al., 2013; Bongaerts et al., 2017; Studivan and Voss, 2018; Eckert et al., 2019). It is important to evaluate symbiont community assemblages as well as coral genetic structure when assessing connectivity of shallow and mesophotic reefs to give insight into the potential barriers to vertical connectivity for depth-generalist scleractinians (Bongaerts et al., 2010).
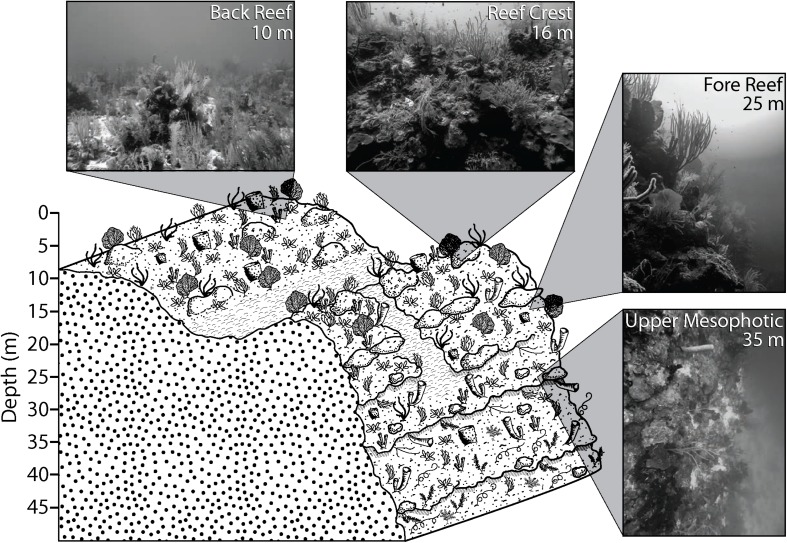
Several studies have examined Symbiodiniaceae associated with scleractinian corals in Belize (Warner et al., 2006; Finney et al., 2010; Baumann et al., 2018), yet none have assessed how Symbiodiniaceae vary across shallow and mesophotic depths. The Belize Barrier Reef surrounding Carrie Bow Cay and the seaward margin of Glover’s Reef Atoll provide abundant mesophotic habitat directly adjacent to shallow coral ecosystems. These reefs exhibit spur and groove structures to a depth of 20–33 m, a near-vertical step from 30–37 m, and a sloping reef wall continuing to >100 m (James and Ginsburg, 1979; Figure 1). Populations of the depth-generalist, broadcast spawning, scleractinian coral Montastraea cavernosa lack gene flow between relatively shallow (10 and 16 m) and deep (25 and 35 m) populations within the South Water Caye and Glover’s Reef Marine Reserves (Eckert et al., 2019). To better understand if the identity of algal endosymbionts associated with M. cavernosa populations in Belize followed similar patterns as observed in genetic structuring of their coral hosts, we characterized the Symbiodiniaceae found within previously genotyped M. cavernosa samples using the high-throughput sequencing of the ITS2 marker and the SymPortal analysis framework.
FIGURE 1.
Generalized reef structure and sampling depths on the Belize Barrier Reef. Inset photographs are representative of typical habitat at sampling depths. Figure from Eckert et al. (2019).
Materials and Methods
Sampling Sites and Sample Collection
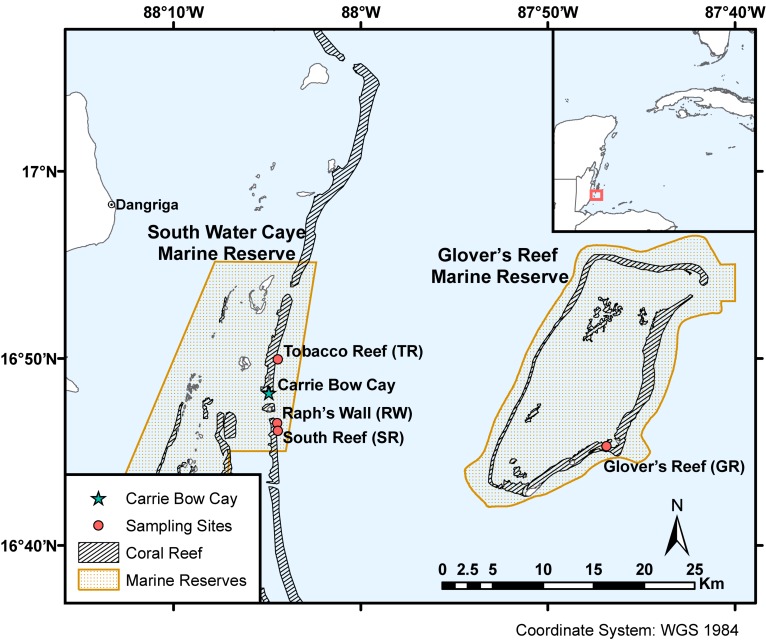
This study examined the algal endosymbionts within M. cavernosa populations found across two marine reserves (South Water Caye and Glover’s Reef Marine Reserves) on the Belize Barrier Reef. Samples were collected from three sites along the barrier reef near Carrie Bow Cay (South Reef, Raph’s Wall, and Tobacco Reef) and one site on Glover’s Reef Atoll (Glover’s Reef), ∼30 km southeast of Carrie Bow Cay (Figure 2). Samples were collected from sites containing sufficiently abundant M. cavernosa across all depth zones. Approximately 15 M. cavernosa colonies were sampled at each of the four depth zones (back reef ∼10 m; reef crest ∼16 m; fore reef ∼25 m; upper mesophotic ∼35 m; Figure 1) per reef site (n = 242). Sample collection and initial processing are detailed in Eckert et al. (2019). After field collection and processing at Carrie Bow Cay, samples were preserved in TRIzol reagent and initially stored at –20°C. Samples were then transported to FAU-HBOI on ice and stored at –80°C until genomic DNA extraction. Samples were collected over two field expeditions (Table 1). All samples were collected in the spring, nearly one year apart, to avoid potential seasonal Symbiodiniaceae community shifts. Samples from 35 m depth zones within South Water Caye Marine Reserve (i.e., Tobacco Reef, Raph’s Wall, and South Reef; n = 45) were collected in March 2016 (Studivan and Voss, 2018), and all remaining samples (n = 137) in South Water Caye Marine Reserve were collected in March 2017. All samples (n = 60) from Glover’s Reef Marine Reserve were collected in March 2017.
FIGURE 2.
Sampling sites within South Water Caye and Glover’s Reef Marine Reserves on the Belize Barrier Reef. Coral reef habitat and MPA shapefiles adapted from Meerman and Clabaugh (2017).
TABLE 1.
Site and sampling information for Montastraea cavernosa samples collected near Carrie Bow Cay, Belize.
| Site name | Latitude | Longitude | Depth zone | n | Sampling date |
| Tobacco Reef (TR) | 16°49.946′ N | 88°4.441′ W | 10 m | 15 | 25 March 2017 |
| 16 m | 15 | 24–25 March 2017 | |||
| 25 m | 15 | 25 March 2017 | |||
| 35 m | 15 | 7–8 March 2016 | |||
| Raph’s Wall (RW) | 16°46.564′ N | 88°4.479′ W | 10 m | 15 | 23 March 2017 |
| 16 m | 15 | 23 March 2017 | |||
| 25 m | 16 | 23 March 2017 | |||
| 35 m | 15 | 6 March 2016 | |||
| South Reef (SR) | 16°46.137′ N | 88°4.433′ W | 10 m | 16 | 24 March 2017 |
| 16 m | 15 | 24 March 2017 | |||
| 25 m | 15 | 24 March 2017 | |||
| 35 m | 15 | 5 and 8 March 2016 | |||
| Glover’s Reef (GR) | 16°45.323′ N | 87°46.875′ W | 10 m | 15 | 27 March 2017 |
| 16 m | 15 | 27 March 2017 | |||
| 25 m | 15 | 27 March 2017 | |||
| 35 m | 15 | 27 March 2017 |
Symbiodiniaceae ITS2 Amplicon Sequencing Library Preparation
Total genomic DNA was extracted using a modified cetyl trimethylammonium bromide (CTAB) extraction (Mieog et al., 2009) as in Eckert et al. (2019). Following DNA extraction, all samples were cleaned with the Zymo Research DNA Clean & Concentrator-5 kit to enhance downstream polymerase chain reaction (PCR) amplification. Cleaned extracts were checked for quality and concentration with a NanoDrop 2000 (Thermo Fisher Scientific) spectrophotometer and sample dilutions were prepared for PCR amplifications.
The ITS2 region of Symbiodiniaceae ribosomal DNA operon was targeted for sequencing using Symbiodiniaceae specific primers its-dino and its2rev2 (Pochon et al., 2001; Stat et al., 2009) modified to include adapter regions for the incorporation of indexed forward and reverse Illumina adapters (Klepac et al., 2015; Supplementary Table S1). Each 30 μL PCR included the following components: 1U Takara HS Taq, 1X Takara Taq Buffer, 0.15 μM each forward and reverse primer, 0.25 mM dNTP mixture, and 20 ng of template genomic holobiont DNA. All PCRs were run with an initial melt of 95°C for 5 min, followed by 22–28 cycles of 95°C for 40 s, 65°C for 2 min, and 72°C for 1 min, and a final extension of 72°C for 10 min. To avoid over-amplification, any samples with only a faint band visible on a 2% agarose gel after 22 cycles received an additional 1–6 PCR cycles following the same PCR profile without the initial melt of 95°C for 5 min (Kenkel et al., 2013; Klepac et al., 2015). Samples that did not amplify after 28 cycles were excluded from further analyses (n = 1; Supplementary Table S2).
PCR products were cleaned with the Thermo Scientific GeneJET PCR Purification Kit according to manufacturer protocols, quantified fluorescently using Qubit (Invitrogen), and diluted for subsequent PCRs. Samples were randomly assigned to one of three sequencing pools (∼80 samples per pool) and a second PCR was run on each sample to incorporate a unique combination of indexed forward and reverse Illumina adapter primers producing a unique dual index (i.e., “barcode”) for each sample in each pool (Klepac et al., 2015; Supplementary Table S1). A 20 μL PCR was run for each sample in each pool with 15 ng initial PCR product and 0.15 μM of each indexed Illumina forward and reverse adapter primer. All other components were identical to the initial ITS2 amplification PCR. Cycling conditions were identical to initial PCRs, but with only 4 cycles required to incorporate indexed adapters.
Indexed PCR products were run on a 2% agarose gel with SYBR Green (Invitrogen). The resulting ∼500 bp amplicon was excised, extracted with the QIAGEN Gel Extraction Kit, and quantified using quantitative PCR (qPCR) on an Eppendorf Realplex4 using Thermo Scientific Maxima SYBR Green qPCR Master Mix with 0.1 μM Illumina adapter primers (Supplementary Table S1). Indexed ITS2 libraries were pooled based on calculated cycling thresholds (CT) to ensure equitable representation among samples in each sequencing pool. Pooled libraries were purified and concentrated through isopropanol precipitation and eluted in nuclease-free water for sequencing. The libraries were loaded and sequenced with 20% phiX on the Illumina MiSeq platform (v3 chemistry) using paired-end 300 bp reads.
Amplicon Sequencing Analysis With SymPortal
Demultiplexed forward and reverse. fastq. gz files were remotely submitted to SymPortal.org for analysis and were subjected to standard sequence quality control protocols implemented with MOTHUR 1.39.5 (Schloss et al., 2009), the BLAST+ suite of executables (Camacho et al., 2009), and minimum entropy decomposition (Eren et al., 2015) to filter non-Symbiodiniaceae and sequencing artifacts from the dataset (Hume et al., 2019). Sequences were grouped by genera and only groups with more than 200 sequences were algorithmically searched. Sequences occurring in a sufficient number of samples within both the dataset being analyzed and the entire database of samples run through SymPortal were identified as DIVs which were then used to characterize ITS2 type profiles (Hume et al., 2019).
Statistical Analysis of SymPortal Results
Subsequent statistical analyses of Symbiodiniaceae diversity were conducted on SymPortal outputs in the R statistical environment (R Core Team, 2019) and PRIMER v7 software package (Clarke and Gorley, 2015). To account for differences in sequencing depth among individual libraries, resulting ITS2 sequences and ITS2 type profile reads were normalized using trimmed mean of M-values (TMM) in the package edgeR (Robinson and Oshlack, 2010), which effectively decreases false discovery rates and increases true positive rates (Pereira et al., 2018). Non-metric multidimensional scaling analyses were conducted in PRIMER v7 using Bray-Curtis dissimilarities of square root-transformed sample read counts to visualize differences in beta diversity of ITS2 sequences and ITS2 type profiles.
Subsequent statistical analyses were carried out on ITS2 type profile sequencing reads, which are representative of putative Symbiodiniaceae taxa. The betadisper function was used in the package vegan to calculate multivariate homogeneity of dispersion (PERMDISP) using Bray-Curtis distances (Oksanen et al., 2019). Pairwise comparisons were calculated with permutation tests using the permutest function in vegan for any significant factors (9,999 permutations). Permutational multivariate analysis of variance (PERMANOVA) was used to test for differences in Symbiodiniaceae ITS2 type profiles, due to the balance of sampling design and the demonstrated lack of sensitivity to heterogeneity of dispersion compared to other multivariate statistical tests (e.g., ANOSIM; Anderson and Walsh, 2013). Depth and sampling site were used as fixed factors in the adonis function in vegan with 9,999 permutations of residuals from Bray-Curtis dissimilarities. After significant PERMANOVA results, pairwise PERMANOVA tests were conducted with the package pairwiseAdonis (Martinez Arbizu, 2017) using false discovery rate (FDR) corrected p-values. PERMANOVA were also run on a subset of the data with 35 m samples removed to test if different sampling times (2016 vs. 2017) influenced the observed results. Finally, similarity percentage (SIMPER) tests were run in PRIMER v7 using an 80% cumulative similarity cutoff to determine which ITS2 type profiles contributed most to significant differences among factors identified by pairwise PERMANOVA.
Results
Symbiodiniaceae ITS2 Sequences and ITS2 Type Profiles
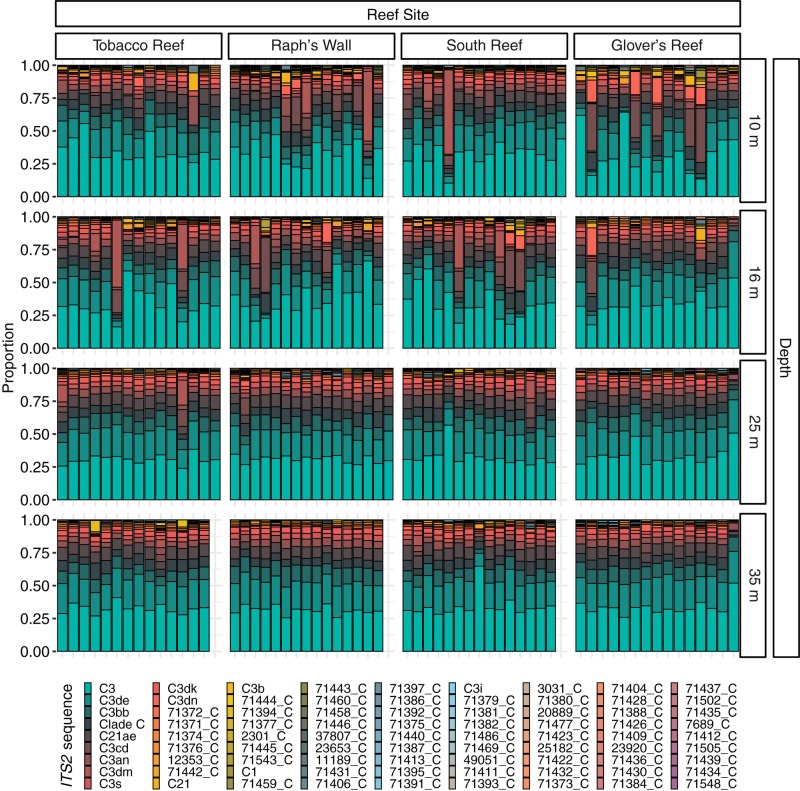
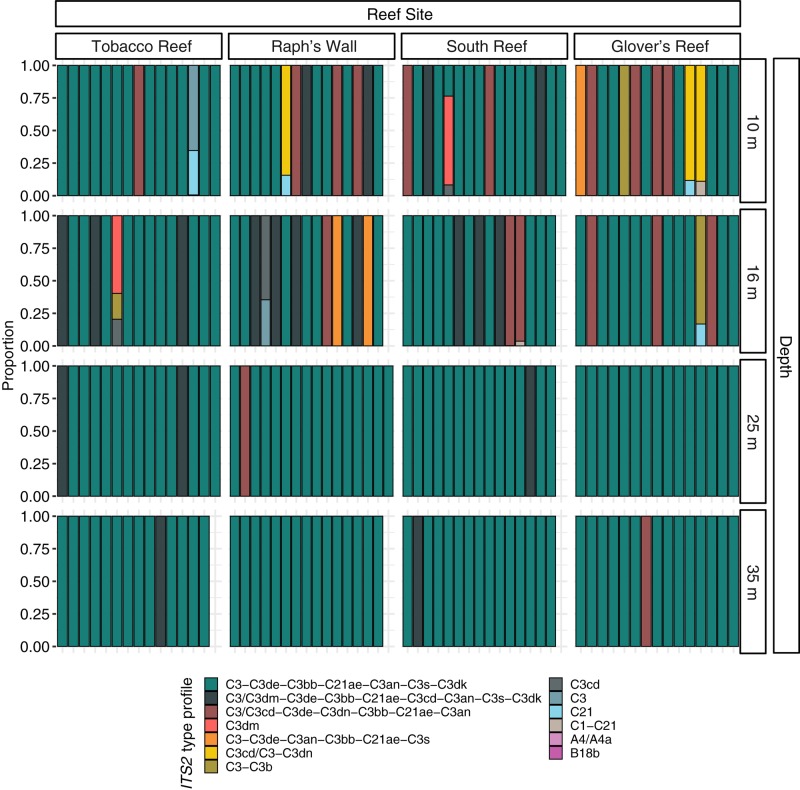
Prior to quality filtering, the 241 samples returned 46,476,815 sequencing reads, 30,858,017 of which passed the described initial quality and sample assignment filters (66.39%). ITS2 sequences from the genera Symbiodinium (formerly Clade A), Breviolum (formerly Clade B), and Cladocopium (formerly Clade C) were used to calculate ITS2 type profiles, with the majority of filtered ITS2 sequences being of the genus Cladocopium (99.99%; Figure 3). Thirteen ITS2 type profiles were identified across all samples, eleven of which were from the genus Cladocopium, with the remaining Symbiodinium and Breviolum profiles comprising 0.00017% of all ITS2 type profiles (Figure 4).
FIGURE 3.
Normalized relative proportion of ITS2 sequences from Montastraea cavernosa samples. Only the most abundant (>0.01% of all reads) sequences are displayed (n = 81). ITS2 sequences are listed in order of overall abundance. Sequences not used in the definition of ITS2 type profiles are named with a unique database ID number followed by a letter which refers to the genus the sequence is from (e.g., 71372_C is a sequence from the genus Cladocopium with the database ID 71372).
FIGURE 4.
Normalized relative proportion of ITS2 type profiles from Montastraea cavernosa samples. ITS2 type profiles are listed in order of overall abundance. ITS2 type profiles are named for the defining intragenomic variants (DIVs) used to characterize them. DIVs in a type profile name are listed in order of abundance with ‘/’ separating DIVs with co-dominance in samples.
Symbiodiniaceae Variation Across Depth
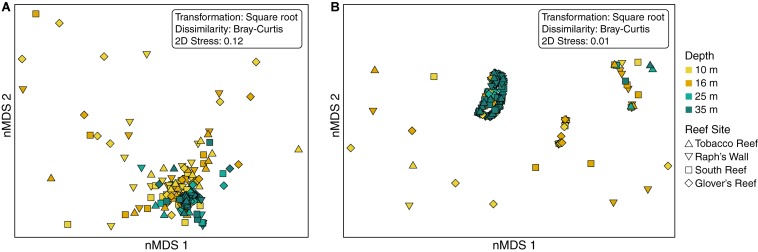
Visualization of beta diversity of ITS2 sequences with nMDS revealed that most samples from 25 and 35 m populations, hereafter referred to as “deep populations,” clustered together while samples from 10 and 16 m, hereafter referred to as “shallow populations,” were much less tightly clustered (Figure 5A). Ordination of ITS2 type profiles with nMDS illustrated strong clustering of the majority of samples (Figure 5B). Nearly all (94.17%) of the deep population samples grouped together in the main cluster, with shallow population samples much more dispersed in comparison (66.12% in the main cluster). Beta diversity of ITS2 type profiles was significantly higher in shallow populations (F = 11.5653, 237, p < 0.0001; Table 2) than deep populations but did not differ significantly across sampling sites.
FIGURE 5.
Non-metric multidimensional scaling (nMDS) biplot based on (A) ITS2 sequences and (B) ITS2 type profiles using Bray-Curtis dissimilarities. Each point represents the Symbiodiniaceae found within a Montastraea cavernosa sample. Color denotes sampling depth and shape denotes reef site.
TABLE 2.
Test results for homogeneity of multivariate dispersions (PERMDISP) and pairwise permutation tests using the betadisper and permutest functions in vegan.
| Test | Comparison | Test statistic | p-value |
| PERMDISP | Site | 0.5837 | ns |
| Depth | 11.565 | 0.0001 | |
| Permutation test | 10 m vs. 16 m | –0.1384 | ns |
| 10 m vs. 25 m | 3.9797 | 0.0004 | |
| 10 m vs. 35 m | 4.2363 | 0.0001 | |
| 16 m vs. 25 m | 4.0923 | 0.0001 | |
| 16 m vs. 35 m | 4.3452 | 0.0001 | |
| 25 m vs. 35 m | 0.3414 | ns |
Pairwise p-values are the permuted p-values generated by the test (9,999 permutations). Non-significant tests are listed as “ns.”
PERMANOVA results indicated that Symbiodiniaceae communities varied significantly by depth (Pseudo-F = 6.8803, 225, p = 0.0001), but that neither site nor the interaction between site and depth had any significant effects on community composition. Pairwise PERMANOVA identified that differences in Symbiodiniaceae communities occurred between shallow populations and deep populations (Table 3). Removing all 35 m samples did not change results of PERMANOVA or pairwise comparisons, demonstrating that time differences between sampling trips did not significantly influence patterns in algal symbiont community structure (Supplementary Table S3). SIMPER tests found shallow and deep populations were 37.46% dissimilar to one another on average. The three most abundant ITS2 type profiles (Figure 4) accounted for over 85% of the dissimilarity between shallow and deep populations (Table 4).
TABLE 3.
Test results from permutational multivariate analysis of variance (PERMANOVA; 9,999 permutations) of Symbiodiniaceae ITS2 type profiles from M. cavernosa colonies and pairwise comparisons between all depth zones (FDR corrected).
| Test | Comparison | Pseudo-F | p-value |
| Overall | Depth | 6.8801 | 0.0001 |
| Site | 1.4559 | ns | |
| Depth:Site | 1.1611 | ns | |
| Depth | 10 m vs. 16 m | 0.9838 | ns |
| 10 m vs. 25 m | 9.0844 | 0.0005 | |
| 10 m vs. 35 m | 10.0260 | 0.0004 | |
| 16 m vs. 25 m | 9.4051 | 0.0004 | |
| 16 m vs. 35 m | 10.6563 | 0.0004 | |
| 25 m vs. 35 m | 0.1196 | ns |
Non-significant tests are listed as “ns.”
TABLE 4.
Similarity percentage (SIMPER) test results.
| Group | Average Dissimilarity | ITS2 type profile | Contribution |
| Shallow vs. Deep | 37.46% | C3-C3de-C3bb-C21ae-C3an-C3s-C3dk | 46.43% |
| C3/C3dm-C3de-C3bb-C21ae-C3cd-C3an-C3s-C3dk | 19.64% | ||
| C3/C3cd-C3de-C3dn-C3bb-C21ae-C3an | 19.13% |
ITS2 type profiles contributions to Average dissimilarity between depth zones are shown in descending order. SIMPER cutoff was set at 80%.
Discussion
Cladocopium was the most abundant algal genus identified across all M. cavernosa samples from all depth zones on the Belize Barrier Reef, the majority of which were C3-related sequences. These results agree with previous characterizations of M. cavernosa algal symbiont communities on the Belize Barrier Reef which all hosted Cladocopium (mainly C3 type) species as their dominant algal symbiont across a depth gradient from 8–25 m (Warner et al., 2006). The genus Cladocopium is one of the most species-rich, ecologically abundant, and diverse genera of Symbiodiniaceae (LaJeunesse et al., 2018). Some members of the genus, C. goreaui for example, are considered host-generalists because they exist in association with many coral species and across broad environmental, geographic, and depth ranges (LaJeunesse, 2001, 2005; Serrano et al., 2014; LaJeunesse et al., 2018). Previous studies across multiple regions have reported M. cavernosa harboring Cladocopium spp. almost exclusively (Warner et al., 2006; Serrano et al., 2014; Bongaerts et al., 2015b; Klepac et al., 2015; Polinski and Voss, 2018). Similar to the results presented here, Cladocopium spp. have also been identified as the predominant symbiont species within M. cavernosa on reefs in Florida, United States Virgin Islands, Barbados, Bermuda, as well as other sites in Belize (Finney et al., 2010; Serrano et al., 2014). Just as Cladocopium spp. are extremely cosmopolitan reef inhabitants, M. cavernosa is one of the most ubiquitous coral species in the TWA, potentially due in part to its association with this common genus of algal symbiont. Many species of Cladocopium are also found associated with coral species at depths >60 m (Lesser et al., 2010; Bongaerts et al., 2015b; Lucas et al., 2016). Members of Cladocopium were found to be more photochemically efficient than members of Durusdinium, especially in temperatures ≤24°C (Silverstein et al., 2017). This may make Cladocopium species more beneficial symbionts at greater depths, including in mesophotic coral ecosystems where temperatures can be lower and light is more limited (Lesser et al., 2009). Niche specialization and coincidental metabolic costs across depth have been demonstrated in the algal symbiont communities of Seriatopora hystrix colonies (Cooper et al., 2011). While Durusdinium spp. were dominant in S. hystrix between 3–23 m, Cladocopium dominated endosymbiont communities were more common between 23–45 m, coinciding with a decline in net photosynthetic production.
Forming symbiotic relationships predominantly with a cosmopolitan and hyper-diverse Symbiodiniaceae genus may be advantageous to high dispersal, allowing M. cavernosa to dominate and persist across most reefs in the TWA. However, some studies examining M. cavernosa Symbiodiniaceae communities have also detected background (≤5%) levels of other genera, including Symbiodinium, Breviolum and Durusdinium (Serrano et al., 2014; Klepac et al., 2015; Polinski and Voss, 2018). All of these genera have been previously identified within other scleractinian species on the Belize Barrier Reef (Baumann et al., 2018), but we only detected minimal abundances (<0.01%) of ITS2 type profiles from Symbiodinium and Breviolum in a minority of our samples (n = 2), demonstrating an affinity for symbioses between M. cavernosa and Cladocopium spp. in this region.
Previous examination of Symbiodiniaceae from mesophotic and shallow M. cavernosa has found significant differences in communities and physiology across depth in multiple regions. In the northwestern Gulf of Mexico (NW GOM), M. cavernosa from MCEs had higher densities of Symbiodiniaceae cells and greater levels of chlorophyll a and chlorophyll c2 per unit area of coral tissue in comparison to shallow conspecifics (Polinski and Voss, 2018). Despite these differences, there was no significant difference among Symbiodiniaceae communities observed across depth in the NW GOM. Other studies have found differences in Symbiodiniaceae communities of M. cavernosa and other scleractinian corals across depth in the Bahamas (Lesser et al., 2010) and Curaçao (Bongaerts et al., 2015a, b). In these instances, significant differences were observed in the lower mesophotic zone (>60 m). However, in Curaçao there was also a significant shift in symbiont community profile observed at 25 m for M. cavernosa (Bongaerts et al., 2015a), the same depth at which we have seen differences in both symbiont communities and M. cavernosa genetic structure in Belize (Eckert et al., 2019). While there is evidence for Symbiodiniaceae community shifts with increasing depth, these patterns do not appear to be universal. Rather, they appear to depend upon both coral species and region (Bongaerts et al., 2010, 2015a, 2017).
Symbiodiniaceae ITS2 type profiles from M. cavernosa on the Belize Barrier Reef are depth-stratified, but not as distinctly as M. cavernosa population genetic structure. The symbiont community “break point” still appears to remain between the geomorphologic transition between reef crest and fore reef (16 and 25 m; Figure 1), but rather than having distinct depth-specialized assemblages, there is a relatively abundant depth-generalist ITS2 type profile in Belize (Figure 4). Shallow communities instead show greater algal diversity across coral samples, characterized by the presence of additional ITS2 type profiles not present in samples beyond 16 m. In Curaçao, multiple depth-generalist corals also harbored more diverse Symbiodiniaceae communities on shallow reefs, but overall, hosted similar algal symbiont compositions between shallow and mesophotic depths, until depths of 50–60 m (Bongaerts et al., 2015a). These results are similar to what we observed in the algal symbiont communities of M. cavernosa across shallow and upper mesophotic depths on the Belize Barrier Reef. There may be extreme depth-specialized communities in Belize among deeper (i.e., 40–60 m) M. cavernosa populations not captured in this study, as reported in other regions (Lesser et al., 2010; Bongaerts et al., 2015a).
On the Belize Barrier Reef, we found the majority (94.17%) of deep populations of M. cavernosa were dominated by a single ITS2 type profile with only 2 additional ITS2 type profiles present in these populations. The majority of all sampled colonies along the depth gradient from 10–35 m had the same Symbiodiniaceae ITS2 type profile (Figure 4; n = 192). Recent studies suggest that there is a high level of intra-genus diversity in physiological tolerances of algal symbionts. For example, a consensus ranking algorithm found that thermal tolerance of different Cladocopium species ranged highly, between the 3rd and 71st percentile (Swain et al., 2017). There is the potential that the ITS2 type profiles which were only present in shallow populations of M. cavernosa are specialized for shallow reef habitats. Previous work has suggested that the taxa of in hospite Symbiodiniaceae in coral can be significantly influenced by the availability and diversity of free-living Symbiodiniaceae present in the surrounding environment (Cunning et al., 2015b; Quigley et al., 2017). However, there is limited information on whether depth is a significant factor in structuring communities of free-living Symbiodiniaceae or if the in hospite Symbiodiniaceae mirror free-living communities across depth. Distinct algal symbiont profiles may also be related to coral skeletal morphology and symbiont photochemistry. In the Gulf of Mexico, Symbiodiniaceae density and chlorophyll measurements changed with host morphology (Polinski and Voss, 2018; Studivan et al., 2019). Observed differences in Symbiodiniaceae communities found in Belize may also occur as a function of host morphology as in the Gulf of Mexico. We are presently unable to assess this hypothesis due to the small fragment sizes sampled (∼ 6 cm2).
While this study implemented a balanced sampling design over depth and site, sampling occurred over two excursions (Table 1). This could introduce variance over time, due to the temporal changes which may occur in the numerically dominant Symbiodiniaceae within a coral colony (Baker, 2003; Berkelmans and van Oppen, 2006; Reich et al., 2017). A total of 13 Symbiodiniaceae ITS2 type profiles were found within M. cavernosa samples taken from 10 and 16 m in March 2017. There were only three ITS2 type profiles found in 25 m samples, even though they were also sampled in March 2017 (Table 1). Additionally, the entire complement of Glover’s Reef samples was sampled during a single day (27 March 2017) and these profiles are indistinguishable from profiles at all other sites (Table 3). Montastraea cavernosa has previously demonstrated stability in its Symbiodiniaceae through temporal sampling of tagged colonies, even in a comparatively variable environment (Klepac et al., 2015). Finally, there were no observed coral bleaching events between sampling events, which is a common impetus for changing of dominant Symbiodiniaceae taxa (Berkelmans and van Oppen, 2006; Silverstein et al., 2015). Based on these combined factors, the data presented here likely represent differences driven by depth rather than any temporal co-factors.
Symbiodiniaceae ITS2 type profiles that dominated deep populations of M. cavernosa in this study were also typically abundant in shallow populations of M. cavernosa. However, a subset of Symbiodiniaceae ITS2 type profiles unique to shallow populations contributed to the significant differences in algal symbiont assemblages reported between the shallow and deep M. cavernosa populations. Physiological differences among putative Symbiodiniaceae species have been previously documented, particularly in terms of thermal tolerance (Hume et al., 2015; Díaz-Almeyda et al., 2017; Silverstein et al., 2017; Swain et al., 2017), which could potentially be a driver of observed depth stratification in Belize. Nonetheless, the ubiquity of the majority of Symbiodiniaceae ITS2 type profiles across both shallow and deep zones suggest that algal symbionts are unlikely to be driving the observed lack of gene flow between shallow and deep M. cavernosa populations on the Belize Barrier Reef (Eckert et al., 2019). Further resolution of Symbiodiniaceae taxonomy, coupled with investigations of environmental tolerances and preferred ranges (e.g., light, temperature, etc.) among individual Symbiodiniaceae species are needed to understand the consequences of various coral-algal symbioses and how these may drive observed variations in Symbiodiniaceae assemblages across depth zones in Belize and elsewhere.
Data Availability Statement
All protocols, including sample preparation and data analysis scripts are available in a GitHub repository (Eckert, 2020). Raw ITS2 amplicon sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the Project No. PRJNA579363, Accession Nos. SAMN13109002 to SAMN13109242.
Author Contributions
JV and RE designed the research. JV, RE, and MS collected the coral samples. AR and RE optimized the DNA extraction and clean up. AR, AS, and RE extracted the DNA. RE prepared the sequencing libraries, performed the data analyses, and created the figures. All authors contributed to the final edited manuscript prepared by RE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank D. Dodge, A. Shatters, and A. Alker for field assistance, Z. Foltz and S. Jones for facilitating access to Smithsonian Carrie Bow Cay Field Station and assisting with permits, and B. Hume for running SymPortal analysis remotely. Samples were sequenced at The University of Texas at Austin’s Genome and Sequencing Analysis Facility. All corals were collected under CITES permits 7123 and 000023-17 from the Belize Fisheries Department. This is contribution 2274 from Harbor Branch Oceanographic Institute and contribution 1040 from Caribbean Coral Reef Ecosystems (CCRE) Program, Smithsonian Institution.
Footnotes
Funding. Funding for this research was awarded to JV through the NOAA Office of Ocean Exploration and Research under awards NA09OAR4320073 and NA14OAR4320260 to the Cooperative Institute for Ocean Exploration, Research and Technology (CIOERT) at Florida Atlantic University’s Harbor Branch Oceanographic Institute. This research was also supported by a donation from the Banbury Fund in memory of John and Andreija Robertson and a donation from the River Branch Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00518/full#supplementary-material
References
- Anderson M. J., Walsh D. C. I. (2013). PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 83 557–574. 10.1890/12-2010.1 [DOI] [Google Scholar]
- Arif C., Daniels C., Bayer T., Banguera-Hinestroza E., Barbrook A., Howe C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23 4418–4433. 10.1111/mec.12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34 661–689. 10.1146/annurev.ecolsys.34.011802.132417 [DOI] [Google Scholar]
- Baker A. C., Starger C. J., McClanahan T. R., Glynn P. W. (2004). Corals’ adaptive response to climate change. Nature 430 741–741. 10.1038/430741a [DOI] [PubMed] [Google Scholar]
- Baumann J. H., Davies S. W., Aichelman H. E., Castillo K. D. (2018). Coral Symbiodinium community composition across the Belize Mesoamerican Barrier Reef System is influenced by host species and thermal variability. Microb. Ecol. 75 903–915. 10.1007/s00248-017-1096-6 [DOI] [PubMed] [Google Scholar]
- Baums I. B., Devlin-Durante M. K., Lajeunesse T. C. (2014). New insights into the dynamics between reef corals and their associated dinoflagellate endosymbionts from population genetic studies. Mol. Ecol. 23 4203–4215. 10.1111/mec.12788 [DOI] [PubMed] [Google Scholar]
- Berkelmans R., van Oppen M. J. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. Lond. B 273 2305–2312. 10.1098/rspb.2006.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank R. J., Trench R. K. (1985). Speciation and symbiotic dinoflagellates. Science 229 656–658. 10.1126/science.229.4714.656 [DOI] [PubMed] [Google Scholar]
- Bongaerts P., Carmichael M., Hay K. B., Tonk L., Frade P. R., Hoegh-Guldberg O. (2015a). Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species. R. Soc. Open Sci. 2:140297 10.1098/rsos.140297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts P., Frade P. R., Hay K. B., Englebert N., Latijnhouwers K. R. W., Bak R. P. M., et al. (2015b). Deep down on a Caribbean reef: lower mesophotic depths harbor a specialized coral-endosymbiont community. Sci. Rep. 5:7652 10.1038/srep07652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts P., Ridgway T., Sampayo E. M., Hoegh-Guldberg O. (2010). Assessing the ‘deep reef refugia’ hypothesis: focus on Caribbean reefs. Coral Reefs 29 309–327. 10.1007/s00338-009-0581-x [DOI] [Google Scholar]
- Bongaerts P., Riginos C., Brunner R., Englebert N., Smith S. R., Hoegh-Guldberg O. (2017). Deep reefs are not universal refuges: reseeding potential varies among coral species. Sci. Adv. 3:e1602373 10.1126/sciadv.1602373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts P., Smith T. B. (2019). “Beyond the ‘Deep Reef Refuge’ hypothesis: a conceptual framework to characterize persistence at depth,” in Mesophotic Coral Ecosystems of the World, eds Loya Y., Puglise K. A., Bridge T. C. L. (Cham: Springer; ), 881–895. 10.1007/978-3-319-92735-0_45 [DOI] [Google Scholar]
- Brazeau D. A., Lesser M. P., Slattery M. (2013). Genetic structure in the coral, Montastraea cavernosa: assessing genetic differentiation among and within mesophotic reefs. PLoS One 8:e65845 10.1371/journal.pone.0065845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. E., Le Tissier M. D. A., Bythell J. C. (1995). Mechanisms of bleaching deduced from histological studies of reef corals sampled during a natural bleaching event. Mar. Biol. 122 655–663. 10.1007/BF00350687 [DOI] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R., Gorley R. N. (2015). PRIMER v7: User Manual/Tutorial. Plymouth: PRIMER-E. [Google Scholar]
- Cooper T. F., Ulstrup K. E., Dandan S. S., Heyward A. J., Kühl M., Muirhead A., et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc. R. Soc. Lond. B 278 1840–1850. 10.1098/rspb.2010.2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A. M. S., Baker A. C. (2009). Understanding diversity in coral-algal symbiosis: a cluster-based approach to interpreting fine-scale genetic variation in the genus Symbiodinium. Coral Reefs 28 81–93. 10.1007/s00338-008-0456-6 [DOI] [Google Scholar]
- Cunning R., Gates R. D., Edmunds P. J. (2017). Using high-throughput sequencing of ITS2 to describe Symbiodinium metacommunities in St. John, US Virgin Islands. PeerJ 5:e3472 10.7717/peerj.3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunning R., Silverstein R. N., Baker A. C. (2015a). Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc. R. Soc. Lond. B 282:20141725 10.1098/rspb.2014.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunning R., Yost D. M., Guarinello M. L., Putnam H. M., Gates R. D. (2015b). Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa. PLoS One 10:e0145099 10.1371/journal.pone.0145099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De’ath G., Fabricius K. E., Sweatman H., Puotinen M. (2012). The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc. Natl. Acad. Sci. U.S.A. 109 17995–17999. 10.1073/pnas.1208909109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Almeyda E. M., Prada C., Ohdera A. H., Moran H., Civitello D. J., Iglesias-Prieto R., et al. (2017). Intraspecific and interspecific variation in thermotolerance and photoacclimation in Symbiodinium dinoflagellates. Proc. R. Soc. Lond. B 284:20171767 10.1098/rspb.2017.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner S. D. (2009). Coping with commitment: projected thermal stress on coral reefs under different future scenarios. PLoS One 4:e5712 10.1371/journal.pone.0005712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. E. (2003). Coral bleaching—how and why? Mar. Pollut. Bull. 46 385–392. 10.1016/s0025-326x(03)00037-7 [DOI] [PubMed] [Google Scholar]
- Eakin C. M., Morgan J. A., Heron S. F., Smith T. B., Liu G., Alvarez-Filip L., et al. (2010). Caribbean corals in crisis: record thermal stress, bleaching, and mortality in 2005. PLoS One 5:e13969 10.1371/journal.pone.0013969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R. J. (2020). RyanEckert/Belize_Mcav_Symbiodiniaceae_ITS2: Depth influences Symbiodiniaceae associations among Montastraea cavernosa populations on the Belize Barrier Reef (Version v1.0.1). 10.5281/zenodo.3675991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R. J., Studivan M. S., Voss J. D. (2019). Populations of the coral species Montastraea cavernosa on the Belize Barrier Reef lack vertical connectivity. Sci. Rep. 9:7200 10.1038/s41598-019-43479-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren A. M., Morrison H. G., Lescault P. J., Reveillaud J., Vineis J. H., Sogin M. L. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9 968–979. 10.1038/ismej.2014.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney J. C., Pettay D. T., Sampayo E. M., Warner M. E., Oxenford H. A., LaJeunesse T. C. (2010). The relative significance of host–habitat, depth, and geography on the ecology, endemism, and speciation of coral endosymbionts in the genus Symbiodinium. Microb. Ecol. 60 250–263. 10.1007/s00248-010-9681-y [DOI] [PubMed] [Google Scholar]
- Gardner T. A., Côté I. M., Gill J. A., Grant A., Watkinson A. R. (2003). Long-term region-wide declines in Caribbean corals. Science 301 958–960. 10.1126/science.1086050 [DOI] [PubMed] [Google Scholar]
- Glynn P. W. (1996). Coral reef bleaching: facts, hypotheses and implications. Glob. Chang. Biol. 2 495–509. 10.1111/j.1365-2486.1996.tb00063.x [DOI] [Google Scholar]
- Glynn P. W., D’Croz L. (1990). Experimental evidence for high temperature stress as the cause of El Niño-coincident coral mortality. Coral Reefs 8 181–191. 10.1007/BF00265009 [DOI] [Google Scholar]
- Goreau T. (1964). Mass expulsion of zooxanthellae from Jamaican reef communities after Hurricane Flora. Science 145 383–386. 10.1126/science.145.3630.383 [DOI] [PubMed] [Google Scholar]
- Hatcher B. G. (1988). Coral reef primary productivity: a beggar’s banquet. Trends Ecol. Evol. 3 106–111. 10.1016/0169-5347(88)90117-6 [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2008). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. 10.1126/science.1152509 [DOI] [PubMed] [Google Scholar]
- Hughes T. P. (1994). Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265 1547–1551. 10.1126/science.265.5178.1547 [DOI] [PubMed] [Google Scholar]
- Hughes T. P., Kerry J. T., Álvarez-Noriega M., Álvarez-Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543 373–377. 10.1038/nature21707 [DOI] [PubMed] [Google Scholar]
- Hume B. C. C., D’Angelo C., Smith E. G., Stevens J. R., Burt J., Wiedenmann J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci. Rep. 5:8562 10.1038/srep08562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume B. C. C., Smith E. G., Ziegler M., Warrington H. J. M., Burt J. A., LaJeunesse T. C., et al. (2019). SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19 1063–1080. 10.1111/1755-0998.13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James N. P., Ginsburg R. N. (1979). The Seaward Margin of Belize Barrier and Atoll Reefs. Oxford: Blackwell Scientific. [Google Scholar]
- Jokiel P. L., Coles S. L. (1977). Effects of temperature on the mortality and growth of Hawaiian reef corals. Mar. Biol. 43 201–208. 10.1007/BF00402312 [DOI] [Google Scholar]
- Kahng S. E., Garcia-Sais J. R., Spalding H. L., Brokovich E., Wagner D., Weil E., et al. (2010). Community ecology of mesophotic coral reef ecosystems. Coral Reefs 29 255–275. 10.1007/s00338-010-0593-6 [DOI] [Google Scholar]
- Kenkel C. D., Bay L. K. (2018). Exploring mechanisms that affect coral cooperation: symbiont transmission mode, cell density and community composition. PeerJ 6:e6047 10.7717/peerj.6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel C. D., Goodbody-Gringley G., Caillaud D., Davies S. W., Bartels E., Matz M. V. (2013). Evidence for a host role in thermotolerance divergence between populations of the mustard hill coral (Porites astreoides) from different reef environments. Mol. Ecol. 22 4335–4348. 10.1111/mec.12391 [DOI] [PubMed] [Google Scholar]
- Klepac C., Beal J., Kenkel C., Sproles A., Polinski J., Williams M., et al. (2015). Seasonal stability of coral-Symbiodinium associations italic in the subtropical coral habitat of St. Lucie Reef, Florida. Mar. Ecol. Prog. Ser. 532 137–151. 10.3354/meps11369 [DOI] [Google Scholar]
- LaJeunesse T. C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a “species” level marker. J. Phycol. 37 866–880. 10.1046/j.1529-8817.2001.01031.x [DOI] [Google Scholar]
- LaJeunesse T. C. (2005). “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol. Biol. Evol. 22 570–581. 10.1093/molbev/msi042 [DOI] [PubMed] [Google Scholar]
- LaJeunesse T. C., Parkinson J. E., Gabrielson P. W., Jeong H. J., Reimer J. D., Voolstra C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol 28 2570–2580. 10.1016/j.cub.2018.07.008 2570–2580.e6. [DOI] [PubMed] [Google Scholar]
- LaJeunesse T. C., Parkinson J. E., Reimer J. D. (2012). A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J. Phycol. 48 1380–1391. 10.1111/j.1529-8817.2012.01217.x [DOI] [PubMed] [Google Scholar]
- LaJeunesse T. C., Pettay D. T., Sampayo E. M., Phongsuwan N., Brown B., Obura D. O., et al. (2010). Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37 785–800. 10.1111/j.1365-2699.2010.02273.x [DOI] [Google Scholar]
- LaJeunesse T. C., Thornhill D. J. (2011). Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS One 6:e29013 10.1371/journal.pone.0029013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichter J. J., Helmuth B., Fischer A. M. (2006). Variation beneath the surface: quantifying complex thermal environments on coral reefs in the Caribbean. Bahamas and Florida. J. Mar. Res. 64 563–588. 10.1357/002224006778715711 [DOI] [Google Scholar]
- Lesser M. P., Mazel C., Phinney D., Yentsch C. S. (2000). Light absorption and utilization by colonies of the congeneric hermatypic corals Montastraea faveolata and Montastraea cavernosa. Limnol. Oceanogr. 45 76–86. 10.4319/lo.2000.45.1.0076 [DOI] [Google Scholar]
- Lesser M. P., Slattery M., Leichter J. J. (2009). Ecology of mesophotic coral reefs. J. Exp. Mar. Biol. Ecol. 375 1–8. 10.1016/j.jembe.2009.05.009 [DOI] [Google Scholar]
- Lesser M. P., Slattery M., Mobley C. D. (2018). Biodiversity and functional ecology of mesophotic coral reefs. Annu. Rev. Ecol. Evol. Syst. 49 49–71. 10.1146/annurev-ecolsys-110617-062423 [DOI] [Google Scholar]
- Lesser M. P., Slattery M., Stat M., Ojimi M., Gates R. D., Grottoli A. (2010). Photoacclimatization by the coral Montastraea cavernosa in the mesophotic zone: light, food, and genetics. Ecology 91 990–1003. 10.1890/09-0313.1 [DOI] [PubMed] [Google Scholar]
- Lucas M. Q., Stat M., Smith M. C., Weil E., Schizas N. V. (2016). Symbiodinium (internal transcribed spacer 2) diversity in the coral host Agaricia lamarcki (Cnidaria: Scleractinia) between shallow and mesophotic reefs in the Northern Caribbean (20–70 m). Mar. Ecol. 37 1079–1087. 10.1111/maec.12367 [DOI] [Google Scholar]
- Martinez Arbizu P. (2017). pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. Available online at: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed February 10, 2019). [Google Scholar]
- Meerman J., Clabaugh J. (2017). Biodiversity and Environmental Resource Data System of Belize. Available online at: http://www.biodiversity.bz (accessed November 17, 2017). [Google Scholar]
- Mieog J. C., van Oppen M. J. H., Berkelmans R., Stam W. T., Olsen J. L. (2009). Quantification of algal endosymbionts (Symbiodinium) in coral tissue using real-time PCR. Mol. Ecol. Resour. 9 74–82. 10.1111/j.1755-0998.2008.02222.x [DOI] [PubMed] [Google Scholar]
- Mumby P. J., Dahlgren C. P., Harborne A. R., Kappel C. V., Micheli F., Brumbaugh D. R., et al. (2006). Fishing, trophic cascades, and the process of grazing on coral reefs. Science 311 98–101. 10.1126/science.1121129 [DOI] [PubMed] [Google Scholar]
- Muscatine L., Cernichiari E. (1969). Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol. Bull. 137 506–523. 10.2307/1540172 [DOI] [PubMed] [Google Scholar]
- Muscatine L., Falkowski P. G., Porter J. W., Dubinsky Z. (1984). Fate of photosynthetic fixed carbon in light- and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc. R. Soc. Lond. Ser. B Biol. Sci. 222 181–202. 10.1098/rspb.1984.0058 [DOI] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). vegan: Community Ecology Package. Available online at: https://cran.r-project.org/package=vegan (accessed February 10, 2019). [Google Scholar]
- Pereira M. B., Wallroth M., Jonsson V., Kristiansson E. (2018). Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genomics 19:274 10.1186/s12864-018-4637-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochon X., Pawlowski J., Zaninetti L., Rowan R. (2001). High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar. Biol. 139 1069–1078. 10.1007/s002270100674 [DOI] [Google Scholar]
- Polinski J. M., Voss J. D. (2018). Evidence of photoacclimatization at mesophotic depths in the coral-Symbiodinium symbiosis at Flower Garden Banks National Marine Sanctuary and McGrail Bank. Coral Reefs 37 779–789. 10.1007/s00338-018-1701-2 [DOI] [Google Scholar]
- Quigley K. M., Bay L. K., Willis B. L. (2017). Temperature and water quality-related patterns in sediment-associated Symbiodinium communities impact symbiont uptake and fitness of juveniles in the genus Acropora. Front. Mar. Sci. 4:401 10.3389/fmars.2017.00401 [DOI] [Google Scholar]
- Quigley K. M., Davies S. W., Kenkel C. D., Willis B. L., Matz M. V., Bay L. K. (2014). Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS One 9:e94297 10.1371/journal.pone.0094297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Available online at: https://www.r-project.org/ (accessed January 01, 2019). [Google Scholar]
- Rahav O., Dubinsky Z., Achituv Y., Falkowski P. G. (1989). Ammonium metabolism in the zooxanthellate coral, Stylophora pistillata. Proc. R. Soc. Lond. B 236 325–337. 10.1098/rspb.1989.0026 [DOI] [Google Scholar]
- Reich H. G., Robertson D. L., Goodbody-Gringley G. (2017). Do the shuffle: changes in Symbiodinium consortia throughout juvenile coral development. PLoS One 12:e0171768 10.1371/journal.pone.0171768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampayo E. M., Dove S., Lajeunesse T. C. (2009). Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus Symbiodinium. Mol. Ecol. 18 500–519. 10.1111/j.1365-294X.2008.04037.x [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541. 10.1128/aem.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano X., Baums I. B., O’Reilly K., Smith T. B., Jones R. J., Shearer T. L., et al. (2014). Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol. Ecol. 23 4226–4240. 10.1111/mec.12861 [DOI] [PubMed] [Google Scholar]
- Silverstein R. N., Correa A. M. S., Baker A. C. (2012). Specificity is rarely absolute in coral-algal symbiosis: implications for coral response to climate change. Proc. R. Soc. Lond. B 279 2609–2618. 10.1098/rspb.2012.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein R. N., Cunning R., Baker A. C. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob. Chang. Biol. 21 236–249. 10.1111/gcb.12706 [DOI] [PubMed] [Google Scholar]
- Silverstein R. N., Cunning R., Baker A. C. (2017). Tenacious D: Symbiodinium in clade D remain in reef corals at both high and low temperature extremes despite impairment. J. Exp. Biol. 220 1192–1196. 10.1242/jeb.148239 [DOI] [PubMed] [Google Scholar]
- Skirving W. J., Heron S. F., Marsh B. L., Liu G., De La Cour J. L., Geiger E. F., et al. (2019). The relentless march of mass coral bleaching: a global perspective of changing heat stress. Coral Reefs 38 547–557. 10.1007/s00338-019-01799-4 [DOI] [Google Scholar]
- Smith E. G., Ketchum R. N., Burt J. A. (2017). Host specificity of Symbiodinium variants revealed by an ITS2 metahaplotype approach. ISME J. 11 1500–1503. 10.1038/ismej.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. B., Gyory J., Brandt M. E., Miller W. J., Jossart J., Nemeth R. S. (2016). Caribbean mesophotic coral ecosystems are unlikely climate change refugia. Glob. Chang. Biol. 22 2756–2765. 10.1111/gcb.13175 [DOI] [PubMed] [Google Scholar]
- Stat M., Gates R. D. (2011). Clade D Symbiodinium in scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011:730715 10.1155/2011/730715 [DOI] [Google Scholar]
- Stat M., Pochon X., Cowie R., Gates R. (2009). Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar. Ecol. Prog. Ser. 386 83–96. 10.3354/meps08080 [DOI] [Google Scholar]
- Studivan M. S., Milstein G., Voss J. D. (2019). Montastraea cavernosa corallite structure demonstrates distinct morphotypes across shallow and mesophotic depth zones in the Gulf of Mexico. PLoS One 14:e0203732 10.1371/journal.pone.0203732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studivan M. S., Voss J. D. (2018). Population connectivity among shallow and mesophotic Montastraea cavernosa corals in the Gulf of Mexico identifies potential for refugia. Coral Reefs 37 1183–1196. 10.1007/s00338-018-1733-7 [DOI] [Google Scholar]
- Swain T. D., Chandler J., Backman V., Marcelino L. (2017). Consensus thermotolerance ranking for 110 Symbiodinium phylotypes: an exemplar utilization of a novel iterative partial-rank aggregation tool with broad application potential. Funct. Ecol. 31 172–183. 10.1111/1365-2435.12694 [DOI] [Google Scholar]
- Thornhill D. J., Lajeunesse T. C., Santos S. R. (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 16 5326–5340. 10.1111/j.1365-294X.2007.03576.x [DOI] [PubMed] [Google Scholar]
- Thornhill D. J., Xiang Y., Fitt W. K., Santos S. R. (2009). Reef endemism, host specificity and temporal stability in populations of symbiotic dinoflagellates from two ecologically dominant Caribbean corals. PLoS One 4:e6262 10.1371/journal.pone.0006262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooidonk R., Maynard J., Tamelander J., Gove J., Ahmadia G., Raymundo L., et al. (2016). Local-scale projections of coral reef futures and implications of the Paris Agreement. Sci. Rep. 6:39666 10.1038/srep39666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen M. J. H., Bongaerts P., Underwood J. N., Peplow L. M., Cooper T. F. (2011). The role of deep reefs in shallow reef recovery: an assessment of vertical connectivity in a brooding coral from west and east Australia. Mol. Ecol. 20 1647–1660. 10.1111/j.1365-294X.2011.05050.x [DOI] [PubMed] [Google Scholar]
- Venn A. A., Loram J. E., Douglas A. E. (2008). Photosynthetic symbioses in animals. J. Exp. Bot. 59 1069–1080. 10.1093/jxb/erm328 [DOI] [PubMed] [Google Scholar]
- Warner M. E., LaJeunesse T. C., Robison J. D., Thur R. M. (2006). The ecological distribution and comparative photobiology of symbiotic dinoflagellates from reef corals in Belize: potential implications for coral bleaching. Limnol. Oceanogr. 51 1887–1897. 10.4319/lo.2006.51.4.1887 [DOI] [Google Scholar]
- Weis V. M. (2008). Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211 3059–3066. 10.1242/jeb.009597 [DOI] [PubMed] [Google Scholar]
- Wham D. C., Ning G., LaJeunesse T. C. (2017). Symbiodinium glynnii sp. nov., a species of stress-tolerant symbiotic dinoflagellates from pocilloporid and montiporid corals in the Pacific Ocean. Phycologia 56 396–409. 10.2216/16-86.1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protocols, including sample preparation and data analysis scripts are available in a GitHub repository (Eckert, 2020). Raw ITS2 amplicon sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the Project No. PRJNA579363, Accession Nos. SAMN13109002 to SAMN13109242.