Summary
Precise measurement of action potentials (APs) is needed to observe electrical activity and cellular communication within cardiac tissue. Voltage-sensitive dyes (VSDs) are traditionally used to measure cardiac APs; however, they require acute chemical addition that prevents chronic imaging. Genetically encoded voltage indicators (GEVIs) enable long-term studies of APs without the need of chemical additions, but current GEVIs used in cardiac tissue exhibit poor kinetics and/or low signal to noise (SNR). Here, we demonstrate the use of Archon1, a recently developed GEVI, in hiPSC-derived cardiomyocytes (CMs). When expressed in CMs, Archon1 demonstrated fast kinetics comparable with patch-clamp electrophysiology and high SNR significantly greater than the VSD Di-8-ANEPPS. Additionally, Archon1 enabled monitoring of APs across multiple cells simultaneously in 3D cardiac tissues. These results highlight Archon1's capability to investigate the electrical activity of CMs in a variety of applications and its potential to probe functionally complex in vitro models, as well as in vivo systems.
Subject Areas: Biotechnology, Bioelectronics, Technical Aspects of Cell Biology, Electronic Materials
Graphical Abstract
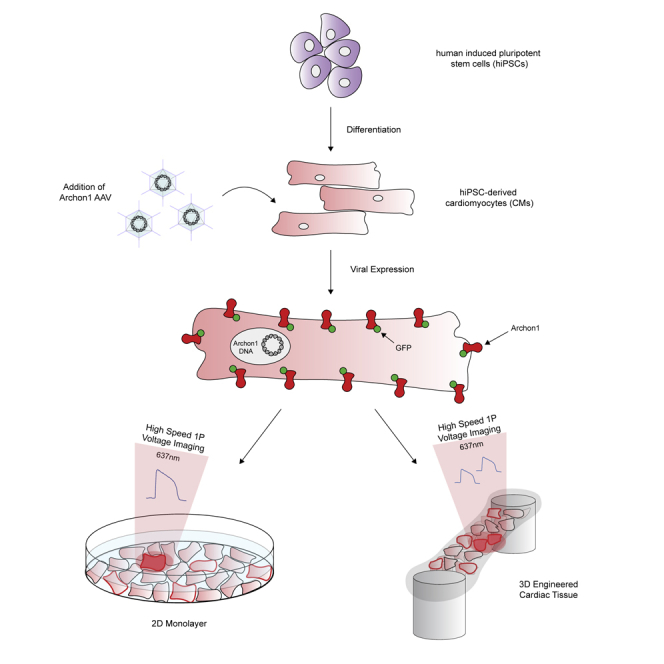
Highlights
-
•
Genetic sensor Archon1 reports membrane voltage in hiPSC-derived cardiomyocytes
-
•
Archon1 monitors action potentials in 2D and 3D cardiac tissue with high sensitivity
-
•
Archon1 repeatedly monitored voltage in the same cells and over extended time periods
-
•
Voltage dynamics of multiple cells were recorded simultaneously with Archon1
Biotechnology; Bioelectronics; Technical Aspects of Cell Biology; Electronic Materials
Introduction
The measurement of electrical potential and conduction in the heart is important for understanding how electrical signals propagate through cardiac tissue in both healthy and diseased conditions. Although numerous methods exist for monitoring whole-heart voltage (EKG, EPS, etc.), few techniques examine the properties at the individual cell level within cardiac tissue. Intracellular calcium release, triggered by action potentials (APs), has long been used to study mechanisms such as excitation-contraction coupling and as a proximal measurement for signal propagation (Bers, 2002, Bers, 2008, Eisner et al., 2017, Hobai and O'Rourke, 2001). However, calcium does not provide insight into membrane voltage dynamics and individual AP waveforms, which may be altered in biological, pharmacological, or pathological states (Leyton-Mange and Milan, 2014, Leyton-Mange et al., 2014, Olivari et al., 1979, Peng et al., 2010, Sanguinetti and Jurkiewicz, 1991, Sanguinetti et al., 1991, Shaheen et al., 2018, Shinnawi et al., 2015, Song et al., 2015). Recent attempts to study cardiac membrane potential have made use of voltage-sensitive dyes (VSDs) such as the ANEPPS series or the PGH series (Entcheva and Bien, 2006, Herron et al., 2012, Hortigon-Vinagre et al., 2016, Panakova et al., 2010, Salama et al., 2005, Salama and Morad, 1976). Although many of these VSDs have the time resolution necessary to capture AP dynamics of cardiac cells, they often suffer from poor signal-to-noise ratio (SNR) and membrane localization and are often cytotoxic (Herron et al., 2012, Hou et al., 2014). In addition, by nature, VSDs are transient and must be reapplied to samples, which hinders long-term studies; they also lack the ability to label specific subpopulations.
Genetically encoded voltage indicators (GEVIs), which have been successfully utilized in a similarly electrically excitable cell population, neurons (Abdelfattah et al., 2019, Adam et al., 2019, Chamberland et al., 2017, Flytzanis et al., 2014, Gong et al., 2014, Gong et al., 2015, Hochbaum et al., 2014, Jin et al., 2012, Lou et al., 2016, Piatkevich et al., 2018, St-Pierre et al., 2014, Villette et al., 2019, Zou et al., 2014), have recently been deployed in cardiac tissue to overcome the shortcomings of VSDs (Hou et al., 2014, Johnston et al., 2018, Leyton-Mange et al., 2014). Although GEVIs such as ArcLight, Mermaid, or the VSFP series have been successfully applied to study arrhythmias, as well as aid in cellular phenotyping, these GEVIs lack the time resolution, sensitivity, and SNR to capture voltage dynamics under various conditions (Chang Liao et al., 2015, Leyton-Mange et al., 2014, Mutoh et al., 2009, Tsutsui et al., 2010, van Opbergen et al., 2018). For example, ArcLight, a commonly used GEVI, has slow kinetics, which hinders its ability to capture the characteristic upstroke morphology of cardiac APs and any perturbations to this upstroke such as those caused by diseases or drugs (Lee et al., 2017, Leyton-Mange et al., 2014, Nakajima et al., 2016, Storace et al., 2016). In contrast, the GEVI VSFP3.1 has relatively fast kinetics (activation time ∼1.3 ms) but displays fluorescence changes of only 0.5% per 100 mV, which impedes monitoring of subthreshold fluctuations and contributes to a very low SNR (Kaestner et al., 2015, Lundby et al., 2008). Many of these GEVIs thus suffer from this dichotomy between superior kinetics and sensitivity.
Recently, Piatkevich et al. developed a high-performance GEVI called Archon1, a far-red fluorescent plasma transmembrane protein evolved from the Arch3 opsin, with fluorescence emission linearly dependent on membrane voltage (Piatkevich et al., 2018). Archon1 exhibits high temporal resolution, sensitivity, and brightness under simple one-photon widefield microscopy, which enabled imaging of neural activity in mouse cortical brain slices, whole zebrafish, and C. elegans (Piatkevich et al., 2018). Archon1's fast kinetics and linear fluorescence-voltage relationship resulted in fluorescence traces that closely matched simultaneously recorded electrophysiology. Since Archon1 does not need chemical cofactors, the sensor showed no phototoxicity and retained photostability for up to 800 s in cultured mouse neurons (Piatkevich et al., 2018). In addition, Archon1 demonstrated excellent membrane localization and did not alter membrane resistance, capacitance, or resting potential as compared with non-expressing cells. Its far-red excitation is also compatible with blue-light-driven optogenetics enabling all-optical electrophysiology (Piatkevich et al., 2018, Piatkevich et al., 2019). These features of Archon1, along with its easily adaptable one-photon widefield imaging setup, make it an ideal GEVI candidate to deploy in other electrically dependent systems. Here, we demonstrate Archon1's utility for monitoring cardiac APs in both 2D cell culture and 3D tissue environments under a variety of biologically relevant conditions.
Results
Characterization of Archon1 in iPSC-Derived Cardiomyocytes
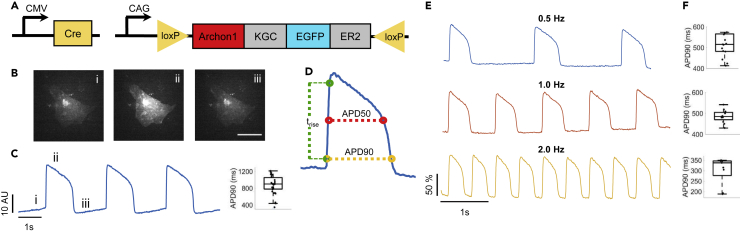
Archon1 was expressed in 2D monolayers of human induced pluripotent stem cell (iPSC)-derived cardiomyocytes (CMs) using both Cre-dependent CAG-Archon1-KGC-EGFP-ER2 (Archon1) and CMV-Cre AAV (Figure 1A) 5–7 days after spontaneous beating was observed. CM membrane potential was recorded optically at 100 Hz using a scientific cMOS camera (lex = 637 nm). Archon1 fluorescence changes exhibited typical cardiac AP waveforms with clearly identifiable phases (Figures 1B and 1C; i: resting potential, ii: peak depolarization, iii: end of repolarization; Video S1). Changes in intensity were not due to motion from cellular contraction as indicated by lack of fluorescence changes in the GFP channel (Video S2), as compared with the Archon1 channel (Video S1). In order to quantify AP duration, we calculated rise time (trise; defined by 10%–90% of the valley-to-peak duration), APD50 (AP duration at 50% repolarization), and APD90 (AP duration at 90% repolarization) (Figure 1D, see Transparent Methods). Without electrical stimulation, APD90 values ranged from 345 to 1,210 ms (n = 31 cells, 880 ± 218 ms, mean ± standard deviation, Figure 1C right). We then evoked APs by electrically pacing CMs at 0.5, 1, and 2 Hz and found that Archon1 fluorescence faithfully followed electrical stimulation (Figure 1E; respective APD90 distributions, Figure 1F). Together, these results demonstrate that Archon1 can robustly capture AP dynamics and waveform morphology, and this capability is maintained at higher frequencies.
Figure 1.
Demonstration of Archon1 for Monitoring Action Potentials of iPSC-Derived Cardiomyocytes
(A) Diagrams of Cre-recombinase AAV system used to infect CMs with Archon1. KGC, linker; ER2, ER export trafficking sequence.
(B) Single-frame images of CM expressing Archon1 at different phases of an AP as shown in (C). Scale bar, 50 μm. See Video S1.
(C) Left, optically recorded raw voltage trace for cell shown in (B). Right, quantification of APD90, as defined in (D), per voltage trace for unpaced CMs (n = 31 cells).
(D) An example AP waveform with APD90, APD50, and rise time (trise) defined.
(E) Representative normalized fluorescence voltage traces from CM electrically paced at 0.5 Hz (blue), 1 Hz (orange), and 2 Hz (yellow).
(F) Corresponding APD90 box plot for each pacing shown in E (from n = 15, 12, 9, cells, respectively).
Box plots represent 25th to 75th percentiles, with whiskers extending 1.5x the interquartile range; horizontal line represents median; each dot represents data from a single cell. All optical traces obtained at image acquisition rate of 100 Hz.
Scale bar, 25 μm
Scale bar, 25 μm.
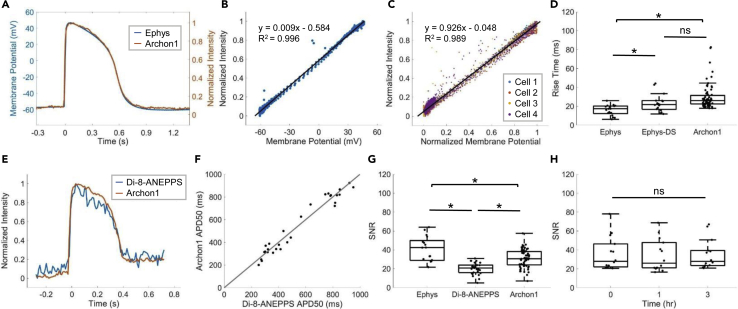
To quantify Archon1 performance, we conducted patch-clamp electrophysiology while simultaneously optically recording Archon1 fluorescence of CMs. We found that membrane voltage recorded via Archon1 fluorescence matched well with that recorded via electrophysiology (representative overlay in Figure 2A). Archon1 fluorescence was linearly correlated with electrically recorded membrane potential (Figure 2B; example corresponding to Figure 2A; least-squares regression line: y = 0.009 x – 0.584; R2 = 0.996) for all simultaneously recorded cells (n = 4 cells, Figure 2C; least-squares regression line: y = 0.926 x – 0.048; R2 = 0.989). In addition, Archon1 expression did not alter CM properties such as resting membrane potential, beat amplitude, membrane resistance, rise time, APD90, and beat period (Figure S1, see statistics in Table S1).
Figure 2.
Characterization of Archon1 in iPSC-derived Cardiomyocyte Monolayers
(A) Representative overlay of single AP waveform simultaneously measured using patch-clamp electrophysiology (blue) and Archon1 fluorescence (orange).
(B) Normalized-intensity Archon1 fluorescence trace versus membrane potential recorded via electrophysiology for cell simultaneously recorded in (A). Trendline equation and R2 value shown on chart.
(C) Normalized intensity Archon1 fluorescence traces versus normalized membrane potential from all simultaneously recorded cells (n = 4). Trendline equation and R2 value shown on chart.
(D) Quantification of rise time, as defined in Figure 1D, per cell for patch-clamp electrophysiology (Ephys; acquired at 10–20kHz), down-sampled electrophysiology (Ephys-DS; same electrophysiology data down-sampled to 100 Hz to match the acquisition rate of Archon1), and Archon1 (Archon1; acquired at 100 Hz) from n = 17, 17, and 67, cells, respectively. ∗: p < 0.05 in two-tailed Student’s t test, ns: p > 0.05 in two-tailed Student’s t test.
(E) Representative overlay of single AP waveform measured with Di-8-ANEPPS (blue) versus Archon1 (orange).
(F) APD50 values per cell for Archon1 versus Di-8-ANEPPS. Trendline equation and R2 value shown on chart.
(G) Quantification of SNR (defined in Transparent Methods) per cell for patch-clamp electrophysiology, Di-8-ANEPPS, and Archon1 (from n = 17, 29, 67, cells, respectively). ∗, p < 0.05 in two-tailed Student’s t test.
(H) Quantification of SNR per cell recorded repeatedly with Archon1 at t = 0, 1, and 3 h (n = 14 cells). ns: p > 0.05 in two-tailed Student's t test. See Figures S2A–S2C for optical traces of representative cell.
Figure S2 Box plots displayed as described in Figure 1. For full statistics see Table S1. All optical traces obtained at image acquisition rate of 100 Hz; patch clamp electrophysiology acquisition rate, 10–20 kHz.
To further characterize the performance of Archon1, rise times were calculated across all CMs recorded with electrophysiology and Archon1 (n = 17 cells for electrophysiology, 67 cells for Archon1; Figure 2D). The rise time of Archon1 (29.8 ± 13.2 ms, mean ± standard deviation) was slightly longer than that of electrophysiology (15.9 ± 5.6 ms, mean ± standard deviation; Ephys ↔ Archon1: p = 5.8 × 10−5; two-tailed Student’s t test; Table S1). To examine how these kinetic measures are influenced by the difference in data acquisition rates, 10–20 kHz for electrophysiology versus 100 Hz for Archon1, we down-sampled the same electrophysiology data to 100 Hz to match the optical imaging frame rate. We obtained a rise time of 23.3 ± 9.4 ms (mean ± standard deviation) for down-sampled electrophysiology traces, which is no longer significantly different from Archon1 (p = 0.06, two-tailed Student’s t test; Figure 2D).
We also compared Archon1 with a widely used red membrane-targeted voltage dye, Di-8-ANEPPS (Iex = 470nm; Figure 2E and Video S3). APD90 values were compared per cell between Di-8-ANEPPS and Archon1 and showed a nearly 1:1 correlation (least-squares regression line: y = 1.001x – 0.003, R2 = 0.954; Figure 2F; n = 29 cells). To compare performances across electrophysiology, Di-8-ANEPPS, and Archon1, we calculated the corresponding SNR (defined as the valley-to-peak amplitude divided by the standard deviation of the noise) for each method (n = 17, 29, 67 cells, respectively; Figure 2G). SNR was significantly different between all groups, with Archon1's SNR (Archon1: 30.7 ± 9.3, mean ± standard deviation) significantly higher than that of Di-8-ANEPPS (Di-8-ANEPPS: 20.2 ± 6.1, mean ± standard deviation; Archon1 ↔ Di-8-ANEPPS: p = 4.87 × 10−8; two-tailed Student’s t test) but significantly lower than that of electrophysiology (Ephys: 40.4 ± 13.5, mean ± standard deviation; Ephys ↔ Archon1: p = 8.3 × 10−4, two-tailed Student’s t test; Table S1). Together, these results demonstrate that Archon1 voltage imaging exhibited comparable kinetics to that achieved via patch-clamp electrophysiology and SNR significantly better than the VSD Di-8-ANEPPS.
Scale bar, 25 μm.
Because Archon1 is genetically encoded and optical voltage imaging does not incur mechanical damage to the cell membrane, as with electrophysiology, Archon1 should allow repeated imaging of the same cells. To demonstrate Archon1's robustness over time, CMs expressing Archon1 were imaged repeatedly at three time points: (t = 0, 1, and 3 h) (n = 14 cells, Figure 2H, example in Figures S2A–S2C). Over repeated recordings, SNR remained constant (36.0 ± 17.7, 34.1 ± 17.1, 35.1 ± 15.6 mean ± standard deviation for t = 0,1,3 h respectively; p = 0.96, repeated measures one-way ANOVA). We also found that Archon1 was highly photostable over 5 min of continuous imaging with SNR remaining stable across the recording (Figures S2D–S2F; SNR: 24.67 ± 0.659, 24.72 ± 0.475, 24.56 ± 0.816, mean ± standard deviation, for t = 0–20 s, 100–120 s, 280–300 s, respectively; p = 0.82, repeated measures one-way ANOVA; full statistics in Table S1). Furthermore, CMs expressing Archon1 retained expression months post-infection (see Video S4 for video of Archon1-expressing CM at day 62 after infection). Archon1 is thus capable of monitoring CM APs over extended periods of time, with high fidelity as compared with currently available methods, in cultured 2D monolayers.
Scale bar, 25 μm.
Archon1 Can Reliably Detect Pharmacologically Induced Changes in AP Waveforms
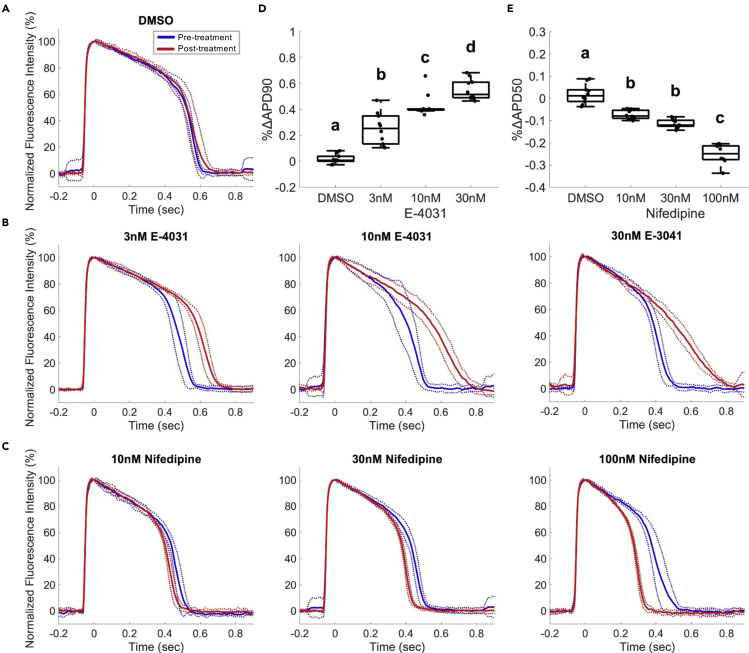
To examine the sensitivity of Archon1 for detecting changes in AP waveforms, we recorded Archon1-expressing CMs upon the application of two well-characterized ion channel inhibitors, E-4031 and Nifedipine. E-4031 is a hERG K+ ion channel inhibitor that prolongs the refractory period of the cardiac AP (Peng et al., 2010, Sanguinetti et al., 1991). Nifedipine, on the other hand, is a Ca2+ ion channel inhibitor that shortens the AP plateau and overall AP duration and is commonly used to treat hypertension and angina (Olivari et al., 1979). Archon1 fluorescence was imaged from the same cells before and after inhibitor treatment. Treated groups were compared with a DMSO control group (Figure 3A). E-4031 treatment of 2D monolayers at increasing concentrations (3, 10, 30 nM) showed concentration-dependent prolongation of APs (Figure 3B). We compared the mean APD90 before and after E-4031 treatment for all concentrations and observed increasing %ΔAPD90 with increasing E-4031 concentration (mean ± standard deviation, n = 10; 3 nM: 25.2 ± 12.2%, 10 nM: 43.2 ± 8.9%, 30 nM: 55.2 ± 8.1%; full statistics in Table S2) in comparison with the %ΔAPD90 of the DMSO control (n = 10, 1.7% ± 3.6%), consistent with previous studies (Faivre et al., 1999, Hortigon-Vinagre et al., 2016, Ma et al., 2011, Maddah et al., 2015, Peng et al., 2010) (Figure 3D). Nifedipine treatment of 2D monolayers at increasing concentrations (10, 30, 100 nM) also showed the expected concentration-dependent shortening of AP length (Figure 3C). Comparison of the mean APD50 before and after treatments for all three concentrations showed an inverse relationship between %ΔAPD50 and Nifedipine concentration (n = 10; 10 nM: −7.3 ± 2.0%, 30 nM: −11.4 ± 1.9%, 100 nM: −25.0 ± 4.4%, n = 10), compared with the DMSO control (n = 10, 1.9 ± 4.3%) (Figure 3E; Table S2), consistent with previous studies that have performed similar measurements (Hortigon-Vinagre et al., 2016, Ma et al., 2011, Maddah et al., 2015, Scheel et al., 2014). Thus, Archon1 is capable of capturing changes in AP waveform morphology in 2D monolayers.
Figure 3.
Archon1 Can Reliably Detect Changes in AP Waveforms Induced by K+ and Ca2+ Ion Channel Inhibitors
E-4031 (K+ Channel Blocker) and Nifedipine (Nif; Ca2+ Channel Blocker) compared with DMSO control; electrically paced at 1 Hz.
(A–C) (A) Average normalized Archon1 fluorescence intensity traces for DMSO control (n = 10 cells), (B) E-4031 (3, 10, 30 nM; n = 10 cells), and (C) Nifedipine (10, 30, 100 nM; n = 10 cells) before (blue) and after (red) drugs, SD (dotted lines).
(D and E) (D) %ΔAPD90 for E-4031 APs from pre-drug condition. (E) %ΔAPD50 for Nifedipine APs from pre-drug condition where a, b, c, and d represent different statistical groups (all pairs Tukey HSD test). For full statistics see Table S2.
Box plots displayed as described in Figure 1. All optical traces obtained at image acquisition rate of 100 Hz.
Archon1 Enables Monitoring of Action Potentials within Cardiac Tissues
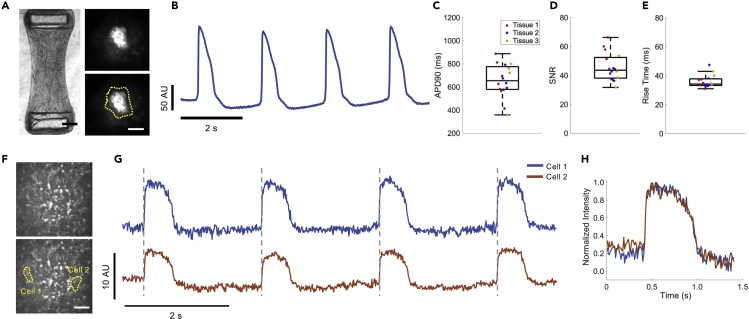
Engineered cardiac tissues can provide an optical, structural, and functional context that is more similar to in vivo tissue. The use of engineered cardiac tissues, particularly with iPSC-derived CMs, has been shown to increase both the structural and functional maturity of CMs, providing better models compared with more simple 2D monolayer systems (Nunes et al., 2013, Ronaldson-Bouchard et al., 2018, Tu et al., 2018, Zimmermann et al., 2002). To demonstrate Archon1's ability to measure the APs of single cells embedded within a tissue, we constructed engineered cardiac tissues containing Archon1-expressing CMs alongside a stromal cell population (Figure 4A left, Video S5; see Transparent Methods). The same Cre-recombinase system (Figure 1A) was used for controlled stochastic labeling of CMs to enable optical isolation of individual cells within the engineered tissues. Single CMs expressing Archon1 within the tissues were optically identified via EGFP fluorescence and their APs recorded in the far-red Archon1 fluorescence channel (Figure 4A right, 4B).
Figure 4.
Archon1 Enables Monitoring of APs in μTUG hMSC-Cardiomyocyte Tissues
(A) Brightfield image of μTUG fibroblast-CM tissues (left). Scale bar, 250 μm. See Video S5. Maximum-minus-minimum projection image of CM within μTUG tissue expressing Archon1 (top right), with corresponding ROI overlaid (bottom right). Scale bar, 20 μm.
(B) Optically recorded voltage trace for the cell shown in (A). Image acquisition rate, 50 Hz.
(C) Quantification of APD90 for CMs within tissues (n = 5, 7, 4, cells for Tissue 1 (red), 2 (blue), 3 (yellow), respectively).
(D) Quantification of SNR for CMs within tissues (n = 5, 7, 4, cells for Tissue 1 (red), 2 (blue), 3 (yellow), respectively).
(E) Quantification of rise time for CMs within tissues (n = 5, 7, 4, cells for Tissue 1 (red), 2 (blue), 3 (yellow), respectively). Box plots displayed as described in Figure 1; image acquisition rate, 50 Hz.
(F) Max projection image of μTUG tissue (top), with cells expressing Archon1 identified in yellow (bottom). Scale bar, 50 μm.
(G) Corresponding optically recorded voltage traces for cells identified in (F), with gray dashed vertical lines demonstrating AP synchrony. Image acquisition rate, 100 Hz.
(H) Overlay of representative normalized AP from two traces shown in (G) highlighting waveform consistency.
Scale bar, 250 μm.
Across three unpaced tissues, APD90 ranged from 360 to 890 ms (mean ± standard deviation: 655 ± 143 ms; range: 530 ms), with cells from individual tissues more closely clustered (range: 321, 379, 442 ms and n = 5, 7, 4 cells for Tissue 1, 2, 3, respectively; Figure 4C) demonstrating expected within-tissue homogeneity compared with between-tissue heterogeneity. The SNR and rise time across the three tissues (mean ± standard deviation: 45.7 ± 9.6, and 35 ± 4.3 ms, for SNR and rise time, respectively; Figures 4D and 4E) were comparable with those of 2D monolayers (Figures 2D and 2G), demonstrating that Archon1 is as capable of measuring high-fidelity APs in complex 3D environments and highlighting its potential for use in future in vivo studies.
In order to investigate how CMs interact and synchronize in the tissue, we simultaneously recorded two cells in the same field of view in an unpaced tissue (Figures 4F and 4G). MYK-461, a cardiac-specific myosin ATPase inhibitor, was used to prevent tissue contraction and associated movement-related optical artifacts (Stern et al., 2016). We found that the two cells' APs were highly synchronized (Figure 4G, gray lines), suggesting that they are likely electrically coupled within the tissue, even in the absence of externally applied electrical pacing. In addition, by overlaying normalized single AP waveforms from these two cells, we found that the AP waveform morphology was largely similar, further highlighting their intercellular synchrony (Figure 4H). The ability to simultaneously record APs from multiple CMs with single-cell precision enables the monitoring of intercellular conductance and/or whole tissue synchrony. Such measurements, when combined with high-speed and ultra-large field of view imaging, could enable future studies to examine how individual cell APs contribute to population dynamics in complex 3D environments and in vivo systems.
Discussion
Here, we demonstrate the use of a high-performance GEVI, Archon1, to monitor cardiac action potentials in iPSC-derived cardiomyocytes in 2D monolayers and engineered 3D tissues. Our results demonstrate that Archon1 is capable of robustly reporting AP dynamics under a variety of conditions, including when exposed to electrical pacing and ion channel inhibitors, and has comparable sensitivity and time resolution to patch-clamp electrophysiology—the technically challenging but current gold standard in the field for measuring membrane potential. In addition, Archon1 outperformed the commonly used VSD, Di-8-ANEPPS, in SNR. In contrast to VSDs, Archon1 is fully genetically encoded, which enabled long-term studies and has the potential for cell-type-specific labeling in in vivo models. Furthermore, the Cre-recombinase system used here enabled optical isolation of CMs within a complex 3D tissue environment, which can be easily translatable to imaging of whole heart slices or cardiac tissue in Cre-transgenic animals. Finally, Archon1 is capable of reporting membrane potential of multiple cells simultaneously, including in 3D tissue environments. This enables population studies, while retaining single-cell precision, allowing measurement of conduction velocity and AP propagation, as well as intercellular phase locking or phase delay. Future studies could examine changes in individual cell voltage dynamics or intercellular propagation around and distal from injury or diseased sites within tissues.
Although we did not take advantage of this utility here, as Archon1 is a far-red indicator, it is compatible with blue-light-driven optogenetic molecules such as channelrhodopsins (Piatkevich et al., 2019). These molecules could be paired to provide simultaneous control and monitoring of cardiac membrane potential. Similarly, Archon1 can be used together with the genetically encoded calcium indicator, GCaMP6, for simultaneous, multicolor imaging of voltage and calcium dynamics (Dempsey et al., 2016, Song et al., 2015). This could enable studies of diseases like Timothy syndrome or heart failure, which affect both voltage dynamics and calcium handling (Beuckelmann et al., 1992, Gwathmey et al., 1987, Kaab et al., 1996, Splawski et al., 2005). Furthermore, the far-red spectrum of Archon1 enables deeper penetration into tissues than those operating at shorter wavelengths, facilitating future in vivo applications.
The genetically encoded nature of Archon1 makes it a useful tool for longer-duration studies of disease mechanisms, such as detecting arrhythmias, which are known to affect AP waveforms or for phenotyping atrial, ventricular, or nodal cells based on waveform morphology (Leyton-Mange et al., 2014, Shaheen et al., 2018, Song et al., 2015). In particular, Archon1 could be used to follow the development of these cell types through differentiation, maturation, etc. or during disease progression (Leyton-Mange et al., 2014, Song et al., 2015). In addition, although we currently transduce CMs with AAVs expressing Archon1, lentiviral particles could additionally be used to create stable cell lines expressing Archon1 for characterization through iPSC differentiation, development, or integration into living tissue.
Finally, Archon1's capability to monitor CMs in both 2D and 3D environments could assist in the development and evaluation of more functionally mature, biologically relevant engineered cardiac tissues.
Limitations of the Study
Photobleaching is often a concern for all optical imaging techniques that deploy fluorescent indicators. However, under 5 min of continuous imaging, Archon1 demonstrated robust monitoring of membrane voltage with no change in SNR, which we expect could extend to tens of minutes. For experiments requiring hours of continuous monitoring, periodic imaging of short durations may be necessary. As GEVIs are engineered to become more sensitive, less light illumination power will be needed to achieve the same SNR and longer imaging studies could be conducted.
In addition, due to current limitations in camera speed, in order to capture an entire CM (typically ∼50–100 μm across) within the field of view, the acquisition rate must be limited to ∼100 Hz. We have previously shown that Archon1 and its variants can follow millisecond timescale events, such as neuron APs, with high temporal precision at acquisition rates up to 1 kHz (Piatkevich et al., 2018, Piatkevich et al., 2019). As camera technology continues to improve, we hope to be able to conduct such high-speed imaging of larger fields of view containing populations of CMs. Like all non-ratiometric GEVIs or VSDs, Archon1 does not report absolute values of membrane potential, and SNR may vary due to expression levels across individual cells. However, relative membrane potential is still valuable for comparing AP dynamics between different biological states.
In conclusion, Archon1, with its diverse functionality and simple one-photon widefield imaging setup, can be easily adapted for investigating a variety of avenues in cardiac biology and relevant pathology.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Kiryl D. Piatkevich and Edward S. Boyden for providing the rAAV2 CAG-FLEX-Archon1 virus used in all experiments. We thank Paige Cloonan for maintenance of the iPSC lines.
S.N.S. acknowledges funding from the NIH/NIGMS T32 Quantitative Biology and Physiology Fellowship (GM008764) through the Boston University Biomedical Engineering department and from the NIH Ruth L. Kirschstein National Research Service Award (1 F31 NS115421-01). S.L.D. acknowledges funding from the National Science Foundation Graduate Research Fellowship (1122374). C.S.C. acknowledges funding from the National Institutes of Health R01 HL080494 and R01 HL127087. S.N.S., S.L.D. H.T., C.S.C., and X.H. acknowledge funding from the National Science Foundation 1647837. The funders had no role in study design, data collection, and analysis; decision to publish; or preparation of the manuscript.
Author Contributions
S.N.S., S.L.D., H.T., C.S.C, and X.H. designed all experiments and interpreted all data. S.L.D performed all cell culture and tissue preparation. S.N.S. and S.L.D. performed all microscopy experiments and ROI selection. J.N. performed optically guided electrophysiology. F.F. and J.A.W. assisted with and supervised the electrophysiology study. H.T. performed voltage trace processing and subsequent analyses. S.N.S. and S.L.D. prepared all figures, and all authors wrote and edited the manuscript. C.S.C. and X.H. oversaw all aspects of the project.
Declaration of Interests
The authors declare no competing financial interests.
Published: April 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.100974.
Contributor Information
Christopher S. Chen, Email: chencs@bu.edu.
Xue Han, Email: xuehan@bu.edu.
Data and Code Availability
The data and codes used in this study are available upon reasonable request from the Lead Contact (Xue Han, xuehan@bu.edu). Plasmids used in this study and their sequences are available at Addgene.org (pAAV-CAG-FLEX-Archon1-KGC-EGFP-ER2, Addgene: 108422; pENN.AAV.CMVs.PI.Cre.rBG, Addgene: 105537-AAV9). For correspondence and requests for materials, please contact corresponding authors, xuehan@bu.edu and chencs@bu.edu.
Supplemental Information
References
- Abdelfattah A.S., Kawashima T., Singh A., Novak O., Liu H., Shuai Y., Huang Y.C., Campagnola L., Seeman S.C., Yu J. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science. 2019;365:699–704. doi: 10.1126/science.aav6416. [DOI] [PubMed] [Google Scholar]
- Adam Y., Kim J.J., Lou S., Zhao Y., Xie M.E., Brinks D., Wu H., Mostajo-Radji M.A., Kheifets S., Parot V. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature. 2019;569:413–417. doi: 10.1038/s41586-019-1166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers D.M. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- Bers D.M. Calcium cycling and signaling in cardiac myocytes. Annu. Rev. Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- Beuckelmann D.J., Nabauer M., Erdmann E. Intracellular calcium handling in isolated ventricular myocytes from patients with terminal heart failure. Circulation. 1992;85:1046–1055. doi: 10.1161/01.cir.85.3.1046. [DOI] [PubMed] [Google Scholar]
- Chamberland S., Yang H.H., Pan M.M., Evans S.W., Guan S., Chavarha M., Yang Y., Salesse C., Wu H., Wu J.C. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife. 2017;6 doi: 10.7554/eLife.25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Liao M.L., de Boer T.P., Mutoh H., Raad N., Richter C., Wagner E., Downie B.R., Unsold B., Arooj I., Streckfuss-Bomeke K. Sensing cardiac electrical activity with a cardiac myocyte--targeted optogenetic voltage indicator. Circ. Res. 2015;117:401–412. doi: 10.1161/CIRCRESAHA.117.306143. [DOI] [PubMed] [Google Scholar]
- Dempsey G.T., Chaudhary K.W., Atwater N., Nguyen C., Brown B.S., McNeish J.D., Cohen A.E., Kralj J.M. Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging. J. Pharmacol. Toxicol. Methods. 2016;81:240–250. doi: 10.1016/j.vascn.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Eisner D.A., Caldwell J.L., Kistamas K., Trafford A.W. Calcium and excitation-contraction coupling in the heart. Circ. Res. 2017;121:181–195. doi: 10.1161/CIRCRESAHA.117.310230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entcheva E., Bien H. Macroscopic optical mapping of excitation in cardiac cell networks with ultra-high spatiotemporal resolution. Prog. Biophys. Mol. Biol. 2006;92:232–257. doi: 10.1016/j.pbiomolbio.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Faivre J.F., Forest M.C., Gout B., Bril A. Electrophysiological characterization of BRL-32872 in canine Purkinje fiber and ventricular muscle. Effect on early after-depolarizations and repolarization dispersion. Eur. J. Pharmacol. 1999;383:215–222. doi: 10.1016/s0014-2999(99)00614-7. [DOI] [PubMed] [Google Scholar]
- Flytzanis N.C., Bedbrook C.N., Chiu H., Engqvist M.K., Xiao C., Chan K.Y., Sternberg P.W., Arnold F.H., Gradinaru V. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat. Commun. 2014;5:4894. doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Huang C., Li J.Z., Grewe B.F., Zhang Y., Eismann S., Schnitzer M.J. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Wagner M.J., Zhong Li J., Schnitzer M.J. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwathmey J.K., Copelas L., MacKinnon R., Schoen F.J., Feldman M.D., Grossman W., Morgan J.P. Abnormal intracellular calcium handling in myocardium from patients with end-stage heart failure. Circ. Res. 1987;61:70–76. doi: 10.1161/01.res.61.1.70. [DOI] [PubMed] [Google Scholar]
- Herron T.J., Lee P., Jalife J. Optical imaging of voltage and calcium in cardiac cells & tissues. Circ. Res. 2012;110:609–623. doi: 10.1161/CIRCRESAHA.111.247494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobai I.A., O'Rourke B. Decreased sarcoplasmic reticulum calcium content is responsible for defective excitation-contraction coupling in canine heart failure. Circulation. 2001;103:1577–1584. doi: 10.1161/01.cir.103.11.1577. [DOI] [PubMed] [Google Scholar]
- Hochbaum D.R., Zhao Y., Farhi S.L., Klapoetke N., Werley C.A., Kapoor V., Zou P., Kralj J.M., Maclaurin D., Smedemark-Margulies N. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortigon-Vinagre M.P., Zamora V., Burton F.L., Green J., Gintant G.A., Smith G.L. The use of ratiometric fluorescence measurements of the voltage sensitive dye Di-4-ANEPPS to examine action potential characteristics and drug effects on human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2016;154:320–331. doi: 10.1093/toxsci/kfw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J.H., Kralj J.M., Douglass A.D., Engert F., Cohen A.E. Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front. Physiol. 2014;5:344. doi: 10.3389/fphys.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Han Z., Platisa J., Wooltorton J.R., Cohen L.B., Pieribone V.A. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C.M., Krafft A.J., Russe M.F., Rog-Zielinska E.A. A new look at the heart-novel imaging techniques. Herzschrittmacherther Elektrophysiol. 2018;29:14–23. doi: 10.1007/s00399-017-0546-7. [DOI] [PubMed] [Google Scholar]
- Kaab S., Nuss H.B., Chiamvimonvat N., O'Rourke B., Pak P.H., Kass D.A., Marban E., Tomaselli G.F. Ionic mechanism of action potential prolongation in ventricular myocytes from dogs with pacing-induced heart failure. Circ. Res. 1996;78:262–273. doi: 10.1161/01.res.78.2.262. [DOI] [PubMed] [Google Scholar]
- Kaestner L., Tian Q., Kaiser E., Xian W., Muller A., Oberhofer M., Ruppenthal S., Sinnecker D., Tsutsui H., Miyawaki A. Genetically encoded voltage indicators in circulation Research. Int. J. Mol. Sci. 2015;16:21626–21642. doi: 10.3390/ijms160921626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Geiller T., Jung A., Nakajima R., Song Y.K., Baker B.J. Improving a genetically encoded voltage indicator by modifying the cytoplasmic charge composition. Sci. Rep. 2017;7:8286. doi: 10.1038/s41598-017-08731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton-Mange J.S., Milan D.J. Pluripotent stem cells as a platform for cardiac arrhythmia drug screening. Curr. Treat. Opt. Cardiovasc. Med. 2014;16:334. doi: 10.1007/s11936-014-0334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton-Mange J.S., Mills R.W., Macri V.S., Jang M.Y., Butte F.N., Ellinor P.T., Milan D.J. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014;2:163–170. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S., Adam Y., Weinstein E.N., Williams E., Williams K., Parot V., Kavokine N., Liberles S., Madisen L., Zeng H. Genetically targeted all-optical electrophysiology with a transgenic cre-dependent optopatch mouse. J. Neurosci. 2016;36:11059–11073. doi: 10.1523/JNEUROSCI.1582-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A., Mutoh H., Dimitrov D., Akemann W., Knopfel T. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS One. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Guo L., Fiene S.J., Anson B.D., Thomson J.A., Kamp T.J., Kolaja K.L., Swanson B.J., January C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M., Heidmann J.D., Mandegar M.A., Walker C.D., Bolouki S., Conklin B.R., Loewke K.E. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Rep. 2015;4:621–631. doi: 10.1016/j.stemcr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H., Perron A., Dimitrov D., Iwamoto Y., Akemann W., Chudakov D.M., Knopfel T. Spectrally-resolved response properties of the three most advanced FRET based fluorescent protein voltage probes. PLoS One. 2009;4:e4555. doi: 10.1371/journal.pone.0004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R., Jung A., Yoon B.J., Baker B.J. Optogenetic monitoring of synaptic activity with genetically encoded voltage indicators. Front. Synaptic Neurosci. 2016;8:22. doi: 10.3389/fnsyn.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Masse S., Gagliardi M., Hsieh A. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari M.T., Bartorelli C., Polese A., Fiorentini C., Moruzzi P., Guazzi M.D. Treatment of hypertension with nifedipine, a calcium antagonistic agent. Circulation. 1979;59:1056–1062. doi: 10.1161/01.cir.59.5.1056. [DOI] [PubMed] [Google Scholar]
- Panakova D., Werdich A.A., Macrae C.A. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature. 2010;466:874–878. doi: 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Lacerda A.E., Kirsch G.E., Brown A.M., Bruening-Wright A. The action potential and comparative pharmacology of stem cell-derived human cardiomyocytes. J. Pharmacol. Toxicol. Methods. 2010;61:277–286. doi: 10.1016/j.vascn.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Piatkevich K.D., Bensussen S., Tseng H.A., Shroff S.N., Lopez-Huerta V.G., Park D., Jung E.E., Shemesh O.A., Straub C., Gritton H.J. Population imaging of neural activity in awake behaving mice. Nature. 2019;574:413–417. doi: 10.1038/s41586-019-1641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich K.D., Jung E.E., Straub C., Linghu C., Park D., Suk H.J., Hochbaum D.R., Goodwin D., Pnevmatikakis E., Pak N. Publisher Correction: a robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 2018;14:901. doi: 10.1038/s41589-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama G., Choi B.R., Azour G., Lavasani M., Tumbev V., Salzberg B.M., Patrick M.J., Ernst L.A., Waggoner A.S. Properties of new, long-wavelength, voltage-sensitive dyes in the heart. J. Membr. Biol. 2005;208:125–140. doi: 10.1007/s00232-005-0826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama G., Morad M. Merocyanine 540 as an optical probe of transmembrane electrical activity in the heart. Science. 1976;191:485–487. doi: 10.1126/science.191.4226.485. [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jurkiewicz N.K. Delayed rectifier outward K+ current is composed of two currents in Guinea pig atrial cells. Am. J. Physiol. 1991;260:H393–H399. doi: 10.1152/ajpheart.1991.260.2.H393. [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jurkiewicz N.K., Scott A., Siegl P.K. Isoproterenol antagonizes prolongation of refractory period by the class III antiarrhythmic agent E-4031 in Guinea pig myocytes. Mechanism of action. Circ. Res. 1991;68:77–84. doi: 10.1161/01.res.68.1.77. [DOI] [PubMed] [Google Scholar]
- Scheel O., Frech S., Amuzescu B., Eisfeld J., Lin K.H., Knott T. Action potential characterization of human induced pluripotent stem cell-derived cardiomyocytes using automated patch-clamp technology. Assay Drug Dev. Technol. 2014;12:457–469. doi: 10.1089/adt.2014.601. [DOI] [PubMed] [Google Scholar]
- Shaheen N., Shiti A., Huber I., Shinnawi R., Arbel G., Gepstein A., Setter N., Goldfracht I., Gruber A., Chorna S.V. Human induced pluripotent stem cell-derived cardiac cell sheets expressing genetically encoded voltage indicator for pharmacological and arrhythmia studies. Stem Cell Rep. 2018;10:1879–1894. doi: 10.1016/j.stemcr.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnawi R., Huber I., Maizels L., Shaheen N., Gepstein A., Arbel G., Tijsen A.J., Gepstein L. Monitoring human-induced pluripotent stem cell-derived cardiomyocytes with genetically encoded calcium and voltage fluorescent reporters. Stem Cell Rep. 2015;5:582–596. doi: 10.1016/j.stemcr.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Awari D.W., Han E.Y., Uche-Anya E., Park S.H., Yabe Y.A., Chung W.K., Yazawa M. Dual optical recordings for action potentials and calcium handling in induced pluripotent stem cell models of cardiac arrhythmias using genetically encoded fluorescent indicators. Stem Cells Transl. Med. 2015;4:468–475. doi: 10.5966/sctm.2014-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I., Timothy K.W., Decher N., Kumar P., Sachse F.B., Beggs A.H., Sanguinetti M.C., Keating M.T. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc. Natl. Acad. Sci. U S A. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086-8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F., Marshall J.D., Yang Y., Gong Y., Schnitzer M.J., Lin M.Z. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern J.A., Markova S., Ueda Y., Kim J.B., Pascoe P.J., Evanchik M.J., Green E.M., Harris S.P. A small molecule inhibitor of sarcomere contractility acutely relieves left ventricular outflow tract obstruction in feline hypertrophic cardiomyopathy. PLoS One. 2016;11:e0168407. doi: 10.1371/journal.pone.0168407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storace D., Sepehri Rad M., Kang B., Cohen L.B., Hughes T., Baker B.J. Toward better genetically encoded sensors of membrane potential. Trends Neurosci. 2016;39:277–289. doi: 10.1016/j.tins.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Higashijima S., Miyawaki A., Okamura Y. Visualizing voltage dynamics in zebrafish heart. J. Physiol. 2010;588:2017–2021. doi: 10.1113/jphysiol.2010.189126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Chao B.S., Wu J.C. Strategies for improving the maturity of human induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2018;123:512–514. doi: 10.1161/CIRCRESAHA.118.313472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opbergen C.J.M., Koopman C.D., Kok B.J.M., Knopfel T., Renninger S.L., Orger M.B., Vos M.A., van Veen T.A.B., Bakkers J., de Boer T.P. Optogenetic sensors in the zebrafish heart: a novel in vivo electrophysiological tool to study cardiac arrhythmogenesis. Theranostics. 2018;8:4750–4764. doi: 10.7150/thno.26108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V., Chavarha M., Dimov I.K., Bradley J., Pradhan L., Mathieu B., Evans S.W., Chamberland S., Shi D., Yang R. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell. 2019;179:1590–1608 e1523. doi: 10.1016/j.cell.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W.H., Schneiderbanger K., Schubert P., Didie M., Munzel F., Heubach J.F., Kostin S., Neuhuber W.L., Eschenhagen T. Tissue engineering of a differentiated cardiac muscle construct. Circ. Res. 2002;90:223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- Zou P., Zhao Y., Douglass A.D., Hochbaum D.R., Brinks D., Werley C.A., Harrison D.J., Campbell R.E., Cohen A.E. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 2014;5:4625. doi: 10.1038/ncomms5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bar, 25 μm
Scale bar, 25 μm.
Scale bar, 25 μm.
Scale bar, 25 μm.
Scale bar, 250 μm.
Data Availability Statement
The data and codes used in this study are available upon reasonable request from the Lead Contact (Xue Han, xuehan@bu.edu). Plasmids used in this study and their sequences are available at Addgene.org (pAAV-CAG-FLEX-Archon1-KGC-EGFP-ER2, Addgene: 108422; pENN.AAV.CMVs.PI.Cre.rBG, Addgene: 105537-AAV9). For correspondence and requests for materials, please contact corresponding authors, xuehan@bu.edu and chencs@bu.edu.