Abstract
Purpose
Bile acids play an important role in Clostridioides difficile life cycle. Deoxycholate (DCA), one of the most abundant secondary bile acids, is known to inhibit vegetative growth and toxin production. However, limited data are available on the role of DCA on C. difficile sporulation. Here, we investigated the phenotypic and genotypic impact of DCA on the growth, toxin production, and sporulation of C. difficile.
Methodology
Four genetically divergent C. difficile strains were cultured in nutrient-rich broth with and without DCA at various concentrations, and growth activity was evaluated for each strain. Cytotoxicity assays using culture supernatants from cells grown in nutrient-rich broth with and without 0.01% DCA were conducted. Sporulation efficiency was determined using sporulation media with and without 0.01% DCA. Transcript levels of tcdB and spo0A were analyzed using quantitative reverse-transcription polymerase chain reaction.
Results
We found that DCA led to growth reduction in a dose-depended manner and regulated toxin production by repressing tcdB expression during vegetative growth. To our knowledge, we have also provided the first evidence that DCA reduces C. difficile sporulation efficiency through the downregulation of spo0A expression during the sporulation stage.
Conclusions
DCA modulates C. difficile sporulation, vegetative growth, and toxin production.
Keywords: Microbiology, Hepatobiliary system, Infectious disease, Medical microbiology, Microbiology epidemiology, Bacteriology, Deoxycholate, Clostridioides difficile, Sporulation, Toxin production, Vegetative growth
Microbiology; Hepatobiliary System; Infectious Disease; Medical Microbiology; Microbiology Epidemiology; Bacteriology; deoxycholate; Clostridioides difficile; sporulation; toxin production; vegetative growth.
1. Introduction
Clostridioides difficile is a Gram-positive, anaerobic, spore-forming bacterium that causes a spectrum of symptoms, ranging from mild diarrhea to severe pseudomembranous colitis and even death [1, 2]. C. difficile infection (CDI) is a well-known cause of health-care-associated infectious diarrhea related to the disruption of the indigenous intestinal microbiota as a result of prolonged drug treatment, including treatment with antibiotics, and it is especially prevalent among hospitalized old adults [2, 3]. Clinical manifestations of CDI are largely triggered by the toxins produced by vegetative cells, such as toxin A (TcdA), toxin B (TcdB), and binary toxin (Cdt) [4]. The incidence of community-acquired CDI has also increased over the past decade [5].
Bile acids are known to play important roles during infection by C. difficile at all stages of its life cycle [6, 7, 8, 9]. Primary bile acids, such as taurocholate (TCA) and cholate, together with glycine, are known to initiate C. difficile spore germination in the distal small intestine [6, 10]. However chenodeoxycholate, another primary bile acid, can inhibit TCA-mediated spore germination [11]. Moreover, previous studies have demonstrated that deoxycholate (DCA), one of the most abundant secondary bile acids derived from cholate, inhibits the vegetative growth of C. difficile and its toxin activity in vitro [6, 9]. Although this inhibition may be strain-dependent and is reported to be triggered by the functional disruption of the cell membrane in some bacteria [12, 13], the mechanisms underlying it are yet to be elucidated. Moreover, in a murine model of CDI, antibiotic treatment induced spore germination, and increased the vegetative growth and toxin activity due to reduced levels of secondary bile acids, including DCA, in the cecum [14]. Therefore, an understanding of how DCA and other secondary bile acids control the life cycle of C. difficile in terms of vegetative cell proliferation and toxin production in the large intestine, may contribute to the prediction of CDI risk and therapeutic response, including response to recurrent CDI treatments.
A previous study has indicated that the human gut bacterium Clostridium scindens expresses 7-alpha-dehydroxylase in the large intestine, which converts the primary bile acids, cholate and chenodeoxycholate, to the secondary bile acids, DCA and lithocholate (LCA), respectively, and inhibits CDI in animal models and human patients [15]. Very recently, Kang et al. [16] reported that secondary bile acids enhance the activity of tryptophan-derived antibiotics secreted by bile acid 7-alpha-dehydroxylating gut bacteria, including C. scindens, resulting in the inhibition of C. difficile growth. Therefore, reduction in the biosynthesis of secondary bile acids due to decreased abundance of gut bacteria secreting 7-alpha-dehydroxylase results in the disruption of CDI resistance [17, 18]. Among the approximately 50 different secondary bile acids found in the large intestine of humans, the physiological concentrations of the most abundant bile acids, DCA, LCA, and ursodeoxcholate, are in the 0.03–0.7, 0.001–0.45, and 0–0.769 mM ranges, respectively [9, 10]. Moreover, Solbach et al. [19] recently reported that the abundance of the baiCD gene cluster, which is required for the conversion of primary bile acids to secondary bile acids in C. scindens, is negatively correlated with CDI. Although several studies have shown that DCA induces the germination of C. difficile [6, 9], the precise role of secondary bile acids in the sporulation of C. difficile still remains unclear due to a lack of relevant information or knowledge. Thus, additional data are needed to comprehend how secondary bile acids cause the re-sporulation of C. difficile. Such data may also contribute to the suppression of vegetative cell proliferation and toxin release and would further advance our knowledge regarding the role of bile acids in both the developmental life cycle of C. difficile, as well as in the onset of CDI.
In the present study, we investigated the phenotypic and genotypic effects of DCA, which is one of the most abundant secondary bile acids in the large intestine, and aimed to investigate the phenotypic and genotypic impacts of DCA on the growth, toxin production, and sporulation of C. difficile.
2. Materials and methods
2.1. Bacteria strains and growth conditions
In this study, we used the clinically relevant strains of C. difficile, American Type Culture Collection (ATCC) 700057 (Ribotype 038; TcdA−, TcdB−, Cdt−) and ATCC BAA-1870 (Ribotype 027; TcdA+, TcdB+, Cdt+) were used in the study. We also used clinical isolates 138 (TcdA+, TcdB+, Cdt+) [20, 21] and B-12-14 (TcdA+, TcdB+, Cdt−) [20], which were previously isolated from stool samples of patients with diarrhea. These genetically divergent strains belong to different multilocus sequence typing-based sequence types (STs): ATCC 700057 belongs to ST 48, ATCC BAA-1870 to ST 1, 138 to ST 97, and B-12-14 to ST 17. The C. difficile strains were grown at 37 °C using brain heart infusion (BHI; Oxoid, Basingstoke, UK) broth or agar plates supplemented with 5 g/L yeast extract and 0.1% (w/v) L-cysteine (BHIS) in an anaerobic bag flushed and filled with N2 gas [22]. For stimulating spore formation, a 70:30 medium consisting of 63 g/L Bacto Peptone, 3.5 g/L Proteose Peptone, 11.1 g/L BHI, 1.5 g/L yeast extract, 1.06 g/L Tris base, 0.7 g/L NH4SO4, and 0.3 g/L L-cysteine, which is a well-established and commonly used medium for inducing C. difficile sporulation [23, 24], was used. Sodium DCA (Wako Pure Chemical Industries, Osaka, Japan) was added to the BHIS broth and 70:30 media as necessary for all assays. For measuring sporulation efficiency, BHIS agar plates supplemented with 0.1% sodium TCA (Nacalai Tesque, Kyoto, Japan), which is well-known as a major germinant [23, 25], were used for the proper germination of spores.
2.2. Growth kinetics
Pre-cultured C. difficile strains grown on BHIS agar plates were inoculated into BHIS broth and incubated anaerobically overnight at 37 °C. The optical density of the overnight culture was measured at 550 nm (OD550), and the culture was subsequently diluted in fresh 10 mL BHIS broth containing different concentrations of DCA (0.001%, 0.01%, or 0.05%), such that the starting OD550 was 0.01 for all the strains. The 0.001%, 0.01%, and 0.05% strengths correspond approximately to 0.024, 0.24, and 1.2 mM DCA, respectively [9, 10]. After 24 h of incubation at 37 °C, the OD550 values of all the cultures were determined. Based on the results obtained for the four strains, we used 0.01% DCA (approximately 0.24 mM, estimated as the physiological concentration) and/or 0.001% DCA (approximately 0.024 mM, estimated to be slightly below the physiological concentration) for analyzing the growth kinetics and for conducting subsequent experiments. The growth kinetics of the four strains used in this study were also determined by measuring the OD550 values of the cultures for up to 12 h, because, in our preliminary experiment, we found that all these strains attained the stationary growth phase by 12 h (data not shown). The procedure for analysis of the growth kinetics was repeated three times.
2.3. Cytotoxicity assay
An in vitro cytotoxicity assay was performed using Vero cells, in which C. difficile strains show potent toxic activity [26]. Vero cells were cultured in D-MEM medium (Wako Pure Chemical Industries, Osaka, Japan), supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin for 72 h. Then, Vero cells (1.0 × 104 cells) were seeded (100 μL) into each of the 96 wells of culture plates and incubated at 37 °C and 5% CO2 for 48 h.
Cells of C. difficile strains in the stationary growth phase, after being cultured at 37 °C for 24 h in BHIS broth with and without 0.01% DCA (estimated as the physiological concentration), were diluted in fresh BHIS broth to an OD550 of 0.6 to adjust cell counts. The C. difficile cultures were centrifuged at 19,000 × g for 30 min, and the filtered supernatant (10 μL) was added to 96-well plates containing Vero cells. After the plates were incubated at 37 °C and 5% CO2 for 48 h, cell viability was determined in triplicate using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), a WST-8 dye-based colorimetric reaction [20, 27]. Optical density was measured at 450 nm using a microplate reader and normalized to the untreated blank samples. The ATCC 700057 and ATCC BAA-1870 strains were used as negative and positive controls, respectively. The results are presented as means ± standard deviations (SD) of percentage cell viability.
2.4. Sporulation efficiency
C. difficile strains grown for 24 h in 70:30 broth with and without 0.001% DCA and 0.01% DCA, were heated at 80 °C for 10 min to kill all the vegetative cells (germinated spores). To quantify heat-resistant spores, appropriate volumes of serially-diluted heated or unheated cultures, which represented heat-resistant cells and total cells, respectively, were plated onto BHIS agar plates supplemented with 0.1% TCA. After 24 h of anaerobic incubation at 37 °C, the colonies were counted. Sporulation efficiency was determined as the ratio of heat-resistant cells and total cells (heat-resistant cells/total cells) for each strain relative to the data from cultures without DCA.
2.5. Morphological analysis using fluorescent staining
To investigate whether DCA affects the sporulation process, we analyzed cell morphology using fluorescence microscopy. C. difficile strains ATCC BAA-1870 and 138 were anaerobically cultured in 10 mL 70:30 broth with and without 0.01% DCA for 24 h. The cells were collected from 1-mL aliquots by centrifugation at 19,000 × g for 3 min and immediately washed with phosphate-buffered saline (PBS) and resuspended in 1 mL PBS. The 5-μL bacterial suspensions were mixed with 1 μL of each 5 μg/mL FM4-64 (Thermo Fisher Scientific, Tokyo, Japan) and 15 μg/mL Hoechst 33342 (Dojindo) for staining the cell membrane and double-stranded DNA, respectively [24], and the stained cells were mounted on 1% agarose-coated glass slides. The stained cells were then observed using fluorescence microscopy (Olympus Model BX5; Olympus, Tokyo, Japan) and classified into four stages based on their morphology (vegetative cells, early stage of spore formation, late stage of spore formation, and mature spore). One thousand cells were counted for each sample from 24-h cultures grown in 70:30 broth with and without 0.01% DCA.
2.6. RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
For the analysis of tcdB transcript levels, total RNA was extracted from cultures of the three toxin-producing strains, BAA-1870, 138, and B-12-14, growing in the exponential growth phase, after incubation for 7 h at 37 °C in BHIS broth with and without 0.01% DCA, using the NucleoSpin RNA kit (Takara Bio, Shiga, Japan). For the analysis of spo0A transcript levels, total RNA extraction was performed using cultures of all the four strains, ATCC 700056, BAA-1870, 138, and B-12-14, growing in the stationary growth phase, after incubation for 24 h at 37 °C in 70:30 broth with and without 0.01% DCA. Complementary DNA (cDNA) was synthesized from 200 ng of total RNA using the PrimeScript RT Master Mix (Takara Bio). The RT-qPCR assays were performed in triplicate using the KAPA SYBR Fast qPCR Kit (NIPPON Genetics, Tokyo, Japan). Each reaction contained 5 μL of master mix, 200 nM of each primer, 1 μL of cDNA template (10-fold dilution), and nuclease-free water in a final volume of 10 μL. The primers used in these reactions are listed in Table 1. Amplification and detection of PCR products were performed using an Eco Real-Time PCR System (Illumina, San Diego, California) as follows: initial denaturation at 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, and 50 °C for 20 s; and melting curve analysis was performed at temperatures from 55 °C to 95 °C. The transcript levels were normalized to that of the housekeeping gene 16S rRNA and quantified using the comparative threshold cycle (Ct) method (2−ΔΔCt).
Table 1.
Primers used in the current study.
| Name | Sequence (5′ to 3′) |
|---|---|
| 16S ribosomal RNA | |
| 16S-F | GATTTACTTCGGTAAAGAGCGG |
| 16S-R |
CCTTACCAACTAGCTAATCAGACG |
|
tcdB | |
| tcdB-F | GGCAAATGTAAGATTTCGTACTCA |
| tcdB-R |
TCGACTACAGTATTCTCTGAC |
|
spo0A | |
| spo0A-F | AGCGCAATAAATCTAGGAGCAGA |
| spo0A-R | TGGTCTAGGTTTTGGCTCAACT |
2.7. Statistical analyses
All assays were performed three times, and the data are shown as means ± SD. In growth and sporulation efficiency studies (Figure 1A and Figure 3A), statistical significance of the differences between controls and other groups was calculated using one-way analysis of variance and Dunnett's multiple comparison post-hoc test. In other experiments, such as those involving the study of toxin production (Figure 2A, B, and Figure 3D), the significance between two different groups was calculated by multiple Student's t-test and Holm–Sidak post-hoc pairwise comparison of means. All statistical analyses were performed using Prism version 7.0 (GraphPad Software, La Jolla, California, USA) and p < 0.05 was considered significant.
Figure 1.
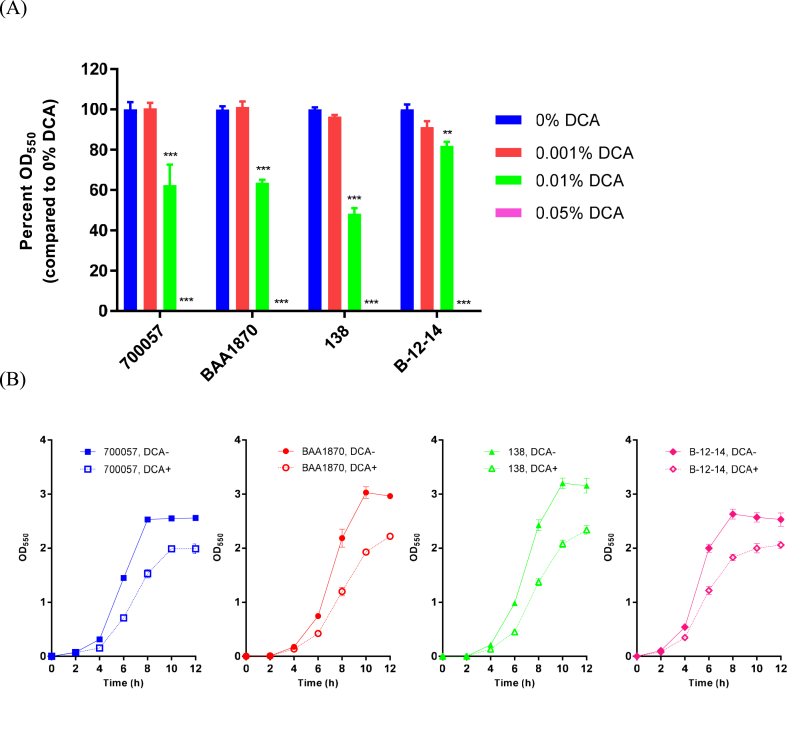
Growth of Clostridioides difficile strains (A) C. difficile strains were cultured in BHIS broth with and without DCA (at 0%, 0.001%, 0.01%, or 0.05%). Optical density at 550 nm (OD550) was measured after 24 h of incubation. The ODs were normalized to relative to that in the presence of 0% DCA (B) Growth kinetics were determined by measuring the OD550 values of f the cultures of all the four strains in the presence or absence of 0.01% DCA for up to 12 h of incubation. Data are means ± standard deviation of triplicate experiments. ∗∗∗p < 0.001, ∗∗p < 0.01.
Figure 3.
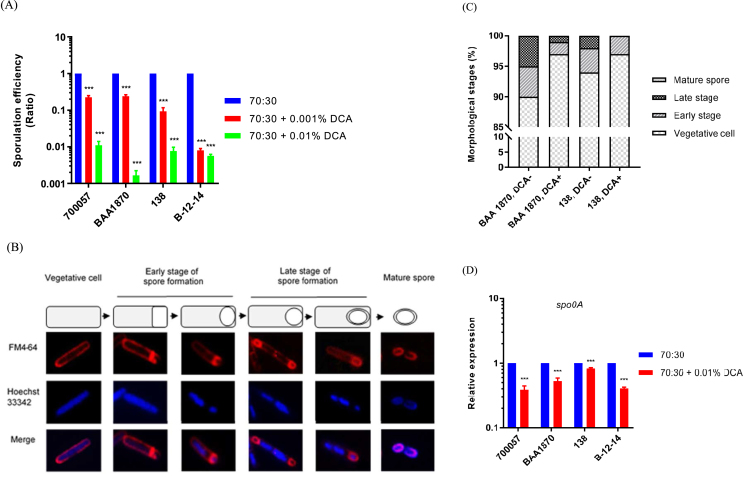
Sporulation efficiency, morphological analysis, and transcript levels of spo0A in Clostridioides difficile strains (A) Sporulation efficiency (based on the reduction of non-germinated spores at 80 °C) was defined as the ratio of heat-resistant cells/total cells of each strain grown in the presence of 0.001% and 0.01% DCA compared to the cells grown without DCA (B) The cell membranes of C. difficile strains grown in the 70:30 broth were stained with FM4-64 and the double-stranded DNA was stained with Hoechst 33342. The samples were then observed under a fluorescence microscope (C) The stained cells were classified into four morphological stages, as shown in panel (B) and the number of each cell type was determined. One thousand cells were counted in each sample (D) The expression of spo0A in C. difficile strains grown for 24 h in 70:30 broth, with and without 0.01% DCA was analyzed using RT-qPCR. Values are relative to that in the absence of DCA (70:30 broth) after normalization to the transcript levels of the housekeeping 16S rRNA gene. Data are means ± standard deviation of triplicate experiments. ∗∗∗p < 0.001.
Figure 2.
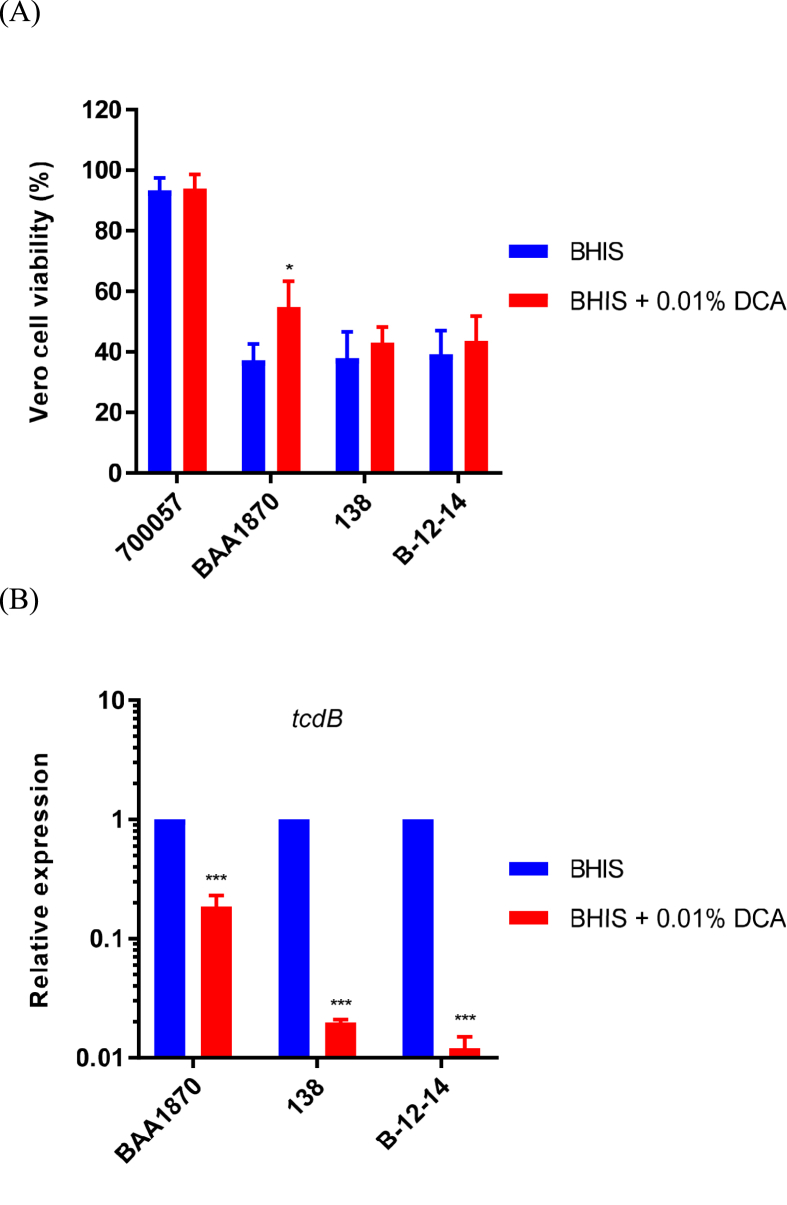
Toxin production and tcdB expression in Clostridioides difficile strains (A) After 24 h of incubation in BHIS broth with and without 0.01% DCA, the final number of C. difficile cells in the stationary growth phase was adjusted, and the culture supernatants were then used for cytotoxicity assays. The viability of Vero cells was determined using the Cell Counting Kit-8. The data are presented as the percentage of Vero cell viability (B) Transcript levels of tcdB in C. difficile cells (in the exponential growth phase) grown for 7 h in BHIS broth with and without 0.01% DCA were analyzed using RT-qPCR. The transcript levels are relative to that in cells grown in the absence of DCA (BHIS) after normalization with respect to the transcript levels of the housekeeping 16S rRNA gene. Data are means ± standard deviation of triplicate experiments. ∗∗∗p < 0.001, ∗p < 0.05.
3. Results
3.1. DCA inhibited C. difficile growth in a dose-dependent manner
Previous studies have demonstrated that DCA reduces C. difficile growth in a strain-dependent manner [6, 9]. Therefore, we investigated the growth of C. difficile strains in the presence of four different DCA concentrations in BHIS broth containing the physiological relevant concentration of DCA (0.01%, corresponding to approximately 0.24 mM) [9, 10]. We found that the growth of the strains examined in the current study was reduced by DCA in a dose-dependent manner (Figure 1A). No proliferation was observed under vegetative growth conditions for 24 h at 37 °C with 0.05% DCA. Furthermore, when ATCC BAA-1870 and 138 strains were grown under vegetative growth conditions for 12 h at 37 °C with 0.01% DCA, both the strains demonstrated slower growth compared to that in the absence of DCA, and the delayed growth involved both a prolonged exponential phase and a decrease in the final cell density (Figure 1B). Taken together, these results indicate that DCA led to a reduction in the vegetative growth of both the strains in a dose-dependent manner, and that the highest concentration of DCA (0.05%) completely inhibited the growth.
3.2. DCA regulated tcdB expression and toxin production
Because DCA is known to affect the toxin activity of C. difficile [9], we conducted cytotoxicity assays using the supernatants of cells in the stationary growth phase, after culture in BHIS broth with and without the addition of 0.01% DCA, to analyze the amount of toxin released from C. difficile cells. When the final cell counts were adjusted based on the optical density of the cultures, Vero cells exposed to the culture supernatants of the ATCC BAA-1870 (TcdA+, TcdB+, Cdt+) strain grown in the presence of 0.01% DCA demonstrated significantly increased cell viability compared to that of Vero cells exposed to the culture supernatants of the ATCC BAA-1870 strain grown without DCA (Figure 2A). Vero cells exposed to the culture supernatants of the 138 (TcdA+, TcdB+, Cdt+) and B-12-14 (TcdA+, TcdB+, Cdt−) strains grown in the presence of 0.01% DCA also tended to demonstrate increased cell viability (Figure 2A). In contrast, Vero cells exposed to culture supernatants of the non-toxin-producing strain, ATCC 700057 grown with and without DCA, had the same level of viability, suggesting that DCA did not directly affect the viability of Vero cells. These results indicate that DCA reduces the production of toxin by C. difficile under vegetative growth conditions.
To confirm these phenotypic findings, we further analyzed the cellular transcript levels of tcdB, which encodes a major virulence factor in C. difficile, at the exponential growth phase using RT-qPCR. Consistent with the phenotypic results, tcdB transcript levels were significantly decreased in all the toxin-producing strains grown in the presence of 0.01% DCA compared to that in the cells grown without DCA (Figure 2B). Collectively, our results indicated that DCA negatively regulates toxin production, at least by repressing tcdB transcription under vegetative growth conditions.
3.3. DCA reduced C. difficile sporulation efficiency by downregulating spo0A expression
While DCA inhibited the vegetative cell growth and toxin production under vegetative growth conditions, it was still unclear whether it also controlled the sporulation under sporulation conditions. Therefore, we also evaluated the relationship between DCA and the ability to form spores. In the stationary growth phase, the sporulation efficiency of C. difficile, based on the reduction in non-germinated spores at 80 °C, was significantly decreased in the presence of both 0.001% and 0.01% DCA compared to that of cells grown without DCA for all the four strains used in this study (Figure 3A). Moreover, we also found that DCA inhibited the sporulation efficiencies of all the four strains in a dose-dependent manner. Because DCA not only suppressed the vegetative growth but also reduced the sporulation efficiency in the same manner in all the four strains, we then investigated whether it directly modulated the sporulation process by assessing its effect of DCA on the overall homogeneity of spore-forming cells using fluorescent staining (Figure 3B). We observed that in ATCC BAA-1870, the sporulation efficiency of which was most strongly reduced in 0.01% DCA, and 138, the sporulation efficiency of which was similar to that of 700057 and B-12-14, the subpopulation (percentage) of cells in the sporulation stage was decreased in the presence of 0.01% DCA compared to that in the absence of DCA (Figure 3C). Thus, our findings suggest that DCA affected the viability of vegetative cells and directly inhibited the sporulation process leading to reduced sporulation efficiency.
Based on a previous finding that the accumulation of Spo0A in C. difficile cells leads to more rapid sporulation [25], we hypothesized that DCA may diminish the sporulation efficiency via inhibiting a Spo0A-dependent sporulation pathway. To investigate this possibility, we evaluated spo0A expression under sporulation conditions using RT-qPCR. As expected, the spo0A transcript levels in ATCC 700057, ATCC BAA-1870, 138, and B-12-14 cells (means ± SD: 0.39 ± 0.06, 0.53 ± 0.06, 0.82 ± 0.03, 0.41 ± 0.02, respectively) in the stationary growth phase were significantly decreased when they were grown for 24 h in the presence of 0.01% DCA relative to spo0A transcript levels observed when these cells were grown without DCA (Figure 3D). This indicated that reduced spo0A transcript levels resulted in lower sporulation efficiency.
4. Discussion
In several bacteria, such as Gram-positive Lactobacilli and Bifidobacteria, the inhibition of growth by DCA is caused by its cell membrane-damaging activities, which lead to the leakage of essential cellular components, and the activities are involved in strong hydrophobicity, which increases the affinity of DCA for the phospholipid bilayer of the bacterial cell membrane [12, 13]. Although the mechanism of inhibition of C. difficile growth remains unclear, previous studies have indicated that DCA inhibits its vegetative growth in C. difficile [6, 9]. In the present study, the findings of the several experiments were consistent and demonstrated that DCA inhibited the vegetative growth of C. difficile in a dose-dependent manner. We also found that the four phylogenetically divergent strains showed the same growth tendencies in the presence of 0.001%–0.05% DCA. However, Thanissery et al. [9] demonstrated that the sensitivity of vegetative growth in DCA-containing broths is strain dependent, because among seven clinically relevant strains, including epidemic R20291 and M68 strains, historic 630 strain and current the Bl-9 strain [28] were unable to grow in the presence of 0.02% DCA. Therefore, the discrepancy between our results and those reported previously [9] might result from the differences in the hydrophobicities of the cell membranes of the vegetative cells of different strains. It is not surprising that DCA inhibited the vegetative growth of C. difficile through cell membrane-damaging activities in agreement with previous findings [12, 13]. Further analysis of vegetative growth through a hydrophobicity assay, using more strains should provide additional insights into the physiological role of DCA during CDI.
It is evidently critical to routinely test the levels of toxin A and/or B secreted by vegetative cells in patients with antibiotic-associated diarrhea and/or pseudomembranous colitis for immediate start of treatment with metronidazole or vancomycin in a clinical setting [2, 3]. A previous study reported that DCA affected the toxin activity depending on the C. difficile strain [9]; however, this statement was based on a semi-quantitative cytotoxicity assay and no accurate data for the amount of toxins was provided. Here, we investigated if DCA modulates the synthesis and/or release of toxin to obtain additional data that would improve our understanding of the relationship between DCA and toxin production using a well-established and commercially available kit based on the index of cell viability. Our current results, coupled with the results of the prior study [9], demonstrate that DCA negatively regulates toxin production in C. difficile cells in the stationary growth phase under vegetative growth conditions. However, because reduced C. difficile growth may lead to reduced toxin production, we further investigated the relationship between DCA and tcdB transcription. In the current study, our findings suggested that the reduced toxin production resulted from the downregulation of tcdB expression, although the transcript levels obtained in cells in the exponential growth phase may be underestimated relative to those in the stationary phase. The expression of tcdB was significantly decreased in strains 138 and B-12-14, but we were unable to detect a significant phenotypic difference between the two strains (Figure 2A and B). One possibility for the contradiction is that the cytotoxicity levels in strains 138 and B-12-14 resulted from other toxins, such as TcdA and Cdt, indicating toxicity to Vero cells [4, 29, 30]. However, in this study, we did not investigate the relationship between the transcript levels of toxins besides TcdB and DCA. Further research is warranted to determine whether DCA controls the production of TcdA and Cdt, as well as that of their regulators, such as TcdR and SigD.
To date, it remains unclear as to how DCA affects the sporulation in C. difficile. Previous studies using a related species Clostridium perfringens strain SM101 demonstrated that DCA induces sporulation through the activation of a Spo0A-dependent signaling pathway [31, 32]. However, in contrast to this finding, our results suggested that DCA reduces the sporulation efficiency in response to repression of spo0A expression. This discrepancy may result from the fact that the sporulation regulatory circuit in C. difficile differs from that in C. perfringens. Moreover, it is possible that a classical two-component system involving several histidine kinases [33] primarily regulates the DCA-mediated sporulation in C. difficile by modulating the phosphorylation of Spo0A. Unfortunately, although we were unable to determine the detailed mechanism through which DCA regulates Spo0A, we believe that our study is the first to elucidate the relationship between DCA and C. difficile sporulation. Further studies evaluating the related histidine kinases would help to determine any possible cross talk involved in DCA-mediated sporulation.
In conclusion, we found that: 1) DCA inhibited efficient vegetative growth in C. difficile in a dose-dependent manner; 2) DCA regulated toxin production under vegetative growth conditions by repressing tcdB expression; and 3) DCA reduced C. difficile sporulation efficiency through the downregulation of spo0A. Further investigations, including transcriptome analyses, are required to clarify the interaction between DCA and the cellular physiology of C. difficile.
Declarations
Author contribution statement
Yukino Usui: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Alafate Ayibieke: Performed the experiments; Analyzed and interpreted the data.
Yuko Kamiichi: Performed the experiments.
Shu Okugawa, Kyoji Moriya, Shuji Tohda: Contributed reagents, materials, analysis tools or data.
Ryoichi Saito: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number JP17K08822 (R.S.).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Rupnik M., Wilcox M.H., Gerding D.N. Clostridium difficileinfection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 2.Smits W.K., Lyras D., Lacy D.B., Wilcox M.H., Kuijper E.J. Clostridium difficile infection. Nature Reviews Disease Primers. 2016;2 doi: 10.1038/nrdp.2016.20. PubMed PMID: WOS:000381363700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reveles K.R., Lee G.C., Boyd N.K., Frei C.R. The rise in Clostridium difficile infection incidence among hospitalized adults in the United States: 2001-2010. Am. J. Infect. Contr. 2014;42(10):1028–1032. doi: 10.1016/j.ajic.2014.06.011. PubMed PMID: WOS:000342385700002. [DOI] [PubMed] [Google Scholar]
- 4.Aktories K., Schwan C., Jank T. Clostridium difficile toxin biology. Annu. Rev. Microbiol. 2017;71:281–307. doi: 10.1146/annurev-micro-090816-093458. Epub 2017/06/28. . PubMed PMID: 28657883. [DOI] [PubMed] [Google Scholar]
- 5.Gupta A., Khanna S. Community-acquired Clostridium difficile infection: an increasing public health threat. Infect. Drug Resist. 2014;7:63–72. doi: 10.2147/IDR.S46780. Epub 2014/03/17. PubMed PMID: 24669194; PubMed Central PMCID: PMCPMC3962320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorg J.A., Sonenshein A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008;190(7):2505–2512. doi: 10.1128/JB.01765-07. Epub 2008/02/01. PubMed PMID: 18245298; PubMed Central PMCID: PMCPMC2293200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darkoh C., Brown E.L., Kaplan H.B., DuPont H.L. Bile salt inhibition of host cell damage by Clostridium difficile toxins. PloS One. 2013;8(11) doi: 10.1371/journal.pone.0079631. Epub 2013/11/11. PubMed PMID: 24244530; PubMed Central PMCID: PMCPMC3823588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorg J.A., Sonenshein A.L. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 2009;191(3):1115–1117. doi: 10.1128/JB.01260-08. Epub 2008/12/05. PubMed PMID: 19060152; PubMed Central PMCID: PMCPMC2632082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thanissery R., Winston J.A., Theriot C.M. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. Epub 2017/03/06. PubMed PMID: 28279860; PubMed Central PMCID: PMCPMC5466893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton J.P., Xie G., Raufman J.P., Hogan S., Griffin T.L., Packard C.A. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293(1):G256–G263. doi: 10.1152/ajpgi.00027.2007. Epub 2007/04/05. PubMed PMID: 17412828. [DOI] [PubMed] [Google Scholar]
- 11.Sorg J.A., Sonenshein A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010;192(19):4983–4990. doi: 10.1128/JB.00610-10. PubMed PMID: WOS:000281866900020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe M., Fukiya S., Yokota A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. JLR (J. Lipid Res.) 2017;58(6):1143–1152. doi: 10.1194/jlr.M075143. PubMed PMID: WOS:000402517000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurdi P., Kawanishi K., Mizutani K., Yokota A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 2006;188(5):1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. PubMed PMID: WOS:000235819200033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koenigsknecht M.J., Theriot C.M., Bergin I.L., Schumacher C.A., Schloss P.D., Young V.B. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun. 2015;83(3):934–941. doi: 10.1128/IAI.02768-14. Epub 2014/12/22. PubMed PMID: 25534943; PubMed Central PMCID: PMCPMC4333439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. Epub 2014/10/22. PubMed PMID: 25337874; PubMed Central PMCID: PMCPMC4354891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J.D., Myers C.J., Harris S.C., Kakiyama G., Lee I.K., Yun B.S. Bile acid 7 alpha-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chemical Biology. 2019;26(1):27. doi: 10.1016/j.chembiol.2018.10.003. PubMed PMID: WOS:000456042100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theriot C.M., Bowman A.A., Young V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1(1) doi: 10.1128/mSphere.00045-15. Epub 2016/01/06. PubMed PMID: 27239562; PubMed Central PMCID: PMCPMC4863611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antunes L.C., Han J., Ferreira R.B., Lolić P., Borchers C.H., Finlay B.B. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob. Agents Chemother. 2011;55(4):1494–1503. doi: 10.1128/AAC.01664-10. Epub 2011/01/31. PubMed PMID: 21282433; PubMed Central PMCID: PMCPMC3067180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solbach P., Chhatwal P., Woltemate S., Tacconelli E., Buhl M., Gerhard M. BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PloS One. 2018;13(5) doi: 10.1371/journal.pone.0196977. Epub 2018/05/08. PubMed PMID: 29738579; PubMed Central PMCID: PMCPMC5940204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwata Y., Tanimoto S., Sawabe E., Shima M., Takahashi Y., Ushizawa H. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34(4):763–772. doi: 10.1007/s10096-014-2290-9. Epub 2014/12/05. PubMed PMID: 25471195. [DOI] [PubMed] [Google Scholar]
- 21.Sawabe E., Kato H., Osawa K., Chida T., Tojo N., Arakawa Y. Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a five-year period. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26(10):695–703. doi: 10.1007/s10096-007-0355-8. PubMed PMID: WOS:000249259500003. [DOI] [PubMed] [Google Scholar]
- 22.Weingarden A.R., Dosa P.I., DeWinter E., Steer C.J., Shaughnessy M.K., Johnson J.R. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PloS One. 2016;11(1) doi: 10.1371/journal.pone.0147210. PubMed PMID: WOS:000368529100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam E.E., Nock A.M., Lawley T.D., Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 2013;195(6):1214–1225. doi: 10.1128/JB.02181-12. Epub 2013/01/04. PubMed PMID: 23292781; PubMed Central PMCID: PMCPMC3592010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fimlaid K.A., Bond J.P., Schutz K.C., Putnam E.E., Leung J.M., Lawley T.D. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 2013;9(8) doi: 10.1371/journal.pgen.1003660. PubMed PMID: WOS:000323830300015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dembek M., Willing S.E., Hong H.A., Hosseini S., Salgado P.S., Cutting S.M. Inducible expression of spo0A as a universal tool for studying sporulation in Clostridium difficile. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01793. PubMed PMID: WOS:000411430600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stabler R.A., He M., Dawson L., Martin M., Valiente E., Corton C. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10(9) doi: 10.1186/gb-2009-10-9-r102. PubMed PMID: WOS:000271425300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larabee J.L., Bland S.J., Hunt J.J., Ballard J.D. Intrinsic toxin-derived peptides destabilize and inactivate Clostridium difficile TcdB. mBio. 2017;8(3) doi: 10.1128/mBio.00503-17. PubMed PMID: WOS:000404733300027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He M., Sebaihia M., Lawley T.D., Stabler R.A., Dawson L.F., Martin M.J. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U.S.A. 2010;107(16):7527–7532. doi: 10.1073/pnas.0914322107. PubMed PMID: WOS:000276892300075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Verstraete I., Peltier J., Dupuy B. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins. 2016;8(5) doi: 10.3390/toxins8050153. Epub 2016/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundriyal A., Roberts A.K., Ling R., McGlashan J., Shone C.C., Acharya K.R. Expression, purification and cell cytotoxicity of actin-modifying binary toxin from Clostridium difficile. Protein Expr. Purif. 2010;74(1):42–48. doi: 10.1016/j.pep.2010.04.014. PubMed PMID: WOS:000281495100006. [DOI] [PubMed] [Google Scholar]
- 31.Yasugi M., Okuzaki D., Kuwana R., Takamatsu H., Fujita M., Sarker M.R. Transcriptional profile during deoxycholate-induced sporulation in a Clostridium perfringens isolate causing foodborne illness. Appl. Environ. Microbiol. 2016;82(10):2929–2942. doi: 10.1128/AEM.00252-16. Epub 2016/03/13.PubMed PMID: 26969700; PubMed Central PMCID: PMCPMC4959072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talukdar P.K., Olguín-Araneda V., Alnoman M., Paredes-Sabja D., Sarker M.R. Updates on the sporulation process in Clostridium species. Res. Microbiol. 2015;166(4):225–235. doi: 10.1016/j.resmic.2014.12.001. Epub 2014/12/23. PubMed PMID: 25541348. [DOI] [PubMed] [Google Scholar]
- 33.Edwards A.N., McBride S.M. Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol. Lett. 2014;358(2):110–118. doi: 10.1111/1574-6968.12499. Epub 2014/06/26. PubMed PMID: 24910370; PubMed Central PMCID: PMCPMC4189817. [DOI] [PMC free article] [PubMed] [Google Scholar]