Abstract
Background
Recent years have witnessed an increasing interest in multi-omics data, because these data allow for better understanding complex diseases such as cancer on a molecular system level. In addition, multi-omics data increase the chance to robustly identify molecular patient sub-groups and hence open the door towards a better personalized treatment of diseases. Several methods have been proposed for unsupervised clustering of multi-omics data. However, a number of challenges remain, such as the magnitude of features and the large difference in dimensionality across different omics data sources.
Results
We propose a multi-modal sparse denoising autoencoder framework coupled with sparse non-negative matrix factorization to robustly cluster patients based on multi-omics data. The proposed model specifically leverages pathway information to effectively reduce the dimensionality of omics data into a pathway and patient specific score profile. In consequence, our method allows us to understand, which pathway is a feature of which particular patient cluster. Moreover, recently proposed machine learning techniques allow us to disentangle the specific impact of each individual omics feature on a pathway score. We applied our method to cluster patients in several cancer datasets using gene expression, miRNA expression, DNA methylation and CNVs, demonstrating the possibility to obtain biologically plausible disease subtypes characterized by specific molecular features. Comparison against several competing methods showed a competitive clustering performance. In addition, post-hoc analysis of somatic mutations and clinical data provided supporting evidence and interpretation of the identified clusters.
Conclusions
Our suggested multi-modal sparse denoising autoencoder approach allows for an effective and interpretable integration of multi-omics data on pathway level while addressing the high dimensional character of omics data. Patient specific pathway score profiles derived from our model allow for a robust identification of disease subgroups.
Keywords: Deep learning, Patient clustering, Multi-omics
Background
Precision medicine aims for the delivery of the right treatment for the right patients. One important goal is therefore to identify molecular sub-types of diseases, which opens the opportunity for a better targeted therapy of malignancies in the future. In that context high throughput omics data has been extensively used. Prominent examples include the gene expression based groupings of breast cancer and glioblastoma multiforme (GBM) into four clusters [1, 2]. However, analysis of one type of omics data alone provides only a very limited view on a complex and systemic disease such as cancer. Correspondingly, parallel analysis of multiple omics data types is needed and employed more and more routinely [3–5]. However, leveraging the full potential of multi-omics data requires statistical data fusion, which comes along with a number of unique challenges, including differences in data types (e.g. numerical vs discrete), scale, data quality and dimension (e.g. hundreds of thousands of SNPs vs few hundred miRNAs), see Ahmad and Fröhlich for a review [6].
Several authors have proposed methods, which allow for integrating multi-modal omics data into one clustering model and thus allow for detection of clusters that are consistently supported by several biological scales [7–10]. Prominent examples include Similarity Network Fusion (SNF) [11], iCluster [12] and multi-view non-negative matrix factorization [13], see [6] for a more complete overview and discussion. A challenge that all these methods face is the high dimensional character of omics data and the large difference of the number of features across different omics modalities.
In this paper we propose an unsupervised multi-modal neural network architecture to learn a low dimensional embedding of omics features from multiple sources, which can be mapped to the same biological pathway. The result of such an embedding is one score per pathway and patient. In a second step we then combine scores of multiple pathways into a profile for each patient, which we use to bi-cluster patients and pathways via consensus sparse non-negative matrix factorization (sNMF) [14], hence providing insights into interpretable and cluster specific pathways of distinct patient subgroups. Furthermore, by leveraging a recently proposed machine learning technique [15], we are also able to disentangle the relevance of individual genes, miRNAs, CNVs and CpG sites for each individual patient subgroup.
We demonstrate the utility of our approach in comparison to conventional sNMF based clustering based on a gene expression leukemia dataset, and in comparison to SNF and iCluster based on four cancer datasets from the GDC Data Portal and cBioPortal, respectively. We specifically show the association of clusters identified by our Pathway based Multi-modal autoEncoder (PathME) method to well-known somatic mutation patterns and a variety of clinical outcomes, including survival. Moreover, we highlight the possibility to interpret features selected by our method for individual patient subgroups in the light of existing disease knowledge.
Methods
Overview
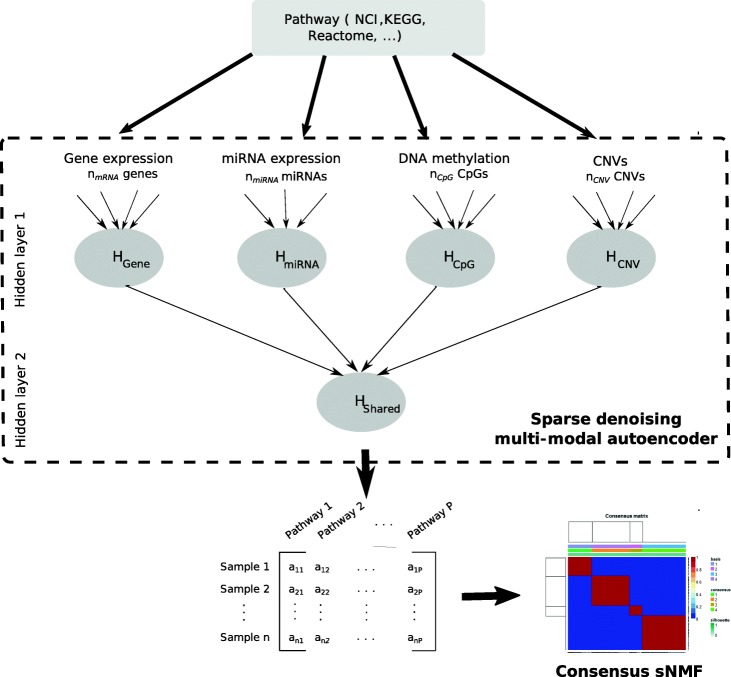
The overall aim of our proposed multi-modal autoencoder framework is to embed k patient-level omics data types mapping to a particular pathway of interest into a common low dimensional latent space. Our method thus compresses hundreds of original features into one score per pathway and patient. Conducting the same embedding for P pathways results into a pathway profile representation, which we use to stratify patients based on sparse NMF in an unsupervised manner [14]. This effectively allows for a bi-clustering of patients and pathways and thus ensures a certain level of interpretation. Overall our proposed PathME method consists of four major steps (Fig. 1):
Mapping of omics features from each data source to pathways.
Estimation of a per-patient score for each pathway using multi-modal sparse denoising autoencoders.
Bi-clustering of patients using consensus sNMF.
Interpretation of clusters and cluster specific pathway scores using recent statistical and game theoretic methods.
Fig. 1.
Conceptual overview about our approach: Multi-omics feature mapping to a specific pathway are summarized into a pathway level score via a sparse denoising multi-modal autoencoder architecture. Hidden layer 1 consists of up to [pj/2] hidden units per omics modality, where p_j is the number of features in omics type j. Hidden units for omics modality j are densely connected to input features of the same omics type, but there are no connections from input features of other data modalities. Hidden layer 2 consists of one hidden unit, which represents the overall multi-omics pathway score. Concatenation of P multi-omics pathway scores for each patient allows for application of consensus sparse NMF clustering in a subsequent step
In the following we describe each of these steps in more detail.
Mapping of Omics features to pathways
To demonstrate the principle of our method in this paper we used combinations of gene expression, miRNA expression, DNA methylation (chip based) and copy number variation (CNV) data. Entrez genes IDs were mapped to NCI pathways [16] using the graphite R-package [17], but of course other pathway databases could be used as well. For DNA methylation data we relied on the annotation by the manifacturer to map individual CpGs to Entrez gene IDs. For assignment of CNVs to genes we relied on the mapping provided by The Cancer Genome Atlas (TCGA), which uses the Genomic Identification of Significant Targets in Cancer (GISTIC2) method [18]. TCGA provides for each patient a list of CNVs mapped to Entrez gene IDs. These are available for download via http://firebrowse.org/. For miRNA data, we considered the predicted miRNA target genes (again as Entrez gene IDs) obtained from miRBase [19]. Overall, CpGs, CNVs and miRNAs were mapped to Entrez gene IDs and Entrez gene IDs to pathways. Hence, all relevant omics modalities considered in this paper could be mapped to pathways.
Estimation of pathway scores via multi-modal sparse Denoising autoencoders
Deep Denoising autoencoders
We start by explaining a standard autoencoder [20]. Briefly, an autoencoder network takes a feature vector x ∈ ℝ d as input and transforms / encodes it to a hidden representation y ∈ ℝ q (typically q < d) via
| 1 |
where s(·) is a non-linear activation function, e.g. tanh or rectified linear unit. Matrix W consists of weights and b is a bias vector. Several encoding steps can be performed sequentially, resulting into a deep encoder.
The latent representation y can be decoded / mapped back via
| 2 |
where W0,b0 are other weights and a bias vector. Again, several decoding steps can be performed sequentially, resulting into a deep decoder. Encoder plus decoder network together form a (deep) autoencoder. In most cases (as well as in our work) the decoder network has a laterally reversed architecture to the encoder. That means the input to the encoder network has the same number of units as the last layer of the decoder (which is at the same time the output of the entire autoencoder). Hidden layer 1 in the encoder has the same architecture as the second last layer of the decoder, and so on.
The objective of training a deep autoencoder is to minimize the reconstruction loss L(·,·) (e.g. the mean squared difference between original input x and reconstructed input, i.e. output of the autoencoder, z) by updating weights W,W0 and biases b,b0:
| 3 |
The training can be conducted via common stochastic gradient descent methods.
To further increase robustness of the representation learning Vincent et al. [21] suggested to add random noise to input features while attempting to reconstruct original data, which results into a denoising autoencoder.
Multi-modal Denoising autoencoders
In our case we have multiple omics data sources X1,X2,···,Xk from the same n patients mapping to a particular pathway of interest. To account for this multimodal character of the data we suggest the architecture shown in Fig. 1: Each data modality j is first encoded separately into up to pj/2 hidden units (using dense connections between inputs and hidden units), where pj is the number of input features for omics type j. In Fig. 1 these units in the first hidden layer are denoted as Hgene, HmiRNA, etc. The exact number of hidden units for each data modality is determined via a hyper-parameter optimization, which is described in Section 1.3.4. In a second step units from the first hidden layer are further encoded (via dense connections) into one pathway score, which is denoted as Hshared. Hshared is the only unit in the second hidden layer. As mentioned before, the decoder has a laterally reversed architecture to the encoder and is thus not explicitly shown in Fig. 1.
Note that our suggested architecture has significantly fewer trainable weights than a fully connected standard autoencoder network, because initially a separate hidden representation within each data modality is learned.
The specific architecture of our multi-modal (denoising) autoencoder induces a modified loss function for training the network compared to a standard autoencoder.
More specifically, let W denote the set of all trainable network parameters. Let denote the reconstruction of feature vector x(i) corresponding to patient i in data modality j at the output layer of the autoencoder. Our training objective is then to minimize
| 4 |
That means we aim to find network parameters W, which jointly minimize the reconstruction error of features within each data modality.
Introducing Sparsity
To control overfitting, we regularize our model by enforcing sparsity of weights W. More specifically, we used two techniques for this purpose: i) random drop-out of input features with predefined probability p [22]; ii) a sparse group lasso penalty for features at the input layer [23]. The sparse group lasso is an extension of the classical lasso algorithm [24], which has originally been introduced in the context of generalized linear models. The sparse group lasso enforces a sparse regression model by jointly pushing coefficients of certain predefined groups of features towards zero, i.e. there is a feature selection at the group level similar to the group lasso [25]. Furthermore, also sparsity within each group of features is promoted.
The idea of the sparse group lasso can also be applied to neural networks. More specifically, by adding a sparse group lasso penalty we modify our training objective as:
| 5 |
where d is the number of layers, sl is the number of units in lth layer, s_l_succ is the number of units in the successor layer of the lth layer, λ is the weight decay parameter, α is a convex combination of the lasso and group lasso penalties. The first penalty in the second line (l2-norm of weights) promotes sparsity at the group (here: omics modality) level. The second penalty in the third line of Eq. (5) (l1-norm of weights) promotes sparsity at the individual feature level within each data modality. Parameter α ∈ [0,1] balances between sparse selection of entire omics data modalities and sparsity within each data modality. If α = 1 we recover the original lasso penalty by Tibshirani [24]. If α = 0 we get the group lasso penalty by Yuan and Lin [25].
Training and optimizing the multi-modal sparse Denoising autoencoder
For training of our muli-modal sparse denoising autoencoder we use mini-batch stochastic gradient descent [26, 27], i.e. the training procedure updates parameters iteratively using randomly chosen small mini-batches of patients instead of the entire training set. We evaluated different variants of the stochastic gradient descent algorithm: momentum [28], RMSProp [29], adam [30] and Nesterov-accelerated Adam (nadam) [31]. To account for differences in numerical ranges of data modalities we employed batch normalization of hidden units [32], i.e. scaled their inputs to mean zero and standard deviation one. The complementary batch de-normalization was performed at the output layer. Note that batch normalization also accounts for the covariance shift of network weights and typically yields a dramatic speed-up of the training procedure [32].
All experiments in this paper were carried out with tanh activation functions, because initial results looked most promising. In addition to the regularization techniques described in last paragraph we incorporated an early stopping mechanism in the training process [26]. More specifically, we stopped training when there was no improvement in the loss function for more than twenty iterations.
Altogether we considered the following hyper-parameters for training or model:
mini-batch size ∈ {4,8,16,32}
sparse group lasso parameters α ∈ [0,1] and λ ∈ [10− 9,···,10− 1]
learning rate ρ ∈ [10− 5,···,10− 1]
probability p of dropout of an input feature p ∈ [0.5,1]
number of units in the first hidden layer in the range [1,···,pj/2], where pj is the input size of the jth omics modality. Note that we used for the decoder network an architecture that was laterally reversed to the encoder.
To deal with the large number of different hyper-parameters we used Bayesian hyper-parameter optimization with 50 evaluations [33]. Within this procedure each selected candidate set of hyper-parameters was assessed via the reconstruction error of the model on unseen test data via a 5-fold cross-validation procedure. That means we randomly split the entire dataset into 5-folds and sequentially left out one of these folds (i.e. 20% of the data) for validation / testing, whereas the rest of the data was used to learn network weights. The entire implementation has been conducted in TensorFlow and is available in the Supplementary files to this paper.
Patient bi-clustering via consensus sparse non-negative matrix factorization
Our suggested multi-modal sparse autoencoder resulted into a patient specific pathway score profile. We used this matrix to jointly identify clusters of patients and cluster specific pathways. We employed sparse non-negative matrix factorization (sNMF) for this purpose [14], which is an extension of the algorithm by Lee et al. [34]. Briefly, this algorithm factorizes a data matrix X (here: P pathways ×n patients) as X ≈ BH, where B is a non-negative P × m matrix containing basis vectors and H is a non-negative m × n matrix containing coefficient vectors. The solution is found by minimizing the reconstruction error between X and BH via multiplicative updates. Sparse NMF extends this approach by additionally enforcing sparsity of matrix B:
| 6 |
where η > 0 and β > 0 are regularization parameters, and H(:,j) is the jth column vector of H. According to Kim et al. [14] η was set to the maximum value of X. Sparse NMF effectively yields a bi-clustering of patients and pathways and thus allows for identifying a cluster specific pathway profile. For doing so we employed the method by Carmona et al. [35]: For each basis component i (i.e. ith column in B) values are first sorted by decreasing magnitude. Then, only the first consecutive features are considered as most descriptive for cluster i. That means PathME allows for assessing the most descriptive pathways per data modality (e.g. gene expression, CNVs) and cluster.
To help the interpretation of these most descriptive pathways we calculated the fraction of genes carrying any form of somatic mutation, resulting into a mutational burden score. We then compared mutational burden across clusters via a Kruskal-Wallis test. P-values were corrected for multiple testing via the Benjamini-Hochberg method [36].
NMF based matrix factorization is performed in an iterative fashion and starting from some initial conditions. The obtained solution is thus sensitive to the initialization. To account for this aspect we performed 500 repeated runs of the entire sNMF algorithm, starting from different random initializations of B and H [14], as implemented in the R-package NMF [37]. One possible strategy is then to look for the individual solutions yielding minimal reconstruction loss, which resembles the approach typically taken in k-means clustering. Another approach, which we followed here, is to conduct consensus clustering over these 500 individual clustering solutions by hierarchical clustering of the consensus matrix [38]. The agreement of the hierarchical clustering with the similarity of patients according to the consensus matrix can be assessed via the cophenetic correlation [39]. The cophenetic correlation can then be used for model selection. More specifically, we here used this measure to tune the regularization beta in the range [0.001,1] and to find the optimal rank of the factorization (i.e. number of clusters) in the range 2,...,9. While doing so we aimed for comparing the cophenetic correlation achieved by our consensus sNMF algorithm with the cophenetic correlation achieved by chance. Since consensus sNMF clustering is computationally quite costly (as it is the result of 500 sparse non-negative matrix factorizations) we limited ourselves to 40 random permutations of the data matrix X here. For each of these random permutations we re-ran the entire consensus sNMF algorithm. We then looked for the smallest number of clusters m, for which the cophenetic correlation achieved on the basis of the original data exceeded the upper bound of the 95% confidence interval achieved on the basis of randomly permuted data. In addition, we recorded also the silhouette index as a widely used distance based clustering index [40]. The silhouette s(i) for data point (here: patient) i assigned to cluster Ci is defined as.
| 7 |
provided that |Ci| > 1, and 0 otherwise. Furthermore, a(i) is the mean distance of data point i to all other data points in cluster Ci, and b(i) is defined as.
| 8 |
where d(i,j) is the distance between data points i and j. The silhouette s(i) ranges from − 1 to 1, where 0 indicates that data point i falls in between two clusters. The silhouette index is defined as the mean s(i) over all data points. A value closer to 1 indicates a more tightly grouping of data points into clusters.
Interpretation of cluster specific pathway scores via SHAP
One of the main criticism of neural network based approaches (here: autoencoders) is the difficulty to interpret them. Recently, Lundberg et al. [15] proposed a game theoretic framework to address this issue. Briefly, the idea behind Shapley Additive exPlanations (SHAP) is that the relevance of feature i for model output f(x) can be regarded as the average weighted difference between f(x) and outputs from all possible models trained on subsets of features, excluding feature i:
| 9 |
where F is the set of all features. The authors propose several local approximation techniques, which can circumvent the exact combinatorial calculation of Φi(x), one which is specifically tailored towards neural networks (Deep SHAP). Deep SHAP effectively combines SHAP values calculated for smaller components of a neural network into SHAP values for the entire network. We refer to Lundberg et al. [15] for details. In this work we used SHAP to understand the impact of individual omics modalities and features on the autoencoded score that we learned for each pathway. SHAP results into a patient specific score that may be positive (feature i increases f(x)) or negative (feature i decreases f(x)). In agreement to [15] we considered the mean of the absolute SHAP values per omics feature to score the overall impact of a variable on the pathway score. Moreover, we investigated the overall mean of absolute SHAP values per omics data modality. Hence, our PathME method allows for interpreting the influence of pathways, omics data modalities and individual markers on dedicated patient sub-groups.
Association to clinical features
An important question is, in how far molecular subgroups might be clinically relevant. For this purpose we investigated for each cancer type a wide range of available clinical features for each disease, including different survival endpoints. More specifically, we considered overall survival (OS), progression free survival (PFS) and disease free survival (DFS), as defined in Liu et al. [41]. We first checked nominal significance of the association to age via a Cox regression model that only contained age as predictor. If this was true, we fitted a Cox regression model that contained a factor “cluster” plus age as predictors. This model was then compared against the “null” Cox regression model that only contained age as predictor. Both models were compared via a likelihood ratio test, and the corresponding p-value was reported as the significance of the age corrected association to the clustering. In case of no significant association with age a conventional log-rank test was used.
Significance of additional categorical clinical variables (e.g. gender) was tested via a χ2 test, including the agreement with existing molecular classification schemes. For numerical variables a one-way ANOVA was applied.
Multiple testing correction of p-values was performed via the Benjamini-Hochberg method to control false discovery rate (FDR) [36].
Compared approaches
We compared our PathME approach against two competing multi-omics clustering methods: 1) SNF and 2) iCluster. For both methods we performed a pre-filtering of features: We selected only features that mapped to the same pathways used by our method. In addition, for the iCluster method we further down-filtered these features to the 100–200 most variable ones (depending on each disease). This was done to increase the robustness of the iCluster method and get better separated clusters.
According to the suggestion by the respective authors of each method the number of clusters for SNF was selected via the eigen-gap method [11] and for iCluster via the proportion of deviation score [12].
Although we motivated the development of our approach by the use of multi-omics data, PathME can in principle also be applied to single omics data. We therefore compared PathME also against sNMF directly applied to individual omics features, which could be mapped to NCI pathways.
Results
Evaluation on gene expression dataset with known grouping
We first used gene expression data of 62 bone marrow samples from leukemia patients (41 Acute Lymphocytic Leukemia (ALL), 21 Acute Myelocytic Leukemia (AML) - [42]) to test PathME against sNMF. The dataset is available as Bioconductor R-package golubEsets (see http://bioconductor.org/packages/release/ data/experiment/html/golubEsets.html) and has been frequently used to test clustering methods [14]. 33 out of the 41 ALL patient samples are from B-cells and 8 from T-cells. So, there are presumably k = 3 clusters. 1729 out of the 7129 omics features could be mapped to 211 NCI pathways.
We compared consensus sNMF applied to individual genes against PathME based consensus clustering in terms of the adjusted rand index [43]. According to that measure PathME outperformed conventional consensus sNMF clustering using individual genes as features (adj. Rand index 0.16 vs 0.02). This suggests that autoencoder based dimensionality reduction yields more statistically stable and coherent clusters, i.e. was successful in capturing relevant signal from the data.
Multi-Omics datasets
We next applied PathME to four diseases with available multi-omics data from The Cancer Genome Atlas (TCGA) [44, 45] available via the cBioPortal (https:// www.cbioportal.org/) and GDC Data Portal (https://portal.gdc.cancer.go), respectively: Colorectal Cancer (CRC), Lung Squamous Cell Carcinoma (LSCC), Glioblastoma Multiforme (GBM) and Breast Cancer (PanCancer BRCA). These cancer types were chosen due to the comparable large number of patients with available multi-omics data. For each of these diseases we selected those patients where three omics data types were jointly available and pre-filtered features that were mapping to NCI cancer pathways, as previously described in Section 1.2. Details are shown in Table 1. Notably, gene expression for GBM and LSCC was based on normalized microarray data, whereas for CRC and BRCA we used RSEM estimates available from Firebrowse (http://firebrowse.org/). CNV and DNA methylation profiles were available as preprocessed microarray data.
Table 1.
Datasets from The Cancer Genome Atlas (TCGA) used for evaluation: colorectal cancer (CRC), glioblastoma multiforme (GBM), lung squamous cell carcinoma (LSCC) and breast cancer (BRCA). Omics features correspond to those mapable to NCI pathways
| Dataset | Patients | Omics types | Features |
|---|---|---|---|
| mRNA | 2295 | ||
| CRC | 294 | miRNA | 264 |
| CNV | 2310 | ||
| mRNA | 2039 | ||
| GBM | 273 | miRNA | 18 |
| DNA methylation | 1798 | ||
| mRNA | 2039 | ||
| LSCC | 106 | miRNA | 150 |
| DNA methylation | 1846 | ||
| mRNA | 2329 | ||
| BRCA | 747 | miRNA | 99 |
| CNV | 2334 |
Training error curves of all autoencoder models for the optimal set of hyperparameters (found according to the procedure described in the Methods part) can be found in the Supplementary material.
We applied PathME, SNF and iCluster to the multi-omics data from each disease. Based on our above defined criteria for determining a good number of clusters PathME arrived at 5 sub-groups for CRC and BRCA, and clusters 4 for GBM and LSCC (Figure S1). Table 2 shows silhouette indices (SI) of our method compared to those obtained for SNF and iCluster. While the significantly higher SI of our method after applying consensus clustering is not unexpected, it is interesting to see that also the SI obtained from the best among 500 individual sNMA runs is clearly better compared to SNF and iCluster for GBM, BRCA and LSCC.
Table 2.
Comparison of PathME vs SNF and iCluster in terms of silhouette index. For PathME we report the silhouette index of the consensus clustering as well as the best individual one among 500 randomly initialized sNMF runs
| Cancer datasets | PathME | SNF | iCluster | |||
|---|---|---|---|---|---|---|
| Disease | Omics type | Cophenetic correlation | Consensus silhouette | Optimal silhouette | Silhouette | Silhouette |
| CRC | Number of clusters | (5) | (2) | (2) | ||
| Multi-omics | 1 | 0.98 | 0.51 | 0.54 | 0.06 | |
| GBM | Number of clusters | (4) | (2) | (3) | ||
| Multi-omics | 1 | 1 | 0.67 | 0.58 | 0.11 | |
| LSCC | Number of clusters | (4) | (4) | (2) | ||
| Multi-omics | 1 | 1 | 0.82 | 0.71 | 0.35 | |
| BRCA | Number of clusters | (5) | (2) | (3) | ||
| Multi-omics | 1 | 0.93 | 0.67 | 0.57 | 0.12 | |
For comparison reasons we also include the results of PathME applied to each individual omics data type (Table 3), indicating that multi-omics based clustering via PathME based consensus clustering for CRC, GBM and LSCC results into at least as well discriminated patient subgroups as consensus clustering of individual omics sources. In agreement to our findings in the previous Section, PathME in most cases also resulted into better SI than sNMF clustering applied to individual omics features (i.e. without autoencoder based representation learning). Notably, SNF and iCluster are not applicable to individual omics types data.
Table 3.
Comparison of PathME vs conventional sNMF consensus clustering of individual omics features in terms of cophenetic correlation and silhouette index
| Cancer datasets | Cophenetic correlation | Consensus silhouette | |||
|---|---|---|---|---|---|
| Disease | Omics type | PathME | sNMF (ind. features) | PathME | sNMF (ind. features) |
|
CRC (5 clusters) |
mRNA | 1 | 1 | 0.99 | 0.87 |
| miRNA | 1 | 1 | 0.97 | 0 | |
| CNV | 1 | 0.99 | 0.99 | 0.93 | |
|
GBM (4 clusters) |
mRNA | 1 | 0.99 | 1 | 1 |
| miRNA | 1 | 1 | 0.93 | 0.86 | |
| Methylation | 1 | 1 | 1 | 1 | |
|
LSCC (4 clusters) |
mRNA | 0.92 | 1 | 0.67 | 1.00 |
| miRNA | 1 | 1 | 0.98 | 0.98 | |
| Methylation | 1 | 0.99 | 1.00 | 0.97 | |
|
BRCA (5 clusters) |
mRNA | 0.98 | 0.99 | 0.88 | 0.94 |
| miRNA | 1 | 0.91 | 0.99 | 0.49 | |
| CNV | 1 | 1 | 1 | 1 | |
PathME allows for interpretable multi-Omics based patient stratification
We next analyzed clinical and biological features of the patient clusters obtained via PathME in the last Section.
Colorectal Cancer (CRC)
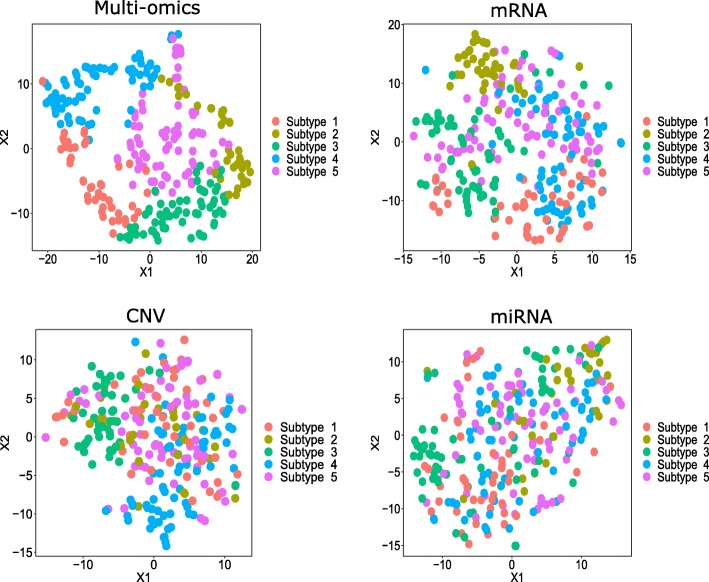
CRC patient clusters identified by PathME differed significantly by tissue site as well as pathological and histological stage (Table S1). Interestingly, our clusters 1-4 were highly enriched for formerly published consensus molecular subtypes (CMS) in CRC [46] (see Table S1, S2). A visual representation of the PathME consensus clustering via t-stochastic neighorhood embedding (t-SNE [47]) can be found in Fig. 2. We here relied on the t-SNE implementation in Bioconductor R-package M3C [48] using default parameters.
Fig. 2.
T-SNE visualization of CRC consensus clustering based on PathME. Data points (= patients) have been colored according to the consensus sNMF clustering of multi-omics pathway scores. T-SNE visualization of individual omics modalities is based on all features map-able to the pathways used by PathME. T-SNE plots for other datasets can be found in the Supplementary material
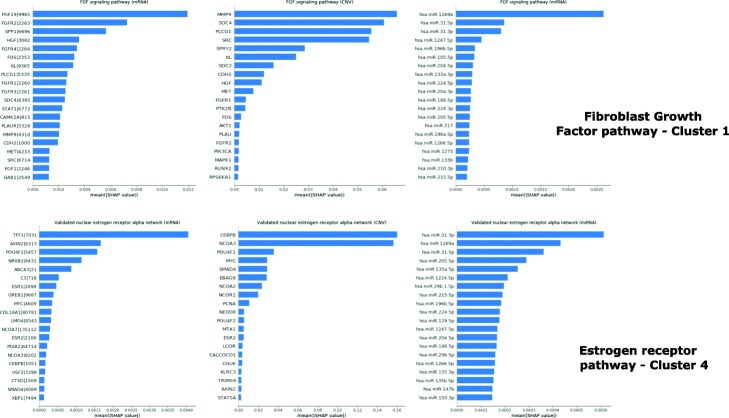
In the following we use examples to further demonstrate, how PathME can be interpreted on molecular level based on the methods described before (Fig. 3, Table S8): For cluster 1 the Fibroblast Growth Factor (FGF) signaling pathway is one of the most contributing molecular features. FGF signaling dysregulation is associated with cancer tumorgenesis and progression [49, 50]. Its components Fibroblast Growth Factor 19 (FGF19) and Fibroblast Growth Factor Receptor 2 (FGFR2) are reported to be expressed in CRC and could be useful therapeutic targets [49, 51]. Their relevance is underlined by high SHAP values. SHAP analysis also demonstrates the impact of miR-31-3p/5p. These are known to be important predictive and prognosis biomarkers in CRC [52, 53]. Altogether CNVs demonstrate the strongest impact on the multi-omics pathway score learned for FGF signaling (Table S8), and CNVs in the metalloproteinase-9 (MMP9) gene are most impactful. The MMP9 level has been suggested as a biological predictor of prognosis in CRC [54] For cluster 4, a high contribution of the nuclear estrogen receptor (ER) activation pathway is determined. ER signaling including ER-α and ER-β is implicated in CRC pathogenesis and progression and it is considered as a potential preventive and therapeutic target [55]. Estrogen Receptor 2 (ESR2), the gene encoding ER-β, is considered as a prognostic marker [56]. Another important pathway related to the CRC adenoma to carcinoma transition is the Ras pathway [57]. Regulation of Ras family activation is associated to cluster 5. The oncogene KRAS is considered as an important component of this pathway, because KRAS gene copy number alteration might be a useful marker to predict treatment response [58]. SHAP values demonstrate the impact of CNVs in that gene on the composite Ras signaling score, which is otherwise mostly influenced by miRNAs. The most influential miR-206 is a known prognostic factor in CRC [59].
Fig. 3.
Example of SHAP analysis results obtained for CRC. For gene expression data gene symbols are shown together with Entrez gene IDs. Further results can be found in the Supplementary material, including Table S8
Glioblastoma Multiforme (GBM)
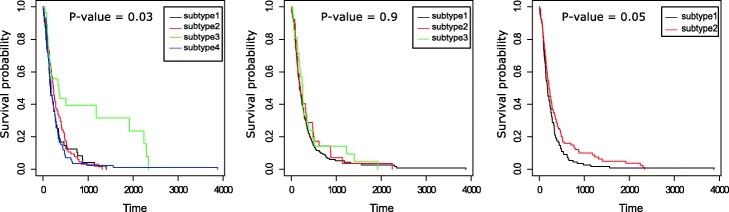
A t-SNE visualization of the PathME consensus clustering can be found in Figure S4. GBM patient clusters found by PathME differed significantly by PFS (FDR = 0.03; iCluster: FDR = 0.9; SNF: FDR = 0.05 – Fig. 4), gender distribution (in agreement with recent observations that gender may impact cancer survival.
Fig. 4.
Progression free survival (days) of patients stratified by PathME (left), iCluster (middle) and SNF (right) for GBM. P-values were corrected for the confounding effect of age and adjusted for multiple testing
[60]), Verhaak’s molecular classification [2] and MGMT methylation status [61] (Tables S3, S4). PathME cluster 3 (green PFS curve) showed the best prognosis of patients and an enrichment of somatic mutations in Isocitrate Dehydrogenase 1 (IDH1), which is a well-known positive prognostic factor in GBM [62]. Notably, this enrichment of somatic mutations was not found in clustering results of SNF and iCluster. Cluster 3 also showed a strong enrichment of the proneural Verhaak subtype, which has been associated to positive prognosis. In contrast, cluster 4 (blue PFS curve) revealed a high enrichment of the mesenchymal subtype that has been associated with poor prognosis [2].
By investigating the sNMF based matrix factorization we can find that cluster 4 is specifically associated to transcriptional targets of c-MYC, which has been associated with disease prognosis [63]. Interestingly, there is also a significant difference in the fraction of mutated genes in that NCI pathway across clusters (FDR = 0.03, Kruskal-Wallis test, Figure S7).
Cluster 2 is associated to Platelet-Derived Growth Factor Receptor-β (PDGFR-β) signaling and Transforming Growth Factor -β (TGF-β) receptor signaling. PDGFR and TGF-β pathways are known to be dysregulated in GBM and they contribute to its pathogenesis and progression [64]. PDGFR and TGF-β are are currently considered as therapeutic targets [64, 65]. In addition, TGF-β activity is associated with differences in prognosis in gliomas, including GBM [65, 66]. Investigation of SHAP values allows for gaining additional insight (Table S9): The most contributing gene to the PDGFR signaling pathway score is transgelin-2, which has recently been found to be expressed at significantly higher levels in IDH1 WT patients. The most influential data modalities on the multi-omics score for TGF-β signaling are miRNAs and DNA methylation, and cg15001381 is the most contributing CpG. This CpG is in the promotor region of AXIN1, a gene that has been reported to be downregulated in GBM [67]. More results can be found in the Supplementary material.
Lung squamous cell carcinoma (LSCC)
LSCC patient clusters identified by PathME showed significant differences in lymph node pathology, race, tissue source site and tumor size (Table S5). A visual representation of the PathME consensus clustering via t-SNE can be found Figure S5.
According to the analysis of the PathME model (Table S10), cluster 1 is associated to α6β4 integrin-ligand interactions. The α6β4 signaling pathway is known to play a role in many cellular processes in human malignancies including LSCC. its alteration induces aggressive behavior [68]. In LSCC, α6/β4 integrin encoded by Integrin Alpha-6 (ITGA6) and Integrin Beta-4 (ITGB4) genes (high SHAP values), respectively, have been found moderately expressed and β4 integrin was highly expressed [69]. MiRNAs are the most influential data modality for the multi-omics score of that pathway according to SHAP, and miR-149 is the most impactful miRNA. It has been associated with the Epithelial-to-Mesenchymal Transition (EMT) phenotype in Non-Small-Cell Lung Cancer (NSLCC) [70]. Moreover, SHAP values show that the Murine Double Minute 2 (MDM2) gene has a high impact on the androgen receptor activity pathway, which was found as a feature for cluster 4. MDM2 was found to strongly correlate with patient survival in NSCLC [71]. For miR-21, associated to the Ataxia Telangiectasia Mutated (ATM) pathway (cluster 4), and miR-155, associated to calcineurin-dependent Nuclear Factor of Activated T-cells (NFAT) signaling in lymphocytes (cluster 2), expression has been described as a prognosis factor in NLSCC, including LSCC [72, 73].
Breast Cancer (BRCA)
A visual representation of the PathME consensus clustering via t-SNE can be found Figure S6. BRCA patient clusters found by PathME showed significant differences in race and histology type (Table S6). Moreover, there was a strong correlation with the common molecular classification into luminal A, luminal B, normal-like, basal-like and HER2 enriched subtypes [74] (Table S7). Cluster 1 and 2 were highly enriched for the luminal B subtype, cluster 3 for the basal-like subtype, cluster 4 for the liminal A subtype and cluster 5 for the normal-like subtype. Cluster 3 showed a strong enrichment of TP53 somatic mutations (p < 1E − 11), and cluster 4 of somatic mutations in Cadherin-1 (CDH1), GATA3 and Phosphatidylinositol4,5-bisphosphate 3-Kinase Catalytic subunit Alpha (PIK3CA). All these are known prognostic factors for BRCA [75, 76].
According to the further inspection of the PathME model (Table S11), cluster 1 is associated to the stabilization and expansion of the E-cadherin adherens junction pathway, which has been associated with tumor survival in several cancers including breast [77]. There is a significant difference in the fraction of genes carrying somatic mutations in this pathway across clusters (Figure S9). According to SHAP analysis data the most influential gene in the pathway is Insulin-like Growth Factor 1 Receptor (IG1FR), which has an important role in tumor growth and survival [78]. MiR-184 is the most impactful miRNA in the same pathway. It has been discussed as a predictive biomarker for breast cancer [79]. Cluster 4 is highly associated to CDC42 (Cell Division Control) signaling. CDC42 has been discussed as a drug target in breast cancer [80]. According to SHAP a CNV in the Ribosomal Protein S6 Kinase B1 (RPS6KB1) has a high impact on the pathway score. Indeed copy number alterations in this gene have been associated to prognosis of breast cancer patients [81].
Once again, these are only examples and more results can be found in the Supplements to this paper. Altogether they indicate the ability to interpret PathME clusters via their association to clinical and molecular features.
Discussion
We could show that PathME is better able to recover known patient leukemia sub-groups than consensus sparse NMF clustering using individual omics features. Additional analysis of 12 individual omics datasets (3 per disease – Table 3) with unknown subgroups in most cases resulted into a higher silhouette index for PathME compared to sNMF.
For multi-omics data we observed higher silhouette indices compared to SNF and iCluster. In contrast to these methods PathME is not limited to multi-omics data, but can also be applied to a single data modality. A further advantage of PathME is that it can effectively address the high dimensionality of omics data due to the proposed multi-modal autoencoder architecture. At the same time this approach yields interpretable results via investigation of sNMF results and SHAP analysis, as demonstrated above.
Of course, PathME is not without limitations: Instead of auto-encoding each pathway separately one could try to take advantage of the fact that pathways are not independent from each other and design the neural network model such that all pathway scores are jointly learned. However, this approach – although theoretically being more elegant – would not only yield a drastic increase of computation time, but also of the number of parameters to be learned, hence raising the risk of overfitting due to the high dimensionality of omics data. It is worthwhile to emphasize that the proposed modular architecture of our multi-modal autoencoder heavily reduces the number of trainable weights compared to a fully connected standard autoencoder architecture. Our approach is therefore in essence a compromise between a realistic consideration of sample sizes in omics data and affordable model complexity in the light of the typical high dimensionality of omics data. In that context we should also mention that we use multiple regularization techniques (drop-out, weight penalties) to further address this point.
Conclusion
The overall contribution of this work is two-fold:
We suggested a multi-modal sparse denoising autoencoder architecture that allows for an effective and interpretable combination of multi-omics data and pathway information. Our specific model addresses the high dimensionality of omics data.
Our second contribution is to demonstrate that patient specific pathway scores derived from our autoencoder model allow for a robust and interpretable disease subtype identification. Disease sub-groups are tighter (i.e. have higher silhouette indices) than the ones produced by SNF and iCluster. Our results based on different cancer datasets demonstrate the association of patient subgroups identified by our method to relevant clinical and biological features.
Altogether we see our proposed model as a step towards a more effective integration of multi-omics data in the context of precision medicine by using modern data science techniques. As a next step we could use pathway profiles in a supervised learning context by changing the loss function of our multi-modal neural network accordingly.
Supplementary information
Additional file 1. Supplementary text, including links to further online material.
Additional file 2. PathME code (python) and additional analysis codes: https://github.com/AminaLEM/PathME.
Acknowledgements
We thank the University of Constantine 2 for supporting AL during her stay at the University of Bonn.
Abbreviations
- PathME
PATHway based Multi-modal sparse autoEncoders sNMF: sparse Non-negative Matrix Factorization
- SNF
Similarity Network Fusion
- SHAP
Shapley additive exPlanations
- SI
Silhouette Index
- TCGA
The Cancer Genome Atlas
- GDC
Genomic Data Commons
- GBM
Glioblastoma Multiforme
- CRC
Colorectal Cancer
- LSCC
Lung Squamous Cell Carcinoma
- BRCA
Breast Cancer
- NSLCC
Non-Small-Cell Lung Cancer
- ALL
Acute Lymphocytic Leukemia
- AML
Acute Myeloacytic Leukemia
- CNV
Copy Number Variation
- GISTC2
Genomic Identification of Significant Targets in Cancer
- FDR
False Discovery Rate
- OS
Overall Survival
- PFS
Progression Free Survival
- DFS
Disease Free Survival
- Nadam
Nesterov-accelerated Adam
- ATM
Ataxia-Telangiectasia Mutated
- CDC42
Cell Division Control 42
- CDH1
Cadherin 1
- EMT
Epithelial-tomesenchymal Transition
- ER
Estrogen Receptor
- ESR2
Estrogen Receptor 2
- FGF
Fibroblast Growth Factor
- FGF19
Fibroblast Growth Factor 19
- FGFR2
Fibroblast Growth Factor Receptor 2
- IDH1
Isocitrate Dehydrogenase 1
- IG1FR
Insulin-like Growth Factor 1 Receptor
- ITGA6
Integrin alpha-6
- ITGB4
Integrin beta-4
- MDM2
Murine Double Minute 2
- NFAT
Nuclear Factor of Activated T-cells
- PDGFR
Platelet-Derived Growth Factor Receptor
- PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-Kinase Catalytic subunit Alpha
- RPS6KB1
Ribosomal Protein S6 Kinase B1 TGF-β: Transforming Growth Factor-β
Authors’ contributions
AL implemented the code. AL and SO conducted the analysis. HF designed and guided the project. HF and AL drafted the manuscript. All authors have read the manuscript and agreed to its content.
Funding
AL was partially supported by the PhD student internship program of the University of Constantine 2. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data used in this work are publicly available via https://www.cbioportal.com, https://www.portal.gdc.cancer.go, http://firebrowse.org and https://www.bioconductor.org/packages/release/data/experiment/html/golubEsets.html. PathME code is available under https://github.com/AminaLEM/PathME.
Ethics approval and consent to participate
Does not apply - we used only published data.
Consent for publication
Not applicable.
Competing interests
Author HF has partially received salaries from UCB Biosciences GmbH during the time that this study was conducted. This was unrelated to this work and no influence on its content.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12859-020-3465-2.
References
- 1.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabrie S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17(1):98. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkins RD, Hon GC, Ren B (2011) Next-generation genomics: an integrative approach. Nature Reviews Genetics , URL http://www.nature.com/doifinder/10.1038/nrg2795. [DOI] [PMC free article] [PubMed]
- 5.Kristensen VN, Lingjærde OC, Russnes HG, Vollan HKM, Frigessi A, Børresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nature Reviews Cancer. 2014;14(5):299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad A, Fröhlich H. Integrating Heterogeneous omics Data via Statistical Inference and Learning Techniques. Genomics and Computational Biol. 2016;2(1):32. doi: 10.18547/gcb.2016.vol2.iss1.e32. [DOI] [Google Scholar]
- 7.Kirk P, Griffin JE, Savage RS, Ghahramani Z, Wild DL. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics. 2012;28(24):3290–3297. doi: 10.1093/bioinformatics/bts595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kormaksson M, Booth JG, Figueroa ME, Melnick A. Integrative model-based clustering of microarray methylation and expression data. Ann Appl Stat. 2012;6(3):1327–1347. doi: 10.1214/11-AOAS533. [DOI] [Google Scholar]
- 9.Serra A, Fratello M, Fortino V, Raiconi G, Tagliaferri R, Greco D. MVDA: a multi-view genomic data integration methodology. BMC Bioinformatics. 2015;16(1):261. doi: 10.1186/s12859-015-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Y, Savage RS, Markowetz F. Patient-specific data fusion defines prognostic Cancer subtypes. PLoS Comput Biol. 2011;7(10):e1002,227. doi: 10.1371/journal.pcbi.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11(3):333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 12.Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, Huse J, Ladanyi M, Sander C (2012) Integrative Subtype Discovery in Glioblastoma Using iCluster. PLOS ONE 7(4):e35,236, , URL http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0035236. [DOI] [PMC free article] [PubMed]
- 13.Liu J, Wang C, Gao J, Han J. Proceedings of the 2013 SIAM international conference on data mining, Proceedings, Society for Industrial and Applied Mathematics. 2013. Multi-view clustering via joint nonnegative matrix factorization; pp. 252–260. [Google Scholar]
- 14.Kim H, Park H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics. 2007;23(12):1495–1502. doi: 10.1093/bioinformatics/btm134. [DOI] [PubMed] [Google Scholar]
- 15.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, Liston DE, Low DKW, Newman SF, Kim J, Lee SI (2018) Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nature Biomedical Engineering 2(10):749–760, , URL https://www.nature.com/articles/s41551-018-0304-0. [DOI] [PMC free article] [PubMed]
- 16.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH (2009) PID: the Pathway Interaction Database. Nucleic Acids Research 37(Database issue):D674–D679, , URL https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2686461/. [DOI] [PMC free article] [PubMed]
- 17.Sales G, Calura E, Cavalieri D, Romualdi C (2012) graphite - a Bioconductor package to convert pathway topology to gene network. BMC Bioinformatics 13(1):20, , URL 10.1186/1471-2105-13-20. [DOI] [PMC free article] [PubMed]
- 18.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. Gistic2. 0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Saini HK, Sv D, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinton GE, Salakhutdinov RR. Reducing the Dimensionality of Data with Neural Networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 21.Vincent P, Larochelle H, Bengio Y, Manzagol PA (2008) Extracting and composing robust features with denoising autoencoders. In: proceedings of the 25th international conference on machine learning, ACM, pp 1096–1103..
- 22.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from Overfitting. J Machine Learning Research 15:1929–1958, URL http://jmlr.org/papers/v15/srivastava14a.html.
- 23.Simon N, Friedman J, Hastie T, Tibshirani R (2013) A Sparse-Group Lasso. Journal of Computational and Graphical Statistics 22(2):231–245, , URL 10.1080/10618600.2012.681250.
- 24.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58(1):267–288. [Google Scholar]
- 25.Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc. 2007;68(1):49–67. doi: 10.1111/j.1467-9868.2005.00532.x. [DOI] [Google Scholar]
- 26.Bengio Y. Practical recommendations for gradient-based training of deep architectures. In: neural networks: tricks of the trade. Heidelberg: Springer; 2012. pp 437–478.
- 27.Masters D, Luschi C (2018) Revisiting small batch training for deep neural networks. arXiv preprint arXiv:180407612.
- 28.Sutskever I, Martens J, Dahl G, Hinton G. International conference on machine learning. 2013. On the importance of initialization and momentum in deep learning; pp. 1139–1147. [Google Scholar]
- 29.Hinton G, Srivastava N, Swersky K. Neural networks for machine learning lecture 6a overview of mini-batch gradient descent. 2012. [Google Scholar]
- 30.Kingma DP, Ba JL (2015) Adam: Amethod for stochastic optimization. In: Proc. 3rd Int. Conf. Learn. Representations.
- 31.Dozat T. Incorporating Nesterov momentum into Adam. In: proceedings of 4th international conference on learning representations, workshop track. 2016. [Google Scholar]
- 32.Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep Network training by reducing internal covariate shift. arXiv:150203167 [cs] URL http://arxiv.org/abs/1502.03167, arXiv: 1502.03167.
- 33.Bergstra J, Yamins D, Cox DD (2013) Hyperopt: a python library for optimizing the hyperparameters of machine learning algorithms. In: proceedings of the 12th Python in science conference, Citeseer, pp 13–20.
- 34.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401(6755):788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 35.Carmona-Saez P, Pascual-Marqui RD, Tirado F, Carazo JM, Pascual-Montano A. Biclustering of gene expression data by non-smooth non-negative matrix factorization. BMC bioinformatics. 2006;7(1):78. doi: 10.1186/1471-2105-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statist Soc Series B. 1995;57:289–300. [Google Scholar]
- 37.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52(1):91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 39.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(12):4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comp and Applied Mathematics. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 41.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 43.Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc. 1971;66(336):846–850. doi: 10.1080/01621459.1971.10482356. [DOI] [Google Scholar]
- 44.Tomczak K, Czerwin’ska P, Wiznerowicz M. The cancer genome atlas (tcga): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Network CGAR, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guinney J, Dienstmann R, Wang X, De Reyni’es A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21(11):1350. [DOI] [PMC free article] [PubMed]
- 47.Maaten Lvd, Hinton G (2008) visualizing data using t-SNE. Journal of machine learning research 9(Nov):2579–2605, URL http://jmlr.org/papers/v9/vandermaaten08a.html.
- 48.John CR, Watson D, Russ D, Goldmann K, Ehrenstein M, Pitzalis C, Lewis M, Barnes M (2019) M3C: Monte Carlo reference-based consensus clustering. bioRxiv p 377002,. [DOI] [PMC free article] [PubMed]
- 49.Matsuda Y, Ueda J. Ishiwata T (2012) fibroblast growth factor receptor 2: expression, roles, and potential as a novel molecular target for colorectal cancer. Pathol Res Int. 2012. [DOI] [PMC free article] [PubMed]
- 50.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 51.Desnoyers L, Pai R, Ferrando R, Hötzel K, Le T, Ross J, Carano R, D’souza A, Qing J, Mohtashemi I, et al. Targeting fgf19 inhibits tumor growth in colon cancer xenograft and fgf19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27(1):85. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- 52.Laurent-Puig P, Paget-Bailly S, Vernerey D, Vazart C, Decaulne V, Fontaine K, Rousseau F, Elliott F, Quirke P, Richman S, et al (2015) Evaluation of mir 31 3p as a biomarker of prognosis and panitumumab benefit in ras-wt advanced colorectal cancer (acrc): analysis of patients (pts) from the piccolo trial.
- 53.Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, Zagatti B, Radova L, Svoboda M, Nemecek R, John S, Kiss I, Vyzula R, et al. Microrna expression profiling identifies mir-31-5p/3p as associated with time to progression in wild-type ras metastatic colorectal cancer treated with cetuximab. Oncotarget. 2015;6(36):38,695. doi: 10.18632/oncotarget.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonsson A, Hjalmarsson C, Falk P, Ivarsson ML. Stability of matrix metalloproteinase-9 as biological marker in colorectal cancer. Med Oncol. 2018;35(4):50. doi: 10.1007/s12032-018-1109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barzi A, Lenz AM, Labonte MJ, Lenz HJ. Molecular pathways: estrogen pathway in colorectal cancer. Clin Cancer Res. 2013;19(21):5842–5848. doi: 10.1158/1078-0432.CCR-13-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevanato Filho PR, Ju’nior SA, Begnami MD, de Oliveira Ferreira F, Nakagawa WT, RMSB S, Bezerra TS, Boggiss PE, Lopes A. Estrogen receptor β as a prognostic marker of tumor progression in colorectal cancer with familial adenomatous polyposis and sporadic polyps. Pathology Oncol Research. 2018;24(3):533–540. doi: 10.1007/s12253-017-0268-5. [DOI] [PubMed] [Google Scholar]
- 57.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14(8):16,365–16,385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mekenkamp LJ, Tol J, Dijkstra JR, de Krijger I, Vink-Bo¨rger ME, van Vliet S, Teerenstra S, Kamping E, Verwiel E, Koopman M, et al (2012) Beyond kras mutation status: influence of kras copy number status and micrornas on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer 12(1):292. [DOI] [PMC free article] [PubMed]
- 59.Sun P, Sun D, Wang X, Liu T, Ma Z, Duan L. miR-206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancer. Cancer Biomarkers: Section A of Disease Markers. 2015;15(4):391–396. doi: 10.3233/CBM-150489. [DOI] [PubMed] [Google Scholar]
- 60.Tian M, Ma W, Chen Y, Yu Y, Zhu D, Shi J, Zhang Y. Impact of gender on the survival of patients with glioblastoma. Bioscience Reports. 2018;38(6):BSR20180,752. doi: 10.1042/BSR20180752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smrdel U, Popovic M, Zwitter M, Bostjancic E, Zupan A, Kovac V, Glavac D, Bokal D, Jerebic J. Long-term survival in glioblastoma: methyl guanine methyl transferase (mgmt) promoter methylation as independent favourable prognostic factor. Radiol Oncol. 2016;50(4):394–401. doi: 10.1515/raon-2015-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 63.Cenci T, Martini M, Montano N, D’alessandris QG, Falchetti ML, Annibali D, Savino M, Bianchi F, Pierconti F, Nasi S, et al. Prognostic relevance of c-myc and bmi1 expression in patients with glioblastoma. Am J Clin Pathol. 2012;138(3):390–396. doi: 10.1309/AJCPRXHNJQLO09QA. [DOI] [PubMed] [Google Scholar]
- 64.Pearson JR, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduction Targeted Therapy. 2017;2:17,040. doi: 10.1038/sigtrans.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seystahl K, Papachristodoulou A, Burghardt I, Schneider H, Hasenbach K, Janicot M, Roth P, Weller M. Biological role and therapeutic targeting of tgf-β3 in glioblastoma. Mol Cancer Ther. 2017;16(6):1177–1186. doi: 10.1158/1535-7163.MCT-16-0465. [DOI] [PubMed] [Google Scholar]
- 66.Bruna A, Darken RS, Rojo F, Ocan˜a A, Pen˜uelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, et al (2007) High tgfβ-smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the pdgf-b gene. Cancer Cell 11(2):147–160. [DOI] [PubMed]
- 67.Pe’cina-Slaus N, Nikuˇseva Marti’c T, Kokotovi’c T, Kuˇsec V, Tomas D, Hraˇs’can R (2011) Axin-1 proteinˇ expression and localization in glioblastoma. Collegium antropologicum 35(1):101–106. [PubMed]
- 68.Stewart RL, O’connor KL. Clinical significance of the integrin α6β4 in human malignancies. Lab Investig. 2015;95(9):976. doi: 10.1038/labinvest.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costantini RM, Falcioni R, Battista P, Zupi G, Kennel SJ, Colasante A, Venturo I, Curcio CG, Sacchi A. Integrin (α6/β4) expression in human lung cancer as monitored by specific monoclonal antibodies. Cancer Res. 1990;50(18):6107–6112. [PubMed] [Google Scholar]
- 70.Ke Y, Zhao W, Xiong J, Cao R. mir-149 inhibits non-small-cell lung cancer cells emt by targeting foxm1. Biochem Res Int. 2013:506731. 10.1155/2013/506731. Epub 2013 May 16. [DOI] [PMC free article] [PubMed]
- 71.Ko JL, Cheng YW, Chang SL, Su JM, Chen CY, Lee H. Mdm2 mrna expression is a favorable prognostic factor in non-small-cell lung cancer. Int J Cancer. 2000;89(3):265–270. doi: 10.1002/1097-0215(20000520)89:3<265::AID-IJC9>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 72.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. Microrna classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu K, Wei L, Chu M. The prognostic value of mir-21 and mir-155 in non-small-cell lung cancer: a meta-analysis. Jpn J Clin Oncol. 2013;43(8):813–820. doi: 10.1093/jjco/hyt084. [DOI] [PubMed] [Google Scholar]
- 74.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van De Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98(19):10,869–10,874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Network CGA, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silwal-Pandit L, Vollan HKM, Chin SF, Rueda OM, McKinney S, Osako T, Quigley DA, Kristensen VN, Aparicio S, Børresen-Dale AL, et al. Tp53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20(13):3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 77.Rubtsova SN, Zhitnyak IY, Gloushankova NA. A novel role of e-cadherin-based adherens junctions in neoplastic cell dissemination. PLoS One. 2015;10(7):e0133,578. doi: 10.1371/journal.pone.0133578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farabaugh SM, Boone DN, Lee AV. Role of igf1r in breast cancer subtypes, stemness, and lineage differentiation. Front Endocrinol. 2015;6:59. doi: 10.3389/fendo.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M, Aldahmash A, Alajez NM. Circulating micrornas in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8(9):e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Li J, Lai XN, Jiao XQ, Xiong JP, Xiong LX. Focus on cdc42 in breast cancer: new insights, target therapy development and non-coding rnas. Cells. 2019;8(2):146. doi: 10.3390/cells8020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van der Hage J, van den Broek L, Legrand C, Clahsen P, Bosch C, Robanus-Maandag E, van de Velde C, Van de Vijver M. Overexpression of p70 s6 kinase protein is associated with increased risk of locoregional recurrence in node-negative premenopausal early breast cancer patients. Br J Cancer. 2004;90(8):1543. doi: 10.1038/sj.bjc.6601741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary text, including links to further online material.
Additional file 2. PathME code (python) and additional analysis codes: https://github.com/AminaLEM/PathME.
Data Availability Statement
All data used in this work are publicly available via https://www.cbioportal.com, https://www.portal.gdc.cancer.go, http://firebrowse.org and https://www.bioconductor.org/packages/release/data/experiment/html/golubEsets.html. PathME code is available under https://github.com/AminaLEM/PathME.