Abstract
The diverse fruit colors of peppers (Capsicum spp.) are due to variations in carotenoid composition and content. Mature fruit color in peppers is regulated by three independent loci, C1, C2, and Y. C2 and Y encode phytoene synthase (PSY1) and capsanthin-capsorubin synthase (CCS), respectively; however, the identity of the C1 gene has been unknown. With the aim of identifying C1, we analyzed two pepper accessions with different fruit colors: Capsicum frutescens AC08-045 and AC08-201, whose fruits are light yellow and white, respectively. Ultra-performance liquid chromatography showed that the total carotenoid content was six times higher in AC08-045 than in AC08-201 fruits, with similar composition of main carotenoids and slight difference in minor components. These results suggest that a genetic factor in AC08-201 may down-regulate overall carotenoid biosynthesis. Analyses of candidate genes related to carotenoid biosynthesis and plastid abundance revealed that both accessions carry non-functional alleles of CCS, golden2-like transcription factor (GLK2), and PSY1. However, a nonsense mutation (C2571T) in PRR2, a homolog of Arabidopsis pseudo response regulator2-like (APRR2), was present in only AC08-201. In a population derived from a cross between AC08-045 and AC08-201, a SNP marker based on the nonsense mutation co-segregated fully with fruit color, implying that the mutation in PRR2 may cause the white color of AC08-201 fruits. Transmission electron microscopy (TEM) of AC08-201 fruit pericarp also showed less developed granum structure in chloroplast and smaller plastoglobule in chromoplast compared to those of AC08-045. Virus-induced gene silencing (VIGS) of PRR2 significantly reduced carotenoid accumulation in Capsicum annuum ‘Micropep Yellow’, which carries non-functional mutations in both PSY1 and CCS. Furthermore, sequence analysis of PSY1, CCS, and PRR2 in other white pepper accessions of C. annuum and Capsicum chinense showed that they commonly have non-functional alleles in PSY1, CCS, and PRR2. Thus, our data demonstrate that the fruit color locus C1 in Capsicum spp. corresponds to the gene PRR2.
Keywords: mature fruit color, pepper (Capsicum spp.), carotenoid, plastid, Pseudo response regulator2-like gene
Introduction
Carotenoids, also called tetraterpenoids, are derived from 8 isoprene units and contain 40 carbons in their polyene backbone. Carotenoids are essential for human nutrition and health as they provide dietary sources of provitamin A and serve as antioxidants that reduce the incidence of many diseases, including age-related eye diseases, cardiovascular diseases, and cancers (Fraser and Bramley, 2004). Carotenoids also play crucial roles in plants, including photosynthesis and photoprotection, and provide precursors for the biosynthesis of phytohormones such as abscisic acid (ABA) and strigolactones (Domonkos et al., 2013; Nambara and Marion-Poll, 2005; Al-Babili and Bouwmeester, 2015). The carotenoid biosynthetic pathway and its associated enzymes have been well elucidated in Solanaceous plants. Carotenoid biosynthesis begins with the formation of phytoene with the aid of phytoene synthase (PSY). Through a series of desaturation and isomerization steps, phytoene is converted into lycopene, which is the major carotenoid component in mature tomato (Solanum lycopersicum). Whereas the red color in tomato is due to lycopene accumulation, the red color in peppers results from the accumulation of capsanthin and capsorubin (Paran and van der Knaap, 2007). This pepper-specific process is regulated by capsanthin-capsorubin synthase (CCS), which converts downstream products of lycopene, antheraxanthin and violaxanthin into capsanthin and capsorubin, respectively (Bouvier et al., 1994).
The distinct colors of mature pepper fruits result from the conversion of chloroplasts to chromoplasts during ripening. Compared to chloroplasts, chromoplasts have a greater capacity to synthesize and sequester carotenoids, and as a result the fruits change from their immature green to display ivory to red colors in maturity (Sun et al., 2018). Based on inheritance studies of pepper fruit color, a three-locus model (C1, C2, and Y) was proposed by Hurtado-Hernandez and Smith (1985). In this model, mature fruit color can be classified into eight groups according to their allelic combinations, ranging from white (c1 c2 y) to red (C1 C2 Y). Candidate gene analyses of carotenoid biosynthetic genes revealed that the C2 and Y loci are the sites of the genes PSY1 and CCS, respectively (Popovsky and Paran, 2000; Huh et al., 2001; Lefebvre et al., 1998), but the nature of the third locus, C1, remained to be discovered. Since the identification of the PSY1 and CCS genes, many researchers have tried to understand fruit-color variation in peppers based on the allelic variations of these two loci. However, since fruit color is not only qualitative, but can also be quantitative, explanations of fruit-color variation based on only PSY1 and CCS are limited.
Other researchers have analyzed quantitative variation in color intensity in Solanaceous fruits. Brand et al. (2012) revealed that the chlorophyll content of immature pepper fruits is mainly controlled by two quantitative trait loci (QTLs) designated pc8 and pc10, respectively. pc10 encodes the golden2-like transcription factor (GLK2), which regulates chloroplast compartment size during the early stages of fruit development (Brand et al., 2014). The identity of pc8 had not been known, although several transcription factor genes have been regarded as candidates, including Arabidopsis pseudo response regulator2-like (APRR2), as the gene of pc8 (Brand et al., 2014). Only recently, it was identified as the pepper homolog of transcription factor LSD ONE LIKE1 (LOL1), which eventually termed pc1 as further mapping showed the QTL in chromosome 1. However, mutation of CcLOL1 showed significant difference in fruit chlorophyll, but not in carotenoid contents (Borovsky et al., 2019). Similar to the case of CcLOL1 mutant pepper, one C. annuum pepper cultivar that is white when immature, a nonsense mutation in APRR2 is strongly associated with color intensity in the immature fruits (Pan et al., 2013). Although both GLK2 and APRR2 affect the color of immature pepper fruits, overexpression of each gene in tomato led to increased levels of carotenoids as well as chlorophyll (Powell et al., 2012; Pan et al., 2013). By contrast, the possible involvement of GLK2 and APRR2 in the color of mature pepper fruits was unknown.
In this study, we performed a candidate analysis to identify the gene in the C1 locus that controls white fruit color. Six carotenoid biosynthetic genes along with two plastid related genes were analyzed. We showed that the white color of C. frutescens AC08-201 is due to a nonsense mutation in PRR2, as well as PSY1 and CCS mutations. Expression analysis of PRR2 showed that this gene is specifically expressed at the immature fruit stage. A genetic analysis demonstrated that the nonsense mutation co-segregated fully with the fruit color in a population derived from a cross between AC08-045 and AC08-201. Virus-induced gene silencing of the PRR2 gene of C. annuum cultivar MY demonstrated that down-regulation of PRR2 resulted in lighter color of both immature and mature fruits. Furthermore, sequence analysis of PRR2 showed that two additional white pepper cultivars, from C. annuum and C. chinense, also had non-functional alleles in PSY1, CCS, and PRR2. In conclusion, we have demonstrated that PRR2 is the gene corresponding to C1.
Materials and Methods
Plant Materials
Capsicum frutescens accessions AC08-045 and AC08-201 were selected from the germplasm of the Horticultural Crops Breeding and Genetics lab (Seoul National University, South Korea). Both accessions have bush-type architecture and set numerous fruit. A total of 127 F2 individuals were obtained from a cross between AC08-045 and AC08-201, and this population was grown in the experimental greenhouse of Seoul National University (Suwon, South Korea). Dominance of light yellow over white fruit color was confirmed in an F1 hybrid.
Capsicum annuum ‘MicroPep Yellow (MY)’ was used for VIGS analysis because of its fast growth rate and short internode length. MY peppers have dark green immature fruits and yellow mature fruits. MY has non-functional mutations in PSY1 and CCS. C. chinense ‘Habanero White’ and ‘White Moruga’ and C. annuum ‘Chupetino White’ were purchased from Mojo Pepper (Italy) and grown in the greenhouse of Seoul National University (Seoul, South Korea). All three cultivars have white mature fruit color.
Nucleic Acid Extraction
Genomic DNA (gDNA) was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Lee et al., 2017). Leaf tissues were homogenized using 3-mm steel beads with the aid of TissueLyserII (Qiagen, Hilden, Germany). The concentration and purity of gDNA were measured with a Nanodrop spectrophotometer (BioTek, Winooski, VT), and the DNA was then diluted to a final concentration of 20 ng/μL in distilled water for further experiments. Total RNA was also extracted from various tissues. For AC08-045 and AC08-201, RNA was extracted from roots, stems, leaves, and three stages of fruit. RNA was also extracted from immature fruits of the virus-inoculated MY used for VIGS analysis, and from young leaves of C. chinense ‘Habanero White’ and ‘White Moruga’ C. annuum ‘Chupetino White.’ Total RNA was extracted using MG RNAzol kit (MGmed, Seoul, South Korea) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 2 μg of RNA using the EasyScript Reverse Transcriptase kit (TransGen, Beijing, China) with oligo (dT) primers. The resulting cDNAs were used for further analyses.
PCR Amplification and Sequencing
PCR was performed in a total volume of 25 μL using PrimeStar GXL DNA polymerase (Takara Bio, Kusatsu, Japan) with 100 ng of each gDNA template and 0.5 μL of 10 pmol gene-specific primers for the carotenogenic genes PSY1, PSY2, Lcyb, CrtZ-2, ZEP, and CCS. Five of these genes were amplified using one primer set to amplify the whole gene, while three sets of primers were used for ZEP amplification because of its large size: a gDNA length of 4,802 bp and a cDNA length of 1,986 bp. Primers for ZEP were designed to cover all exons of the gene and to overlap each amplicon. For GLK2 and PRR2, cDNAs from immature fruits were used as amplification templates, as both genes contain long introns (Table 1). For reverse-transcription PCR (RT-PCR), 2 μL of 4 × -diluted cDNA was used as a template using primers identical to those used for gDNA PCR with following reaction mixture: 0.3 μL EX Taq (Takara, Japan), 2.5 μL 10 × PCR buffer, 2 μL 2.5 mM dNTPs, 0.5 μL 10 pmol primers, and 17.2 μL triple distilled water. The RT-PCR conditions were 28 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 90 s. The resulting amplicons were separated on a 1% agarose gel and DNA was recovered using a LaboPass PCR clean-up kit (Cosmo Genetech, Seoul, South Korea). Elution products were either directly sequenced or cloned into a modified T-blunt vector (SolGent, Daejeon, South Korea) prior to sequencing. Plasmid DNA was extracted using an AccuPrep plasmid mini extraction kit (Bioneer, Daejeon, South Korea). Sanger sequencing was performed at Macrogen (Seoul, South Korea) and the nucleotide sequences were analyzed with Lasergene’s SeqMan program (DNASTAR, Madison, WI).
TABLE 1.
List of primers used to amplify fruit-color-related genes.
| Gene | Gene no. | gDNA (bp) | CDS (bp) | Primer sequence (5′ to 3′) |
| PSY1 | CA04g04080 | 2,844 | 1,260 | F: ATGTCTGTTGCCTTGTTATGG R: ATCCTGATTTCATGTTCTTGTAGAAG |
| PSY2 | CA02g20350 | 2,985 | 1,299 | F: ATGTCTGTTGCTTTGTTGTGG R: CAACTTCATTCATGTCTTTGTTAGTG |
| Lcyb | CA05g00080 | 1,497 | 1,497 | F: ATGGATACGCTCTTGAGAACC R: TCATTCTTTATCCTGTAACAAATTG |
| CrtZ-2 | CA03g25820 | 2,025 | 948 | F: ATGGCTGCTGAAATTTCAAT R: CTTTGATCATAATCTCTTCGAAC |
| ZEP (fragment 1) | CA02g10990 | 1,760 | - | F: TCCTTTCACTTCCTTTGGCCT R: AGCTTCACTGTGTCCGAACA |
| ZEP (fragment 2) | 1,952 | - | F: GAATGGACAACGGTTTACAGGT R: CAAACCACAGGATATCAACTTCC | |
| ZEP (fragment 3) | 1,709 | - | F: GGACTTGGGAATGCCTCTAATG R: ATGCTGTACAAATTTCCCGTTT | |
| CCS | CA06g22860 | 1,497 | 1,497 | F: ATGGAAACCCTTCTAAAGCCT R: TCAAAGGCTCTCTATTGCTAG |
| GLK2 | CA10g02900 | 5,859 | 942 | F: ATGATGCTTGTTGTATCTACACCA R: GAGGTATTTTTGTAATCCCTTGAC |
| PRR2 | Capana01g000809 | 5,588 | 1,764 | F: ATGATTTGCATTGAGGATGAA R: TCATCTCCAACATCGAGAGC |
Carotenoid Extraction and Saponification of AC08-045 and AC08-201
Mature fruits were harvested and the pericarp tissue was sectioned and collected without placenta for carotenoid extraction. Three biological replicates of fruits for each AC08-045 and AC08-201 were used. The carotenoid pigments were extracted according to method of Collera-Zúñiga et al. (2005) but using different volumes of chemicals. Acetone (20 mL) was added to 1 g of freeze-dried pericarp and incubated at 4°C for 20 h to extract the pigments. The extracts were dried by evaporation and incubated with 3 mL acetone, 3 mL methanol, and 1 mL 30% potassium hydroxide/methanol at room temperature for 2 h 30 min in dark condition to avoid carotenoid degradation. After saponification, the extracts were transferred to a separatory funnel with 20 mL diethyl ether, shaken, and left to settle. The extracts were combined with 2 mL of 10% NaCl to separate the phases and to transfer the pigments to the ether. Subsequently, 2 mL of 2% Na2SO4 was added to remove all water in the ether phase. The ether phase was collected and dried using vacuum concentrator. The resulting dried residue was dissolved in 2 mL acetone and filtered through an Acrodisc syringe filter (13 mm, 0.2 μm) (Pall Corporation, New York, NY).
Carotenoid Analysis by Ultra-Performance Liquid Chromatography
Carotenoids extracted from mature fruit pericarps of the two AC08 accessions were analyzed using an Acquity UPLC-H-Class system (Waters, Milford, MA). Separation was performed using an Acquity UPLC HSS T3 column (2.1 × 100, 1.8 μm) at 35°C. The mobile phase was a binary solvent system consisting of phase A (acetonitrile/methanol/methylene chloride, 65/25/10, v/v/v) and phase B (distilled water). The gradients were programmed as previously described (Kim et al., 2016). The UV wavelength was set to 450 nm. For qualitative and quantitative analysis of carotenoids, 11 standards were purchased from Sigma-Aldrich (St. Louis, MO): antheraxanthin, capsanthin, capsorubin, lutein, neoxanthin, violaxanthin, zeaxanthin, α-carotene, α-cryptoxanthin, β-carotene, and β-cryptoxanthin.
High-Resolution Melting Analysis
A pair of primers was designed to amplify the short genomic region (<300 bp) centered on the target PRR2 target SNP. Genotyping analysis was performed using a Rotor-Gene 6000 real-time PCR (Qiagen, Hilden, Germany). The reaction mixture was prepared in a total volume of 20 μL containing 80 ng of DNA, 0.3 μL of R Taq (Takara Bio), 2 μL of 10 × PCR buffer, 2 μL of 2.5 mM dNTPs, 0.6 μL of SYTO 9 (Thermo Fisher Scientific, Waltham, MA), and 0.5 μL of 10 pmol primers. Sequences of forward and reverse primers used in the study were as following: 5′-TTTGAAAGAGGAGAATGGTTCA-3′ (forward) and 5′-TGAGCTATGGGGACCAGAAG-3′ (reverse). The PCR conditions consisted of 95°C for 5 min; 55 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s; and 60°C for 1 min. For high-resolution melting (HRM) analysis, the temperature was increased 0.1°C per every minute from 65°C to 90°C. Melting curve and HRM-normalized graphs were analyzed using Rotor-Gene Q series software 2.1.0.
Transmission Election Microscopy (TEM)
TEM analysis was conducted using immature and mature fruits of AC08-045 and AC08-201. Both immature and mature fruit pericarps of AC08 accessions were longitudinally sliced to a thickness of 1 mm. Specimen preparation for TEM was done following the method described by Seo et al. (2014). Polymerized samples were than trimmed, sliced to a thickness of 500 nm, stained with toluene blue, and examined under a light microscope. After confirming plastid existence and localization, regions with plastids were sliced to a thickness of 80 nm and observed using a JEM-1400 Flash (JEOL, Tokyo, Japan) (120 kV) transmission electron microscope. Processes from trimming to TEM were kindly done by the Dental Research Institute (Seoul National University, Seoul).
Virus-Induced Gene Silencing of PRR2
Ligation-independent cloning (LIC) for pTRV2-PRR2 construction was conducted as described by Kim et al. (2017). The partial coding sequence of PRR2 was amplified with the following primers containing 15-bp adapter sequences (underlined): 5′-CGACGACAAGACCCTTAAGTCC TCCTGGGCAACAA-3′ (forward) and 5′- GAGGAGAA GAGCCCTGGTTCAGCAGAATGACTAATGC-3′ (reverse). Purified PCR product was treated with T4 DNA polymerase (Enzymatics, Beverly, MA) along with 5 × blue buffer and 10 mM dATP. PstI digested pTRV2-LIC vector was also treated with T4 DNA polymerase and 5 × blue buffer with 10 mM dTTP. Both reaction mixtures were incubated at 22°C for 30 min followed by 75°C for 20 min. A total of 30 ng of PCR product and 400 ng of TRV2-LIC vector were mixed and incubated at room temperature for 15 min. The mixture was transformed into Escherichia coli DH5α competent cells (TransGen). Plasmids were extracted from transformants and sequenced by Macrogen. Plasmids with intact sequences were introduced into Agrobacterium tumefaciens strain GV3101 through electroporation at 2.0 kV. For a negative control for VIGS, the construct pTRV2-GFP was used. The pTRV2-LIC and pTRV2-GFP constructs were kindly provided by Dr. Doil Choi at Seoul National University.
Agrobacterium carrying pTRV1, pTRV2-GFP, and pTRV2-PRR2 were grown overnight at 30°C in 5 mL LB medium containing rifampicin (50 μg/mL) and kanamycin (50 μg/mL). 4 mL of Agrobacterium cultures was harvested by centrifugation and resuspended in 10 mM MES, 10 mM MgCl2, and 200 μM acetosyringone solution to a final OD600 of 0.7. Cell suspensions were incubated in a rocking incubator at room temperature for 3 h. Cell cultures containing pTRV1 and each pTRV2 construct were mixed at a 1:1 ratio and infiltrated into the abaxial side of both cotyledons of MY seedlings. Inoculated plants were incubated at 16°C in the dark for 1 day and then grown at 25°C with a 16/8-h light/dark photoperiod.
Expression Analysis of PRR2 in VIGS-Treated Fruit Using RT-PCR
Mature fruits were harvested from pTRV2-GFP- or pTRV2-PRR2-inoculated MY plants four months after inoculation. Segments of the mature pericarp displaying lighter colors were excised and their RNA was extracted. RT-PCR was conducted to verify the gene silencing using the same reaction mixture and primers to the method above for PCR and sequencing. The RT-PCR conditions were 28 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 90 s. The Actin was used for internal control with following primers: 5’-ATTCTCACCTTGAAGTATCCCA-3’ (forward) and 5’-ATAGCAACATACATGGCAGG-3’ (reverse). Three biological replicates were used for reproducibility.
Carotenoid Extraction and Saponification of VIGS-Treated Fruits
Carotenoids were extracted as described by Yoo et al. (2017), with some modifications. The samples were put on ice, and the whole procedure was done under dim light to prevent carotenoid oxidation and degradation. The lighter-colored pTRV2-PRR2-inoculated pericarps and the pTRV2-GFP-inoculated pericarps were diced, immediately frozen in liquid nitrogen, and freeze-dried using a vacuum freezer.
The freeze-dried tissues were ground into fine powder, and about 50 mg of tissue powder was aliquoted into a 2-mL tube with two glass beads. The upper phases of saponified samples were collected and concentrated using a vacuum concentrator until they were fully dried. The dried samples were dissolved with 500 μL HPLC-grade acetone (Honeywell) with sonication, filtered using a 0.2-μm syringe filter (Acrodisc LC 13 mm syringe filter, PVDF membrane; Pall, NY, United States), and bottled in a 1.5-mL HPLC amber vial to prevent light exposure. HPLC analysis of carotenoids was performed by the NICEM chromatography lab (Seoul National University). Seven carotenoids were used as standards: capsanthin, capsorubin, lutein, zeaxanthin, α-carotene, β-carotene, and β-cryptoxanthin.
Results
The White-Fruited Accession AC08-201 Accumulates Minimal Levels of Carotenoids
Capsicum frutescens AC08-045 bears dark green immature fruit, which change to yellow and then light yellow as they ripe (Figure 1A). AC08-201 fruits start out light green and change to off-white and finally white at maturity (Figure 1B). F1 hybrids obtained from a cross between AC08-045 and AC08-201 showed dominance of light yellow over white (Figure 1C); consistently with this, the F2 population showed a roughly 3:1 ratio of dark green/light yellow to light green/white.
FIGURE 1.
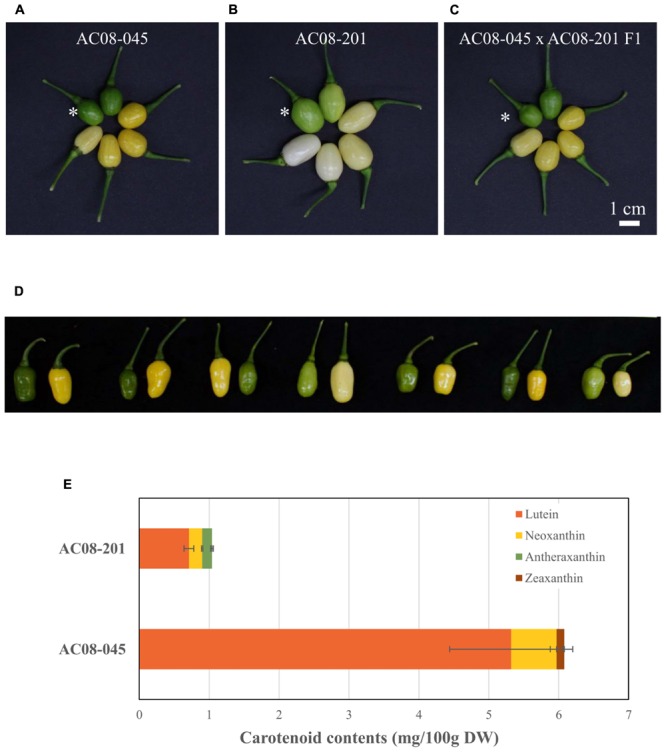
Pepper fruits and carotenoid profiles of AC08-201 and AC08-201 showed less carotenoid accumulation in AC08-201 fruit. (A–C) Pepper fruits are displayed in a clockwise di ection according to the ripening stages. An asterisk indicates fully expanded immature fruits. (A) In AC08-045, fruit color changes from dark green to yellow and then light yellow. (B) In AC08-201, fruit color starts out light green and becomes light yellow to white. (C) Fruit color of an F1 hybrid is similar to that of AC08-045. Scale bar, 1 cm. (D) F2 individuals from cross between AC08-045 and AC08-201. Seven pairs of immature and mature fruit from each F2 individuals were selected from F2 population and shown in the figure. (E) Carotenoid profiles of AC08-201 and AC08-201. Carotenoids extracted from mature fruit pericarp. Levels of four major carotenoids, lutein, neoxanthin, antheraxanthin, and zeaxanthin, are shown in the graph. Horizontal bars indicate standard deviations of each carotenoid content.
To investigate the differences in carotenoid composition and levels between the two accessions, we analyzed the carotenoid contents of their mature fruits by UPLC. Among 11 carotenoids, only three were detected in each accession; lutein and neoxanthin in both accessions, zeaxanthin only in AC08-045 and antheraxanthin only in AC08-201 (Figure 1E and Supplementary Table S1). Lutein was the major carotenoid in both accessions. In AC08-045, it accounted for 88% of total carotenoid content, followed by neoxanthin and zeaxanthin. In AC08-201, lutein accounted for 70% of total carotenoids, followed by neoxanthin and antheraxathin. The main differences between two accessions were total carotenoids content and the ratio of zeaxanthin and antheraxanthin in each accession: the total carotenoid content was six times higher in AC08-045 (6.07 ± 0.97 mg/100 g DW) than in AC08-201 (1.02 ± 0.08 mg/100 g DW). Although the colors of zeaxanthin and antheraxanthin are slightly different, these two pigments were least in each accession. These results suggest that the color difference between the two accessions is due to their abundance rather than composition of carotenoids.
AC08-201 Has Mutations in CCS, GLK2, PRR2, and PSY1
To identify the gene(s) controlling the carotenoid levels in AC08-201, we amplified fruit-color-related genes and sequenced the resulting amplicons. We analyzed six carotenoid biosynthetic genes, PSY1, PSY2, Lcyb, CrtZ-2, ZEP, and CCS, along with two genes regulating plastid compartment size, GLK2 and PRR2. Amplicons of the genes except GLK2 showed the sizes expected based on the reference genome (CM334 v1.55), and no size differences between the two accessions were observed in any of the candidate genes, although for GLK2, the amplicons of both accessions were slightly smaller than expected based on the reference genome (Supplementary Figure S1).
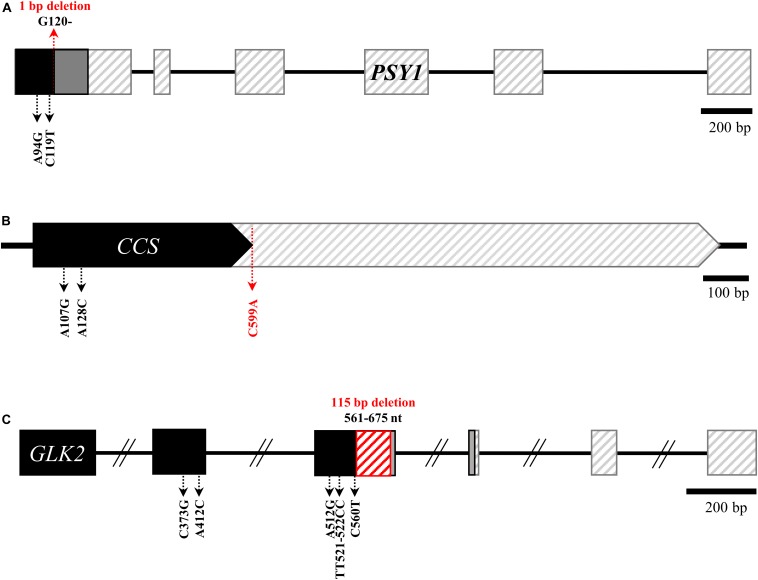
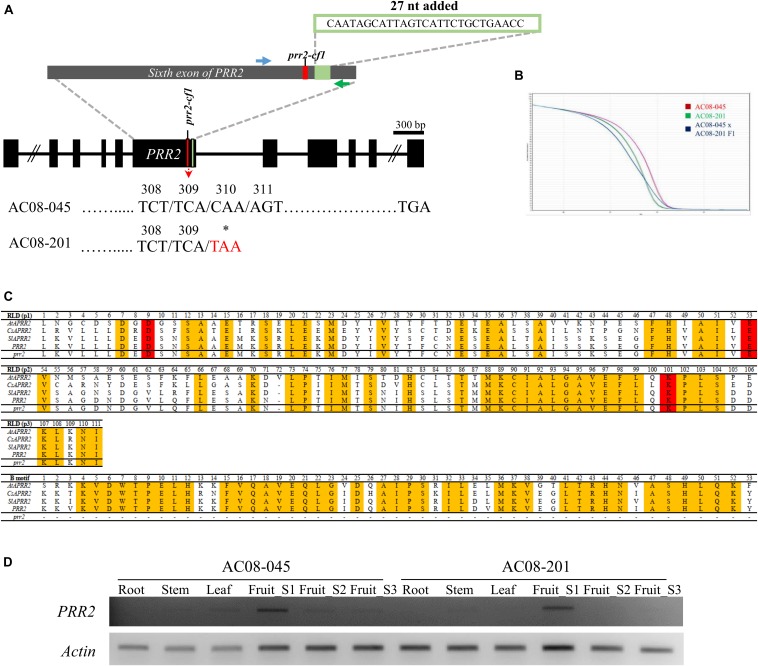
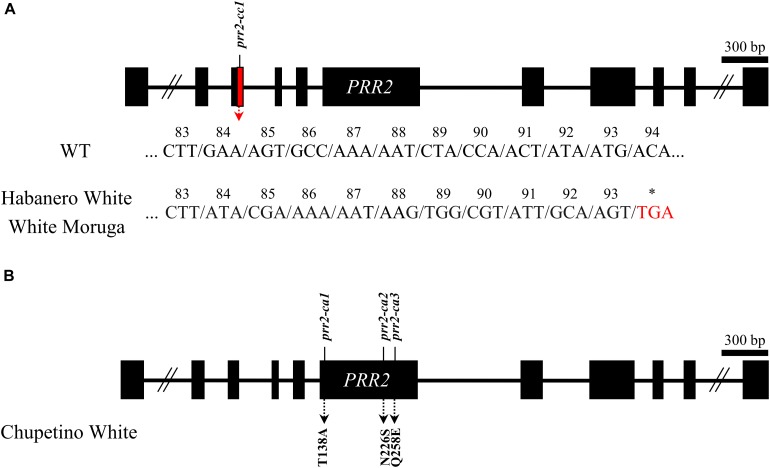
Sequence analyses revealed no mutations causing amino acid changes in PSY2, Lcyb, CrtZ-2, or ZEP. By contrast, we identified the same structural mutations of GLK2, PSY1, and CCS in both accessions, and a mutation in PRR2 in AC08-201 only. The three mutations in both accessions were a frameshift mutation due to a 1-bp deletion at nucleotide (nt) position 120 in PSY1 (Figure 2A); a nonsense mutation (serine to stop codon) in CCS due to a cytosine-to-adenine substitution at nt 599 (Figure 2B); and a 115-bp deletion in GLK2 from nt 561 to nt 675, which causes premature translational termination (Figure 2C). The alignment of the PSY1, CCS, and GLK2 coding sequences between the AC08 accessions and CM334 is shown in Supplementary Material S1. The PRR2 mutation in AC08-201 was a nonsense mutation (cytosine to thymine) at nt 928 (Figure 3A). We designed a HRM marker to discriminate this mutation from the wild-type allele of AC08-045 and used this in our further studies (Figures 3A,B).
FIGURE 2.
Common background mutations in PSY1, CCS, and GLK2 of AC08-045 and AC08-201. (A–C) Common mutations in the genes PSY1, CCS, and GLK2 of AC08. In alleles with early stop codons, non-synonymous mutations downstream from those stop codons are omitted. Major mutation of each gene are highlighted in red letters. Black region indicates normal transcription, and gray regions indicate frameshifted regions. Regions after early translation termination are indicated with diagonal lines. The 115-bp deletion in GLK2 is indicated with red diagonal lines.
FIGURE 3.
Mutation and HRM markers of PRR2 in AC08-045 and AC08-201, alignment of PRR2 domain between Arabidopsis and three horticultural crops, and expression analysis of PRR2. (A) Comparison of PRR2 variation between AC08-045 and AC08-201. Variation in the nucleotide sequence and resulting codon change are highlighted in red. A premature stop codon due to a 1-bp substitution was detected in AC08-201. The sixth exon of PRR2 is expanded in the gray exon. Nucleotide sequence of 27-nt insertion observed in this study compared to Zunla v2.0 (Capana01g000809) is shown above. The positions of the designed HRM marker are shown in blue (forward primer) and green (reverse primer). Asterisks indicate stop codons. (B) HRM genotyping of PRR2. Melting curves are differentiated according to the accessions. Red, green, and blue lines indicate maternal, paternal, and heterozygous type of PRR2 genotype. x axis shows temperature and y axis shows normalized fluorescence. (C) Alignment of PRR2 domain between Arabidopsis and three horticultural crops. AtAPRR2, CsAPRR2, SlAPRR2, and PRR2 refer to the PRR2 genes of Arabidopsis thaliana, Cucumis sativus, Solanum lycopersicum, and Capsicum frutescens, respectively. RLD for receiver-like domain; B-motif for DNA-binding domain. (D) Expression pattern of PRR2 in various tissues. Results of RT-PCR amplification of PRR2. Roots, stems, leaves, and three stages of fruits were analyzed. In both AC08-045 and AC08-201, PRR2 was exclusively expressed in immature stage (Fruit_S1). Actin served as a control. S stands for stage.
We compared the truncated protein sequences of AC08-201 to previously reported sequences from species including Arabidopsis, cucumber (Cucumis sativus), and tomato (Figure 3C). Sequence alignment analysis revealed that the deleted region is crucial for the protein’s enzymatic activity, as it contains a DNA-binding motif conserved in transcription factors and a functional domain for nuclear localization (Jiao et al., 2017).
White Color in Peppers Is Controlled by a Single Recessive Gene
To test whether the nonsense mutation in PRR2 resulted in the lighter fruit color in AC08-201, we prepared a population segregating for white fruit color by crossing AC08-045 and AC08-201. All fruits from the F1 progeny showed a light yellow color indicating that light yellow is dominant over white color. In the F2 population, light yellow and white fruits were observed in a ratio of 170:57, which fit a 3:1 ratio with an χ2 value of 0.001468 (P-value 0.9694) (Table 2). During phenotyping, we observed an interesting trend between immature fruit color and mature fruit color. Fruits with a dark green immature color became light yellow when ripe, whereas fruits with light green immature color turn white when fully mature. This trend of immature fruits with more intense color (dark green) becoming more dark mature fruits (light yellow) indicates that the white fruit-color phenotype is related to plastid development. To test the co-segregation of the mature fruit phenotype and the PRR2 genotype, we used the HRM marker targeting the nonsense mutation (C928T in cDNA, C2571T in gDNA) (Figure 3B). The genotypes co-segregated completely with the colors, which makes PRR2 a strong candidate gene for controlling the white fruit color in AC08-201.
TABLE 2.
Segregation of phenotypes in a F2 population.
| Population | Size | Phenotype | Expected ratio | p-value | |
| Dark green then light yellow | Light green then white | ||||
| AC08-045 × AC08-201 F2 | 227 | 170 | 57 | 3:1 | 0.97 |
PRR2 Is Expressed in Immature Fruit Pericarps
To evaluate the expression of PRR2, we performed RT-PCR using fruit pericarps harvested at different ripening stages, including the immature (S1), breaker (S2), and mature stage (S3). We also analyzed the expression of PRR2 in root, stem, and leaf tissues (Figure 3D). In both accessions, we observed transcripts of the expected size predominantly at the immature stage (S1), whereas they were almost undetectable in more mature fruits (stages S2 to S3) and in the vegetative tissues. The relative transcript levels of PRR2 were slightly less than that of actin, which was used as a control. This result demonstrates the expression pattern of PRR2, which expressed specifically in immature fruit.
PRR2 Regulates Plastid Development in Pepper Fruits
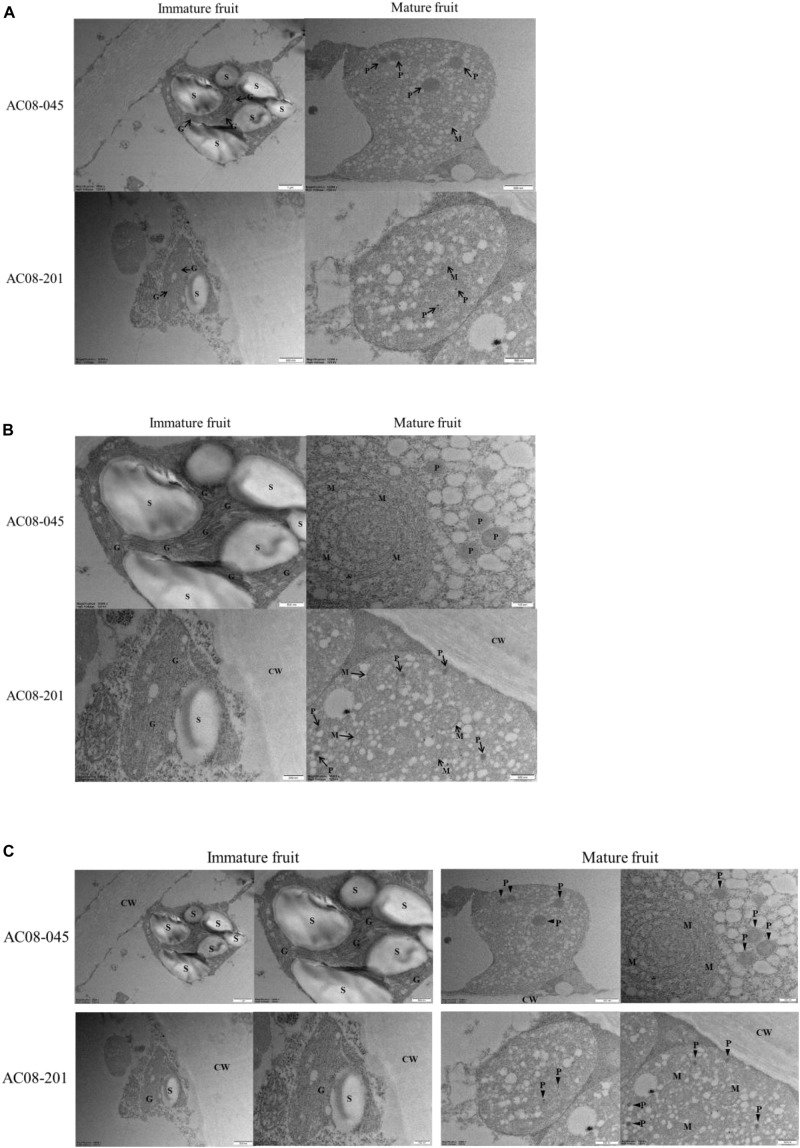
To study how PRR2 control fruit color, we conducted TEM analysis of fully expanded immature and mature fruits of AC08-045 and AC08-201, using a light microscope and toluene blue-stained samples. We observed a much smaller number of plastid-like structures in AC08-201 than in AC08-045 in both mature and immature fruits. All the cell organelles were displaced toward the cell wall, possibly due to the large tonoplast that develops in the fruit pericarp during fruit maturation (Supplementary Figure S2). We also observed the inner structures of the plastids using TEM. A single plastid from each sample is shown in Figure 4A for size comparison. Chloroplasts were observed in immature fruits of both accessions, with the characteristic thylakoid membrane structure and starch grains. AC08-201 had much smaller chloroplasts than AC08-045, almost half the size, as indicated by the scale bar. The starch grains in the chloroplasts were also larger and more numerous in AC08-045 than in AC08-201. Chromoplasts, which we observed in mature fruit pericarp samples, showed no significant size difference between the accessions. Enlarged images of the inner structures of each plastid are shown in Figure 4B. In immature fruits, chloroplasts in AC08-045 had much thicker thylakoids than those of AC08-201, which were almost undetectable even in magnified images. Moreover, the thylakoids in AC08-045 were highly stacked into well-developed granum, in contrast to those of AC08-201. The plastoglobuli in the chromoplasts were much smaller in mature AC08-201 fruits compared than mature AC08-045 fruits, although the number of plastoglobuli did not differ significantly. The plastoglobule is the inner structure of the plastid, which mainly accumulates carotenoids and contains the carotenoid biosynthetic enzymes of chromoplasts; thus, the number and size of plastoglobuli are closely related to the chromoplast carotenoid content (Oleszkiewicz et al., 2018). Images of a single plastid and its inner structures are collected in Figure 4C to provide a comprehensive comparison.
FIGURE 4.
Plastid structures of AC08-045 and AC08-201 observed by TEM. (A–C) Chloroplast of immature fruits and chromoplast of mature fruits were observed. (A) Images of single plastids of immature and mature fruits of AC08-045 and AC08-201, shown in wide field. (B) Magnified images showing inner structures of each plastid. (C) Combined image of A and B for comparison. S, starch grain; CW, cell wall; G, granum; P, plastoglobule; M, internal membrane structure. Scale bars for each micrograph are located at the bottom right.
Silencing of PRR2 Leads to Lighter Yellow Fruit Color in MY
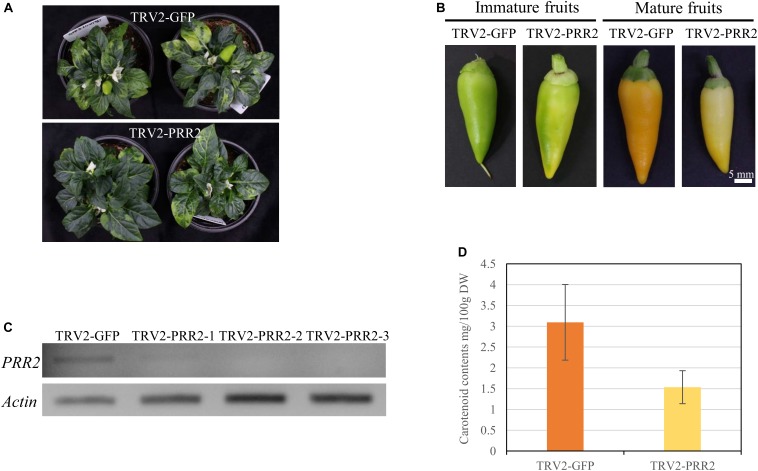
We evaluated the function of PRR2 using C. annuum ‘MY,’ which has non-functional mutations in both PSY1 and CCS, via TRV-mediated VIGS. Agrobacterium carrying pTRV2-GFP served as a negative control. Neither TRV2-GFP- nor TRV2-PRR2-inoculated plants displayed any significant differences from the un-inoculated wild-type MY plants, except for the slight discoloration due to the TRV symptoms (Figure 5A). The flowers and fruits of silenced plants developed at about 60 and 75 DAI, respectively. There were no visible changes in the flower colors in either group, nor in the colors of the leaves (data not shown). In the pTRV2-GFP-inoculated plants, the immature fruits were dark green in color, whereas the immature fruits in the PRR2-silenced plants showed patches of ivory to pale green color. The mature fruit color showed a similar trend: the pTRV2-GFP-inoculated fruits were yellow, whereas the PRR2-silenced fruits were ivory to light yellow (Figure 5B). The fruit-specific effect of TRV2-PRR2 inoculation support our hypothesis that PRR2 is C1, as C1 does not affect the vegetative tissue phenotype.
FIGURE 5.
VIGS of PRR2 in MY. (A) Phenotypes of TRV2-GFP-inoculated MY (top) and TRV2-PRR2-inoculated MY (bottom) plants at 60 days after inoculation (DAI). (B) Effect of the VIGS of GFP and PRR2 on the color of immature and mature fruit. Scale bar, 5 mm. (C) Expression analysis of PRR2 using RT-PCR. Three biological replicates of PRR2-silenced fruits were used. Actin served as a control. (D) Total carotenoid contents of TRV2-GFP- or TRV2-PRR2-inoculated MY fruits. The standard deviation of carotenoid contents of TRV2-GFP- or TRV2-PRR2-inoculated MY fruits were 0.909484 and 0.396838, respectively, with the p-value of 0.09078 using Student’s t-test. TRV2-GFP was used as a vector control.
We conducted RT-PCR to test the expression levels of PRR2 in the pTRV2-GFP-inoculated and PRR2-silenced plants. The cDNA of immature fruits was used for RT-PCR, as PRR2 is predominantly expressed in immature fruits and barely expressed in mature fruits (Figure 3D). Whereas the fruits of the pTRV2-GFP-inoculated plants showed PRR2 expression, there was almost no detectable PRR2 expression in the fruits from the PRR2-silenced plants, indicating the successful post-transcriptional gene silencing (PTGS) of the target gene (Figure 5C).
We used HPLC analysis to obtain a quantitative and qualitative measurement of the carotenoid contents of the mature fruits from the TRV-GFP- and TRV2-PRR2-inoculated plants. The total carotenoid contents were 3.09 mg/100 g DW and 1.54 mg/100 g DW, respectively, with non-significant P-value according to Student’s t-test. The mature fruits of PRR2-silenced plants had less than half the carotenoid content of those from the control plants. Lutein was the main component in fruits from both the TRV2-GFP- and TRV2-PRR2-inoculated plants, and α-carotene and β-carotene were also detected in the former (Figure 5D and Supplementary Figure S3).
Several White Pepper Accessions Carry PRR2 Mutations
To validate PRR2 mutations are commonly observed in other Capsicum spp., we analyzed the cDNA sequences of PRR2, along with PSY1 and CCS, in three other white pepper accessions: C. chinense ‘Habanero White’ and ‘White Moruga’ and C. annuum ‘Chupetino White.’ Non-functional alleles of PSY1, CCS, and PRR2 were present in all three accessions, although the types of mutation were slightly different between accessions.
In PSY1, Habanero White and Chupetino White had an early stop codon in the cDNA sequence at 679 nt, along with some non-synonymous mutations (N16S and A40V in Chupetino White; A40V in Habanero White). White Moruga had a 28-nt deletion in the coding sequence at nt 1,103, which also leads to early translation termination of the PSY1 gene, along with one non-synonymous mutation resulting in a G62R substitution in the amino acid sequence. In CCS, all three accessions have several nucleotide substitutions (resulting in the amino acid alterations K36R and Y43S), including the common SNP leading to an early stop codon at nt 599 (data not shown). Along with the early stop codons of PSY1 and CCS, these accessions also had structural mutations in PRR2. Habanero White and White Moruga had the same structural mutation of PRR2, which results in an early stop codon at amino acid 93 of the protein (Figure 6A). Chupetino White had three non-synonymous mutations in the sixth exon of PRR2 (Figure 6B). Taken together, these results indicate that pepper accessions with white mature fruits commonly carry mutations in PSY1, CCS, and PRR2. Taken together, our results suggest that PRR2 might be the gene corresponding to C1, the last unidentified locus governing pepper mature fruit color.
FIGURE 6.
Structural mutations of PRR2 in pepper cultivars with white mature fruit. (A) C. chinense ‘Habanero White’ and ‘White Moruga’ had same structural mutation leading to a premature stop codon. Consecutive non-synonymous mutations are shown in the figure. (B) C. annuum ‘Chupetino White’ had three non-synonymous substitutions. The amino acid substitutions are shown in the figure.
Discussion
In Capsicum, a three-locus model (C1, C2, and Y) has been proposed to explain fruit-color inheritance (Hurtado-Hernandez and Smith, 1985). The genes at C2 and Y were previously reported to encode phytoene synthase and capsanthin-capsorubin synthase, respectively. In this study, we verified that the C1 locus contains the gene pseudo response regulator2-like (PRR2), and the white color of AC08-201 is due to triple mutations in PSY1 (C2), CCS (Y), and PRR2 (C1). According to this hypothesis, the fruits of plants with a functional C1 allele are expected to be more intensely colored than those of plants with a non-functional C1 allele: red (C1, PSY1, CCS) vs. light red (c1, PSY1, CCS); orange (C1, psy1, CCS) vs. pale orange (c1, psy1, CCS); orange-yellow (C1, PSY1, ccs) vs. lemon-yellow (c1, PSY1, ccs); pale orange-yellow (C1, psy1, ccs) vs. white (c1, psy1, ccs). As the C1 genotype does not affect the overall color category (red, orange, yellow, or white), C1 was proposed to be a regulatory gene of carotenoid biosynthesis rather than a structural gene. In accordance with this hypothesis, our results strongly suggest that C1 corresponds to PRR2.
In the previous study, Pan et al. (2013) showed that APRR2 overexpression in tomato largely enhanced the levels of chlorophyll in immature fruits and slightly enhanced the levels of carotenoids in red fruits by increasing plastid number and area. The same group also reported a premature stop codon due to a guanine-to-adenine substitution at 1,404 nt in PRR2 (based on the coding sequence of Capana01g000809) in sweet peppers with white immature fruits. In cucumber, PRR2 is reported to control the green immature fruit color, and its mutated allele, prr2, causes a white color. prr2 encodes a truncated 101-amino-acid protein due to a frameshift mutation that creates a premature stop codon (Liu et al., 2016). All previous studies of the function of PRR2 have focused mainly on the immature stage of the fruits. However, in this study, we showed that PRR2 affects the color of mature fruits. Given that chromoplasts, the differentiated form of proplastids that are the primary sites of carotenoid accumulation, derive from chloroplasts (Lu et al., 2006; Jarvis and López-Juez, 2013), a larger chloroplast compartment size at the immature stage could result in a larger chromoplast compartment at the mature stage, which could accumulate more carotenoids. However, a contrasting report indicates that increased chlorophyll content is not always statistically linked to increased carotenoid content in peppers (Brand et al., 2012), and thus more studies will be required to clarify the function of PRR2.
During sequencing analysis, we observed that the sequences of PRR2 in the accessions we sequenced are different from that of the reference genome. Pan et al. (2013) reported that the coding sequence of PRR2 consisted of 1,683 bp (GenBank accession no. KC175445). In this study, however, we performed RT-PCR using primers that cover the full length of the gene and identified bigger amplicons (total length 1,764 bp). The resulting sequences were very similar to the sequence of this gene derived from the C. annuum Zunla cultivar genome, v2.0 (Capana01g000809), which is differed by 54 bp in sixth exon of the gene compared to KC175445 sequence, although the resulting sequence also differed in length, by 27 bp (total length 1,737 bp) compared to the Zunla reference. For verification, we sequenced PRR2 from other accessions, including C. annuum. The sequencing results showed the same extra 27 bp of sequence, located in the sixth exon of the gene (Figure 3A). The coding sequences of PRR2 in AC08-045 and AC08-201 are shown in Supplementary Material S2.
We classified the ripening stages of each accession into three stages. In AC08-045, fruit color changes from dark green to yellow and then light yellow. In AC08-20l, fruits become light green, ivory, and then white. However, if PRR2 is the only candidate for regulating color intensity, fruits of both accessions would be expected to have only two ripening stages. In other words, once the plastid compartment size is determined by PRR2 and then the chloroplasts transition into chromoplasts during ripening, the fruit color must become yellowish, and the color would be expected to be maintained. It was assumed that the original classification of ripening into more than two stages might have resulted from degradation of carotenoids during ripening. Hurtado-Hernandez and Smith (1985) also reported that some mature pepper fruits lose pigment as they move from physiological maturity to over-maturity. Ha et al. (2007) studied the carotenoid profiles and genotype of C. annuum IT158782, with white mature fruit. They observed that PSY1 and CCS could not be amplified, indicating the presence of structural mutations abolishing the functions of those genes. In this context, IT158782 and AC08-201 can both be considered to have non-functional alleles of PSY1 and CCS, although the alleles may be different. In IT158782 fruits, however, no carotenoids were detected, including lutein, the major carotenoid component of AC08-201. This might be due to the difference in harvest time. Indeed, IT158782 fruits accumulated 1.28 μg/g (0.13 mg/100 g DW) of lutein before they were fully mature. If the carotenoid extraction of AC08-201 were performed with more fully ripened fruits, no carotenoids might be detected.
In this study, we observed the PRR2 gene was mainly expressed in immature fruits indicating that carotenoid accumulation in mature fruit is affected by the early expression of PRR2. To understand the discrepancy between PRR2 expression and carotenoid accumulation, we observed the plastid structures of both immature and mature fruit pericarps of AC08 accessions as the PRR2 is reported to regulate the plastid number and area. Immature fruits of AC08-201 had smaller chloroplasts and less developed thylakoid structures, which were stacked only one or twice into each granum. Although the chromoplast sizes in mature fruits were not significantly different, the plastoglobuli were larger in AC08-045, with a normal PRR2 gene. As the plastoglobule, the inner structure of the plastid, is where the carotenoids mainly accumulate, it is plausible that its size could be closely related to carotenoid content in chromoplast (Oleszkiewicz et al., 2018). Considering the fact that conversion from chloroplast to chromoplast is one of the most important developmental changes in fruit ripening, thylakoid structures of chloroplast are dissembled and plastoglobuli are formed in chromoplast (Klee and Giovannoni, 2011), indicating that expression of PRR2 in immature fruit might be connected to the carotenoid accumulation in mature fruit. This also supports our theory that PRR2 is a carotenoid-content-regulating gene, as carotenoid accumulation could be limited by the smaller compartment space due to the PRR2 mutation. However, the mechanism regulating plastid development still needs to be studied.
We also performed VIGS analysis of PRR2, using the accession C. annuum ‘MicroPep Yellow (MY),’ which has non-functional PSY1 and CCS alleles and a functional PRR2 gene. Therefore, MY and AC08-045 belong to the same group according to the three-locus model. Silencing of PRR2 resulted in lighter color of immature and mature fruits in MY plants, similar to those of AC08-201 with non-functional PRR2. This result supports our hypothesis about the role of PRR2 in controlling fruit color in peppers that produce pale orange-yellow and white fruits.
We also analyzed the coding sequences of PSY1, CCS, and PRR2 in other pepper accessions with white mature fruit (Habanero White, Chupetino White, and White Moruga) and identified several significant mutations in all three genes. All three cultivars carried non-synonymous mutations and an early stop codon in PSY1 and CCS, respectively. In PRR2, Habanero White and White Moruga had an early stop codon, and Chupetino White had three non-synonymous mutations (Figure 6). The portion of PRR2 deleted in Habanero White and White Moruga contained a receiver-like domain (RLD) and DNA-binding domain (B-motif), and the mutations in Chupetino White were mostly located in the part of the gene encoding the golden-2 like transcription factor domain, which is critical for the normal function of APRR2 (Jiao et al., 2017). To sum up, all white accessions used in this study had prr2/psy1/ccs genotypes. However, PRR2 mutations do not appeared to be conserved within species as mutations found in this study were different from those reported by Pan et al. (2013) and Borovsky et al. (2019) in C. annuum and C. chinense. Furthermore, the mutations in PSY1 and CCS are also not species specific as the various allelic variations were previously reported even in same species (Ha et al., 2007; Jeong et al., 2018). However, to confirm that PRR2 is the gene corresponding to the C1 locus, more study is still needed in PSY1/CCS, psy1/CCS, and PSY1/ccs pepper of other background genotypes.
Borovsky et al. (2019) comprehensively demonstrated the effects of CcLOL1, GLK2, and PRR2 on chloroplast biogenesis. In fact, CcLOL1 is closely located very closely to PRR2 and has similar function. Therefore, we cannot fully rule out the effect of LOL1 in mature color development of white pepper accessions. However, the VIGS study using MY supports our hypothesis that the gene determining white mature color in these accessions is PRR2 as we used PRR2 specific sequence to induce gene silencing. In the further study, the relationship between GLK2, LOL1, and PRR2 on chromoplast biogenesis needs to be revealed as chromoplast can be converted from chloroplast.
In conclusion, in this study we have demonstrated the function of PRR2, the regulator of color intensity in both PSY1- and CCS-mutated genotypes. Silencing of PRR2 by VIGS resulted in lighter fruit color. Moreover, although there were some differences in the specific mutations, non-functional PRR2 alleles are common in white pepper accessions of the three major Capsicum species, along with non-functional alleles of PSY1 and CCS. This is the first report revealing the role of PRR2 in mature fruit color and the identity of the gene at the C1 locus.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
H-BJ conducted the VIGS, UPLC, and sequence analysis of genes in the AC08 cultivar, while S-JJ conducted VIGS, microscopy, expression analysis, and sequence analysis of other cultivars. Both wrote the manuscript and made the figures. M-YK supervised VIGS experiment. J-KK won supervised TEM microscopy, and SK kindly lent machine for UPLC for carotenoid analysis. B-CK supervised the overall processes and revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Agriculture, Food, and Rural Affairs Research Center Support Program (Vegetable Breeding Research Center, 710011-03), funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA). This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01322901), Rural Development Administration, Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00399/full#supplementary-material
References
- Al-Babili S., Bouwmeester H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66 161–186. 10.1146/annurev-arplant-043014-114759 [DOI] [PubMed] [Google Scholar]
- Borovsky Y., Monsonego N., Mohan V., Shabtai S., Kamara I., Faigenboim A., et al. (2019). The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J. 99 41–55. 10.1111/tpj.14305 [DOI] [PubMed] [Google Scholar]
- Bouvier F., Hugueney P., d’harlingue A., Kuntz M., Camara B. (1994). Xanthophyll biosynthesis in chromoplasts: Isolation and molecular cloning of an enzyme catalyzing the conversion of 5, 6-epoxycarotenoid into ketocarotenoid. Plant J. 6 45–54. 10.1046/j.1365-313x.1994.6010045.x [DOI] [PubMed] [Google Scholar]
- Brand A., Borovsky Y., Hill T., Rahman K. A., Bellalou A., Van Deynze A., et al. (2014). CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 127 2139–2148. 10.1007/s00122-014-2367-y [DOI] [PubMed] [Google Scholar]
- Brand A., Borovsky Y., Meir S., Rogachev I., Aharoni A., Paran I. (2012). pc8.1, a major QTL for pigment content in pepper fruit, is associated with variation in plastid compartment size. Planta 235 579–588. 10.1007/s00425-011-1530-9 [DOI] [PubMed] [Google Scholar]
- Collera-Zúñiga O., Jiménez F. G., Gordillo R. M. (2005). Comparative study of carotenoid composition in three mexican varieties of Capsicum annuum L. Food Chem. 90 109–114. 10.1016/j.foodchem.2004.03.032 [DOI] [Google Scholar]
- Domonkos I., Kis M., Gombos Z., Ughy B. (2013). Carotenoids, versatile components of oxygenic photosynthesis. Prog. Lipid. Res. 52 539–561. 10.1016/j.plipres.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Bramley P. M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid. Res. 43 228–265. 10.1016/j.plipres.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Ha S. H., Kim J. B., Park J. S., Lee S. W., Cho K. J. (2007). A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J. Exp. Bot. 58 3135–3144. 10.1093/jxb/erm132 [DOI] [PubMed] [Google Scholar]
- Huh J. H., Kang B. C., Nahm S. H., Kim S., Ha K. S., Lee M. H., et al. (2001). A candidate gene approach identified Phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp.). Theor. Appl. Genet. 102 524–530. 10.1007/s001220051677 [DOI] [Google Scholar]
- Hurtado-Hernandez H., Smith P. G. (1985). Inheritance of mature fruit color in Capsicum annuum L. J. Hered. 76 211–213. 10.1093/oxfordjournals.jhered.a110070 [DOI] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14 787–802. 10.1038/nrm3702 [DOI] [PubMed] [Google Scholar]
- Jeong H., Kang M., Jung A., Han K., Lee J., Jo J., et al. (2018). Single-molecule real-time sequencing revelas diverse allelic variations in carotenoid biosynthetic genes in pepper (Capsicum spp.). Plant Biotechnol. J. 17 1081–1093. 10.1111/pbi.13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Liu H., Liu J., Cui M., Xu J., Meng H., et al. (2017). Identification and functional characterization of APRR2 controlling green immature fruit color in cucumber (Cucumis sativus L.). Plant Growth Regul. 83 233–243. 10.1007/s10725-017-0304-1 [DOI] [Google Scholar]
- Kim J., Park M., Jeong E. S., Lee J. M., Choi D. (2017). Harnessing anthocyanin-rich fruit: a visible reporter for tracing virus-induced gene silencing in pepper fruit. Plant Methods 13:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., An C. G., Park J. S., Lim Y. P., Kim S. (2016). Carotenoid profiling from 27 types of paprika (Capsicum annuum L.) with different colors, shapes, and cultivation methods. Food Chem. 201 64–71. 10.1016/j.foodchem.2016.01.041 [DOI] [PubMed] [Google Scholar]
- Klee H. J., Giovannoni J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45 41–59. 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- Lee J. H., An J. T., Siddique M. I., Han K., Choi S., Kwon J. K., et al. (2017). Identification and molecular genetic mapping of Chili veinal mottle virus (ChiVMV) resistance genes in pepper (Capsicum annuum). Mol. Breed. 37:121. [Google Scholar]
- Lefebvre V., Kuntz M., Camara B., Palloix A. (1998). The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol. Biol. 36 785–789. [DOI] [PubMed] [Google Scholar]
- Liu H., Jiao J., Liang X., Liu J., Meng H., Chen S., et al. (2016). Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 129 1247–1256. 10.1007/s00122-016-2700-8 [DOI] [PubMed] [Google Scholar]
- Lu S., Eck J. V., Zhou X., Lopez A. B., O’Halloran D. M., Cosman K. M., et al. (2006). The cauliflower Or gene encodes a DnaJ cystein-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18 3594–3605. 10.1105/tpc.106.046417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- Oleszkiewicz T., Klimek-Chodacka M., Milewska-Hendel A., Zubko M., Stró Ź, Kurczyńska E., et al. (2018). Unique chromoplast organization and carotenoid gene expression in carotenoid-rich carrot callus. Planta 248 1455–1471. 10.1007/s00425-018-2988-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Bradley G., Pyke K., Ball G., Lu C., Fray R., et al. (2013). Network inference analysis identifies an APRR2-Like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161 1476–1485. 10.1104/pp.112.212654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I., van der Knaap E. (2007). Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 58 3841–3852. 10.1093/jxb/erm257 [DOI] [PubMed] [Google Scholar]
- Popovsky S., Paran I. (2000). Molecular genetics of the y locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit color. Theor. Appl. Genet. 101 86–89. 10.1007/s001220051453 [DOI] [Google Scholar]
- Powell A. L., Nguyen C. V., Hill T., Cheng K. L., Figueroa-Balderas R., Aktas H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336 1711–1715. 10.1126/science.1222218 [DOI] [PubMed] [Google Scholar]
- Seo Y., Park J., Kim Y. S., Park Y., Kim Y. H. (2014). Screening and histopathological characterization of Korean carot lines for resistance to the root-knot nematode Meloidogyne incognita. Plant Pathol. J. 30 75–81. 10.5423/ppj.oa.08.2013.0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., Li L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11 58–74. 10.1016/j.molp.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Yoo H. J., Park W. J., Lee G., Oh C., Yeam I., Won D., et al. (2017). Inferring the genetic determinants of fruit colors in tomato by carotenoid profiling. Molecules 22:764 10.3390/molecules22050764 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.