Abstract
Vaccination is the most effective method of disease prevention and control. Many viruses and bacteria that once caused catastrophic pandemics (e.g., smallpox, poliomyelitis, measles, and diphtheria) are either eradicated or effectively controlled through routine vaccination programs. Nonetheless, vaccine manufacturing remains incredibly challenging. Viruses exhibiting high antigenic diversity and high mutation rates cannot be fairly contested using traditional vaccine production methods and complexities surrounding the manufacturing processes, which impose significant limitations. Virus‐like particles (VLPs) are recombinantly produced viral structures that exhibit immunoprotective traits of native viruses but are noninfectious. Several VLPs that compositionally match a given natural virus have been developed and licensed as vaccines. Expansively, a plethora of studies now confirms that VLPs can be designed to safely present heterologous antigens from a variety of pathogens unrelated to the chosen carrier VLPs. Owing to this design versatility, VLPs offer technological opportunities to modernize vaccine supply and disease response through rational bioengineering. These opportunities are greatly enhanced with the application of synthetic biology, the redesign and construction of novel biological entities. This review outlines how synthetic biology is currently applied to engineer VLP functions and manufacturing process. Current and developing technologies for the identification of novel target‐specific antigens and their usefulness for rational engineering of VLP functions (e.g., presentation of structurally diverse antigens, enhanced antigen immunogenicity, and improved vaccine stability) are described. When applied to manufacturing processes, synthetic biology approaches can also overcome specific challenges in VLP vaccine production. Finally, we address several challenges and benefits associated with the translation of VLP vaccine development into the industry.
Keywords: capsomere, computational, omics technologies, synthetic biology, vaccine, virus‐like particle
Vaccination is the most effective method of disease prevention and control. Many viruses and bacteria that once caused catastrophic pandemics (e.g. smallpox, poliomyelitis, measles, and diphtheria) are either eradicated or effectively controlled through routine vaccination programs. Nonetheless, vaccine manufacturing remains incredibly challenging.
1. INTRODUCTION
Virus‐like particles (VLPs) are self‐assembling complexes of capsid proteins that mimic the overall structure of their parental virus. Void of viral genetic material, these noninfectious particles possess biologically desirable traits that are attributed to the particulate viral structure (Grgacic & Anderson, 2006; Pattenden, Middelberg, Niebert, & Lipin, 2005). Of particular interest is their efficient recognition, cellular uptake, and processing by host immune systems. VLP technology is a rapidly expanding field, which aims to embrace such features to achieve specific biological outcomes. VLPs are amenable to a broad range of modifications including encapsulation, chemical conjugation, and genetic manipulation (Roldão, Mellado, Castilho, Carrondo, & Alves, 2010). This versatility of VLPs, combined with the natural ability to package and deliver nucleic acids has prompted their use as biodegradable delivery agents for gene therapy (Takamura et al., 2004; Tegerstedt, Franzen, et al., 2005). The successful packaging of peptides, proteins, and synthetic drugs into VLPs has revealed a prospective role in drug delivery (Kaczmarczyk, Sitaraman, Young, Hughes, & Chatterjee, 2011; Zdanowicz & Chroboczek, 2016). In the diagnostic field, VLPs containing gadolinium have shown potential as molecular imaging contrasting agents (Schwarz & Douglas, 2015) while the application of VLPs as research surrogates to study the clearance of live viruses has also been demonstrated (Johnson, Brorson, Frey, Dhar, & Cetlin, 2017; Loisy et al., 2005). At the forefront of VLP technology impact, however, is vaccinology.
Licensed prophylactic VLP vaccines, such as Gardasil®, Cervarix®, Hecolin®, and Porcilis PCV®, demonstrate that VLP vaccines are safe and effective. VLP technology can overcome numerous drawbacks associated with traditional methods of vaccine production; specifically, the infectious nature associated with live and inactivated vaccines and the lengthy production time. Synthetic biology, the redesign and construction of novel artificial biological organisms, pathways or processes, is revolutionizing vaccine production. When applied to VLPs, synthetic biology allows for more precise and predictable control over the composition and assembly of the viral capsid. This, in turn, widens the range of unrelated antigenic modules that can be incorporated. Consequently, vaccines can be engineered for prophylactic or therapeutic means (Jennings & Bachmann, 2009) to match specific viral strains (Schwartzman et al., 2015) or to generate multivalent (Pushko et al., 2011) or broadly cross‐protective (Ben‐Yedidia, 2011) vaccines. Moreover, new bioprocessing modalities including in vitro assembly (Chuan, Fan, Lua, & Middelberg, 2010) and cell‐free expression (Bundy, Franciszkowicz, & Swartz, 2008), open the way to reimagined vaccine production processes.
2. DESIGN TOOLS FOR MODERN VACCINES
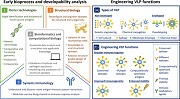
With the ability to integrate biological data and computational analysis, synthetic biology has significantly contributed to move vaccine development beyond the constraints of Pasteur, assisting in the design of novel biological systems with enhanced efficacy and safety as well as reducing vaccine production times (Ruder, Lu, & Collins, 2011). With today's focus gearing towards using VLPs, not only as vaccines of unmodified viral assemblies against parental viruses but also as scaffolds for display of heterologous antigens (Lua et al., 2014; Roldão et al., 2010), synthetic biology has become a centerpiece in VLP vaccine engineering. Seconded by a multitude of tools, such as omics technologies, structural biology, system immunology, and bioinformatics and computational biology, one can now screen for pathogen‐specific antigens with high immunogenic potential and apply that information to rationally design modern VLP vaccines (Figure 1).
Figure 1.
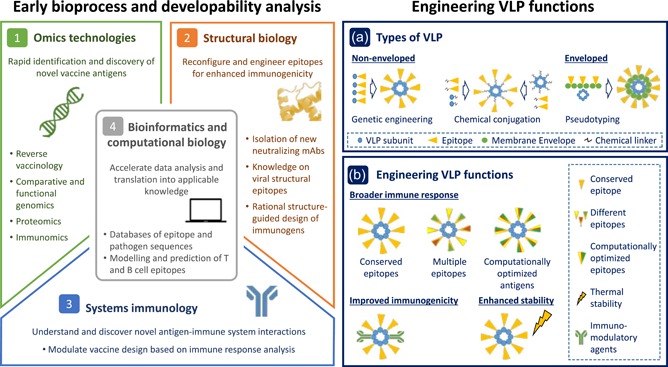
Design tools for VLP vaccine engineering. Multitude of tools and recent advances in synthetic biology enable screening for pathogen‐specific antigens with high immunogenic potential and engineering of VLP function. (1) Omics technologies enable rapid identification and discovery of novel/potential vaccine antigens. (2) Structural biology and (3) system immunology assist rational reconfiguration and engineering of epitopes/VLPs for enhanced immunogenicity. (4) Bioinformatics and computational biology accelerate data analysis and translation into applicable knowledge. (a) Engineering VLP function on different types of VLP. While nonenveloped VLPs are commonly engineered using genetic engineering or chemical conjugation, enveloped VLPs rely on pseudotyping for function engineering. (b) VLPs can be engineered to offer broader immunogenicity, improved immunogenicity, or enhanced stability. Broadly immunogenic VLPs can be obtained by displaying multiple antigenically distinct epitopes (Pushko et al., 2011; Schwartzman et al., 2015), highly conserved epitopes (Krammer, 2015; Wiersma et al., 2015), or computationally optimized epitopes (Carter et al., 2016) within a single VLP. Improving VLP immunogenicity can be achieved by incorporating immunomodulatory agents, such as dendritic cells targeting antibodies into particles structure (Rosenthal et al., 2014). VLP stability can be enhanced by modulating particles formulation (Collins et al., 2017; Lua et al., 2014). VLP: virus‐like particle [Color figure can be viewed at wileyonlinelibrary.com]
2.1. Omics technologies
Influenza is one of the best examples on how omics, more precisely genomics, is revolutionizing vaccine design. Dedicated databases detailing complete and accurate influenza genomic information have been created and can be readily accessed worldwide (McHardy & Adams, 2009). In the last decade, this vast wealth of data has been translated into knowledge, which, in conjunction with synthetic biology, has aided the design of VLP vaccine candidates (Prabakaran et al., 2010; Pushko et al., 2011; Pushko et al., 2017). These novel biological entities are undoubtedly safer with the potential to be more broadly reactive than their previous counterparts (i.e., traditional, commercially available live attenuated influenza vaccines).
Several genomic applications aid in the identification of novel antigens (Liao et al., 2017), namely, reverse vaccinology and comparative and functional genomics. Reverse vaccinology combines genomics with proteomics and bioinformatics to identify virtually all potentially protective antigens from coding regions within the genome (Bambini & Rappuoli, 2009; Rappuoli, 2007). Comparative genomics is primarily used to design broadly protective vaccines as it allows comparison of conserved and variable open reading frames within the same species. Functional genomics allows the identification of protein function based on reverse genetic evaluation (mutations and knockouts) or gene expression analysis (transcriptomics; Bagnoli et al., 2011; Bambini & Rappuoli, 2009; Rappuoli, 2007; Sette & Rappuoli, 2010). Although genomics per se has contributed significantly to the design of many vaccine candidates, such as influenza (Prabakaran et al., 2010; Pushko et al., 2011; Pushko et al., 2017), malaria (Draper et al., 2015; Y. Wu, Narum, Fleury, Jennings, & Yadava, 2015), and human immunodeficiency virus (HIV; Calazans, Boggiano, & Lindsay, 2017), its full potential for vaccine development can only be realized when integrated with proteomics and immunomics (Bagnoli et al., 2011). Moreover, the coordinated deployment of these omics technologies along with miniaturized bioprocess screening in, for example, microbioreactor and microfluidic cell culture systems can aid developability assessment leading to accelerated manufacture (Bhambure, Kumar, & Rathore, 2011; Gong & Lei, 2014; Hemmerich, Noack, Wiechert, & Oldiges, 2018).
Immunomics specifically addresses the interface between the host immune system and the pathogen proteome. It studies the subset of pathogen‐derived proteins or epitopes that are recognized by the host immune system. This information can be used to validate antigens identified from in silico and/or in vitro approaches by evaluating whether they are targets of clinically relevant immune responses (i.e., they stimulate the production of specific cytokines or activate specific cell types; Kuleš et al., 2016). An example of immunomics application to VLP vaccine design is GlaxoSmithKline's RTS,S vaccine, the most advanced malaria vaccine candidate. RTS,S is a chimeric VLP in which a large portion of the C‐terminal of Plasmodium falciparum‐derived circumsporozoite protein (CSP) is displayed on a protein scaffold (hepatitis B surface antigen; Y. Wu et al., 2015). In combination with systems biology (genomics and proteomics), immunomics was used to highlight regions within CSP that were immunogenic and protective in humans and, thus, indispensable to incorporate into a chimeric VLP. From the many candidates identified, only one proved to be successful in Phase IIa/b efficacy trials (Draper et al., 2015; Kazmin et al., 2017).
2.2. Structural biology
Structural information can be used to reconfigure and engineer conserved epitopes to expose areas that exhibit high immunogenicity or to insert multiple immunodominant epitopes within the same VLP platform (Liljeroos, Malito, Ferlenghi, & Bottomley, 2015). These strategies can broaden the immune response or enhance the existing response to weak immunogenic antigens. Identifying conformational epitopes and studying their interactions with the immune system can provide significant information for rational antigen design (Anggraeni et al., 2013; Mulder et al., 2012).
Encouraged by recent results from HIV‐1 VLPs (C. Zhao, Ao, & Yao, 2016), structural biology is emerging as a powerful tool to assist in the rational design of a modern HIV VLP vaccine. Novel protective epitopes can now be identified in conformational epitope mapping studies via structural biology (Liljeroos et al., 2015; Malito, Carfi, & Bottomley, 2015). In addition, broad and potent HIV antibodies discovered in the pool of antigen‐specific memory B cells using structural biology, highlight novel sites of vulnerability on HIV envelope glycoprotein epitope (J. Huang et al., 2014). When incorporated into HIV‐1 VLPs, these antigens can be strategically modified to insert the extended 2F5, 4E10 epitope and membrane proximal external region (MPER) of HIV‐1 gp41 (Zhai, Zhong, Zariffard, Spear, & Qiao, 2013) providing a better display of the conserved CD4 binding site and capturing broadly neutralizing antibodies (Ingale et al., 2014).
Structural biology is equally informative for VLP engineering as the capsid proteins forming each VLP need to be correctly folded to ensure functionality, that is, induce a protective humoral immune response. The morphological characteristics of VLPs can be assessed through imaging technologies, such as cryoelectron microscopy, atomic force microscopy, and dynamic light scattering. These imaging methods provide essential data to solve the three‐dimensional structure of a VLP, therefore, help to identify optimal VLP insertion sites for display of any given antigen. Many VLP platforms have been developed so far using this structure‐based design approach. The most commonly reported are those derived from viral structures of hepatitis core antigen (HBcAg), murine polyomavirus (MuPyV), human papillomavirus (HPV), and bacteriophages Qβ and AP250 (Table 1).
Table 1.
Nonenveloped virus‐like particle platforms for the display of unrelated antigens
| Platforms | Targets | Antigens | References |
|---|---|---|---|
| Bacteriophage AP205 | HIV‐1 | HIV‐1 gp41 epitopes | Pastori et al. (2012) |
| Influenza | M2e | Tissot et al. (2008) | |
| Malaria | Circumsporozoite | Janitzek et al. (2016) | |
| Malaria | Pfs25 and VAR2CSA proteins | Thrane et al. (2016) | |
| Tuberculosis | Ag58A | Thrane et al. (2016) | |
| West Nile virus | Domain III of E glycoprotein | Spohn et al. (2010) | |
| Bacteriophage Qβ | Allergenic | Allergen Der p 1 | Kundig et al. (2006) |
| Alzheimer's disease | Aβ1–6 (amyloid peptide) | Wiessner et al. (2011) | |
| HIV‐1 | CCR5 coreceptor | Hunter, Smyth, Durfee, and Chackerian (2009) | |
| Hypertension | Angiotensin II | Tissot et al. (2008) | |
| Influenza | Hemagglutinin (globular head) | Jegerlehner et al. (2013) | |
| Influenza | M2e | Bessa et al. (2008) | |
| Nicotine dependence | Nicotine | Maurer et al. (2005) | |
| Type 2 diabetes | Interleukin‐1β | Spohn et al. (2010) | |
| Bovine Papillomavirus | Alzheimer's disease | Amyloid β peptide | Li et al. (2004) |
| HIV‐1 | CCR5 peptide | Chackerian, Lowy, and Schille (1999) | |
| HIV‐1 | V3 loop of HIV‐1 gp120 | X. S. Liu et al. (2002) | |
| HIV‐1 | HIV‐1 gp41 neutralizing epitopes | Zhai et al. (2013) | |
| Human papillomavirus (HPV) | HPV 16 L2 neutralizing epitopes | Slupetzky et al. (2007) | |
| Cowpea mosaic virus | Canine parvovirus | VP2 capsid protein | Langeveld et al. (2001) |
| HIV‐1 | Glycoprotein 41 peptide | McLain, Porta, Lomonossoff, Durrani, and Dimmock (1995) | |
| Pseudomonas aeruginosa | CPMV‐PAE5 peptide | Brennan et al. (1999) | |
| Staphylococcus aureus | Truncated D2‐domain | Rennermalm et al. (2001) | |
| Cucumber mosaic virus | Alzheimer's disease | Amyloid β peptides | Vitti et al. (2010) |
| Hepatitis C virus | HCV‐derived R9 and R10 mimotopes | Nuzzaci et al., (2007) | |
| Newcastle disease virus | Neutralizing epitopes | Y. Zhao and Hammond (2005) | |
| Flock House virus | Anthrax | Von Willebrand A domain of ANTXR2 cellular receptor/protective antigen | Manayani et al. (2007) |
| Hepatitis B and hepatitis C virus | Epitopes of hepatitis C virus and hepatitis B surface antigen | Chen et al. (2006) | |
| HIV‐1 | V3 loop of HIV‐1 gp120 protein | Scodeller (1995) | |
| Influenza | A‐helix epitope of HA2 | Schneemann et al. (2012) | |
| Hepatitis B core | Anthrax | Domain 4 epitope of the protective antigen (PA) of anthrax toxin | Bandurska et al. (2008) |
| Anthrax | 2β2–2β3 loop of PA | Yin et al. (2014) | |
| Dengue virus type 2 | Envelope domain III | Arora, Tyagi, Swaminathan, and Khanna (2012); Arora, Tyagi, Swaminathan, and Khanna (2013) | |
| Enterovirus 71 | SP55 and SP70 epitopes of enterovirus 71 | Ye et al. (2014) | |
| Hepatitis C virus (HCV) | B‐ and T‐cell epitopes of HCV | Mihailova et al. (2006) | |
| Influenza | M2e | De Filette et al. (2008); Fu et al. (2009) | |
| Lyme disease | OspA and variants of OspC | Nassal et al. (2008), (2005) | |
| Lyme disease | tHRF, Salp15, and Iric‐1 | Kolb, Wallich, and Nassal (2015) | |
| Malaria | CSP‐specific B and T cell epitopes | Sällberg, Hughes, Jones, Phillips, and Milich (2002) | |
| Tuberculosis | CFP‐10 | Dhanasooraj, Kumar, and Mundayoor (2013) | |
| Human papillomavirus | Human respiratory syncytial virus | Neutralizing epitopes | Murata, Lightfoote, Rose, and Walsh (2009) |
| Murine polyomavirus | Cancer (Breast) | Her2 | Tegerstedt, Lindencrona, et al. (2005) |
| Cancer (Prostate) | Prostate‐specific antigen | Eriksson et al. (2011) | |
| Group A Streptoccocus | J8 peptide | Middelberg et al. (2011) | |
| Influenza | M2e | Wibowo, Chuan, Lua, and Middelberg (2013) | |
| Influenza | Helix 190 antigen | Anggraeni et al. (2013) | |
| Influenza | Hemagglutinin (globular head) | Waneesorn, Wibowo, Bingham, Middelberg, and Lua (2016) | |
| Rotavirus | VP8 antigen | Tekewe et al. (2015) | |
| Tobacco mosaic virus | Foot and mouth disease | Foot and mouth disease peptides | L. G. Wu et al. (2003) |
| Murine hepatitis | Murine hepatitis coronavirus neutralizing epitope | Koo et al. (1999) | |
| Poliovirus | Poliovirus type 3 epitope | Haynes et al. (1986) | |
| Pseudomonas aeruginosa | Peptide of outer membrane protein F | Staczek, Bendahmane, Gilleland, Beachy, and Gilleland (2000) | |
| Rabbit papillomavirus | L2 epitopes | Palmer et al. (2006) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.3. Systems immunology
Systems immunology is an emerging area of research that uses a broad and integrated, multilevel approach to study the immune system to identify immune correlates of protection or immunogenicity signatures (Davis, Tato, & Furman, 2017). Combined with recent technological advances in human immunology, systems immunology can provide guidance for rational vaccine design. Systems immunology allows the assessment of most cell types of the immune system (including specialized B and T white blood cells), their state, function, signaling molecules, and encoding genes (Bird, 2017). This accumulation of data captures a snapshot of the human immune system, providing valuable information for the creation of human immune response models that may later be translated into improved vaccine design (Davis et al., 2017). A systems immunology approach was previously undertaken to investigate the immune response to the live attenuated yellow fever vaccine YF‐17D (Hou et al., 2017; Muyanja et al., 2014). Following immunization, comprehensive characterization of the immune system was performed providing insights into the vaccine's mechanism of action. A similar method was later applied to the malaria vaccine RTS,S, where a systems‐level approach led to the identification of molecular and cellular signatures associated with protection and immunogenicity unique to this VLP‐based vaccine (Kazmin et al., 2017). Systems immunology may also play an important role in designing vaccines capable of stimulating parts of the immune system not addressed by current vaccines (Davis et al., 2017). VLPs are ideal testing candidates for systems immunology approach due to their ability to stimulate B‐cell‐mediated immune responses, as well as CD4 proliferative responses and cytotoxic T lymphocyte responses (Gause et al., 2017).
The correct identification of epitopes that stimulate an immune response is crucial for the design of novel immunogens. These epitopes are regions within the antigen that are recognized by B‐ and T‐cell receptors (Patronov & Doytchinova, 2013). Potential B‐ and T‐cell epitopes can be mapped using bioinformatics and computational biology tools, however, without recent improvements in immunological characterization methods, the identification of epitopes that optimally stimulate the human immune response becomes challenging (Rappuoli, Bottomley, D'Oro, Finco, & De Gregorio, 2016). The ability to isolate memory B cells through high‐throughput FACS followed by in vitro culture enables secretion of sufficient amounts of recombinant monoclonal antibodies or antigen‐binding fragments to allow for antigen screening in binding and functional assays (J. Huang et al., 2013). The ultimate goal being the incorporation of specific antigenic regions on a VLP scaffold. Thus, improving vaccine specificity for generation of effective vaccines. An example is the above‐mentioned novel vulnerability sites on HIV envelope glycoprotein that is used for HIV VLP vaccine design (J. Huang et al., 2014; Zhai et al., 2013).
2.4. Bioinformatics and computational biology
Bioinformatics and computational biology tools have the potential to accelerate data analysis and translate results into applicable knowledge, fostering the discovery of new lead antigens by reducing the number of empirical experiments (He & Xiang, 2013). Vaccine design is an inherently complex and laborious process but software, algorithms, and databases outlined below have the potential to streamline vaccine development via identification of candidate antigens that may otherwise have been overlooked.
Epitope mapping is essential for designing vaccines capable of mounting a robust T‐ and B‐cell response. The discovery of such epitopes relies upon immunological prediction software, such as netMHC, SYFPEITHI, EpiJen, or Epivax, as described in other studies (Soria‐Guerra, Nieto‐Gomez, Govea‐Alonso, & Rosales‐Mendoza, 2015). For T‐cell epitope prediction, as are many software available that can reach up to 95% of positive predictive values (Soria‐Guerra et al., 2015). In addition, there are interface platforms, such as MHCBench (Salomon & Flower, 2006) allowing direct evaluation of various Major Histocompatibility Complex (MHC)‐binding peptide prediction algorithms. Interface platforms prove valuable to users unsure of which prediction model best suits their needs. For B‐cell epitope prediction, available software is supported by algorithms that can reach up to 25% for discontinuous B‐cell antigens (e.g., COBEpro [Sweredoski & Baldi, 2009], BCPRed and FBCPred [El‐Manzalawy, Dobbs, & Honavar, 2008]) or up to 95% for continuous B‐cell antigens (e.g., EPMeta [Liang et al., 2010]) of positive predictive values.
Bioinformatics and computational biology can also assist in the discovery of conserved epitopes through sequence variability analysis. This is particularly relevant when dealing with pathogens capable of evading the immune system due to their high mutation rates. The Protein Variability Server (PVS) is a valuable tool for the identification of such conserved epitopes, which can facilitate the development of broadly protective vaccines (Garcia‐Boronat, Diez‐Rivero, Reinherz, & Reche, 2008). In this study, PVS was used to identify a conserved fragment in the ectodomain of HIV‐1‐gp41. Several databases retain classified and well‐curated data from experimentally verified vaccines and/or vaccine components and, thus, are useful tools for vaccine design. The Immune Epitope Database and Analysis Resource (Peters et al., 2005; https://www.iedb.org) collects all published experimental data characterizing immune epitopes and the context in which the molecular structure is recognized as an epitope by the immune receptors. IEDB has over 88,382 epitopes and multiple tools to identify B‐ and T‐cell epitopes (Vita et al., 2015). The Syfpeithi database (Schuler, Nastke, & Stevanovikc, 2007; http://www.syfpeithi.de) has information on MHC classes I and II anchor motifs and binding specificity. The Conformational Epitope Database offers information about protein conformation. The AntigenDB (http://www.imtech.res.in/raghava/antigendb/) stores sequences, structures, origins, and epitopes of pathogen antigens (Ansari, Flower, & Raghava, 2010). The Computationally Optimized Broadly Reactive Antigen (COBRA) methodology was recently developed to overcome the challenges associated with antigenic diversity in influenza subtypes (Carter et al., 2016). This in silico approach, which uses consensus building to generate a number of antigen candidates termed COBRA antigens, was used to identify HA antigens that were broadly protective against H1N1 strains. Display of these antigens on VLPs demonstrated immunogenicity and efficacy in a murine model validating this technology.
3. ENGINEERING VLP FUNCTIONS
With the advent of novel technologies for screening and discovery of immunogenic antigen targets and the advances in synthetic biology one can now engineer VLP function and design vaccines with increased or broader immunogenicity and/or improved stability (cold‐chain free; Figure 1; Collins, Snaith, Cottingham, Gilbert, & Hill, 2017; Lua et al., 2014). VLPs can be engineered to enhance and tune the immune response to vaccination. Using synthetic biology tools, one can harbor immunogens inside or outside the VLP that upon vaccination are capable of triggering an early innate immune response that enhances vaccine effectiveness by increasing vaccine's uptake (Rosenthal, Chen, Baker, Putnam, & DeLisa, 2014). These immunomodulatory agents (e.g., pattern recognition receptor ligands, dendritic cells targeting antibodies, and endoplasmic reticulum targeting peptides) boost the immune response mainly through expansion of specific CD8+ T cells and the production of cytokines (without any long‐lasting effects).
VLPs are divided into two main groups, enveloped and nonenveloped. Enveloped VLPs are self‐assembling capsids, which acquire a lipid layer when budding from their host cells. This layer is absent in nonenveloped VLPs (Mateu, 2011). Insertion of heterologous antigens (otherwise known as modularization) into nonenveloped VLPs is mainly achieved through genetic fusion or chemical conjugation (Peacey, Wilson, Baird, & Ward, 2007). The size of the insert has implications for VLP assembly and correct presentation of the antigen. Small peptide epitopes are easily inserted into VLP structures without affecting VLP assembly, which can often occur when modularizing whole or large protein domains. Genetic fusion is the most popular method despite being time‐consuming and error‐prone (Mateu, 2011). Chemical conjugation supports the insertion of large antigens in preformed VLPs and these modifications are performed on naturally occurring conjugation sites using chemical crosslinkers or enzymes (Patel & Swartz, 2011; Tang, Xuan, Ye, Huang, & Qian, 2016). The downside of chemical conjugation is the incurred cost as it requires the production of both the VLP and the epitope(s) as well as conducting the chemical conjugation (Chackerian, 2007; Smith, Hawes, & Bundy, 2013). Recently developed technologies have started to address the cost and technological challenges. An example is the “Plug and Display” system, a technology based on two proteins, “the tag” and “the catcher,” which react irreversibly when in close proximity of each other. When “the catcher” is fused to a VLP and “the Tag” is fused with a vaccine target, these form a two‐component VLP vaccine ready to use (Brune et al., 2016). Examples of nonenveloped VLPs used to display foreign antigens are HBcAg (Chu et al., 2016), HPV L1 protein (Slupetzky et al., 2001), and Qβ (O'Rourke, Peabody, & Chackerian, 2015).
Enveloped VLPs can present heterologous membrane proteins (i.e., glycoproteins) in their native configuration on top of a self‐assembling capsid protein(s) without the need to engineer both epitope and capsid structures in a process called pseudotyping (Chua et al., 2013; Kirchmeier et al., 2014). With pseudotyping, one can alter the VLP stability or even its tropism (Cronin, Zhang, & Reiser, 2005; K. Palomares et al., 2013). In addition, transmembrane domains within foreign viral envelope proteins can be replaced with transmembrane domains of specific viruses (e.g., vesicular stomatitis virus) to improve pseudotyping efficiency and immunogenicity (Kirchmeier et al., 2014). Enveloped VLPs, such as retro‐ and lenti‐VLPs, have shown promise as vaccines candidates against diseases, such as Influenza, Malaria, or Dengue virus (Chua et al., 2013; Pitoiset, Vazquez, & Bellier, 2015).
The engineering of VLP has long been a complex process and often unsuccessful, as the insertion of small peptides can disrupt the VLP structure. The field has evolved and now VLP chimeras are used in both fundamental and applied research (Mateu, 2011; Murata et al., 2009). A successful example of the insertion of large peptides is the VLP derived from flock house virus that was engineered to carry a receptor domain and could be used as an anthrax antitoxin as well as a vaccine (Manayani et al., 2007).
Circulating viruses that exhibit high antigenic variability and high mutation rates, such as influenza, pose substantial challenges for current vaccination strategies (Wibowo et al., 2014). Novel strategies are, therefore, under development to create broadly cross‐protective vaccines. Highly conserved antigens have been identified between divergent influenza viruses. These broadly reactive antigens, which are located within the membrane proximal stalk domain of the hemagglutinin protein have been inserted into VLPs and demonstrate success as vaccines candidates (Ben‐Yedidia, 2011; Krammer, 2015; Wiersma, Rimmelzwaan, & de Vries, 2015). Broad protection is also possible through the simultaneous display of multiple, antigenically distinct (Prabakaran et al., 2010; Pushko et al., 2011; Schwartzman et al., 2015), or chimeric (Carter et al., 2016) HA antigens from different influenza subtypes within the same VLP.
4. VLP‐BASED PLATFORM TECHNOLOGY
The engineering of viral structural proteins to display heterologous antigens presents an opportunity to manufacture vaccines against unrelated viruses, as well as pathogens from other sources, that is, bacteria and parasitic protozoans. This versatile nature, coupled with the suite of VLPs amenable to modularization, makes VLPs ideal candidates for the development of vaccine platforms. Noninfectious, generic VLP platforms offer the potential for streamlined bioprocesses, parallel infrastructure, predictable biosafety, and regulatory practices (Charlton Hume & Lua, 2017), which, in turn, would significantly reduce vaccine production times. Given the lack of preparedness during recent global outbreaks of influenza A (H1N1) (Fineberg, 2014), Zika (Boeuf, Drummer, Richards, Scoullar, & Beeson, 2016), and Ebola virus (Coltart, Lindsey, Ghinai, Johnson, & Heymann, 2017), as well as Middle East Respiratory Syndrome (Okba, Raj, & Haagmans, 2017), VLP platform technologies are pertinent to address the rapid spread of emergent viruses.
4.1. Platform capabilities
Several viral structural proteins and bacteriophages are currently under development as vaccine platforms to target a variety of pathogens (Tables 1, 2). Synthetic biology approaches have widened the scope of potential antigens for modularization and show that platforms can be modified to overcome specific challenges associated with VLP vaccine manufacturing (Figure 2). Nonenveloped VLPs are structurally less complex than their enveloped counterparts and can be produced in prokaryotic and lower order eukaryotic systems making them easily scalable, cost‐effective, and rapid to manufacture. The presence of a lipid bilayer in enveloped VLPs necessitates the use of eukaryotic hosts for expression, which increases the overall production time and cost. Nonetheless, unlike nonenveloped VLPs, enveloped VLPs permit presentation of antigenic modules that require membrane‐association.
Table 2.
Enveloped virus‐like particle platforms for pseudotyping
| Platforms | Targets | Antigens | References |
|---|---|---|---|
| BIV Gag | Influenza | Hemagglutinin (HA) (subtypes) and neuraminidase (NA) | Pushko et al. (2017); Tretyakova et al. (2016) |
| HBsAg | Dengue virus | Envelope domain III | Harahap‐Carrillo, Ceballos‐Olvera, and Valle (2015) |
| Malaria | Circumsporozoite protein | Stoute et al. (1997) | |
| HIV1‐Gag | Dengue virus | Envelope domain III | Chua et al. (2013) |
| West Nile virus | Glycoprotein E | ||
| Influenza | HA and NA | Carter et al. (2016) | |
| Influenza M1 | Influenza | HA subtypes | Schwartzman et al. (2015) |
| Influenza | M2 | Song et al. (2011) | |
| Influenza | NA | Ben‐Yedidia (2011); Wiersma et al. (2015) | |
| Respiratory syncytial virus | RSV A2 fusion | Kim et al. (2015) | |
| Murine leukemia virus Gag | Cancer | Melanoma antigens | Kurg et al. (2016) |
| Human cytomegalovirus | Glycoprotein B | Kirchmeier et al. (2014) | |
| Rift Valley fever virus | Glycoproteins GN, GC, and nucleoprotein N | Mandell et al. (2010) | |
| Influenza | HA and NA | Haynes et al. (1986) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 2.
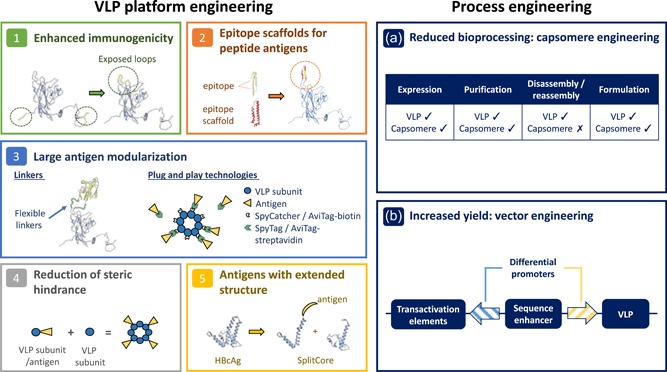
Application of synthetic biology to VLP vaccine platforms. (1) Enhanced immunogenicity of peptides is achieved through their insertion into exposed loops of viral capsid proteins (Murata et al., 2009; Slupetzky et al., 2007; Ye et al., 2014). (2) The structural properties of complex peptides are maintained through the incorporation of epitope scaffolds into exposed loops (Schneemann et al., 2012). (3) Large antigens are modularized onto VLP vaccine platforms using long flexible linkers to maintain structural separation between the viral capsid protein and the antigen (Kratz et al., 1999); or onto preformed VLPs using plug and play technologies, such as SpyCatcher/SpyTag (Brune et al., 2016) and AviTag (Thrane et al., 2015). (4) Dual expression of modified and unmodified viral capsid proteins reduces steric hindrance and permits VLP assembly (Tekewe et al., 2017). (5) The SplitCore system permits modularization of antigens with an extended structure through the coexpression of modified and unmodified HBcAg core fragments (Walker, Skamel, and Nassal, 2011). (a) Synthetic production of capsomeres minimizes host cell contaminants reducing required bioprocessing steps (Chuan et al., 2010). (b) Synthetic engineering of baculovirus vectors can increase VLP expression yield (Gómez‐Sebastián et al., 2014; Y. K. Liu et al., 2015; Y. V. Liu et al., 2015). VLP: virus‐like particle [Color figure can be viewed at wileyonlinelibrary.com]
High immunogenicity is a desirable vaccine attribute that potentially translates to protectivity against target infections. To enhance immunogenicity, modularized antigens must be strategically inserted to maximize their presentation to the immune system. Modularization of peptides to the N‐ or C‐termini of viral proteins has been previously reported (De Filette et al., 2008; Haynes et al., 1986). However, low surface expression or the inability of peptides to adopt native conformation can result in weak immunogenicity at these insertion sites (Schödel et al., 1992). Platforms based upon HBcAg, papillomavirus (bovine and human), Flock House virus (FHV), and others, are thus engineered to take advantage of surface‐exposed loops that demonstrate enhanced immunogenicity (Murata et al., 2009; Slupetzky et al., 2007; Ye et al., 2014). Peptides inserted into exposed loops protrude from the surface of VLPs making them more accessible to the immune system. Short peptides displaying relatively simple structures pose little problem for modularization within exposed loops, yet those with more complex structures require further platform engineering (Anggraeni et al., 2013). Incorporation of epitope scaffolds into exposed loops of VLPs maintain the structural properties of complex peptides as shown with the presentation of a 20aa A‐helix of the influenza HA2 chain on the FHV platform (Schneemann et al., 2012). This strategy, however, requires detailed structural knowledge of the chosen peptide and the identification of a suitable scaffold fragment. Antigens inserted into the immunodominant loops of the HBcAg platform require closely juxtaposed N‐ and C‐termini to maintain VLP integrity (Walker et al., 2011). This imposes considerable limitations upon antigen choice. To support the modularization of antigens that possess an extended structure, a SplitCore system has thus been devised (Walker et al., 2011). Dividing the HBcAg core protein within the immunodominant c/e1 loop yields two fragments, which are able to form VLPs when coexpressed. Fusion of antigen to the c/e1 termini of either fragment before coexpression enables modularization of antigens that may otherwise have been structurally incompatible.
Display of whole protein domains on the surface of VLPs allows presentation of multiple antigenic epitopes and increases the likelihood that epitopes will adopt their native conformation. Their large size, however, can cause steric hindrance resulting in compromised VLP assembly (Lua, Fan, Chang, Connors, & Middelberg, 2015). As such, several strategies have been developed to specifically modularize large antigens. Long, flexible glycine‐rich linkers were engineered to flank the antigen in the c/e1 loop of the HBcAg platform, allowing spatial separation and enabling proteins of up to 238aa to be inserted (Kratz, Bottcher, & Nassal, 1999). Though effective, optimal linker length for different antigens needs to be empirically determined. Steric hindrance can also be addressed in some cases by reducing the antigen content on the surface of the VLP. For example, Peyret et al. (2015) engineered a single polypeptide composed of an unmodified HBcAg fused to a chimeric HBcAg. A dual expression construct was also devised to coexpress unmodified murine polyomavirus (MuPyV) VP1, with antigen‐modularized VP1 displaying an 18 kDa rotavirus antigen (Lua et al., 2015; Tekewe, Fan, Tan, Middelberg, & Lua, 2017). Although such methods can lead to successful VLP assembly, it is possible that increasing the antigen mass, whereas reducing the antigen number may be reflected in lower immunogenicity, as is seen with the synthetically derived RTS,S malaria vaccine (Pitoiset et al., 2015). Nevertheless, this trade‐off is inevitable as the mass of the antigen increases relative to the carrier, necessitating an understanding of the optimization domain.
Display of large antigens has been reported using novel platforms based upon the HPV 16 L1 protein and the Acinetobacter phage, AP205 (Thrane et al., 2015; Thrane et al., 2016; Brune et al., 2016). The AviTag and SpyTag/SpyCatcher platforms allow the conjugation of antigens post VLP assembly through biotin‐streptavidin reactions (AviTag) or spontaneous formation of an irreversible isopeptide bond (SpyTag/SpyCatcher). With linkers already predefined, platforms remain constant permitting high throughput screening of vaccine candidates. In addition, independent production of the antigen in a different expression system is possible and of particular interest, if posttranslational modifications are critical for immunogenicity, provided these do not sterically inhibit attachment.
Encapsulation of host‐cell proteins and nucleic acid during cell‐based VLP assembly can alter immunogenicity and contribute significantly to the batch‐to‐batch variation of vaccine preparations. Specific limitations regarding host contaminants are clearly stipulated in licensing regulations. Host contaminants can be removed by VLP disassembly and ex vivo reassembly, as necessitated for licensing of HPV L1 VLPs (McCarthy, White, Palmer‐Hill, Koenig, & Suzich, 1998). Unfavorably, this complicates downstream processing and adds to manufacturing costs. Synthetic modification of the HPV L1 and MuPyV VP1 proteins has led to the development of capsomere platforms (Middelberg et al., 2011; Schadlich et al., 2009). Removal of the carboxyl termini from each protein yields capsomeres incapable of forming VLPs in vivo. MuPyV capsomeres display increased stability, although less immunogenic than VLPs when administered with adjuvant they are just as effective (Middelberg et al., 2011). Furthermore, a quantitative process study reported that when produced in E. coli, the MuPyV capsomere platform is capable of producing 320 million vaccine doses in 2.3 days at low cost highlighting its suitability as a rapid response and low‐cost vaccine platform (Chuan, Wibowo, Lua, & Middelberg, 2014).
The safe and robust influenza M1 platform further illustrates the aforementioned, broadly immunoprotective capabilities of VLP platforms. Multiple subtypes of HA antigens have been displayed on M1‐VLPs either individually (Schwartzman et al., 2015) or simultaneously (Pushko et al., 2011; Sequeira et al., 2017; Tretyakova, Pearce, Florese, Tumpey, & Pushko, 2013). M1‐VLP based vaccines afford protection against virus challenge in multiple species (Liu, Massare, et al., 2015; Pyo et al., 2012) and have been the subject of recent influenza phases I and II clinical trials (NCT01897701 and NCT02078674). The vast majority of studies producing M1‐VLPs utilize either mammalian cell lines (transient or stable expression) or insect cell lines (mainly transient expression using the baculovirus expression system, IC‐BEVS). Synthetic biology has recently been applied to the IC‐BEVS platform to overcome process‐related drawbacks. When produced in IC‐BEVS, VLPs and baculovirus are not easily separated owing to current limitations in purification and analytical techniques. A recently developed biorthogonal labeling strategy enables distinction between baculovirus and HA containing M1‐VLPs. Thus, allowing greater control over vaccine contaminants (Carvalho et al., 2016). This system involves fluorescent labeling of an azide‐tagged noncanonical amino acid, which is incorporated into the HA protein within the enveloped VLP. Combinatorial analysis using size exclusion chromatography (SEC), confocal microscopy, and flow cytometry demonstrated the possibility of obtaining VLPs independently of baculovirus. At present, this synthetic modification holds promise as an analytical tool at the laboratory level. However, given the use of fluorescent reagents and their cost, further development is required to address practicality at the industrial scale.
5. CONSIDERATIONS FOR VLP PRODUCTION
The choice of expression host and culture parameters can greatly influence expression yield (Kushnir, Streatfield, & Yusibov, 2012; Lua et al., 2014; Roldão et al., 2010; Vicente, Roldão, Peixoto, Carrondo, & Alves, 2011). E. coli is far superior to other expression systems in terms of cost, yield, and speed making it a popular choice for the production of single capsid protein nonenveloped VLPs (X. Huang, Wang, Zhang, Xia, & Zhao, 2017). The inability of E. coli to perform posttranslational modifications means that complexity is minimized. In cases where posttranslational complex modification is required, IC‐BEVS is used to manufacture licensed VLP vaccines, such as Cervarix® and Porcilis PCV®, yet vaccine costs are high, owing partly to low production yield. Synthetic engineering of baculovirus vectors using combinations of transcriptional or translational elements can increase yield in this system (Gómez‐Sebastián, López‐Vidal, & Escribano, 2014; Liu, Zhang, et al., 2015). Similarly, a synthetically designed expression cassette, containing rearranged genetic regulatory elements, transcription factors IE1 and IE0 and a transcription enhancer sequence linked to differential promoters, increased both expression and cell viability of porcine circovirus VLPs and rabbit hemorrhagic disease VLPs in IC‐BEVS (López‐Vidal et al., 2015).
Maintaining VLP integrity is another key consideration. In multiprotein capsids, specific protein ratios are required to prevent aggregation of unassembled VLPs (L. A. Palomares & Ramirez, 2009; Roldão et al., 2012). Protein stoichiometry can be controlled in IC‐BEVS and mammalian expression systems through manipulating multiplicity of infection, transfection ratios, or thermodynamics (Arevalo, Wong, & Ross, 2016; L. A. Palomares & Ramirez, 2009; Roldão et al., 2012) though it involves increased process cost. Different strength promoters can also be synthetically integrated into multicistronic vectors resulting in differential expression of individual proteins (Jere, O'Neill, Potgieter, & van Dijk, 2014). However, given that optimal ratios vary depending on the specific VLP, individual optimization is required. To surpass the drawbacks of traditional coinfection strategies and/or larger, unstable viral vectors, a modular expression system in which the number of genes to be expressed is rationally distributed between a recombinant viral construct and a stable cell line can be adopted. Such a strategy has proven already to be successful for the production of multivalent influenza VLPs using an insect High Five cell‐based platform (Sequeira et al., 2017).
Vaccine production scale, cost, and purity vary depending on VLP purification processes. Centrifugation, depth, and tangential flow filtration, used for initial clarification and concentration steps, are easily scaled. At the industrial level, chromatographic methods are used to remove host cell and DNA impurities as density gradients and ultracentrifugation are not easily scalable. Anion exchange and SEC are time‐consuming and costly, highlighting the need for more efficient and cost‐effective methods. Nonchromatographic strategies based upon aqueous two‐phase systems (ATPS), a process presently used for enzyme production at the industrial level, are being developed for VLP purification. High yields of rotavirus VLPs have been purified from insect cell supernatant using ATPS, although purity was relatively poor (Benavides et al., 2006). More recently, single and multistep ATPS were used to purify human B19 parvovirus‐like particles from insect cell lysates (Effio et al., 2015). Purity levels were greater than 90%, yet this appeared to be at the expense of yield. Monolith technology presents a rapid and scalable method that offers distinct advantages over the classical packed‐bed chromatography (Vicente et al., 2011). HBsAg VLPs from yeast homogenate was effectively purified using a hydroxyl derivatized monolith (Burden, Jin, Podgornik, & Bracewell, 2012) and when compared to density gradient centrifugation, an anion‐exchange monolith yielded 220‐fold more HIV‐1 gag VLPs from Chinese hamster ovary (CHO) cell supernatant (Steppert et al., 2016). Sulfated cellulose membrane absorbers offer significant improvements over conventional ion exchangers membrane absorbers (Carvalho, Fortuna, et al., 2017). As they are easily scaled and reduce the number of required processing steps they may qualify as a generic purification platform for VLP‐based vaccines. These technologies hold promise for large‐scale VLP purification, warranting further investigation.
VLP characterization is a critical step in analyzing VLP stability and integrity. Additional information can be obtained regarding host‐ and product‐derived impurities, both of which have significant impacts on vaccine efficacy and biosafety. Robust analytical tools are used in tandem to provide comprehensive characterization data. Traditional methods, such as western blot analysis, SEC, and ultracentrifugation are time‐consuming. Others, such as transmission electron microscopy demand high investment costs, extensive preparation work, and specific expertize. Methods allowing swift analysis are currently under development. Tekewe et al. (2015) developed a high throughput method based upon dynamic light scattering technology that permits rapid analysis of capsomere stability based on their hydrodynamic radius. A size exclusion ultrahigh performance liquid chromatography method was also developed in which VLP samples were analyzed in 3.1 min using an interlaced injection technique, allowing accurate quantitation of VLP aggregates (Effio, Oelmeier, & Hubbuch, 2016). More recently, a universal label‐free analytical tool for influenza VLPs quantification based on biolayer interferometry technology applied on an octet platform was developed (Carvalho, Moleirinho, et al., 2017). Overall, these analytical methods support the rapid characterization of large sample sets aiding more efficient vaccine development.
6. VLP VACCINE TRANSLATION
From R&D to licensing, new vaccines can take up to two decades to come to fruition and are estimated to cost from USD 500 million to over 1 billion (S. A. Plotkin, Mahmoud, & Farrar, 2015). VLP vaccines have been under development for >30 years (Lua et al., 2014). Today there are six commercially licensed VLP vaccines and over 110 VLP vaccine trials (completed and current) documented at http://www.clinicaltrials.gov. Over 50% of these trials are designed to assess VLP vaccine candidates against various cancers (where Merck Sharp & Dohme Corp. and GlaxoSmithKline plc. are the leading sponsors) and over 20% against influenza (where Novavax, Inc. and Medicago Inc. dominate the trials). VBI Vaccines Inc. and Takeda Pharmaceutical Co. Ltd. are currently conducting trials for vaccines against cytomegalovirus and norovirus, respectively. India‐based manufacturers, CPL Biologicals Pvt. Ltd. have developed Cadiflu‐S, the world's first VLP influenza vaccine to successfully complete phase 3 trials (http://cadilapharma.com). These multistage clinical trials assess safety and efficacy and determine the suitability of vaccine candidates for licensing approval. Successful translation of vaccines, however, depends upon viable vaccine manufacturing processes to ensure reliable vaccine supply and to minimize production costs.
VLP vaccine platforms provide opportunities to overcome several limitations of traditional vaccine manufacturing. Substantial variation between antigen characteristics of traditional vaccines and their infectious nature adds significantly to production cost as they demand specialized production facilities and tailored manufacturing processes (S. Plotkin, Robinson, Cunningham, Iqbal, & Larsen, 2017). The standardized production of noninfectious, generic vaccine platforms (used to display various antigens) would reduce process variation (i.e., platform remains constant) and provide grounds to establish multiproduct facilities. Streamlining of upstream and downstream processes is likely to increase efficiency, reduce laboratory waste, and permit cost savings through bulk buying of equipment and reagents (Konstantinov & Cooney, 2015). Predictable bioprocesses may allow the standardization of facility infrastructure and the use of single‐use technologies, already in use for some VLP vaccines (Roldão et al., 2010). Disposable technologies offer several advantages over traditional stainless steel equipment including lower initial investment and operating costs, elimination of cross‐contamination, and flexibility in terms of scale (Lopes, 2015; Shukla & Gottschalk, 2013). Combining disposable technologies with VLP platforms offer advantages on multiple levels and would facilitate VLP translation into the industry.
6.1. Regulation and economics
The increased cost and regulatory uncertainty associated with producing new vaccines have long been challenges for vaccine manufacturers (Ulmer, Valley, & Rappuoli, 2006). The intricacies of vaccine manufacturing demand a substantial amount of expertize and contribute significantly to cost (S. Plotkin et al., 2017). Expenses associated with vaccine R&D, testing and manufacturing as well as a poor profit margin compared with current drug markets, have led to a dramatic fall in funding from profit‐driven pharmaceutical companies (Offit, 2005). However, this decline is thought to be premature. As demonstrated throughout this review, VLP technologies are advancing rapidly with a strong focus on developing platform capabilities and improving VLP production yields to enable rapid production of safer and cheaper vaccines, particularly when combined with single‐use bioprocessing systems. This progress is set to continue with financial investment provided from “not‐for‐profit,” sources, such as national governments, international organizations, and philanthropic bodies. In particular, Gavi, the vaccine Alliance (Balaji, 2004) and the Bill & Melinda Gates Foundation (http://www.gatesfoundation.org) are motivated to deliver affordable vaccines to third world countries that are the world's biggest vaccine market. To ensure vaccine purity, safety, efficacy, and stability, regulatory authorities govern every stage of vaccine manufacturing from raw materials and production processes to clinical trials and beyond. This includes the assessment and certification of safe and viable manufacturing processes (S. Plotkin et al., 2017). Meeting regulatory requirements can be a complicated process, particularly when manufacturing vaccines for overseas markets or utilizing newly developed bioprocesses. By streamlining upstream and downstream processes and utilizing a generic VLP base, VLP platform technology is expected to reduce the regulatory load of individual vaccines given that regulations for the base and its purification will become well characterized. VLP platforms may even fast track vaccine delivery in response to pandemic circumstances.
7. FUTURE PERSPECTIVE/CONCLUDING REMARKS
Vaccinology is experiencing an impressive technological revolution, enabling to move vaccines development beyond the rules of Pasteur (empirical approach), using data‐rich disciplines, such as systems biology, immunology, or computational biology to assist rational vaccines design (modern approach). Indeed, the last decade has witnessed a trend toward the use of alternative vaccine designs to attenuated pathogens, having VLPs emerged as a powerful and versatile platform for their production. Today, VLPs are being used not only as vaccines of unmodified viral assemblies against parental viruses but also as scaffolds for displaying heterologous antigens. In addition, tremendous investments have been made to develop new technologies capable of (a) deciphering pathogen biology and vaccine mechanistic responses, and (b) storing and curating extracted data into biological references databases. Machine learning algorithms can then use this information for epitope prediction and structure‐based modular design. Complemented with synthetic biology, this information will provide the basis for (a) engineering VLP functions and (a) developing generic VLP platforms offering not only the potential for streamlined bioprocesses, parallel infrastructure, and predictable biosafety but also the ability to manufacture vaccines against unrelated viruses or pathogens from other sources (i.e., bacteria and parasitic protozoans). Although unable to deliver any marketable product to date, these technologies have massive potential to provide, in near future, solutions against untargeted infectious agents (e.g., antibiotic‐resistant bacteria, HIV, malaria, or tuberculosis), and most importantly to reduce vaccine development times and manufacturing cost associated with current vaccine platforms.
CONFLICTS OF INTEREST
The University of Queensland (UQ) filed patents on the use of MuPyV as a vaccine platform. L. H. L. L. and A. P. J. M. contributed to those patents and, through their employment with UQ, hold an indirect interest in this intellectual property.
ACKNOWLEDGMENTS
The authors acknowledge the support from Australian Research Council (ARC Discovery DP160102915), European Commission (Project EDUFLUVAC, Grant No. 602640), and Portuguese “Fundação para a Ciência e a Tecnologia” through the following programs: FCT Investigator Starting Grant (IF/01704/2014), Exploratory Research and Development Project EXPL/BBB‐BIO/1541/2013, and PhD fellowship SFRH/BD/90564/2012. iNOVA4Health Research Unit (LISBOA‐01‐0145‐FEDER‐007344), which is co‐funded by Fundação para a Ciência e Tecnologia / Ministério da Ciência e do Ensino Superior, through national funds, and by FEDER under the PT2020 Partnership Agreement, is also acknowledged.
Charlton Hume HK, Vidigal J, Carrondo MJT, Middelberg APJ, Roldão A, Lua LHL. Synthetic biology for bioengineering virus‐like particle vaccines. Biotechnology and Bioengineering. 2019;116:919–935. 10.1002/bit.26890
References
REFERENCES
- Anggraeni, M. R. , Connors, N. K. , Wu, Y. , Chuan, Y. P. , Lua, L. H. L. , & Middelberg, A. P. J. (2013). Sensitivity of immune response quality to influenza helix 190 antigen structure displayed on a modular virus‐like particle. Vaccine, 31(40), 4428–4435. [DOI] [PubMed] [Google Scholar]
- Ansari, H. R. , Flower, D. R. , & Raghava, G. P. S. (2010). AntigenDB: An immunoinformatics database of pathogen antigens. Nucleic Acids Research, 38, D847–D853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo, M. T. , Wong, T. M. , & Ross, T. M. (2016). Expression and purification of virus‐like particles for vaccination. Jove‐Journal of Visualized Experiments, 10.3791/54041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, U. , Tyagi, P. , Swaminathan, S. , & Khanna, N. (2012). Chimeric hepatitis B core antigen virus‐like particles displaying the envelope domain III of dengue virus type 2. Journal of Nanobiotechnology, 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, U. , Tyagi, P. , Swaminathan, S. , & Khanna, N. (2013). Virus‐like particles displaying envelope domain III of dengue virus type 2 induce virus‐specific antibody response in mice. Vaccine, 31(6), 873–878. [DOI] [PubMed] [Google Scholar]
- Bagnoli, F. , Baudner, B. , Mishra, R. P. N. , Bartolini, E. , Fiaschi, L. , Mariotti, P. , … Rappuoli, R. (2011). Designing the next generation of vaccines for global public health. OMICS, 15(9), 545–566. [DOI] [PubMed] [Google Scholar]
- Balaji, K. A. (2004). GAVI and the vaccine Fund—A boon for immunization in the developing world. Indian Journal of Public Health, 48(2), 45–48. [PubMed] [Google Scholar]
- Bambini, S. , & Rappuoli, R. (2009). The use of genomics in microbial vaccine development. Drug Discovery Today, 14(5‐6), 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandurska, K. , Brodzik, R. , Spitsin, S. , Kohl, T. , Portocarrero, C. , Smirnov, Y. , … Golovkin, M. (2008). Plant‐produced hepatitis B core protein chimera carrying anthrax protective antigen domain‐4. Hybridoma (2005), 27(4), 241–247. [DOI] [PubMed] [Google Scholar]
- Benavides, J. , Mena, J. A. , Cisneros‐Ruiz, M. , Ramirez, O. T. , Palomares, L. A. , & Rito‐Palomares, M. (2006). Rotavirus‐like particles primary recovery from insect cells in aqueous two‐phase systems. Journal of Chromatography B, Analytical Technologies in the Biomedical and Life Sciences, 842(1), 48–57. [DOI] [PubMed] [Google Scholar]
- Ben‐Yedidia, T. (2011). Progress towards a universal influenza vaccine. Future Virology, 6(2), 237–248. [Google Scholar]
- Bessa, J. , Schmitz, N. , Hinton, H. J. , Schwarz, K. , Jegerlehner, A. , & Bachmann, M. F. (2008). Efficient induction of mucosal and systemic immune responses by virus‐like particles administered intranasally: Implications for vaccine design. European Journal of Immunology, 38(1), 114–126. [DOI] [PubMed] [Google Scholar]
- Bhambure, R. , Kumar, K. , & Rathore, A. S. (2011). High‐throughput process development for biopharmaceutical drug substances. Trends in Biotechnology, 29(3), 127–135. [DOI] [PubMed] [Google Scholar]
- Bird, L. (2017). Immune regulation: Immune cell social networks. Nature Reviews Immunology, 17(4), 216–216. [DOI] [PubMed] [Google Scholar]
- Boeuf, P. , Drummer, H. E. , Richards, J. S. , Scoullar, M. J. L. , & Beeson, J. G. (2016). The global threat of Zika virus to pregnancy: Epidemiology, clinical perspectives, mechanisms, and impact. BMC Medicine, 14(1), 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, F. R. , Gilleland, L. B. , Staczek, J. , Bendig, M. M. , Hamilton, W. D. O. , & Gilleland, H. E., Jr. (1999). A chimaeric plant virus vaccine protects mice against a bacterial infection. Microbiology, 145(Pt 8), 2061–2067. [DOI] [PubMed] [Google Scholar]
- Brune, K. D. , Leneghan, D. B. , Brian, I. J. , Ishizuka, A. S. , Bachmann, M. F. , Draper, S. J. , … Howarth, M. (2016). Plug‐and‐display: Decoration of virus‐like particles via isopeptide bonds for modular immunization. Scientific Reports, 6, 19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy, B. C. , Franciszkowicz, M. J. , & Swartz, J. R. (2008). Escherichia coli‐based cell‐free synthesis of virus‐like particles. Biotechnology and Bioengineering, 100(1), 28–37. [DOI] [PubMed] [Google Scholar]
- Burden, C. S. , Jin, J. , Podgornik, A. , & Bracewell, D. G. (2012). A monolith purification process for virus‐like particles from yeast homogenate. Journal of Chromatography B, Analytical Technologies in The Biomedical and Life Sciences, 880(1), 82–89. [DOI] [PubMed] [Google Scholar]
- Calazans, A. , Boggiano, C. , & Lindsay, R. (2017). A DNA inducing VLP vaccine designed for HIV and tested in mice. PLOS One, 12(8), e0183803. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carter, D. M. , Darby, C. A. , Lefoley, B. C. , Crevar, C. J. , Alefantis, T. , Oomen, R. , … Ross, T. M. (2016). Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. Journal of Virology, 90(9), 4720–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, S. B. , Fortuna, A. R. , Wolff, M. , Peixoto, C. , Alves, P. M. , Reichl, U. , & Carrondo, M. J. T. (2018). Purification of influenza virus‐like particles using sulfated cellulose membrane adsorbers. Journal of Chemical Technology & Biotechnology, 93, 1988–1996. 10.1002/jctb.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, S. B. , Freire, J. M. , Moleirinho, M. G. , Monteiro, F. , Gaspar, D. , Castanho, M. A. R. B. , … Peixoto, C. (2016). Bioorthogonal strategy for bioprocessing of specific‐site‐functionalized enveloped influenza‐virus‐like particles. Bioconjugate Chemistry, 27, 2386–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, S. B. , Moleirinho, M. G. , Wheatley, D. , Welsh, J. , Gantier, R. , Alves, P. M. , … Carrondo, M. J. T. (2017). Universal label‐free in‐process quantification of influenza virus‐like particles. Biotechnology Journal, 12(8) 10.1002/biot.201700031 [DOI] [PubMed] [Google Scholar]
- Chackerian, B. (2007). Virus‐like particles: Flexible platforms for vaccine development. Expert Review of Vaccines, 6(3), 381–390. [DOI] [PubMed] [Google Scholar]
- Chackerian, B. , Lowy, D. R. , & Schiller, J. T. (1999). Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proceedings of the National Academy of Sciences, 96(5), 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton Hume, H. K. , & Lua, L. H. L. (2017). Platform technologies for modern vaccine manufacturing. Vaccine, 35, 4480–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Xiong, X. , Liu, X. , Li, J. , Wen, Y. , Chen, Y. , … Yu, W. (2006). Immunoreactivity of HCV/HBV epitopes displayed in an epitope‐presenting system. Molecular Immunology, 43(5), 436–442. [DOI] [PubMed] [Google Scholar]
- Chu, X. , Li, Y. , Long, Q. , Xia, Y. , Yao, Y. , Sun, W. , … Ma, Y. (2016). Chimeric HBcAg virus‐like particles presenting a HPV 16 E7 epitope significantly suppressed tumor progression through preventive or therapeutic immunization in a TC‐1‐grafted mouse model. International Journal of Nanomedicine, 11, 2417–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, A. J. , Vituret, C. , Tan, M. L. , Gonzalez, G. , Boulanger, P. , Ng, M. L. , & Hong, S. S. (2013). A novel platform for virus‐like particle‐display of flaviviral envelope domain III: Induction of Dengue and West Nile virus neutralizing antibodies. Virology Journal, 10, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuan, Y. P. , Fan, Y. Y. , Lua, L. H. L. , & Middelberg, A. P. J. (2010). Virus assembly occurs following a pH‐ or Ca2+‐triggered switch in the thermodynamic attraction between structural protein capsomeres. Journal of the Royal Society, Interface, 7(44), 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuan, Y. P. , Wibowo, N. , Lua, L. H. L. , & Middelberg, A. P. J. (2014). The economics of virus‐like particle and capsomere vaccines. Biochemical Engineering Journal, 90, 255–263. [Google Scholar]
- Collins, K. A. , Snaith, R. , Cottingham, M. G. , Gilbert, S. C. , & Hill, A. V. S. (2017). Enhancing protective immunity to malaria with a highly immunogenic virus‐like particle vaccine. Scientific Reports, 7, 46621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltart, C. E. M. , Lindsey, B. , Ghinai, I. , Johnson, A. M. , & Heymann, D. L. (2017). The Ebola outbreak, 2013–2016: Old lessons for new epidemics. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 372, 1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin, J. , Zhang, X. Y. , & Reiser, J. (2005). Altering the tropism of lentiviral vectors through pseudotyping. Current Gene Therapy, 5(4), 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filette, M. , Martens, W. , Smet, A. , Schotsaert, M. , Birkett, A. , Londoño‐Arcila, P. , … Saelens, X. (2008). Universal influenza A M2e‐HBc vaccine protects against disease even in the presence of pre‐existing anti‐HBc antibodies. Vaccine, 26(51), 6503–6507. [DOI] [PubMed] [Google Scholar]
- Davis, M. M. , Tato, C. M. , & Furman, D. (2017). Systems immunology: Just getting started. Nature Immunology, 18(7), 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasooraj, D. , Kumar, R. A. , & Mundayoor, S. (2013). Vaccine delivery system for tuberculosis based on nano‐sized hepatitis B virus core protein particles. International Journal of Nanomedicine, 8, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, S. J. , Angov, E. , Horii, T. , Miller, L. H. , Srinivasan, P. , Theisen, M. , & Biswas, S. (2015). Recent advances in recombinant protein‐based malaria vaccines. Vaccine, 33(52), 7433–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Manzalawy, Y. , Dobbs, D. , & Honavar, V. (2008). Predicting linear B‐cell epitopes using string kernels. Journal of Molecular Recognition, 21(4), 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M. , Andreasson, K. , Weidmann, J. , Lundberg, K. , Tegerstedt, K. , Dalianis, T. , & Ramqvist, T. (2011). Murine polyomavirus virus‐like particles carrying full‐length human PSA protect BALB/c mice from outgrowth of a PSA expressing tumor. PLOS One, 6(8), e23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg, H. V. (2014). Pandemic preparedness and response—Lessons from the H1N1 influenza of 2009. New England Journal of Medicine, 370(14), 1335–1342. [DOI] [PubMed] [Google Scholar]
- Fu, T. M. , Grimm, K. M. , Citron, M. P. , Freed, D. C. , Fan, J. , Keller, P. M. , … Joyce, J. G. (2009). Comparative immunogenicity evaluations of influenza A virus M2 peptide as recombinant virus like particle or conjugate vaccines in mice and monkeys. Vaccine, 27(9), 1440–1447. [DOI] [PubMed] [Google Scholar]
- Garcia‐Boronat, M. , Diez‐Rivero, C. M. , Reinherz, E. L. , & Reche, P. A. (2008). PVS: A web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Research, 36, W35–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause, K. T. , Wheatley, A. K. , Cui, J. , Yan, Y. , Kent, S. J. , & Caruso, F. (2017). Immunological principles guiding the rational design of particles for vaccine delivery. ACS Nano, 11(1), 54–68. [DOI] [PubMed] [Google Scholar]
- Gong, C. S. A. , & Lei, K. F. (2014). Advances in miniaturized instruments for genomics. BioMed Research International, 2014, 734675–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grgacic, E. V. L. , & Anderson, D. A. (2006). Virus‐like particles: Passport to immune recognition. Methods, 40(1), 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Sebastián, S. , López‐Vidal, J. , & Escribano, J. M. (2014). Significant productivity improvement of the baculovirus expression vector system by engineering a novel expression cassette. PLOS One, 9(5), e96562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harahap‐Carrillo, I. S. , Ceballos‐Olvera, I. , & Valle, J. R. (2015). Immunogenic subviral particles displaying domain III of dengue 2 envelope protein vectored by measles virus. Vaccines, 3(3), 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, J. R. , Cunningham, J. , Von Seefried, A. , Lennick, M. , Garvin, R. T. , & Shen, S. H. (1986). Development of a genetically‐engineered, candidate polio vaccine employing the self‐assembling properties of the tobacco mosaic‐virus coat protein. Bio‐Technology, 4(7), 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, J. R. , Dokken, L. , Wiley, J. A. , Cawthon, A. G. , Bigger, J. , Harmsen, A. G. , & Richardson, C. (2009). Influenza‐pseudotyped Gag virus‐like particle vaccines provide broad protection against highly pathogenic avian influenza challenge. Vaccine, 27(4), 530–541. [DOI] [PubMed] [Google Scholar]
- He, Y. , & Xiang, Z. (2013). Databases and in silico tools for vaccine design. Methods in Molecular Biology, 993, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich, J. , Noack, S. , Wiechert, W. , & Oldiges, M. (2018). Microbioreactor systems for accelerated bioprocess development. Biotechnology Journal, 13(4), 1700141. [DOI] [PubMed] [Google Scholar]
- Hou, J. , Wang, S. , Jia, M. , Li, D. , Liu, Y. , Li, Z. , … Shao, Y. (2017). A systems vaccinology approach reveals temporal transcriptomic changes of immune responses to the yellow fever 17D vaccine. Journal of Immunology, 199(4), 1476–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Doria‐Rose, N. A. , Longo, N. S. , Laub, L. , Lin, C. L. , Turk, E. , … Connors, M. (2013). Isolation of human monoclonal antibodies from peripheral blood B cells. Nature Protocols, 8(10), 1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Kang, B. H. , Pancera, M. , Lee, J. H. , Tong, T. , Feng, Y. , … Connors, M. (2014). Broad and potent HIV‐1 neutralization by a human antibody that binds the gp41‐gp120 interface. Nature, 515(7525), 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Wang, X. , Zhang, J. , Xia, N. , & Zhao, Q. (2017). Escherichia coli‐derived virus‐like particles in vaccine development. NPJ Vaccines, 2(3), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, Z. , Smyth, H. D. , Durfee, P. , & Chackerian, B. (2009). Induction of mucosal and systemic antibody responses against the HIV coreceptor CCR5 upon intramuscular immunization and aerosol delivery of a virus‐like particle based vaccine. Vaccine, 28(2), 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingale, J. , Tran, K. , Kong, L. , Dey, B. , McKee, K. , Schief, W. , … Wyatt, R. T. (2014). Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies. Journal of Virology, 88(24), 14002–14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janitzek, C. M. , Matondo, S. , Thrane, S. , Nielsen, M. A. , Kavishe, R. , Mwakalinga, S. B. , … Sander, A. F. (2016). Bacterial superglue generates a full‐length circumsporozoite protein virus‐like particle vaccine capable of inducing high and durable antibody responses. Malaria Journal, 15(1), 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegerlehner, A. , Zabel, F. , Langer, A. , Dietmeier, K. , Jennings, G. T. , Saudan, P. , & Bachmann, M. F. (2013). Bacterially produced recombinant influenza vaccines based on virus‐like particles. PLOS One, 8(11), e78947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, G. T. , & Bachmann, M. F. (2009). Immunodrugs: Therapeutic VLP‐based vaccines for chronic diseases. Annual Review of Pharmacology and Toxicology, 49, 303–326. [DOI] [PubMed] [Google Scholar]
- Jere, K. C. , O'Neill, H. G. , Potgieter, A. C. , & van Dijk, A. A. (2014). Chimaeric virus‐like particles derived from consensus genome sequences of human rotavirus strains co‐circulating in Africa. PLOS One, 9(9), e105167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. , Brorson, K. A. , Frey, D. D. , Dhar, A. K. , & Cetlin, D. A. (2017). Characterization of non‐infectious virus‐like particle surrogates for viral clearance applications. Applied Biochemistry and Biotechnology, 183, 318–331. [DOI] [PubMed] [Google Scholar]
- Kaczmarczyk, S. J. , Sitaraman, K. , Young, H. A. , Hughes, S. H. , & Chatterjee, D. K. (2011). Protein delivery using engineered virus‐like particles. Proceedings of the National Academy of Sciences of the United States of America, 108(41), 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmin, D. , Nakaya, H. I. , Lee, E. K. , Johnson, M. J. , van der Most, R. , van den Berg, R. A. , … Pulendran, B. (2017). Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proceedings of the National Academy of Sciences of the United States of America, 114(9), 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. H. , Lee, Y. T. , Hwang, H. S. , Kwon, Y. M. , Kim, M. C. , Ko, E. J. , … Kang, S. M. (2015). Virus‐like particle vaccine containing the f protein of respiratory syncytial virus confers protection without pulmonary disease by modulating specific subsets of dendritic cells and effector T cells. Journal of Virology, 89(22), 11692–11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmeier, M. , Fluckiger, A. C. , Soare, C. , Bozic, J. , Ontsouka, B. , Ahmed, T. , … Anderson, D. E. (2014). Enveloped virus‐like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity. Clinical and Vaccine Immunology, 21(2), 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, P. , Wallich, R. , & Nassal, M. (2015). Whole‐chain tick saliva proteins presented on hepatitis b virus capsid‐like particles induce high‐titered antibodies with neutralizing potential. PLOS One, 10(9), e0136180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov, K. B. , & Cooney, C. L. (2015). White paper on continuous bioprocessing. May 20‐21, 2014 continuous manufacturing symposium. Journal of Pharmaceutical Sciences, 104(3), 813–820. [DOI] [PubMed] [Google Scholar]
- Koo, M. , Bendahmane, M. , Lettieri, G. A. , Paoletti, A. D. , Lane, T. E. , Fitchen, J. H. , … Beachy, R. N. (1999). Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proceedings of the National Academy of Sciences of the United States of America, 96(14), 7774–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer, F. (2015). Emerging influenza viruses and the prospect of a universal influenza virus vaccine. Biotechnology Journal, 10(5), 690–701. [DOI] [PubMed] [Google Scholar]
- Kratz, P. A. , Bottcher, B. , & Nassal, M. (1999). Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proceedings of the National Academy of Sciences of the United States of America, 96(5), 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleš, J. , Horvatić, A. , Guillemin, N. , Galan, A. , Mrljak, V. , & Bhide, M. (2016). New approaches and omics tools for mining of vaccine candidates against vector‐borne diseases. Molecular BioSystems, 12(9), 2680–2694. [DOI] [PubMed] [Google Scholar]
- Kundig, T. , Senti, G. , Schnetzler, G. , Wolf, C. , Prinzvavricka, B. , Fulurija, A. , … Bachmann, M. (2006). Der p 1 peptide on virus‐like particles is safe and highly immunogenic in healthy adults. Journal of Allergy and Clinical Immunology, 117(6), 1470–1476. [DOI] [PubMed] [Google Scholar]
- Kurg, R. , Reinsalu, O. , Jagur, S. , Õunap, K. , Võsa, L. , Kasvandik, S. , … Ustav, M. (2016). Biochemical and proteomic characterization of retrovirus Gag based microparticles carrying melanoma antigens. Scientific Reports, 6, 29425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir, N. , Streatfield, S. J. , & Yusibov, V. (2012). Virus‐like particles as a highly efficient vaccine platform: Diversity of targets and production systems and advances in clinical development. Vaccine, 31(1), 58–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd Effio, C. , Oelmeier, S. A. , & Hubbuch, J. (2016). High‐throughput characterization of virus‐like particles by interlaced size‐exclusion chromatography. Vaccine, 34(10), 1259–1267. [DOI] [PubMed] [Google Scholar]
- Ladd effio, C. , Wenger, L. , Ötes, O. , Oelmeier, S. A. , Kneusel, R. , & Hubbuch, J. (2015). Downstream processing of virus‐like particles: Single‐stage and multi‐stage aqueous two‐phase extraction. Journal of Chromatography A, 1383, 35–46. [DOI] [PubMed] [Google Scholar]
- Langeveld, J. P. M. , Brennan, F. R. , Martínez‐Torrecuadrada, J. L. , Jones, T. D. , Boshuizen, R. S. , Vela, C. , … Hamilton, W. D. O. (2001). Inactivated recombinant plant virus protects dogs from a lethal challenge with canine parvovirus. Vaccine, 19(27), 3661–3670. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Cao, C. , Chackerian, B. , Schiller, J. , Gordon, M. , Ugen, K. E. , & Morgan, D. (2004). Overcoming antigen masking of anti‐amyloidbeta antibodies reveals breaking of B cell tolerance by virus‐like particles in amyloidbeta immunized amyloid precursor protein transgenic mice. BMC Neuroscience, 5(1), 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S. , Zheng, D. , Standley, D. M. , Yao, B. , Zacharias, M. , & Zhang, C. (2010). EPSVR and EPMeta: Prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinformatics, 11, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W. , Zhang, T. T. , Gao, L. , Lee, S. S. , Xu, J. , Zhang, H. , … Li, W. (2017). Integration of novel materials and advanced genomic technologies into new vaccine design. Current Topics in Medicinal Chemistry, 17(20), 2286–2301. [DOI] [PubMed] [Google Scholar]
- Liljeroos, L. , Malito, E. , Ferlenghi, I. , & Bottomley, M. J. (2015). Structural and computational biology in the design of immunogenic vaccine antigens. Journal of Immunology Research, 2015, 156241–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. S. , Liu, W. J. , Zhao, K. N. , Liu, Y. H. , Leggatt, G. , & Frazer, I. H. (2002). Route of administration of chimeric BPV1 VLP determines the character of the induced immune responses. Immunology and Cell Biology, 80(1), 21–29. [DOI] [PubMed] [Google Scholar]
- Liu, Y. K. , Zhang, Y. Y. , Yao, L. G. , Hao, H. F. , Fu, X. J. , Yang, Z. Q. , & Du, E. Q. (2015). Enhanced production of porcine circovirus type 2 (PCV2) virus‐like particles in Sf9 cells by translational enhancers. Biotechnology Letters, 37(9), 1765–1771. [DOI] [PubMed] [Google Scholar]
- Liu, Y. V. , Massare, M. J. , Pearce, M. B. , Sun, X. , Belser, J. A. , Maines, T. R. , … Tumpey, T. M. (2015). Recombinant virus‐like particles elicit protective immunity against avian influenza A(H7N9) virus infection in ferrets. Vaccine, 33(18), 2152–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisy, F. , Atmar, R. L. , Le Saux, J. C. , Cohen, J. , Caprais, M. P. , Pommepuy, M. , & Le Guyader, F. S. (2005). Use of rotavirus virus‐like particles as surrogates to evaluate virus persistence in shellfish. Applied and Environmental Microbiology, 71(10), 6049–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, A. G. (2015). Single‐use in the biopharmaceutical industry: A review of current technology impact, challenges and limitations. Food and Bioproducts Processing, 93, 98–114. [Google Scholar]
- Lua, L. H. L. , Connors, N. K. , Sainsbury, F. , Chuan, Y. P. , Wibowo, N. , & Middelberg, A. P. J. (2014). Bioengineering virus‐like particles as vaccines. Biotechnology and Bioengineering, 111(3), 425–440. [DOI] [PubMed] [Google Scholar]
- Lua, L. H. L. , Fan, Y. , Chang, C. , Connors, N. K. , & Middelberg, A. P. J. (2015). Synthetic biology design to display an 18 kDa rotavirus large antigen on a modular virus‐like particle. Vaccine, 33(44), 5937–5944. [DOI] [PubMed] [Google Scholar]
- López‐Vidal, J. , Gómez‐Sebastián, S. , Bárcena, J. , Nuñez, M. C. , Martínez‐Alonso, D. , Dudognon, B. , … Escribano, J. M. (2015). Improved production efficiency of virus‐like particles by the baculovirus expression vector system. PLOS One, 10(10), e0140039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malito, E. , Carfi, A. , & Bottomley, M. J. (2015). Protein crystallography in vaccine research and development. International Journal of Molecular Sciences, 16(6), 13106–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manayani, D. J. , Thomas, D. , Dryden, K. A. , Reddy, V. , Siladi, M. E. , Marlett, J. M. , … Schneemann, A. (2007). A viral nanoparticle with dual function as an anthrax antitoxin and vaccine. PLOS Pathogens, 3(10), e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell, R. B. , Koukuntla, R. , Mogler, L. J. K. , Carzoli, A. K. , Freiberg, A. N. , Holbrook, M. R. , … Flick, R. (2010). A replication‐incompetent Rift Valley fever vaccine: Chimeric virus‐like particles protect mice and rats against lethal challenge. Virology, 397(1), 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu, M. G. (2011). Virus engineering: Functionalization and stabilization. Protein Engineering, Design & Selection, 24(1‐2), 53–63. [DOI] [PubMed] [Google Scholar]
- Maurer, P. , Jennings, G. T. , Willers, J. , Rohner, F. , Lindman, Y. , Roubicek, K. , … Bachmann, M. F. (2005). Frontline: A therapeutic vaccine for nicotine dependence: Preclinical efficacy, and phase I safety and immunogenicity. European Journal of Immunology, 35(7), 2031–2040. [DOI] [PubMed] [Google Scholar]
- McCarthy, M. P. , White, W. I. , Palmer‐Hill, F. , Koenig, S. , & Suzich, J. A. (1998). Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. Journal of Virology, 72(1), 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy, A. C. , & Adams, B. (2009). The role of genomics in tracking the evolution of influenza A virus. PLOS Pathogens, 5(10), e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLain, L. , Porta, C. , Lomonossoff, G. P. , Durrani, Z. , & Dimmock, N. J. (1995). Human immunodeficiency virus type 1‐neutralizing antibodies raised to a glycoprotein 41 peptide expressed on the surface of a plant virus. AIDS Research and Human Retroviruses, 11(3), 327–334. [DOI] [PubMed] [Google Scholar]
- Middelberg, A. P. J. , Rivera‐Hernandez, T. , Wibowo, N. , Lua, L. H. L. , Fan, Y. , Magor, G. , … Batzloff, M. R. (2011). A microbial platform for rapid and low‐cost virus‐like particle and capsomere vaccines. Vaccine, 29(41), 7154–7162. [DOI] [PubMed] [Google Scholar]
- Mihailova, M. , Boos, M. , Petrovskis, I. , Ose, V. , Skrastina, D. , Fiedler, M. , … Viazov, S. (2006). Recombinant virus‐like particles as a carrier of B‐ and T‐cell epitopes of hepatitis C virus (HCV). Vaccine, 24(20), 4369–4377. [DOI] [PubMed] [Google Scholar]
- Mulder, A. M. , Carragher, B. , Towne, V. , Meng, Y. , Wang, Y. , Dieter, L. , … Zhao, Q. (2012). Toolbox for non‐intrusive structural and functional analysis of recombinant VLP based vaccines: A case study with hepatitis B vaccine. PLOS One, 7(4), e33235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, Y. , Lightfoote, P. M. , Rose, R. C. , & Walsh, E. E. (2009). Antigenic presentation of heterologous epitopes engineered into the outer surface‐exposed helix 4 loop region of human papillomavirus L1 capsomeres. Virology Journal, 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyanja, E. , Ssemaganda, A. , Ngauv, P. , Cubas, R. , Perrin, H. , Srinivasan, D. , … Trautmann, L. (2014). Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. Journal of Clinical Investigation, 124(7), 3147–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal, M. , Skamel, C. , Vogel, M. , Kratz, P. A. , Stehle, T. , Wallich, R. , & Simon, M. M. (2008). Development of hepatitis B virus capsids into a whole‐chain protein antigen display platform: New particulate Lyme disease vaccines. International Journal of Medical Microbiology, 298(1‐2), 135–142. [DOI] [PubMed] [Google Scholar]
- Nassal, M. , Skamel, C. , Kratz, P. A. , Wallich, R. , Stehle, T. , & Simon, M. M. (2005). A fusion product of the complete Borrelia burgdorferi outer surface protein A (OspA) and the hepatitis B virus capsid protein is highly immunogenic and induces protective immunity similar to that seen with an effective lipidated OspA vaccine formula. European Journal of Immunology, 35(2), 655–665. [DOI] [PubMed] [Google Scholar]
- Nuzzaci, M. , Piazzolla, G. , Vitti, A. , Lapelosa, M. , Tortorella, C. , Stella, I. , … Piazzolla, P. (2007). Cucumber mosaic virus as a presentation system for a double hepatitis C virus‐derived epitope. Archives of Virology, 152(5), 915–928. [DOI] [PubMed] [Google Scholar]
- Offit, P. A. (2005). Why are pharmaceutical companies gradually abandoning vaccines? Health Affairs, 24(3), 622–630. [DOI] [PubMed] [Google Scholar]
- Okba, N. M. , Raj, V. S. , & Haagmans, B. L. (2017). Middle East respiratory syndrome coronavirus vaccines: Current status and novel approaches. Current Opinion in Virology, 23, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'rourke, J. P. , Peabody, D. S. , & Chackerian, B. (2015). Affinity selection of epitope‐based vaccines using a bacteriophage virus‐like particle platform. Current Opinion in Virology, 11, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, K. E. , Benko, A. , Doucette, S. A. , Cameron, T. I. , Foster, T. , Hanley, K. M. , … Christensen, N. D. (2006). Protection of rabbits against cutaneous papillomavirus infection using recombinant tobacco mosaic virus containing L2 capsid epitopes. Vaccine, 24(26), 5516–5525. [DOI] [PubMed] [Google Scholar]
- Palomares, K. , Vigant, F. , Van Handel, B. , Pernet, O. , Chikere, K. , Hong, P. , … Lee, B. (2013). Nipah virus envelope‐pseudotyped lentiviruses efficiently target ephrinB2‐positive stem cell populations in vitro and bypass the liver sink when administered in vivo. Journal of Virology, 87(4), 2094–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares, L. A. , & Ramírez, O. T. (2009). Challenges for the production of virus‐like particles in insect cells: The case of rotavirus‐like particles. Biochemical Engineering Journal, 45(3), 158–167. [Google Scholar]
- Pastori, C. , Tudor, D. , Diomede, L. , Drillet, A. S. , Jegerlehner, A. , Röhn, T. A. , … Lopalco, L. (2012). Virus like particle based strategy to elicit HIV‐protective antibodies to the alpha‐helic regions of gp41. Virology, 431(1‐2), 1–11. [DOI] [PubMed] [Google Scholar]
- Patel, K. G. , & Swartz, J. R. (2011). Surface functionalization of virus‐like particles by direct conjugation using azide‐alkyne click chemistry. Bioconjugate Chemistry, 22(3), 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patronov, A. , & Doytchinova, I. (2013). T‐cell epitope vaccine design by immunoinformatics. Open Biology, 3(1), 120139–120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattenden, L. K. , Middelberg, A. P. J. , Niebert, M. , & Lipin, D. I. (2005). Towards the preparative and large‐scale precision manufacture of virus‐like particles. Trends in Biotechnology, 23(10), 523–529. [DOI] [PubMed] [Google Scholar]
- Peacey, M. , Wilson, S. , Baird, M. A. , & Ward, V. K. (2007). Versatile RHDV virus‐like particles: Incorporation of antigens by genetic modification and chemical conjugation. Biotechnology and Bioengineering, 98(5), 968–977. [DOI] [PubMed] [Google Scholar]
- Peters, B. , Sidney, J. , Bourne, P. , Bui, H. H. , Buus, S. , Doh, G. , … Sette, A. (2005). The immune epitope database and analysis resource: From vision to blueprint. PLOS Biology, 3(3), e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret, H. , Gehin, A. , Thuenemann, E. C. , Blond, D. , El Turabi, A. , Beales, L. , … Rowlands, D. J. (2015). Tandem fusion of hepatitis B core antigen allows assembly of virus‐like particles in bacteria and plants with enhanced capacity to accommodate foreign proteins. PLOS One, 10(4), e0120751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoiset, F. , Vazquez, T. , & Bellier, B. (2015). Enveloped virus‐like particle platforms: Vaccines of the future? Expert Review of Vaccines, 14(7), 913–915. [DOI] [PubMed] [Google Scholar]
- Plotkin, S. , Robinson, J. M. , Cunningham, G. , Iqbal, R. , & Larsen, S. (2017). The complexity and cost of vaccine manufacturing—An overview. Vaccine, 35(33), 4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin, S. A. , Mahmoud, A. A. F. , & Farrar, J. (2015). Establishing a global vaccine‐development fund. New England Journal of Medicine, 373(4), 297–300. [DOI] [PubMed] [Google Scholar]
- Prabakaran, M. , He, F. , Meng, T. , Madhan, S. , Yunrui, T. , Jia, Q. , & Kwang, J. (2010). Neutralizing epitopes of influenza virus hemagglutinin: Target for the development of a universal vaccine against H5N1 lineages. Journal of Virology, 84(22), 11822–11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushko, P. , Pearce, M. B. , Ahmad, A. , Tretyakova, I. , Smith, G. , Belser, J. A. , & Tumpey, T. M. (2011). Influenza virus‐like particle can accommodate multiple subtypes of hemagglutinin and protect from multiple influenza types and subtypes. Vaccine, 29(35), 5911–5918. [DOI] [PubMed] [Google Scholar]
- Pushko, P. , Tretyakova, I. , Hidajat, R. , Zsak, A. , Chrzastek, K. , Tumpey, T. M. , & Kapczynski, D. R. (2017). Virus‐like particles displaying H5, H7, H9 hemagglutinins and N1 neuraminidase elicit protective immunity to heterologous avian influenza viruses in chickens. Virology, 501, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo, H. M. , Masic, A. , Woldeab, N. , Embury‐Hyatt, C. , Lin, L. , Shin, Y. K. , … Zhou, Y. (2012). Pandemic H1N1 influenza virus‐like particles are immunogenic and provide protective immunity to pigs. Vaccine, 30(7), 1297–1304. [DOI] [PubMed] [Google Scholar]
- Rappuoli, R. (2007). Bridging the knowledge gaps in vaccine design. Nature Biotechnology, 25(12), 1361–1366. [DOI] [PubMed] [Google Scholar]
- Rappuoli, R. , Bottomley, M. J. , D'oro, U. , Finco, O. , & De Gregorio, E. (2016). Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. Journal of Experimetnal Medicine, 213(4), 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennermalm, A. , Li, Y. H. , Bohaufs, L. , Jarstrand, C. , Brauner, A. , Brennan, F. R. , & Flock, J. I. (2001). Antibodies against a truncated Staphylococcus aureus fibronectin‐binding protein protect against dissemination of infection in the rat. Vaccine, 19(25‐26), 3376–3383. [DOI] [PubMed] [Google Scholar]
- Roldão, A. , Mellado, M. C. M. , Castilho, L. R. , Carrondo, M. J. , & Alves, P. M. (2010). Virus‐like particles in vaccine development. Expert Review of Vaccines, 9(10), 1149–1176. [DOI] [PubMed] [Google Scholar]
- Roldão, A. , Mellado, M. C. M. , Lima, J. C. , Carrondo, M. J. T. , Alves, P. M. , & Oliveira, R. (2012). On the effect of thermodynamic equilibrium on the assembly efficiency of complex multi‐layered virus‐like particles (VLP): The case of rotavirus VLP. PLOS Computational Biology, 8(2), e1002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, J. A. , Chen, L. , Baker, J. L. , Putnam, D. , & DeLisa, M. P. (2014). Pathogen‐like particles: Biomimetic vaccine carriers engineered at the nanoscale. Current Opinion in Biotechnology, 28, 51–58. [DOI] [PubMed] [Google Scholar]
- Ruder, W. C. , Lu, T. , & Collins, J. J. (2011). Synthetic biology moving into the clinic. Science, 333(6047), 1248–1252. [DOI] [PubMed] [Google Scholar]
- Salomon, J. , & Flower, D. R. (2006). Predicting class II MHC‐peptide binding: A kernel based approach using similarity scores. BMC Bioinformatics, 7, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadlich, L. , Senger, T. , Gerlach, B. , Mucke, N. , Klein, C. , Bravo, I. G. , … Gissmann, L. (2009). Analysis of modified human papillomavirus type 16 L1 capsomeres: The ability to assemble into larger particles correlates with higher immunogenicity. Journal of Virology, 83(15), 7690–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneemann, A. , Speir, J. A. , Tan, G. S. , Khayat, R. , Ekiert, D. C. , Matsuoka, Y. , & Wilson, I. A. (2012). A virus‐like particle that elicits cross‐reactive antibodies to the conserved stem of influenza virus hemagglutinin. Journal of Virology, 86(21), 11686–11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M. M. , Nastke, M. D. , & Stevanović, S. (2007). SYFPEITHI: Database for searching and T‐cell epitope prediction. Methods in Molecular Biology, 409, 75–93. [DOI] [PubMed] [Google Scholar]
- Schwartzman, L. M. , Cathcart, A. L. , Pujanauski, L. M. , Qi, L. , Kash, J. C. , & Taubenberger, J. K. (2015). An intranasal virus‐like particle vaccine broadly protects mice from multiple subtypes of influenza A virus. mBio, 6(4), e01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, B. , & Douglas, T. (2015). Development of virus‐like particles for diagnostic and prophylactic biomedical applications. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 7(5), 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel, F. , Moriarty, A. M. , Peterson, D. L. , Zheng, J. A. , Hughes, J. L. , Will, H. , … Milich, D. R. (1992). The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. Journal of Virology, 66(1), 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scodeller, E. A. T. , Sergio, G. , Porro, F. , Schiappacassi, M. , De Rossi, A. , Chiecco‐Bianchi, L. , … Francisco, E. (1995). A new epitope presenting system displays a HIV‐1 V3 loop sequence and induces neutralizing antibodies. Vaccine, 13(13), 1233–1239. [DOI] [PubMed] [Google Scholar]
- Sequeira, D. P. , Correia, R. , Carrondo, M. J. , Roldao, A. , Teixeira, A. P. , & Alves, P. M. (2017). Combining stable insect cell lines with baculovirus‐mediated expression for multi‐HA influenza VLP production. Vaccine, 36(22), 3112–3123. 10.1016/j.vaccine.2017.02.043 [DOI] [PubMed] [Google Scholar]
- Sette, A. , & Rappuoli, R. (2010). Reverse vaccinology: Developing vaccines in the era of genomics. Immunity, 33(4), 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, A. A. , & Gottschalk, U. (2013). Single‐use disposable technologies for biopharmaceutical manufacturing. Trends in Biotechnology, 31(3), 147–154. [DOI] [PubMed] [Google Scholar]
- Slupetzky, K. , Shafti‐Keramat, S. , Grassauer, A. , Lenz, P. , Brandt, S. , Grassauer, A. , & Sara, M. (2001). Chimeric papillomavirus‐like particles expressing a foreign epitope on capsid surface loops. Journal of General Virology, 82(Pt 11), 2799–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupetzky, K. , Gambhira, R. , Culp, T. D. , Shafti‐Keramat, S. , Schellenbacher, C. , Christensen, N. D. , … Kirnbauer, R. (2007). A papillomavirus‐like particle (VLP) vaccine displaying HPV16 L2 epitopes induces cross‐neutralizing antibodies to HPV11. Vaccine, 25(11), 2001–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. T. , Hawes, A. K. , & Bundy, B. C. (2013). Reengineering viruses and virus‐like particles through chemical functionalization strategies. Current Opinion in Biotechnology, 24(4), 620–626. [DOI] [PubMed] [Google Scholar]
- Song, J. M. , Wang, B. Z. , Park, K. M. , Van Rooijen, N. , Quan, F. S. , Kim, M. C. , … Kang, S. M. (2011). Influenza virus‐like particles containing M2 induce broadly cross protective immunity. PLOS One, 6(1), e14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria‐Guerra, R. E. , Nieto‐Gomez, R. , Govea‐Alonso, D. O. , & Rosales‐Mendoza, S. (2015). An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. Journal of Biomedical Informatics, 53, 405–414. [DOI] [PubMed] [Google Scholar]
- Spohn, G. , Jennings, G. T. , Martina, B. E. , Keller, I. , Beck, M. , Pumpens, P. , … Bachmann, M. F. (2010). A VLP‐based vaccine targeting domain III of the West Nile virus E protein protects from lethal infection in mice. Virology Journal, 7, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohn, G. , Schori, C. , Keller, I. , Sladko, K. , Sina, C. , Guler, R. , … Bachmann, M. F. (2014). Preclinical efficacy and safety of an anti‐IL‐1β vaccine for the treatment of type 2 diabetes. Molecular Therapy. Methods & Clinical Development, 1, 14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staczek, J. , Bendahmane, M. , Gilleland, L. B. , Beachy, R. N. , & Gilleland, H. E. (2000). Immunization with a chimeric tobacco mosaic virus containing an epitope of outer membrane protein F of Pseudomonas aeruginosa provides protection against challenge with P. aeruginosa . Vaccine, 18(21), 2266–2274. [DOI] [PubMed] [Google Scholar]
- Steppert, P. , Burgstaller, D. , Klausberger, M. , Berger, E. , Aguilar, P. P. , Schneider, T. A. , … Jungbauer, A. (2016). Purification of HIV‐1 gag virus‐like particles and separation of other extracellular particles. Journal of Chromatography A, 1455, 93–101. [DOI] [PubMed] [Google Scholar]
- Stoute, J. A. , Slaoui, M. , Heppner, D. G. , Momin, P. , Kester, K. E. , Desmons, P. , … Cohen, J. D. (1997). A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. New England Journal of Medicine, 336(2), 86–91. [DOI] [PubMed] [Google Scholar]
- Sweredoski, M. J. , & Baldi, P. (2009). COBEpro: A novel system for predicting continuous B‐cell epitopes. Protein Engineering, Design & Selection, 22(3), 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sällberg, M. , Hughes, J. , Jones, J. , Phillips, T. R. , & Milich, D. R. (2002). A malaria vaccine candidate based on a hepatitis B virus core platform. Intervirology, 45(4‐6), 350–361. [DOI] [PubMed] [Google Scholar]
- Takamura, S. , Niikura, M. , Li, T. C. , Takeda, N. , Kusagawa, S. , Takebe, Y. , … Yasutomi, Y. (2004). DNA vaccine‐encapsulated virus‐like particles derived from an orally transmissible virus stimulate mucosal and systemic immune responses by oral administration. Gene Therapy, 11(7), 628–635. [DOI] [PubMed] [Google Scholar]
- Tang, S. , Xuan, B. , Ye, X. , Huang, Z. , & Qian, Z. (2016). A modular vaccine development platform based on sortase‐mediated site‐specific tagging of antigens onto virus‐like particles. Scientific Reports, 6, 25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegerstedt, K. , Franzen, A. V. , Andreasson, K. , Joneberg, J. , Heidari, S. , Ramqvist, T. , & Dalianis, T. (2005). Murine polyomavirus virus‐like particles (VLPs) as vectors for gene and immune therapy and vaccines against viral infections and cancer. Anticancer Research, 25(4), 2601–2608. [PubMed] [Google Scholar]
- Tegerstedt, K. , Lindencrona, J. A. , Curcio, C. , Andreasson, K. , Tullus, C. , Forni, G. , … Ramqvist, T. (2005). A single vaccination with polyomavirus VP1/VP2Her2 virus‐like particles prevents outgrowth of HER‐2/neu‐expressing tumors. Cancer Research, 65(13), 5953–5957. [DOI] [PubMed] [Google Scholar]
- Tekewe, A. , Connors, N. K. , Sainsbury, F. , Wibowo, N. , Lua, L. H. L. , & Middelberg, A. P. J. (2015). A rapid and simple screening method to identify conditions for enhanced stability of modular vaccine candidates. Biochemical Engineering Journal, 100, 50–58. [Google Scholar]
- Tekewe, A. , Fan, Y. , Tan, E. , Middelberg, A. P. J. , & Lua, L. H. L. (2017). Integrated molecular and bioprocess engineering for bacterially produced immunogenic modular virus‐like particle vaccine displaying 18 kDa rotavirus antigen. Biotechnology and Bioengineering, 114(2), 397–406. [DOI] [PubMed] [Google Scholar]
- Thrane, S. , Janitzek, C. M. , Agerbæk, M. Ø. , Ditlev, S. B. , Resende, M. , Nielsen, M. A. , … Sander, A. F. (2015). A novel virus‐like particle based vaccine platform displaying the placental malaria antigen VAR2CSA. PLOS One, 10(11), e0143071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane, S. , Janitzek, C. M. , Matondo, S. , Resende, M. , Gustavsson, T. , de Jongh, W. A. , … Sander, A. F. (2016). Bacterial superglue enables easy development of efficient virus‐like particle based vaccines. Journal of Nanobiotechnology, 14, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot, A. C. , Maurer, P. , Nussberger, J. , Sabat, R. , Pfister, T. , Ignatenko, S. , … Bachmann, M. F. (2008). Effect of immunisation against angiotensin II with CYT006‐AngQb on ambulatory blood pressure: A double‐blind, randomised, placebo‐controlled phase IIa study. Lancet, 371(9615), 821–827. [DOI] [PubMed] [Google Scholar]
- Tissot, A. C. , Renhofa, R. , Schmitz, N. , Cielens, I. , Meijerink, E. , Ose, V. , … Bachmann, M. F. (2010). Versatile virus‐like particle carrier for epitope based vaccines. PLOS One, 5(3), e9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova, I. , Hidajat, R. , Hamilton, G. , Horn, N. , Nickols, B. , Prather, R. O. , … Pushko, P. (2016). Preparation of quadri‐subtype influenza virus‐like particles using bovine immunodeficiency virus gag protein. Virology, 487, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova, I. , Pearce, M. B. , Florese, R. , Tumpey, T. M. , & Pushko, P. (2013). Intranasal vaccination with H5, H7 and H9 hemagglutinins co‐localized in a virus‐like particle protects ferrets from multiple avian influenza viruses. Virology, 442(1), 67–73. [DOI] [PubMed] [Google Scholar]
- Ulmer, J. B. , Valley, U. , & Rappuoli, R. (2006). Vaccine manufacturing: Challenges and solutions. Nature Biotechnology, 24(11), 1377–1383. [DOI] [PubMed] [Google Scholar]
- Vicente, T. , Roldão, A. , Peixoto, C. , Carrondo, M. J. T. , & Alves, P. M. (2011). Large‐scale production and purification of VLP‐based vaccines. Journal of Invertebrate Pathology, 107, S42–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita, R. , Overton, J. A. , Greenbaum, J. A. , Ponomarenko, J. , Clark, J. D. , Cantrell, J. R. , … Peters, B. (2015). The immune epitope database (IEDB) 3.0. Nucleic Acids Research, 43, D405–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti, A. , Piazzolla, G. , Condelli, V. , Nuzzaci, M. , Lanorte, M. T. , Boscia, D. , … Tortorella, C. (2010). Cucumber mosaic virus as the expression system for a potential vaccine against Alzheimer's disease. Journal of Virological Methods, 169(2), 332–340. [DOI] [PubMed] [Google Scholar]
- Walker, A. , Skamel, C. , & Nassal, M. (2011). SplitCore: An exceptionally versatile viral nanoparticle for native whole protein display regardless of 3D structure. Scientific Reports, 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waneesorn, J. , Wibowo, N. , Bingham, J. , Middelberg, A. , & Lua, L. (2016). Structural‐based designed modular capsomere comprising HA1 for low‐cost poultry influenza vaccination. Vaccine, 36, 3064–3071. [DOI] [PubMed] [Google Scholar]
- Wibowo, N. , Chuan, Y. P. , Lua, L. H. L. , & Middelberg, A. P. J. (2013). Modular engineering of a microbially‐produced viral capsomere vaccine for influenza. Chemical Engineering Science, 103, 12–20. [Google Scholar]
- Wibowo, N. , Hughes, F. K. , Fairmaid, E. J. , Lua, L. H. L. , Brown, L. E. , & Middelberg, A. P. J. (2014). Protective efficacy of a bacterially produced modular capsomere presenting M2e from influenza: Extending the potential of broadly cross‐protecting epitopes. Vaccine, 32(29), 3651–3655. [DOI] [PubMed] [Google Scholar]
- Wiersma, L. C. , Rimmelzwaan, G. F. , & de Vries, R. D. (2015). Developing universal influenza vaccines: Hitting the nail, not just on the head. Vaccines, 3(2), 239–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiessner, C. , Wiederhold, K. H. , Tissot, A. C. , Frey, P. , Danner, S. , Jacobson, L. H. , … Staufenbiel, M. (2011). The second‐generation active A β immunotherapy CAD106 reduces amyloid accumulation in APP transgenic mice while minimizing potential side effects. Journal of Neuroscience, 31(25), 9323–9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. G. , Jiang, L. B. , Zhou, Z. A. , Fan, J. H. , Zhang, Q. Q. , Zhu, H. H. , … Xu, Z. K. (2003). Expression of foot‐and‐mouth disease virus epitopes in tobacco by a tobacco mosaic virus‐based vector. Vaccine, 21(27–30), 4390–4398. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Narum, D. L. , Fleury, S. , Jennings, G. , & Yadava, A. (2015). Particle‐based platforms for malaria vaccines. Vaccine, 33(52), 7518–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X. , Ku, Z. , Liu, Q. , Wang, X. , Shi, J. , Zhang, Y. , … Huang, Z. (2014). Chimeric virus‐like particle vaccines displaying conserved enterovirus 71 epitopes elicit protective neutralizing antibodies in mice through divergent mechanisms. Journal of Virology, 88(1), 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y. , Zhang, S. , Cai, C. , Zhang, J. , Dong, D. , Guo, Q. , … Chen, W. (2014). Deletion modification enhances anthrax specific immunity and protective efficacy of a hepatitis B core particle‐based anthrax epitope vaccine. Immunobiology, 219(2), 97–103. [DOI] [PubMed] [Google Scholar]
- Zdanowicz, M. , & Chroboczek, J. (2016). Virus‐like particles as drug delivery vectors. Acta Biochimica Polonica, 63(3), 469–473. [DOI] [PubMed] [Google Scholar]
- Zhai, Y. , Zhong, Z. , Zariffard, M. , Spear, G. T. , & Qiao, L. (2013). Bovine papillomavirus‐like particles presenting conserved epitopes from membrane‐proximal external region of HIV‐1 gp41 induced mucosal and systemic antibodies. Vaccine, 31(46), 5422–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Ao, Z. , & Yao, X. (2016). Current advances in virus‐like particles as a vaccination approach against HIV infection. Vaccines, 4(1),2. 10.3390/vaccines4010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , & Hammond, R. W. (2005). Development of a candidate vaccine for Newcastle disease virus by epitope display in the Cucumber mosaic virus capsid protein. Biotechnology Letters, 27(6), 375–382. [DOI] [PubMed] [Google Scholar]


